Abstract
Human herpesvirus 8 (HHV-8) is associated with Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease; in all of these diseases, interleukin-6 (IL-6) has been implicated as a likely mitogenic and/or angiogenic factor. HHV-8 encodes a homologue of IL-6 (viral IL-6 [vIL-6]) that has been shown to be biologically active in several assays and whose activities mirror those of its mammalian counterparts. Like these proteins, vIL-6 mediates its effects through the gp130 signal transducer, but signaling is not dependent on the structurally related IL-6 receptor (IL-6R; gp80) subunit of the receptor-signal transducer complex. However, as we have shown previously, IL-6R can enhance vIL-6 signal transduction and can enable signaling through a gp130 variant (gp130.PM5) that is itself unable to support vIL-6 activity, indicating that IL-6R can form part of the signaling complex. Also, our analysis of a panel of vIL-6 mutants in transfection experiments in Hep3B cells (that express IL-6R and gp130) showed that most were able to function normally in this system. Here, we have used in vitro vIL-6–receptor binding assays to demonstrate direct binding of vIL-6 to both gp130 and IL-6R and vIL-6-induced gp130–IL-6R complex formation, and we have extended our functional analyses of the vIL-6 variants to identify residues important for IL-6R-independent and IL-6R-dependent signaling through native gp130 and gp130.PM5, respectively. These studies have identified residues in vIL-6 that are important for IL-6R-independent and IL-6R-mediated functional complex formation between vIL-6 and gp130 and that may be involved directly in binding to gp130 and IL-6R.
Human herpesvirus 8 (HHV-8) has been associated with all forms of Koposi's sarcoma (KS) and primary effusion lymphoma (PEL) and is found in a high proportion of affected lymph nodes of patients with Multicentric Castleman's disease (MCD). The roles of interleukin-6 (IL-6) in KS and MCD have long been suspected; IL-6 promotes the growth of KS cells in culture, and IL-6 is present at elevated levels in KS and MCD lesions. Recent reports have also implicated human IL-6 (hIL-6) in the growth of HHV-8 latently infected PEL cells in soft agar and in inoculated nude mice, and hIL-6 and viral IL-6 (vIL-6) enhance tumor formation in mice (1, 2, 9). vIL-6 has been shown to enhance the growth of PEL cells in culture, although IL-10 and other factors also play a role, and to be a mitogenic factor for human T-cell and murine hybridoma cell lines (5, 13, 20, 23). Since vIL-6 has been shown to be produced by uninduced PEL cell lines, in freshly isolated PEL tissue, and in MCD and at least some KS lesions, it is possible that vIL-6, in addition to hIL-6, can play a role in the development and progression of these diseases (6, 20, 23, 26, 34). In fact, because vIL-6 signaling is not restricted to cells expressing IL-6R (also called gp80, the α-subunit of the IL-6 receptor), which is required for hIL-6 signaling through the gp130 signal transducer, it is possible that vIL-6 plays a disproportionate or distinct role in the disease process (19, 22, 24, 35).
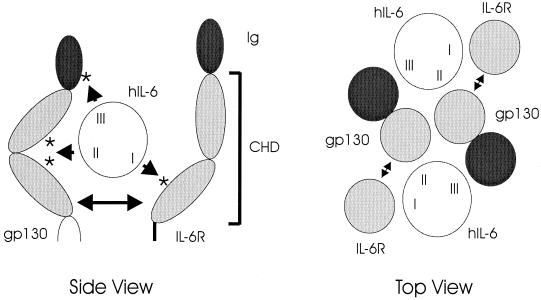
Over the past few years, extensive work has been undertaken to characterize IL-6 and its receptor subunits, IL-6R and gp130. These studies include mutagenesis of ligand and receptors and their use in binding and functional experiments to try to elucidate ligand-receptor contact sites and the nature of the functional receptor-ligand complex (see, e.g., references 3, 7, 8, 11, 14, 27, 28, 29, 30, and 37). More recently, the structures of hIL-6 and the “cytokine receptor homology domain” (CHD) of human gp130 (hgp130) have been elucidated through X-ray crystallography (4, 33). Taken together, this large body of data has provided a fairly detailed picture of how the ligand and receptor subunits interact to form a complex that then mediates, through phosphorylation of cytoplasmic tyrosine residues of gp130, signal transduction via the Jak/STAT and mitogen-activated protein kinase pathways. Dimerization of gp130, the centrally important event that initiates signal transduction, occurs on formation of a hexameric complex between two molecules of IL-6 and two molecules of each of the receptor subunits (36). IL-6 can bind to IL-6R in the absence of gp130; binding to gp130 requires prior binding of IL-6 to IL-6R. Three receptor-binding regions (sites I, II, and III) have been identified; site I interacts with IL-6R, whereas sites II and III are involved in IL-6 interactions with gp130 (17, 25, 32, 33) (Fig. 1). Site II interacts with the CHD region of gp130, while site III interacts with the more proximal immunoglobulin homology domain. For IL-6R, several residues have been shown to be important for IL-6 interaction and/or functional complex formation. Some of these residues are conserved in gp130 (which is related structurally to IL-6R) (Fig. 1), and resolution of the structure of the CHD region of gp130 has shown that some occur in two interstrand loops (E-F and B2-C2) that are positioned appropriately for interactions with ligand (via site II of hIL-6) (4). We have shown previously that alteration of three adjacent residues within one of these loops (E-F) in the gp130 variant gp130.PM5 abrogates IL-6R-independent signaling by vIL-6 (35).
FIG. 1.
Diagrammatic representations of the cytokine receptor homology and immunoglobulin-like domains (CHD and Ig, respectively) of gp130 and IL-6R, their interactions with hIL-6, and complex formation resulting from these interactions. The figure on the left is a side view that indicates the general locations (asterisks) of receptor residues known to interact with hIL-6. Sites II and III of hIL-6 interact with distinct gp130 molecules to form, in association with IL-6R, the functional hexameric complex, as shown schematically in the right panel (top view); for IL-6R, only the hIL-6 site I-interacting CHD region is depicted (individual, lightly shaded circle), with CHD (site II-interacting) and Ig (site III-interacting) regions of gp130 indicated as adjoined lightly and darkly shaded circles, respectively. There is strong experimental evidence for an IL-6R–gp130 dimerization interface (27, 37), here indicated by double-headed arrows.
In this report, we have used in vitro ligand-receptor binding assays to demonstrate binding of vIL-6 to gp130 and IL-6R both independently and as a complex, and we have measured the signaling activities of our previously reported panel of vIL-6 mutants (35) in IL-6R-independent and IL-6R-dependent assays (employing transfected gp130 or gp130.PM5 plus IL-6R) to identify residues within vIL-6 that potentially are important for interactions with gp130 and/or IL-6R. Two variants that were found to be defective in signaling through gp130 alone were able to abrogate native vIL-6 activity, indicating competitive interactions with receptor. Our data demonstrate that vIL-6 is indeed able to bind directly and independently to IL-6R and gp130 and that vIL-6 can induce IL-6R–gp130 complexing, and we identify vIL-6 residues that are important for vIL-6–gp130 and vIL-6–gp130–IL-6R functional complex formation.
MATERIALS AND METHODS
Cell culture and transfections.
