Abstract
During latent infections of sensory neurons, herpes simplex virus type 1 gene expression is restricted to the latency-associated transcripts (LATs). The association of the stable 2.0-kb LAT intron with polysomes has suggested that it might represent a novel mRNA. In this work, we investigated expression of 2.0-kb LAT open reading frames (ORFs) by inserting the gene for green fluorescent protein (GFP) within the 2.0-kb LAT sequence, both within a LAT expression plasmid and in the context of the virus. Upon transient transfection of cells of both neuronal and nonneuronal origin with LAT-GFP expression vectors, low-level GFP fluorescence was distributed over the cell cytoplasm and likely resulted from infrequent initiation at a GFP AUG codon, on either unspliced or alternately spliced LAT RNAs. A second nucleolar GFP expression pattern which resulted from fusion of GFP to a conserved ORF in exon 1 of the LAT gene was also observed. However, the abundant expression of this fusion protein was dependent upon an artificially added translation initiation codon. Expression was much reduced and restricted to a small subset of transfected cells when this initator codon was removed. Neither the 2.0-kb LAT-GFP intron itself nor transcripts originating from the latency-associated promoter 2 (LAP2) were responsible for GFP expression. Abundant alternate splicing involving the 1.5-kb LAT splice acceptor and including splicing between the 1.5-kb LAT splice donor and acceptor, was observed in the nonneuronal Cos-1 cell line. Contrary to the results of our transfection studies, GFP expression could not be detected from a LAT-GFP virus at any stage of the infection cycle. Our results suggest that the inhibition of LAT ORF expression during viral infection occurred primarily at the level of translation.
Herpes simplex virus type 1 (HSV-1) is a neurotropic virus with an infection cycle characterized by two distinct phases. The virus replicates in a conventional lytic cycle in epithelial tissue but may also establish a latent infection in sensory ganglia that innervate the initial replication site (17, 35, 43). During latent infection, viral transcription is restricted to a family of RNAs known as the latency-associated transcripts (LATs) (see Fig. 1a) (8, 39, 45). The most abundant LAT is 2.0 kb long, while a second transcript of 1.5 kb is also routinely detected in latently infected neurons (33, 39, 40, 44, 45, 51). These transcripts are partially colinear and are considered to be differentially spliced products of a large precursor RNA (41, 51). Such a precursor RNA, identified by in situ hybridization and known as the minor hybridized LAT (mLAT), has been proposed to span the 2.0-kb LAT sequence and extend 8.3 kb from the major LAT promoter LAP1, to the first downstream polyadenylation site (26).
FIG. 1.
HSV-1 LAT genetic locus and 2.0-kb LAT-GFP expression vectors. (a) A schematic representation of the HSV-1 genome with an expanded view of the LAT transcription unit and the LAT RNAs is shown. (b) The LAT expression vector pcDNA-Pst/Mlu contains a PstI-MluI fragment of the HSV-1 genome, which spans the 2.0-kb LAT and includes LAT exon 1 and part of LAT exon 2. Transcription is driven by a CMV promoter, which directly replaces the LAT promoter. GFP was inserted into pcDNA-Pst/Mlu in both orientations (sense [+] and antisense [−]) at a HpaI site, directly upstream of conserved ORFs 1 and 2 but downstream of the 1.5-kb splice acceptor site (SA) to produce plasmids pLAT(H)GFP+ and pLAT(H)GFP−. GFP was also inserted at the HpaI site of an XcmI deletion variant of pcDNA-Pst/Mlu to create the plasmids pLAT(X)GFp+ and pLAT(X)GFP−. SD, splice donor site.
The 2.0-kb LAT is not capped or poladenylated (10, 39, 49) and is located 600 bp downstream of the major LAT promoter. Several lines of evidence now indicate that the 2.0-kb LAT is an intron. This RNA is circular (34, 54) and is excised both from the context of the mLAT (55) and from within the β-galactosidase (β-Gal) gene (13). In addition, consensus RNA processing sites flank the 2.0-kb LAT sequence (13, 41), are spliced together within processed LAT RNA (55), and are required for 2.0-kb LAT production in the context of the virus (1). RNA processing is also responsible for the removal of 500 bp from within the 2.0-kb LAT sequence and results in the production of the 1.5-kb LAT (1, 41). In contrast to the 2.0-kb LAT, which is detectable both in productive and latent infection, the 1.5-kb LAT is detectable only in latently infected neurons (39, 40, 50).
While the mLAT is present at low levels (56) and the spliced exons of this RNA have yet to be detected in HSV-1-infected cells, the steady-state levels of the 2.0-kb LAT are strikingly high during both productive and latent infection (11, 13). This abundance has been attributed to the unusual stability of the 2.0-kb LAT intron (34), which appears to result from an atypical branch point (53, 55) and/or from secondary structure at the 3′ terminus of the RNA (20). In latently infected cells, the 2.0-kb LAT is retained in the nucleus (39, 45, 46), while in productively infected or transfected tissue culture cells and in mouse brain stems, this RNA is also found in the cytoplasm (28, 55).
Some HSV-1 LAT-negative mutants display a reduced efficiency in the establishment or maintenance of latency (37) and/or a delayed reactivation phenotype in animal models (3, 16, 23, 29, 42, 48). Several mechanisms to account for the latency or reactivation function in molecular terms have been envisaged, for example, the antisense suppression of immediate-early gene expression (45) and an active role of the 2.0-kb LAT in suppression of macromolecular synthesis (28). Recently, the LAT gene has been demonstrated to encode an antiapoptosis function both in vitro and in vivo (30), which may contribute to the survival of infected neurons and hence may affect both establishment of latency and subsequent reactivation. The product of the LAT gene responsible for enhanced neuron survival and the mechanism by which it acts have yet to be addressed.
In addition to the possibility that the LAT RNA itself is active, it is thought that the HSV-1 LAT gene may encode a protein with a latency-related function, as has been demonstrated for bovine herpesvirus (38). Three open reading frames (ORFs) located within the HSV-1 2.0-kb LAT (Fig. 1a) have been extensively investigated. These ORFs are well conserved between HSV-1 strains 17, F, and KOS, although in the latter strain the larger two ORFs are fused (41, 49, 52; GenBank accession no. X14112). Recently, the largest of the 2.0-kb LAT ORFs has been placed directly under the control of a powerful cytomegalovirus (CMV) promoter and expressed from an ectopic site within the HSV-1 genome (47). A protein product of this ORF confers a significant growth advantage to the recombinant virus and is able to complement viruses deficient in immediate-early genes. Whether this ORF can be expressed from its normal context within the LAT gene remains unresolved. There has been one report of a LAT-encoded protein in latently infected primary neuron cultures (12), but it has not been confirmed. In this case the protein had an apparent molecular mass of 80 kDa, which far exceeds the capacity of any of the 2.0-kb LAT ORFs, but may reflect fusion of LAT ORFs as a result of an alternate splicing event. It remains possible that the large 2.0-kb LAT ORF is expressed at very low levels or in a highly regulated manner during a particular phase of the infection cycle.
In this work, we investigated the possibility that ORFs within the 2.0-kb LAT are translated by introducing the gene for the green fluorescent protein (GFP) directly into the 2.0-kb LAT sequence. In transient-transfection studies, we observed two types of GFP signal. The first, a weak cytoplasmic signal, is most likely produced by low-frequency translation initiation upon a LAT mRNA and occurs at a GFP AUG codon located far from the mRNA cap site. The second signal is localized to the nucleus and results from translation of alternately spliced LAT mRNAs encoding an exon 1 ORF-GFP fusion protein. The majority of this fusion protein is artificially expressed, as a result of the unintentional addition of an extraneous AUG initiation codon. However, residual expression can still be detected when this initiator is removed, albeit only in a subset of cells. Contrary to previous predictions (15), we have demonstrated that the excised 2.0-kb LAT intron does not act as a mRNA for the inserted GFP gene in transfected cells of either neuronal or nonneuronal origin. In contrast to our findings in vitro, insertion of the GFP gene into the 2.0-kb LAT region of HSV-1 does not result in GFP expression in either productively infected tissue culture cells or latently infected trigeminal ganglia (TGs). The implications of our results with respect to LAT ORF expression during the viral life cycle is discussed.
MATERIALS AND METHODS
Cell culture and viruses.
Cos-1, HeLa, and Vero cells were grown in Eagle's minimal essential medium supplemented with 5% calf serum at 37°C, and SY5Y neuroblastomas were grown in RPMI medium supplemented with 10% fetal calf serum. High-titer stocks of HSV-1 strain F and LAT-GFP recombinant viruses were produced by infection of Vero cells at a low multiplicity (0.1) followed by concentration from infected-cell lysates as previously described (8, 9). Kos-GFP (kindly provided by J. Baillet and P. A. Schaffer) is a Kos strain recombinant with a CMV promoter-GFP cassette inserted between viral genes UL26 and UL27.
Plasmid construction.
The plasmid pcDNA-Pst/Mlu (55) containing the PstI-MluI restriction fragment of the HSV-1 strain F LAT gene (Fig. 1) served as the parental construct from which all other clones were derived. A NheI-EcoRI GFP fragment from pEGFP-C1 (Clontech, Palo Alto, Calif.) was cloned directly using standard methods into the HpaI site of pcDNA-Pst/Mlu and a variant (pΔXcm) (20) containing an XcmI deletion (36) to produce plasmids pLAT(H)GFP+ and pLAT(X)GFP+, respectively. Subclones with the GFP gene inserted in an antisense direction [pLAT(H)GFP− and pLAT(X)GFP−] were also isolated. The cons mutation, which has been previously described (20) and contains a LAT branch point mutation, was subcloned from plasmid pCONS and inserted as a SalI-XbaI restriction fragment into pLAT(H)GFP+ and as a BbsI-EcoRI fragment into pLAT(X)GFP+. An NruI-HindIII fragment containing the CMV promoter was deleted from pLAT(H)GFP+ to generate the plasmid pΔCMV(H)GFP. All further manipulation was performed on the background of pLAT(X)GFP+. Plasmids pΔPA(X)GFP and pΔPB(X)GFP were obtained by deleting PpuMI-BsmI and PpuMI-AgeI restriction fragments from pLAT(X)GFP+, respectively. An SphI site, located in the pLAT(X)GFP+ polylinker region between the CMV promoter and the PstI site of the LAT gene insert and which contains an AUG codon, was removed by inserting an oligonucleotide adapter (5′AGCTTCGCGCTGCA3′) into the HindIII and PstI sites on either side of the SphI site to generate pΔSph(X)GFP. Plasmid pΔSph(X)FS+1 was produced from pΔSph(X)GFP by shifting the entire GFP gene one reading frame in a 5′-to-3′ direction with respect to the 1.5-kb splice acceptor. This was accomplished by linearizing pΔSph(X)GFP with AgeI, filling in the blunt ends with the Klenow fragment of DNA polymerase I, and religating. A second frameshift mutant [pΔSph(X)FS+2 (GFP gene shifted two reading frames in a 5′-to-3′ direction)] was produced by ligating an AgeI adapter into the AgeI site of pΔSph(X)GFP. The AgeI adapter consisted of annealed primers GFPF2-1 (5′CCGGAGGATCCGCTAGCGGATCC3′) and GFPF2-2 (5′CCGGGGATCCGCTAGCGGATCCT3′). All introduced mutations were verified by double-strand DNA sequencing using an ABI Prism automated sequencer.
