Abstract
Influenza viruses pose a threat to public health as evidenced by severe morbidity and mortality in humans on a yearly basis. Given the constant changes in the viral glycoproteins owing to antigenic drift, seasonal influenza vaccines need to be updated periodically and effectiveness often drops due to mismatches between vaccine and circulating strains. In addition, seasonal influenza vaccines are not protective against antigenically shifted influenza viruses with pandemic potential. Here, we have developed a highly immunogenic vaccination regimen based on live-attenuated influenza vaccines (LAIVs) comprised of an attenuated virus backbone lacking non-structural protein 1 (ΔNS1), the primary host interferon antagonist of influenza viruses, with chimeric hemagglutinins (cHA) composed of exotic avian head domains with a highly conserved stalk domain, to redirect the humoral response towards the HA stalk. In this study, we showed that cHA-LAIV vaccines induce robust serum and mucosal responses against group 1 stalk and confer antibody-dependent cell cytotoxicity activity. Mice that intranasally received cH8/1-ΔNS1 followed by a cH11/1-ΔNS1 heterologous booster had robust humoral responses for influenza A virus group 1 HAs and were protected from seasonal H1N1 influenza virus and heterologous highly pathogenic avian H5N1 lethal challenges. When compared with mice immunized with the standard of care or cold-adapted cHA-LAIV, cHA-ΔNS1 immunized mice had robust antigen-specific CD8+ T-cell responses which also correlated with markedly reduced lung pathology post-challenge. These observations support the development of a trivalent universal influenza vaccine for the protection against group 1 and group 2 influenza A viruses and influenza B viruses.
Subject terms: Live attenuated vaccines, Influenza virus
Introduction
Seasonal influenza virus infections are estimated to cause globally 290,000 to 650,000 deaths annually1. Given that vaccines are the best form of control measure against influenza virus infections, the US Advisory Committee on Immunization Practices (ACIP) recommends the use of seasonal influenza vaccines for individuals aged 6 months and older2. Although seasonal influenza vaccines can mitigate the disease burden, variable effectiveness is observed annually, especially when vaccine strains mismatch circulating strains. In addition, despite the availability of seasonal vaccines, influenza A (IAV) and influenza B (IBV) viruses continue to circulate in humans on a global scale3. The latter issue is due to the constant antigen drift of the viral hemagglutinin (HA) which is the primary target for seasonal vaccines4. Furthermore, occasional antigenic shift due to zoonotic outbreaks of IAV strains containing “novel” HA genes derived from non-human IAV can result in human influenza pandemics4–7. In the past, influenza pandemics were observed in 1918 (‘Spanish Flu’; >50 million deaths), 1957 (‘Asian Flu’; 1–2 million deaths), 1968 (H3N2; >700,000 deaths) and the most recent pandemic which took place in 2009 (‘swine flu’; >300,000 deaths)8.
Currently, licensed seasonal influenza virus vaccines are formulated as inactivated influenza virus vaccines (IIV), live-attenuated influenza virus vaccines (LAIV) based on cold-adapted influenza viruses with limited replication in the lower respiratory tract, or HA-based vaccines based on recombinant protein technology9. IIVs and recombinant HA-protein-based vaccines have been shown to induce a suboptimal immune response in adults against antigenically mismatched viruses (especially in the absence of adjuvants), and these categories of vaccines are known not to induce a mucosal immune response which might better protect from respiratory infection by influenza viruses10. Conversely, LAIVs which are recommended for the use of individuals aged between 2 - 49 years, can induce robust humoral and cell-mediated immune responses given that they mimic a natural infection by intranasal administration11. Studies have indicated that some of the key protective immunological factors are IgA responses in the upper respiratory tract, IgG-dominant serum responses that readily transude, and tissue-resident memory CD8+ T-cells that confer heterosubtypic protection in subsequent exposures. In addition, the longevity and potency of the immune responses induced by LAIVs are greatly dependent on the activation of T-follicular helper cells (Tfh)12,13. However, these have not been identified as correlates of protection by regulatory agencies11,14–17. Moreover, the vaccine loses efficacy in older adults due to decreased vaccine virus replication and low immune responses in this segment of the human population, which includes a major risk group for influenza virus-mediated severe disease.
The diverse antigenic HA groups (19) and NA groups (11) circulating in animal reservoirs along with the time frame (~6 months) to develop all the required doses of an influenza vaccine once a pandemic outbreak is detected highlight the need for a vaccine that would protect against all influenza viruses regardless of the antigenic make-up, while inducing long-lasting immunity18–20. These features will also eliminate the need for annual reformulations of influenza vaccines. Accordingly, the National Institute of Allergy and Infectious Diseases (NIAID) initiated a directive to develop a “universal influenza vaccine” (UIV) that would protect against antigenically drifted and shifted influenza viruses while also curbing symptomatic seasonal influenza infections21. Scientists around the world have focused on conserved epitopes such as the HA stalk, viral nucleoprotein (NP), matrix 1 (M1), and the ectodomain of the M2 ion channel (M2e) as target antigens for the basis of UIVs22–24. HA stalk-directed antibodies have been demonstrated to possess neutralizing capacity at specific stages of the virus life cycle, such as inhibition of fusion, uncoating, and viral budding. In addition, these broadly cross-reactive antibodies are known to induce Fc-Fcγ receptor-mediated effector functions such as antibody-dependent cell cytotoxicity (ADCC), antibody-dependent cellular phagocytosis (ADCP), and complement-mediated lysis, which also contribute to in vivo protection from influenza disease24.
In this study, we have developed a prime-boost vaccination regimen using an in vivo replication incompetent IAV (ΔNS1) containing chimeric HAs (cHA) for the induction of HA stalk antibodies able to confer robust protection against group 1 IAV. Prime-boost approaches using cHA immunogens previously have been shown to induce protective levels of HA stalk antibodies against influenza A and B viruses in several animal models, as well as in human clinical trials25–29. In a ferret model, we have previously shown that a prime-boost regimen using cold-adapted temperature-sensitive cHA-LAIVs induced broadly cross-reactive humoral responses that protected the animals from an IAV with pandemic potential30. In a follow-up study, the durability of the generated immune response in a ferret model was assessed indicating that sequential immunization of cHA-LAIV-cHA-LAIV and cHA-LAIV-cHA-IIV induced long-lived HA-stalk specific and broadly reactive humoral, and cross-reactive T-cell mediated responses. When compared to other immunization schemes, the cHA-LAIV-cHA-LAIV vaccination provided superior protection against an infection by a 2009 pandemic H1N1 (pH1N1) IAV31. Nevertheless, in a human clinical trial, the cold-adapted LAIV was poorly immunogenic. This prompted us to evaluate a different live attenuation approach known to be safe and immunogenic32,33. Specifically, we used a modified virus backbone that is devoid of the main viral interferon antagonist, the non-structural protein 1 (NS1)34. NS1 has been shown to inhibit the host interferon (IFN) response by sequestering viral factors such as dsRNA which trigger pathogen-associated molecular patterns (PAMPS)35. Conversely, NS1 is known to directly interact with host factors involved in innate immunity such protein kinase R (PKR), retinoic acid-inducible gene I (RIG-I), and oligoadenylate synthetase 1 (OAS1), resulting in inhibition of the host-induced antiviral innate response36–38. In a previous report, we showed that influenza viruses lacking NS1 (ΔNS1) are highly immunogenic in vivo, while conferring a broad interferon-mediated antiviral state that results in both short-lived but immediate innate protection, followed by long-lasting adaptive immune protection39,40. In fact, ΔNS1 IAV are inherently more immunogenic than wild-type IAV. Several clinical trials have shown that ΔNS1-LAIVs are immunogenic and well tolerated in humans, leading to robust humoral and cellular immune responses.
In the present study, we have generated ΔNS1 viruses expressing cHAs corresponding to the head of H8 and the stalk of H1 (cH8/1) and to the head of H11 and the stalk of H1 (cH11/1). We then show that mice that were immunized with a cH8/1-N1-ΔNS1 prime followed by cH11/1-N1-ΔNS1 boost had robust levels of humoral immunity across a multitude of group 1 IAV HAs, including stalk-specific responses. Consequently, effector functions conferred by the stalk antibodies induced by this immunization regimen were superior compared to other immunization regimens assessed in the present study. Furthermore, mucosal IgA HA stalk responses for this group were superior to their cold-adapted temperature-sensitive cHA-LAIV counterparts. The cHA-ΔNS1 vaccine regimen also induced high levels of tissue-resident antigen-specific memory CD8+ T-cells and splenic antigen-specific IFN-γ+-CD8+ responses. Challenges with heterologous highly pathogenic avian influenza (HPAI) virus further demonstrated the efficacy of the vaccine-induced immunity indicating the suitability of this vaccine platform for a universal influenza vaccine candidate. Additionally, given the in vivo replication-deficient nature of ΔNS1 based backbones, a strategy as such could further increase the safety and immunogenicity of an LAIV universal vaccine.
Results
Comparative In vitro characterization of cHA-LAIV vaccine strains
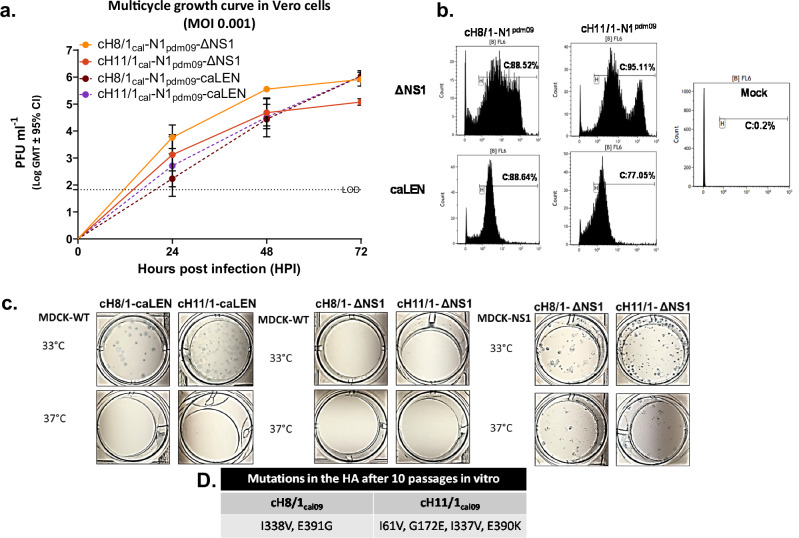
The cHA-ΔNS1-LAIVs viruses were rescued using reverse genetics and were passaged ten times at 33 °C for 72 h. The resulting viruses were plaque purified and were assessed for growth with plaque purified cell culture grown cHA-caLEN viruses. Figure 1a indicates that cHA-ΔNS1-LAIV strains replicate in a multi-cycle growth curve in an IFN-incompetent Vero cell line during 72 h at 33 °C. The cHA-caLEN viruses showed growth competence but had relatively slower kinetics than their counterparts. One way ANOVA tests indicated that the cH8/1-ΔNS1-LAIV grew to a significantly higher level in comparison to the other viruses at both 24 h and 48 h timepoints. By 72 h, cH8/1-ΔNS1-LAIV grew significantly more than the cH11/1-ΔNS1-LAIV, while the other assessed viruses caught up with the titers of cH8/1-ΔNS1-LAIV. Thus, the data indicated that cH11/1-ΔNS1-LAIV demonstrated slower replication kinetics. Given that ΔNS1-LAIV viruses were passaged over ten times to ensure genetic stability, we were interested in assessing any antigenic changes in the HA stalk. Amino-acid changes observed in the HA stalk of the ΔNS1-LAIVs are indicated in Fig. 1d and might be related to selection for good compatibility with the new HA heads. To assess whether these changes impact antigenicity of the H1 stalk, we used flow cytometric analysis with a broadly neutralizing HA stalk antibody (Fig. 1b) and observed a high level of reactivity with infected Vero cells, although cHA-ΔNS1-LAIVs displayed more heterogenicity than cHA-caLAIV with respect to levels of HA staining of infected cells.
