Abstract
Relative to wild-type herpes simplex virus type 1 (HSV-1), ICP0-null mutant viruses reactivate inefficiently from explanted, latently infected mouse trigeminal ganglia (TG), indicating that ICP0 is not essential for reactivation but plays a central role in enhancing the efficiency of reactivation. The validity of these findings has been questioned, however, because the replication of ICP0-null mutants is impaired in animal models during the establishment of latency, such that fewer mutant genomes than wild-type genomes are present in latently infected mouse TG. Therefore, the reduced number of mutant viral genomes available to reactivate, rather than mutations in the ICP0 gene per se, may be responsible for the reduced reactivation efficiency of ICP0-null mutants. We have recently demonstrated that optimization of the size of the ICP0 mutant virus inoculum and transient immunosuppression of mutant-infected mice with cyclophosphamide can be used to establish wild-type levels of ICP0-null mutant genomes in latently infected TG (W. P. Halford and P. A. Schaffer, J. Virol. 74:5957–5967, 2000). Using this procedure to equalize mutant and wild-type genome numbers, the goal of the present study was to determine if, relative to wild-type virus, the absence of ICP0 function in two ICP0-null mutants, n212 and 7134, affects reactivation efficiency from (i) explants of latently infected TG and (ii) primary cultures of latently infected TG cells. Although equivalent numbers of viral genomes were present in TG of mice latently infected with either wild-type or mutant viruses, reactivation of n212 and 7134 from heat-stressed TG explants was inefficient (31 and 37% reactivation, respectively) relative to reactivation of wild-type virus (KOS) (95%). Similarly, n212 and 7134 reactivated inefficiently from primary cultures of dissociated TG cells plated directly after removal from the mouse (7 and 4% reactivation, respectively), relative to KOS (60% reactivation). The efficiency and kinetics of reactivation of KOS, n212, and 7134 from cultured TG cells (treated with acyclovir to facilitate the establishment of latency) in response to heat stress or superinfection with a nonreplicating HSV-1 ICP4− mutant, n12, were compared. Whereas heat stress induced reactivation of KOS from 69% of latently infected TG cell cultures, reactivation of n212 and 7134 was detected in only 1 and 7% of cultures, respectively. In contrast, superinfection with the ICP4− virus, which expresses high levels of ICP0, resulted in the production of infectious virus in nearly 100% of cultures latently infected with KOS, n212, or 7134 within 72 h. Thus, although latent mutant viral genome loads were equivalent to that of wild-type virus, in the absence of ICP0, n212 and 7134 reactivated inefficiently from latently infected TG cells during culture establishment and following heat stress. Collectively, these findings demonstrate that ICP0 is required to induce efficient reactivation of HSV-1 from neuronal latency.
Unlike productive infection, which is initiated by the activities of virion-associated proteins, including the transcriptional activator VP16, reactivation of herpes simplex virus type 1 (HSV-1) from neuronal latency is thought to occur in the absence of any preexisting viral proteins. Because ICP0 is the only HSV-1 protein expressed at very early times during productive infection and is capable of activating expression of all classes of viral genes (immediate-early [IE], early, and late) (4), it has been suggested that low-level expression of ICP0 in neurons may be responsible for the initiation of productive-phase gene expression during reactivation. This possibility is supported by the following observations. (i) In three independent studies, ICP0-null mutants reactivated from explanted mouse trigeminal ganglia (TG) with significantly reduced efficiencies relative to wild-type virus (2, 5, 16). Thus, although not essential for reactivation, ICP0 enhances reactivation efficiency significantly. (ii) Expression of ICP0 by adenoviral vectors or by temperature-sensitive mutants with mutations in the ICP4 gene at the nonpermissive temperature induces reactivation of HSV-2 and productive-phase HSV-2 gene expression from quiescent, nonreplicating viral genomes in human embryonic lung cells (12, 28). (iii) In the absence of VP16, ICP0 enhances the ability of transfected viral DNA to initiate productive infection in Vero cells by 10,000-fold, which resembles reactivation from neuronal latency in that it occurs in the absence of other viral proteins (3).
Central to the goals of this study are the following. In the reactivation studies mentioned above, all of which utilized the mouse ocular model of HSV latency, it was subsequently shown that the number of genomes in TG latently infected with ICP0-null mutants was significantly lower than the number of genomes in TG latently infected with wild-type virus. Thus, in none of these studies was it possible to determine definitely whether the impaired reactivation efficiency of ICP0-null mutants was a consequence of (i) reduced numbers of latent ICP0-null mutant genomes available to reactivate (5 to 20% of the number of wild-type genomes) or (ii) a requirement for ICP0 function during the reactivation process.
We have recently demonstrated that a reduction in the size of the inoculum of ICP0-null mutants from 2 × 106 to 2 × 105 PFU/eye coupled with transient immunosuppression of mutant-infected mice can be used to enable HSV-1 ICP0-null mutant genomes to reach wild-type levels in the TG of latently infected mice (11). Utilizing this procedure to equalize mutant and wild-type genome loads, the goal of the present study was to determine if loss of ICP0 function impairs HSV-1 reactivation from latency. For this purpose, the reactivation efficiencies of two ICP0-null mutants, n212 and 7134, were compared to the reactivation efficiency of wild-type virus in four different assay systems: (i) explanted, latently infected TG subjected to heat stress, (ii) latently infected TG cells subjected to the stress associated with dissociation and establishment of primary cultures (10), (iii) acyclovir (ACV)-treated, latently infected TG cell cultures subjected to heat stress (10), and (iv) ACV-treated, latently infected TG cell cultures superinfected with a replication-defective, ICP4− mutant which overexpresses ICP0. In the absence of ICP0 function, n212 and 7134 reactivated inefficiently relative to wild-type virus. In contrast, when ICPs 0, 22, 27, and 47 were provided in trans from a replication-defective ICP4− virus, reactivation occurred in nearly 100% of TG cell cultures latently infected with n212 or 7134. Collectively, the results of this study demonstrate that ICP0 is required for the efficient reactivation of HSV-1 from neuronal latency.
MATERIALS AND METHODS
Cells and viruses.
Vero, L7 (an ICP0-expressing Vero cell line [22]), and E5 (an ICP4−-expressing Vero cell line [6]) cells were propagated in complete Dulbecco modified Eagle medium (DMEM) containing 0.375% HCO3− and supplemented with 10% fetal bovine serum, penicillin G (100 U/ml), streptomycin (100 mg/ml), and 2 mM l-glutamine as previously described (15). The viruses used in this study were wild-type HSV-1 strain KOS (passage 12 from human isolation [24]), the KOS-derived ICP0-null mutants n212 and 7134 (2), and the KOS-derived ICP4-null mutant n12 (7). Viruses were propagated in Vero cells (wild-type, n212, and 7134) or in E5 cells (n12) as described previously (2, 7, 24). Wild-type viral titers were determined in Vero cells, n212 and 7134 titers were determined in L7 cells, and n12 titers were determined in E5 cells. The deletion in 7134 that removed the ICP0 gene also removed ∼1 kb of the 3′ end of the latency-associated transcript (LAT) gene (8); consequently, 7134 is an ICP0− LAT− double mutant. In contrast, n212 produces full-length LAT and ICP0 transcripts but contains a mutation in codon 212 of the 775-codon ICP0 open reading frame that introduces stop codons in all three reading frames (2).
Infection and transient immunosuppression of mice.
Male ICR mice (6 to 8 weeks, 29 ± 2 g) were obtained from Harlan-Sprague Dawley (Indianapolis, Ind.) and handled in accordance with Guide for the Care and Use of Laboratory Animals (14). Mice were anesthetized by intraperitoneal administration of xylazine (6.6 mg/kg) and ketamine (100 mg/kg). Following corneal scarification with a 26-gauge needle, tear film was blotted from eyes with tissue, and 3 μl of viral inoculum containing 2 × 105 PFU of KOS, n212, or 7134 virus was placed on each eye. Virus suspensions used to infect mice were titrated immediately after inoculation to confirm the size of the viral inoculum. Transient immunosuppression of mice was achieved by intraperitoneal administration of 18 mg of cyclophosphamide (CyP) (Pharmacia/Upjohn Co., Kalamazoo, Mich.) per ml in a volume of 0.25 ml of phosphate-buffered saline to achieve a dose of 150 mg/kg/day 1 day before and 1 and 3 days after inoculation.
