Abstract
Reovirus induces apoptosis in cultured cells and in vivo. Genetic studies indicate that the efficiency with which reovirus strains induce apoptosis is determined by the viral S1 gene, which encodes attachment protein ς1. However, the biochemical properties of ς1 that influence apoptosis induction are unknown. To determine whether the capacity of ς1 to bind cell surface sialic acid determines the magnitude of the apoptotic response, we used isogenic reovirus mutants that differ in the capacity to engage sialic acid. We found that T3SA+, a virus capable of binding sialic acid, induces high levels of apoptosis in both HeLa cells and L cells. In contrast, non-sialic-acid-binding strain T3SA− induces little or no apoptosis in these cell types. Differences in the capacity of T3SA− and T3SA+ to induce apoptosis are not due to differences in viral protein synthesis or production of viral progeny. Removal of cell surface sialic acid with neuraminidase abolishes the capacity of T3SA+ to induce apoptosis. Similarly, incubation of T3SA+ with sialyllactose, a trisaccharide comprised of lactose and sialic acid, blocks apoptosis. These findings demonstrate that reovirus binding to cell surface sialic acid is a critical requirement for the efficient induction of apoptosis and suggest that virus receptor utilization plays an important role in regulating cell death.
Many viruses are capable of inducing the genetically programmed death pathway that leads to apoptosis of infected cells (36, 50, 52). In some cases, apoptosis triggered by virus infection may serve as a host defense mechanism to limit viral replication (48). In other instances, induction of apoptosis may enhance viral infection by facilitating virus spread or allowing the virus to evade host inflammatory or immune responses (10, 36, 52). Although apoptosis is a common mechanism of cell death for many viruses, little is known about the biochemical pathways that lead to this cellular response. Such knowledge is of critical importance to an understanding of viral disease mechanisms and has the potential to lead to the development of novel antiviral therapeutics capable of apoptosis blockade.
Mammalian reoviruses have served as a useful experimental system for studies of viral pathogenesis. Reoviruses are nonenveloped viruses with a genome consisting of 10 segments of double-stranded RNA (33). Infection of newborn mice with reovirus results in injury to a variety of tissues, including the central nervous system, liver, and heart (54). Both in cultured cells (11, 41, 56) and in vivo (13, 35), reovirus infection induces the biochemical and morphological features of apoptosis. In cultured cells, reovirus infection triggers activation of nuclear factor kappa B (NF-κB) (11), a transcription factor known to play an important role in regulating cellular stress responses (37). Apoptosis induced by reovirus is significantly reduced in cells expressing a transdominant inhibitor of NF-κB and in cells deficient in the expression of NF-κB subunits p50 and RelA (p65) (11), indicating that NF-κB activation is required for reovirus-induced apoptosis.
Mechanisms by which reovirus infection triggers activation of NF-κB and apoptosis are unknown. However, the segmented nature of the reovirus genome has enabled identification of viral gene segments that determine the efficiency with which reovirus strains induce apoptosis. Reovirus strain type 3 Dearing (T3D) induces high levels of apoptosis in both murine L929 (L) fibroblast cells and Madin-Darby canine kidney (MDCK) epithelial cells, whereas strain type 1 Lang (T1L) induces low levels of apoptosis in both of these cell types (41, 55, 56). Analysis of reassortant viruses containing mixtures of gene segments from T1L and T3D indicate that the viral S1 gene is the primary determinant of differences in the efficiency of apoptosis induction exhibited by these strains. The S1 gene encodes two proteins, viral attachment protein ς1 and nonstructural protein ς1s (17, 23, 47). A ς1s-null reovirus mutant induces apoptosis with an efficiency equivalent to that of its ς1s-expressing parental strain (42). Therefore, the influence of the S1 gene in apoptosis induction is mediated by either the ς1 protein or the s1 RNA. Currently, there is no experimental evidence to distinguish between these two possibilities.
Although apoptosis efficiency differs between T1L and T3D in both L cells and MDCK cells, these strains produce equivalent yields in L cells (56). In MDCK cells, however, infection with T1L results in higher yields than infection with T3D. Differences in the growth of T1L and T3D in MDCK cells segregate with the viral L1 gene (41), which encodes λ3, a protein involved in viral RNA-dependent RNA polymerase activity (15, 30, 51). These results demonstrate that strain-specific differences in apoptosis efficiency are not linked to viral growth and suggest that the attachment step is a critical determinant of apoptosis induction.
To better understand mechanisms by which reovirus induces apoptosis, we investigated whether specific biochemical properties of ς1 influence apoptosis induction. The ς1 protein forms an elongated fibrous tail and a globular head (18, 20). Type 3 (T3) ς1 recognizes two cell surface receptors using discrete receptor-binding domains (16, 32, 61). Sequences in the T3 ς1 head bind junction adhesion molecule (JAM), a recently identified reovirus receptor (4). Sequences in the T3 ς1 tail bind sialic acid (8, 9, 14). Using isogenic reovirus mutants that differ by a single point mutation in the sialic acid-binding domain of ς1, we conducted experiments to determine whether the capacity of ς1 to bind sialic acid influences the efficiency of apoptosis induction. We found that sialic acid-binding reovirus strains induce high levels of apoptosis, whereas non-sialic-acid-binding reovirus strains induce little or no apoptosis. Differences in apoptosis induction are mediated by the capacity to engage cell surface sialic acid and are independent of viral protein synthesis and viral replication. These results indicate that reovirus binding to cell surface sialic acid is required to achieve maximal levels of apoptosis and provide the first direct evidence linking a ς1 function to modulation of the apoptotic response.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
HeLa cells were grown in Dulbecco's modified Eagle medium (Gibco BRL, Gaithersburg, Md.) supplemented with 10% fetal bovine serum (Intergen, Purchase, N.Y.), 2 mM l-glutamine, 100 U of penicillin per ml, 100 μg of streptomycin (Sigma-Aldrich Chemical Co., St. Louis, Mo.) per ml and 0.25 μg of amphotericin (Irvine Scientific, Santa Ana, Calif.) per ml. L cells were maintained as previously described (42).
T1L and T3D are laboratory stocks. T1L × T3D reassortant viruses were grown from stocks originally obtained from Kevin Coombs, Bernard Fields, and Max Nibert (6, 12, 59). Non-sialic-acid-binding reovirus strains T3 clone 43 (T3C43), T3 clone 44 (T3C44), and T3 clone 84 (T3C84) were obtained from the collection of L. Rosen (14, 43–45). Isolation and characterization of murine erythroleukemia (MEL) cell-adapted (MA) mutants T3C43-MA, T3C44-MA, and T3C84-MA (9) and isogenic ς1 point mutants T3/C44-SA− (T3SA−) and T3/C44MA-SA+ (T3SA+) have been previously described (3). Purified virion preparations were made using second- or third-passage L-cell lysate stocks of twice-plaque-purified reovirus (20).
Polyclonal rabbit antiserum raised against poly(ADP-ribose) polymerase (PARP) was obtained from Roche Molecular Biochemicals (Indianapolis, Ind.). Polyclonal rabbit anti-T1L antiserum was obtained by inoculating a New Zealand White rabbit with 100 μg of purified T1L virions in complete Freund's adjuvant, followed by booster doses of 100 μg of purified T1L virions in incomplete Freund's adjuvant (Cocalico, Reamstown, Pa.) at 2, 3, and 7 weeks after the initial inoculation. Antiserum was heat inactivated by incubation at 56°C for 60 min prior to use.
Quantitation of apoptosis by acridine orange staining.
Cells (5 × 104) grown in 24-well tissue culture plates (Costar, Cambridge, Mass.) were infected with reovirus virions at a multiplicity of infection (MOI) of 10 to 1,000 PFU per cell or treated with 10 μg of cycloheximide (Sigma-Aldrich) per ml alone or in combination with 10 ng of human recombinant tumor necrosis factor alpha (TNF-α; Sigma-Aldrich) per ml. The percentage of apoptotic cells was determined by acridine orange staining as previously described (56). For each experiment, 200 to 300 cells were counted, and the percentage of cells exhibiting condensed chromatin was determined by epi-illumination fluorescence microscopy using a fluorescein filter set (Zeiss Photomicroscope III; Carl Zeiss, Oberkochen, Germany).
Immunoblot for PARP cleavage.
Cells (5 × 106) grown in 75-cm2 tissue culture flasks (Costar) were infected with reovirus at an MOI of 100 PFU per cell. After viral adsorption for 1 h, cells were incubated at 37°C for 6 to 48 h. Nuclear extracts were prepared as previously described (11). Extracts (50 μg of total protein) were electrophoresed in 7% polyacrylamide gels (27) and transferred to nitrocellulose membranes. Immunoblots were performed as previously described (42), using PARP-specific primary antibody (Roche) followed by horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Amersham Pharmacia Biotech, Piscataway, N.J.) each diluted 1:2,000 in Tris-buffered saline containing 0.05% Tween 20 and 5% low-fat dry milk.
