Summary
Mitochondria (mt) represent the vital hub of the molecular physiology of the cell, being decision-makers in cell life/death and information signaling, including major redox regulations and redox signaling. Now we review recent advances in understanding mitochondrial redox homeostasis, including superoxide sources and H2O2 consumers, i.e., antioxidant mechanisms, as well as exemplar situations of physiological redox signaling, including the intramitochondrial one and mt-to-cytosol redox signals, which may be classified as acute and long-term signals. This review exemplifies the acute redox signals in hypoxic cell adaptation and upon insulin secretion in pancreatic β-cells. We also show how metabolic changes under these circumstances are linked to mitochondrial cristae narrowing at higher intensity of ATP synthesis. Also, we will discuss major redox buffers, namely the peroxiredoxin system, which may also promote redox signaling. We will point out that pathological thresholds exist, specific for each cell type, above which the superoxide sources exceed regular antioxidant capacity and the concomitant harmful processes of oxidative stress subsequently initiate etiology of numerous diseases. The redox signaling may be impaired when sunk in such excessive pro-oxidative state.
Keywords: Mitochondrial superoxide formation, Redox regulations, Redox signaling, Pancreatic β-cells, β-oxidation, Peroxiredoxins
Mitochondrial reactive oxygen species (ROS) sources
Primary sources of mitochondrial superoxide
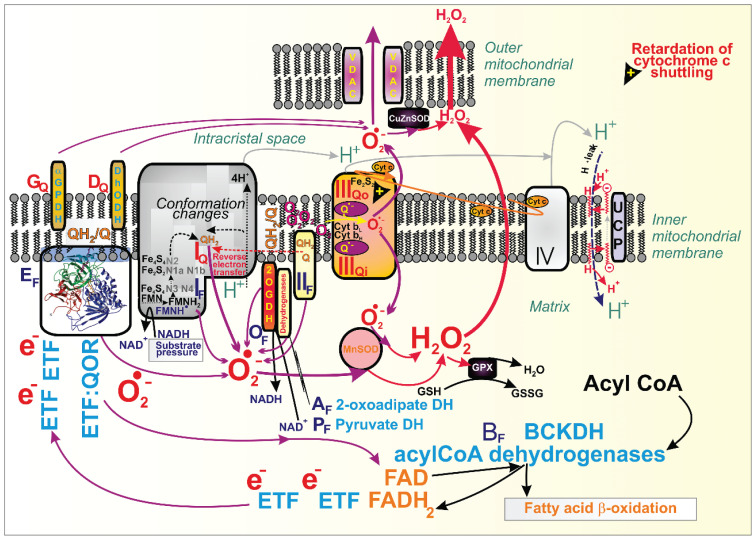
According to the classification of M. Brand [1–4], flavin (F) and ubiquinone (Q) containing binding sites, typically within structures of respiratory chain (RC) complexes, belong to the most critical superoxide formation sites in mitochondria [5–7], besides particular loci of dehydrogenases [1,2] (Fig. 1). Recent progress in understanding mechanisms involved in proton-coupled electron transfer via the RC and in resolving supercomplexes formation and single crista architecture [7–9] then calls for reconsiderations of these mechanisms and more precise determination of superoxide formation sites within the given and already resolved protein structures.
Fig. 1.
Sites of superoxide formation in mitochondria. Schema depicts locations for the identified sites of superoxide formation, acting at the ~280 mV redox potential of the NADH/NAD+ iso-potential pool (index F, flavin; dark blue capitalized fonts) and sites acting at the ~20 mV redox potential of the ubiquinol/ubiquinone (QH2/Q) iso-potential pool (index Q; red capitalized fonts), according to the Brand’s nomenclature introduced by Martin Brand [1]. Thus the major sites IF, IQ and IIIQo exists for Complex I and III, respectively; at certain circumstances also Complex II/succinate dehydrogenase forms superoxide at site IIF; and sites for probable superoxide formation by dehydrogenases (DH) are shown (GQ for α-glycerolphosphate dehydrogenase, GPDH; DQ for dihydroorotate DH; AF for 2-oxoadipated DH; BF for branched-chain ketaoacid DH, BCKDH; OF for 2-oxoglutarate DH, 2OGDH; PF for pyruvate DH. In case of acylCoA dehydrogenases acting in fatty acid β-oxidation, two electron transfer flavoproteins (ETF) are required to transfer electrons to the membrane-attached ETF:ubiquionone oxidoraductase (ETF:QOR, depicted by its structure) containing a site EF (though a site EQ also potentially exists). The scheme also depicts a situation when retardation fo cytochrome c shuttling induces superoxide formation on site IIIQo; as well as attenuation of superoxide formation by uncoupling proteins (UCP) based on the fatty acid-cycling mechanism [119]. Finally, formed superoxide is converted to H2O2 by MnSOD in the matrix or by CuZnSOD within the intermembrane space. H2O2 may readily penetrate to the cell cytosol (for special relations of such diffusion, see Reference [7].
A general requirement for superoxide (O2•−; and its conjugated acid – hydroperoxyl radical, HO2•, pKa 4.9) to be formed is a local retardation of the electron transfer or an enzyme reaction process so that intermediate radicals have enough lifetime to react with oxygen. These intermediates are typically semiquinone anion radical (Q•−; or semiquinol QH•) for Q sites and flavosemiquinone radical FMNH• for F sites. Thus, the flavin site on Complex I (termed IF) can produce superoxide at a higher NADH/NAD+ ratio after the direct H− transfer between NADH and FMN [10]. When an excessive electron cannot pass through the existing FeS chain within the Complex I matrix arm, the NAD+ binding is interrupted, and the pairing of FMNH− and NADH form FMNH•. At lower NADH, indeed, NAD+ can pair with FMNH•, and superoxide cannot be formed [11].
Complex I, as an H+-pumping NADH:quinone oxidoreductase, possesses a Q-tunnel structure, where an ongoing inhibition by a product (ubiquinol, QH2) can form superoxide at the phenomenologically defined site IQ [1] (Fig. 1). This typically occurs when the whole Complex I runs backward during so-called reversed electron transfer (RET). Disputes still exist whether under conditions of e.g. reperfusion after ischemic accumulation of succinate, superoxide is formed at IQ or IF site [12,13]. Due to the suppression of the electron leak to oxygen at site IQ by a specific antioxidant S1QEL, the site IQ is more plausible to act in RET-derived superoxide formation [1].
We have also revealed that the maximum superoxide was formed only when electron transport and H+ pumping were retarded [14,15]. H+ pumping may be attenuated by a high electrochemical gradient of protons established at the inner mitochondrial membrane (IMM), termed protonmotive force, Δp (when expressed in mV units) [16–18]. In pathologies this can be induced by mutations of the ND5 subunit (or other mitochondrion-encoded subunits) of the Complex I membrane arm.
Complex III, a ubiquinol-cytochrome c reductase, contributes to O2•− generation by autooxidation of the semiquinone anion radical (Q•−) within the so-called Q cycle [1,5,7,19,20], while it releases O2•− about equally to both sides of IMM [20,21]. Typically, when cytochrome c turnover is delayed for some reason, then a feedback inhibition of the Q-cycle within CIII is induced, causing the superoxide formation at the Complex III site, termed IIIQo („o“ for outer, which is located in proximity to the intracristal space, [1]) (Fig. 1). This is because of the increased lifetime of QH• and oxygen diffusion into this site [22]. In vivo, a physiological delay of the Q-cycle occurs at hypoxia or pathological one with specific mutations in Complex IV [23]. Retardation of the cytochrome c cycling automatically exists at the escape of cytochrome c from the cristae lumen during initiation of mitochondria-related apoptosis.
Also, Complex II (succinate dehydrogenase, SDH) may form superoxide under specific conditions but not at high succinate concentrations [24,25]. But superoxide is formed when the flavin site IIF within the SDHA subunit is less occupied, such as when succinate concentrations approach to Km of 100–500 μM [26–28]. Pathologically, with the blocked SDHD subunit and hence interrupted electron transfer to Q, Complex II/SDH produces H2O2 (with a 70 % capacity) directly due to the ability of existing three FeS clusters to provide two-electron transfer to oxygen [29]. The 3Fe-4S cluster may also theoretically provide superoxide [30].
Evidence was also reported for superoxide formation within the dehydrogenase (DH) complexes in isolated mitochondria when excessive particular substrates for given DHs were used. Hence with excessive 2-oxoglutarate (2OG, 2-ketoglutarate) for OGDH, pyruvate for pyruvate dehydrogenase (PDH) and substrates of branched-chain 2-ketoacid (2-oxoacid) dehydrogenase (BCKDH) superoxide/H2O2 formation in skeletal muscle mitochondria was eightfold, fourfold, and twofold higher, respectively, than that one ascribed to the site IF [31]. Phenomenological sites were termed as site OF, PF, and BF, respectively, but mechanisms and occurrence in vivo must be further investigated.
Also, isolated mitochondria respiring with glycerol-3-phosphate partly produced superoxide at site GQ of the glycerol-3-phosphate dehydrogenase [4,26, 32–34]. Analogously, dihydroorotate dehydrogenase was reported to form superoxide at side DQ [1,4,33,35]. Moreover, ongoing fatty acid (FA) β-oxidation also produces superoxide. Its portion may originate from the site EF of the electron-transferring flavoprotein – ubiquinone oxido reductase (ETFQOR) [27] (Fig. 1).
Superoxide dismutation into H2O2
Manganese superoxide dismutase (MnSOD or SOD2) is localized in the mitochondrial matrix, whereas CuZnSOD (SOD1) localizes to the mitochondrial intermembrane space, besides residing in the cell cytosol. MnSOD dismutates the majority of superoxide released to the matrix into H2O2. It is not known whether CuZnSOD is exclusively located between the outer mitochondrial membrane (OMM) and the inner boundary membrane (IBM, the unfolded part of IMM) in the so-called intermembrane space peripheral (IMSp), or whether in also resides in the intracristal space (ICS). In the latter case, it could more effectively convert superoxide therein [36], namely the part released from the site IIIQo.
MnSOD activity was found to be regulated. Rather fast posttranslational modifications (PTMs) were reported for NAD+-dependent sirtuin-3- (SIRT3−) mediated deacetylation of MnSOD, activating the enzyme [37–40]. These results and derived conclusions need to be validated since not always a large population of MnSOD molecules in the matrix is acetylated/deacetylated. Only a fraction is affected, therefore, one should expect that this particular MnSOD molecule fraction is activated and, for example, only the resulting fraction of H2O2 may thus participate in intramitochondrial redox signaling or a weak redox signal directed to the cytosol. MnSOD regulations rather proceeded within an hour-time frame and could be regarded as chronic regulations [41–45]. Thus a subtle change in H2O2 release, accumulated during sufficient time, can be effective.
Ultrastructure of mitochondrion vs. H2O2 diffusion to the cell cytosol
We have published several reviews on how mitochondrial morphology and ultrastructure affect the diffusion of H2O2 into the cell cytosol and/or other organelles and up to the plasma membrane or even diffusion into the extracellular space [5–9,46–48]. Hence, let’s briefly summarize 3D architecture of mitochondria (Fig. 2). Actually the plural is adequate for isolated fragments of the original mitochondrial network, the mitochondrion [6–8,49–51]. Note that such a network also exists in skeletal muscle and the heart [52,53]. Small fragments are constantly separated from the main mitochondrial network by fission machinery (Fig. 2E), while at the same time, fragments join the main network by fusion (Fig. 2D), which is aided by the pro-fusion proteins [54,55]. This process is important, since mitochondrial-specific autophagy, termed mitophagy, eliminates those fragments that do not possess a sufficient IMM membrane potential (or Δp) as a result of local predominance of the mutated mt-DNA-encoded RC and ATP synthase subunits.
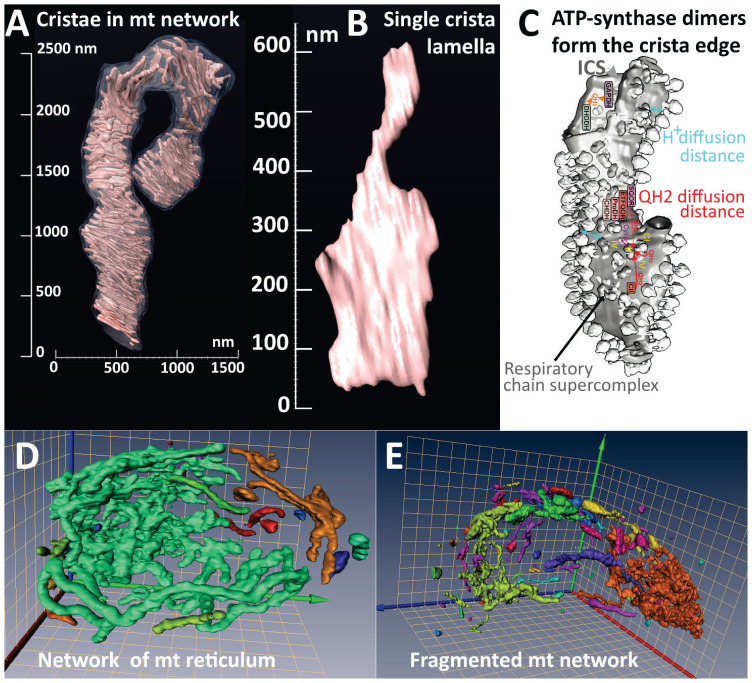
Fig. 2.
Cristae in mitochondrial network of pancreatic islet β-cells. (A) An exemplar 3D image of crista lamellae within a 4-μm segment of the mitochondrial tubule, obtained by the focused ion beam/scanning electron microscopy (FIB/SEM); (B) detail of the single crista lamella from A). (C) Comparison with the 3D image of a single crista with resolved ATP-synthase dimers at the lamella edge, adapted from Ref. [67]. Also, structures of respiratory chain supercomplexes are visible on a lamella flank, which represents the crista membrane lipid bilayer leaflet oriented toward the matrix. The distances are marked for a minimum path of proton diffusion (mild blue arrows), providing a substantial coupling between the respiratory chain proton pumping and the ATP-synthase. The purple arrow indicates a shuttling of cytochrome c at the supercomplex surface. The distances are also marked for a short ubiquinol QH2 (or ubiquionone Q) diffusion between Complex I (CI) and Complex III (CIII) around supercomplexes (red arrow) and a much longer diffusion path from Complex II (red arrow) to CIII or from oxidoreductases and dehydrogenases to CIII (dashed red arrows). Inside the broken portion of crista lamella at the inner (intracristal space) surface, a QH2-diffusion path is indicated by orange arrows. This path must be followed by the flip across the membrane to CIII. (D, E) Mitochondrial reticular network in pancreatic islet β-cells of Wistar (D) and diabetic Goto-Kakizaki rats (E) in 3D images adopted from Ref. [49]. Note the nearly continuous mitochondrial network in intact β-cells (D), but the fragmented network in diabetic β-cells (E).
