Abstract
Background
In the era of tyrosine kinase inhibitor (TKI) treatment, the progression of chronic myeloid leukemia (CML) remains a significant clinical challenge, and genetic biomarkers for the early identification of CML patients at risk for progression are limited. This study explored whether essential circular RNAs (circRNAs) can be used as biomarkers for diagnosing and monitoring CML disease progression and assessing CML prognosis.
Methods
Peripheral blood (PB) samples were collected from 173 CML patients (138 patients with chronic phase CML [CML-CP] and 35 patients with accelerated phase/blast phase CML [CML-AP/BP]) and 63 healthy controls (HCs). High-throughput RNA sequencing (RNA-Seq) was used to screen dysregulated candidate circRNAs for a circRNA signature associated with CML disease progression. Quantitative real-time PCR (qRT-PCR) was used for preliminary verification and screening of candidate dysregulated genes, as well as subsequent exploration of clinical applications. Receiver operating characteristic (ROC) curve analysis, Spearman’s rho correlation test, and the Kaplan-Meier method were used for statistical analysis.
Results
The aberrant expression of hsa_circ_0006010 and hsa_circ_0002903 during CML progression could serve as valuable biomarkers for differentiating CML-AP/BP patients from CMP-CP patients or HCs. In addition, the expression levels of hsa_circ_0006010 and hsa_circ_0002903 were significantly associated with the clinical features of CML patients but were not directly related to the four scoring systems. Furthermore, survival analysis revealed that high hsa_circ_0006010 expression and low hsa_circ_0002903 expression indicated poor progression-free survival (PFS) in CML patients. Finally, PB hsa_circ_0006010 and hsa_circ_0002903 expression at diagnosis may also serve as disease progression surveillance markers for CML patients but were not correlated with PB BCR-ABL1/ABL1IS.
Conclusions
Our study demonstrated that PB levels of hsa_circ_0006010 and hsa_circ_0002903 may serve as novel diagnostic, surveillance, and prognostic biomarkers for CML disease progression and may contribute to assisting in the diagnosis of CML patients at risk for progression and accurate management of advanced CML patients.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-024-12943-x.
Keywords: Hsa_circ_0006010, Hsa_circ_0002903, Chronic myeloid leukemia, Disease progression, Diagnosis, Surveillance, Prognostic biomarkers
Introduction
Chronic myeloid leukemia (CML) is a clonal myeloproliferative tumor characterized by balanced chromosomal ectopic t(9; 22)(q34; q11.2), leading to the production of the breakpoint cluster region-Abelson leukemia (BCR-ABL1) fusion gene [1]. CML has traditionally been divided into three phases: the chronic phase (CP), accelerated phase (AP), and blast phase (BP). The application of tyrosine kinase inhibitors (TKIs) has significantly improved treatment outcomes in CML patients, particularly disease duration and quality of life [2, 3]. However, a proportion of CML-CP patients still progress to the advanced (AP or BP) phase and often have poor outcomes [4]. Unfortunately, the exact mechanisms driving progression to the advanced stage of CML are complex and poorly understood [5, 6]. The management of advanced CML remains a pivotal clinical challenge even in the era of TKI treatment [4, 7]. Therefore, early and accurate detection of CML progression and disease monitoring are essential for timely intervention guidance and directly affect patient survival.
At present, four main traditional CML risk scoring systems are used: the Sokal score, Hasford score, EURO score, and the newer EUTOS long-term survival (ELTS) score [1]. These systems use different patient and clinical parameters, excluding cytogenetic and genomic variables. Importantly, these risk scoring systems are primarily used to predict the long-term survival of CML patients rather than the risk of progression to advanced stages [8–10]. The ELTS score more accurately predicts CML-related death, which is strongly associated with the transition to advanced disease [11, 12]. The risk factors for progression to advanced CML vary among different risk scoring tools and include genomic, epigenetic, and other intracellular and extracellular changes [4]. In particular, gene acquisition mutations play a crucial role in the progression of CML [13]. Genomic research has the potential to lay the foundation for the classification of diagnostic risks based on clinical parameters and genomic changes [14]. Owing to the lack of reliable predictors for the risk of disease progression [7], identifying additional novel diagnostic and prognostic biomarkers for CML disease progression is urgently needed.
Recently, studies have shown that circular RNAs (circRNAs) may play a vital role in the development, progression, recurrence, and resistance of diverse human cancers, including leukemia, that are often accompanied by dysregulation of circRNAs [15]. CircRNAs are evolutionarily conserved, are abundant, have global and extracellular stability, and exhibit tissue or pathological condition-specific expression [16, 17]. Due to their prominent characteristics, circRNAs may play important roles as diagnostic and prognostic markers for leukemia [18]. Some circRNAs related to CML resistance and treatment have recently been identified. For example, Pan Y et al. reported that circ-BA9.3 is involved in TKI resistance and may be a potential target for treating TKI-resistant CML patients [19]. Similarly, circ_0080145 promotes imatinib resistance in CML by regulating the miR-326/PPFIA1 axis, which may provide a new approach for CML therapy [20]. According to another study, hsa_circ_0058493 may be a novel biomarker for imatinib-resistant CML [21]. Other studies have identified the carcinogenic effect of F-circBA1 on CML cells [22] and circHIPK3 as a prognostic marker that promotes CML progression [23]. Studies have shown that circRNAs are associated with the development of CML resistance and CML progression, which suggests new opportunities for CML treatment [24]. However, the role of circRNAs as potential biomarkers for the early detection and monitoring of CML progression and for predicting progression-free survival (PFS) during CML treatment follow-up is still unclear.
During the follow-up period, peripheral blood (PB) is used to evaluate the effectiveness of TKIs in treating CML patients [25, 26]. Studies have also shown that PB is a better sample source than bone marrow (BM), and lnc-LOC in PB can serve as a novel noninvasive biomarker for treating and monitoring acute promyelocytic leukemia [27]. Thus, we hypothesize that dysregulated circRNAs associated with disease progression in PB could be ideal noninvasive biomarkers for diagnosing and monitoring CML disease progression and evaluating CML prognosis.
Based on previous studies, we systematically analyzed the circRNA signature in advanced CML using RNA sequencing (RNA-Seq) and identified six candidate circRNAs associated with CML disease progression (GEO accession: GSE212254). Using quantitative real-time PCR (qRT-PCR), we identified PB hsa_circ_0006010 and hsa_circ_0002903 as the most significantly upregulated and downregulated circRNAs, respectively, in CML patients with disease progression. We further evaluated the clinical value of PB hsa_circ_0006010 and hsa_circ_0002903 in diagnosing, monitoring, and determining the prognosis of CML disease progression. This study aimed to identify novel diagnostic, surveillance, and prognostic biomarkers for disease progression in CML patients, contributing to the precise management of advanced CML.
Materials and methods
Patient profiles
In this study, 173 CML patients (comprising 35 CML-AP/BP patients and 138 CML-CP patients) who met the European LeukemiaNet (ELN) 2013 criteria [28] were recruited from January 2009 to December 2022 at The Second Affiliated Hospital and Yuying Children’s Hospital of Wenzhou Medical University. Patients were over 18 years of age and received any TKI as initial therapy. The stage of CML was assessed according to the ELN 2013 criteria [28]. CML patients who received induction treatment, follow-up treatment, or post-TKI resistance therapy and those who achieved molecular complete remission (CR), were defined according to the ELN recommendations [29]. Patients with prior blood transfusion and hematopoietic stem cell transplantation were excluded from the study. Moreover, 63 age- and sex-matched individuals who were free of hematologic diseases, other types of malignancies and any chronic diseases were recruited as healthy controls (HCs).
