Abstract
Mutations in parkin and PINK1 cause early-onset Parkinson’s disease (EOPD). The ubiquitin ligase parkin is recruited to damaged mitochondria and activated by PINK1, a kinase that phosphorylates ubiquitin and the ubiquitin-like domain of parkin. Activated phospho-parkin then ubiquitinates mitochondrial proteins to target the damaged organelle for degradation. Here, we present the mechanism of activation of a new class of small molecule allosteric modulators that enhance parkin activity. The compounds act as molecular glues to enhance the ability of phospho-ubiquitin (pUb) to activate parkin. Ubiquitination assays and isothermal titration calorimetry with the most active compound (BIO-2007817) identify the mechanism of action. We present the crystal structure of a closely related compound (BIO-1975900) bound to a complex of parkin and two pUb molecules. The compound binds next to pUb on RING0 and contacts both proteins. Hydrogen-deuterium exchange mass spectrometry (HDX-MS) experiments confirm that activation occurs through release of the catalytic Rcat domain. In organello and mitophagy assays demonstrate that BIO-2007817 partially rescues the activity of parkin EOPD mutants, R42P and V56E, offering a basis for the design of activators as therapeutics for Parkinson’s disease.
Subject terms: Structure-based drug design, X-ray crystallography, Solution-state NMR, Molecular modelling, Mitophagy
Parkin is a ubiquitin ligase that protects against early-onset Parkinson’s disease. Here, the authors show a molecular glue that promotes binding of phosphorylated ubiquitin to parkin and rescues the mitophagy defect of mutations in the parkin ubiquitin-like domain.
Introduction
Mitochondrial homeostasis is a critical process for neuronal survival. Mutations in several mitochondrial proteins that regulate the dynamics and turnover of this organelle lead to specific neurodegenerative diseases1. In particular, mutations in parkin or PINK1 cause early-onset Parkinson’s disease (EOPD), a movement disorder arising from the loss of specific neurons including (but not exclusively) dopaminergic neurons in the midbrain that are responsible for fine motor control2,3. Parkin and PINK1 work together to mediate selective turnover of damaged mitochondria via macroautophagy (a process called mitophagy) or through formation of mitochondria-derived vesicles that deliver damaged cargo to lysosomes for degradation4. Defects in these pathways lead to mitochondria-linked inflammation and autoimmune responses that lead to neuronal death5–8. Furthermore, loss of soluble parkin in the human brain correlates with age9,10 and impaired mitophagy is associated with sporadic PD11. While the details of the pathways leading to cell loss remain to be elucidated, it is clear that parkin and PINK1 suppress these deleterious responses by targeting mitochondrial compartments for degradation.
The molecular mechanisms of parkin/PINK1 have been elucidated in detail over the last two decades12. Parkin is a member of the RING-In-Between-RING (RBR) family of E3 ubiquitin ligases, which carry a RING1 domain that binds E2 ubiquitin-conjugating enzymes, as well as a catalytic Rcat (or RING2) domain that transfers ubiquitin to a substrate via a thioester intermediate13. Parkin also has two zinc-finger domains called RING0 and IBR, as well as an N-terminal ubiquitin-like (Ubl) domain with a regulatory phosphorylation site (Fig. 1a)14,15. In the basal state, parkin adopts a conformation with three features that maintain auto-inhibition: (1) the Rcat domain binds to RING0, which blocks access to the active site Cys431 in Rcat, (2) the Repressor Element of Parkin (REP), located upstream of Rcat, binds to RING1 and occludes the E2-binding site, (3) the Ubl binds to RING1, which prevents Ubl phosphorylation by PINK1 at Ser6516–19. Upon mitochondrial damage, PINK1 accumulates on the outer mitochondrial membrane (OMM), where its cytosolic kinase domain phosphorylates Ser65 in ubiquitin20–25. Phospho-ubiquitin (pUb) recruits parkin to the OMM through a high-affinity site on the RING1 domain, which induces the release and phosphorylation of the Ubl domain19,26–28. In turn, the phosphorylated Ubl domain (pUbl) binds to RING0 and displaces the Rcat domain and REP, allowing the formation of the active thioester transfer complex with a charged E2~Ub conjugate29–31. This complex is further stabilized by the activation element (ACT) of parkin, located in the Ubl-RING0 linker, which makes contacts with both the pUbl and RING0 domains30. Active parkin then ubiquitinates OMM proteins such as Mfn2 and Miro, which leads to selective mitochondrial turnover32–34.
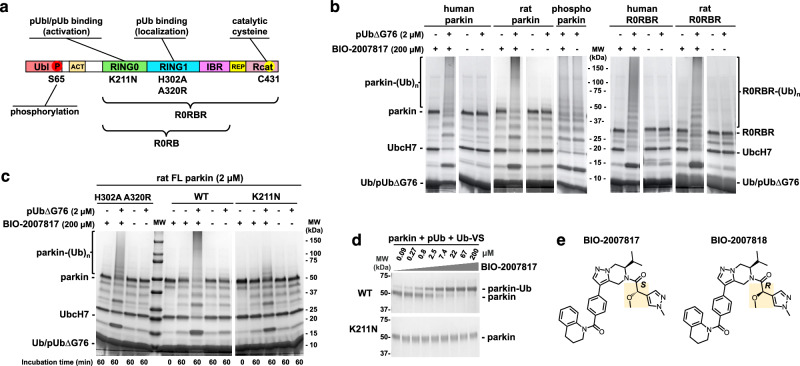
Fig. 1. Activation of parkin by the tetrahydropyrazolo-pyrazine (THPP) BIO-2007817 in the presence of pUb.
a Domain structure of parkin. b Autoubiquitination assays show full-length parkin and the Ubl-ACT deletion (R0RBR) are activated by BIO-2007817 and phosphorylated ubiquitin (pUb). pUb without a C-terminal glycine (pUb∆G76) was used to prevent its incorporation by E1. No effect was observed on activated, phosphorylated human parkin (n = 1 assay). c Effect of mutations in the pUb-binding sites on RING1 (H302A A320R) and RING0 (K211N) on the parkin activation by BIO-2007817 (n = 1 assay). d Ubiquitin vinyl sulfone (Ub-VS) assay of parkin activation by BIO-2007817 in the presence of pUb. Formation of the parkin-Ub crosslink measures release of the Rcat domain from RING0 (n = 1 assay). e Chemical structures of BIO-2007817 and inactive diastereomer BIO-2007818.
Pathogenic missense EOPD mutations can be found throughout parkin domains. Those have been categorized into 5 functional groups based on their normalized abundance and impact on mitophagy35. Groups 1 and 2 are clearly or likely pathogenic and have the greatest impact on mitophagy. Group 1 mutants affect protein folding and stability and include mutations in zinc-coordinating cysteines, as well as R275W in the RING1 domain. Group 2 mutants reduce mitophagy without affecting protein stability. Those include mutations in the active site (e.g., C431F), as well as mutations that prevent pUb or pUbl binding (e.g., K211N). Mutations that affect the PINK1-mediated activation such as Ubl mutations R42P and V56E, or K211N in RING0 which impairs pUbl-binding, could all be rescued by synthetic activating mutations that disrupt auto-inhibitory interactions. Those activating mutations include F146A in RING0 (Rcat and ACT interface), and W403A in the REP (RING1 interface), which can also rescue the parkin S65A mutation or Ubl deletion19,36. In a recent survey of synthetic activation mutants, we found that mutations that robustly increase parkin’s activity and can rescue the S65A mutation form a cluster at the junction of the REP-RING1 and RING0-Rcat interfaces, suggesting this is a hotspot for parkin activation37.
Activation of the parkin/PINK1 pathway is an attractive therapeutic strategy for the treatment of EOPD as well as sporadic PD38. Building on the discovery of kinetin as a PINK1 activator39, Mitokinin (acquired by Abbvie in 2023) developed potent small-molecules that stimulate PINK1-dependent mitophagy in the presence of mitochondrial damage (EC50 ~ 500 nM) and promote the clearance of α-synuclein aggregates, which are a hallmark of sporadic PD40. Inhibitors of USP30, a deubiquitinating enzyme on the OMM that counters parkin’s activity41, are also being developed by several companies and are currently under clinical evaluation. Other groups used phenotypic screens or computational approaches to identify small molecules that stimulate parkin/PINK1-mediated mitophagy, but the targets of these molecules remain unknown42,43. Parkin-activating compounds have also been pursued by several groups, with three patents reporting small-molecules that activate parkin-mediated mitophagy44–46. However, it is unclear how these compounds increase parkin activity, and whether they mediate their effect through parkin directly47. In 2022, our group at Biogen reported a class of small-molecule allosteric modulators that increase parkin’s E3 ubiquitin ligase activity in biochemical assays48. These tetrahydropyrazolo-pyrazine (THPP) compounds activate parkin in a pUb-dependent manner, demonstrate stereospecificity, and have submicromolar EC50 values. However, in the absence of structural data, the mechanism of action of these compounds remained unknown.
