Abstract
Autosomal Dominant Polycystic Kidney Disease (ADPKD) is the most prevalent kidney genetic disorder, producing structural abnormalities and impaired function. This research investigates its evolution on mouse models, utilizing a combination of histology imaging, Computed Tomography (CT) and Magnetic Resonance Imaging (MRI) to evaluate its progression thoroughly. ADPKD has been induced in mice via PKD2 gene knockout, followed by image acquisition at different stages. Histology data provides two-dimensional details, like the cystic area ratio, whereas CT and MRI facilitate three-dimensional temporal monitoring. Our approach allows to quantify the affected tissue at different disease stages through multiple quantitative metrics. A pivotal point is shown at approximately ten weeks after induction, marked by a swift acceleration in disease advancement, and leading to a notable increase in cyst formation. This multimodal strategy augments our comprehension of ADPKD dynamics and suggests the possibility of employing higher-resolution imaging in the future for more accurate volumetric analyses.
Subject terms: Image processing, Experimental models of disease
Studies of ADPKD progression in mouse models using histology, CT, and MRI to monitor disease evolution reveals a significant acceleration in cyst formation around ten weeks post-induction, underscoring the utility of multimodal imaging for detailed analysis of disease stages.
Introduction
Polycystic Kidney Disease (PKD) is the most prevalent hereditary kidney disorder1. It is a condition that impairs renal function and can ultimately lead to mortality. Characterized by the emergence of fluid-filled cysts within the kidneys, these cysts cause structural distortions, enlargement, and dysfunction. Autosomal Dominant Polycystic Kidney Disease (ADPKD) is the most frequent form of PKD, where cysts predominantly form in the distal nephron segments and collecting ducts2. This hereditary disorder is normally diagnosed in humans through visual inspection of images such as ultrasound or magnetic resonance imaging (MRI), which identifies a multitude of cysts throughout the kidney. This method generally detects PKD in its advanced stages, where the manifestations are more pronounced3,4. ADPKD is only detected at an early stage (1 or 2 cysts) through echography in children when they are screened due to their parents presenting the disease. Otherwise, it normally goes unnoticed until a later stage. The impact of PKD on human kidneys has been extensively studied, often involving the extraction of the kidney’s total volume from segmented Computed Tomography (CT) or MRI scans to assess the disease-induced deformations across the entire organ5–7. The late detection of PKD complicates effective treatment, and a succesful cure has not yet been found, with tolvaptan being able to delaying to an extent the disease development. This has driven in-depth research of the disease in animal models to understand its earlier steps in evolution. This allows the evaluation of PKD on more controllable specimens, opening the doors for different types of analysis. These include the use of histological imaging to measure the impact of the disease in mice at different stages8–11. These images allow to visualize small-scale modifications in the kidney, such as the initial stages in cyst formation that would not be visible with the previous techniques in as much detail. However, histology imaging does not allow for longitudinal monitoring, as it necessitates organ extraction. Despite advancements in integrating histology with techniques like MRI12, the primary focus of 3D imaging has been on assessing the total kidney volume. Therefore, we advocate for a comprehensive study using mouse models, incorporating histology, CT, and MRI to evaluate PKD’s effects at multiple development stages. Our method aims for a more subtle segmentation of affected kidney areas, enhancing the histological data. Combining the information provided by diverse imaging modalities, we aim to elucidate the dynamics of ADPKD and delineate the key stages of structural changes triggered by the disease.
In this study, we conducted a comprehensive analysis of Autosomal Dominant Polycystic Kidney Disease (ADPKD) progression in a mouse model using a combination of histology, Computed Tomography (CT), and Magnetic Resonance Imaging (MRI). The validation of ADPKD induction was confirmed through significant reductions in PKD2 expression and corresponding increases in kidney-to-body weight ratio, cystic index, and serum urea nitrogen levels. Our results highlight a critical acceleration in cyst formation starting at 10 weeks post-induction, with pronounced changes observed at 12 and 16 weeks. CT provided detailed volumetric insights into kidney and diseased region growth, while MRI allowed for the quantification of individual cyst numbers and areas, capturing the rapid progression of the disease. These findings validate the successful induction of ADPKD and demonstrate the value of multimodal imaging in tracking disease progression, particularly during the crucial 8 to 12-week window, thus offering potential for early therapeutic interventions.
Results
This section presents the findings from our investigation into the progression of ADPKD in a mouse model using a combination of histological analysis, CT, and MRI imaging techniques. We aimed to elucidate the temporal development of ADPKD, focusing on the onset and growth of cysts within the kidney tissue, changes in kidney volume, and the overall impact of these alterations on renal function. The results are organized to first detail the histological observations, which provide a microscopic view of the impact of the disease on kidney structure over time. Subsequently, we explore the contributions of CT and MRI imaging in offering a macroscopic perspective on the progression of the disease, including the evolution of kidney volume and the dynamics of cyst formation. Through this multifaceted approach, we aim to offer a comprehensive understanding of the evolution of ADPKD and its implications for kidney health.
Critical increase in cyst formation and impact on renal function post-induction
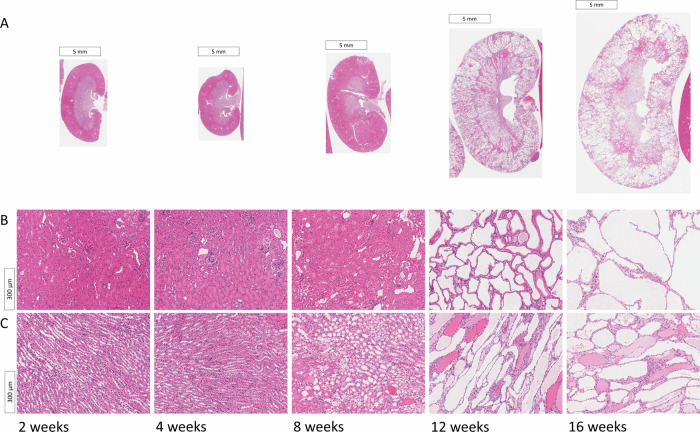
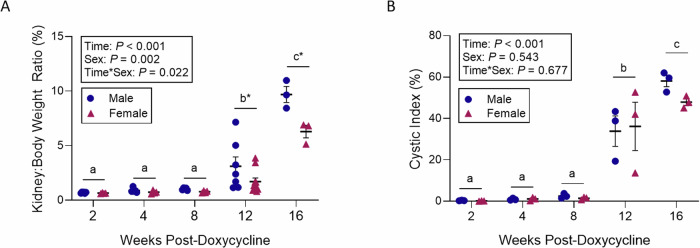
Histological examination of PKD2 KO mice, sacrificed at 2, 4, 8, 12, and 16 weeks following PKD2 gene deletion via doxycycline treatment, show cysts starting to be noticeable in the images at around 8 weeks, but a marked increase in cystogenesis was revealed at 12 weeks post-induction. Hematoxylin and eosin (H&E) stained sections of the kidneys displayed a pronounced cystic phenotype from the 12-week mark, with observable cyst enlargement continuing through to the 16-week point (Fig. 1). This trend in cyst development was quantitatively supported by analyzes of the kidney-to-body weight ratio (Fig. 2A) and the cystic index (Fig. 2B). A significant interaction was noted between the time post-induction and the sex of the mice concerning the kidney-to-body weight ratio (p = 0.022), with significant differences due to both time (p < 0.001) and sex (p = 0.002). The ratio remained consistent at 2, 4, and 8 weeks, then escalated to roughly threefold and tenfold at 12 and 16 weeks, respectively, in comparison to the initial time points (Fig. 2A). Notably, sex-specific variations in the kidney-to-body weight ratio were only apparent at 12 and 16 weeks, showing an 85% increase at 12 weeks (p < 0.001) and a 54% increase at 16 weeks (p < 0.001) in male mice relative to female mice (Fig. 2A). The cystic index showed no significant change during the first 8 weeks; however, it surged by approximately 35-fold and 53-fold at 12 and 16 weeks, respectively, compared to the baseline measurements (Fig. 2B).
