Abstract
Aberrant activation of IL-18 signaling regulates tumor immune evasion and progression. However, the underlying mechanism remains unclear. Here, we report that long-chain acyl-CoA synthase 6 (ACSL6) is highly expressed in liver cancer and correlated with poor prognosis. ACSL6 promotes tumor growth, metastasis, and immune evasion mediated by IL-18, independent of its metabolic enzyme activity. Mechanistically, upon IL-18 stimulation, ACSL6 is phosphorylated by ERK2 at S674 and recruits IL-18RAP to interact with IL-18R1, thereby reinforcing the IL-18R1–IL-18RAP heterodimer and triggering NF-κB–dependent gene expression to facilitate tumor development. Furthermore, the up-regulation of CXCL1 and CXCL5 by ACSL6 promotes tumor-associated neutrophil and tumor-associated macrophage recruitment, thereby inhibiting cytotoxic CD8+ T cell infiltration. Ablation or S674A mutation of ACSL6 potentiated anti–PD-1 therapeutic efficacy by increasing the effector activity of intertumoral CD8+ T cells. We revealed that ACSL6 is a potential adaptor that activates IL-18–NF-κB axis–mediated tumor immune evasion and provides valuable insights for developing effective immunotherapy strategies for cancer.
Ablation or S674A mutation of ACSL6 inhibits IL-18–mediated tumor progression and enhances the therapeutic efficacy of anti–PD-1.
INTRODUCTION
Liver cancer is the sixth most common cancer worldwide and has the third highest cancer-related mortality rate (1). Despite the availability of clinical treatments, the overall survival rate of patients with liver cancer remains discouraging, mainly due to the high recurrence and metastasis rates of liver cancer (2). Therefore, identifying potential driving molecules of liver cancer is needed to develop more effective therapeutic targets and improve the prognosis of patients with liver cancer.
Immune checkpoint inhibitors (ICIs) have emerged as promising primary therapeutic agents for advanced liver cancer (3). In 2020, atezolizumab [targeting programmed cell death ligand 1 (PD-L1)] combined with bevacizumab (targeting vascular endothelial growth factor) was approved as a first-line treatment for advanced liver cancer (4). However, only 20 to 30% of patients with liver cancer respond to immunotherapy. Various factors contribute to decreased ICI efficacy, such as the generation of an immunosuppressive tumor microenvironment (TME) through the inhibition of cytotoxic CD8+ T cells or recruitment of myeloid-derived suppressor cells (MDSCs) and macrophages (5), which release inflammatory cytokines, such as interleukin-12 (IL-12) and IL-18, to inhibit the antitumor response (6). Recent studies have indicated that IL-18 switches on immunosuppressive TME through the generation of functional MDSCs, which allows the immune evasion of multiple myeloma cells (7). Therefore, developing combination strategies involving inhibitors of cytokine signaling and ICIs for liver cancer treatment has great significance.
Long-chain acyl–coenzyme A (CoA) synthases (ACSLs), which comprise five isoforms—ACSL1, ACSL3, ACSL4, ACSL5, and ACSL6—are pivotal enzymes that catalyze the first step of fatty acid activation for subsequent fatty acid catabolism and anabolism (8), Recently, numerous studies have suggested that ACSL up-regulation is associated with tumor progression. High expression of ACSL1 is observed in liver cancer and is correlated with a poor prognosis (9, 10). ACSL3 protects melanoma cells from ferroptosis and promotes metastasis (11). ACSL4 up-regulation suppresses chemotherapy-induced apoptosis in triple-negative breast cancer cells by increasing mitochondrial membrane lipid levels (12). However, those studies focused on the contribution of ACSL metabolic enzyme activity to tumor progression. Whether ACSLs play a role in liver cancer through non-metabolic enzymatic mechanisms is unclear.
Here, we identified ACSL6 as a potentially critical oncogenic factor in liver cancer pathogenesis. We showed that IL-18 promotes the phosphorylation of ACSL6 and its localization to the cell membrane, where it interacts with IL-18 receptor 1 (IL-18R1), IL-18 receptor accessory protein (IL-18RAP), and the nuclear factor κB (NF-κB) signaling proteins TNF (tumor necrosis factor) receptor–associated factor 6 (TRAF6) and transforming growth factor β–activated kinase 1 (TAK1), ultimately contributing to liver cancer immune evasion and progression.
RESULTS
ACSL6 is up-regulated and associated with the prognosis of liver cancer
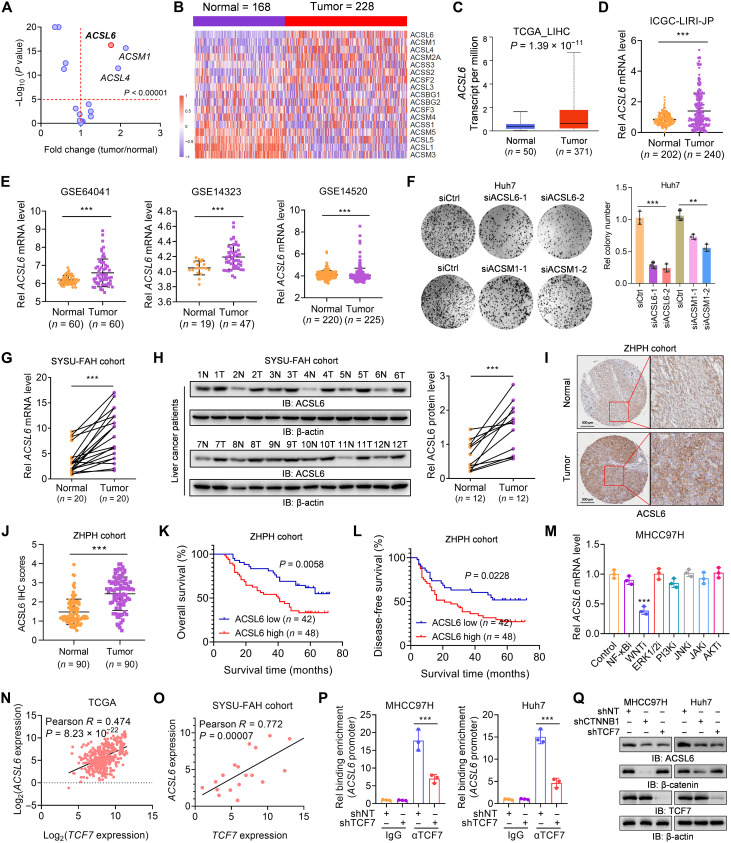
To define the function of ACSs in liver cancer, we analyzed the expression of 17 identified ACSs in liver cancer tissues and adjacent nontumor tissues in the Gene Expression Omnibus (GEO) database GSE63898 (table S1) (13). The results showed that six genes were significantly up-regulated in liver cancer tissues compared to those in normal liver tissues (Fig. 1, A and B). However, among the six genes, only three genes (ACSL4, ACSL6, and ACSM1) exhibited an average increase >1.5-fold (Fig. 1A). Moreover, ACSL4, ACSL6, and ACSM1 were up-regulated in The Cancer Genome Atlas (TCGA), International Cancer Genome Consortium (ICGC), and other GEO datasets (Fig. 1, C to E, and fig. S1, A to F). ACSL4 is reportedly related to liver cancer progression through its enzymatic activity (14, 15), but the roles of ACSL6 and ACSM1 in liver cancer have not been well elucidated. We then individually silenced ACSL6 and ACSM1 in Huh7 cells (fig. S1, G and H) and found that ACSL6 knockdown had a greater inhibitory effect on cell growth (Fig. 1F). To further confirm the results from bioinformatics analyses of online databases, the mRNA and protein levels of ACSL6 were detected from patients with liver cancer. The results demonstrated that the mRNA and protein levels of ACSL6 were higher in tumor tissues than those in paired adjacent nontumor tissues (Fig. 1, G and H). Immunohistochemistry (IHC) confirmed higher ACSL6 expression in liver cancer tissues than in paired normal tissues (Fig. 1, I and J). Moreover, overall survival and disease-free survival analyses revealed that higher expression of ACSL6 was correlated with a worse prognosis for patients with liver cancer (Fig. 1, K and L).
Fig. 1. ACSL6 is up-regulated in liver cancer and is associated with prognosis.
(A and B) Expression analyses (A) and heatmap (B) of 17 ACSs in liver tumor and nontumor tissues from GSE63898. (C to E) ACSL6 expression in liver tumor and nontumor tissues from The Cancer Genome Atlas (TCGA) database (C), International Cancer Genome Consortium (ICGC) database (D), and Gene Expression Omnibus (GEO) database (E). (F) Colony formation of Huh7 cells transfected with small interfering RNA control (siCtrl) or small interfering RNA (siRNA) targeting ACSL6 or ACSM1. (G and H) qPCR analyses of ACSL6 mRNA expression (G) and immunoblotting (IB) analyses of protein levels (H) in paired liver tumor and adjacent nontumor tissues from The First Affiliated Hospital, Sun Yat-sen University (SYSU-FAH). (I and J) Immunohistochemistry (IHC) analyses of ACSL6 expression in liver tumor and paired nontumor tissues from Zhuhai People’s Hospital (ZHPH). (K and L) Overall survival analyses (K) and disease-free survival analyses (L) based on ACSL6 expression in liver tumors. (M) Quantitative polymerase chain reaction (qPCR) analyses of ACSL6 expression in MHCC97H cells stimulated with inhibitors targeting NF-κB, WNT, extracellular signal–regulated kinase 1/2 (ERK1/2), phosphatidylinositol 3-kinase (PI3K), c-Jun N-terminal kinase (JNK), Janus kinase (JAK), and AKT. (N) A correlation analysis was conducted to assess the correlation between ACSL6 and TCF7 expression in TCGA liver cancer database. (O) A correlation analysis was performed to assess the correlation between ACSL6 and TCF7 expression in liver cancer tissue samples from SYSU-FAH cohort. (P) Chromatin immunoprecipitation (ChIP)–qPCR analyses of TCF7 binding to the ACSL6 promoter in shNT and short hairpin RNA targeting TCF7 (shTCF7) MHCC97H or Huh7 cells. (Q) Immunoblotting analyses were performed with the indicated antibodies in shNT, short hairpin RNA targeting CTNNB1 (shCTNNB1), and shTCF7 MHCC97H or Huh7 cells. **P < 0.01 and ***P < 0.001. Student’s t test [(C) to (E), (G), (H), (J), and (P)], one-way analysis of variance [ANOVA; (F) and (M)], and log-rank test [(K) and (L)].
We then investigated the molecular regulation of high ACSL6 expression in liver cancer. By treating cells with inhibitors targeting multiple signaling pathways, we observed that inhibitors of the WNT pathway significantly decreased ACSL6 expression (Fig. 1M and fig. S1, I and J). Because T cell factor 7 (TCF7) is a central transcription factor in WNT signaling, we next investigated whether ACSL6 is a direct target gene of TCF7 and identified the TCF7 binding motif in the ACSL6 promoter (fig. S1K). In addition, an analysis of expression data from the TCGA database corroborated a positive correlation between ACSL6 and TCF7 expression in liver cancer (Fig. 1N). This correlation was further validated in liver cancer samples from The First Affiliated Hospital, Sun Yat-sen University (SYSU-FAH) cohort (Fig. 1O). Chromatin immunoprecipitation (ChIP) quantitative polymerase chain reaction (qPCR) analysis confirmed the binding of TCF7 to the ACSL6 promoter, and this interaction was notably reduced in TCF7-depleted cells (Fig. 1P). Moreover, the construction of a reporter plasmid containing the ACSL6 promoter (ACSL6-luc) revealed that TCF7 knockdown attenuated ACSL6-luc activity, whereas TCF7 overexpression enhanced ACSL6-luc activity (fig. S1, L and M). Further experiments involving individual knockdown of the signal transduction proteins in the WNT pathway, namely, β-catenin (encoded by the CTNNB1 gene) and TCF7, revealed that both catenin beta 1 (CTNNB1) and TCF7 knockdown markedly down-regulated ACSL6 expression (Fig. 1Q and fig. S1N). Conversely, the overexpression of β-catenin and TCF7 increased ACSL6 expression (fig. S1O). These findings suggest that the WNT/β-catenin/TCF7 signaling pathway up-regulates ACSL6 expression in liver cancer cells. Overall, we showed that ACSL6 is up-regulated in liver cancer and that its high expression is associated with a poor prognosis for patients with liver cancer.
