Abstract
CoQ10 (Coenzyme Q10) is an essential fat-soluble metabolite that plays a key role in cellular metabolism. A less-known function of CoQ10 is whether it may act as a plasma membrane-stabilizing agent and whether this property can affect cancer development and progression. Here, we show that CoQ10 and its biosynthetic enzyme UBIAD1 play a critical role in plasmamembrane mechanical properties that are of interest for breast cancer (BC) progression and treatment. CoQ10 and UBIAD1 increase membrane fluidity leading to increased cell stiffness in BC. Furthermore, CoQ10 and UBIAD1 states impair ECM (extracellular matrix)-mediated oncogenic signaling and reduce ferroptosis resistance in BC settings. Analyses on human patients and mouse models reveal that UBIAD1 loss is associated with BC development and progression and UBIAD1 expression in BC limits CTCs (circulating tumor cells) survival and lung metastasis formation. Overall, this study reveals that CoQ10 and UBIAD1 can be further investigated to develop therapeutic interventions to treat BC patients with poor prognosis.
Subject terms: Breast cancer, Mechanisms of disease, Cell death
The roles of CoQ10 and producing enzymes in cancer have not been well explored. Here, the authors identify that the CoQ10-biosynthetic enzyme UBIAD1 is a tumor suppressor in breast cancer. CoQ10 induced alterations in plasma membrane mechanical properties lead to increased cellular stiffness and impaired tumorigenic signaling, as well as enhanced sensitivity to ferroptosis.
Introduction
CoQ10 (Coenzyme Q10, also known as ubiquinone) represents an essential fat-soluble compound present in the plasma and inner membranes of most eukaryotic cells1,2. CoQ10 consists of a benzoquinone ring and a lipophilic isoprenoid side chain. CoQ10 plays a role in several aspects of cellular metabolism, including mitochondrial respiration and shielding membrane lipids from oxidative stress3. A less-known function of CoQ10 is its capacity to act as a membrane-stabilizing agent that alters the mechanical properties of phospholipid membranes. Indeed, the role of CoQ10 in plasma membrane homeostasis influences cholesterol-poor membranes such as in mitochondria and aerobic gram-negative bacteria4,5. However, whether increased plasma membrane CoQ10 levels affect cholesterol-dependent signaling in cholesterol-rich membranes and pathological settings is poorly understood. Such CoQ10 studies would be critical also from a clinical point of view since genetic mutations, disease, and aging can affect Q10 levels in the cells and consequently result in serious health issues. How and whether CoQ10 contents can affect cancer development and progression has thus far been poorly investigated6,7.
The biosynthesis of CoQ10 involves distinct steps: production of the aromatic group that forms the quinone head, production of the isoprene tail, attachment of the quinone head to the isoprene tail, followed with slight biochemical modifications that lead to the formation of the final CoQ10 product8,9. Several metabolic pathways contribute to CoQ10 biosynthesis by providing precursors that generate CoQ10 molecules: the quinone head derives from tyrosine catabolism in higher eukaryotic cells, and the isoprene tail derives from acetyl-CoA in the mevalonate pathway10. There are different rate-limiting steps to synthesizing CoQ10. The first one is the availability of isopentenyl pyrophosphate (IPP) since the production of the quinone head comes from an abundant source of tyrosine, and the second is the condensation of the isoprenoid tail to the quinone group mediated by UbiA enzymes11. In mammals, two UbiA enzymes act as polyprenyl-4-hydroxybenzoate transferase, called COQ2 and UBIAD1, located in the mitochondria and Golgi/ER apparatus. Indeed, mitochondria and Golgi/ER apparatus are proposed sites for CoQ10 synthesis in mammals12,13.
Ferroptosis is an iron-dependent form of programmed cell death mediated by the accumulation of lipid reactive oxygen species (ROS), inducing oxidative damage and disrupting antioxidant defense balance14. Cancer cells are more prone to undergo ferroptosis, as they have a more vigorous metabolism and accumulate more ROS in comparison to normal cells15. Recent studies reveal that ferroptosis induction holds great promise for cancer therapy, especially for therapeutic-resistant tumors or those with high metastasizing tendencies15. However, cancer cells can also achieve lower susceptibility to ferroptosis through additional mechanisms like the glutathione (GSH) system, CoQ10 system, and thioredoxin (TXN) system detoxify lipid hydroperoxides and suppress ferroptosis16. Induction of ferroptosis is an explorable therapeutic strategy as triple-negative breast cancer (TNBC) is an iron-rich and lipid-rich tumor17,18. Recently it has been shown that TNBCs display ferroptosis heterogeneity, and the luminal androgen receptor (LAR) subtype of TNBC shows hypersensitive to ferroptosis inducers proposing that the combination of GPX4 inhibitors and immune checkpoint inhibitors could represent a treatment strategy for LAR TNBCs19. Resistance to ferroptosis is also associated with the development of some refractory cancers. Ferroptosis is a process regulated by multiple metabolic pathways, but a clear picture of ferroptosis in TNBC remains poorly characterized. In this context we investigate whether UBIAD1 and CoQ10 affect the survival and aggressiveness of the TNBC subtype that strongly relies on survival and invasion signaling and ferroptosis protection for the progression of the disease. Overall, we wonder whether UBIAD1 and CoQ10 dysfunction cause the acquisition of aggressive phenotypes in BC being the function of the mevalonate pathway intermediates in BC far from being completely understood.
In this work, we discover a function for CoQ10 and UBIAD1 in controlling membrane fluidity properties that is associated with a reduced metastatic potential and ferroptosis resistance in BC settings and patients.
Results
CoQ10 regulates membrane fluidity and cell stiffness in BC
CoQ10 (or ubiquinone) is a potent lipid-soluble antioxidant enriched not only in mitochondria but also in plasma membranes2,20. Whether plasma membrane (PM) CoQ10 levels might have redox-independent functions such as altering cell membrane lipid homeostasis and acting as a membrane-stabilizing agent in pathological settings remains not characterized. To this purpose, we treated the TNBC cell lines MDA-MB-231 with different formulations of CoQ10 (CoQ10 and CoQ10H2) and we evaluated changes in PM cholesterol-rich lipid rafts, microdomains that are known to regulate signal transduction and spatial organization of the PM. CoQ10 treatment of TNBC cells leads to a significant increase of CoQ10 contents in the membrane of these cells (Supplementary Fig. 1a, b) without affecting overall survival. By using the cholera toxin B-subunit staining we detected a decrease in lipid rafts microdomains upon CoQ10 treatments (Fig. 1a, b). These effects are independent of the redox state of CoQ10 and are evident also in a different TNBC cell line such as MDA-MB-436 (Supplementary Fig. 1c).
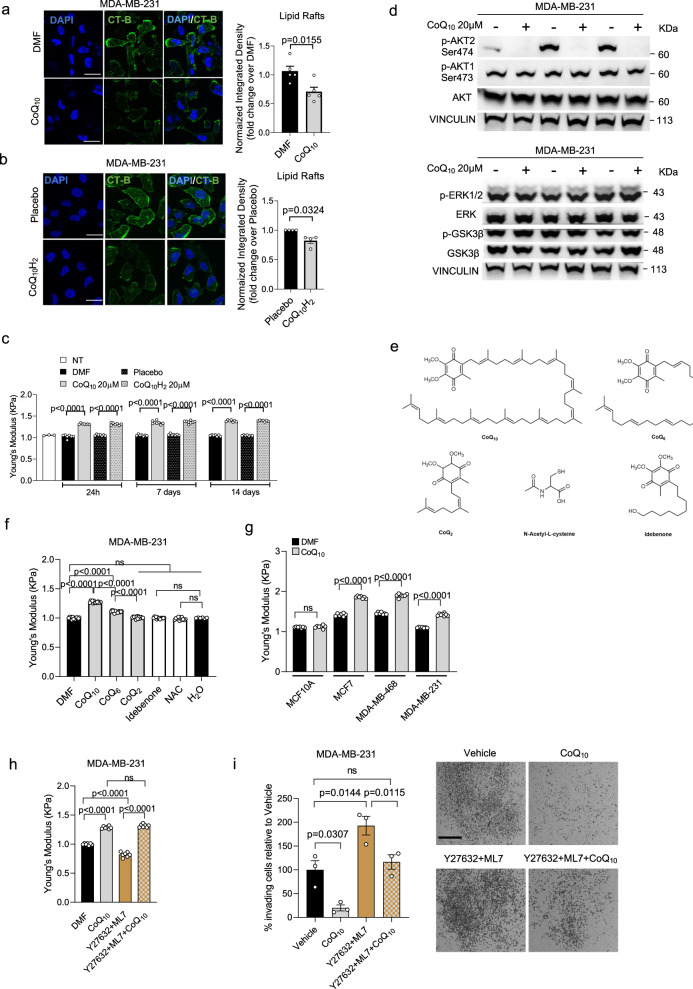
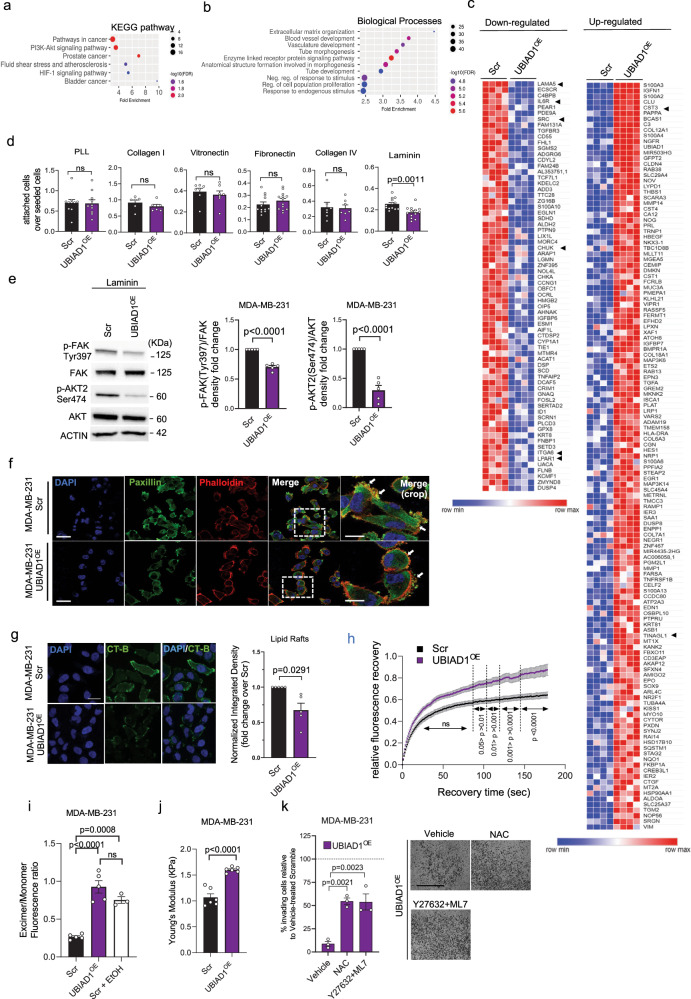
Fig. 1. CoQ10 alters BC cell stiffness and impairs AKT2 activation.
a, b Representative images of lipid rafts (CT-B) in MDA-MB-231 cells treated with 20 μM CoQ10 (a) or 20 μM CoQ10H2 (b). Scale bar, 25 μm. DAPI and CT-B/lipid rafts. Integrated density normalized to cell number, expressed as fold change over Di-Methyl-Formamide (a) or Placebo (b). Mean ± SEM from n = 5 for (a); n = 4 for b independent experiments, analyzed with a two-tailed t-test. c Stiffness of MDA-MB-231 cell after 20 μM CoQ10 or CoQ10H2 treatment. Each dot is the average of 3 measurements per cell from 2 independent experiments (n = 3 for Not-Treated, n = 8 for CoQ10 and CoQ10H2 at 7 days, n = 7 for others). P-values from two-tailed t-test. d Western blot for AKT1, AKT2, ERK1/2, and GSK3β phosphorylation to assess PI3K-AKT, MEK/ERK, and Wnt pathway activation. VINCULIN as control. Data from independent replicates (n = 3). e Chemical structures of CoQ compounds and antioxidants, including N-Acetyl-L-Cysteine (NAC) and Idebenone. f Stiffness of MDA-MB-231 cell after 24-h treatment with 20 μM CoQ10, CoQ6, CoQ2, Idebenone, or 10 mM NAC. Vehicles: Di-Methyl-Formamide or H2O. Each dot is the average of 3 measurements per cell from 3 independent experiments (n = 6 for H2O, n = 15 for CoQ2, n = 14 for others). Mean ± SEM, adjusted p-values from one-way ANOVA and Tukey’s test. g Stiffness in MCF10A and breast cancer cell lines after 1 week of 20 μM CoQ10 treatment. Each dot is the average of 3 measurements per cell from 2 independent experiments (n = 8 for MDA-MB-231 CoQ10, n = 7 for others). Mean ± SEM, p-values from two-tailed t-test. h Stiffness of MDA-MB-231 cells treated with 20 μM CoQ10, ROCK inhibitor Y27632 (20 μM), MLCK inhibitor ML7 (20 μM), or their combination. Each dot is the average of 3 measurements per cell from 2 independent experiments (n = 7). Mean ± SEM, adjusted p-values from one-way ANOVA and Tukey’s test. i Invasion assay of MDA-MB-231 cells through Matrigel-coated membranes, treated for 24 h with Di-Methyl-Formamide, 20 μM CoQ10, Y27632 (20 μM) + ML7 (20 μM), or their combination. Data are percentage of invading cells relative to Vehicle (100%). Each dot is the average of two wells from 3 independent experiments. Mean ± SEM, adjusted p-values from one-way ANOVA and Tukey’s test. Source data are provided in Source data file.
It has been reported that plasma membrane cholesterol depletion rises cellular stiffness by increasing cortical rigidity21–23. To examine whether increased membrane CoQ10 level influences BC cell mechanical properties, we directly measured single-cell cortical stiffness using Atomic Force Microscopy (AFM)24. We found that CoQ10-supplemented BC cells exhibited higher cortical stiffness, compared with the untreated or control cells (Fig. 1c). This is true for both formulations of CoQ10 and independent of the timing of treatments.
Next, we evaluated whether such alteration of PM homeostasis induced by CoQ10 treatment may lead to changes in the way how cells regulate signal transduction25. Different signaling pathways related to BC development were analyzed (Fig. 1d). While most of them remained unchanged, specific inhibition of the PI3K/AKT pathway has been detected as the impairment of the phosphorylation status of the AKT2 isoform. Interestingly, this specific isoform has been previously associated with worse breast cancer patient outcomes26. Considering the biochemical properties of membrane-bound CoQ10, these data suggest that treatment with CoQ10 affects PM homeostasis and PI3K/AKT oncogenic signaling in BC cells.
To support our hypothesis regarding the unique dual functionality of CoQ10—combining antioxidant properties through the redox cycling of its quinone ring and mechano-properties associated with the PM through its polyprenyl tail (isoprenoid moiety)—we conducted a comprehensive investigation utilizing AFM. Our experimental approach involved the examination of diverse molecules, including structural analogs of CoQ10 featuring a shortened isoprenoid moiety, such as CoQ6 and CoQ2, alongside counterparts possessing antioxidant function but lacking the structural features of CoQ10, such as idebenone and NAC.
This analysis aims to contribute valuable insights into the multifaceted roles of CoQ10, providing a better understanding of its potential applications in both antioxidant and mechano-property domains. Interestingly, we observed that antioxidant compounds such as idebenone and NAC do not alter cellular stiffness. CoQ2 which contains a short side chain of 2 isoprenoid units also does not alter cell stiffness, while CoQ6 which contains a side chain of 6 isoprenoid units significantly promotes stiffness although not at the same levels of CoQ10 (Fig. 1e, f). Therefore, these data showed that it is conceivable to separate the lipid antioxidant role of CoQ10 (linked to the redox ubiquinone ring) from its plasma membrane mechanical role (linked to the polyprenyl tail).
Next, we analyzed whether CoQ10 can equally exert its mechano-properties in different BC cell lines (Fig. 1g). CoQ10 does increase stiffness in MCF7, MDA-MB-468, and MDA-MB-231, but not in non-tumorigenic MCF10A suggesting that the effect of CoQ10 is possibly related to the tumorigenic state of breast cancer cells or rather a different PM composition. To further confirm the ability of CoQ10 to regulate cellular stiffness, we treated MDA-MB-231 cells with CoQ10 in combination with the ROCK inhibitor Y27632 and the MLCK inhibitor ML7 known to make BC softer27 (Fig. 1h). Here we show that Y27632 + ML7 inhibitors promote softness in MDA-MB-231 cells as expected. However, in presence of CoQ10, the two inhibitors fail to promote cellular softness and the cells present the same stiffness as with CoQ10 treatment alone.
Moreover, we examined whether CoQ10 could interfere with BC proliferation and/or alter invasion properties (Fig. 1i and Supplementary Fig. 1d). CoQ10 treatment does not alter cell proliferation of MDA-MB-231, while significantly impairs their ability to invade Matrigel-coated membranes. On the other hand, in the presence of the ROCK inhibitor Y27632 and the MLCK inhibitor ML7, cell invasiveness increases, but such effect is significantly reduced in the presence of CoQ10 (Fig. 1i).
Finally, we tested whether CoQ10 treatment could alter cellular morphology by examining structures containing Paxillin and FAK proteins as crucial molecular components that play a central role in connecting ECM signaling with actin filaments. They regulate the dynamics of the cytoskeleton and cellular morphology during cell invasion28. MDA-MB-231 cells treated with CoQ10 show different cellular protrusions compared to controls. Notably, these alterations manifest as multiple blebs at the plasma membrane where FAK and Paxillin are localized. In contrast, control cells exhibit a more elongated morphology with visible lamellipodia, characteristic of mesenchymal-like cells (Supplementary Fig. 1e–h). To confirm that CoQ10 acts by altering cortical contractility, we examined cytoskeletal changes in cells that have been treated with the ROCK inhibitor Y27632 and the MLCK inhibitor ML7 (Supplementary Fig. 1i–k). Y27632 + ML7-treated cells showed pronounced lamellipodia but no blebs and, interestingly, such morphological features were significantly rescued by the co-treatment with CoQ10, confirming that CoQ10 acts by altering cortical contractility.
Altogether, these findings indicate an unforeseen role for CoQ10 in regulating PM mechano-properties, cellular morphology and PI3K/AKT signaling pathways in BC cells.