Here, we report the structural basis for the activation of parkin by THPP compounds. Using autoubiquitination assays and isothermal calorimetry (ITC), we show that binding of the most potent THPP compound BIO-2007817 (EC50 = 150 nM) is dependent on the binding of pUb or exogenous pUbl to RING0. Indeed, we recently discovered that pUb can directly activate parkin through binding the RING0 domain49,50. We built on this to determine the crystal structure of a related THPP compound BIO-1975900 bound to the complex of parkin RING0-RING1-IBR domains (R0RB) and two pUb molecules. The structure, confirmed by nuclear magnetic resonance (NMR) and mutagenesis, reveals that the THPP compounds bind to the RING0 domain and make contacts with the pUb molecule in the RING0 site (pUbl binding site), mimicking the ACT element. Structure-activity relationship (SAR) analysis of two subseries of THPP compounds establishes the importance of binding pockets in RING0 (near W183 and P180) and pUb (near K48). Based on this structural model, we hypothesized that the THPP compounds would rescue Ubl mutations. Indeed, we demonstrate that BIO-2007817 can partially rescue EOPD mutations R42P, V56E, as well S65A and ΔUbl using in organello ubiquitination assays and mitoKeima-based cellular mitophagy assays. These results show that RBR ligases can be modulated pharmacologically and highlight the clinical potential of THPP compounds in the treatment of EOPD.
Results
Ubl is not essential for parkin activation by BIO-2007817
Previous experiments showed that BIO-2007817 could trigger autoubiquitination activity of unphosphorylated parkin in the presence of pUb48. We confirmed those results with both human and rat parkin, which share 86% identity (Fig. 1b). At 200 µM, BIO-2007817 induced activity similar to phosphorylated (activated) parkin but the compound did not further stimulate the activity of phosphorylated parkin. As previously reported48, the activation was strictly dependent on the presence of pUb. To avoid charging of the E2 enzyme with pUb, a C-terminal deletion (pUbΔG76) was used. Activation could also be detected in ubiquitin-vinyl sulfone (Ub-VS) assays51 that measure the accessibility of Rcat and are not dependent on the upstream components of the ubiquitination cascade (Fig. 1d). We next asked if the parkin Ubl domain was required for activation. We observed equal autoubiquitination activity of full-length and R0RBR parkin (lacking the Ubl domain) in both the human and rat proteins (Fig. 1b). Thus, the Ubl domain is not implicated in the mechanism of activation.
BIO-2007817 requires pUb bound to RING0 domain
Phospho-ubiquitin can bind two distinct sites in parkin. The high-affinity site on RING1 mediates the recruitment of parkin to the mitochondria19. With an affinity of around 20 nM, it is formed by residues H302 and R305 and promotes detachment and phosphorylation of the Ubl domain52. The second site is located on the RING0 domain and regulates parkin activity. While the second site typically binds pUbl (when parkin is phosphorylated), we previously demonstrated that it can also bind pUb49. The site is formed by residues K211, R163 and K161 and with an affinity of around 400 nM for pUb and 1 µM for pUbl49,50. This binding forces the dissociation of Rcat domain and the release of parkin ligase activity.
We investigated which of the two sites is needed to promote activation by comparing the effects of a RING1 double mutation (H302A A320R) and a RING0 mutation (K211N). In the absence of compound, none of the parkin variants—wild-type, H302A A320R, or K211N—can autoubiquitinate in the presence of pUb (Fig. 1c). Upon addition of BIO-2007817, the double mutant H302A A320R was activated to close to wild-type levels as judged by the disappearance of unmodified parkin and appearance of a high molecular weight smear of autoubiquitinated parkin. In contrast, the K211N parkin mutant was strongly impaired with an absence of high molecular weight parkin adducts (Fig. 1c). Thus, pUb binding to RING0 but not RING1 is required for activation by BIO-2007817. We confirmed this result in the Ub-VS assay that measures Rcat release. At 2.5 µM BIO-2007817, half of wild-type parkin was modified by the Ub-VS reagent while the K211N mutation completely prevented parkin modification even at 200 µM (Fig. 1d). Taken together, these results demonstrate that activation by BIO-2007817 requires the binding of an exogenous pUb molecule to the RING0 binding site of parkin.
BIO-2007817 acts a molecular glue for RING0 and pUb/pUbl
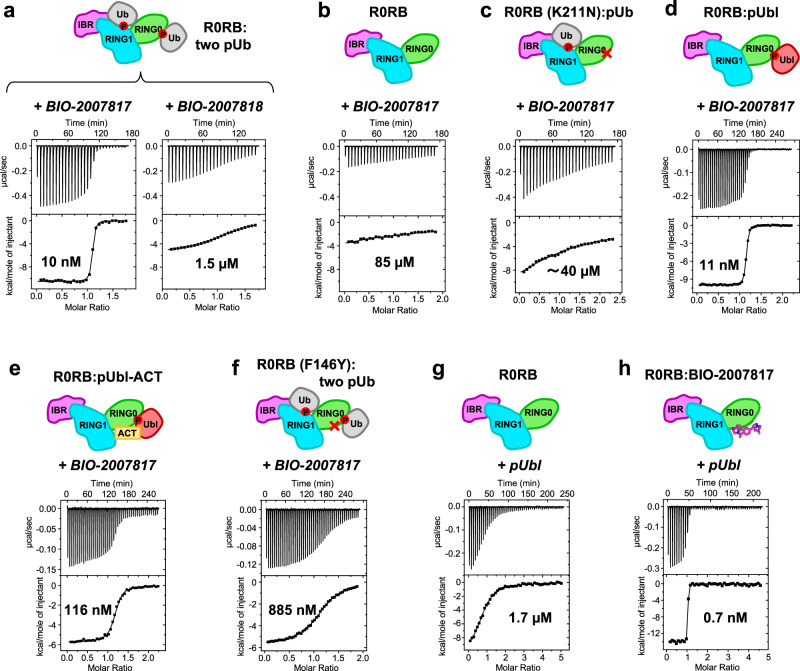
We measured the affinity of BIO-2007817 for parkin using isothermal titration calorimetry (ITC) experiments with a parkin construct R0RB that lacks both the Ubl domain and the Rcat domain (Fig. 2, Supplementary Table 1). Since pUb and Rcat compete for binding to RING0, deleting the Rcat domain favors pUb binding. The initial experiment measured BIO-2007817 binding to R0RB with pUb bound to both the RING1 and RING0 sites (R0RB + two pUb). BIO-2007817 bound the complex with 10 nM affinity and one-to-one stoichiometry (Fig. 2a). The interaction was highly specific as the inactive diastereomer BIO-200781848 (Fig. 1e) bound with 150-fold less affinity (Fig. 2a).
Fig. 2. BIO-2007817 binds to the complex of parkin and pUb or pUbl.
Isothermal titration calorimetry measurements with different parkin complexes and titrants (in italics). a BIO-2007817 binds to the R0RB:2×pUb complex with 150-fold higher affinity than the diastereomer BIO-2007818. b High-affinity binding requires the presence of pUb. c The K211N mutation in the RING0 pUb/pUbl-binding site prevents high-affinity binding. d BIO-2007817 also binds to the R0RB:pUbl complex. e Inclusion of ACT element with pUbl decreases the affinity of BIO-2007817 10-fold. f The F146Y mutation in RING0 ACT-binding site decreases the affinity 80-fold. g pUbl binds weakly to R0RB49,50. h Inclusion of BIO-2007817 increases the affinity of pUbl binding by 2400-fold. Protein concentrations and binding parameters are given in Supplementary Table 1.
We next asked if pUb was required for BIO-2007817 binding (Fig. 2b). In the absence of pUb, the affinity was decreased over 5000-fold (to the limit of detection), mirroring the requirement for pUb observed in autoubiquitination experiments. Loss of the RING0 pUb-binding site (due to the K211N mutation) similarly prevented binding (Fig. 2c). Although BIO-2007817 had no effect on phosphorylated parkin, surprisingly, the phosphorylated Ubl domain (pUbl) added in trans could replace pUb. In ITC experiments, BIO-2007817 bound to the R0RB:pUbl complex with the same affinity as the pUb complex (Fig. 2d). As the R0RB fragment does not undergo any conformational rearrangements and pUbl binds only to RING0, this suggests that BIO-2007817 binds at the RING0/pUb (or RING0/pUbl) interface.
