Abstract
An alignment of cardiovirus sequences led to the prediction of three conserved stem-loops in the 3′ untranslated region (UTR) of mengovirus. Deletions of each stem were engineered in mengovirus cDNAs and also in mengovirus replicons, in which part of the viral capsid sequences were replaced with the firefly luciferase gene. The effect of deletion on RNA infectivity and plaque phenotype was evaluated after transfection of viral transcripts into HeLa cells or by luciferase assays of cellular extracts after transfection with RNA replicons. Stem I (mengovirus bases 7666 to 7687) was found to be dispensable for viral growth or exponential luciferase expression. Deletion of stem III (bases 7711 to 7721) was lethal to the virus, and the replicons were incapable of RNA synthesis. Deletion of stem II (ΔII; bases 7692 to 7705) produced an intermediate phenotype, in that replicons had marginal RNA synthesis activity but transfection with genomic RNA usually failed to produce plaques after normal incubation times (31 h, 37°C). In a few of the ΔII transfections, however, plaques were observed after long incubation, especially if the cells received large amounts of RNA (3 μg per 3 × 106 cells). Viruses from two ΔII-derived plaques were isolated and amplified. Their RNAs were converted into cDNA, sequenced, and mapped for genotype. Each maintained the ΔII deletion and, in addition, had one or two reversion mutations, which were characterized by reverse genetics as responsible for the phenotypes. One reversion caused an amino acid change in the polymerase (3Dpol), and the other was localized to the 3′ UTR, upstream of stem I.
Picornavirus polymerase enzymes (3Dpol) are capable of copying almost any RNA template in vitro if provided with a suitable RNA or DNA primer (6). An apparent lack of template selectivity during elongation contrasts with the enzymes' apparent specificity during natural viral RNA synthesis, in that viral, but not cellular, RNAs are copied in infected cells. Although the precise biological functions of the untranslated regions (UTRs) of picornaviruses are only poorly understood, the proximity of the 3′ UTR to the genetically encoded poly(A) tail, the normal site of RNA synthesis initiation, has led to proposals that these segments may work in cis with other portions of the RNA, the polymerase, other viral proteins (2B, 2C, 3A, 3B, 3C, etc), and perhaps unidentified cellular factors, to bring about template selection or a timely regulation of RNA replication processes (1, 22, 27, 28, 38, 39, 41).
The 3′ UTR segments of picornaviruses are generally quite short, ranging from 40 bases (rhinoviruses) to 126 bases (cardioviruses), are located between the polyprotein termination codon(s) and 3′-terminal poly(A), and consist of purine-rich heteropolymeric sequences (37, 42, 43). Computer-aided predictions, combined with enzymatic and chemical probing, have led to partial models for the 3′ UTRs of several enteroviruses (19, 27, 29, 41). Within members of this genus, the bases encoding two short stems, designated X and Y (bases 7376 to 7417 and 7423 to 7445 in poliovirus 1M, respectively), are reasonably well conserved. The terminal loops of X and Y typically contain additional short complementary sequences capable of generating higher-order tertiary structures by base pairing between the terminal loops (29). When the putative X-Y interactions were tested via site-directed mutagenesis in coxsackievirus A9 and coxsackievirus B3 cDNAs, the resultant genomic RNAs were noninfectious, consistent with the idea that loop-loop interactions may be involved in the mechanism of viral RNA synthesis (27, 29, 41).
Protein recognition of these or related motifs could conceivably play roles in viral template recognition or the initiation of minus-strand RNA synthesis. Poliovirus RNA probes containing the 3′-terminal 108 bases and the poly(A) tail undergo demonstrably specific reactions in vitro with recombinant poliovirus protein 3AB or with complexes of recombinant proteins 3AB and 3CD (14). Such interactions are consistent with a predicted early step in the 3Dpol-dependent initiation of minus-strand RNA synthesis or with the addition of VPg (protein 3B) to the 5′ end of the minus strand (30). Although RNA structure mapping experiments on genome RNAs have been reasonably successful (39, 40), they are technically quite difficult, and the binding segments that are specifically responsible for 3AB and 3CD interactions have yet to be more narrowly mapped within the enteroviral 3′ UTR or to be correlated more directly with the putative loop-loop tertiary structure (14). Moreover, these binding complexes have not yet been rigorously tested for in vivo biological activity by mutagenesis mapping within the context of viral genomes or within the context of replication-competent recombinant RNAs (replicons) in assays that use reporter gene expression as a quantitative indicator of viral RNA synthesis. Indeed, recent deletion and substitution experiments with poliovirus and rhinovirus cDNAs suggest that specific 3′ UTR segments or structures may not even be uniquely required for negative-strand RNA synthesis, because they can be replaced with fragments from heterologous picornaviruses (33) or removed entirely from certain replication-competent RNAs (39, 40).
Furthermore, as compelling as it is, the enterovirus X-Y model for the binding of viral protein is neither common nor consistent with the 3′-terminal sequences for other picornavirus genera. In rhinovirus 14 (HRV-14) for example, a different, albeit specific, interaction with a 34- to 36-kDa cellular protein has been documented in UV cross-linking reactions with a rhinovirus-specific 3′ UTR stem (38). Recombinant HRV-14 deletions in the 3′ UTR resulting in reduced ability to bind the cellular protein produced defective-replication phenotypes when engineered into genomic sequences. Also, in contrast to the poliovirus experiments, parallel attempts to demonstrate stable rhinovirus protein interactions (e.g., 3Dpol or 3CD) with the 3′ UTR have yet to be successful (38). With encephalomyocarditis virus (EMCV), a cardiovirus, only the poly(A) tail and a contiguous, short U-rich sequence located 38 to 49 bases upstream are reportedly required for interactions with homologous recombinant 3Dpol, as determined by RNA mobility shift assays (4, 5). Again, this result is different from that for poliovirus. Therefore, if particular 3′ structural elements are indeed involved in picornaviral RNA synthesis, it is conceivable that they might participate through broader, more-general template recognition or membrane binding mechanisms (40) or perhaps share only subtle commonalities with those of other members of this extended virus family.
To address these questions in more depth for the cardioviruses, we have begun a more detailed sequence and structural mapping of the mengovirus 3′ UTR. Phylogenetic analyses and computer modeling now suggest a genus-specific secondary structure consisting of three conserved stem-loop motifs in this region. We report here the development of a cDNA replicon assay for cardioviruses, using firefly luciferase as a reporter gene, in a system analogous to those described for enteroviruses (1) and the use of this assay to test the specific steps in the mengovirus life cycle that might be disrupted by deletional modification of the 3′ UTR. Parallel experiments with genome-length recombinant RNAs have also been carried out; these confirm the replicon results and implicate two of the three newly described 3′ stem regions as being required for minus-strand RNA synthesis in cardioviruses.
MATERIALS AND METHODS
Virus and cells.
H1 HeLa cell (ATCC-CRL1958) growth in suspension cultures and cardiovirus plaque development on plated cells under agar were performed as described previously (35). Virus was clonally propagated by picking an isolated plaque and inoculating a fresh cell monolayer (60-mm-diameter plates). After the development of cytopathic effect (24 h at 37°C), infected cells were lysed by three cycles of freezing and thawing, the cell debris was removed by centrifugation for 10 min at 2,000 × g, and the supernatant was used to infect another monolayer (100-mm-diameter plates) for virus amplification. Subsequent cell lysis (after 24 h at 37°C) and virus purification by sedimentation through sucrose cushions have been described (10).