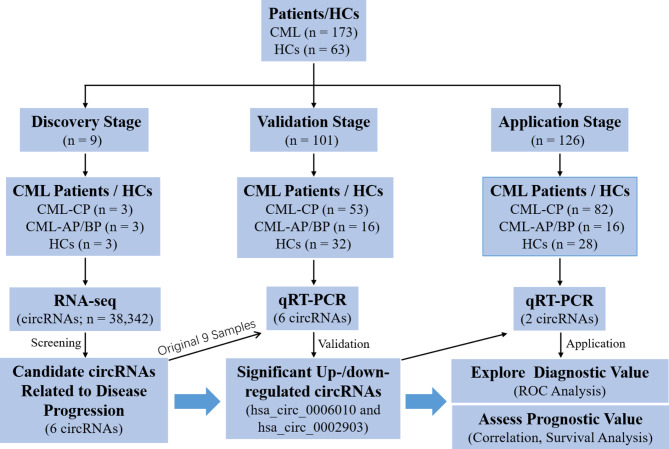
PB samples were collected from all CML-CP patients at diagnosis, CML-AP/BP patients either at diagnosis or during therapy, and HCs. At the discovery phase, 9 PB samples (3 each from CML-CP patients, CML-AP/BP patients, and HCs) were subjected to high-throughput RNA-Seq. In addition, 101 and 126 PB samples were allocated to the validation and application phases, respectively (Fig. 1). The characteristics of the CML patients in the validation and application phases are presented in Table 1, and the clinical characteristics of HCs are presented in Supplementary Table 1. Informed consent was obtained from all participants, and approval was obtained from the Ethics Committee of The Second Affiliated Hospital and Yuying Children’s Hospital of Wenzhou Medical University.
Fig. 1.
Flow chart of the study design
Table 1.
Characteristics of CML patients in the validation and application cohorts
| Characteristics | Total (n = 167) | Validation cohort (n = 69) | Application cohort (n = 98) | p value |
|---|---|---|---|---|
| Ages, median (range) | 51 (13–81) | 52 (13–77) | 54 (34–81) | 0.453 |
| Gender male (n), (%) | 90 (53.89) | 38 (55.07) | 52 (53.06) | 0.875 |
| WBC counts (×109/L), median (range) | 101.45 (13.19-726.73) | 98.67 (13.19-666.88) | 106.82 (13.69-726.73) | 0.296 |
| Hemoglobin (g/L), median (range) | 109 (54–163) | 108 (54–163) | 110.50 (55–159) | 0.345 |
| Platelet counts (×109/L), median (range) | 321 (67-2300) | 251.50 (67-2300) | 348 (138–2107) | 0.256 |
| Blast (%) (PB), median (range) | 3 (0–42) | 3 (0–42) | 4 (0–35) | 0.197 |
| Eosinophil (%) (PB), median (range) | 3 (0–11) | 3 (0–9) | 3 (0–11) | 0.678 |
| Basophil (%) (PB), median (range) | 4 (0–21) | 4 (0–21) | 3 (0–19) | 0.732 |
| RDW-CV (%) | 14.90 (11.80–19.80) | 14.75 (12.80–19.80) | 15.10 (11.80–19.40) | 0.488 |
| Splenomegaly (n), (%) | 103 (60.48) | 46 (66.67) | 55 (56.12) | 0.200 |
| Phase at diagnosis | 0.319 | |||
| CP (n), (%) | 135 (80.84) | 53 (76.81) | 82 (83.67) | |
| AP/BP (n), (%) | 32 (19.16) | 16 (23.19) | 16 (16.33) | |
| Initial treatment | < 0.001 | |||
| Imatinib (n), (%) | 144 (86.23) | 69 (100%) | 75 (76.53%) | |
| Other TKIs (n), (%) | 23 (13.77) | 0 | 23 (23.47%) |
Sample collection and processing
Blood samples were collected and treated as follows: 2 ml PB samples were extracted from the median cubital vein of each subject and placed in an EDTA-K2 anticoagulant vacuum container. The PB mononuclear cells (PBMCs) were prepared by Ficoll Hypaque gradient centrifugation. Subsequently, PBMCs were resuspended in 1 ml of TRIzol® (Invitrogen, USA) and stored at -70 °C for subsequent analysis.
RNA-Seq
RNA-Seq of circRNAs was performed by Novogene Co., Ltd. (Beijing, China). The operating procedure was briefly described as follows: total RNA was extracted from nine samples using TRIzol® (Invitrogen, USA) reagent and purified using a Ribo-ZeroTM rRNA Removal Kit (Epicenter, Madison, WI, USA). Single-stranded cDNA and double-stranded cDNA were synthesized successively by reverse transcription. Subsequently, the double-stranded cDNA was purified, and the terminal was repaired. According to the RNA species, sequence amplification and purification were conducted through PCR for library construction by adding primers. A quality inspection of the library was carried out, and nine samples were subsequently sequenced using the Illumina HiSeqTM 2500 sequencing platform (Illumina, San Diego, CA, USA). Bioinformatics analysis of the raw sequence data was performed, and the results were annotated using an integrated transcript database.
RNA-Seq data analysis
Fast QC software (v0.11.2) was used to evaluate the quality of the raw data. Low-quality data were filtered using NGSQC software (v2.3.2). High-quality clean reads were compared with the reference genome using TopHat (v2.0.9). The read codes of each gene were counted using HTSeq software (v0.5.3p9). DESeq (v1.12.0) was used for differential expression analysis between the two groups. Genes with a p value < 0.05 were considered differentially expressed. GOseq, topGO, hmmscan (release 2.12), and KOBAS (v2.0) were used for functional annotation and pathway enrichment analysis of differentially expressed genes. Finally, the circRNA signature associated with disease progression in CML patients was annotated in the NCBI GEO database (GSE212254).
RNA reverse transcription and qRT-PCR validation
Total RNA was extracted and eluted with 50 µL of RNase-free water and then evaluated and quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, USA). Total RNA was reverse transcribed into cDNA using the PrimeScript™ RT reagent kit (Takara, Japan). qRT-PCR was performed using TB Green® Premix Ex Taq™ II (Takara, Japan) on an ABI7500 qRT-PCR system (Applied Biosystems, USA). The comparative cycle threshold (Ct) method was used to analyze gene expression levels. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control. The primer sequences are shown in Supplementary Table 2.
Assessment of the four risk scoring systems
Four main risk scoring systems, including the Sokal score, Hasford score, EURO score, and ELTS score, are used to evaluate CML prognosis. Recording the components of the four risk scoring systems was mandatory at the validation stage. The demographic and clinical information required for the risk scoring systems was extracted from medical records. CML patients in the application stage were grouped according to different scoring systems, and the Sokal score [10], Hasford score [8], EUTOS score [30], and ELTS score [31] at diagnosis were calculated as described previously.
Statistical analysis
The statistical analysis was performed with SPSS V.26.0 (SPSS, USA), and figures were produced using GraphPad Prism 8.0.1 (GraphPad Software, USA). Significant differences in sequencing data between the two groups were estimated by t tests. CircRNAs with a |fold change (FC)| > 2 and a p value < 0.05 were considered differentially expressed. If the data were normally distributed, a t test was used for comparisons between groups. If the data exhibited a nonnormal distribution, a nonparametric Mann-Whitney U test was used to analyze the measurement data. Receiver operating characteristic (ROC) curve analysis was used to assess the diagnostic value of the circRNAs. PFS was calculated as the time from TKI therapy initiation to progression, death from any cause, or censoring at the last follow-up using the Kaplan-Meier (K-M) estimator and log-rank tests. Moreover, Spearman’s rho correlation test and chi-square test were applied to analyze the correlation among circRNA expression, risk score, and clinical characteristics in CML patients. A p value < 0.05 was considered to indicate statistical significance.
