Abstract
The gp120 envelope glycoprotein of primary human immunodeficiency virus type 1 (HIV-1) promotes virus entry by sequentially binding CD4 and the CCR5 chemokine receptor on the target cell. Previously, we adapted a primary HIV-1 isolate, ADA, to replicate in CD4-negative canine cells expressing human CCR5. The gp120 changes responsible for CD4-independent replication were limited to the V2 loop-V1/V2 stem. Here we show that elimination of a single glycosylation site at asparagine 197 in the V1/V2 stem is sufficient for CD4-independent gp120 binding to CCR5 and for HIV-1 entry into CD4-negative cells expressing CCR5. Deletion of the V1/V2 loops also allowed CD4-independent viral entry and gp120 binding to CCR5. The binding of the wild-type ADA gp120 to CCR5 was less dependent upon CD4 at 4°C than at 37°C. In the absence of the V1/V2 loops, neither removal of the N-linked carbohydrate at asparagine 197 nor lowering of the temperature increased the CD4-independent phenotypes. A CCR5-binding conformation of gp120, achieved by CD4 interaction or by modification of temperature, glycosylation, or variable loops, was preferentially recognized by the monoclonal antibody 48d. These results suggest that the CCR5-binding region of gp120 is occluded by the V1/V2 variable loops, the position of which can be modulated by temperature, CD4 binding, or an N-linked glycan in the V1/V2 stem.
Human immunodeficiency virus types 1 and 2 (HIV-1 and HIV-2) are the etiologic agents of AIDS in humans (5, 12, 30). AIDS is associated with the depletion of CD4-positive T lymphocytes, which are the major target cells of viral infection in vivo (26).
The entry of primate immunodeficiency viruses into target cells is mediated by the viral envelope glycoproteins, gp120 and gp41, which are organized into trimeric complexes on the virion surface (2, 53). Viral entry usually requires the binding of the exterior envelope glycoprotein, gp120, to the primary receptor CD4 (14, 36, 42). gp120 is heavily glycosylated and contains protruding variable loops (38, 40), features that are thought to decrease the susceptibility of the virus to host immune responses (73, 75). The interaction between gp120 and CD4 promotes a series of conformational changes in gp120 that result in the formation or exposure of a binding site for particular members of the chemokine receptor family that serve as coreceptors (68, 72). The chemokine receptor CCR5 is the major coreceptor for primary HIV-1 isolates (1, 10, 16, 19, 20) and can be utilized by HIV-2 and simian immunodeficiency virus (SIV) isolates as well (9, 43). Binding of gp120 to the coreceptor is thought to induce additional conformational changes that lead to activation of the transmembrane glycoprotein gp41 and subsequent fusion of the viral and cellular membranes (8, 61, 69).
In addition to anchoring and orienting the viral envelope glycoproteins with respect to the target cell membrane, binding to CD4 initiates changes in the conformation of the envelope glycoproteins (3, 4, 17, 22, 55–57, 66, 70, 74). Some of these conformational changes allow high-affinity interaction with CCR5 (68, 72). CD4-induced movement of the V1/V2 loops results in the exposure of conserved, discontinuous structures on the HIV-1 gp120 glycoprotein recognized by the monoclonal antibodies 17b and 48d (66, 74). Analysis of a panel of gp120 mutants suggested that this conformational change is functionally important for virus entry (64). The close physical relationship between the 17b and 48d epitopes and conserved gp120 structures shown to be important for CCR5 binding (52) supports a model in which conformational changes in the V1/V2 stem-loop structure induced by CD4 binding create and/or expose a high-affinity binding site for the CCR5 coreceptor.
Insights into the molecular basis for receptor binding by the primate immunodeficiency virus gp120 glycoproteins have been obtained from analysis of antibody binding, mutagenesis, and X-ray crystallography (39, 48–52, 54, 60, 75). These studies suggest that the major variable loops are well exposed on the surface of gp120, whereas the more conserved regions fold into a core structure. This HIV-1 gp120 core has been crystallized in a complex with fragments of the CD4 glycoprotein and the monoclonal antibody 17b (39, 75). The gp120 core is composed of an inner and an outer domain and a four-stranded β-sheet (the “bridging sheet”). Elements of both domains and the bridging sheet contribute to CD4 binding (39). Thermodynamic analysis of the gp120-CD4 interaction suggests that core elements of gp120 undergo significant conformational changes upon CD4 binding (50a). Alteration of the relationships among the gp120 domains by CD4 binding may be relevant to the induction of CCR5 binding. CCR5 binding apparently involves a conserved gp120 element (39, 52, 52a) and the third variable (V3) loop, which determines the choice of a particular chemokine receptor (10, 13, 60). The conserved element is located on two gp120 strands that connect the gp120 domains (52, 52a) and therefore is potentially modified by CD4-induced changes in gp120 interdomain relationships.
Infection by primate immunodeficiency viruses is generally more efficient when CD4 is expressed on the surface of the target cells. However, some viral isolates are able to achieve reasonably efficient infection of cells lacking CD4. For example, some HIV-2 isolates have been shown to enter CD4-negative cells by using CXCR4 (11, 24). Some SIV strains can infect CD4-negative brain capillary endothelial cells or other cell types by using CCR5 as a primary receptor (23, 57). The gp120 glycoproteins of some SIV isolates can efficiently bind rhesus monkey CCR5 in the absence of soluble CD4 (sCD4) (44). Naturally occurring, CD4-independent HIV-1 isolates appear to be less common, but CXCR4-using HIV-1 isolates have been derived by passage on CD4-negative cultured cells (21, 32a, 39a). We have previously derived a CD4-independent variant of the ADA HIV-1 strain that utilizes the CCR5 coreceptor and demonstrated that changes in the gp120 V2 loop and/or V1/V2 stem region were responsible for both CD4-independent entry into cells and gp120 binding to CCR5 in the absence of CD4 (36a). Here we show that elimination of a single N-linked glycosylation site in the ADA gp120 glycoprotein is responsible for both of these CD4-independent phenotypes. Moreover, deletion of the gp120 V1/V2 loops was also sufficient for CD4-independent virus entry and CCR5 binding. CD4-independent binding of the wild-type gp120 to CCR5 was enhanced at 4°C. These observations allow us to propose a mechanistic model for CD4 independence.
MATERIALS AND METHODS
Cell lines.
293T and Cf2Th cells were obtained from the American Type Culture Collection and maintained as previously described (10). Cf2Th-CCR5 and Cf2Th-CCR5-CD4 cells were derived and maintained as previously described (36a).
Site-directed mutagenesis.
