Abstract
We have previously described two replication-competent adenovirus vectors, named KD1 and KD3, for potential use in cancer gene therapy. KD1 and KD3 have two small deletions in the E1A gene that restrict efficient replication of these vectors to human cancer cell lines. These vectors also have increased capacity to lyse cells and spread from cell to cell because they overexpress the adenovirus death protein, an adenovirus protein required for efficient cell lysis and release of adenovirus from the cell. We now describe a new vector, named KD1-SPB, which is the KD1 vector with the E4 promoter replaced by the promoter for surfactant protein B (SPB). SPB promoter activity is restricted in the adult to type II alveolar epithelial cells and bronchial epithelial cells. Because KD1-SPB has the E1A mutations, it should replicate within and destroy only alveolar and bronchial cancer cells. We show that KD1-SPB replicates, lyses cells, and spreads from cell to cell as well as does KD1 in H441 cells, a human cancer cell line where the SPB promoter is active. KD1-SPB replicates, lyses cells, and spreads only poorly in Hep3B liver cancer cells. Replication was determined by expression of the E4ORF3 protein, viral DNA accumulation, fiber synthesis, and virus yield. Cell lysis and vector spread were measured by lactate dehydrogenase release and a “vector spread” assay. In addition to Hep3B cells, KD1-SPB also did not express E4ORF3 in HT29.14S (colon), HeLa (cervix), KB (nasopharynx), or LNCaP (prostate) cancer cell lines, in which the SPB promoter is not expected to be active. Following injection into H441 or Hep3B tumors growing in nude mice, KD1-SPB caused a three- to fourfold suppression of growth of H441 tumors, similar to that seen with KD1. KD1-SPB had only a minimal effect on the growth of Hep3B tumors, whereas KD1 again caused a three- to fourfold suppression. These results establish that the adenovirus E4 promoter can be replaced by a tissue-specific promoter in a replication-competent vector. The vector has three engineered safety features: the tissue-specific promoter, the mutations in E1A that preclude efficient replication in nondividing cells, and a deletion of the E3 genes which shield the virus from attack by the immune system. KD1-SPB may have use in treating human lung cancers in which the SPB promoter is active.
Human adenovirus serotype 5 (Ad5) offers great promise in cancer gene therapy. It primarily causes asymptomatic or mild respiratory infections in young children, followed by long-term effective immunity (24). Fatalities are extremely rare, except when the patient is immunocompromised. Ad5 is very well understood, can be grown in culture to high-titered stocks that are stable, and can replicate in most human cancer cell types (46). Its genome can be manipulated by site-directed mutagenesis and insertion of foreign sequences.
Most Ad vectors are replication defective (RD) because they lack the genes in the E1A and E1B transcription units (7). The E1A proteins induce transcription of other Ad genes, and in nontransformed cells, they deregulate the cell cycle, induce or repress a variety of cellular genes, and force cells from G0 into S phase (57, 62). The E1B proteins inhibit cellular apoptosis (57, 62). In RD vectors, the E1A and E1B regions are replaced by an anticancer therapeutic gene, e.g., p53, expressed from a foreign promoter-enhancer (65). Since they cannot replicate, RD vectors express their therapeutic protein only in the cells which they initially infect, and that can be only a small fraction of the cells in the tumor. Some of these therapeutic proteins or their products exhibit a bystander effect where cells proximal to the infected cell are destroyed.
Some Ad vectors are replication competent (RC) (22). When these vectors infect a cell, they go through their natural cycle of replication, lysis of the cell, vector release from the cell, and reinfection of other cells. These vectors should be capable of destroying more cells in the tumor than RD vectors. In order to prevent RC vectors from damaging normal tissues and causing disseminated viremia, it is important that they have some feature that limits their replication to cancer cells. The RC vector ONYX-015, which was originally named dl1520 (2), has a deletion in the E1B transcription unit that removes the gene for E1B-55K (2, 4, 20, 21). E1B-55K is multifunctional: it is required for efficient cytoplasmic accumulation and translation of Ad mRNAs, it binds the tumor suppressor p53 and represses p53-responsive promoters, it (acting together with the Ad E4ORF6 protein) induces degradation of p53, and it inhibits p53-induced apoptosis, thereby enabling cells to enter S phase (11, 17, 23, 29, 30, 36, 41, 43, 49, 50, 59, 66, 67). Replication of ONYX-015 was originally reported to be restricted to tumor cells lacking wild-type p53 (20, 56), but it is now known that it can replicate in cells with wild-type p53 (15, 17, 19, 42, 55). ONYX-015 partially suppresses growth of p53-negative xenotransplants (20, 56) and has shown safety and suggestive clinical benefit in phase I and II clinical trials of head and neck cancer (13, 26). Efforts have been made to improve the specificity and efficacy of ONYX-015. For example, addition of a polylysine tail to the fiber gene caused the vector to be targeted to gliomas (48). In another approach, the combined use of ONYX-015 with cisplatin and 5-fluorouracil (chemotherapeutic drugs) (20, 68) or radiation (39, 40) improved the ability of ONYX-015 to retard the growth of xenotransplants. ONYX-015 also synergized with chemotherapy in cytolysis of primary cultures from human lung cancers (68). In other E1B-55K deletion vectors, introduction of the herpes simplex virus type 1 thymidine kinase (TK) cancer therapeutic gene (60, 61) or a fusion gene between TK and Escherichia coli cytosine deaminase (10, 60) into the vector increased the potency of the vector. Further, vector-mediated expression of interleukin 2 potentiated immune responses against vector-infected tumors in mice (33).
Another strategy for RC vector improvement has been to place replication under the control of tissue-specific promoters. One group replaced the basal E1A promoter with a modified promoter for α-fetoprotein (AFP) (16). AFP is expressed in the liver during development, but it is not expressed in adults. However, it is expressed in 70 to 80% of patients with hepatocellular carcinoma. Growth of this vector was limited to AFP-expressing cells, and the vector showed some suppression of xenotransplants (16). A number of RC vectors have been developed that have expression of the E1A and E1B genes dependent on the prostate tumor-specific prostate-specific antigen and kallikrein (38, 69, 70) promoters-enhancers.
We have described two RC vectors, named KD1 and KD3 (Fig. 1), which have unique features (8). First, KD1 and KD3 are more cytolytic than Ad5 or the Ad5 mutant dl309, resulting in faster spread of vector from cell to cell in culture. KD1 and KD3 are more cytolytic because they overexpress the adenovirus death protein (ADP), an 11.6-kDa Ad membrane glycoprotein synthesized at very late stages of infection when virus has assembled in the cell nucleus, which mediates efficient cell lysis and release of Ad from cells (44, 52–54). Second, KD1 and KD3 contain two small deletions in the E1A gene that abolish the ability of E1A to deregulate the cell cycle and cause cells to move from G0 to S phase (25). These deletions leave intact the ability of E1A to transactivate Ad genes. Replication of KD1 and KD3 is very poor in human quiescent or primary cells, but it is very good in many cancer cell lines. KD1 and KD3 suppress the growth of human A549 (lung cancer) and Hep3B (hepatocellular carcinoma) tumors in immunodeficient nude mice (8). Third, KD1 and KD3 lack the E3 region genes that protect Ad-infected cells from destruction by the immune system (62, 63); this feature could mitigate against possible runaway viremia by the vector (8).
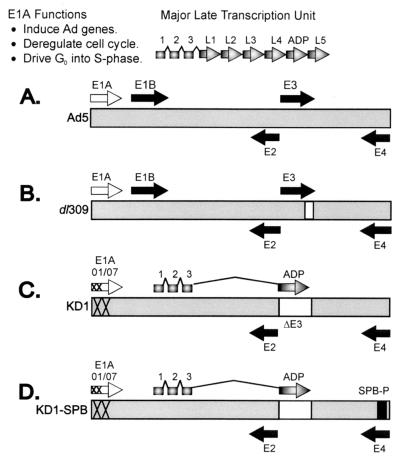
FIG. 1.
Schematic of vectors. (A) Ad5. The gray bar indicates the DNA genome of 36 kbp. The open arrow indicates the immediate-early E1A transcription unit, and the black arrows are the delayed-early E1B, E2, E3, and E4 transcription units. The upper shaded arrows indicate the five families of major late mRNAs and also the ADP mRNA, which is synthesized as part of the major late transcription unit. Each major late mRNA has a tripartite leader (leaders 1, 2, and 3) spliced to its 5′ terminus. (B) dl309. dl309 is identical to Ad5, except that it has the E3-RID and E3-14.7K genes deleted. dl309 expresses ADP at levels similar to those of Ad5. (C) KD1. KD1 has two small deletions (indicated by “X” marks) in the E1A gene that abolish binding of the E1A proteins to pRB or p300/CBP. It lacks all E3 genes except adp. ADP is expressed earlier in infection and in greater abundance than is ADP from Ad5 or dl309 (8). (D) KD1-SPB. KD1-SPB is identical to KD1, except that it has the E4 promoter replaced by the promoter for SPB (SPB-P).
