Abstract
In 1997 and 1998, H9N2 influenza A viruses were isolated from the respiratory organs of Indian ring-necked parakeets (Psittacula Krameri manillensis) that had been imported from Pakistan to Japan. The two isolates were closely related to each other (>99% as determined by nucleotide analysis of eight RNA segments), indicating that H9N2 viruses of the same lineage were maintained in these birds for at least 1 year. The hemagglutinins and neuraminidases of both isolates showed >97% nucleotide identity with those of H9N2 viruses isolated from humans in Hong Kong in 1999, while the six genes encoding internal proteins were >99% identical to the corresponding genes of H5N1 viruses recovered during the 1997 outbreak in Hong Kong. These results suggest that the H9N2 parakeet viruses originating in Pakistan share an immediate ancestor with the H9N2 human viruses. Thus, influenza A viruses with the potential to be transmitted directly to humans may be circulating in captive birds worldwide.
Phylogenetic analysis of influenza A viruses, together with the presence of all 15 hemagglutinin (HA) and 9 neuraminidase (NA) subtypes of viruses in aquatic birds, suggests that these hosts are natural reservoirs of such viruses (31). Influenza A viruses have also been isolated from psittacines, among other avian species (3, 17), although little is known about their roles in the ecology, epizootiology, and epidemiology of these viruses. Because they are captured in the wild and distributed worldwide for sale as pets, psittacines might serve as biological vectors in the spread of influenza A viruses to animals in other countries. In fact, a Newcastle disease virus originating from imported psittacines caused outbreaks of the disease among domestic poultry in southern California in the early 1970s (30).
In recent years, H9N2 viruses have caused influenza outbreaks in poultry worldwide, including Europe, Pakistan, and Asia (3, 4, 10, 16). H9N2 viruses are genetically distinct in Asia, where at least three lineages defined by the nucleotide sequence of the NP gene are circulating (10). Some of these Asian H9N2 viruses have been transmitted to pigs and humans (10) in mainland China, as well as Hong Kong (13, 21, 22). For example, an H9N2 virus, A/quail/Hong Kong/G1/97(G1), that was genetically related to the H9N2 viruses from humans was isolated during surveillance for influenza A viruses in Hong Kong birds (9). Moreover, the genes encoding the internal proteins (PA, PB1, PB2, NP, M, and NS) of the H9N2 viruses isolated from two children in Hong Kong were genetically closely related to those of the H5N1 viruses that had been directly transmitted from birds to humans, killing 6 of the 18 people infected (6, 11, 13, 26, 27, 28, 29).
During routine virologic diagnosis of birds imported to Japan, influenza A viruses were isolated in embryonated eggs from Indian ring-necked parakeets (Psittacula Krameri manillensis) imported from Pakistan. The first virus, A/parakeet/Chiba/1/97, was isolated from the respiratory organ (trachea) of a bird that died at a pet shop within 10 days of importation in March 1997. The second virus, A/parakeet/Narita/92A/98, also isolated from respiratory organs (mixture of trachea and lung), came from a bird that died at the animal quarantine station at the Narita airport in Japan in June 1998. Both isolates were identified as influenza A viruses of the H9N2 subtype by conventional hemagglutination inhibition and neuraminidase inhibition assays (2, 14). The viruses were plaque purified in primary chicken kidney cells, and stock viruses were prepared by inoculation into the allantoic cavities of 10-day-old chicken embryos. To our knowledge, these are the first H9N2 influenza viruses isolated from psittacine birds.
To determine the genetic relationship of these isolates with other H9N2 viruses, all eight genes of both isolates were sequenced. Viral RNA was extracted with a commercial kit (ISOGEN; Nippongene, Tokyo, Japan) from allantoic fluids containing virus. After reverse transcription with Superscript II (Life Technologies, Gaithersburg, Md.) using random 9-mers, cDNAs were amplified by PCR. PCR amplification of the coding regions of the viral gene segments was performed with gene-specific primer sets (sequences of the primers are available on request). PCR-derived double-stranded DNA was used as a template for sequencing on an Applied Biosystems 373S automated DNA sequencer using cycle sequencing dye terminator chemistry (Perkin-Elmer/Applied Biosystems, Foster City, Calif.). The nucleotide sequences were analyzed using version 10.0 of the sequence analysis software package GENETYX-MAC (Software Development, Tokyo, Japan). Identical results were obtained when the isolates were resequenced.