TG explants.
Latently infected mice were sacrificed on day 30 postinfection (p.i.), TG were removed, each TG was cut into eight equal sized pieces, and all eight pieces were placed in one well of a 24-well plate (on ice) containing 0.5 ml of complete DMEM. Once processing of all TG was complete, the 24-well plates were removed from ice and heat stressed by transfer to a 43°C, 5% CO2 incubator for 3 h. After heat stress, explants were transferred to a 37°C, 5% CO2 incubator. Twenty-four hours later, TG pieces and culture medium were transferred to 24-well plates seeded 6 h earlier with 2 × 104 Vero or L7 cells per well, TG obtained from mice latently infected with KOS were cocultured with Vero cells, and TG obtained from mice latently infected with n212 and 7134 were cocultured with L7 cells. Daily, on days 2 through 12 postexplant, 100 μl of culture supernatant was transferred from each TG explant culture to a freshly seeded culture of the appropriate indicator cells (i.e., Vero or L7 cells). These cultures were observed 4 and 6 days later for the appearance of cytopathic effects. After 12 days in culture, TG explants that exhibited no evidence of reactivation were homogenized, frozen and thawed once, sonicated, and centrifuged, and aliquots of the clarified supernatant fluids were placed on monolayers of the appropriate indicator cells to test for the presence of infectious (reactivated) virus.
Infectious-center assay.
On day 30 p.i., the left and right TG were removed from each latently infected mouse and combined. Both TG were teased apart longitudinally with a scalpel and incubated in 100 μl of 0.1% collagenase in Ca2+- and Mg2+-free Hanks' balanced salt solution (HBSS) at 37°C for 40 min. After 20 and 35 min, TG were gently pipetted up and down to mechanically dissociate individual cells from pieces of tissue. Collagenase was removed from TG cells by performing three sequential washes with 1 ml of complete DMEM. Dissociated cells were resuspended in 2 ml of complete DMEM, transferred to a single well of collagen-coated six-well dishes (i.e., cells from each TG pair were placed in one well), and centrifuged at 1,000 rpm for 5 min in a Sorvall RT7 centrifuge (Kendro Laboratory Products, Newtown, Conn.) to facilitate rapid cell adhesion. After incubation at 37°C for 12 h, culture medium was gently aspirated from wells and replaced with 300 μl of DMEM containing 107 PFU of n12 to achieve a multiplicity of infection of ∼10 PFU/cell, n12 is a replication-defective ICP0-expressing ICP4− mutant. After 1 h at 37°C, the viral inoculum was aspirated, wells were rinsed gently with 1 ml of complete DMEM, and 2 ml of complete DMEM containing 2 × 105 Vero or L7 indicator cells was added per well. After 3 h of incubation at 37°C, medium was removed and 3 ml of complete DMEM containing 1.5% methylcellulose was added to each well. After 4 days at 37°C, the methylcellulose overlay was removed, monolayers were fixed and stained with 20% methanol–0.1% crystal violet, and plaques initiated by infectious centers (single infected cells) were counted.
Establishment and maintenance of TG cell cultures.
Primary cultures of TG cells were prepared by a procedure described previously (10), with the following modifications. On day 30 p.i., TG were collected from mice latently infected with KOS, n212, or 7134 viruses (n = 8 mice or 16 TG per virus). Pooled TG were placed on ice in TG cell culture medium, which consists of minimal essential medium containing 0.075% HCO3− and supplemented with 10% fetal bovine serum, penicillin G (100 U/ml), streptomycin (100 mg/ml), 2 mM l-glutamine, and 2.5S nerve growth factor (10 ng/ml; Becton Dickinson, Chicago, Ill.). After collection of all 16 TG, the medium was decanted and pooled TG were rinsed twice with 5 ml of Ca2+- and Mg2+-free HBSS. The TG were then teased apart longitudinally with a scalpel, transferred to 1 ml of HBSS containing 0.1% collagenase type I (Sigma Chemical Co., St. Louis, Mo.), and incubated at 37°C. After 20 and 35 min of incubation, pooled TG were gently triturated with a 1-ml serological pipette. After 40 min, dissociated TG cells and tissue pieces were resuspended in 10 ml of TG cell medium and centrifuged at 1,000 rpm for 5 min in a Sorvall RT7 centrifuge (Kendro Laboratory Products). Cell pellets were rinsed two additional times by resuspension and centrifugation. After the final rinse, cells were resuspended in 32 ml of TG cell medium. From each 32-ml preparation of TG cell suspension (prepared from 16 TG), (i) 4 1-ml aliquots were frozen at −70°C thawed, sonicated, and tested immediately for the presence of infectious virus, (ii) total DNA was extracted from an additional 4 1-ml aliquots to measure viral genome load by competitive PCR, and (iii) the remaining 24 1-ml aliquots were cultured in a 24-well plate.
To facilitate neuronal survival during the establishment of TG cell cultures, 24-well plates were prepared 1 day in advance by thin-coating wells with rat tail collagen per the instructions of the manufacturer (Becton Dickinson). Each well was then seeded with 1.5 × 104 primary, uninfected feeder mouse TG cells in a volume of 0.5 ml of TG cell culture medium and incubated overnight at 37°C. One milliliter of TG cell suspension from mice latently infected with KOS, n212, or 7134 was then added to 24-well plates and centrifuged at 1,000 rpm for 5 min in a Sorvall RT7 centrifuge (Kendro Laboratory Products) to facilitate rapid adhesion of TG cells to the feeder layer in collagen-coated wells. In experiments in which reactivation was induced from quiescent, latently infected cells (see below), TG cell cultures latently infected with KOS, n212, or 7134 were established in the presence of 200 μM ACV (44 μg/ml) to suppress viral replication (reactivation) during culture establishment. After 7 days in culture, the ACV-containing medium was removed, monolayers were rinsed with 1 ml of culture medium, and 1.5 ml of new TG cell culture medium was added per well.
Analysis of reactivation in TG cell cultures.
Reactivation induced in response to the combined stress associated with dissociation of TG cells and culture establishment was measured as follows. Daily from day 2 to 12 following culture establishment, 100 μl of medium was transferred from each TG cell culture to a freshly seeded culture of the appropriate indicator cells (i.e., Vero or L7 cells). These indicator cultures were observed 4 and 6 days later for the appearance of cytopathic effects. After 12 days in culture, TG cell cultures were frozen at −70°C and thawed, and homogenates were placed on appropriate indicator cells to test for the presence of infectious virus by the appearance of cytopathic effects.
Reactivation in response to either heat stress or superinfection with an ICP0-expressing ICP4− mutant virus (n12) was carried out as follows. Cultures were transferred to a 43°C, 5% CO2 incubator for 3 h and then returned to a 37°C, 5% CO2 incubator. Viral superinfection was achieved by replacing the culture medium in each well of a 24-well plate with 200 μl containing 107 PFU/ml (multiplicity of infection, ∼10 PFU/cell) of the ICP4− virus, n12. After 1 h of adsorption, the viral inoculum was aspirated, wells were rinsed with 0.5 ml of culture medium, and 1.5 ml of new TG cell culture medium was added per well. Control cultures were superinfected with 0.2 ml of a 107-PFU/ml solution of UV-inactivated n12. UV inactivation was achieved by exposing 35-mm-diameter dishes containing 1.5-ml aliquots of n12 in complete DMEM to UV radiation (1 J/cm2) in a Stratalinker (Stratagene, La Jolla, Calif.). This treatment reduced the viral titer from 107 PFU/ml to less than 4 PFU/ml. To test for infectious virus in induced-reactivation experiments, 100 μl of TG cell culture medium was transferred to freshly seeded cultures of the appropriate indicator cells (i.e., Vero or L7 cells) on day 4 and on days 7 to 21. On day 21, heat-stressed TG cell cultures were immunocytochemically stained for HSV antigens as described below.
Measurement of viral DNA loads in TG by competitive PCR.
