Abstract
Murine cells do not support human immunodeficiency virus type 1 (HIV-1) replication because of blocks to virus entry, proviral expression, and virion assembly. In murine 3T3 fibroblasts, the block to HIV-1 entry is relieved by the introduction of human CD4 and CCR5 or CXCR4, and proviral expression is increased by the introduction of the Tat cofactor, human cyclin T1; however, because of the assembly block, virus fails to spread. A panel of rodent cell lines expressing human CD4, CCR5, and cyclin T1 was established and studied for the ability to support virus replication. Mus musculus lymphoid cell lines EL4 and L1-2 and Mus dunni fibroblasts supported only low levels of virus assembly and released small amounts of infectious virus. CHO and Rat2 cell lines produced more infectious virus, but this production was still 40-fold lower than production in human cells. Only CHO cells expressing the three human cofactors were partially permissive for HIV-1 replication. To investigate the basis of the block to HIV-1 assembly, mouse-human heterokaryons were tested for ability to assemble and release virus. Fusion of human cells to HIV-1-infected mouse cells expressing CD4, CCR5, and cyclin T1 caused a 12-fold increase in virion release and a 700-fold increase in infectious virus production. Fusion of HIV-1-infected M. dunni tail fibroblasts to uninfected human cells caused a similar increase in virus release. More efficient virus release was not caused by increased proviral transcription or increased synthesis of virion components. Analysis of reciprocal heterokaryons suggested the absence of an inhibitor of virus assembly. Taken together, the results suggested that murine fibroblasts lack a cofactor that is required for efficient virus assembly and release.
Mouse cells, regardless of whether they express human CD4 (hu-CD4) and chemokine receptor, do not support human immunodeficiency virus type 1 (HIV-1) replication (8, 44). In contrast, activated human cells expressing endogenous or transfected hu-CD4 and coreceptor are generally permissive (3). Mouse cells fail to support HIV-1 replication as a result of blocks at various steps of the virus replication cycle. Virus fails to enter because murine CD4 and CCR5 do not interact with the SU subunit of the envelope glycoprotein (although murine CXCR4 is functional) (8). Expression of hu-CD4-CCR5 or hu-CD4-CXCR4 on mouse cells at least partially relieves the entry block, but replication fails due to subsequent blocks (3, 44).
In addition to the entry block, Tat transactivation of the HIV-1 long terminal repeat (LTR) is inefficient in murine cells, resulting in low-level provirus expression (2, 14, 17, 34, 41). The defect in Tat function was complemented in human-rodent somatic cell lines containing human chromosome 12 (2, 17, 34), a finding that suggested the presence of a species-specific Tat cofactor. This cofactor was identified as cyclin T1 (47), a component of the pTEFb transcription factor complex (16, 27, 49). Human cyclin T1 (hu-cyclin T1), in association with the cyclin-dependent kinase CDK9, binds to Tat to form a heterodimer with high affinity for the TAR stem-loop at the 5′ ends of nascent HIV-1 mRNA transcripts. The complex mediates hyperphosphorylation of the carboxy-terminal domain of RNA polymerase II, causing increased transcriptional processivity (14). Mouse cyclin T1 and hu-cyclin T1 differ at several amino acids, accounting for inefficient Tat transactivation of the HIV-1 LTR in mouse cells. A single amino acid change, the replacement of Tyr261 with Cys, in the mouse protein prevents it from interacting with Tat (5, 15, 22). Transfection of CHO or 3T3 cells with hu-cyclin T1 expression vector restores transactivation activity (5, 15, 22, 47).
Expression of hu-cyclin T1 together with hu-CD4 and hu-CCR5 or hu-CXCR4 in 3T3 cells restores Tat function, yet the cells remain refractory to HIV-1 replication (15, 28). Entry, reverse transcription, integration, and proviral expression are efficient in 3T3 cells (23, 28, 36), although some reduction of entry and RNA synthesis in comparison to a control human cell line was noted (28). Upon infection of the mouse cells with vesicular stomatitis virus G (VSV-G) pseudotypes, abundant Gag precursor polyprotein, Pr55gag, is synthesized; however, this is inefficiently converted to processed p24 CA. Electron microscopy showed that the cells contained electron-dense accumulations in intracellular vesicles of what appeared to be aggregates of Gag. In spite of the block to assembly and release, a small number of virions were observed budding from the cell membrane, and low levels of virions could be detected in the supernatant of the infected cells. These virions were infectious but were apparently not present in sufficient quantity to allow spread through the culture. A small reduction in CCR5 coreceptor function on the mouse cells was noted; however, the major block to HIV-1 replication appeared to be at assembly and release (28).
The restricted host range of HIV-1 has limited the development of small animal models for AIDS. Various rodent species have been inoculated, but virus replication has not been detected (32). Rabbits have been investigated as a potential AIDS model, as CD4-transgenic animals were found to support low-level virus replication (9, 12, 21) and a rabbit cell line expressing transfected CD4 and CCR5 was fully permissive for HIV-1 replication (44). Mice have also been investigated as a possible model system because of the experimental utility of this species. CD4-CCR5- and CD4-CXCR4-transgenic mice were established; however, these were found to support only low levels of HIV-1 replication (6, 40).
The restriction of the HIV-1 host range to human cells results from the requirement for viral proteins to interact with specific host cofactors. In the case of murine CD4-gp120 (24) and murine cyclin T1-Tat interactions (5, 15), species-specific alterations in the host cofactors prevents their association with the cognate virus protein. Similarly, the failure of the mouse fibroblasts expressing hu-CD4, hu-CCR5, and hu-cyclin T1 to support HIV-1 replication could have been caused by the absence in the mouse cells of a cellular cofactor required for virus assembly. Alternatively, it could have been caused by the presence of an inhibitory factor in the mouse cells. Here, we used human-mouse heterokaryons to investigate this question. The results demonstrated that the assembly defect could be complemented by fusion of infected mouse cells to uninfected human cells, suggesting that the human cells provided an assembly cofactor that was lacking in the mouse cells. In addition, we extended our earlier study to additional rodent species and cell types. This showed that various rodent species and cell lines shared the assembly block that was first observed in 3T3 cells, although a rat and hamster cell line were able to produce significant quantities of infectious virus. Interestingly, the hamster cell line supported a low level of virus replication.
MATERIALS AND METHODS
Cell lines.
Adherent cell lines derived from HOS, 3T3, 293T, Mus dunni tail fibroblasts (MDTF), the Chinese hamster ovary cell line (CHO; ATCC CCL-61) (37), and the rat fibroblast cell line Rat2 (46) were cultured in DMEM–10% fetal bovine serum. Nonadherent cell lines derived from CEMx174, EL4 (38), and L1-2 (19) were cultured in RPMI–10% fetal bovine serum. Cultures were maintained in 5% CO2 at 37°C.
Retroviral vector transduction was used to introduce hu-CD4 and hu-CCR5 into rodent and human cell lines and hu-cyclin T1 into rodent cell lines. L1-2, MDTF, CHO, and Rat2 cells were infected with pBABE-CCR5 retroviral vector stock produced in 293T packaging cells and selected 2 days later in medium containing 0.2 μg of puromycin/ml. Puromycin-resistant cells were stained with anti-CCR5 monoclonal antibody 2D7 (Pharmingen), and high-CCR5-expressing cells were sorted by flow cytometry. The cells were then infected with the retroviral vector pMX-CD4, which was derived from pMX (35), a retroviral vector that contains no selectable marker. CD4-expressing cells were enriched on anti-CD4-coated magnetic beads (Dynal) and then plated at a limiting dilution. Individual clones were analyzed for CD4 expression by fluorescence-activated cell sorting, and a single clone of each was chosen based on uniform high CD4 expression. L1-2.CD4.R5, MDTF.CD4.R5, and CHO.CD4.R5 cells were infected with pBABE.CyT(hygro), a hu-cyclin T1 expression vector derived from pBABE.hygro (29). The infected cells were selected in medium containing 200 μg of hygromycin/ml. Individual drug-resistant clones were expanded and tested for their response to transfected Tat and LTR-luciferase plasmids. GHOST.R5 (7) cells are HOS cells that contain a transactivatable HIV-2 LTR-enhanced green fluorescent protein (EGFP) cassette and express hu-CD4 and hu-CCR5. The CEMx.R5.GFP cell line that was used to determine virus titers by endpoint dilution was described previously (28).
