Summary
Metabolomics and lipidomics have emerged as tools in understanding the connections of metabolic syndrome (MetS) with cardiovascular diseases (CVD), type 1 and type 2 diabetes (T1D, T2D), and metabolic dysfunction-associated steatotic liver disease (MASLD). This review highlights the applications of these omics approaches in large-scale cohort studies, emphasizing their role in biomarker discovery and disease prediction. Integrating metabolomics and lipidomics has significantly advanced our understanding of MetS pathology by identifying unique metabolic signatures associated with disease progression. However, challenges such as standardizing analytical workflows, data interpretation, and biomarker validation remain critical for translating research findings into clinical practice. Future research should focus on optimizing these methodologies to enhance their clinical utility and address the global burden of MetS-related diseases.
Keywords: Metabolomics, Lipidomics, Mass spectrometry, Metabolic syndrome, Cardiovascular diseases, Type 1 diabetes, Type 2 diabetes, Metabolic dysfunction-associated steatotic liver disease
Introduction
Metabolic syndrome (MetS), also known as insulin resistance syndrome, is defined as a cluster of risk factors for cardiovascular disease and diabetes. The main risk factors include raised blood pressure, visceral obesity, hyperglycemia, and dyslipidemia (reduced high-density lipoprotein cholesterol or raised triacylglycerols) [1–3]. These features are often related to insulin resistance, which can lead to prediabetes or type 2 diabetes [4]. Recent studies have shown that even non-obese patients may suffer from insulin resistance, with visceral adiposity being considered the primary contributor to MetS pathology. Visceral adiposity is strongly associated with hepatic fatty infiltration, indicating that the amount of fatty acids in the liver is indirectly linked with MetS, both as a cause and a consequence of the syndrome [5]. Furthermore, in recent decades, MetS has become a significant health concern with a high prevalence worldwide [4,6–8]. To properly understand MetS metabolism and the relationships between the aforementioned risk factors [9,10], metabolomics and lipidomics can be applied.
Metabolite profiling is conducted using either untargeted or targeted approaches, applied to biological samples through various analytical methods and platforms [11]. Large-scale metabolomics and lipidomics studies, which involve extensive populations or numerous samples (over 1000), have demonstrated their effectiveness in various scientific fields. These studies have defined individual phenotypes and shown the effects of genetic, environmental, intervention, or aging factors. They have also discovered biomarkers and validated metabolite patterns associated with specific biological states [11]. Integrating newly identified metabolite biomarkers with clinical characteristics can potentially enhance the prediction of disease development [12].
In this review, we examine metabolomics and lipidomics human cohort studies and their application in MetS research. We introduce the analytical workflow and provide examples of recent MetS studies on cardiovascular diseases, type 1 and type 2 diabetes, and metabolic dysfunction-associated steatotic liver disease.
Metabolomics and lipidomics in large cohort studies
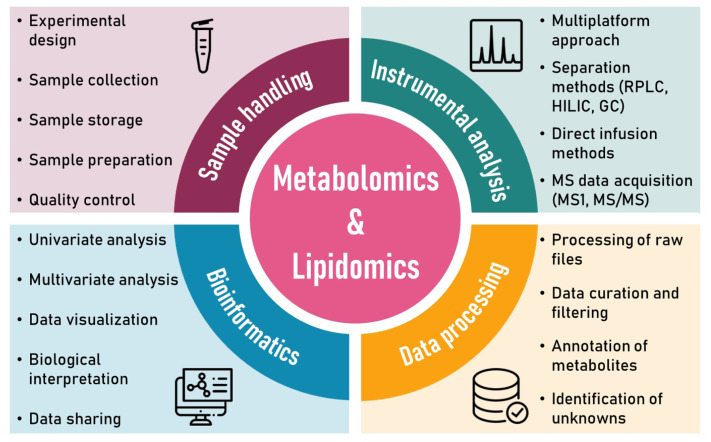
Large-scale metabolomics and lipidomics studies analyze hundreds to thousands of human samples containing thousands of metabolites. These samples are often processed in multiple batches over several weeks or months. No single analytical platform can cover all metabolites in a biological sample due to the complexity, diversity, and size of the human metabolome and lipidome. Therefore, multiple analytical platforms are employed to increase metabolite coverage [13]. Figure 1 shows metabolomics and lipidomics workflow, consisting of sample handling, instrumental analysis, data processing, and bioinformatics.
Fig. 1.
Metabolomics and lipidomics workflow.
Sample handling
The first step in metabolomics and lipidomics studies is creating a proper experimental design, including sample size, sample collection and storage, sample preparation, quality control, and analytical techniques [14].
Determining the appropriate sample size, both overall and for each group, is essential. Insufficient sample size can lead to errors and lack of precision. Conversely, even small, insignificant differences might appear statistically significant with a larger sample size, while clinically important effects might seem statistically non-significant with a small sample size [15]. A high sample size may also waste resources for minimal information gain [16]. The minimal sample size is calculated using power analysis, taking into account the significance level (e.g., α=0.05), statistical power (e.g., 0.8), and effect size (d=0.8, 0.5, 0.2 for large, medium, small effect size, respectively) [17]. To this end, freely available software such as G*Power can be used [18]. However, for untargeted metabolomics and lipidomics studies with a priori unknown number of measured metabolites [19], alternative strategies have become available, such as the Data-driven Sample size Determination (DSD) algorithm for MATLAB and GNU Octave [20], MetSizeR [21], or the online tool SSizer (idrblab.org/ssizer) [22].
Generally, at least 20–30 samples per group are advised for human studies, although the number of samples can range from hundreds to even thousands to achieve reasonable statistical power. On the other hand, for cell and animal studies with tightly controlled conditions, 3–6 and 5–10 samples per group, respectively, are recommended [23–25].
Another crucial aspect to consider is sample collection and storage. These steps must be decided during preanalytical processing to ensure reliable results [26]. Collection procedures differ based on the type of samples and planned analysis. For human cohort studies, samples typically consist of plasma or serum. The selection of a specific anticoagulant for plasma (e.g., EDTA, citrate, heparin) should be decided in advance and maintained consistently throughout the study. Inaccurate sample collection or improper storage may cause metabolite degradation, increased variability, or interference with instrumentation [27].
It is important to quench the metabolism of samples as soon as possible prior to their storage. Quenching should stop all enzymatic and chemical activities and maintain the current metabolite levels during harvesting [28]. The recommended method for quenching is to rapidly freeze the samples using liquid nitrogen, dry ice, or freeze clamping. After that, samples should be stored at -80 °C [29].
The next step is sample extraction to capture as many metabolites as possible in the sample. Various sample preparation techniques are available [30]. Minimal sample preparation methods, such as dilution, are sufficient for some matrices like urine. Water is a suitable diluent for reversed-phase liquid chromatography platforms, which start with a high percentage of water in the mobile phase. On the other hand, acetonitrile as a diluent is preferred for hydrophilic interaction chromatography, which begins with a high percentage of organic solvent (acetonitrile). Additionally, normalization to creatinine or osmolality values is a common strategy for urine due to its high variability in concentration, which correlates with metabolite composition [31]. On the other hand, plasma and serum, often used in large human cohort studies, contain many interfering proteins and require an extraction step to remove these before instrumental analysis. Common preparation methods like buffering, dilution, evaporation, and centrifugation may lead to metabolite losses and issues such as high salt concentration and instrument disruption, which can be reduced by adding an extraction step [32].
Extraction techniques in metabolomics and lipidomics commonly include organic solvent-based protein precipitation, liquid–liquid extraction (LLE), or solid-phase extraction (SPE). Isolation can also be performed in single or multiple fractions [33]. Single-phase extraction uses methanol, acetonitrile, isopropanol, a mixture of isopropanol/acetonitrile/water, acetonitrile/methanol, butanol/methanol [27,34,35]. This method enables simultaneous extraction of lipids and polar metabolites, but such extracts are very complex and can be challenging during instrumental analysis.
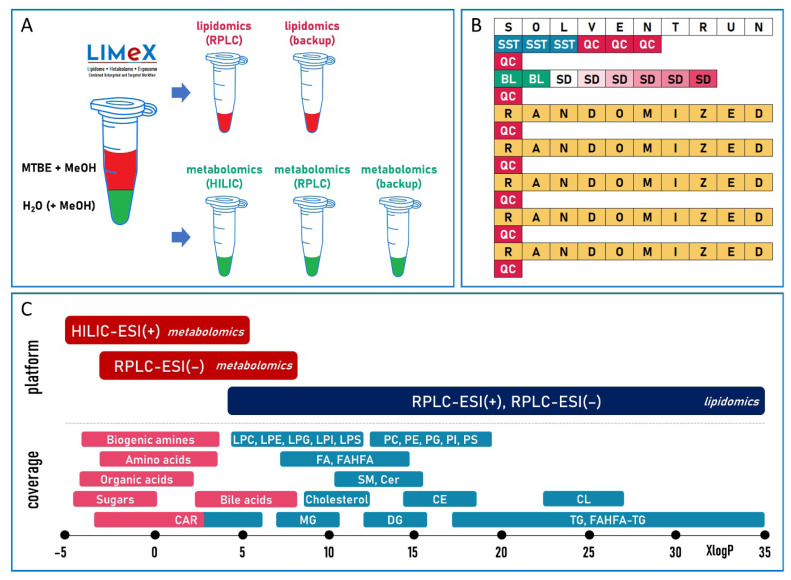
The most utilized method for reducing extract complexity is two-phase liquid extraction, where the separate phases are created by combining immiscible solvents: methyl tert-butyl ether (MTBE)/methanol/water [36], chloroform/methanol/water [37], and dichloromethane/methanol/water [38]. After centrifugation, the organic phase primarily contains nonpolar metabolites, such as lipids, while the polar (water) phase mainly consists of polar metabolites (Fig. 2A). In 2019, Vale et al. [39] introduced three-phase extraction using hexane, methyl acetate, acetonitrile, and water. After centrifugation, the upper organic phase is enriched with neutral lipids such as triacylglycerols and cholesteryl esters; the middle organic phase contains the glycerophospholipids, and the bottom aqueous phase contains polar metabolites.
