Abstract
Chronic myelomonocytic leukemia (CMML) is a rare blood cancer of older adults (3 in every 1,000,000 persons) characterized by poor survival and lacking effective mutation-specific therapy. Mutations in the ubiquitin ligase Cbl occur frequently in CMML and share biological and molecular features with a clonal disease occurring in children, juvenile myelomonocytic leukemia (JMML). Here we analyzed the clinical presentations, molecular features and immunophenotype of CMML patients with CBL mutations enrolled in a prospective Phase II clinical trial stratified according to molecular markers. Clinically, CBL mutations were associated with increased bone marrow blasts at diagnosis, leukocytosis and splenomegaly, similar to patients harboring NRAS or KRAS mutations. Interestingly, 64% of patients presented with more than one CBL variant implying a complex subclonal architecture, often with co-occurrence of TET2 mutations. We found CBL mutations in CMML frequently clustered in the RING domain in contrast to JMML, where mutations frequently involve the linker helix region (P<0.0001). According to our comparative alignment of available X-ray structures, mutations in the linker helix region such as Y371E give rise to conformational differences that could be exploited by targeted therapy approaches. Furthermore, we noted an increased percentage of CMML CD34+ stem and progenitor cells expressing CD116 and CD131 in all CBL mutant cases and increased CD116 receptor density compared to healthy controls, similar to CMML overall. In summary, our data demonstrate that CBL mutations are associated with distinct molecular and clinical features in CMML and are potentially targetable with CD116-directed immunotherapy.
Introduction
Monocytes give rise to tissue macrophages that can perform a myriad of biological functions ranging from innate immune activation to phagocytosis and wound healing. Chronic myelomonocytic leukemia (CMML) is a rare blood cancer of older adults (3 in every 1,000,000 persons) [1, 2] characterized by an increase in clonal CD14+ CD16– classical monocytes and their precursors in the blood and bone marrow [3, 4]. Because of its subtle presentation, the disease is often diagnosed late, but is likely to rise in prevalence due to routine uptake of next-generation sequencing, lower threshold monocytosis criteria by the World Health Organization and increasing recognition by physicians, especially in persons previously treated with cytotoxic therapy (therapy-related CMML) [5, 6]. Many patients present with autoinflammatory features such as vasculitis, polychondritis, Sweet syndrome and pleural/pericardial effusions [7–10] but the mechanism and intersection of clonal monocytes and innate vs. adaptive immunity is not understood.
The molecular pathogenesis of CMML is only beginning to be characterized. Though 70% of CMML patients present without any cytogenetic abnormalities [11, 12] they harbor somatic mutations in genes that influence epigenetic regulation (TET2, DNMT3A, ASXL1, EZH2, IDH1, IDH2), mRNA splicing (SRSF2, U2AF1), genome stability (SETBP1, TP53), transcription regulation (RUNX1, CEBPA, NPM1) and cell signaling pathways (KRAS, NRAS, CBL, PTPN11, JAK2, MPL) [13–17]. Interestingly, CMML shares some biological and morphological features with a clonal disease occurring in young children, juvenile myelomonocytic leukemia (JMML). Around 90% of cases of JMML are associated with mutations in the RAS signaling pathway (PTPN11, NRAS, KRAS, NF1 and CBL) [18–21]. Notably, while the overall molecular patterns of CMML and JMML are distinct, mutations in CBL are found with equal frequency, at approximately 15% [17, 18, 22, 23], in both diseases. Both CMML and JMML display hypersensitivity to the pro-inflammatory cytokine granulocyte-macrophage colony-stimulating factor (GM-CSF) which promote the differentiation of classical pro-inflammatory monocytes [24, 25]. Emerging reports suggest that CBL mutations are associated with inferior survival in both CMML and JMML [15, 26].
The CBL gene is located on 11q23.3 and encodes an E3 ubiquitin ligase (c-Cbl or Cbl) that acts as both a positive and negative regulator in the signal transduction of activated receptor tyrosine kinases (RTKs) and cytokine receptors. Cbl relays signals downstream of activated RTKs by functioning as an adaptor [27–30], and at the same time, attenuates signaling by promoting the ubiquitination of RTKs through its E3 ligase activity, marking them for degradation by the proteasome or via endocytosis [31–33]. Cbl recognizes phosphorylated tyrosines on active RTKs through its Src homology 2 domain within the tyrosine kinase binding domain (TKBD), and binds E2 ubiquitin-conjugating enzymes though its conserved RING domain. The TKBD and RING domain are connected by a 28 amino acid sequence, referred to as the linker helix region (LHR). A proline-rich region, serine-rich region, several C-terminal principal phosphorylation sites (Y674, Y700, Y731, Y774) and a ubiquitin-association domain (UBA) complete the structure of Cbl. Several studies on the role of the LHR and RING domain have shown that mutation of these domains, could lead to loss of activity and/or gain of oncogenicity [34–42].
To date, there has not been significant mutation-specific therapy developed for CMML, unlike chronic myeloid leukemia, and standard-of-care with hypomethylating agents azacitidine or decitabine is not curative [16, 43–47]. While allogeneic hematopoietic stem cell transplantation may be potentially curative, stem cell transplantation is not a viable option for most older CMML patients [48, 49]. Currently, survival is estimated at a median of 31 months, with even shorter life expectancy for patients with CMML-2 (more than 10% bone marrow blasts/promonocytes). Transformation to acute myeloid leukemia (AML), an aggressive cancer with poor long-term survival, occurs in up to 20% of CMML patients within 5 years [50–52].
In this study, we analyzed the clinical presentation of CMML patients with CBL mutations enrolled in a prospective clinical trial. Strikingly, 7 out of 11 (64%) patients were found to have more than one CBL clone, implying a complex clonal architecture. Clinically, CBL mutations were associated with a more proliferative phenotype evidenced by increased bone marrow blasts, leukocytosis and splenomegaly, similar to other RAS pathway mutations such as KRAS, NRAS and PTPN11. We also found that CMML CBL mutations often co-occurred with TET2 mutations and were enriched in the RING domain compared to the LHR (P<0.0001). Furthermore, we noted an increased percentage of CD116 and CD131-expressing CMML CD34+ progenitors compared to healthy controls. In summary, our data suggest that CBL mutants are associated with distinct clinical and molecular features in CMML.
Materials and methods
Patients
Between 1 October 2021 and 30 September 2023, 24 patients with CMML, diagnosed according to the 2016 WHO Classification of Myeloid Neoplasms [53], who met the eligibility criteria (untreated CMML with high white cell count, cytopenia or constitutional symptoms) were enrolled in the trial with written, informed consent. Detection of TET2, KRAS, NRAS or CBL mutation at a variant allele frequency (VAF) percentage of ≥3% was a key inclusion criteria. Of these patients, 13 were male and 11 were female. The median age was 73 years (range 56−86 years). Written informed consent for genetic analysis and use of laboratory results and samples for scientific research were obtained during trial enrolment. The trial was approved in multiple centers across Australia including the Royal Adelaide Hospital, Royal Brisbane and Women’s Hospital and Austin Health. The trial was conducted with approval from the Central Adelaide Local Health Network Human Research Ethics Committee (2021/HRE00017) and registered on the Australian and New Zealand Clinical Trials Registry (ANZCTR) (Registration number ACTRN12621000223831, Acronym PREACH-M).
Clinical presentation
All clinical and laboratory data were acquired at screening, prior to commencement of the therapy protocol outlined in the trial. Complete blood examination and bone marrow (BM) analyses were performed for each patient. The spleen craniocaudal length was determined by ultrasonography.
Mutation screening
Targeted enrichment of selected coding exons and flanking intronic regions of 46 genes was performed using a custom-designed hematological neoplasms capture panel (Integrated DNA Technologies; HaemV1) and analyzed by next-generation sequencing (NGS) (Illumina NextSeq sequencing system). Variant calling was performed using Vardict and Mutect2 where variants with VAF <5% were reported where clinically significant. These assays were performed by accredited pathology laboratories across Australia.
Cord blood and peripheral blood mononuclear cells from healthy donors
Umbilical cord blood was collected with written informed consent from scheduled cesarean section deliveries at the Women’s Health Unit, Lyell-McEwin Hospital (Adelaide, South Australia) between 12 November 2020 to 31 December 2023 with approval from the Women’s and Children’s Health Network Human Research Ethics Committee (HREC/20/WCHN/65; 2020/HRE01664). Peripheral blood buffy coat samples obtained with written informed consent was retrieved on 29 May 2023 and studies were approved by the Central Adelaide Local Health Network Human Research Ethics Committee (HREC/15/RAH/448) and conducted in accordance with the Declaration of Helsinki. Samples were processed by density gradient centrifugation using LymphoprepTM (Stemcell Technologies, USA) to isolate mononuclear cells (MNCs).