Additional insight into the site of BIO-2007817 binding came from the observation that inclusion of the ACT region with the pUbl domain (pUbl-ACT) decreased the affinity of BIO-2007817 binding 10-fold (Fig. 2e). In contrast, in the absence of compound, the ACT improves the affinity of pUbl binding to R0BR three-fold50. This suggests they compete for a common binding site. Identification of the binding site was confirmed by a mutation in ACT-binding site. Addition of a single oxygen atom (F146Y) decreased BIO-2007817 binding 10-fold (Fig. 2f). As F146 is surface-exposed and does not make contacts with pUb or pUbl in crystal structures of parkin, this strongly suggests that F146 directly binds BIO-2007817.
To directly test if BIO-2007817 behaves as a molecular glue, we measured the affinity of the pUbl domain binding to RING0 in the R0RB construct. As previously reported49,50, pUbl in trans has low micromolar affinity (Fig. 2g). This contrasts with the situation in intact phosphorylated parkin when the pUbl domain is present in cis. Inclusion of a saturating concentration (200 µM) of BIO-2007817 increased the pUbl binding affinity by more than three orders of magnitude, demonstrating that the compound acts as a molecular glue (Fig. 2h).
Mechanism of activation by THPP compounds
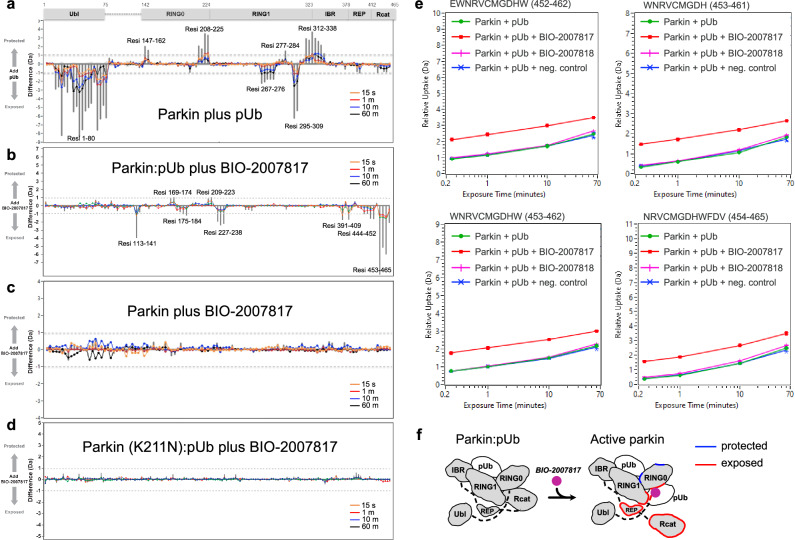
Hydrogen-deuterium exchange mass spectrometry (HDX-MS) has been instrumental for characterizing the conformational changes during parkin activation29–31. HDX-MS measures solvent exposure which provides information on protein structure and interdomain contacts. We performed a series of HDX-MS experiments to resolve the mechanism of parkin activation by THPP compounds (Fig. 3). As previously observed, addition of pUb to parkin increased deuteration of the Ubl domain (Fig. 3a). This reflects the release of the Ubl from its binding site on RING1, which facilitates its phosphorylation by PINK129–31. There was no increase in deuteration of Rcat domain as pUb binding to RING1 is not sufficient to activate parkin. In contrast, addition of BIO-2007817 in the presence of pUb, increased deuteration of the catalytic Rcat domain, reflecting its release from RING0 (Fig. 3b). The effect of BIO-2007817 was strictly dependent on the presence of pUb; no changes in parkin deuteration were observed upon addition of BIO-2007817 alone (Fig. 3c). The increase in Rcat exposure was also completely blocked by the K211N mutation that prevents pUb binding to RING0 (Fig. 3d) or by substitution of BIO-2007818 with the inactive diastereomer (Fig. 3e). Thus, as observed in the autoubiquitination assays (Fig. 1), the combined action of pUb and BIO-2007817 is required to activate parkin. This occurs through the release of the Rcat domain from its site on RING0 in a manner analogous to activation by parkin phosphorylation29–31 (Fig. 3f).
Fig. 3. HDX-MS analysis of conformational changes in parkin upon pUb and BIO-2007817 binding.
a Changes in solvent exposure of the parkin complex upon addition of pUb at four time points (15 s, 1 min, 10 min, 60 min) and the sum of the four time points (gray bars). The largest changes occur in the Ubl domain, which is released by pUb binding, and RING1, which is the site of pUb binding. b Changes in solvent exposure upon addition of BIO-2007817 in the presence of pUb. The largest changes occur in the Rcat domain. c, d Addition of BIO-2007817 does not change solvent exposure in the absence of pUb or in the parkin K211N mutant. e HDX-MS analysis of individual peptides from the parkin Rcat domain show solvent exposure in the presence of BIO-2007817 but not the inactive diastereomer BIO-2007818. f Mapping of increased (red) and decreased (blue) exchange upon BIO-2007817 addition to parkin:pUb (b).
Crystal structure of parkin R0RB:2×pUb complex with BIO-1975900
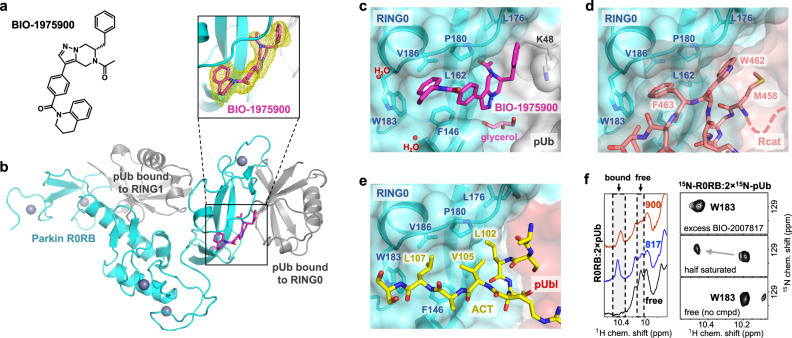
We turned to crystallography to understand how BIO-2007817 acts as a molecular glue. As crystallization trials with BIO-2007817 were unsuccessful, we expanded the screen to include related compounds with different ethanone substituents. We obtained crystals with the compound BIO-1975900 bound to rat parkin R0RB with two pUb and solved the structure by molecular replacement (Fig. 4; Supplementary Table 2). BIO-1975900 is composed of a central tetrahydropyrazolo-pyrazine core, decorated by a tetrahydroquinoline head and a benzyl and ethanone tail (Fig. 4a). It has roughly 20-fold lower affinity with an EC50 of 2.9 µM in in vitro TR-FRET activity assays compared to 0.15 µM for BIO-200781748.
Fig. 4. Crystal structure of BIO-1975900 bound to parkin R0RB:2×pUb complex.
a Chemical structure of BIO-1975900. b Crystal structure of rat R0RB:2×pUb with BIO-1975900. Inset shows the 2.5 Å omit map of BIO-1975900 contoured at 3 σ. c Details of the BIO-1975900 (magenta) bound to a hydrophobic groove formed by residues from RING0 and a lysine from pUb. A glycerol and two water molecules were observed at the binding site. d Overlay of the inactive parkin structure (PDB 5N2W)26. The C-terminus of the Rcat domain (salmon) occupies the BIO-1975900 binding site. e Overlay of the active, phosphorylated parkin:pUb structure (PDB 6GLC). The ACT element (yellow) binds the same site as BIO-1975900. f NMR spectra of THPP binding. 1D NMR spectra (left panel) of the downfield region upon addition of BIO-1975900 or BIO-2007817. 2D 1H-15N correlation NMR spectra (right panel) of the tryptophan W183 indole amide titrated with BIO-2007817.
Overlay of the electron density of the unliganded structure50 (PDB 7US1) showed additional electron density at the RING0-pUb interface corresponding to BIO-1975900 (Fig. 4b). The compound binds a hydrophobic groove composed of three regions: a RING0 W183 pocket, a P180 pocket, and a pUb K48 pocket (Fig. 4c). The tetrahydroquinoline group contacts F146, P180, W183, and V186. Replacement of F146 by tyrosine introduces a polar group and disrupts binding as observed in ITC experiments. The ethanone group inserts into a hydrophobic pocket formed by parkin RING0 residues L162, V164, L176, P180, and F208. The benzyl moiety extends to sit in a pocket sandwiched between the L176 residues of RING0 and residue K48 of the pUb molecule bound to RING0 (Fig. 4c). In the difference density, we also identified a glycerol molecule from the cryoprotectant bound near the THPP central ring. The glycerol bridges the backbone amide of L162 and the guanidino group of R163 in the RING0 phosphoserine binding site.