ACSL6 promotes cell proliferation and migration in vitro and facilitates liver cancer growth and metastasis in vivo
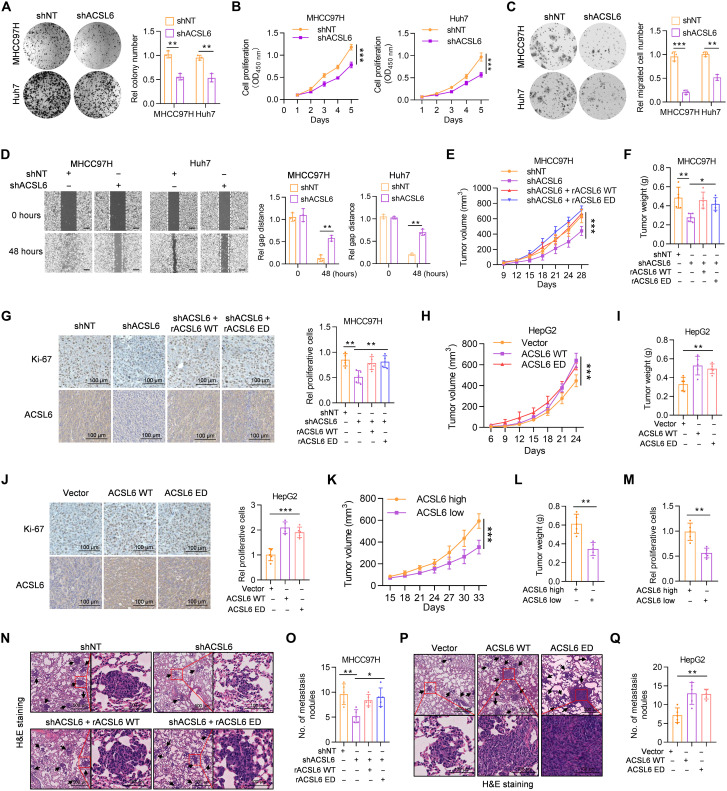
To investigate the role of ACSL6 in liver cancer progression, we detected the expression of ACSL6 in liver cancer cell lines, and the results revealed high endogenous ACSL6 expression in most liver cancer cell lines (fig. S2A). We then depleted endogenous ACSL6 using two independent short hairpin RNAs (shRNAs) in MHCC97H and Huh7 cells (fig. S2B). ACSL6 depletion markedly decreased cell colony formation and growth rates (Fig. 2, A and B). We also overexpressed ACSL6 in HepG2 and Sk-Hep1 cells, which exhibited relatively low ACSL6 expression (fig. S2C). ACSL6 overexpression notably increased cell proliferation (fig. S2, D to F). The high expression of ACSL6 in cells with high metastatic potential (MHCC97H) indicated that ACSL6 may function in cell metastasis. As expected, ACSL6 depletion greatly inhibited tumor cell migration (Fig. 2, C and D). In contrast, the overexpression of ACSL6 strongly enhanced tumor cell metastasis ability (fig. S2, G and H). To explore whether the promoting effect of ACSL6 on liver cancer progression depends on its metabolic activity, we restored ACSL6 expression with a synonymously mutated shRNA-resistant ACSL6 wild-type (rACSL6 WT) and rACSL6 enzymatically dead (ED; Y344A) (16) in ACSL6-depleted MHCC97H and Huh7 cells (fig. S2I). The results showed that rACSL6 WT and ED restored ACSL6 depletion inhibited cell growth and metastasis, excluding the possibility of off-target effects of short hairpin RNA targeting ACSL6 (shACSL6), which indicated that ACSL6 metabolic enzyme activity is not essential for tumor cell growth and metastasis (fig. S2, J to L).
Fig. 2. ACSL6 promotes cell proliferation and migration in vitro and facilitates liver cancer growth and metastasis in vivo.
(A and B) Colony formation (A) and CCK-8 assays (B) of shNT or shACSL6 expressed MHCC97H and Huh7 cells. OD450 nm, optical density at 450 nm. (C and D) Transwell (C) and wound healing assays (D) in MHCC97H and Huh7 cells expressing shNT or shACSL6. (E to G) Subcutaneous injection of shNT, shACSL6, and shACSL6 rescued with resistant ACSL6 wild-type (rACSL6 WT) or enzymatically dead (ED) MHCC97H cells into nude mice. Tumor volumes (E), tumor weights (F), and Ki-67 staining (G) in the xenograft model. (H to J) Subcutaneous injection of vector, ACSL6 WT–, or ED-overexpressing HepG2 cells into nude mice. Tumor volumes (H), tumor weights (I), and Ki-67 staining (J) in the xenograft model. (K to M) Assessment of the effect of high or low ACSL6 expression in tumors obtained from patients with liver cancer on PDX mouse models. Tumor volumes (K), tumor weights (L), and relative proliferative cells (M) in the PDX mouse model. (N and O) Representative images of hematoxylin and eosin (H&E) staining (N) and statistical analysis (O) of metastatic lung nodules from mice injected with shNT, shACSL6, and shACSL6 rescued with rACSL6 WT or ED MHCC97H cells via the tail vein. (P and Q) Representative images of H&E staining (P) and statistical analysis (Q) of metastatic lung nodules from mice injected with vector, ACSL6 WT–, or ED-overexpressed HepG2 cells via the tail vein. [(E) to (Q)] n = 5. *P < 0.05, **P < 0.01, and ***P < 0.001. Student’s t test [(A), (C), (D), (L), and (M)], two-way ANOVA [(B), (E), (H), and (K)], one-way ANOVA [(F), (G), (I), (J), (O), and (Q)].
To further study the effect of ACSL6 on liver cancer progression in vivo, we constructed a subcutaneous xenograft tumor model. ACSL6 knockdown decreased MHCC97H cell–derived tumor growth in mice. However, the decrease in tumor growth was rescued when rACSL6 WT and ED were reconstituted in tumor cells (Fig. 2, E to G). Consistently, both ACSL6 WT and ED overexpression promoted tumor growth (Fig. 2, H to J). Furthermore, in patient-derived xenograft (PDX) mouse models with tumors from patients with liver cancer, tumors with high ACSL6 expression exhibited accelerated growth (Fig. 2, K to M, and fig. S2M). Similar phenomena were observed in in vivo metastasis models (Fig. 2, N to Q). Collectively, these in vitro and in vivo findings support that ACSL6 promotes tumor growth and metastasis in liver cancer independent of its enzyme activity.
ACSL6 activates the IL-18–IL-18R1–NF-κB pathway by forming a complex with IL-18R1 and consolidating the IL-18R1–IL-18RAP heterodimer
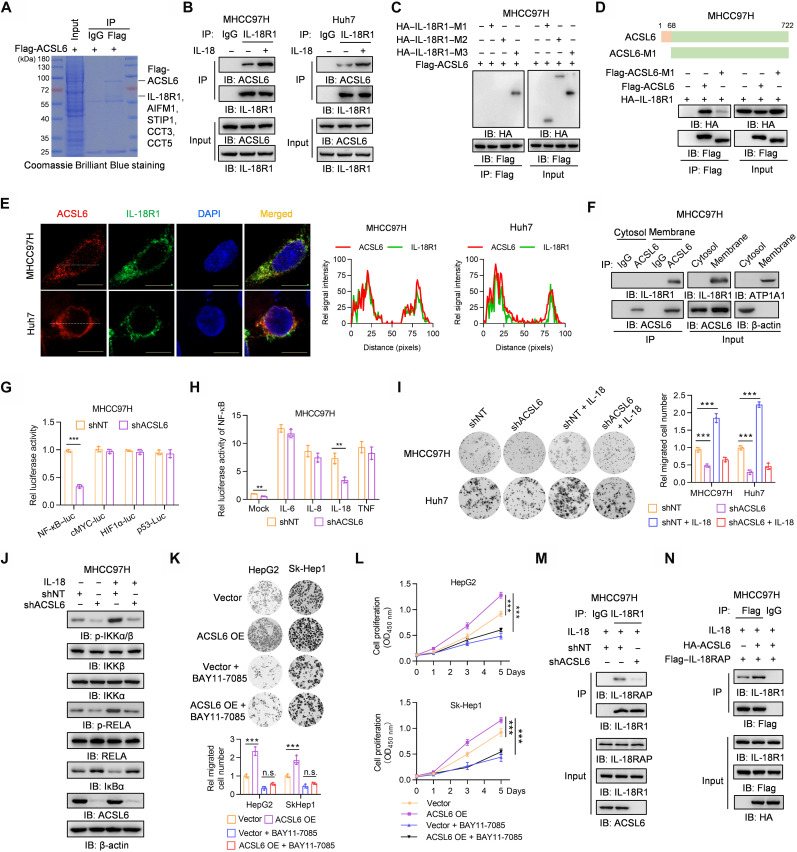
To investigate the mechanism underlying ACSL6-promoted liver cancer progression, we performed a mass spectrometry (MS) analysis and identified five proteins that potentially interact with ACSL6 (Fig. 3A). Subsequent co-immunoprecipitation (co-IP) experiments confirmed the binding of four of the five identified proteins to ACSL6, with a particularly strong interaction observed between IL-18R1 and ACSL6 (fig. S3A). We individually knocked down each of the four proteins that bind to ACSL6 to further understand the specific role. We found that, only when IL-18R1 was depleted, the ability of ACSL6 to promote liver cancer cell proliferation was heavily inhibited (fig. S3B). On the basis of these findings, we focused further research on IL-18R1 (fig. S3C). IL-18 is a member of the IL-1 superfamily, and its receptor contains IL-18R1 and IL-18RAP (17). To validate this interaction, we performed exogenous and endogenous co-IP assays and demonstrated that IL-18R1 interacted with ACSL6 in liver cancer cells and that this interaction was reinforced upon IL-18 stimulation (Fig. 3B and fig. S3, D and E) and was not depended on ACSL6 enzymatic activity (fig. S3F). Moreover, the interaction between IL-18R1 and ACSL6 barely occurred in nontumorigenic THLE2 liver cells (fig. S3G). To study the mechanism by which ACSL6 interacted with IL-18R1, we constructed three IL-18R1 truncations (fig. S3H) and found that ACSL6 interacted with IL-18R1–M3, which contains the transmembrane domain responsible for IL-18R1 membrane localization (Fig. 3C). Furthermore, IL-18R1 interacted with ACSL6 but not with the ACSL6-M1 truncation, which lacks the transmembrane domain (Fig. 3D), indicating that ACSL6 and IL-18R1 interacted with each other in a manner dependent on their transmembrane domains. The computational model of the structure predicted by ZDOCK and PyMOL software also showed that ACSL6 and IL-18R1 potentially interact with each other (fig. S3I). Immunofluorescence (IF) and co-IP assays showed that ACSL6 and IL-18R1 colocalized and interacted on the membrane (Fig. 3, E and F, and fig. S3J). In addition, IL-18 stimulation induced ACSL6 translocation to the membrane to form a complex with IL-18R1 (fig. S3K).
Fig. 3. ACSL6 activates the IL-18–IL-18R1–NF-κB pathway by forming a complex with IL-18R1.
(A) Flag-ACSL6 was immunoprecipitated from MHCC97H cells expressing Flag-ACSL6, and then SDS–polyacrylamide gel electrophoresis (PAGE) followed by Coomassie Brilliant Blue staining and mass spectrometry (MS) analysis was performed. (B) MHCC97H and Huh7 cells were treated with or without IL-18 (20 ng ml−1) for 1 hour. (C) MHCC97H cells expressing Flag-ACSL6 were transfected with hemagglutinin (HA)–tagged vector, IL-18R1–M1, IL-18R1–M2, or IL-18R1–M3. (D) MHCC97H cells expressing HA–IL-18R1 were transfected with vector, Flag-ACSL6 WT, or M1. (E) Immunofluorescence (IF) analyses of ACSL6 and IL-18R1 colocalization in MHCC97H and Huh7 cells. DAPI, 4′,6-diamidino-2-phenylindole. (F) Membrane and cytosol fractions were prepared from MHCC97H cells. (G) Luciferase analyses in shNT and shACSL6 MHCC97H cells. HIF1α, hypoxia-inducible factor 1α. (H) Luciferase analyses of NF-κB–luc in shNT and shACSL6 MHCC97H cells treated with IL-6, IL-8, IL-18, and TNF (20 ng ml−1) for 12 hours. (I) Transwell assays in shNT or shACSL6 MHCC97H and Huh7 cells treated with or without IL-18 (20 ng ml−1). (J) Immunoblotting analyses in shNT and shACSL6 MHCC97H cells treated with or without IL-18 (20 ng ml−1) for 1 hour. (K and L) Transwell (K) and CCK-8 assays (L) in vector or ACSL6-overexpressed (OE) HepG2 and Sk-Hep1 cells treated with or without 10 μM BAY11-7085. n.s., not significant. (M) MHCC97H cells expressing shNT or shACSL6 were treated with IL-18 (20 ng ml−1) for 1 hour. (N) MHCC97H cells expressing Flag–IL-18RAP were infected with HA-ACSL6 and then treated with IL-18 (20 ng ml−1) for 1 hour. [(B) to (D), (F), (M), and (N)] Immunoprecipitation and immunoblotting analyses were performed with indicated antibodies. **P < 0.01 and ***P < 0.001. Student’s t test [(G) and (H)], one-way ANOVA [(I) and (K)], and two-way ANOVA (L).
IL-18R1 acts as a receptor for IL-18, transferring IL-18 signaling from the extracellular to intracellular space and activating the NF-κB pathway (17). Because ACSL6 interacts with IL-18R1, we wondered whether ACSL6 regulates IL-18R1–NF-κB activation. ACSL6 depletion significantly decreased NF-κB activity without affecting other transcription factors, such as p53, hypoxia-inducible factor 1α, and MYC proto-oncogene bHLH transcription factor (cMYC), which are known regulators of tumor progression (Fig. 3G) (18). We then performed a luciferase assay following stimulation with IL-18 or other cytokines that activate NF-κB, including, IL-6, IL-8, and TNF. The results showed that ACSL6 depletion decreased NF-κB activity upon IL-18 treatment but not upon treatment with other cytokines (Fig. 3H), which was independent of ACSL6 metabolic activity (fig. S3L). Furthermore, the ability of IL-18 to promote tumor cell migration and proliferation was significantly reduced in ACSL6-depleted cells (Fig. 3I and fig. S3M). Moreover, phosphorylated inhibitor of nuclear factor kappa B kinase subunit alpha/beta (p-IKKα/β) and phosphorylated RELA proto-oncogene (p-RELA) levels were decreased, and IκBα expression was increased in ACSL6-depleted MHCC97H cells upon IL-18 stimulation (Fig. 3J), while the opposite effect was observed in ACSL6-overexpressed cells (fig. S3N). We then treated ACSL6-overexpressed cells with NF-κB inhibitor BAY11-7085 (fig. S3O). The results showed that BAY11-7085 treatment significantly inhibited the ability of ACSL6 overexpression to promote the cell proliferation and migration of HepG2 and Sk-Hep1 cells (Fig. 3, K and L). Together, these results indicate that ACSL6 activates the NF-κB signaling pathway upon IL-18 stimulation.
IL-18RAP heterodimerizes with IL-18R1 during activation after IL-18 stimulation (19). Co-IP revealed that the interaction between IL-18R1 and IL-18RAP increased upon IL-18 stimulation (fig. S3P). Given that ACSL6 interacts with IL-18R1, we wondered whether ACSL6 is involved in regulating the formation of IL-18R1 and IL-18RAP heterodimers. The results showed that ACSL6 depletion weakened the interaction between IL-18R1 and IL-18RAP (Fig. 3M and fig. S3Q). Conversely, ACSL6 overexpression increased the interaction between IL-18R1 and IL-18RAP (Fig. 3N). Moreover, ACSL6 interacted with IL-18RAP, and this interaction increased upon IL-18 stimulation (fig. S3R). These results suggest that ACSL6 strengthens the interaction between IL-18R1 and IL-18RAP by recruiting IL-18RAP, thereby activating IL-18R1–NF-κB signaling to promote liver cancer cell proliferation and migration.
