Abstract
Cross-talk between hematopoietic stem and progenitor cells (HSPCs) and bone marrow (BM) cells is critical for homing and sustained engraftment after transplantation. In particular, molecular and physical adaptation of sinusoidal endothelial cells (ECs) promote HSPC BM occupancy; however, signals that govern these events are not well understood. Extracellular vesicles (EVs) are mediators of cell-cell communication crucial in shaping tissue microenvironments. Here, we demonstrate that integrin α4β7 on murine HSPC EVs targets uptake into ECs. In BM ECs, HSPC EVs induce up-regulation of C-C motif chemokine receptor 2 (CCR2) ligands that synergize with CXCL12-CXCR4 signaling to promote BM homing. In nonirradiated murine models, marrow preconditioning with HSPC EVs or recombinant CCR2 ligands improves homing and early graft occupancy after transplantation. These findings identify a role for HSPC EVs in remodeling ECs, newly define CCR2-dependent graft homing, and inform novel translational conditioning strategies to improve HSPC transplantation.
Reciprocal cross-talk with endothelial cells reveals a CCR2-dependent homing axis for hematopoietic stem cells.
INTRODUCTION
Hematopoietic stem and progenitor cells (HSPCs) maintain hematopoiesis through continued self-renewal and differentiation into mature blood lineages. After migration from the fetal liver during development, adult HSPCs predominantly reside in the bone marrow (BM), a highly vascularized microenvironment (1). Under steady-state conditions, marrow HSPCs proliferate and differentiate into mature progeny, which exit across the sinus endothelium to circulate into the blood. During steady-state hematopoiesis, low levels of noncycling HSPCs also continuously egress and enter the systemic circulation, likely contributing to host immunity, before migrating back to the BM (2–4).
Physiologic homing to the BM occurs partly along the CXCL12-CXCR4 axis and is exploited during HSPC transplantation (HSPCT), where donor HSPCs migrate to the BM and assume niche occupancy for sustained hematopoiesis. Increasingly used for otherwise incurable hematopoietic disorders, and especially in combination with innovations in gene editing, HSPCT is a powerful platform to treat malignancy, BM failure, or monogenic diseases, including hemoglobinopathies and metabolic storage disorders (5). Despite its curative potential and expanding access, patient morbidity and mortality remain major concerns in HSPCT, partly attributable to delayed or insufficient levels of engraftment (6). Conditioning regimens to boost engraftment have improved long-term (LT) patient outcomes; however, current strategies are limited by toxicity. A better understanding of the mechanisms guiding HSPC homing will provide potential to develop selective approaches to maximizing early engraftment and de-escalating morbidity.
During adoptive transfer, HSPC migration from circulation to the BM relies upon interaction with sinusoidal endothelial cells (ECs), where adhesion and tethering allow subsequent transmigration through the endothelium to the niche (7–10). In the fetal liver, HSPC colonization triggers physical adaptation of ECs to sustain HSPC occupancy in a term called “cuddling” (11). However, a critical gap in knowledge exists with regard to the dynamic roles that HSPCs take in shaping their BM niche. Extracellular vesicles (EVs) have emerged as important mediators of endocrine and paracrine communication by delivery of bioactive proteins, lipids, and nucleic acids (12). HSPC-derived EVs remain largely undefined, but parallel studies of tumor EVs show that vesicles display organotropism through integrin-ligand interactions, aiding in the establishment of premetastatic niches by remodeling the tissue microenvironment (13, 14).
Here, we demonstrate that HSPC EVs target the BM vasculature through analogous integrin-mediated uptake and induce the secretion of chemoattractant ligands that promote CCR2-dependent homing. Moreover, we show for the first time that CCR2 cooperates with CXCR4 to boost HSPC homing. These studies unveil a novel mechanism of niche cross-talk by which HSPC vesicles reshape niche function and improve engraftment during adoptive transfer.
RESULTS
Integrin α4β7 promotes HSPC vesicle uptake into ECs
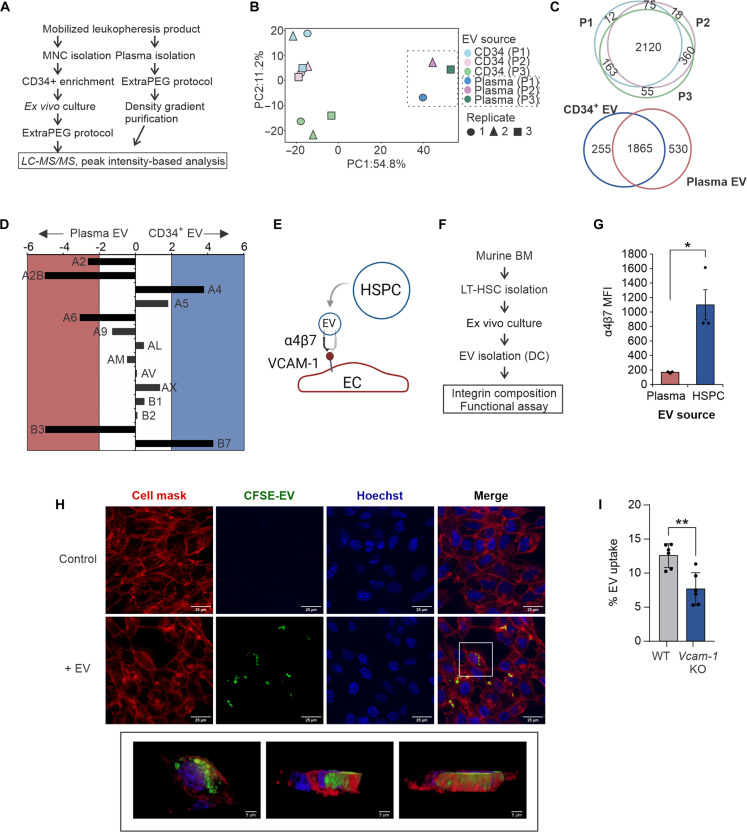
EVs represent a core component of the cellular secretome; however, only a few studies have investigated HSPC vesicle cargo (15–18). To profile the composition of HSPC-derived EVs and better understand the contribution of secreted HSPC cargo in graft products, we performed comparative mass spectrometry of secreted vesicles purified from short-term (ST) ex vivo cultures of leukapheresis-derived CD34+ cells or donor-matched plasma EVs (Fig. 1A; fig. S1, A to C; and data S1). This initial study was conducted using human samples due to limitations in scalability for in-depth proteomic profiling. HSPC EVs contained distinct cargo compared to plasma EVs, with enrichment of chaperone and binding proteins involved in cell migration and motility (Fig. 1, B and C, and fig. S1, D to E). Compared to circulating plasma EVs, human HSPC EVs revealed significant enrichment of the integrin α4 and β7 subunits (Fig. 1D). Heterodimerized, integrin α4β7 forms a receptor for vascular cell adhesion molecule–1 (VCAM-1), highly expressed on ECs, and with known functions in lymphocyte homing and adhesion (19, 20). Together with existing evidence for integrins in directing cell type–specific vesicle uptake (14, 21), these data suggested a role for integrin α4β7 in targeting HSPC EVs to ECs (Fig. 1E). To facilitate further mechanistic study at scale, we adopted a polyvinyl alcohol–based ex vivo expansion protocol for murine HSPCs (Fig. 1F) (18, 22). Murine HSPC EVs showed similar enrichment of integrin α4β7 (Fig. 1G and fig. S1F). Using a well-established model of Akt1-expressing primary murine BM ECs (23, 24), we demonstrate robust uptake of HSPCs into ECs that is reduced after neutralization with an integrin α4β7 inhibitor, TR-14035 (Fig. 1H, fig. S1G, and movie S1). HSPCs also trigger increased surface expression of VCAM-1 that is unaltered by TR-14035 treatment (fig. S1H). CRISPR-Cas9–directed knockout (KO) of Vcam-1 in ECs also mitigated HSPC EV uptake, nearly comparable to inhibition of dynamin-dependent endocytosis using dynasore (Fig. 1I and fig. S1I). Together, these findings demonstrate a role for integrin α4β7 in promoting HSPC EV uptake into BM ECs.
Fig. 1. Enriched integrin α4β7 targets HSPC EV uptake into ECs.
(A) Workflow for isolation and purification of HSPC-derived vesicles and donor-matched plasma EV controls (n = 3 donors). (B) Principle components analysis of protein cargo identified in HSPC and plasma EVs. (C) Overlap of unique proteins in HSPC EVs from donors 1 to 3 (P1 to P3) and plasma EVs. (D) Enrichment analysis of integrin subunits in HSPC EVs compared to plasma EVs. (E) Proposed model of HSPC EV uptake into ECs; created with BioRender. (F) Workflow for isolation and analysis of murine HSPC-derived EVs. (G) Flow cytometric analysis of integrin α4β7 in murine EVs. (H) Representative confocal microscopy images of HSPC EV uptake into BM ECs. (I) Flow cytometric measurement of CFSE-dyed EV uptake into Vcam-1 KO ECs. LC-MS/MS, liquid chromatography tandem mass spectrometry; DC, differential centrifugation; MFI, mean fluorescence intensity; LT-HSC, long-term HSC. *P < 0.05 and **P < 0.01.
HSPC EVs induce CCR2 ligand up-regulation in ECs through NF-κB signaling
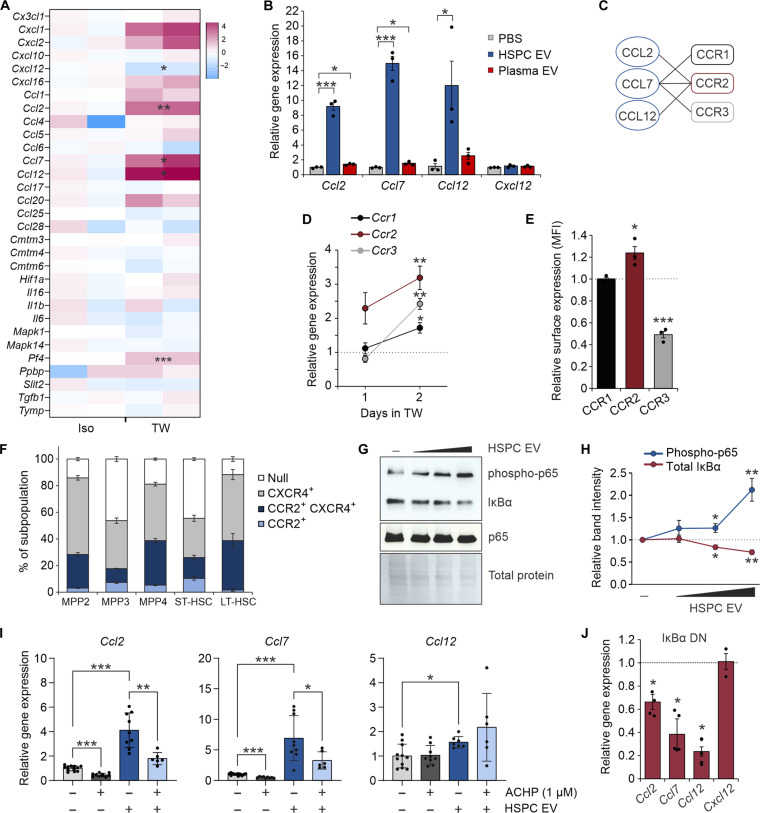
Secreted EVs from immune cells not only harbor encapsulated cytokines (25) but may also induce inflammatory signaling downstream of uptake into recipient cells (26). To determine the impact of the HSPC secretome on BM ECs in the absence of cell-cell contact, we adopted a 1-μm porous transwell coculture system. ECs cultured in transwell culture with HSPCs showed transcriptional up-regulation of several cytokines and chemokines, including Ccl2, Ccl7, and Ccl12 (Fig. 2A). Similar gene expression induction was observed after addition of purified HSPC EVs, but not plasma-derived EVs, into EC cultures (Fig. 2B). Notably, Ccl2, Ccl7, and Ccl12 mRNA was minimal to undetectable in HSPC-derived EVs, arguing against a mechanism of direct transfer. CCL2, CCL7, and CCL12 bind a family of C-C motif chemokine receptors (CCRs) that include CCR1 to CCR3, with CCR2 acting as a common receptor (Fig. 2C). Of these receptors, Ccr2 showed predominant reciprocal up-regulation on HSPCs in EC transwell coculture (Fig. 2D). Corresponding increases in CCR2 surface expression was also seen after a 2-day coculture (Fig. 2E). Neither Cxcr4 nor Cxcl12 were increased in gene or protein expression in cocultured HSPCs and ECs, respectively (Fig. 2B and fig. S2, A to C). However, subsets of stem and progenitor cell subpopulations were noted to coexpress CCR2 and CXCR4 (Fig. 2F).
Fig. 2. HSPC EVs remodel the EC secretome through NF-κB activation.
