Abstract
The K8 locus in Kaposi's sarcoma-associated herpesvirus (KSHV) is syntenic with the Epstein-Barr virus (EBV) BZLF (Z) locus and expresses three alternatively spliced transcripts. The fully spliced transcript encodes K-bZIP, the KSHV homologue of the EBV immediate-early transcriptional transactivator Z. Here we show that despite the presence of alternatively spliced transcripts, the protein from the fully spliced RNA, K-bZIP, is the principal product detectable in KSHV-infected B cells. The protein is detected only in lytically infected cells and is localized to the nucleus. We further characterized K-bZIP by determining its phosphorylation status. Phosphoamino acid analysis revealed phosphorylation on serine and threonine. Analysis of the sites of K-bZIP phosphorylation by tandem mass spectrometry revealed that K-bZIP was phosphorylated on Thr 111 and Ser 167. These phosphorylation sites are contained within cyclin-dependent kinase (CDK) recognition sites with the consensus sequence (S/T)PXR, suggesting that K-bZIP could be phosphorylated by CDKs. We tested this hypothesis using an in vitro kinase reaction performed in whole-cell extracts that resemble in vivo conditions more closely than standard in vitro kinase reactions. We found that the three CDK-cyclin complexes we tested phosphorylated K-bZIP but not the control ORF 73 protein, which contains four (S/T)PXR sites. Ectopic expression of K-bZIP cannot reactivate KSHV from latency, and single and double mutants of K-bZIP in which alanines replaced the phosphorylated serine and/or threonine also failed to induce lytic replication. These studies indicate that K-bZIP is a substrate for CDKs and should inform further functional analyses of the protein.
Kaposi's sarcoma (KS) was first described as a rare and indolent neoplasm of elderly Mediterranean men and was later found to be more frequent in African men. With the onset of the AIDS epidemic, another, much more aggressive, form of KS emerged (4). The epidemiology of AIDS-associated KS strongly suggested that a transmissible agent caused KS (5). The search for such an agent led to the discovery in 1994 of a new human herpesvirus, Kaposi's sarcoma-associated herpesvirus (KSHV), or human herpesvirus 8 (12). Subsequent studies indicated that KSHV infection is central to KS pathogenesis but that other cofactors (for example, immunosuppression) are also required for the development of the lesion (8, 33). In addition to KS, KSHV is associated with the B-cell lymphoproliferative diseases primary effusion lymphoma (PEL; formerly known as body cavity-based lymphoma) and Castleman's disease (10, 38). The observation that KSHV infects B cells is consistent with sequence analysis indicating that KSHV is a member of the gamma-2 (lymphotropic) subfamily of herpesviruses.
KS lesions consist primarily of spindle cells, presumably of endothelial origin, and are permeated with neovascular structures. Most of the spindle cells in a KS tumor are latently infected with KSHV, and the latency program is likely to play a key role in spindle cell survival and expansion. However, in KS lesions some of the cells also express markers for lytic replication, and several lines of evidence suggest that KSHV lytic replication also contributes to the formation of a KS lesion. For example, ganciclovir, a drug that inhibits lytic replication of herpesviruses, can decrease the incidence of KS in high-risk AIDS patients (27). Moreover, many viral genes that play roles in angiogenesis and inflammation—key features of KS histology—are expressed predominantly in the lytic cycle (1, 6, 7). For these and other reasons we have been investigating the control of the lytic cascade of gene expression.
Extensive work has been done in the other human gammaherpesvirus, Epstein-Barr virus (EBV), on the mechanisms of activation and control of lytic replication. In EBV, two immediate-early genes are capable of reactivating EBV from latent viral infection: Z (Zta, ZEBRA, EB1, or BZLF1) and R (Rta or BRLF1) (14, 28). Both of these genes are transcriptional transactivators, and ectopic expression of either can induce latently infected cells to undergo lytic replication. Apart from its function as a transcription factor, Z also associates with helicase-primase replication proteins and may be involved in the formation of the EBV DNA replication complex (18). Additionally, Z expression can cause G0/G1 cell cycle arrest, suggesting that it helps redirect cellular metabolism to aid viral replication (9, 30).
KSHV codes for apparent homologues of R and Z called ORF 50 (RTA) and K-bZIP, respectively. The location of the corresponding genes is syntenic to EBV R and Z, and the transcription and splicing pattern at this locus is somewhat similar to that at the EBV R/Z locus. KSHV ORF 50 can reactivate latently infected B cells and induce the lytic cascade of gene expression (26, 39). In addition, ORF 50 can transactivate the promoters of K-bZIP, ORF 57, PAN (nut-1), thymidine kinase, and DNA binding protein (25). K-bZIP is encoded by a spliced mRNA in which sequences from the genomic ORF K8 (exon 1 in the cDNA diagram of Fig. 1C) are spliced to downstream exons (E2 to E4) bearing a basic region (encoded by E2) and a leucine zipper (from E3) which together form a bZIP domain (19, 24). bZIP domains are DNA binding and oligomerization motifs, and the bZIP domain of EBV Z is necessary for its function as an activator of lytic replication. Several other mRNAs can also be generated from the locus via alternative splicing (Fig. 1C), and these could potentially encode other isoforms of K-bZIP (24).
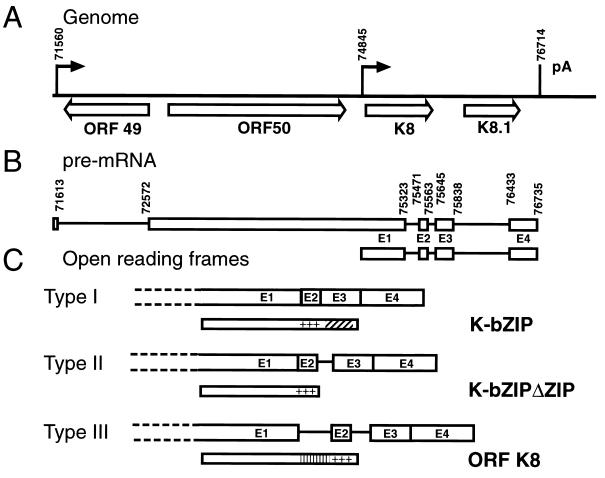
FIG. 1.
The ORF 50 (K8) locus. (A) Genomic organization. Nucleotide numbering and ORF designations (open arrows) are according to the work of Russo et al.(32). The closed arrows indicate the transcriptional start site, and the vertical line indicates the polyadenylation site. (B) Pre-mRNAs from the ORF 50 (K8) locus. The lines represent introns; the open boxes represent exons. The numbers indicate the locations of splice sites. (C) Potential protein products for the alternatively spliced RNAs. The narrow open boxes represent the protein products of the RNAs. The diagonally striped box represents the leucine zipper, and the plus signs represent the basic region. The vertically striped box represents the unique region of K8. The dashed lines indicate that exon 1 can originate from the monocistronic or bicistronic transcript.