IL-18 induces ACSL6 pS674 to promote its translocation, thereby activating IL-18R1–NF-κB signaling
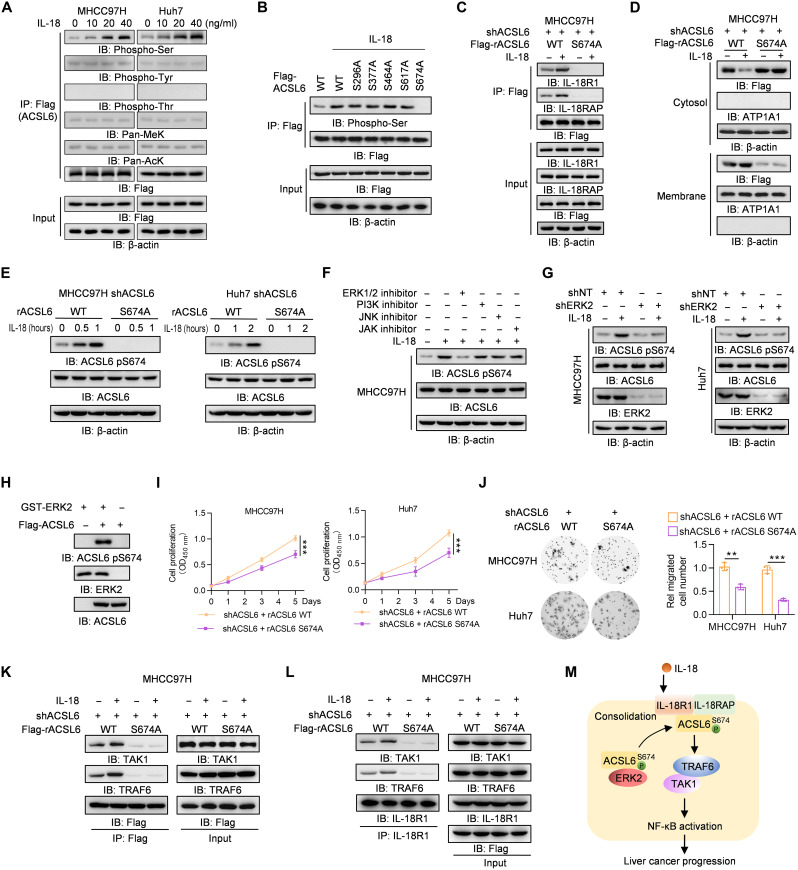
The mechanism by which IL-18 stimulation leads to ACSL6 and IL-18R1 interaction and ACSL6 membrane localization is unclear. Phosphorylation is an important posttranslational modification that regulates protein function or subcellular location in signaling cascades (20). To determine whether ACSL6 undergoes posttranslational modification upon IL-18 stimulation, ACSL6 phosphorylation, pan-lysine methylation, and pan-lysine acetylation were detected. The results showed that only serine phosphorylation of ACSL6 increased upon IL-18 stimulation (Fig. 4A). We performed MS analysis to validate this finding, and the results revealed that five serine residues may be phosphorylated upon IL-18 stimulation, including S296, S377, S464, S617, and S674 (fig. S4A). We constructed five phospho-deficient ACSL6 mutants in which S296, S377, S464, S617 and S674 were mutated to alanine (A) and found that only S674A significantly decreased IL-18–induced ACSL6 phosphorylation (Fig. 4B and fig. S4B). We then rescued ACSL6-depleted cells with Flag-ACSL6 WT or S674A expression. The co-IP results showed that, compared to ACSL6 WT, ACSL6 S674A hardly interacted with IL-18R1 and IL-18RAP (Fig. 4C) and attenuated IL-18R1–IL-18RAP heterodimer upon IL-18 treatment (fig. S4C). Furthermore, ACSL6 S674A was predominantly localized in the cytosol even after IL-18 stimulation (Fig. 4D). These results suggest that ACSL6 S674 phosphorylation (pS674) is essential for ACSL6 function as an adaptor to regulate IL-18 signaling by promoting IL-18R heterodimer formation.
Fig. 4. IL-18 induces ACSL6 pS674 to activate NF-κB signaling.
(A) Flag-ACSL6–overexpressed MHCC97H and Huh7 cells were treated with different concentrations of IL-18 for 1 hour. Immunoprecipitation and immunoblotting analyses were performed. (B) Flag-ACSL6 WT and mutant overexpressed cells were treated with or without IL-18 (20 ng ml−1) for 1 hour. Immunoprecipitation and immunoblotting were performed. (C and D) ACSL6-depleted MHCC97H cells were infected with Flag-ACSL6 WT or S674A and then treated with or without IL-18 (20 ng ml−1) for 1 hour. Immunoprecipitation (C) and subcellular fractionation detection (D) were performed. (E) Immunoblotting analyses in ACSL6-depleted MHCC97H and Huh7 cells reconstituted with Flag-rACSL6 WT or S674A and treated with or without IL-18 (20 ng ml−1) for indicated times. (F) Immunoblotting analyses in MHCC97H cells treated with or without indicated inhibitors for 6 hours, followed by treatment with IL-18 (20 ng ml−1) for 1 hour. (G) Immunoblotting analyses in shNT or shERK2 MHCC97H and Huh7 cells treated with or without IL-18 (20 ng ml−1) for 1 hour. (H) In vitro kinase assay was performed by mixing GST-ERK2 and Flag-ACSL6. (I and J) CCK-8 (I) and transwell assays (J) in ACSL6-depleted MHCC97H and Huh7 cells reconstituted with rACSL6 WT or S674A. (K and L) ACSL6-depleted MHCC97H cells were infected with Flag-rACSL6 WT or S674A and then treated with or without IL-18 (20 ng ml−1) for 1 hour. Immunoprecipitation analyses were performed using anti-Flag (K) or anti–IL-18R1 antibodies (L). (M) Mechanism through which ACSL6 activates the IL-18–IL-18R1–NF-κB pathway by forming a complex with IL-18R1 and consolidating the IL-18R1–IL-18RAP heterodimer. **P < 0.01 and ***P < 0.001. Two-way ANOVA (I) and one-way ANOVA (J).
We next investigated the kinase responsible for ACSL6 pS674. An antibody specifically against ACSL6 pS674 was generated, and its specificity was validated by analyzing in ACSL6-depleted MHCC97H cells rescued with Flag-rACSL6 WT or S674A, which showed that S674A abrogated ACSL6 pS674 (fig. S4D). In addition, IL-18 induced pS674 in the rACSL6 WT cells but not in rACSL6 S674A cells (Fig. 4E). We then treated cells with several kinase inhibitors, including the inhibitors of extracellular signal–regulated kinase 1/2 (ERK1/2), phosphatidylinositol 3-kinase, c-Jun N-terminal kinase, and Janus kinase. The results showed that the ERK1/2 inhibitor attenuated IL-18–induced ACSL6 pS674 (Fig. 4F and fig. S4E). In addition, ACSL6 bound to ERK2 but not to ERK1 (fig. S4F), and ERK2 depletion markedly decreased ACSL6 pS674 (Fig. 4G), while ERK1 knockdown did not block ACSL6 pS674 (fig. S4G). Furthermore, we found that IL-18 treatment promoted ERK2 phosphorylation in MHCC97H and Huh7 cells (fig. S4H). Moreover, ERK2 directly phosphorylated ACSL6 pS674 by in vitro kinase assay (Fig. 4H). These results indicate that ERK2 is required for ACSL6 pS674. ACSL6 S674A decreased cell growth and migration (Fig. 4, I and J), highlighting the importance of ACSL6 pS674 for ACSL6 function. Together, these results indicate that ACSL6 pS674 is indispensable for the consolidation of the IL-18R1–IL-18RAP heterodimer to promote liver cancer progression.
We then determined whether ACSL6 serves as an adaptor to link IL-18R1 and NF-κB signaling key protein activation, including TRAF6, IKKα, TAK1, TGF-β activated kinase 1 binding protein 2 (TAB2), TAB3, p50, and RELA. The results showed that ACSL6 strongly interacted with TRAF6 and TAK1 and weakly interacted with TAB2 and TAB3 (fig. S4I). However, ACSL6 did not exhibit binding affinity for key proteins associated with the nonclassical NF-κB signaling pathway, including TRAF2, TRAF3, NF-κB–inducing kinase, IKKα, RELB proto-oncogene (RELB), and p52 (fig. S4J), and ACSL6 depletion did not alter the NF-κB luciferase activity induced by nonclassical NF-κB proteins (fig. S4K), which suggests that ACSL6 does not modulate the nonclassical NF-κB pathway. The interaction between ACSL6 WT and TRAF6 and TAK1 was significantly enhanced upon stimulation with IL-18, but ACSL6 S674A hardly interacted with TAK1 or TRAF6 (Fig. 4K). Furthermore, ACSL6 S674A did not promote the binding of IL-18R1 to TRAF6 or TAK1 upon IL-18 stimulation (Fig. 4L). Collectively, these results indicate that ACSL6 serves as an adaptor to linking IL-18 signaling and NF-κB activation to promote liver cancer progression (Fig. 4M).
ACSL6 promotes NF-κB signaling to drive NF-κB–dependent gene expression
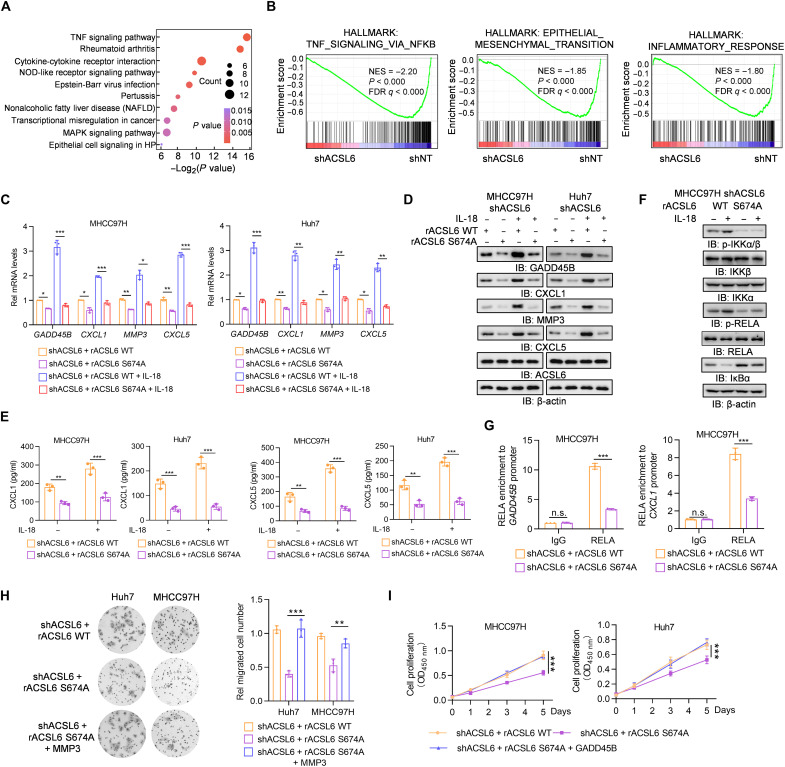
To examine whether and how the oncogenic role of IL-18 in liver cancer progression is regulated by ACSL6, we performed RNA sequencing (RNA-seq) analysis in MHCC97H cells with endogenous ACSL6 depletion. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed that the TNF signaling pathway was the most significantly altered pathway (Fig. 5A), with genes whose expression decreased after ACSL6 depletion. Gene set enrichment analysis (GSEA) also showed that NF-κB signaling, epithelial-mesenchymal transition, and inflammatory response gene sets were enriched among the down-regulated genes in ACSL6-depleted cells (Fig. 5B). We assessed the expression of the top seven NF-κB target genes and observed notable reductions in GADD45B, CXCL1, MMP3, and CXCL5 expression in ACSL6-depleted cells (fig. S5A). Moreover, ACSL6 depletion or overexpression decreased or increased the expression of these genes, respectively, in subcutaneous xenograft tumors (fig. S5B). These data suggest that ACSL6 promotes NF-κB target gene expression.
Fig. 5. ACSL6 promotes NF-κB signaling to drive NF-κB–dependent gene expression.
(A and B) Kyoto Encyclopedia of Genes and Genomes (A) and gene set enrichment analysis (GSEA) analyses (B) of RNA-seq data from shNT and shACSL6 MHCC97H cells. MAPK, mitogen-activated protein kinase; FDR, false discovery rate; NES, normalized enrichment score. (C to E) ACSL6-depleted MHCC97H and Huh7 cells were infected with rACSL6 WT or S674A and then treated with or without IL-18 (20 ng ml−1) for 12 hours. The mRNA (C) and protein expression (D) levels of the indicated genes and concentrations of CXCL1 and CXCL5 (E) in the medium were detected. (F) ACSL6-depleted MHCC97H cells were infected with rACSL6 WT or S674A and then treated with or without IL-18 (20 ng ml−1) for 1 hour. Immunoblotting analyses were performed with indicated antibodies. (G) ChIP-qPCR analyses were performed with the indicated antibodies, and DNA was amplified with primers targeting positive sites in the GADD45B or CXCL1 gene in ACSL6-depleted MHCC97H cells with forced expression of rACSL6 WT and S674A. (H) Transwell assays of the migration ability of ACSL6-depleted MHCC97H or Huh7 cells with forced expression of rACSL6 WT, S674A, or S674A and MMP3. (I) CCK8 analyses of ACSL6-depleted MHCC97H or Huh7 cells with forced expression of rACSL6 WT, S674A, or S674A with GADD45B. *P < 0.05, **P < 0.01, and ***P < 0.001. One-way ANOVA [(C), (E), (G), and (H)] and two-way ANOVA (I).
