Abstract
Human immunodeficiency virus type 1 (HIV-1) variants resistant to protease (PR) and reverse transcriptase (RT) inhibitors may display impaired infectivity and replication capacity. The individual contributions of mutated HIV-1 PR and RT to infectivity, replication, RT activity, and protein maturation (herein referred to as “fitness”) in recombinant viruses were investigated by separately cloning PR, RT, and PR-RT cassettes from drug-resistant mutant viral isolates into the wild-type NL4-3 background. Both mutant PR and RT contributed to measurable deficits in fitness of viral constructs. In peripheral blood mononuclear cells, replication rates (means ± standard deviations) of RT recombinants were 72.5% ± 27.3% and replication rates of PR recombinants were 60.5% ± 33.6% of the rates of NL4-3. PR mutant deficits were enhanced in CEM T cells, with relative replication rates of PR recombinants decreasing to 15.8% ± 23.5% of NL4-3 replication rates. Cloning of the cognate RT improved fitness of some PR mutant clones. For a multidrug-resistant virus transmitted through sexual contact, RT constructs displayed a marked infectivity and replication deficit and diminished packaging of Pol proteins (RT content in virions diminished by 56.3% ± 10.7%, and integrase content diminished by 23.3% ± 18.4%), a novel mechanism for a decreased-fitness phenotype. Despite the identified impairment of recombinant clones, fitness of two of the three drug-resistant isolates was comparable to that of wild-type, susceptible viruses, suggestive of extensive compensation by genomic regions away from PR and RT. Only limited reversion of mutated positions to wild-type amino acids was observed for the native isolates over 100 viral replication cycles in the absence of drug selective pressure. These data underscore the complex relationship between PR and RT adaptive changes and viral evolution in antiretroviral drug-resistant HIV-1.
Nucleoside analog reverse transcriptase inhibitors and protease inhibitors effectively impair human immunodeficiency virus type 1 (HIV-1) replication and protein maturation. However, viral variants resistant to these drugs can be selected in patients in the course of treatment (16). Mutations in viral reverse transcriptase (RT) and protease (PR) leading to drug resistance are rarely detected in viruses not previously exposed to antiretroviral pressure, suggesting a selective in vivo growth disadvantage of mutant variants in the absence of treatment (10).
Mutations selected by RT and PR inhibitors generally involve changes of enzyme active-site residues. Reduction in primer extension activity by purified mutant RT has been demonstrated in vitro (2, 3). Mutant PR enzymes exhibit diminished catalytic efficiency in the processing of Gag and Gag-Pol polyproteins (8, 11, 29, 31, 39). These modifications in enzyme processivity may translate into in vitro or in vivo changes in viral infectivity or replication capacity (generally referred to as “fitness”) due to accumulation of immature viral particles (reviewed in reference 33).
Because of the remarkable plasticity of the HIV-1 genome, selection of compensatory mutations leading to improving enzyme function takes place. This process involves structurally relevant amino acid substitutions in target enzymes (e.g., in the hinge or flap regions of the viral protease) (6, 31), changes in the enzyme substrate which improve processing (e.g., Gag cleavage sites) (8, 13, 39), or modifications at as-yet-unrecognized sites elsewhere in the genome (26). However, in vitro data indicate that the pathways of compensation may not provide an immediate or full remediation of viral fitness deficits (22).
A number of issues related to fitness determinants and the natural evolution of mutant viruses have yet to be explored in depth. One issue is the respective contributions of PR and RT to viral fitness. Information to date suggests that PR rather than RT deficits are key to the infectivity and replicative impairment of mutant viruses (33, 34). A second issue is the extent of compensation of viral deficits and thus the potential for stability or reversion of the multimutated genome in the absence of selective pressure (6). This latter aspect will have consequences for the future of patients infected with multidrug-resistant HIV-1.
In the present study, we analyzed (i) the fitness phenotype of clinical isolates and their stability in the absence of drug selective pressure and (ii) the extent to which constructs incorporating mutant PR and RT, together or individually, reproduce the behavior of parental isolates. Our work proposes a standardized assessment of determinants of viral fitness through detailed correlation of various aspects of viral phenotype—infectivity, replication, RT activity, protein processing, and maturation—in the context of specific resistant genotypes and in various cellular testing systems.
MATERIALS AND METHODS
Isolates and viral culture conditions.
Native wild-type susceptible (n = 5) and drug-resistant HIV-1 isolates (n = 3) were cultured from blood cells (N2 to N5, B495, and B497) or plasma (N1 and B670) from HIV-1 infected patients (Table 1) using donor peripheral blood mononuclear cells (PBMCs) stimulated with phytohemagglutinin (2 μg/ml during 24 to 72 h). One drug-resistant isolate (B670) was isolated from a patient presenting with a primary infection after transmission of a multidrug-resistant HIV-1 strain. B495 and B497 are representative of the spectrum of replicative capacities of drug-resistant viruses (18).
TABLE 1.
Clinical characteristics and amino acid mutations in PR and RT of resistant isolatesa
| Isolate | Current treatment | Prior treatment | No. of CD4+ cells/μl | Viremia (log copies/ml) | Protein and mutation(s) | Comments |
|---|---|---|---|---|---|---|
| B495 | d4T, ritonavir, saquinavir | AZT, ddC, ddI, indinavir | 230 | 5.5 | PR: M46I, V77I, I84V, L90M RT: M41L, L210W, T215Y CL: P453L | Long-term failure to achieve viral suppression and slow immunological deterioration |
| B497 | d4T, ritonavir, saquinavir | AZT, 3TC, ddI, indinavir | 411 | 5.2 | PR: L10I, K20M, I54V, A71V, G73S, I84V, L90M RT: K65R, V75I, F77L, Y115F, F116Y, Q151M, K219Q CL: P453L | Clinical and immunological stability despite lack of viral suppression |
| B670 | None | None | 773 | 3.4 | PR: L10I, A71T, V77I, V82A, L90M RT: M41L, E44D, D67N, M184V, L210W, T215A CL: WT | Horizontal multidrug-resistant-HIV-1 transmission |
PBMCs were cultured in RPMI 1640 supplemented with Glutamax (2 mM) (Life Technologies, GIBCO BRL), gentamicin (50 μg/ml), fetal calf serum (FCS; 20% [vol/vol]), and interleukin 2 (10 U/ml). CEM T cells were cultured in RPMI 1640 supplemented with Glutamax (2 mM), gentamicin (50 μg/ml), and FCS (10% [vol/vol]). HeLa, COS-7, and GHOST cells (stably transduced with the chemokine receptor CCR5 or CXCR4 and with the green fluorescence protein [GFP] linked to the HIV-1 long terminal repeat; provided by D. Littman and V. K. Ramani, AIDS Research and Reference Reagent Program) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with Glutamax (2 mM), gentamicin (50 μg/ml), and FCS (10% [vol/vol]). For GHOST cells, selective medium also contained puromycin (1 μg/ml; Sigma), hygromycin (100 μg/ml; GIBCO), and G-418 (500 μg/ml; GIBCO).
