Abstract
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV) (also known as human herpesvirus 8) latently infects KS tumors, primary effusion lymphomas (PELs), and PEL cell lines. In latently infected cells, KSHV DNA is maintained as circularized, extrachromosomal episomes. To persist in proliferating cells, KSHV episomes must replicate and efficiently segregate to progeny nuclei. In uninfected B-lymphoblastoid cells, KSHV latency-associated nuclear antigen (LANA1) is necessary and sufficient for persistence of artificial episomes containing specific KSHV DNA. In previous work, the cis-acting sequence required for episome persistence contained KSHV terminal-repeat (TR) DNA and unique KSHV sequence. We now show that cis-acting KSHV TR DNA is necessary and sufficient for LANA1-mediated episome persistence. Furthermore, LANA1 binds TR DNA in mobility shift assays and a 20-nucleotide LANA1 binding sequence has been identified. Since LANA1 colocalizes with KSHV episomes along metaphase chromosomes, these results are consistent with a model in which LANA1 may bridge TR DNA to chromosomes during mitosis to efficiently segregate KSHV episomes to progeny nuclei.
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV) or human herpesvirus 8 is a gamma-2 herpesvirus tightly linked to KS, primary effusion lymphoma (PEL), and multicentric Castleman's disease, an aggressive lymphoproliferative disorder (8, 9, 35, 47). KSHV infection in tumors and PEL cell lines is predominantly latent. Latently infected cells have multiple copies of extrachromosomal, circularized KSHV DNA (episomes) (8, 13). To persist in proliferating cells, viral episomes must first replicate and then segregate to progeny cells.
The latency-associated nuclear antigen (LANA1), encoded by KSHV open reading frame 73, is one of a limited number of KSHV genes expressed during latent infection (23, 24, 40). Confocal microscopy using immunofluorescence and fluorescent in situ hybridization demonstrated that LANA1 colocalized with KSHV episomes in PEL cell nuclei in interphase and along mitotic chromosomes (4, 12, 22–24, 48). Furthermore, in KSHV-uninfected lymphoblastoid cells, LANA1 mediates extrachromosomal persistence of artificial KSHV episomes containing specific KSHV DNA (4). The KSHV Z6 cosmid, which includes KSHV terminal-repeat (TR) elements and the left end of the KSHV genome, persisted as an episome in LANA1-expressing cells. In contrast, the Z8 cosmid, which contains sequence from near the center of the KSHV genome, did not persist as an episome in LANA1-expressing cells. Further, DNA containing the left end of Z6, but not other Z6 segments, persisted as episomes in LANA1-expressing cells (4, 43). Here we show that KSHV TR sequence is necessary and sufficient for LANA1-mediated episome persistence.
The ability of LANA1 to bind to TR DNA was also investigated since such an interaction would be compatible with a role in episome maintenance. Consistent with this possibility, LANA1 bound a segment of Z6 DNA which includes the TR elements (12). Both the Epstein-Barr virus (EBV) nuclear antigen 1 (EBNA1) and bovine papillomavirus E2 protein directly bind DNA and are hypothesized to tether cognate viral DNA to chromosomes in order to mediate episome segregation to progeny cells (5, 21, 30, 41, 46, 50, 52). For instance, EBNA1 binds to multiple sites within a cis-acting 1.8-kb EBV DNA sequence termed origin of plasmid replication (oriP) to mediate episome persistence (6, 41, 50, 52). Here we define a 20-nucleotide LANA1 binding site corresponding to nt 603 to 622 of the KSHV TR (43). Since LANA1 colocalizes with KSHV episomes along mitotic chromosomes and binds TR DNA, these results are consistent with a model in which LANA1 directly binds KSHV episomes to mediate episome persistence.
MATERIALS AND METHODS
Plasmids.
pRepCK has the sequence between ClaI and KpnI of pRep9 (Invitrogen) deleted. Plasmid Z6-BE contains the BglII-EcoRI fragment from the Z6-13 plasmid and has about eight copies of the TR unit and unique KSHV sequence cloned into the BamHI-XhoI sites of the pRepCK vector (4). Z6-BE was digested with NotI, and the 0.8- and ∼0.6-kb KSHV DNA fragments were purified and ligated into the NotI site of the pRepCK vector to generate Z6-3TRA, Z6-2TR, Z6-1TR, and Z6-A. Z6-3TRA contains three copies of the 0.8-kb TR unit and one copy of the 0.6-kb sequence. Z6-2TR and Z6-1TR contain two copies and one copy, respectively, of the 0.8-kb TR unit. Z6-A contains one copy of the 0.6-kb KSHV DNA (termed A), which was sequenced at a core facility by automated sequencing.
Selection of G418-resistant cells and Gardella gel analysis.
BJAB (KSHV-uninfected) B-lymphoblastoid cells or hygromycin (200 U/ml) (Calbiochem)-resistant BJAB cells stably expressing FLAG epitope-tagged LANA1 (BJAB/F-LANA1) (4) were transfected in 400 μl at 200V and 960 μF in a 0.4-cm-gap cuvette using a Bio-Rad Electroporator with Z6-BE, Z6-3TRA, Z6-2TR, Z6-1TR, or Z6-A. After 48 h, the cells were plated in 96-well microtiter plates (1,000 cells/well) in medium containing G418 (600 μg/ml) (Gibco). G418 resistance is conferred by the plasmid vector. After selection of G418-resistant cell lines, Gardella gel analysis was performed by in situ lysis of cells in gel-loading wells with pronase and sodium dodecyl sulfate and electrophoresis in 1× Tris-borate-EDTA (TBE) (17). DNA was transferred to a nylon membrane, and KSHV DNA was detected by Southern blot analysis using a 32P-labeled TR probe. Signal was captured with a Molecular Dynamics PhosphorImager and analyzed with ImageQuant software.
DNA binding assay.
The 535-bp NotI-AscI and 266-bp NotI-AscI TR fragments were gel purified after digestion of the Z6-2TR plasmid. After digestion of the 535-bp fragment with AvaII, the 297-bp NotI-AvaII fragment was isolated. The 535-bp fragment was also digested with Sau3A, and the 370-bp fragment was gel purified. Fragments were 32P radiolabeled by Klenow fill-in. Probes were purified on a Sepharose G-50 column (Stratagene).