We next investigated the function of ACSL6 pS674 in regulating the expression of GADD45B, CXCL1, MMP3, and CXCL5. Compared to ACSL6 WT, ACSL6 S674A significantly reduced the expression of these genes (Fig. 5, C and D). We then examined the soluble CXCL1 and CXCL5 levels and found that ACSL6 S674A reduced the concentration of CXCL1 and CXCL5 secreted in the medium (Fig. 5E). Moreover, ACSL6 S674A significantly inhibited NF-κB activity (fig. S5C). Consistent with this finding, compared with ACSL6 WT, IL-18 hardly promoted IKKα/β and RELA phosphorylation and decreased IκBα levels in cells rescued with ACSL6 S674A (Fig. 5F and fig. S5D). ChIP assays showed that ACSL6 S674A decreased the recruitment of RELA to the GADD45B, CXCL1, MMP3, and CXCL5 promoters (Fig. 5G and fig. S5E), and ACSL6 S674A blocked the luciferase activity of the GADD45B, CXCL1, MMP3, and CXCL5 promoters (fig. S5F). Growth arrest and DNA damage inducible beta (GADD45B) and matrix metalloproteinase 3 (MMP3) promote the proliferation and metastasis of tumor cells, respectively (21, 22). We next overexpressed exogenous GADD45B and MMP3 in ACSL6-depleted cells, respectively (fig. S5G). The results showed that overexpression of MMP3 or GADD45B in ACSL6-depleted cells almost restored the decreases in tumor cell migration and proliferation caused by ACSL6 depletion (Fig. 5, H and I). Together, these results suggest that ACSL6 up-regulates GADD45B and MMP3 by activating NF-κB signaling, thereby promoting tumor cell proliferation and migration, respectively.
ACSL6 pS674 promotes TANs and TAMs recruitment thereby promoting IL-18–mediated tumor immune escape
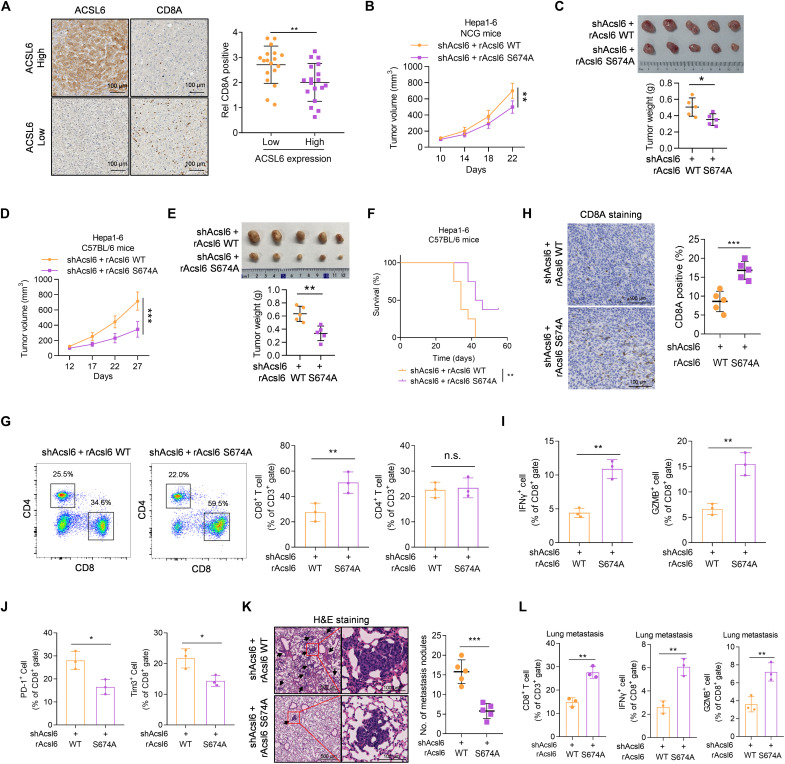
IL-18 signaling facilitates immune evasion in tumors and promotes tumor growth and metastasis (7). However, the exact underlying mechanisms are incompletely understood. Our results suggest that ACSL6 activates IL-18R1–NF-κB to promote the expression of CXCL1 and CXCL5, which are associated with the immunotherapy response (23, 24). On the basis of these observations, it is reasonable to hypothesize that ACSL6 may contribute to IL-18–mediated immune evasion. To further test this hypothesis, we first interrogated public transcriptome data of patients with liver cancer in TCGA database to define the connection between ACSL6 and the immune microenvironment (25). The results showed that patients with liver cancer with higher expression of ACSL6 exhibited lower immune scores, decreased cytotoxicity scores, CD8+ T cell infiltration, and lower CD3E and CD8A levels (fig. S6, A to C). We further assessed ACSL6 expression and T cell marker levels in liver cancer tissues and showed that tumors with low ACSL6 expression exhibited high CD3E and CD8A levels (fig. S6D) and a high CD8A intensity (Fig. 6A), suggesting that ACSL6 inversely correlates with CD8+ T cell levels and may function to promote immune evasion in liver cancer.
Fig. 6. ACSL6 pS674 drives liver cancer immune evasion.
(A) IHC staining and quantification showing the inverse correlation between ACSL6 expression and CD8A levels in patients with liver cancer. (B and C) Subcutaneous injection of Acsl6-depleted Hepa1-6 cells with forced expression of rAcsl6 WT and S674A into NCG mice. Tumor volumes (B) and tumor images and weights (C) in the xenograft model. (D to J) Subcutaneous injection of Acsl6-depleted Hepa1-6 cells with forced expression of rAcsl6 WT or S674A into C57BL/6 mice. Tumor volumes (D), tumor images and weights (E), and survival rates (F) are shown. Flow cytometry analyses of tumor-infiltrating CD8+ and CD4+ T cells (G). IHC staining analyses of CD8A levels (H). Flow cytometry analyses of tumor-infiltrating IFN-γ+CD8+ and GZMB+CD8+ T cells (I) and tumor-infiltrating PD-1+CD8+ and Tim3+CD8+ T cells (J) in indicated tumors. (K and L) Representative images of H&E staining and statistics of metastatic lung nodules (K) and flow cytometry analyses of tumor-infiltrating CD8+ T cells, IFN-γ+CD8+ T cells, and GZMB+CD8+ T cells (L) from C57BL/6 mice injected with Acsl6-depleted Hepa1-6 cells with forced expression of rAcsl6 WT and S674A. [(B) to (E)] n = 5; (F) n = 10. *P < 0.05, **P < 0.01, and ***P < 0.001. Student’s t test [(A), (C), (E), and (G) to (L)], two-way ANOVA [(B) and (D)], and log-rank test (F).
Our results indicated that IL-18 promoted ACSL6-mediated activation of NF-κB signaling by inducing ACSL6 pS674. We further analyzed the sequence of ACSL6 among different species and found that ACSL6 S674 is conserved in multiple species (fig. S6E). To investigate whether ACSL6 pS674 affects the possible tumor cell–extrinsic function of ACSL6 within the microenvironment, we generated Acsl6-depleted Hepa1-6 cells rescued with rAcsl6 WT or S674A (fig. S6F). NF-κB signaling and Cxcl1 and Cxcl5 expression were inhibited in the rAcsl6 S674A Hepa1-6 cells compared to those in rAcsl6 WT Hepa1-6 cells (fig. S6, G and H). Although rAcsl6 S674A moderately impaired Hepa1-6 tumor growth in immune-deficient NOD-SCID gamma (NCG) mice (Fig. 6, B and C), this impairment was much more pronounced in immune-competent C57BL/6 mice (Fig. 6, D and E). Moreover, C57BL/6 mice bearing tumors expressing rAcsl6 S674A had a higher survival rate (Fig. 6F). Consistently, we found a significant increase in CD8+ T cells but a small increase in CD4+ T cells from rAcsl6 S674A tumors (Fig. 6, G and H). In addition, increased interferon-γ (IFN-γ)+CD8+ and granzyme B (GZMB)+CD8+ T cells (Fig. 6I), decreased programmed cell death protein 1 (PD-1)+CD8+ and Tim3+CD8+ T cells (Fig. 6J), and increased IFN-γ levels were observed in the rAcsl6 S674A tumors (fig. S6I). The lung metastasis model showed that rAcsl6 S674A markedly inhibited tumor metastasis in C57BL/6 mice (Fig. 6K), followed by increases in CD8+ T cells, IFN-γ+CD8+, and GZMB+CD8+ T cells and decreases in PD-1+CD8+ and Tim3+CD8+ T cells (Fig. 6L and fig. S6J). These results suggest that ACSL6 inhibits the infiltration and activation of CD8+ T cells.
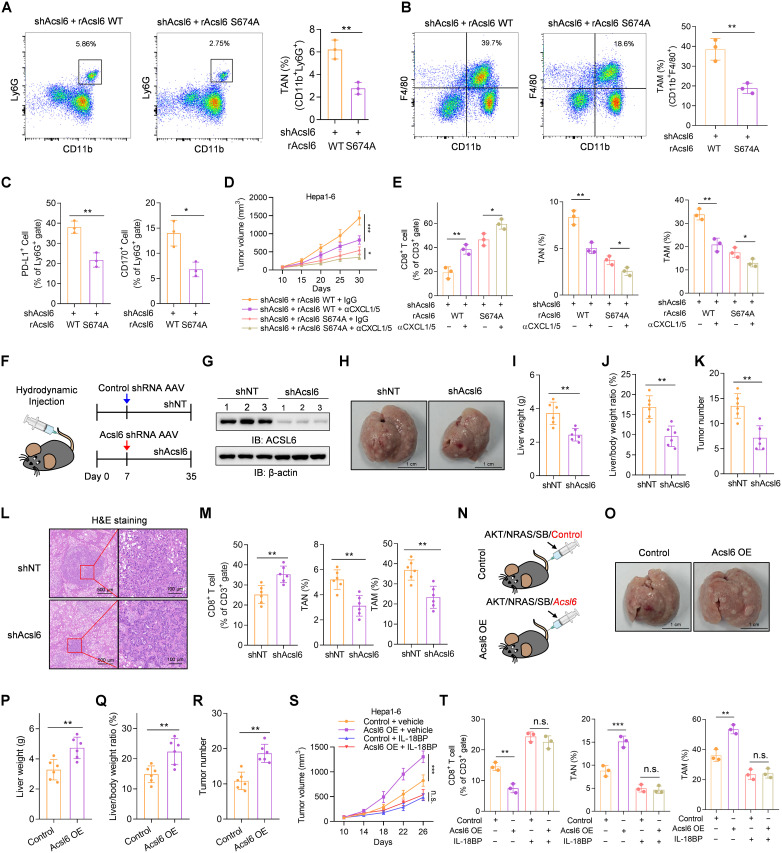
Immunosuppressive cells, such as tumor-associated neutrophils (TANs) and tumor-associated macrophages (TAMs), have been reported to induce immunosuppressive effects and promote tumor progression by suppressing CD8+ T cell infiltration and activation (26). The chemokines CXCL1 and CXCL5 promote cancer progression by recruiting neutrophils and macrophages (27). We next investigated the infiltration of TANs and TAMs in the TME. Compared with rAcsl6 WT tumors, rAcsl6 S674A tumors exhibited reduced TAN and TAM recruitment and a decrease in CXCL1 and CXCL5 secretion (Fig. 7, A and B, and fig. S7, A and B). Notably, PD-L1+ TANs, CD170+ TANs, PD-L1+ TAMs, and CD206+ TAMs were significantly decreased (Fig. 7C and fig. S7C). Moreover, the expression levels of immunosuppressive molecules, such as Il10, Agr-1, and Tgfβ, in TANs and TAMs were reduced in rAcsl6 S674A tumors (fig. S7D). The lung metastasis model also showed rAcsl6 S674A markedly decreased TAN and TAM infiltration in metastatic tumors (fig. S7, E and F). To further investigate the roles of CXCL1 and CXCL5 in ACSL6 pS674–promoted immunosuppression, we neutralized CXCL1 and CXCL5 by pretreatment with anti-CXCL1 and anti-CXCL5 antibodies to reshape ACSL6–NF-κB–regulated immune functions. The neutralization of CXCL1 and CXCL5 strongly hampered cancer cell growth in the rAcsl6 WT tumors but had a relatively weak effect on the rAcsl6 S674A tumors (Fig. 7D). We also showed that neutralizing CXCL1 and CXCL5 significantly promoted CD8+ T cell infiltration and decreased TANs and TAMs recruitment in rAcsl6 WT tumors (Fig. 7E). These data indicate that ACSL6 pS674 is required for TANs and TAMs recruitment, which promotes immune escape in liver cancer and contributes to tumor progression.
Fig. 7. ACSL6 pS674 promotes TANs and TAMs recruitment.
(A to C) Flow cytometry analyses of tumor-infiltrating TANs (A), TAMs (B), PD-L1+ TANs, and CD170+ TANs (C) in indicated tumors from C57BL/6 mice. (D and E) Acsl6-depleted Hepa1-6 cells with forced expression of rAcsl6 WT or S674A were subcutaneously injected into C57BL/6 mice, which were then treated with or without neutralizing antibodies against CXCL1 and CXCL5. Tumor volumes were recorded (D). Flow cytometry analyses of tumor-infiltrating CD8+ T cells, TANs, and TAMs (E). (F to M) Liver cancer mouse model was constructed by hydrodynamic transfection of AKT/NRAS/SB plasmids before the injection of AAV8-TBG-miR30-Acsl6-shRNA-GFP or AAV8-TBG-GFP-miR30-shRNA. Schematic diagram and injection timeline (F). Immunoblotting analyses were performed with indicated antibodies (G). Representative liver images are shown (H). Liver weight (I), liver weight/body weight ratio (J), and tumor numbers (K) were measured. Representative image of H&E staining of mouse liver sections are shown (L). Flow cytometry analyses of tumor-infiltrating CD8+ T cells, TANs, and TAMs in indicated tumors (M). (N to R) Liver cancer mouse model was generated by hydrodynamic transfection of indicated plasmids (N). Representative liver images are shown (O). Liver weights (P), liver weight/body weight ratio (Q), and tumor numbers (R) were calculated. (S and T) Control or Acsl6-OE Hepa1-6 cells were subcutaneously injected into C57BL/6 mice, which were then treated with or without IL-18BP. Tumor volumes (S) and tumor-infiltrating CD8+ T cells, TANs, and TAMs (T) were analyzed. [(D) and (S)] n = 5; [(F) to (R)] n = 6. *P < 0.05, **P < 0.01, and ***P < 0.001. Student’s t test [(A) to (C), (I) to (K), (M), and (P) to (R)], two-way ANOVA [(D) and (S)], and one-way ANOVA [(E) and (T)].