Construction of deletion vector pNL4-3Δ series.
To perform stepwise cloning of RNA-derived PR and RT, three restriction sites were introduced into the proviral clone pNL-NF (a pNL4-3 lacking the cellular flanking regions 10353 to 11286 and 13708 to 14772; provided by T. Klimkait) by site-directed mutagenesis (Stratagene): an XbaI site 10 codons upstream of the PR sequence (primers pH7 and pH9) (Table 2) (38), a ClaI site 6 codons downstream of the PR sequence (primers pH8 and pH10) (38), and an MluI site in the RT sequence, 286 codons inside the RT coding region (primers pH11 and pH12).
TABLE 2.
Oligonucleotides used in this study
| Primer | Primer sequencea | Positions |
|---|---|---|
| Vector construction | ||
| pH7 | 5′GGAGCCTCTAGACAAGGAACTGTATCCT3′ | 2217–2244 |
| pH8 | 5′GTACAGTATCGATAGGACTAATGGGAAA3′ | 2547–2574 |
| pH9 | 5′AGGATACAGTTCCTTGTCTAGAGGCTCC3′ | 2217–2244 |
| pH10 | 5′TTTCCCATTAGTCCTATCGATACTGTAC3′ | 2547–2574 |
| pH11 | 5′GTAAACTTCTTAGGGGAACGCGTGCACTAAC3′ | 3388–3418 |
| pH12 | 5′GTTAGTGCACGCGTTCCCCTAAGAAGTTTAC3′ | 3388–3418 |
| pH13 | 5′GGAATTGGAGGTTTTATCTAGATAGGAC3′ | 2397–2424 |
| pH14 | 5′CAAATACGCGTGTATTGTATGGATTTTCAG3′ | 2704–2733 |
| PR amplification | ||
| PR1849 | 5′GATGACAGCATGTCAGGGAGTA3′ | 1827–1847 |
| PR2796 | 5′CTTCCCAGAAGTCTTGAGTTCT3′ | 2796–2817 |
| PR1937 | 5′ATGCTGCAGAGAGGCAATTT3′ | 1918–1937 |
| PR2716 | 5′GGCAAATACTCGAGTATTGT3′ | 2716–2735 |
| RT amplification | ||
| RTA1 | 5′AATTTTCCCATTAGTCCTATT3′ | 2544–2564 |
| Gag3 | 5′TAAGTCTTTTGATGGGTCATAATA3′ | 3501–3524 |
| RTNNA | 5′AAGCCAGGAATGGATGGCCCA3′ | 2586–2606 |
| RTNE1 | 5′TATGTCATTGACAGTCCAGCT3′ | 3300–3320 |
Restriction sites used for cloning are underlined, and modified nucleotides are in bold. Nucleotide numbers correspond to the pNL4-3 sequence (GenBank accession number M19921).
The series of cloning vectors was rendered nonfunctional by targeted PR and/or RT deletions, so that only correct cloning of viral PR and RT would allow production of viable virions. Deletion vectors pNL-ΔPR, -ΔRT, and -ΔPRΔRT were constructed by cloning a pNL4-3 fragment containing amino acids 55 to 99 of PR and 1 to 58 of RT (amplified by PCR with primers pH13 and pH14) into the XbaI/MluI restriction sites of the modified vector (to construct pNL-ΔPRΔRT) and by stepwise reinsertion of wild-type PR (for pNL-ΔRT) or RT sequences (for pNL-ΔPR). This series of vectors allows cloning from amino acids D47 to P159 of Pol (complete PR), from P159 to G440 (RT P1 to G285), and the PR-RT fragment (Pol D47 to G440).
Recombinant clones.
Mutant PR and RT from viral isolates were amplified from extracted viral RNA (RNeasy kit; Qiagen) by reverse transcription-PCR (primer pairs PR1849-PR2796 for PR and RTA1-Gag3 for RT) (Table 2), followed by a nested PCR with primer pairs pH7-pH8 (for PR) and pH10-pH12 (for RT). PCR products were digested with XbaI and ClaI (PR) and ClaI and MluI (RT) and cloned into the series of pNL-Δ vectors. All constructs were confirmed by sequencing. Recombinant viruses were obtained by HeLa cell transfection (Geneporter transfection reagent; Axon Labs, Baden, Switzerland).
Single-cycle infectivity assay.
GHOST/CCR5 cells in 48-well plates (4 × 104 cells/well) were infected in triplicate with clinical isolates (1,500 pg of p24 antigen) in 300 μl of supplemented DMEM in the presence of 20 μg of Polybrene/ml by a spinoculation technique: 3 h of centrifugation at 1,500 × g and 22°C (4). Cells were trypsinized 24 h postinfection, harvested in 700 μl of phosphate-buffered saline–5% FCS–2 mM EDTA, and resuspended in 250 μl of cell-fixing solution (Becton Dickinson, Erembodegem, Belgium). The infectious titer was determined by fluorescence-activated cell sorting (FACS) analysis as the proportion of GFP-positive cells. The single-cycle infectivity titer of recombinant viruses was determined in GHOST/CXCR4 cells upon infection with 3,500 pg of p24 antigen as described above.
Replication kinetic assay.
PBMCs (3 × 106 cells) were infected with native or recombinant virus (1,500 pg of p24 antigen) in 1 ml of supplemented RPMI 1640 for 3 h. Thereafter, the residual inoculum was removed by washing, and cells were maintained in culture for 10 days at 106 cells/ml. Aliquots of culture supernatant were collected to monitor viral replication using an HIV-1 p24 antigen enzyme-linked immunosorbent assay (HIVAG-1 Monoclonal; Abbott). CEM cells (106) were infected with recombinant virus (3,000 pg of p24 antigen) in 1 ml of supplemented RPMI 1640 for 3 h. Thereafter, the residual inoculum was removed by washing, and cells were maintained in culture for 21 days at 0.5 × 106 cells/ml. Input virus concentrations were also normalized by 50% tissue culture infectious dose values. Relative replication rates were estimated, after logarithmic transformation of p24 values, by comparison of p24 antigen slopes between recombinant clones and NL4-3. Student's t test was used for statistical analysis.
