Abstract
Early HIV-1 reverse transcription can be separated into initiation and elongation phases. Here we show, using PCR analysis of negative-strand strong-stop DNA [(−)ssDNA] synthesis in intact virus, that different reverse transcriptase (RT) inhibitors affect distinct phases of early natural endogenous reverse transcription (NERT). The effects of nevirapine on NERT were consistent with a mechanism of action including both specific and nonspecific binding events. The nonspecific component of this inhibition targeted the elongation reaction, whereas the specific effect seemed principally to be directed at very early events (initiation or the initiation-elongation switch). In contrast, foscarnet and the nucleoside analog ddATP inhibited both early and late (−)ssDNA synthesis in a similar manner. We also examined compounds that targeted other viral proteins and found that Ro24-7429 (a Tat antagonist) and rosmarinic acid (an integrase inhibitor) also directly inhibited RT. Our results indicate that NERT can be used to identify and evaluate compounds that directly target the reverse transcription complex.
Human immunodeficiency virus type 1 (HIV-1), like all retroviruses, uses a virally encoded reverse transcriptase (RT) to convert its positive-strand RNA genome into double-stranded DNA (2, 56). Synthesis of the first product of reverse transcription, 181 nucleotides (nt) of single-stranded DNA called negative-strand strong-stop DNA [(−)ssDNA], is subject to complex regulation by both cellular and viral factors. A ribonucleoprotein complex composed of (at least) RT and a cell-derived tRNA molecule initiates reverse transcription from the primer binding site (PBS) (54), an 18-nt viral genomic sequence complementary to the 3′ end of tRNA
molecule initiates reverse transcription from the primer binding site (PBS) (54), an 18-nt viral genomic sequence complementary to the 3′ end of tRNA . A specific reverse transcription initiation complex (RTIC) is thought to form as a result of intrastrand base pairing between the viral A-rich loop sequences located upstream of the PBS and the tRNA
. A specific reverse transcription initiation complex (RTIC) is thought to form as a result of intrastrand base pairing between the viral A-rich loop sequences located upstream of the PBS and the tRNA anticodon loop sequences, together with intermolecular interactions between tRNA
anticodon loop sequences, together with intermolecular interactions between tRNA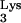 , RT, and viral genomic RNA (23, 25).
, RT, and viral genomic RNA (23, 25).
Many viral factors, including Nef (1), Vif (12, 51, 61), matrix protein (MA) (28), nucleocapsid protein (NCp7) (36, 49), integrase (IN) (40, 66), and Tat (17), affect the efficiency of reverse transcription. Viruses mutated or deleted in the nef, vif, or matrix genes showed decreased reverse transcription efficiency as a result of defective virus formation and/or postentry capsid uncoating. NCp7 greatly facilitates strand transfer and reduced pausing of RT at RNA stem-loop structures during reverse transcription (14, 26). Viruses lacking IN or Tat are defective for initiation of reverse transcription, but this defect can be rescued by trans complementation in the virus-infected cell (60, 66). Analysis of mutated IN and tat genes has shown that their roles in reverse transcription are distinct from their other well-characterized roles in virus replication, but the mechanisms by which IN and Tat affect reverse transcription are not known.
Lanchy et al. (34) and Thrall et al. (57) have described the kinetics of HIV-1 reverse transcription. A general mechanism of DNA synthesis by RT includes binding of RT to the template, binding of the appropriate nucleotide, chemical synthesis (phosphodiester bond formation), and release of pyrophosphate. Pre-steady-state kinetic measurements indicate that the rate-limiting step during the incorporation of a single nucleotide is the conformational change of the RT complex from an inactive to an active form (63), which precedes covalent bond synthesis. In addition, the RTIC, which forms around an RNA-RNA duplex, must alter its conformation to accommodate RNA-DNA hybrids during RNA-dependent synthesis of (−)ssDNA (27). The requirement for a conformational change in RT and the contacts in the narrow minor groove around the DNA-tRNA junction are major factors responsible for early (+1 to +5) pause sites observed in reverse transcription in vitro (reviewed in reference 13). Virion-derived tRNA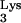 placed on the RNA genome is found both in an unextended form and with the first two bases of (−)ssDNA added (22), suggesting that reverse transcription initiation is somehow restricted in intact viruses obtained from tissue culture supernatants. In other respects, DNA synthesis by HIV-1 RT is kinetically similar to the actions of other polymerases, although HIV-1 RT is particularly susceptible to pausing caused by RNA stem-loop structures that can dislodge it from the template (9, 18, 34, 55).
placed on the RNA genome is found both in an unextended form and with the first two bases of (−)ssDNA added (22), suggesting that reverse transcription initiation is somehow restricted in intact viruses obtained from tissue culture supernatants. In other respects, DNA synthesis by HIV-1 RT is kinetically similar to the actions of other polymerases, although HIV-1 RT is particularly susceptible to pausing caused by RNA stem-loop structures that can dislodge it from the template (9, 18, 34, 55).
Intact HIV-1 can carry out reverse transcription of at least part of its genome in physiological milieux, without the mild detergent treatment used to permeabilize virions in classical endogenous reverse transcription (ERT) assays (39, 58). Intravirion DNA synthesis in the absence of permeabilizing agents has been termed natural ERT (NERT) to distinguish it from the somewhat artificial process which takes place in standard ERT assays (69). NERT is made possible by the amphipathic domains of the gp41 transmembrane protein, which render the HIV-1 envelope permeable to a range of small molecules (68). In vivo, NERT is an active process which is responsive to the virion microenvironment. Virus isolated from seminal plasma, which contains high levels of deoxynucleoside triphosphates (dNTPs), contained much higher levels of full-length or nearly full-length intravirion reverse transcripts than did virus isolated from the blood of the same patients (69). Moreover, the ability of purified virions to infect initially quiescent T cells and nonproliferating cells such as macrophages was significantly increased by preincubation of the virions with seminal plasma (69), indicating that NERT may be an integral part of the viral life cycle and play an important role in the infection of nondividing cells. NERT is also susceptible to inhibition in vivo: the levels of intravirion reverse transcripts in virus isolated from the blood of HIV-infected patients dropped dramatically after commencement of nevirapine (NVP) therapy and rebounded to pretreatment levels concomitant with the development of NVP resistance in the virus (70). Virion infectivity also decreased dramatically in response to NVP therapy and returned almost to pretreatment levels with the development of NVP resistance (70), indicating that the antiviral effects of NVP begin with cell-free virions.
In the present study, we used a PCR-based assay to measure NERT in order to investigate the effects of RT inhibitors on early reverse transcription in intact virions. Compounds that target RT, Tat, or IN were tested for their ability to inhibit both NERT and the activity of recombinant HIV-1 RT in vitro. Our data indicate that although all of the compounds tested, including those with anti-Tat and anti-IN activities, could directly inhibit RT, their effect on early and late (−)ssDNA synthesis produced strikingly different inhibition profiles. Analysis of the relationship between inhibition and drug concentration showed that nucleotide or pyrophosphate (PPi) analogs had similar effects on both early and late (−)ssDNA synthesis, a pattern consistent with inhibition of the elongation reaction. In contrast, nonnucleoside inhibitors of RT had different effects on early and late (−)ssDNA synthesis, which were consistent with inhibition of very early reverse transcription, the most likely targets being initiation or early pausing associated with the transition to elongation (24, 37). Since the use of intact virus preserves all of the viral and cellular factors that contribute to efficient reverse transcription, these results demonstrate the utility of the NERT-PCR assay as a probe to examine the mechanisms of drug action and to identify novel antiretroviral compounds with unique mechanisms of action.
MATERIALS AND METHODS
Cells and virus.
Virus was grown in stably transfected 293 cells and Jurkat T cells. The isolation and characterization of a 293 cell line stably transfected with wild-type (HXB2-neo) HIV-1 is described elsewhere (16). 293 cells were cultured in Iscove's modified Dulbecco's medium supplemented with 5% newborn calf serum, 2% fetal bovine serum, 1% penicillin-streptomycin (pen/strep; Life Technologies), and 0.5 mg of Geneticin per ml. Jurkat cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum, 1% pen/strep, and 1 mg of Geneticin per ml. Cell culture supernatants were filtered through polyethersulfone membranes with a pore size of 0.45 μm (Nalgene) and stored in aliquots at −80°C for use as virus stocks. Aliquots were used only once after thawing, and the stocks were not stored for more than 6 months.
Drugs.