Fig. 1. Characterization of cHA-LAIV vaccine strains in vitro.
a The final purified viral stocks were used to assess the comparative replication competence in Vero-CCL81 cells at a multiplicity of infection (MOI – 0.001) in triplicate and all viral supernatants were assessed by plaque assay. b Vero cells were infected at an MOI – 5 for 16 h for a single replication cycle and cells were stained using the broadly neutralizing antibody CR9114 for flow cytometric analysis. Infection percentages are shown for each virus. c cHA-LAIVs were plaqued in MDCK and MDCK-NS1 (only for ΔNS1 viruses) at 33 or 37 °C for 48 h. Plaques were immunostained using HT103 mAb (anti-NP) to assess temperature sensitive and attenuated phenotypes. d Amino acid changes noted in the HAs of cHA-ΔNS1 after passaging 10 times after virus rescue. Statistics were done using one way-ANOVA conducting multiple comparison against cH8/1-ΔNS1. **P < 0.083, ***P < 0.0009 and ****P < 0.0001. Data are shown as mean ± SE.
We next investigated the ability of all these viruses to plaque in IFN-competent MDCK cells at 33 °C or 37 °C for 48 h. In addition, we also used MDCK cell constitutively expressing NS1 (MDCK-NS1) to plaque ΔNS1 viruses. As expected, a temperature-sensitive phenotype was observed for cHA-caLEN viruses as indicated by the lack of plaque formation at 37 °C. ΔNS1-LAIV viruses were able to infect at both temperatures in MDCK-NS1 cells indicating reduced sensitivity to higher temperature. However, the attenuated nature of these viruses was evident by their inability to form plaques in wild-type MDCK cell monolayers at either temperature (Fig. 1c).
cHA-ΔNS1-LAIV prime-boost vaccination regimen induces robust serum and mucosal HA stalk responses and confers ADCC reporter assay activity
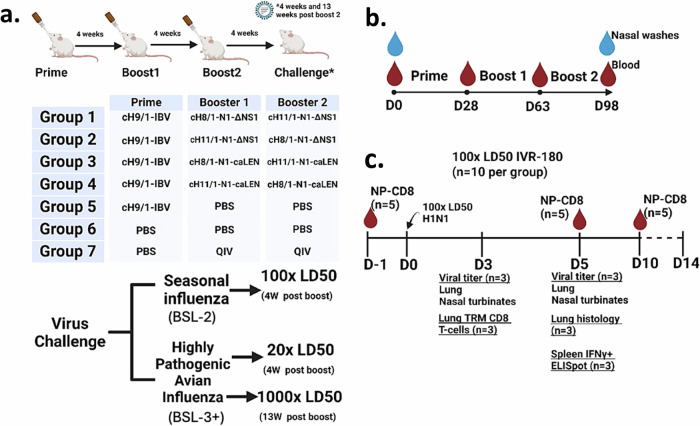
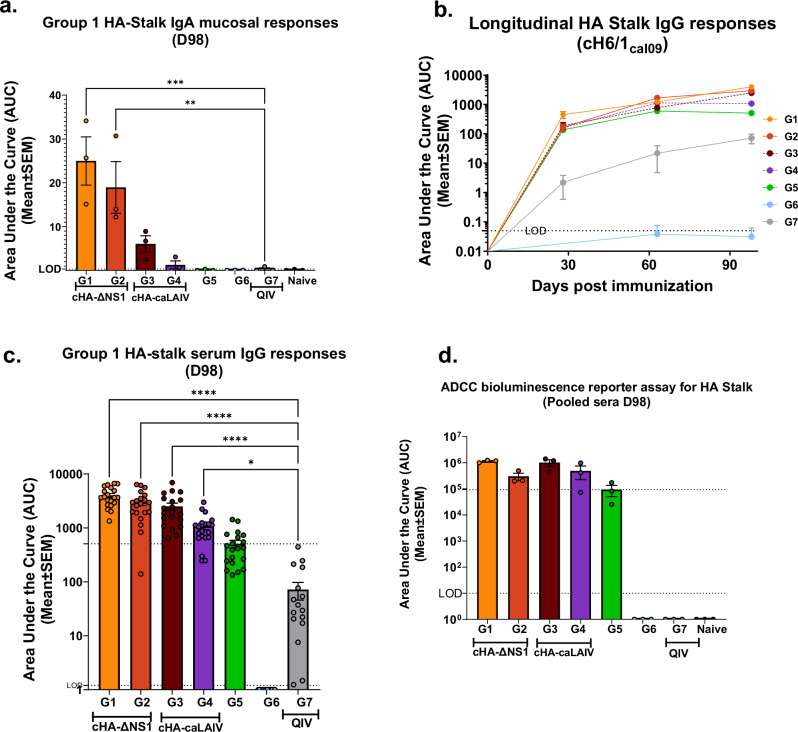
We next immunized groups of mice according to the scheme in Fig. 2a. Mice immunized with cHA viruses were primed with an influenza B virus expressing a cH9/1 HA to simulate a low level of pre-existing immunity against the group 1 HA stalk present in the human population. As indicated in Fig. 2b, mice were bled between and after immunization regimens to assess the serum-derived humoral responses. LAIVs are known inducers of a mucosal response11,41. Therefore, we assessed the mucosal IgA response against the HA stalk derived from clarified nasal washes to compare the differences between QIV as well as the different regimens of LAIVs used in this study. The area under the curve values (AUCs) derived from endpoint titers for nasal washes indicated that ΔNS1-LAIV vaccinated mice had a higher mucosal IgA response in comparison to responses to cold-adapted counterparts (caLEN-LAIV). As expected, the intramuscularly administered standard of care failed to induce mucosal HA stalk responses (Fig. 3a). As expected, all groups (except group 6) showed increased group1 HA stalk titers as a function of time with boosting (Fig. 3b). All cHA-LAIVs had higher responses compared to the HA-stalk prime only (group 5) and nonadjuvanted standard of care (QIV) (Fig. 3c). When considering individual responses at the end of the immunization regimen, ΔNS1-LAIV vaccinated groups showed comparatively higher responses than the caLEN-LAIV groups. Interestingly, mice that received a cH8/1-LAIV first had better responses than groups that received cH11/1-LAIV first. When compared to the standard of care, groups 1-4 had statistically significant higher responses of HA stalk titers while group 1 had the highest titers from groups 1-4. Given that HA stalk antibodies are known to induce ADCC activity, we used a modified reporter assay using an in-house generated MDCK-HA cell-line harboring a cH6/1 (same HA as the cH6/1 protein) to test this hypothesis. While groups that received cHA-LAIVs had higher ADCC activity in sera compared to other vaccination regimens, group 1 had the highest ADCC reporter activity (Fig. 3d). As anticipated, the standard of care failed to mount ADCC activity likely due to its bias towards an IgG1 serological phenotype (Fig. 3d, group 7).
Fig. 2. cHA-LAIV study design and sample collection.
a Animals were randomly assigned into seven groups (n = 35). Groups 1-5 were intranasally infected with a sub-lethal dose of purified cH9/1-IBV (2 × 105 PFU). Mice were immunized in 4-week intervals. Group 1 received cH8/1-N1-ΔNS1 and then cH11/1-N1-ΔNS1, group 2 received the vaccine doses in reverse order (105 PFU per dose). Groups 3 and 4 followed the regimens of groups 1 and 2 except that the strains had a cold-adapted temperature-sensitive LAIV backbone (with a full length, functional NS1). Group 5 and 6 were mock immunized with identical volumes of sterile 1x PBS. Group 7 was intramuscularly given 50 µl of non-adjuvanted QIV per dose (~1.5 µg per dose). We used Group 7 as a “standard-of-care” vaccine comparator. Although this group does not include a cH9/1-IBV, previous experiments demonstrated that even in the presence of priming, QIV vaccination did not confer good protection against heterologous challenges26. Animals were intranasally challenged 4 weeks post final-boost either with IVR-180 (BSL-2; 100x LD50) or influenza A/Vietnam/1203/04 virus (BSL-3 + ; 20x LD50) or 13 weeks post final-boost for a separate BSL-3+ challenge (1000x LD50). b Animals were bled (submandibular), and nasal washes were taken at D0 (naïve) and D98 animals. Mice were bled between immunization doses at approximately 28-day intervals up until D98 since the first dose. c Sample collection for BSL2 100 LD50 challenge included submandibular bleeds at D1, D5 and D10 (n = 5) for pooled circulating tetramer specific CD8+ T-cell analysis. Lungs (left lobes) and nasal turbinates were harvested on D3 and D5 for viral titration. Right lobes of lungs on D3 were used for analysis of tissue-resident memory CD8+ T-cell analysis and on D5 for histopathology. Spleens were harvested on D5 for enzyme-linked immunosorbent spot (ELISpot) and intracellular staining assays. For BSL-3+ studies, whole-lung and nasal turbinates were harvested on D3 and D5 for viral titration. Image was created using BioRender.
Fig. 3. cHA-LAIV vaccines induce robust serum and mucosal responses against group 1 stalk and confer ADCC reporter assay activity.
Longitudinal area-under-the-curve values (AUCs) were calculated using endpoint titers from serum/nasal washes derived from mice before, between and after immunization. a Mucosal total IgA titers depicted as AUC values for group 1 HA stalk derived from endpoint titers of (n = 3) from animal nasal washes (undiluted starting dilution) on D98. b Longitudinal group 1 HA stalk total IgG titers from pooled sera. c Individual values of group 1 HA stalk total IgG titers from serum on D98. d ADCC reporter assay activity from serially diluted serum samples (1:20 starting dilutions) in which endpoint titers were used to calculate area under the curve values (AUCs) plotted from 3 sets of pooled animal serum from each respective group. In (a), significance established by comparisons against the group with the highest values (Group 1) while all other panels had comparisons to QIV standard of care group (Group 7) using one way-ANOVA for multiple comparisons corrected and adjusted as per Dunnett’s correction. ****P- < 0.0001, ***P-0.001, **P-0.002, *P-0.02. Data are shown as mean ± SEM. Limits of detection (LOD) is shown in dotted lines. Upper dotted line indicates HA-Stalk Prime only (Group 5) mean.
In comparison to the LAIV vaccination groups the standard of care (QIV) group failed to mount robust responses for the antigens that were tested here. Data indicated here suggests that ΔNS1-LAIV vaccination regimens induced comparatively higher responses for group 1 stalk, H18, and H2 antigens when compared to the cHA-caLEN vaccination regimen (Fig. 4). In general, all LAIV groups had significantly higher serological readouts (except for ADCC) as compared to baseline HA-stalk primed mice. In all cases, ΔNS1-LAIV groups had statistically significant higher titers when compared to the QIV group by D98 as measured by a multiple-comparison one-way ANOVA test for all used antigens (Fig. 4a–c bottom panels). Animals from group 5 induced an HA-specific immune response conferred by the cH9/1-IBV (Influenza B virus) sublethal infection to induce pre-existing immunity while group 6 failed to develop titers as expected, given that these animals were mock immunized. In summary group 1 of the ΔNS1-LAIV had the highest HA stalk humoral response (although not significant between LAIV groups).
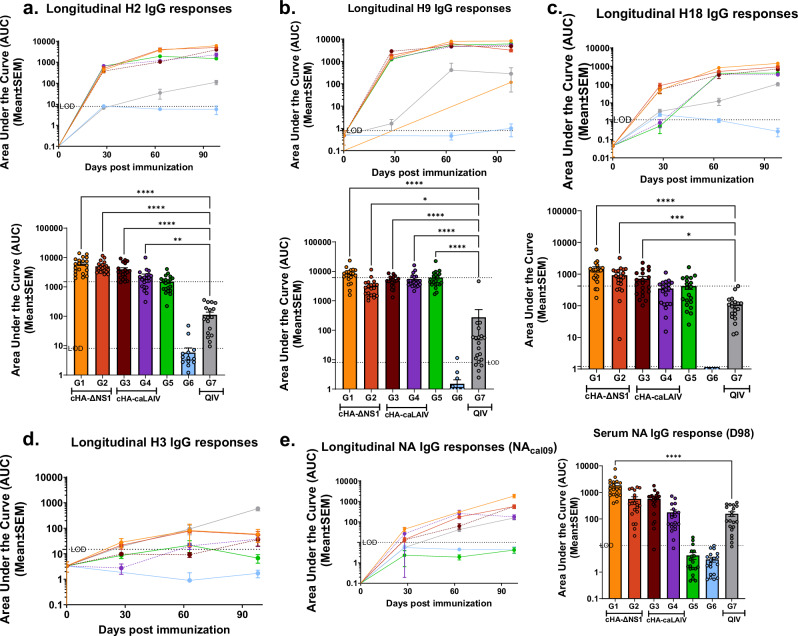
Fig. 4. cHA-LAIV vaccines induce broad group specific HA and NA total IgG serum responses.
