Abstract
Background: TLR7, the receptor accountable for immune response to RNA viruses, has been studied extensively to identify its variants related to the severity of Covid-19 in different populations worldwide. However, the genotype of Pakistani population is still unknown. This study aimed to determine the TLR7 genotypes and their relation with severity in our population. Methods: This cross sectional study collected data on 151 Covid-19 positive patients (aged 18-80 years), from June 2022 to May 2023, after an informed consent, from Ziauddin University and Hospital. Prior to that approval from ethics review committee was taken. The demographic variables and comorbidities were recorded along with health status till LAMA (Leave Against Medical Advise), recovery or death. The DNA was extracted from collected blood samples, PCR and Sanger sequencing was done for identification of TLR7 variants. SPSS was used for data analyses and Chi-Square for categorical variables. P-values of <0.05 was considered significant. Results: Out of 151 patients’ sequencing was done for 59 samples. The restriction site, rs864058 of TLR7 gene, identified G/A and G/G variants. This missense variant of TLR7 identified at rs864058 of TLR7 gene, has not been previously reported in population control databases. The genotype G/G was main variant of 49 (83%) patients, whereas, G/A was found in 10 (17%). Majority, 25 (51%) of patients with mild covid-19 had GG genotype but results were not significant (P=0.684). Among female patients the main genotype was GA 8 (80%) while male had G/G 29 (59.2%) with significant results (P=0.024). Since G/G genotype was the major genotype, high percentage was found in hypertensives [20 (40.8%)], Diabetics [13 (26.5%)], depression [24 (49%)] and pneumonia patients [20 (40.8%)]. However, significant association (P=0.023) was only found with pneumonia. Males, in majority had severe [17 (68%)] infection and death [40 (26.4%)], whereas, females had mild [14 (25%)] with [12 (7.9%)] deaths. Conclusion: A variant rs864058 “G/A” of TLR7, in relation to covid-19 were found in our population. Males were found more at risk of morbidity and mortality due to covid-19. Larger studies are required to further confirm these results.
Keywords: COVID-19, Toll-like receptors, PCR, polymorphism, single nucleotide, TLR7, mRNA, RNA viruses, risk factors
Introduction
Considering host resistance and susceptibility, an immuno-polymorphism database was set up to study genetic aspects of infectious diseases. Covid 19 necessitated the need to explore the role of Toll-like receptors (TLRs) especially TLR7 which plays the main role in inducing immune response against RNA Viruses. Thus TLR7 protein and its regulators plays a fundamental role in pathogen recognition and activation of innate immunity through direct and indirect signaling, regulating cell mediated immunity [1]. TLR7 encodes for an endosomal innate immune sensor which detects single-stranded RNA (ssRNA) of the virus, including covid 19 (SARS-CoV-2) and gets activated by viral products, generating an immune response by stimulating the production of tumor necrosis factors (TNF) and interleukin-12 (IL12) cytokines and activation of NFKB, inducing the production of type 1 IFN and the release of other proinflammatory cytokines [2,3].
The gene for TLR7 is approximately 23 kb present on the X chromosome and has 3 exons. The codon for initiation, “methionine” is present on exon 2, whereas, exon 3, encodes other TLR7 proteins [4]. Chuang et al. analyzed the TLR7 sequence and found that it is a type I transmembrane protein, a signal peptide made of 1,049 amino acid with, multiple leucine-rich repeats (LRRs) and a cysteine-rich region. Predominantly TLR7 has been detected in lung, placenta, brain, spleen, small intestine, stomach and spleen [5].
Polymorphism even in a single nucleotide in the TLR7 gene can result in mutation in the TLR7 receptor which may not provide the required immune response, has been reported by various studies. A significant association of the polymorphisms ‘T/T’ genotypes and the ‘T’ alleles of TLR7 at rs179008 was found with Covid-19 pneumonia [6]. Whereas, another study found GG genotype of the TLR7 rs3853839, a genetic risk factor for Covid-19 infection, severe illness and poor clinical outcome [7].
These studies show that TLR7 genes due to allelic polymorphisms, have genetic variations which results in several immune-pathological consequences in the form of various infections due to RNA viruses [2]. A number of studies reported associations of genetic polymorphism in the TLR7 receptor genes which resulted in untoward morbidity and mortality. The findings, however, are controversial in various infectious diseases depending also on the size of patients and controls studied. This could also be related to differences in the ethnicity and genetic variations of the populations [6]. Thus, we designed this study of TLR7 gene polymorphisms hypothesizing that it will enable us to find out important clues on the susceptibility and clinical outcomes of covid 19 infection in our population. Therefore, it was required to study and investigate the variations in the TLR7 genes and find out its association with Covid 19 in our setting. The purpose of this study was to find out polymorphism in the TLR7 gene and to investigate the function of the TLR7 variant as a consequence of this polymorphism in the targeted Pakistani population.