Heterokaryon formation.
Three days prior to fusion, MGT5.CyT, MDTF.CD4.R5.CyT, and GHOST.R5 (106) were infected with NL4-3(VSV-G) at a multiplicity of infection (MOI) of 1. After 24 h, the cells were washed three times with medium to remove input virus, and the next day the cells were cocultured with 106 uninfected human 293T or murine 3T3 cells in a six-well culture plate. After 48 h, the cells were fused by adding 500 μl of 50% polyethylene glycol (PEG) 3350 (Sigma) for 5 min at 37°C. PEG was thoroughly removed by several washes with phosphate-buffered saline (PBS), and the heterokaryons were cultured for another 24 h. Heterokaryons were fixed with 4% paraformaldehyde–PBS for 15 min at room temperature, washed with PBS, and then stained with 100 μg of Hoechst 33258 (Sigma)/ml in PBS–0.2% Triton X-100. The efficiency of heterokaryon formation was analyzed by counting heterokaryons under UV fluorescence microscopy. Mouse and human nuclei could be distinguished morphologically, and only heterokaryons containing both nuclei were counted. Culture supernatant was collected before the heterokaryons were fixed, and virions were pelleted in a Beckman L-70 ultracentrifuge at 35,000 rpm in a SW55 Ti rotor for 1 h at 4°C. Pelleted virions were resuspended in 100 μl of lysis buffer (100 mM NaCl, 10 mM EDTA, 20 mM Tris [pH 7.5], 1% Triton X-100, 1% sodium deoxycholate), and 10 μl was used for Western analysis. The number of 50% tissue culture infective doses (TCID50) was determined by adding 100 μl of 10-fold serial dilutions of virus-containing supernatant to the eight-well columns of 96-well microtiter dishes containing 104 CEMx.R5.GFP cells per well. After 3 days of incubation, wells were scored microscopically for the presence or absence of fluorescent cells. The TCID50 per milliliter was calculated as the reciprocal of the dilution at which 50% of the wells scored positive.
To test for reporter gene activity in mouse-human heterokaryons, cells were fused by an Env-mediated mechanism. MGT5.CyT and GHOST.R5 (106) cells were infected with VSV-G pseudotyped NL4-3 env− luciferase virus. On the same day, 293T and 3T3 (2 × 106) cells were cotransfected with 20 μg of JR.FL Env expression vector or control empty vector and pRSV-Rev expression vector to promote efficient Env synthesis. Two days later, the infected and transfected cells were mixed at a 1:1 ratio. Luciferase activity was detected 24 to 48 h postfusion using luciferase detection reagents (Promega).
Viruses and infections.
NL4-3(VSV-G) pseudotypes were produced by transfecting 293T cells with an equal amount of pNL4-3 and VSV-G expression vector. Culture supernatants were harvested 48 h postinfection, filtered, and frozen at −80°C in aliquots. Viruses were quantitated by p24gag enzyme-linked immunosorbent assay and, for titer, by limiting dilution on CEMx.R5.GFP cells, as described above. Luciferase reporter viruses were produced by cotransfecting 293T cells with NL-Luc-E−R− and Env expression vectors for JR.FL or VSV-G as previously described (11). For dual luciferase infections, NL-Ren.Luc-E−R− reporter vector containing the Renilla luciferase gene in place of the firefly luciferase gene was pseudotyped with VSV-G (28). Cultures were infected with an equal mixture of the two different reporter viruses, and each virus was quantitated independently using dual luciferase reporter assay reagents that specifically detect each enzyme (Promega). HXB.BaL was a replication-competent virus consisting of HXB2 and BaL env (10). NL.BaL was similarly constructed using NL4-3 and substituting the BaL env. HIV.YU2.GFP, provided by M. Muesing (Aaron Diamond AIDS Research Center), was based on the HXB2 derivative R7 and contained EGFP in place of nef and the env of HIV YU2.
Immunoblot analysis.
Cells were infected with NL4-3(VSV-G), and 3 days later virions and cell lysates were prepared. Virions were pelleted by centrifuging 10 ml of filtered culture supernatant in an SW40 Ti rotor for 1 h and lysed in lysis buffer (100 mM NaCl, 10 mM EDTA, 20 mM Tris [pH 7.5], 1% Triton X-100, 1% sodium deoxycholate). Cell lysates were prepared by removing medium from the infected cells, washing with PBS, and lysing in 500 μl of lysis buffer. Protein in the lysates was quantitated by Coomassie blue reagent (Bio-Rad) and stored at −80°C before use. Lysate containing 10 μg of protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride filters, and probed with AIDS patient serum diluted 1:500 (provided by D. Richman, University of California, San Diego [UCSD]) followed by horseradish peroxidase-conjugated rabbit anti-human immunoglobulin (BioSource International). Filters were developed using enhanced-chemiluminescence reagents (Amersham).
Electron microscopy.
CHO.CD4.R5.CyT and MDTF.CD4.R5.CyT cells were seeded on large glass coverslips in 10-cm dishes (106 per dish) and 1 day later were infected with NL4-3(VSV-G) at an MOI of 0.5. Infected cells were analyzed 3 days after infection. Cells were washed twice and fixed in situ for 30 min with ice-cold 2.5% glutaraldehyde in 100 mM PIPES [piperazine-1,4-bis(2-ethanesulfonic acid), pH 6.9]. Fixed cells were washed three times in PIPES, scraped from the plate with a rubber policeman, and collected by low-speed centrifugation. Cell pellets were embedded in low-melting-temperature agarose and cut into small blocks. Pellets were postfixed in 1% osmium tetroxide in PIPES for 30 min at 4°C. After extensive washing in PIPES, the blocks were treated with 1% tannic acid in water, washed again, dehydrated in ethanol, and embedded in ERL resin (45) in gelatin capsules. Silver-gray sections were stained with lead citrate and uranyl acetate and examined under a Philips CM120 electron microscope at 60 kV.
For immunoelectron microscopy, the cells were washed twice and fixed in situ for 30 min in cold 4% formaldehyde–0.25% glutaraldehyde–0.2% picric acid in 100 mM PIPES (pH 6.9). Cell pellets were embedded in low-melting-temperature agarose and cooled to 0°C, and small blocks were cut and stained for 30 min at 4°C with 1% uranyl acetate in water. Subsequently, the blocks were dehydrated in ethanol and embedded in London resin white in gelatin capsules (33). Polymerization was initiated by addition of 0.5% benzoinmethylether and continued under UV light for 2 days at 4°C. Blocks were cut using a Reichert Ultracut ultratome, and silver sections were mounted on nickel grids without supporting film. For immunolabeling, grids were placed in a Pelco grid storage box converted as described by Sparkman and White (42) with a 1.5-mm hole drilled through each of the diamond-shaped grid slots. Grids were etched for 15 min with 1% sodium periodate in water, washed three times with water, and blocked for 30 min in 0.5% casein–0.2% gelatin (CaGe) in Tris-buffered salt solution (150 mM NaCl, 20 mM Tris-HCl [pH 7.5]). Grids were then incubated overnight at 4°C with rabbit anti-HIV-1 CA diluted 1:1,000 or with normal rabbit serum in CaGe, washed five times with CaGe, and treated with colloidal gold-labeled goat anti-rabbit immunoglobulin (12-nm diameter; Dianova, Hamburg, Germany) in CaGe for 1 h at room temperature. Labeled grids were washed with CaGe and distilled H2O and subsequently stained with 4% aqueous uranyl acetate and Reynold's lead citrate, air dried, and viewed at 60 kV. Ultrathin sections of uninfected cells were processed as described above for comparison.