The competitive PCR analysis used in these studies has been described in detail elsewhere (11). In brief, total DNA was isolated from the pooled left and right TG of individual mice or from aliquots of dissociated TG cells by standard phenol-chloroform DNA extraction (25). Oligonucleotide primers specific for the HSV-1 genome, RR-a (5′-ATGCCAGACCTGTTTTTCAA) and RR-b (5′-gtctttgaacatgacgaagg), which amplified 243- and 322-bp fragments from each template, respectively, together with competitor templates were used in these tests. Relative yields of the two PCR products were determined by hybridization of radiolabeled HSV-specific and competitor-specific probes to duplicate slot blots of the completed PCR products. A standard curve generated from samples containing known amounts of viral DNA was included in each competitive PCR and used to define the relationship between the logarithm of the yield with the primers/HSV RR competitor product yield and the logarithm of the viral genome copy number. The viral genome load per TG was calculated as the number of genomes per 100 ng of TG cell culture DNA × 70, because each 1-ml aliquot of TG cell culture suspension contained ∼7 μg of DNA.
Southern blot analysis.
L7 cells were mock infected or infected with a 0.1-PFU/cell concentration of (i) n212 reactivation isolates, (ii) 7134 reactivation isolates, or (iii) the viral stocks of KOS, n212, and 7134 used to infect mice in these studies. Total DNA was harvested from infected L7 cells at 24 h p.i. DNA from L7 cells infected with n212 reactivation isolates (1 μg) was digested with NcoI and SpeI (New England Biolabs, Beverly. Mass.), and DNA from 7134 reactivation isolates (1 μg) was digested with NotI (New England Biolabs). Following electrophoresis on 0.8% agarose gels, restriction digests were blotted on Zeta-probe GT nylon membranes (Bio-Rad Laboratories, Inc., Richmond, Calif.) with a vacuum blotter (Bio-Rad Laboratories), and blots were irradiated with 0.2 J/cm2 in a UV cross-linker (Stratagene). A LAT-specific oligonucleotide probe (complementary to nucleotides 120744 to 120766; 5′-ACCACACACCAGCGGGTCTTTTG-3′) was end labeled with [α-32P]dATP using terminal transferase (Promega Corp.), and the probe was hybridized to Southern blots at 45°C for 16 h in hybridization solution containing 2 ng of labeled probe per ml, 7% sodium dodecyl sulfate, 120 mM NaH2PO4, and 250 mM NaCl. Excess probe was removed from membranes by sequential rinses in 0.1× standard saline citrate (SSC) (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% sodium dodecyl sulfate. Southern blots were exposed in phosphor screens, scanned with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.), and analyzed with ImageQuant 3.3 software (Molecular Dynamics).
Detection of HSV antigens by immunocytochemistry.
TG cell cultures were fixed in 4% phosphate-buffered formalin for 5 min, rinsed with phosphate-buffered saline, and incubated with a 1:100 dilution of horseradish peroxidase-conjugated polyclonal rabbit antibody to HSV-1 infected cells (DAKO Corporation, Carpenteria, Calif.) for 1 h. Excess antibody was removed, cultures were washed three times with phosphate-buffered saline, and cells were stained with aminoethylcarbazole (Vector Laboratories, San Francisco, Calif.).
Statistics.
Numerical data are presented as the mean ± standard error of the mean (SEM). The significance of differences in reactivation efficiency was determined by Fisher's exact test. The significance of differences in the time of reactivation (i.e., time at which reactivation was first detected and mean time to reactivation) was determined by one-way analysis of variance (ANOVA) followed by Tukey's post hoc t test. The equivalence of measurements of viral genome load and reactivation efficiency in TG explants and TG cell cultures was compared by a two-sided, paired t test.
RESULTS
Reactivation of KOS, n212, and 7134 from explanted latently infected TG.
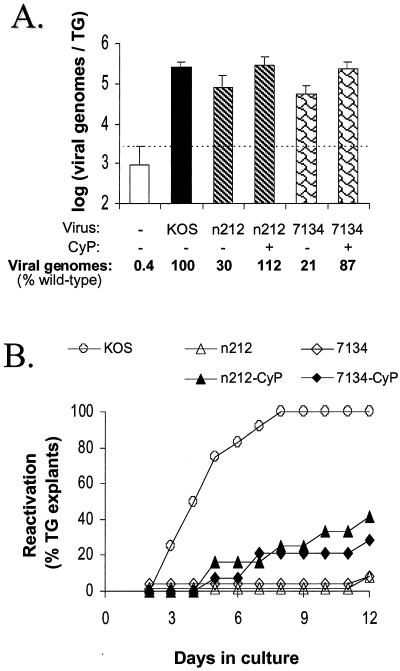
Administration of the immunosuppressive drug CyP to mice on days −1, 1, and 3 p.i. enables the ICP0− mutants n212 and 7134 to establish wild-type levels of latent genomes in mouse TG (11). Using this approach to equalize viral genome loads, the reactivation efficiencies of n212 and 7134 were compared to that of the wild-type virus KOS to determine if ICP0 is necessary for the efficient reactivation of HSV-1 from TG explants. As shown by the results of a representative experiment, competitive PCR demonstrated that wild-type levels of n212 and 7134 genomes were present in TG of CyP-treated mice on day 30 p.i. (Fig. 1A). In contrast, the numbers of n212 and 7134 genomes in TG of untreated mice were only 30 and 21% of wild-type levels, respectively (Fig. 1A). On day 30 p.i., the reactivation kinetics and efficiencies of KOS, n212, and 7134 in latently infected TG in response to the combined stimuli of explant and heat stress were compared. Latent KOS reactivated from 12 of 12 TG by day 8 postexplant (Table 1, experiment 1), and the mean time required to detect new infectious (reactivated) virus in culture medium was 4.8 ± 0.5 days (Fig. 1B). In contrast, and despite the presence of wild-type levels of mutant genomes in TG, reactivation of n212 and 7134 was detected in only 5 of 14 and 4 of 12 TG obtained from CyP-treated mice (Fig. 1B and Table 1, experiment 1). In addition to the reduced reactivation efficiency of n212 and 7134, the mean time required to detect reactivation of these mutants was significantly longer than that for KOS (8.0 ± 1.5 and 7.8 ± 1.7 days, respectively). As anticipated, reactivation of n212 and 7134 was detected in only 1 of 12 TG obtained from untreated mice (Fig. 1B and Table 1, experiment 1), in which the numbers of genomes were significantly lower than the number of genomes in TG latently infected with wild-type virus (Fig. 1A).
FIG. 1.
Viral genome loads and reactivation efficiencies of KOS, n212, and 7134 from latently infected TG explants. (A) Viral genome loads in TG latently infected with KOS, n212, or 7134 and derived from untreated or CyP-treated mice (n = 5 TG per group), as determined by competitive PCR assay. The dashed line indicates the lower limit of the quantitative range of the competitive PCR. Measurements below this line are not significantly different from background. Error bars indicate SEMs. (B) Efficiencies of reactivation from explanted TG latently infected with KOS, n212, or 7134 (n = 12 TG per group) and heat stressed at the time of explantation. On days 2 to 11 postexplant, reactivation was assessed by the presence or absence of infectious virus in cell culture medium. On day 12, reactivation was detected by the presence of infectious virus in homogenates of TG explants.
TABLE 1.
Viral genome loads and reactivation efficiencies of KOS, n212, and 7134 from explanted TG
| Expt | Reactivation frequency (viral genome load)a
|
||||
|---|---|---|---|---|---|
| KOS | n212-VEH | n212-CyP | 7134-VEH | 7134-CyP | |
| 1 | 12/12 (2.6 × 105) | 1/12 (7.9 × 104) | 4/12 (2.9 × 105) | 1/12 (5.5 × 104) | 4/14 (2.3 × 105) |
| 2 | 15/17 (3.0 × 105) | 1/12 (8.5 × 104) | 2/12 (2.1 × 105) | 0/12 (7.4 × 104) | 1/12 (2.8 × 105) |
| 3 | 12/12 (2.8 × 105) | NDb | 5/12 (3.1 × 105) | ND | 9/12 (2.7 × 105) |
| Sum of reactivation efficiencies | 39/41 [96% ± 5%] | 2/24** [12% ± 9%] | 11/36** [31% ± 9%] | 1/24** [4% ± 4%] | 14/38** [37% ± 24%] |
| Avg no. of viral genomes/TG | 2.8 × 105 | 8.2 × 104* | 2.7 × 105 | 6.4 × 104* | 2.6 × 105 |
The frequency of reactivation in TG explants is based on detection of infectious virus in culture supernatants or in homogenates of TG and indicator cells collected on day 12 postexplant. The viral genome load is expressed as the number of viral genomes per TG, as determined by competitive PCR of TG derived from mice (n = 5 per determination) in the same inoculation group used to establish TG explant cultures. This value is calculated as viral genomes per 100 ng of TG DNA (parameter measured by competitive PCR) × 300, because there is ∼30 μg of DNA in a mouse TG. *, P < 0.05 (H0: treatment group = KOS), based on comparison of viral genome loads by ANOVA and Tukey's post hoc t test. **, P < 0.001 (H0: treatment group = KOS), based on comparison of reactivation frequencies by Fisher's exact test. VEH, vehicle.