Transfections and fluorescence microscopy.
Cell monolayers were grown directly on coverslips in six-well plates to 90% confluence, and plasmids were transfected overnight using Lipofectamine 2000 reagent (Gibco-BRL) as indicated by the manufacturer. Forty-eight hours following transfection or 24 h post-HSV-1 infection, cells were fixed with 4% paraformaldehyde. For direct observation of GFP fluorescence, monolayers were washed with phosphate-buffered saline, mounted with Fluoromount-G (Southern Biotechnology Associates), excited with a krypton-argon laser, and visualized using a Nikon Microphot FXA microscope and fluorescein isothiocyanate filters. For immunofluorescence studies, fixed monolayers were incubated twice at room temperature for 5 min each in 100 mM glycine, permeabilized for 2 min with 0.1% Nonidet P-40, and then blocked for 30 min in 3% bovine serum albumin–0.1% Nonidet P-40. Anti-Rev ascitic fluid (courtesy of Michael Malim, University of Pennsylvania) or anti-HSV-1 polyclonal antibody (American Qualex Antibodies, San Clemente, Calif.) was diluted appropriately in blocking buffer and applied to monolayers for 1 h at room temperature. Coverslips were washed three times and incubated in Texas Red-conjugated anti-mouse immunoglobulin G (IgG) (Amersham) or rhodamine-conjugated anti-rabbit IgG (Chemicon) for 30 min at room temperature. After three final washes, coverslips were mounted and observed as described above for GFP fluorescence, except standard rhodamine filters were employed.
RNA analysis.
Total RNA extracts of transfected cells and snap-frozen TGs were made using Trizol reagent (GIBCO-BRL) according to the manufacturer's instructions. Northern analysis was performed as previously described (55) with minor modifications. Following DNase treatment and phenol-chloroform extraction, 10-μg aliquots of glyoxylated RNA were resolved on 1.2% agarose gels, vacuum blotted to nylon membranes (GeneScreen Plus; NEN), and UV cross-linked. To synthesize probes, various DNA restriction fragments were isolated from pcDNA-Pst/Mlu and 32P radiolabeled using a RadPrime DNA labeling system (LTI). Heat-denatured probes were hybridized to immobilized RNAs overnight, and blots were washed six times at 65°C as detailed previously (55) followed by autoradiography. For PCR analysis, 1 μg of DNase-treated RNA was reverse transcribed using primers d(N)6 and/or oligo(dT) according to the instructions supplied with the Superscript preamplification system (Gibco-BRL). Following RNase H treatment, 10% of the cDNA reaction mixture was PCR amplified with primers exon 1 (5′TATCGGTACCGCTCCATCGCCTTTCTGT3′) and GFP5P (5′ATAGTCTAGACTTGTGCCCCAGGATGTGC3′). The amplification products were subcloned into pBluescript (Stratagene) and sequenced on an automated sequencer (ABI) using a T7 promoter primer.
Protein analysis.
Total protein extracts were generated by scraping cell monolayers into radioimmunoprecipitation assay (RIPA) buffer (0.15 M NaCl, 10 mM Tris [pH 7.4], 1% sodium deoxycholate, 1% Triton X-100, 0.1% sodium dodecyl sulfate). Extracts were freeze-thawed three times and clarified at 600 × g for 10 min, and residual genomic DNA was broken up by passing through a 30-gauge needle. Fifty-microgram aliquots of protein were resolved by sodium dodecyl sulfate–11% polyacrylamide gel electrophoresis and transferred to Immobilon P membranes (Millipore). Blots were blocked in 5% dried milk and probed with a polyclonal GFP antibody (Clontech), and bands were visualized by enhanced chemiluminescence (Amersham).
Recombinant virus production and viral DNA analysis.
LAT-GFP recombinant viruses were obtained by cotransfection of purified viral F strain DNA and the plasmid pΔCMV(H)GFP. The screening procedure for GFP-positive plaques has been described previously (19). Briefly, plaques were transferred from agarose overlays to nitrocellulose filters, denatured, and then screened by plaque hybridization using a random-primed 32P-labeled GFP probe. Positive plaques were rescreened by this technique (five rounds) until all plaques were GFP positive. The correct insertion of GFP in the final isolates was verified by Southern blot analysis. Total infected-cell DNA was obtained by standard methods (36), restricted with AgeI and/or BstEII and separated on 1% agarose gels. DNA was vacuum transferred under alkaline conditions (1.0 N NaOH) to GeneScreen Plus nylon membrane (NEN), followed by hybridization to a 32P-labeled 2.0-kb LAT-specific probe. High-stringency washes were performed as detailed by the membrane manufacturer, and bands were visualized by autoradiography.
Animal infections and tissue analysis.
BALB/c mice were infected bilaterally via an ocular route with 106 PFU of various virus stocks per scarified eye. At 4 days pastinfection (PI) (acute) or 40 to 50 days PI (latent), TGs were excised. For positive controls, BALB/c mice were infected intracranially with 2 × 106 PFU of Kos-GFP. At 1 day PI (acute), the brains were excised. Ganglia were immediately snap-frozen in liquid nitrogen for RNA analysis, while for fluorescence microscopy and immunohistochemistry, the animals were perfused with 4% paraformaldehyde, and the TGs and brains were removed and incubated in 4% sucrose for 2 h, transferred to 20% sucrose overnight, frozen in OCT (Tissue-Tek), and then cryostat sectioned. Sections were observed directly using long-pass emission filters (catalog no. 41018; Chroma). The standard methods for tissue processing and light microscopic immunohistochemical analysis were used. Antibodies used were anti-HSV-1 polyclonal rabbit antibody (American Qualex Antibodies) and anti-GFP A-V peptide rabbit antibody (Clontech). Cells were detected by an indirect avidin-biotin immunoperoxidase method (Vectastain ABC kit; Vector Labs, Burlingame, Calif.) as specified by the manufacturer with slight modifications. Briefly, tissue sections were rehydrated, quenched in peroxide (H2O2), and blocked in 3.5% goat serum (Sigma Chemical Co., St. Louis, Mo.). Tissue sections were incubated with the primary antibody (1:10). Next, the tissues were incubated with biotinylated goat anti-rabbit IgG, the avidin-biotin horseradish peroxidase complex, and finally Vector red (Vector Labs) as the chromagen. Sections were counterstained with hematoxylin and examined under the light microscope. Sections were washed twice with 0.01 M Tris-HCl (pH 7.9) and: then twice with 0.01 M Tris-HCl containing 5% goat serum between every step except for the chromagen addition step when sections were washed twice with 0.01 M Tris-HCl only. As an additional control for the specificity of immunostaining, tissues were processed as described above, except that nonimmune goat serum was substituted for the primary antibody. Quantification of positive stained cells was done using the Phase 3 Imaging program (Phase 3 Imaging, Glen Mills, Pa.).
RESULTS
In vitro system for assessing protein expression from within the HSV-1 2.0-kb LAT.
Previously we described a LAT minigene expression vector containing a PstI-MluI fragment of the mLAT gene (Fig. 1b) (55). Transcripts originating from the CMV promoter of this vector are efficiently spliced in cultured cells to yield both the 2.0-kb LAT intron and spliced LAT exons (55). We have utilized this transient-transfection system to ask whether the 2.0-kb LAT can function as a mRNA for a reporter protein sequence placed within it and to explore alternate expression strategies for this protein. In this way we hoped to throw light upon expression strategies for endogenous 2.0-kb LAT ORFs. A GFP gene was placed directly into the 2.0-kb LAT sequence of the minigene construct such that the GFP AUG initiation codon was in the vicinity of the initiation codons of the two major 2.0-kb LAT ORFs. Specifically, the plasmids pLAT(H)GFP+ and pLAT(H)GFP− (Fig. 1b) were created by inserting GFP (EGFP; Clontech) in both orientations into the 2.0-kb LAT region of pcDNA-Pst/Mlu at a HpaI restriction site just downstream of the 1.5-kb LAT splice acceptor and upstream of two conserved 2.0-kb LAT ORFs (41). To minimize the size difference between the 2.0-kb LAT intron bearing GFP and the wild-type 2.0-kb LAT intron, a second set of constructs [pLAT(X)GFP+ and pLAT(X)GFP− (Fig. 1b)] were created by deleting an 805-bp region between two XcmI sites within the pLAT(H)GFP plasmids. The resulting expression vectors were transfected into both nonneuronal (Cos-1) and neuronal (SY5Y) cells where GFP fluorescence was recorded.
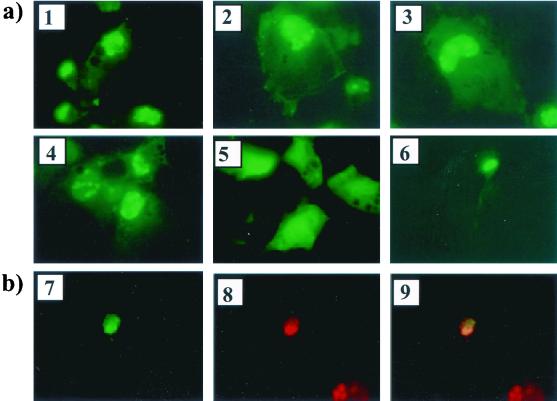
Two distinct patterns of fluorescence are detected when LAT-GFP expression vectors are transfected into cultured cells. Transient transfection of the plasmid pEGFP-C1 (Clontech), which contains the GFP gene under the control of the CMV promoter and which has no LAT sequences present, resulted in a typical pattern of GFP fluorescence over the whole cell (Fig. 2a, panel 5). In contrast, transient transfection of either one of the 2.0-kb LAT-GFP expression vectors pLAT(H)GFP+ and pLAT(X)GFP+ resulted in two different patterns of fluorescence within the same cell. A weak GFP fluorescent signal was clearly detectable throughout the cytoplasm of both SY5Y, a cell line of neuronal origin and nonneuronal Cos-1 cells, transfected with either LAT-GFP vector (Fig. 2a, panels 1, 3, and 6). A second more noticeable fluorescence pattern in which GFP accumulated in the nuclei of both SY5Y and Cos-1 cells (panels 1, 3 and 6) was also observed. Moreover, the GFP fluorescence appeared to be localized to specific areas within the nucleus which were reminiscent of nucleoli. To determine if the punctate nuclear GFP signal observed did in fact reflect nucleolar targeting of GFP, the vector pLAT(X)GFP+ and vector pcREV, which expresses the human immunodeficiency virus type 1 (HIV-1) Rev protein known to localize specifically to the nucleolus, were cotransfected. GFP colocalized with Rev in this experiment (Fig. 2b, panels 7 to 9), demonstrating that LAT sequences present in the LAT-GFP expression vectors are capable of targeting GFP to the nucleolus. Such targeting is not a function of the conserved 2.0-kb LAT ORFs 1 and 2 (Fig. 1a), since with the exception of 16 residues at the carboxy terminus of ORF 1, these ORFs are absent in the plasmid pLAT(X)GFP.
