Abstract
Aim:
A systematic review was conducted to summarize the methylated circulating tumor DNA (ctDNA) markers reported over the last decade for early detection of colorectal cancer (CRC) and to identify the main technical challenges that are impeding their clinical implementation.
Background:
CRC is a major cause of cancer deaths worldwide, but early detection is key for successful treatment. Non-invasive methods such as methylated ctDNA testing show promise for improving detection and monitoring of CRC.
Methods:
A comprehensive search was performed using Web of Science, PubMed, and Scopus up to December 30, 2023, limited to articles published in the last 10 years (after 2012), while including advanced adenoma/stage 0 or stage I/II samples in biomarker validation.
Results:
After identifying 694 articles, removing duplicates and screening titles, abstracts, and full texts, a total of 62 articles were found to meet the inclusion criteria. Among the single biomarkers, MYO1-G, SEPT9, SDC2, and JAM3 revealed the highest sensitivity for polyps and stage I/II CRC. For multi-biomarkers with suitable sensitivity, combinations of SFRP1, SFRP2, SDC2, PRIMA1, or ALX4, BMP3, NPTX2, RARB, SDC2, SEPT9, VIM or ZFHX4, ZNF334, ELOVL2, UNC5C, LOC146880, SFMBT2, GFRA1 were identified for polyps and stage I/II CRC.
Conclusion:
Enhancing sensitivity and specificity of molecular screening methods is crucial for improving CRC detection. Identifying a select few valuable biomarkers is key to reducing costs, despite challenges posed by low ctDNA levels in plasma, particularly in early-stage cancers.
Key Words: Colorectal cancer, Circulating tumor DNA, DNA methylation, Liquid biopsy, Cancer biomarker, Early detection
Introduction
Despite advances in detection and treatment methods, colorectal cancer (CRC) is the second most deadly cancer and the third most common malignant tumor among men and women worldwide (1, 2). CRC survival rates are closely linked to early detection, which ranges from a 90% five-year survival rate in stage I disease to 10% five-year survival in stage IV disease (3). Approximately 60% of CRC patients at diagnosis present with either locally advanced lesions or distant metastases. This advanced stage unfortunately precludes surgical intervention, significantly impacting treatment options and prognosis (4, 5). Since developing malignant lesions from precancerous lesions (adenomas) is a long process (6), it allows CRC to be detected early by screening (7). Early detection of CRC leads to successful treatment, and proactive screening helps reduce its incidence.
Colonoscopy is widely considered the gold standard for diagnosing CRC, but it has limitations such as invasiveness, cost, and patient discomfort, and may not always detect early-stage lesions (8). Molecular biomarkers for CRC detection are currently limited, and non-invasive stool-based tests such as fecal occult blood testing (FOBT) and fecal immunochemical tests (FIT), as well as blood-based protein biomarkers like CEA and CA19, have shown reduced sensitivity and specificity (9). Most patients (83%) prefer blood-based tests over stool-based tests (10). Liquid biopsies, especially blood-based ones, are gaining interest and are seen as the future of cancer screening (11, 12).
The development of CRC is linked to genetic mutations, epigenetic changes, environmental factors, and non-modifiable risk factors such as age, family history, and personal medical history. Liquid biopsies can detect these cancer-related progressive build-up of genetic and epigenetic alterations in plasma DNA (13). However, the identification of mutations for early cancer diagnosis is limited by the frequency and diversity of mutations gained throughout cancer development in CRC patients (14, 15). Epigenetic alterations, particularly DNA methylation, are mostly constant, occur before gene mutations in tumor development (16), with blood/tissue paired studies in CRC showing good tissue and tumor type specificity (17). These aberrant DNA methylations, as a most prevalent epigenetic alteration, can be effectively detected in numerous types of biological samples such as blood, tissue, and stool. Thus, Methylated circulating tumor DNA (mctDNA) can be a valuable source for tumor DNA in the diagnosis, prognosis, and surveillance of tumors (18), with potential applications in therapeutic interventions (19).
Cell-free DNA (cfDNA), generally at approximately 167 bp in length, is fragmented double helix DNA found in the blood circulation. It comprises nucleic acids released into body fluids through active release, apoptosis, necrosis, or other cell death processes. CfDNA in blood circulation has a short half-life of about 15 minutes to 2.5 hours, which allows ongoing surveillance of tumors (20), where the concentration of cfDNA in plasma is 1-10 ng/mL (often <20ng/ml) in healthy individuals, mostly from the hematopoietic system (21, 22). This concentration can be higher in patients with cancer (23) and also in diseases other than cancer (20). CtDNA is a subset of cfDNA that originates from tumor cells and contains unique genetic as well as epigenetic markers of the tumor (24). Compared to cfDNA, which primarily reflects apoptotic cell debris with a characteristic length of ~150-200 bp (coinciding with nucleosome size), ctDNA exhibits substantial heterogeneity. Its fragment size often exceeds 200 bp and can even reach >1000 bp, likely due to impaired apoptotic pathways in cancer cells (25). Further, the ctDNA fraction within cfDNA varies widely between 0.05% and 93% (20, 26).
Currently, several epigenetic in vitro diagnosis (IVD) tests are being added to the market for CRC screening (Table 1). The Cologuard® (27) is the first stool DNA-based test, and the Epi proColon® (28) is the first blood DNA-based test approved by the FDA for early CRC detection. Beyond commercially available kits, researchers have reported a diverse array of methylation-specific PCR assays for CRC detection. Primarily, these encompass potential biomarkers for diagnosis, yet they also include markers aimed at evaluating tumor burden, detecting disease relapse, and estimating patient prognosis. Due to the importance and fast-growing knowledge of blood-based biomarkers, in this article, we review all investigated blood DNA methylation-based biomarkers over the last 10 years to have an updated overview of improvements and directions for research. Although CRC-related epigenetic biomarkers were reviewed in several articles with different points of view (8, 17, 29-31), we aim to gather only blood-based epigenetic biomarkers.
Table 1.
Commercially available Epigenetic IVD/RUO tests for CRC
| Tests | Sample | Biomarker target | Sensitivity (%) | SPE (%) | Company | ||||
|---|---|---|---|---|---|---|---|---|---|
| CRC | I/II | III/IV | HGD | AA | |||||
| Cologuard (27, 68, 69) | Stool | NDRG4, BMP3, 7 KRAS mutations | 92 | 69 | 42 | 87 | Exact Sciences | ||
| Epi proColon (28, 70) | Blood | SEPT9 | 75–81 | 71-77 | 22 | 96–99 | Epigenomics | ||
| Colosafe (63, 71-73) | Stool | SDC2 | 81- 91 | 87 | 95 | 80 | 42-58 | 86 - 98 | Creative Biosciences |
| EarlyTect (74, 75) | Stool | SDC2 | 90 | 89 | 67 | 90 | EarlyTect | ||
| Colodefense (45-47) | Blood | SEPT9, SDC2 | 89 | 48 | 93 | VersaBio | |||
| Stool | 88-92 | 55-67 | 93 | ||||||
| ColoSure™ (76) | Stool | Vimentin | 72-77 | 83-94 | Labcorp | ||||
| ColonSecure (64) | Blood | 149 markers | 86 | 90% | - | ||||
AA: advanced adenoma, HGD: polyps with high-grade dysplasia, CRC: colorectal cancer
Methods
Search strategy
A comprehensive search up to December 30, 2023, limited to articles published in the last 10 years (after 2012), which included AA/stage 0 or stage I/II samples in biomarker validation, written in English, was performed exploiting three main electronic libraries: Web of Science® Core Collection (Clarivate Analytics, Philadelphia/London, USA/GB), PubMed® (National Library of Medicine’s, Bethesda, MD, USA), and Scopus® (Elsevier, Amsterdam, NL, USA). The employed keywords were “colorectal cancer,” “colorectal neoplasms,” “circulating tumor DNA,” “cfDNA,” “Cell-free DNA,” “biomarker,” “DNA methylation,” “cancer screening,” “cancer detection,” “diagnosis,” “early detection,” “adenoma,” “plasma,” “serum,” or “liquid biopsy.” The investigation utilized these specific terms to navigate through the keywords, titles, and abstracts of scholarly articles. Additionally, the bibliographies of all accessed full-text articles and significant reviews were meticulously examined to uncover further relevant research. The study was conducted based on Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) (32).
Study selection
The articles were initially screened based on their titles and abstracts to eliminate any irrelevant studies. They were then categorized into three groups: include, exclude, or unclear. For the "unclear" articles, the full-text versions were examined and placed into one of the other two categories: include or exclude. All full-text manuscripts were evaluated to determine if they met the eligibility criteria, which included being written in English, focusing on human subjects, methylation testing at least one ctDNA biomarker in CRC patients, and offering a detailed description of the patients and detection rate of assays. Studies that used animal models, reviews, congress abstracts, or articles in languages other than English were excluded. If needed, the corresponding authors of the selected published reports were contacted.
Data extraction
Two reviewers independently extracted data from the included studies, collecting information such as publication year, number of patients, demographic details, and clinical and laboratory data for methylation evaluation. Duplicate cases were identified and eliminated, and the medical records from all papers were compiled and consolidated. In the event of any disagreements between the two reviewers, a third author was consulted.