The CoQ10 biosynthetic enzyme, UBIAD1, is associated with breast cancer (BC) patient outcome
Membrane-bound CoQ10 is synthesized by the UBIAD1 enzyme that is specifically localized in the Golgi compartment29,30. Therefore, we investigate the function of the UBIAD1 enzyme during BC tumorigenesis. At first, we analyzed the expression of the CoQ10 biosynthetic enzyme UBIAD1 in human patients with BC. We interrogated clinical and genomic data of ~2000 BC patients with well-annotated clinicopathological information and long-term follow-up, comprised in the METABRIC (Molecular Taxonomy of Breast Cancer Consortium) dataset31. Copy number alterations (CNA) analysis of this database revealed that, compared to the normal tissue, breast tumors show frequent copy number aberrations at the level of the UBIAD1 locus, consisting in most cases in deep or shallow deletions, with only a minority of cases (~1%) showing a copy gain of UBIAD1 (Fig. 2a). In the analysis of the METABRIC cohort, UBIAD1 deletion appears to be highly significantly enriched in Luminal B, HER2+ and Basal vs. Luminal A patients (Fig. 2b). To confirm the possible alterations of UBIAD1 expression in BC patients, we performed immunohistochemistry (IHC) analyses on a large retrospective consecutive cohort of ~2000 BC patients with long-term complete follow-up (median 14.1 years), available in a tissue microarray32,33. The availability of matched normal mammary tissue samples for a subset of patients also enabled us to compare UBIAD1 expression in normal vs. tumor tissue. UBIAD1 appeared uniformly and proficiently expressed in the normal mammary gland, with comparatively higher expression levels in the luminal compared to the basal epithelial layer, arguing for a functional role of UBIAD1 in homeostatic conditions (Fig. 2c). This observation strikingly contrasted with the heterogeneous pattern of UBIAD1 staining across breast tumors, with a vast majority of tumors displaying a remarkable downregulation or even total loss of UBIAD1 expression, compared to the normal tissue (Fig. 2c). Then, we categorized patients of the cohort in three groups based on UBIAD1 protein levels in the primary tumor (see also Materials and Methods): UBIAD1High, tumors with UBIAD1 protein levels undistinguishable from normal (9.2% of patients); UBIAD1Intemediate, tumors with residual UBIAD1 expression (67.0% of patients); UBIAD1Low, tumors with almost complete or total loss of UBIAD1 (23.8% of patients). Remarkably, we found clinical significance, with UBIAD1Low BC patients showing a significantly shorter overall survival (OS) compared to UBIAD1High patients (HRHigh vs. Low = 0.57, CI, 0.34–0.94; P = 0.03) (Fig. 2d). The behavior of UBIAD1 as a marker of poor prognosis was independent of all the standard clinicopathological parameters, as assessed in a multivariable analysis adjusted for estrogen and progesterone receptor expression, nodal status, tumor size, proliferative index, age, and grade. We also found that UBIAD1Low tumors, compared to UBIAD1High tumors, are associated with a more malignant clinicopathological status, as witnessed by an increase in tumor size, poorer degree of differentiation, loss of estrogen and progesterone receptor, and higher proliferation rate (Supplementary Table 1). While no significant associations were detected for nodal status, HER2 expression and Age (Supplementary Table 2). Consistently, UBIAD1Low tumors also significantly correlated with triple-negative BC (TNBC) (ORLow vs. High = 4.08, p = 0.0012), which represents the most biologically and clinically aggressive BC subtype (Fig. 2e). Similar evidence was obtained for basal tumors when examining the UBIAD1 mRNA expression levels in the METABRIC cohort (Supplementary Table 3).
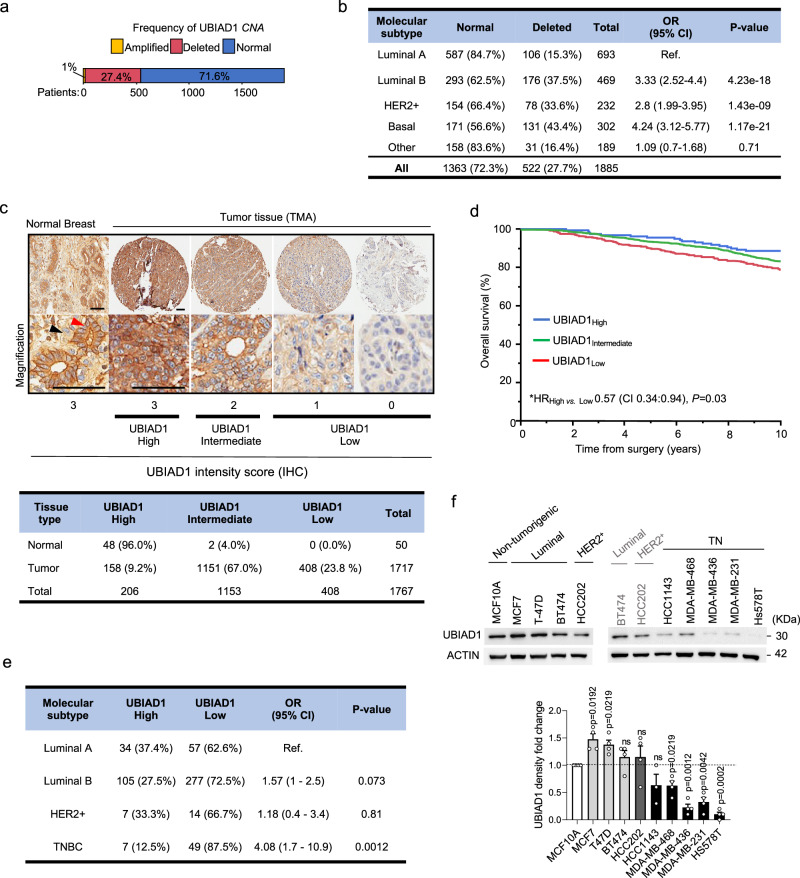
Fig. 2. Loss of the CoQ10-biosynthetic enzyme UBIAD1 is associated to a worse prognosis in BC patients.
a Distribution of UBIAD1 copy number alterations (CNA) in the METABRIC cohort of breast cancer patients (see Methods). Percentages of patients with no CNA (Normal), amplifications (Amplified), or deep and shallow deletions (Deleted) are indicated. b Association between UBIAD1 status (Normal vs. Deleted) and PAM50 BC subtypes in the METABRIC cohort. HER2 +, HER2-positive. OR, odds ratio; CI, confidence intervals. P-value from two-sided Fisher’s exact test, with Luminal A subtype as the reference (ref.). The number and percentage of patients in each group are shown. c Representative images showing heterogeneous UBIAD1 expression in a cohort 1767 breast samples available as TMA (1717 tumor plus 50 normal mammary gland samples) by IHC. Red and black arrowheads: luminal and basal cells, respectively. Tumors are classified as High, Intermediate, or Low based on UBIAD1 intensity scores. Scale bars, 100 µm. Quantitative analysis of UBIAD1 expression by IHC in normal and tumor tissues, with the number and percentage of samples in each UBIAD1 category (High, Intermediate, Low) shown. d Kaplan-Meier analysis of overall survival in the TMA cohort, stratified by UBIAD1 status as in (c). *HR, multivariate hazard ratio for UBIAD1High vs. UBIAD1Low groups. HR is calculated with 95% CI using multivariate Cox proportional hazards regression, adjusted for ER/PGR, pT, pN, Ki-67, HER2, age, and grade. CI and P (p-value) are indicated. P-value from two-sided Wald test. e Association between UBIAD1 status (High, Low) and BC molecular subtypes. HER2 +, HER2-positive; TNBC, triple-negative. OR, odds ratio; CI, confidence intervals. P-value from Fisher’s exact test with Luminal A subtype as reference (ref.). The number and percentage of patients in each group are shown. P-value from two-sided Fisher’s exact test. f Western blot analysis of UBIAD1 protein expression in human BC cell lines, grouped by molecular subtype. HER2+, HER2-positive; TN, triple-negative. Densitometric analysis of UBIAD1 protein expression with each dot representing n = 4 independent seedings and treatments. Data normalized over ACTIN and expressed relative to the non-tumorigenic breast line MCF10A. Mean ± SEM. P-value calculated using one-sample (two-tailed) t-test to compare the mean of each sample with 1.0. ns, not significant. Source data are provided as a Source data file.
In agreement with patients, we obtained similar evidence by analyzing UBIAD1 expression both at mRNA and protein levels in a panel of human BC cell lines. We found that UBIAD1 expression is reduced particularly in TNBC lines (Fig. 2f and Supplementary Fig. 2a), especially in the most aggressive MDA-MB-231, MDA-MB-436, and Hs578T lines. Interestingly, also the levels of total CoQ10 are significantly higher in MCF10A cells compared to other tumorigenic cells lines (Supplementary Fig. 2b).
To support the role of UBIAD1 in breast tissues, we evaluated the estrogen-mediated regulation of UBIAD1. Thus, we treated with the Estrogen receptor-inhibitor 4-OH-Tamoxifen (4OHT) three ERα-positive lines (Supplementary Fig. 2c) and we evaluated the changes in the expression of the ERα-targets genes such as BCL2, MYC and CCND1 and UBIAD1. We detected a reduction in the expression of all these ER-target genes as well as of UBIAD1 (Supplementary Fig. 2d, e). To confirm this regulation, we treated the ERα-positive lines with 17β-Estradiol to activate the ER-mediated transcription. Accordingly, we detect an increase of UBIAD1 expression upon estrogen treatment (Supplementary Fig. 2f). These data show the regulation of UBIAD1 expression through ERα signaling and support the involvement of the CoQ10-producing enzyme UBIAD1 in BC.
In summary, we propose a function of the CoQ10-biosynthetic enzyme UBIAD1 as a tumor suppressor gene in mammary tumorigenesis, with an intrinsic clinical value as an independent biomarker for the prognostic stratification of BC patients.
Ubiad1 genetic loss impairs CoQ9 synthesis and promotes tumor development in mouse BC models
To validate the functional role of UBIAD1 and CoQ10 in BC development, we generated a Ubiad1-deficient mouse model by disrupting exon1 through the insertion of a β-Gal/Neo Cassette (Supplementary Fig. 3a). Ubiad1−/− mice were not viable (Supplementary Fig. 3b) starting to die around E10.5. Protein lysates from E9.5 embryos showed a strong reduction in UBIAD1 protein level compared to both Ubiad1+/− and Ubiad1+/+ littermates at the same embryonic stage (Supplementary Fig. 3c). We observed that almost 40% of Ubiad1+/− embryos at E10.5/E11.5 show a hemorrhagic phenotype with cranial and heart bleeding, while the rest of the embryos show normal appearance (Supplementary Fig. 3d). These results suggested that part of Ubiad1+/− embryos die during embryogenesis due to cardiovascular failure and explain the reduced number of Ubiad1+/− pups observed at birth. Similar results were previously obtained in the zebrafish model30. Next, we measured by MS the level of CoQ9 in whole Ubiad1+/− embryos and we found a reduced amount of CoQ9 in Ubiad1+/− embryos compared to Ubiad1+/+ siblings (Supplementary Fig. 3e). To test whether CoQ9 is the limiting factor responsible for the embryonic lethality of Ubiad1 KO mice, we injected CoQ10 or Vitamin K2 in Ubiad1+/− pregnant females and analyzed the number of pups for each genotype at P0. We found that CoQ10, but not Vitamin K2, was able to increase the percentage of Ubiad1+/− embryos at P0, while both failed to rescue embryonic lethality of Ubiad1−/− genotype (Supplementary Fig. 3f). Moreover, to validate the contribution of enhanced oxidative stress in determining the cardiovascular system failure observed in CoQ9-deficient embryos, we showed that injection of the antioxidant NAC rescues the embryonic lethality at similar extent as CoQ10 treatments (Supplementary Fig. 3f).
Next, we crossed Ubiad1+/− mice with two mouse genetic models of BC: (i) MMTV-NeuT, where tumor formation is driven by the overexpression of an active form of the HER2 oncogene34; (ii) MMTV-PyMT, in which the oncogenic polyomavirus middle T antigen (PyMT) expressed under the control of the mammary gland specific promoter MMTV, and drives the spontaneous development of multifocal mammary adenocarcinoma35. In both models, Ubiad1+/− mice, compared to Ubiad1+/+ controls, showed a significantly accelerated tumorigenesis, with a mean tumor formation interval of 31 weeks instead of 37 weeks for MMTV-NeuT mice and 15 weeks instead of 16.5 weeks for MMTV-PyMT mice (Fig. 2a, b and Supplementary Fig. 4a, b). Moreover, we found an accelerated growth rate in Ubiad1+/− vs. Ubiad1+/+ control mice by monitoring tumor growth kinetics after the tumors had reached a palpable size (Fig. 3c and Supplementary Fig. 4c). Accordingly, we detected an increased number of proliferating Ki67+ cells in MMTV-PyMT; Ubiad1+/− tumors compared to controls (Supplementary Fig. 4d). At the sacrifice, we observed an increase in the number of primary tumors developed per mouse and the total tumor weight per mouse in Ubiad1+/− compared to Ubiad1+/+ control for both models (Fig. 3d, e, and Supplementary Fig. 4e, f). We then analyzed Ubiad1 expression in tumors spontaneously generated in MMTV-NeuT; Ubiad1+/− and MMTV-PyMT; Ubiad1+/− mice and we found a significant reduction of Ubiad1 mRNA expression in Ubiad1+/−-derived tumors compared to controls, supporting the association between the loss of Ubiad1 and increased tumorigenicity (Fig. 3f and Supplementary Fig. 4g). We did also measure the level of CoQ9 in these tumors showing a reduction in Ubiad1+/− mice (Fig. 3g).
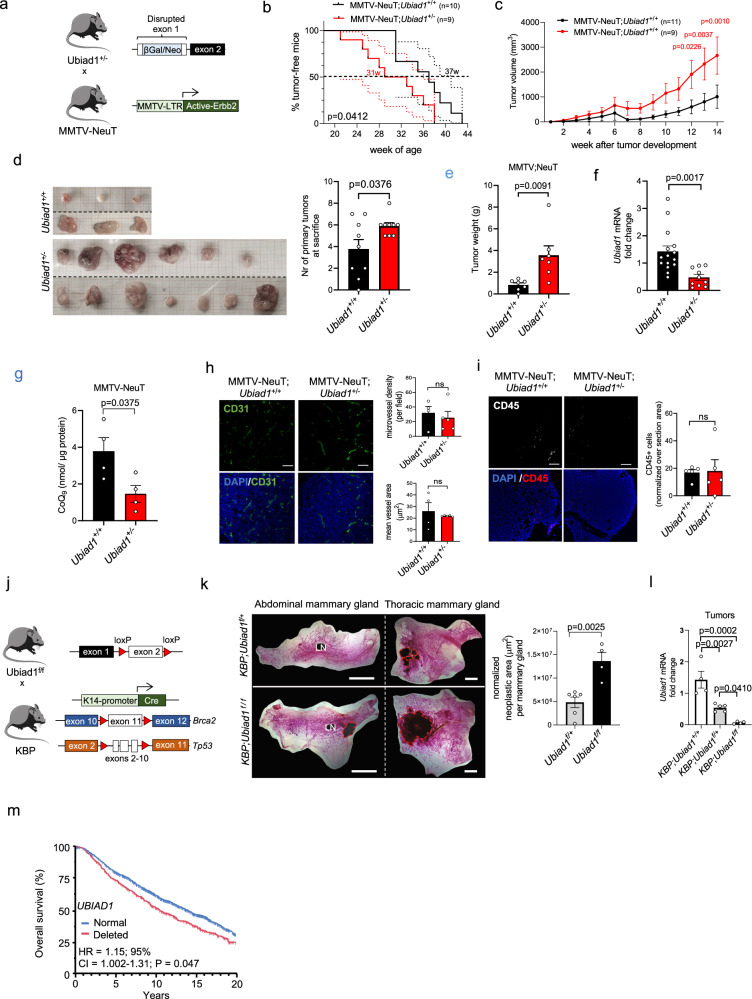
Fig. 3. Deletion of Ubiad1 gene accelerates BC development in mouse models.
a Generation of MMTV-NeuT BC model with exon 1 disruption of the Ubiad1 gene. b Tumor-free survival from birth to tumor detection. Dashed lines show 95% Confidence Interval (CI). Group mean survival is reported. Analysis used the Gehan-Breslow-Wilcoxon test (1 degree-of-freedom). c Tumor growth from first palpable tumor (week 0). Data for n = 11 MMTV-NeuT;Ubiad1+/+ and n = 9 MMTV-NeuT;Ubiad1+/− mice shown as mean ± SEM. Adjusted p-value from two-way ANOVA and Sidak’s test. d Tumors from two MMTV-NeuT;Ubiad1+/+ and two MMTV-NeuT;Ubiad1+/− mice. Number of primary tumors per mouse (n = 9 mice) shown as mean ± SEM. P-value from a two-tailed t-test. e Total tumor weight per n = 7 mice/group shown as mean ± SEM. P-value from a two-tailed t-test. f Ubiad1 mRNA levels in n = 15 MMTV-NeuT;Ubiad1+/+ and n = 10 MMTV-NeuT;Ubiad1+/− tumors. Mean ± SEM. P-value from a two-tailed t-test. g CoQ9 levels in tumor lysates from n = 4 mice/group. Data shown as mean ± SEM. P-value from a two-tailed t-test. h CD31 staining in tumors. Each dot represents a tumor from n = 4 MMTV-NeuT;Ubiad1+/+ and n = 5 MMTV-NeuT;Ubiad1+/− mice. Scale bar, 100 µm. Micro-vessel density and area shown as mean ± SEM. P-value from a two-tailed t-test. i CD45 staining in tumors. Each dot represents a tumor from n = 5 mice/group. Scale bar, 100 µm. CD45+ cells shown as mean ± SEM. P-value from a two-tailed t-test. j Generation strategy for the KBP BC model with exon 2 deletion of Ubiad1 in basal mammary gland cells. k Whole mount mammary gland staining to visualize lesions in heterozygous (Ubiad1f/+) or homozygous (Ubiad1f/f) Ubiad1 knockout KBP females. Red dashed lines highlight lesions. Each dot represents a mouse (n = 6 KBP;Ubiad1f/+ and n = 4 KBP;Ubiad1f/f). Data shown as mean ± SEM. P-value from a two-tailed t-test. LN, Lymph node. l Ubiad1 mRNA levels in n = 4 KBP, n = 5 KBP;Ubiad1f/+, and n = 4 KBP;Ubiad1f/f tumors. Mean ± SEM. Adjusted p-value from one-way ANOVA and Tukey’s test. m Kaplan-Meier analysis of survival in METABRIC BC patients by UBIAD1 status (522 for UBIAD1 deleted and 1363 for UBIAD1 normal. *HR, multivariate hazard ratio adjusted for clinicopathological parameters. HR and 95% Confidence Interval from Cox regression after adjustment. CI and P-value indicated. P-value from two-sided Wald test without adjustments for multiple comparisons. Source data are provided in Source data file.
Considering the essential role of UBIAD1 in endothelial cells30 and the frequency of the hemorrhagic phenotypes observed in Ubiad1+/− embryos, we investigated the presence of blood vessels inside the tumor by analyzing vessel density and mean vessel area (Fig. 3h and Supplementary Fig. 4h). The analysis revealed no significant differences in both parameters between Ubiad1+/+ and Ubiad1+/− mice. In parallel, we analyzed immune cell infiltration (CD45+ cells) in these tumors with no significant difference between Ubiad1+/− vs Ubiad1+/+ animals (Fig. 3i and Supplementary Fig. 4i) arguing in favor of a tumor cell-autonomous function of UBIAD1.
To better check the tissue-specific function of Ubiad1 in breast cancer onset, we generated a flox allele model of Ubiad1f/f mice, carrying the second exon of Ubiad1 gene flanked by two lox-P sites (Supplementary Fig. 4j). We then crossed these Tg animals with the BC model K14-Cre; Brca1f/f; Tp53f/f (Fig. 3j36); in which deletion of Tp53, Brca1, and Ubiad1 occurs concomitantly and specifically in the basal cells of the mammary gland. We verified the specific deletion of exon 2 only in the mammary gland and in the breast cancer cells of KBP; Ubiad1f/+ and KBP; Ubiad1f/f mice by PCR on the genomic DNA (Supplementary Fig. 4j, k). In line with our previous results, deep deletion of the Ubiad1 gene in KBP; Ubiad1f/f mice leads to the development of larger neoplastic areas in the mammary glands, compared to KBP; Ubiad1f/+ littermates (Fig. 3k). We then confirmed that Ubiad1 expression was absent in KBP; Ubiad1f/f tumor compared to KBP; Ubiad1+/+ and KBP; Ubiad1Δ/+ conditions (Fig. 3l). Interestingly, using the METABRIC dataset we confirmed these findings in humans where deep deletion of UBIAD1 gene in BC patients is associated with reduced overall survival (Fig. 3m). Moreover, as previously demonstrated (Fig. 2), absence of UBIAD1 is more frequent in TN and HER2 + BC subtypes, that are known to be the most clinically aggressive (Fig. 2e).