Fig. 2.
(A) Example of sample extraction using MTBE, methanol, and water [41], leading to two phases for subsequent metabolomics and lipidomics platforms. (B) Example of a typical LC-MS sequence during metabolomics and lipidomic analysis, consisting of solvent injection for general platform equilibration, followed by a system suitability test (SST), platform equilibration using pooled QC samples, analysis of method blanks (BL), a diluted series of QC samples (SD), randomized study samples with regular QC sample injections after every 10 study samples. (C) Example of different LC-MS platforms [41,42] for metabolomic and lipidomic analysis in relation to the XlogP (predicted octanol/water partition coefficient) range of subgroups of polar metabolites and complex lipids.
While organic solvent-based protein precipitation and LLE methods are typically used for untargeted methods, SPE is the first choice for targeted methods, usually covering trace concentrations of metabolites [40].
The success of any research study also depends on an effective quality control (QC) process. Using internal standards in the extraction and resuspension solvents helps control the method’s performance. These standards verify that aliquots are collected correctly from all extracts, the autosampler injects the correct volume, chromatographic and mass accuracy drifts are monitored, signal intensity fluctuations are tracked, and the quality of generated data is assessed during data processing [29]. They can also be used for quantification using a single-point calibration approach if added during the extraction step. Internal standards are essential because they represent true positives in the sample.
QC samples are crucial for obtaining high-quality data in high-throughput analytical chemistry laboratories [43]. They help assess the precision and stability of the analysis. QC samples are used to equilibrate the analytical platform, monitor signals for precision (within and between days), correct signals (normalization), and standardize methods. QC data can also help indicate random errors or fluctuations during the analytical run [44].
QC samples can be created by pooling aliquots of each study sample, reflecting the composition of all samples during analysis. Another option is to employ external QC using a matrix that matches the study samples, which can be useful in large-scale studies where pooling is challenging. In such studies, pooling QC samples can be simplified by using pooled aliquots from only a portion of the samples. Additionally, commercially accessible QC samples (e.g., human plasma NIST SRM 1950 standard reference material [45]) can be applied, though there is a risk of missing some metabolites compared to pooled QC samples [46]. These approaches can be combined; however, they should be planned in advance and not modified during the study.
As Figure 2B shows, a typical metabolomics and lipidomics sequence consists of pre-injection steps (injection of solvents, QC sample) to equilibrate a particular platform, followed by a system suitability test (e.g., a mixture of selected metabolites or biological samples with known composition), analysis of method blanks, a diluted series of QC samples, randomized samples, and regular injection of QC samples [42]. All these steps are essential to generate reliable metabolomics and lipidomics data.
Instrumental analysis
A multiplatform approach using various analytical techniques and platforms is necessary due to the diversity and complexity of the metabolome and lipidome. This approach can improve the overall coverage and reliability of detected metabolites [47]. Liquid chromatography-mass spectrometry (LC-MS) dominates metabolomics and lipidomics. Other commonly applied platforms are gas chromatography-mass spectrometry (GC-MS), capillary electrophoresis-mass spectrometry (CE-MS), and nuclear magnetic resonance (NMR). However, NMR does not offer as broad metabolite coverage as MS-based approaches [29].
LC-MS separates metabolites with a wide range of polarities due to its versatility in stationary phases, column dimensions, mobile phase modifiers, and solvents [48]. Commonly used LC-MS separation platforms are reversed-phase LC (RPLC) and hydrophilic interaction chromatography (HILIC). RPLC separates polar to semi-polar metabolites using C18, C8, or C30 columns, whereas HILIC separates highly polar metabolites using silica, alkyl amide, aminopropylsilane, or sulfobetaine groups as the stationary phase [48]. Efficient chromatographic separation enhances the sensitivity of MS detection, while background noise reduction improves the quality of MS data [49]. For the analysis of polar metabolites (Fig. 2C), RPLC and HILIC are preferred, with mobile phases containing water, acetonitrile, and methanol. On the other hand, for RPLC-based lipidomics, stronger mobile phases are needed, typically containing a high percentage of isopropanol [41,42]. The column formats vary around 50–150 mm in length, with an internal diameter of 2.1 mm, packed with sub-2 μm particles. The separation process takes between 10 and 30 min [50]. However, fast, high-throughput LC-MS methods (<5 min), combined with 96-well plate sample preparation, are preferred for large cohort studies since they allow for hundreds of injections to be performed daily [29,42].
Once separated, analytes are ionized in an ion source to create charged particles. In LC-MS, electrospray ionization (ESI) is typically used, allowing ion formation for small molecules (<2,000 Da) and large molecules, such as peptides and proteins. Due to the chemical diversity of the metabolome and lipidome, ESI is usually applied in both positive and negative modes for more efficient coverage. ESI is a soft ionization technique, minimizing the fragmentation of molecular ions compared to electron ionization (EI) in GC-MS. However, ESI is sensitive to non-volatile salts, leading to limited use of only volatile mobile phase modifiers (e.g., formic acid, acetic acid, ammonium formate, ammonium acetate) in the chromatography part of the method. Due to the possible occurrence of ion suppression, metabolites with lower affinity for electrons or protons can be masked or undetected when competing for ionization [51].
MS techniques used for analyte detection can be either in a simple MS system with a single mass analyzer or in a tandem MS/MS system with multiple analyzers. These systems fall into low-resolution (LRMS) and high-resolution (HRMS) techniques. The main difference between LRMS and HRMS is their mass accuracy, i.e., the precision in determining the mass. HRMS can reach the accurate mass and increase confidence during metabolite annotation, whereas LRMS can only differentiate compounds based on nominal mass, which can cause false positives for compounds that share mass but are structurally unrelated [52]. Therefore, untargeted metabolomics and lipidomics rely on HRMS and HR-MS/MS using time-of-flight or orbital ion trap analyzers and operating in data-dependent acquisition (DDA) or data-independent acquisition (DIA) modes. In DDA mode, precursor ions above a pre-set threshold are selected using a narrow isolation window, making connecting product and precursor ions easier. However, low-abundance ions can be missed, and the settings are more complex than in DIA, which may lead to errors [53,54]. Conversely, in DIA mode, all precursor ions within the wide isolation window are fragmented, covering more low-abundance ions; however, this results in more complex spectra that are harder to interpret [55]. Tools like MS-DIAL [56], DecoMetDIA [57], and DecoID [58] help to deconvolute these complex MS/MS spectra. For the targeted LC-MS method, LRMS triple-quadrupole (QqQ) and quadrupole/linear ion trap (QLIT) are used, usually operating in a multiple reaction monitoring (MRM) mode to improve sensitivity and selectivity of monitored ions [50].
In general, untargeted methods provide semi-quantitative data, meaning that the results are reported as peak areas or heights in arbitrary units within the linear dynamic range of the detector. In contrast, targeted methods report quantitative data in molar concentrations [29]. Although quantification is often requested, it is not necessary for many studies since both semi-quantitative and quantitative data can be used for statistical analysis. However, the advantage of quantitative data is that it allows for the immediate distinction between major and minor metabolites and enables direct comparisons of results between laboratories and studies.
Data processing
Properly handling complex datasets produced by metabolomics and lipidomics experiments is crucial, as this process significantly impacts metabolite annotation and quantification, consequently affecting the biological interpretation of results [59]. A standard untargeted metabolomics and lipidomics study can generate hundreds of annotated metabolites and numerous unknown features characterized by retention time and mass-to-charge ratio (m/z). Data handling can be divided into data processing and data analysis. Data processing uses signal processing methods to refine the raw data and combine them between measurements, converting data into a format that is easier for further analysis. This includes feature detection, chromatogram building, deisotoping, peak alignment, and gap-filling. Data analysis involves examining and interpreting processed data from previous steps, using methods like clustering metabolic profiles or finding key differences between sample groups [59]. Over the last decade, numerous processing tools have been introduced, such as MarkerLynx, MarkerView, MassHunter Profiling, Compound Discoverer, MS-DIAL (Fig. 3A), MZmine, XCMS, MetAlign, GeneDATA, Matlab and R scripts [29].
Fig. 3.
(A) Example of MS-DIAL software [56] used for processing lipidomics data acquired using the RPLC-ESI(–)-MS [41], with annotated PC 16:0_18:2 in human serum. Using ammonium acetate and acetic acid as mobile phase modifiers led to the detection PC 34:2 as an acetate adduct ([M+CH3COO]−) (m/z 816.576). The MS/MS spectrum of PC 34:2 provided a fragment ion [M–CH3]− (m/z 742.539) and a series of fragments for elucidating fatty acyl chains (e.g., m/z 255.233 for 16:0 and m/z 279.233 for 18:2). The use of the underscore “_” indicates certainty in the composition of the fatty acyl constituents but not their specific placement on the glycerol backbone. (B) Example of MS-FINDER software [60] used for the structure elucidation of an unknown compound (m/z 189.1597, retention time 4.57 min) in human serum acquired using the HILIC-ESI(+)-MS platform [41], with tentative annotation as N6,N6,N6-trimethyl-L-lysine.
Due to the structural variability and diversity of metabolites, detecting and annotating metabolites can be challenging. On average, successful annotation occurs for only approximately 10 % of the molecules, underscoring the importance of accurately identifying most molecular structures [61]. It should also be noted that in LC-MS, each metabolite can be detected in multiple ion forms, which can be annotated if present in spectral libraries or based on accurate mass differences. For instance, phosphatidylcholines (PC) can be detected during lipidomics profiling in positive ESI as [M+H]+ (major peak) and [M+Na]+ (minor peak), while negative ESI provides [M+CH3COO]− (major peak in the presence of ammonium acetate in the mobile phase [41]) and [M+Cl]− (minor peak). Depending on the data processing workflow, various options are possible for reporting these ion forms, such as providing all annotated species separately, species from one ionization mode only (e.g., the one with a lower relative standard deviation in QC samples), or combining adducts (summing peak intensities) for each ionization mode.
