Abstract
Mouse hepatitis virus (MHV), a member of the Coronaviridae, contains a polyadenylated positive-sense single-stranded genomic RNA which is 31 kb long. MHV replication and transcription take place via the synthesis of negative-strand RNA intermediates from a positive-strand genomic template. A cis-acting element previously identified in the 3′ untranslated region binds to trans-acting host factors from mouse fibroblasts and forms at least three RNA-protein complexes. The largest RNA-protein complex formed by the cis-acting element and the lysate from uninfected mouse fibroblasts has a molecular weight of about 200 kDa. The complex observed in gel shift assays has been resolved by second-dimension sodium dodecyl sulfate-polyacrylamide gel electrophoresis into four proteins of approximately 90, 70, 58, and 40 kDa after RNase treatment. Specific RNA affinity chromatography also has revealed the presence of a 90-kDa protein associated with RNA containing the cis-acting element bound to magnetic beads. The 90-kDa protein has been purified from uninfected mouse fibroblast crude lysates. Protein microsequencing identified the 90-kDa protein as mitochondrial aconitase. Antibody raised against purified mitochondrial aconitase recognizes the RNA-protein complex and the 90-kDa protein, which can be released from the complex by RNase digestion. Furthermore, UV cross-linking studies indicate that highly purified mitochondrial aconitase binds specifically to the MHV 3′ protein-binding element. Increasing the intracellular level of mitochondrial aconitase by iron supplementation resulted in increased RNA-binding activity in cell extracts and increased virus production as well as viral protein synthesis at early hours of infection. These results are particularly interesting in terms of identification of an RNA target for mitochondrial aconitase, which has a cytoplasmic homolog, cytoplasmic aconitase, also known as iron regulatory protein 1, a well-recognized RNA-binding protein. The binding properties of mitochondrial aconitase and the functional relevance of RNA binding appear to parallel those of cytoplasmic aconitase.
The coronaviruses belong to the newly established order Nidovirales (5) and have enveloped virions containing the largest known RNA virus genome. Mouse hepatitis virus (MHV) is a prototypic member of the Coronaviridae family and contains a single-stranded, positive-sense RNA genome approximately 31 kb in length (28, 29, 33, 44). Viral proteins are translated from six to seven subgenomic mRNAs as well as from the genome. These RNAs are produced in different quantities, and their molar ratios remain constant during MHV replication. The virus-specific subgenomic and genomic RNAs make up a 3′-coterminal nested set (33, 62) and contain a leader sequence of approximately 70 nucleotides (nt) at the 5′ end (27, 60). Coronaviruses perform their entire replication program in the cytoplasm of infected cells. Following uncoating, coronaviruses express the largest known replicase polyproteins, which in turn are proteolytically processed to yield a large number of mature proteins, including RNA-dependent RNA polymerase. The RNA-dependent RNA polymerase, perhaps in association with host proteins, directs the synthesis of negative-sense full-length and subgenomic RNA from the 3′ end of the viral genome (40). Several alternative models have been described to explain the mechanism of MHV RNA synthesis (25, 54, 61). In all of these models, the initial step in MHV RNA replication is the synthesis of negative-sense RNA from a positive-strand genomic template.
Analysis of the structure of defective interfering RNAs indicated that approximately 470 nt at the 5′ terminus, 436 nt at the 3′ terminus, and about 135 internal nt were required for defective interfering RNA replication in MHV-infected cells and suggested that these sequences retain signals necessary for RNA replication (21, 22, 35, 36). The cis-acting signals for the synthesis of negative-strand RNA have been shown to be contained within the last 55 nt plus the poly(A) tail at the 3′ end of the MHV genome (34). Evidence supporting the involvement of host proteins in the replication of a number of RNA viruses has been reported previously (for reviews, see reference 26). Recently, the specific binding of host cellular proteins to two distinct sites within the 3′ untranslated region (3′-UTR) of MHV-JHM genomic RNA was reported (68). One site, the 3′(+)42 protein-binding element [(3′(+)42], was mapped within the 3′-most 42 nt of the genomic RNA (68), and the other was mapped between nt 154 to 129 upstream from the 3′ end of the viral genome (38).
In the current work, we have attempted to identify and characterize the host proteins that interact with the 3′-most protein-binding element of 3′-UTR in MHV-JHM. Our results indicate that the RNA-protein complex formed at this region contains at least four proteins of 90, 70, 58, and 40 kDa. The 90-kDa protein has been identified as mitochondrial aconitase (m-aconitase), the counterpart of a well-known RNA-binding protein, the iron-regulatory protein (IRP), which is also known as cytoplasmic aconitase. Although m-aconitase structural data have been used extensively to model the RNA-binding site in cytoplasmic aconitase, this is the first report to identify a target RNA for m-aconitase. We present data suggesting that binding of m-aconitase to the MHV 3′-UTR substantially increases the production of infectious virus and the expression of MHV proteins during the early stages of infection.
MATERIALS AND METHODS
Virus and cells.
Murine 17Cl-1 cells were cultured with Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (DMEM-10) at 37°C in a 5% CO2 atmosphere (33). The origin and growth of MHV-JHM virus used in this study have been described (33).
Preparation of cytoplasmic lysates and subcellular fractionation.
Cytoplasmic extracts from uninfected and MHV-infected 17Cl-1 cells were prepared by a modification of a previously described method (68). Nearly confluent monolayers were washed twice with phosphate-buffered saline (PBS), removed from the plastic substrate by scraping into PBS, and then pelleted by centrifugation. The cell pellets were resuspended in an isotonic buffer containing 25 mM Tris(pH 7.6), 1.5 mM MgCl2, 10 mM KCl, 250 mM sucrose, and protease inhibitors (10 μg/ml each of leupeptin and aprotinin plus 1 mM phenylmethylsulfonyl fluoride [PMSF]) and phosphatase inhibitors (10 μM NaVO4 and 1 mM NaF) and disrupted with 20 strokes of a Dounce homogenizer. Unlysed cells and nuclei were removed by centrifugation at 750 × g for 10 min. The supernatant was centrifuged at 10,000 × g for 30 min at 4°C, and the resulting pellet was washed once with the same buffer and stored as the mitochondrial fraction. The 10,000 × g supernatant was stored as the postmitochondrial fraction. The protein concentration in each sample was determined by the Bradford method (Bio-Rad, Richmond, Calif.).
Fractionation of cytoplasmic lysates by ion-exchange matrix.
Ten milliliters of Macro-Prep High Q support (Bio-Rad, Hercules, Calif.) were equilibrated at 4°C with buffer A (10 mM Tris [pH 7.6], 5 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol [DTT], 5% glycerol, 1 mM PMSF, plus 1 μg of leupeptin, 1 μg of aprotinin, and 0.5 μg of pepstatin per ml). Cytoplasmic lysates (50 ml) were mixed with the High Q matrix and incubated at 4°C for 1 h. Proteins which bound to the High Q matrix were removed by low-speed centrifugation. The supernatant was collected and incubated for 1 h with Macro-Prep High S matrix (Bio-Rad) preequilibrated with buffer B (identical to buffer A except pH 6.8). The beads were washed four times with buffer B containing 100 mM KCl. The bound proteins were then eluted from the High S matrix with 4 ml of 150 mM KCl in buffer B. The eluate was concentrated, desalted, exchanged into 1 ml of buffer A using a Centricon 10 filtration unit (Millipore Corp., Bedford, Mass.), and kept frozen in aliquots labeled Q/S tandem eluate.
Heparin-agarose affinity purification.
Five hundred microliters of heparin-agarose matrix type I (Sigma, St. Louis, Mo.) was equilibrated with buffer A. The concentrated, desalted Q/S tandem eluate (500 μl) was incubated with heparin-agarose matrix for 1 h at 4°C. The beads were washed four times with buffer A (pH 7.4), and the bound proteins were eluted with 500 μl of buffer A containing 100 mM KCl. The eluted material was concentrated and desalted with a Centricon 10 and frozen in aliquots.
At each step of the purification procedure, a sample (0.1 to 2 μl) of each fraction was assayed for MHV-JHM 3′(+)42 RNA-binding activity by RNase protection and gel mobility shift assays as described (68). Protein concentrations were determined with the Bradford reagent (Bio-Rad). Fractions with MHV-JHM 3′(+)42 RNA-binding activity were also analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE).
Specific RNA affinity purification.
A biotinylated synthetic RNA (5′-AGUAAAUGAAUGAAGUUGAUCAUGGCCAAUUGGAAGA-3′) corresponding to nt 42 to 5 at the 3′ end of the MHV genome [counting the first nucleotide upstream from the 3′ poly(A) tail as position 1] was purchased from Dharmacon Research (Boulder, Colo.). Aliquots of biotinylated RNA were cleaved and deprotected as per the manufacturer's guidelines. Briefly, 40 μl of 200 mM acetic acid (pH 3.0) was added to the synthetic RNA. The cleavage reaction was incubated at 60°C for 10 min and centrifuged briefly, then 40 μl of 300 mM Tris (pH 8.7) was added, and the incubation was continued for 15 min at 60°C. The concentration of the RNA was determined by UV absorbance, and the RNA solution was adjusted to 100 mM KCl, 5 mM MgCl2, and 1 mM DTT. One milligram of BioMag Streptavidin beads (PerSeptive Biosystems, Framingham, Mass.) was equilibrated with buffer C (10 mM Tris [pH 7.4], 100 mM KCl, 5 mM MgCl2, 1 mM EDTA, 10% glycerol, plus PMSF, leupeptin, aprotinin, and pepstatin at the concentrations used in buffer A). RNA was added to the beads and incubated at room temperature for 15 min, and the adsorption of RNA to the beads was monitored by UV absorbance. After washing the beads, the eluate from the heparin-agarose matrix was bound to the beads for 2 h at 4°C. The beads were washed four times with buffer C, and the bound proteins were eluted by boiling in 1× SDS-PAGE loading buffer.