HEK293-T cells were grown in minimal essential medium supplemented with 5% fetal calf serum. Cells were passaged 12 to 24 h before transfection to produce monolayers of 40 to 60% confluency in 25-cm2 flasks (for vIL-6 functional assays) or 100-mm dishes (for generation of secreted proteins for receptor-ligand binding assays). Transfections were carried out by calcium phosphate-DNA coprecipitation with HEPES-buffered saline; cells or media were harvested 24 to 48 h posttransfection. For functional analyses of vIL-6 variants, gp130 or gp130.PM5 plus IL-6R expression constructions (1 μg) were used together with 3 μg of each vIL-6 expression vector (35) or pSG5 (negative control) and 1 μg of pα2MCAT reporter plasmid. Where necessary, total amounts of transfected DNA were made up to a standard amount using pEF-BOS (receptor expression vector) DNA. For wild-type–variant vIL-6 competition assays, 2 μg of each effector was used.
Plasmids and oligonucleotides.
The pSG5-based eukaryotic expression plasmids for vIL-6 and specifically altered derivatives have been described previously (35). The hIL-6 open reading frame was cloned as a BamHI fragment into the BamHI site of pSG5 to generate an hIL-6 expression plasmid. The hIL-6R and hgp130 expression vectors pEF-BOS–hIL-6R and pEF-BOS–hgp130 were provided by M. Narazaki and T. Kishimoto; they comprise the receptor coding sequences cloned between the XbaI (IL-6R) or SacI and BamHI sites of the pEF-BOS eukaryotic expression vector (18). The pα2MCAT reporter plasmid comprises α2-macroglobulin promoter sequences cloned upstream of the chloramphenicol acetyltransferase (CAT) gene (31). The generation of the signaling-defective gp130 variant, gp130.PM5, has been described previously (35). Extracellular regions of gp130 and IL-6R were cloned into a pSG5-based expression vector containing human immunoglobulin G (IgG) Fc coding sequences to generate contiguous soluble gp130 (sgp130)-Fc and sIL-6R–Fc coding sequences for the expression of these fusion proteins by transfected cells. For expression and purification of vIL-6 protein from bacteria, the vIL-6 open reading frame was cloned as a BamHI-XhoI fragment into the expression vector pTrcHisB (Invitrogen, San Diego, Calif.).
Western blotting.
vIL-6 and sgp130 proteins derived from transfected cell media were size fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoretically transferred to nitrocellulose membranes. These were blocked in TBS-T (10 mM Tris-HCl, pH 8.0; 150 mM NaCl; 0.05% Tween 20) containing 5% nonfat milk prior to the addition of primary antibody (0.1 to 1.0 μg/ml). Antibody used for the detection of gp130 was obtained from PharMingen (Catalog no. 33831; San Diego, Calif.); vIL-6 peptide antiserum used for the detection of vIL-6 has been described previously (35); hIL-6 antibody was obtained from R&D System (catalog no. AB-206-NA; Minneapolis, Minn.). After washing in TBS-T containing 5% nonfat milk, horseradish peroxidase (HRP)-conjugated anti-mouse (gp130), anti-rabbit (vIL-6), or anti-goat IgG secondary antibody (Santa Cruz Biotechnology [Santa Cruz, Calif.] catalog numbers sc-2005, sc-2004, and sc-2020, respectively) diluted 1:2,000 was used to detect filter-bound primary antibody. HRP on TBS-washed filters was visualized by the chemiluminescence assay.
Bacterially produced vIL-6.
His6–vIL-6 fusion protein was extracted from pTrcHisB– vIL-6-transformed bacteria and purified by Ni2+ affinity chromatography. For this, 250-ml cultures of bacteria in log-phase growth were induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 1 mM and incubated at 18°C overnight. Cells were harvested by centrifugation, resuspended in 10 ml lysis buffer (50 mM sodium phosphate, pH 8.0; 300 mM NaCl), and sonicated to disrupt the cells. Following centrifugation (10,000 × g, 20 min), the cleared lysate was loaded onto a Ni2+ affinity column (Hi-Trap; Pharmacia Biotech, Piscataway, N.J.), which was then washed with lysis buffer containing 10 mM imidazole. Bound His6–vIL-6 fusion protein was eluted in lysis buffer containing 250 mM imidazole.
vIL-6-receptor binding assays.
Binding of vIL-6 to purified gp130 and IL-6R was determined by using a microplate binding assay in which wells were coated with either sgp130 or sIL-6R (75 ng/well) in NaHCO3 at pH 9.6 (overnight, 4°C) prior to blocking with 10% fetal calf serum in phosphate-buffered saline (PBS; 2 h, room temperature), application of vIL-6 protein (4 h, room temperature), and detection of bound vIL-6 by vIL-6 antiserum (35)–HRP-conjugated α-rabbit-IgG secondary antibody (1-h incubations at room temperature for each). Visualization of bound HRP was effected by application of 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) (ABTS)–citric acid (2 μg of ABTS/ml; 100 mM citric acid), and the relative amounts of bound HRP were quantified by measuring the optical density at 450 nm (OD450) in a microplate spectrophotometer. Extensive washing with PBS–0.05% Tween 20 was performed between each step of the assay. Purified sgp130 and sIL-6R proteins were obtained from R&D Systems and detection reagents from Santa Cruz Biotechnology. Purified His6–vIL-6 protein was used at doses ranging from 25 to 200 ng (in 100 μl of PBS). Analogous experiments were undertaken with rhIL-6 (R&D Systems catalog no. 206-IL) using 100 ng of the cytokine per binding assay, and immunological detection was achieved using HRP-conjugated α-hIL-6 antibody (R&D Systems catalog no. AB-206-NA). Assays of vIL-6 binding to sgp130-Fc, gp130.PM5-Fc, or sIL-6R-Fc fusion proteins, vIL-6.24 binding to sgp130-Fc, hIL-6 binding to IL-6R-Fc, and ligand-induced gp130 associations with IL-6R-Fc were carried out using the component proteins derived from growth media of HEK293-T cells transfected with the appropriate expression vectors. Next, 400-μl aliquots of ligand and receptor samples (or pSG5 controls) were incubated with protein A-Sepharose (Pharmacia Biotech catalog no. 170974) at 4°C overnight, and the matrix and bound material were then subjected to repeated centrifugation and washing in PBS. Denatured samples were applied to SDS-polyacrylamide gels and transferred to nitrocellulose for Western analysis to detect vIL-6, hIL-6, or gp130.
RESULTS
Interactions of vIL-6 with gp130 and IL-6R.
Previously published data have shown that vIL-6, unlike hIL-6, can signal through gp130 alone, in the absence of IL-6R (19, 22, 35). However, it has also been reported that IL-6R neutralizing antibodies can partially inhibit vIL-6 activity and that IL-6R can enhance vIL-6 signaling through gp130 or enable signaling through an otherwise nonfunctional variant, gp130.PM5, in cotransfection assays, indicating that IL-6R can form part of the signaling complex induced by vIL-6 (5, 35). To identify direct interactions of vIL-6 with gp130 and IL-6R, we used two distinct in vitro binding assays, one employing purified receptor subunits in enzyme-linked immunosorbent assay (ELISA) and the other utilizing receptor-Fc fusion proteins in coprecipitation experiments.