In summary, these in vivo data provide genetic evidence that loss of the CoQ9/10-biosynthetic enzyme UBIAD1 cooperates with the oncogene-driven tumorigenesis to increase spontaneous breast tumor formation, pointing to a fundamental tumor suppressor function of UBIAD1 and possibly CoQ9/10, in BC development.
UBIAD1 expression limits metastasis formation in aggressive BC lines
To better characterize the role of CoQ10 and UBIAD1 during BC development and progression, we took advantage of the metastatic human BC cell line MDA-MB-231 that showed a remarkable reduction of UBIAD1 expression. We then (re)expressed UBIAD1 (UBIAD1OE) in MDA-MB-231 cells (Supplementary Fig. 5a) and measured the level of different UBIAD1-derived metabolites through HPLC (Supplementary Fig. 5b). CoQ10 amount was increased while free and esterified cholesterol did not change. Vitamin K2 was not detectable by MS in both control and UBIAD1OE cells (data not shown). Interestingly, UBIAD1 expression stabilizes HMGCR (Supplementary Fig. 5c), the rate-limiting enzyme of the mevalonate pathway, suggesting a metabolic response due to the CoQ10 increase. Then, we measured whether UBIAD1 expression might alter the tumorigenic properties of MDA-MB-231. While UBIAD1OE cells do not show alteration in cell proliferation rate, they do show impairment in cell invasiveness compared to Scramble control cells (Supplementary Fig. 5d, e, respectively). Moreover, expression of UBIAD1 reduced the ability of MDA-MB-231 cells to grow and form colonies in anchorage-free conditions (Supplementary Fig. 5f). These results indicated a decreased cell ability to escape anoikis, which is an essential process also during the dissemination of circulating tumor cells (CTCs)37,38. Consistently, restoration of UBIAD1 expression in MDA-MB-436 has similar effects (Supplementary Fig. 5g, h).
Next, we tested the in vivo tumorigenic ability of UBIAD1-expressing MDA-MB-231 cells using xenografts assays in immunodeficient mice39. After labeling these cells with Luciferase-eGFP, we injected UBIAD1OE or control cells in the fat pad of the fourth mammary gland of NSG mice (Fig. 4a, b). We measured tumor growth weekly, and we found a decreased growth rate of UBIAD1OE compared to control cells (Fig. 4b). Moreover, we evaluated the number of circulating tumor cells (CTCs) and the metastasis formation at different sites (Fig. 4c–f). The number of CTCs sorted from the whole blood is dramatically reduced in mice injected intracardiacally with UBIAD1OE cells compared to controls (Fig. 4d), suggesting a decreased ability of UBIAD1OE cells to survive in the bloodstream. Moreover, the lung metastasis formed by UBIAD1OE cells was reduced both by analyzing the whole organs (Fig. 4e) or sorting the GFP+ cells in dissociated lungs (Fig. 4f). No significant differences were detected in the other whole organs analyzed. We confirmed through qPCR that UBIAD1 expression was maintained high in the UBIAD1OE cells sorted from the primary tumor, blood, and different metastatic sites (Supplementary Fig. 5i). In line with our experimental evidence, we found that UBIAD1 expression is specifically turned off in MDA-MB-231-derived CTCs and lung metastasis but not in metastasis formed in other sites (i.e., brain and liver), supporting the idea that low level of UBIAD1 could be beneficial specifically for the survival of CTCs and the growth of pulmonary metastasis (Fig. 4g, h).
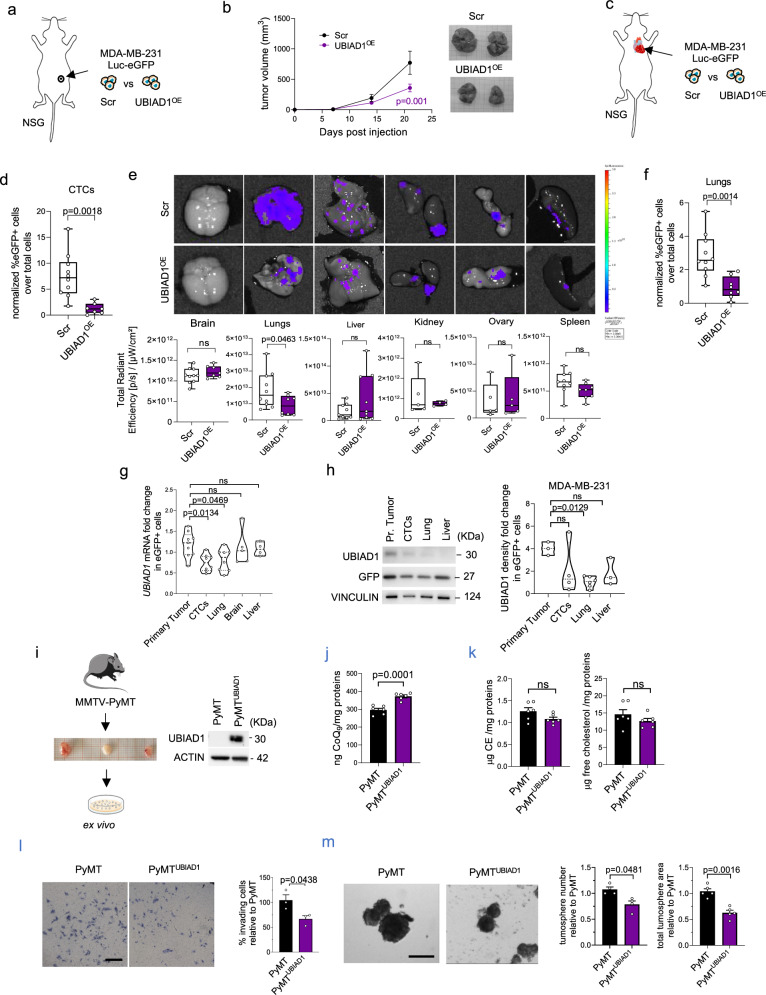
Fig. 4. UBIAD1 expression increases CoQ10 level and reduces BC dissemination.
a, b Tumor growth in NSG tumor-bearing mice engrafted in the mammary fat pad with Scramble and UBIAD1-overexpressing MDA-MB-231 cells. Data are mean ± SEM, with n = 8 mice per group from two independent experiments. Adjusted p-value from two-way ANOVA and Sidak’s test. c–f Scramble and UBIAD1-overexpressing MDA-MB-231 cells injected intracardially into NSG mice. d–f Data shown as box plots with median, 25th, and 75th percentiles and with whiskers at minimum and maximum values. P-value from two-tailed unpaired t-test. d Percentage of Circulating Tumor Cells (CTCs) in whole blood. Each dot represents a mouse (n = 10 MDA-MB-231 Scr and n = 7 MDA-MB-231 UBIAD1OE). Gating: eGFP+ cells from Single_cell_2 population. e eGFP signal of MDA-MB-231-derived metastasis in different organs. Each dot represents a mouse. f Percentage of eGFP+ cells in dissociated lungs. Each dot represents a mouse (n = 10 MDA-MB-231 Scr and n = 8 MDA-MB-231 UBIAD1OE). g, h UBIAD1 mRNA and protein expression in eGFP+ cells from whole blood, organs, or primary tumors of MDA-MB-231 Scr-injected mice. Each dot represents a single organ pooled from 3 independent experiments. Data are mean ± SEM. Adjusted p-value from one-way ANOVA and Sidak’s test. i–m Generation and characterization of Scramble (PyMT) and UBIAD1-overexpressing (PyMTUBIAD1) ex vivo cultures from MMTV-PyMT-derived tumors. i UBIAD1 protein expression. j Total CoQ9 in mouse ex vivo cultures. Each dot represents an independent seeding (n = 6). Data shown as mean ± SEM. P-value from two-tailed unpaired t-test. k Cholesterol esters (CE) and free cholesterol levels in mouse ex vivo cultures. Each dot represents an independent seeding (n = 6). Data shown as mean ± SEM. P-value from two-tailed unpaired t-test. l Invading mouse ex vivo PyMT and PyMTUBIAD1 cells. Data expressed as percentage of invading cells compared to PyMT (set at 100%). Each dot represents an independent seeding (n = 3). Data shown as mean ± SEM. P-value from two-tailed unpaired t-test. m PyMT tumorspheres. Quantification of total tumorspheres and total area relative to PyMT cells. Each dot represents an independent seeding (n = 4 for number, n = 5 for area). Data shown as mean ± SEM. P-value from two-tailed unpaired t-test. Source data are provided in Source data file.
To verify if our findings might be extended to other aggressive BC lines independently by UBIAD1 basal expression, we performed similar experiments using the JIMT-1 cell line, a HER2+ human breast cancer cell line with high UBIAD1 expression (Supplementary Fig. 5j, k). Thus, we overexpressed UBIAD1 in JIMT-1 and injected them in the fat pad of the fourth mammary gland of NSG mice or into the bloodstream through intracardiac injection (Supplementary Fig. 5l–r). Similarly, to MDA-MB-231, we found that increased UBIAD1 expression led to a decreased primary tumor growth (Supplementary Fig. 5m, n). Moreover, we found a reduction in the number of CTCs isolated from mice blood at 35 days post-intracardiac injection of JIMT-1 UBIAD1OE cells and in the colonization of different distal sites, like lung, brain, and bone (Supplementary Fig. 5o–r)40.
To validate the ability of UBIAD1 expression to impair BC tumorigenesis, we took advantage of MMTV-PyMT tumors. We isolated primary PyMT tumor cells and we (re)expressed UBIAD1 by lentiviral infections (Fig. 4i). We confirm that UBIAD1 expression increases CoQ10 without affecting free cholesterol or CE levels (Fig. 4j, k). Then, we tested their invasion properties on Matrigel-coated membranes as well as their capacity to generate tumorspheres (Fig. 4l, m). In agreement with previous results in human BC cell lines, UBIAD1 expression in ex vivo PyMT cells impaired both their invasion properties and cancer stem cell-like features evaluated in the tumorsphere assay.
Altogether, our data showed a role of the metabolic enzyme UBIAD1 in decreasing BC primary tumor growth and metastasis formation with specificities related to the metastatic site.
CoQ10 and UBIAD1 expression impairs ECM-mediated signaling and AKT2 pathway in BC cells
To investigate the molecular mechanisms driven by UBIAD1 in BC, we performed RNA-seq analyses on control and UBIAD1OE MDA-MB-231 cells (Fig. 5a–c, Supplementary Tables 4–5, and Supplementary Fig. 6a). KEGG pathway and Gene Ontology (GO) enrichment analyses identified the PI3K-AKT pathway as one of the most affected oncogenic signaling pathways, and the Extracellular matrix organization as the most significant biological process affected. Several genes that have been pointed out as oncogenes in breast cancer were shown to be downregulated in UBIAD1OE cells, i.e., LAMA5, IL6R, SRC, CHUK/IKKα, ITGA6, and LPAR141–46, and likely oncosuppressor genes were upregulated, i.e., CST3 and TINAGL147,48 (Fig. 5c and Supplementary Tables 4–5, and Supplementary Fig. 6a).
Fig. 5. UBIAD1 expression impairs AKT oncogenic pathway and alter cytoskeletal stiffness of BC cells.
a–c RNA-seq analysis of MDA-MB-231 Scramble (Scr) and UBIAD1-overexpressing (UBIAD1OE) cells. a KEGG pathway and b Gene Ontology (GO) enrichment analysis on differentially expressed genes. c Downregulated and upregulated genes in UBIAD1OE cells. Arrowheads: main genes listed in the GO analysis’ top rank. d Cell adhesion assay in MDA-MB-231. Data shown as ratio of attached cells over total seeded cells. Mean ± SEM of n = 10 for PLL, n = 6 for Collagen I, n = 7 for Vitronectin, n = 12 for Collagen IV, and n = 13 for Laminin independent seedings. P-value from two-tailed unpaired t-test. ns, not significant. e Phospho-FAK and phospho-AKT2 in laminin-attached cells. Densitometric analysis of phosphorylated protein over total protein. Each dot represents an independent seeding (n = 5). Data are mean ± SEM. P-value from two-tailed one-sample t-test comparing the mean of UBIAD1OE with 1.0. f Paxillin, DAPI, and Phalloidin staining. Arrows: cell adhesions or blebbing structures. Scale bar, 60 μm; crop, 30 μm. Results of three independent cell seedings, stainings, and analyses. g Lipid rafts (CT-B). Scale bar, 25 μm. Integrated density normalized over number of cells. Data as fold change over Scr. Each dot represents the average of five pictures for n = 5 independent seedings. P-value from two-tailed one-sample t-test comparing the mean of UBIAD1OE with 1.0. h FRAP analysis on MDA-MB-231 cells. Data shown as relative fluorescence intensity over recovery time. Each dot represents a single cell (n = 25 Scr and n = 27 UBIAD1OE) analyzed across three independent seedings, stainings, and analyses. Adjusted p-value from two-way ANOVA and Sidak’s test. ns, not-significant. i Membrane fluidity analysis. EtOH as a positive control. Each dot represents an independent seeding (n = 5 for Scr and UBIAD1OE, and n = 3 for Scr+EtOH). Data are mean ± SEM. Adjusted p-value from one-way ANOVA and Tukey’s test. j Cellular stiffness. Each dot represents the average of 3 repeated measurements for a single cell (n = 6 cells/group). P-value from two-tailed unpaired t-test. k Percentage of invading cells compared to Vehicle-treated Scr (set at 100%). Scale bar, 100 µm. N-Acetyl-L-Cysteine (NAC). Each dot represents an independent seeding (n = 3). Data are mean ± SEM. Adjusted p-value from one-way ANOVA and Dunnett’s test. Source data are provided in Source Data file.
The most significant downregulated gene is LM-511 (aka LAMA5). LM-511 has been recently shown to play a crucial role in the luminal mammary epithelial cells highlighting its function in mammary gland remodeling and luminal differentiation49. LM-511 expression correlates with breast tumor grade and is linked to metastatic potential in experimental models making an interesting target of our study. Indeed, it has been proposed that LM511 contributes to the metastatic progression of breast cancer and its stemness in a mechano-dependent manner50,51. Thus, we investigated the ability of MDA-MB-231OE cells to bind different extracellular matrix components by the engagement of integrin receptors (Fig. 5d). Cell attachment assays revealed that compared to controls, UBIAD1OE cells had a reduced capacity to bind to laminin-coated plates, while no differences were detected for other integrin-ligands, like Collagen I and IV, Vitronectin, or Fibronectin-coated surface.
Laminin-binding integrins are recruited in BC cells to activate AKT through the activation of the Focal Adhesion Kinase (FAK)25. Thus, we led the cells to adhere on a laminin-coated surface to confirm that both FAK and AKT signaling were more active compared to cells in suspension (BSA treated-surface) or cells attached to a PLL-coated plate (Supplementary Fig. 6b). Given that, we compared the level of the activating phosphorylation Tyr397 on FAK and Ser474 on AKT2 in MDA-MB-231 vs UBIAD1OE let adhere on a laminin-coated plate (Fig. 5e). As expected, UBIAD1 expression significantly decreased both FAK and AKT2 activation. We previously showed the effect of an extended CoQ10 treatment on AKT pathway activation (Fig. 1d). We then decided to investigate AKT2 activation in MDA-MB-231 upon UBIAD1 overexpression in standard culture conditions. Again, we detected a decrease in AKT2 phosphorylation in UBIAD1OE cells, in line with a CoQ10 overload (Supplementary Fig. 6c). We obtained similar results using PyMTUBIAD1 ex vivo cell line being PyMTUBIAD1 cells impaired in their ability to bind laminin (Supplementary Fig. 6d).
To establish a functional role for AKT2 in this context, we overexpressed wild-type AKT2 or its constitutively active form, AKT2-Myr, in UBIAD1-expressing cells (Supplementary Fig. 6e). We found that AKT2 did not alter UBIAD1-overexpressing cell invasion ability, while the constitutively active form is able to restore the invasion properties of these cells. Next, we have analyzed the behavior of UBIAD1 mRNA levels in the METABRIC cohort upon conducting a subgroup analysis by stratifying patients according to the status of PIK3CA/PTEN alterations (Supplementary Fig. 6f). We observed a significantly worse prognosis associated with low UBIAD1 mRNA expression exclusively in the subgroup without PIK3CA/PTEN alterations. These data lead to finding that the presence of constitutive active PI3K-AKT2 pathway overrides UBIAD1 functions and tumor suppressor status.
Overall, we conclude that CoQ10 treatment or UBIAD1 overexpression leads BC cells to lose their capacity to bind to laminins and engaged integrin-dependent FAK/AKT oncogenic signaling pathway.
Lastly, we examined cellular protrusions and adhesions as integrin-containing, multi-protein structures that form mechanical links between intracellular actin bundles and the extracellular substrate in many cell types. Confocal imaging analyses of FAK-containing structures clearly showed alterations in membrane morphology in MDA-MB-231 overexpressing UBIAD1 compared to controls (Fig. 5f and Supplementary Fig. 6g–i). Notably, these alterations manifest as multiple blebs at the plasma membrane resembling previous conditions in MDA-MB-231 cells treated with CoQ1052 (Supplementary Fig. 1e–k). Similarly, we observed that MDA-MB-231 overexpressing UBIAD1 were less responsive to the cytoskeletal changes induced by the ROCK inhibitor Y27632 and the MLCK inhibitor ML7 (Supplementary Fig. 6j–l).
It has been reported that the oncogenic PI3K/AKT pathway sustains cancer cell survival, invasion, and metastasis formation via activation of integrin receptors localized on the plasma membrane53. Among the integrin analyzed, laminin-binding integrins require a proper PM lipid raft assembly to recruit the transduction machinery and activate the downstream pathways54. Considering the ability of CoQ10 treatment to increase membrane fluidity, reduce lipid rafts contents and increase cortical stiffness (Fig. 1a–c), we analyzed whether a similar phenotype was induced by UBIAD1OE. Indeed, we detected a decrease of lipid rafts in UBIAD1OE cells (Fig. 5g). In line with this evidence, using the fluorescent probe Dil-C16 to perform a Fluorescence Recovery After Photobleaching (FRAP) analysis, we found an increased recovery of fluorescence in UBIAD1OE MDA-MB-231 cells compared to controls. Indeed Dil-C16 has more affinity for disordered cholesterol-poor membrane domains (where it can freely diffuse) than for cholesterol-enriched lipid rafts (Fig. 5h55). Knowing that alteration of PM lipid raft structures is associated to changes in membrane fluidity56, we tested the effect of UBIAD1 overexpression on membrane fluidity of MDA-MB-231 cells. The analysis showed an increased membrane fluidity of UBIAD1OE cells compared with control conditions that are similar to what was observed after treatment with EtOH, used as a positive control (Fig. 5i).
Modifications in membrane fluidity are important also in the context of the organization of the actin cytoskeleton that can control the activity of receptors exposed on cell surface25,56. To confirm whether this is the case, we analyzed the single-cell cortical stiffness of MDA-MB-231 cells upon UBIAD1 overexpression. Using AFM, we showed that UBIAD1OE cells are stiffer than control cells (Fig. 5j) as for CoQ10 treatment (Fig. 1c) suggesting that BC adapts to increased plasma membrane fluidity, impairment of cholesterol-rich lipid rafts with increasing cortical cytoskeletal stiffness.
