Abstract
Because lentiviruses are able to infect nondividing cells, these viruses might be utilized in gene therapy applications where the target cell does not divide. However, it has been suggested that the introduction of primate lentivirus sequences, particularly those of human immunodeficiency virus, into human cells may pose a health risk for the patient. To avoid this concern, we have constructed gene transfer systems based on a nonprimate lentivirus, bovine immunodeficiency virus. A panel of vectors and packaging constructs was generated and analyzed in a transient expression system for virion production and maturation, vector expression and encapsidation, and envelope protein pseudotyping. Virion preparations were also analyzed for transduction efficiency in a panel of human and nonhuman primary cells and immortalized cell lines. The virion preparations transduced most of the target cell types, with efficiencies up to 90% and with titers of unconcentrated virus up to 5 × 105 infectious doses/ml. In addition, infection of nondividing human cells, including unstimulated hematopoietic stem cells and irradiated endothelial cells, was observed.
One method of transferring therapeutic genes into human cells for disease applications involves inserting the gene into a virus genome and letting the modified virus infect the cells. Because retroviruses integrate their genomes into the target cell chromosomes, retrovirus-mediated gene transfer theoretically provides for long-term expression of the therapeutic gene in the transduced cell. However, the retroviral preintegration complex does not traverse the nuclear membrane pore, requiring target cell division in order for the viral genome to be integrated (27, 44). Lentiviruses also integrate their genomes but do not require target cell division, since the lentivirus preintegration complex can traverse the nuclear membrane pore (8, 26). Therefore, lentiviruses are attractive vehicles for gene therapy applications requiring long-term expression of a therapeutic gene in nondividing cells (for reviews, see references 7 and 47).
Gene transfer systems based on human immunodeficiency virus type 1 (HIV-1) are by far the most developed lentivirus systems, with documented in vivo transduction of rat brain (5, 37, 38), retina (34, 36), muscle and liver (23), and mouse trachea (22). Mouse pancreatic islets were transduced ex vivo and transplanted in vivo, with stable expression of the transgene (17). In addition, HIV-1-derived vectors have transduced human corneal tissue ex vivo (50). Moreover, unstimulated human hematopoietic stem cells (HSCs) transduced in vitro have developed into mature T (14) and B cells (35) in in vivo models of lymphocyte maturation. Gene transfer systems have been derived from other lentiviruses as well, including HIV-2, simian immunodeficiency virus, feline immunodeficiency virus, (13a, 42), equine infectious anemia virus (33, 40), and visna virus (4a).
Although HIV-1 vectors are the most well studied, some authors have suggested that HIV-based vectors pose safety risks for human clinical applications (7, 47). The possibility has been raised that if the HIV-1 vector recombines with endogenous human retroviruses present in the cells (29, 32) or with exogenous viruses present during transient infections, there is a chance of generating replication-competent HIV-1 or transferring the vector to other cells in the patient. Such a recombination event is less likely to occur for nonhuman or even nonprimate vectors. In addition, even if the nonprimate lentivirus were to become replication competent, it may not be as destructive in humans as HIV-1, although the point has been debated (47).
Bovine immunodeficiency virus (BIV) is a lentivirus which infects cows and causes AIDS-like disease after a variable asymptomatic phase (19). Although the virus has a set of accessory genes similar to those of HIV and simian immunodeficiency virus, it is the most ancient known lentivirus and does not readily infect T cells. Instead, the virus is found predominantly in monocytes and splenic macrophages in vivo (19). In this report we describe the generation and characterization of gene transfer systems based on BIV clone 127 (18). The vectors were observed to transduce proliferating and nondividing human and nonhuman cell lines and primary cells, in some instances with virus titers almost as high as that of an HIV-1 gene transfer system (14).
MATERIALS AND METHODS
Plasmids.
All restriction endonucleases were purchased from Roche Molecular Biochemicals (Indianapolis, Ind.). Plasmid pBIV, containing BIV proviral clone 127 (18), was obtained from the National Institutes of Health, Rockville, Md. Plasmid pCI was obtained from Promega (Madison, Wis.). Plasmid pCIGL contains the vesicular stomatitis virus glycoprotein (VSV-G) cDNA (9, 51) in the pCI polylinker, downstream of the human cytomegalovirus (CMV) immediate-early promoter and chimeric intron and upstream of the simian virus 40 (SV40) late polyadenylation signal. Plasmid pCrev, containing the HIV-1 rev cDNA under control of the CMV promoter, has been described previously (30). Plasmid pBH1 was generated by sequential insertion of two pBIV segments into the pCI polylinker using standard techniques: a 5.5-kb SmaI-XbaI fragment containing the gag, pol, vif, vpw, and vpy genes, as well as the first coding exons of the tat and rev genes; and a 1.3-kb DraIII-PvuII fragment containing the putative rev-response element (RRE) and the second coding exons of the tat and rev genes. Plasmids pBH2 and pBH3 were constructed in the same way, but the chimeric intron was deleted; moreover, the pBH2 5′ BIV fragment also contained the 3′ 70 bp of the leader, including the major splice donor site. Plasmids pBH1 and pBH3 were also modified by insertion of (1) an approximately 500-bp fragment of HIV-1 containing the RRE, (2) an internal ribosome entry site (IRES) from encephalomyocarditis virus (21), and (3) the puromycin N-acetyltransferase cDNA (49). Plasmid pBBB was generated by digesting plasmid pBIV with BfrI and BglII to remove most of the coding region and inserting into the gap a short polylinker created by annealing oligonucleotides BB5 (5′-TTAAGATTTAAATACGCGTGCGGCCGCA-3′) and BB3 (5′-GATCTGCGGCCGCACGCGTATTTAAATC-3′).
Packaging construct BH2 and an HIV-1 packaging construct (14) were each modified by insertion of two hemagglutinin (HA) oligonucleotides immediately upstream of the gag stop codon and deletion of most of the pol coding region as follows. First, a small fragment containing the gag stop codon of each packaging construct (290-bp ApaI-AccI fragment of BIV, 360-bp ApaI-BstXI fragment of HIV-1) was ligated to ApaI/EcoRV-digested pBluescript SK(+) (Stratagene, La Jolla, Calif.). Next, each subclone was subjected to reverse PCR amplification using primers containing the HA tags HHAS (5′-TATCCATACGATGTTCCAGATTATGCTTAAAGATAGGGGGGCAATTAAAG-3′) and HHAA(5′-AGCATAATCTGGAACATCGTATGGATATTGTGACGAGGGGTCGCTG-3′) for HIV-1 and BHAS (5′-TATCCATACGATGTTCCAGATTATGCTTAGACAAACAGCCTTTTATAAAG-3′) and BHAA (5′-AGCATAATCTGGAACATCGTATGGATAATCTAATATAAGAGGGGGTGC-3′) for BIV. Each PCR product was circularized, and then the modified gag fragment was excised with ApaI/SmaI and ligated to the parental packaging construct deleted between the ApaI site in gag and the 3′ end of pol (Asp718 for HIV-1, NsiI for BIV).
The CMV immediate-early enhancer-promoter was used to replace the BIV promoter in the 5′ LTR in plasmids pBIV and pBBB as follows. First, the CMV region upstream of the TATA box was subjected to PCR amplification with primers CB5 (5′-CGGGATCCCGTAGTTATTAATAGTAATCAATTACGG-3′) and CMVBIV3 (5′ AGATATGGTTTATATAGACCTCCCACCGTACA-3′) while the BIV region downstream of the TATA box was subjected to PCR amplification with primers CMVBIV5 (5′-GGGAGGTCTATATAAACCATATCTTCACTCTGT-3′) and Bgag3 (5′-GCCGTTTCTGTACTCTCTGGT-3′). Second, the two amplified products were mixed and subjected to amplification using primers CB5 and Bgag3. The final product was digested with XmaCI and ligated to plasmids pBIV and pBBB, previously digested with NruI and XmaCI, generating plasmids pBIVC and pBBBC. Plasmid pBIVC was then digested with SmaI and AflIII to remove most of the coding region and blunt-end ligated to a DNA segment containing either the CMV promoter or the mouse phosphoglycerate kinase (PGK) promoter (1) linked to the enhanced green fluorescent protein (eGFP) cDNA (Promega) generating plasmids pBCCG and pBCPG, respectively. Plasmid pBIVC was also digested with BfrI and BglII to remove a smaller segment of the coding region and then blunt-end ligated to the CMV-eGFP and PGK-eGFP cassettes, as well as a DNA segment containing eGFP linked to MND, a modified version of the myeloid proliferative sarcoma virus long terminal repeat (LTR) (43), generating plasmids pBC2CG, pBC2PG, and pBC2MG, respectively. Plasmid pBBBC was digested with MunI and HpaI to remove BIV sequences between the putative RRE and the 3′ LTR and then ligated to the MND-eGFP and PGK-eGFP cassettes to generate plasmids pBC3MG and pBC3PG, respectively. The CMV-eGFP cassette was also inserted into the BstEII site of plasmid pBIV to generate pBCG.