FLAG epitope-tagged LANA1 (F-LANA1) (4) and control immune precipitates were assayed for the ability to bind radiolabeled TR DNA. BJAB cells (2 × 107) stably expressing F-LANA1 (BJAB/F-LANA1 cells [4]) were lysed in lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% NP-40, 2 mM EDTA, 25 μg of aprotinin per ml, 25 μg of leupeptin per ml, 1 mM phenylmethylsulfonyl fluoride). Cell lysates were precleared with protein G beads at 4°C for 1 h and incubated at 4°C for 2 h with M2 anti-FLAG monoclonal antibody-conjugated beads (Sigma) or isotype-matched control antibody bound to protein G beads. The beads were washed twice in lysis buffer and then washed three times in 1× binding buffer (25 mM Tris [pH 7.5], 100 mM KCl, 5 mM EDTA, 10 mM MgCl2, 0.1% NP-40, 5% glycerol, 0.1 mM dithiothreitol). Radiolabeled probe was incubated with the beads and 2 μg of poly (dI-dC) in 1× binding buffer for 30 min at room temperature. The beads were then washed five times with 1× binding buffer, and bound DNA was eluted from the beads at 95°C for 5 min with TE containing 0.1% sodium dodecyl sulfate. Eluted DNA fragments were detected by autoradiography after resolution on a nondenaturing 5% polyacrylamide gel. The signal was captured with a PhosphorImager and analyzed with ImageQuant software. Western blotting detected F-LANA1 only in M2 immunoprecipitates.
EMSA.
Oligonucleotides with 5′-GATC overhangs were annealed and 32P radiolabeled by Klenow fill-in. For electrophoretic mobility shift assays (EMSAs), in vitro-translated F-LANA1 (3 μl of the reaction mixture) (TNT coupled reticulocyte lysate systems [Promega]) was incubated in 1× reaction buffer [20 mM Tris (pH 7.5), 10% glycerol, 50 mM KCl, 0.1 mM dithiothreitol, 10 mM MgCl2, 1 mM EDTA, 1 μg (experiments in Fig. 5 and 6A and B) or 20 μg (experiments in Fig. 6C and 7) of poly(dI-dC) per ml] with 50,000 cpm of probe in 20 μl for 30 min at room temperature with or without excess unlabeled competitor oligonucleotides. For supershift assays, M2 or an irrelevant, murine isotype matched control antibody (Southern Biotechnology Associates) was added to the incubation mixture for 15 min prior to the addition of radiolabeled probe. Bound and unbound probes were resolved by electrophoresis in 3.5% nondenaturing polyacrylamide gels in 1× TBE. The gels were dried, and the signal was detected by autoradiography. Nuclear extracts were made by modified Dignam method (3). For supershift assays with nuclear extracts, a rat monoclonal antibody to LANA1 (ABI Biotechnology) or an irrelevant rat immunoglobulin G control antibody (Southern Biotechnology Associates) was used.
FIG. 5.
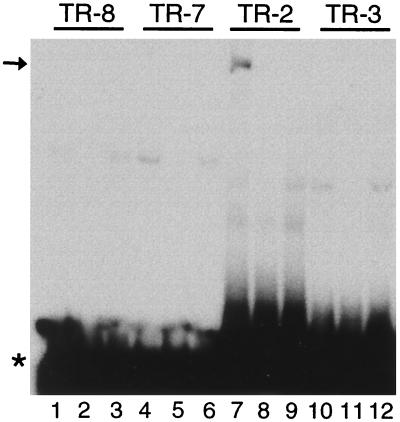
In vitro-translated F-LANA1 gel shifts TR-2. TR-8 (lanes 1 to 3), TR-7 (lanes 4 to 6), TR-2 (lanes 7 to 9) and TR-3 (lanes 10 to 12) 32P-labeled oligonucleotides were each incubated with in vitro-translated F-LANA1 (lanes 1, 2, 4, 5, 7, 8, 10, and 11) or RBP-Jκ (lanes 3, 6, 9, and 12), and EMSA was performed. A 50-fold excess of unlabeled TR-8 (lane 2), TR-7 (lane 5), TR-2 (lane 8), or TR-3 (lane 11) oligonucleotide was included in the incubation. The arrow indicates the specific gel shift in lane 7, and the asterisk indicates free probe. Data shown are representative of two experiments.
FIG. 6.
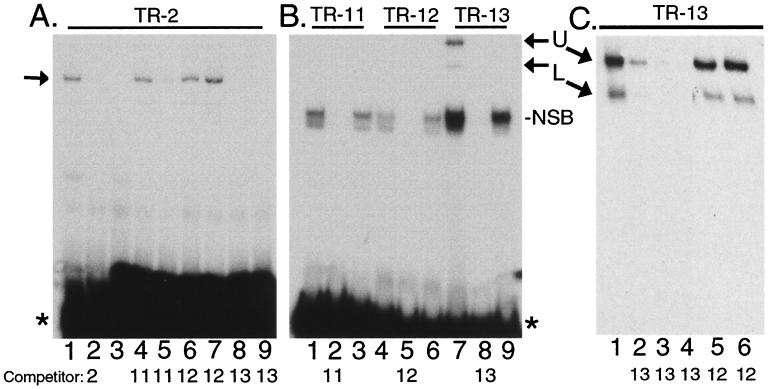
Delineation of F-LANA1 binding within TR-2. (A) Competition for TR-2 binding to F-LANA1 with oligonucleotides comprising TR-2. In vitro-translated F-LANA1 was incubated with TR-2 prior to EMSA in all lanes except lane 3, where RBP-Jκ was used. A 50-fold molar excess of unlabeled TR-2 was incubated with F-LANA1 for 5 min before addition of TR-2 probe (lane 2). A 50-fold (lanes 4, 6, and 8) or 100-fold (lanes 5, 7, and 9) molar excess of unlabeled TR-11, TR-12, or TR-13 was incubated with F-LANA 1 before the addition of TR-2 probe. The arrow indicates specific gel shifts, and the asterisk indicates free probe. (B) F-LANA1 specifically gel shifts TR-13. In vitro-translated F-LANA1 (lanes 1, 2, 4, 5, 7, and 8) or RBP-Jκ (lanes 3, 6, and 9) was incubated with radiolabeled TR-11 (lanes 1 to 3), TR-12 (lanes 4 to 6), or TR-13 (lanes 7 to 9) (indicated at the top). A 50-fold excess of unlabeled oligonucleotide was included in the incubation (lanes 2, 5, and 8). Asterisk indicates unbound probe. NSB, nonspecific band. (C) Excess nonradiolabeled TR-13 specifically competes the TR-13 gel shift. In vitro-translated F-LANA1 was incubated with radiolabeled TR-13. A 10-, 50-, or 100-fold excess of unlabeled TR-13 was included in the incubation in lanes 2, 3, and 4 respectively. A 50- or 100-fold excess of unlabeled TR-12 was included in the incubations in lanes 5 and 6, respectively. Free probe was run off the gel. U, upper F-LANA1 TR-13 gel-shifted complex; L, lower F-LANA1 TR-13 gel-shifted complex. Unlabeled competitor oligonucleotides are indicated at the bottom. Data shown are representative of two experiments.
FIG. 7.
