Abstract
Christensenella minuta DSM 22607 has recently been suggested as a potential microbiome-based therapy for inflammatory bowel disease (IBD) because it displays strong anti-inflammatory effects both in vitro and in vivo. Here, we aimed to decipher the mechanism(s) underlying the DSM 22607-mediated beneficial effects on the host in a mouse model of chemically induced acute colitis. We observed that C. minuta plays a key role in the preservation of the epithelial barrier and the management of DNBS-induced inflammation by inhibiting interleukin (IL)-33 and Tumor necrosis factor receptor superfamily member 8 (Tnfrsf8) gene expression. We also showed that DSM 22607 abundance was positively correlated with Akkermansia sp. and Dubosiella sp. and modulated microbial metabolites in the cecum. These results offer new insights into the biological and molecular mechanisms underlying the beneficial effects of C. minuta DSM 22607 by protecting the intestinal barrier integrity and regulating inflammation.
Subject terms: Microbiome, Bacteria
Introduction
Over the last decade, several studies have highlighted the use of microbiome-derived therapies using a single bacterium as a promising approach to treat chronic inflammatory disorders, such as necrotizing enterocolitis1 and inflammatory bowel diseases (IBD)2–5. IBD is a multifactorial diseases6 characterized by chronic and abnormal activation of the gut immune system, leading to an uncontrolled and deleterious inflammatory response7. Symptoms associated with IBD include abdominal pain, diarrhea, hematochezia, and weight loss8. Moreover, IBD is associated with a depletion of mucus-secreting goblet cells (GC) and a reduction in the intestinal mucus layer9, which is known to negatively impact intestinal homeostasis10. An increase in intestinal permeability has also been observed, which triggers an immunological response that drives intestinal inflammation11. These are also associated with a gut microbiome imbalance characterized by decreases in bacterial richness and diversity12, features that have been suggested to play an important role in the establishment and maintenance of mucosal inflammation13. In fact, an increase in some commensal opportunistic pathogenic bacteria, such as members of the Enterobacteriaceae and Bacteroidaceae families14–18 and a decrease in members of the Lactobacillaceae family and the butyrate-producing genus Faecalibacterium19–22 have been observed in patients suffering from IBD. Clinical studies have also highlighted the strong depletion of the Christensenellaceae family in the gut microbiota of patients with Crohn’s disease (CD) and ulcerative colitis (UC), the two main types of IBD23–25. Of particular interest, a drop in Christensenellaceae has been shown to be predictive of flares in patients with CD, indicating a possible causal relationship between the presence of Christensenellaceae in the gut community and the maintenance of immune homeostasis in the intestinal barrier23.
Modulating the gut microbiota with members of the Christensenellaceae family could be considered as a therapeutic option. For example, it has been shown that in people with a low body mass index (BMI), the Christensenellaceae family was significantly more abundant compared to adults with a hight BMI26. Moreover, Chistensenella minuta DSM 33407, one member of Christensenellaceae family, represents a potential anti-obesity treatment by its capacity to decrease glycemia and free fatty acids in serum and liver respectively in vivo27. C. minuta has been suggested to be a key member of the healthy human gut community28 and has been recently identified as a key component of healthy microbiota29. Keystone species were defined in 1969 as “a specie which has a disproportionately large effect on its natural environment relative to its abundance”30. They play a critical role in maintaining the structure of ecological communities. These taxa, which are highly connected within the ecosystem, play a unique and crucial role in microbial communities, carrying essential functions to support the growth of other species. Their reduction can cause a dramatic shift in microbiome structure and functioning31 as Christensenellaceae family has been described in co-occurrence network analysis as the central member of a hub of microorganisms that includes other heritable taxa26. Maintaining physiological levels of Christensenellaceae in the gut microbiota of patients thus represents an interesting therapeutic strategy for the management of the disease. We have previously demonstrated that C. minuta DSM 22607 exerts anti-inflammatory effects by inhibiting the NF-κB signaling pathway in vitro. It also reduced colonic inflammation in vivo in two murine models of induced-colitis32.
Here, we aimed to elucidate the mechanism(s) underlying the immunomodulatory effects of C. minuta DSM 22607 on the host. For this purpose, we analyzed the impact of the administration of C. minuta in a chemical-induced mouse model of colitis (dinitrobenzene sulfonic acid (DNBS)). We focused on the intestinal barrier, including mucus production and permeability, and immunomodulatory effects at local and systemic levels. These effects on the host were complemented by RNA-seq transcriptomic analysis of intestinal tissues and an investigation of microbiota modifications at the taxonomic and functional levels. This study provides new insights into the mechanism of action underlying the anti-inflammatory effects of C. minuta, supporting the multiple benefits of this bacterium in IBD management.
Results
Christensenella minuta DSM 22607 attenuates colitis severity in a DNBS-induced mouse model
To decipher the mechanism of action of C. minuta, we induced colitis in mice using dinitrobenzene sulfonic acid (DNBS). Mice were administered DSM 22607 by gavage 14 days before the induction of inflammation. We validated C. minuta protective effects of C. minuta, as it reduced DNBS-induced weight loss and improved weight recovery (p < 0.001, Supplementary Fig. 1a). Furthermore, DSM 22607 treatment decreased DNBS-induced macroscopic and microscopic damage, as observed by the macroscopic (p < 0.0001) and histological (p < 0.001) scores (Supplementary Fig. 1b, c), and decreased myeloperoxidase (MPO) activity (p = 0.06, Supplementary Fig. 1d), which confirmed the anti-inflammatory effects observed in our previous study32.
DSM 22607 improves intestinal barrier integrity by modulating paracellular pathways and tissue resistance in the colon
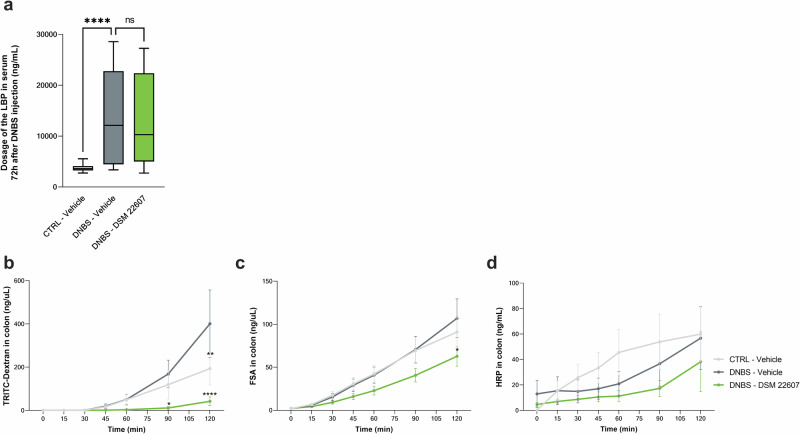
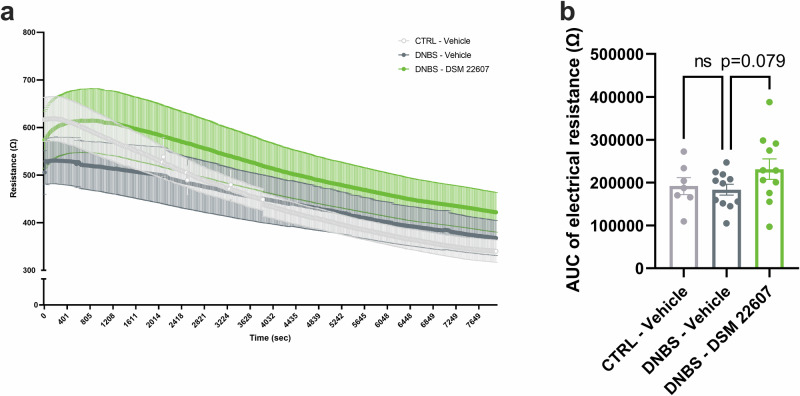
We assessed intestinal permeability by measuring bacterial translocation using serum lipopolysaccharide (LPS)-Binding Protein (LBP) as a proxy marker (Fig. 1a). As expected, we observed an increase in LBP in the DNBS-Vehicle group compared with that in the CTRL-Vehicle group (p < 0.0001), reflecting an increase in intestinal permeability. However, no improvements were observed in the DNBS-DSM 22607 group. To better evaluate local intestinal permeability, colon, ileum, and cecum biopsies collected at the endpoint were mounted in Ussing chambers for 120 min. We analyzed both paracellular (fluorescein-5-(and-6)-sulfonic acid (FSA) and isothiocyanate (TRITC)-dextran) and transcellular peroxidase (horseradish peroxidase (HRP)) permeabilities. A significant decrease in the passage of TRITC-dextran (at 90 min, p < 0.05; at T120 min, p < 0.001) and FSA (at 120 min, p < 0.05) was observed in the colon of DSM 22607-treated mice compared to that in inflamed control mice (Fig. 1b, c), indicating differences in paracellular permeability. In contrast, no significant effects were observed in HRP measurements, indicating no differences in transcellular permeability (Fig. 1d). No significant differences were observed in ileum or cecum sections (Supplementary Fig. 2). Transepithelial electrical resistance (TEER) was recorded across the colon samples (Fig. 2a, b). As expected, TEER was lower in DNBS-treated inflamed mice than in non-inflamed mice. Interestingly, this was partially restored in DSM 22607-treated biopsies compared to the untreated inflamed group (p = 0.079), confirming the potential of DSM 22607 to limit tissue alteration of permeability.
Fig. 1. C. minuta maintained intestinal barrier integrity by modulating permeability.
Lipopolysaccharide (LPS) binding protein (LBP) in the serum (a). CTRL-Vehicle (N = 20; white); DNBS-Vehicle (N = 19; dark gray); DNBS-DSM 22607 (N = 19; green). Ussing chamber kinetics of tetramethylrhodamine isothiocyanate (TRITC)-dextran (b), fluorescein-5-(and-6)-Sulfonic Acid (FSA) (c), and horseradish peroxidase (HRP) (d) in colon. CTRL-Vehicle (N = 9; light gray); DNBS-Vehicle (N = 12; dark gray); DNBS-DSM 22607 (N = 12; green). Data are means and standard error of the mean (SEM). Results of the Original FDR method of Benjamini and Hochberg multiple comparison tests comparing the DNBS-Vehicle group to the other three groups. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Fig. 2. Colonic permeability tended to improve following C. minuta treatment.