To better understand the function of K-bZIP, we have characterized the protein products of the K-bZIP locus. In most of this work we used BCBL-1 cells, which are derived from a PEL and can be induced to lytic replication using phorbol esters. Here we show that despite the presence of alternatively spliced RNAs the K-bZIP protein is the predominant protein isoform detectable in infected B cells. We also find that K-bZIP is phosphorylated at two cyclin-dependent kinase (CDK) recognition sites and that K-bZIP can be phosphorylated by CDK-cyclin complexes in vitro. Unlike EBV Z, K-bZIP cannot activate lytic viral replication when expressed in latently infected B cells. Single and double mutants of K-bZIP in which an alanine(s) replaced the phosphorylated serine and/or threonine also failed to induce lytic replication.
MATERIALS AND METHODS
Plasmids.
The bacterial expression vector pRSET-K-bZIPSE contains the SmaI/EcoRI fragment of pBS SKII-ORF50 cDNA 12 inserted into the PvuII/EcoRI sites of pRSETA (Invitrogen). This clone expresses all but 13 N-terminal amino acids of K-bZIP and has a six-His tag. pBS-K-bZIP was created by PCR amplification of the K-bZIP open reading frame from pBS SKII-ORF50 cDNA 12 (26) with primers that contained BamHI and EcoRI restriction sites and insertion of the digested PCR product into the BamHI/EcoRI sites of pBluescript SK(+) (Promega). The sequences of the primers used to clone the open reading frame were as follows: K-bZIP5′BamHI, GATCGGATCCCCCAGAATGAAGGACATA, and K-bZIP3′EcoRI, GATCGAATTCAACATGGTGGGAGTGG. pcDNA3.1-K-bZIP was created by digestion of pBS SKII-ORF50 cDNA 12, with SalI filling in of the overhanging end by extension with Klenow fragment and further digestion with XhoI to generate a fragment that was inserted in to the EcoRV/XhoI sites of pcDNA3.1 (Invitrogen). pcDNA3.1-K-bZIPΔZIP was created using the Stratagene Quickchange kit to change the codon for Ala 190 (GCA) to a Val codon (GTA) and the codon for Leu 191 (TTA) to a stop codon (TGA). This results in the same open reading frame that is present in K-bZIP splice variant II. pcDNA3.1-K8 was created by digestion of pcDNA3 gZ (25) with NruI and SalI and ligation of the resultant insert into pcDNA3.1 that had been prepared by digesting with ApaI, blunting of the resultant ends with T4 DNA polymerase, and digestion with XhoI. pcDNA3.1-HIS-K-bZIP contains the EcoRI/BamHI fragment of pBS-K-bZIP inserted into pcDNA3.1-HIS-C (Invitrogen) to produce K-bZIP with a six-His tag and an Xpress tag. pcDNA3.1-K-bZIP T111A, pcDNA3.1-K-bZIP S167A, and pcDNA3.1-K-bZIP T111A S167A were made by site-directed mutagenesis of pcDNA3.1-K-bZIP using the Stratagene Quickchange kit.
Cell lines and transfections.
BCBL-1, BC-1, BC-3, and BCP-1 cells were maintained and induced with 12-O-tetradecanoylphorbol-13-acetate (TPA) and/or ionomycin as previously described (2, 11, 17, 29). SLK and Cos-7 cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum and penicillin-streptomycin. Transfections were performed with Fugene 6 (Boehringer Mannheim) as described by the manufacturer.
Antibodies.
An anti-K-bZIP rabbit serum was raised against the protein product of pRSET-K-bZIPSE by Animal Farm Services (Healdsberg, Calif.). To produce K-bZIPSE BL21(DE3)-pLysS, cells were transformed with pRSET-K-bZIPSE and grown at 37°C in 2 liters of Luria-Bertani medium until the cells reached an optical density at 600 nm of 0.6. Cells were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and harvested by centrifugation 4 h later. Cells were resuspended in 200 ml of urea binding buffer (500 mM NaCl, 20 mM Tris [pH 8.0], 5 mM imidazole, 50 mM phenylalanine, 50 mM isoleucine, 6 M urea) supplemented with 0.25% Nonidet P-40 and a 1:500-diluted protease inhibitor cocktail for eukaryotes (Sigma). The extract was sonicated, and the insoluble material was removed by centrifugation for 30 min at 20,000 × g and filtration through a 0.45-μm-pore-size bottle-top filter. The extract was loaded onto a 5-ml HiTrap metal-chelating column (Pharmacia) charged with NiSO4. The column was washed with 100 ml of binding buffer, and the bound protein was eluted with urea elution buffer (500 mM NaCl, 20 mM Tris [pH 8.0], 500 mM imidazole, 50 mM phenylalanine, 50 mM isoleucine, 6 M urea) and collected in 2.5-ml fractions. The fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the fractions containing the bulk of the protein were pooled and dialyzed against phosphate-buffered saline (PBS).
Anti-K8 antibodies were made by conjugation of 10 mg of the peptide acetyl-H2N-AVPAAPITPPSPRVQRPAC-CO2H to 5 mg of keyhole limpet hemocyanin using sulfo-sulfosuccinimidyl-4-[N-maleimido-methyl]cyclohexane-1-carboxylate and subsequent immunization of a rabbit (Animal Farm Services). ORF 73 antisera were made by conjugation of peptides that correspond to the repeats in ORF 73 (H2N-CDDEEDDEEDDEEDDEEEDEEED-CO2H, H2N-CQQQEPQQQEPQQREPQQREP-CO2H, and H2N-LEEQEQELEEQEQELEEQEQEC-CO2H) as described above and immunization of one rabbit with all three peptides.
Purification of His-tagged K-bZIP from Cos-7 cells.
Twenty 10-cm plates of Cos-7 cells were transfected with 8 μg of pcDNA3.1-HIS-K-bZIP. Forty-eight hours after transfection, cells were lysed in a total of 20 ml of urea binding buffer supplemented with 5 mM sodium orthovanadate, 25 mM NaF, and 1 mM sodium pyrophosphate. The extract was sonicated, and the insoluble material was removed by centrifugation for 30 min at 20,000 × g and filtered through a Millipore 0.45-μm-pore-size syringe filter. The extract was loaded onto a 5-ml HiTrap metal-chelating column (Pharmacia) charged with NiSO4. The column was washed with 100 ml of binding buffer, and the bound protein was eluted with urea elution buffer (500 mM NaCl, 20 mM Tris [pH 8.0], 500 mM imidazole, 50 mM phenylalanine, 50 mM isoleucine, 6 M urea) and collected in 2.5-ml fractions. The fractions were then precipitated with 2.7 ml of acetone, and the pellets were resuspended in SDS-PAGE sample buffer. The fractions were run on an SDS–10% PAGE gel and silver stained according to reference 36, and the His-tagged K-bZIP bands were cut out and stored at 4°C.
Western analysis.
To make extracts, nonadherent cells were washed twice with PBS, resuspended in radioimmunoprecipitation (RIPA) buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 0.5% Triton X-100, 0.5% deoxycholic acid, 25 mM NaF, 1 mM sodium pyrophosphate, 1 mM sodium orthophosphate, and 1:500 protease inhibitor cocktail set III [Calbiochem]) at 107 cells per ml and vortexed, and the insoluble material was removed by centrifugation. Cos-7 cell extracts were made by transfecting cells on 10-cm plates with 8 μg of plasmid using Fugene 6 and harvesting the confluent cells 48 h after transfection. The cells were then washed twice with PBS and scraped into 1 ml of RIPA buffer, and the insoluble material was removed by centrifugation. In vitro translations were performed with the TNT system (Promega) as described by the manufacturer. Western analysis was performed as previously described (25). The anti-K-bZIP sera were used at a 1:20,000 dilution, and the anti-K8 sera were used at a 1:4,000 dilution.