The pSVIIIenv-ADA plasmids expressing ADA envelope glycoproteins with the individual V2 loop-V1/V2 stem changes depicted in Fig. 1 were created by PCR mutagenesis. Complementary pairs of primers were used to introduce the following mutations into pSVIIIenv-ADA by the QuikChange protocol (Stratagene). Only one primer of each pair is given here: for 190 S/R–197 N/S, primer ADARSf (5′-CCAATAGATAATGATAATACTAGGTATCGAT TGATAAAT TG TAGTACC TCAACCAT TACACAGG-3′); for 190 S/N–197 N/S, primer ADANSf (5′-GTACCAATAGATAATGATAATACTAACTATCGATTGATAAATTGTAGCACCTCAACCATTACACAGGC-3′); for 197 N/K, primer a-Knewf (5′-CCAATAGATAATGATAATACTAGCTATCGATTGATAAATTGTAAGACCTCAACCATTACACAGG-3′); for 190 S/R, primer ADAR-f (5′-CCAATAGATAATGATAATACTAGGTATCGATTGATAAATTGTAATACCTCAACCATTACACAGG-3′); for 197 N/S, primer ADA-Sf (5′-CCAATAGATAATGATAATACTAGCTATCGATTGATAAATTGTAGTACCTCAACCATTACACAGG-3′); for 188 N/Q, primer ADAQ-f (5′-CCAATAGATAATGATCAGACTAGCTATCGATTGATAAATTGTAATACCTCAACCATTACACAGG-3′); for 197 N/Q, primer ADA-Qf (5′-CCAATAGATAATGATAATACTAGCTATCGATTGATAAATTGTCAGACCTCAACCATTACACAGG-3′); and for 188 N/Q–197 N/Q, primer ADAQQf (5′-CCAATAGATAATGATCAGAC TAGC TATCGAT TGATAAAT TG TCAGACCTCAACCATTACACAGG-3′).
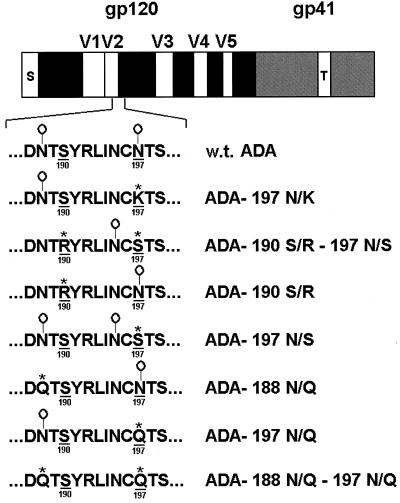
FIG. 1.
HIV-1 ADA envelope glycoprotein mutants. The V2 loop-V1/V2 stem sequences of the wild-type (w.t.) and mutant ADA envelope glycoproteins are shown beneath the representation of the gp120 and gp41 envelope glycoprotein sequences. The ADA envelope glycoprotein residues altered in the CD4-independent virus are underlined and numbered according to the prototypic HXBc2 sequence (37). In the mutants, the altered residues are marked with an asterisk. Sites of potential N-linked glycosylation are indicated (O|). S, signal peptide; V1 to V5, gp120 variable regions; T, gp41 transmembrane region.
To generate V1/V2 deletion-containing envelopes, the following primers were used: AdeltaB (5′-ACAATTGCCGGCGCCAACACAGAGTGGGGTTAATTTTACACATGG-3′), AdeltaF (5′-CTCTGTGTTggcgccggcAATTGTAATACCTCAACCATTACACAGGCCTGTCC-3′), SdeltaF (5′-CTCTGTGTTggcgccggcAATTGTAgTACCTCAACCATTACACAGGCCTGTCC-3′), QdeltaF (5′-CT CTGTGTTggcgccggcAATTGTcAgACCTCAACCATTACACAGGCCTGTC C-3′), ADACD4f (5′-GAAAGAGCAGAAGAGAGTGGCAATGAGAGTG-3′), and ADACD4b (5′-GCCATCCAATCACACTAC-3′) (lowercase letters indicate changes). Using a wild-type ADA envelope-encoding construct, primers AdeltaB and ADACD4f were used to amplify fragment A by PCR. Primers AdeltaF and ADACD4b were used to PCR amplify fragment B from the same template. Fragments A and B were then gel purified using a Bio-Rad Freeze n' Spin column and were combined into one PCR that produced a fragment encoding an envelope glycoprotein containing a Gly-Ala-Gly linker in place of the gp120 V1/V2 loops. This larger PCR fragment was gel purified and cloned into the original pSVIIIenv template plasmid using KpnI and BamHI. To make plasmids encoding the 197 N/S and 197 N/Q envelope glycoproteins with V1/V2 deletions, primers SdeltaF and QdeltaF, respectively, were substituted for AdeltaF in the scheme above.
To generate secreted, soluble versions of particular gp120 envelope glycoproteins, the primer ADA stop (5′-GAAGAGTGGTGCAGAGAGAAAAAAGATAAGTGGGAACGATAGGAGCTATGTTCC-3′) was used to introduce a stop codon at a position corresponding to the natural gp120-gp41 cleavage site. All constructs were sequenced using the set of GBK96 primers described previously (7a).
env complementation assay.
Recombinant reporter viruses (32) were generated by transfecting 293T cells by the calcium phosphate method with 2 μg of an env-expressing pSVIIIenv plasmid, 5 μg of a packaging plasmid, and 15 μg of a vector plasmid expressing firefly luciferase. The ADA envelope glycoprotein variants were expressed equivalently in the transfected cells (data not shown). Seventy-two hours after transfection, the virus-containing supernatant was harvested and cleared by low-speed centrifugation. The virus in the supernatant was quantitated by measuring reverse transcriptase as described previously (32).
Target cells were seeded at a density of 6,000 cells/well in 96-well, luminometer-compatible tissue culture plates (Dynex). Twenty-four hours later, the medium was removed from the wells containing the target cells, and 20,000 reverse transcriptase units of virus was added to the cells. After 8 h of virus-cell incubation, the supernatant was removed and fresh medium was added (200 μl). Seventy-two hours after infection, the medium was removed from each well and the cells were lysed with agitation in 20 μl of passive lysis buffer (Promega). Luciferase activity was measured using an EG&G Berthold LB 96V microplate luminometer in accordance with the luciferase assay system technical bulletin from Promega.
Immunoprecipitations.
One hundred microliters of cell supernatants containing radiolabeled gp120 was preincubated with shaking at 37°C for 1 h with or without sCD4 (30 μg/ml). Approximately 40 μl of protein A-Sepharose beads suspended in phosphate-buffered saline (50% of bead volume) were added together with 0.5 μg of monoclonal antibody 48d to the gp120 solutions. The mixture was incubated with rocking for 2 to 24 h at 37°C or 4. Beads were washed once with NP-40 buffer and once with phosphate-buffered saline. Beads were resuspended in 40 μl of 2× sodium dodecyl sulfate (SDS) sample buffer, boiled, and centrifuged briefly. The samples were run under denaturing conditions on an SDS–10% polyacrylamide gel and then analyzed on a phosphorimager. In parallel, 100 μl of cell supernatants was precipitated by a mixture of sera from HIV-1-infected individuals. To control for the small degree of variation in the amounts of the gp120 glycoprotein variants, the values obtained for 48d antibody recognition were normalized based on the amount of gp120 precipitated by the mixture of sera.
CCR5 binding assay.
Radiolabeled gp120 glycoproteins were produced by transfection of 293T cells as previously described (36a). The amounts of the gp120 variants were adjusted prior to binding to Cf2Th-CCR5 cells, as previously described (36a). Bound gp120 was detected by precipitation with the C11 antibody, followed by SDS-polyacrylamide gel electrophoresis (PAGE) and quantitation with a phosphorimager (36a).