(A) Transcriptional analysis of murine BM ECs by RT2 Profiler PCR Array. ECs were cultured in isolation or in noncontact 1.0-μm transwell dishes with murine HSPCs for comparative analysis. (B) Relative gene expression of target chemokines following transfer of HSPC or plasma-derived EVs (103 to 104 EV per cell). (C) Known receptor-ligand binding partners of CCR1 to CCR3. (D) Relative gene expression of CCR2 ligand (CCR2L) receptors on HSPCs after 1 to 2 days in transwell culture with ECs. Dotted line denotes isolated HSPCs. (E) Relative surface expression of CCR1 to CCR3 on HSPCs after 2 days in transwell EC culture. Dotted line denotes isolated HSPCs. (F) Surface expression of CCR2 and CXCR4 on HSPC subpopulations. (G) Representative immunoblots and (H) quantitative analysis demonstrating a dose-dependent activation of canonical NF-κB signaling in ECs after HSPC EV uptake. (I) Relative gene expression of CCR2L in ECs after HSPC EV transfer and treatment with the IκB kinase inhibitor, ACHP. (J) Relative gene expression of CCR2L and Cxcl12 after HSPC EV uptake into WT ECs (dotted line) or cells transduced with an IκBα dominant-negative (DN) mutant. Iso, isolated; TW, transwell. *P < 0.05, **P < 0.01, and ***P < 0.001.
Although common transcriptional regulation of CCR2 ligands (CCR2Ls) has not been well described, a prominent role exists for nuclear factor κB (NF-κB) in modulating the expression of angiogenic chemokines (27). Addition of HSPC EVs showed dose-dependent activation of canonical NF-κB signaling through enhanced inhibitor of nuclear factor κBα (IκBα) degradation and p65 phosphorylation (Fig. 2, G and H). Inhibition of canonical NF-κB signaling using an IκB kinase inhibitor, ACHP (28, 29), or expression of a dominant negative IκBα construct largely blocked CCR2L and Vcam-1 up-regulation induced by HSPC EVs (Fig. 2, I and J, and fig. S2, D to G). These data support a mechanism by which HSPC EVs remodel the NF-κB–responsive EC secretome and likely promote a chemoattractant state.
CCR2-ligand and CXCL12 interactions drive HSPC chemotaxis
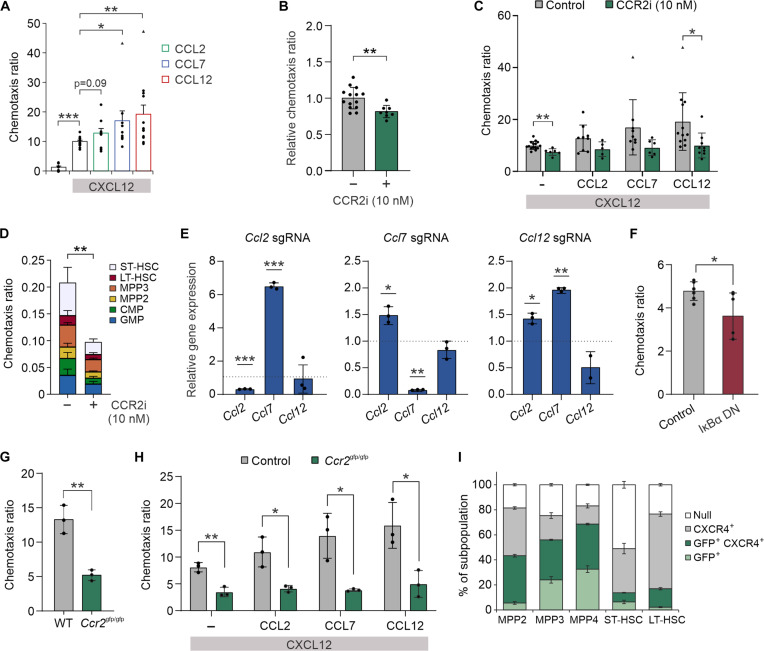
To test whether CCR2-ligand interactions actively promote HSPC chemotaxis, we established CCR2L gradients in the bottom chamber of a 5-μm porous transwell dish. Addition of individual CCR2Ls did not affect HSPC chemotaxis (fig. S3A); however, CCL7 and CCL12 synergistically enhanced migration in the presence of CXCL12 with no effect on HSPC CXCR4 surface expression (Fig. 3A and fig. S3B). A small, although statistically insignificant, augmentation of migration was also seen with the addition of CCL2. No significant differences in migration toward chemokine gradients were observed across stem and progenitor cells in vitro (fig. S3, C and D). Pharmacologic inhibition of CCR2 (CCR2i) reduced migration toward BM ECs (Fig. 3B and fig. S3E) and recombinant CXCL12/CCR2L gradients (Fig. 3C; control data depicted from Fig. 3A). CCR2 inhibition was also noted to significantly decrease CXCL12-mediated chemotaxis in the absence of CCR2L, suggesting a possible cooperativity between CXCR4 and CCR2. Integrin α4β7 blockade similarly decreased HSPC migration toward ECs (fig. S3F). Proportional decreases across stem and progenitor cell subpopulation migration toward ECs were seen after CCR2i, with no significant differences in subpopulation frequency during coculture (Fig. 3D and fig. S3G).
Fig. 3. CCR2 receptor-ligand interactions promote HSPC chemotaxis toward ECs.
(A) Chemotaxis ratio of murine HSPCs through 5-μm pores toward CCR2L chemokine gradients (200 ng/ml). Chemotaxis assays were performed in the presence of CXCL12 (200 ng/ml). Chemotaxis of HSPCs toward (B) BM ECs or (C) CXCL12/CCR2L gradients after CCR2i. (D) Flow cytometric analysis of HSPC subpopulation-specific migration toward ECs after CCR2i. (E) Relative gene expression of all CCR2L in BM ECs harboring individual gene KO by CRISPR-Cas9 after HSPC EV uptake (WT, dotted line), demonstrating compensatory up-regulation of residual CCR2L after EV exposure. (F) Chemotaxis ratio of HSPCs toward WT ECs or ECs transduced with an IκBα dominant-negative mutant. Chemotaxis ratio of Ccr2gfp/gfp HSPCs compared to WT HSPCs toward (G) BM ECs or (H) chemokine gradients. (I) GFP and surface CXCR4 expression across subpopulations in Ccr2gfp/gfp marrows. *P < 0.05, **P < 0.01, and ***P < 0.001. Triangle points denote statistically significant outliers.
CRISPR-Cas9 directed KO of each individual CCR2L in BM ECs revealed notable compensatory up-regulation of the remaining CCR2Ls during HSPC coculture or after HSPC EV uptake, suggesting a critical function of these genes in BM ECs (Fig. 3E and fig. S3H). Given the functional redundancy, individual CCR2L KOs in ECs had little effect on chemotaxis (fig. S3I). However, suppression of all CCR2Ls by expression of dominant-negative IκBα in ECs significantly reduced HSPC chemotaxis (Figs. 2J and 3F).
In addition, ex vivo cultured HSPCs derived from Ccr2gfp/gfp KO/knock-in(KI) mice similarly demonstrated reduced chemotaxis toward BM ECs and CXCL12/CCR2L gradients (Fig. 3, G and H). This mouse strain contains an enhanced green fluorescent protein (GFP) sequence followed by a polyadenylation signal inserted into the translation site of the Ccr2 gene, abolishing gene expression and resulting in GFP positivity in a subpopulation of cells normally expressing CCR2 (fig. S3, J and K). The migrated cell compartment in EC transwell culture was enriched in GFP+ cells, indicating that other factors typically enriched in CCR2+ HSPCs may also play a role in migration (fig. S3L). Relative increases in GFP+ and GFP+ CXCR4+ progenitor cell subsets were noted in Ccr2 KO BM, likely reflective of added intracellular signal compared to CCR2 surface expression measurement (Figs. 2F and 3I).
The EC secretome promotes CXCR4 colocalization to CCR2-rich membrane domains
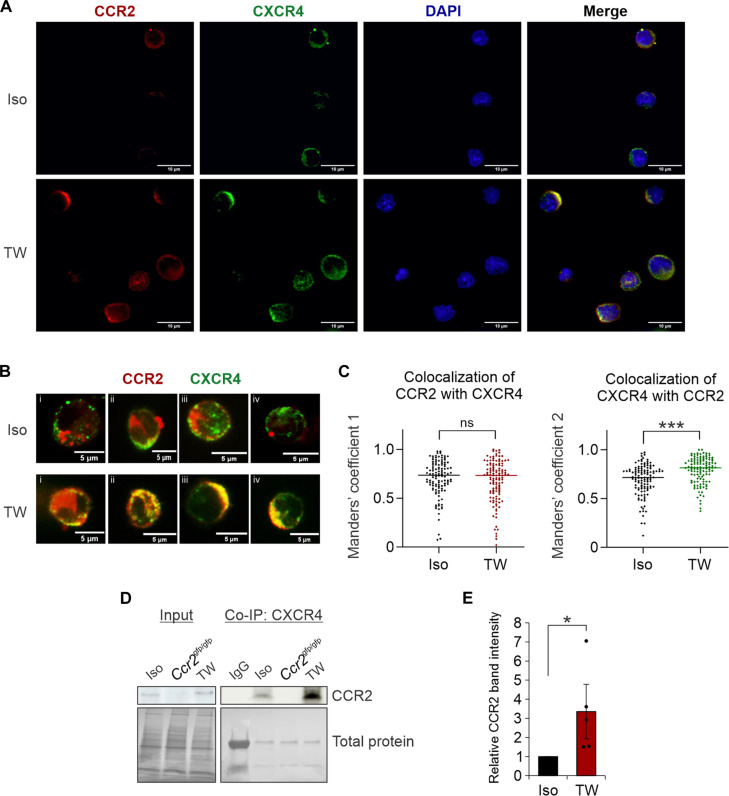
The observed functional synergy of CCR2L and CXCL12 (Fig. 3A) raised the possibility of CCR2 and CXCR4 receptor cooperativity in driving HSPC migration. To assess the membrane distribution of CCR2 on HSPCs exposed to the EC secretome, immunofluorescence analysis was performed on HSPCs in isolation compared to cells in transwell EC coculture. CCR2 demonstrated a polarized membrane distribution on HSPCs with overall increased expression following transwell EC coculture (Fig. 4A and fig. S4, A and B). CXCR4 similarly showed a polar, membranous distribution; however, in isolated HSPCs, CXCR4hi regions were predominantly distinct from CCR2hi domains, with no change in CXCR4 expression after coculture (fig. S4C). Measurements of CCR2 and CXCR4 colocalization in isolated cells (Fig. 4C) are congruent with prior reports that a significant proportion of each receptor exists in a mutually heterodimerized formation (30–32). Following EC transwell culture, increased colocalization of CXCR4 with CCR2 was noted (Fig. 4, B and C, and fig. S4, D to G). Immunoprecipitation of CXCR4 confirmed increased levels of CXCR4-CCR2 heterodimers in cocultured HSPCs (Fig. 4, D and E). These data support a model whereby the EC secretome promotes CCR2 and CXCR4 receptor interaction and cooperativity.
Fig. 4. EC-activated HSPCs show CXCR4 and CCR2 cooperation.
(A) Representative confocal images showing CCR2 and CXCR4 localization in isolated HSPCs versus those grown in EC cocultures. (B) Representative high-power images of individual cells analyzed. (C) Calculation of Manders’ overlap coefficients (n = 118 to 120 cells per condition). (D) Representative immunoblot analyses and (E) quantitative analysis of CCR2 in HSPC lysates after CXCR4 immunoprecipitation. *P < 0.05 and ***P < 0.001. ns, not significant; Co-IP, coimmunoprecipitation; DAPI, 4′,6-diamidino-2-phenylindole; IgG, immunoglobulin G.
CCR2 regulates HSPC BM niche occupancy
To directly measure CCR2-dependent BM homing after transplantation, we performed competitive transplantations with donor BM derived from wild-type (WT) versus Ccr2gfp/gfp mice (33). For these experiments, recipient mice were conditioned with a sublethal irradiation dose (5.2 Gy), as we noted a radiation dosage-dependent up-regulation of Ccl2 and Ccl7 (fig. S5A). With only some cellular subsets harboring GFP positivity in Ccr2gfp/gfp mice (fig. S3K), single-cell suspensions of whole BM were stained with carboxyfluorescein succinimidyl ester (CFSE) to evaluate comparative global BM homing efficiency (fig. S5B). No significant differences in proportions of native stem and progenitor cells were seen at baseline in CCR2-deficient animals (fig. S5C). Overall, total cellular homing was reduced from CCR2-deficient grafts, although this effect was predominantly driven by decreased homing of mature lineage-positive subpopulations (fig. S5D).