RT activity assay.
The RT activity assay was carried out according to the manufacturer's protocol (Lenti RT activity assay; Cavidi Tech, Uppsala, Sweden). Briefly, 50 μl of filtered culture supernatant was serially diluted (1/5, 1/25, and 1/125) and added to a 96-well plate with poly(rA) (enzyme template) bound to the bottom of the wells and with 150 μl of reaction solution containing bromodeoxyuridine triphosphate as the enzyme substrate. Polymerization was allowed to proceed for 3 h at 33°C. Immunological product detection with alkaline phosphatase (AP)-conjugated antibromodeoxyuridine antibody was carried out at 33°C for 90 min. The level of bound antibody was determined colorimetrically with an AP substrate, para-nitrophenyl phosphate, in a standard microtiter plate reader (405 nm) at 0.5, 1, 2, 4, and 6 h after addition of the AP substrate. RT activity was determined at the time of replicative peak in PBMC cultures, normalized to p24 antigen content, and expressed as picograms of RT per nanogram of p24.
Protein maturation analysis.
Subconfluent COS-7 cells were transfected with 10 μg of the different recombinant clones in a 100-mm petri dish. At 24 h posttransfection, cells were metabolically labeled for 12 to 17 h with 5 ml of DMEM (methionine-cysteine-free, 10% dialyzed FCS) containing 40 μCi of [35S]methionine-[35S]cysteine (35S : Easy Tag EXPRESS; NEN, Life Science Products, Boston, Mass.)/ml (38).
To analyze particle-associated proteins, virions in the culture supernatant were concentrated through a 20% sucrose cushion by ultracentrifugation (90 min with a centrifugal force of 100,000 × g at 4°C) and lysed in 200 μl of radioimmunoprecipitation assay buffer (1% Nonidet P-40, 0.5% deoxycholic acid, 0.9% sodium dodecyl sulfate [SDS], 2 mM EDTA, 150 mM NaCl, 50 mM Tris-HCl [pH 8]), supplemented with complete protease inhibitor cocktail (Roche Diagnostics).
To analyze cell-associated viral proteins, transfected cells were washed with phosphate-buffered saline, lysed in 500 μl of radioimmunoprecipitation assay buffer, and centrifuged for 30 min at 21,000 × g and 4°C. Viral proteins were immunoprecipitated from lysates using anti-HIV human immunoglobulin G (NIH AIDS Research & Reference Reagent Program) and protein A-Sepharose CL-4B beads (Amersham Pharmacia Biotech). Twenty microliters of immunoprecipitated radiolabeled proteins per sample was separated by SDS-polyacrylamide gel electrophoresis (PAGE) (5-to-15% gradient gel), and radioactivity content was quantified using Instant Imager (Packard Instruments). Values were normalized to the quantity of p24 and gp160/120 and compared to NL4-3 protein content.
In vitro analysis of genetic stability of drug-resistant viral isolates.
PBMCs (3 × 106) were infected with resistant viral isolates (104 pg of p24 antigen) and cultured for 2 weeks at 106 cells/ml in supplemented RPMI 1640. At day 14 postinfection, aliquots of culture supernatant were removed for p24 antigen measurement. An equivalent of 104 pg of p24 antigen from each passage was used to infect fresh cultures of donor PBMCs. Isolates were passaged up to seven times, corresponding to 100 viral life cycles.
At the end of each passage, amplification and sequencing of PR, RT, and Gag cleavage sites of the viral isolates were performed by using primer pairs PR1849-PR2796 (for Gag cleavage sites and PR) and RTA1-Gag3 (for RT) for reverse transcription and the first round of amplification. Primer pairs for the nested PCR were PR1937-PR2716 (for Gag cleavage sites and PR) and RTNNA-RTNE1 (for RT) (Table 2).
Nucleotide sequence accession numbers.
Nucleotide sequences have been submitted to GenBank under accession no. AF316831 to AF316837 (B495 isolate and recombinant clones), AF316838 to AF316844 (B497 isolate and recombinant clones), and AF316845 to AF316851 (B670 isolate and recombinant clones).
RESULTS
Infectivity and replication efficiency of wild-type and mutant resistant isolates.
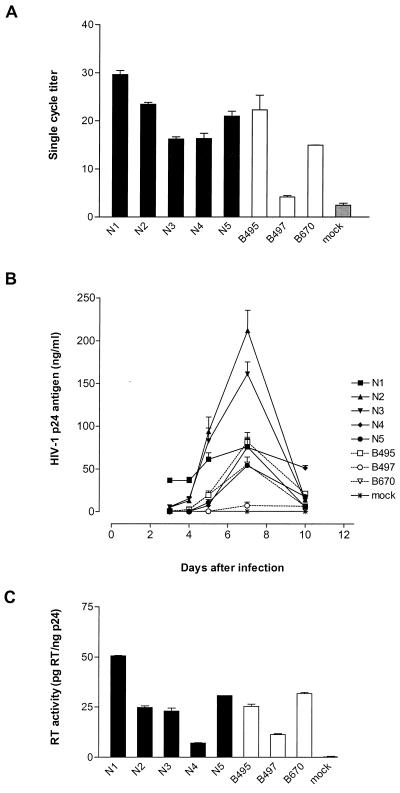
Wild-type and mutant viral isolates (all obtained using CCR5 cells) displayed a range of infectivity as determined in GHOST/CCR5 cells (Fig. 1A). While isolates B495 and B670 presented an infectious titer comparable to that of wild-type isolates, B497 displayed a 40 to 85% reduction in infectivity compared to wild-type isolates.
FIG. 1.
Infectivity, replication, and RT activity of clinical isolates. (A) Single-cycle infectivity of wild-type (N1 to N5) and mutant (B495, B497, and B670) viral isolates. The single-cycle titer was determined by FACS analysis of GHOST/CCR5 cell fluorescence as the percent GFP-positive cells. Error bars indicate the ranges of values obtained from triplicate data points. (B) Replication kinetics of wild-type (solid lines) and mutant (dotted lines) viral isolates. PBMCs were infected with p24-normalized amounts of particles, and virus production was monitored by measuring p24 antigen concentration in the culture supernatant. The data are representative of three independent experiments in which comparable results were obtained. Error bars reflect triplicate data points from a single experiment. (C) RT activity of wild-type and mutant viral isolates. Values were normalized to p24 antigen concentration in supernatant. Error bars represent the ranges of values obtained in two independent assays. RT activity was determined in PBMC culture supernatants at the peak of replication.