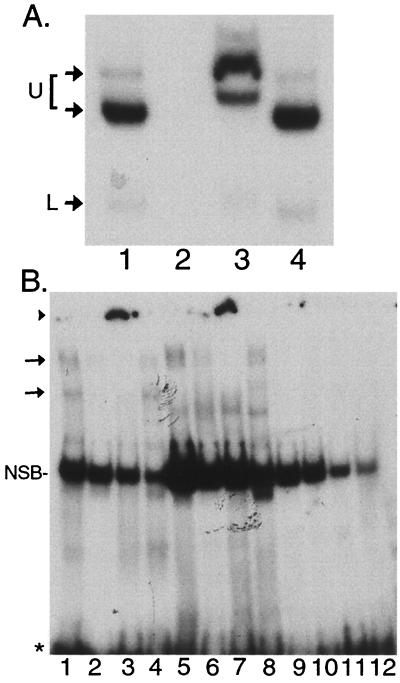
Supershift analyses of F-LANA1 and PEL cell nuclear extracts with TR-13. (A) Anti-FLAG antibody supershifts F-LANA1 TR-13 complexes. In vitro-translated F-LANA1 was incubated with TR-13 probe for all lanes. A 50-fold excess of unlabeled TR-13 oligonucleotide (lane 2), monoclonal anti-FLAG antibody (lane 3), or isotype-matched control antibody (lane 4) was added 15 min prior to the addition of 50,000 cpm of TR-13. Arrows indicate specific gel shifts. U, upper F-LANA1 shifted complex seen in Fig. 6B and C, which is resolved into two complexes here after a longer gel run; L, lower F-LANA1 gel-shifted complex. Data shown are representative of three experiments. Free probe was run off the gel. (B) LANA1 from PEL cells gel shifts TR-13. EMSA was performed using TR-13 probe and nuclear extracts from BCBL-1 (lanes 1 to 4), BC-1 (lanes 5 to 8) or (uninfected) BJAB cells (lanes 9 to 12). A 50-fold molar excess of unlabeled TR-13 (lanes 2, 6, and 10), anti-LANA1 monoclonal antibody (lanes 3, 7, and 11), or control antibody (lanes 4, 8, and 12) were included in incubations prior to the addition of 50,000 cpm of TR-13 probe. The arrows indicate specific gel shifts, and the arrowhead indicates supershifted probe near the gel origin (lanes 3 and 7). NSB, nonspecific band. Free probe is indicated by an asterisk. Data shown are representative of three experiments.
RESULTS
LANA1 mediates efficient persistence of DNA containing the KSHV TR sequence.
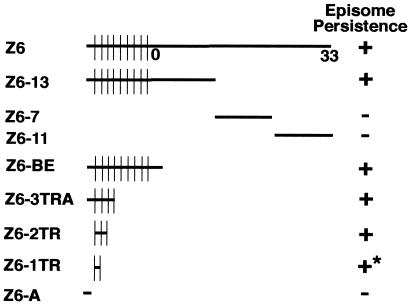
Experiments were performed to localize the cis-acting sequence upon which LANA1 acts to mediate episome persistence. Previous work demonstrated that LANA1 mediated the episome persistence of Z6, Z6-13, and Z6-3TRA but not Z6-7 and Z6-11 (Fig. 1) (4). Since Z6-3TRA contains only TR elements and ∼0.6 kb of KSHV sequence from the far left end of Z6 (termed A) (Fig. 1), either the TR elements or A is critical for LANA1-mediated episome persistence. Sequencing of the ∼0.6-kb A segment demonstrated that it is composed of KSHV nt 74931 to 75144 fused to a partial TR. The 5′ end of the partial TR sequence is TR nt 801, which is fused to TR nt 1 to 322, which are in turn fused to TR nt 348 to 383 (43). (In contrast to other reported KSHV TR sequences, A has GA in place of CC at TR at 351 and 352 [28, 38, 43].) nt 74931 to 75144 in A result from an insertion of this unique sequence into the BC-1 TRs (41). Plasmids were generated to assay TR and A for LANA1-mediated episome persistence. Z6-2TR and Z6-1TR contain two and one TR element, respectively. Z6-A contains the ∼0.6-kb A element (Fig. 1).
FIG. 1.
Schematic diagram of KSHV DNAs assayed for episome persistence. Approximate KSHV genome coordinates (in kilobases) are shown for Z6 (43). Vertical lines separate the ∼0.8-kb TR units. The diagrams are not drawn to scale. Episome persistence in LANA1-expressing cells is indicated by +, and lack of episome persistence is indicated by −. Z6, Z6-13, Z6-7, and Z6-11 were assayed in earlier experiments for episome persistence (4). The asterisk indicates that initial G418-resistant outgrowth of BJAB/F-LANA1 cells transfected with Z6-1TR was slower than after transfection of BJAB/F-LANA1 cells with other DNAs that scored positive for episome persistence.
To define the cis-acting element upon which LANA1 acts to mediate episome persistence, Z6-BE, Z6-3TRA, Z6-2TR, Z6-1TR, and Z6-A (Fig. 1) were each transfected into BJAB/F-LANA1 cells (4) or BJAB cells and selected for G418 resistance (conferred by the plasmid vector). Z6-BE, Z6-3TRA, and Z6-2TR DNA efficiently persisted in BJAB/F-LANA1 cells, and over 95% of microtiter wells had easily detectable macroscopic cell clumps, each containing at least several hundred cells, by ∼10 days posttransfection. Z6-1TR also persisted in BJAB/F-LANA1 cells in over 95% of microtiter wells, although the wells had smaller macroscopic cell clumps at ∼10 days posttransfection and hence the cells took longer to acidify the medium than after transfection with Z6-BE, Z6-3TRA, or Z6-2TR. However, after several weeks in culture there was no obvious difference between the growth of G418-resistant BJAB/F-LANA1 cells transfected with Z6-1TR and the growth of those transfected with Z6-BE, Z6-3TRA, or Z6-2TR. Efficient outgrowth was F-LANA1 dependent since only ∼20% of microtiter wells were positive for outgrowth after transfection of Z6-BE, Z6-3TRA, Z6-2TR, or Z6-1TR into BJAB cells and G418 selection. Although Z6-A contains truncated TR sequence, it lacks a cis-acting component necessary for efficient persistence. In contrast to the efficient G418-resistant outgrowth (in over 95% of microtiter wells) after transfection of DNA containing unit-length TR sequence into BJAB/F-LANA1 cells, only ∼20% of microtiter wells were positive for outgrowth after Z6-A transfection into BJAB/F-LANA1 cells and G418 selection. A similar low level of G418-resistant outgrowth was observed after transfection of Z6-A into BJAB cells. The Z6-A-transfected, G418-resistant BJAB/F-LANA1 and BJAB cells contain integrated Z6-A DNA (see below). These results indicate that unit-length TR DNA, but not the truncated TR sequence in A, efficiently persists in F-LANA1-expressing cells. Of note, the truncated TR sequence in A lacks the 20-nt LANA1 binding sequence defined below.
LANA1 acts in trans on KSHV TR DNA to mediate long-term episome persistence.