Plasmid pBC3MG was digested with BfrI and BglII to remove the short polylinker and then (i) ligated to an ∼500-bp BglII fragment of the pBIV pol gene (containing the putative central polypurine tract) to produce plasmid pBC3MGppt; (ii) ligated to an ∼1-kb BfrI fragment of pBIV (containing the 3′ end of the BIV gag gene) to produce plasmid pBC3MGgag; or (iii) ligated to an ∼800-bp fragment containing the human beta interferon scaffold attachment region (SAR) (2) to produce plasmid pBC3MGsar. Plasmid pBC3MGgag was linearized with BfrI and ligated to the SAR and central polypurine tract (cPPT) fragments to produce plasmids pBC3MGgagSAR and pBC3MGgagppt, respectively. Plasmid pBC3MGppt was linearized with BfrI and ligated to the SAR fragment to produce plasmids pBC3MGpptsar. Plasmids pBC3MP and pBC3MPsar were generated from plasmids pBC3MG and pBC3MGsar, respectively, by replacement of the eGFP cDNA with the puromycin N-acetyltransferase cDNA (49).
Plasmids pBC4MG and pBC4MGppt, in which the 3′ LTR contains a large deletion in the U3 region and an insertion of the SV40 late polyadenylation signal upstream enhancer element (USE) (45), were generated from plasmids pBC3MG and pBC3MGppt, respectively, as follows. First, the 5′ portion of the LTR and SV40 regions was subjected to PCR amplification with primers GFP5 (5′-GAGGACGGCAACATCCTGG-3′) and BSINSV3 (5′-AGCAATAGCATCACAAATTTCACAAATAAACACATATGGGAAGTCCGGG-3′) while the 3′ portion was subjected to PCR amplification with primers BSINSV5 (5′-GTG AAATTTGTGATGCTATTGCTTTATTTGTAATCTTGTACTTCAGCTCGT GTAG-3′) and BIV3 (5′-TCGCCGACATCACCGATGG-3′). Second, the two amplified products were mixed and subjected to amplification using primers GFP5 and BIV3. The final product was digested with SspBI and SphI and ligated to plasmids pBC3MG and pBC3MGppt previously digested with SspBI and SphI. The resulting plasmids, pBC4MG and pBC4MGppt, contain the 40-bp SV40 USE in place of 332 bp of the U3 region.
For the relative assessment of the BIV gene transfer system, HIV-1 and murine leukemia virus (MLV) gene transfer systems were used. Details of the construction of the packaging constructs and vectors have been published elsewhere (14).
Immortalized cells.
293T cells were obtained from Gary Nolan (Stanford University, Palo Alto, Calif.). CEMSS cells were obtained from the AIDS Reagent Program (Rockville, Md.). A-10 and D-17 cells were obtained from the American Tissue Type Collection (Manassas, Va.). MN9D cells (13) were obtained from Rainer Ortman (Novartis, Basel, Switzerland). Embryonal rabbit epithelial (EREp) cells (39) were obtained from the National Institutes of Health. Human umbilical vein endothelial cells (HUVEC) (20) and primary rat aorta (smooth muscle) cells were obtained from Clonetics (San Diego, Calif.). HUVEC were cultured in EBM basal medium with the FGM bullet kit containing 0.1% human epidermal growth factor, 2% fetal calf serum (FCS), 0.4% bovine brain extract with heparin, 0.1% gentamicin-amphotericin B, and 0.1% hydrocortisone. Rat aorta cells were maintained in EBM basal medium with the EGM-MV bullet kit containing 0.1% human epidermal growth factor, 5% FCS, 0.4% bovine brain extract with heparin, 0.1% gentamicin-amphotericin B, 0.1% hydrocortisone and cultured in Primaria tissue culture flasks (Becton Dickinson Biosciences, San Jose, Calif.). CEMSS cells were cultured in RPMI 1640 medium supplemented with 10% FCS. All other cell lines were cultured in Dulbecco's modified Eagle medium supplemented with 10% FCS. HUVEC (1.25 × 105) were suspended in 5 ml of medium and irradiated with 8,000 rads from a 137Cs source irradiator (J. L. Shepherd, San Fernando, Calif.) and then transferred to a six-well dish and incubated for 2 days to allow for synchronization in the G2/M phase of the cell cycle.
Virus production.
293T cells (4 × 106 to 10 × 106 293T cells) were seeded into 10-cm-diameter dishes overnight and transfected the next day with 20 to 30 μg of plasmid DNA by the calcium phosphate method (Clontech, Palo Alto, Calif.). Typically, 20 μg of the vector, 10 μg of the packaging construct, and 3 μg of the VSV-G plasmid were used. In cases where the HIV-1 rev protein was required, 4 μg of plasmid pCrev was added. After 24 to 72 hr, the cells or the virus-containing medium was collected and analyzed in a variety of ways (see below). To assess the efficiency of transfection, a portion of the cells were analyzed for eGFP expression by flow cytometry, using a FACScan apparatus (Becton Dickinson Biosciences). To measure the amount of virus shed into the medium, the medium was cleared of cellular debris by low-speed centrifugation, and then 10 μl was lysed and analyzed for reverse transcriptase (RT) activity using a commercial kit (Roche Molecular Biochemicals).
Analysis of RNA levels: Northern blotting.
Transfected cells were lysed and cytoplasmic RNA was prepared using a commercial kit (Qiagen, Valencia, Calif.). In addition, virus-containing medium was collected, subjected to low-speed centrifugation to remove cellular debris, and then subjected to high-speed centrifugation (50,000 × g for 90 min at 4°C) to collect the virus particles. The viral pellet was lysed and the viral RNA was prepared using a commercial kit (Qiagen). A fixed amount of cytoplasmic RNA (10 μg) or viral RNA (one-third of the RNA preparation, not quantitated) was subjected to 1% agarose gel electrophoresis and transferred to a nylon filter (Bio-Rad, Hercules, Calif.). The filter was exposed to 40 × 106 cpm of a DNA fragment random primed with [32P]dCTP using a commercial kit (Ambion, Austin, Tex.) and then washed and analyzed for bound probe with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). The probes included a BIV gag fragment and an HIV-1 gag fragment (both ∼1 kb), an ∼330-bp BIV U3 fragment, and an ∼800-bp eGFP fragment.
Analysis of protein levels: Western blotting assay.
Transfected cells were lysed on ice for 1 h in a buffer containing 1% NP-40, 150 mM NaCl, 10 mM Tris-Cl (pH 7.4), 1 mM EDTA, and Pefabloc protease inhibitor (Roche Molecular Biochemicals). The lysate was subjected to centrifugation at 8,000 × g for 20 min at 4°C to remove precipitated proteins and other debris. Alternatively, virus-containing medium was collected, subjected to brief centrifugation to remove cellular debris, and then subjected to high-speed centrifugation (50,000 × g for 90 min at 4°C) to collect the virus particles. The viral pellet was lysed directly in sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer (Novex, San Diego, Calif.). A fixed amount of cell or viral lysate was subjected to acrylamide gel electrophoresis and transferred to a nitrocellulose filter. The filter was exposed to rabbit serum specific for BIV Gag protein (obtained from the National Institutes of Health), then to horseradish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin (Ig) antibody (Zymed, South San Francisco, Calif.), and then to the HRP substrate o-phenylenediamine dihydrochloride (OPD; Sigma, St. Louis, Mo.). Prestained molecular weight standards (Bio-Rad) were used to determine the approximate molecular weight of the BIV Gag bands. For the HIV Western blot, the filter was exposed to biotinylated anti-HIV Gag antibody (Beckman Coulter, Fullteron, Calif.) and then to steptavidin-conjugated HRP (Beckman Coulter) before the OPD reaction. For the HA Western blot, the filter was exposed to a biotinylated anti-HA antibody (Roche Molecular Biochemicals) and then to the steptavidin-conjugated HRP before the OPD reaction. For the VSV-G Western blot, the filter was exposed to mouse anti-VSV-G monoclonal antibody (MAb) (Sigma) and then to an HRP-conjugated goat anti-mouse Ig antibody (Zymed) before the OPD reaction.
Analysis of transduction.
To transduce cells, the virus-containing medium was subjected to brief centrifugation to remove cellular debris and then 1 ml was added to fresh cells in polypropylene tubes (5 × 105 CEMSS cells) or in six-well dishes (3 × 105 to 6 × 105 adherent cells seeded per well the previous day). Protamine sulfate (Sigma) was added to the wells at a final concentration of 8 μg/ml and the tubes or dishes were subjected to centrifugation (“spinoculation”) at 1,500 × g for 2 to 3 h at 32 to 37°C. In later experiments, the tubes or wells were also supplemented with 10 mM HEPES buffer prior to spinoculation to prevent the pH of the medium from rising while the cells were in the centrifuge. After spinoculation, the tubes and dishes were processed in different ways: supernatant was aspirated from the tubes, and the CEMSS cells were suspended in fresh medium and transferred to six-well dishes. In contrast, the spinoculated dishes were placed back into the incubator for 30 to 60 min, and then the medium was removed and fresh medium was added to the wells. At 2 to 3 days postspinoculation, a portion of the cells were removed from the plate and analyzed for eGFP expression by flow cytometry, using a FACScan apparatus (Becton Dickinson Biosciences). For virus preparations containing the vectors pBC3MP and pBC3MPsar, 293T cells were subjected to spinoculation with serial dilutions of the virus-containing medium, and fresh medium supplemented with puromycin (5 μg/ml) was added to the cells 24 h postspinoculation. Seven to ten days later, colonies were counted by direct visualization.