The Metabolomics Standardization Initiative (MSI) describes community-based guidelines for reporting and performing metabolomics workflows, proposing four confidence levels [62]: Level 1 – matching based on retention time, MS1, and MS/MS spectrum; Level 2 – matching based on MS1 and MS/MS spectrum; Level 3 – annotation based on matching MS1 accurate mass only; Level 4 – unknown compound characterized by retention time and m/z. However, multiple researchers have suggested revisions and modifications [63–65]. The Lipidomics Standards Initiative (LSI) has recently been introduced to create standardized lipid species annotations and unify community efforts [66].
The most reliable approach for metabolite annotation represents the use of spectral libraries containing retention time, m/z (MS1 accurate mass), and MS/MS fragmentation spectra (MSI – Level 1). However, it is virtually impossible to obtain all three pieces of information for every possible metabolite. Thus, commercial or open-access MS/MS libraries (with MS1 precursor ions and MS/MS spectra) are crucial in confident compound annotation in metabolomics and lipidomics (MSI – Level 2). In recent years, spectral libraries and databases have grown in both coverage and diversity [67]. METLIN Gen2 is the most extensive spectral library (metlin.scripps.edu), containing over 900,000 molecular standards and MS/MS data, comprising over 4 million tandem spectra [68]. Other extensive MS/MS libraries include the National Institute of Standards and Technology (NIST) MS/MS library (chemdata.nist.gov) and MassBank of North America (MoNA, massbank.us). Additional resources include MassBank (massbank.jp), ReSpect (spectra.psc.riken.jp), RIKEN PlaSMA (plasma.riken.jp), mzCloud (mzcloud.org), GNPS (gnps.ucsd.edu), MSforID (msforid.com), and HMBD (hmdb.ca).
Furthermore, numerous software and tools have been developed to help annotate unknown compounds, such as MS-FINDER (Fig. 3B), CFM-ID, MetFrag, ChemDistiller, and CSI:FingerID. These tools convert mass data into molecular fragments using combinatorial structure generation techniques and search against existing structures in various databases. Potential candidates can be filtered using additional orthogonal filters based on retention time prediction [69] or hydrogen/deuterium exchange mass spectrometry (HDX-MS) [70,71]. Nevertheless, confirmation should always follow by analyzing an analytical standard under identical instrumental conditions [72].
Bioinformatics
Statistical analysis is essential to properly extract relevant information from the obtained data. Statistical analyses can be categorized as univariate and multivariate methods. Univariate statistical methods include t-test, ANOVA, and fold-change analysis to compare different sets of samples. These methods are used for sets of tens to hundreds of metabolites, which increases the chances of false positives [73]. Therefore, correction methods such as Bonferroni correction [74] or the Benjamini-Hochberg [75] false discovery rate should be applied. These corrections have been addressed in multiple studies [74–76]. Commonly used multivariate methods include principal component analysis (PCA), partial least squares discriminant analysis (PLS-DA), and hierarchical cluster analysis (HCA) [77]. A routinely employed web-based platform for comprehensive metabolomics and lipidomics data analysis and interpretation is MetaboAnalyst (metaboanalyst.ca) [78].
Next, the biological relevance of the measured metabolites is interpreted using pathway and enrichment analysis. Enrichment analysis identifies functionally relevant metabolites and links their changes to biological contexts, suggesting key pathways or disease conditions for further study. Pathway analysis, on the other hand, finds pathways that significantly affect specific biological processes [79]. Both analyses are performed using various software tools such as MetaMapR, MetabNet, GNPS, MS2LDA, MetaboAnalyst, or MetFlow to map the metabolic pathways. New tools, such as an ontology database and enrichment analysis (LION, lipidontology.com) and lipid over-representation analysis (LORA, lora.metabolomics.fgu.cas.cz), are also available to interpret complex lipids [80].
An important part of every experiment is data sharing. Data should be shared following the Findable, Accessible, Interoperable, and Reusable (FAIR) Guiding Principles for scientific data management and stewardship [81]. Public repositories such as Metabolomics Workbench (metabolomicsworkbench.org), MetaboLights (ebi.ac.uk/metabolights), and MassIVE (massive.ucsd.edu/ProteoSAFe/static/massive.jsp) enable data sharing. A newly introduced dynamic checklist (lipidomicstandards.org/reporting_checklist) summarizing key details of lipidomic analyses can be stored or shared in the supporting materials of papers or at a general-purpose open repository Zenodo (zenodo.org).
Recently introduced metabolomics and lipidomics atlases should also serve as open-access resources [29]. These atlases monitor the quantities and relationships of metabolites in different biological matrices, highlighting the importance of reusing and sharing data [82].
Metabolomics and lipidomics for studying metabolic syndrome
In recent years, MetS has become a major health risk with its increasing prevalence, reaching pandemic proportions [83]. The disease affects around 25 % of the global population, making prevention and management essential [84]. Understanding its pathophysiology is crucial in this effort. Metabolomics and lipidomics have been employed to investigate various diseases by identifying diagnostic biomarkers. Recently, research efforts have focused on cardiovascular diseases (CVD), type 2 diabetes (T2D), and metabolic dysfunction-associated steatotic liver disease (MASLD), all of which are associated with MetS. Wishart’s comprehensive review in 2019 further underscored the significance of metabolomics studies in understanding physiological and pathophysiological processes [19]. Supplementary Tables S1–S3 overview metabolomics and lipidomics large-cohort studies focusing on CVD, T1D/T2D, and MASLD. Next, we briefly highlight some of these studies to elucidate key findings and advancements, emphasizing how metabolomic and lipidomic profiles have provided deeper insights into disease mechanisms and potential therapeutic targets.
Cardiovascular diseases
CVDs are the leading cause of death globally. In 2022, CVDs caused approximately 19.8 million deaths, accounting for about one-third of all global mortality that year. Major contributors to this toll were ischemic heart disease (9.2 million deaths) and ischemic stroke (3.5 million deaths) [85]. More than three-quarters of CVD deaths occur in low- and middle-income countries, compared to high-income countries, where the CVD death rate has declined [86,87].
CVDs are disorders of the heart and blood vessels, including coronary heart disease, cerebrovascular disease, peripheral arterial disease, rheumatic heart disease, congenital heart disease, deep vein thrombosis, and pulmonary embolism. Heart attacks and strokes are usually considered acute events, primarily resulting from a blockage that obstructs blood flow to the heart or brain [86]. The risk factors for cardiovascular diseases often include an unhealthy diet, physical inactivity, tobacco use, and harmful use of alcohol. These factors can be controlled, reducing the risk of CVD occurrence [86].
One area in CVD research involves exploring the role of different metabolites in disease promotion and progression. For instance, amino acids (alanine, glutamine, glycine, histidine, isoleucine, leucine, lysine, valine, phenylalanine, and tyrosine) have been identified as predictors of incident CVD risks [88–92]. Other discovered biomarkers of CVD are choline, trimethylamine N-oxide (TMAO), and betaine [93–96]. Similarly, compounds such as trimethyllysine [97], phenylacetyl glutamine [98], and niacin metabolites (N1-methyl-2-pyridone-5-carboxamide and N1-methyl-4-pyridone-3-carboxamide) [99] have been linked to CVD risks. Moreover, in recent studies, the endogenous sugar alcohols erythritol and xylitol were both clinically and mechanistically linked to CVD [100,101].
The association of diet-linked metabolites with CVD has also been explored. Fu et al. [102] investigated metabolites connected with a healthy lifestyle and their effect on CVD incidence. They identified and validated 111 metabolites associated with overall lifestyle, 65 of which were related to CVD risk. Healthy lifestyle-linked metabolites were also studied by Lu et al. [90]. Diabetes patients free of CVD were divided into groups based on the healthy level of five lifestyle factors and observed. Adherence to healthy lifestyle factors was associated with 44 plasma metabolites (e.g., 3-hydroxybutyrate, alanine, glutamine, glycine, branched-chain amino acids), and approximately half of them mediated between at least one lifestyle factor and CVD risk. Both studies suggest that a healthy diet positively affects the incidence of CVD.
Additionally, the effects of legume [103] and walnut [104] consumption on CVD risk were researched. Walnut consumption was found to lower the risk of incident CVD and T2D, while legume consumption was associated with a lower risk of T2D but not CVD. Furthermore, gut microbiome-derived metabolites such as p-cresol sulfate and indoxyl sulfate have garnered attention [105]. This study shows that these abundant microbiome-derived metabolites have a greater impact on CVD than previously thought. It also suggests targeting the gut microbial pathways that produce p-cresol and indole as a potential strategy for treating CVD.
Lipidomics profiling also reveals characteristic lipid signatures associated with increased CVD risk. Harm et al. [106] focused on the platelet lipidome of coronary artery disease patients and found alterations in the lipid composition of patients with adverse cardiovascular events. The results showed that the platelet lipidome of CVD patients with increased cardiovascular risk is changed, and specific platelet lipids may indicate adverse events. These findings may help discriminate the individual risk of patients with coronary artery disease. Eichelmann et al. [88] investigated associations of plasma lipid alterations with incident cardiometabolic diseases and studied the effect of dietary fat modulation on discovered risk-associated lipids. The results suggest that dietary fat intervention can alter lipids, which may serve as a potential tool for primary disease prevention. Furthermore, Seah et al. [107] suggested that certain classes of sphingolipids may also affect CVD risk.
Type 1 & 2 diabetes
As of 2021, the global prevalence of diabetes was estimated at 10.5 % (537 million people), projected to rise to 12.2 % (783 million people) by 2045. Diabetes was responsible for approximately 6.7 million deaths worldwide in 2021, with global healthcare expenditures amounting to approximately USD 966 billion [108]. However, the majority of these cases are attributed to T2D, while T1D affected approximately 8.4 million individuals globally in 2021 [109]. In the future, access to and affordability of insulin may become challenging, particularly in underdeveloped and developing countries, due to the increasing prevalence and incidence of T1D [110].