We used NMR spectroscopy to confirm that BIO-1975900 and BIO-2007817 bind at the same location. The proton NMR spectrum shows similar chemical shift changes in the downfield region upon addition of the two compounds (Fig. 4f, left panel). 2D NMR with 15N-labeled R0RB:2×pUb protein confirmed that the shifting signal was from W183, which is the only tryptophan present in R0RB:2×pUb (Fig. 4f, right panel). Addition of half an equivalent of BIO-2007817 to 15N-labeled R0RB:2×pUb split the W183 side chain signal at 10.2 ppm into two peaks, indicative of slow-exchange kinetics typical for nanomolar binding affinity. Addition of excess of BIO-2007817 shifted the signal to 10.5 ppm, as observed in the 1D spectra (Fig. 4f, left panel). The downfield chemical shift change arises from the interaction between the tetrahydroquinoline moiety of either BIO-2007817 or BIO-1975900 and the W183 side chain. This confirms that BIO-2007817 binds at the RING0/pUb interface in the same manner as BIO-1975900.
The crystal structure highlights the importance of the hydrophobic groove that binds BIO-1975900 for controlling parkin activity. In autoinhibited structures of parkin, Rcat residues bind the groove: W462 forms hydrophobic interactions with RING0 while F463 contacts RING0 W183 (Fig. 4d). The F463 is positioned almost identically to the tetrahydroquinoline group of BIO-1975900. BIO-2007817 and Rcat compete for binding the same site on RING0.
The structure also explains the competition observed in ITC experiments between ACT and BIO-2007817 binding. The ACT, formed by residues 102–109 in the Ubl-R0RBR linker, binds to the same hydrophobic groove than BIO-1975900 (Fig. 4e), as observed in the active, phosphorylated parkin:pUb structure (PDB 6GLC). ACT L107 contacts W183, L102 contacts F208, and V105 stacks against RING0 P180. The molecular mimicry between the compound and the hydrophobic residues of the ACT element explains, in part, why the compound is unable to increase the activity of phosphorylated parkin.
The importance of the hydrophobic groove in parkin activation has been observed before. The mutation F146A weakens the hydrophobic interaction with Rcat F463 and, consequently, promotes parkin activation16. Cells transfected with the F146A mutant showed increased recruitment of parkin to mitochondria and mitophagy36. The F146A mutation also rescues mitochondrial recruitment and mitophagy in parkin mutants with defective Ubl (R42P, V56E, S65A, and ∆Ubl) or RING0 pUbl/pUb-binding sites (K161N and K211N)35,36.
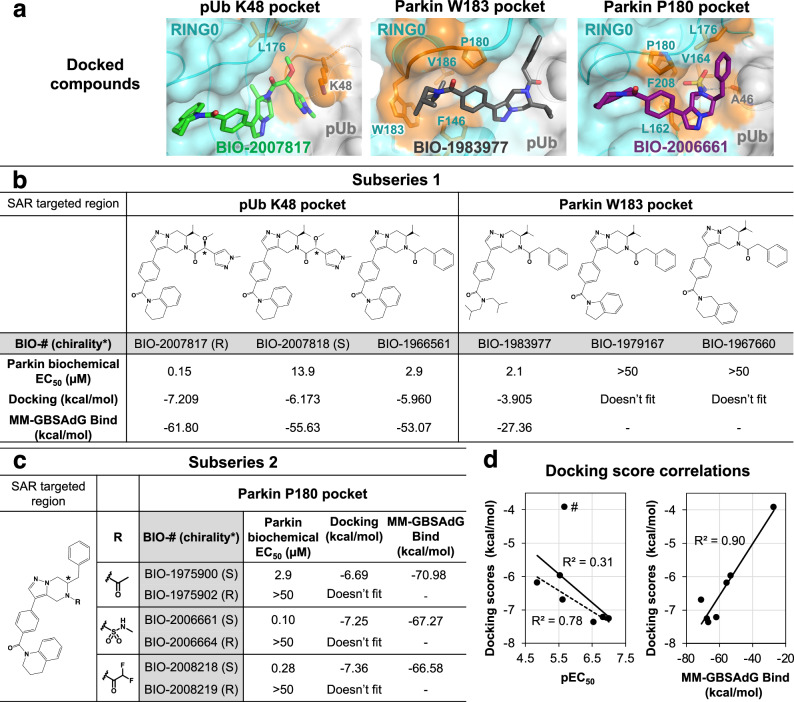
Structure-activity relationship of THPP compounds
We used molecular docking to explore the structure-activity relationship of related THPP compounds. Parkin activity was previously determined for compounds from two subseries: BIO-2007817-related subseries 1, which has an isopropyl group attached to its pyrazolo-pyrazine moiety, and BIO-1975900-related subseries 2, in which the isopropyl is replaced by a benzyl group46. Additional compounds from both subseries were selected to rationalize how chemical modifications in the three interaction regions affect their activity (Fig. 5a, Supplementary Fig. 1). To validate the protocol, BIO-1975900 was first docked into the R0RB:2×pUb crystal structure using the standard precision ligand docking module in Glide. The docked compound had favorable energies (Fig. 5c) and superimposed well (less than 0.3 Å deviation) with the experimentally determined structure.
Fig. 5. Structure-activity relationships based on the crystal structure predict potencies of two subseries of THPP compounds.
a Docking-based positioning of BIO-1975900 related-compounds. Comparison of biochemical potency (EC50 in parkin autoubiquitination assays) and the docking scores for subseries 1 (b) and subseries 2 (c) compounds. Autoubiquitination activity profiles are shown in Supplementary Fig. 1. d Correlation between docking scores, biochemical potency, and MM-GBSA scores. BIO-1983977 lacks the THPP group and is an outlier: removing it (marked #) improves the correlation between the docking score and pEC50 (dashed line).
Subseries 1-compound BIO-2007817 was docked into the prepared R0RB:2×pUb protein grid (Fig. 5a left panel, Supplementary Fig. 2). NMR experiments with BIO-2007817 revealed hindered rotation with the methoxy-methylpyrazine-ethanone moiety adopting cis and trans conformations (Supplementary Fig. 3). In the docked structure, BIO-2007817 is in the cis conformation, suggesting a conformationally locked compound could likely improve potency further. The isopropyl moiety points towards parkin V164 and L176 and the methoxy-methylpyrazine-ethanone groups are adjacent to the positively charged side chain of pUb K48, forming a cation-pi interaction. The pi-stacking interactions between tetrahydroquinoline group and W183 and F146 are similar to BIO-1975900. In the weakly binding BIO-2007818 diastereomer, the oxygen of the methoxy group points away from pUb K48. The loss of the electrostatic interaction is reflected in lower docking and MM-GBSA scores compared to BIO-2007817 (Fig. 5b). This agrees with the limited effect of BIO-2007818 on parkin activation48 and its reduced binding in ITC experiments (Fig. 2a). Replacement of the methoxy and methyl-pyrazolyl groups with a phenyl moiety (BIO-1966561) also decreased the docking score and EC50 (Fig. 5b).
Docking of BIO-1983977 (Fig. 5a, middle panel) demonstrates that the hydrophobic diisobutylamine functional group fits in the W183 pocket. The predicted binding is weaker than compound BIO-1966561 (identical except for the tetrahydroquinoline head group) but BIO-1983977 was similar in biochemical assays (Fig. 5b). The inactive compounds BIO-1967660 and BIO-1979167 could not be docked due to steric clashes with the sides of the W183 pocket.
Modulation of compound interactions with the parkin P180 pocket was investigated by docking subseries 2 compounds which are most closely related to the crystallized ligand BIO-1975900 (Fig. 5c). In contrast to the ethanone group of BIO-1975900, which showed limited interactions with the P180 pocket (Fig. 4c), the methylsulfonamide moiety of BIO-2006661 formed a hydrogen bond with the backbone carbonyl of pUb A46 (Fig. 5a, right panel). The additional interaction would explain the higher potency of BIO-2006661 relative to BIO-1975900 (EC50 of 0.1 µM and 2.9 µM). BIO-2008218, which contains a difluoro-ethanone moiety, showed a similar increase in potency. Positioning of the subseries 2 benzyl group was critical for binding. BIO-1975902, BIO-2006664, and BIO-2008219, R-enantiomers of active S-compounds, were inactive in biochemical assays and could not be docked. In general, the calculated binding interactions correlated well with the biochemical EC50, suggesting that docking could be used for shape-based screening of new parkin activators (Fig. 5d).