Fig. 1. ADPKD visual evolution in histology images.
Cystic formations, identifiable as white areas, exhibit progressive enlargement correlating with disease advancement. A Representative kidney sections stained with hematoxylin and eosin (H&E) across longitudinal studies at 2, 4, 8, 12, and 16 weeks post-doxycycline treatment. B cortex. C medulla.
Fig. 2. Kidney to weight ratio and cystic index.
Quantitative assessment of cyst progression in Pax8rtTA; TetO-Cre; PKD2fl/fl mice following PKD2 gene knockout. A Kidney to body weight ratio (n = 3–10) and B cystic index (n = 3) in Pax8rtTA; TetO-Cre; PKD2fl/fl at 2, 4, 8, 12 and 16 weeks post-doxycycline treatment. Mean values were evaluated for statistically significant differences with the use of a two-factor ANOVA, and all pairwise comparisons were run with a Bonferroni adjustment. Data are expressed as mean values ± SEM. Distinct lowercase letters denote differences across time points, while asterisks highlight differences between sexes at specific times (P < 0.05).
Survival analysis was conducted for PKD2 KO mice using the Kaplan–Meier method to assess the survival probabilities of male and female mice treated with oral doxycycline between 4 and 6 weeks of age. The median survival time was found to be 16 weeks for male mice and 15 weeks for female mice, with no significant differences in survival rates between both sexes (p = 0.980) as shown in Fig. 3.
Fig. 3. Survival analysis results.
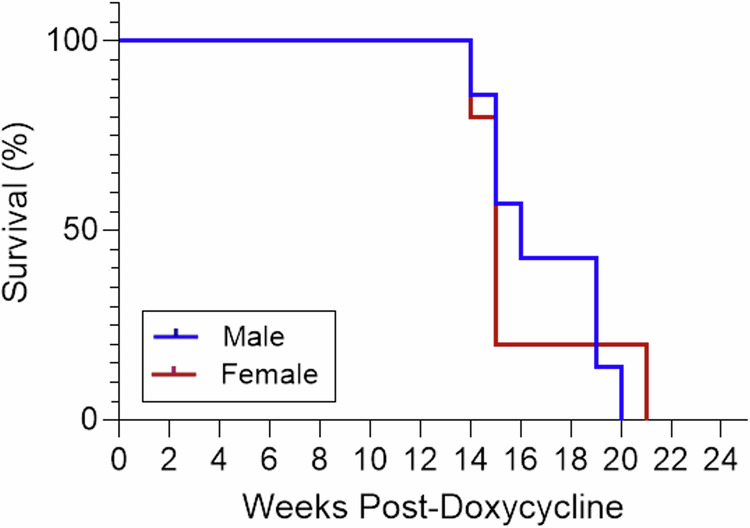
Kaplan–Meier survival analysis for male and female Pax8rtTA; TetO-Cre; PKD2fl/fl mice administered 2 mg/mL doxycycline from ages 4 to 6 weeks. Median survival times were 16 weeks for males (n = 7) and 15 weeks for females (n = 4), respectively.
Renal PKD2 Expression
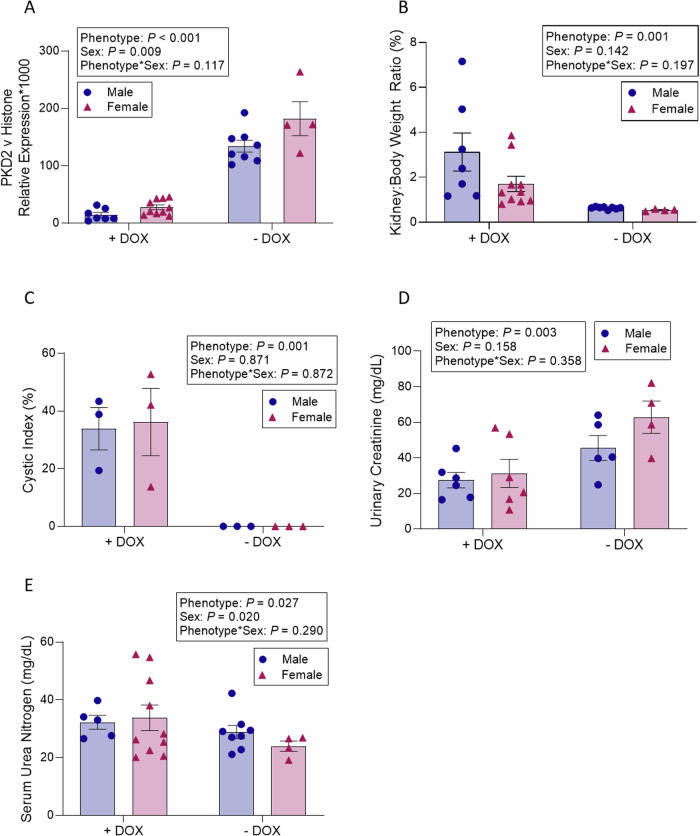
Consistent with expectations, PKD2 expression in the kidneys was found to be 87% lower in PKD2 KO mice relative to their age-matched controls (p < 0.001). Furthermore, male mice exhibited a 29% reduction in renal PKD2 expression compared to female mice (p = 0.009), irrespective of the phenotype, indicating sex-related variations in baseline renal PKD2 expression (Fig. 4A).
Fig. 4. Comparisons of APKD-induced mice vs control mice.
Analysis of Pax8rtTA; TetO-Cre; PKD2fl/fl mice administered oral doxycycline versus untreated, age-matched controls 12 weeks post-doxycycline treatment. A Shows PKD2 expression levels (n = 4–10), B details the kidney-to-body weight ratio (n = 4–10), C presents the cystic index (n = 3), D lists urinary creatinine (n = 4–6) and E outlines serum urea nitrogen levels (n = 4–10). Mean values were evaluated for statistically significant differences with the use of a two-factor ANOVA, and all pairwise comparisons were run with a Bonferroni adjustment. Data are expressed as mean values ± SEM. DOX denotes doxycycline.
Regarding the analysis of cyst progression and renal biomarkers in PDK2 KO mice compared to their age-matched controls 12 weeks after doxycycline treatment, control mice not treated with doxycycline remained healthy and exhibited no cystic development. Twelve weeks post-doxycycline treatment, the kidney-to-body weight ratio in PKD2 KO mice increased fourfold relative to their age-matched controls (p = 0.001) (Fig. 4B). Similarly, the cystic index significantly rose in PKD2 KO mice compared to the controls (p = 0.001) (Fig. 4C). No significant differences were noted between male and female mice in terms of the kidney-to-body weight ratio (p = 0.142) or the cystic index (p = 0.871). A decline in kidney function was evident in PKD2 KO mice, indicated by a 46% reduction in urinary creatinine excretion (p = 0.158) (Fig. 4D). However, serum urea nitrogen levels were 44% higher in male mice than in female mice, irrespective of their phenotype (p = 0.020) (Fig. 4E).