Interestingly, the inhibitory effect of UBIAD1 expression on blocking BC invasion is attenuated either by inducing softness using Y27632 + ML7 inhibitors or by supplying antioxidants such as NAC (Fig. 5k). This suggests that the increased stiffness and potential alteration of redox homeostasis caused by UBIAD1-synthesized CoQ10 are likely responsible for decreasing tumorigenic activities in these cells.
Taken together our data demonstrate that UBIAD1 and CoQ10 alter plasma membrane mechano-properties that in turn impair ECM-mediated oncogenic AKT pathway activation during BC progression and decrease their tumorigenic properties.
UBIAD1 and CoQ10 enhance BC sensitivity to ferroptosis inducers via FSP1
Modification of PM lipid properties is associated with the alteration of cell mechanical properties and signaling. However, whether it might also modulate cell sensitivity to lipid peroxidation (e.g., ferroptosis and lipid-peroxidation-triggered death upon treatment with ferroptosis inducers) is currently poorly understood. Resistance to ferroptosis has been recently associated with the development of some refractory cancers, in particular BC19. Thus, we treated MDA-MB-231 UBIAD1OE cells with the lipid peroxidation inducers Erastin and RSL3 and we evaluated the cell sensitivity to lipid peroxidation. We found that in vehicle-treated cells, the level of Malondialdehyde (MDA), a bioproduct derived from lipid peroxidation, was not affected by UBIAD1 overexpression, while UBIAD1OE cells showed high redox stress upon Erastin treatment compared to controls (Fig. 6a). Similar evidence was obtained evaluating the oxidative status of BODIPY C11, a fluorescent lipid peroxidation probe that shifts the emission spectrum when oxidized. Indeed, UBIAD1OE showed a higher level of oxidized BODIPY compared to control cells after RSL3 treatment (Fig. 6b). Interestingly, we found that while treatment with the ROCK inhibitor Y27632 and the MLCK inhibitor ML7 tends to protect cells from RSL3, lipid peroxidation is significantly higher in MDA-MB-231 UBIAD1OE cells treated with the same inhibitors. This suggests that changes in PM cortical properties might influence ferroptosis sensitivities (Fig. 6c).
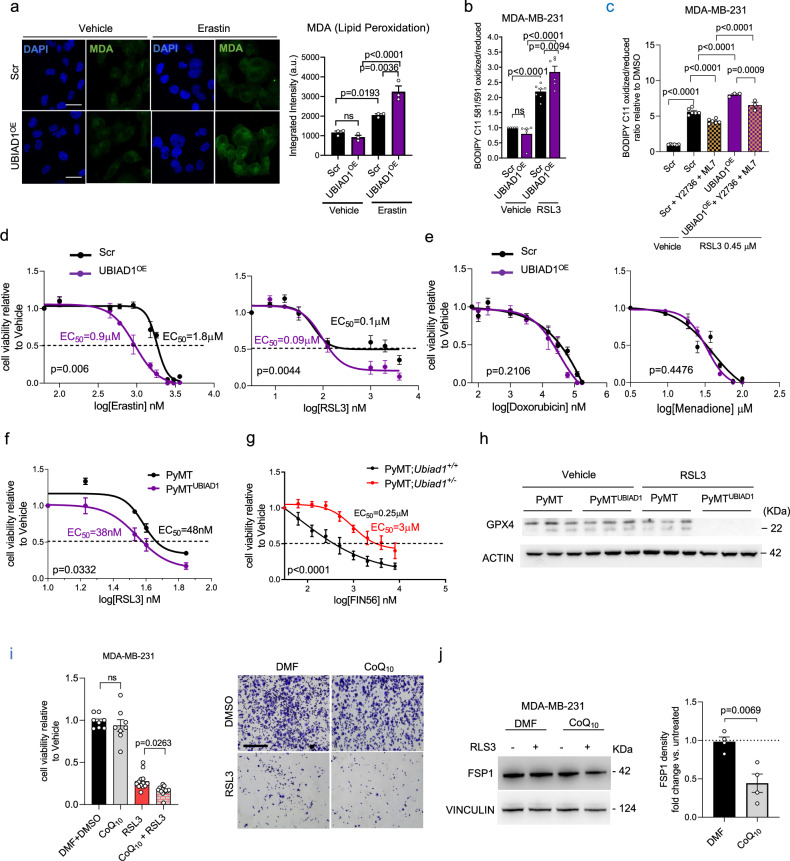
Fig. 6. UBIAD1 affects ferroptosis resistance in BC cells.
a Malondialdehyde (MDA) staining in MDA-MB-231 cells treated with Vehicle (DMSO) or 2 μM Erastin for 24 h. Scale bar, 100 μm. MDA signal intensity normalized over cell number. Mean ± SEM of three independent experiments. Adjusted p-value from one-way ANOVA and Sidak’s test. b Lipid peroxidation in cells treated with vehicle (DMSO) or 0.45 μM RSL3 for 24 h. Data expressed as fold change over vehicle-treated cells. Each dot represents an independent experiment (n = 6 for Vehicle-treated Scramble (Scr), n = 6 for RSL3-treated Scr and Scr+Y2736 + ML7, and n = 3 for RSL3-treated UBIAD1OE and UBIAD1OE + Y2736 + ML7). Mean ± SEM. Adjusted p-value from one-way ANOVA followed by Sidak’s test. c Lipid peroxidation in cells treated with vehicle (DMSO), 0.45 μM RSL3 for 12 h, in combination with 20 μM Y27632 and 20 μM ML7 inhibitors. Gating: P1-Live_Cells-FITC/PE. Data expressed as fold change over vehicle-treated cells. Each dot represents an independent experiment (n = 5 for Vehicle-treated and n = 6 for RSL3-treated). Mean ± SEM. Adjusted p-value from one-way ANOVA and Tukey’s test. d, e Cell viability assay with Erastin and RSL3 (d), Doxorubicin (e), and Menadione (e). Cells treated for 24 h. Mean ± SEM of n = 5 independent replicates. P-value from two-way ANOVA. f Cell viability assay with RSL3 in MMTV-PyMT ex vivo cultures. Mean ± SEM of n = 3 independent replicates. P-value calculated using two-way ANOVA. EC50 reported. g Cell viability assay with FIN56 in PyMT;Ubiad1+/+ and PyMT;Ubiad1+/− cells. Mean ± SEM of n = 5 independent replicates. P-value from two-way ANOVA. EC50 reported. h GPX4 protein in ex vivo cells treated with vehicle (DMSO) or 50 nM RSL3 for 24 h. ACTIN used as loading control. Each column represents an independent replicate (n = 3). i Cell viability assay in cells were treated for 24 h with 0.45 μM RSL3 alone, 20 μM CoQ10 alone or in combination. Each dot represents an independent seeding, treatment and analysis (n = 8 for DMF + DMSO (Vehicle) and CoQ10 only, and n = 13 for RSL3 only and CoQ10 + RSL3). Mean ± SEM. Adjusted p-value from one-way ANOVA and Sidak’s test. j FSP1 protein with DMF (Vehicle) or RSL3 0.45 μM for 24 h with or without CoQ10 20 μM. VINCULIN used as loading control. Each dot represents an independent experiment (n = 4). Mean ± SEM. P-value from two-tailed unpaired t-test. Source data are provided in Source data file.
We then evaluated cell viability at increasing concentrations of Erastin and RSL3 and we found that MDA-MB-231 UBIAD1OE cells showed reduced viability at lower concentrations of both drugs (Fig. 6d). Similar evidence was obtained for the two TNBC cell lines MDA-MB-436 and Hs578T (Supplementary Figs. 5g and 7a–c). Such effect was specific for ferroptosis-inducers since cell sensitivity to general oxidative stress inducers (e.g., Menadione) or chemotherapeutic drug treatment (e.g., Doxorubicin) was not affected by UBIAD1 expression (Fig. 6e and Supplementary Fig. 7b, c). In line with these results, we found that also ex vivo PyMTUBIAD1 cells were more sensitive to RSL3 than controls (Fig. 6f). To strengthen our results, we tested also tumor-derived cells from MMTV-PyMT-Ubiad1+/+ and MMTV-PyMT-Ubiad1+/− mice. We then treated with increasing concentrations of the ferroptosis inducers FIN56 (Fig. 6g) and found that MMTV-PyMT-Ubiad1+/− cancer cells with low UBIAD1 levels (Supplementary Fig. 4g) were more resistant to ferroptosis. GPX4, the central regulator that drives ferroptosis resistance, was analyzed in PyMT cells after treatment with RSL3 (Fig. 6h) and we found that UBIAD1 expression in PyMT cancer cells decreases importantly GPX4 levels and, therefore, their resistance to ferroptosis. Similar outcomes were obtained for MDA-MB-231 cells where both Erastin and RLS3 promote GPX4 level increase in control cells but not in UBIAD1-overexpressing cells (Supplementary Fig. 7d). Furthermore, we quantified Acsl4 mRNA expression in PyMT control and PyMTUBIAD1 cells. Indeed, ACSL4 functions to catalyze the ligation of CoA to free PUFAs to generate PUFA-CoA that predispose for ferroptosis onset. ACSL4 expression is tuned down by activation of integrin-mediated signaling, especially in cells detached by the ECM57. Interestingly, we found that in detached PyMTUBIAD1 cells, Acsl4 mRNA expression is significantly increased compared to control cells (Supplementary Fig. 7e). The same results were achieved in tumorspheres obtained by PyMTUBIAD1 cells (Supplementary Fig. 7f). Similarly, ACSL4 mRNA expression increases more in detached MDA-MB-231 UBIAD1OE cells compared to Scr (Supplementary Fig. 7g). These data suggest that UBIAD1 levels can modulate the sensitivity of BC cells to ferroptosis inducers through different anti-ferroptosis mechanisms58.
Lastly, we tested whether UBIAD1 increases ferroptosis sensitivity in BC cells through its biosynthetic product CoQ10. Therefore, we treated MDA-MB-231 cells with CoQ10 with or without the ferroptosis inducers RSL3 and tested cell viability (Fig. 6i). While CoQ10 treatment alone does not affect MDA-MB-231 cell viability, the co-treatment with RSL3 significantly enhanced cell death. To explain these results, we investigate FSP1 expression in MDA-MB-231 compared to other BC cell lines since the oxidized form CoQ10 needs to be reduced by the NADH-dependent oxidoreductases FSP1 to CoQ10H2, and only CoQ10H2 acts as a radical-trapping antioxidant to suppress ferroptosis59. We examined FSP1 expression by Western blot analysis in different BC cell lines and scored that MDA-MB-231 and other TNBC lines express very low amounts of FSP1 compared to, for instance, the luminal BC cell line MCF7 (Supplementary Fig. 7h). We therefore tested whether FSP1 expression correlates with ferroptosis sensitivity of BC cells by comparing RSL3-mediated lipid peroxidation between MCF7 and MDA-MB-231 (Supplementary Fig. 7i). MDA-MB-231 are more sensitive to RSL3 compared MCF7. This biological tendency was evident also analyzing cell viability after RSL3 treatment with or without CoQ10; indeed, CoQ10 treatment made MDA-MB-231 (but not MCF7) significantly more sensitive to RSL3 (Supplementary Fig. 7j). Next, to explain the increased sensitivity of the CoQ10-treated cells for ferroptosis inducers we analyzed FSP1 levels in CoQ10-treated cells after ferroptosis inductions (Fig. 6j). RSL3 stimulation reduces FSP1 protein levels only in CoQ10 treated cells suggesting the possibility that during ferroptosis CoQ10 act by impairing the stability of FSP1.
Overall, we propose that treatment with CoQ10 or UBIAD1 expression sensitizes breast cancer cells to ferroptosis (and consequently reduces ferroptosis resistance) by altering PM mechanical properties and, possibly, the effectiveness of the FSP1 shield system.
Discussion
While it is known that CoQ10 acts as a redox-cycling agent in cellular membrane59,60, its previously hypothesized role as a membrane-stabilizing agent remains elusive5. Here we demonstrate that CoQ10 can alter membrane mechanical properties by increasing PM fluidity and possibly cortical stiffness in BC. Such biological changes can be achieved either by expressing Golgi/ER-localized producing enzyme UBIAD1 or treating cells with the metabolite itself, an effect that is independent of its redox state. Such CoQ10-mediated changes in PM homeostasis leads to an impairment of lipid rafts content and ECM-mediated signaling in BC. Interestingly, CoQ10-dependent impairment of integrin signaling has been previously described during atherosclerosis61. Aggressive BC cells, such as TNBC cells, display higher levels of cholesterol-rich lipid rafts62. Therefore, it is conceivable that UBIAD1 and CoQ10 affect the survival and aggressiveness of the TNBC subtype that strongly relies on lipid raft integrity and integrin/ECM-mediated signal transduction for invasion/survival and ferroptosis protection63,64. Mechanistically, we foresee that such effect depends on the ability of CoQ10 to intercalate within the lipid bilayer and altering cholesterol-dependent PM rigidity and cortical cell softness vs. stiffness23,65,66. Cholesterol is known for its stabilizing effect in biological lipid membranes, therefore, it is tempting to speculate that CoQ10 might increase the fluidity state of cellular PM by directly competing with the function of cholesterol in membrane stabilization, causing opposite effects (cholesterol→rigidity vs. CoQ10→fluidity). Alternatively, CoQ10 might exert a membrane-destabilizing effect in certain types of biological membranes, independently of cholesterol67. We can speculate that these membrane alterations might be responsible for the spatial organization of membrane-associated contractile proteins, potentially altering their function and interactions with the plasma membrane. This could interfere with the effect of inhibitors of MLCK and ROCK such as ML7 and Y27632 limiting their effect on cellular softness. Regardless, the sum of results of this study clearly point to a direct role for the UBIAD1/CoQ10 axis in membrane homeostasis, with its dysfunction being causal to the acquisition of aggressive phenotypes in BC, thus highlighting alterations in cancer cell membrane fluidity and mechano-properties as potential therapeutically actionable targets for specific treatments in BC. Supporting this view, clinical trials showed a decrease of both plasma and tumor tissue levels of CoQ10 in breast cancer patients, suggesting a statistically inverse relationship between the plasma CoQ10 level and breast cancer prognosis68,69. Of note, reduced plasma concentrations of CoQ10 have also been reported for other types of human cancers, such as melanoma, lymphoma and lung cancer70,71 and correlated, in particular in one study in melanoma patients, with increased risk of metastasis72.
Another peculiar aspect of our studies is the correlation of UBIAD1 and CoQ10 expression with the regulation of ferroptosis resistance during BC spreading and CTCs survival. It has been recently shown that melanoma cells exit the primary site of tumor growth through the bloodstream; the oxidative stress that they encounter makes them prone to undergo a type of cell death called ferroptosis that in theory limits the formation of distant metastases. However, melanoma cancer cells develop specific mechanisms to counteract ferroptosis and survive in the bloodstream leading to metastasis73. Our data show that UBIAD1 expression limits significantly CTCs and metastasis in xenograft models of BC and that this is possibly related to its ability to enhance sensitivity to ferroptosis. We were able to recapitulate these findings using CoQ10 treatments suggesting that an increased concentration of plasma CoQ10 could be therapeutic for BC patients. Mechanistically, we demonstrate that altering PM function (e.g., cortical actin, lipid raft contents) CoQ10 and UBIAD1 impair laminin/α6 integrin activation and oncogenic functions in BC. This also limits integrin-dependent ferroptosis protection function. These data are consistent with previous data showing that laminin (e.g., LAMA5) and integrin (e.g., α6β4) are essential to protect BC cells by sustaining GPX4 expression51,57. This would be of particular importance for the luminal androgen receptor (LAR) subtypes of TNBC characterized by an upregulation of the GPX4 ferroptosis defense mechanism19.
Some questions remain unanswered such as why we can detect UBIAD1 effects on GPX4 levels only under a ferroptosis-inducing condition, but not under normal basal situations. It has been shown that cancer cells use the mevalonate pathway (MVA) and its intermediates, such IPP and DMAPP, to support selenoproteins and, thus, GPX4 synthesis74,75. We could therefore speculate that when UBIAD1 is low in TNBC, MVA intermediates are used to support the DMAPP-tRNASelCys-GPX4 branch while when UBIAD1 is high MVA intermediates are needed to support CoQ10 synthesis explaining the reduced levels of GPX4 and higher sensibility to ferroptosis in UBIAD1 high condition (Fig. 7). It would be interesting to metabolically test the hypothesis whether ferroptosis inducers heavily engaged the DMAPP-tRNASelCys-GPX4 branch response that would explain the higher sensitivity to RSL3 of cells expressing UBIAD1, being the metabolic mevalonate pathway of the cells fully rewired to synthetize CoQ10 instead of GPX4. In agreement with this hypothesis, it has been shown that soft conditions in BC increase the mevalonate IDI gene required to synthesize DMAPP to support selenoprotein/GPX4 synthesis27. FSP1 is also an alternative suppressor of ferroptosis. Interestingly, It has been shown that myristoylation of FSP1 mediates the recruitment of this protein to the plasma membrane and leads to ferroptosis inhibition60,76. Therefore, we can not exclude that by reducing the number of lipids rafts and altering PM cortical properties UBIAD1/CoQ10 also act to reduce the stability of FSP1 making cells, with already low levels of FSP1, more sensitive to ferroptosis.
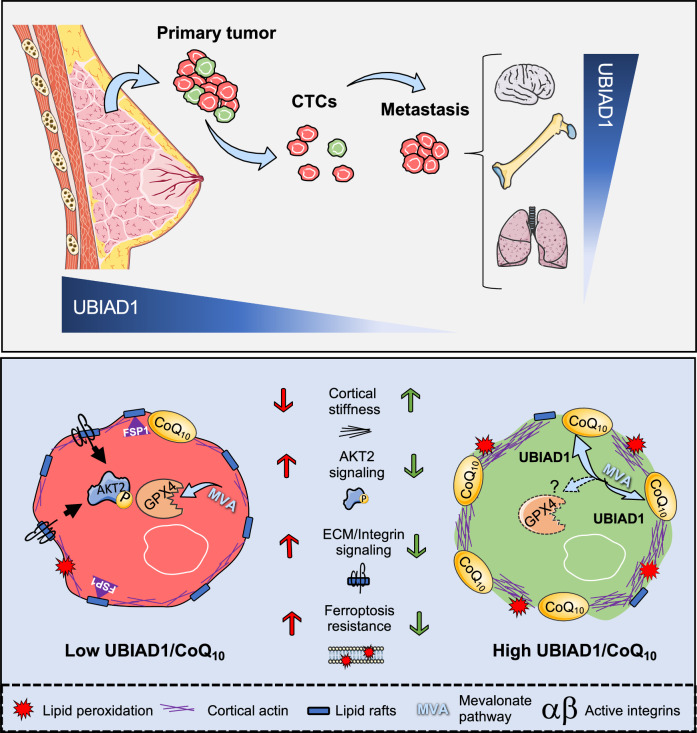
Fig. 7. CoQ10 and UBIAD1 expression impair BC spreading and survival via modification of plasma membrane-dependent signaling.
Expression of the CoQ10-biosynthetic enzyme UBIAD1 is reduced in highly aggressive BC, especially during the metastatic cascade. Moreover, the ability of colonize specific distant sites is associated to specific UBIAD1 expression levels. High levels of CoQ10 or expression of UBIAD1 in BC alters lipid raft plasma membrane fluidity that leads to increase cellular stiffness, reduced integrin activity and associated oncogenic AKT2 signaling. Furthermore, BC cells with high UBIAD1/CoQ10 levels show increased sensitivity to ferroptosis and lipid peroxidation possibly by decreasing GPX4 synthesis and FSP1 levels contributing to the impaired blood circulating cancer cell survival and reduced lung colonization. The images were generated using Smart Servier Medical Art under license CC BY4.0.