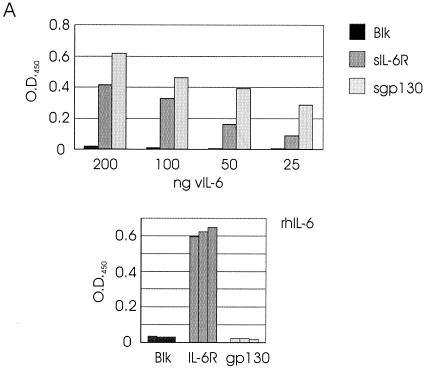
In the first approach, sIL-6R and sgp130 proteins were used to coat the wells of assay plates, to which purified, bacterially expressed His6–vIL-6 protein or recombinant hIL-6 was applied; wells were subsequently washed, and bound ligands were detected and quantified with α-vIL-6–α-rabbit-IgG-HRP or α-hIL-6-HRP reagents and ABTS-based colorimetric analysis (see Materials and Methods). The results of these assays (Fig. 2A) demonstrated binding by vIL-6 to both sIL-6R and sgp130; hIL-6 bound sIL-6R but not sgp130 (in isolation), as has been shown previously. No significant binding was detected when IL-6 protein was added to wells containing no receptor protein (Fig. 2A), nor in controls in which no α-vIL-6 detection antibody was used prior to the addition of α-rabbit-IgG-HRP (data not shown). Furthermore, no binding of His6-DHFR protein (50 ng) to wells coated with either sIL-6R or sgp130 was detected in separate experiments giving positive results for vIL-6 binding, thus ruling out the possibility of nonspecific binding of the His6 tag to the receptor subunits (data not shown).
FIG. 2.
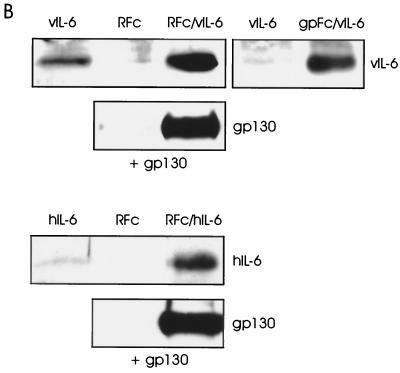
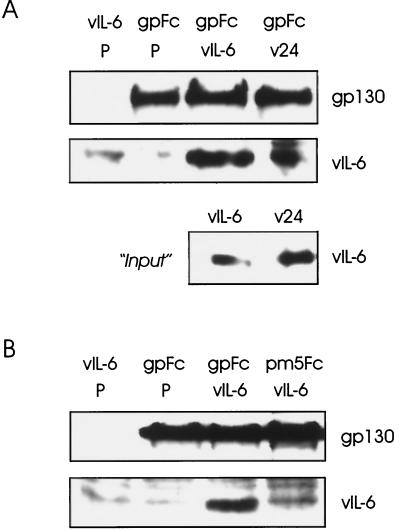
In vitro binding of vIL-6 to IL-6R and gp130. (A) ELISA techniques (see Materials and Methods) were used for the detection of binding by vIL-6 and hIL-6 of sgp130 and sIL-6R. For vIL-6, various amounts (25 to 200 ng) of bacterially produced, purified His6–vIL-6 fusion protein were applied to microassay plate wells coated with sIL-6R or sgp130 or to untreated wells (Blk). After incubation and washing, bound ligand was detected with vIL-6 rabbit antiserum (35) and HRP-conjugated α-rabbit-IgG secondary antibody. Visualization and quantitation was carried out using ABTS reagent and the determination of the OD450 (top panel). Analogous assays were undertaken with rhIL-6 using HRP-conjugated α-hIL-6 as the detection antibody. Experiments were performed in triplicate using 100 ng of ligand (bottom panel). (B) vIL-6 binding to IL-6R and gp130 was determined also by coprecipitation assays using transfected cell media containing sIL-6R–Fc (RFc) or sgp130-Fc (gpFc) together with media containing vIL-6 (see Materials and Methods). Protein A-Sepharose-precipitated material was analyzed by Western blot for the detection of vIL-6. Analogous experiments were undertaken with hIL-6. Inclusion of sgp130 in the sIL-6R–Fc/ligand-binding assays, followed by Western analysis to detect coprecipitated sgp130, demonstrated that vIL-6, in addition to hIL-6, could induce gp130–IL-6R complexing. Experiments using vIL-6 or hIL-6 in the absence of Fc fusion protein or sIL-6R-Fc plus sgp130 in the absence of ligand were included to control for nonspecific binding of proteins to protein A-Sepharose.
To confirm the results of our plate assays, we used coprecipitation assays to detect associations of vIL-6 with gp130 and IL-6R. The soluble receptor subunits were expressed as IgG Fc fusion proteins by transfection of HEK293-T cells with appropriate expression vectors (see Materials and Methods). These proteins, vIL-6 and hIL-6, were derived from transfected cell media for use in the protein A-Sepharose-mediated coprecipitation assays; precipitated ligands were detected by Western analysis. As shown in Fig. 2B (top panels), both sgp130-Fc and sIL-6R–Fc fusion proteins were able to precipitate vIL-6, whereas only low background levels of vIL-6 were detected in the absence of receptor fusion protein. Similar results were obtained using sIL-6R–Fc and hIL-6 in this assay (Fig. 2B, top panel of bottom pair). To determine whether vIL-6 could induce gp130–IL-6R complexing, similar experiments were undertaken in which IL-6R–Fc- and sgp130-containing media were mixed either in the presence or in the absence of vIL-6, and subsequent Western analysis was used for the detection of precipitated sgp130. Analogous experiments were undertaken with hIL-6 to provide a positive control. The results of these assays (Fig. 2B) demonstrate that vIL-6, like hIL-6, can induce interactions between gp130 and IL-6R, thereby providing direct evidence of vIL-6–gp130–IL-6R complexing.
The combined data from our in vitro binding assays demonstrate that vIL-6 can interact with both IL-6R and gp130 and that vIL-6 can induce interactions between gp130 and IL-6R. These data are consistent with our previously reported findings from vIL-6 functional assays of enhancement of gp130 signaling and “rescue” of gp130.PM5 signaling by IL-6R (35).
IL-6R-independent signaling by vIL-6 variants through gp130.
We previously reported on the generation, by targeted mutagenesis, of a series of specifically altered vIL-6 proteins that were assayed for function by reporter-based transfection assays conducted in Hep3B cells, which naturally express gp130 and IL-6R (35). Only two (variants 15 and 24, “site III” mutants) of a panel of 28 altered proteins (Table 1) were found to be defective for signaling (although appropriately expressed) in this experimental system, despite the targeting for mutagenesis of residues whose equivalents in hIL-6 were shown to affect hIL-6-receptor binding or that were highly conserved between vIL-6 and IL-6 proteins from several different species. We decided to investigate the possibility that vIL-6 variants other than 15 and 24 might be abrogated in their ability to signal through to gp130 alone. We used cotransfection assays to overexpress gp130 and each of the vIL-6 variants in turn, together with the STAT-inducible reporter pα2MCAT, in HEK293-T cells (see Materials and Methods).
TABLE 1.