Results
Profile of circRNA expression in advanced CML patients
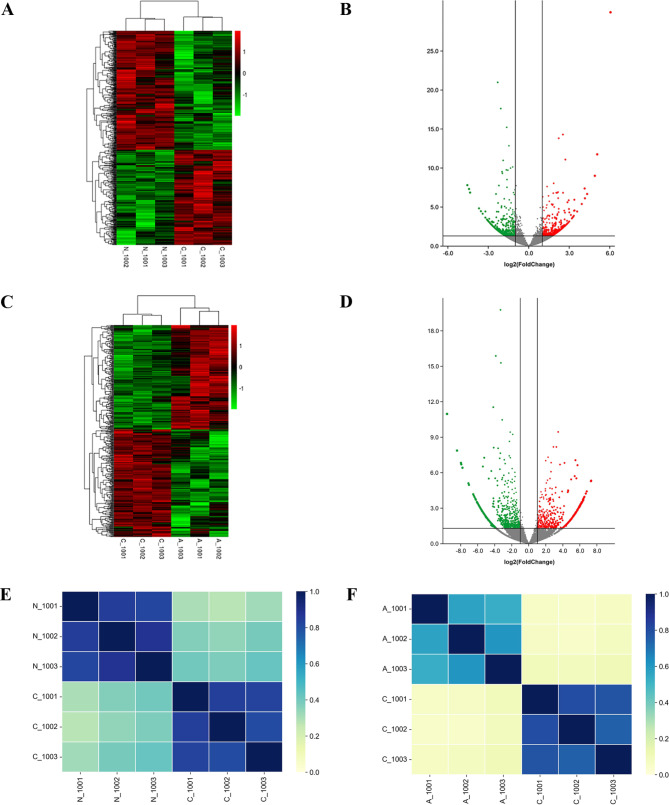
To characterize CML-related circRNAs, CML patients and HCs were subjected to RNA-Seq analysis. The detailed clinical data of the patients are shown in Supplementary Table 3. A total of 38,342 distinct circRNAs were detected. The raw RNA-Seq data were submitted to the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE212254). To determine the specific expression of circRNAs under pathological conditions, differential expression analysis was performed for different stages of CML. First, we selected 1154 differentially expressed circRNAs between CML-CP patients and HCs by FC filtering (|FC| > 2) with a p value < 0.05. Compared with those in HCs, there were 459 upregulated circRNAs and 695 downregulated circRNAs in CML-CP patients (Fig. 2A, B). The analysis also identified 1848 differentially expressed circRNAs between the CML-AP/BP and CML-CP groups according to the same statistical inclusion criteria. Compared with those in CML-CP, there were 581 upregulated circRNAs and 1267 downregulated circRNAs in CML-AP/BP (Fig. 2C, D). Then, we evaluated the relevant heatmaps for samples in the different subgroups. The results showed that all comparison groups had strong classification properties, but the intragroup differences were slight (Fig. 2E, F). The differential expression results indicated that there were more changes in the expression of circRNAs between CML-AP/BP and CML-CP patients than between CML-CP patients and HCs.
Fig. 2.
Profile of circRNA expression in CML. a Differentially expressed circRNA heatmap between CML-CP patients and HCs. b Volcano plot of differentially expressed circRNAs between CML-CP patients and HCs. c Differentially expressed circRNA heatmap between CML-CP and CML-AP/BP patients. d Volcano plot of differentially expressed circRNAs between CML-CP and CML-AP/BP patients. e Relevant heatmap results between CML-CP patients and HCs. f Relevant heatmap results between CML-CP and CML-AP/BP patients. Note: a, c On the left, circRNAs are clustered according to expression similarity. At the top of the figure, each sample is clustered according to the similarity of the expression spectrum, with the clustering intensity increasing from green to red. b, d The two vertical black lines represent twofold up- and downregulation, and the horizontal black lines symbolize a p value of 0.05. e, f The relative intensity increased from yellow to blue. N_1001, N_1002, and N_1003 represent 3 HCs; C_1001, C_1002, and C_1003 represent 3 CML-CP patients; and A_1001, A_1002, and A_1003 represent 3 CML-AP/BP patients
Selection of a circRNA signature related to CML disease progression
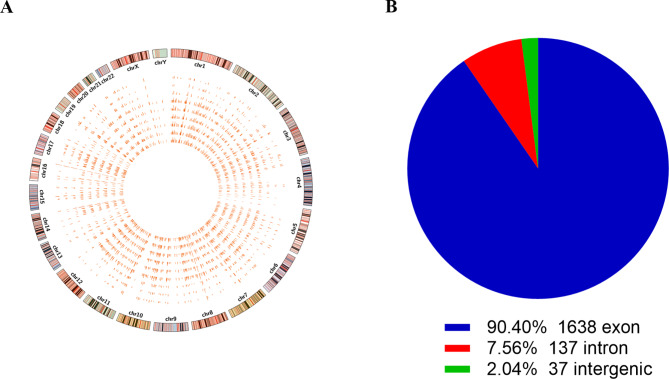
Considering that circRNA changes between CML-AP/BP and CML-CP patients were more prominent than those between CML-CP patients and HCs, we further focused on the circRNA signature associated with disease progression. A Circos plot revealed that these circRNAs were widely distributed on all chromosomes (Fig. 3A). Most circRNAs were derived from exons (90.4%) (Fig. 3B). circRNAs with a high raw signal intensity of expression were selected first. Based on the inclusion criteria (|FC| >2, p value < 0.001, and potential coding function), three upregulated and three downregulated circRNAs were chosen for further validation. Information on the six candidate circRNAs is shown in Table 2.
Fig. 3.
Selection of candidate circRNAs related to CML disease progression. a Circos plot displaying the distribution of significantly dysregulated circRNAs between CML-AP/BP and CML-CP patients. The outermost layer is the human chromosome map. The inner 9 circles represent each sample analyzed by RNA-seq. The bar chart shows the expression levels of circRNAs. b Classification of dysregulated circRNAs according to genomic origin
Table 2.
Information on the six candidate circRNAs
| circRNA | Regulation in CML | r value | Fold change | circRNA type | Chrom | Best transcript | Gene symbol |
|---|---|---|---|---|---|---|---|
| hsa_circ_0001523 | Up | 0.00005 | 3.668 | exon | Chr5 | NM_020747 | ZNF608 |
| hsa_circ_0066971 | Up | 0.00004 | 2.627 | exon | Chr3 | NM_018456 | EAF2 |
| hsa_circ_0006010 | Up | 0.00061 | 2.204 | exon | Chr7 | NR_036680 | DPY19L1P1 |
| hsa_circ_0000095 | Down | < 0.0001 | -3.873 | exon | Chr1 | NM_001199691 | TMEM56RWDD3 |
| hsa_circ_0001801 | Down | < 0.0001 | -2.181 | exon | Chr8 | NM_052937 | PCMTD1 |
| hsa_circ_0002903 | Down | 0.0002 | -2.348 | exon | Chr21 | NM_006031 | PCNT |
Validation of differentially expressed circRNAs in CML-AP/BP patients
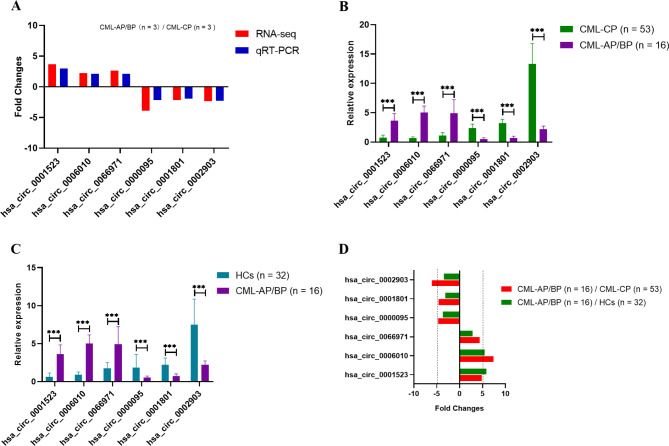
To verify the RNA-Seq data, qRT-PCR was performed on identical samples (n = 9) from CML patients and HCs. The six candidate circRNAs were dysregulated and exhibited the same trend of dysregulation as that shown in the RNA-Seq results (Fig. 4A). Subsequently, we performed qRT-PCR on a validation cohort (n = 101) of CML patients and HCs. PB hsa_circ_0006010 and hsa_circ_0002903 in CML-AP/BP patients were the most significantly downregulated and upregulated circRNAs, respectively, among the six candidates compared with the levels in CML-CP patients or HCs (Fig. 4B, C). Similarly, the FC values calculated by the ratio of the mean or median relative expression of hsa_circ_0006010 and hsa_circ_0002903 were also the highest or lowest in the two comparison groups (CML-AP/BP /CML-CP and CML-AP/BP /HCs) (Fig. 4D). Meanwhile, we explored the expression levels of the six candidate circRNAs in both CML-CP and HCs groups. The six candidate circRNAs were slightly dysregulated (|FC| < 2) and exhibited the same trend of dysregulation as the RNA-Seq results in the CML-CP /HCs group (Supplementary Fig. 1A). In addition, although qRT-PCR validation results showed that 4 of the 6 candidate circRNAs were differentially expressed in the comparison between CML-CP and HCs (Supplementary Fig. 1B), the FC values calculated by the ratio of the mean or median relative expression of the six circRNAs were |FC| < 2 (Supplementary Fig. 1C). Therefore, we selected the two most differentially expressed circRNAs (hsa_circ_0006010 and hsa_circ_0002903) and investigated whether they could serve as new biomarkers for the diagnosis of CML progression and the assessment of CML prognosis.
