Abstract
The nucleoside triphosphatase (NTPase)/helicase associated with nonstructural protein 3 of West Nile (WN) virus was purified from cell culture medium harvested from virus-infected Vero cells. The purification procedure included sequential chromatography on Superdex-200 and Reactive Red 120 columns, followed by a concentration step on an Ultrogel hydroxyapatite column. The nature of the purified protein was confirmed by immunoblot analysis using a WN virus-positive antiserum, determination of its NH2 terminus by microsequencing, and a binding assay with 5′-[14C]fluorosulfonylbenzoyladenosine. Under optimized reaction conditions the enzyme catalyzed the hydrolysis of ATP and the unwinding of the DNA duplex with kcat values of 133 and 5.5 × 10−3 s−1, respectively. Characterization of the NTPase activity of the WN virus enzyme revealed that optimum conditions with respect to the Mg2+ requirement and the monovalent salt or polynucleotide response differed from those of other flavivirus NTPases. Initial kinetic studies demonstrated that the inhibition (or activation) of ATPase activity by ribavirin-5′-triphosphate is not directly related to changes in the helicase activity of the enzyme. Further analysis using guanine and O6-benzoylguanine derivatives revealed that the ATPase activity of WN virus NTPase/helicase may be modulated, i.e., increased or reduced, with no effect on the helicase activity of the enzyme. On the other hand the helicase activity could be modulated without changing the ATPase activity. Our observations show that the number of ATP hydrolysis events per unwinding cycle is not a constant value.
West Nile virus (WN virus), a member of the family of Flaviviridae, is a small enveloped single-stranded RNA positive-strand virus. The viral genome encodes a monocistronic polyprotein of 3,430 amino acids that is processed into three structural proteins, protein M, capsid protein C, and glycoprotein E, and seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (10, 11, 52). The processing of the polyprotein is carried out by the host signal peptidase associated with the endoplasmic reticulum and viral proteases. The polyprotein of WN virus and its processing are similar to those of the pestivirus- and hepatitis C virus (HCV)-related viruses (36, 44, 55). Sequence analysis of the nonstructural region of WN virus polyprotein revealed numerous conserved motifs specific for serine proteases, RNA helicase with intrinsic RNA-stimulated nucleoside triphosphatase (NTPase) localized in the NS3 protein, and RNA-directed RNA polymerase associated with the NS5 protein (3, 16, 17). These predictions were partially confirmed by verifying the enzymatic properties of a COOH-terminal segment of NS3 released from a membrane fraction of infected cells by subtilisin (54). Further information about the interactions and functions of the viral proteins was obtained by using synthesized recombined proteins of Flaviviridae or HCV-related viruses (19, 23, 47, 49, 50).
Due to multiple enzymatic and biological activities associated with NS3, this protein appears to be the most promising target for antiviral agents. The protease activity of NS3 is the subject of numerous studies and has been well characterized previously (24, 31). However, despite the importance of enzymes modulating RNA structures in diverse metabolic processes and their critical role in the life cycles of viruses whose genomes are composed of RNA, only limited information on the viral helicases or helicase-like enzymes is available.
Helicases are capable of enzymatically unwinding duplex DNA or RNA structures by disrupting the hydrogen bonds that keep the two strands together (18, 21). The unwinding reaction is accomplished by the hydrolysis of γ-phosphate of nucleotide triphosphate (NTP). Based on sequence comparisons, the viral helicases have been divided into three superfamilies. The WN virus helicase is a member of superfamily II (SFII), which includes helicases from bymovirus, potyvirus, pestivirus, herpesvirus, poxvirus, HCV, and other Flaviviridae (22). All of the helicases contain seven highly conserved amino acid sequences (motifs I to VII) that are located on the surfaces of domains 1 and 2 of the three-domain enzymes. The involvement of the motifs in NTP binding, NTP hydrolysis, and the binding of polynucleotide(s) was well explained by resolving the crystal structures of several enzymes (25, 57). However, these structures did not elucidate the mechanisms coupling ATP hydrolysis to the unwinding reaction. Although numerous studies about the quantification of the interaction of SFII helicases with NTP and polynucleotides were performed, uniform results were not obtained (1, 25, 57). This challenged us to perform a study in which the regulation of WN virus helicase activity was investigated by using compounds that are potent modulators (activators or inhibitors) of NTPase activity. Our data presented here demonstrated a dissociation of both activities of the enzyme. Consequently, the number of the ATP hydrolysis events per unwinding reaction was not a constant value.
MATERIALS AND METHODS
Materials.
Human WN virus-positive antisera were kindly provided by G. Rietdorf (Bernhard-Nocht-Institute, Hamburg, Germany). Oligonucleotide synthesis was performed by M. Schreiber (Bernhard-Nocht-Institute). [γ-33P]ATP (110 TBq/mmol) and [γ-32P]ATP (220 TBq/mmol) were purchased from Hartmann Analytic. 125I-protein A (1.11 GBq/mmol) and 5′-[14C]fluorosulfonylbenzoyladenosine ([14C]FSBA) (1.7 GBq/mmol)were obtained from NEN/Du Pont. All other chemicals were obtained from Sigma.
Cell culture.
Vero E6 cells were cultivated in RPMI 1640 medium containing 10% fetal calf serum (Gibco), 100 μg of ampicillin/ml, and 80 μg of gentamicin/ml. The cells in the log phase of growth were infected with WN virus (ATCC strain VR-82) as described previously (53). The medium used as starting material for the enzyme purification was harvested 5 days postinfection.
Purification of WN virus NTPase/helicase.
Portions of 200 ml of medium were supplemented with protease inhibitors (5 U of aprotinin/ml, 1 mM N-tosyl-l-phenylalanine chloromethyl ketone, 1 mM N-p-tosyl-l-lysine chloromethyl ketone, 10 μg of leupeptin/ml, 5 mM phenylmethylsulfonyl fluoride, 5 mM EDTA, and 5 mM EGTA) and with 0.5% Triton X-100 and concentrated to 6 ml by ultrafiltration on a 30-kDa membrane (Amicon). Aliquots of 2 ml of the concentrated material (approximately 600 mg of protein) were loaded onto a Superdex-200 column (Hi-Load; Amersham Pharmacia Biotech). The column was equilibrated with TGT buffer (20 mM Tris-HCl[pH 7.5], 10% Glycerol, 0.05% Triton X-100, 1 mM EDTA, 1 mM β-mercaptoethanol) and calibrated with reference proteins as described in the legend to Fig. 2A. The fractions (fractions 10, 11, 14, and 15; each 3.2 ml) containing the ATPase and helicase activities were pooled and directly applied to a Reactive Red 120 column (12-ml bed volume, 0.8 cm in diameter). The column was washed with TGT buffer and developed with a discontinuous KCl gradient (0 to 1 M). Fractions 16 to 20 were pooled, concentrated 20-fold by ultrafiltration, diluted with 10 volumes of TGT buffer, and chromatographed again on the Reactive Red 120 column. Pooled fractions with the ATPase and helicase activities (fractions 16 to 20) were applied to a hydroxyapatite (HA-Ultrogel) column (2-ml bed volume, 0.8 cm in diameter) preequilibrated with TGT buffer. The loaded column was washed with 10 ml of TGT buffer, and the bound protein was eluted with 1 ml of 50 mM KH2PO4 in the same buffer.
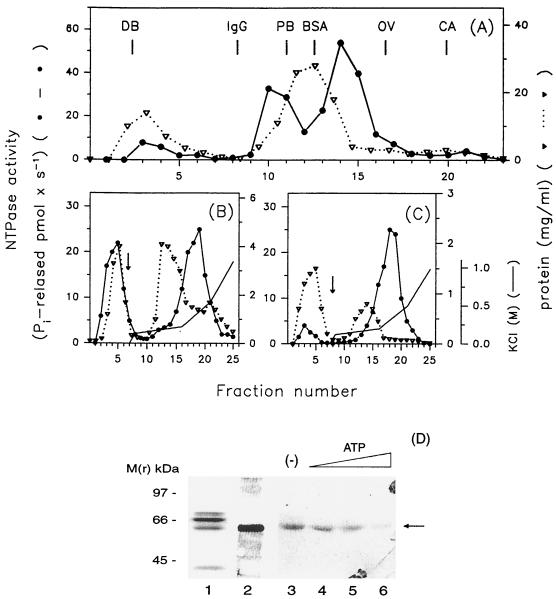
FIG. 2.