Mutated residues in vIL-6
| Position(s) | Residue(s) | Change(s) | Mutant no. | Comments |
|---|---|---|---|---|
| 13 | G | W | 1 | Conserved |
| 41, 43 | W, L | F, G | 19 | Site II |
| 54 | C | D | 2 | Conserved |
| G | 3 | |||
| 60 | C | S | 4 | Conserved |
| L | 5 | |||
| 75 | P | S | 7 | Conserved |
| 83 | C | G | 8 | Conserved |
| 88 | F | L | 20 | Site I |
| 93 | C | G | 9 | Site I |
| W | 10 | |||
| 103–105 | EFE | EGG | 22 | Site I |
| GVA | 23 | |||
| 129 | T | L | 11 | Site II |
| A | 12 | |||
| 151 | P | F | 13 | Conserved |
| S | 14 | |||
| 167 | W | G | 15 | Site III |
| R | 16 | |||
| 167, 172 | W, A | R, P | 24 | Site III |
| 183 | F | W | 17 | Conserved |
| G | 18 | |||
| E | 29 | |||
| L | 31 | |||
| 189 | R | L | 25 | Site I |
| S | 26 | |||
| C | 27 | |||
| 195 | P | Stop | 28 | C-tail deletion |
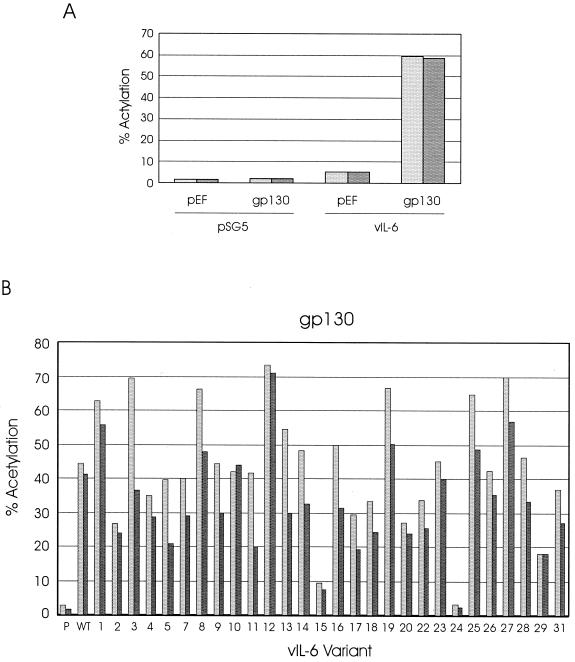
Initial experiments were performed to confirm that high level induction by vIL-6 of the pα2MCAT reporter was dependent on cotransfection of pEFBOS-hgp130 and that cotransfection of the gp130 expression vector alone had no effect on CAT expression (Fig. 3A). The profile of signal transduction by vIL-6 and its derivatives through overexpressed gp130 alone generally paralleled that obtained previously in Hep3B cells (35) (Fig. 3B). It is notable, however, that several of the vIL-6 variants showed somewhat reduced or enhanced activities relative to wild-type vIL-6, while being functionally equivalent to vIL-6 in the Hep3B system (35). For those proteins with reduced activity, such as variants 2, 17, 18, 20, 22, and 29, this could indicate that IL-6R (expressed in Hep3B cells) can stabilize otherwise lower-affinity interactions between some of the vIL-6 variants and gp130, a situation akin to the stabilization of hIL-6–IL-6R interactions by complex formation with gp130.
FIG. 3.
Signaling by vIL-6 variants through overexpressed gp130. (A) Dependence on overexpressed gp130 of high-level activation of the STAT binding site-containing pα2MCAT reporter plasmid was tested by comparing the levels of CAT expression in HEK293-T cells cotransfected with pSG5–vIL-6 plus pα2MCAT and either pEF-BOS–hgp130 or the empty pEF-BOS expression vector. Similar transfection experiments were performed in which pSG5 was substituted for pSG5–vIL-6 to ensure that gp130 overexpression alone did not influence pα2MCAT expression. (B) pSG5-cloned vIL-6 (WT) and vIL-6 variants (see Table 1), or pSG5 (P, negative control), were cotransfected into HEK293-T cells, together with pEF-BOS–hgp130 and pα2MCAT. Cells were harvested 2 days after transfection, and CAT activities present in cell extracts were assayed to determine the relative activities of each of the vIL-6 variants relative to wild-type vIL-6. The results of duplicate experiments are shown.
Functional competition and receptor binding by gp130 signaling-defective vIL-6 variants.
To assess whether the vIL-6 variants 15 and 24 that were essentially inactive in the gp130 transfection assay might be able to bind to gp130, further transfection experiments were undertaken using each of the variants together with wild-type vIL-6. We hypothesized that if inhibition of vIL-6 by the nonsignaling variants occurred, this would most likely be because of competition for receptor binding. The transfection experiments used equal amounts of wild-type, vIL-6.15, and vIL-6.24 expression vectors cotransfected, either alone or in combination, with pEFBOS-hgp130 and the pα2MCAT reporter plasmid. The results of these experiments are shown in Fig. 4. When transfected individually, the vIL-6 effectors led to levels of signaling similar to those obtained in previous experiments, with native vIL-6 leading to over 10-fold activation, relative to the pSG5 control, and the vIL-6 variants being functionally inactive. When vIL-6 was transfected with vIL-6 variants 15 or 24, activation by the wild-type protein was almost completely inhibited. These data suggest that vIL-6 variants 15 and 24 are able to bind to gp130 and to inhibit gp130–vIL-6 interactions and/or vIL-6-induced gp130–gp130 functional dimerization.
FIG. 4.
Inhibition of gp130-mediated vIL-6 signaling by functionally negative vIL-6 variants. Plasmids expressing vIL-6 variants 15 or 24, or pSG5 (P), were cotransfected with pEF-BOS–hgp130 and pα2MCAT and either pSG5–vIL-6 or pSG5. Results with pSG5 confirmed those obtained previously (Fig. 3B); these altered vIL-6 proteins are unable to signal through gp130. Coexpression of variants 15 and 24 with vIL-6 resulted in almost complete inhibition of vIL-6 signaling. The results of duplicate experiments are shown.
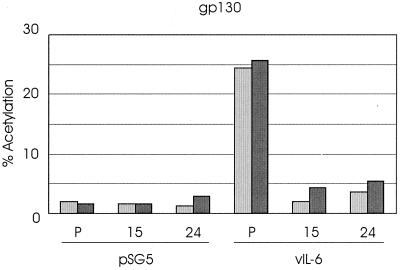
To determine directly the gp130 binding activity of vIL-6 variant 24 relative to wild-type vIL-6, we used sgp130-Fc fusion protein together with vIL-6.24 or vIL-6 proteins in protein A-Sepharose-mediated coprecipitation assays (see above and Materials and Methods). Detection of precipitated sgp130-Fc (to control for efficient and uniform precipitation) and vIL-6 proteins was achieved by Western analysis using peptide antisera specific for these proteins. The expression and relative amounts of vIL-6 and vIL-6.24 in the transfected cell media used in the binding assays were determined by Western analysis of samples of each. The results of a representative experiment are shown in Fig. 5A and indicate that vIL-6 variant 24 binds with reduced affinity to sgp130. Whether this level of binding is sufficient to account for the inhibition of wild-type vIL-6 activity by vIL-6.24 through direct competition for binding is uncertain, but it is possible that within the context of cell-expressed gp130, vIL-6.24 is able to bind (nonfunctionally) with high affinity to gp130 monomers or to gp130 in higher-order complexes (e.g., those involving gp130 dimers). Clearly, gp130 binding by vIL-6.24 is incompatible with the view that this vIL-6 variant is functionally negative because it cannot recognize gp130.