RESULTS
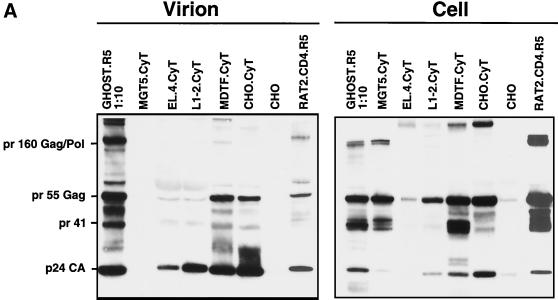
Analysis of HIV-1 assembly and release in Mus musculus, M. dunni, and hamster cell lines. Our initial study demonstrated a block to HIV-1 assembly and release in MGT5.CyT, a derivative of the murine fibroblast cell line 3T3 that expresses hu-CD4, hu-CCR5, and hu-cyclin T1 and contains an LTR-EGFP cassette (28). To determine how generalized this phenomenon is and with the hope of identifying rodent cell lines that might be more permissive for HIV-1 replication, we tested several additional rodent cell lines. These included the M. musculus T-cell lymphoma EL4 and B-cell lymphoma L1-2 lines, the CHO line, MDTF, and Rat2. Hu-cyclin T1-expressing derivatives of the cell lines were generated by retroviral vector transduction, except for Rat2, which, in preliminary experiments, expressed high levels of viral proteins without exogenous hu-cyclin T1 (data not shown). The cells were infected with NL4-3(VSV-G), and 3 days postinfection, viral protein in the cell lysates and virions pelleted from the culture medium were processed for immunoblot analysis. L1-2, MDTF, and Rat2 cell lysates were found to contain readily detectable Pr55gag but relatively low levels of processed p24gag compared to infected GHOST.R5 control cells (Fig. 1A). The block to processing was not as pronounced in these cell lines as in MGT5.CyT where p24gag was barely detectable, but processing was clearly reduced from that of the human cell controls. Interestingly, the CHO.CD4.R5.CyT supported processing with an efficiency comparable to that of the human cells as judged by the Pr55gag/p24gag ratio. Gag levels in EL4 were much lower than in the other cell lines, perhaps reflecting rapid degradation or release of viral proteins. Some variability in Pr160gag-pol, the Gag-Pol precursor, was also apparent. In the MDTF and CHO derivatives, Pr160gag-pol seemed to be replaced by a higher-molecular-weight band (Fig. 1A, right panel). The composition of this protein is unknown, but it could be a modified form of the Gag-Pol precursor.
FIG. 1.
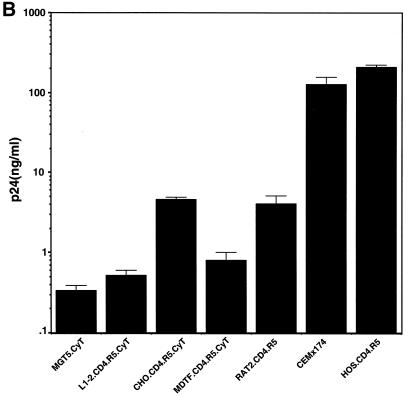
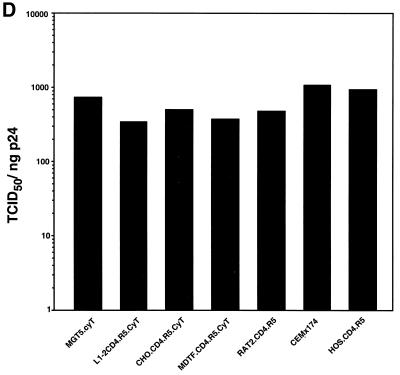
Processing ability of various rodent cell lines. M. musculus (MGT5.CyT, EL4.CyT, and L1-2.CyT), M. dunni (MDTF.CyT), hamster (CHO.CyT), or rat (Rat2.CD4.R5) cell lines with or without transfected hu-cyclin T1 were infected with NL4-3(VSV-G) at an MOI of 1. (A) Immunoblot analysis of infected cell lysates and virions pelleted from supernatant with anti-HIV serum. (B) Virion production of the infected cell lines. (C) Infectious titer of virus produced by the infected cell lines. Results are the average of triplicates. (D) Calculated ratio of TCID50 to supernatant p24gag from panels B and C.
Analysis of the viral protein pelleted from the culture supernatants of the infected cells showed the presence of p24gag and some partially processed Gag, suggesting that each of the cell lines had released at least low levels of virions (Fig. 1A). Quantitation of the supernatant p24gag showed that all of the rodent cells produced considerably fewer virions than the two control human cell lines. The best rodent producer cell lines were the CHO and Rat2 derivatives, which produced about 40-fold less p24gag than the two human cell lines (Fig. 1B). The 3T3-, L1-2-, and MDTF-derived cell lines produced about 10- to 20-fold less p24gag than the hamster or rat cells. Virus titers, as measured by TCID50 limiting dilution analysis, were consistent with the relative p24gag levels. CHO and Rat2 produced the highest virus titers (Fig. 1C), although these were still nearly 100-fold below those of the two human cell lines. The titer of virus produced by the 3T3, L1-2, and MDTF derivatives was another 10-fold lower. We previously reported that the infectivity, defined as TCID50 per nanogram of p24gag, of the virus produced by MGT5.CyT was only slightly reduced from that produced by GHOST.R5 (28). Consistent with our earlier findings, the infectivity of virus produced by the additional rodent cell lines was only two- to threefold reduced from that produced by the two human cell lines (Fig. 1D). Thus, the rodent cell lines produced much less virus than human cells, but that which was released was nearly as infectious.
Electron microscopic analysis of infected MDTF and CHO-derived cell lines.