Methodology
Sampling
This cross-sectional analytical study was conducted by recruiting 151 COVID-19 PCR-positive patients using Convenient sampling from OPD, wards, and ICU at Ziauddin Hospital from June 2022 to May 2023. Patients Prior to collection approval from the Ziauddin University Ethics Review Committee (Ref code: 5360522BKBC), was taken.
Inclusion criteria: All PCR positive adult patients who were treated by the Pulmonologist at Ziauddin Hospital Clifton and those patients who already had COVID-19.
Exclusion criteria: Patient with post chemotherapy and radiotherapy, or any malignant condition.
The participants comprised both males and females, aged between 18-80. Informed consent was taken from the Patient. All Parameters, including demographic variables such as age, weight, BMI, socioeconomic status, laboratory investigations, past medical history, family history, etc. were recorded through a questionnaire. The patients were classified into mild, moderate and severe by the Pulmonologist according to international criteria of SpO2<94%, PaO2/FiO2<300 mm Hg, suffocation or a respiratory rate >30 breaths/min, or lung infiltrates >50%. The Patients were followed up for their clinical condition in terms of other co-morbidities and health status, till recovery, death or LAMA (leave against medical advice).
DNA extraction and PCR
Blood samples collected from 82 patients were stored at 4°C. DNA was extracted from the whole blood and the TLR7 gene was amplified by PCR using primers forward 5’TGGGCTCAAATCTTTCAGTTG3’, Reverse 5’GATCACACTTTGGCCCTTGT3’. Amplification products were observed using 1% agarose gel electrophoresis.
Sequencing and alignment
Sanger’s sequencing was performed at the Lab. Genetic Lahore, Pakistan for identification of polymorphic sites. Samples sent for sequencing were 59 which included 29 mild, 12 moderate and 18 severe cases. For alignment and trimming of sequences, MEGA X was used.
Statistical analyses were carried out using SPSS version 21. Frequency and percentages were used to present qualitative data, whereas, mean and standard deviation were used to present quantitative data. The chi-Square test was applied to categorical variables. P-values of <0.05 were considered significant... SPSS version 21 was used for statistical analysis.
Results
Covid 19 infection’s among patients
Out of total 151 Covid 19 patients, the majority of cases in males had severe [17 (68%)] infection, compared to females [13 (23%)]. Whereas, females mostly suffered a mild [14 (25%)] infection (Figure 1). It was found that from total 151 patients, 82 (54.3%) recovered, 52 (34.4%) passed away and 17 (11.2%) were LAMA (Leave Against Medical Advice). Deaths in males 40 (26.4%), were higher, compared to females 12 (7.9%).
Figure 1.
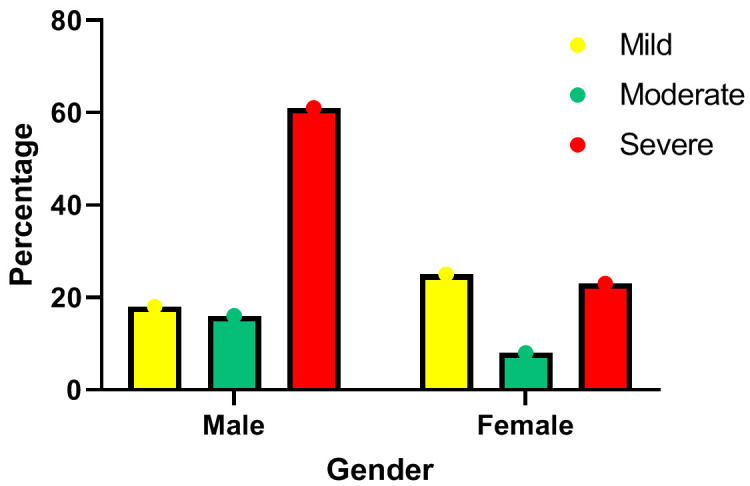
Distribution of severity of COVID among males and females.
Sequence analysis of TLR7 gene
Further the recovered patients were followed and sequence analysis of 59 samples were studied thouroghly for multiple mutations reported worldwide for their extreme virulence. Our samples showed two synonymous mutations, GA and GG, at restriction site rs864058. The GG genotype was the major genotype in 49 samples whereas, GA was in 10 samples (Figure 2).