FIG. 2.
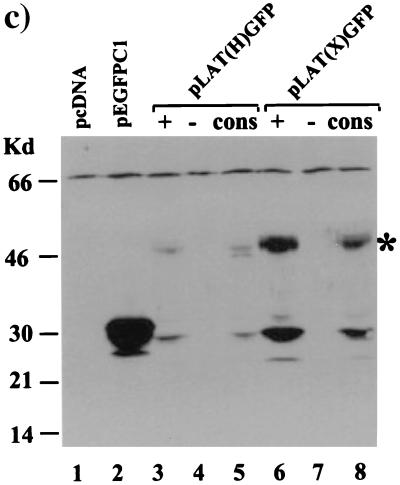
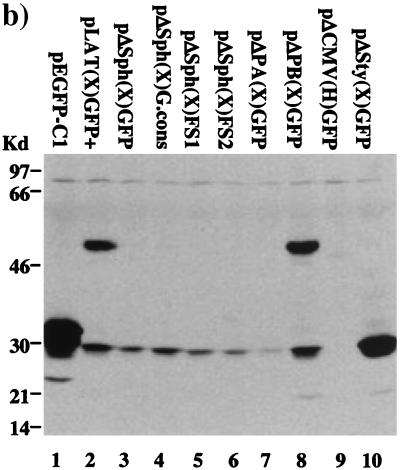
GFP fluorescence in LAT-GFP vector-transfected cells. (a) Expression plasmids pLAT(H)GFP+ (panel 1), pLAT(X)GFP+ (panel 3), and pEGFP-Cl (panel 5) were transfected into Cos-1 cells, and GFP fluorescence images were captured at a magnification of ×45. GFP expression from 2.0-kb LAT-GFP plasmids containing a conserved intron branch point [pLAT(H)G.cons (panel 2) and pLAT(X)G.cons (panel 4)] was also analyzed. GFP fluorescence from pLAT(X)GFP+ in SY5Y, a cell line of neuronal origin, is shown in panel 6. (b) Plasmids expressing GFP from 2.0-kb LAT sequences [pLAT(X)GFP+] and the HIV-1 Rev protein (pcREV) were cotransfected into Cos-1 cells. GFP fluorescence (panel 7) and Rev expression (panel 8) from cotransfected cells are shown, and the images are superimposed in panel 9. The photographs in panel b were taken at a lower resolution than those in panel a in order to demonstrate colocalization in the nucleus; hence, the weak cytoplasmic GFP signal from pLAT(X)GFP+ cannot be seen. (c) Whole-cell extracts from Cos-1 cells transfected with 2.0-kb LAT-GFP expression vectors were analyzed on Western blots using a polyclonal anti-GFP antibody (Clontech). The plasmids transfected are indicated above the lanes, with GFP oriented in the sense (+) and antisense (−) directions or with a consensus branch point (cons), and a band representing the 48-kDa LAT-GFP fusion protein (∗) is marked.
A LAT-GFP fusion protein is produced from 2.0-kb LAT-GFP expression vectors.
One explanation for the nuclear localization pattern conferred upon GFP by the surrounding LAT sequences was that a LAT-encoded polypeptide bearing a nuclear localization signal was fused to the reporter protein. The other signal observed in LAT-GFP-transfected cells, a weak cytoplasmic fluorescence, probably reflected either low-frequency GFP translation or incomplete localization of the fusion protein to the nuclear compartment. To examine these possibilities further, protein extracts from cells transfected with LAT-GFP expression vectors were separated on polyacrylamide gels and subjected to Western blot analysis using a polyclonal antibody specific for GFP (Clontech). As shown in Fig. 2c, extracts from cells transiently transfected with pEGFP-C1 express high levels of a 32-kDa protein, corresponding to full-length GFP (lane 2). In contrast, a 48-kDa protein is detected in extracts from cells transfected with pLAT(H)GFP+ and pLAT(X)GFP+ (lanes 3 and 6), albeit at lower levels. A second polypeptide (30 kDa) is also detected in these extracts and may represent an independently initiated protein. These data suggest that the 48-kDa protein is a fusion between GFP and a LAT polypeptide, which directs GFP to the nucleolus, while the 30-kDa protein is responsible for the observed cytoplasmic GFP signal.
Alternately spliced transcripts are found in cells transfected with LAT-GFP expression vectors.
We were interested in identifying the LAT polypeptide fused to GFP, since it contained a signal responsible for the accumulation of GFP within the nucleolus and might have significance for both the biology and latency of HSV-1. The conserved 2.0-kb LAT ORFs lying downstream of the GFP insertion point are not candidates for the LAT-encoded polypeptide fused to GFP, since their deletion in the expression vector pLAT(X)GFP+ has no effect on the nucleolar accumulation of GFP observed in cells transfected with this plasmid. A third 2.0-kb LAT ORF (ORF 3 [Fig. 1b]) lies partially upstream and is in frame with the GFP ORF inserted into both LAT-GFP expression vectors. However, it is unlikely that the 48-kDa protein results from a fusion between this ORF and GFP, since this fusion would produce a protein with a lower expected molecular mass (38 kDa).
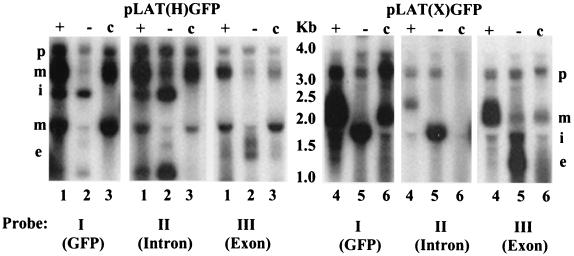
Another way in which a fusion between a LAT ORF and GFP may have occurred is through an RNA splicing event other than that responsible for the excision of the 2.0-kb LAT intron. This possibility was addressed by Northern blot analysis of total cell RNA prepared from Cos-1 cells, transiently transfected with various LAT-GFP expression vectors (Fig. 3). Probes specific for GFP (probe I), the 2.0-kb LAT intron (probe II), and exon 1 of the LAT gene (probe III) were used. Cells transfected with pLAT(H)GFP+ and pLAT(H)GFP− both produced a 2.7-kb transcript (labeled i in Fig. 3, lanes 1 and 2), the predicted size of the 2.0-kb LAT-GFP intron, which contains both GFP and 2.0-kb LAT sequences (Fig. 3, probes I and II, lanes 1 and 2). This 2.7-kb transcript was shown to be the excised LAT-GFP intron, since it failed to hybridize to exon-specific probe III (lanes 1 and 2). Novel transcripts of 3.0 kb, 1.9 kb [pLAT(H)GFP+], and 2.2 kb [pLAT(X)GFP+] were also detected on Northern blots (Fig. 3, probes I, II, and III, lanes 1 and 4, labeled m). Interestingly, these transcripts were absent (lane 5) or markedly less abundant (lane 2) when GFP was inserted into the 2.0-kb LAT in the reverse orientation. The disappearance of these transcripts coincided with increases in the levels of the 2.0-kb LAT intron and spliced LAT exon RNA and suggested that certain sequence elements within the GFP gene can influence the choice of splice sites, resulting in the production of novel, alternately spliced transcripts. This scenario is consistent with the previously published observation that the orientation of phage lambda DNA inserted at the HpaI site within the 2.0-kb LAT region strongly affected splicing of the primary LAT transcript (20).
FIG. 3.
Northern blot analysis of LAT-GFP RNA. Total transfected Cos-1 cell RNA was blotted to a nylon membrane and probed sequentially with each of three probes. The probes used were a 0.65-kb NheI-EcoRI GFP fragment (probe I), a 1.0-kb BstEII fragment specific for the 2.0-kb LAT (probe II), and a 0.37-kb StyI fragment specific for LAT exon 1) (probe III). The positions of precursor RNAs (p), introns (i), spliced exons (e), and novel transcripts (m) are indicated. The plasmids transfected were pLAT(H)GFP and pLAT(X)GFP with GFP oriented in the sense (+) and antisense (−) directions or pLAT(H)GFP+ and pLAT(X)GFP+ bearing a consensus intron branch point (c).