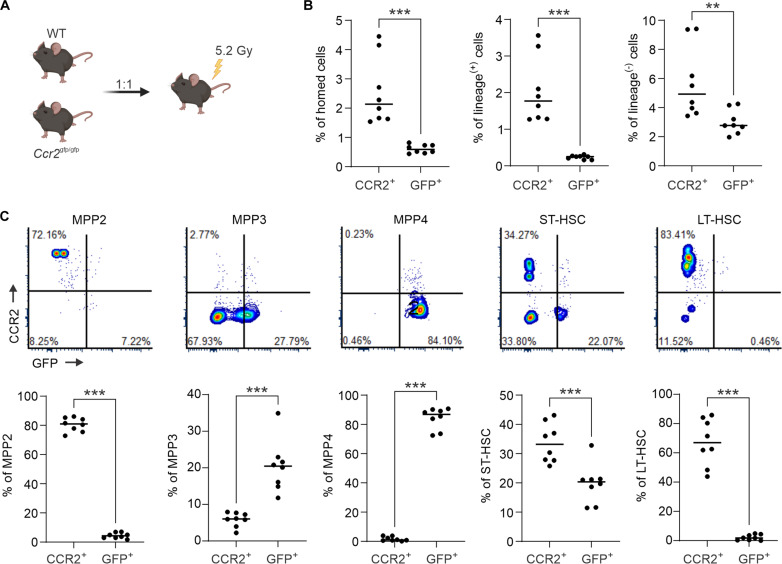
To specifically evaluate CCR2+ population homing frequencies, WT BM was competitively transplanted with unstained Ccr2gfp/gfp marrow (Fig. 5A). In this experiment, only mutually exclusive CCR2+ or GFP+ (CCR2−) cells were analyzed (fig. S5E). Here, CCR2-deficient grafts demonstrated deficits in homing of both mature and immature hematopoietic cells (Fig. 5B). However, marked differences in subpopulation-specific homing was noted, with Ccr2 deletion resulting in a reduction in myeloid-biased multipotent progenitor 2 (MPP2) and HSC populations and enrichment in lymphoid-biased MPP3 and MPP4 progenitors (Fig. 5C). Notably, substantial contributions of MPP3 and MPP4 populations to the lineage-negative compartment may explain the modest, but insignificant, increases in overall precursor homing (fig. S5D). Strikingly, although only a subset of HSCs and MPP2 show CCR2 positivity (Fig. 2F), the majority of cells that occupy the marrow and spleen after transplantation are positive for CCR2 (Fig. 5C and fig. S5F), suggesting a critical role for this receptor in guiding niche homing.
Fig. 5. CCR2 is necessary for optimal HSPC homing to the BM.
(A) Experimental schematic of competitive homing using WT and Ccr2gfp/gfp BM (n = 8). (B) Flow cytometric analysis measuring the relative frequency of CCR2+ versus GFP+ (CCR2−) cells, including lineage-positive and lineage-negative cells within the CD45.2 cell compartment 20 hours after transplantation. (C) Representative flow cytometry plots and quantitative analysis of CD45.2+ CCR2+ versus CD45.2+ GFP+ cells across each subpopulation. Experimental schematics created using BioRender. **P < 0.01 and ***P < 0.001.
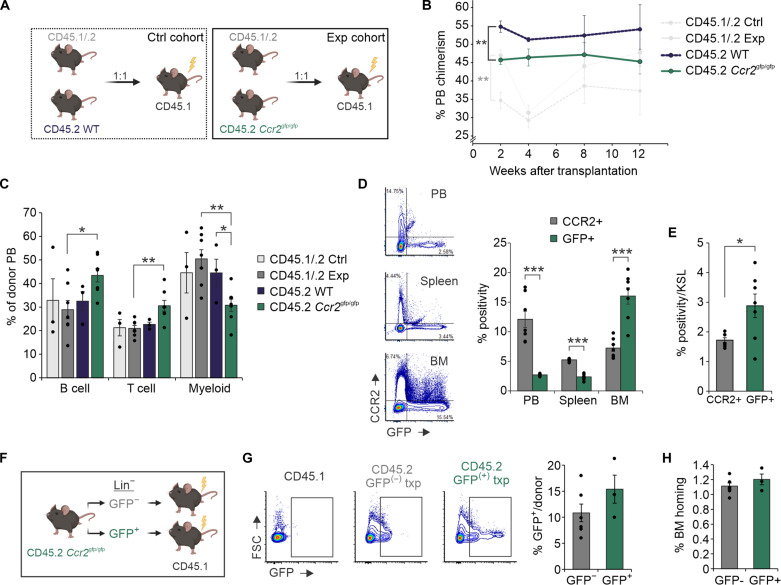
To further assess the impact of CCR2 depletion on overall ST reconstitution, Ccr2gfp/gfp (CD45.2) marrow was transplanted against competitor CD45.1/.2 marrow into irradiated recipients (Fig. 6A). To account for known advantages of CD45.2 grafts (34), a control cohort was included. WT CD45.2 outperformed WT CD45.1/.2 marrow as expected; however, relative impairment of Ccr2gfp/gfp engraftment was seen at 2 weeks after transplantation (Fig. 6B). At 4 weeks onward, no significant differences in Ccr2gfp/gfp and WT peripheral reconstitution were seen, suggesting possible additional roles of CCR2 in regulating HSPC differentiation and marrow retention. To this point, relative chimerism levels of Ccr2gfp/gfp grafts were modestly higher in peripheral blood (PB) and spleens compared to BM (fig. S6A). Myeloid output in the PB and spleen and MPP3 frequency in the BM were diminished from Ccr2gfp/gfp grafts at 12 weeks; these findings are consistent with prior evidence supporting a myeloid differentiation bias of CCR2+ HSC (Fig. 6C and fig. S6, B to D) (35). In the experimental cohort, notable differences in chimerism were seen in CCR2+ versus GFP+ populations in the PB and spleen versus BM, including specific increases in GFP+ BM c-Kit+ Sca-1+ lineage- (KSL) cells, suggesting CCR2+ cells harbor are later prone to marrow egress (Fig. 6, D and E).
Fig. 6. Dynamic CCR2 signaling regulates BM niche occupancy.
(A) Experimental schematic of competitive transplantations (1:1 donor ratio) using CD45.1/.2 WT versus Ccr2gfp/gfp BM (n = 7). Recipient CD45.1 mice were lethally irradiated 24 hours before transplantation. A control cohort (n = 3) using CD45.1/CD45.2 WT versus CD45.2 WT BM was also analyzed. (B) PB chimerism of WT and Ccr2gfp/gfp grafts in individual mice after transplantation. (C) PB lineage analysis at 12 weeks after transplantation. (D) Representative flow cytometric plots and quantitative analysis of CCR2+ and GFP+ donor cells and (E) BM KSL in experimental cohort animals 12 weeks after transplantation. (F) Experimental schematic of noncompetitive Ccr2gfp/gfp GFP− versus GFP+ graft transplantations. (G) Representative flow cytometric plots and quantitative analysis of GFP positivity in BM 20 hours after transplantation from initial GFP− versus GFP+ grafts. (H) Flow cytometry measurement of overall donor cell BM frequency. Experimental schematics created using BioRender. *P < 0.05, **P < 0.01, and ***P < 0.001.
Chemokine receptor surface expression may fluctuate in response to inflammatory stimuli (36). We hypothesized that CCR2 expression would increase on HSPCs during myeloablative adoptive transfer. To test this hypothesis, lineage-depleted GFP+ or GFP− cells from Ccr2gfp/gfp marrows were independently transplanted into irradiated recipients (Fig. 6F). GFP levels on homed cells from initially GFP− and GFP+ grafts normalized to baseline levels, suggesting physiologic calibration of CCR2 expression during homing (Fig. 6G). In particular, >50% of initially GFP- KSL that homed to the BM were noted to activate GFP expression (fig. S6E). Functionally, noncell autonomous HSPC adjustment to the local niche environment rapidly neutralizes differences in BM homing between GFP− and GFP+ grafts (Fig. 6H). Overall, these findings indicate that dynamic CCR2 signaling adds subpopulation specificity to BM homing in a CXCR4-CXCL12 replete system and further provokes a role for CCR2 in fine-tuning marrow retention.
Marrow conditioning with CCR2 ligands enhances HSPC homing
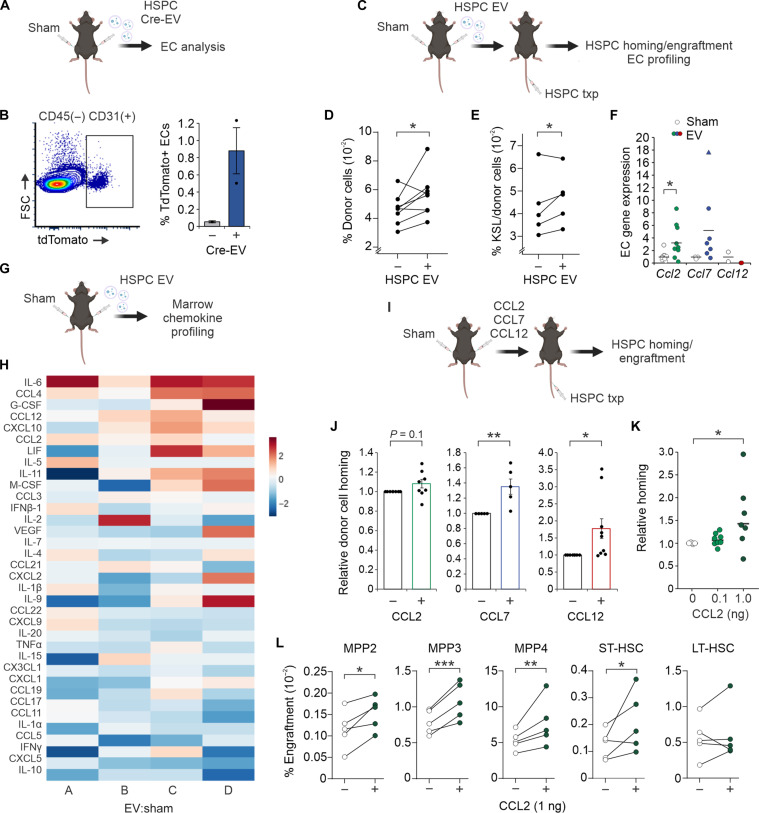
Pharmacologic targeting of the CXCR4 surface receptor is widely used in clinical settings to improve peripheral mobilization of HSPCs for pre-transplantation leukapheresis (37). More recent investigations into CXCR4 activation and expression on hematopoietic cells have explored the utility of this axis in alternatively promoting BM homing (38, 39). We hypothesized that CCR2-ligand signaling via HSPC-EC cross-talk may additionally be exploited to enhance marrow homing. To first measure rates of in vivo EV uptake into BM ECs, we injected HSPC EVs carrying Cre recombinase mRNA into Ai9 reporter mice (Fig. 7A). Intracellular expression of Cre recombinase protein following EV uptake in these mice results in tdTomato fluorescence (40, 41). Accordingly, in vivo uptake of HSPC EVs induced measurable tdTomato fluorescence into BM ECs (Fig. 7B).
Fig. 7. Preconditioning with HSPC EVs or CCR2 ligands improves HSPC homing to the BM.
(A) Schematic detailing intrafemoral injection of HSPC EVs containing Cre mRNA into tdTomato (Ai9) mice. (B) Representative flow plot and quantitative analysis of tdTomato fluorescence in BM ECs isolated from EV-injected marrows. (C) Schematic detailing intrafemoral injection of HSPC EVs before systemic cell transplantation. (D) Flow cytometric quantification of HSPC homing (20 hours after transplantation) toward EV versus sham conditioned marrows (n = 7). (E) Quantification of c-Kit+ Sca-1+ lineage− (KSL) progenitors engrafted in EV versus sham conditioned marrows after 4 weeks (n = 5). (F) Relative gene expression analysis of BM ECs isolated from EV versus sham conditioned marrows (n = 7). (G) Schematic detailing intrafemoral injection of EVs for BM cytokine array profiling (n = 4). (H) Heatmap of fold change (FC; log 2) abundance of cytokines in EV versus sham-conditioned BM. Each column corresponds to the FC of EV:sham-injected marrows in an individual mouse. (I) Schematic detailing intrafemoral injection of individual CCR2L prior to cell transplantation. (J) Flow measurement of HSPC homing toward BM conditioned with individual CCR2L (0.1 ng) versus contralateral sham-injected marrows (n = 4 to 8 per cohort). (K) Relative homing of HSPCs following dose escalation (n = 8) of CCL2 conditioning compared to contralateral sham-injected marrows. (L) Subpopulation-specific engraftment in CCL2-conditioned marrows compared to contralateral sham-injected marrows 4 weeks after transplantation (n = 5). Schematics created using BioRender. *P < 0.05, **P < 0.01, and ***P < 0.001.