During 2 h, raw transepithelial electrical resistance recording (a) and associated aera under the curve (b) were obtained using Ussing chambers. CTRL-Vehicle (N = 9; light gray); DNBS-Vehicle (N = 12; dark gray); DNBS-DSM 22607 (N = 12; green). Data are means and SEM. Results of Original FDR method of Benjamini and Hochberg multiple comparison tests comparing the DNBS-Vehicle group to the two other groups. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
DSM 22607 improves intestinal barrier structure and function by hampering GC depletion and increasing mucin 2 (MUC2) expression
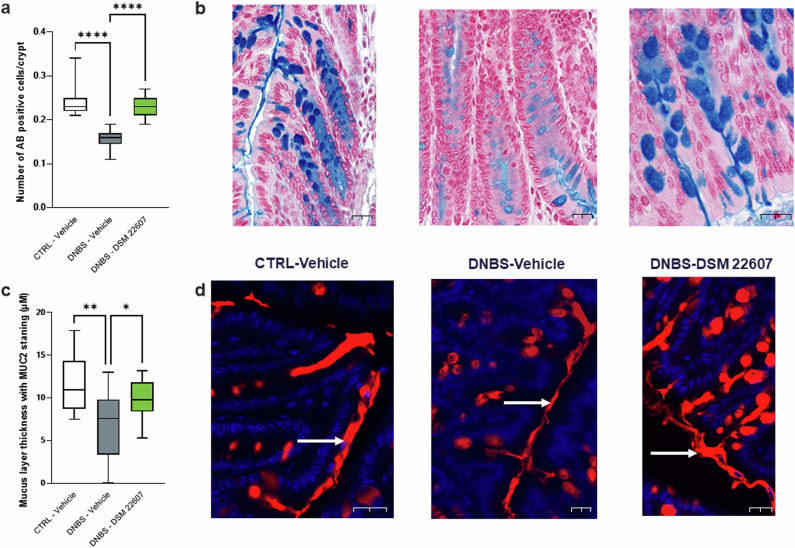
To evaluate the effects of DSM 22607 on the intestinal barrier, we decided to further analyze the epithelial structure with a focus on the GC and mucus layer. First, as GC depletion is characteristic of DNBS-induced colitis33, we quantified GC populations in the colon using alcian blue (AB) staining. As expected, a decrease in AB-positive cells was observed following DNBS injection (p < 0.0001), whereas C. minuta DSM 22607 prevented the loss of mucus-producing cells (p < 0.0001; Fig. 3a). To determine mucus layer thickness, MUC2 staining, one of the major mucins in the mucus in the colon, was performed. We observed that DSM 22607 treatment significantly improved the thickness of this layer compared to that in the DNBS-Vehicle group (p < 0.05, Fig. 3b).
Fig. 3. DSM 22607 improves mucus production by decreasing goblet cell depletion.
AB positive cells/Crypt (a), goblet cell staining (b), thickness of the mucus layer (c) and MUC2 staining showed with white arrows (d) in the colon. CTRL-Vehicle (N = 26; white); DNBS-Vehicle (N = 29; dark gray); DNBS-DSM 22607 (N = 31; green). Data are means and SEM. Length of scale bar measured 20 µm. Results of Original FDR method of Benjamini and Hochberg multiple comparison tests comparing the DNBS-Vehicle group to the two other groups. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
C. minuta DSM 22607 displayed anti-inflammatory effects at local and systemic levels
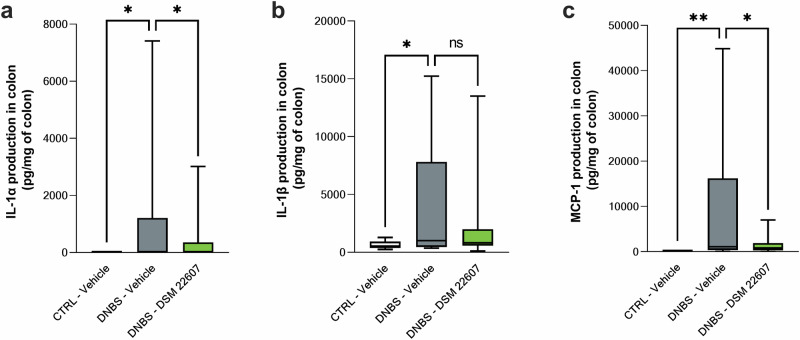
In the colon, DNBS injection stimulated the protein expression of interleukin (IL)-1α (Fig. 4a), IL-1β (Fig. 4b), and monocyte chemoattractant protein (MCP)-1 (Fig. 4c). DSM 22607 showed anti-inflammatory properties through a significant reduction in IL-1α (p < 0.05) and MCP-1 (p < 0.01) levels, whereas no effects were observed for IL-1β. No significant effects were observed for tumor necrosis factor (TNF)-α, interferon (IFN), γ and IL-23 (data not shown). IL-12p70, IL-10, IL-6, IL-27, IL-17A, and IFNβ and granulocyte-macrophage colony-stimulating factor (GM-CSF) were not detected.
Fig. 4. Anti-inflammatory properties of Christensenella minuta at local level.
Determination of IL-1α (a), IL-1β (b) and MCP-1 (c) cytokines in the colon. CTRL-Vehicle (N = 20; white); DNBS-Vehicle (N = 19; dark gray); DNBS-DSM 22607 (N = 19; green). Data are means and SEM. Results of Original FDR method of Benjamini and Hochberg multiple comparison tests comparing the DNBS-Vehicle group to the three other groups.*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
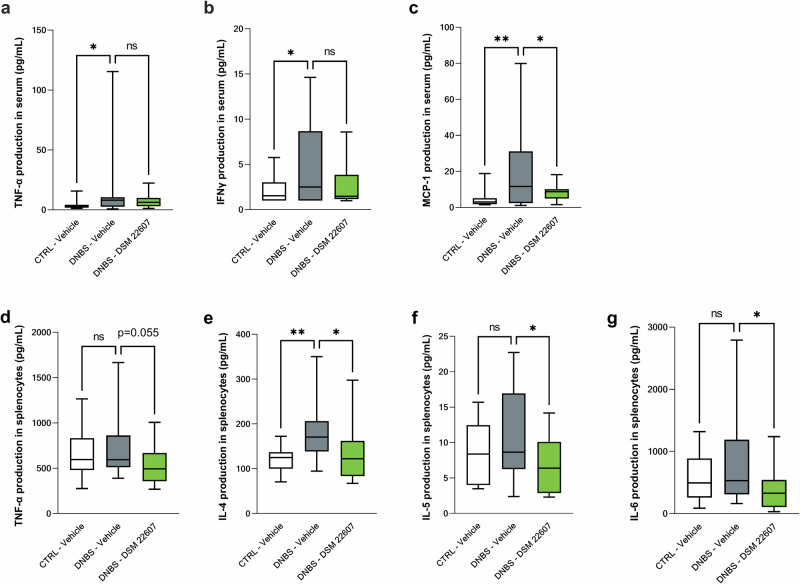
To evaluate the immunomodulatory effects of DSM 22607 at the systemic level, we measured cytokine levels in serum samples and in PMA-IONO-stimulated splenocytes, which activate Th cells. We observed a significant increase in the serum levels of TNF-α (Fig. 5a), IFNγ (Fig. 5b), and MCP-1 (Fig. 5c) following DNBS injection (p < 0.05), which tended to be counterbalanced by DSM 22607 treatment for TNF-α and IFNγ (p = 0.08) and decreased MCP-1 (p < 0.05). No significant effects were observed for IL-1α and IL-23 (data not shown). IL-12p70, IL-1b, IL-10, IL-6, IL-27, IL-17A, and GM-CSF were not detected in these samples. Cytokine measurement of the supernatant of stimulated splenocytes showed that DSM 22607 decreased TNF-α (p < 0.05; Fig. 5d), IL-4 (p < 0.05; Fig. 5e), IL-5 (p < 0.05; Fig. 5f), and IL-6 (p < 0.01; Fig. 5g) levels compared with the DNBS-vehicle group. No differences were observed in IFNγ, IL-2, IL-10, IL-17A, and IL-22 cytokines between the groups (data not shown). IL-9, IL-17F, and IL-13 were not detected (data not shown).
Fig. 5. Anti-inflammatory properties of Christensenella minuta at systemic levels in serum and spleen.
Determination of TNF-α (a); IFNγ (b); and MCP-1 (c) cytokines in serum. Following splenocyte stimulation, TNF-α (d), IL-4 (e), IL-5 (f) and IL-6 (g) were measured. CTRL-Vehicle (N = 20; white); DNBS-Vehicle (N = 19; dark gray); DNBS-DSM 22607 (N = 19; green). Data are means and SEM. Results of Original FDR method of Benjamini and Hochberg multiple comparison tests comparing the DNBS-Vehicle group to the three other groups. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
DSM 22607 modulates gut microbiota composition
To assess the modulation of the gut microbiota following C. minuta administration and DNBS injection, we collected feces at time (T) T0 (before any treatment), T14 (14 days after gavage with C. minuta or controls), and fecal content at T17 (3 days post-injection and final time point). After quality control, there were a total of 4,678,378 reads, and a mean of 27,358 reads per sample (min = 20,132 and max = 34,377). A total of 982 amplicon sequence variants (ASVs) were identified. The assessment of microbiota diversity indices at each time point revealed no significant differences between T0 and T14 (Supplementary Fig. 3a, b, d, and e) between groups. Neither alpha nor beta diversity was impacted. Conversely, significant changes were observed at the end of the experiment (T17). The most significant differences were detected between the inflamed (DNBS-vehicle and DNBS-DSM 22607) and non-inflamed (CTRL-vehicle) groups. The inflamed groups showed a significantly lower alpha diversity (Supplementary Fig. 3c), and increased dissimilarity compared to that in the non-inflamed groups (Supplementary Fig. 3f). This result indicates that inflamed gut microbial communities are less diverse and more heterogeneous, displaying the most dissimilar communities among individuals of the same group.