Phosphonoformic acid (PFA; foscarnet), phosphonoacetic acid (PAA), ddATP, ddTTP, rosmarinic acid (RA), and curcumin were purchased from Sigma Aldrich (Sydney, Australia). The benzodiazepins Ro 5-3335 (Ro5) and Ro 24-7429 (Ro24) were kindly provided by Roche Products. NVP was obtained from commercial drug preparations (Viramune, 200 mg; Boehringer, Ingelheim, Germany).
NERT assay.
Virus stocks were assayed for total RT activity on a synthetic homopolymer template in the presence of detergent, using a commercial enzyme-linked immunosorbent assay (ELISA) kit (Roche) as specified by the manufacturer. Aliquots of virus (typically 0.25 mU of RT) were incubated for 30 to 60 min at 37°C, with or without inhibitors, in a final volume of 100 μl of Iscove's modified Dulbecco's medium supplemented with 50 U of DNase I (Worthington Biochemical) and 10 mM MgCl2. Enzymatic activity was then terminated in controls by the addition of 150 μl of stop solution (10 mM Tris-HCl [pH 7.4], 10 mM EDTA, 20 μg of sheared salmon sperm DNA per ml, 50 μg of proteinase K per ml) followed by incubation for 10 min at 37°C and then a further 10 min in a boiling-water bath. The remaining assay mixtures were supplemented with 200 μM dNTPs and incubated at 37°C for 90 min, and the reactions were then terminated as described above. Samples of each stopped reaction mixture were serially diluted in twofold steps, and each dilution was assayed for HIV-1 DNA by quantitative PCR using combinations of the following HIV-1-specific oligonucleotides (Fig. 1): S1 (5′-CAA GTA GTG TGT GCC CGT CTG TT-3′), P1 (5′-TAG AGA TCC CTC AGA CCC TTT-3′), P2 (5′-CTG CTA GAG ATT TTT CCA CAC TGA C-3′), D1 (5′-GGT CTC TCT GGT TAG ACC A-3′), and D2 (5′-AAG CAG TGG GTT CCC TAG TT AG-3′). One primer in each pair was labeled with 32P using T4 polynucleotide kinase, and the reaction products were resolved on 6% polyacrylamide–Tris-borate-EDTA (TBE) gels and detected using a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). PhosphorImager data were analyzed with ImageQuant software (Molecular Dynamics). PCR standard curves were generated using proviral plasmid DNA serially diluted in mock stop solution containing MgCl2 (MMS) (6 mM Tris-HCl [pH 7.4], 6 mM EDTA, 12 μg of sheared salmon sperm DNA per ml, 4 mM MgCl2), and samples were diluted in MMS to keep all signals within the linear range of the assay (approximately 75 to 2,500 copies). At least two (typically three or four) dilutions of every sample were assayed, and data sets in which the linear correlation coefficient of the standard curve was less than 0.98 were not included in further analysis.
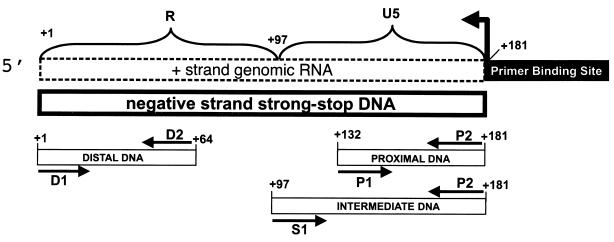
FIG. 1.
Schematic of the PCR strategy to detect and quantitate (−)ssDNA. Regions corresponding to R (+1 to +97) and U5 (+98 to +181) were detected by PCR using oligonucleotide probes as shown. Probes were used to detect and quantitate different regions of (−)ssDNA designated proximal (oligonucleotides P1 and P2, nt +132 to +181), intermediate (oligonucleotides S1 and P2, nt +96 to +181), and distal (oligonucleotides D1 and D2, nt +1 to +64) according to their positions relative to the site of reverse transcription initiation. The site of initiation and the direction of DNA synthesis by RT are indicated by the bent arrow.
RESULTS
NERT.
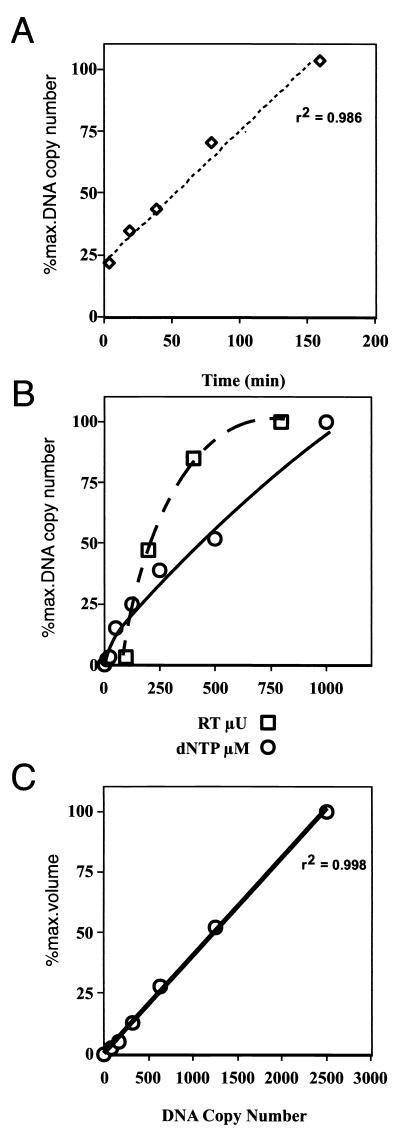
Intact virus was incubated with 200 μM total dNTPs for 90 min at 37°C in the presence of DNase I, and the reactions were terminated by EDTA addition and boiling. Reverse transcription products were detected by PCR using oligonucleotides specific for early, intermediate, or late (−)ssDNA (Fig. 1, probes P1 and P2, S1 and P2, and D1 and D2, respectively). The products of these PCRs were separated on polyacrylamide gels and analyzed on a PhosphorImager. Virions incubated without dNTPs did not synthesize appreciable levels of (−)ssDNA, and (−)ssDNA levels increased roughly proportionally with increasing virus and dNTP concentrations (Fig. 2B). The amount of (−)ssDNA detected in NERT-PCR assays reached ∼20% of maximum in the first 5 min and then increased in an almost linear fashion during the first 180 min (Fig. 2A). This long temporal linear range may be due to complex differences within the virus population, such as differences in virion maturity. Control PCR using a plasmid containing the target DNA sequences demonstrated that the PCR response was linear over a 32-fold range (Fig. 2C).
FIG. 2.
Characterization of the NERT-PCR assay. (A) Virus supernatants containing 0.25 mU of total RT activity were incubated with 200 μM total dNTPs at 37°C for the indicated times and assayed for (−)ssDNA by PCR using both proximal (P1 and P2) and distal (D1 and D2 [not shown]) primer pairs (Fig. 1). NERT reaction mixtures were serially diluted, where necessary, to conform to the linear range of the PCR assay. All experiments were performed at least three times with similar results, and results of representative experiments are shown. (B) NERT assays were carried out using 200 μM total dNTPs and increasing quantities of virus supernatant (0.1, 0.2, 0.4, and 0.8 mU of RT activity, measured using a commercial homopolymer template RT assay), or using virus supernatants containing 0.25 mU of RT activity and increasing total concentrations of dNTPs (0, 5, 10, 50, 100, 250, 500, and 1000 μM); these assay mixtures were incubated for 90 min at 37°C, and (−)ssDNA was measured using primers P1 and P2 (Fig. 1). (C) Analysis of control DNAs, used to quantify the PCR results (r2 = 0.998).
Effects of nucleoside and nonnucleoside RT inhibitors on NERT.