Diabetes is a complex chronic metabolic disease characterized by high prevalence and mortality, encompassing T1D, T2D, and gestational diabetes occurring during pregnancy. T1D results from insufficient insulin production by the pancreas, necessitating daily insulin administration. T2D arises from inadequate insulin secretion and the body’s ineffective use of insulin, leading to elevated blood sugar levels. T2D impacts the metabolism of glucose, lipids, and amino acids [111,112].
Metabolomics and lipidomics studies of T1D aim to identify biomarkers for predicting T1D risk and aiding in early disease detection. Orešič et al. [113] analyzed the lipidome profile of cord serum samples to investigate associations between lipid profile changes and β-cell autoimmunity development or clinical T1D. Their study found that progression to T1D correlated with decreased concentrations of major choline-containing phospholipids (sphingomyelins and phosphatidylcholines) in cord blood. The study also indicated that phospholipid reduction is associated explicitly with T1D progression rather than general β-cell autoimmunity.
La Torre [89] and Tapia [90] also studied cord blood samples. La Torre et al. [114] discovered that decreased levels of phospholipids at birth, especially phosphatidylcholines and phosphatidylethanolamines, may contribute to early induction of islet autoimmunity and increased T1D risk. Conversely, Tapia et al. [115] focused more on changes in the metabolome profile than lipidome alterations. However, the research showed no strong associations of selected polar metabolites with T1D. Nevertheless, Webb-Robertson et al. [116] identified multiple metabolites associated with T1D progression by age 6, primarily comprising sugar metabolism compounds such as fructose, levoglucosan, glycerol-α-phosphate, and xylulose.
Recent studies have explored metabolomics’ potential in predicting T2D risk based on dietary patterns and corresponding biomarkers. One study involving nearly 6,000 participants identified 29 plasma metabolites associated with inflammatory and insulinemic dietary patterns [117]. The top five biomarkers included PE 36:4, CAR 5:0, PC 34:4, 1-methylguanosine, and N4-acetyl-cytidine. Additionally, investigations into the lipid profile of lean and obese individuals with T2D revealed significant lipidome changes (lyso-, diacyl- and ether-phospholipids, and 1-deoxyceramides), aiding in T2D diagnosis [118].
Lipid profiles containing 69 odd-chain saturated fatty acids (OCFA) among 15 lipid subclasses were also examined for their potential as T2D biomarkers [119], revealing variations dependent on lipid class and sex, correlating with food consumption. Sun et al. [120] investigated plasma acylcarnitines’ role in early T2D prediction, identifying long-chain acylcarnitines as significantly linked to future T2D risk.
Moreover, interventions targeting weight loss have shown promise in altering metabolite signatures associated with T2D. Studies have noted positive associations between changes in branched-chain amino acids (valine, leucine, isoleucine) and branched-chain ketoacids (α-ketoisovalerate, α-ketoisocaproate, α-keto-β-methylvalerate) with glycated hemoglobin (HbA1c) levels following weight loss [121]. Branched-chain amino acids are frequently studied due to their association with increased T2D risk [122–127]. 3-Hydroxybutyrate is another frequently studied metabolite, often alongside branched-chain amino acids [122–126,128]. Similar to its association with CVD, TMAO has also been investigated in relation to T2D [129]. Lemaitre et al. [129] explored the connections of TMAO, carnitine, crotonobetaine, and γ-butyrobetaine with insulin resistance, and betaine and choline with enhanced insulin sensitivity. However, they did not establish a definitive association.
Metabolic dysfunction-associated steatotic liver disease
MASLD is the latest term used to describe steatotic liver disease associated with MetS, encompassing various metabolic risk factors and often coexisting with other chronic liver conditions [130]. Historically, the term nonalcoholic fatty liver disease (NAFLD) was used. In 2020, Eslam et al. [131] proposed the term metabolic dysfunction-associated fatty liver disease (MAFLD), which was further modified to MASLD in 2023 [132]. Both MAFLD and MASLD identify patients with hepatic steatosis and metabolic dysfunction [133]. There are slight differences in the definitions of MASLD and MAFLD, which have been discussed in several articles [130,132–134]. Notably, MAFLD encompasses patients with fatty liver regardless of alcohol consumption pattern or amount [132], whereas MASLD introduces the term MetALD for patients who meet alcohol-related fatty liver disease criteria [134]. MASLD diagnosis requires meeting one of five cardiometabolic risk factors [132], while MAFLD requires meeting two out of seven metabolic dysfunction parameters [131]. De et al. [135] suggest that MASLD and SLD (steatotic liver disease) criteria may better suit lean patients with NAFLD than MAFLD criteria. Consequently, both MASLD and MAFLD terms are used in literature to classify liver diseases associated with metabolic dysfunction, although NAFLD remains prevalent in many studies since the new nomenclature’s introduction.
The global prevalence of NAFLD was estimated to be approximately 30 % between 1990 and 2019, with a continuing upward trend [136]. This increasing prevalence of NAFLD is likely associated with rising rates of diabetes and obesity. However, the global mortality rate declined from 2.39 per 100,000 population in 1990 to 2.09 per 100,000 population in 2019 [137].
MASLD includes a range of steatotic liver conditions, from isolated hepatic steatosis to metabolic dysfunction-associated steatohepatitis (MASH), with varying levels of liver fibrosis that can potentially lead to cirrhosis. MASLD is associated with a higher risk of liver complications (e.g., cirrhosis), end-stage liver disease, and hepatocellular carcinoma, as well as an increased risk of developing extrahepatic issues such as cardiovascular disease (CVD), chronic kidney disease, and certain extrahepatic cancers [138].
Recent large-scale cohort studies aim to identify risk factors and biomarkers for MASLD, aiding in its challenging diagnosis. Commonly identified biomarkers include amino acids, particularly aromatic amino acids (tyrosine, tryptophan) and branched-chain amino acids (isoleucine, leucine, valine) [139–143]. Studies by Hirata [142] and Martínez-Arranz [144] examined the association of NAFLD with cardiovascular risk, identifying metabolomic signatures aligning with known CVD risk factors. Hirata et al. [142] found that NAFLD was positively associated with the cardio-ankle vascular index (CAVI), an indicator of subclinical atherosclerosis, and identified ten metabolites involved in both NAFLD and CAVI: branched-chain amino acids (valine, leucine, and isoleucine), aromatic amino acids (tyrosine and tryptophan), alanine, proline, glutamic acid, glycerophosphorylcholine, and 4-methyl-2-oxopenta-noate. Martínez-Arranz et al. [144] investigated lipidomic profile changes, particularly in triacylglycerols, phosphatidylcholines, and sphingomyelins, providing evidence of distinct metabolic mechanisms associated with NAFLD progression that vary between subtypes.
McGlinchey et al. [145] observed lipidomic and metabolomic profile changes across different stages of NAFLD progression, highlighting unique metabolites and 27 common metabolites across all stages, including significant alterations in cholesteryl esters, ceramides, lysophosphatidylcholines, phosphatidylcholines, phosphatidylethanolamine, sphingomyelins, and triacylglycerols. Hu et al. [146] discovered correlations between NAFLD and uric acid, as well as oleic acid-hydroxy oleic acid (OAHOA), identifying OAHOA as a novel biomarker for NAFLD prevalence in a cohort of 1,479 patients (aged 18–80 years). Other studies have explored potential biomarkers, such as anandamide [147] or taurochloric acid [148].
Conclusions
Metabolomics and lipidomics represent effective tools for studying MetS and related disorders. The comprehensive multiplatform-based profiling of polar metabolites and complex lipids in large cohorts has enabled the identification of novel biomarkers and enhanced our understanding of disease mechanisms. Key advancements include the discovery of metabolic signatures associated with CVD, T1D, T2D, and MASLD.
Regarding polar metabolites, branched-chain amino acids (valine, leucine, isoleucine), TMAO, betaine, choline, and 3-hydroxybutyrate have been identified in multiple studies as promising biomarkers. For complex lipids, a panel or combination of affected lipids is expected to be useful as biomarkers, including acylcarnitines, phospholipids, sphingomyelins, and triacylglycerols as key lipid subclasses.
Further research is needed to validate these reported biomarkers in diverse populations and clinical settings, ensuring their robustness and clinical utility. Standardization of experimental protocols and data analysis methods will be critical to facilitate data comparability and reproducibility across studies. Based on a review of multiple studies, we also advocate for the inclusion of authoritative identifiers such as InChI keys or identifiers from bioinformatics resources such as the Human Metabolome Database (hmdb.ca) and LIPID MAPS (lipidmaps.org). This will expedite the comparison of potential biomarkers within studies, making the process faster and more effective.
In addition, further advances in analytical technologies and computational tools will continue to drive innovation in metabolomics and lipidomics, offering new opportunities for early disease detection and personalized therapeutic interventions.
Supplementary Information
Acknowledgements
This research was funded by the Czech Health Research Council (NU20-01-00186), the Ministry of Education, Youth and Sport of the Czech Republic (LUAUS24169), and the National Institute for Research of Metabolic and Cardiovascular Diseases (Programme EXCELES, ID Project No. LX22NPO5104) funded by the European Union - Next Generation EU. Examples of LC-MS chromatographic records (Vanquish UHPLC & Q Exactive Plus, Vanquish UHPLC & Orbitrap Exploris 480) processed in MS-DIAL and MS-FINDER software were provided by the Metabolomics Core Facility at the Institute of Physiology of the Czech Academy of Sciences (metabolomics.fgu.cas.cz).
Footnotes
Conflict of Interest: There is no conflict of interest.