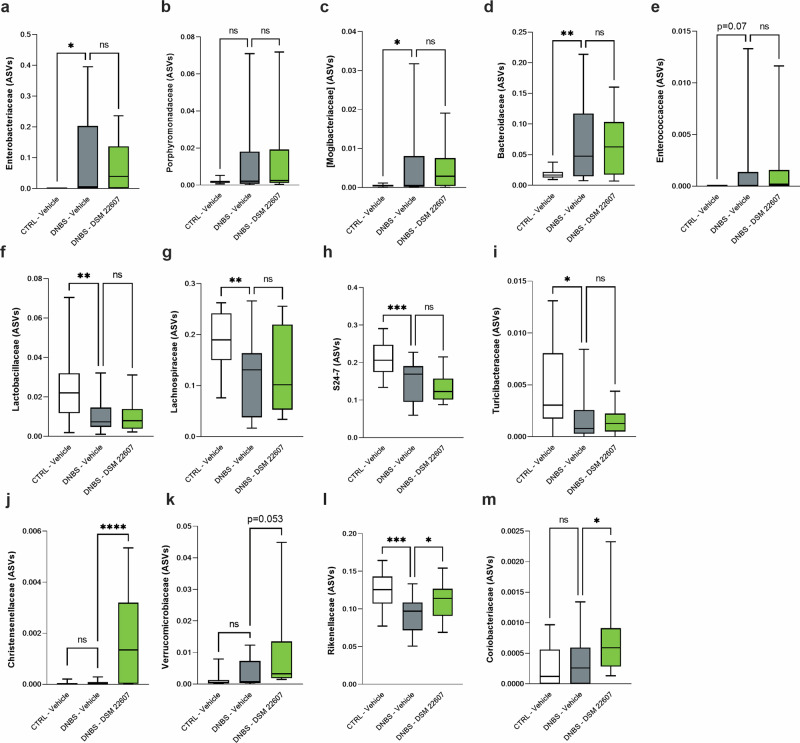
A differential abundance analysis that identified significant differences in the gut microbiota composition at the family level was performed. Members of Enterobacteriaceae (p < 0.05; Fig. 6a), Porphyromonadaceae (Fig. 6b), Mogibacteriaceae (p < 0.05; Fig. 6c), Bacteroidaceae (p < 0.01; Fig. 6d), and Enterococcaceae (p = 0.07; Fig. 6e) were higher in the inflamed group than in the control group. In contrast, Lactobacillaceae (p < 0.01; Fig. 6f), Lachnospiraceae (p < 0.01; Figs. 6g), S24–6 (p < 0.001; Fig. 6h), and Turicibacteraceae (p < 0.05; Fig. 6i) decreased in the DNBS-vehicle group. Members of the Christensenellaceae family were exclusively present in the DNBS-DSM 22607 group (p < 0.0001; Fig. 6j), where they were present in a concentration higher of the detection limit in 10 of 16 animals. By using a specific set of primers, we confirmed the presence of C. minuta at the end of the experiment (D21) on the same gDNA by PCR in all the mice (data not shown). We also detected a higher abundance of members of the families Verrucomicrobiaceae (p = 0.053; Fig. 6k), Rikenellaceae (p < 0.05; Fig. 6l), and Coriobacteriaceae (p < 0.05; Fig. 6m) in the DNBS-DSM 22607 group.
Fig. 6. Amplicon sequence variant (ASV) family diversity among groups at the experiment end.
Enterobacteriaceae (a), Prophyromonadaceae (b) Mogibacteriaceae (c), Bacteroidaceae (d), Enterococcaceae (e), Lactobacillaceae (f), Lachnospiraceae (g), S24-7 (h), Rikenellaceae (i), Turibacteraceae (j), Christensenellaceae (k), Verrucomicrobiaceae (l) and Coriobacteriaceae (m) ASVs at the end of the experiment. CTRL-Vehicle (N = 20; white); DNBS-Vehicle (N = 19; dark gray); DNBS-DSM 22607 (N = 19; green). Data are means and SEM. Results of Original FDR method of Benjamini and Hochberg multiple comparison tests comparing the DNBS-Vehicle group to the three other groups. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
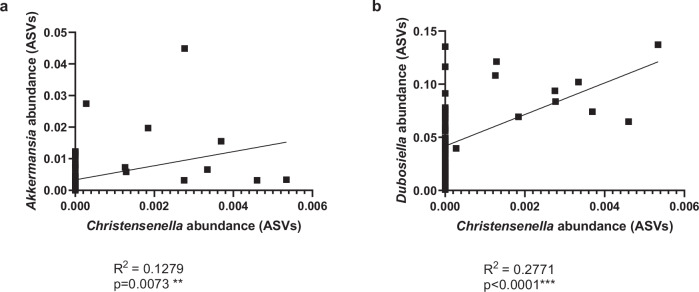
We further examined the correlation between the relative abundances of C. minuta and other genera in the DNBS-DSM 22607 group. The high relative abundances of Akkermansia sp. and Dubosiella sp. were slightly positively correlated with the presence of the Christensenella genus (Figs. 7a and 7b), as illustrated by the correlation coefficients of R2 = 0.1269 (p = 0.0073) and R2 = 0.2661 (p < 0.0001), respectively. Interestingly, an increase in these two genera, compared to DNBS-Vehicle, was only observed in mice in which we detected the Christensenellaceae family in the differential abundance analysis (data not shown).
Fig. 7. Positive correlation between Christensenella and Akkermansia and Dubosiella genus.
Akkermansia (a) and Dubosiella (b) amplicon sequence variant (ASV) abundances versus Christensenella. Results correlation with Pearson r test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Modulation of microbial metabolites by DSM 22607
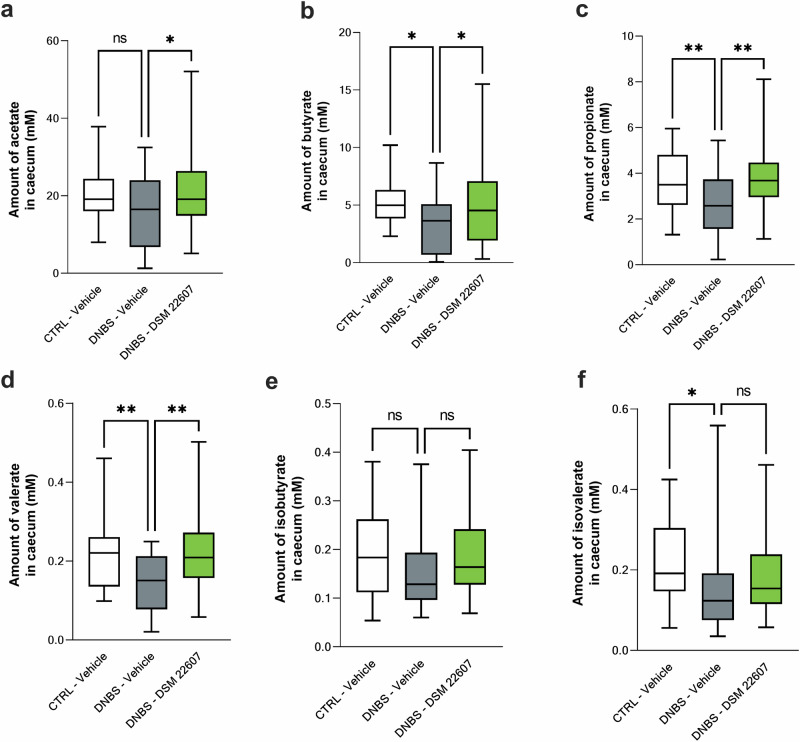
Modulation of short chain fatty acids (SCFA) production in the cecum was measured using gas chromatography. The three main SCFAs found in the gut are acetate (Fig. 8a), butyrate (Fig. 8b), and propionate (Fig. 8c). DNBS induced a decrease in these three SCFAs compared to that in the CTRL-vehicle group. In contrast, an increase in SCFAs was observed in the C. minuta DSM 22607 group (p < 0.05), mitigating the effects of DNBS-induced inflammation on these metabolites. A similar effect was observed for valerate (p < 0.01; Fig. 8d). For both branched chain fatty acids (BCFA), isobutyrate (Fig. 8e), and isovalerate (Fig. 8f), no effects were observed following treatment with C. minuta.
Fig. 8. C. minuta increases cecal short chain fatty acid (SCFA) production.
Assessment of acetate (a); butyrate (b); propionate (c); valerate (d); isobutyrate (e) and isovalerate (f) production in the cecum. CTRL-Vehicle (N = 26; white); DNBS-Vehicle (N = 29; dark gray); DNBS-DSM 22607 (N = 31; green). Data are means and SEM. Results of the Original FDR method of Benjamini and Hochberg multiple comparison tests comparing the DNBS-Vehicle group to the other three groups. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
DSM 22607 modulates inflammatory and functional genes in the colon
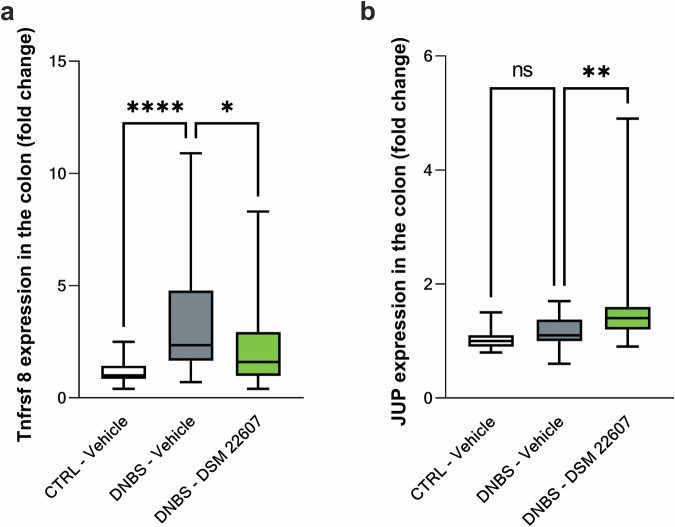
RNA samples (CTRL-Vehicle (N = 20); DNBS-Vehicle (N = 19)); DNBS-DSM 22607 (N = 19) were sequenced with a yield of 50 million reads per sample. Multivariate discriminant analysis using an sPLS-DA approach enabled the identification of the top 100 genes (Table 1) that discriminated the inflamed groups DNBS-DSM 22607 and DNBS-Vehicle: 82 downregulated and 18 upregulated by DSM 22607. We observed that our treatment reduced the expression of genes involved in inflammation (Tnfrsf8 and Atg5), mitochondrial membrane (Tomm7 and Tmem126a), cell migration, and tumorigenesis (Sn x 16). To validate the transcriptomic analysis, we performed RT-qPCR analysis of two genes that were found to be modulated and associated with the observed phenotype: one involved in inflammation, a member of the TNF receptor family (Tnfrsf8), and the other linked to cell-cell adhesion (Jup, a plakoglobin protein that is a component of adherent junctions and desmosomes). We performed this quantification in the colon samples. As expected, Tnfrsf8 was significantly decreased (p < 0.05; Fig. 9a), and Jup was significantly increased (p < 0.01; Fig. 9b) in the DNBS-DSM 22607 group. These results validated the results of transcriptome analysis and led us to continue investigating the mechanisms of action of C. minuta.
Table 1.