Electrophoretic mobility shift assay.
Cells (5 × 106) grown in 75-cm2 tissue culture flasks were adsorbed with reovirus at an MOI of 100 PFU per cell. After incubation at 37°C for 10 h, nuclear extracts were prepared as previously described (11). Nuclear extracts (10 μg total protein) were assayed for NF-κB activation by electrophoretic mobility shift assay using a 32P-labeled oligonucleotide consisting of the NF-κB consensus binding sequence (Santa Cruz Biotechnology, Santa Cruz, Calif.) as previously described (11).
Quantitation of reovirus growth.
Cells (105) grown in 24-well plates were infected with reovirus at an MOI of 10 to 1,000 PFU per cell. After viral adsorption for 1 h, the inoculum was removed, cells were washed once with phosphate-buffered saline (PBS), 1.0 ml of fresh medium was added, and cells were incubated at 37°C for 0, 24, or 48 h. After incubation, cells and culture media were frozen (−70°C) and thawed twice, and viral titers in cell lysates were determined by plaque assay using L-cell monolayers (57).
Immunoprecipitation of viral proteins.
Cells (2 × 106) grown in 25-cm2 tissue culture flasks (Costar) were infected with reovirus at an MOI of 10 to 1,000 PFU per cell. After viral adsorption for 1 h, the inoculum was removed, and cell culture medium containing 100 μCi of [35S]methionine-[35S]cysteine (DuPont NEN Research Products, Boston, Mass.) per ml was added. Following incubation at 37°C for 24 h, the medium was removed and cells were washed once with PBS. Cells were lysed by adding 300 μl of radioimmunoprecipitation (RIPA) buffer (150 mM NaCl, 10 mM Tris [pH 7.4], 1% lGEPAL, 1% deoxycholate, 1% sodium dodecyl sulfate [SDS]) and passed through a syringe with a 25-gauge needle to shear cells and DNA. Lysates (10 to 150 μl) were precleared by adding 800 μl of low-stringency RIPA buffer (150 mM NaCl, 10 mM Tris [pH 7.4], 1% lgepal, 1% deoxycholate, 0.1% SDS), 1 μl of normal rabbit serum (Vector Laboratories, Burlingame, Calif.), and 100 μl of a 10% protein A-Sepharose (Amersham) bead slurry (25 mg of protein A-Sepharose resuspended in 1 ml of low-stringency RIPA buffer). Samples were rotated at 4°C for 1 h and centrifuged at 13,000 × g for 5 min. Supernatants were transferred to tubes containing 300 μl of a 10% protein A-Sepharose bead slurry, followed by addition of 3 μl of rabbit polyclonal T1L-specific serum. Samples were rotated overnight at 4°C and centrifuged at 13,000 × g for 5 min. Supernatants were removed, and beads were washed six times in high-stringency RIPA buffer (1 M NaCl, 10 mM Tris [pH 7.4], 1% lGEPAL, 0.5% deoxycholate, 0.1% SDS). Following washes, beads were boiled in sample buffer for 5 min, subjected to electrophoresis in SDS–10% polyacrylamide gels, dried under vacuum, and exposed to Biomax MR film (Kodak) or a phosphorimaging plate (Fuji, Edison, N.J.). Total immunoprecipitated viral protein was quantitated with a phosphorimager (Fuji BAS2000).
Treatment with neuraminidase.
Cells (5 × 104) grown in 24-well plates were incubated in gel saline (HeLa cells) (57) or incomplete cell culture medium (L cells) containing 1 or 40 mU of Arthrobacter ureafaciens neuraminidase (Sigma-Aldrich) per ml at 37°C for various intervals. Following incubation, neuraminidase was removed, and cells were adsorbed with reovirus. After adsorption at 4°C for 1 h, the inoculum was removed, cells were washed once with PBS, and fresh medium was added. Neuraminidase-treated cells were incubated at 37°C and used in apoptosis and gel shift assays.
Treatment with SLL.
Virus was incubated with 0.1 to 10 mM α-sialyllactose (SLL; Sigma-Aldrich), 10 mM lactose (Sigma-Aldrich) as a control, or PBS at 37°C for 30 min. Following incubation, cells (5 × 104) were adsorbed with PBS containing virus and either SLL or lactose at 4°C for 1 h. After adsorption, the inoculum was removed, and cells were washed once with PBS and incubated at 37°C for 48 h. Cells were then processed for apoptosis assays.
RESULTS
The reovirus S1 gene is the primary genetic determinant of differences in apoptosis induction in HeLa cells.
T3 reovirus strains are significantly more efficient inducers of apoptosis than T1 strains in both L cells (55, 56) and MDCK cells (41). This difference in apoptosis induction efficiency is determined primarily by the S1 gene (41, 56), which encodes viral attachment protein ς1 (28, 59). To determine whether the S1 gene also segregates with strain-specific differences in the efficiency of reovirus-induced apoptosis in HeLa cells, and to validate the use of HeLa cells for studies of the role of the ς1 protein in reovirus-induced apoptosis, we tested previously characterized T1L × T3D reassortant viruses (6, 12, 59) for the capacity to induce apoptosis. HeLa cells were infected with either T1L, T3D, or 1 of 11 T1L × T3D reassortant viruses at an MOI of 100 PFU per cell. This MOI was chosen to produce a synchronous infection and to ensure maximum levels of apoptosis (11, 41, 56). Apoptosis was quantitated by acridine orange staining 48 h after infection (Table 1). Reassortant viruses containing a T3D S1 gene induced higher levels of apoptosis than reassortant viruses containing a T1L S1 gene. The association of apoptosis efficiency and the T3D S1 gene was highly statistically significant (Mann-Whitney test, P < 0.01). No other reovirus genes were significantly associated with the efficiency of apoptosis induction, which indicates that the ς1-encoding S1 gene is the primary determinant of apoptosis efficiency in HeLa cells.
TABLE 1.
Capacities of T1L × T3D reassortants to induce apoptosis in HeLa cells
| Virus strain | Origin of gene segmenta
|
% Apoptotic cellsb | Rank | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L1 | L2 | L3 | M1 | M2 | M3 | S1 | S2 | S3 | S4 | |||
| Parental | ||||||||||||
| T1L | L | L | L | L | L | L | L | L | L | L | 4 | |
| T3D | D | D | D | D | D | D | D | D | D | D | 38 | |
| Reassortant | ||||||||||||
| EB138 | D | L | L | D | D | L | D | D | L | L | 64 | 1 |
| KC150 | D | L | L | L | D | L | D | D | L | D | 52 | 2 |
| 1HA3 | L | L | L | L | L | L | D | L | L | L | 51 | 3 |
| EB39 | L | D | D | L | D | D | D | D | D | D | 50 | 4 |
| H9 | D | D | L | D | L | L | D | D | D | D | 29 | 5 |
| G2 | L | D | L | L | L | L | D | L | L | L | 22 | 6 |
| EB1 | L | D | L | L | D | L | L | L | D | L | 21 | 7 |
| EB18 | D | D | L | D | D | D | L | L | D | L | 7 | 8 |
| H41 | D | D | L | L | L | D | L | D | D | L | 6 | 9 |
| EB145 | D | D | D | D | D | L | L | D | D | D | 5 | 10 |
| EB121 | D | D | L | D | L | D | L | D | D | D | 4 | 11 |
L, gene segment derived from T1L; D, gene segment derived from T3D.
HeLa cells (5 × 104) were infected with the strains shown at an MOI of 100 PFU per cell, and percent apoptotic cells was determined by acridine orange staining 48 h after viral infection. Shown are the means for three independent experiments.
Reovirus strains selected for the capacity to bind sialic acid are potent inducers of apoptosis.
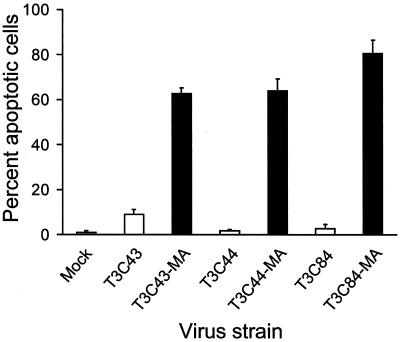
To determine whether the capacity of T3 strains to bind cell surface sialic acid influences the efficiency of apoptosis induction, we used three sialic acid-binding T3 reovirus isolates, T3C43-MA, T3C44-MA, and T3C84-MA, that were derived by serial passage of three non-sialic-acid-binding viruses, T3C43, T3C44, and T3C84, respectively, in MEL cells (9). HeLa cells were infected with either sialic-acid-binding or non-sialic-acid-binding strains at an MOI of 100 PFU per cell, and apoptosis was assessed 48 h after infection by acridine orange staining (Fig. 1). Infection with the sialic acid-binding strains resulted in a high percentage (approximately 60 to 80%) of apoptotic cells; however, the non-sialic acid-binding strains induced low levels of apoptosis, with approximately 2 to 9% of cells apoptotic. These results suggest that the capacity to bind sialic acid on the surface of HeLa cells significantly enhances the efficiency of reovirus-induced apoptosis.
