Abstract
We isolated a revertant virus after prolonged culturing of a replication-impaired human immunodeficiency virus type 1 (HIV-1) mutant of which the Rev open reading frame was inactivated by mutation of the AUG translation initiation codon. Sequencing of the tat-rev region of this revertant virus identified a second-site mutation in tat that restored virus replication in the mutant background. This mutation activated a cryptic 5′ splice site (ss) that, when used in conjunction with the regular HIV 3′ ss #5, fuses the tat and rev reading frames to encode a novel T-Rev fusion protein that rescues Rev function. We also demonstrate an alternative route to indirectly activate this cryptic 5′ ss by mutational inactivation of an adjacent exon splicing silencer element.
Human immunodeficiency virus type 1 (HIV-1) transcripts contain multiple splicing signals that are utilized by the cellular splicing machinery to produce a large set of subgenomic mRNAs. More than 30 mRNA species are produced by differential usage of the 5′ and 3′ splice sites (ss) that are present in the HIV pre-mRNA (reviewed in reference 20). This splicing process is highly regulated by mechanisms that control the efficiency by which these splicing signals are recognized by components of the splicing machinery (19). For instance, suboptimal branch point sequences are used for lariat formation (12), and splicing enhancer and silencer elements regulate the activity of splice signals (1, 2, 22). The viral Rev protein stimulates the nuclear export of unspliced and partially spliced HIV-1 transcripts through its binding to a structured RNA motif termed the Rev-responsive element (RRE) located in the env gene (20). At low Rev levels, primary HIV transcripts will be multiply spliced, producing transcripts that encode the early viral proteins Tat, Nef, and Rev itself. Upon accumulation of Rev protein in the nucleus, unspliced and singly spliced transcripts appear in the cytoplasm. The latter mRNAs encode the Vif, Vpr, Vpu, and Env proteins. The full-length, unspliced HIV RNA serves two functions. As mRNA, it produces Gag and Gag-Pol polyproteins, and as viral genome it is packaged in virus particles. A balanced and properly timed expression of viral transcripts and proteins is required to ensure efficient production of new, infectious HIV virions. The Rev protein is critical for this regulation and is therefore an essential viral function.
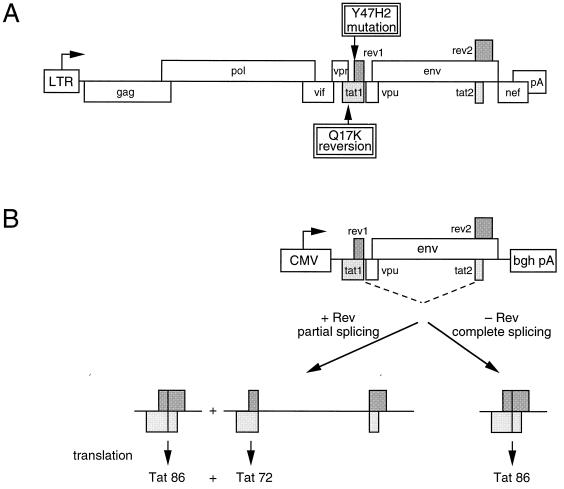
This study describes how a replication-impaired virus mutant with an inactivated Rev start codon can be repaired by activation of a cryptic 5′ ss in the tat gene. This leads to the synthesis of a functional Tat-Rev fusion protein. A large set of mutants was previously generated in a study on the structure and function of the HIV-1 Tat protein (26). The mutant proteins were tested for transcriptional activity in transfections with reporter constructs and in virus replication assays. Some of the mutations that target the tyrosine (Y) residue at codon 47 also affect the overlapping Rev translation initiation codon. For instance, the same Tat amino acid substitution is present in Y47H1 and Y47H2, but only the latter mutation destroys the overlapping Rev start codon (26). The Y47H1 virus replicates suboptimally due to a partially active Tat protein. The Y47H2 virus has the same Tat problem, but this mutant is replication impaired because expression of the essential Rev protein is abolished. We tried to select for virus revertants through prolonged culturing of this replication-impaired Y47H2 virus (25). Despite multiple attempts, we generated only one candidate Y47H2 revertant. Sequencing of the Tat-Rev region identified a second-site mutation (Q17K) in the tat gene, upstream of the original Y47H2 mutation that was maintained (Fig. 1A). The revertant tat gene, with the point mutation that alters Tat codon 17 from CAG (glutamine; Q) to AAG (lysine; K), was subsequently cloned in a eukaryotic expression vector. Transient transfection of this plasmid with a long terminal repeat-chloramphenicol acetyltransferase reporter construct in SupT1 T cells revealed that the Q17K second-site mutation does not improve Tat function (data not shown and reference 25).