RESULTS
Effects of changes in gp120 glycosylation sites on CCR5 binding.
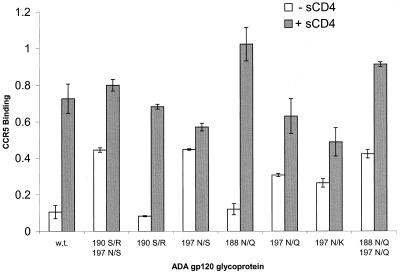
The ADA gp120 glycosylation site variants used in this study are shown in Fig. 1. The CD4-independent ADA viruses contained two sets of changes, one involving an N-linked glycosylation site at asparagine 188 in the V2 loop and the other involving an N-linked glycosylation site at asparagine 197 in the V1/V2 stem. In addition, the substitution of positively charged residues near the modified N-glycosylation sites was commonly observed (36a). We created several ADA gp120 variants that contained these or related changes individually or in combination. The ability of these variants to bind CCR5 in the absence or presence of sCD4 was examined. When cell supernatants containing equivalent amounts of the different radiolabeled gp120 glycoproteins were incubated with CCR5-expressing Cf2Th cells in the presence of 10 μg of sCD4/ml, all of the gp120 variants bound efficiently to the cells (Fig. 2). However, in the absence of sCD4, only the gp120 glycoproteins altered at residue 197 were able to bind to the CCR5-expressing cells more efficiently than the wild-type ADA gp120. For example, the 197 N/Q and 197 N/K mutants exhibited the ability to bind CCR5-expressing cells in the absence of sCD4. In the absence of sCD4, the 197 N/S glycoprotein bound to Cf2Th-CCR5 cells even more efficiently than the 197 N/Q and 197 N/K mutants. The relative migration of these gp120 variants on SDS-polyacrylamide gels (data not shown) is consistent with the expectation that the latter two mutants would exhibit a loss of an N-linked glycosylation site, whereas the 197 N/S substitution allows, in addition, modification of asparagine 195 by carbohydrates. The results indicate that the removal of the complex sugar moiety at asparagine 197 is sufficient to allow CD4-independent binding of ADA gp120 to CCR5. Removal of this carbohydrate in conjunction with the introduction of a site of N-linked glycosylation at asparagine 195 allows for even more efficient CD4-independent CCR5 binding.
FIG. 2.
CCR5-binding ability of wild-type (w.t.) and mutant ADA glycoproteins. The binding of radiolabeled, soluble gp120 glycoproteins to Cf2Th-CCR5 cells in the absence and presence of 10 μg of sCD4/ml at 37°C is shown. Cells were washed and lysed, and the bound gp120 was precipitated by the anti-gp120 antibody C11. Proteins were analyzed using SDS-PAGE and then quantitated with a phosphorimager. The averages and standard deviations of values obtained from a binding assay performed in triplicate are shown.
To determine whether monomeric gp120 binding to CCR5 was achieving equilibrium in the experiment described in Fig. 2, we repeated the CCR5-binding assay and measured the amount of gp120 bound at time points between 20 min and 4 h of incubation. The results showed that maximal binding to CCR5-Cf2Th cells occurred between 20 min and 1 h after supernatant was added to cells (data not shown).
Effects of changes in gp120 glycosylation sites on virus infection.
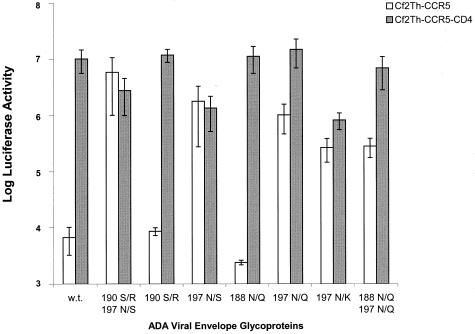
To examine the ability of the mutant envelope glycoproteins to support HIV-1 entry into CCR5-expressing cells, in either the presence or absence of CD4, an env complementation assay was used. This assay measures the ability of HIV-1 envelope glycoproteins to complement a single round of infection of an env-defective HIV-1 provirus expressing firefly luciferase. The target cells used in this assay were Cf2Th-CCR5 cells and Cf2Th-CCR5-CD4 cells. The level of CCR5 expression in the Cf2Th-CCR5 cells is substantially higher than that in the Cf2Th-CCR5-CD4 cells (data not shown); thus, for a particular envelope glycoprotein, any observed differences in the infection of these two cell lines may be due to differences in cellular expression of both CD4 and CCR5. Figure 3 shows that all ADA envelope glycoproteins lacking the N-linked glycosylation site at asparagine 197 were able to mediate infection of the CD4-negative Cf2Th-CCR5 cells more efficiently than the wild-type ADA glycoproteins. In some cases, changes in V2 loop residue 190 modulated the efficiency of infection of CD4-positive or -negative Cf2Th-CCR5 cells in conjunction with the altered glycosylation site at asparagine 197. In no case, however, were the changes in residue 190 sufficient to confer entry into CD4-negative cells above the level seen for the wild-type ADA envelope glycoproteins.
FIG. 3.
Influence of envelope glycoprotein changes on infection of CD4-negative and CD4-positive cells. An env-deficient, luciferase-expressing HIV-1 provirus was complemented by the wild-type (w.t.) ADA or mutant envelope glycoproteins. The recombinant viruses were used to infect either Cf2Th-CCR5 or Cf2Th-CCR5-CD4 cells. Luciferase activities observed for equivalent amounts of cell lysates are shown. The assay was done in quadruplicate, with average values and standard deviations shown.
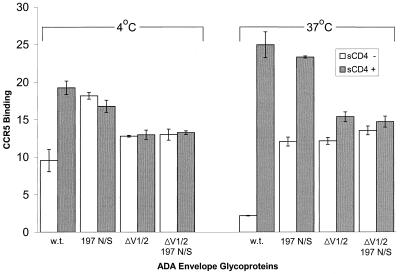
Effects of V1/V2 loop deletion on gp120 binding to CCR5.
Deletion of the V1/V2 variable loops has been shown to result in the exposure of CD4-induced gp120 epitopes thought to be proximal to the chemokine receptor-binding site (73, 74). Examination of the current structural models of gp120 revealed that the carbohydrate at asparagine 197 is unlikely to occlude the chemokine receptor-binding surface directly (39, 75). Hypothesizing that changes in the sugar at position 197 might be mediating the CD4-independent phenotypes through repositioning of the V1/V2 loops, we tested the effects of deleting the V1/V2 loops on CCR5-binding. The right panel of Fig. 4 shows the relative CCR5 binding levels of wild-type and mutant gp120 glycoproteins at 37°C. The deletion of the V1/V2 loops was sufficient to allow gp120 to bind CCR5 efficiently in the absence of CD4. The ΔV1/V2 gp120 glycoprotein bound CCR5 in the absence of CD4 at a level comparable to that seen for 197 N/S gp120. Alteration of asparagine 197 did not affect the level of CD4-independent CCR5 binding exhibited by the ΔV1/V2 gp120. Differences in the migration of the ΔV1/V2 and ΔV1/V2 197 N/Q glycoproteins on SDS-polyacrylamide gels suggest that asparagine 197 is modified in the former protein (data not shown). Thus, with respect to CCR5 binding in the absence of CD4, deletion of the V1/V2 variable loops is functionally equivalent to removal of the sugar moieties at asparagine 197.
