Abstract
Previous work has demonstrated that the V protein of simian virus 5 (SV5) targets STAT1 for proteasome-mediated degradation (thereby blocking interferon [IFN] signaling) in human but not in murine cells. In murine BF cells, SV5 establishes a low-grade persistent infection in which the virus fluxes between active and repressed states in response to local production of IFN. Upon passage of persistently infected BF cells, virus mutants were selected that were better able to replicate in murine cells than the parental W3 strain of SV5 (wild type [wt]). Viruses with mutations in the Pk region of the N-terminal domain of the V protein came to predominate the population of viruses carried in the persistently infected cell cultures. One of these mutant viruses, termed SV5 mci-2, was isolated. Sequence analysis of the V/P gene of SV5 mci-2 revealed two nucleotide differences compared to wt SV5, only one of which resulted in an amino acid substitution (asparagine [N], residue 100, to aspartic acid [D]) in V. Unlike the protein of wt SV5, the V protein of SV5 mci-2 blocked IFN signaling in murine cells. Since the SV5 mci-2 virus had additional mutations in genes other than the V/P gene, a recombinant virus (termed rSV5-V/P N100D) was constructed that contained this substitution alone within the wt SV5 backbone to evaluate what effect the asparagine-to-aspartic-acid substitution in V had on the virus phenotype. In contrast to wt SV5, rSV5-V/P N100D blocked IFN signaling in murine cells. Furthermore, rSV5-V/P N100D virus protein synthesis in BF cells continued for significantly longer periods than that for wt SV5. However, even in cells infected with rSV5-V/P N100D, there was a late, but significant, inhibition in virus protein synthesis. Nevertheless, there was an increase in virus yield from BF cells infected with rSV5-V/P N100D compared to wt SV5, demonstrating a clear selective advantage to SV5 in being able to block IFN signaling in these cells.
Alpha/beta and gamma interferons (IFN-α/β and IFN-γ) mediate their biological activities, including the induction of an antiviral state within cells, by inducing the expression of IFN-responsive genes. To survive in nature, it seems likely that all viruses must have some means of circumventing the IFN response (for a review on IFNs, cell signaling, immune modulation, antiviral responses, and virus countermeasures, see references 9 and 33). Many Paramyxoviridae at least partially circumvent the IFN response by inhibiting IFN-induced signal transduction pathways (2, 8, 16, 38), although they achieve this by distinct mechanisms (40). Thus, for example, we have shown that simian virus 5 (SV5) blocks IFN signaling by targeting STAT1, an essential component of IFN-α/β and IFN-γ signaling cascades, for proteasome-mediated degradation (3). However, the ability of SV5 to block IFN signaling is species dependent. Thus, wild-type (wt) SV5 blocks IFN signaling in human, monkey, and canine cells but fails to block IFN signaling in murine cells. Indeed, the ability to block IFN signaling in a particular species may be one factor that limits the host range of a given virus (2).
SV5 is a prototype member of the Rubulavirus genus within the Paramyxoviridae family (19). Like other paramyxoviruses, SV5 has a nonsegmented, negative-sense, single-stranded RNA genome. Its genome is encapsidated within a helical nucleocapsid which is surrounded by a pleomorphic, lipid-containing envelope containing the HN and F virus glycoproteins. (For a review of the Paramyxoviridae, see reference 17.) Over the last few years, techniques have been developed to manipulate the genomes of many Paramyxoviridae, including SV5 (11, 12, 24, 27). Thus, it is now possible to manipulate the genome of SV5 and to correlate genotypes with phenotypes. We report here that a single amino acid substitution in the V protein confers on SV5 the ability to block IFN signaling in murine cells, and we demonstrate protracted virus protein synthesis in murine cells infected with a recombinant virus which blocks IFN signaling in these cells compared to cells infected with wt virus.
MATERIALS AND METHODS
Cells, viruses, and IFN.
Murine BF cells (cloned from a primary cell culture of a BALB/c mouse embryo) and human 2fTGH cells (22) were grown as monolayers in 25- or 75-cm2 tissue culture flasks in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (growth medium). Monolayer cultures of A549, BHK-21, MDBK, and CV-1 cells were maintained in DMEM and 10% fetal calf serum as described previously (26). Chick embryo fibroblasts (CEFs) were grown as described previously (18). All cell lines were negative for mycoplasmas as screened by DAPI (4′,6′-diamidino-2-phenylindole) staining. Human and mouse cells were treated with recombinant human αA/D (rHuIFN-αA/D) IFN (31) at 1,000 IU/ml in medium containing 2% bovine serum (maintenance medium). The wt (strain W3A) (1), rSV5-V/P N100D, and SV5 mci-2 strains of SV5 were grown and titrated under appropriate conditions in Vero cells using maintenance medium. High-multiplicity infections of BF cells were carried out at 50 to 100 PFU/cell and of human cells at 5 to 10 PFU/cell. Modified vaccinia virus Ankara (MVA) expressing bacteriophage T7 RNAP (34, 37) was grown in CEFs and was kindly provided by Bernard Moss, National Institutes of Health, Bethesda, Md. Plaque assays were performed in BHK-21 or CV-1 cells as described earlier (25).
Generation of a rSV5 containing a mutation of N to D at residue 100 of the V and P proteins.
The N-to-D mutation in the V and P proteins at residue 100 was introduced into the SV5 V/P gene cDNA (36) by four-primer PCR. The resulting PCR DNA product was digested with the restriction enzymes ClaI and StuI and cloned into the SV5 infectious cDNA clone, pSV5 (11) by four successive cloning steps. Details of the oligonucleotide primers used and the cloning steps are available on request. The nucleotide sequence of the region amplified by PCR was determined and shown to contain only the desired mutation.
