Abstract
We analyzed genomic data from the prostate cancer of African- and European American men to identify differences contributing to racial disparity of outcome. We also performed FISH-based studies of Chromodomain helicase DNA-binding protein 1 (CHD1) loss on prostate cancer tissue microarrays. We created CHD1-deficient prostate cancer cell lines for genomic, drug sensitivity and functional homologous recombination (HR) activity analysis. Subclonal deletion of CHD1 was nearly three times as frequent in prostate tumors of African American than in European American men and it associates with rapid disease progression. CHD1 deletion was not associated with HR deficiency associated mutational signatures or HR deficiency as detected by RAD51 foci formation. This was consistent with the moderate increase of olaparib and talazoparib sensitivity with several CHD1 deficient cell lines showing talazoparib sensitivity in the clinically relevant concentration range. CHD1 loss may contribute to worse disease outcome in African American men.
Subject terms: Prognostic markers, Cancer, Computational biology and bioinformatics
Introduction
Despite an improving trend, African American (AA) men with PCa still have a significantly worse outcome with a 2.2-fold higher mortality rate compared with men of European ancestry (EA)1. Recent studies demonstrated that AA men are at higher risk of progression after radical prostatectomy, even in equal access settings and when accounting for socioeconomic status2,3. While the reasons underlying these disparities are multifactorial, these data strongly argue that germline and/or somatic genetic differences between AA and EA men may in part explain these differences.
Comparative analysis of AA and EA prostate tumors have identified several genomic differences. PTEN deletions, ERG rearrangements and consequent ERG over-expression are more frequent in PCas of EA men4–6. In contrast, LSAMP and ETV3 deletions, ZFHX3 mutations, MYC and CCND1 amplifications and KMT2D truncations are more frequent in PCas of AA men7–9. ERF, an ETS transcriptional repressor, also showed an increased mutational frequency in AA prostate cancer cases with probable functional consequences such as increased anchorage independent growth10. Additionally, SPINK1 expression is also enriched in African American PCa11.
Chromodomain helicase DNA-binding protein 1 (CHD1) deletion is frequently present in prostate cancer. Deletions are associated with increased Gleason score and faster biochemical recurrence12, activation of transcriptional programs that drive prostate tumorigenesis13 and enzalutamide resistance14. Mechanistically, CHD1 loss influences prostate cancer biology in at least two ways. CHD1, an ATPase-dependent chromatin remodeler, contributes to a specific distribution of androgen receptor (AR) binding in the genome of prostate tissue. When lost, the AR cistrome redistributes to HOXB13 enriched sites and thus alters the transcriptional program of prostate cancer cells13. CHD1 may also contribute to genome integrity. It is required for the recruitment of CtIP, an exonuclease, to DNA double-strand breaks (DSB) to initiate end-resection. Impairing this important step of DSB repair upon CHD1 loss was proposed to lead to homologous recombination deficiency15,16. The functional impact of CHD1 loss is likely further influenced by the frequent co-occurrence of SPOP mutations, which were reported to be associated with the suppression of DNA repair17.
CHD1 loss is frequently subclonal18 (present only in a subset of cells), which makes its detection by next-generation sequencing more challenging19 and it may go undetected depending on the fraction of cells harboring this aberration. Therefore, the true proportion of PCa cases with CHD1 loss may be underestimated. Thus, we decided to investigate the frequency of CHD1 loss in EA and AA PCa by methods more sensitive to detecting subclonal deletions including evaluations of multiple tumor foci present in each prostatectomy specimen.
Results
Subclonal CHD1 deletion is more frequent in African American prostate cancers and associated with worse clinical outcome
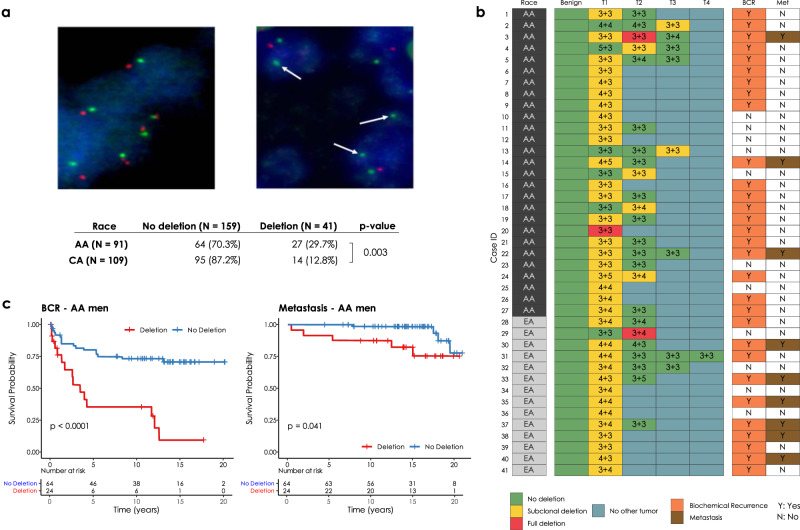
CHD1 is frequently subclonally deleted in prostate cancer18. Our initial analysis on the SNP array data from TCGA comparing AA and EA PCa cases suggested that the subclonal loss of CHD1 may be a more frequent event in AA men (Supplementary Figs. 1 and 2). To independently validate this observation, we assessed CHD1 copy number by FISH (for probe design see Supplementary Fig. 3) in tissue microarrays (TMAs) sampling multiple tissue cores from each tumor focus. Sampling included index tumors and non-index tumors per whole mounted radical prostatectomy sections in a matched cohort of 91 AA and 109 EA patients from the equal-access military healthcare system (Fig. 1a). Key clinico-pathological features including age at the diagnosis, serum PSA levels at diagnosis, pathological T-stages, Gleason sums, Grade groups, margin status, biochemical recurrence (BCR) and metastasis had no significant differences between AA and EA cases (Supplementary Table 1a). Consistent with the cohort design and long-term follow up (median: 14.5 years), we observed a 40% biochemical recurrence (BCR) and 16% metastasis rate20. For each case up to four cancerous foci were analyzed, each sampled by two TMA punch cores on average (for details see “Methods” and Supplementary Table 1a–c). We detected monoallelic CHD1 loss in 27 out of 91 AA cases (29.7%), and 14 out of 109 (11%) EA cases indicating that CHD1 deletion is about three times more frequent in prostate tumors of AA men. Our FISH data showed only 3 (2 AA cases and 1 EA case) cases where all TMA punch cores in a single tumor focus harbored CHD1 deletion in the entire samples areas of a given tumor (Fig. 1b and see the “Methods and materials” “FISH assay part” for details.) In most cases CHD1 deletion was present in only a subset of tumor glands within a 1 mm TMA punch, which further confirmed the subclonal nature of CHD1 deletion in prostate cancer. As a control, we performed FISH staining for PTEN deletion and immunohistochemistry (IHC) staining for ERG overexpression in a subset of the cohort (42 AA and 59 EA prostate cancer cases) confirming previously described frequency differences between AA and EA PCa4,5 (Supplementary Table 1e). There was a frequent exclusivity between CHD1 deletion, PTEN deletion and ERG expression both when individual tumor cores or when all tumor cores from a given patient were considered (Supplementary Fig. 4a, b). In general, the genomic defects including CHD1 deletion, PTEN deletion and ERG expression were mainly detected in index tumors.
Fig. 1. CHD1 copy number by FISH in tissue microarrays.
a Prostate cancer cells with wild type (diploid) CHD1 (upper left) vs. prostate cancer cells harboring mono-allelic deletion for CHD1 (upper right) are visualized by FISH assay. Orange signal: CHD1 probe; green signal: human chromosome 5 short arm probe; blue color: DAPI nuclear stain. Arrows are representing the lack of CHD1. Representative view fields capture 3-3 cell nuclei at ×60 magnification. Inset table summarizes the higher frequency of CHD1 deletion in prostatic carcinoma of AA vs. EA patients (The p value is from a one-sided Fisher exact test). b CHD1 deletion is a subclonal event in prostate cancer. Multiple tumor samples from 200 patients were assessed by FISH assay that identified 41 patients with CHD1 deletion (left panel). The heatmap depicts the sampled largest tumor 1 (T1), second largest tumor (T2), and so on. Numbers denote pathological Gleason grade for each tumor. BCR biochemical recurrence (orange), Met metastasis (brown). c Deletion of CHD1 (clonal or subclonal in any of the nodes) is strongly associated with disease progression in AA prostate cancer patients (N = 91). BCR univariable Kaplan–Meier curve, Metastasis univariable Kaplan–Meier curve.