Assessing PKD Progression Through CT and MRI Imaging Techniques
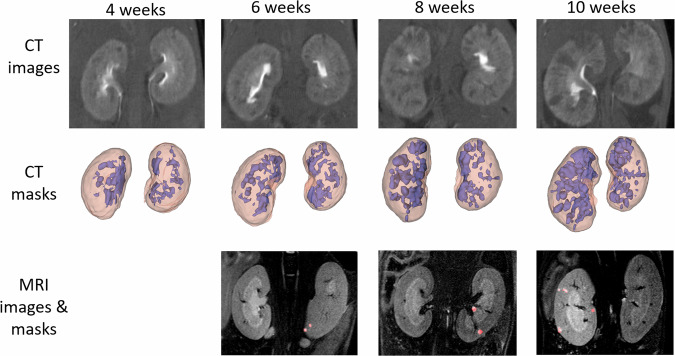
Figure 5 illustrates the visual progression of the disease captured through CT (3D) and MRI (2D) imaging.
Fig. 5. MRI and CT PKD evolution of 3D CT masks and and 2D MRI masks.
Kidney volumes in pink and diseased volumes in purple in the CT images. Cysts marked in red in the MRI images.
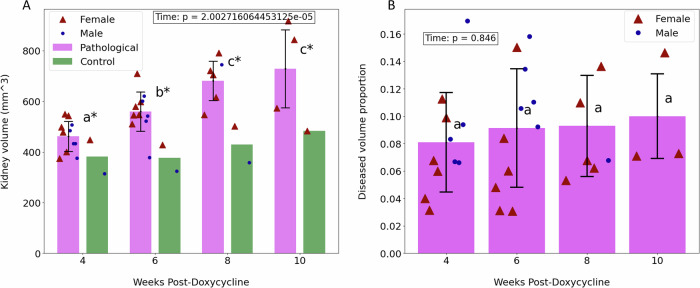
The CT images provided three-dimensional insights into the entire volume impacted by the disease, enabling us to track its progression at each time interval. Initially, comprehensive kidney segmentations were derived from the imaging volumes. Subsequently, the volumes of the diseased regions, identified as the darkest areas, were isolated to examine the extent of organ deterioration over time. Calculations were made for the total volumes of the kidneys (combining both kidneys) as well as for the volumes of diseased tissue at various stages. Figure 6A illustrates the progression of the total kidney volume across four-time points. It is evident in all pathological specimens that the total kidney volume incrementally increases as the disease advances, in contrast to the control group. This graph demonstrates that the average kidney volume of the diseased mice, initially similar to that of the control group, experienced a 60% increase by the final time point. A statistically significant difference was observed in the mean total kidney volume over the time points (p = 1.886 × 10−5).
Fig. 6. Resuls on CT images.
Findings from CT scans obtained at 4, 6, 8, and 10 weeks post-ADPKD induction. Distinct lowercase letters indicate significant changes over time between groups, while an asterisk (*) denotes a significant difference between pathological and control mice. Data are expressed as mean values ± standard deviation. A Illustrates the total kidney volume over time for both pathological mice,in pink, and control mice, in green. B Depicts the ratio of diseased volume to total kidney volume over time.
In addition to the overall increase in kidney size, the growth of cysts was also considered. Figure 6B displays the ratio of diseased volume to total kidney volume at four distinct time points for all pathological mice. There is a general trend of increase in these ratios, although the rise is not markedly pronounced. The comparison of average ratios across different time points yielded a p-value of 0.05588. This value does not provide sufficient statistical evidence to conclude significant differences between the means at each time point.
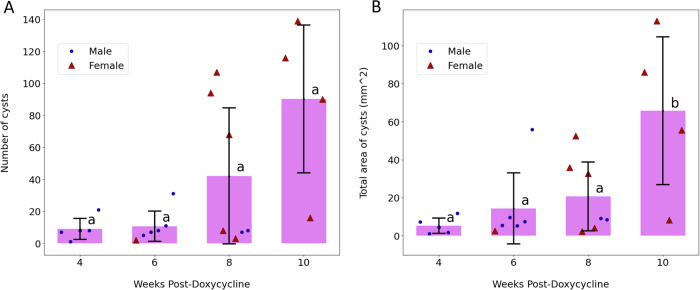
The MRI scans provided distinct insights compared to CT images. While CT imaging captured the entire volume impacted by the disease, MRI scans highlighted individual cysts. Due to the lower z-axis resolution in MRI than in CT, a volume-based analysis was deemed less reliable. Consequently, the number of cysts was quantified as an alternative measure to track ADPKD progression. These individual cysts could also be differentiated better in MRI, since it allows for a more detailed visualization of soft tissues. The data depicted in Fig. 7A suggest a gradual increase in cyst numbers over time, aligning with expectations and mirroring the trend in diseased volume proportion observed in CT scans for the pathological subjects. No statistically significant difference was observed in cyst counts (p > 0.05), which showed only a moderate increase. Additionally, the evolution of the total cyst area was monitored, with Fig. 7B illustrating a notable growth in this area between 8 and 10 weeks (p = 0.0426)
Fig. 7. Resuls on MRI images.
Analysis of MRI scans conducted at 4, 6, 8, and 10 weeks following PKD induction. Distinct lowercase letters indicate significant changes over time across groups. Data are expressed as mean values ± standard deviation. A Displays the count of cysts over time. B Shows the total area occupied by cysts over time, highlighting differences between pathological and control groups.
Discussion
This study presents a comprehensive workflow for assessing the progression of cyst-affected regions in PKD across various stages, utilizing histology, CT, and MRI on mouse models post-ADPKD induction. The methodology integrates several innovative approaches to enhance our understanding of ADPKD.
One of the key strengths of this study is the ability to track cyst development in vivo through CT and MRI imaging. This allows for continuous observation of kidney changes in individual specimens, closely reflecting the progression of the disease in a real-life scenario. Particularly noteworthy is the analysis of the entire kidney in CT scans, where the proportion of diseased tissue is calculated across the whole three-dimensional volume. This approach provides a detailed representation of the extent to which PKD affects kidney function in mice. Histological analysis complements these findings by offering a microscopic view of the structural changes of the organ, thus enriching our understanding of the impact of the disease.
In recent studies on ADPKD development in mouse models, image analysis usually consists on cyst evaluation through histology imaging10,13,14 or employing alternative microscopy techniques, such as confocal microscopy, to gather additional structural data15. Our study introduces a novel approach by combining the widely used animal histology analysis with both CT and MRI imaging of the complete organ, techniques more commonly applied in human patient analysis16,17. The integration of these three-dimensional modalities allows us to provide a comprehensive view of cyst formation throughout the entire kidney, complementing large-scale data with high-resolution details obtained from histology. We have aimed to extract additional information from these three-dimensional images beyond overall kidney volume, observing the inner structure of the organ and quantifying the affected tissue within it. This combined approach, along with molecular and urinary biomarker analysis, has provided us a deeper insight into ADPKD development in the animal model, that can serve as a foundation for further research into the mechanisms of the disease.