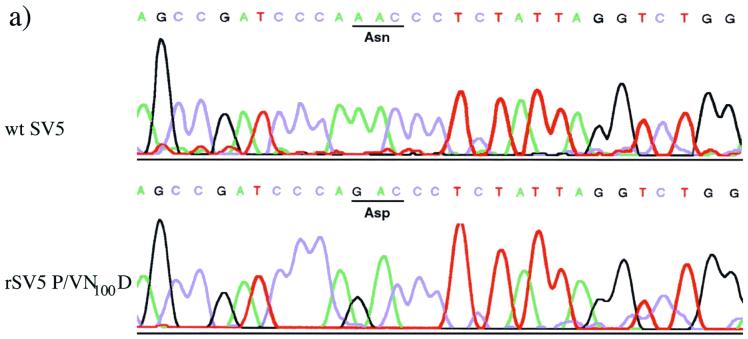
The rSV5-V/P N100D was obtained essentially as described previously (11, 12). Briefly, A549 cells at 95% confluency in a 3.5-cm-diameter plate were infected with MVA at a multiplicity of infection (MOI) of 3 in phosphate-buffered saline (PBS) containing 1% bovine serum albumin for 1 h and then the plasmids pSV5 and those encoding NP, P, and L (11) were transfected into cells with Lipofectin (Life Technologies Town Co.). The amounts of plasmids were as follows: 1 μg of pSV5, 2 μg of pUC19 NP3A, 0.2 μg of pGEM 2-P, and 2 μg of pGEM 3-L. After incubation for 24 h the transfection medium was changed to DMEM containing 10% fetal calf serum. After 48 h at 37°C, the media were harvested, the cell debris was pelleted by low-speed centrifugation (1,250 × g, 5 min), and the supernatant was filtered through a 0.45-μm (pore-size) filter to remove the MVA. This virus stock was used to generate rSV5-V/P N100D stocks by infection of MDBK cells. The virus was plaque purified on BHK cells, and the nucleotide sequence of the V/P gene of rSV5-V/P N100D was confirmed by reverse transcription (RT)-PCR by using an ABI 310 sequence analyzer (see Fig. 4A).
FIG. 4.
(a) The nucleotide sequence of the V/P gene of wt SV5 and rSV5-V/P N100D (N > D) as confirmed by RT-PCR. (b) Comparison of the ability of wt SV5 and rSV5-V/P N100D to block IFN signaling in human and murine cells. Human 2fTGH or murine BF cells were transfected with 0.1 μg of pJATlacZ, 0.1 μg of the IFN-α/β-responsive plasmid, and 0.3 μg of pEFlink2 (control plasmid). At 24 h posttransfection the cells were infected with either wt SV5 or rSV5-V/P N100D. At 44 h posttransfection, the culture medium was supplemented with IFN (+IFN) or left untreated (−IFN). After 4 h, the luciferase and β-galactosidase activities in cellular lysates were measured. The luciferase activity, expressed in relative light units, was normalized to the β-galactosidase activity. (c) Immunoblot analysis showing the relative levels of STAT1 in total cell extracts made from BF cells that had been mock infected or infected with wt or rSV5-V/P N100D Cells were harvested at 24, 48, or 72 h p.i. as indicated in subpanels a to c. In addition, extracts were made from cells that were harvested at 72 h p.i. but to which exogenous IFN-α/β was added at 24 h p.i. (subpanel d). The lower band (arrow) is an unidentified host cell protein.
Immunofluorescence.
Cells to be stained for immunofluorescence were grown on multispot microscope slides (C. A. Hendley Ltd., Essex, United Kingdom) or 10-mm-diameter coverslips (General Scientific Co., Ltd.). The cells were treated and stained with specific monoclonal antibodies (MAbs) as described previously (29). Antibody binding was detected by indirect immunofluorescence using a secondary goat anti-mouse immunoglobulin Texas red-conjugated antibody (Seralab, catalog number SBA 1010-02). The primary antibodies were SV5-P-e, SV5-P-k, and SV5-NP-a (30). After staining for immunofluorescence, the monolayers of cells were examined using a Nikon Microphot-FXA immunofluorescence microscope.
Preparation of radiolabeled antigen extracts, immunoprecipitation, and SDS-PAGE.
BF cell monolayers in 25-cm2 tissue culture flasks were infected with 50 PFU of SV5 per cell. After an adsorption period of 1 h at 37°C, the inoculum was removed and replaced with maintenance medium. At various times postinfection (p.i.) the cells were metabolically labeled with l-[35S]methionine (500 Ci/mmol; Amersham International, Ltd.) in tissue culture medium containing 1/10 the normal concentration of methionine (i.e., 1.5 mg/liter). At the end of the labeling interval, the cells were washed in ice-cold PBS and lysed into immune precipitation buffer (10 mM Tris-HCl, pH 7.8; 5 mM EDTA; 0.5% Nonidet P-40; 0.65 M NaCl; 0.1% sodium dodecyl sulfate [SDS]; with 4 × 106 to 6 × 106 cells per ml of buffer) by sonication with an ultrasonic probe. Soluble antigen extracts were obtained after pelleting the particulate material from the total cell antigen extracts by centrifugation at 400,000 × g for 30 min. Immune complexes were formed by incubating (for 2 h at 4°C) 1-ml samples of the soluble antigen extracts with an excess of anti-SV5 MAbs to the HN, F, P, M, and NP proteins (1 μl of concentrated tissue culture fluid of MAbs SV5-HN-4a, NP-a, P-e, M-h, and F-1a [30]). The immune complexes were isolated (13) using an excess of a fixed suspension of the Cowan A strain of Staphylococcus aureus (20 μl of a 10% [wt/vol] suspension per μl of concentrated tissue culture fluid for 30 min at 4°C). The proteins in the immune complexes were dissociated by heating (100°C for 5 min) in polyacrylamide gel electrophoresis (PAGE) sample buffer (0.05 M Tris-HCl, pH 7.0; 0.2% SDS; 5% 2-mercaptoethanol; 5% glycerol) and analyzed by SDS-PAGE using an 11% gel. After electrophoresis the gels were fixed, stained, and dried. Labeled polypeptides were then visualized by phosphorimager analysis.