Further analyses revealed a significant association between CHD1 deletion and pathologic stages and Gleason sum. Higher frequency of CHD1 deletion was detected in T3-4 pathological stage compared to T2 stage (p = 0.043, Supplementary Table 1d). Prostate cancer cases with higher Gleason sum scores (3 + 4, 4 + 3, 8–10) were seen more frequently in the CHD1 deletion group than in the non-deletion group (p < 0.001). In contrast, lower Gleason sum score (3 + 3) was more often seen in non-deletion cases (p < 0.001, Supplementary Table 1d). The CHD1 deletion was more commonly detected in the cases with higher Gleason sum score (3 + 4, 4 + 3, 8–10) (p = 0.024, Supplementary Table 1d). CHD1 deletion was more strongly associated with rapid biochemical recurrence in AA cases (p < 0.0001, Fig. 1c) than in EA cases (p = 0.051, Supplementary Fig. 5b). The univariable survival analysis was conducted to determine the association of the clinical features including CHD1 deletion to BCR and metastasis for further multivariable model analysis (Supplementary Fig. 5a, c, respectively). The multivariate Cox model analysis showed that CHD1 deletion was an independent predictor of BCR (p = 0.012 and p = 0.032, Supplementary Fig. 5b) after adjusting for age at diagnosis, PSA at diagnosis, race, pathological tumor stage, grade group and surgical margins. Moreover, a significant correlation between CHD1 deletion and metastasis was also detected in both AA (p = 0.0055, Fig. 1d) and EA (p = 0.023, Supplementary Fig. 5d) patients with Kaplan–Meier analysis. Following multivariable adjustment in the Cox proportional hazards model, CHD1 deletion was significantly associated with metastasis (p = 0.032 and p = 0.048, Supplementary Fig. 5d). Taken together, our data strongly support the association of CHD1 deletions with aggressive prostate cancer and worse clinical outcomes in AA PCa.
Estimating the frequency of subclonal CHD1 loss in next-generation sequencing data of AA and EA prostate cancer
Previous publications characterizing the genome of AA prostate cancer cases10,21 did not report an increased frequency of CHD1 loss as we observed in the FISH-based analysis presented above. Methods to detect copy number variations from whole genome sequencing (WGS) or whole exome sequencing (WES) data have at least two major limitations. First, subclonal copy number variations (sCNV) can be missed if they are present in fewer than 30%, of the sampled cells19. Second, copy number loss can be underestimated with smaller deletions (e.g., <10 kb). Although various tools are available for inferring sCNVs from WES, WGS or SNP array data, such as TITAN19, THetA22, and Sclust23, they are designed to work on the entire genome, and likely miss small (~1–10 kb) CNVs during the data segmentation process. To maximize the accuracy of our analysis we performed a gene focused analysis of the copy number loss in CHD1. We considered several factors such as the change in the normalized coverage in the tumors relative to their normal pairs’, the cellularity of the tumor genome, and the approximate proportion of tumor cells exhibiting the loss. We also evaluated whether the deletion was heterozygous or homozygous using a statistical method designed for calling subclonal loss of heterozygosity (LOH) events within a confined genomic region (details are available in the “Methods”).
Using this approach in a large cohort (N = 530 cases; 59 AA WES, 18AA WGS, 408 EA WES and 45 EA WGS, Supplementary Figs. 6–25), we observed that CHD1 is more frequently deleted in AA tumors (N = 20; 26%) than in EA tumors (N = 73 EA; 16%). Taken together, when next-generation sequencing based copy number variations were analyzed with a more sensitive method, on the combined cohorts of whole exomes and whole genomes, CHD1 loss was detected more frequently in AA cases than in EA cases (p = 0.029, Fisher exact test), which is consistent with our observations with FISH method in the TMA cohort.
Subclonal CHD1 loss is present in a significant subset of prostate cancer cases without SPOP mutations
SPOP mutations and CHD1 deletions often occur together in prostate cancer, with SPOP mutation as an early event and CHD1 loss is a later, subclonal event during tumor progression18. However, as we pointed out above, subclonal CHD1 loss is often missed by routine next-generation sequencing analysis. Therefore, we reanalyzed the next-generation sequencing cohorts for SPOP mutations and found that CHD1 loss and SPOP mutations frequently occur independently from each other as well. In the 530 cases analyzed, we identified 61 SPOP mutant cases and 95 subclonal CHD1 deletions, but only 42 cases (about 68% of SPOP mutants and 44% of CHD1 deleted cases) had both genomic aberration present. CHD1 deletions were mutually exclusive with PTEN deletions and TP53 mutations in AA PCa cases (Supplementary Fig. 24).
CHD1 loss is not associated with genomic aberration features that are usually observed in HR-deficient cancers
CHD1 loss was proposed to be associated with reduced HR competence in cell line model systems15,24. Detecting and quantifying HR deficiency in tumor biopsies is currently best achieved by analyzing next-generation sequencing data for specific HR deficiency associated mutational signatures. Those mutational signatures include: (1) A single nucleotide variation based mutational signature (“COSMIC signatures 325 and SBS326); (2) a short insertions/deletions based mutational profile, often dominated by deletions with microhomology, a sign of alternative repair mechanisms joining double-strand breaks in the absence of HR, which is also captured by COSMIC indel signatures ID6 and ID826; (3) large-scale rearrangements such as non-clustered tandem duplications in the size range of 1–100 kb (mainly associated with BRCA1 loss of function)27. Some of these signatures can be efficiently induced by the inactivation of BRCA1, BRCA2 or several other key downstream HR genes (Supplementary Figs. 26–44 and Supplementary Data 1 and 2)28.
HR deficiency is also assessed in the clinical setting by a large-scale genomic aberration based signature, namely the HRD score29, which is also approved as companion diagnostic for PARP inhibitor therapy. A composite mutational signature, HRDetect30, combining several of the mutational features listed above was also evaluated as an alternative method to detect HR deficiency in prostate adenocarcinoma31. In order to investigate whether an association between CHD1 loss and HR deficiency exists in prostate cancer biopsies, we performed a detailed analysis on the mutational signature profiles of CHD1 deficient prostate cancer.
We analyzed whole exome and whole genome sequencing data of several prostate adenocarcinoma cohorts containing samples both from AA (52 WES and 18 WGS cases) and EA (387 WES and 45 WGS) individuals in order to determine whether CHD1 loss is associated with the HRD mutational signatures.
We divided the cohorts into three groups: (1) BRCA2 deficient cases that served as positive controls for HR deficiency, (2) CHD1 deleted cases without mutations in HR genes, and (3) cases without BRCA gene aberration or CHD1 deletion.
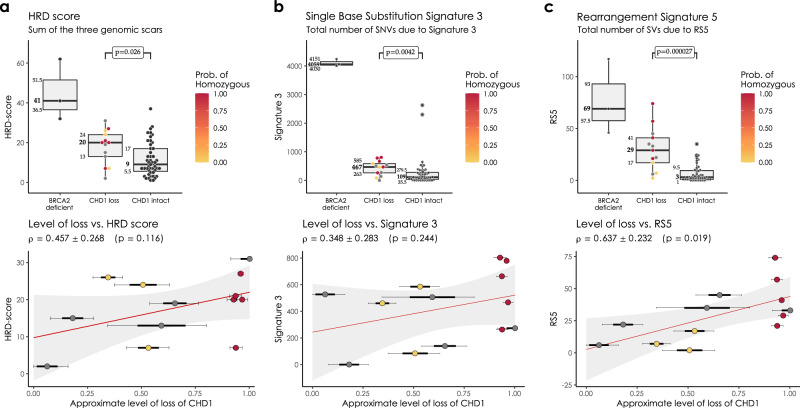
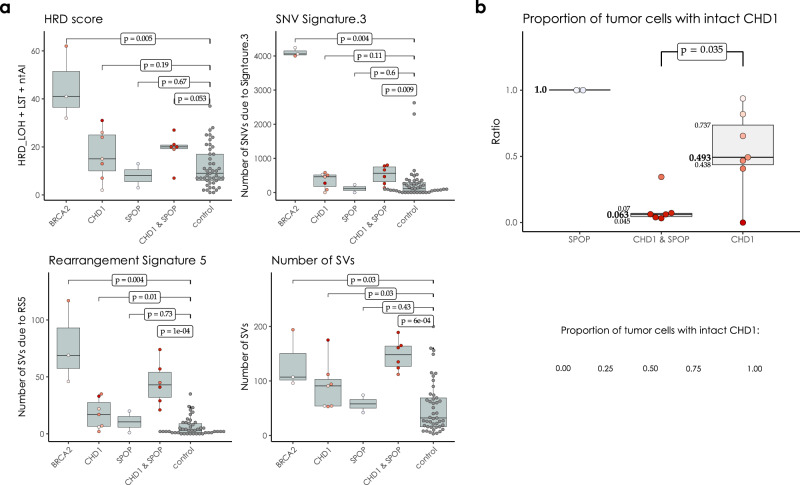
In the WGS cohorts CHD1 deficient cases showed a limited increase of the HRD score relative to the control cases but significantly lower than the BRCA2 deficient cases and none of the CHD1 deficient cases had an HRD score above the threshold currently accepted in the clinic as an indicator of HR deficiency (Fig. 2a). Since CHD1 deletions tend to be subclonal, we investigated whether the low levels of HRD score is due to a “dilution” effect, where the HR proficient regions without CHD1 deletion reduce the intensity of the HRD score. The HRD score did not show a statistically significant correlation with the estimated fraction of the subclonal loss of CHD1 (Fig. 2a and Supplementary Figs. 26 and 27), and even cases where all cells had CHD1 deletion did not have a high enough HRD score indicating HR deficiency. Similarly, the most characteristic HRD associated single nucleotide variation signature (signature 3, SBS3), was significantly increased in the BRCA2 deficient cases but only slightly increased in the CHD1 deficient cases (Fig. 2b).