FIG. 1.
(A) Genetic organization of the HIV-1 provirus. The position of the Y47H2 mutation at the tat-rev border and the Q17K second-site reversion in the tat gene are indicated. The construction of the Y47H2 virus and the selection of the Y47H2 Q17K revertant were described previously (25). (B) The pcDNA3-Tat expression vector can be used as a Rev reporter construct. Both one-exon Tat (72-amino acid form) and two-exon Tat (86-amino acid form) are expressed if there is partial splicing in the presence of Rev. In the absence of Rev translation, there are complete splicing and exclusive synthesis of the extended Tat form.
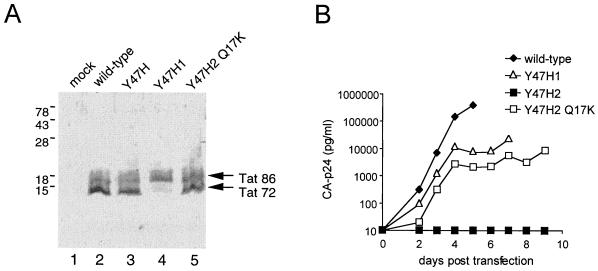
Steady-state expression of the wild-type, mutant, and revertant Tat protein was verified by Western blot analysis of lysates from COS cells that were transiently transfected with a Tat/Rev expression vector (Fig. 1B). Expression of wild-type Tat from this subgenomic expression construct produces two bands (Fig. 2A, lane 2). The upper band corresponds to the full-length, 86-amino acid Tat protein that is expressed from the spliced transcript (Tat 86). The lower band represents a truncated form of Tat (Tat 72) that is translated from unspliced mRNA. This latter Tat form predominates, due to the Rev-mediated nuclear export of the unspliced mRNA (the RRE is also encoded by this construct). The wild-type Tat pattern is observed for the Y47H1 mutant (lane 3) but not for Y47H2 (lane 4). This result suggests that the Y47H2 mutant cannot express the Rev protein. However, the normal Tat pattern is obtained with the Y47H2 Q17K double mutant (lane 5), indicating that Rev expression is restored.
FIG. 2.
(A) Western blot analysis of the Tat protein forms expressed by the different constructs in an indirect assay for Rev activity. COS cells were transfected with 10 μg of the indicated Tat expression plasmids, cell lysates were prepared at 3 days posttransfection, and Western blotting was performed with a monoclonal anti-Tat antibody (Ab #2 in reference 26). (B) Growth kinetics of wild-type HIV-1 LAI, the Y47H1 and Y47H2 mutants, and the Y47H2 Q17K revertant virus variants. SupT1 T cells were electroporated with 4 μg of the HIV-1 molecular clones containing the indicated mutations. Replication was monitored by measuring CA-p24 levels in the culture supernatant by enzyme-linked immunosorbent assay.
We next sought to determine whether the Q17K mutation is able to restore replication of a virus containing the inactivating Y47H2 mutation. To test this, we cloned the revertant tat gene, containing the Y47H2 and Q17K mutations, back into the pLAI molecular clone of HIV-1 and measured virus replication after transfection of SupT1 cells (Fig. 2B). Several appropriate control constructs were transfected in parallel, and CA-p24 production in the culture supernatant was monitored for several days. Wild-type LAI virus spreads rapidly, and this culture was stopped at 5 days posttransfection because of massive, virus-induced cell death. In contrast, we did not measure any virus replication for the Rev-minus Y47H2 virus, which encodes a partially active Tat protein. Delayed virus replication was measured for the Y47H1 virus, which encodes the same Tat mutant but with normal Rev expression. The Y47H2 Q17K double mutant replicated to almost the same extent as the Y47H1 mutant, which is consistent with the finding that Rev expression is restored.