Results
Study characteristics
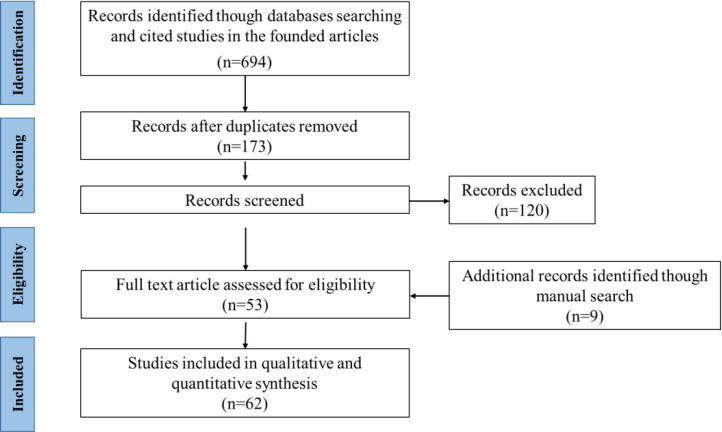
A comprehensive literature search identified 694 articles. Following deduplication, 173 articles were screened by title and abstract, leading to the exclusion of 120. Hand-searching of full-text articles and major reviews yielded an additional 9 articles. As illustrated in Figure 1, 61 articles ultimately met the inclusion criteria and proceeded to full-text assessment.
Figure 1.
Study selection flow chart.
Based on the nature of the study, the collected studies are presented in three categories. The "Genome-wide methylation studies" part discusses high throughput studies that were centered on biomarker identification, whereas the "Single methylation-based ctDNA biomarker" and "Multi-methylated ctDNA biomarker" sections deal with single or multi-biomarker validation.
Genome-wide methylation studies
Genome-wide methylation studies provide the most valuable and straightforward solution for de novo biomarker discovery. Most de novo biomarker-finding studies were accomplished on tissue, cell lines, or fecal samples (Table 2). Twenty-one studies used different sample sources and methods to discover novel methylated ctDNA (mctDNA) biomarkers in CRC for diagnosis, prognosis, and metastasis or therapy monitoring. Some of them used previously reported data from TCGA and GSE databases (15, 33-36) on cell lines and tissue samples. CpG microarray analysis is the most prevalent method for genome-wide methylation analysis. Nevertheless, recently sequencing-based methods have provided more hope to find suitable biomarkers for CRC. Methylation-specific quantitative PCR (MS-qPCR), digital PCR (dPCR), or droplet digital PCR (ddPCR) mostly have been used for the verification of selected biomarkers. Most of the studies used CRC tissues and paired matched normal tissue adjacent to the tumor (PNAT/MNAT) to discover biomarkers; however, it seems the best strategy is to simultaneously array or sequence both tissue and blood samples. Wang et al. (37) conducted a study on the overlapping markers between tissue DNA and cfDNA in advanced adenoma (AA) and early-stage CRC. They identified seven DNA methylation biomarkers that can be used to monitor the malignant progression from AA to CRC. However, it is important to note that their tissue and blood samples were not matched. Although high-throughput analysis such as simultaneously analyzing fragment length and methylation profiles gives a high AUC (0.989) with good sensitivity (96.8) and specificity (97%) in detecting CRC (38) and could provide high accuracy for early-stage CRC detection, it is expensive and computational analysis is also required. Hence, it is necessary to achieve a cost-effective set of biomarkers that possess the appropriate level of sensitivity and specificity for regular use in CRC screening and monitoring.
Table 2.
Biomarker discovery through genome-wide methylation studies
| Year/ Ref |
Serum/ Plasma Samples | Method of biomarker Discovery / Verification | Selected biomarker | AUC/ SEN, SPE% | AUC/ SEN, SPE% in stages | |
|---|---|---|---|---|---|---|
| CRC/ Adenoma | Normal | |||||
| Tissue | ||||||
| 2013 (77) | S:32 (6I/II, 26III/IV), 26A | 161 | Illumina Goldengate array (Tissue, stool, Serum, I-IV) / MS-qPCR | NPY, PENK, WIF1 | SEN:87, SPE:80 | |
| 2013 (78) | S:131(94I, 5II, 17III,15IV) | 125 | Discovery: MeDIA with microarray (Tissue I-IV, PNAT), Verification: MS-qPCR | SDC2 | SEN:87, SPE:95.2 | I: SEN:92 |
| 2015 (79) | P:353(42I, 140II, 108III, 63IV) | Discovery: HM450 array (Tissue/PNAT, 23 MSI/MSS CRC), Verification: MS-qPCR | AGBL4, FLI1, TWIST1 | SEN:93 any gene | SEN for I & II: 90 any gene | |
| 2018 (80) | P:45 | Discovery: HM450 array of tissue (18 C, 21 A, 7 N), Verification: Pyrosequencing | GRIA4, SLC8A1 and SYN3 | - | - | |
| 2019 (81) | P: 256 (41I, 143II, 55III, IV17) | 178 | Discovery: HM450 array (Tissue) Verification: ddPCR |
C9orf50, KCNQ5, CLIP4 | SEN:85, SPE:99 | I:80, II: 85, III: 89, IV: 88 |
| 2020 (82) | P:22 | 20 | Discovery: SureSelectXT Methyl-Seq (Tissue/PNAT), Verification: ddPCR | CLDN1, INHBA SLC30A10 | SEN:41, SPE:100 | |
| 2021 (83) | 44 P | 44 | Discovery: Targeted array (Tissue/PNAT) Verification: MS-qPCR & MSRE-qPCR |
WT1, PENK, SPARC, GDNF, TMEFF2, DCC |
AUC: >0.80 any gene | - |
| Plasma | ||||||
| 2014 (84) | P: 30 (11I, 19II) | 30 | Discovery: microarray (56 gene, Plasma) Verification: MS-qPCR |
CYCD2, HIC, VHL | AUC: 0.93, SEN:83, SPE:94 |
|
| 2018 (85) | S:20 (7I,13II), 20 AA | 20 | Discovery: MethylationEPIC (Plasma) | 1384 CpG sites | - | - |
| 2021 (86) | 13 P | 16 | Discovery: MBD-seq (Plasma) | CLIP4, LONRF2, RNF217 | ||
| 2021 (87) | P: 248 (66I, 86II, 62III, 34IV), 40A, 68AA | 133 | Discovery: Enrichment & HiSeq Sequencing of Plasma | 11 markers | AUC:0.92(88-0.96) CRC |
0.77 A, 0.85AA, 0.9 I |
| 2022 (88) | P: 5 (I), 5 A | Discovery: MethylationEPIC (HM850) | 1865 differently methylated CpG sites | - | - | |
| 2022 (89) | 4 P (2III, 2IV) | 3 | Discovery: MeDIP‑seq | PRDM14, RALYL, ELMOD1, TMEM132E | - | - |
| 2023 (44) | P: 590, 182 AA | 366 | Discovery: targeted bisulfite sequencing (ColonES assay) | 191 regions | AA SEN: 79, CRC SEN: 87, SPE: 88 | 0.903AA, 0.937 CRC |
| Bioinformatics | ||||||
| 2018 (34) | P: 182 | 50 | Discovery: TCGA and GEO cell/tissue data (HM450 array), Verification: dPCR | EYA4, GRIA4, ITGA4, MAP3K14-AS1, MSC | AUC: 0.86 | - |
| 2020 (15) | P: 801 | 1021 | Discovery: TCGA and GSE data Verification: ddPCR |
cg10673833 | AUC:0.90 SEN:90, SPE: 87 |
- |
| 2020 (35) | P:117 (17I,24II,33III,23IV) | 60 | Discovery: TCGA and GEO data Verification: ddPCR |
FAM123A, GLI3, PPP1R16B, SLIT3, TMEM90B |
SEN:58, SPE:95 | SEN:50 I-III, 96 IV, SPE: 95 |
| 2022 (36) | 47 (7III, 30IV, 6), 41AA | 81 | Discovery: TCGA and GEO tissue data Verification: MS-qPCR |
LINC00473 | AUC; CRC:0.88, AA: 0.84 | |
| Cell line | ||||||
| 2014 (90, 91) | P: 120 (12I, 30II, 12III, 66IV) | 96 | Discovery: MeDIP (cell lines) Verification: MS-PCR, pyrosequencing |
PPP1R3C, EFHD1 | SEN:90, SPE:64 | |
| 2017 (92) | P: 95 (10I, 22II, 48III, 15IV) | 47 | Discovery: MeDIP‑seq in NCM460 cell line | CBS | - | - |
| Tissue, Plasma | ||||||
| 2022 (37) | P: 218 (43I, 56II, 50III, 69IV), 88AA | Discovery: AnchorIRIS™ sequencing (Tissue, Plsama) | ZFHX4, ZNF334, ELOVL2, UNC5C, LOC146880, SFMBT2, GFRA1 |
AUC: 0.92, SEN:90, SPE:90 CRC VS AA |
||
CRC: Colorectal cancer; NA: Not ascertained; P/MNAT: paired,matched normal tissue adjacent to the tumor; MS-RE: methylation-sensitive restriction enzyme–based; MeDIP‑seq: immunoprecipitation coupled with high‑through‑put sequencing; P: plasma; S: serum; SEN: sensitivity, MBD-seq: methyl-CpG-binding domain sequencing, MeDIA: methylated DNA isolation assay; SPE: specificity,
Single methylation-based ctDNA biomarker
Totally, 22 studies examined single methylated ctDNA biomarkers in blood-based samples (plasma, serum, or a combination of them) which included AA/stage 0 or stage I/II. SEPT9 biomarker was the most studied biomarker and methylation-specific-qPCR is the method of choice for most of the methylation analysis. Twelve studies included polyp (adenoma or advanced adenoma) samples, 9 studies included polyps and stage I-III samples, and 8 studies included stage I-III without polyps’ samples (Table 3). Still, SEPT9 totally had the highest sensitivity and specificity among biomarkers (39) for CRC, while if we consider polyp and stage I/II CRC, MYO1-G (40), SEPT9 (39, 41), JAM3 (42) showed the highest sensitivity. MYO1-G revealed 74-86% sensitivity and 94% specificity in 50 patients with stage I/II CRC (40). SEPT9 indicated 23-31% (24 patients) sensitivity for advanced adenoma, 40% (12 patients) sensitivity for polyps, and 64-88% (67 patients) sensitivity for stage I/II CRC (39, 41). JAM3 showed AUC=0.8611 (P<0.001) and interestingly methylation rate in stage I/II CRC (81.96%) was greater than in stage III/IV CRC (Table 3).