To validate the function of ACSL6 in tumor immune evasion, we established a liver cancer mouse model via hydrodynamic transfection (28) before the injection of adeno-associated virus (AAV) expressing shAcsl6 or short hairpin RNA non-targeting (shNT) (Fig. 7, F and G). Injection of AAV expressing shAcsl6 after hydrodynamic injection blocked tumor progression, as indicated by decreases in liver weights, liver weight/body weight ratios, and tumor numbers (Fig. 7, H to L). Acsl6 knockdown facilitated the infiltration of CD8+ T cells and suppressed the recruitment of TANs and TAMs (Fig. 7M). Subsequently, a mouse liver cancer model generated via hydrodynamic transfection of Acsl6 overexpression showed that mice coinjected with Acsl6 developed substantial tumor burdens compared to control mice (Fig. 7, N to R, and fig. S7, G and H). Acsl6 overexpression significantly decreased CD8+ T cell infiltration and increased TANs and TAMs recruitment (fig. S7, I and J), suggesting that ACSL6 promotes tumor immune evasion. We further established a liver cancer orthotopic mouse model derived from Hepa1-6 cells. The results illustrated a significant reduction in the growth of orthotopic tumors upon Acsl6 depletion (fig. S7, K to M). To confirm the role of IL-18 in tumor immune regulation via the IL-18R–ACSL6–NF-κB pathway in vivo, we blocked IL-18 signaling with IL-18 binding protein (IL-18BP), which binds to IL-18 and inhibits the binding of IL-18 to IL-18R1 (29), in mice bearing tumors overexpressing Acsl6 or control. The results showed that IL-18BP treatment inhibited tumor growth and eliminated the differences in tumor burden between control and Acsl6 tumors (Fig. 7S and fig. S7N) and increased CD8+ T cells and decreased TANs and TAMs (Fig. 7T). Subsequently, we established an orthotopic liver cancer mouse model using Il18r1-depleted Hepa1-6 cells. The results revealed a significant reduction in the growth of orthotopic tumors upon Il18r1 depletion (fig. S7, O to Q). Overall, our data demonstrate the strong function of Acsl6 response to IL-18 in accelerating liver cancer tumorigenesis and progression in vivo.
ACSL6 pS674 level negatively correlates with ICI efficacy and the prognosis of patients with liver cancer
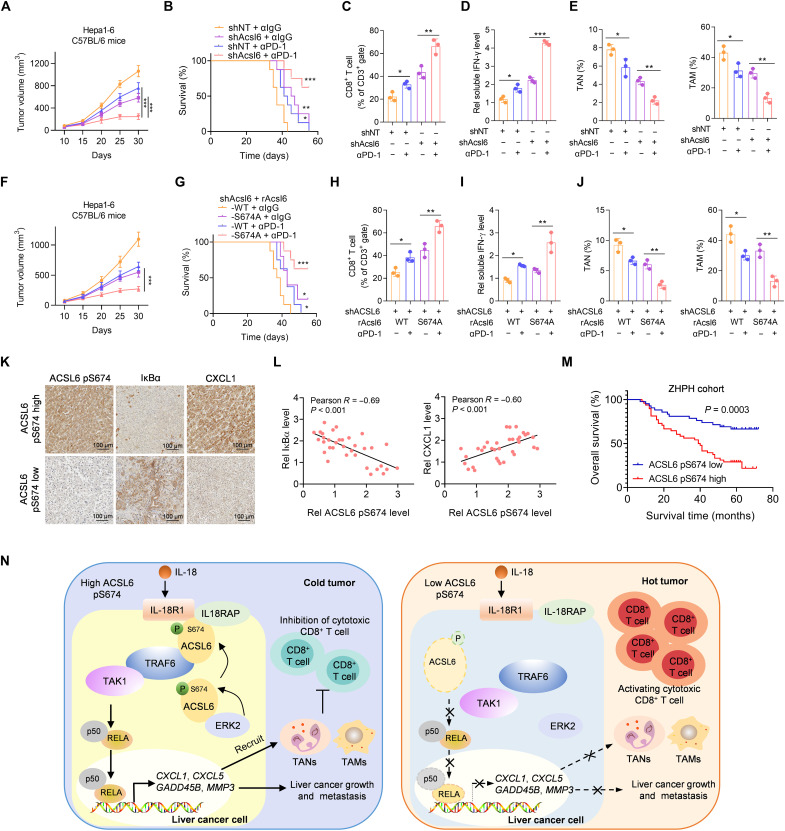
We next examined whether ACSL6 affects ICI efficacy. We treated mice bearing Acsl6-knockdown or control tumors with an anti–PD-1 antibody. The results indicated that Acsl6 depletion significantly enhanced the antitumor efficacy of anti–PD-1, as indicated by the slower tumor growth and higher survival rate in this group (Fig. 8, A and B). We also found that the levels of tumor-infiltrating CD8+ T cells and secreted T cell–derived IFN-γ were significantly increased in mice bearing Acsl6 knockdown tumors and treated with anti–PD-1, compared with those bearing control tumors (Fig. 8, C and D). Moreover, TANs and TAMs were reduced in the Acsl6 knockdown group treated with anti–PD-1 (Fig. 8E). We further investigated the correlation between the level of ACSL6 pS674 and ICI efficacy. Compared to rAcsl6 WT tumors, rAcsl6 S674A tumors showed a remarkable response to anti–PD-1, as indicated by slower tumor growth and higher survival rate (Fig. 8, F and G). Consistently, rAcsl6 S674A tumors treated with anti–PD-1 showed increased infiltrating CD8+ T cells and secreted IFN-γ levels (Fig. 8, H and I) and decreased TANs and TAMs (Fig. 8J). These results suggest that ACSL6 knockdown or the ACSL6 S674A mutation facilitates antitumor immunity and ICI efficacy.
Fig. 8. The level of ACSL6 pS674 negatively correlates with the efficacy of ICIs therapy and the prognosis of patients with liver cancer.
(A to E) Mice with established Acsl6 depletion or control Hepa1-6 tumors were treated with or without anti–PD-1. Tumor volumes (A) and survival rates (B) were recorded. Flow cytometry analyses of tumor-infiltrating CD8+ T cells (C). Measurement of the concentration of IFN-γ (D). Flow cytometry analyses of TANs and TAMs (E). (F to J) Acsl6 WT or S674A was reconstituted into Acsl6-depleted Hepa1-6 cells, which were then subcutaneously injected into C57BL/6 mice, and mice were subsequently treated with anti–PD-1. Tumor volumes (F) and survival rates (G) were recorded. Flow cytometry analyses of tumor-infiltrating CD8+ T cells (H). Measurement of the concentration of IFN-γ (I). Flow cytometry analyses of TANs and TAMs (J). (K and L) IHC staining with anti–ACSL6 pS674, anti-IκBα, and anti-CXCL1 antibodies in tumors from patients with liver cancer from ZHPH (K) and correlation analyses (L). (M) Kaplan-Meier analyses of overall survival according to the ACSL6 pS674 level in patients with liver cancer from ZHPH cohort. (N) Proposed model of the mechanism by which ACSL6 activates IL-18–NF-κB signaling to promote immune evasion and tumor progression in liver cancer. [(A) and (E)] n = 5; [(B) and (G)] n = 10. *P < 0.05, **P < 0.01, and ***P < 0.001. Two-way ANOVA [(A) and (F)], log-rank test [(B), (G), and (M)], and one-way ANOVA [(C) to (E) and (H) to (J)].
To validate the clinical significance of ACSL6 pS674, we quantified ACSL6 pS674 levels in liver cancer samples. IHC staining showed that ACSL6 pS674 was negatively correlated with IκBα expression and positively correlated with CXCL1 expression, suggesting that ACSL6 pS674 positively regulates NF-κB signaling and CXCL1 expression in the clinic (Fig. 8, K and L). Furthermore, patients with high ACSL6 pS674 levels had poorer prognoses than patients with low ACSL6 pS674 levels (Fig. 8M). These data support our finding that ACSL6 pS674 negatively correlates with ICI efficacy and the prognosis of patients with liver cancer.
DISCUSSION
Here, we discovered that ACSL6 plays an important role in IL-18–mediated immune escape and progression in liver cancer. Moreover, ACSL6 pS674 determines the progression of liver cancer and predicts the suitability of PD-1 blockade immunotherapy for patients with liver cancer. Specifically, upon IL-18 stimulation, ERK2 phosphorylates ACSL6 at S674, leading to its cell membrane translocation. Membrane-localized ACSL6 recruits IL-18RAP to interact with IL-18R1, thus consolidating the IL-18R1–IL-18RAP heterodimer and activating the expression of NF-κB target genes, including GADD45B, CXCL1, MMP3, and CXCL5. GADD45B and MMP3 have been shown to play critical roles in promoting tumor growth and metastasis (21, 22), while CXCL1 and CXCL5 are secreted by tumor tissues and generally recruit TANs and TAMs, which remodel the TME and decrease ICI efficacy (Fig. 8N).
Immunotherapy is a promising anticancer strategy that enhances the specificity and intensity of the anticancer immune response (30). PD-1/PD-L1 plays a crucial role in inhibiting T cell activity (31) and promoting cancer cell evasion through immune surveillance and killing (32). By blocking PD-L1 binding to PD-1, ICIs disrupt this negative feedback loop and restore T cell function, enabling them to effectively target and kill cancer cells. The first clinical study on ICIs for liver cancer was CheckMate 040, which evaluated the use of nivolumab in patients after the failure of sorafenib (33). The study reported a total effective rate of 20% and a disease control rate of 64%, which indicated the potential of ICIs to trigger long-term responses. However, the TME of liver cancer contains many immunosuppressive cells, including TANs and TAMs, which greatly impair ICI efficacy (34). Moreover, TANs have been shown to promote tumorigenesis, proliferation, metastasis, and immunosuppression (35). Robust TAN infiltration in solid tumors often correlates with poor clinical outcomes (36). Similarly, TAMs are involved in tumor initiation, progression, angiogenesis, and metastasis. The abundant infiltration of TAMs is linked to adverse clinical outcomes and a reduced response to standard treatments, including radiotherapy, chemotherapy, and ICIs (37, 38). Immunosuppressive cells are the major barrier to maximizing the clinical potential of immunotherapies.
Here, we found that ACSL6 promotes liver cancer cell immune escape in response to IL-18. IL-18 induces ACSL6 pS674 to promote ACSL6 translocation and activate NF-κB signaling, thereby promoting the expression of CXCL1 and CXCL5, which recruit TANs and TAMs to inhibit CD8+ T cell infiltration. Furthermore, ACSL6 pS674 was positively correlated with CXCL1 expression in human liver cancer tissues. Patients with liver cancer with high ACSL6 pS674 levels exhibited worse overall survival. Our study revealed a mechanism through which ACSL6 functions in immune regulation by acting as an adaptor to link IL-18R and NF-κB, induce NF-κB–dependent gene expression, and inhibit CD8+ T cell accumulation and ICI efficacy, which facilitates liver cancer immune escape and progression.
NF-κB signaling is widely recognized to be closely involved in immunosuppression. This pathway can promote the expression of various immunosuppressive factors, including CXCL1 and CXCL5, which, in turn, recruit immunosuppressive cells and contribute to the formation of an immunosuppressive microenvironment (27, 39). Acharyya et al. (40) reported that TNF increases CXCL1/2 expression in cancer cells by activating NF-κB, leading to myeloid cell recruitment and promoting immunosuppression, thereby enhancing cancer cell survival and contributing to chemoresistance. Notably, many types of cancer exhibit constitutive NF-κB activation, and this heightened NF-κB activity is largely attributed to increased production of cytokines such as IL-1β and IL-18 (41, 42). NF-κB activation is also involved in regulating cytokine and growth factor secretion in macrophages through the interaction of repressor activator protein 1 (Rap1) with IKK and that promotes NF-κB activation in proinflammatory macrophages (43). Ding et al. (44) showed that Rap1 deficiency reduces NF-κB activity and impairs cytokine production, which affects the immunosuppressive potency of mesenchymal stem cells. Notably, in addition to the classical NF-κB pathway, the nonclassical NF-κB pathway also plays an important role in various diseases, including tumors, and in maintaining a functional immune system (45). In the present study, we found that ACSL6 specifically activates the classical NF-κB pathway induced by IL-18 without affecting nonclassical NF-κB signaling, revealing the important role of ACSL6 in regulating the IL-18 signaling through classical NF-κB signaling.
As a proinflammatory/immunostimulatory cytokine, IL-18 regulates various immune cells, including natural killer, T helper 1, and cytotoxic T cell 1 (Tc1) (46, 47). Increasing evidence suggests that IL-18 also promotes tumorigenesis. Higher IL-18 levels promote pancreatic cancer invasion and metastasis via the activation of NF-κB pathway and are correlated with a poor prognosis (48). However, the mechanism linking IL-18 signaling and NF-κB activation to promote tumor progression remains largely unclear. Here, we revealed a protumor role for IL-18 in liver cancer and demonstrated that IL-18 induces ACSL6 pS674 by ERK2 and promotes the ACSL6–IL-18R1 interaction. First, ACSL6 pS674 plays a role in IL-18RAP recruitment to consolidate the IL-18R heterodimer, thereby facilitating IL-18 signaling. Moreover, ACSL6 acts as a scaffold protein to bridge IL-18 signaling and NF-κB pathway activation, resulting in liver cancer immune evasion and progression. Our study identified ACSL6 as a driver of IL-18–NF-κB signaling, providing a target for inhibiting the IL-18 pathway to inhibit tumor progression.