Fig. 4.
qRT-PCR validation of differentially expressed circRNAs in CML patients. a qRT-PCR validation and RNA-Seq analysis of dysregulated genes. b qRT-PCR validation of six circRNAs between CML-AP/BP and CML-CP patients. c qRT-PCR validation of six circRNAs between CML-AP/BP patients and HCs. d FC values of hsa_circ_0006010 and hsa_circ_0002903 expression when comparing CML-AP/BP/CML-CP and CML-AP/BP/HCs. *** p < 0.001
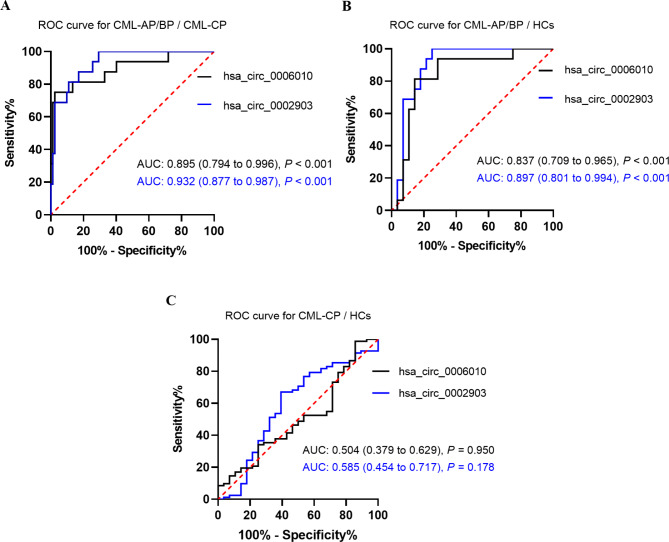
PB hsa_circ_0006010 and hsa_circ_0002903 as biomarkers for the diagnosis of CML disease progression
We further validated another sample cohort to explore the diagnostic value of PB hsa_circ_0006010 and hsa_circ_0002903. The expression of the two candidate circRNAs was determined by qRT-PCR in an application cohort of 82 CML-CP patients, 16 CML-AP/BP patients, and 28 HCs. ROC curve analysis revealed that hsa_circ_0006010 and hsa_circ_0002903 had high accuracy in discriminating CML-AP/BP patients from CML-CP patients (cutoff = 2.851 and 1.837; area under the curve (AUC) = 0.895 and 0.932, respectively, both P < 0.001; Fig. 5A). Similarly, hsa_circ_0006010 and hsa_circ_0002903 had high accuracy in discriminating CML-AP/BP patients from HCs (cutoff = 1.924 and 1.931; AUC = 0.837 and 0.897, respectively; both P < 0.001; Fig. 5B). However, hsa_circ_0006010 and hsa_circ_0002903 had low accuracy in discriminating CML-CP patients from HCs (cutoff = 1.008 and 3.349; AUC = 0.504 and 0.585, respectively; both P > 0.05; Fig. 5C). ROC analysis suggested that PB hsa_circ_0006010 and hsa_circ_0002903 had high diagnostic efficiency and promising potential as biomarkers for CML disease progression.
Fig. 5.
Diagnostic value of PB hsa_circ_0006010 and hsa_circ_0002903 for CML disease progression. a ROC curve analysis of PB hsa_circ_0006010 and hsa_circ_0002903 for discriminating CML-AP/BP patients from CML-CP patients. b ROC curve analysis of PB hsa_circ_0006010 and hsa_circ_0002903 for discriminating CML-AP/BP patients from HCs. c ROC curve analysis of PB hsa_circ_0006010 and hsa_circ_0002903 for discriminating CML-CP patients from HCs
Expression of PB hsa_circ_0006010 and hsa_circ_0002903 is independent of the four risk scoring systems in CML patients.
To explore whether the expression of the two circRNAs is correlated with the four risk scoring systems for CML, we divided the patients with CML-CP (n = 82) into four groups according to the risk scoring system (Sokal score, Hasford score, EURO score, and ELTS score). The correlation between the expression of PB hsa_circ_0006010 and PB hsa_circ_0002903 and the scoring system was analyzed for each corresponding risk group. The results showed that in any of the four different scoring systems, there were no significant differences in hsa_circ_0006010 or hsa_circ_0002903 expression among the different risk groups (all P > 0.05) (Table 3). Therefore, PB hsa_circ_0006010 and hsa_circ_0002903 expression may not be directly related to the four scoring systems of CML patients, suggesting that they may be independent of the four scoring systems.
Table 3.
Correlations between hsa_circ_0006010, hsa_circ_0002903 and different scoring systems
| Risk score | N (%) | hsa_circ_0006010 | hsa_circ_0002903 | |||||
|---|---|---|---|---|---|---|---|---|
| Relative expression | r value | p value | Relative expression | r value | p value | |||
| Sokal | -0.078 | 0.489 | -0.022 | 0.844 | ||||
| Low | 54 (65.86%) | 0.92 (0.13–2.77) | 2.91 (0.41–16.18) | |||||
| Intermediate | 14 (17.07%) | 0.70 (0.31–2.97) | 2.42 (1.06–15.31) | |||||
| High | 14 (17.07%) | 0.91 (0.01–1.83) | 2.33 (1.54–4.87) | |||||
| Hasford | 0.053 | 0.638 | 0.022 | 0.847 | ||||
| Low | / | / | / | |||||
| Intermediate | 62 (75.61%) | 0.79 (0.13–2.77) | 2.69 (0.42–16.18) | |||||
| High | 20 (24.39%) | 0.92 (0.10–2.97) | 2.44 (1.06–6.73) | |||||
| EUOTS | 0.121 | 0.279 | -0.100 | 0.373 | ||||
| Low | 46 (56.10%) | 0.78 (0.10–2.63) | 2.66 (0.42–16.18) | |||||
| High | 36 (49.90%) | 0.87 (0.15–2.97) | 2.31 (1.02–15.32) | |||||
| ELTS | 0.036 | 0.749 | -0.018 | 0.871 | ||||
| Low | 64 (78.05%) | 0.83 (0.13–2.77) | 2.76 (0.42–16.18) | |||||
| Intermediate | 14 (17.07%) | 0.78 (0.10–2.97) | 2.44 (1.06–6.72) | |||||
| High | 4 (4.88%) | 1.45 (0.83–1.83) | 1.87 (1.55–4.87) | |||||
Expression of PB hsa_circ_0006010 and hsa_circ_0002903 is associated with adverse clinical prognostic indicators
We performed correlation analysis to assess the correlation between the expression of the two circRNAs and the clinical features of CML-CP patients in the application phase. As shown in Table 4, PB hsa_circ_0006010 was positively correlated with splenomegaly, red blood cell distribution width (RDW), and blast counts in PB but negatively correlated with hemoglobin. In contrast, PB hsa_circ_0002903 was negatively correlated with blast counts in PB and positively related to RDW and basophil count. However, the expression of these two circRNAs was not correlated with sex or age (Table 4). Consequently, PB hsa_circ_0006010 and hsa_circ_0002903 expression may be associated with adverse clinical prognostic indicators (such as splenomegaly, blast counts, basophil counts, hemoglobin, and RDW) in CML patients.
Table 4.