Table 3.
Individual biomarker studies
| Year/ref | marker | Serum/ Plasma Samples | Method | Polyp | Stage I/II | Stage III/IV | Total | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CRC | Polyp | Normal | SPE % | SEN % | AUC | SPE % | SEN % | AUC | SPE % | SEN% | AUC | SEN % |
SPE % | AUC | ||||
| 1 | 2022 (36) | LINC00473 | P:47 | AA:41 | 81 | qMSP ddPCR |
0,84 | |||||||||||
| 2 | 2022 (93) | RASSF1A | P: 92 (15I, 24II, 22III, 31IV) | 67 CRP | - | 0.75 | I(0.83) II(0.87) |
III(0.87) IV(0.86) |
||||||||||
| 3 | 2022 (94) | SEPT9 | EORC:27(4I,1II,5III,12IV) | 87 | 90.8 | 96.3 | ||||||||||||
| 4 | 2021 (95) | SHISA3 | P: 30 (8I, 10II, 10III, 2IV) | - | 9 | BSP | - | - | - | - | I(4.78) II(5.96) |
- | - | III(7.02) IV(5.2) |
- | - | - | 0.50 |
| 5 | 2021 (40) | MYO1-G | P: 305 (3I, 32II, 85III, 185IV) | - | 307 | ddPCR | - | - | - | 95.4 | I(85.7) II(74.4) |
- | 95.4 | III(83.2) IV(86.8) |
- | 84.3 | 95.4 | 0.94 |
| 6 | 2021 (41) | SEPT9 | P: 53 (14I,16II,9III,14IV) | 48AA, 30A | 48 | qMSP | 88.2 | 83.3 | 84.9 | |||||||||
| 7 | 2020 (39) | SEPT9 | P: 90 (18I, 27II, 26III, 10IV) | 13AA | 81 | qMSP | AA (96.3) |
AA (30.8) |
- | - | I (77.8) II (85.2) |
- | - | III(92.3) IV(80.0) |
- | 85.6 | 90.1 | 0.88 |
| 8 | 2020 (96) | NEUROG1 | S:16 (6I, 4II, 6III) | 89AA, 17A | 33 | qMSP | - | AA (32.8) | - | - | - | - | - | - | - | 33.33 | 90.6 | - |
| 9 | 2019 (42) | JAM3 | P:18 (1I, 4II, 7III, 3IV,3) | 18 | qMSP | - | I/II (81.96) | III/IV (73.36) |
61 | |||||||||
| 10 | 2019 (97) | SFRP2 | S: 62 (13I,27II,17III,5IV) | 7AA | 55 | qMSP | AA (87.3) | AA (42.9) | - | - | I (46.2) II (74.1) |
- | - | III(70.6) IV(100) |
- | 69.4 | 87.3 | 0.82 |
| 11 | 2019 (98) | RUNX3 | S:85 (9I, 39II, 34III, 3IV) | 40A | 40 | qMSP | 17.5 | - | - | - | - | - | - | - | - | - | 82.5 | 60 |
| 12 | 2019 (98) | SFRP1 | S:85 (9I, 39II, 34III, 3IV) | 40A | 40 | qMSP | 30 | - | - | - | - | - | - | - | - | - | 70 | 77.6 |
| 13 | 2018 (99) | BMP3 | P:50 | 40A | 50 | qMSP | 40 | 94 | ||||||||||
| 14 | 2018 (100) | MGMT | S: 30 (4I, 17II, 5III, 1IV) | 40 | MSP | 90 | ||||||||||||
| 15 | 2017 (101) | SEPT9 | P:85 | 364 A, 216 AA | 324 | qMSP | A (38.7), AA (47.0-62.5) |
|||||||||||
| 16 | 2016 (102) | SST | P: 165 (26I, 62II, 62III, 15IV) | - | qMSP | |||||||||||||
| 17 | 2015 (103) | NDRG4 | P: 154 (43I, 44II, 46III, 21IV) | _ | 444 | qMSP | - | - | - | 95 | I (16) II (11) |
- | 95 | III (35) IV (62) |
- | 27 | 95 | 0.61 |
| 18 | 2015 (103) | FOXE1 | P: 154 (43I, 44II, 46III, 21IV) | _ | 444 | qMSP | - | - | - | 93 | I (35) II (43) |
- | 93 | III (50) IV (67) |
- | 46 | 93 | 0.70 |
| 19 | 2015 (103) | SYNE1 | P: 154 (43I, 44II, 46III, 21IV) | _ | 444 | qMSP | - | - | - | 96 | I (28) II (52) |
- | 96 | III (76) IV (47) |
- | 47 | 96 | 0.72 |
| 20 | 2015 (103) | GATA5 | P: 154 (43I, 44II, 46III, 21IV) | _ | 444 | qMSP | - | - | - | 99 | I (14) II (9) |
- | 99 | III (48) IV (18) |
- | 18 | 99 | 0.59 |
| 21 | 2014 (104) | SEPT9 | P: 34 (6I, 11II, 11III, 5IV) | 26A | 24 | qMSP | - | 30.8 | - | - | - | - | - | - | - | 88.2 | - | - |
| 22 | 2014 (105) | VIM | S: 242 (7 0, 36I, 73II, 74III, 49IV) | 25 | qMSP | - | 0 (57.1) | - | - | I (30.6) II (28.8) |
- | - | III(35.1) IV(32.7) |
- | 32.6 | - | - | |
| 23 | 2014 (106) |
SEPT9 | P:44 (39 I-III, 5 IV) | 621AA | 444 | qMSP | - | AA (22) |
- | - | I/II (64) | - | - | III(64) IV(100) |
- | 68 | 78.8 | - |
| 24 | 2014 (107) |
CAHM | P: 73 (12I, 21II, 23III, 12IV) | 73A | 74 | qMSP | - | 4 | - | - | I (42) II (52) |
- | - | III(52) IV(75) |
- | 55 | 93 | - |
| 25 | 2014 (108) |
SEPT9 | P: 53 (22I, 14II, 12III, 5IV) | 209A 314AA |
1457 | qMSP | - | A(7.7) AA(9.6) |
- | - | I (36.4) II (57.1) |
- | - | III(58.3) IV(80.0) |
- | 50.9 | 91.5 | - |
| 26 | 2013 (109) | PCDH10 | S:63(12I,26II, 17III, 8IV) | qMSP | - | - | - | - | 62.7 | - | - | - | - | |||||
CRC: Colorectal cancer; NA: Not ascertained; P: plasma; S: serum; SEN: sensitivity, SPE: specificity; A: Adenoma; AA: Advanced adenoma; MSP: Methylation-specific PCR; qMSP: Methylation-specific qPCR, CRP: colorectal polyp, BSP: bisulfite sequencing PCR, EOCRC:early-onset colorectal cancer
Multi-methylated ctDNA biomarker
We considered a biomarker panel any evaluation of two or more methylated genes. Totally, 17 studies investigated multitarget methylated ctDNA in blood-based samples (plasma or serum). Two studies used 32 (43) and 191 (44) CpG sites to evaluate CRC samples. The most used method was qMSP; also ddPCR, MS-HRM, and NGS were used to methylation profiles. Most studies have included mSEPT9 and/or mSDC2 markers whose combination has been produced as a ColoDefense® kit. Colodefense is a stool/blood-based DNA hypermethylation screening test for CRC with SEPT9 and SDC2 assessment that has been approved by the China Food and Drug Administration. ColoDefense® showed 89% sensitivity for CRC and 48% sensitivity for advanced adenoma with 93% specificity (45-47).
The highest sensitivity for detection of polyps (adenoma, advanced adenoma) was reported in 4 biomarkers, SFRP1, SFRP2, SDC2, and PRIMA1 (48). Bartak et al, in a small sample size with 37 advanced adenoma tissues, showed 89% sensitivity. Another study with 191 CpG sites (44) in a large sample with 182 AA revealed 79% sensitivity. For the detection of stage I/II CRC, 100% sensitivity and 78% specificity were reported for SEPT9 and SDC2 biomarkers (49). Seven biomarkers (50), ALX4, BMP3, NPTX2, RARB, SDC2, SEPT9, and VIM, showed 89% sensitivity, and another seven DNA-Methylation Marker (37) with unpublished biomarkers indicated 87% sensitivity.