Supplementary Materials: The supporting information (Table S1–S3) can be downloaded at:
https://www.biomed.cas.cz/physiolres/pdf/2024/Rakusanova_Supl_Tables_1-3.pdf
References
- 1.Alberti KG, Eckel RH, Grundy SM, Zimmet PZ, Cleeman JI, Donato KA, Fruchart JC, et al. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009;120:1640–1645. doi: 10.1161/CIRCULATIONAHA.109.192644. [DOI] [PubMed] [Google Scholar]
- 2.Alberti KG, Zimmet P, Shaw J. The metabolic syndrome - a new worldwide definition. Lancet. 2005;366:1059–1062. doi: 10.1016/S0140-6736(05)67402-8. [DOI] [PubMed] [Google Scholar]
- 3.Han TS, Lean MEJ. Metabolic syndrome. Medicine. 2015;43:80–87. doi: 10.1016/j.mpmed.2014.11.006. [DOI] [Google Scholar]
- 4.Grundy SM. Metabolic Syndrome Pandemic. Arterioscler Thromb Vasc Biol. 2008;28:629–636. doi: 10.1161/ATVBAHA.107.151092. [DOI] [PubMed] [Google Scholar]
- 5.Radu F, Potcovaru C-G, Salmen T, Filip PV, Pop C, Fierbințeanu-Braticievici C. The link between NAFLD and metabolic syndrome. Diagnostics. 2023;13:614. doi: 10.3390/diagnostics13040614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noubiap JJ, Nansseu JR, Lontchi-Yimagou E, Nkeck JR, Nyaga UF, Ngouo AT, Tounouga DN, Tianyi F-L, Foka AJ, Ndoadoumgue AL, et al. Geographic distribution of metabolic syndrome and its components in the general adult population: A meta-analysis of global data from 28 million individuals. Diabetes Res Clin Pr. 2022;188:109924. doi: 10.1016/j.diabres.2022.109924. [DOI] [PubMed] [Google Scholar]
- 7.Reisinger C, Nkeh-Chungag BN, Fredriksen PM, Goswami N. The prevalence of pediatric metabolic syndrome-a critical look on the discrepancies between definitions and its clinical importance. Int J Obes. 2021;45:12–24. doi: 10.1038/s41366-020-00713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cameron AJ, Shaw JE, Zimmet PZ. The metabolic syndrome: prevalence in worldwide populations. Endocrinol Metab Clin North Am. 2004;33:351–375. doi: 10.1016/j.ecl.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Allam-Ndoul B, Guénard F, Garneau V, Cormier H, Barbier O, Pérusse L, Vohl MC. Association between metabolite profiles, metabolic syndrome and obesity status. Nutrients. 2016;8:324. doi: 10.3390/nu8060324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong L-L, Yang S, Zhang W, Han F-F, Lv Y-L, Xuan L-L, Liu H, Liu L-H. Discovery of metabolite profiles of metabolic syndrome using untargeted and targeted LC-MS based lipidomics approach. J Pharm Biomed. 2020;177:112848. doi: 10.1016/j.jpba.2019.112848. [DOI] [PubMed] [Google Scholar]
- 11.Hajjar G, Barros Santos MC, Bertrand-Michel J, Canlet C, Castelli F, Creusot N, Dechaumet S, et al. Scaling-up metabolomics: Current state and perspectives. Trends Analyt Chem. 2023;167:117225. doi: 10.1016/j.trac.2023.117225. [DOI] [Google Scholar]
- 12.Pujos-Guillot E, Brandolini M, Pétéra M, Grissa D, Joly C, Lyan B, Herquelot É, et al. Systems metabolomics for prediction of metabolic syndrome. J Proteome Res. 2017;16:2262–2272. doi: 10.1021/acs.jproteome.7b00116. [DOI] [PubMed] [Google Scholar]
- 13.Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, Brown M, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2011;6:1060–1083. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- 14.Salek R, Emery L, Beisken S. Metabolomics: an introduction. EMBL-EMI. 2014 doi: 10.6019/TOL.MBS.2014.00001.1. [DOI] [Google Scholar]
- 15.Gardner MJ, Altman DG. Confidence intervals rather than P values: Estimation rather than hypothesis testing. Br Med J (Clin Res Ed) 1986;292:746–750. doi: 10.1136/bmj.292.6522.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vinaixa M, Samino S, Saez I, Duran J, Guinovart JJ, Yanes O. A guideline to univariate statistical analysis for LC/MS-based untargeted metabolomics-derived data. Metabolites. 2012;2:775–795. doi: 10.3390/metabo2040775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serdar CC, Cihan M, Yücel D, Serdar MA. Sample size, power and effect size revisited: simplified and practical approaches in pre-clinical, clinical and laboratory studies. Biochem Med. 2021;31:010502. doi: 10.11613/BM.2021.010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faul F, Erdfelder E, Buchner A, Lang AG. Statistical power analyses using G*Power 3.1: Tests for correlation and regression analyses. Behav Res Methods. 2009;41:1149–1160. doi: 10.3758/BRM.41.4.1149. [DOI] [PubMed] [Google Scholar]
- 19.Zhao X, Niu L, Clerici C, Russo R, Byrd M, Setchell KDR. Data analysis of MS-based clinical lipidomics studies with crossover design: A tutorial mini-review of statistical methods. Clin Mass Spectrom. 2019;13:5–17. doi: 10.1016/j.clinms.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Billoir E, Navratil V, Blaise BJ. Sample size calculation in metabolic phenotyping studies. Brief Bioinform. 2015;16:813–819. doi: 10.1093/bib/bbu052. [DOI] [PubMed] [Google Scholar]
- 21.Nyamundanda G, Gormley IC, Fan Y, Gallagher WM, Brennan L. MetSizeR: Selecting the optimal sample size for metabolomic studies using an analysis based approach. BMC Bioinform. 2013;14:338. doi: 10.1186/1471-2105-14-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li F, Zhou Y, Zhang X, Tang J, Yang Q, Zhang Y, Luo Y, et al. SSizer: Determining the sample sufficiency for comparative biological study. J Mol Biol. 2020;432:3411–3421. doi: 10.1016/j.jmb.2020.01.027. [DOI] [PubMed] [Google Scholar]
- 23.Ivanisevic J, Want EJ. From samples to insights into metabolism: Uncovering biologically relevant information in LC-HRMS metabolomics data. Metabolites. 2019;9:308. doi: 10.3390/metabo9120308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lenth RV. Some practical guidelines for effective sample size determination. Am Stat. 2001;55:187–193. doi: 10.1198/000313001317098149. [DOI] [Google Scholar]
- 25.Arifin WN, Zahiruddin WM. Sample size calculation in animal studies using resource equation approach. Malays J Med Sci. 2017;24:101–105. doi: 10.21315/mjms2017.24.5.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunn WB, Wilson ID, Nicholls AW, Broadhurst D. The importance of experimental design and QC samples in large-scale and MS-driven untargeted metabolomic studies of humans. Bioanalysis. 2012;4:2249–2264. doi: 10.4155/bio.12.204. [DOI] [PubMed] [Google Scholar]
- 27.Smith L, Villaret-Cazadamont J, Claus SP, Canlet C, Guillou H, Cabaton NJ, Ellero-Simatos S. Important considerations for sample collection in metabolomics studies with a special focus on applications to liver functions. Metabolites. 2020;10:104. doi: 10.3390/metabo10030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alseekh S, Aharoni A, Brotman Y, Contrepois K, D’Auria J, Ewald J, Ewald JC, et al. Mass spectrometry-based metabolomics: A guide for annotation, quantification and best reporting practices. Nat Methods. 2021;18:747–756. doi: 10.1038/s41592-021-01197-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rakusanova S, Fiehn O, Cajka T. Toward building mass spectrometry-based metabolomics and lipidomics atlases for biological and clinical research. Trends Anal Chem. 2023;158:116825. doi: 10.1016/j.trac.2022.116825. [DOI] [Google Scholar]
- 30.Mushtaq MY, Choi YH, Verpoorte R, Wilson EG. Extraction for metabolomics: Access to the metabolome. Phytochem Anal. 2014;25:291–306. doi: 10.1002/pca.2505. [DOI] [PubMed] [Google Scholar]
- 31.Khamis MM, Holt T, Awad H, El-Aneed A, Adamko DJ. Comparative analysis of creatinine and osmolality as urine normalization strategies in targeted metabolomics for the differential diagnosis of asthma and COPD. Metabolomics. 2018;14:115. doi: 10.1007/s11306-018-1418-9. [DOI] [PubMed] [Google Scholar]
- 32.Álvarez-Sánchez B, Priego-Capote F, Castro MDL. Metabolomics analysis II. Preparation of biological samples prior to detection. Trends Anal Chem. 2010;29:120–127. doi: 10.1016/j.trac.2009.12.004. [DOI] [Google Scholar]
- 33.Dunn WB. Mass spectrometry in systems biology an introduction. Methods Enzymol. 2011;500:15–35. doi: 10.1016/B978-0-12-385118-5.00002-5. [DOI] [PubMed] [Google Scholar]
- 34.Showalter MR, Nonnecke EB, Linderholm AL, Cajka T, Sa MR, Lönnerdal B, Kenyon NJ, Fiehn O. Obesogenic diets alter metabolism in mice. PLoS One. 2018;13:e0190632. doi: 10.1371/journal.pone.0190632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lepoittevin M, Blancart-Remaury Q, Kerforne T, Pellerin L, Hauet T, Thuillier R. Comparison between 5 extractions methods in either plasma or serum to determine the optimal extraction and matrix combination for human metabolomics. Cell Mol Biol Lett. 2023;28:43. doi: 10.1186/s11658-023-00452-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res. 2008;49:1137–1146. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. doi: 10.1016/S0021-9258(18)64849-5. [DOI] [PubMed] [Google Scholar]
- 38.Southam AD, Haglington LD, Najdekr L, Jankevics A, Weber RJM, Dunn WB. Assessment of human plasma and urine sample preparation for reproducible and high-throughput UHPLC-MS clinical metabolic phenotyping. Analyst. 2020;145:6511–6523. doi: 10.1039/D0AN01319F. [DOI] [PubMed] [Google Scholar]
