Abstract
We screened phage display libraries of porcine reproductive and respiratory syndrome virus (PRRSV) protein fragments with sera from experimentally infected pigs to identify linear B-cell epitopes that are commonly recognized during infection in vivo. We identified 10 linear epitope sites (ES) 11 to 53 amino acids in length. In the replicase polyprotein, a total of eight ES were identified, six of which localized to the Nsp2 replicase polyprotein processing end product. In the structural proteins, a total of two ES were identified, in the ORF3 and ORF4 minor envelope glycoproteins. The ORF4 ES was previously identified by monoclonal antibody mapping (J. J. M. Meulenberg, A. P. van Nieuwstadt, A. van Essen-Zandenbergen, and J. P. M. Langeveld, J. Virol. 71:6061–6067, 1997), but its immunogenicity had not been examined in pigs. We found that six experimentally PRRSV-infected pigs consistently had very high antibody titers against the ORF4 ES. In some animals, sera diluted 1:62,500 still gave weak positive enzyme immunoassay reactivity against the ORF4 ES. This hitherto unrecognized immunodominance likely caused phages displaying the ORF4 ES to outcompete phages displaying other ES during library screening with porcine sera and accounted for our failure to identify more than two ES in the structural genes of PRRSV. Genetic analysis showed that variable ES were also the most immunogenic in vivo. Serological analysis indicated differences in the immunoglobulin A responses between short-term and longer-term viremic pigs towards some ES. The implications of these findings for PRRSV diagnostics and immunopathogenesis are discussed.
Porcine reproductive and respiratory syndrome virus (PRRSV) is a recently emerged pathogen of domesticated swine. The virus, which belongs to the Arteriviridae family, has a 15-kb positive-sense, single-stranded RNA genome. PRRSV encodes an approximately 4,000-amino-acid large replicase polyprotein (open reading frame [ORF] 1a and 1b) and six structural proteins of 130 to 265 amino acids (ORFs 2 to 7) (reviewed in references 6, 36, and 45). The replicase polyprotein is processed by autoproteolytic cleavage into nonstructural protein fragments (Nsps). The replicase polyprotein processing cascade has recently been reviewed by Ziebuhr et al. (45), and the Nsp and protease domain nomenclature suggested by Ziebuhr et al. is used throughout this article. Two main PRRSV genotypes exist, the American (US) and European (EU) types, which are only approximately 60% identical at the nucleotide level. For reasons currently not understood, these two distantly related PRRSV types emerged virtually simultaneously on their respective continents in the late 1980s. Since then, intermingling of the genotypes has occurred through the use of a live, US-type PRRSV vaccine in Europe (2, 18).
PRRSV infection poses a challenge to current serodiagnostic and vaccination strategies. Although live PRRSV vaccines provide protection against homologous challenge, the genetic diversity of field PRRSV isolates is very high, and vaccine effect against heterologous challenge may be limited (39). Also, live PRRSV vaccines have been observed to revert to virulence (2, 38), and the safer, killed vaccines have so far proved less effective (28). Finally, PRRSV can persist in some animals despite high levels of antiviral antibodies; in boars, this is associated with long-term, intermittent seminal excretion of the virus (3, 13). Serological tests cannot discriminate between seropositive animals which have cleared PRRSV infection and carrier animals. Addressing these problems might involve improving the design of antigens for vaccines and diagnostic tests. For example, PRRSV envelope glycoproteins are candidates for use in subunit vaccines (11), and the nonstructural ORF 1 polyprotein could be a possible candidate antigen for the development of serological tests to identify carrier animals (24). Production of full-length recombinant proteins to explore such potential applications may be hampered by the presence of hydrophobic regions and, for the ORF 1 polyprotein, a very large size and the ability to undergo autoproteolytic cleavage. This could be overcome by expression of protein subunits, guided by prior knowledge of naturally antigenic regions. However, no knowledge currently exists about epitopes in the PRRSV ORF 1 nonstructural polyprotein, and only two epitopes have been mapped in the envelope glycoproteins (23, 30), one of which was mapped using monoclonal antibodies (MAbs) and thus is of unknown significance for PRRSV infection in vivo (23). To address these questions, we have in the present study screened 97% of the PRRSV protein mass for linear porcine B-cell epitopes using phage display.
MATERIALS AND METHODS
Construction of phage libraries of random PRRSV fragments.
Routine M13 methods, such as phage amplification in liquid cultures of F′ Escherichia coli, purification of phages from E. coli culture supernatants by polyethylene glycol (PEG)-NaCl precipitation, titration of phages, and preparation of phage DNA for sequencing were done as described in reference 35 and the Ph.D-7 phage display kit manual from New England Biolabs (Hitchin, Hertfordshire, U.K.).
PRRSV 111/92, a Danish European-type isolate (1), was used for library construction. Long reverse transcription (RT)-PCR was performed using the primers detailed in Table 1 and previously described protocols (27). The three PCR fragments described in Table 1 were cloned in the pCR-XL TOPO vector (Invitrogen, Groningen, The Netherlands). To prepare random PRRSV fragments, 6 μg of pCR-XL construct was digested at 15°C in 95-μl reactions containing 100 μg of bovine serum albumin per ml–50 mM Tris (pH 7.6)–1 mM MnCl2–0.3 U of DNase I (Pharmacia, Allerød, Denmark). DNase digestion times were adjusted to produce fragments with an average of 70 to 100 nucleotides (nt), estimated by agarose gel electrophoresis. The random fragments were blunt-ended with T4 DNA polymerase (Novagen, Madison, Wis.), and size fractionated on Chroma Spin 30 gel filtration columns (Clontech, Basingstoke, Hampshire, U.K.).
TABLE 1.
RT-PCR primers used to amplify three fragments of the PRRSV 111/92 genome for the manufacture of ORF 1, ORF 2-3, and ORF 4-7 phage display librariesa
| RT primer | PCR primers | Size and characteristics of amplified PRRSV genome segment |
|---|---|---|
| 5′CTTTCCAGACCATTAG3′ (nt 11275–11290) | Forward (nt 194–214): 5′GATCTCCACCCTTTAACCATG3′ | ORF 1 library: 11.1-kb PCR fragment containing the whole ORF 1a, and 89% of ORF 1b (nonstructural proteins); lacks 485 nt of the 3′ end of ORF 1b |
| Reverse (nt 11272–11290): 5′CTTTCCAGACCATTAGTCG8AC3′ | ||
| poly(dT) | Forward (nt 11269–11290): 5′GT8CGACTAATGGTCTGGAAAG3′ | ORF 2-3 library: 2-kb PCR fragment containing the whole ORF 2 and ORF and ORF 3 (minor structural glycoproteins) |
| Reverse (nt 13234–13253): 5′GCACGCAGAAAGCATCAGCA3′ | ||
| poly(dT) | Forward (nt 12784–12803): 5′GTGTCGCGCGTCTTCGTGGA3′ | ORF 4-7 library: 2.2-kb PCR fragment containing whole ORFs 4 (minor structural glycoprotein), 5 (major structural glycoprotein), 6 (major structural nonglycosylated protein), and 7 (nucleocapsid protein) |
| Reverse (nt 15020–15040): 5′TCGCCCTAATTGAATAGGTGA3′ |
The three RT-PCR amplicons were cloned and used for the construction of three separate phage-display libraries. The 8 in primer sequences denotes inosine. The nucleotide numbering is according to the complete Lelystad PRRSV sequence (GenBank accession no. M96262; total, 15,101 nt).
To prepare M13 vector, 1 μg of M13KE gIII replicative-form DNA (New England Biolabs) was digested with 15 U Eag-I (New England Biolabs) for 5 h at 37°C in a 20-μl reaction, blunt ended with Klenow DNA polymerase (New England Biolabs), and dephosphorylated with calf intestinal phosphatase (Pharmacia).
Approximately 10 to 100 ng of random PRRSV fragments were ligated overnight at 16°C to 100 ng of M13 vector in 20-μl reactions containing 400 U of T4 DNA ligase (New England Biolabs). Following chloroform extraction, ethanol precipitation, and a 70% ethanol wash, ligation reactions were used to electroporate 80 μl of TOP10F′ E. coli (Invitrogen) in a single 0.1-cm cuvette (Bio-Rad, Hemel Hempstead, Hertfordshire, U.K.). Electroporated bacteria were incubated in 500 μl of SOC medium for 20 min at 37°C and 250 rpm, added to 100 ml of a 100-fold-diluted overnight culture of TOP10F′ E. coli in Luria-Bertani (LB) medium, and grown at 250 rpm and 37°C for 5 h. M13 phages were prepared from culture supernatants by two rounds of PEG-NaCl precipitation. Phage pellets were resuspended in 400 μl of 44% glycerol in phosphate-buffered saline (PBS, pH 7.6), and stored at −20°C.
To characterize libraries, PCR was done across the EagI site with primers 5′GTGGTACCTTTCTATTCTCAC3′ and 5′GACGTTAGTAAATGAATTTTCTG3′, which produced a 70-nt band from phages without the insert. In initial experiments, libraries were characterized in terms of frequencies of insert-bearing phage as well as insert sizes by PCR performed on 10 to 20 individual plaques. In a later approach, a sample of 106 phages taken from a library were used as the template for a single PCR. When run on 3% agarose gels, such whole-library PCRs produced a discrete 70-nt band from phages without inserts and a >70-nt smear from phages with inserts of different sizes. The intensity and size distribution of the >70-nt smear, judged by eye from Polaroid photographs, proved an excellent indicator of library quality.
Typically, three to six ligation reactions with different insert-vector ratios were set up for each preparation of random PRRSV fragments, and the phage library containing the highest proportion of insert-bearing phages was used for selection (see below). Thus, altogether, three different phage libraries based on the cloned PCR fragments described in Table 1 were constructed: an ORF 1 (nonstructural protein) library, an ORF 2-3 (structural proteins) library, and an ORF 4-7 (structural proteins) library. From 40 to 60% of the phages in these libraries contained inserts.
Experimental infections and characterization of PRRSV-immune sera.