Infection of primary T cells.
Human peripheral blood mononuclear cells (PBMC) were isolated from adult peripheral whole blood by Ficoll density gradient centrifugation, rinsed in phosphate-buffered saline, and suspended in RPMI 1640 medium supplemented with 10% FCS. A portion of the PBMC were activated by adding interleukin 2 (Peprotech, Rocky Hill, N.J.) to the medium at a final concentration of 200 U/ml and culturing the cells for 3 days in 12-well dishes (3 × 106 cells per well) precoated as follows: the dishes were incubated with 1 μg of goat anti-mouse Ig Fc (Pierce, Rockford, Ill.) per ml for 3 h, rinsed with phosphate-buffered saline, incubated with a mixture of MAbs (1 μg of anti-CD3 [OKT3] per ml and 10 ng of anti-CD28 [BD PharMingen, San Diego, Calif.] per ml) for 1 h and rinsed with medium. Activated or unstimulated PBMC (5 × 105) were spinoculated with viral supernatant in polypropylene tubes similar to CEMSS cells (see above); after spinoculation, the cells were rinsed and suspended in medium containing interlenkin-2 and cultured in 24-well dishes precoated with the anti-CD3 and anti-CD28 MAbs. Four days and 2 weeks later, a portion of the cells was removed from the well and analyzed for eGFP expression by flow cytometry, using a FACScan apparatus (Becton Dickinson Biosciences). T cells were identified by light scatter properties and by expression of CD4 and CD8, using allophycocyanin (APC)-conjugated anti-CD4 and PerCP-conjugated anti-CD8 MAbs (Becton Dickinson Biosciences).
Infection of HSCs.
Human CD34+ cells were isolated from granulocyte colony-stimulating factor-mobilized peripheral whole blood from healthy donors using Isolex 300SA (Baxter Healthcare, Deerfield, Ill.). The cells (80 to 90% pure CD34+) were aliquoted (107) and frozen in medium consisting of 45% Iscove's modified Eagle medium, 45% FCS and 10% dimethyl sulfoxide. Prior to transduction, the frozen cells were first thawed in buffer containing 2% FCS, 1% HEPES, and 10 U of heparin per ml. After thawing, the cells (5 × 105) were either spinoculated (see above) with viral supernatant in the absence of cytokines or cultured for 48 h in cytokine-containing medium (X-vivo15 medium [BioWhittaker, Walkersville Md.], thrombopoietin [tpo] mimetic [50 ng/ml; Novartis], flt3 ligand [100 ng/ml], and c-kit ligand [100 ng/ml; both from Systemix, Palo alto, Calif.]), and then spinoculated with viral supernatant. After infection, the cells were cultured in cytokine-containing medium. Three days or 2 weeks later, a portion of the cells was stained with APC-conjugated anti-CD34 antibody (Becton Dickinson Biosciences) and analyzed for eGFP on the CD34+ cells on a FACScan apparatus (Becton Dickinson Biosciences).
RESULTS
Can BIV transduce human cells?
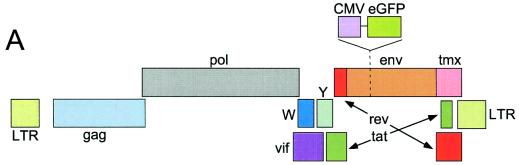
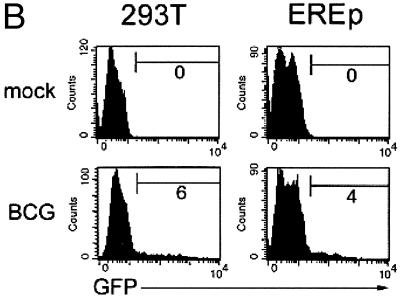
To determine whether BIV could transduce human cells, the wild-type BIV genome (Fig. 1A) was modified by insertion of the eGFP marker gene, producing construct BCG. The eGFP gene was inserted into an interior position within the viral envelope gene so as not to affect viral rev or tat expression or RRE function. The human kidney carcinoma cell line 293T was cotransfected with construct BCG and a plasmid encoding VSV-G; 2 days later, the virus-containing medium was collected and exposed to fresh 293T cells. In addition, the medium was added to the embryonal rabbit epithelial cell line EREp, which supports wild-type BIV replication (39). Three days later, the exposed cells were assayed for eGFP expression by flow cytometry (Fig. 1B). A subset of the 293T cells (approximately 5%) expressed eGFP, indicating that BIV could carry out all of the functions (including reverse transcription and integration) required for transduction of human cells. Similar transduction efficiencies were noted for the EREp cell line (Fig. 1B).
FIG. 1.
(A) Wild-type BIV genome. Viral coding regions are arranged in three horizontal lines representing the different reading frames. The rev and tat genes are composed of two coding regions each. The first coding region of rev and the tmx coding region are in the same reading frame as env. The approximate position of the insertion of CMV-eGFP (in construct BCG) is indicated above the top line. (B) BIV can transduce human cells. VSV-G-pseudotyped construct BCG was produced in 293T cells and exposed to EREp (right column) and fresh 293T (left column) cells; 3 days later, the cells were analyzed for eGFP expression by flow cytometry. The percentage of eGFP+ cells is indicated inside each histogram.
BIV packaging constructs.
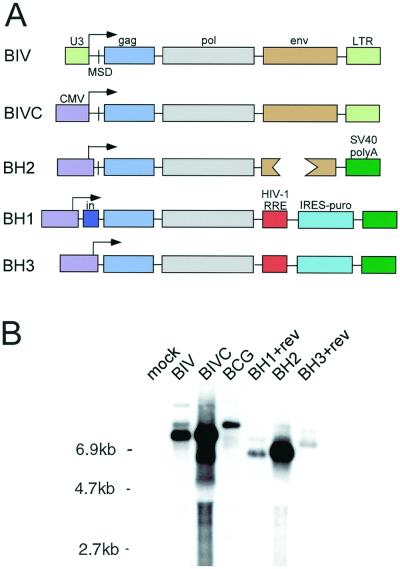
As a first step towards setting up a BIV-based gene transfer system, in which the eGFP gene and viral genes are encoded on separate plasmids, packaging constructs (containing the viral genes) were generated (Fig. 2A) and assayed for gag mRNA expression (Fig. 2B) and virus production (Table 1) in the transient expression system. One construct, BIVC, is identical to wild-type BIV except for a precise replacement of the 5′ LTR U3 region with the human CMV immediate-early promoter. The junction between the two segments is located at the identical TATA boxes to maximize the chances that the viral RNA would contain the proper 5′ end for infection of target cells. The three other BIV packaging constructs contain the CMV promoter and transcription initiation site linked to the BIV leader near of the gag gene, deleting the 5′ LTR and primer binding site and—in the case of constructs BH1 and BH3—the major splice donor. Construct BH1 contains a small chimeric intron inserted between the CMV and BIV segments. Downstream of the pol gene, construct BH2 contains all viral coding sequences except for a deletion (approximately 1 kb) of the interior of env (not predicted to affect tat and rev expression or RRE function), while constructs BH1 and BH3 contain BIV sequences terminating approximately 250 bp downstream of the pol gene. Since BH1 and BH3 lack the rev gene and the RRE, the HIV-1 RRE was inserted downstream of the BIV sequences and HIV-1 rev protein was provided in trans when the constructs were characterized. In addition, these two constructs contain the puromycin N-acetyltransferase cDNA (49) coupled to an IRES from the encephalomyocarditis virus (21), for selecting cell lines stably producing the packaging construct.
FIG. 2.
BIV packaging constructs and expression in transfected 293T cells. (A) wild-type BIV, CMV-driven BIV, and three packaging constructs (BH1 to -3) are depicted for their gag, pol, and env genes; accessory genes are not shown. Also depicted are the viral major splice donor (MSD), a small chimeric intron (in), the HIV-1 RRE, the puromycin N-acetyltransferase cDNA linked to an IRES from encephalomyocarditis virus (IRES-puro), and the SV40 late polyadenylation signal (SV40 polyA). (B) Northern analysis of the constructs' steady-state cytoplasmic expression levels. Cytoplasmic RNA from transfected 293T cells was probed for gag sequences; the high-molecular weight band in each lane corresponds to the construct's gag mRNA.
TABLE 1.
Virus production by BIV constructsa
| Construct | Virus produced (pg of RT/ml of medium) in expt no.:
|
||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| HIV | 132 | 121 | 115 | 123 | 121 | ||
| BIV | 0 | 1 | 4 | ||||
| BIVC | 1 | 5 | |||||
| BH2 | 39 | 137 | 79 | 276 | 464 | 624 | |
| BH1 | 0 | 14 | |||||
| BH3 | 0 | 0 | |||||
Virus-containing medium was collected from 293T cells transfected with the indicated construct and analyzed by RT assay. Seven transfections were performed.