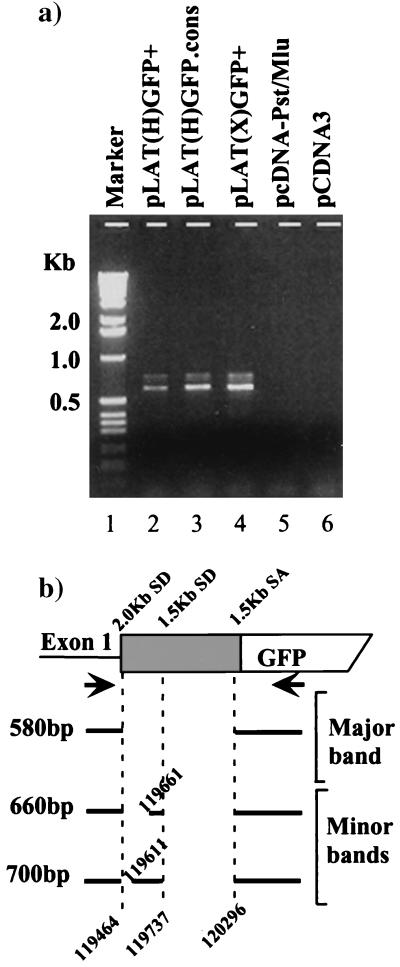
In order to verify that alternate splicing was occurring and to identify the splice donor and acceptor sites utilized, DNase-treated total RNA from cells transfected with LAT-GFP expression vectors was reverse transcribed using oligo(dT) as the primer. cDNAs were PCR amplified using primers exon 1 and GFP5P, annealing upstream of the 2.0-kb LAT splice donor and within the GFP gene, respectively (Fig. 4a). A major band of 580 bp and two slightly slower-migrating, less-abundant bands were amplified from cells transfected with pLAT(H)GFP+ and pLAT(X)GFP+ (lanes 2 and 4). These bands were specific for LAT-GFP transcripts, since they were not amplified from RNA from cells transfected with either pcDNA-Pst/Mlu or an unrelated plasmid (lanes 5 and 6). The reverse transcription-PCR (RT-PCR) products were subcloned and sequenced, and a schematic representation of the sequencing results is shown in Fig. 4b. The major RT-PCR product represents a spliced transcript with an intron excised between the 2.0-kb LAT splice donor site (nucleotides [nt] 119464) and the 1.5-kb LAT splice acceptor site (nt 120296). The minor RT-PCR products detected involve splicing both between the 1.5-kb LAT splice donor (nt 119737) and acceptor (nt 120296) sites and between the 2.0-kb LAT splice donor and two other splice acceptor sites (nt 119611 and 119661). Notably, splicing between the 1.5-kb LAT splice donor and acceptor occurs in nonneuronal cells, whereas previously these splice sites have been characterized as “neuron specific” (41). In summary, alternate splicing, predominantly between the 2.0-kb LAT splice donor and the 1.5-kb LAT splice acceptor, is abundant in the transient-expression system and is influenced by the orientation in which GFP is inserted into the 2.0-kb LAT intron. The alternately spliced transcripts produced are mRNA candidates for the LAT-GFP fusion protein.
FIG. 4.
RT-PCR mapping of LAT-GFP RNA splice junctions. (a) Total transfected cell RNA was reverse transcribed and PCR amplified using primers exon 1 and GFP5P. RT-PCR products from cells transfected with LAT-GFP expression plasmids, as indicated above the lanes, were resolved on a 1% agarose gel. (b) Splicing patterns of transfected cell RNAs as determined by sequencing RT-PCR products. A truncated LAT gene containing the GFP coding sequence is represented by shaded and open boxes, respectively. The positions of 2.0- and 1.5-kb LAT splice acceptor and donor sites (SD and SA) and of the amplification primers exon 1 and GFP5P (arrows) are also shown. Spliced exons are represented by solid lines, and excised introns are shown as gaps. The corresponding HSV-1 genome positions are indicated (in nucleotides).
Nucleolar GFP expression is mediated by alternate mRNA splicing.
To assess whether the GFP fluorescence and polypeptides observed in LAT-GFP-transfected cells resulted from alternate splicing involving the 1.5-kb LAT splice acceptor site, a pair of deletion mutants [pΔPA(X)GFP and pΔPB(X)GFP] were generated from pLAT(X)GFP+ (Fig. 5a). In pΔPA(X)GFP, the 5′ half of the 2.0-kb LAT is deleted, including the 1.5-kb LAT splice acceptor, while the deletion in pΔPB(X)GFP overlaps with that in pΔPA(X)GFP but does not include the splice acceptor site. Transfection of pΔPB(X)GFP into Cos-1 cells gives a pattern of nuclear fluorescence identical to that seen upon transfection of pLAT(X)GFP+ (Fig. 5a, compare panels 1 and 7) and leads to the production of both the 48- and 30-kDa proteins typically seen in cells transfected with pLAT(X)GFP+ (Fig. 5b, compare lanes 2 and 8). In contrast, when pΔPA(X)GFP was transfected into Cos-1 cells, nucleolar GFP expression was not detectable by fluorescence microscopy (Fig. 5a, panel 6) and the 48-kDa protein was not seen on Western blots (Fig. 5b, lane 7). Detection of the weak cytoplasmic GFP signal was unaffected by this mutation, although a decrease in the level of the 30-kDa protein was observed (Fig. 5a, panel 6, and b, lane 7). Taken together, these results demonstrate that alternate splicing involving the 1.5-kb LAT splice acceptor site is critical for expression of the 48-kDa LAT-GFP fusion protein and for nucleolar localization of GFP, but not for 30-kDa GFP polypeptide expression or for the weak cytoplasmic GFP fluorescence pattern. Since we have shown that GFP sequences affect alternate splicing (Fig. 3, lanes 1 and 2), it should be noted that the production of the 48-kDa protein through alternate splicing may be an artificial outcome. The 30-kDa protein is apparently a weakly expressed unfused GFP polypeptide.
FIG. 5.
Characterization of LAT-GFP expression products. (a) Various deletions and mutations were introduced onto the background of pLAT(X)GFP+ as described in Materials and Methods and indicated in the figure. The SphI deletion [pΔSph(X)GFP] removes an artificially introduced AUG initiation codon in the vector polylinker, while the cons mutation (solid circle) [pΔSph(X)G.cons] destabilizes the 2.0-kb LAT-GFP intron. In constructs pΔSph(X)FS+1 and pΔSph(X)FS+2, the GFP gene is shifted one and two reading frames in a 5′-to-3′ direction (arrows). The PpuMI-AgeI deletion [pΔPA(X)GFP] deletes both the 1.5-kb LAT splice acceptor site (SA) and a leader sequence lying between the 2.0-kb LAT splice donor site (SD) and the 1.5-kb LAT splice acceptor, while the PpuMI-BsmI deletion [pΔPB(X)GFP] deletes only the leader. The CMV promoter is removed from the vector pLAT(H)GFP+ to produce the pΔCMV(H)GFP plasmid, while a 370-bp StyI [pΔSty(X)GFP] deletion removes two-thirds of LAT exon 1. The fluorescence patterns shown upon transfection of Cos-1 cells are shown to the right of the schematics. (b) Western blot analysis of cells transfected with various mutant constructs as indicated above the lanes. The blot was probed with a polyclonal antibody specific for GFP.
An artificially initiated ORF in LAT exon 1 is fused to GFP and contains a nuclear localization signal.
The sequences of the RT-PCR products and the requirement of the 1.5-kb LAT splice acceptor site for the production of the 48-kDa protein strongly suggested that this protein was produced by splicing of an ORF in LAT exon 1 to GFP. In support of this theory, deletion of the StyI fragment from pLAT(X)GFP+ which includes 370 bases of the 600-bp exon 1 [pΔSty(X)GFP (Fig. 5a)] resulted in the loss of the 48-kDa protein (Fig. 5b, lane 10) and of the nucleolar GFP expression pattern (Fig. 5a, panel 9). This result demonstrates not only that an exon 1 LAT ORF is fused to GFP but also that this ORF contains a nuclear localization signal. Interestingly, GFP is strongly expressed from pΔSty(X)GFP as a 30-kDa protein (Fig. 5b, lane 10) that localizes in a wild-type pattern throughout the cell (Fig. 5a, panel 9). The data suggest that removal of the StyI fragment from exon 1 enhances GFP translation, possibly by moving the GFP ORF closer to the CMV promoter or by deleting a sequence inhibitory to translation. Shifting the entire GFP coding region one or two reading frames [pΔSph(X)FS-1 and -FS-2] in a 3′ direction with respect to the 1.5-kb splice acceptor also disrupted nucleolar localization (Fig. 5a, panels 4 and 5) but had no effect on either the weak cytoplasmic fluorescence pattern or expression of the 30-kDa GFP polypeptideide (Fig. 5b, lanes 5 and 6). Since the frameshift mutations were made on the background of pLAT(X)GFP where the 2.0-kb LAT ORFs are deleted, they specifically address fusions with LAT ORFs upstream of the GFP insertion site. Thus, together with the pΔSty(X)GFP result, the frameshift mutations confirm that GFP is fused to an ORF in LAT exon 1. In addition, it appears that other ORFs lying upstream of GFP in different reading frames are not fused to GFP, either directly or by alternate splicing, since no new polypeptides were detected on Western blots (Fig. 5b, lanes 5 and 6).
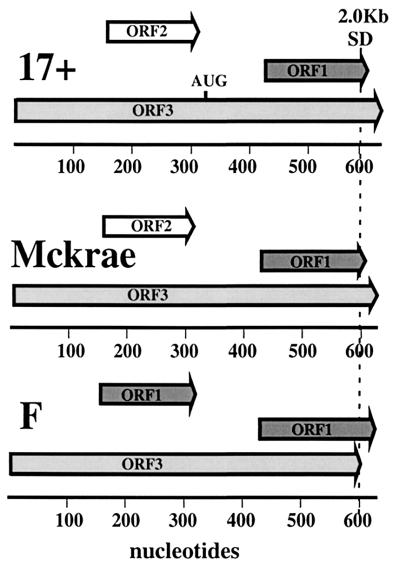
Analysis of the LAT exon 1 sequence (Fig. 6) revealed that only a single ORF in reading frame 3 spanning the entire length of the exon was sufficient in length to account for the 16- to 18-kDa sequence fused to GFP to create the 48-kDa protein. In the HSV-1 strain F sequence, this ORF contains no in-frame AUG initiation codons downstream of the transcriptional start site, although putative non-AUG initiation codons are present. In our expression vectors, an AUG codon in the same reading frame as exon 1 ORF 3 was present in the SphI restriction site of the polylinker. Deletion of this SphI site [pΔSph(X)GFP] led to a loss of the LAT-GFP fusion protein expression, as determined by Western blotting (Fig. 5b, lane 3). Interestingly, however, although nucleolar fluorescence was absent in most cells, a small percentage of cells still exhibited this phenotype (Fig. 5a, panel 2). We conclude therefore that ORF 3 in LAT exon 1 is fused to GFP and contains a nuclear localization signal. However, the translation of this fusion protein is artificially initiated due to the unintentional addition of an AUG initiator codon. In the absence of this initiator, very low-level expression occurs, detectable only by fluorescence microscopy in a minor subset of cells, in which permissive conditions appear to exist. This expression is most likely mediated by non-AUG translational initiation. The identity of the non-AUG initiation codon and the conditions required for its use are not known.
FIG. 6.
LAT exon 1 ORFs. Schematic ORF analysis for bases 1 to 650 of the LAT gene of HSV-1 strains 17+ (GenBank accession no. X14112), McKrae (GenBank accession no. AF041142), and F (M. Lock and N. W. Fraser, unpublished data). The 5′-terminal base of the PstI site is designated the first base of exon 1 due to a lack of sequence information upstream of this site for strain F. ORFs are shown as arrows and are numbered accordingly. The AUG codon that is present in ORF 3 of strain 17+, but not in strains McKrae and F, is indicated. The position of the 2.0-kb LAT splice donor site (SD) is shown.