Immunoblotting.
At the time of harvest, cells were washed twice with PBS, disrupted into SDS-gel loading buffer, sonicated, and boiled for 5 min. Polypeptides were separated by SDS-PAGE using a 7% gel and transferred to nitrocellulose membranes, and STAT1 was detected with a polyclonal anti-STAT1 antibody raised against the N-terminal 194 amino acids (Transduction Laboratories catalog number G16930). All protein-antibody interactions were detected by enhanced chemiluminescence using horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin G (Amersham International, Ltd.).
Plasmid DNAs and transfections.
The IFN-α/β-responsive plasmid [termed p(9–27)4tkΔ(−39)lucter] contains four tandem repeat sequences of the ISRE from the IFN-inducible gene, 9–27, fused to the firefly luciferase gene (15). pJATlacZ, a plasmid used as a transfection standard, contains a β-galactosidase gene under the control of the rat β-actin promoter (23). The construction of the plasmid pEF.SV5-V/wt has been reported elsewhere (3). pEF.SV5-V/SV5 mci-2 was constructed by first generating a PCR copy of the V gene of the SV5 mci-2 virus. The gene was then subcloned between the NcoI and XhoI sites of the EF1a promoter vector, pEFlink2 (a kind gift of R. H. Treisman, Imperial Cancer Research Fund).
Monolayers of BF cells or 2fTGH cells grown in 24-well plates to 50 to 70% confluence were transfected with 0.5 μg of DNA and 2 μl of Lipofectamine (Life Technologies, Inc.) according to the manufacturer's instructions. At various times posttransfection, cells were or were not induced with 1,000 IU of rHuIFN-αA/D per ml for 4 h immediately prior to harvesting. Luciferase and β-galactosidase activities were measured as described previously (14). The relative expression levels were calculated by dividing the luciferase values by the β-galactosidase values.
RESULTS
Passage of persistently infected BF cells and selection of mutant viruses.
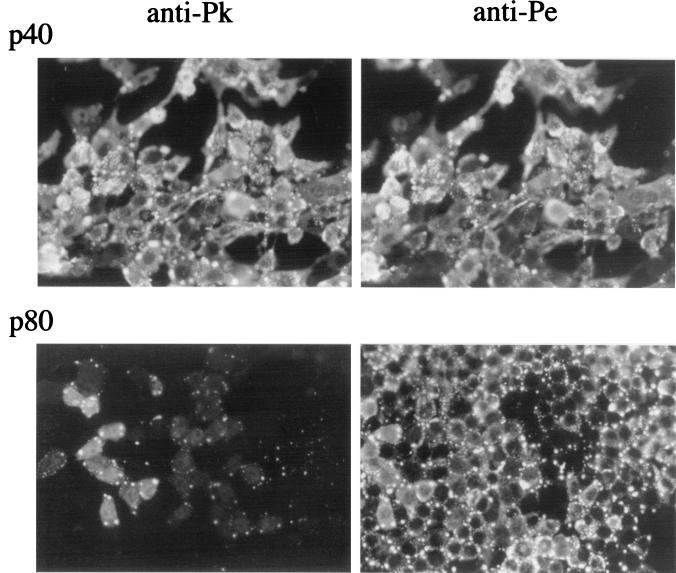
We have shown previously that SV5 infects murine cells but fails to block IFN signaling. Thus, early in infection normal patterns of virus protein synthesis are observed. However, once the cells begin to produce and respond to IFN, there is a rapid reduction in virus protein synthesis (2). Despite this, SV5 establishes persistent infections in murine cells (5, 39). Following high-MOI infections of murine BF cells, the majority of cells, although they initially synthesize high levels of virus proteins, survive the infection. Furthermore, by 8 to 15 days p.i. most, but not all, cells initially infected clear the infection. Upon continued passage of these cells, virus variants were selected that are better able to replicate in murine cells (39). The first mutant virus isolated (which was originally termed W3-f but has been renamed SV5 mci-1, for mouse cell isolate 1), although remaining sensitive to IFN, was highly fusogenic and could spread more rapidly in BF cells than wt SV5 when IFN levels dropped, for example, during cell passage. (The property of this virus and the early passage history of the persistently infected BF cells have been reported elsewhere [39].) On further passage of the persistently infected cells, other mutant viruses came to dominate the pool of viruses being carried. As shown in Fig. 1, the majority of passage 40 (p40) cells reacted with both the anti-Pk and anti-Pe MAbs, whereas by passage 80 (p80) the majority of infected cells within the culture no longer reacted with the anti-Pk MAb, indicating the accumulation of mutations that result in a loss of the epitope for the Pk MAb.
FIG. 1.
Immunofluorescence analysis of BF cells persistently infected with wt SV5 that had been passaged 40 (p40) or 80 (p80) times. The cells were stained with either the anti-Pk or the anti-Pe MAbs, which recognize epitopes in either the N-terminus P/V common domain of the P and V proteins (anti-Pk) or the P-unique C-terminal domain (anti-Pe).