Northern analysis of transfected cell cytoplasmic RNA (Fig. 2B) indicated that construct BIVC produced higher steady-state levels of gag mRNA (816 cpm) than did wild-type BIV (249 cpm) or construct BCG (113 cpm) (Fig. 1A), suggesting that the CMV promoter was more active than the BIV LTR in 293T cells. Packaging constructs BH1 (70 cpm) and BH3 (39 cpm) expressed low levels of gag mRNA, but BH2 (861 cpm) expressed high levels comparable to construct BIVC. For comparison, a CMV-driven HIV-1-derived packaging construct (14) was observed to express even higher levels of gag mRNA (1,120 cpm).
RT assay (Table 1) and Western blot analysis (data not shown) of virus collected from the transfected cells indicated that BIVC produced more virus particles than did wild-type BIV, BH1, and BH3, but much less than BH2 or the HIV-1 packaging construct. The low amount of BIVC virus may have been due to toxicity in the transfected cells resulting from BIVC's high-level expression of the wild-type BIV envelope protein, as cytopathic effects and small syncytia were observed in the culture. Western blot analysis indicated that both the BIVC and BH2 virus preparations had undergone maturation, i.e., the Gag polyprotein was almost entirely cleaved (data not shown and Fig. 3, see below).
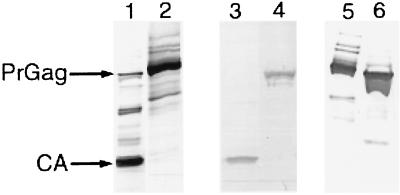
FIG. 3.
BIV produces levels of virus similar to HIV-1. BIV packaging construct BH2 and an HIV-1 packaging construct were each modified by insertion of HA tags at the C terminus of Gag and deletion of pol (see text). Parental and modified viruses were produced in 293T cells, collected and subjected to anti-HIV-1 Gag (lanes 1 and 2), anti-BIV Gag (lanes 3 and 4), or anti-HA (lanes 5 and 6) Western blotting. Lane 1, parental HIV-1 packaging construct. Most of the Gag precursor polyprotein (PrGag) has been cleaved. Lane 2, modified HIV-1 packaging construct. The amount of uncleaved Gag polyprotein is similar to the amount of capsid protein (CA) in the parental virus preparation, indicating that the modifications did not alter virus production substantially. Lane 3, parental BIV packaging construct. Lane 4, modified BIV packaging construct. As with the case of the HIV-1 packaging construct, the modification did not alter virus production. Lane 5, modified HIV-1 packaging construct. Lane 6, modified BIV packaging construct, after normalization by RT assay of the parental constructs. The amount of Gag precursor polyprotein is similar to the amount in lane 5, indicating that the RT assay has comparable sensitivity for BIV and HIV-1 virus and hence, that the packaging constructs produce similar levels of virus.
The RT assay indicated that the amount of virus produced by BH2 was similar to that produced by the HIV-1 packaging construct. To verify that the amounts of virus were similar, these two packaging constructs were modified by insertion of two consecutive HA tags immediately upstream of the gag stop codon. The HA-tagged constructs, which had further deletions of most of the pol gene to block Gag polyprotein cleavage, were introduced into 293T cells alongside the parental constructs. The modified virus was collected 2 days later and compared to the parental virus by Western blot assay using antibodies specific for Gag protein: both modified constructs produced amounts of virus similar to the parental constructs (Fig. 3). The modified viruses were then normalized by RT assay of the parental constructs (649 pg for HIV-1, 205 pg for BH2) and analyzed for HA tag content by Western blot assay. The amounts of HA-tagged HIV-1 Gag polyprotein and HA-tagged BIV Gag polyprotein were similar (Fig. 3), corroborating the RT assay: the BH2 packaging construct produced levels of virus roughly similar to the HIV-1 packaging construct.
Western blot analysis was also performed on the BH2 and HIV-1 virus preparations to assess the efficiency of VSV-G incorporation. Virus was collected from 293T cells transfected with each packaging construct and the VSV-G construct, then normalized by RT assay and subjected to Western blot analysis using a MAb specific for VSV-G. The amounts of VSV-G detected in the BH2 and HIV-1 samples were similar (data not shown), indicating that BIV incorporated VSV-G as efficiently as did HIV-1.
BIV vectors.
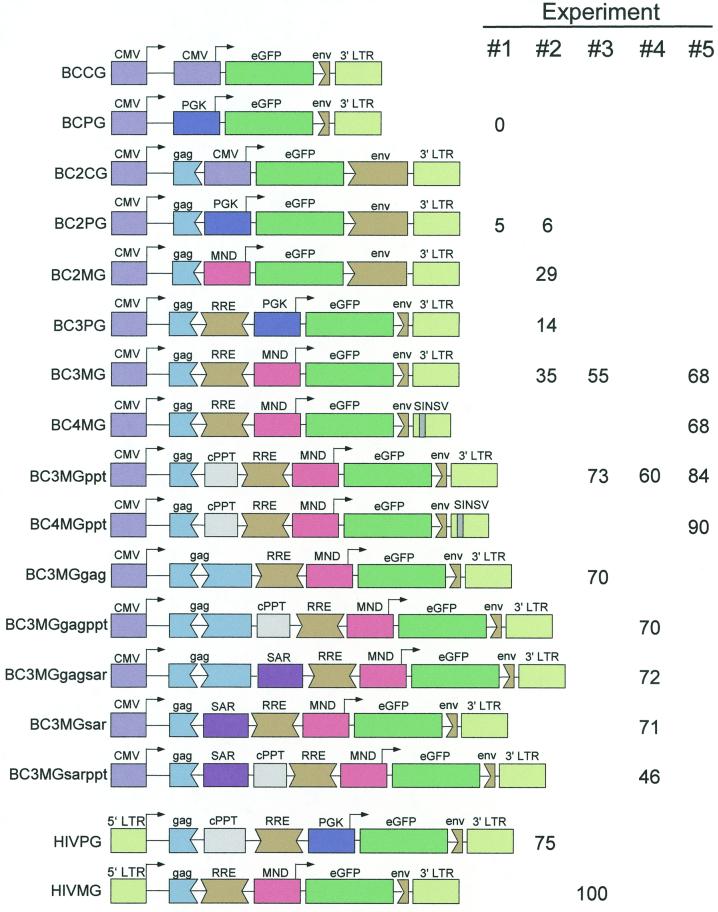
Next, BIV-derived vectors were generated, containing the eGFP marker gene and all viral elements required in cis for transfer into target cells. A panel of vectors was generated (Fig. 4), differing in (i) the amount of gag and env sequences (BC versus BC2 prefix), (ii) the internal promoter driving eGFP expression: CMV (constructs with a CG suffix) versus PGK (1) (constructs with a PG suffix) versus MND (modified myeloid proliferative sarcoma virus promoter [43]) (constructs with an MG suffix); (iii) the placement of eGFP within the vector (i.e., upstream or downstream of the putative BIV RRE) (BC2 versus BC3 prefix); and (iv) additional segments inserted downstream of the putative BIV packaging signal (constructs with gag, ppt, and sar suffixes). In addition, two vectors contained a modified 3′ LTR in which a large portion of U3 (including the TATA box) was replaced by a small SV40 segment containing the late polyadenlyation signal USE (45) (BC3 versus BC4 prefix).
FIG. 4.
BIV vectors and transduction efficiencies. All vectors contain the CMV immediate-early promoter, ending in the TATA box, linked to the BIV 5′ LTR starting immediately after the TATA box. BIV sequences terminate at the gag start codon (BCCG and BCPG) or approximately 510bp into the gag coding region (all others). All vectors contain the eGFP cDNA linked to a heterologous, “internal” promoter: CMV, PGK, or MND (see text). Some vectors contain one or more insertions between the BIV 5′ segment and the internal promoter: these insertions include a potential BIV cPPT, the 3′ segment of the gag gene, the beta interferon SAR, and the putative BIV RRE. Downstream of the eGFP cDNA lies the BIV 3′ LTR and approximately 130 bp (BCCG and BCPG), 1.2 kb (BC2CG, BC2PG, and BC2MG) or 80 bp (all others) of adjacent env sequences. Two vectors (BC4MG and BC4MGppt) contain modified 3′ LTRs in which most of the 3′ LTR U3 region has been replaced by the SV40 late polyadenylation signal enhancer element (SINSV). Transcription start sites and directions are indicated with arrows. At the bottom are depicted two HIV-1 control vectors used in this study. At the right are the transduction efficiencies of the vectors in 293T cells 3 days postinfection, using packaging construct BH2 and VSV-G, in a series of experiments. Infections from experiments 3 to 5 were performed in the presence of an additional buffer to retard pH elevation during spinoculation. As a result, transduction efficiencies were elevated.