Figure 2.
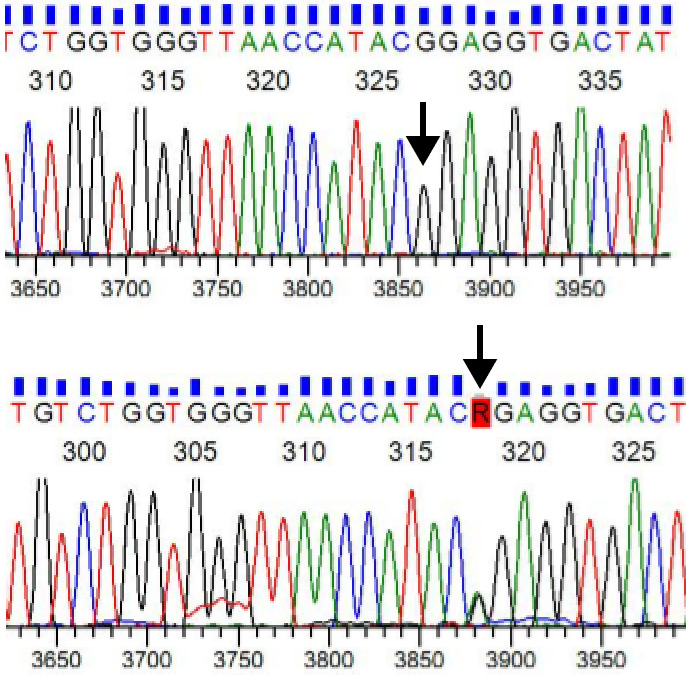
Electropherogram showing heterozygous (G/A) and homozygous (G/G) genotype at the restriction site rs864058.
The polymorphism of restriction sites rs200553089 (G/T) and rs3853839 (C/G) were not found in any sample. The majority of patients with mild COVID had TLR7 genotype GG 25 (51%) (P=0.684). However, in GA genotype 40% of patients had mild levels of disease symptoms and 60% had moderate to severe levels of disease status (Table 1). It can be stated that patients who had Genotype GA experienced relatively moderate to severe levels of disease symptoms compared to GG genotype patients. The variant GG was seen approximately the same in each age group while younger age patients were in higher proportion, 4 (40%) in GA group (P-value =0.903). The majority of patients in GA group were females 8 (80%) while in GG group were males 29 (59.2%) with significant results (P=0.024). In ethnicity, Punjabi and Sindhi had GG genotypes 8 (16.3%), 7 (14.3%). Vaccinated patients [41 (83.7%)] had GG genotype (Table 1).
Table 1.
Association of demographic characteristics with TLR7 genotype in covid-19 patients
| Study variables | TLR7 Genotype (n=59) | Total | P value | ||
|---|---|---|---|---|---|
|
| |||||
| GA (n=10) | GG (n=49) | ||||
| Severity of COVID | Mild | 4 (40%) | 25 (51%) | 29 (49.2%) | 0.684 |
| Moderate | 3 (30%) | 9 (18.4%) | 12 (20.3%) | ||
| Severe | 3 (30%) | 15 (30.6%) | 18 (30.5%) | ||
| Age groups | <25 | 4 (40%) | 16 (32.7%) | 20 (33.9%) | 0.903 |
| 25-50 | 3 (30%) | 16 (32.7%) | 19 (32.2%) | ||
| >50 | 3 (30%) | 17 (34.7%) | 20 (33.9%) | ||
| Gender | Female | 8 (80%) | 20 (40.8%) | 28 (47.5%) | 0.024* |
| Male | 2 (20%) | 29 (59.2%) | 31 (52.5%) | ||
| Ethnicity | Others | 0 (0%) | 2 (4.1%) | 2 (3.4%) | 0.708 |
| Pathan | 1 (10%) | 1 (2%) | 2 (3.4%) | ||
| Punjabi | 2 (20%) | 8 (16.3%) | 10 (16.9%) | ||
| Sindhi | 1 (10%) | 7 (14.3%) | 8 (13.6%) | ||
| Urdu Speaking | 6 (60%) | 31 (63.3%) | 37 (62.7%) | ||
| Occupation | Employed | 4 (40%) | 25 (51%) | 29 (49.2%) | 0.815 |
| House Wife | 1 (10%) | 6 (12.2%) | 7 (11.9%) | ||
| Student | 4 (40%) | 16 (32.7%) | 20 (33.9%) | ||
| Unemployed | 1 (10%) | 2 (4.1%) | 3 (5.1%) | ||
| Vaccination | No | 2 (20%) | 8 (16.3%) | 10 (16.9%) | 0.778 |
| Yes | 8 (80%) | 41 (83.7%) | 49 (83.1%) | ||
| Tobacco History | No | 10 (100%) | 36 (73.5%) | 46 (78%) | 0.065 |
| Yes | 0 (0%) | 13 (26.5%) | 13 (22%) | ||
Chi square test applied; Significance level set at 0.05.
