Abstract
Detection and identification of microorganisms are the first steps to guide susceptibility testing and enable clinicians to confirm diseases and guide therapy. The faster the pathogen identification is determined, the quicker the appropriate treatment can be started. In the clinical microbiology laboratory, multiple methodologies can be used to identify organisms, such as traditional biochemical testing or more recent methods like MALDI TOF MS and nucleic acid detection/identification assays. Each of these techniques has advantages and limitations, and clinical laboratories need to determine which methodology is best suited to their particular setting in terms of clinical needs, availability of technical expertise and cost. This article presents a concise review of the history, utilization, advantages and limitations of the main methods used for identifying microorganisms in microbiology laboratories.
Introduction
Since the first description of microorganisms by Antonie van Leeuwenhoek, the identification and taxonomy of microorganisms have been evolving; today, we can detect and identify organisms directly from patient samples in less than an hour using molecular testing, or give an identification in a few minutes directly from pure colonies with MALDI-TOF MS.1
One of the main functions of the microbiology laboratory is the rapid, sensitive and accurate identification of microorganisms responsible for infections. Knowing an organism's identity allows the microbiologist to correctly set up the appropriate susceptibility testing and give the correct susceptibility interpretation according to established breakpoints by regulatory bodies. Detection and identification of microorganisms in the clinical specimens are the first steps that enable clinicians to confirm the aetiology of an infection and guide the choice of empirical therapy.2
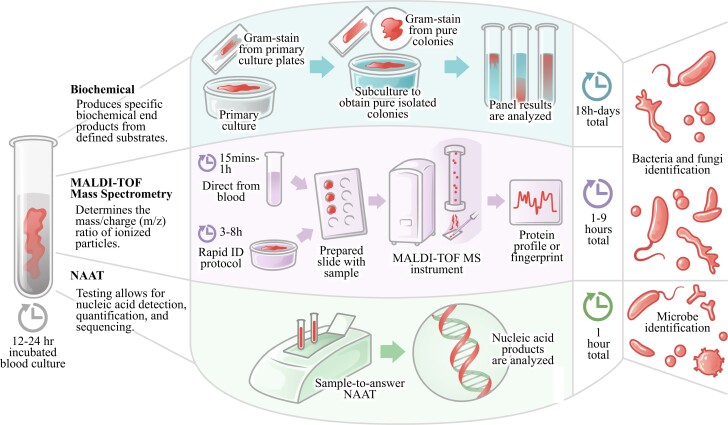
A wide variety of methods are used for identification, as reflected in Figure 1 for blood cultures. The most frequently encountered organisms can easily be identified with typical phenotypic profiles using morphology and rapid tests that are easy to perform and relatively inexpensive, or semi-automated/automated methods that include a battery of biochemical tests. Usually, the time required for identification by these phenotypic conventional methods is estimated to be 2–5 days. MALDI-TOF MS instruments can identify organisms from pure colonies at very low cost in a few minutes. They have extensive databases approved for in vitro diagnostic use (IVD) and research use only (RUO) that allow the identification of common and unusual organisms. Molecular assays can detect the genetic components of single or multiple unknown microorganisms directly from patient samples, like throat swabs or stool specimens, in 1 h or less.3 The variety of approaches the laboratory uses for identification depends on various factors such as the size of the laboratory, the volume of testing, the experience of technical staff, access to new technologies, and cost.
Figure 1.
Biochemical assays, MALDI-TOF MS, and NAAT identification processes and performance turnaround times. NAAT, nucleic acid amplification test. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Biochemical identification techniques
Overview of biochemical identification techniques
Regardless of the changing trends in diagnostic microbiology, the performance of well-characterized biochemical tests for the identification of medically important bacteria is still relevant in the clinical microbiology laboratory.4–6
The contributions of Fannie Hesse7 in 1881, who proposed using agar in culture media, and Richard Petri8 in 1887, who developed the Petri dish (plate), permitting the isolation and propagation of pure cultures, were pivotal in advancing microbial identification. The use of agar-based media spurred the development of various selective, supplemented and enriched culture media, enhancing the recovery of fastidious organisms from clinical specimens, enabling the characterization (morphology patterns, staining and biochemical), identification and taxonomic classification of bacteria.9
During the last two decades of the 19th and the early 20th centuries, as biochemical tests were developed to differentiate among bacterial genera and species, flow charts, dichotomous keys and diagnostic tables were published to guide organism identification.10 Various serological and immunological assays (e.g. serotyping, latex agglutination) were also introduced in the microbiology laboratories to detect microbial antigens or antibodies in bacterial cultures and clinical samples for the purpose of microbial identification. Simultaneously, software to analyse large amounts of data and miniaturized versions of conventional systems were developed to shorten and automate the identification process.