Discussion
CRC has high mortality rates, underscoring the need for effective screening tools to enhance early detection and improve curability. While colonoscopy is the gold standard with sensitivity rates exceeding 95% for CRC and 88-98% for precancerous lesions (AA), its drawbacks, such as invasiveness, discomfort, sedation requirements, risks of bowel damage and infection, and cost contribute to low patient adherence. Sigmoidoscopy and CT colonography offer high sensitivity rates (95% and 90% for CRC and precancerous lesions, respectively) but also require uncomfortable bowel preparation (51, 52). Fecal tests such as FOBT, guaiac FOBT (gFOBT) and FIT have lower sensitivity rates (33–75%) and are more suitable for detecting advanced colorectal abnormalities. In addition, current tumor markers such as CEA and CA19-9 have limitations in diagnosing CRC, highlighting the need for novel, non-invasive, and highly sensitive detection methods (52). Recent progress in circulating DNA methylation analysis shows promise in identifying both CRC and precancerous lesions.
Tumor-specific methylation changes in peripheral blood mononuclear cells (PBMCs), particularly in blood leukocytes, may reflect those seen in cancerous tissues, indicating their potential as novel early cancer biomarkers. However, the clinical significance of PBMC methylation in cancer diagnosis and patient prognosis remains uncertain (53). Studies have shown that a substantial portion of plasma cfDNA comes from hematopoietic lineages, particularly leukocyte genomic DNA, in individuals with or without cancer (22). This poses a challenge as it raises the risk of false-positive results in detecting mutations and methylation markers. The high prevalence of mutations in both normal and cancerous tissue limits their usefulness as early-stage cancer biomarkers due to insufficient specificity (54, 55). However, recently, Chen et al. (2021) demonstrated that the leukocyte’s gDNA will not affect the performance of the plasma methylated-ctDNA test, and methylation markers effectively distinguish CRC from benign tumors and healthy controls, while leukocyte levels of these markers lacked discriminatory power (49). They verified the performance of the ColoDefense test, consisting of methylated SEPT9 (mSEPT9) and methylated SDC2 (mSDC2), in plasma and the paired leucocyte fraction of 213 blood sample from CRC patients, adenomatous polyps’ patients, hyperplastic polyp patients, and control subjects. It is also worth noting that, when compared to tissue, cfDNA has a lower methylation quantity, which may be useful in their differentiation (56).
Compared to plasma, serum demonstrates a marked increase in the total quantity of cfDNA and displays significantly higher integrity. This observation suggests the presence of contaminating genomic DNA (gDNA) potentially introduced during serum separation via clotting processes. Thus, plasma is favored for cfDNA analyses since serum cfDNA has elevated gDNA (57). Despite variations in ctDNA abundance, several studies demonstrate equivalent sensitivity for detecting KRAS, TP53, BRAF, and SMAD4 mutations in both plasma and serum of breast cancer patients (58).
In ctDNA assays, plasma is the preferred biofluid over serum due to reduced contamination with non-tumor cfDNA. Serum preparation involves blood clotting, which induces leukocyte lysis and release of their cfDNA. This non-tumor cfDNA dilutes ctDNA, particularly those with low allele fraction mutations, potentially hindering their detection and compromising assay sensitivity (59). Therefore, current National Cancer Institute (NCI) Biorepositories and Biospecimen Research Branch Biospecimen Evidence-Based Practices (BEBP) guidelines for cfDNA analysis in biospecimens recommend shorter durations of pre-analytical storage at room temperature. Specifically, they suggest a maximum of 2–4 hours for EDTA tubes and up to 3 days for preservative tubes before initiating plasma isolation and subsequent storage at −80 °C (60).
Bisulfite sequencing (BS-seq) has been widely regarded as the gold standard for quantifying DNA methylation at a base-level resolution. MS-qPCR is the most commonly employed method, followed by MS-PCR (Tables 3 and 4). Methods for profiling methylation at specific genomic locations have progressed to encompass a broader genomic scope through the use of high-throughput sequencing or array-based approaches. This expansion enables a more comprehensive analysis of the genome and the discovery of aberrations that had not been detected before (61) (Table 2). Several detection methods, including ddPCR and next-generation sequencing (NGS), have expanded the options for detecting genetic alterations in trace amounts of cfDNA. However, the sensitivity and specificity of the markers used remain critical factors for accurate detection, particularly in early cancer screening tests. NGS technologies, capable of local deep BS-seq, offer the highest sensitivity for detecting DNA methylation at the single-molecule level; however, when analyzing a limited number of samples, qPCR remains the simplest and most frequently utilized method for DNA methylation detection (20).
Table 4.
Multi-biomarker studies
| Year/ref | Marker | Serum/ Plasma Samples | Method | Polyp | Stage I/II | Stage III/IV | Total | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CRC | Polyp | Normal | SEN % |
SPE % |
AUC | SEN % |
SPE % |
AUC | SEN % |
SPE % |
AUC | AUC | SPE% | SEN% | |||||
| 1 | 2023 (110) | SDC2, NPY, IKZF1, SEPT9 | P: 124 (5 0, 36I, 34II, 45III, 3IV) | 137AA | 164 | ddPCR | AA (7) | _ | _ | _ | _ | _ | _ | _ | _ | _ | 92 | 44 | |
| 2 | 2023 (111) | GALNT9/UPF3A | 27 | 21 | NGS | 78.6 | I (87.5) II (100) |
III(100) IV(33.3) |
100 | 78.8 | |||||||||
| 3 | 2023 (44) | 191 CpG sites | 590 | 182AA | 366 | NGS | AA (79.0) | _ | AA (0.90) | I (85.1) II (79.3) |
_ | _ | IV(91.8) | _ | _ | 0.93 | 88.1 | 86.6 | |
| 4 | 2022 (37) | Seven DNA-Methylation Marker | P: 218 (43I, 56II, 50III, 69IV) | 88AA | _ | NGS | _ | AA (89.66) | I (87.5) II (93.75) |
_ | _ | III(88) IV(92.8) |
_ | _ | 0.86 | 97 | |||
| 5 | 2022 (43) | 32 CpG sites | P: 20 | 4 | NGS | _ | _ | _ | _ | _ | _ | _ | _ | _ | 92 | 85 | |||
| 6 | 2022 (112) | FBN1, SPG20 | P:62 | 8 | 50 | MS-HRM | _ | _ | _ | _ | _ | _ | _ | _ | _ | 0.94 | 97 | 91.1 | |
| 7 | 2021 (83) | 35 CpG sites | 16 | 40 | MSRE- qPCR | 63.0 | 88.0 | 0.80 | |||||||||||
| 8 | 2021 (49) | SEPT9, SDC2 | P: 91 (4 0, 9I, 31II, 29III, 4IV) | 49A, 27AA | 38 | qMSP | 0 (50) | _ | I (100) II (76.9) |
_ | III(85.7) IV(100) |
_ | 0.97 | 86.8 | 85.7 | ||||
| 9 | 2021 (113) | C9orf50, TWIST1, KCNJ12, ZNF132 | P:35 | 2A, 22AA | 32 | qMSP | 0.91 | 97 | 80 | ||||||||||
| 10 | 2021 (114) | SEPT9, SDC2, BCAT1 | P: 104 | 83A,47AA | 60 | qMSP | Polyps (4.4) |
_ | _ | _ | _ | _ | _ | _ | _ | 0.91 | 96.9 | 82.7 | |
| 11 | 2018 (115) |
Uc160, Uc283 and Uc346 | P: 50 | 59A | 40 | qMSP | _ | _ | _ | _ | _ | _ | _ | _ | _ | 0.63 | 74.3 | 45 | |
| 12 | 2017 (50) |
ALX4, BMP3, NPTX2, RARB, SDC2, SEPT9, VIM | P:193 | 33A | 102 | qMSP | 88.7 | 73.5 | 0.85 | _ | _ | _ | 0.86 | 72.5 | 90.7 | ||||
| 13 | 2017 (48) |
SFRP1, SFRP2, SDC2, PRIMA1 | P: 7 | 37A | 47 | qMSP | 89.2 | 86.5 | 0.93 | _ | _ | _ | _ | _ | _ | 0.97 | 97.3 | 91.5 | |
| 14 | 2015 (116) |
BCAT1, IKZF1 | P:129 (29I, 42II, 40III, 16IV) | 346A, 338AA | 1291 | qMSP | AA (6) | _ | I /II (56) | _ | 79 | _ | 94 | 66 | |||||
| 15 | 2015 (103) |
SYNE1 and FOXE1 | P:66 (27I, 15II, 20III, 4IV) | _ | 240 | qMSP | _ | _ | _ | I (37) II (87) |
_ | III (55) IV (100) |
_ | 91 | 58 | ||||
| 16 | 2015 (117) |
GATA5, SFRP2 | P: 57 | 30A | 47 | MSP | 26.77 | 91.49 | _ | _ | _ | _ | _ | _ | _ | _ | 91.49 | 42.86 | |
| 17 | 2013 (118) |
TAC1, SEPT9 | S:26(I) | 26 | qMSP | _ | _ | _ | I (73.1) | I (92.3) | I (82.1) | _ | _ | _ | _ | _ | _ | ||
CRC: Colorectal cancer; NA: Not ascertained; P: plasma; S: serum; SEN: sensitivity, SPE: specificity; A: Adenoma; AA: Advanced adenoma; MSP: Methylation-specific PCR; qMSP: Methylation-specific qPCR , MethylLight Droplet Digital PCR (ML-ddPCR), Methylation-Sensitive Restriction Enzyme (MSRE)
There is a limited report of paired evaluation of stool/plasma samples in patients. Meanwhile, patients greatly preferred noninvasive testing, and most of them (83%) preferred a blood-based test over a stool-based test (10). However, normally stool samples showed higher sensitivity in comparison to plasma samples (44). The mSEPT9 showed higher sensitivity in stool samples vs. plasma samples while having a similar specificity (39). Similarly, mSFRP2 demonstrated greater sensitivity in tissue and stool samples compared to plasma samples. However, the methylation of SFRP2 in serum revealed markedly higher specificity for distinguishing CRCs from benign adenomas, as opposed to the methylation levels of SFRP2 found in tumor and fecal DNA (62). The methylation test for SDC2 in stool samples outperformed the mSEPT9 blood test in the detection of nonmetastatic CRC and adenomas (63). Epi proColon, with its improved edition Epi proColon 2.0, was developed using the methylation status of SEPT9 in plasma ctDNA and received FDA approval (28). However, the U.S. Preventive Services Task Force (USPSTF) does not endorse it as an initial screening approach due to its low sensitivity for CRC (48%) including early-stage CRC (35%) and AA (11.2%). Fortunately, a more advanced formulation of this biomarker, Epi proColon 2.0, has been recently introduced, exhibiting enhanced sensitivity and specificity. This development has heightened expectations for the discovery of an optimal blood-based biomarker. Colodefense (45-47) with detection of SEPT9, and SDC2 biomarkers, and ColonSecure (64) with 149 biomarkers are other kits introduced for blood-based CRC screening. Present commercial blood-based tests have a low sensitivity for AA ranging from 22% (Epi proColon) to 48% (Colodefense) and for I/II stage ranging within 71-77% (Epi proColon) (Table 1).