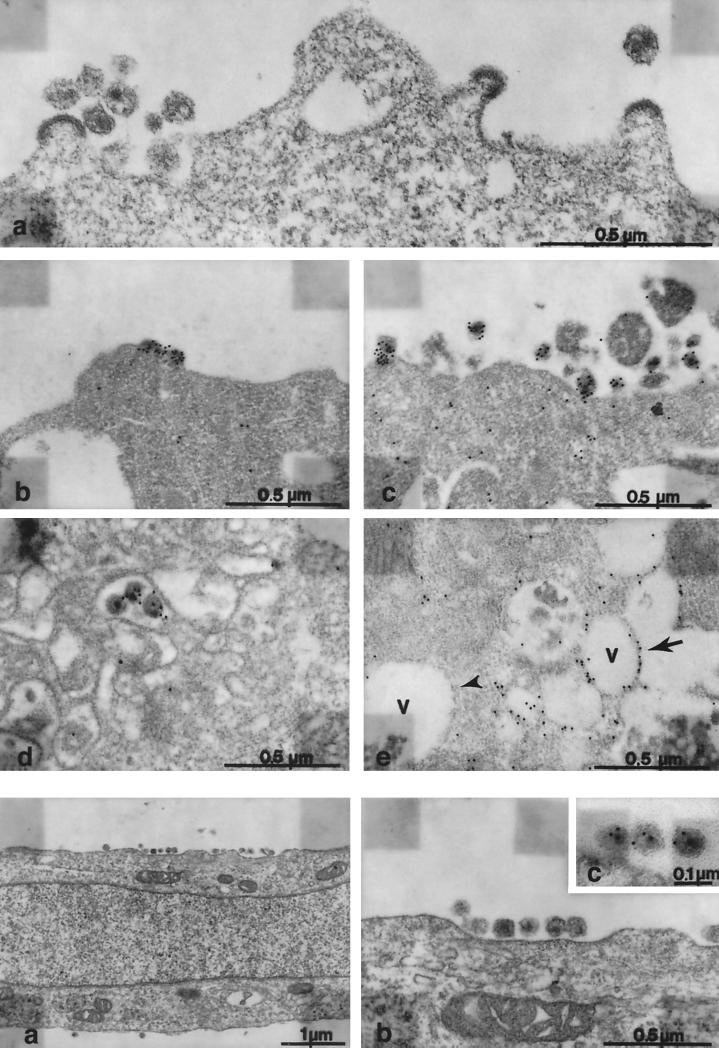
Electron microscopic analysis of infected CHO.CD4.R5.CyT showed budding structures at the plasma membrane and abundant extracellular immature and mature HIV-1 particles of normal morphology (Fig. 2a [upper]). The extracellular particles (Fig. 2b and c [upper]) but not the cytoplasm were labeled by immunogold staining with anti-CA antibody. Some labeled intracellular particles appeared to have budded into the endoplasmic reticulum (ER) cisternae (Fig. 2d). Some of the cytoplasmic vacuoles were labeled, while others in the immediate vicinity remained negative (Fig. 2e). The ER cisternae appeared enlarged and cytoplasmic vesiculation was apparent late in infection compared to uninfected controls (not shown), while the Golgi, mitochondria, and lysosomal structures appeared to be unaffected and the number of multivesicular bodies and lamellar lysosomes was normal (not shown). The cells did not have the electron-dense thickened ER membranes present in infected MGT5.CyT (28). Uninfected cells and infected cells stained with irrelevant serum contained very few gold particles, demonstrating the specificity of the staining (data not shown). In infected MDTF.CD4.R5.CyT cells, viral particles were present but less abundant than in the CHO-derived cells. Extracellular immature and mature particles were observed (Fig. 2a and b [lower]) and these were labeled by anti-CA antibody (Fig. 2c [lower]). Labeled intracellular structures were not detected, and no morphologic alterations were observed in comparison to uninfected cells, perhaps as a result of the reduced virus expression.
FIG. 2.
Electron microscopy of HIV-1 infected CHO and MDTF derivatives. (Upper panels) (a) Ultrathin sections were processed by glutaraldehyde-osmium fixation and ERL embedding. Budding structures (a) and extracellular HIV-1 particles are present. (b and c) Immunoelectron microscopy with anti-CA antibody of HIV-1-infected CHO derivative shows labeled budding structures (b) and adjacent extracellular particles (c). Gold particles are scarce in the cytoplasm and nearly absent in the extracellular space. (d and e) Gold-labeled HIV-like particles in the ER cisternae. Some cytoplasmic vacuoles (V) were labeled (arrow), while adjacent vacuoles remained unlabeled (arrowhead). (Lower panels) (a) Ultrathin sections of infected, fixed, and embedded MDTF.CD4.R5.CyT cells at low magnification showed virus particles at the plasma membrane. Morphological alterations were not observed. (b) Higher magnification revealed mature extracellular HIV-1 particles adjacent to the plasma membrane. (c) The extracellular particles were immunolabeled.
Inefficient virus replication in rodent cell lines.
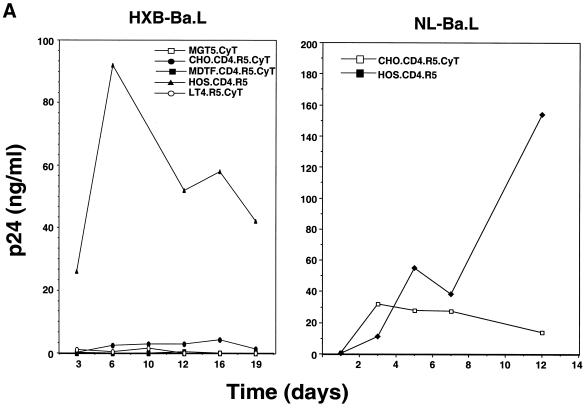
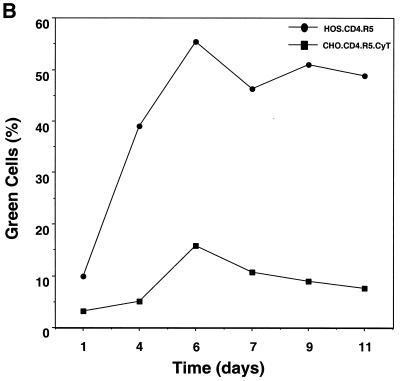
In light of the ability of some of the rodent cell lines to produce significant quantities of infectious virus, we tested whether they would support spreading infection by M-tropic HIV-1. HOS.CD4.R5 cells infected with either HXB.BaL or NL.BaL generated high levels of p24gag (Fig. 3A). MGT5.CyT, MDTF.CD4.R5.CyT, and L1-2.CD4.R5.CyT did not support detectable virus replication. A low level of virus replication was detected in CHO.CD4.R5.CyT, as reflected by a small increase in p24gag up to day 3, after which the level remained fairly constant over 9 days. This finding was confirmed by infecting the cells with HIV.YU2.GFP. On day 6, 15% of cells had become infected as detected by flow cytometry (Fig. 3B). The proportion of infected cells decreased over the next 5 days, suggesting that limited virus spread had occurred but that the infected cells were lost from the culture over time.
FIG. 3.
CHO.CD4.R5.CyT but not other rodent cells support low-level HIV-1 replication. (A) MGT5.CyT, CHO.CD4.R5.CyT, MDTF.CD4.R5.CyT, LT4.R5.CyT, and HOS.CD4.R5 cells were infected with HXB.BaL and CHO.CD4.R5.CyT and HOS.CD4.R5 cells were also infected with NL-BaL at an MOI of 0.05, and p24gag production was measured over time. (B) CHO.CD4.R5.CyT and HOS.CD4.R5 were infected with HIV.YU2.GFP at an MOI of 0.05, and the percentage of green cells was scored by fluorescence-activated cell sorting analysis.
Efficient coreceptor function in CHO.CD4.R5.CyT.
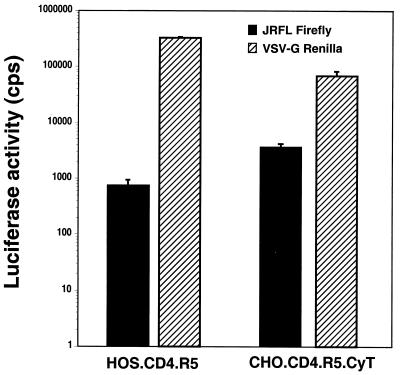
We previously found that entry of JR.FL-pseudotyped luciferase virus into MGT5.CyT was 30% as efficient as entry into HOS.CD4.R5 cells (28) and suggested that coreceptor function in 3T3 cells was partially reduced. To determine whether a similar phenomenon might limit replication of M-tropic HIV-1 in the CHO-derived cell line, a dual luciferase reporter system was used. JR.FL pseudotyped firefly luciferase reporter virus and VSV-G pseudotyped Renilla luciferase reporter viruses were mixed at a 10:1 ratio, and the mixture was used to infect HOS.CD4.R5 and CHO.CD4.R5.CyT cells. The VSV-G pseudotype served to control for postentry effects. Three days later, firefly and Renilla luciferase levels were measured using commercial reagents that separately detect each enzyme. The results showed that the JR.FL pseudotype generated about threefold more firefly luciferase activity in the hamster cells than in the HOS.CD4.R5 cells (Fig. 4). The VSV-G pseudotype generated about threefold more activity in HOS.CD4.R5 than in CHO.CD4.R5.CyT. Thus, the hamster cells were nearly as efficient as the human cells with respect to the early events of the virus life cycle (reverse transcription, integration, and expression). After normalization to VSV-G-mediated entry, JR.FL entry was found to be about ninefold more efficient in the hamster than in the human cells, probably as a result of increased cell surface CCR5 (data not shown). Thus, unlike in 3T3-derived cell lines, coreceptor function is efficient in the hamster cell line.