In vivo phosphorylation analysis.
BCBL-1 cells were labeled according to the method described in reference 3. Briefly, 107 BCBL-1 cells or BJAB cells that had been induced with ionomycin and TPA 48 h before labeling were collected by centrifugation and washed once with RPMI 1640 medium without sodium phosphate (Gibco BRL). Cells were resuspended in 20 ml of labeling medium (RPMI 1640 without sodium phosphate supplemented as described previously [29] except that dialyzed fetal bovine serum [Gibco BRL] was used); then 5 mCi of orthophosphate was added, and cells were incubated for 4 h. Subsequently, cells were washed once with PBS and resuspended in 1 ml of RIPA buffer, and the insoluble material was removed by a high-speed spin in a microcentrifuge. Immunoprecipitation was performed by incubating 200 μl of extract, 250 μl of RIPA buffer, and 3 μl of anti-K-bZIP rabbit sera for 3 h at room temperature followed by the addition of 40 μl of a 50% slurry of protein A-Sepharose CL-4B beads (Sigma) in RIPA buffer for 1 h. The beads were pelleted, washed once with RIPA buffer, and resuspended in SDS-PAGE sample buffer. The proteins were separated by SDS-PAGE, blotted onto Immobilon (Millipore), and then hydrolyzed and released from the membrane with 6 M HCl (22). The phosphoamino acids were separated by two-dimensional thin-layer electrophoresis (21). The first dimension was run in pH 1.9 in acidic acid-formic acid-water (78:25:897 [vol/vol/vol]) at 1,000 V for 1 h. The second dimension was run at pH 3.5 in acidic acid-pyridine-water (50:5:945 [vol/vol/vol]) at 1,000 V for 45 min. The plates were dried, the marker phosphoamino acids were visualized with 0.1% ninhydrin in 95% ethanol, and the plates were exposed to film.
Protein in-gel digestion.
Protein bands were excised and digested as described in reference 20. Briefly, minced gel pieces were first washed with 25 mM NH4HCO3 in 50% acetonitrile, dried in Speedvac, rehydrated in 25 mM NH4HCO3 solution containing trypsin, and digested overnight at 37°C. Peptides were then extracted by washing with high-pressure liquid chromatography-grade water followed by three washes with 60% acetonitrile–0.1% formic acid. The combined supernatants were dried down under vacuum and redissolved in 0.1% formic acid for desalting using Ziptip(C18). The cleaned digest was analyzed by mass spectrometry.
Mass spectrometric analysis of tryptic peptides.
Molecular masses of all tryptic peptides were determined by analyzing 1/10 of desalted unseparated digest using matrix-assisted laser desorption ionization–time of flight (MALDI-TOF; Voyager DESTR mass spectrometer; Perseptive Biosystems). Peptides were cocrystallized with equal volumes of matrices, either α-cyano-4-hydroxycinnamic acid or 2,5-dihydroxylbenzonic acid. MALDI spectra were internally calibrated using trypsin autolysis products to get accurate monoisotopic masses of all the tryptic peptides (<20 ppm). Peptide masses were submitted for protein mass database searching using the MS-Fit program (13).
Peptide sequencing by quadrupole-TOF mass spectrometry.
Tandem mass spectra of peptides were obtained on a quadrupole reflector TOF mass spectrometer with a quadrupole collision cell (QSTAR PE Sciex, Toronto, Canada). A nanoelectrospray ion source (Protana A/S, Odense, Denmark) was used for all experiments. Mass resolution was routinely obtained in the range of 7,000 to 12,000 (for both conventional MS and MS/MS modes of operation), allowing unambiguous charge state determination for ions at least up to m/z 5000. The mass accuracy obtained for peptides and peptide fragment ions (with external calibration) was typically about 50 ppm or better, which is usually sufficient to distinguish between Gln and Lys, between Phe and oxidized Met, and between several combinations of two amino acid residues with the same nominal masses. Therefore, interpretation of the ion series becomes much easier due to the increased accuracy and sensitivity. Cleaned digest mixture was loaded into nanoelectrospray tip. Conventional mass spectra for parent ions of digest mixture were first obtained. Tandem mass spectra for selected parent ions were then collected. The spectra were interpreted with the aid of the MS-Tag program (13).
Preparation of cyclin-CDK complexes.
Sf9 cells were infected with baculoviruses encoding human CDK2F80G, CDC2F80G, cyclin A, cyclin E, or cyclin B. CDKs were tagged at the carboxy terminus with a hemagglutinin epitope tag, and cyclins contained amino-terminal six-His tags. Cyclin E-CDK2F80G, cyclin A-CDK2F80G, and cyclin B-CDC2F80G complexes were assembled by mixing the appropriate Sf9 extracts. Activation of cyclin-CDK complexes was accomplished by the addition of 1 mM ATP, 10 mM MgCl2, and a small amount of Sf9 extract containing baculovirus-expressed human CDK-activating kinase. Kinase complexes were purified by iminodiacetic acid (IDA)-Co2+ affinity chromatography and stored in 150 mM NaCl–20 mM HEPES [pH 7.4]–10% glycerol.
Production of N6-benzyl-[γ-35S]ATP.
N6-benzyl-[γ-35S]ATP was produced by a method to be described elsewhere (J.D.B. and D.M., unpublished data). Briefly, purified 10-His-tagged nucleoside-diphosphate kinase was bound to IDA-Co2+ agarose beads in a miniature column and washed with [γ-35S]ATP, yielding a population of autophosphorylated nucleoside-diphosphate kinase. The column was washed with buffer to remove residual ADP and then washed with N6-benzyl-ADP to yield an eluted mixture of N6-benzyl-ADP and N6-benzyl-[γ-35S]ATP. Following quantitation, the product was used in kinase assays without further purification.
In vitro phosphorylation analysis.
RIPA buffer extracts were prepared from BJAB cells, uninduced BCBL-1 cells, and BCBL-1 cells that had been induced with TPA for 48 h. Samples of each extract (750 μg of protein) were labeled with CDK2F80G-cyclin E, CDK2F80G-cyclin A, or CDC2F80G-cyclin B by the addition of 30 μCi of N6-benzyl-[γ-35S]ATP (1,250 Ci/mmol) and kinase activities equal to approximately 12 μg of active kinase, in total reaction volumes of 338 μl. Kinase activities were normalized by their relative abilities to phosphorylate histone H1. After 20 min at 24°C, reactions were stopped with 10 μl of 0.5 M EDTA. Reaction products were then immunoprecipitated as described above, separated by SDS-PAGE, blotted onto nitrocellulose, and analyzed by autoradiography.
Lytic reactivation assay.