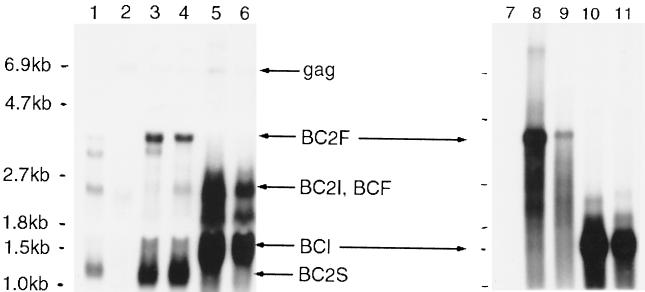
First, the two vectors (BCCG and BCPG) containing the full leader, but no gag sequences, and minimal env sequences (i.e., no RRE) were compared to the analogous vectors (BC2CG and BC2PG) containing approximately 500 bp of gag sequences and 1.2 kb of env sequences (including the putative RRE). The latter vectors were generated because previous studies with other retroviruses have indicated that the 5′ end of the gag gene increases the extent of vector RNA encapsidation in the virus particles, either by containing packaging elements or by stabilizing packaging elements further upstream (3, 6, 41). In addition, studies with HIV-1 have indicated that the gag gene contains sequences which block RNA export from the nucleus and that the RRE removes this block in the presence of the rev protein (31, 46). The two vectors containing the PGK internal promoter were analyzed for transduction in 293T cells, using the BH2 packaging construct pseudotyped with VSV-G: BC2PG transduced 5% of the cells, while BCPG transduced none of the cells (Fig. 4, experiment 1). All four vectors were then assessed for cytoplasmic RNA expression in transfected 293T cells and in collected BH2 virions by Northern blot analysis. Each of the vectors produced high levels of full-length vector RNA in the transfected cell cytoplasm, but only the BC2CG and BC2PG full-length RNAs were encapsidated efficiently by the BH2 virions (Fig. 5). Therefore, the 5′ end of the BIV gag gene is likely to contain sequences directly or indirectly required for viral RNA encapsidation. Interestingly, without those sequences on the BCPG and BCCG full-length RNAs, subgenomic vector mRNAs (presumably internally initiated, based on expected mobility) were encapsidated, suggesting that other packaging elements reside in the 3′ portion of the BIV genome (4).
FIG. 5.
Northern analysis of BIV RNA in transfected 293T cells (left) and in virus particles shed from the transfected cells (right). Cytoplasmic RNA was probed with BIV sequences at the 3′ end of the genome, to detect all packaging construct and vector RNAs. Viral RNA was probed with eGFP sequences, to detect full-length vector RNA and RNA initiating at the internal promoter. Lane 1, vector BC2PG; lanes 2 and 7, packaging construct BH2; lanes 3 and 8, BH2 and BC2PG; lanes 4 and 9, BH2 and BC2CG; lanes 5 and 10, BH2 and BCPG; lanes 6 and 11, BH2 and BCCG. The positions of certain RNAs are indicated: gag, the gag mRNA encoded by BH2; BC2F, full-length BC2PG and BC2CG vector mRNA; BC2I, internally initiated BC2PG and BC2CG vector mRNA; BCF, full-length BCPG and BCCG vector RNA; BCI, internally initiated BCPG and BCCG vector mRNA; BC2S, spliced BC2PG and BC2CG vector mRNA. Note that this last RNA is only present in the BC2 vectors, since the rev splice acceptor is present in the BC2 vectors but not the BC vectors.
Next, the PGK promoter was compared to the MND promoter in two contexts, i.e., with the promoter-eGFP cassette upstream (BC2PG and BC2MG) or downstream (BC3PG and BC3MG) of the putative BIV RRE. In 293T cells, the latter context produced slightly higher transduction efficiencies than the former (14 versus 6% for the PGK vectors and 35 versus 29% for the MND vectors), and in both contexts the MND promoter performed better than the PGK promoter (Fig. 4, experiment 2). However, all vectors transduced substantially fewer cells than did an HIV-1 virus containing an analogous PGK-eGFP vector (75% transduction).
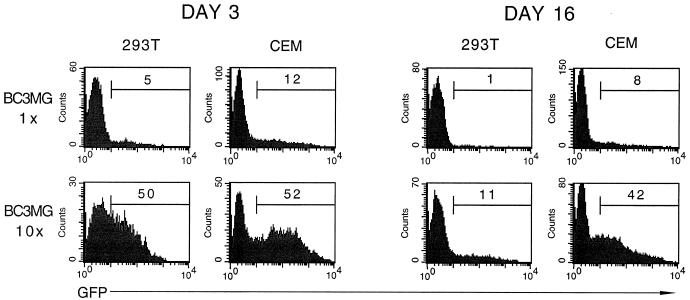
Virus containing the optimal vector, BC3MG, was then produced and analyzed for its ability to be concentrated by centrifugation, for its ability to transduce a lymphoid cell line, and for the change in the frequency of eGFP expression over 2 weeks postinfection (Fig. 6). A portion of the virions were collected by centrifugation, suspended in a volume 100-fold lower than the original volume, and then diluted either 10-fold (to generate “10×” virus) or 100-fold (“1×” virus). Although the 1× virus exhibited low transduction efficiencies, the T-lymphoid cell line CEMSS was transduced more efficiently (12%) than the 293T cells (5%). In addition, the 10× virus transduced a 10-fold-higher percentage of 293T cells and a 4.5-fold-higher percentage of CEM cells. Moreover, the percentage of eGFP+ cells decreased approximately fivefold over 2 weeks in the 293T line and to a much lesser extent in the CEMSS line. Subsequent infections with new, unconcentrated virus preparations indicated that the fold reduction in the percentage of eGFP+ 293T cells over time correlated inversely with the initial percentage of eGFP+ cells. For example, 293T cells which were 73% eGFP+ 2 days postinfection were found to be 43% eGFP+ 2 weeks postinfection and 28% eGFP+ 4 weeks postinfection (data not shown).
FIG. 6.
Analysis of virus concentration and the duration of transgene expression in transduced human cell lines. A BIV preparation consisting of packaging construct BH2, VSV-G, and vector BC3MG was collected by centrifugation, then suspended in the original volume (1× virus) or in a 10-fold-lower volume (10× virus) and exposed to fresh 293T or CEMSS cells. eGFP expression was measured at 3 and 16 days postinfection by flow cytometry; the percentage of transduced cells is indicated in each histogram.
The higher transduction efficiencies exhibited by the BC3 vectors relative to the BC2 vectors might have been due to an increased stability of the packaging signal, due to its positioning farther away from non-viral sequences, i.e., the internal promoter (24). Additional viral segments were therefore inserted immediately downstream of the gag region in vector BC3MG, including an approximately 500-bp segment of the pol gene, containing polypurine tracts that might function in an analogous manner to the cPPT region of HIV-1 (11, 12). In addition, the 3′ portion (approximately 1 kb) of the gag gene was inserted: this vector (BC3MGgag) contains the entire gag gene except for approximately 200 bp in the capsid domain. The two new vectors were found to transduce slightly higher frequencies of 293T cells (70 and 73%) than the parental vector BC3MG (55%) when used with BH2 and VSV-G (Fig. 4, experiment 3). For comparison, an HIV-1 strain containing an MND-eGFP vector transduced 100% of the cells.
The beta interferon SAR, which has been shown to potentiate vector expression from integrated proviral DNA (2), was also inserted immediately downstream of the packaging signal. This vector, BC3MGsar, transduced a slightly higher frequency of 293T cells than did the BC3MGppt vector (71 versus 60% [Fig. 4, experiment 4]). In addition, vectors containing pairwise combinations of the SAR segment, the 3′ gag segment, and the cPPT segment were generated; however, none of these vectors exhibited higher transduction efficiencies than the vectors containing the individual segments alone (Fig. 4, experiment 4).
The BC3MG and BC3MGppt vectors were then compared to their SIN counterparts, BC4MG and BC4MGppt. These SIN vectors had deletions in the interior 322 bp of the U3 region of the 3′ LTR, retaining 55 bp at the 5′ end and 7 bp at the 3′ end; as a result, the TATA box and most of the promoter elements were removed. Vectors with deletions in this area are termed self-inactivating, or SIN, because the integrated vector in the target cell possesses a 5′ LTR incapable of directing transcription (34, 52). This effect not only increases the safety of the vector, but in cases where transcription from the 5′ LTR interferes with transcription from the vector's internal promoter, the SIN deletion may also increase transgene expression in the transduced cell. It has also previously been reported that read-through transcription occurs from an integrated HIV-1 provirus (15), suggesting that HIV-1 transcripts do not always terminate at the 3′ LTR. Since this phenomenon might also occur in the BIV vectors and might decrease the titer of the gene transfer system (10), the SV40 late polyadenylation signal USE (45) was inserted into the gap created by the SIN deletion. The two new vectors (BC4MG and BC4MGppt) were analyzed for transduction efficiency with packaging construct BH2 and VSV-G: BC4MG transduced 68% of the 293T cells, while BC4MGppt transduced 90% (Fig. 4, experiment 5). However, since the parental, non-SIN vectors exhibited similar transduction efficiencies (68 and 84%, respectively), the SIN deletion and SV40 USE insertion did not substantially increase the titer of the BIV gene transfer system.