Next, nonmyeloablated recipient animals were preconditioned with intrafemoral injection of HSPC EVs 4 hours before intravenous HSPCT (Fig. 7C). Significant increases in cell homing to femurs conditioned with EVs were seen compared to contralateral sham [phosphate-buffered saline (PBS)] injected femurs (Fig. 7D). Engraftment of KSL progenitors was significantly increased in most animals 4 weeks after transplantation (Fig. 7E). Similar to ex vivo conditions (Fig. 2B), ECs isolated from EV-conditioned marrows showed up-regulation of Ccl2 and Ccl7, although Ccl12 expression was notably undetectable in vivo (Fig. 7F). To understand the impact of HSPC EVs on the global marrow secretome, chemokine profiling was performed on femoral BM after EV exposure (Fig. 7G). Significantly differentially secreted proteins included increased IL-6, CCL4, CCL12, and CCL2, while IL-10, CXCL5, CCL5, IL-1α, CX3CL1, IL-20 were decreased in marrows following EV exposure (Fig. 7H and fig. S7A).
Last, we tested whether nonirradiated recipients conditioned in situ with recombinant CCR2L could improve HSPC homing and ST engraftment (Fig. 7I). HSPCs demonstrated significant improvements in homing to BM conditioned with 0.1 ng of CCL7 and CCL12 compared to contralateral sham-injected femurs (Fig. 7J). Dose escalation of CCL2 conditioning overall offered additional improvement in homing efficiency (Fig. 7K). After 4 weeks, significant increases in engraftment of MPP and ST-HSC populations were seen in CCL2-conditioned marrows (Fig. 7L). Together, these findings suggest that HSPC EVs act on ECs to enhance chemoattraction to the BM and that exploitation of this pathway by CCR2L conditioning may offer avenues to improve homing and engraftment after therapeutic transplantation.
DISCUSSION
Therapeutic applications of HSPCT continue to expand and evolve, including as a platform for personalized gene therapy. Beyond conventional chemotherapy and graft engineering, clinical interest has risen in developing targeted preconditioning strategies to improve HSPC BM occupancy by nongenotoxic cell depletion (42–45). Alternative or compounded approaches to increase marrow homing will likely further enhance graft outcomes and decrease morbidity through improving early count recovery and polyclonal reconstitution. In addition, a deeper understanding of molecular mechanisms involved in BM homing may serve as a source of insight to improve mobilization strategies. For instance, canonical homing and retention of HSPCs occurs through the CXCR4-CXCL12 axis (46–48); this signaling pathway has been leveraged clinically by use of a CXCR4 antagonist plerixafor during HSPC mobilization and collection. Substantial evidence has demonstrated a role for CCL2-CCR2–driven chemotaxis of mature granulocytes and monocytes (49–52). More recent studies have detailed a role for this axis in driving HSPCs to sites of inflammation, including in models of peritonitis and myocardial infarction (35, 53).
Building on these studies, here, we describe a mechanism of CCR2-dependent chemotaxis that synergizes with CXCR4 signaling to promote BM homing after HSPCT. We demonstrate that this signaling axis may be driven not only by CCL2 but also by additional CCR2 ligands CCL7 and CCL12. A likely explanation toward the migratory synergy provoked with CXCL12 and CCR2L is our observation that activated HSPCs demonstrate increased interaction of CXCR4 and CCR2 that may partly be driven by an increase in CCR2 expression in response to inflammatory signals or by membrane redistribution of one or both receptors. Existing data have shown that CCR2 and CXCR4 form functional heterodimers in native T cells (30, 54, 55). Although specificity of CXCL12 and CCR2 ligands for their respective receptors exist, an allosteric negative binding cooperativity between CXCR4 and CCR2 has been reported, whereby activation of CCR2 may desensitize CXCR4 (30, 55, 56). In this study, we observe evidence supporting a positive CCR2-CXCR4 cooperativity on HSPCs. Our data support increased heterodimer formation in activated HSPCs, which may act in a more potent capacity than respective homodimers to drive migration (57). Notably, a counter balance between CXCR4 and CCR2 signaling has been described to modulate mature immune cell retention and egress from the marrow, where CCR2 up-regulation drives efflux of cells to sites of inflammation (51, 56, 58, 59). In the context of conventional preconditioning regimens and donor cell infusion, the BM itself serves as a site of potent acute inflammation (60), and therefore, CCR2 activation may drive homing and retention along with CXCR4.
A role for CCR2 in fine-tuning marrow homing and retention is supported by results from our competitive transplantation studies between WT and Ccr2-deficient grafts. Here, we observed initial deficits in marrow homing from Ccr2gfp/gfp grafts and impaired myeloid output during early reconstitution; these findings are congruent with previous studies suggesting that CCR2+ HSCs are myeloid-biased and contribute to emergency myelopoiesis (35). CCR2 dependency for BM homing varied significantly across stem and progenitor subpopulations, with impaired homing of HSC and myeloid-biased MPP2 after Ccr2 KO, and relative enrichment of lymphoid-biased MPP3 and MPP4 populations. Despite homing heterogeneity, supraphysiologic CCL2 conditioning resulted in homogeneous increases in progenitor cell frequency in the BM at early engraftment time points; this may partly be due to enhanced CCR2+ HSC homing and early progenitor reconstitution. Our data further indicate that CCR2 expression on HSPCs is dynamically up-regulated during chemotaxis and BM homing. At later time points after transplantation and likely during homeostatic hematopoiesis, CCR2 continues to regulate niche occupancy. This is evident in the observed enrichment of CCR2− HSPCs in the BM 12 weeks after transplantation, with CCR2+ cells alternatively enriched in the PB and spleen. These findings support a role for CCR2 in facilitating marrow egress after resolution of marrow inflammation induced by conventional conditioning.
Notably, limited studies of CCR2-deficient patient BM composition has recently been reported (61); these suggest overall relatively normal mature cell counts, although stem and progenitor cell composition has not been detailed. Our studies likewise show that HSPC subpopulation frequency in native Ccr2 KO animals is preserved; however, important functional differences during transplantation are evident. These findings stress the importance of further investigation into CCR2 as a regulator of marrow retention, egress, and clonogenic reconstitution.
An undesirable side effect of the pro-inflammatory microenvironments provoked by conditioning chemotherapy or radiotherapy is the significant toxicity including marrow suppression (62, 63). Here, we show that the HSPC vesicle secretome engages a more targeted inflammatory signal through NF-κB–driven expression and secretion of chemotactic CCR2L. Radiation-induced production of C-C chemokines from ECs has been previously described (64). Although the impact of sustained NF-κB signaling has been shown to compromise LT HSPC self-renewal and differentiation capacity (65, 66), the role of acute signaling in promoting marrow homing during transplantation is relatively unknown. We propose targeted marrow inflammation, such as induced by HSPC EVs, as a strategy to improve homing. Future studies will determine key vesicle cargo driving EC activation. It is possible that EV exposure represents only one mechanism of up-regulating CCR2L and that alternative signals including pathogen-associated molecular pattern molecules and damage-associated molecular pattern molecules (DAMPs) promote similar effects. The release of DAMPs after toxic marrow irradiation may be key to driving nonspecific elevations of these chemokines.
In this study, we show that enrichment of integrin α4β7 targets secreted vesicles from HSPCs to VCAM-1+ ECs. Recent evidence points to an additional mechanism of selectin-dependent HSPC EV uptake (67). These findings may partly explain our observations of reduced, but not abolished, EV uptake into ECs after integrin α4β7 blockade or Vcam-1 KO. Moreover, we have seen transcriptional up-regulation of EC selectin-E following HSPC EV treatment, data that suggest possible positive feedback driving EV binding and uptake and further substantiate the functional impact of HSPC EVs on a multistep process guiding HSPC chemoattraction and tethering to the endothelium.
Although physiologic EV cross-talk shapes tissue microenvironments and particularly promoting EC remodeling (15, 17), therapeutic delivery of EVs harbors many challenges, including scalability, purity, and batch-to-batch reproducibility (68). Here, we provide the first proof-of-concept studies demonstrating how BM conditioning with CCR2L may leverage physiologic chemotactic mechanisms underlying HSPC homing to the BM vasculature and provide a novel nontoxic strategy to improve HSPC niche occupancy. This may be a promising approach in cases of limited donor cell quantity or requirements for reduced intensity conditioning. In addition, enhancing homing efficiency of infused HSPCs could likely lead to meaningful outcomes in cases of low-level residual leukemia, where healthy donor cells and leukemic stem cells (LSCs) compete for niche occupancy in the BM (69). In contrast, a role for CCR2 in leukemic cell migration and niche occupancy warrants additional examination (70, 71).
Further studies investigating a noninvasive approach to initiate therapeutic EC remodeling events including the delivery of small molecules or lipid nanoparticles carrying key biologically active EV cargo are the focus of our future efforts. This study is limited by its focus on HSPC:EC cross-talk in the murine model system. Differences in chemokine-receptor expression and interactions exist between mice and humans (72); these will need to be addressed in humanized models of marrow homing. For example, while human CCL2 and CCL7 bind CCR2, a human homolog for CCL12 does not exist, whereas additional chemokines CCL8, CCL13, and CCL16 share CCR2 binding capacity. Future studies will need to evaluate the translational relevance of individual signaling components. Overall, these findings add to our understanding of the functional HSPC secretome during transplantation and provide evidence of a novel cross-talk mechanism between HSPCs and BM ECs that originates from secreted HSPC-derived vesicles. We propose that there are opportunities to leverage these signals to improve CCR2-dependent mobilization and homing.
MATERIALS AND METHODS
Animals
C57BL/6J (the Jackson Laboratory strain #000664), B6.SJL-Ptprca Pepcb/BoyJ (Pep Boy; the Jackson Laboratory strain #002014), B6(C)-Ccr2tm1.1Cln/J (Ccr2gfp/gfp KI/KO; Jackson Laboratory strain #027619), B6.Cg-Tg(VAV1-cre)1Graf/MdfJ (VavCre; Jackson Laboratory strain #035670), and B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J (tdTomato; Jackson Laboratory strain #007914) mice were purchased from the Jackson Laboratories or bred in-house. Animals were housed under standard conditions and kept on a 12-hour light/12-hour dark cycle with ad libitum access to food and water. Our study examined male and female animals, and similar findings are reported for both sexes.
HSPC enrichment and culture
Human HSPCs were enriched from frozen leukapheresis products containing granulocyte colony-stimulating factor mobilized stem and progenitor cells and cultured as detailed previously (18). For mouse HSPCs, ex vivo expansion cultures were initiated from murine LT-HSCs flushed from hindlegs of 6- to 12-week-old mice and cultured as previously described in detail (18). Stem and progenitor cell populations were analyzed by flow cytometry as previously described in detail (18). A list of antibodies used in this study is listed in data S2.
BM EC culture
Akt1-activated murine BM-derived ECs were provided by J. Butler (University of Florida). Cells were cultured in 1:1 Dulbecco’s modified Eagle medium (low glucose)/Ham’s F-12 media with 20% (v/v) heat-inactivated fetal bovine serum (FBS), 20 mM Hepes, penicillin G (100 U/ml), streptomycin sulfate (100 μg/ml), 1X minimum essential medium nonessential amino acids (Corning, 25-025-CI), heparin sodium salt (100 μg/ml; Sigma-Aldrich, H3149), EC growth supplement (50 μg/ml; Corning, 356006). Heat-inactivated FBS was centrifuged at 100,000g for 20 hours at 4°C to deplete bovine serum EVs. Complete medium was filtered through a 0.22-μm filter and stored at 4°C for up to 1 month. Sterile plates and flasks for cell culture were coated with 0.0001% fibronectin (Sigma-Aldrich, F0895) diluted in PBS for 20 min and then aspirated off before culturing. To detach ECs, an Accutase cell detachment solution (BioLegend, 423201) was pipetted onto cells and incubated at 37°C for 5 min or until cells were no longer adherent.
EV enrichment
Human HSPC EV samples for proteomic analysis were enriched using the ExtraPEG method, as previously described and thoroughly characterized (73–75). Plasma EVs were further purified using an iodixanol density gradient (75–77). Final EV pellets were lysed in strong urea-containing lysis buffer [5% SDS, 10 mM EDTA, 120 mM tris-HCl (pH 6.8), and 8 M urea] with the addition of a protease inhibitor cocktail (Thermo Fisher Scientific). Vesicle protein was quantified using an EZQ Protein Quantitation Kit (Thermo Fisher Scientific). Murine HSPC EVs from serum-free cultures were isolated by differential centrifugation as described (18) and resuspended in particle-free PBS for functional assays. Single-particle analysis, including transmission electron microscopy and tunable resistance pulse sensing, was performed according to the minimal information for studies of EVs guidelines (18, 78).