The mitochondrial tubular network possesses a complex ultrastructural organization of mt cristae, i.e., rich invaginations of IMM from the IBM, which shrink or inflate according to the metabolic performance or other reasons [7] and might exhibit dynamics in a short time scale [56]. To understand mitochondrial compartments, one must recognize their three-dimensional (3D) architecture. The cristae form rather lamellae with bottleneck connections to the IBM, where ICS meets IMSp (Fig. 2A, B). The mitochondrial cristae organization system (MICOS) complex is attached to the OMM SAM complex and thus forms crista junctions (CJs) [57–59] around the crista outlets [7,60]. Note that at the edges of single crista lamella, the ATP-synthase dimers form rows or arrays [61–67] (Fig. 2C), the dynamic of which may also affect the cristae morphology [68–70]. Small MICOS subunits may intercalate between ATP-synthase dimers, such as Mic10, bound to the ATP-synthase membrane subunit e [71]; or Mic27 [72].
RC supercomplexes (typically CI CIII2 CIV1 [73–75] then reside at flanks of crista lamellae [61,67] (Fig. 2C). Just below CJs (crista outlets) other cristae-shaping proteins reside within cristae membranes (CM) facing ICS (crista lumen), such as various oligomers of OPA1 [76,77] or even filaments of OPA1 ortholog MGM1 [78]; and scaffolding proteins prohibitins, forming hetero-oligomeric 20–27 nm rings [79]. Positive curvature of 90° bends of the crista outlets, when IBM meets CM, is provided by oligomers of MICOS subunit MIC10 [80,81], while the negative curvature of crista lamellae is established by FAM92A1 protein, which binds cardiolipin and phosphatidylinositol 4,5-bisphosphate [82].
The rich lamellar cristae organization affects H2O2 diffusion into the cytosol [6]. If even H2O2 released into ICS (crista lumen) diffuses across the crista membrane, it will still reach the mt matrix in ~99 % of CM surface. Only at the proximity of CJs the ICS-located H2O2 might escape into the IMSp, across CM or IBM or through the crista outlet; and subsequently to the cytosol via the OMM. So, taking into account the mitochondrion ultrastructure, we see the limitations of H2O2 diffusion from ICS to the cytosol. On the contrary, such H2O2 diffusion is allowed upon initiation of mt-related apoptosis, when CJs (or crista outlets) are widened or broken, and this is accelerated by the cytochrome c escape [76,77,83] and concomitant increased superoxide formation.
Nevertheless, cristae lamellae inflate physiologically under hypoxic conditions due to partial losses of MIC60/mitofilin subunit of MICOS complex [60] or in pancreatic β-cells at low glucose (insulin non-stimulating) [46]. On the contrary, with a sudden excess of respiration substrate, the inflated cristae shrink. Such a narrowing of cristae was observed after dimethyl-2-oxoglutarate addition to hypoxia-preadapted HEPG2 cells [63] or upon glucose-stimulated insulin secretion (GSIS) in pancreatic β-cells, i.e., when high glucose was set [46].
Mitochondrial redox buffers and/or anti-oxidant systems
Mitochondrial glutathione reductase & glutathione peroxidase system
Redox buffers and antioxidant enzymes detoxify the produced ROS and may exert specific roles in redox signaling. Similarly to the cell cytosol, in addition to small antioxidant molecules such as vitamin E (α-tocopherol), ascorbate, and uric acid, enzyme systems of glutathione peroxidase (GPX) and peroxiredoxin represent the most critical intracellular antioxidants and a primary defense system. Catalase is absent in mitochondria except in the heart [5,9].
Glutathione is present in a reduced (GSH) or oxidized (GSSG, glutathione disulfide) form. Glutathione reductase (GRX; EC 1.8.1.7) catalyzes the NADPH-dependent reduction of GSSG to GSH [84], and hence oxidized glutathione is regenerated. Glutathione provides/GRX a major mt matrix redox buffer in numerous cells [85]. On the contrary, pancreatic β-cells exhibit a less abundant glutathione/GRX system [6,18,86–88].
GSH is also a cofactor of enzymes of the glutathione peroxidase (GPX) family. These enzymes reduce H2O2 to water, and some isoforms (e.g. GPX4) also reduce lipid hydroperoxides to their corresponding alcohols. The GPX family contains five enzymes with seleno-cysteine active sites (GPX1 to 4, and GPX6) and three other enzymes, acting as redox sensors (GPX5, GPX7, GPX8) [89,90]. The latter possess cysteine residues in their active sites and modest peroxidase activity [91]. The cytosolic and mt-residing GPX1 and plasma membrane and cytosolic GPX4 are abundant in all tissues and cell types. GPX1, GPX2, and GPX3 are homotetrameric proteins. GPX4 has a monomeric structure.
Mitochondrial vs. cytosolic peroxiredoxin system
Peroxiredoxins (PRDX) are hydroperoxide reductases, either of the 2-Cys type (cytosolic peroxiredoxins PRDX1 and PRDX2 (Fig. 3); PRDX3 residing in the mt matrix; and PRDX4 of the endoplasmic reticulum) or 1-Cys type (PRDX6) [92–98]. The second mitochondrial PRDX, PRDX5, also contains two cysteines in the monomer [99] but allows an atypical mechanism, while forming the intra-subunit S-S bridge within the single monomeric subunit [92–96,98]. However, PRDX5 is located also in the cytosol and peroxisomes and prefers lipid peroxides and peroxynitrite over H2O2. Artificial PRDX5 expression in IMSp attenuated hypoxic transcriptome reprogramming [100] and cancerogenesis [101].
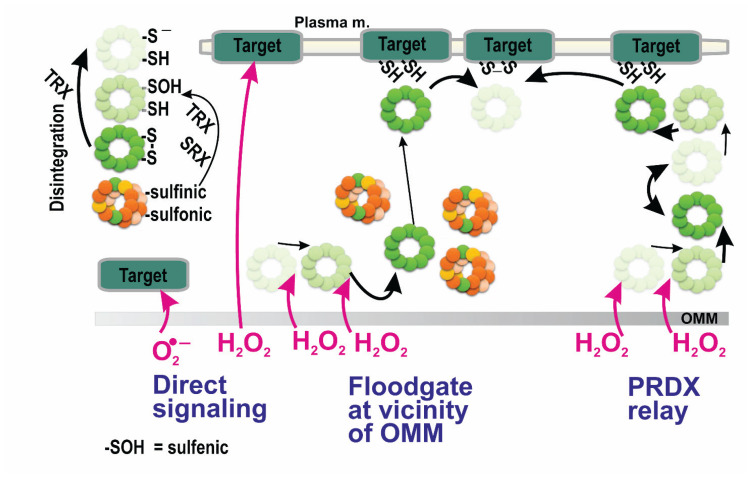
Fig. 3.
Possible modes of peroxiredoxin participation in redox signaling. Possible ways of redox signal spreading from vicinity of the outer mitochondrial membrane (OMM) to the plasma membrane – from left to right: i) direct superoxide diffusion (range only in OMM proximity); ii) direct H2O2 diffusion; iii) peroxiredoxin-mediated redox signal transfer, including diffusion of peroxiredoxin decamers allowed by the flood-gate model mechanism; iv) hypothetical redox relay via an array of peroxiredoxins. Note, that according to the flood-gate model, H2O2 oxidizes PRDX to higher states than a sulfenic state, allowing distant decamers in a S-S state to migrate to the target and exchange the two target sulfhydryls for PRDX S-S bridge. In (iv) the target is the PRDX itself. Left inset: color coding of PRDX monomers: green – PRDX S-S bridge; the very light green – basic reduced state (sulfhydryl and thiolate anionic form); the light green – the first-degree of oxidation, i.e., sulfenic state; yellow – oxidation into the second degree, i.e., sulfinic state; orange – oxidation into the third degree, irreversible, sulfonic state. Note, thioredoxin (TRX) converts either disassembled S-S state dodecamers to the basic sate; and, together with sulfiredoxin (SRX), TRX regenerates PRDX in sulfinic state into the sulfenyl state.
PRDX6 is a 1-cys-PRDX, which can also be recruited to mitochondria (probably to OMM) [102–104]. PRDX6 forms only homodimers, cannot form disulfide bonds, and is not reduced by sulfinyl reductase (SRX). PRDX6 reduces oxidized phospholipids. The sulfenic moiety of PRDX6 is subsequently reduced with GSH/GRX system but not with thioredoxins. PRDX6 also exerts Ca2+-independent phospholipase A2 activity.
Cytosolic peroxiredoxins are decameric, containing five homodimers. Mitochondrial PRDX3 is dodecameric, consisting of six homodimers. Thus, PRDX1, PRDX2, and PRDX3 form a toroid (doughnut-like) structure of five (six) homodimers, which can split from the toroid in an unstable disulfide conformation (see below). Such mechanism allows disulfide regeneration.
Peroxiredoxin catalytic cycle
PRDX monomers of the 2-Cys type contain the peroxidatic cysteine, CP and the resolving cysteine, CR. After reaction with H2O2, the peroxidatic cysteine CP of the first monomer within a homodimer forms an inter-subunit disulfide bond with the resolving cysteine CR of the second homodimer subunit [92–98]. As an intermediate, sulfenic acid (R-SOH) is first formed by two-electron reversible oxidation of the CP. Sub-sequently, the disulfide (S-S bridge) between CP and CR is formed. Interestingly, the PRDX ring is destabilized when such disulfide bonds are formed [92,94], thus allowing homodimers (monomers) to interact with their regenerating enzyme systems, completing the cycle. Such regeneration is catalyzed either by a couple of thioredoxin (TRX) plus NADPH-dependent TRX reductase (TRXR) [105] or by glutathione (GSH)/glutaredoxin (GRX) [92–98]. The disulfide bonds of homodimers are thus converted back to two cysteines.
PRDXs react with H2O2 faster than other peroxidases (catalases and GPX); hence they outcompete them and serve as the primary regulators of cytosolic H2O2 and in specific tissues also of the mt matrix H2O2. The latter is valid for pancreatic β-cells. Therefore PRDXs have been considered major players in cancerogenesis [93,97] and are promising targets for therapies of cardiovascular [106] or neurodegenerative diseases [107] and for defense against oxidative stress in pancreatic β-cells [108].
Peroxiredoxins enable redox signaling
There are two other unique PRDX properties, making them essential players in the redox homeostasis regulations and even redox signaling. The first such property is the formation of stacks of decamers/dodecamers, thus establishing high molecular weight complexes (HMW), which can even form filaments with chaperone function [109]. Since this formation happens only with PRDXs oxidized into higher oxidation state (sulfinyl and sulfonyl), the HMW formation effectively withdraws PRDX molecules from their entire population. This instantly leads to a higher local H2O2 concentration in the HMW loci.
The second property lies in the specific interaction of PRDXs with other proteins containing the two proximal cysteines, which enables their direct redox regulation (targeting the redox signal). The PRDX disulfides (S-S bridges) oxidize those proximal cysteines of the target protein into the S-S bridge between them, while PRDX homodimer become reduced back into two cysteines, CP and CR.
Indeed, when sulfenyls of PRDX1,2 and PRDX3 are oxidized into higher oxidized states, i.e., sulfinyls or sulfonyls, HMW complexes are formed [109]. The sulfinyls within HMW complexes or filaments can still be reduced by ATP-dependent sulfinyl reductase (SRX) enzymes [109,110]. However, hyperoxidation into sulfonyls is irreversible and can be regarded as a sign of oxidative stress. Mitochondrial PRDX3 underlies hyperoxidation about twice as slower when compared to PRDX2 [109].
In summary, the redox signal can be initiated by peroxiredoxins, by two mechanisms, which can even be considered as the two different interpretations of the same phenomenon. So we can point out that the formation of HMW complexes withdraws PRDX molecule from the catalytic cycle reaction, otherwise consuming H2O2. Despite that, the prerequisite for such a withdrawal is oxidation into sulfinyls or sulfonyls. Due to their formation, still consuming H2O2, the local PRDX molecules can no longer react with the local H2O2 after a certain time period. Hence, if such a locus still contains the H2O2 source, the local H2O2 concentration is elevated.
An alternative interpretation considers the so-called floodgate model (Fig. 3). This model has been predicted to describe the shift of the oxidation from the original to distant locations [6,9,111,112]. According to the floodgate model, the HMWs formed in the original locations upon a sustained H2O2 flux allow oxidation in the distant loci, proximal to target proteins, enabling execution of the redox signal. This can happen simply by the direct interaction of H2O2 with target proteins or via oxidation of distant PRDX molecules, which subsequently oxidize proximal cysteines in the target protein. The latter mechanism can be regarded literally as a “redox kiss”. Cytosolic peroxiredoxins convey their oxidation by H2O2 to the terminal target proteins (Fig. 3), typically phosphatases or transcription factors [92,94,113, 114–118]. It still remains to be investigated whether mitochondrial PRDX3 exhibits its own specific mitochondrial targets.
Mitochondrial redox signaling enabled by peroxiredoxins
However, sulfinyls in PRDX decamers/dodecamers or HMW complexes can still be slowly reduced back to cysteines after their reduction by the SRX system. Mitochondrial SRXs were reported to act even in the transfer of circadian rhythms to the mt matrix. This is enabled by periodically enhanced SRX expression intermittent with enhanced SRX degradation by LON-protease under control of the clock genes in the adrenal gland, brown adipose tissue and heart [109,119,120]. It should be further investigated whether such elegant circadian regulation of the mitochondrial matrix redox homeostasis exists in pancreatic β-cells.
Mild uncoupling attenuates mitochondrial ROS generation at intact mtDNA
Oxidative phosphorylation (OXPHOS) represents an ATP synthesis by the mt ATP-synthase (Complex V), which is driven by the protonmotive force, Δp. Δp is formed by the respiratory chain H+ pumping at Complex I, III, and IV [7,16–18,119]. The IMM domain (membrane domain) of the ATP-synthase (FOATPase) consumes an adequate Δp portion in a state, historically termed state-3, for isolated mitochondria with an ADP excess. In vivo, cellular respiration is governed by the metabolic state and/or availability of substrates. Hence, a finely tuned spectrum of various states-3 can be established, depending on the substrate load (e.g., increasing glucose). A state-4, is then given by zero ATP synthesis, when zero H+ backflux via the FOATPase exists, while respiration and H+ pumping are given by so-called H+ leak, mediated by mitochondrial carrier proteins, as their side-function and by the native H+ permeability of IMM. Since Δp exists predominantly in the form of Ψm (IMM electrical potential), Ψm is maximum at state-4 with the maximum substrate load.
Besides other proteins, such as the ADP/ATP carrier, Δp dissipation by a protonophoric short-circuit, termed uncoupling, can be physiologically provided by mitochondrial uncoupling proteins (UCPs) [119], frequently in synergy with mitochondrial phospholipases cleaving nascent fatty acids [120–123]. A mild uncoupling exists when carrier-mediated protonophore activity plus the native IMM H+ leak do not overwhelm the FOATPase protonophoric activity, and hence, ATP synthesis still takes place. This contrasts to a complete uncoupling when Δp approaches zero, such as established by agents termed uncouplers. The mild uncoupling is able to decrease mitochondrial O2•− formation at Complex I [62,63] and Complex III [124]. In cell types where such mitochondrial ROS source predominates, even redox homeostasis in the cytosol may be more pro-oxidant. However, oxidative stress originating from irreversible changes, such as stress due to mutated subunits encoded by mitochondrial DNA (mtDNA), cannot be counteracted by mild uncoupling [62].