FIG. 1.
Apoptosis induced by reovirus strains that differ in sialic acid binding. HeLa cells (5 × 104) were either mock infected or infected with non-sialic-acid-binding strains T3C43, T3C44, and T3C84 (white bars) or their sialic acid-binding MA variants (black bars) at an MOI of 100 PFU per cell. Cells were incubated at 37°C for 48 h and stained with acridine orange. The results are expressed as the mean percentage of cells undergoing apoptosis for three independent experiments. Error bars indicate standard deviations of the means.
Isogenic reovirus mutants that differ in the capacity to bind sialic acid also differ in the capacity to induce apoptosis and activate NF-κB.
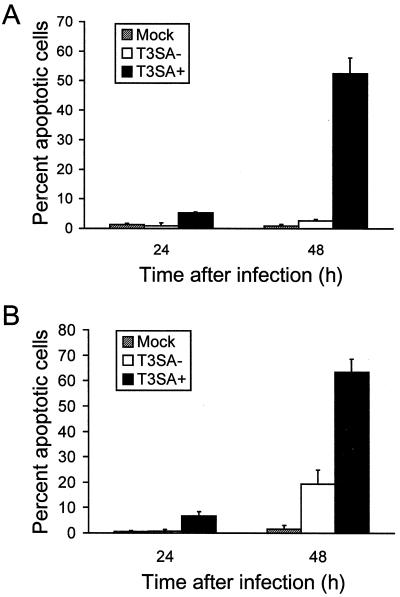
We thought it possible that adaptation of non-sialic-acid-binding strains to growth in MEL cells may have resulted in mutations in gene segments other than S1 that influence apoptosis induction. Therefore, to more conclusively assess the role of sialic acid binding in reovirus-induced apoptosis, we used two isogenic viruses, T3SA− and T3SA+, that differ genetically by a single point mutation in the S1 gene and phenotypically by the capacity to bind sialic acid (3). T3SA− contains the S1 gene from non-sialic-acid-binding strain T3C44 and all other genes from T1L, while T3SA+ contains the S1 gene from sialic acid-binding strain T3C44-MA and all other genes from T1L. HeLa cells were either mock infected or infected with T3SA− or T3SA+ at an MOI of 100 PFU per cell, and apoptosis was assessed 24 and 48 h after infection by acridine orange staining (Fig. 2A). Infection with T3SA+ induced approximately 5 and 55% of cells to undergo apoptosis at 24 and 48 h after infection, respectively. However, apoptosis was not detected in cells infected with T3SA− above the level of mock-infected cells at either time point. To exclude the possibility that the difference in apoptosis efficiency observed between T3SA− and T3SA+ is specific to HeLa cells, we tested the capacity of these viruses to induce apoptosis in L cells. L cells were either mock infected or infected with T3SA− or T3SA+ at an MOI of 100 PFU per cell, and apoptosis was quantitated 24 and 48 h after infection by acridine orange staining (Fig. 2B). In L cells, infection with T3SA+ induced apoptosis of approximately 60% of cells at 48 h, whereas T3SA− induced apoptosis of approximately 20% of cells at this time point. These results suggest that sialic acid binding enhances reovirus-induced apoptosis in both HeLa cells and L cells.
FIG. 2.
Apoptosis induced by T3SA− and T3SA+ in HeLa cells (A) and L cells (B). Cells (5 × 104) were either mock infected or infected with T3SA− or T3SA+ at an MOI of 100 PFU per cell. Cells were incubated at 37°C for 48 h and stained with acridine orange. The results are expressed as the mean percentage of cells undergoing apoptosis for three independent experiments. Error bars indicate standard deviations of the means.
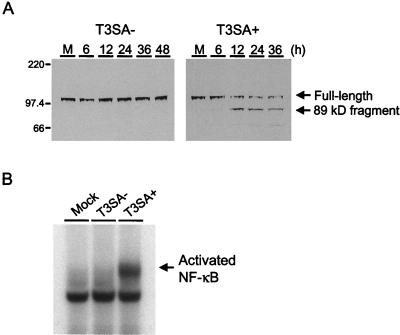
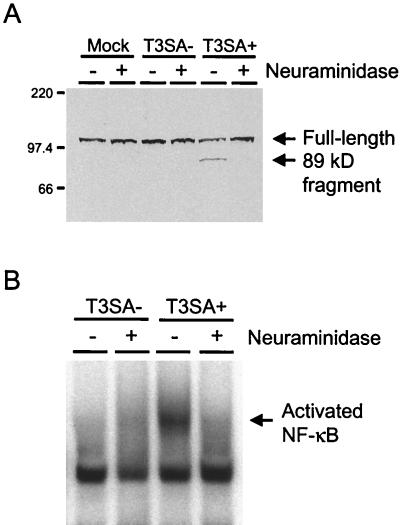
To corroborate our findings using acridine orange staining, infected cells were assayed for cleavage of PARP, a protein involved in DNA repair (31). PARP serves as a substrate for the proapoptotic enzyme caspase-3, which is activated during programmed cell death (34, 46). HeLa cells were either mock infected or infected with T3SA− or T3SA+ at an MOI of 100 PFU per cell, and nuclear extracts were prepared either 6 to 48 h following virus infection or 24 h after mock infection and used in immunoblots for detection of full-length PARP and the 89-kDa cleavage fragment of PARP (Fig. 3A). PARP cleavage was first detected by 12 h postinfection in cells infected with T3SA+ and was evident as late as 36 h after infection with this strain. By 48 h, the extent of cell death induced by T3SA+ precluded the isolation of nuclear protein for immunoblot analysis. In contrast, PARP cleavage was not detected in mock-infected cells or in cells infected with T3SA− at any time point. Together, these results provide both morphological and biochemical evidence that binding to sialic acid enhances reovirus-induced apoptosis.
FIG. 3.
PARP cleavage and NF-κB activation induced by T3SA− and T3SA+. (A) Immunoblot for PARP cleavage. HeLa cells (5 × 106) were either mock infected (M) or infected with T3SA− or T3SA+ at an MOI of 100 PFU per cell and incubated at 37°C. Nuclear extracts were prepared 6 to 48 h following virus infection or 24 h after mock infection, and 50 μg of total protein was resolved by acrylamide gel electrophoresis, transferred to nitrocellulose, and blotted with PARP-specific antiserum. The positions of full-length PARP and the 89-kDa PARP cleavage product are indicated. Molecular size markers are given in kilodaltons. (B) NF-κB gel shift activity. HeLa cells (5 × 106) were either mock infected or infected with T3SA− or T3SA+ at an MOI of 100 PFU per cell and incubated at 37°C for 10 h. Nuclear extracts were prepared and incubated with a 32P-labeled oligonucleotide comprised of the NF-κB consensus binding sequence. Incubation mixtures were resolved by acrylamide gel electrophoresis, dried, and exposed to film. The activated NF-κB complex is indicated.
We have previously demonstrated that apoptosis induced by reovirus infection requires activation of the transcription factor NF-κB (11). To determine whether the capacity to bind sialic acid also influences NF-κB activation, HeLa cells were either mock infected or infected with T3SA− or T3SA+ at an MOI of 100 PFU per cell, and nuclear extracts were prepared 10 h after infection. Extracts were incubated with a 32P-labeled oligonucleotide consisting of the NF-κB consensus binding sequence and resolved in a nondenaturing polyacrylamide gel (Fig. 3B). We found that NF-κB was activated following infection with T3SA+ but not T3SA−. Using antisera specific for NF-κB subunits p50 and p65, we determined that infection with T3SA+ led to the activation of a heterodimeric NF-κB complex containing p50 and p65, while all samples contained a constitutive level of nuclear p50 homodimers (data not shown). This pattern is identical to that previously observed in HeLa cells infected with T3D (11). These results suggest that the capacity to bind sialic acid significantly enhances the capacity of reovirus to activate NF-κB, which is a proximal biochemical signal required for induction of apoptosis by reovirus.
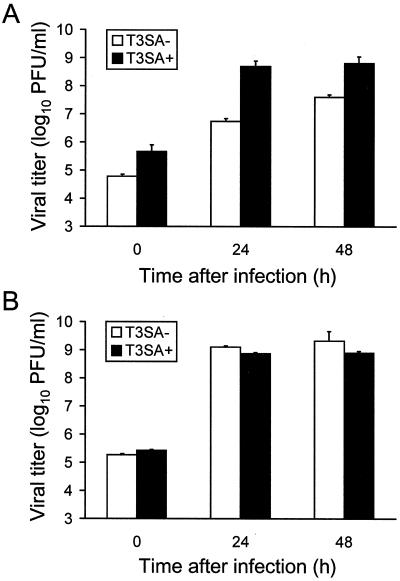
T3SA+ produces higher yields than T3SA− in HeLa cells, but T3SA− and T3SA+ grow equivalently in L cells.