EV uptake
To measure uptake, HSPC EVs were dyed with 40 μM CFSE for 10 min, then washed with PBS, and repelleted by ultracentrifugation at 100,000g for 2 hours. On the basis of our prior data supporting steady-state secretion of EVs at a ratio of 1 × 104 to 1 × 106 EVs per cell across numerous cell and tissue origins (74), doses for all EV uptake experiments were maintained at 104 to 105 EVs per cell. Four hours after addition of HSPC EVs to EC cultures, ECs were pelleted, resuspended in 0.01% trypsin, incubated for 1 min at 37°C, and washed with PBS. To further detach any surface-bound (but not endocytosed) vesicles, cells were resuspended in acid-wash buffer (0.5 M NaCl and 0.2 M acetic acid) for 1 min, pelleted, and resuspended in 1× PBS for fluorescence-activated cell sorting (FACS). Flow cytometric analysis was performed using BD FACSCanto II flow cytometer. For integrin α4β7 inhibition studies, HSPC EVs were incubated with 80 nM TR-14035 for either 30 min at room temperature or 2 hours at 4°C before adding to BM EC cultures. Endocytosis inhibition using dynasore (80 μM) was performed by pretreating ECs for 30 min before addition of HSPC EVs. For dose-dependent studies, increasing concentrations of HSPC EVs (2500 vesicles per cell, 7500 vesicles per cell, and 22,500 vesicles per cell) were added to ECs for 24 hours before cells were harvested, washed with PBS, and lysed for immunoblot analysis. For gene expression analyses, HSPC EVs (104 per cell) were added to ECs for 4 hours before washing cells in PBS for RNA extraction. To pharmacologically inhibit IKKα/IKKβ, ECs were treated with 1 μM ACHP for 30 min before the addition of HSPC EVs.
Confocal microscopy
BM ECs were seeded into a glass eight-well chamber slide and allowed to adhere for 2 days. For EV uptake visualization, HSPC EVs were dyed with 40 μM CFSE (Abcam, #ab113853) for 10 min, then washed with PBS, and repelleted by ultracentrifugation at 100,000g for 2 hours. EVs were transferred onto ECs and incubated at 37°C for 2 hours. Cells were washed with PBS and then stained with 1X CellMask Deep Red Plasma Membrane Stain (Invitrogen, #C10046) for 10 min at 37°C. Cells were then stained with Hoechst stain (2 μg/ml; Thermo Fisher Scientific #62249) for 20 min and washed three times before imaging. Imaging was performed on a CrestOptics X-Light V3 Spinning Disk Confocal using the 100× oil immersion objective lens (numerical aperture, 1.45) and 405-, 488-, and 640-nm lasers. Each image covered 133 μm by 133 μm per field. Three-dimensional (3D) images were constructed with the Fiji 3D viewer plugin set at 55% transparency.
For colocalization experiments, before staining, cells were blocked with 3% bovine serum albumin (BSA) in PBS for 1 hour. Cells were stained as follows: 1:50 with purified anti-mouse CD184 (CXCR4) antibody (BioLegend, #146502) in 3% BSA for 1 hour, 1:50 with chicken anti-rat IgG (H+L) cross-adsorbed secondary antibody, Alexa Fluor 488 (Invitrogen, #A-21470) in 3% BSA for 30 to 60 min, 1:25 with allophycocyanin anti-mouse CD192 (CCR2) antibody (BioLegend, #150628) in PBS for 30 min, and 4′,6-diamidino-2-phenylindole (1 μg/ml; BioLegend, #422801) for 5 min. Alternatively, cells were costained as follows: 1:50 with CCR2 polyclonal antibody (Invitrogen, #PA5-23043) and purified anti-mouse CD184 (CXCR4) antibody (BioLegend, #146502) for 1 hour in 3% BSA, followed by 1:50 donkey anti-rabbit secondary antibody Alexa Fluor 488 (Invitrogen, #A-21206), and goat anti-rat secondary antibody Alexa Fluor 647 (Invitrogen, #A-21247) for 1 hour in 3% BSA. Cells were washed with PBS three times in between stains. Imaging was performed on a CrestOptics X-Light V3 Spinning Disk Confocal using the 100× oil immersion objective lens (numerical aperture, 1.45) and 405-, 488-, and 640-nm lasers. Each image covered 133 μm by 133 μm per field. Using Fiji software, CCR2 intensity was measured using the CXCR4 channel to create regions of interest and overlay them onto the CCR2 channel. To measure co-occurrence of CXCR4 and CCR2 fluorescence, background signal was uniformly subtracted from all images before analysis. The Fiji plugin PTBIOP JACoP was used to generate thresholded area measurements. Manders’ and Pearson correlation coefficients of the two channels were generated using the Moments thresholding algorithm (79). Area overlap analysis was performed as described (80).
Transwell coculture assays
BM ECs were plated in fibronectin-coated tissue culture–treated plates in complete EC media and incubated overnight to allow adhesion of cells. A 1-μm pore transwell was placed in each well, and murine HSPCs were placed in the top chamber with additional media. Cells were cocultured without direct cell-cell contact in transwell dishes for 1 to 3 days at 37°C before resuspending HSPCs or scraping ECs for downstream analyses.
Chemotaxis assays
For transwell cell-to-cell chemotaxis assays, ECs were plated in fibronectin-coated tissue culture–treated wells and incubated overnight to allow adhesion of cells. The following day, HSPCs were dyed with 40 μM CFSE for 10 min at room temperature. A 5-μm pore transwell was placed in each well, and the HSPCs were placed in the top chamber with additional media. For cell-to-chemokine gradient chemotaxis assays, recombinant chemokines CXCL12, CCL2, CCL7, and CCL12 were added to HSPC ex vivo expansion medium lacking insulin-transferrin-selenium-ethanolamine, stem cell factor, and thrombopoietin for a final concentration of 200 ng/ml. HSPCs were dyed with 40 μM CFSE for 10 min at room temperature. A 5-μm pore transwell was placed in each well, and the HSPCs were placed in the top chamber with additional media. For inhibitor studies, HSPCs were treated with indicated inhibitor (CCR2 inhibitor, Sigma-Aldrich, 227016; integrin inhibitor, TR-14035, Tocris, 6908) or equal volume dimethyl sulfoxide before seeding into top chambers. Following an overnight incubation at 37°C, top and bottom chambers were harvested for flow cytometry and 10 μl of compensation beads was added to each sample as an internal control for quantification. Flow cytometric analysis was performed using a BD FACSCanto II flow cytometer. Chemotaxis ratio was calculated as: (Bottom chamber cell events/bead events)/[(Top chamber cell events/bead events) + (Bottom chamber events/bead events)].
Co-immunoprecipitation
Ex vivo cultured HSPCs grown in isolation, or in a 2-day EC transwell coculture were pelleted and washed with PBS three times before resuspension in ice-cold radioimmunoprecipitation assay (RIPA) buffer with protease inhibitor. Samples were incubated on ice for 10 min, sonicated three times for 5-s pulses each, and then centrifuged at 13,000g for 10 min at 4°C to clear supernatants. Equal volumes of lysates were aliquoted as input samples and boiled in 5X Laemmli sample buffer before loading equal volume into an SDS page gel for immunoblot analysis. Remaining lysates were incubated with anti-CXCR4 or control anti-IgG antibody for 1 hour at room temperature with gentle agitation. Pierce Protein A/G Magnetic Beads (Thermo Fisher Scientific, 88802) were precleared by washing four times with RIPA buffer, collecting magnetized beads after every step. Equal volume washed beads were added to immunocomplexes and incubated for 45 min at room temperature with gentle agitation. Beads were magnetized to separate from supernatant, and washed four times with RIPA buffer before resuspending in 2X reducing Laemmli sample buffer and boiling for 10 min. Beads were magnetized, and equal volume of supernatants was loaded into an SDS page gel for immunoblot analysis.
Immunoblot analysis
Cell and vesicle lysate preparation, protein quantification, and immunoblot analysis were performed as detailed (18). Ponceau S stain was used for total protein quantification. Primary and secondary antibodies used are listed in data S2. Gel images were taken using an Amersham ImageQuant 800 instrument and processed and quantitated using ImageQuant TL8.2.0.
Enzyme-linked immunosorbent assay
To measure secreted CXCL12 levels, HSPC EVs were added to BM EC cultures at a final dose of 104 to 105 HSPC EVs per cell. Five hours after EV treatment, supernatants from EC cultures were collected and analyzed by CXCL12 ELISA assay (Thermo Fisher Scientific, EMCXCL12) according to the manufacturer’s instructions.
Vesicle mass spectrometry
In-gel digestion and trypsinization of vesicle protein, mass spectrometry data acquisition, and data processing were performed as previously detailed (18). Data were filtered out at a false discovery rate (FDR) of 1% at precursor, peptide, and protein level and are available in data S1. Differential expression and enrichment analysis were performed in the DEP package (v3.16) in R (81) and ShinyGO v0.77 (82), respectively. All integrin subunits identified in the dataset were analyzed for fold change expression in HSPC EVs.
Generation of CRISPR-Cas9 KO ECs
To produce stable CRISPR-Cas9–mediated BMEC KO cell lines, single guide RNAs (sgRNAs) were designed using Synthego’s Knockout Guide Design tool (Ccl2, Ccl12, and Vcam-1) and IDT’s Custom Alt-R CRISPR-Cas9 guide RNA design tool (Ccl7). The sgRNAs were cloned into the lentiCRISPRv2GFP vector (Addgene, #82416) at the Bsm BI restriction site. Target sgRNA 5′ to 3′ sequences were as follows: Vcam-1 (AGCAATCGTTTTGTATTCAG), Ccl2 (AGTGGGGCGTTAACTGCATC), Ccl7 (TTCCCAGGGACACCGACTAC), and Ccl12 (ATTATAACAGCACGTGACTG). For lentiviral production, human embryonic kidney 293T cells were seeded in T-75 flasks and cultured to 80% confluency at the time of transfection. For each flask, 5 μg of lenti-sgRNA plasmid was cotransfected with 1.18 μg of pRSV-Rev (Addgene, #12253), 2.39 μg of pMDLg/pRRE (Addgene, #12251), and 1.42 μg of pCMV-VSV-G (Addgene, #8454) using CalFectin (SignaGen, #SL100478S) per the manufacturer’s instructions. Cell culture medium was replaced 24 hours after transfection. Viral particles were harvested from the supernatant 48 and 72 hours after transfection. The supernatant was centrifuged at 1000g for 10 min to pellet cells, filtered through a 0.45-μm filter, and stored at −80°C until transduction.
BM ECs were seeded in a six-well plate and cultured to 80% confluency at the time of transduction. Viral stocks were warmed to 37°C and transferred onto ECs for 24 hours before removing viral media and replacing with fresh complete EC media. ECs were then cultured for an additional 24 to 48 hours before passage to a T-25 flask. At confluency, GFP+ ECs were sorted on a BD FACSAria Fusion flow cytometer and then cultured in complete EC media. Gene KO was confirmed via RT-qPCR (see data S3 for primer sequences).
Electroporation
Transient overexpression of RelA or IκBα dominant-negative mutant was performed by electroporation of ECs using an Amaxa 4D-Nucleofector (Lonza) in the presence of RelA cFlag pcDNA3 (Addgene, #20012), pLXSN IkB alpha M (Addgene, #12330), or control vector c-Flag pcDNA3 (Addgene, #20011). Cells were then transferred to a six-well plate and incubated at 37°C overnight. Following incubation, ECs were harvested for immunoblot or gene expression analysis or seeded into bottom chambers for chemotaxis assays.
Gene expression analyses
Total RNA was extracted using the RNeasy Plus Micro Kit (QIAGEN) according to manufacturer’s instructions and quantified using a NanoPhotometer (Implen). RNA was converted into cDNA using SuperScript IV VILO Master Mix (Invitrogen). All RT-PCR reactions were performed using PowerUp SYBR Green Master Mix (Applied Biosystems) on a ViiA 7 Real-Time PCR System (Applied Biosystems). See data S3 for primer sequences. Chemokine transcriptional array analyses were performed using an RT2 Profiler PCR Array Mouse Cytokines & Chemokines (QIAGEN) according to the manufacturer’s instructions.
BM chemokine profiling
HSPC-derived EVs were injected into the left (ipsilateral) femur and PBS into the right (contralateral) femur of WT mice (n = 4). Twenty hours after injections, bilateral femurs were harvested and flushed into 100 μl of PBS. Cells were pelleted by centrifugation at 400g for 5 min, and frozen supernatants were analyzed by a Cytokine/Chemokine 44-Plex Discovery Assay Array (MD44) (Eve Technologies).