Longitudinal area under the curve values (AUCs) were calculated using endpoint titers from serum derived from mice before, between and after immunization (A-C top panels). Respective individual AUC values on D98 (A-C bottom panels) are also shown. a Total serum IgG responses for full-length H2. b Total serum IgG responses for full-length H9. c Total serum IgG responses for full-length H18. d Total serum IgG responses for full-length H3 (group 2 HA). e Longitudinal serum IgG responses against N1 NA and individual values on D98 for N1-NA responses (right panel). Significance was calculated compared to the QIV standard of care group (Group 7) using one way-ANOVA for multiple comparisons corrected and adjusted as per Dunnett’s correction. ****P- < 0.0001, ***P-0.0001, **P-0.001, *P-0.01. Data are shown as mean ± SEM. Limits of detection (LOD) is shown in dotted lines. Upper dotted line indicates HA-Stalk Prime only (Group 5) mean.
cHA-ΔNS1-LAIV prime-boost vaccination regimen induces robust humoral responses against diverse group 1 specific HAs and N1 NA antigens
Given the importance of a broad humoral response in developing a universal influenza vaccine, we focused on the breadth of serum IgG responses by testing against a panel of diverse group specific HAs, a group 2 HA and N1 NA. All cHA-LAIV vaccination platforms showed immune responses towards the group specific HAs (H2, H9, and H18) in a longitudinal manner (Fig. 4a–c, top panels). QIV immunized mice demonstrated a similar trend longitudinally, albeit to a lesser extent.
When considering individual values for each of these group specific HAs on D98, all cHA-LAIV vaccination regimens had significantly higher titers for H2 and H18 when compared with mice that received the standard of care. Furthermore, all groups except group 6 had significantly higher responses against the full length H9 coinciding with the initial intranasal administration of cH9/1-IBV virus. For all group 1 specific HAs tested here, the data from D98 derived serum indicated that mice from group 1 had the highest responses, all of which were statistically significant when compared to QIV (Fig. 4a–c, bottom panels). Like the HA stalk responses observed in Fig. 3, mice immunized first with cH8/1-LAIVs tend to demonstrate higher responses when compared to mice that received first a cH11/1-LAIV for H2 and H18 HA antigens. The group 1 specific responses towards the HA were observed by the inability of the cHA-LAIV immunized mice to confer a boosting effect towards a group 2 HA antigen (H3) tested here. In contrast, mice that received QIV vaccinations were able to boost H3-specific HA responses by the end of the immunization regime (Fig. 4d), consistent with the presence of H3-inactivated viruses in QIV. NA as an immunogen has been also shown to induce protection from influenza virus infections42. Therefore, we assessed N1-NA serum total IgG responses via ELISA and the data suggested that immunized mice from group 1 had higher NA responses compared to caLEN-LAIVs (Fig. 4e, left panel). By the end of the immunization regimen group 1 mice had significantly higher titers compared to mice that received QIV, and mice immunized first with cH8/1 had higher responses within their respective groups (Fig. 4e, right panel).
cHA-LAIV vaccination protects mice from a lethal challenge with seasonal-like influenza virus
To test whether the immune responses conferred by cHA-LAIV regimens protected against high-dose lethal challenge from a seasonal influenza virus, mice were challenged with a 100x LD50 of the QIV-matched IVR-180 (H1N1) strain.
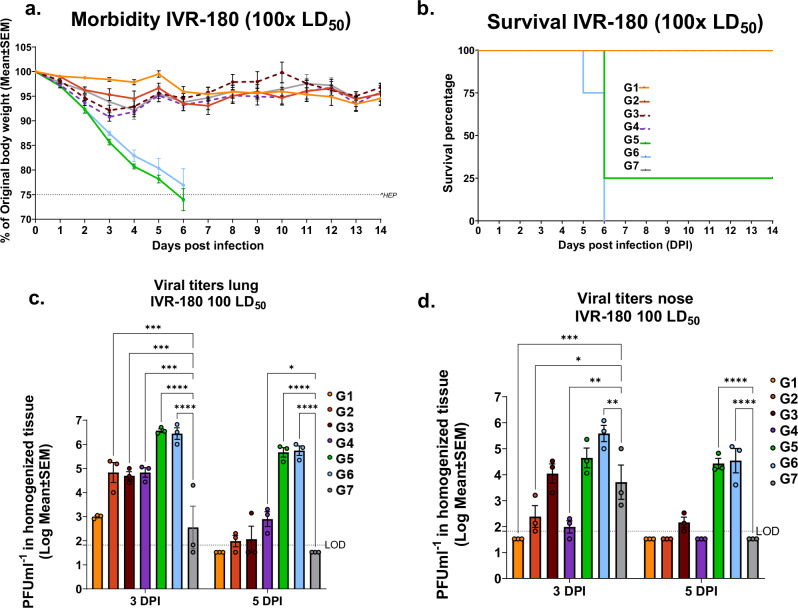
Weight loss induced by the seasonal influenza virus challenge was minimal between the immunized groups (Fig. 5a). Furthermore, all the vaccinated animals had 100% survival and animals that had pre-existing HA stalk immunity due to prior infection with the influenza B virus expressing a cHA9/1 had 25% survival in this lethal challenge (Fig. 6b) while mock immunized animals succumbed to death.
Fig. 5. cHA-LAIV vaccination protects mice from a lethal high-dose seasonal influenza challenge and confers superior upper respiratory tract protection.
Four weeks post-final-boost, mice were challenged with a QIV-matched IVR-180 H1N1 virus using a lethal dose of 100x LD50. a Morbidity was assessed by monitoring weight loss. b Survival of challenged animals. c Viral titers in lungs assessed by plaque assays on day 3 and day 5 post-infection. d Viral titers in nasal turbinates on day 3 and 5 post-infection. Each dot represents one animal. Data are shown as mean ± SEM. Statistical significance was compared to QIV standard of care group (Group 7) using 2 way-ANOVA with Dunnett multiple comparisons. ****P- < 0.0001, ***P-0.0001, **P-0.001, *P-0.01. Limits of detection (LOD) is shown in dotted lines.
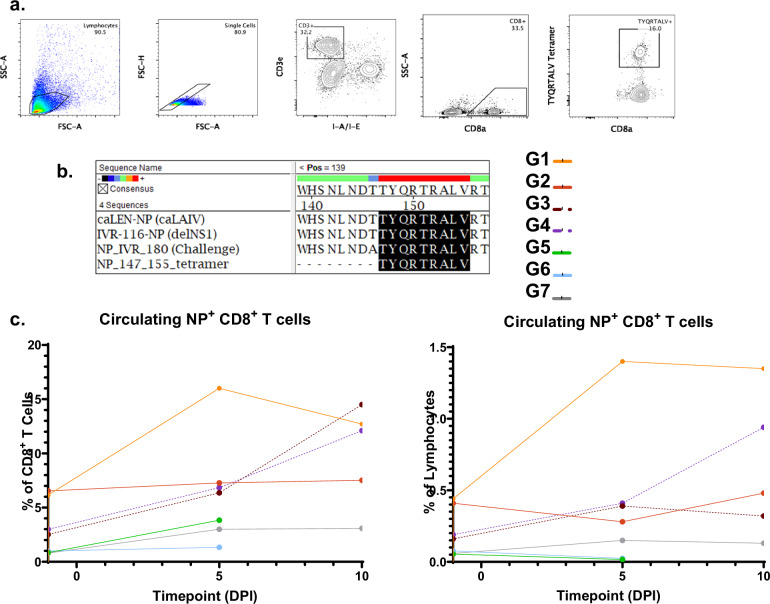
Fig. 6. cHA-ΔNS1-LAIV vaccination induces potent antigen-specific cytotoxic CD8 T-cell responses during virus challenge.
Four weeks post-final-boost, mice (n = 5 per group) were challenged with a QIV-matched IVR-180 using a lethal dose of 100x LD50. a A representative image of the gating strategy used to probe NP-tetramer+ CD8+ T-cell responses of pooled blood derived from mice on D1, D5, and D10 post-challenge. (The same animals were followed throughout the study). The following markers were utilized for the gating strategy; CD3e – T cell marker, CD8a – CD8 T cell marker. b Amino-acid sequence of the NP tetramer used in this study corresponds to all the NPs of the viruses used in this study. c Longitudinal assessment of IAV-NP+CD8+ circulating T-cells expressed as a percentage of total CD8+ T-cells (left) and percentage of total lymphocytes (right).
Viral replication in lungs was highest in groups 5 and 6 (IBV cH9/1 infection only and mock immunized) on both days. Group 1 that received the cH8/1-ΔNS1 and cH11/1-ΔNS1 boost had lower levels of viral titers on D3 as compared to the other groups and had complete clearance by 5 days post-infection. The strain-matched QIV group had a similar trend where virus clearance was apparent by day 5, although the titers were comparatively higher on D3 (Fig. 5c). As indicated in Fig. 5d, group 1 immunized mice had no detectable levels of virus in nasal turbinates on any of the sampled days. In general, all the LAIV groups (except group 3) had lower viral titers in nasal turbinates as compared to controls. Group 7 also showed clearance by 5 days post-infection (Fig. 5d).
cHA-ΔNS1-LAIV vaccination induces potent circulating antigen-specific cytotoxic T-cell responses during the virus challenge
By using NP-tetramers specific for the highly conserved TYQRTRALV epitope (Fig. 6b) we used flow cytometry to assess the level of IAV-specific circulating cytotoxic T-cell (CTL; CD8+) responses after IAV challenge of immunized mice (Fig. 6a includes the gating scheme). We used the well-characterized immunodominant (conserved) T-cell epitope (TYQRTRALV) expressed from NP as a model antigen to gauge T-cell responses.
In general, cHA-ΔNS1-LAIV vaccinated mice (groups 1 and 2) had a high starting baseline (pre-challenge) for IAV-NP specific CTL responses (Fig. 6c). When considering this population of immune cells as a percentage of all lymphocytes, a dramatic expansion was observed by day 5 for the cHA-ΔNS1-LAIV immunized group 1, which then started to plateau by day 10 (Fig. 6c; right panel). The effect was similar when the data were assessed as a percentage of total CD8+ T cells (Fig. 6c; left panel).
In contrast, mice that were immunized with the unadjuvanted-QIV prime-boost regimen, failed to mount a robust CTL response. Conversely, cHA-caLEN immunized mice demonstrated high amounts of CTL activation, although the starting baseline for these groups was lower in comparison to mice that received LAIVs lacking a full-length NS1. When comparing the increase of trends with time, cHA-caLEN immunized mice demonstrated a slower rate of CTL expansion in comparison to their cHA-ΔNS1-LAIV counterpart from group 1. Interestingly group 2 (cHA-ΔNS1 group that received cH11/1 first) did not show expansion after challenge despite a high baseline, while group 1 or the cHA-caLEN groups did (Fig. 6c left and right panels).