- 39.Vale G, Martin SA, Mitsche MA, Thompson BM, Eckert KM, McDonald JG. Three-phase liquid extraction: a simple and fast method for lipidomic workflows. J Lipid Res. 2019;60:694–706. doi: 10.1194/jlr.D090795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gasperotti M, Masuero D, Guella G, Mattivi F, Vrhovsek U. Development of a targeted method for twenty-three metabolites related to polyphenol gut microbial metabolism in biological samples, using SPE and UHPLC-ESI-MS/MS. Talanta. 2014;128:221–230. doi: 10.1016/j.talanta.2014.04.058. [DOI] [PubMed] [Google Scholar]
- 41.Cajka T, Hricko J, Rudl Kulhava L, Paucova M, Novakova M, Kuda O. Optimization of mobile phase modifiers for fast LC-MS-based untargeted metabolomics and lipidomics. Int J Mol Sci. 2023;24:1987. doi: 10.3390/ijms24031987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hricko J, Rudl Kulhava L, Paucova M, Novakova M, Kuda O, Fiehn O, Cajka T. Short-term stability of serum and liver extracts for untargeted metabolomics and lipidomics. Antioxidants. 2023;12:986. doi: 10.3390/antiox12050986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Broadhurst D, Goodacre R, Reinke SN, Kuligowski J, Wilson ID, Lewis MR, Dunn WB. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics. 2018;14:72. doi: 10.1007/s11306-018-1367-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Begou O, Gika HG, Theodoridis GA, Wilson ID. Quality control and validation issues in LC-MS metabolomics. Methods Mol Biol. 2018;1738:15–26. doi: 10.1007/978-1-4939-7643-0_2. [DOI] [PubMed] [Google Scholar]
- 45.Bowden JA, Heckert A, Ulmer CZ, Jones CM, Koelmel JP, Abdullah L, Ahonen L, et al. Harmonizing lipidomics: NIST interlaboratory comparison exercise for lipidomics using SRM 1950-Metabolites in frozen human plasma. J Lipid Res. 2017;58:2275–2288. doi: 10.1194/jlr.M079012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dudzik D, Barbas-Bernardos C, García A, Barbas C. Quality assurance procedures for mass spectrometry untargeted metabolomics. A review. J Pharm Biomed Anal. 2018;147:149–173. doi: 10.1016/j.jpba.2017.07.044. [DOI] [PubMed] [Google Scholar]
- 47.Jeppesen MJ, Powers R. Multiplatform untargeted metabolomics. Magn Reson Chem. 2023;61:628–653. doi: 10.1002/mrc.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harrieder EM, Kretschmer F, Böcker S, Witting M. Current state-of-the-art of separation methods used in LC-MS based metabolomics and lipidomics. J Chromatogr B. 2022;1188:123069. doi: 10.1016/j.jchromb.2021.123069. [DOI] [PubMed] [Google Scholar]
- 49.Zhou B, Xiao JF, Tuli L, Ressom HW. LC-MS-based metabolomics. Mol Biosyst. 2012;8:470–81. doi: 10.1039/C1MB05350G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cajka T, Fiehn O. Toward merging untargeted and targeted methods in mass spectrometry-based metabolomics and lipidomics. Anal Chem. 2016;88:524–545. doi: 10.1021/acs.analchem.5b04491. [DOI] [PubMed] [Google Scholar]
- 51.Gowda GAN, Djukovic D. Overview of mass spectrometry-based metabolomics: Opportunities and challenges. In: RAFTERY D, editor. Mass Spectrometry in Metabolomics: Methods and Protocols. New York, NY: 2014. pp. 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zarrouk E, El Balkhi S, Saint-Marcoux F. Low-resolution or high-resolution MS for clinical and forensic toxicology: some considerations from two real cases. LCGC Supplements. 2023;41:24–27. doi: 10.56530/lcgc.na.ez3089i6. [DOI] [Google Scholar]
- 53.Defossez E, Bourquin J, von Reuss S, Rasmann S, Glauser G. Eight key rules for successful data-dependent acquisition in mass spectrometry-based metabolomics. Mass Spectrom Rev. 2023;42:131–143. doi: 10.1002/mas.21715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nash WJ, Dunn WB. From mass to metabolite in human untargeted metabolomics: Recent advances in annotation of metabolites applying liquid chromatography-mass spectrometry data. Trends Anal Chem. 2019;120:115324. doi: 10.1016/j.trac.2018.11.022. [DOI] [Google Scholar]
- 55.Guo J, Huan T. Comparison of full-scan, data-dependent, and data-independent acquisition modes in liquid chromatography-mass spectrometry based untargeted metabolomics. Anal Chem. 2020;92:8072–8080. doi: 10.1021/acs.analchem.9b05135. [DOI] [PubMed] [Google Scholar]
- 56.Tsugawa H, Ikeda K, Takahashi M, Satoh A, Mori Y, Uchino H, Okahashi N, et al. A lipidome atlas in MS-DIAL 4. Nat Biotechnol. 2020;38:1159–1163. doi: 10.1038/s41587-020-0531-2. [DOI] [PubMed] [Google Scholar]
- 57.Yin Y, Wang R, Cai Y, Wang Z, Zhu Z-J. DecoMetDIA: Deconvolution of multiplexed MS/MS spectra for metabolite identification in SWATH-MS-based untargeted metabolomics. Anal Chem. 2019;91:11897–11904. doi: 10.1021/acs.analchem.9b02655. [DOI] [PubMed] [Google Scholar]
- 58.Stancliffe E, Schwaiger-Haber M, Sindelar M, Patti GJ. DecoID improves identification rates in metabolomics through database-assisted MS/MS deconvolution. Nat Methods. 2021;18:779–787. doi: 10.1038/s41592-021-01195-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Katajamaa M, Oresic M. Data processing for mass spectrometry-based metabolomics. J Chromatogr A. 2007;1158:318–28. doi: 10.1016/j.chroma.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 60.Tsugawa H, Kind T, Nakabayashi R, Yukihira D, Tanaka W, Cajka T, Saito K, Fiehn O, Arita M. Hydrogen rearrangement rules: Computational MS/MS fragmentation and structure elucidation using MS-FINDER software. Anal Chem. 2016;88:7946–7958. doi: 10.1021/acs.analchem.6b00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jonge NF, Mildau K, Meijer D, Louwen JJR, Bueschl C, Huber F, van der Hooft JJJ. Good practices and recommendations for using and benchmarking computational metabolomics metabolite annotation tools. Metabolomics. 2022;18:103. doi: 10.1007/s11306-022-01963-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TWM, et al. Proposed minimum reporting standards for chemical analysis. Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Creek DJ, Dunn WB, Fiehn O, Griffin JL, Hall RD, Lei Z, Mistrik R, et al. Metabolite identification: Are you sure? And how do your peers gauge your confidence? Metabolomics. 2014;10:350–353. doi: 10.1007/s11306-014-0656-8. [DOI] [Google Scholar]
- 64.Spicer RA, Salek R, Steinbeck C. A decade after the metabolomics standards initiative it’s time for a revision. Sci Data. 2017;4:170138. doi: 10.1038/sdata.2017.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salek RM, Steinbeck C, Viant MR, Goodacre R, Dunn WB. The role of reporting standards for metabolite annotation and identification in metabolomic studies. Gigascience. 2013;2:13. doi: 10.1186/2047-217X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liebisch G, Fahy E, Aoki J, Dennis EA, Durand T, Ejsing CS, Fedorova M, et al. Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS-derived lipid structures. J Lipid Res. 2020;61:1539–1555. doi: 10.1194/jlr.S120001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kind T, Tsugawa H, Cajka T, Ma Y, Lai Z, Mehta SS, Wohlgemuth G, et al. Identification of small molecules using accurate mass MS/MS search. Mass Spectrom Rev. 2018;37:513–532. doi: 10.1002/mas.21535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xue J, Guijas C, Benton HP, Warth B, Siuzdak G. METLIN MS2 molecular standards database: a broad chemical and biological resource. Nat Methods. 2020;17:953–954. doi: 10.1038/s41592-020-0942-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Witting M, Böcker S. Current status of retention time prediction in metabolite identification. J Sep Sci. 2020;43:1746–1754. doi: 10.1002/jssc.202000060. [DOI] [PubMed] [Google Scholar]
- 70.Damont A, Legrand A, Cao C, Fenaille F, Tabet J-C. Hydrogen/deuterium exchange mass spectrometry in the world of small molecules. Mass Spectrom Rev. 2023;42:1300–1331. doi: 10.1002/mas.21765. [DOI] [PubMed] [Google Scholar]
- 71.Cajka T, Hricko J, Rakusanova S, Brejchova K, Novakova M, Rudl Kulhava L, Hola V, et al. Hydrophilic interaction liquid chromatography-hydrogen/deuterium exchange-mass spectrometry (HILIC-HDX-MS) for untargeted metabolomics. Int J Mol Sci. 2024;25:2899. doi: 10.3390/ijms25052899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blaženović I, Kind T, Ji J, Fiehn O. Software tools and approaches for compound identification of LC-MS/MS data in metabolomics. Metabolites. 2018;8:31. doi: 10.3390/metabo8020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saccenti E, Hoefsloot HCJ, Smilde AK, Westerhuis JA, Hendriks MMWB. Reflections on univariate and multivariate analysis of metabolomics data. Metabolomics. 2014;10:361–374. doi: 10.1007/s11306-013-0598-6. [DOI] [Google Scholar]
- 74.Sedgwick P. Multiple significance tests: the Bonferroni correction. BMJ. 2012;344:e509. doi: 10.1136/bmj.e509. [DOI] [Google Scholar]