Six specific-pathogen-free pigs, 4 weeks of age, were intranasally inoculated with 105.4 50% tissue culture-infective doses (TCID50) of PRRSV 111/92, the same Danish European-type isolate used for phage library construction. Serum samples were prepared prior to infection (0 days postinfection [dpi]) and at 3- to 7-day intervals thereafter for up to 56 days. The pigs were euthanized at 42 dpi (pigs 9 and 10) or 56 dpi (pigs 11, 12, 13, and 14), at which time tonsil homogenates, lung homogenates, and lung washings were tested for PRRSV by RT-PCR (29).
In-house routine diagnostic assays were used to characterize the serum samples. Antibodies to PRRSV were detected by blocking enzyme-linked immunosorbent assay (ELISA) as well as immunoperoxidase monolayer assay (1, 38). Viremia was detected by culturing serum samples on primary cultures of porcine pulmonary alveolar macrophages (1). Neutralizing antibodies were determined by mixing 100 TCID50 of PRRSV 111/92 with serial dilutions of heat-inactivated test serum, followed by inoculation on MARC-145 cells (15), essentially as described (43).
Selection of phage libraries with porcine sera and characterization of selected phages by sequencing.
Serum to be used for selection of phage libraries was filtered (0.45-μm pore size), sodium azide was added to 0.02%, and UV-inactivated M13KO7 helper phage (New England Biolabs) was added to 1012 particles/ml. Sera were stored at 4°C. Prior to being used in the selection described below, protein A- and G-Sepharose particles (Pharmacia) were blocked for 30 min at room temperature (or 4°C overnight) in bovine serum albumin (BSA) (100 mg/m). The selection protocol below was modified from the PhD-7 kit manual (New England Biolabs). First, to remove nonspecifically binding phage, 10 μl of phage library (typically 1011 PFU) was added to 190 μl of PBS–0.5% Tween 20–20% protein A-Sepharose in a polypropylene tube and incubated for 20 min at room temperature with vigorous shaking. The Sepharose particles were centrifuged for 20 s in a tabletop microcentrifuge and discarded. The supernatant was transferred to a new polypropylene tube, 0.25 to 2 μl of whole serum was added, and the phage-serum mix was incubated for 20 min at room temperature with shaking. To isolate antibody-bound phages, protein A-Sepharose was added to 20%, and the tubes were incubated for a further 15 min at room temperature with shaking. The Sepharose particles were washed 15 times with 1 ml of PBS–0.5% Tween 20 and once with 1 ml of PBS without detergent. During washing, the Sepharose suspensions were twice transferred to new polypropylene tubes to reduce the background of nonspecifically binding or adsorbing phage. Finally, phages were eluted through a 7-min incubation of the Sepharose in 1 ml of 200 mM glycine–1 mg of BSA per ml (pH 2.2). The Sepharose particles were centrifuged for 60 s in a table-top microcentrifuge and discarded. The eluate was neutralized by adding 150 μl 1 M Tris (pH 9.1) and stored at 4°C. At this stage, the phage content of the eluate (the output from the selection) and of the phage library (the input to the selection) were determined by titration on E. coli ER2537 (New England Biolabs). Eluates were amplified in 30 ml of 100-fold-diluted overnight culture of E. coli ER2537 in LB medium. Eluates thus amplified were subjected to further cycles of selection with porcine sera, as described above. Typically, a total of three or four cycles of selection and amplification were carried out. To reduce the occurrence of background phage, protein A and protein G-Sepharose particles were used alternately in successive selection cycles.
For sequencing, plaques were amplified in 1 ml of 100-fold-diluted overnight culture of E. coli ER2537 in LB medium for 5 h at 37°C and 250 rpm. Only plaques from nonamplified eluates were used to reduce bias caused by differences in replication of phages displaying inserts of different sizes. Cycle sequencing was done using the BigDye kit (PE Biosystems, Allerød, Denmark), and a reverse sequencing primer (5′ GAC GTT AGT AAA TGA ATT TTC TG 3′), which anneals 30 nt downstream from the M13KE gIII EagI site.
The sequences of the phage-displayed peptides were read using the SeqMan-II software (DNASTAR Inc.) and matched to PRRSV using the dot plot function of the Omiga software (Oxford Molecular Ltd., Oxford, U.K.). Inserts displayed by phages selected from the ORF 2-3 and ORF 4-7 structural protein libraries were matched to the ORF 2-7 sequence of the 111/92 PRRSV isolate, which was used for library construction and experimental infection (GenBank accession no. AJ223078). Inserts displayed by phages selected from the ORF 1 (nonstructural protein) library were matched to the ORF 1 sequence of the Lelystad PRRSV isolate (GenBank accession no. M96262), since ORF 1 of 111/92 has not yet been sequenced.
Phage ELISA.
Phages for ELISA were plaque purified and underwent two rounds of PEG-NaCl precipitation. The particle content of phage preparations was determined spectrophotometrically by measurement of the absorbance at 269 nm (5). A total of 4 × 1010 phage particles in 100 μl of PBS were added per well to Maxisorp U-96 ELISA plates (Nunc, Life Technologies, Tåstrup, Denmark) and incubated at 4°C overnight. Prior to addition of the antibody layers, plates were blocked for 1 h at room temperature with 10% dried skimmed milk in PBS (Blotto). In the following, reagents were used at 100 μl/well, incubations, unless otherwise stated, were for 1 h at room temperature (with shaking), and washes between antibody layers were done with 0.5 M NaCl–15 mM Na2HPO4–2 mM KH2PO4–0.05% Tween 20 (ELISA buffer).
The plates were first incubated with swine serum diluted in Blotto (1:100, unless otherwise stated) with 1011 particles of insertless M13KE gIII library phage per ml. The insertless phage was a background library phage from the selection process described above which was shown by sequencing not to have any insert (i.e., did not display any foreign epitope) at the EagI site. To determine total immunoglobulin (Ig) levels against the phage-displayed epitopes (total Ig ELISA), horseradish peroxidase (HRP)-conjugated rabbit anti-swine Ig (Dako, Glostrup, Denmark) diluted 1:5,400 in PBS–0.05% Tween 20 containing 1011 particles of insertless M13KE gIII library phage per ml and 1% normal rabbit serum (HRP conjugate buffer) was used as the secondary antibody. To determine IgA levels against the phage-displayed epitopes (IgA ELISA), a monoclonal antibody against porcine IgA (murine IgG1 isotype; ID-Lelystad, The Netherlands) (40) was used at 10 μg/ml in Blotto, followed by an HRP-conjugated rabbit anti-mouse Ig (Dako) diluted 1:1,000 in HRP conjugate buffer. For a negative isotype control, an irrelevant MAb (anti-Aspergillus niger glucose oxidase, murine IgG1 isotype; Dako) was used, also at 10 μg/ml. The ELISA plates were developed with 0.42 mM 3,3′,5,5′-tetramethylbenzidine–0.007% H2O2–35 mM citric acid–67 mM Na2HPO4 (pH 5.0) for 5 to 15 min at room temperature. The reaction was stopped with 1 M sulfuric acid, and the absorbance at 450 (tetramethylbenzidine substrate product absorbance) and 620 nm (background absorbance from dirt and scratches on the ELISA plates) was determined in a microplate reader.
To avoid background problems due to the common anti-M13 reactivity found in animal sera, each serum was tested in parallel on wells coated with peptide-displaying phage (test antigen), and a well coated with the same insertless library phage used as the adsorbent in the antibody dilution buffers above (negative antigen). The specific reactivity of the serum against the phage-displayed peptide was calculated as a ratio between the reactivity against test and negative antigen: (OD450test antigen − OD620test antigen)/(OD450negative antigen − OD620negative antigen). This ratio is referred to in Fig. 1 and is hereafter called the OD450 ratio. For titration of sera, we examined twofold serum dilutions in the range from 1:50 to 1:3,200 and fivefold serum dilutions in the range from 1:100 to 1:1,562,500 (Table 3 and Fig. 5). The specific reactivity of the serum against the phage-displayed peptide was calculated as a difference between the reactivity against test and negative antigen: (OD450test antigen − OD620test antigen)−(OD450negative antigen − OD620negative antigen), and the titer was expressed as the serum dilution range which spanned half-maximal specific ELISA reactivity.
FIG. 1.
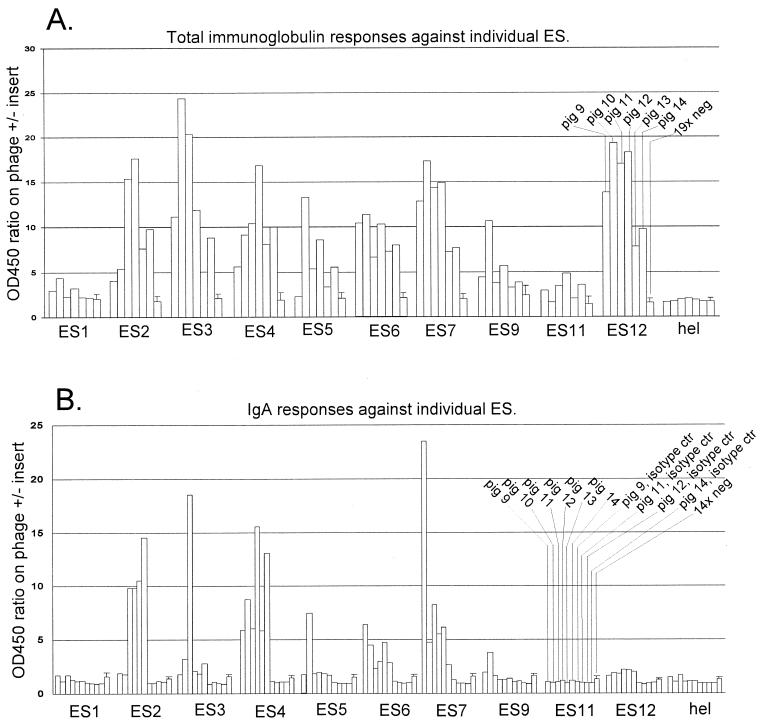
Serological confirmation of ES by ELISA. ELISA to detect total porcine Ig or porcine IgA directed against the different ES was done as described in Material and Methods. Hel is a phage displaying a 14-amino-acid fragment from the PRRSV helicase which is not an epitope, i.e., a negative-control antigen. To estimate the specific reactivity of porcine sera against the phage-displayed PRRSV peptides and to correct for any unspecific reactivity against the M13 virion, the OD450 ratio (y axis) was calculated as described in Materials and Methods. The 19× neg (A) and 14× neg (B) columns show the mean reaction of negative serum panels, with bars indicating 1 standard deviation. Experimental sera (pigs 9, 10, 11, 12, 13, and 14) were from 42-dpi PRRSV 111/92 infection. (B) Isotype control bars indicate the reactivity of the indicated pig serum using an isotype-matched MAb instead of the anti-IgA MAb. Sera from pigs 10 and 12 were used for phage library selection, as described in Table 2, footnote a.