Activation and expansion of tissue-resident memory (TRM) IAV-NP+CD8+ T cells of mice that were intranasally immunized with cHA-ΔNS1-LAIV vaccines
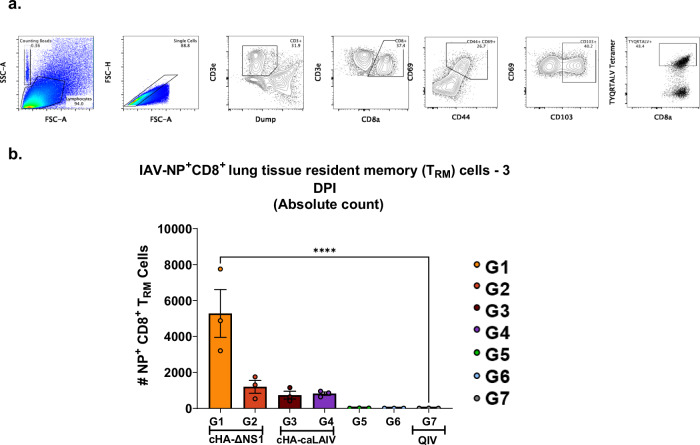
Influenza virus antigen-specific T cells that reside in the lungs (tissue-resident memory T cells, TRM) have been shown to be reactivated upon homologous and heterologous virus challenge. These T cells can reduce disease burden and associated pathology to confer improved survival43. Universal influenza vaccine candidates are characterized by their ability to induce heterologous and heterosubtypic immunity. Given that these T-cells target epitopes in conserved viral proteins such as NP, we used NP tetramers to assess antigen-specific TRM CD8+ CTLs derived from immunized mice after challenge with 100x LD50 of IVR-180 (3 days post-infection). We utilized flow cytometry for the processed lung tissues and assessed the absolute values of CD8+ TRMs as shown in Fig. 7a.
Fig. 7. cHA-ΔNS1-LAIV vaccination activates tissue-resident memory (TRM) IAV-NP+CD8+ T-cells on day 3 post-infection.
Four weeks post-final-boost, mice were challenged with a QIV matched IVR-180 using a lethal dose of 100 LD50. a A representative image of the gating strategy used to probe lung resident NP-tetramer+ CD8+ T-cell responses of animals on day 3 post-infection. The following markers were utilized for the gating strategy; CD3e—T cell marker, CD8a—CD8 T cell marker, CD69—Activation marker, CD44—Memory marker, CD103—Tissue resident marker (absent in CD4+) b Absolute count of TRM IAV-NP+CD8+ T-cells 3 days post infection. Each dot is one animal and data are shown as mean ± SEM Statistical significance was compared to QIV standard of care group (Group 7) using one-way ANOVA using Dunnett’s correction. **** P- <0.0001.
The data indicated the rapid expansion of IAV-specific CD8+ TRM (CD3ε+ CD8+ CD44+ CD69+ CD103+ IAV-NP+) in group 1 that received cH8/1-ΔNS1 prime followed by cH11/1-ΔNS1 boost, suggesting efficient priming by our intranasal vaccine regimen. Even though the rest of the LAIV-immunized animals showed the presence of IAV-specific TRMs, they did not have comparable amounts to those in group 1. In comparison to mice that received QIV, the TRM levels of group 1 were statistically significantly higher as compared to the HA-stalk prime only mice (Fig. 7b).
Mice intranasally immunized with cHA-ΔNS1-LAIV vaccines induce robust splenic IFN-γ+-CD8+ T-cell responses upon subsequent infection
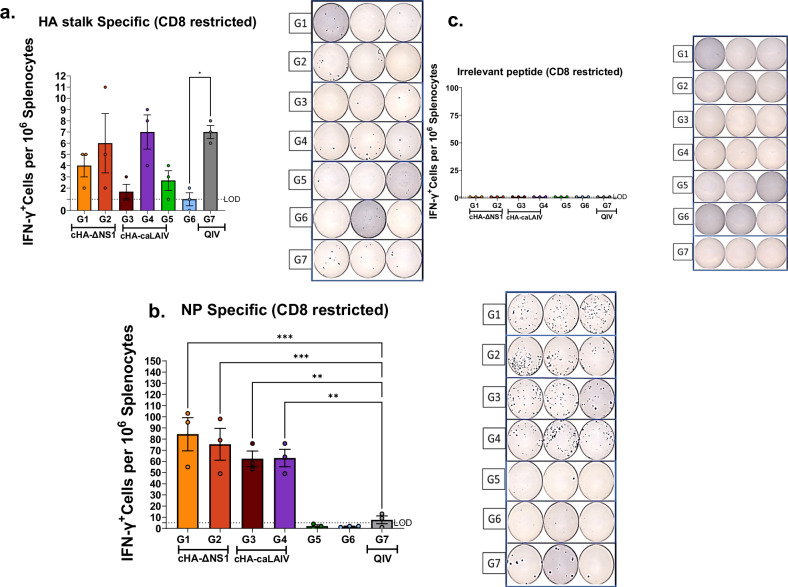
Given that IFN-γ is a cytokine that is readily produced by both CD4+ and CD8+ in order to exert the biological function of activating other immune cells such as NK cells, macrophages and other CD8+T-cells43,44, we used an established IFN-γ ELISpot assay to readily detect CD8 restricted peptide stimulated T-cells of splenic origin. Cells stimulated with HA stalk peptides (IYSTVASSL) indicated groups 2, 4 and 7 to be the highest responders for this assay (Fig. 8a). Interestingly groups 1 and 3 which received the same immunization sequence with different backbones showed reduced amounts in comparison to all the other immunized groups, although this was not statistically significant due to high variability. When considering the absolute numbers with Fig. 8b, it is apparent that HA stalk-based T-cell responses are less pronounced as compared to those induced by NP peptide stimulation. NP peptide stimulated splenic T-cells showed the highest responses to groups 1 and 2, with group 1 being the most pronounced, mirroring results seen in Figs. 6 and 7. This indicates that immune dominance of NP over HA stalk T cell responses was not changed due to the vaccination protocol. As expected QIV immunized mice mounted suboptimal T-cell responses against NP. In parallel, Respiratory syncytial virus fusion protein RSV-F was used as a negative control and yielded no response in any group (Fig. 8c).
Fig. 8. ELISpot assays for IFNγ+CD8+ T-cells from splenocytes indicate robust T-cell responses against viral NP and HA stalk, induced by cHA-LAIV vaccinations.
Four weeks post-final-boost, mice were challenged with a QIV-matched IVR-180 using a lethal dose of 100x LD50. On day 5 post-infection, spleens were harvested and processed for ELISpot assays. Splenocytes (105 cells) were stimulated with indicated CD8 restricted peptides for 16 h. a HA stalk-peptide stimulated. b NP peptide stimulated and c. RSV-F peptide stimulated (irrelevant peptide). Each dot represents one animal. Side panels indicate representative ELISpot images form each animal. Data present as mean ± SEM. Statistical significance was compared to QIV standard of care group (Group 7) using one-way ANOVA using Dunnett’s-correction. ****P- <0.0001, ***P-0.0002, **P-0.0011, *P-0.01. Limits of detection (LOD) are shown in dotted lines.
Intranasal immunization of mice with cHA-ΔNS1-LAIV vaccines minimizes challenge virus-induced interstitial damage and prevents airway epithelial injury
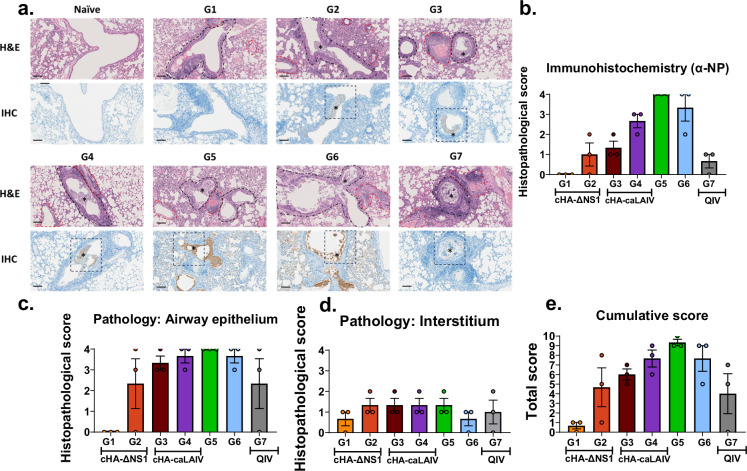
Immune protective parameters induced by vaccination can reduce lung pathology in vivo and therefore, we examined the pathological features of mice that received the different vaccination regimens45,46. Lung tissues were taken from mice 5 days post-infection with a lethal dose of 100x LD50 seasonal influenza virus. We used IHC targeting NP as an antigen to assess the effect of the challenge infection in the lung on day 5. The data indicated that groups 1 and 7 (matched) had the lowest amounts of viral antigen present in the lungs. For group 1 mice, failure to detect viral antigens by day 5 coincided with a lack of viral titers (Figs. 9a, b, and 5c). Although titers were not detected for group 7, IHC indicated the presence of low levels of NP antigen (Figs. 9a, b, and 5c). In contrast, groups that received cH11/1-LAIVs as the prime tended to have higher IHC scores, indicating similar patterns in agreement with the other data sets shown.
Fig. 9. cHA-ΔNS1-LAIV vaccination protects mice from severe bronchointerstitial histopathology after seasonal virus challenge.
Four weeks post-final-boost, mice were challenged with a QIV-matched IVR-180 using a lethal dose of 100 LD50. On day 5 post infection left lung lobes were fixed in formalin and were processed for H&E staining and IHC. Lungs were scored by a blinded independent veterinary pathologist to assess pathology features. a Representative images from an animal derived from each group are shown as H&E staining (top panels) and NP IHC immunostaining (bottom panels). Peribronchiolar (black hashed outlines) and perivascular inflammation were observed to variable degrees in all influenza virus-inoculated animals regardless of vaccination group. Normal naïve lungs are represented for comparison. Scale bar = 100 microns. All images acquired at 200x total magnification. b Scoring for IHC (α-NP), c scoring for interstitial pathology, d. scoring for airway epithelial integrity and e. cumulative scores based on the assessed parameters. Scoring for individual parameters range from 0–5: 0 no pathology and 5 being severe pathology. Each dot is derived from a single animal and data is shown as mean ± SEM.
Mock immunized animals and IBV HA-stalk only primed animals had the highest IHC scores, with some variability in group 6.
Influenza-mediated pulmonary damage initially occurs in the airways and then subsequently disseminates to the respiratory zones of the alveoli as disease progresses. Tissue necrosis derived from the viral infection results in lung pathology and can be readily detected47. Scoring indicated no airway epithelial damage in group 1 whereas all the other groups had measurable amounts of tissue damage, coinciding with the IHC data (Fig. 9a, b and d). Group 2 had the second lowest pathology scoring of all the vaccinated groups together with the standard of care, although groups 2-4 had similar trends in viral titers (Fig. 5c). Group 7 had no detectable virus by day 5 but had similar pathological scoring for the airway epithelia (Figs. 5c and 9b).
Given the vital role in air diffusion mediated by the interstitium, thickening of the interstitium due to influenza virus-induced damage and affiliated immune cell infiltrates contributes to clinicopathological outcomes of viral pneumonia46. To assess this parameter, histopathological analysis of challenged mice was conducted (Fig. 9c). Pathological scoring indicated a general trend of mild-to-moderate interstitial pathology across all animals. Group 1 however, showed comparatively low pathological scores as compared with most of the other groups. However, interstitial pathology scores at day 5 were too low in all groups to make any comparative conclusions.
Taken together, the cumulative scores indicate an overview of pathology induced by the virus challenge. Group 1 showed the lowest cumulative pathology scores of all the vaccinated mice. Interestingly group 2 and the standard of care had similar levels of overall pathology, while the cHA-caLEN LAIV vaccinated groups had moderate levels of pathology (Fig. 9d). The effects of pathology were not completely mirrored by weight loss (Fig. 5a).