Correlations between hsa_circ_0006010, hsa_circ_0002903 and clinical indicators
| Features | N/r | hsa_circ_0006010 | hsa_circ_0002903 |
|---|---|---|---|
| Gender | |||
| male | 44 | 0.97 (0.13–2.97) | 2.32 (0.42–16.18) |
| female | 38 | 0.72 (0.10–2.77) | 2.66 (1.06–15.32) |
| p value | 0.323 | 0.712 | |
| Splenomegaly | |||
| yes | 39 | 0.88 (0.15–2.97) | 2.29 (0.67–15.32) |
| no | 43 | 0.78 (0.10–2.63) | 2.89 (0.42–16.18) |
| p value | < 0.001 | 0.207 | |
| Age (years) | r value | -0.070 | 0.147 |
| p value | 0.494 | 0.189 | |
| Blast counts (×10 9 /L) | r value | 0.381 | -0.303 |
| p value | < 0.001 | < 0.001 | |
| Eosinophils (×10 9 /L) | r value | 0.060 | 0.044 |
| p value | 0.591 | 0.694 | |
| Basophils (×10 9 /L) | r value | 0.039 | 0.461 |
| p value | 0.725 | < 0.001 | |
| Hemoglobin (g/L) | r value | -0.385 | 0.051 |
| p value | < 0.001 | 0.651 | |
| RDW-CV (%) | r value | 0.569 | 0.631 |
| p value | < 0.001 | < 0.001 |
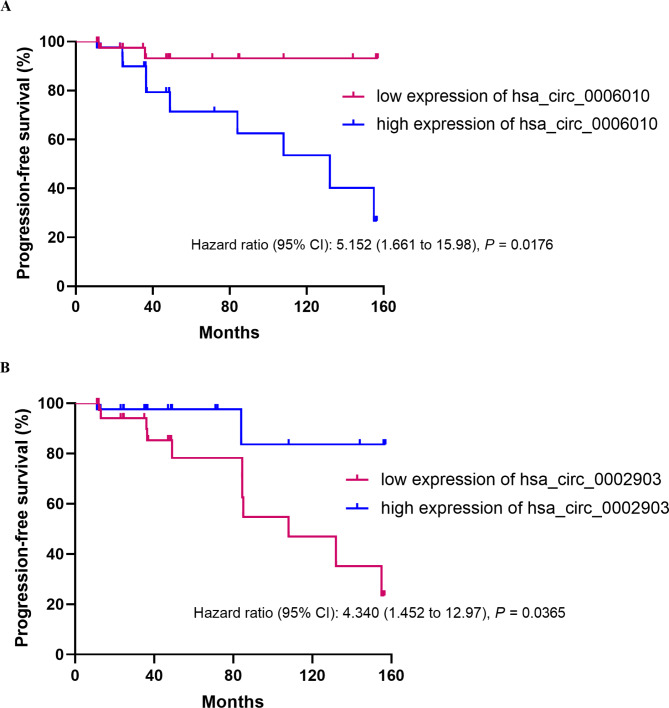
High PB hsa_circ_0006010 expression and low PB hsa_circ_0002903 expression indicate worse PFS in CML patients
We analyzed the PFS of patients to evaluate the correlation between PB hsa_circ_0006010 and hsa_circ_0002903 and the prognosis of CML-CP patients in the application phase. High and low expressions of hsa_circ_0006010 and hsa_circ_0002903 were defined by median expression. Survival analysis revealed that patients with high hsa_circ_0006010 expression had significantly shorter PFS than patients with low hsa_circ_0006010 expression (HR = 5.152, 95% CI = 1.661–15.98, P = 0.0176; Fig. 6A). In addition, the PFS of patients with low hsa_circ_0002903 expression was significantly shorter than that of patients with high hsa_circ_0002903 expression (HR = 4.340, 95% CI = 1.452–12.97, P = 0.0365; Fig. 6B). Therefore, our results showed that PB hsa_circ_0006010 and hsa_circ_0002903 expression significantly affected patient PFS, specifically that high hsa_circ_0006010 expression and low hsa_circ_0002903 might indicate worse PFS in CML patients.
Fig. 6.
Correlations between PB hsa_circ_0006010 and hsa_circ_0002903 expression and PFS in CML-CP patients. a K-M curve of PB hsa_circ_0006010. b K-M curve of PB hsa_circ_0002903
PB hsa_circ_0006010 and hsa_circ_0002903 as disease progression surveillance markers for CML patients
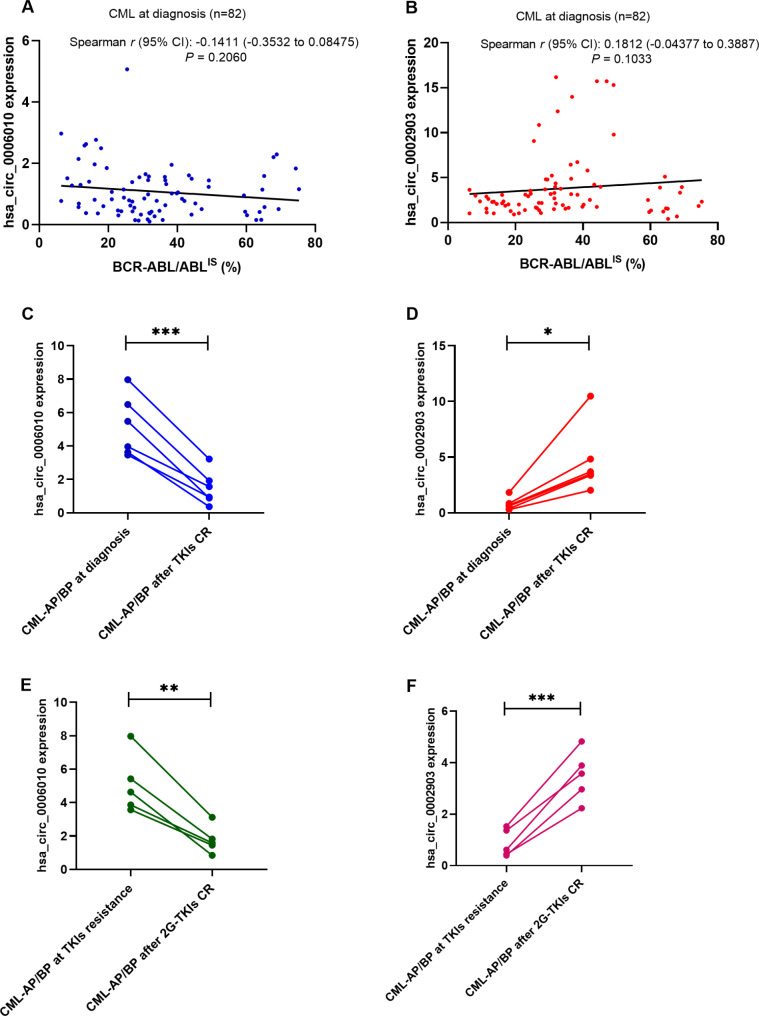
To investigate whether PB hsa_circ_0006010 or hsa_circ_0002903 expression is related to PB BCR-ABL1 quantification on the international scale (IS), we analyzed the correlation between BCR-ABL1/ABL1IS and the two circRNAs in 82 CML patients (82/98) diagnosed at the application stage. The results showed that PB hsa_circ_0006010 expression at diagnosis (Spearman r = -0.1411, P = 0.2060; Fig. 7A) and PB hsa_circ_0002903 expression at diagnosis were not correlated with PB BCR-ABL1/ABL1IS (Spearman r = 0.1812, P = 0.1033; Fig. 7B). These results indicated that PB hsa_circ_0006010 and hsa_circ_0002903 might be independent of BCR-ABL1 expression. We continued to explore whether PB hsa_circ_0006010 or hsa_circ_0002903 can be used to monitor CML disease progression. We collected six pairs of PB samples from CML-AP/BP patients at diagnosis and at CR after TKI treatment at the application stage. Compared with that in CML-AP/BP patients at diagnosis, PB hsa_circ_0006010 expression in patients who achieved CR decreased significantly (P < 0.001, Fig. 7C), and hsa_circ_0002903 expression increased significantly (P = 0.0118, Fig. 7D). Furthermore, we collected five pairs of PB samples from CML-CP patients who developed disease progression due to TKI resistance and who achieved CR after second-generation (2G)-TKI treatment at the application stage. Similarly, compared with that in CML-AP/BP patients who developed TKI resistance, PB hsa_circ_0006010 expression in patients who achieved CR decreased significantly (P < 0.01, Fig. 7E), and hsa_circ_0002903 expression increased significantly (P < 0.001, Fig. 7F). Taken together, these results suggested that PB hsa_circ_0006010 and hsa_circ_0002903 could reflect the disease progression status of CML patients and can be utilized as disease progression surveillance markers for CML patients, which may be beneficial for the clinical management of advanced CML patients.
Fig. 7.
