Abstract
Alzheimer’s disease (AD) is one of the main causes of dementia worldwide, whereby neuronal death or malfunction leads to cognitive impairment in the elderly population. AD is highly prevalent, with increased projections over the next few decades. Yet current diagnostic methods for AD occur only after the presentation of clinical symptoms. Evidence in the literature points to potential mechanisms of AD induction beginning before clinical symptoms start to present, such as the formation of amyloid beta (Aβ) extracellular plaques and neurofibrillary tangles (NFTs). Biomarkers of AD, including Aβ40, Aβ42, and tau protein, amongst others, show promise for early AD diagnosis. Additional progress is made in the application of biosensing modalities to measure and detect significant changes in these AD biomarkers within patient samples, such as cerebral spinal fluid (CSF) and blood, serum, or plasma. Herein, a comprehensive review of the emerging nano-biomaterial approaches to develop biosensors for AD biomarkers’ detection is provided. Advances, challenges, and potential of electrochemical, optical, and colorimetric biosensors, focusing on nanoparticle-based (metallic, magnetic, quantum dots) and nanostructure-based biomaterials are discussed. Finally, the criteria for incorporating these emerging nano-biomaterials in clinical settings are presented and assessed, as they hold great potential for enhancing early-onset AD diagnostics.
Keywords: Alzheimer’s disease, biomarkers, colorimetric biosensors, electrochemical biosensors, nano-biomaterials, nanoparticles, optical biosensors
1. Introduction
Alzheimer’s disease (AD) is a neurodegenerative disease that accounts for more than 78% of dementia cases in elderly individuals, with 55 million people affected by the disease worldwide. This number is expected to increase to 135 million by 2050.[1] AD is the most common cause of dementia, an umbrella term that is used to describe various conditions and diseases that occur when neurons, the nerve cells in the brain, die or fail to function normally.[2] This neuronal death or malfunction subsequently affects cognitive function, such as memory and behavior.[2] In its advanced stages, dementia also has a significant impact on basic daily activities, such as walking and swallowing food.[2] Unfortunately, discernible AD symptoms only begin to appear 10 to 20 years after cellular degeneration begins. For this reason, novel strategies for early diagnosis of the disease are essential to allow successful medical intervention. To this end, scholars continue to strive to develop effective diagnosis strategies that detect AD before symptoms start to appear, allowing for both the development and application of interventions that could halt the expansion of the disease.[2,3]
Currently, AD is diagnosed based on the clinical presentation of the disease, including patient history, cognitive tests, and medical examination, which can be followed by imaging and fluid biomarker tests. Clinicians will first evaluate criteria for patients experiencing mild cognitive impairment based on the Diagnostic and Statistical Manual of Mental Disorders, fifth edition (DSM-5).[4] Tests for AD pathology and biomarkers will then be conducted once this preliminary diagnosis is given. Several imaging techniques are used to diagnose AD. For example, magnetic resonance imaging (MRI) is utilized to identify brain atrophies, while positron emission tomography (PET) is used to detect the accumulation of Aβ or tau in the brain.[5] Biomarkers of AD can be detected in biofluids, most commonly in cerebrospinal fluid (CSF). However, CSF is collected via lumbar puncture, an invasive and painful procedure. Instead, blood samples – in particular, serum samples – are gaining significant attention as a more cost-effective and non-invasive approach compared to the aforementioned diagnostic techniques.[6] Researchers have demonstrated that AD biomarkers can be detected in blood as well as CSF, and it has been shown that the concentration of target proteins significantly increases in patients with AD compared to healthy people.[6]
Moving beyond the delineation of the disease based on the presentation of symptoms to detect AD in the pre-symptomatic stage, there is a high demand for a new generation of diagnostic strategies that are non-invasive, cost-effective, reliable, and highly accurate.[5] A novel approach gaining attention in diagnostic techniques is the use of nanoparticles (NPs) in nano-biomaterial biosensing. The unique properties of NPs, including the flexibility to synthesize different sizes and shapes, functionalization of the outer surface with several ligands, and their high surface area to volume ratio, has fueled significant interest in applying NPs in the diagnosis of multifactorial diseases such as AD.[7] NPs can be employed to contend with many of the limitations of conventional diagnostic techniques (e.g., high-cost and invasiveness), providing a key solution to the current traditional diagnostic methods in AD. With ease-of-use, NPs can be incorporated into miniaturized diagnostic platforms in the form of nano-biomaterials, which makes it possible to create preventative diagnostics for AD available at the point of care. This literature review will explore early detection methods that can be used to target a range of biomarkers known to be involved or associated with the development of AD, as shown in Figure 1. First, we will explore biomarkers that have been identified for AD, expanded beyond the traditional detection of beta-amyloid (Aβ) or tau proteins. We then explore optical, electrochemical, and colorimetric forms of biosensors that leverage NP-based strategies to diagnose AD. Finally, we explore the use of NPs as contrast agents (CAs) to improve diagnostic imaging.