FIG. 4.
CHO cells are competent for coreceptor function but fail to support efficient virus replication. CHO.CD4.R5.CyT and HOS.CD4.R5 cells were infected with JR.FL pseudotyped firefly luciferase or VSV-G pseudotyped Renilla luciferase single-cycle reporter viruses. Luciferase activity was measured in cell lysates 3 days postinfection. The results are the averages of triplicates and are representative of three experiments.
Virus production in mouse-human heterokaryons.
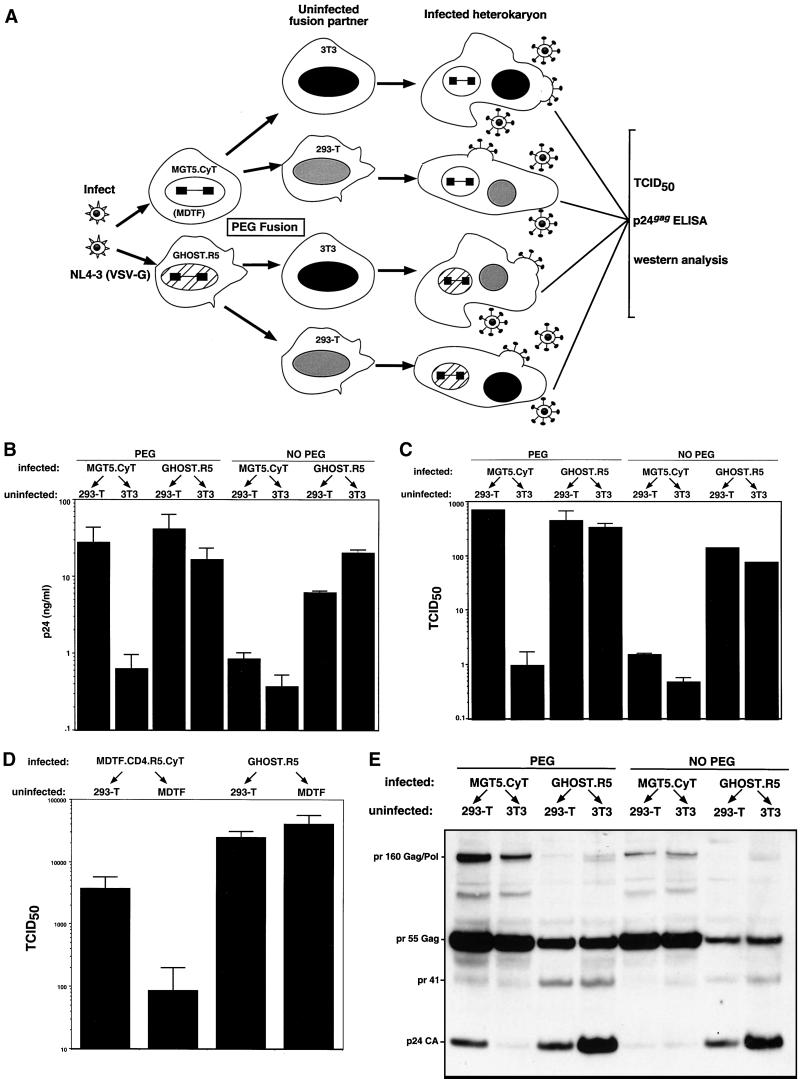
The inability of murine 3T3 cells to assemble and release virions could result from the absence of a required cofactor or the presence of an inhibitory activity. To distinguish between these possibilities, human-mouse heterokaryons were formed and tested for their ability to release virus, as diagrammed in Fig. 5A. The cells were infected with NL4-3(VSV-G) at a high MOI, the input virus was thoroughly removed the next day, and then 2 days postinfection the cells were fused to uninfected human 293T or 3T3 cells. To test for inhibitory activity in the murine cells, reciprocal heterokaryons were prepared in which GHOST.R5 was infected and then fused to 293T or 3T3. Virus assembly and release were evaluated by measuring supernatant p24gag and infectious titer and by immunoblot analysis of viral protein in the cell lysates.
FIG. 5.
The assembly defect in 3T3 cells is complemented by fusion to human cells. (A) Strategy for testing HIV assembly and production in mouse-human heterokaryons. MGT5.CyT, GHOST.R5, and MDTF.CD4.R5.CyT cells were infected at an MOI of 1 with NL4-3(VSV-G), washed three times, and fused 3 days later to 293T, 3T3, or MDTF cells. After 24 h postfusion, the cells were lysed for immunoblot analysis, and virions were collected for p24gag and TCID50 measurement. To control for input virus contamination, mock fusions in which PEG was omitted were analyzed in parallel. (B) p24gag concentration in heterokaryon supernatants. (C) TCID50 measurement of virus in heterokaryon supernatants. (D) TCID50 measurement of virus in M. dunni-human heterokaryon supernatants. (E) Immunoblot analysis of heterokaryon lysates using anti-HIV serum.
Heterokaryons formed by the fusion of infected MGT5.CyT to 293T produced 12-fold more supernatant p24gag than control heterokaryons formed by fusion of infected MGT5.CyT to 3T3 (Fig. 5B). A potential artifactual explanation for this result was that the increase in virus release was caused by carryover of input NL4-3(VSV-G) that had remained in the heterokaryon culture despite extensive washing 24 h earlier. Carryover would have led to direct infection of the human cell fusion partner, resulting in increased virus production independent of heterokaryon formation. To test for the contribution of input virus carryover, the infected and uninfected cells were cocultivated but the PEG was omitted. Omission of the PEG prevented the increase in p24gag (Fig. 5B), arguing against carryover as an explanation for the increase in p24gag release and further demonstrating the requirement for cell-cell fusion. We also considered whether the PEG itself had stimulated virus release in the heterokaryons; however, if this had occurred, increased virus release would have been apparent in the mouse-mouse cell fusions. To test for an inhibitory activity in the murine cell line, infected GHOST.R5 was fused to 3T3 and to 293T. These heterokaryons released large amounts of p24gag, and these levels did not differ between the two fusion partners (Fig. 5B). Omission of the PEG did not affect the p24gag levels. As determined by counting mouse and human nuclei following staining with Hoechst dye, the efficiency of heterotypic fusion was 70 to 80%. Thus, the contribution to virus production by unfused human cells was small.
Measurement of the virus titers showed more pronounced differences. Heterokaryons formed by the fusion of infected MGT5.CyT to 293T released virus with 700-fold higher TCID50 than those in which infected MGT5.CyT was fused to 3T3 (Fig. 5C). Heterokaryons composed of infected GHOST.R5 fused to 3T3 produced virus with titer as high as that produced by GHOST.R5 fused to 293T. Omitting the PEG from the infected mouse-human heterokaryons prevented the increase in TCID50 and had no effect on the infected human-mouse heterokaryon. Taken together, the p24gag and TCID50 data suggested that the human cell line contained a species-specific factor that complemented the assembly defect in the mouse cells. The results did not support the presence of an inhibitor in the mouse cells, although it is conceivable that such an inhibitor was present but that its activity was suppressed by a factor in the human cells.