FIG. 4.
Independent and combined effects of the 197 N/S change, V1/V2 loop deletion, and temperature on CCR5 binding of gp120. Supernatants containing radiolabeled wild-type (w.t.) and mutant gp120 glycoproteins were preincubated with or without sCD4 (30 μg/ml) at 37°C for 1 h and then added to Cf2Th-CCR5 cells at 4 or 37°C for 2 h. Cells were washed and lysed, and the bound gp120 was analyzed as described in the legend to Fig. 2. The mean values and ranges of duplicate experiments are shown.
The gp120 glycoproteins with the V1/V2 loop deleted did not exhibit significant increases in CCR5 binding following preincubation with sCD4. Although V1/V2 loop deletion results in a twofold decrease in the affinity of the ADA gp120 glycoprotein for CD4 (data not shown), because the CCR5-binding assays were performed in an excess of sCD4 (30 μg/ml), decreases in affinity for CD4 do not account for the lack of induction of CCR5 binding of the V1/V2 deletion-containing gp120 glycoproteins by sCD4.
Effects of V1/V2 loop deletion on the recognition of gp120 by monoclonal antibody 48d.
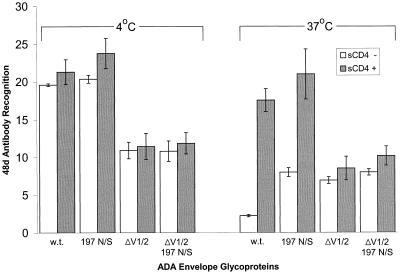
Monoclonal antibody 48d recognizes a conserved, CD4-induced epitope on HIV-1 gp120 near the proposed CCR5-binding region (52, 66, 75). We therefore examined the effects of deletion of the V1/V2 loops on the ability of 48d to precipitate the gp120 variants. At 37°C in the absence of CD4, the recognition of the 197 N/S mutant and the V1/V2 loop deletion-containing variants by 48d was equivalent and was higher than that of the wild-type ADA gp120 glycoprotein (Fig. 5, right panel). Although sCD4 increased the precipitation of the wild-type and 197 N/S gp120 glycoproteins by 48d, insignificant increases were observed for the V1/V2 loop deletion-containing glycoproteins. Thus, the recognition of this panel of gp120 variants by 48d at 37°C closely parallels the pattern observed for CCR5 recognition (Fig. 4).
FIG. 5.
Independent and combined effects of the 197 N/S change, V1/V2 loop deletion, and temperature on 48d recognition of gp120. Approximately 100 μl of supernatants containing radiolabeled wild-type and mutant gp120 glycoproteins were preincubated with or without sCD4 (30 μg/ml) at 37°C for 1 h. The supernatants were incubated with 48d (5 μg/ml) and protein A-Sepharose at 4 or 37°C for 2 h. Precipitated proteins were analyzed using SDS-PAGE and a phosphorimager. The results were normalized for the relative abundance of gp120 in the supernatants, which was determined by a parallel precipitation with a mixture of sera from HIV-1-infected individuals. The mean values and ranges for duplicate experiments are shown.
The effects of lower temperature on gp120 binding to CCR5 and antibodies.
To examine the effects of lower temperature on the gp120 binding characteristics, the interaction of gp120 variants with CCR5 and monoclonal antibodies (including 48d) was examined at 4°C. Compared with CCR5 binding at 37°C, the wild-type ADA gp120 bound better to CCR5 at 4°C in the absence of sCD4 (Fig. 4, left). The binding of wild-type gp120 to CCR5 at 4°C in the absence of sCD4 approached that of the 197 N/S gp120 at 37°C in the absence of sCD4. At 4°C, the 197 N/S change still resulted in an increase in CCR5 binding of full-length gp120 in the absence of sCD4. Thus, the effect of a low temperature on CD4-independent CCR5 binding is not completely equivalent to the effect of the removal of the carbohydrate at asparagine 197. As was seen at 37°C, both V1/V2 loop deletion-containing gp120 glycoproteins bound CCR5 efficiently at 4°C in the absence of sCD4. In the absence of the V1/V2 loops, no additional CCR5-binding phenotype was observed for the 197 N/S change. Binding of the gp120 variants to Cf2Th cells not expressing CCR5 was negligible at 4 and 37°C (data not shown), indicating that all of the observed gp120 binding was CCR5 dependent.
At 4°C, the addition of sCD4 increased the binding of wild-type ADA gp120 to CCR5 but did not increase the binding of the other gp120 variants. This lack of increased binding was attributable neither to the lack of unbound gp120 nor to the saturation of available CCR5 on the target cells (data not shown). Thus, at 4°C, the loss of the carbohydrate at asparagine 197 results in a CCR5-binding affinity comparable to that achieved by the wild-type gp120 in the presence of CD4. As was seen at 37°C, the enhancing effect of CD4 binding on CCR5 binding at 4°C was not observed for the gp120 variants lacking the V1/V2 loops.
At 4°C in the absence of CD4, the recognition of wild-type gp120 by 48d was identical to that of the 197 N/S mutant (Fig. 5). The V1/V2 loop deletion-containing variants were efficiently recognized by 48d at 4°C, although not as well as the glycoproteins with intact variable loops. The addition of sCD4 only minimally increased the precipitation of the wild-type and mutant gp120 glycoproteins at 4°C. The addition of more gp120 or more 48d to these mixtures resulted in increased amounts of precipitated gp120 (data not shown), indicating that the lack of observed increase in 48d binding after sCD4 treatment was due neither to saturation of the 48d antibody nor to lack of free gp120. The ability of CD4-Ig and the monoclonal anti-gp120 antibodies F105 and 17b to precipitate wild-type ADA gp120 did not differ appreciably at 4°C versus 37°C (data not shown). Thus, the binding of one CD4-induced antibody, 48d, parallels CCR5 binding closely, whereas the binding of another CD4-induced antibody, 17b, resembles that of CCR5 less closely. This indicates that differences exist in the way that individual CD4-induced antibodies bind HIV-1 gp120.
Effects of V1/V2 loop deletion and temperature on virus infection.
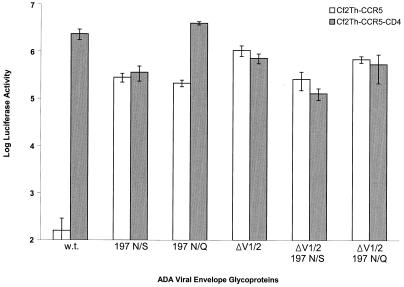
The above results indicated that deletion of the ADA gp120 V1/V2 variable loops confers a degree of CD4-independent CCR5 binding equivalent to that seen for the 197 N/S mutant. To examine the effect of V1/V2 loop deletion on virus infection, recombinant luciferase-encoding viruses containing the ADA HIV-1 envelope glycoprotein variants were used to infect Cf2Th-CCR5 and Cf2Th-CCR5-CD4 cells. The results indicate that the wild-type envelope glycoproteins did not allow efficient infection of Cf2Th-CCR5 cells, whereas all of the other envelope glycoproteins, including those lacking the V1/V2 loops, did (Fig. 6). The efficient infection of these CD4-negative target cells by the viruses with the ΔV1/V2 envelope glycoproteins was not further enhanced by the absence of glycosylation of asparagine 197. All of the viruses efficiently infected the Cf2Th-CCR5-CD4 cells.