Flow cytometry
Peripheral blood mononuclear cells (PB-MNCs) were collected at baseline from patients in the PREACH-M trial. 2–3 CBL mutant and 2–3 CBL wildtype CMML samples (n = 4–6) were immunophenotyped by spectral flow cytometry (Cytek Aurora, USA). Control samples include cord blood mononuclear cells (CB-MNCs) (n = 2) and PB-MNCs from healthy donors (n = 1). All MNCs were incubated with Human TruStain FcXTM (BioLegend, USA) and True-Stain Monocyte BlockerTM (BioLegend, USA) prior to antibody staining. Antibody panel included mouse anti-human CD45 (HI30) BV421, CD14 (M5E2) PE-Cy7, CD16 (3G8) PE-Cy5, CD34 (8G12) APC, CD114 (LMM741) PE, CD116 (hGMCSFR-M1) PE, CD131 (1C1) PE; CD131 (3D7) BV421 and rat anti-human CD115 (9-4D2-1E4) PE. Viability was determined using Zombie AquaTM Fixable Dye (BioLegend, USA). A list of catalogue numbers and clones of antibodies can be found in S1 Table.
Hotspot and protein structure analysis
Data from the Catalogue of Somatic Mutations in Cancer (COSMIC) were analyzed (last accessed on 15 February 2024). Analysis of CBL mutations were filtered as follows: Primary site hematopoietic and lymphoid, histology hematopoietic neoplasm; sub-histology CMML or JMML. 120 variants were found for CMML, 46 for JMML. Variants were stratified to include nonsense or missense substitutions, frameshift insertion or deletions, and in-frame insertions or deletions, with the exclusion of synonymous mutations, within the coding sequence. 17 coding region variants were detected in the PREACH-M cohort, making the total variants for CMML 137. The six most common mutations for CMML and JMML were identified. Protein structures were sourced from the Protein Data Bank (PDB) and analyzed using Maestro 13.8 (Schrodinger, USA) and UCSF ChimeraX (UCSF Resource for Biocomputing, Visualization and Information, USA). RMSD calculations were performed without rejection of any atoms during fit and distance was measured between alpha carbons of residue 227 and 371 of Cbl.
Statistical analysis
Values between unique samples were presented as mean ± standard error of margin (S.E.M.) or as mean ± standard deviation (S.D.) between technical replicates. For comparisons between groups, Student’s t-test or Mann-Whitney test was applied to analyze measurement (continuous) data and Fisher’s exact test for enumeration (categorical) data. All statistical analyses were performed using GraphPad Prism 10. P-values for Student’s t-tests were two-tailed, Mann-Whitney tests were one-tailed, and Fisher’s exact tests were two-tailed. P<0.05 was considered statistically significant.
Results
CBL mutations are associated with increased marrow blasts, leukocytosis and splenomegaly, consistent with RAS pathway activation
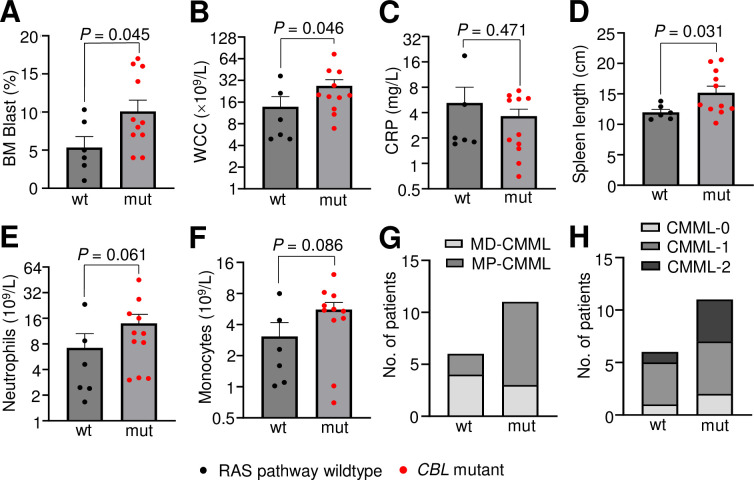
Targeted NGS was performed on 24 de novo CMML baseline patient samples from the PREACH-M trial. Overall, RAS pathway (KRAS, NRAS, PTPN11, CBL) mutations were detected in 18/24 (75%) patients with CBL mutations detected in 11/24 (46%) patients (Tables 1, 2 and S2). Consistent with previous findings where RAS pathway mutations were linked to the proliferative variant of CMML [54, 55], patients with RAS pathway mutations had increased BM blast percentage (9.2 ± 1.1 vs. 5.3. ± 1.4%, P = 0.05), white cell count (WCC) (30.4 ± 6.0 vs. 13.8 ± 5.3×109/L, P = 0.02), neutrophil absolute count (15.2 ± 3.3 vs. 7.2 ± 3.4×109/L, P = 0.03), monocyte absolute count (7.8 ± 1.7 vs. 3.1 ± 1.1×109/L, P = 0.03) and spleen length (15.2 ± 0.9 vs. 12.0 ± 0.5 cm, P = 0.04) compared to patients wildtype for any RAS pathway mutations (Table 1). This finding led us to further stratify our cohort and indeed, patients with CBL mutations were also noted to have increased BM blast percentage (10.1 ± 1.5 vs. 5.3 ± 1.4%, P = 0.05), WCC (26.8 ± 5.9 vs. 13.8 ± 5.2×109/L, P = 0.05), and spleen length (15.2 ± 1.1 vs. 12.0 ± 0.5 cm, P = 0.03) compared to CBL wildtype patients without RAS pathway mutations (Table 2 and Fig 1A–1F). Comparisons between CBL mutant/RAS pathway wildtype vs. RAS pathway mutant/CBL wildtype cases revealed consistent trends (S1 Fig). 10/11 (90.9%; P = 0.03) patients with CBL variants presented with splenomegaly (S3 Table). This underscores the strong proliferative phenotype conferred by mutations in CBL in CMML. Importantly, 8/11 (73%) CBL variants were in cases classified as myeloproliferative-CMML (MP-CMML) based on WCC (Fig 1G), and 9/11 (82%) classified as CMML-1 or -2 based on BM blast percentage (Fig 1H) according to the 2016 WHO classification, linking CBL mutations not just to a proliferative phenotype but to more advanced stages of the disease, and therefore, to increased risk of progression to AML [50].
Table 1. Clinical characteristics, complete blood examination and bone marrow analyses of CMML patients in the PREACH-M trial stratified as RAS pathway (KRAS, NRAS, PTPN11, CBL) mutant vs. wildtype.
| Variable | Total (n = 24) | RAS pathway wildtype (n = 6) | RAS pathway mutant (n = 18) | P-value |
|---|---|---|---|---|
| Gender | ||||
| Male, n (%) | 13 (54%) | 2 (15%) | 11 (85%) | 0.3572 |
| Female, n (%) | 11 (46%) | 4 (36%) | 7 (64%) | |
| Age (years) | ||||
| Mean (range) | 72 (56–86) | 73 (56−86) | 70 (56−79) | 0.1424 |
| WHO classification | ||||
| CMML-0, n (%) | 4 (17%) | 1 (17%) | 3 (17%) | 0.8215 |
| CMML-1, n (%) | 13 (54%) | 4 (67%) | 9 (50%) | |
| CMML-2, n (%) | 7 (29%) | 1 (17%) | 6 (33%) | |
| MD-CMML, n (%) | 9 (38%) | 4 (67%) | 5 (28%) | 0.1501 |
| MP-CMML, n (%) | 15 (63%) | 2 (33%) | 13 (72%) | |
| BM Blast (%) | ||||
| Mean (range) | 7.3 (1.0−17.0) | 5.3 (1.0−10.3) | 9.2 (2.0−17.0) | 0.0493 |
| WCC (×109/L) | ||||
| Mean (range) | 22.1 (4.9−103.3) | 13.8 (4.9−36.8) | 30.4 (6.9−103.3) | 0.0224 |
| Hb (g/L) | ||||
| Mean (range) | 107 (79−143) | 104 (79−124) | 109 (82−143) | 0.3426 |
| PLT (×109/L) | ||||
| Mean (range) | 82 (7−219) | 91 (7−219) | 73 (18−192) | 0.3235 |
| Neutrophils (109/L) | ||||
| Mean (range) | 11.2 (1.67−47.5) | 7.2 (1.7−23.3) | 15.2 (3.0–47.5) | 0.0329 |
| Monocytes (109/L) | ||||
| Mean (range) | 5.4 (0.7−32.8) | 3.1 (1.0−8.0) | 7.8 (0.7−32.8) | 0.0267 |
| CRP (mg/L) | ||||
| Mean (range) | 4.9 (0.6−18.9) | 5.2 (1.7−18.9) | 4.5 (0.6−17) | 0.4796 |
| Spleen craniocaudal length (cm) | ||||
| Mean (range) | 13.6 (9.8−20.6) | 12.0 (10.8−13.8) | 15.2 (9.8−20.6) | 0.0400 |
n number of patients; BM bone marrow; WCC white blood cell count; Hb haemoglobin; PLT platelet; CRP C-reactive protein; MD-CMML myelodysplastic CMML; MP-CMML myeloproliferative CMML.