In this communication, we describe the construction and characterization of a version of KD1 that is not only tumor selective but also tissue specific. Accordingly, this vector should display increased safety. In this vector, the Ad E4 promoter is deleted and replaced with the promoter for surfactant protein B (SPB) (Fig. 1). The new vector is named KD1-SPB. The regulation of the SPB promoter is cell type specific: it is active nearly exclusively in type II alveolar epithelial cells and bronchial epithelial cells (Clara cells) of the lung (5, 64). The E4 region is required for Ad DNA replication and late mRNA maturation (28). Because of the E4 promoter substitution and the E1A mutation, KD1-SPB is expected to replicate efficiently in lung tumors, where both the SPB promoter and the E1A proteins are active, but not in normal lung tissues or in other types of tissues. Indeed, we show here that KD1-SPB replicates well in H441 papillary lung adenocarcinoma cells but very poorly in Hep3B cells. In addition, KD1-SPB suppresses the growth of H441 tumors but not the growth of Hep3B tumors in nude mice.
MATERIALS AND METHODS
Cells and viruses.
Human cancer cell lines A549 (human lung carcinoma), Hep3B (human hepatocellular carcinoma), and LNCaP (prostate carcinoma) were obtained from the American Type Culture Collection. H441 (papillary lung carcinoma) cells were obtained from Jeffrey Whitsett (University of Cincinnati), HeLa (cervical carcinoma) cells were from Eileen White (Rutgers University), HT29.14S (colon carcinoma) cells were from Jeff Browning (Biogen, Cambridge, Mass.), and KB cells were from Maurice Green (St. Louis University). HEK 293 cells were obtained from Microbix (Toronto, Ontario, Canada). VK10-9 cells are 293 cells that in addition to E1 contain and express E4 and pIX (27). We will refer to these cells as 293-E4 cells. A549, Hep3B, HeLa, 293, and 294-E4 cells were grown in Dulbecco's modified essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS). H441 and LNCaP cells were grown in RPMI medium 1640 (HyClone) plus 10% FBS. HT29.14S cells were grown in McCoy's 5A medium (Gibco/BRL) plus 10% FBS.
The Ad KD1 and KD3 vectors overexpressing ADP and restricted to tumor cells by mutations in the E1A region have been previously described (8). The E1A deletions were derived from the Ad5 mutant dl1101/1107 (obtained from Stanley Bayley, McMaster University). Ad KD1-SPB is a modification of KD1 with the E4 promoter replaced by the lung-specific SPB promoter. KD1-SPB was constructed starting with plasmid p491, which is based on the pCRII (Invitrogen) backbone and contains Ad5 sequences from BamHI (21,595 bp in Ad5) to the end of the viral genome (35,935 bp). p491 has modifications in the E3 region as follows: deletion of Ad5 bp 27858 to 27860, TAA inserted; deletion of Ad5 bp 27982 to 28134; deletion of Ad5 bp 28395 to 29397, CCTTAATTAAA inserted; deletion of Ad5 bp 29783 to 30883, TTAATTAAGG inserted. p491 also has a modification of the E4 region as follows: deletion of Ad5 E4 promoter, bp 35623 to 35775, and insertion of the SPB B-500 promoter (64) (obtained from Jeffrey Whitsett) flanked by Bst1107I sites.
dl1101/1107 DNA was digested with EcoRI and then was cotransfected into 293-E4 cells together with p491 DNA using the calcium phosphate technique. The resulting plaques were screened by PCR for the presence of recombinant versions of E1A, E3, and E4. One plaque was purified three times and expanded into a large-scale CsCl stock, and the titer was determined on 293-E4 cells (51). Viral DNA from the CsCl preparation was screened for the presence of recombinant sequences and absence of wild-type sequences by PCR, restriction enzyme digestion, and DNA sequencing. The ratios of particles to PFU, as determined by Mittereder et al. (32), were 1,750 for KD1-SPB and 1,380 for KD1.
Plaque development assay.
The plaque development assay was performed as described previously (51, 53). In brief, serial dilutions of the viruses were prepared and plaques were assayed on monolayers of 293-E4, 293, and A549 cells. Visible plaques were counted at 1- to 3-day intervals. The number of plaques on a given day was plotted as a percentage of plaques seen on the final day of the assay.
Virus spread assay.
Hep3B or H441 cells were seeded in 48-well plates and were mock infected or infected with 10-fold serial dilutions of the indicated viruses, ranging from 101 to 10−4 PFU/cell. Monolayers were fixed and stained with crystal violet at 5 or 8 days postinfection (p.i.), as described previously (8).
Western blots.
H441 or Hep3B cells (in 60-mm-diameter dishes) were infected with 10 PFU of KD1 or KD1-SPB/cell. At 24 h p.i., the cells were washed three times with phosphate-buffered saline (PBS) and harvested by scraping. The cells were lysed by radioimmunoprecipitation assay buffer. The protein concentration was measured by the Bio-Rad DC protein assay kit (Bio-Rad Laboratories, Hercules, Calif.), and 10 μg of each sample was electrophoresed on sodium dodecyl sulfate–15% polyacrylamide gels. The gels were electroblotted onto polyvinylidene difluoride membranes (Immobilon; Millipore, Bedford, Mass.). The membranes were blocked in TBST (50 mM Tris-Cl [pH 7.6], 150 mM NaCl, 0.2% Tween 20) containing 10% dry milk (Carnation) overnight at 4°C. After blocking, the membranes were incubated with a rabbit polyclonal antiserum against E4ORF3 (gift of Gary Ketner) or ADP (54) or with M73, a monoclonal antibody against E1A (18). The secondary antibodies were goat anti-rabbit immunoglobulin G (IgG)-horseradish peroxidase or goat anti-mouse IgG-horseradish peroxidase. The blots were developed using the ECL protocol (Amersham Pharmacia, Arlington Heights, Ill.).
Lactate dehydrogenase release assay.
For the lactate dehydrogenase release assay for cell lysis (53), H441 cells (7.7 × 105 cells per 35-mm dish) and Hep3B cells (9.0 × 105 cells per 35-mm dish) were infected at 20 PFU/cell in 1 ml of serum-free DMEM. After an adsorption period of 1 h, 3 ml of DMEM (10% FBS) was added (final FBS concentration of 7.5%). Cells were incubated at 37°C with 6% CO2. At daily intervals, supernatants were collected and microcentrifuged to remove floating cells, and cell-free supernatants were frozen at −70°C until assayed. Total lysis samples were prepared by addition of 10× lysis buffer included in the Cyto Tox 96 kit (Promega, Madison, Wis.). After all samples were collected, 20-μl samples were assayed in triplicate using the lactate dehydrogenase assay kit Cyto Tox 96 and read on an EL340 microplate reader (BioTec Instruments, Inc.) at 490 nm.
Immunofluorescence.
In the experiment shown in Fig. 6, H441 and Hep3B cells were plated on Corning no. 1 coverslips in 35-mm dishes. H441 (1.5 × 106 cells/35-mm dish) and Hep3B (9.0 × 105 cells/35-mm dish) cells were infected with 20 PFU of the indicated viruses/cell in 1 ml of serum-free DMEM. After 1 h, 1 ml of DMEM–20% FBS was added (final FBS concentration of 10%). At the indicated times (48 h or 6 days p.i.), cells were fixed for 10 min in 3.7% paraformaldehyde in PBS and then permeabilized for 6 min in methanol (−20°C) and rehydrated in PBS. Coverslips were stained with rabbit antiserum to E4ORF3 (1:250 dilution; gift of Gary Ketner), followed by goat anti-rabbit IgG-fluorescein isothiocyanate (FITC) conjugate (1:50 dilution; Cappel/ICN). In the experiment shown in Fig. 7, H441, Hep3B, HeLa, KB, LNCaP, or HT29.14S cells were plated onto poly-l-lysine-coated coverslips in 35-mm dishes. The cells were infected with 10 PFU of KD1-SPB or KD3/cell. At 44 h p.i., cells were fixed and permeabilized as described above. Cells were double immunostained using E4ORF3 rabbit antiserum and a pool of E1A-specific monoclonal antibodies (gift of Ed Harlow) (18), followed by goat anti-rabbit IgG-FITC conjugate and goat anti-mouse IgG-rhodamine (RITC) conjugate. All antibodies were diluted in PBS containing 1% bovine serum albumin and 0.1% sodium azide. Photographs were taken on a Nikon epifluorescence microscope using a 100× Planapo lens and Tmax 400 film (Kodak). The film was developed in a Diafine developer.