BIO-2007817 rescues ubiquitination and mitophagy by parkin Ubl mutants
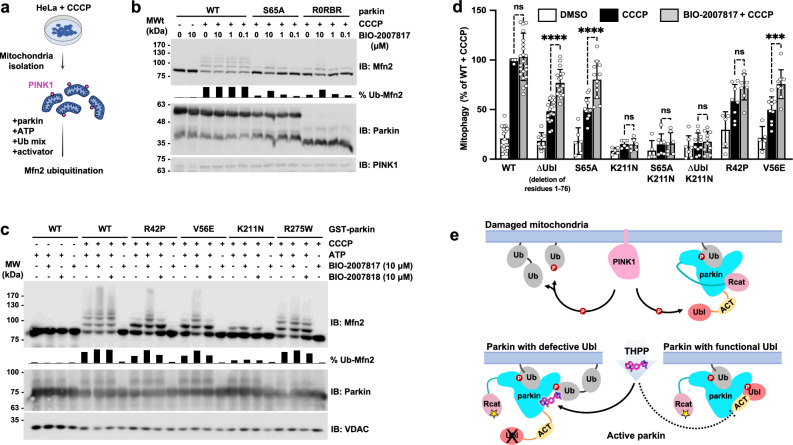
The observation that BIO-2007817 can activate parkin in the presence of pUb suggested that it might rescue the activity of parkin variants without a functional Ubl domain. We tested this hypothesis using an in organello assay that measures parkin ubiquitination of the mitochondrial protein mitofusin-2 (Mfn2)36. As substrate ubiquitination and autoubiquitination rates can differ, it was important to test the activity of the compounds on a physiologically relevant substrate. In the assay, HeLa cells are stimulated by treatment with mitochondria depolarizer CCCP and then the mitochondria are isolated and added ex vivo to a ubiquitination reaction mix including parkin with or without activator compounds (Fig. 6a).
Fig. 6. BIO-2007817 rescues parkin Ubl mutations in organello and in cells.
a Schematic of in organello ubiquitination assay of parkin. PINK1 expression is induced in cultured cells by CCCP and parkin activity measured by ubiquitination of mitofusin-2 (Mfn2). b Immunoblots of in organello assays showing the rescue of ubiquitination activity of the non-phosphorylatable parkin mutants, S65A and R0RBR, by BIO-2007817. Quantification of the percentage of ubiquitinated Mfn2 is shown as chart bar. c Immunoblots of in organello assays showing the rescue of ubiquitination activity of Parkinson’s disease parkin mutants, R42P and V56E, by BIO-2007817, but not by the diastereomer, BIO-2007818. Quantification of the percentage of ubiquitinated Mfn2 is shown as chart bar. d Quantification of average percentage of mitophagy detected by fluorescence‐activated cell sorting (FACS) of mitochondrially targeted mitoKeima U2OS cells expressing transient WT or mutant GFP‐parkin. Cells were treated with DMSO (white), CCCP (black), or pre-treated with BIO-2007817 followed by CCCP treatment (gray). Percentages were normalized to WT Parkin pre-treated with DMSO followed by CCCP treatment. A partial rescue by BIO-2007817 was observed in cells expressing parkin with defective Ubl. Error bars indicate s.e.m. For statistical analysis, a one‐way ANOVA with Tukey’s post‐test was performed. The number of independent, biological replicates varied between 5 and 23. ****P < 0.0001; ***P = 0.0004; ns, not significant. e Model for THPP activation of parkin with a defective Ubl domain.
Without CCCP, there was no ubiquitination of the Mfn2 substrate due to the absence of PINK1 and pUb (Fig. 6b). In contrast, CCCP-treatment led to an accumulation of PINK1, which led to parkin recruitment, phosphorylation/activation, and Mfn2 ubiquitination. In both conditions, addition of BIO-2007817 had no effect, in agreement with autoubiquitination assays with phospho-parkin (Fig. 1) and previous mitochondrial translocation and mitophagy assays48. BIO-2007817 was able to partially rescue Mfn2 ubiquitination by non-phosphorylatable parkin variants, either S65A or R0RBR, which lacks the Ubl domain (Fig. 6b). At 10 µM BIO-2007817, the fraction of total ubiquitination for Mfn2 was roughly 50% that of wild-type parkin.
We next asked if BIO-2007817 could rescue pathological mutations that impair parkin activity. R42P and V56E both affect the Ubl domain to destabilize its folding53,54 and impair mitophagy in cells in response to CCCP treatment35. BIO-2007817 at 10 µM effectively suppressed the ubiquitination defect of both mutations (Fig. 6c). The suppression was limited to Ubl mutations: neither R275W nor K211N showed enhanced activity in the presence of BIO-2007817. R275W occurs in the RING1 domain, disrupting parkin stability and possibly pUb binding35,55. The lack of effect on the K211N mutant recapitulates the conclusions of autoubiquitination and Ub-VS assays that pUb-binding is essential for activation by BIO-2007817 (Figure 1). The rescue of Mfn2 ubiquitination was selective. We did not observe rescue of either R42P and V56E by the diastereomer BIO-200781848 that differs at only one chiral center (Fig. 1e). Taken together, the in organello results show that mutations (ΔUbl, R42P, V56E, S65A) that affect the parkin Ubl domain can be rescued by BIO-2007817.
We turned to mitoKeima assays to confirm this result and assess the ability of BIO-2007817 to derepress non-phosphorylatable parkin mutants which do not normally induce mitophagy in cells. The assay measures mitophagy using mitochondrially targeted fluorescent protein (mitoKeima) that shifts its excitation spectrum when mitochondria enter the acidic environment of lysosomes. Depolarization of mitochondria by CCCP led to robust mitophagy in cells expressing wild-type parkin with or without 1 h pre-exposure to BIO-2007817 (Fig. 6d). Cells expressing non-phosphorylatable S65A parkin or ∆Ubl parkin showed only partial mitophagy that was enhanced by BIO-2007817. PD mutations destabilizing parkin Ubl (R42P and V56E) and reducing mitophagy response to CCCP, could be rescued by pre-treatment with BIO-2007817. In agreement with our previous findings, the importance of RING0 pUb/pUbl binding site in BIO-2007817 activation of parkin was confirmed by the complete loss of mitophagy response in K211N mutant which could not be rescued by the compound.
Based on our results, we propose a mechanism of activation of parkin by BIO-2007817 or THPP compounds (Fig. 6e). In the case of parkin with impaired Ubl, the THPP compound can allosterically promote the binding of pUb onto RING0 through bridging between parkin and pUb. When parkin Ubl can be phosphorylated by PINK1, the THPP binding site is already occupied by the ACT following pUbl relocalization to RING0. This explains the lack of activation effect of THPP on wild-type parkin observed in in organello and mitophagy assays, as well as in previously reported cell assays48.
Discussion
Boosting the activity of parkin using small molecule drugs has been proposed as a potential treatment for Parkinson’s disease. To do so, the molecules need to shift the inactive/active equilibrium toward the active conformation by rearrangement of the Ubl and Rcat domains. Several point mutations that increase parkin activity have been suggested as models for the design of parkin activators. The mutations W403A and F146A increase the activity of unphosphorylated parkin by disrupting inhibitory interactions. W403A weakens the REP-RING1 interaction while F146A disrupts the RING0-Rcat interface. Both are strongly activating in in vitro and cellular experiments and can suppress some naturally occurring EOPD parkin mutations16,36,37. Unfortunately, no small molecule activators mimicking those mutations have been identified so far. One possible explanation is that many of the parkin surfaces are multifunctional which creates additional constraints for small molecule binders. A compound facilitating the release of Ubl from RING1 could accelerate the phosphorylation of Ubl by PINK1 and its relocalization to RING0; however, it cannot interfere with the E2-binding site, which overlaps with the Ubl domain. It also cannot inhibit the Ubl domain’s ability to act as a substrate for PINK1 or hinder the phosphorylated domain from binding to RING0. Similarly, a compound facilitating the release of Rcat from RING0 shouldn’t compete with the physiological pathways of activation. If binding RING0, it shouldn’t block pUb or pUbl binding. If binding Rcat, it shouldn’t prevent the transthiolation step of ubiquitin transfer from the E2 to the Rcat cysteine or the subsequent step of ubiquitin transfer to the substrate lysine. THPP compounds were identified in a biochemical screen with unphosphorylated parkin and pUb, not with phosphorylated parkin. As such, the compounds circumvent these problems by acting as a molecular glue to promote a minor activation mechanism in which pUb substitutes for pUbl. This avoids interfering with the activation and catalytic steps necessary for parkin function.
An additional consideration for the design of parkin activators is avoiding excessive or inappropriate activation. Phosphorylated parkin is highly active and ubiquitinates proteins indiscriminately, including itself. Compound BIO-2007817 amplifies an existing activation mechanism to obtain activation in a regulated way. BIO-2007817 is not a direct activator of parkin since it has a low affinity for parkin in the absence of pUb (Fig. 1d). The pUb-dependent activation is only functional upon parkin translocation to damaged mitochondria displaying pUb. In in vitro ubiquitination assays, activity was detected only in the presence of pUb and functional pUb-binding site (Fig. 1).