Figure 1.
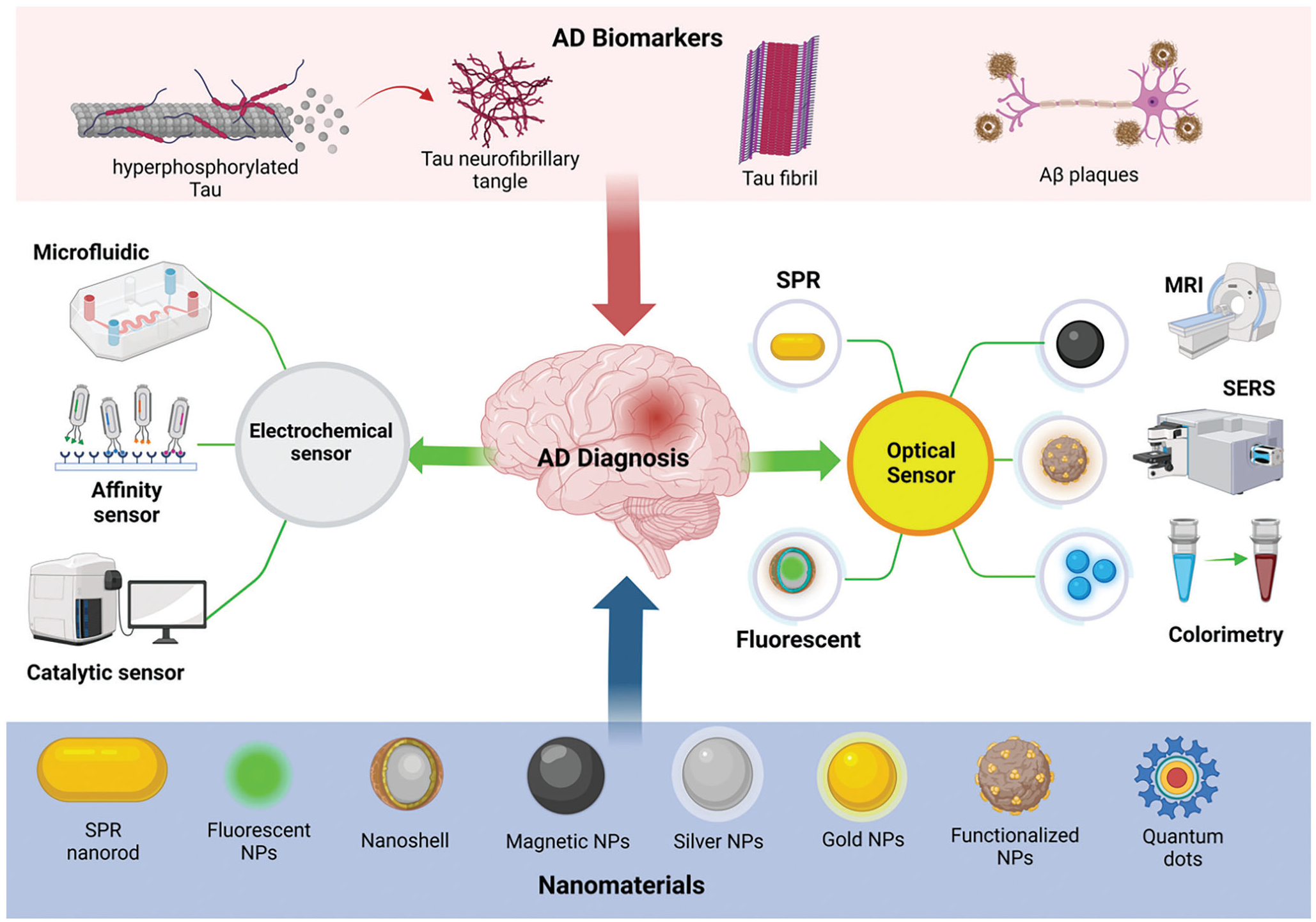
A schematic showing the major biomarkers of AD and various nanoparticles used to design advanced ultra-sensitive diagnostic tools, including optical and electrochemical biosensors for AD detection in the early stage.