Mann-Whitney test was applied to continuous and Fisher’s exact test to categorical data for statistical analysis where P<0.05 was statistically significant.
2016 WHO Classification:
(i) Based on BM blast %: CMML-0 PB <2%, BM <5%; CMML-1 PB 2–4%, BM 5–9%, CMML-2 PB>5%, BM 10–19%.
(ii) Based on WCC: MD-CMML WCC<13×109/L, MP-CMML WCC>13×109/L
Table 2. Clinical characteristics, complete blood examination and bone marrow analyses of CMML patients in the PREACH-M trial stratified as CBL mutant vs. RAS pathway wildtype.
| Variable | Total (n = 17) | RAS pathway wildtype (n = 6) | CBL mutant (n = 11) | P-value |
|---|---|---|---|---|
| Gender | ||||
| Male, n (%) | 8 (47%) | 2 (33%) | 6 (55%) | 0.6199 |
| Female, n (%) | 11 (65%) | 4 (67%) | 5 (45%) | |
| Age (years) | ||||
| Mean (range) | 72 (56−86) | 73 (56−86) | 71 (56−79) | 0.2539 |
| WHO classification | ||||
| CMML-0, n (%) | 3 (18%) | 1 (17%) | 2 (18%) | 0.7964 |
| CMML-1, n (%) | 9 (53%) | 4 (67%) | 5 (45%) | |
| CMML-2, n (%) | 5 (29%) | 1 (17%) | 4 (36%) | |
| MD-CMML, n (%) | 7 (41%) | 4 (67%) | 3 (27%) | 0.1618 |
| MP-CMML, n (%) | 10 (59%) | 2 (33%) | 8 (73%) | |
| BM Blast (%) | ||||
| Mean (range) | 7.7 (1.0−17.0) | 5.3 (1.0−10.3) | 10.1 (4.0−17.0) | 0.0457 |
| WCC (×109/L) | ||||
| Mean (range) | 20.3 (4.9−74.1) | 13.8 (4.9−36.8) | 26.8 (6.9−74.1) | 0.0462 |
| Hb (g/L) | ||||
| Mean (range) | 106 (79−128) | 104 (79−124) | 107 (91−128) | 0.4893 |
| PLT (×109/L) | ||||
| Mean (range) | 89 (7−219) | 91 (7−219) | 88 (27−192) | 0.5000 |
| Neutrophils (109/L) | ||||
| Mean (range) | 10.6 (1.7−45.2) | 7.2 (1.7−23.3) | 14.0 (3.0−45.2) | 0.0608 |
| Monocytes (109/L) | ||||
| Mean (range) | 4.3 (0.7−12.2) | 3.1 (1.0−8.0) | 5.6 (0.7−12.2) | 0.0859 |
| CRP (mg/L) | ||||
| Mean (range) | 4.4 (1.7−18.9) | 5.2 (1.7−18.9) | 3.6 (0.7−7.3) | 0.4708 |
| Spleen craniocaudal length (cm) | ||||
| Mean (range) | 13.6 (10.2−20.6) | 12.0 (10.8−13.8) | 15.2 (10.2−20.6) | 0.0308 |
n number of patients; BM bone marrow; WCC white blood cell count; Hb haemoglobin; PLT platelet; CRP C-reactive protein; MD-CMML myelodysplastic CMML; MP-CMML myeloproliferative CMML
Mann-Whitney test was applied to continuous and Fisher’s exact test to categorical data for statistical analysis where P<0.05 was statistically significant.
2016 WHO Classification:
Based on BM blast %: CMML-0 PB <2%, BM <5%; CMML-1 PB 2–4%, BM 5–9%, CMML-2 PB>5%, BM 10–19%.
Based on WCC: MD-CMML WCC<13×109/L, MP-CMML WCC>13×109/L
Fig 1. CBL mutants are associated with proliferative features, increased BM blast percentage, leukocytosis and splenomegaly.
(A-F) Clinical characteristics of PREACH-M cohort at baseline, stratified based on the detection of CBL mutation. (G) MD-, MP-CMML classification based on WCCα (H) CMML-0, -1, -2 classifications based on BM blast percentageβ. 2016 WHO Classification: αBased on WCC: MD-CMML WCC<13×109/L, MP-CMML WCC>13×109/L. β Based on BM blast %: CMML-0 PB <2%, BM <5%; CMML-1 PB 2–4%, BM 5–9%, CMML-2 PB>5%, BM 10–19%. Bars represent mean ± standard error of mean. Mann-Whitney test used to determine statistical significance, where P<0.05 was statistically significant. [BM bone marrow; WCC white cell count; CRP C-reactive protein; MD-CMML myelodysplastic-CMML; MP-CMML myeloproliferative-CMML, wt wildtype, mut mutant].
CBL mutations co-occur frequently with TET2
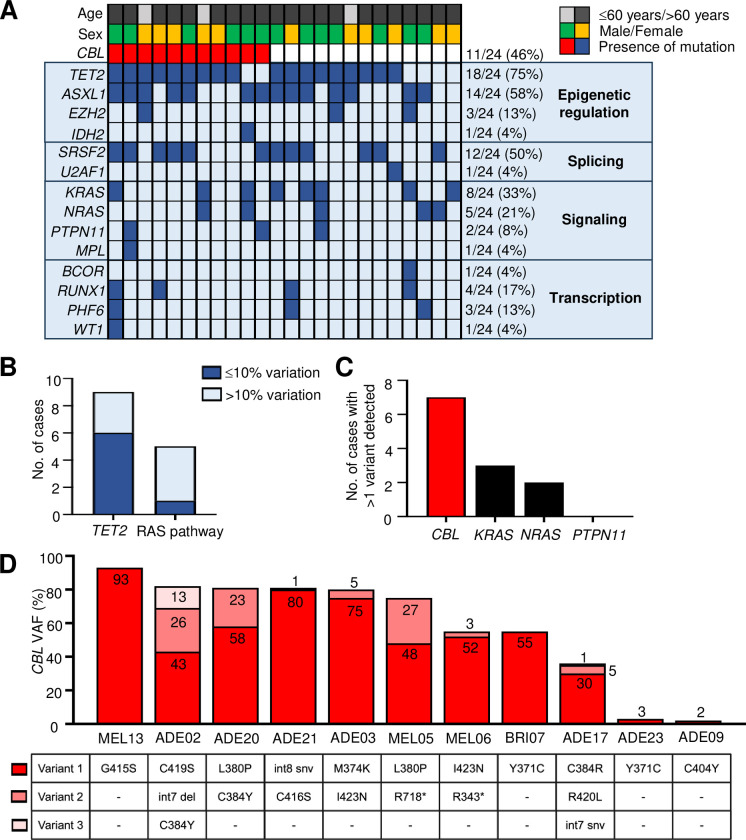
Of the 24 patients in the study, 75% (18/24) were detected to have TET2 mutation, 58% (14/24) ASXL1, 50% (12/24) SRSF2 and 46% (11/24) CBL (Fig 2A). Of the 11 patients with CBL mutation, 9 (82%) had co-occurring TET2 mutation. In 67% (6/9) of instances when these mutations are detected in the same patient, the difference in VAF magnitude between CBL and TET2 were ≤10% (MEL13, ADE02, ADE20, MEL05, MEL06, BRI07) (Figs 2B and S2), indicating these mutations may co-occur within the same clone. On the contrary, mutations in CBL and other RAS pathway genes (KRAS, NRAS, PTPN11) are not only infrequently found in the same patient (5/11; 45%), the VAF of the dominant CBL or RAS pathway mutant clone tend to be discordant, with VAF differences >10% (MEL13, BRI07, ADE17, ADE09), except in one case where CBL and PTPN11 VAF were ≤3% (Figs 2B and S2).
Fig 2. CBL mutants frequently co-occur with TET2 mutants and are associated with a complex subclonal architecture.
(A) Oncoplot for the PREACH-M cohort (n = 24). Mutation groups are shown in rows with each individual patient represented by a column. The presence of a mutation is indicated by the red or blue colored bars. Age category of the patients indicated by the black and grey bars and sex of patients by the green and gold bars. (B) Number of CBL mutant cases where TET2 mutations (n = 9) and other RAS pathway mutations (n = 5) were detected, where variation in the VAF of CBL vs. TET2 or RAS pathway mutant clones were ≤10% (dark blue) or >10% (light blue) (C) Number of cases where more than one variant of CBL, NRAS, KRAS or PTPN11 mutation was detected. (D) Details of CBL variants detected in each patient with CBL mutation. [VAF variant allele frequency].