Fig. 2. HRD markers in the PRAD WGS cohorts.
a HRD-score, the sum of the three genomic scars, HRD-LOH, LST, and ntAI, b number of somatic mutations due to single-base substitution signature 3, c number of structural variants due to rearrangement signature 5. The significance of the difference between the means of the “CHD1 loss” and “control” groups were assessed with Wilcoxon ranked sum tests. Below the box plots are the correlations between the approximate levels of loss in CHD1 and the HRD measures are visualized. The standard errors and the corresponding p values of the correlation coefficients (Pearson) are also indicated. Horizontal lines indicate the uncertainty in the level of loss in each sample. Thick black lines correspond to the 66%, thin black error-bars to the 95% percentile intervals.
The increase of the relative contribution of short indel signatures ID6 and ID8 to the total number of indels, which is characteristic of loss of function on BRCA2 biallelic mutants, was not observed in the CHD1 loss cases (Supplementary Figs. 32–34). This suggests, that the alternative end-joining repair pathways do not dominate the repair of DSBs in CHD1 deleted tumors.
In the WGS cohort we also determined the number of structural variants (SVs) as previously defined (Supplementary Fig. 35)32. The SV signature associated with HR deficiency (SV3) was not elevated in the CHD1 deficient tumors. Interestingly, an SV signature characterized by an increase in the number of non-clustered 1kb-1Mb deletions (termed RS527) was significantly increased both in the BRCA2 mutant and CHD1 deficient cases (Fig. 2c), with the latter showing a less significant increase. Notably, this signature also displayed a strong subclonal dilution. This signature was described to be associated with BRCA2 deficiency previously27,32 but it is also present in tumors without BRCA2 deficiency and the current version of this signature, SV5 (https://cancer.sanger.ac.uk/signatures/sv/sv5/) is not associated with HR deficiency.
Finally, the BRCA2 deficient cases showed high HRDetect scores (Supplementary Figs. 36–38). However, since the HRDetect scores arise from a logistic regression, which involves the non-linear transformation of the weighted sum of its attributes, even slightly lower linear sums in the CHD1 loss cases compared to the BRCA2 mutant cases can result in substantially lower HRDetect scores (Supplementary Fig. 38).
We have previously processed WES prostate adenocarcinoma data for the various HR deficiency associated mutational signatures31. When the CHD1 deficient cases were compared to the BRCA1/2 deficient and BRCA1/2 intact cases we obtained results that were consistent with the WGS based results outlined above (Supplementary Figs. 39–44).
Deleting CHD1 in prostate cancer cell lines does not induce homologous recombination deficiency as detected by the RAD51 foci formation assay or mutational signatures
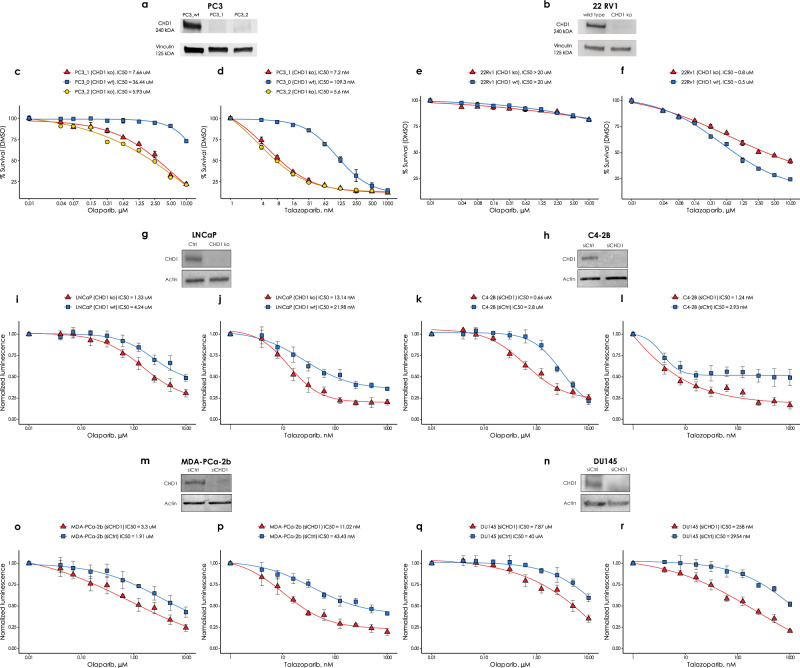
To investigate the functional impact of the biallelic loss of CHD1 we created several CRISPR-Cas9 edited clones of the AR− PC-3 and AR+ 22Rv1 cell lines (Fig. 4a and Supplementary Fig. 47a). RAD51 foci formation was induced by 4 Gy irradiation. The CHD1 deficient prostate cancer cell lines did not show reduction of RAD51 foci formation (Fig. 3a). Non-irradiated cells were used as controls (Supplementary Fig. 46).
Fig. 4. The effect of CHD1 loss on olaparib and talazoparib sensitivity in six prostate cancer cell lines.
CHD1 was either knocked out CRISPR-Cas9-mediated editing in the PC-3, 22Rv1 and LNCaP cell lines or suppressed by shRNA in the C4-2b, Du145 and MDA-PCa-2b cell lines. Western immunoblots show the successful elimination CHD1 in a PC-3, b 22Rv1, g LNCaP, h C4-2b, m MDA-PCa-2b and n Du145 cell lines. Sensitivity assays of parental wt and CHD1 eliminated clones to PARP inhibitor Olaparib and talazoparib in PC-3 cells (c, d), in 22Rv1 cells (e, f), in LNCaP cells (i, j), in C4-2b cells (k, l), in MDA-PCa-2b cells (o, p), and in Du145 cells (q, r). Cells viability was measured the PrestoBlueTM reagent. SD of triplicates are shown.
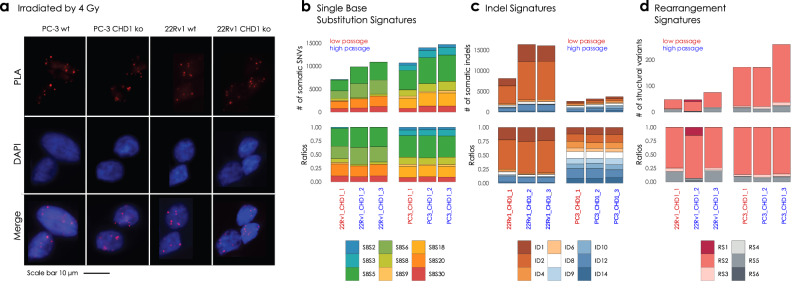
Fig. 3. PC-3 and 22Rv1 CHD1 ko cell line experiment and somatic signature extraction.
a RAD51 foci formation. Examples of the most common staining patterns in WT and CHD1 ko 22Rv1 and PC-3 cell lines. Cells were fixed by 4% PFA 3 h after irradiation (IR = 4 Gy) PLA was carried out using antibodies against γH2Ax and RAD51 proteins. b Single Nucleotide Substitution (SBS) signatures, c Indel signatures, d Rearrangement signatures. The number of mutations indicated originate from the reconstructed mutational spectra.
DNA repair pathway aberration induced mutational signatures can also be detected in cell lines by whole genome sequencing28,33. We grew single cell clones from the PC-3 and 22Rv1 cell lines for 45 generations to accumulate the genomic aberrations induced by CHD1 loss (Supplementary Fig. 45). Two of such late passage clones and one early passage clone were subjected to WGS analysis. All the clones retained the BRCA2 wild type background of their parental clone.
Furthermore, CHD1 elimination did not induce any of the mutational signatures commonly associated with HR deficiency (Fig. 3b–d).
Taken together, CHD1 loss in prostate cancer cell line models did not induce any signs of HR deficiency.
CHD1 deficient cell lines show limited sensitivity to the PARP inhibitors olaparib and talazoparib
CHD1 deficient cancer cells were reported to have moderately increased sensitivity to the PARP inhibitor olaparib15, which is consistent with the lack of observed HR deficiency described in the previous section. PARP inhibitors were initially thought to exert their therapeutic activity by inhibiting the enzymatic activity of PARP, but it was later revealed that trapped PARP on DNA may have a more significant contribution to cytotoxicity (reviewed in ref. 34). Therefore, in addition to olaparib, we also determined the efficacy of the strong PARP trapping agent talazoparib in several prostate cancer cell lines in which CHD1 was either knocked out or suppressed. In addition to the PC-3, 22Rv1 and LNCaP cells with CRISPR-Cas9-mediated CHD1 deletion we also suppressed CHD1 by shRNA in the C4-2b, Du145 and MDA-PCa-2b prostate cancer cell lines, the last one is one of the few AA derived prostate cancer cell line models. Our goal was to assess in several CHD1 deficient prostate cancer cell lines whether their PARP inhibitor sensitivity is in the therapeutically achievable concentration range.