Previously, an antioxidant role for UCP2 has been demonstrated in vivo [123,125,126]. For example, Duval et al. [127] have shown that UCP2-mediated uncoupling in endothelial cells is able to decrease extracellular ROS in co-incubated low-density-lipoproteins (LDL). Mice with deleted LDL receptors exhibited extensive diet-induced atherosclerotic plaques when they received bone marrow transplanted from UCP2 (−/−) mice, and the appearance of these plaques was prevented when they received bone marrow transplants from UCP2 (+/+) mice [128]. We have also demonstrated that UCP2 function suppresses mitochondrial superoxide production in vitro [121,123, 129,130].
Physiological redox signaling vs. oxidative stress
Oxidative stress
In principle, there exists no net oxidative stress without the other consequences, such as proteinaceous stress due to the disrupted turnover of intact proteins, concomitantly impaired autophagy and/or mitochondria-specific authophagy, i.e., mitophagy, or without the endoplasmic reticulum stress. Oxidative stress cannot be separated from the possible initiation of apoptosis, ferroptosis, or other forms of the cell death, as well as from impaired mitochondrial biogenesis. Moreover, all these phenomena are projected to an abnormal mt network morphology, frequently also to an abnormal cristae morphology (e.g., apoptosis). Redox-sensitive transcriptomic reprogramming sets altered metabolism and changes in epigenetics, which may further accelerate pathogenesis. In addition, the internal causes should be distinguished from the external ones, such as macrophage attacks and other immune system stimuli. That is why oxidative-stress-related pathologies must always be analyzed in a complex way, and frequently it is difficult to establish the primary cause. The reason is that under oxidative stress, cellular constituents are oxidized, i.e., covalently modified, deteriorating function and/or quality with serious consequences. To avoid the encyclopedic description, next, we will only briefly describe the oxidative stress of mitochondrial origin and how we can distinguish it from the redox signaling.
Moreover, recently, mitochondria are regarded as signaling organelles when signals of different origins are produced, not only the redox signals [55,130,131]. The evoked signals affects not only cells and tissues but also the systemic levels of the organism. The latter exists with the metabokine/mitokine signaling [133–135], mt-nuclear crosstalk [136–139], and mt-initiated epigenome remodeling [140–143].
Oxidative stress of mitochondrial origin
Theoretically, when an overly excessive superoxide/H2O2 formation exceeds the mitochondrial redox buffering, i.e., antioxidant capacity in the mt matrix, local oxidative stress in the matrix takes place. When concomitant H2O2 diffusion into the cytosol and/or other cell constituents exceeds the cellular antioxidant buffers and defense mechanisms, cellular oxidative stress is developed. We should admit that the frequent causes of such disequilibria are consequences of certain mutations in mtDNA and changes resulting from the impaired mt network morphology and/or cristae architecture. The extracellular origins or cytosolic oxidative stress acting on mitochondrial constituents also belong to frequently occurring pathologies.
The typical example of oxidative stress of mt origin is RET due to a previous accumulation of succinate, such as during heart reperfusion after ischemia [12]. Artificially induced mt oxidative stress resulted in chromatin release into the cytosol when mediated by MAPK/JNK signaling [144]. Similar manipulations induced telomere damage [145] or altered nuclear DNA methylation [146]. Senescent signaling due to increased mt ROS production activating NFκB pathway belongs to other examples [147–149].
Typically, proteinaceous stress and impairments of mitophagy and/or induction of all distinct types of cell death are developed when these thresholds are overcome. These mechanisms are out the scope of this review. We exemplified these phenomena in cases of normal physiology of pancreatic β-cells and the effects of lipotoxicity, glucotoxicity and glucolipotoxicity in the etiology of type 2 diabetes [150].
Mitochondrial redox signaling
Mitochondrial redox signaling of any time range was previously reviewed in References [151,152]. Here, we deal specifically with the acute redox signals. The triggering of redox-sensitive gene-regulatory processes (e.g. [153,154]) is beyond the scope of this review. We will discuss in detail redox signals in pancreatic β-cells in the next chapter. Now, we will list a few examples of rather acute redox signals of mitochondrial origin.
The uncoupling protein UCP1 was reported to be activated in order to switch on heat production and, therefore, nonshivering thermogenesis in brown adipose tissue (BAT) by oxidation of its Cys253 due to the elevated mt superoxide/H2O2 [155]. We speculated that H2O2-activated mt phospholipase iPLA2γ can also participate in this process by providing free fatty acids required for the UCP1-mediated uncoupling (thermogenic and not mild one) [156]. Mitochondrial superoxide/H2O2 may influence the local synaptic activity of neurons [157], and increased ROS upon mt fission provided a repair signal [158].
Mitochondrial redox signaling at hypoxia
One would not expect increasing ROS with a lowering oxygen. This paradox has been investigated, and mitochondrial contribution to oxygen sensing and hypoxic transcriptome reprogramming is still debated [159,160]. The central cytosolic mechanism of oxygen sensing is based on prolyl hydroxylases (PHD1 to 3, or Egl nine homolog 1 proteins, GLNs)[159,161–163], which catalyze hydroxylation of hypoxia-induced factor HIF-1α, -2α or -3α in a ferrous iron- (FeII-) plus 2OG- plus O2-dependent manner. The resulting hydroxylation promotes its constant proteasome degradation after ubiquitination by pVHL ubiquitin ligase (Von Hippel-Lindau tumor suppressor protein) [164–166]. Therefore, by decreasing O2, by lowering 2OG and by oxidation of FeII to FeIII due to increasing cytosolic ROS, PHDs are inhibited, and HIF-1α stabilization occurs. Also, another O2− and 2OG-dependent dioxygenase, termed factor inhibiting HIF (FIH), hydroxylates HIFα, but at asparagines. This blocks the binding of the coactivators CBP (CREB-binding protein) and p300, and thereby disables HIF-1-mediated transcription. Upon hypoxia, both PHD and FIH are inhibited; hence, HIF-α is stabilized and binds HIF-β plus coactivators VBP and p300, which allows activation of transcription of >400 genes [167–172]. In this way, the HIF system is activated, resulting in transcriptome reprogramming important in cancerogenesis and numerous physiological and pathological situations.
Since both PHD and FIH are affected by ROS, both can be targets of redox signaling. Various ROS may oxidize FeII of PHD to FeIII [173]. Still, also reactive cysteines were recognized in PHD2, which, after oxidation, inactivate the enzyme (probably inactive PHD homodimers are formed due to S-S bridges) and initiate HIF-response upon oxidation [174–176]. Cytosolic peroxiredoxins may also be involved. In any case, PHDs sense oxygen independently of mitochondria, however, mitochondrial metabolism and mt redox signaling may also independently participate. Since PHDs are also inhibited by the lack of 2OG-related substrates, such as fumarate, succinate, malate isocitrate, and lactate [177,178], suppression of mitochondrial metabolism may stimulate HIF system.
Moreover, participation of mt redox signaling linked to HIF-1α stabilization was suggested by the ΔΨm restoration, which returned a higher mt superoxide formation in cells with deleted mtDNA polymerase when respiration and hence Krebs cycle turnover was largely abolished [179]. After an instant hypoxia switch-on, a hypoxic burst of mt matrix superoxide release was observed [180] but delayed by several hours [181,182]. Other reports described an instant hypoxic ROS burst in endothelial, HeLa, and HK2 cells [183]. Originally, the Complex III site IIIQo was considered as the superoxide source for the mt hypoxic ROS burst [180,184–188]. A similar hypoxic mt ROS burst was also detected for normoxic HIF activation [189]. The emanated mt H2O2 to the cytosol was suggested to oxidize FeII in PHDs. When certain Complex III subunits, such as Rieske iron-sulfur protein [190] or others were ablated, HIF-1α was stabilized [180], unlike in anoxia [187]. Moreover, suppressors of site IIIQo electron leak (S3QELs) prevented the HIF response [34]. The key evidence for mt redox signal participation in HIF-system signaling was provided by PRDX5 overexpression in the mt intermembrane space, which abolished HIF-1α stabilization [100].
HIF strikes back on mitochondria
The chicken-and-egg problem of the steady-state established upon HIF-signaling can be solved by precisely time-resolved events. This is because the execution of HIF-mediated transcriptome reprogramming affects redox homeostasis, which is then different than that one allowing HIF-system initiation. Indeed, HIF activates transcription for the expression of proteins, decreasing ROS formation or scavenging ROS [171].
Mitochondrial cristae inflate with dormant ATP synthesis in hypoxic cells and shrink with its restoration
We have also encountered that mitochondrial cristae inflate after adaptation of HepG2 cells to hypoxia [60] (Fig. 4). It is recognized as cristae widening in transmission electron microscopic (TEM) images (Fig. 4A) and as inflation (widening in 2D projections) of 3D super-resolution images of mitochondrial cristae stained with Eos-Lactamase-β (Fig. 4B,D). Due to the HIF transcriptome reprogramming, hypoxic HepG2 cells exhibited a low-intensity (dormant) ATP synthesis and respiration [60,182]. Partial degradation of mitofilin/MIC60 protein led to the decrease of crista junctions and the widening of crista outlets from the inflated crista to the intermembrane space [60,63].
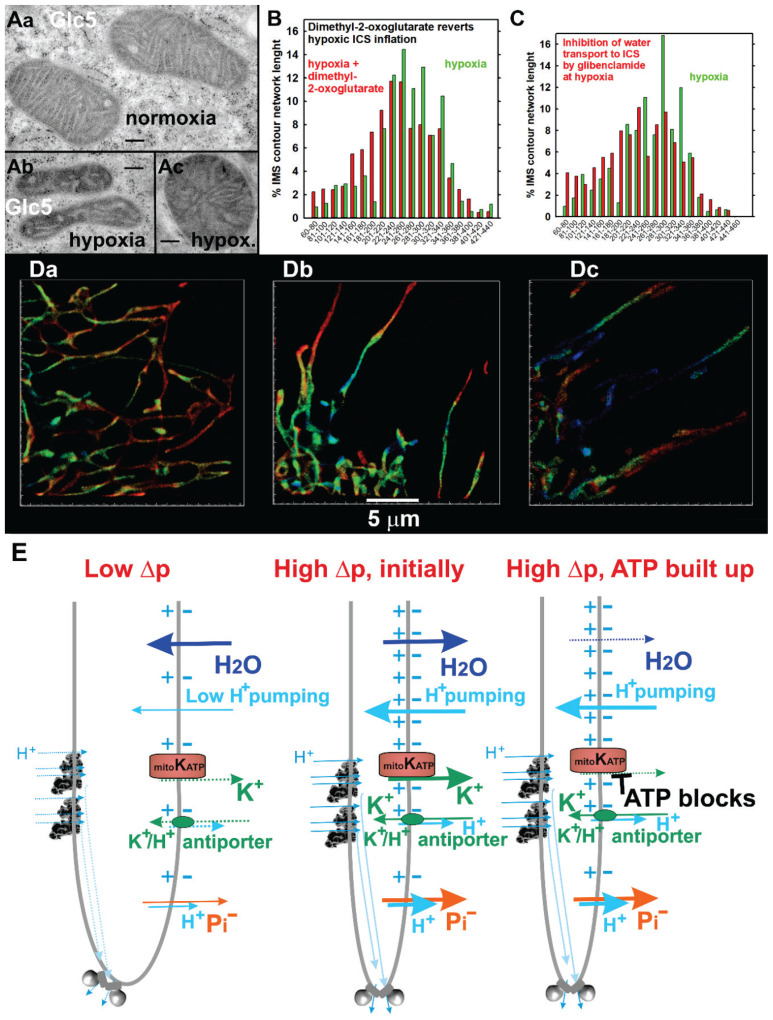
Fig. 4.
Hypoxic cristae inflation, its reversal at restored respiration and ATP synthesis and possible osmotic mechanism involving mitochondrial ATP-sensitive K+ channel. (A) Illustration of mitochondrial cristae widening after adaptation of HepG2 cells to hypoxia in transmission electron microscopic (TEM) sections (adapted from Ref. [60]). (B–D) Mitochondrial cristae widening indicated by the Eos-Lactamase-β 3D-superresolution fluorescence microscopy (adapted from Ref. [60]). Since Lactamase-β stains the intracristal space, an apparent width of its fluorescence contour on 2D sections of 3D images (panels Da for normoxia, Db, Dc for hypoxia) semi-quantifies the volume of crista lamellae. Thus, panel (B) illustrates on histograms of such width the existence of bulky cristae after hypoxic adaptation and their shrinkage after the addition of respiratory substrate, dimethyl-2-oxoglutarate. In contrast, panel (C) shows inhibition of the cristae lamellae shrinkage by glibenclamide, a known inhibitor of ATP-sensitive K+ channel, including its mitochondrial form (mtKATP). (E) Osmotic hypothesis for participation of mtKATP in cristae shrinkage. For an explanation, see text (Chapter Mitochondrial cristae inflate with dormant ATP synthesis in hypoxic cells and shrink with its restoration).
In contrast, after addition of respiratory substrate to the hypoxia-adapted HepG2 cells, a sudden narrowing of cristae in 2D projections (shrinkage of crista lamellae in a space) resulted from the restored respiration and ATP-synthesis [63,188] (Fig. 4B). We have observed similar changes in rat pancreatic β-cells, INS-1E, after the addition of a substrate, i.e. glucose (which stimulates secretion of insulin) [46]. This observation led us to a hypothesis assuming that strengthening and ordering the ATP-synthase dimers at the crista lamellar edges leads to sharpening of these edges and that the two lamellae flanks mechanistically come close together [46,63]. When metabolic conditions and signaling allow disordering of the ATP-synthase dimers at the crista lamellar edges, this allows a more flat edge, which mechanistically puts apart the two lamellae flanks of the crista, resulting in cristae inflation [7,46,63].
However, considering that individual mechanistic tension within the single crista is responsible for the lamella inflation and shrinkage seems insufficient. Hence, recently, we came up with a novel hypothesis [7], which may be valid simultaneously, that the osmotic forces are the real engines of the crista lamellae inflation and shrinkage (Fig. 4E). The hypothesis expects that at low ATP-synthesis and low Δp, ion fluxes allow the salt to be extruded from the matrix (cations such as K+ and Na+ and anions such as phosphate and Krebs cycle intermediates). After a switch-on of the ATP synthesis and before the built-up of the high ATP concentration, i.e., at the beginning of cristae morphology changes, the open mitochondrial ATP-sensitive K+ channel (mtKATP) allows an influx of K+ from the intracristal space to the matrix. Since simultaneous phosphate uptake to the matrix diminishes a salt content in the ICS and enriches it in the matrix, water uptake to the matrix occurs concomitantly to the salt influx. As a result, this osmotic force shrinks the ICS. However, to prevent an infinite cristae shrinkage, a built-up of ATP closes the mtKATP, stopping salt leakage from the ICS and water transport to the matrix. Also, mt K+/H+ and Na+/H+ antiporters, being driven by Δp, prevent the infinite shrinkage of cristae.
We have already obtained the first evidence supporting the relevance of the osmotic hypothesis of cristae morphology changes. In hypoxia-adapted HepG2 cells where the addition of dimethyl-2-oxoglutarate initiated respiration and cristae shrinkage, glibenclamide, an inhibitor of the mtKATP, blocked such shrinkage [60] (Fig. 4C). Further investigations are required to reveal whether the mechanistic or osmotic hypothesis is relevant, or whether both are relevant; as well as to describe possible regulations which transfer metabolic changes to the activation of relevant proteins which initiate cristae morphology changes.