Top 100 genes modulated by DSM 22607 compared to DNBS-vehicle via sPLS-DA approach
| GeneID | SYMBOL | logFC | adj.P.Val | GeneID | SYMBOL | logFC | adj.P.Val |
|---|---|---|---|---|---|---|---|
| ENSMUSG00000028602 | Tnfrsf8 | −0,99743626 | 0,29510731 | ENSMUSG00000002580 | Mien1 | −0,22417223 | 0,30891771 |
| ENSMUSG00000047854 | Stx19 | −0,72030651 | 0,27035889 | ENSMUSG00000024293 | Esco1 | −0,22040676 | 0,20828067 |
| ENSMUSG00000023919 | Cenpq | −0,46702204 | 0,30891771 | ENSMUSG00000025967 | Eef1b2 | −0,21902614 | 0,28500164 |
| ENSMUSG00000114540 | NA | −0,44988898 | 0,34684554 | ENSMUSG00000078970 | Wdr92 | −0,21818105 | 0,30760988 |
| ENSMUSG00000096010 | H4f16 | −0,43470579 | 0,30891771 | ENSMUSG00000037531 | Mrpl47 | −0,2173443 | 0,30891771 |
| ENSMUSG00000114456 | H2bc9 | −0,42550252 | 0,29950651 | ENSMUSG00000022370 | Mrpl13 | −0,21664537 | 0,29951729 |
| ENSMUSG00000078974 | Sec61g | −0,41581979 | 0,25057889 | ENSMUSG00000063362 | Alg11 | −0,21499792 | 0,19625406 |
| ENSMUSG00000030521 | Mphosph10 | −0,38416676 | 0,30760988 | ENSMUSG00000026568 | Mpc2 | −0,21294554 | 0,27611749 |
| ENSMUSG00000036781 | Rps27l | −0,37678571 | 0,22844889 | ENSMUSG00000001891 | Ugp2 | −0,21110044 | 0,30891771 |
| ENSMUSG00000052033 | Pfdn4 | −0,37455825 | 0,28500164 | ENSMUSG00000031696 | Vps35 | −0,21053581 | 0,28118045 |
| ENSMUSG00000032330 | Cox7a2 | −0,36291934 | 0,20828067 | ENSMUSG00000020076 | Ddx50 | −0,21023024 | 0,20828067 |
| ENSMUSG00000028175 | Depdc1a | −0,3620709 | 0,33499303 | ENSMUSG00000054302 | Eapp | −0,20672131 | 0,23936558 |
| ENSMUSG00000062328 | Rpl17 | −0,32598048 | 0,27035889 | ENSMUSG00000034800 | Zfp661 | −0,20642286 | 0,3849899 |
| ENSMUSG00000028295 | Smim8 | −0,3238612 | 0,27755867 | ENSMUSG00000020397 | Med7 | −0,20403306 | 0,29510731 |
| ENSMUSG00000069208 | Zfp825 | −0,31927764 | 0,27035889 | ENSMUSG00000029462 | Vps29 | −0,20114252 | 0,30891771 |
| ENSMUSG00000030469 | Zfp719 | −0,31692917 | 0,19305783 | ENSMUSG00000026500 | Cox20 | −0,19770674 | 0,34568622 |
| ENSMUSG00000058291 | Zfp68 | −0,30754312 | 0,25057889 | ENSMUSG00000046603 | Tcaim | −0,1971252 | 0,29969993 |
| ENSMUSG00000003198 | Zfp959 | −0,30357571 | 0,29142092 | ENSMUSG00000040044 | Orc3 | −0,19574769 | 0,25900506 |
| ENSMUSG00000032673 | Prorsd1 | −0,29841183 | 0,25057889 | ENSMUSG00000037075 | Rnf139 | −0,19447024 | 0,27035889 |
| ENSMUSG00000027534 | Snx16 | −0,29345952 | 0,27575137 | ENSMUSG00000026577 | Blzf1 | −0,18959363 | 0,30891771 |
| ENSMUSG00000074521 | NA | −0,29060099 | 0,39568849 | ENSMUSG00000051469 | Zfp24 | −0,18958995 | 0,19305783 |
| ENSMUSG00000031347 | Cetn2 | −0,28992334 | 0,25406939 | ENSMUSG00000025979 | Mob4 | −0,18572347 | 0,29950651 |
| ENSMUSG00000027835 | Pdcd10 | −0,28964885 | 0,26112428 | ENSMUSG00000027937 | Jtb | −0,18161537 | 0,29510731 |
| ENSMUSG00000048495 | Tyw5 | −0,28875924 | 0,27035889 | ENSMUSG00000066880 | Zfp617 | −0,18004659 | 0,28118045 |
| ENSMUSG00000021290 | Atp5mpl | −0,28574474 | 0,20828067 | ENSMUSG00000031479 | Vps36 | −0,17721061 | 0,27040179 |
| ENSMUSG00000041153 | Osgin2 | −0,27926591 | 0,29510731 | ENSMUSG00000041124 | Msantd4 | −0,17512831 | 0,30891771 |
| ENSMUSG00000068882 | Ssb | −0,27668082 | 0,25057889 | ENSMUSG00000021023 | Prorp | −0,17440299 | 0,34376946 |
| ENSMUSG00000070394 | Tmem256 | −0,27331311 | 0,26728859 | ENSMUSG00000071267 | Zfp942 | −0,17016226 | 0,4057334 |
| ENSMUSG00000042670 | Immp1l | −0,27226643 | 0,30891771 | ENSMUSG00000036299 | BC031181 | −0,16428556 | 0,34376946 |
| ENSMUSG00000021711 | Trappc13 | −0,26365975 | 0,19305783 | ENSMUSG00000027088 | Phospho2 | −0,16215153 | 0,35840453 |
| ENSMUSG00000078994 | Zfp429 | −0,26043082 | 0,30760988 | ENSMUSG00000037860 | Aim2 | −0,16038052 | 0,40282073 |
| ENSMUSG00000073448 | NA | −0,25843035 | 0,34906971 | ENSMUSG00000017686 | Rhot1 | −0,15711633 | 0,2254286 |
| ENSMUSG00000028998 | Tomm7 | −0,25808785 | 0,27035889 | ENSMUSG00000001552 | Jup | 0,308949158 | 0,19625406 |
| ENSMUSG00000078862 | Gm14326 | −0,25441179 | 0,32043191 | ENSMUSG00000039910 | Cited2 | 0,388552298 | 0,25900506 |
| ENSMUSG00000026021 | Sumo1 | −0,25411878 | 0,19625406 | ENSMUSG00000002763 | Pex6 | 0,389786576 | 0,19305783 |
| ENSMUSG00000021022 | Ppp2r3c | −0,25270256 | 0,28118045 | ENSMUSG00000000631 | Myo18a | 0,42506165 | 0,19305783 |
| ENSMUSG00000030615 | Tmem126a | −0,25162666 | 0,25057889 | ENSMUSG00000020747 | Tmem94 | 0,434966894 | 0,20828067 |
| ENSMUSG00000045624 | Esf1 | −0,25147634 | 0,30891771 | ENSMUSG00000037686 | Aspg | 0,453762602 | 0,27035889 |
| ENSMUSG00000116802 | Gm5165 | −0,24678902 | 0,33565587 | ENSMUSG00000044707 | Ccnjl | 0,471308593 | 0,29510731 |
| ENSMUSG00000028563 | Tm2d1 | −0,24532911 | 0,30891771 | ENSMUSG00000022199 | Slc22a17 | 0,4767096 | 0,25057889 |
| ENSMUSG00000039270 | Megf9 | −0,24455218 | 0,30891771 | ENSMUSG00000022096 | Hr | 0,540875898 | 0,21432634 |
| ENSMUSG00000040374 | Pex2 | −0,24419174 | 0,25057889 | ENSMUSG00000048916 | NA | 0,554931734 | 0,28996021 |
| ENSMUSG00000024145 | Pigf | −0,2437491 | 0,30891771 | ENSMUSG00000097336 | Fendrr | 0,585339795 | 0,20828067 |
| ENSMUSG00000053289 | Ddx10 | −0,24056409 | 0,28255213 | ENSMUSG00000045348 | Nyap1 | 0,598431945 | 0,29510731 |
| ENSMUSG00000033983 | Coil | −0,23576517 | 0,28118045 | ENSMUSG00000064345 | ND2 | 0,64893192 | 0,28527504 |
| ENSMUSG00000002728 | Naa20 | −0,23397621 | 0,28118045 | ENSMUSG00000038473 | Nos1ap | 0,66887166 | 0,28118045 |
| ENSMUSG00000038160 | Atg5 | −0,22954702 | 0,22844889 | ENSMUSG00000110772 | NA | 0,701002337 | 0,28500164 |
| ENSMUSG00000074781 | Ube2n | −0,22927426 | 0,25057889 | ENSMUSG00000090561 | NA | 0,739140612 | 0,36375605 |
| ENSMUSG00000038628 | Polr3k | −0,22897652 | 0,23936558 | ENSMUSG00000109325 | NA | 0,75294619 | 0,30891771 |
| ENSMUSG00000062931 | Zfp938 | −0,22658542 | 0,35840453 | ENSMUSG00000108365 | NA | 1,005271021 | 0,27035889 |
Fig. 9. C. minuta significantly modulated Tnfrsf8 and Jup gene expressions.
Tnfrsf8 (a) and Jup (b) gene expression in the colon. CTRL-Vehicle (N = 26); DNBS-Vehicle (N = 29); DNBS-DSM 22607 (N = 31). Data are means and SEM. Results of the Original FDR method of Benjamini and Hochberg multiple comparison tests comparing the DNBS-Vehicle group with the other two groups. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
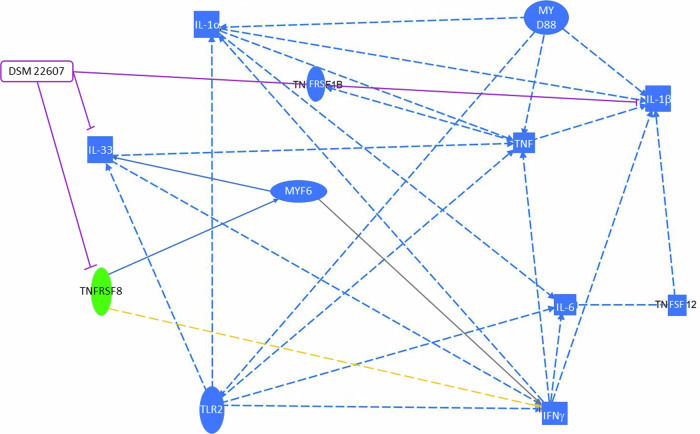
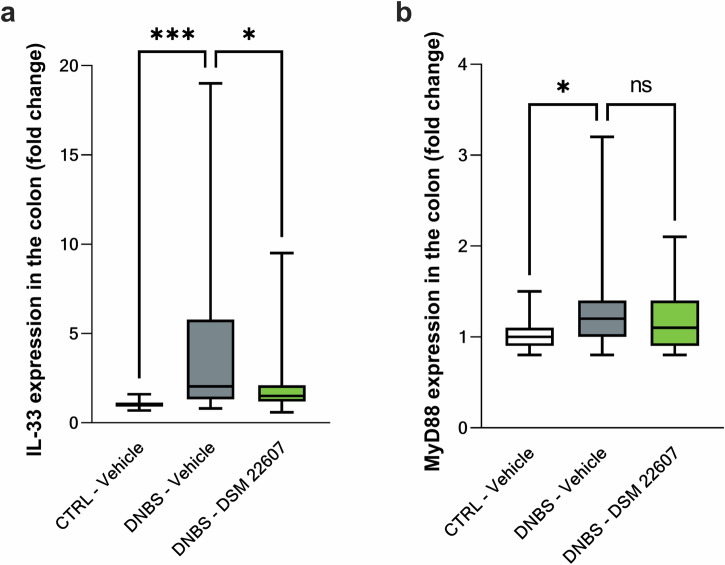
Finally, transcriptomic results were analyzed using ingenuity pathway analysis (IPA) software. Analysis of upstream regulators and networks showed inhibition of il-33 and siglec8 following DSM 22607 administration. Interestingly, il-33 and tfnrsf8 expression depends on the same transcription regulator, MYF6, and is linked to the MyD88 pathway (Fig. 10). Therefore, we quantified il-33 and myd88 gene expression in the colon by qPCR, which is linked to the activation of the pro-inflammatory NF-κB pathway34,35. We observed that DSM 22607 decreased il-33 (p < 0.05; Fig. 11a) without modulating myd88 gene expression (Fig. 11b).