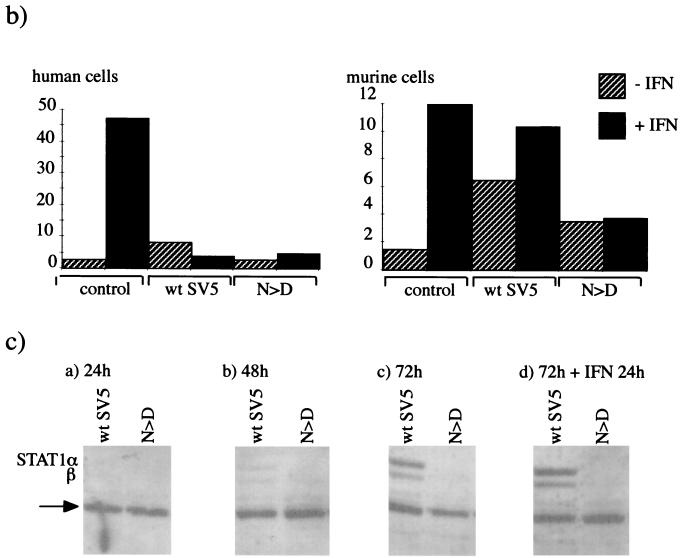
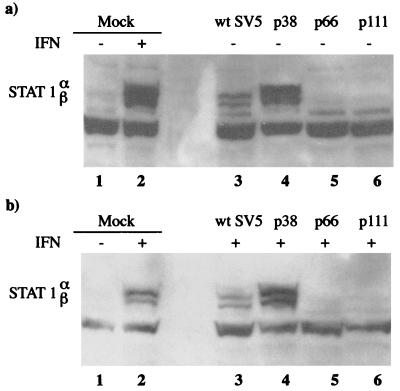
Since we had shown previously that SV5 failed to block IFN signaling in murine cells (2), it was of interest to determine if viruses selected upon prolonged passage of the persistently infected cells had acquired the ability to block IFN signaling. To determine whether this was the case, the levels of STAT1 in the persistently infected cultures were examined. In uninfected BF cells the levels of STAT1 were below the levels of detection using immunoblot analysis as described above. However, the levels of STAT1 could be increased to readily detectable levels by treating the cells with IFN (i.e., the induction of STAT1 can be used as a marker for IFN signaling; Fig. 2, compare lanes 1 and 2). Examination of STAT1 levels in the persistently infected cells showed that during early passages of the persistently infected cells STAT1 could readily be detected irrespective of the presence or absence of exogenous IFN in the culture medium (STAT1 can be detected in the absence of exogenous IFN because infected cells produce and respond to IFN). However, by passage 66 (a point at which viruses with mutations in the Pk epitope dominated the population of virus genomes carried by the cells) STAT1 could not be detected either in the presence or absence of exogenous IFN, suggesting that IFN signaling was being blocked in these cells.
FIG. 2.
Immunoblot analysis showing the relative levels of STAT1 in mock-infected, wt SV5-infected (48 h p.i.), or persistently infected BF cells that had been passaged 38 (p38), 66 (p66), or 111 (p111) times. Cells were (+; panel b) or were not (−; panel a) treated with IFN-α/β for 24 h prior to harvest.
A single point mutation in the V gene open reading frame (ORF) confers the ability to block IFN signaling in murine cells.
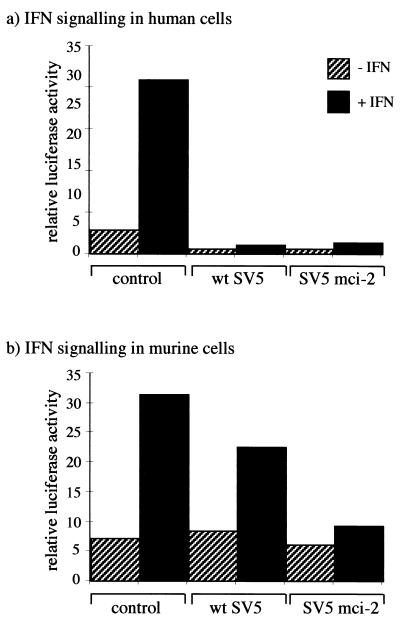
The anti-Pk MAb binds to a short epitope of contiguous amino acid residues within the N-terminal common domain of the V and P proteins (4, 32). Since the V protein targets STAT1 for proteasome-mediated degradation in human cells, it appeared likely that mutation(s) in the V/P gene had been selected which conferred on SV5 the ability to block IFN signaling in murine cells. To determine whether this was the case, a mutant virus was isolated from p80 cells which was no longer recognized by the anti-Pk MAb; this virus is referred to here as SV5 mci-2. The V/P gene from SV5 mci-2 was cloned and inserted into a eukaryotic expression vector such that the V protein could be expressed. The vector was cotransfected into human and murine cells, together with an IFN-α/β-responsive reporter plasmid, and the activation of the reporter (luciferase) gene in response to the addition of IFN was measured. Whereas the V proteins from both wt SV5 and SV5 mci-2 blocked IFN signaling in human cells, only the SV5 mci-2 V protein efficiently blocked IFN signaling in murine cells (Fig. 3). Nevertheless, in this assay the wt SV5 V protein reproducibly inhibited approximately 30% of the activity of IFN-responsive promoter, suggesting that it may be weakly active in murine cells. Furthermore, very low levels of IFN signaling were reproducibly observed in BF cells transfected with the SV5 mci-2V gene (Fig. 3b).
FIG. 3.
Comparison of the ability of the V proteins from wt SV5 and SV5 mci-2 to block IFN signaling in human and murine cells. Human 2fTGH (a) or murine BF (b) cells were transfected with 0.1 μg of pJATlacZ, 0.1 μg of the IFN-α/β-responsive plasmid, and either 0.3 μg of pEFlink2 (control plasmid), pEF.SV5-V/wt (that encodes the V protein of wt SV5), or pEF.SV5-V/SV5 mci-2 (that encodes the V protein of SV5 mci-2). At 40 h posttransfection, the culture medium was supplemented with IFN (+IFN) or left untreated (−IFN). After 4 h, the luciferase and β-galactosidase activities in cellular lysates were measured. The luciferase activity, expressed in relative light units, was normalized to the β-galactosidase activity.
Nucleotide sequence analysis of the V/P gene isolated from the SV5 mci-2 virus revealed a single A-to-G nucleotide change in the V gene ORF at genome position 2147 (GenBank AF052755). This resulted in an asparagine (N)-to-aspartic-acid (D) substitution at amino acid position 100 (N100D) within the N-terminal V/P common domain. There was an additional A-to-G nucleotide change within the P ORF (at genome position 2587), but outside the V ORF, which resulted in a glutamine-to-arginine substitution at amino acid position 247 in the P-unique C-terminal domain. Sequence analysis of other genes, including HN, isolated from the SV5 mci-2 virus revealed additional mutations which also resulted in amino acid substitutions (data not shown). Thus, to determine what effect the asparagine-to-aspartic-acid substitution had on the biological properties of wt SV5, the A-to-G mutation at position 2147 was introduced into an infectious cDNA clone of SV5 (11). A recombinant virus, rSV5-V/P N100D, was recovered that showed no significant difference in infectious particle yield from wt SV5 in infected Vero cells. The nucleotide sequence of the rSV5-V/P N100D and wt SV5 genome in the region covering the mutation confirmed the presence of the mutation in the recovered virus (Fig. 4a).