Previous studies using reassortant viruses indicate that the capacity of reovirus to induce apoptosis is not linked to viral growth (41). However, we thought it possible that the higher levels of apoptosis induced by T3SA+ might be due to an increase in viral replication. To test this possibility, we infected HeLa cells and L cells with T3SA− or T3SA+ at an MOI of 100 PFU per cell and determined viral titers at 0, 24, and 48 h by plaque assay on L-cell monolayers (Fig. 4). In HeLa cells, titers of T3SA+ were approximately 100- and 10-fold, respectively, greater than titers of T3SA− at 24 and 48 h following adsorption. This difference in viral titer is partially explained by the approximately 10-fold higher titer of T3SA+ relative to T3SA− at 0 h, as yields (calculated by dividing titer at 24 or 48 h by titer at 0 h) of T3SA+ are approximately 10- and 2-fold, respectively, greater than yields of T3SA− at 24 and 48 h. In contrast, growth of T3SA− and T3SA+ in L cells did not significantly differ at either time point tested, indicating that differences in the capacity of these strains to induce apoptosis are independent of viral growth in this cell type.
FIG. 4.
Growth of T3SA− and T3SA+ in HeLa cells (A) and L cells (B). Cells (105) were infected with T3SA− or T3SA+ at an MOI of 100 PFU per cell and incubated at 37°C. Viral titers at 0, 24, and 48 h were determined by plaque assay. The results are expressed as the mean viral titer for three independent experiments. Error bars indicate standard deviations of the means.
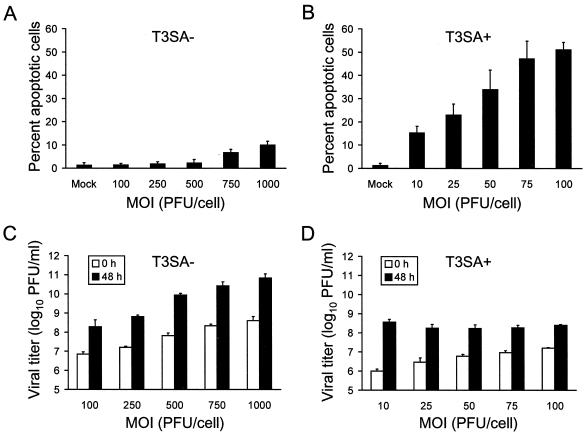
Levels of apoptosis induced by T3SA+ are dependent on viral dose but independent of viral growth in HeLa cells.
Since T3SA+ binding and growth are more efficient than binding and growth of T3SA− in HeLa cells, we next quantitated apoptosis and viral growth following infection over a range of viral doses to determine whether T3SA− could overcome the block to apoptosis by increasing the inoculum. HeLa cells were infected with 100 to 1,000 PFU of T3SA− or 10 to 100 PFU of T3SA+ per cell, and apoptosis (Fig. 5A and B) and viral growth (Fig. 5C and D) were assessed 48 h after infection. In cells infected with T3SA+, apoptosis was detected at each dose of virus tested, and levels of apoptosis increased in relationship to MOI. However, T3SA− induced levels of apoptosis in excess of mock-infected cells only when cells were infected at an MOI of 750 or 1,000 PFU per cell. Strikingly, the percentage of cells undergoing apoptosis following infection with 10 PFU of T3SA+ per cell was greater than the percentage of apoptotic cells following infection with 1,000 PFU of T3SA− per cell, which more than compensates for the differences exhibited by these strains in viral attachment and growth. In infectivity assays, an increase in viral titer as a function of MOI was observed with both viruses at the 0-h time point, as expected. Titers of T3SA− also increased at 48 h with increasing MOI; however, titers of T3SA+ at 48 h were equivalent at each dose. Importantly, 48 h titers of T3SA− were equal to or greater than titers of T3SA+ at each dose tested. Therefore, even at MOIs of T3SA− that result in more virus bound at 0 h and higher viral titers at 48 h relative to T3SA+, T3SA− still induces substantially less apoptosis than T3SA+. In additional experiments, we observed no detectable apoptosis of HeLa cells inoculated with T3SA− at an MOI of 100 PFU per cell over a 4-day observation period, by which time titers of T3SA− were greater than titers of T3SA+ (data not shown). These results, combined with those obtained using L cells, in which viral yield is clearly independent of virus-induced apoptosis, indicate that differences in the capacity of T3SA− and T3SA+ to induce apoptosis are not attributable to increased virus binding or growth but rather due to the capacity of T3SA+ to bind sialic acid.
FIG. 5.
Quantitation of apoptosis and viral growth following adsorption with increasing doses of T3SA− and T3SA+. (A and B) Apoptosis induced by T3SA− and T3SA+. HeLa cells (5 × 104) were either mock infected or infected with T3SA− at MOIs of 100 to 1,000 PFU per cell (A) or T3SA+ at MOIs of 10 to 100 PFU per cell (B). Cells were incubated at 37°C for 48 h and stained with acridine orange. (C and D) Growth of T3SA− and T3SA+. HeLa cells (105) were infected with T3SA− at MOIs of 100 to 1,000 PFU per cell (C) or T3SA+ at MOIs of 10 to 100 PFU per cell (D). After adsorption for 1 h, the inocula were removed and cells were incubated at 37°C. Viral titers at 0 and 48 h were determined by plaque assay. The results are expressed as the mean percentage of cells undergoing apoptosis or mean viral titer for three independent experiments. Error bars indicate standard deviations of the means.
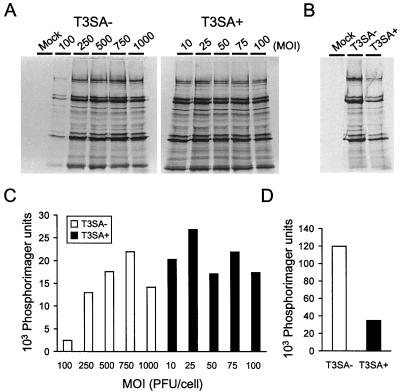
Differences in the capacity of T3SA− and T3SA+ to induce apoptosis are not attributable to differences in viral protein synthesis.
To exclude the possibility that T3SA+ is a more potent inducer of apoptosis than T3SA− due to increased levels of viral protein synthesis, we quantitated total viral protein present in cell extracts prepared following infection by either strain. HeLa cells were adsorbed with T3SA− at MOIs of 100 to 1,000 PFU per cell or T3SA+ at MOIs of 10 to 100 PFU per cell and incubated for 24 h in medium containing [35S]methionine-[35S]cysteine. Radiolabeled viral proteins in cell lysates were immunoprecipitated using a T1L-specific polyclonal serum, which recognizes all T3SA− and T3SA+ proteins with the exception of ς1, and resolved by SDS-polyacrylamide gel electrophoresis (Fig. 6A). Total immunoprecipitated protein was quantitated with a phosphorimager (Fig. 6C). With the exception of cells infected with T3SA− at an MOI of 100 PFU per cell, levels of viral proteins produced by T3SA− and T3SA+ differed by less than twofold at each MOI. Furthermore, levels of viral protein synthesis do not correlate with levels of apoptosis. Supernatants were immunoprecipitated a second time using an excess of protein A-Sepharose beads to confirm that the majority (>90%) of viral proteins were removed from cell lysates (data not shown). Levels of viral protein synthesis also were quantitated following infection of L cells with 100 PFU of either T3SA− or T3SA+ per cell (Fig. 6B and D). T3SA− produced approximately threefold more viral protein than T3SA+, which provides additional evidence that levels of viral protein synthesis are independent of levels of apoptosis.
FIG. 6.
Quantitation of viral protein synthesis in cells infected with T3SA− and T3SA+. (A and C) HeLa cells infected with increasing doses of T3SA− and T3SA+. Cells (2 × 106) were either mock infected or infected with T3SA− at MOIs of 100 to 1,000 PFU per cell or T3SA+ at MOIs of 10 to 100 PFU per cell. Cells were incubated at 37°C for 24 h in the presence of [35S]methionine-[35S]cysteine. Viral proteins were immunoprecipitated from cell lysates, resolved by acrylamide gel electrophoresis, dried, and exposed to film (A). Total viral protein was quantitated by phosphorimager analysis (C). (B and D) L cells infected with T3SA− and T3SA+. Cells (2 × 106) were either mock infected or infected with T3SA− or T3SA+ at an MOI of 100 PFU per cell. Cells were incubated at 37°C for 24 h in the presence of [35S]methionine-[35S]cysteine. Viral proteins were immunoprecipitated from cell lysates, resolved by acrylamide gel electrophoresis, dried, and exposed to film (B). Total viral protein was quantitated by phosphorimager analysis (D).