Peptide sequencing and amino acid sequence analysis.
The partially purified 90-kDa protein was resolved from other proteins by SDS-PAGE, located by staining with Gelcode Blue staining reagent (Pierce, Rockford, Ill.), and the band containing this protein was excised from the gel. In-gel digestion of the protein was carried out with 1% trypsin (Promega). The resulting tryptic peptides were purified by high-performance liquid chromatography (HPLC) and analyzed by matrix-assisted laser desorption-ionization (MALDI)-mass spectrometry. Identification of the protein was attempted from peptide fingerprints, taking into account several parameters: the accuracy of fragment mass determination, the number of masses submitted for query, the mass distribution of query masses, the number of masses matching between sample and database protein, the size of the sequence database, and the kind and number of modifications considered (3). A peptide mass fingerprinting tool (http://prospector.ucsf.edu/ucsfhtml3.2/msfit.htm) from the University of California–San Francisco Mass Spectrometry Facility was used to fit the mass spectrometry data obtained for the tryptic digests to a protein sequence in an existing database. Two peptides were subjected to sequential Edman degradation at the Protein Chemistry Laboratory at Texas A&M University. The amino acid sequences obtained were used to search the Swiss-Prot protein sequence database with the BLAST sequence similarity searching program (default filtering) at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov).
In vitro transcription reactions.
Templates for transcribing RNA probes were prepared by PCR, and transcription reactions were carried out with [α-32P]UTP to generate radiolabeled probes as described earlier (68). The synthesized RNA was extracted with phenol-chloroform and purified by electrophoresis on a 6% polyacrylamide–urea gel. The gel was autoradiographed, and RNA was eluted from excised gel slices by overnight incubation at 4°C in 10 mM Tris (pH 7.6)–1 mM EDTA (TE). The eluted RNA was filtered through a 0.45-μm cellulose filter (VWR) to remove gel pieces and then loaded onto Sep-Pak light C18 cartridges (Waters Corporation, Milford, Mass.) preequilibrated with methanol. The cartridges were thoroughly washed with TE, and the RNA was then eluted with 60% methanol. The eluted material was concentrated in a Speedvac. 32P-labeled MHV 3′ plus-strand RNA probe produced by in vitro transcription generally had a specific activity of approximately 108 cpm/μg. Unlabeled competitor RNAs were synthesized in 50-μl transcription reactions carried out as described previously (67). Competitor RNAs were purified through Sep-Pak light C18 cartridges and quantitated by UV absorbance.
RNase protection-gel mobility shift assay.
RNA-protein binding reactions were performed in a volume of 10 μl as described previously (68). Briefly, cytoplasmic extracts or purified lysates containing 1 to 3 μg of total protein, 2 to 5 ng of one of the 32P-labeled MHV 3′ plus-strand RNA probes, 10 μg of heparin (Sigma), 500 ng of poly(I)-poly(C) (Sigma), and 6% glycerol were incubated at 22°C for 20 min in binding buffer (10 mM Tris [pH 7.6], 5 mM MgCl2, 1 mM DTT, 100 mM KCl) and then digested at 22°C for 20 min with RNase T1 (Calbiochem). For competition assays, various amounts of unlabeled RNA were added to the binding reaction prior to addition cytoplasmic lysate. tRNA and a mutant RNA designated mG1, with only 9% of wild-type binding activity (67), were used as nonspecific competitors. Unlabeled RNA identical to the radioactive probe was used as a specific competitor. RNA-protein complexes were subjected to nondenaturing electrophoresis, dried, and autoradiographed as described (68). In some experiments RNA-protein complexes were quantitated with a phosphoimager (Molecular Dynamics Inc.).
UV-induced and glutaraldehyde cross-linking assays.
UV cross-linking was performed as described previously (68) with the modification that 2 to 5 ng of labeled RNA and cytoplasmic extract or purified lysate containing approximately 10 μg of protein were used. Labeled proteins were separated by SDS-PAGE and visualized by autoradiography.
Glutaraldehyde cross-linking studies were performed by incubating the contents of UV cross-linking reactions with 0.1% glutaraldehyde (Sigma) for 1 to 15 min at room temperature, followed by SDS-PAGE and autoradiography.
Northwestern blot analysis.
Protein samples were resolved by SDS-PAGE (10% polyacrylamide gel) and transferred to a nitrocellulose membrane (Protan nitrocellulose; Schleicher & Schuell) by semidry blotting. The transferred proteins were renatured overnight in PBS containing 0.01% Nonidet P-40 at 40°C. Prehybridization and hybridization were carried out as described (53). Briefly, the membrane was prehybridized for 1 h in 25 ml of hybridization buffer (10 mM Tris [pH 7.6], 100 mM KCl, 0.1% [wt/vol] Ficoll 400-DL, 0.1% [wt/vol] polyvinyl pyrrolidone, 0.01% [vol/vol] Nonidet P-40, 0.1 mM MnCl2, 0.1 mM ZnCl2, 0.1 mM EDTA, 1 mM DTT), supplemented with 0.1 mg of yeast tRNA per ml. After the addition of the 32P-labeled RNA probe (106 cpm/ml in 5 ml of hybridization buffer), the membrane was hybridized for 2 h at room temperature. The membrane was washed four times for exactly 5 min in 100 ml of hybridization buffer. The membranes were completely dried and autoradiographed.
In situ UV cross-linking and two-dimensional electrophoresis.
In situ UV cross-linking was carried out as described by Garcia-Blanco et al. (13). After the binding reaction, RNA protein complexes were resolved by nondenaturing PAGE. The wet polyacrylamide gel was placed on ice at a distance of 10 cm from the UV source (254 nm; Hoefer UVC 1000) for 30 min. After UV cross-linking, the polyacrylamide gel was wrapped in Saran wrap, and RNA protein complexes were visualized by autoradiography at 4°C. The band containing the RNA protein complexes was excised, placed in a tube with TE (10 mM Tris [pH 8], 1 mM EDTA) containing RNase A (330 μg/ml) and RNase T1 (50 U/ml) and incubated at 37°C for 1 h. The RNase solution was replaced with 2 × SDS-PAGE sample buffer and incubated at 37°C for 1 h and then at 65°C for 15 min. The gel slice was embedded into the stacking portion (4 cm) of an SDS–10% polyacrylamide gel (10-cm resolving gel) by layering low-melting-temperature agarose below and above the gel slice. After electrophoresis, the gel was fixed, dried, and autoradiographed.
Alternatively, binding reactions were carried out with or without UV cross-linking. The RNA-protein complexes were resolved after being loaded into multiple lanes to ensure good resolution and maximum signals. The native gel was dried between cellophane sheets (Bio-Rad) after electrophoresis. The region containing the RNA-protein complex was cut out from the gel after brief exposure to X-ray film and incubated at room temperature for 30 min with 1× SDS loading buffer with or without prior RNase treatment (30 min at 37°C). The cellophane was removed, and the contents were collected after being boiled for 5 min at 95°C. The eluted cross-linked proteins were separated on 4 to 20% gradient gels. After electrophoresis, either the gel was dried and autoradiographed or the proteins were transferred from the gel to a nitrocellulose membrane for Western blotting.
Antibodies.
The 1.16.1 monoclonal antibody directed against the MHV-JHM N protein has been described (32). Antiserum recognizing the putative helicase (anti-Hel) subunits of MHV replicase was a gift from Mark R. Denison, Vanderbilt University, Knoxville, Tenn. (9). Anti-m-aconitase antibody was kindly provided by Richard S. Eisenstein, University of Wisconsin, Madison (6).
Indirect immunofluorescence assays.
17Cl-1 cells were grown on coverslips to 50% confluence, infected with MHV-JHM at a multiplicity of infection (MOI) of 1 PFU/cell, and fixed at various times postinfection (p.i.) with 3% paraformaldehyde in PBS. After being washed with 10 mM Tris-HCl and 50 mM NH4Cl, cells were permeabilized for 10 min with cold acetone. Overnight blocking was carried out with PBS–Tween 20 (0.1%) containing 5% normal goat serum. Antibodies were diluted with PBS containing 0.1% Tween 20; all washes were performed with the same solution. After sequential primary antibody incubations (60 min each), the cells were washed four times. Secondary antibodies were added together and incubated for 45 min, followed by four washes, and cells were embedded in ProLong Antifade kit (Molecular Probes).
Laser confocal microscopy was performed in the confocal imaging center at Texas A&M University System Health Science Center. Cells were analyzed using a Meridian Ultima-Z laser confocal microscope system, operating with blue line excitation from an argon laser for Oregon green and yellow line excitation from an krypton laser for rhodamine red X. Scanning parameters were adjusted to negative control slides to eliminate nonspecific fluorescence collection. Fixed contrast, brightness, and exposure times were used for all data collection and image analysis. The thickness of each digital section obtained by the microscope was 0.1 μm at a pinhole size of 200 μm. Each experiment was performed at least five times, and approximately 10 fields were examined to ensure that the results were representative. Only cells that were not in syncytia and did not show any visible cytopathic effects were selected for colocalization analysis. Two or more individual cell images were recorded for each experiment.
Iron supplementation.