Mice intranasally immunized with cHA-ΔNS1-LAIV vaccines are protected from a heterologous challenge with a highly pathogenic avian (HPAI) H5N1 influenza virus
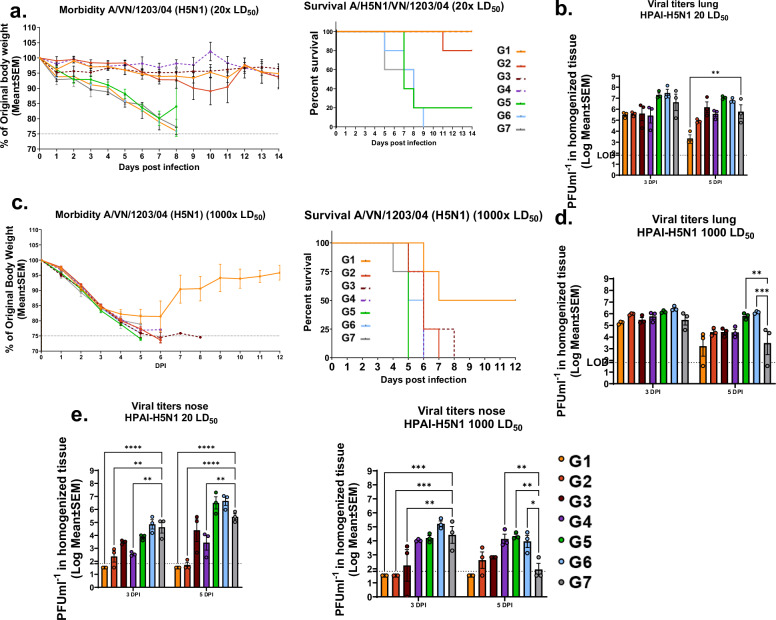
Universal influenza vaccines should protect from any influenza virus regardless of the antigenic make-up. The present study evaluates IAV group 1-specific vaccine candidates that should confer protection against heterologous viruses derived from the same group23. To assess the level of protection, we conducted two BSL-3+ level studies with wild-type H5N1 as challenge virus, that would stringently assess the protective capacity of the proposed immunization regimen. In the first study, we challenged mice with a low lethal dose (20 LD50) 4 weeks post-final boost.
All LAIV immunized groups showed minimal weight loss except group 2 for which weight loss initiated on day 8. By day ten 100% survival was observed for all cHA-LAIV groups while the standard of care group 7 (QIV immunized mice) and group 5 (mice pre-existing HA-stalk based immunity) had 20% survival (Fig. 10a left and right panels). Interestingly, by the end of the study, group 2 had 80% survival. Viral titers in lungs indicated that group 1 was able to reduce replication from day 3 to day 5, showing the lowest amounts of virus in comparison to other vaccinated groups with significance observed at 5 DPI. While cHA-LAIV immunized animals had lower lung viral titers, QIV immunized mice lung viral titers were higher and were almost as high as those for mock immunized animals and HA-stalk primed mice (Fig. 10b). Viral titers in nasal turbinates indicated that cHA-ΔNS1 immunized mice had the lowest amount of virus, particularly in group1 with no viral titers detectable (limit of detection 66.67 PFUml-1; Fig. 10e left panel). cHA-caLEN immunized mice showed intermediate levels of reduction, while the standard-of-care group had only slight reductions on both days in comparison to the naïve control group.
Fig. 10. cHA-LAIV vaccination protects mice from a high dose heterologous highly pathogenic avian influenza challenge and confers superior upper respiratory tract protection.
Foue weeks post final-boost, mice were challenged with an A/VN/1203/04/H5N1 virus using a lethal dose of 20x LD50.Thirteen weeks post final-boost the mice were challenged with high lethal dose 1000x LD50 challenge. a Morbidity (left) and mortality (right) were assessed by monitoring weight loss (20x LD50). b Viral titers in lungs (left) and nasal turbinates (right) on day 3 and 5 post-infection of challenged animals (20x LD50). c Morbidity (left) and mortality (right) were assessed by monitoring weight loss (1000x LD50). d Viral titers in lungs (left) and nasal turbinates (right) on day 3 and 5 post-infection of challenged animals (1000x LD50). Each dot represents one animal. Data is shown as mean ± SEM. Statistical significance was compared to QIV-Standard of care group (Group 7) using one-way ANOVA using Dunnett’s-correction. ****P-<0.0001, ***P-0.0002, **P-0.0011, *P-0.02. Limits of detection (LOD) are shown in dotted lines.
Mice were then challenged 13 weeks post final boost with a high dose of (1000x LD50) HPAI-H5N1 to assess the durability and the protective capacity of this vaccination regimen. Only group 1 had 50% survival and the rest of the mice succumbed to death by day 9 (Fig. 10c left and right panels). Viral titers in this lethal challenge indicated similar levels of viral replication in lungs, although group 1 had slightly lower titers at both sampled days (Fig. 10d). Group 1 did not have any detectable amounts of virus in nasal turbinates on either day (detection limit 666.67 PFUml-1; Fig. 10e right panel).
Discussion
Despite the availability of seasonal vaccines, influenza viruses continue to circulate globally. To mitigate the need for annual vaccine formulations, a universal influenza vaccine that would protect against any influenza virus is currently being pursued worldwide. Many scientists focus on viral proteins containing conserved epitopes that would induce broadly protective immune responses. Such epitopes include the HA stalk, NA, M2e, M1 and NP48.
In the present study we compared two different universal influenza vaccine platforms that use the format of an intranasally administered LAIV. A growing body of evidence shows that natural infections are able to provide appropriate mucosal responses that are driven by secretory IgA in the upper respiratory tract as well as cell-mediated immune responses within the respiratory tract43,49,50. These features of an LAIV would be highly beneficial in a universal influenza virus vaccine.
Another characteristic of an effective universal influenza virus vaccine is the ability to induce protective immune responses which are long-lasting. A balanced mix of humoral and cell-mediated immunity against influenza virus has been shown to reduce symptomatic disease, transmission, tissue pathology, and overall influenza virus infection-induced disease burden5,10. Needle-free administration is another benefit that LAIVs offer, which may also increase vaccine accessibility. The LAIVs in this study were produced in cell culture, which omits the need for an egg-based vaccine pipeline.
To achieve universal protection, we focused on the HA stalk which has been demonstrated to induce broadly neutralizing antibodies that can confer neutralizing and effector function-mediated protection at different stages of the viral life cycle51. Data suggest that the HA stalk antigenically drifts at a much slower rate in comparison to the globular HA head, and therefore is less likely to allow immune escape52,53. To induce HA stalk-specific immune responses, we devised – based on LAIV constructs - a prime-boost immunization regimen composed of two different chimeric HAs (cHA) that harbor exotic avian heads to which humans are naïve, while maintaining the same group-specific HA stalk, with the expectation of boosting antibodies against the stalk. This principle has been tested in various animal models and data suggest that cHA-LAIV vaccination platforms provide superior levels of protection against diverse influenza viruses54. However, a recent clinical trial using the cold-adapted LenLAIV bearing chimeric HAs found that this LAIV vaccine platform did not increase antibodies against the HA stalk in healthy adult individuals, while an adjuvanted IIV vaccine did. To potentially improve an LAIV vaccine approach, we focused on removing the main interferon antagonist NS1 to increase the immunogenicity of the LAIV platform. Our group reported the self-adjuvanting capacity of ΔNS1-LAIV viruses by creating a broad antiviral state in IFN-competent hosts39. ΔNS1-LAIVs are replication-deficient when compared to their WT counterparts due to the induction of local IFN responses. However, it has been shown mice intranasally inoculated with ΔNS1 viruses induced IAV-NP reactive spleen cells while inactivated viruses failed to confer a similar phenotype. This suggests that ΔNS1 viruses go through “abortive replication” resulting in induction of B and T cell responses55. Furthermore, ΔNS1-LAIVs have been shown to be highly immunogenic when given as a seasonal vaccine and have been tested in clinical trials (NCT00724997, NCT03745274)32,41.
In the present study, we combined a cHA regimen with a ΔNS1-LAIV backbone to assess whether a prime-boost immunization strategy would confer robust humoral, and cell-mediated responses, and we compared this to the cold-adapted Len-LAIV backbone. We showed that the cHA-ΔNS1-LAIVs were able to grow in IFN-incompetent cells while maintaining antigenicity. Nevertheless, these viruses were unable to replicate in IFN-competent cells in vitro further indicating their replication-deficient phenotype (Fig. 1). We then showed that a prime-boost regimen of cHA-ΔNS1-LAIVs in mice conferred superior levels of serum-based IgG titers against group specific diverse HAs, N1 NA, as well as high titers of HA stalk antibodies, which conventional seasonal vaccines are unable to induce robustly (Fig. 3). While HA stalk antibodies can neutralize viruses during viral fusion and budding, they also can induce effector functions such as ADCC, antibody dependent cellular phagocytosis (ADCP) and complement mediated lysis (CML) by interacting with other innate immune cells such as NK cells, macrophages, neutrophils, and the complement system56,57. Using an in-house developed MDCK-cH6/1 cell line, we showed that the serum IgG induced by the ΔNS1-LAIVs were readily able to induce ADCC in vitro (Fig. 3d), in agreement with our previous cHA vaccine studies54,58. We noticed a considerably high levels of HA-stalk serological readouts (IgG and ADCC) in the HA-prime only group, although by itself was not protective.
Given the importance of mucosal secretory IgA, we were interested in seeing whether our vaccines can induce robust IgA responses49. Although the inductions were low, cHA-ΔNS1-LAIV vaccination induced higher levels compared to vaccination with the Leningrad-based backbones, further supporting the idea that vaccine constructs lacking a functional full-length NS1 increase immunogenicity. (Fig. 3a). To test whether our cHA vaccine preparations can protect from a seasonal influenza lethal dose infection, we challenged mice with 100 LD50 using the QIV matched IVR-180 strain. Although none of the vaccinated mice showed signs of morbidity by weight loss and death, differences in viral replication in the lungs and nasal turbinates were observed. Group 1 mice that received cH8/1-ΔNS1 followed by cH11/1-ΔNS1 had no detectable virus titers in the upper respiratory tract (URT) in agreement with the IgA responses we measured by ELISAs (Figs. 3a and 5). During the challenge, we followed a group of animals to assess their circulating CTL responses towards ongoing influenza infection. We selected NP as an antigen given its wide use for in vivo studies as well its known contributions for T-cell mediated immunity in both humans and preclinical animal models59. While both cHA-ΔNS1-LAIV vaccine regimens had high baseline levels of NP-specific CTLs, only group 1 mice were able to readily expand these cells by day 5 post-infection, and this level was superior to those induced by the caLEN immunized groups (Fig. 6). The higher starting baseline is due to the cellular immunity induced by vaccination. Tissue-resident memory (TRM) CTLs are known to be induced by natural infections as well as intranasal vaccines10. The importance of these is exemplified by the fact that these cells can readily expand locally upon subsequent infections. Furthermore, these cells are known to confer heterologous immunity. Using tetramer-staining and flow cytometry, we demonstrated the presence of NP-specific TRM-CTLs in ΔNS1-LAIV immunized groups but not in QIV immunized mice. Group 1 mice showed the most robust response indicative of a trend across several immunogenic and protective assays we have performed.