FIG. 5.
Interactions of vIL-6 and derivatives with gp130 and gp130.PM5. Media from cells transfected independently with vIL-6, vIL-6.24 (v24), sgp130-Fc (gpFc), and sgp130.PM5-Fc (pm5Fc) expression plasmids or pSG5 (negative control; P) were mixed, as indicated, precipitated with protein A-Sepharose, and size fractionated by SDS-PAGE, and component proteins were detected by Western blot analysis (see Materials and Methods). (A) The relative gp130 binding activities of vIL-6 and vIL-6.24 were determined from the amounts of coprecipitated vIL-6 protein. The amounts of sgp130-Fc protein precipitated were equivalent (top panel). The first lane shows no cross-reaction of the gp130 antibody with proteins binding (nonspecifically) to the protein A-Sepharose; there was a protein species, migrating slightly slower than vIL-6, that cross-reacted with the vIL-6 antiserum. The relative amounts of the vIL-6 proteins present in samples of the transfected cell media used in the binding assays are indicated (bottom panel). (B) Similar experiments using sgp130.PM5-Fc showed that vIL-6 was not coprecipitated with the altered receptor subunit but was coprecipitated with sgp130-Fc.
IL-6R-dependent signaling by vIL-6 variants through gp130.PM5.
To examine the effects of the various vIL-6 alterations on signaling involving IL-6R-mediated complex formation, we made use of the IL-6R-dependent CHD E-F loop-altered gp130 protein, gp130.PM5, described previously (35). Since this gp130 variant is unable to signal independently of IL-6R, we used it as a means of testing, by cotransfection assays, the vIL-6 variants for their ability to signal through gp130.PM5–IL-6R complexes and so identify amino acid residues in vIL-6 that might be important for IL-6R recognition and/or functional vIL-6–gp130–IL-6R complex formation. Initially, however, we sought to determine the likely effect of the PM5 mutation (residues Y190FV192 changed to AAA) on vIL-6 binding, as determined by coprecipitation assays using gp130.PM5-Fc (see Materials and Methods). The results of these assays are shown in Fig. 5B and demonstrate the reduced ability of gp130.PM5 to bind vIL-6. This supports our earlier hypothesis (35) that gp130.PM5 cannot form stable interactions with vIL-6 and that IL-6R complexes with gp130.PM5 and vIL-6 to allow signaling through gp130.PM5.
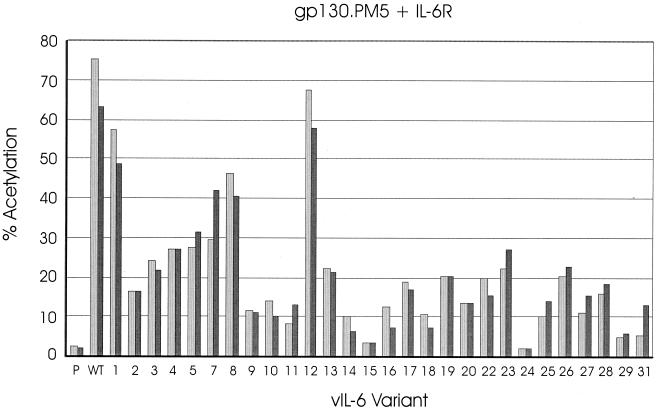
In cotransfections of the vIL-6 variant expression constructions with gp130.PM5 and IL-6R, the results were very different from those obtained in Hep3B cells (35) and in the gp130 cotransfection studies (Fig. 3B). The data from the gp130.PM5–IL-6R cotransfection experiments (Fig. 6) identified several vIL-6 variants (i.e., variants 2, 9, 10, 11, 14, 16, 17, 18, 20, 22, 25, 27, 28, 29, and 31), in addition to variants 15 and 24, that were greatly abrogated in their ability to signal (less than 25% of wild-type). The amino acid changes in these proteins occurred in positions corresponding to site I (variants 9, 10, 20, 22, 25, and 27), site II (variant 11), a conserved F183 residue previously reported to be important for high-affinity cell surface binding by hIL-6 (16) (variants 17, 18, 29, and 31), loci containing interspecies-conserved residues (variants 2 and 14), and the C-terminal 10 amino acids of vIL-6 (deleted in variant 28) representing an “extension” relative to endogenous IL-6 proteins. It is likely that these residues occur in regions that are important for direct interactions of vIL-6 with IL-6R and/or gp130. Multiple receptor interaction points in vIL-6 would allow “bridging” between neighboring IL-6R and gp130 proteins to allow complex formation between vIL-6 and IL-6R–gp130 dimers; disruption of protein-protein interaction interfaces in gp130.PM5 (vIL-6–gp130) and the signaling-defective vIL-6 variants (vIL-6–IL-6R or vIL-6–gp130) would thus affect the formation of stable vIL-6–IL-6R–gp130 complexes and signaling. For the vIL-6 variants that are specifically defective in the gp130.PM5–IL-6R-type transfections, it is uncertain from these data whether they are defective for binding to gp130, IL-6R, or both receptor subunits or, indeed, whether they bind to the receptor subunits but are impaired in their ability to form signaling complexes.
FIG. 6.
IL-6R-dependent signaling of vIL-6 variants through gp130.PM5. Transfection assays similar to those outlined in Fig. 3 were carried out to investigate the profile of activities of the vIL-6 variants, relative to pSG5 (P) and vIL-6 (WT) signaling through overexpressed gp130.PM5 and IL-6R. The results of duplicate experiments are shown.
Signaling-defective vIL-6 variants do not inhibit vIL-6 effects through gp130.PM5–IL-6R.
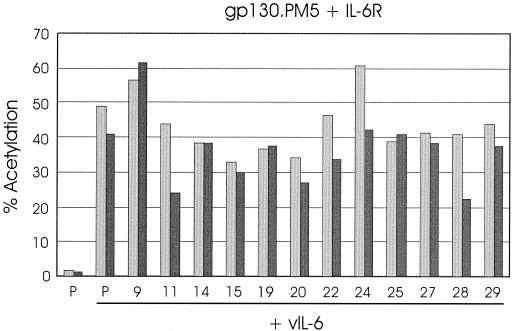
Competition experiments similar to those undertaken with vIL-6, gp130, and the variants 15 and 24 (Fig. 4) were performed to determine whether vIL-6 proteins showing reduced activity through cotransfected gp130.PM5–IL-6R could inhibit IL-6R-dependent signaling by wild-type vIL-6 through gp130.PM5. Several of the functionally defective variants, including 15 and 24, were cotransfected with native vIL-6, gp130.PM5, and IL-6R, and the levels of CAT activity in the cell extracts were compared with those from cells transfected with pSG5 instead of competitor. The results of these experiments are shown in Fig. 7. None of the vIL-6 variants was able to substantially inhibit vIL-6 activity, indicating a lack of competitive binding to gp130.PM5 and IL-6R. For variants 15 and 24, the results in this system contrast sharply with those obtained in gp130 transfected cells (Fig. 4), suggesting that gp130 binding, abrogated by the PM5 mutation (Fig. 5), is essential for the inhibitory effects of vIL-6.15 and vIL-6.24 on vIL-6 signaling through gp130.