Binding both parkin and pUb, BIO-2007817 belongs to a growing class of molecules known as molecular glues56. Chemically diverse, they share the property of directing a protein-protein interaction often with therapeutic goal. Cyclosporin, rapamycin, and thalidomide are among the best-known examples. Targeted protein degradation through the linking of ubiquitin ligases and substrate proteins is a particularly exciting application57, although molecular glues can also act to inhibit ligase activity58. Here, we observed the compounds act as activators through linking pUb and the RING0 domain to shift parkin toward its active conformation (Fig. 6e). The binding site of the compounds in hydrophobic groove between pUb and RING0 is a strategic location. In the inactive conformation, the groove is occupied by elements of the Rcat domain, while in the phosphorylated active conformation, it is occupied by the ACT element (Fig. 4d, e).
The identification of a hot spot for parkin activators lays the basis for further development. BIO-2007817 exists in two conformations in solution so constraining it to the bound conformation should improve its potency. The BIO-1975900 crystal structure points to other improvements. BIO-1975900 mimics the shape of the hydrophobic groove but does not fully occupy the region between RING0 and pUb. Notably, the chiral methoxy-methylpyrazine group, which makes important contributions to the binding affinity, is not present in the crystal structure. Instead, there is a well-ordered glycerol molecule that interacts with L162 and R163 of RING0 (Fig. 4c). The glycerol could be linked to the methyl-pyrazolyl moiety to boost the solubility and affinity of BIO-1975900. More generally, the small number of hydrogen bonds in hydrophobic groove suggests that higher affinity molecules are possible by enhancing salt bridge and H-bond interactions.
Finally, while THPP compounds are not universal activators of parkin, we note that their selectivity in rescuing parkin Ubl mutations could be an advantage for patients with those specific mutations. BIO-2007817 increases mitophagy in cells transfected with parkin mutants with defective Ubl domains (Fig. 6d). Generalized activation of parkin is unlikely to be a viable therapeutic strategy due to parkin’s lack of substrate selectivity. As selective inducers, THPP compounds are promising leads for personalized medicine for patients with certain naturally occurring parkin mutations.
Methods
Cloning, expression, and purification of recombinant proteins in E. coli
Single-point mutations and deletions were generated using PCR mutagenesis (Agilent) and proteins expressed in BL21 (DE3) E. coli. Purification of full-length parkin, R0RBR, R0RB:2×pUb, pUb∆G76, pUbl, pUbl-ACT, UbcH7, Tc-PINK1, and human His-E1 were done using methods previously described19,59. Briefly, proteins were purified by glutathione-Sepharose (Cytiva) or Ni-NTA agarose (Qiagen) affinity chromatography, followed by either 3C protease cleavage to remove the GST tag or Ulp protease cleavage to remove the His-SUMO tag. Size-exclusion chromatography was used as a last step. 15N-labeled rat R0RB and pUb were produced in M9 minimal medium supplemented with 15NH4Cl. Phosphorylated Ub and Ubl were produced and purified according to published procedures28,52. Purified proteins were verified using SDS-PAGE analysis. Protein concentrations were determined using UV absorbance.
Ubiquitination assays
The autoubiquitination assays of full-length and R0RBR parkin were performed at 22 °C for 60 min by adding 2 µM full-length WT, K211N or H302A A320R parkin or R0RBR, to 50 nM human His-Ube1, 2 µM UbcH7, 100 µM ubiquitin in 50 mM Tris pH 8.0, 150 mM NaCl, 1 mM TCEP, 0.4% DMSO, 5 mM ATP and 10 mM MgCl2. Where indicated, pUb without the C-terminal glycine (pUbΔG76) was used. When stated, 2 µM of pUbΔG76 and/or 200 µM BIO-2007817 were added to the sample. pUbΔG76 was used to prevent incorporation of pUb by E1 into the ubiquitination reaction. Reactions were stopped by the addition of reducing SDS-PAGE loading buffer and the level of ubiquitination was analyzed on SDS-PAGE gels stained with Coomassie blue. Phosphorylated full-length parkin in complex with pUb was used as positive control.
Ubiquitin vinyl sulfone assays
To assess the effect of compounds on Ub-VS-charging of parkin at a fixed pUb concentration, 2 µM parkin (either WT or K211N mutant) were incubated at 25 °C for 2 h in the presence of 10 µM Ub-VS (R&D Systems), 3 µM pUb, at the following compound concentrations of BIO-2007817 (0 µM, 0.09 µM, 0.27 µM, 0.8 µM, 2.5 µM, 7.4 µM, 22.2 µM, 66.7 µM, and 200 µM) in 50 mM HEPES, 150 mM NaCl, 1 mM TCEP, pH 8.0 containing 2%(v/v) DMSO. Reactions were stopped by addition of reducing SDS-PAGE loading buffer and the level of cross-linking was analyzed on SDS-PAGE gels (Biorad) stained with Coomassie blue.
Isothermal titration calorimetric assays
ITC measurements were carried out at 20 °C using VP-ITC (Microcal). The composition of the cell and syringe components are given in Supplementary Table 1. Titrations were performed with either 28 injections of 10 μl or 42 or 48 injections of 6 μl in 50 mM Tris-HCl, 150 mM NaCl, 1 mM TCEP, 0.33% DMSO, pH 7.4. Data were modeled using a single set of identical sites using Origin v7 software.
HDX-MS
Parkin (15 µM) with additions of 15 µM pUb and 600 µM THPP compounds (diluted from 10 mM in 100% DMSO) were incubated at RT for 30 min in 25 mM Tris, 150 mM NaCl, and 1 mM TCEP-HCl, pH 8. Deuterium labeling was initiated by diluting the solution 7-fold (v/v) with D2O buffer (25 mM Tris, 150 mM NaCl, and 1 mM TCEP-HCl, pD 7.6) at room temperature. After different time intervals, labeling was quenched by addition of an equal volume of buffer containing 2 M guanidine hydrochloride, 0.6% formic acid, pH 2.6. Inline digestion and peptide separation were conducted on a Waters nanoACQUITY system with HDX sample manager. 30 to 40 pmol of sample were injected on a pepsin immobilized column (2.1 mm × 30 mm, Waters, Milford, MA) with a flow rate of 100 µl/min in 0.1% formic acid at 15 °C. The proteolytic peptides were trapped on a VanGuard Pre-Column (2.1 mm × 5 mm, ACQUITY UPLC BEH C18, 1.7 µm, Waters) for 2 min. The trap was then placed in line with an ACQUITY UPLC BEH C18 column (1.7 µm, 1.0 × 100 mm, Waters), and an 3–35% gradient of acetonitrile over 11 min at a flow rate of 50 µl/min was used to separate the peptides at 0.4 °C. Formic acid (0.1%) was added to both mobile phases to maintain a pH of 2.5. Mass accuracy was maintained through continuous lock-mass correction using a Glu-fibrinogen B peptide standard (Sigma). Mass determination was performed on a Waters Synapt G2S Q-Tof mass spectrometer and the amounts of deuterated and undeuterated peptides analyzed using Waters DynamX 3.0. The data were not corrected for back-exchange.
Crystallization and structure determination
Crystals of R0RB:2×pUb in complex with BIO-1975900 were grown using hanging drop vapor diffusion method by mixing 0.3 µl of protein complex at 10 mg/ml (230 µM) supplemented with 400 µM of BIO-1975900 in 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.7 mM TCEP, 4% (v/v) DMSO with 0.3 µl of 0.2 M NaCl, 0.1 M HEPES pH 7.5, 25% (w/v) PEG 3350 at 4 °C. Crystals appeared after 9 days. After 3 weeks, crystals were cryoprotected in mother liquor supplemented with 25% (v/v) glycerol before being cryo-cooled in liquid nitrogen.
Diffraction data was collected at the CMCF beamline 08ID-1 at the Canadian Light Source. A total of 1800 images were collected with an oscillation angle of 0.2° at 0.953 Å. Reflections were integrated and scaled using XDS package60. The diffraction data were phased by molecular replacement using R0RB and two pUb molecules from PDB deposition 7US1 as search model using Phaser implemented in PHENIX61–63. The structure was refined using PHENIX package61 and the model building was performed using COOT64. Calculated Ramachandran values for the final structure are 98.1% of residues in the favored region and 0% outliers (Supplementary Table 2).