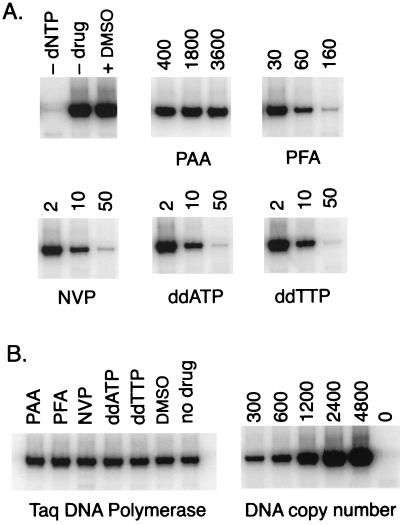
We tested the effects on (−)ssDNA synthesis by NERT of various reverse transcriptase inhibitors, including PAA, PFA, NVP, ddATP, and ddTTP (Fig. 3). These compounds were selected to represent a range of different mechanisms of RT inhibition. The PPi analog PFA inhibits RT by occupying the PPi binding pocket (7, 43). NVP is a specific noncompetitive inhibitor of RT that can bind directly to a hydrophobic pocket within the enzyme (31), causing a conformational change that disrupts the catalytic site and blocks its DNA polymerase activity. Both ddATP and ddTTP are nucleoside analogs which, when incorporated into a nascent DNA chain by RT, result in the termination of DNA synthesis. PAA, which is structurally closely related to PFA, had no effect on NERT even at millimolar concentrations. In contrast, PFA at 60 and 160 μM decreased total (−)ssDNA levels (measured using PCR probes S1 and P2) by 75% and by more than 98%, respectively, compared to uninhibited virus. NVP at 10 to 50 μM inhibited NERT to a similar degree, in agreement with studies of virus isolated from patients taking NVP (67). Both ddATP at 10 μM and ddTTP at 50 μM also effectively inhibited NERT. All of the observed reductions in PCR signals could be attributed to inhibition of NERT, since none of the drugs tested showed any inhibitory activity against Taq DNA polymerase (Fig. 3B). We also compared the effects of the various drugs in our NERT-PCR assay to their effects in a commercial reverse transcriptase ELISA (Table 1). In general, the two assays showed similar results, although some compounds were more effective against in vitro reverse transcription on a homopolymer template by recombinant HIV-1 RT than against NERT in intact virions. This observation may reflect the different nucleotide concentrations used in each assay, particularly with respect to nucleoside analog inhibitors, and/or the effect of membrane permeability on intravirion drug concentration.
FIG. 3.
NERT is inhibited by nucleoside analogs and nonnucleoside RT inhibitors. (A) NERT reaction mixtures containing 0.25 mU of virion RT activity were preincubated for 30 min with RT inhibitors (PAA, PFA, NVP, ddATP, and ddTTP) at the concentrations indicated (micromolar), or with dimethyl sulfoxide (DMSO) (solvent vehicle for NVP). Then 200 μM dNTP was added to each NERT reaction mixture except as indicated (panel A, -dNTP), and all the mixtures were incubated at 37°C for 90 min. The reactions were terminated, and the products were assayed for (−)ssDNA by PCR using primers P1 and P2 (Fig. 1). (B) RT inhibitors or DMSO was added to PCR mixtures containing 1,000 copies of proviral plasmid DNA at concentrations slightly higher than were carried over from any of the corresponding NERT assays. All PCR results were within the linear range of the assay (r2 > 0.99). Experiments were performed at least three times, and typical results are shown.
TABLE 1.
Comparison of inhibition in NERT and homopolymer RT assays
| Inhibitor | Inhibitor concn (μM) | % Inhibitiona by:
|
|
|---|---|---|---|
| ELISA | NERT | ||
| NVP | 10 | 87 ± 12 | 77 ± 11 |
| 100 | 99 ± 2 | 100 | |
| PFA | 30 | 98 ± 3 | 45 ± 5 |
| 300 | 98 ± 2 | 100 | |
| PAA | 70 | 0 ± 1 | 0 |
| 700 | 9 ± 2 | 0 | |
| 2,800 | 42 ± 4 | 0 | |
| ddATP | 10 | NTb | 71 ± 6 |
| 100 | NT | 100 | |
| ddTTP | 10 | 99 ± 1 | 70 ± 5 |
| 100 | 100 | 100 | |
| Ro 5-3335 | 20 | NT | 0 |
| 25 | 6 ± 5 | NT | |
| 200 | NT | 0 | |
| 250 | 19 ± 8 | NT | |
| Ro 24-7429 | 20 | NT | 32 ± 9 |
| 25 | 62 ± 16 | NT | |
| 200 | NT | 56 ± 5 | |
| 250 | 84 ± 19 | NT | |
| RA | 10 | 16 ± 3 | 26 ± 9 |
| 100 | 30 ± 7 | 44 ± 13 | |
| 200 | 50 ± 4 | 62 ± 4 | |
| Curcumin | 12.5 | 18 ± 10 | 0 |
| 100 | 29 ± 12 | 0 | |
| 200 | 29 ± 13 | 0 | |
| DMSO | ∼1% (vol/vol) | 0 | 0 |
Data are shown as average ± standard deviation from at least three separate experiments. Percent inhibition was calculated as 100(1 − Sn/S0), where Sn and S0 represent the PCR signals in the presence of n and 0 μM drug, respectively. Proximal or intermediate PCR primer pairs were used in all cases.
NT, not tested.
Tat and IN inhibitors down regulate NERT.
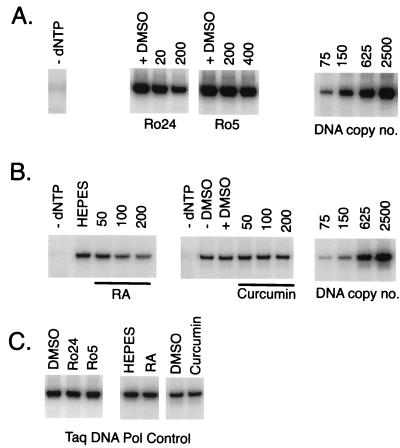
Previous studies have shown that the drugs Ro24 and Ro5 inhibit Tat-induced HIV-1 gene expression (20). Since Tat plays a role in reverse transcription, we tested the effects of these Tat antagonists on NERT (Fig. 4) and on recombinant RT in vitro (Table 1). Ro24 decreased (−)ssDNA synthesis in NERT assays (measured using PCR probes S1 and P2) by ∼55% at 200 μM and inhibited recombinant RT activity in vitro by ∼60 and ∼85% at 25 and 250 μM, respectively. It therefore appeared that the observed inhibition of NERT by Ro24 was mediated by direct activity against RT and was unrelated to Tat antagonist activity. Ro5 had no effect on NERT and little effect on recombinant RT in vitro (less than 20% inhibition at 250 μM).
FIG. 4.
NERT is inhibited by anti-Tat and anti-IN compounds. (A and B) NERT reaction mixtures containing 0.25 mU of virion RT activity were preincubated for 30 min with solvent (DMSO) or with Ro24, Ro5, RA, or curcumin at the concentrations indicated (micromolar). The reaction mixtures were supplemented with 200 μM dNTP, except as indicated (panel A, -dNTP) and incubated at 37°C for 90 min, the reactions were terminated, and the products were assayed for (−)ssDNA by PCR using primers P1 and P2 (Fig. 1). (C) RT inhibitors or DMSO was added to PCR mixtures containing 1,000 copies of proviral plasmid DNA at concentrations slightly higher than were carried over from any of the corresponding NERT assays. All PCR results were within the linear range of the assay (r2 > 0.99). Experiments were performed at least three times, and typical results are shown.
Like Tat, HIV-1 IN plays a role in reverse transcription which is separate from its eponymous role (66). We therefore tested the effects on reverse transcription of two related IN inhibitors, curcumin and RA (Fig. 4; Table 1). In in vitro assays of IN activity, the 50% inhibitory concentration of RA is ∼15-fold lower against 3′-processing activity and ∼35-fold lower against strand transfer activity than the 50% inhibitory concentration of curcumin (41). RA inhibited the activity of both NERT and recombinant HIV-1 RT in commercial ELISAs by ∼30 to 40% at 100 μM and by ∼50 to 60% at 200 μM. This indicates that the effect of RA on NERT is probably attributable to direct inhibition of RT. We also found that Moloney murine leukemia virus (M-MLV) RT, in a standard RT assay using homopolymer template and primer, was inhibited by high (200 μM) but not by low (≤50 μM) concentrations of RA (data not shown). Curcumin inhibited recombinant RT by only ∼30% at 100 and 200 μM and had no effect on NERT.
RT inhibitors display differential inhibition of proximal and distal (−)ssDNA synthesis.