To test whether the boost in virion production in the heterokaryons could be detected in murine cells other than 3T3, heterokaryons were formed with MDTF.CD4.R5.CyT. Infected MDTF.CD4.R5.CyT fused to 293T produced virus titers about 40-fold greater than those fused to MDTF. (Fig. 5D). The increase was not as pronounced as that in the 3T3 cells, not because of reduced complementation but because these cells are not as tightly blocked for virion assembly and release (Fig. 1A). Fusion of infected GHOST.R5 to MDTF did not reduce virus production, arguing against an inhibitory activity in the mouse cell line.
The biochemical basis for the increase in virus release in the interspecies heterokaryons was studied by immunoblot analysis of the viral structural proteins. Infected heterokaryons were lysed 24 h postfusion, and the viral proteins in the lysates were detected using AIDS patient serum. Immunoblot analysis showed that the heterokaryons contained Pr160gag-pol, Pr55gag, fully processed p24gag, and the MA-CA processing intermediate pr41gag (Fig. 5E). Heterokaryons formed by fusing infected MGT5.CyT to 293T contained threefold more p24gag than those fused to 3T3. Heterokaryons formed by fusing infected GHOST.R5 to 3T3 contained about twofold more p24gag than those fused to 293T. Thus, the human cells stimulated processing of the Gag polyprotein and the murine cells did not inhibit processing. Fusion of the infected MGT5.CyT to 293T also caused a small increase in the overall amount of virion protein in the cells. This is likely due to the presence of additional activating factors in the human cells, as described below. Consistent with the viral release measurements, omission of the PEG blocked the increase in p24gag levels in the infected mouse-human heterokaryons. A small increase in virion protein production was reproducibly detected in infected GHOST.R5 fused to 3T3. The increase did not require fusion, as it occurred without PEG. The basis for the increase is not clear, but it could be due to a soluble factor released from 3T3 that modestly stimulates HIV-1 assembly.
Reporter gene expression in mouse-human heterokaryons.
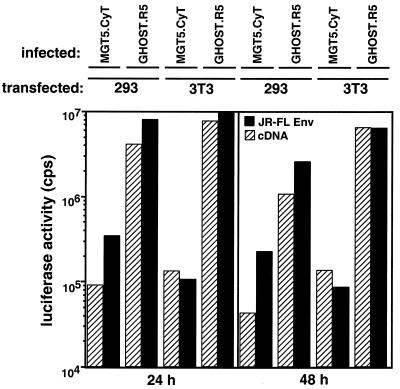
The increase in virion release that occurred in the infected MGT5.CyT upon fusion to 293T could have been due to more efficient virus assembly, to increased proviral transcription, to increased synthesis of virion components or to a combination of these factors. To gauge the contribution of increased proviral expression, MGT5.CyT or GHOST.R5 cells were infected with reporter virus containing the firefly luciferase gene in place of nef. The infected cells were then fused to 3T3 or 293T cells that had been previously transfected with JR.FL Env expression vector or control pcDNAI/amp plasmid. Mixing of the infected MGT5.CyT or GHOST.R5 cells that express CD4-CCR5 with transfected 293T or 3T3 cells that express JR.FL Env leads to fusion of the two cell types, as observed by the massive syncytia that form 24 h later (data not shown). Env-mediated fusion was used to restrict fusion to heterotypic rather than homotypic reactions. To control for nonspecific effects, 3T3 and 293T were transfected with pcDNAI/amp in place of JR.FL Env vector to prevent fusion. Luciferase activity in the heterokaryons was measured 24 and 48 h later. The results showed that reporter gene expression from proviruses integrated in the murine genome increased threefold upon fusion to 293T (Fig. 6). A similar increase occurred when reporter virus-infected GHOST.R5 was fused to 293T. The increase could have been due to activating factors provided by the 293T that are limiting in 3T3 and GHOST.R5. Alternatively, the increase may have been caused by an increased synthetic capacity of the larger cells that form during fusion. Fusion of the infected GHOST.R5 cells to mouse cells did not diminish luciferase activity, arguing against a murine inhibitor of proviral transcription. The MGT5.CyT cells expressed 30-fold less luciferase activity than the GHOST.R5, but this comparison probably reflects differences in luciferase enzyme detection sensitivity between the two cell types rather than absolute differences in provirus expression. Thus, the increase in virus production in the heterokaryons could not be attributed to increased proviral transcription but was the result of complementation of the assembly defect.
FIG. 6.
Fusing human cells to infected murine cells that express hu-cyclin T1 increases LTR transactivation. Murine or human cells were infected with luciferase reporter virus. Three days postinfection, the cells were cocultivated with JR.FL Env-expressing or control cells for an additional 24 or 48 h to allow Env-mediated fusion between the two cell types. Luciferase activity in the cell lysates was then measured. Results are the averages of triplicates and are representative of three experiments.
DISCUSSION
Murine 3T3 cells expressing hu-CD4, hu-CCR5, and hu-cyclin T1 fail to support HIV-1 replication as a result of the failure of the cells to support virus assembly and release (15, 28). Here we show that the defect can be complemented by fusing the infected mouse cells to uninfected human cells. Upon their formation, the heterokaryons rapidly released large quantities of infectious virus. The increase was not due to augmented proviral transcription or to greater synthesis of virion components in the heterokaryons. Reciprocal heterokaryons in which infected human cells were fused to mouse cells continued to release large amounts of virus, arguing against an inhibitor of HIV-1 assembly or release in the mouse cells. The detection of an inhibitor in the mouse cells could have been missed if the fusion efficiency with the human cells had been low; however, fusion efficiency was at least 70%, suggesting that an inhibitor would have been detected. It remains possible that the murine cells contained an inhibitor that was diluted in the human cytoplasm upon heterokaryon formation and therefore inactive; however, this seems unlikely, as fusion would have diluted the murine cytoplasm about twofold and the increase in virus titer in the mouse-human heterokaryon was 700-fold. Taken together, the findings suggest that the human cells contain an assembly cofactor that is nonfunctional, inefficient, or expressed at low levels in the various rodent cell lines.
To determine whether the block to assembly first described for 3T3 cells was a generalized property of rodent cells, additional cell types and species were studied. As with the 3T3 cells, all of the rodent cell lines synthesized abundant virus protein upon infection but none supported a high level of virus replication. The rodent cell lines produced 30- to 500-fold fewer virions than comparable human cell lines, and with the exception of the CHO-derived cell line, each had reduced levels of Gag processing. Also consistent with earlier findings (28), the infectivity of the virus released by the rodent cells was only slightly reduced (two- to threefold) from that of the virus produced by human cells. Interestingly, the CHO cells supported a low-level HIV-1 replication.
It is conceivable that some of the differences in the ability of the various rodent cell lines to assemble and release virus were due to cell type differences and not species-specific differences. However, human cells that express CD4 and a coreceptor, regardless of cell type, are generally permissive for virus replication (1, 3, 8). Postentry effects, with a few exceptions (30, 31), are not a major determinant of virus replication. An inability to support virus assembly or release has not been described for any activated human cell type. Thus, it seems likely that the blocks to replication described here were primarily due to species-specific differences.