Fig. 10. Casual network involved in DSM 22607 effects.
Legend: Leads to an inhibition (blue); findings inconsistent with state of downstream molecule (yellow); effect not predicted (gray); inhibition induced by DSM 22607 (purple). Activation, causation or expression (entire arrow); indirect interaction (streaked arrows). Cytokine (square); transmembrane receptor (oval); transcription regulator (elongated oval); other (circle). Decreased measurement (green oval); predicted inhibition (blue oval).
Fig. 11. C. minuta inhibited IL-33 pathway independently of MyD88 gene expression.
IL-33 (a) and MyD88 (b) gene expression in the colon. CTRL-Vehicle (N = 26; white); DNBS-Vehicle (N = 29; dark gray); DNBS-DSM 22607 (N = 31; green). Data are means and SEM. Results of Original FDR method of Benjamini and Hochberg multiple comparison tests comparing the DNBS-Vehicle group to the two other groups. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Discussion
One promising innovative approach for the management of IBD is the use of microbiome-based therapies. Thus, the use of a single keystone bacterium from the human intestinal microbiota, such as C. minuta is an interesting therapeutic option to complement existing treatments that fail to address underlying gut microbial dysbiosis. Consequently, following the discovery of C. minuta anti-inflammatory properties of C. minuta in both in vitro and in vivo models32, our objective was to further explore its mechanisms of action.
First, we determined the effects of C. minuta extract on the structure of the intestinal barrier. In a previous study, C. minuta DSM 22607 partially restored permeability in an in vitro model of intestinal cells, whereas no improvements were observed in DNBS-induced systemic hyperpermeability in vivo32. To assess local intestinal permeability in vivo, we tested colon, ileum, and cecum permeability in Ussing chambers after DNBS challenge. Two permeability pathways were measured: (i) paracellular, which can result from dysregulated tight junction (TJ) protein expression and (ii) transcellular, which is defined as passive diffusion through cells36. We observed a significant effect of C. minuta DSM 22607 on the paracellular pathway but not on the transcellular pathway. IBD is associated with dysfunction of the paracellular pathway, increasing permeability due to TJs dysfunction (i.e., Claudin or Occludin) and other junctional complexes such as desmosomes11,37. In addition, colonic TEER was increased after DSM 22607 administration, confirming the improvement of paracellular permeability, since 90% of the total TEER in the colonic tissue is due to the paracellular pathway38. It has recently been reported that probiotics such as Bifidobacterium bifidum could play a role in TJs regulation in animal models of colitis in a TLR-2-dependent manner, compensating for the deleterious effects induced by DNBS39. Interestingly, we observed upregulation of Jup expression, a gene involved in the regulation of cell-cell adhesion, such as desmosomes and E-cadherin adhesion complexes (UniProtKB, Q02256), which are essential proteins regulating the paracellular pathway. In addition, it has been demonstrated that desmosome adhesion during IBD plays a major role to stabilize and reverse intestinal permeability40. The mucus layer is another essential component of the intestinal barrier structure41. DSM 22607 treatment prevented the depletion of mucus-producing cells induced by DNBS, a trait also observed in IBD patients33. MUC2 staining confirmed that C. minuta DSM 22607 limited mucus thinning in the colon. These results corroborate that DSM 22607 can limit intestinal damage during IBD by protecting the intestinal barrier.
The intestinal epithelial barrier is also composed of an inner layer known as the lamina propria which hosts numerous immune cells42. Previously, we observed that DSM 22607 has anti-inflammatory effects in vitro by decreasing IL-8 cytokine production in TNF–α stimulated HT-29 cells and in vivo by reducing MPO activity in a murine colitis model32. Thus, we monitored the immunomodulatory effects of DSM 22607 at local and systemic levels. At the local level, we observed a decrease in colonic IL-1α and MCP-1 levels, two cytokines involved in IBD43,44. MCP-1 is induced by TNF-α and is involved in macrophage recruitment45. Interestingly, macrophages are known to release IL-1α, which suggests that the observed decrease in the colon may be due to lower macrophage infiltration46. This finding is consistent with the beneficial ability of some bacteria to modulate inflammation. For example, Akkermansia muciniphila has been shown to decrease inflammation in the colon through the reduction of MCP-1 release in a DSS-induced colitis model47. Other probiotics have also demonstrated the ability to prevent the synthesis of pro-inflammatory cytokines, such as IL-8, TNF-, α and MCP-1, in UC patients48. At a systemic level, we observed that C. minuta treatment led to a decrease in MCP-1 and tended to decrease IFNγ and TNF-α cytokines in the serum, indicating a systemic immunomodulatory effect. Thus, to characterize the adaptive immune response, which is known to play an important role in IBD49, we evaluated cytokine release by activated lymphocytes in the spleen. We observed a decrease in TNFα and, interestingly, a decrease in IL-4, IL-5, and IL-6. The spleen is a secondary lymphoid tissue involved in the main immune response, and its function is affected during IBD50. TNF-α, produced by M1 macrophages, is known to be a major Th1 cytokine in IBD, with pro-inflammatory effects51. To manage inflammation, anti-TNF-α therapy has been developed to hamper the pro-inflammatory response in IBD52. Moreover, the reduction of TNF-α, an inducer of the NF-κB inflammatory pathway53 following C. minuta administration is consistent with the previous observation of the inhibition of the NF-κB pathway32. Researchers have also highlighted a high prevalence of allergic diseases in patients with CD54, associated with Th2 cytokines, such as IL-4, IL-5, and IL-655. The decrease in IL-4 suppresses the development of colitis in mice56 and the inhibition of IL-6 induces immunosuppression in patients57. These results showed that DSM 22607 displayed strong anti-inflammatory properties and confirmed its immunomodulatory potential.
Gut microbiota, a component of the intestinal barrier, is altered during IBD12. Therefore, we hypothesized that C. minuta DSM 22607 could exert some protective effects through the modulation of the gut microbiota and observed that DNBS induced a decrease in gut microbiota richness and diversity, including an increase in some bacterial families such as Enterobacteriaceae and Bacteroidaceae and a decrease in Lactobacillaceae family. All these modulations have also been previously reported in IBD in humans and mouse colitis models16–18,22. Interestingly, C. minuta administration was correlated with the abundance of Akkermansia spp., a genus widely documented for its positive action on the gut barrier58 and Dubosiella sp., shown to be positively correlated with the increase of butyric acid and negatively correlated with IL-1b and IL-6 mRNA expressions in a DSS mouse colitis model59. Recently, the presence of these two species was reported to be positively correlated: following oral administration of pasteurized A. muciniphila, the authors observed a significant increase in Dubosiella sp., thereby alleviating intestinal damage induced by DSS in mice60. These microbial modulations are associated with functional modifications, as illustrated by the changes in SCFAs production. DSM 22607 treatment increased the levels of butyrate, acetate, propionate, and valerate in the cecum. DSM 22607 is a high acetate and low butyrate producer32 and a recent study showed that the addition of C. minuta DSM 33407 was sufficient to stimulate the production of acetate, butyrate and propionate, known as the three main SCFAs27. The increase in Akkermansia and Dubosiella could also help explain the restoration of SCFA levels as butyrate, acetate, and propionate by modulating microbial metabolism and downregulating Th1 cytokines in the colon61. Interestingly, acetate and butyrate were associated with MUC2 expression in a dose-dependent manner62. Acetate is also involved in GC differentiation through the activation of the KLF4 transcription factor63; butyrate is well known to have anti-inflammatory effects, notably by inhibiting the NF-κB pathway64. Therefore, it is possible that the increased production of these SCFAs largely contributes to other protective effects on the intestinal barrier observed following C. minuta treatment. Christensenellaceae is considered as a gut microbiota keystone species known to held a role as a hub in a co-occurrence network that includes heritable taxa26. Multiple studies indicate that Akkermensia spp., Dubosiella spp., Christensenellaceae and other families as Rikenellaceae are often positively correlated. For instance, it has been sown that a combination of baicalin and berberine hydrochloride ameliorates Dextran Sulfate Sodium-Induced Colitis by increasing Christensenellaceae and Erysipelotrichaceae (Dubosiella family) and modulating inflammation in a DSS colitis model65. Moreover in 2023, polysaccharides from the seeds Gleditsia sinensis stimulated the abundance of Akkermansia spp. and Christensenella spp. which could be linked to the improvement of intestinal barrier by increasing Muc2 expression, tight junction protein and SCFA amount66. All these information established that C. minuta administration induced several shifts with beneficial effects in our mouse model confirming the importance of this bacterium.
Finally, Tnfrsf8 levels were significantly decreased in the C. minuta-treated group. Tnfrsf8 plays a major role in the management of inflammatory diseases by activating innate and adaptive immune cells34. For example, in a TNBS-induced colitis model, Tnfrsf8 KO mice were resistant to TNBS-associated effects67. Moreover, network analyses revealed that DSM 22607 inhibits IL-33 and SIGLEC 8, both of which are associated with pro-inflammation. IL-33, which activates SIGLEC 8-mediated apoptosis68, is a member of the IL-1 family that induces T cell differentiation into Th1 cells, maturation of dendritic cells, upregulation of natural killer (NK) cells, and CD8+ cells35 and drives type 2 innate lymphoid cells in allergy69. However, here, IL-33 expression in the colon was not regulated via Myd88. Other studies have observed a role of IL-33 due to microbiota modulation during inflammation. The administration of A. muciniphila has been found to restore the diversity of the intestinal microbiota depleted during abdominal aortic aneurysms and reduced markers of inflammation such as IL-3370. In contrast, Arifuzzaman and co workers showed recently that inulin-induced gut microbiota modifications, which altered microbiota-derived metabolites as bile acids and enhanced the expression of IL-3371. More studies should be performed to determine which is the mechanism that allow C. minuta to decrease IL-33 production.
In conclusion, our research confirmed the protective effects of C. minuta DSM 22607 on the intestinal epithelial barrier and its anti-inflammatory effects in a rodent model of IBD. This study also provides new insights into the mechanisms of action of C. minuta DSM 22607 and further supports the need for a clinical trial to validate the use of C. minuta strains as a microbiome-based therapy to manage IBD.