BCBL-1 cells (107) were electroporated at 960 μF and 210 V with 12 μg of test expression vector and 8 μg of pCMV-GFP expression vector and incubated for 48 h in 20 ml of complete RPMI 1640 (29). The cells were then washed twice in PBS, fixed in 2 ml of 4% paraformaldehyde for 20 min at room temperature, washed three times in PBS and once in fluorescence-activated cell sorting (FACS) buffer–saponin (PBS–1% bovine serum albumin–0.02% saponin), and then incubated in 100 μl of mouse anti-ORF 59 antibody diluted in FACS buffer-saponin on ice for 30 min. The cells were then washed three times with FACS buffer-saponin, incubated with 100 μl of goat anti-mouse immunoglobulin-phycoerythrin diluted in FACS buffer-saponin for 30 min on ice, and washed three times with FACS buffer-saponin and once with PBS. The cells were then analyzed by flow cytometry gating only on intact cells. As controls, cells transfected without green fluorescent protein (GFP) were used to mark the limits of GFP-negative (untransfected) cells.
RESULTS AND DISCUSSION
K-bZIP is the predominant isoform detectable in induced BCBL-1 cells.
Figure 1 summarizes the genomic organization of the K-bZIP region and the structure and coding potential of its RNAs. Lin et al. (24) have previously detected three differentially spliced transcripts in BCBL-1 cells from the K-bZIP locus, termed types I, II, and III, and found them to occur in a ratio of 16:4:1. Type I RNA, the predominant transcript, encodes the leucine zipper-containing K-bZIP protein that is the homologue of EBV Z. Type II RNA encodes a protein that is almost identical to K-bZIP except that it lacks the leucine zipper domain (K-bZIPΔZIP) by omitting the splicing of E2 to E3. Type III RNA, the least abundant form, fails to splice E1 to E2 and can encode only a protein that corresponds to the K8 ORF originally recognized in the genomic DNA sequence of KSHV (32) (Fig. 1). That ORF also includes coding sequences that are depicted as the intron between E1 and E2 in Fig. 1C.
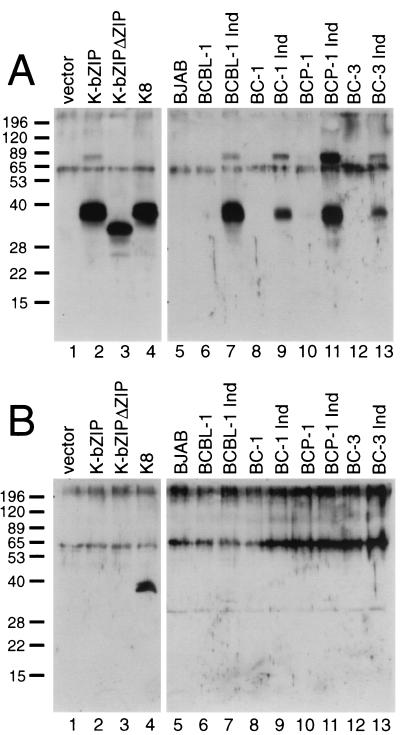
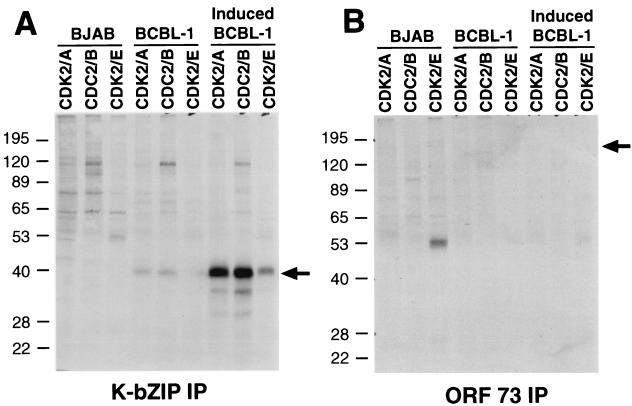
To see which of these potential protein products are expressed in KSHV-infected cells, we generated polyclonal antibodies against recombinant K-bZIP protein; since this protein includes E1 sequences common to all potential isoforms (Fig. 1), the antiserum should recognize all three potential protein products of the locus. Figure 2A (lanes 1 to 4) confirms that this is so. Proteins corresponding to K-bZIP, K-bZIPΔZIP, and K8 were generated by in vitro translation, fractionated by SDS-PAGE, and examined by immunoblotting using this antibody. As shown in Fig. 2A, the antiserum recognized each of these proteins (lanes 2 to 4). Next, we used this antibody to probe Western blots of extracts from an uninfected B-cell line (BJAB) and several uninduced and lytically induced PEL cell lines (BCBL-1, BC-1, BC-3, and BCP-1) (Fig. 2A, lanes 5 to 13). As expected, no immunoreactive material was seen prior to lytic induction. Following induction, a band of 38 kDa was seen in all four PEL lines. At best, only traces of immunoreactive material that comigrates with the smaller K-bZIPΔZIP protein were observed. Additionally, there is a ∼80-kDa band that is recognized by the anti-K-bZIP antibody in the infected cell lines and from the K-bZIP translated in vitro. We speculate that this is a dimeric form of K-bZIP; K-bZIP is known to form homodimers (19, 24).
FIG. 2.
K-bZIP is the predominant isoform expressed from the K8 region in PEL cells. For lanes 1 to 4, protein products were generated by coupled in vitro transcription and translation from the empty pcDNA3.1 vector, pcDNA3.1-K-bZIP, pcDNA3.1-K-bZIPΔZIP, and pcDNA3.1-K8, respectively. Samples were examined by SDS-PAGE and immunoblotting with anti-K-bZIP antibody (A) or anti-K8 antibody (B). For lanes 5 to 13, extracts from the indicated B-cell lines either before or after induction with TPA were examined by SDS-PAGE and immunoblotting with anti-K-bZIP (A) or anti-K8 (B) antibody. The molecular weights of the protein markers are shown on the left, in thousands.
Because the K-bZIP and K8 proteins comigrate on SDS-PAGE (Fig. 2A, lanes 2 and 4), the identity of the 38-kDa species could not be rigorously assigned from this immunoblot. Accordingly, we prepared an antiserum to a synthetic peptide derived from the K8-specific coding sequences lying between E1 and E2 (Fig. 1C). As expected, in control experiments using in vitro translation products, this antibody recognized only the K8 protein and not the K-bZIP protein (Fig. 2B, lanes 1 to 4). When used to probe immunoblots of PEL cell extracts, however, this antibody detected no K8 protein in these samples (Fig. 2B, lanes 5to 13). Although we cannot exclude the presence of low levels of K8 and K-bZIPΔZIP in lytically infected cells, it is clear that the predominant protein isoform generated by KSHV in vivo is K-bZIP itself.