Prior to infection for single-step growth assays, the HeLa cells from suspension cultures were collected, washed once in phosphate-buffered saline (PBS), gently pelleted, and then resuspended at a concentration of 4 × 107 cells/ml. Mengovirus samples were added at a multiplicity of infection of 10 PFU per cell, and attachment was carried out for 30 min at room temperature under constant rotation. After another gentle pelleting, unattached virus was removed from the cells by washing with PBS (10 ml). The infected cultures were resuspended to a concentration of 4 × 106 cells/ml in medium A (35) and incubated at 37°C in a shaking water bath. Aliquots (0.5 ml) were removed at the required times (0 to 12 h), buffered with 15 μl of 1.25 M HEPES, pH 7.4, and then frozen in an ethanol-dry ice bath before storage at −80°C. After two subsequent freeze-thaw cycles, virus titers (PFU per cell) were determined by standard plaque assays at 37°C for 30 h.
Recombination procedures.
Standard cDNA procedures were used (2, 36). Restriction endonucleases, T7 polymerase, and Moloney murine leukemia virus reverse transcriptase were obtained from commercial sources (New England Biolabs, Boehringer Mannheim Biochemicals, Gibco BRL, and Takara). DNA sequencing was performed by the alkaline denaturation method for double-stranded DNA, using Sequenase (version 2.0; U.S. Biochemicals). PCR amplifications used cloned Pfu polymerase (Stratagene), reaction conditions recommended by the manufacturer, and the primers (Integrated DNA Technologies, Inc.) listed in Table 1. Denaturation cycles were performed at 92°C for 45 s, annealing cycles were for 45 s at temperatures 5°C below the melting temperature of the least-stable primer, and elongation cycles were at 72°C for 2 min per kb of expected product.
TABLE 1.
Mutagenesis primers
| Location | Viral basea | Primer
|
|
|---|---|---|---|
| Name | Sequence | ||
| 5′ UTR | 209 (5′) | P74 | TCACATTACTGGCCGAAG |
| 1D/2A | 3450 (5′) | P85 | CTTATGCTTGAAAGCCCCAACCCT |
| 3D | 6563 (5′) | P88 | GGACCCTATGGACAAGAACAC |
| 3D | 7416 (3′) | P90 | AGGTCGATATAGAGGGCCCTC |
| lucc | NAb (3′) | P93 | CCCCACTAGTTTCCAAAGGGTTGGTCAATTTGGACTTTCCGCCC |
| luc | NA (5′) | P94 | CCCCACTAGTAATGGAAGACGCCAAAAAC |
| 2B | 4012 (3′) | P113 | GTTGGCATCGAGCAAAGCCG |
| 3′ UTR | 7690 (5′) | P162 | CACGGTCGTCATACCGCAG |
| 3′ UTR | 7706 (5′) | P163 | CAGACAGGGTTCTTCTACTTTG |
| 3′ UTR | 7723 (5′) | P164 | CTTTGCAAGATAAACTAGAG |
| 3′ UTR | 7650 (3′) | P165 | CGGTATGACGACCGTGTAACGCGTTGTGCCAGTG |
| 3′ UTR | 7676 (3′) | P166 | GTAGAAGAACCCTGTCTGTGTACACCCGGTTGGC |
| 3′ UTR | 7694 (3′) | P167 | CTAGTTTATCTTGCAAAGTGTCTGCGGTATGACGAC |
| Vector | 7987 (3′) | P168 | CGGGCAGTGAGCGCAACG |
| 3′poly(A) | 7757 (3′) | P179 | TTTTTTTTTTTTTTTAAAAC |
| 3D | 7143 (3′) | P208 | TAACATTGAGGTCGCTGC |
| 3C | 5825 (5′) | P212 | CCACACCCGCTCCTCAGT |
| 3D | 6664 (5′) | P251 | TTGCAGCAGAGAGACTAG |
| 3′ UTR | 7650 (5′) | P263 | CACTGGCACACCGCGTTACCC |
| 3D | 6710 (5′) | P265 | CATTGTCTATCGGACATTCCT |
| 3′ UTR | 7650 (3′) | P267 | GGGTAACGCGGTGTGCCAGTG |
| 3D | 6710 (3′) | P268 | GAGGAATGTCCGATAGACAAT |
Mengovirus genome base encoded by the 5′- or 3′ (reverse complement)-most nucleotide of the primer.
NA, not applicable.
luc, luciferase gene.
Engineered genomes.
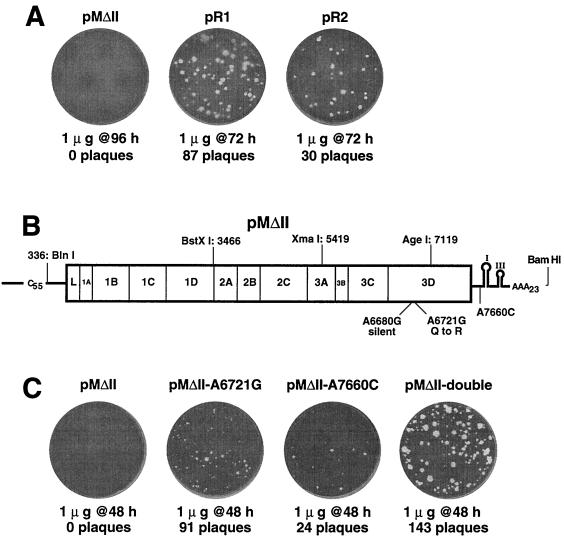
Mengovirus cDNA contained in plasmid pMwt has been described (8). The viral sequence encoded by this plasmid may be accessed from GenBank (accession no. L22089). Amplicons containing precise deletion of stem I, II, or III were generated by four sequential PCR steps. First, fragments upstream of each stem's sequence were amplified from pMwt templates. The primer pairs were P88 and P165 (for deletion of stem I [ΔI]), P88 and P166 (for ΔII), and P88 and P167 (for ΔIII). Next, downstream fragments from pMwt were synthesized using P162 and P168 (for ΔI), P163 and P168 (for ΔII), and P164 and P168 (for ΔIII). For each segment, the primers were designed to give upstream and downstream products that overlapped by 16 nucleotides and contained the desired deletion. The appropriate unfractionated amplicons were combined and assembled into linked fragments by an additional 10 cycles of PCR. Each sample was then supplemented with primers P88 and P168 before a final amplification. After gel purification, the sized fragments were introduced into the EcoRV site of pBluescript SK +/− (Stratagene) and transformed into Escherichia coli (strain MV1190; Bio-Rad). Resultant AgeI-HindIII cDNA segments containing the sequenced deletions were substituted into pMwt, and, after sequence confirmation, the new plasmids were designated pMΔI, pMΔII, and pMΔIII.
Similar procedures engineered two putative reversion mutations in a site-specific manner. The A-to-G substitution at base 6721 (A6721G) was created by PCR using primer combinations P265-P168 and P212-P268, followed by amplicon linkage with primers P212-P208. The product was ligated into pMΔII using BspDI and AgeI sites to generate pMΔII-A6721G. The parallel A7660C mutation used primer sets P263-P168 and P251-P267, with product amplification by primers P251-P168. This fragment was ligated into pMΔII using AgeI and HindIII sites to generate pMΔII-A7660C. These mutations were combined into pMΔII-double by substituting the BspDI-AgeI fragment from pMΔII-A6721G for the analogous fragment of pMΔII-A7660C.