Bacterial biochemical characteristics refer to the ability of individual genera or species of bacteria to produce specific biochemical end products from defined substrates. The difference in protein and fat metabolism, carbohydrate metabolism, enzyme production and compound utilization of each organism has enough specificity to provide information for classifying bacteria into different groups based on their reactions.11
Processes and techniques involved
Traditionally, rapid tests are performed selectively and individually according to Gram staining and colonial morphology following the recommendations of standard guides for rapid bacterial identification. Some of the most rapid biochemical tests used to identify a few organisms commonly encountered in clinical microbiology are catalase, coagulase, leukocyte alkaline phosphatase, oxidase, indole, pyrrolidonyl arylamidase (PYR) and bile solubility test. For example, Escherichia coli can rapidly be identified by determining that the organism is Gram-negative, spot-indole-positive, oxidase-negative and β-haemolytic on the blood agar plate or lactose-positive and PYR-negative.12 Streptococcus pneumoniae occurs as Gram-positive lancet-shaped cocci in pairs that are ‘catalase negative’, have a sharp zone of α-haemolysis on a blood agar plate with a colony ≥0.5 mm in diameter, and are bile-solubility positive.12 These rapid tests take only a few minutes and permit the rapid identification of a few commonly encountered pathogens. However, the number of pathogens able to be identified with rapid tests is small, limiting their use to only a few commonly seen organisms.
Identification relies on matching the characteristics of an unknown organism to well-characterized established entities.13 Gram staining, microscopic morphology, colonial morphology, and growth on specific media are some of the first key characteristics used for classifying organisms into specific genera or species and dictate which additional key biochemical data are needed to provide more in-depth identification to the species level. Until manual miniaturized identification systems were introduced in the late 1960s, each biochemical reaction was set up manually in tubes.5 Using miniaturized, multi-test identification kits, like the API system (bioMérieux), was the first step towards rapid bacterial identification. These identification kits carefully preselected biochemical tests for different groups of organisms, for example, Gram-negative versus Gram-positive, improving efficiency. The demand for quicker turnaround time, efficiency and increased testing capacity pushed the introduction of automation and computers into clinical laboratories throughout the 1970s. Commercial automated systems, such as the bioMérieux VITEK 2, the BD Phoenix and the Beckman Coulter MicroScan, use identification cards or plates to simultaneously test many biochemical reactions, increasing identification accuracy. The instrument performs the incubation, analysis and interpretation of the biochemical reactions to produce an identification. The automated systems further developed their algorithm to shorten their time to identification, and some systems can give an identification within 4 h.14,15
Advantages and limitations of biochemical identification
The development of data analysis software and the expansion of the databases of semi-automated or fully automated biochemical identification systems have increased the accuracy of identifying the most commonly encountered microorganisms in routine clinical microbiology laboratories.3,16 The fully automated systems have larger capacities and can accommodate laboratories with significant workloads. They require minimal hands-on time to set up and generally can generate an identification in less than 24 h. Their ability to simultaneously perform susceptibility testing increases their efficiencies.
These identification systems generally work very well for common and non-fastidious pathogens but have difficulty differentiating closely related and metabolically inert organisms. Not all microorganisms are reliably identified by biochemical methods. The time needed for microorganism identification based on the traditional manual test-tube approach, which includes morphology, physiology, chemistry and biochemical characterization, is estimated to be at least 2–5 days. To stay accurate and up to date, the databases associated with the commercially available system need to be updated regularly. The accuracy of any system is limited to the manufacturer’s claims and the current edition of the database.
Manual or automated conventional identification methods in clinical microbiology have been the only means of bacterial isolation and identification for nearly 150 years and are still used widely in clinical microbiology. However, the introduction of more accurate and faster identification technologies, like nucleic acid–based molecular testing and MALDI-TOF MS, has gradually replaced the conventional biochemical test identification methods. They are still being used as a second line of identification in laboratories that have adopted molecular and/or MALDI-TOF as primary means of identification when identification needs confirmation for a particular isolate or during repairs of the primary instrument. Most fully automated conventional systems can perform automated susceptibility testing and thus are required in laboratories, explaining their continued use for identification when needed.
Overview of MALDI-TOF MS
Historically, MALDI-TOF MS was used in research to characterize proteins.17 However, it was discovered in 1996 that it could also be used for microorganism identification from whole-cell preparations without the need for pretreatment and then was used for identification in the clinical microbiology laboratory environment in the early 2000s.18 There are several MALDI-TOF MS instruments on the market now, but the two most prevalent for clinical microbiology laboratories are manufactured by bioMérieux (VITEK MS or VITEK MS PRIME) and Bruker (Biotyper and Sirius). These instruments have clinical laboratory-friendly workflows and databases for bacteria and fungi that are approved for IVD as well as RUO. Additionally, these instruments provide rapid and accurate identification of bacteria frequently identified in the clinical laboratory.19,20
MALDI-TOF MS identifies organisms by analysing their protein profile. A sample is placed on a target slide, and matrix, α-cyano-4-hydroxycinnamic acid, is added and allowed to dry. In some cases, formic acid is added and allowed to dry prior to the addition of matrix. The target slide with the sample is then placed in the instrument and the laser is activated to strike the sample spot. Sample particles are ionized and transformed into a gas phase. The ionized particles make their way through the flight tube and the mass/charge (m/z) ratio is calculated from the speed of travel of the ions through the vacuum tube; this creates a unique spectrum that is the microorganism’s protein profile or fingerprint. The protein profile can then be compared with a database to determine the identification of the organism. This process is quite rapid, with the potential for an identification in minutes.