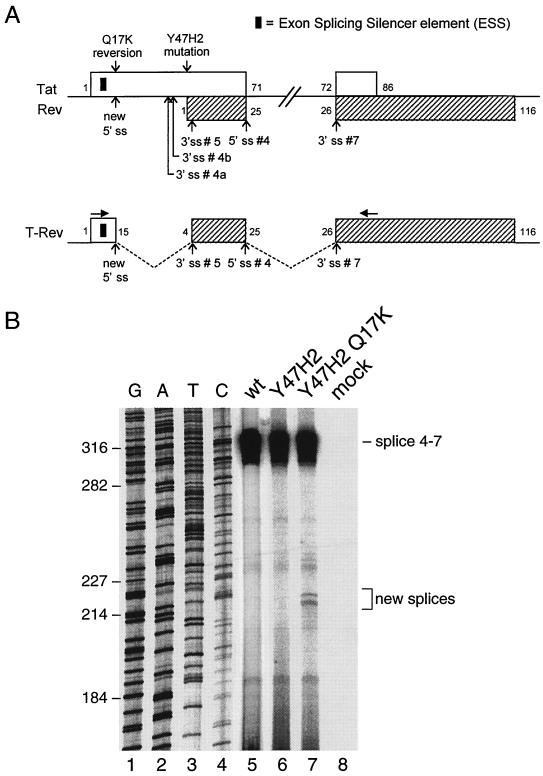
It is not immediately obvious how the Q17K mutation in Tat can rescue Rev expression, as the mutation does not simply create a new translational start codon for the Rev open reading frame. We therefore analyzed the wild-type and revertant sequences for differences in splicing potential. The sequence around codon 17 of both the wild-type and revertant tat gene was entered into a computer program that predicts 5′ and 3′ ss (http://www.fruitfly.org). Whereas the wild-type sequence was scored unlikely to be recognized as a splicing signal (P < 0.10; . .AAGUCAGC. .; with the putative 5′ ss core nucleotides underlined), the revertant sequence scored as a high probability 5′ ss (P = 0.91; …AAGUAAGC, altered nucleotide in bold). Several downstream 3′ ss can be used as acceptors to combine with this putative 5′ ss to generate novel mRNA species (Fig. 3A, top). In fact, usage of the putative 5′ ss in combination with the downstream 3′ ss #5 that is normally used to express the Env-Nef genes will fuse the Tat and Rev open reading frames to encode a new Tat-Rev fusion protein (Fig. 3A, bottom). In this way, translation of the functional Rev domains would be under control of the Tat AUG start codon and thus bypass the inactivated Rev start codon.
FIG. 3.
(A) Schematic of the known splicing signals in the tat-rev coding regions of the HIV-1 genome. The putative 5′ ss (new 5′ ss) generated by the Q17K mutation is indicated. This new 5′ ss, in combination with 3′ ss #5, generates a Tat-Rev fusion open reading frame that encodes the T-Rev protein. Several amino acid positions in the respective open reading frames are indicated. The black rectangle in the tat gene is the ESS element (see also Fig. 4). The primers used in RT-PCR analysis hybridize at the positions indicated by the horizontal arrows. (B) RT-PCR analysis of spliced HIV transcripts produced from infected SupT1 T cells. Total RNA was isolated, DNase I treated, and used as template in RT-PCR with the HIV-specific 3′ Tat1 primer (5′ TTTGAATTCTAATCGAATGGATCTGTCTC 3′) and Tat-AUG (5′ ATGGAGCCAGTAGATCCTAG 3′) in a standard 35-cycle PCR. Products were resolved on a sequencing gel (a standard dideoxy sequencing reaction was run in parallel as a marker). Several new, low-abundance splice signals, specific for the Y47H2 Q17K revertant, are indicated. These products were purified from gel, cloned, and sequenced using standard techniques.