In conclusion, our study shows that UBIAD1 loss as well as reduced CoQ10 plasma membrane levels promote BC development and metastatic spreading and, possibly contribute to BC resistance to ferroptosis19. Therapeutic interventions able to stabilize UBIAD1 expression and/or increase CoQ10 levels (e.g., based on chronic CoQ10 supplementations) would act to increase the fluidity of the plasma membrane of tumor cells/CTCs and counteract ECM-mediated oncogenic signaling, representing an interesting therapeutic level of action in BC patients with aggressive disease course.
Methods
Ethics statement
All procedures involving animals for research purposes were approved by the Organismo preposto al benessere degli animali (O.P.B.A.) of the University of Padua and by the Italian Ministry of Health (Permit Numbers: 336/2019-PR).
Mouse model
We generated constitutive Ubiad1 Knockout mice by cloning a neomycin-resistance cassette into the ATG start codon in the first exon of Ubiad1, transfected by electroporation in mouse ES cells. Southern blotting identified 5 clones that carried the targeted allele, 2 of them were injected into C57BL/6J mouse blastocysts. We obtained heterozygous Ubiad1+/− mice using chimeras. Transgenic male C57BL/6J mice expressing the polyoma virus middle T antigen under the control of the mouse mammary tumor virus (MMTV) long terminal repeat (LTR) promoter (MMTV-PyMT, The Jackson Laboratory JAX #022974) were crossed with the Ubiad1+/− females to generate MMTV-PyMT; Ubiad1+/+ and MMTV-PyMT; Ubiad1+/− females. We monitored the female mice from birth to week 28. Similarly, transgenic male C57BL/6J mice expressing an activated rat c-neu oncogene (Erbb2) under the control of the mouse mammary tumor virus (MMTV) long terminal repeat (LTR) promoter (MMTV-NeuT/Erbb277), were crossed with the Ubiad1+/− females to generate MMTV-NeuT; Ubiad1+/+ and NeuT; Ubiad1+/− females. We monitored the female mice from birth to week 50. We obtained MMTV-NeuT mice in BALB/c background and were backcrossed in C57BL/6J for more than 10 generations before proceeding with the experiments. Ubiad1f/f mice carrying the second exon of Ubiad1 flanked by two Lox-P sites were custom-made from Cyagen in C57BL/6J background. C57BL/6J K14-Cre; Brca1 f/f; Tp53 f/f mice, alias KBP36, were kindly obtained from Prof. Bouwman at The Netherlands Cancer Institute. We crossed KBP mice with Ubiad1f/f mice to generate the KBP; Ubiad1f/f used in this study. Only males K14-Cre-positive were used for breeding78, while only females were used in the experiments. 6–8-week-old NOD/SCID common γ chain knockout (NSG; Charles River) female mice were used for xenografts experiments.
Mouse genotyping began with whole mouse embryos or from a small piece of the ear that was digested for 16 h at 56 °C with 0.2 mg/ml Proteinase K (Thermo Fisher Cat. #EO0492). After heat inactivation of the Proteinase K, samples were used for genotyping with the following primers: Ubiad1+/− FW 5′-CACGGGGCCGGCAATTTGGTCAAG-3′, RV 5′-GGGGGTCTCCAGCCCCACAGTAAGAGA-3′ and Neo-Cassette 5′-CAGCTCGGGCTCGACTAGAG CTTGC-3′ (400 bp KO allele and 500 bp WT allele); MMTV-PyMT IMR-15 5′- CAAATGTTGCTTGTCTGGTG-3′, IMR-16 5′-GTCAGTCGAGTGCACAGTTT-3′, IMR-384 5′-GGAAGCAAGTACTTCACAAGGG-3′ and IMR-385 5′-GGAAAGTCACTAGGAGCAGGG-3′ (200 bp internal CTRL and 556 bp PyMT allele); MMTV-NeuT/Erbb2 FW 5′-ATCGGTGATGTCGGCGATAT-3′, RV 5′-GTAACACAGGCAGATGTAGGA-3′, β-cas-FW 5′-GATGTGCTCCAGGCTAAAGTT-3′ and β-cas-RV 5′-AGAAACGGAATGTTGTGGAGT-3′ (525 bp internal CTRL and 225 bp NeuT allele); Ubiad1f/f LoxP1_FW 5′-CTCCTGAAGACCATTGTCACCTGTC-3′, LoxP1_RV 5′-GAACACAGAGCCACAGTAACAAACAC-3′, LoxP2_FW 5′-CACAAGGAGCAAGCAAGCCAGTA-3′ and LoxP2_RV 5′-GGGAGGATAGAGGGAAATGAGGTTA-3′; K14-Cre FW5′-CGATGCAACGAGTGATGAGGTTC-3′, RV5′-GCACGTTCACCGGCATCAAC-3′, oIMR8744_Int_ctrl5′-CAAATGTTGCTTGTCTGGTG-3′ and oIMR8745_Int_ctrl5′-GTCAGTCGAGTGCACAGTTT-3′; Brca1f/f LoxP1_FW 5′-TATCACCACTGAATCTCTACCG-3′, LoxP1_RV 5′-GACCTCAAACTCTGAGATCCAC-3′, LoxP2_FW 5′-TATTCTTACTTCGTGGCACATC-3′ and LoxP2_RV 5′-TCCATAGCATCTCCTTCTAAAC-3′; Tp53f/f LoxP1_FW 5′-CACAAAAACAGGTTAAACCCAG-3′, LoxP1_RV 5′-AGCACATAGGAGGCAGAGAC-3′, LoxP2_FW 5′-AAGGGGTATGAGGGACAAGG-3′ and LoxP2_RV 5′-GAAGACAGAAAAGGGGAGGG-3′. HOT FIREPol® Blend Master Mix (Solis BioDyne) was used for genotyping and Tannealing was set at 58 °C for all the reactions, except for Ubiad1+/− in which Tannealing was set at 62 °C.
Tissue-specific deletion of exon 2 of Ubiad1 was verified using the following primers: Ubiad1f/f LoxP1_FW 5′-CTCCTGAAGACCATTGTCACCTGTC-3′, LoxP2_FW 5′-CACAAGGAGCAAGCAAGCCAGTA-3′ and LoxP2_RV 5′-GGGAGGATAGAGGGAAATGAGGTTA-3′. HOT FIREPol® Blend Master Mix (Solis BioDyne) was used, Tannealing was set at 63 °C and time of extension was set at 20 s.
Mice were housed under conventional conditions in individual cages in the controlled room at 22 °C and 12 h light/dark cycle with ad libitum access to food (Mucedola S.r.l., 4RF21 standard diet) and water and were regularly monitored for weight and food consumption. The maximal permitted tumor volume of 4000 mm³ per single tumor has not been exceeded. Only female mice were used for the experiments as the gold standard model to study breast cancer.
Primary cells
Ex vivo primary BC PyMT (alias PyMT; Ubiad1+/+) or PyMT; Ubiad1+/− cells were isolated from mammary gland tumors of MMTV-PyMT (alias MMTV-PyMT; Ubiad1+/+) or MMTV-PyMT;Ubiad1+/− transgenic C57BL/6J female mice, respectively. The collected tumor was placed on ice in DMEM/F12 medium (Thermo Fisher Scientific Cat. #31331028) and processed in less than 1 h. A piece of 1 g was dissected from the tumor, cut into small pieces (<4 mm) and dissociated using Tumor Dissociation Kit (Miltenyi Biotec Cat. #130-096-730) according to manufacturer’s instructions (program for soft tumors). The suspension was applied to a 70 μm and a 40 μm cell strainer, then centrifuged at 300 × g for 10 min. Cells were resuspended in DMEM/F12 GlutaMAX™ medium containing 10% FBS (Carlo Erba Reagents Cat. #FA30WS1810500), 5 ng/ml EGF (PeproTech Cat. #AF10015), 1 µg/ml Hydrocortisone (Sigma Cat. #H0888), 100 ng/ml Cholera Toxin (Sigma Cat. #C8052), 5 µg/ml Insulin (Sigma Cat. #I6634), 100 µg/ml penicillin and 100 µg/mL streptomycin (Thermo Fisher Scientific Cat. #15140122) and placed in culture. The medium was changed regularly and any fibroblasts were removed by several passages of differential trypsinization79. Then, PyMT cells were maintained in DMEM/F12 GlutaMAX™ medium supplemented with 10% FBS, 100 µg/ml penicillin and 100 µg/mL streptomycin in a 37 °C incubator with a humidified atmosphere of 5% CO2. The cells were tested for mycoplasma infection using N GARDE Mycoplasma PCR Reagent Set (EuroClone Cat. #EMK090020) prior to use for experiments.
Cell lines
MCF7 (ATCC Cat. #HTB-22™), T-47D (ATCC Cat. #HTB-133™), BT474 (ATCC Cat. #HTB-20™), HCC202 (ATCC Cat. #CRL-2316™), HCC1143 (ATCC Cat. #CRL-2321™), MDA-MB-438 (ATCC Cat. #HTB-132™), MDA-MB-436 (ATCC Cat. #HTB-130™), MDA-MB-231 (ATCC Cat. #HTB-26™), Hs578T (ATCC Cat. #HTB-126™), JIMT-1 (Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures, Cat. #ACC-589) and HEK293T (ATCC Cat. #CRL-11268) cells were cultured in DMEM, high glucose, GlutaMAX™ Supplement, pyruvate (Thermo Fisher Scientific Cat. #31966047) containing 10% FBS (Carlo Erba Reagents Cat. #FA30WS1810500) and 100 µg/ml penicillin and 100 µg/mL streptomycin (Thermo Fisher Scientific Cat. #15140122). MCF10A (ATCC Cat. #CRL-10317™) cells were cultured in DMEM/F12 GlutaMAX™ medium (Thermo Fisher Scientific Cat. #31331028) supplemented with 5% Horse Serum (Sigma Cat. #H1138), 20 ng/ml human EGF (PeproTech Cat. #AF10015), 0.5 µg/ml Hydrocortisone (Sigma Cat. #H0888), 100 ng/ml Cholera Toxin (Sigma Cat. #C8052), 10 µg/ml Insulin (Sigma Cat. #I6634), 100 µg/ml penicillin and 100 µg/mL streptomycin. Cells were maintained in a 37 °C incubator with humidified atmosphere of 5% CO2. All cell lines were regularly tested for mycoplasma infection using N GARDE Mycoplasma PCR Reagent Set (EuroClone Cat. #EMK090020).
Public dataset analysis
Publicly available Copy Number Alteration (CNA), UBIAD1 mRNA level, PIK3CA/PTEN mutational status, associated clinicopathological, and survival data of 1904 breast cancer patients enrolled in the METABRIC cohort were retrieved from cBioPortal (http://www.cbioportal.org/).
Patients were stratified according to UBIAD1 CNA in Normal (without CNA), Amplified (with amplifications), and Deleted (with deep or shallow deletions).
The 19 patients carrying UBIAD1 amplifications were not included in the survival analysis as they represented a rare subpopulation not of primary interest.
Inferred ER and HER2 status from RNA expression status were used for the molecular classification of breast cancer to compensate for the lack of IHC information in a relevant fraction of patients.
Human breast cancer immunohistochemical analysis
IHC analysis was performed on formalin-fixed paraffin-embedded tissue microarrays (TMA) of breast cancer specimens from a retrospective consecutive cohort of ~2400 BC patients enrolled at the European Institute of Oncology, Milan, from 1997 to 2000, with complete long-term follow-up (~15 years)80. Expression of UBIAD1 was assessed using rabbit anti-human UBIAD1 custom-made from ProteoGenix (peptide sequence: cys-FRSQAFNKLPQRTAK), which was used at final concentration of 0.17 µg/ml following an antigen retrieval procedure in Citrate pH 6.0. Immunocomplexes were visualized by the EnVision+ HRP Rabbit (DAB+) kit (DAKO Cat. #K4007), acquired with the Aperio ScanScope system (Leica Biosystems). Informed consent was obtained from all subjects. For correlation analysis with clinical and pathological parameters, patients were stratified based on the intensity of UBIAD1 staining, as follows: UBIAD1Low (IHC score <2), UBIAD1Intermediate (IHC score >2 and <3) and UBIAD1High (IHC score ≥3).
Spontaneous tumor models
2-month-old mice were monitored once a week for palpable tumors. When tumors became palpable, the volumes of all the mammary tumors in each mouse were measured once a week till week 28 for MMTV-PyMT and week 50 for MMTV-NeuT mice. Tumor volume was measured with a caliper using the formula Tumor volume = (length × width2)/2, where length indicates the largest tumor diameter and width indicates the perpendicular tumor diameter. The total tumor volume per mouse was calculated as the sum of the volumes of all the tumors developed from the single mouse. At the experimental endpoint, tumors were counted, weighed, and collected for histological examination or quantitative PCR/Western blot analysis. Total tumor weight was calculated as the sum of the weights of all the tumors developed from a single mouse. 4-month-old KBP mice were monitored once a week for palpable tumors. As soon as a tumor became palpable in one female of the litter (between 29 and 32 weeks of age), all siblings were sacrificed, the 2nd−3rd (thoracic) and the 4th (abdominal) mammary glands were dissected and stained for whole mammary gland staining. Alternatively, tumor lesions were collected and dissociated in TRIzol for RNA extraction or digested with Proteinase K for DNA extraction. Genomic DNA was purified through phenol/chloroform method prior to proceed with PCR.
Xenografts tumor models
MDA-MB-231 and JIMT-1 cScr and UBIAD1OE cells were transduced with a lentivirus carrying the plasmid for Firefly Luciferase and eGFP expression81. Only eGFP+ cells were selected with FACS Aria™ IIIu Cell Sorting and expanded in vitro. 6–8-week-old NSG female mice were anesthetized and 5 × 105 eGFP+ MDA-MB-231 or 4 × 105 JIMT-1 cells resuspended in 100 μL of equal amount PBS: Matrigel (Growth Factor Reduced Matrigel, Corning Cat. #354230) were injected into the fat pad of one 4th mammary gland using a syringe with a 26G needle. Tumor growth was monitored once a week using a caliper till 21 days post-injection for MDA-MB-231 and 35 days post-injection for JIMT-1 cells. For intracardiac injection, 2 × 105 eGFP+ MDA-MB-231 or JIMT-1 were resuspended in 100 μL of PBS and injected into the left ventricle of anesthetized NSG females using a syringe with 30G needle82. Metastasis formation was evaluated once a week using bioluminescence imaging (BLI) till 21 days post-injection for MDA-MB-231 and 35 days post-injection for JIMT-1 cells. Bioluminescence images were collected with the IVIS Spectrum Imaging System (PerkinElmer). Ten minutes before imaging, animals were anesthetized with isoflurane/oxygen and administered i.p. with 150 mg/kg of D-luciferin (PerkinElmer) in PBS. Ventral and dorsal images were obtained for each animal and quantified through the region of interest (ROI). Living Image Software (PerkinElmer) was used to acquire and quantify the bioluminescence imaging data sets. At sacrifice, mouse blood was collected via cardiac puncture using a syringe with 25G needle containing 50 μL of 500 U/mL Heparin (Sigma). After centrifugation at 1100 × g for 5 min at RT, the upper layer of plasma was discarded and the underlying buffy coat layer containing WBC and Circulating Tumor Cells was collected. The buffy coat was then washed with Red Bood Cell Lysis Buffer (Sigma, Cat. #11814389001) according to manufacturer instructions to remove any remaining red blood cells. WBC and CTCs were resuspended in 3%BSA in PBS and sorted with FACSAria™ IIIu Cell Sorting to isolate only CTC-eGFP+ cells. After blood collection, the primary tumor and several organs were collected and the eGFP signal was evaluated in the whole organ using Xenogen IVIS Lumina II (Perkin Elmer) or using Leica Stereomicroscope connected to a fluorescent lamp. Organs and tumors were then cut into 2 mm pieces, resuspended in 1 mg/mL Collagenase A (Sigma, Cat. #11088793001) in DMEM (Thermo Fisher), and dissociated at 37 °C for 30 min. After red blood cells were lysed, cells were resuspended in 3%BSA in PBS and sorted with FACSAria™ IIIu Cell Sorting to isolate only eGFP+ cancer cells. Isolated cells were used either for protein extraction using home-made RIPA supplemented with 1x halt protease inhibitor cocktail (Roche Cat. #04693116001) and phosphatase inhibitor (Sigma Cat. #4906845001) or for RNA extraction. Depending on the number of eGFP+ cells isolated, either TRIzol™ Reagent (ThermoFisher Scientific Cat. #15596018) or NucleoSpin RNA XS Kit (Carlo Erba, Cat. #FC140902L) was used according to manufacturer instructions.
Mouse treatment
N-Acetyl-L-cysteine (NAC; Sigma Cat. #A9165) and Vitamin K2 (Sigma Cat. #V9378) were diluted in PBS while CoQ9 (Sigma Cat. #C9538) was diluted in sunflower oil. NAC, Vitamin K2 and CoQ10 were injected at a dose of 150 mg/kg. Pregnant females were injected daily between E2.5 and E9.5 intraperitoneally. Pups were genotyped at P0.
CoQ9 measurement from mouse embryos and tumors
Samples for HPLC-MS analysis were prepared as described previously30. CoQ9 levels were detected by HPLC-MS using CoQ6 (Avanti Cat. #900150 O) as an internal standard. Protein concentration in each sample was measured by Pierce™ BCA Protein Assay (ThermoFisher Scientific Cat. #23225) and the quantity of CoQ9 was normalized on this protein quantity.
Whole mount mammary gland staining
Mouse mammary gland whole mount preparation and staining was performed as previously described83. In particular, 2nd−3rd (thoracic) and 4th (abdominal) mammary glands from mice aged 29 to 32 weeks were fixed in Carnoy’s solution for 24 h, and staining was performed using a Carmine dye solution (Sigma, C1022) and aluminum potassium (Sigma, Cat. #A7167) in deionized H2O for 3 days. Slides were kept in Xylene while pictures were taken using a Leica MZ16F stereomicroscope. The area of neoplastic lesions was calculated with ImageJ and normalized over different mouse litters.
Cell viability assay
1 × 104 MDA-MB-231, MCF7, MDA-MB-436, Hs578T cells/well, and 5 × 103 ex vivo PyMT cells/well were seeded in 96-well plates and allowed to adhere for 4 h or O/N. Cells were treated for 24 h with indicated compounds at the indicated concentrations. In the case of combinatory treatment, cells were allowed to adhere for 4 h and then, simultaneously treated for 24 h with medium containing 20 μM CoQ10 (Sigma) and 0.45 μM RSL3 (Sigma Cat. #SML2234). Erastin (Sigma Cat. #329600), RSL3, and Doxorubicin hydrochloride (Sigma Cat. #D2975000) were solved in DMSO (Sigma Cat. #D2650), Menadione (Sigma Cat. #M5625) in 96% ethanol and CoQ10 in DMF (Sigma Cat. #227056). Then, the medium was removed and cells were washed 2 times with tap water. Then, 50 µl of 0.5% (w/v) Crystal Violet staining solution (Sigma Cat. #C6158) in 20% methanol was added to each well and incubated for 20 min at room temperature on a bench rocker with a frequency of 20 oscillations per minute. The plates were washed four times with tap water and allowed to dry. Crystal Violet was then dissolved by adding 200 µl of methanol to each well and incubating for 20 min at room temperature on a bench rocker with a frequency of 20 oscillations per minute. Next, the absorbance at 595 nm was analyzed with a plate reader. Alternatively, pictures of each well were taken using a Leica DMI4000 or Nikon AZ100 microscope, and crystal violet-stained cells were counted with ImageJ software.