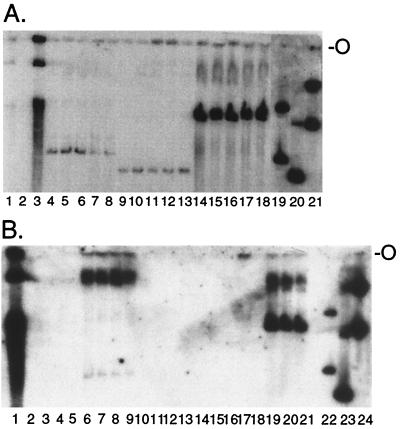
Since efficient outgrowth of F-LANA1-expressing cells transfected with TR DNA is consistent with episome persistence, Gardella gel analysis was performed on G418-resistant cells to assay for episomes. In Gardella gels, live cells are lysed in situ in the gel wells at the start of the gel run. Episomal DNA (as large as 200 kb) migrates into the gel, whereas chromosomal DNA is unable to migrate into the gel (17). As expected, BC-1 (Fig. 2A, lane 1) and BCBL-1 (Fig. 2A, lane 3; Fig. 2B, lane 1) KSHV-infected PEL cells had episomal DNA whereas uninfected BJAB cells did not (Fig. 2, lanes 2). After 2 weeks of G418 selection, BJAB/F-LANA1 cells that had been transfected with Z6-3TRA (Fig. 2A, lanes 4 to 8), Z6-2TR (lanes 9 to 13), or Z6-BE (lanes 14 to 18) had episomal DNA. Because of their slower initial outgrowth in microtiter plates, BJAB/F-LANA1 cells transfected with Z6-1TR and Z6-A and BJAB cells transfected with Z6-3TRA, Z6-2TR, Z6-1TR, Z6-BE, and Z6-A could not be assayed for extrachromosomal DNA after 2 weeks of G418 selection.
FIG. 2.
LANA1 acts on cis-acting unit-length KSHV TR DNA to mediate episome persistence. (A) G418-resistant BJAB/F-LANA1 cells (2 × 106) transfected with Z6-BE, Z6-3TRA, or Z6-2TR were lysed in situ in wells of Gardella gels, electrophoresis was performed, DNA was transferred to a nylon membrane, and KSHV TR DNA was detected. Lanes: 1, BC-1; 2, BJAB; 3, BCBL-1; 4 to 18, G418-resistant BJAB/F-LANA1 cells transfected with Z6-3TRA (lanes 4 to 8), Z6-2TR (lanes 9 to 13) or Z6-BE (lanes 14 to 18); 19, Z6-3TRA plasmid; 20, Z6-2TR plasmid; 21, Z6-BE plasmid. The upper bands in lanes 19 to 21 are from nicked plasmid DNA. The lower bands in lanes 1 and 3 are from linear and degraded virus DNA. The data shown are representative of two experiments. (B) G418-resistant BJAB or BJAB/F-LANA1 cells transfected with Z6-BE, Z6-3TRA, or Z6-A were analyzed in Gardella gels. Lanes: 1 BCBL-1; 2, BJAB; 3 to 5, Z6-3TRA-transfected BJAB cells; 6 to 9, Z6-3TRA-transfected BJAB/F-LANA1 cells; 10 to 12, Z6-A-transfected BJAB cells; 13 to 15, Z6-A-transfected BJAB/F-LANA1 cells; 16 to 18, Z6-BE-transfected BJAB cells; 19 to 21, Z6-BE-transfected BJAB/F-LANA1 cells; 22, Z6-3TRA plasmid; 23, Z6-A plasmid; 24, Z6-BE plasmid. The upper bands in lanes 22 to 24 are from nicked plasmid DNA. The lower band and smear in lane 1 is from linear and degraded DNA. The data shown are representative of two experiments. O, well origins.
Gardella gel assays for episomes were repeated after 1 month of G418 selection. Z6-3TRA (Fig. 2B, lanes 6 to 9) and Z6-BE (lanes 19 to 21), continued to persist as episomes in BJAB/F-LANA1 cells, and BJAB/F-LANA1 cells transfected with Z6-2TR or Z6-1TR also had episomes (data not shown). Episomes persisted for as long as 7 months in BJAB/F-LANA1 cells (data not shown). In contrast, BJAB/F-LANA1 cells that had grown out as G418 resistant after transfection with Z6-A (Fig. 2B, lanes 13 to 15) did not have extrachromosomal DNA. After transfection of LANA1-negative BJAB cells and G418 selection, Z6-3TRA (Fig. 2B, lanes 3 to 5), Z6-A (lanes 10 to 12), Z6-BE (lanes 16 to 18), Z6-2TR, and Z6-1TR (data not shown) did not have episomal DNA. These cells had transfected DNA as demonstrated by PCR, and upon longer exposures of Southern blots of Gardella gels, DNA was sometimes detected at the loading wells, consistent with the presence of integrated DNA in these cells (data not shown). Since episomal DNA persisted in BJAB/F-LANA1 cells only after transfection of DNA containing one or more complete TR elements, these data demonstrate that F-LANA1 acts on unit-length KSHV TR DNA to mediate episome persistence. The truncated TR sequence in Z6-A (which lacks the LANA1 binding sequence defined below) is insufficient for F-LANA1-mediated episome maintenance.
A decrease in episome migration rate in Gardella gels, consistent with an increase in episome size, was observed over time. For instance, after 2 weeks of G418 selection, most extrachromosomal Z6-3TRA (Fig. 2A, lanes 4 to 8) and Z6-BE (lanes 14 to 18) DNA comigrated with covalently closed circular plasmid (lanes 19 and 21, respectively), but after 1 month of selection, almost all Z6-3TRA (Fig. 2B, lanes 6 to 9) and much of Z6-BE (lanes 19 to 21) extrachromosomal DNA migrated at a rate similar to that of ∼170-kb (42) viral BCBL-1 episomes (lane 1). Similar decreases in migration rates were observed with other artificial KSHV episomes. Initial Southern blot analyses of Hirt-extracted DNA (20) from BJAB/F-LANA1 cells containing large episomes and restriction enzyme analyses of plasmids transferred from BJAB/F-LANA1 cells to bacteria indicate that the slower migration is due to TR duplication and arrangement of input plasmids into multimers (data not shown). These findings are reminiscent of multimerization of input DNA containing partial EBV oriP sequences in EBNA1-expressing cells, with a resultant increase in EBNA1 binding sites per episome (11, 51).
Immunoprecipitated F-LANA1 binds KSHV TR restriction fragments.