Notably, the role of IL-18 in liver cancer progression remains controversial. Although elevated levels of IL-18 in the serum of patients with liver cancer and higher levels of circulating IL-18 correlate with a worse prognosis (49), conflicting experimental results regarding the protumor or antitumor functions of IL-18 have been reported (50). Markowitz et al. (51) reveal that IL-18 exerts inflammation-dependent tumor-suppressive effects by promoting the differentiation, activity, and survival of tumor-infiltrating T cells using mouse models with systemic knockout of the IL-18R1. In our study, we focused on the function of IL-18 in liver cancer cells and found that the IL-18–IL-18R1–ACSL6 signaling axis plays a pro-oncogenic role in liver cancer. Therefore, our study, along with the study of Markowitz et al., investigated the function of IL-18 in tumor cells and immune cells, respectively. Together, these findings complement each other and provide a more comprehensive understanding of the contradictory results for the role of IL-18 in the occurrence and progression of liver cancer. However, different types of tumors have distinct microenvironments and signaling pathways, which can influence tumor behavior and response to various factors. Therefore, further investigation is necessary to determine whether the impact of ACSL6 observed in liver cancer also holds true for other tumor types.
MATERIALS AND METHODS
Cell culture
THLE-2 cells were donated by S. Lin (The First Affiliated Hospital, Sun Yat-sen University, China) and cultured in complete THLE-2 medium (MeisenCTCC, CTCC-002-046). The human cell lines Huh7, Sk-Hep1, MHCC97L, MHCC97H, Hep3B, HepG2, SNU423, SNU449, and human embryonic kidney (HEK) 293T and the mouse cell line Hepa1-6 were purchased from the American Type Culture Collection or maintained in our laboratory. Cells were cultured at 37°C with 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) or RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), penicillin (100 μg ml−1), and streptomycin (100 μg ml−1). All cell lines tested negative for mycoplasma.
Plasmid construction and transfection
To generate a constitutive human ACSL6 overexpression plasmid, ACSL6 was amplified from human cDNA and cloned into pCDH-Flag, pCDH-SFB-hygro, pCDH-HA, pcDNA4.0, and pXJ40.HA. PCR-amplified mouse Acsl6 was cloned into PCDH-SFB-hygro. To generate a constitutive human IL18R1 overexpression plasmid, IL18R1 was amplified from human cDNA and cloned into pXJ40.HA. PCR-amplified IL18RAP was cloned into the pCDH-Flag vector. The plasmids of TRAF6, TAK1, TAB2, TAB3, IKKα, RELA, and P50 were cloned into the pCDH-Flag vector and were gifts from the Cancer Center of Sun Yat-sen University. shRNAs targeting ACSL6, Acsl6, IL18R1, and ERK2 were cloned into the pLKO.1 vector. For plasmid or small interfering RNA (siRNA) transfection, HEK293T and liver cancer cells were transfected with EZ Trans (Shanghai Life iLab Biotechnology, AC04L092) and Lipo3000 (Invitrogen, L3000-015), respectively, in Opti-DMEM according to the manufacturer’s instructions and harvested 48 hours after transfection. The sequences of the shRNAs and siRNAs used are listed in table S2.
Patients
The study was approved by the Ethics Committee of The First Affiliated Hospital, Sun Yat-sen University, and Zhuhai People’s Hospital, Zhuhai Hospital Affiliated with Jinan University. The studies were conducted in accordance with the recognized ethical guidelines of the Declaration of Helsinki. qPCR and immunoblotting analyses of ACSL6 expression were performed using tumor tissues and paired adjacent normal tissues from patients with liver cancer obtained from The First Affiliated Hospital, Sun Yat-sen University, with the consent of the patients ([2024]108). IHC analyses of ACSL6 or ACSL6 pS674 levels were performed in tumor tissues and paired adjacent normal tissues from 90 patients (ZHPH2022053) with liver cancer obtained from Zhuhai People’s Hospital, Zhuhai Hospital Affiliated with Jinan University, with the consent of the patients.
Mice
All mice, including female BALB/c nude mice, NCG mice, and C57BL/6 mice, were purchased from Guangdong GemPharmatech. All animal experiments were conducted in accordance with the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals. The animals were maintained under specific pathogen–free conditions. The experimental procedures were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University (approval number SYSU-IACUC-2022-001685). For the BALB/c nude mouse subcutaneous xenograft model, 6-week-old female BALB/c nude mice were randomly allocated to several groups. Briefly, 2 × 106 MHCC97H cells stably expressing shNT, shACSL6, or shACSL6 reconstituted with rACSL6 WT or ED were subcutaneously injected into the dorsal flank of BALB/c nude mice. After 9 days of inoculation, the tumor volumes were measured every 3 days. After injection for 28 days, the mice were euthanized, and the tumors were excised and weighed. A total of 2 × 106 HepG2 cells stably expressing EV, ACSL6 WT, or ED were subcutaneously injected into the dorsal flank of BALB/c nude mice. Tumor volumes were measured every 3 days after injection for 6 days, the mice were euthanized, and the tumors were excised and weighed after injection for 24 days.
For the NCG mouse or C57BL/6 mouse subcutaneous mouse liver cancer model, 2 × 106 Hepa1-6 cells stably expressing shAcsl6 reconstituted with Acsl6 WT or S674A were subcutaneously injected into the dorsal part of 6-week-old female NCG mice or C57BL/6 mice. Tumor volumes were monitored every 4 to 5 days following inoculation. Mice were euthanized after inoculation for about 30 days. For PD-1 antibody treatments, mice were treated with 200 μg of anti–PD-1 antibody (BioXcell, no. BE0273) or immunoglobulin G (IgG) control via intraperitoneal injection twice a week for 2 weeks beginning on day 15 after subcutaneous tumor implantation. For the IL-18 blockade assay, mice were treated with mouse IL-18BP (25 μg/kg; MedChemExpress, no. HY-P75841) or vehicle phosphate-buffered saline (PBS) via intratumoral injection for three cycles, with 3 days per cycle and a 2-day interval between cycles. For the CXCL1 and CXCL5 antibody neutralization model, 3 × 106 Hepa1-6 cells stably expressing shAcsl6 reconstituted with Acsl6 WT or S674A were subcutaneously injected into the dorsal flank of 6-week-old female C57BL/6 mice. After injection for 8 days, the tumor-bearing mice were treated by intraperitoneally injecting 100 μg of anti-CXCL1 antibody (R&D Systems, no. MAB453) and 100 μg of anti-CXCL5 antibody (Novus Biologicals, no. NBP2-22026) or IgG control twice a week for 2 weeks beginning on day 8 after subcutaneous tumor implantation.
For the tail vein metastasis model, 6-week-old female BALB/c nude or C56BL/6J mice were randomly divided into several groups. Then, 2 × 106 stable MHCC97H, HepG2, or Hepa1-6 cells were injected into the mice via the tail vein. Six to 8 weeks after the injection, the mice were euthanized, and the lungs were removed and fixed with 4% paraformaldehyde (Biosharp Life Science, no. BL539A). Lungs were embedded in paraffin and subjected to hematoxylin and eosin staining.
The hydrodynamic injection liver cancer model was established in mice as previously described (28, 52). Briefly, 20 μg of pT3-EF1αH-myr-AKT-HA, 20 μg of pT/Caggs-NRASV12, and sleeping beauty (SB) transposase (1 of the 25 total plasmids) were diluted in 2 ml of saline, and the mixture was subsequently injected into the lateral tail vein within 7 s. For the Acsl6 overexpression mouse model (Acsl6 OE), 20 μg of pT3-EF1αH-myr-AKT-HA, 20 μg of pT/Caggs-NRASV12, and 20 μg of pT3-EF1α-Acsl6 plasmids, along with 2.4 μg of SB, were injected into 6-week-old female C57BL/6 mice via the same protocol. In the control group, pT3-EF1αH-myr-AKT-HA, pT/Caggs-NRASV12, pT3-EF1α-empty, and SB were hydrodynamically injected. After 4 weeks, the mice were euthanized, and the livers were collected to evaluate liver cancer development. For the Acsl6 knockdown model, 20 μg of pT3-EF1αH-myr-AKT-HA, 20 μg of pT/Caggs-NRASV12, and 1.6 μg of SB were injected via the same protocol to induce liver cancer. After hydrodynamic injection for 1 week, the mice were randomly assigned six mice per group. Then, AAV8-TBG-miR30-Acsl6-shRNA-GFP or AAV8-TBG-GFP-miR30-shRNA (1.5 × 1012 genome copies per mouse) AAV (WZ Biosciences Inc.) was injected via tail vein. After AAV injection for 4 weeks, mouse livers were collected to evaluate the effect of Acsl6 knockdown on liver cancer progression.
For the PDX mouse model, xenografts were generated from patients with orthotopic liver cancer. Tumor tissues resected from patients were isolated, cut into pieces, and implanted into the flanks of NCG mice. The tissues were divided into groups on the basis of ACSL6 expression levels. After reaching a tumor diameter of 10 mm, the tumor tissues were harvested, isolated, and cut into blocks. Subsequently, these blocks were implanted into the left liver lobe of tumor-free NCG mice. After 33 days, the mice were euthanized, and the xenografts were removed, weighed, and subjected to pathological analysis. For the liver cancer orthotopic mouse model, a microinjector delivered a mixture of Hepa1-6 cells in PBS into the left liver lobe of C56BL/6 mice. The needle was inserted 2 to 3 mm into the liver and withdrawn slightly to ensure that it was not in a blood vessel. The cell suspension was then injected slowly, with the needle remaining in place for 1 min after injection. Sterile cotton swabs were applied to the needle entry site for 30 s after injection to prevent bleeding. Last, the surgical incision was closed with sutures. Mice were euthanized after 4 to 6 weeks for analyses.
Cytokine measurements
CXCL1 and CXCL5 protein levels in the conditioned medium from indicated tumor cells treated with IL-18 (20 ng ml−1) for 24 hours were measured using enzyme-linked immunosorbent assay (ELISA; R&D Systems, nos. DGR00B and DX000) according to the manufacturer’s protocol. Mouse CXCL1, CXCL5, and IFN-γ levels in tumor tissue supernatants from C56BL/6J mice were measured using ELISAs (R&D Systems, no. MKC00B; eBioscience, nos. EMCXCL5 and 88-7314-22) according to the manufacturer’s protocol.
Isolation of tumor-infiltrating cells and flow cytometry
The mice were euthanized at the indicated time points, and the tumors were cut into small pieces in PBS. Tumors were digested in RPMI medium supplemented with 2% FBS, collagenase type IV (50 U ml−1; Invitrogen, no. 17104019), and deoxyribonuclease (20 U ml−1; Roche, no. 10104159001) at 37°C for 1 hour. Suspensions were washed with cold PBS and then filtered through a 70-μm strainer. The cells were centrifuged, and the cell pellet was resuspended in 200 μl of PBS supplemented with 2% FBS. For tumor-infiltrating CD4+ and CD8+ T cell analysis, the cells were stained with fixable viability stain 700 (FVS700, BD Biosciences, no. 564997), mouse anti-CD45 (eBioscience, no. 47-0451-82), mouse anti-CD3 (eBioscience, no. 11-0031-82), mouse anti-CD4 (eBioscience, no. 48-0041-82), mouse anti-CD8 (BD Pharmingen, no. 552877), mouse anti–PD-1 (BioLegend, no. 109103), and mouse anti–T cell immunoglobulin and mucin domain-containing protein 3 (TIM-3) (BD Biosciences, no. 747623) antibodies. For tumor-infiltrating IFN-γ+ and GZMB+CD8 T cell analysis, the cells were stained with FVS700, mouse anti-CD45, mouse anti-CD3, and mouse anti-CD8 antibodies. Then, the cells were fixated and permeabilized using the Fixation/Permeabilization Kit (BD Biosciences, no. 554714) and stained with mouse anti–IFN-γ (eBioscience, no. 12-7311-82) or mouse anti-GZMB (eBioscience, no. 17-8898-82). For tumor-infiltrating TANs and TAMs analysis, the cells were stained with FVS700, mouse anti-CD45, mouse anti-CD11b (BD Pharmingen, no. 557396), mouse anti-Ly6G (BD Pharmingen, no. 560602), mouse anti-Ly6C (Biolegend, no. 128033), mouse anti-PD-L1 (BD Pharmingen, no. 558091), mouse anti-CD170 (BD Pharmingen, no. 562681), mouse anti-F4/80 (BD Pharmingen, no. 566787), and mouse anti-CD206 (BD Pharmingen, no. 141720) antibodies. Flow cytometry was performed (BD Pharmingen), and the data were analyzed using FlowJo v10.8 software.
Detection of cell proliferation using the CCK-8 assay
A total of 1000 cells per well were seeded into 96-well plates. The plates were preincubated with 5% CO2 at 37°C. Cell Counting Kit-8 (CCK-8) solution (Good Laboratory Practice Bioscience, no. XYM-GLP-GK10001-500) was added to the wells at specific time points (1 to 5 days). Then, the plate was incubated in an incubator for 1 to 4 hours. Last, the absorbance was measured at 450 nm using an Infinite 200 PRO microplate reader (M PLEX, Tecan).
Colony formation assay
A total of 2000 cells per well were seeded in six-well plates and cultured in an incubator (37°C, 5% CO2). After 8 to 14 days of culture, the colonies were gently washed with PBS, fixed with 4% paraformaldehyde for 30 min, and stained with crystal violet (Beyotime, no. C0121) for 30 min. The stained colonies were photographed and counted.
Transwell assay
Cells (8 × 104 to 10 × 104) in serum-free medium were seeded in the upper chamber of a transwell insert with 8-μm pores and cultured for 24 to 48 hours. Cells on the basal surface of the membrane were then fixed with 4% paraformaldehyde for 30 min and stained with crystal violet for 30 min. The non-migrated cells on the apical surface of the membrane were removed by wiping with a cotton swab. Images were acquired with an inverted light microscope and analyzed with ImageJ software.
Wound healing assay
Stable MHCC97H, Huh7, HepG2, or Sk-Hep1 cells were seeded in six-well plates and allowed to grow to confluence. When the cells had adhered to the plate, a sterile plastic pipette tip was used to scratch one linear wound in each well. Then, the cells were washed with PBS twice to remove cellular debris, and medium supplemented with 1% FBS was added. After 0 and 48 hours, images of the cells were captured with an inverted light microscope. The area of the scratch wound was compared between the two time points, and the cell migration/wound healing rate was calculated.