Following submission of this article, Bieniasz and Cullen (4) reported findings consistent with those presented here obtained with a panel of rodent cell lines. One notable difference between the two studies was that they found that virus produced by the rodent cell lines was about 50-fold lower than that produced by human cells while we noted only a two- to threefold reduction. The reason for this difference is not clear but could be related to the use of different methods for measuring infectious titer. In that study, infectious titer was determined by quantitating infectivity of the virus on CHO cells, while human CEM T cells were used here. It is possible that virus produced by rodent cells more readily infects human than rodent cells, although the mechanism by which this could occur is not obvious.
The relationship of intracellular Gag processing to virus assembly and release is not clear. The Rat2-derived cell line released an amount of virions similar to that released by the infected CHO-derived cells but by immunoblot analysis appeared not to support Gag processing. Thus, the extent of intracellular processing does not always reflect the ability of a cell to produce virus. HIV-1 assembles at the cell membrane, and Gag processing appears to initiate at or after virus budding (13, 20). Intracellular p24gag presumably results from several sources: virions that are in the process of budding, released virions that have attached to the cell surface, virions in which the proteinase was prematurely activated, and virions that have inappropriately budded into intracellular compartments. It is possible that in Rat2 cells, nonproductive pathways such as inappropriate targeting are minimized and that this results in more efficient virus release and less accumulation of intracellular p24gag. Also contributing to the relatively efficient virion release in Rat2 is the overall increased abundance of Gag in the cells.
Infected 3T3-derived cells contained abundant dense material in intracytoplasmic vesicles (28). Similar material was not present in infected CHO- or MDTF-derived cells. The absence of this material may reflect more efficient assembly and Gag processing in these two cell lines. This would be consistent with the greater abundance of budding particles at the surfaces of cells of both lines apparent by electron microscopy analysis. The budding structures on the surfaces of the cells appeared to have normal morphology, consistent with their high infectivity. Some intracytoplasmic vesicles were labeled by immunogold staining for CA, suggesting that intracellular trapping or mistargeting of Gag may also occur in CHO and MDTF but to a lesser extent than in 3T3. Whether the assembly defect in MDTF has the same molecular basis as that in 3T3 is not clear; however, the defect appeared to be caused by a missing cofactor, since it can be complemented in heterokaryons.
The nature of the species-specific factor that facilitates HIV-1 assembly and release that was detected in the heterokaryon analysis is a matter of speculation. The accumulation of Gag in inappropriate intracellular vesicles (28) could be caused by mistargeting of Pr55gag. Plasma membrane targeting of Gag is mediated by the MA portion of Pr55gag (13). MA contains an amino-terminal myristate and a basic patch that appear to participate in cell membrane association (13, 43, 48). It is conceivable that in the murine cells, the MA targeting does not function appropriately. As a result, only the Gag molecules that traffic fortuitously to the cell membrane are assembled and released. In such a model the cofactor could be a chaperone that facilitates intracellular transport (25), a cytoskeleton component on which virus travels, or a facilitator of protein folding (26, 39). Alternatively, the factor could be a lipid component of the cell membrane that interacts with MA (18). Whether the factor is controlled by a single or multiple genes is not clear. Analysis of human-mouse somatic-cell hybrids may be useful for determining this.
Establishing a rodent model for HIV-1 replication will require overcoming the block to HIV-1 assembly. This could be accomplished by identifying the genes that complement the assembly defect in the murine cells or by altering the virus such that appropriate membrane targeting of the Gag precursor is restored in the mouse cells. Repeated attempts to adapt HIV-1 in culture to replicate in the rodent cell lines have not resulted in selection for more fit virus variants (data not shown), suggesting that the required alteration is not minor. The ability of the various rodent cells to release significant quantities of infectious virus and, in one example, to support a low level of virus replication suggests that a such model may be achievable.
ACKNOWLEDGMENTS
We thank Susan Hedrick for technical assistance, Douglas Richman and Richard Kornbluth for anti-HIV-1 serum, Julie Overbaugh for MDTF, Ge Wei for NL.BaL, Mark Muesing for HIV.YU2.GFP, and Lynn Artale for administrative assistance.
The work was funded by NIH grants AI43252 and CA7214, the Advanced Technology Program of NIST, Elisabeth Glaser Pediatric AIDS Foundation grant 77328 to R.M., UCSD CFAR developmental grant AI36214 to R.M., and the J. B. Pendleton Charitable Trust. N.R.L. is an Elizabeth Glaser Scientist of the Elizabeth Glaser Pediatric AIDS Foundation.
REFERENCES
- 1.Alkhatib G, Broder C C, Berger E A. Cell type-specific fusion cofactors determine human immunodeficiency virus type 1 tropism for T-cell lines versus primary macrophages. J Virol. 1996;70:5487–5494. doi: 10.1128/jvi.70.8.5487-5494.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso A, Derse D, Peterlin B M. Human chromosome 12 is required for optimal interactions between Tat and TAR of human immunodeficiency virus type 1 in rodent cells. J Virol. 1992;66:4617–4621. doi: 10.1128/jvi.66.7.4617-4621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger E A, Murphy P M, Farber J M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 4.Bieniasz P D, Cullen B R. Multiple blocks to human immunodeficiency virus type 1 replication in rodent cells. J Virol. 2000;74:9868–9877. doi: 10.1128/jvi.74.21.9868-9877.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieniasz P D, Grdina T A, Bogerd H P, Cullen B R. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 1998;17:7056–7065. doi: 10.1093/emboj/17.23.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browning J, Horner J W, Pettoello-Mantovani M, Raker C, Yurasov S, DePinho R A, Goldstein H. Mice transgenic for human CD4 and CCR5 are susceptible to HIV infection. Proc Natl Acad Sci USA. 1997;94:14637–14641. doi: 10.1073/pnas.94.26.14637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cecilia D, KewalRamani V N, O'Leary J, Volsky B, Nyambi P, Burda S, Xu S, Littman D R, Zolla-Pazner S. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J Virol. 1998;72:6988–6996. doi: 10.1128/jvi.72.9.6988-6996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chesebro B, Wehrly K, Maury W. Differential expression in human and mouse cells of human immunodeficiency virus pseudotyped by murine retroviruses. J Virol. 1990;64:4553–4557. doi: 10.1128/jvi.64.9.4553-4557.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho S, Kindt T J, Zhao T M, Sawasdikosol S, Hague B F. Replication of HIV type 1 in rabbit cell lines is not limited by deficiencies in tat, rev, or long terminal repeat function. AIDS Res Hum Retrovir. 1995;11:1487–1493. doi: 10.1089/aid.1995.11.1487. [DOI] [PubMed] [Google Scholar]
- 10.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:936–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 11.Deng D, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, DiMarzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 12.Dunn C S, Mehtali M, Houdebine L M, Gut J P, Kirn A, Aubertin A M. Human immunodeficiency virus type 1 infection of human CD4-transgenic rabbits. J Gen Virol. 1995;76:1327–1336. doi: 10.1099/0022-1317-76-6-1327. [DOI] [PubMed] [Google Scholar]