FIG. 7.
Effects of functionally altered vIL-6 variants on vIL-6 signaling through gp130.PM5–IL-6R. Cotransfections of selected vIL-6 variants or pSG5 (P) with wild-type vIL-6 and gp130.PM5, IL-6R, and pα2MCAT were undertaken (see Materials and Methods). The CAT activities present in cell extracts from duplicate experiments are shown.
DISCUSSION
Numerous studies, using a variety of approaches, including X-ray crystallography, mutagenesis, and in vitro and in vivo binding assays, have been undertaken to determine the structures of hIL-6, IL-6R, and gp130 and the basis of their interactions. It has been determined that there are three main receptor-interaction sites of hIL-6 (sites I, II, and III) that are involved in interactions with IL-6R (site I) and in hIL-6–IL-6R associations with gp130 (sites II and III). However, the precise nature of the hIL-6 interactions with IL-6R and gp130 and the conformational structure of the hexameric signaling complex formed between ligand and receptor subunit dimers remain to be determined. Binding assays using specifically altered hIL-6 or receptor subunit molecules have been extremely useful for the identification of residues likely to be involved in ligand-receptor interactions, but it has not been possible to examine hIL-6–gp130 interactions in the absence of IL-6R. The fact that vIL-6 can bind directly to gp130 affords the opportunity to examine vIL-6–gp130 interactions more directly.
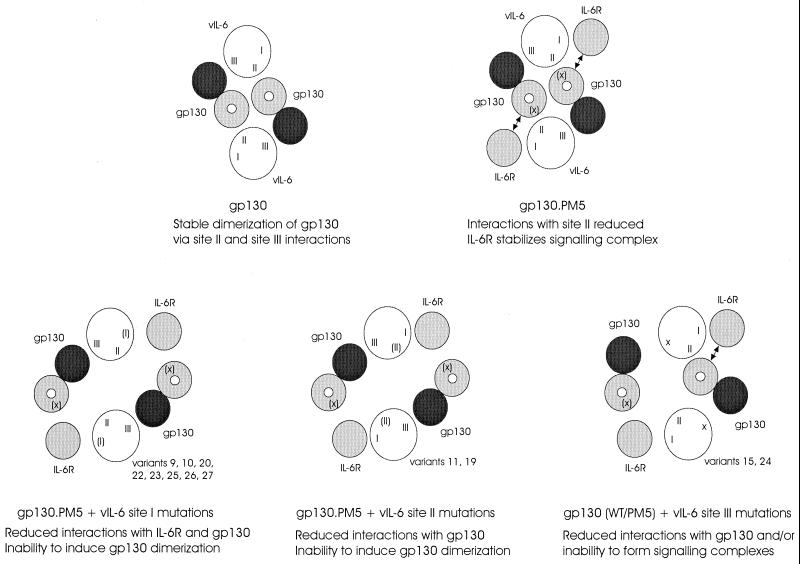
In the present study we have established that vIL-6 interacts directly with gp130 and IL-6R and induces gp130–IL-6R complex formation and have utilized our panel of vIL-6 variants in cotransfections with gp130 and gp130.PM5–IL-6R to examine residues important for IL-6R-independent and IL-6R-dependent signaling. The main findings from our functional assays are summarized in Table 2, and models for their interpretation are shown in Fig. 8. Of relevance to interactions of vIL-6 with gp130 was the finding that vIL-6 residues W167 and A172, occurring in site III and altered in variants 15 and 24 and that we previously found were unable to signal in Hep3B cells (which express gp130 and IL-6R), were also functionally defective in the gp130 and gp130.PM5–IL-6R cotransfection studies presented here and vIL-6.24 showed reduced in vitro binding to gp130. This suggests that the site III variants have altered interactions with gp130. However, both acted as dominant negatives in competition with vIL-6 in gp130 cotransfection assays, indicating that they can form stable, possibly higher-order (but nonfunctional) complexes with gp130 within the context of the cell. This conclusion is supported by the finding that variants 15 and 24 failed to influence signaling by wild-type vIL-6 through gp130.PM5, in the presence of IL-6R; gp130 binding is therefore necessary for inhibition of vIL-6 signaling. These data indicate that the functional deficiencies of vIL-6 variants 15 and 24 are likely to be due to their inabilities to induce higher-order signaling complexes with gp130 (involving appropriately conformed gp130 dimers) rather than their failure to recognize gp130.
TABLE 2.
Summary of transfection data for functionally altereda vIL-6 variants
| vIL-6 variant | Hep3Bb | gp130 transfection | gp130–IL-6R transfection | Comments |
|---|---|---|---|---|
| WT | +++ | +++ | +++ | |
| 9 | +++ | +++ | + | Site I, C93G |
| 10 | +++ | +++ | + | Site I, C93W |
| 20 | ++++ | ++ | + | Site I, F88L |
| 22 | +++ | ++ | + | Site I, FE105GG |
| 23 | +++ | +++ | ++ | Site I, EFE105GVA |
| 25 | +++ | ++++ | + | Site I, R189L |
| 26 | +++ | +++ | ++ | Site I, R189S |
| 27 | +++ | ++++ | + | Site I, R189C |
| 11 | +++ | ++ | + | Site II, T129L |
| 19 | +++ | ++++ | ++ | Site II, W41F + L43G |
| 15 | + | + | − | Site III, W167G; dominant negativec |
| 16 | +++ | +++ | + | Site III, W167R |
| 24 | + | − | − | Site III, W167R + A172P; dominant negativec |
| 2 | +++ | ++ | + | C54D |
| 3 | +++ | +++ | ++ | C54G |
| 13 | +++ | +++ | ++ | P151F |
| 14 | +++ | +++ | + | P151S |
| 17 | +++ | ++ | + | F183W |
| 18 | +++ | ++ | + | F183G |
| 29 | +++ | ++ | − | F183E |
| 31 | +++ | ++ | + | F183L |
| 28 | +++ | +++ | + | C-terminal deletion (residues 195 to 204) |
Signaling efficiencies relative to wild type (WT) vIL-6 are indicated: ++++, >125%; +++, 75 to 125%; ++, 25 to 75%; +, 10 to 25%; −, 0 to 5%. Scoring less than 35% wild-type vIL-6 activity in at least one assay.
Data from transfections of IL-6R+ gp130+ Hep3B cells (35).
Determined by cotransfection with vIL-6 and gp130 (Fig. 4).
FIG. 8.
Models of vIL-6 interactions with gp130 and IL-6R, formation of functional signaling complexes, and effects of vIL-6 mutations on complex formation. Functional complexes, in which gp130 molecules (marked by white dots) are brought into close proximity, result from interactions of vIL-6 (e.g., via sites I and II) with different gp130 molecules. Amino acid changes affecting but not destroying interaction sites (e.g., the PM5 mutation in gp130, which occurs in one of two loci predicted to interact with vIL-6 site II) are indicated by parentheses [(X), (I), (II)]; site III changes in vIL-6 variants 15 and 24 (indicated by an “X” at the appropriate position in vIL-6) are presumed to abolish receptor interactions through this site.