RNA extraction, reverse transcription, and qPCR
Total RNA extraction reagent (Vazyme, no. R401-01) was used to isolate total RNA according to the manufacturer’s instructions, and 1 μg of total RNA was used to synthesize cDNA using HiScript II Reverse Transcriptase for qPCR from Vazyme (no. RL201-01). qPCR was performed using ChamQ SYBR qPCR Master Mix from Vazyme (no. Q712) according to the manufacturer’s instructions. Relative gene expression was normalized to that of the control gene actin beta (ACTB). The qPCR primers used are listed in table S3.
RNA-seq and data analysis
Total RNA was extracted with TRIzol reagent (Vazyme, no. R401-01) and sequenced on the Illumina HiSeq 2500 platform (Shanghai Jingfang Biotechnology Co. Ltd., China). The R programming language was used to calculate P values and plot heatmaps. GSEA was performed using GSEA software (53).
Immunoblotting and co-IP
For immunoblotting analyses, cultured cells were lysed using radioimmunoprecipitation assay lysis buffer (Cell Signaling Technology, no. 9806) supplemented with protease inhibitors (MedChemExpress, no. HY-K0010) and phosphatase inhibitors (Biosharp Life Science, no. BL615A). Protein concentrations were measured with a bicinchoninic acid (BCA) protein assay kit (Bestbio, no. BB-3401-500 T), and samples containing equal amounts of protein were used for immunoblotting analyses. To determine the relative ACSL6 protein levels in liver cancer tissues and paired adjacent tissues, the intensities of ACSL6 and β-actin bands were analyzed using ImageJ software, and the band intensity of ACSL6 was divided by that of β-actin (ACSL6/β-actin). Subsequently, the relative ACSL6 level was calculated by dividing the ACSL6/β-actin value for each sample by the ACSL6/β-actin value for the first sample. For the co-IP analysis, Pierce IP lysis buffer (Thermo Fisher Scientific, no. 87787) containing protease and phosphatase inhibitors was used to lyse cells. Anti-Flag M2 beads (Sigma-Aldrich, no. A2220) or the indicated antibodies with Protein A/G beads (Santa Cruz Biotechnology, no. sc-2003) were added to the cell lysates, which were incubated at 4°C for 6 to 12 hours. IP buffer was used to wash the protein bead complexes three times. The immunoprecipitated proteins were used for immunoblotting analyses. The antibodies used are listed in table S4.
MS analysis
For the IP-MS analysis, we performed IP on MHCC97H cells stably expressing Flag-ACSL6 using anti-Flag M2 beads or an IgG control. The precipitated complexes were then boiled at 95°C for 10 min and separated via SDS–polyacrylamide gel electrophoresis (PAGE). After staining with Coomassie Brilliant Blue (Beyotime, no. P0017), we excised bands with significant differences after precipitation using anti-Flag M2 beads and the IgG control for MS identification, and IL-18R1 was identified. For the identification of ACSL6 phosphorylation sites, Flag-ACSL6 proteins were immunoprecipitated from MHCC97H cells stably expressing Flag-ACSL6, and the precipitated complexes were boiled at 95°C for 10 min and separated via SDS-PAGE. After staining with Coomassie Brilliant Blue, we excised the ACSL6 bands for MS identification. MS analysis was conducted at the Institutional Technology Center of the Shanghai Institute of Materia Medica. The peptides were analyzed using liquid chromatography–tandem MS on a Q Exactive mass spectrometer (Thermo Fisher Scientific, no. 0726090). Proteins were identified by searching the fragment spectra against the universal SWISS-PROT protein database (European Bioinformatics Institute, EBI) using Mascot Server 2.4. The phospho-peptide matches were analyzed using MaxQuant v.1.5.2.8 implemented in Proteome Discoverer and manually curated.
Computational structure prediction
The three-dimensional structures of ACSL6 (AF-Q9UKU0-F1) and IL-18R1 (AF-Q13478-F1) were obtained from the AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk/) (54). The structure of ACSL6 in complex with IL-18R1 was predicted using the ZDOCK protein-protein docking web server (https://zdock.umassmed.edu/) based on the pose with the best docking score (55).
In vitro kinase assay
Flag-ACSL6 was immunoprecipitated from MHCC97H cells using anti-Flag M2 beads. Flag-ACSL6 was incubated with or without bacterially purified recombinant GST-ERK2 in kinase assay buffer (Cell Signaling Technology, no. 9802) supplemented with 200 μM adenosine 5′-triphosphate (Cell Signaling Technology, no. 9804) for 30 min at 30°C. The kinase assay was stopped with 20 μl of 3× SDS sample buffer, and the protein expression was measured using the indicated antibody.
Subcellular fractionation analysis
The cells were washed and collected with ice-cold PBS. Cytoplasmic and membrane proteins were extracted using a Mem-PER Plus kit (Thermo Fisher Scientific, no. 89842) according to the manufacturer’s instructions. The protein concentration was measured using a BCA protein assay kit, and samples containing equal amounts of protein were used for immunoblot analysis.
Immunohistochemistry
IHC staining was performed as previously described (56). Briefly, tissue sections from paraffin-embedded human or mouse liver cancer specimens were stained with anti-ACSL6, anti-Ki67 (Invitrogen, no. PA5-114437), mouse anti-CD8A (Cell Signaling Technology, no. 98941), human anti-CD8A (Cell Signaling Technology, no. 85336), anti–ACSL6 pS674, anti-IκBα (Cell Signaling Technology, no. 4814), or anti-CXCL1 (Proteintech, no. 12335-1-AP) antibodies. IHC was used to score the tissue sections. On the basis of the patient’s survival status and time, we generated overall survival graphs using GraphPad Prism software (v.8.0) and used the Kaplan-Meier method to calculate P values.
IF staining
The cells were fixed and incubated with primary antibodies, Alexa Fluor–conjugated secondary antibodies, and 4′,6-diamidino-2-phenylindole (Sigma-Aldrich, no. D9542) according to the manufacturer’s instructions. An LSM 900 laser confocal microscope (Zeiss) was used to acquire images of the cells and determine the locations of ACSL6 and IL-18R1. ImageJ was used to analyze the fluorescence intensity.
Chromatin immunoprecipitation
ChIP was performed as previously described (57). Briefly, 37% formaldehyde (Sangon Biotech, no. A501912-0500) was added to the culture medium to fix the cells. Cross-linking was performed at 37°C for 6 min and quenched by adding glycine (Sigma-Aldrich, no. V900144) for 6 min. Chromatin from the lysed cells was digested by sonication and enzymatic methods. Two percent of sonicated DNA not subjected to immunoprecipitation was used as the input. Chromatin was then immunoprecipitated using rabbit antibodies against RELA and IgG (negative control). Reverse transcription qPCR was used to evaluate the enrichment of RELA at the GADD45B, CXCL1, MMP3, and CXCL5 promoters.
Dual-luciferase reporter assay
The promoters of GADD45B, CXCL1, MMP3, and CXCL5 were cloned and inserted separately into the pGL3 basic vector. Cells were transfected with the reporter plasmid and thymidine kinase (TK) plasmid. After 48 hours, the cells were lysed, and the supernatant was analyzed using the TransDetect Double-Luciferase Reporter Assay Kit (TransGen Biotech, no. RF201) according to the manufacturer’s instructions.
Database analyses
We compared ACSL4, ACSL6, and ACSM1 transcript levels between liver cancer tissues and adjacent normal tissues using the GSE63898 (13), GSE64041 (58), GSE14323 (59), GSE14520 (60), and ICGC-LIRI-JP datasets. TCGA database was used to compare the expression of ACSL6 between normal tissues and liver adenocarcinoma tissues and to analyze the survival rate of patients according to ACSL6 expression.
Immune cell infiltration estimates for TCGA and GEO database expression profiles
To investigate the relationship between ACSL6 expression and the immune microenvironment, we used the immune estimation function in TIMER 2.0 (http://timer.cistrome.org) (25). First, we uploaded expression profiles from TCGA and GEO databases (GSE63898 and GSE14520) and selected “human” as the species and “liver hepatocellular carcinoma” as the cancer type. After running the analysis on the TIMER 2.0 website, immune infiltration estimates, such as immune scores, cytotoxicity scores, and CD8+ T cell infiltration levels, were calculated.
Statistical analysis
All in vitro experiments were repeated at least three times, and the animal experiments were repeated at least twice. All quantitative data are presented as individual data points and as the means ± SD. GraphPad Prism software (v.8.0) was used for statistical analysis. The statistical methods used for each experiment are described in the figure legends. P < 0.05 was considered to indicate statistical significance.
Acknowledgments
We would like to thank the Institutional Technology Center of Shanghai Institute of Materia Medica for MS technology support. We thank J. Wang and S. Lin (The First Affiliated Hospital, Sun Yat-sen University, China) for comments and suggestions, S. Chen (Sun Yat-sen University Cancer Center, China) for providing hydrodynamic plasmids, and H. Zhang (Southern Medical University, China) for assistance in flow cytometry analysis.
Funding: This project was supported by grants from the National Natural Science Foundation of China (82303614, 82203152, 21933010, 82230067, and 92359302), National Key Research and Development Plan (2022YFC3401000), Guangdong Basic and Applied Basic Research Foundation (2022A1515111079 and 2021B1515230009), Guangdong Provincial Key Areas R&D Programs of “Precision medicine and stem cells” (2023B1111020005), Strategic Priority Research Program of Chinese Academy of Sciences (XDB 37000000), and China Postdoctoral Science Foundation (2022 T150760 and 2023 T160750).
Author contributions: Conceptualization: X.Wa., Y.D., Z.W., and L.L. Methodology: X.Wa., Y.D., Z.W., and L.L. Investigation: Y.D., Z.W., L.L., and J.X. Validation: Y.D., Z.W., X.Z., L.Y., X.We., J.X., and L.L. Sources: W.H., Y.D., Z.W., L.L., and J.X. Formal analysis: Y.D., Z.W., X.Z., and L.L. Visualization: X.Wa., Y.D., Z.W., and L.L. Data curation: Z.W. and J.Q. Funding acquisition: W.H., Y.D., Z.W., and L.L. Supervision: X.Wa., Z.W., W.H., and L.L. Writing—original draft: X.Wa., Y.D., Z.W., and L.L. Writing—review and editing: X.Wa., Y.D., Z.W., W.H., X.Z., L.L., and J.X. Project administration: X.Wa., Z.W., Y.D., W.H., and L.L.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq data have been submitted to the GEO with accession number GSE240768.