Pancreatic β-cells as an exemplar mitochondrial redox system
Oxidative phosphorylation and NADPH-oxidase-4-mediated redox signaling as essential determinant of glucose-stimulated insulin secretion
Previously, an effect of antioxidants upon exhausted glutathione in pancreatic β-cells has been reported as an unspecified link between glucose-stimulated insulin secretion (GSIS) and external H2O2 [191]. Recently, it has been established that the essential conditions for GSIS involve the elevated OXPHOS and consequent ATP/ADP elevation in the peri-plasma membrane space [87,192,193] plus essential redox signaling, mediated by the NADPH-oxidase 4 (NOX4) [18,88,194] (Fig. 5). Together with ATP, the cytosolic redox (H2O2) signal closes the plasma membrane ATP-sensitive K+ channels (KATP), together with the elevated ATP [18,88,194]. For a closing of the entire KATP population and setting a threshold membrane potential to −50 mV, opening of other non-specific calcium channels (NSCCs, such as TRMP2 channels, [195]) or Cl− channels is required [87]. At −50 mV an intermittent opening of voltage-dependent Ca2+ channels (CaV) is initiated, being instantly counteracted by voltage-dependent K+ channels (KV). This leads to a pulsatile Ca2+ entry into the cytosol and in-phase exocytosis of insulin granule vesicles (IGV) [18,87].
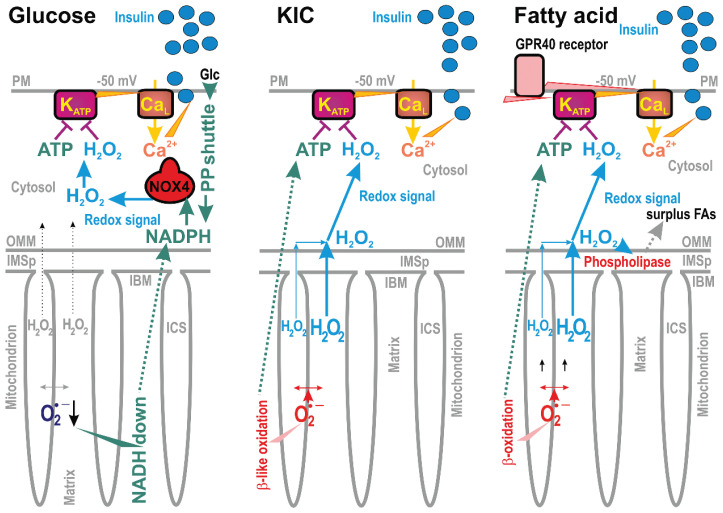
Fig. 5.
Redox signaling upon insulin secretion stimulated with glucose or ketoisocaproate (KIC) or fatty acid. Redox signaling is depicted for insulin secretion stimulated with three distinct secretagogues. In all cases, the plasma membrane ATP-sensitive K+ channel (KATP) is synergically closed only when both ATP and H2O2 (redox signaling) are elevated [194]. This predetermines plasma membrane depolarization to -50 mV and concomitant opening of the voltage-dependent Ca2+ channels (typically CaL), allowing the Ca2+ entry and exocytosis of insulin granule vesicles [88]. For the glucose-stimulated insulin secretion (GSIS) the constitutively expressed NADPH-oxidase isoform 4 (NOX4) substantiates cytosolic redox signaling, while NADPH is supplied by pentose phosphate (PP) shuttle [194] and by redox pyruvate transport shuttles (causing matrix NADH to be down and increased cytosolic NADPH, [197]). For ketoisocaproate stimulation of insulin secretion, KIC oxidation (termed β-like oxidation) generates both ATP and H2O2, which now originates from the mt-matrix-formed superoxide/H2O2 [194]. For fatty acid, stimulating insulin secretion even at low glucose [123], fatty acid β-oxidation also provides both ATP and H2O2 [88]. Similarly, as for KIC, H2O2 substantiates the redox signal from the mitochondrial matrix directed to the plasma membrane. Simultaneously, H2O2 also activates mitochondrial phospholipase iPLA2γ (“phospholipase”), which adds a surplus of mitochondrial fatty acids for both β-oxidation and the metabotropic GPR40 receptor on the plasma membrane [123]. The downstream pathways of the GPR40 receptor further stimulate insulin secretion.
Upon GSIS, an NADPH supply to the constitutively expressed NOX4 originates from the two enzymes of the pentose phosphate pathway (PPP), producing NADPH, i.e., glucose-6-phosphate dehydrogenase (G6PDH) 6-phosphogluconate dehydrogenase (6PGDH) [196]; plus from so-called (pyruvate redox) shuttles [18,88,193,197]. Interestingly, these shuttles do not allow synthesis of one NADH molecule in the mt matrix, but instead, NADPH is formed in the cytosol after a few transport steps and enzyme reactions [197]. As a result, production of superoxide released to the mt matrix is slowed down, most probably due to the decreased NADH/NAD+ ratio affecting the Complex I superoxide formation site IF [7,197]. We have linked two redox shuttles with the decreasing superoxide formation upon GSIS, the pyruvate-malate shuttle and the pyruvate-isocitrate shuttle [197]. Other shuttles were also reported [193,198]. The pyruvate-malate shuttle is allowed by the pyruvate carboxylase (PC). Such a bypass of pyruvate dehydrogenase makes possible a reverse reaction of malate dehydrogenase (MDH2), consuming NADH. A concomitant malate export from the mt matrix enables the cytosolic malic enzyme (ME1) to convert malate into pyruvate while yielding NADPH. The pyruvate-isocitrate shuttle stems from a truncated Krebs cycle after citrate synthase so that isocitrate dehydrogenase 3 (IDH3) does not form NADH. Instead, matrix NADPH is converted by IDH2 together with 2-oxoglutarate (2OG) into isocitrate, allowing its export from the mt matrix and subsequent reaction of cytosolic IDH1, transforming cytosolic isocitrate back to 2OG and synthesizing NADPH. 13C-glutamine-assisted isotope tracing enabled to verify the existence of this redox shuttle [197,199].
Relative easy spread of redox changes in pancreatic β-cells is possible due to a rather weak antioxidant defense system and low capacity of redox buffers [200,201]. Such a delicate redox homeostasis is then disturbed by a rather weak insult. Expression and activity of antioxidant enzymes is low in rodent β-cells as compared to other organs [202].
Oxidative phosphorylation and mitochondrial redox signaling as essential determinant of branched-chain-ketoacid- and fatty-acid-stimulated insulin secretion
Insulin is also stimulated by other metabolites, collectively termed secretagogues [7,88] (Fig. 5). Thus, a leucine metabolite, keto-isocaproate (KIC) was demonstrated to stimulate insulin secretion, while its oxidation (termed commonly as β-like oxidation) provides both elevated ATP and redox (H2O2) signal [194]. The mitochondrial origin of this redox signal was suggested by the blockage of KIC-stimulated insulin secretion with mt-matrix-targeted antioxidant SkQ1 [194]. This excludes the previous hypothesis that leucine itself stimulate IGV exocytosis.
Also, free fatty acids (FAs) have been regarded to augment GSIS [203], meaning that insulin secretion in the presence of FAs required certain higher glucose concentrations [204–206]. Nevertheless, we and others have demonstrated that the net FA-stimulated insulin secretion (FASIS) exists [18,88,123,207–210], i.e., insulin secretion stimulated by FAs at low glucose concentration, which otherwise does not stimulate insulin release alone. Similarly to KIC, FA β-oxidation [123] provides both elevated ATP and increased mt superoxide formation transformed into the redox (H2O2) signal, which is subsequently spread up to the plasma membrane [9]. Previously, mitochondrial ROS resulting from the addition of monooleoyl-glycerol [211] have been suggested to modulate insulin secretion, and mt-derived ROS were regarded as obligatory signals for insulin secretion [212].
FASIS is more complex than the KIC-stimulated insulin secretion (Fig. 5), since also metabotropic GPR40 receptors, residing presumably on the plasma membrane, sense FAs and initiate a complex downstream signaling. When this proceeds via Gαq/11 heterotrimeric G-proteins, followed by the Ca2+-dependent phospholipase-C-(PLC)-mediated hydrolysis of phosphatidylinositol-4,5-bisphosphate (PIP2) into diacyl-glycerol (DAG) and inositol-3-phosphate (IP3), a non-metabolizable GPR40 agonist can stimulate insulin secretion even at low glucose and ATP (Jezek et al., unpublished). Indeed, PIP2 was known to stabilize the open KATP, hence its degradation by PLC-hydrolysis facilitates the KATP closure [213]. The reaction product DAG stimulates protein kinase-C (PKC) iso-enzymes, some of which phosphorylate TRPM4 and TRPM5 [214], which opens these channels, enabling them to activate CaV channels, similarly to the TRPM2 action upon GSIS.
Moderately elevated cytosolic [Ca2+] should be required for PLC activity, however, DAG could also originate from the so-called glycerol/FA cycle [203], if it exists at “fasting” glucose. Interestingly, novel PKCs (nPKCs) are activated by DAG alone, but not by Ca2+ [215]. Hence, GPR40-signaling to final targets via nPKCs, promoting IGV-exocytosis, could exist even at low glucose.
The other product IP3 acts in the Ca2+-induced Ca2+-release from endoplasmic reticulum, enabled by the IP3-receptor (IP3R), functioning as a Ca2+ channel. Note also that in vivo, FASIS is not separated but acts in parallel with the signaling by monoacyl-glycerols (MAG) via the GPR119-Gαs-PKA(EPAC2) pathway. The PKA and EPAC2 pathways also serve as a biased pathways for certain GPR40 agonists. The protein kinase A (PKA) phosphorylates glucose transporter GLUT2 to facilitate glucose entry. PKA also phosphorylates KATP and CaV to ease action potential triggering. The EPAC pathway acts similarly by phosphorylating TRPM2, releasing PIP2 from KATP and affecting Rim2a interaction with SNARE proteins, thus facilitating IGV exocytosis [18,87,88].
Yet another phenomenon is concomitant to FASIS. Interestingly not the extracellular FAs, but FAs cleaved from the mt phospholipids by the redox-activated mt phospholipase A2, isoform γ (iPLA2γ) stimulate the GPR40 receptors [123]. Silencing of iPLA2γ led to a profound decrease of FASIS [123] despite the redox signaling up to the plasma membrane was not attenuated (Jabůrek et al., unpublished). There was a paradox encountered which has to be resolved. Due to the antioxidant synergy provided by a couple of iPLA2γ and uncoupling protein 2 (UCP2), the FA addition to pancreatic β-cells first attenuates mitochondrial superoxide formation released to the matrix. This mechanism exists since the redox-activated iPLA2γ provides nascent free FAs for UCP2 to initiate a mild uncoupling and thus reduce the mt superoxide formation [123]. However, when H2O2 is monitored in the cell cytosol or extracellularly at the same time, it is elevated (MJ, unpublished data). This paradox could be speculatively explained by MnSOD activation or by the involvement of the peroxiredoxin system [9]. Indeed PRDX3 silencing in INS-1E cells partly inhibited FASIS (MJ, unpublished data).
Future perspectives
It is an experimental challenge to track or monitor acute redox signaling by observing changes in particular reactive oxygen species in a given compartment [9]. Any event has to be studied to reliably identify the source, the path, and the target of redox signaling. Tracking changes in the surrounding proteins, e.g., in cysteine oxidation, cannot frequently distinguish the real target from those collateral ones. Correlations must be found for the initiation of the signal with the initiation of the effect and particular molecular changes in the effector proteins. It has to be investigated, for example, whether the changes in mitochondrial cristae morphology are governed by certain redox signals. With already identified redox signals, such as NADPH-oxidase 4 essential participation in glucose-stimulated insulin secretion (GSIS), it must be further studied, whether a cytosol-targeted antioxidant therapy would not rather promote a certain harm. Strong artificial suppression of redox signals could inevitably suppress GSIS and even redox signals, which otherwise contribute to the fitness of pancreatic β-cells [194]. Thus, instead of healing, this would amplify symptoms of prediabetes. In contrast, mitochondria-targeted antioxidants would not harm physiological redox signaling (except that of oxoacids and fatty acids) and might avoid the premature oxidative stress in the matrix of β-cells at the prediabetes stage. In conclusion, future studies of redox signaling should answer not only the basic questions of molecular physiology, but will also lead to novel translational aspects.
Acknowledgements
This work has been supported by Grant Agency of the Czech Republic, grant No. 21-01205S and No. 24-10132S to PJ, 23-05798S to HE, 22-02203S to AD, 22-17173S to MJ and by the project National Institute for Research of Metabolic and Cardiovascular Diseases (Programme EXCELES, ID Project No. LX22NPO5104) – Funded by the European Union – Next Generation EU.
Footnotes
Conflict of Interest: There is no conflict of interest.