With regard to signaling by vIL-6 variants through gp130.PM5 in the presence of IL-6R, many of the vIL-6 alterations led to significant decreases in signal transduction. These signaling deficiencies could be due to effects of the mutations on vIL-6 binding to IL-6R and/or gp130 or on the formation or conformation of higher-order signaling complexes. For hIL-6, experimental and structural data have indicated that distinct regions of hIL-6 interact with IL-6R (site I), gp130 cytokine receptor homology domain (site II), and gp130 immunoglobulin-like domain (site III) (10, 12, 15, 21, 25, 36). By analogy, our functional data for vIL-6 variants 9, 10, 20, 22, 23, 25, 26, and 27 (altered in site I and showing reduced IL-6R-dependent signaling activities) and variants 15 and 24 (altered in site III and being negative in both IL-6R-independent and IL-6R-dependent functional assays) can be explained by effects of the amino acid alterations on IL-6R and gp130 (immunoglobulin domain) binding, respectively (Fig. 8). Decreased activities of site II variants 11 and 19 in IL-6R-dependent signaling through the gp130 CHD E-F loop variant gp130.PM5 are likely to reflect decreased interactions with distinct site II-interacting gp130 CHD residues (Fig. 8). Other vIL-6 variants (i.e., variants 2, 3, 13, 14, 17, 18, 28, 29, and 31) that showed reduced IL-6R-dependent signaling through gp130.PM5 may be altered in their abilities to bind IL-6R or gp130.
The data presented here have established that HHV-8-encoded vIL-6 can bind directly and independently to IL-6R (reported by others not to be involved in vIL-6 signaling [19, 22]) and gp130 and can induce gp130–IL-6R complexing, and we have identified residues in vIL-6 that affect vIL-6 signaling through gp130 alone or through IL-6R-dependent gp130.PM5 and that may represent receptor subunit interaction sites. Results from the in vitro binding assays are in agreement with our previous functional data from gp130.PM5–IL-6R cotransfections (35) indicating that vIL-6, like other IL-6 proteins, can complex with gp130 and IL-6R, although clearly they also are consistent with the previously reported ability of vIL-6 to bind to and signal through gp130 in the absence of IL-6R (19, 22, 35). Data from our IL-6R–gp130.PM5 cotransfection assays demonstrate that residues in vIL-6 that correspond to site I and site III regions of hIL-6 are important for IL-6R-dependent signaling through gp130.PM5 and may represent analogous receptor-binding interfaces. However, we have also identified additional residues, namely C54, P151, F183, and the C-terminal region of vIL-6, that also are important for IL-6R–gp130.PM5 signaling and that may include additional receptor binding residues. Future delineation of differences in vIL-6 and hIL-6 receptor recognition and signaling complex formation could potentially be exploited to specifically block the action of vIL-6, a cytokine that may be important in HHV-8-induced pathogenesis.
ACKNOWLEDGMENT
This work was supported by NIH grant R01-CA76445.
REFERENCES
- 1.Aoki Y, Jaffe E S, Chang Y, Jones K, Teruya-Feldstein J, Moore P S, Tosato G. Angiogenesis and hematopoiesis induced by Kaposi's sarcoma-associated herpesvirus-encoded interleukin-6. Blood. 1999;93:4034–4043. [PubMed] [Google Scholar]
- 2.Asou H, Said J W, Yang R, Munker R, Park D J, Kamada N, Koeffler H P. Mechanisms of growth control of Kaposi's sarcoma-associated herpes virus-associated primary effusion lymphoma cells. Blood. 1998;91:2475–2481. [PubMed] [Google Scholar]
- 3.Brakenhoff J P J, de Hon F D, Fontaine V, Ten Boekel E, Schooltink H, Rose-John S, Heinrich P C, Content J, Aarden L A. Development of a human interleukin-6 receptor agonist. J Biol Chem. 1994;269:86–93. [PubMed] [Google Scholar]
- 4.Bravo J, Staunton D, Heath J K, Jones E Y. Crystal structure of a cytokine-binding region of gp130. EMBO J. 1998;17:1665–1674. doi: 10.1093/emboj/17.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burger R, Neipel F, Fleckenstein B, Savino R, Ciliberto G, Kalden J R, Gramatzki G. Human herpesvirus type 8 interleukin-6 homologue is functionally active on human melanoma cell lines. Blood. 1998;91:1858–1863. [PubMed] [Google Scholar]
- 6.Cannon J S, Nicholas J, Ornstein J M, Mann R B, Murray P G, Browning P J, DiGiuseppe J A, Cesarman E, Hayward G S, Ambinder R F. Heterogeneity of viral IL-6 expression in HHV-8-associated diseases. J Infect Dis. 1999;180:824–828. doi: 10.1086/314956. [DOI] [PubMed] [Google Scholar]
- 7.Ehlers M, Grötzinger J, de Hon F D, Müllberg J, Brakenhoff J P J, Liu J, Wollmer A, Rose-John S. Identification of two novel regions of human IL-6 responsible for receptor binding and signal transduction. J Immunol. 1994;153:1744–1753. [PubMed] [Google Scholar]
- 8.Fontaine V, Savino R, Arcone R, De Wit L, Brakenhoff J P, Content J, Ciliberto G. Involvement of the Arg179 in the active site of human IL-6. Eur J Biochem. 1993;211:749–755. doi: 10.1111/j.1432-1033.1993.tb17605.x. [DOI] [PubMed] [Google Scholar]
- 9.Foussat A, Wijdenes J, Bouchet L, Gaidano G, Neipel F, Balabanian K, Galanaud P, Couderc J, Emilie D. Human interleukin-6 is in vivo an autocrine growth factor for human herpesvirus-8-infected malignant B lymphocytes. Eur Cyt Netw. 1999;10:501–508. [PubMed] [Google Scholar]
- 10.Hammacher A, Richardson R T, Layton J E, Smith D K, Angus L J, Hilton D J, Nicola N A, Wijdenes J, Simpson R J. The immunoglobulin-like module of gp130 is required for signaling by interleukin-6, but not by leukemia inhibitory factor. J Biol Chem. 1998;273:22701–22707. doi: 10.1074/jbc.273.35.22701. [DOI] [PubMed] [Google Scholar]
- 11.Hammacher A, Ward L D, Weinstock J, Treutlein H, Yasukawa K, Simpson R J. Structure-function analysis of human IL-6: identification of two distinct regions that are important for receptor binding. Protein Sci. 1994;3:2280–2293. doi: 10.1002/pro.5560031213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horsten U, Müller-Newen G, Gerhartz C, Wollmer A, Wijdenes J, Heinrich P C, Grötzinger J. Molecular modeling-guided mutagenesis of the extracellular part of gp130 leads to the identification of contact sites in the interleukin-6 (IL-6).IL-6 receptor.gp130 complex. J Biol Chem. 1997;272:23748–23757. doi: 10.1074/jbc.272.38.23748. [DOI] [PubMed] [Google Scholar]