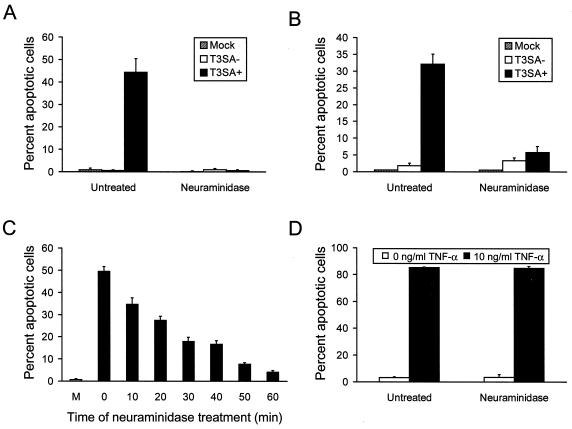
Removal of cell surface sialic acid with neuraminidase blocks apoptosis and NF-κB activation induced by T3SA+.
To test specifically whether sialic acid binding is required to achieve the high levels of apoptosis detected following T3SA+ infection, cells were treated with A. ureafaciens neuraminidase to remove cell surface sialic acid. HeLa cells or L cells were treated with neuraminidase for 1 h and then either mock infected or infected with 100 PFU of T3SA− or T3SA+ per cell. Apoptosis was assessed by acridine orange staining 48 h after infection (Fig. 7A and B). T3SA+ efficiently induced apoptosis in untreated HeLa cells and untreated L cells; however, apoptosis was abolished in HeLa cells treated with neuraminidase and substantially diminished in neuraminidase-treated L cells. Treatment of HeLa cells with neuraminidase also blocked the capacity of T3SA+ to induce cleavage of PARP (Fig. 8A). To determine whether levels of apoptosis induced by T3SA+ could be altered by the degree of sialic acid removal from the cell surface, HeLa cells were treated with a lower concentration of neuraminidase over a time course ranging from 0 to 60 min (Fig. 7C). Levels of apoptosis induced by T3SA+ decreased with increasing time of neuraminidase treatment, confirming the specificity of the neuraminidase effect on apoptosis induction by reovirus. To confirm that treatment with neuraminidase does not alter the capacity of HeLa cells to undergo apoptosis, untreated cells and cells treated with neuraminidase were incubated with 10 ng of TNF-α per ml and assessed for apoptosis by acridine orange staining 24 h after addition of TNF-α (Fig. 7D). TNF-α induced high levels of apoptosis in both untreated and neuraminidase-treated cells, indicating that the apoptotic machinery in HeLa cells is not altered by treatment with neuraminidase.
FIG. 7.
Apoptosis induced by T3SA− and T3SA+ in neuraminidase-treated cells. (A and B) HeLa cells (A) and L cells (B) infected with T3SA− and T3SA+. Cells (5 × 104) were either untreated or treated with 40 mU of A. ureafaciens neuraminidase per ml for 1 h. Following treatment, neuraminidase was removed, and cells were either mock infected or infected with T3SA− or T3SA+ at an MOI of 100 PFU per cell. Cells were incubated at 37°C for 48 h and stained with acridine orange. (C) Time course of neuraminidase treatment in HeLa cells. Cells (5 × 104) were either untreated or treated with 1 mU of neuraminidase per ml for the times shown. Following treatment, neuraminidase was removed, and cells were either mock infected (M) or infected with T3SA+ at an MOI of 100 PFU per cell. Cells were incubated at 37°C for 48 h and stained with acridine orange. (D) HeLa cells treated with TNF-α. Cells (5 × 104) were either untreated or treated with 40 mU of neuraminidase per ml for 1 h. Following treatment, neuraminidase was removed, and cells were incubated with 10 μg of cycloheximide per ml either alone or in combination with 10 ng of TNF-α per ml. Cells were incubated at 37°C for 24 h and stained with acridine orange. The results are expressed as the mean percentage of cells undergoing apoptosis for three independent experiments. Error bars indicate standard deviations of the means.
FIG. 8.
PARP cleavage and NF-κB activation induced by T3SA− and T3SA+ in neuraminidase-treated HeLa cells. (A) Immunoblot for PARP cleavage. HeLa cells (5 × 106) were either untreated or treated with 40 mU of A. ureafaciens neuraminidase per ml for 1 h. Following treatment, neuraminidase was removed, and cells were either mock infected or infected with T3SA− or T3SA+ at an MOI of 100 PFU per cell. Nuclear extracts were prepared after incubation at 37°C for 24 h, and 50 μg of total protein was resolved by acrylamide gel electrophoresis, transferred to nitrocellulose, and blotted with PARP-specific antiserum. The positions of full-length PARP and the 89-kDa PARP cleavage product are indicated. Molecular size markers are given in kilodaltons. (B) NF-κB gel shift activity. HeLa cells (5 × 106) were either untreated or treated with 40 mU of A. ureafaciens neuraminidase per ml for 1 h. Following treatment, neuraminidase was removed, and cells were either mock infected or infected with T3SA− or T3SA+ at an MOI of 100 PFU per cell. Nuclear extracts were prepared following incubation at 37°C for 10 h and incubated with a 32P-labeled oligonucleotide comprised of the NF-κB consensus binding sequence. Incubation mixtures were resolved by acrylamide gel electrophoresis, dried, and exposed to film. The activated NF-κB complex is indicated.
To determine whether neuraminidase treatment of HeLa cells alters the capacity of T3SA+ to activate NF-κB, nuclear extracts were prepared from untreated or neuraminidase-treated cells that were either mock infected or infected with 100 PFU of T3SA− or T3SA+ per cell and used in a gel shift assay (Fig. 8B). We found that T3SA+ activated NF-κB in untreated cells; however, activation did not occur in neuraminidase-treated cells. As expected, all samples contained a constitutive complex comprised of p50 homodimers. These results indicate that removal of cell surface sialic acid blocks both NF-κB activation and apoptosis induced by infection with T3SA+.
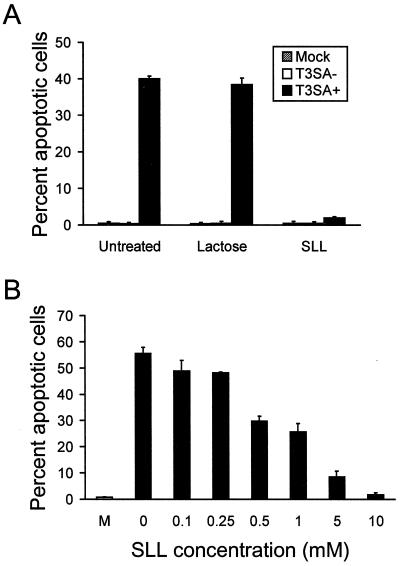
Incubation with SLL blocks apoptosis induced by T3SA+.
Sialic acid-binding reovirus strains bind terminal sialic acid residues in α2,3 and α2,6 linkages (38). To confirm results obtained using neuraminidase treatment, we tested the effect of SLL, a trisaccharide comprised of lactose and α-linked terminal sialic acid residues, on apoptosis induction by T3SA− and T3SA+. Using surface plasmon resonance, we have shown that binding of T3SA+ to sialic acid residues on glycophorin is inhibited in a dose-dependent manner by the addition of SLL and is completely abolished by 10 mM SLL (3). T3SA− and T3SA+ were preincubated with either 10 mM lactose as a control or 10 mM SLL prior to adsorption to cells at an MOI of 100 PFU per cell. Apoptosis was assessed by acridine orange staining 48 h following infection (Fig. 9A). Incubation of T3SA+ with lactose did not alter the capacity of T3SA+ to induce apoptosis; however, incubation with SLL completely blocked apoptosis induced by this strain. To confirm that the effect of SLL on T3SA+-induced apoptosis correlates with the dose-dependent inhibition of T3SA+ binding to sialic acid, we preincubated T3SA+ with 0.1 to 10 mM SLL and assessed apoptosis by acridine orange staining 48 h following infection (Fig. 9B). Apoptosis decreased with increasing concentrations of SLL, demonstrating that the effect of SLL on apoptosis efficiency parallels the inhibition of sialic acid binding (3). Therefore, these results indicate that the capacity of T3SA+ to engage cell surface sialic acid significantly enhances virus-induced apoptosis.
FIG. 9.
Apoptosis induced by T3SA− and T3SA+ following incubation with SLL. (A) Incubation of T3SA− and T3SA+ with SLL. No virus (Mock), T3SA−, or T3SA+ was incubated with either PBS (Untreated), 10 mM lactose, or 10 mM SLL at 37°C for 30 min prior to adsorption to HeLa cells. Cells were incubated at 37°C for 48 h and stained with acridine orange. (B) Incubation of T3SA+ with increasing concentrations of SLL. No virus (M) or T3SA+ were incubated with either PBS or 0.1 to 10 mM SLL at 37°C for 30 min prior to adsorption to HeLa cells. Cells were incubated at 37°C for 48 h and stained with acridine orange. The results are expressed as the mean percentage of cells undergoing apoptosis for three independent experiments. Error bars indicate standard deviations of the means.