The 2.0-kb LAT intron does not function as a mRNA for GFP expression in vitro.
Our transfection experiments indicated that expression of GFP occurs from both 2.0-kb LAT-GFP vectors where GFP was inserted in the sense direction. We demonstrated above that nucleolar fluorescence resulted from translation of an alternately spliced mRNA. However, it remained unclear whether the spliced intron was functioning as a mRNA for the 30-kDa protein and consequently the weak cytoplasmic fluorescence or whether a polyadenylated RNA was responsible. To distinguish between these possibilities, a previously described mutation cons, which changes the 2.0-kb LAT intron branch point into a consensus branch point and destabilizes the 2.0-kb LAT RNA (20), was introduced into both LAT-GFP expression vectors to yield pLAT(H)G.cons and pLAT(X)G.cons. Analysis of RNA from transfected cells demonstrates that as expected, the cons mutation destabilizes the LAT-GFP intron produced from pLAT(H)G.cons, since the 2.7-kb RNA present in cells transfected with LAT(H)GFP+ was absent in cells transfected with pLAT(H)G.cons (Fig. 3, probes I and II, compare lanes 1 and 2). However, loss of the LAT-GFP intron does not affect the GFP fluorescence patterns seen in cells transfected with pLAT(H)GFP+ (Fig. 2a, panel 2, and 5a, panel 3) or production of either the 48-kDa fusion protein or the 30-kDa polypeptide (Fig. 2c, lane 4, and 5b, lane 4). Furthermore, following transfection of pLAT(X)GFP+, both types of GFP fluorescence and both the 48- and 30-kDa GFP proteins were observed when the GFP gene was present in the sense orientation (Fig. 2a, panel 3, and c, lane 6). In this orientation, the production of the LAT-GFP intron is inhibited, since there was negligible hybridization to an intron-specific probe at the expected size for the LAT-GFP intron (1.9 kb) (Fig. 3a, probe III, lane 4). Thus, as is the case for pLAT(H)GFP+, neither nucleolar nor cytoplasmic GFP expression observed upon transfection of pLAT(X)GFP+ originates from an excised 2.0-kb LAT-GFP intron.
Another possibility is that the RNA responsible for ORF expression is a mRNA initiated at the promoter mapped just upstream of the 2.0-kb LAT splice donor site (LAP2) (Fig. 1) (14). However, a construct in which the CMV promoter was removed but in which LAP2 was left unaltered [pΔCMV(H).GFP] (Fig. 1) failed to express both GFP, as measured by fluorescence microscopy and Western blot analysis (Fig. 5a, panel 8, and b, lane 9) and LAT-specific transcripts (not shown) in transiently transfected cells. Clearly, in the transient-transfection system described, LAP2 is not operational in isolation and may require cooperation with LAP1 or other viral factors for activity. Taking all our data into account, we conclude that the 30-kDa GFP protein observed in transfected cells is translated from CMV promoter-driven mRNAs and not the LAT-GFP intron.
GFP is not expressed from a 2.0-kb LAT-GFP virus during either productive or latent infection.
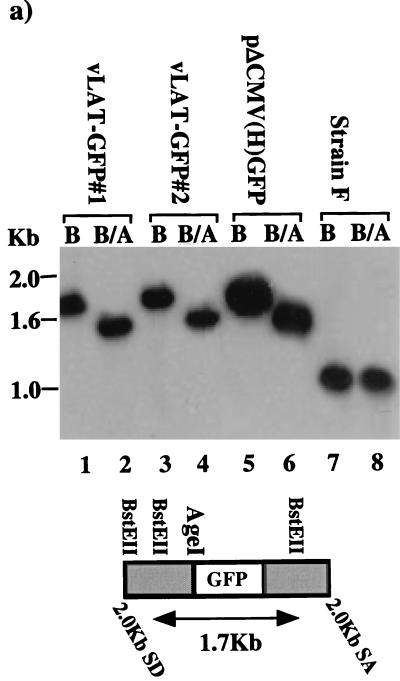
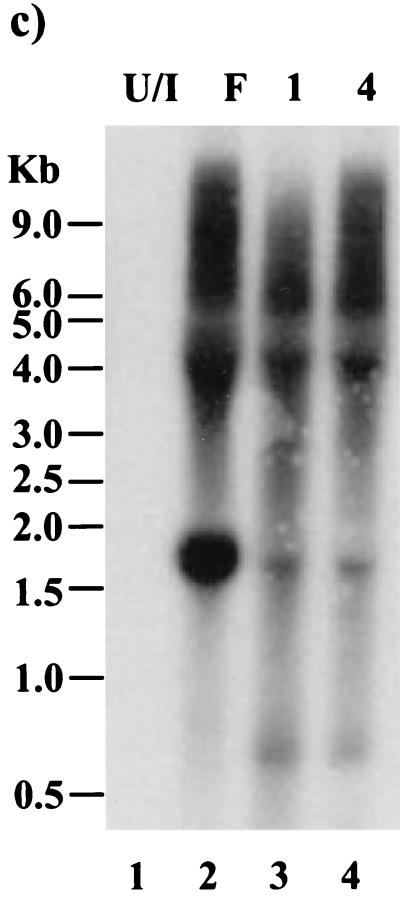
Our finding that GFP can be expressed from within the 2.0-kb LAT DNA sequence suggested that under optimal conditions, endogenous 2.0-kb LAT ORFs might also be expressed from viral LAT transcripts. To address this possibility, a recombinant virus, vLAT-GFP, was produced by cotransfection of the plasmid pΔCMV(H)GFP and parental F strain DNA, followed by several rounds of plaque purification as described in Materials and Methods. This process also has the effect of removing the SphI site which encodes the artificial AUG codon at the amino terminus of exon 1 ORF 3, since it is deleted in the pΔCMV(H)GFP construct and lies outside the region of homology between the transfer vector and the virus. The correct insertion of GFP into the viral LAT gene was determined by Southern blot analysis of vLAT-GFP DNA. A LAT-specific BstEII band of 1.7 kb (Fig. 7a, lanes 1 and 3) was observed with both isolates of the vLAT-GFP virus, while the parental F strain gave only a 1.0-kb band (lane 7). This mobility shift and the introduction of an AgeI site (compare lanes 2 and 5) confirm that GFP is present in the 2.0-kb LAT locus of the recombinant viruses. However, when LAT-GFP virus-infected cells were observed for fluorescence, GFP expression could not be detected in either fibroblasts (Fig. 7b, panels 7 and 8) or an SY5Y neuroblastoma cell line (not shown), despite high-level expression of other HSV-1 antigens (panels 3 and 4). Confirmation that the viruses were altered with respect to LAT expression was obtained by Northern analysis of infected-cell RNA (Fig. 7c). A band of 2.7 kb representing the LAT-GFP intron could not be detected in RNA from Cos-1 cells infected with either LAT-GFP virus isolate using an intron-specific probe (lanes 3 and 4). A minor band of 1.9 kb was seen in LAT-GFP virus-infected cells migrating alongside the F strain 2.0-kb LAT intron (lane 2). The 1.9-kb RNA is also detected by an exon 1-specific probe (data not shown) and is possibly the same alternately spliced RNA detected previously in Cos-1 cells transfected with pLAT(H)GFP+ (Fig. 3, lanes 1 and 3). The presence of alternately spliced exon 1-GFP transcripts in LAT-GFP virus-infected cells was assessed by RT-PCR analysis, as outlined above for transiently transfected cells (Fig. 4), and the results are shown in Fig. 7d. The 580-bp major band was amplified from LAT-GFP virus-infected Cos-1 cells (lanes 4 and 5) and from TGs excised on day 5 (lanes 8 and 9) and day 40 (lanes 12 and 13) PI with LAT-GFP virus isolates. These time points represent acute infection/establishment of latency and latent infection, respectively. In addition, at higher amplification levels, the faster-migrating minor bands previously amplified from transiently transfected cells were also observed (lanes 4, 5, and 13). Thus, alternate splicing of LAT transcripts involving the 2.0-kb splice donor and the 1.5-kb splice acceptor is occurring in cells both productively and latently infected with LAT-GFP viruses, while production of a 2.7-kb LAT-GFP intron (involving 2.0-kb LAT splice donor and splice acceptor sites) is apparently suppressed.
FIG. 7.
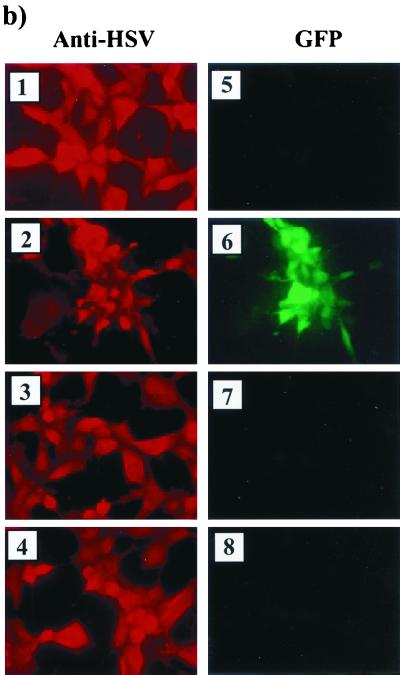
Construction and analysis of a LAT-GFP virus. (a) Southern blot of LAT-GFP recombinant virus DNA. Viral DNA was isolated from infected cells and digested with either BstEII (B) alone or BstEII and AgeI (B/A). Two isolates of the plaque-purified recombinant virus (lanes 1 to 4) were compared with the donor plasmid (lanes 5 and 6) and with the parental virus strain (lanes 7 and 8). The blot was hybridized with a 1.0-kb BstEII LAT probe, which detects a 0.7-kb increase in the size of the BstEII viral fragment (1.0 kb) due to the insertion of GFP. SA and SD, LAT splice acceptor and splice donor sites, respectively. (b) Fluorescence analysis of vLAT-GFP-infected cells. Cells infected with F strain (panels 1 and 5) or Kos-GFP (panels 2 and 6) or with two isolates of the LAT-GFP virus (panels 3 and 7 and panels 4 and 8) were examined both by immunofluorescence using an anti-HSV-1 polyclonal antibody and directly for GFP expression. (c) Northern blot analysis of total RNA from Cos-1 cells either uninfected (U/I) or infected with strain F or LAT-GFP viral isolates (isolates 1 and 4) as indicated. The blot was hybridized with a 1.0-kb BstEII LAT probe. (d) RT-PCR analysis of total RNA isolated from infected Cos-1 cells and from both productively infected (day 5) and latently infected (day 40) TGs. Cells or animals were either left uninfected (U) or infected with HSV-1 strain F (F) or with two isolates of the LAT-GFP virus (isolates 1 and 4) as indicated above the lanes. The reverse transcription primer for Cos-1 and day 5 TG was oligo(dT), and for latently infected TGs, it was a mixture of oligo(dT) and p(N)6. PCR was performed with primers exon 1 and GFP5P as indicated in Fig. 4. Lane M contains molecular size markers. (e) Immunohistochemistry and fluorescence of acutely infected LAT-GFP and KOS-GFP tissue. Animals were infected and sacrificed at day 5 (LAT-GFP [panels 1, 2, 4, and 5]) or day 1 (Kos-GFP [panels 3 and 6]). Immunohistochemistry sections were stained with both anti-HSV (panel 1) and anti-GFP antibodies (panel 4), and images were captured at a magnification of ×7. LAT-GFP rhodamine (anti-HSV) and GFP fluorescence images were captured at a magnification of ×14 (panels 2 and 5), while KOS-GFP rhodamine (anti-HSV) and GFP fluorescence images were captured at a magnification of ×32 (panels 3 and 6).