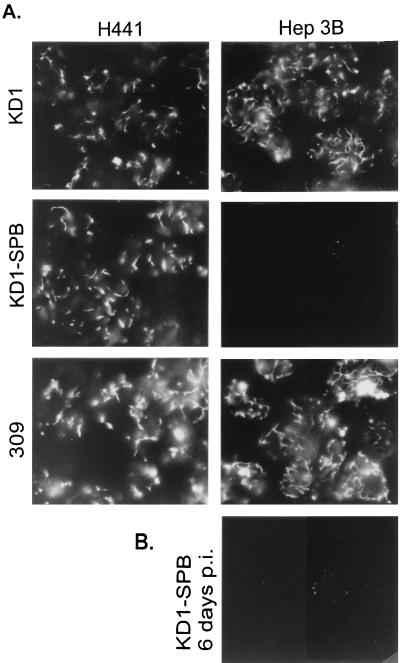
FIG. 6.
Immunofluorescence assay results showing that KD1-SPB expresses E4ORF3 in H441 but not Hep3B cells. Cells growing on coverslips were infected with 20 PFU of KD1, KD1-SPB, or dl309 (wild type) per cell. At 48 h (A) or 6 days (B), cells were fixed and stained with a rabbit polyclonal antipeptide antiserum against E4ORF3. Each panel shows about eight nuclei.
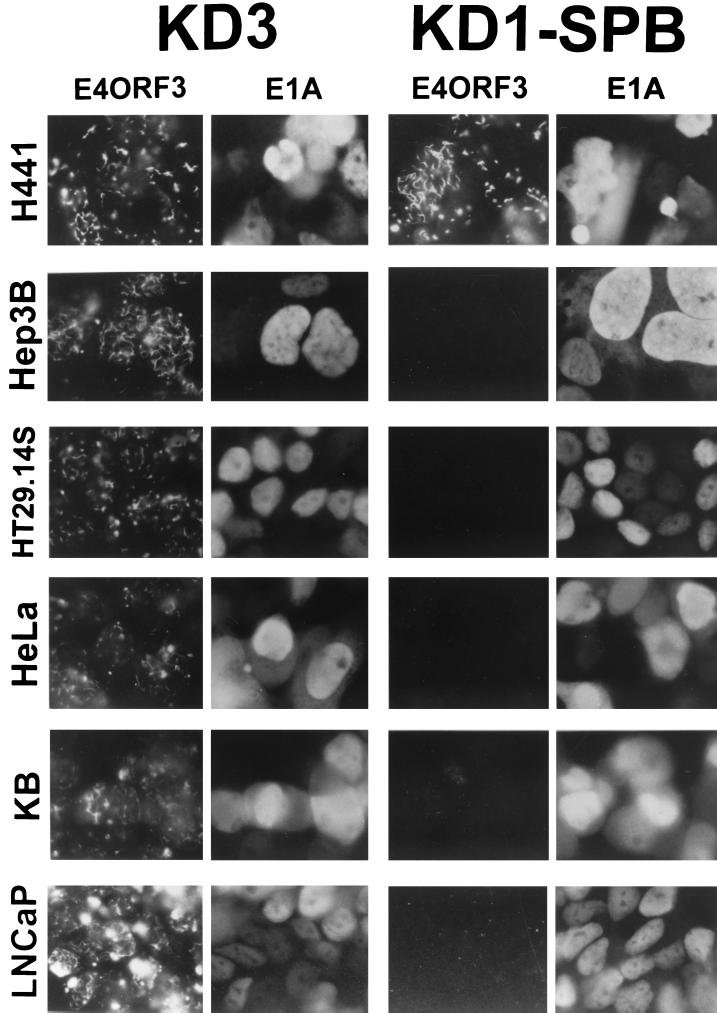
FIG. 7.
Coimmunofluorescence assay results showing that, whereas KD1-SPB infects a variety of human cancer cell types, it expresses E4ORF3 efficiently only in H441 cells. Cells were infected with 10 PFU of KD1-SPB or KD3/cell. At 44 h p.i., cells were fixed and double stained using a rabbit polyclonal antibody to E4ORF3 and a pool of mouse monoclonal antibodies to E1A. The same fields are shown for E4ORF3 and E1A.
Southern blot analysis of viral DNA replication.
H441 and Hep3B cells were grown in 60-mm dishes in DMEM supplemented with 10% FBS. Cells were infected at 70% confluency with 10 PFU of KD1 or KD1-SPB/cell. Dishes were incubated in a humidified 5% CO2 atmosphere at 37°C. Total genomic DNAs were isolated at 5, 24, 48, 72, and 96 h p.i. Equal amounts of total genomic DNAs were digested with HindIII and resolved on a 1% agarose gel prior to transfer onto membranes. A random primed 32P-labeled pBHG10 plasmid probe (3) was used for hybridization, and the blots were autoradiographed. DNA fragments were quantitated on a Molecular Dynamics PhosphorImager.
Virus yields.
Hep3B cells or H441 cells grown as monolayers in 35-mm dishes were infected with 10 PFU of KD1 or KD1-SPB/cell. At days 0 to 4 (for H441) or days 0 to 9 (for Hep3B) p.i., cells and culture medium were frozen at −70°C. Samples were frozen and thawed three times to release the virus from the cells, and total virus yields were determined by plaque assay on 293-E4 monolayers.
Tumors and intratumor injections.
A nude mouse model (8) was used to examine the effect of KD1-SPB and KD1 on H441 and Hep3B tumors. Tumor cells (107 cells in 200 μl of DMEM–50% Matrigel [Becton Dickinson Labware, Bedford, Mass.] for H441 cells or 107 cells in 200 μl of DMEM–10% Matrigel for Hep3B cells) were injected into flanks of 5- to 6-week-old athymic nude mice and allowed to grow for 3 weeks to about 100 (H441)- or 150 (Hep3B)-μl volumes. Preestablished tumors (n = 10) were injected with 50 μl of DMEM or 5 × 107 PFU of indicated viruses in DMEM. Injections of the viruses were repeated twice weekly for 3 weeks to the total dose of 3.0 × 108 PFU per tumor. Tumor size measurements were taken twice per week for H441 cells or weekly for Hep3B cells using Sylvac digital calipers. Tumor volumes were calculated according to the formula length × width2/2. Data are represented as the mean increase in tumor size relative to the tumor size at the initial injection.
RESULTS
Construction of KD1-SPB.
As described in Materials and Methods, the plasmid p491 was constructed, containing Ad5 sequences from the BamHI site at map position 60 (position 21595 in Ad5) to the right end of the genome of the vector KD1 (8) with the E4 promoter replaced by the B-500 SPB promoter (64). The deletion in E4, Ad5 bp 35623 to 35775, removes the E4F and ATF-2 transcription factor binding sites and the E4 TATA box. The deletion in E3 of 2,260 bp changes the translation initiation codon for the 12,500-molecular-weight (12.5K) protein to a stop codon, removes sequences between the pVIII gene and the L4 polyadenylation signal, removes sequences between the L4 polyadenylation signal and a position upstream of the adp gene, and removes sequences from the end of the adp gene to the end of the E3-14.7K gene. As such, all E3 genes are deleted except the adp gene. There are no methionine codons upstream of the adp gene in what remains of the E3 transcription unit. L4 mRNAs are expected to undergo polyadenylation at their natural site.
Plasmid p491 was cotransfected into 293-E4 cells together with the EcoRI-digested DNA from the Ad5 mutant dl1101/1107 (8). The EcoRI-A fragment of dl1101/1107, map positions 0 to 76, contains two deletions in the E1A gene. One deletion (amino acids 112 to 122), in conserved region 2 of the E1A proteins, removes the binding site for pRB. The other deletion (amino acids 5 to 24), in the NH2-terminal region and in conserved region 1, removes the binding site for p300/CBP. Following transfection, the complete genome of KD1-SPB forms by overlap recombination of the EcoRI-A fragment from dl1101/1107 and plasmid p491. 293-E4 cells were used for transfection because they express E4 proteins and therefore offer the recombinant genomes an increased opportunity to replicate. Plaques were picked, examined by PCR for the expected genome structure, plaque purified three times, and expanded into CsCl stocks. The titers were determined on 293-E4 cells. These stocks were confirmed by PCR, restriction enzyme digestion, and DNA sequence analysis.
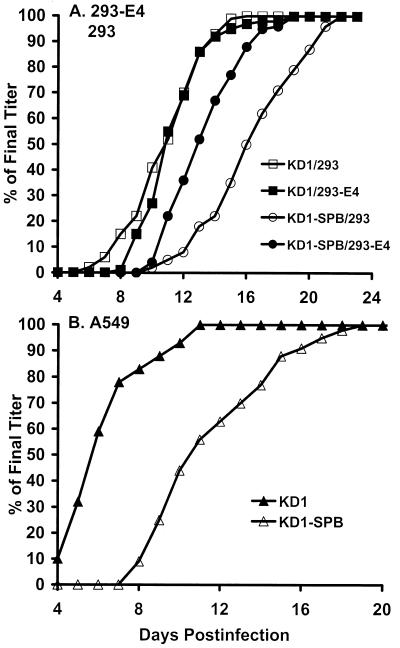
In subsequent experiments, the properties of KD1-SPB were compared to those of its “parent,” KD1. Figure 2 shows the plaque development properties of these vectors on 293-E4, 293, and A549 cells. The data are plotted as the number of plaques seen on any day of the plaque assay as a percentage of the number of plaques seen at the end of the assay (i.e., when new plaques cease to appear) (53). This assay is an indicator of the size of the plaques. KD1 formed plaques equally well on 293-E4 and 293 cells (Fig. 2A). With KD1-SPB, plaques were observed about 3 to 4 days sooner on 293-E4 cells than on 293 cells (Fig. 2A). On A549 cells, KD1 formed plaques 4 to 6 days sooner than did KD1-SPB (Fig. 2B).