DISCUSSION
Many viruses, including reovirus (21, 38), adenovirus (2), influenza virus (60), polyomavirus (5, 29), rotavirus (19), Theiler's murine encephalomyelitis virus (65, 66), and transmissible gastroenteritis coronavirus (26), are capable of binding sialic acid. For these viruses, sialic acid binding plays an important role in attachment (19, 29, 60), internalization (2, 65, 66), or tropism (5). However, it is not known whether interaction of these viruses with cell surface sialic acid activates signaling pathways or leads to alterations in cellular homeostasis. Here, we demonstrate that the capacity of reovirus to bind sialic acid enhances the activation of NF-κB and leads to apoptosis, which indicates that engagement of sialic acid by reovirus activates cellular signaling pathways that result in apoptosis of infected cells.
Carbohydrate-binding proteins, or lectins, have been shown to play important roles in many cellular processes, including cell adhesion (39, 63), cell proliferation (25, 39, 63), and cell death (7, 22, 39, 40, 49, 62, 64). There are several studies that report the capacity of sialic acid-binding lectins to induce apoptosis of lymphocytes (7, 22, 40, 64) and tumor cell lines (49, 62). In thymocytes, lectin binding can lead to mitogenic stimulation (25), secretion of interleukin-2 (22), increased intracellular [Ca2+] (58), and induction of apoptosis (22, 40, 64). The level of sialylation of T- and B-cell surface proteins appears to influence the susceptibility of these cells to apoptosis, suggesting that sialic acid-binding proteins may influence selection of lymphocytes in vivo (1, 24). In support of this idea, galectin-1, a carbohydrate-binding protein produced by thymic epithelial cells, induces apoptosis of lymphocytes, particularly CD4lo and CD8lo T cells, raising the possibility that galectin-1 plays a role in negative selection (40). Furthermore, certain subsets of T lymphocytes are more susceptible to lectin-induced apoptosis, suggesting that carbohydrate moieties on specific cellular receptors are involved in the apoptotic response (25, 58). Similarly, it is possible that sialic acid-binding strains of reovirus function like lectins to trigger signaling pathways that lead to cell death.
We used a panel of T1L × T3D reassortant viruses to confirm that the ς1-encoding S1 gene is the primary genetic determinant of apoptosis induction in HeLa cells, which had previously been demonstrated for both L cells (55, 56) and MDCK cells (41). To gather evidence to support a functional role for ς1 in apoptosis induction by reovirus, we assessed apoptosis of HeLa cells and L cells following infection by reovirus isolates that encode ς1 molecules that differ only in the capacity to bind sialic acid. We found that sialic acid-binding strain T3SA+ induces significantly higher levels of apoptosis than non-sialic-acid-binding strain T3SA− in both cell types. Enzymatic removal of sialic acid with neuraminidase or blockade of virus binding to cell surface sialic acid by SLL abolished apoptosis induced by T3SA+, which demonstrates that apoptosis induction is tightly coupled to sialic acid binding. In fact, the SLL concentration for 50% inhibition of apoptosis induced by T3SA+ is identical to that for inhibition of T3SA+ binding to sialoglycophorin (3). Thus, these findings support the linkage of the ς1 protein, and not the s1 RNA, to the efficiency of apoptosis induction, and they demonstrate that sialic acid binding plays an important role in this process.
It seemed possible to us that the enhanced capacity of T3SA+ to induce apoptosis in HeLa cells might be due to an increase in T3SA+ binding relative to T3SA− binding to this cell type. We found that when 100 PFU of T3SA− or T3SA+ per cell was adsorbed to HeLa cells for 1 h, approximately 10-fold more PFU of T3SA+ than of T3SA− was detectable, as determined by plaque titration on L-cell monolayers. This result raises the possibility that PFU of T3SA− and T3SA+ on L cells may not be equivalent to PFU on HeLa cells. Using a fluorescent-focus assay to directly assess the infectivity of T3SA− and T3SA+ on HeLa cells, we found that the numbers of HeLa cells infected by T3SA− and T3SA+ were equivalent when the dose of T3SA− was 10-fold greater than the dose of T3SA+ (data not shown). Our experiments assessing levels of apoptosis induction in response to viral dose demonstrate that greater than 100-fold more T3SA− is required to induce levels of apoptosis equivalent to those induced by T3SA+, indicating that differences in the capacity of T3SA− and T3SA+ to bind HeLa cells do not account for the difference in apoptosis induction efficiency observed for these strains.
To exclude the possibility that T3SA+ is a more efficient inducer of apoptosis as a result of increased viral protein synthesis or growth, we used increasing doses of both T3SA− and T3SA+ in apoptosis, protein synthesis, and growth assays. The results demonstrate that the dramatic increase in apoptosis observed with sialic acid-binding strain T3SA+ is not due to increased synthesis of viral proteins or production of viral progeny. Importantly, T3SA− and T3SA+ produce equivalent yields in L cells, and T3SA− produces threefold more viral protein in L cells than T3SA+. However, T3SA+ is significantly more efficient at inducing apoptosis than T3SA− in this cell line. Therefore, the capacity of reovirus to bind sialic acid does not enhance apoptosis by simply increasing viral attachment or replication.
We conclude that ς1 binding to sialic acid activates a signaling pathway that results in nuclear translocation of NF-κB and induction of apoptosis. However, it is clear that binding to cell surface sialic acid is not sufficient to trigger these events. Sialic acid-binding reovirus strains must also engage ς1 head receptor JAM to activate NF-κB and induce apoptosis (4). JAM contains two potential N-linked glycosylation sites in the membrane-proximal immunoglobulin-like domain. It is possible that ς1 binding to sialic acid moieties present on JAM induces conformational changes in JAM or results in prolonged receptor activation, either of which could lead to initiation of signals required for reovirus-induced apoptosis. Alternatively, it is possible that the sialic acid residues bound by reovirus are conjugated to other cell surface proteins in close proximity to JAM. In this scenario, the simultaneous ligation of both molecules induces the requisite activation events. To formally test these possibilities, we must determine whether alteration of the JAM glycosylation sites influences the efficiency with which reovirus induces apoptosis.
Although receptor ligation is required for reovirus to activate NF-κB and induce apoptosis, there is evidence to suggest that additional steps in reovirus replication are necessary to elicit these cellular responses. Activation of NF-κB following reovirus infection of HeLa cells is first detectable 4 h after viral adsorption and peaks at 10 h (11). However, when activated solely in response to receptor-ligand interactions, such as the binding of TNF-α to its receptor, peak activation of NF-κB typically occurs with more rapid kinetics (53). Therefore, we suspect that aspects of reovirus entry or replication are required to potentiate signals induced by sialic acid and JAM binding to activate the host NF-κB pathway and induce apoptosis. However, our data clearly indicate that these postattachment steps are not sufficient to induce NF-κB activation and apoptosis, as T3SA− completes all steps of viral replication in HeLa cells and L cells yet is incapable of activating the signaling pathway that leads to apoptotic cell death.
Does the capacity to bind sialic acid offer a selective advantage to reovirus? Sialic acid binding clearly enhances apoptosis, which would inhibit host inflammatory responses triggered by reovirus infection in vivo, potentially aiding in viral dissemination within the infected host (52). However, as MOIs of T3SA+ increase, viral yields decrease, perhaps as a result of increased apoptosis. Therefore, the enhancement of reovirus-induced apoptosis mediated by sialic acid binding is likely beneficial for viral replication and spread at early stages of infection when levels of infectious virus are low but may limit viral growth at later stages of infection when viral titers are high. The capacity of reovirus to bind sialic acid appears to be a balanced polymorphism (9, 14). Of 11 T3 reovirus field isolate strains characterized thus far, 8 are capable of binding sialic acid and 3 are not (14). This observation suggests that selection pressures exist to maintain both sialic acid-binding and non-sialic-acid-binding reovirus strains in nature.
Our results demonstrate that virus-receptor utilization is not only an important determinant of infectivity and tropism but also an important mediator of virus-induced cell death. Sequence variations in the reovirus attachment protein that confer the capacity to bind sialic acid dictate the efficiency with which reovirus strains induce apoptosis. Utilization of a strategy in which virus-receptor interactions are linked to cell death pathways suggests that reovirus actively promotes this cellular response. Our ongoing studies of the mechanisms and pathogenic role of apoptosis pathways induced by reovirus binding will contribute to an enhanced understanding of host injury mediated by neurotropic viruses.
ACKNOWLEDGMENTS
We thank Michelle Becker for advice about immunoprecipitation of viral proteins.
This work was supported by Public Health Service award AI38296 from the National Institute of Allergy and Infectious Diseases, the National Science Foundation, the Vanderbilt University Research Council (E.S.B.), and the Elizabeth B. Lamb Center for Pediatric Research. Additional support was provided by Public Health Service awards CA68485 for the Vanderbilt Cancer Center and DK20593 for the Vanderbilt Diabetes Research and Training Center.