Processes and techniques involved
Though quite rapid, MALDI-TOF MS identifications still require an organism first to be grown on an agar plate for at least 18 h. As a result, for time-critical samples, such as positive blood cultures, laboratories have investigated methods to decrease the time needed prior to identification. Two specific approaches that have shown success are short-incubation cultures (‘smudge’ or ‘scum’ plates) or direct from blood culture preparation methods.
For short-incubation, laboratories subculture a positive blood culture and then incubate plates for a shorter timeframe before setting up MALDI-TOF MS from the small amount of growth available. Several studies have been performed on this off-label method using various media, incubation times, and both the VITEK MS and Bruker Biotyper. Overall, performance reported for both instruments in multiple different studies with incubation times ranging from 3 to 6 h showed 78.3%–91% and 61.8%–88% identification to the species level, respectively, for VITEK MS and Bruker Biotyper, with higher percent identification to the genus level and as high as 93.4% for Gram-negative bacteria.21–26 These results are not surprising, because Gram-negative bacteria generally grow faster than Gram-positive bacteria and yeast, and inherently short-incubation culture will work better for organisms that grow faster.
For even more rapid identification, many studies have investigated MALDI-TOF MS identifications directly from positive blood culture. Before placement on the target slide, samples must first be processed. Multiple approaches have been used for sample processing, including centrifugation, centrifugation plus lysis, and centrifugation plus lysis plus additional extraction methods, with varying success.27,28 Some commercially available processing kits include the BACpro II kit (Nittobo Medical Co., Ltd), the FAST™ System (Qvella), the VITEK® MS Blood Culture Kit (RUO) (bioMérieux) and the Rapid MBT Sepsityper® IVD Kit (Bruker Daltonics). The Sepsityper is the most commonly used, and there are many reports in the literature for performance, so focus will be placed on recent studies with larger sample numbers. Two recent large studies, one with over 2000 Gram-negative positive blood cultures and the other with over 6000 Gram-positive and Gram-negative positive blood cultures, both showed good performance using the Sepsityper with the Bruker Biotyper, with 92.1% identification to the species level for the Gram-negative study, and 86.3% overall identification and 97% identification for Enterobacterales for the larger study.29,30 Two other studies with 443 and 227 monomicrobial positive blood cultures, both using the Rapid Sepsityper method and Bruker Biotyper, respectively showed similar results, with 87.6% and 89.4% identification to the species level and better performance for Gram-negative (98.6% and 95.2% identification) than for Gram-positive (85.9% and 84.4%).31,32 However, in contrast to these, a recent report showed lower performance using the Rapid Sepsityper protocol combined to the MBT-Sepsityper module, with 65.4%, 78.9% and 62% identification to the species level of monomicrobial positive blood culture growing respectively Gram-positive bacteria, Gram-negative bacteria or yeast.27 And in another study with 560 positive blood cultures, 69.3% identification to the species level was observed, with 85.8% for Gram-negative and 59.7% for Gram-positive.33 For the BacPro II kit using the Bruker instrument, rates of identification reported are 80.8%, 87.4% and 80% to species level, with 92.3% for Gram-negative and 72.4% for Gram-positive.33–35 And for VITEK MS Blood Culture Kit (RUO) combined with the RUO VITEK MS and SARAMIS database, 78.2% of positive blood cultures were identified to the species level.36 A newer preparation kit on the market, the FAST System, has shown 87.7% species identification with VITEK MS, 89.5% identification with the Bruker Biotyper, and 93.8% identification with Bruker Biotyper.37–39
Advantages and limitations of MALDI-TOF MS
Some key advantages of MALDI-TOF MS are the ease of target slide preparation, rapid identification once the target slide has been loaded on the instrument, and the ability to potentially identify any organism that is in the database. However, when working with direct specimens such as direct from positive blood culture, additional processing is needed prior to loading the sample on the slide, increasing the sample preparation time. Additionally, reports for both direct from blood culture and short-incubation cultures have shown that MALDI-TOF MS performs much better for Gram-negative bacteria and often struggles with the identification of Gram-positive microorganisms. Finally, one major limitation is related to the database itself; though there are comprehensive IVD and RUO databases available, the chance of identification of an organism is only as good as the database used. Moving forwards, there are opportunities for continued expansion of IVD databases as well as a need for improved direct from positive blood culture processing methods for identification of Gram-positive microorganisms.