FIG. 2.
Plaque development assay showing that KD1-SPB plaques form more slowly in 293 and A549 cells than do KD1 plaques. The titers of CsCl-banded stocks of KD1-SPB and KD1 were determined using standard methods (51) on 293-E4 or 293 cells (A) or on A549 cells (B). The data are plotted as the number of plaques seen on any day of the plaque assay as a percentage of the number of plaques seen on the final day of the assay (53). The final titers were as follows (PFU per milliliter): KD1 on A549 cells, 4.1 × 1010; KD1-SPB on A549 cells, 5.7 × 109; KD1 on 293 cells, 2.8 × 109; KD1-SPB on 293 cells, 3.5 × 108; KD1 on 293-E4 cells, 2.1 × 109; KD1-SPB on 293-E4 cells, 1.9 × 108.
KD1-SPB grows as well as KD1 in H441 lung carcinoma cells but much more poorly than KD1 in Hep3B hepatocellular carcinoma cells.
The properties of KD1-SPB versus KD1 were characterized in detail in H441 cells, a human papillary lung adenocarcinoma cell line known to express the TTF1 transcription factor and in which the SPB promoter is active (64). Hep3B cells, a human hepatocellular carcinoma line in which the SPB promoter should not be active, were used as a negative control. H441 and Hep3B monolayers were infected with 10 PFU of KD1 or KD1-SPB/cell and photographed at 4 and 7 days p.i. KD1 produced a cytopathic effect (CPE) on both cell lines at 4 and 7 days p.i. (data not shown). KD1-SPB caused CPE on H441 cells but not on Hep3B cells. Since CPE in Ad-infected cells is usually an indicator of virus growth, these results suggest that KD1-SPB grows in H441 but not in Hep3B cells.
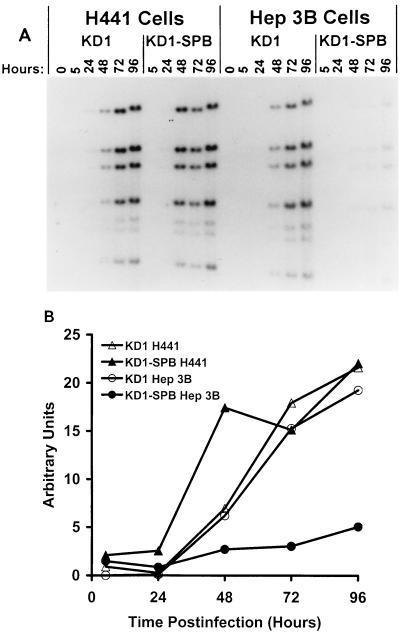
To examine viral DNA replication, H441 and Hep3B cells were infected with 10 PFU of KD1 or KD1-SPB/cell, and then the accumulation of viral DNA was determined by DNA blotting. With H441 cells, KD1 and KD1-SPB DNAs were readily detected at similar levels at 48 to 96 h p.i. (Fig. 3A). With Hep3B cells, KD1 DNA levels were similar to those in H441 cells, but KD1-SPB DNA was barely detectable. This was confirmed by PhosphorImager analysis of the DNA bands (Fig. 3B).
FIG. 3.
KD1-SPB DNA is synthesized efficiently in H441 but not Hep3B cells. H441 or Hep3B cells were infected with 10 PFU of KD1 or KD1-SPB/cell. Total genomic DNA was isolated at 0, 5, 24, 48, 72, and 96 h p.i.; digested with HindIII; resolved by agarose gel electrophoresis; blotted; and hybridized with 32P-labeled Ad DNA. (A) Autoradiogram. (B) PhosphorImager quantitation of the DNA bands in panel A.
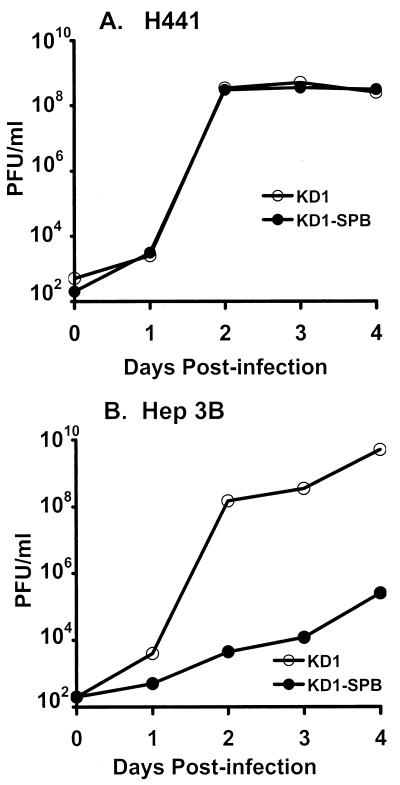
Growth of KD1-SPB and KD1 in H441 and Hep3B cells was determined by a single-step growth assay. Cells were infected with 10 PFU of vector/cell, and then total vector yield was determined by plaque assay. Total yields of both vectors were similar in H441 cells, reaching a plateau after 2 days (Fig. 4A). KD1 yield plateaued in Hep3B cells after 2 to 4 days p.i. (Fig. 4B). However, KD1-SPB levels were about 5 logs lower in Hep3B cells after 2 to 4 days, and even by 9 days, they had not achieved the levels of KD1. We conclude that KD1-SPB grows with significant specificity on H441 versus Hep3B cells. Further, KD1-SPB grows as well as does KD1 on H441 cells, indicating that the E4 promoter can be replaced by a tissue-specific promoter in an RC vector.
FIG. 4.
Single-step growth curve showing that KD1-SPB grows well in H441 but not Hep3B cells. Cells were infected with 10 PFU of KD1 or KD1-SPB/cell. Vectors were extracted at the indicated days p.i., and titers were determined by plaque assay.
The E4-encoded protein E4ORF3 is expressed well by KD1-SPB in H441 cells but not in Hep3B, HT29.14S, HeLa, KB, or LNCaP cells.
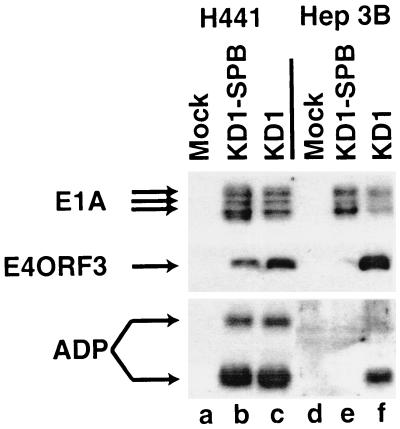
To obtain further details on the replication of KD1-SPB compared with that of KD1 in H441 and Hep3B cells, the expression of representative Ad proteins by KD1-SPB and KD1 was examined. H441 or Hep3B cells were mock infected or infected with 10 PFU of KD1 or KD1-SPB/cell, and then at 24 h p.i., the proteins were extracted and the E1A and E4ORF3 proteins and ADP were examined by immunoblotting. E4ORF3 is one of the six proteins encoded by the E4 transcription unit (28). As anticipated, KD1-SPB expressed E4ORF3 well in H441 cells but at only trace levels in Hep3B cells (Fig. 5). KD1-SPB expressed the E1A proteins in Hep3B cells. Synthesis of E1A proteins by KD1-SPB in Hep3B cells is expected because E1A expression does not require E4 proteins; it also indicates that the block to infection with KD1-SPB is downstream of E1A. KD1 expressed E1A in both cell lines (Fig. 5). KD1-SPB expressed ADP as well as did KD1 in H441 cells, but it did not make detectable ADP in Hep3B cells. ADP is primarily a late protein, and so this result is consistent with the relative lack of E4 protein expression, DNA replication, and growth of KD1-SPB in Hep3B cells.
FIG. 5.
Immunoblots showing that KD1-SPB expresses E4ORF3 and ADP in H441 but not Hep3B cells. Cells were infected with 10 PFU of KD1 or KD1-SPB/cell. At 24 h p.i., protein extracts were analyzed for E1A, E4ORF3, and ADP using specific antisera. The E1A proteins appear as multiple bands. ADP appears as two bands; the upper band is glycosylated, and the lower band is a proteolytically cleaved species (44, 54).