Elution profiles of ATPase activity of WN virus NTPase/helicase from consecutive chromatographic columns and analysis of the purified protein obtained from the last purification step by SDS-PAGE. Aliquots (20 μl) of the tested fractions were assayed for NTPase activity and protein content as described in Materials and Methods. (A) Superdex-200 column chromatography of the concentrated medium harvested from an infected-cell culture. The column was calibrated with dextran blue (DB; 2,000 kDa) and with the following marker proteins: immunoglobulin G (IgG; 160 kDa), phosphorylase b (PB; 97 kDa), BSA (66 kDa), ovalbumin (OV; 45 kDa), and carbonic anhydrase (CA; 30 kDa). (B) Reactive Red 120 chromatography of pooled Superdex-200 fractions 10, 11, 14, and 15. (C) Reactive Red 120 rechromatography of pooled fractions 16 to 20 obtained from panel B. Arrows (B and C), start of the salt gradients. (D) Aliquots of the final enzyme preparation (10 μg of protein) were separated by SDS-PAGE and visualized by staining with Coomassie blue (lane 1) or transferred onto nitrocellulose and immunoblotted with the NS3/HEL(1&2) antibody followed by incubation with rabbit anti-human antiserum and 125I-protein A (lane 2). The blot was exposed for 12 h. Aliquots (20 μg) of the final enzyme preparation were incubated with [14C]FSBA in the absence (lane 3) or presence of ATP added at 10 μM (lane 4), 100 μM (lane 5), or 1 mM (lane 6). The samples were subjected to SDS-PAGE, and the gel was dried and exposed for 4 days. The molecular masses of the investigated proteins were estimated by reference to protein standards (left). Arrow, position of the WN virus NTPase/helicase.
ATPase and helicase assays.
The ATPase activity of the WN virus NTPase/helicase was determined under conditions described previously (6, 7) with slight modifications. Briefly, the assay was performed with 2 pmol of WN virus NTPase/helicase incubated in a reaction mixture (final volume, 25 μl) that contained 20 mM Tris-HCl, pH 7.5, 2 mM MgCl2, 1 mM β-mercaptoethanol, 10% glycerol, 0.01% Triton X-100, 0.1 mg of bovine serum albumin (BSA)/ml, and 9.5 μM [γ-33P]ATP (0.025 μCi). The reaction proceeded for 30 min at 30°C and was terminated by the addition of 0.5 ml of activated charcoal (2 mg/ml). After centrifugation at 10,000 × g for 10 min, aliquots (100 μl) of the supernatant were subjected to scintillation counting.
The substrate for the helicase assays was obtained by annealing the two complementary DNA oligonucleotides, which were synthesized with a sequence corresponding to the deoxynucleotide version of the RNA strands described previously (15). The release strand (26-mer) with sequence 5′-CAAACTCTCTCTCTCTCAACAAAAAA-3′ was 5′ end labeled with [γ-32P]ATP by using T4 polynucleotide kinase (MBI; Fermentas) as recommended by the manufacturer. For the annealing reaction the labeled oligonucleotide was combined at a molar ratio of 1:10 with the template strand (40-mer) with sequence 5′-AGAGAGAGAGGTTGAGAGAGAGAGAGTTTGAGAGAGAGAG-3′, denatured for 5 min at 96°C, and slowly renatured as described previously (15). The duplex DNA was electrophoresed on a 15% native Tris-borate-EDTA (TBE)-polyacrylamide gel, visualized by autoradiography, and extracted as described previously (46). The helicase activity, unless otherwise specified, was tested with 2 pmol of enzyme incubated in reaction mixture (final volume, 25 μl) containing 20 mM Tris-HCl, pH 7.5, 2 mM MgCl2, 1 mM β-mercaptoethanol, 10% glycerol, 0.01% Triton X-100, 0.1 mg of BSA/ml, 9.5 μM ATP, and substrate at concentrations indicated in the figure legends. The reaction proceeded for 30 min at 30°C and was stopped by the addition of 5 μl of termination buffer (100 μM Tris-HCl [pH 7.5], 20 mM EDTA, 0.5% sodium dodecyl sulfate (SDS), 0.1% Triton X-100, 25% glycerol, 0.1% bromophenol blue, 0.1% xylene cyanol). The samples were separated on a 15% polyacrylamide–TBE gel containing 0.1% SDS (30). The gels were dried and exposed to Kodak X-ray films at −70°C. Subsequently, the parts of the gels corresponding to the released strand and to the not-unwound substrate were cut out and 32P radioactivity was measured. Alternatively, the films were scanned and the radioactivity associated with the released strand and with the not-unwound substrate was quantified with Gellmage software (Amersham Pharmacia Biotech). The efficacy of the helicase reaction was calculated as a total amount of nucleotide bases of the unwound substrate. Kinetic parameters were determined by nonlinear regression analysis using ENZFITTER (BioSoft) and SIGMA PLOT (Jandel Corp.).
Affinity labeling with [14C]FSBA.
The labeling of ATP-binding site of the WN virus NTPase/helicase with [14C]FSBA was carried out according to the Woodford and Pardee method (56) with modifications described previously (6). The labeling reaction was terminated by addition of SDS sample buffer and boiling. The proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE), and the gels were dried and exposed to Kodak BioMax MR film supplemented by an intensifying screen (BioMax TranScreen). In control experiments the incubation was performed in the presence of ATP as indicated in the legend to Fig. 2. The same reaction was performed to monitor the inactivation of the NTPase and helicase activities of the enzyme by FSBA. Aliquots of the enzyme preparation obtained from each purification step corresponding to 200 μg of protein were incubated with the nucleoside in the absence or presence of ATP added to the reaction mixture in a 10-fold excess. The nucleoside not incorporated was removed by dialysis or gel filtration on the Superdex-200 column, and the enzymatic activities of the protein sample were investigated as described above. The incorporation of the [14C]FSBA was monitored by autoradiography of SDS-PAGE-separated proteins.
Synthesis of nucleosides.
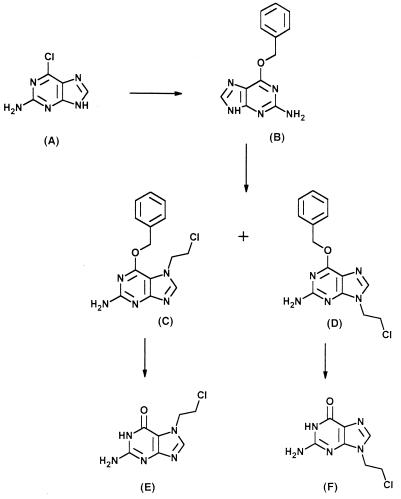
Ribavirin-5′-triphosphate (ribavirin-TP) was synthesized according to the method of Ludwig, Mishra and Broom, and Yoshikawa et al. (33, 37, 58) with some modifications (7). Guanine derivatives were synthesized according to methods reported previously. Briefly, O6-benzylguanine (Fig. 6B) was prepared from 6-chloro-2-aminopurine (Fig. 6A) by the method of Bowles et al. (9). N-alkylation of O6-benzylguanine gave a mixture of N7- and N9-substituted guanine derivatives (Fig. 6C and D) in almost quantitative yield. The mixture of the regioisomers was separated on a silica gel column by flash chromatography to give individual isomers. These isomers were hydrolyzed in aqueous hydrochloric acid to give the corresponding N7- and N9-chloroethylguanines (Fig. 6E and F) in high yield. The UV and nuclear magnetic resonance spectra were identical to those reported by Ramzaeva et al. (42). The synthetic procedure is schematically presented in Fig. 6. 5-Fluoro-2-selenocytosine was synthesized as described previously (14). All compounds were dissolved in water.
FIG. 6.
Structures of guanine and O6-benzylguanine derivatives used in this study. The synthesis and purification of the compounds were performed according to procedures presented in Materials and Methods.