2. Biomarkers of AD as Targets for Diagnostic Platforms
Research has established that the pathophysiological process of AD starts decades before symptoms begin to appear.[8] Unfortunately, most diagnoses only occur upon the clinical presentation of symptoms through a combination of clinical history, medical examinations, and imaging.[9] Accordingly, researchers are striving to identify biomarkers that can be used for early diagnosis of AD to facilitate more effective interventions.[10] The following discussion will briefly provide an overview of potential CSF and blood biomarkers to diagnose AD. Here, we intend to provide an overview of the literature to provide context for the subsequent discussion of biosensor platforms rather than a critical evaluation of the clinical validity of each biomarker.
2.1. Protein Biomarkers in AD Neurodegeneration Pathways
AD is a multifactorial disease that is caused by the presence and aggregation of different types of proteins that work synergistically to promote neuronal apoptosis and synaptic dysfunction. A few hypotheses have been suggested involving the dysfunction and loss of synapses and neurons, based on the two hallmarks of AD: extracellular plaque deposits of the Aβ peptide,[11] and neurofibrillary tangles (NFTs) of the microtubule-binding protein, tau.[12] These pathological changes have respectively led to the Aβ hypothesis and tau hypothesis.[13] However, it has been proposed that the production and accumulation of the Aβ peptide play a more significant role in the pathogenesis of AD. Thus, understanding and identification of mechanisms that are involved in the etiology of AD will play an essential role in finding biomarkers for AD. Currently, isoforms of Aβ and tau proteins are the most studied among these biomarkers.
Aβ is a small protein made up of 38–43 amino acids.[14] Aβ peptides are formed via the cleavage of a membrane-bound protein known as an amyloid precursor protein (APP).[14] APP can undergo amyloidogenic or non-amyloidogenic cleavage.[14] When APP cleaves through the latter method, the result will produce soluble forms of Aβ that are involved in several essential biological processes in the brain, such as neuroprotection and synaptic plasticity.[14] However, the amyloidogenic cleavage process of APP, shown in Figure 2A, produces insoluble isoforms of Aβ, such as Aβ40 and Aβ42, that have the potential to aggregate with time and create Aβ plaques.[14] It has been shown that the Aβ42 isoform is more insoluble than the former.[13] Aβ plaques accumulate when the APP is cut by specific secretase enzymes called β-secretase and γ-secretase.[4] This cleavage leads to the production of insoluble types of Aβ that conjugate together and form toxic irregularly shaped plaques that can potentially position between synapses and interrupt their connection.[13] Because Aβ protein is directly involved in AD pathology, intensive research has been done to study the ability to utilize Aβ and its derived isoforms as a biomarker for AD. In 1995, Motter et al. published the first paper that examined changes in Aβ42 expression in CSF and its correlation with AD, where a significant decrease in Aβ42 in CSF samples collected from individuals with AD was shown.[15] Several studies since showed that the expression levels of Aβ42 in CSF of AD patients were significantly reduced compared to healthy controls.