Multiple CBL mutant subclones found in CMML
Strikingly, in 7/11 (64%) patients with CBL mutation, more than one CBL variant can be detected (Fig 2C). Of these, 2 patients had 3 variants while 5 patients had 2 variants (Fig 2D). In comparison, multiple subclones within a patient are more uncommon in KRAS (3/8; 38%), NRAS (2/5; 40%) and PTPN11 (0/3; 0%) mutant cases (Fig 2C).
CMML linked to high CD116 and CD131 in the progenitor subpopulation
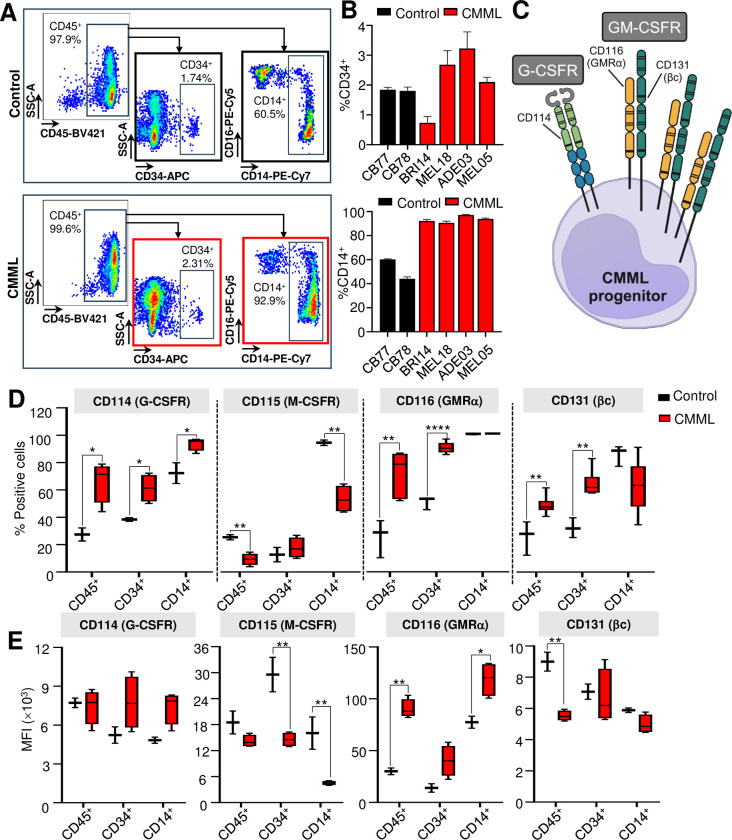
As CMML is a disease characterized by upregulation of inflammatory cytokines [56, 57] and expansion of pro-inflammatory granulocyte-macrophage-like progenitor cells and monocytes with enhanced cytokine receptor signaling [58, 59], we analyzed the immunophenotype of primary patient samples (n = 2–3 CBL mutant; n = 2–3 CBL wildtype) focusing on cytokine receptor expression, including granulocyte colony-stimulating factor receptor (G-CSFR, CD114), macrophage colony-stimulating factor receptor (M-CSFR, CD115) and the heterodimeric granulocyte-macrophage colony-stimulating factor receptor (GM-CSFR) comprising the alpha subunit (GMRα, CD116) and the beta common subunit (βc, CD131), in the CD45+ MNCs, CD34+ hematopoietic stem and progenitor cells and CD14+ monocytes (gating strategy outlined in Fig 3A). CMML patient samples had higher percentage of CD14+ cells compared to normal donors, consistent with expansion of monocytes and clinical presentation of the disease (Fig 3B).
Fig 3. CMML have an increased percentage of CD116 and CD131 positive CD34+ stem and progenitor cells.
(A) Flow cytometry analysis of a representative CMML sample and healthy control stained for CD45, CD34, CD14 and CD16, and gating strategy used to define CD45+ mononuclear cells, CD34+ stem and progenitor cells and CD14+ monocytes. (B) Percentage of CD34+ progenitors and CD14+ monocytes in CMML samples (n = 4) vs. healthy control (n = 2). (C) Illustration of the cluster of differentiation (CD) markers where CD114 is a marker for G-CSFR, CD116 GMRα and CD131 βc. G-CSFR is homodimeric while GM-CSFR is heterodimeric receptor consisting of GMRα and βc. The expression of CD114, CD115, CD116 and CD131 in CMML samples (n = 4–6) vs. control (n = 2–3) in CD45+, CD34+ and CD14+ subpopulations, expressed as percentage of positively stained cells (D) and MFI (E) compared to control (cord blood or peripheral blood mononuclear cells from healthy donors). Bars represent mean ± standard deviation in (B). Box and whiskers graphs were plotted with min and max in (C) and (D). Unpaired Student’s t-test between CMML vs. healthy control used to determine statistical significance, where P<0.05 was statistically significant. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. [MFI mean fluorescence intensity; G-CSFR granulocyte-colony stimulating factor receptor; GMRα granulocyte-macrophage colony stimulating factor receptor subunit α; βc beta common subunit].
Our data revealed that the percentage of cells in CMML patient samples expressing CD116 (GMRα) was significantly higher in the total MNC population compared to control, and this was most pronounced in the CD34+ progenitor subpopulation (89.7 ± 1.6 vs. 50.3 ± 2.7%; P = 0.000003) (Fig 3C and 3D). In contrast, the percentage of CD116 expressing cells was similar between healthy and CMML-derived CD14+ monocytes (Fig 3D). Interestingly, the density of CD116 expression represented by mean fluorescence intensity (MFI) was upregulated in our CMML cohort vs. healthy controls (CD45+ 90.3 ± 4.6×103 vs. 30.0 ± 3.2×103, P = 0.001; CD34+ 40.2 ± 7.4×103 vs. 13.9 ± 4.1×103, P = 0.08; CD14+ 118.9×103 ± 8.1×103 vs. 77.4×103 ± 5.8×103; P = 0.03) (Fig 3E). We also noted an increase in the percentage of CD131 expression in the MNCs, particularly in the CD34+ progenitors in CMML compared to control (64.3 ± 3.8 vs. 32.1 ± 4.1%; P = 0.001), although not in terms of MFI (Fig 3D and 3E). We did not observe a difference in receptor expressions between CBL mutant and CBL wildtype CMML patient samples (S3 Fig).
We also noted increases in the percentage of CD114+ cells across all CMML cell populations (CD45+ P = 0.03; CD34+ P = 0.04; CD14+ P = 0.02), with no difference in the MFI (Fig 3C and 3D). In contrast, the percentage of CD115+ cells was notably lower in the CMML CD45+ MNCs (P = 0.01) and CD14+ monocytes (P = 0.005), with reductions in MFI seen in both CD34+ (P = 0.005) and CD14+ (P = 0.007) populations (Fig 3D and 3E).
CBL mutations are enriched in the RING domain in CMML compared to JMML
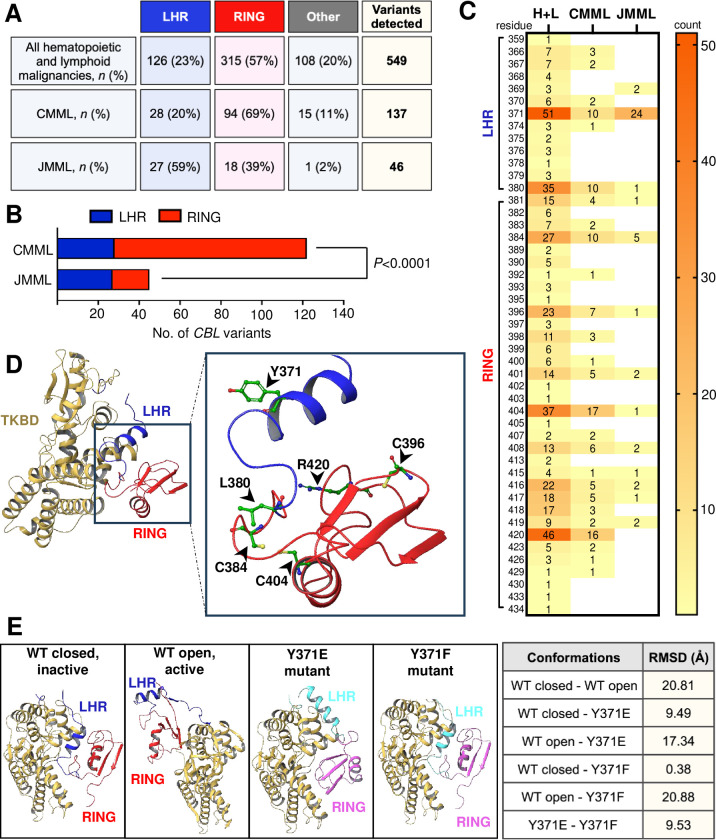
We then combined the new mutation data from our cohort with the publicly available data in COSMIC to assess which domains of Cbl were commonly perturbed. We found that CBL mutations in CMML and JMML are concentrated within the coding sequence of the LHR and RING domain of Cbl (Fig 4A). Furthermore, we noted that in CMML, mutations most frequently occur within the RING domain (amino acid residues 381–435) contrary to JMML, where mutations within the LHR (amino acid residues 353–380) are most common (P<0.0001) (Fig 4B).