Affinity purification of antibody.
The sequence encoding a fragment of NS3 of HCV consisting of amino acid residues 1189 to 1525 (corresponding to domains I and II of the HCV NTPase/helicase [25]) was expressed downstream of the glutathione S-transferase gene in Escherichia coli (4). The polypeptide was purified on a glutathione-Sepharose 4B column and separated from the fusion protein by limited proteolysis with thrombin. The NS3 fragment was immobilized on the CNBr-activated Sepharose 4B (Sigma) as recommended by manufacturer and used for purification of the affinity antibody [NS3/HEL(1&2)] from the pooled human WN virus-positive antiserum. The purification was performed according to methods reported previously (20). The antibody recognized NTPases/helicases of Flaviviridae in immunoblots using the respective proteins produced in E. coli (data not shown).
Immunoblotting.
Cells or cell culture medium was precipitated with cold trichloroacetic acid (TCA) (20%), washed with TCA and twice with acetone, and dissolved in SDS sample buffer (27). After SDS-PAGE the proteins were transferred on nitrocellulose filters (BA 85; 0.45-μm pore size; Schleicher & Schüll) (48). The filters were incubated for 1 h with 1 mg of BSA/ml in 25 mM Tris-HCl, pH 7.5– 150 mM NaCl–0.05% Tween 80 (TTBS buffer) and for 2 h with NS3/HEL(1&2) antiserum (1:500 in TTBS containing 10% glycerol and 3 mg of BSA/ml). The filters were washed again with 1 mg of BSA/ml in TTBS, and the bound antibody was detected with rabbit anti-human antibodies followed by 125I-protein A. The nitrocellulose filters were dried and subjected to autoradiography.
Other assays.
For end sequencing, the 60-kDa protein was purified as described above. The final enzyme preparation was subjected to SDS-PAGE and electroblotted onto an Immobilon-PSQ polyvinylidene difluoride membrane (Millipore) essentially as described by Matsudaira (34). The NH2-terminal sequencing was performed by F. Buck (University of Hamburg) and by D. McCourt (Midwest Analytical, Inc., St. Louis, Mo.). The amount of the DNA duplex used as the substrate for the WN virus NTPase/helicase was determined by the ethidium bromide fluorescence quantitation method described previously (43). The protein concentration was measured by the method of Lowry et al. (32).
RESULTS
Source of WN virus NTPase/helicase.
The investigations presented here were carried out with an enzyme produced by WN virus-infected Vero E6 cells. For the detection of the protein we used an antibody obtained from WN virus-seropositive individuals purified on an affinity column containing immobilized domains 1 and 2 of the related HCV NTPase/helicase [NS3/HEL(1&2)].
An immunoblot analysis of the total cellular proteins of WN virus-infected cells performed with this antibody revealed three polypeptides with molecular masses of 80, 60, and 47 kDa and several minor protein bands (Fig. 1, lane 2). In contrast, when the proteins of the cell culture medium were immunoblotted, the NS3/HEL(1&2) antibody recognized only the 60-kDa protein. Analyses of the medium aliquots removed after various times of cultivation revealed considerable enrichment of the protein in the course of the infection (Fig. 1, lanes 4 to 6). This accumulation of the 60-kDa protein was accompanied by an increase of ATPase activity in the investigated samples. The cell culture medium collected 5 days after the infection was therefore used as starting material for the purification of the NTPase/helicase.
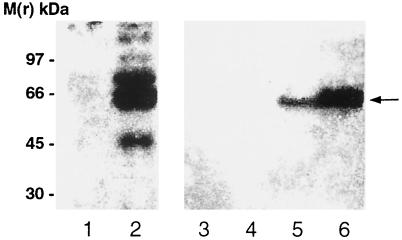
FIG. 1.
Analysis of SFII and WN virus immunoreactivity of cells and cell culture medium infected with WN virus. Vero E6 cells were infected with WN virus as described in Materials and Methods. Five days after the infection the cells were precipitated with TCA and an aliquot (10 μg) of the collected proteins was subjected to SDS-PAGE. The separated proteins were transferred onto a nitrocellulose filter and reacted with NS3/HEL(1&2) antibody. Simultaneously, portions of the cell culture medium were removed immediately (lane 4), 2 days (lane 5), or 5 days (lane 6) after infection. The proteins were precipitated with TCA, and aliquots (50 μg of protein) were separated by SDS-PAGE followed by immunoblotting with NS3/HEL(1&2) antibody. Immunoblots were prepared using uninfected cells (lane 1), infected cells (lane 2), and cell culture supernatant from uninfected cells (lane 3). The nitrocellulose filters were autoradiographed for 14 h. Molecular mass markers are indicated at the left. Arrow, position of WN virus NTPase/helicase.
When the NS3/HEL(1&2) antibody was used, no immunoreactivity was detected in the materials obtained from the uninfected-cell culture (Fig. 1, lanes 1 and 3).
Purification protocol.
The medium was supplemented with Triton X-100 at 0.5%, extensively concentrated, and subjected to size exclusion chromatography on Superdex-200. The fractions obtained were tested for ATPase activity in the standard assay described in Materials and Methods. As shown in Fig. 2A the activity migrated in three peaks: the first protein peak eluted at the void volume of the column (fractions 3 to 5), the second contained proteins with molecular masses of 120 to 110 kDa (fractions 10 and 11), and the third corresponded to proteins migrating at 65 to 55 kDa (fractions 14 and 15) (Fig. 2A). In all these fractions containing ATPase activity antibody NS3/HEL(1&2) recognized a single protein with a molecular mass of 60 kDa (data not shown).
Pooled fractions 10, 11, 14, and 15 were subjected to chromatography on a Reactive Red 120 column. The column was intentionally overloaded so that the ATPase activity migrated in two approximately equal peaks: the first one was found in not-retained fractions, and the second eluted with 0.2 to 0.5 M KCl (fractions 16 to 20) (Fig. 2B). The fractions eluted with salt containing ATPase activity were concentrated, desalted, and rechromatographed on the Reactive Red column. Fractions containing ATPase activity (fractions 16 to 20) (Fig. 2C) were pooled, applied to a HA-Ultrogel column, and, after being washed, eluted with 50 mM KH2PO4. The final enzyme preparation eluted with the phosphate was used in the study. When the fractions obtained during the purification procedure were monitored in the helicase assay the same profile of distribution of activity was observed. The analysis of the proteins present in the final preparation of the enzyme in the Coomassie blue-stained SDS-polyacrylamide gel revealed four proteins with molecular masses of 75, 66, 60, and 40 kDa (Fig. 2D, lane 1). Of these proteins only the one with a molecular mass of 60 kDa was recognized by the NS3/HEL(1&2) antibody (Fig. 2D, lane 2). The densitometric scan of the stained gel revealed that the 60-kDa polypeptide constitutes 25% of the total protein. The purification protocol of the enzyme is summarized in Table 1. The NTPase/helicase nature of the 60-kDa protein was further confirmed by NH2-terminal sequencing. The amino acid sequences, Xaa-Xaa-Leu-Asp-Pro-Tyr and Asp-Pro-Tyr-Trp, that were found within NS3 protein of WN virus defined the 60-kDa protein as the COOH-terminal part of NS3.
TABLE 1.
Purification of the NTPase/helicase from WN virus-infected cell culture mediuma
| Step | Amt. of protein (mg) | Sp. act (nmol/s/mg) | Purification (fold) |
|---|---|---|---|
| Cell culture medium | 600 | 0.0032 | |
| Superdex-200 | 110 | 2.19 | |
| Reactive Red 120 (I) | 15 | 121.22 | 1 |
| Reactive Red 120 (II) | 1.4 | 1,914.3 | 15.8 |
| HA Ultrogel | 0.8 | 2,680 | 22.1 |
The estimation of the ATPase activity was performed under conditions described in Materials and Methods. The NTPase activity and recovery of the cell culture medium, Superdex-200, and Reactive Red 120 (I) steps could not be estimated because of the presence of other enzymes hydrolyzing ATP and because not-whole WN virus NTPase/helicase-mediated ATPase activity obtained in a given step was used as the starting material in the next purification step. Consequently, the salt-eluted activity obtained from chromatography on Reactive Red 120 (I) was more accurate for the estimation of the fold purification. Thus, the purification factors shown are significantly underestimated.