Cell treatments
Cells were treated with 20 μM CoQ10 (Sigma Cat. #C9538), CoQ6 (Avanti Polar Lipids, Inc Cat. #900150), CoQ2 (Sigma Cat. #C8081) or DMF (vehicle, Sigma Cat. #227056), 20 μM water-dispersible CoQ10H2 (alias ShiroQ™, 40% Kaneka Ubiquinol water-soluble powder) or Placebo (vehicle), Idebenone (Tocris Bioscience Cat. #3002), Y27632 (Adooq Bioscience Cat. #A11001), ML7 (Tocris Bioscience Cat. #4310), NAC (Sigma Cat. # A9165) or equal volume of ddH2O. Cells were treated for 14 days (unless specified otherwise) changing medium every two days.
4-OHT-treated cells were seeded in complete medium in 6-well plates and let adhere overnight. The following day, cells were treated with 10 μM 4-OH-Tamoxifen (Sigma Cat. #H7904) in DMSO or with Vehicle (DMSO) for 48 h before protein and RNA collection.
Before treatment with 17β-Estradiol, cells were seeded and grown for 48 h in DMEM without phenol-red (Thermo Fisher Scientific, Cat. #A1443001) supplemented with 4.5 g/L D-glucose (Sigma, Cat. #G7021), 1 mM Sodium Pyruvate (Thermo Fisher Scientific, Cat. #11360070), 2 mM L-Glutamine (Thermo Fisher Scientific, Cat. #25030024), 10% Fetal Bovine Serum charcoal stripped (Thermo Fisher Scientific Cat. #A3382101) and 100 µg/ml penicillin and 100 µg/mL streptomycin (Thermo Fisher Scientific Cat. #15140122). In the same medium composition, cells were treated for 24 h with 10 nM 17β-Estradiol (Merk, Cat. #2001128945) in EtOH or with Vehicle (EtOH), before proceeding with RNA extraction.
Cell proliferation assay
3000 cells per well were seeded in 96-well plates and stained after 24 h, 48 h, 72 h, 96 h, and 120 h intervals using Crystal Violet staining solution (Sigma Cat. #C6158). The medium was removed, and cells were washed twice with tap water at each interval. Then, 50 µl of 0.5% (w/v) Crystal Violet staining solution in 20% methanol was added to each well and incubated for 20 min at room temperature on a bench rocker with a frequency of 20 oscillations per minute. The plates were washed four times with tap water and allowed to dry. Crystal Violet was then dissolved by adding 200 µl of methanol to each well and incubating for 20 min at room temperature on a bench rocker at a frequency of 20 oscillations per minute. Next, we determined the value of absorbance at 595 nm with a plate reader. For cell proliferation assay in presence of CoQ10, 10,000 cells per well were seeded in 96-well plates and treated with DMF (Vehicle) or 20 μM CoQ10 (Sigma Cat. #C9538). Medium with treatment was replaced every 48 h.
Viral transduction and generation of stable lines
The coding sequence of the human UBIAD1 gene (NM_013319.3) was cloned into the pLenti-CMV-Neo-DEST vector (Addgene Cat. #17392) using the Gateway cloning system. Recombinant lentiviruses carrying the pLenti-CMV-Neo-DEST-UBIAD1, pLenti-CMV-Neo-GFP-DEST (Addgene Cat. #19732) or pLenti-CMV-Neo-DEST-empty were produced by co-transfecting HEK293T cells with a mixture of plasmid DNA consisting of pMD2.G (VSV-G envelope; Addgene Cat. #12259), pMDLg/pRRE (Gag/Pol; Addgene Cat. #12251), and pRSV-Rev (Rev; Addgene Cat. #12253) using Lipofectamine™ 2000 Transfection Reagent (Life Technologies Cat. #11668019) according to the manufacturer’s recommendations. Supernatants containing lentivirus were collected, passed through 0.45-µm filters, and stored at −80 °C. Lentiviral particles were quantified using Lenti-X™ p24 Rapid Titer Kit (Takara Cat. #632200). Similarly, lentiviruses carrying lentiMPH v2 plasmid (Addgene Ref.# 89308) and lenti_SAM_v2 plasmid (Addgene Ref.# 75112) containing scramble-gRNA (5′-GGCACTCACATCGCTACATCAGTTT-3′) or UBIAD1-gRNA (5′-GGCCAGAGGCGGGGAGAAGGGGTTT-3′) for the generation of CRISPR-edited lines were produced.
Stable lines overexpressing UBIAD1 were generated by transducing MDA-MB-231, MDA-MB-436, Hs578T and ex vivo PyMT cells with 5 MOI of lentivirus. Transduced cells were selected with 1.5 mg/ml G418 (Gibco™ Cat. #10131019) until the complete death of MOCK control (not transduced cells). All the following experiments were performed in culture medium without G418. Alternatively, MDA-MB-231 stable UBIAD1 overexpressing cells were generating using CRSIPR-dCas9 technology: first cells were transduced with lentiMPH v2 and selected with 100 μg/mL Hygromycin b (Santa Cruz Cat. #sc-29067) and second with lenti_SAM_v2_UBIAD1-gRNA or lenti_SAM_v2_scamble-gRNA and selected with 2 μg/mL Blasticidine (Sigma, Cat. #15205)84.
For the generation of the Luc-eGFP lentivirus, the following lentiviral packaging vectors were used: pMDLg/pRRE, pRSV-Rev, pMD2.VSVG, and pADVantage. The transfer vector #945.pCCL.sin.cPPT.SV40ployA.eGFP.- nCMV.hPGK.deltaLNGFR.Wpre is a self-inactivating (SIN) HIV-derived vector85, which carries a minCMVPGK divergent bidirectional promoter driving the simultaneous expression of two genes in antisense orientation. The transfer vectors used in this study were modified to express two reporter genes: the Luciferase sequence (fluc) under the control of the hPGK promoter, and the eGFP (enhanced Green Fluorescent Protein) under the control of minCMV. fluc sequence were synthetized by GeneArt, Life Technologies (Regensburg, Germany).
AKT2 overexpression
Cells were transfected via reverse-transfection using Lipofectamine 2000 (Invitrogen, Cat. #11668). Briefly, 1.5 × 106 cells were mixed with Lipofectamine and 2 μg of pcDNA3-empty, pcDNA3-AKT2-wt, and pcDNA3-Myr-AKT2 in Pen/Strep- and FBS-free medium and seeded in 10 cm petri-dish86. After 6 h, cells were washed with 1x PBS and full culture medium was added. 48 h-post transfection, cells were trypsinized and seeded for invasion assay. AKT2 overexpression was confirmed by Western blot analysis using anti-human AKT2 antibody (1:1000, GeneTex Cat. #GTX128457, RRID:AB_2750992).
Fluorescence recovery after photobleaching (FRAP)
Cells were seeded in µ-Dish 35 mm, high (Ibidi, Cat. #81156) in complete medium and observed overnight. The following day, cells were washed with PBS and incubated with 1 μg/mL DiI-C16(3) solution (ThermoFisher Scientific Cat. #D-384) in PBS for 1 min at RT. After three washes with PBS, cells were maintained in warm HBSS (ThermoFisher Scientific, Cat. #14025050). Cells were kept at 37 °C using a heating chamber for the entire experiment. FRAP was performed using a 63× oil-immersion objective in a confocal microscope (Leica SP8). 60–80% of photobleaching of a defined area (3 × 3 μm2) of the labeled membrane was obtained with four 100% 552 nm laser scans for a total duration of 4 s. The fluorescence intensity was recorded before photobleaching and after taking pictures every 1 sec over a period of 3 min. After background normalization, the fluorescence was plotted over time to generate a fluorescence recovery curve. Unwanted photobleaching during imaging was negligible with the laser power set.
Membrane fluidity assay using lipophilic pyrene probes
Cells were seeded in 96-well Black/Clear Flat Bottom TC-treated Imaging Microplates (Corning Cat. #353219) in standard medium. When reached 80–90% confluency, cells were stained using the Membrane Fluidity Kit (Abcam, Cat. #ab189819) according to manufacturer’s instructions. In brief, a solution of 5 μM Fluorescent Lipid Reagent with 0.08% Pluronic F127 in Perfusion Buffer was used to stain the cells at 25 °C for 1 h in the dark. Cells were then washed two times with PBS. In PBS, the fluorescence emission between 390 nm and 510 nm (5 nm step size) was recorded from the bottom of the plate after an excitation at 350 nm (±10 nm) using a Tecan Plate reader Infinite 200 PRO. The mean of fluorescent intensity at 390–395 nm or 475–485 nm was considered for probe monomers or excimer, respectively. After background (cells without staining) subtraction, the ratio of excimers over monomers fluorescence intensity was calculated as a measure of membrane fluidity. Increased membrane fluidity leads to pyrene probes forming excimers instead of monomers87. Treatment for 48 h with 400 mM EtOH was used as the positive control.
Cell adhesion assay
Cell adhesion assay was performed to analyze integrin-mediated binding to different extracellular matrix components. Fibronectin (Sigma Cat. #ECM001), Collagen I (Roche Cat. #11179179001), Collagen IV (Sigma Cat. #C7521), Vitronectin (Sigma Cat. #SRP3186), Laminin from human placenta (Sigma Cat. #L6274), and Poly-L-lysine (PLL; Sigma Cat. #P4707) were solubilized according to manufacturer’s instructions and 100 μl/well were used to coat 96-well plates for 1 h at 37 °C. Plates were coated with 1.25 μg/ml Fibronectin, 0.4 μg/ml Collagen I, 0.2 μg/ml Collagen IV, 4 μg/ml Vitronectin, and 2.5 μg/ml Laminin. 12.5 μg/ml Poly-L-lysine (PLL) was used as integrin-independent attachment control. After coating, unspecific binding was reduced by blocking the plate with 2.5 mg/ml of heat-denatured BSA (Sigma Cat. #10735094001) in water for 2 h at 37 °C. The blocking solution was removed and the wells were washed twice with serum-free DMEM. Cells were detached using an enzyme-free buffer containing 2 mM EDTA in PBS pH 7.4 and were seeded 5 × 104 cells/well in 100 μl of serum-free DMEM. Cells were allowed to adhere for 30 min in a 37 °C incubator with a humidified atmosphere of 5% CO2 before staining using Crystal Violet staining protocol. Briefly, cells were washed twice with tap water before adding 50 µl of 0.5% (w/v) Crystal Violet staining solution (Sigma) in 20% methanol to each well. The plates were incubated for 20 min at room temperature on a bench rocker with a frequency of 20 oscillations per minute. Wells were washed four times with tap water and allowed to dry. Crystal Violet was then dissolved by adding 200 µl of methanol to each well and incubating for 20 min at room temperature on a bench rocker with a frequency of 20 oscillations per minute. Next, the absorbance at 595 nm was analyzed with a plate reader.
Similarly, 6-well plates were coated with 7.5 μg/ml Laminin from human placenta (Sigma) or 7.5 μg/ml Poly-L-lysine (Sigma) and blocked as described above. 5 × 105 cells were seeded in 2 ml of serum-free medium and allowed to adhere for 30 min in a 37 °C incubator with a humidified atmosphere of 5% CO2. Then, samples were prepared for Western blotting as described in “Methods” details.
Staining of lipid rafts
To visualize lipid rafts in living cells we used Vybrant™ Alexa Fluor™ 555 Lipid Raft Labeling Kit (Thermo Fisher Scientific Cat. #V34404) following the manufacturer’s instructions. Briefly, 1.5 × 104 cells/well were seeded on glass coverslips placed in 24-well plates with DMEM GlutaMAX™ medium (Thermo Fisher Scientific) supplemented with 10%FBS and allowed to adhere overnight. Then, the medium was removed and cells were incubated with 350 μl/well of CT-B conjugate in medium w/o FBS for 10 min at +4 °C with gentle shaking. After the incubation, cells were washed twice with PBS and incubated with 350 μl/well of anti-CT-B antibody and Hoechst 33342 (1:2000, Thermo Fisher Scientific Cat. #62249) in medium w/o FBS for 15 min at +4 °C with gentle shaking. Cells were washed three times with PBS and fixed with 4% formaldehyde at room temperature for 15 min. After three additional washes with PBS, the glass coverslips were mounted with Mowiol® 4-88 (Sigma Cat. #81381). Samples were visualized using Leica SP8 DLS microscopy and images were processed using Leica LASX software (https://www.leica-microsystems.com/). The fluorescence intensity was measured with ImageJ software (https://imagej.nih.gov/ij/).
Real time q-PCR
Harvested tumors were washed in PBS and radially divided into four pieces. One piece was resuspended in TRIzol™ Reagent (Thermo Fisher Scientific Cat. #15596018) and homogenized using IKA T-10 Basic Homogenizer. Cells were rinsed with ice-cold PBS and either TRIzol™ Reagent or NucleoSpin RNA XS Kit (Carlo Erba, Cat. #FC140902L) was directly added to the plate. After sample collection, RNA isolation was performed according to the manufacturer’s instructions. RNA concentration and purity were determined using a NanoDrop spectrophotometer at A260 and A280/260, respectively. RNA was reverse transcribed to cDNA using a High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific Cat. #4368814). Quantitative PCR (q-PCR) was carried out with 5x HOT FIREPol® EvaGreen® qPCR Supermix (Solis Cat. #08-36-00020) on QuantStudio™ 5 Real-Time PCR System (Thermo Fisher Scientific). Transcripts levels were normalized to β-Actin and reported as relative to a reference sample. All primers used in q-PCR have an efficiency close to 100%, and their sequences are listed below. QuantStudio™ Design & Analysis Software (www.thermofisher.com) was used for performing q-PCR data processing. Mouse Ubiad1 FW: 5′-CACAGGAGGAATTGGATTCAAGT-3′; Mouse Ubiad1 RV: 5′-GGATGGCGTAGATTAGAGGGAAG-3′; Mouse β-Actin FW 5′-GTACTCTGTGTGGATCGGTGG-3′; Mouse β-Actin RV: 5′-AAACGCAGCTCAGTAACAGTCC-3′; Human UBIAD1 FW: 5′-CACTTGGCTCTTATCTACTTTGGA-3′; Human UBIAD1 RV: 5′-GTCTCCCAGAGCCACGTACTTG-3′; Human β-ACTIN FW: 5′-GATGGAGTTGAAGGTAGTTTCGT-3′; Human β-ACTIN RV: 5′-GCGGGAAATCGTGCGTAGCATT-3′; Mouse Acsl4 FW: 5′-GGCTGACAGAATCATGTGGTG-3′; Mouse Acsl4 RV: 5′-GAACTGTATAACCACCTTCCTGC-3′; Human MYC FW: 5′-CTCTGAAAGGCTCTCCTTG-3′; Human MYC RV: 5′-CGTAGTCGAGGTCATAGTTC-3′; Human BCL2 FW: 5′-TGGATGACTGAGTACCTGAA-3′; Human BCL2 RV: 5′-AGGAGAAATCAAACAGAGGC-3′; Human CCND1 FW: 5′-TCGGTGTCCTACTTCAAATG-3′; Human CCND1 RV: 5′-CTCGCACTTCTGTTCCTC-3′.
Western blotting
Cells were rinsed with ice-cold PBS and collected from culture plates by scraping. Cells were lysed in ice-cold homemade RIPA Lysis Buffer (20 mM Tris-HCl, pH 7.4, 200 mM NaCl2, 2% sodium deoxycholate, 0.2% SDS, 2% NP40) supplemented with 1x halt protease inhibitor cocktail (Roche Cat. #04693116001) and phosphatase inhibitor (Sigma Cat. #4906845001) for 20 min, and soluble lysate fractions were clarified by centrifugation at 20,000 × g for 10 min at 4 + C.
Mouse embryos were resuspended in ice-cold RIPA Lysis and Extraction Buffer (Life Technologies) supplemented with 1x halt protease and phosphatase inhibitor cocktail (Roche and Sigma), gently homogenized with a pestle and incubated for 30 min on ice. Soluble lysate fractions were clarified by centrifugation at 20,000 × g for 10 min.
Protein concentrations were determined with Bradford (Bio-Rad Cat. #5000006). Twenty micrograms of protein per well were loaded in SDS-PAGE (4–12% pre-cast gels Invitrogen) and transferred to nitrocellulose membrane or 100% methanol-activated PVDF membrane. To check the proper transfer, membranes were stained with Ponceau (EuroClone Cat. #EMR194500). After Ponceau removal with several washes in 1x TBS-T, membranes were blocked with 5% milk in 1x TBS-T for 1 h and then incubated with primary antibodies overnight at 4 °C in 1% BSA. For anti-human UBIAD1 (Santa Cruz Biotechnology), membrane was blocked with 5% BSA and then incubated with primary antibody 3 h at RT. Following the incubation, all membranes were washed three times before incubation with the appropriate horseradish peroxidase (HRP)-conjugated anti-rabbit immunoglobulin G (IgG) (Sigma Cat. #A6154, RRID: AB_258284), HRP-conjugated anti-mouse IgG (Sigma Cat. #A4416, RRID: AB_258167). The bands were visualized and acquired using ChemiDoc™ Imaging System (Bio-Rad). The band intensities on developed films were quantified using Image Lab software (Bio-Rad Laboratories; www.bio-rad.com). Data were normalized on loading control (ACTIN or VINCULIN). The following primary antibodies were used: anti-human and mouse UBIAD1 (1:1000, Atlas Antibodies Cat. #HPA044862, RRID: AB_2679117), anti-human VINCULIN (1:5000, Sigma Cat. #V9131, RRID:AB_477629), anti-human and mouse β-ACTIN (1:5000, Sigma Cat. #A5316, RRID:AB_476743), anti-human p-FAK (Tyr397) (1:1000, GeneTex Cat. #GTX129840, RRID:AB_2886103), anti-human FAK (1:200, Santa Cruz Cat. #sc-558, RRID:AB_2300502), anti-human AKT(pan) (1:1000, Cell Signaling Cat. #4691, RRID:AB_915783), anti-human p-AKT1 (Ser473) (1:1000, Cell Signaling Cat. #9018, RRID:AB_2629283), anti-human p-AKT2 (Ser474) (1:1000, Cell Signaling Cat. #8599, RRID:AB_2630347), anti-human GSK-3β (1:1000, Cell Signaling Cat. #9832, RRID:AB_10839406), anti-human p-GSK-3β (Ser9) (1:1000, Cell Signaling Cat. #9323, RRID:AB_2115201), anti-human ERK1/2 (1:1000, Santa Cruz Cat. #sc-514302, RRID:AB_2571739), anti-human p-ERK1/2 (Thr202/Tyr204) (1:1000, Cell Signaling Cat. #9101, RRID:AB_331646), anti-human FSP1/AMID (1:1000, Santa Cruz Cat. #sc-377120), anti-human GPX4 (1:1000, Abcam Cat. #41787, RRID:AB_941790), anti-human UBIAD1 (1:250, Santa Cruz Biotechnology Cat. #sc-377013, clone H8, Lot. #I2313), anti-human HMGCR (1:500, Santa Cruz Biotechnology Cat. #sc-271595, clone C-1, RRID: AB_10650274) and anti-human ERα (1:500, Santa Cruz Biotechnology Cat. #sc-542, clone MC-20, RRID:AB_631470).
For Western blot analysis, human cell lines and mouse ex vivo cells were seeded at 80% confluency and allowed to adhere for 4 h. Then, cells were treated for 24 h with Vehicle or 0.9 μM ERASTIN, 0.45 μM RSL3 for human cells and 50 nM RSL3 for mouse ex vivo cells, before proceeding with protein sample collection.