The ability of rSV5-V/P N100D to block IFN signaling in human and murine cells was compared to that of wt SV5. Human (2fTGH) and murine (BF) cells were transfected with the IFN-α/β-responsive reporter plasmid and at 24 h posttransfection were infected with wt SV5 and rSV5-V/P N100D. At 44 h posttransfection, exogenous IFN was or was not added to the culture media, and 4 h later the relative activation of the IFN-α/β reporter (luciferase) gene was estimated by measuring the luciferase activity in the cells. As shown in Fig. 4b, rSV5-V/P N100D blocked IFN signaling in both human and murine cells, whereas wt SV5 blocked IFN signaling in human but not in murine cells. The ability of rSV5-V/P N100D to block IFN signaling in murine cells was confirmed by demonstrating that STAT1 levels did not increase in BF cells infected with rSV5-V/P N100D, even upon the addition of exogenous IFN to the culture medium (Fig. 4c).
Prolonged virus protein synthesis in murine cells infected with a recombinant virus capable of blocking IFN signaling.
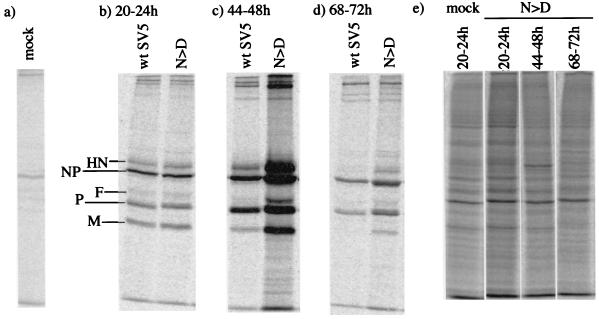
To determine what effect the ability to block IFN signaling had on virus protein synthesis in murine cells, BF cells were infected at a high MOI with either wt SV5 or rSV5-V/P N100D. The cells were metabolically labeled with [35S]methionine at various times p.i. prior to immunoprecipitation of virus proteins with a panel of MAbs specific for the HN, NP, F, P, and M proteins. As shown in Fig. 5, similar levels of virus protein synthesis were found in cells infected with wt SV5 or rSV5-V/P N100D at between 20 and 24 h p.i. However, by 44 to 48 h p.i. the cells infected with rSV5-V/P N100D synthesized significantly more viral proteins than the cells infected with wt SV5, but by 68 to 72 h p.i., although cells infected with the rSV5-V/P N100D were still making larger amounts of viral proteins than cells infected with wt SV5, the relative levels of rSV5-V/P N100D virus protein synthesis were significantly less than at 44 to 48 h p.i. This was not due to a reduction in the overall levels of protein synthesis in these cells, since the relative levels of total cell protein synthesis was similar at all of the time points examined (Fig. 5d).
FIG. 5.
Analysis of 35S-labeled polypeptides present in immune precipitates formed by the reaction of a pool of MAbs specific for the HN, NP, F, M, and P/V proteins with soluble antigen extracts made from BF cells that had been mock infected (a) or infected with wt SV5 or rSV5-V/P N100D (N > D) (b to d). The cells were radioactively labeled from 20 to 24, 44 to 48, or 68 to 72 h prior to harvesting as indicated. (e) 35S-labeled polypeptides present in the total cell extracts from rSV5-V/P N100D-infected cells.
wt SV5 is cleared from BF cells more rapidly than rSV5-V/P N100D.
As described above, following infection with wt SV5 the majority of BF cells clear the infection by 14 days p.i. due to the cells producing and responding to IFN. However, 5 to 10% of the cells remain infected and, on passage, these cells give rise to persistently infected cultures. Given that the rSV5-V/P N100D virus blocked IFN signaling but that there was a reduction in virus protein synthesis at late times p.i., it was of interest to determine the percentage of cells that remained infected with rSV5-V/P N100D upon cell passage. Monolayers of BF cells were infected at high MOI with either wt SV5 or rSV5-V/P N100D. Despite the fact that rSV5-V/P N100D blocked IFN signaling, the majority of BF cells infected with rSV5-V/P N100D (and wt SV5) survived the infection and could be passaged. After three passages the cells were stained for fluorescence using anti-NP MAbs. As shown in Fig. 6, the vast majority of cells infected with wt SV5 had cleared the infection whereas, in contrast, all of the cells infected with the rSV5-V/P N100D remained infected.
FIG. 6.
BF cells were infected at a high MOI with wt SV5 or rSV5-V/P N100D and passaged three times over a period of 2 weeks. At 24 h p.i. (a) and after the third passage (b), cells growing on coverslips in the tissue culture dishes were fixed and stained for immunofluorescence using an anti-NP MAb (NP-a).
To determine whether the increase in the relative amounts of virus-specific protein synthesis in BF cells infected with rSV5-V/P N100D compared to the amount of SV5 virus-specific protein synthesis was reflected in virus production, the culture media were harvested at various times p.i., and the amount of infectious virus was quantified (Table 1). Although similar levels of virus were produced by cells infected with wt SV5 and rSV5-V/P N100D for up to 2 days p.i., thereafter cells infected with rSV5-V/P N100D produce more virus, indicating that the ability to block IFN signaling in murine cells has a clear selective advantage for SV5 in terms of its ability to replicate in BF cells.
TABLE 1.