Methods
Bacteria growth conditions
Christensenella minuta DSM 22607 was grown in GAM broth modified medium (HyServe, Germany) as described by Kropp et al.32. Briefly, after revivification of a stock for 4 days at 37 °C under anaerobic conditions, a preculture was obtained from a single colony grown in GAM for 2 days. Finally, to prepare gavage aliquots, this preculture was used to inoculate the final culture under the same conditions. The culture was centrifuged at 2500 g during 20 min, washed in phosphate-buffered saline 1X (PBS) (Gibco, Thermo Fisher Scientific, USA), resuspended in PBS with 16% glycerol (PBS/glycerol) at 5 *109 CFU/mL, and stored at -80°C until use.
Mouse model experimental design
Eighty-eight 7-week-old male C56BL/6 J mice were obtained from Janvier Lab and maintained under specific pathogen-free (SPF) conditions in the animal facilities of the National Research Institute for Agriculture, Food and Environment (IERP, INRAE, Jouy-en-Josas, France). The experiments were performed in two separate batches, and the mice were housed in cages of 4 or 5. All animal experiments were performed in accordance with the European Community Guidelines for Animal Care. Animal care and work protocols were approved by the local regional ethical committee Comethea according to EU Directive 2010/63/EU. After 7 days of acclimation, the mice were divided into three groups: control non-inflamed mice which received 150 µL of PBS/glycerol solution (vehicle) by oral gavage (CTRL-Vehicle, n = 26); control inflamed mice that received 150 µL of vehicle by oral gavage (DNBS-Vehicle, n = 30), treated mice that received C. minuta DSM 22607 at 109 CFU/mL by oral gavage (DNBS-DSM 22607, n = 32). Two weeks after the start of daily force-feeding, the mice were anesthetized by intraperitoneal injection of 0.1% ketamine and 0.06% xylazine and received by intra rectal injection a solution of 165 mg/kg DNBS dissolved in ethanol 30%. The control group without inflammation (CTRL-Vehicle) received an intra rectal injection with a solution of ethanol 30%. Three days after DNBS injection, the mice were euthanized by cervical dislocation just after taking a terminal blood sample from the submandibular vein. Body weight variation was monitored daily, and macroscopic and microscopic scores were obtained as previously described32. Briefly, a 1 cm piece of colon was collected and fixed for 24 h in a 4% paraformaldehyde (PFA) solution before dipping it in 70% ethanol. Samples were embedded in paraffin using a tissue embedding system (Leica), cut into 5 μm sections using a microtome (UC6, Reicher E - Leica UC6), and then stained with hematoxylin and eosin (HE) for histological scoring using an automated staining system (Leica). Tissue sections were evaluated to characterize alterations in mucosal architecture and the degree of immune cell infiltration72. DNBS-induced chronic inflammation is also visible at the macroscopic level, and inflammation intensity can be evaluated by measuring different parameters as damage in colon tissue and stool consistency, using Wallace’s score73 with the following modifications: tissue sections from each mouse were scored by evaluating ulcerations (score of 0–5), adhesions (presence/absence: 0/1), hyperemia (presence/absence: 0/1), altered transit with constipation (presence/absence: 0/1), and increases in colon wall thickness (presence/absence: 0/1; measured using an electronic caliper, Control Company, WVR, United States).
To observe the effect of C. minuta on local permeability, Ussing chamber experiments were performed. Thirty-three 7-week-old C56BL/6 J male mice were obtained from Janvier Lab, maintained under SPF conditions in IERP animal facilities, and housed in cages of 3. Mice were divided into three groups (CTRL-Vehicle: n = 9, DNBS-Vehicle: n = 12, and DNBS-DSM 22607: n = 12) and the same protocol as for the DNBS experiment was followed.
MPO activity determinations
1 cm section of colon was placed in liquid nitrogen and stored at -80°C until MPO dosage to determine inflammation74. Briefly, after adding 300 µl of hexadecyl trimethyl ammonium bromide (HTAB) (Sigma-Aldrich, USA) buffer, tissues were homogenized for three cycles of 30 s at 8000 rpm at 4°C with Precellys® evolution coupled to Cryolys® evolution (Ozyme, France). The samples were centrifuged at 14,000 x g for 5 min, and the supernatants were collected. o-Dianisidine 1% H202 solution (200 µL, Sigma-Aldrich, USA) was added to 6 µL of the samples in a 96-well plate. One hour later, the plates were read using a spectrophotometer at 450 nm (TECAN, Switzerland). The results were normalized to the sample weight.
Measurement of general intestinal permeability in vivo
Blood was collected on the day of necropsy in microtest Kima tubes (Labellians, France) and processed immediately by centrifugation at 2000 g for 10 min at room temperature. Serum was stored at -80°C until use. Lipopolysaccharide (LPS)-binding protein (LBP) (Hycult Biotech, Netherlands) in serum was measured by ELISA according to the manufacturer’s specifications.
Ex vivo local permeability measurement
Three days after DNBS injection, mice were euthanized and the colon, ileum, and cecum were collected; content samples were removed by flushing with cold PBS 1X and immediately mounted in Ussing chambers as described previously75. To assess the para- and trans-cellular permeability, we measured the mucosal-to-serosal flux of three different molecules over 120 min: HRP (40 kDa, type II, Merck, Germany), FSA (500 Da, Thermo Fisher Scientific, USA), and TRITC-dextran (4 kDa, TdB Labs, Sweden). Molecules were added to the mucosal side of the chamber at final concentrations of 200 µg/mL, 40 µg/mL, and 400 µg/mL. The electrical part was assessed by clamping the voltage and recording the change in conductance (G) and resistance (Ω) over 2 h. The results are expressed as conductivity and resistance per second and represented by the area under the curve of these measurements.
Staining of mucus producing cells and MUC2 determinations
One centimeter section of the colon was fixed for at least 3 weeks in water-free methacarn solution76. The tissues were then embedded in paraffin (VWR, USA), and sections were cut using a microtome (Leica, USA). AB staining was performed to color the mucins present in GC, according to standard protocols. To avoid bias, the % of positive AB cells in each crypt was normalized to the total cell count. For MUC2 determination, immunohistochemistry was performed according to standard methods using Leica BOND RXm. The protocol included a deparaffinization and rehydration step, an acidic antigen retrieval step (20 min at 100°C in BondTM Epitope Retrieval 1), and blocking with 1% BSA and 10% Normal Goat Serum for 30 min at room temperature. The primary antibody anti-MUC2 (dilution 1/500, Novus Biological, USA) was applied to the slides for 1 h at room temperature, and anti-rabbit secondary antibody ((dilution 1/200, Goat anti-Rabbit IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 568, Invitrogen, USA) with DAPI) was added for 30 min at room temperature. The slides were mounted using Fluoromount® (Cliniscience, France). All slides were scanned using a Pannoramic SCAN II automated slide scanner (3D HISTECH, Hungary) at 20X and analysed using Caseviewer software.
Cytokine determinations
Cytokines were measured locally in the colon and at systemic levels in serum samples using the LEGENDplex mouse inflammation panel IL-1α, IL-1β, IL-6, IL-10, IL-12p60, IL-16A, IL-23, IL-26, MCP-1, IFN-β, IFN-γ, TNF-α, and GM-CSF (BioLegend, USA) according to the manufacturer’s instructions. The samples were analyzed using a flow cytometer Accuri C6 (BD Biosciences, USA). Samples were analyzed using LEGENDplex software according to the data analysis software suite manual (legendplex.qognit.com; Supplementary Figure 5). For colonic concentrations, the results were normalized based on tissue weight.
Spleen cells stimulation
Spleens were collected in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Thermo Fisher Scientific, USA) and kept on ice. Spleens were smashed in a 40 µm sterile cell strainer (Falcon®), and cells were collected in 2 mL mL DMEM supplemented with 10% fetal calf serum (FCS) (Eurobio Scientific, France) and 1% penicillin/streptomycin cocktail (Sigma-Aldrich, USA). The spleens were centrifuged at 300 g for 10 min at 4°C. Supernatants were removed and 1 mL of 1X red blood cell lysis buffer was added to lyse the erythrocytes. After 5 min, complete DMEM was added to stop the reaction and the samples were centrifuged under the same conditions. Finally, the supernatant was removed, and the cell pellets were resuspended in 3 mL of complete DMEM. Cells were counted on an Accuri C6 cytometer, and 1 million lymphocytes per mouse were deposed in 24 well-plates. cell stimulation cocktail (PMA/ionomycin, Invitrogen, USA) was added to stimulate immune cells, and plates were incubated at 37°C for 48 h under 10% of CO2. At The end, supernatants were collected and stored at -80°C until use. Cytokines were determined using a T helper cytokine panel LEGENDPLEX assay (IL-2, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12p70, IL-13, IL-17A, IL-17F, IL-22, IFN-γ, and TNF-α; BioLegend, USA) according to the manufacturer’s instructions. The samples were read and analyzed as explained above.
SCFA determination in cecum samples
The day of the sacrifice, cecal samples were collected and placed in liquid nitrogen at −80 °C. SCFA were extracted from the samples with water (wt g/vol) and centrifuged for 15 min at 15000xg. Supernatants were collected and deproteinized overnight at 4°C by the addition of phosphotungstic acid (10%, Sigma-Aldrich, USA). The samples were processed and analyzed as described by Kropp et al.32.
DNA extraction in feces
Mouse feces were sampled at three time points: day 0 (T0), 14 (T14), and 17 (T17) (Supplementary Fig. 4) and stored at −80 °C until total genomic DNA extraction using Godon’s protocol77. Feces were suspended in 250 µL of 4 M guanidine thiocyanate (Sigma-Aldrich, USA) and 40 µL of 10% N-lauroyl sarcosine (Sigma-Aldrich, USA) solution. Samples were vortexed and incubated for 10 min at room temperature. Then, 500 µL of 5% N-lauroyl sarcosine was added, and samples were incubated 1 h at 60°C under agitation. 0.1 mm beads were used for bacterial cell lysis, and feces were homogenized three cycles of 30 s at 10,000 rpm at 4 °C with Precellys® evolution coupled with Cryolys® (Ozyme, France) evolution. Subsequently, 20 mg polyvinylpyrrolidone (PVPP) (Sigma-Aldrich, USA) was added, and the suspension was centrifuged at 20,000 × g at 4 °C for 5 min. The supernatants were recovered in a separate tube. Pellets were washed twice with 650 μL of a 6,5 pH 1 M Tric-Cl, 0,5 M EDTA, 5 M NaCl, 1% PVPP, and mQ water solution (TENP buffer), centrifuged, and supernatants were recovered in the same tube. The pooled supernatants were centrifuged at 20,000 × g and 4 °C for 1 min. Nucleic acids were precipitated using isopropanol. Following 10 min incubation at room temperature, the pool was centrifuged at 20,000 × g at 4 °C for 10 min, and the pellet was resuspended in 450 µL of a pH 8 0,1 M phosphate buffer. Fifty µL of M potassium acetate 5 M solution was added, and samples were left at 4 °C overnight. Finally, samples were centrifuged at 20,000xg at 4 °C for 30 min. supernatants were collected, and 2 μL of RNAse (10 mg/mL, Fisher Scientific) was added and incubated at 36 °C for 30 min. DNA was precipitated using 50 μL of 3 M sodium acetate and 1000 µL ice-cold 100% ethanol. Samples were centrifuged at 20,000 x g for 10 min at 4 °C the pellets were washed in 60% ethanol two times, dried, and stored at −20 °C in TE buffer.