As with the infectivity testing, there was a range in replication capacity in PBMCs for both wild-type and mutant viral isolates (Fig. 1B). B497, the least infectious virus, displayed a 1- to 2-log replication impairment compared to wild-type isolates.
RT activity (Fig. 1C), analyzed on PBMC culture supernatants, correlated with infectivity: R2 = 0.625, P = 0.03.
Genome stability of resistant isolates.
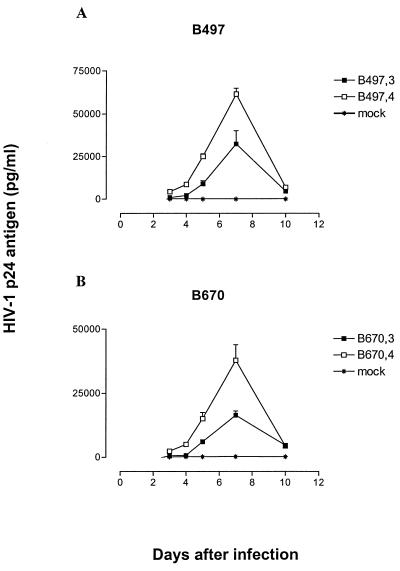
The stability of viral Gag, PR, and RT sequences in the absence of drug pressure was assessed in PBMCs for up to 100 viral life cycles (20). Limited genetic evolution in terms of new polymorphisms, putative compensatory amino acid changes, or reversion of mutations associated with drug resistance was observed. For the region sequenced, encompassing from the last 96 amino acids of Gag to the first 232 amino acids of RT, amino acid variation over time ranged from 0 to 4%. With two exceptions, sequence analysis at the end of each passage revealed stability for resistance-associated mutations (16) in PR and RT as well as for cleavage site adaptations in Gag (5). At the third passage (≈40 viral life cycles), isolate B497 reverted RT residue 65 from arginine to wild-type lysine, and isolate B670 reverted RT residue 184 from valine to wild-type methionine. Comparison of replication capacity of B497 and B670 from passage 3 to 4 revealed, in both instances, an improvement in viral growth (Fig. 2).
FIG. 2.
Replication kinetics of B497 and B670 in PBMCs before and after reversion of RT codons 65 from arginine (B497,3) to wild-type lysine (B497,4) and 184 from valine (B670,3) to wild-type methionine (B670,4). PBMCs were infected with p24-normalized amounts of particles, and virus production was monitored by measuring p24 antigen concentration in the culture supernatant. The data are representative of two independent experiments in which comparable results were obtained. Error bars reflect triplicate data points from a single experiment.
PR and RT recombinant viruses.
For each drug-resistant isolate, three sets of recombinant viruses were constructed in a wild-type pNL4-3 background: clones comprising only the mutated PR (PR clones), only the mutated RT (RT clones), or both mutated PR and RT (PR-RT clones). Two individual clones per construct were analyzed. Mutations introduced in the vector pNL4-3 for the purpose of cloning (Pol D47S, E161D, and K442R) had no impact on infectivity or replication (results not shown).
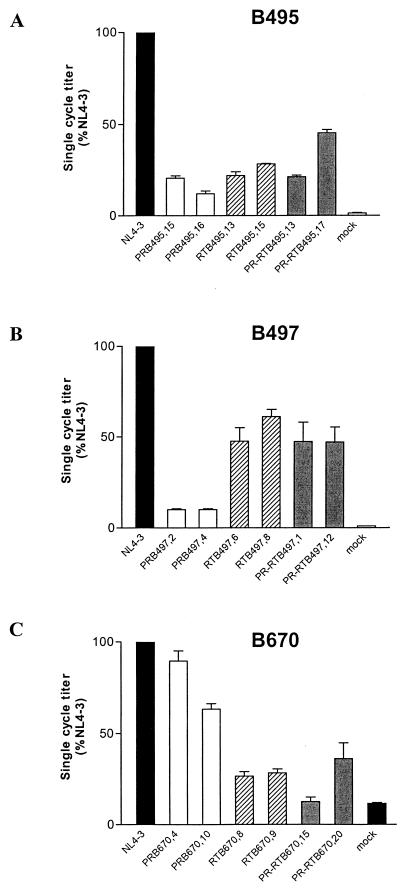
Infectivity of PR, RT, and PR-RT constructs.
The infectivity of various constructs in GHOST/CXCR4 is shown in Fig. 3. PR and RT recombinants of B495 displayed a 55 to 88% decrease in infectivity with respect to wild-type NL4-3 (Fig. 3A). For B497, PR constructs displayed a 10% infectivity, RT constructs displayed a 48 to 61% infectivity; and PR-RT cognate constructs compensated for the PR deficit and exhibited a 47% infectivity (Fig. 3B). For B670, PR constructs displayed a 60 to 90% infectivity, RT constructs displayed a 27% infectivity, and infectivity of PR-RT constructs followed the behavior of the corresponding RT clones, with 12 to 36% infectivity (Fig. 3C).
FIG. 3.
Single-cycle infectivity of recombinant clones of B495, B497, and B670. The single-cycle titer of each PR (white bars)-, RT (hatched bars)-, and PR-RT (grey bars)-mutated virus was determined by FACS analysis of GHOST/CXCR4 cell fluorescence and expressed as a percentage of the wild-type NL4-3 value (black bars). Error bars indicate the ranges of values obtained from triplicate data points in a single experiment.
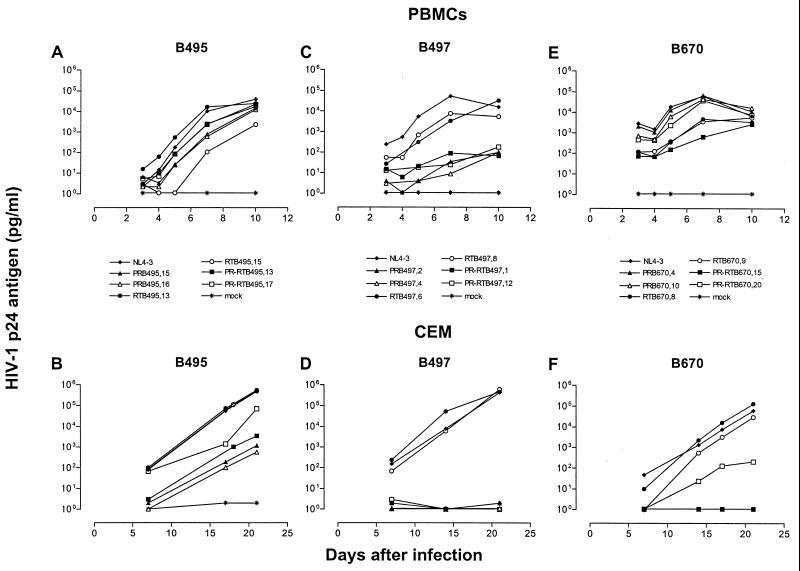
Replication of PR, RT, and PR-RT constructs.