TABLE 3.
ELISA titers against ES.
| Epitope sitea | Length of phage-displayed PRRSV sequence (amino acids)b | No. of pigs (n = 6) with 42-dpi total Ig ELISA titerc:
|
||||||
|---|---|---|---|---|---|---|---|---|
| 0 (seronegative) | 50–200 | 200–400 | 400–800 | 800–1,600 | 2,500–12,500 | 12,500–62,500 | ||
| ES1 | 17 | 3 | 3 | |||||
| ES2 | 14 | 2 | 1 | 2 | 1 | |||
| ES3 | 32 | 4 | 1 | 1 | ||||
| ES4 | 47 | 2 | 1 | 2 | 1 | |||
| ES5 | 11 | 1 | 5 | |||||
| ES6 | 28 | 5 | 1 | |||||
| ES7 | 24 | 2 | 3 | 1 | ||||
| ES9 | 26 | 6 | ||||||
| ES11 | 17 | 2 | 4 | |||||
| ES12 | 24 | 5 | 1 | |||||
| Hel | 14 | 6 | ||||||
Hel, phage displaying a 14-amino-acid fragment from the PRRSV helicase which is not an epitope.
In all cases, the phage displaying the longest insert was used to represent an ES in ELISA (see also Fig. 2).
Discrimination between seropositive and seronegative ELISA reactions was done as described in Materials and Methods. Sera from six pigs at 42 dpi were examined. The titer was examined on twofold serum dilutions in the range from 1:50 to 1:3,200 and fivefold serum dilutions in the range from 1:100 to 1:1,562,500 (ES12) and is expressed as the dilution range which spanned the half-maximal ELISA reaction.
FIG. 5.
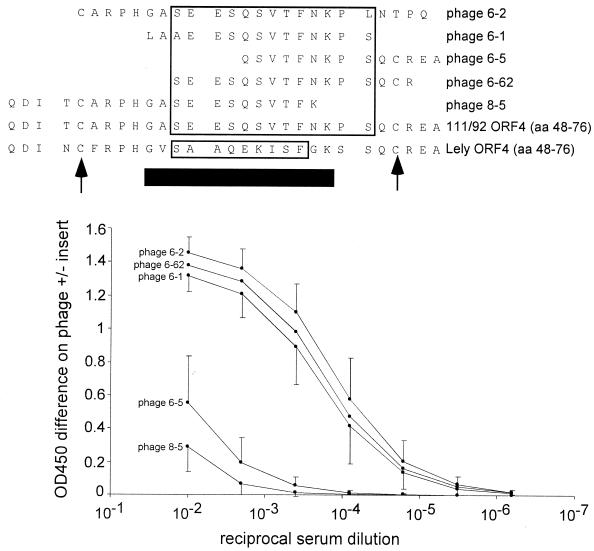
Fine mapping of the immunodominant ES12. Serum samples taken at 42 dpi from six experimentally infected pigs were titrated in ELISA on each of the five phage clones defining the ORF 4 epitope site (ES12). Each data point represents the average ELISA reactivity of the six experimental sera, and bars indicate 1 standard deviation. The boxed residues in the Lelystad PRRSV ORF 4 sequence indicate a core epitope previously determined by Meulenberg et al. using murine MAbs and Pepscan (23). The boxed residues in the 111/92 ORF 4 sequence indicate a core epitope defined by the sequence overlap between the three most strongly reacting phage clones (6-1, 6-2, and 6-62). Arrows indicate two highly conserved cysteine residues, and the heavy horizontal black bar indicates PRRSV sequence which is hypervariable and deletion prone in field isolates (30). The five C-terminal residues in phage 6-2, the three N-terminal residues in phage 6-1, and the C-terminal residue in phage 8-05 do not match the 111/92 sequence and likely represent a ligation or cloning artifact during library construction.
To score sera as seropositive or seronegative (Fig. 4 and 6), the OD450 ratio for the test serum was compared to the maximal OD450 ratio for a panel of 19 (total Ig ELISA) or 14 (IgA ELISA) known PRRSV-negative field sera. A test serum was considered PRRSV seropositive if it exhibited an OD450 ratio above the maximum OD450 ratio for the negative serum panel.
FIG. 4.
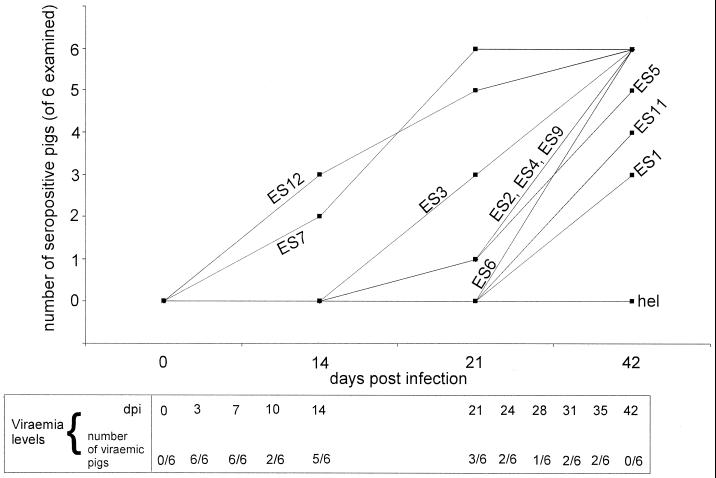
Seroconversion kinetics of six experimentally infected pigs against phage-displayed PRRSV ES. Serum samples taken at 0, 14, 21, and 42 dpi from six experimentally PRRSV-infected pigs were examined in total Ig ELISA against all the ES identified in this study. Seropositive/seronegative scoring was done as described in Materials and Methods. Hel is a phage displaying a 14-amino-acid fragment from the PRRSV helicase which is not an epitope. No pigs seroconverted to this negative-control antigen. Viremia levels were examined on more closely spaced serum samples than seroconversion, as indicated.
FIG. 6.
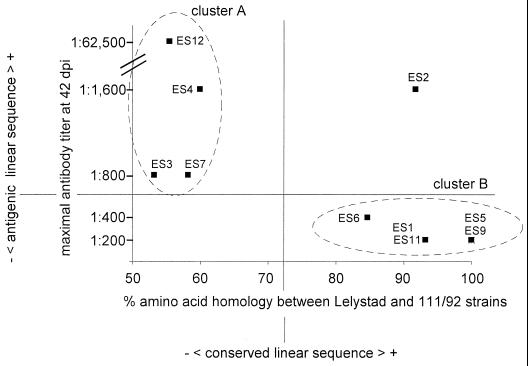
For linear ES in PRRSV, antigenicity in vivo correlates with genetic variability. The amino acid homology values (x axis) are from Fig. 2, and the maximal antibody titers (y axis) are from Table 3.
RESULTS
Selection with PRRSV-immune sera enriches for phages displaying PRRSV epitopes.
Three segments of the PRRSV genome were RT-PCR amplified (Table 1), cloned, fragmented by DNase treatment, and used to construct three separate phage display libraries. The RT-PCR amplicon used for the ORF 1 library contained 94% of the ORF 1 coding sequence, lacking only 485 nt of the 3′ end of ORF 1b. The two RT-PCR amplicons used for ORF 2-3 and ORF 4-7 library construction provided complete, overlapping coverage of all the structural genes of PRRSV. The cloning strategy described in Materials and Methods resulted in the surface display of random PRRSV fragments of 10 to 50 amino acids, fused to the N-terminus of the phage pIII protein, which is present in three to five copies at one end of the filamentous M13 virion. The libraries were selected in parallel with porcine sera taken at 42 days post-PRRSV infection (42-dpi sera) and with preinfection sera from the same animals (0-dpi sera). The 42-dpi sera used for selection had high (>1:1,000) antibody titers against PRRSV in routine diagnostic assays and contained neutralizing antibodies. In 0-dpi sera, no anti-PRRSV antibodies could be detected. The phage display libraries underwent three or four rounds of selection, where the eluted phage population from one selection was amplified and used for a successive selection (Table 2). To bias the screening towards commonly recognized epitopes, sera from two individual pigs were used alternately in successive selection cycles. Also, to bias the screening towards identifying epitopes which induce higher-affinity antibodies, progressively lower levels of serum were used for each selection (Table 2). For each selection, we measured the total amount of phage added and the total amount of phage recovered and calculated an output-input ratio by dividing these two values. Reproducibly, the output-input ratio increased through successive selections with 42-dpi sera, indicating that the phage population was increasingly enriched for antibody binders (Table 2). To assess how much of this enrichment was due to nonspecific reaction of porcine antibodies against M13 virions, the output-input ratios obtained with 42-dpi sera were divided by the output-input ratios obtained in parallel selections with 0-dpi sera to yield specific enrichment ratios. The specific enrichment ratios also increased through successive selections (Table 2), indicating that our selection strategy was effective in specifically enriching the libraries for phages that reacted with anti-PRRSV antibodies, ie, phages that likely expressed PRRSV epitopes. After three or four rounds of selection, the specific enrichment ratios were at least 10, which indicated that the majority of phages selected with 42-dpi sera might express PRRSV epitopes. At this stage, the inserts displayed by individual phages were determined by DNA sequencing.