The two H9N2 isolates were genetically closely related to each other (>99% identity by nucleotide analysis of all eight RNA segments) (Table 1), indicating that they belong to the same lineage. Since the viruses were identified 1 year apart, their lineage must have been established in Pakistan for at least a year.
TABLE 1.
Nucleotide and amino acid differences between the two parakeet isolates
| Segment | No. of different:
|
|
|---|---|---|
| Nucleotides (% identity) | Amino acids (% identity) | |
| PB2 | 7 (99.7) | 3 (99.6) |
| PB1 | 17 (99.3) | 4 (99.5) |
| PA | 9 (99.6) | 1 (99.9) |
| HA | 14 (99.2) | 9 (98.4) |
| NP | 10 (99.3) | 3 (99.4) |
| NA | 9 (99.4) | 5 (98.9) |
| M | 5 (99.5) | 1 (99.6) (M1), 0 (100) (M2) |
| NS | 3 (99.6) | 1 (99.6) (NS1), 0 (100) (NS2) |
The entire coding regions of the HA and NA genes of the H9N2 parakeet viruses showed >97% identity with those of the H9N2 viruses isolated from humans in 1999 and from quail in 1997 (G1 virus), but there were appreciable differences in the PA, HA, NP, NA, M, and NS genes compared with a chicken isolate (A/chicken/Hong Kong/G9/97) and in all genes compared with A/duck/Hong Kong/Y439/97 (Table 2). Potential HA N-glycosylation sites with the N-X-T/S motif (in which X may be any amino acid except proline) are shown in Table 3. Unlike most other H9 HAs, those of the two parakeet viruses had a glycosylation site at Asn-87, consistent with findings for the H9N2 human and G1 quail viruses. However, the potential glycosylation site at Asn-188 in the three Hong Kong isolates is not represented in the parakeet viruses or other H9N2 viruses. The A/parakeet/Narita/92A/98 virus lost a glycosylation site at Asn-200 that is conserved in all other H9 HAs. The significance of this variability in glycosylation sites remains unknown. Both of the parakeet viruses possessed an R-S-S-R sequence at the HA cleavage site (Table 4). It is identical to that found in the H9N2 human and G1 quail viruses but differs from that found in other H9 viruses.
TABLE 2.
Sequence comparison of A/parakeet/Chiba/1/97 with H9N2 Hong Kong influenza virusa
| Segment | No. of nucleotides sequenced | % Homology with A/parakeet/Chiba/1/97
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| A/Hong Kong/1073/99
|
A/quail/Hong Kong/G1/97
|
A/chicken/Hong Kong/G9/97
|
A/duck/Hong Kong/Y439/97
|
||||||
| Nucleotides | Amino acids | Nucleotides | Amino acids | Nucleotides | Amino acids | Nucleotides | Amino acids | ||
| PB2 | 2,280 | 98.5 | 99.1 | 98.5 | 98.7 | 98.2 | 98.3 | 87.6 | 96.4 |
| PB1 | 2,277 | 98.9 | 99.7 | 98.9 | 99.6 | 98.4 | 99.1 | 90.6 | 97.3 |
| PA | 2,151 | 97.6 | 98.7 | 98.4 | 99.2 | 89.0 | 94.2 | 89.9 | 96.1 |
| HA | 1,683 | 97.7 | 97.5 | 98.0 | 97.4 | 92.5 | 92.9 | 85.1 | 88.1 |
| NP | 1,497 | 98.9 | 99.4 | 99.3 | 99.4 | 90.6 | 96.3 | 94.7 | 97.0 |
| NA | 1,410 | 97.9 | 97.7 | 98.1 | 97.4 | 94.0 | 93.4 | 89.7 | 89.3 |
| M | 982 | 99.1 | 99.6 (M1), 96.9 (M2) | 99.1 | 99.6 (M1), 97.9 (M2) | 96.4 | 98.0 (M1), 96.9 (M2) | 92.9 | 95.6 (M1), 93.8 (M2) |
| NS | 838 | 98.2 | 96.1 (NS1), 98.3 (NS2) | 98.6 | 96.5 (NS1), 99.2 (NS2) | 93.3 | 92.5 (NS1), 95.7 (NS2) | 91.1 | 89.5 (NS1), 97.5 (NS2) |
Nucleotide and amino acid sequences were compared, and identity was determined by GenBank searches.