[16] The decreased CSF level of Aβ42 in individuals with AD can be explained by the deposition of Aβ42 molecules as senile plaques in AD patients, decreasing the level of the free Aβ monomers (AβMs) in the CSF.[17] However, testing CSF is not convenient, especially for elderly people, since it is collected through a painful method known as a lumbar puncture.[18] Thus, researchers are shifting to detect AD biomarkers in other biofluidic samples, such as blood and saliva. For example, Janelidze et al. measured plasma Aβ42 and Aβ40 levels using a single-molecule array (Simoa) assay.[19] It was found that levels of plasma Aβ40 and Aβ42 decreased in AD patients compared to healthy controls, and there is a significant correlation in the levels of Aβ40 and Aβ42 found in the plasma and CSF samples of AD patients.[19] On the other hand, many studies have reported no significant difference in plasma Aβ40 and Aβ42 between AD and healthy controls;[20] Therefore, the relationship between the brain and peripheral Aβ levels is still unclear. Besides Aβ40 and Aβ42, all proteins involved in the amyloidogenic process could be promising biomarkers for AD, including APP (see Figure 2A)[21] and beta-site amyloid precursor protein cleaving enzyme (BACE).[22] Tau is another protein that plays an essential role in the pathology of AD.[23] Tau is a major component of microtubules that has a pivotal role in the intracellular transportation of molecules.[23] In AD, tau proteins undergo hyperphosphorylation and bind to form intracellular NFTs,[23] as shown in Figure 2B. The phosphorylation of tau occurs at multiple sites on the protein, with at least 30 different phosphorylation sites found to date.[24] NFTs are abnormal accumulations of the tau protein, that, in healthy physiologic conditions, play a significant role in stabilizing microtubules that act as paths for the intracellular transport of several molecules and nutrients.[7a] Similar to Aβ, the expression of total tau (T-tau) and phosphorylated tau (p-tau) proteins are utilized as diagnostic biomarkers for AD. In 2016, Mattson et al. found that the expression of tau protein increased in plasma and CSF from AD patients compared to healthy controls.[25] Also, tau was not only identified as a biomarker but as a prognostic marker since its expression was proportionally correlated with the severity of cognition decline and atrophy.[25] In other words, more tau expression was correlated with more atrophy and reduced cognitive abilities. Several isoforms of p-tau have been discovered to have the potential to act as diagnostic biomarkers for AD, for example, p-T181-tau,[26] p-T217-tau,[26] and p-s396-tau.[27] In 2022, Gonzalez-Ortiz et al. designed an anti-tau antibody, TauJ.5H3 antibody, that can specifically target and capture brain-derived tau (BD-tau) proteins from plasma d serum samples.[28] Besides the proteins that are included in the amyloidogenic and tau phosphorylation processes, researchers found other proteins that also have a major role in the pathology of the disease and can be used as a biomarker for AD. For example, Lanni et al. found that conformationally altered p53 has the potential to act as a biomarker for AD since it was detected in the fibroblasts of individuals diagnosed with AD in a high concentration.[29]
Figure 2.