This observation is noteworthy because K-bZIPΔZIP RNA (type II) is only fourfold less abundant than K-bZIP mRNA (type I). One possible explanation for this seeming discrepancy between mRNA levels and protein levels comes from the way in which the amounts of the spliced variants were determined in an earlier study (24). As summarized in Fig. 1B, the K8-K8.1 region is not only transcribed as such but is also represented in ORF 50 mRNAs that read through this region and are polyadenylated downstream. The RNase protection probe previously used to detect the spliced RNAs could not distinguish the RNA initiating from the K8 promoter from those initiating from the upstream ORF 50 promoter. Thus, the type II (K-bZIPΔZIP) and type III (K8 splice) variants could have derived primarily from the ORF 50 mRNAs that traverse this region. If so, they would be expected to be poorly translated relative to the monocistronic K-bZIP mRNA (type I).
To further characterize K-bZIP, we performed indirect immunofluorescence with the anti-K-bZIP antisera on TPA-induced BCBL-1 cells and SLK cells (an endothelial cell line) transfected with a K-bZIP expression vector and showed that K-bZIP was localized to the nucleus (data not shown). This observation is consistent with published results (23) and K-bZIP's putative role as a transcription factor.
K-bZIP is phosphorylated on threonine and serine residues.
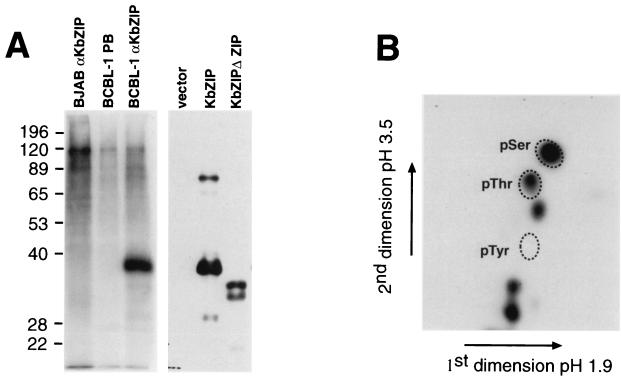
EBV Z is phosphorylated on Ser 186, which is in the basic domain of the bZIP region. This phosphorylation is required for the ability of Z to induce lytic reactivation (15, 16). Therefore, we were interested to see if K-bZIP was phosphorylated. We labeled BCBL-1 in vivo with [32P]orthophosphate, made extracts of the labeled cells, immunoprecipitated the extracts with anti-K-bZIP polyclonal sera, separated the proteins by SDS-PAGE, and blotted them onto an Immobilon membrane. Exposure of the blot showed that only one band was present and that the band comigrated with K-bZIP (Fig. 3A). Immunoprecipitation of BCBL-1 extract with preimmune sera or immunoprecipitation from extracts of BJAB cells (an uninfected B-cell line) with anti-K-bZIP sera did not show a band of the correct size (Fig. 3A). These data indicate that K-bZIP is phosphorylated in KSHV-infected cells. To identify the phosphoamino acids of K-bZIP, we cut out the piece of membrane containing K-bZIP, hydrolyzed the protein with 6 M HCl, and performed two-dimensional thin-layer electrophoresis (Fig. 3B). This analysis showed that K-bZIP is phosphorylated on serine and threonine residues. These data are also consistent with antiphosphotyrosine immunoblots that suggested that K-bZIP contains no phosphotyrosine (data not shown).
FIG. 3.
K-bZIP is phosphorylated at serine and threonine residues. (A) Induced BCBL-1 or BJAB cells were labeled with orthophosphate, and extracts of the labeled cells were immunoprecipitated with anti-K-bZIP polyclonal sera or preimmune sera (PB) as indicated. Immunoprecipitated proteins were separated by SDS-PAGE, blotted onto Immobilon membranes, and exposed to film. Western analysis of extracts of Cos cells transfected with empty vector or vectors expressing K-bZIP or K-bZIPΔZIP as indicated and probed with anti-K-bZIP antibody was also carried out. (B) Two-dimensional thin-layer electrophoresis of K-bZIP phosphoamino acids. The immunoprecipitated K-bZIP was excised from the Immobilon and digested to amino acids. The amino acids were separated by two-dimensional thin-layer electrophoresis with unlabeled marker phosphoamino acids that were visualized with ninhydrin.
K-bZIP is phosphorylated at CDK recognition site motifs.
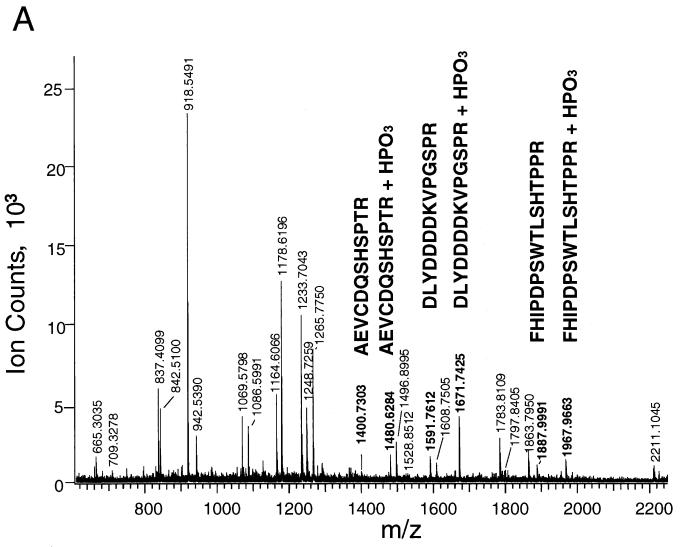
We used mass spectrometry to determine the location of the phosphorylation sites in K-bZIP. K-bZIP was purified by expression of His-tagged K-bZIP (HISK-bZIP) in Cos-7 cells followed by fractionation of the cell extract on a nickel-IDA column and excision of the HISK-bZIP from a silver-stained SDS-PAGE gel of the nickel column fractions. The HISK-bZIP was then digested with trypsin, and the molecular masses of the tryptic fragments were determined by MALDI-TOF mass spectrometric analysis. We found three pairs of fragments which have molecular masses corresponding to the calculated masses of K-bZIP tryptic peptides with and without the presence of a HPO3 moiety (Fig. 4A). To determine the location of the phosphorylation sites, we sequenced the phosphorylated tryptic fragments using tandem mass spectrometry (Fig. 4B and C).
FIG. 4.