NMR experiments
For 1D NMR spectra, 60 μM of either BIO-2007817 or BIO-1975900 was added to 50 μM unlabeled rat R0RB:2×pUb in 20 mM Tris-HCl, 120 mM NaCl, 0.5 mM TCEP, 4% DMSO, pH 7.4 and spectra acquired for 8 h. For 2D NMR spectra, 14 μM and 32 μM of BIO-2007817 were added to 28 μM 15N-labeled rat R0RB:2×pUb in 20 mM Tris-HCl, 120 mM NaCl, 0.5 mM TCEP, 1% DMSO, pH 7.4. 1H-15N correlation spectra were acquired for 16 h. For the characterization of the cis-trans isomerization of BIO-2007817, 0.5 mM of the compound was dissolved in a buffer containing 1% DMSO, 10% D2O, 150 mM NaCl, and 20 mM Tris-HCl pH 7.4. Resonances were assigned using 2D 1H-1H DQF-COSY (ns 32, 256 t1 increments, SW1 13 ppm, 5 h) and 13C-1H HSQC spectra in the aromatic and aliphatic regions (ns 64, 180 t1 increments, SW1 80 ppm, 1 h 16 min). The EXSY experiment was acquired on the same sample using a 2D 1H-1H NOESY pulse sequence (noesygpph19) with a 500 msec mixing time (ns 16, 256 t1 increments, SW1 10 ppm, 3 h). All acquisitions were done at 25 °C at 600 MHz on a Bruker spectrometer equipped with a triple-resonance (1H, 13C, 15N) cryoprobe.
Compound synthesis and characterization
Synthesis of compounds BIO-1966561 (44 mg), BIO-2007817 (19.1 mg), and BIO-2007818 (30.9 mg) was described previously48. See Supplementary Methods in the Supplementary Information for the synthesis of compounds: BIO-1983977, BIO-1979167, BIO-1967660, BIO-1975900, BIO-1975902, BIO-2008218, BIO-2008219, BIO-2006661, and BIO-2006664. Compounds were >95% pure as determined by LC/MS analysis.
Docking
The co-crystal structure of BIO-1975900 was used as a starting point for docking. The protein was prepared using the Protein Preparation Wizard accessible from Maestro interface of the Schrodinger 2019-3 suite at a pH of 7.065. The hydrogen atom positions were added and optimized using OPLS3e force field66–68. The optimization was done in the absence of the ligand. Crystal waters, not interacting with the ligand were removed from the receptor structure. Only hydrogen atoms were minimized while the heavy atoms were kept fixed. Ligands were prepared using LigPrep utility in Schrodinger using OPLS3e force field as well. Protein grid was generated using Receptor Grid Generation utility in Glide for assigning the ligand position in the binding pocket. Docking of ligands was performed using standard precision (SP) ligand docking module in Glide. In the advanced setting of the docking panel, we performed enhanced conformational sampling for ligand conformer generation by increasing it to 4 times. We also checked the expanded sampling option during selection of initial poses so that more poses pass through the initial Glide screens.
The Molecular Mechanics-Generalized Born/Surface Area (MM-GBSA) method is slightly more complex than docking calculations as it computes ligand strain and solvation and therefore adds more accuracy to the protein-ligand binding prediction. MM-GBSA calculations were performed directly from Maestro interface using Prime. The solvation model used for the ligand was VSGB 2.1 with OPLS4 force field. Protein residues were kept rigid for the calculation and the default minimization was used for the protein sampling method.
In organello assays
Assays were performed as described previously36. HeLa cells treated with 10 mM CCCP or DMSO for 3 h were suspended in mitochondrial isolation buffer (MIB) (20 mM HEPES-KOH pH 7.4, 220 mM mannitol, 10 mM KAc and 70 mM sucrose) on ice. Cells were disrupted by nitrogen cavitation, and cell homogenates were centrifuged at 500 × g for 5 min at 4 °C to obtain a post-nuclear supernatant. Cytosolic fractions were collected by two further centrifugation steps for 15 min at 4 °C at 12,000 × g. Mitochondria pellets were suspended in MIB to a concentration of 2 mg/ml and stored at −80 °C.
Five micrograms of CCCP- or DMSO-treated mitochondria were supplemented with a ubiquitination reaction mix (100 nM E1, 200 nM of UbcH7, 5 mM Ub, 1 mM ATP, 5 mM MgCl2 and 50 μM TCEP in MIB), 200 nM of recombinant WT or mutant human parkin and 10 µM of BIO2007817, BIO2007818 or some MIB buffer with equivalent DMSO concentration into a final 10 μl reaction volume. After a 10-min incubation at 37 °C, reactions were stopped with reducing SDS-PAGE loading buffer and separated by SDS-PAGE gels. Proteins were transferred to PVDF membrane for Western blotting analysis. Membranes were blocked with 5% BSA in TBS-T (0.1% Tween 20) and incubated with rabbit anti-Mfn2 (1:2000, mAb D2D10, Cell Signaling), mouse anti-parkin (1:40,000, mAb Prk8, Cell Signaling), or rabbit anti-VDAC (1:5000, mAb D73D12, Cell Signaling) diluted in TBS-T with 3% BSA. Membranes were washed with TBS-T and incubated with HRP-coupled horse anti-mouse or goat anti-rabbit IgG antibodies (1:10,000, Cell Signaling). Detection was performed with Clarity Lightning ECL (Bio-Rad) and images acquired with a ImageQuant LAS 500 (GE Healthcare). The fraction of Mfn2 ubiquitination was quantified from western blot signals using Fiji69, according to the formula f = (total ubiquitination)/(unmodified + total ubiquitination).
Mitophagy assay
Mitophagy was assessed using flow cytometry analysis of mitochondrially targeted mitoKeima as previously described19,36. Briefly, U2OS cells stably expressing an ecdysone-inducible mitoKeima were induced with 10 µM ponasterone A, transfected with GFP-parkin or parkin mutants for 20 h, and treated (or not) with 20 µM CCCP for 4 h. Where indicated, cells were pre-treated with DMSO or 2.5 µM BIO-2007817 1 h prior to addition of CCCP. Cells were trypsinized, washed and resuspended in PBS prior to fluorescence‐activated cell sorting (FACS) analysis on a Thermo Attune NxT cytometer (NeuroEDDU Flow Cytometry Facility, McGill University). Lysosomal mitoKeima was measured ratiometrically using excitation at 405 nm and 561 nm (Supplementary Fig. 4). For each sample, 100,000 events were collected, and single GFP-parkin-positive cells were gated for mitoKeima. Data were analyzed using FlowJo v10.7.2 (Tree Star). The percentage of cells with an increase in the 405:561 nm ratio (i.e., cells within the upper gate) was quantified and normalized to the percentage observed in GFP-WT-parkin CCCP treated samples for each replicate.
Supplementary information
Source data
Acknowledgements
We thank Hao Liu for assistance with HDX-MS experiments. This work was supported by a Michael J. Fox Foundation grant (MJFF-019029) to K.G., Canadian Institutes of Health Research grants (FDN 154301) to E.A.F., (PJT 186189) to J.F.T. and (FDN 159903) to K.G. and Canada Research Chairs awards to E.A.F., J.F.T., and K.G. X-ray data was collected at beamline CMCF-ID at the Canadian Light Source, which is supported by the Canada Foundation for Innovation, the Natural Sciences and Engineering Research Council, the National Research Council, the Canadian Institutes of Health Research, the Government of Saskatchewan, and the University of Saskatchewan.
Author contributions
V.S. performed autoubiquitination and ITC assays. J.F.T. performed NMR analysis. A.H., J.F., and S.C. designed and performed the Ub-VS and HDX-MS experiments. V.S. and R.F. screened crystals and V.S. solved the X-ray crystal structure. E.S. prepared compounds and structure-activity relationship analysis. P.M. conducted autoubiquitination assays for structure-activity relationships. N.B. performed docking and energy analysis. N.C. performed in organello assays. T.G. performed and analyzed the mitophagy assays. V.S., J.F.T., and K.G. wrote the paper. E.A.F., W.D.H., L.F.S., J.F.T., and K.G. oversaw the research.
Peer review
Peer review information
Nature Communications thanks Shourya Roy Burman and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Source data are provided with this manuscript as a Source data file. A reporting summary for this article is available as a Supplementary Information file. Uncropped and unprocessed scans of gels and blots are available in the Source data file. Atomic coordinates and structure factors for X-ray crystal structures are available from the Protein Data Bank: 5N2W, 6GLC, 8W31. Source data are provided with this paper.
Competing interests
Eric Stefan, Nupur Bansal, Adelajda Hadzipasic, Jing Fang, Paramasivam Murugan, Shimin Chen, Warren D. Hirst, and Laura F. Silvian are or were employees and shareholders of Biogen Inc. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Laura F. Silvian, Email: laura.silvian@biogen.com
Kalle Gehring, Email: kalle.gehring@mcgill.ca.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51889-3.