ELISpot assays were performed targeting splenic IFN-γ + CD8 T-cells against HA stalk and NP. We found that the level of HA stalk-specific T-cell responses to be low across all groups, albeit highly variable. This is arguably due to the poor immunogenicity of the antigen in the context of T-cell immunity. In contrast, NP specific IFN-γ + CD8 T-cells had a higher response in agreement with its T-cell-specific immunogenicity. Once again, group 1 mice that received cHA-ΔNS1-LAIV prime-boost vaccination had the best response, consistent with the other NP based T-cell data we presented here (Figs. 6, 7 and 8). We believe that a synergistic effect of strong serological responses at serum and mucosal levels together with circulating and tissue-resident T-cell responses are at play in the observed protective effect. When already present in the tissue (tissue-resident memory T cells, TRM), these effector-type lymphoid cells can respond very fast and contribute to the first line of defense during the early phases of infection by elimination of infected cells60. Inactivated virus-based vaccines such as the currently licensed QIV or recombinant protein vaccines typically are poor inducers of T cell responses in the periphery, and do not particularly contribute to mucosal T cell induction. Live attenuated virus vaccines like the one tested here, on the contrary, are potent inducers of T cell responses at mucosal sites, and therefore can be considered a more optimal way to activate cellular vaccine responses61. T cell responses also allow to target highly conserved epitopes in vaccine antigens, which allows to further broaden the protective effect of vaccination to protect against antigenically drifted and shifted influenza viruses. Influenza virus infections can be characterized by their ability to confer respiratory tissue damage inducing necrosis in airway epithelial cells as well as interstitial pneumonia46,47. We showed that group 1 mice had the best outcome based on pathology scoring; this is in line with the serum and mucosal responses which control virus replication. The CTL response may have also contributed to controlling of viral replication by clearing infected cells. Moreover, group 1 mice had no detectable pathology in the alveolar epithelia, coinciding with the lack of infectious virus in lungs at day 5 (Figs. 5 and 9). Overall, cHA-ΔNS1-LAIV (specifically group 1) mice immunized with cH8/1-ΔNS1 followed by cH11/1-ΔNS1 were able to control virus induced tissue damage, likely owing to the superior humoral and T-cell responses. The reason why this order of immunization is more efficacious than a regimen of cH11/1-ΔNS1 virus followed by the cH8/1-ΔNS1 is not clear.
Finally, we challenged groups of mice with a highly pathogenic avian influenza virus (H5N1) in a BSL-3+ setting, to test whether our vaccine preparation would protect mice from a heterosubtypic lethal challenge. We observed 100% survival in a 20x LD50 low lethal dose challenge 4 weeks post challenge for group 1 mice; the standard of care group had 20% survival. However, we did notice onset of sudden weight loss and subsequent death in group 2 on day 9, probably due to the neurotropic nature of the virus (viral titers in URT and lower respiratory tract (LRT) were modest). Another group of mice was challenged 13-weeks post final boost with a high lethal dose (1000x LD50) to better understand the potency of the immune response induced by the vaccinations. Group 1 was the only group that had survivors (50%) while all the other groups succumbed to death. The survival likely can be attributed to the robust TRM as well as mucosal responses. Upon assessing viral replication by plaque assays, we found that the same group had no detectable levels of viral replication in URT (Fig. 10d). While the vaccine-induced immune response may have certainly assisted in this observation, we acknowledge the possibility that the levels were below the limit of detection which was relatively high in this assay (666.67 PFUml-1). Furthermore, avian viruses such as the one used in this study have been shown to preferentially replicate in cell types expressing α-2,3 sialic acid moieties that are highly expressed in lungs compared to the URT62. The breadth and the strength of the immune responses to LAIVs have been shown to be dependent on the induction of robust Tfh CD4+ T-cell responses. It has been shown that LAIV-induced CXCR5 dependent Tfh cells aid cognate B cells in producing antibodies with high affinity and contributing to long-lasting memory12,13. In the current study we were unable to look at these immune parameters and future studies will be performed to decipher the immunology in the context of cHA-ΔNS1-LAIVs.
In summary, we demonstrated the potential of a cHA-ΔNS1-based LAIV vaccination platform to provide robust humoral and cellular immune responses. The vaccine regimen given in a specific order provided optimal protective immune responses. However, when the order was reversed, we observed that the immune responses and protection outcomes were reduced (group 2). Although the mechanism behind this discrepancy is not known, it highlights the importance of the immunization sequence in a prime boost vaccination regime. This also indicates the need for further studies to assess the impact of vector immunity in the context of repeated dosing. Future work will focus on understanding the effect of pre-existing immunity by modeling the vaccination strategy in pre-immune ferrets. Taken together, the results of this study emphasize the importance of antigen selection and vaccine delivery platforms to develop a universal influenza vaccine and their impacts on a multidose-live attenuated influenza vaccination strategy.
Methods
Ethics statement
The present animal study was conducted upon approval of an animal protocol by the Institutional Animal Care and Use Committee (IACUC) of the Icahn School of Medicine (ISMMS). All mice were housed in a BSL-2 barrier facility which was temperature and humidity controlled and all procedures were conducted by trained personnel and were designed to minimize animal suffering. Challenge studies with select agents were done by trained personnel with approved protocols performed within a certified ABSL3+ facility at the Icahn School of Medicine at Mount Sinai.
Cells, viruses, and recombinant proteins
Madin-Darby canine kidney (MDCK) cells (ATCC® CCL-34), an in-house generated MDCK cell line stably expressing NS1 (MDCK-NS1) from A/Puerto Rico/8/1934(H1N1) and Vero-CCL81 (ATCC® CRL-81™) cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% heat inactivated-FBS and penicillin/streptomycin at 37 °C with 5% CO2. Plaque assays in BSL-2 were done as previously described39. Virus titrations were done using standard plaque assays to obtain the plaque forming unit (PFU) titer. For plaque assays within the BSL3+, an overlay composed of 2.1% Avicel (microcrystalline cellulose) was used63. Plaques were visualized by immunostaining using an α-Nucleoprotein NP HT103 mAb64 at a dilution of 1:1000. The cH6/1 (containing H6 head domain from A/mallard/Sweden/81/02 combined with an H1 stalk domain of A/California/04/09 (Cal/09)), H2 (A/mallard/Netherlands/5/99), and H18 (A/flat-facedbat/Peru/33/10) recombinant proteins were expressed using a baculovirus expression system as described previously65. For the BSL-2 challenge, A/Singapore/GP1908/15 H1N1 (IVR-180) (matched H1N1 component for the quadrivalent inactivated vaccine (QIV)) was obtained from the National Institute for Biological Standards and Control (NIBSC). For the BSL-3+ challenge, the highly pathogenic avian influenza (HPAI) H5N1 strain A/Vietnam/1203/2004 (H5N1) virus was rescued using reverse genetics within BSL-3+ setting as previously described66.
cHA-universal vaccine preparations and standard-of-care vaccine preparations
cHA-ΔNS1-LAIV were generated as described previously32. The external glycoproteins of these viruses are cH8/1 or cH11/1 with N1 of the 09-pandemic origin virus (A/California/7/09 (H1N1)). The viruses were passaged 10 times and were sequenced to confirm the genetic stability. The cold-adapted temperature sensitive Leningrad backbone-based viruses (caLEN) had identical HAs and NAs to the ΔNS1 vaccine strains and have been described previously30. These viruses were sequenced, and all vaccine strains were assessed for infectivity using multi-cycle growth curves and fluorescence-activated cell sorting (FACS) sorting using mAb CR911467 and Galios FACS sorter. An influenza B virus (B/Yamagata/16/88 backbone) harboring a cH9/1 HA was generated as previously described25. As a standard of care, a QIV, Flucelvax Quadrivalent Influenza Vaccine (2018-2019 season), was obtained from BEI Resources (NR-51698). All vaccine viruses were propagated in cell culture (MDCK-WT or MDCK-NS1 cells) and were purified through a 30% sucrose cushion using ultra centrifugation (2 rounds in SW28 Beckman rotor at 25,000 RPM for 2 h at 4 °C). Viral pellets were suspended in sterile 1X PBS (pH 7.0).
Mouse immunizations, sample collection and virus challenges
Female, 6–8-week-old BALB/c mice were purchased from Taconic Biosciences. Animals were randomly assigned to groups and study layout is shown in Fig. 2a. Anesthetized animals (ketamine and xylazine diluted in PBS administered via intraperitoneal injection; 100 mg/kg ketamine and 10 mg/kg xylazine IP) were intranasally infected using 30 µl of appropriately diluted viruses or PBS. Intramuscular vaccinations were administered using 50 µl undiluted, unadjuvanted QIV (standard of care). Sample collection setup is shown in Fig. 2b, c.
Mice were subjected to submandibular bleeds and nasal washes (terminal procedure) pre-prime and were also bled between immunizations (approximately at 28-day intervals) as indicated in Fig. 2b. A total of 5 animals from each group were bled D1, D5, and D10 for circulating CD8+ T-cell analysis and the same animals were followed for this experimental procedure. The terminal procedure for tissue harvesting was facilitated by humanly sacrificing animals via an IP injection of pentobarbital (250 mg/kg). Left lung lobes and nasal turbinates were harvested for viral titration via plaque assays for D3 and D5 post-challenge. Right lobes were used for flow cytometric analysis of tissue-resident memory CD8+ T-cells on D3 and left lung lobes were used for histology on D5 post-infection. Spleens were harvested on D5 for peptide-stimulated IFN-γ+-CD8+ T-cells ELISpot assays. Median lethal doses (LD50) were calculated as described before using groups of mice (n = 5 per group) which were infected with ten-fold serially diluted virus (of known titer)68. IVR-180 had an LD50 of 180 PFU while the HPAI-H5N1 had an LD50 of ~2 PFU.
Enzyme-linked immunosorbent assays (ELISA)
ELISA for serum IgG (Rabbit α-mouse IgG-HRP; Sigma A9044) and nasal wash IgA (goat α-mouse IgA-HRP; Bethyl Laboratories A90-103P) were established. Serum ELISAs were done with heat-inactivated serum at a starting dilution of 1:30 and nasal washes were studied using clarified undiluted washes. The format for ELISA was described previously25. In brief, high-binding 96-well ELISA plates (Thermo Scientific) were coated with 2 µg/ml of in the indicated recombinant proteins in PBS overnight at 4 °C. Plates were washed three times using PBS-T (0.1% Tween-20) and were incubated for 1.5 h at room temperature with blocking buffer composed of PBS-T with 3% goat serum (Life Technologies) and 0.5% milk powder. Plates were then incubated with serially diluted (three-fold) serum/nasal wash samples. Plates were then washed again three times and incubated with the appropriate secondary antibody (1:3000 for IgG and 1:2000 for IgA) for an hour. Plates were then washed four times with PBS-T and were developed by using SigmaFast OPD (o-Phenylenediamine dihydrochloride) substrate followed by 3 M HCL, and plates were read at 490 nm using a Biotek Synergy H1 reader. The mean + 3X standard deviations (SD) of blank wells were calculated as a cut-off endpoint value.
Antibody dependent cellular cytotoxicity (ADCC) reporter assays
ADCC reporter assays were done using a modified protocol as described previously58. Briefly, MDCK-HA (cH6/1) cells were seeded (2.5 ×104 cells per well) in white polystyrene 96-well assay plates (Costar Corning) and were incubated overnight. Heat-inactivated serum samples with a starting dilution of 1:20 (25 µl) were three-fold serially diluted in RPMI 1640 (Life Technologies) and were incubated for 7 h at 37 °C and 5% CO2 with modified Jurkat cells expressing the murine FcγRIV receptor which activates the luciferase under the control of NFAT promotor (7.5 × 104 cells per well; Promega). The cross-linking of Fcγ receptors by immune complexes results in luciferase activity. This activity was assessed by using Bio-Glo luciferase substrate and the plates were read after 15 minutes using a Synergy H1 hybrid multimode microplate reader (BioTek). Background luminescence (average + 3 x SD) of wells without Ab was subtracted, and the area under the curve was calculated.
Circulating NP-specific T cells in the blood
Blood was collected from the submandibular vein on 1 day before, 5 days after, and 10 days after challenge (n = 5 per group) into tubes containing 20 μl of 0.5 M EDTA to prevent coagulation and pooled per group. Whole blood was stained with H-2K(d) TYQRTALV tetramer in PE (1:75 dilution, Cat. No. 44053, NIH Tetramer Core Facility, Atlanta, GA, USA) for 40 min at room temperature in the dark, then the following antibodies were spiked in: purified rat anti-mouse CD16/CD32 Fc Block (1:100 dilution, Clone 2.4G2, Cat. No. 553142, BD), CD3 Alexa Fluor 700 (1:100 dilution, Clone 17A2, Cat. No. 100216, Biolegend, San Diego, CA, USA), CD8a PerCP (1:100 dilution, Clone 53-6.7, Cat. No. 553036, BD), and MHC Class II eFluor 450 (1:200 dilution, Clone M5/114.15.2, Cat. No. 48-5321-82, ThermoFisher). Samples were incubated for 20 minutes at room temperature in the dark. After incubation, samples were centrifuged at 400g for 5 min at room temperature and supernatants were discarded. Cells were resuspended in 200 μl of eBioscience Foxp3 Fixation/Permeabilization Buffer (Cat. No. 00-5523-00, ThermoFisher), diluted as recommended by the manufacturer. Samples were incubated for 10 minutes at room temperature in the dark. After fixation, cells were washed twice with 1 ml of staining buffer. After the second wash, cells were resuspended in 200 μl of staining buffer. Samples and compensation controls were acquired and analyzed as described above.