Overview of molecular testing identification
The introduction of molecular testing identification has transformed the field of clinical microbiology, permitting rapid and accurate detection and identification of microorganisms, allowing for early targeted management of patients and improving their subsequent clinical outcomes.40,41 The history of molecular testing identification can be traced back to the development of the PCR by Kary Mullis and the team at Cetus Corporation in the 1980s, which subsequently led Mullis to earn the Nobel Prize in Chemistry in 1993.42–44 Since this time, the routine use of PCR and similar molecular testing technology has exploded in clinical microbiology as streamlining and automation of nucleic acid processing, amplification and detection techniques have evolved in addition to progressive decreases in costs.45
Molecular testing allows for nucleic acid detection, quantification and sequencing from specific DNA or RNA genetic markers, enabling both qualitative determination of organisms and quantitative assessment of abundance depending on the technique employed.45 For most applications, this generally requires nucleic acid extraction and amplification, then detection, allowing for species determinations Clinical laboratories employed early non-amplified probe technologies such as fluorescent in situ hybridization to detect isolates from culture, for example, blood cultures, which became more routinely used in the early 2000s. Whereas single-target PCR has been employed since the 1980s for HIV, the use of multiplex PCR for detection of multiple targets in a patient presenting with a potential infectious disease clinical syndrome has more recently developed with the earliest device approved in 2012 by the FDA.46 Similarly, routine and broad use of sequencing-based approaches such as metagenomics allowing for the detection of all possible pathogens is still evolving as the processing requirements, data analysis infrastructure and cost continue to improve. With continued improvement, the impact of molecular technologies on clinical microbiology and patient outcomes will continue to progress.
Processes and techniques involved
To begin the process of PCR testing, a clinical sample (e.g. blood, stool, swab, other body fluid) is collected for preparation.45 The preparation includes lysis of microbial cells for the extraction of nucleic acids—both RNA and DNA. After extraction, the amplification step occurs, which includes denaturing, annealing and elongation of the nucleic acids; in many applications this requires thermocycling for the primers to attach and detach with each cycle. These cycles are repeated 25–40 times, increasing the copies of nucleic acid products with each cycle. These products are detected and sometimes quantified with probes such as fluorescent probes. The design of primers and probes is critical to the performance of PCR testing. Also critical to testing is ensuring a quality specimen and processing to avoid contamination as both can lead to erroneous results.
Akin to PCR, next-generation sequencing (NGS) starts with extraction of nucleic acids from the clinical specimen; this also requires a quality specimen and processing to ensure quality results.47 NGS often additionally includes a step for fragmentation of the DNA to create shorter fragments that are appropriate for sequencing. After this step is the library preparation stage in which adapter sequences are added that enable the DNA fragments to bind the sequencing platform. An enrichment process occurs next to amplify the DNA to guarantee there is enough DNA for sequencing. In contrast to Sanger sequencing, which sequences a single fragment at a time, NGS then sequences millions of fragments at the same time in a parallel manner, which substantially increases the throughput of sequencing. Finally, after sequencing is finished, data analysis is completed with bioinformatics. This involves quality assurance, alignment of sequence outputs to a reference database, and determining an explanation of results. The bioinformatics steps require substantial expertise and computational resources.
Advantages and limitations of molecular testing identification
Advantages and limitations of molecular testing can be divided among pre-analytical, analytical and post-analytical stages of testing. The pre-analytical stage of testing includes the processes prior to testing (sample collection, transport, preparation). Advantages of molecular testing include the eligibility of broad sample types for testing. Additionally, testing is sensitive enough often to require minimal sample in addition to allowing for utility of samples that have had antibiotic pre-exposure.48 Disadvantages of molecular technology include being prone to contamination and the need for careful sample preparation and handling. Additionally, with increased sensitivity, selecting the right patient for testing becomes increasingly important.49 In the absence of this, detections may be clinically unclear, as in the example of Clostridiodes difficile PCR testing.50,51 Solutions to this clinical ambiguity have focused around diagnostic stewardship—ensuring the right test for the right patient at the right time. For C. difficile PCR testing, guidelines have recommended multistep algorithms for combining sensitive and specific tests to increase the overall clinical utility. Alternatively, guidelines have recommended increasing the pre-test probability of disease with pre-analytical strategies by only testing patients without recent laxative use but with unexplained and new onset of three or more unformed stools in 24 h. Similar approaches have been more recently suggested with PCR testing of respiratory samples due to the increased sensitivity and diagnostic yield compared with the historical gold standard of culture.52