BM homing
WT homing assays were performed following 14-day ex vivo culture of murine HSPCs. Before injection, donor cells were labeled with 40 μM CFSE dye (Abcam, ab113853) for 10 min and washed with PBS by centrifugation at 400g for 5 min. For homing experiments with EV preconditioning, 109 to 1010 HSPC-derived EVs were injected into the left (ipsilateral) femur and PBS into the right (contralateral) femur. Four hours after the intrafemoral injection, 6 × 106 to 10 × 106 total murine CFSE-labeled HSPCs were injected intravenously through the tail vein. For homing experiments with CCR2 ligands, mice were preconditioned via intrafemoral injection of either recombinant mouse CCL2 (0.1 or 1 ng), CCL7 (0.1 ng), or CCL12 (0.1 ng). Immediately following the intrafemoral injection, 6 × 106 to 10 × 106 total murine CFSE-labeled HSPCs were injected intravenously through the tail vein. For all homing experiments, femurs were collected and flushed 20 hours after transplantation. Relative proportions of CFSE-positive cells and HSPC sub-populations were analyzed using a BD LSRFortessa flow cytometer. For EC collection, CD45(−) CD31(+) ECs were sorted using FACSAria Fusion Sorter. In competitive homing experiments, fresh whole BM (WBM) from CD45.2 WT and Ccr2gfp/gfp KI/KO mice (with or without CFSE labeling) was injected intravenously (1:1 ratio, 107 total cells) following red blood cell lysis into sublethally irradiated (5.2 Gy) CD45.1 mice. For homing assays using GFP− versus GFP+ cells, BM from Ccr2gfp/gfp was flushed and lineage-depleted before FACS sorting for GFP- and GFP+ cells. Sublethally irradiated (5.2 Gy) recipient mice were injected with 2 × 106 total cells before BM analysis 20 hours later. At end-point analyses, BM from leg bones and spleens were harvested from recipient mice for analysis of relative chimerism and subpopulation frequency by a BD LSRFortessa flow cytometer.
Short-term engraftment
For competitive engraftment of CD45.1/0.2 WT versus CD45.2 Ccr2gfp/gfp marrow, a total of 1 × 106 WBM cells were transplanted (1:1) into lethally irradiated (10.4 Gy) CD45.1 recipient mice. To account for CD45 allele variation, a control cohort was included where CD45.1/0.2 WT was competed against CD45.2 WT (1:1). PB was collected by retro-orbital bleed at 2 weeks to assess chimerism levels. At 12 weeks after transplantation, PB (cardiac puncture), spleens, and BM were collected and single-cell suspensions were analyzed by flow cytometry. For EV/chemokine conditioning engraftment experiments, non-myeloablated CD45.1 mice were preconditioned via intrafemoral injection of either 108 to 109 HSPC EVs or 1 ng of recombinant CCL2 (ipsilateral femur) versus a sham PBS injection (contralateral femur). A total of 107 BM cells from CD45.2 mice were injected intravenously (via tail vein) immediately following CCL2 conditioning or 4 hours after EV conditioning. After 4 weeks, BM was harvested from bilateral femurs separately. Proportions of CD45.2 donor cells and subpopulations were analyzed using a BD LSRFortessa flow cytometer.
In vivo Cre-TdTomato recombination
Cre mRNA containing EVs were harvested from conditioned media of ex vivo cultures of murine Vav1-Cre HSPCs. Ai14 (tdTomato) mice received intrafemoral injection of 108 HSPC Cre EVs into the ipsilateral femur and a sham PBS injection into the contralateral femur. After 11 days, BM was harvested from bilateral leg bones separately. Flow cytometry was used to detect tdTomato-positive CD45(−)CD31(+) ECs.
Statistics
Statistical significance between two groups was measured by unpaired Student’s t test and by ANOVA with Bonferroni correction for experiments with three or more groups. Normal distribution was tested by Kolmogorov-Smirnov test before parametric testing. A Grubb’s test (extreme Studentized deviate method) was used to evaluate statistical outliers. Statistical analyses were conducted using R version 4.2.2, GraphPad, or Microsoft Excel. Bar plots reflect the means ± SEM.
Study approval
Animal experiments were approved by the Children’s Hospital of Philadelphia Institutional Animal Care and Use Committee (IACUC) and conducted in accordance with the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals. Human HSPCs were enriched from deidentified remnant human leukapheresis products obtained from the University of Pennsylvania (UPenn) Hematopoietic Stem Cell Laboratory.
Acknowledgments
We thank the Children’s Hospital of Philadelphia (CHOP) Flow Cytometry core laboratory, CHOP Department of Veterinary Resources, the CHOP-PENN Proteomics Core Facility, and the Penn Cell and Developmental Biology (CDB) Microscopy Core. We additionally thank the University of Pennsylvania Hematopoietic Stem Cell Laboratory for donation of de-identified donor-derived samples (IRB exempt).
Funding: This work was supported by the National Institutes of Health grants R01HL164633 (P.K.), 4-R38-HL143613-04 (to S.N.H.), and T32 HL-07439 (to S.N.H.).
Author contributions: Conceptualization: S.N.H., J.M.B., and P.K. Methodology: S.N.H., D.R.K., S.K.J., and P.K. Investigation: S.N.H., D.R.K., S.K.J., and J.J.C. Formal analysis: S.N.H., D.R.K., and S.K.J. Visualization: D.R.K. and S.N.H. Supervision: S.N.H. and P.K. Writing—original draft: S.N.H. Writing—review and editing: S.N.H., D.R.K., S.K.J., J.J.C., J.M.B., and P.K.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Proteomic data are available in the ProteomeXchange Consortium via the PRIDE (83) partner repository.
Supplementary Materials
The PDF file includes:
Figs. S1 to S7
Legend for movie S1
Legends for data S1 to S3
Other Supplementary Material for this manuscript includes the following:
Movie S1
Data S1 to S3
REFERENCES AND NOTES
- 1.Nombela-Arrieta C., Pivarnik G., Winkel B., Canty K. J., Harley B., Mahoney J. E., Park S.-Y., Lu J., Protopopov A., Silberstein L. E., Quantitative imaging of haematopoietic stem and progenitor cell localization and hypoxic status in the bone marrow microenvironment. Nat. Cell Biol. 15, 533–543 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wright D. E., Wagers A. J., Gulati A. P., Johnson F. L., Weissman I. L., Physiological migration of hematopoietic stem and progenitor cells. Science 294, 1933–1936 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Massberg S., Schaerli P., Knezevic-Maramica I., Köllnberger M., Tubo N., Moseman E. A., Huff I. V., Junt T., Wagers A. J., Mazo I. B., von Andrian U. H., Immunosurveillance by hematopoietic progenitor cells trafficking through blood, lymph, and peripheral tissues. Cell 131, 994–1008 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katayama Y., Battista M., Kao W.-M., Hidalgo A., Peired A. J., Thomas S. A., Frenette P. S., Signals from the sympathetic nervous system regulate hematopoietic stem cell egress from bone marrow. Cell 124, 407–421 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Watts K. L., Adair J., Kiem H.-P., Hematopoietic stem cell expansion and gene therapy. Cytotherapy 13, 1164–1171 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simione A. J., das Neves H. R. A., da Silva C. C., da Silva Sabaini P. M., da Silva Santos Geraldo B. L., Pasquini M. C., Vigorito A. C., Ammi M., Colturato V. A. R., Nabhan S. K., Seber A., Silvério A., Moreira M. C. R., Barros G. M. N., Astigarraga C. C., Daudt L. E., de Almeida Macedo M. C. M., Chiattone R., Novis Y. A. S., Fernandes J. F., Vilela V. A. L., Lerner D., de Almeida Soares R. D., Scheinberg P., Teixeira G. M., Arrais-Rodrigues C., Colella M. P., da Silva R. L., Funke V. A. M., Arcuri L. J., Hamerschlak N., Filho J. S., de Molla V. C., de Holanda Farias J. S., Pasquini R., Bonfim C. M. S., Neto A. E. H., Calixto R. F., Bouzas L. F., Duarte F. B., Current use and outcome of hematopoietic stem cell transplantation: CIBMTR summary slides. Biol. Blood Marrow Transplant. 3, 171 (2022). [Google Scholar]
- 7.Frenette P. S., Subbarao S., Mazo I. B., von Andrian U. H., Wagner D. D., Endothelial selectins and vascular cell adhesion molecule-1 promote hematopoietic progenitor homing to bone marrow. Proc. Natl. Acad. Sci. U.S.A. 95, 14423–14428 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Buul J. D., Voermans C., van den Berg V., Anthony E. C., Mul F. P. J., van Wetering S., van der Schoot C. E., Hordijk P. L., Migration of human hematopoietic progenitor cells across bone marrow endothelium is regulated by vascular endothelial cadherin. J. Immunol. 168, 588–596 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Rademakers T., Goedhart M., Hoogenboezem M., García Ponce A., van Rijssel J., Samus M., Schnoor M., Butz S., Huveneers S., Vestweber D., Nolte M. A., Voermans C., van Buul J. D., Hematopoietic stem and progenitor cells use podosomes to transcellularly cross the bone marrow endothelium. Haematologica 105, 2746–2756 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalafati L., Chavakis T., Hematopoietic stem and progenitor cells take the route through the bone marrow endothelium. Haematologica 105, 2700–2701 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamplin O. J., Durand E. M., Carr L. A., Childs S. J., Hagedorn E. J., Li P., Yzaguirre A. D., Speck N. A., Zon L. I., Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell 160, 241–252 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salomon C., Das S., Erdbrügger U., Kalluri R., Lim S. K., Olefsky J. M., Rice G. E., Sahoo S., Tao W. A., Vader P., Wang Q., Weaver A. M., Extracellular vesicles and their emerging roles as cellular messengers in endocrinology: An endocrine society scientific statement. Endocr. Rev. 43, 441–468 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costa-Silva B., Aiello N. M., Ocean A. J., Singh S., Zhang H., Thakur B. K., Becker A., Hoshino A., Mark M. T., Molina H., Xiang J., Zhang T., Theilen T.-M., García-Santos G., Williams C., Ararso Y., Huang Y., Rodrigues G., Shen T.-L., Labori K. J., Lothe I. M. B., Kure E. H., Hernandez J., Doussot A., Ebbesen S. H., Grandgenett P. M., Hollingsworth M. A., Jain M., Mallya K., Batra S. K., Jarnagin W. R., Schwartz R. E., Matei I., Peinado H., Stanger B. Z., Bromberg J., Lyden D., Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 17, 816–826 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoshino A., Costa-Silva B., Shen T.-L., Rodrigues G., Hashimoto A., Tesic Mark M., Molina H., Kohsaka S., Di Giannatale A., Ceder S., Singh S., Williams C., Soplop N., Uryu K., Pharmer L., King T., Bojmar L., Davies A. E., Ararso Y., Zhang T., Zhang H., Hernandez J., Weiss J. M., Dumont-Cole V. D., Kramer K., Wexler L. H., Narendran A., Schwartz G. K., Healey J. H., Sandstrom P., Labori K. J., Kure E. H., Grandgenett P. M., Hollingsworth M. A., de Sousa M., Kaur S., Jain M., Mallya K., Batra S. K., Jarnagin W. R., Brady M. S., Fodstad O., Muller V., Pantel K., Minn A. J., Bissell M. J., Garcia B. A., Kang Y., Rajasekhar V. K., Ghajar C. M., Matei I., Peinado H., Bromberg J., Lyden D., Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahoo S., Klychko E., Thorne T., Misener S., Schultz K. M., Millay M., Ito A., Liu T., Kamide C., Agrawal H., Perlman H., Qin G., Kishore R., Losordo D. W., Exosomes from human CD34+ stem cells mediate their proangiogenic paracrine activity. Circ. Res. 109, 724–728 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ratajczak J., Kucia M., Mierzejewska K., Marlicz W., Pietrzkowski Z., Wojakowski W., Greco N. J., Tendera M., Ratajczak M. Z., Paracrine proangiopoietic effects of human umbilical cord blood-derived purified CD133+ cells—Implications for stem cell therapies in regenerative medicine. Stem Cells Dev. 22, 422–430 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathiyalagan P., Liang Y., Kim D., Misener S., Thorne T., Kamide C. E., Klyachko E., Losordo D. W., Hajjar R. J., Sahoo S., Angiogenic mechanisms of human CD34+ stem cell exosomes in the repair of ischemic hindlimb. Circ. Res. 120, 1466–1476 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurwitz S. N., Jung S. K., Kobulsky D. R., Fazelinia H., Spruce L. A., Baltasar-Pérez E., Groen N., Mesaros C., Kurre P., Neutral sphingomyelinase blockade enhances hematopoietic stem cell fitness through an integrated stress response. Blood 142, 1708–1723 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erle D. J., Briskin M. J., Butcher E. C., Garcia-Pardo A., Lazarovits A. I., Tidswell M., Expression and function of the MAdCAM-1 receptor, integrin alpha 4 beta 7, on human leukocytes. J. Immunol. 153, 517–528 (1994). [PubMed] [Google Scholar]