TABLE 3.
Potential glycosylation sites on HA proteins of parakeet and closely related H9N2 viruses
| Virus | Glycosylation sitea at position:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 11 | 87 | 123 | 188 | 200 | 280 | 287 | 474 | |
| A/parakeet/Chiba/1/97 | + | + | + | − | + | + | + | + |
| A/parakeet/Narita/92A/98 | + | + | + | − | − | + | + | + |
| A/quail/Hong Kong/G1/97 | + | + | + | + | + | + | + | + |
| A/Hong Kong/1073/99 | + | + | + | + | + | + | + | + |
| A/Hong Kong/1074/99 | + | + | + | + | + | + | + | + |
| A/chicken/Beijing/1/94 | + | − | + | − | + | + | + | + |
| A/chicken/Hong Kong/739/94 | + | − | + | − | + | + | + | + |
| A/chicken/Hong Kong/G9/97 | + | − | + | − | + | + | + | + |
| A/duck/Hong Kong/Y439/97 | + | − | + | − | + | + | + | + |
| A/quail/Hong Kong/AF157/93 | + | − | + | − | + | + | + | + |
| A/turkey/California/189/66 | + | − | + | − | + | + | + | + |
| A/turkey/Minnesota/1/95 | + | − | + | − | + | + | + | + |
+, glycosylation site present; −, glycosylation site absent.
TABLE 4.
HA connecting peptide sequences of H9N2 viruses
| Virus | Connecting peptide amino acid sequence | Reference |
|---|---|---|
| A/turkey/Wisconcin/66 | P A V S S R | 19 |
| A/quail/Arizona/29209/93 | P A A S N R | 9 |
| A/duck/Hong Kong/168/77 | P A A S G R | 9 |
| A/duck/Hong Kong/784/79 | P A A S D R | 9 |
| A/chicken/Beijing/1/94 | P A R S S R | 9 |
| A/quail/Hong Kong/G1/97 | P A R S S R | 9 |
| A/chicken/Hong Kong/G9/97 | P A R S S R | 9 |
| A/Hong Kong/1073/99 | P A R S S R | 13 |
| A/parakeet/Chiba/1/97 | P A R S S R | This study |
| A/parakeet/Narita/92A/98 | P A R S S R | This study |
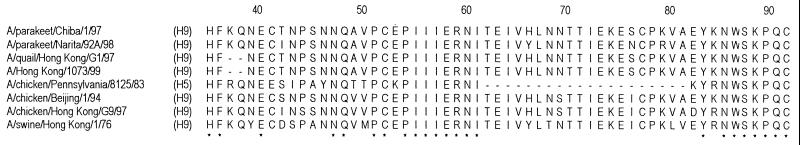
The parakeet viruses also differed from the H9N2 human and G1 quail viruses in the length of the NA stalk. In contrast to the latter viruses, whose NA stalks are two amino acid residues shorter than other N2 NA stalks, the parakeet virus NAs had the same number of amino acid residues in this region as other N2 NAs (Fig. 1). Interestingly, the Hong Kong H5N1 viruses also have a shorter NA stalk than do other N1 viruses (28, 32). Although the biological significance of this finding is uncertain, a shorter NA stalk has been associated with high virulence in poultry (8).
FIG. 1.
Comparison of NA stalks of representative N2 influenza A viruses. Identical amino acids are shown by asterisks, and the positions of deletions are shown by dashes. The HA subtype of each virus is shown in parentheses.