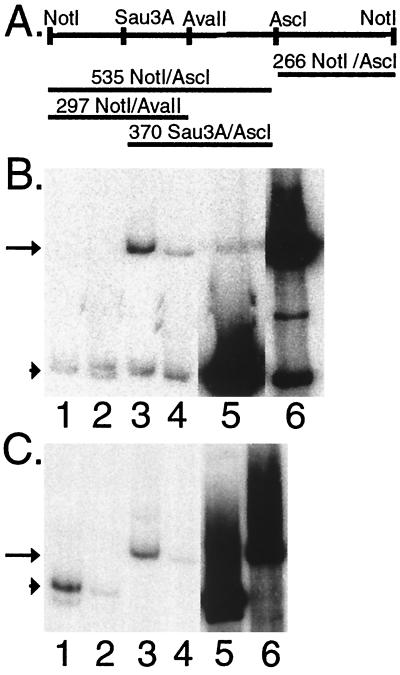
Since KSHV TR DNA is necessary and sufficient for LANA1-mediated episome persistence, we investigated whether LANA1 associates with TR DNA. TR restriction fragments (Fig. 3A) were assayed for the ability to bind anti-FLAG M2 or isotype-matched control antibody immunoprecipitates from BJAB/F-LANA1 cells. F-LANA1 immunoprecipitate (Fig. 3B, lane 3) bound sixfold more NotI-AscI 535-nt TR fragment than did control immunoprecipitate (lane 4, arrow) (input NotI-AscI 535-nt TR probe [Fig. 3B, lane 6]). In contrast, there was no significant difference between the low level of binding of F-LANA1 (lane 1) and control (lane 2, arrowhead) immunoprecipitates to the NotI-AscI 266-nt TR fragment (input NotI-AscI 266-nt TR probe [Fig. 3B, lane 5]). Since F-LANA1 immunoprecipitate specifically bound the NotI-AscI 535-nt fragment, binding to this fragment was further investigated by assaying the NotI-AvaII 297-nt and Sau3A-AscI 370-nt restriction fragments (Fig. 3A) for F-LANA1 binding. F-LANA1 immunoprecipitate (Fig. 3C, lane 1) bound 12-fold more NotI-AvaII 297-nt TR fragment than did control immunoprecipitate (lane 2, arrowhead) (input NotI-AvaII 297-nt TR probe [Fig. 3C, lane 5]). F-LANA1 immunoprecipitate (Fig. 3C, lane 3) bound 17-fold more Sau3A-AscI 370-nt TR fragment than did control immunoprecipitate (lane 4, arrow) (input Sau3A-AscI 370-nt TR probe [Fig. 3C, lane 6]). Since both the the NotI-AvaII 297-nt and Sau3A-AscI 370-nt fragments specifically bound F-LANA1 immunoprecipitates, these findings are consistent with the possibility that F-LANA1 binds to a site(s) within the 132-bp overlapping region between these fragments.
FIG. 3.
KSHV TR DNA restriction fragments are bound by immunoprecipitated F-LANA1. Radiolabeled KSHV TR restriction fragments were incubated with F-LANA1 or control immunoprecipitates from BJAB/F-LANA1 cells. Bound restriction fragments were eluted and resolved on 5% nondenaturing polyacrylamide gels. (A) Restriction map of the KSHV TR. Numbers indicate the the length of restriction fragments in nucleotides. (The NotI restriction site is at TR nt 383 [43].) Only the nonmethylated AvaII site is shown. (B) F-LANA1 (lanes 1 and 3) or control (lane 2 and 4) immunoprecipitates were incubated with the 266-bp NotI-AscI probe (lanes 1 and 2) or the 535-bp NotI-AscI probe (lanes 3 and 4); input 266-bp NotI-AscI probe (lane 5) and input 535-bp NotI-AscI probe (lane 6) are also shown. The arrow indicates the 535-bp NotI-AscI probe, and the arrowhead indicates the 266-bp NotI-AscI probe. All lanes are from the same gel and have the same exposure time. (C) F-LANA1 (lanes 1 and 3) or control (lane 2 and 4) immunoprecipitates were incubated with the 297-bp NotI-AvaII probe (lanes 1 and 2) or the 370-bp Sau3A-AscI probe (lanes 3 and 4); the input 297-bp NotI-AvaII probe (lane 5) and the input 370-bp Sau3A-AscI probe (lane 6) are also shown. All lanes are from the same gel and have the same exposure time. The arrow indicates the 370-bp Sau3A-AscI probe, and the arrowhead indicates the 297-bp NotI-AvaII probe. The data shown are representative of three experiments.
F-LANA1 binds specific TR sequence in EMSA analysis.
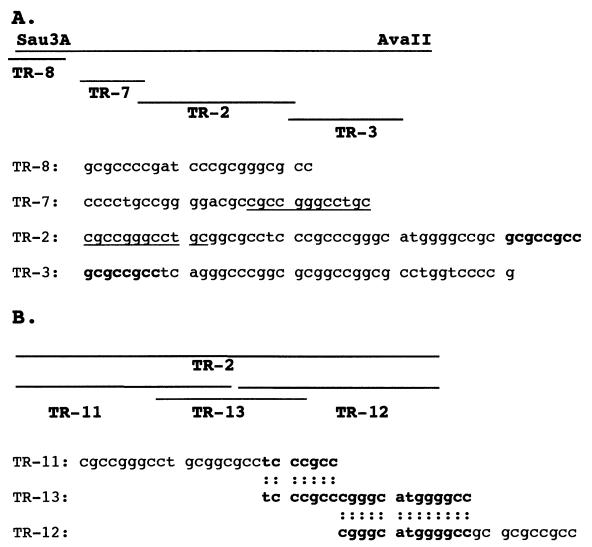
EMSA experiments with oligonucleotides TR-8, TR-7, TR-2, and TR-3 (Fig. 4A) were performed to assay F-LANA1 binding within the 132-nt overlapping region between the NotI-AvaII 297-nt and Sau3A-AscI 370-nt TR fragments. In vitro translated F-LANA1 (Fig. 5, lanes 1, 2, 4, 5, 7, 8, 10 and 11) or RBP-Jκ (lanes 3, 6, 9, and 12) was incubated with radiolabeled TR-8, TR-7, TR-2, or TR-3, and EMSA was performed. RBP-Jκ is a DNA binding protein that lacks a cognate sequence in TR-8, TR-7, TR-3, and TR-2 (18, 19). F-LANA1 (Fig. 5, lanes 1, 4, and 10) and RBP-Jκ (lanes 3, 6, and 12) gel shifts were faint and similar for TR-8, TR-7, and TR-3 probes, indicating only nonspecific gel shifts. However, F-LANA1 (lane 7, arrow) generated a specific TR-2 gel shift not observed with RBP-Jκ (lane 9). A 50-fold excess of nonradiolabeled oligonucleotide of the same sequence as the radiolabeled probe competed the specific gel shift (lane 8). These results indicate that F-LANA1 specifically gel shifts TR-2 but not TR-3, TR-7, or TR-8.
FIG. 4.
KSHV TR oligonucleotides. (A) The positions of TR-8, TR-7, TR-2, and TR-3 within the 132-bp Sau3A-AvaII fragment of the KSHV TR are shown schematically, TR-8 extends 7 nt 5′ to the Sau3A site. The sequence between TR-7 and TR-8 and the sequence 3′ to TR-3 was not synthesized. The sequences of TR-8, TR-7, TR-2 and TR-3 are shown. Sequences common to TR-7 and TR-2 are underlined, and the overlapping sequence between TR-2 and TR-3 is shown in bold type. (B) The positions of TR-11, TR-12, and TR-13 within TR-2 are shown schematically. Identical sequence is aligned.
Localization of a 20-nt LANA1 binding sequence in TR-2.