ND, not determined.
The combined results of several independent experiments are summarized in Table 1. Although KOS reactivated from 96% ± 5% of heat-stressed TG explants, reactivation of n212 and 7134 was detected in only 31% ± 9% and 37% ± 24% of heat-stressed TG explants derived from CyP-treated mice (Table 1). Therefore, in the absence of ICP0 and despite the presence of equal genome loads, the potent combined stimuli of TG explanation and heat stress failed to induce reactivation of n212 and 7134 as efficiently as reactivation of wild-type virus from latently infected TG.
Infectious-center assays of cells from TG latently infected with KOS, n212, or 7134.
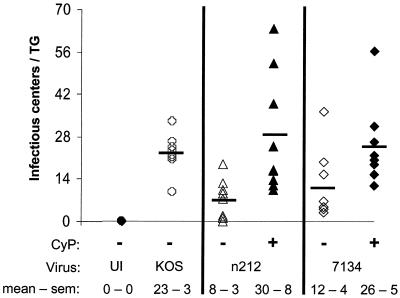
The infectious-center assay initially developed by Leib et al. (16) was used in the present study to determine the relative number of reactivatable, latently infected cells in TG of mice that were (i) uninfected, (ii) KOS infected, (iii) n212 infected, (iv) n212-infected and CyP treated, (v) 7134 infected, or (vi) 7134 infected and CyP treated (Fig. 2). Briefly, this ex vivo assay involves superinfecting dissociated TG cells with a replication-defective HSV-1 ICP4− mutant that expresses four functional IE proteins (ICPs 0, 22, 27, and 47), overlaying superinfected cells with permissive indicator cells, and counting the number of plaques that develop.
FIG. 2.
Numbers of KOS, n212, and 7134 infectious centers detected in latently infected TG. Infectious centers were initially determined in pairs of TG derived from untreated and CyP-treated mice that were either uninfected (UI) (n = 3 TG pairs) or latently infected with KOS, n212, or 7134 (n = 8 TG pairs per group), and the total numbers were divided by 2. The horizontal bars in each column indicate the average number of infectious centers observed in individual TG of each treatment group.
As expected, infectious centers were not detected in cultures derived from uninfected TG following superinfection with the ICP4− virus (Fig. 2). Cultures derived from TG latently infected with KOS, however, contained an average of 23 infectious centers per TG (Fig. 2). Cultures derived from TG of untreated mice latently infected with n212 and 7134 contained an average of 8 and 12 infectious centers per TG, respectively (Fig. 2). Cultures derived from TG of CyP-treated mice, however, contained an average 29 and 26 infectious centers per TG, respectively. Thus, although considerable animal-to-animal variation was observed, CyP treatment during acute infection increased the number of n212 and 7134 infectious centers established in each TG significantly (P < 0.05; two-sided t test). Therefore, CyP treatment produced an increase in the number of latently infected cells (i.e., infectious centers) per TG as well as an increase in the number of viral genomes per TG, both of which contributed to the increased reactivation frequency of ICP0− mutants.
Reactivation of KOS, n212, and 7134 during establishment of primary TG cell cultures.
Primary cultures of dissociated, latently infected TG cells can be used as an ex vivo model of HSV-1 reactivation (10, 18). TG cell cultures have an important advantage over TG explants, however, in that viable neurons are present in monolayers of dividing support cells such that drugs (10, 18) and viral expression vectors can be delivered efficiently to neurons and other cell types in culture. Before utilizing the model for these purposes, however, the reactivation efficiencies of KOS, n212, and 7134 in response to the transient, reactivation-inducing stimuli that accompany the establishment of TG cell cultures were compared (10, 18).
Using the pooled TG from eight mice latently infected with each virus as starting material, suspensions of dissociated TG cells were used to (i) test for the presence of infectious virus in freeze-thawed homogenates of TG cells, (ii) measure viral genome loads in DNA extracted from TG cells, and (iii) establish cultures of TG cells in the absence of ACV.
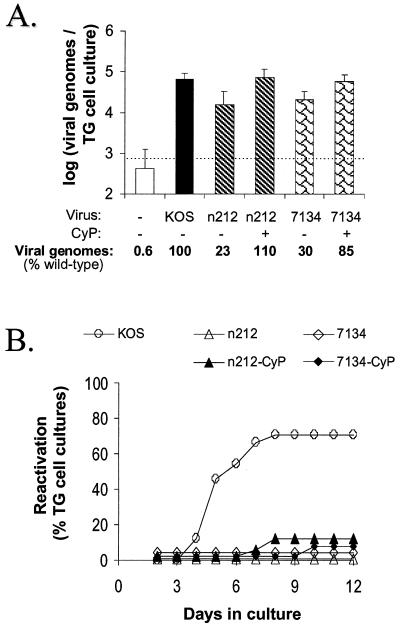
Notably, no infectious virus was detected in any TG cell suspension at the time of culture establishment in any of the experiments described below. In a representative experiment, competitive PCR demonstrated that wild-type levels of n212 and 7134 genomes were present in TG cells derived from CyP-treated mice (Fig. 3A). In contrast, the numbers of n212 and 7134 genomes in TG cells derived from untreated mice were only 23 and 30% of wild-type levels, respectively (Fig. 3A).
FIG. 3.
Viral genome loads and reactivation efficiencies of KOS, n212, and 7134 from primary cultures of latently infected TG cells. (A) Viral genome loads in aliquots of TG cells latently infected with KOS, n212, or 7134 derived from untreated or CyP-treated mice (n = 4 1-ml aliquots per group), as determined by competitive PCR. The dashed line indicates the lower limit of the quantitative range of the competitive PCR. Measurements below this line are not significantly different from background. Error bars indicate SEMs. (B) Efficiencies of reactivation from TG cells latently infected with KOS, n212, or 7134 (n = 24 TG cell cultures per group). On days 2 to 11 after culture establishment, reactivation was assessed by the presence or absence of infectious virus in culture medium. On day 12, reactivation was detected by the presence of infectious virus in TG cell culture suspensions prepared by sequential freeze-thawing and sonication.
Reactivation of KOS was detected in 17 of 24 TG cell cultures with a mean reactivation time of 5.5 ± 1.5 days postplating (Fig. 3B and Table 2, experiment 1). In contrast, and despite wild-type levels of ICP0− mutant genomes in TG cell cultures derived from CyP-treated mice (Fig. 3A), reactivation of n212 and 7134 was detected only in 2 of 24 and 1 of 24 cultures, respectively (Fig. 3B). Although reactivation of n212 and 7134 was not detected in TG cell cultures derived from untreated mice (Fig. 3B and Table 2, experiment 1), the numbers of n212 and 7134 genomes in these TG cell cultures were significantly lower than that of wild-type virus (Fig. 3A).
TABLE 2.