Before transfection of HeLa cells, viral genome-containing cDNAs were linearized by digestion with BamHI, transcribed into RNA by T7 polymerase (8), and incorporated into liposomes (34), as described previously (12).
Revertant cDNAs.
Two well-isolated viral plaques, vMΔII.1 and vMΔII.2, that resulted after pMΔII-derived RNA was transfected into HeLa cells were picked, and the viruses were amplified by passage through HeLa cells and then purified by sedimentation through sucrose cushions as described above. Genomic RNAs were extracted by sodium dodecyl sulfate-phenol and collected by ethanol precipitation. Moloney murine leukemia virus reverse transcriptase (Gibco BRL) was used for reverse transcription with primer P179, and then PCR using primers P88-P179, P85-P90, and P74-P113 converted the viral sequences into overlapping cDNA sets (covering ∼7,700 viral bases), which were subsequently ligated together, inserted into plasmid vectors (pBluescript SK +/−), and used to transform E. coli. Defined restriction fragments from these plasmids, pR1 and pR2, were purified on agarose gels and then substituted for the analogous fragments of pMΔII.
Mengovirus replicons.
A PCR fragment containing the complete firefly luciferase cDNA was amplified from pGL2 (Promega) using primers P93-P94. Engineered restriction sites (BstXI in P93 and SpeI in P94) facilitated the in-frame substitution of this gene for the SpeI-BstXI fragment in the capsid-coding region of pMwt. The SacII-AgeI fragment from the resulting plasmid, pMluz, was then transferred into the analogous sites of pMΔI, pMΔII, and pMΔIII, creating pMLuzΔI, pMluzΔII, and pMluzΔIII, respectively. To monitor the luciferase induced by these sequences, 60-mm-diameter plates of confluent HeLa cell monolayers were transfected with cDNA-derived T7 RNA transcripts (2.5 μg) incorporated into liposomes (34). After 30 min at room temperature, 5 ml of P5 medium (35) was added to each plate, and the incubation was continued at 37°C. At the required times (0 to 14 h later), plates were washed with 5 ml of PBS and then reacted with luciferase lysis buffer (250 μl; Promega) for 10 min. The materials were collected into small tubes and subjected to centrifugation for 1 min at 16,000 × g in a microcentrifuge. The supernatants were decanted and stored at −80°C until the enzyme activities (relative light units) could be determined in standard luciferin assays (15) with a luminometer (Moonlight; model 2010).
Murine inoculations.
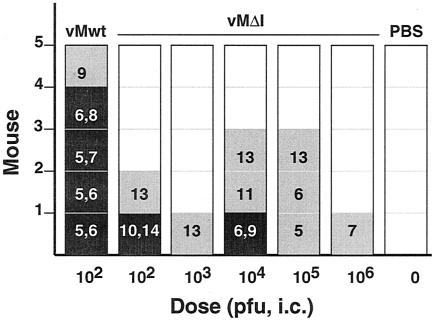
ICR Swiss mice (4-week-old females; Charles River) were randomly divided into seven groups of five animals. Members of five groups were inoculated intracranially with sucrose-purified vMΔI virus (30 μl of PBS containing 102, 103, 104, 105, or 106 PFU). A sixth group received vMwt virus (30 μl; containing 102 PFU in PBS), and the remaining group received PBS alone. Mortality was recorded for 14 days postinoculation, and the 50% lethal dose for vMΔI (>106 PFU) was estimated from these data by standard methods (32).
RESULTS
Model of 3′ UTR.
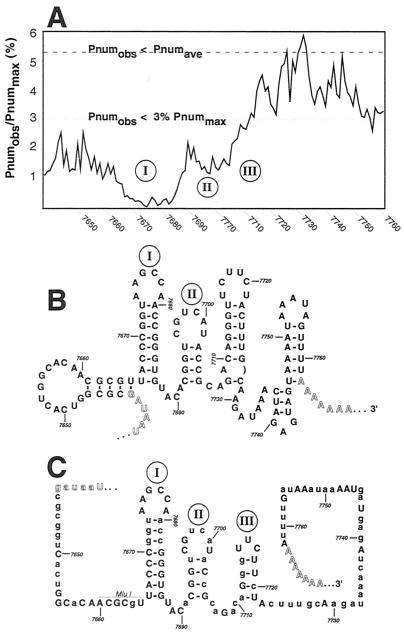
The complete genomic sequence for mengovirus M was analyzed for optimal and suboptimal minimum-free-energy secondary structures by the program mFOLD (44). This data set, as well as many others for various picornavirus folds, sequences, and alignments, is available on the Internet (www.bocklabs.wisc.edu/acp). The described findings (31) predict that most RNA virus sequences are in constant flux among local energy minima and that many bases can adopt multiple, energetically similar, configurations with diverse pairing partners throughout the genome. The picornavirus 5′ and 3′ UTR segments are special, however, in that they tend to fold as independent segments over a range of suboptimal energies (−12 kcal/mol), without alternative contacts from the interior portions of the genomes (31). For mengovirus, a plot of each base's pairing number (Pnum), as determined from the genomic fold (Fig. 1A), highlighted a particular short segment of the 3′ UTR (bases 7664 to 7687) with exceptionally low Pnum values, indicative of one or more well-determined stems with high pairing fidelity (17, 45). Compared to the rest of this genome (average Pnum is 5.35% of maximum Pnum [Pnummax]), the segments flanking this trough are also composed of relatively low-Pnum bases (defined as <3% of Pnummax), and indeed, among the (several) low-energy configurations for this region (Fig. 1B), two short unbranched stems containing bases 7666 to 7687 (I) and 7692 to 7705 (II) were consistently observed.
FIG. 1.
Mengovirus 3′ UTR models. (A) The observed pairing number (Pnumobs) for each base in the family of suboptimal folds within 12 kcal of the minimum free energy of the mengovirus M genomic fold was determined. The values for the 3′ UTR were normalized as percentages of Pnummax that might have been observed at infinite energy (31). On this scale, the average Pnumobs (Pnumave) for all genomic bases was 5.348% +/− 3.57% (standard deviation). Values <3% are considered low, and the inherent structures are considered well determined (45). (B) The high-road minimum-free-energy structure for the mengovirus M 3′ UTR, as determined from the complete genomic fold (31). The polyprotein termination codons, the poly(A) tail, and stems I and II of the model are highlighted. (C) Thirteen cardiovirus genomes, including Theiler's murine encephalomyelitis virus (TMEV) strain BeAn, TMEV-GDVII, TMEV-GDVII variant, TMEV-Da, EMCV-Rueckert, EMCV-B, EMCV-B variant, EMCV-D, EMCV-D variant, EMCV-DV1, EMCV-PV1, EMCV-IP, and mengovirus M were aligned and analyzed for potentially conserved, minimum-free-energy secondary structures (31). Within the 3′ UTR, only mengovirus stems I, II, and III meet these criteria. Uppercase letters indicate conserved bases in all sequences of the alignment. Nucleotide numbering corresponds to the mengovirus M genome sequence (GenBank accession no. L22089).