Since TGs both acutely and latently infected with the LAT-GFP virus produced alternately spliced transcripts, we were interested to know if the specific environment of productively or latently infected TGs might permit GFP expression. Frozen tissue sections were examined directly for GFP fluorescence with long-pass GFP filters, which have previously been used to discriminate between background fluorescence from the tissue section and a true GFP signal (31). Employing both this technique and GFP antibody-based immunohistochemistry, we were unable to detect GFP expression in TGs harvested at day 5 (Fig. 7e) or day 40 PI or at 24 h postreactivation (data not shown). Therefore, despite expression of LAT transcripts, which in a transient-transfection system mediate GFP expression, GFP is not expressed from the 2.0-kb LAT locus in the context of the virus at any stage of infection of peripheral nervous system tissue.
DISCUSSION
Expression of the LAT gene in infected cells is a highly regulated process, involving a tightly controlled promoter (2, 11, 56), a downstream regulatory region (24), and splicing of a LAT precursor RNA (41, 51). An ORF contained within the 2.0-kb LAT intron has recently been shown to encode a protein with considerable biological activity in terms of viral growth enhancement (47). However, this activity was detected from a recombinant gene where all LAT regulatory elements were removed and it is presently unclear if this ORF might be expressed from the natural context within the LAT gene or indeed which expression strategies might be used. With these issues in mind, we assessed the expression of a reporter gene placed within the 2.0-kb LAT DNA sequence, initially in an in vitro system from the powerful CMV immediate-early promoter and subsequently in the context of the virus. In the in vitro system we observed two patterns of GFP expression, which suggest that 2.0-kb LAT ORFs and perhaps an exon 1 ORF have the potential to be expressed in the viral life cycle. The 2.0-kb LAT intron (15) and alternately spliced RNAs or polyadenylated RNAs initiating from LAP1 and LAP2, respectively, have been previously postulated as mRNA candidates mediating 2.0-kb LAT ORF expression (47). Our data indicate that the only potential mRNAs for the expression of 2.0-kb LAT ORFs are alternately spliced and unspliced RNAs originating from LAP1. However, despite the production of alternately spliced transcripts from a recombinant LAT-GFP virus in productively and latently infected cells, no GFP signal could be detected.
The GFP gene placed within the 2.0-kb LAT DNA sequence can be expressed in transfected cells.
Previous attempts to visualize HSV-1 LAT ORF expression using polyclonal antibodies against LAT peptides (J. G. Spivak and N. W. Fraser, unpublished observations) or by an epitope tagging approach (22, 32) have been largely unsuccessful. In the latter studies, two ORFs towards the 3′ terminus of the mLAT were expressed from tagged viruses (ORF O and ORF P [Fig. 1a]) but only under conditions where the viral regulatory protein ICP4 was not expressed. Current evidence suggests that both these proteins are translated from an overlapping set of transcripts known as the LS/Ts, rather than from a LAT RNA (4, 54a). In our transfection studies, any viral regulation of LAT ORF expression has been largely circumvented by removing the LAT gene from the context of the virus and by replacing the LAT promoter with the constitutive immediate-early CMV promoter. In this system we observe two patterns of GFP expression, one of which is low level and resembles the normal cytoplasmic distribution pattern of GFP and another that localizes to the nucleolus (Fig. 2 and 5). Previously, Coffin et al. (6) inserted a chloramphenicol acetyltransferase (CAT) reporter gene between two XcmI sites within the 2.0-kb LAT in a construct driven by the CMV promoter but were unable to demonstrate CAT expression in transfected cells. This contradictory result may reflect the different choice of gene insertion sites used in the two studies and the relative effects of the two inserted sequences on alternate splicing involving the 1.5-kb LAT splice acceptor site. In our hands, GFP insertion at the HpaI site affects usage of the 1.5-kb LAT splice acceptor site and results in nucleolar GFP expression. However, GFP expression is not completely dependent on alternate splicing, since a weak cytoplasmic GFP signal and a 30-kDa GFP protein are consistently observed, even after deleting the 1.5-kb splice acceptor site [pΔPA(X)GFP (Fig. 5)]. Our data indicate that a polyadenylated RNA originating from LAP1 serves as a mRNA for expression of the 30-kDa GFP protein, despite the fact that the GFP coding sequence is located 1,500 bp downstream of the RNA cap site. Previously it has been demonstrated that the “leader” sequence between the 5′ terminus of the 2.0-kb LAT and the 1.5-kb LAT splice acceptor acts to enhance translation of 2.0-kb LAT ORFs (6). The presence of this element might help explain the initiation of translation so far downstream of the cap site of the LAT mRNA. Interestingly, deletion of this leader sequence [PpuMI-AgeI; pΔPA(X)GFP] leads to a decrease in expression of the 30-kDa GFP protein (Fig. 5b), while deletion of the PpuMI-BsmI sequence [pΔPB(X)GFP], which leaves 150 bp of the leader sequence intact just upstream of the splice acceptor, has no effect on 30-kDa GFP expression. These results are consistent with the existence of an element just upstream of the 1.5-kb LAT splice acceptor that acts to enhance translation but are complicated by the fact that the PpuMI-AgeI deletion also removes the 1.5-kb LAT splice acceptor which may negatively affect expression. In a separate study using a wheat germ cell-free translation system, the 2.0-kb LAT leader sequence was demonstrated to inhibit rather than promote translation of 2.0-kb LAT ORFs (41). It is possible that this result reflected a lack of appropriate trans-acting factors in the wheat germ extract required for leader-mediated enhancement of translation.
Alternate splicing and LAT ORF expression.
The demonstration that alternate splicing of LAT RNAs occurs in transfected cells (Fig. 3 and 4) (20) suggests novel strategies for LAT ORF expression. Alternate splicing would bring downstream LAT ORFs into closer proximity to LAP1 and would eliminate the small ORFs and secondary structure lying in between. In addition, splicing between the 2.0-kb LAT splice donor and alternate acceptor sites may lead to the fusion of ORFs in exon 1 with other LAT ORFs: Our results are consistent with the first scenario, since the 30-kDa GFP polypeptideide appears to be translated despite being located far downstream of the mRNA cap site. Interestingly, deletion of the 1.5-kb splice acceptor [pΔPA(X)GFP (Fig. 5)] appears to decrease the level of GFP expression, although as mentioned above, this deletion does not differentiate between disruption of the leader sequence and the 1.5-kb splice acceptor. It is also relevant to note that deletion of a large fragment from exon 1 [pΔSty(X)GFP] markedly increases 30-kDa GFP expression and probably reflects the closer proximity of GFP to the cap site, brought about by a combination of the deletion and alternate splicing involving the 1.5-kb LAT splice acceptor.
In our transfection studies, fusion occurred between an ORF in LAT exon 1 and GFP and leads to the production of a 48-kDa fusion protein and nucleolar localization of GFP. In this case, however, introduction of an artificial initiation codon to the exon 1 ORF was required to detect expression of the fusion protein to any major extent. In addition, the insertion of GFP into the HpaI site just downstream of the 1.5-kb splice acceptor favors the use of this site and hence the production of the fusion protein. Thus, the expression of fusions between exon 1 ORF 3 (Fig. 6) and downstream LAT ORFs is unlikely to occur in the context of wild-type HSV-1. In the absence of an artificially introduced AUG at the N terminus, the exon 1-GFP fusion protein is translated sporadically in transfected cells, a phenotype consistent with the requirement of a specific set of cellular conditions for translation initiation. Such conditional protein expression has been demonstrated for other proteins (for example, the cell cycle-dependent expression of p58pitslre through an internal ribosome entry sequence [IRES] element located within the mRNA coding region [7]). Therefore, it remains possible that HSV-1 LAT exon 1 ORF 3 may be translated in specific cells in vivo. Expression of the ORF 3-GFP fusion protein, however, was not readily detectable from the LAT-GFP virus where the artifical initiation codon was not present.
Finally, with regard to alternate splicing, we show that 1.5-kb LAT splice sites are used in nonneuronal cells (Fig. 4). Thus, the production of the 1.5-kb LAT in HSV-1 latently infected neurons (41, 51) may not be due to neuron-specific recognition of these sites but rather to cellular conditions which influence their utilization.
The 2.0-kb LAT does not function as a mRNA in an in vitro expression system.
The presence of the 2.0-kb LAT in the cytoplasm of infected cells and the association with polysomes have led to the proposal that the 2.0-kb LAT is translated (15). In our experiments, transfection of the 2.0-kb LAT-GFP expression plasmids into both neuronal and nonneuronal cells did indeed result in GFP expression (Fig. 2). However, we have shown here that this expression is not due to direct translation of the 2.0-kb LAT, since the destabilization of the LAT-GFP intron by the introduction of the cons mutation has no effect on either nucleolar or cytoplasmic GFP expression in transfected cells (Fig. 2 and 3). The conclusion that the 2.0-kb LAT is not directly translated is consistent with results obtained previously by Coffin et al. (6) where a CAT gene inserted between the XcmI sites within the 2.0-kb LAT could not be expressed in transient-transfection assays. We cannot formally rule out the possibility that translation of the 2.0-kb LAT occurs if the correct viral factors are present, since a LAT-GFP intron was not produced from the recombinant virus vLAT-GFP. However, preliminary experiments in which cells were transfected with pLAT(H)GFP+ are subsequently infected with HSV-1 F strain (not shown) do not demonstrate a significant increase in GFP expression over that of cells transfected with pLAT(H)G.cons, as might be expected if viral factors indeed influence 2.0-kb LAT translation. We are therefore inclined to believe that if the 2.0-kb LAT has a functional role in the viral life cycle, it is probably at the level of an RNA-ribosome interaction which is unrelated to 2.0-kb LAT translation, as has been previously proposed (28).