The examination of mice 12 weeks after doxycycline treatment revealed a significant decrease in PKD2 expression in the treated group, confirming the effectiveness of the PKD2 inactivation method. Significant changes were observed in several key indicators, including the kidney-to-body weight ratio, cystic index, and serum urea nitrogen levels, with variations between sexes also noted. This fact correlates with previous studies in mice and humans where it has been described that Male gender is a risk factor for progression of autosomal-dominant polycystic kidney disease18. The results collectively validate the induction of ADPKD and its pronounced effects on kidney parameters.
The use of various imaging modalities provides a multifaceted view of ADPKD progression. The kidney-to-body weight ratio analysis over time highlights a critical increase in organ mass at 12 weeks, following a period of relative stability, with a more pronounced effect in males, matching again with the results of18. This increase correlates with the surge in cyst development, as indicated by the cystic index, suggesting a significant contribution of cyst growth to the overall kidney enlargement. This pattern is consistent across sexes, supported by survival analysis findings.
CT imaging reveals a gradual enlargement of the kidney volume over time, hinting at ongoing cyst formation and organ deformation before the 12-week mark. However, the proportion of diseased to total kidney volume remains relatively stable, suggesting that the growth of diseased regions is proportional to overall kidney enlargement. In contrast, MRI data indicates a sharp increase in cyst numbers and total area between 8 and 10 weeks, aligning with the accelerated phase of disease progression observed in histological analysis post-12 weeks.
This study successfully demonstrates the induction of ADPKD in mice and provides detailed documentation of the progression of the disease using a combination of imaging techniques. The collective evidence points to a pivotal moment in ADPKD development around the 10-week mark, where the progression of the disease accelerates, leading to significant renal deterioration. This critical phase is clearly captured in the histological images, showcasing the rapid overtaking of kidney tissue by cysts from this stage onward, highlighting the critical nature of this phase in ADPKD progression. Interestingly, this analysis allowed us to identify ADPKD progression several weeks before observing biomarkers that define renal failure providing a powerful tool to assess disease progression in mice and a sensitve method to study therapeutical treatments before any sign of the disease can be detected in blood and urine.
Our study encountered several limitations that warrant consideration. One of them was the number of samples per group. Given time and resource constraints, only a certain number of animals were able to be breed for the study. For histology imaging, animals needed to be sacrificed at the specific time points in order to harvest their kidneys. Therefore, the decision was taken to only process the necessary amount to obtain balanced groups of a few individuals for histology and molecular and urinary biomarker analysis. This allowed for a separate evaluation of male and female individuals, to assess the effects of sex in the observed parameters. The study could have benefited from larger sample sizes, which will be taken into account for future developments. This would provide more robust conclusions to the analyzes.
For CT and MRI imaging, changing the animal’s location from their initial breeding facility seemed to negatively influence their survival, resulting in an earlier mortality. Given our lower total sample size for this second type of analyzes due to this, we decided to observe together individuals from both sexes, taking also into account that disease quantification in the histology images through cystic index had shown no clear differences between male and female. This way, we mitigated the low number of specimens, aiming for more representative groups. A larger number of samples per time point would be desirable to obtain more robust estimations on disease development. This is, however, much more feasible for human scans, while mouse models need to be precisely controlled, since ADPKD development is notably fast.
CT acquisitions provided an overall volumetric view of the mouse body, from where the kidneys were specifically segmented. The available resolution allowed to segment the full kidney and the damaged area, which was sufficient to track ADPKD development in the whole volume. For this, manual checking of all segmentations was performed to ensure the measurements were as accurate as possible. More precise volumetric cyst segmentation could be obtained using high resolution μCT, but this would most likely imply extracting the kidneys, as shown in19. For the purposes of our study, the focus was on longitudinal tracking, being able to use the same animals for different acquisitions, and obtaining overall measurements.
MRI imaging was added to complement the CT volumes. These images were commissioned as a service with a limited amount of time. Given volumetric data was already present in the form of CT volumes, a lower number of slices was acquired in this case, which did not allow for accurate volumetric reconstruction. This led to the acquisition of non-volumetric parameters like number of cysts. A more detailed 3D MRI reconstruction could be able to provide more detailed measurements for the cysts moving forward.
While the PC2 (polycystin-2) protein levels are an important factor in the pathophysiology of ADPKD, their analysis was not included in our current study. The observation of PKD2 low expression was the main factor to determine that ADPKD had been correctly induced on the animals. This was complemented by low creatinine levels, and high serum urea nitrogen, as well as kidney to body weight ratios. These, together with the image analysis measurements, covered a multitude of aspects of ADPKD effects. PC2 levels would have added an additional molecular analysis that provides more information, and will be considered for further studies.
The temporal scope of CT and MRI imaging was restricted to a maximum of 10 weeks, beyond which the survival rate of the subjects declined significantly. This constraint prevented the alignment of imaging data with the final histological and biomarker analyzes conducted across all data sets at identical time points. Extending the observation period could facilitate more accurate correlations between the various modalities, enhancing the comprehensiveness of our findings.
Another potential limitation to consider is the variability in disease progression among individual subjects, which may introduce variability in the imaging and histological data. This heterogeneity can complicate the interpretation of results and the establishment of definitive timelines for disease progression. Future studies may benefit from larger sample sizes to mitigate the impact of individual variability and improve the robustness of the findings.
Additionally, the reliance of the study on a single genetic model of ADPKD may limit the generalizability of the results. Different genetic strains or models might exhibit variations in disease progression and response to treatment, suggesting that findings from this study may not be universally applicable across all forms of the disease. Expanding the research to include multiple models could provide a more comprehensive understanding of their various manifestations.
Overall, in this research we conducted a comprehensive study on the effects of ADPKD in mice after inducting a PKD2 gene knockout. Initial assessments confirmed the successful onset of ADPKD through various kidney parameter measurements. We then utilized histology, CT, and MRI to observe and document the structural changes in the kidneys over time. Our findings reveal a gradual progression of ADPKD up to the 10-week mark post-induction, after which the severity of the disease escalates significantly, leading to rapid and substantial alterations in kidney structure and function.
This study provided an in-depth look at the damaging effects of ADPKD, leveraging multiple imaging modalities to offer a comprehensive perspective on disease progression. Through the analysis of kidney condition at different stages and employing various metrics, we have edged closer to unraveling the complex mechanisms driving ADPKD progression.
For future endeavors, the integration of advanced high-resolution 3D imaging techniques, such as Light Sheet Fluorescence Microscopy, could enhance the precision of volumetric analyzes of cysts, enabling deeper exploration of their shapes and distribution patterns. Additionally, the use of higher-resolution MRI could provide a more granular temporal and volumetric view of the kidneys in specific mouse models, significantly improving our capacity to monitor disease progression with increased detail and accuracy. For new studies, the focus could be shifted to a higher temporal resolution assessment of the stage starting from 8 weeks after induction. This could provide valuable information about the detailed evolution along this period, as it has been observed that most of ADPKD evolution occurs there. Another interesting direction to be explored is the analysis of fibrotic areas, besides cysts. A similar multimodal evaluation to the one performed during this project could help understand the behavior of the surrounding interstitium, although it would require higher resolution volumetric imaging.