Reactivation of KOS, n212, and 7134 during establishment of TG cell cultures
| Expt | Reactivation frequency (input no. of viral genomes/culture)a
|
||||
|---|---|---|---|---|---|
| KOS | n212-VEH | n212-CyP | 7134-VEH | 7134-CyP | |
| 1 | 17/24 (6.7 × 104) | 0/24 (1.6 × 104) | 2/24 (7.3 × 104) | 0/24 (2.0 × 104) | 1/24 (5.7 × 104) |
| 2 | 11/24 (5.2 × 104) | 0/24 (1.4 × 104) | 2/24 (5.8 × 104) | 2/24 (1.3 × 104) | 1/24 (6.4 × 104) |
| 3 | 15/24 (5.4 × 104) | NDb | 1/24 (4.0 × 104) | ND | 1/24 (3.8 × 104) |
| Sum of reactivation frequencies | 43/72 [60% ± 9%] | 0/48** [<5%] | 5/72** [7% ± 2%] | 2/48** [<5%] | 3/72** [4% ± 0%] |
| Avg no. of viral genomes/TG | 5.8 × 104 | 1.5 × 104* | 5.7 × 104 | 1.7 × 104* | 5.3 × 104 |
The frequency of reactivation in TG cell cultures is based on detection of infectious virus in culture supernatants or in freeze-thawed TG cells collected on day 12 after culture establishment. The input number of viral genomes per TG cell culture was determined by competitive PCR of TG cells (n = 4 aliquots per determination) at the time of culture establishment. This value is calculated as viral genomes per 100 ng of TG DNA (parameter measured by competitive PCR) × 70, because aliquots of TG cells contained ∼7 μg of DNA. *, P < 0.05 (H0: treatment group = KOS), based on comparison of viral genome loads by ANOVA and Tukey's post hoc t test. **, P < 0.001 (H0: treatment group = KOS), based on comparison of reactivation frequencies by Fisher's exact test. VEH, vehicle.
ND, not determined.
The combined results of several independent experiments are summarized in Table 2. Although KOS reactivated in 60% ± 9% of TG cell cultures, reactivation of n212 and 7134 was detected in only 7% ± 2% and 4% ± 0% of cultures derived from CyP-treated mice (Table 2). Thus, in the absence of ICP0, the transient stress associated with the establishment of TG cell cultures did not induce wild-type levels of reactivation of n212 or 7134.
Comparison of ex vivo reactivation models: TG explants versus TG cell cultures.
To date, the TG cell culture model of HSV-1 reactivation has not been characterized extensively (10, 18). Therefore, it was not known how well measurements of viral genome loads and reactivation efficiencies made in TG explants would compare with similar measurements made in TG cell cultures. To compare the two models, the TG explant experiments described in Table 1 were performed in parallel with the TG cell culture experiments described in Table 2 using mice from the same inoculation groups. The results of these comparisons are presented in Table 3.
TABLE 3.
Comparison of measurements obtained from TG explants and TG cell cultures
| Virus | Exptg | Log viral genomes per 100 ng of TG DNAab
|
Viral genomes per culturecd
|
% Reactivationef
|
|||
|---|---|---|---|---|---|---|---|
| Explant | Cells | Explant | Cells | Explant | Cells | ||
| KOS | 1 | 2.94 | 2.98 | 2.6 × 105 | 6.7 × 104 | 100 | 71 |
| 2 | 3.00 | 2.87 | 3.0 × 105 | 5.2 × 104 | 88 | 46 | |
| 3 | 2.97 | 2.89 | 2.8 × 105 | 5.4 × 104 | 100 | 62 | |
| n212 | 1 | 2.42 | 2.35 | 7.9 × 104 | 1.6 × 104 | 8 | 0 |
| 2 | 2.45 | 2.30 | 8.5 × 104 | 1.4 × 104 | 8 | 0 | |
| n212-CyP | 1 | 2.99 | 3.02 | 2.9 × 105 | 7.3 × 104 | 33 | 8 |
| 2 | 2.85 | 2.92 | 2.1 × 105 | 5.8 × 104 | 17 | 8 | |
| 3 | 3.02 | 2.76 | 3.1 × 105 | 4.0 × 104 | 42 | 4 | |
| 7134 | 1 | 2.26 | 2.46 | 5.5 × 104 | 2.0 × 104 | 8 | 0 |
| 2 | 2.39 | 2.27 | 7.4 × 104 | 1.3 × 104 | 0 | 8 | |
| 7134-CyP | 1 | 2.88 | 2.91 | 2.3 × 105 | 5.7 × 104 | 29 | 4 |
| 2 | 2.97 | 2.96 | 2.8 × 105 | 6.4 × 104 | 8 | 4 | |
| 3 | 2.95 | 2.73 | 2.7 × 105 | 3.8 × 104 | 75 | 4 | |
The logarithm of the concentration of viral genomes, as measured by competitive PCR, in individual TG explants and TG cell cultures at the time of culture establishment.
The mean ± standard error of the fold differences in viral genome concentration and total genome load in TG explants versus TG cells is 1.02 ± 0.01.
The total number of viral genomes in individual TG explants and TG cell cultures was calculated by multiplying the concentration of viral genomes by the total amount of DNA in a TG explant (∼30 μg) or an aliquot of dissociated TG cells (∼7 μg).
The mean ± standard error of the fold differences in viral genome concentration and total genome load in TG explants versus TG cells is 4.37 ± 0.06.
Percent reactivation in latently infected TG explants (n = 12 to 17 per determination) and TG cell cultures (n = 24 per determination).
The mean ± standard error of differences in reactivation efficiency observed in TG explants versus TG cells is 23 ± 4 (P < 0.05 [H0: difference = 0], based on comparison of measurements of [i] log viral genome concentration and [ii] percent reactivation in TG explants and TG cell cultures by two-sided paired t tests).
Competitive PCR was used to measure the concentrations of KOS, n212, and 7134 genomes in DNA isolated from (i) individual TG or (ii) 1-ml aliquots of dissociated TG cell suspension. As shown in Table 3, competitive PCR measurements of viral genome concentrations in total DNA were equivalent in individual TG and in dissociated TG cells when TG were obtained from the same inoculation group of mice. Although the total number of latent viral genomes was approximately 4.5-fold higher in individual TG than in TG cell cultures, this simply reflects the fact that the total amount of DNA in an individual TG is approximately 4.5-fold greater than the total amount of DNA in a 1-ml aliquot of dissociated TG cell suspension (Table 3). Comparison of reactivation efficiencies from TG explants and TG cell cultures demonstrated that KOS, n212, and 7134 reactivated with significantly higher efficiencies from heat-stressed TG explants than from primary cultures of dissociated TG cells not subjected to heat stress (Table 3). Although heat stress and other factors likely play a role in the enhanced reactivation frequency from TG explants, the ∼4.5-fold-greater number of latent viral genomes in individual TG is a likely explanation for the increased frequency of reactivation of all three viruses from TG explants relative to TG cell cultures.
Reactivation of KOS, n212, and 7134 from ACV-treated, latently infected TG cell cultures following induction by heat stress or viral superinfection.
The capacities of KOS, n212, and 7134 to reactivate from latently infected TG cell cultures in response to heat stress or superinfection with an ICP4− virus that overexpresses ICP0 were compared. Cultures were established from TG of mice latently infected with KOS, n212 (CyP treated only), or 7134 (CyP treated only) in the presence of 200 μM ACV. As previously noted, antiviral drugs such as ACV inhibit reactivation during the establishment of TG cell cultures (10, 18).
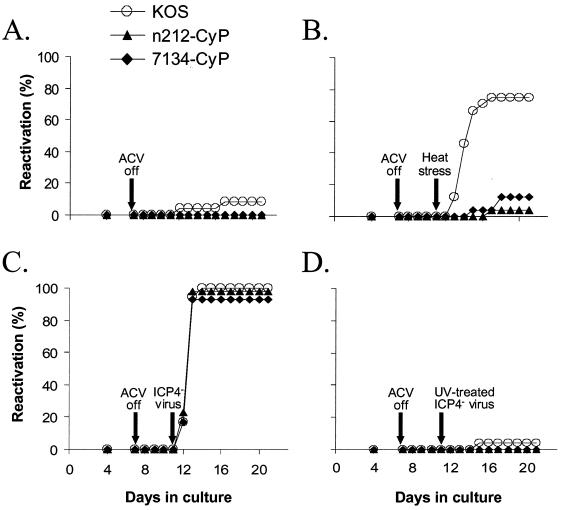
At the time of plating (day 0), competitive PCR revealed no significant differences in KOS, n212, or 7134 genome loads in total DNA from TG cell suspensions (one-way ANOVA, P = 0.43) (data not shown). On day 7 postplating, ACV-containing medium was replaced with ACV-free medium, and on day 11, cultures were either (i) not treated (Fig. 4A), (ii) heat stressed (Fig. 4B), (iii) superinfected with the ICP4− virus n12, (Fig. 4C), or (iv) superinfected with the same number of PFU of UV-inactivated ICP4− virus (Fig. 4D).