Deleting CHD1 induced a maximum of approximately 5-fold increase in olaparib sensitivity with minimal or no change in some cell lines (Fig. 4c, e, i, k, o, q)15. Three cell lines (LNCaP, C4-2H and MDA-PCa-2B) without deleting CHD1 showed olaparib sensitivity of low micromolar concentrations, which is in the therapeutic concentration range for this agent. This sensitivity was further increased by 1.5-3 fold by CHD1 deletion. The increase in talazoparib sensitivity was similar to that of olaparib for most cell lines with a few notable exceptions. Talazoparib sensitivity increased by about 15–20-fold in the CHD1 deficient PC-3 cells (Fig. 4d), and, notably in the CHD1 deficient AA derived cell line (MDA-PCa-2b), talazoparib sensitivity increased by 4-fold (Fig. 4p), while the increase in olaparib sensitivity was approximately 1.5-fold for the same cell line (Fig. 4o). In summary, in four of the six cell lines (Fig. 4d, j, l, p), CHD1 suppression was associated with a talazoparib sensitivity consistent with therapeutically achievable concentrations (around 10 nM or less.)
These data suggest that despite the lack of inducing HR deficiency, CHD1 deletion may lead to PARP inhibitor sensitivity with likely clinical benefit. Supplementary Figs. 45 and 46 provide the uncropped immunoblot images.
The impact of SPOP mutations on the clonality of CHD1 deletions and HR deficiency associated mutational signatures
Although less frequent, SPOP mutations and CHD1 deletions may co-exist in a subset of prostate cancer35 and SPOP mutations have been shown to suppress key HR genes17. Therefore, we investigated whether the presence of SPOP mutation in a CHD1 deficient prostate cancer is associated with a further increase of HR deficiency associated mutational signatures. We identified cases with SPOP mutations or CHD1 deletions only, cases with both SPOP mutations and CHD1 deletions and cases without either of those aberrations (Fig. 5a). Cases with both mutations showed significantly higher levels of signature SBS3, RS5 and the total number of large-scale structural rearrangements relative to cases with either mutation alone. It should be noted, however, that the proportion of cells in a given tumor with CHD1 deletions tended to be significantly higher in SPOP mutant cases than those with CHD1 deletions without SPOP mutations. Therefore, it is possible that the presence of SPOP will intensify HR deficiency associated mutational signatures by enhancing the proportion of CHD1 deficient cells in a tumor (Fig. 5b).
Fig. 5. CHD1 loss and SPOP mutation in the WGS cohorts.
a HRD-related markers and total number of structural variants in samples with mutations in SPOP, BRCA2 and loss in CHD1 versus the controls. Samples that simultaneously harbor mutations in SPOP and a loss in CHD1 tend to have higher markers. p values were estimated using non-parametric Wilcoxon signed-rank tests. b Proportion of cells with intact CHD1 in SPOP mutants and samples identified with CHD1 loss. While the deletion in CHD1 in SPOP mutants is mostly clonal, in samples with wild type SPOP background it is mostly subclonal. The color-code for points in both (a, b) is illustrated in the bottom right corner of the figure.
Finally, we investigated whether adding SPOP mutations to a CHD1 deficient background increases PARP inhibitor sensitivity. We overexpressed the SPOP mutant SPOPF102C in the CHD1 deleted PC3 cells (Supplementary Figs. 47–55), but we could not detect a further increase in sensitivity to either olaparib or talazoparib.
Discussion
The presence of functionally relevant subclonal mutations in various solid tumor types is well documented36,37. Deletions present only in a minority of tumor cells are difficult to detect unless more targeted analytical approaches are applied. Here we present one example of such detection bias with significant functional relevance. We used a FISH-based approach to detect CHD1 deletion in PCa. Consistent with the previously described subclonal nature of CHD1 loss, we found that while this gene is often deleted in prostate cancer, it is rarely deleted in every tumor core or tumor focus. When we took the subclonal nature of CHD1 loss into consideration a significant racial disparity emerged, with an approximately 3-fold increase in the frequency of CHD1 deletion in AA PCa patients vs. EA patients. This loss was also significantly associated with rapid disease progression to biochemical recurrence and metastasis. Since CHD1 loss is associated with a more malignant phenotype, the significantly higher frequency of CHD1 loss in AA PCa may account for the diverging clinical course observed in PCa between men of African and European Ancestry. It is possible that CHD1 loss is in fact more frequent in EA PCa as well but with a lower focal density than in AA cases. This is certainly a limitation of our bioinformatics approach. However, CHD1 single cell-level deletions have not been observed in our high-resolution FISH assay in tumors of EA patients.
Several studies pointed out a potential link between CHD1 loss and homologous recombination deficiency15,16,24. Interestingly, CHD1 null cells showed only a modest (3-fold) increase in sensitivity to PARP inhibitor or platinum-based therapy15,16,24. This suggested that CHD1 loss may not lead to a significant level of HR deficiency. Our results support this assumption since CHD1 deficient tumors did not display increased levels of the verified HR deficiency associated mutational signatures and CHD1 loss in cell lines did not induce HR deficiency as detected by functional assays either. The moderate increase in PARP inhibitor sensitivity may be caused by other mechanisms such as an interaction between nucleosome remodeling factors, such as ACL1 and PARP38,39.
Consequently, the limited sensitivity of CHD1 deficient cell lines to PARP inhibitors suggests that this treatment may be less effective than in bona fide HR deficient, such as inactivated BRCA2 cases. Nevertheless, the facts that talazoparib is effective in some of the CHD1 deficient cell line models and the that CHD1 suppression induces enzalutamide sensitivity14 may explain some of the unexpected results of the TALAPRO-2 study40. In this trial, patients without mutations in the DNA damage pathway (BRCA2 etc.) also benefitted from a combination of talazoparib and enzalutamide. We are hypothesizing that talazoparib, perhaps by eliminating CHD1 deficient cells, may delay the emergence of enzalutamide resistance, which may define an effective therapy in a significant subset, those with CHD1 deficiency, of AA PCa cases. Taken together, our cell line sensitivity data suggest, that despite the lack of CHD1 induced HR deficiency PARP inhibitors may still provide clinical benefit by targeting CHD1 deficient prostate cancer.
CHD1 was also reported to be associated with altered immunogenic phenotype in prostate cancer41. These results coupled with the demonstrated differences of tumor immunity between EA and AA prostate cancer cases42 raises the possibility that CHD1 deficiency may make the AA PCa population sensitive to targeted immunotherapy.
Finally, the somewhat increased genomic instability of CHD1 deficient cases, as reflected by the moderately elevated HRD scores, may also indicate that it is the genomic instability rather than the CHD1 loss that is responsible for the significantly worse outcome of CHD1 deficient cases detected in our AA PCa cohort. Separating these two effects will require further studies.
Methods
Institutional Review Board—Center for Prostate Disease Research (CPDR)
The Uniformed Services University of the Health Sciences’ (Department of Defense) Institutional Review Board (OHRP #IRB00000968; FWA #FWA00005897) reviewed the work in this study and was “determined to be considered research not involving human subjects as defined by 32 CFR 219.102(e) because the research involves the use of de-identified specimens and data not collected specifically for this study.” (IRB protocol #910230). 32 CFR 219 is the Department of Defense’s adoption of the Common Rule (45 CFR 46) and also adheres to DoD Instruction 3216.02 titled, “Protection of human subjects and adherence to ethical standards in DoD-conducted and -supported research”. An informed consent form was not utilized for this study. A full HIPAA Waiver was granted for the use of the data in the Center for Prostate Disease Research (CPDR) Database Repository (IRB protocol #GT90CM). This study used already banked specimens and data from consented participants who agreed to the future use of their specimens and data from the CPDR’s repositories:
CPDR Biospecimen Bank (IRB protocol number #393738) at the Walter Reed National Military Medical Center IRB (OHRP #IRB00008418; FWA #FWA00017749).
CPDR Database Repository (IRB protocol number #GT90CM) at the Uniformed Services University of the Health Sciences IRB (OHRP #IRB00000968; FWA #FWA00005897).
Cohort selection and tissue microarray (TMA) generation
The aggregate cohort was composed of 2 independently selected cohort samples from Bio-specimen bank of Center for Prostate Disease Research and the Joint Pathology Center. Whole-mount prostates were collected from 1996 to 2008 with minimal follow-up time of 10 years. Self-reported race was validated by genomic ancestry analysis showing an 95% accuracy43. The first cohort of 42 AA and 59 EA cases was described before7,43. Similarly, the second cohort of 50 AA and 50 EA cases was selected based on the tissue availability (>1.0 cm tumor tissue) and tissue differentiation status (1/3 well differentiated, 1/3 moderately differentiated and 1/3 poorly differentiated).