Tissue-resident CD8+ T cells in the lung
To assess elicitation and activation of tissue-resident memory CD8+ T cells, lungs were collected at 3 days post-challenge (n = 3 mice per group) into supplemented RPMI-1640 (5% FBS, 1X Pen-Strep). Lungs were minced into small pieces, roughly 4 mm3 in size, then digested with collagenase D (Cat. No. 11088866001, Roche Diagnostics, Basel, Switzerland) diluted to a working concentration of 2 mg/ml in supplemented RPMI-1640 for 15 min at 37 °C, shaking. Digested lung pieces were then forced through a 70 μm cell strainer (Cat. No. 352350, Corning Inc., Corning, NY, USA) with the plunger of a 1 ml syringe (Cat. No. 309628, BD) to generate single-cell suspensions. After cells were centrifuged at 400g for 5 min at 4 °C, media was aspirated from the pellet and cells were resuspended in 5 ml of ammonium chloride red blood cell lysis buffer and incubated at room temperature for 5 min. After centrifugation at 400g for 5 min at 4 °C supernatant was aspirated and cells were resuspended in 50 μl of staining buffer containing Fc Block (Clone 2.4G2, Cat. No. 553142, BD) diluted 1:100. Cells were incubated with diluted Fc Block for 5 minutes at room temperature, before 50 μl of surface stain cocktail was added on top. The surface stain cocktail consisted of the following: CD3ε FITC (1:100 dilution, Clone 145-2C11, Cat. No. 553061, BD), CD44 PE-CF594 (1:100 dilution, Clone IM7, Cat. No. 562464, BD), CD8a PerCP (1:100 dilution, Clone 53-6.7, Cat. No. 553036, BD), CD69 PE-Cy7 (1:100 dilution, Clone H1.2F3, Cat. No. 561930, BD), MHC Class II (I-A/I-E) eFluor 450 (1:200 dilution, Clone M5/114.15.2, Cat. No. 48-5321-82, ThermoFisher), Fixable Viability Dye eFluor 450 (1:100 dilution, Cat. No. 65-0863-14, ThermoFisher), and H-2K(d) TYQRTALV tetramer in PE (1:75 dilution, Cat. No. 44053, NIH Tetramer Core Facility) diluted in staining buffer. Cells were surface stained for 20 min at room temperature in the dark. Cells were then washed by adding 1 ml fresh staining buffer on top and centrifuged at 400g for 5 min at room temperature, then the supernatant was decanted before proceeding to the next step. Cells were resuspended in 100 μl of BD Cytofix (Cat. No. 554714, BD) and incubated for 5 min at room temperature. Cells were washed with 1 ml staining buffer and resuspended in 200 μl of staining buffer. Five μl of CountBright Absolute Counting Beads (Cat. No. C36950, ThermoFisher) were added to each tube. Samples and compensation controls were acquired and analyzed as described above.
Flow cytometry
After samples were processed and stained as described below, samples were measured using a Gallios Flow Cytometer (Beckman Coulter, Brea, CA, USA) with Kaluza for Gallios Version 1.0 software. For all compensation controls, 1 μl of antibody was added to 1 drop of Invitrogen UltraComp eBeads Plus Compensation Beads (Cat. No. 01-3333-42, ThermoFisher, Waltham, MA, USA) then vortexed. 150 μl of staining buffer (2% bovine serum albumin and 200 mM EDTA in 1X PBS) was added to the compensation controls after incubating at room temperature for 15 minutes. Data analysis was performed using FlowJo Version 10.8 (BD, Franklin Lakes, NJ, USA). Data was compensated using the AutoSpill algorithm in FlowJo. Exported cell counts and population frequencies were visualized using GraphPad Prism Version 9.2.0 (GraphPad Software, San Diego, CA, USA).
ELISpot assays
ELISpot assays were done using murine IFN-γ ELISpot kit (R&D systems EL485) using processed splenocytes as mentioned above. Briefly, 105 processed splenocytes were stimulated with HA peptide (IYSTVASSL, Cat. No. RP20284, Genescript), NP peptide (TYQRTALV, Cat. No. RP20260 Genescript), or RSV-F glycoprotein (KYKNAVTEL; MBL international) peptides diluted according to manufacturer’s instructions and were incubated for 16 h at 37 °C and 5% CO2. The plates were then washed with wash buffer and incubated in detection antibody for 2 h a room temperature and developed with BCIP/NBT substrates, as per manufacturer recommendations. The plates were then washed with deionized water and were dried before imaging using an ELISpot imaging device (Cellular Technology 2018). The images were then manually counted. A total of three animals in duplicate for each group were assessed.
Histopathology
Left lung lobes were inflated with 4% formaldehyde in PBS and fixed for a week. These were embedded in paraffin blocks and 5μm sections were cut on a microtome (Microm, Thermo Scientific). Sections were subjected to staining by hematoxylin and eosin (H&E) by the Biorepository and Pathology Core (N.A.C., Icahn School of Medicine at Mount Sinai). Sections were mounted (Histomount Solution; Life Technologies) and analyzed by a veterinary pathologist blinded to the treatment groups. Lung H&E slides were evaluated using a pathological scoring system to assess three parameters: percentage of lung parenchyma immunoreactive to influenza A virus NP, severity of epithelial degeneration/necrosis of bronchioles, and alveolar interstitial inflammation. A cumulative score of all three parameters was also utilized to evaluate differences in severity amongst experimental cohorts. For the area affected, a scale of 0–5 was used where 0 = not affected, 1 = 5–10% 2 = 10–25%, 3 = 25–50%, 4 = 50–75%, and 5 = 75–100% of lung affected. For histopathological parameters a score of 0–5 was used: 0 = not affected, 1 = minimal, 2 = mild, 3 = moderate, 4 = marked and 5 = severe.
Brightfield immunohistochemistry
A Ventana Discovery Ultra (Roche, Basel, Switzerland) tissue autostainer was used for brightfield chromogenic immunohistochemistry (IHC). Lungs from uninfected (negative control) and infected mice with mock vaccinated (group 6) (positive control) were used as internal controls for assay optimization. Antigen retrieval was conducted using a Tris-based buffer-Cell Conditioning 1 (CC1)-Catalog # 950-124(Roche) and retrieval was done at 94 °C for 32 minutes. An influenza A nucleoprotein antibody (Catalog #PA5-32242 Invitrogen, Waltham, MA) was used at a dilution of 1:2000 diluted in casein buffer and incubations were done for 64 minutes at 37 °C. The anti-influenza A primary antibody was of rabbit origin, and thus was developed with a pre-diluted (1:5000) secondary goat anti-rabbit HRP-polymer antibody-Catalog #MP-7451-50 (Vector Laboratories, Burlingame, CA). A ChromoMap DAB (3,3′-Diaminobenzidine) Kit-Catalog #760-159 (Roche) was applied to slides to form a brown precipitate at the site of primary antibody complexes containing HRP with hematoxylin nuclear counterstain. IHC scores ranged from 0 to 4 and represented the following: 0-no antigen detected; 1-minimal antigen detected, 2-mild to regionally moderate antigen detected, 3-moderate to regionally marked antigen detected, 4-marked to severe antigen detected.
Acknowledgements
The authors thank Ricard Cadagan for excellent technical support. This work was partly supported by NIAID grant R01AI141226 to A.G.-S. and T.M. and by NIAID grant P01AI097092, by CRIPT (Center for Research in Influenza Pathogenesis and Transmission), an NIAID-funded Center of Excellence for Influenza Research and Response (CEIRR, contract # 75N93021C00014, subcontract to P.P.) and by CIVICs (NIAID contract # 75N93019C00051 subcontract to P.P.) to A.G.-S. Influenza research in the M.S. laboratory is supported by R21AI176069 and R21AI151229. This work used NIH S10 Shared Instrumentation Grants S10-OD026983 and SS10-OD030269 to N.A.C. The enhanced Emerging Pathogens Facility (EPF) is a NIHBSL3/BSL3/ABSL3 facility that is part of the BSL-3 Biocontainment CoRE. This core is supported by subsidies from the ISMMS Dean’s Office and by investigator support through a cost recovery mechanism. Research reported in this publication was supported by the National Institute of Allergy And Infectious Diseases of the National Institutes of Health under Award Number G20AI174733 (R.A. Albrecht). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
R.R., P.P, F.K., M.S., T.M., and A.G.-S. planned the experiments and analyzed the data. R.R., R.P., A.A., A.H., and R.A.A. generated the vaccine and challenge viruses. R.R., L.A.C., R.P., S.J., S.Y., I.M., W.S., M.L., S.A., and R.A.A. conducted the experiments. N.A.C., H.P.G., and A.E.T. conducted the pathological analysis of the tissue samples. R.R. and M.S. generated the figures. R.R., M.S., and A.G.-S. wrote the manuscript. All authors read and edited the manuscript.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Competing interests
The A.G.-S. laboratory has received research support from GSK, Pfizer, Senhwa Biosciences, Kenall Manufacturing, Blade Therapeutics, Avimex, Johnson & Johnson, Dynavax, 7Hills Pharma, Pharmamar, ImmunityBio, Accurius, Nanocomposix, Hexamer, N-fold LLC, Model Medicines, Atea Pharma, Applied Biological Laboratories and Merck. A.G.-S. has consulting agreements for the following companies involving cash and/or stock: Castlevax, Amovir, Vivaldi Biosciences, Contrafect, 7Hills Pharma, Avimex, Pagoda, Accurius, Esperovax, Farmak, Applied Biological Laboratories, Pharmamar, CureLab Oncology, CureLab Veterinary, Synairgen, Paratus, Pfizer and Prosetta. A.G.-S. has been an invited speaker in meeting events organized by Seqirus, Janssen, Abbott and AstraZeneca. A.G.-S. and P.P. are inventors on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections and cancer, owned by the Icahn School of Medicine at Mount Sinai, New York. The M.S. laboratory has received unrelated funding support in sponsored research agreements from Phio Pharmaceuticals, 7Hills Pharma, ArgenX, and Moderna. The Icahn School of Medicine at Mount Sinai has filed patent applications regarding influenza virus vaccines on which F.K. is listed as inventor. The Krammer laboratory has received support for influenza virus research in the past from GSK and is currently receiving support from Dynavax. The Icahn School of Medicine at Mount Sinai has filed patent applications relating to influenza virus vaccines and therapeutics vaccines, which lists F.K. as co-inventor. Several of these patents have been licensed and F.K. has received royalty payments from commercial entities. F.K. has consulted from Merck, Pfizar, Seqirus, GSK and Curevac and is currently consulting for Gritstone, 3rd Rock Ventures and Avimex and he is co-founder and scientific advisory board member of Castlevax. The F.K. laboratory is also collaborating with Dynavax on influenza virus vaccine development and with VIR on influenza therapeutics. T.M. is inventor on patents and patent applications on vaccines and immunotherapies, owned by Vivaldi Biosciences and BlueSky Immunotherapies. T.M. owns stocks and options from BlueSky Immunotherapies, Nuvonis Technologies and Vivaldi Biosciences. A.A. is inventor on patents and patent applications on vaccines and immunotherapies, owned by Vivaldi Biosciences and BlueSky Immunotherapies. A.A. owns stock options from Vivaldi Biosciences. All other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Macias, A. E. et al. The disease burden of influenza beyond respiratory illness. Vaccine39, A6–A14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grohskopf, L. A. Prevention and control of seasonal influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices—United States, 2019–20 influenza season. MMWR Recomm. Rep. 68 (2019). [DOI] [PMC free article] [PubMed]
- 3.Kennedy, R. B., Ovsyannikova, I. G. & Poland, G. A. Update on Influenza Vaccines: needs and progress. J. Allergy Clin. Immunol. Pract.9, 3599–3603 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez, K. J. & Strauch, E. M. Decreased vaccine protection of egg-based influenza vaccine in the elderly and nonhemagglutinin-focused immunity. J. Clin. Invest. 131 (2021). [DOI] [PMC free article] [PubMed]