Replication profiles of the recombinant clones were investigated in two cell systems, PBMCs and the CEM T-cell line. Distinct replication patterns were observed for the same constructs when the were propagated in these two cell types (Fig. 4). While replication capacity in PBMCs generally paralleled the respective patterns of GHOST cell infectivity, CEM cells favored the growth of RT recombinants in the cases of five of six RT clones (RTB495.13 already displayed a replication rate comparable to that of wild-type NL4-3 in PBMCs). Overall, relative replication rates (mean ± standard deviation) of RT recombinants improved by 28%, increasing from 72.5% ± 27.3% of the NL4-3 replicative rate in PBMCs to 100.2% ± 7.1% in CEM cells (P = 0.04). PR recombinants exhibited various degrees of compromised replication profiles in PBMCs and exhibited a profound impairment in CEM cells. Overall, relative replication rates of PR recombinants worsened by 45%, decreasing from 65.5% ± 33.6% of the NL4-3 replicative rate in PBMCs to 15.8% ± 23.5% in CEM (P = 0.02).
FIG. 4.
Replication kinetics of recombinant clones of B495, B497, and B670. PBMCs and CEM cells were infected with p24-normalized amounts of viral particles. Virus production was monitored by measuring p24 antigen concentration in the culture supernatant. The data are from one of two independent experiments in which comparable results were obtained.
While relative replication rates of the most impaired recombinants improved from 54.7% ± 38.0% to 64.3% ± 24.9% in PBMCs and from 15.8% ± 23.5% to 31.3% ± 34.8% in CEM cells upon cloning of the cognate enzyme, differences for this small collective were not statistically significant (P > 0.05). Two recombinants of B495 (Fig. 4B) increased relative replication rates in CEM cells from 43 and 49% in PR-only clones to 59 and 79%, and one clone of B670 increased the relative replication rate from 0 to 47% (Fig. 4F). In an alternatively interpretation of the data, the introduction of the mutant PR could result in an overall reduction of replication rate of RT constructs if the PR growth phenotype was dominant.
Normalization of viral input by 50% tissue culture infectious doses did not alter the outcome of replication kinetics (data not shown). Differences in the behavior of one of the two recombinants per category (PR, RT, or PR-RT) might be explained by observed variations in sequence of 0.3 to 2%, reflecting the original pool of quasispecies in patients' plasma. Importantly, all the resistance mutations of the parental virus were present in the recombinants.
RT activity of PR, RT, and PR-RT constructs.
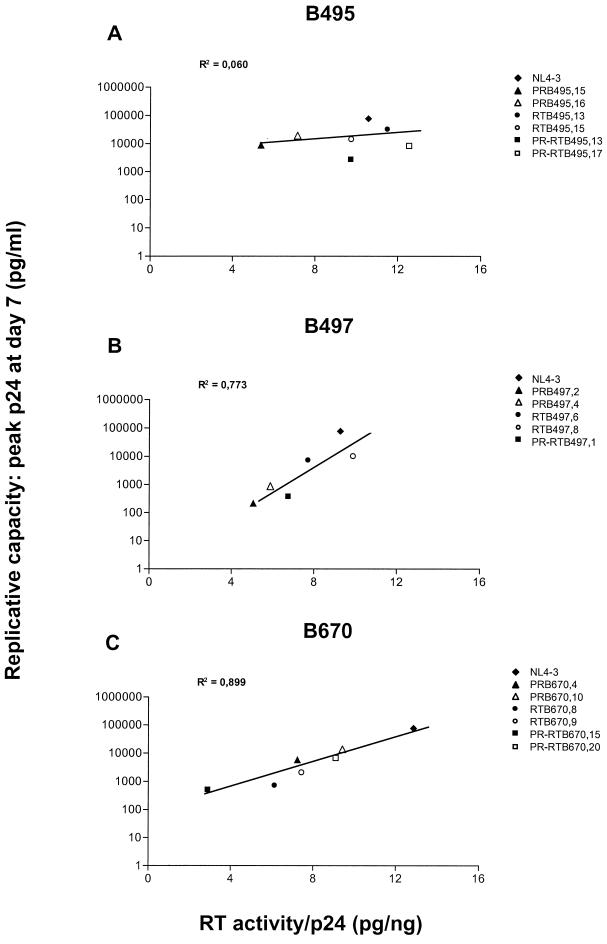
For B497 and B670, RT activity of recombinant virions matched their replication capacity in PBMCs (Fig. 5B and C). No correlation was observed for B495, where RT activities for mutant clones ranged from 5.37 to 12.54 pg of RT/ng of p24, compared to 10.57 pg of RT/ng of p24 for the control NL4-3 (Fig. 5A).
FIG. 5.
RT activity of recombinant clones of B495, B497, and B670. RT activity was determined in PBMC culture supernatants at the peak of replication and normalized to p24 antigen concentration. Regression lines show the correlation of viral RT activity with replicative capacity. PR-RTB497,12 is not represented due to limited replication and RT activity outside the assay range. The data are from one of two independent experiments in which comparable results were obtained.