SEPT9 and Syndecan-2 (SDC2) are the two most studied biomarkers in CRC detection. Here we discuss these two biomarkers specifically. MSEPT9 methylation in plasma/serum demonstrated variable sensitivity (47-87%) and high specificity (89-98%) across diverse studies. While sensitivity increased with advanced CRC stages (reaching 100% in some stage IV reports), it remained suboptimal in the early stages (I-II) (17). Furthermore, mSEPT9's sensitivity for detecting adenomas and polyps is suboptimal in most studies, ranging from 8 to 40%. Notably, combining FOBT and mSEPT9 assessment achieved a remarkable 100% sensitivity for stage I CRC identification (65). While SDC2 methylation shows promise as a biomarker for early CRC detection, its stage specificity appears limited. Assay sensitivity is demonstrably higher for advanced (stage III/IV) disease compared to early (stage I/II) stages, suggesting a potential need for further optimization for improved sensitivity in early detection applications (8). It seems co-analyzing of SEPT9 and SDC2 showed a superior detection rate for stage I/II (49). Another single biomarkers with high sensitivity for polyp and stage I/II CRC include MYO1-G (40), SEPT9 (39, 41), and JAM3 (42) showed the highest sensitivity. Multi-biomarkers with suitable sensitivity for polyp and stage I/II CRC include SFRP1, SFRP2, SDC2, PRIMA1 combination (48), 191 CpG sites (44), ALX4, BMP3, NPTX2, RARB, SDC2, SEPT9, VIM combination (50), and another seven markers (37).
Logically, the detection rate of CRC will grow with increasing the number of biomarkers for analysis. However, there is a discrepancy within studies (Table 1 and 2) which may be related to sample size, heterogeneity of samples, and even the location of CpG island in biomarkers (66). Although the studies and reported biomarkers showed the invaluable potential of blood-based methylation assays for CRC early detection even before dysplasia, identifying potential biomarkers and combinational strategy is an urgent need today.
Conclusion and future perspective
Compared to immunoaffinity tests such as FIT, PCR-based DNA tests offer superior sensitivity for detecting molecular markers, particularly in early-stage CRC. Despite the higher costs associated with these tests, their increased sensitivity may outweigh this factor due to the significant potential for early diagnosis to save lives and reduce costs. In this study, we presented promising biomarkers such as MYO1-G and JAM3, in addition to SEPT9 and SDC2. Multi-biomarker assays, while offering increased sensitivity, may also lead to higher costs. Combinations of biomarkers such as SFRP1, SFRP2, SDC2, PRIMA1, or ALX4, BMP3, NPTX2, RARB, SDC2, SEPT9, VIM, or ZFHX4, ZNF334, ELOVL2, UNC5C, LOC146880, SFMBT2, and GFRA1 have been identified for polyps and stage I/II CRC. However, these molecular screening methods still require enhancements in sensitivity and specificity to maximize their clinical utility.
Early tumor detection using ctDNA methylation faces critical limitations. Low ctDNA abundance in plasma, particularly in early-stage cancers, hinders efficient capture for methylation analysis. Innovations such as Droplet digital PCR or deep sequencing methods utilizing larger volumes of plasma may help overcome this challenge.
Despite numerous studies indicating tumor-specific methylation changes, their validity remains uncertain. Most of these alterations are limited to individual studies lacking independent sample validation. So only a small fraction has advanced through clinical trials and commercialization for CRC detection. Another limitation of current research is the small number of patients included, hindering the discovery of clinically significant biomarker candidates. Genome-wide methylome profiling offers a powerful solution, providing comprehensive and reproducible data to unlock the true potential of methylation biomarkers. Future large-scale studies or the integration of existing methylome-level data will be necessary to identify biomarkers that are reliable enough for clinical application.
Conflict of interests
The authors declare no conflict of interest.
References
- 1.Xie YH, Chen YX, Fang JY. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct Target Ther. 2020;5:1–30. doi: 10.1038/s41392-020-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao W, Chen HD, Yu YW, Li N, Chen WQ. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J. 2021;134:783–91. doi: 10.1097/CM9.0000000000001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, et al. Colorectal cancer. Nat Rev Dis Primers. 2015;1:15065. doi: 10.1038/nrdp.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Pool AE, Damhuis RA, Ijzermans JN, de Wilt JH, Eggermont AM, Kranse R, Verhoef C. Trends in incidence, treatment and survival of patients with stage IV colorectal cancer: a population-based series. Colorectal Dis. 2012;14:56–61. doi: 10.1111/j.1463-1318.2010.02539.x. [DOI] [PubMed] [Google Scholar]
- 5.Kow AWC. Hepatic metastasis from colorectal cancer. J Gastrointest Oncol. 2019;10:1274–98. doi: 10.21037/jgo.2019.08.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen LH, Goel A, Chung DC. Pathways of Colorectal Carcinogenesis. Gastroenterology. 2020;158:291–302. doi: 10.1053/j.gastro.2019.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Provenzale D, Ness RM, Llor X, Weiss JM, Abbadessa B, Cooper G, et al. NCCN Guidelines Insights: Colorectal Cancer Screening, Version 2 2020. J Natl Compr Canc Netw. 2020;18:1312–20. doi: 10.6004/jnccn.2020.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anghel SA, Ioniță-Mîndrican CB, Luca I, Pop AL. Promising epigenetic biomarkers for the early detection of colorectal cancer: a systematic review. Cancers. 2021:13. doi: 10.3390/cancers13194965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaukat A, Levin TR. Current and future colorectal cancer screening strategies. Nat Rev Gastroenterol Hepatol. 2022;19:521–31. doi: 10.1038/s41575-022-00612-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adler A, Geiger S, Keil A, Bias H, Schatz P, deVos T, et al. Improving compliance to colorectal cancer screening using blood and stool based tests in patients refusing screening colonoscopy in Germany. BMC Gastroenterol. 2014;14:183. doi: 10.1186/1471-230X-14-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic - implementation issues and future challenges. Nat Rev Clin Oncol. 2021;18:297–312. doi: 10.1038/s41571-020-00457-x. [DOI] [PubMed] [Google Scholar]
- 12.Razzaghi H, Khabbazpour M, Heidary Z, Heiat M, Moghaddam ZS, Derogar P, et al. Emerging role of tumor-educated platelets as a new liquid biopsy tool for colorectal cancer. Arch Iran Med. 2023:26. doi: 10.34172/aim.2023.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hossain MS, Karuniawati H, Jairoun AA, Urbi Z, Ooi J, John A, et al. Colorectal cancer: a review of carcinogenesis, global epidemiology, current challenges, risk factors, preventive and treatment strategies. Cancers. 2022:14. doi: 10.3390/cancers14071732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–9. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo H, Zhao Q, Wei W, Zheng L, Yi S, Li G, et al. Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Sci Transl Med. 2020:12. doi: 10.1126/scitranslmed.aax7533. [DOI] [PubMed] [Google Scholar]
- 16.Sprang M, Paret C, Faber J. CpG-Islands as Markers for Liquid Biopsies of Cancer Patients. Cells. 2020:9. doi: 10.3390/cells9081820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nassar FJ, Msheik ZS, Nasr RR, Temraz SN. Methylated circulating tumor DNA as a biomarker for colorectal cancer diagnosis, prognosis, and prediction. Clin Epigenetics. 2021;13:111. doi: 10.1186/s13148-021-01095-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Summers T, Langan RC, Nissan A, Brücher BL, Bilchik AJ, Protic M, et al. Serum-based DNA methylation biomarkers in colorectal cancer: potential for screening and early detection. J Cancer. 2013;4:210–6. doi: 10.7150/jca.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulis M, Esteller M. 2 - DNA Methylation and Cancer. In: In: Herceg Z, Ushijima T., editors. Advances in Genetics. Academic Press; 2010. pp. 27–56. [DOI] [PubMed] [Google Scholar]
- 20.Hu Z, Chen H, Long Y, Li P, Gu Y. The main sources of circulating cell-free DNA: Apoptosis, necrosis and active secretion. Crit Rev Oncol Hematol. 2021;157:103166. doi: 10.1016/j.critrevonc.2020.103166. [DOI] [PubMed] [Google Scholar]