FIG. 4.
Reactivation efficiencies of KOS, n212, and 7134 from latently infected TG cell cultures that were heat stressed or superinfected with an ICP4− mutant virus. At the time of cell culture preparation, cultures were incubated in medium containing 200 μM ACV to inhibit virus replication. On day 7, ACV-containing medium was replaced with ACV-free medium. On day 11, cultures were left untreated (A), subjected to heat stress (B), superinfected with an ICP4− virus (C), or superinfected with a UV-inactivated ICP4− virus (D). On day 4 and days 7 to 21 after culture establishment, reactivation was assessed by the presence or absence of infectious virus in the culture medium.
In the absence of a specific stressor, reactivation of latent KOS, n212, and 7134 was detected in 8, 0, and 0% of TG cell cultures, respectively, following ACV removal (Fig. 4A). Following removal of ACV and the application of heat stress, reactivation was detected in 75% of KOS-infected TG cell cultures (Fig. 4B) but in only 4 and 13% of n212- and 7134-infected cultures, respectively (Fig. 4B). When cultures derived from the same groups of latently infected mice were superinfected with a replication-defective ICP4− virus, reactivation was detected in 100% of TG cell cultures latently infected with KOS, n212, or 7134 by 72 h postsuperinfection (Fig. 4C), indicating that superinfection with n12 constitutes a highly efficient means of inducing reactivation. As expected, infectious virus was not recovered following ICP4− virus infection of 12 uninfected TG cell cultures (data not shown). Also as expected, UV irradiation destroyed the capacity of the ICP4− virus to induce reactivation in cultures of TG cells latently infected with any of the three viruses (Fig. 4D), demonstrating that expression of one or more IE protein (ICPs 0, 22, 27, and 47) is essential to induce reactivation in these tests.
The combined results of several independent experiments are summarized in Table 4. Although KOS reactivated in 69% ± 7% of heat-stressed TG cell cultures, reactivation of n212 and 7134 was detected in only 1% ± 2% and 7% ± 4% of cultures treated with this potent reactivation-inducing stimulus. The failure of heat stress to induce n212 and 7134 reactivation was not due to the absence of latent genomes, because superinfection with an ICP4− virus induced reactivation in nearly 100% of TG cell cultures latently infected with n212 or 7134. Therefore, in the absence of ICP0, heat stress failed to induce reactivation of n212 and 7134 efficiently in latently infected TG cell cultures.
TABLE 4.
Induced reactivation of KOS, n212, and 7134 from TG cell cultures
| Stressor | % Reactivation frequency (n)a
|
||
|---|---|---|---|
| KOS | n212-CyP | 7134-CyP | |
| None | 13 ± 5 (36) | 2 ± 2 (56) | 0 ± 0 (36) |
| Heat stress | 69 ± 7** (72) | 1 ± 2 (76) | 7 ± 4 (72) |
| ICP4− virus | 100 ± 0** (53) | 95 ± 5** (53) | 100 ± 0** (41) |
| UV-treated ICP4− virus | 2 ± 2 (41) | 0 ± 0 (24) | 0 ± 0 (24) |
Mean ± standard error of reactivation frequency observed in two or three independent experiments, based on the detection of infectious virus in culture supernatants within 10 days after stress (i.e., day 21 after culture establishment). n, total number of cultures tested per treatment group. **, P < 0.001, for each virus (H0: treated cultures = unstressed cultures), based on comparison of reactivation frequencies (i.e., expressed as fractions) by Fisher's exact test.
Detection of HSV-1 antigens in TG cell cultures latently infected with KOS, n212, or 7134.
Once reactivation of KOS was detected in a given culture well, infectious virus was detected daily thereafter. Unlike reactivation of KOS, however, reactivation of n212 and 7134 from TG cell cultures was difficult to verify by daily transfer of culture medium to L7 indicator cells. Thus, when infectious n212 or 7134 was detected in a given well, infectious virus was often undetectable in the same well on the remaining 4 to 6 days of an experiment. Therefore, a second method was employed to assess reactivation of n212 and 7134. Specifically, 10 days after heat stress, cultures were fixed, stained immunocytochemically, and examined for the presence of viral antigens by light microscopy.
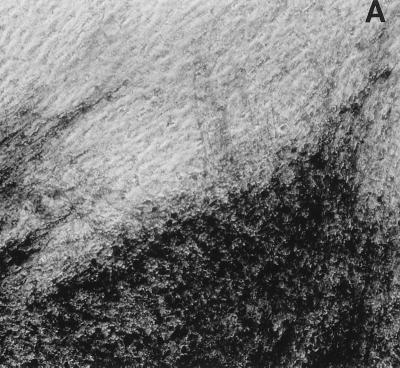
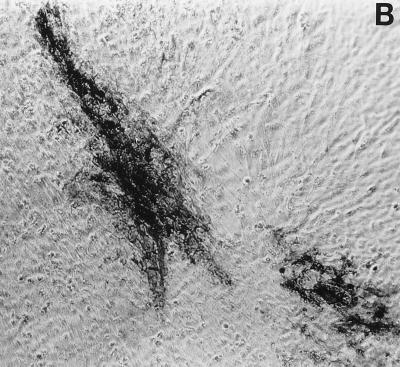
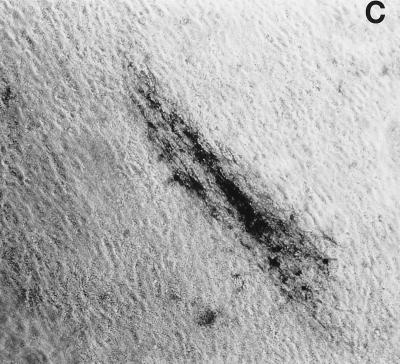
Immunocytochemical staining of TG cell cultures latently infected with KOS demonstrated an absolute correlation between the detection of HSV antigens in TG cells on day 10 post-heat stress (Fig. 5A) and the earlier detection of infectious KOS in culture medium. As expected, HSV antigens were not detected in cultures in which infectious KOS had not been detected. In n212- and 7134- infected cultures, however, the frequency of detection of HSV antigens (Fig. 5B and C) was twice as high as the frequency of detection of infectious virus in culture media (2 of 76 and 10 of 72, respectively). Moreover, foci of HSV antigen-positive cells in n212- and 7134-infected cultures were noticeably smaller than those observed in KOS-infected cultures (Fig. 5). Collectively, these results indicate that relative to KOS, the ICP0− mutant viruses are markedly impaired in their ability to express viral antigens, synthesize new infectious virus, and spread from cell to cell in TG cell cultures. Thus, even if one used the presence of detectable HSV antigen in TG cell cultures as a measure of reactivation, n212 and 7134 produced fewer, smaller foci in heat-stressed TG cell cultures (3 and 14%, respectively) than wild-type virus (69%).
FIG. 5.
Foci of HSV antigen expression in TG cell cultures infected with KOS (A), n212 (B), or 7134 (C) on day 10 after heat stress. Magnification, ×10. Viral antigens were visualized using horseradish peroxidase-conjugated polyclonal rabbit antibody to HSV-1 and the substrate aminoethylcarbazole.
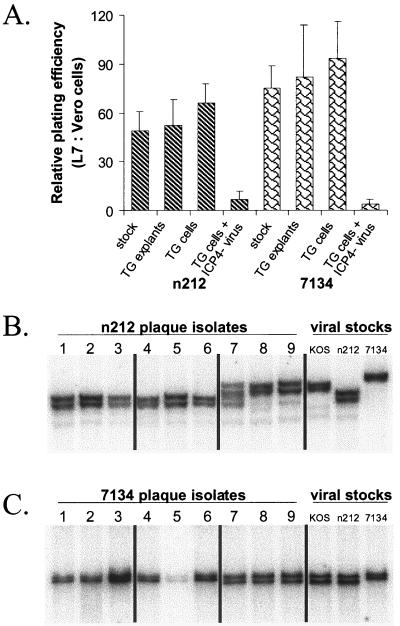
Stability of the mutations in n212 and 7134 in vivo.