Next, we investigated whether the measured NTPase and helicase activities were specific for the WN virus NS3 protein. To do this, we tested if the antibodies reacting with the 60-kDa protein in the immunoblot were capable of inhibiting the enzymatic activities associated with the enzyme. The assays were performed with the NS3/HEL(1&2) antibody and WN virus- or HCV-positive human sera (diluted 1:10). All antibodies used completely inhibited the NTPase and helicase activities, while WN virus- and HCV-negative sera failed to affect these enzymatic activites.
Previous FSBA-binding studies using HCV NTPase/helicase or its proteolytically generated ATP-binding domain revealed the specific covalent binding of the nucleotide (4, 6). Therefore, aliquots of the final enzyme preparation were labeled in a reaction with [14C]FSBA in the presence or absence of ATP. The autoradiography of the samples separated by SDS-PAGE showed only one 14C-labeled 60-kDa protein. The binding was specific, since the ATP added reduced the 14C labeling in a concentration-dependent manner (Fig. 2D, lanes 3 to 6). As a consequence of the FSBA binding, the NTPase and helicase activities of the enzyme disappeared. The loss of the activity was irreversible and could not be restored by removing the unbound nucleoside. The inactivating effect did not appear when the FSBA-binding reaction was performed in the presence of ATP. Therefore, it was assured that the 60-kDa FSBA-binding protein with immunoreactivity to NS3 and WN virus is the only NTPase/helicase present in the enzyme preparation.
NTPase and helicase activities of the enzyme.
The ATPase activity was monitored in a standard assay by determination of the 33Pi released from [γ-33P]ATP due to the enzyme-mediated hydrolysis. The experimental curve of the Mg2+ requirement was relatively flat (150% of the control at an optimum Mg2+ concentration of 1 to 3 mM); higher concentrations (>10 mM) of Mg2+ were inhibitory. In the presence of 2 mM Mg2+ the apparent kcat for ATP was 133 mol of ATP hydrolyzed/s/mol of enzyme. At ATP concentrations up to 85 μM the Lineweaver-Burk plot was linear and yielded an apparent Km of 9.5 μM. At higher ATP concentrations (>0.5 mM), however, reduction of the ATPase activity rather than a saturable experimental curve was observed (Fig. 3A). The deviation from Michaelis-Menten behavior suggested substrate or product inhibition of the reaction. When the ATPase activity was determined as a function of increasing concentrations of ADP, inhibition of the reaction was observed. The extent of the inhibition was dependent on the ATP concentration in the reaction mixture. When the graphical method described by Dixon (reciprocal velocity of the reaction versus inhibitor concentration) (12) was used, the analysis of the kinetic parameters revealed a competitive inhibition modus, thus indicating that inhibition by the product occurred (Fig. 3B).
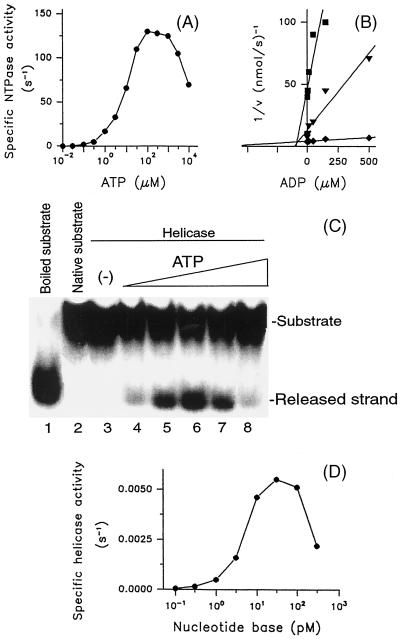
FIG. 3.
Substrate and product inhibition of the NTPase and helicase reactions mediated by WN virus NTPase/helicase. (A) The ATPase reaction as a function of increasing concentrations of ATP was investigated as shown. The product of the reaction (33Pi) was quantified using the charcoal adsorption method described in Materials and Methods. (B) The plots, performed as described by Dixon, demonstrate the competitive type of inhibition of the ATPase activity by ADP. The reaction was carried out in the presence of ATP adjusted to concentrations equal to 1/10 (▪), 1 (▾), or 10 (⧫) times of the Km value (9.5 μM) and the indicated ADP concentrations. (C) The strand-displacing activity of the WN virus NTPase/helicase was determined with a 4.7 pM concentration of the nucleotide bases of the DNA duplex as the substrate in the absence (lane 3) or presence of ATP adjusted to 0.95 μ (lane 4), 9.5 μM (lane 5), 95 μM (lane 6), 950 μM (lane 7), and 9.5 mM (lane 8). Lanes 1 and 2, boiled and native substrates, respectively. The samples were separated in a TBE-polyacrylamide gel, and the levels of 32P radioactivity associated with the substrate and the released strand were visualized by exposition of the dried gels for 10 h. (D) The helicase activity of the WN virus NTPase/helicase as a function of increasing concentrations of the DNA duplex as the substrate was determined at the saturating ATP concentration (90 μM). The samples were separated in a TBE-polyacrylamide gel, and the levels of 32P radioactivity associated with the substrate and the released strand were quantified as described in Materials and Methods. The data obtained were presented as the sum of the nucleotide bases of the unwound DNA duplex. The results shown are representative of three independent experiments.
The influence of monovalent cations Na+ and K+ on the ATPase activity was tested. Both ions influenced the ATPase activity in a similar manner: added at 100 to 300 mM they activated the enzyme (200 to 220% of the control), and added at higher concentrations (>500 mM) they were inhibitory (data not shown).
In order to monitor and characterize the helicase reaction mediated by the enzyme, we prepared a substrate consisting of two annealed DNA oligonucleotides as described in Materials and Methods. The unpairing activity was demonstrated by the release of the shorter labeled strand of the DNA duplex (Fig. 3C). The enzyme required ATP and Mg2+ for the strand-displacing reaction. The maximum velocity of the reaction was observed at 0.3 to 5 mM Mg2+ (data not shown). Analogous to the ATPase reaction, an optimum value for the ATP concentration (90 μM) rather than a saturation curve was obtained (Fig. 3C). At optimum Mg2+ and saturating ATP concentrations 1 pmol of the enzyme unwound 5.5 fmol (given as nucleotide bases) of the DNA duplex per s. The helicase activity reached maximum at a 30 pM concentration of nucleotide bases and yielded a saturable curve at concentrations of up to 120 pM. At higher concentrations of the substrate the reaction declined (Fig. 3D). However, as estimated in a separate helicase assay, the number of single-stranded oligonucleotides that are produced in the course of the helicase reaction was not inhibitory. Thus, for the DNA duplex the deviation from the Michaelis-Menten model resulted from substrate inhibition. When the ATP was adjusted to a concentration at which the half-maximal velocity of the unwinding reaction was measured (10 μM) the Km value for the DNA duplex was a 4.7 pM concentration of nucleotide bases.
The helicase activity remained unchanged in the presence of monovalent ions Na+ and K+ up to concentrations of 500 mM; higher concentrations of the salts were inhibitory.
Regulation of the NTPase and helicase activities by polynucleotides.
The effects of various polynucleotides, known as modulators of NTPase activity of SFII helicases, were investigated. The ATPase activity was determined as a function of increasing concentrations of homopolymeric polynucleotides (poly[U], poly[A], poly[G], poly[C], poly[dT], poly[dA], poly[dG], and poly[dC]), RNA, and DNA at ATP concentrations equal to its Km value. In contrast to those for NTPases/helicases described previously (15, 26, 29, 39, 45, 47, 50, 54), the kinetic parameters of our enzyme were only marginally influenced by the tested compounds. The highest activation was observed with poly(dA) (170 to 180% of control in a concentration range of 1.7 to 3.3 mM for nucleotide bases). Other polynucleotides had no effect or were inhibitory (data not shown).