To test this hypothesis, we transfected SupT1 T cells with molecular clones of wild-type, mutant, and revertant genotype and isolated viral RNA at 2 days posttransfection. Sequences encompassing the tat gene were amplified in a reverse transcription (RT)-PCR (primer positions are indicated in Fig. 3A). A prominent, slowly migrating RT-PCR product was observed for all virus variants (Fig. 3B). The size of this DNA fragment corresponds to the expected size of the regular tat exon 1-2 splice product (using 5′ ss #4 and 3′ ss #7), and this was verified by sequence analysis. For the revertant virus, several additional, faster-migrating RT-PCR products were observed (new splices; lane 7). These DNA fragments were excised from gel, amplified, and cloned. Sequence analysis of several clones revealed that the novel 5′ ss, generated by the Q17K change in the tat gene, was utilized in all cases. The heterogeneity in size of the cDNA fragments stems from the combination of this new 5′ ss with alternative 3′ ss 4a, 4b, or 5. Of the 4 clones that were sequenced, the new 5′ ss was combined twice with 3′ ss #4b and once with #4a and #5. The splices with 3′ ss #4a/4b yield mRNAs that encode an internally deleted Tat protein that is likely to be inactive due to the absence of the cysteine-rich domain. The splicing event with 3′ ss #5 is special in that an mRNA is produced that encodes a Tat-Rev fusion protein that we have coined T-Rev (Fig. 3A). The transcript encodes the first 15 amino acids of Tat, an Arg codon that is generated at the splice, and the nearly complete Rev protein (residues 4 to 116). The T-Rev protein lacks the functional Tat domains but is predicted to have Rev function. T-Rev contains all essential Rev domains: the arginine-rich domain that is required for nuclear localization and binding to the RRE (residues 35 to 50), sequences that mediate Rev multimerization, and the leucine-rich domain (residues 75 to 83) that functions as an activation domain-nuclear export sequence.
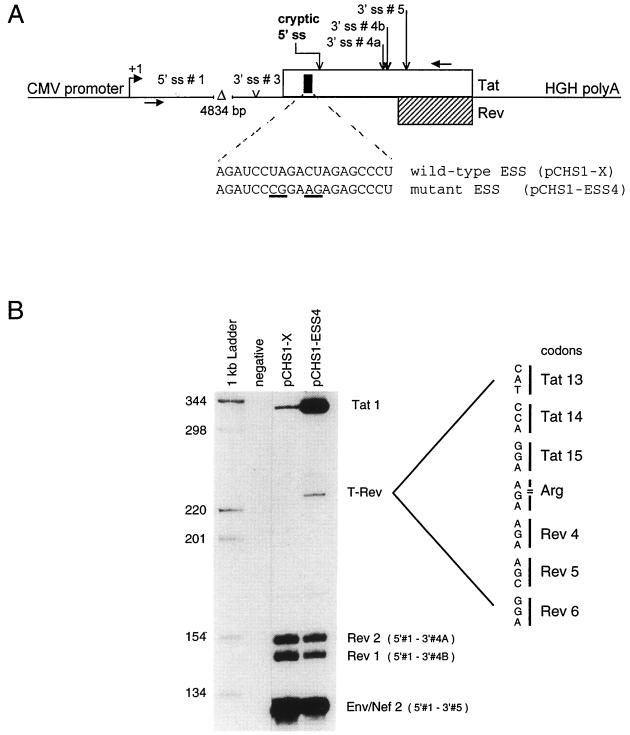
These results indicate that a point mutation in the 5′ part of the tat gene creates a novel splicing signal that rescues Rev expression in the context of the Y47H2 mutant. The wild-type HIV-1 Tat sequence is not utilized as 5′ ss. This may in part be due to the presence of an exon splicing silencer (ESS) element immediately upstream from this site (Fig. 4A) (1). To test this possibility, we inactivated the ESS element by four base substitutions as indicated in Fig. 4A. Two effects were observed upon inactivation of the ESS enhancer (Fig. 4B). First, there is a selective increase in splicing to the Tat ss, as evidenced by the increase in Tat1 RNA product. Second, a novel splice product was observed by RT-PCR. Cloning and sequencing of the unique cDNA product revealed that the GU dinucleotide at tat codon 16 had served as 5′ ss in combination with 3′ ss #5, thereby creating the T-Rev open reading frame.
FIG. 4.