Next, we modified the PCR step in the NERT-PCR assay to distinguish between early and late (−)ssDNA synthesis. We assayed NERT DNA products over two short regions proximal (Fig. 1, probes P1 and P2) and distal (Fig. 1, probes D1 and D2) to the PBS and compared the levels of proximal and distal DNA generated by wild-type virus in the presence of increasing concentrations of various RT inhibitors. At least three serial dilutions of each NERT reaction mixture were assayed by PCR, and at least two dilutions with signals that were within the linear range of the assay were used to generate each data point. Only data sets for which the standard curve showed a correlation coefficient (r2) greater than 0.98 were included in the final analyses. The relative levels of proximal and distal DNA observed in the absence of drug varied between virus stocks, with distal DNA copy numbers ranging from 30 to 50% of proximal DNA copy numbers (Table 2). This intrinsic termination has not been previously described in NERT or any other system, but several studies have observed that HIV-1 RT is susceptible to pausing along adenosine-rich RNA sequences or at stable RNA secondary structures (9, 18, 34, 55). As expected, increasing concentrations of inhibitor resulted in increased total inhibition of (−)ssDNA, with the greatest inhibitory effect on distal DNA levels. Table 2 shows representative data for ddATP and NVP; PFA and RA gave similar total inhibition patterns, and four to six separate experiments with each compound gave simular results (data not shown).
TABLE 2.
Inhibition of NERT
| Compound | PCR probesa | Concn (μM) of compound | % Uninhibited copy numberb |
|---|---|---|---|
| ddATP | Proximal | 0 | 100 ± 1.90 |
| 0.5 | 96.5 ± 2.51 | ||
| 1 | 67.3 ± 3.84 | ||
| 2 | 68.0 ± 1.50 | ||
| 4 | 40.5 ± 0.53 | ||
| 6 | 37.4 ± 1.75 | ||
| 8 | 30.0 ± 1.35 | ||
| Distal | 0 | 32.2 ± 1.64cd | |
| 0.5 | 18.3 ± 0.64c | ||
| 1 | 13.3 ± 0.59c | ||
| 2 | 8.72 ± 0.50c | ||
| 4 | 4.1 ± .013c | ||
| 6 | 3.8 ± .017c | ||
| 8 | 1.49 ± 0.15c | ||
| NVP | Proximal | 0 | 100 ± 3.6 |
| 0.5 | 73.3 ± 1.7 | ||
| 1 | 63.9 ± 4.5 | ||
| 2 | 53.5 ± 0.75 | ||
| 4 | 41.8 ± 0.37 | ||
| 6 | 36.8 ± 2.2 | ||
| Distal | 0 | 42.7 ± 2.7cd | |
| 0.5 | 23.0 ± 1.5c | ||
| 1 | 18.2 ± 0.56c | ||
| 2 | 13.4 ± 0.67c | ||
| 4 | 5.7 ± 0.41c | ||
| 6 | 2.7 ± 0.51c |
Primers shown in Fig. 1.
Data are shown as average ± standard deviation from at least two PCR amplifications of serially diluted NERT assays. Copy numbers are expressed as percentages of the copy number measured in the absence of inhibitor with proximal primers (P1 and P2). These data are representative of four (ddATP) or five (NVP) independent NERT-PCR assays.
These values, like %T, include intrinsic termination effects.
Independent virus stocks with different, but internally consistent, levels of intrinsic termination were used for ddATP and NVP inhibition assays.
It is clear from these observations that there exists an intrinsic termination rate (ITR = d0/p0, where d0 is the distal copy number in the absence of drug [0 μM drug] and p0 is the proximal copy number in the absence of drug [0 μM drug]), which describes premature termination in the absence of an inhibitor and must be taken into account in order to calculate the degree of inhibition of the distal signal that can be attributed to drug action. We defined the expected distal value (edvn = pn × ITR, where edvn is the expected distal copy number at a given drug concentration [n μM drug] and pn is the proximal copy number at a given drug concentration [n μM drug]) as the distal copy number that would be expected in the absence of any drug-related termination. This value can then be used to define the percent inhibition of the distal signal which is due to drug activity [%D = 100(1 − dn/edvn), where dn is the distal copy number at a given drug concentration (n μM drug)]. Plots of the percent inhibition of the proximal signal [%P = 100 (1 − pn/p0)] and %D against drug concentration (inhibition curves) revealed differences between the anti-RT compounds tested (Fig. 5). The inhibitors were examined over concentration ranges which resulted in similar total levels of inhibition [%T = 100(1 − dn/p0)] of NERT, from ∼70 to 98%. The proximal and distal inhibition curves of both PFA and ddATP showed a regular hyperbolic concentration dependence consistent with a single binding site {I = Imax [c/(c + kd)], where I is the percent inhibition [%P or %D], Imax is the maximal percent inhibition expressed as a percentage, c is the micromolar drug concentration and kd is the micromolar dissociation coefficient}. Regression analysis showed that the PFA and ddATP inhibition curves fit this expression with correlation coefficients (r2) of ∼0.98 to 0.99 in all cases. Consistent with its known activity as an inhibitor of the RT elongation reaction, ddATP effected greater inhibition of late than of early DNA synthesis (Fig. 5A). A similar but even more marked effect was observed for PFA: maximal inhibition (Imax) of ddATP proximal and distal inhibition curves and of PFA distal inhibition curves was approximately 100%, but Imax of PFA proximal inhibition curves ranged from 50 to 70% in five independent assays (Fig. 5B and data not shown). These data distinguish the effects of PFA from inhibition by dideoxynucleotides and suggest that PFA is a relatively weak inhibitor of RT which requires many rounds of nucleotide addition to achieve complete termination of reverse transcription.
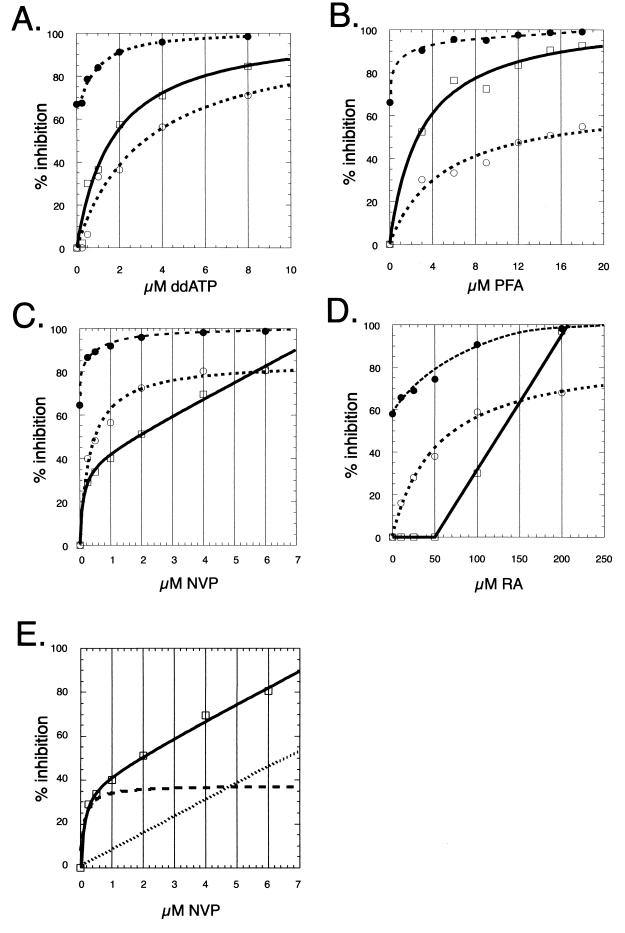
FIG. 5.
NERT inhibition curves for nucleoside analogs and nonnucleoside compounds. (A to D) NERT assays were carried out in the presence of increasing concentrations of four different inhibitors: ddATP at 0.25, 0.5, 1, 2, 4, and 8 μM (A); PFA at 3, 6, 9, 12, 15, and 18 μM (B); NVP at 0.25, 0.5, 1, 2, 4, and 6 μM (C); and RA at 10, 25, 50, 100, and 200 μM (D). Each NERT reaction product was assayed for (−)ssDNA by PCR using either the proximal (P1 and P2) or the distal (D1 and D2) DNA primer pair (Fig. 1). Total inhibition (%T; dotted line, solid circles), proximal inhibition (%P; dotted line, open circles), and distal inhibition (%D; solid line, open squares) were calculated and plotted against drug concentrations. All of the proximal inhibition curves were accurately described by regular hyperbolic expressions (see the text), as were the distal inhibition curves for ddATP and PFA. The expression which best fit the NVP distal inhibition curve, however, included an additional component describing nonspecific interaction (see the text). (E) The specific (hyperbolic, dashed line) and nonspecific (linear, dotted line) components of the NVP distal inhibition curve (solid line, open squares). Three to five independent experiments were performed with each drug, and representative results are shown.