GFP is not expressed from within 2.0-kb LAT DNA in the context of the virus and implications for expression of 2.0-kb LAT ORFs.
A caveat of the in vitro expression system used in this work is that any potential role of viral factors in the expression of 2.0-kb LAT ORFs is not assessed. This concern was addressed by the construction of a virus containing GFP inserted at the HpaI site of the 2.0-kb LAT intron. Expression of GFP from this virus, in either neuronal or nonneuronal cells in culture or in infected TGs at any stage of the infection cycle was not observed, despite the production of alternately spliced LAT-GFP RNAs in both acutely and latently infected TGs (Fig. 7). It has been suggested that the putative promoter or regulatory element LAP2 (14), which abuts the 5′ terminus of the 2.0-kb LAT and which has been reported to be active during productive infection (5), may also be involved in the expression of 2.0-kb LAT ORFs (47). Based upon the observation that LAT transcripts can initiate just upstream of the 2.0-kb LAT (5), it was envisioned that LAP2 might drive the transcription of a polyadenylated, unspliced 2.0-kb LAT. The lack of GFP expression from our LAT-GFP virus during productive infection implies that this scenario is not likely. One possibility is that since LAP2-initiated RNA is efficiently spliced to form the 2.0-kb LAT intron (5), the unspliced form is never present in sufficient abundance to act as a mRNA.
The major LAT promoter, LAP1, was initially reported to be active during both productive infection and latency, while a second promoter was active in tissue culture cells, (27). More recently, it has been claimed that LAP1 is active mainly in latently infected cells, while the 2.0-kb LAT-proximal LAP2 is predominantly active during productive infection (5). Whichever the case, transcripts originating from LAP1 in the context of the LAT-GFP virus did not express detectable levels of GFP either in infected-cell cultures or in productively or latently infected TGs. Our results from cell culture transfections indicate that LAP1 initiated transcripts, either alternately spliced or unspliced, should indeed act as mRNAs for GFP expression. This discrepancy suggests that translation of LAP1-initiated transcripts is specifically affected in infected cells. In previous studies, the LAT HpaI site has been used successfully for expression of foreign gene sequences from the LAT promoter of HSV-1 vectors (21). However, some modifications were made to achieve expression, which occurred weakly during productive infection but more strongly during latency. Specifically, a cassette containing the lacZ gene under the control of an IRES and terminated with a CMV poly(A) signal was inserted at the HpaI site. Since LAT-GFP alternately spliced transcripts observed in our studies are polyadenylated during productive infection (Fig. 7d), it appears that the important difference between this study and our own in terms of latent reporter gene expression is the inclusion of the IRES element. Previously, we have observed that expression of the β-galactosidase (β-Gal) gene from the LAT promoter differs at the transcriptional and translational levels (18). In this study, 89-fold-more neurons from TGs acutely infected with a LAT promoter-LacZ virus, were positive for β-Gal RNA than for enzyme activity. It was suggested that the 5′ untranslated region (UTR) between the LAT cap site and the AUG codon of the lacZ gene was responsible for the inhibition of translation, since substituting a neurofilament 5′ UTR restored β-Gal enzyme expression. Margolis et al. (25) also noticed a similar low level of β-Gal expression from the LAT promoter during productive ganglionic infection, although in this case, the relationship between β-Gal enzyme and transcript expression on a single neuron basis was not investigated. Interestingly, in the LAT-GFP expression vectors used in the transfection assays, cloning of the LAT gene resulted in the deletion of a large portion of the 100-bp 5′ UTR element. The difference in translation between the plasmid-produced transcripts and viral transcripts may therefore reflect the presence or absence of the LAT 5′ UTR and suggests that viral LAP-1-initiated RNAs are blocked at the level of translation.
Previously, our group has been unable to demonstrate a protein product of 2.0-kb LAT ORF 1 in either productively infected cells or in infected-mouse brains, using a polyclonal antibody directed against an ORF 1 peptide (Spivak and Fraser, unpublished). Together with the results of the current work, this lack of detection leads us to believe that the biologically active ORF 1 product (47) may not be expressed from its natural context within the 2.0-kb LAT intron during viral infection. However, we cannot formally exclude the possibility that expression does occur but is limited to a particular window of the infection process not covered by either the current study or by previous work. Such expression might require cellular factors or activation signals unique to the specific environment of infected neurons.
ACKNOWLEDGMENTS
We are indebted to John Baillet and Priscilla Schaeffer for supplying the Kos-GFP virus. We also thank Michael Malim for the kind gift of the anti-Rev ascitic fluid, and we are grateful for the insightful comments of participants in the Herpes Latency Program Project biweekly meetings.
This work was supported by Public Health Service Program Project grant NS33768 from the National Institute of Health. In addition, M.L. was supported by National Institute of Health postdoctoral traineeship T32 A107324.
REFERENCES
- 1.Alvira M R, Goins W F, Cohen J B, Glorioso J C. Genetic studies exposing the splicing events involved in herpes simplex type 1 latency-associated transcript production during lytic and latent infection. J Virol. 1999;73:3866–3876. doi: 10.1128/jvi.73.5.3866-3876.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batchelor A H, O'Hare P. Regulation and cell-type-specific activity of a promoter located upstream of the latency-associated transcript of herpes simplex virus type 1. J Virol. 1990;64:3269–3279. doi: 10.1128/jvi.64.7.3269-3279.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Block T M, Deshmane S L, Masonis J, Maggioncalda J, Valyi-Nagy T, Fraser N W. An HSV LAT null mutant reactivates slowly from latent infection and makes small plaques on CV-1 monolayers. Virology. 1992;192:618–630. doi: 10.1006/viro.1993.1078. [DOI] [PubMed] [Google Scholar]
- 4.Bohenzky R A, Papavassiliou A G, Gelman I H, Silverstein S. Identification of a promoter mapping within the reiterated sequences that flank the herpes simplex virus type 1 UL region. J Virol. 1993;67:632–642. doi: 10.1128/jvi.67.2.632-642.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X, Schmidt M, Goins W, Glorioso J. Two herpes simplex virus type 1 latency-active promoters differ in their contributions to latency-associated transcript expression during lytic and latent infections. J Virol. 1995;69:7899–7908. doi: 10.1128/jvi.69.12.7899-7908.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coffin R S, Thomas S K, Thomas D P, Latchman D S. The herpes simplex virus 2 kb latency associated transcript (LAT) leader sequence allows efficient expression of downstream proteins which is enhanced in neuronal cells: possible function of LAT ORFs. J Gen Virol. 1998;79:3019–3026. doi: 10.1099/0022-1317-79-12-3019. [DOI] [PubMed] [Google Scholar]
- 7.Cornelis S, Bruynooghe Y, Denecker G, Van Huffel S, Tinton S, Beyaert R. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol Cell. 2000;5:597–605. doi: 10.1016/s1097-2765(00)80239-7. [DOI] [PubMed] [Google Scholar]
- 8.Deatly A M, Spivack J G, Lavi E, Fraser N W. RNA from an immediate early region of the HSV-1 genome is present in the trigeminal ganglia of latently infected mice. Proc Natl Acad Sci USA. 1987;84:3204–3208. doi: 10.1073/pnas.84.10.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deatly A M, Spivack J G, Lavi E, O'Boyle II D R, Fraser N W. Latent herpes simplex virus type 1 transcripts in peripheral and central nervous system tissues of mice map to similar regions of the viral genome. J Virol. 1988;62:749–756. doi: 10.1128/jvi.62.3.749-756.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devi-Rao G B, Goodart S A, Hecht L M, Rochford R, Rice M A, Wagner E K. Relationship between polyadenylated and nonpolyadenylated herpes simplex virus type 1 latency-associated transcripts. J Virol. 1991;65:2179–2190. doi: 10.1128/jvi.65.5.2179-2190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobson A T, Sederati F, Devi-Rao G, Flanagan J, Farrell M J, Stevens J G, Wagner E K, Feldman L T. Identification of the latency-associated transcript promoter by expression of rabbit beta-globin mRNA in mouse sensory nerve ganglia latently infected with a recombinant herpes simplex virus. J Virol. 1989;63:3844–3851. doi: 10.1128/jvi.63.9.3844-3851.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doerig C, Pizer L I, Wilcox C L. An antigen encoded by the latency-associated transcript in neuronal cell cultures latently infected with herpes simplex virus type 1. J Virol. 1991;65:2724–2727. doi: 10.1128/jvi.65.5.2724-2727.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farrell M J, Dobson A T, Feldman L T. Herpes simplex virus latency-associated transcript is a stable intron. Proc Natl Acad Sci USA. 1991;88:790–794. doi: 10.1073/pnas.88.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goins W F, Sternberg L R, Croen K D, Krause P R, Hendricks R L, Fink D J, Straus S E, Levine M, Glorioso J C. A novel latency-active promoter is contained within the herpes simplex virus type 1 UL flanking repeats. J Virol. 1994;68:2239–2252. doi: 10.1128/jvi.68.4.2239-2252.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldenberg D, Mador N, Ball M J, Panet A, Steiner I. The abundant latency-associated transcripts of herpes simplex virus type 1 are bound to polyribosomes in cultured neuronal cells and during latent infection in mouse trigeminal ganglia. J Virol. 1997;71:2897–2904. doi: 10.1128/jvi.71.4.2897-2904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hill J M, Sederati F, Javier R T, Wagner E K, Stevens J G. Herpes simplex virus latent phase transcription facilitates in vivo reactivation. Virology. 1990;174:117–125. doi: 10.1016/0042-6822(90)90060-5. [DOI] [PubMed] [Google Scholar]
- 17.Hill T J. Herpes simplex virus latency. In: Roizman B, editor. The herpes viruses. Vol. 4. New York, N.Y: Plenum Publishing Corp.; 1985. pp. 175–240. [Google Scholar]
- 18.Huang Q S, Valyi-Nagy T, Kesari S, Fraser N W. beta-Gal enzyme activity driven by the HSV LAT promoter does not correspond to beta-gal RNA levels in mouse trigeminal ganglia. Gene Ther. 1997;4:797–807. doi: 10.1038/sj.gt.3300476. [DOI] [PubMed] [Google Scholar]
- 19.Johnson P, Friedmann T. Replication defective recombinant herpes simplex virus vectors. Methods Cell Biol. 1994;43:211–230. doi: 10.1016/s0091-679x(08)60605-6. [DOI] [PubMed] [Google Scholar]