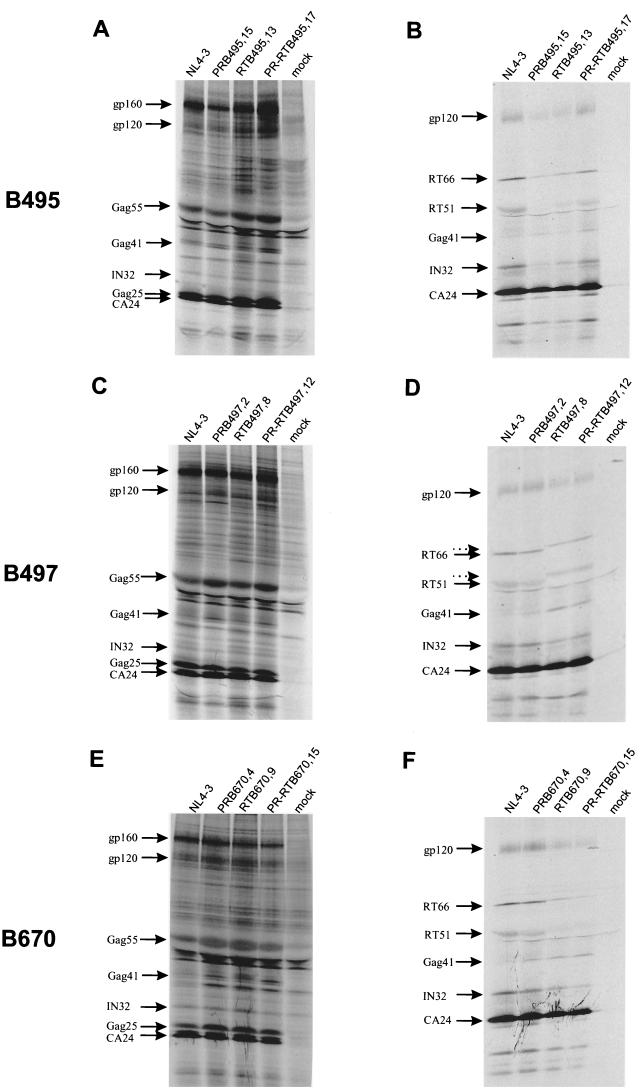
Viral protein maturation.
Maturation was assessed by immunoprecipitation of cell-associated and supernatant viral proteins after metabolic labeling in transfected COS-7 cells (Fig. 6). Assessment of RT constructs identified various maturation abnormalities. B497 RT recombinants exhibited an upward shift in RT66 and RT51 migration, likely resulting from conformational or charge changes in the mutated enzyme. B670 and B497 RT recombinants showed an associated Gag processing defect, as demonstrated by up to 2.5-fold accumulation of Gag41 intermediates in virions (Fig. 6D and F). A second defect in B670 RT recombinants involved a failure to package Pol products into virions: RT particle content diminished by 56.3% ± 10.7% and integrase content diminished by 23.3% ± 18.4% compared to that of NL4-3 (Fig. 6F). PR was not evaluable by the SDS-PAGE strategy used.
FIG. 6.
Protein processing and maturation profiles in cell lysates (A, C, and E) and supernatant virions (B, D, and F) of recombinant clones of B495, B497, and B670. COS-7 cells were transfected with recombinant clones and metabolically labeled with [35S]methionine-[35S]cysteine. Proteins in cell-associated particles and virions were immunoprecipitated with anti-HIV human immunoglobulin G and analyzed by SDS-PAGE and fluorography. Precursor and processed viral proteins are indicated. Dotted arrows (D) indicate upward shifts in migration of RT66/RT51.
The extent of precursor processing defects due to mutant PR was not pronounced. Only in recombinants of B670 did Gag 41 accumulate up to threefold in cell lysate material compared to NL4-3. (Fig. 6E).
DISCUSSION
Experiences with viral mutant clones have generally linked replicative capacity to the functionality of the mutant PR (8, 11, 13, 23, 29, 34, 39). In contrast, drug resistance mutations in RT have been associated with a broad spectrum of in vitro and in vivo fitness (15, 32, 33), frequently with minor defects or no alteration in RT enzyme processivity (7, 30). While fitness of a virus is best defined by its replicative capacity in the course of competition kinetics (21, 23), most published literature extends the concept of fitness to various other measurements of viral infectivity and replicative phenotype.
In the present study, detailed analysis of recombinant viruses shows that defects in RT, in PR, or in both may contribute to the infectivity and replicative capacity phenotype of a clone. These defects can be expressed differently depending on the cellular system used for testing. Results of single-cycle infectivity in GHOST cells indicate a good correlation with replication kinetics and RT activities of recombinant clones tested in PBMCs. In contrast, testing in the CEM T-cell line accentuated the defects linked to mutant PR, while improving the replicative capacity of clones carrying a mutant RT. Technical as well as biological events may explain the discrepant behavior of mutant constructs: cell lines with a large intracellular pool of deoxynucleoside triphosphates may improve RT defects (2), while cell line culture manipulation through greater removal of infected cells and substitution with fresh media, as done for CEM cells, may limit expansion of viruses with late cycle defects linked to PR mutations. However, we do not exclude cellular factors leading to deepening of PR mutant replication defects in specific cell lines. Back et al. have recommended that primary cell types should be used in fitness assays with drug-resistant variants (3). Clearly, analysis and reporting of biological behavior of mutants should take into account the cellular system used for testing (33).
Despite the impaired fitness of mutant PR clones, we could not identify more than moderate defects in HIV-1 protein maturation profiles. Although more pronounced defects have been previously described (9, 38), resistant clones derived from clinical isolates in the present study might have undergone significant compensation to preserve PR function in vivo. Profound defects of maturation and any cleavage site processing rate lower than 61% are predicted to result in noninfectious, nonviable immature virions (28).
RT-determined impairment was identified in recombinant clones from two resistant isolates. Clones from a particular isolate, a multidrug-resistant virus transmitted through sexual contact (B670), exhibited a novel characteristic: their behavior in GHOST cells and PBMCs was determined by the deficits imposed by the mutant RT rather than by the PR. The specific defect was manifested by a packaging failure of Pol products. Virions carried 56% less RT than the parental NL4-3 isolate. In addition, they exhibited a moderate defect in maturation, as demonstrated by the presence of Gag41 intermediates in viral particles. RT activity paralleled the fitness of the clones. This phenomenon could be explained by the alteration of specific RT domains involved in packaging, or by structural constraints imposed by the mutant RT leading to unstable Gag-Pol intermediates (9). Although assembly and function of RT can be unlinked (35), mutations of the RT primer grip (L234D and W239A) (36) and integrase mutants (1, 27) result in defective maturation and failure to package Pol, possibly through premature cleavage of the Gag-Pol precursors (36).
Fitness of two of the three drug-resistant clinical isolates studied was, despite the deficits of recombinant clones, comparable to that of wild-type drug-susceptible controls. This suggests that extensive compensatory changes take place not only at the intramolecular (PR or RT) level, through accumulation of secondary mutations, but also through intermolecular adaptation, as underscored by the fitness improvement from cloning of the cognate RT into PR constructs shown by us and others (9), or by compensation at a distance, at characterized (13, 39) or uncharacterized (24, 26) genomic sites. Potential differences in behavior between native isolates and their reconstructed clones should be taken into account when data from newly available fitness assays that use recombinant viruses are analyzed (34).