TABLE 2.
Selection of PRRSV phage-display libraries with porcine sera: description of a simple method to monitor the specific enrichment for epitope-displaying phagesa
| Library | Selection no. and Sepharose type | Pig no. | Amt of 0-dpi serum used (μl) | Titer (PFU/ml)
|
Output-input ratio | Pig no. | Amt of 42-dpi serum used (μl) | Titer (PFU/ml)
|
Output-input ratio | Specific enrichment ratio | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Input | Output | Input | Output | |||||||||
| ORF 1 | 1, A | 12 | 2 | 1.0 × 1013 | 1.2 × 105 | 1.4 × 10−6 | 12 | 2 | 1.0 × 1013 | 6.0 × 105 | 6.9 × 10−6 | 4.6 |
| 2, G | 10 | 0.5 | 3.6 × 1013 | 1.6 × 106 | 5.1 × 10−6 | 10 | 0.5 | 3.7 × 1013 | >4 × 107 | >1.2 × 10−4 | >24 | |
| 3, A | 12 | 0.25 | 5.3 × 1013 | 2.5 × 105 | 5.3 × 10−7 | 12 | 0.25 | 1.7 × 1013 | 1.8 × 107 | 1.3 × 10−4 | 245 | |
| ORF 2-3 | 1, A | 12 | 2 | 1.3 × 1013 | 1.5 × 105 | 1.4 × 10−6 | 12 | 2 | 1.3 × 1013 | 1.6 × 105 | 1.4 × 10−6 | 1 |
| 2, G | 10 | 0.5 | 7.1 × 1013 | 4.8 × 105 | 7.8 × 10−7 | 10 | 0.5 | 8.8 × 1013 | 1.2 × 106 | 1.6 × 10−6 | 2 | |
| 3, A | 12 | 0.25 | 8.6 × 1013 | 3.2 × 106 | 4.3 × 10−6 | 12 | 0.25 | 8.3 × 1013 | 2.7 × 107 | 3.7 × 10−5 | 9 | |
| 4, G | 10 | 0.25 | 9.0 × 1013 | 1.0 × 106 | 1.3 × 10−6 | 10 | 0.25 | 8.0 × 1013 | 1.5 × 107 | 2.1 × 10−5 | 16 | |
| ORF 4-7 | 1, A | 12 | 2 | 4.6 × 1012 | 1.3 × 105 | 3.3 × 10−6 | 12 | 2 | 4.6 × 1012 | 1.0 × 105 | 2.5 × 10−6 | 1 |
| 2, G | 10 | 0.5 | 3.7 × 1013 | 2.0 × 105 | 6.2 × 10−7 | 10 | 0.5 | 1.1 × 1013 | ND | ND | ND | |
| 3, A | 12 | 0.25 | 1.7 × 1012 | 2.1 × 105 | 1.4 × 10−5 | 12 | 0.25 | 3.5 × 1012 | 4.2 × 106 | 1.4 × 10−4 | 10 | |
PRRSV phage display libraries underwent parallel selections with 0-dpi (free for PRRSV antibodies) and 42-dpi (containing high titers of anti-PRRSV antibodies) porcine sera. Serum from two pigs and two types of Sepharose (protein A or protein G) were used in alternate selection rounds, as indicated. Three or four cycles of selection were performed using successively lower serum levels. For all selection rounds, the input was 10 μl of library with the indicated titer, and the output was 1.15 ml of eluate with the indicated titer. The output-input ratio was calculated as (titer of eluate × 1.15)/(titer of input library × 0.01). The specific enrichment ratio was calculated by dividing the output-input ratio obtained with the 42-dpi serum with the output-input ratio obtained with the 0-dpi serum. ND, not done.
A total of 79 phages selected with 0-dpi sera were sequenced, and 73 of these phages either carried no inserts at the Eag-I site or displayed peptides which did not match any known PRRSV protein. These non-PRRSV peptides were derived from the pCR-XL vector sequence, the negative-sense PRRSV sequence, and alternate reading frames in the PRRSV positive-sense sequence. Only 6 of the 79 phages selected with 0-dpi sera displayed peptides which matched a known PRRSV protein (not shown). In contrast, of 213 phages selected with 42-dpi sera (57 from the ORF 1 library, 95 from the ORF 2-3 library, and 61 from the ORF 4-7 library), 135 displayed peptides of 10 to 47 amino acids which matched a known PRRSV protein. This difference in the frequency of PRRSV-displaying phages between populations selected with 0-dpi and 42-dpi sera (8 and 63%, respectively) was in agreement with the specific enrichment observed during selection (Table 2); together, these results suggested that PRRSV-displaying phages identified putative epitope sites in the viral proteins.
The 135 PRRSV-displaying phages which were selected with 42-dpi sera were not all unique, i.e., did not display 135 different peptide sequences; repetitions of the same phage clone were common. Also, several unique peptides sometimes clustered at the same site in a PRRSV protein. For these reasons, the 135 PRRSV-displaying phages altogether identified 12 putative epitope sites (ES), 10 in the ORF 1 nonstructural protein and one each in the ORF 3 and 4 minor envelope glycoproteins.
A total of 83% of putative epitope sites identified by sequencing are real and are not unique to the sera used for library selection.
Plaque-purified phage clones representing each putative ES were used as the antigen in total Ig ELISA (Fig. 1A). If more than one phage clone identified an ES, the phage clone displaying the longest insert was used for ELISA in all cases. Of the 12 putative ES identified by sequencing, 10 could be verified serologically (Fig. 1A). Experimental pig sera (42-dpi sera) reacted with these ES, whereas a panel of known negative sera did not. Also, experimental 0-dpi sera did not react with the ES (not shown in Fig. 1A, but see Fig. 4). A phage representing one of the epitope sites that could not be verified serologically (ES10) was used as a negative-control antigen. Experimental 42-dpi sera did not react with this phage, which displayed a 14-amino-acid nonepitope fragment of the PRRSV helicase (Fig. 1A, hel). Importantly, the serologically confirmed 10 ES reacted not only with the two pig sera that were used for library selection but also with one or more of the other experimental sera (Fig. 1A), indicating that our selection procedure (Table 2) had been successful in isolating B-cell epitopes that are commonly recognized by pigs. For most ES (except the weak ES1 and ES11), an IgA response could also be detected (Fig. 1B). Unsurprisingly, a strong reaction in total Ig ELISA generally correlated with a strong reaction in IgA ELISA. More remarkably, the opposite was also seen, with ES12 exhibiting a strong reaction with all experimental sera in total Ig ELISA but a very weak (albeit positive) reaction with all experimental sera in IgA ELISA (Fig. 1, compare A and B).
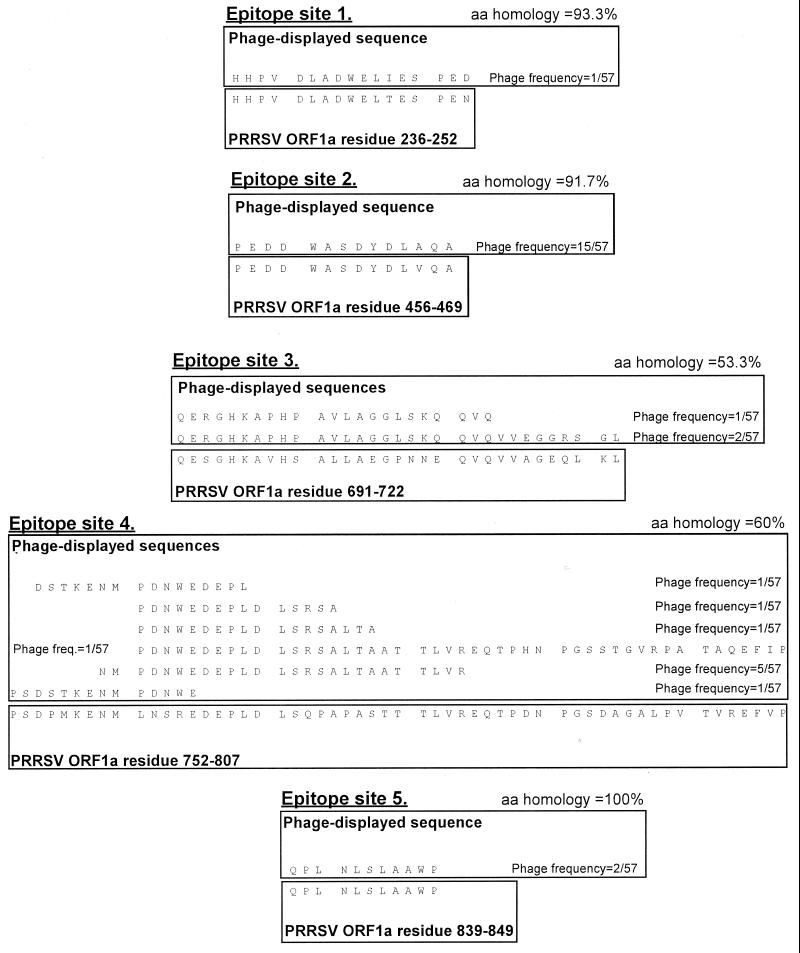
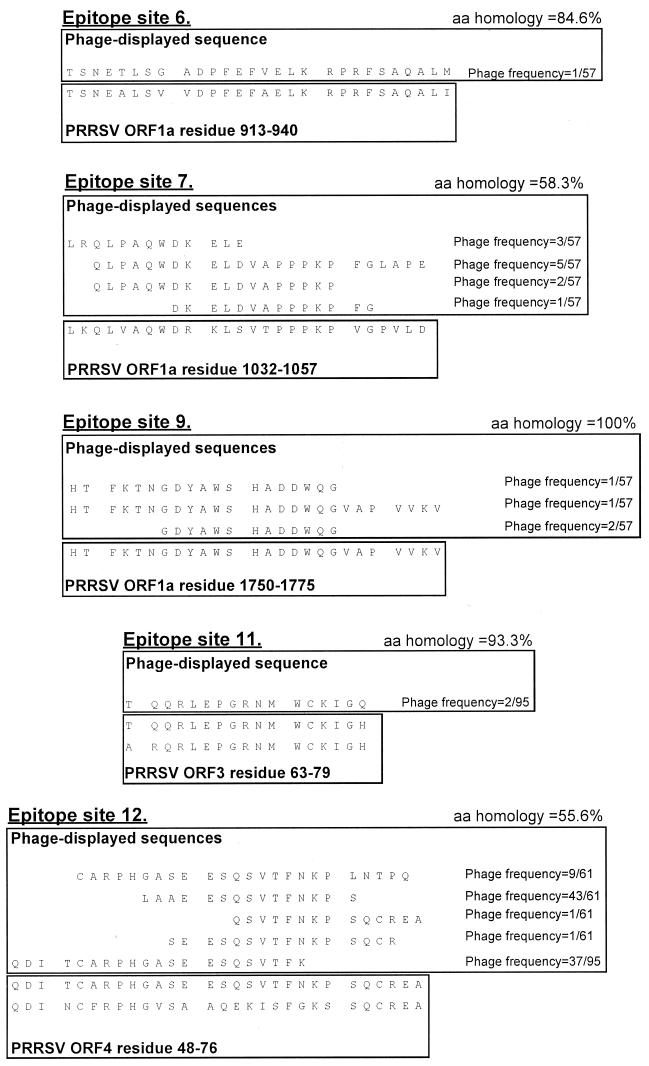
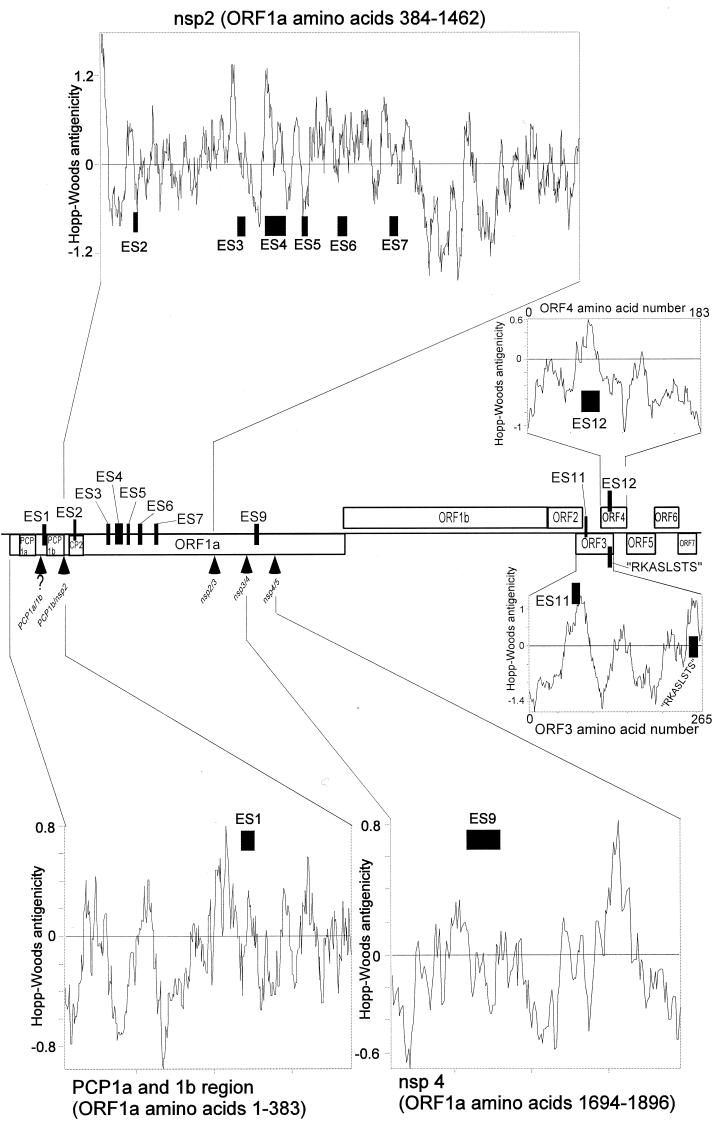
The sequences of the 10 serologically confirmed ES are presented in Fig. 2, and the positions of the confirmed ES in the PRRSV genome are shown in Fig. 3.