To stimulate translation of m-aconitase (55), 50% confluent 17Cl-1 monolayers had their medium replaced with DMEM-10 containing 60 to 100 μg of ferric ammonium citrate (FAC) (Sigma) per ml and incubated overnight. The next day, the cells were either mock infected or infected with MHV-JHM at an MOI of 1 in the presence of FAC. The cells were harvested at various times p.i. Either the cells were frozen for later virus titration or postmitochondrial lysates were prepared as described above. To measure the effect of FAC supplementation on MHV plaquing efficiency, L2 cells were grown to confluence in the presence or absence of FAC (100 μg/ml), and then a standard aliquot of MHV was counted with the continued presence (or absence) of FAC in the agarose overlay.
Western blot analysis.
Equal amounts of postmitochondrial lysates were added to SDS lysis buffer heated to 100°C and boiled for 5 min. Proteins were separated on 10% (wt/vol) SDS-polyacrylamide gels and transferred onto Hybond polyvinylidene difluoride membranes (Amersham Pharmacia Biotech) by semidry blotting. Membranes were blocked overnight in TBST (25 mM Tris-HCl [pH 8], 150 mM NaCl, 0.1% [vol/vol] Tween 20) containing 5% (wt/vol) powdered dried milk. The membranes were incubated for 1 h with primary antibodies diluted in TBST–5% (wt/vol) powdered dried milk, washed in TBST, and incubated with a second antibody conjugated to horseradish peroxidase for 1 h. Membranes were developed using an ECL detection kit (Amersham Pharmacia Biotech). Exposed films were scanned on a flat-bed scanner, and the signals were quantified using NIH Image software (version 1.60).
RNA extraction and Northern blot analysis.
Total RNA from 3 × 106 cells was extracted using the Trizol method (Life Technologies). RNA was denatured and electrophoresed through a 0.8% formaldehyde–agarose gel and transferred to a nitrocellulose membrane. Hybridization was carried out using a 32P-labeled negative-sense RNA which was generated by in vitro transcription of a cDNA which extended from the 3′ poly(A) tail to nt 119 of the MHV-JHM N gene (31). The blot was washed four times for 15 min in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% (wt/vol) SDS at 70°C and processed for autoradiography.
RESULTS
Characteristics of cytosolic protein binding to the MHV-JHM 3′(+)42-containing RNA.
To identify, purify, and characterize proteins interacting with the 3′(+)42 protein-binding element, we employed the RNase T1 protection-gel mobility shift assay that we originally used to identify this protein-binding element (68). Cytosolic protein extracts from murine 17Cl-1 cells were assayed using a 68-nt 32P-labeled transcript corresponding to nt 16 to 84 upstream from the 3′ end of the MHV-JHM genome. This probe contains the 3′(+)42 protein-binding element, and we have previously shown that this RNA possesses host cell protein-binding activity (68). As shown in Fig. 1A, three RNA-protein complexes formed, with the slowest-migrating and presumably largest complex, complex 1, being the most abundant. Identical results were obtained using a probe, designated 3′(+)42, corresponding to the last 42 nt upstream of the poly(A) tail (not shown). These results are similar to those obtained previously (68). The specificity of the RNA-protein binding was established by competition experiments. The formation of the T1-resistant complexes was almost completely blocked by 25-, 50-, and 100-fold molar excesses of unlabeled specific competitor RNAs but not by unlabeled nonspecific competitor RNAs (not shown).
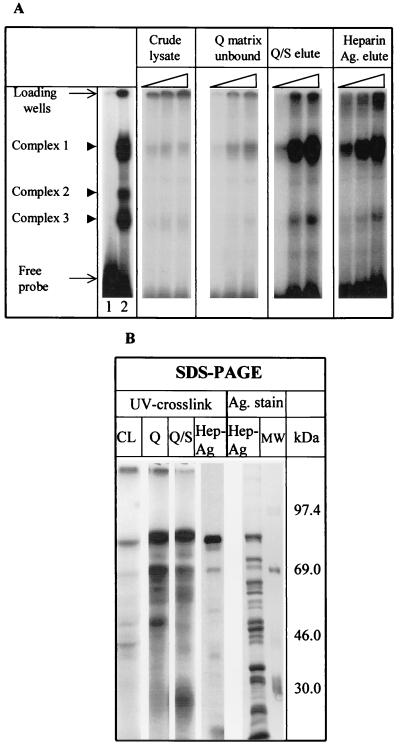
FIG. 1.
Purification of proteins recognizing the MHV 3′(+)42 protein-binding element. (A) Increasing amounts of 17Cl-1 cytoplasmic extracts (0.5, 1.5, and 2.5 μg) or partially purified fractions were assayed for their ability to bind to a 32P-labeled RNA (nt 16 to 84) by RNase protection-gel mobility shift assays. RNA-protein complexes were resolved by nondenaturing PAGE. The far left panel represents an assay which has been overexposed to clearly demonstrate complexes 2 and 3. Lane 1 represents free probe assayed in the absence of cytoplasmic lysate. Lane 2 represents the probe after incubation with cellular lysate. (B) 32P-labeled RNA (nt 16 to 84) was incubated with 5 μg of either uninfected 17Cl-1 extract (CL), the fraction which failed to bind to High Q matrix (Q), the Q/S tandem eluate (Q/S), or proteins purified through the heparin-agarose step (Hep-Ag), and the RNA-protein complexes formed were irradiated with UV light for 30 min. The samples were digested with RNase A (20 μg/μl) and directly analyzed by SDS-PAGE (10% gel) and autoradiography. A silver stain of the heparin-agarose-eluted material and molecular size markers is also shown (Ag. stain). The sizes of the markers are indicated on the right-hand side (in kilodaltons).
The molecular masses of the cytoplasmic proteins that bind to the MHV-JHM 3′(+)42 protein-binding element were estimated by UV-induced cross-linking assays. Proteins to which 32P label was transferred by cross-linking were analyzed by SDS-PAGE. Four proteins of approximately 90, 70, 58, and 40 kDa were consistently detected (Fig. 1B), with the 90-kDa protein being the most prominent. No bands were observed when cytoplasmic extracts were omitted. Similarly, 17Cl-1 cytoplasmic extracts probed in Northwestern blot assays contained four proteins of 90, 70, 58, and 40 kDa which bound 32P-labeled MHV 3′(+)42 RNA (Fig. 2, lane 7). The apparent molecular masses of three of these four proteins (90, 70, and 55 kDa), although not identical, are consistent with the molecular masses of three of the proteins that we had identified previously in UV cross-linking experiments (68), considering the different markers used with different gel systems in experiments run years apart. The 40-kDa signal that we observe here was noted previously as a faint band (68) which was not reproducibly present at that time. We did not detect the 120-kDa protein that we reported previously. The reason for this is unclear.
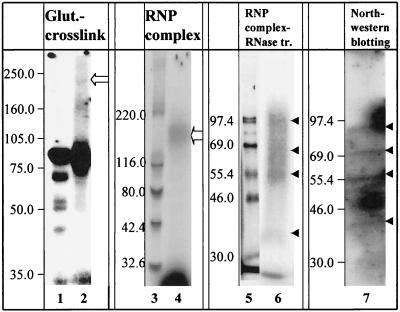
FIG. 2.
Analysis of RNA-protein complex 1. RNA-protein complexes were UV cross-linked in solution and then either treated with glutaraldehyde as described in Materials and Methods (lane 2) or untreated (lane 1). The position of a complex of about 200 kDa is marked by an open arrow. In situ UV cross-linking of RNA-protein (RNP) complexes was performed after native gel electrophoresis as described in Materials and Methods. Complex 1 was then eluted from the native gel and either treated with RNase (lane 6) or untreated (lane 4). The samples were then subjected to SDS-PAGE and autoradiography in parallel with 14C-labeled molecular size markers (lanes 3 and 5). The open arrow in lane 4 indicates the position of a band of approximately 200 kDa. The solid arrowheads in lane 6 indicate the positions of four proteins of approximately 90, 70, 58, and 40 kDa. Lane 7 contains an autoradiograph of a Northwestern blot of 17Cl-1 cytoplasmic extract (10 μg) probed with 32P-labeled MHV 3′(+)42 RNA (2 × 106 cpm). The positions of four proteins of approximately 90, 70, 58, and 40 kDa are indicated by solid arrowheads.
To probe for protein-protein interactions among the four proteins present in the RNA-protein complex, lysates were incubated with labeled 16–84 RNA probe, UV cross-linked to the RNA by UV irradiation in solution, treated with glutaraldehyde as described in Materials and Methods, and analyzed by SDS-PAGE and autoradiography. A band with an apparent mass of 200 kDa (Fig. 2, lane 2) was visible along with a band of intermediate mass (approximately 160 kDa) which was apparent only after glutaraldehyde cross-linking, suggesting that there were protein-protein interactions as well as RNA-protein interactions in the RNA-protein complex.
The proteins comprising the most prominent complex, complex 1, were further characterized by in situ cross-linking and two-dimensional gel electrophoresis. After the binding reaction and electrophoresis for mobility shift detection, the gel was exposed to UV light to covalently cross-link the radiolabeled RNA to proteins at sites of direct RNA-protein contact. The RNA-protein complex was then excised from the gel, treated with RNase to trim all but the short stretch of RNA that is cross-linked to or protected by the protein, and layered onto an SDS-polyacrylamide gel for electrophoresis in the second dimension (13, 45). Four major radiolabeled protein species of approximately 90, 70, 58, and 40 kDa were present in complex 1 (Fig. 2). The sizes of the RNA-binding proteins detected by in situ cross-linking of complex 1, by solution UV cross-linking (Fig. 1B), and by Northwestern blot (Fig. 2, lane 7) are identical.
Partial purification of the MHV-JHM 3′(+)42 RNA-binding proteins.