- 75.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 76.Storey JD. A direct approach to false discovery rates. J R Stat Soc Ser B Stat Method. 2002;64:479–498. doi: 10.1111/1467-9868.00346. [DOI] [Google Scholar]
- 77.Bartel J, Krumsiek J, Theis FJ. Statistical methods for the analysis of high-throughput metabolomics data. Comput Struct Biotechnol J. 2013;4:e201301009. doi: 10.5936/csbj.201301009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang L, Barrette M, Gauthier C, et al. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021;49:W388–W396. doi: 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen Y, Li EM, Xu LY. Guide to metabolomics analysis: a bioinformatics workflow. Metabolites. 2022;12:357. doi: 10.3390/metabo12040357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vondrackova M, Kopczynski D, Hoffmann N, Kuda O. LORA, Lipid Over-Representation Analysis based on structural information. Anal Chem. 2023;95:12600–12604. doi: 10.1021/acs.analchem.3c02039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Witting M. (Re-)use and (re-)analysis of publicly available metabolomics data. Proteomics. 2023;23:2300032. doi: 10.1002/pmic.202300032. [DOI] [PubMed] [Google Scholar]
- 83.Rus M, Crisan S, Andronie-Cioara FL, Indries M, Marian P, Pobirci OL, Ardelean AI. Prevalence and risk factors of metabolic syndrome: a prospective study on cardiovascular health. Medicina. 2023;59:1711. doi: 10.3390/medicina59101711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Madan K, Paliwal S, Sharma S, Kesar S, Chauhan N, Madan M. Metabolic syndrome: the constellation of co-morbidities, a global threat. Endocr Metab Immune Disord Drug. 2023;23:1491–1504. doi: 10.2174/1871530323666230309144825. [DOI] [PubMed] [Google Scholar]
- 85.Mensah GA, Fuster V, Murray CJL, Roth GA, Mensah GA, Abate YH, Abbasian M, et al. Global burden of cardiovascular diseases and risks, 1990–2022. J Am Coll Cardiol. 2023;82:2350–2473. doi: 10.1016/j.jacc.2023.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.World Health Organization. Cardiovascular diseases (CVDs) 2021. May 28, 2024. https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds)
- 87.Di Cesare M, Perel P, Taylor S, Kabudula C, Bixby H, Gaziano TA, McGhie DV, et al. The Heart of the World. Glob Heart. 2024;19:11. doi: 10.5334/gh.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eichelmann F, Sellem L, Wittenbecher C, Jäger S, Kuxhaus O, Prada M, Cuadrat R, et al. Deep lipidomics in human plasma: Cardiometabolic disease risk and effect of dietary fat modulation. Circulation. 2022;146:21–35. doi: 10.1161/CIRCULATIONAHA.121.056805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang Z, Klaric L, Krasauskaite J, Khalid W, Strachan MWJ, Wilson JF, Price JF. Combining serum metabolomic profiles with traditional risk factors improves 10-year cardiovascular risk prediction in people with type 2 diabetes. Eur J Prev Cardiol. 2023;30:1255–1262. doi: 10.1093/eurjpc/zwad160. [DOI] [PubMed] [Google Scholar]
- 90.Lu Q, Chen J, Li R, Wang Y, Tu Z, Geng T, Liu L, Pan A, Liu G. Healthy lifestyle, plasma metabolites, and risk of cardiovascular disease among individuals with diabetes. Atherosclerosis. 2023;367:48–55. doi: 10.1016/j.atherosclerosis.2022.12.008. [DOI] [PubMed] [Google Scholar]
- 91.Tzoulaki I, Castagné R, Boulangé CL, Karaman I, Chekmeneva E, Evangelou E, Ebbels TMD, et al. Serum metabolic signatures of coronary and carotid atherosclerosis and subsequent cardiovascular disease. Eur Heart J. 2019;40:2883–2896. doi: 10.1093/eurheartj/ehz235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Würtz P, Havulinna AS, Soininen P, Tynkkynen T, Prieto-Merino D, Tillin T, Ghorbani A, et al. Metabolite profiling and cardiovascular event risk: A prospective study of 3 population-based cohorts. Circulation. 2015;131:774–85. doi: 10.1161/CIRCULATIONAHA.114.013116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, DuGar B, Feldstein AE, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Benson TW, Conrad KA, Li XS, Wang Z, Helsley RN, Schugar RC, Coughlin TM, et al. Gut microbiota-derived trimethylamine N-oxide contributes to abdominal aortic aneurysm through inflammatory and apoptotic mechanisms. Circulation. 2023;147:1079–1096. doi: 10.1161/CIRCULATIONAHA.122.060573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tang WHW, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368:1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhu W, Gregory JC, Org E, Buffa JA, Gupta N, Wang Z, Li L, et al. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell. 2016;165:111–124. doi: 10.1016/j.cell.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li XS, Wang Z, Cajka T, Buffa JA, Nemet I, Hurd AG, Gu X, et al. Untargeted metabolomics identifies trimethyllysine, a TMAO-producing nutrient precursor, as a predictor of incident cardiovascular disease risk. JC Insight. 2018;3:e99096. doi: 10.1172/jci.insight.99096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nemet I, Saha PP, Gupta N, Zhu W, Romano KA, Skye SM, Cajka T, et al. A cardiovascular disease-linked gut microbial metabolite acts via adrenergic receptors Cell 2020180862–877..e22 10.1016/j.cell.2020.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ferrell M, Wang Z, Anderson JT, Li XS, Witkowski M, DiDonato JA, Hilser JR, et al. A terminal metabolite of niacin promotes vascular inflammation and contributes to cardiovascular disease risk. Nat Med. 2024;30:424–434. doi: 10.1038/s41591-023-02793-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Witkowski M, Nemet I, Alamri H, Wilcox J, Gupta N, Nimer N, Haghikia A, et al. The artificial sweetener erythritol and cardiovascular event risk. Nat Med. 2023;29:710–718. doi: 10.1038/s41591-023-02223-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Witkowski M, Nemet I, Li XS, Wilcox J, Ferrell M, Alamri H, Gupta N, et al. Xylitol is prothrombotic and associated with cardiovascular risk. Eur Heart J. 2024;45:2439–2452. doi: 10.1093/eurheartj/ehae244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fu Z, Liu Q, Liang J, Weng Z, Li W, Xu J, Zhang X, Xu C, Gu A. Association between NMR metabolomic signatures of healthy lifestyle and incident coronary artery disease. Eur J Prev Cardiol. 2022;30:243–253. doi: 10.1093/eurjpc/zwac252. [DOI] [PubMed] [Google Scholar]
- 103.Margara-Escudero HJ, Paz-Graniel I, García-Gavilán J, Ruiz-Canela M, Sun Q, Clish CB, Toledo E, et al. Plasma metabolite profile of legume consumption and future risk of type 2 diabetes and cardiovascular disease. Cardiovasc Diabetol. 2024;23:38. doi: 10.1186/s12933-023-02111-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Guasch-Ferré M, Hernández-Alonso P, Drouin-Chartier JP, Ruiz-Canela M, Razquin C, Toledo E, Li J, et al. Walnut consumption, plasma metabolomics, and risk of type 2 diabetes and cardiovascular disease. J Nutr. 2021;151:303–311. doi: 10.1093/jn/nxaa374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nemet I, Funabashi M, Li XS, Dwidar M, Sangwan N, Skye SM, Romano KA, et al. Microbe-derived uremic solutes enhance thrombosis potential in the host mBio 202314e0133123 10.1128/mbio.01331-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Harm T, Dittrich K, Brun A, Fu X, Frey M, Petersen Uribe A, Schwarz F-J, et al. Large-scale lipidomics profiling reveals characteristic lipid signatures associated with an increased cardiovascular risk. Clin Res Cardiol. 2023;112:1664–1678. doi: 10.1007/s00392-023-02260-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Seah JYH, Chew WS, Torta F, Khoo CM, Wenk MR, Herr DR, Choi H, Tai ES, van Dam RM. Plasma sphingolipids and risk of cardiovascular diseases: a large-scale lipidomic analysis. Metabolomics. 2020;16:89. doi: 10.1007/s11306-020-01709-8. [DOI] [PubMed] [Google Scholar]
- 108.Sun H, Saeedi P, Karuranga S, Pinkepank M, Ogurtsova K, Duncan BB, Stein C, et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res Clin Pract. 2022;183:109119. doi: 10.1016/j.diabres.2021.109119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gregory GA, Robinson TIG, Linklater SE, Wang F, Colagiuri S, de Beaufort C, Donaghue KC, et al. Global incidence, prevalence, and mortality of type 1 diabetes in 2021 with projection to 2040: a modelling study. Lancet Diabetes Endocrinol. 2022;10:741–760. doi: 10.1016/S2213-8587(22)00218-2. [DOI] [PubMed] [Google Scholar]
- 110.Mobasseri M, Shirmohammadi M, Amiri T, Vahed N, Hosseini Fard H, Ghojazadeh M. Prevalence and incidence of type 1 diabetes in the world: a systematic review and meta-analysis. Health Promot Perspect. 2020;10:98–115. doi: 10.34172/hpp.2020.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rakusanova S, Cajka T. Current analytical methods to monitor type 2 diabetes medication in biological samples. Trends Anal Chem. 2023;158:116831. doi: 10.1016/j.trac.2022.116831. [DOI] [Google Scholar]
- 112.World Health Organization. Diabetes. 2023. Apr 5, 2023. https://www.who.int/news-room/fact-sheets/detail/diabetes .