References
- 1.Brand MD. Mitochondrial generation of superoxide and hydrogen peroxide as the source of mitochondrial redox signaling. Free Radic Biol Med. 2016;100:14–31. doi: 10.1016/j.freeradbiomed.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Brand MD. Riding the tiger - physiological and pathological effects of superoxide and hydrogen peroxide generated in the mitochondrial matrix. Crit Rev Biochem Mol Biol. 2020;55:592–661. doi: 10.1080/10409238.2020.1828258. [DOI] [PubMed] [Google Scholar]
- 3.Fang J, Wong HS, Brand MD. Production of superoxide and hydrogen peroxide in the mitochondrial matrix is dominated by site IQ of complex I in diverse cell lines. Redox Biol. 2020;37:101722. doi: 10.1016/j.redox.2020.101722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quinlan CL, Perevoshchikova IV, Hey-Mogensen M, Orr AL, Brand MD. Sites of reactive oxygen species generation by mitochondria oxidizing different substrates. Redox Biol. 2013;1:304–312. doi: 10.1016/j.redox.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ježek P, Hlavatá L. Mitochondria in homeostasis of reactive oxygen species in cell, tissues, and organism. Int J Biochem Cell Biol. 2005;37:2478–2503. doi: 10.1016/j.biocel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 6.Ježek P, Holendová B, Plecitá-Hlavatá L. Redox Signaling from Mitochondria: Signal Propagation and Its Targets. Biomolecules. 2020;10:93. doi: 10.3390/biom10010093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ježek P, Jabůrek M, Holendová B, Engstová H, Dlasková A. Mitochondrial Cristae Morphology Reflecting Metabolism, Superoxide Formation, Redox Homeostasis, and Pathology. Antioxid Redox Signal. 2023;39:635–683. doi: 10.1089/ars.2022.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawano I, Bazila B, Ježek P, Dlasková A. Mitochondrial dynamics and cristae shape changes during metabolic re-programming. Antioxid Redox Signal. 2023;39:684–707. doi: 10.1089/ars.2023.0268. [DOI] [PubMed] [Google Scholar]
- 9.Ježek P. Pitfalls of Mitochondrial Redox Signaling Research. Antioxidants (Basel) 2023;12:1696. doi: 10.3390/antiox12091696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohnishi ST, Shinzawa-Itoh K, Ohta K, Yoshikawa S, Ohnishi T. New insights into the superoxide generation sites in bovine heart NADH-ubiquinone oxidoreductase (Complex I): the significance of protein-associated ubiquinone and the dynamic shifting of generation sites between semiflavin and semiquinone radicals. Biochim Biophys Acta. 2010;1797:1901–1909. doi: 10.1016/j.bbabio.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 11.Hirst J, Roessler MM. Energy conversion, redox catalysis and generation of reactive oxygen species by respiratory complex I. Biochim Biophys Acta. 2016;1857:872–883. doi: 10.1016/j.bbabio.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chouchani ET, Pell VR, Gaude E, Aksentijević D, Sundier SY, Robb EL, Logan A, Nadtochiy SM, Ord ENJ, Smith AC, Eyassu F, Shirley R, Hu CH, Dare AJ, James AM, Rogatti S, Hartley RC, Eaton S, Costa ASH, Brookes PS, Davidson SM, Duchen MR, Saeb-Parsy K, Shattock MJ, Robinson AJ, Work LM, Frezza C, Krieg T, Murphy MP. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515:431–435. doi: 10.1038/nature13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robb EL, Hall AR, Prime TA, Eaton S, Szibor M, Viscomi C, James AM, Murphy MP. Control of mitochondrial superoxide production by reverse electron transport at complex I. J Biol Chem. 2018;293:9869–9879. doi: 10.1074/jbc.RA118.003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dlasková A, Hlavatá L, Ježek P. Oxidative stress caused by blocking of mitochondrial complex I H(+) pumping as a link in aging/disease vicious cycle. Int J Biochem Cell Biol. 2008;40:1792–1805. doi: 10.1016/j.biocel.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Dlasková A, Hlavatá L, Ježek J, Ježek P. Mitochondrial Complex I superoxide production is attenuated by uncoupling. Int J Biochem Cell Biol. 2008;40:2098–2109. doi: 10.1016/j.biocel.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Ježek P, Žáčková M, Růžička M, Škobisová E, Jabůrek M. Mitochondrial uncoupling proteins--facts and fantasies. Physiol Res. 2004;53(Suppl 1):S199–S211. doi: 10.33549/physiolres.930000.53.S199. [DOI] [PubMed] [Google Scholar]
- 17.Ježek P, Olejár T, Smolková K, Ježek J, Dlasková A, Plecitá-Hlavatá L, Zelenka J, Špaček T, Engstová H, Pajuelo Reguera D, Jabůrek M. Antioxidant and regulatory role of mitochondrial uncoupling protein UCP2 in pancreatic beta-cells. Physiol Res. 2014;63(Suppl 1):S73–S91. doi: 10.33549/physiolres.932633. [DOI] [PubMed] [Google Scholar]
- 18.Ježek P, Holendová B, Jabůrek M, Dlasková A, Plecitá-Hlavatá L. Contribution of Mitochondria to Insulin Secretion by Various Secretagogues. Antioxid Redox Signal. 2022;36:920–952. doi: 10.1089/ars.2021.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brand MD, Affourtit C, Esteves TC, Green K, Lambert AJ, Miwa S, Pakay JL, Parker N. Mitochondrial superoxide: production, biological effects, and activation of uncoupling proteins. Free Radic Biol Med. 2004;37:755–767. doi: 10.1016/j.freeradbiomed.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 20.Muller FL, Roberts AG, Bowman MK, Kramer DM. Architecture of the Qo site of the cytochrome bc1 complex probed by superoxide production. Biochemistry. 2003;42:6493–6499. doi: 10.1021/bi0342160. [DOI] [PubMed] [Google Scholar]
- 21.Muller FL, Liu Y, Van Remmen H. Complex III releases superoxide to both sides of the inner mitochondrial membrane. J Biol Chem. 2004;279:49064–49073. doi: 10.1074/jbc.M407715200. [DOI] [PubMed] [Google Scholar]
- 22.Husen P, Nielsen C, Martino CF, Solov’yov IA. Molecular Oxygen Binding in the Mitochondrial Electron Transfer Flavoprotein. J Chem Inf Model. 2019;59:4868–4879. doi: 10.1021/acs.jcim.9b00702. [DOI] [PubMed] [Google Scholar]
- 23.Reichart G, Mayer J, Zehm C, Kirschstein T, Tokay T, Lange F, Baltrusch S, Tiedge M, Fuellen G, Ibrahim S, Köhling R. Mitochondrial complex IV mutation increases reactive oxygen species production and reduces lifespan in aged mice. Acta Physiol (Oxf) 2019;225:e13214. doi: 10.1111/apha.13214. [DOI] [PubMed] [Google Scholar]
- 24.Grivennikova VG, Kozlovsky VS, Vinogradov AD. Respiratory complex II: ROS production and the kinetics of ubiquinone reduction. Biochim Biophys Acta. 2017;1858:109–117. doi: 10.1016/j.bbabio.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 25.St-Pierre J, Buckingham JA, Roebuck SJ, Brand MD. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J Biol Chem. 2002;277:44784–44790. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- 26.Perevoshchikova IV, Quinlan CL, Orr AL, Gerencser AA, Brand MD. Sites of superoxide and hydrogen peroxide production during fatty acid oxidation in rat skeletal muscle mitochondria. Free Radic Biol Med. 2013;61:298–309. doi: 10.1016/j.freeradbiomed.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quinlan CL, Treberg JR, Perevoshchikova IV, Orr AL, Brand MD. Native rates of superoxide production from multiple sites in isolated mitochondria measured using endogenous reporters. Free Radic Biol Med. 2012;53:1807–1817. doi: 10.1016/j.freeradbiomed.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trewin AJ, Bahr LL, Almast A, Berry BJ, Wei AY, Foster TH, Wojtovich AP. Mitochondrial Reactive Oxygen Species Generated at the Complex-II Matrix or Intermembrane Space Microdomain Have Distinct Effects on Redox Signaling and Stress Sensitivity in Caenorhabditis elegans. Antioxid Redox Signal. 2019;31:594–607. doi: 10.1089/ars.2018.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siebels I, Dröse S. Q-site inhibitor induced ROS production of mitochondrial complex II is attenuated by TCA cycle dicarboxylates. Biochim Biophys Acta. 2013;1827:1156–1164. doi: 10.1016/j.bbabio.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 30.Manhas N, Duong QV, Lee P, Richardson JD, Robertson JD, Moxley MA, Bazil JN. Computationally modeling mammalian succinate dehydrogenase kinetics identifies the origins and primary determinants of ROS production. J Biol Chem. 2020;295:15262–15279. doi: 10.1074/jbc.RA120.014483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinlan CL, Goncalves RL, Hey-Mogensen M, Yadava N, Bunik VI, Brand MD. The 2-oxoacid dehydrogenase complexes in mitochondria can produce superoxide/hydrogen peroxide at much higher rates than complex I. J Biol Chem. 2014;289:8312–8325. doi: 10.1074/jbc.M113.545301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDonald AE, Pichaud N, Darveau CA. “Alternative” fuels contributing to mitochondrial electron transport: Importance of non-classical pathways in the diversity of animal metabolism. Comp Biochem Physiol B Biochem Mol Biol. 2018;224:185–194. doi: 10.1016/j.cbpb.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 33.Orr AL, Quinlan CL, Perevoshchikova IV, Brand MD. A refined analysis of superoxide production by mitochondrial sn-glycerol 3-phosphate dehydrogenase. J Biol Chem. 2012;287:42921–42935. doi: 10.1074/jbc.M112.397828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orr AL, Vargas L, Turk CN, Baaten JE, Matzen JT, Dardov VJ, Attle SJ, Li J, Quackenbush DC, Goncalves RL, Perevoshchikova IV, Petrassi HM, Meeusen SL, Ainscow EK, Brand MD. Suppressors of superoxide production from mitochondrial complex III. Nat Chem Biol. 2015;11:834–836. doi: 10.1038/nchembio.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boukalova S, Hubackova S, Milosevic M, Ezrova Z, Neuzil J, Rohlena J. Dihydroorotate dehydrogenase in oxidative phosphorylation and cancer. Biochim Biophys Acta. 2020;1866:165759. doi: 10.1016/j.bbadis.2020.165759. [DOI] [PubMed] [Google Scholar]
- 36.Valentine JS, Doucette PA, Zittin Potter S. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu Rev Biochem. 2005;74:563–593. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- 37.Ozden O, Park SH, Kim HS, Jiang H, Coleman MC, Spitz DR, Gius D. Acetylation of MnSOD directs enzymatic activity responding to cellular nutrient status or oxidative stress. Aging. 2011;3:102–107. doi: 10.18632/aging.100291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park SH, Ozden O, Jiang H, Cha YI, Pennington JD, Aykin-Burns N, Spitz DR, Gius D, Kim HS. Sirt3, mitochondrial ROS, ageing, and carcinogenesis. Int J Mol Sci. 2011;12:6226–6239. doi: 10.3390/ijms12096226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salvatori I, Valle C, Ferri A, Carrì MT. SIRT3 and mitochondrial metabolism in neurodegenerative diseases. Neurochem Int. 2017;109:184–192. doi: 10.1016/j.neuint.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 40.Tao R, Vassilopoulos A, Parisiadou L, Yan Y, Gius D. Regulation of MnSOD enzymatic activity by Sirt3 connects the mitochondrial acetylome signaling networks to aging and carcinogenesis. Antioxid Redox Signal. 2014;20:1646–1654. doi: 10.1089/ars.2013.5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anamika Roy A, Trigun SK. Hippocampus mitochondrial MnSOD activation by a SIRT3 activator, honokiol, correlates with its deacetylation and upregulation of FoxO3a and PGC1α in a rat model of ammonia neurotoxicity. J Cell Biochem. 2023;124:606–618. doi: 10.1002/jcb.30393. [DOI] [PubMed] [Google Scholar]
- 42.Gao E, Sun X, Thorne RF, Zhang XD, Li J, Shao F, Ma J, Wu M. NIPSNAP1 directs dual mechanisms to restrain senescence in cancer cells. J Transl Med. 2023;21:401. doi: 10.1186/s12967-023-04232-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X, Xie X, Li D, Liu Z, Zhang B, Zang Y, Yuan H, Shen C. Sirt3-dependent regulation of mitochondrial oxidative stress and apoptosis contributes to the dysfunction of pancreatic islets after severe burns. Free Radic Biol Med. 2023;198:59–67. doi: 10.1016/j.freeradbiomed.2023.01.027. [DOI] [PubMed] [Google Scholar]
- 44.Ma C, Sun Y, Pi C, Wang H, Sun H, Yu X, Shi Y, He X. Sirt3 Attenuates Oxidative Stress Damage and Rescues Cellular Senescence in Rat Bone Marrow Mesenchymal Stem Cells by Targeting Superoxide Dismutase 2. Front Cell Dev Biol. 2020;8:599376. doi: 10.3389/fcell.2020.599376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mohan MS, Aswani SS, Aparna NS, Boban PT, Sudhakaran PR, Saja K. Effect of acute cold exposure on cardiac mitochondrial function: role of sirtuins. Mol Cell Biochem. 2023;478:2257–2270. doi: 10.1007/s11010-022-04656-1. [DOI] [PubMed] [Google Scholar]
- 46.Dlasková A, Engstová H, Špaček T, Kahancová A, Pavluch V, Smolková K, Špačková J, Bartoš M, Hlavatá L, Ježek P. 3D super-resolution microscopy reflects mitochondrial cristae alternations and mtDNA nucleoid size and distribution. Biochim Biophys Acta. 2018;1859:829–844. doi: 10.1016/j.bbabio.2018.04.013. [DOI] [PubMed] [Google Scholar]
- 47.Ježek P, Plecitá-Hlavatá L. Mitochondrial reticulum network dynamics in relation to oxidative stress, redox regulation, and hypoxia. Int J Biochem Cell Biol. 2009;41:1790–1804. doi: 10.1016/j.biocel.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 48.Plecitá-Hlavatá L, Ježek P. Integration of superoxide formation and cristae morphology for mitochondrial redox signaling. Int J Biochem Cell Biol. 2016;80:31–50. doi: 10.1016/j.biocel.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 49.Dlasková A, Špaček T, Šantorová J, Plecitá-Hlavatá L, Berková Z, Saudek F, Lessard M, Bewersdorf J, Ježek P. 4Pi microscopy reveals an impaired three-dimensional mitochondrial network of pancreatic islet beta-cells, an experimental model of type-2 diabetes. Biochim Biophys Acta. 2010;1797:1327–1341. doi: 10.1016/j.bbabio.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 50.Dlasková A, Engstová H, Plecitá-Hlavatá L, Lessard M, Alán L, Reguera DP, Jabůrek M, Ježek P. Distribution of mitochondrial DNA nucleoids inside the linear tubules vs. bulk parts of mitochondrial network as visualized by 4Pi microscopy. J Bioenerg Biomembr. 2015;47:255–263. doi: 10.1007/s10863-015-9610-3. [DOI] [PubMed] [Google Scholar]