The immunoblot data in Fig. 5 depict E4ORF3, E1A, and ADP synthesis in the bulk cell culture. To examine the synthesis of E4ORF3 in individual cells, infected cells were characterized by immunofluorescence. H441 or Hep3B cells were infected with 20 PFU/cell. At 48 h or 6 days p.i., cells were fixed and immunostained. E4ORF3 was detected in the nuclei of H441 cells at 48 h p.i. with KD1, KD1-SPB, or dl309 (Fig. 6A) (dl309 is an Ad5 mutant that has wild-type E1A, expresses Ad5 levels of ADP, and lacks the E3-RID and E3–14.7K genes). The staining pattern for E4ORF3 was indistinguishable between KD1-SPB and KD1, suggesting that E4ORF3 (and other E4 proteins) function normally in H441 cells infected with KD1-SPB. E4ORF3 could not be detected in the vast majority of Hep3B cells infected with KD1-SPB at 48 h (Fig. 6A) or even at 6 days (Fig. 6B) p.i. Thus, KD1-SPB expresses E4ORF3 well in H441 cells but not in Hep3B cells.
The host range of KD1-SPB was further examined by immunofluorescence using four additional human cancer cell lines, HT29.14S (colon), HeLa (cervix), KB (nasopharynx), and LNCaP (prostate) (Fig. 7). As is the case with Hep3B cells, the SPB promoter is not expected to be active in these cells. Whereas KD1 and dl309 were used as positive controls in Fig. 6, KD3 was used as the positive control in Fig. 7; the properties of KD3 are very similar to those of KD1 (8). Cells in monolayers were infected with 10 PFU of KD1-SPB or KD3/cell, and then at 44 h p.i. they were fixed and double immunostained for E4ORF3 and E1A. With H441 cells, E4ORF3 was expressed well by both KD1-SPB and KD3 (Fig. 7, top). Most of the same cells that expressed E4ORF3 also expressed E1A strongly in the nucleus, the known location of E1A. Nuclei were identifiable because they were stained with 4′,6′-diamidino-2-phenylindole (DAPI; data not shown). The staining for E4ORF3 was confined largely to the nucleus, consistent with the previous finding that E4ORF3 localizes to promyelocytic leukemia oncogenic domains and causes them to be reorganized (9, 35). With all the other cell lines that were infected with KD3, there was strong staining of both E4ORF3 and E1A. In contrast, with these cell lines infected with KD1-SPB, there was very little staining for E4ORF3, even though there was strong staining for E1A in the same cells (Fig. 7). These results demonstrate that, although KD1-SPB can infect a variety of human cancer cell lines efficiently (as measured by E1A expression), it can replicate efficiently (as measured by E4ORF3 expression) only in the cell line where the SPB promoter is predicted and known to be active.
KD1-SPB lyses cells and spreads efficiently from cell to cell in H441 but not Hep3B cells.
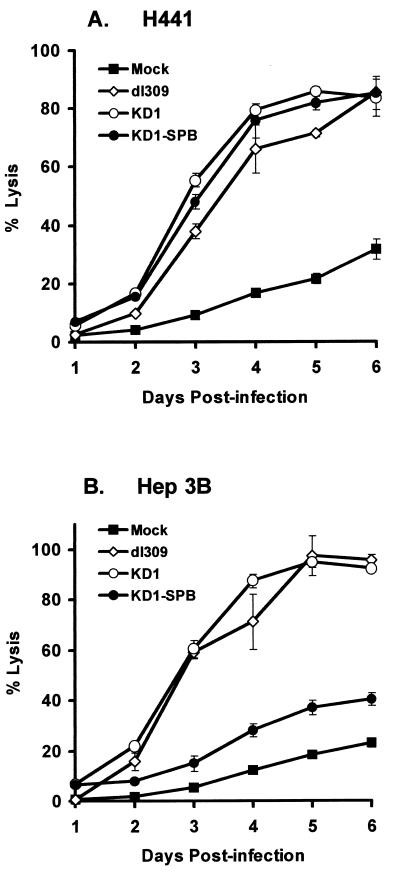
At the culmination of replication, Ad-infected cells are lysed and the virus spreads to other cells; this process is mediated in large part by ADP (52, 53). To examine vector-induced cell lysis, H441 and Hep3B cells were mock infected or infected with 20 PFU of KD1, KD1-SPB, or dl309/cell, and cell lysis was determined by release of lactate dehydrogenase (53). All vectors lysed H441 cells beginning at 2 to 3 days p.i. (Fig. 8A). KD1 and dl309 also lysed Hep3B cells in the same time period; however, KD1-SPB caused only minimal cell lysis (Fig. 8B).
FIG. 8.
KD1-SPB lyses H441 but not Hep3B cells as efficiently as does KD1. H441 or Hep3B cells were mock infected or infected with 20 PFU of dl309, KD1, or KD1-SPB/cell. Cell lysis was determined by release of lactate dehydrogenase from the cells into the medium.
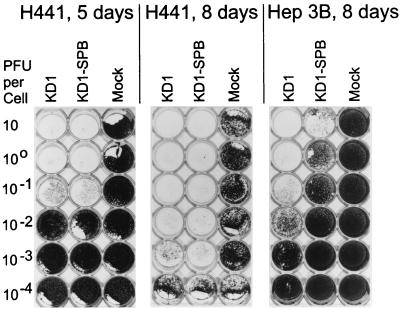
Ad replication and cell lysis were also examined in a vector spread assay (8). H441 or Hep3B cells were mock infected or infected with 101, 100, 10−1, 10−2, 10−3, or 10−4 PFU of KD1 or KD1-SPB/cell, and then cell lysis was determined by crystal violet staining at 5 and 8 days p.i. (Fig. 9). At low PFU per cell, the vectors must go through two or more rounds of replication and cell lysis in order to infect and destroy every cell in the monolayer. With H441 cells at 5 days p.i., both KD1 and KD1-SPB eliminated the monolayer at 10−1 PFU/cell (Fig. 9). At 8 days, the monolayer was destroyed at 10−3 PFU/cell. With Hep3B cells at 8 days, KD1 destroyed nearly all the monolayer at 10−1 PFU/cell, much of it at 10−2 PFU/cell, and some of it at 10−3 PFU/cell. KD1-SPB caused some destruction of Hep3B cells, but about 102- to 103-fold-more KD1-SPB than KD1 was required (Fig. 9).
FIG. 9.
Vector spread assay showing that KD1-SPB spreads from cell to cell as well as does KD1 in H441 but not Hep3B cells. H441 or Hep3B cells were mock infected or infected with KD1 or KD1-SPB at multiplicities of infection ranging from 101 to 10−4 PFU/cell. At 5 and 8 days p.i., cells remaining on the dish were stained with crystal violet and photographed.
KD1-SPB and KD1 suppress the growth of H441 tumors in nude mice, but only KD1 suppresses the growth of Hep3B tumors.
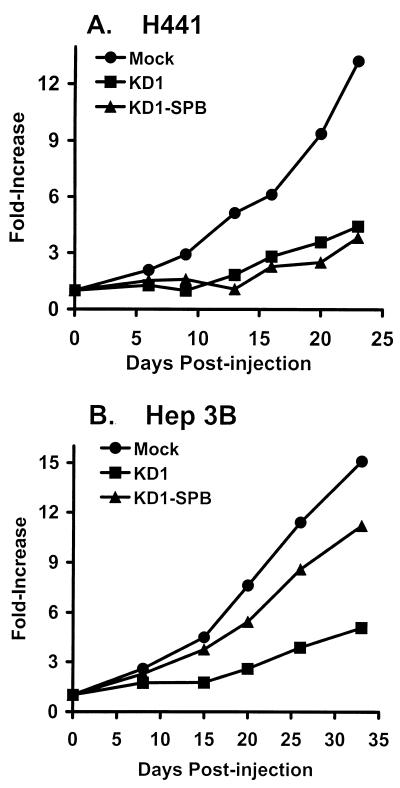
An experiment was conducted to determine whether KD1-SPB or KD1 would suppress H441 tumors in nude mice. H441 or Hep3B cells were injected into each hind flank. When tumors had grown to about 100 μl (H441) or 150 μl (Hep3B), they were injected twice weekly for 3 weeks with DMEM (mock) or 5 × 107 PFU of test virus in 50 μl of DMEM (3.0 × 108 total PFU). Ten tumors (five mice) were used for each virus. Growth of H441 tumors was suppressed similarly by KD1-SPB and KD1 (Fig. 10A). KD1 suppressed growth of Hep3B tumors, whereas KD1-SPB caused only minimal suppression (Fig. 10B). These results show that KD1-SPB is as effective as KD1 in suppressing tumors when the SPB promoter is active. Further, the cell type specificity observed with KD1-SPB in vitro is maintained in vivo.
FIG. 10.
KD1-SPB suppresses growth of H441 tumors in nude mice equally as well as does KD1. Tumor cells were injected into flanks of nude mice and allowed to grow to about 100-μl (H441) or 150-μl (Hep3B) volumes. Tumors (n = 10) were injected with DMEM (mock) or with 5 × 107 PFU of KD1 or KD1-SPB. Injections of the viruses were repeated twice weekly for 3 weeks to a total dose of 3.0 × 108 PFU per tumor. Tumors were measured, and the mean fold increase in tumor size was calculated. Significance was as follows for H441 tumors: mock versus KD1, P = 0.03; mock versus KD1-SPB, P = 0.02; KD1-SPB versus KD1, P = 0.53.