- 13.Jones K D, Aoki Y, Chang Y, Moore P S, Yarchoan R, Tosato G. Involvement of interleukin-10 (IL-10) and viral IL-6 in the spontaneous growth of Kaposi's sarcoma herpesvirus-associated infected primary effusion lymphoma cell lines. Blood. 1999;94:2871–2879. [PubMed] [Google Scholar]
- 14.Kalai M, Montero-Julian F A, Grötzinger J, Fontaine V, Vandenbussche P, Deschuyteneer R, Wollmer A, Brailly H, Content J. Analysis of the human interleukin-6/human interleukin-6 receptor binding interface at the amino acid level: proposed mechanism of interaction. Blood. 1997;89:1319–1333. [PubMed] [Google Scholar]
- 15.Kurth I, Horsten U, Pflanz S, Dahmen H, Kuster A, Grotzinger J, Heinrich P C, Müller-Newen G. Activation of the signal transducer glycoprotein 130 by both IL-6 and IL-11 requires two distinct binding epitopes. J Immunol. 1999;162:1480–1487. [PubMed] [Google Scholar]
- 16.Li X, Rock F, Chong P, Cockle S, Keating A, Ziltner H, Klein M. Structure-function analysis of the C-terminal segment of human interleukin-6. J Biol. 1993;268:22377–22384. [PubMed] [Google Scholar]
- 17.Menziani M C, Fanelli F, Benedetti P G. Theoretical investigations of IL-6 multiprotein receptor assembly. Proteins. 1997;29:528–544. [PubMed] [Google Scholar]
- 18.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molden J, Chang Y, You Y, Moore P S, Goldsmith M A. A Kaposi's sarcoma-associate herpesvirus-encoded cytokine homolog (vIL-6) activates signaling through the shared gp130 receptor subunit. J Biol Chem. 1997;272:19625–19631. doi: 10.1074/jbc.272.31.19625. [DOI] [PubMed] [Google Scholar]
- 20.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 21.Moritz R L, Ward L D, Tu G F, Fabri L J, Ji H, Yasukawa K, Simpson R J. The N-terminus of gp130 is critical for the formation of the high affinity interleukin-6 receptor complex. Growth Factors. 1999;16:265–278. doi: 10.3109/08977199909069145. [DOI] [PubMed] [Google Scholar]
- 22.Müllberg J, Geib T, Jostock T, Hoischen S H, Vollmer P, Voltz N, Heinz D, Galle P R, Klouche M, Rose-John S. IL-6 receptor independent stimulation of human gp130 by viral IL-6. J Immunol. 2000;164:4672–4677. doi: 10.4049/jimmunol.164.9.4672. [DOI] [PubMed] [Google Scholar]
- 23.Nicholas J, Ruvolo V R, Burns W H, Sandford G, Wan X, Ciufo D, Hendrickson S B, Guo H-G, Hayward G S, Reitz M S. Kaposi's sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat Med. 1997;3:287–292. doi: 10.1038/nm0397-287. [DOI] [PubMed] [Google Scholar]
- 24.Osborne J, Moore P S, Chang Y. KSHV-encoded viral IL-6 activates multiple human IL-6 signaling pathways. Hum Immunol. 1999;60:921–927. doi: 10.1016/s0198-8859(99)00083-x. [DOI] [PubMed] [Google Scholar]
- 25.Paonessa G, Graziani R, De Serio A, Savino R, Ciapponi L, Lahm A, Salvati A L, Toniatti C, Ciliberto G. Two distinct and independent sites on IL-6 trigger gp130 dimer formation and signalling. EMBO J. 1995;14:1942–1951. doi: 10.1002/j.1460-2075.1995.tb07186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parravicini C, Chandran B, Corbellino M, Berti E, Paulli M, Moore P S, Chang Y. Differential viral protein expression in Kaposi's sarcoma-associated herpesvirus-infected diseases. Am J Pathol. 2000;156:743–749. doi: 10.1016/S0002-9440(10)64940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salvati A L, Lahm A, Paonessa G, Ciliberto G, Toniatti C. Interleukin-6 (IL-6) antagonism by soluble IL-6 receptor α mutated in the predicted gp130-binding interface. J Biol Chem. 1995;270:12242–12249. doi: 10.1074/jbc.270.20.12242. [DOI] [PubMed] [Google Scholar]
- 28.Savino R, Ciapponi L, Lahm A, Demartis A, Cabibbo A, Toniatti C, Delmastro P, Altamura S, Ciliberto G. Rational design of a receptor super-antagonist of human interleukin-6. EMBO J. 1994;13:5863–5870. doi: 10.1002/j.1460-2075.1994.tb06931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savino R L, Lahm A, Giorgio M, Tramontano A, Ciliberto G. Saturation mutagenesis of the human interleukin-6 receptor-binding site: implications for its three-dimensional structure. Proc Natl Acad Sci USA. 1993;90:4067–4071. doi: 10.1073/pnas.90.9.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savino R L, Lahm A, Salvati A L, Ciapponi L, Sporeno E, Altamura S, Paonessa G, Toniatti C, Ciliberto G. Generation of interleukin-6 receptor antagonists by molecular-modeling guided mutagenesis of residues important for gp130 activation. EMBO J. 1994;13:1357–1367. doi: 10.1002/j.1460-2075.1994.tb06389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaefer T S, Sanders L K, Nathans D. Cooperative transcriptional activity of Jun and Stat3β, a short form of Stat3. Proc Natl Acad Sci USA. 1995;92:9097–9101. doi: 10.1073/pnas.92.20.9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simpson R J, Hammacher A, Smith D K, Matthews J M, Ward L D. Interleukin-6: structure-function relationships. Protein Sci. 1997;6:929–955. doi: 10.1002/pro.5560060501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Somers W, Stahl M, Seehra J S. 1.9 Å crystal structure of interleukin-6: implications for a novel mode of receptor dimerization and signaling. EMBO J. 1997;16:989–997. doi: 10.1093/emboj/16.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staskus K A, Sun R, Miller G, Racz P, Jaslowski A, Metroka C, Brett-Smith H, Haase A T. Cellular tropism and viral interleukin-6 expression distinguish human herpesvirus 8 involvement in Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. J Virol. 1999;73:4181–4187. doi: 10.1128/jvi.73.5.4181-4187.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan X, Wang H, Nicholas J. Human herpesvirus 8 interleukin-6 (vIL-6) signals through gp130 but has structural and receptor-binding properties distinct from those of human IL-6. J Virol. 1999;73:8268–8278. doi: 10.1128/jvi.73.10.8268-8278.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward L D, Howlett G J, Discolo G, Yasukawa K, Hammacher A, Moritz R L, Simpson R J. High affinity interleukin-6 receptor is a hexameric complex consisting of two molecules of interleukin-6, interleukin-6 receptor and gp130. J Biol Chem. 1994;269:23286–23289. [PubMed] [Google Scholar]
- 37.Yawata H, Yasukawa K, Natsuka S, Murakami M, Yamasaki K, Hibi M, Taga T, Kishimoto T. Structure-function analysis of human IL-6 receptor: dissociation of amino acid residues required for IL-6 binding and for IL-6 signal transduction through gp130. EMBO J. 1993;12:1705–1712. doi: 10.1002/j.1460-2075.1993.tb05815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]