- 20.Yu Y., Zhu J., Mi L.-Z., Walz T., Sun H., Chen J., Springer T. A., Structural specializations of α4β7, an integrin that mediates rolling adhesion. J. Cell Biol. 196, 131–146 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu M., Kenific C. M., Boudreau N., Lyden D., Tumor-derived nanoseeds condition the soil for metastatic organotropism. Semin. Cancer Biol. 93, 70–82 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkinson A. C., Ishida R., Nakauchi H., Yamazaki S., Long-term ex vivo expansion of mouse hematopoietic stem cells. Nat. Protoc. 15, 628–648 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poulos M. G., Crowley M. J. P., Gutkin M. C., Ramalingam P., Schachterle W., Thomas J.-L., Elemento O., Butler J. M., Vascular platform to define hematopoietic stem cell factors and enhance regenerative hematopoiesis. Stem Cell Rep. 5, 881–894 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poulos M. G., Ramalingam P., Gutkin M. C., Llanos P., Gilleran K., Rabbany S. Y., Butler J. M., Endothelial transplantation rejuvenates aged hematopoietic stem cell function. J. Clin. Invest. 127, 4163–4178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fitzgerald W., Freeman M. L., Lederman M. M., Vasilieva E., Romero R., Margolis L., A system of cytokines encapsulated in extracellular vesicles. Sci. Rep. 8, 8973 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buzas E. I., The roles of extracellular vesicles in the immune system. Nat. Rev. Immunol. 23, 236–250 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richmond A., NF-κB, chemokine gene transcription and tumour growth. Nat. Rev. Immunol. 2, 664–674 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murata T., Shimada M., Sakakibara S., Yoshino T., Kadono H., Masuda T., Shimazaki M., Shintani T., Fuchikami K., Sakai K., Inbe H., Takeshita K., Niki T., Umeda M., Bacon K. B., Ziegelbauer K. B., Lowinger T. B., Discovery of novel and selective IKK-beta serine-threonine protein kinase inhibitors. Bioorg. Med. Chem. Lett. 13, 913–918 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Sanda T., Iida S., Ogura H., Asamitsu K., Murata T., Bacon K. B., Ueda R., Okamoto T., Growth inhibition of multiple myeloma cells by a novel IκB kinase inhibitor. Clin. Cancer Res. 11, 1974–1982 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Sohy D., Parmentier M., Springael J.-Y., Allosteric transinhibition by specific antagonists in CCR2/CXCR4 heterodimers*. J. Biol. Chem. 282, 30062–30069 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Manders E. M. M., Verbeek F. J., Aten J. A., Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 169, 375–382 (1993). [DOI] [PubMed] [Google Scholar]
- 32.Dunn K. W., Kamocka M. M., McDonald J. H., A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 300, C723–C742 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Satpathy A. T., Briseño C. G., Lee J. S., Ng D., Manieri N. A., Kc W., Wu X., Thomas S. R., Lee W.-L., Turkoz M., McDonald K. G., Meredith M. M., Song C., Guidos C. J., Newberry R. D., Ouyang W., Murphy T. L., Stappenbeck T. S., Gommerman J. L., Nussenzweig M. C., Colonna M., Kopan R., Murphy K. M., Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat. Immunol. 14, 937–948 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waterstrat A., Liang Y., Swiderski C. F., Shelton B. J., Van Zant G., Congenic interval of CD45/Ly-5 congenic mice contains multiple genes that may influence hematopoietic stem cell engraftment. Blood 115, 408–417 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dutta P., Sager H. B., Stengel K. R., Naxerova K., Courties G., Saez B., Silberstein L., Heidt T., Sebas M., Sun Y., Wojtkiewicz G., Feruglio P. F., King K., Baker J. N., van der Laan A. M., Borodovsky A., Fitzgerald K., Hulsmans M., Hoyer F., Iwamoto Y., Vinegoni C., Brown D., Di Carli M., Libby P., Hiebert S., Scadden D., Swirski F. K., Weissleder R., Nahrendorf M., Myocardial infarction activates CCR2+ hematopoietic stem and progenitor cells. Cell Stem Cell 16, 477–487 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medina-Ruiz L., Bartolini R., Wilson G. J., Dyer D. P., Vidler F., Hughes C. E., Schuette F., Love S., Pingen M., Hayes A. J., Fu J., Stewart A. F., Graham G. J., Analysis of combinatorial chemokine receptor expression dynamics using multi-receptor reporter mice. eLife 11, e72418 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liles W. C., Broxmeyer H. E., Rodger E., Wood B., Hübel K., Cooper S., Hangoc G., Bridger G. J., Henson G. W., Calandra G., Dale D. C., Mobilization of hematopoietic progenitor cells in healthy volunteers by AMD3100, a CXCR4 antagonist. Blood 102, 2728–2730 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Biondi M., Tettamanti S., Galimberti S., Cerina B., Tomasoni C., Piazza R., Donsante S., Bido S., Perriello V. M., Broccoli V., Doni A., Dazzi F., Mantovani A., Dotti G., Biondi A., Pievani A., Serafini M., Selective homing of CAR-CIK cells to the bone marrow niche enhances control of the acute myeloid leukemia burden. Blood 141, 2587–2598 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levy E., Reger R., Segerberg F., Lambert M., Leijonhufvud C., Baumer Y., Carlsten M., Childs R., Enhanced bone marrow homing of natural killer cells following mRNA transfection with gain-of-function variant CXCR4R334X. Front. Immunol. 10, 1262 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ridder K., Keller S., Dams M., Rupp A.-K., Schlaudraff J., Del Turco D., Starmann J., Macas J., Karpova D., Devraj K., Depboylu C., Landfried B., Arnold B., Plate K. H., Höglinger G., Sültmann H., Altevogt P., Momma S., Extracellular vesicle-mediated transfer of genetic information between the hematopoietic system and the brain in response to inflammation. PLOS Biol. 12, e1001874 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zomer A., Maynard C., Verweij F. J., Kamermans A., Schäfer R., Beerling E., Schiffelers R. M., de Wit E., Berenguer J., Ellenbroek S. I. J., Wurdinger T., Pegtel D. M., van Rheenen J., In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell 161, 1046–1057 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Czechowicz A., Palchaudhuri R., Scheck A., Hu Y., Hoggatt J., Saez B., Pang W. W., Mansour M. K., Tate T. A., Chan Y. Y., Walck E., Wernig G., Shizuru J. A., Winau F., Scadden D. T., Rossi D. J., Selective hematopoietic stem cell ablation using CD117-antibody-drug-conjugates enables safe and effective transplantation with immunity preservation. Nat. Commun. 10, 617 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uchida N., Stasula U., Demirci S., Germino-Watnick P., Hinds M., Le A., Chu R., Berg A., Liu X., Su L., Wu X., Krouse A. E., Linde N. S., Bonifacino A., Hong S. G., Dunbar C. E., Lanieri L., Bhat A., Palchaudhuri R., Bennet B., Hoban M., Bertelsen K., Olson L. M., Donahue R. E., Tisdale J. F., Fertility-preserving myeloablative conditioning using single-dose CD117 antibody-drug conjugate in a rhesus gene therapy model. Nat. Commun. 14, 6291 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saha A., Hyzy S., Lamothe T., Hammond K., Clark N., Lanieri L., Bhattarai P., Palchaudhuri R., Gillard G. O., Proctor J., Riddle M. J., Panoskaltsis-Mortari A., MacMillan M. L., Wagner J. E., Kiem H.-P., Olson L. M., Blazar B. R., A CD45-targeted antibody-drug conjugate successfully conditions for allogeneic hematopoietic stem cell transplantation in mice. Blood 139, 1743–1759 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Breda L., Papp T. E., Triebwasser M. P., Yadegari A., Fedorky M. T., Tanaka N., Abdulmalik O., Pavani G., Wang Y., Grupp S. A., Chou S. T., Ni H., Mui B. L., Tam Y. K., Weissman D., Rivella S., Parhiz H., In vivo hematopoietic stem cell modification by mRNA delivery. Science 381, 436–443 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGrath K. E., Koniski A. D., Maltby K. M., McGann J. K., Palis J., Embryonic expression and function of the chemokine SDF-1 and its receptor, CXCR4. Dev. Biol. 213, 442–456 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Möhle R., Bautz F., Rafii S., Moore M. A., Brugger W., Kanz L., The chemokine receptor CXCR-4 is expressed on CD34+ hematopoietic progenitors and leukemic cells and mediates transendothelial migration induced by stromal cell-derived factor-1. Blood 91, 4523–4530 (1998). [PubMed] [Google Scholar]
- 48.Wright D. E., Bowman E. P., Wagers A. J., Butcher E. C., Weissman I. L., Hematopoietic stem cells are uniquely selective in their migratory response to chemokines. J. Exp. Med. 195, 1145–1154 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnston B., Burns A. R., Suematsu M., Issekutz T. B., Woodman R. C., Kubes P., Chronic inflammation upregulates chemokine receptors and induces neutrophil migration to monocyte chemoattractant protein-1. J. Clin. Invest. 103, 1269–1276 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reichel C. A., Khandoga A., Anders H.-J., Schlöndorff D., Luckow B., Krombach F., Chemokine receptors Ccr1, Ccr2, and Ccr5 mediate neutrophil migration to postischemic tissue. J. Leukoc. Biol. 79, 114–122 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Fujimura N., Xu B., Dalman J., Deng H., Aoyama K., Dalman R. L., CCR2 inhibition sequesters multiple subsets of leukocytes in the bone marrow. Sci. Rep. 5, 11664 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feterowski C., Mack M., Weighardt H., Bartsch B., Kaiser-Moore S., Holzmann B., CC chemokine receptor 2 regulates leukocyte recruitment and IL-10 production during acute polymicrobial sepsis. Eur. J. Immunol. 34, 3664–3673 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Si Y., Tsou C.-L., Croft K., Charo I. F., CCR2 mediates hematopoietic stem and progenitor cell trafficking to sites of inflammation in mice. J. Clin. Invest. 120, 1192–1203 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Percherancier Y., Berchiche Y. A., Slight I., Volkmer-Engert R., Tamamura H., Fujii N., Bouvier M., Heveker N., Bioluminescence resonance energy transfer reveals ligand-induced conformational changes in CXCR4 homo- and heterodimers. J. Biol. Chem. 280, 9895–9903 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Sohy D., Yano H., de Nadai P., Urizar E., Guillabert A., Javitch J. A., Parmentier M., Springael J.-Y., Hetero-oligomerization of CCR2, CCR5, and CXCR4 and the protean effects of “Selective” antagonists. J. Biol. Chem. 284, 31270–31279 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jung H., Mithal D. S., Park J. E., Miller R. J., Localized CCR2 activation in the bone marrow niche mobilizes monocytes by desensitizing CXCR4. PLOS One 10, e0128387 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mellado M., Rodríguez-Frade J. M., Vila-Coro A. J., Fernández S., Martín de Ana A., Jones D. R., Torán J. L., Martínez-A C., Chemokine receptor homo- or heterodimerization activates distinct signaling pathways. EMBO J. 20, 2497–2507 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pereira da Costa M., Minutti C. M., Piot C., Giampazolias E., Cardoso A., Cabeza-Cabrerizo M., Rogers N. C., Lebrusant-Fernandez M., Iliakis C. S., Wack A., Sousa C. R. E., Interplay between CXCR4 and CCR2 regulates bone marrow exit of dendritic cell progenitors. Cell Rep. 42, 112881 (2023). [DOI] [PubMed] [Google Scholar]