Within the remainder of the 3′ UTR (bases 7710 to 7761), the pairing became more promiscuous as there were many alternative optimal and suboptimal structures. To discriminate among the elements with the highest probability of occurrence, an alignment of 13 cardiovirus genomic sequences, including 4 Theiler's murine encephalomyelitis viruses and 9 EMCV variants, was probed with the program mFOLD-Phylo, as previously reported (31), to give a new minimum-free-energy model of the complete mengovirus genome, consistent with possible folds for all other cardioviruses. This program reports base pairs only if they can form in identical locations among every member of the alignment (31; M. Zuker, personal communication). In addition to stems I and II, the phylogenetic model predicted that a putative third stem, III, was common to all cardioviruses. This new model could be formed with an energetic stability at or very near the minimum free energy of each individual genome when folded independently (Fig. 1C). No other stems, motifs, or obvious tertiary elements were found to be otherwise common to the 3′ UTRs of these viruses. On the basis of the collective folding data, and as a first approximation for mapping experiments, the regions of stems I, II and III were chosen for further genetic analysis.
RNA transcript infectivity.
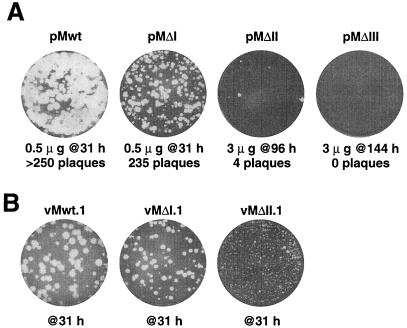
Individual deletions of stems I, II, and III were engineered by PCR and introduced into mengovirus full-length cDNAs (pMΔI, pMΔII, and pMΔIII, respectively). As an initial test of engineering consistency, RNA transcripts from these cDNAs were used to program cell-free reactions in rabbit reticulocyte lysates. All samples translated with equivalent efficiencies, and the resultant protein processing patterns could not be distinguished from wild-type patterns (data not shown), indicating that the polyprotein open reading frames were intact and that the function of the internal ribosome entry sites within the 5′ UTRs was not affected. Similar viral transcripts were then transfected into HeLa cell monolayers and assayed for the ability to produce infectious progeny. The sequence from pMΔI had a specific infectivity of ∼500 PFU/μg of RNA, similar to that obtained from pMwt, although this deletion always gave plaques (at 31 h) that were somewhat smaller than those from pMwt sequences (Fig. 2A). RNAs from pMΔII and pMΔIII were much less infectious. After transfection, pMΔII RNA gave only an occasional plaque and these typically required at least 96 h to develop visibly, even when the monolayers were transfected with large amounts of transcript (3 μg per plate). RNA from pMΔIII failed at every attempt to produce plaque-forming virus. Even consecutive blind passages (three times) of lysed transfected cells (3 μg per plate) and long incubation times (96 h per passage) were unsuccessful in yielding any evidence of plaques (or revertants), and we subsequently considered ΔIII to be a lethal mutation.
FIG. 2.
Plaque phenotype after transfection and infection. (A) HeLa monolayers were transfected with the indicated quantities of transcript RNA and then incubated at 37°C for 31, 96, or 144 h before crystal violet staining and an assay for plaque development. (B) Two individual, well-separated plaques from panel A were picked, amplified, and then used to reinfect HeLa monolayers. After 31 h at 37°C, the plates were stained and assayed for plaque development. The viruses derived from vMΔI.2 and vMΔII.2 plaques (not shown) gave plaque phenotypes indistinguishable from those of their parallel counterparts, vMΔI.1 and vMΔII.1, respectively.
Viral infectivity.
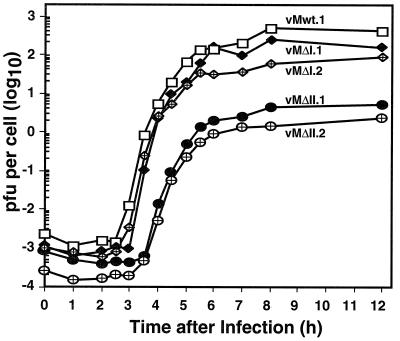
Isolated plaques (two each) from ΔI, ΔII, and wild-type transfections were picked and amplified in HeLa cells. The maintenance of the relevant 3′ deletions was reconfirmed in every case by reverse transcription-PCR and sequencing. When the viruses were replated, the plaque sizes from both of the ΔI isolates were again somewhat smaller than those from wild type and those from vMΔII were even smaller, with plaques having minute-plaque phenotypes (Fig. 2b). These effects were better quantitated in single-step growth experiments, where the cells were infected synchronously with the amplified isolates and progeny titers were determined at defined intervals (Fig. 3). The two wild-type isolates gave typical mengovirus growth patterns, with eclipse periods of ∼2.5 h and progeny yields of 2 to 3 log10 PFU/cell. The viruses with ΔI deletions (vMΔI.1 and vMΔI.2) showed marginal delays in the eclipse phase (2.5 to 3 h), and, by 12 h postinfection, neither had produced quite as many progeny as the wild type. In addition, their overall yields were two- to fourfold lower per cell. The ΔII viruses (vMΔII.1 and vMΔII.2) were much more impaired than ΔI or wild-type viruses, with average eclipse times (3.5 h) and progeny yields (76- to 170-fold lower) indicative of strong replication delays or other impediments to normal growth. Collectively, the infectivity data suggest that each of the engineered 3′ UTR deletions had a measurably negative impact on overall mengovirus growth, and moreover that these impacts could be clearly weighted as ΔIII ≫ ΔII ≫ ΔI > wild type.
FIG. 3.
Single-step viral growth. Viruses from amplified plaques (Fig. 2) were used to infect HeLa cell cultures (multiplicity of infection of 10) as described in Materials and Methods. Each point represents the viral titer determined by serial dilution for a sample removed at that time.
Mengovirus replicons and RNA synthesis assays.
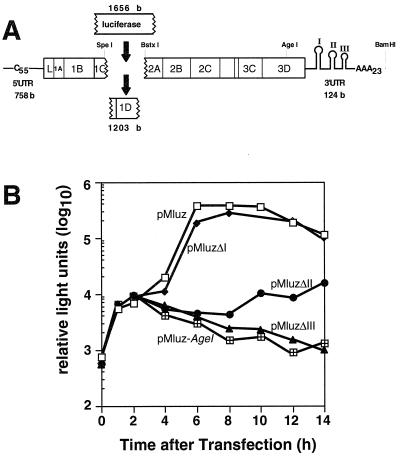
Efficient viral growth is a cumulative phenotype reliant on many steps in the infectious cycle. To separate some of these events, mengovirus-luciferase replicon pMluz, in which a portion of the capsid-coding region (1,203 bases) was replaced with an in-frame luciferase reporter gene (1,656 bases; Fig. 4A), was constructed. Upon translation, the luciferase sequence was expressed as a fusion protein linked to 109 amino-terminal residues from viral protein 1C and 141 carboxyl-terminal residues of viral protein 2A. The remainder of the mengovirus polyprotein was left intact and yielded a leader protein, 1A, 1B, and RNA synthesis functions (2B, 2C, 3A, 3B, 3C, and 3D). Also intact were both viral UTRs, including the wild-type-length 5′ poly(C) tract (C44UC10) and the 3′ poly(A) tail (of length A23). During multiple experiments with these replicons, transfection of HeLa cells with pMluz transcripts always produced luciferase activity in two distinct phases (Fig. 4B), as also has been reported for analogous poliovirus replicons (1). Translation of the input transcripts peaked after 2.5 to 3 h, before synthesis resumed in a secondary, exponential burst (4 to 8 h). Since replication-defective replicon controls, linearized within 3Dpol (pMluz-Agel), only showed the first phase of input translation, the 200- to 500-fold activity difference relative to pMluz (8 to 10 h) could be attributed to new plus-strand RNA synthesis by replication-competent sequences. At later times (>10 h), detectable luciferase tended to diminish, even in high-activity samples, as further RNA synthesis presumably declined and the proteins degraded within the cells.