Methods
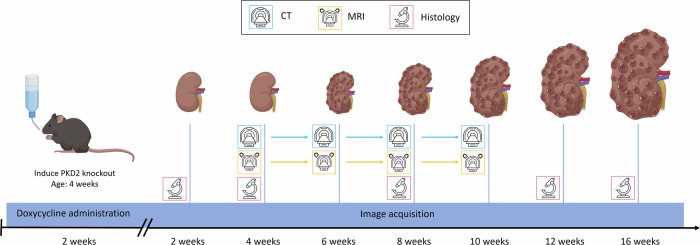
Figure 8 shows a temporal diagram including all the image acquisitions performed during the study.
Fig. 8. Temporal diagram for image acquisitions.
Diagram illustrating the timeline for acquiring in vivo CT and MRI images, along with ex vivo histological specimens and their corresponding images, following PKD induction in mouse models. CT, magnet and microscope models extracted from Vecteezy.com. Kidneys and mouse models created with BioRender.com.
We have complied with all relevant ethical regulations for animal use. Experimental procedures conducted during this project adhered to the European Communities Council Directive (2010/63/EU) and national rules (RD53/2013, ECC/566/2015) for care of laboratory animals. All protocols received approval from the Ethics Committees for Animal Experimentation of each center, “Comité de Ética de Experimentación Animal, CEEA" from Hospital General Universitario Gregorio Maranon and “Comité de Ética de Experimentación Animal, CEEA" from Centro de Investigacion Medica Aplicada (CIMA) Universidad de Navarra, and followed the ARRIVE guidelines (10.1371/journal.pbio.3000410). All efforts were made to minimize the number of animals used, following the 3Rs principle and based in previous studies of the group.
Sample sizes were limited as well by the laboratory capacity to breed the amount of animals and the survivability of the individuals. For the histology analysis, no less than 6 mice per time step were included, attempting to maintain a balanced proportion of males and females, which provides a sufficient series of specimens so that statistical measures can be extracted. For CT and MRI, no less than 4 pathological mice were imaged at each time for statistical measures. Sample numbers were lower at later stages of the disease, due to mortality. 2 additional control specimens were allocated to the CT study to provide a baseline comparison.
Generation and housing of PKD2 knockout mice
Throughout the project, we utilized Pax8rtTA20; TetO-Cre21; PKD2fl/fl mice to generate a conditional inducible knockout (KO) of the polycystic kidney disease 2 (PKD2) gene. These mice were obtained from the Baltimore PKD Research and Clinical Core Center. The transgenic mice possess loxP sites flanking exons 11–13 of the PKD2 gene, which are recognized by Cre recombinase under the control of a tetracycline-responsive promoter element (tetO). Administration of the tetracycline analog, doxycycline, results in the expression of the reverse tetracycline-controlled transactivator protein (rtTA), leading to Cre-mediated recombination and excision of the floxed PKD2 sequence. In this system, the Pax8 promoter directs gene expression to the proximal and distal tubules and collecting ducts of the kidney, allowing for tissue-specific control of PKD2 gene deletion.
The animals were bred in the CIMA Universidad de Navarra animal facilities. All of them were housed in ventilated plastic cages of up to 6 mice per cage in a temperature-controlled room with a 12-hour light-dark cycle. They received a standard laboratory chow diet, and water was supplied ad libitum. PKD2 was deleted in the mice when they reached four weeks of age. This was achieved by providing the mice with drinking water containing 2 mg/mL doxycycline (Sigma D9891-25G), supplemented with 2.5% sucrose, over a duration of 2 weeks. Littermates were randomly assigned to doxycycline-treated and untreated groups, and they were monitored by personnel who were blinded to the treatment assignments.
When the welfare of the animals was compromised, they were sacrificed by using CO2. End point was considered when a weight loss of 20% was observed.
Samples for biomarker analyzes, histological imaging and survival analysis
A first set of pathological mice was selected for histology imaging, survival analysis, and molecular and urinary biomarker analyzes at CIMA Universidad de Navarra. This consisted on 26 male and 26 female pathological Pax8rtTA; TetO-Cre PKD2fl/fl mice, with doxycycline administration triggering PKD2 knockout. Additionally, 8 male and 4 female littermates were used as wild-type controls.
Most of the mice were euthanized at various stages of disease progression (2, 4, 8, 12, and 16 weeks) for histology imaging. Prior to euthanasia and other measurements. Mice were anesthetized with a combination of ketamine and xylazine (90:10 mg/kg body weight) administered via a single intraperitoneal injection. The kidneys were then excised, and selected sections were fixed in 4% formaldehyde for a period of 48 h. They were then embedded in paraffin, sectioned to a thickness of 3 μm, and stained with hematoxylin and eosin (H&E). The stained sections were imaged using an Aperio CS2 slide scanner (Leica Biosystems) at 20× optical magnification.
For kindey to body weight ratio measurements the following specimens were used. At 2 weeks: 3 male and 3 female; at 4 weeks: 3 male and 3 female; at 8 weeks: 3 male and 3 female; at 12 weeks: 7 male and 10 female, and at 16 weeks: 3 male and 3 female
For cystic index calculations, the following specimens were used. 3 male and 3 female for each of the 2, 4, 8, 12 and 16 weeks points.
To later assess urinary parameters, the specimens (both pathological and control) that would be later used for the 12 week point histology images were housed at that moment of time in metabolic cages for 24 h (1–3 mice per cage) for urine collection, with unrestricted access to food and water. The collected urine samples were then frozen at −80 °C until further analysis. Concurrently, whole blood was drawn at the 12-week euthanasia point via retro-orbital bleeding, followed by centrifuging to separate the serum, which was then stored in new tubes at −80 °C.
The specimens used for the PKD2 vs histone expression analysis were as follows. Pathological mice: 7 male and 10 female; control mice: 8 male and 4 female.
The specimens used for the urinary creatinine analyzes were the following. Pathological mice: 6 male and 6 female; control mice: 5 male and 4 female.
Some of the pathological mice that were not used for histopathology collection were separated for a survival analysis (7 males and 4 females).
Samples for CT and MRI Imaging
A second set of mice was used for CT and MRI image acquisition at Hospital General Universitario Gregorio Marañón. This set included 11 pathological animals (5 females, 6 males) and 2 controls (1 female, 1 male). Images were acquired at 4, 6, 8, and 10 weeks after the disease induction.
For both CT and MRI, the female and male mice were observed together as a single group. With these techniques we were looking to extract overall trends of our mouse population, for which this arrangement would be adequate. The number of images in some of the time points was also not enough to perform two separate analyzes.
For the CT study, Iopamiro (300 mg/ml, Iopamidol, Bracco Imaging S.p.A, Italy) was administered intravenously via tail vein (0.25 ml). After 5 minutes of its distribution, CT images were acquired under sevoflurane-inhaled anesthesia using a small-animal ARGUS PET/CT scanner (SEDECAL, Spain), with the following parameters: 340 mA, 40 kV, 360 projections, 8 shots and 200 μm of resolution. Image acquisition was performed at the nephrographic phase of the contrast agent, aiming to enhance the visualization of kidney structures. CT images were reconstructed using a Feldkamp algorithm. The resolution was close to identical in all three dimensions, resulting in images of 514 x 514 x 469 voxels, with a voxel size of 0.00187 cubic mm.
For MRI, animals were scanned using a 7-Tesla Biospec 70/20 scanner (Bruker, Germany) under sevoflurane inhaled anesthesia. Axial T2-weighted images of the abdomen were acquired with a Fast Spin-Echo sequence with TE = 29.73 ms, TR = 1500 ms, RARE factor = 8, averages = 6, a slice thickness of 0.5 mm (15 slices), and the matrix size was 256 × 256 pixels. The acquired MRI images were centered around the kidney area, with a lower number of coronal planes. The pixel resolutions for the X and Y dimensions were almost the same, resulting in approximately 0.01 squared mm per pixel when considering these two dimensions.