FIG. 2.
Sequences of serologically confirmed ES. For each ES, phage-displayed sequences are shown in the top box. The phage-displayed sequences were derived from PRRSV isolate 111/92, which was used for library construction. The sequences shown were fused to the phage pIII protein, in the context MKKLLFAIPLVVPFYSHS∗A?AQTVQSCLA, where the pIII leader peptide is underlined, ∗ is the predicted leader peptidase cleavage site, ? is the phage-displayed PRRSV sequence shown in the figure, and naturally phage-encoded residues immediately neighboring the displayed sequence in the mature pIII protein are in italics. The published PRRSV sequence is shown in the bottom box, annotated and aligned to the phage-displayed sequences. For ES in ORF 1, only published Lelystad PRRSV sequence is shown. For ES in the structural genes (sites 11 and 12), published 111/92 and Lelystad PRRSV sequences are shown (111/92 top, Lelystad bottom). The sequences for two ES which could not be confirmed by ELISA (ES8 and ES10) are not shown. Amino acid (aa) homologies were calculated between the phage-displayed sequences and Lelystad PRRSV, in all cases omitting the first and last of the phage-displayed residues, which might have been corrupted during the cloning procedure. For ES12, the 5 C-terminal residues in the first phage sequence and the three N-terminal residues in the second sequence likely represented a ligation artifact and were omitted from the homology calculation. For each phage clone, the number of times the clone was observed (numerator) in the total number of sequenced phages from a given library (denominator) is indicated. Altogether 57, 95, and 61 phage clones were sequenced from the ORF 1, ORF 2-3 and ORF 4-7 libraries, respectively. Where more than one phage identified an ES, the phage displaying the longest PRRSV sequence was used as the ELISA antigen in all cases.
FIG. 3.
Positions of serologically confirmed ES in the PRRSV genome. The positions of the serologically confirmed ES are shown by black boxes. Box width is proportional to the total length of the ES, as defined in Fig. 2. Only ORF 1 features of relevance to the identified ES are shown, using the protease domain and Nsp nomenclature suggested by Ziebuhr et al. (45). Arrowheads indicate predicted autoproteolytic cleavage sites in the ORF 1 replicase polyprotein. For the PCP1 α/β cleavage site, the question mark indicates that the exact position is not known. PCP1α, PCP1β, and CP2 denote accessory protease domains; the Nsp4 fragment is the main arteriviral protease. RKASLSTS is a previously identified ORF 3 epitope (30). Hopp-Woods antigenicity predicitions were made using the built-in option of the Omiga software and a 17-amino-acid sliding window.
Examination of seroconversion kinetics and titers.
Pigs seroconverted to the 10 ES at different times postinfection in what appeared to be a consistent pattern (Fig. 4). Seroconversion was first noted towards ES12 (ORF 4 envelope glycoprotein) and ES7 (Nsp2). Very late seroconversion was seen towards ES1 (ORF1a) and ES11 (ORF 3 envelope glycoprotein). Thus, the kinetics of seroconversion did not discriminate between ES in structural and nonstructural parts of the viral genome. This is the first report of ES in the PRRSV replicase polyprotein, and it is noteworthy that some of the nonstructural ES (ES7, Fig. 4) were targeted very rapidly by the pig antibody response. To examine the relationship between the speed and the magnitude of the porcine antibody response, total Ig titers towards all ES were determined at 42 dpi (Table 3). We found that the speed with which ES were recognized during infection in vivo (Fig. 4) was not generally predictive of the 42-dpi titers (Table 3), yet the 42-dpi titers in Table 3 are unlikely to represent a “final” antibody response; it is possible that ES exhibiting very late seroconversion such as ES1, ES5, and E11 (Fig. 4), attained titers higher than those shown in Table 3 at later times postinfection. In support of this, we observed that seroconversion to ES11 did not occur untill 56 dpi in one pig (not shown). It is an intriguing possibility that the seroconversion and antibody titer towards such very late epitopes might be informative about late phases of PRRSV replication.
Fine mapping of ES12.
ES12 corresponded to an epitope identified by murine MAbs and peptide scan in the Lelystad virus ORF 4 protein by Meulenberg et al. (23). Based on the reaction of 15 of 15 MAbs raised against recombinant ORF 4 protein with this site, the authors noted that the region was immunodominant in mice but did not examine its relevance in pigs. The anti-ORF 4 MAbs made by Meulenberg et al. were neutralizing in vitro (23). We observed that high anti-ES12 titers coexisted with viremia in individual animals (not shown) as well as in the experimental group as a whole (Table 3). Thus, the protective effect of anti-ES12 antibodies appeared to be limited in vivo. Interestingly, a recent study showed that anti-ORF 4 MAbs are inefficient at neutralization in vitro compared to anti-ORF 5 MAbs (42). Six of six experimentally infected pigs consistently produced very high antibody titers towards ES12 (Table 3 and Fig. 5). In one pig, half-maximal ELISA reaction was not reached untill a serum dilution of 1:12,500 to 1:62,500 (Table 3), and in two pigs, weak positive ELISA reactivity was still evident at a 1:62,500 serum dilution (not shown). Thus, our study confirmed the immunodominance of ES12 in pigs. In addition to inducing high antibody titers, ES12 was special in that it appeared to be deficient in IgA induction (Fig. 1, compare A and B). It remains to be determined whether this deficiency is one of IgA amount, affinity, or both. The deficiency might also be influenced by host factors, because serum from 1-year-old experimentally PRRSV-infected boars in some cases produces a stronger anti-ES12 signal in the IgA ELISA (M. B. Oleksiewicz, unpublished).