Microbiota determinations by 16S rRNA sequencing from fecal content
The 16S rRNA gene segments spanning the variable V3 and V4 regions were amplified from fecal DNA using the Herculase II Fusion DNA Polymerase Nextera XT Index Kit V2 with the primers Bakt_341F 5′- CCTACGGGNGGCWGCAG -3′ and Bakt_805R 5′- GACTACHVGGGTATCTAATCC -3′78. The V3-V4 amplicons were prepared according to the 16S Metagenomic Sequencing Library Preparation Part #15044223 Rev. B and sequenced on an Illumina MiSeq platform. Sequences were processed using version 2 of the Quantitative Insights into Microbial Ecology (QIIME2) pipeline79. Cleaned sequencing reads were obtained after removing the barcode sequences, primers, and low-quality reads (Q < 30). The cleaned 16S rRNA gene sequences were processed using amplicon sequence variants (ASVs). Taxonomic assignment of the ASVs was performed using the primer-specific trained version of the GreenGenes2 Database80.
Polymerase chain reactions (PCR) to detect C. minuta
To confirm the results obtained after 16S sequencing of the fecal content regarding the presence of C. minuta, we decided to perform a PCR with primers specific for the amplification of C. minuta according to Jinthas and co workers81 on the gDNA samples of day 21 used for the microbiota analysis. Negative controls were also included (mouse gDNA, water and another bacterium).
RNA isolation and colon transcriptome analysis
One centimeter section of the distal colon (n = 80) was placed in RNA later (ThermoFisher Scientific, USA), frozen in liquid nitrogen, and stored at -80°C before RNA extraction using RNeasy mini kit (Qiagen, Germany) according to manufacturer’s specifications. RNA samples were checked for quality control, and qualified samples were used for library construction using the TruSeq Stranded Total RNA LT Sample Prep Kit (Human, Mouse, Rat, Illumina). The ribo-depleted fractions of the prepared libraries were paired-end sequenced (2 × 150 bp) on a NovaSeq6000 Illumina platform with a target sequencing yield of 50 million reads per sample. Cleaned sequencing reads were obtained after removing low-quality sections (Phred score < 33), reads shorter than 36 bp, and reads adapter. Contaminants (humans and microorganisms) were also eliminated, following the guidelines recommended by Kraken 2 (Wood, Lu, and Langmead 2019) recommended guidelines. The processed reads were then aligned and annotated based on the GRCm39 assembly version using STAR v2.6.9a (Dobin et al. 2013).
Lowly expressed genes were filtered out following the best practice recommended in edgeR v.3.14.1, using the filterByExpr function82. Prior to gene expression analysis, we examined potential outliers using a Mahalanobis distance83 and applied Bonferroni correction for multiple test comparisons.
Transcriptomic analysis was uploaded to Qiagen’s IPA system for core analysis and then overlaid with the global molecular network in the Ingenuity Pathway Knowledge Base (IPKB). IPA was performed to identify canonical pathways and gene networks that were differentially expressed after DSM 22607 administration compared with DNBS-Vehicle.
Quantitative reverse transcription (RT)-qPCR
Total RNA from each sample was reverse-transcribed using a high-capacity RNA-to-cDNA kit (ThermoFisher Scientific, USA). qPCR reactions were performed with the ROX probe 1X MasterMix dTTP blue (Takyon, Eurogentec, Belgium), primers with probe (Thermo Fisher Scientific, USA), and 125 ng of cDNA. The genes analyzed were TNF receptor superfamily member 8 (Tnfrsf8), junction plakoglobin (jup), interleukine-33 (il-33), myeloid differentiation primary response 88 (myd88), and the housekeeping gene TATA-box-binding protein (tbp). Amplification was performed using the StepOneTM Real-Time qPCR system (Applied Biosystems, USA). Cycle threshold (Ct) values were used to calculate fold-change values, according to Rao et al.84.
Statistical analysis
All results are expressed as the mean ± standard error of the mean (SEM). Statistical analyses were performed using GraphPad Prism version 9.4.0 (GraphPad Software, La Jolla, CA, USA). The observed differences were analyzed for statistical significance using one-way analysis of variance with the Original FDR method of Benjamini and Hochberg multiple comparison post-hoc tests or Kruskal–Wallis test with Dunn’s multiple comparisons as post-hoc tests. The differences were considered significant at *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Supplementary information
Acknowledgements
This work benefited from the facilities and expertize of @BRIDGe (Université Paris-Saclay, INRAE, AgroParisTech, GABI, 78350 Jouy-en-Josas, France). We wish to thank the staff of the INRAE Infectiology of Fishes and Rodents Facility (IERP-UE907, Jouy-en-Josas Research Center, France) for performing animal experiments. The IERP Facility belongs to the National Distributed Research Infrastructure for the Control of Animal and Zoonotic Emerging Infectious Diseases through an In Vivo Investigation. We also wanted to thank Carla Dos ANjos De Souza from DataOmics for the support on the bioinformatic analysis.
Author contributions
C.K., K.T., S.C. and R.M. performed experiments. P.L., S.P.C., and R.M. conceived and supervised the study. C.K. wrote the first draft of the manuscript. P.L., S.P.C., and R.M. corrected the manuscript. All authors have read and accepted the manuscript.
Data availability
The 16S RNA raw reads and transcriptomic raw data are available at 10.57745/XXAXFA and 10.57745/02ODSE respectively.
Competing interests
C.K. and S.C. were employees of Ysopia Bioscience. Founders participated in the study design. P.L. reports lecture fee, board membership, or consultancy from Biose, Biostime, Boiron, Bonduelle, BMS, Bromatech, IPSEN, iTaK, Lallemand, Lesaffre, L’Oréal, Mayoli, Merck, Procter and Gamble, Second Genome, Therascience and URGO and is co-founder of Exeliom Biosciences. RM declares no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41522-024-00540-6.
References
- 1.Patel, R. M. & Underwood, M. A. Probiotics and necrotizing enterocolitis. Semin. Pediatr. Surg.27, 39–46 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filidou, E. & Kolios, G. Probiotics in intestinal mucosal healing: a new therapy or an old friend? Pharmaceuticals (Basel)14, 1181 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simon, E., Călinoiu, L. F., Mitrea, L. & Vodnar, D. C. Probiotics, prebiotics, and synbiotics: implications and beneficial effects against irritable bowel syndrome. Nutrients13, 2112 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jakubczyk, D., Leszczyńska, K. & Górska, S. The effectiveness of probiotics in the treatment of inflammatory bowel disease (IBD)—a critical review. Nutrients12, 1973 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheykhsaran, E. et al. Gut microbiota and obesity: an overview of microbiota to microbial-based therapies. Postgrad. Med. J.99, 384–402 (2022). [DOI] [PubMed]
- 6.Lee, M. & Chang, E. B. Inflammatory bowel diseases (IBD) and the microbiome-searching the crime scene for clues. Gastroenterology160, 524–537 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abraham, C. & Cho, J. H. Inflammatory bowel disease. N. Engl. J. Med.361, 2066–2078 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stokkers, P. C. F. & Hommes, D. W. New cytokine therapeutics for inflammatory bowel disease. Cytokine28, 167–173 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Fernández-Tomé, S., Ortega Moreno, L., Chaparro, M. & Gisbert, J. P. Gut microbiota and dietary factors as modulators of the mucus layer in inflammatory bowel disease. Int. J. Mol. Sci.22, 10224 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang, S. & Yu, M. Role of goblet cells in intestinal barrier and mucosal immunity. J. Inflamm. Res.14, 3171–3183 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michielan, A. & D’Incà, R. Intestinal permeability in inflammatory bowel disease: pathogenesis, clinical evaluation, and therapy of leaky gut. Mediators Inflamm.2015, 628157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong, D., Gong, X., Wang, L., Yu, X. & Dong, Q. Involvement of reduced microbial diversity in inflammatory bowel disease. Gastroenterol. Res. Pract.2016, 6951091 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishida, A. et al. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin. J. Gastroenterol.11, 1–10 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Seksik, P. Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut52, 237–242 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sokol, H. et al. Prominence of ileal mucosa-associated microbiota to predict postoperative endoscopic recurrence in Crohn’s disease. Gut69, 462–472 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Baldelli, V., Scaldaferri, F., Putignani, L. & Del Chierico, F. The role of enterobacteriaceae in gut microbiota dysbiosis in inflammatory bowel diseases. Microorganisms9, 697 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nomura, K. et al. Bacteroidetes species are correlated with disease activity in ulcerative colitis. J. Clin. Med.10, 1749 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gkouskou, K. K., Deligianni, C., Tsatsanis, C. & Eliopoulos, A. G. The gut microbiota in mouse models of inflammatory bowel disease. Front Cell Infect. Microbiol.4, 28 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sokol, H. et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl Acad. Sci. USA105, 16731–16736 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halfvarson, J. et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat. Microbiol.2, 17004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rehman, A. et al. Geographical patterns of the standing and active human gut microbiome in health and IBD. Gut65, 238–248 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Sultan, S. et al. Metabolic influences of gut microbiota dysbiosis on inflammatory bowel disease. Front Physiol.12, 715506 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braun, T. et al. Individualized dynamics in the gut microbiota precede Crohnʼs disease flares. Am. J. Gastroenterol.114, 1142–1151 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Teofani, A. et al. Intestinal taxa abundance and diversity in inflammatory bowel disease patients: an analysis including covariates and confounders. Nutrients14, 260 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pittayanon, R. et al. Differences in gut microbiota in patients with vs without inflammatory bowel diseases: a systematic review. Gastroenterology158, 930–946.e1 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Goodrich, J. K. et al. Human genetics shape the gut microbiome. Cell159, 789–799 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazier, W. et al. A new strain of christensenella minuta as a potential biotherapy for obesity and associated metabolic diseases. Cells10, 823 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alard, J. et al. New probiotic strains for inflammatory bowel disease management identified by combining in vitro and in vivo approaches. Beneficial. Microbe.9, 317–331 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Mancabelli, L. et al. Identification of universal gut microbial biomarkers of common human intestinal diseases by meta-analysis. FEMS Microbiol. Ecol. 93, fix153 (2017). [DOI] [PubMed]