RNA-binding proteins were purified from cytoplasmic lysates by sequential ion-exchange chromatography and affinity chromatography steps. Enrichment of the RNA-binding proteins from the cytoplasmic extract was assessed by gel mobility shift-RNase protection assay, UV cross-linking, and silver staining after SDS-PAGE (Fig. 1). RNA-binding activity was not removed from cytoplasmic lysates by a High Q matrix, a strong anion-exchange resin. Virtually all of the RNA-binding activity was detected in the unbound fraction. This fraction was then directly applied to a High S matrix, a strong cation-exchange resin, which bound the activity. After a series of preliminary experiments which determined the conditions for optimal elution from the High S matrix, the RNA-binding proteins were eluted from the High S resin with 150 mM KCl. Fractions having maximum RNA-binding activity were pooled and subjected to nonspecific affinity chromatography with heparin-agarose. UV cross-linking assays and SDS-PAGE analysis followed by silver staining (43) and autoradiography demonstrated that this purification scheme resulted in the enrichment of RNA-binding proteins. In particular, the 90-kDa protein present in material purified through the heparin-agarose affinity step appeared to be well resolved from other proteins in this preparation and was strongly labeled by UV cross-linking to our RNA probe (Fig. 1B). Two-dimensional gel electrophoresis (first dimension being isoelectric focusing) of the heparin-agarose-purified material followed by silver staining indicated that the 90-kDa band seen on SDS-PAGE was a single protein (not shown).
Identification of the MHV-JHM 3′ plus-strand RNA-binding protein as m-aconitase by MALDI-mass spectrometry and peptide sequencing.
A large-scale preparation of heparin-agarose affinity matrix-purified material was reduced and carboxymethylated, and the 90-kDa protein was resolved from the other proteins in the preparation by SDS-PAGE. The band containing this protein was excised and digested with trypsin. After fractionation of the tryptic digest by HPLC, the molecular masses of the peptides were determined by MALDI-mass spectrometry. Mass spectrometry data were used for an MS-Fit search of the SwissProt.r06.03.99 database. In rank order, the three best fits with the data were m-aconitases from bovine (Swiss-Prot AC P20004), swine (Swiss-Prot AC P16276), and human (Swiss-Prot AC Q09259) species, with 63% (7 of 11), 54% (6 of 11), and 54% (6 of 11) masses matched, respectively. Two peptides well resolved by HPLC were chosen for sequencing, and complete sequence information was obtained for both. The resulting peptide sequences, IVYGHLDDPANQEIER and LTIQGLK, were used to search the Swiss Protein sequence data bank. The search revealed 100% sequence identity with amino acids 69 to 83 and 724 to 730 in human m-aconitase (Swiss-Prot AC Q09259).
Evidence for the presence of m-aconitase in the RNA-protein complex.
Proteins binding to the MHV 3′(+)42 protein-binding element were further purified by affinity chromatography employing a biotinylated RNA corresponding to nt 42 to 5 at the 3′ end of the MHV genome as the affinity reagent. Sypro-Orange staining of the proteins eluted from the specific RNA affinity matrix demonstrated four proteins of 90, 70, 58, and 40 kDa (Fig. 3A, lane 2). The prominent signal observed around 30 kDa is due to the bromphenol blue tracking dye.
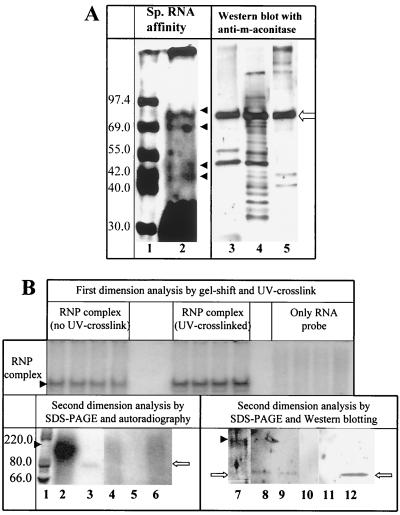
FIG. 3.
Western blot analyses with anti-m-aconitase antibody. (A) Biotinylated RNAs representing nt 42 to 5 at the 3′ end of the MHV genome were immobilized on magnetic beads. Heparin-agarose eluates were incubated with the beads, and the RNA-binding proteins were eluted as described in Materials and Methods, subjected to SDS-PAGE, and stained with the fluorescent dye Sypro-Orange (lane 2) in parallel to molecular size markers (lane 1). The image shown in lanes 1 and 2 is a negative of the image obtained after UV illumination of the gel. The positions of proteins of 90, 70, 50, and 40 kDa are marked by solid arrowheads. We believe that the 50-kDa band corresponds to the approximately 58-kDa band observed in Fig. 1, lane 4, and Fig. 2, lanes 6 and 7. The portion of the gel containing affinity-purified protein (lane 3), crude cytoplasmic lysate (lane 4), and purified m-aconitase (lane 5) was transferred to nitrocellulose and probed with an anti-m-aconitase antibody. The position of m-aconitase is marked with an open arrow. (B) 32P-radiolabeled 3′(+)42 MHV RNA was incubated with a heparin-agarose-eluted protein preparation and analyzed on a 6% nondenaturing polyacrylamide gel. A portion of the gel containing the majority but not all of the lanes was exposed to UV light, and RNA-protein (RNP) complex 1 was visualized by autoradiography (upper panel). Complex 1 was excised from the gel, and the extracted protein (as described in Materials and Methods) was subjected to electrophoresis in an SDS–10% polyacrylamide gel with and without prior RNase treatment. One set of lanes (lanes 1 to 6) was autoradiographed. Lanes 7 to 12 were transferred to nitrocellulose and probed with anti-m-aconitase antibodies. Molecular size markers are shown in lane 1. Lanes 2 and 7 contain complex 1 which was UV cross linked but not treated with RNase. Lanes 3 and 8 represent complex 1 which was UV cross linked and treated with RNase prior to SDS-PAGE. Lanes 4 and 9 represent complex 1 which was not UV cross-linked but was treated with RNase prior to SDS-PAGE. Lanes 5 and 10 represent samples excised from the region of the nondenaturing gel corresponding to complex 1 from binding reactions containing only labeled RNA in the absence of protein. Lanes 6 and 11 represent extracts from a region of the nondenaturing gel containing no RNA-protein complex. Lane 12 represents purified m-aconitase. The position of the 200-kDa complex is indicated by solid arrowheads; the position of m-aconitase is indicated by the open arrows.
When specific-affinity-purified protein was analyzed by SDS-PAGE followed by immunoblotting with an anti-m-aconitase antibody, a 90-kDa protein which coelectrophoresed with purified bovine m-aconitase (kindly provided by M. Claire Kennedy, Medical College of Wisconsin, Milwaukee) was detected (Fig. 3A). The lower-molecular-weight bands present in the Western blot in lanes 3 to 5 are not detected with greater dilutions of the primary antiserum, and, thus we believe that these bands represent nonspecific immunoreactivity. However, we cannot completely rule out the possibility that some of these bands are proteolytic breakdown products of m-aconitase. Furthermore, RNA-protein complex 1 isolated by native gel electrophoresis contained a 90-kDa protein which was labeled by 32P after UV cross-linking (Fig. 3B, lane 3) and was also recognized by anti-m-aconitase antibody (Fig. 3B, lane 8). This antibody also reacted with the complex of 200 kDa demonstrated when the RNase treatment step is omitted in UV cross-linking reactions (Fig. 3B, lane 7). This immunoreactive band corresponds in size to the 32P-labeled band observed when this complex is autoradiographed rather than probed by immunoblotting (Fig. 3B, lane 2). When the UV cross-linking step was omitted prior to isolation of complex 1 and RNase digestion (Fig. 3B, lane 4), a radioactive signal was not detected at either 200 or 90 kDa. However, a 90-kDa band which reacted with the anti-m-aconitase antibody was observed in these complexes (Fig. 3B, lane 9). When binding reactions performed in the absence of protein were handled similarly (lanes 5 and 10) and when samples were isolated from a region of the nondenaturing gel which did not contain an RNA-protein complex (lanes 6 and 11), no radioactive or immunoreactive signal was detected. These results strongly support the identification of the 90-kDa protein in RNA-protein complex 1 as m-aconitase.
RNA-binding activity of purified m-aconitase is dependent on Fe-S state.
To determine if purified m-aconitase binds to the MHV-JHM 3′(+)42 RNA in the absence of other proteins, RNase protection-gel mobility shift reactions were carried out with purified bovine mitochondrial holo- and apo-aconitase. Both forms of the enzyme were tested since it is well known that the binding of the cytoplasmic homolog of m-aconitase (IRP1) to RNA is dependent on the Fe-S state of the protein (see Discussion). Binding activity was not detected with either form of the protein in this assay. However, when a UV cross-linking step was added prior to electrophoresis, the mitochondrial apo-aconitase formed an RNA-protein complex. This complex coelectrophoresed with complex 2 formed when the same RNA probe was reacted with cytoplasmic lysate (Fig. 4A). Binding of the mitochondrial holoenzyme to the MHV 3′(+)42 RNA could not be detected by this method (not shown). Binding assays with bovine serum albumin failed to demonstrate any RNA binding activity (data not shown). The specificity of this binding of mitochondrial apo-aconitase to the MHV 3′(+)42 RNA was verified by competition with specific and nonspecific unlabeled competitors. At a 50-fold excess of self-competitor RNA, the complex was not visible, whereas the complex remained unchanged at even a 100-fold excess of mutant competitor RNA mG1 and there was a slightly higher level of binding activity with increasing amounts of tRNA (Fig. 4B). We have noted this slightly higher binding activity in the presence of tRNA competitor on several occasions. All of the RNA-binding activity was detected in the postmitochondrial lysate (Fig. 4C), although the vast majority of m-aconitase fractionates into the mitochondrial lysate (Fig. 4D). The amount of m-aconitase detected by Western blot does not reflect the Fe-S status of the protein. The m-aconitase present in the mitochondrial fraction is likely to reflect the active enzyme containing a [4Fe-4S] iron sulfur cluster.