Immunodominant envelope protein regions have been implicated in the pathogenesis of other RNA virus infections (4). Therefore, to map ES12 more precisely, each of the five phage clones identifying ES12 were tested in an ELISA (Fig. 5). The termini of some of the phage-displayed peptides were corrupted, most likely by the blunt-ending or ligation steps during library construction (Fig. 5). Nevertheless, based on the sequence overlap between the three clones that reacted strongly in ELISA (Fig. 5, phages 6-1, 6-2, and 6-62), the porcine ES12 epitope mapped to a 13-amino-acid segment of the 111/92 ORF 4 protein (Fig. 5, large box in the 111/92 virus sequence). This region was slightly larger than the 9-amino-acid murine epitope identified by Meulenberg et al. (23) (Fig. 5, small box in the Lelystad virus sequence). The extra residues in the porcine epitope site appeared to be important for B-cell recognition in vivo, since a phage clone which displayed the whole murine site but lacked three of the four residues unique to the porcine site exhibited strongly reduced ELISA reactivity (Fig. 5, phage 8-5).
The mechanisms governing immunodominance are not well understood (4). For example, the high anti-ES12 titers at 42 dpi (Table 3 and Fig. 5) could not be explained by an early seroconversion towards ES12, because ES7 induced similar early seroconversion (Fig. 4) but never reached titers above 1:1,600 (Table 3). Also, the Hopp-Woods antigenicity of ES12 was not particularly high (Fig. 3). Finally, the high anti-ES12 titers were not due to a high level of antigenic stimulation, because the ORF 4 protein is a minor component of PRRSV virions (36). Interestingly, the ES12 core epitope is flanked by two cysteine residues which are conserved in EU-type PRRSV and even in US-type PRRSV (Fig. 5 and data not shown). Based on the results in Fig. 5, the flanking cysteines were clearly not required for the reaction of mature Ig with ES12; however, disulfide bridging might enhance presentation of ES12 to developing B cells.
In PRRSV, the antigenicity of linear epitopes correlates with genetic variability.
We and others have previously found, based on the analysis of two epitope sites, that linear epitopes in the envelope glycoproteins of PRRSV may exhibit a very high level of genetic variability (10, 14, 23, 30). In the present study, we were able to examine the variability-antigenicity relationship on a larger panel of epitope sequences and to extend the analysis to the replicase polyprotein. The amino acid homology between the Lelystad and 111/92 PRRSV strains was plotted against the maximal antibody titer at 42 dpi for each of the 10 ES (Fig. 6). Interestingly, in this plot two main clusters of ES were apparent: one cluster exhibited a low level of sequence conservation and induced higher levels of antibodies (cluster A, Fig. 6), and one cluster exhibited a high level of sequence conservation and induced lower levels of antibodies (cluster B, Fig. 6).
Antibody responses of short-term and longer-term viremic pigs are different.
It is well known that pigs may exhibit various periods of viremia following PRRSV infection, but the mechanisms responsible are unknown. Since our pigs were experimentally infected by the nasopharyngeal route and the initial viremic peak was likely to be derived from acute PRRSV replication in alveolar pulmonary macrophages (16, 17), we examined whether local mucosal humoral responses might have influenced viremia levels. We estimated viremia duration by the number of viremia-positive samples among 11 serum samples taken at 3, 7, 10, 14, 17, 21, 24, 28, 31, 35, and 42 dpi (all pigs were viremia negative at 0 dpi). Two pigs had only three positive samples, the latest at 14 dpi, and remained viremia negative thereafter. One pig had four positive samples, the latest at 14 dpi, and remained viremia negative thereafter. One pig had four viremic samples, the latest at 21 dpi, and remained viremia negative therafter. The remaining two pigs had seven and eight viremic samples, respectively, the latest at 35 dpi, and were viremia negative at 42 dpi (summarized for the group as a whole in Fig. 4). Thus, our group of animals contained short-term as well as longer-term viremic pigs. For analysis of IgA responses, the ES were grouped as follows: ES11 was excluded from the analysis because seroconversion in IgA ELISA was not consistently seen against this weak epitope (Fig. 1 and not shown). Also, owing to large interanimal variability in which the ORF 1 ES were targeted by the IgA response (not shown in detail, but see Fig. 1B), the ORF 1 ES (ES1 to ES9) were analyzed collectively. Finally, ES12 was analyzed separately because, in comparison to the ORF 1 ES, ES12 appeared to be deficient in IgA induction relative to total Ig induction (Fig. 1, compare ES12 in A and B). Interestingly, viremia duration correlated with the kinetics of the IgA response in individual pigs: short-term viremic pigs also exhibited the quickest IgA response to the “virtual antigen” consisting of the combined ORF 1 epitopes (Fig. 7). For ES12, the kinetics of IgA production did not correlate with viremia duration (Fig. 7).
FIG. 7.
Short-term and longer-term viremic pigs exhibit differences in humoral immune responses. Serum samples taken from six experimentally infected pigs at 0, 14, 21, 31, and 42 dpi were examined in IgA ELISA. The time of seroconversion (the earliest time when ELISA reactivity exceeded the cutoff determined by examining a negative serum panel, as described in Materials and Methods) was plotted against the duration of viremia for ES12 as well as for the ORF 1 ES (ES1 to ES9) collectively. Each data point represents a single animal. All six experimentally infected pigs, irrespective of viremia duration, were positive for PRRSV by RT-PCR on lung or tonsil material at euthanasia at 42 to 56 dpi.
DISCUSSION
In this study, we describe an approach based on long RT-PCR and phage display for the screening of whole viral genomes for linear B-cell epitopes. We show that, given an effective library selection strategy and an appropriate monitoring of enrichment ratios (Table 2), sequencing of phage clones is more than 80% accurate in identifying B-cell epitopes. Sequencing phages is technically simple and well suited to high-throughput formats. In contrast, downstream serological confirmation of epitopes is more labor intensive and requires larger panels of characterized sera. Thus, although the phage display method identifies only linear epitopes and may exhibit residue bias in the displayed peptides (32), the relatively high degree of accuracy in epitope identification based on sequencing alone makes it attractive for rapid first-pass epitope screening of viral genomes using minimal amounts of immune sera and pathogen material. The phage display method identified 10 ES in total, 8 of which were in the replicase polyprotein. The results from the serological characterization of the 10 ES appeared to have implications for the diagnostics as well as immunopathology of PRRSV infection, as discussed below.
Underestimate of the total number of linear ES in the structural genes.
Although seven ES were identified in ORF 1a, none were identified in ORF 1b (Fig. 3). In ORF 1a, six unique phage clones sometimes clustered at an ES (Fig. 2), indicating that the phage display library provided at least a sixfold coverage of ORF 1. Also, by using appropriate ligands, we have isolated phage displaying ORF 1b functional domains from the ORF 1 library (unpublished data). Thus, we believe that the failure to identify linear ES in ORF 1b was not due to poor library quality. The paucity of ES in ORF 1b may reflect a low level of protein production and hence poor stimulation of porcine B cells: translation of ORF 1b is thought to require a ribosomal frameshift event at a “slippery knot” RNA feature just upstream of the ORF 1a stop codon. The frameshift efficiency has been estimated at 15 to 20% in vitro (8), suggesting that significantly more ORF 1a protein than ORF 1ab protein may be produced in vivo. Additionally, the lack of linear ES in ORF 1b may reflect the antigenicity-conservation relationship for short linear sequences described in this study (Fig. 6). ORF 1b is generally more conserved than ORF 1a and contains several very highly conserved functional domains (22, 25). Interestingly, the Nsp2 fragment found to contain most linear ES (Fig. 3) has been noted by others to be the most variable part of the PRRSV genome, not excluding the structural genes (6, 34). In short, we believe that the absence of linear ES from ORF 1b and the clustering of linear ES in Nsp2 represent meaningful rather than artifactual results (Fig. 3). The biological mechanisms behind the clustering of ES in Nsp2 are unknown, but the protein is produced in quantity in Arterivirus-infected cells, where it carries out several functions, such as participating in the induction of double lipid membrane vesicles, as well as acting as a membrane anchor for the assembly of multiprotein viral replication complexes on the surface of the vesicles (31, 41).
Only two ES were identified in the structural genes (Fig. 3). This low level of ES was surprising, and also surprising was the failure to identify any ES in the ORF 7 nucleocapsid protein, which is known to contain linear ES (21) and to stimulate a strong antibody response in pigs. However, in agreement with the very high titers observed against ES12, ES12-representing phages constituted a very high proportion of the clones sequenced from the structural libraries (compare Table 3 and Fig. 2). Also, due to the location of ES12 at the ORF 3-4 overlap, ES12-displaying phages were present in ORF 2-3 as well as ORF 4-7 phage display libraries (Fig. 3 and Table 1). Thus, the most likely explanation for the low level of ES identified from the structural libraries is that phages representing the very strong ES12 outcompeted phages representing other structural-gene ES during the screening of the ORF 2-3 and ORF 3-7 libraries. Finally, the ORF 7 nucleocapsid protein is rich in positive residues, and it is possible that residue bias (32) prevented identification, especially of ORF 7 ES. Work is in progress to construct libraries without ES12, to perform a more sensitive screen for ES in the structural genes of PRRSV.
Antigenicity-variability relationship for linear ES in PRRSV.
We observed that the ES exhibited an unexpectedly marked clustering in antigenicity-variability plots (Fig. 6). Only one of the 10 ES did not exhibit clustering (Fig. 6, ES2). The very strong ES12 localized to cluster A (Fig. 6). ES12 induced very high antibody levels in vivo (Table 3), is known to mediate neutralization in vitro (23), and is hypervariable in field PRRSV isolates (10, 14, 23, 30). Thus, although our results suggested that the protective effect of anti-ES12 antibodies may be limited in vivo (see results above and Fig. 4), it nevertheless seems likely that the porcine antibody response exerts a selective pressure, which results in the sequence diversity at the ES12 site, as has also been suggested by others (10, 14, 23, 30). The genetic mechanisms regulating ES12 sequence diversity may be quite complex: the stretch of the viral genome which encodes ES12 in ORF 4 also codes for an RKASLSTS epitope in the overlapping ORF 3 (Fig. 3) (30). Nucleotide changes in the first codon position in ORF 4 result in third-codon position changes in the overlapping ORF 3. This suggests the presence of a selective pressure amplifying and quenching mechanism, where radical (first codon position) changes in ES12 are linked with more conserved (third codon position) changes in the RKASLSTS ORF 3 epitope. This mechanism, if it exists, is not essential; we have previously reported that whereas the genetic locus encoding ES12 and the ORF3 RKASLSTS epitope was intact in 70% of Danish field PRRSV isolates from 1992, it is deleted in 63% of present-day Danish field PRRSV isolates (30).