PB hsa_circ_0006010 and hsa_circ_0002903 as potential disease progression surveillance markers for CML patients. a PB hsa_circ_0006010 expression was not correlated with PB BCR-ABL1/ABL1IS in CML patients at diagnosis. b PB hsa_circ_0002903 expression was not correlated with PB BCR-ABL1/ABL1IS in CML patients at diagnosis. c PB hsa_circ_0006010 expression in 6 pairs of samples from CML-AP/BP patients at diagnosis and after achieving CR through TKI treatment. d hsa_circ_0002903 expression in 6 pairs of samples from CML-AP/BP patients at diagnosis and after achieving CR through TKI treatment. e PB hsa_circ_0006010 expression in 5 pairs of PB samples from CML-CP patients who developed disease progression due to TKI resistance and who achieved CR after 2G-TKI treatment. f PB hsa_circ_0006010 expression in 5 pairs of PB samples from CML-CP patients who developed disease progression due to TKI resistance and after 2G-TKI treatment
Discussion
In this study, we systematically explored the PB circRNA signature associated with CML disease progression, using RNA-Seq to identify dysregulated candidate circRNAs, which were subsequently validated via qRT-PCR in an independent cohort of CML patients. We confirmed that PB hsa_circ_0006010 is a significantly upregulated circRNA and that PB hsa_circ_0002903 is a downregulated circRNA in advanced CML patients. ROC analysis revealed that both hsa_circ_0006010 and hsa_circ_0002903 have excellent diagnostic efficacy for detecting CML disease progression. Furthermore, we found that the expression of PB hsa_circ_0006010 and hsa_circ_0002903 was not associated with the four scoring systems of CML patients but was correlated with CML patients’ critical clinical indicators. Moreover, high hsa_circ_0006010 and low hsa_circ_0002903 expression were associated with poor PFS in CML patients. Finally, we confirmed that PB hsa_circ_0006010 and hsa_circ_0002903, which are independent of BCR-ABL1 expression, could be used for monitoring CML disease progression at initial diagnosis or follow-up. Therefore, our study demonstrated that PB hsa_circ_0006010 and hsa_circ_0002903 might serve as novel diagnostic, surveillance, and prognostic biomarkers for CML disease progression, contributing to the precise management of advanced CML.
Disease progression is often fatal for CML patients and remains a major clinical challenge today [4]. In recent years, the role of circRNAs as diagnostic and prognostic markers of leukemia has gradually been revealed [32]. However, the existing monitoring methods for CML disease progression have limitations and need to be enhanced to detect disease progression at an early stage [33]. For example, diagnosing additional chromosomal abnormalities (ACAs) and drug-resistant mutation points is achieved through invasive bone marrow puncture. Moreover, the sensitivity of ACA detection is limited by the number of mid-stage cells, and the resistance mutation point is limited by the specific mutation type. Therefore, it is imperative to use noninvasive diagnostic and monitoring methods to detect disease progression at an early stage. circRNAs, which are expressed in a developmental stage- and tissue-specific manner, are superior to linear RNAs because of their high abundance and stability and can even be stably present in blood [34], suggesting that circRNAs have promise as biomarkers for disease diagnosis and treatment. However, there are still unknowns in the study of PB circRNAs as early diagnostic, monitoring and prognostic markers for CML progression.
The progression of CML from CP to AP/BP involves gene expression changes associated with progression [35] and is often accompanied by an increase in genomic instability [36]. Therefore, identifying the genes that are significantly altered in CML patients may identify novel biomarkers to improve early diagnosis and personalized treatment [36]. The circRNA expression profiles in CML patients identified in this study showed that CML-AP/BP patients may undergo more significant circRNA changes than CML-CP patients or HCs. We subsequently identified six candidate circRNAs associated with CML disease progression. Considering the potential errors that may result from an insufficient sample size for RNA-Seq, we performed qRT-PCR on a large validation cohort. PB hsa_circ_0006010 and hsa_circ_0002903 were the most significantly upregulated and downregulated circRNAs, respectively, in CML-AP/BP patients. In an independent application cohort, we found that PB hsa_circ_0006010 and hsa_circ_0002903 had high accuracy in discriminating CML-AP/BP patients from CML-CP patients or HCs (Fig. 5), suggesting that they may enable early differentiation of patients who will likely progress from follow-up CML-CP patients or HCs via regular medical examinations of CML-AP/BP patients. Furthermore, this study also showed that PB hsa_circ_0006010 and hsa_circ_0002903 could be used for monitoring CML disease progression at initial diagnosis or follow-up, suggesting their role in the early identification of CML disease progression.
Some scholars have researched other markers and detection methods related to CML disease progression, such as lipids, proteins, and histone chaperones, which may expand the types of disease progression markers beyond PB circRNAs. For example, a new study showed that bioactive lipids could be potential biomarkers for disease progression and response to TKIs in CML patients [37]. Similarly, in a previous study, cancerous inhibitor of protein phosphatase 2 A (CIP2A) was validated as a biomarker of disease progression and treatment failure in CML [38], which may aid in the planning of CML treatment strategies. However, detecting bioactive lipids requires specialized and expensive mass spectrometry equipment and trained personnel, and detecting the CIP2A protein requires expensive flow cytometry equipment and specialized personnel, which may limit its application. Furthermore, another study confirmed that the histone chaperone ASF1A accelerates CML cell crisis by activating Notch signaling in BM samples from CML-BP patients [39], suggesting that it may not be suitable for the early detection of CML-AP or active CML using PB. However, specimen availability and method simplicity are essential factors for effective long-term monitoring of CML progression. As shown in the present study, PB hsa_circ_0006010 and hsa_circ_0002903 detection by qRT-PCR has the advantages of convenient specimen collection and simple methodologies. These biomarkers may be more suitable for diagnosing and monitoring patients with CML disease progression.
Previous studies have revealed that CML patients who initially present with AP and BP have a better prognosis than those who progress to these stages during TKI therapy [4, 40, 41] and often respond to TKI therapy like CML-CP patients, especially when 2G-TKIs are used. Achieving a stable deep molecular response and discontinuing medication for treatment-free remission (TFR) are considered some of the main goals for most CML-CP patients [29, 42]. Therefore, it is imperative to monitor CML-CP patients that achieve TFR during discontinuation, with particular attention given to CML-AP/BP reversal of CML-CP due to TKI resistance. Our study revealed that when CML-AP/BP patients at diagnosis and TKI-resistant CML-AP/BP patients achieved CR, the expression of PB hsa_circ_0006010 and PB hsa_circ_0002903 was significantly downregulated and upregulated, respectively (Fig. 7), which suggested that the expression of PB hsa_circ_0006010 and hsa_circ_0002903 could be used as surveillance biomarkers for CML disease progression, especially in patients with CML-CP or CML-AP/BP who achieve TFR. In addition, we found that neither hsa_circ_0006010 nor hsa_circ_0002903 expression at diagnosis was correlated with BCR-ABL1/ABL1IS in PB samples.
There is evidence that in CML-CP patients treated with TKIs, especially those treated with 2G-TKIs, the ELTS score is more accurate than the Sokal score, and patients with moderate-risk ELTS scores may benefit from 2G-TKIs compared to imatinib [43]. In this study, we analyzed the correlation between hsa_circ_0006010 and hsa_circ_0002903 expression and the respective scoring systems in CML-CP patients at the application stage. We found that PB hsa_circ_0006010 and hsa_circ_0002903 expression may not be directly related to any of the four scoring systems used for CML patients, suggesting that the use of PB hsa_circ_0006010 and hsa_circ_0002903 as prognostic biomarkers may be independent of the four scoring systems.
Our previous study demonstrated that in CML-CP patients treated with TKIs, a higher RDW at diagnosis suggested a poor prognosis [44]. Similarly, other studies have shown that lower hemoglobin concentrations and higher white blood cell (WBC) counts are associated with more unsatisfactory responses and outcomes [45, 46]. Surprisingly, in the present study, we further demonstrated that the expression of PB hsa_circ_0006010 and hsa_circ_0002903 was associated with other adverse clinical prognostic indicators of CML, such as splenomegaly, blast counts, basophil counts, hemoglobin level, and RDW, suggesting that these indicators might play essential roles in the progression of CML and could be necessary complements to existing prognostic indicators of CML, including RDW. Our study also showed that high hsa_circ_0006010 expression and low hsa_circ_0002903 expression predicted worse PFS in CML patients, indicating that both hsa_circ_0006010 and hsa_circ_0002903 may be independent prognostic factors for CML patients at risk of disease progression.