Fig 4. CBL mutation hotspots in CMML cluster in the RING domain, unlike in JMML where they more commonly occur within the LHR.
(A) Table of CBL variants detected in our PREACH-M cohort combined with data sourced from COSMIC. Variants include nonsense or missense substitutions, frameshift and in-frame insertions or deletions within the coding sequence of CBL, filtered for all hematopoietic and lymphoid malignancies including CMML and JMML (n = 549), and CMML only (n = 137) or JMML only (n = 46) (B) Contingency analysis of CBL mutation hotspots within the LHR and RING domain of Cbl in CMML and JMML (C) Heat map representation of all sites within the LHR (amino acid residues 353–380) and RING domain (amino acid residues 381–435) where mutations have been reported. Numbers within the figure and on the scale depict counts (D) Tertiary protein structure of native wildtype Cbl (PDB ID 2Y1M) in inactive, closed conformation. The TKBD is colored beige, LHR blue and RING domain red. Amino acid residues of the top 6 mutation hotspots are indicated in inset; Tyrosine 371 (Y371), Leucine 380 (L380), Cysteine 384 (C384), Cysteine 396 (C396), Cysteine 404 (C404) and Arginine 420 (R420). (E) X-ray structures of wildtype Cbl in unphosphorylated, inactive state and in closed conformation (PDB ID 2Y1M), wildtype Cbl in Y371 phosphorylated, active state and in open conformation (PDB ID 4A4C), mutant Cbl Y371E (PDB ID 5HKX) and mutant Cbl Y371F (PDB ID 5J3X). The TKBD is colored beige, LHR of wildtype blue, LHR of mutant cyan, RING domain of wildtype red, RING domain of mutant pink. RMSD values between various Cbl conformations are shown in table. Statistical analysis was performed using two-sided Fisher’s exact test, where P<0.05 was statistically significant. [TKBD tyrosine kinase binding domain; LHR linker helix region; RING RING domain; H+L all hematopoietic and lymphoid malignancies; WT wildtype; RMSD root mean square deviation (distanced-based measure of protein structure similarity)].
The 6 most common CBL mutations in all hematopoietic and lymphoid malignancies occur in codons affecting amino acid residues 371, 380, 384, 396, 404, 420 (Fig 4C and 4D). In the PREACH-M cohort in particular, mutations in residue 384 were detected 3 patients, 371 in 2 patients, 380 in 2 patients, 404 and 420 in 1 patient, respectively. In CMML, missense substitutions cysteine 404 to tyrosine (C404Y) (13/137; 10%) and arginine 420 to glutamine (R420Q) (12/137; 9%) were most common, while in JMML, tyrosine 371 to histidine (Y371H) substitution was most common (21/46; 46%).
Mutations at residue 371 within the LHR can result in novel conformational change
Finally, we performed comparative structural alignments of available mutant Cbl structures resolved by X-ray diffraction publicly available via the Protein Data Bank (PDB). Comparison of the LHR of wildtype Cbl protein in the closed, inactive conformation (Y371 unphosphorylated) (PDB 2Y1M) [40] against the open, active conformation (Y371 phosphorylated) (PDB 4A4C) [40] (Fig 4E), revealed a root mean square deviation (RMSD) of 20.81Å, indicating that a significant conformational change takes place when Cbl becomes activated by phosphorylation. Further, we also compared PDB structures 2Y1M and 4A4C with structures comprising tyrosine 371 to glutamic acid (Y371E) (PDB 5HKX) and tyrosine 371 to phenylalanine (Y371F) (PDB 5J3X) [41] LHR mutations. Interestingly, we inferred that the Y371E mutant Cbl possesses an entirely different conformation to either closed, inactive or open, active wildtype Cbl (RMSD 9.49Å and 17.34Å, respectively). We noted that replacement of the polar, bulky tyrosine with the negatively charged glutamic acid (Y371E) resulted in perturbation of the LHR-TKBD interface and subsequent total displacement of the LHR and RING domain compared to both inactive and active wildtype Cbl (Fig 4E). In contrast, when tyrosine was replaced with a structurally similar residue phenylalanine (Y371F), the LHR-TKBD interface was unperturbed, and the mutant closely mimicked the native, inactive state of wildtype Cbl (RMSD 0.38Å) and not the active state (RMSD 20.88Å) (Fig 4E). Additional measurements relating to the structural differences between wildtype and mutant Cbl can be found in S4 Table.
Discussion
Our data, obtained from patients enrolled in a prospective multicenter interventional study, highlight several clinical and molecular features of CBL mutants in CMML. Our results are generally consistent with previous studies that performed next generation sequencing in CMML patients but we note a higher frequency of CBL variants (11 of 24 patients, 46%) than others report (12.8%) [60, 61], possibly due to strict trial eligibility criteria (higher white cell count or cytopenia). CBL variants were associated with a myeloproliferative phenotype, including higher white cell count and splenomegaly with many patients having increased blasts at diagnosis, similar to patients with other RAS pathway mutations. Notably, many patients harbored multiple CBL subclones (intrapatient molecular heterogeneity) that was not observed to the same extent for other RAS pathway mutations. This may be significant because subclonal abundance, especially a branched pattern of clonal evolution, is associated with a favorable outcome in AML [62].
We observed a strong overlap and clonal correlation between CBL mutations and TET2 mutation. We noted another study that has found a modest association of TET2 with CBL mutations (r<0.25; P<0.1) [60], despite the high frequency of TET2 mutation overall. A number of in vivo murine studies have highlighted a role for TET2 in suppressing innate immune signaling in monocytes, with TET2 mutant monocytes showing enhanced pro-inflammatory responses to stimuli such as lipopolysaccharide [63, 64]. It is plausible that the CBL mutation serves to further amplify innate immune signaling by preventing ubiquitination and turnover of cytokine and Toll-like receptors but the exact cellular compartments within which this occurs is not defined. Thus, it is significant that a high percentage of CBL mutant CMML CD34+ progenitors express GMRα (CD116, 89.7%) and its partner subunit, βc (CD131, 64.3%), suggesting that a substantial proportion of CD34+ cells are primed to respond to the cytokine GM-CSF (Fig 3C–3E). Indeed, we know from previous studies that CMML display hypersensitivity to GM-CSF akin to JMML, especially in cases that have RAS pathway mutations [24, 25]. In contrast, the percentage expression of the receptor for M-CSF (CD115) was decreased in CMML progenitors compared to controls, indicating this alternate monocyte cytokine is unlikely to be driving the disease. We also noted increased percentage of G-CSFR (CD114)-expressing CMML compared to controls, although not to the same extent as CD116, indicating CMML progenitors may also respond to G-CSF.
Previous studies with GM-CSF neutralizing antibody [25] and our own work with the GM-CSF E21R antagonist and successful engraftment of CMML patient samples in mice transgenic for human GM-CSF [24] provide strong evidence that GM-CSF is an essential growth factor for CMML in vitro and in vivo. Our findings that CD116 is upregulated in the CD34+ progenitors is interesting as it raises the question of the effect of GM-CSF on the leukemia-initiating cell population. This is in agreement with a recent study using single cell RNA-seq to map the differentiation trajectories of CD34+ progenitors in CMML primary patient samples [59] where the authors also showed the upregulation of CD116 in a cell cluster enriched for granulocyte/monocyte progenitor-like inflammatory hematopoietic stem and progenitor cells that may have self-renewal capacity in CMML patients with a monocyte-biased differentiation trajectory. Recently, we found that interleukin-3 (IL-3) receptor stoichiometry is a critical determinant in cell fate and IL-3 receptor overexpression in leukemia stem cells leads to biased activation of distinct transcriptional programs and signaling pathways to drive stemness programs vs. cell differentiation [65]. This propels us to hypothesize that although GM-CSF has been mostly associated with the proliferation and differentiation of hematopoietic progenitors and mature cells, it is possible that aside from the cytokine hypersensitivity previously shown [25], GM-CSF may also have unique signaling and effects on stem cell maintenance and function in CD116-overexpressing CD34+ progenitors for disease initiation and generation of the pro-inflammatory phenotype associated with this disease. Future studies should examine the kinetics of receptor turnover and phosphorylation peak and attenuation in this primary population, the signaling pathways involved and determine whether CD116 can be used to distinguish between CBL mutant leukemia vs. healthy stem cells. Indeed, the effects of anti-CD116 or anti-GM-CSF therapies (or in combination), on these populations warrant further investigation.