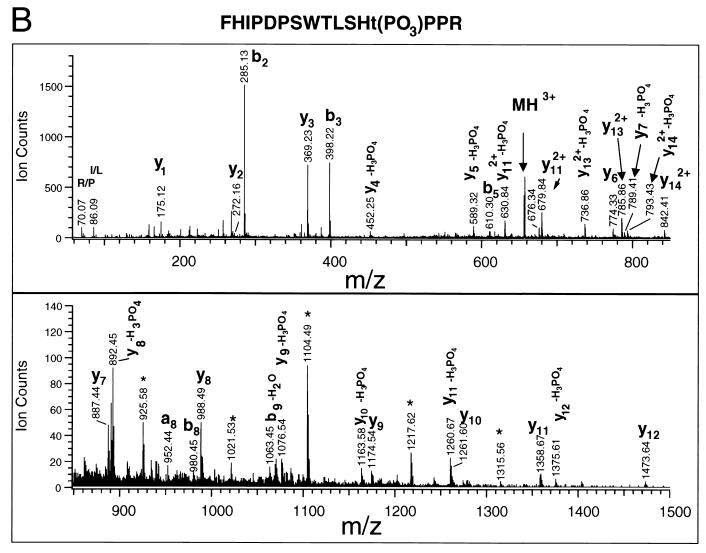
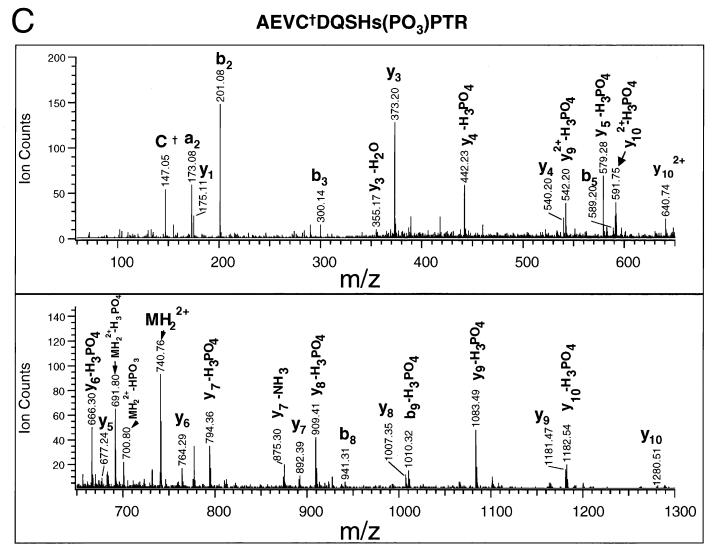
K-bZIP is phosphorylated at T111 and S167 (A) MALDI-TOF of HIS-K-bZIP tryptic fragments. His-tagged K-bZIP was expressed in Cos-7 cells and purified by fractionation on a nickel column and excision from a silver stained SDS-PAGE gel of the nickel column fractions. The K-bZIP was then digested with trypsin, and the molecular masses of the tryptic fragments were determined by MALDI-TOF mass spectrometric analysis. The peaks whose molecular masses match those of HIS-K-bZIP and phosphoHIS-K-bZIP are indicated in bold. (B) QSTAR tandem mass spectra of the phosphopeptide FHIPDPSWTLSHTPPR. The major y series and b series and the molecular ion (MH3+), internal ions (∗), acrylamide adducts (†), and immonium ions corresponding to specific amino acids (R/P and I/L) are indicated. Some the ionic masses correspond to a fragment of the parent ion with loss of a phosphate (−H3PO4 or −H2O). (C) QSTAR tandem mass spectra of the phosphopeptide AEVCDQSHSPTR. The spectra are labeled as described above.
The presence of y6 (m/z 774.330) and y6-H3PO4 (m/z 676.34) ions in the mass spectrum of phosphopeptide FHIPDPSWTLSHTPPR (MH+ = 1967.9) indicates that either T111 or S109 is phosphorylated. It is difficult to distinguish between these two possibilities because the ions at m/z 589.32 and 452.25 can be interpreted as either y5-H3PO4 and y4-H3PO4 or y5-H2O and y4-H2O, respectively. However, an internal ion, m/z 279.07, was observed that can be attributed to the phosphodipeptidyl moiety T(PO3)P, supporting the phosphorylation at T111. The sequencing of the phosphopeptide AEVCDQSHSPTR establishes that S167 is phosphorylated. As shown in the spectrum (Fig. 4C), starting from y4, all the y ion series contain both y and y-H3PO4 ions, indicating that the phosphorylation occurred at position 4 (i.e., S167) from the C terminus.
The peptide DLYDDDKVPGSPR contained only one serine and no threonines, and sequencing confirmed that the serine residue was phosphorylated (data not shown). This serine residue is part of the His tag constituent and therefore not relevant to the function of K-bZIP. The mass spectrometric data are summarized in Fig. 5. We noted that all three phosphorylation sites were located in consensus CDK recognition motifs: the sites within K-bZIP itself are found in the recognition motif (S/T)PXR, suggesting that K-bZIP may be a substrate for CDKs in vivo.
FIG. 5.
Summary of the mass spectrometry data. The numbering is based on the location of the amino acid in wild-type K-bZIP; the N-terminal methionine of K-bZIP was removed when the His tag was added. The regions of HIS-K-bZIP that were identified by the MALDI-TOF analysis are shown in bold; the leucine zipper, basic region, and HIS tag are underlined; the CDK recognition site is overlined; and the phosphorylated amino acids within K-bZIP are indicated by an asterisk.
K-bZIP is phosphorylated in vitro.
Since K-bZIP is phosphorylated at CDK recognition sites in vivo, we hypothesized that K-bZIP is a CDK target. To test this hypothesis, we performed kinase reactions in vitro using a method first developed for the tyrosine kinase Src by Shah et al. (35). This method involves mutation of the kinase ATP binding site so that it will accept N6-benzyl-substituted ATP, which is a very poor substrate for most wild-type kinases. With this method, kinase substrates can be analyzed in crude lysates that provide a more physiological environment than is normally possible with reaction mixtures containing purified kinase and substrate. To further increase specificity, we used N6-benzyl-[γ-35S]ATP, because ATP containing thiophosphate in the gamma position is a poor substrate for many kinases but an effective substrate for human CDC2(CDK1) and CDK2. Addition of mutant CDK and N6-benzyl-[γ-35S]ATP to cell extracts results in specific radiolabeling of protein substrates with negligible background phosphorylation due to endogenous kinase activity.
For our labeling reactions we used mutant CDK2-cyclin E, which is normally active in late G1 phase of the cell cycle, mutant CDK2-cyclin A, which is active in S phase, and CDK1-cyclin B, which is required for entry into mitosis. The kinase complexes were incubated with extracts from BJAB cells, uninduced BCBL-1 cells, and BCBL-1 cells that had been induced with TPA for 48 h. K-bZIP or LANA (ORF 73) was immunoprecipitated from the labeled extracts, separated by SDS-PAGE, blotted onto nitrocellulose, and analyzed by autoradiography. We used ORF 73 as a control because it is found in all BCBL-1 cells and contains four potential CDK recognition motifs: SPER, TPMR, SPPR, and TPPR (which is also found in K-bZIP). Under these conditions, ORF 73 was not radioactively labeled (Fig. 6B), even though Western blot analysis of the kinase reaction immunoprecipitates demonstrated a strong ORF 73 signal. In contrast, K-bZIP from induced BCBL-1 lysates was phosphorylated by all three of the CDK-cyclin pairs, to various extents (CDK1-cyclin B > CDK2-cyclin A > CDK2-cyclin E) (Fig. 6A). The differences in the extent of phosphorylation could reflect differences in kinetics and/or site usage. Since we normalized the amount of CDK activity added to the reaction mixtures, we believe that this preference order probably reflects K-bZIP reactivity in vitro. The fact that ORF 73 was not phosphorylated under these conditions despite the presence of multiple CDK consensus sites argues strongly that the phosphorylation of K-bZIP is not due simply to opportunistic kinase activity in vitro. However, we note that while our in vitro results confirm that K-bZIP is an effective substrate for CDKs, they do not allow us to determine which CDKs are the principal in vivo catalysts of K-bZIP phosphorylation.