NVP concentrations as low as 0.25 μM inhibited reverse transcription by more than 80%, and ∼95% inhibition was observed at NVP concentrations of 4 μM or higher (%T [Fig. 5C; Table 2]). Like ddATP and PFA, NVP proximal inhibition curves were accurately (r2 > 0.99 for two separate experiments and ∼0.96 for a third data set) described by a hyperbolic binding curve, suggesting a single binding site (see above). In contrast, NVP distal curves were most accurately (r2 > 0.99 in all cases) described by the addition of an expression describing nonspecific inhibition {I = Imax [c/(c + kd)] + mc, where m is a nonspecific inhibition coefficient} (Fig. 5E). The nonspecific component was most apparent at concentrations of NVP (1 to 8 μM) sufficient to saturate the well-characterized hydrophobic binding pocket (kd = 0.025 μM) which is believed to mediate the specific effect of NVP (53), suggesting that the nonspecific inhibition does not involve this site. The nonspecific component was not apparent in the proximal data, probably because primer P1 is less than 60 bp from the PBS: the observed effect of nonspecific interference with NERT would be expected to increase with distance from the PBS. Whereas NVP proximal-inhibition curves showed maximal theoretical inhibition levels (Imax) of 80 to 90% in each of five different experiments (Fig. 5 and data not shown), the Imax of the portion of distal inhibition which was described by a regular hyperbolic expression (see above) was only 30 to 40% (Fig. 5E, dashed line). These experiments suggest that in NERT, the elongation reaction is somewhat resistant to inhibition by NVP. For example, NVP at 0.5 μM inhibited proximal DNA synthesis by ∼50%, but drug action inhibited further DNA synthesis by only ∼33%. In other words, once proximal DNA was completed, ∼33% of the elongation reactions was further inhibited by NVP (Fig. 5C). Both ddATP and PFA had much greater downstream effects, inhibiting further DNA synthesis by ∼67 and ∼86%, respectively, at concentrations resulting in ∼50% inhibition of proximal DNA synthesis (Fig. 5A and B). A single binding event which affected elongation would be expected to result in greater inhibition of late than of early (−)ssDNA synthesis, as was observed in ddATP and PFA inhibition curves. The two hyperbolic NVP inhibition curves (proximal and distal specific) therefore appear to represent different inhibitory effects and are consistent with a model in which NVP exerts a strong inhibitory effect on very early events (initiation or the initiation-elongation switch) and a weaker effect on the elongation reaction, in addition to nonspecific interference with enzyme activity.
The level of inhibition of NERT by RA, whose described antiviral target is HIV-1 IN, was lower than that observed for the other RT inhibitors (Fig. 5D, %T). Total NERT inhibition (%T) was 75 and 98% at 50 and 200 μM RA, respectively. Inhibition of proximal DNA synthesis showed hyperbolic concentration dependence (see above), with Imax values of 87 and 91% in two independent assays (r2 = 0.99) and 73% in a third assay (r2 = 0.97). RA at concentrations up to 50 μM inhibited proximal DNA synthesis by up to 40% but had no additional inhibitory effect on distal DNA synthesis (indicating that these concentrations of RA did not affect the RT elongation reaction) in three independent assays. In two other assays, however, we observed up to 20% inhibition of distal DNA synthesis in addition to the proximal inhibition. Higher concentrations of RA caused variable levels of inhibition, but 200 μM RA inhibited distal DNA synthesis by 86 to 100% in five independent NERT-PCR assays, and this concentration of RA also inhibited Moloney murine leukemia virus RT in a standard RT assay using homopolymer template and primer (data not shown). These results are similar to those observed for NVP and indicate that RT which completed proximal (−)ssDNA was nearly resistant to further inhibition by RA at concentrations up to 50 μM. While these experiments cannot precisely map termination, the data are consistent with direct inhibition of either reverse transcription initiation or the switch to elongation.
DISCUSSION
DNA synthesized within intact HIV-1 virions is the product of partial reverse transcription that can be influenced by physiological environments (39, 58). For example, seminal fluid, which contains high levels of dNTPs, stimulates negative-strand DNA synthesis, while virus isolated from infected patients taking NVP contains diminished levels of intravirion DNA compared to virus obtained before therapy (69, 70). In the present study, we used a quantitative NERT-PCR assay that directly measured the effects of RT inhibitors on early reverse transcription in intact virus particles. We have provided experimental evidence, for the first time using intact virus, that both NVP and RA (a previously identified HIV-1 IN inhibitor) can inhibit very early reverse transcription, with the most likely targets being initiation or the switch from initiation to elongation.
In the present study, we have not examined whether the effects of NVP and RA during NERT are identical to the effects of these drugs during cell infections by HIV-1. Difficulties in accurately controlling the many additional variables in a cellular system, particularly intracellular concentrations of RT inhibitors, render such experiments technically difficult, especially with respect to detailed analysis of inhibitory effects on early and late (−)ssDNA synthesis. Others have shown, however, that endogenous RT assays can accurately reveal synergies of drug-resistant RT genotypes previously observed in vivo (8, 45). Our comparisons of reverse transcription initiation defects observed in viruses carrying mutations in either the tat gene (60) or the TAR RNA element (15) have demonstrated that NERT provides an accurate model of early reverse transcription.
Biochemical analysis of in vitro reverse transcription has shown that the initiation of reverse transcription can be distinguished from the subsequent elongation reaction (24, 33). Initiation of reverse transcription is regulated by complex interactions between the cellular tRNA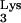 , RT (3, 44), NCp7 (35, 36), and the RNA genome containing the PBS and 3′-flanking RNA sequences, U5 (21, 23), and the TAR RNA element (6, 15). The initiation reaction is characterized by early pausing (18, 29, 33), which is more prominent at low concentrations of dNTPs (37), so that reverse transcription elongation appears to be induced under conditions that favor the completion of proviral DNA synthesis. NVP, which binds within a hydrophobic pocket near the catalytic site of RT (32), inhibits the initial “burst” of polymerization in vitro (53); in the presence of bound NVP, dNTPs bind tightly but nonproductively to RT, resulting in a decreased rate of polymerization. In the present study, we compared the effects of different antiviral compounds on (−)ssDNA synthesis in intact virus. Induction of reverse transcription with 200 μM dNTP in the absence of any inhibitors revealed a high level of intrinsic termination, ranging from 50 to 70% in different virus stocks. To our knowledge, this phenomenon has not been previously described. Pause sites in (−)ssDNA have been mapped in vitro and are enhanced at adenosine-rich RNA regions (18) and stable RNA stem-loop structures (65), such as the TAR element and the poly(A) stem-loop (29), that can dislodge RT from the RNA template. Less efficient elongation in NERT could be due to several reasons, such as a postinitiation reorganization of the genomic RNA-tRNA
, RT (3, 44), NCp7 (35, 36), and the RNA genome containing the PBS and 3′-flanking RNA sequences, U5 (21, 23), and the TAR RNA element (6, 15). The initiation reaction is characterized by early pausing (18, 29, 33), which is more prominent at low concentrations of dNTPs (37), so that reverse transcription elongation appears to be induced under conditions that favor the completion of proviral DNA synthesis. NVP, which binds within a hydrophobic pocket near the catalytic site of RT (32), inhibits the initial “burst” of polymerization in vitro (53); in the presence of bound NVP, dNTPs bind tightly but nonproductively to RT, resulting in a decreased rate of polymerization. In the present study, we compared the effects of different antiviral compounds on (−)ssDNA synthesis in intact virus. Induction of reverse transcription with 200 μM dNTP in the absence of any inhibitors revealed a high level of intrinsic termination, ranging from 50 to 70% in different virus stocks. To our knowledge, this phenomenon has not been previously described. Pause sites in (−)ssDNA have been mapped in vitro and are enhanced at adenosine-rich RNA regions (18) and stable RNA stem-loop structures (65), such as the TAR element and the poly(A) stem-loop (29), that can dislodge RT from the RNA template. Less efficient elongation in NERT could be due to several reasons, such as a postinitiation reorganization of the genomic RNA-tRNA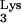 complex, uncoating, or other viral and cellular factors. While others have shown that truncated (−)ssDNA can complete the first strand jump of reverse transcription, the efficiency of such strand transfers is reduced relative to that of transfers involving full-length or near-full-length (−)ssDNA (30, 38, 46, 64). Together, these observations suggest that postentry events are probably required for optimal (−)ssDNA synthesis and complete reverse transcription.
complex, uncoating, or other viral and cellular factors. While others have shown that truncated (−)ssDNA can complete the first strand jump of reverse transcription, the efficiency of such strand transfers is reduced relative to that of transfers involving full-length or near-full-length (−)ssDNA (30, 38, 46, 64). Together, these observations suggest that postentry events are probably required for optimal (−)ssDNA synthesis and complete reverse transcription.