Patients who have donated tissue for this study also contributed to the long-term follow-up data (the mean follow-up time was 14.5 years). TMA block was assigned as 10 cases each slide and each case with 2 benign tissue cores, 2 Prostatic intraepithelial neoplasia (PIN) cores if available and 4–10 tumor cores covering the index and non-index focal tumors from formalin fixed paraffin embedded (FFPE) whole-mount blocks. The description of numbers of patients, tumors and tumor cores of combined cohort was in Supplementary Table 1d. All the blocks were sectioned into 8 µM tissue slides for FISH staining.
Fluorescence in situ hybridization (FISH) assay
A gene-specific FISH probe for CHD1 was generated by selecting a combination of bacterial artificial chromosome (BAC) clones (Thermo Fisher Scientific, Waltham, MA) within the region of observed deletions near 5q15-q21.1, resulting in a probe matching ca. 430 kbp covering the CHD1 gene as well as some upstream and downstream adjacent genomic sequences including the complete repulsive guidance molecule B (RGMB) gene. Due to the high degree of homology of chromosome 5-specific alpha satellite centromeric DNA to the centromere repeat sequences on other chromosomes, and the resulting potential for cross-hybridization to other centromere sequences, particularly on human chromosomes 1 and 19, a control probe matching a stable genomic region on the short arm of chromosome 5—instead of a centromere 5 probe—was used for chromosome 5 counting (Supplementary Fig. 1e). The FISH assay of CHD1 was performed on TMA as previously described7. The green signal was from probe detecting control chromosome 5 short arm and the red signal was from probe detecting CHD1 gene copy. The FISH-stained TMA slides were scanned with Leica Aperio VERSA digital pathology scanner for further evaluation. The criteria for CHD1 deletion was that in over 50% of counted cancer cells (with at least 2 copies of chromosome 5 short arm detected in one tumor cell) more than one copy of CHD1 gene had to be undetected. Examining tumor cores, deletions were called when more than 75% of evaluable tumor cells showed loss of allele. Focal deletions were called when more than 25% of evaluable tumor cells showed loss of allele or when more than 50% evaluable tumor cells in each gland of a cluster of two or three tumor glands showed loss of allele. Benign prostatic glands and stroma served as built-in control.
The sub-clonality of CHD1 deletion was presented with a heatmap showing CHD1 deletion status in all the given tumors sampled from whole-mount sections of each patient. The color designations were denoted as: red color (full deletion) meaning all the tumor cores carrying CHD1 deletion within a given tumor, yellow color (subclonal deletion) meaning only partial tumor cores carrying CHD1 deletion within a given tumor and green color (no deletion) meaning no tumor core carry CHD1 deletion (Supplementary Table 1b).
Statistics analysis
The correlations of CHD1 deletion and clinic-pathological features, including pathological stages, Gleason score sums, Grade groups, margin status, and therapy status were calculated using an unpaired t-test or chi-square test. Gleason Grade Groups were derived from the Gleason patterns for cohort from Grade group 1 to Grade group 5. Due to the small sample sizes within each Grade group, Grade group 1 through Grade group 3 were categorized as one level as well as Grade group 4 through Grade group 5. A BCR was defined as either two successive post-RP PSAs of ≥0.2 ng/mL or the initiation of salvage therapy after a rising PSA of ≥0.1 ng/mL. A metastatic event was defined by a review of each patient’s radiographic scan history with a positive metastatic event defined as the date of a positive CT scan, bone scan, or MRI in their record. The associations of CHD1 deletion and clinical outcomes with time to event outcomes, including BCR and metastasis, were analyzed by a Kaplan–Meier survival curves and tested using a log-rank test. Multivariable Cox proportional hazards models were used to estimated hazard ratios (HR) and 95% confidence intervals (Cis) to adjust for age at diagnosis, PSA at diagnosis, race, pathological tumor stage, grade group, and surgical margins. We checked the proportional hazards assumption by plotting the log-log survival curves. A p value < 0.05 was considered statistically significant. Analyses were performed in R version 4.0.2.
Immunohistochemistry for ERG
ERG immunohistochemistry was performed as previously described44. Briefly, four μm TMA sections were dehydrated and blocked in 0.6% hydrogen peroxide in methanol for 20 min. and were processed for antigen retrieval in EDTA (pH 9.0) for 30 min in a microwave followed by 30 min of cooling in EDTA buffer. Sections were then blocked in 1% horse serum for 40 min and were incubated with the ERG-MAb mouse monoclonal antibody developed at CPDR (9FY, Biocare Medical Inc.) at a dilution of 1:1280 for 60 min at room temperature. Sections were incubated with the biotinylated horse anti-mouse antibody at a dilution of 1:200 (Vector Laboratories) for 30 min followed by treatment with the ABC Kit (Vector Laboratories) for 30 min. The color was developed by VIP (Vector Laboratories,) treatment for 5 min, and the sections were counter stained by hematoxylin. ERG expression was reported as positive or negative. ERG protein expression was correlated with clinico-pathologic features.
TCGA SNP-array data
We analyzed data from 495 TCGA patients using the Affymetrix SNP Array 6.0 and preprocessed it with the AROMA affymetrix R package. They calculated principal components from the B-allele frequencies, finding that PC2 and PC3 distinguished samples by ancestry. The DBSCAN algorithm identified 251 Caucasian and 46 African American patients, excluding outliers (Supplementary Fig. 1). The analysis revealed a notable depletion near the centromere of chromosome 5, with a more significant loss in African American patients, particularly around the CHD1 gene (Supplementary Fig. 2). This discovery prompted further investigation into the observed genetic differences.
Next-generation sequencing data
Whole exomes from 498 patients were downloaded from the GDC data portal and aligned to the GRCh38 reference genome. The samples included the following self-declared ancestries: 52 African American (nAA = 52), 387 European American (nEA = 387), 12 Asian American (nAS = 12), 1 American Native, and 46 not reported (Supplementary Fig. 6). Additionally, whole genome normal-tumor pairs from 63 patients were obtained from various sources. We acquired 20 sample pairs (nAA = 2, nEA = 18) from the ICGC data portal (TCGA PRAD-US cohort), 19 sample pairs (nAA = 9, nEA = 10) from the Dana Farber Cancer Institute, 14 sample pairs (nAA = 7, nEA = 7) from the Center for Prostate Disease Research (CPDR), and 10 sample pairs (nAA = 0, nEA = 10) from the Decker et al. study45.
Evaluation of the self-declared ancestries
To identify the ancestries of the 46 unreported cases in the TCGA whole exome cohort, we sought to determine the genotypes at key genomic SNP coordinates, which are significantly more prevalent in the three common ancestry groups (European American—EA, African American—AA, and Asian—AS)46. A Bayes Classifier was used to identify the most probable ancestries of the “not reported” cases and to detect outliers among cases with self-declared ancestries. Variants more prevalent in the three ancestries were chosen from the Exome Aggregation Consortium (ExAC) database, emphasizing those supported by at least 4000 African American donors and 10,000 Asian and European American donors. The top 1000 most common variants in each ancestry group, which were nearly absent in the other two groups, were selected as predictors. (Supplementary Figs. 7–9).
The collected 3000 SNPs were used to create a single genotype matrix (G) with 498 rows (patients) and 3000 columns (genotypes). In this matrix, an element (G[i, j]) was set to 0 for REF/REF genotypes, 1 for heterozygous ALT/REF variants, and 2 for ALT/ALT homozygotes. Singular value decomposition was performed on matrix G to determine its singular values and their corresponding singular vectors, representing the principal components (PCs). The projections onto 2-dimensional planes formed by the first few principal components showed that the first principal component accounted for the largest proportion of variance and best separated African American patients from European American patients. The second principal component, while representing a smaller fraction of the variance, differentiated the Asian samples from the other two ancestries (Supplementary Figs. 10 and 11). We identified and filtered outliers based on their distances in the PC1-PC2 space, focusing on the mean distance from their 10 closest neighbors of the same ancestry. These outliers were reclassified and treated similarly to samples with “not reported” ancestries (Supplementary Figs. 12 and 13).
Our approach involved training a model to learn the distribution of ancestry points in the PC1-PC2 space. We used these learned distributions to predict the likely ancestry of ‘not reported’ and ‘outlier’ cases based on their genotypes. Ancestry classes were encoded as follows; European American: 0, African American: 1, Asian American: 2.
The columns of the G matrix were standardized according to:
| 1 |
The probability that sample [i,•] belongs to ancestry group was calculated as the following:
| 2 |
The likelihood is modeled using a multivariate normal density , with the maximum a posteriori (MAP) estimates of the parameters obtained from the classifier algorithm. The prior probability is determined by the relative sample size of ancestry , calculated as , where represents the number of patients from ancestry . The 441 samples from AA, EA, and AS patients were randomly split into training (ntraining = 352, 80%) and test (ntest = 89, 20%) sets. The classifier was implemented in R, and its accuracy was estimated to be within the range of [0.949, 0.999] (Supplementary Figs. 14–19). Additionally, the model trained on TCGA whole exomes was evaluated on the full cohort of whole genomes, achieving a 100% agreement between the predicted ancestries and the self-reported ancestries (Supplementary Figs. 20–22).