To further define the binding domain of F-LANA1 within TR-2, the oligonucleotides which comprise TR-2 (TR-11, TR-12, and TR-13) (Fig. 4B) were each assayed for the ability to compete the F-LANA1 TR-2 gel shift. F-LANA1 (Fig. 6A, lane 1, left arrow) again specifically gel shifted TR-2 compared to RBP-Jκ (lane 3), and the complex was competed with a 50-fold excess of unlabeled TR-2 (lane 2). Both 50- and 100-fold excesses of unlabeled TR-13 competed the F-LANA1 TR-2 gel shift (lanes 8 and 9). TR-11 was intermediate in its ability to compete the F-LANA1 TR-2 gel shift, since a 100-fold excess (lane 5) but not a 50-fold excess (lane 4) of TR-11 competed the shift. TR-12 did not compete the F-LANA1 gel shift at any concentration (lanes 6 and 7). Therefore, TR-13 competed the TR-2 gel shift more efficiently than did TR-11, and TR-12 did not compete the F-LANA1 shift at all. Since TR-11 and TR-13 overlap by 7 nt (Fig 4B), this result indicates a role for these nucleotides in competing with TR-2 for F-LANA1 binding.
The ability of F-LANA1 to gel shift TR-11, TR-12, and TR-13 was assayed. Consistent with the inability of TR-12 to compete the TR-2 gel shift (Fig. 6A), F-LANA1 did not specifically gel shift TR-12 (Fig. 6B, lane 4 [compare to RBP-Jκ in Fig. 6B, lane 6]). Despite the finding that a 100-fold excess of TR-11 competed the TR-2 gel shift (Fig. 6A, lane 5), F-LANA1 did not specifically shift TR-11 (Fig. 6B, lane 1 [compare to RBP-Jκ in Fig. 6B, lane 3]). Consistent with the efficient competition by TR-13 of the TR-2 gel shift (Fig. 6A), F-LANA1 specifically gel shifted TR-13 (Fig. 6B, lane 7, right arrows [compare to RBP-Jκ in Fig. 6B, lane 9]). Excess nonradiolabeled competitor oligonucleotides competed all gel shifts (Fig. 6B, lanes 2, 5, and 8). The nonspecific band in Fig. 6B (lanes 1, 3, 4, 6, 7 and 9) was greatly diminished when poly(dI-dc) at 20 μg/ml (Fig. 6C) rather than 1 μg/ml (Fig. 6B) was included in the EMSA binding buffer. Further, anti-FLAG antibody supershifted only the specific F-LANA1 gel shifts (see below) and not the nonspecific band (data not shown). Competition of the F-LANA1 TR-13 gel shifts by excess, unlabeled TR-13 was very efficient. A 10-fold excess of unlabeled TR-13 competed most of the TR-13 gel shifts (Fig. 6C, lane 2) and 50- and 100-fold excesses of TR-13 (lanes 3 and 4, respectively) competed all the TR-13 gel shifts. In contrast, a 50- or 100-fold excess of unlabeled TR-12 did not compete the TR-13 gel shifts (lanes 5 and 6). The upper TR-13 gel shift (Fig 6B, lane 7; Fig. 6C) was routinely resolved into two complexes on longer gel runs (Fig. 7A, lanes 1 and 4).
Supershift analyses with F-LANA1 and LANA1 from PEL cells.
To confirm the presence of F-LANA1 in the TR-13 gel-shifted complexes, supershift assays with anti-FLAG antibody were performed. Anti-FLAG (Fig. 7A, lane 3), but not an isotype-matched control antibody (lane 4), induced supershifts of both the upper and lower F-LANA1/TR-13 complexes (lane 1). Excess unlabeled TR-13 competed the gel shifts (lane 2). These results demonstrate that these complexes contain F-LANA1.
Since KSHV-infected PEL cells express LANA1, PEL cell nuclear extracts were assayed for the ability to gel shift TR-13 probe (Fig. 7B). Specific LANA1 complexes were identified with anti-LANA1 monoclonal antibody. EMSA with nuclear extract from BCBL-1 PEL cells (Fig. 7B, lane 1, arrows) gel shifted two complexes which were supershifted with LANA1-specific antibody (lane 3, arrowhead) but not control antibody (lane 4). BC-1 PEL cell nuclear extract (lane 5) gel shifted a complex which comigrated with the upper BCBL-1 complex (lane 1, upper arrow) and which also supershifted with antibody to LANA1 (lane 7, arrowhead) but not control antibody (lane 8). Excess unlabeled TR-13 competed the LANA1 complexes (lanes 2 and 6). Incubation of TR-13 probe with BJAB (uninfected) nuclear extract (lanes 9 to 12) did not result in specific gel shifts. These results indicate that BCBL-1 and BC-1 nuclear extracts gel shift TR-13 in complexes that contain LANA1.
DISCUSSION
This work demonstrates that LANA1 acts in trans on cis-acting KSHV TR DNA to mediate episome persistence and that LANA1 specifically binds KSHV TR DNA. Plasmids containing one or more TR units persisted as episomes in F-LANA1-expressing cells but not in cells lacking F-LANA1. Immunoprecipitated F-LANA1 but not control immune precipitates specifically bound certain TR restriction fragments. Specific TR oligonucleotides were gel shifted by in vitro-translated F-LANA1, and a 20-nt LANA1 binding site (TR-13) was defined. Nuclear extracts from BCBL-1 and BC-1 PEL cells specifically gel shifted TR-13, and the presence of LANA1 in these complexes was confirmed by supershift analyses. Z6-A DNA contains truncated TR sequence but lacks the LANA1 binding site defined here (TR-13) and did not persist as an episome in F-LANA1-expressing cells. Medveczky and colleagues have also shown that LANA1 binds to KSHV TR sequence (Medveczky et al., Third International Workshop on KSHV and Related Agents, abstr. 21, 2000). These data support a model in which LANA1 directly binds KSHV episomes via TR DNA to efficiently mediate episome persistence.
The high TR copy number (∼40 copies) (28, 43) in genomic KSHV may enhance LANA1-mediated episome persistence. The EBV oriP contains 24 EBNA1 binding sites distributed among 20 tandem repeats and a dyad symmetry element (29, 37, 41, 50, 52). More than one EBNA1 binding site is required for efficient EBNA1 mediated episome retention (11, 34, 51). Plasmids lacking the full complement of EBNA1 binding sites may dimerize or form tetramers in EBNA1-expressing cells, increasing the number of EBNA1 binding sites per episome (11, 47). Although LANA1 may bind to additional sites in the TR unit, the tandemly repeated TR elements in genomic KSHV provide a large number of LANA1 binding sites. Multimerization of input plasmid DNA and TR duplication (Fig. 2 and data not shown) in artificial KSHV episomes increases the number of LANA1 binding sites and suggests that this increase improves the efficiency of LANA1-mediated episome maintenance.