FIG. 4.
Mengovirus replicon pMluz. (A) Within this cDNA, the 3′ half of the viral 1C region, the complete 1D region, and the first two codons of the viral 2A region were replaced in-frame by the firefly luciferase gene. Additional cDNAs differed from pMluz in that they contained separate deletions of 3′ UTR stems I, II, or III. (B) Luciferase activity in transfected HeLa cells was determined as described in Materials and Methods. All samples (20 μl of lysate) were prepared and assayed in parallel.
The three 3′ deletion sequences were transferred into pMluz, and their transcripts were assayed in parallel with the parental sequence (Fig. 4B). Initially, all samples showed equivalent luciferase activities, confirming that all the RNAs were translated equivalently and assuring that the deletions did not impinge on viral translational efficiency. As expected from the infectivity data, the pMluzΔI samples proved replication competent, as their curves resembled that for pMluz throughout the secondary translation phase. Still, none of these samples ever quite achieved the wild-type level of enzyme activity, even after 8 to 10 h, suggesting that overall RNA synthesis was slightly less than optimal for this mutation. At the other extreme, the pMluzΔIII samples paralled the negative controls and showed no evidence of new RNA synthesis. The pMluzΔII samples had intermediate phenotypes. They produced low, albeit continuous, levels of luciferase after the initial translation phase, which marginally kept pace with protein turnover. Clearly, this mutant was significantly impaired in RNA synthesis relative to the wild type or pMluzΔI, but viral translation was nevertheless sustained for at least 14 h after transfection.
Revertant mapping.
The low specific infectivity of pMΔII viral transcripts, the long time required for plaque development (96 h), and the marginal fecundity of replicons carrying this defect suggested that vMΔII.1 and vMΔII.2 might contain pseudoreversions. Both viruses maintained the ΔII deletions, but viral sequences, even those with minute-plaque phenotypes, should have performed better in the replicon assays, and we suspected that these genomes might have second-site changes as well. The two viral RNAs were cloned into cDNAs that spanned ∼7,700 bases, from the 5′ poly(C) tract through the 3′ poly(A) tail. Since native cardiovirus poly(C) tracts are notoriously refractory to cDNA conversion (24), each isolated sequence was made full length by linkage to a pMwt cassette containing the 5′-most mengovirus segment (336 bases), including a wild-type poly(C). Transfection with RNAs (1 μg) derived from the new constructs, pR1 and pR2, produced 104 and 34 medium plaques, respectively, after 72 h, a significant improvement in number and size relative to parental pMΔII transcripts (Fig. 5A). Restriction fragments spanning these revertant genomes were then swapped back into pMΔII until the enhanced growth phenotypes were reconstructed. In both cases, the similar fragments containing the 3Dpol gene and portions of the 3′ UTR restored viability in the presence of the ΔII deletion (not shown). When sequenced in their entirety, all of these fragments showed the same point mutation, A6721G, in the 3Dpol coding region, which would cause a Gln-to-Arg substitution at amino acid 155 in the 3D protein (Fig. 5B). Additionally, the pR2 fragment contained a silent A-to-G substitution at base 6680. The pR1 fragment had a different, secondary substitution of A7660C in the 3′ UTR sequence just upstream of stem I.
FIG. 5.
Revertant identification and cloning. (A) Putative revertant viruses vMΔII.1 and vMΔII.2 were cloned into cDNAs pR1 and pR2, respectively. RNA transcripts were transfected into HeLa cells and assayed for plaque development relative to that of parental pMΔII RNA. (B) Large restriction fragments (indicated) from pR1 and pR2 were substituted into pMΔII until the plaquing phenotypes in panel A were reconstructed. Locations of identified sequence changes are indicated. (C) Point mutations A6721G and A7660C were engineered separately and together into pMΔII cDNA. RNA transcripts were transfected into HeLa cells, and the plates were assayed after 48 h for plaque development.
Confirmation of pseudoreversion.
The effects of the polymerase (A6721G) and 3′ UTR (A7660C) reversion mutations were confirmed experimentally by recreating the sequences synthetically and then engineering them separately or in combination into pMΔII cDNA. Transcripts from each of these constructs (pMΔII-A6721G, pMΔII-A7660C, and pMΔII-double) had improved infectivity compared to that of the parental pMΔII (Fig. 5C). Though all of these plaques still took longer to develop than the wild type (48 versus 31 h) and were small in size, both reversions, singly or preferably, in combination, were clearly responsible for the enhanced viability of mengovirus genomes carrying the ΔII deletion. The reversion activities were also evident when the engineered mutations were transferred into pMluzΔII replicons. Again, either alone or, preferably, in combination, the 3Dpol and 3′ UTR point mutations increased the ability of the ΔII sequences to produce new RNAs and boosted the consequent luciferase activity 6 h after transfection (not shown).
Murine lethality.
Engineered cardioviruses can be unexpectedly attenuated for murine pathogenicity even though their growth potential in tissue culture may be comparable to that for the wild type (7). The surprising dispensability of the 3′ UTR stem I for viral growth in HeLa cells or RNA synthesis activity in replicons, despite its structural conservation among viruses and low Pnum values, suggested a phenotype that might be more apparent if tested in animals. Groups of outbred Swiss mice were inoculated intracranially with vMwt, vMΔI, or buffer and then monitored for disease (Fig. 6). As expected (24), most animals receiving vMwt (102 PFU) became sick (5 to 7 days) and died quickly (6 to 8 days). Only one animal from this group, paralyzed at day 9, survived the experiment (30 days). The data are consistent with the previously reported 50% lethal dose for vMwt of 102 to 103 PFU (24). When inoculated with vMΔI, the mice seemed more resistant to this virus, and, regardless of doses of up to 106 PFU, 15 of 25 never showed symptoms of disease. Only two in these groups died, and, surprisingly, these had received low (102 PFU) or medium doses (104 PFU). An eight additional survivors were scattered among the groups, each eventually showing the development of hind limb paralysis typical of cardiovirus disease. Generally, detectable disease onset was delayed in the low-dose groups (10 to 13 days) compared to that in the high-dose groups (5 to 13 days), but none of these animals' conditions seemed to further degenerate after the first 2 or 3 days. Based on these data, we assess vMΔI as much less pathogenic than vMwt.
FIG. 6.
Murine pathogenicity. Mice (groups of five) were infected with the indicated doses of virus or buffer (PBS). Animals were monitored for 3 weeks postinfection (PI) and scored for death (black boxes), paralysis (grey boxes), or lack of symptoms (white boxes). The dates PI of first detectable paralysis and death (black boxes only) are indicated for all appropriate animals. i.c. intracranially.
DISCUSSION
The thermodynamics of base pairing predicts that large segments of most viral RNA genomes fluctuate through multiple topologies according to variation in environmental temperatures and ionic conditions that are prevalent during viral infections (16, 31, 46). Accordingly, it is not always reliable to assign a single, unilateral minimum-free-energy model to these genomes, because such a model does not always convey alternative or equivalent secondary or tertiary structures of the RNA as a whole (46). Chemical and enzymatic mapping methods are very useful tools if the local structures are stable enough or populous enough within the family of molecules to allow reproducible targeting. Sometimes whole RNA genomes may be stable enough to map (39), but isolating an RNA fragment from its genome always has the potential to obscure alternative pairing opportunities that could occur for these bases and that indeed may occur within the context of a larger sequence (16). Instead, as a first step in RNA model building, we prefer to search for consistent viral RNA secondary structures by querying large families of related suboptimal genomic folds for regions, motifs, or elements with low Pnum and then to compare these regions for structural conservation in phylogenetically related viral sequences. This method was originally developed and validated for the structures of procaryotic and eucaryotic ribosomal RNAs and several RNA phage genomes (18, 20, 21, 25, 45, 46). We have also applied it successfully to multiple picornavirus genomes for the prediction of structurally important 5′ UTR internal ribosome entry site elements (31) and cis-acting replication elements (23, 26). Model building is then followed by genetic and biochemical probing according to the base pairing predictions.