REFERENCES
- 1.Alvarez G, Lascurain R, Perez A, Degand P, Montano L F, Martinez-Cairo S, Zenteno E. Relevance of sialoglycoconjugates in murine thymocytes during maturation and selection in the thymus. Immunol Investig. 1999;28:9–18. doi: 10.3109/08820139909022719. [DOI] [PubMed] [Google Scholar]
- 2.Arnberg N, Edlund K, Kidd A H, Wadell G. Adenovirus type 37 uses sialic acid as a cellular receptor. J Virol. 2000;74:42–48. [PMC free article] [PubMed] [Google Scholar]
- 3.Barton E S, Connolly J L, Forrest J C, Chappell J D, Dermody T S. Utilization of sialic acid as a coreceptor enhances reovirus attachment by multi-step adhesion-strengthening. J Biol Chem. 2001;276:2200–2211. doi: 10.1074/jbc.M004680200. [DOI] [PubMed] [Google Scholar]
- 4.Barton E S, Forrest J C, Connolly J L, Chappell J D, Liu Y, Schnell F J, Nusrat A, Parkos C A, Dermody T S. Junction adhesion molecule is a receptor for reovirus. Cell. 2001;104:441–451. doi: 10.1016/s0092-8674(01)00231-8. [DOI] [PubMed] [Google Scholar]
- 5.Bauer P H, Cui C, Stehle T, Harrison S C, DeCaprio J A, Benjamin T L. Discrimination between sialic acid-containing receptors and pseudoreceptors regulates polyomavirus spread in the mouse. J Virol. 1999;73:5826–5832. doi: 10.1128/jvi.73.7.5826-5832.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown E G, Nibert M L, Fields B N. The L2 gene of reovirus serotype 3 controls the capacity to interfere, accumulate deletions and establish persistent infection. In: Compans R W, Bishop D H L, editors. Double-stranded RNA viruses. New York, N.Y: Elsevier Biomedical; 1983. pp. 275–287. [Google Scholar]
- 7.Bussing A, Multani A S, Pathak S, Pfuller U, Schietzel M. Induction of apoptosis by the N-acetyl-galactosamine-specific toxic lectin from Viscum album L. is associated with a decrease of nuclear p53 and Bcl-2 proteins and induction of telomeric associations. Cancer Lett. 1998;130:57–68. doi: 10.1016/s0304-3835(98)00124-4. [DOI] [PubMed] [Google Scholar]
- 8.Chappell J D, Duong J, Wright B W, Dermody T S. Identification of carbohydrate-binding domains in the attachment proteins of type 1 and type 3 reoviruses. J Virol. 2000;74:8472–8479. doi: 10.1128/jvi.74.18.8472-8479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chappell J D, Gunn V L, Wetzel J D, Baer G S, Dermody T S. Mutations in type 3 reovirus that determine binding to sialic acid are contained in the fibrous tail domain of viral attachment protein ς1. J Virol. 1997;71:1834–1841. doi: 10.1128/jvi.71.3.1834-1841.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen J J. Programmed cell death in the immune system. Adv Immunol. 1991;50:55–85. doi: 10.1016/s0065-2776(08)60822-6. [DOI] [PubMed] [Google Scholar]
- 11.Connolly J L, Rodgers S E, Clarke P, Ballard D W, Kerr L D, Tyler K L, Dermody T S. Reovirus-induced apoptosis requires activation of transcription factor NF-κB. J Virol. 2000;74:2981–2989. doi: 10.1128/jvi.74.7.2981-2989.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coombs K M, Fields B N, Harrison S C. Crystallization of the reovirus type 3 Dearing core. Crystal packing is determined by the λ2 protein. J Mol Biol. 1990;215:1–5. doi: 10.1016/s0022-2836(05)80089-0. [DOI] [PubMed] [Google Scholar]
- 13.DeBiasi R, Edelstein C, Sherry B, Tyler K. Calpain inhibition protects against virus-induced apoptotic myocardial injury. J Virol. 2001;75:351–361. doi: 10.1128/JVI.75.1.351-361.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dermody T S, Nibert M L, Bassel-Duby R, Fields B N. A ς1 region important for hemagglutination by serotype 3 reovirus strains. J Virol. 1990;64:5173–5176. doi: 10.1128/jvi.64.10.5173-5176.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drayna D, Fields B N. Activation and characterization of the reovirus transcriptase: genetic analysis. J Virol. 1982;41:110–118. doi: 10.1128/jvi.41.1.110-118.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duncan R, Horne D, Strong J E, Leone G, Pon R T, Yeung M C, Lee P W K. Conformational and functional analysis of the carboxyl-terminal globular head of the reovirus cell attachment protein. Virology. 1991;182:810–819. doi: 10.1016/0042-6822(91)90622-i. [DOI] [PubMed] [Google Scholar]
- 17.Ernst H, Shatkin A J. Reovirus hemagglutinin mRNA codes for two polypeptides in overlapping reading frames. Proc Natl Acad Sci USA. 1985;82:48–52. doi: 10.1073/pnas.82.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraser R D, Furlong D B, Trus B L, Nibert M L, Fields B N, Steven A C. Molecular structure of the cell-attachment protein of reovirus: correlation of computer-processed electron micrographs with sequence-based predictions. J Virol. 1990;64:2990–3000. doi: 10.1128/jvi.64.6.2990-3000.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukudome K, Yoshie O, Konno T. Comparison of human, simian, and bovine rotaviruses for requirement of sialic acid in hemagglutination and cell adsorption. Virology. 1989;172:196–205. doi: 10.1016/0042-6822(89)90121-9. [DOI] [PubMed] [Google Scholar]
- 20.Furlong D B, Nibert M L, Fields B N. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J Virol. 1988;62:246–256. doi: 10.1128/jvi.62.1.246-256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gentsch J R, Pacitti A F. Differential interaction of reovirus type 3 with sialylated receptor components on animal cells. Virology. 1987;161:245–248. doi: 10.1016/0042-6822(87)90192-9. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh S, Majumder M, Majumder S, Ganguly N K, Chatterjee B P. Saracin: a lectin from Saraca indica seed integument induces apoptosis in human T-lymphocytes. Arch Biochem Biophys. 1999;371:163–168. doi: 10.1006/abbi.1999.1418. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs B L, Atwater J A, Munemitsu S M, Samuel C E. Biosynthesis of reovirus-specified polypeptides. The S1 mRNA synthesized in vivo is structurally and functionally indistinguishable from in vitro-synthesized S1 mRNA and encodes two polypeptides, ς1a and ς1bNS. Virology. 1985;147:9–18. doi: 10.1016/0042-6822(85)90222-3. [DOI] [PubMed] [Google Scholar]
- 24.Keppler O T, Peter M E, Hinderlich S, Moldenhauer G, Stehling P, Schmitz I, Schwartz-Albiez R, Reutter W, Pawlita M. Differential sialylation of cell surface glycoconjugates in a human B lymphoma cell line regulates susceptibility for CD95 (APO-1/Fas)-mediated apoptosis and for infection by a lymphotropic virus. Glycobiology. 1999;9:557–569. doi: 10.1093/glycob/9.6.557. [DOI] [PubMed] [Google Scholar]
- 25.Kilpatrick D C. Mechanisms and assessment of lectin-mediated mitogenesis. Mol Biotechnol. 1999;11:55–65. doi: 10.1007/BF02789176. [DOI] [PubMed] [Google Scholar]
- 26.Krempl C, Ballesteros M-L, Zimmer G, Enjuanes L, Klenk H-D, Herrler G. Characterization of the sialic acid binding activity of transmissible gastroenteritis coronavirus by analysis of haemagglutination-deficient mutants. J Gen Virol. 2000;81:489–496. doi: 10.1099/0022-1317-81-2-489. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Lee P W, Hayes E C, Joklik W K. Protein sigma 1 is the reovirus cell attachment protein. Virology. 1981;108:156–163. doi: 10.1016/0042-6822(81)90535-3. [DOI] [PubMed] [Google Scholar]
- 29.Liu C K, Wei G, Atwood W J. Infection of glial cells by the human polyomavirus JC is mediated by an N-linked glycoprotein containing terminal α(2–6)-linked sialic acids. J Virol. 1998;72:4643–4649. doi: 10.1128/jvi.72.6.4643-4649.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morozov S Y. A possible relationship of reovirus putative RNA-polymerase to polymerases of positive-strand RNA viruses. Nucleic Acids Res. 1989;17:5394. doi: 10.1093/nar/17.13.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murcia G d, Murcia J M d. Poly (ADP-ribose) polymerase: a molecular nick-sensor. Trends Biochem Sci. 1994;19:172–176. doi: 10.1016/0968-0004(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 32.Nagata L, Masri S A, Pon R T, Lee P W K. Analysis of functional domains on reovirus cell attachment protein sigma 1 using cloned S1 gene deletion mutants. Virology. 1987;160:162–168. doi: 10.1016/0042-6822(87)90056-0. [DOI] [PubMed] [Google Scholar]