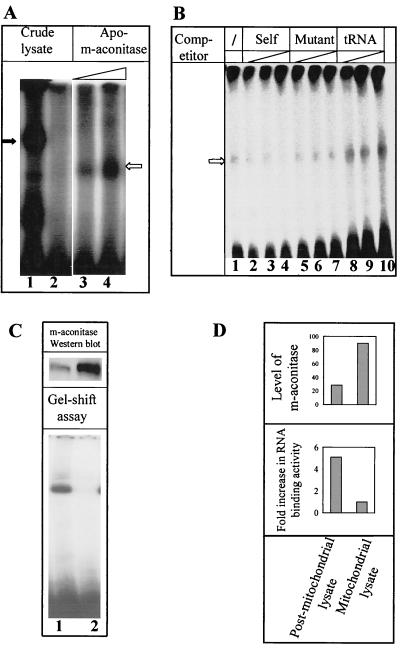
FIG. 4.
Interaction of purified apo-m-aconitase with MHV 3′(+)42 RNA is specific. (A) Purified apo-m-aconitase (7 and 14 μg in lanes 3 and 4, respectively) and cytoplasmic lysates (lanes 1 and 2) were incubated with 5 ng of 32P-labeled MHV 3′(+)42 RNA for 20 min at 22°C, exposed to UV light for 30 min, digested with RNase, and analyzed on a nondenaturing polyacrylamide gel. The positions of complex 1 and complex 2 are indicated by solid and open arrows, respectively. (B) Various amounts of unlabeled RNA competitors were added along with 32P-labeled MHV 3′(+)42 RNA prior to the addition of purified apo-m-aconitase. The RNA-protein complexes formed were then analyzed by gel mobility shift assays after UV cross-linking. Reactions displayed in lane 1 contained no competitor; lanes 2 to 4 contained 50, 125, and 250 ng, respectively, of unlabeled specific competitor RNA; lanes 5 to 7 contained 50, 125, and 250 ng, respectively, of mutant (mG1) competitor RNA; lanes 8 to 10 contained increasing amounts (50, 125, and 250 ng, respectively) of unlabeled yeast tRNA. (C) Cytoplasmic extracts of 17Cl-1 cells were separated into mitochondrial (lane 2) and postmitochondrial (lane 1) fractions. Equal amounts of protein from each fraction were used in either RNase protection-gel shift assays or resolved by SDS-PAGE and blotted onto nitrocellulose membranes. (D) Quantitation of the Western blot and RNA-binding assays was performed as described in Materials and Methods.
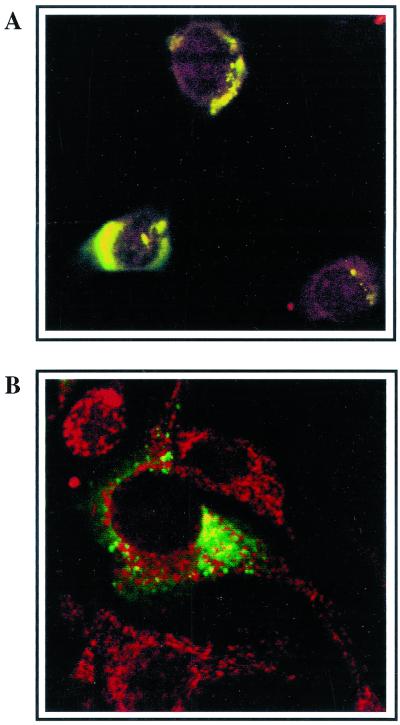
Partial colocalization of nucleocapsid protein and m-aconitase early in infection.
Dual-labeling experiments were carried out to visualize the localization of MHV replication in relation to m-aconitase. The MHV helicase protein (Hel) has been shown to localize to the site of newly synthesized MHV RNA (9). However, the antibody available for the visualization of the Hel protein was raised in rabbits, as was the antibody to m-aconitase. It has been reported that the MHV nucleocapsid (N) protein colocalized with helicase containing MHV replication complexes (9, 65). We first confirmed the colocalization of N protein with the Hel protein at early time points of MHV infection (3 h p.i.) by dual immunofluorescent labeling with an N protein-specific mouse monoclonal antibody and rabbit anti-Hel antibody (Fig. 5A). Consistent with the previously published work, we observed almost complete colocalization of the two labels, which gave a punctate pattern of staining. Therefore, we used the dual labeling of N protein and m-aconitase to visualize their proximity inside MHV-infected 17Cl-1 cells. Partial overlap of the fluorescent signal from these two proteins was observed in double labeling experiments with anti-N and anti-m-aconitase antibodies (Fig. 5B). Similarly, we have seen isolated foci of m-aconitase staining when cells were labeled with the anti-m-aconitase antibody and Mito Tracker Red CMXRos, a mitochondrion-selective dye (Molecular Probes) (data not shown). However, these experiments did not allow us to definitively determine if N protein and extramitochondrial m-aconitase colocalize.
FIG. 5.
Confocal immunofluorescence of MHV-infected 17Cl-1. At 3 h p.i., MHV-infected 17Cl-1 cells were fixed, labeled with an anti-N monoclonal antibody (green) and with either an anti-Hel (red) polyclonal rabbit antiserum (A) or with rabbit anti-m-aconitase (red) antibody (B). Images were captured by dual laser beam scanning. Colocalization of green (oregon green) and red (rhodamine red X) signals in a single pixel produces yellow, while separated signals remain green or red.
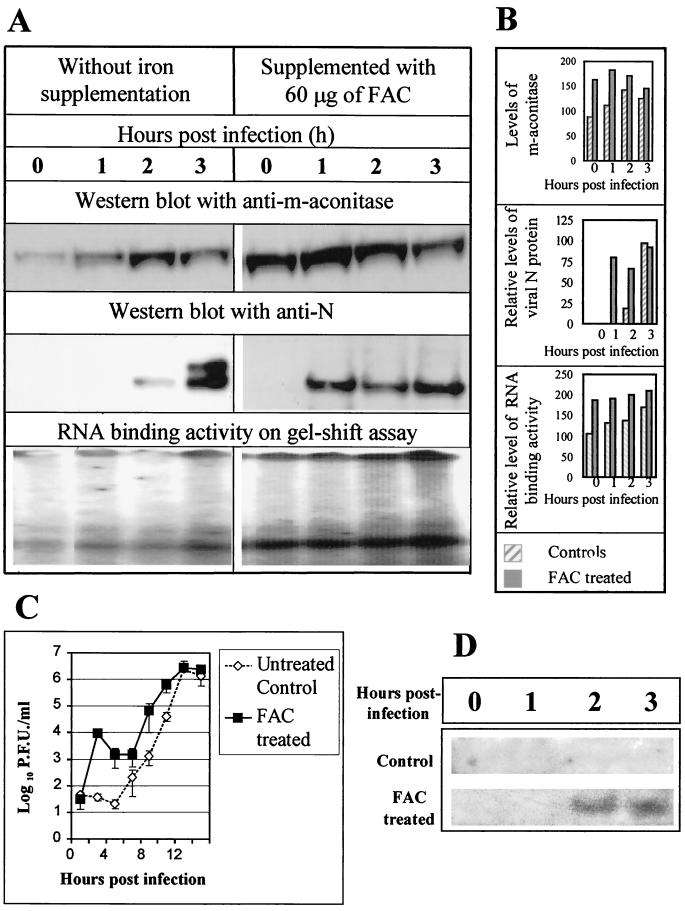
Iron treatment increases m-aconitase expression, RNA-binding activity, and MHV replication early in infection.
The level of m-aconitase is regulated in part by intracellular iron concentration (55). We thus attempted to manipulate m-aconitase levels and MHV 3′(+)42 RNA binding activity by treating cells with FAC prior to and during infection. Postmitochondrial lysates prepared from FAC-treated 17Cl-1 cells had increased RNA-binding activity relative to lysates prepared from untreated cultures, as judged by an RNase protection-gel mobility shift assay (Fig. 6A). We also observed a slight increase in binding activity during the course of infection (last time point, 3 h p.i.). Western blotting of these lysates with anti-m-aconitase antibody indicated an increase in the level of m-aconitase in the FAC-treated samples, confirming the effect of iron supplementation (Fig. 6A). An increase in the level of m-aconitase which paralleled the increase in RNA-binding activity was also observed during the course of infection.
FIG. 6.
Modulation of m-aconitase synthesis and viral replication by iron supplementation. 17Cl-1 cells were either untreated or supplemented with FAC (60 μg/ml) and infected with MHV at an MOI of 1 as described in Materials and Methods. At various times p.i., the cells were harvested for analysis. (A) Equal amounts of protein were electrophoresed, blotted to a nitrocellulose membrane, and probed with either anti-m-aconitase polyclonal rabbit serum or an anti-N monoclonal antibody. RNA-binding activity was assessed by RNase protection-gel mobility shift assay. (B) Quantitation of the Western blot and RNA-binding assays was performed as described in Materials and Methods. (C) Replicate cultures of 17Cl-1 cells were either treated with FAC (60 μg/ml) or untreated and infected with MHV-JHM at an MOI of 0.1. Cultures were harvested at various times p.i., and virus was counted by plaque assay. The data are plotted as virus titer, and each point represents the mean of three experiments, with each experiment containing duplicate or triplicate samples for each time point. Standard errors are indicated by the error bars. (D) Replicate cultures of 17Cl-1 cells were either treated with FAC (60 μg/ml) or untreated and infected with MHV-JHM at an MOI of 0.1. At various times p.i., RNA was extracted and analyzed by Northern blot hybridzation as described in Materials and Methods. The region of the gel containing RNA 7 is shown.
