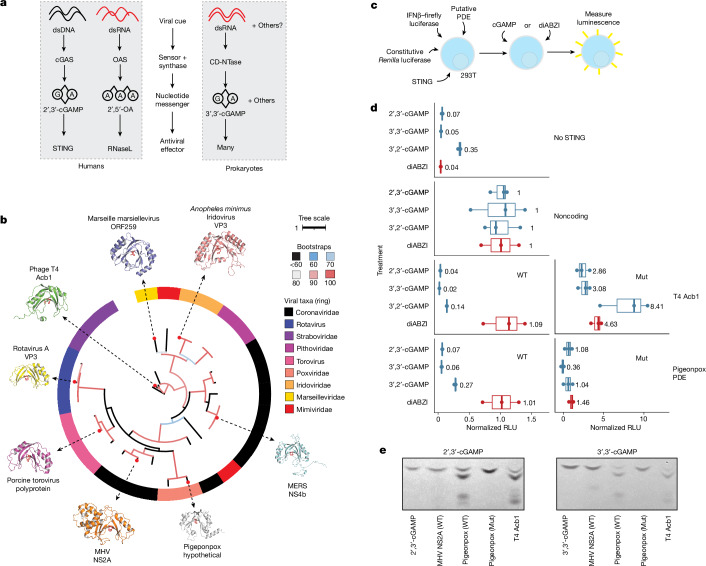
Fig. 4. LigT-like PDEs are frequently used to subvert host immunity.
a, Some innate immune pathways in eukaryotes and prokaryotes rely on a viral synthase sensor that detects virus-associated molecular patterns such as dsDNA or dsRNA and generates a nucleotide second messenger that stimulates an antiviral effector. b, A phylogenetic tree showing the polyphyletic lineages of LigT-like PDEs. Shaded boxes indicate viral taxa. The red residues in each protein structure are the conserved catalytic histidines. Units are substitutions per residue. The tree is coloured according to bootstrap values. NCBI Protein accessions: YP_008798230, YP_002302228, YP_009021100, YP_003406995, NP_049750, YP_009047207, YP_009046269 and YP_009824980. c, HEK 293T cells were transfected with constructs encoding STING, firefly luciferase driven by an IFNB promoter, a constitutively expressed Renilla luciferase, and a transgene. After 5 h, cells were treated with 10 μg ml−1 cGAMP or 0.1 μM diABZI. Around 24 h after the first transfection, luminescence of the firefly and Renilla luciferases was measured. d, Pigeonpox PDE prevents STING activation by cGAMP isomers. On the x axis, luminescence in relative luminescence units (RLU) is normalized to the RLU from cells transfected with noncoding vector and treated with the same STING agonist. RLUs were initially normalized as firefly RLU/Renilla RLU. Mut indicates mutations of the catalytic histidines. In box plots, the centre line is the median, box edges delineate 25th and 75th percentiles, and whiskers extend to the highest or lowest point up to 1.5 times the inter-quartile range. Data are from one biological replicate and three wells per condition. e, 2′,3′-cGAMP or 3′,3′-cGAMP was incubated with indicated wild-type or catalytic histidine mutant PDE proteins. Degradation of each cGAMP isomer was visualized by TLC. Uncropped TLC images are presented in Supplementary Fig. 1.