For the analytical phase, there are several advantages to molecular testing. PCR has mostly replaced viral cultures based on the technical challenges and time to result for those approaches.45 Additionally, the decreased turnaround time—often results come within 1 h for PCR—has allowed for earlier appropriate therapy decisions, which has translated to decreased length of stay (meningitis/encephalitis, acute respiratory tract infections and bloodstream infection) and mortality (bloodstream infections).40,53,54 Moreover for NGS, the broad detections allow for determination of atypical aetiologies of disease.55 One disadvantage of molecular testing with PCR is non-standardized quantification in most use cases leading to inability to create consensus standards and evidence for management decisions.45 Additionally, PCR may not detect all aetiologies of an illness, which may be limiting in certain patient populations with broader differentials of disease.56 In contrast, NGS testing allows very broad detection of potential aetiologies of disease, which may often require expert knowledge and interpretation for the clinical relevance of the detections.55 Finally, NGS currently has prolonged turnaround times, increased costs, large infrastructure requirements, and a need for significant expertise for use, which are all challenges to widespread implementation.55
Finally, for the post-analytical phase, newer technologies have the advantage over manual biochemical identifications by generally being connected to laboratory information systems, allowing for ease of data capture without manual entry. The resultant connectivity can also aid in tracking antimicrobial resistance and outbreaks at the local, regional, national and international levels, which can facilitate antimicrobial stewardship, infection control and public health efforts.57 A disadvantage of post-analytical molecular techniques is that many newer technologies may require education or guidance for their results.58 Opportunities in standardizing reporting results due to current heterogeneity have been reported and are critical as generally in microbiology these variations in reporting have been shown to significantly impact antimicrobial prescribing.59–61
Conclusion
In the practice of clinical microbiology, there has been a trajectory towards advanced diagnostic testing, transforming the field. Firstly, the identification of bacteria has evolved considerably in clinical microbiology from historical techniques of biochemical testing to modern approaches with the protein profiling with MALDI-TOF and the accuracy of molecular testing. Secondly, although biochemical testing continues to provide results for many specific applications and settings, MALDI-TOF and molecular testing have transformed clinical microbiology with the speed and accuracy of their results. Thirdly, it is important to keep in mind that each testing approach has differing advantages and disadvantages, with their collective contribution allowing for fast treatment decisions and improved clinical outcomes.
Clinical microbiologists and bedside practitioners should anticipate future enhancements in approaches. These include: (i) the increasing role of automation and bioinformatics (including machine learning and artificial intelligence); (ii) routine clinical practice use of sequencing for typical bacterial infections for management and infection prevention activities; and (iii) introduction of next-generation host response technologies incorporating transcriptomics and/or proteomics. As these technologies advance, there is little doubt that further evolution in rapid identification of microorganisms and improved treatment of patients will continue.
Acknowledgements
We would like to thank Justin Klein of MitoPop for the commissioned medical illustration of the included conceptual framework.
Contributor Information
Sophie S Arbefeville, Microbiology & Molecular Pathology, Marshfield Clinic Health System, 1000 N. Oak Ave., Marshfield, WI 54449, USA.
Tristan T Timbrook, Department of Global Medical Affairs, St Louis, MO, USA; Department of Pharmacotherapy, University of Utah College of Pharmacy, Salt Lake City, UT, USA.
Cherilyn D Garner, Department of Global Medical Affairs, St Louis, MO, USA.
Funding
This paper was published as part of a supplement financially supported by bioMérieux.
Transparency declarations