(A) Schematic of a subgenomic cytomegalovirus-driven cassette expressing the untranslated HIV-1 leader RNA, containing the major 5′ ss, and coupled to the Tat-Rev coding region. The various 5′ and 3′ splice signals in this construct are indicated, as well as a well-characterized ESS element (black rectangle in 5′ half of the tat gene). Mutant ESS4 contains specific mutations in this element that abolish the silencing function. Horizontal arrows indicate the 5′ to 3′ direction of the primers used for the analysis of spliced products in the Tat-Rev coding regions. (B) RT-PCR analysis of HIV-1-specific transcripts from transfected cells. The RT-PCRs were performed essentially as described elsewhere(9). The sequence around the novel splice junction and the new T-Rev open reading frame is indicated.
The analysis of spontaneous revertant viruses that appear in long-term cultures of replication-impaired HIV-1 mutants is a powerful genetic approach to dissect complex regulatory mechanisms and to study nucleic acid and protein motifs that control virus replication (7). We constructed an HIV-1 mutant with a single amino acid substitution in the Tat protein (Y47H2) that partially inactivates Tat function. However, this same mutation inactivates the start codon of the overlapping rev gene (AUG to ACG), thereby causing a severe replication defect. The Tat-Rev coding region of a revertant virus was sequenced. Surprisingly, no repair of the Rev start codon was apparent, but instead a second-site mutation was observed in the upstream Tat sequences, changing codon position 17 in Tat (Q17K). No improved Tat function was measured in transient assays, but we could nevertheless demonstrate that this mutation in the tat gene was responsible for restored virus replication. Furthermore, the reversion mutation was able to restore Rev expression and Rev function as measured in sensitive reporter assays. Interestingly, we detected novel spliced transcripts by RT-PCR, and sequence analysis revealed that a cryptic 5′ ss was activated in the revertant. The reversion mutation improves the sequence of the cryptic 5′ ss (AA/GUCAGC to AA/GUAAGC; the consensus 5′ ss is AG/GUAAGC). This new 5′ ss is linked to common 3′ ss further downstream in the HIV-1 genome. Combination with the vpu-env 3′ ss (3′ ss #5) produces a new open reading frame in which the N terminus of Tat (amino acids 1 to 15) is fused to most of Rev (amino acids 4 to 116). Thus, Rev expression is restored by a novel splicing event that places the Rev open reading frame in register with the Tat translational start codon, generating a novel fusion protein that we termed T-Rev. Another Tat-Rev fusion protein has been described previously and was termed Tnv (21) or Tev (4). This Tev-Tnv fusion protein is much different from T-Rev because it fuses the first coding exon of Tat and the second coding exon of Rev, joined together by sequences derived from the env gene. The Tnv-Tev protein exhibits both Tat and Rev activity and can be detected in cells that are latently infected with HIV-1 as well as in T cells that have been transfected with HIV-1 molecular clones.
The T-Rev fusion protein could not be detected directly by Western blotting because the new 5′ ss is used very inefficiently (approximately 2 to 6% of the regular Tat splice). For instance, we used a monoclonal antibody directed against the Tat N-terminal 15 amino acids, which readily detects the Tat protein itself but not the T-Rev protein (data not shown). Thus, a low level of T-Rev splicing is apparently sufficient to rescue virus replication. It seems likely that more frequent use of the new splice pattern will have a negative impact on virus replication because it will compete with the synthesis of Tat-encoding mRNAs. These results indicate that very little Rev protein is needed, although it is possible that T-Rev translation is more efficient due to the stronger translation start signal provided by the tat AUG than by the usual rev AUG.
We described activation of the cryptic 5′ ss by a single nucleotide substitution. The cryptic splicing signal is too weak in the wild-type HIV-1 context. This is due in part to the presence of an adjacent ESS element, because the cryptic 5′ ss can also be triggered by inactivation of this ESS element. Interestingly, a recent study of pseudoexons indicated that inactivation of cryptic ss by ESS elements may be a more general mechanism (24).