Significant.
Clinical evaluation of genotypes of TLR7
In clinical features, there was found no significant mean difference between temperature, pulse rate, respiratory rate, etc., and genotypes (P≥0.05) (Table 2). Further association of TLR7 Genotype with covid 19 patients’ symptoms was performed and it was found that all the symptoms reported more in GA when compared to the GG genotype patients like Cough 10 (100%), Sore Throat 9 (90%), chest congestion 8 (80%), headache 8 (80%), fatigue and muscle ache or joint pain 10 (100%) were the symptoms (P>0.05). Whereas, other symptoms fatigue or weakness 9 (90%) and rashes 16 (32.7%) were reported as having GG genotype (Table 3).
Table 2.
Comparison of clinical features between TLR7 genotypes
| Study variables | TLR7 genotype GA (n=10) | TLR7 genotype GG (n=49) | P value |
|---|---|---|---|
|
| |||
| Mean ± SD | Mean ± SD | ||
| Temperature | 101.6±1.1 | 101.7±1.2 | 0.655 |
| Systolic Blood Pressure | 130.9±18.9 | 130.7±22.4 | 0.886 |
| Diastolic Blood Pressure | 73.9±9.4 | 77.8±11.7 | 0.296 |
| Pulse rate | 94±13.7 | 86.1±12.8 | 0.052 |
| Respiratory Rate | 20.1±1.7 | 20.1±2.4 | 0.803 |
| Oxygen Saturation % | 90±10.8 | 92.7±6.8 | 0.640 |
Mann Whiteny U test applied.
Table 3.
Association of symptoms with TLR7 genotype in covid-19 patients
| Covid-19 Symptoms | TLR7 Genotype (n=59) | Total | P value | |
|---|---|---|---|---|
|
| ||||
| GA (n=10) | GG (n=49) | |||
| Shortness of breath | 5 (50%) | 23 (46.9%) | 28 (47.5%) | 0.860 |
| Cough | 10 (100%) | 38 (77.6%) | 48 (81.4%) | 0.097 |
| Sore Throat | 8 (80%) | 26 (53.1%) | 34 (57.6%) | 0.116 |
| Chest Congestion | 9 (90%) | 27 (55.1%) | 36 (61%) | 0.039 |
| Chest Pain | 4 (40%) | 19 (38.8%) | 23 (39%) | 0.942 |
| Palpitation | 2 (20%) | 15 (30.6%) | 17 (28.8%) | 0.499 |
| Headache | 8 (80%) | 28 (57.1%) | 36 (61%) | 0.177 |
| Fatigue/Weakness | 9 (90%) | 48 (98%) | 57 (96.6%) | 0.205 |
| Loss of Taste | 6 (60%) | 23 (46.9%) | 29 (49.2%) | 0.451 |
| Loss of Smell | 6 (60%) | 26 (53.1%) | 32 (54.2%) | 0.688 |
| Rashes | 3 (30%) | 16 (32.7%) | 19 (32.2%) | 0.870 |
| Hoarse Voice | 1 (10%) | 7 (14.3%) | 8 (13.6%) | 0.718 |
| Nausea/Vomiting | 5 (50%) | 19 (38.8%) | 24 (40.7%) | 0.510 |
| Diarrhea | 4 (40%) | 20 (40.8%) | 24 (40.7%) | 0.962 |
| Muscle aches/Joint pain | 10 (100%) | 37 (75.5%) | 47 (79.7%) | 0.080 |
Chi square test applied; Significance level set at 0.05.
TLR7 genotypes and co-morbidities
Similarly, the association of TLR7 genotype with co-morbidities was evaluated. The high percentage of GG genotype was reported in patients with hypertension 20 (40.8%), diabetes 13 (26.5%) and pneumonia 20 (40.8%). Only depression 24 (49%) showed the association with genotype (P=0.023). Heart disease had a similar prevalence (10%) in both genotypes. Whereas, COPD and Asthma were seen with high GA genotypes 2 (20%), 4 (40%) respectively (Table 4).