S.S.A. reports no relevant potential conflicts of interest. T.T.T. and C.D.G. are employees of bioMérieux.
References
- 1. Isenberg HD. Clinical microbiology: past, present, and future. J Clin Microbiol 2003; 41: 917–8. 10.1128/JCM.41.3.917-918.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Procop GW. Koneman's Color Atlas and Textbook of Diagnostic Microbiology. Wolters Kluwer Health, 2017. [Google Scholar]
- 3. Altheide ST, Butina M, Chatterjee U et al. Evolution of the clinical microbiology laboratory. Am Soc Clin Lab Sci 2020. 10.29074/ascls.119.001867 [DOI] [Google Scholar]
- 4. Aslanzadeh S. Biochemical profile-based microbial identification systems. In: Advanced Techniques in Diagnostic Microbiology. Springer, 2006. 10.1007/0-387-32892-0_6 [DOI] [Google Scholar]
- 5. Janda JM, Abbott SL. Bacterial identification for publication: when is enough enough? J Clin Microbiol 2002; 40: 1887–91. 10.1128/JCM.40.6.1887-1891.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sandle T. Pharmaceutical Microbiology: Essentials for Quality Assurance and Quality Control. Woodhead Publishing, 2016. [Google Scholar]
- 7. Hesse W. Walther and Angelina Hesse: early contributors to bacteriology. ASM News 1992; 58: 425–8. [Google Scholar]
- 8. Petri RJ. Eine Kleine Modification des Koch'schen Plattenverfahrens. Centralblatt fur Bakteriologie und Parasitenkunde 1887; 1: 279–80. [Google Scholar]
- 9. Bonnet M, Lagier JC, Raoult D et al. Bacterial culture through selective and non-selective conditions: the evolution of culture media in clinical microbiology. New Microbes New Infect 2020; 34: 100622. 10.1016/j.nmni.2019.100622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wainwright M, Lederberg J. Encyclopedia of Microbiology. Academic Press Inc., 1992. [Google Scholar]
- 11. O'Hara CM. Manual and automated instrumentation for identification of Enterobacteriaceae and other aerobic gram-negative bacilli. Clin Microbiol Rev 2005; 18: 147–62. 10.1128/CMR.18.1.147-162.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. CLSI . Abbreviation Identification of Bacteria and Yeast; Approved Guideline—Second Edition: M35-A2. 2008. [Google Scholar]
- 13. Franco-Duarte R, Cernakova L, Kadam S et al. Advances in chemical and biological methods to identify microorganisms—from past to present. Microorganisms 2019; 7: 130. 10.3390/microorganisms7050130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Spanu T, Sanguinetti M, Ciccaglione D et al. Use of the VITEK 2 system for rapid identification of clinical isolates of staphylococci from bloodstream infections. J Clin Microbiol 2003; 41: 4259–63. 10.1128/JCM.41.9.4259-4263.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barman P, Chopra S, Thukral T. Direct testing by VITEK((R)) 2: a dependable method to reduce turnaround time in gram-negative bloodstream infections. J Lab Physicians 2018; 10: 260–4. 10.4103/JLP.JLP_11_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Monteiro AC, Fortaleza CM, Ferreira AM et al. Comparison of methods for the identification of microorganisms isolated from blood cultures. Ann Clin Microbiol Antimicrob 2016; 15: 45. 10.1186/s12941-016-0158-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tanaka K, Waki H, Ido Y et al. Protein and polymer analyses up to m/z 100 000 by laser ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 1988; 2: 151–3. 10.1002/rcm.1290020802 [DOI] [Google Scholar]
- 18. Holland RD, Wilkes JG, Rafii F et al. Rapid identification of intact whole bacteria based on spectral patterns using matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 1996; 10: 1227–32. [DOI] [PubMed] [Google Scholar]
- 19. Grohs P, Remaud E, Lath C et al. Comparison of the new VITEK MS PRIME system with the matrix-assisted laser desorption ionization Biotyper Microflex LT for the identification of microorganisms. Ann Lab Med 2023; 43: 574–84. 10.3343/alm.2023.43.6.574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bardelli M, Padovani M, Fiorentini S et al. A side-by-side comparison of the performance and time-and-motion data of VITEK MS. Eur J Clin Microbiol Infect Dis 2022; 41: 1115–25. 10.1007/s10096-022-04472-x [DOI] [PubMed] [Google Scholar]
- 21. Bhatti MM, Boonlayangoor S, Beavis KG et al. Rapid identification of positive blood cultures by matrix-assisted laser desorption ionization-time of flight mass spectrometry using prewarmed agar plates. J Clin Microbiol 2014; 52: 4334–8. 10.1128/JCM.01788-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Altun O, Botero-Kleiven S, Carlsson S et al. Rapid identification of bacteria from positive blood culture bottles by MALDI-TOF MS following short-term incubation on solid media. J Med Microbiol 2015; 64: 1346–52. 10.1099/jmm.0.000168 [DOI] [PubMed] [Google Scholar]
- 23. Curtoni A, Cipriani R, Marra ES et al. Rapid identification of microorganisms from positive blood culture by MALDI-TOF MS after short-term incubation on solid medium. Curr Microbiol 2017; 74: 97–102. 10.1007/s00284-016-1161-2 [DOI] [PubMed] [Google Scholar]