- 30.Banerjee, S., Schlaeppi, K. & van der Heijden, M. G. A. Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol.16, 567–576 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Tudela, H., Claus, S. P. & Saleh, M. Next generation microbiome research: identification of keystone species in the metabolic regulation of host-gut microbiota interplay. Front Cell Dev. Biol.9, 719072 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kropp, C. et al. The Keystone commensal bacterium christensenella minuta DSM 22607 displays anti-inflammatory properties both in vitro and in vivo. Sci. Rep.11, 11494 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morampudi, V. et al. DNBS/TNBS colitis models: providing insights into inflammatory bowel disease and effects of dietary fat. J. Vis. Exp.27, e51297 (2014). [DOI] [PMC free article] [PubMed]
- 34.Croft, M. et al. TNF superfamily in inflammatory disease: translating basic insights. Trends Immunol.33, 144–152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu, L. et al. IL33 activates CD8+T and NK cells through MyD88 pathway to suppress the lung cancer cell growth in mice. Biotechnol. Lett.42, 1113–1121 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Nagpal, R. & Yadav, H. Bacterial translocation from the gut to the distant organs: an overview. ANM71, 11–16 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Hu, J.-C. E. et al. Expression of tricellular tight junction proteins and the paracellular macromolecule barrier are recovered in remission of ulcerative colitis. BMC Gastroenterol.21, 141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clarke, L. L. A guide to Ussing chamber studies of mouse intestine. Am. J. Physiol. Gastrointest. Liver Physiol.296, G1151–G1166 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al-Sadi, R. et al. Bifidobacterium bifidum enhances the intestinal epithelial tight junction barrier and protects against intestinal inflammation by targeting the toll-like receptor-2 pathway in an NF-κB-independent manner. Int. J. Mol. Sci.22, 8070 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlegel, N., Boerner, K. & Waschke, J. Targeting desmosomal adhesion and signalling for intestinal barrier stabilization in inflammatory bowel diseases-lessons from experimental models and patients. Acta. Physiol. (Oxf)231, e13492 (2021). [DOI] [PubMed] [Google Scholar]
- 41.Cornick, S., Tawiah, A. & Chadee, K. Roles and regulation of the mucus barrier in the gut. Tissue Barriers3, e982426 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vancamelbeke, M. & Vermeire, S. The intestinal barrier: a fundamental role in health and disease. Expert Rev. Gastroenterol. Hepatol.11, 821–834 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murata, Y., Ishiguro, Y., Itoh, J., Munakata, A. & Yoshida, Y. The role of proinflammatory and immunoregulatory cytokines in the pathogenesis of ulcerative colitis. J. Gastroenterol.30, 56–60 (1995). [PubMed] [Google Scholar]
- 44.Singh, S., Anshita, D. & Ravichandiran, V. MCP-1: Function, regulation, and involvement in disease. Int. Immunopharmacol.101, 107598 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe, S., Kawamoto, S., Ohtani, N. & Hara, E. Impact of senescence-associated secretory phenotype and its potential as a therapeutic target for senescence-associated diseases. Cancer Sci.108, 563–569 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pyrillou, K., Burzynski, L. C. & Clarke, M. C. H. Alternative pathways of IL-1 activation, and its role in health and disease. Front Immunol.11, 613170 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qu, S. et al. Akkermansia muciniphila alleviates dextran sulfate sodium (DSS)-induced acute colitis by NLRP3 activation. Microbiol. Spectr.9, e0073021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghyselinck, J. et al. A 4-strain probiotic supplement influences gut microbiota composition and gut wall function in patients with ulcerative colitis. Int. J. Pharm.587, 119648 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Geremia, A., Biancheri, P., Allan, P., Corazza, G. R. & Di Sabatino, A. Innate and adaptive immunity in inflammatory bowel disease. Autoimmun. Rev.13, 3–10 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Kawashima, K. et al. Evaluation of the relationship between the spleen volume and the disease activity in ulcerative colitis and Crohn disease. Medicine (Baltimore)101, e28515 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shapouri-Moghaddam, A. et al. Macrophage plasticity, polarization, and function in health and disease. J. Cell Physiol.233, 6425–6440 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Peyrin-Biroulet, L. Anti-TNF therapy in inflammatory bowel diseases: a huge review. Minerva. Gastroenterol. Dietol.56, 233–243 (2010). [PubMed] [Google Scholar]
- 53.Hayden, M. S. & Ghosh, S. Regulation of NF-κB by TNF family cytokines. Semin. Immunol.26, 253–266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chudnow, M. L., Levy, M. B., Kelly, K. J. & Binion, D. G. Increased prevalence of environmental allergy in patients with Crohn’s disease. J. Allergy Clin. Immunol.113, S276 (2004). [Google Scholar]
- 55.León, B. & Ballesteros-Tato, A. Modulating Th2 cell immunity for the treatment of asthma. Front. Immunol.12, 637948 (2021). [DOI] [PMC free article] [PubMed]
- 56.Mizoguchi, A., Mizoguchi, E. & Bhan, A. K. The critical role of interleukin 4 but not interferon gamma in the pathogenesis of colitis in T-cell receptor α mutant mice. Gastroenterology116, 320–326 (1999). [DOI] [PubMed] [Google Scholar]
- 57.Schreiber, S. et al. Therapeutic interleukin-6 trans-signaling inhibition by olamkicept (sgp130Fc) in patients with active inflammatory bowel disease. Gastroenterology160, 2354–2366.e11 (2021). [DOI] [PubMed] [Google Scholar]
- 58.Collado, M. C., Derrien, M., Isolauri, E., de Vos, W. M. & Salminen, S. Intestinal integrity and akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl. Environ. Microbiol.73, 7767–7770 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wan, F. et al. Dihydroquercetin supplement alleviates colonic inflammation potentially through improved gut microbiota community in mice. Food Funct.12, 11420–11434 (2021). [DOI] [PubMed] [Google Scholar]
- 60.Xue, L. et al. The effects of live and pasteurized akkermansia muciniphila on DSS-induced ulcerative colitis, gut microbiota, and metabolomics in mice. Food Funct.14, 4632–4646 (2023). [DOI] [PubMed] [Google Scholar]
- 61.Zhai, R. et al. Strain-specific anti-inflammatory properties of two akkermansia muciniphila strains on chronic colitis in mice. Front. Cell. Infect. Microbiol.9, 239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burger-van Paassen, N. et al. The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: implications for epithelial protection. Biochem. J.420, 211–219 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Pérez-Reytor, D., Puebla, C., Karahanian, E. & García, K. Use of short-chain fatty acids for the recovery of the intestinal epithelial barrier affected by bacterial toxins. Front Physiol.12, 650313 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berni Canani, R. et al. The potential therapeutic efficacy of lactobacillus GG in children with food allergies. Pharmaceuticals5, 655–664 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan, Y. et al. A combination of baicalin and berberine hydrochloride aeliorates dextran sulfate sodium-induced colitis by modulating colon gut microbiota. J. Med. Food25, 853–862 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qin, N. et al. Polysaccharides from the seeds of Gleditsia sinensis Lam. attenuate DSS-induced colitis in mice via improving gut barrier homeostasis and alleviating gut microbiota dybiosis. Food Funct.14, 122–132 (2023). [DOI] [PubMed] [Google Scholar]
- 67.Sun, X. et al. A critical role of CD30 ligand/CD30 in controlling inflammatory bowel diseases in mice. Gastroenterology134, 447–458 (2008). [DOI] [PubMed] [Google Scholar]
- 68.Na, H. J., Hudson, S. A. & Bochner, B. S. IL-33 enhances siglec-8 mediated apoptosis of human eosinophils. Cytokine57, 169–174 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ngo Thi Phuong, N. et al. IL-33 drives expansion of type 2 innate lymphoid cells and regulatory T cells and protects mice from severe, acute colitis. Front. Immunol.12, 669787 (2021). [DOI] [PMC free article] [PubMed]
- 70.He, X., Bai, Y., Zhou, H. & Wu, K. Akkermansia muciniphila alters gut microbiota and immune system to improve cardiovascular diseases in murine model. Front Microbiol.13, 906920 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arifuzzaman, M. et al. Inulin fibre promotes microbiota-derived bile acids and type 2 inflammation. Nature611, 578–584 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ameho, C. K. et al. Prophylactic effect of dietary glutamine supplementation on interleukin 8 and tumour necrosis factor α production in trinitrobenzene sulphonic acid induced colitis. Gut41, 487–493 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wallace, J. L., MacNaughton, W. K., Morris, G. P. & Beck, P. L. Inhibition of leukotriene synthesis markedly accelerates healing in a rat model of inflammatory bowel disease. Gastroenterology96, 29–36 (1989). [DOI] [PubMed] [Google Scholar]
- 74.Barone, M. et al. A versatile new model of chemically induced chronic colitis using an outbred murine strain. Front. Microbiol. 9, 565 (2018). [DOI] [PMC free article] [PubMed]
- 75.Tomas, J. et al. Early colonizing Escherichia coli elicits remodeling of rat colonic epithelium shifting toward a new homeostatic state. ISME J.9, 46–58 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Howat, W. J. & Wilson, B. A. Tissue fixation and the effect of molecular fixatives on downstream staining procedures. Methods70, 12–19 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Godon, J. J., Zumstein, E., Dabert, P., Habouzit, F. & Moletta, R. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl. Environ. Microbiol.63, 2802–2813 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sinclair, L., Osman, O. A., Bertilsson, S. & Eiler, A. Microbial community composition and diversity via 16S rRNA gene amplicons: evaluating the illumina platform. PLoS ONE10, e0116955 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Almeida, A., Mitchell, A. L., Tarkowska, A. & Finn, R. D. Benchmarking taxonomic assignments based on 16S rRNA gene profiling of the microbiota from commonly sampled environments. GigaScience7, giy054 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McDonald, D. et al. Greengenes2 unifies microbial data in a single reference tree. Nat. Biotechnol.42, 715–718 (2023). [DOI] [PMC free article] [PubMed]
- 81.Jinatham, V., Kullawong, N., Kespechara, K., Gentekaki, E. & Popluechai, S. Comparison of gut microbiota between lean and obese adult Thai individuals. 46, 277–287 (2018).
- 82.Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Suzuki, H., Sota, M., Brown, C. J. & Top, E. M. Using mahalanobis distance to compare genomic signatures between bacterial plasmids and chromosomes. Nucleic Acids Res.36, e147 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rao, X., Huang, X., Zhou, Z. & Lin, X. An improvement of the 2ˆ(–delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinforma. Biomath.3, 71–85 (2013). [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S RNA raw reads and transcriptomic raw data are available at 10.57745/XXAXFA and 10.57745/02ODSE respectively.