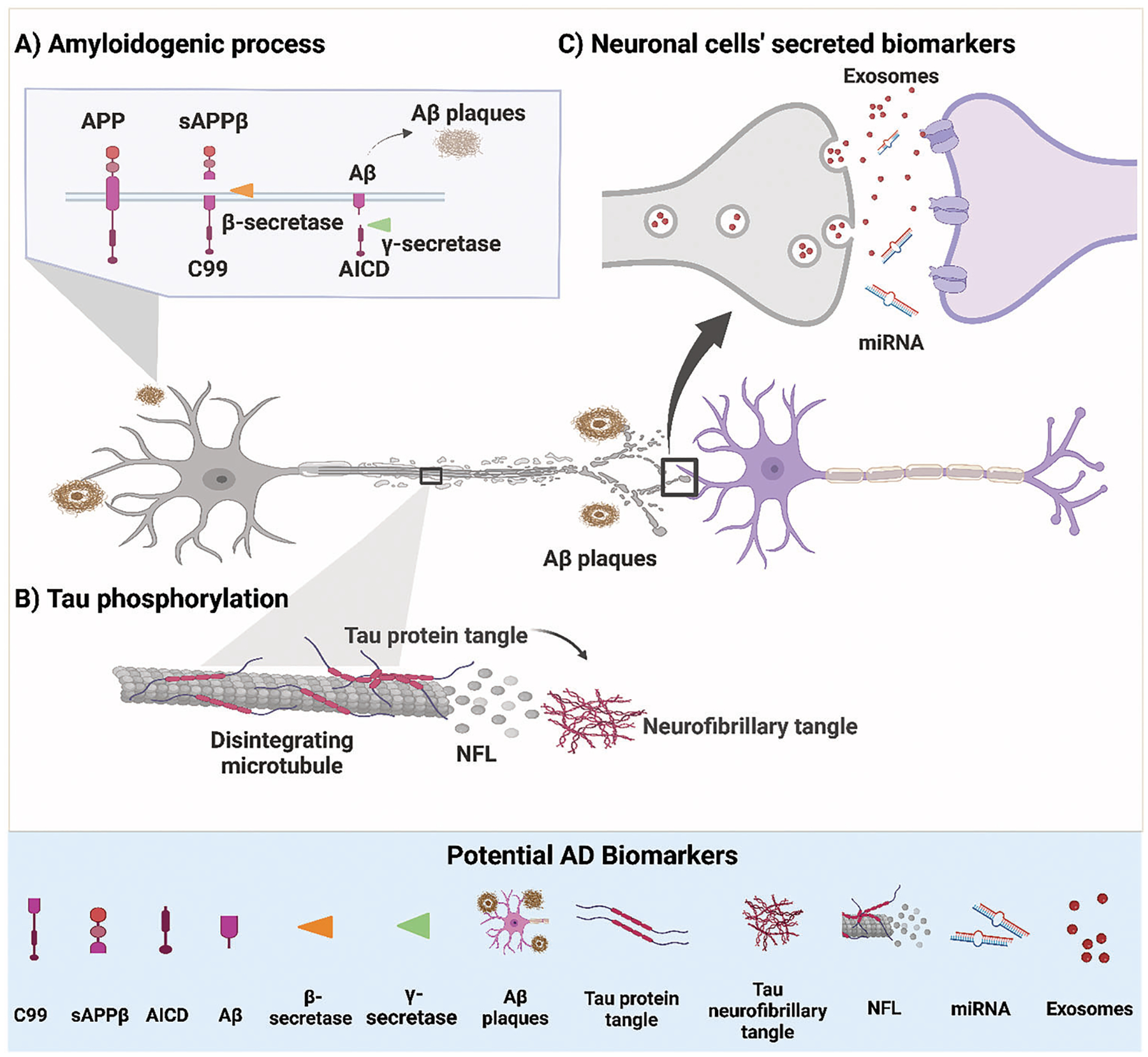
A schematic representation showing potential biomarkers for Alzheimer’s disease (AD). A) Shows the amyloidogenic process of amyloid precursor protein (APP); the protein is cut by a β-secretase enzyme and produces a membrane-bound protein (C99) and soluble peptide APPβ (sAPPβ). Then, the C99 protein is further cut by another enzyme, γ-secretase, that produces Aβ monomers (AβMs) and amyloid precursor intracellular domain (AICD). AβMs aggregate to form Aβ-plaques. B) Shows tau phosphorylation: tau is phosphorylated at multiple sites. The phosphorylation of tau protein causes the protein to dissociate from the microtubule structure, which leads to the disruption of microtubules and the release of its content to the extracellular space, such as the neurofilaments light chain (NFL). Moreover, the phosphorylated tau (p-tau) proteins tend to aggregate and form NFTs. C) Potential biomarkers secreted from neuronal cells, including miRNA and exosomes containing biomarkers.