- 24. Ha J, Hong SK, Han GH et al. Same-day identification and antimicrobial susceptibility testing of bacteria in positive blood culture broths using short-term incubation on solid medium with the MicroFlex LT, Vitek-MS, and Vitek2 systems. Ann Lab Med 2018; 38: 235–41. 10.3343/alm.2018.38.3.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mitchell SL, Alby K. Performance of microbial identification by MALDI-TOF MS and susceptibility testing by VITEK 2 from positive blood cultures after minimal incubation on solid media. Eur J Clin Microbiol Infect Dis 2017; 36: 2201–6. 10.1007/s10096-017-3046-0 [DOI] [PubMed] [Google Scholar]
- 26. Verroken A, Defourny L, Lechgar L et al. Reducing time to identification of positive blood cultures with MALDI-TOF MS analysis after a 5-h subculture. Eur J Clin Microbiol Infect Dis 2015; 34: 405–13. 10.1007/s10096-014-2242-4 [DOI] [PubMed] [Google Scholar]
- 27. Ponderand L, Pavese P, Maubon D et al. Evaluation of rapid Sepsityper(R) protocol and specific MBT-Sepsityper module (Bruker Daltonics) for the rapid diagnosis of bacteremia and fungemia by MALDI-TOF-MS. Ann Clin Microbiol Antimicrob 2020; 19: 60. 10.1186/s12941-020-00403-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nomura F, Tsuchida S, Murata S et al. Mass spectrometry-based microbiological testing for blood stream infection. Clin Proteomics 2020; 17: 14. 10.1186/s12014-020-09278-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Comini S, Bianco G, Boattini M et al. Evaluation of a diagnostic algorithm for rapid identification of gram-negative species and detection of extended-spectrum beta-lactamase and carbapenemase directly from blood cultures. J Antimicrob Chemother 2022; 77: 2632–41. 10.1093/jac/dkac230 [DOI] [PubMed] [Google Scholar]
- 30. Cordovana M, Zignoli A, Ambretti S. Rapid Sepsityper in clinical routine: 2 years’ successful experience. J Med Microbiol 2020; 69: 1398–404. 10.1099/jmm.0.001268 [DOI] [PubMed] [Google Scholar]
- 31. Pranada AB, Cordovana M, Meyer M et al. Identification of micro-organism from positive blood cultures: comparison of three different short culturing methods to the Rapid Sepsityper workflow. J Med Microbiol 2022; 71. 10.1099/jmm.0.001571 [DOI] [PubMed] [Google Scholar]
- 32. Watanabe N, Koyama S, Taji Y et al. Direct microorganism species identification and antimicrobial susceptibility tests from positive blood culture bottles using rapid Sepsityper kit. J Infect Chemother 2022; 28: 563–8. 10.1016/j.jiac.2021.12.030 [DOI] [PubMed] [Google Scholar]
- 33. Oviano M, Ingebretsen A, Steffensen AK et al. Multicenter evaluation of rapid BACpro((R)) II for the accurate identification of microorganisms directly from blood cultures using MALDI-TOF MS. Diagnostics (Basel) 2021; 11: 2251. 10.3390/diagnostics11122251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tsuchida S, Murata S, Miyabe A et al. Application of the biocopolymer preparation system, rapid BACpro(R) II kit, for mass-spectrometry-based bacterial identification from positive blood culture bottles by the MALDI Biotyper system. J Microbiol Methods 2018; 152: 86–91. 10.1016/j.mimet.2018.07.017 [DOI] [PubMed] [Google Scholar]
- 35. Kayin M, Mert B, Aydemir S et al. Comparison of rapid BACpro(R) II, Sepsityper(R) kit and in-house preparation methods for direct identification of bacteria from blood cultures by MALDI-TOF MS with and without Sepsityper(R) module analysis. Eur J Clin Microbiol Infect Dis 2019; 38: 2133–43. 10.1007/s10096-019-03654-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fothergill A, Kasinathan V, Hyman J et al. Rapid identification of bacteria and yeasts from positive-blood-culture bottles by using a lysis-filtration method and matrix-assisted laser desorption ionization-time of flight mass spectrum analysis with the SARAMIS database. J Clin Microbiol 2013; 51: 805–9. 10.1128/JCM.02326-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ugaban K, Pak P, She RC. Direct MALDI-TOF MS and antimicrobial susceptibility testing of positive blood cultures using the FASTTM system and FAST-PBC prep cartridges—performance evaluation in a clinical microbiology laboratory serving high-risk patients. Microorganisms 2022; 10: 2076. 10.3390/microorganisms10102076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Verroken A, Hajji C, Bressant F et al. Performance evaluation of the FAST system and the FAST-PBC prep cartridges for speeded-up positive blood culture testing. Front Microbiol 2022; 13: 982650. 10.3389/fmicb.2022.982650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bonaiuto C, Baccani I, Chilleri C et al. Evaluation of the liquid colony produced by the FAST system for shortening the time of bacterial identification and phenotypic antimicrobial susceptibility testing and detection of resistance mechanisms from positive blood cultures. Diagnostics (Basel) 2023; 13: 1849. 10.3390/diagnostics13111849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Timbrook TT, Morton JB, McConeghy KW et al. The effect of molecular rapid diagnostic testing on clinical outcomes in bloodstream infections: a systematic review and meta-analysis. Clin Infect Dis 2017; 64: 15–23. 10.1093/cid/ciw649 [DOI] [PubMed] [Google Scholar]