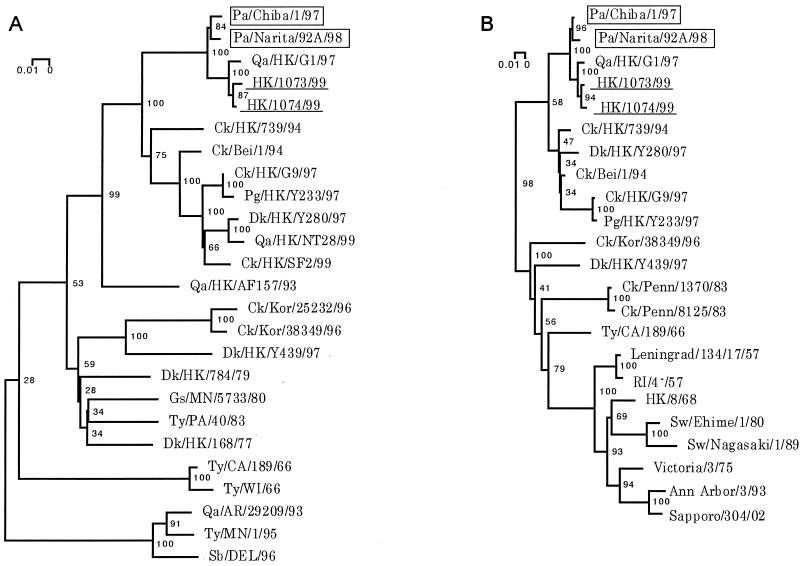
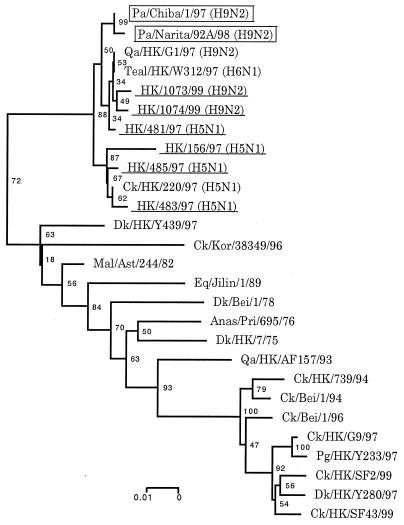
Phylogenetic trees for the HA and NA genes constructed by the neighbor-joining method (25) reinforced viral RNA sequencing results, suggesting that the parakeet viruses share an immediate ancestor with the H9N2 human and G1 quail viruses (Fig. 2). Phylogenetic analysis of the entire coding regions of the NP, PB2, PB1, PA, M, and NS genes showed that the parakeet viruses cluster with the H9N2 human and G1 quail viruses as well as human H5N1 Hong Kong viruses. A phylogenetic tree of the NP gene is shown as an example (Fig. 3).
FIG. 2.
Phylogenetic trees of the H9 HA (A) and N2 NA (B) genes of influenza A viruses. Nucleotides 46 to 1091 (1,046 bp) of the H9 HAs and nucleotides 1 to 1386 (1,386 bp) of the N2 NAs were used for phylogenetic analysis. Horizontal distances are proportional to the minimum number of nucleotide differences required to join nodes and sequences. Viruses isolated from parakeets or humans are boxed or underlined, respectively. Numbers at the nodes indicate confidence levels of bootstrap analysis with 1,000 replications as a percentage value. Abbreviations: Pa, parakeet; Qa, quail; HK, Hong Kong; Ck, chicken; Bei, Beijing; Pg, pigeon; Dk, duck; Kor, Korea; Gs, goose; MN, Minnesota; Ty, turkey; PA, Pavia; CA, California; WI, Wisconcin; AR, Arizona; Sb, shorebird; DEL, Delaware; Penn, Pennsylvania; Sw, swine.
FIG. 3.
Evolutionary relationship of influenza A virus NP genes. Nucleotides 741 to 1398 (658 bp) of the NP genes were used for phylogenetic analysis. All viruses underlined were isolated from humans. Horizontal distances are proportional to the minimum number of nucleotide differences required to join nodes and sequences. Viruses isolated from parakeets or humans are boxed or underlined, respectively. Numbers at the nodes indicate confidence levels of bootstrap analysis with 1,000 replications as a percentage value. Abbreviations: Pa, parakeet; Qa, quail; HK, Hong Kong; Ck, chicken; Dk, duck; Kor, Korea; Mal, mallard; Ast, Astrakhan; Eq, equine; Bei, Beijing; Anas, Anas acuta; Pri, Primorje; Pg, pigeon.