FIG. 6.
K-bZIP is phosphorylated by CDK in vitro. Extracts from BJAB cells, uninduced BCBL-1 cells, and BCBL-1 cells that had been induced with TPA for 48 h were labeled by addition of CDK2F80G-cyclin E, CDK2F80G-cyclin A, or CDC2F80G-cyclin B and of N6-benzyl-[γ-35S]ATP. The extracts were incubated at room temperature and immunoprecipitated with anti-K-bZIP sera (A) or anti-ORF 73 (B). The immunoprecipitate (IP) was analyzed on SDS-PAGE gels followed by electroblotting onto Immobilon and exposure to film. Arrows indicate the position of the K-bZIP protein (A) and the ORF 73 (LANA) protein (B).
Ectopic expression of K-bZIP phosphorylation site mutants does not reactivate KSHV from latency.
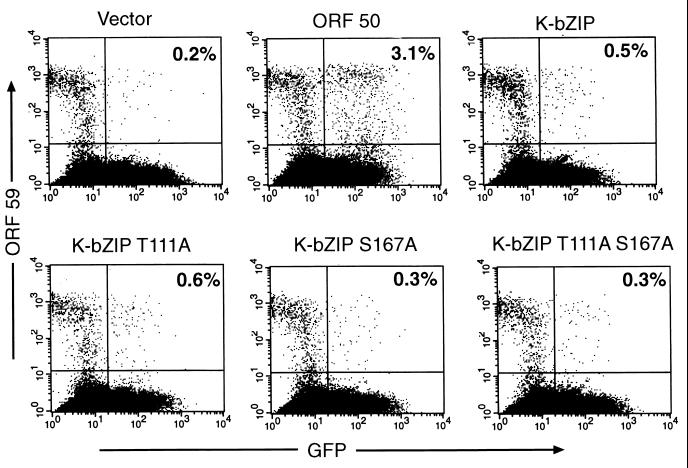
EBV Z and EBV R proteins are known to reactivate EBV from latency when ectopically expressed. KSHV ORF 50 (the EBV R homologue) can reactivate KSHV in infected B cells (26, 39); however, initial studies of the K8 genomic locus suggested that protein products from this region were unable to reactivate KSHV (39). We wondered if the phosphorylation of K-bZIP could be inhibiting the ability of K-bZIP to reactivate KSHV from latency. To test this possibility we made single and double mutants of K-bZIP in which alanines replaced the phosphorylated serine and/or threonine and tested their ability to reactivate BCBL-1 cells from latency. (In parallel studies [results not shown], both wild-type and mutant proteins were shown to be expressed with comparable efficiencies in transfected cells.) BCBL-1 cells were transfected with empty vector or with ORF 50 or K-bZIP expression plasmids and with a GFP expression plasmid as a marker of transfection. Forty-eight hours after transfection, the cells were assayed by flow cytometry for expression of GFP and KSHV ORF 59 (a delayed early gene). This allows us to score which cells were transfected and, of those cells, which had entered the lytic cycle. In agreement with earlier studies (39), wild-type K-bZIP expression could not induce lytic KSHV reactivation; moreover, it could not enhance ORF50's ability to do so (Fig. 7 and data not shown). Similarly, expression of the phosphorylation site mutants also failed to reactivate KSHV lytic replication (Fig. 7). This finding excludes the possibility that a switch protein activity intrinsic to K-bZIP is masked by negative regulatory phosphorylation events mediated by CDKs.
FIG. 7.
Ectopically expressed K-bZIP does not reactivate BCBL-1 cells from latency. BCBL-1 cells were electroporated with empty vector (pcDNA3.1) or vector expressing the indicated protein (pcDNA3.1-K-bZIP, pcDNA3.1-K-bZIP T111A, pcDNA3.1-K-bZIP S167A, and pcDNA3.1-K-bZIP T111A S167A) and a vector expressing GFP (CMV-GFP). Cells were assayed 48 h after electroporation by FACS for the presence of GFP, indicating transfection (x axis), and for the marker ORF 59, indicating lytic replication (y axis). The number in the FACS plot indicates the percentage of transfected cells that are positive for ORF 59.
These data suggest that, despite its similarities to EBV Z, K-bZIP does not function as an activator of lytic replication. Several other lines of evidence suggest that the role of K-bZIP in the lytic cycle of KSHV may be different from that of EBV Z. (i) While EBV Z is an immediate-early gene, the monocistronic K-bZIP transcript displays delayed early kinetics. (ii) The sequence similarity between K-bZIP and EBV Z is modest and limited to the bZIP domain itself. (iii) K-bZIP expression does not activate either the ORF 50 promoter or its own promoter in reporter gene assays in transiently transfected cells (34; A. G. Polson and D. Ganem, data not shown). While none of these data are in themselves definitive, together they suggest that K-bZIP does not act as a lytic switch protein like EBV Z.
Although K-bZIP does not appear to be an activator of lytic replication, the observation that K-bZIP is phosphorylated by CDKs suggests several other possibilities. Other transcription factors that are phosphorylated by CDKs include nucleolar transcription factor, UBF, MyoD, and human estrogen receptor α (31, 37, 40). In these cases the phosphorylation of the transcription factor is used to link its activity to the cell cycle. The phosphorylation state of K-bZIP could serve as a viral sensor of host cell cycle progression; in addition, cell cycle-dependent changes in the phosphorylation state of K-bZIP could be used to modulate its function, for example, triggering differential activation of host or viral genes at different stages. Alternatively, K-bZIP phosphorylation could be used to link KSHV DNA replication to the cell cycle. In addition to its role as an activator of lytic gene expression, EBV Z associates with helicase-primase replication proteins and may be involved in the formation of the EBV replication complex. If K-bZIP plays a similar role in KSHV DNA replication, then its phosphorylation by CDKs could be used to link viral DNA synthesis to a specific cell cycle stage.
ACKNOWLEDGMENTS
We are grateful to Joseph Lin for excellent technical help, Elizabeth Prescott for great advice on two-dimensional thin-layer electrophoresis, and Michael Lagunoff for help with flow cytometry assays.
A.G.P. was supported by grant PF-98–161-01 form the American Cancer Society. Financial support for the mass spectrometry was provided by NIH NcRR grant PR01614 (to A.L.B.).