- 51.Plecitá-Hlavatá L, Lessard M, Šantorová J, Bewersdorf J, Ježek P. Mitochondrial oxidative phosphorylation and energetic status are reflected by morphology of mitochondrial network in INS-1E and HEP-G2 cells viewed by 4Pi microscopy. Biochim Biophys Acta. 2008;1777:834–846. doi: 10.1016/j.bbabio.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 52.Glancy B, Hartnell LM, Malide D, Yu ZX, Combs CA, Connelly PS, Subramaniam S, Balaban RS. Mitochondrial reticulum for cellular energy distribution in muscle. Nature. 2015;523:617–620. doi: 10.1038/nature14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eisner V, Cupo RR, Gao E, Csordás G, Slovinsky WS, Paillard M, Cheng L, Ibetti J, Chen SR, Chuprun JK, Hoek JB, Koch WJ, Hajnóczky G. Mitochondrial fusion dynamics is robust in the heart and depends on calcium oscillations and contractile activity. Proc Natl Acad Sci U S A. 2017;114:E859–E868. doi: 10.1073/pnas.1617288114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Picard M, Shirihai OS. Mitochondrial signal transduction. Cell Metab. 2022;34:1620–1653. doi: 10.1016/j.cmet.2022.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kondadi AK, Anand R, Hänsch S, Urbach J, Zobel T, Wolf DM, Segawa M, Liesa M, Shirihai OS, Weidtkamp-Peters S, Reichert AS. Cristae undergo continuous cycles of membrane remodelling in a MICOS-dependent manner. EMBO Rep. 2020;21:e49776. doi: 10.15252/embr.201949776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bohnert M, Wenz LS, Zerbes RM, Horvath SE, Stroud DA, von der Malsburg K, Müller JM, Oeljeklaus S, Perschil I, Warscheid B, Chacinska A, Veenhuis M, van der Klei IJ, Daum G, Wiedemann N, Becker T, Pfanner N, van der Laan M. Role of mitochondrial inner membrane organizing system in protein biogenesis of the mitochondrial outer membrane. Mol Biol Cell. 2012;23:3948–3956. doi: 10.1091/mbc.e12-04-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pfanner N, van der Laan M, Amati P, Capaldi RA, Caudy AA, Chacinska A, Darshi M, Deckers M, Hoppins S, Icho T, Jakobs S, Ji J, Kozjak-Pavlovic V, Meisinger C, Odgren PR, Park SK, Rehling P, Reichert AS, Sheikh MS, Taylor SS, Tsuchida N, van der Bliek AM, van der Klei IJ, Weissman JS, Westermann B, Zha J, Neupert W, Nunnari J. Uniform nomenclature for the mitochondrial contact site and cristae organizing system. J Cell Biol. 2014;204:1083–1086. doi: 10.1083/jcb.201401006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zerbes RM, Bohnert M, Stroud DA, von der Malsburg K, Kram A, Oeljeklaus S, Warscheid B, Becker T, Wiedemann N, Veenhuis M, van der Klei IJ, Pfanner N, van der Laan M. Role of MINOS in mitochondrial membrane architecture: cristae morphology and outer membrane interactions differentially depend on mitofilin domains. J Mol Biol. 2012;422:183–191. doi: 10.1016/j.jmb.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 60.Plecitá-Hlavatá L, Engstová H, Alán L, Špaček T, Dlasková A, Smolková K, Špačková J, Tauber J, Strádalová V, Malínský J, Lessard M, Bewersdorf J, Ježek P. Hypoxic HepG2 cell adaptation decreases ATP synthase dimers and ATP production in inflated cristae by mitofilin down-regulation concomitant to MICOS clustering. FASEB J. 2016;30:1941–1957. doi: 10.1096/fj.201500176. [DOI] [PubMed] [Google Scholar]
- 61.Davies KM, Strauss M, Daum B, Kief JH, Osiewacz HD, Rycovska A, Zickermann V, Kühlbrandt W. Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc Natl Acad Sci U S A. 2011;108:14121–14126. doi: 10.1073/pnas.1103621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davies KM, Anselmi C, Wittig I, Faraldo-Gómez JD, Kühlbrandt W. Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc Natl Acad Sci U S A. 2012;109:13602–13607. doi: 10.1073/pnas.1204593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dlasková A, Špaček T, Engstová H, Špačková J, Schröfel A, Holendová B, Smolková K, Plecitá-Hlavatá L, Ježek P. Mitochondrial cristae narrowing upon higher 2-oxoglutarate load. Biochim Biophys Acta. 2019;1860:659–678. doi: 10.1016/j.bbabio.2019.06.015. [DOI] [PubMed] [Google Scholar]
- 64.Dudkina NV, Oostergetel GT, Lewejohann D, Braun HP, Boekema EJ. Row-like organization of ATP synthase in intact mitochondria determined by cryo-electron tomography. Biochim Biophys Acta. 2010;1797:272–277. doi: 10.1016/j.bbabio.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 65.Gu J, Zhang L, Zong S, Guo R, Liu T, Yi J, Wang P, Zhuo W, Yang M. Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1. Science. 2019;364:1068–1075. doi: 10.1126/science.aaw4852. [DOI] [PubMed] [Google Scholar]
- 66.Guo H, Bueler SA, Rubinstein JL. Atomic model for the dimeric FO region of mitochondrial ATP synthase. Science. 2017;358:936–940. doi: 10.1126/science.aao4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nesterov S, Chesnokov Y, Kamyshinsky R, Panteleeva A, Lyamzaev K, Vasilov R, Yaguzhinsky L. Ordered Clusters of the Complete Oxidative Phosphorylation System in Cardiac Mitochondria. Int J Mol Sci. 2021;22:1462. doi: 10.3390/ijms22031462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blum TB, Hahn A, Meier T, Davies KM, Kühlbrandt W. Dimers of mitochondrial ATP synthase induce membrane curvature and self-assemble into rows. Proc Natl Acad Sci U S A. 2019;116:4250–4255. doi: 10.1073/pnas.1816556116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Daum B, Walter A, Horst A, Osiewacz HD, Kühlbrandt W. Age-dependent dissociation of ATP synthase dimers and loss of inner-membrane cristae in mitochondria. Proc Natl Acad Sci U S A. 2013;110:15301–15306. doi: 10.1073/pnas.1305462110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spikes TE, Montgomery MG, Walker JE. Structure of the dimeric ATP synthase from bovine mitochondria. Proc Natl Acad Sci U S A. 2020;117:23519–23526. doi: 10.1073/pnas.2013998117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rampelt H, Bohnert M, Zerbes RM, Horvath SE, Warscheid B, Pfanner N, van der Laan M. Mic10, a Core Subunit of the Mitochondrial Contact Site and Cristae Organizing System, Interacts with the Dimeric F1Fo-ATP Synthase. J Mol Biol. 2017;429:1162–1170. doi: 10.1016/j.jmb.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 72.Eydt K, Davies KM, Behrendt C, Wittig I, Reichert AS. Cristae architecture is determined by an interplay of the MICOS complex and the F1FO ATP synthase via Mic27 and Mic10. Microb Cell. 2017;4:259–272. doi: 10.15698/mic2017.08.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lenaz G, Tioli G, Falasca AI, Genova ML. Complex I function in mitochondrial supercomplexes. Biochim Biophys Acta. 2016;1857:991–1000. doi: 10.1016/j.bbabio.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 74.Letts JA, Fiedorczuk K, Sazanov LA. The architecture of respiratory supercomplexes. Nature. 2016;537:644–648. doi: 10.1038/nature19774. [DOI] [PubMed] [Google Scholar]
- 75.Lobo-Jarne T, Ugalde C. Respiratory chain supercomplexes: Structures, function and biogenesis. Semin Cell Dev Biol. 2018;76:179–190. doi: 10.1016/j.semcdb.2017.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Giacomello M, Pyakurel A, Glytsou C, Scorrano L. The cell biology of mitochondrial membrane dynamics. Nat Rev Mol Cell Biol. 2020;21:204–224. doi: 10.1038/s41580-020-0210-7. [DOI] [PubMed] [Google Scholar]
- 77.Pernas L, Scorrano L. Mito-Morphosis: Mitochondrial Fusion, Fission, and Cristae Remodeling as Key Mediators of Cellular Function. Annu Rev Physiol. 2016;78:505–531. doi: 10.1146/annurev-physiol-021115-105011. [DOI] [PubMed] [Google Scholar]
- 78.Faelber K, Dietrich L, Noel JK, Wollweber F, Pfitzner AK, Mühleip A, Sánchez R, Kudryashev M, Chiaruttini N, Lilie H, Schlegel J, Rosenbaum E, Hessenberger M, Matthaeus C, Kunz S, von der Malsburg A, Noé F, Roux A, van der Laan M, Kühlbrandt W, Daumke O. Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1. Nature. 2019;571:429–433. doi: 10.1038/s41586-019-1372-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tatsuta T, Langer T. Prohibitins. Curr Biol. 2017;27:R629–R631. doi: 10.1016/j.cub.2017.04.030. [DOI] [PubMed] [Google Scholar]
- 80.Barbot M, Jans DC, Schulz C, Denkert N, Kroppen B, Hoppert M, Jakobs S, Meinecke M. Mic10 oligomerizes to bend mitochondrial inner membranes at cristae junctions. Cell Metab. 2015;21:756–763. doi: 10.1016/j.cmet.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 81.Bohnert M, Zerbes RM, Davies KM, Mühleip AW, Rampelt H, Horvath SE, Boenke T, Kram A, Perschil I, Veenhuis M, Kühlbrandt W, van der Klei IJ, Pfanner N, van der Laan M. Central role of Mic10 in the mitochondrial contact site and cristae organizing system. Cell Metab. 2015;21:747–755. doi: 10.1016/j.cmet.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 82.Wang L, Yan Z, Vihinen H, Eriksson O, Wang W, Soliymani R, Lu Y, Xue Y, Jokitalo E, Li J, Zhao H. FAM92A1 is a BAR domain protein required for mitochondrial ultrastructure and function. J Cell Biol. 2019;218:97–111. doi: 10.1083/jcb.201806191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T, Bartoli D, Polishuck RS, Danial NN, De Strooper B, Scorrano L. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 84.Deponte M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim Biophys Acta. 2013;1830:3217–3266. doi: 10.1016/j.bbagen.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 85.Sies H, Jones DP. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat Rev Mol Cell Biol. 2020;21:363–383. doi: 10.1038/s41580-020-0230-3. [DOI] [PubMed] [Google Scholar]
- 86.Merrins MJ, Corkey BE, Kibbey RG, Prentki M. Metabolic cycles and signals for insulin secretion. Cell Metab. 2022;34:947–968. doi: 10.1016/j.cmet.2022.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rorsman P, Ashcroft FM. Pancreatic β-Cell Electrical Activity and Insulin Secretion: Of Mice and Men. Physiol Rev. 2018;98:117–214. doi: 10.1152/physrev.00008.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ježek P, Holendová B, Jabůrek M, Tauber J, Dlasková A, Plecitá-Hlavatá L. The Pancreatic β-Cell: The Perfect Redox System. Antioxidants. 2021;10:197. doi: 10.3390/antiox10020197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brigelius-Flohe R, Maiorino M. Glutathione peroxidases. Biochim Biophys Acta. 2013;1830:3289–3303. doi: 10.1016/j.bbagen.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 90.Ighodaro OM, Akinloye OA. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and gluta-thione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alex J Med. 2018;54:287–293. doi: 10.1016/j.ajme.2017.09.001. [DOI] [Google Scholar]
- 91.Herbette S, Roeckel-Drevet P, Drevet JR. Seleno-independent glutathione peroxidases. More than simple antioxidant scavengers. FEBS J. 2007;274:2163–2180. doi: 10.1111/j.1742-4658.2007.05774.x. [DOI] [PubMed] [Google Scholar]
- 92.Bolduc J, Koruza K, Luo T, Malo Pueyo J, Vo TN, Ezeriņa D, Messens J. Peroxiredoxins wear many hats: Factors that fashion their peroxide sensing personalities. Redox Biol. 2021;42:101959. doi: 10.1016/j.redox.2021.101959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu Y, Wang P, Hu W, Chen D. New insights into the roles of peroxiredoxins in cancer. Biomed Pharmacother. 2023;164:114896. doi: 10.1016/j.biopha.2023.114896. [DOI] [PubMed] [Google Scholar]
- 94.Rhee SG. Overview on Peroxiredoxin. Mol Cells. 2016;39:1–5. doi: 10.14348/molcells.2016.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rhee SG, Kang SW, Chang TS, Jeong W, Kim K. Peroxiredoxin, a novel family of peroxidases. IUBMB Life. 2001;52:35–41. doi: 10.1080/15216540252774748. [DOI] [PubMed] [Google Scholar]
- 96.Rhee SG, Woo HA. Multiple functions of 2-Cys peroxiredoxins, I and II, and their regulations via post-translational modifications. Free Radic Biol Med. 2020;152:107–115. doi: 10.1016/j.freeradbiomed.2020.02.028. [DOI] [PubMed] [Google Scholar]
- 97.Thapa P, Jiang H, Ding N, Hao Y, Alshahrani A, Wei Q. The Role of Peroxiredoxins in Cancer Development. Biology (Basel) 2023;12:666. doi: 10.3390/biology12050666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Villar SF, Ferrer-Sueta G, Denicola A. The multifaceted nature of peroxiredoxins in chemical biology. Curr Opin Chem Biol. 2023;76:102355. doi: 10.1016/j.cbpa.2023.102355. [DOI] [PubMed] [Google Scholar]
- 99.Knoops B, Goemaere J, Van der Eecken V, Declercq JP. Peroxiredoxin 5: structure, mechanism, and function of the mammalian atypical 2-Cys peroxiredoxin. Antioxid Redox Signal. 2011;15:817–829. doi: 10.1089/ars.2010.3584. [DOI] [PubMed] [Google Scholar]
- 100.Sabharwal SS, Waypa GB, Marks JD, Schumacker PT. Peroxiredoxin-5 targeted to the mitochondrial intermembrane space attenuates hypoxia-induced reactive oxygen species signalling. Biochem J. 2013;456:337–346. doi: 10.1042/BJ20130740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sabharwal SS, Dudley VJ, Landwerlin C, Schumacker PT. H2O2 transit through the mitochondrial intermembrane space promotes tumor cell growth in vitro and in vivo. J Biol Chem. 2023;299:104624. doi: 10.1016/j.jbc.2023.104624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.López-Grueso MJ, Lagal DJ, García-Jiménez ÁF, Tarradas RM, Carmona-Hidalgo B, Peinado J, Requejo-Aguilar R, Bárcena JA, Padilla CA. Knockout of PRDX6 induces mitochondrial dysfunction and cell cycle arrest at G2/M in HepG2 hepatocarcinoma cells. Redox Biol. 2020;37:101737. doi: 10.1016/j.redox.2020.101737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ma S, Zhang X, Zheng L, Li Z, Zhao X, Lai W, Shen H, Lv J, Yang G, Wang Q, Ji J. Peroxiredoxin 6 Is a Crucial Factor in the Initial Step of Mitochondrial Clearance and Is Upstream of the PINK1-Parkin Pathway. Antioxid Redox Signal. 2016;24:486–501. doi: 10.1089/ars.2015.6336. [DOI] [PubMed] [Google Scholar]