Cbl adopts a closed and open conformation dependent on Y371 phosphorylation to allow for the binding of ubiquitin-conjugating enzyme E2 [40]. Thus, it follows that the loss of this key tyrosine residue in position 371 within the LHR could have dramatic implications for the conformation and activity of Cbl. Tyrosine 371 is in a buried environment, where it forms a hydrogen bond with threonine 227 (T227) and makes several van der Waals interactions with residues in the hydrophobic pocket of the TKBD [36, 40], playing a structural role in maintaining the integrity of the LHR-TKBD interface, and importantly, in keeping Cbl in a closed conformation, autoinhibited state [40]. When Y371 is phosphorylated, an open conformation is adopted, and autoinhibition is abolished leading to Cbl becoming a more active ligase [35, 38, 40, 66, 67]. Our analysis of the X-ray structures demonstrates that Cbl conformation is sensitive to the amino acid residue at position 371. Indeed, our inference following comparative structural alignments is that, depending on the nature of the substituting residue, mutation at position 371 (such as Y371E, PDB 5HKX) not only results in the impairment of phosphorylation-dependent activation, but can also yield an entirely novel conformation of Cbl, different from both the inactive (unphosphorylated, closed) and active (phosphorylated, open) conformations. With this conformational drift, the RING domain and E2 enzyme cannot be in sufficient proximity to the substrate binding site of the TKBD for effective ubiquitination. Thus, ubiquitination and degradation of activated RTKs would be predicted to occur less efficiently, resulting in sustained downstream signaling that may contribute to oncogenicity and disease progression. In contrast, mutations that do not perturb the LHR-TKBD interaction (such as Y371F, PDB 5J3X [41]) would mimic the conformation of native, wildtype Cbl, albeit no longer capable of increased catalytic efficiency due to loss of the phosphorylation site. This is consistent with early evidence that Cbl Y371 mutants can exist in different states of activity depending on the chemical nature of the amino acid substitution [41].
In CMML, CBL mutations were found in both the LHR and RING domains of the protein but with significant enrichment for mutations in the RING domain compared to JMML. The RING domain determines the specificity of Cbl E3 for its cognate E2 enzyme, recognizes lysines to be ubiquitinated and serves as a scaffold for optimal orientation for ubiquitin transfer between E2 and its substrate RTK [36] but the structure of Cbl RING domain mutations has not been determined. Elucidation of distinct mutant Cbl conformations is significant because new drug development strategies could employ proteolysis targeting chimera (PROTAC) technology for targeted protein degradation of mutant Cbl with conformations different from wildtype. Conversely, a new Cbl-b inhibitor C7683 currently in phase I clinical trials for advanced solid tumor malignancies, designed to keep wildtype Cbl-b locked in an inactive state [68], may partially mimic LHR Cbl mutations and thus should be used with caution in patients with clonal hematopoiesis, CMML or JMML.
A limitation of our study is the relatively small number of CBL mutant positive cases, reflecting the rarity of CMML. Nevertheless, certain clinical and molecular features are consistent across the cohort and are congruent with data available in COSMIC, implying that CBL mutant CMML may have a characteristic phenotype. Studies with larger cohorts are required to distinguish CBL mutation CMML from other RAS pathway mutations such as NRAS, KRAS and PTPN11. To date, one study using serial VAF measurements did not show significant change in clone size for CMML patients, including CBL clones, treated with azacitidine alone suggesting epigenetic effects, rather than mutation-specific effects, were linked to therapeutic benefit [16]. In a phase I study, quizartinib inhibition of the receptor tyrosine kinase FLT3, a Cbl target for ubiquitination and internalization [69], did not appear to impact CMML with CBL mutations, suggesting phosphorylated FLT3 is not a critical substrate of Cbl in CMML [70]. Future work should examine the effect of CD116-targeted immunotherapy on the clonal dynamics of CBL RING domain vs. LHR mutants.
Supporting information
(PDF)
(PDF)
(PDF)
(PDF)
(TIF)
(TIF)
Cytokine receptor CD114, CD115, CD116 and CD131 expression of CBL mutant and wildtype compared to healthy control by (A) percentage positive cells and (B) mean fluorescence intensity (MFI).
(TIF)
(XLSX)
Acknowledgments
The authors thank all patients and their families for donating specimens to research. The authors thank Verity Saunders and Kiralee Vuglar for isolation and cryopreservation of MNCs following blood collection from patients in the PREACH-M trial and Professor Gus Dekker and the Northern Adelaide Local Health Network for provision of fresh cord blood. Flow cytometry was performed at the Adelaide Health and BioMedical Precinct Cytometry Facility. Illustrations were created with BioRender.com.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
D. Thomas is supported by a Commonwealth Serum Laboratories (2020 Centenary Fellowship), Australian Medical Research Future Fund (1201012, 2024427, 2008972) and National Health and Medical Research Council (2023/GNT2029809) and The Leukemia and Lymphoma Society (LLS 6619-21; LLS 6650-23; CMML Special Initiative) (www.lls.org), Snowdome Foundation (co-funder LLS 6619-21, LLS 6650-23) (www.snowdome.org.au) and the Leukaemia Foundation (co-funder LLS 6619-21, LLS 6650-23) (www.leukaemia.org.au). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rollison DE, Howlader N, Smith MT, Strom SS, Merritt WD, Ries LA, et al. Epidemiology of myelodysplastic syndromes and chronic myeloproliferative disorders in the United States, 2001–2004, using data from the NAACCR and SEER programs. Blood. 2008;112(1):45–52. doi: 10.1182/blood-2008-01-134858 [DOI] [PubMed] [Google Scholar]
- 2.Guru Murthy GS, Dhakal I, Mehta P. Incidence and survival outcomes of chronic myelomonocytic leukemia in the United States. Leukemia & Lymphoma. 2017;58(7):1648–54. doi: 10.1080/10428194.2016.1258700 [DOI] [PubMed] [Google Scholar]
- 3.Selimoglu-Buet D, Wagner-Ballon O, Saada V, Bardet V, Itzykson R, Bencheikh L, et al. Characteristic repartition of monocyte subsets as a diagnostic signature of chronic myelomonocytic leukemia. Blood. 2015;125(23):3618–26. doi: 10.1182/blood-2015-01-620781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Talati C, Zhang L, Shaheen G, Kuykendall A, Ball M, Zhang Q, et al. Monocyte subset analysis accurately distinguishes CMML from MDS and is associated with a favorable MDS prognosis. Blood. 2017;129(13):1881–3. doi: 10.1182/blood-2016-12-753210 [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K, Pemmaraju N, Strati P, Nogueras-Gonzalez G, Ning J, Bueso-Ramos C, et al. Clinical characteristics and outcomes of therapy-related chronic myelomonocytic leukemia. Blood. 2013;122(16):2807–11. doi: 10.1182/blood-2013-03-491399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patnaik MM, Vallapureddy R, Yalniz FF, Hanson CA, Ketterling RP, Lasho TL, et al. Therapy related-chronic myelomonocytic leukemia (CMML): Molecular, cytogenetic, and clinical distinctions from de novo CMML. Am J Hematol. 2018;93(1):65–73. doi: 10.1002/ajh.24939 [DOI] [PubMed] [Google Scholar]
- 7.Saffie M, Sun D, Hsia C. Sweet’s syndrome in chronic myelomonocytic leukemia. American Journal of Hematology. 2013;88(7):630-. doi: 10.1002/ajh.23415 [DOI] [PubMed] [Google Scholar]
- 8.Peker D, Padron E, Bennett JM, Zhang X, Horna P, Epling-Burnette PK, et al. A close association of autoimmune-mediated processes and autoimmune disorders with chronic myelomonocytic leukemia: observation from a single institution. Acta Haematol. 2015;133(2):249–56. doi: 10.1159/000365877 [DOI] [PubMed] [Google Scholar]