- 5.Krammer, F. The human antibody response to influenza A virus infection and vaccination. Nat. Rev. Immunol.19, 383–397 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Bouvier, N. M. & Palese, P. The biology of influenza viruses. Vaccine26, D49–D53 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Treanor, J. Influenza vaccine—outmaneuvering antigenic shift and drift. N. Engl. J. Med.350, 218–220 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Taubenberger, J. K., Kash, J. C. & Morens, D. M. The 1918 influenza pandemic: 100 years of questions answered and unanswered. Sci. Transl. Med.11, eaau5485 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamayoshi, S. & Kawaoka, Y. Current and future influenza vaccines. Nat. Med.25, 212–220 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Cox, R., Brokstad, K. & Ogra, P. Influenza virus: immunity and vaccination strategies. Comparison of the immune response to inactivated and live, attenuated influenza vaccines. Scand. J. Immunol.59, 1–15 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Sridhar, S., Brokstad, K. A. & Cox, R. J. Influenza vaccination strategies: comparing inactivated and live attenuated influenza vaccines. Vaccines3, 373–389 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lartey, S. et al. Live-attenuated influenza vaccine induces tonsillar follicular T helper cell responses that correlate with antibody induction. J. Infect. Dis.221, 21–32 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aljurayyan, A. et al. Activation and induction of antigen-specific T follicular helper cells (TFH) play a critical role in LAIV-induced human mucosal anti-influenza antibody response. J. Virol.92, e00114–e00118 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clements, M., Betts, R., Tierney, E. & Murphy, B. Serum and nasal wash antibodies associated with resistance to experimental challenge with influenza A wild-type virus. J. Clin. Microbiol.24, 157–160 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner, D. L. & Farber, D. L. Mucosal resident memory CD4 T cells in protection and immunopathology. Front. Immunol.5, 103608 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown, L. E. & Kelso, A. Prospects for an influenza vaccine that induces cross‐protective cytotoxic T lymphocytes. Immunol. Cell Biol.87, 300–308 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Sridhar, S. et al. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat. Med.19, 1305–1312 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Medina, R. A. & García-Sastre, A. Influenza A viruses: new research developments. Nat. Rev. Microbiol.9, 590–603 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hampson, A. et al. Improving the selection and development of influenza vaccine viruses–Report of a WHO informal consultation on improving influenza vaccine virus selection, Hong Kong SAR, China, 18–20 November 2015. Vaccine35, 1104–1109 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weir, J. P. & Gruber, M. F. An overview of the regulation of influenza vaccines in the United States. Influenza Other Respir. Viruses10, 354–360 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erbelding, E. J. et al. A universal influenza vaccine: the strategic plan for the National Institute of Allergy and Infectious Diseases. J. Infect. Dis.218, 347–354 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pica, N. & Palese, P. Toward a universal influenza virus vaccine: prospects and challenges. Annu. Rev. Med.64, 189–202 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Nachbagauer, R. & Palese, P. Is a universal influenza virus vaccine possible? Annu. Rev. Med.71, 315–327 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Krammer, F., García-Sastre, A. & Palese, P. Is it possible to develop a “universal” influenza virus vaccine? Toward a universal influenza virus vaccine: potential target antigens and critical aspects for vaccine development. Cold Spring Harb. Perspect. Biol. a028845 (2017). [DOI] [PMC free article] [PubMed]
- 25.Nachbagauer, R. et al. A chimeric haemagglutinin-based influenza split virion vaccine adjuvanted with AS03 induces protective stalk-reactive antibodies in mice. Npj Vaccines1, 1–10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krammer, F., Natalie, P., Hai, R., Margine, I. & Palese, P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J. Virol. 87, 6542–6550 (2013). [DOI] [PMC free article] [PubMed]
- 27.Krammer, F. et al. H3 stalk-based chimeric hemagglutinin influenza virus constructs protect mice from H7N9 challenge. J. Virol.88, 2340–2343 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ermler, M. E. et al. Chimeric hemagglutinin constructs induce broad protection against influenza b virus challenge in the mouse model. J. Virol.91, e00286–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernstein, D. I. et al. Immunogenicity of chimeric haemagglutinin-based, universal influenza virus vaccine candidates: interim results of a randomised, placebo-controlled, phase 1 clinical trial. Lancet Infect. Dis.20, 80–91 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, W.-C. et al. Sequential immunization with live-attenuated chimeric hemagglutinin-based vaccines confers heterosubtypic immunity against influenza A viruses in a preclinical Ferret model. Front. Immunol.10, 756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, W.-C. et al. Chimeric hemagglutinin-based live-attenuated vaccines confer durable protective immunity against influenza A viruses in a preclinical Ferret model. Vaccines9, 40 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicolodi, C. et al. Safety and immunogenicity of a replication-deficient H5N1 influenza virus vaccine lacking NS1. Vaccine37, 3722–3729 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Wacheck, V. et al. A novel type of influenza vaccine: safety and immunogenicity of replication‐deficient influenza virus created by deletion of the interferon antagonist NS1. J. Infect. Dis.201, 354–362 (2010). [DOI] [PubMed] [Google Scholar]
- 34.García-Sastre, A. et al. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology252, 324–330 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Hatada, E. & Fukuda, R. Binding of influenza A virus NS1 protein to dsRNA in vitro. J. Gen. Virol.73, 3325–3329 (1992). [DOI] [PubMed] [Google Scholar]
- 36.Krug, R. M. Functions of the influenza A virus NS1 protein in antiviral defense. Curr. Opin. Virol.12, 1–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hale, B. G., Albrecht, R. A. & García-Sastre, A. Innate immune evasion strategies of influenza viruses. Future Microbiol5, 23–41 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hale, B. G., Randall, R. E., Ortín, J. & Jackson, D. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol.89, 2359–2376 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Rathnasinghe, R. et al. Interferon mediated prophylactic protection against respiratory viruses conferred by a prototype live attenuated influenza virus vaccine lacking non-structural protein 1. Sci. Rep.11, 22164 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fernandez-Sesma, A. et al. Influenza Virus Evades Innate and Adaptive Immunity via the NS1 Protein. J. Virol.80, 6295–6304 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morokutti, A., Muster, T. & Ferko, B. Intranasal vaccination with a replication-deficient influenza virus induces heterosubtypic neutralising mucosal IgA antibodies in humans. Vaccine32, 1897–1900 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Krammer, F. et al. NAction! How Can Neuraminidase-Based Immunity Contribute to Better Influenza Virus Vaccines? mBio9, e02332–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zens, K. D., Chen, J. K. & Farber, D. L. Vaccine-generated lung tissue–resident memory T cells provide heterosubtypic protection to influenza infection. JCI Insight1 (2016). [DOI] [PMC free article] [PubMed]
- 44.Tewari, K., Nakayama, Y. & Suresh, M. Role of direct effects of IFN-γ on T cells in the regulation of CD8 T cell homeostasis. J. Immunol.179, 2115–2125 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Mackenzie, C. D., Taylor, P. M. & Askonas, B. A. Rapid recovery of lung histology correlates with clearance of influenza virus by specific CD8+ cytotoxic T cells. Immunology67, 375–381 (1989). [PMC free article] [PubMed] [Google Scholar]
- 46.Guo, H. H., Sweeney, R. T. & Regula, D., Leung, A. & Fatal, N. 2009 Influenza A (H1N1) infection, complicated by acute respiratory distress syndrome and pulmonary interstitial emphysema. RadioGraphics30, 327–333 (2010). [DOI] [PubMed]
- 47.Herold, S., Becker, C., Ridge, K. M. & Budinger, G. R. S. Influenza virus-induced lung injury: pathogenesis and implications for treatment. Eur. Respir. J.45, 1463–1478 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Paules, C. I., Sullivan, S. G., Subbarao, K. & Fauci, A. S. Chasing seasonal influenza—the need for a universal influenza vaccine. N. Engl. J. Med.378, 7–9 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Van Riet, E., Ainai, A., Suzuki, T. & Hasegawa, H. Mucosal IgA responses in influenza virus infections; thoughts for vaccine design. Vaccine30, 5893–5900 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Arulanandam, B. P. et al. IgA immunodeficiency leads to inadequate Th cell priming and increased susceptibility to influenza virus infection. J. Immunol.166, 226–231 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Krammer, F. & Palese, P. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr. Opin. Virol.3, 521–530 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kirkpatrick, E., Qiu, X., Wilson, P. C., Bahl, J. & Krammer, F. The influenza virus hemagglutinin head evolves faster than the stalk domain. Sci. Rep.8, 10432 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roubidoux, E. K. et al. Mutations in the hemagglutinin stalk domain do not permit escape from a protective, stalk-based vaccine-induced immune response in the mouse model. mBio12, e03617–e03620 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi, A. et al. Chimeric hemagglutinin-based influenza virus vaccines induce protective stalk-specific humoral immunity and cellular responses in mice. ImmunoHorizons3, 133–148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Talon, J. et al. Influenza A and B viruses expressing altered NS1 proteins: A vaccine approach. Proc. Natl Acad. Sci.97, 4309–4314 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mallajosyula, V. V. A. et al. Influenza hemagglutinin stem-fragment immunogen elicits broadly neutralizing antibodies and confers heterologous protection. Proc. Natl. Acad. Sci. 111 (2014). [DOI] [PMC free article] [PubMed]
- 57.Laursen, N. S. & Wilson, I. A. Broadly neutralizing antibodies against influenza viruses. Antivir. Res.98, 476–483 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chromikova, V. et al. Activity of human serum antibodies in an influenza virus hemagglutinin stalk-based ADCC reporter assay correlates with activity in a CD107a degranulation assay. Vaccine38, 1953–1961 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grant, E. et al. Nucleoprotein of influenza A virus is a major target of immunodominant CD8 + T‐cell responses. Immunol. Cell Biol.91, 184–194 (2013). [DOI] [PubMed] [Google Scholar]
- 60.Lee, S., Yeung, K. K. & Watts, T. H. Tissue-resident memory T cells in protective immunity to influenza virus. Curr. Opin. Virol.65, 101397 (2024). [DOI] [PubMed] [Google Scholar]
- 61.Mohn, K. G.-I., Smith, I., Sjursen, H. & Cox, R. J. Immune responses after live attenuated influenza vaccination. Hum. Vaccines Immunother.14, 571–578 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ito, T. Interspecies transmission and receptor recognition of influenza A viruses. Microbiol. Immunol.44, 423–430 (2000). [DOI] [PubMed] [Google Scholar]
- 63.Matrosovich, M., Matrosovich, T., Garten, W. & Klenk, H.-D. New low-viscosity overlay medium for viral plaque assays]. Virol. J.3, 63 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Breen, M., Nogales, A., Baker, S. F., Perez, D. R. & Martínez-Sobrido, L. Replication-competent influenza A and B viruses expressing a fluorescent dynamic timer protein for in vitro and in vivo studies. PLoS ONE11, e0147723 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Margine, I., Palese, P. & Krammer, F. Expression of functional recombinant hemagglutinin and neuraminidase proteins from the novel H7N9 influenza virus using the baculovirus expression system. J. Vis. Exp. 81, 51112 (2013) [DOI] [PMC free article] [PubMed]
- 66.Pleschka, S. et al. A plasmid-based reverse genetics system for influenza A virus. J. Virol.70, 4188–4192 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dreyfus, C. et al. Highly conserved protective epitopes on influenza B viruses. Science337, 1343–1348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ramakrishnan, M. & Muthuchelvan, D. Influence of reed-muench median dose calculation method in virology in the millennium. Antivir. Res.28, 16–17 (2018). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.