Identification of local subclonal loss of CHD1 in prostate adenocarcinoma
The paired germline and tumor BAM files were analyzed to determine their mean sequencing depths using bedtools genomecov47 and samtools48. The coverage data around the CHD1 gene (chr5:98,853,485–98,930,272 in GRCh38 and chr5:98,190,408–98,262,740 in GRCh37) was collected in 50 bp wide bins, resulting in m-dimensional vectors (m_GRCh37 = 1447, m_GRCh38 = 1536). These vectors were normalized based on their respective mean sequencing depths. The linear relationship between the paired germline-tumor coverages were determined in the following form:
| 3 |
where is the normalized depth of the germline sample and is the normalized depth of its corresponding tumor pair. The intercept () was used to ensure that the data was free of outliers, and the slope () was used as a raw measure of the observable loss in the tumor. Similar slopes were calculated for 14 housekeeping genes (G6PD, IPO8, PGK1, PP1A, HMBS, GUSB, UBC, YWHAZ, GAPDH, HPRT1, ACTB, B2M, TBP, and TFRC) in each sample-pair to assess the significance of the loss. The 14 estimated slopes were standardized into z-scores using their mean and standard deviation. The estimated slopes for CHD1 were also converted into z-scores based on previously determined parameters from their donors, and p values were calculated. Samples with p values greater than 0.1 for whole genomes or 0.05 for whole exomes were labeled as “CHD1 intact,” while those with lower p values were classified as “CHD1 loss”.
The cellularity (c) of the tumors were estimated using sequenza49 with the most reliable cellularity-ploidy pair selected from the tool’s alternative solutions. To account for the uncertainty in the reported cellularity values, a beta distribution was fitted on the grid-approximated marginal posterior densities of c. These were used to simulate random variables to determine the proportion of the approximate loss of CHD1 in the tumors using the following formula:
| 4 |
which was derived from:
| 5 |
The ∼ operator in Eq. (4) indicates that the true level of CHD1 can only be determined with a certain degree of accuracy, which depends on the uncertainties in β0 and c. The uncertainty in β0 arises from the fitted linear model itself:
| 6 |
Sequenza provides the joint posterior distribution of ploidy and cellularity using a grid approximation. We sampled from the peak of the cellularity’s discretized marginal posterior, which matched the final copy number segments. To convert these discrete values to a continuous scale, a beta distribution was fitted to the cellularity samples: . Using the distributions of β₀ and cellularity, we estimated the uncertainty in the true level of CHD1 loss, calculated as
Genotyping
Variant and copy number calling were conducted in the same manner as described by Sztupinszki et al.31. Genotypes were categorized as follows: wild type (+|+) if no pathogenic or likely pathogenic variants were found in the gene, monoallelic (+|−) if at least one pathogenic germline or somatic variant or a loss of heterozygosity (LOH) was identified, and biallelic (−|−) if a pathogenic variant was present along with an LOH or a deep deletion was observed (Supplementary Fig. 24).
Local subclonal LOH-calling
The SNP variant allele frequencies (VAF) at CHD1 in the tumor were collected with GATK HaplotypeCaller50. The coverage and VAF data were carefully analyzed to ensure a strict focus on regions that have suffered the most serious loss (e.g., if only a part of the gene was lost, the unaffected regions were excluded from the analysis). Using the tumor cellularity (c) and the estimated level of loss in the tumor (), we evaluated whether a heterozygous or homozygous subclonal deletion was more likely responsible for the observed frequency pattern.
The observed distribution of SNP ALT allele frequencies in the tumor sample () were considered as stochastic variables generated by the following process:
| 7 |
Here, c represents the cellularity of the tumor sample (a stochastic random variable approximated by a beta process, as described earlier), and is the proportion of cancer cells with intact CHD1 in the sample, not accounting for normal contamination. is the distribution of allele frequencies for heterozygous SNPs in the normal sample, which is also modeled using a beta process:
| 8 |
centered on 0.5, i.e.,
When the loss in the tumor is homozygous, all the reads come from either the normal cells or the tumor cells that still have the normal phenotype, meaning they have intact CHD1. To ensure that , we assume that in this case, . This means the observed allele frequency in the homozygous loss scenario is the same as the normal allele frequency (specifically in the vicinity of the target gene), expressed as:
| 9 |
In cases where there is no deletion in the targeted gene, the same allele frequency distribution is observed. The only indication of a loss in this scenario is a decrease in coverage in the tumor.
A heterozygous deletion (LOH) can occur through the loss of either the ALT allele (resulting in ) or the REF allele (resulting in ), and the distribution of the observable allele frequencies becomes bimodal. Equation(7) can be simplified to the following formula:
| 10 |
where and are stochastic variables that depend only on the cellularity and the estimated level of CHD1 loss, subject to the constraint In a heterozygous model, the observable allele frequencies will be generated by the following stochastic process:
| 11 |
The left-hand side will produce variants with higher AFs, while the right-hand side will produce lower AFs. The distance between the two modes is influenced by . The larger is, the closer the modes will be to and .
The likelihoods that the data were produced by either a homozygous or heterozygous process are:
| 12 |
and
| 13 |
The probability that the deletion affects only one of the alleles (i.e., it is heterozygous) can be calculated from the likelihoods:
| 14 |
This process is illustrated in Supplementary Fig. 25.
Mutational signatures
Somatic point-mutational signatures were estimated with the deconstructSigs R package51. The list of considered mutational processes whose signatures’ linear combination could lead to the final mutational catalogs (a.k.a. mutational spectra) were extracted in a dynamic process in which every single signature components were investigated one by one in an iterative manner and only those were kept that have improved the cosine similarity between the reconstructed and original spectra by a considerable margin (>0.001).
HRD-scores
The calculation of the genomics scar scores (loss-of-heterozygosity: LOH, large-scale transitions: LST and number of telomeric allelic imbalances: ntAI) was performed using the scarHRD R package52. The allele-specific segmentation data of the samples were provided by sequenza49.
Cell culture models
PC-3, 22Rv1, C4-2B and DU-145 prostate cell lines were purchased from ATCC® and grown in RPMI 1640 (Gibco) supplemented with 10% FBS (Gibco). MDA-PCa-2b cells were grown in BRFF-HPC1 media (Athena Enzyme Systems #0403) supplemented with 20% FBS (Gibco) and growing surface was coated with FNC coating mix (Athena Enzyme Systems #0407). All the cell lines were grown at 37 °C in 5% CO2, and regularly tested negative for Mycoplasma spp. contamination. The CRISPR edited CHD1 deficient LNCaP cell lines were generously shared by the authors13.
Stable CRISPR-Cas9 expressing isogenic PC-3 cell line generation
Full length SpCas9 ORF was introduced in PC-3 cell population by Lentiviral transduction using lentiCas9-Blast (Addgene #52962) construction. After antibiotics (blasticidin) selection, survival populations were single cell cloned, isogenic cell lines were generated and tested for Cas9 activity by cleavage assay.
Gene knock-out induction
CHD1 was targeted in CRISPR-Cas9 expressing PC-3 cell line using guide RNA CHD1_ex2_g1 (gCTGACTGCCTGATTCAGATC), resulted PC-3 CHD1 ko 1, and CHD1 ko 2 homozygous knock out cell lines. The same guide RNA was used to transiently knock out CHD1 gene in the 22Rv1 parental cell line.
Transfection
Cells were transiently transfected by Nucleofector® 4D device (Lonza) by using supplemented, Nucleofector® SF solution and 20 μl Nucleocuvette® strips following the manufacturer’s instructions. Following transfection, cells were resuspended in 100 μl culturing media and plated in 1.5 ml pre-warmed culturing media in a 24 well tissue culture plate. Cells were subjected to further assays 72 h post transfection.
In vitro T7 endonuclease I (T7E1) assay
Templates used for T7E1 were amplified by PCR using CGTCAACGATGTCACTAGGC forward and ATGATTTGGGGCTTTCTGCT reverse oligos generating a 946 bp amplicon. In total, 500 ng PCR products were denatured and reannealed in 1x NEBuffer 2.1 (New England Biolabs) using the following protocol: 95 °C, 5 min; 95–85 °C at −2 °C/s; 85–25 °C at −0.1 °C/s; hold at 4 °C. Hybridized PCR products were then treated with 10 U of T7E1 enzyme (New England Biolabs) for 30 min in a reaction volume of 30 μl. Reactions were stopped by adding 2 μl 0.5 M EDTA, fragments were visualized by agarose gel electrophoresis.
Generating of SPOPF102C mutant overexpressing PC cell lines. SPOPF102C ORF was previously cloned into pInducer20 (Addgene #44012)53 vector and overexpressed in PC-3 and 22Rv1 wt and CHD1 knock out cells by lentiviral transduction. After G418 (500 ug/ml) antibiotics selection survival populations were propagated and utilized for further assays. Using 48 h doxycycline (0.5 ug/ul) induction, olaparib sensitivity assay was performed. Endogenous wt SPOP and mutant SPOPF102C protein levels were determined SPOP specific (Abcam) and HA-tag (Sigma-Aldrich) antibodies, respectively.