To determine the titer of the BIV viruses with precision, the eGFP cDNA was removed from vectors BC3MG and BC3MGsar and replaced with the puromycin N-acetyltransferase cDNA (49), generating vectors BC3MP and BC3MPsar. The same was done for the HIV-1 vector containing the MND-eGFP cassette. The three viruses were prepared in 293T cells, and serial dilutions were exposed to fresh 293T cells; after 2 days, the cells were treated with puromycin to kill the nontransduced cells. After a week in culture, the number of colonies growing in the dishes was determined and used to calculate the titer of the original viruses. The HIV titer was 1.2 × 107 per ml, the BC3MP titer was 3 × 105 per ml, and the BC3MPsar titer was 4.5 × 105 per ml, nearly 30-fold lower than the HIV titer.
Transduction of other cell lines and nondividing cells.
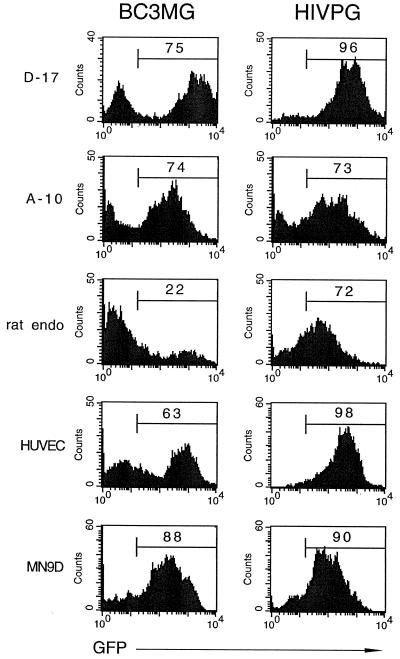
Concentrated BIV, prepared in 293T cells using packaging construct BH2, vector BC3MG, and VSV-G, was used to transduce a panel of cell lines: D-17, a dog osteosarcoma line; A-10, a rat smooth muscle cell line; HUVEC, a human endothelial cell line; and MN9D, a mouse neuronal cell line. In each of these cell lines, the BIV preparation transduced a large percentage (63 to 88%) of the cells, even 2 weeks postinfection (Fig. 7). In addition, primary rat endothelial cells were transduced by BIV, albeit not as efficiently as the immortalized lines (22%).
FIG. 7.
BIV can transduce a variety of cell lines and primary cells. A 1× (D-17 cells) or 10× (all others) BIV preparation consisting of packaging construct BH2, VSV-G, and vector BC3MG was exposed to the indicated cell lines or primary cells (see text for details on each cell type). Two weeks later, the cells were analyzed for eGFP expression by flow cytometry; the percentage of transduced cells is indicated in each histogram. For comparison, the HIVPG histograms indicate the cells exposed to an unconcentrated HIV-1 preparation consisting of a packaging construct, VSV-G, and a vector containing the PGK internal promoter.
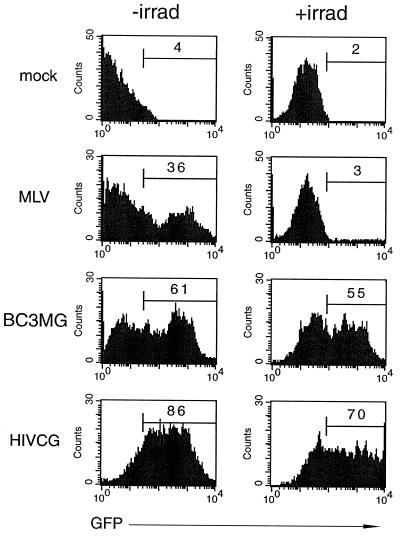
To assess the ability of BIV to transduce nondividing cells, the concentrated BIV (10×) was assayed for transduction of irradiated HUVEC and resting human peripheral blood lymphocytes (PBLs). The HUVEC line was irradiated 2 days before exposure to BIV, to synchronize the cells at the G2/M phase of the cell cycle. As expected, an MLV preparation was observed to readily transduce the untreated cells but not the irradiated cells (Fig. 8). In contrast, both BIV and HIV-1 transduced the untreated and irradiated cells with similar efficiencies, indicating that each lentivirus was able to transduce the nondividing cells efficiently.
FIG. 8.
BIV can transduce nondividing cells. A 10× BIV virus preparation consisting of packaging construct BH2, VSV-G, and vector BC3MG was exposed to HUVEC 2 days after the cells were irradiated to abrograte cell division (right column) or to nonirradiated HUVEC (left column). Two days later, the cells were analyzed for eGFP expression by flow cytometry; the percentage of transduced cells is indicated in the histogram (right column). For comparison, the HIVCG histograms indicate cells exposed to an unconcentrated HIV-1 preparation consisting of a packaging construct, VSV-G, and a vector containing the CMV internal promoter. In addition, the MLV histograms indicate cells exposed to an MLV preparation consisting of a packaging construct, VSV-G, and an LTR-driven eGFP vector.
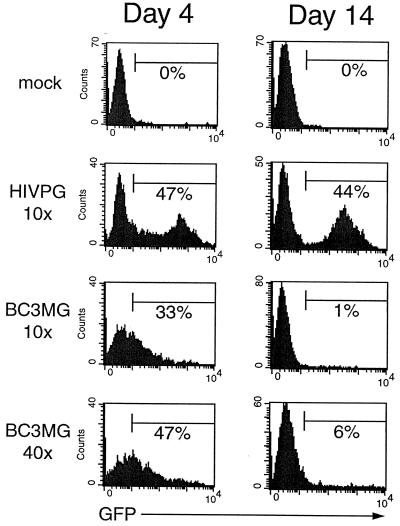
In unstimulated human PBLs, BIV exhibited transduction efficiencies similar to those of HIV-1 when the cells were assayed 4 days after infection, although most of the BIV-transduced cells expressed very low levels of eGFP (Fig. 9). Two weeks postinfection, however, these dim cells were nearly absent from the population. As a result, the percentage of transduced cells was low: 1% for the 10× virus and 6% for the 40× virus. However, the virus also exhibited low transduction efficiencies in preactivated (i.e., proliferating) PBLs, as the 10× virus transduced only 5% of the cells (data not shown). In contrast, 10× HIV-1 transduced 81% of the preactivated cells and 44% of the unstimulated cells, while 10× MLV transduced 43% of the preactivated cells but only 1% of the unstimulated cells.
FIG. 9.
BIV can transduce resting human lymphocytes. Unstimulated human PBLs were infected with 10× and 40× BIV consisting of packaging construct BH2, VSV-G, and vector BC3MG. After infection, the cells were activated and cultured for 4 days (left column) or 2 weeks (right column) in the presence of antibodies specific for CD3 and CD28, after which a portion of the cells were analyzed for eGFP expression by flow cytometry. The percentage of transduced cells is indicated in each histogram. For comparison, the cells were infected with the 10× HIV-1 PGK preparation.
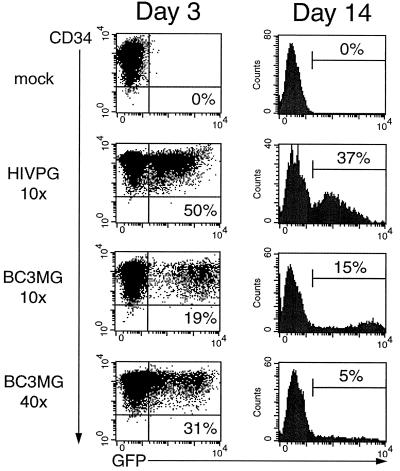
Finally, the concentrated BIV was used to transduce unstimulated mobilized peripheral CD34+ HSCs, most of which are quiescent (25, 48). The 10× BIV preparation transduced 19% of the HSCs 3 days postinfection, while the 40× BIV virus transduced 31% of the cells (Fig. 10). Interestingly, almost all of the cells transduced with the 10× BIV virus expressed high levels of eGFP, and these cells did not disappear when the cells were analyzed 11 days later. The cells infected with the 40× BIV virus contained two eGFP+ populations: one (18% of the cells) which expressed very low levels of eGFP and one (13% of the cells) which expressed higher levels of eGFP. Both populations exhibited sixfold reductions in frequency over the next 11 days.
FIG. 10.
BIV can transduce resting HSCs Unstimulated human HSCs were infected with 10x and 40x BIV consisting of packaging construct BH2, VSV-G, and vector BC3MG. After infection, the cells were activated and cultured for 3 days (left column) or 2 weeks (right column) in the presence of tpo mimetic, flt3 ligand, and c-kit ligand, after which a portion of the cells were analyzed for eGFP expression by flow cytometry. The percentage of transduced cells is indicated in each histogram. For comparison, the cells were infected with the 10× HIV-1 PGK preparation.