In contrast to ES12, the remaining three ES in cluster A and four of the five ES in cluster B localized to a nonstructural PRRSV protein (Fig. 3 and 6), and selective pressure from the porcine antibody response therefore appears to be a less immediate explanation for the observed diversity-antigenicity relationship. A possible factor other than the host immune response which could link antigenicity and sequence conservation is tertiary protein structure. Tertiary structure-rich regions might impede the presentation of strictly linear ES to porcine B cells and might predominate in, for example, functional protein domains, which are in turn more likely to be composed of conserved than nonconserved linear sequence. This hypothesis would not fully explain our data, because while ES2 and ES9 both localized to functional protein domains (Fig. 3, ES2 in the CP2 accessory protease and ES9 in the Nsp4 main protease), ES2 induced high and ES9 induced low antibody titers. Alternatively, nonstructural PRRSV protein(s) may be the target of selective pressure from the porcine antibody response due to a hitherto unrecognized localization on the surface of infected cells or as a trace component of PRRSV virions. Trace amounts of replicase protein in the particle of a positive-stranded RNA virus is not without precedent (26), and the enveloping mechanism for PRRSV may be sufficiently leaky to allow incorporation of “bystander” proteins into PRRSV particles (6). Finally, IgA might exert a selective pressure on nonstructural PRRSV proteins inside infected cells (12, 19, 20).
Although the mechanisms behind the observed diversity-antigenicity relationship (Fig. 6) can at present only be guessed at, the interplay between hypervariable epitope sites in viral envelope glycoproteins and the host antibody response is thought to be of importance for the pathogenesis of, for example, human immunodeficiency virus and hepatitis C virus infections. For those viruses, linear strongly antigenic envelope glycoprotein sequences have in some cases been shown to be decoy epitopes, which lure the host antibody response away from critical, neutralizing epitope sites (4, 33). ES12 exhibited some features expected of a decoy epitope, such as the ability to induce very high antibody levels rapidly and consistently in all infected animals (Table 3 and Fig. 4), hypervariability (10, 14, 23, 30), poor correlation between seroconversion and clearance of viremia in vivo (Fig. 4), and apparent polarization in the type of Ig induced (Fig. 1, compare ES12 in A and B). Thus, examination of the in vivo protective effect of anti-ES12 antibodies may be of value for vaccine development.
Implications of the identified ES for PRRSV diagnostics.
ELISA using phage-displayed ES as the antigen exhibited characteristics which might be attractive for diagnostic tests. (i) The wide range in seroconversion times against the different ES in effect made seroconversion predictive of the occurrence of viremia in the group of animals (Fig. 4, compare serology and virus isolation data). This might be exploited to develop serological tests with improved ability to measure disease progression at the herd level. (ii) Since preliminary experiments indicate that the ES described in Fig. 2 are not recognized by antibodies from animals infected with US-type PRRSV (unpublished data), they may be applicable for highly discriminatory serodiagnostics in areas where US- and EU-type PRRSV coexist, such as Denmark (2, 18, 38) and Canada (9). (iii) M13 phage particles are resistant to proteolysis, allow very simple and reproducible coating of ELISA plates with peptide antigen, and combine multivalent display with a good presentation of peptides for antibody binding (46). All of these features may be attractive for detection of low-level antibodies in protease-containing sources such as boar semen or meat juice. In fact, ELISA using phage-displayed ES as the antigen can be used to detect antibodies in boar semen (M. B. Oleksiewicz, submitted for publication).
More generally, the wide variation in ES sequence conservation (Fig. 2) and serological characteristics (Fig. 4 and Table 3) may allow the ES catalogue generated in the present study (Fig. 2 and 3) to act as a guide for the development of new test antigens, particularly from the hitherto serologically uncharacterized replicase polyprotein. In fact, our results predicted interesting properties for ELISAs using the N-terminal part of Nsp2 as the antigen, be it in the form of short ES (Fig. 4) or as a whole (Fig. 7). However, prior to diagnostic exploitation, more information is needed about the relationship between the genetic variability and serological cross-reactivity between different PRRSV isolates at the epitope sites, as well as about the interpig variability in anti-ES responses.
Implications of the identified ES for PRRSV immunopathology.
For PRRSV, the demonstration of antibody-dependent enhancement of infection in macrophages in vitro has caused interest in the effect of humoral responses on viral replication in vivo (44). A recent study showed a correlation between the humoral immune response and quenching of PRRSV replication in the lungs of pigs (16). In our study, we observed a correlation between the kinetics of the IgA response towards ES in the replicase polyprotein and the duration of viremia (Fig. 7). Although the presence of neutralizing ES in the replicase polyprotein cannot be excluded, as mentioned above, it seems more likely at present to assume that the IgA response per se, rather the IgA response specifically against ORF 1 ES, might be of importance in shortening viremia duration. Also, the IgA levels at late times postinfection did not correlate with viremia duration (not shown). This indicates that the early IgA response may directly affect viral replication, as opposed to IgA being a marker for another, unknown protective immune parameter. Importantly, in all six experimentally infected pigs, PRRSV could be detected by RT-PCR on lung or tonsil material at euthanasia at 42 to 56 dpi (not shown). Thus, the IgA response did not correlate with the clearance of virus from solid tissues. It is also noteworthy that no correlation existed between the IgA response kinetics and viremia duration for ES12 (Fig. 7). While further examination of the correlation between virus replication and serological parameters is warranted, it appears that the use of “precise” antigen, such as the ES identified in the present study, may have inherent advantages in examining the interactions between PRRSV and the humoral immune system.
ACKNOWLEDGMENTS
We thank H. S. Nielsen for assistance with long RT-PCR and cloning, B. Strandbygaard for providing panels of characterized sera, and R. Forsberg and Å. Uttenthal for critical reading and discussion of the manuscript.
REFERENCES
- 1.Bøtner A, Nielsen J, Bille-Hansen V. Isolation of porcine reproductive and respiratory syndrome (PRRS) virus in a Danish swine herd and experimental infection of pregnant gilts with the virus. Vet Microbiol. 1994;40:351–360. doi: 10.1016/0378-1135(94)90122-8. [DOI] [PubMed] [Google Scholar]
- 2.Bøtner A, Strandbygaard B, Sørensen K J, Have P, Madsen K G, Smedegaard Madsen E, Alexandersen S. Appearance of acute PRRS-like symptoms in sow herds after vaccination with a modified live PRRS vaccine. Vet Rec. 1997;141:497–499. doi: 10.1136/vr.141.19.497. [DOI] [PubMed] [Google Scholar]
- 3.Christopher-Hennings J, Nelson E A, Hines R J, Nelson J K, Swenson S L, Zimmerman J J, Chase C L, Yaeger M J, Benfield D A. Persistence of porcine reproductive and respiratory syndrome virus in serum and semen of abult boars. J Vet Diagn Investig. 1995;7:456–464. doi: 10.1177/104063879500700406. [DOI] [PubMed] [Google Scholar]
- 4.Cleveland S M, Buratti E, Jones T D, North P, Baralle F, McLain L, McInerney T, Durrani Z, Dimmock N J. Immunogenic and antigenic dominance of a nonneutralizing epitope over a highly conserved neutralizing epitope in the gp41 envelope glycoprotein of human immunodeficiency virus type 1: its deletion leads to a strong neutralizing response. Virology. 2000;266:66–78. doi: 10.1006/viro.1999.0041. [DOI] [PubMed] [Google Scholar]
- 5.Day L A. Conformations of single-stranded DNA and coat protein in fd bacteriophage as revealed by ultraviolet absorption spectroscopy. J Mol Biol. 1969;39:265–277. doi: 10.1016/0022-2836(69)90316-7. [DOI] [PubMed] [Google Scholar]
- 6.Dea S, Gagnon C A, Mardassi H, Pirzadeh B, Rogan D. Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the North American and European isolates. Arch Virol. 2000;145:659–688. doi: 10.1007/s007050050662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Haan C A, Vennema H, Rottier P J. Assembly of the coronavirus envelope: homotypic interactions between the M proteins. J Virol. 2000;74:4967–4978. doi: 10.1128/jvi.74.11.4967-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Den Boon J A, Snijder E J, Chirnside E D, de Vries A A, Horzinek M C, Spaan W J. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dewey C, Charbonneau G, Carman S, Hamel A, Nayar G, Friendship R, Eernisse K, Swenson S. Lelystad-like strain of porcine reproductive and respiratory syndrome virus (PRRSV) identified in Canadian swine. Can Vet J. 2000;41:493–494. [PMC free article] [PubMed] [Google Scholar]
- 10.Drew T, Lowings J P, Yapp F. Variation in open reading frames 3, 4 and 7 among porcine reproductive and respiratory syndrome isolates in the UK. Vet Microbiol. 1997;55:209–221. doi: 10.1016/s0378-1135(96)01328-4. [DOI] [PubMed] [Google Scholar]
- 11.Durand J P, Climent I, Sarraseca J, Urinza A, Cortes E, Vela C, Casal I. Baculovirus expression of proteins of porcine reproductive and respiratory syndrome virus strain olot/91: involvement of ORF3 and ORF5 proteins in protection. Virus Genes. 1997;14:19–29. doi: 10.1023/a:1007931322271. [DOI] [PubMed] [Google Scholar]