Table 1 shows the number of CT and MRI images taken at different time points, both for pathological and control specimens.
Table 1.
Number of CT and MRI images at each time point
| Time((weeks) | 4 | 6 | 8 | 10 |
|---|---|---|---|---|
| Pathological (CT) | 11 | 11 | 7 | 4 |
| Control (CT) | 2 | 2 | 2 | 1 |
| Pathological (MRI) | 6 | 6 | 7 | 4 |
| Control (MRI) | 1 | 1 | 2 | 1 |
Molecular and urinary biomarker analysis in PKD2 knockout mice
We conducted an analysis in the first set of mice at CIMA Universidad de Navarra to compare the expression levels of PKD2 against Histone and assess the impact of our disease induction protocol. Kidney tissue samples from the 12 week euthanized specimens were processed to isolate RNA using the Maxwell® RSC simplyRNA Tissue Kit (Promega; AS1340), with the yield and purity of the extracted RNA being quantified through spectrophotometry (Nanodrop ND 1000). Complementary DNA (cDNA) was synthesized from 1 μg of total RNA in a 21 μL reaction volume using M-MLV Reverse Transcriptase (Invitrogen; 28025-013). We then performed real-time quantitative PCR (qPCR) to measure PKD2 expression, utilizing iQTM SYBR® Green Supermix (BioRad; 1708882) on a CFX96 Real-Time System (BioRad), with PKD2 primer sets referenced against Histone levels.
Additionally, DNA was extracted from ear tissue samples of this set of specimens for genotyping PCR, conducted with the KAPA HotStart Mouse Genotyping Kit (Kapa Biosystems; KK7352). The primer sets designed for amplifying PKD2, rtTA, and Cre recombinase sequences are detailed in Table 2. Electrophoresis of the PCR products was performed on 2% agarose gels for PKD2 and 1.5% for rtTA and Cre, with the resulting bands visualized using the GelDoc XR Imaging System (Bio-Rad). Serum urea nitrogen levels and urinary creatinine concentrations were determined from the available samples at the 12 week time point using an autoanalyzer (Cobas C311, Roche).
Table 2.
List of primers
| Gene | Forward Primer | Reverse Primer | Use |
|---|---|---|---|
| PKD2 | GGGGTTTCCTATGAAGAGTTCCAAG | CTGACAGGCACCTACAGAACAGTG | Genotyping |
| Cre | GCATTACCGGTCGATGCAACGAGTGATGAG | GAGTGAACGAACCTGGTCGAAATGACTGCG | Genotyping |
| rtTA | CCATGTCTAGACTGGACAAGA | CTCCAGGCCACATATGATTAG | Genotyping |
| rtTA | CTAGGCCACAGAATTGAAAGATCT | GTAGGTGGAAATTCTAGCATCATCC | Genotyping Internal Control |
| PKD2 | CTCTTTACCACGTCCGATGA | GGAAACGATGCTGCCAAT | RT-qPCR |
| Histone | AAAGCCGCTCGCAAGAGTGCG | ACTTGCCTCCTGCAAAGCAC | RT-qPCR |
Histological analysis of ADPKD progression and cyst quantification
The histology images acquired were analyzed using Aperio ImageScope software (Leica Biosystems) at 2, 4, 8, 12, and 16 weeks post-doxycycline administration. Figure 1 showcases selected histological images from these intervals, featuring detailed views to illustrate the textural changes in kidney architecture as ADPKD advances.
Representative images of H&E stained longitudinal kidney sections were analyzed to determine the cystic index, which reflects the proportion of the total kidney parenchyma occupied by cysts. The quantification of the cysts was conducted using CystAnalyser software22, an automated tool designed for cyst recognition. For each time point (2, 4, 8, 12, and 16 weeks) and each sex, 3 different histology cuts were chosen. The cystic index of each cut was derived from the average measurements of four randomly chosen fields per sample, with two fields each from the cortex and medulla. In these fields, the total area occupied by cysts was measured and expressed as a percentage of the overall kidney tissue area to calculate the cystic index.
CT and MRI Imaging Techniques for ADPKD Progression Analysis
CT and MRI scans were obtained in vivo from the second set of mice, with a focus on segmenting both kidneys and specifically the areas impacted by the disease. In the course of ADPKD, our CT images reveal the progression of small cysts merging into larger affected regions, disrupting normal kidney functions. This is caused by groups of nearby cysts that grow adjacent to each other, creating areas in the kidney that are formed by only cysts, separated by thin walls. These are detected as homogeneous areas in our CT analysis, which is also supported by observations in histology images of cystic walls breaking so that several nearby cysts become combined to form a larger one (Supplementary Fig. 1). Given the expansive nature of these diseased areas across the kidney, quantifying the volume of affected tissue provides a more comprehensive indicator of the impact of the disease rather than concentrating solely on individual cysts. Therefore, the analysis of CT scans yields the proportion of the volume of the kidney that is diseased, offering a measure of the extent of the effects of ADPKD on the organ.
In recent years, the application of automatic methods for full kidney segmentation has experienced a significant increase, especially regarding deep learning algorithms. This stems from the aim to avoid completely manual segmentation methods, which can be very time-consuming and less reproducible compared to their automatic counterparts. For instance, the application of convolutional networks trained on 2D slices of human abdominal CT images has been proven as an accurate method to segment full PKD-affected kidneys5. Training three-dimensional networks for this task has also provided promising results. For instance,23 presents a 3D fully convolutional neural network with pyramid pooling, which applies pooling operations at different levels of the network to capture local features. Other approaches include the SE-ResNeXT U-Net (SERU)24, a combination of the U-net encoder-decoder main structure25 with SE-Net blocks26, used to recalibrate channel-wise feature responses, and ResNeXT layers27, which use aggregated residual transformations to improve feature extraction. In28, a U-net is trained on 2D CT cuts to segment the kidneys and mark the Region of Interest (ROI), after which a second U-net is applied to segment Renal Masses (RM) inside them. Similarly,29 presents the initial application of an AlexNet classification network30 to keep only the relevant slices from an abdominal CT, to be later segmented using a 2D U-net architecture. All of these have been applied on human CT scans.
Regarding mouse CT processing,19 presents a comparison of U-net derived methods for kidney and cyst segmentation on very high resolution μCT images depicting ex vivo ADPKD rat kidneys. On a more comparable scale to our images,31 shows a pipeline for full organ segmentation on full body mouse CT, based again on a U-net-like network applied slice by slice. On the other hand, in32 a semiautomatic method is presented as a way to compensate for the lower resolution of mouse full body CT when compared to human CT. In general, most of the examples mentioned show the prevalent use of different variations of the U-net or similar arquitectures throughout the literature for kidney segmentation.