Table 4.
Association of co-morbidities and outcomes with TLR7 genotype in covid-19 patients
| Co-morbidities & Outcomes | TLR7 Genotype (n=59) | Total (n=59) | P value | |
|---|---|---|---|---|
|
| ||||
| GA (n=10) | GG (n=49) | |||
| Hypertension | 3 (30%) | 20 (40.8%) | 23 (39%) | 0.523 |
| Diabetes | 2 (20%) | 13 (26.5%) | 15 (25.4%) | 0.666 |
| Heart Disease | 1 (10%) | 5 (10.2%) | 6 (10.2%) | 0.984 |
| COPD | 2 (20%) | 5 (10.2%) | 7 (11.9%) | 0.383 |
| Asthma | 4 (40%) | 12 (24.5%) | 16 (27.1%) | 0.315 |
| Bronchitis | 0 (0%) | 7 (14.3%) | 7 (11.9%) | 0.203 |
| Pneumonia | 3 (30%) | 20 (40.8%) | 23 (39%) | 0.523 |
| Depression | 1 (10%) | 24 (49%) | 25 (42.4%) | 0.023 |
| Stroke | 0 (0%) | 2 (4.1%) | 2 (3.4%) | 0.516 |
| Dengue | 2 (20%) | 4 (8.2%) | 6 (10.2%) | 0.259 |
Chi square test applied; Significance level set at 0.05.
Discussion
While exploring and investigating the challenges to overcome the covid-19 pandemic, the Toll-like receptor (TLR7) and its gene driving its expression were studied extensively in different populations. We matched our TLR7 sequences at NCBI with MEGA X alignment comparing with the previously reported variants responsible for severe covid-19 worldwide. None of the restriction sites, such as TLR7 rs179008, rs3853839, rs200553089 (G/T) and rs3853839 (C/G) were detected in our samples.
These studies by other researchers showed a significant association between the ‘T/T’ genotype and the ‘T’ allele of TLR7 rs179008 to increased risk of COVID-19 pneumonia, similarly, the TLR7 genetic site rs3853839 and the G allele and GG genotype were significantly associated with COVID-19 cases, whereas, the CC genotype and C allele with healthy volunteers [6,7]. However, polymorphism at these restiction sites were not detected in our patients.
A new TLR7 polymorphism at restriction site rs864058 (GA), was identified in our patients which has not been reported by any other populations in the world for covid-19. This genotype GA and GG in connection with covid-19, is being reported for the first time.
These two variants of TLR7 gene in our population were GG in 49 (59.8%) and GA in 10 (12.2%) patients at rs864058. Previously, this restriction site has been found associated with allergic Rhinitis in Swedish as well as Chinese populations [8] but not with covid 19 disease. A study reported the same restriction site but a different polymorphism, “CT” for allergic Rhinitis. Willie et al. reported GA genotype related to HIV at rs864058 SNP 2403G>A of TLR7 on X chromosome at position 12906030 [9]. Thus this TLR7 gene was found by other other studies, to have a very strong association with other disorders such as asthma, rhinitis, as well as atopic dermatitis but at another restriction site, rs179008 [10].
Although the SNPs studied were found to be silent mutation, differences were found between patients of these genotypes. It can be stated that patients who had Genotype GA experienced relatively more moderate to severe levels of disease symptoms compared to GG genotype patients. The majority of patients in the GA group were females 8 (80%) while in GG group were males 29 (59.2%) with significant results (P=0.024). Symptoms observed showed that more severity in GA when compared to the GG genotype patients like Cough 10 (100%), Sore Throat 9 (90%), chest congestion 8 (80%), headache 8 (80%), fatigue and muscle ache or joint pain 10 (100%) with significant results (P>0.05). A report published in Science has toppled the silent mutation theory. The report states that “silent mutations” can, under certain circumstances, determine the performance of a protein. The researchers hypothesized that this may cause a slight alteration in the three-dimensional shape which could slow down the cell’s protein-making machinery [11].
Researchers around the world have studied many TLR7 SNPs, investigating their associations with various different conditions. In Moroccan patients, rs179008 has been found linked to the progression of hepatic illness due to HCV [12]. In the Finnish population, in children 5 to 7 years of age, it was found associated with insufficiency of post-bronchiolitis lung function deficiency [13]. Similarly, TLR7 rs3853839, has also been found linked to many diseases in different populations. It was found significantly linked to HIV-1 infection in Chinese Han patients as well as HCV [14]. Further, its association with hand, foot, and mouth infections mediated by enterovirus-71 has also been reported [15]. In the Indian population (rs3853839 and rs179008) were found significantly related to Dengue virus infection [16] and to chikungunya virus [17].