The limitations of our study should be acknowledged. Due to our focus on CML disease progression, we studied only PFS, an important prognostic factor associated with disease progression, in CML patients, not overall survival. To avoid introducing data bias, we did not combine CML patients at the validation stage with those at the application stage, which resulted in relatively few cases of CML-AP/BP. In addition, the molecular mechanisms by which hsa_circ_0006010 and hsa_circ_0002903 are involved in CML disease progression and their interaction were not explored in this study, and further studies are needed. Furthermore, this was a small-sample, single-center study, and a large-sample, multicenter study is required to verify the results of this study before the results can be applied clinically.
Conclusion
In summary, the present study identified a unique circRNA signature in advanced CML patients, of which two candidate circRNAs, hsa_circ_0006010 and hsa_circ_0002903, were demonstrated to be associated with CML disease progression and identified as potential diagnostic biomarkers for CML disease progression. Furthermore, we found that hsa_circ_0006010 and hsa_circ_0002903 expression was associated with adverse clinical features but was independent of the four scoring systems used for CML patients. Moreover, high hsa_circ_0006010 expression and low hsa_circ_0002903 expression indicated poor PFS in CML patients. Our study provided the first evidence that PB hsa_circ_0006010 and hsa_circ_0002903 could serve as novel diagnostic, surveillance, and prognostic biomarkers for disease progression in CML patients, contributing to assisting in the diagnosis of CML patients at risk for progression and the accurate management of patients with advanced CML.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank all the participants for donating blood samples and all the researchers for their contributions to this work.
Abbreviations
- CML
Chronic myeloid leukemia
- BCR-ABL1
Breakpoint cluster region-Abelson leukemia
- CP
Chronic phase
- AP
Accelerated phase
- BP
Blast phase
- TKIs
Tyrosine kinase inhibitors
- ELTS
EUTOS long-term survival
- circRNAs
Circular RNAs
- PFS
Progression-free survival
- PB
Peripheral blood
- BM
Bone marrow
- RNA-Seq
RNA sequencing
- qRT-PCR
Quantitative real-time PCR
- ELN
European LeukemiaNet
- CR
Complete remission
- HCs
Healthy controls
- PBMCs
Peripheral blood mononuclear cells
- Ct
Cycle threshold
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- FC
Fold change
- ROC
Receiver operating characteristic
- K-M
Kaplan-Meier
- GEO
Gene Expression Omnibus
- AUC
Area under the curve
- RDW
Red blood cell distribution width
- IS
International scale
- 2G
Second-generation
- ACAs
Additional chromosomal abnormalities
- CIP2A
Cancerous inhibitor of protein phosphatase 2 A
- TFR
Treatment-free remission
- WBC
White blood cell
Author contributions
JWZ, GRW, and GLY analyzed and drafted the manuscript. GRW and JWZ performed the laboratory work for this study. MTZ and HSL collected the samples. YYB provided technical support. ZGC critically revised the manuscript for important intellectual content. ZGC and XQZ were responsible for the conception and design of this study and revised the manuscript. All authors approved the final manuscript submitted for publication.
Funding
This study was supported by the Medical and Health Research Science and Technology Plan Project of Zhejiang Province (2021KY216), the Basic Public Welfare Technology Research Project of Zhejiang Province (LGF20H200005), and the Basic Scientific Research Project of Wenzhou City (Y20220123 and Y20220744).
Data availability
The RNA-seq data were annotated on the NCBI GEO database (GSE212254), and other datasets generated and analyzed during the current study are not publicly available for patient privacy reasons but are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
The Ethics Committee of The Second Affiliated Hospital and Yuying Children’s Hospital of Wenzhou Medical University approved the study. All participants provided written informed consent. Research involving human participants was conducted following the Declaration of Helsinki. All methods were carried out under relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jingwei Zhao and Guiran Wang contributed equally to this work.
Contributor Information
Xiaoqun Zheng, Email: jszhengxq@163.com.
Zhanguo Chen, Email: steve0577@126.com.
References
- 1.Cortes J, Pavlovsky C, Saußele S. Chronic myeloid leukaemia. Lancet. 2021;398(10314):1914–26. 10.1016/s0140-6736(21)01204-6. [DOI] [PubMed] [Google Scholar]
- 2.Zhou T, Medeiros LJ, Hu S. Chronic myeloid leukemia: beyond BCR-ABL1. Curr Hematol Malig Rep. 2018;13(6):435–45. 10.1007/s11899-018-0474-6. [DOI] [PubMed] [Google Scholar]
- 3.Iezza M, Cortesi S, Ottaviani E, Mancini M, Venturi C, Monaldi C, et al. Prognosis in chronic myeloid leukemia: baseline factors, dynamic risk assessment and novel insights. Cells. 2023;12(13):1703. 10.3390/cells12131703. [DOI] [PMC free article] [PubMed]
- 4.Senapati J, Jabbour E, Kantarjian H, Short NJ. Pathogenesis and management of accelerated and blast phases of chronic myeloid leukemia. Leukemia. 2023;37(1):5–17. 10.1038/s41375-022-01736-5. [DOI] [PubMed] [Google Scholar]
- 5.Chandran RK, Geetha N, Sakthivel KM, Kumar RS, Krishna K, Sreedharan H. Differential gene expression changes and their implication on the disease progression in patients with chronic myeloid leukemia. Blood Cells Mol Dis. 2019;77:51–60. 10.1016/j.bcmd.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Iqbal Z, Absar M, Akhtar T, Aleem A, Jameel A, Basit S, et al. Integrated genomic analysis identifies ANKRD36 gene as a novel and common biomarker of disease progression in chronic myeloid leukemia. Biology (Basel). 2021;10(11):1182. 10.3390/biology10111182. [DOI] [PMC free article] [PubMed]
- 7.Bonifacio M, Stagno F, Scaffidi L, Krampera M, Di Raimondo F. Management of chronic myeloid leukemia in advanced phase. Front Oncol. 2019;9:1132. 10.3389/fonc.2019.01132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hasford J, Pfirrmann M, Hehlmann R, Allan NC, Baccarani M, Kluin-Nelemans JC, et al. A new prognostic score for survival of patients with chronic myeloid leukemia treated with interferon alfa. Writing Committee for the Collaborative CML Prognostic Factors Project Group. J Natl Cancer Inst. 1998;90(11):850–8. 10.1093/jnci/90.11.850. [DOI] [PubMed] [Google Scholar]
- 9.Pfirrmann M, Clark RE, Prejzner W, Lauseker M, Baccarani M, Saussele S, et al. The EUTOS long-term survival (ELTS) score is superior to the Sokal score for predicting survival in chronic myeloid leukemia. Leukemia. 2020;34(8):2138–49. 10.1038/s41375-020-0931-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sokal JE, Cox EB, Baccarani M, Tura S, Gomez GA, Robertson JE, et al. Prognostic discrimination in good-risk chronic granulocytic leukemia. Blood. 1984;63(4):789–99. [PubMed] [Google Scholar]
- 11.Sato E, Iriyama N, Tokuhira M, Takaku T, Ishikawa M, Nakazato T, et al. The EUTOS long-term survival score predicts disease-specific mortality and molecular responses among patients with chronic myeloid leukemia in a practice-based cohort. Cancer Med. 2020;9(23):8931–9. 10.1002/cam4.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang XS, Gale RP, Huang XJ, Jiang Q. Is the Sokal or EUTOS long-term survival (ELTS) score a better predictor of responses and outcomes in persons with chronic myeloid leukemia receiving tyrosine-kinase inhibitors? Leukemia. 2022;36(2):482–91. 10.1038/s41375-021-01387-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osman AEG, Deininger MW. Chronic myeloid leukemia: modern therapies, current challenges and future directions. Blood Rev. 2021;49:100825. 10.1016/j.blre.2021.100825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Branford S, Kim DDH, Apperley JF, Eide CA, Mustjoki S, Ong ST, et al. Laying the foundation for genomically-based risk assessment in chronic myeloid leukemia. Leukemia. 2019;33(8):1835–50. 10.1038/s41375-019-0512-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Yang L, Chen LL. The Biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–42. 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 16.Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One. 2015;10(10):e0141214. 10.1371/journal.pone.0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–85. 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 18.Li Q, Ren X, Wang Y, Xin X. CircRNA: a rising star in leukemia. PeerJ. 2023;11:e15577. 10.7717/peerj.15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan Y, Lou J, Wang H, An N, Chen H, Zhang Q, et al. CircBA9.3 supports the survival of leukaemic cells by up-regulating c-ABL1 or BCR-ABL1 protein levels. Blood Cells Mol Dis. 2018;73:38–44. 10.1016/j.bcmd.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Che H, Ding H, Jia X. circ_0080145 enhances Imatinib resistance of chronic myeloid leukemia by regulating miR-326/PPFIA1 axis. Cancer Biother Radiopharm. 2020. 10.1089/cbr.2020.3600. Epub ahead of print [DOI] [PubMed] [Google Scholar]