- 59.Wang Y., Cui L., Gonsiorek W., Min S.-H., Anilkumar G., Rosenblum S., Kozlowski J., Lundell D., Fine J. S., Grant E. P., CCR2 and CXCR4 regulate peripheral blood monocyte pharmacodynamics and link to efficacy in experimental autoimmune encephalomyelitis. J. Inflamm. 6, 32 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Melenhorst J. J., Tian X., Xu D., Sandler N. G., Scheinberg P., Biancotto A., Scheinberg P., McCoy J. P., Hensel N. F., McIver Z., Douek D. C., Barrett A. J., Cytopenia and leukocyte recovery shape cytokine fluctuations after myeloablative allogeneic hematopoietic stem cell transplantation. Haematologica 97, 867–873 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neehus A.-L., Carey B., Landekic M., Panikulam P., Deutsch G., Ogishi M., Arango-Franco C. A., Philippot Q., Modaresi M., Mohammadzadeh I., Berndt M. C., Rinchai D., Le Voyer T., Rosain J., Momenilandi M., Martin-Fernandez M., Khan T., Bohlen J., Han J. E., Deslys A., Bernard M., Gajardo-Carrasco T., Soudée C., Le Floc’h C., Migaud M., Seeleuthner Y., Jang M.-S., Nikolouli E., Seyedpour S., Begueret H., Emile J.-F., Le Guen P., Tavazzi G., Colombo C. N. J., Marzani F. C., Angelini M., Trespidi F., Ghirardello S., Alipour N., Molitor A., Carapito R., Mazloomrezaei M., Rokni-Zadeh H., Changi-Ashtiani M., Brouzes C., Vargas P., Borghesi A., Lachmann N., Bahram S., Crestani B., Pahari S., Schlesinger L. S., Marr N., Bugonovic D., Boisson-Dupuis S., Béziat V., Abel L., Borie R., Young L. R., Deterding R., Shahrooei M., Rezaei N., Parvaneh N., Craven D., Gros P., Malo D., Sepulveda F. E., Nogee L. M., Aladjidi N., Trapnell B. C., Casanova J.-L., Bustamante J., Human inherited CCR2 deficiency underlies progressive polycystic lung disease. Cell 187, 390–408.e23 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meng A., Wang Y., Brown S. A., Zant G. V., Zhou D., Ionizing radiation and busulfan inhibit murine bone marrow cell hematopoietic function via apoptosis-dependent and -independent mechanisms. Exp. Hematol. 31, 1348–1356 (2003). [DOI] [PubMed] [Google Scholar]
- 63.Cachaço A. S., Carvalho T., Santos A. C., Igreja C., Fragoso R., Osório C., Ferreira M., Serpa J., Correia S., Pinto-do-Ó P., Dias S., TNF-α regulates the effects of irradiation in the mouse bone marrow microenvironment. PLOS One 5, e8980 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang L., Jiang J., Chen Y., Jia Q., Chu Q., The roles of CC chemokines in response to radiation. Radiat. Oncol. 17, 63 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poulos M. G., Ramalingam P., Gutkin M. C., Kleppe M., Ginsberg M., Crowley M. J. P., Elemento O., Levine R. L., Rafii S., Kitajewski J., Greenblatt M. B., Shim J.-H., Butler J. M., Endothelial-specific inhibition of NF-κB enhances functional haematopoiesis. Nat. Commun. 7, 13829 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pietras E. M., Mirantes-Barbeito C., Fong S., Loeffler D., Kovtonyuk L. V., Zhang S., Lakshminarasimhan R., Chin C. P., Techner J.-M., Will B., Nerlov C., Steidl U., Manz M. G., Schroeder T., Passegué E., Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat. Cell Biol. 18, 607–618 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Isaioglou I., Aldehaiman M. M., Li Y., Lahcen A. A., Rauf S., Al-Amoodi A. S., Habiba U., Alghamdi A., Nozue S., Habuchi S., Salama K. N., Merzaban J. S., CD34+ HSPCs-derived exosomes contain dynamic cargo and promote their migration through functional binding with the homing receptor E-selectin. Front. Cell Dev. Biol. 11, 1149912 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meng W., He C., Hao Y., Wang L., Li L., Zhu G., Prospects and challenges of extracellular vesicle-based drug delivery system: Considering cell source. Drug Deliv. 27, 585–598 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boyd A. L., Campbell C. J. V., Hopkins C. I., Fiebig-Comyn A., Russell J., Ulemek J., Foley R., Leber B., Xenocostas A., Collins T. J., Bhatia M., Niche displacement of human leukemic stem cells uniquely allows their competitive replacement with healthy HSPCs. J. Exp. Med. 211, 1925–1935 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jacamo R. O., Mu H., Zhang Q., Chachad D., Zhiqiang W., Ma W., Zhang M., Mak P. Y., Mak D., Ruvolo P., McQueen T., Lowe S., Zuber J., Eulberg D., Kruschinski A., Konopleva M., Davis R. E., Andreeff M., Effects of CCL2/CCR2 blockade in acute myeloid leukemia. Blood 126, 1348 (2015). [Google Scholar]
- 71.Macanas-Pirard P., Quezada T., Navarrete L., Broekhuizen R., Leisewitz A., Nervi B., Ramírez P. A., The CCL2/CCR2 axis affects transmigration and proliferation but not resistance to chemotherapy of acute myeloid leukemia cells. PLOS One 12, e0168888 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zlotnik A., Yoshie O., Nomiyama H., The chemokine and chemokine receptor superfamilies and their molecular evolution. Genome Biol. 7, 243 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rider M. A., Hurwitz S. N., Meckes D. G., ExtraPEG: A polyethylene glycol-based method for enrichment of extracellular vesicles. Sci. Rep. 6, 23978 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hurwitz S. N., Conlon M. M., Rider M. A., Brownstein N. C., Meckes D. G., Nanoparticle analysis sheds budding insights into genetic drivers of extracellular vesicle biogenesis. J. Extracell. Vesicles 5, 31295 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hurwitz S. N., Meckes D. G., An adaptable polyethylene glycol-based workflow for proteomic analysis of extracellular vesicles. Methods Mol. Biol. 1660, 303–317 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Kowal J., Arras G., Colombo M., Jouve M., Morath J. P., Primdal-Bengtson B., Dingli F., Loew D., Tkach M., Théry C., Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. U.S.A. 113, E968–E977 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hurwitz S. N., Cheerathodi M. R., Nkosi D., York S. B., Meckes D. G. Jr., Tetraspanin CD63 bridges autophagic and endosomal processes to regulate exosomal secretion and intracellular signaling of epstein-barr virus LMP1. J. Virol. 92, e01969-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Théry C., Witwer K. W., Aikawa E., Alcaraz M. J., Anderson J. D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G. K., Ayre D. C., Bach J.-M., Bachurski D., Baharvand H., Balaj L., Baldacchino S., Bauer N. N., Baxter A. A., Bebawy M., Beckham C., Zavec A. B., Benmoussa A., Berardi A. C., Bergese P., Bielska E., Blenkiron C., Bobis-Wozowicz S., Boilard E., Boireau W., Bongiovanni A., Borràs F. E., Bosch S., Boulanger C. M., Breakefield X., Breglio A. M., Brennan M. Á., Brigstock D. R., Brisson A., Broekman M. L., Bromberg J. F., Bryl-Górecka P., Buch S., Buck A. H., Burger D., Busatto S., Buschmann D., Bussolati B., Buzás E. I., Byrd J. B., Camussi G., Carter D. R., Caruso S., Chamley L. W., Chang Y.-T., Chen C., Chen S., Cheng L., Chin A. R., Clayton A., Clerici S. P., Cocks A., Cocucci E., Coffey R. J., Cordeiro-da-Silva A., Couch Y., Coumans F. A., Coyle B., Crescitelli R., Criado M. F., D’Souza-Schorey C., Das S., Chaudhuri A. D., de Candia P., De Santana E. F., De Wever O., Del Portillo H. A., Demaret T., Deville S., Devitt A., Dhondt B., Di Vizio D., Dieterich L. C., Dolo V., Rubio A. P. D., Dominici M., Dourado M. R., Driedonks T. A., Duarte F. V., Duncan H. M., Eichenberger R. M., Ekström K., El Andaloussi S., Elie-Caille C., Erdbrügger U., Falcón-Pérez J. M., Fatima F., Fish J. E., Flores-Bellver M., Försönits A., Frelet-Barrand A., Fricke F., Fuhrmann G., Gabrielsson S., Gámez-Valero A., Gardiner C., Gärtner K., Gaudin R., Gho Y. S., Giebel B., Gilbert C., Gimona M., Giusti I., Goberdhan D. C., Görgens A., Gorski S. M., Greening D. W., Gross J. C., Gualerzi A., Gupta G. N., Gustafson D., Handberg A., Haraszti R. A., Harrison P., Hegyesi H., Hendrix A., Hill A. F., Hochberg F. H., Hoffmann K. F., Holder B., Holthofer H., Hosseinkhani B., Hu G., Huang Y., Huber V., Hunt S., Ibrahim A. G.-E., Ikezu T., Inal J. M., Isin M., Ivanova A., Jackson H. K., Jacobsen S., Jay S. M., Jayachandran M., Jenster G., Jiang L., Johnson S. M., Jones J. C., Jong A., Jovanovic-Talisman T., Jung S., Kalluri R., Kano S.-I., Kaur S., Kawamura Y., Keller E. T., Khamari D., Khomyakova E., Khvorova A., Kierulf P., Kim K. P., Kislinger T., Klingeborn M., Klinke D. J., Kornek M., Kosanović M. M., Kovács Á. F., Krämer-Albers E.-M., Krasemann S., Krause M., Kurochkin I. V., Kusuma G. D., Kuypers S., Laitinen S., Langevin S. M., Languino L. R., Lannigan J., Lässer C., Laurent L. C., Lavieu G., Lázaro-Ibáñez E., Le Lay S., Lee M.-S., Lee Y. X. F., Lemos D. S., Lenassi M., Leszczynska A., Li I. T., Liao K., Libregts S. F., Ligeti E., Lim R., Lim S. K., Linē A., Linnemannstöns K., Llorente A., Lombard C. A., Lorenowicz M. J., Lörincz Á. M., Lötvall J., Lovett J., Lowry M. C., Loyer X., Lu Q., Lukomska B., Lunavat T. R., Maas S. L., Malhi H., Marcilla A., Mariani J., Mariscal J., Martens-Uzunova E. S., Martin-Jaular L., Martinez M. C., Martins V. R., Mathieu M., Mathivanan S., Maugeri M., McGinnis L. K., McVey M. J., Meckes D. G., Meehan K. L., Mertens I., Minciacchi V. R., Möller A., Jørgensen M. M., Morales-Kastresana A., Morhayim J., Mullier F., Muraca M., Musante L., Mussack V., Muth D. C., Myburgh K. H., Najrana T., Nawaz M., Nazarenko I., Nejsum P., Neri C., Neri T., Nieuwland R., Nimrichter L., Nolan J. P., Hoen E. N. M. N., Hooten N. N., O’Driscoll L., O’Grady T., O’Loghlen A., Ochiya T., Olivier M., Ortiz A., Ortiz L. A., Osteikoetxea X., Østergaard O., Ostrowski M., Park J., Pegtel D. M., Peinado H., Perut F., Pfaffl M. W., Phinney D. G., Pieters B. C., Pink R. C., Pisetsky D. S., von Strandmann E. P., Polakovicova I., Poon I. K., Powell B. H., Prada I., Pulliam L., Quesenberry P., Radeghieri A., Raffai R. L., Raimondo S., Rak J., Ramirez M. I., Raposo G., Rayyan M. S., Regev-Rudzki N., Ricklefs F. L., Robbins P. D., Roberts D. D., Rodrigues S. C., Rohde E., Rome S., Rouschop K. M., Rughetti A., Russell A. E., Saá P., Sahoo S., Salas-Huenuleo E., Sánchez C., Saugstad J. A., Saul M. J., Schiffelers R. M., Schneider R., Schøyen T. H., Scott A., Shahaj E., Sharma S., Shatnyeva O., Shekari F., Shelke G. V., Shetty A. K., Shiba K., Siljander P. R.-M., Silva A. M., Skowronek A., Snyder O. L., Soares R. P., Sódar B. W., Soekmadji C., Sotillo J., Stahl P. D., Stoorvogel W., Stott S. L., Strasser E. F., Swift S., Tahara H., Tewari M., Timms K., Tiwari S., Tixeira R., Tkach M., Toh W. S., Tomasini R., Torrecilhas A. C., Tosar J. P., Toxavidis V., Urbanelli L., Vader P., van Balkom B. W., van der Grein S. G., Van Deun J., van Herwijnen M. J., Van Keuren-Jensen K., van Niel G., van Royen M. E., van Wijnen A. J., Vasconcelos M. H., Vechetti I. J., Veit T. D., Vella L. J., Velot É., Verweij F. J., Vestad B., Viñas J. L., Visnovitz T., Vukman K. V., Wahlgren J., Watson D. C., Wauben M. H., Weaver A., Webber J. P., Weber V., Wehman A. M., Weiss D. J., Welsh J. A., Wendt S., Wheelock A. M., Wiener Z., Witte L., Wolfram J., Xagorari A., Xander P., Xu J., Yan X., Yáñez-Mó M., Yin H., Yuana Y., Zappulli V., Zarubova J., Žėkas V., Zhang J.-Y., Zhao Z., Zheng L., Zheutlin A. R., Zickler A. M., Zimmermann P., Zivkovic A. M., Zocco D., Zuba-Surma E. K., Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7, 1535750 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsai W.-H., Moment-preserving thresolding: A new approach. Comput. Vis. Graph. Image Process. 29, 377–393 (1985). [Google Scholar]
- 80.Aaron J. S., Taylor A. B., Chew T.-L., Image co-localization—Co-occurrence versus correlation. J. Cell Sci. 131, jcs211847 (2018). [DOI] [PubMed] [Google Scholar]
- 81.Zhang X., Smits A. H., van Tilburg G. B., Ovaa H., Huber W., Vermeulen M., Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat. Protoc. 13, 530–550 (2018). [DOI] [PubMed] [Google Scholar]
- 82.Ge S. X., Jung D., Yao R., ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Perez-Riverol Y., Bai J., Bandla C., García-Seisdedos D., Hewapathirana S., Kamatchinathan S., Kundu D. J., Prakash A., Frericks-Zipper A., Eisenacher M., Walzer M., Wang S., Brazma A., Vizcaíno J. A., The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Legend for movie S1
Legends for data S1 to S3
Movie S1
Data S1 to S3