Studies around the world have shown a very diverse role of various TLR7 SNP in different infections and pathogenesis in different ethnic groups (Table 5).
Table 5.
Studies showing the role of various TLR7 SNP in viral infection and pathogenesis in different ethnic groups worldwide
| Study | TLR7 Restriction site | Pathogen | Disease | Population |
|---|---|---|---|---|
| Fakhir 2017 [18] | rs179008 | HCV | Mediated hepatic illness | Moroccan patients |
| Lauhkonen et al. 2016 [19] | rs179008 | Respiratory Syncytial Virus (RSV) | Post-bronchiolitis lung function deficiency | Finnish children 5 to 7 yrs of age |
| Mukherjee et al. 2019 [16] | rs3853839 & rs179008 | Dengue virus infection | Significant ssociation with dengue | Indian |
| Zhang et al. 2020 [14] | rs179010 | HIV | HIV-1 infection | Chinese Han |
| Dutta et al. 2017 [17] | rs3853839 | Chikungunya virus | Chikungunya infection | Indians |
| Yue et al. 2014 [15] | rs3853839 | HCV persistence and predisposition | Enterovirus-71-mediated hand, foot and mouth infection | Chinese |
Higher severity and mortality of infections among males compared to females have been observed worldwide, across all age groups [20]. The majority of our severe cases were males [17 (68%)] with more deaths [40 (26.4%)], compared to females [12 (7.9%)]. Many hypotheses regarding advocating underlying causes have been suggested [21], but the exact sex-related clinicopathological reason has yet to be confirmed. However, based on female anatomy some actions can be explained. The functional receptor of SARS-CoV-1 and 2 is influenced by sex hormones and estrogen has a protective role [22]. At the same time, estrogens indirectly control the immune cells which are richly provided by estrogen receptors (ER-alpha and ER-beta) [23]. Estrogen upregulates the pro-inflammatory cytokine such as TNFα, improving the immune system. On the contrary, in males, testosterone upregulates anti-inflammatory cytokines such as interleukin 10, serving as an immune suppressor [24]. This could probably be the reason for cytokine storm in males as the cause of death due to covid 19.
The female genetic makeup, regarding sex chromosome differences, may further explain this gender disparity [25]. The gene of RNA receptor, TLR7, is present on chromosome X, therefore, biological sex has wide-ranging impacts on RNA viruses like HIV, HCV or Covid 19 [26]. Females with XX chromosomes have double copies of key immune genes compared to men who have one X, single copy of the gene, making females more equipped to combat infections.
Regarding co-morbidities in a study of more than 1.3 million PCR-proven COVID-19 cases in the United States and China, they found that individuals with past co-morbidities had a greater rate of hospitalization, ICU admission and mortality than those without previous co-morbidities [27-29]. Comparing genotypes with co-morbidities we found a high percentage of GG genotype was reported in patients with hypertension 20 (40.8%), Diabetes 13 (26.5%) and pneumonia 20 (40.8%). Only in depression, GG genotype 24 (49%) showed a significant association (P=0.023). A study from Kenya of 913 patients in which 80.8% were of African origin showed a high incidence of Diabetes, followed by hypertension co-morbidities in patients infected with COVID-19 [30].
Overall, according to www.worldometers.info [31] the intensity of illness and deaths due to covid-19 in Pakistan were much less (total deaths =30,664) compared to its neighboring countries India (533,570) and Iran (146,811). This proves that the host’s genetic background plays a major role in the intensity of the illness of any disease. GenOMICC investigators found loci 3p21.31, 9q34.2, 12q24.13 and 21q22.1, all with involvement of multiple genes associated with severity of covid 19. The substantial variation could be due to host selection in the past during evolutionary growth [32].
Conclusion
Silent mutation due to synonymous variants of TLR7 at restriction site rs864058 in relation to Covid-19 was found in our targeted population. Males were found more at risk of morbidity and mortality due to Covid-19. Larger studies are required to further confirm these results as well as work on other loci.
Acknowledgements
I ackhnowledge the hospital staff who helped in the collection of data.
Disclosure of conflict of interest
None.