- 21.Yao W, Mei C, Nan X, Hui L. Evaluation and comparison of in vitro degradation kinetics of DNA in serum, urine and saliva: a qualitative study. Gene. 2016;590:142–8. doi: 10.1016/j.gene.2016.06.033. [DOI] [PubMed] [Google Scholar]
- 22.Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA comprises an In Vivo nucleosome footprint that informs its tissues-of-origin. Cell. 2016;164:57–68. doi: 10.1016/j.cell.2015.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zill OA, Banks KC, Fairclough SR, Mortimer SA, Vowles JV, Mokhtari R, et al. The Landscape of actionable genomic alterations in cell-free circulating tumor DNA from 21,807 advanced cancer patients. Clin Cancer Res. 2018;24:3528–38. doi: 10.1158/1078-0432.CCR-17-3837. [DOI] [PubMed] [Google Scholar]
- 24.Wu Z, Yu L, Hou J, Cui L, Huang Y, Chen Q, et al. Plasma cfDNA for the diagnosis and prognosis of colorectal cancer. J Oncol. 2022;2022:9538384. doi: 10.1155/2022/9538384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giacona MB, Ruben GC, Iczkowski KA, Roos TB, Porter DM, Sorenson GD. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas. 1998;17:89–97. doi: 10.1097/00006676-199807000-00012. [DOI] [PubMed] [Google Scholar]
- 26.Luo H, Wei W, Ye Z, Zheng J, Xu RH. Liquid biopsy of methylation biomarkers in Cell-Free DNA. Trends Mol Med. 2021;27:482–500. doi: 10.1016/j.molmed.2020.12.011. [DOI] [PubMed] [Google Scholar]
- 27.Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287–97. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 28.Lamb YN, Dhillon S. Epi proColon((R)) 2 0 CE: a blood-based screening test for colorectal cancer. Mol Diagn Ther. 2017;21:225–32. doi: 10.1007/s40291-017-0259-y. [DOI] [PubMed] [Google Scholar]
- 29.Müller D, Győrffy B. DNA methylation-based diagnostic, prognostic, and predictive biomarkers in colorectal cancer. Biochim Biophys Acta Rev Cancer. 2022;1877:188722. doi: 10.1016/j.bbcan.2022.188722. [DOI] [PubMed] [Google Scholar]
- 30.Galoș D, Gorzo A, Balacescu O, Sur D. Clinical applications of liquid biopsy in colorectal cancer screening: current challenges and future perspectives. Cells. 2022:11. doi: 10.3390/cells11213493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fatemi N, Tierling S, Es HA, Varkiani M, Mojarad EN, Aghdaei HA, et al. DNA methylation biomarkers in colorectal cancer: Clinical applications for precision medicine. Int J Cancer. 2022;151:2068–81. doi: 10.1002/ijc.34186. [DOI] [PubMed] [Google Scholar]
- 32.Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Br Med J. 2021;372:n71. doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lange CP, Campan M, Hinoue T, Schmitz RF, van der Meulen-de Jong AE, Slingerland H, et al. Genome-scale discovery of DNA-methylation biomarkers for blood-based detection of colorectal cancer. PLoS One. 2012;7:50266. doi: 10.1371/journal.pone.0050266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barault L, Amatu A, Siravegna G, Ponzetti A, Moran S, Cassingena A, et al. Discovery of methylated circulating DNA biomarkers for comprehensive non-invasive monitoring of treatment response in metastatic colorectal cancer. Gut. 2018;67:1995–2005. doi: 10.1136/gutjnl-2016-313372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho NY, Park JW, Wen X, Shin YJ, Kang JK, Song SH, et al. Blood-Based Detection of Colorectal Cancer Using Cancer-Specific DNA Methylation Markers. Diagnostics. 2020:11. doi: 10.3390/diagnostics11010051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruiz-Bañobre J, Rodriguez-Casanova A, Costa-Fraga N, Bao-Caamano A, Alvarez-Castro A, Carreras-Presas M, et al. Noninvasive early detection of colorectal cancer by hypermethylation of the LINC00473 promoter in plasma cell-free DNA. Clin Epigenetics. 2022;14:86. doi: 10.1186/s13148-022-01302-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang W, Zhang X, Zhu X, Cui W, Ye D, Tong G, et al. Seven DNA methylation biomarker prediction models for monitoring the malignant progression from advanced adenoma to colorectal cancer. Front Oncol. 2022;12:827811. doi: 10.3389/fonc.2022.827811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen HT, Khoa Huynh LA, Nguyen TV, Tran DH, Thu Tran TT, Khang Le ND, et al. Multimodal analysis of ctDNA methylation and fragmentomic profiles enhances detection of nonmetastatic colorectal cancer. Future Oncol. 2022;18:3895–3912. doi: 10.2217/fon-2022-1041. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Zhao G, Miao J, Li H, Ma Y, Liu X, et al. Performance comparison between plasma and stool methylated SEPT9 tests for detecting colorectal cancer. Front Genet. 2020;11:324. doi: 10.3389/fgene.2020.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin WH, Xiao J, Ye ZY, Wei DL, Zhai XH, Xu RH, et al. Circulating tumor DNA methylation marker MYO1-G for diagnosis and monitoring of colorectal cancer. Clin Epigenetics. 2021;13:232. doi: 10.1186/s13148-021-01216-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin S, Zhu D, Shao F, Chen S, Guo Y, Li K, et al. Efficient detection and post-surgical monitoring of colon cancer with a multi-marker DNA methylation liquid biopsy. Proc Natl Acad Sci USA. 2021;118:2017421118. doi: 10.1073/pnas.2017421118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou D, Tang W, Zhang Y, An HX. JAM3 functions as a novel tumor suppressor and is inactivated by DNA methylation in colorectal cancer. Cancer Manag Res. 2019;11:2457–70. doi: 10.2147/CMAR.S189937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gouda MA, Duose DY, Lapin M, Zalles S, Huang HJ, Xi Y, et al. Mutation-agnostic detection of colorectal cancer using liquid biopsy-based methylation-specific signatures. Oncologist. 2023;28:368–372. doi: 10.1093/oncolo/oyac204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mo S, Dai W, Wang H, Lan X, Ma C, Su Z, et al. Early detection and prognosis prediction for colorectal cancer by circulating tumour DNA methylation haplotypes: A multicentre cohort study. EClinicalMedicine. 2023;55:101717. doi: 10.1016/j.eclinm.2022.101717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao G, Liu X, Liu Y, Li H, Ma Y, Li S, et al. Aberrant DNA Methylation of SEPT9 and SDC2 in Stool specimens as an integrated biomarker for colorectal cancer early detection. Front Genet. 2020;11:643. doi: 10.3389/fgene.2020.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao G, Li H, Yang Z, Wang Z, Xu M, Xiong S, et al. Multiplex methylated DNA testing in plasma with high sensitivity and specificity for colorectal cancer screening. Cancer Med. 2019;8:5619–28. doi: 10.1002/cam4.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dai Y, Zhao G, Yang J, Zhou X, Xiong S, Lu X, et al. A simplified multiplex methylated DNA testing for early detection of colorectal cancer in stool DNA. BMC Gastroenterol. 2022;22:428. doi: 10.1186/s12876-022-02512-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barták BK, Kalmár A, Péterfia B, Patai Á V, Galamb O, Valcz G, et al. Colorectal adenoma and cancer detection based on altered methylation pattern of SFRP1, SFRP2, SDC2, and PRIMA1 in plasma samples. Epigenetics. 2017;12:751–63. doi: 10.1080/15592294.2017.1356957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Z, Zhao G, Wang K, Wang X, Ma Y, Xiong S, et al. Blood leukocytes methylation levels analysis indicate methylated plasma test is a promising tool for colorectal cancer early detection. J Cancer. 2021;12:3678–85. doi: 10.7150/jca.57114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rasmussen SL, Krarup HB, Sunesen KG, Johansen MB, Stender MT, Pedersen IS, et al. Hypermethylated DNA, a circulating biomarker for colorectal cancer detection. PLoS One. 2017;12:0180809. doi: 10.1371/journal.pone.0180809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simon K. Colorectal cancer development and advances in screening. Clin Interv Aging. 2016;11:967–76. doi: 10.2147/CIA.S109285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swiderska M, Choromanska B, Dabrowska E, Konarzewska-Duchnowska E, Choromanska K, Szczurko G, et al. The diagnostics of colorectal cancer. Contemp Oncol. 2014;18:1–6. doi: 10.5114/wo.2013.39995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y, Fan Z, Meng Y, Liu S, Zhan H. Blood-based DNA methylation signatures in cancer: A systematic review. Biochim Biophys Acta Mol Basis Dis. 2023;1869:166583. doi: 10.1016/j.bbadis.2022.166583. [DOI] [PubMed] [Google Scholar]