To seek possible functional effects of the interaction of m-aconitase with the MHV-JHM 3′(+)42 protein-binding element, we examined the effect of FAC supplementation on plaque formation and virus production. The ability of a virus inoculum to form plaques on cells treated with FAC and on untreated control cultures was nearly equal (data not shown). As shown in Fig. 6C, one-step growth curves indicated that FAC supplementation increased the amount of infectious virus in the cultures at early times p.i. However, the FAC-treated and control cultures ultimately produced similar amounts of virus by 12 to 14 h p.i. In a control experiment, FAC supplementation had no effect on the kinetics of growth of mengovirus in the same 17Cl-1 cell line. To determine if iron supplementation altered the amount of viral protein produced during the infection, we monitored the accumulation of N protein in the FAC-treated cells during the course of infection. Western blot analysis demonstrated increased N protein production at early time points of infection in the FAC-treated cells compared to the untreated cultures (Fig. 6A and B). Two forms of N protein which differ in their level of phosphorylation have been observed (42). The increased N protein found at early time points (1 to 3 h p.i.) of infection observed in the FAC-treated cells was largely the lesser-phosphorylated form of the N protein. Northern blot hybridization detected a corresponding acceleration in the appearence of MHV RNA 7 (Fig. 6D). These results strongly support the hypothesis that modulation of m-aconitase expression led to increases in infectious virus production and increased viral protein synthesis, probably through increased RNA-binding activity present in infected cells.
DISCUSSION
Two protein-binding elements, conserved among MHV strains (46), have been mapped within the MHV 3′-UTR and are implicated in viral replication (38, 67, 68). By direct peptide sequence analysis, affinity purification, Northwestern blotting, UV cross-linking–second-dimension analysis, and Western blotting, we have demonstrated that there are at least four proteins in the complex formed at the 3′-most cis element of the MHV-JHM 3′-UTR and identified the 90-kDa component of this complex as m-aconitase. The binding activity is not limited to murine cells, as we have observed similar binding activity in HeLa cell extracts by gel shift and UV cross-linking assays (data not shown). The stoichiometry of the four proteins is not detectably different in second-dimension analysis after UV cross-linking and in Northwestern blotting assays. However, the relative signal strength for m-aconitase after in-solution UV cross-linking and its recovery during specific RNA affinity purification is severalfold greater than that of the other three proteins. Although this is not a direct measurement of protein abundance, m-aconitase can be considered the dominant constituent of the RNA-protein complex, at least in terms of the relative abilities of the four proteins to bind to the MHV RNA. This is the first report to identify an RNA-binding activity for m-aconitase. Purified m-aconitase specifically binds to the MHV-JHM 3′-UTR RNA despite the absence of a consensus ribonucleoprotein (RNP)-binding domain (4). The lack of a consensus RNP-binding domain is not unique to m-aconitase, since several proteins binding to viral RNAs, including calreticulin to rubella virus RNA (58) and glyceraldehyde-3-phosphate dehydrogenase to hepatitis C virus (47) and human parainfluenza virus RNAs (7), do not possess such domains. The IRP (23) and some of the small nuclear RNPs (51) also belong to this category of RNA-binding proteins.
Mammalian cells have two aconitases encoded by separate nuclear genes: the mitochondrial enzyme (m-aconitase), a component of the citric acid cycle (citrate [isocitrate] hydrolyase, EC 4.2.1.3), and cytoplasmic aconitase, better known as the IRP. Aconitase catalyzes stereospecific interconversion of citrate into isocitrate through a cis-aconitate intermediate in the Krebs cycle (2). Activity of the enzyme critically depends on the presence of an iron-sulfur [4Fe-4S] cluster in the catalytic center that is highly sensitive to oxygen. Molecular cloning of IRP revealed 30% amino acid identity with m-aconitase (19) and ∼56% overall amino acid sequence similarity. The highest degree of sequence similarity is in the vicinity of the active center; 18 active-site residues of m-aconitase are conserved in IRP, including three of the four cysteines that coordinate the [4Fe-4S] cluster (48, 52). IRP is a conditional cytoplasmic mRNA-binding protein which interacts with iron-responsive elements (IREs) located in the 5′-UTR of ferritin mRNA and the 3′-UTR of transferrin receptor (TfR) mRNA (23, 64) and functions to coordinate posttranscriptional regulation of cellular iron metabolism (17, 24). Disassembly of the IRP iron-sulfur cluster upon iron starvation yields the active RNA-binding form which binds to IREs of target RNAs. Thus, when iron becomes limiting inside the cell, the binding of IRPs to the IRE in the 5′-UTR of ferritin mRNA blocks translation, whereas an association of IRPs with IREs in the 3′-UTR of TfR mRNA stabilizes this transcript and increases its subsequent translation. Folded m-aconitase has a centrally located active-site cleft and conserved substrate-binding residues (50, 52). When the identified IRE contact sites are projected onto the m-aconitase structure, it appears that the RNA is most likely to bind at the cleft between N-terminal domains 1 to 3 and the C-terminal globular domain (domain 4). The variable opening of this cleft was proposed to explain the mutually exclusive enzymatic and RNA-binding functions of IRP1 (23). In the enzymatically active closed conformation, the IRE-binding sites would remain inaccessible. Only after [4Fe-4S] cluster and/or substrate removal would the cleft open to allow access to the RNA-binding site. In iron-replete cells, IRP is active as aconitase, whereas in iron-depleted cells, IRP has no enzyme activity but is able to bind IREs. Site-directed mutagenesis replacing the cluster-coordinating cysteine residues with serines has also demonstrated the role of the [Fe-S] cluster in RNA binding (18, 48). Overexpression of IRP in baculovirus-infected insect cells yields the apo-form of IRP having IRE-binding activity even though it lacks the [Fe-S] cluster. We have clearly demonstrated that only the m-aconitase apoprotein binds directly to the MHV-JHM 3′(+)42 RNA, and this binding is specific for this RNA. Although the stability of the interaction of purified m-aconitase with the MHV 3′(+)42 RNA is low, it seems likely that the other proteins detected in the RNA-protein complex stabilize the interaction and lead to the formation of complex 1 in the gel mobility shift assay. The m-aconitase, like its cytoplasmic homolog IRP, is a bifunctional protein whose dynamic (and perhaps labile) [4Fe-4S] cluster determines if it functions as an enzyme or as a posttranscriptional regulator (2, 23).
It is surprising that m-aconitase, a mitochondrial matrix protein, is involved in binding to a viral RNA that replicates in the cytoplasm, notably on the late endosomal vesicles at early times of infection (56, 65). During rubella virus infection, P32, another mitochondrial matrix protein, has been shown to interact with its capsid protein (1). Moreover, rubella virus replication sites are modified endosomes or lysosomes located in the perinuclear region (30). Dual labeling of m-aconitase and the MHV N protein, a protein that colocalizes with MHV replication complexes (9, 56, 65), suggests that the interaction of MHV RNA and m-aconitase is possible in vivo. Although breakage of mitochondria during the preparation of postmitochondrial lysate could be attributed as the source of m-aconitase in that fraction, the RNA-binding activity did not partition with the majority of m-aconitase detected in mitochondrial lysates. We believe that the m-aconitase interacts with MHV RNA prior to importation into mitochondria. We have observed a few foci of extramitochondrial m-aconitase in cells stained with MitoTracker Red CMXRos and immunofluorescently labeled with anti-m-aconitase antibody (data not shown). Immunoelectron microscopy studies are needed to confirm this result. Furthermore, RNA specific-affinity Western blotting and supershift assays with antibody to mitochondrial heat shock protein 70 (mtHSP70) suggest that the 70-kDa protein present in the four-protein–RNA complex is mtHSP70 (unpublished), a finding which is consistent with this idea. The mtHSP70 is also predominantly located inside the mitochondrial inner membrane and is implicated in the importation of mitochondrial proteins through its chaperone activity. Indeed, the presence of extramitochondrial mtHSP70 has been established through biochemical fractionation (8) and immunoelectron microscopic studies (57). Numerous other mitochondrial proteins have been shown to leave this organelle under certain conditions (59). It has also been demonstrated previously that mitochondrial stress proteins mtHSP70 and HSP58 transiently interact with newly synthesized mitochondrial proteins (41). These data are consistent with our notion that the assembly of the MHV RNA–m-aconitase RNA-protein complex occurs outside the mitochondria in the absence of any reported observation of involvement of this organelle in MHV infection.
The identification of an IRE in the 5′-UTR of m-aconitase indicated that IRPs might regulate m-aconitase synthesis (69). Translational arrest via the IRE-IRP interaction in the 5′-UTR has been demonstrated in in vitro translation systems (16, 20) and in intact cells (63), even in the context of a heterologous RNA. A link between cellular iron status and m-aconitase expression has also been established; 5 h of iron exposure results in a 2- to 2.4-fold increase in m-aconitase synthesis compared to untreated cells (20, 55). We hypothesized an effect on viral replication in iron-replete cells through the regulated expression of m-aconitase. However, there was no striking difference in virus plaque formation (plaque number, morphology, or size) with either iron supplementation or depletion (data not shown). Iron supplementation resulted in increased virus production at early times postinfection, although virus production was equivalent in supplemented and control cultures by 12 h p.i. Iron supplementation increased the expression of the 57-kDa form of the N protein as early as 1 h p.i., accompanied by a corresponding increase in MHV-specific mRNA levels compared to the control cultures. We believe that m-aconitase binding to the 3′-UTR likely increases the stability of the viral mRNAs and hence increases translation of viral proteins, similar to the role of IRP in regulating TfR (24). The relatively modest effect of iron supplementation on MHV replication was not unexpected, since dramatic changes in m-aconitase levels cannot be achieved by iron depletion or repletion, as it is involved in key physiological functions. Moreover, other proteins which are components of the complex which binds to the 3′-UTR may be limiting.