References
- 1.Moehlman, A. T. & Youle, R. J. Mitochondrial quality control and restraining innate immunity. Annu. Rev. Cell Dev. Biol.36, 265–289 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Agarwal, S. & Muqit, M. M. K. PTEN-induced kinase 1 (PINK1) and Parkin: unlocking a mitochondrial quality control pathway linked to Parkinson’s disease. Curr. Opin. Neurobiol.72, 111–119 (2022). [DOI] [PubMed] [Google Scholar]
- 3.Hattori, N. & Mizuno, Y. Twenty years since the discovery of the parkin gene. J. Neural Transm.124, 1037–1054 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Konig, T. & McBride, H. M. Mitochondrial-derived vesicles in metabolism, disease, and aging. Cell Metab.36, 21–35 (2024). [DOI] [PubMed] [Google Scholar]
- 5.Sliter, D. A. et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature561, 258–262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matheoud, D. et al. Intestinal infection triggers Parkinson’s disease-like symptoms in Pink1(−/−) mice. Nature571, 565–569 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Mouton-Liger, F. et al. Parkin deficiency modulates NLRP3 inflammasome activation by attenuating an A20-dependent negative feedback loop. Glia66, 1736–1751 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panicker, N. et al. Neuronal NLRP3 is a parkin substrate that drives neurodegeneration in Parkinson’s disease. Neuron110, 2422–2437.e9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pawlyk, A. C. et al. Novel monoclonal antibodies demonstrate biochemical variation of brain parkin with age. J. Biol. Chem.278, 48120–48128 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Tokarew, J. M. et al. Age-associated insolubility of parkin in human midbrain is linked to redox balance and sequestration of reactive dopamine metabolites. Acta Neuropathol.141, 725–754 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsieh, C. H. et al. Functional impairment in miro degradation and mitophagy is a shared feature in familial and sporadic Parkinson’s disease. Cell Stem Cell19, 709–724 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trempe, J. F. & Gehring, K. Structural mechanisms of mitochondrial quality control mediated by PINK1 and Parkin. J. Mol. Biol.435, 168090 (2023). [DOI] [PubMed] [Google Scholar]
- 13.Wenzel, D. M., Lissounov, A., Brzovic, P. S. & Klevit, R. E. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature474, 105–108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondapalli, C. et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol.2, 120080 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaugule, V. K. et al. Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J.30, 2853–2867 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trempe, J. F. et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science340, 1451–1455 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Riley, B. E. et al. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat. Commun.4, 1982 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wauer, T. & Komander, D. Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J.32, 2099–2112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sauve, V. et al. A Ubl/ubiquitin switch in the activation of Parkin. EMBO J.34, 2492–2505 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koyano, F. et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature510, 162–166 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Kane, L. A. et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol.205, 143–153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vives-Bauza, C. et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl Acad. Sci. USA107, 378–383 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narendra, D. P. et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol.8, e1000298 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuda, N. et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol.189, 211–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lazarou, M., Jin, S. M., Kane, L. A. & Youle, R. J. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev. Cell22, 320–333 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar, A. et al. Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis. EMBO J.34, 2506–2521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kazlauskaite, A. et al. Binding to serine 65-phosphorylated ubiquitin primes Parkin for optimal PINK1-dependent phosphorylation and activation. EMBO Rep.16, 939–954 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ordureau, A. et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell56, 360–375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sauvé, V. et al. Mechanism of parkin activation by phosphorylation. Nat. Struct. Mol. Biol.25, 623–630 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Gladkova, C., Maslen, S. L., Skehel, J. M. & Komander, D. Mechanism of parkin activation by PINK1. Nature559, 410–414 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Condos, T. E. et al. Synergistic recruitment of UbcH7~Ub and phosphorylated Ubl domain triggers parkin activation. EMBO J.37, e100014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narendra, D., Tanaka, A., Suen, D. F. & Youle, R. J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol.183, 795–803 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLelland, G. L., Soubannier, V., Chen, C. X., McBride, H. M. & Fon, E. A. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J.33, 282–295 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vincow, E. S. et al. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc. Natl Acad. Sci. USA110, 6400–6405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yi, W. et al. The landscape of Parkin variants reveals pathogenic mechanisms and therapeutic targets in Parkinson’s disease. Hum. Mol. Genet28, 2811–2825 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang, M. Y. et al. Structure-guided mutagenesis reveals a hierarchical mechanism of Parkin activation. Nat. Commun.8, 14697 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevens, M. U. et al. Structure-based design and characterization of Parkin-activating mutations. Life Sci. Alliance6, e202201419 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silvian, L. F. PINK1/Parkin pathway activation for mitochondrial quality control - which is the best molecular target for therapy? Front Aging Neurosci.14, 890823 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hertz, N. T. et al. A neo-substrate that amplifies catalytic activity of Parkinson’s-disease-related kinase PINK1. Cell154, 737–747 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chin, R. M. et al. Pharmacological PINK1 activation ameliorates Pathology in Parkinson’s Disease models. Preprint at bioRxiv10.1101/2023.02.14.528378 (2023).
- 41.Bingol, B. et al. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature510, 370–375 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Moskal, N. et al. An AI-guided screen identifies probucol as an enhancer of mitophagy through modulation of lipid droplets. PLoS Biol.21, e3001977 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shiba-Fukushima, K. et al. A cell-based high-throughput screening identified two compounds that enhance PINK1-Parkin signaling. iScience23, 101048 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garofalo, A. W., Johnston, J. & Fatheree, P. R. Triazole Benzamide Derivatives and the Compositions and Methods of Treatment Regarding the Same (ed. Office, U.S.P.a.T.) (2017).
- 45.Johnston, J. & Garofalo, A. W. Pyradazinone Derivatives and the Compositions and Methods of Treatment Regarding the Same (ed. Office, U.S.P.a.T.) (2017).
- 46.Springer, W., Fiesel, F. C. & Caulfield, T. R. Small Molecule Activators of Parkin Enzyme Function (ed. Office, U.S.P.a.T.) (2018).
- 47.Traynor, R. et al. Design and high-throughput implementation of MALDI-TOF/MS-based assays for Parkin E3 ligase activity. Cell Rep. Methods4, 100712 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shlevkov, E. et al. Discovery of small-molecule positive allosteric modulators of Parkin E3 ligase. iScience25, 103650 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sauvé, V. et al. Structural basis for feedforward control in the PINK1/Parkin pathway. EMBO J.41, e109460 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fakih, R., Sauvé, V. & Gehring, K. Structure of the second phosphoubiquitin-binding site in parkin. J. Biol. Chem.298, 102114 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borodovsky, A. et al. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J.20, 5187–5196 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wauer, T., Simicek, M., Schubert, A. & Komander, D. Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature524, 370–374 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Safadi, S. S. & Shaw, G. S. A disease state mutation unfolds the parkin ubiquitin-like domain. Biochemistry46, 14162–14169 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Safadi, S. S., Barber, K. R. & Shaw, G. S. Impact of autosomal recessive juvenile Parkinson’s disease mutations on the structure and interactions of the parkin ubiquitin-like domain. Biochemistry50, 2603–2610 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Broadway, B. J. et al. Systematic functional analysis of PINK1 and PRKN coding variants. Cells11, 2426 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schreiber, S. L. The rise of molecular glues. Cell184, 3–9 (2021). [DOI] [PubMed] [Google Scholar]
- 57.Bekes, M., Langley, D. R. & Crews, C. M. PROTAC targeted protein degraders: the past is prologue. Nat. Rev. Drug Discov.21, 181–200 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.St-Cyr, D. et al. Identification and optimization of molecular glue compounds that inhibit a noncovalent E2 enzyme-ubiquitin complex. Sci. Adv.7, eabi5797 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berndsen, C. E. & Wolberger, C. A spectrophotometric assay for conjugation of ubiquitin and ubiquitin-like proteins. Anal. Biochem.418, 102–110 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kabsch, W. Xds. Acta Crystallogr D. Biol. Crystallogr66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. Biol. Crystallogr66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D. Biol. Crystallogr68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr D. Biol. Crystallogr66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sastry, G. M., Adzhigirey, M., Day, T., Annabhimoju, R. & Sherman, W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des.27, 221–234 (2013). [DOI] [PubMed] [Google Scholar]
- 66.Shivakumar, D. et al. Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J. Chem. Theory Comput.6, 1509–1519 (2010). [DOI] [PubMed] [Google Scholar]
- 67.Harder, E. et al. OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput.12, 281–296 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Jorgensen, W. L., Maxwell, D. S. & Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc.118, 11225–11236 (1996). [Google Scholar]
- 69.Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data are provided with this manuscript as a Source data file. A reporting summary for this article is available as a Supplementary Information file. Uncropped and unprocessed scans of gels and blots are available in the Source data file. Atomic coordinates and structure factors for X-ray crystal structures are available from the Protein Data Bank: 5N2W, 6GLC, 8W31. Source data are provided with this paper.