Invasion assay
Cell invasion was assessed using a permeable support for a 24-well plate with an 8.0 µm pore size transparent PET membrane (Falcon Cat. #353097). The membrane was coated with 70 μl of ice-cold Growth Factor Reduced Matrigel (Corning Cat. #354230) diluted 1 mg/ml in DMEM w/o FBS and incubated overnight at 37 °C to allow the Matrigel to solidify. The next day, 2 × 105 cells were added to the upper chamber in 200 μl of medium without FBS and the lower chamber was filled with 650 μl of medium supplemented with 10%FBS to create a gradient of FBS. In the case of treatment with Vehicle, NAC, CoQ10, Y27632, and ML7, compounds were added at the indicated concentrations to the upper chamber. Cells were allowed to invade for 24 h in a 37 °C incubator with humidified atmosphere of 5% CO2. The chambers were then removed, washed twice in PBS and fixed with 4% PFA for 2 min at RT. After two additional washes in PBS, the membranes were stained with 0.5% (w/v) Crystal Violet staining solution (Sigma) in 20% methanol for 20 min at RT. Finally, the excess staining solution was removed with several washes in PBS and the not invading cells were gently removed from the upper surface of the membrane using a cotton swab. Membranes were let to air-dry before proceeding with imaging. Invading cells were photographed using Leica DFC300 FX digital camera (software Leica Application Suite V4.13) connected to Nikon AZ100 microscope and counted with ImageJ software.
Soft agar assay
12-well plates were coated with a layer of 600 µl phenol red- and serum-free DMEM (Thermo Fisher Scientific Cat. #11594416) containing 1% low-gelling temperature agarose (Sigma Cat. #A9414). Upon solidification, 1 × 104 cells/well were seeded in 400 µl phenol red-free DMEM containing 0.6% agarose and 4% FBS. Upon solidification, 1 ml DMEM containing 10% FBS was added on top of the agarose layers. The medium was replaced once a week and after 4 weeks the medium was substituted with a solution containing 0.5% (w/v) Crystal Violet staining solution (Sigma) in 20% methanol diluted 1:100 in PBS for 2 hrs at RT. The staining solution was removed, and the agarose layers were washed twice with PBS before imaging with a Leica MZ16F stereomicroscope. The number and area of the colonies were calculated with ImageJ software (https://imagej.nih.gov/ij/).
Tumorsphere formation assay
Tumorsphere formation assay was performed as previously described88. Cells were trypsinized and centrifuged at 200 × g for 5 min at RT. The supernatant was removed and cells were resuspended in 5 ml of tumorsphere medium: DMEM/F12 GlutaMAX™ medium (Thermo Fisher Scientific) containing 100 µg/ml penicillin, 100 µg/mL streptomycin, 20 ng/ml recombinant human epidermal growth factor (EGF; PeproTech Cat. #AF10015), 10 ng/ml recombinant human basic fibroblast growth factor (bFGF; Sigma Cat. #F3685) and 1x B27 supplement (Gibco Cat. #7504044). The cell suspension was passed through a 40 µm cell strainer to obtain a single-cell suspension. Cells were counted and 2 × 104 cells/well in 500 µl of tumorsphere medium were seeded in ultra-low attachment 24-well plates (Corning Cat. #3473). After 5 days, tumorspheres were photographed using Leica DMI4000 microscope and counted with ImageJ software. Only tumorspheres greater than 100 µm were considered.
Immunofluorescence
Tumor cryosections (10 µm-thick) were fixed in 4% PFA for 10 min, blocked in PBS containing 5% goat serum (Merk Millipore Cat. #S26), 1% BSA and 0.3 M glycine for 30 min. Sections were then incubated with anti-mouse CD31 (1:20, Dianova Cat. #DIA-310, RRID:AB_2631039) and anti-mouse Ki-67 (1:100, Cell Signaling Cat. #9129-S, RRID:AB_2687446) primary antibodies and APC-Cy7 anti-mouse CD45 (1:100, BD Biosciences Cat. #557659, RRID:AB_396774) antibody overnight at 4 °C. After three washes with PBS, sections were incubated for 1 h at RT with the corresponding Alexa Fluor conjugated secondary antibodies (5 µg/ml, Thermo Fisher Scientific) and 300 nM DAPI (Thermo Fisher Scientific Cat. #D1306) for nuclear staining. Sections were mounted and then analyzed using Leica SP8 DLS microscopy. For micro-vessel density and mean vessel area measurements, CD31 staining was evaluated in four randomly chosen fields per tumor sample. Quantification of Ki-67+ and CD45+ cells was assessed by counting all the stained cells of the tumor section and normalizing the number for the total tumor section area.
For cultured cell immunofluorescence, cells were seeded either on glass coverslips placed in a 24-wells plate or in 15 μ-Slide 8 well (Ibidi Cat. #80826). For the analysis of blebs and lamellipodia, cells were seeded after coating with 2.5 μg/ml of Laminin from human placenta (Sigma Cat. #L6274) and 3 h after seeding, samples were processed as follows. Cells were washed three times with PBS and fixed with 4% paraformaldehyde for 15 min at RT. Only for MDA staining, cells were not permeabilized and the use of detergents was avoided also in the following steps. The fixed cells were then permeabilized with 0.2% Triton X-100 in 1× PBS for 10 min at RT and blocked with 3% goat serum + 3% BSA + 1.5 mg/ml glycine in PBS for 1 h at room temperature. Cells were incubated overnight at +4 °C with anti-human MDA (1:100, Abcam Cat. #ab6463, RRID: AB_305484), anti-human Paxillin (1:400, Transduction Laboratories Cat. #P13520), anti-human FAK (1:50, Santa Cruz Cat. #sc-558, RRID:AB_2300502) diluted in PBS + 3%BSA. Filamentous actin (F-actin) was stained with Phalloidin (1:300, Sigma-Aldrich Cat. #P1951, RRID:AB_2315148) diluted in PBS for 10 min at RT. After three washes with PBS, cells were incubated for 1 h at room temperature with the corresponding Alexa Fluor conjugated secondary antibodies (0.75–2 µg/ml, Thermo Fisher Scientific) in 0.2% BSA/PBS and 300 nM DAPI (Thermo Fisher Scientific) for nuclear staining. After three additional washes with PBS, coverslips were mounted with Mowiol® 4-88 (Sigma) and then visualized using Leica SP8 DLS microscopy. Images were processed using Leica LASX software and the fluorescence intensity was measured with ImageJ software. The percentage of cells without any protrusions, with lamellipodia only, or with bleb structures was calculated using samples of cells stained for PAX and Phalloidin, or FAK and Phalloidin, by manually counting the number of cells in each group. Cells were considered to have blebs if two or more bleb structures were visible along the plasma membrane.
Lipid peroxidation measurement
Lipid peroxidation was evaluated in adherent living cells using cholesteryl BODIPY® 576/589 C11 (Thermo Fisher Scientific Cat. # C12680). 1.5 × 104 cells/well were seeded in the standard medium in 96-well Black/Clear Flat Bottom TC-treated Imaging Microplates (Corning Cat. #353219) and allowed to adhere for 4 h. Alternatively, 5 × 105 cells/well were seeded in 6-well plates and allowed to adhere overnight. Then, cells were treated with 0.45 μM RSL3 (Sigma) or 0.1% DMSO (NT) for 12 or 24 h. The medium was removed and cells were incubated for 15 min with 3 μM BODIPY® 576/589 C11 and Hoechst 33342 (1:2000, Thermo Fisher Scientific Cat. #62249) at 37 °C in the dark. The staining solution was removed and wells were gently washed three times with PBS. The fluorescence signal was analyzed (1) using an Infinite 200 PRO plate reader (Tecan) with the Hoechst Ex360/Em465, Oxidized BODIPY Ex485/Em535, and Reduced BODIPY Ex535/Em590 filters and data were expressed in Oxidized/Reduced BODIPY normalized on Hoechst intensity signal, or (2) by flow cytometry analysis using BD FACSCanto™ II Cell Analyzer (BD Biosciences) with non-treated cells without Bodipy C11 staining used as blank and the ratio between oxidized (FITC-A) and reduced (PE-A) Bodipy C11 was measured (a minimum of 20,000 events were obtained per sample).
To evaluate the lipid peroxidation in fixed cells, immunofluorescence staining using anti-MDA an antibody (Abcam) was performed as described in the relative section. Lipid peroxidation was induced by treating the cells for 24 h with 2 μM ERATIN (Sigma) or 0.1%DMSO (NT) before fixation.
Growth in attachment-free condition
6-well plates were coated with 1 ml of 1% low-gelling temperature agarose (Sigma Cat. #A9414) in standard medium w/o FBS and the agarose was let solidify for 1 h at +4 °C. Then, 1 × 105 cells/well were seeded on top of the agarose layer in 2 ml of complete growth medium and allowed to grow in suspension for 2–6 days. Before resuspending the cells in TRIzol™ Reagent (Thermo Fisher Scientific Cat. #15596018), the cell suspension was collected, cells were pelleted by centrifugation at 300 × g for 5 min at +4 °C and the pellet was washed three times with cold PBS. Then, we proceeded with RNA extraction as described.
Atomic force microscopy (AFM)
MDA-MB-231, MCF10A, MCF7, and MDA-MB-468 cells were seeded in a complete growth medium in 35 mm dishes (Falcon Cat. #353001) to have 30–40% confluency the day after. Immediately before the analysis, the culture medium was replaced with 3 mL of PBS, after 2 previous washes with PBS. AFM force indentation measurements were performed using a XE Bio - AFM (Park Systems, South Korea) equipped with an inverted microscope (Nikon Eclipse Ti).
The force-displacement curves were acquired using PPP-CONTSCR-10 pyramidal tips mounted on Si3N4 cantilevers with a nominal spring constant of ~0.2 N/m (NanoSensors, Neuchatel, Switzerland). Values for the cantilever spring constants were obtained by the manufacturer before use and were used in all calculations. Before each test, the sensitivity of the AFM photodetector (optical lever sensitivity) was calculated by measuring the slope of the force-distance curve acquired on a silicon standard. All experiments were performed at room temperature.
Indentation curves were acquired by approaching the cell surface at a rate of 5 μm/s and producing an indentation with a depth of 0.5 μm. For each cell, at least two-force curves were collected.
Young’s modulus was calculated by applying a fit of the Hertz model to each force-distance curve, assuming a Poisson ratio of 0.5.
RNA-seq analysis
MDA-MB-231 cells stably transduced with lenti_SAM_v2_UBIAD1-gRNA or lenti_SAM_v2_scamble-gRNA were seeded in a complete medium to obtain 70% confluency the day after. Cells were washed three times with autoclaved PBS and resuspended in TRIzol™ Reagent (Thermo Fisher Scientific Cat. #15596018). Four biological replicates per condition were collected. Total RNA was extracted according to TRIzol™ Reagent manufacturer’s instructions. 1 μL of DNase I (Thermo Fisher Scientific, Cat. #18068015) was used for every 1 μg of isolated RNA to digest any remaining genomic DNA. After DNase I inactivation, RNA Clean & Concentrator-5 kit (Euroclone, Cat. #R1015) was used to purify the RNA samples. RNA concentration and purity were determined using a NanoDrop spectrophotometer at A260 and A280/260, respectively, while integrity was verified by Tapestation System (AGILENT). Only samples with RIN > 8 were used for the following RNA-seq analysis. For library preparation, total RNA was quantified using the Qubit 4.0 fluorimetric Assay (Thermo Fisher Scientific). Libraries were prepared from 125 ng of total RNA using the NEGEDIA Digital mRNA-seq research grade sequencing service (Next Generation Diagnostic srl)89 which included library preparation, quality assessment, and sequencing on a NovaSeq 6000 sequencing system using a single-end, 100 cycle strategy (Illumina Inc.).
The raw data were analyzed by Next Generation Diagnostic srl proprietary NEGEDIA Digital mRNA-seq pipeline (v2.0) which involves a cleaning step by quality filtering and trimming, alignment to the reference genome, and counting by gene90,91. The raw expression data were normalized, analyzed, and visualized by Rosalind HyperScale architecture92 (OnRamp BioInformatics, Inc.) Only genes with −1.3 ≥ fold-change ≤ 1.3 and p-Adj ≤ 0.05 were considered differentially expressed and shown as a heatmap or volcano plot.
Starting from the differentially expressed genes between Scramble and UBIAD1OE a Gene Ontology (GO) and KEGG pathway enrichment analysis were performed using ShinyGO 0.76.3 (http://bioinformatics.sdstate.edu/go/) with FDR cutoff 0.05 and minimum pathway size 2. Enrichment was calculated relative to a set of background genes relevant to the experiment.
HPLC analysis of CoQs level
Cells were resuspended in standard medium and seeded in a 100 mm petri dish to reach 70% of confluency the next day. After three washes with ice-cold PBS, cells were scraped and resuspended in 250 µl ice-cold PBS. 10 µl of cells/PBS suspension were put aside to quantify the protein content to normalize the quantification of CoQ10 or CoQ9. The remaining sample was immediately frozen in liquid nitrogen and stored at −80 °C. To extract CoQs from cells, 100 µl of cell suspension in PBS containing 30ug of protein was transferred to 300 µl of ice-cold extraction solution (ethanol/hexane 1:2) containing 0.2 µM of CoQ9 (Sigma Cat. #27597) or CoQ6 (Avanti Cat. #900150 O) as an internal standard for human and mouse cells, respectively, followed by vortexing. The upper, CoQ containing, the hexane layer was separated by centrifugation (5 min, 13,200 × g, +4 °C) and added to a glass mass spectrometry vial. The extraction procedure was repeated twice. The hexane was dried down under a stream of nitrogen and dried samples were then resuspended in 100 µl of methanol. The calibration standards were prepared by serial dilutions of stock solution of CoQ10 or CoQ9 in methanol within the range from 2 to 1000 nM. Internal standard (CoQ9 or CoQ6) was spiked in each calibration standard at a concentration of 0.2 µM. The UHPLC-MS/MS analysis was performed on a Hybrid quadrupole-Orbitrap mass spectrometer (Q Exactive, Thermo Scientific) coupled to a UHPLC system (Ultimate 3000, Thermo Dionex) via a heated electrospray ionization source. Chromatographic separation of targeted analytes and internal standard was achieved on an Accucore C18 100 × 2.1 (2.6 µ particle size) column with a gradient elution of 10 mM of ammonium formate in water (phase A) and 10 mM of ammonium formate in methanol/2-propanol 80:20 (phase B) at a flow rate of 250 µl/min. The sample injection volume was 4 µl.
Detection and determination were performed in full MS/ddMS2 mode with positive electrospray ionization mode. The mass spectrometer was calibrated before analysis using a commercial calibration solution to maintain mass accuracy below 5 ppm. CoQs were detected as [M + H] + and [M + NH4] + adducts ions. CoQs concentration was quantified relative to the internal standard by an external calibration curve. Results are expressed as ng of CoQ10 or CoQ9 normalized over mg of total proteins.
Quantification of lipid classes
Cells were resuspended in standard medium (10% FBS) and seeded in a 100 mm petri dish to reach 70% of confluency the next day. After three washes with ice-cold PBS, cells were scraped and resuspended in 250 µl ice-cold PBS. 10 µl of cells/PBS suspension were put aside to quantify the protein content to normalize the quantification of lipid classes. The remaining sample was immediately frozen in liquid nitrogen and stored at −80 °C.
For lipid class quantification the following internal standard was used: trinonanoylglycerol (Sigma Cat. #92909) and tripentadecanoylglycerol (Sigma Cat. #T4217) for triacylglycerols (TG), 5-α-cholestane (Sigma Cat. #C8003) for cholesteryl esters (CE), (3α, 5β)- cholestan-3-ol (epicoprostanol) (Steraloids Inc. Cat. #C5050-000) for free cholesterol (CO).
The trimethylsilylating agents, N, O-Bis(trimethylsilyl)trifluoroacetamide (BSTFA) with trimethylchlorosilane (Cat. #15238) and HCl methanol (Cat. #90964) were purchased from Merk. All organic solvents were of analytical grade and were purchased from Merk. The fatty acid reference standard mixture (GLC 461) was supplied by Nuchek Prep (Cat. #GLC-461).
Lipids were extracted from 100 µl of a sample using a chloroform-methanol solution by the Folch method93. Lipid extracts were resolved in classes by thin layer chromatography as previously reported94.
The TG fraction was hydrolyzed with 2 ml of HCl-methanol (5%) and the resulting free fatty acids were trans-esterified and extracted with hexane. Hexane-containing fatty acid methyl esters (FAME) was directly taken for gas chromatographic analysis.
In brief, CO and CE was eluted from the silica with 5 ml of chloroform-methanol 2:1. The solvent was collected and evaporated to dryness under a stream of nitrogen. CE were hydrolyzed by saponification and derived free CO was extracted with hexane. Finally, both dried residues containing CO were derivatized in 50 µl of BSTFA for 30 min at room temperature, and 1 µl of the resulting mixture was injected for GC-MS analysis.
The sterols were quantified by gas chromatography-mass spectrometry (GC-MS). The instrument used was Agilent 6890 GC coupled with Agilent 5973 inert MS on the single ion monitoring (SIM) mode. Samples were injected in splitless mode at 270 °C and separated through an HP-5ms column (30 m × 0.25 mm internal diameter × 0.25 µm film thickness; Agilent Technologies, Folsom, CA, USA). The oven temperature was held initially at 200 °C for 1 min, ramped to 275 °C at 10 °C/min, increased to 277 °C at 0.1 °C/min, and finally increased to 290 °C at 10 °C/min and held for 3 min. Peak identification was achieved by comparing the retention time and matching the height ratios of the characteristic ions. The internal standard method was used for quantitative analysis.
Gas chromatography analysis was performed with an Agilent 5890 gas chromatograph (GC) equipped with a flame-ionization detector. FAMEs derived from TG were resolved with an Omegawax column (30 m × 0.25 mm internal diameter × 0.25 µm film thickness; Supelco) with 1 µl injection volume running in on-column mode. The oven temperature was programmed as follows: 60 °C for 3 min, increased 20 °C/min to 205 °C, which remained constant for 15 min. The temperature then increased 0.4 °C/min up to 213 °C, which was maintained for 10 min and finally increased to 240 °C at 5.0 °C/min and held for 8 min. Peaks were identified to a reference standard mixture (GLC 461, Nuchek Prep). The internal standard method was used for quantitative analysis. Results are expressed as pg of TGs, CE, or CO per mg of proteins.
Quantification and statistical analysis
Statistical analysis was performed in GraphPad Prism 8.0 software and R (version 4.1). The values reported in the figures represent ± SEM unless otherwise stated. The exact value of sample size (n) is given in the figure legends or represented as single dots in the figures. Three or more groups were analyzed with one-way ANOVA and Dunnett’s post-test or using Sidak’s multiple comparisons tests. Two-way ANOVA was used to compare data with two variables. Two-tailed unpaired t-test was used to compare the two groups. One-sample t-test was used to compare a group to a hypothetical value when values were expressed as relative to the control and the control was set at 1.0 or 100. Multivariate Hazard ratio (HR) was calculated with 95% confidence interval using multivariate Cox proportional hazards regression after adjustment for age, pT (Tumor size), pN (Regional lymph nodes), ER (Estrogen receptor), HER2 (Erb-B2 Receptor Tyrosine Kinase 2/HER2), tumor grade, patient age at diagnosis. Fisher’s exact test was used to determine statistical significance in contingency tables. Statistical significance is reported as exact p-value, adjusted-p-value or n.s., when not significant (p-value or adjusted-p-value > 0.05). Additional statistical details can be found in the figure legends.