As might be expected of RT elongation inhibitors, the nucleoside analog ddATP and the PP; analog PFA both effected greater inhibition of distal DNA synthesis than of synthesis near the reverse transcription initiation site. In contrast, portions of the inhibition curves for both NVP and RA showed greater effects at or near the initiation site than downstream. RA exhibited little or no inhibition of distal DNA synthesis (in excess of the inhibitory effect observed on proximal DNA) at concentrations up to 50 μM. The specific effect of NVP on distal DNA synthesis was relatively weak and appeared to reflect a different inhibitory effect from that which was evident in NVP proximal inhibition curves. These results are consistent with inhibition of very early events in NERT, such as reverse transcription initiation or the switch from initiation to elongation, and suggest that both RA and NVP act on targets within the RTIC. Binding of NVP to RT is known to induce conformational change in the enzyme (32) in addition to inhibiting the elongation reaction (52). This conformational change may also reduce the ability of RT to function within the RTIC. Recently, it was shown that zidovudine (AZT), a known elongation inhibitor, has different inhibitory effects on initiation and elongation reactions (48). This was attributed to an apparent resistance to removal of incorporated AZT by pyrophosphorolysis during the initiation, but not the elongation, reaction. Our data suggest that NVP and RA also have distinct inhibitory effects during these phases of reverse transcription.
In these experiments, nonspecific inhibition of the elongation reaction by NVP, which has not been previously described, was observed in five of five independent NERT assays. The inhibition of distal DNA synthesis by NVP (Fig. 5C) gave a characteristic concentration dependence pattern, a regular hyperbolic binding curve on a sloping baseline, that has been previously observed (reviewed in reference 62). When mathematically separated, the nonspecific component is represented by a straight line that passes through zero and the remaining curve is a regular hyperbola (Fig. 5E). Inhibition of proximal DNA synthesis by NVP probably arises from the binding of NVP to the previously characterized hydrophobic binding pocket adjacent to the active site of RT (32, 47). The noncompetitive, specific proximal inhibition never reached 100% (Imax < 90%), indicating that the nonspecific mechanism described here contributes significantly to the total inhibition by NVP during NERT, particularly at concentrations above the kd of the known binding site. It is possible that NVP increased the intrinsic termination rate in NERT reactions. NVP does not block nucleotide binding to RT but makes this an unproductive event that could lead to increased pausing (53). Such a mechanism may in fact account for the relatively weak (Imax = 30 to 40%) specific component of distal inhibition, but the mechanism underlying the nonspecific component remains unclear and is the subject of a new investigation.
The nature of the RTIC is not fully understood. Many viral factors affect reverse transcription; Nef (1, 50) and Vif (11, 12, 42, 51) seem to affect reverse transcription during virus particle formation or uncoating or by affecting RNA structure (10, 71), while IN (40, 59, 66) and Tat (17, 60) are required for efficient initiation of reverse transcription, and NCp7 greatly increases the efficiency of the initiation reaction (36, 49) and improves transcript elongation by destabilizing RNA secondary structures (14, 26). Following entry into the cytoplasm, a number of viral proteins including MA, Vpr, IN, and RT form large, dynamic nucleoprotein complexes that carry out reverse transcription and direct the nuclear import of the newly synthesized provirus (5, 19). Whether these or other factors cooperate in an RTIC, which may be distinct from an elongation complex, is under investigation. For example, virus lacking the Tat protein (Δtat) shows no obvious structural or biochemical defects but is defective (relative to wild-type virus) for early (−)ssDNA synthesis by NERT (17). Interestingly, Ro24, which has been reported to inhibit transactivation by Tat of RNA polymerase II-mediated transcription (4, 20), directly inhibited RT in the present study. Recent studies revealed that IN-defective HIV-1 failed to initiate reverse transcription efficiently (66); as has also been observed in Δtat HIV-1, this function could be rescued by supply of the missing protein in trans. Our data show that the IN inhibitor RA can inhibit NERT. Although RA directly inhibited RT in both ELISA and a NERT assay, experiments are in progress to determine whether RA and IN interact with a common domain of RT.
In the present study, NERT-PCR assays discriminated between the anti-initiation and antielongation effects on viral DNA synthesis of PFA, ddATP, NVP, and RA and indicated that both NVP and RA directly targeted very early reverse transcription in intact virions. In addition, we describe for the first time a significant nonspecific component, whose mechanism of action is unknown, of the inhibition of RT by NVP in NERT assays. The NERT-PCR method has distinct advantages over in vitro systems in the testing of antiviral compounds, since all of the cell-derived and viral factors and structural elements that contribute to initiation are present in the intact virus. It seems likely that novel compounds which target either RT or other components of the RTIC could be identified and their activities could be optimized using this strategy.
Footnotes
Manuscript 125 from The Sir Albert Sakzewski Virus Research Centre.
REFERENCES
- 1.Aiken C, Trono D. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J Virol. 1995;69:5048–5056. doi: 10.1128/jvi.69.8.5048-5056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970;226:1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 3.Barat C, Lullien V, Schatz O, Keith G, Nugeyre M T, Gruninger-Leitch F, Barre-Sinoussi F, LeGrice S F, Darlix J L. HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J. 1989;8:3279–3285. doi: 10.1002/j.1460-2075.1989.tb08488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braddock M, Cannon P, Muckenthaler M, Kingsman A J, Kingsman S M. Inhibition of human immunodeficiency virus type 1 Tat-dependent activation of translation in Xenopus oocytes by the benzodiazepine Ro24–7429 requires trans-activation response element loop sequences. J Virol. 1994;68:25–33. doi: 10.1128/jvi.68.1.25-33.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukrinsky M I, Sharova N, McDonald T L, Pushkarskaya T, Tarpley W G, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clever J L, Eckstein D A, Parslow T G. Genetic dissociation of the encapsidation and reverse transcription functions in the 5′ R region of human immunodeficiency virus type 1. J Virol. 1999;73:101–109. doi: 10.1128/jvi.73.1.101-109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crumpacker C S. Mechanism of action of foscarnet against viral polymerases. Am J Med. 1992;92:3S–7S. doi: 10.1016/0002-9343(92)90329-a. [DOI] [PubMed] [Google Scholar]
- 8.Debyser Z, Vandamme A M, Pauwels R, Baba M, Desmyter J, De Clercq E. Kinetics of inhibition of endogenous human immunodeficiency virus type 1 reverse transcription by 2′,3′-dideoxynucleoside 5′-triphosphate, tetrahydroimidazo-[4,5,1-jk][1,4]-benzodiazepin-2(1H)-thione, and 1-[(2-hydroxyethoxy)methyl]-6-(phenylthio)thymine derivatives. J Biol Chem. 1992;267:11769–11776. [PubMed] [Google Scholar]
- 9.DeStefano J J, Buiser R G, Mallaber L M, Fay P J, Bambara R A. Parameters that influence processive synthesis and site-specific termination by human immunodeficiency virus reverse transcriptase on RNA and DNA templates. Biochim Biophys Acta. 1992;1131:270–280. doi: 10.1016/0167-4781(92)90025-u. [DOI] [PubMed] [Google Scholar]