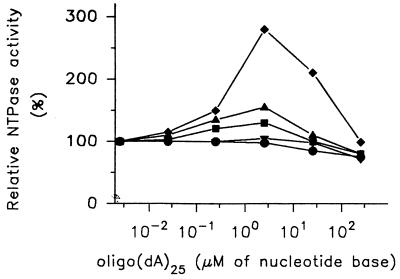
It has been documented that the most-activating effect is exerted by polynucleotides with a chain length greater than 18 to 25 bases (40). In assays performed at standard conditions no significant increase of ATPase activity by synthesized oligo(dA)25 was measured. However, in assays performed at increased ATP concentrations (>100 × Km), oligo(dA)25 exerted an activating effect. The activation reached a maximum at a 2.5 μM concentration of the nucleotide bases of the oligonucleotide and correlated closely to the ATP concentration (Fig. 4). Oligo(dA)25 and other polynucleotides tested did not stimulate the helicase activity and at concentrations higher than 0.5 mM for nucleotide bases inhibited the unwinding reaction (data not shown).
FIG. 4.
Modulation of the ATPase activity of the WN virus NTPase/helicase by oligo(dA)25. The ATPase reaction was investigated as a function of increasing concentrations of oligo(dA)25. The reaction was performed at ATP concentrations equal to 1/100 (ℑ), 1/10 (▾), 1 (▪), 10 (∓), and 100 (⧫) times the Km value (9.5 μM). The ATPase activity measured for each ATP concentration in the absence of oligo(dA)25 was taken as 100%. The results shown are representative of three independent experiments.
Correlation between the NTPase and helicase activities of the WN virus NTPase/helicase.
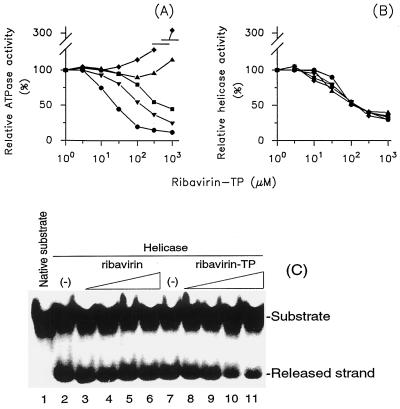
In our previous work we have reported that the reduction of the accessibility of the ATP-binding site of HCV NTPase/helicase for ATP resulted in a decline of ATPase activity of the enzyme (6). Our next interest was to find out whether the reduction of the NTPase activity could lead to an adequate inhibition of unwinding activity. To answer this question, we made use of ribavirin-TP, a competitive inhibitor of the ATPase activity of the related HCV NTPase/helicase (7). Similar to what was found for the HCV enzyme, the susceptibility of the ATPase activity of WN virus NTPase/helicase toward ribavirin-TP was strongly dependent on the ATP concentration. At an ATP concentration equal to Km a half-maximum inhibition (IC50) of the NTPase reaction was observed at 400 μM ribavirin-TP. The reduction of the ATP concentration in the reaction mixture resulted in an amplification of the inhibitory effect. The inhibition was not complete and reached 10 to 15% of control. Graphic analysis of the data obtained revealed that ribavirin-TP acts as a classical competitive inhibitor with regard to ATP (data not shown). On the other hand, when the ATP concentration was enhanced (>100 × Km), a significant increase of the ATPase activity in the presence of ribavirin-TP was measured (Fig. 5A). Interestingly, ribavirin was capable of activating the ATPase activity with similar efficacy, without displaying any inhibitory potential (data not shown).
FIG. 5.
Inhibition of the ATPase and helicase activities of WN virus NTPase/helicase by ribavirin and ribavirin-TP. The ATPase (A) and helicase (B) reactions were performed at ATP concentrations equal to 1/100 (ℑ), 1/10 (▾), 1 (▪), 10 (∓), and 100 (⧫) times of the Km value (9.5 μM) as a function of increasing amounts of ribavirin-TP as the inhibitor. The ATPase activity of the enzyme was determined by the charcoal adsorption method described in Materials and Methods. The helicase activity was assayed at a 4.7 pM concentration of nucleotide bases of the DNA duplex as the substrate. The samples were separated in a TBE-polyacrylamide gel, and the 32P radioactivity associated with the released strand was quantified as described in Materials and Methods. The activity of the enzyme measured for each ATP concentration in the absence of the ribavirin-TP was taken as 100%. (C) Comparison of the inhibitory effects of ribavirin and ribavirin-TP on the helicase activity of WN virus NTPase/helicase. The reaction took place in the absence (lanes 2 and 7) or presence of ribavirin (lanes 3 to 6) or ribavirin-TP (lanes 8 to 11). The concentrations of both compounds were adjusted to 5 μM (lanes 3 and 8), 50 μM (lanes 4 and 9), 500 μM (lanes 5 and 10), and 5 mM (lanes 6 and 11). The assay was performed at 9.5 μM ATP and a DNA duplex concentration equal to the Km value (4.7 pM concentration of nucleotide bases). The substrate and released strand were separated in a TBE-polyacrylamide gel and visualized by exposition of dried gel onto X-ray film for 14 h. The results shown are representative of three independent experiments.
The ribavirin- and ribavirin-TP-mediated changes of the ATPase activity were, however, only partially accompanied by respective alterations of the helicase activity. At ATP and DNA duplex concentrations corresponding to their Km values an IC50 of 120 μM for ribavirin-TP was measured. The inhibition reached a maximum of 30% of the control at 450 μM and was not competitive with regard to the ATP. Under these experimental conditions ribavirin did not affect the helicase activity (Fig. 5B and C). The reduction or increase of the ATP or DNA duplex did not lead to an alteration of the inhibitory potential of ribavirin or ribavirin-TP. The juxtaposition of both the ATPase and the helicase activities as a function of ribavirin-TP concentration at selected ATP concentrations is presented in Fig. 5A and B.
Experiments performed with some cytosine and uracil derivatives revealed that the inhibitory behavior of ribavirin-TP was not without precedent. For example, 5-fluoro-2-selenocytosine reduced the NTPase activity with an IC50 of 75 μM and did not influence the helicase activity up to a concentration of >500 μM (A. Niebuhr, unpublished data).
Action of guanine and O6-benzylguanine derivatives.
Evidence for a divergence in the NTPase and the helicase activities of WN virus NTPase/helicase was further provided by experiments using derivatives of O6-benzylguanine, an inhibitor of O6-alkylguanine-DNA alkyltransferase with enhanced specificity toward human and rodent enzymes (13).
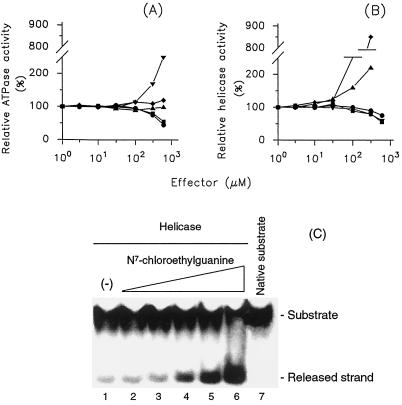
The parent substance, O6-benzylguanine (Fig. 6B), was a weak inhibitor of the ATPase activity of WN virus NTPase/helicase (IC50 > 500 μM). Interesting observations were made, however, with derivatives of O6-benzylguanine in which a chloroethyl moiety was added at position 7 or 9 (Fig. 6C and D). While the O6-benzyl-N7-chloroethylguanine conserved the inhibitory properties of the parent compound, the O6-benzyl-N9-chloroethylguanine proved to be a stimulator of NTPase activity, with a maximum effect of 350% of control at 650 μM. Both chloroethyl derivatives in which the O6-benzyl moiety was omitted (N7-chloroethylguanine and N9-chloroethylguanine; Fig. 6E and F) did not influence the ATPase activity up to concentrations of 500 μM (Fig. 7A).
FIG. 7.