DISCUSSION
In this study, we have demonstrated that gene transfer systems derived from BIV can transduce a variety of cell types from a variety of organisms in vitro. Transduction was observed in cell lines and primary cells, including resting human HSCs, as well as irradiated, nondividing human endothelial cells. In several cell types, transduction efficiencies approached those of an HIV-1 gene transfer system (14), and in most cell types, transgene expression was stable for at least 2 weeks. In addition, the titer of VSV-G-pseudotyped BIV preparations could be increased by centrifugation of the virus. One of the unconcentrated VSV-G-pseudotyped BIV preparations possessed a titer of approximately 5 × 105 infectious units (IU) per ml in the human kidney epithelial cell line 293T. With virus concentration, the titer could be raised much higher.
With these results, BIV is the third nonprimate lentivirus to be reported to efficiently transduce nondividing cells in vitro. VSV-G-pseudotyped gene transfer preparations derived from feline immunodeficiency virus were observed to transduce G1/S-arrested human and nonhuman cell lines, as well as postmitotic human monocyte-derived macrophages and neurons; viral concentration yielded titers (in growing HeLa cells) of 1.8 × 107 IU/ml (42). VSV-G-pseudotyped gene transfer preparations derived from equine infectious anemia virus were observed to transduce G1/S-arrested human (40) and canine cell lines, as well as postmitotic primary rat neurons, with titers in the canine cells as high as 5 × 106 IU/ml (33). In contrast, VSV-G-pseudotyped gene transfer preparations derived from visna virus transduce human and sheep cells poorly, due to blocks in reverse transcription and integration (4a).
Although the VSV-G-pseudotyped BIV gene transfer preparations transduced all of the human cells tested, the relative transduction efficiencies varied greatly. The epithelial cell line 293T, the T lymphoid cell line CEMSS and the endothelial cell line HUVEC were all transduced efficiently, although the percentage of eGFP+ cells declined over time in the 293T cells. In addition, although CEMSS cells were efficiently transduced, the same was not true for primary lymphocytes: BIV preparations transduced 16-fold fewer proliferating PBLs and 29 to 44-fold fewer unstimulated PBLs than did an HIV-1 preparation. This relative inefficiency is not surprising, in light of the BIV's lack of tropism for lymphocytes in vitro and in vivo (19). In contrast, BIV transduced HSCs moderately well, as a BIV preparation transduced only 2.5-fold fewer unstimulated HSCs than did the HIV-1 preparation.
In unstimulated human PBLs and unstimulated human HSCs, BIV transduced a portion of the cells in such a way as to express very low levels of eGFP, and only for a short duration (i.e., less than 2 weeks). One possible explanation for this phenomenon is pseudotransduction: eGFP protein expressed in the transfected cells is associated with the virus particles and transferred to the target cells during infection and then is diluted out of the cells as they divide. Pseudotransduction can occur with other transgenes besides eGFP and has been observed for VSV-G-pseudotyped MLV vectors (16, 28) and spleen necrosis virus vectors (J. Douglas and S. Tamaki, unpublished data). Other possible explanations for the transient eGFP phenomenon include eGFP expression from unintegrated forms of the vector which are degraded over time and transgene promoter inactivation occurring on integrated forms of the vector.
The HSCs infected with the 40× BIV contained, in addition to the population expressing very low levels of eGFP, a population which expressed high levels of eGFP. Despite being exposed to more virions, this population expressed slightly lower levels of eGFP per cell and was less stable over 2 weeks than the cells transduced with the 10× BIV. Since VSV-G protein is toxic to cells, it is possible that the 40× BIV contained levels of VSV-G that damaged the HSCs during infection.
The lower transduction efficiencies exhibited by BIV, relative to HIV-1, could be due to deficiencies during infection. For example, BIV RT or integrase might interact with human proteins less efficiently than do the HIV-1 counterparts. However, it is also possible that the BIV preparations contained fewer infectious virus particles than the HIV-1 preparations. Although the two viruses' packaging constructs were often found to produce comparable numbers of virus particles and to incorporate VSV-G with comparable efficiency, the extent of virion encapsidation of the full-length vector RNA and primer tRNA was not analyzed thoroughly. Preliminary experiments indicate that HIV-1 preparations contain up to 3.5-fold more full-length vector RNA than do BIV preparations (R. D. Berkowitz, unpublished data). In any regard, the transduction efficiencies exhibited by BIV should certainly rise as the titer of infectious virus is increased, i.e. by increasing virus production or the efficiency of vector RNA encapsidation. Such increases in titer should be particularly valuable for cell types that require a high multiplicity of infection for efficient transduction or high-level transgene expression.
In addition to increasing viral titer, future studies on BIV gene transfer systems should evaluate the possibilities of potentially harmful side effects of such systems, i.e., generation of replication-competent retrovirus and propagation by or recombination with HIV virions during natural HIV infection. BIV does not infect human cells, presumably due to a number of blocks including the lack of a competent viral envelope, but nevertheless BIV-transduced cells must be monitored closely with a sensitive replication-competent retrovirus detection assay. In addition, if an individual containing BIV-transduced cells is infected with HIV, it is theoretically possible that the HIV virions could spread the BIV vector to new cells and/or recombine with the vector. Although it is not known if such an event would be harmful, the likelihood of the event could be measured by in vitro cross-packaging (13a) and recombination studies.
ACKNOWLEDGMENT
CEMSS cells were obtained from the AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Adra C N, Boer P H, McBurney M W. Cloning and expression of the mouse pgk-1 gene and the nucleotide sequence of its promoter. Gene. 1987;60:65–74. doi: 10.1016/0378-1119(87)90214-9. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal M, Austin T W, Morel F, Chen J, Böhnlein E, Plavec I. Scaffold attachment region-mediated enhancement of retroviral vector expression in primary T cells. J Virol. 1998;72:3720–3728. doi: 10.1128/jvi.72.5.3720-3728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender M A, Palmer T D, Gelinas R E, Miller A D. Evidence that the packaging signal of Moloney murine leukemia virus extends into the gag region. J Virol. 1987;61:1639–1646. doi: 10.1128/jvi.61.5.1639-1646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkowitz R D, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 4a.Berkowitz R D, Ilves H, Plavec I, Veres G. Gene transfer systems derived from Visna virus: analysis of virus production and infectivity. Virology. 2001;279:116–129. doi: 10.1006/viro.2000.0659. [DOI] [PubMed] [Google Scholar]
- 5.Blomer U, Naldini L, Kafri T, Trono D, Verma I M, Gage F H. Highly efficient and sustained gene transfer in adult neurons with a lentivirus vector. J Virol. 1997;71:6641–6649. doi: 10.1128/jvi.71.9.6641-6649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchschacher G L, Panganiban A T. Human immunodeficiency virus vectors for inducible expression of foreign genes. J Virol. 1992;66:2731–2739. doi: 10.1128/jvi.66.5.2731-2739.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchschacher G L, Wong-Staal F. Development of lentiviral vectors for gene therapy for human disease. Blood. 2000;95:2499–2504. [PubMed] [Google Scholar]
- 8.Bukrinsky M, Sharova N, Dempsey M P, Stanwick T L, Bukrinskaya A G, Haggerty S, Stevenson M. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc Natl Acad Sci USA. 1992;89:6580–6584. doi: 10.1073/pnas.89.14.6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burns J C, Friedmann T, Driever W, Burrascano M, Yee J K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carswell S, Alwine J C. Efficiency of utilization of the simian virus 40 late polyadenylation site: effects of upstream sequences. Mol Cell Biol. 1989;9:4248–4258. doi: 10.1128/mcb.9.10.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charneau P, Alizon M, Clavel F. A second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. J Virol. 1992;66:2814–2820. doi: 10.1128/jvi.66.5.2814-2820.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charneau P, Mirambeau R G, Paulous P S, Buc H, Clavel F. HIV-1 reverse transcription. A termination step at the center of the genome. J Mol Biol. 1994;241:651–662. doi: 10.1006/jmbi.1994.1542. [DOI] [PubMed] [Google Scholar]
- 13.Choi H K, Won L A, Kontur P J, Hammond D N, Fox A P, Wainer B H, Hoffmann P C, Heller A. Immortalization of embryonic mesencephalic dopaminergic neurons by somatic cell fusion. Brain Res. 1991;552:67–76. doi: 10.1016/0006-8993(91)90661-e. [DOI] [PubMed] [Google Scholar]