It has been reported that RNA fragments containing the 3′ UTR of EMCV, a cardiovirus serotypically indistinguishable from mengovirus, bind to viral polymerase 3Dpol in reactions that require only an intact 3′ poly(A) tail and a short U-rich sequence nearby (4, 5). Because of these data, we began our investigations with mengovirus with the expectation that cardioviruses, like the enteroviruses and rhinoviruses, might have a few well-defined structure elements within the required portions of their 3′ UTRs to help guide the polymerase in the initiation of minus-strand synthesis. Our computer analyses of the intact genomic sequence were encouraging, as two nodes of particularly low Pnum values were apparent in the 3′ region. These nodes correlated well with small stems I and II, common to 50 randomly selected minimum-free-energy structures of mengovirus. These stems were also conserved and of low Pnum in 12 independent folds with related sequences from different cardiovirus strains. Neither stem, however, included the U-rich segment (mengovirus bases 7715 to 7726), of which only a portion (mengovirus bases 7715 to 7721) is actually common among sequences from the other cardioviruses. Instead, the higher Pnum values in the distal portion of the 3′ UTR predicted a significant degree of structural disorder, with these bases accepting many different local pairing partners and also distant partners from the middle and 5′ UTRs of the genome, suggesting that bases 3′ to the stem I and stem II segments do not routinely adopt a definable conformation. Yet when the genomic sequence was analyzed phylogenetically, a third putative stem emerged (III) from the models that could form in each individual cardiovirus sequence at slightly suboptimal energies. Moreover, the putative location of stem III exactly coincided with the conserved portion of the U-rich sequence implicated in polymerase binding experiments (4, 5). As a place to start with our genetic experiments, we engineered precise deletions of the stem I, II, and III sequences into mengovirus cDNAs and tested the resulting mutants for phenotype within the context of full-length genomes and replication-competent RNA replicons.
Surprisingly, stem I, the motif of lowest Pnum and highest sequence conservation, proved virtually dispensable for viral growth in tissue culture. Deleted ΔI transcripts had infectivity similar to that of pMwt RNAs, and the resultant viruses grew with nearly the same single-cycle kinetics. The replicon assays confirmed that this stem was not required for viral translation or effective RNA synthesis activities. Except for resulting in modest diminutions in plaque size and ultimate viral titer, deletion of stem I actually seemed relatively innocuous. In mice, however, we found an unusual phenotype. Both vMwt and vMΔI viruses contain the same 5′ poly(C) tract, C44UC10, which should have made them extremely lethal to animals (7). The wild-type virus killed or crippled all recipients at a dose of only 100 PFU. But most animals receiving vMΔI, even at doses up to 10,000 times that amount, survived or were only partially paralyzed. This course of disease is very unusual for mengoviruses, as paralysis is usually the harbinger of serious neurological damage and subsequent inevitable death. That so many animals survived unscathed or survived (and recovered) after virus-induced paralysis suggests that the stem I region of the 3′ UTR may play a previously unrecognized role in cardiovirus neurovirulence, cell tropism, or host immune regulatory induction. The strong conservation of stem I in sequence and structure among all cardioviruses may mark this site as a potential host protein recognition site. We now plan further investigations along these lines.
In contrast to stem I, stem III was modeled as part of an indeterminant, shifting conformation within the 3′ UTR. This is the only stable stem that can commonly form within this distal region of all cardioviruses, but, for any particular virus, there are many other structural arrangements with nearly equivalent energies. Yet deletion of this segment conferred absolute lethality to the mengovirus genome. Transfection with very high RNA doses, long incubation times, and multiple forced passage of transfected-cell lysates gave no evidence of infectivity or viable revertants. Nor was there overt evidence of a deletion-dependent change in RNA stability (not shown). When assayed within a replicon context, the input pMluzΔIII sequences were translated with an efficiency similar to that for the wild type replicons but were incapable of new mRNA synthesis and the luciferase decay rate was the same as in control samples (pMluz-AgeI) with inactive 3Dpol. The absence of new mRNA synthesis and the complete lack of detectable reversion point to the stemIII sequence as an absolute requirement for cardiovirus RNA synthesis. We assume that the requisite step probably involves the initiation of minus-strand synthesis. The stem III segment contains the conserved portion of the U-rich sequence identified in EMCV as a putative 3Dpol binding region for that virus (4). Our phylogenetic folding data also suggest that this element probably is universally required as a replication component in all cardioviruses.
No amount of energy concession or forced pairings allowed us to manipulate the 3′-most half of this UTR segment into more-plausible, alternative secondary (or tertiary) models consistent with a majority of the sequences. If cardioviruses share a sequence or structural commenality (other than stem III) in the region between stem II and the poly(A) tail which confers polymerase recognition to these RNAs, we have yet to find it. Rather, we now recognize that the conserved, low-Pnum, upstream stem II is probably another important part of the polymerase recognition motif. When our RNA genomes with the ΔII deletion first developed minute plaques in HeLa cells, we assumed that this sequence region, like stem I, might prove peripheral to the RNA synthesis pathways. The progeny viruses were found to maintain the ΔII deletion, and although they grew more slowly than ΔI or wild-type viruses, they were still quite viable. However, the replicon assays suggested otherwise. Transfection with pMluzΔII RNA gave the expected input translational signal, but thereafter luciferase synthesis barely kept pace with turnover. This slow rate of viral RNA synthesis from a replicon was inconsistent with a viable infectious sequence, and we began to suspect that the ΔII virus phenotypes might have acquired additional, second-site reversions.
When converted into cDNA, the pR1 and pR2 sequences grew faster and gave larger plaques than the parental (ΔII) sequences. These phenotypes were very similar to those of the vMΔII.1 and vMΔII.2 viruses from which pR1 and pR2 had been derived, and while not completely wild type in vigor, these cloned progeny clearly had incorporated some genetic improvements that helped them synthesize more viral RNA despite their ΔII deletions. In both cases, restriction fragment replacement experiments localized the responsible genomic segments to the 3′-terminal 1,100 bases. Sequencing then highlighted three nucleotide changes. Both revertants contained the same A6721G mutation that caused a Q155R substitution in the 3Dpol protein. A second mutation in pR1 changed a single base (A7660C) in the 3′ UTR, just upstream of stem I. An additional silent mutation in pR2 (A6680G) was not investigated further because it caused no protein changes and because its context was in a very-high-Pnum region of the genome. The two most likely candidate reversions were engineered separately or as a pair into pMΔII. Restoration of the original plaque phenotypes allowed definitive assignment of the rescue mechanism to these particular mutations.