DISCUSSION
Tumor specificity is one of the biggest challenges facing RC and RD Ad vector-mediated cancer gene therapy. Two main strategies that confer specificity have been described: targeting the vector to infect only the tumor cells, e.g., by genetically modifying the Ad fiber protein that interacts with a specific receptor on cells (1, 58), and targeting transcription such that vector replication and/or transgene expression will occur only in tumors. Transcriptional targeting utilizes tumor- and tissue-specific promoters (31). In RC vectors, insertion of the tissue- or tumor-specific promoter-enhancer into the E1A promoter-enhancer region has been used exclusively (16, 38, 69, 70). The rationale behind these vectors is that expression of E1A and therefore the whole Ad transcription program will depend on these tissue- or tumor-specific promoters. However, as a generic approach, there may be difficulties. The E1A enhancer-promoter is very complex. The enhancer controls not only the E1A promoter but also distant promoters such as the E4 promoter (46). In addition, it has been shown that the E1A enhancer in the inverted terminal repeat region changes the tissue specificity of cellular promoters (47). Also, the E1A enhancer-promoter is partially embedded within the signals required to package the Ad genome into virions (45), and it may be problematic to remove all the E1A enhancer elements without impairing virus production. Accordingly, we chose to replace the E4 promoter with a tissue-specific promoter. E4 genes are essential for Ad replication, and therefore, the replication of the recombinant virus should be dependent on tissue-specific regulatory elements.
To construct the KD1-SPB RC vector, ca. 150 bp of the E4 promoter were deleted and the B-500 version (ca. 500 bp) of the SPB promoter (64) was inserted (Fig. 1D). We selected the SPB promoter because of its strict tissue specificity: it is active exclusively in type II alveolar cells and bronchial epithelial cells of the lung (5). Since the parental virus KD1 contains and expresses two E1A mutations that restrict virus replication to tumor cells (8), we anticipated that the vector would selectively replicate in cells derived from lung tumors. Thus, H441 cells, a papillary lung carcinoma cell line, were used to characterize the replication, gene expression, and functional profile of KD1-SPB. KD1-SPB was originally isolated on 293-E4 cells, a 293 cell line that constitutively expresses E4 proteins.
KD1-SPB formed plaques 3 to 4 days sooner on 293-E4 cells than on 293 cells, whereas KD1 formed plaques with the same kinetics on both cell lines. These data are in accord with the expectation that KD1-SPB would not grow well in 293 cells and that it would be complemented by the E4 proteins expressed in 293-E4 cells. It is not clear why KD1-SPB forms plaques at all on 293 cells; it could be because these cells, which are derived from human embryonic kidney, express hepatocyte nuclear factor 3, a transcription factor that regulates the SPB promoter (5). It is also possible that TTF1, the master regulatory factor of SPB expression, is minimally active in 293 cells. In A549 cells, KD1-SPB plaques were delayed 4 to 6 days compared to KD1 plaques. Since A549 cells are derived from an alveolar adenocarcinoma, it is possible that they express small amounts of TTF1.
Importantly, KD1-SPB replicated efficiently, equally well as KD1, in H441 cells, in which the SPB promoter is active. Replication was determined by viral DNA accumulation, viral early and late protein expression, CPE, lactate dehydrogenase release (an indicator of replication-induced cell lysis), and production of infectious virions. In contrast to KD1, KD1-SPB replicated very poorly in Hep3B cells, a liver cancer cell line in which the SPB promoter is not expected to be active. At 2 days p.i. of Hep3B cells, KD1-SPB synthesized only small amounts of DNA and E4ORF3 (as detected by immunoblotting), and the yield of infectious virus was 4 to 5 logs lower than that of KD1.
The ability of KD1-SPB to infect and express E4ORF3 was examined with four additional cell lines in which the SPB promoter is not predicted to be active. These lines are HT29.14S (colon carcinoma), HeLa (human papillomavirus-positive cervical carcinoma), KB (nasopharyngeal carcinoma), and LNCaP (prostate carcinoma). Ad5 is well known to infect these cells. Consistent with this, these cells were efficiently infected with KD1-SPB and KD3 as indicated by immunofluorescence staining for the E1A proteins. Most of the cells on the coverslip were infected, as judged by comparing E1A-positive nuclei with DAPI-stained nuclei. However, in contrast to all cell types infected with KD3 and to H441 cells infected with KD1-SPB, there was very little staining for E4ORF3. Presumably, other E4 proteins also were not expressed. Considering that E4 proteins are required for Ad replication, these results indicate that KD1-SPB does not replicate in these cell lines. These findings support the conclusion that a cell-type-specific promoter inserted in place of the E4 promoter can drive replication in a cell-type-specific manner.
Although there was very weak immunostaining for E4ORF3, a few speckles were seen (e.g., in LNCaP cells in Fig. 7) in a small fraction of cells. This may mean that the cell lines are heterogeneous and a few cells support the SPB promoter. Alternatively, perhaps general transcription factors and specific transcription factors in the AP1, Sp1, CAAT box-binding protein, or hepatocyte nuclear factor families (5, 6) can assemble inefficiently on the promoter, and once a threshold is reached, the promoter fires to allow E4 protein synthesis and a normal lytic infection. This would result in a low rate of replication but nevertheless a gradual increase in virus accumulation in culture, as was observed for KD1-SPB-infected Hep3B cells (Fig. 4). This scenario would be analogous to the replication observed at high multiplicities of infection and at several days p.i. with the E1A-negative mutant dl312 (14) and with E1− first-generation RD vectors (34).
Of great interest, the cell type specificity in cultured H441 and Hep3B cells was also observed when these cells were growing as tumors in nude mice. Growth of H441 tumors was suppressed by KD1-SPB and KD1 at similarly high efficiencies (Fig. 10). Unlike KD1, KD1-SPB had only a minimal effect on the growth of Hep3B tumors (Fig. 10). These observations raise the possibility that KD1-SPB could be used to treat lung cancer. The E1A mutation in KD1-SPB should spare normal lung tissue, and the E1A mutation and E4 promoter substitution should spare other tissues. Although the lung ranks as the second most common cancer site for both men and women in the United States (37), lung cancer has not been a major target for cancer vector gene therapy, since intratumoral injection of virus is generally not feasible in the lungs. However, there has been a recent report of intratumoral injection of an RD Ad vector into a lung tumor (12), and such an approach could be attempted with KD1-SPB. It may also be feasible to administer KD1-SPB systemically in the lung.
In addition to the increased tumoricidal capacity afforded by overexpression of ADP (8), KD1-SPB has three safety features. First, because of the SPB promoter, it can replicate well in lung tissues only where this promoter is active. Second, because of the E1A mutation, it should replicate only in cancer cells of the lung, not in normal lung cells. We have not tested this idea directly with KD1-SPB, but we did show previously that KD1 did not induce CPE in human primary bronchial epithelial cells and small airway epithelial cells (8). Third, KD1 lacks all the E3 genes (except adp). E3 genes protect Ad-infected cells from being destroyed by killer cells of the immune system (62, 63). Thus, as discussed earlier (8), the immune system may be able to control a disseminated KD1-SPB infection more easily than a wild-type Ad5 infection.
In summary, we have provided proof of the principle that replacement of the E4 promoter with a tissue-specific promoter allows highly tissue-specific replication of Ad vectors. Further, replication in the target tissue is as efficient as the replication of the parental virus.
ACKNOWLEDGMENTS
K.D., M.K., and K.T. contributed equally.
We thank Meribeth Broadway and Chris Wells for technical assistance; Jeffrey A. Whitsett for the SPB promoter and H441 cells; Stanley Bayley and Thomas Shenk for viruses; Jeff Engler, Maurice Green, and Gary Ketner for antisera; Pat Farrar for advice; and Sue Sulkey and Dawn Schwartz for preparation of the figures and manuscript.
This work was supported by grants CA71704 and CA81829 from the National Cancer Institute.