- 54.Razavi P, Li BT, Brown DN, Jung B, Hubbell E, Shen R, et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med. 2019;25:1928–37. doi: 10.1038/s41591-019-0652-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu Y, Ulrich BC, Supplee J, Kuang Y, Lizotte PH, Feeney NB, et al. False-positive plasma genotyping due to clonal hematopoiesis. Clin Cancer Res. 2018;24:4437–43. doi: 10.1158/1078-0432.CCR-18-0143. [DOI] [PubMed] [Google Scholar]
- 56.Bedin C, Enzo MV, Del Bianco P, Pucciarelli S, Nitti D, Agostini M. Diagnostic and prognostic role of cell-free DNA testing for colorectal cancer patients. Int J Cancer. 2017;140:1888–98. doi: 10.1002/ijc.30565. [DOI] [PubMed] [Google Scholar]
- 57.Trigg RM, Martinson LJ, Parpart-Li S, Shaw JA. Factors that influence quality and yield of circulating-free DNA: A systematic review of the methodology literature. Heliyon. 2018;4:e00699. doi: 10.1016/j.heliyon.2018.e00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mijnes J, Tiedemann J, Eschenbruch J, Gasthaus J, Bringezu S, Bauerschlag D, et al. SNiPER: a novel hypermethylation biomarker panel for liquid biopsy based early breast cancer detection. Oncotarget. 2019;10:6494–508. doi: 10.18632/oncotarget.27303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pittella-Silva F, Chin YM, Chan HT, Nagayama S, Miyauchi E, Low SK, Nakamura Y. Plasma or serum: which is preferable for mutation detection in liquid biopsy? Clin Chem. 2020;66:946–57. doi: 10.1093/clinchem/hvaa103. [DOI] [PubMed] [Google Scholar]
- 60.Greytak SR, Engel KB, Parpart-Li S, Murtaza M, Bronkhorst AJ, Pertile MD, Moore HM. Harmonizing cell-free dna collection and processing practices through evidence-based guidance. Clin Cancer Res. 2020;26:3104–9. doi: 10.1158/1078-0432.CCR-19-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Y, Tollefsbol TO. DNA methylation detection: bisulfite genomic sequencing analysis. Methods Mol Biol. 2011;791:11–21. doi: 10.1007/978-1-61779-316-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tang D, Liu J, Wang DR, Yu HF, Li YK, Zhang JQ. Diagnostic and prognostic value of the methylation status of secreted frizzled-related protein 2 in colorectal cancer. Clin Invest Med. 2011;34:88–95. doi: 10.25011/cim.v34i1.15105. [DOI] [PubMed] [Google Scholar]
- 63.Zhan Y, Wang S, Yuan Z, Zhao X, Ni K, Xin R, et al. The stool syndecan2 methylation test is more robust than blood tests for methylated septin9, CEA, CA19-9 and CA724: a diagnostic test for the early detection of colorectal neoplasms. Transl Cancer Res. 2023;12:65–77. doi: 10.21037/tcr-22-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao F, Bai P, Xu J, Li Z, Muhammad S, Li D, et al. Efficacy of cell-free DNA methylation-based blood test for colorectal cancer screening in high-risk population: a prospective cohort study. Mol Cancer. 2023;22:157. doi: 10.1186/s12943-023-01866-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xie L, Jiang X, Li Q, Sun Z, Quan W, Duan Y, et al. Diagnostic value of methylated Septin9 for colorectal cancer detection. Front Oncol. 2018;8:247. doi: 10.3389/fonc.2018.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Massen M, Lommen K, Wouters KAD, Vandersmissen J, van Criekinge W, Herman JG, et al. Technical considerations in PCR-based assay design for diagnostic DNA methylation cancer biomarkers. Clin Epigenetics. 2022;14:56. doi: 10.1186/s13148-022-01273-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clebak KT, Nickolich S, Mendez-Miller M. Multitarget stool DNA testing (Cologuard) for colorectal cancer screening. Am Fam Physician. 2022;105:198–200. [PubMed] [Google Scholar]
- 68.Stürzlinger H, Conrads-Frank A, Eisenmann A, Invansits S, Jahn B, Janzic A, et al. Stool DNA testing for early detection of colorectal cancer: systematic review using the HTA Core Model(®) for rapid relative effectiveness assessment. Ger Med Sci. 2023;21:06. doi: 10.3205/000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shirley M. Epi proColon(®) for colorectal cancer screening: a profile of its use in the USA. Mol Diagn Ther. 2020;24:497–503. doi: 10.1007/s40291-020-00473-8. [DOI] [PubMed] [Google Scholar]
- 70.Wang J, Liu S, Wang H, Zheng L, Zhou C, Li G, et al. Robust performance of a novel stool DNA test of methylated SDC2 for colorectal cancer detection: a multicenter clinical study. Clin Epigenetics. 2020;12:162. doi: 10.1186/s13148-020-00954-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Niu F, Wen J, Fu X, Li C, Zhao R, Wu S, et al. Stool DNA test of Methylated Syndecan-2 for the early detection of colorectal neoplasia. Cancer Epidemiol Biomarkers Prev. 2017;26:1411–9. doi: 10.1158/1055-9965.EPI-17-0153. [DOI] [PubMed] [Google Scholar]
- 72.Wang X, Xu P, Shen H, Tao Y, Liu X, Cao H, et al. A novel stool-based SDC2 DNA methylation test is more robust than FIT and plasma CEA in detecting colorectal neoplasia in China. Am J Transl Med. 2021;5:37–50. [Google Scholar]
- 73.Beltran-Garcia J, Osca-Verdegal R, Mena-Molla S, Garcia-Gimenez JL. Epigenetic IVD tests for personalized precision medicine in cancer. Front Genet. 2019;10:621. doi: 10.3389/fgene.2019.00621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim CW, Kim H, Kim HR, Kye BH, Kim HJ, Min BS, et al. Colorectal cancer screening using a stool DNA-based SDC2 methylation test: a multicenter, prospective trial. BMC Gastroenterol. 2021;21:173. doi: 10.1186/s12876-021-01759-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ned RM, Melillo S, Marrone M. Fecal DNA testing for colorectal cancer screening: the ColoSure™ test. PLoS Curr. 2011;3:Rrn1220. doi: 10.1371/currents.RRN1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roperch JP, Incitti R, Forbin S, Bard F, Mansour H, Mesli F, et al. Aberrant methylation of NPY, PENK, and WIF1 as a promising marker for blood-based diagnosis of colorectal cancer. BMC Cancer. 2013;13:566. doi: 10.1186/1471-2407-13-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oh T, Kim N, Moon Y, Kim MS, Hoehn BD, Park CH, et al. Genome-wide identification and validation of a novel methylation biomarker, SDC2, for blood-based detection of colorectal cancer. J Mol Diagn. 2013;15:498–507. doi: 10.1016/j.jmoldx.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 78.Lin PC, Lin JK, Lin CH, Lin HH, Yang SH, Jiang JK, et al. Clinical relevance of plasma DNA methylation in colorectal cancer patients identified by using a genome-wide high-resolution array. Ann Surg Oncol. 2015;22:1419–27. doi: 10.1245/s10434-014-4277-2. [DOI] [PubMed] [Google Scholar]
- 79.Fadda A, Gentilini D, Moi L, Barault L, Leoni VP, Sulas P, et al. Colorectal cancer early methylation alterations affect the crosstalk between cell and surrounding environment, tracing a biomarker signature specific for this tumor. Int J Cancer. 2018;143:907–20. doi: 10.1002/ijc.31380. [DOI] [PubMed] [Google Scholar]
- 80.Jensen S, Øgaard N, Ørntoft MW, Rasmussen MH, Bramsen JB, Kristensen H, et al. Novel DNA methylation biomarkers show high sensitivity and specificity for blood-based detection of colorectal cancer-a clinical biomarker discovery and validation study. Clin Epigenetics. 2019;11:158. doi: 10.1186/s13148-019-0757-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pulverer W, Kruusmaa K, Schönthaler S, Huber J, Bitenc M, Bachleitner-Hofmann T, et al. Multiplexed DNA methylation analysis in colorectal cancer using liquid biopsy and its diagnostic and predictive value. Curr Issues Mol Biol. 2021;43:1419–35. doi: 10.3390/cimb43030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Melson J, Li Y, Cassinotti E, Melnikov A, Boni L, Ai J, et al. Commonality and differences of methylation signatures in the plasma of patients with pancreatic cancer and colorectal cancer. Int J Cancer. 2014;134:2656–62. doi: 10.1002/ijc.28593. [DOI] [PubMed] [Google Scholar]