Comparison of the modulating effects of the guanine and O6-benzylguanine derivatives on the ATPase and helicase activities of the WN virus NTPase/helicase. The ATPase (A) and helicase (B) activities of the enzyme were investigated as a function of increasing amounts of O6-benzylguanine (ℑ), O6-benzyl-N9-chloroethylguanine (▾), O6-benzyl-N7-chloroethylguanine (▪), N9-chloroethylguanine (∓), and N7-chloroethylguanine (⧫). The ATPase reaction was performed in the presence of ATP adjusted to a concentration equal to the Km value, and the activity of the enzyme was determined by using the charcoal adsorption method described in Materials and Methods. The helicase activity of the enzyme was investigated at 9.5 μM ATP and a 4.7 pM concentration of nucleotide bases of DNA duplex as the substrate. The 32P radioactivity of the released strand was quantified as described in Materials and Methods. The ATPase and helicase activities measured in the absence of the inhibitors were taken as 100%. (C) Autoradiography of a TBE-polyacrylamide gel demonstrating the activating effect of N7-chloroethylguanine on the helicase activity of WN virus NTPase/helicase. The reaction took place in the absence (lane 1) or presence of the compound added at 2.2 (lane 2), 6.6 (lane 3), 22 (lane 4), 66 (lane 5), or 220 μM (lane 6). The native substrate is presented in lane 7. The dried gel was exposed for 14 h without an intensifying screen. The results shown are representative of three independent experiments.
Subsequently the helicase activity of the enzyme as a function of the increasing concentrations of the O6-benzylguanine derivatives was investigated. While compounds containing the O6-benzyl moiety displayed comparably weak inhibitory potentials, N7-chloroethylguanine and N9-chloroethylguanine significantly enhanced the unwinding activity of the enzyme. The activation reached maxima of 850 and 220% of control for N7-chloroethylguanine and N9-chloroethylguanine, respectively, at a concentration of 200 to 250 μM and a 50% effective dose equal to 150 μM (Fig. 7B and C). The effect of the derivatives on the ATPase activity juxtaposed to the corresponding changes of the helicase activity is shown in Fig. 7A and B. In summary, the data indicated that both activities of the WN virus NTPase/helicase could be modulated (inhibited or activated) independently of each other.
DISCUSSION
NTPase/helicase activity associated with a 60-kDa protein was purified from medium obtained from a WN virus-infected cell culture. The WN virus and helicase SFII immunoreactivity together with the presence of the ATP-binding site confirmed the nature of the protein. Sequencing experiments revealed two NH2 termini of the protein: at Gly1564 and Asp1567. Assuming an average molecular mass of 110 to 120 Da per amino acid, we could calculate the COOH terminus of our enzyme corresponding to the boundary of NS3 (Arg2120).
This finding appears to be in concordance with the data obtained by Wengler and Wengler (54). The authors have obtained a fully active WN virus NTPase by treatment of the particulate fraction of infected cells with subtilisin. The 50-kDa fragment of NS3 started at Gly1668 (numbered according to the position within the WN virus polyprotein [10, 11, 52]) and, similar to our enzyme, contained the complete COOH-terminal part of the NS3 protein (54). A structural comparison with the NS3 protein of HCV allowed the localization of the start region of the reported 50-kDa enzyme within the putative interdomain “hinge” connecting the protease and NTPase/helicase domains (6). Our previous study revealed a high susceptibility of these interdomain links to proteolytic attack (5, 6). In this context the localization of the NH2 termini of the 60-kDa protein within the compact protease domain is rather surprising. The presence of the closely neighboring NH2 termini may suggest the presence of a hinge region within the protease domain of the WN virus NS3 protein. This region could be used by different proteases as for other hinge regions present within the NS3 of HCV characterized by us previously (5, 6). Our immunoblot data, however, strongly suggest that the 60-kDa form of the enzyme is produced intracellularly during virus replication and does not result from artificial proteolysis, e.g., after cell disruption. The cause for the enrichment of the 60-kDa protein in the cell culture medium remains unclear. It is tempting to suggest that the secretion of the viral protein or subviral particles may be a mechanism responsible for the appearance of large amounts of the 60-kDa protein in the medium. A secretion of large amounts of NS1 or subviral particles was previously observed in tick-borne encephalitis or Japanese encephalitis (JE) virus (2, 35, 38, 41).
An interesting property of the NTPase/helicase was associated with the multimerization status of the enzyme. When investigated by size exclusion gel chromatography, the 60-kDa protein, accompanied by NTPase and helicase activities, migrated in two peaks: at 120 to 110 kDa and at 65 to 55 kDa. This chromatographic behavior did not result from the formation of a heteromer with other proteins. When the protein migrating at 65 to 55 kDa was rechromatographed on the gel filtration column, the NTPase and helicase activities were distributed between the two peaks: 110 to 120 kDa and 65 to 55 kDa (data not shown). The tendency to polymerization among the SFII helicases is not without precedent. In a recent report Levin and Patel demonstrated the formation of HCV NTPase/helicase oligomers and postulated that the oligomeric state is the active form of the enzyme (28). In contrast, the migration of the 60-kDa protein at the void volume of the Superdex-200 column may result from a formation of heteromers with other viral proteins. An immunoblot analysis of the proteins comigrating in fractions 3 to 5, performed with WN virus-positive human sera, revealed numerous viral proteins that were not detectable with affinity antibody NS3/HEL(1&2). Indeed, there is evidence concerning complex formation and a direct interaction between NS3 and NS5 for the related dengue virus (23).
Similar to the WN virus enzyme purified by Wengler and Wengler (54) or dengue and yellow fever virus helicases described by Li et al. (29) and Warrener et al. (50), respectively, our NTPase/helicase preferred poly(dA) and poly(A) as activators. It is noteworthy that the strongest stimulator of the NS3-associated NTPase activity of JE virus or of other members of the family Flaviridae such as HCV and bovine viral diarrhea pestivirus (BVDV) was poly(U) (26, 45, 47). However, compared to that for enzymes described previously the activation was only modest and was measured at higher ATP concentrations. Further kinetic analyses of the modulation of the NTPase activity by polynucleotides are under way; nevertheless our preliminary study may suggest a mechanism based on an interesting property of poly(dA): as a regulator of the capacity of the NTP-binding domain for ATP, described recently for NTPase/helicase of HCV. The binding of the polynucleotide increased the affinity of the ATP-binding domain for ATP. On the other hand ATP enhanced the affinity of this domain for poly(dA) (6). The extent of the interaction was dependent on poly(dA) and ATP concentrations. Whether this mechanism occurs on the level of the holoenzyme and for WN virus NTPase/helicase remains to be elucidated.
The cause of the limited response of the enzyme to polynucleotide has not been addressed in this report. In a study mentioned above Kuo et al. compared the poly(U)-induced NTPase activities of full-length NS3 and of the NTPase/helicase domain of JE virus and demonstrated that the NH2-terminal part of NS3 is sufficient to suppress the sensitivity to polynucleotide stimulation (26). Similar observations were made with full-length and NH2-terminally truncated forms of dengue virus NS3 (29). In the light of these observations it appears conceivable that the fragment of the protease domain present in the molecule of our enzyme is responsible for the reduced stimulatory effect of the polynucleotide.
Kinetic data published elsewhere revealed significant differences between basal and polynucleotide-induced NTPase activities. In the presence of polynucleotides the NTPase activities of NS3 of HCV, BVDV, JE virus, yellow fever virus, and dengue virus displayed strict dependency on Mg2+ or Mn2+ and high sensitivity to monovalent ions. In the absence of the polynucleotide these enzymatic activities were not significantly altered by these ions (29, 45, 47, 54). Thus, the rather insignificant effect on ATPase activity by monovalent ions and the flat Mg2+ requirement curve of our enzyme, observed also in the presence of the polynucleotide, were not surprising.