Mitochondria may be a critical target during virus infection in view of their central roles as regulators of cell death and survival. Aconitase activity in mitochondria has been reported to be a sensitive redox sensor of reactive oxygen and nitrogen species in cells (12, 14). Reaction between m-aconitase and superoxide plays a major role in mitochondrial oxidative damage (15, 37, 66). The activity of m-aconitase, an iron-sulfur enzyme, is inhibited in activated macrophages (10, 11) and correlates with the appearance of iron-dinitrosyl complexed with protein thiols as a consequence of the cytotoxic effects of NO through its reactivity with iron (39). Macrophages from MHV-resistant A/J mice treated with IFN-γ produce higher levels of inducible nitric oxide synthase mRNA transcripts and higher levels of nitrite than macrophages from MHV-susceptible BALB/c mice. In vivo inhibition of NO also resulted in loss of resistance to MHV-3 in A/J mice. The relative defect in the production of NO in macrophages from susceptible BALB/c mice may contribute to their susceptibility to lethal MHV-3 infection (49). It is tempting to speculate that inhibition of MHV replication in resistant A/J mice, which produce more NO in response to IFN-γ, might be involved in increased IRE-binding activity of IRP, thus downregulating m-aconitase expression.
ACKNOWLEDGMENTS
This work was supported in part by National Multiple Sclerosis Society grant RG2203-B-6 and a generous gift from the Stearman family.
We thank Larry Dangott, Director of the Texas A&M University Protein Chemistry Laboratory, for performing the MS and sequencing analyses and Nancy Dawson for performing the confocal microscopy. We gratefully acknowledge Claire Kennedy and Richard Eisenstein for providing purified aconitase and antiaconitase antibodies. We also thank Qi Liu, Reed Johnson, and Elena Belyavskaya for help and encouragement.
REFERENCES
- 1.Beatch M D, Hobman T C. Rubella virus capsid associates with host cell protein p32 and localizes to mitochondria. J Virol. 2000;74:5569–5576. doi: 10.1128/jvi.74.12.5569-5576.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beinert H, Kennedy M C. Aconitase, a two-faced protein: enzyme and iron regulatory factor. FASEB J. 1993;7:1442–1449. doi: 10.1096/fasebj.7.15.8262329. [DOI] [PubMed] [Google Scholar]
- 3.Berndt P, Hobohm U, Langen H. Reliable automatic protein identification from matrix-assisted laser desorption/ionization mass spectrometric peptide fingerprints. Electrophoresis. 1999;20:3521–3526. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3521::AID-ELPS3521>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 4.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 5.Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- 6.Chen O S, Schalinske K L, Eisenstein R S. Dietary iron intake modulates the activity of iron regulatory proteins and the abundance of ferritin and mitochondrial aconitase in rat liver. J Nutr. 1997;127:238–248. doi: 10.1093/jn/127.2.238. [DOI] [PubMed] [Google Scholar]
- 7.Choudhary S, De B P, Banerjee A K. Specific phosphorylated forms of glyceraldehyde 3-phosphate dehydrogenase associate with human parainfluenza virus type 3 and inhibit viral transcription in vitro. J Virol. 2000;74:3634–3641. doi: 10.1128/jvi.74.8.3634-3641.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dahlseid J N, Lill R, Green J M, Xu X, Qiu Y, Pierce S K. PBP74, a new member of the mammalian 70-kDa heat shock protein family, is a mitochondrial protein. Mol Biol Cell. 1994;5:1265–1275. doi: 10.1091/mbc.5.11.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denison M R, Spaan W J, van der Meer Y, Gibson C A, Sims A C, Prentice E, Lu X T. The putative helicase of the coronavirus mouse hepatitis virus is processed from the replicase gene polyprotein and localizes in complexes that are active in viral RNA synthesis. J Virol. 1999;73:6862–6871. doi: 10.1128/jvi.73.8.6862-6871.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drapier J C, Hibbs J B., Jr Differentiation of murine macrophages to express nonspecific cytotoxicity for tumor cells results in l-arginine-dependent inhibition of mitochondrial iron-sulfur enzymes in the macrophage effector cells. J Immunol. 1988;140:2829–2838. [PubMed] [Google Scholar]
- 11.Drapier J C, Hibbs J B., Jr Murine cytotoxic activated macrophages inhibit aconitase in tumor cells: inhibition involves the iron-sulfur prosthetic group and is reversible. J Clin Investig. 1986;78:790–797. doi: 10.1172/JCI112642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenstein R S, Kennedy M C, Beinert H. The iron responsive element (IRE), the iron regulatory protein (IRP), and cytosolic aconitase: posttranscriptional regulation of mammalian iron metabolism. In: Silver S, Walden W, editors. Metal ions in gene regulation. New York, N.Y: Chapman and Hill, Inc.; 1998. pp. 157–216. [Google Scholar]
- 13.Garcia-Blanco M A, Jamison S F, Sharp P A. Identification and purification of a 62,000-dalton protein that binds specifically to the polypyrimidine tract of introns. Genes Dev. 1989;3:1874–1886. doi: 10.1101/gad.3.12a.1874. [DOI] [PubMed] [Google Scholar]
- 14.Gardner P R, Nguyen D D, White C W. Aconitase is a sensitive and critical target of oxygen poisoning in cultured mammalian cells and in rat lungs. Proc Natl Acad Sci USA. 1994;91:12248–12252. doi: 10.1073/pnas.91.25.12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gardner P R, Raineri I, Epstein L B, White C W. Superoxide radical and iron modulate aconitase activity in mammalian cells. J Biol Chem. 1995;270:13399–13405. doi: 10.1074/jbc.270.22.13399. [DOI] [PubMed] [Google Scholar]
- 16.Gray N K, Pantopoulous K, Dandekar T, Ackrell B A, Hentze M W. Translational regulation of mammalian and Drosophila citric acid cycle enzymes via iron-responsive elements. Proc Natl Acad Sci USA. 1996;93:4925–4930. doi: 10.1073/pnas.93.10.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hentze M W, Kuhn L C. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc Natl Acad Sci USA. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirling H, Henderson B R, Kuhn L C. Mutational analysis of the [4Fe-4S]-cluster converting iron regulatory factor from its RNA-binding form to cytoplasmic aconitase. EMBO J. 1994;13:453–461. doi: 10.1002/j.1460-2075.1994.tb06280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennedy M C, Mende-Mueller L, Blondin G A, Beinert H. Purification and characterization of cytosolic aconitase from beef liver and its relationship to the iron-responsive element binding protein Proc. Natl Acad Sci USA. 1992;89:11730–11734. doi: 10.1073/pnas.89.24.11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim H Y, LaVaute T, Iwai K, Klausner R D, Rouault T A. Identification of a conserved and functional iron-responsive element in the 5′-untranslated region of mammalian mitochondrial aconitase. J Biol Chem. 1996;271:24226–24230. doi: 10.1074/jbc.271.39.24226. [DOI] [PubMed] [Google Scholar]
- 21.Kim Y, Makino S. Characterization of a murine coronavirus defective interfering RNA internal cis-acting replication signal. J Virol. 1995;69:4963–4971. doi: 10.1128/jvi.69.8.4963-4971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y-N, Jeong Y S, Makino S. Analysis of cis-acting sequences essential for coronavirus defective interfering RNA replication. Virology. 1993;197:53–63. doi: 10.1006/viro.1993.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klausner R D, Rouault T A, Harford J B. Regulating the fate of mRNA: the control of cellular iron metabolism. Cell. 1993;72:19–28. doi: 10.1016/0092-8674(93)90046-s. [DOI] [PubMed] [Google Scholar]
- 24.Kuhn L C, Hentze M W. Coordination of cellular iron metabolism by post-transcriptional gene regulation. J Inorg Biochem. 1992;47:183–195. doi: 10.1016/0162-0134(92)84064-t. [DOI] [PubMed] [Google Scholar]
- 25.Lai M M C. Coronavirus: organization, replication and expression of genome. Annu Rev Microbiol. 1990;44:303–333. doi: 10.1146/annurev.mi.44.100190.001511. [DOI] [PubMed] [Google Scholar]
- 26.Lai M M C, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai M M C, Patton C D, Stohlman S A. Further characterization of mRNAs of mouse hepatitis virus: presence of comon 5′-end nucleotides. J Virol. 1982;41:557–565. doi: 10.1128/jvi.41.2.557-565.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai M M C, Stohlman S A. The RNA of mouse hepatitis virus. J Virol. 1978;26:236–242. doi: 10.1128/jvi.26.2.236-242.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee H J, Shieh C K, Gorbalenya A E, Koonin E V, La Monica N, Tuler J, Bagdzhadzhyan A, Lai M M C. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J Y, Bowden D S, Marshall J A. Membrane junctions associated with rubella virus infected cells. J Submicrosc Cytol Pathol. 1996;28:101–108. [PubMed] [Google Scholar]
- 31.Leibowitz J L, DeVries J R. Synthesis of virus-specific RNA in permeabilized murine coronavirus-infected cells. Virology. 1988;166:66–75. doi: 10.1016/0042-6822(88)90147-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leibowitz J L, DeVries J R, Rodriguez M. Increased hepatotropism of mutants of MHV, strain JHM, selected with monoclonal antibodies. Adv Exp Med Biol. 1987;218:321–331. doi: 10.1007/978-1-4684-1280-2_41. [DOI] [PubMed] [Google Scholar]