References
- 1.Duan T, Du Y, Xing C, Wang HY, Wang RF. Toll-like receptor signaling and its role in cell-mediated immunity. Front Immunol. 2022;13:812774. doi: 10.3389/fimmu.2022.812774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mukherjee S, Huda S, Sinha Babu SP. Toll-like receptor polymorphism in host immune response to infectious diseases: a review. Scand J Immunol. 2019;90:e12771. doi: 10.1111/sji.12771. [DOI] [PubMed] [Google Scholar]
- 3.Hemmi H, Kaisho T, Takeuchi O, Sato S, Sanjo H, Hoshino K, Horiuchi T, Tomizawa H, Takeda K, Akira S. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 4.Du X, Poltorak A, Wei Y, Beutler B. Three novel mammalian toll-like receptors: gene structure, expression, and evolution. Eur Cytokine Netw. 2000;11:362–71. [PubMed] [Google Scholar]
- 5.Chuang TH, Ulevitch RJ. Cloning and characterization of a sub-family of human toll-like receptors: hTLR7, hTLR8 and hTLR9. Eur Cytokine Netw. 2000;11:372–8. [PubMed] [Google Scholar]
- 6.Alseoudy MM, Elgamal M, Abdelghany DA, Borg AM, El-Mesery A, Elzeiny D, Hammad MO. Prognostic impact of toll-like receptors gene polymorphism on outcome of COVID-19 pneumonia: a case-control study. Clin Immunol. 2022;235:108929. doi: 10.1016/j.clim.2022.108929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El-Hefnawy SM, Eid HA, Mostafa RG, Soliman SS, Omar TA, Azmy RM. COVID-19 susceptibility, severity, clinical outcome and Toll-like receptor (7) mRNA expression driven by TLR7 gene polymorphism (rs3853839) in middle-aged individuals without previous comorbidities. Gene Rep. 2022;27:101612. doi: 10.1016/j.genrep.2022.101612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nilsson D, Andiappan AK, Halldén C, De Yun W, Säll T, Tim CF, Cardell LO. Toll-like receptor gene polymorphisms are associated with allergic rhinitis: a case control study. BMC Med Genet. 2012;13:66. doi: 10.1186/1471-2350-13-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willie B, Hall NB, Stein CM, Jurevic RJ, Weinberg A, Mehlotra RK, Zimmerman PA. Association of Toll-like receptor polymorphisms with HIV status in North Americans. Genes Immun. 2014;15:569–77. doi: 10.1038/gene.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moller-Larsen S, Nyegaard M, Haagerup A, Vestbo J, Kruse TA, Borglum AD. Association analysis identifies TLR7 and TLR8 as novel risk genes in asthma and related disorders. Thorax. 2008;63:1064–1069. doi: 10.1136/thx.2007.094128. [DOI] [PubMed] [Google Scholar]
- 11.Shen X, Song S, Li C, Zhang J. Synonymous mutations in representative yeast genes are mostly strongly non-neutral. Nature. 2022;606:725–731. doi: 10.1038/s41586-022-04823-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fakhir FZ, Lkhider M, Badre W, Alaoui R, Meurs EF, Pineau P, Ezzikouri S, Benjelloun S. Genetic variations in toll-like receptors 7 and 8 modulate natural hepatitis C outcomes and liver disease progression. Liver Int. 2018;38:432–442. doi: 10.1111/liv.13533. [DOI] [PubMed] [Google Scholar]
- 13.Lauhkonen E, Koponen P, Vuononvirta J, Teräsjärvi J, Nuolivirta K, Toikka JO, Helminen M, He Q, Korppi M. Gene polymorphism of toll-like receptors and lung function at five to seven years of age after infant bronchiolitis. PLoS One. 2016;11:e0146526. doi: 10.1371/journal.pone.0146526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang T, Zhu J, Su B, Cao L, Li Z, Wei H, Huang X, Zheng K, Li A, Chen N, Liu L, Xia W, Wu H, He Q. Effects of TLR7 polymorphisms on the susceptibility and progression of HIV-1 infection in Chinese MSM population. Front Immunol. 2020;11:589010. doi: 10.3389/fimmu.2020.589010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yue M, Feng L, Tang SD, Wang JJ, Xue XX, Ding WL, Zhang Y, Deng XZ. Sex-specific association between X-linked Toll-like receptor 7 with the outcomes of hepatitis C virus infection. Gene. 2014;548:244–50. doi: 10.1016/j.gene.2014.07.040. [DOI] [PubMed] [Google Scholar]
- 16.Mukherjee S, Tripathi A. Contribution of Toll like receptor polymorphisms to dengue susceptibility and clinical outcome among eastern Indian patients. Immunobiology. 2019;224:774–785. doi: 10.1016/j.imbio.2019.08.009. [DOI] [PubMed] [Google Scholar]