- 10.Dettenhofer M, Cen S, Carlson B A, Kleiman L, Yu X F. Association of human immunodeficiency virus type 1 Vif with RNA and its role in reverse transcription. J Virol. 2000;74:8938–8945. doi: 10.1128/jvi.74.19.8938-8945.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dornadula G, Yang S, Pomerantz R J, Zhang H. Partial rescue of the Vif-negative phenotype of mutant human immunodeficiency virus type 1 strains from nonpermissive cells by intravirion reverse transcription. J Virol. 2000;74:2594–2602. doi: 10.1128/jvi.74.6.2594-2602.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goncalves J, Korin Y, Zack J, Gabuzda D. Role of Vif in human immunodeficiency virus type 1 reverse transcription. J Virol. 1996;70:8701–8709. doi: 10.1128/jvi.70.12.8701-8709.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gotte M, Li X, Wainberg M A. HIV-1 reverse transcription: a brief overview focused on structure-function relationships among molecules involved in initiation of the reaction. Arch Biochem Biophys. 1999;365:199–210. doi: 10.1006/abbi.1999.1209. [DOI] [PubMed] [Google Scholar]
- 14.Guo J, Henderson L E, Bess J, Kane B, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J Virol. 1997;71:5178–5188. doi: 10.1128/jvi.71.7.5178-5188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrich D, Hooker C W, Parry E. The human immunodeficiency virus type 1 TAR RNA upper stem-loop plays distinct roles in reverse transcription and RNA packaging. J Virol. 2000;74:5639–5646. doi: 10.1128/jvi.74.12.5639-5646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrich D, Hsu C, Race E, Gaynor R B. Differential growth kinetics are exhibited by human immunodeficiency virus type 1 TAR mutants. J Virol. 1994;68:5899–5910. doi: 10.1128/jvi.68.9.5899-5910.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrich D, Ulich C, Garcia-Martinez L F, Gaynor R B. Tat is required for efficient HIV-1 reverse transcription. EMBO J. 1997;16:1224–1235. doi: 10.1093/emboj/16.6.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison G P, Mayo M S, Hunter E, Lever A M. Pausing of reverse transcriptase on retroviral RNA templates is influenced by secondary structures both 5′ and 3′ of the catalytic site. Nucleic Acids Res. 1998;26:3433–3442. doi: 10.1093/nar/26.14.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinzinger N K, Bukinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu M C, Dhingra U, Earley J V, Holly M, Keith D, Nalin C M, Richou A R, Schutt A D, Tam S Y, Potash M J, et al. Inhibition of type 1 human immunodeficiency virus replication by a tat antagonist to which the virus remains sensitive after prolonged exposure in vitro. Proc Natl Acad Sci USA. 1993;90:6395–6399. doi: 10.1073/pnas.90.14.6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y, Khorchid A, Gabor J, Wang J, Li X, Darlix J L, Wainberg M A, Kleiman L. The role of nucleocapsid and U5 stem/A-rich loop sequences in tRNA(3Lys) genomic placement and initiation of reverse transcription in human immunodeficiency virus type 1. J Virol. 1998;72:3907–3915. doi: 10.1128/jvi.72.5.3907-3915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Y, Wang J, Shalom A, Li Z, Khorchid A, Wainberg M A, Kleiman L. Primer tRNA3Lys on the viral genome exists in unextended and two-base extended forms within mature human immunodeficiency virus type 1. J Virol. 1997;71:726–728. doi: 10.1128/jvi.71.1.726-728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isel C, Ehresmann C, Keith G, Ehresmann B, Marquet R. Initiation of reverse transcription of HIV-1: secondary structure of the HIV-1 RNA/tRNA(3Lys) (template/primer) J Mol Biol. 1995;247:236–250. doi: 10.1006/jmbi.1994.0136. [DOI] [PubMed] [Google Scholar]
- 24.Isel C, Lanchy J M, Le Grice S F, Ehresmann C, Ehresmann B, Marquet R. Specific initiation and switch to elongation of human immunodeficiency virus type 1 reverse transcription require the post-transcriptional modifications of primer tRNA3Lys. EMBO J. 1996;15:917–924. [PMC free article] [PubMed] [Google Scholar]
- 25.Isel C, Marquet R, Keith G, Ehresmann C, Ehresmann B. Modified nucleotides of tRNA(3Lys) modulate primer/template loop-loop interaction in the initiation complex of HIV-1 reverse transcription. J Biol Chem. 1993;268:25269–25272. [PubMed] [Google Scholar]
- 26.Ji X, Klarmann G J, Preston B D. Effect of human immunodeficiency virus type 1 (HIV-1) nucleocapsid protein on HIV-1 reverse transcriptase activity in vitro. Biochemistry. 1996;35:132–143. doi: 10.1021/bi951707e. [DOI] [PubMed] [Google Scholar]
- 27.Kerr S G, Anderson K S. RNA dependent DNA replication fidelity of HIV-1 reverse transcriptase: evidence of discrimination between DNA and RNA substrates. Biochemistry. 1997;36:14056–14063. doi: 10.1021/bi971385+. [DOI] [PubMed] [Google Scholar]
- 28.Kiernan R E, Ono A, Englund G, Freed E O. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J Virol. 1998;72:4116–4126. doi: 10.1128/jvi.72.5.4116-4126.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klasens B I, Huthoff H T, Das A T, Jeeninga R E, Berkhout B. The effect of template RNA structure on elongation by HIV-1 reverse transcriptase. Biochim Biophys Acta. 1999;1444:355–370. doi: 10.1016/s0167-4781(99)00011-1. [DOI] [PubMed] [Google Scholar]
- 30.Klaver B, Berkhout B. Premature strand transfer by the HIV-1 reverse transcriptase during strong-stop DNA synthesis. Nucleic Acids Res. 1994;22:137–144. doi: 10.1093/nar/22.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohlstaedt L A, Steitz T A. Reverse transcriptase of human immunodeficiency virus can use either human tRNA(3Lys) or Escherichia coli tRNA(2Gln) as a primer in an in vitro primer-utilization assay. Proc Natl Acad Sci USA. 1992;89:9652–9656. doi: 10.1073/pnas.89.20.9652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 33.Lanchy J M, Ehresmann C, Le Grice S F, Ehresmann B, Marquet R. Binding and kinetic properties of HIV-1 reverse transcriptase markedly differ during initiation and elongation of reverse transcription. EMBO J. 1996;15:7178–7187. [PMC free article] [PubMed] [Google Scholar]
- 34.Lanchy J M, Keith G, Le Grice S F, Ehresmann B, Ehresmann C, Marquet R. Contacts between reverse transcriptase and the primer strand govern the transition from initiation to elongation of HIV-1 reverse transcription. J Biol Chem. 1998;273:24425–24432. doi: 10.1074/jbc.273.38.24425. [DOI] [PubMed] [Google Scholar]
- 35.Lapadat-Tapolsky M, De Rocquigny H, Van Gent D, Roques B, Plasterk R, Darlix J L. Interactions between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle. Nucleic Acids Res. 1993;21:831–839. doi: 10.1093/nar/21.4.831. . (Erratum, 21:2024.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Quan Y, Arts E J, Li Z, Preston B D, de Rocquigny H, Roques B P, Darlix J L, Kleiman L, Parniak M A, Wainberg M A. Human immunodeficiency virus type 1 nucleocapsid protein (NCp7) directs specific initiation of minus-strand DNA synthesis primed by human tRNA(Lys3) in vitro: studies of viral RNA molecules mutated in regions that flank the primer binding site. J Virol. 1996;70:4996–5004. doi: 10.1128/jvi.70.8.4996-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang C, Rong L, Gotte M, Li X, Quan Y, Kleiman L, Wainberg M A. Mechanistic studies of early pausing events during initiation of HIV-1 reverse transcription. J Biol Chem. 1998;273:21309–21315. doi: 10.1074/jbc.273.33.21309. [DOI] [PubMed] [Google Scholar]
- 38.Lobel L I, Goff S P. Reverse transcription of retroviral genomes: mutations in the terminal repeat sequences. J Virol. 1985;53:447–455. doi: 10.1128/jvi.53.2.447-455.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lori F, di Marzo Veronese F, de Vico A L, Lusso P, Reitz M S, Jr, Gallo R C. Viral DNA carried by human immunodeficiency virus type 1 virions. J Virol. 1992;66:5067–5074. doi: 10.1128/jvi.66.8.5067-5074.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masuda T, Planelles V, Krogstad P, Chen I S Y. Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J Virol. 1995;69:6687–6696. doi: 10.1128/jvi.69.11.6687-6696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazumder A, Neamati N, Sunder S, Schulz J, Pertz H, Eich E, Pommier Y. Curcumin analogs with altered potencies against HIV-1 integrase as probes for biochemical mechanisms of drug action. J Med Chem. 1997;40:3057–3063. doi: 10.1021/jm970190x. [DOI] [PubMed] [Google Scholar]