The genotypes of resistant clinical isolates proved stable in vitro in the absence of drug pressure. While Borman and Clavel described the appearance of secondary PR mutations in resistant isolates cultured in drug-free medium (6), minimal genome evolution and only two instances of reversion of a RT mutation associated with resistance were documented in the present study. Interestingly, the isolate identified in the setting of sexual transmission reverted RT residue 184 from valine to the wild-type residue methionine both in vitro and in vivo in the absence of antiretroviral therapy (S. Yerly and L. Perrin, personal communication). In vitro this was associated with an improvement of replicative capacity of the isolate. While we ascribe the viral evolution to a limited reversion of resistance-associated mutations, we cannot exclude a gradual overgrowth with a more fit but less resistant viral variant.
The genome stability observed in drug-free culture over 6 months could be explained by a significant interlocking of primary and compensatory mutations that limits reversion through unfit intermediate amino acid variants. This observation has relevance to the current debate on structured treatment interruption in individuals experiencing virological failure due to multidrug resistance (17). Treatment interruption will generally lead to rapid overgrowth by archival, more fit wild-type virus rather than reversion of the multidrug-resistant variant (12, 14). In contrast, viruses in patients primarily infected by multidrug-resistant viruses may evolve slowly towards more susceptible variants whenever the diversity of transmitted quasispecies is restricted to resistant variants through a founder effect.
This study included only a limited number of representative clinical isolates. However, it serves to underscore the fact that the fitness phenotype of drug-resistant HIV-1 strains represents a complex interplay of cellular and virological factors. A better knowledge of mechanisms of viral infectivity and replication capacity impairment and compensation is relevant to the analysis of the clinical impact and pathogenesis of multidrug-resistant HIV-1 infection and to the analysis of evolutionary and adaptive limits of HIV-1 (24, 37). The immune stability displayed by a subgroup of individuals infected with drug-resistant viruses can be explained in part by a reduced fitness phenotype of resistant mutants under continued drug pressure (18, 19, 25).
ACKNOWLEDGMENTS
We thank Sabine Yerly and Luc Perrin for viral isolate B670, Thomas Klimkait for the HIV molecular clone pNL-NF, Brendan Larder for helpful discussions, and Solange Peters, Gilbert Greub, and Raquel Martinez for assistance.
Support for this work was provided by the Swiss National Science Foundation (grants 3346–62092.99 and 33–61321.00) and by the Santos-Suarez Foundation.
REFERENCES
- 1.Ansari-Lari A M, Donehower L A, Gibbs R A. Analysis of human immunodeficiency virus type 1 integrase mutants. Virology. 1995;211:332–335. doi: 10.1006/viro.1995.1412. [DOI] [PubMed] [Google Scholar]
- 2.Back N K T, Berkhout B. Limiting deoxynucleoside triphosphate concentrations emphasize the processivity defect of lamivudine-resistant variants of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1997;41:2484–2491. doi: 10.1128/aac.41.11.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Back N K T, Nijhuis M, Keulen W, Boucher C A B, Oude Essink B B, van Kuilenburg A B P, van Gennip A H, Berkhout B. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 1996;15:4040–4049. [PMC free article] [PubMed] [Google Scholar]
- 4.Bahnson A. Centrifugal enhancement of retroviral mediated gene transfer. J Virol Methods. 1995;54:131–134. doi: 10.1016/0166-0934(95)00035-s. [DOI] [PubMed] [Google Scholar]
- 5.Bally F, Martinez R, Peters S, Sudre P, Telenti A. Polymorphism of HIV-1 Gag p7/p1 and p1/p6 cleavage sites. Clinical significance and implications for resistance to protease inhibitors. AIDS Res Hum Retrovir. 2000;16:1209–1213. doi: 10.1089/08892220050116970. [DOI] [PubMed] [Google Scholar]
- 6.Borman A M, Clavel F. Resistance to human immunodeficiency virus type 1 to protease inhibitors: selection of resistance mutations in the presence and absence of the drug. J Gen Virol. 1996;77:419–426. doi: 10.1099/0022-1317-77-3-419. [DOI] [PubMed] [Google Scholar]
- 7.Caliendo A M, Savara A, An D, DeVore K, Kaplan J C, D'Aquila R T. Effects of zidovudine-selected human immunodeficiency virus type 1 reverse transcriptase amino acid substitutions on processive DNA synthesis and viral replication. J Virol. 1996;70:2146–2153. doi: 10.1128/jvi.70.4.2146-2153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrillo A, Stewart K D, Sham H L, Norbeck D W, Kohlbrenner W E, Leonard J M, Kempf D J, Molla A. In vitro selection and characterization of human immunodeficiency virus type 1 variants with increased resistance to ABT-378, a novel protease inhibitor. J Virol. 1998;72:7532–7541. doi: 10.1128/jvi.72.9.7532-7541.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carron de la Carriere L, Paulous S, Clavel F, Mammano F. Effects of human immunodeficiency virus type 1. Resistance to protease inhibitors on reverse transcriptase processing, activity, and drug sensitivity. J Virol. 1999;73:3455–3459. doi: 10.1128/jvi.73.4.3455-3459.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 11.Croteau G, Doyon L, Thibeault D, McKercher G, Pilote L, Lamarre D. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J Virol. 1997;71:1089–1096. doi: 10.1128/jvi.71.2.1089-1096.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devereux H L, Youle M, Johnson M A, Loveday C. Rapid decline in detectability of HIV-1 resistance mutations after stopping therapy. AIDS. 1999;13:F123–F127. [PubMed] [Google Scholar]
- 13.Doyon L, Croteau G, Thibeault D, Poulin F, Pilote L, Lamarre D. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J Virol. 1996;70:3763–3769. doi: 10.1128/jvi.70.6.3763-3769.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Essajee S M, Kim M, Gonzalez C, Rigaud M, Kaul A, Chandwani S, Hoover W, Lawrence R, Spiegel H, Pollack H, Krasinski K, Borkowsky W. Immunological and virological responses to HAART in severely immunocompromised HIV-1 infected children. AIDS. 1999;13:2523–2532. doi: 10.1097/00002030-199912240-00005. [DOI] [PubMed] [Google Scholar]