A mutation at the initiation codon of the NP gene of A/parakeet/Narita/92A/98 virus (CUG instead of AUG) was detected. Sequencing analysis of this region was repeated five times with identical results. The CUG initiation codon has not been observed in the genes of any influenza virus; however, it is the most efficient non-AUG initiation codon (15) and is used in the synthesis of both eukaryotic and viral proteins (7, 15, 24). Thus, we suggest that this CUG codon is used as an initiation codon for NP translation, although we cannot speculate on its biologic significance in the NP gene of A/parakeet/Narita/92A/98.
G1 quail virus replicated in chickens and ducks without producing any disease signs, but it spread to the brain in mice (10). We therefore examined the pathogenicities of our H9N2 parakeet viruses in chickens and mice by intranasally inoculating 6-week-old specific-pathogen-free chickens (approximately 107 50% egg infective doses/chicken; eight chickens/virus). Viruses were recovered from tracheal and cloacal swabs, but none of the infected chickens showed any disease signs. When 5-week-old female BALB/c mice (Japan Clea) were intranasally inoculated with virus (approximately 107 50% egg infective doses; 12 mice/virus), all survived without signs of disease. Of all organs tested, the lung was the only site from which virus was recovered at 3 and 6 days postinfection (data not shown). These results indicate that the parakeet viruses differ from G1 quail virus in both tissue tropism and virulence in mice and hence contain amino acid alterations that would account for the discrepant phenotypes.
Taken together, our findings indicate that H9N2 viruses, genetically closely related to those transmitted to humans (13, 21, 22), were circulating in psittacines in Pakistan in 1997 to 1998 and were introduced into Japan during this period. Thus, viruses with the potential for bird-to-human transmission may still be circulating in countries other than China and may be spreading across geographical boundaries through the importation and sale of wild psittacine birds as pets.
An H9N2 influenza virus caused an influenza outbreak in poultry in Pakistan in 1999 (16). Although the causative virus was never fully characterized, the HA cleavage site sequence was identical to that of our viruses. Efforts to clarify the relationship between the Pakistan virus and ours will depend on more complete molecular characterization of the former.
Avian influenza A viruses are genetically divided into two geographically based lineages: Eurasian and American. The G1 quail strain and the 1997 H5N1 Hong Kong virus belong to the Eurasian lineage. Recent surveillance studies indicate that Eurasian avian viruses can be subdivided further and that the NP genes of the Hong Kong H5N1 and H9N2 genes belong to one of these sublineages (10, 20). Although earlier experimental attempts to infect human volunteers with avian influenza viruses had failed (5), the Hong Kong H5N1 and H9N2 viruses were clearly able to infect humans. Thus, the H5N1 and H9N2 Hong Kong viruses seem unique in their ability to replicate efficiently in humans. Although amino acid differences were found between these Hong Kong and other avian viruses, it is still not clear which differences account for the direct transmissibility of the former viruses to humans or, alternatively, which simply represent unique traits of the different avian lineages in nature. Thus, further genetic studies of Eurasian avian influenza viruses are needed to evaluate the potential of avian viruses to cross host range barriers.
The PB2, PA, NP, and M proteins of the 1997 H5N1 viruses contained human virus-like amino acid residues that might have been responsible for direct transmission of the avian virus to humans (32). Some of these residues are also found in the H9N2 parakeet viruses: position 661 (Ala [avian] and Thr [human]) in PB2, the residue located in the region responsible for interaction with other polymerase components (23), and position 136 (Leu [avian] and Met [human]) in NP, the residue in the RNA binding domain of the protein (1). To unequivocally determine the contributions of these amino acid residues to the viruses' replicative capacity in mammals, one needs to generate viruses with specific mutations, using reverse genetics (12, 18).
In conclusion, the international trade of exotic pet birds carrying influenza A viruses may pose a serious health threat to humans. Previously, psittacine birds have not been thought to play a major role in the epizootiology and epidemiology of influenza A viruses. Since the trading of these birds across regional and international boundaries is extensive (over 400,000 nonpoultry birds [50,000 from Pakistan] were imported to Japan each year for the last 5 years), the risk of worldwide dissemination of potentially virulent influenza A virus is considerable. Thus, adequate quarantine and surveillance systems should be established in countries engaging in such trade.
Nucleotide sequence accession numbers.
All sequences used in this study were sent to DDBJ, and the accession numbers are AB049153 to AB049168.