- 33.Leibowitz J L, Wilhelmsen K C, Bond C W. The virus-specific intracellular RNA species of two murine coronaviruses: MHV-A59 and MHV-JHM. Virology. 1981;114:39–51. doi: 10.1016/0042-6822(81)90250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin Y, Liao C, Lai M M C. Identification of the cis-acting signal for minus-strand RNA synthesis of a murine coronavirus: implications for the role of minus-strand RNA in RNA replication and transcription. J Virol. 1994;68:8131–8140. doi: 10.1128/jvi.68.12.8131-8140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin Y J, Zhang X, Wu R C, Lai M M C. The 3′ untranslated region of coronavirus RNA is required for subgenomic mRNA transcription from a defective interfering RNA. Virol. 1996;70:7236–7240. doi: 10.1128/jvi.70.10.7236-7240.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin Y-J, Lai M M C. Deletion mapping of a mouse hepatitis virus defective interfering RNA reveals the requirement of an internal and discontinous sequence for replication. J Virol. 1993;67:6110–6118. doi: 10.1128/jvi.67.10.6110-6118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liochev S L. The role of iron-sulfur clusters in in vivo hydroxyl radical production. Free Radical Res. 1996;25:369–384. doi: 10.3109/10715769609149059. [DOI] [PubMed] [Google Scholar]
- 38.Liu Q, Yu W, Leibowitz J L. A specific host cellular protein binding element near the 3′ end of mouse hepatitis virus genomic RNA. Virology. 1997;232:74–85. doi: 10.1006/viro.1997.8553. [DOI] [PubMed] [Google Scholar]
- 39.MacMicking J, Xie Q W, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 40.Maeda A, An S, Makino S. Importance of coronavirus negative-strand genomic RNA synthesis prior to subgenomic RNA transcription. Virus Res. 1998;57:35–42. doi: 10.1016/S0168-1702(98)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mizzen L A, Kabiling A N, Welch W J. The two mammalian mitochondrial stress proteins, grp 75 and hsp 58, transiently interact with newly synthesized mitochondrial proteins. Cell Regul. 1991;2:165–179. doi: 10.1091/mbc.2.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohandas D V, Dales S. Endosomal association of a protein phosphatase with high dephosphorylating activity against a coronavirus nucleocapsid protein. FEBS Lett. 1991;282:419–24. doi: 10.1016/0014-5793(91)80528-B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nesterenko M V, Tilley M, Upton S J. A simple modification of Blum's silver stain method allows for 30 minute detection of proteins in polyacrylamide gels. J Biochem Biophys Methods. 1994;28:239–242. doi: 10.1016/0165-022x(94)90020-5. [DOI] [PubMed] [Google Scholar]
- 44.Pachuk C J, Breedenbeek P J, Zoltick P W, Spaan W J M, Weiss S R. Molecular cloning of the gene encoding the putative polymerase of mouse hepatitis coronavirus strain A59. Virology. 1989;171:141–148. doi: 10.1016/0042-6822(89)90520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park Y W, Katze M G. Translational control by influenza virus. Identification of cis-acting sequences and trans-acting factors which may regulate selective viral mRNA translation. J Biol Chem. 1995;270:28433–28439. doi: 10.1074/jbc.270.47.28433. [DOI] [PubMed] [Google Scholar]
- 46.Parker M M, Masters P S. Sequence comparison of the N genes of five strains of the coronavirus mouse hepatitis virus suggests a three domain structure for the nucleocapsid protein. Virology. 1990;179:463–468. doi: 10.1016/0042-6822(90)90316-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petrik J, Parker H, Alexander G J. Human hepatic glyceraldehyde-3-phosphate dehydrogenase binds to the poly(U) tract of the 3′ non-coding region of hepatitis C virus genomic RNA. J Gen Virol. 1999;80:3109–3113. doi: 10.1099/0022-1317-80-12-3109. [DOI] [PubMed] [Google Scholar]
- 48.Philpott C C, Haile D, Rouault T A, Klausner R D. Modification of a free Fe-S cluster cysteine residue in the active iron-responsive element-binding protein prevents RNA binding. J Biol Chem. 1993;268:17655–17658. [PubMed] [Google Scholar]
- 49.Pope M, Marsden P A, Cole E, Sloan S, Fung L S, Ning Q, Ding J W, Leibowitz J L, Phillips M J, Levy G A. Resistance to murine hepatitis virus strain 3 is dependent on production of nitric oxide. J Virol. 1998;72:7084–7090. doi: 10.1128/jvi.72.9.7084-7090.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robbins A H, Stout C D. Structure of activated aconitase: formation of the [4Fe-4S] cluster in the crystal. Proc Natl Acad Sci USA. 1989;86:3639–3643. doi: 10.1073/pnas.86.10.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rokeach L A, Haselby J A, Hoch S O. Molecular cloning of a cDNA encoding the human Sm-D autoantigen. Proc Natl Acad Sci USA. 1988;85:4832–4836. doi: 10.1073/pnas.85.13.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rouault T A, Stout C D, Kaptain S, Harford J B, Klausner R D. Structural relationship between an iron-regulated RNA-binding protein (IRE-BP) and aconitase: functional implications. Cell. 1991;64:881–883. doi: 10.1016/0092-8674(91)90312-m. [DOI] [PubMed] [Google Scholar]
- 53.Sagesser R, Martinez E, Tsagris M, Tabler M. Detection and isolation of RNA-binding proteins by RNA-ligand screening of a cDNA expression library. Nucleic Acids Res. 1997;25:3816–3822. doi: 10.1093/nar/25.19.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sawicki S G, Sawicki D L. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J Virol. 1990;64:1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schalinske K L, Chen O S, Eisenstein R S. Iron differentially stimulates translation of mitochondrial aconitase and ferritin mRNAs in mammalian cells. Implications for iron regulatory proteins as regulators of mitochondrial citrate utilization. J Biol Chem. 1998;273:3740–3746. doi: 10.1074/jbc.273.6.3740. [DOI] [PubMed] [Google Scholar]
- 56.Sims A C, Ostermann J, Denison M R. Mouse hepatitis virus replicase proteins associate with two distinct populations of intracellular membranes. J Virol. 2000;74:5647–5654. doi: 10.1128/jvi.74.12.5647-5654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh B, Soltys B J, Wu Z C, Patel H V, Freeman K B, Gupta R S. Cloning and some novel characteristics of mitochondrial Hsp70 from Chinese hamster cells. Exp Cell Res. 1997;234:205–216. doi: 10.1006/excr.1997.3609. [DOI] [PubMed] [Google Scholar]
- 58.Singh N K, Atreya C D, Nakhasi H L. Identification of calreticulin as a rubella virus RNA binding protein. Proc Natl Acad Sci USA. 1994;91:12770–12774. doi: 10.1073/pnas.91.26.12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soltys B J, Gupta R S. Mitochondrial-matrix proteins at unexpected locations: are they exported? Trends Biochem Sci. 1999;24:174–177. doi: 10.1016/s0968-0004(99)01390-0. [DOI] [PubMed] [Google Scholar]
- 60.Spaan W, Delius H, Skinner M, Armstrong J, Rottier P, Smeekens S, van der Zeijst B A, Siddell S G. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2:1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spaan W J M, Cavanagh D, Horzinek M C. Coronaviruses: structure and genome expression. J Gen Virol. 1988;69:2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- 62.Spaan W J M, Rottier P J M, Horzinek M C, van der Zeijst B A M. Sequence relationships between the genome and the intracellular RNA species 1, 3, 6, and 7 of mouse hepatitis virus strain A59. J Virol. 1982;42:432–439. doi: 10.1128/jvi.42.2.432-439.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stripecke R, Oliveira C C, McCarthy J E, Hentze M W. Proteins binding to 5′ untranslated region sites: a general mechanism for translational regulation of mRNAs in human and yeast cells. Mol Cell Biol. 1994;14:5898–5909. doi: 10.1128/mcb.14.9.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Theil E C, McKenzie R A, Sierzputowska-Gracz H. Structure and function of IREs, the noncoding mRNA sequences regulating synthesis of ferritin, transferrin receptor and (erythroid) 5-aminolevulinate synthase. Adv Exp Med Biol. 1994;356:111–118. doi: 10.1007/978-1-4615-2554-7_12. [DOI] [PubMed] [Google Scholar]
- 65.Van der Meer Y, Snijder E J, Dobbe J C, Schleich S, Denison M R, Spaan W J M, Locker J K. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J Virol. 1999;73:7641–7657. doi: 10.1128/jvi.73.9.7641-7657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vasquez-Vivar J, Kalyanaraman B, Kennedy M C. Mitochondrial aconitase is a source of hydroxyl radical: an electron spin resonance investigation. J Biol Chem. 2000;275:14064–14069. doi: 10.1074/jbc.275.19.14064. [DOI] [PubMed] [Google Scholar]
- 67.Yu W, Leibowitz J L. A conserved motif at the 3′ end of mouse hepatitis virus genomic RNA required for host protein binding and viral RNA replication. Virology. 1995;214:128–138. doi: 10.1006/viro.1995.9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu W, Leibowitz J L. Specific binding of host cellular proteins to multiple sites within the 3′ end of mouse hepatitis virus genomic RNA. J Virol. 1995;69:5033–5038. doi: 10.1128/jvi.69.4.2016-2023.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng L, Kennedy M C, Blondin G A, Beinert H, Zalkin H. Binding of cytosolic aconitase to the iron responsive element of porcine mitochondrial aconitase mRNA. Arch Biochem Biophys. 1992;229:356–360. doi: 10.1016/0003-9861(92)90287-7. [DOI] [PubMed] [Google Scholar]