REFERENCES
- 1.Alemany R, Balague C, Curiel D T. Replicative adenoviruses for cancer therapy. Nat Biotechnol. 2000;18:723–727. doi: 10.1038/77283. [DOI] [PubMed] [Google Scholar]
- 2.Barker D D, Berk A J. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology. 1987;156:107–121. doi: 10.1016/0042-6822(87)90441-7. [DOI] [PubMed] [Google Scholar]
- 3.Bett A J, Haddara W, Prevec L, Graham F L. An efficient and flexible system for construction of adenovirus vectors with insertions or deletions in early regions 1 and 3. Proc Natl Acad Sci USA. 1994;91:8802–8806. doi: 10.1073/pnas.91.19.8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischoff J R, Kirn D H, Williams A, Heise C, Horn S, Muna M, Ng L, Nye J A, Sampson-Johannes A, Fattaey A, McCormick F. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science. 1996;274:373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- 5.Bohinski R J, Di Lauro R, Whitsett J A. The lung-specific surfactant protein B gene promoter is a target for thyroid transcription factor 1 and hepatocyte nuclear factor 3, indicating common factors for organ-specific gene expression along the foregut axis. Mol Cell Biol. 1994;14:5671–5681. doi: 10.1128/mcb.14.9.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohinski R J, Huffman J A, Whitsett J A, Lattier D L. Cis-active elements controlling lung cell-specific expression of human pulmonary surfactant protein B gene. J Biol Chem. 1993;268:11160–11166. [PubMed] [Google Scholar]
- 7.Crystal R G. In vivo and ex vivo gene therapy strategies to treat tumors using adenovirus gene transfer vectors. Cancer Chemother Pharmacol. 1999;43(Suppl.):S90–S99. doi: 10.1007/s002800051105. [DOI] [PubMed] [Google Scholar]
- 8.Doronin K, Toth K, Kuppuswamy M, Ward P, Tollefson A E, Wold W S M. Tumor-specific, replication-competent adenovirus vectors overexpressing the adenovirus death protein. J Virol. 2000;74:6147–6155. doi: 10.1128/jvi.74.13.6147-6155.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doucas V, Ishov A M, Romo A, Juguilon H, Weitzman M D, Evans R M, Maul G G. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 10.Freytag S O, Rogulski K R, Paielli D L, Gilbert J D, Kim J H. A novel three-pronged approach to kill cancer cells selectively: concomitant viral, double suicide gene, and radiotherapy. Hum Gene Ther. 1998;9:1323–1333. doi: 10.1089/hum.1998.9.9-1323. [DOI] [PubMed] [Google Scholar]
- 11.Gabler S, Schutt H, Groitl P, Wolf H, Shenk T, Dobner T. E1B 55-kilodalton-associated protein: a cellular protein with RNA-binding activity implicated in nucleocytoplasmic transport of adenovirus and cellular mRNAs. J Virol. 1998;72:7960–7971. doi: 10.1128/jvi.72.10.7960-7971.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gahery-Segard H, Molinier-Frenkel V, Le Boulaire C, Saulnier P, Opolon P, Lengagne R, Gautier E, Le Cesne A, Zitvogel L, Venet A, Schatz C, Courtney M, Le Chevalier T, Tursz T, Guillet J-G, Farace F. Phase I trial of recombinant adenovirus gene transfer in lung cancer. J Clin Investig. 1997;100:2218–2226. doi: 10.1172/JCI119759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganly I, Kirn D, Eckhardt S G, Rodriguez G I, Soutar D S, Otto R, Robertson A G, Park O, Gulley M L, Heise C, Von Hoff D D, Kaye S B. A phase I study of Onyx-015, an E1B attenuated adenovirus, administered intratumorally to patients with recurrent head and neck cancer. Clin Cancer Res. 2000;6:798–806. [PubMed] [Google Scholar]
- 14.Gaynor R B, Berk A J. Cis-acting induction of adenovirus transcription. Cell. 1983;33:683–693. doi: 10.1016/0092-8674(83)90011-9. [DOI] [PubMed] [Google Scholar]
- 15.Goodrum F D, Ornelles D A. p53 status does not determine outcome of E1B 55-kilodalton mutant adenovirus lytic infection. J Virol. 1998;72:9479–9490. doi: 10.1128/jvi.72.12.9479-9490.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallenbeck P L, Chang Y-N, Hay C, Golightly D, Stewart D, Lin J, Phipps S, Chiang Y L. A novel tumor-specific replication-restricted adenoviral vector for gene therapy of hepatocellular carcinoma. Hum Gene Ther. 1999;10:1721–1733. doi: 10.1089/10430349950017725. [DOI] [PubMed] [Google Scholar]
- 17.Harada J N, Berk A J. p53-independent and -dependent requirements for E1B–55K in adenovirus type 5 replication. J Virol. 1999;73:5333–5344. doi: 10.1128/jvi.73.7.5333-5344.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harlow E, Franza B R J, Schley C. Monoclonal antibodies specific for adenovirus early region 1A proteins: extensive heterogeneity in early region 1A products. J Virol. 1985;55:533–546. doi: 10.1128/jvi.55.3.533-546.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hay J G, Shapiro N, Sauthoff H, Heitner S, Phupakdi W, Rom W N. Targeting the replication of adenoviral gene therapy vectors to lung cancer cells: the importance of the adenoviral E1B–55kD gene. Hum Gene Ther. 1999;10:579–590. doi: 10.1089/10430349950018652. [DOI] [PubMed] [Google Scholar]
- 20.Heise C, Sampson-Johannes A, Williams A, McCormick F, Von Hoff D D, Kirn D H. ONYX-015, an E1B gene-attenuated adenovirus, causes tumor-specific cytolysis and antitumoral efficacy that can be augmented by standard chemotherapeutic agents. Nat Med. 1997;3:639–645. doi: 10.1038/nm0697-639. [DOI] [PubMed] [Google Scholar]
- 21.Heise C C, Williams A M, Xue S, Propst M, Kirn D H. Intravenous administration of ONYX-015, a selectively replicating adenovirus, induces antitumoral efficacy. Cancer Res. 1999;59:2623–2628. [PubMed] [Google Scholar]
- 22.Hermiston T, Heise C, Kirn D H. The discovery and development of selectively replicating adenoviruses as anticancer agents. Tumor Targeting. 2000;4:218–224. [Google Scholar]
- 23.Horridge J J, Leppard K N. RNA-binding activity of the E1B 55-kilodalton protein from human adenovirus type 5. J Virol. 1998;72:9374–9379. doi: 10.1128/jvi.72.11.9374-9379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horwitz M S. Adenoviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2149–2171. [Google Scholar]
- 25.Howe J A, Mymryk J S, Egan C, Branton P E, Bayley S T. Retinoblastoma growth suppressor and a 300-kDa protein appear to regulate cellular DNA synthesis. Proc Natl Acad Sci USA. 1990;87:5883–5887. doi: 10.1073/pnas.87.15.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirn D, Hermiston T W, McCormick F. ONYX-015: clinical data are encouraging. Nat Med. 1998;4:1341–1342. doi: 10.1038/3902. [DOI] [PubMed] [Google Scholar]
- 27.Krougliak V, Graham F L. Development of cell lines capable of complementing E1, E4, and protein IX defective adenovirus type 5 mutants. Hum Gene Ther. 1995;6:1575–1586. doi: 10.1089/hum.1995.6.12-1575. [DOI] [PubMed] [Google Scholar]
- 28.Leppard K N. E4 gene function in adenovirus, adenovirus vector and adeno-associated virus infections. J Gen Virol. 1997;78:2131–2138. doi: 10.1099/0022-1317-78-9-2131. [DOI] [PubMed] [Google Scholar]
- 29.Martin M E, Berk A J. Corepressor required for adenovirus E1B 55,000-molecular-weight protein repression of basal transcription. Mol Cell Biol. 1999;19:3403–3414. doi: 10.1128/mcb.19.5.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin M E D, Berk A J. Adenovirus E1B 55K represses p53 activation in vitro. J Virol. 1998;72:3146–3154. doi: 10.1128/jvi.72.4.3146-3154.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maxwell I H, Cripe T P. Transcriptional targeting. Tumor Targeting. 2000;4:189–209. [Google Scholar]
- 32.Mittereder N, March K L, Trapnell B C. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J Virol. 1996;70:7498–7509. doi: 10.1128/jvi.70.11.7498-7509.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Motoi F, Sunamura M, Ding L, Duda D G, Yoshida Y, Zhang W, Matsuno S, Hamada H. Effective gene therapy for pancreatic cancer by cytokines mediated by restricted replication-competent adenovirus. Hum Gene Ther. 2000;11:223–235. doi: 10.1089/10430340050015978. [DOI] [PubMed] [Google Scholar]