Acknowledgments
Support for this work came from the Ministry of Education and Culture of Japan and NIAID Public Health Service research grants and from the Japan Health Science Foundation.
We thank John Gilbert for scientific editing.
REFERENCES
- 1.Albo C, Valencia A, Portela A. Identification of an RNA binding region within the N-terminal third of the influenza A virus nucleoprotein. J Virol. 1995;69:3799–3806. doi: 10.1128/jvi.69.6.3799-3806.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander D J, Allan W H, Parsons G. Characterisation of influenza viruses isolated from turkeys in Great Britain during 1963–1977. Res Vet Sci. 1979;26:17–20. [PubMed] [Google Scholar]
- 3.Alexander D J. A review of avian influenza in different bird species. Vet Microbiol. 2000;74:3–13. doi: 10.1016/s0378-1135(00)00160-7. [DOI] [PubMed] [Google Scholar]
- 4.Banks J, Speidel E C, Harris P A, Alexander D J. Phylogenetic analysis of influenza A viruses of H9 haemagglutinin subtype. Avian Pathol. 2000;29:353–359. doi: 10.1080/03079450050118485. [DOI] [PubMed] [Google Scholar]
- 5.Beare A S, Webster R G. Replication of avian influenza viruses in humans. Arch Virol. 1991;119:37–42. doi: 10.1007/BF01314321. [DOI] [PubMed] [Google Scholar]
- 6.Claas E C, de Jong J C, van Beek R, Rimmelzwaan G F, Osterhaus A D. Human influenza virus A/HongKong/156/97 (H5N1) infection. Vaccine. 1998;16:977–978. doi: 10.1016/s0264-410x(98)00005-x. [DOI] [PubMed] [Google Scholar]
- 7.Dasso M C, Jackson R J. Efficient initiation of mammalian mRNA translation at a CUG codon. Nucleic Acids Res. 1989;17:6485–6497. doi: 10.1093/nar/17.16.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deshpande K L, Naeve C W, Webster R G. The neuraminidases of the virulent and avirulent A/Chicken/Pennsylvania/83 (H5N2) influenza A viruses: sequence and antigenic analyses. Virology. 1985;147:49–60. doi: 10.1016/0042-6822(85)90226-0. [DOI] [PubMed] [Google Scholar]
- 9.Guan Y, Shortridge K F, Krauss S, Webster R G. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci USA. 1999;96:9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo Y J, Krauss S, Senne D A, Mo I P, Lo K S, Xiong X P, Norwood M, Shortridge K F, Webster R G, Guan Y. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology. 2000;267:279–288. doi: 10.1006/viro.1999.0115. [DOI] [PubMed] [Google Scholar]
- 11.Hiromoto Y, Yamazaki Y, Fukushima T, Saito T, Lindstrom S E, Omoe K, Nerome R, Lim W, Sugita S, Nerome K. Evolutionary characterization of the six internal genes of H5N1 human influenza A virus. J Gen Virol. 2000;81:1293–1303. doi: 10.1099/0022-1317-81-5-1293. [DOI] [PubMed] [Google Scholar]
- 12.Horimoto T, Kawaoka Y. Reverse genetics provides direct evidence for a correlation of hemagglutinin cleavability and virulence of an avian influenza A virus. J Virol. 1994;68:3120–3128. doi: 10.1128/jvi.68.5.3120-3128.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin Y P, Shaw M, Gregory V, Cameron K, Lim W, Klimov A, Subbarao K, Guan Y, Krauss S, Shortridge K, Webster R, Cox N, Hay A. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc Natl Acad Sci USA. 2000;97:9654–9658. doi: 10.1073/pnas.160270697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madeley C R, Allan W H, Kendal A P. Studies with avian influenza A viruses: serological relations of the haemagglutinin and neuraminidase antigens of ten virus isolates. J Gen Virol. 1971;12:69–78. doi: 10.1099/0022-1317-12-2-69. [DOI] [PubMed] [Google Scholar]
- 15.Mehdi H, Ono E, Gupta K C. Initiation of translation at CUG, GUG, and ACG codons in mammalian cells. Gene. 1990;91:173–178. doi: 10.1016/0378-1119(90)90085-6. [DOI] [PubMed] [Google Scholar]