- 9.Grignano E, Mekinian A, Braun T, Liozon E, Hamidou M, Decaux O, et al. Autoimmune and inflammatory diseases associated with chronic myelomonocytic leukemia: A series of 26 cases and literature review. Leuk Res. 2016;47:136–41. doi: 10.1016/j.leukres.2016.05.013 [DOI] [PubMed] [Google Scholar]
- 10.Zahid MF, Barraco D, Lasho TL, Finke C, Ketterling RP, Gangat N, et al. Spectrum of autoimmune diseases and systemic inflammatory syndromes in patients with chronic myelomonocytic leukemia. Leuk Lymphoma. 2017;58(6):1488–93. doi: 10.1080/10428194.2016.1243681 [DOI] [PubMed] [Google Scholar]
- 11.Onida F, Kantarjian HM, Smith TL, Ball G, Keating MJ, Estey EH, et al. Prognostic factors and scoring systems in chronic myelomonocytic leukemia: a retrospective analysis of 213 patients. Blood. 2002;99(3):840–9. doi: 10.1182/blood.v99.3.840 [DOI] [PubMed] [Google Scholar]
- 12.Such E, Cervera J, Costa D, Solé F, Vallespí T, Luño E, et al. Cytogenetic risk stratification in chronic myelomonocytic leukemia. Haematologica. 2011;96(3):375–83. doi: 10.3324/haematol.2010.030957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grossmann V, Kohlmann A, Eder C, Haferlach C, Kern W, Cross NC, et al. Molecular profiling of chronic myelomonocytic leukemia reveals diverse mutations in >80% of patients with TET2 and EZH2 being of high prognostic relevance. Leukemia. 2011;25(5):877–9. [DOI] [PubMed] [Google Scholar]
- 14.Patnaik MM, Tefferi A. Cytogenetic and molecular abnormalities in chronic myelomonocytic leukemia. Blood Cancer J. 2016;6(2):e393. doi: 10.1038/bcj.2016.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itzykson R, Kosmider O, Renneville A, Gelsi-Boyer V, Meggendorfer M, Morabito M, et al. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J Clin Oncol. 2013;31(19):2428–36. doi: 10.1200/JCO.2012.47.3314 [DOI] [PubMed] [Google Scholar]
- 16.Merlevede J, Droin N, Qin T, Meldi K, Yoshida K, Morabito M, et al. Mutation allele burden remains unchanged in chronic myelomonocytic leukaemia responding to hypomethylating agents. Nat Commun. 2016;7:10767. doi: 10.1038/ncomms10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel BJ, Przychodzen B, Thota S, Radivoyevitch T, Visconte V, Kuzmanovic T, et al. Genomic determinants of chronic myelomonocytic leukemia. Leukemia. 2017;31(12):2815–23. doi: 10.1038/leu.2017.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stieglitz E, Taylor-Weiner AN, Chang TY, Gelston LC, Wang YD, Mazor T, et al. The genomic landscape of juvenile myelomonocytic leukemia. Nature genetics. 2015;47(11):1326–33. doi: 10.1038/ng.3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caye A, Strullu M, Guidez F, Cassinat B, Gazal S, Fenneteau O, et al. Juvenile myelomonocytic leukemia displays mutations in components of the RAS pathway and the PRC2 network. Nature genetics. 2015;47(11):1334–40. doi: 10.1038/ng.3420 [DOI] [PubMed] [Google Scholar]
- 20.Lipka DB, Witte T, Toth R, Yang J, Wiesenfarth M, Nollke P, et al. RAS-pathway mutation patterns define epigenetic subclasses in juvenile myelomonocytic leukemia. Nat Commun. 2017;8(1):2126. doi: 10.1038/s41467-017-02177-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murakami N, Okuno Y, Yoshida K, Shiraishi Y, Nagae G, Suzuki K, et al. Integrated molecular profiling of juvenile myelomonocytic leukemia. Blood. 2018;131(14):1576–86. doi: 10.1182/blood-2017-07-798157 [DOI] [PubMed] [Google Scholar]
- 22.Lasho T, Patnaik MM. Juvenile myelomonocytic leukemia ‐ A bona fide RASopathy syndrome. Best Pract Res Clin Haematol. 2020;33(2):101171. doi: 10.1016/j.beha.2020.101171 [DOI] [PubMed] [Google Scholar]
- 23.Loh ML, Sakai DS, Flotho C, Kang M, Fliegauf M, Archambeault S, et al. Mutations in CBL occur frequently in juvenile myelomonocytic leukemia. Blood. 2009;114(9):1859–63. doi: 10.1182/blood-2009-01-198416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramshaw HS, Bardy PG, Lee MA, Lopez AF. Chronic myelomonocytic leukemia requires granulocyte-macrophage colony-stimulating factor for growth in vitro and in vivo. Exp Hematol. 2002;30(10):1124–31. doi: 10.1016/s0301-472x(02)00903-7 [DOI] [PubMed] [Google Scholar]
- 25.Padron E, Painter JS, Kunigal S, Mailloux AW, McGraw K, McDaniel JM, et al. GM-CSF–dependent pSTAT5 sensitivity is a feature with therapeutic potential in chronic myelomonocytic leukemia. Blood. 2013;121(25):5068–77. doi: 10.1182/blood-2012-10-460170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hecht A, Meyer JA, Behnert A, Wong E, Chehab F, Olshen A, et al. Molecular and phenotypic diversity of CBL-mutated juvenile myelomonocytic leukemia. Haematologica. 2022;107(1):178–86. doi: 10.3324/haematol.2020.270595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baumann CA, Ribon V, Kanzaki M, Thurmond DC, Mora S, Shigematsu S, et al. CAP defines a second signalling pathway required for insulin-stimulated glucose transport. Nature. 2000;407(6801):202–7. doi: 10.1038/35025089 [DOI] [PubMed] [Google Scholar]
- 28.Ueno H, Sasaki K, Honda H, Nakamoto T, Yamagata T, Miyagawa K, et al. c-Cbl is tyrosine-phosphorylated by interleukin-4 and enhances mitogenic and survival signals of interleukin-4 receptor by linking with the phosphatidylinositol 3’-kinase pathway. Blood. 1998;91(1):46–53. [PubMed] [Google Scholar]
- 29.Grishin A, Sinha S, Roginskaya V, Boyer MJ, Gomez-Cambronero J, Zuo S, et al. Involvement of Shc and Cbl-PI 3-kinase in Lyn-dependent proliferative signaling pathways for G-CSF. Oncogene. 2000;19(1):97–105. doi: 10.1038/sj.onc.1203254 [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Guzman M, Larsen E, Vuori K. The proto-oncogene c-Cbl is a positive regulator of Met-induced MAP kinase activation: a role for the adaptor protein Crk. Oncogene. 2000;19(35):4058–65. doi: 10.1038/sj.onc.1203750 [DOI] [PubMed] [Google Scholar]
- 31.Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286(5438):309–12. doi: 10.1126/science.286.5438.309 [DOI] [PubMed] [Google Scholar]
- 32.Schmidt MHH, Dikic I. The Cbl interactome and its functions. Nature Reviews Molecular Cell Biology. 2005;6(12):907–19. doi: 10.1038/nrm1762 [DOI] [PubMed] [Google Scholar]
- 33.Swaminathan G, Tsygankov AY. The Cbl family proteins: ring leaders in regulation of cell signaling. J Cell Physiol. 2006;209(1):21–43. doi: 10.1002/jcp.20694 [DOI] [PubMed] [Google Scholar]
- 34.Blake TJ, Shapiro M, Morse HC, 3rd, Langdon WY. The sequences of the human and mouse c-cbl proto-oncogenes show v-cbl was generated by a large truncation encompassing a proline-rich domain and a leucine zipper-like motif. Oncogene. 1991;6(4):653–7. [PubMed] [Google Scholar]
- 35.Andoniou CE, Thien CB, Langdon WY. Tumour induction by activated abl involves tyrosine phosphorylation of the product of the cbl oncogene. EMBO J. 1994;13(19):4515–23. doi: 10.1002/j.1460-2075.1994.tb06773.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102(4):533–9. doi: 10.1016/s0092-8674(00)00057-x [DOI] [PubMed] [Google Scholar]
- 37.Thien CBF, Walker F, Langdon WY. RING Finger Mutations that Abolish c-Cbl-Directed Polyubiquitination and Downregulation of the EGF Receptor Are Insufficient for Cell Transformation. Molecular Cell. 2001;7(2):355–65. doi: 10.1016/s1097-2765(01)00183-6 [DOI] [PubMed] [Google Scholar]
- 38.Kassenbrock CK, Anderson SM. Regulation of ubiquitin protein ligase activity in c-Cbl by phosphorylation-induced conformational change and constitutive activation by tyrosine to glutamate point mutations. Journal of Biological Chemistry. 2004;279(27):28017–27. doi: 10.1074/jbc.M404114200 [DOI] [PubMed] [Google Scholar]