Amount of infectious virus released from BF cells infected at a high MOI with wt SV5 or rSV5-V/P N100Da
| Passage and collection point (day) | Amt of virus (log10 PFU/ml) released from BF cells infected with:
|
|
|---|---|---|
| wt SV5 | rSV5-V/P N100D | |
| p0 | ||
| 1 | 5.7 | 5.9 |
| 2 | 5.3 | 5.7 |
| 3 | 5.2 | 6.7 |
| 4 | 5.0 | 6.5 |
| p1 (3) | 2.0 | 6.3 |
| p2 | ||
| 1 | 2.0 | 4.1 |
| 2 | 3.2 | 4.7 |
| 3 | 3.4 | 6.3 |
Samples of culture media were collected at 1, 2, 3, and 4 days p.i. On day 4 p.i., the cells were passaged (p1), and media were collected 3 days later (p1, day 3). The following day, the cells were again passaged (p2), and media were collected at 1, 2, and 3 days postpassage as indicated. Cells were infected in suspension, and approximately 2 × 106 cells in 10 ml were plated into 25-cm2 flasks; 106 cells in 10 ml of medium were passaged into 25-cm2 flasks. The amount of infectious virus in the medium samples was estimated by plaque assays on Vero cells.
DISCUSSION
The asparagine-to-aspartic-acid mutation at amino acid position 100 within the V protein conferred on SV5 the ability to block IFN signaling in murine cells while retaining its ability to block IFN signaling in human cells. Furthermore, rSV5-V/P N100D was clearly able to synthesize greater amounts of viral proteins and for longer periods than wt SV5 in murine BF cells. However, when making single amino acid substitutions in an RNA virus, there is always a danger that other unknown mutations may arise which will affect the virus phenotype. Whereas this cannot be completely ruled out without sequencing the entire genome of the rSV5-V/P N100D, during the reverse genetics procedure a recombinant “wild type” virus was recovered which did not have the N100D mutation. This virus behaved exactly the same as wt SV5 with regard to its sensitivity to IFN and the kinetics of protein synthesis in BF cells (data not shown). Taken together, it is thus reasonable to conclude that the prolonged virus protein synthesis observed in BF cells infected with the rSV5-V/P N100D is a direct consequence of its ability to block IFN signaling. Thus, a single point mutation within the V gene differentially affects the ability of SV5 to block IFN signaling in cells from different species. For Sendai virus the P/V gene encodes, in addition to P and V, another set of proteins termed the C proteins (C′, C, Y1, and Y2) and for Sendai virus it is the C proteins, rather than V, which block IFN signaling and the induction of an antiviral state by IFN (2, 7, 8, 10, 16). Furthermore, a single amino acid substitution (Phe 170 to Ser) in the Sendai virus C proteins adversely affects the ability of the proteins to block IFN signaling, and the presence of this mutation in Sendai virus correlates with an attenuated phenotype (6).
The reason for the late inhibition of rSV5-V/P N100D virus protein synthesis in BF cells remains unclear. It may be that a low level of IFN signaling occurs in BF cells infected with rSV5-V/P N100D that eventually induces an antiviral state within these cells. Consistent with this idea was the observation that there is never a complete block in IFN signaling in reporter assays using BF cells tranfected with the V gene isolated from SV5 mci-2. Furthermore, a low level of signaling also appeared to be occurring in BF cells infected with rSV5-V/P N100D (Fig. 4b). It is also possible that the accumulation of virus proteins at late times p.i. may lead to an inhibition of virus transcription. Alternatively, other cellular transcription factors may induce an antiviral state in cells in which IFN signaling has been inhibited. Indeed, given that blocking IFN signaling seems an obvious strategy for viruses, it seems likely that cells may have other compensatory strategies for inducing antiviral responses. This may be an important function of IRF-1, a cellular transcription factor which can bind to and activate many of the promoters normally activated by IFN-α/β (9).
The anti-Pk MAb binds to a sequence of eight amino acid residues within the N-terminus common domain of the V and P proteins (4, 32). The asparagine residue within this domain is critical for antibody binding (4). Substitution of asparagine with aspartic acid in the SV5 mci-2 virus was thus clearly responsible for the loss of binding of anti-Pk MAb to the SV5 mci-2 virus. The exact molecular details of how V blocks IFN signaling and targets STAT1 for proteasome-mediated degradation have yet to be determined, but it seems likely that the Pk epitope may be critical for the interaction of V with the host cell protein(s) involved in this process. However, given that the Pk epitope is critical for V function but is also present in P, it is also unclear why P fails to block IFN signaling (3). Presumably, since P (unlike V) is a tetramer (35) and has a unique C terminus, exposed epitopes on V will either be hidden or altered on P. It thus seems likely that although part of the specificity for targeting STAT1 for degradation resides in the N-terminal domain of V, the C terminus will also contribute to the functionality and/or specificity of this interaction. It has been reported that V interacts with the host cell UV DNA damage-binding protein (UV DDB) (20), although the role, if any, that this interaction has in V-induced inhibition of IFN signaling remains unclear. Indeed, the interaction of V with UV DDB may be more concerned with slowing the progression of the cell cycle in virus infected cells (21). V also binds soluble, but not polymeric, NP and may thus have a role in keeping NP soluble prior to encapsidation (28). Thus, V is clearly a multifunctional protein with a variety of roles in the life cycle of SV5, and further detailed structural and/or functional analysis of V needs to be undertaken to elucidate the relationship(s) of these functions.