- 41. Hanson KE, Azar MM, Banerjee R et al. Molecular testing for acute respiratory tract infections: clinical and diagnostic recommendations from the IDSA's diagnostics committee. Clin Infect Dis 2020; 71: 2744–51. 10.1093/cid/ciaa508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mullis K, Faloona F, Scharf S et al. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb Symp Quant Biol 1986; 51: 263–73. 10.1101/SQB.1986.051.01.032 [DOI] [PubMed] [Google Scholar]
- 43. Saiki RK, Scharf S, Faloona F et al. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 1985; 230: 1350–4. 10.1126/science.2999980 [DOI] [PubMed] [Google Scholar]
- 44. Wrublewski DT. Analysis for science librarians of the 2018 Nobel Prize in chemistry: directed evolution of enzymes and phage display of peptides and antibodies. Sci Technol Libraries 2019; 38: 51–69. 10.1080/0194262X.2019.1579159 [DOI] [Google Scholar]
- 45. Schmitz JE, Stratton CW, Persing DH et al. Forty years of molecular diagnostics for infectious diseases. J Clin Microbiol 2022; 60: e0244621. 10.1128/jcm.02446-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ramanan P, Bryson AL, Binnicker MJ et al. Syndromic panel-based testing in clinical microbiology. Clin Microbiol Rev 2018; 31: e00024-17. 10.1128/CMR.00024-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yohe S, Thyagarajan B. Review of clinical next-generation sequencing. Arch Pathol Lab Med 2017; 141: 1544–57. 10.5858/arpa.2016-0501-RA [DOI] [PubMed] [Google Scholar]
- 48. Achermann Y, Vogt M, Leunig M et al. Improved diagnosis of periprosthetic joint infection by multiplex PCR of sonication fluid from removed implants. J Clin Microbiol 2010; 48: 1208–14. 10.1128/JCM.00006-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hueth KD, Prinzi AM, Timbrook TT. Diagnostic stewardship as a team sport: interdisciplinary perspectives on improved implementation of interventions and effect measurement. Antibiotics (Basel) 2022; 11: 250. 10.3390/antibiotics11020250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Crobach MJT, Planche T, Eckert C et al. European Society of Clinical Microbiology and Infectious Diseases: update of the diagnostic guidance document for Clostridium difficile infection. Clin Microbiol Infect 2016; 22: S63–81. 10.1016/j.cmi.2016.03.010 [DOI] [PubMed] [Google Scholar]
- 51. McDonald LC, Gerding DN, Johnson S et al. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis 2018; 66: e1–48. 10.1093/cid/cix1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Falsey AR, Branche AR, Croft DP et al. Real-life assessment of Biofire Filmarray(r) pneumonia panel in adults hospitalized with respiratory illness. J Infect Dis 2024; 229: 214–22. 10.1093/infdis/jiad221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hueth KD, Thompson-Leduc P, Totev TI et al. Assessment of the impact of a meningitis/encephalitis panel on hospital length of stay: a systematic review and meta-analysis. Antibiotics (Basel) 2022; 11: 1028. 10.3390/antibiotics11081028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Clark TW, Lindsley K, Wigmosta TB et al. Rapid multiplex PCR for respiratory viruses reduces time to result and improves clinical care: results of a systematic review and meta-analysis. J Infect 2023; 86: 462–75. 10.1016/j.jinf.2023.03.005 [DOI] [PubMed] [Google Scholar]
- 55. Azar MM, Turbett S, Gaston D et al. A consensus conference to define the utility of advanced infectious disease diagnostics in solid organ transplant recipients. Am J Transplant 2022; 22: 3150–69. 10.1111/ajt.17147 [DOI] [PubMed] [Google Scholar]
- 56. Nguyen A, Chen J, Isaza E et al. Biofire pneumonia panel in lung donors: faster detection but limited pathogens. Transpl Infect Dis 2023; 25: e14091. 10.1111/tid.14091 [DOI] [PubMed] [Google Scholar]
- 57. Timbrook TT, Olin KE, Spaulding U et al. Epidemiology of antimicrobial resistance among blood and respiratory specimens in the United States using genotypic analysis from a cloud-based population surveillance network. Open Forum Infect Dis 2022; 9: ofac296. 10.1093/ofid/ofac296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Foster RA, Kuper K, Lu ZK et al. Pharmacists’ familiarity with and institutional utilization of rapid diagnostic technologies for antimicrobial stewardship. Infect Control Hosp Epidemiol 2017; 38: 863–6. 10.1017/ice.2017.67 [DOI] [PubMed] [Google Scholar]
- 59. Prinzi AM, Wattier RL, Curtis DJ et al. Impact of organism reporting from endotracheal aspirate cultures on antimicrobial prescribing practices in mechanically ventilated pediatric patients. J Clin Microbiol 2022; 60: e0093022. 10.1128/jcm.00930-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Musgrove MA, Kenney RM, Kendall RE et al. Microbiology comment nudge improves pneumonia prescribing. Open Forum Infect Dis 2018; 5: ofy162. 10.1093/ofid/ofy162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Simner PJ, Dien Bard J, Doern C et al. Reporting of antimicrobial resistance from blood cultures, an antibacterial resistance leadership group survey summary: resistance marker reporting practices from positive blood cultures. Clin Infect Dis 2023; 76: 1550–8. 10.1093/cid/ciac952 [DOI] [PMC free article] [PubMed] [Google Scholar]