- 104.Pacifici F, Della-Morte D, Capuani B, Coppola A, Scioli MG, Donadel G, Andreadi A, Ciccosanti F, Fimia GM, Bellia A, Orlandi A, Lauro D. Peroxiredoxin 6 Modulates Insulin Secretion and Beta Cell Death via a Mitochondrial Dynamic Network. Front Endocrinol. 2022;13:842575. doi: 10.3389/fendo.2022.842575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bachnoff N, Trus M, Atlas D. Alleviation of oxidative stress by potent and selective thioredoxin-mimetic peptides. Free Radic Biol Med. 2011;50:1355–1367. doi: 10.1016/j.freeradbiomed.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 106.Jeong SJ, Park JG, Oh GT. Peroxiredoxins as Potential Targets for Cardiovascular Disease. Antioxidants. 2021;10:1244. doi: 10.3390/antiox10081244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Szeliga M. Peroxiredoxins in Neurodegenerative Diseases. Antioxidants. 2020;9:1203. doi: 10.3390/antiox9121203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stancill JS, Corbett JA. The Role of Thioredoxin/Peroxiredoxin in the β-Cell Defense Against Oxidative Damage. Front Endocrinol. 2021;12:718235. doi: 10.3389/fendo.2021.718235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rhee SG, Kil IS. Mitochondrial H2O2 signaling is controlled by the concerted action of peroxiredoxin III and sulfiredoxin: Linking mitochondrial function to circadian rhythm. Free Radic Biol Med. 2016;100:73–80. doi: 10.1016/j.freeradbiomed.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 110.Mishra M, Jiang H, Wu L, Chawsheen HA, Wei Q. The sulfiredoxin-peroxiredoxin (Srx-Prx) axis in cell signal transduction and cancer development. Cancer Lett. 2015;366:150–159. doi: 10.1016/j.canlet.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Reczek CR, Chandel NS. ROS-dependent signal transduction. Curr Opin Cell Biol. 2015;33:8–13. doi: 10.1016/j.ceb.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Woo HA, Yim SH, Shin DH, Kang D, Yu DY, Rhee SG. Inactivation of peroxiredoxin I by phosphorylation allows localized H(2)O(2) accumulation for cell signaling. Cell. 2010;140:517–528. doi: 10.1016/j.cell.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 113.Heo S, Kim S, Kang D. The Role of Hydrogen Peroxide and Peroxiredoxins throughout the Cell Cycle. Antioxidants. 2020;9:280. doi: 10.3390/antiox9040280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jarvis RM, Hughes SM, Ledgerwood EC. Peroxiredoxin 1 functions as a signal peroxidase to receive, transduce, and transmit peroxide signals in mammalian cells. Free Radic Biol Med. 2012;53:1522–1530. doi: 10.1016/j.freeradbiomed.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 115.Rhee SG, Woo HA, Kang D. The Role of Peroxiredoxins in the Transduction of H2O2 Signals. Antioxid Redox Signal. 2018;28:537–557. doi: 10.1089/ars.2017.7167. [DOI] [PubMed] [Google Scholar]
- 116.Rhee SG. A catalytic career: Studies spanning glutamine synthetase, phospholipase C, peroxiredoxin, and the intracellular messenger role of hydrogen peroxide. J Biol Chem. 2019;294:5169–5180. doi: 10.1074/jbc.X119.007975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sobotta MC, Liou W, Stocker S, Talwar D, Oehler M, Ruppert T, Scharf AN, Dick TP. Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat Chem Biol. 2015;11:64–70. doi: 10.1038/nchembio.1695. [DOI] [PubMed] [Google Scholar]
- 118.Stocker S, Van Laer K, Mijuskovic A, Dick TP. The Conundrum of Hydrogen Peroxide Signaling and the Emerging Role of Peroxiredoxins as Redox Relay Hubs. Antioxid Redox Signal. 2018;28:558–573. doi: 10.1089/ars.2017.7162. [DOI] [PubMed] [Google Scholar]
- 119.Ježek P, Holendová B, Garlid KD, Jabůrek M. Mitochondrial Uncoupling Proteins: Subtle Regulators of Cellular Redox Signaling. Antioxid Redox Signal. 2018;29:667–714. doi: 10.1089/ars.2017.7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jabůrek M, Průchová P, Holendová B, Galkin A, Ježek P. Antioxidant Synergy of Mitochondrial Phospholipase PNPLA8/iPLA2γ with Fatty Acid-Conducting SLC25 Gene Family Transporters. Antioxidants. 2021;10:678. doi: 10.3390/antiox10050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Průchová P, Gotvaldová K, Smolková K, Alán L, Holendová B, Tauber J, Galkin A, Ježek P, Jabůrek M. Antioxidant Role and Cardiolipin Remodeling by Redox-Activated Mitochondrial Ca2+-Independent Phospholipase A2γ in the Brain. Antioxidants. 2022;11:198. doi: 10.3390/antiox11020198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fedorenko A, Lishko PV, Kirichok Y. Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell. 2012;151:400–413. doi: 10.1016/j.cell.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ježek J, Dlasková A, Zelenka J, Jabůrek M, Ježek P. H2O2-Activated Mitochondrial Phospholipase iPLA2γ Prevents Lipotoxic Oxidative Stress in Synergy with UCP2, Amplifies Signaling via G-Protein-Coupled Receptor GPR40, and Regulates Insulin Secretion in Pancreatic β-Cells. Antioxid Redox Signal. 2015;23:958–972. doi: 10.1089/ars.2014.6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Korshunov SS, Skulachev VP, Starkov AA. High protonic potential actuates a mechanism of production of reactive oxygen species in mitochondria. FEBS Lett. 1997;416:15–18. doi: 10.1016/S0014-5793(97)01159-9. [DOI] [PubMed] [Google Scholar]
- 125.Arsenijevic D, Onuma H, Pecqueur C, Raimbault S, Manning BS, Miroux B, Couplan E, Alves-Guerra MC, Goubern M, Surwit R, Bouillaud F, Richard D, Collins S, Ricquier D. Disruption of the uncoupling protein-2 gene in mice reveals a role in immunity and reactive oxygen species production. Nat Genet. 2000;26:435–439. doi: 10.1038/82565. [DOI] [PubMed] [Google Scholar]
- 126.Nègre-Salvayre A, Hirtz C, Carrera G, Cazenave R, Troly M, Salvayre R, Pénicaud L, Casteilla L. A role for uncoupling protein-2 as a regulator of mitochondrial hydrogen peroxide generation. FASEB J. 1997;11:809–815. doi: 10.1096/fasebj.11.10.9271366. [DOI] [PubMed] [Google Scholar]
- 127.Duval C, Nègre-Salvayre A, Dogilo A, Salvayre R, Pénicaud L, Casteilla L. Increased reactive oxygen species production with antisense oligonucleotides directed against uncoupling protein 2 in murine endothelial cells. Biochem Cell Biol. 2002;80:757–764. doi: 10.1139/o02-158. [DOI] [PubMed] [Google Scholar]
- 128.Blanc J, Alves-Guerra MC, Esposito B, Rousset S, Gourdy P, Ricquier D, Tedgui A, Miroux B, Mallat Z. Protective role of uncoupling protein 2 in atherosclerosis. Circulation. 2003;107:388–390. doi: 10.1161/01.CIR.0000051722.66074.60. [DOI] [PubMed] [Google Scholar]
- 129.Ježek J, Jabůrek M, Zelenka J, Ježek P. Mitochondrial phospholipase A2 activated by reactive oxygen species in heart mitochondria induces mild uncoupling. Physiol Res. 2010;59:737–747. doi: 10.33549/physiolres.931905. [DOI] [PubMed] [Google Scholar]
- 130.Jabůrek M, Ježek J, Zelenka J, Ježek P. Antioxidant activity by a synergy of redox-sensitive mitochondrial phospholipase A2 and uncoupling protein-2 in lung and spleen. Int J Biochem Cell Biol. 2013;45:816–825. doi: 10.1016/j.biocel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 131.Chandel N. Evolution of mitochondria as signaling organelles. Cell Metab. 2015;22:204–206. doi: 10.1016/j.cmet.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 132.Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Forsstrom S, Jackson CB, Carroll CJ, Kuronen M, Pirinen E, Pradhan S, Marmyleva A, Auranen M, Kleine IM, Khan NA, Roivainen A, Marjamäki P, Liljenbäck H, Wang L, Battersby BJ, Richter U, Velagapudi V, Nikkanen J, Euro L, Suomalainen A. Fibroblast growth factor 21 drives dynamics of local and systemic stress responses in mitochondrial myopathy with mtDNA deletions. Cell Metab. 2019;30:1040–1054.e7. doi: 10.1016/j.cmet.2019.08.019. [DOI] [PubMed] [Google Scholar]
- 135.Tyynismaa H, Carroll CJ, Raimundo N, Ahola-Erkkila S, Wenz T, Ruhanen H, Guse K, Hemminki A, Peltola-Mjosund KE, Tulkki V, Oresic M, Moraes CT, Pietiläinen K, Hovatta I, Suomalainen A. Mitochondrial myopathy induces a starvation-like response. Hum Mol Genet. 2010;19:3948–3958. doi: 10.1093/hmg/ddq310. [DOI] [PubMed] [Google Scholar]
- 136.Cardamone MD, Tanasa B, Cederquist CT, Huang J, Mahdaviani K, Li W, Rosenfeld MG, Liesa M, Perissi V. Mitochondrial retrograde signaling in mammals is mediated by the transcriptional cofactor GPS2 via direct mitochondria-to-nucleus translocation. Mol Cell. 2018;69:757–772.e7. doi: 10.1016/j.molcel.2018.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kim KH, Son JM, Benayoun BA, Lee C. The mitochondrial-encoded peptide MOTS-c translocates to the nucleus to regulate nuclear gene expression in response to metabolic stress. Cell Metab. 2018;28:516–524.e7. doi: 10.1016/j.cmet.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337:587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, Kaech SM, Smiley JR, Means RE, Iwasaki A, Shadel GS. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520:553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lozoya OA, Martinez-Reyes I, Wang T, Grenet D, Bushel P, Li J, Chandel N, Woychik RP, Santos JH. Mitochondrial nicotinamide adenine dinucleotide reduced (NADH) oxidation links the tricarboxylic acid (TCA) cycle with methionine metabolism and nuclear DNA methylation. PLoS Biol. 2018;16:e2005707. doi: 10.1371/journal.pbio.2005707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Picard M, Zhang J, Hancock S, Derbeneva O, Golhar R, Golik P, O’Hearn S, Levy S, Potluri P, Lvova M, Davila A, Lin CS, Perin JC, Rappaport EF, Hakonarson H, Trounce IA, Procaccio V, Wallace DC. Progressive increase in mtDNA 3243A>G heteroplasmy causes abrupt transcriptional reprogramming. Proc Natl Acad Sci U S A. 2014;111:E4033–E4042. doi: 10.1073/pnas.1414028111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Smiraglia D, Kulawiec M, Bistulfi GL, Ghoshal S, Singh KK. A novel role for mitochondria in regulating epigenetic modifications in the nucleus. Cancer Biol Ther. 2008;7:1182–1190. doi: 10.4161/cbt.7.8.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tian Y, Garcia G, Bian Q, Steffen K, Joe L, Wolff S, Meyer B, Dillin A. Mitochondrial stress induces chromatin reorganization to promote longevity and UPR(mt) Cell. 2016;165:1197–1208. doi: 10.1016/j.cell.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Vizioli MG, Liu T, Miller KN, Robertson NA, Gilroy K, Lagnado AB, Perez-Garcia A, Kiourtis C, Dasgupta N, Lei X, Kruger PJ, Nixon C, Clark W, Jurk D, Bird TG, Passos JF, Berger SL, Dou Z, Adams PD. Mitochondria-to-nucleus retrograde signaling drives formation of cytoplasmic chromatin and inflammation in senescence. Genes Dev. 2020;34:428–445. doi: 10.1101/gad.331272.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Qian W, Kumar N, Roginskaya V, Fouquerel E, Opresko PL, Shiva S, Watkins SC, Kolodieznyi D, Bruchez MP, Van Houten B. Chemoptogenetic damage to mitochondria causes rapid telomere dysfunction. Proc Natl Acad Sci U S A. 2019;116:18435–18444. doi: 10.1073/pnas.1910574116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lozoya OA, Xu F, Grenet D, Wang T, Grimm SA, Godfrey V, Waidyanatha S, Woychik RP, Santos JH. Single nucleotide resolution analysis reveals pervasive, long-lasting DNA methylation changes by developmental exposure to a mitochondrial toxicant. Cell Rep. 2020;32:108131. doi: 10.1016/j.celrep.2020.108131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Correia-Melo C, Marques FD, Anderson R, Hewitt G, Hewitt R, Cole J, Carroll BM, Miwa S, Birch J, Merz A, Rushton MD, Charles M, Jurk D, Tait SW, Czapiewski R, Greaves L, Nelson G, Bohlooly-Y M, Rodriguez-Cuenca S, Vidal-Puig A, Mann D, Saretzki G, Quarato G, Green DR, Adams PD, von Zglinicki T, Korolchuk VI, Passos JF. Mitochondria are required for pro-ageing features of the senescent phenotype. EMBO J. 2016;35:724–742. doi: 10.15252/embj.201592862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nelson G, Kucheryavenko O, Wordsworth J, von Zglinicki T. The senescent bystander effect is caused by ROS-activated NF-kB signalling. Mech Ageing Dev. 2018;170:30–36. doi: 10.1016/j.mad.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.da Silva PFL, Ogrodnik M, Kucheryavenko O, Glibert J, Miwa S, Cameron K, Ishaq A, Saretzki G, Nagaraja-Grellscheid S, Nelson G, von Zglinicki T. The bystander effect contributes to the accumulation of senescent cells in vivo. Aging Cell. 2019;18:e12848. doi: 10.1111/acel.12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ježek P, Jabůrek M. Plecitá-Hlavatá L. Contribution of oxidative stress and impaired biogenesis of pancreatic beta-cells to type 2 diabetes. Antioxid Redox Signal. 2019;31:722–751. doi: 10.1089/ars.2018.7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 152.Shadel G, Horvath T. Mitochondrial ROS signaling in organismal homeostasis. Cell. 2015;163:560–569. doi: 10.1016/j.cell.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Al-Mehdi AB, Pastukh VM, Swiger BM, Reed DJ, Patel MR, Bardwell GC, Pastukh VV, Alexeyev MF, Gillespie MN. Perinuclear mitochondrial clustering creates an oxidant-rich nuclear domain required for hypoxia-induced transcription. Sci Signal. 2012;5:ra47. doi: 10.1126/scisignal.2002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Desai R, East DA, Hardy L, Faccenda D, Rigon M, Crosby J, Alvarez MS, Singh A, Mainenti M, Hussey LK, Bentham R, Szabadkai G, Zappulli V, Dhoot GK, Romano LE, Xia D, Coppens I, Hamacher-Brady A, Chapple JP, Abeti R, Fleck RA, Vizcay-Barrena G, Smith K, Campanella M. Mitochondria form contact sites with the nucleus to couple prosurvival retrograde response. Sci Adv. 2020;6:eabc9955. doi: 10.1126/sciadv.abc9955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chouchani ET, Kazak L, Jedrychowski MP, Lu GZ, Erickson BK, Szpyt J, Pierce KA, Laznik-Bogoslavski D, Vetrivelan R, Clish CB, Robinson AJ, Gygi SP, Spiegelman BM. Mitochondrial ROS regulate thermogenic energy expenditure and sulfenylation of UCP1. Nature. 2016;532:112–116. doi: 10.1038/nature17399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ježek P, Jabůrek M, Porter RK. Uncoupling mechanism and redox regulation of mitochondrial uncoupling protein 1 (UCP1) Biochim Biophys Acta. 2019;1860:259–269. doi: 10.1016/j.bbabio.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 157.Fu ZX, Tan X, Fang H, Lau PM, Wang X, Cheng H, Bi GQ. Dendritic mitoflash as a putative signal for stabilizing long-term synaptic plasticity. Nat Commun. 2017;8:31. doi: 10.1038/s41467-017-00043-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Horn A, Raavicharla S, Shah S, Cox D, Jaiswal JK. Mitochondrial fragmentation enables localized signaling required for cell repair. J Cell Biol. 2020;219:e201909154. doi: 10.1083/jcb.201909154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Fuhrmann DC, Brune B. Mitochondrial composition and function under the control of hypoxia. Redox Biol. 2017;12:208–215. doi: 10.1016/j.redox.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pan Y, Mansfield KD, Bertozzi CC, Rudenko V, Chan DA, Giaccia AJ, Simon MC. Multiple factors affecting cellular redox status and energy metabolism modulate hypoxia-inducible factor prolyl hydroxylase activity in vivo and in vitro. Mol Cell Biol. 2007;27:912–925. doi: 10.1128/MCB.01223-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Appelhoff RJ, Tian YM, Raval RR, Turley H, Harris AL, Pugh CW, Ratcliffe PJ, Gleadle JM. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J Biol Chem. 2004;279:38458–38465. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- 162.Lu H, Dalgard CL, Mohyeldin A, McFate T, Tait AS, Verma A. Reversible inactivation of HIF-1 prolyl hydroxylases allows cell metabolism to control basal HIF-1. J Biol Chem. 2005;280:41928–41939. doi: 10.1074/jbc.M508718200. [DOI] [PubMed] [Google Scholar]