FIG. 6.
Effects of gp120 V1/V2 loop deletion on virus infection of CD4-negative and CD4-positive cells. An env-deficient, luciferase-expressing HIV-1 provirus was complemented by the wild-type ADA or mutant envelope glycoproteins. The recombinant viruses were used to infect either Cf2Th-CCR5 or Cf2Th-CCR5-CD4 cells. Luciferase activity observed for equivalent amounts of cell lysates is shown. The assay was done in quadruplicate, with average values and standard deviations shown.
To examine whether CD4-independent infection might be enhanced by low temperature, recombinant viruses with the wild-type ADA envelope glycoproteins were incubated with Cf2Th-CCR5 cells at 4°C for various periods of time prior to incubation at 37°C. The preincubation at 4°C, however, did not result in an increase in infection of these CD4-negative target cells (data not shown).
DISCUSSION
We have investigated the structural changes in the envelope glycoproteins responsible for the conversion of a primary, CCR5-using HIV-1 isolate from a virus dependent on the presence of CD4 on the target cells to a virus that no longer requires CD4 for efficient infection. Remarkably, the absence of N-linked glycosylation at a single site, asparagine 197, in the ADA gp120 glycoprotein is sufficient for CD4-independent binding to CCR5 and for CD4-independent infection of CCR5-expressing cells. Shifting the potential glycosylation site two residues in the N-terminal direction also resulted in CD4 independence. Complex carbohydrates are known to be added to asparagine 197 in mammalian cells (40). Although changes in gp120 glycosylation have been observed in CXCR4-using HIV-1 strains that have been selected for the ability to infect CD4-negative cells (21, 39a), in no instance has the alteration of a single site been shown to be sufficient for CD4 independence. The simplicity of the changes involved in our example provided an opportunity to understand the mechanism underlying CD4 independence. Our data indicate that the absence of carbohydrate modification of asparagine 197 results in an increase in affinity of the ADA gp120 for CCR5 in the absence of CD4. How might this affinity increase arise? Asparagine 197 resides in the conserved V1/V2 stem, which consists of two antiparallel strands linked at both ends by disulfide bonds. The V1 and V2 variable loops, which have been suggested to undergo conformational changes upon CD4 binding (64, 74), project from the conserved V1/V2 stem. The X-ray crystal structure of a ternary complex of the HIV-1 gp120 core, CD4, and a neutralizing antibody (39) provides further insights into the possible consequences of the loss of asparagine 197 glycosylation on CD4 independence. In the ternary complex, the asparagine 197 side chain and associated N-acetylglucosamine project away from the putative chemokine receptor-binding surface of gp120. This makes direct steric masking of the chemokine receptor-binding site of gp120 by the complex carbohydrate residues associated with asparagine 197 unlikely. Indeed, our results demonstrate that the modulation of CCR5-binding affinity by the removal of carbohydrate from asparagine 197 is dependent on the presence of the V1/V2 variable loops. For CCR5 binding in the absence of CD4 and for infection of CD4-negative cells, deletion of the V1/V2 loops of the ADA gp120 glycoprotein resulted in phenotypes that were almost identical to those seen for mutants lacking the asparagine 197 carbohydrate.
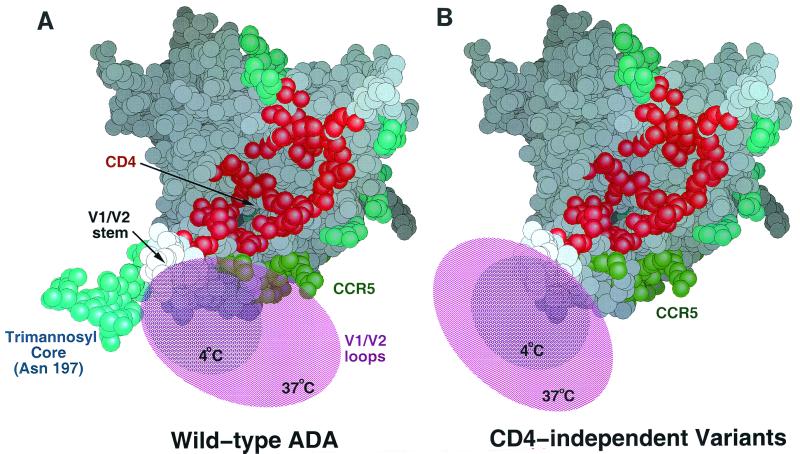
Movement of the V1/V2 loops from their native position is likely to be required for exposure of the CCR5-binding region (52, 64, 74). We propose that the carbohydrate added to asparagine 197 sterically impedes this movement, thereby restricting the flexible loops to a region of space overlapping the CCR5-binding site (Fig. 7A). In the absence of carbohydrate on asparagine 197, the V1/V2 loops are free to assume positions that result in exposure of the gp120 CCR5-binding site (Fig. 7B). Shifting the carbohydrate to asparagine 195 would likewise abrogate its ability to influence the positions of the V1/V2 loops. Our data suggest that the ability of the V1/V2 loops to mask the CCR5-binding region and the 48d epitope on gp120 is more effective at 37°C than at 4°C. An increase in the conformational flexibility of these surface-exposed loops at higher temperatures might contribute to this effect.
FIG. 7.
Proposed model for the mechanism of CD4 independence. (A) The HIV-1 gp120 core, in the CD4-bound conformation (39), is shown. In this orientation, the viral membrane would be at the top of the figure and the target cell membrane would be at the bottom. The gp120 atoms that contact CD4 (center-to-center distance, less than 5 Å) are in red. Carbohydrates are in cyan. The trimannosyl core added to asparagine 197 was modeled based upon the position and orientation of the N-acetylglucosamine, which was resolved in the structure (39). Residues important for CCR5 binding (52, 52a) are in green. Hypothetical spheres of influence of the V1/V2 loops at 4 and 37°C in the absence of CD4 are depicted. (B) The proposed shift in the locations of the V1/V2 loops of HIV-1 ADA gp120 glycoproteins lacking the glycan at asparagine 197 is illustrated. Note the resulting exposure of the gp120 region important for CCR5 binding.
The recognition of the gp120 variants by one of the CD4-induced monoclonal antibodies, 48d, exhibited many parallels with CCR5 binding. By contrast, gp120 binding by another CD4-induced antibody, 17b, did not exhibit the same temperature dependence as did CCR5 and 48d binding. Of the CD4-induced-antibody binding activities examined, 48d binding to HIV-1 gp120 is most effectively competed by sulfated peptides corresponding to the CCR5 N terminus (25a). Like CCR5 binding, recognition of the HIV-1 envelope glycoproteins by 48d is disrupted by deletion of the gp120 V3 loop, even in the presence of sCD4 (73). These observations suggest that 48d and CCR5 bind to closely related gp120 structures. Compared with the CD4-dependent parent virus, a CD4-independent CXCR4-using HIV-1 isolate was demonstrated to exhibit an increase in the binding of the antibody 17b (32a). Exposure of the gp120 surface recognized by the CD4-induced antibodies and by the chemokine receptors may be a common feature of CD4-independent HIV-1 isolates. However, differences in the specific gp120 epitopes that become exposed as a consequence of CD4 independence may relate to the different chemokine receptors utilized or to other virus strain-dependent factors.