- 20.Krummenacher C, Zabolotny J M, Fraser N W. Selection of a nonconsensus branch point is influenced by an RNA stem-loop structure and is important to confer stability to the herpes simplex virus 2-kilobase latency-associated transcript. J Virol. 1997;71:5849–5860. doi: 10.1128/jvi.71.8.5849-5860.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lachmann R, Efstathiou S. Utilization of the herpes simplex virus type 1 latency-associated regulatory region to drive stable reporter gene expression in the nervous system. J Virol. 1997;71:3197–3207. doi: 10.1128/jvi.71.4.3197-3207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lagunoff M, Roizman B. Expression of a herpes simplex virus 1 open reading frame antisense to the g134.5 gene and transcribed by an RNA 3′ coterminal with the unspliced latency-associated transcript. J Virol. 1994;68:6021–6028. doi: 10.1128/jvi.68.9.6021-6028.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leib D A, Bogard C L, Kosz-Vnenchak M, Hicks K A, Coen D M, Knipe D M, Schaffer P A. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J Virol. 1989;63:2893–2900. doi: 10.1128/jvi.63.7.2893-2900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lokensgard J R, Berthomme H, Feldman L T. The latency-associated promoter of herpes simplex virus type 1 requires a region downstream of the transcription start site for long-term expression during latency. J Virol. 1997;71:6714–6719. doi: 10.1128/jvi.71.9.6714-6719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Margolis T P, Sedarati F, Dobson A T, Feldman L T, Stevens J G. Pathways of viral gene expression during acute neuronal infection with HSV-1. Virology. 1992;189:150–160. doi: 10.1016/0042-6822(92)90690-q. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell W J, Lirette R P, Fraser N W. Mapping of low abundance latency associated RNA in the trigeminal ganglia of mice latently infected with herpes simplex virus type 1. J Gen Virol. 1990;71:125–132. doi: 10.1099/0022-1317-71-1-125. [DOI] [PubMed] [Google Scholar]
- 27.Nicosia M, Deshmane S L, Zabolotny J M, Valyi-Nagy T, Fraser N W. Herpes simplex virus type 1 latency-associated transcript (LAT) promoter deletion mutants can express a 2-kilobase transcript mapping to the LAT region. J Virol. 1993;67:7276–7283. doi: 10.1128/jvi.67.12.7276-7283.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicosia M, Zabolotny J M, Lirette R P, Fraser N W. The HSV-1 2-kb latency-associated transcript is found in the cytoplasm comigrating with ribosomal subunits during productive infection. Virology. 1994;204:717–728. doi: 10.1006/viro.1994.1587. [DOI] [PubMed] [Google Scholar]
- 29.Perng G-C, Ghiasi H, Slanina S M, Nesburn A B, Wechsler S L. The spontaneous reactivation function of the herpes simplex virus type 1 LAT gene resides completely within the first 1.5 kilobases of the 8.3-kilobase primary transcript. J Virol. 1996;70:976–984. doi: 10.1128/jvi.70.2.976-984.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perng G C, Jones C, Ciacci-Zanella J, Stone M, Henderson G, Yukht A, Slanina S M, Hofman F M, Ghiasi H, Nesburn A B, Wechsler S L. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science. 2000;287:1500–1503. doi: 10.1126/science.287.5457.1500. [DOI] [PubMed] [Google Scholar]
- 31.Perng G C, Slanina S M, Yukht A, Ghiasi H, Nesburn A B, Wechsler S L. The latency-associated transcript gene enhances establishment of herpes simplex virus type 1 latency in rabbits. J Virol. 2000;74:1885–1891. doi: 10.1128/jvi.74.4.1885-1891.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Randall G, Lagunoff M, Roizman B. The product of ORF O located within the domain of herpes simplex virus 1 genome transcribed during latent infection binds to and inhibits in vitro binding of infected cell protein 4 to its cognate DNA site. Proc Natl Acad Sci USA. 1997;94:10379–10384. doi: 10.1073/pnas.94.19.10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rock D L, Nesburn A B, Ghiasi H, Ong J, Lewis T L, Lokensgard J R, Wechsler S M. Detection of latency-related viral RNAs in trigeminal ganglia of rabbits latently infected with herpes simplex virus type 1. J Virol. 1987;61:3820–3826. doi: 10.1128/jvi.61.12.3820-3826.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodahl E, Haarr L. Analysis of the 2-kilobase latency-associated transcript expressed in PC12 cells productively infected with herpes simplex virus type 1: evidence for a stable, nonlinear structure. J Virol. 1997;71:1703–1707. doi: 10.1128/jvi.71.2.1703-1707.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roizman B, Sears A E. Herpes simplex viruses and their replications. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2231–2296. [Google Scholar]
- 36.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Sawtell N M, Thompson R L. Herpes simplex virus type 1 latency-associated transcription unit promotes anatomical site-dependent establishment and reactivation from latency. J Virol. 1992;66:2157–2169. doi: 10.1128/jvi.66.4.2157-2169.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schang L M, Hossain A, Jones C. The latency-related gene of bovine herpesvirus 1 encodes a product which inhibits cell cycle progression. J Virol. 1996;70:3807–3814. doi: 10.1128/jvi.70.6.3807-3814.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spivack J G, Fraser N W. Detection of herpes simplex virus type 1 transcripts during latent infection in mice. J Virol. 1987;61:3841–3847. doi: 10.1128/jvi.61.12.3841-3847.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spivack J G, Fraser N W. Expression of herpes simplex virus type 1 latency-associated transcripts in the trigeminal ganglia of mice during acute infection and reactivation of latent infection. J Virol. 1988;62:1479–1485. doi: 10.1128/jvi.62.5.1479-1485.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spivack J G, Woods G M, Fraser N W. Identification of a novel latency-specific splice donor signal within the herpes simplex virus type 1 2.0-kilobase latency-associated transcript (LAT): translation inhibition of LAT open reading frames by the intron within the 2.0-kilobase LAT. J Virol. 1991;65:6800–6810. doi: 10.1128/jvi.65.12.6800-6810.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steiner I, Spivack J G, Lirette R P, Brown S M, MacLean A R, Subak-Sharpe J, Fraser N W. Herpes simplex virus type 1 latency-associated transcripts are evidently not essential for latent infection. EMBO J. 1989;8:505–511. doi: 10.1002/j.1460-2075.1989.tb03404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stevens J G. Herpetic latency and reactivation. In: Rapp F, editor. Oncogenesis and herpesviruses. Vol. 2. Boca Raton, Fla: CRC Press; 1980. pp. 1–12. [Google Scholar]
- 44.Stevens J G, Haar L, Porter D, Cook M L, Wagner E K. Prominence of the herpes simplex virus latency associated transcript in trigeminal ganglia from seropositive humans. J Infect Dis. 1988;158:117–123. doi: 10.1093/infdis/158.1.117. [DOI] [PubMed] [Google Scholar]
- 45.Stevens J G, Wagner E K, Devi-Rao G B, Cook M L, Feldman L T. RNA complementary to a herpes virus gene mRNA is prominent in latently infected neurons. Science. 1987;235:1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- 46.Stroop W G, Rock D L, Fraser N W. Localization of herpes simplex virus in the trigeminal and olfactory systems of the mouse central nervous system during acute and latent infections by in situ hybridization. Lab Investig. 1984;51:27–38. [PubMed] [Google Scholar]
- 47.Thomas S K, Gough G, Latchman D S, Coffin R S. Herpes simplex virus latency-associated transcript encodes a protein which greatly enhances virus growth, can compensate for deficiencies in immediate-early gene expression, and is likely to function during reactivation from virus latency. J Virol. 1999;73:6618–6625. doi: 10.1128/jvi.73.8.6618-6625.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trousdale M, Steiner I, Spivack J G, Deshmane S L, Brown S M, MacLean A S, Subak-Sharpe J H, Fraser N W. Evidence that the herpes simplex virus type 1 latency-associated transcripts play a role in reactivation of latent infection in vivo. J Virol. 1991;65:6989–6993. doi: 10.1128/jvi.65.12.6989-6993.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner E K, Devi-Rao G, Feldman L T, Dobson A T, Zhang Y F, Hill J M, Flanagan W M, Stevens J G. Physical characterization of the herpes simplex virus latency-associated transcript in neurons. J Virol. 1988;62:1194–1202. doi: 10.1128/jvi.62.4.1194-1202.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner E K, Flanagan W M, Devi-Rao G, Zhang Y F, Hill J M, Anderson K P, Stevens J G. The herpes simplex virus latency-associated transcript is spliced during the latent phase of infection. J Virol. 1988;62:4577–4585. doi: 10.1128/jvi.62.12.4577-4585.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wechsler S L, Nesburn A B, Watson R, Slanina S M, Ghiasi H. Fine mapping of the latency-related gene of herpes simplex virus type 1: alternate splicing produces distinct latency-related RNAs containing open reading frames. J Virol. 1988;62:4051–4058. doi: 10.1128/jvi.62.11.4051-4058.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wechsler S L, Nesburn J, Zaagstra N, Ghiasi H. Sequence of the latency related gene of herpes simplex virus type 1. Virology. 1989;168:168–172. doi: 10.1016/0042-6822(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 53.Wu T, Su Y, Block T, Taylor J. Atypical splicing of the latency-associated transcript of herpes simplex type 1. Virology. 1998;243:140–149. doi: 10.1006/viro.1998.9036. [DOI] [PubMed] [Google Scholar]
- 54.Wu T T, Su Y H, Block T M, Taylor J M. Evidence that two latency-associated transcripts of herpes simplex virus type 1 are nonlinear. J Virol. 1996;70:5962–5967. doi: 10.1128/jvi.70.9.5962-5967.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54a.Yeh L, Schaffer P A. A novel class of transcripts expressed with late kinetics in the absence of ICP4 spans the junction between the long and short segments of the herpes simplex virus type 1 genome. J Virol. 1993;67:7373–7382. doi: 10.1128/jvi.67.12.7373-7382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zabolotny J M, Krummenacher C, Fraser N W. The herpes simplex virus type 1 2.0-kilobase latency-associated transcript is a stable intron which branches at a guanosine. J Virol. 1997;71:4199–4208. doi: 10.1128/jvi.71.6.4199-4208.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zwaagstra J, Ghiasi H, Slanina S M, Nesburn A B, Wheatley S C, Lillycrop K, Wood J, Latchman D S, Patel K, Wechsler S L. Activity of herpes simplex virus type 1 latency-associated transcript (LAT) promoter in neuron-derived cells: evidence for neuron specificity and for a large LAT transcript. J Virol. 1990;64:5019–5028. doi: 10.1128/jvi.64.10.5019-5028.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]