Immunoblot analysis
Freshly harvested cells were lysed in RIPA buffer. Protein concentrations were determined by Pierce BCATM Protein Assay Kit (Pierce). Proteins were separated via Mini Protean TGX stain free gel 4–15% (BioRad) and transferred to polyvinylidene difluoride membrane by using iBlot 2 PVDF Regular Stacks (Invitrogen) and iBlot system transfer system (Life Technologies).
Membranes were blocked in 5% BSA solution (Sigma). Primary antibodies were diluted following the manufacturer’s instructions: anti-Vinculin antibody (Cell Signaling) (1:1000) and antiCHD1 (Novus Biologicals) (1:2000).
Signals were developed by using Clarity Western ECL Substrate (BioRad) and Image Quant LAS4000 System (GE HealthCare).
Proximity ligation assay (PLA)
Cells were seeded in μ-slide 8 well chambers (Ibidi GmbH, Germany) and incubated overnight. Next day, cells were subjected to irradiation (4 Gy). Irradiated and control cells (0 Gy) were recovered for 3 h, then fixed with 4% PFA and permeabilized with 0.3% Triton X-100.
Duolink® Proximity Ligation Assay (Sigma) was carried out using antibodies against γH2Ax and RAD51(Cell Signaling) according to the manufacturer’s instruction. Signals were detected by fluorescent microscopy (Nikon Ti2-e Live Cell Imaging System). Quantification of fluorescent signals were carried out by using the Fiji-ImageJ software.
Sample preparation for whole genome sequencing (WGS)
DNA was extracted from 22Rv1 and PC-3 CHD1 knock out isogenic cell lines at low passage number of the cells (22Rv1_1, PC-3_1). Following 45 passages, CHD1 knock out isogenic cell line was single cell cloned, and two colonies per cell line (22Rv1_2, 22Rv1_3, PC-3_2, PC-3_3) were propagated for DNA isolation.
DNA was extracted by using QIAamp DNA Mini Kit (QIAGENE). Whole Genome Sequencing of the DNA samples was carried out at Novogene service company.
Viability cell proliferation assays
Exponentially growing PC-3 cell lines WT, CHD1 ko1, CHD1 ko2, and 22Rv1 WT and chd1 ko respectively, were seeded in 96-well plates (1500 PC-3 cells/well, and 3000 22 Rv1 cells/well) and incubated for 36 h to allow cell attachment. Identical cell numbers of seeded parallel isogenic lines were verified by the Celigo Imaging Cytometer after attachment. C4-2B, MDA-PCa-2b and DU145 cells were transiently transfected with Ctrl siRNA (5’-CGUACGCGGAAUACUUCGAUUUU-3’) and CHD1 siRNA (5’-CACAAGAGCUGGAGGUCUAUU-3’) using RNAiMAX (Invitrogen, 13778-150) according to the manufacturer’s instructions. Cells were exposed to talazoparib (Selleckchem) and olaparib (MedChemExpress) for 24 h, then kept in drug-free fresh media for 5 days until cell growth was determined by the addition of PrestoBlueTM (Invitrogen) and incubated for 2.5 h or with CellTiter-Glo (Promega, #G7572). Cell viability was determined by using the BioTek plate reader system. Fluorescence was recorded at 560 nm/590 nm, and values were calculated based on the fluorescence intensity. IC50 values were determined by using the AAT Bioquest IC50 calculator tool. p values were calculated using Student’s t test. p values < 0.05 were considered statistically significant.
NGS analysis of the PC-3 and 22Rv1 whole genomes sequences
The reads of the six WGS (3 PC-3 and 3 22Rv1) were aligned to the grch37 reference genome using the bwa-mem54 aligner. The resulting bam files were post-processed according to the GATK best-practices guidelines. Novel variants were called using Mutect2 (v4.1.0) by using CHD1 intact WGS references downloaded from the Sequence Read Archive (SRA, with accession IDs; PC-3: SRX5466646, 22Rv1: SRX5437595) as “normal” and the CHD1 ko clones as “tumor” specimens50. These vcfs were converted into tab-delimited files and further analyzed in R. Annotation was performed via Intervar55.
Supplementary information
Acknowledgements
The authors thank Zita Bratu for technical assistance, Alimamy Bundu and Treissy Soares for FISH probe preparation and testing, Dr. Hua Zou, Audrey Flores and Safaa Khairi for valuable experimental support and Orsolya Pipek and Aimilia Schina for the technical support. This work was supported by the Research and Technology Innovation Fund (KTIA_NAP_13-2014-0021 and NAP2-2017-1.2.1-NKP-0002); Breast Cancer Research Foundation (BCRF-17-156 to Z. Szallasi) and the Novo Nordisk Foundation Interdisciplinary Synergy Program Grant (NNF15OC0016584), Det Fri Forskningsrad (award number #7016-00345B; to Z. Szallasi); Department of Defense through the Prostate Cancer Research Program (award number is W81XWH-18-2-0056; to Z. Szallasi, A. Dobi and M.L.F.); and the National Cancer Institute (P01CA228696, to A. D’Andrea, Z. Szallasi, M.L.F.). Z. Szallasi, Z. Sztupinszki and J.B. were supported by Velux Foundation 00018310 grant. P.S. was supported by the Finnish Cultural Foundation, Sigrid Jusélius Foundation and Instrumentarium Science Foundation. S.K. is supported by the Prostate Cancer Foundation (18YOUN09 and 19CHAL07). This research was also supported by National Institute of Health grant R01CA273696 (S.P.) and U54 pilot project grant to S.P and Z.S. (parent grant: 5U54 CA156734-13 to Macoska and Colon-Carmona, PI). The contents of this publication are the sole responsibility of the author(s) and do not necessarily reflect the views, opinions or policies opinions of Uniformed Services University of the Health Sciences (USUHS), the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., the Department of Defense (DoD) or the Departments of the Army, Navy, or Air Force. Mention of trade names, commercial products, or organizations does not imply endorsement by the U.S. Government.
Author contributions
Conception and design: Miklos Diossy, V. Tisza, H. Li, P. Sahgal, J. Zhou, Zs. Sztupinszki, S. Spisak, G. Valcz, P. V. Nuzzo, D. Ribli, T. Ried, S. Kaochar, K. Rizwan, S. Pathania, A. D’Andrea, I. Csabai, S. Srivastava, A. Dobi, M. L. Freedman, and Z. Szallasi. Development of methodology: M. Diossy, V. Tisza, H. Li, P. Sahgal, J. Zhou, Zs. Sztupinszki, M. Krzystanek, A. Dobi, and Z. Szallasi. TMA analysis: H. Li, D. Young, D. Nousome, C. Kuo, Y. Chen, R. Ebner, I. A. Sesterhenn, Gy. Petrovics, and G. Valcz. Acquisition of data: M. Diossy, V. Tisza, H. Li, P. Sahgal, J. Zhou, Zs. Sztupinszki, D. Young, D. Nousome, C. Kuo, J. Jiang, G. Valcz, and D. Ribli. Cell line experiments: M. Diossy, V. Tisza, P. Sahgal, J. Zhou, G. T. Klus, S. Spisak, T. Ried, and Z. Szallasi. Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): M. Diossy, V. Tisza, H. Li, P. Sahgal, Zs. Sztupinszki, D. Nousome, J. Börcsök, A. Prosz, I. Csabai, S. Srivastava, M. L. Freedman, and Z. Szallasi. Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): M. Diossy, V. Tisza, H. Li, J. T. Moncur, G. T. Chesnut, S. Srivastava, A. Dobi, and Z. Szallasi. Study supervision: A. Dobi, S Spisak, and Z. Szallasi. All authors were involved in the preparation of the manuscript and the Supplementary Materials.
Data availability
Whole exome and whole genome TCGA data presented in this study are available from the GDC (https://portal.gdc.cancer.gov/) and ICGC (https://dcc.icgc.org/) data portals respectively. The whole genomes from the Mayo clinic are available from dbGap (phs001105.v1.p1), while whole genomes from DFCI and CPDR are available upon request.
Code availability
All analysis was done using standard R (v4.1) codes with the help of the following packages: ggplot2, data.table, deconstructSigs, lsa, ggbeeswarm, RColorBrewer, sequenza, copy number, and cluster. In particular, standard variant files were converted to tab-delimited tables using GATK (v3.8) VariantsToTable and manipulated using the data.table package in R. Figures were created using ggplot2. Every tool mentioned in the “Methods” section were used with default parameters unless stated otherwise.
Competing interests
Z. Szallasi is listed as a co-inventor on a patent to quantify homologous recombination deficiency, which is owned by Children’s Hospital Boston and licensed to Myriad Genetics. No potential conflicts of interest were disclosed by the other authors.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Miklos Diossy, Viktoria Tisza, Hua Li.