CD4 induction of 48d and CCR5 binding to the ADA gp120 envelope glycoprotein was dependent on the presence of the V1/V2 loops. This observation suggests that most of the effects of sCD4 binding on the recognition of the ADA gp120 glycoprotein by these ligands involve adjustments of V1/V2 loop conformation. In the presence of sCD4, the binding of gp120 variants with intact variable loops to the 48d and CCR5 proteins was better than that of gp120 variants lacking the V1/V2 loops. This suggests that sCD4 not only moves the V1/V2 loops from locations that occlude the gp120 binding sites for 48d and CCR5 but also positions the V1/V2 loops to allow them to contribute to the ultimate affinity for ligands that is achieved. Because this CD4-induced, V1/V2 loop-dependent affinity increase was observed for two different ligands, 48d and CCR5, it is less likely to result from direct contact between the V1/V2 variable loops and the ligand. Rather, CD4 binding may allow the newly positioned V1/V2 loops to modulate the conformation of gp120 regions that directly contact CCR5 and 48d.
Our results also suggest an explanation for the appearance of changes in the V2 loop glycosylation site at asparagine 188 in CD4-independent isolates. The CD4-independent gp120 variants with only an alteration in asparagine 197 exhibited decreased CCR5 binding in the presence of sCD4 compared with the wild-type ADA gp120. The CCR5-binding ability of these mutants in the presence of sCD4 could be restored by changes that resulted in the loss of glycosylation of asparagine 188. Apparently, in the absence of the sugar moieties associated with asparagine 197, the asparagine 188 carbohydrate can exert negative effects either on CD4 induction of a CCR5-binding conformation or on the CCR5-binding process per se. These effects were more apparent in the gp120 binding assay than in the virus infectivity assay, but they may have contributed to some advantage for viruses during the selection process, which involved mixtures of CD4-positive and CD4-negative, CCR5-expressing target cells (36a).
Our results underscore the importance of the V1/V2 stem-loop structure in the induction of chemokine receptor-binding conformations by CD4. They also emphasize the contribution of some of the carbohydrate structures on the HIV-1 envelope glycoproteins to the virus entry process. A better understanding of the functionally relevant interactions among protein and carbohydrate components of the HIV-1 envelope should assist intervention.
ACKNOWLEDGMENTS
We acknowledge Raymond Sweet for reagents. We thank Sheri Farnum and Yvette McLaughlin for manuscript preparation.
This work was supported by NIH grants AI24755 and AI41851 and by Center for AIDS Research grant AI28691. We also acknowledge the support of the G. Harold and Leila Mathers Foundation, The Friends 10, Douglas and Judith Krupp, and the late William F. McCarty-Cooper.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC-CKR5: a RANTES, MIP-1a, MIP-1b receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Allan J, Lee T H, McLane M F, Sodroski J, Haseltine W, Essex M. Identification of the major envelope glycoprotein product of HTLV-III. Science. 1983;228:1322–1323. [Google Scholar]
- 3.Allan J S. Receptor-mediated activation of immunodeficiency viruses in viral fusion. Science. 1991;252:1322–1323. doi: 10.1126/science.1925547. [DOI] [PubMed] [Google Scholar]
- 4.Allan J S, Strauss J, Buck D W. Enhancement of SIV infection with soluble receptor molecules. Science. 1990;247:1084–1088. doi: 10.1126/science.2309120. [DOI] [PubMed] [Google Scholar]
- 5.Barre-Sinoussi F, Chermann J C, Rey F, Nugeyre M T, Chamaret S, Gruest J, Daguet C, Axler-Bin C, Vezinet-Brun F, Rouzioux C, Rozenbaum W, Montagnier L. Isolation of a T-lymphocyte retrovirus from a patient at risk for acquired immunodeficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 6.Bieniasz P D, Fridell R, Aramori I, Ferguson S, Caron M, Cullen B. HIV-1-induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR5 co-receptor. EMBO J. 1997;16:2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bugelski P J, Ellens H, Hart T K, Kirsh R L. Soluble CD4 and dextran sulfate mediate release of gp120 from HIV-1: implications for clinical trials. J Acquir Immune Defic Syndr. 1991;4:923–924. [PubMed] [Google Scholar]
- 7a.Cayabyab M, Karlsson G, Etemad-Moghadam B, Hofmann W, Steenbeke T, Halloran M, Fanton J, Axthelm M, Letvin N, Sodroski J. Changes in human immunodeficiency virus type 1 envelope glycoproteins responsible for the pathogenicity of a multiply passaged simian-human immunodeficiency virus (SHIV-HXBc2) J Virol. 1999;73:976–984. doi: 10.1128/jvi.73.2.976-984.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan D C, Fass D, Berger J M, Kim P. Core structure of gp41 from HIV envelope glycoprotein. Cell. 1997;73:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z, Zhou P, Ho D D, Landau N, Marx P. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 11.Clapham P R, McKnight A, Weiss R A. Human immunodeficiency virus type 2 infection and fusion of CD4-negative human cell lines: induction and enhancement by soluble CD4. J Virol. 1992;66:3531–3537. doi: 10.1128/jvi.66.6.3531-3537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clavel F. HIV-2, the West African AIDS virus. AIDS. 1987;1:135–140. [PubMed] [Google Scholar]
- 13.Cocchi F, DeVico A, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 14.Dalgleish A G, Beverly P C L, Clapham P R, Crawford D H, Greaves M F, Weiss R A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 15.Deen K, McDougal J S, Inacker R, Folena-Wasserman G, Arthos J, Rosenberg J, Maddon P, Axel R, Sweet R. A soluble form of CD4 (T4) protein inhibits AIDS virus infection. Nature. 1988;331:82–84. doi: 10.1038/331082a0. [DOI] [PubMed] [Google Scholar]
- 16.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 17.Denisova G, Raviv D, Mondor I, Sattentau Q J, Gershoni J M. Conformational transitions in CD4 due to complexation with HIV-envelope glycoprotein gp120. J Immunol. 1997;158:1157–1164. [PubMed] [Google Scholar]
- 18.Desrosiers R C. The simian immunodeficiency viruses. Annu Rev Immunol. 1990;8:557–578. doi: 10.1146/annurev.iy.08.040190.003013. [DOI] [PubMed] [Google Scholar]
- 19.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Permentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 20.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 21.Dumonceaux J, Nisole S, Chanel C, Quivet L, Amara A, Baleux F, Briand P, Hazan U. Spontaneous mutations in the env gene of the human immunodeficiency virus type 1 NDK isolate are associated with a CD4-independent phenotype. J Virol. 1998;72:512–519. doi: 10.1128/jvi.72.1.512-519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebenbichler C, Westervelt P, Carrillo A, Henkel T, Johnson D, Ratner L. Structure-function relationships of the HIV-1 envelope V3 loop tropism determinant: evidence for two distinct conformations. AIDS. 1993;7:639–646. [PubMed] [Google Scholar]