To our knowledge there are only few reports demonstrating the unwinding activity associated with the NS3 proteins of arthropod-borne viruses (29, 49). The common feature of these enzymes is suspiciously low specific helicase activity in comparison with those of HCV or BVDV (40, 46, 51). It is well established that during NTP hydrolysis the energy released is utilized in helicase translocation along the RNA strand; however, the number of ATP hydrolysis events per measured unwinding reaction cycle remains controversial (25, 57). When the activities of the enzyme were tested as a function of the ribavirin-TP concentration, the experimental inhibitory curves fitted for NTPase and helicase activities did not go in parallel. The activities were inhibited with different efficacies and by different mechanisms. For example, at ATP and ribavirin-TP concentrations at which the NTPase concentration was reduced to 20% of the control the helicase activity remains unaffected. It could not be ruled out that at strongly decreased ATP concentrations (<1/1,000 of the Km value), at which ribavirin-TP displays a significantly enhanced inhibitory potential toward NTPase activity (7), the helicase activity begins to be affected by the lowered NTPase activity. Such an effect of strongly reduced ATP concentrations, in the presence of competitive NTPase inhibitors, on the helicase activity of HCV NTPase/helicase was observed by us recently (P. Borowski, A. Niebuhr, and N. Schmitz, unpublished data). Nevertheless, for the WN virus enzyme the reduced ATP concentrations were not sufficient to provide a measurable level of unwinding activity (Fig. 3C). On the other hand in view of the relatively low specificity toward NTP of the viral NTPases/helicases (40, 47, 51), it is possible that the investigated enzyme catalyzed the hydrolysis of the ribavirin-TP to derivatives that are less potent regarding helicase activity (8). We have found, however, numerous chemically unrelated compounds that exerted a similar effect, i.e., inhibition of ATPase reaction, at least to a certain extent, without influencing the helicase activity.
Taken together these results indicate that the NTPase activity is not directly coupled to the unwinding reaction. Consequently, the number of ATP hydrolysis events per unwinding cycle is not any constant value. This was ascertained by using guanine and O6-benzylguanine derivatives. Our experiments indicated that the ATPase activity of WN virus NTPase/helicase may be modulated, i.e., increased or reduced, without affecting the helicase activity of the enzyme. Furthermore, N7- and N9-chloroethyl derivatives of guanine stimulate helicase activity without enhancing the requirement for ATP of the enzyme. Similar observations were made when effects of the guanine derivatives were verified with the respective enzymatic activities of the holo-NS3 protein of JE and dengue viruses or with the NTPase/helicase domain of HCV. Interestingly, under our standard assay conditions the HCV enzyme exhibited a specific unwinding activity higher by one or more orders of magnitude than that exhibited by the WN virus NTPase/helicase at comparable specific ATPase activities. It is noteworthy that even the high helicase activity could be further stimulated (approximately 5- and 10-fold for N9- and N7-chloroethylguanine, respectively; data not shown). Consequently, the regulation of the helicase activity in vivo might involve compounds or cofactors that are capable of coupling the unwinding reaction to ATP hydrolysis.
A further unanswered question is how the enzymes recognize the compounds. Previous experimental data (7) together with kinetic analyses of Porter (39) suggest strongly the presence of a further NTP-binding site within the SFII NTPase/helicase molecule. Thus, it is possible that the compounds tested exerted their effect(s) by occupation of this site. On the basis of the data presented above one could speculate that in the course of the NTPase reaction more ATP will be hydrolyzed than directly necessary for the unwinding reaction.
REFERENCES
- 1.Ali J A, Lohman T M. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science. 1997;275:377–380. doi: 10.1126/science.275.5298.377. [DOI] [PubMed] [Google Scholar]
- 2.Allison S L, Stadler K, Mandl C W, Kunz C, Heinz F X. Synthesis and secretion of recombinant tick-borne encephalitis virus protein E in soluble and particulate form. J Virol. 1995;69:5816–5820. doi: 10.1128/jvi.69.9.5816-5820.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bazan J F, Fletterick R. Detection of a trypsin-like serine protease domain in flaviviruses and pestiviruses. Virology. 1989;171:637–639. doi: 10.1016/0042-6822(89)90639-9. [DOI] [PubMed] [Google Scholar]
- 4.Borowski P, Heiland M, Oehlmann K, Becker B, Kornetzky L, Feucht H-H, Laufs R. Non-structural protein 3 of hepatitis C virus inhibits phosphorylation mediated by cAMP-dependent protein kinase. Eur J Biochem. 1996;237:611–618. doi: 10.1111/j.1432-1033.1996.0611p.x. [DOI] [PubMed] [Google Scholar]
- 5.Borowski P, Kühl R, Laufs R, Schulze zur Wiesch J, Heiland M. Identification and characterization of a histone binding site of nonstructural protein 3 of hepatitis C virus. J Clin Virol. 1999;13:61–69. doi: 10.1016/s1386-6532(99)00007-4. [DOI] [PubMed] [Google Scholar]
- 6.Borowski P, Kuehl R, Mueller O, Hwang L-H, Schulze zur Wiesch J, Schmitz H. Biochemical properties of a minimal functional domain with ATP-binding activity of the NTPase/helicase of hepatitis C virus. Eur J Biochem. 1999;266:715–723. doi: 10.1046/j.1432-1327.1999.00854.x. [DOI] [PubMed] [Google Scholar]
- 7.Borowski P, Mueller O, Niebuhr A, Kalitzky M, Hwang L-H, Schmitz H, Siwecka M, Kulikowski T. ATP-binding domain of NTPase/helicase as a target for hepatitis C antiviral therapy. Acta Biochim Pol. 2000;47:173–180. [PubMed] [Google Scholar]
- 8.Borowski, P., M. Lang, A. Niebuhr, A. Haag, H. Schmitz, J. Schulze zur Wiesch, J. Choe, M. A. Siwecka, and T. Kulikowski. Inhibition of the helicase activity of HCV NTPase/helicase by 1-β-d-ribofuranosyl-1,2,4-triazole-3-carboxamide-5′-triphosphate (ribavirin-TP). Acta Biochim. Pol., in press. [PubMed]
- 9.Bowles W A, Schneider F H, Lewis L, Robins R K. Synthesis and antitumor activity of 9-(tetrahydro-2-furyl)purine analogs of biologically important deoxynucleosides. J Med Chem. 1963;6:471–480. doi: 10.1021/jm00341a002. [DOI] [PubMed] [Google Scholar]
- 10.Castle E, Nowak T, Leidner U, Wengler G, Wengler G. Sequence analysis of the viral core protein and the membrane-associated proteins V1 and NV2 of the flavivirus West Nile virus and of the genome sequence for these proteins. Virology. 1985;145:227–236. doi: 10.1016/0042-6822(85)90156-4. [DOI] [PubMed] [Google Scholar]
- 11.Castle E, Leidner U, Nowak T, Wengler G, Wengler G. Primary structure of the West Nile flavivirus genome region coding for all nonstructural proteins. Virology. 1986;149:10–26. doi: 10.1016/0042-6822(86)90082-6. [DOI] [PubMed] [Google Scholar]
- 12.Dixon M. The determination of enzyme inhibitor constants. Biochem J. 1952;55:170–171. doi: 10.1042/bj0550170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elder R, Margison G, Rafferty J. Differential inactivation of mammalian and Escherichia coli O6-alkylguanine-DNA alkyltransferases by O6-benzylguanine. Biochem J. 1994;298:231–235. doi: 10.1042/bj2980231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felczak K, Miazga A, Golos B, Rode W, Shugar D, Kulikowski T. Synthesis, conformation and biological properties of selenonucleosides. Nucleosides Nucleotides. 1999;18:635–636. [Google Scholar]