- 12.Fujioka H, Emancipator S N, Aikawa M, Huang D S, Blatnik F, Karban T, DeFife K, Mazanec M B. Immunocytochemical colocalization of specific immunoglobulin A with sendai virus protein in infected polarized epithelium. J Exp Med. 1998;188:1223–1229. doi: 10.1084/jem.188.7.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gradil C, Dubuc C, Eaglesome M D. Porcine reproductive and respiratory syndrome virus: seminal transmission. Vet Rec. 1996;138:521–522. doi: 10.1136/vr.138.21.521. [DOI] [PubMed] [Google Scholar]
- 14.Katz J B, Schafer A L, Eernisse K, Landgraf J G N E A. Antigenic differences between European and American isolates of porcine reproductive and respiratory syndrome virus (PRRSV) are encoded by the carboxyterminal portion of viral open reading frame 3. Vet Microbiol. 1995;44:65–76. doi: 10.1016/0378-1135(94)00113-B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim H S, Kwang J, Yoon I J, Joo H S, Frey M L. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch Virol. 1993;133:477–483. doi: 10.1007/BF01313785. [DOI] [PubMed] [Google Scholar]
- 16.Labarque G G, Nauwynck H J, Van Reeth K, Pensaert M B. Effect of cellular changes and onset of humoral immunity on the replication of porcine reproductive and respiratory syndrome virus in the lungs of pigs. J Gen Virol. 2000;81:1327–1334. doi: 10.1099/0022-1317-81-5-1327. [DOI] [PubMed] [Google Scholar]
- 17.Lawson S R, Rossow K D, Collings J E, Benfield D A, Rowland R R R. Porcine reproductive and respiratory syndrome virus infection of gnotobiotic pigs: sites of virus replication and co-localization with MAC-387 staining at 21 days post-infection. Virus Res. 1997;51:105–113. doi: 10.1016/s0168-1702(97)00086-5. [DOI] [PubMed] [Google Scholar]
- 18.Madsen K G, Hansen C M, Madsen E S, Strandbygaard B, Bøtner A, Sørensen K J. Detection of porcine reproductive and respiratory syndrome virus of the American type in Danish swine herds. Arch Virol. 1998;143:1683–1700. doi: 10.1007/s007050050409. [DOI] [PubMed] [Google Scholar]
- 19.Mazanec M B, Coudret C L, Fletcher D R. Intracellular neutralization of influenza virus by immunoglobulin A antihemagglutinin monoclonal antibodies. J Virol. 1995;69:1339–1343. doi: 10.1128/jvi.69.2.1339-1343.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazanec M B, Kaetzel C S, Lamm M E, Fletcher D, Nedrud J G. Intracellular neutralization of virus by immunoglobulin A antibodies. Proc Natl Acad Sci USA. 1992;89:6901–6905. doi: 10.1073/pnas.89.15.6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meulenberg J J, Van Nieuwstadt A P, Essen-Zandbergen A, Bos-de Ruijter J N, Langeveld J P, Meloen R H. Localization and fine mapping of antigenic sites on the nucleocapsid protein N of porcine reproductive and respiratory syndrome virus with monoclonal antibodies. Virology. 1998;252:106–114. doi: 10.1006/viro.1998.9436. [DOI] [PubMed] [Google Scholar]
- 22.Meulenberg J J M, Hulst M M, De Meijer E J, Moonen P L J M, Den Besten A, De Kluyver E P, Wensvoort G, Moormann R J M. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology. 1993;192:62–72. doi: 10.1006/viro.1993.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meulenberg J J M, Van Nieuwstadt A P, Van Essen-Zandbergen A, Langeveld J P M. Posttranslational processing and identification of a neutralization domain of the GP4 protein encoded by ORF4 of Lelystadt virus. J Virol. 1997;71:6061–6067. doi: 10.1128/jvi.71.8.6061-6067.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neitzert E, Beck E, de Mello P A, Gomes I, Bergmann I E. Expression of the aphthovirus RNA polymerase gene in Escherichia coli and its use together with other bioengineered nonstructural antigens in detection of late persistent infections. Virology. 1991;184:799–804. doi: 10.1016/0042-6822(91)90456-l. [DOI] [PubMed] [Google Scholar]
- 25.Nelsen C J, Murtaugh M P, Faaberg K S. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J Virol. 1999;73:270–280. doi: 10.1128/jvi.73.1.270-280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newman J F, Brown F. Foot-and-mouth disease virus and poliovirus particles contain proteins of the replication complex. J Virol. 1997;71:7657–7662. doi: 10.1128/jvi.71.10.7657-7662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen H S, Storgaard T, Oleksiewicz M B. Analysis of ORF 1 in European porcine reproductive and respiratory syndrome virus by long RT-PCR and restriction fragment length polymorphism (RFLP) analysis. Vet Microbiol. 2000;76:221–228. doi: 10.1016/s0378-1135(00)00258-3. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen T L, Nielsen J, Have P, Baekbo P, Hoff J R, Botner A. Examination of virus shedding in semen from vaccinated and from previously infected boars after experimental challenge with porcine reproductive and respiratory syndrome virus. Vet Microbiol. 1997;54:101–112. doi: 10.1016/s0378-1135(96)01272-2. [DOI] [PubMed] [Google Scholar]
- 29.Oleksiewicz M B, Botner A, Madsen K G, Storgaard T. Sensitive detection and typing of porcine reproductive and respiratory syndrome virus by RT-PCR amplification of whole viral genes. Vet Microbiol. 1998;64:7–22. doi: 10.1016/S0378-1135(98)00254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oleksiewicz M B, Botner A, Toft P, Grubbe T, Nielsen J, Kamstrup S, Storgaard T. Emergence of porcine reproductive and respiratory syndrome virus deletion mutants: correlation with the porcine antibody response to a hypervariable site in the ORF 3 structural glycoprotein. Virology. 2000;267:135–140. doi: 10.1006/viro.1999.0103. [DOI] [PubMed] [Google Scholar]
- 31.Pedersen K W, van der Meer Y, Roos N, Snijder E J. Open reading frame la-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J Virol. 1999;73:2016–2026. doi: 10.1128/jvi.73.3.2016-2026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peters E A, Schatz P J, Johnson S S, Dower W J. Membrane insertion defects caused by positive charges in the early mature region of protein pIII of filamentous phage fd can be corrected by prlA suppressors. J Bacteriol. 1994;176:4296–4305. doi: 10.1128/jb.176.14.4296-4305.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ray S C, Wang Y M, Laeyendecker O, Ticehurst J R, Villano S A, Thomas D L. Acute hepatitis C virus structural gene sequences as predictors of persistent viremia: hypervariable region 1 as a decoy. J Virol. 1999;73:2938–2946. doi: 10.1128/jvi.73.4.2938-2946.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen S, Kwang J, Liu W, Liu D X. Determination of the complete nucleotide sequence of a vaccine strain of porcine reproductive and respiratory syndrome virus and identification of the nsp2 gene with a unique insertion. Arch Virol. 2000;145:871–883. doi: 10.1007/s007050050680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith G P, Scott J K. Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 1993;217:228–257. doi: 10.1016/0076-6879(93)17065-d. [DOI] [PubMed] [Google Scholar]
- 36.Snijder E J, Meulenberg J J M. The molecular biology of arteriviruses. J Gen Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- 37.Sørensen K J, Strandbygaard B, Bøtner A, Madsen E S, Nielsen J, Have P. Blocking ELISA's for the distinction between antibodies against European and American strains of porcine reproductive and respiratory syndrome (PRRS) virus. Vet Microbiol. 1998;60:169–177. doi: 10.1016/s0378-1135(98)00159-x. [DOI] [PubMed] [Google Scholar]
- 38.Storgaard T, Oleksiewicz M, Bøtner A. Examination of the selective pressures on a live PRRS vaccine virus. Arch Virol. 1999;144:2389–2401. doi: 10.1007/s007050050652. [DOI] [PubMed] [Google Scholar]
- 39.van Woensel P A M, Liefkens K, Demaret S. Effect on viraemia of an American and a European serotype PRRSV vaccine after challenge with European wild-type strains of the virus. Vet Rec. 1998;142:510–512. doi: 10.1136/vr.142.19.510. [DOI] [PubMed] [Google Scholar]
- 40.Van Zaane D, Hulst M M. Monoclonal antibodies against porcine immunoglobulin isotypes. Vet Immunol Immunopathol. 1987;16:23–36. doi: 10.1016/0165-2427(87)90171-1. [DOI] [PubMed] [Google Scholar]
- 41.Wassenaar A L, Spaan W J, Gorbalenya A E, Snijder E J. Alternative proteolytic processing of the arterivirus replicase ORF 1a polyprotein: evidence that Nsp2 acts as a cofactor for the Nsp4 serine protease. J Virol. 1997;71:9313–9322. doi: 10.1128/jvi.71.12.9313-9322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiland E, Wieczorek-Krohmer M, Kohl D, Conzelmann K K, Weiland F. Monoclonal antibodies to the GP5 of porcine reproductive and respiratory syndrome virus are more effective in virus neutralization than monoclonal antibodies to the GP4. Vet Microbiol. 1999;66:171–186. doi: 10.1016/s0378-1135(99)00006-1. [DOI] [PubMed] [Google Scholar]
- 43.Yoon I J, Joo H, Goyal S, Molitor T. A modified serum neutralization test for the detection of antibody to porcine reproductive and respiratory syndrome virus in swine sera. J Vet Diagn Investig. 1994;6:289–292. doi: 10.1177/104063879400600326. [DOI] [PubMed] [Google Scholar]
- 44.Yoon K J, Wu L L, Zimmerman J J, Hill H T, Platt K B. Antibody-dependent enhancement (ADE) of porcine reproductive and respiratory syndrome virus (PRRSV) infection in pigs. Viral Immunol. 1996;9:51–63. doi: 10.1089/vim.1996.9.51. [DOI] [PubMed] [Google Scholar]
- 45.Ziebuhr J, Snijder E J, Gorbalenya A E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol. 2000;81 Pt 4:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 46.Zwick M B, Bonnycastle L L, Noren K A, Venturini S, Leong E, Barbas III C F, Noren C J, Scott J K. The maltose-binding protein as a scaffold for monovalent display of peptides derived from phage libraries. Anal Biochem. 1998;264:87–97. doi: 10.1006/abio.1998.2793. . (Erratum, 266:240, 1999.) [DOI] [PMC free article] [PubMed] [Google Scholar]