- 23.Edinger A L, Mankowski J L, Doranz B J, Margulies B J, Lee B, Rucker J, Sharron M, Hoffman T L, Berson J F, Zink M C, Hirsch V M, Clements J E, Doms R W. CD4-independent, CCR5-dependent infection of brain capillary endothelial cells by a neurovirulent simian immunodeficiency virus strain. Proc Natl Acad Sci USA. 1997;94:14742–14747. doi: 10.1073/pnas.94.26.14742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Endres J M, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 25.Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard N, Gerard C, Sodroski J, Choe H. Tyrosine sulfation of the amino-terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96:667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 25a.Farzan M, Vasilieva N, Schnitzler C, Chung S, Robinson J, Gerard N, Gerard C, Choe H, Sodroski J. A tyrosine-sulfated peptide based on the N-terminus of CCR5 interacts with a CD4-enhanced epitope of the HIV-1 gp120 envelope glycoprotein and inhibits HIV-1 entry. J Biol Chem. 2000;275:33516–33521. doi: 10.1074/jbc.M007228200. [DOI] [PubMed] [Google Scholar]
- 26.Fauci A, Macher A, Longo D, Lane H C, Rook A, Masur H, Gelmann E. Acquired immunodeficiency syndrome: epidemiologic, clinical, immunologic, and therapeutic considerations. Ann Intern Med. 1984;100:92–106. doi: 10.7326/0003-4819-100-1-92. [DOI] [PubMed] [Google Scholar]
- 27.Feng Y, Davey R A, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 28.Fisher R, Bertonis J, Meier W, Johnson V, Costopoulos D, Liu T, Tizard R, Walder B, Hirsch M, Schooley R, Flavell R. HIV infection is blocked in vitro by recombinant soluble CD4. Nature. 1988;331:76–78. doi: 10.1038/331076a0. [DOI] [PubMed] [Google Scholar]
- 29.Fu Y K, Hart T K, Jonak Z L, Bugelski P J. Physicochemical dissociation of CD4-mediated syncytium formation and shedding of human immunodeficiency virus type 1 gp120. J Virol. 1993;67:3818–3825. doi: 10.1128/jvi.67.7.3818-3825.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallo R C, Salahuddin S Z, Popovic M, Shearer G M, Kaplan M, Haynes B F, Palker T J, Redfield R, Oleske J, Safai B, White G, Foster P, Markham P D. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science. 1984;224:500–503. doi: 10.1126/science.6200936. [DOI] [PubMed] [Google Scholar]
- 31.Hart T K, Kirsh R, Ellens H, Sweet R W, Lambert D M, Petteway S R, Jr, Leary J, Bugelski P J. Binding of soluble CD4 proteins to human immunodeficiency virus type 1 and infected cells induces release of envelope glycoprotein gp120. Proc Natl Acad Sci USA. 1991;88:2189–2193. doi: 10.1073/pnas.88.6.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Helseth E, Kowalski M, Gabuzda D, Olshevsky U, Haseltine W, Sodroski J. Rapid complementation assays measuring replicative potential of human immunodeficiency virus type 1 envelope glycoprotein mutants. J Virol. 1990;64:2416–2420. doi: 10.1128/jvi.64.5.2416-2420.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Hoffman T, LaBranche C, Zhang W, Canziani G, Robinson J, Chaiken I, Hoxie J, Doms R. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope glycoprotein. Proc Natl Acad Sci USA. 1999;96:6359–6364. doi: 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hussey R, Richardson N, Kowalski M, Brown N, Change H, Siliciano R, Dorfman T, Walker B, Sodroski J, Reinherz E. A soluble CD4 protein selectively inhibits HIV replication and syncytium formation. Nature. 1988;331:78–81. doi: 10.1038/331078a0. [DOI] [PubMed] [Google Scholar]
- 34.Kanki P, McLane M, King N, Essex M. Serological identification and characterization of a macaque T-lymphotropic retrovirus closely related to HTLV-III. Science. 1985;228:1199–1422. doi: 10.1126/science.3873705. [DOI] [PubMed] [Google Scholar]
- 35.Kirsh R, Hart T, Ellens H, Miller J, Petteway S, Lambert D, Leary B, Bugelski P. Morphometric analysis of recombinant soluble CD4-mediated release of the envelope glycoprotein gp120 from HIV-1. AIDS Res Hum Retrovir. 1990;6:1209–1212. doi: 10.1089/aid.1990.6.1209. [DOI] [PubMed] [Google Scholar]
- 36.Klatzmann D, Champagne E, Charmaret S, Gruest J, Guetard D, Hercend T, Glueckman J-C, Montagnier L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 36a.Kolchinsky P, Mirzabekov T, Farzan M, Kiprilov E, Cayabyab M, Mooney L J, Choe H, Sodroski J. Adaptation of a CCR5-using, primary human immunodeficiency virus type 1 isolate for CD4-independent replication. J Virol. 1999;73:8120–8126. doi: 10.1128/jvi.73.10.8120-8126.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korber B, Foley B, Kuiken C, Pillai S, Sodroski J. Numbering positions in HIV relative to HXB2. In: Korber B, Kuiken C, Foley B, Hahn B, McCutchan F, Mellors J, Sodroski J, editors. Human retroviruses and AIDS 1998. Los Alamos, N.Mex: Los Alamos National Laboratory; 1998. pp. III102–III111. [Google Scholar]
- 38.Korber B, Foley B, Leitner T, McCutchan F, Hahn B, Mellors J, Myers G, Kuiken C, editors. Human retroviruses and AIDS 1997. Los Alamos, N.Mex: Los Alamos National Laboratory; 1997. [Google Scholar]
- 39.Kwong P D, Wyatt R, Robinson J, Sweet R, Sodroski J, Hendrickson W. Structure of an HIV-1 gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.LaBranche C, Hoffmann T, Romano J, Haggarty B, Edwards T, Matthews T, Doms R, Hoxie J. Determinants of CD4 independence for a human immunodeficiency virus type 1 variant map outside regions for coreceptor specificity. J Virol. 1999;73:10310–10319. doi: 10.1128/jvi.73.12.10310-10319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leonard C, Spellman M, Riddle L, Harris R, Thomas J, Gregory T. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem. 1990;265:10373–10382. [PubMed] [Google Scholar]
- 41.Letvin N L, Daniel M D, Sehgal P K, Desrosiers R C, Hunt R D, Waldron L M, MacKey J J, Schmidt D K, Chalifoux L V, King N W. Induction of AIDS-like disease in macaque monkeys with T-cell tropic retrovirus STLV-III. Science. 1985;230:71–73. doi: 10.1126/science.2412295. [DOI] [PubMed] [Google Scholar]
- 42.Maddon P J, Dalgleish A G, McDougal J S, Clapham P R, Weiss R A, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 43.Marcon L, Choe H, Martin K A, Farzan M, Ponath P D, Wu L, Newman W, Gerard N, Gerard G, Sodroski J. Utilization of C-C chemokine receptor 5 by the envelope glycoproteins of a pathogenic simian immunodeficiency virus (SIVmac239) J Virol. 1997;71:2522–2527. doi: 10.1128/jvi.71.3.2522-2527.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin K A, Wyatt R, Farzan M, Choe H, Marcon L, Desjardins E, Robinson J, Sodroski J, Gerard C, Gerard N P. CD4-independent binding of SIV gp120 to rhesus CCR5. Science. 1997;278:1470–1473. doi: 10.1126/science.278.5342.1470. [DOI] [PubMed] [Google Scholar]