- 42.Nascimbeni M, Bouyac M, Rey F, Spire B, Clavel F. The replicative impairment of Vif-mutants of human immunodeficiency virus type 1 correlates with an overall defect in viral DNA synthesis. J Gen Virol. 1998;79:1945–1950. doi: 10.1099/0022-1317-79-8-1945. [DOI] [PubMed] [Google Scholar]
- 43.Oberg B. Antiviral effects of phosphonoformate (PFA, foscarnet sodium) Pharmacol Ther. 1989;40:213–285. doi: 10.1016/0163-7258(89)90097-1. [DOI] [PubMed] [Google Scholar]
- 44.Oude Essink B B, Das A T, Berkhout B. Structural requirements for the binding of tRNA Lys3 to reverse transcriptase of the human immunodeficiency virus type 1. J Biol Chem. 1995;270:23867–23874. doi: 10.1074/jbc.270.40.23867. [DOI] [PubMed] [Google Scholar]
- 45.Quan Y, Gu Z, Li X, Li Z, Morrow C D, Wainberg M A. Endogenous reverse transcription assays reveal high-level resistance to the triphosphate of (−)2′-dideoxy-3′-thiacytidine by mutated M184V human immunodeficiency virus type 1. J Virol. 1996;70:5642–5645. doi: 10.1128/jvi.70.8.5642-5645.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramsey C A, Panganiban A T. Replication of the retroviral terminal repeat sequence during in vivo reverse transcription. J Virol. 1993;67:4114–4121. doi: 10.1128/jvi.67.7.4114-4121.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ren J, Esnouf R, Garman E, Somers D, Ross C, Kirby I, Keeling J, Darby G, Jones Y, Stuart D, et al. High resolution structures of HIV-1 RT from four RT-inhibitor complexes. Nat Struct Biol. 1995;2:293–302. doi: 10.1038/nsb0495-293. [DOI] [PubMed] [Google Scholar]
- 48.Rigourd M, Lanchy J M, Le Grice S F, Ehresmann B, Ehresmann C, Marquet R. Inhibition of the initiation of HIV-1 reverse transcription by 3′-azido-3′-deoxythymidine. Comparison with elongation. J Biol Chem. 2000;275:26944–26951. doi: 10.1074/jbc.M003262200. [DOI] [PubMed] [Google Scholar]
- 49.Rong L, Liang C, Hsu M, Kleiman L, Petitjean P, de Rocquigny H, Roques B P, Wainberg M A. Roles of the human immunodeficiency virus type 1 nucleocapsid protein in annealing and initiation versus elongation in reverse transcription of viral negative-strand strong-stop DNA. J Virol. 1998;72:9353–9358. doi: 10.1128/jvi.72.11.9353-9358.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz O, Marechal V, Danos O, Heard J M. Human immunodeficiency virus type 1 Nef increases the efficiency of reverse transcription in the infected cell. J Virol. 1995;69:4053–4059. doi: 10.1128/jvi.69.7.4053-4059.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sova P, Volsky D J. Efficiency of viral DNA synthesis during infection of permissive and nonpermissive cells with vif-negative human immunodeficiency virus type 1. J Virol. 1993;67:6322–6326. doi: 10.1128/jvi.67.10.6322-6326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spence R A, Anderson K S, Johnson K A. HIV-1 reverse transcriptase resistance to nonnucleoside inhibitors. Biochemistry. 1996;35:1054–1063. doi: 10.1021/bi952058+. [DOI] [PubMed] [Google Scholar]
- 53.Spence R A, Kati W M, Anderson K S, Johnson K A. Mechanism of inhibition of HIV-1 reverse transcriptase by nonnucleoside inhibitors. Science. 1995;267:988–993. doi: 10.1126/science.7532321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Starcich B, Ratner L, Josephs S F, Okamoto T, Gallo R C, Wong-Staal F. Characterization of long terminal repeat sequences of HTLV-III. Science. 1985;227:538–540. doi: 10.1126/science.2981438. [DOI] [PubMed] [Google Scholar]
- 55.Suo Z, Johnson K A. Effect of RNA secondary structure on the kinetics of DNA synthesis catalyzed by HIV-1 reverse transcriptase. Biochemistry. 1997;36:12459–12467. doi: 10.1021/bi971217h. [DOI] [PubMed] [Google Scholar]
- 56.Temin H M, Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970;226:1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 57.Thrall S H, Krebs R, Wohrl B M, Cellai L, Goody R S, Restle T. Pre-steady-state kinetic characterization of RNA-primed initiation of transcription by HIV-1 reverse transcriptase and analysis of the transition to a processive DNA-primed polymerization mode. Biochemistry. 1998;37:13349–13358. doi: 10.1021/bi981102t. [DOI] [PubMed] [Google Scholar]
- 58.Trono D. Partial reverse transcripts in virions from human immunodeficiency and murine leukemia viruses. J Virol. 1992;66:4893–4900. doi: 10.1128/jvi.66.8.4893-4900.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsurutani N, Kubo M, Maeda Y, Ohashi T, Yamamoto N, Kannagi M, Masuda T. Identification of critical amino acid residues in human immunodeficiency virus type 1 IN required for efficient proviral DNA formation at steps prior to integration in dividing and nondividing cells. J Virol. 2000;74:4795–4806. doi: 10.1128/jvi.74.10.4795-4806.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ulich C, Dunne A, Parry E, Hooker C W, Gaynor R B, Harrich D. Functional domains of Tat required for efficient human immunodeficiency virus type 1 reverse transcription. J Virol. 1999;73:2499–2508. doi: 10.1128/jvi.73.3.2499-2508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.von Schwedler U, Song J, Aiken C, Trono D. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J Virol. 1993;67:4945–4955. doi: 10.1128/jvi.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Winzor D, Sawyer W. Quantitative characterization of ligand binding. New York, N.Y: Wiley-Liss, Inc.; 1995. [Google Scholar]
- 63.Wohrl B M, Krebs R, Goody R S, Restle T. Refined model for primer/template binding by HIV-1 reverse transcriptase: pre-steady-state kinetic analyses of primer/template binding and nucleotide incorporation events distinguish between different binding modes depending on the nature of the nucleic acid substrate. J Mol Biol. 1999;292:333–344. doi: 10.1006/jmbi.1999.3057. [DOI] [PubMed] [Google Scholar]
- 64.Wu W, Blumberg B M, Fay P J, Bambara R A. Strand transfer mediated by human immunodeficiency virus reverse transcriptase in vitro is promoted by pausing and results in misincorporation. J Biol Chem. 1995;270:325–332. doi: 10.1074/jbc.270.1.325. [DOI] [PubMed] [Google Scholar]
- 65.Wu W, Henderson L E, Copeland T D, Gorelick R J, Bosche W J, Rein A, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J Virol. 1996;70:7132–7142. doi: 10.1128/jvi.70.10.7132-7142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu X, Liu H, Xiao H, Conway J A, Hehl E, Kalpana G V, Prasad V, Kappes J C. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J Virol. 1999;73:2126–2135. doi: 10.1128/jvi.73.3.2126-2135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang H, Bagasra O, Niikura M, Poiesz B J, Pomerantz R J. Intravirion reverse transcripts in the peripheral blood plasma on human immunodeficiency virus type 1-infected individuals. J Virol. 1994;68:7591–7597. doi: 10.1128/jvi.68.11.7591-7597.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang H, Dornadula G, Alur P, Laughlin M A, Pomerantz R J. Amphipathic domains in the C terminus of the transmembrane protein (gp41) permeabilize HIV-1 virions: a molecular mechanism underlying natural endogenous reverse transcription. Proc Natl Acad Sci USA. 1996;93:12519–12524. doi: 10.1073/pnas.93.22.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang H, Dornadula G, Pomerantz R J. Endogenous reverse transcription of human immunodeficiency virus type 1 in physiological microenvironments: an important stage for viral infection of nondividing cells. J Virol. 1996;70:2809–2824. doi: 10.1128/jvi.70.5.2809-2824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang H, Dornadula G, Wu Y, Havlir D, Richman D D, Pomerantz R J. Kinetic analysis of intravirion reverse transcription in the blood plasma of human immunodeficiency virus type 1-infected individuals: direct assessment of resistance to reverse transcriptase inhibitors in vivo. J Virol. 1996;70:628–634. doi: 10.1128/jvi.70.1.628-634.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang H, Pomerantz R J, Dornadula G, Sun Y. Human immunodeficiency virus type 1 Vif protein is an integral component of an mRNP complex of viral RNA and could be involved in the viral RNA folding and packaging process. J Virol. 2000;74:8252–8261. doi: 10.1128/jvi.74.18.8252-8261.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]