Although SV5 mci-2 was able to block IFN signaling in murine cells, it has acquired additional mutations which adversely affected its ability to spread and replicate efficiently in BF cells (data not shown). The reasons for the selection of these additional mutations remains unclear, but there are clearly very different selection pressures on viruses in persistently infected cells than on viruses which are propagated by passage through uninfected cells. Nevertheless, it was surprising that it took multiple passages to select for viruses capable of blocking IFN signaling in murine cells. One possible reason for this may be that, due to constraints imposed by other functions of V and P, there are only a restricted number of possible mutations in the P/V gene that result in a block of IFN signaling in murine cells and are compatible with the generation of infectious virus. However, the ability to block IFN signaling is also clearly not the only factor which affects the efficiency of SV5 replication in BF cells. Thus, for example, the time at which BF cells first express SV5 proteins (as judged by immunofluorescence) is significantly later, even in the absence of IFN, than that observed in other cells, including human 2fTGH cells. Furthermore, there was a 10- to 100-fold reduction in the amount of infectious virus produced by BF cells infected with rSV5-V/P N100D compared to 2fTGH or Vero cells infected with either wt SV5 or rSV5-V/P N100D. These additional constraints on SV5 may partially explain why rSV5-V/P N100D remains nonpathogenic in normal mice (unpublished observations). However, by selecting other variants that are more pathogenic in mice and using reverse genetics to associate genotypes with phenotypes, a more detailed understanding of the molecular pathogenesis of paramyxoviruses may be gained.
ACKNOWLEDGMENTS
D. F. Young is supported by the BBSRC, and N. Chatziandreou is supported by a Ph.D. studentship from the Maitland Ramsay Trust. S. Goodbourn and R. E. Randall are also indebted to the Wellcome Trust for support. This research was also supported in part by Public Health Service Research Award AI-23173 from the National Institute of Allergy and Infectious Diseases. B. He is an Associate and R. A. Lamb is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Choppin P W. Multiplication of a myxovirus (SV5) with minimal cytopathic effects and without interference. Virology. 1964;23:224–233. doi: 10.1016/0042-6822(64)90286-7. [DOI] [PubMed] [Google Scholar]
- 2.Didcock L, Young D F, Goodbourn S, Randall R E. Sendai virus and simian virus 5 block activation of interferon-responsive genes: importance for virus pathogenesis. J Virol. 1999;73:3125–3133. doi: 10.1128/jvi.73.4.3125-3133.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Didcock L, Young D F, Goodbourn S, Randall R E. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J Virol. 1999;73:9928–9933. doi: 10.1128/jvi.73.12.9928-9933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunn C, O'Dowd A M, Randall R E. Fine mapping of the binding sites of monoclonal antibodies raised against the Pk tag. J Immunol Methods. 1999;224:141–150. doi: 10.1016/s0022-1759(99)00017-4. [DOI] [PubMed] [Google Scholar]
- 5.Fearns R, Young D, Randall R E. The paramyxovirus, simian virus 5, can remain inactive in cytoplasmic inclusion bodies in persistent infections. J Gen Virol. 1994;75:3525–3539. doi: 10.1099/0022-1317-75-12-3525. [DOI] [PubMed] [Google Scholar]
- 6.Garcin D, Itoh M, Kolakofsky D. A point mutation in the Sendai virus accessory proteins attenuates virulence for mice, but not virus growth in cell culture. Virology. 1997;238:424–431. doi: 10.1006/viro.1997.8836. [DOI] [PubMed] [Google Scholar]
- 7.Garcin D, Latorre P, Kolakofsky D. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J Virol. 1999;73:6559–6565. doi: 10.1128/jvi.73.8.6559-6565.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcin D, Curran J, Kolakofsky D. Sendai virus C proteins must interact directly with cellular components to interfere with interferon action. J Virol. 2000;74:8823–8830. doi: 10.1128/jvi.74.19.8823-8830.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodbourn S, Didcock L, Randall R E. Interferons: cell signalling, immune modulation, antiviral responses and virus countermeasures. J Gen Virol. 2000;81:2341–2364. doi: 10.1099/0022-1317-81-10-2341. [DOI] [PubMed] [Google Scholar]
- 10.Gotoh B, Takeuchi K, Komatsu T, Yokoo J, Kimura Y, Kurotani A, Kato A, Nagai Y. Knockout of the Sendai virus C gene eliminates the viral ability to prevent the interferon-alpha/beta-mediated responses. FEBS Lett. 1999;459:205–210. doi: 10.1016/s0014-5793(99)01241-7. [DOI] [PubMed] [Google Scholar]
- 11.He B, Paterson R G, Ward C D, Lamb R A. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology. 1997;237:249–260. doi: 10.1006/viro.1997.8801. [DOI] [PubMed] [Google Scholar]
- 12.He B, Leser G P, Paterson R G, Lamb R A. The paramyxovirus SV5 small hydrophobic (SH) protein is not essential for virus growth in tissue culture cells. Virology. 1998;250:30–40. doi: 10.1006/viro.1998.9354. [DOI] [PubMed] [Google Scholar]
- 13.Kessler S W. Rapid isolation of antigens from cells with a staphylococcus protein A-antibody absorbent: parameters of the interaction of antibody-antigen complexes with protein A. J Immunol. 1975;115:1617–1627. [PubMed] [Google Scholar]
- 14.King P, Goodbourn S. The β-interferon promoter responds to priming through multiple independent regulatory elements. J Biol Chem. 1994;269:30609–30615. [PubMed] [Google Scholar]
- 15.King P, Goodbourn S. STAT1 is inactivated by a caspase. J Biol Chem. 1998;273:8699–8704. doi: 10.1074/jbc.273.15.8699. [DOI] [PubMed] [Google Scholar]