- 45.Moore J, McKeating J, Weiss R, Sattentau Q. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science. 1990;250:1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 46.Moore J P, Klasse P J. Thermodynamic and kinetic analysis of sCD4 binding to HIV-1 virions and of gp120 dissociation. AIDS Res Hum Retrovir. 1992;8:443–450. doi: 10.1089/aid.1992.8.443. [DOI] [PubMed] [Google Scholar]
- 47.Moore J P, McKeating J A, Norton W A, Sattentau Q J. Direct measurement of soluble CD4 binding to human immunodeficiency virus type 1 virions: gp120 dissociation and its implications for virus-cell binding and fusion reactions and their neutralization by soluble CD4. J Virol. 1991;65:1133–1140. doi: 10.1128/jvi.65.3.1133-1140.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore J P, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J Virol. 1996;70:1863–1872. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore J P, Sattentau Q J, Wyatt R, Sodroski J. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J Virol. 1994;68:469–484. doi: 10.1128/jvi.68.1.469-484.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morrison H, Kirchhoff F, Desrosiers R. Effects of mutations in constant regions 3 and 4 of envelope of simian immunodeficiency virus. Virology. 1995;210:448–455. doi: 10.1006/viro.1995.1361. [DOI] [PubMed] [Google Scholar]
- 50a.Myszka D, Sweet R, Hensley P, Brigham-Burke M, Kwong P, Hendrickson W, Wyatt R, Sodroski J, Doyle M. Energetics of the HIV gp120-CD4 binding reaction. Proc Natl Acad Sci USA. 2000;97:9026–9031. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olshevsky U, Helseth E, Furman C, Li J, Haseltine W, Sodroski J. Identification of individual human immunodeficiency virus type 1 gp120 amino acids important for CD4 receptor binding. J Virol. 1990;64:7025–7031. doi: 10.1128/jvi.64.12.5701-5707.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rizzuto C, Wyatt R, Hernández-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 52a.Rizzuto C, Sodroski J. Fine definition of a conserved CCR5-binding region on human immunodeficiency virus type 1 gp120 glycoprotein. AIDS Res Human Retrovir. 2000;16:741–749. doi: 10.1089/088922200308747. [DOI] [PubMed] [Google Scholar]
- 53.Robey W G, Safai B, Oroszlan S, Arthur L, Gonda M, Gallo R, Fischinger P J. Characterization of envelope and core structural gene products of HTLV-III with sera from AIDS patients. Science. 1985;228:593–595. doi: 10.1126/science.2984774. [DOI] [PubMed] [Google Scholar]
- 54.Sattentau Q J, Zolla-Pazner S, Poignard P. Epitope exposure on functional, oligomeric HIV-1 gp41 molecules. Virology. 1995;206:713–717. doi: 10.1016/s0042-6822(95)80094-8. [DOI] [PubMed] [Google Scholar]
- 55.Sattentau Q J, Moore J P, Vignaux F, Traincard F, Poignard P. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J Virol. 1993;67:7383–7393. doi: 10.1128/jvi.67.12.7383-7393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sattentau Q J, Moore J P. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schenten D, Marcon L, Karlsson G, Parolin C, Kodama T, Gerard N, Sodroski J. Effects of soluble CD4 on simian immunodeficiency virus infection of CD4-positive and CD4-negative cells. J Virol. 1999;73:5373–5380. doi: 10.1128/jvi.73.7.5373-5380.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schutten M, Andeweg A C, Bosch M L, Osterhaus A D. Enhancement of infectivity of a non-syncytium inducing HIV-1 by sCD4 and by human antibodies that neutralize syncytium inducing HIV-1. Scand J Immunol. 1995;41:18–22. doi: 10.1111/j.1365-3083.1995.tb03528.x. [DOI] [PubMed] [Google Scholar]
- 59.Smith D, Byrn R, Marsters S, Gregory T, Groopman J, Capon D. Blocking of HIV-1 infectivity by a soluble secreted form of the CD4 antigen. Science. 1987;238:1704–1707. doi: 10.1126/science.3500514. [DOI] [PubMed] [Google Scholar]
- 60.Speck R, Wehrly K, Platt E, Atchison R, Charo I, Kabat D, Chesebro B, Goldsmith M. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stein B S, Gouda S, Lifson J, Penhallow R, Bensch K, Engelman E. pH-independent HIV entry into CD4-positive T cells via virus envelope fusion to the plasma membrane. Cell. 1987;49:659–668. doi: 10.1016/0092-8674(87)90542-3. [DOI] [PubMed] [Google Scholar]
- 62.Strebel K, Daugherty D, Clouse K, Cohen D, Folks T, Martin M. The HIV ‘A’ (sor) gene product is essential for virus infectivity. Nature. 1987;328:728–730. doi: 10.1038/328728a0. [DOI] [PubMed] [Google Scholar]
- 63.Sullivan N, Sun Y, Li J, Hofmann W, Sodroski J. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J Virol. 1995;69:4413–4422. doi: 10.1128/jvi.69.7.4413-4422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore J, Sodroski J. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J Virol. 1998;72:4694–4703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thali M, Furman C, Helseth E, Repke H, Sodroski J. Lack of correlation between soluble CD4-induced shedding of the human immunodeficiency virus type 1 exterior envelope glycoprotein and subsequent membrane fusion events. J Virol. 1992;66:5516–5524. doi: 10.1128/jvi.66.9.5516-5524.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thali M, Moore J P, Furman C, Charles M, Ho D D, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Traunecker A, Luke W, Karjalainen K. Soluble CD4 molecules neutralize human immunodeficiency virus type 1. Nature. 1988;331:84–86. doi: 10.1038/331084a0. [DOI] [PubMed] [Google Scholar]
- 68.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its coreceptor CCR5. Nature. 1996;384:184–186. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 69.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 70.Werner W, Levy J. Human immunodeficiency virus type 1 envelope gp120 is cleaved after incubation with recombinant soluble CD4. J Virol. 1993;67:2566–2574. doi: 10.1128/jvi.67.5.2566-2574.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Willey R L, Martin M A, Peden K W. Increase in soluble CD4 binding to and CD4-induced dissociation of gp120 from virions correlates with infectivity of human immunodeficiency virus type 1. J Virol. 1994;68:1029–1039. doi: 10.1128/jvi.68.2.1029-1039.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 73.Wyatt R, Sullivan N, Thali M, Repke H, Ho D, Robinson J, Posner M, Sodroski J. Functional and immunological characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J Virol. 1993;67:4557–4565. doi: 10.1128/jvi.67.8.4557-4565.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wyatt R, Moore J, Accola M, Desjardins E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wyatt R, Kwong P D, Desjardins E, Sweet R, Robinson J, Hendrickson W, Sodroski J. The antigenic structure of the human immunodeficiency virus gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]