- 34.Nelson J E, Kay M A. Persistence of recombinant adenovirus in vivo is not dependent on vector DNA replication. J Virol. 1997;71:8902–8907. doi: 10.1128/jvi.71.11.8902-8907.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nevels M, Tauber B, Kremmer E, Spruss T, Wolf H, Dobner T. Transforming potential of the adenovirus type 5 E4orf3 protein. J Virol. 1999;73:1591–1600. doi: 10.1128/jvi.73.2.1591-1600.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Querido E, Marcellus R C, Lai A, Charbonneau R, Teodoro J G, Ketner G, Branton P E. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J Virol. 1997;71:3788–3798. doi: 10.1128/jvi.71.5.3788-3798.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reis L A, Wingo P A, Miller D S, Howe H L, Weir H K, Rosenberg H M, Vernon S W, Cronin K, Edwards B K. The annual report to the nation on the status of cancer, 1973–1997, with a special section on colorectal cancer. Cancer Res. 2000;88:2398–2424. doi: 10.1002/(sici)1097-0142(20000515)88:10<2398::aid-cncr26>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez R, Schuur E R, Lim H Y, Henderson G A, Simons J W, Henderson D R. Prostate attenuated replication competent adenovirus (ARCA) CN706: a selective cytotoxic for prostate-specific antigen-positive prostate cancer cells. Cancer Res. 1997;57:2559–2563. [PubMed] [Google Scholar]
- 39.Rogulski K R, Freytag S O, Zhang K, Gilbert J D, Paielli D L, Kim J H, Heise C C, Kirn D H. In vivo antitumor activity of ONYX-015 is influenced by p53 status and is augmented by radiotherapy. Cancer Res. 2000;60:1193–1196. [PubMed] [Google Scholar]
- 40.Rogulski K R, Wing M S, Paielli D L, Gilbert J D, Kim J H, Freytag S O. Double suicide gene therapy augments the antitumor activity of a replication-competent lytic adenovirus through enhanced cytotoxicity and radiosensitization. Hum Gene Ther. 2000;11:67–76. doi: 10.1089/10430340050016166. [DOI] [PubMed] [Google Scholar]
- 41.Roth J, Konig C, Wienzek S, Weigel S, Ristea S, Dobbelstein M. Inactivation of p53 but not p73 by adenovirus type 5 E1B 55-kilodalton and E4 34-kilodalton oncoproteins. J Virol. 1998;72:8510–8516. doi: 10.1128/jvi.72.11.8510-8516.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rothmann T, Hengstermann A, Whitaker N J, Scheffner M, zur Hausen H. Replication of ONYX-015, a potential anticancer adenovirus, is independent of p53 status in tumor cells. J Virol. 1998;72:9470–9478. doi: 10.1128/jvi.72.12.9470-9478.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarnow P, Ho Y S, Williams J, Levine A J. Adenovirus E1B–58kd tumor antigen and SV40 large tumor antigen are physically associated with the same 54 kd cellular protein in transformed cells. Cell. 1982;28:387–394. doi: 10.1016/0092-8674(82)90356-7. [DOI] [PubMed] [Google Scholar]
- 44.Scaria A, Tollefson A E, Saha S K, Wold W S M. The E3-11.6K protein of adenovirus is an Asn-glycosylated integral membrane protein that localizes to the nuclear membrane. Virology. 1992;191:743–753. doi: 10.1016/0042-6822(92)90250-s. [DOI] [PubMed] [Google Scholar]
- 45.Schmid S I, Hearing P. Cellular components interact with adenovirus type 5 minimal DNA packaging domains. J Virol. 1998;72:6339–6347. doi: 10.1128/jvi.72.8.6339-6347.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shenk T. Adenoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2111–2148. [Google Scholar]
- 47.Shi Q, Wang Y, Worton R. Modulation of the specificity and activity of a cellular promoter in an adenoviral vector. Hum Gene Ther. 1997;8:403–410. doi: 10.1089/hum.1997.8.4-403. [DOI] [PubMed] [Google Scholar]
- 48.Shinoura N, Yoshida Y, Tsunoda R, Ohashi M, Zhang W, Asai A, Kirino T, Hamada H. Highly augmented cytopathic effect of a fiber-mutant E1B-defective adenovirus for gene therapy of gliomas. Cancer Res. 1999;59:3411–3416. [PubMed] [Google Scholar]
- 49.Steegenga W T, Riteco N, Jochemsen A G, Fallaux F J, Bos J L. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene. 1998;16:349–357. doi: 10.1038/sj.onc.1201540. [DOI] [PubMed] [Google Scholar]
- 50.Teodoro J G, Branton P E. Regulation of apoptosis by viral gene products. J Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tollefson A E, Hermiston T W, Wold W S M. Preparation and titration of CsCl-banded adenovirus stocks. In: Wold W S M, editor. Adenovirus methods and protocols. Totowa, N.J: Humana Press, Inc.; 1998. pp. 1–9. [Google Scholar]
- 52.Tollefson A E, Ryerse J S, Scaria A, Hermiston T W, Wold W S M. The E3-11.6kDa adenovirus death protein (ADP) is required for efficient cell death: characterization of cells infected with adp mutants. Virology. 1996;220:152–162. doi: 10.1006/viro.1996.0295. [DOI] [PubMed] [Google Scholar]
- 53.Tollefson A E, Scaria A, Hermiston T W, Ryerse J S, Wold L J, Wold W S M. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J Virol. 1996;70:2296–2306. doi: 10.1128/jvi.70.4.2296-2306.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tollefson A E, Scaria A, Saha S K, Wold W S M. The 11,600-Mw protein encoded by region E3 of adenovirus is expressed early but is greatly amplified at late stages of infection. J Virol. 1992;66:3633–3642. doi: 10.1128/jvi.66.6.3633-3642.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turnell A S, Grand R J, Gallimore P H. The replicative capacities of large E1B-null group A and group C adenoviruses are independent of host cell p53 status. J Virol. 1999;73:2074–2083. doi: 10.1128/jvi.73.3.2074-2083.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vollmer C M, Ribas A, Butterfield L H, Dissette V B, Andrews K J, Eilber F C, Montejo L D, Chen A Y, Hu B, Glaspy J A, McBride W H, Economou J S. p53 selective and nonselective replication of an E1B-deleted adenovirus in hepatocellular carcinoma. Cancer Res. 1999;59:4369–4374. [PubMed] [Google Scholar]
- 57.White E. Regulation of apoptosis by adenovirus E1A and E1B oncogenes. Semin Virol. 1998;8:505–513. [Google Scholar]
- 58.Wickham T J. Targeting adenovirus. Gene Ther. 2000;7:110–114. doi: 10.1038/sj.gt.3301115. [DOI] [PubMed] [Google Scholar]
- 59.Wienzek S, Roth J, Dobbelstein M. E1B 55-kilodalton oncoproteins of adenovirus types 5 and 12 inactivate and relocalize p53, but not p51 or p73, and cooperate with E4orf6 proteins to destabilize p53. J Virol. 2000;74:193–202. doi: 10.1128/jvi.74.1.193-202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wildner O, Blaese R M, Morris J C. Therapy of colon cancer with oncolytic adenovirus is enhanced by the addition of herpes simplex virus-thymidine kinase. Cancer Res. 1999;59:410–413. [PubMed] [Google Scholar]
- 61.Wildner O, Morris J C, Vahanian N N, Ford H J, Ramsey W J, Blaese R M. Adenoviral vectors capable of replication improve the efficacy of HSVtk/GCV suicide gene therapy of cancer. Gene Ther. 1999;6:57–62. doi: 10.1038/sj.gt.3300810. [DOI] [PubMed] [Google Scholar]
- 62.Wold W S M, Chinnadurai G. Adenovirus proteins that regulate apoptosis. In: Cann A J, editor. DNA virus replication: frontiers in molecular biology. Oxford, United Kingdom: Oxford University Press; 2000. pp. 200–232. [Google Scholar]
- 63.Wold W S M, Doronin K, Toth K, Kuppuswamy M, Lichtenstein D L, Tollefson A E. Immune responses to adenoviruses: viral evasion mechanisms and their implications for the clinic. Curr Opin Immunol. 1999;11:380–386. doi: 10.1016/S0952-7915(99)80064-8. [DOI] [PubMed] [Google Scholar]
- 64.Yan C, Sever Z, Whitsett J A. Upstream enhancer activity in the human surfactant protein B gene is mediated by thyroid transcription factor 1. J Biol Chem. 1995;270:24852–24857. doi: 10.1074/jbc.270.42.24852. [DOI] [PubMed] [Google Scholar]
- 65.Yang C, Cirielli C, Capogrossi M C, Passaniti A. Adenovirus-mediated wild-type p53 expression induces apoptosis and suppresses tumorigenesis of prostatic tumor cells. Cancer Res. 1995;55:4210–4213. [PubMed] [Google Scholar]
- 66.Yew P R, Berk A J. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 67.Yew P R, Liu X, Berk A J. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 1994;8:190–202. doi: 10.1101/gad.8.2.190. [DOI] [PubMed] [Google Scholar]
- 68.Young L, Yang C-T, Jablons D M. ONYX-015 works synergistically with chemotherapy in lung cancer cell lines and primary cultures freshly made from lung cancer patients. Cancer Res. 2000;60:1009–1013. [PubMed] [Google Scholar]
- 69.Yu D-C, Chen Y, Seng M, Dilley J, Henderson D R. The addition of adenovirus type 5 region E3 enables Calydon virus 787 to eliminate distant prostate tumor xenografts. Cancer Res. 1999;59:4200–4203. [PubMed] [Google Scholar]
- 70.Yu D C, Sakamoto G T, Henderson D R. Identification of the transcriptional regulatory sequences of human kallikrein 2 and their use in the construction of Calydon virus 764, an attenuated replication competent adenovirus for prostate cancer therapy. Cancer Res. 1999;59:1498–1504. [PubMed] [Google Scholar]