During acute infection of mice (days 1 to 9 p.i.), no evidence of reversion of the ICP0− mutants to wild type was detected, based on the fact that isolates of n212 and 7134 from tear film consistently produced far more plaques on ICP0-complementing L7 cells than on the parental Vero cell line (data not shown; more than 50 independent plaque isolates per virus were tested). Similarly, isolates of n212 and 7134 derived from reactivating TG explants or TG cell cultures exhibited the ICP0− phenotype, based on their ∼50- to 100-fold-greater plating efficiency on L7 than on Vero cells (Fig. 6A). As anticipated, viral isolates obtained from TG cell cultures latently infected with n212 and 7134 and superinfected with the ICP4− virus contained a large proportion of ICP0+ recombinant virus, based on the fact that the titers of individual plaque isolates were only two- to threefold higher on L7 cells than on Vero cells (Fig. 6A). Because high-frequency recombination occurs in the joint region between the ICP0 and ICP4 genes (20, 23, 27) these results demonstrate that coreplication of latent ICP0− genomes (n212 or 7134) and superinfecting ICP4− genomes (n12) leads to the production of ICP0+ ICP4+ recombinant viruses which have a strong selective advantage over both parental viruses.
FIG. 6.
Stability of phenotypes and genotypes of n212 and 7134 reactivation isolates relative to viral stocks used for ocular inoculation. (A) Ratios of relative plating efficiencies (L7 versus Vero cells) of the n212 and 7134 viral stocks used to inoculate mice (n = 3 independent tests per virus), of isolates obtained from reactivating TG latently infected with n212 and 7134 (n = 6 isolates per group), of isolates obtained from TG cell cultures that reactivated during culture establishment or following heat stress (n = 6 isolates per group), and of isolates obtained from latently infected TG cell cultures superinfected with ICP4− virus (n = 17 isolates per group). Error bars indicate SEMs. (B and C) Southern blot analysis of viral DNAs from individual reactivation isolates of n212 (B) and 7134 (C) compared with n212 and 7134 virus stocks used for ocular inoculation. Total DNA (3 μg) from n212-infected L7 cells was digested with NcoI and SpeI, and total DNA from 7134-infected L7 cells was digested with NotI. Following eletrophoretic separation of DNA fragments on a 0.8% agarose gel and transfer of separated fragments to nitrocellulose, blots were hybridized to an ICP0-specific oligonucleotide. From left to right, the samples on each blot include DNAs of three independent reactivation isolates obtained from latently infected TG explants (lanes 1 to 3), latently infected TG cells that reactivated during culture establishment or following heat stress (lanes 4 to 6), or latently infected TG cell cultures superinfected with ICP4− virus (lanes 7 to 9). The controls were total DNAs obtained from L7 cells infected with viral stocks of KOS, n212, or 7134.
Southern blot analysis was used as a second test to determine if mutations in the ICP0 gene were retained in n212 and 7134 reactivation isolates. NcoI-SpeI digests confirmed that the SpeI linker insertion in codon 212 was present in all plaque isolates from TG explants and TG cell cultures latently infected with n212 (Fig. 6B, lanes 1 to 6). In contrast, all three isolates obtained from cultures of TG cells latently infected with n212 and superinfected with the ICP4− mutant contained either a mixture of n212 and wild-type alleles in the ICP0 locus (Fig. 6B, lane 7), or predominantly wild-type ICP0 (Fig. 6B, lanes 8 and 9). Likewise, NotI digestion confirmed that all six isolates obtained from TG explants and TG cell cultures latently infected with 7134 were genotypically identical to the 7134 stock used to inoculate mice (Fig. 6B, lanes 1 to 6). In contrast, all three isolates obtained from cultures of TG cells latently infected with 7134 and superinfected with ICP4− virus contained predominantly wild-type ICP0 (Fig. 6C, lanes 7 to 9). Therefore, reversion or repair of the n212 and 7134 mutations did not occur in TG explants or in TG cell cultures that were not superinfected with an ICP4− virus.
DISCUSSION
ICP0 is necessary for efficient reactivation from latency.
The results of the present study provide conclusive evidence that expression of ICP0 is necessary for the efficient reactivation of HSV-1 from neuronal latency. Previous attempts to establish a role for ICP0 in reactivation have been inconclusive because ICP0− mutants failed to establish wild-type levels of viral genomes in latently infected TG (2, 5, 16). In the present study, transient immunosuppression with CyP was used to eliminate differences in viral genome load, and thus the comparison was restricted to latent HSV-1 genomes that either did or did not express ICP0 during reactivation. In multiple tests, reactivation of n212 and 7134 was significantly and reproducibly less efficient than that of wild-type virus.
With regard to the ICP0− mutants used in this study, a previously characterized rescuant of 7134, 7134R, replicates as efficiently as wild-type virus in vivo and reactivates from latently infected TG with wild-type kinetics and efficiency (2). Therefore, the phenotypes of 7134 described herein can be ascribed to the deletion in the ICP0 gene (which also removes the portion of the LAT gene that lies on the complementary strand). Because a rescuant of n212 has not yet been constructed, it remains a formal possibility that the phenotypes of n212 may be influenced by secondary mutations acquired during construction of the virus. It should be noted, however, that the in vitro replication defects of n212 are (i) virtually indistinguishable from those of 7134 (2, 19) and are (ii) fully reversed when ICP0 is provided in trans (unpublished observations). It remains to be formally proven that ICP0 provided in trans would be sufficient to complement the reactivation deficiencies of n212 and 7134.
Superinfection of TG cell cultures with an ICP4− virus.
Despite its potency as a reactivation stimulus, heat stress did not induce reactivation of n212 and 7134 efficiently in latently infected TG cell cultures. Superinfection with an ICP4− virus, however, induced reactivation in nearly 100% of cultures latently infected with n212 or 7134 derived from the same preparation of dissociated TG cells. Because UV irradiation destroyed this activity, reactivation was dependent on expression of at least one of the IE genes (i.e., ICPs 0, 22, 27, and 47). At a minimum, the results confirmed that biologically competent n212 and 7134 genomes were present in nearly 100% of the TG cell cultures. Thus, inefficient reactivation of n212 and 7134 following heat stress cannot be attributed to defects in the establishment or maintenance of latent ICP0− mutant genomes.
The precise mechanism by which the ICP4− mutant induces reactivation of n212 and 7134 from TG cell cultures has yet to be determined. The production of infectious virus was dependent on the presence of latent n212 and 7134 genomes in TG cells, but the majority of the progeny of these reactivation events lacked the mutant ICP0 alleles present in n212 and 7134. This is not surprising, because high-frequency recombination occurs between the L (location of the ICP0 gene) and S (location of the ICP4 gene) regions of the HSV genome during viral replication (20, 23, 27). At this writing, it remains to be determined whether expression of ICP0 is critical to the capacity of an ICP4− virus to trigger HSV reactivation in latently infected TG cell cultures.
Other potential effects of CyP treatment.
Transient administration of CyP was used to increase both the number of latent n212 and 7134 genomes and the number of latently infected cells in mouse TG. In theory, CyP treatment on days −1, 1, and 3 p.i. may also have affected other parameters of the establishment of latency in mice. Unfortunately, it is not possible to control for this variable, because KOS infection is uniformly lethal to mice treated with the CyP regimen used in this study (11). The drug likely had no direct effect on the outcome of reactivation experiments, because CyP is metabolized within hours following in vivo administration (1). Likewise, all known parameters of immune function have been shown to return to normal within 10 days after termination of CyP treatment (17, 26). In the present study, it was evident that reactivation of n212 and 7134 occurred more efficiently in TG and TG cell cultures derived from CyP-treated mice than in those obtained from non-drug-treated mice. Although other possibilities cannot be excluded, the simplest interpretation of this observation is that TG of CyP-treated mice contained ∼3 to 4 times as many latent ICP0− mutant genomes and 2 to 4 times as many latently infected cells as those of untreated mice.
What role does ICP0 play in the HSV-1 reactivation process?