- 13a.Curran M A, Kaiser S M, Achacoso P L, Nolan G P. Efficient transduction of non-dividing cells by optimised feline immunodeficiency virus vectors. Mol Ther. 2000;1:31–38. doi: 10.1006/mthe.1999.0007. [DOI] [PubMed] [Google Scholar]
- 14.Douglas, J., W.-Y. Lin, M. Panis, and G. Veres. Efficient HIV-based vector transduction of unstimulated human CD34+ cells in the SCID-hu Thy/Liv model of human T cell lymphopoiesis. Gene Ther., in press. [DOI] [PubMed]
- 15.Dron M, Hameau L, Benboudjema L, Guymarho J, Cajean-Feroldi C, Rizza P, Godard C, Jasmin C, Tovey M G, Lang M C. Cloning of a long HIV-1 readthrough transcript and detection of an increased level of early growth response protein-1 (Egr-1) mRNA in chronically infected U937 cells. Arch Virol. 1999;144:19–28. doi: 10.1007/s007050050482. [DOI] [PubMed] [Google Scholar]
- 16.Gallardo H F, Tan C, Ory D, Sadelain M. Recombinant retroviruses pseudotyped with the vesicular stomatitis virus G glycoprotein mediate both stable gene transfer and pseudotransduction in human peripheral blood lymphocytes. Blood. 1997;90:952–957. [PubMed] [Google Scholar]
- 17.Gallichan W S, Kafri T, Krahl T, Verma I M, Sarvetnick N. Lentivirus-mediated transduction of islet grafts with interleukin 4 results in sustained gene. Hum Gene Ther. 1998;10:2717–2726. doi: 10.1089/hum.1998.9.18-2717. [DOI] [PubMed] [Google Scholar]
- 18.Garvey K J, Oberste M S, Elser J E, Braun M J, Gonda M A. Nucleotide sequence and genome organization of biologically-active proviruses of the bovine immunodeficiency-like virus. Virology. 1990;175:391–409. doi: 10.1016/0042-6822(90)90424-p. [DOI] [PubMed] [Google Scholar]
- 19.Gonda M A, Luther D G, Fong S E, Tobin G J. Bovine immunodeficiency virus: molecular biology and virus-host interactions. Virus Res. 1994;32:155–181. doi: 10.1016/0168-1702(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 20.Hoshi H, McKeehan W L. Brain- and liver cell-derived factors are required for growth of human endothelial cells in serum-free culture. Proc Natl Acad Sci USA. 1984;81:6413–6417. doi: 10.1073/pnas.81.20.6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jang S K, Davies M V, Kaufman R J, Wimmer E. Initiation of protein synthesis by internal entry of ribosomes into 5′ nontranslated region of encephalomyocarditis virus RNA in vivo. J Virol. 1989;63:1651–1660. doi: 10.1128/jvi.63.4.1651-1660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson L G, Olsen J C, Naldini L, Boucher R C. Pseudotyped human lentiviral vector-mediated gene transfer to airway epithelia in vivo. Gene Ther. 2000;7:568–574. doi: 10.1038/sj.gt.3301138. [DOI] [PubMed] [Google Scholar]
- 23.Kafri T, Blomer U, Peterson D A, Gage F H, Verma I M. Sustained expression of genes delivered directly into liver and muscle by lentiviral vectors. Nat Genet. 1997;17:314–317. doi: 10.1038/ng1197-314. [DOI] [PubMed] [Google Scholar]
- 24.Kaye J F, Richardson J H, Lever A M L. cis-acting sequences involved in human immunodeficiency virus type 1 RNA packaging. J Virol. 1995;69:6588–6592. doi: 10.1128/jvi.69.10.6588-6592.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knaan-Shanzer F, Valerio D, van Beusechem V W. Cell cycle state, response to hemopoietic growth factors and retroviral vector-mediated transduction of human hemopoietic stem cells. Hum Gene Ther. 1996;3:323–333. [PubMed] [Google Scholar]
- 26.Lewis P, Hensel M, Emerman M. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 1992;11:3053–3058. doi: 10.1002/j.1460-2075.1992.tb05376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis P F, Emerman M. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J Virol. 1994;68:510–516. doi: 10.1128/jvi.68.1.510-516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu M L, Winther B L, Kay M A. Pseudotransduction of hepatocytes by using concentrated pseudotyped vesicular stomatitis virus G glycoprotein (VSV-G)-Moloney murine leukemia virus-derived retrovirus vectors: comparison of VSV-G and amphotropic vectors for hepatic gene transfer. J Virol. 1996;70:2497–2502. doi: 10.1128/jvi.70.4.2497-2502.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lower R, Lower J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malim M H, Hauber J, Fenrick R, Cullen B R. Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature. 1988;335:181–183. doi: 10.1038/335181a0. [DOI] [PubMed] [Google Scholar]
- 31.Malim M H, Hauber J, Le S Y, Maizel J V, Cullen B R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 32.Mayer J, Sauter M, Racz A, Scherer D, Mueller-Lantzsch N, Meese E. An almost-intact human endogenous retrovirus K on human chromosome 7. Nat Genet. 1999;21:257–258. doi: 10.1038/6766. [DOI] [PubMed] [Google Scholar]
- 33.Mitrophanous K, Yoon S, Rohll J, Patil D, Wilkes F, Kim V, Kingsman S, Kingsman A, Mazarakis N. Stable gene transfer to the nervous system using a non-primate lentiviral vector. Gene Ther. 1999;6:1808–1818. doi: 10.1038/sj.gt.3301023. [DOI] [PubMed] [Google Scholar]
- 34.Miyoshi H, Blomer U, Takahashi M, Gage F H, Verma I M. Development of a self-inactivating lentivirus vector. J Virol. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyoshi H, Smith K A, Mosier D E, Verma I M, Torbett B E. Transduction of human CD34+ cells that mediate long-term engraftment of NOD/SCID mice by HIV vectors. Science. 1999;283:682–686. doi: 10.1126/science.283.5402.682. [DOI] [PubMed] [Google Scholar]
- 36.Miyoshi H, Takahashi M, Gage F H, Verma I M. Stable and efficient gene transfer into the retina using an HIV-based lentiviral vector. Proc Natl Acad Sci USA. 1997;94:10319–10323. doi: 10.1073/pnas.94.19.10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naldini L, Blomer U, Gage F H, Trono D, Verma I M. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci USA. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 39.Oberste M S, Williamson J C, Greenwood J D, Nagashima K, Copeland T D, Gonda M A. Characterization of bovine immunodeficiency virus rev cDNAs and identification and subcellular localization of the rev protein. J Virol. 1993;67:6395–6405. doi: 10.1128/jvi.67.11.6395-6405.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olsen J C. Gene transfer vectors derived from equine infectious anemia virus. Gene Ther. 1998;5:1481–1487. doi: 10.1038/sj.gt.3300768. [DOI] [PubMed] [Google Scholar]
- 41.Parolin C P, Dorfman T, Palu G, Gottlinger H, Sodroski J. Analysis in human immunodeficiency virus type 1 vectors of cis-acting sequences that affect gene transfer into human lymphocytes. J Virol. 1994;68:3888–3895. doi: 10.1128/jvi.68.6.3888-3895.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poeschla E, Wong-Staal F, Looney D. Efficient transduction of nondividing human cells by feline immunodeficiency virus lentiviral vectors. Nat Med. 1998;4:354–357. doi: 10.1038/nm0398-354. [DOI] [PubMed] [Google Scholar]
- 43.Robbins P B, Yu X J, Skelton D M, Pepper K A, Wasserman R M, Zhu L, Kohn D B. Increased probability of expression from modified retroviral vectors in embryonal stem cells and embryonal carcinoma cells. J Virol. 1997;71:9466–9474. doi: 10.1128/jvi.71.12.9466-9474.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roe T, Reynolds T C, Yu G, Brown P O. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schek N, Cooke C, Alwine J C. Definition of the upstream efficiency element of the simian virus 40 late polyadenylation signal by using in vitro analyses. Mol Cell Biol. 1992;12:5386–5393. doi: 10.1128/mcb.12.12.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartz S, Felber B K, Pavlakis G N. Distinct RNA sequences in the gag region of human immunodeficiency virus type 1 decrease RNA stability and inhibit expression in the absence of Rev protein. J Virol. 1992;66:150–159. doi: 10.1128/jvi.66.1.150-159.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trono D. Lentiviral vectors: turning a deadly foe into a therapeutic agent. Gene Ther. 2000;7:20–23. doi: 10.1038/sj.gt.3301105. [DOI] [PubMed] [Google Scholar]
- 48.Uchida N, He D, Friera A, Reitsma M, Sasaki D, Chen B, Tsukamoto A. The unexpected G0/G1 cell cycle status of mobilized hematopoietic stem cells from peripheral blood. Blood. 1997;89:465–472. [PubMed] [Google Scholar]
- 49.Vara J A, Portela A, Ortin J, Jimenez A. Expression in mammalian cells of a gene from Streptomyces alboniger conferring puromycin resistance. Nucleic Acids Res. 1986;14:4617–4624. doi: 10.1093/nar/14.11.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Appukuttan B, Ott S, Pate I R, Irvine J, Song J, Park J H, Smith R, Stout J T. Efficient and sustained transgene expression in human corneal cells mediated by a lentiviral vector. Gene Ther. 2000;7:196–200. doi: 10.1038/sj.gt.3301075. [DOI] [PubMed] [Google Scholar]
- 51.Yee J K, Miyanohara A, Laporte P, Bouic K, Burns J C, Friedmann T. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc Natl Acad Sci USA. 1994;91:9564–9568. doi: 10.1073/pnas.91.20.9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zufferey R, Dull T, Mandel R J, Bukovsky A, Quiroz D, Naldini L, Trono D. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]