- 13.Freed E O. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 14.Garber E M, Jones K A. HIV-1 Tat: coping with negative elongation factors. Curr Opin Immunol. 1999;11:460–465. doi: 10.1016/S0952-7915(99)80077-6. [DOI] [PubMed] [Google Scholar]
- 15.Garber E M, Wei P, KewalRamani V N, Mayall T P, Herrmann C H, Rice A P, Littman D R, Jones K A. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998;12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gold M, Yang X, Herrmann C, Rice A. PITALRE, the catalytic subunit of TAK, is required for human immunodeficiency virus Tat transactivation in vivo. J Virol. 1997;72:4448–4453. doi: 10.1128/jvi.72.5.4448-4453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hart C E, Ou C Y, Galphin J C, Moore J, Bacheler L T, Wasmuth J J, Petteway S R J, Schochetman G. Human chromosome 12 is required for elevated HIV-1 expression in human-hamster hybrid cells. Science. 1989;246:488–491. doi: 10.1126/science.2683071. [DOI] [PubMed] [Google Scholar]
- 18.Hermida-Matsumoto L, Resh M D. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J Virol. 2000;74:8670–8679. doi: 10.1128/jvi.74.18.8670-8679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jutila A M, Bargatze R F, Kurk S, Warnock R A, Ehsani N, Watson S R, Walcheck B. Cell surface P- and E-selectin support shear-dependent rolling of bovine gamma/delta T cells. J Immunol. 1994;153:3917–3928. [PubMed] [Google Scholar]
- 20.Krausslich H G, Welker R. Intracellular transport of retroviral capsid components. Curr Top Microbiol Immunol. 1996;214:25–63. doi: 10.1007/978-3-642-80145-7_2. [DOI] [PubMed] [Google Scholar]
- 21.Kulaga H, Folks T M, Rutledge R, Kindt T J. Infection of rabbit T-cell and macrophage lines with human immunodeficiency virus. Proc Natl Acad Sci USA. 1988;85:4455–4459. doi: 10.1073/pnas.85.12.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwak Y T, Ivanov D, Guo J, Nee E, Gaynor R B. Role of the human and murine cyclin T proteins in regulating HIV-1 tat-activation. J Mol Biol. 1999;288:57–69. doi: 10.1006/jmbi.1999.2664. [DOI] [PubMed] [Google Scholar]
- 23.Landau N R, Page K A, Littman D R. Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range. J Virol. 1991;65:162–169. doi: 10.1128/jvi.65.1.162-169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landau N R, Warton M, Littman D R. The envelope glycoprotein of the human immunodeficiency virus binds to the immunoglobulin-like domain of CD4. Nature. 1988;334:159–162. doi: 10.1038/334159a0. [DOI] [PubMed] [Google Scholar]
- 25.Lingappa J R, Martin R L, Wong M L, Ganem D, Welch W J, Lingappa V R. A eukaryotic cytosolic chaperonin is associated with a high molecular weight intermediate in the assembly of hepatitis B virus capsid, a multimeric particle. J Cell Biol. 1994;125:99–111. doi: 10.1083/jcb.125.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu B, Dai R, Tian C J, Dawson L, Gorelick R, Yu X F. Interaction of the human immunodeficiency virus type 1 nucleocapsid with actin. J Virol. 1999;73:2901–2908. doi: 10.1128/jvi.73.4.2901-2908.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mancebo H S, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, Flores O. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mariani R, Rutter G, Harris M E, Hope T J, Krausslich H G, Landau N R. A block to human immunodeficiency virus type 1 assembly in murine cells. J Virol. 2000;74:3859–3870. doi: 10.1128/jvi.74.8.3859-3870.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mori K, Ringler D J, Desrosiers R C. Restricted replication of simian immunodeficiency virus strain 239 in macrophages is determined by env but is not due to restricted entry. J Virol. 1993;67:2807–2814. doi: 10.1128/jvi.67.5.2807-2814.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moriuchi H, Moriuchi M, Arthos J, Hoxie J, Fauci A S. Promonocytic U937 subclones expressing CD4 and CXCR4 are resistant to infection with and cell-to-cell fusion by T-cell-tropic human immunodeficiency virus type 1. J Virol. 1997;71:9664–9671. doi: 10.1128/jvi.71.12.9664-9671.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrow W J, Wharton M, Lau D, Levy J A. Small animals are not susceptible to human immunodeficiency virus infection. J Gen Virol. 1987;68:2253–2257. doi: 10.1099/0022-1317-68-8-2253. [DOI] [PubMed] [Google Scholar]
- 33.Newman G R, Jasani B, Williams E D. A simple post-embedding system for the rapid demonstration of tissue antigens under the electron microscope. Histochem J. 1983;15:543–555. doi: 10.1007/BF01954145. [DOI] [PubMed] [Google Scholar]
- 34.Newstein M, Stanbridge E J, Casey G, Shank P R. Human chromosome 12 encodes a species-specific factor which increases human immunodeficiency virus type 1 tat-mediated trans activation in rodent cells. J Virol. 1990;64:4565–4567. doi: 10.1128/jvi.64.9.4565-4567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Onishi M, Kinoshita S, Morikawa Y, Shibuya A, Phillips J, Lanier L L, Gorman D M, Nolan G P, Miyajima A, Kitamura T. Applications of retrovirus-mediated expression cloning. Exp Hematol. 1996;24:324–329. [PubMed] [Google Scholar]
- 36.Page K A, Landau N R, Littman D R. Construction and use of a human immunodeficiency virus vector for analysis of virus infectivity. J Virol. 1990;64:5270–5276. doi: 10.1128/jvi.64.11.5270-5276.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puck T T. Genetics of somatic mammalian cells. III. Long-term cultivation of eupoid cells from human and animal subjects. J Exp Med. 1958;108:945–956. doi: 10.1084/jem.108.6.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ralph P, Nakoinz I. Inhibitory effects of lectins and lymphocyte mitogens on murine lymphomas and myelomas. J Natl Cancer Inst. 1973;51:883–890. doi: 10.1093/jnci/51.3.883. [DOI] [PubMed] [Google Scholar]
- 39.Rey O, Canon J, Krogstad P. HIV-1 Gag protein associates with F-actin present in microfilaments. Virology. 1996;220:530–534. doi: 10.1006/viro.1996.0343. [DOI] [PubMed] [Google Scholar]
- 40.Sawada S, Gowrishankar K, Kitamura R, Suzuki M, Suzuki G, Tahara S, Koito A. Disturbed CD4+ T cell homeostasis and in vitro HIV-1 susceptibility in transgenic mice expressing T cell line-tropic HIV-1 receptors. J Exp Med. 1998;187:1439–1449. doi: 10.1084/jem.187.9.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seigel L J, Ratner L, Josephs S F, Derse D, Feinberg M B, Reyes G R, O'Brien S J, Wong-Staal F. Transactivation induced by human T-lymphotropic virus type III (HTLV III) maps to a viral sequence encoding 58 amino acids and lacks tissue specificity. Virology. 1986;148:226–231. doi: 10.1016/0042-6822(86)90419-8. [DOI] [PubMed] [Google Scholar]
- 42.Sparkman D R, White C L., III A simple apparatus for processing large numbers of specimens for colloidal gold immunoelectron microscopy: application to paired helical filaments of Alzheimer's disease. J Electron Microsc Techniques. 1989;13:152–153. doi: 10.1002/jemt.1060130207. [DOI] [PubMed] [Google Scholar]
- 43.Spearman P, Horton R, Ratner L, Kuli-Zade I. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J Virol. 1997;71:6582–6592. doi: 10.1128/jvi.71.9.6582-6592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Speck R F, Penn M L, Wimmer J, Esser U, Hague B F, Kindt T J, Atchison R E, Goldsmith M A. Rabbit cells expressing human CD4 and human CCR5 are highly permissive for human immunodeficiency virus type 1 infection. J Virol. 1998;72:5728–5734. doi: 10.1128/jvi.72.7.5728-5734.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spurr A. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969;26:31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- 46.Topp W C. Normal rat cell lines deficient in nuclear thymidine kinase. Virology. 1981;113:408–411. doi: 10.1016/0042-6822(81)90168-9. [DOI] [PubMed] [Google Scholar]
- 47.Wei P, Garber M E, Fang S M, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 48.Zhou W, Parent L J, Wills J W, Resh M D. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews M B, Price D H. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]