Activation of a cryptic 5′ ss has also been reported upon inactivation of the major 5′ ss in the untranslated leader (10, 19). It thus appears that the HIV-1 genome is riddled with spare ss that may help the virus to overcome deleterious mutations. It has been proposed that the cryptic splice signals may have an active regulatory function. These elements may act as a cis-acting repressor sequence to establish Rev dependence of the viral RNA (10). These combined results underscore both the complexity and plasticity of the HIV-1 genome. Coordinated viral gene expression results from the interplay of multiple regulatory signals in the retroviral genome, but complex gene expression strategies are also likely to be important for the more simple animal retroviruses (3, 11, 15). Simple retroviruses have certainly evolved strategies for maintaining appropriate levels of splicing. In avian cells, it has been shown that the two 3′ ss of Rous sarcoma virus (env and src) are both suboptimal because of nonconsensus polypyrimidine tracts or branch point sequences (15, 27). In addition, other cis elements upstream and downstream of the 3′ ss, as well as within the gag gene, are necessary to control the levels of splicing (3, 6, 13, 14, 18, 23). In mammalian cell types, a cryptic 5′ ss within the env gene is activated in the formation of a double-spliced mRNA containing sequences from both the env and src genes (5, 17). Although the intricate gene expression circuit of HIV-1 is vulnerable to mutational inactivation, the plasticity of the HIV-1 genome allows subsequent repair strategies by acquisition of spontaneous reverse transcriptase errors and subsequent selection of beneficial second-site mutations. This plasticity is underscored by the forced evolution experiments presented in this and other in vitro studies (8, 16), but the plasticity is also apparent from inspection of naturally evolved HIV-1 isolates that belong to the different subtypes (9). In fact, inspection of the Los Alamos HIV-1 sequence database (http://hiv-web.lanl.gov/) indicates that activation of the 5′ ss may also occur infrequently in vivo. Among 776 viral sequences, we identified four isolates with the Q17K mutation in the tat gene (UG273, subtype A; 98SE-MP1213, subtype AG; and MVP5180 and BCF06, both group O viruses).
Acknowledgments
We thank Wim van Est for artwork.
This study was sponsored by the Dutch AIDS Fund (AIDS Fonds, Amsterdam) and by U.S. Public Health Service grant AI36073 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Amendt B A, Hesslein D, Chang L-J, Stoltzfus C M. Presence of negative and positive cis-acting RNA splicing elements within and flanking the first Tat coding exon of human immunodeficiency virus type 1. Mol Cell Biol. 1994;14:3960–3970. doi: 10.1128/mcb.14.6.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amendt B A, Si Z-H, Stoltzfus C M. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: evidence for inhibition mediated by cellular factors. Mol Cell Biol. 1995;15:4606–4615. doi: 10.1128/mcb.15.8.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrigo S, Beemon K. Regulation of Rous sarcoma virus RNA splicing and stability. Mol Cell Biol. 1988;8:4858–4867. doi: 10.1128/mcb.8.11.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benko D M, Schwartz S, Pavlakis G N, Felber B K. A novel human immunodeficiency virus type 1 protein, Tev, shares sequences with Tat, Env, and Rev proteins. J Virol. 1990;64:2505–2518. doi: 10.1128/jvi.64.6.2505-2518.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berberich S L, Macias M, Zhang L, Turek L P, Stoltzfus C M. Comparison of Rous sarcoma virus RNA processing in chicken and mouse fibroblasts: evidence for double-spliced RNA in nonpermissive mouse cells. J Virol. 1990;64:4313–4320. doi: 10.1128/jvi.64.9.4313-4320.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berberich S L, Stoltzfus C M. Mutations in the regions of the Rous sarcoma virus 3′ splice sites: implications for regulation of alternative splicing. J Virol. 1991;65:2640–2646. doi: 10.1128/jvi.65.5.2640-2646.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkhout B, Das A T. Functional analysis of RNA signals in the HIV-1 genome by forced evolution. In: Barciszewski J, Clark B F C, editors. RNA biochemistry and biotechnology. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 249–275. [Google Scholar]