The terms ‘independent experiment’ and ‘biological replicate’ present in the figure legends refer to experiments in which seedings, treatments, stainings, and analyses were conducted independently, using different cell preparations and working solutions.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank Dr. Raj Sewduth for antioxidant rescue experiments on UBIAD1 KO mice, Dr. Chiara Gorrini for advice on breast explant experiments and mouse models, Dr. Marianna Spizzotin for mouse handling and Prof. Federica Cavallo for NeuT mice. Prof. Pier Paolo DI Fiore, Dr. Giovanna Jodice, Dr. Francesca Montani and Prof. Francesca Sanguedolce at the Molecular and Digital Pathology Unit of the European Institute of Oncology (IEO) for support, tissue processing, IHC staining and analysis. Dr. M. Takahashi and Dr. Peter Lambrecht for ShiroQTM Kaneka Ubiquinol water-soluble powder and placebo solutions. Prof. Enzo Calautti for Akt2 constructs. MMS’s laboratory is supported by the European Research Council (ERC) Grants (ERC-CoG 647057 and ERC-PoC 963865) and AIRC (Associazione Italiana Ricerca sul Cancro) IG Grant 20119, Telethon Grant GGP20003. Grants AIRC MultiUnit -5×1000 MCO 10.000, IG 11904, IG 23049, the Italian Ministry of University and Scientific Research (MIUR)-PRIN 20177E9EPY_002 and MIUR-PRIN 202032AZT3_004, the Italian Ministry of Health-Ricerca Corrente and Fondazione Umberto Veronesi (FUV), PSR-University of Milan support S.P. We thank Ellen Jane Corcoran for editorial editing.
Author contributions
M.M.S. and G.T. managed the planning and execution of this study. G.T., A.P., and G.Z. performed and analyzed in vivo mice experiments. A.R. provided supervision for NGS mice experiments. G.T. and A.P. performed and analyzed AFM and cell culture experiments. E.T. and M.M.S. generated the UBIAD1 knock-out mice. M.S. and P.C. performed mass spectrometry analyses. M.G. and N.E. performed and supervised AFM analysis. S.P., D.T., and F.T. performed and analyzed UBIAD1 expression in human breast cancer samples. G.T., A.P., and P.L. performed cell culture experiments analyses. G.T. and M.M.S. wrote the manuscript. MMS conceived the study and the experimental design, provided supervision. All authors agreed on the final version of the manuscript.
Peer review
Peer review information
Nature Communications thanks Luis Carlos Lopez, Ghassan Mouneimne and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The raw and processed RNA sequencing data generated in this study have been deposited in the GEO database under accession code GSE229792. The RNA expression and associated clinicopathological data from the METABRIC cohort used in this study are available in cBioPortal under accession id brca_metabric [https://www.cbioportal.org/study/summary?id=brca_metabric]. Human GRCh38 and mouse GRCm39 reference genomes (primary genome assembly) used for the alignment of RNA sequencing data are available on GENCODE. Publicly available RNA expression and associated clinicopathological data of 1904 patients from the METABRIC (Molecular Taxonomy of Breast Cancer Consortium) dataset 3 were retrieved from cBioPortal (http://www.cbioportal.org/). All data are available in the manuscript, in the Supplementary Information or in the Source data files. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52523-y.
References
- 1.Morton, R. A. Ubiquinone. Nature182, 1764–1767 (1958). [DOI] [PubMed] [Google Scholar]
- 2.Crane, F. L. Discovery of ubiquinone (coenzyme Q) and an overview of function. Mitochondrion7, S2–S7 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Navas, P., Villalba, J. M. & de Cabo, R. The importance of plasma membrane coenzyme Q in aging and stress responses. Mitochondrion7, S34–S40 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Agmo Hernández, V., Eriksson, E. K. & Edwards, K. Ubiquinone-10 alters mechanical properties and increases stability of phospholipid membranes. Biochim. Biophys. Acta (BBA) - Biomembr.1848, 2233–2243 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Sévin, D. C. & Sauer, U. Ubiquinone accumulation improves osmotic-stress tolerance in Escherichia coli. Nat. Chem. Biol.10, 266–272 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Thapa, M. & Dallmann, G. Role of coenzymes in cancer metabolism. Semin Cell Dev. Biol.98, 44–53 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Martínez-Reyes, I. et al. Mitochondrial ubiquinol oxidation is necessary for tumour growth. Nature585, 288–292 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee, S. Q. E., Tan, T. S., Kawamukai, M. & Chen, E. S. Cellular factories for coenzyme Q10 production. Microb. Cell Fact.16, 39 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guerra, R. M. & Pagliarini, D. J. Coenzyme Q biochemistry and biosynthesis. Trends Biochem. Sci.48, 463–476 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Göbel, A., Riffel, R. M., Hofbauer, L. C. & Rachner, T. D. The mevalonate pathway in breast cancer biology. Cancer Lett.542, 215761 (2022). [DOI] [PubMed] [Google Scholar]
- 11.George, D. M., Ramadoss, R., Mackey, H. R. & Vincent, A. S. Comparative computational study to augment UbiA prenyltransferases inherent in purple photosynthetic bacteria cultured from mangrove microbial mats in Qatar for coenzyme Q10 biosynthesis. Biotechnol. Rep.36, e00775 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang, Y. & Hekimi, S. Molecular genetics of ubiquinone biosynthesis in animals. Crit. Rev. Biochem. Mol. Biol.48, 69–88 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, W. Bringing bioactive compounds into membranes: the UbiA superfamily of intramembrane aromatic prenyltransferases. Trends Biochem Sci.41, 356–370 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang, X., Stockwell, B. R. & Conrad, M. Ferroptosis: mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol.22, 266–282 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lei, G., Zhuang, L. & Gan, B. Targeting ferroptosis as a vulnerability in cancer. Nat. Rev. Cancer22, 381–396 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng, J. & Conrad, M. The metabolic underpinnings of ferroptosis. Cell Metab.32, 920–937 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Doll, S. et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol.13, 91–98 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao, Y. et al. Comprehensive metabolomics expands precision medicine for triple-negative breast cancer. Cell Res.32, 477–490 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang, F. et al. Ferroptosis heterogeneity in triple-negative breast cancer reveals an innovative immunotherapy combination strategy. Cell Metab.35, 84–100.e108 (2023). [DOI] [PubMed] [Google Scholar]
- 20.Bentinger, M. et al. Stimulation of coenzyme Q synthesis. Biofactors32, 99–111 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Byfield, F. J., Aranda-Espinoza, H., Romanenko, V. G., Rothblat, G. H. & Levitan, I. Cholesterol depletion increases membrane stiffness of aortic endothelial cells. Biophys. J.87, 3336–3343 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khatibzadeh, N., Gupta, S., Farrell, B., Brownell, W. E. & Anvari, B. Effects of cholesterol on nano-mechanical properties of the living cell plasma membrane. Soft Matter8, 8350–8360, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lei, K. et al. Cancer-cell stiffening via cholesterol depletion enhances adoptive T-cell immunotherapy. Nat. Biomed. Eng.5, 1411–1425 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vahabikashi, A. et al. Probe sensitivity to cortical versus intracellular cytoskeletal network stiffness. Biophys. J.116, 518–529 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanchanawong, P. & Calderwood, D. A. Organization, dynamics and mechanoregulation of integrin-mediated cell–ECM adhesions. Nat. Rev. Mol. Cell Biol.24, 142–161 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinz, N. & Jücker, M. Distinct functions of AKT isoforms in breast cancer: a comprehensive review. Cell Commun. Signal17, 154 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romani, P. et al. Extracellular matrix mechanical cues regulate lipid metabolism through Lipin-1 and SREBP. Nat. Cell Biol.21, 338–347 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Wu, C. Focal adhesion: a focal point in current cell biology and molecular medicine. Cell Adh Migr.1, 13–18 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, X. et al. A novel Golgi retention signal RPWS for tumor suppressor UBIAD1. PLoS ONE10.1371/journal.pone.0072015 (2013). [DOI] [PMC free article] [PubMed]
- 30.Mugoni, V. et al. Ubiad1 is an antioxidant enzyme that regulates eNOS activity by CoQ10 synthesis. Cell152, 504–518 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Curtis, C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature486, 346–352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veronesi, U. et al. A randomized comparison of sentinel-node biopsy with routine axillary dissection in breast cancer. N. Engl. J. Med.349, 546–553 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Filippone, M. G. et al. CDK12 promotes tumorigenesis but induces vulnerability to therapies inhibiting folate one-carbon metabolism in breast cancer. Nat. Commun.13, 2642 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taneja, P. et al. MMTV mouse models and the diagnostic values of MMTV-like sequences in human breast cancer. Expert Rev. Mol. Diagn.9, 423–440 (2009). [DOI] [PMC free article] [PubMed]
- 35.Fluck, M. M. & Schaffhausen, B. S. Lessons in signaling and tumorigenesis from polyomavirus middle T antigen. Microbiol Mol. Biol. Rev.73, 542–563 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jonkers, J. et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet.29, 418–425 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Borowicz, S. et al. The soft agar colony formation assay. J. Vis. Exp.10.3791/51998 (2014). [DOI] [PMC free article] [PubMed]
- 38.Paoli, P., Giannoni, E. & Chiarugi, P. Anoikis molecular pathways and its role in cancer progression. Biochim. Biophys. Acta.1833, 3481–3498 (2013). [DOI] [PubMed]
- 39.Capaci, V. et al. Mutant p53 induces Golgi tubulo-vesiculation driving a prometastatic secretome. Nat. Commun.11, 3945 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao, L., Wu, X., Li, T., Luo, J. & Dong, D. ctcRbase: the gene expression database of circulating tumor cells and microemboli. Database (Oxf.)2020, baaa020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang, C. et al. A laminin 511 matrix is regulated by TAZ and functions as the ligand for the alpha6Bbeta1 integrin to sustain breast cancer stem cells. Genes Dev.29, 1–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geraldo, L. H. M. et al. Role of lysophosphatidic acid and its receptors in health and disease: novel therapeutic strategies. Signal Transduct. Target Ther.6, 45 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mezquita, B. et al. All-trans-retinoic acid activates the pro-invasive Src-YAP-Interleukin 6 axis in triple-negative MDA-MB-231 breast cancer cells while cerivastatin reverses this action. Sci. Rep.8, 7047 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song, L. et al. Proto-oncogene Src links lipogenesis via lipin-1 to breast cancer malignancy. Nat. Commun.11, 5842 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weng, Y. S. et al. MCT-1/miR-34a/IL-6/IL-6R signaling axis promotes EMT progression, cancer stemness and M2 macrophage polarization in triple-negative breast cancer. Mol. Cancer18, 42 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, W. et al. A NIK-IKKalpha module expands ErbB2-induced tumor-initiating cells by stimulating nuclear export of p27/Kip1. Cancer Cell23, 647–659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen, M. et al. Tinagl1 suppresses triple-negative breast cancer progression and metastasis by simultaneously inhibiting integrin/FAK and EGFR signaling. Cancer Cell35, 64–80.e67 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Song, F. et al. Mast cells inhibit colorectal cancer development by inducing ER stress through secreting Cystatin C. Oncogene42, 209–223 (2023). [DOI] [PubMed] [Google Scholar]
- 49.Englund, J. I. et al. Laminin alpha 5 regulates mammary gland remodeling through luminal cell differentiation and Wnt4-mediated epithelial crosstalk. Development148, dev199281 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chia, J. et al. Evidence for a role of tumor-derived laminin-511 in the metastatic progression of breast cancer. Am. J. Pathol.170, 2135–2148 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang, C. et al. A laminin 511 matrix is regulated by TAZ and functions as the ligand for the α6Bβ1 integrin to sustain breast cancer stem cells. Genes Dev.29, 1–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ghosh, I., Singh, R. K., Mishra, M., Kapoor, S. & Jana, S. S. Switching between blebbing and lamellipodia depends on the degree of non-muscle myosin II activity. J. Cell Sci.134, jcs248732 (2021). [DOI] [PubMed] [Google Scholar]
- 53.Leitinger, B. & Hogg, N. The involvement of lipid rafts in the regulation of integrin function. J. Cell Sci.115, 963–972 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Lietha, D. & Izard, T. Roles of membrane domains in integrin-mediated cell adhesion. Int J. Mol. Sci.21, 5531 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baumgart, T. et al. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc. Natl Acad. Sci. USA104, 3165–3170 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sezgin, E., Levental, I., Mayor, S. & Eggeling, C. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol.18, 361–374 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown, C. W., Amante, J. J., Goel, H. L. & Mercurio, A. M. The α6β4 integrin promotes resistance to ferroptosis. J. Cell Biol.216, 4287–4297 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doll, S. et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature575, 693–698 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Santoro, M. M. The antioxidant role of non-mitochondrial CoQ10: mystery solved! Cell Metab.31, 13–15 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Bersuker, K. et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature575, 688–692 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ya, F. et al. Coenzyme Q10 attenuates platelet integrin αIIbβ3 signaling and platelet hyper-reactivity in ApoE-deficient mice. Food Funct.11, 139–152 (2020). [DOI] [PubMed] [Google Scholar]
- 62.Ehmsen, S. et al. Increased cholesterol biosynthesis is a key characteristic of breast cancer stem cells influencing patient outcome. Cell Rep.27, 3927–3938.e3926 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Li, Y. C., Park, M. J., Ye, S. K., Kim, C. W. & Kim, Y. N. Elevated levels of cholesterol-rich lipid rafts in cancer cells are correlated with apoptosis sensitivity induced by cholesterol-depleting agents. Am. J. Pathol.168, 1107–1118 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Badana, A. et al. Lipid raft integrity is required for survival of triple negative breast cancer cells. J. Breast Cancer19, 372–384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grzybek, M. et al. ESR and monolayer study of the localization of coenzyme Q10 in artificial membranes. Gen. Physiol. Biophys.24, 449–460 (2005). [PubMed] [Google Scholar]
- 66.Garg, S. et al. CoQ10 selective miscibility and penetration into lipid monolayers with lower lateral packing density. Biochim. Biophys. Acta (BBA) - Biomembr.1859, 1173–1179 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Clarke, C. F., Rowat, A. C. & Gober, J. W. Is CoQ a membrane stabilizer? Nat. Chem. Biol.10, 242–243 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Jolliet, P. et al. Plasma coenzyme Q10 concentrations in breast cancer: prognosis and therapeutic consequences. Int. J. Clin. Pharm. Ther.36, 506–509 (1998). [PubMed] [Google Scholar]
- 69.Portakal, O. et al. Coenzyme Q10 concentrations and antioxidant status in tissues of breast cancer patients. Clin. Biochem.33, 279–284 (2000). [DOI] [PubMed] [Google Scholar]
- 70.Folkers, K. Relevance of the biosynthesis of coenzyme Q10 and of the four bases of DNA as a rationale for the molecular causes of cancer and a therapy. Biochem. Biophys. Res. Commun.224, 358–361 (1996). [DOI] [PubMed] [Google Scholar]
- 71.Folkers, K., Osterborg, A., Nylander, M., Morita, M. & Mellstedt, H. Activities of vitamin Q10 in animal models and a serious deficiency in patients with cancer. Biochem. Biophys. Res. Commun.234, 296–299 (1997). [DOI] [PubMed] [Google Scholar]
- 72.Rusciani, L. et al. Low plasma coenzyme Q10 levels as an independent prognostic factor for melanoma progression. J. Am. Acad. Dermatol.54, 234–241 (2006). [DOI] [PubMed] [Google Scholar]
- 73.Ubellacker, J. M. et al. Lymph protects metastasizing melanoma cells from ferroptosis. Nature585, 113–118 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hassannia, B., Vandenabeele, P. & Berghe, T. V. Targeting ferroptosis to iron out cancer. Cancer Cell35, 830–849 (2019). [DOI] [PubMed] [Google Scholar]
- 75.Juarez, D. & Fruman, D. A. Targeting the mevalonate pathway in cancer. Trends Cancer7, 525–540 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jin, D. Y. et al. A genome-wide CRISPR-Cas9 knockout screen identifies FSP1 as the warfarin-resistant vitamin K reductase. Nat. Commun.14, 828 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Quaglino, E., Mastini, C., Forni, G. & Cavallo, F. ErbB2 transgenic mice: a tool for investigation of the immune prevention and treatment of mammary carcinomas. Curr. Protoc. Immunol.82, 20.29.21–20.29.10 (2008). [DOI] [PubMed] [Google Scholar]
- 78.Hafner, M. et al. Keratin 14 Cre transgenic mice authenticate keratin 14 as an oocyte-expressed protein. Genesis38, 176–181 (2004). [DOI] [PubMed] [Google Scholar]
- 79.Saxena, M., Kalathur, R. K. R., Neutzner, M. & Christofori, G. PyMT-1099, a versatile murine cell model for EMT in breast cancer. Sci. Rep.8, 12123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pece, S. et al. Identification and clinical validation of a multigene assay that interrogates the biology of cancer stem cells and predicts metastasis in breast cancer: a retrospective consecutive study. EBioMedicine42, 352–362 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Porcù, E. et al. BMP9 counteracts the tumorigenic and pro-angiogenic potential of glioblastoma. Cell Death Differ.25, 1808–1822 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Campbell, J. P., Merkel, A. R., Masood-Campbell, S. K., Elefteriou, F. & Sterling, J. A. Models of bone metastasis. J. Vis. Exp. 10.3791/4260 (2012). [DOI] [PMC free article] [PubMed]
- 83.Tolg, C., Cowman, M. & Turley, E. A. Mouse mammary gland whole mount preparation and analysis. Bio Protoc.8, e2915 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wiel, C. et al. BACH1 stabilization by antioxidants stimulates lung cancer metastasis. Cell178, 330–345.e322 (2019). [DOI] [PubMed] [Google Scholar]
- 85.Amendola, M., Venneri, M. A., Biffi, A., Vigna, E. & Naldini, L. Coordinate dual-gene transgenesis by lentiviral vectors carrying synthetic bidirectional promoters. Nat. Biotechnol.23, 108–116 (2005). [DOI] [PubMed] [Google Scholar]
- 86.Saoncella, S. et al. Nuclear Akt2 opposes limbal keratinocyte stem cell self-renewal by repressing a FOXO-mTORC1 signaling pathway. Stem Cells32, 754–769 (2014). [DOI] [PubMed] [Google Scholar]
- 87.Zhao, W. et al. Candidate antimetastasis drugs suppress the metastatic capacity of breast cancer cells by reducing membrane fluidity. Cancer Res.76, 2037–2049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lombardo, Y., de Giorgio, A., Coombes, C. R., Stebbing, J. & Castellano, L. Mammosphere Formation Assay from Human Breast Cancer Tissues and Cell Lines. J. Vis. Exp.97, e52671 (2015). [DOI] [PMC free article] [PubMed]
- 89.Xiong, Y. et al. A comparison of mRNA sequencing with random primed and 3’-directed libraries. Sci. Rep.7, 14626 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Folch, J., Lees, M. & Sloane Stanley, G. H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem.226, 497–509 (1957). [PubMed] [Google Scholar]
- 94.Correani, A. et al. The maternal-fetal gradient of free and esterified phytosterols at the time of delivery in humans. Clin. Nutr.37, 2107–2112 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw and processed RNA sequencing data generated in this study have been deposited in the GEO database under accession code GSE229792. The RNA expression and associated clinicopathological data from the METABRIC cohort used in this study are available in cBioPortal under accession id brca_metabric [https://www.cbioportal.org/study/summary?id=brca_metabric]. Human GRCh38 and mouse GRCm39 reference genomes (primary genome assembly) used for the alignment of RNA sequencing data are available on GENCODE. Publicly available RNA expression and associated clinicopathological data of 1904 patients from the METABRIC (Molecular Taxonomy of Breast Cancer Consortium) dataset 3 were retrieved from cBioPortal (http://www.cbioportal.org/). All data are available in the manuscript, in the Supplementary Information or in the Source data files. Source data are provided with this paper.