- 15.Galinari P, Brennan D, Nardi C, Brunetti M, Tomei L, Steinkühler C, De Francesco R. Multiple enzymatic activities associated with recombinant NS3 of hepatitis C virus. J Virol. 1998;72:6758–6769. doi: 10.1128/jvi.72.8.6758-6769.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorbalenya A E, Donchenko A P, Koonin E V, Blinov V M. N-terminal domains of putative helicase of flavi- and pestiviruses may be serine proteases. Nucleic Acids Res. 1989;17:3889–3897. doi: 10.1093/nar/17.10.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Two related superfamilies of putative helicases involved in replication, recombination, repair, and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorbalenya A E, Koonin E V. Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- 19.Grakoui A, Wychowski C, Lin C, Feinstone S, Rice C M. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 21.Hodgman T C. A new superfamily of replicative proteins. Nature. 1988;333:22–23. doi: 10.1038/333022b0. [DOI] [PubMed] [Google Scholar]
- 22.Kadare G, Haenni A. Virus-encoded RNA helicase. J Virol. 1997;71:2583–2590. doi: 10.1128/jvi.71.4.2583-2590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner K E, Padmanabhan R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem. 1995;270:19100–19106. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- 24.Kim J, Morgenstern K, Lin C, Fox T, Dwyer M, Landro J A, Chambers S P, Markland W, Lepre C A, O'Malley E T, Harbeson S L, Rice C M, Murcko M, Caron P, Thomson J A. Crystal structure of the hepatitis C virus NS3 protease domain complexed with synthetic NS4A cofactor peptide. Cell. 1996;87:343–355. doi: 10.1016/s0092-8674(00)81351-3. [DOI] [PubMed] [Google Scholar]
- 25.Kim J, Morgenstern K, Griffith J, Dwyer M, Thomson J, Murcko M, Lin C, Caron P. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 26.Kuo M-D, Chin C, Hsu S-L, Shiao J-Y, Wang T-M, Lin J-H. Characterization of the NTPase activity of Japanese encephalitis virus NS3 protein. J Gen Virol. 1996;77:2077–2084. doi: 10.1099/0022-1317-77-9-2077. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Levin M, Patel S. The helicase from hepatitis C virus is active as an oligomer. J Biol Chem. 1999;274:31839–31846. doi: 10.1074/jbc.274.45.31839. [DOI] [PubMed] [Google Scholar]
- 29.Li H, Clum S, You S, Ebner K, Padmanabhan R. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J Virol. 1999;73:3108–3116. doi: 10.1128/jvi.73.4.3108-3116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin C, Kim J. Structure-based mutagenesis study of hepatitis C virus NS3 helicase. J Virol. 1999;73:8798–8807. doi: 10.1128/jvi.73.10.8798-8807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love R A, Parge H E, Wickersham J A, Hostomsky Z, Habuka N, Moomaw E W, Adachi T, Hostomska Z. The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and structural zinc binding site. Cell. 1996;87:331–342. doi: 10.1016/s0092-8674(00)81350-1. [DOI] [PubMed] [Google Scholar]
- 32.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 33.Ludwig J. Chemical synthesis of nucleoside triphosphates. Acta Biochim Biophys Acad Sci Hung. 1981;16:131–133. [PubMed] [Google Scholar]
- 34.Matsudaira P T. A practical guide to protein and peptide purification for microsequencing. San Diego, Calif: Academic Press; 1989. [Google Scholar]
- 35.Matveeva V A, Bugrysheva V, Bakhvalova V N, Morozova O V. Secretion of tick-borne encephalitis virus glycoproteins E and NS1 heterocomplex in the late stage of infection. Vopr Virusol. 1997;42:179–182. [PubMed] [Google Scholar]
- 36.Miller H, Purcell R. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc Natl Acad Sci USA. 1990;87:2057–2061. doi: 10.1073/pnas.87.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mishra N C, Broom A D. A novel synthesis of nucleoside 5′-triphosphates. J Chem Soc Chem Commun. 1991;1991:1276–1277. [Google Scholar]
- 38.Muylaert I R, Galler R, Rice C M. Genetic analysis of the yellow fever virus NS1 protein: identification of a temperature-sensitive mutation which blocks RNA accumulation. J Virol. 1997;71:291–298. doi: 10.1128/jvi.71.1.291-298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Porter D. A kinetic analysis of the oligonucleotide-modulated ATPase activity of the helicase domain of the NS3 protein from hepatitis C virus. J Biol Chem. 1998;273:14247–14253. doi: 10.1074/jbc.273.23.14247. [DOI] [PubMed] [Google Scholar]
- 40.Preugschat F, Averett D, Clarke B, Porter D. A steady-state and pre-steady-state kinetic analysis of the NTPase activity associated with the hepatitis C virus NS3 helicase domain. J Biol Chem. 1996;271:24449–24459. doi: 10.1074/jbc.271.40.24449. [DOI] [PubMed] [Google Scholar]
- 41.Pugachev K V, Mason P W, Frey T K. Sindbis vectors suppress secretion of subviral particles of Japanese encephalitis virus from mammalian cells infected with SIN-JEV recombinants. Virology. 1995;209:155–166. doi: 10.1006/viro.1995.1239. [DOI] [PubMed] [Google Scholar]
- 42.Ramzaeva N, Alksnis E, Goldberg A, Lidaks M. Alkylation of 6-methylthio-and 6-benzyloxyguanine under phase-transfer conditions. Synth Commun. 1989;19:3121–3128. [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Speight G, Westaway G. Positive identification of NS4A, the last of the hypothetical nonstructural proteins of flaviviruses. Virology. 1989;170:299–301. doi: 10.1016/0042-6822(89)90383-8. [DOI] [PubMed] [Google Scholar]
- 45.Suzich J, Tamura J, Palmer-Hill F, Warrener P, Grakoui A, Rice C M, Feinstone S, Collett M. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J Virol. 1993;67:6152–6158. doi: 10.1128/jvi.67.10.6152-6158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tai C-L, Chi W-K, Chen D-S, Hwang L-H. The helicase activity associated with hepatitis C virus nonstructural protein 3 (NS3) J Virol. 1996;70:8477–8484. doi: 10.1128/jvi.70.12.8477-8484.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamura J, Warrener P, Collett M. RNA-stimulated NTPase activity associated with the p80 protein of the pestivirus bovine viral diarrhea virus. Virology. 1993;193:1–10. doi: 10.1006/viro.1993.1097. [DOI] [PubMed] [Google Scholar]
- 48.Towbin H, Staehlin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Utama A, Shimizu H, Morikawa S, Hasebe F, Morita K, Igarashi A, Hatsu M, Takamizawa K, Miyamura T. Identification and characterization of the RNA helicase activity of Japanese encephalitis virus NS3 protein. FEBS Lett. 2000;465:74–78. doi: 10.1016/s0014-5793(99)01705-6. [DOI] [PubMed] [Google Scholar]
- 50.Warrener P, Tamura J K, Collett M. RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J Virol. 1993;67:989–996. doi: 10.1128/jvi.67.2.989-996.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warrener P, Collett M. Pestivirus NS3 (p80) protein possesses RNA helicase activity. J Virol. 1995;69:1720–1726. doi: 10.1128/jvi.69.3.1720-1726.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wengler G, Castle E, Leidner U, Nowak T, Wengler G. Sequence analysis of the membrane protein V3 of the flavivirus West Nile virus and of its gene. Virology. 1986;147:264–274. doi: 10.1016/0042-6822(85)90129-1. [DOI] [PubMed] [Google Scholar]
- 53.Wengler G, Nowak T, Castle E. Description of a procedure which allows isolation of viral nonstructural proteins from BHK vertebrate cells infected with the West Nile flavivirus in a state which allows their direct chemical characterization. Virology. 1990;177:795–801. doi: 10.1016/0042-6822(90)90552-3. [DOI] [PubMed] [Google Scholar]
- 54.Wengler G, Wengler G. The carboxy-terminal part of the NS3 protein of the West Nile flavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology. 1991;184:707–715. doi: 10.1016/0042-6822(91)90440-m. [DOI] [PubMed] [Google Scholar]
- 55.Westaway E G, Brinton M A, Gaidamovich S, Horzinek M C, Igrashi A, Käariäinen L, Lvov D K, Porterfield J S, Russell P K, Trent D W. Flaviviridae. Intervirology. 1985;24:183–192. doi: 10.1159/000149642. [DOI] [PubMed] [Google Scholar]
- 56.Woodford T, Pardee A B. Histone H1 kinase in exponential and synchronous populations of Chinese hamster fibroblasts. J Biol Chem. 1986;261:4669–4676. [PubMed] [Google Scholar]
- 57.Yao N, Hesson T, Cable M, Hong Z, Kwong A, Le H, Weber P. Structure of the hepatitis C virus RNA helicase domain. Nat Struct Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- 58.Yoshikawa M, Kato T, Takenishi T. A novel method of phosphorylation of nucleosides to 5′-nucleotides. Tetrahedron Lett. 1967;50:5065–5068. doi: 10.1016/s0040-4039(01)89915-9. [DOI] [PubMed] [Google Scholar]