- 16.Naeem K, Ullah A, Manvell R J, Alexander D J. Avian influenza A subtype H9N2 in poultry in Pakistan. Vet Rec. 1999;145:560. doi: 10.1136/vr.145.19.560. [DOI] [PubMed] [Google Scholar]
- 17.Nestorowicz A, Kawaoka Y, Bean W J, Webster R G. Molecular analysis of the hemagglutinin genes of Australian H7N7 influenza viruses: role of passerine birds in maintenance or transmission? Virology. 1987;160:411–418. doi: 10.1016/0042-6822(87)90012-2. [DOI] [PubMed] [Google Scholar]
- 18.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez D R, Donis R, Hoffmann E, Hobom G, Kawaoka Y. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci USA. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nobusawa E, Aoyama T, Kato H, Suzuki Y, Tateno Y, Nakajima K. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology. 1991;182:475–485. doi: 10.1016/0042-6822(91)90588-3. [DOI] [PubMed] [Google Scholar]
- 20.Okazaki K, Takada A, Ito T, Imai M, Takakuwa H, Hatta M, Ozaki H, Tanizaki T, Nagano T, Ninomiya A, Demenev V A, Tyaptirganov M M, Karatayeva T D, Yamnikova S S, Lvov D K, Kida H. Precursor genes of future pandemic influenza viruses are perpetuated in ducks nesting in Siberia. Arch Virol. 2000;145:885–893. doi: 10.1007/s007050050681. [DOI] [PubMed] [Google Scholar]
- 21.Peiris M, Yuen K Y, Leung C W, Chan K H, Ip P L, Lai R W, Orr W K, Shortridge K F. Human infection with influenza H9N2. Lancet. 1999;354:916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 22.Peiris M, Yam W C, Chan K H, Ghose P, Shortridge K F. Influenza A H9N2: aspects of laboratory diagnosis. J Clin Microbiol. 1999;37:3426–3427. doi: 10.1128/jcm.37.10.3426-3427.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perales B, de la Luna S, Palacios I, Ortin J. Mutational analysis identifies functional domains in the influenza A virus PB2 polymerase subunit. J Virol. 1996;70:1678–1686. doi: 10.1128/jvi.70.3.1678-1686.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prats A C, De Billy G, Wang P, Darlix J L. CUG initiation codon used for the synthesis of a cell surface antigen coded by the murine leukemia virus. J Mol Biol. 1989;205:363–372. doi: 10.1016/0022-2836(89)90347-1. [DOI] [PubMed] [Google Scholar]
- 25.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 26.Shortridge K F, Zhou N N, Guan Y, Gao P, Ito T, Kawaoka Y, Kodihalli S, Krauss S, Markwell D, Murti K G, Norwood M, Senne D, Sims L, Takada A, Webster R G. Characterization of avian H5N1 influenza viruses from poultry in Hong Kong. Virology. 1998;252:331–342. doi: 10.1006/viro.1998.9488. [DOI] [PubMed] [Google Scholar]
- 27.Shortridge K F, Gao P, Guan Y, Ito T, Kawaoka Y, Markwell D, Takada A, Webster R G. Interspecies transmission of influenza viruses: H5N1 virus and a Hong Kong SAR perspective. Vet Microbiol. 2000;74:141–147. doi: 10.1016/s0378-1135(00)00174-7. [DOI] [PubMed] [Google Scholar]
- 28.Suarez D L, Perdue M L, Cox N, Rowe T, Bender C, Huang J, Swayne D E. Comparisons of highly virulent H5N1 influenza A viruses isolated from humans and chickens from Hong Kong. J Virol. 1998;72:6678–6688. doi: 10.1128/jvi.72.8.6678-6688.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 30.Utterback W W, Schwartz J H. Epizootiology of velogenic viscerotropic Newcastle disease in southern California, 1971–1973. J Am Vet Med Assoc. 1973;163:1080–1088. [PubMed] [Google Scholar]
- 31.Webster R G, Bean W J, Gorman O T, Chambers T M, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou N N, Shortridge K F, Claas E C J, Krauss S L, Webster R G. Rapid evolution of H5N1 influenza viruses in chickens in Hong Kong. J Virol. 1999;73:3366–3374. doi: 10.1128/jvi.73.4.3366-3374.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]