Contributor Information
Matthew L. Freedman, Email: freedman@broadinstitute.org
Albert Dobi, Email: adobi@cpdr.org.
Sandor Spisak, Email: Spisak.sandor@ttk.hu.
Zoltan Szallasi, Email: Zoltan.Szallasi@childrens.harvard.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41698-024-00705-8.
References
- 1.DeSantis, C. E., Miller, K. D., Goding Sauer, A., Jemal, A. & Siegel, R. L. Cancer statistics for African Americans, 2019. CA Cancer J. Clin.69, 211–233 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Gaines, A. R. et al. The association between race and prostate cancer risk on initial biopsy in an equal access, multiethnic cohort. Cancer Causes Control25, 1029–1035 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu, D. I. et al. Effect of race and socioeconomic status on surgical margins and biochemical outcomes in an equal-access health care setting: results from the Shared Equal Access Regional Cancer Hospital (SEARCH) database. Cancer118, 4999–5007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khani, F. et al. Evidence for molecular differences in prostate cancer between African American and Caucasian men. Clin. Cancer Res.20, 4925–4934 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosen, P. et al. Differences in frequency of ERG oncoprotein expression between index tumors of Caucasian and African American patients with prostate cancer. Urology80, 749–753 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sedarsky, J., Degon, M., Srivastava, S. & Dobi, A. Ethnicity and ERG frequency in prostate cancer. Nat. Rev. Urol.15, 125–131 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Petrovics, G. et al. A novel genomic alteration of LSAMP associates with aggressive prostate cancer in African American men. EBioMedicine2, 1957–1964 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koga, Y. et al. Genomic profiling of prostate cancers from men with African and European ancestry. Clin. Cancer Res.26, 4651–4660 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahal, B. A. et al. Racial differences in genomic profiling of prostate cancer. N. Engl. J. Med.383, 1083–1085 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang, F. W. et al. Exome sequencing of African-American prostate cancer reveals loss-of-function ERF mutations. Cancer Discov.7, 973–983 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faisal, F. A. et al. SPINK1 expression is enriched in African American prostate cancer but is not associated with altered immune infiltration or oncologic outcomes post-prostatectomy. Prostate Cancer Prostatic Dis.22, 552–559 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Burkhardt, L. et al. CHD1 is a 5q21 tumor suppressor required for ERG rearrangement in prostate cancer. Cancer Res.73, 2795–2805 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Augello, M. A. et al. CHD1 loss alters AR binding at lineage-specific enhancers and modulates distinct transcriptional programs to drive prostate tumorigenesis. Cancer Cell35, 817–819 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Zhang, Z. et al. Loss of CHD1 promotes heterogeneous mechanisms of resistance to AR-targeted therapy via chromatin dysregulation. Cancer Cell37, 584–598.e11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou, J. et al. Human CHD1 is required for early DNA-damage signaling and is uniquely regulated by its N terminus. Nucleic Acids Res.46, 3891–3905 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kari, V. et al. Loss of CHD1 causes DNA repair defects and enhances prostate cancer therapeutic responsiveness. EMBO Rep.17, 1609–1623 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hjorth-Jensen, K. et al. SPOP promotes transcriptional expression of DNA repair and replication factors to prevent replication stress and genomic instability. Nucleic Acids Res.46, 9484–9495 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baca, S. C. et al. Punctuated evolution of prostate cancer genomes. Cell153, 666–677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ha, G. et al. TITAN: inference of copy number architectures in clonal cell populations from tumor whole-genome sequence data. Genome Res.24, 1881–1893 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han, M. et al. Biochemical (prostate specific antigen) recurrence probability following radical prostatectomy for clinically localized prostate cancer. J. Urol.169, 517–523 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Yuan, J. et al. Integrative comparison of the genomic and transcriptomic landscape between prostate cancer patients of predominantly African or European genetic ancestry. PLoS Genet.16, e1008641 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oesper, L., Mahmoody, A. & Raphael, B. J. THetA: inferring intra-tumor heterogeneity from high-throughput DNA sequencing data. Genome Biol.14, R80 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cun, Y., Yang, T.-P., Achter, V., Lang, U. & Peifer, M. Copy-number analysis and inference of subclonal populations in cancer genomes using Sclust. Nat. Protoc.13, 1488–1501 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Shenoy, T. R. et al. CHD1 loss sensitizes prostate cancer to DNA damaging therapy by promoting error-prone double-strand break repair. Ann. Oncol.28, 1495–1507 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature500, 415–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alexandrov, L. B. et al. The repertoire of mutational signatures in human cancer. Nature578, 94–101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nik-Zainal, S. et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature534, 47–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Póti, Á. et al. Correlation of homologous recombination deficiency induced mutational signatures with sensitivity to PARP inhibitors and cytotoxic agents. Genome Biol.20, 240 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Telli, M. L. et al. Homologous recombination deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin. Cancer Res.22, 3764–3773 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davies, H. et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat. Med.23, 517–525 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sztupinszki, Z. et al. Detection of molecular signatures of homologous recombination deficiency in prostate cancer with or without BRCA1/2 mutations. Clin. Cancer Res.26, 2673–2680 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, Y. et al. Patterns of somatic structural variation in human cancer genomes. Nature578, 112–121 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zámborszky, J. et al. Loss of BRCA1 or BRCA2 markedly increases the rate of base substitution mutagenesis and has distinct effects on genomic deletions. Oncogene36, 746–755 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murai, J. & Pommier, Y. PARP trapping beyond homologous recombination and platinum sensitivity in cancers. Annu. Rev. Cancer Biol.3, 131–150 (2019). [Google Scholar]
- 35.Barbieri, C. E. et al. The mutational landscape of prostate cancer. Eur. Urol.64, 567–576 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yates, L. R. et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat. Med.21, 751–759 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Linch, M. et al. Intratumoural evolutionary landscape of high-risk prostate cancer: the PROGENY study of genomic and immune parameters. Ann. Oncol.28, 2472–2480 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hewitt, G. et al. Defective ALC1 nucleosome remodeling confers PARPi sensitization and synthetic lethality with HRD. Mol. Cell81, 767–783.e11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verma, P. et al. ALC1 links chromatin accessibility to PARP inhibitor response in homologous recombination-deficient cells. Nat. Cell Biol.23, 160–171 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agarwal, N. et al. Talazoparib plus enzalutamide in men with first-line metastatic castration-resistant prostate cancer (TALAPRO-2): a randomised, placebo-controlled, phase 3 trial. Lancet402, 291–303 (2023). [DOI] [PubMed] [Google Scholar]
- 41.Calagua, C. et al. A subset of localized prostate cancer displays an immunogenic phenotype associated with losses of key tumor suppressor genes. Clin. Cancer Res.27, 4836–4847 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minas, T. Z. et al. Serum proteomics links suppression of tumor immunity to ancestry and lethal prostate cancer. Nat. Commun.13, 1759 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merseburger, A. S. et al. Limitations of tissue microarrays in the evaluation of focal alterations of bcl-2 and p53 in whole mount derived prostate tissues. Oncol. Rep.10, 223–228 (2003). [PubMed] [Google Scholar]
- 44.Furusato, B. et al. ERG oncoprotein expression in prostate cancer: clonal progression of ERG-positive tumor cells and potential for ERG-based stratification. Prostate Cancer Prostatic Dis.13, 228–237 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Decker, B. et al. Biallelic BRCA2 mutations shape the somatic mutational landscape of aggressive prostate tumors. Am. J. Hum. Genet.98, 818–829 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karczewski, K. J. et al. The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic Acids Res.45, D840–D845 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Favero, F. et al. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann. Oncol.26, 64–70 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res.20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosenthal, R., McGranahan, N., Herrero, J., Taylor, B. S. & Swanton, C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol.17, 31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sztupinszki, Z. et al. Migrating the SNP array-based homologous recombination deficiency measures to next generation sequencing data of breast cancer. NPJ Breast Cancer4, 16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Geng, C. et al. SPOP regulates prostate epithelial cell proliferation and promotes ubiquitination and turnover of c-MYC oncoprotein. Oncogene36, 4767–4777 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li, Q. & Wang, K. InterVar: clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am. J. Hum. Genet.100, 267–280 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole exome and whole genome TCGA data presented in this study are available from the GDC (https://portal.gdc.cancer.gov/) and ICGC (https://dcc.icgc.org/) data portals respectively. The whole genomes from the Mayo clinic are available from dbGap (phs001105.v1.p1), while whole genomes from DFCI and CPDR are available upon request.
All analysis was done using standard R (v4.1) codes with the help of the following packages: ggplot2, data.table, deconstructSigs, lsa, ggbeeswarm, RColorBrewer, sequenza, copy number, and cluster. In particular, standard variant files were converted to tab-delimited tables using GATK (v3.8) VariantsToTable and manipulated using the data.table package in R. Figures were created using ggplot2. Every tool mentioned in the “Methods” section were used with default parameters unless stated otherwise.