- 21.Zhong AN, Yin Y, Tang BJ, Chen L, Shen HW, Tan ZP, et al. CircRNA microarray profiling reveals hsa_circ_0058493 as a novel biomarker for imatinib-resistant CML. Front Pharmacol. 2021;12:728916. 10.3389/fphar.2021.728916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan Y, Huang Z, Wang X, Dai H, Jiang G, Feng W. A novel fusion circular RNA F-circBA1 derived from the BCR-ABL fusion gene displayed an oncogenic role in chronic myeloid leukemia cells. Bioengineered. 2021;12(1):4816–27. 10.1080/21655979.2021.1957749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng XQ, Nie SM, Huang JX, Li TL, Zhou JJ, Wang W, et al. Circular RNA circHIPK3 serves as a prognostic marker to promote chronic myeloid leukemia progression. Neoplasma. 2020;67(1):171–7. 10.4149/neo_2018_181129N908. [DOI] [PubMed] [Google Scholar]
- 24.Wu Z, Sun H, Li J, Jin H. Circular RNAs in leukemia. Aging. 2019;11(13):4757–71. 10.18632/aging.102091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deininger MW, Shah NP, Altman JK, Berman E, Bhatia R, Bhatnagar B, et al. Chronic myeloid leukemia, Version 2.2021, NCCN Clinical Practice guidelines in Oncology. J Natl Compr Canc Netw. 2020;18(10):1385–415. 10.6004/jnccn.2020.0047. [DOI] [PubMed] [Google Scholar]
- 26.Li T, Li X, Chen H, Huang KZ, Xie Q, Ge HY, et al. Higher red blood cell distribution width is a poor prognostic factor for patients with chronic myeloid leukemia. Cancer Manag Res. 2021;13:1233–43. 10.2147/CMAR.S288589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang G, Yan G, Sang K, Yang H, Sun N, Bai Y, et al. Circulating lnc-LOC as a novel noninvasive biomarker in the treatment surveillance of acute promyelocytic leukaemia. BMC Cancer. 2022;22(1):481. 10.1186/s12885-022-09621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baccarani M, Deininger MW, Rosti G, Hochhaus A, Soverini S, Apperley JF, et al. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood. 2013;122(6):872–84. 10.1182/blood-2013-05-501569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hochhaus A, Baccarani M, Silver RT, Schiffer C, Apperley JF, Cervantes F, et al. European LeukemiaNet 2020 recommendations for treating chronic myeloid leukemia. Leukemia. 2020;34(4):966–84. 10.1038/s41375-020-0776-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uz B, Buyukasik Y, Atay H, Kelkitli E, Turgut M, Bektas O, et al. EUTOS CML prognostic scoring system predicts ELN-based ‘event-free survival’ better than Euro/Hasford and Sokal systems in CML patients receiving front-line imatinib mesylate. Hematology. 2013;18(5):247–52. 10.1179/1607845412y.0000000071. [DOI] [PubMed] [Google Scholar]
- 31.Pfirrmann M, Baccarani M, Saussele S, Guilhot J, Cervantes F, Ossenkoppele G, et al. Prognosis of long-term survival considering disease-specific death in patients with chronic myeloid leukemia. Leukemia. 2016;30(1):48–56. 10.1038/leu.2015.261. [DOI] [PubMed] [Google Scholar]
- 32.Deng W, Chao R, Zhu S. Emerging roles of circRNAs in leukemia and the clinical prospects: an update. Immun Inflamm Dis. 2023;11(1):e725. 10.1002/iid3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dominy K, Mokretar K, Reid AG, Khorashad JS. Molecular monitoring of chronic myeloid leukemia. Methods Mol Biol. 2020;2065:153–73. 10.1007/978-1-4939-9833-3_12. [DOI] [PubMed] [Google Scholar]
- 34.Salzman J, Circular RNA, Expression. Its potential regulation and function. Trends Genet. 2016;32(5):309–16. 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Radich JP, Dai H, Mao M, Oehler V, Schelter J, Druker B, et al. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proc Natl Acad Sci U S A. 2006;103(8):2794–9. 10.1073/pnas.0510423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdulmawjood B, Costa B, Roma-Rodrigues C, Baptista PV, Fernandes AR. Genetic biomarkers in chronic myeloid leukemia: what have we learned so far? Int J Mol Sci. 2021;22(22):12516. 10.3390/ijms222212516. [DOI] [PMC free article] [PubMed]
- 37.de Almeida FC, Berzoti-Coelho MG, Toro DM, Cacemiro MDC, Bassan VL, Barretto GD, et al. Bioactive lipids as chronic myeloid leukemia’s potential biomarkers for disease progression and response to tyrosine kinase inhibitors. Front Immunol. 2022;13:840173. 10.3389/fimmu.2022.840173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clark RE, Basabrain AA, Austin GM, Holcroft AK, Loaiza S, Apperley JF, et al. Validation of CIP2A as a biomarker of subsequent disease progression and treatment failure in chronic myeloid leukaemia. Cancers (Basel). 2021;13(9):2155. 10.3390/cancers13092155. [DOI] [PMC free article] [PubMed]
- 39.Yin X, Zhou M, Zhang L, Fu Y, Xu M, Wang X, et al. Histone chaperone ASF1A accelerates chronic myeloid leukemia blast crisis by activating notch signaling. Cell Death Dis. 2022;13(10):842. 10.1038/s41419-022-05234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jain P, Kantarjian HM, Ghorab A, Sasaki K, Jabbour EJ, Nogueras Gonzalez G, et al. Prognostic factors and survival outcomes in patients with chronic myeloid leukemia in blast phase in the tyrosine kinase inhibitor era: cohort study of 477 patients. Cancer. 2017;123(22):4391–402. 10.1002/cncr.30864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohanian M, Kantarjian HM, Quintas-Cardama A, Jabbour E, Abruzzo L, Verstovsek S, et al. Tyrosine kinase inhibitors as initial therapy for patients with chronic myeloid leukemia in accelerated phase. Clin Lymphoma Myeloma Leuk. 2014;14(2):155–62.e1. 10.1016/j.clml.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atallah E, Sweet K. Treatment-free remission: the new goal in CML therapy. Curr Hematol Malig Rep. 2021;16(5):433–9. 10.1007/s11899-021-00653-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X-S, Gale RP, Huang X-J, Jiang Q. Is the Sokal or EUTOS long-term survival (ELTS) score a better predictor of responses and outcomes in persons with chronic myeloid leukemia receiving tyrosine-kinase inhibitors? Leukemia. 2021;36(2):482–91. 10.1038/s41375-021-01387-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mao XL, Xi YM, Li ZJ, Jia MF, Li M, Wang LN, et al. Higher red blood cell distribution width at diagnose is a simple negative prognostic factor in chronic phase-chronic myeloid leukemia patients treated with tyrosine kinase inhibitors: a retrospective study. Medicine (Baltimore). 2021;100(10):e24003. 10.1097/MD.0000000000024003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bewersdorf JP, Zeidan AM. Hyperleukocytosis and leukostasis in acute myeloid leukemia: can a better understanding of the underlying molecular pathophysiology lead to novel treatments? Cells. 2020;9(10):2310. 10.3390/cells9102310 [DOI] [PMC free article] [PubMed]
- 46.Dou X, Zheng F, Zhang L, Jin J, Zhang Y, Liu B, et al. Adolescents experienced more treatment failure than children with chronic myeloid leukemia receiving imatinib as frontline therapy: a retrospective multicenter study. Ann Hematol. 2021;100(9):2215–28. 10.1007/s00277-021-04544-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data were annotated on the NCBI GEO database (GSE212254), and other datasets generated and analyzed during the current study are not publicly available for patient privacy reasons but are available from the corresponding author upon reasonable request.