- 8.Berkhout B, Klaver B, Das A T. Forced evolution of a regulatory RNA helix in the HIV-1 genome. Nucleic Acids Res. 1997;25:940–947. doi: 10.1093/nar/25.5.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bilodeau P S, Domsic J K, Stoltzfus C M. Splicing regulatory elements within tat exon 2 of human immunodeficiency virus type 1 (HIV-1) are characteristic of group M but not group O HIV-1 strains. J Virol. 1999;73:9764–9772. doi: 10.1128/jvi.73.12.9764-9772.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borg K T, Favaro J P, Arrigo S J, Schmidt M. Activation of a cryptic splice donor in human immunodeficiency virus type-1. J Biomed Sci. 1999;6:45–52. doi: 10.1007/BF02256423. [DOI] [PubMed] [Google Scholar]
- 11.Déjardin J, Bompard-Maréchal G, Audit M, Hope T J, Sitbon M, Mougel M. A novel subgenomic murine leukemia virus RNA transcript results from alternative splicing. J Virol. 2000;74:3709–3714. doi: 10.1128/jvi.74.8.3709-3714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dyhr-Mikkelsen H, Kjems J. Inefficient spliceosome assembly and abnormal branch site selection in splicing of an HIV-1 transcript in vitro. J Biol Chem. 1995;270:24060–24066. doi: 10.1074/jbc.270.41.24060. [DOI] [PubMed] [Google Scholar]
- 13.Fu X-D, Katz R A, Skalka A M, Maniatis T. The role of branchpoint and 3′-exon sequences in the control of balanced splicing of avian retrovirus RNA. Genes Dev. 1991;5:211–220. doi: 10.1101/gad.5.2.211. [DOI] [PubMed] [Google Scholar]
- 14.Guo W, Winistorfer S C, Stoltzfus C M. Selective inhibition of splicing at the avian sarcoma virus src 3′ splice site by direct-repeat posttranscriptional elements. J Virol. 2000;74:8513–8523. doi: 10.1128/jvi.74.18.8513-8523.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katz R A, Skalka A M. Control of retroviral RNA splicing through maintenance of suboptimal processing signals. Mol Cell Biol. 1990;10:696–704. doi: 10.1128/mcb.10.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klaver B, Berkhout B. Evolution of a disrupted TAR RNA hairpin structure in the HIV-1 virus. EMBO J. 1994;13:2650–2659. doi: 10.1002/j.1460-2075.1994.tb06555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knight J B, Stinski M F, Stoltzfus C M. Avian sarcoma virus RNA synthesis, RNA splicing and virus production in human fibroblasts: effect of coinfection with human cytomegalovirus. J Gen Virol. 1993;74:2629–2636. doi: 10.1099/0022-1317-74-12-2629. [DOI] [PubMed] [Google Scholar]
- 18.McNally M T, Beemon K. Intronic sequences and 3′ splice sites control Rous sarcoma virus RNA splicing. J Virol. 1992;66:6–11. doi: 10.1128/jvi.66.1.6-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purcell D F J, Martin M A. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol. 1993;67:6365–6378. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rabson A B, Graves B J. Synthesis and processing of viral RNA. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 205–262. [PubMed] [Google Scholar]
- 21.Salfeld J, Gottlinger H G, Sia R A, Park R E, Sodroski J G, Haseltine W A. A tripartite HIV-1 tat-env-rev fusion protein. EMBO J. 1990;9:965–970. doi: 10.1002/j.1460-2075.1990.tb08195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Staffa A, Cochrane A. Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol Cell Biol. 1995;15:4597–4605. doi: 10.1128/mcb.15.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoltzfus C M, Fogarty S J. Multiple regions in the Rous sarcoma virus src gene intron act in cis to affect accumulation of unspliced RNA. J Virol. 1989;63:1669–1676. doi: 10.1128/jvi.63.4.1669-1676.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun H, Chasin L A. Multiple splicing defects in an intronic false exon. Mol Cell Biol. 2000;20:6414–6425. doi: 10.1128/mcb.20.17.6414-6425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verhoef K, Berkhout B. A second-site mutation that restores replication of a Tat-defective human immunodeficiency virus. J Virol. 1999;73:2781–2789. doi: 10.1128/jvi.73.4.2781-2789.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verhoef K, Koper M, Berkhout B. Determination of the minimal amount of Tat activity required for human immunodeficiency virus type 1 replication. Virology. 1997;237:228–236. doi: 10.1006/viro.1997.8786. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L, Stoltzfus C M. A suboptimal src 3′ splice site is necessary for efficient replication of Rous sarcoma virus. Virology. 1995;206:1099–1107. doi: 10.1006/viro.1995.1033. [DOI] [PubMed] [Google Scholar]