- 39.Kales SC, Ryan PE, Nau MM, Lipkowitz S. Cbl and human myeloid neoplasms: the Cbl oncogene comes of age. Cancer Res. 2010;70(12):4789–94. doi: 10.1158/0008-5472.CAN-10-0610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dou H, Buetow L, Hock A, Sibbet GJ, Vousden KH, Huang DT. Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nat Struct Mol Biol. 2012;19(2):184–92. doi: 10.1038/nsmb.2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buetow L, Tria G, Ahmed SF, Hock A, Dou H, Sibbet GJ, et al. Casitas B-lineage lymphoma linker helix mutations found in myeloproliferative neoplasms affect conformation. BMC Biol. 2016;14(1):76. doi: 10.1186/s12915-016-0298-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nadeau SA, An W, Mohapatra BC, Mushtaq I, Bielecki TA, Luan H, et al. Structural Determinants of the Gain-of-Function Phenotype of Human Leukemia-associated Mutant CBL Oncogene. J Biol Chem. 2017;292(9):3666–82. doi: 10.1074/jbc.M116.772723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Braun T, Itzykson R, Renneville A, de Renzis B, Dreyfus F, Laribi K, et al. Molecular predictors of response to decitabine in advanced chronic myelomonocytic leukemia: a phase 2 trial. Blood. 2011;118(14):3824–31. doi: 10.1182/blood-2011-05-352039 [DOI] [PubMed] [Google Scholar]
- 44.Aribi A, Borthakur G, Ravandi F, Shan J, Davisson J, Cortes J, et al. Activity of decitabine, a hypomethylating agent, in chronic myelomonocytic leukemia. Cancer. 2007;109(4):713–7. doi: 10.1002/cncr.22457 [DOI] [PubMed] [Google Scholar]
- 45.Wijermans PW, Rüter B, Baer MR, Slack JL, Saba HI, Lübbert M. Efficacy of decitabine in the treatment of patients with chronic myelomonocytic leukemia (CMML). Leuk Res. 2008;32(4):587–91. doi: 10.1016/j.leukres.2007.08.004 [DOI] [PubMed] [Google Scholar]
- 46.Costa R, Abdulhaq H, Haq B, Shadduck RK, Latsko J, Zenati M, et al. Activity of azacitidine in chronic myelomonocytic leukemia. Cancer. 2011;117(12):2690–6. doi: 10.1002/cncr.25759 [DOI] [PubMed] [Google Scholar]
- 47.Ades L, Sekeres MA, Wolfromm A, Teichman ML, Tiu RV, Itzykson R, et al. Prognostic Factors of Response and Survival in CMML Patients Treated with Azacitidine (AZA). Blood. 2011;118(21):1726. [Google Scholar]
- 48.Solary E, Itzykson R. How I treat chronic myelomonocytic leukemia. Blood. 2017;130(2):126–36. doi: 10.1182/blood-2017-04-736421 [DOI] [PubMed] [Google Scholar]
- 49.Eissa H, Gooley TA, Sorror ML, Nguyen F, Scott BL, Doney K, et al. Allogeneic hematopoietic cell transplantation for chronic myelomonocytic leukemia: relapse-free survival is determined by karyotype and comorbidities. Biol Blood Marrow Transplant. 2011;17(6):908–15. doi: 10.1016/j.bbmt.2010.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hunter AM, Al Ali N, Sallman DA, Sweet KL, Kuykendall AT, Talati C, et al. WHO-Defined Chronic Myelomonocytic Leukemia-2 (CMML-2) Patients Rapidly Progress to AML Suggesting This Entity Represents a Transitory Clinical State. Blood. 2019;134(Supplement_1):1717-. [Google Scholar]
- 51.Germing U, Kundgen A, Gattermann N. Risk assessment in chronic myelomonocytic leukemia (CMML). Leuk Lymphoma. 2004;45(7):1311–8. doi: 10.1080/1042819042000207271 [DOI] [PubMed] [Google Scholar]
- 52.Bacher U, Haferlach T, Schnittger S, Kreipe H, Kroger N. Recent advances in diagnosis, molecular pathology and therapy of chronic myelomonocytic leukaemia. Br J Haematol. 2011;153(2):149–67. doi: 10.1111/j.1365-2141.2011.08631.x [DOI] [PubMed] [Google Scholar]
- 53.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–405. doi: 10.1182/blood-2016-03-643544 [DOI] [PubMed] [Google Scholar]
- 54.Ricci C, Fermo E, Corti S, Molteni M, Faricciotti A, Cortelezzi A, et al. RAS mutations contribute to evolution of chronic myelomonocytic leukemia to the proliferative variant. Clin Cancer Res. 2010;16(8):2246–56. doi: 10.1158/1078-0432.CCR-09-2112 [DOI] [PubMed] [Google Scholar]
- 55.Carr RM, Vorobyev D, Lasho T, Marks DL, Tolosa EJ, Vedder A, et al. RAS mutations drive proliferative chronic myelomonocytic leukemia via a KMT2A-PLK1 axis. Nature Communications. 2021;12(1):2901. doi: 10.1038/s41467-021-23186-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Everson MP, Brown CB, Lilly MB. Interleukin-6 and granulocyte-macrophage colony-stimulating factor are candidate growth factors for chronic myelomonocytic leukemia cells. Blood. 1989;74(5):1472–6. [PubMed] [Google Scholar]
- 57.Niyongere S, Lucas N, Zhou JM, Sansil S, Pomicter AD, Balasis ME, et al. Heterogeneous expression of cytokines accounts for clinical diversity and refines prognostication in CMML. Leukemia. 2019;33(1):205–16. doi: 10.1038/s41375-018-0203-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Franzini A, Pomicter AD, Yan D, Khorashad JS, Tantravahi SK, Than H, et al. The transcriptome of CMML monocytes is highly inflammatory and reflects leukemia-specific and age-related alterations. Blood Adv. 2019;3(20):2949–61. doi: 10.1182/bloodadvances.2019000585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ferrall-Fairbanks MC, Dhawan A, Johnson B, Newman H, Volpe V, Letson C, et al. Progenitor Hierarchy of Chronic Myelomonocytic Leukemia Identifies Inflammatory Monocytic-Biased Trajectory Linked to Worse Outcomes. Blood Cancer Discov. 2022;3(6):536–53. doi: 10.1158/2643-3230.BCD-21-0217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nie Y, Shao L, Zhang H, He CK, Li H, Zou J, et al. Mutational landscape of chronic myelomonocytic leukemia in Chinese patients. Exp Hematol Oncol. 2022;11(1):32. doi: 10.1186/s40164-022-00284-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han W, Zhou F, Wang Z, Hua H, Qin W, Jia Z, et al. Mutational landscape of chronic myelomonocytic leukemia and its potential clinical significance. International Journal of Hematology. 2022;115(1):21–32. doi: 10.1007/s12185-021-03210-x [DOI] [PubMed] [Google Scholar]
- 62.Benard BA, Leak LB, Azizi A, Thomas D, Gentles AJ, Majeti R. Clonal architecture predicts clinical outcomes and drug sensitivity in acute myeloid leukemia. Nature Communications. 2021;12(1):7244. doi: 10.1038/s41467-021-27472-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Q, Zhao K, Shen Q, Han Y, Gu Y, Li X, et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature. 2015;525(7569):389–93. doi: 10.1038/nature15252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cai Z, Kotzin JJ, Ramdas B, Chen S, Nelanuthala S, Palam LR, et al. Inhibition of Inflammatory Signaling in Tet2 Mutant Preleukemic Cells Mitigates Stress-Induced Abnormalities and Clonal Hematopoiesis. Cell Stem Cell. 2018;23(6):833–49.e5. doi: 10.1016/j.stem.2018.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kan WL, Dhagat U, Kaufmann KB, Hercus TR, Nero TL, Zeng AGX, et al. Distinct Assemblies of Heterodimeric Cytokine Receptors Govern Stemness Programs in Leukemia. Cancer Discov. 2023;13(8):1922–47. doi: 10.1158/2159-8290.CD-22-1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ryan PE, Sivadasan-Nair N, Nau MM, Nicholas S, Lipkowitz S. The N terminus of Cbl-c regulates ubiquitin ligase activity by modulating affinity for the ubiquitin-conjugating enzyme. J Biol Chem. 2010;285(31):23687–98. doi: 10.1074/jbc.M109.091157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell. 1999;4(6):1029–40. doi: 10.1016/s1097-2765(00)80231-2 [DOI] [PubMed] [Google Scholar]
- 68.Kimani SW, Perveen S, Szewezyk M, Zeng H, Dong A, Li F, et al. The co-crystal structure of Cbl-b and a small-molecule inhibitor reveals the mechanism of Cbl-b inhibition. Communications Biology. 2023;6(1):1272. doi: 10.1038/s42003-023-05655-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sargin B, Choudhary C, Crosetto N, Schmidt MHH, Grundler R, Rensinghoff M, et al. Flt3-dependent transformation by inactivating c-Cbl mutations in AML. Blood. 2007;110(3):1004–12. doi: 10.1182/blood-2007-01-066076 [DOI] [PubMed] [Google Scholar]
- 70.Montalban-Bravo G, Jabbour E, Chien K, Hammond D, Short N, Ravandi F, et al. Phase 1 study of azacitidine in combination with quizartinib in patients with FLT3 or CBL mutated MDS and MDS/MPN. Leuk Res. 2024;142:107518. doi: 10.1016/j.leukres.2024.107518 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(TIF)
(TIF)
Cytokine receptor CD114, CD115, CD116 and CD131 expression of CBL mutant and wildtype compared to healthy control by (A) percentage positive cells and (B) mean fluorescence intensity (MFI).
(TIF)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.