- 17.Dutta SK, Tripathi A. Association of toll-like receptor polymorphisms with susceptibility to chikungunya virus infection. Virology. 2017;511:207–213. doi: 10.1016/j.virol.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Fakhir FZ, Lkhider M, Badre W, Alaoui R, Meurs EF, Pineau P, Ezzikouri S, Benjelloun S. Genetic variations in toll-like receptors 7 and 8 modulate natural hepatitis C outcomes and liver disease progression. Liver Int. 2018;38:432–442. doi: 10.1111/liv.13533. [DOI] [PubMed] [Google Scholar]
- 19.Lauhkonen E, Koponen P, Vuononvirta J, Teräsjärvi J, Nuolivirta K, Toikka JO, Helminen M, He Q, Korppi M. Gene polymorphism of toll-like receptors and lung function at five to seven years of age after infant bronchiolitis. PLoS One. 2016;11:e0146526. doi: 10.1371/journal.pone.0146526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dudley JP, Lee NT. Disparities in age-specific morbidity and mortality from SARS-CoV-2 in China and the Republic of Korea. Clin Infect Dis. 2020;71:863–865. doi: 10.1093/cid/ciaa354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer J, Jung N, Robinson N, Lehmann C. Sex differences in immune responses to infectious diseases. Infection. 2015;43:399–403. doi: 10.1007/s15010-015-0791-9. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Jerkic M, Slutsky AS, Zhang H. Molecular mechanisms of sex bias differences in COVID-19 mortality. Crit Care. 2020;24:405. doi: 10.1186/s13054-020-03118-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein SL, Marriott I, Fish EN. Sex-based differences in immune function and responses to vaccination. Trans R Soc Trop Med Hyg. 2015;109:9–15. doi: 10.1093/trstmh/tru167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.vom Steeg LG, Klein SL. SeXX matters in infectious disease pathogenesis. PLoS Pathog. 2016;12:e1005374. doi: 10.1371/journal.ppat.1005374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei XY, Xiao YT, Wang J, Chen R, Zhang W, Yang Y, Lv DJ, Qin C, Gu D, Zhang B, Chen WD, Hou JQ, Song NH, Zeng GH, Ren SC. Sex differences in severity and mortality among patients with COVID-19: evidence from pooled literature analysis and insights from integrated bioinformatic analysis. arXiv:200313547. 2020. https://doi.org/10.48550/arXiv.2003.13547.
- 26.Ziegler SM, Beisel C, Sutter K, Griesbeck M, Hildebrandt H, Hagen SH, Dittmer U, Altfeld M. Human pDCs display sex-specific differences in type I interferon subtypes and interferon α/β receptor expression. Eur J Immunol. 2017;47:251–256. doi: 10.1002/eji.201646725. [DOI] [PubMed] [Google Scholar]
- 27.Stokes EK, Zambrano LD, Anderson KN, Marder EP, Raz KM, El Burai Felix S, Tie Y, Fullerton KE. Coronavirus disease 2019 case surveillance - United States, January 22-May 30, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:759–765. doi: 10.15585/mmwr.mm6924e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrison SL, Fazio-Eynullayeva E, Lane DA, Underhill P, Lip GYH. Comorbidities associated with mortality in 31,461 adults with COVID-19 in the United States: a federated electronic medical record analysis. PLoS Med. 2020;17:e1003321. doi: 10.1371/journal.pmed.1003321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shah R, Shah J, Kunyiha N, Ali SK, Sayed S, Surani S, Saleh M. Demographic, clinical, and co-morbidity characteristics of COVID-19 patients: a retrospective cohort from a Tertiary Hospital in Kenya. Int J Gen Med. 2022;15:4237–4246. doi: 10.2147/IJGM.S361176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. https://www.worldometers.info/coronavirus/country/pakistan/; https://www.worldometers.info/coronavirus/country/india/; https://www.worldometers.info/coronavirus/country/iran/
- 32.Kwok AJ, Mentzer A, Knight JC. Host genetics and infectious disease: new tools, insights and translational opportunities. Nat Rev Genet. 2021;22:137–153. doi: 10.1038/s41576-020-00297-6. [DOI] [PMC free article] [PubMed] [Google Scholar]