- 16.Komatsu T, Takeuchi K, Yokoo J, Tanaka Y, Gotoh B. Sendai virus blocks alpha interferon signaling to signal transducers and activators of transcription. J Virol. 2000;74:2477–2480. doi: 10.1128/jvi.74.5.2477-2480.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamb R A, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields Virology. 3rd ed. New York, N.Y: Lippincott-Raven Press; 1996. pp. 1177–1204. [Google Scholar]
- 18.Lamb R A, Mahy B W, Choppin P W. The synthesis of Sendai virus polypeptides in infected cells. Virology. 1976;69:116–131. doi: 10.1016/0042-6822(76)90199-9. [DOI] [PubMed] [Google Scholar]
- 19.Lamb R A, Collins P L, Kolakofsky D, Melero J A, Nagai Y, Oldstone M B A, Pringle C R, Rima B K. Paramyxoviridae. In: van Reggenmortel M H V, Fauquet C M, Bishop D H L, Carstens E B, Estes M K, Lemon S M, Maniloff J, Mayo M A, McGeoch D J, Pringle C R, Wickner R B, editors. Report of the International Committee on Taxonomy of Viruses. San Diego, Calif: Academic Press, Inc.; 2000. pp. 549–561. [Google Scholar]
- 20.Lin G Y, Lamb R A. The paramyxovirus simian virus 5 V protein slows progression of the cell cycle. J Virol. 2000;74:9152–9166. doi: 10.1128/jvi.74.19.9152-9166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin G Y, Paterson R G, Richardson C D, Lamb R A. The V protein of the paramyxovirus SV5 interacts with damage-specific DNA binding protein. Virology. 1998;249:189–200. doi: 10.1006/viro.1998.9317. [DOI] [PubMed] [Google Scholar]
- 22.McKendry R, John J, Flavell D, Muller M, Kerr I M, Stark G R. High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both alpha and gamma interferons. Proc Natl Acad Sci USA. 1991;88:11455–11459. doi: 10.1073/pnas.88.24.11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masson N, Ellis M, Goodbourn S, Lee K A W. Cyclic-AMP response-binding protein and the catalytic subunit of protein kinase A are present in F9 embryonal carcinoma cells but are unable to activate the somatostatin promoter. Mol Cell Biol. 1992;12:1096–1102. doi: 10.1128/mcb.12.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagai Y, Kato A. Paramyxovirus reverse genetics is coming of age. Microbiol Immunol. 1999;43:613–624. doi: 10.1111/j.1348-0421.1999.tb02448.x. [DOI] [PubMed] [Google Scholar]
- 25.Paterson R G, Lamb R A. The molecular biology of influenza viruses and paramyxoviruses. In: Davidson A, Elliott R M, editors. Molecular virology: a practical approach. Oxford, England: IRL Oxford University Press; 1993. pp. 35–73. [Google Scholar]
- 26.Paterson R G, Harris T J R, Lamb R A. Fusion protein of the paramyxovirus simian virus 5: nucleotide sequence of mRNA predicts a highly hydrophobic glycoprotein. Proc Nat Acad Sci USA. 1984;81:6706–6710. doi: 10.1073/pnas.81.21.6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pekosz A, He B, Lamb R A. Reverse genetics of negative-strand RNA viruses: closing the circle. Proc Natl Acad Sci USA. 1999;96:8804–8806. doi: 10.1073/pnas.96.16.8804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Randall R E, Bermingham A. NP:P and NP:V interactions of the paramyxovirus simian virus 5 examined using a novel protein:protein capture assay. Virology. 1996;224:121–138. doi: 10.1006/viro.1996.0513. [DOI] [PubMed] [Google Scholar]
- 29.Randall R E, Dinwoodie N. Intranuclear localisation of herpes simplex virus immediate-early and delayed-early proteins. Evidence that ICP4 is associated with progeny virus DNA. J Gen Virol. 1986;67:2163–2177. doi: 10.1099/0022-1317-67-10-2163. [DOI] [PubMed] [Google Scholar]
- 30.Randall R E, Young D, Goswami K K A, Russell W C. Isolation and characterisation of monoclonal antibodies to simian virus 5 and their use in revealing antigenic differences between human, canine and simian isolates. J Gen Virol. 1987;68:2769–2780. doi: 10.1099/0022-1317-68-11-2769. [DOI] [PubMed] [Google Scholar]
- 31.Rehberg E, Kelder B, Hoal E G, Pestka S. Specific molecular activities of recombinant and hybrid interferons. J Biol Chem. 1982;257:11497–11502. [PubMed] [Google Scholar]
- 32.Southern J A, Young D F, Heaney F, Baumgartner W, Randall R E. Identification of an epitope on the P & V proteins of simian virus 5 that distinguishes between two isolates with different biological characteristics. J Gen Virol. 1991;72:1551–1557. doi: 10.1099/0022-1317-72-7-1551. [DOI] [PubMed] [Google Scholar]
- 33.Stark G R, Kerr I M, Williams B R G, Silverman R H, Schreiber R D. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 34.Sutter G, Ohlmann M, Erfle V. Non-replicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 1995;371:9–12. doi: 10.1016/0014-5793(95)00843-x. [DOI] [PubMed] [Google Scholar]
- 35.Tarbouriech N, Curran J, Ebel C, Ruigrok R W, Burmeister W P. On the domain structure and the polymerization state of the Sendai virus P protein. Virology. 2000;266:99–109. doi: 10.1006/viro.1999.0066. [DOI] [PubMed] [Google Scholar]
- 36.Thomas S M, Lamb R A, Paterson R G. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell. 1988;54:891–902. doi: 10.1016/S0092-8674(88)91285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wyatt L S, Moss B, Rozenblatt S. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression inmammalian cells. Virology. 1995;210:202–205. doi: 10.1006/viro.1995.1332. [DOI] [PubMed] [Google Scholar]
- 38.Yokosawa N, Kubota T, Fujji N. Poor induction of interferon induced 2′, 5′-oligoadenylate synthetase (2–5 AS) in cells persistently infected with mumps virus is caused by decrease of STAT-1 alpha. Arch Virol. 1998;143:1985–1992. doi: 10.1007/s007050050434. [DOI] [PubMed] [Google Scholar]
- 39.Young D F, Didcock L, Randall R E. Isolation of highly fusogenic variants of simian virus 5 from persistently infected cells that produce and respond to interferon. J Virol. 1997;71:9333–9342. doi: 10.1128/jvi.71.12.9333-9342.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young D F, Didcock L, Goodbourn S, Randall R E. Paramyxoviruses utilize different molecular mechanisms to circumvent the interferon response. Virology. 2000;269:383–390. doi: 10.1006/viro.2000.0240. [DOI] [PubMed] [Google Scholar]