Latency in the herpesvirus field has long been defined as the absence of infectious virus in cells or tissues that contain viral genomes, whereas reactivation has been defined as the de novo production of new infectious virus from cells containing latent viral genomes (13, 21). Based on these definitions, the present study demonstrates that ICP0 is necessary for the efficient reactivation of HSV-1 from latency. Production of wild-type levels of infectious virus from latently infected cells is dependent on the capacity of virus-infected cells to (i) initiate lytic-phase gene expression, (ii) initiate HSV DNA replication, (iii) produce new infectious virus, and (iv) transport virus to nonneuronal cells at the site of primary infection. Therefore, the inconsistent recovery of infectious n212 and 7134 from heat-stressed TG cell cultures suggests that the impaired reactivation of ICP0− mutants could, in theory, occur at any of these four levels. Based on the relative sensitivity of immunocytochemical staining (i.e., groups of 10 HSV antigen-positive cells are readily detected), the failure to detect n212 and 7134 antigens in 97 and 86% of heat-stressed TG cell cultures, respectively, suggests that ICP0 was required very early in the reactivation process.
Given ICP0's demonstrated role as a global transcriptional activator during productive infection (4, 9, 15), we favor the hypothesis that ICP0 contributes to efficient reactivation either by initiating lytic-phase gene expression in latently infected neurons or by sustaining viral gene expression once it has been initiated by cellular (or other viral) factors. As a first step towards addressing this hypothesis, studies to determine if ICP0 is sufficient to trigger HSV-1 reactivation in latently infected neurons are in progress.
ACKNOWLEDGMENTS
This investigation was supported by Public Health Service Program Project grant P01 NS 35138 from the National Institute of Neurological Disorders and Stroke. W.P.H. was the recipient of individual National Research Service Award F32 AI 10147 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Berenbaum M C, Cope W A, Double J A. The effect of microsomal enzyme inhibition in the immunosuppressive and toxic effects of cyclophosphamide. Clin Exp Immunol. 1973;14:257–270. [PMC free article] [PubMed] [Google Scholar]
- 2.Cai W, Astor T L, Liptak L M, Cho C, Coen D M, Schaffer P A. The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J Virol. 1993;67:7501–7512. doi: 10.1128/jvi.67.12.7501-7512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai W, Schaffer P A. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J Virol. 1989;63:4579–4589. doi: 10.1128/jvi.63.11.4579-4589.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai W, Schaffer P A. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J Virol. 1992;66:2904–2915. doi: 10.1128/jvi.66.5.2904-2915.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clements G B, Stow N D. A herpes simplex virus type 1 mutant containing a deletion within immediate early gene 1 is latency-competent in mice. J Gen Virol. 1989;70:2501–2506. doi: 10.1099/0022-1317-70-9-2501. [DOI] [PubMed] [Google Scholar]
- 6.DeLuca N A, McCarthy A M, Schaffer P A. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J Virol. 1985;56:558–570. doi: 10.1128/jvi.56.2.558-570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLuca N A, Schaffer P A. Activities of herpes simplex virus type 1 (HSV-1) ICP4 genes specifying nonsense peptides. Nucleic Acids Res. 1987;15:4491–4511. doi: 10.1093/nar/15.11.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devi-Rao G B, Goodart S A, Hecht L M, Rochford R, Rice M A, Wagner E K. Relationship between polyadenylated and nonpolyadenylated herpes simplex virus type 1 latency-associated transcript. J Virol. 1991;65:2179–2190. doi: 10.1128/jvi.65.5.2179-2190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everett R D, Orr A, Preston C M. A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J. 1998;17:7161–7169. doi: 10.1093/emboj/17.24.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halford W P, Gebhardt B M, Carr D J J. Mechanisms of herpes simplex virus type 1 reactivation. J Virol. 1996;70:5051–5060. doi: 10.1128/jvi.70.8.5051-5060.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halford W P, Schaffer P A. Optimized viral dose and transient immunosuppression enable herpes simplex virus ICP0-null mutants to establish wild-type levels of latency in vivo. J Virol. 2000;74:5957–5967. doi: 10.1128/jvi.74.13.5957-5967.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris R A, Everett R D, Zhu X X, Silverstein S, Preston C M. Herpes simplex virus type 1 immediate-early protein Vmw110 reactivates latent herpes simplex virus type 2 in an in vitro latency system. J Virol. 1989;63:3513–3515. doi: 10.1128/jvi.63.8.3513-3515.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill T J. Herpes simplex virus latency. In: Roizman B, editor. The herpesviruses. Vol. 3. New York, N.Y: Plenum Press; 1985. pp. 175–240. [Google Scholar]
- 14.Institute of Laboratory Animal Resources. Guide for the care and use of laboratory animals. Washington, D.C.: National Academy Press; 1996. [Google Scholar]
- 15.Jordan R, Schaffer P A. Activation of gene expression by herpes simplex virus type 1 ICP0 occurs at the level of mRNA synthesis J. Virol. 1997;71:6850–6862. doi: 10.1128/jvi.71.9.6850-6862.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leib D A, Coen D M, Bogard C L, Hicks K A, Yager D R, Knipe D M, Tyler K L, Schaffer P A. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J Virol. 1989;63:759–768. doi: 10.1128/jvi.63.2.759-768.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marbrook J, Baguley B C. The recovery of immune responsiveness after treatment with cyclophosphamide. Int Arch Allergy. 1971;41:802–812. doi: 10.1159/000230572. [DOI] [PubMed] [Google Scholar]
- 18.Moriya A, Yoshiki A, Kita M, Fushiki S, Imanishi J. Heat shock-induced reactivation of herpes simplex virus type 1 in latently infected mouse trigeminal ganglion cells in dissociated culture Arch. Virol. 1994;135:419–425. doi: 10.1007/BF01310025. [DOI] [PubMed] [Google Scholar]
- 19.Mossman K L, Saffran H A, Smiley J R. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J Virol. 2000;74:2052–2056. doi: 10.1128/jvi.74.4.2052-2056.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parris D S, Dixon R A, Schaffer P A. Physical mapping of herpes simplex virus type 1 ts mutants by marker rescue: correlation of the physical and genetic maps. Virology. 1980;100:275–287. doi: 10.1016/0042-6822(80)90519-x. [DOI] [PubMed] [Google Scholar]
- 21.Preston C M. Repression of viral transcription during herpes simplex virus latency. J Gen Virol. 2000;81:1–19. doi: 10.1099/0022-1317-81-1-1. [DOI] [PubMed] [Google Scholar]
- 22.Samaniego L A, Wu N, DeLuca N A. The herpes simplex virus immediate-early protein ICP0 affects transcription from the viral genome and infected-cell survival in the absence of ICP4 and ICP27. J Virol. 1997;71:4614–4625. doi: 10.1128/jvi.71.6.4614-4625.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaffer P A, Tevethia M J, Benyesh-Melnick M. Recombination between temperature-sensitive mutants of herpes simplex virus type 1. Virology. 1974;58:219–228. doi: 10.1016/0042-6822(74)90156-1. [DOI] [PubMed] [Google Scholar]
- 24.Smith K O. Relationship between the envelope and the infectivity of herpes simplex virus. Proc Soc Exp Biol Med. 1964;115:814–816. doi: 10.3181/00379727-115-29045. [DOI] [PubMed] [Google Scholar]
- 25.Treco D A. Preparation and analysis of DNA. In: Ausubel F M, editor. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1990. pp. 2.0.3–2.2.3. [Google Scholar]
- 26.Turk J L, Poulter L W. Effect of cyclophosphamide on lymphoid tissue labeled with 5-iodo-2-deoxyuridine- 125I and 51Cr. Int Arch Allergy. 1972;43:620–629. doi: 10.1159/000230874. [DOI] [PubMed] [Google Scholar]
- 27.Wilkie N M, Stow N D, Marsden H S, Preston V, Cortini R, Timbury M C, Subak-Sharpe J H. Physical mapping of herpes simplex virus-coded functions and polypeptides by marker rescue and analysis of HSV-1/HSV-2 intertypic recombinants. IARC Sci Publ. 1978;24:11–31. [PubMed] [Google Scholar]
- 28.Zhu X, Chen J, Young C J H, Silverstein S. Reactivation of latent herpes simplex virus by adenovirus recombinants encoding mutant IE-0 gene products. J Virol. 1990;64:4489–4498. doi: 10.1128/jvi.64.9.4489-4498.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]