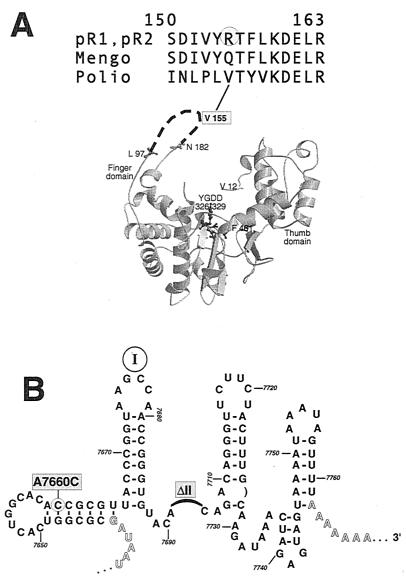
Mengovirus 3Dpol amino acid Q155 is the sequence homolog, and presumably structural analog, of poliovirus V155 (Fig. 7A). The reported atomic structure of poliovirus 3Dpol is disordered in this “fingers domain” (13), but more highly refined structures of human immunodeficiency virus reverse transcriptase and RNA polymerase assign key template binding and positioning functions to the middle, or motif I, portion of this region (3, 11). Logically, this assignment fits our reversion too, if the Q-to-R substitution somehow improved the mengovirus protein's interaction with the defective (ΔII) 3′ UTR and thus enhanced minus-strand initiation. The second reversion, A7660C, also fits this hypothesis by potentially creating two new base pairing opportunities in a predicted minimum-free-energy stem just upstream of stem I (Fig. 7B). In wild-type mengovirus, the bases of this stem are somewhat promiscuous, with low but flexible Pnum values. The reversion mutation would reduce the energy of this stem by 4.4 kcal/mol and additionally could diminish the number of attractive alternative pairing partners elsewhere in the genome. While obviously not a perfect substitution for the deleted stem II, this newly stabilized 3′ motif, in synergy with the altered 3Dpol contact loop, seems to restore sufficient vitality to the viral RNA synthesis pathways to allow these genomes to plaque. This hypothesis predicts that stem II in addition to stem III makes direct contact with the mengovirus 3Dpol, in a reaction mediated at least in part, by the fingers domain of the protein. The lethal ΔIII genotype is obviously more catastrophic to this putative interaction than the ΔII genotype, because reversion fixation indicates at least a minimum level of successful RNA synthesis and because no revertants could be isolated from the ΔIII sequences. When the atomic structure of this or a related viral polymerase is resolved more completely, we hope to identify the contacts that may contribute to the initiation of RNA synthesis within this 3′ UTR and understand more clearly why cardiovirus replication seems to differ in several respects from that of the entero- and rhinoviruses.
FIG. 7.
Pseudoreversion locations. (A) Structural model of poliovirus 3Dpol protein from reference 13 is oriented to show the catalytic YGDD motif and other landmarks. Mengovirus mutation A6721G creates a Q155R substitution in 3Dpol. The amino acid analog in poliovirus, V155, is in the structurally disordered fingers region of the model. (B) A minimum-free-energy structure of the mengovirus 3′ UTR (ΔII) shows that mutation A7660C could stabilize a putative stem motif upstream of stem I by allowing two additional base pairs relative to the wild-type sequence (Fig. 1B), perhaps compensating for the physical loss of stem II.
Our mengovirus analyses, combined with the previous EMCV 3Dpol binding studies, have already highlighted several genus-specific distinctions. Relative to those of the poliovirus and rhinoviruses, the 3′ UTRs of cardioviruses are dissimilar in length, sequence, and apparent structure. Also in contrast to that of poliovirus, where proteins 3AB and 3CD are both required for stable 3′ complexes (14), the 3Dpol of EMCV alone seems able to recognize its plus-strand template in a sequence-specific manner (4, 5), a result consistent with our ΔII revertant mapping to a probable RNA binding loop within the enzyme. Moreover, we have recently discovered that, unlike what was found in reported experiments with poliovirus and rhinovirus sequences (39, 40), the 3′ UTR of mengovirus cannot be deleted in its entirety from cDNAs without complete loss of transcript infectivity or replicon RNA synthesis functions. The required sequence elements center on stems II and III (H. Duque, M. S. McBride, and A. C. Palmenberg, unpublished data). Our future experiments now will aim at extending our 3′ genome analyses to the bases farther downstream of stem III and also at the introduction of less-obtrusive point mutations into stems II and III. Like poliovirus replicon assays (1), our mengovirus replicon assays seem able to distinguish the input from subsequent RNA translation, and we should be able to assign replication-specific phenotypes to individual mutations. We have cloned and expressed the mengovirus 3Dpol (9) and additionally intend to probe the binding properties of this protein with our various sequences.
ACKNOWLEDGMENTS
This work was supported by NIH grant AI-17331 to A.C.P.
We thank Marchel Hill for excellent technical support and Jean-Yves Sgro for invaluable help with the RNA-folding analyses.
REFERENCES
- 1.Andino R, Rieckhof G E, Achacoso P L, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baltimore D, Franklin R M. Preliminary data on a virus-specific enzyme system responsible for the synthesis of viral RNA. Biochem Biophys Res Commun. 1962;9:388–392. doi: 10.1016/0006-291x(62)90021-9. [DOI] [PubMed] [Google Scholar]
- 3.Boyer P L, Ferris A L, Hughes S H. Mutational analysis of the fingers domain of human immunodeficiency virus type 1 reverse transcriptase. J Virol. 1992;66:7533–7537. doi: 10.1128/jvi.66.12.7533-7537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui T, Porter A G. Localization of binding site for encephalomyocarditis virus RNA polymerase in the 3′-noncoding region of the viral RNA. Nucleic Acids Res. 1995;23:377–382. doi: 10.1093/nar/23.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui T, Sankar S, Porter A G. Binding of encephalomyocarditis virus RNA polymerase to the 3′-noncoding region of the viral RNA is specific and requires the 3′-poly(A) tail. J Biol Chem. 1993;268:1–6. [PubMed] [Google Scholar]
- 6.Dasgupta A, Baron M H, Baltimore D. Poliovirus replicase: a soluble enzyme able to initiate copying of poliovirus RNA. Proc Natl Acad Sci USA. 1979;76:2679–2683. doi: 10.1073/pnas.76.6.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duke G M, Osorio J E, Palmenberg A C. Attenuation of mengovirus through genetic engineering of the 5′ noncoding poly(C) tract. Nature. 1990;343:474–476. doi: 10.1038/343474a0. [DOI] [PubMed] [Google Scholar]
- 8.Duke G M, Palmenberg A C. Cloning and synthesis of infectious cardiovirus RNAs containing short, discrete poly(C) tracts. J Virol. 1989;63:1822–1826. doi: 10.1128/jvi.63.4.1822-1826.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duque H, Palmenberg A C. Epitope mapping of monoclonal antibodies raised to recombinant Mengo 3D protein. Virus Genes. 1996;13:159–168. doi: 10.1007/BF00568908. [DOI] [PubMed] [Google Scholar]
- 10.Frolov V G, Duque H, Palmenberg A C. Quantitation of endogenous viral polymerase, 3Dpol, in preparations of Mengo and encephalomyocarditis viruses. Virology. 1999;260:148–155. doi: 10.1006/viro.1999.9808. [DOI] [PubMed] [Google Scholar]
- 11.Georgiadis M M, Jessen S M, Ogata C M, Telesnitsky A, Goff S P, Herndrickson W A. Mechanistic implications from the structure of a catalytic fragment of Moloney murine leukemia virus reverse transcriptase. Structure. 1995;3:879–892. doi: 10.1016/S0969-2126(01)00223-4. [DOI] [PubMed] [Google Scholar]