- 84.Gallardo-Gómez M, Moran S, Páez de la Cadena M, Martínez-Zorzano VS, Rodríguez-Berrocal FJ, Rodríguez-Girondo M, et al. A new approach to epigenome-wide discovery of non-invasive methylation biomarkers for colorectal cancer screening in circulating cell-free DNA using pooled samples. Clin Epigenetics. 2018;10:53. doi: 10.1186/s13148-018-0487-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang J, Soupir AC, Schlick BD, Teng M, Sahin IH, Permuth JB, et al. Cancer detection and classification by CpG island hypermethylation signatures in plasma cell-free DNA. Cancers. 2021:13. doi: 10.3390/cancers13225611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu X, Zhang Y, Hu T, He X, Zou Y, Deng Q, et al. A novel cell-free DNA methylation-based model improves the early detection of colorectal cancer. Mol Oncol. 2021;15:2702–14. doi: 10.1002/1878-0261.12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu YL, Jiang T, Huang W, Wu XY, Zhang PJ, Tian YP. Genome-wide methylation profiling of early colorectal cancer using an Illumina Infinium Methylation EPIC BeadChip. World J Gastrointest Oncol. 2022;14:935–46. doi: 10.4251/wjgo.v14.i4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang X, Li T, Niu Q, Qin CJ, Zhang M, Wu GM, et al. Genome-wide analysis of cell-Free DNA methylation profiling with MeDIP-seq identified potential biomarkers for colorectal cancer. World J Surg Oncol. 2022;20:21. doi: 10.1186/s12957-022-02487-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Takane K, Midorikawa Y, Yagi K, Sakai A, Aburatani H, Takayama T, Kaneda A. Aberrant promoter methylation of PPP1R3C and EFHD1 in plasma of colorectal cancer patients. Cancer Med. 2014;3:1235–45. doi: 10.1002/cam4.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yagi K, Akagi K, Hayashi H, Nagae G, Tsuji S, Isagawa T, et al. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res. 2010;16:21–33. doi: 10.1158/1078-0432.CCR-09-2006. [DOI] [PubMed] [Google Scholar]
- 91.Xue G, Lu CJ, Pan SJ, Zhang YL, Miao H, Shan S, et al. DNA hypomethylation of CBS promoter induced by folate deficiency is a potential noninvasive circulating biomarker for colorectal adenocarcinomas. Oncotarget. 2017;8:51387–401. doi: 10.18632/oncotarget.17988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li J, Li H, Run ZC, Wang ZL, Jiang T, An Y, Li Z. RASSF1A methylation as a biomarker for detection of colorectal cancer and hepatocellular carcinoma. World J Gastrointest Oncol. 2022;14:1574–84. doi: 10.4251/wjgo.v14.i8.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Loomans-Kropp HA, Song Y, Gala M, Parikh AR, Van Seventer EE, Alvarez R, et al. Methylated Septin9 (m SEPT9): a promising blood-based biomarker for the detection and screening of early-onset colorectal cancer. Cancer Res Commun. 2022;2:90–8. doi: 10.1158/2767-9764.CRC-21-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tang SH, Hsiao CW, Chen WL, Wu LW, Chang JB, Yang BH. Hypermethylation of SHISA3 DNA as a blood-based biomarker for colorectal cancer. Chin J Physiol. 2021;64:51–6. doi: 10.4103/CJP.CJP_89_20. [DOI] [PubMed] [Google Scholar]
- 95.Otero-Estévez O, Gallardo-Gomez M, Cadena MP, Rodríguez-Berrocal FJ, Cubiella J, Ramirez VH, et al. Value of serum NEUROG1 methylation for the detection of advanced adenomas and colorectal cancer. Diagnostics. 2020:10. doi: 10.3390/diagnostics10070437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li H, Wang Z, Zhao G, Ma Y, Chen Y, Xue Q, et al. Performance of a MethyLight assay for methylated SFRP2 DNA detection in colorectal cancer tissue and serum. Int J Biol Markers. 2019;34:54–9. doi: 10.1177/1724600818820536. [DOI] [PubMed] [Google Scholar]
- 97.Pasha HF, Radwan MI, Yehia AM, Toam MM. Circulating methylated RUNX3 and SFRP1 genes as a noninvasive panel for early detection of colorectal cancer. Eur J Gastroenterol Hepatol. 2019;31:1342–9. doi: 10.1097/MEG.0000000000001532. [DOI] [PubMed] [Google Scholar]
- 98.Rokni P, Shariatpanahi AM, Sakhinia E, Kerachian MA. BMP3 promoter hypermethylation in plasma-derived cell-free DNA in colorectal cancer patients. Genes Genom. 2018;40:423–8. doi: 10.1007/s13258-017-0644-2. [DOI] [PubMed] [Google Scholar]
- 99.Alizadeh Naini M, Kavousipour S, Hasanzarini M, Nasrollah A, Monabati A, Mokarram P. O6-Methyguanine-DNA Methyl Transferase (MGMT) promoter methylation in serum DNA of Iranian patients with colorectal cancer. Asian Pac J Cancer Prev. 2018;19:1223–7. doi: 10.22034/APJCP.2018.19.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Song L, Peng X, Li Y, Xiao W, Jia J, Dong C, et al. The SEPT9 gene methylation assay is capable of detecting colorectal adenoma in opportunistic screening. Epigenomics. 2017;9:599–610. doi: 10.2217/epi-2016-0146. [DOI] [PubMed] [Google Scholar]
- 101.Liu Y, Chew MH, Tham CK, Tang CL, Ong SY, Zhao Y. Methylation of serum SST gene is an independent prognostic marker in colorectal cancer. Am J Cancer Res. 2016;6:2098–108. [PMC free article] [PubMed] [Google Scholar]
- 102.Melotte V, Yi JM, Lentjes MH, Smits KM, Van Neste L, Niessen HE, et al. Spectrin repeat containing nuclear envelope 1 and forkhead box protein E1 are promising markers for the detection of colorectal cancer in blood. Cancer Prev Res. 2015;8:157–64. doi: 10.1158/1940-6207.CAPR-14-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tóth K, Wasserkort R, Sipos F, Kalmár A, Wichmann B, Leiszter K, et al. Detection of methylated septin 9 in tissue and plasma of colorectal patients with neoplasia and the relationship to the amount of circulating cell-free DNA. PLoS One. 2014;9:115415. doi: 10.1371/journal.pone.0115415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shirahata A, Hibi K. Serum vimentin methylation as a potential marker for colorectal cancer. Anticancer Res. 2014;34:4121–5. [PubMed] [Google Scholar]
- 105.Potter NT, Hurban P, White MN, Whitlock KD, Lofton-Day CE, Tetzner R, et al. Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin Chem. 2014;60:1183–91. doi: 10.1373/clinchem.2013.221044. [DOI] [PubMed] [Google Scholar]
- 106.Pedersen SK, Mitchell SM, Graham LD, McEvoy A, Thomas ML, Baker RT, et al. CAHM, a long non-coding RNA gene hypermethylated in colorectal neoplasia. Epigenetics. 2014;9:1071–82. doi: 10.4161/epi.29046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Church TR, Wandell M, Lofton-Day C, Mongin SJ, Burger M, Payne SR, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63:317–25. doi: 10.1136/gutjnl-2012-304149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Danese E, Minicozzi AM, Benati M, Montagnana M, Paviati E, Salvagno GL, et al. Epigenetic alteration: new insights moving from tissue to plasma - the example of PCDH10 promoter methylation in colorectal cancer. Br J Cancer. 2013;109:807–13. doi: 10.1038/bjc.2013.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Petit J, Carroll G, Williams H, Pockney P, Scott RJ. Evaluation of a multi-gene methylation blood-test for the detection of colorectal cancer. Med Sci. 2023:11. doi: 10.3390/medsci11030060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gallardo-Gómez M, Rodríguez-Girondo M, Planell N, Moran S, Bujanda L, Etxart A, et al. Serum methylation of GALNT9, UPF3A, WARS, and LDB2 as noninvasive biomarkers for the early detection of colorectal cancer and advanced adenomas. Clin Epigenetics. 2023;15:157. doi: 10.1186/s13148-023-01570-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Alizadeh-Sedigh M, Fazeli MS, Mahmoodzadeh H, Sharif SB, Teimoori-Toolabi L. Methylation of FBN1, SPG20, ITF2, RUNX3, SNCA, MLH1, and SEPT9 genes in circulating cell-free DNA as biomarkers of colorectal cancer. Cancer Biomark. 2022;34:221–50. doi: 10.3233/CBM-210315. [DOI] [PubMed] [Google Scholar]
- 112.Zhang Y, Wu Q, Xu L, Wang H, Liu X, Li S, et al. Sensitive detection of colorectal cancer in peripheral blood by a novel methylation assay. Clin Epigenetics. 2021;13:90. doi: 10.1186/s13148-021-01076-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xu F, Yu S, Han J, Zong M, Tan Q, Zeng X, Fan L. Detection of Circulating Tumor DNA Methylation in Diagnosis of Colorectal Cancer. Clin Transl Gastroenterol. 2021;12:00386. doi: 10.14309/ctg.0000000000000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kottorou AE, Antonacopoulou AG, Dimitrakopoulos FD, Diamantopoulou G, Sirinian C, Kalofonou M, et al. Deregulation of methylation of transcribed-ultra conserved regions in colorectal cancer and their value for detection of adenomas and adenocarcinomas. Oncotarget. 2018;9:21411–28. doi: 10.18632/oncotarget.25115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pedersen SK, Symonds EL, Baker RT, Murray DH, McEvoy A, Van Doorn SC, et al. Evaluation of an assay for methylated BCAT1 and IKZF1 in plasma for detection of colorectal neoplasia. BMC Cancer. 2015;15:654. doi: 10.1186/s12885-015-1674-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang X, Song YF, Lu HN, Wang DP, Zhang XS, Huang SL, et al. Combined detection of plasma GATA5 and SFRP2 methylation is a valid noninvasive biomarker for colorectal cancer and adenomas. World J Gastroenterol. 2015;21:2629–37. doi: 10.3748/wjg.v21.i9.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu Y, Tham CK, Ong SY, Ho KS, Lim JF, Chew MH, et al. Serum methylation levels of TAC1 SEPT9 and EYA4 as diagnostic markers for early colorectal cancers: a pilot study. Biomarkers. 2013;18:399–405. doi: 10.3109/1354750X.2013.798745. [DOI] [PubMed] [Google Scholar]