- 113.Sen P, Hyötyläinen T, Orešič M. 1-Deoxyceramides - Key players in lipotoxicity and progression to type 2 diabetes? Acta Physiol. 2021;232:e13635. doi: 10.1111/apha.13635. [DOI] [PubMed] [Google Scholar]
- 114.La Torre D, Seppänen-Laakso T, Larsson HE, Hyötyläinen T, Ivarsson SA, Lernmark Å, Orešič M Group atDS. Decreased cord-blood phospholipids in young age-at-onset type 1 diabetes. Diabetes. 2013;62:3951–3956. doi: 10.2337/db13-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tapia G, Suvitaival T, Ahonen L, Lund-Blix NA, Njølstad PR, Joner G, Skrivarhaug T, et al. Prediction of type 1 diabetes at birth: Cord blood metabolites vs genetic risk score in the Norwegian Mother, Father, and Child cohort. J Clin Endocrinol Metab. 2021;106:e4062–e4071. doi: 10.1210/clinem/dgab400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Webb-Robertson B-JM, Nakayasu ES, Frohnert BI, Bramer LM, Akers SM, Norris JM, Vehik K, et al. Integration of infant metabolite, genetic, and islet autoimmunity signatures to predict type 1 diabetes by age 6 years. J Clin Endocrinol Metab. 2022;107:2329–2338. doi: 10.1210/clinem/dgac225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee DH, Jin Q, Shi N, Wang F, Bever AM, Liang L, Hu FB, et al. The metabolic potential of inflammatory and insulinaemic dietary patterns and risk of type 2 diabetes. Diabetologia. 2024;67:88–101. doi: 10.1007/s00125-023-06021-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hannich JT, Loizides-Mangold U, Sinturel F, Harayama T, Vandereycken B, Saini C, Gosselin P, et al. Ether lipids, sphingolipids and toxic 1-deoxyceramides as hallmarks for lean and obese type 2 diabetic patients. Acta Physiol. 2021;232:e13610. doi: 10.1111/apha.13610. [DOI] [PubMed] [Google Scholar]
- 119.Prada M, Wittenbecher C, Eichelmann F, Wernitz A, Drouin-Chartier J-P, Schulze MB. Association of the odd-chain fatty acid content in lipid groups with type 2 diabetes risk: A targeted analysis of lipidomics data in the EPIC-Potsdam cohort. Clin Nutr. 2021;40:4988–4999. doi: 10.1016/j.clnu.2021.06.006. [DOI] [PubMed] [Google Scholar]
- 120.Sun L, Liang L, Gao X, Zhang H, Yao P, Hu Y, Ma Y, Wang F, Jin Q, Li H, et al. Early prediction of developing type 2 diabetes by plasma acylcarnitines: A population-based study. Diabetes Care. 2016;39:1563–1570. doi: 10.2337/dc16-0232. [DOI] [PubMed] [Google Scholar]
- 121.Thaker VV, Kwee LC, Chen H, Bahnson J, Ilkayeva O, Muehlbauer MJ, Wolfe B, et al. Metabolite signature of diabetes remission in individuals with obesity undergoing weight loss interventions. Obesity. 2024;32:304–314. doi: 10.1002/oby.23943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ahola-Olli AV, Mustelin L, Kalimeri M, Kettunen J, Jokelainen J, Auvinen J, Puukka K, et al. Circulating metabolites and the risk of type 2 diabetes: a prospective study of 11,896 young adults from four Finnish cohorts. Diabetologia. 2019;62:2298–2309. doi: 10.1007/s00125-019-05001-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Delgado-Velandia M, Gonzalez-Marrachelli V, Domingo-Relloso A, Galvez-Fernandez M, Grau-Perez M, Olmedo P, Galan I, et al. Healthy lifestyle, metabolomics and incident type 2 diabetes in a population-based cohort from Spain. Int J Behav Nutr Phys Act. 2022;19:8. doi: 10.1186/s12966-021-01219-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu J, Semiz S, van der Lee SJ, van der Spek A, Verhoeven A, van Klinken JB, Sijbrands E, et al. Metabolomics based markers predict type 2 diabetes in a 14-year follow-up study. Metabolomics. 2017;13:104. doi: 10.1007/s11306-017-1239-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Menni C, Fauman E, Erte I, Perry JR, Kastenmüller G, Shin SY, Petersen AK, et al. Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes. 2013;62:4270–4276. doi: 10.2337/db13-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Peddinti G, Cobb J, Yengo L, Froguel P, Kravić J, Balkau B, Tuomi T, Aittokallio T, Groop L. Early metabolic markers identify potential targets for the prevention of type 2 diabetes. Diabetologia. 2017;60:1740–1750. doi: 10.1007/s00125-017-4325-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rebholz CM, Yu B, Zheng Z, Chang P, Tin A, Köttgen A, Wagenknecht LE, Coresh J, Boerwinkle E, Selvin E. Serum metabolomic profile of incident diabetes. Diabetologia. 2018;61:1046–1054. doi: 10.1007/s00125-018-4573-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ferrannini E, Natali A, Camastra S, Nannipieri M, Mari A, Adam KP, Milburn MV, et al. Early metabolic markers of the development of dysglycemia and type 2 diabetes and their physiological significance. Diabetes. 2013;62:1730–1737. doi: 10.2337/db12-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lemaitre RN, Jensen PN, Wang Z, Fretts AM, McKnight B, Nemet I, Biggs ML, Sotoodehnia N, de Oliveira Otto MC, Psaty BM, et al. Association of Trimethylamine N-Oxide and Related Metabolites in Plasma and Incident Type 2 Diabetes: The Cardiovascular Health Study. JAMA Netw Open. 2021;4:e2122844–e2122844. doi: 10.1001/jamanetworkopen.2021.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Song R, Li Z, Zhang Y, Tan J, Chen Z. Comparison of NAFLD, MAFLD and MASLD characteristics and mortality outcomes in United States adults. Liver Int. 2024;44:1051–1060. doi: 10.1111/liv.15856. [DOI] [PubMed] [Google Scholar]
- 131.Eslam M, Newsome PN, Sarin SK, Anstee QM, Targher G, Romero-Gomez M, Zelber-Sagi S, et al. A new definition for metabolic dysfunction-associated fatty liver disease: An international expert consensus statement. J Hepatol. 2020;73:202–209. doi: 10.1016/j.jhep.2020.03.039. [DOI] [PubMed] [Google Scholar]
- 132.Rinella ME, Lazarus JV, Ratziu V, Francque SM, Sanyal AJ, Kanwal F, Romero D, et al. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. Hepatology. 2023;78:1966–1986. doi: 10.1097/HEP.0000000000000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ramírez-Mejía MM, Jiménez-Gutiérrez C, Eslam M, George J, Méndez-Sánchez N. Breaking new ground: MASLD vs. MAFLD-which holds the key for risk stratification? Hepatol Int. 2024;18:168–178. doi: 10.1007/s12072-023-10620-y. [DOI] [PubMed] [Google Scholar]
- 134.Chen L, Tao X, Zeng M, Mi Y, Xu L. Clinical and histological features under different nomenclatures of fatty liver disease: NAFLD, MAFLD, MASLD and MetALD. J Hepatol. 2024;80:e64–e66. doi: 10.1016/j.jhep.2023.08.021. [DOI] [PubMed] [Google Scholar]
- 135.De A, Bhagat N, Mehta M, Taneja S, Duseja A. Metabolic dysfunction-associated steatotic liver disease (MASLD) definition is better than MAFLD criteria for lean patients with NAFLD. J Hepatol. 2024;80:e61–e62. doi: 10.1016/j.jhep.2023.07.031. [DOI] [PubMed] [Google Scholar]
- 136.Younossi ZM, Golabi P, Paik JM, Henry A, Van Dongen C, Henry L. The global epidemiology of nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH): a systematic review. Hepatology. 2023;77:1335–1347. doi: 10.1097/HEP.0000000000000004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang D, Xu Y, Zhu Z, Li Y, Li X, Li Y, Shen H, et al. Changes in the global, regional, and national burdens of NAFLD from 1990 to 2019: A systematic analysis of the global burden of disease study 2019. Front Nutr. 2022;9:1047129. doi: 10.3389/fnut.2022.1047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Miao L, Targher G, Byrne CD, Cao YY, Zheng MH. Current status and future trends of the global burden of MASLD. Trends Endocrinol Metab. 2024 doi: 10.1016/j.tem.2024.02.007. S1043–2760(24)00036–5. [DOI] [PubMed] [Google Scholar]
- 139.Gagnon E, Manikpurage HD, Mitchell PL, Girard A, Gobeil É, Bourgault J, Bégin F, et al. Large-scale metabolomic profiling and incident non-alcoholic fatty liver disease. iScience. 2023;26:107127. doi: 10.1016/j.isci.2023.107127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gobeil É, Maltais-Payette I, Taba N, Brière F, Ghodsian N, Abner E, Bourgault J, et al. Mendelian randomization analysis identifies blood tyrosine levels as a biomarker of non-alcoholic fatty liver disease. Metabolites. 2022;12:440. doi: 10.3390/metabo12050440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hartley A, Santos Ferreira DL, Anderson EL, Lawlor DA. Metabolic profiling of adolescent non-alcoholic fatty liver disease [version 2; peer review: 2 approved] Wellcome Open Res. 2019;3:166. doi: 10.12688/wellcomeopenres.14974.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hirata A, Harada S, Iida M, Kurihara A, Fukai K, Kuwabara K, Kato S, et al. Association of Nonalcoholic Fatty Liver Disease with Arterial Stiffness and its Metabolomic Profiling in Japanese Community-Dwellers. J Atheroscler Thromb. 2024;31:1–17. doi: 10.5551/jat.64616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Khusial R, Cioffi C, Caltharp S, Krasinskas A, Alazraki A, Knight-Scott J, Cleeton R, et al. Development of a plasma screening panel for pediatric nonalcoholic fatty liver disease using metabolomics. Hepatol Commun. 2019;3:1311–1321. doi: 10.1002/hep4.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Martínez-Arranz I, Bruzzone C, Noureddin M, Gil-Redondo R, Mincholé I, Bizkarguenaga M, Arretxe E, et al. Metabolic subtypes of patients with NAFLD exhibit distinctive cardiovascular risk profiles. Hepatology. 2022;76:1121–1134. doi: 10.1002/hep.32427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.McGlinchey AJ, Govaere O, Geng D, Ratziu V, Allison M, Bousier J, Petta S, et al. Metabolic signatures across the full spectrum of non-alcoholic fatty liver disease. JHEP Rep. 2022;4:100477. doi: 10.1016/j.jhepr.2022.100477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hu X-Y, Li Y, Li L-Q, Zheng Y, Lv J-H, Huang S-C, Zhang W, Liu L, Zhao L, Liu Z, et al. Risk factors and biomarkers of non-alcoholic fatty liver disease: an observational cross-sectional population survey. BMJ Open. 2018;8:e019974. doi: 10.1136/bmjopen-2017-019974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kimberly WT, O’Sullivan JF, Nath AK, Keyes M, Shi X, Larson MG, Yang Q, et al. Metabolite profiling identifies anandamide as a biomarker of nonalcoholic steatohepatitis. JCI Insight. 2017;2:e92989. doi: 10.1172/jci.insight.92989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zeng F, Su X, Liang X, Liao M, Zhong H, Xu J, Gou W, Zhang X, Shen L, Zheng J-S, et al. Gut microbiome features and metabolites in non-alcoholic fatty liver disease among community-dwelling middle-aged and older adults. BMC Med. 2024;22:104. doi: 10.1186/s12916-024-03317-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.