REFERENCES
- 1.Arvanitakis L, Geras-Raaka E, Varma A, Gershengorn M C, Cesarman E. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature. 1997;385:347–350. doi: 10.1038/385347a0. [DOI] [PubMed] [Google Scholar]
- 2.Arvanitakis L, Mesri E A, Nador R G, Said J W, Asch A S, Knowles D M, Cesarman E. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood. 1996;88:2648–2654. [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley and Sons; 1997. [Google Scholar]
- 4.Beral V. Epidemiology of Kaposi's sarcoma. Cancer Surv. 1991;10:5–22. . (Erratum, 12:225, 1992.) [PubMed] [Google Scholar]
- 5.Beral V, Peterman T A, Berkelman R L, Jaffe H W. Kaposi's sarcoma among persons with AIDS: a sexually transmitted infection? Lancet. 1990;335:123–128. doi: 10.1016/0140-6736(90)90001-l. [DOI] [PubMed] [Google Scholar]
- 6.Boshoff C. Coupling herpesvirus to angiogenesis. Nature. 1998;391:24–25. doi: 10.1038/34054. [DOI] [PubMed] [Google Scholar]
- 7.Boshoff C, Endo Y, Collins P D, Takeuchi Y, Reeves J D, Schweickart V L, Siani M A, Sasaki T, Williams T J, Gray P W, Moore P S, Chang Y, Weiss R A. Angiogenic and HIV-inhibitory functions of KSHV-encoded chemokines. Science. 1997;278:290–294. doi: 10.1126/science.278.5336.290. [DOI] [PubMed] [Google Scholar]
- 8.Boshoff C, Weis R. Kaposi's sarcoma-associated herpesvirus. Adv Cancer Res. 1998;75:57–86. doi: 10.1016/s0065-230x(08)60739-3. [DOI] [PubMed] [Google Scholar]
- 9.Cayrol C, Flemington E K. The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 1996;15:2748–2759. [PMC free article] [PubMed] [Google Scholar]
- 10.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 11.Cesarman E, Moore P, Rao P, Inghirami G, Knowles D, Chang Y. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 12.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 13.Clauser K R, Baker P, Burlingame A L. Role of accurate mass measurement (+/- 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal Chem. 1999;71:2871–2882. doi: 10.1021/ac9810516. [DOI] [PubMed] [Google Scholar]
- 14.Countryman J, Miller G. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc Natl Acad Sci USA. 1985;82:4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francis A, Ragoczy T, Gradoville L, Heston L, El-Guindy A, Endo Y, Miller G. Amino acid substitutions reveal distinct functions of serine 186 of the ZEBRA protein in activation of early lytic cycle genes and synergy with the Epstein-Barr virus R transactivator. J Virol. 1999;73:4543–4551. doi: 10.1128/jvi.73.6.4543-4551.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francis A L, Gradoville L, Miller G. Alteration of a single serine in the basic domain of the Epstein-Barr virus ZEBRA protein separates its functions of transcriptional activation and disruption of latency. J Virol. 1997;71:3054–3061. doi: 10.1128/jvi.71.4.3054-3061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao S J, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo C R, Saah A, Phair J, Detels R, Chang Y, Moore P S. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat Med. 1996;2:925–928. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- 18.Gao Z, Krithivas A, Finan J E, Semmes O J, Zhou S, Wang Y, Hayward S D. The Epstein-Barr virus lytic transactivator Zta interacts with the helicase-primase replication proteins. J Virol. 1998;72:8559–8567. doi: 10.1128/jvi.72.11.8559-8567.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gruffat H, Portes-Sentis S, Sergeant A, Manet E. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) encodes a homologue of the Epstein-Barr virus bZip protein EB1. J Gen Virol. 1999;80:557–561. doi: 10.1099/0022-1317-80-3-557. [DOI] [PubMed] [Google Scholar]
- 20.Huang L, Shen M, Chernushevich I, Burlingame A L, Wang C C, Robertson C D. Identification and isolation of three proteasome subunits and their encoding genes from Trypanosoma brucei. Mol Biochem Parasitol. 1999;102:211–223. doi: 10.1016/s0166-6851(99)00096-1. [DOI] [PubMed] [Google Scholar]
- 21.Hunter T, Sefton B M. Transforming gene product of Rous sarcoma virus phosphorylates tyrosine. Proc Natl Acad Sci USA. 1980;77:1311–1315. doi: 10.1073/pnas.77.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamps M P. Determination of phosphoamino acid composition by acid hydrolysis of protein blotted to Immobilon. Methods Enzymol. 1991;201:21–27. doi: 10.1016/0076-6879(91)01005-m. [DOI] [PubMed] [Google Scholar]
- 23.Katano H, Sato Y, Kurata T, Mori S, Sata T. Expression and localization of human herpesvirus 8-encoded proteins in primary effusion lymphoma, Kaposi's sarcoma, and multicentric Castleman's disease. Virology. 2000;269:335–344. doi: 10.1006/viro.2000.0196. [DOI] [PubMed] [Google Scholar]
- 24.Lin S-F, Robinson D R, Miller G, Kung H-J. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J Virol. 1999;73:1909–1917. doi: 10.1128/jvi.73.3.1909-1917.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukac D M, Kirshner J R, Ganem D. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J Virol. 1999;73:9348–9361. doi: 10.1128/jvi.73.11.9348-9361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lukac D M, Renne R, Kirshner J R, Ganem D. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology. 1998;252:304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- 27.Martin D F, Kuppermann B D, Wolitz R A, Palestine A G, Li H, Robinson C A. Oral ganciclovir for patients with cytomegalovirus retinitis treated with a ganciclovir implant. Roche Ganciclovir Study Group. N Engl J Med. 1999;340:1063–1070. doi: 10.1056/NEJM199904083401402. [DOI] [PubMed] [Google Scholar]
- 28.Ragoczy T, Heston L, Miller G. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J Virol. 1998;72:7978–7984. doi: 10.1128/jvi.72.10.7978-7984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez A, Armstrong M, Dwyer D, Flemington E. Genetic dissection of cell growth arrest functions mediated by the Epstein-Barr virus lytic gene product, Zta. J Virol. 1999;73:9029–9038. doi: 10.1128/jvi.73.11.9029-9038.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogatsky I, Trowbridge J M, Garabedian M J. Potentiation of human estrogen receptor alpha transcriptional activation through phosphorylation of serines 104 and 106 by the cyclin A-CDK2 complex. J Biol Chem. 1999;274:22296–22302. doi: 10.1074/jbc.274.32.22296. [DOI] [PubMed] [Google Scholar]
- 32.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schulz T. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) J Gen Virol. 1998;79:1573–1591. doi: 10.1099/0022-1317-79-7-1573. [DOI] [PubMed] [Google Scholar]
- 34.Seaman W T, Ye D, Wang R X, Hale E E, Weisse M, Quinlivan E B. Gene expression from the ORF50/K8 region of Kaposi's sarcoma-associated herpesvirus. Virology. 1999;263:436–449. doi: 10.1006/viro.1999.9963. [DOI] [PubMed] [Google Scholar]
- 35.Shah K, Liu Y, Deirmengian C, Shokat K M. Engineering unnatural nucleotide specificity for Rous sarcoma virus tyrosine kinase to uniquely label its direct substrates. Proc Natl Acad Sci USA. 1997;94:3565–3570. doi: 10.1073/pnas.94.8.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 37.Song A, Wang Q, Goebl M G, Harrington M A. Phosphorylation of nuclear MyoD is required for its rapid degradation. Mol Cell Biol. 1998;18:4994–4999. doi: 10.1128/mcb.18.9.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 39.Sun R, Lin S F, Gradoville L, Yuan Y, Zhu F, Miller G. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1998;95:10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voit R, Hoffmann M, Grummt I. Phosphorylation by G1-specific cdk-cyclin complexes activates the nucleolar transcription factor UBF. EMBO J. 1999;18:1891–1899. doi: 10.1093/emboj/18.7.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]