- 15.Goudsmit J, de Ronde A, de Rooij E, de Boer R. Broad spectrum of the in vivo fitness of human immunodeficiency virus type 1 subpopulations differing at reverse transcriptase codons 41 and 215. J Virol. 1997;71:4479–4484. doi: 10.1128/jvi.71.6.4479-4484.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirsch M S, Brun-Vezinet F, D'Aquila R T, Hammer S M, Johnson V A, Kuritzkes D R, Loveday C, Mellors J W, Clotet B, Conway B, Demeter L M, Vella S, Jacobsen D M, Richman D D. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society-USA panel. JAMA. 2000;283:2417–2426. doi: 10.1001/jama.283.18.2417. [DOI] [PubMed] [Google Scholar]
- 17.Katlama C. Structured antiretroviral treatment interruption in heavily-experienced HIV-infected patients. AIDS Rev. 2000;2:9–14. [Google Scholar]
- 18.Kaufmann D E, Munoz M, Bleiber G, Fleury S, Lotti B, Martinez R, Piechler W, Meylan P, Telenti A. Virological and immunological characteristics of HIV treatment failure. AIDS. 2000;14:1767–1774. doi: 10.1097/00002030-200008180-00012. [DOI] [PubMed] [Google Scholar]
- 19.Kaufmann D E, Pantaleo G, Sudre P, Telenti A for the Swiss HIV Cohort Study. CD4-cell count in HIV-1 infected individuals remaining viraemic with highly active antiretroviral therapy (HAART) Lancet. 1998;351:723–724. doi: 10.1016/s0140-6736(98)24010-4. [DOI] [PubMed] [Google Scholar]
- 20.Klotman M E, Kim S, Buchbinder A, DeRossi A, Baltimore D, Wong-Staal F. Kinetics of expression of multiply spliced RNA in early human immunodeficiency virus type 1 infection of lymphocytes and monocytes. Proc Natl Acad Sci USA. 1991;8:5011–5015. doi: 10.1073/pnas.88.11.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kosalaraksa P, Kavlick M F, Maroun V, Le R, Mitsuya H. Comparative fitness of multi-dideoxynucleoside-resistant human immunodeficiency syndrome HIV-1 in an in vitro competitive replication assay. J Virol. 1999;73:5356–5363. doi: 10.1128/jvi.73.7.5356-5363.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mammano F, Petit C, Clavel F. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and Gag coevolution in protease inhibitor-treated patients. J Virol. 1998;72:7632–7637. doi: 10.1128/jvi.72.9.7632-7637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez-Picado J, Savara A V, Sutton L, D'Aquila R T. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J Virol. 1999;73:3744–3752. doi: 10.1128/jvi.73.5.3744-3752.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nijhuis M, Schuurman R, de Jong D, Erickson J W, Gustchina E, Albert J, Schipper P J, Boucher C A B. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS. 1999;13:2349–2359. doi: 10.1097/00002030-199912030-00006. [DOI] [PubMed] [Google Scholar]
- 25.Perrin L, Telenti A. HIV treatment failure: testing for HIV resistance in clinical practice. Science. 1998;280:1871–1873. doi: 10.1126/science.280.5371.1871. [DOI] [PubMed] [Google Scholar]
- 26.Peters S, Munoz M, Martinez R, Meylan P, Telenti A. Polymorphism of HIV p6 Gag under selective pressure. Antivir Ther. 2000;5(Suppl. 3):38–39. [Google Scholar]
- 27.Quillent C, Borman A M, Paulous S, Dauguet C, Clavel F. Extensive regions of pol are required for efficient human immunodeficiency virus polyprotein processing and particle maturation. Virology. 1996;219:29–36. doi: 10.1006/viro.1996.0219. [DOI] [PubMed] [Google Scholar]
- 28.Rasnick D. Kinetics analysis of consecutive HIV proteolytic cleavages of the Gag-Pol polyprotein. J Biol Chem. 1997;272:6348–6353. [PubMed] [Google Scholar]
- 29.Rayner M M, Cordova B, Jackson D A. Population dynamic studies of wild-type and drug-resistant mutant HIV in mixed infections. Virology. 1997;236:85–94. doi: 10.1006/viro.1997.8620. [DOI] [PubMed] [Google Scholar]
- 30.Schmit J C, Cogniaux J, Hermans P, Van Vaeck C, Sprecher S, Van Remoortel B, Witvrouw M, Balzarini J, Desmyter J, De Clercq E, Vandamme A M. Multiple drug resistance to nucleoside analogues and nonnucleoside reverse transcriptase inhibitors in an efficiently replicating human immunodeficiency virus type 1 patient strain. J Infect Dis. 1996;174:962–968. doi: 10.1093/infdis/174.5.962. [DOI] [PubMed] [Google Scholar]
- 31.Schock H B, Garsky V M, Kuo L C. Mutational anatomy of an HIV-1 protease variant conferring cross-resistance to protease inhibitors in clinical trials. J Biol Chem. 1996;271:31957–31963. doi: 10.1074/jbc.271.50.31957. [DOI] [PubMed] [Google Scholar]
- 32.Sharma P L, Crumpacker C D. Attenuated replication of human immunodeficiency virus type 1 with a didanosine-selected reverse transcriptase mutation. J Virol. 1997;71:8846–8851. doi: 10.1128/jvi.71.11.8846-8851.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Telenti A, Munoz M, Bleiber G, Ledergerber B, Kaufmann D E. Heterogeneity of response to antiretroviral therapy. AIDS Rev. 1999;1:147–155. [Google Scholar]
- 34.Wrin T, Gamarnik A, Whitehurst N, Huang W, Ziermann R, Whitcomb J, Petropoulos C J. Drug resistance is associated with impaired protease and reverse transcriptase function and reduced replication activity: characterization of recombinant viruses from 150 HIV-1 infected patients. Antivir Ther. 2000;5(Suppl. 3):92–93. [Google Scholar]
- 35.Wu X, Liu H, Xiao H, Conway J A, Hunter E, Kappes J C. Functional RT and IN incorporated into HIV-1 particles independently of the Gag/Pol precursor protein. EMBO J. 1997;16:5113–5122. doi: 10.1093/emboj/16.16.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu Q, Ottmann M, Pechoux C, le Grice S, Darlix J-L. Mutations in the primer grip of human immunodeficiency virus type 1 reverse transcriptase impair proviral DNA synthesis and virion maturation. J Virol. 1998;72:7676–7680. doi: 10.1128/jvi.72.9.7676-7680.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuste E, Sanchez-Palomino S, Casado C, Domingo E, Lopez-Galindez C. Drastic fitness loss in human immunodeficiency virus type 1 upon serial bottleneck events. J Virol. 1999;73:2745–2751. doi: 10.1128/jvi.73.4.2745-2751.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zennou V, Mammano F, Paulous S, Mathez D, Clavel F. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J Virol. 1998;72:3300–3306. doi: 10.1128/jvi.72.4.3300-3306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y-M, Imamichi H, Imamichi T, Lane H C, Falloon J, Vasudevachari M B, Salzman N P. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J Virol. 1997;71:6662–6670. doi: 10.1128/jvi.71.9.6662-6670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]