In the case of our study, a semi-automatic approach was selected. The first step was the use of a 2D U-net for full kidney segmentation25,33, as it was deemed a reliable method for this task. More complicated architectures, as presented in the previous examples, were normally aimed at human CTs, while the U-net would suit our aim to evaluate kidney volume evolution at the available resolution. The implemented network was based on the ZeroCostDL4Mic Colab Notebooks34, and was utilized to generate preliminary kidney masks. This U-net was trained on manually segmented 2D slices from CT scans of five distinct mice, selecting only those slices featuring visible kidney regions. The training dataset comprised 624 images, with a subset reserved for validation. The model underwent training over 257 epochs, with a batch size of 4 and an initial learning rate set at 0.0003. Its performance was assessed on manually segmented 2D sections of a CT scan from another mouse, achieving an average Intersection over Union (IoU) score of approximately 0.7. The trained model was then applied to all 2D slices from new CT scans to create 3D kidney masks. The scripts for this automatic processing are available online:35 (https://github.com/BSEL-UC3M/CT-PKD-processing). After obtaining these masks, the two largest connected components (corresponding to the kidneys) were kept, and morphological opening and closing operations were performed to remove small oversegmented areas and fill holes. Since our dataset was not overly extensive, we decided to manually check all the produced masks after processing to correct any remaining mis-segmentation and ensure the masks were as accurate as possible, providing a human component to the process, as in32.
Regarding the diseased area segmentation, some automatic methods have been proposed for similar tasks on human CT images, like cortex/medulla segmentation in36 or RM and tumor segmentations in23,24,28. However, to our knowledge, the segmentation of the ADPKD damaged area in mouse CT has not been widely studied. In our case, the observable effects of the disease in our CT images were depicted as low-intensity areas (cysts) growing through the organ and covering a larger area as ADPKD progressed. Therefore, the use of a U-net or similar network was deemed unnecessary. Instead, we applied a nested Otsu filter37. Within the voxels marked by these full kidney masks, two Otsu thresholding steps were implemented to delineate the damaged regions and track their progression over time. Otsu’s method finds the optimal threshold value that maximizes inter-class variance between both classes of voxels, separated by being smaller than or larger or equal than that specific threshold. First, Otsu’s method was applied to differentiate the high-intensity areas, marked by the contrast agent mainly in the renal pelvis. These high intensity areas were discarded. On the remaining voxels, a second Otsu was applied to isolate the darkest parts of the image, which corresponded to the cystic areas, damaged by the disease. This process facilitates the quantification of the evolving affected volume. The accuracy of the segmented diseased regions was further verified and adjusted manually to ensure precise measurement of disease progression. Th analysis encompassed the changes in both the overall kidney volume and the volume of the diseased areas over time in both the pathological and control mice. This approach allowed for an assessment of the rate and magnitude of ADPKD’s impact on the kidneys.
Given the sparse z-slices in MRI scans, volumetric assessments were not possible through this technique. Instead, the focus was on manually identifying individual cysts, which were more discernible in MRI than in CT scans, appearing as distinct, bright, rounded structures that sometimes merged together. This enabled the counting of individual cysts at various stages, offering an alternative metric to track ADPKD progression.
Kidney cyst segmentation in MRI has also been studied, mostly on human patients, by training convolutional neural networks on a set of manual annotations38,39. Previous studies, however, show the use of semiautomatic approaches to evaluate specific MRI slices40. As for CT, these methods are normally developed for human MRI scans. In our case, the limited number of slices and cysts per slice allowed us to manually mark the cysts without the need of performing training, ensuring a more accurate result that would otherwise be acquired with alternative methods.
The 3DSlicer software41 was employed for generating manual segmentations and for the manual correction of 3D masks.
Statistics and reproducibility
Statistical analyzes were applied to all histological data to identify significant differences, with a significance threshold set at (p < 0.05). Survival rates for male and female PKD2 KO mice were compared using Kaplan–Meier survival analysis and the log-rank (Mantel-Cox) test. Disease progression in PKD2 KO and control mice at 12 weeks was assessed with a two-factor ANOVA (phenotype x sex). When the interaction term was not significant, a main effects analysis was conducted. Significant interactions led to a simple effects analysis, with Bonferroni adjustments for pairwise comparisons. Outliers were identified using Grubb’s test, the normality of residuals was checked with the Shapiro-Wilk test, and variance equality was verified using Levene’s test. For data with unequal variances, a logarithmic transformation was applied, and tests were performed on both original and transformed data to ensure consistent results. In instances of significant outliers, analyzes were conducted with and without these data points to ascertain their impact on the results. Outliers were excluded only if their removal improved normality and had a significant effect on the statistical outcomes. For the survival analysis, n = 6 for males and n = 5 for females; for PKD2 expression levels, n = 4–10; for kidney to body weight ratio, n = 3–10; for cystic index, n = 3; for urinary creatinine, n = 4–6, and for serum urea nitrogen, n = 4–10.
Total kidney volume and ratios of diseased volume over total kidney volume from CT images were assessed via a one-factor ANOVA with significance threshold (p < 0.05), with pathological n = 4–11. The evolution of the number of cysts and total cystic area in MRI images were evaluated using a Welch test due to the differences in variance across different time points, with the same significance threshold (p < 0.05) and with n = 4–7.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Materials
Acknowledgements
This work was partially funded by Ministerio de Ciencia, Innovación y Universidades, Agencia Estatal de Investigación, under Grants FPU19/02854 and PID2019-109820RB-I00, MCIN/AEI/10.13039/501100011033/, co-financed by European Regional Development Fund (ERDF), “A way of making Europe" and the Program Retos-colaboración RTC-2017-6600-1. MLSM was supported by project PID2021-128862OB-I00 funded by MCIN /AEI /10.13039/501100011033 / FEDER, UE; CIBER de Salud Mental - ISCIII (project number CB07/09/0031); and Delegación del Gobierno para el Plan Nacional sobre Drogas correspondiente a fondos del Mecanismo de Recuperación, Transformación y Resiliencia de la Unión Europea (EXP2022/008917). Grant 0011-1411- 2019-000074 from proyectos de I+D estratégicos. RIS3 Navarra (Departamento de Desarrollo Económico, Gobierno de Navarra).
Author contributions
P.D.R. performed experiments on the CT and MRI data and wrote the manuscript. N.L.R. provided the CT and MRI images. C.S. performed experiments on biomarkers, obtained the histology images, and analyzed them. R.A. provided the mice and secured funding. M.L.S.M. provided the CT and MRI images, supervised in vivo imaging studies, secured funding. A.M.B. supervised the project, provided computational resources, secured funding, and wrote the manuscript. All the authors revised and edited the manuscript.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Aylin Bircan. A peer review file is available.
Data availability
The numerical data supporting all graphs can be found in Supplementary Data 1. The images used for this project are available upon request to the authors.
Code availability
The Python scripts for the initial steps of CT, automatic processing, are available at35 (https://github.com/BSEL-UC3M/CT-PKD-processing).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-06868-1.