It is likely that tandemly repeated TR elements mediate the focal concentration of LANA1 to dots seen by immunofluorescence microscopy in KSHV-infected cells (23, 24, 48). LANA1 localizes to dots in BJAB/F-LANA1 cells with artificial KSHV episomes containing tandemly repeated TR elements but is diffusely distributed in the nucleus in the absence of KSHV episomal DNA (4). Localization of LANA1 to dots was also observed in cells with episomal Z6-1TR, Z6-2TR, Z6-3TRA, or Z6-BE DNA (data not shown). LANA1 concentration to dots in cells with these artificial episomes may be dependent on increased numbers of LANA1 binding sites from TR duplication and multimerization of input plasmids, as occurred when cells proliferated over time. Also consistent with the idea that DNA mediates the focal LANA1 concentration, DNase but not RNase treatment of BCBL-1 nuclei eliminated the punctate nuclear immunofluorescent staining pattern characteristic of LANA1 in PEL cells (49). LANA1 forms dimers in the absence of KSHV DNA (44) and may also dimerize and oligomerize at binding sites, as suggested by the presence of three different F-LANA1 gel-shifted TR-13 complexes in EMSA experiments (Fig. 7A, lane 1). By analogy, EBNA1 binds to its cognate DNA sequence as a dimer, and dimerization is required for binding (6, 10).
In order to persist, episomes must replicate at least once per cell cycle in addition to efficiently segregating to progeny cells. DNA replication initiates at or near the EBNA1 binding region of dyad symmetry in the EBV oriP and at other sites in the EBV genome during latency (16, 31). The dyad symmetry element, which contains four EBNA1 binding sites, is also a site of termination of DNA replication (14, 16). Although EBNA1 is necessary for long-term persistence of EBV oriP episomes, its role in EBV DNA replication remains controversial (1, 7, 10, 29, 32, 45, 51, 52). Whether LANA1 plays a role in KSHV DNA replication and how the TR functions as an origin of replication await further investigation.
Open reading frame 73 genes of other gamma-2 herpesviruses may also act on cognate TR DNA to mediate episome persistence. A cis-acting oriP sequence has been defined for herpesvirus saimiri (HVS), although the trans-acting factor has not been described (27). Despite the HVS oriP location within the long unique region of the genome, it is likely that HVS has an additional region(s) involved in episome maintenance, perhaps in the TR elements, since a strain of HVS lacking the described oriP sequence persists as an episome (27, 33). Similar to KSHV, HVS and other gamma-2 herpesviruses also have high TR copy numbers, which may reflect a TR role in episome persistence.
The model of LANA1 tethering TR DNA to chromosomes to mediate episome persistence requires simultaneous association of LANA1 with episomes and chromosomes, and this work demonstrates that LANA1 directly binds KSHV TR DNA. With respect to the association of LANA1 with chromosomes, LANA1 may bind chromosomal DNA or chromosome-associated proteins to mediate episome persistence. In this regard, LANA1 binds to histone H1, RING3, p53, members of the mSin3 corepressor complex, and retinoblastoma protein (12, 15, 26, 36, 39), and it is possible that any of these proteins may mediate its attachment to chromosomes. The DNA binding domain of the EBV EBNA1 protein is different from the domain involved in chromosome association (2, 10, 25, 29, 32, 45). Further investigation should elucidate the functional domains of LANA1 and how they coordinate to mediate episome maintenance.
ACKNOWLEDGMENTS
We thank Elliott Kieff, Siu Chun Hung, Eric Johanssen, and Stephanie Shauer for helpful discussions.
This work was supported by grants 5T32CA09031 and CA85751 (to M.E.B.) and CA67380 and CA82036 (to K.M.K.) from the National Cancer Institute.
REFERENCES
- 1.Aiyar A, Tyree C, Sugden B. The plasmid replicon of EBV consists of multiple cis-acting elements that facilitate DNA synthesis by the cell and a viral maintenance element. EMBO J. 1998;17:6394–6403. doi: 10.1093/emboj/17.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambinder R F, Mullen M A, Chang Y N, Hayward G S, Hayward S D. Functional domains of Epstein-Barr virus nuclear antigen EBNA-1. J Virol. 1991;65:1466–1478. doi: 10.1128/jvi.65.3.1466-1478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1998. [Google Scholar]
- 4.Ballestas M E, Chatis P A, Kaye K M. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 5.Bastien N, McBride A A. Interaction of the papillomavirus E2 protein with mitotic chromosomes. Virology. 2000;270:124–134. doi: 10.1006/viro.2000.0265. [DOI] [PubMed] [Google Scholar]
- 6.Bochkarev A, Barwell J A, Pfuetzner R A, Bochkareva E, Frappier L, Edwards A M. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein, EBNA1, bound to DNA. Cell. 1996;84:791–800. doi: 10.1016/s0092-8674(00)81056-9. [DOI] [PubMed] [Google Scholar]
- 7.Ceccarelli D F, Frappier L. Functional analyses of the EBNA1 origin DNA binding protein of Epstein-Barr virus. J Virol. 2000;74:4939–4948. doi: 10.1128/jvi.74.11.4939-4948.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cesarman E, Moore P S, Rao P H, Inghirami G, Knowles D M, Chang Y. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 9.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 10.Chen M R, Middeldorp J M, Hayward S D. Separation of the complex DNA binding domain of EBNA-1 into DNA recognition and dimerization subdomains of novel structure. J Virol. 1993;67:4875–4885. doi: 10.1128/jvi.67.8.4875-4885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chittenden T, Lupton S, Levine A J. Functional limits of oriP, the Epstein-Barr virus plasmid origin of replication. J Virol. 1989;63:3016–3025. doi: 10.1128/jvi.63.7.3016-3025.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotter M A, Jr, Robertson E S. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology. 1999;264:254–264. doi: 10.1006/viro.1999.9999. [DOI] [PubMed] [Google Scholar]
- 13.Decker L L, Shankar P, Khan G, Freeman R B, Dezube B J, Lieberman J, Thorley-Lawson D A. The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. J Exp Med. 1996;184:283–288. doi: 10.1084/jem.184.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhar V, Schildkraut C L. Role of EBNA-1 in arresting replication forks at the Epstein-Barr virus oriP family of tandem repeats. Mol Cell Biol: 1991;11:6268–6278. doi: 10.1128/mcb.11.12.6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friborg J, Jr, Kong W, Hottiger M O, Nabel G J. p53 inhibition by the LANA1 protein of KSHV protects against cell death. Nature. 1999;402:889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- 16.Gahn T A, Schildkraut C L. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989;58:527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- 17.Gardella T, Medveczky P, Sairenji T, Mulder C. Detection of circular and linear herpesvirus DNA molecules in mammalian cells by gel electrophoresis. J Virol. 1984;50:248–254. doi: 10.1128/jvi.50.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grossman S R, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc Natl Acad Sci USA. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henkel T, Ling P D, Hayward S D, Peterson M G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 20.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 21.Ilves I, Kivi S, Ustav M. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J Virol. 1999;73:4404–4412. doi: 10.1128/jvi.73.5.4404-4412.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones D, Ballestas M E, Kaye K M, Gulizia J M, Winters G L, Fletcher J, Scadden D T, Aster J C. Primary-effusion lymphoma and Kaposi's sarcoma in a cardiac-transplant recipient. N Engl J Med. 1998;339:444–449. doi: 10.1056/NEJM199808133390705. [DOI] [PubMed] [Google Scholar]