Supplementary Materials
The PDF file includes:
Figs. S1 to S8
Tables S1 to S4
Legend for data S1
Other Supplementary Material for this manuscript includes the following:
Data S1
REFERENCES AND NOTES
- 1.Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., Bray F., Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Vogel A., Meyer T., Sapisochin G., Salem R., Saborowski A., Hepatocellular carcinoma. Lancet 400, 1345–1362 (2022). [DOI] [PubMed] [Google Scholar]
- 3.Rizvi S., Wang J., El-Khoueiry A. B., Liver cancer immunity. Hepatology 73, 86–103 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finn R. S., Qin S. K., Ikeda M., Galle P. R., Ducreux M., Kim T. Y., Kudo M., Breder V., Merle P., Kaseb A. O., Li D. N., Verret W., Xu D. Z., Hernandez S., Liu J., Huang C., Mulla S., Wang Y. L., Lim H. Y., Zhu A. X., Cheng A. L., Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N. Engl. J. Med. 382, 1894–1905 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Gordon S. R., Aute R. L. M., Dulken B. W., Hutter G., George B. M., Ccracken M. N. M., Gupta R., Tsai J. M., Sinha R., Corey D., Ring A. M., Connolly A. J., Weissman I. L., PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature 545, 495–499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z. N., Li B. H., Li S., Lin W. L., Wang Z., Wang S. D., Chen W. D., Shi W., Chen T., Zhou H., Yinwang E., Zhang W. K., Mou H. C., Chai X. P., Zhang J. H., Lu Z. M., Ye Z. M., Metabolic control of CD47 expression through lat2-mediated amino acid uptake promotes tumor immune evasion. Nat. Commun. 13, 6308 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakamura K., Kassem S., Cleynen A., Chrétien M. L., Guillerey C., Putz E. M., Bald T., Förster I., Vuckovic S., Hill G. R., Masters S. L., Chesi M., Bergsagel P. L., Avet-Loiseau H., Martinet L., Smyth M. J., Dysregulated IL-18 is a key driver of immunosuppression and a possible therapeutic target in the multiple myeloma microenvironment. Cancer Cell 33, 634–648.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Tang Y., Zhou J., Hooi S. C., Jiang Y. M., Lu G. D., Fatty acid activation in carcinogenesis and cancer development: Essential roles of long-chain acyl-CoA synthetases. Oncol. Lett. 16, 1390–1396 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui M., Xiao Z. L., Wang Y., Zheng M. Y., Song T. Q., Cai X. L., Sun B. D., Ye L. H., Zhang X. D., Long noncoding RNA hulc modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 75, 846–857 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Dongiovanni P., Crudele A., Panera N., Romito I., Meroni M., De Stefanis C., Palma A., Comparcola D., Fracanzani A. L., Miele L., Valenti L., Nobili V., Alisi A., β-Klotho gene variation is associated with liver damage in children with NAFLD. J. Hepatol. 72, 411–419 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Ubellacker J. M., Tasdogan A., Ramesh V., Shen B., Mitchell E. C., Martin-Sandoval M. S., Gu Z. M., McCormick M. L., Durham A. B., Spitz D. R., Zhao Z. Y., Mathews T. P., Morrison S. J., Lymph protects metastasizing melanoma cells from ferroptosis. Nature 585, 113–118 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y. J., Fahrmann J. F., Aftabizadeh M., Zhao Q., Tripathi S. C., Zhang C., Yuan Y., Ann D., Hanash S., Yu H., Fatty acid oxidation protects cancer cells from apoptosis by increasing mitochondrial membrane lipids. Cell Rep. 39, 111044 (2022). [DOI] [PubMed] [Google Scholar]
- 13.Villanueva A., Portela A., Sayols S., Battiston C., Hoshida Y., Mendez-Gonzalez J., Imbeaud S., Letouze E., Hernandez-Gea V., Cornella H., Pinyol R., Sole M., Fuster J., Zucman-Rossi J., Mazzaferro V., Esteller M., Llovet J. M., Consortium H., DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology 61, 1945–1956 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Chen J., Ding C., Chen Y., Hu W., Yu C., Peng C., Feng X., Cheng Q., Wu W., Lu Y., Xie H., Zhou L., Wu J., Zheng S., ACSL4 reprograms fatty acid metabolism in hepatocellular carcinoma via c-Myc/SREBP1 pathway. Cancer Lett. 502, 154–165 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Li H., Song J., He Y., Liu Y., Liu Z., Sun W., Hu W., Lei Q. Y., Hu X., Chen Z., He X., CRISPR/Cas9 screens reveal that hexokinase 2 enhances cancer stemness and tumorigenicity by activating the ACSL4-fatty acid β-oxidation pathway. Adv. Sci. 9, e2105126 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soupene E., Dinh N. P., Siliakus M., Kuypers F. A., Activity of the acyl-CoA synthetase ACSL6 isoforms: Role of the fatty acid gate-domains. BMC Biochem. 11, 18 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yasuda K., Nakanishi K., Tsutsui H., Interleukin-18 in health and disease. Int. J. Mol. Sci. 20, 649 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X. D., Shao S. X., Jiang H. P., Cao Y. W., Wang Y. H., Yang X. C., Wang Y. L., Wang X. S., Niu H. T., Warburg effect or reverse Warburg effect? A review of cancer metabolism. Oncol. Res. Treat. 38, 117–122 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Myhr C. B., Hulme M. A., Wasserfall C. H., Hong P. J., Lakshmi P. S., Schatz D. A., Haller M. J., Brusko T. M., Atkinson M. A., The autoimmune disease-associated SNP rs917997 of IL18RAP controls IFNγ production by PBMC. J. Autoimmun. 44, 8–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M., Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Sternlicht M. D., Lochter A., Sympson C. J., Huey B., Rougier J. P., Gray J. W., Pinkel D., Bissell M. J., Werb Z., The stromal proteinase MMP3/stromelysin-1 promotes mammary carcinogenesis. Cell 98, 137–146 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papa S., Zazzeroni F., Fu Y. X., Bubici C., Alvarez K., Dean K., Christiansen P. A., Anders R. A., Franzoso G., Gadd45β promotes hepatocyte survival during liver regeneration in mice by modulating JNK signaling. J. Clin. Invest. 118, 1911–1923 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seifert L., Werba G., Tiwari S., Giao Ly N. N., Alothman S., Alqunaibit D., Avanzi A., Barilla R., Daley D., Greco S. H., Torres-Hernandez A., Pergamo M., Ochi A., Zambirinis C. P., Pansari M., Rendon M., Tippens D., Hundeyin M., Mani V. R., Hajdu C., Engle D., Miller G., The necrosome promotes pancreatic oncogenesis via CXCL1 and mincle-induced immune suppression. Nature 532, 245–249 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mollaoglu G., Jones A., Wait S. J., Mukhopadhyay A., Jeong S., Arya R., Camolotto S. A., Mosbruger T. L., Stubben C. J., Conley C. J., Bhutkar A., Vahrenkamp J. M., Berrett K. C., Cessna M. H., Lane T. E., Witt B. L., Salama M. E., Gertz J., Jones K. B., Snyder E. L., Oliver T. G., The lineage-defining transcription factors SOX2 and NKX2–1 determine lung cancer cell fate and shape the tumor immune microenvironment. Immunity 49, 764–779.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li T., Fu J., Zeng Z., Cohen D., Li J., Chen Q., Li B., Liu X. S., TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 48, W509–W514 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franklin R. A., Li M. O., Ontogeny of tumor-associated macrophages and its implication in cancer regulation. Trends Cancer 2, 20–34 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kemp S. B., Carpenter E. S., Steele N. G., Donahue K. L., Nwosu Z. C., Pacheco A., Velez-Delgado A., Menjivar R. E., Lima F., The S., Espinoza C. E., Brown K., Long D., Lyssiotis C. A., Rao A., Zhang Y. Q., di Magliano M. P., Crawford H. C., Apolipoprotein E promotes immune suppression in pancreatic cancer through NF-κB–mediated production of CXCL1. Cancer Res. 81, 4305–4318 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X., Calvisi D. F., Hydrodynamic transfection for generation of novel mouse models for liver cancer research. Am. J. Pathol. 184, 912–923 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dinarello C. A., Targeting interleukin 18 with interleukin 18 binding protein. Ann. Rheum. Dis. 59, i17–i20 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Q. Z., Wu X., Wang Z. M., Chen X. Y., Wang L. S., Lu Y. J., Xiong D., Liu Q., Tian Y. H., Lin H. Y., Guo J. Y., Wen S. Q., Dong W., Yang X. F., Yuan Y. C., Yue Z. L., Lei S., Wu Q., Ran L., Xie L. Y. Z., Wang Y. F., Gao L. Q., Tian Q., Zhou X. Y., Sun B. C., Xu L. F., Tang Z. H., Ye L. L., The primordial differentiation of tumor-specific memory CD8+T cells as bona fide responders to PD-1/PD-L1 blockade in draining lymph nodes. Cell 185, 4049–4066.e25 (2022). [DOI] [PubMed] [Google Scholar]
- 31.Kleffel S., Posch C., Barthel S. R., Mueller H., Schlapbach C., Guenova E., Elco C. P., Lee N., Juneja V. R., Zhan Q., Lian C. G., Thomi R., Hoetzenecker W., Cozzio A., Dummer R., Mihm M. C. Jr., Flaherty K. T., Frank M. H., Murphy G. F., Sharpe A. H., Kupper T. S., Schatton T., Melanoma cell-intrinsic PD-1 receptor functions promote tumor growth. Cell 162, 1242–1256 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Efremova M., Rieder D., Klepsch V., Charoentong P., Finotello F., Hackl H., Hermann-Kleiter N., Lower M., Baier G., Krogsdam A., Trajanoski Z., Targeting immune checkpoints potentiates immunoediting and changes the dynamics of tumor evolution. Nat. Commun. 9, 32 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El-Khoueiry A. B., Sangro B., Yau T., Crocenzi T. S., Kudo M., Hsu C., Kim T. Y., Choo S. P., Trojan J., Welling T. H. R., Meyer T., Kang Y. K., Yeo W., Chopra A., Anderson J., Dela Cruz C., Lang L., Neely J., Tang H., Dastani H. B., Melero I., Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): An open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 389, 2492–2502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou S. L., Zhou Z. J., Hu Z. Q., Huang X. W., Wang Z., Chen E. B., Fan J., Cao Y., Dai Z., Zhou J., Tumor-associated neutrophils recruit macrophages and t-regulatory cells to promote progression of hepatocellular carcinoma and resistance to sorafenib. Gastroenterology 150, 1646–1658.e17 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Ogawa R., Yamamoto T., Hirai H., Hanada K., Kiyasu Y., Nishikawa G., Mizuno R., Inamoto S., Itatani Y., Sakai Y., Kawada K., Loss of SMAD4 promotes colorectal cancer progression by recruiting tumor-associated neutrophils via the CXCL1/8-CXCR2 axis. Clin. Cancer Res. 25, 2887–2899 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Shaul M. E., Fridlender Z. G., Tumour-associated neutrophils in patients with cancer. Nat. Rev. Clin. Oncol. 16, 601–620 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Ruffell B., Coussens L. M., Macrophages and therapeutic resistance in cancer. Cancer Cell 27, 462–472 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mantovani A., Marchesi F., Malesci A., Laghi L., Allavena P., Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 14, 399–416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taki M., Abiko K., Baba T., Hamanishi J., Yamaguchi K., Murakami R., Yamanoi K., Horikawa N., Hosoe Y., Nakamura E., Sugiyama A., Mandai M., Konishi I., Matsumura N., Snail promotes ovarian cancer progression by recruiting myeloid-derived suppressor cells via CXCR2 ligand upregulation. Nat. Commun. 9, 1685 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Acharyya S., Oskarsson T., Vanharanta S., Malladi S., Kim J., Morris P. G., Manova-Todorova K., Leversha M., Hogg N., Seshan V. E., Norton L., Brogi E., Massague J., A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell 150, 165–178 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karin M., Greten F. R., NFκB: Linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol. 5, 749–759 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Capece D., Verzella D., Flati I., Arboretto P., Cornice J., Franzoso G., NF-κB: Blending metabolism, immunity, and inflammation. Trends Immunol. 43, 757–775 (2022). [DOI] [PubMed] [Google Scholar]
- 43.Cai Y., Sukhova G. K., Wong H. K., Xu A. M., Tergaonkar V., Vanhoutte P. M., Tang E. H. C., Rap1 induces cytokine production in pro-inflammatory macrophages through NFκB signaling and is highly expressed in human atherosclerotic lesions. Cell Cycle 14, 3580–3592 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding Y., Liang X. T., Zhang Y. L., Yi L., Shum H. C., Chen Q. L., Chan B. P., Fan H. M., Liu Z. M., Tergaonkar V., Qi Z. Q., Tse H. F., Lian Q. Z., Rap1 deficiency-provoked paracrine dysfunction impairs immunosuppressive potency of mesenchymal stem cells in allograft rejection of heart transplantation. Cell Death Dis. 9, 386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morgan D., Garg M., Tergaonkar V., Tan S. Y., Sethi G., Pharmacological significance of the non-canonical NF-κB pathway in tumorigenesis. Biochim. Biophys. Acta Rev. Cancer 1874, 188449 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Voet S., Srinivasan S., Lamkanfi M., van Loo G., Inflammasomes in neuroinflammatory and neurodegenerative diseases. EMBO Mol. Med. 11, e10248 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tarhini A. A., Millward M., Mainwaring P., Kefford R., Logan T., Pavlick A., Kathman S. J., Laubscher K. H., Dar M. M., Kirkwood J. M., A phase 2, randomized study of SB-485232, rhIL-18, in patients with previously untreated metastatic melanoma. Cancer 115, 859–868 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Guo X. J., Zheng L., Jiang J. X., Zhao Y., Wang X., Shen M., Zhu F., Tian R., Shi C. J., Xu M., Li X., Peng F., Zhang H., Feng Y. C., Xie Y., Xu X. D., Jia W., He R. Z., Xie C. C., Hu J., Ye D. W., Wang M., Qin R. Y., Blocking NF-κB is essential for the immunotherapeutic effect of recombinant il18 in pancreatic cancer. Clin. Cancer Res. 22, 5939–5950 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Tangkijvanich P., Thong-Ngam D., Mahachai V., Theamboonlers A., Poovorawan Y., Role of serum interleukin-18 as a prognostic factor in patients with hepatocellular carcinoma. World J. Gastroenterol. 13, 4345–4349 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li S. N., Sun R., Chen Y. Y., Wei H. M., Tian Z. G., TLR2 limits development of hepatocellular carcinoma by reducing IL18-mediated immunosuppression. Cancer Res. 75, 986–995 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Markowitz G. J., Yang P., Fu J., Michelotti G. A., Chen R., Sui J., Yang B., Qin W. H., Zhang Z., Wang F. S., Diehl A. M., Li Q. J., Wang H., Wang X. F., Inflammation-dependent il18 signaling restricts hepatocellular carcinoma growth by enhancing the accumulation and activity of tumor-infiltrating lymphocytes. Cancer Res. 76, 2394–2405 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ho C., Wang C. M., Mattu S., Destefanis G., Ladu S., Delogu S., Armbruster J., Fan L. L., Lee S. A., Jiang L. J., Dombrowski F., Evert M., Chen X., Calvisi D. F., AKT (v-akt murine thymoma viral oncogene homolog 1) and N-Ras (neuroblastoma ras viral oncogene homolog) coactivation in the mouse liver promotes rapid carcinogenesis by way of mTOR (mammalian target of rapamycin complex 1), FOXM1 (forkhead box M1)/SKP2, and c-Myc pathways. Hepatology 55, 833–845 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zídek A., Potapenko A., Bridgland A., Meyer C., Kohl S. A. A., Ballard A. J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A. W., Kavukcuoglu K., Kohli P., Hassabis D., Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pierce B. G., Wiehe K., Hwang H., Kim B. H., Vreven T., Weng Z., ZDOCK Server: Interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 30, 1771–1773 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Di Y., Jing X., Hu K., Wen X., Ye L., Zhang X., Qin J., Ye J., Lin R., Wang Z., He W., The c-MYC-WDR43 signalling axis promotes chemoresistance and tumour growth in colorectal cancer by inhibiting p53 activity. Drug Resist. Updat. 66, 100909 (2023). [DOI] [PubMed] [Google Scholar]
- 57.Wang Z., Sheng C., Kan G., Yao C., Geng R., Chen S., RNAi screening identifies that TEX10 promotes the proliferation of colorectal cancer cells by increasing NF-κB activation. Adv. Sci. 7, 2000593 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Makowska Z., Boldanova T., Adametz D., Quagliata L., Vogt J. E., Dill M. T., Matter M. S., Roth V., Terracciano L., Heim M. H., Gene expression analysis of biopsy samples reveals critical limitations of transcriptome-based molecular classifications of hepatocellular carcinoma. J. Pathol. Clin. Res. 2, 80–92 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mas V. R., Maluf D. G., Archer K. J., Yanek K., Kong X., Kulik L., Freise C. E., Olthoff K. M., Ghobrial R. M., McIver P., Fisher R., Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma. Mol. Med. 15, 85–94 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roessler S., Jia H. L., Budhu A., Forgues M., Ye Q. H., Lee J. S., Thorgeirsson S. S., Sun Z., Tang Z. Y., Qin L. X., Wang X. W., A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 70, 10202–10212 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Tables S1 to S4
Legend for data S1
Data S1