References
- 1.Bergmann, C. et al. Polycystic kidney disease. Nature Reviews Disease Primers 2018 4:14, 1–24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cornec-Le Gall, E., Alam, A. & Perrone, R. D. Autosomal dominant polycystic kidney disease. The Lancet393, 919–935 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Colbert, G. B., Elrggal, M. E., Gaur, L. & Lerma, E. V. Update and review of adult polycystic kidney disease. Disease-a-Month66, 100887 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Odedra, D. et al. Autosomal dominant polycystic kidney disease: role of imaging in diagnosis and management. RadioGraphics43, e220126 (2023). [DOI] [PubMed] [Google Scholar]
- 5.Sharma, K. et al. Automatic segmentation of kidneys using deep learning for total kidney volume quantification in autosomal dominant polycystic kidney disease. Scientific Reports7, 2049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shin, T. Y. et al. Expert-level segmentation using deep learning for volumetry of polycystic kidney and liver. Precision Medicine in Urology61, 555–564 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharbatdaran, A. et al. Deep learning automation of kidney, liver, and spleen segmentation for organ volume measurements in autosomal dominant polycystic kidney disease. Tomography8, 1804–1819 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang, X. et al. Ferroptosis promotes cyst growth in autosomal dominant polycystic kidney disease mouse models. Journal of the American Society of Nephrology32, 2759–2776 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, M. Y. et al. Effects of Suramin on polycystic kidney disease in a mouse model of polycystin-1 deficiency. International Journal of Molecular Sciences23, 8499 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pastor-Soler, N. M. et al. Metformin improves relevant disease parameters in an autosomal dominant polycystic kidney disease mouse model. American Journal of Physiology-Renal Physiology322, F27–F41 (2022). [DOI] [PubMed] [Google Scholar]
- 11.Tham, M. S. et al. Deletion of Aurora kinase A prevents the development of polycystic kidney disease in mice. Nature Communications 2024 15:115, 1–18 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arroyo, J. et al. The genetic background significantly impacts the severity of kidney cystic disease in the Pkd1RC/RC mouse model of autosomal dominant polycystic kidney disease. Kidney International99, 1392–1407 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Talbi, K. et al. The chloride channel cftr is not required for cyst growth in an adpkd mouse model. The FASEB Journal35, e21897 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Dagorn, P. G. et al. A novel direct adenosine monophosphate kinase activator ameliorates disease progression in preclinical models of autosomal dominant polycystic kidney disease. Kidney International103, 917–929 (2023). [DOI] [PubMed] [Google Scholar]
- 15.Li, X.-w. et al. 1-indanone retards cyst development in adpkd mouse model by stabilizing tubulin and down-regulating anterograde transport of cilia. Acta Pharmacologica Sinica44, 406–420 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caroli, A. et al. Diffusion magnetic resonance imaging for kidney cyst volume quantification and non-cystic tissue characterisation in adpkd. European Radiology33, 6009–6019 (2023). [DOI] [PubMed] [Google Scholar]
- 17.Yoo, J. et al. Non-contrast low-dose ct can be used for volumetry of adpkd. BMC nephrology24, 317 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Talbi, K., Cabrita, I., Schreiber, R. & Kunzelmann, K. Gender-dependent phenotype in polycystic kidney disease is determined by differential intracellular ca2+ signals. International Journal of Molecular Sciences22, 6019 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rombolotti, M., Sangalli, F., Cerullo, D., Remuzzi, A. & Lanzarone, E. Automatic cyst and kidney segmentation in autosomal dominant polycystic kidney disease: Comparison of u-net based methods. Computers in Biology and Medicine146, 105431 (2022). [DOI] [PubMed] [Google Scholar]
- 20.Traykova-Brauch, M. et al. An efficient and versatile system for acute and chronic modulation of renal tubular function in transgenic mice. Nature medicine14, 979 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uemura, S., Nagaoka, T., Yokoyama, M., Igarashi, M. & Kishi, M. A simple and highly efficient method to identify the integration site of a transgene in the animal genome. Neuroscience Research80, 91–94 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Cordido, A., Cernadas, E., Fernández-Delgado, M. & García-González, M. A. CystAnalyser: a new software tool for the automatic detection and quantification of cysts in Polycystic Kidney and Liver Disease, and other cystic disorders. PLOS Computational Biology16, e1008337 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang, G. et al. Automatic segmentation of kidney and renal tumor in ct images based on 3d fully convolutional neural network with pyramid pooling module. In 2018 24th International Conference on Pattern Recognition (ICPR), 3790–3795 (IEEE, 2018).
- 24.Xie, X., Li, L., Lian, S., Chen, S. & Luo, Z. Seru: a cascaded se-resnext u-net for kidney and tumor segmentation. Concurrency and Computation: Practice and Experience32, e5738 (2020). [Google Scholar]
- 25.Ronneberger, O., Fischer, P. & Brox, T. U-net: convolutional networks for biomedical image segmentation. Lecture Notes in Computer Science9351, 234–241 (2015). [Google Scholar]
- 26.Hu, J., Shen, L. & Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE conference on computer vision and pattern recognition, 7132–7141 (2018).
- 27.Xie, S., Girshick, R., Dollár, P., Tu, Z. & He, K. Aggregated residual transformations for deep neural networks. In Proceedings of the IEEE conference on computer vision and pattern recognition, 1492–1500 (2017).
- 28.Fatemeh, Z., Nicola, S., Satheesh, K. & Eranga, U. Ensemble u-net-based method for fully automated detection and segmentation of renal masses on computed tomography images. Medical physics47, 4032–4044 (2020). [DOI] [PubMed] [Google Scholar]
- 29.da Cruz, L. B. et al. Kidney segmentation from computed tomography images using deep neural network. Computers in Biology and Medicine123, 103906 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Krizhevsky, A., Sutskever, I. & Hinton, G. E. Imagenet classification with deep convolutional neural networks. Communications of the ACM60, 84–90 (2017). [Google Scholar]
- 31.Schoppe, O. et al. Deep learning-enabled multi-organ segmentation in whole-body mouse scans. Nature communications11, 5626 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen, Z., Wang, H., Cong, F. & Kettunen, L. Low-dose mouse micro-ct image segmentation based on multi-resolution multi-organ shape prior knowledge model. In 2022 International Conference on Computers, Information Processing and Advanced Education (CIPAE), 349–353 (IEEE, 2022).
- 33.Falk, T. et al. U-net: deep learning for cell counting, detection, and morphometry. Nature methods16, 67–70 (2019). [DOI] [PubMed] [Google Scholar]
- 34.von Chamier, L. et al. Democratising deep learning for microscopy with ZeroCostDL4Mic. Nature Communications (2021). [DOI] [PMC free article] [PubMed]
- 35.Delgado-Rodriguez, P. CT-PKD-processing. https://github.com/BSEL-UC3M/CT-PKD-processing.
- 36.Korfiatis, P. et al. Automated segmentation of kidney cortex and medulla in ct images: a multisite evaluation study. Journal of the American Society of Nephrology33, 420–430 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otsu, N. et al. A threshold selection method from gray-level histograms. Automatica11, 23–27 (1975). [Google Scholar]
- 38.Kline, T. L. et al. Automatic semantic segmentation of kidney cysts in mr images of patients affected by autosomal-dominant polycystic kidney disease. Abdominal Radiology46, 1053–1061 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anari, P. Y. et al. Automatic segmentation of clear cell renal cell tumors, kidney, and cysts in patients with von hippel-lindau syndrome using u-net architecture on magnetic resonance images. ArXiv (2023).
- 40.Bae, K. T. et al. Novel methodology to evaluate renal cysts in polycystic kidney disease. American journal of nephrology39, 210–217 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kikinis, R., Pieper, S. D. & Vosburgh, K. G. 3D Slicer: A Platform for Subject-Specific Image Analysis, Visualization, and Clinical Support. Intraoperative Imaging and Image-Guided Therapy 277–289 (2014).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Materials
Data Availability Statement
The numerical data supporting all graphs can be found in Supplementary Data 1. The images used for this project are available upon request to the authors.
The Python scripts for the initial steps of CT, automatic processing, are available at35 (https://github.com/BSEL-UC3M/CT-PKD-processing).