- 23.Kedes D H, Lagunoff M, Renne R, Ganem D. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J Clin Investig. 1997;100:2606–2610. doi: 10.1172/JCI119804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kellam P, Boshoff C, Whitby D, Matthews S, Weiss R A, Talbot S J. Identification of a major latent nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J Hum Virol. 1997;1:19–29. [PubMed] [Google Scholar]
- 25.Kirchmaier A L, Sugden B. Dominant-negative inhibitors of EBNA-1 of Epstein-Barr virus. J Virol. 1997;71:1766–1775. doi: 10.1128/jvi.71.3.1766-1775.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krithivas A, Young D B, Liao G, Greene D, Hayward S D. Human herpesvirus 8 LANA1 interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J Virol. 2000;74:9637–9645. doi: 10.1128/jvi.74.20.9637-9645.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kung S-H, Medveczky P G. Identification of a herpesvirus saimiri cis-acting DNA fragment that permits stable replication of episomes in transformed T cells. J Virol. 1996;70:1738–1744. doi: 10.1128/jvi.70.3.1738-1744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagunoff M, Ganem D. The structure and coding organization of the genomic termini of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) Virology. 1997;236:147–154. doi: 10.1006/viro.1997.8713. [DOI] [PubMed] [Google Scholar]
- 29.Lee M A, Diamond M E, Yates J L. Genetic evidence that EBNA-1 is needed for efficient, stable latent infection by Epstein-Barr virus. J Virol. 1999;73:2974–2982. doi: 10.1128/jvi.73.4.2974-2982.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehman C W, Botchan M R. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc Natl Acad Sci USA. 1998;95:4338–4343. doi: 10.1073/pnas.95.8.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Little R D, Schildkraut C L. Initiation of latent DNA replication in the Epstein-Barr virus genome can occur at sites other than the genetically defined origin. Mol Cell Biol. 1995;15:2893–2903. doi: 10.1128/mcb.15.5.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marechal V, Dehee A, Chikhi-Brachet R, Piolot T, Coppey-Moisan M, Nicolas J C. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J Virol. 1999;73:4385–4392. doi: 10.1128/jvi.73.5.4385-4392.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medveczky M M, Geck P, Sullivan J L, Serbousek D, Djeu J Y, Medveczky P G. IL-2 independent growth and cytotoxicity of herpesvirus saimiri-infected human CD8 cells and involvement of two open reading frame sequences of the virus. Virology. 1993;196:402–412. doi: 10.1006/viro.1993.1495. [DOI] [PubMed] [Google Scholar]
- 34.Middleton T, Sugden B. Retention of plasmid DNA in mammalian cells is enhanced by binding of the Epstein-Barr virus replication protein EBNA1. J Virol. 1994;68:4067–4071. doi: 10.1128/jvi.68.6.4067-4071.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore P S, Chang Y. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection. N Engl J Med. 1995;332:1181–1185. doi: 10.1056/NEJM199505043321801. [DOI] [PubMed] [Google Scholar]
- 36.Platt G M, Simpson G R, Mittnacht S, Schulz T F. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J Virol. 1999;73:9789–9795. doi: 10.1128/jvi.73.12.9789-9795.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Platt T H, Tcherepanova I Y, Schildkraut C L. Effect of number and position of EBNA-1 binding sites in Epstein-Barr virus oriP on the sites of initiation, barrier formation, and termination of replication. J Virol. 1993;67:1739–1745. doi: 10.1128/jvi.67.3.1739-1745.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poole L J, Zong J C, Ciufo D M, Alcendor D J, Cannon J S, Ambinder R, Orenstein J M, Reitz M S, Hayward G S. Comparison of genetic variability at multiple loci across the genomes of the major subtypes of Kaposi's sarcoma-associated herpesvirus reveals evidence for recombination and for two distinct types of open reading frame K15 alleles at the right-hand end. J Virol. 1999;73:6646–6660. doi: 10.1128/jvi.73.8.6646-6660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radkov S A, Kellam P, Boshoff C. The latent nuclear antigen of kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene hras transforms primary rat cells. Nat Med. 2000;6:1121–1127. doi: 10.1038/80459. [DOI] [PubMed] [Google Scholar]
- 40.Rainbow L, Platt G M, Simpson G R, Sarid R, Stoiber S-J G, Herrington C S, Moore P S, Schulz T F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rawlins D R, Milman G, Hayward S D, Hayward G S. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985;42:859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- 42.Renne R, Lagunoff M, Zhong W, Ganem D. The size and conformation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J Virol. 1996;70:8151–8154. doi: 10.1128/jvi.70.11.8151-8154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russo J J, Bohenzky R A, Chien M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Eman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwam D R, Luciano R L, Mahajan S S, Wong L, Wilson A C. Carboxy terminus of human herpesvirus 8 latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies. J Virol. 2000;74:8532–8540. doi: 10.1128/jvi.74.18.8532-8540.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shire K, Ceccarelli D F, Avolio-Hunter T M, Frappier L. EBP2, a human protein that interacts with sequences of the Epstein-Barr virus nuclear antigen 1 important for plasmid maintenance. J Virol. 1999;73:2587–2595. doi: 10.1128/jvi.73.4.2587-2595.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skiadopoulos M H, McBride A A. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J Virol. 1998;72:2079–2088. doi: 10.1128/jvi.72.3.2079-2088.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M-F, Clauvel J-P, Raphael M, Degos L, Sigaux F. Kaposi's sarcoma-associated herpesvirus-Like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 48.Szekely L, Chen F, Teramoto N, Ehlin-Henriksson B, Pokrovskaja K, Szeles A, Manneborg-Sandlund A, Lowbeer M, Lennette E T, Klein G. Restricted expression of Epstein-Barr virus (EBV)-encoded, growth transformation-associated antigens in an EBV- and human herpwevirus type 8-carrying body cavity lymphoma line. J Gen Virol. 1998;79:1445–1452. doi: 10.1099/0022-1317-79-6-1445. [DOI] [PubMed] [Google Scholar]
- 49.Szekely L, Kiss C, Mattsson K, Kashuba E, Pokrovskaja K, Juhasz A, Holmvall P, Klein G. Human herpesvirus-8-encoded LNA-1 accumulates in heterochromatin-associated nuclear bodies. J Gen Virol. 1999;80:2889–2900. doi: 10.1099/0022-1317-80-11-2889. [DOI] [PubMed] [Google Scholar]
- 50.Yates J, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci USA. 1984;81:3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yates J L, Camiolo S M, Bashaw J M. The minimal replicator of Epstein-Barrr virus oriP. J Virol. 2000;74:4512–4522. doi: 10.1128/jvi.74.10.4512-4522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yates J L, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]