- 33.Nibert M L, Schiff L A, Fields B N. Reoviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1557–1596. [Google Scholar]
- 34.Nicholson D W, Ali A, Thornberry N A, Vaillancourt J P, Ding C K, Gallant M, Gareau Y, Griffin P R, Labelle M, Lazebnik Y A, Munday N A, Raju S M, Smulson M E, Yamin T, Yu V L, Miller D K. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 35.Oberhaus S M, Smith R L, Clayton G H, Dermody T S, Tyler K L. Reovirus infection and tissue injury in the mouse central nervous system are associated with apoptosis. J Virol. 1997;71:2100–2106. doi: 10.1128/jvi.71.3.2100-2106.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Brien V. Viruses and apoptosis. J Gen Virol. 1998;79:1833–1845. doi: 10.1099/0022-1317-79-8-1833. [DOI] [PubMed] [Google Scholar]
- 37.Pahl H L. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 38.Paul R W, Choi A H, Lee P W K. The α-anomeric form of sialic acid is the minimal receptor determinant recognized by reovirus. Virology. 1989;172:382–385. doi: 10.1016/0042-6822(89)90146-3. [DOI] [PubMed] [Google Scholar]
- 39.Perillo N L, Marcus M E, Baum L G. Galectins: versatile modulators of cell adhesion, cell proliferation, and cell death. J Mol Med. 1998;76:402–412. doi: 10.1007/s001090050232. [DOI] [PubMed] [Google Scholar]
- 40.Perillo N L, Uittenbogaart C H, Nguyen J T, Baum L G. Galectin-1, an endogenous lectin produced by thymic epithelial cells, induces apoptosis of human thymocytes. J Exp Med. 1997;185:1851–1858. doi: 10.1084/jem.185.10.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodgers S E, Barton E S, Oberhaus S M, Pike B, Gibson C A, Tyler K L, Dermody T S. Reovirus-induced apoptosis of MDCK cells is not linked to viral yield and is blocked by Bcl-2. J Virol. 1997;71:2540–2546. doi: 10.1128/jvi.71.3.2540-2546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodgers S E, Connolly J L, Chappell J D, Dermody T S. Reovirus growth in cell culture does not require the full complement of viral proteins: identification of a ς1s-null mutant. J Virol. 1998;72:8597–8604. doi: 10.1128/jvi.72.11.8597-8604.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosen L. Serologic grouping of reovirus by hemagglutination-inhibition. Am J Hyg. 1960;71:242–249. doi: 10.1093/oxfordjournals.aje.a120107. [DOI] [PubMed] [Google Scholar]
- 44.Rosen L, Abinanti F R. Natural and experimental infection of cattle with human types of reovirus. Am J Hyg. 1960;71:424–429. doi: 10.1093/oxfordjournals.aje.a120108. [DOI] [PubMed] [Google Scholar]
- 45.Rosen L, Abinanti F R, Hovis J F. Further observations on the natural infection of cattle with reoviruses. Am J Hyg. 1963;77:38–48. doi: 10.1093/oxfordjournals.aje.a120294. [DOI] [PubMed] [Google Scholar]
- 46.Salvensen G S, Dixit V M. Caspases: intracellular signaling by proteolysis. Cell. 1997;91:443–446. doi: 10.1016/s0092-8674(00)80430-4. [DOI] [PubMed] [Google Scholar]
- 47.Sarkar G, Pelletier J, Bassel-Duby R, Jayasuriya A, Fields B N, Sonenberg N. Identification of a new polypeptide coded by reovirus gene S1. J Virol. 1985;54:720–725. doi: 10.1128/jvi.54.3.720-725.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwarz E M, Badorff C, Hiura T S, Wessely R, Badorff B, Verma I M, Knowlton K U. NF-κB-mediated inhibition of apoptosis is required for encephalomyocarditis virus virulence: a mechanism of resistance in p50 knockout mice. J Virol. 1998;72:5654–5660. doi: 10.1128/jvi.72.7.5654-5660.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwarz R E, Wojciechowicz D C, Picon A I, Schwarz M A, Paty P B. Wheatgerm agglutinin-mediated toxicity in pancreatic cancer cells. Br J Cancer. 1999;80:1754–1762. doi: 10.1038/sj.bjc.6690593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen Y, Shenk T E. Viruses and apoptosis. Curr Opin Genet Dev. 1995;5:105–111. doi: 10.1016/s0959-437x(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 51.Starnes M C, Joklik W K. Reovirus protein λ3 is a poly(C)-dependent poly(G) polymerase. Virology. 1993;193:356–366. doi: 10.1006/viro.1993.1132. [DOI] [PubMed] [Google Scholar]
- 52.Teodoro J G, Branton P E. Regulation of apoptosis by viral gene products. J Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Traenckner E B, Pahl H L, Henkel T, Schmidt K N, Wilk S, Baeuerle P A. Phosphorylation of human I kappa B-alpha on serines 32 and 36 controls I kappa B-alpha proteolysis and NF-kappa B activation in response to diverse stimuli. EMBO J. 1995;14:2876–2883. doi: 10.1002/j.1460-2075.1995.tb07287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tyler K L, Fields B N. Pathogenesis of viral infections. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Press; 1996. pp. 173–218. [Google Scholar]
- 55.Tyler K L, Squier M K T, Brown A L, Pike B, Willis D, Oberhaus S M, Dermody T S, Cohen J J. Linkage between reovirus-induced apoptosis and inhibition of cellular DNA synthesis: role of the S1 and M2 genes. J Virol. 1996;70:7984–7991. doi: 10.1128/jvi.70.11.7984-7991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tyler K L, Squier M K T, Rodgers S E, Schneider B E, Oberhaus S M, Grdina T A, Cohen J J, Dermody T S. Differences in the capacity of reovirus strains to induce apoptosis are determined by viral attachment protein ς1. J Virol. 1995;69:6972–6979. doi: 10.1128/jvi.69.11.6972-6979.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Virgin H W, IV, Bassel-Duby R, Fields B N, Tyler K L. Antibody protects against lethal infection with the neurally spreading reovirus type 3 (Dearing) J Virol. 1988;62:4594–4604. doi: 10.1128/jvi.62.12.4594-4604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walzel H, Blach M, Hirabayashi J, Kasai K I, Brock J. Involvement of CD2 and CD3 in galectin-1 induced signaling in human Jurkat T-cells. Glycobiology. 2000;10:131–140. doi: 10.1093/glycob/10.2.131. [DOI] [PubMed] [Google Scholar]
- 59.Weiner H L, Ramig R F, Mustoe T A, Fields B N. Identification of the gene coding for the hemagglutinin of reovirus. Virology. 1978;86:581–584. doi: 10.1016/0042-6822(78)90099-5. [DOI] [PubMed] [Google Scholar]
- 60.Weis W, Brown J H, Cusack S, Paulson J C, Skehel J J, Wiley D C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 61.Williams W V, Guy H R, Rubin D H, Robey F, Myers J N, Kieber-Emmons T, Weiner D B, Greene M I. Sequences of the cell-attachment sites of reovirus type 3 and its anti-idiotypic/antireceptor antibody: modeling of their three-dimensional structures. Proc Natl Acad Sci USA. 1988;85:6488–6492. doi: 10.1073/pnas.85.17.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoon T J, Yoo Y C, Kang T B, Shimazaki K, Song S K, Lee K H, Kim S H, Park C H, Azuma I, Kim J B. Lectins isolated from Korean mistletoe (Viscum album coloratum) induce apoptosis in tumor cells. Cancer Lett. 1999;136:33–40. doi: 10.1016/s0304-3835(98)00300-0. [DOI] [PubMed] [Google Scholar]
- 63.Zanetta J P, Badache A, Maschke S, Marschal P, Kuchler S. Carbohydrates and soluble lectins in the regulation of cell adhesion and proliferation. Histol Histopathol. 1994;9:385–412. [PubMed] [Google Scholar]
- 64.Zhou J, Shao H, Cox N R, Baker H J, Ewald S J. Gangliosides enhance apoptosis of thymocytes. Cell Immunol. 1998;183:90–98. doi: 10.1006/cimm.1998.1247. [DOI] [PubMed] [Google Scholar]
- 65.Zhou L, Lin X, Green T J, Lipton H L, Luo M. Role of sialyloligosaccharide binding in Theiler's virus persistence. J Virol. 1997;71:9701–9712. doi: 10.1128/jvi.71.12.9701-9712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou L, Luo Y, Wu Y, Tsao J, Luo M. Sialylation of the host receptor may modulate entry of demyelinating persistent Theiler's virus. J Virol. 2000;74:1477–1485. doi: 10.1128/jvi.74.3.1477-1485.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]