- 12.Hahn H, Palmenberg A C. Encephalomyocarditis viruses with short poly(C) tracts are more virulent than their mengovirus counterparts. J Virol. 1995;69:2697–2699. doi: 10.1128/jvi.69.4.2697-2699.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen J L, Long A M, Schultz S C. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure. 1997;5:1109–1122. doi: 10.1016/s0969-2126(97)00261-x. [DOI] [PubMed] [Google Scholar]
- 14.Harris K S, Xiang W, Alexander L, Paul A V, Lane W S, Wimmer E. Interaction of the poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome: identification of viral and cellular cofactors necessary for efficient binding. J Biol Chem. 1994;269:27004–27014. [PubMed] [Google Scholar]
- 15.Hoffman M A, Palmenberg A C. Mutational analysis of the J-K stem-loop region of the encephalomyocarditis virus IRES. J Virol. 1995;69:4399–4406. doi: 10.1128/jvi.69.7.4399-4406.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobson A B, Arora R, Zuker M, Priano C, Lin C H, Mills D R. Structural plasticity in RNA and its role in the regulation of protein translation in coliphage Q-beta. J Mol Biol. 1998;275:589–600. doi: 10.1006/jmbi.1997.1472. [DOI] [PubMed] [Google Scholar]
- 17.Jacobson A B, Zuker M. Structural analysis by energy dot plot of a large mRNA. J Mol Biol. 1993;233:261–269. doi: 10.1006/jmbi.1993.1504. [DOI] [PubMed] [Google Scholar]
- 18.Jacobson A B, Zuker M, Hirashima A. Molecular biology of RNA: new perspectives. New York, N.Y: Academic Press, Inc.; 1987. Comparative studies on the secondary structure of the RNAs of related RNA coliphages; pp. 331–354. [Google Scholar]
- 19.Jacobson S J, Konings D A M, Sarnow P. Biochemical and genetic evidence for a pseudoknot structure at the 3′ terminus of the poliovirus RNA genome and its role in viral RNA amplification. J Virol. 1993;67:2961–2971. doi: 10.1128/jvi.67.6.2961-2971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaeger J A, Turner D H, Zuker M. Improved predictions of secondary structure for RNA. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le S-Y, Zuker M. Predicting common folds of homologous RNAs. J Biomol Struct Dyn. 1991;8:1027–1044. doi: 10.1080/07391102.1991.10507863. [DOI] [PubMed] [Google Scholar]
- 22.Lloyd R E, Jense H G, Ehrenfeld E. Restriction of translation of capped mRNA in vitro as a model for poliovirus-induced inhibition of host cell protein synthesis: relationship to p220 cleavage. J Virol. 1987;61:2480–2488. doi: 10.1128/jvi.61.8.2480-2488.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lobert P-E, Escriou N, Ruelle J, Michiels T. A coding RNA sequence acts as a replication signal. Proc Natl Acad Sci USA. 1999;96:11560–11565. doi: 10.1073/pnas.96.20.11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin L R, Duke G M, Osorio J E, Hall D J, Palmenberg A C. Mutational analysis of the mengovirus poly(C) tract and surrounding heteropolymeric sequences. J Virol. 1996;70:2027–2031. doi: 10.1128/jvi.70.3.2027-2031.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathews D H, Sabina J, Zuker M, Turner D H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 26.McKnight K L, Lemon S M. Capsid coding sequence is required for efficient replication of human rhinovirus 14 RNA. J Virol. 1996;70:1941–1952. doi: 10.1128/jvi.70.3.1941-1952.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melchers W J G, Hoenderop J G J, Bruins Slot H J, Pleij C W A, Pilipenko E V, Agol V I, Galama J M D. Kissing of the two predominant hairpin loops in the coxsackie B virus 3′ untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J Virol. 1997;71:686–696. doi: 10.1128/jvi.71.1.686-696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mellits K H, Meredith J M, Rohll J B, Evans D J, Almond J W. Binding of a cellular factor to the 3′ untranslated region of the RNA genomes of entero- and rhinoviruses plays a role in virus replication. J Gen Virol. 1998;79:1715–1723. doi: 10.1099/0022-1317-79-7-1715. [DOI] [PubMed] [Google Scholar]
- 29.Mirmomeni M H, Hughes P J, Stanway G. An RNA tertiary structure in the 3′ untranslated region of enteroviruses is necessary for efficient replication. J Virol. 1997;71:2363–2370. doi: 10.1128/jvi.71.3.2363-2370.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nomoto A, Detjen B M, Pozzatti R, Wimmer E. Location of the polio genome protein in viral RNAs and its implication for RNA synthesis. Nature. 1977;268:208–213. doi: 10.1038/268208a0. [DOI] [PubMed] [Google Scholar]
- 31.Palmenberg A C, Sgro J-Y. Topological organization of picornaviral genomes: statistical prediction of RNA structural signals. Semin Virol. 1997;8:231–241. [Google Scholar]
- 32.Reed L J, Muench H A. A simple method of estimating fifty percent end points. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 33.Rohll J B, Moon D H, Evans D J, Almond J W. The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J Virol. 1995;69:7835–7844. doi: 10.1128/jvi.69.12.7835-7844.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rose J K, Buonocore L, Whitt M A. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques. 1991;10:520–525. [PubMed] [Google Scholar]
- 35.Rueckert R R, Pallansch M A. Preparation and characterization of encephalomyocarditis virus. Methods Enzymol. 1981;78:315–325. [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Spector D H, Baltimore D. Requirement of 3′-terminal polyadenylic acid for the infectivity of poliovirus RNA. Proc Natl Acad Sci USA. 1974;71:2983–2987. doi: 10.1073/pnas.71.8.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Todd S, Nguyen J H C, Semler B L. RNA-protein interactions directed by the 3′ end of human rhinovirus genomic RNA. J Virol. 1995;69:3605–3614. doi: 10.1128/jvi.69.6.3605-3614.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Todd S, Semler B L. Structure-infectivity analysis of the human rhinovirus genomic RNA 3′ noncoding region. Nucleic Acids Res. 1996;24:2133–2142. doi: 10.1093/nar/24.11.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Todd S, Towner J S, Brown D M, Semler B L. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J Virol. 1997;71:8668–8874. doi: 10.1128/jvi.71.11.8868-8874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J, Bakkers J M J E, Galama J M D, Bruins Slot H J, Pilipenko E V, Agol V I, Melchers W J G. Structural requirements of the higher order RNA kissing element in the enteroviral 3′ UTR. Nucleic Acids Res. 1999;27:485–490. doi: 10.1093/nar/27.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yogo Y, Wimmer E. Polyadenylic acid at the 3′ terminus of poliovirus RNA. Proc Natl Acad Sci USA. 1972;69:1877–1882. doi: 10.1073/pnas.69.7.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yogo Y, Wimmer E. Sequence studies of poliovirus RNA. III. Polyuridylic acid and polyadenylic acid as components of the purified poliovirus replicative intermediate. J Mol Biol. 1975;92:467–477. doi: 10.1016/0022-2836(75)90292-2. [DOI] [PubMed] [Google Scholar]
- 44.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 45.Zuker M, Jacobson A B. Well-determined regions in RNA secondary structure prediction: analysis of small subunit ribosomal RNA. Nucleic Acids Res. 1995;23:2791–2798. doi: 10.1093/nar/23.14.2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuker M, Jacobson A B. Using reliability information to annotate RNA secondary structures. RNA. 1998;4:669–679. doi: 10.1017/s1355838298980116. [DOI] [PMC free article] [PubMed] [Google Scholar]