- 163.Samanta D, Semenza GL. Maintenance of redox homeostasis by hypoxia-inducible factors. Redox Biol. 2017;13:331–335. doi: 10.1016/j.redox.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 165.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 166.Semenza GL. Regulation of Erythropoiesis by the Hypoxia-Inducible Factor Pathway: Effects of Genetic and Pharmacological Perturbations. Annu Rev Med. 2023;74:307–319. doi: 10.1146/annurev-med-042921-102602. [DOI] [PubMed] [Google Scholar]
- 167.Brocato J, Chervona Y, Costa M. Molecular responses to hypoxia-inducible factor 1alpha and beyond. Mol Pharmacol. 2014;85:651–657. doi: 10.1124/mol.113.089623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Schodel J, Oikonomopoulos S, Ragoussis J, Pugh CW, Ratcliffe PJ, Mole DR. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood. 2011;117:e207–e217. doi: 10.1182/blood-2010-10-314427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Semenza GL. The Genomics and Genetics of Oxygen Homeostasis. Annu Rev Genomics Hum Genet. 2020;21:183–204. doi: 10.1146/annurev-genom-111119-073356. [DOI] [PubMed] [Google Scholar]
- 171.Wicks EE, Semenza GL. Hypoxia-inducible factors: cancer progression and clinical translation. J Clin Invest. 2022;132:e159839. doi: 10.1172/JCI159839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zepeda AB, Pessoa A, Jr, Castillo RL, Figueroa CA, Pulgar VM, Farias JG. Cellular and molecular mechanisms in the hypoxic tissue: Role of HIF-1 and ROS. Cell Biochem Funct. 2013;31:451–459. doi: 10.1002/cbf.2985. [DOI] [PubMed] [Google Scholar]
- 173.Gerald D, Berra E, Frapart YM, Chan DA, Giaccia AJ, Mansuy D, Pouyssegur J, Yaniv M, Mechta-Grigoriou F. JunD reduces tumor angiogenesis by protecting cells from oxidative stress. Cell. 2004;118:781–794. doi: 10.1016/j.cell.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 174.Briggs KJ, Koivunen P, Cao S, Backus KM, Olenchock BA, Patel H, Zhang Q, Signoretti S, Gerfen GJ, Richardson AL, Witkiewicz AK, Cravatt BF, Clardy J, Kaelin WG., Jr Paracrine Induction of HIF by Glutamate in Breast Cancer: EglN1 Senses Cysteine. Cell. 2016;166:126–139. doi: 10.1016/j.cell.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Chowdhury R, Flashman E, Mecinovic J, Kramer HB, Kessler BM, Frapart YM, Boucher JL, Clifton IJ, McDonough MA, Schofield CJ. Studies on the reaction of nitric oxide with the hypoxia-inducible factor prolyl hydroxylase domain 2 (EGLN1) J Mol Biol. 2011;410:268–279. doi: 10.1016/j.jmb.2011.04.075. [DOI] [PubMed] [Google Scholar]
- 176.Lee G, Won HS, Lee YM, Choi JW, Oh TI, Jang JH, Choi DK, Lim BO, Kim YJ, Park JW, Puigserver P, Lim JH. Oxidative Dimerization of PHD2 is Responsible for its Inactivation and Contributes to Metabolic Reprogramming via HIF-1alpha Activation. Sci Rep. 2016;6:18928. doi: 10.1038/srep18928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hewitson KS, Lienard BM, McDonough MA, Clifton IJ, Butler D, Soares AS, Oldham NJ, McNeill LA, Schofield CJ. Structural and mechanistic studies on the inhibition of the hypoxia-inducible transcription factor hydroxylases by tricarboxylic acid cycle intermediates. J Biol Chem. 2007;282:3293–3301. doi: 10.1074/jbc.M608337200. [DOI] [PubMed] [Google Scholar]
- 178.Koivunen P, Hirsila M, Remes AM, Hassinen IE, Kivirikko KI, Myllyharju J. Inhibition of hypoxia-inducible factor (HIF) hydroxylases by citric acid cycle intermediates: Possible links between cell metabolism and stabilization of HIF. J Biol Chem. 2007;282:4524–4532. doi: 10.1074/jbc.M610415200. [DOI] [PubMed] [Google Scholar]
- 179.Martínez-Reyes I, Diebold LP, Kong H, Schieber M, Huang H, Hensley CT, Mehta MM, Wang T, Santos JH, Woychik R, Dufour E, Spelbrink JN, Weinberg SE, Zhao Y, DeBerardinis RJ, Chandel NS. TCA Cycle and Mitochondrial Membrane Potential Are Necessary for Diverse Biological Functions. Mol Cell. 2016;61:199–209. doi: 10.1016/j.molcel.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bell EL, Klimova TA, Eisenbart J, Moraes CT, Murphy MP, Budinger GR, Chandel NS. The Qo site of the mitochondrial complex III is required for the transduction of hypoxic signaling via reactive oxygen species production. J Cell Biol. 2007;177:1029–1036. doi: 10.1083/jcb.200609074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Nguyen LK, Cavadas MA, Scholz CC, Fitzpatrick SF, Bruning U, Cummins EP, Tambuwala MM, Manresa MC, Kholodenko BN, Taylor CT, Cheong A. A dynamic model of the hypoxia-inducible factor 1alpha (HIF-1alpha) network. J Cell Sci. 2013;126:1454–1463. doi: 10.1242/jcs.119974. [DOI] [PubMed] [Google Scholar]
- 182.Plecitá-Hlavatá L, Ježek J, Ježek P. Aglycemia keeps mitochondrial oxidative phosphorylation under hypoxic conditions in HepG2 cells. J Bioenerg Biomembr. 2015;47:467–476. doi: 10.1007/s10863-015-9628-6. [DOI] [PubMed] [Google Scholar]
- 183.Hernansanz-Agustín P, Ramos E, Navarro E, Parada E, Sánchez-López N, Peláez-Aguado L, Cabrera-García JD, Tello D, Buendia I, Marina A, Egea J, López MG, Bogdanova A, Martínez-Ruiz A. Mitochondrial complex I deactivation is related to superoxide production in acute hypoxia. Redox Biol. 2017;12:1040–1051. doi: 10.1016/j.redox.2017.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, Schumacker PT. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc Natl Acad Sci U S A. 1998;95:11715–11720. doi: 10.1073/pnas.95.20.11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Comito G, Calvani M, Giannoni E, Bianchini F, Calorini L, Torre E, Migliore C, Giordano S, Chiarugi P. HIF-1alpha stabilization by mitochondrial ROS promotes Met-dependent invasive growth and vasculogenic mimicry in melanoma cells. Free Radic Biol Med. 2011;51:893–904. doi: 10.1016/j.freeradbiomed.2011.05.042. [DOI] [PubMed] [Google Scholar]
- 186.Guzy RD, Hoyos B, Robin E, Chen H, Liu L, Mansfield KD, Simon MC, Hammerling U, Schumacker PT. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1:401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 187.Schroedl C, McClintock DS, Budinger GR, Chandel NS. Hypoxic but not anoxic stabilization of HIF-1alpha requires mitochondrial reactive oxygen species. Am J Physiol. 2002;283:L922–L931. doi: 10.1152/ajplung.00014.2002. [DOI] [PubMed] [Google Scholar]
- 188.Ježek J, Plecitá-Hlavatá L, Ježek P. Aglycemic HepG2 cells switch from aminotransferase glutaminolytic pathway of pyruvate utilization to complete Krebs cycle at hypoxia. Front Endocrinol. 2018;9:637. doi: 10.3389/fendo.2018.00637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Patten DA, Lafleur VN, Robitaille GA, Chan DA, Giaccia AJ, Richard DE. Hypoxia-inducible factor-1 activation in nonhypoxic conditions: The essential role of mitochondrial-derived reactive oxygen species. Mol Biol Cell. 2010;21:3247–3257. doi: 10.1091/mbc.e10-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Brunelle JK, Bell EL, Quesada NM, Vercauteren K, Tiranti V, Zeviani M, Scarpulla RC, Chandel NS. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab. 2005;1:409–414. doi: 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 191.Pi J, Bai Y, Zhang Q, Wong V, Floering LM, Daniel K, Reece JM, Deeney JT, Andersen ME, Corkey BE, Collins S. Reactive oxygen species as a signal in glucose-stimulated insulin secretion. Diabetes. 2007;56:1783–1791. doi: 10.2337/db06-1601. [DOI] [PubMed] [Google Scholar]
- 192.Ashcroft FM, Rorsman P. Diabetes mellitus and the β cell: the last ten years. Cell. 2012;148:1160–1171. doi: 10.1016/j.cell.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ho T, Potapenko E, Davis DB, Merrins MJ. A plasma membrane-associated glycolytic metabolon is functionally coupled to KATP channels in pancreatic α and β cells from humans and mice. Cell Rep. 2023;42:112394. doi: 10.1016/j.celrep.2023.112394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Plecitá-Hlavatá L, Jabůrek M, Holendová B, Tauber J, Pavluch V, Berková Z, Cahová M, Schröder K, Brandes RP, Siemen D, Ježek P. Glucose-Stimulated Insulin Secretion Fundamentally Requires H2O2 Signaling by NADPH Oxidase 4. Diabetes. 2020;69:1341–1354. doi: 10.2337/db19-1130. [DOI] [PubMed] [Google Scholar]
- 195.Yosida M, Dezaki K, Uchida K, Kodera S, Lam NV, Ito K, Rita RS, Yamada H, Shimomura K, Ishikawa SE, Sugawara H, Kawakami M, Tominaga M, Yada T, Kakei M. Involvement of cAMP/EPAC/TRPM2 activation in glucose- and incretin-induced insulin secretion. Diabetes. 2014;63:3394–3403. doi: 10.2337/db13-1868. [DOI] [PubMed] [Google Scholar]
- 196.Spégel P, Sharoyko VV, Goehring I, Danielsson AP, Malmgren S, Nagorny CL, Andersson LE, Koeck T, Sharp GW, Straub SG, Wollheim CB, Mulder H. Time-resolved metabolomics analysis of β-cells implicates the pentose phosphate pathway in the control of insulin release. Biochem J. 2013;450:595–605. doi: 10.1042/BJ20121349. [DOI] [PubMed] [Google Scholar]
- 197.Plecitá-Hlavatá L, Engstová H, Holendová B, Tauber J, Špaček T, Petrásková L, Křen V, Špačková J, Gotvaldová K, Ježek J, Dlasková A, Smolková K, Ježek P. Mitochondrial Superoxide Production Decreases on Glucose-Stimulated Insulin Secretion in Pancreatic β Cells Due to Decreasing Mitochondrial Matrix NADH/NAD+ Ratio. Antioxid Redox Signal. 2020;33:789–815. doi: 10.1089/ars.2019.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Jitrapakdee S, Wutthisathapornchai A, Wallace JC, MacDonald MJ. Regulation of insulin secretion: role of mitochondrial signalling. Diabetologia. 2010;53:1019–1032. doi: 10.1007/s00125-010-1685-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Smolková K, Dvořák A, Zelenka J, Vítek L, Ježek P. Reductive carboxylation and 2-hydroxyglutarate formation by wild-type IDH2 in breast carcinoma cells. Int J Biochem Cell Biol. 2015;65:125–133. doi: 10.1016/j.biocel.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 200.Lenzen S, Drinkgern J, Tiedge M. Low antioxidant enzyme gene expression in pancreatic islets compared with various other mouse tissues. Free Radic Biol Med. 1996;20:463–466. doi: 10.1016/0891-5849(96)02051-5. [DOI] [PubMed] [Google Scholar]
- 201.Ježek P, Dlasková A, Plecitá-Hlavatá L. Redox Homeostasis in Pancreatic β Cells. Oxid Med Cell Longev. 2012;2012:932838. doi: 10.1155/2012/932838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lenzen S. Oxidative stress: the vulnerable beta-cell. Biochem Soc Trans. 2008;36:343–347. doi: 10.1042/BST0360343. [DOI] [PubMed] [Google Scholar]
- 203.Prentki M, Matschinsky FM, Madiraju SR. Metabolic signaling in fuel-induced insulin secretion. Cell Metab. 2013;18:162–185. doi: 10.1016/j.cmet.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 204.Carpinelli AR, Picinato MC, Stevanato E, Oliveira HR, Curi R. Insulin secretion induced by palmitate-a process fully dependent on glucose concentration. Diabetes Metab. 2002;28(3):S37–44. [PubMed] [Google Scholar]
- 205.Gehrmann W, Elsner M, Lenzen S. Role of metabolically generated reactive oxygen species for lipotoxicity in pancreatic β-cells. Diab Obes Metab. 2010;12(Suppl 2):149–158. doi: 10.1111/j.1463-1326.2010.01265.x. [DOI] [PubMed] [Google Scholar]
- 206.Graciano MF, Valle MM, Kowluru A, Curi R, Carpinelli AR. Regulation of insulin secretion and reactive oxygen species production by free fatty acids in pancreatic islets. Islets. 2011;3:213–223. doi: 10.4161/isl.3.5.15935. [DOI] [PubMed] [Google Scholar]
- 207.Cen J, Sargsyan E, Bergsten P. Fatty acids stimulate insulin secretion from human pancreatic islets at fasting glucose concentrations via mitochondria-dependent and -independent mechanisms. Nutr Metab (Lond) 2016;13:59. doi: 10.1186/s12986-016-0119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Fernandez J, Valdeolmillos M. Increased levels of free fatty acids in fasted mice stimulate in vivo beta-cell electrical activity. Diabetes. 1998;47:1707–1712. doi: 10.2337/diabetes.47.11.1707. [DOI] [PubMed] [Google Scholar]
- 209.Hauke S, Keutler K, Phapale P, Yushchenko DA, Schultz C. Endogenous Fatty Acids Are Essential Signaling Factors of Pancreatic β-Cells and Insulin Secretion. Diabetes. 2018;67:1986–1998. doi: 10.2337/db17-1215. [DOI] [PubMed] [Google Scholar]
- 210.Ježek P, Jabůrek M, Holendová B, Plecitá-Hlavatá L. Fatty Acid-Stimulated Insulin Secretion vs. Lipotoxicity. Molecules. 2018;23:1483. doi: 10.3390/molecules23061483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Saadeh M, Ferrante TC, Kane A, Shirihai O, Corkey BE, Deeney JT. Reactive oxygen species stimulate insulin secretion in rat pancreatic islets: studies using mono-oleoyl-glycerol. PLoS One. 2012;7:e30200. doi: 10.1371/journal.pone.0030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Leloup C, Tourrel-Cuzin C, Magnan C, Karaca M, Castel J, Carneiro L, Colombani AL, Ktorza A, Casteilla L, Pénicaud L. Mitochondrial reactive oxygen species are obligatory signals for glucose-induced insulin secretion. Diabetes. 2009;58:673–681. doi: 10.2337/db07-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Yang HQ, Martinez-Ortiz W, Hwang J, Fan X, Cardozo TJ, Coetzee WA. Palmitoylation of the K(ATP)channel Kir6.2 subunit promotes channel opening by regulating PIP(2) sensitivity. Proc Natl Acad Sci U S A. 2020;117:10593–10602. doi: 10.1073/pnas.1918088117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Shigeto M, Ramracheya R, Tarasov AI, Cha CY, Chibalina MV, Hastoy B, Philippaert K, Reinbothe T, Rorsman N, Salehi A, Sones WR, Vergari E, Weston C, Gorelik J, Katsura M, Nikolaev VO, Vennekens R, Zaccolo M, Galione A, Johnson PR, Kaku K, Ladds G, Rorsman P. GLP-1 stimulates insulin secretion by PKC-dependent TRPM4 and TRPM5 activation. J Clin Invest. 2015;125:4714–4728. doi: 10.1172/JCI81975. [DOI] [PMC free article] [PubMed] [Google Scholar]