Neurofilament light chain (NFL) is another potential protein biomarker for AD since its expression is highly impacted by the presence of the disease. NFL is a cylindrical protein that is a subunit of a group of proteins known as neurofilaments which have an essential role in maintaining the radial structure of the neuronal axons.[30] NFL proteins are expressed in all regions of neuronal cells, namely the soma, axons, and dendritic shaft.[31] In AD, the senile plaques and NFTs caused by Aβ and tau protein, respectively, result in shifting neuronal cells into apoptosis. The apoptosis of neuronal cells leads to the secretion of NFL proteins from the neuronal axon to the interstitial fluid and increases the NFL concentration in biofluids, such as CSF and plasma.[30] Mattson et al. designed longitudinal research to study the ability of NFL to differentiate AD patients from healthy controls and whether the plasma NFL expression correlates with other AD hallmarks, such as tau and Aβ.[32] The expression of NFL was measured in plasma samples of 855 participants with mild cognitive impairment (MCI), 327 with AD dementia, and 401 individuals with no cognitive impairment (controls).[32] The researchers found that the concentration of NFL was higher in plasma collected from the AD dementia group compared to MCI and control groups, proving that NFL can be used to detect AD and also can differentiate between individuals with different stages of the disease.[32] Indeed, a meta-regression analysis of 38 studies found a strong correlation between the expression of NFL and other AD biomarkers, including T-tau, p-tau, and neurogranin (NRGN).[33] This correlation shows that NFL is directly affected by AD pathologic proteins and can be a promising biomarker that reflects the progression of AD.
2.2. Nucleic Acids and Exosomes as AD Diagnostic Biomarkers
MicroRNAs (miRNAs) may be another promising source of biomarkers for the diagnosis of AD.[37] miRNAs are small non-coding RNAs that have an essential role in regulating several physiological and pathophysiological processes, such as differentiation, proliferation, and apoptosis (see Figure 2C).[34] Several studies have shown that the expression of specific types of miRNAs is highly affected by the existence and progression of AD.[35] For example, Tan et al. found that the expression of miR-181c and miR-125b decreased while miR-9 increased in AD patients compared to healthy controls.[36] Moreover, the authors found that the expression of miR-125b alone was able to differentiate AD patients from healthy controls with selectivity and specificity of 80.8% and 68.3%, respectively.[36] Another study found a significant upregulation in miR-210-3p, miR-92a-3p, and miR-181c-5p in the plasma samples obtained from AD patients compared to healthy controls.[37] A recent systematic review evaluated the results of 20 articles that studied the change in the expression of several miRNAs in blood samples derived from individuals with AD and healthy controls and 12 articles that studied the deregulation of miRNA in CSF.[38] From the 20 articles that studied the deregulation of miRNAs in blood samples collected from individuals with AD, the expression of 102 miRNAs was found to be upregulated or downregulated compared to that from healthy controls.[38] According to articles that contain data for miRNA in the CSF, the expression of 153 different types of miRNAs was found to be altered in CSF samples obtained from individuals with AD to healthy controls.[38]
Exosomes are biological vesicles ~30 to 200 nm in size and have an essential role in the crosstalk between different types of cells in the brain (see Figure 2C).[39] These vesicles comprise a phospholipid bilayer that carries many functional biomolecules, such as proteins, DNA, RNA, and miRNA.[39] Exosomes carry their content between different cells in the brain and across the blood-brain barrier (BBB).[39] Therefore, isolating exosomes from CSF or blood samples and studying their content might be a novel promising strategy to diagnose AD. Dong et al. studied the expression of several types of exosomal miRNAs via next-generation sequencing from plasma samples derived from 8 individuals with AD and 8 healthy controls.[40] The authors found 207 different types of exosomal miRNAs that were variably expressed in samples from individuals with AD compared to healthy controls.[40] 21 miRNAs out of the 207 showed a significant difference in expression, with more than ± 2.0-fold change.[40] Besides exosomal miRNA, exosome-associated proteins have also been studied for their ability to differentiate individuals with AD from healthy controls. For example, Winston et al. isolated exosomes from the plasma of AD patients and healthy controls and measured the expression of several AD pathogenic proteins, including Aβ42, P-S396-tau, P-T181-tau, repressor element 1-silencing transcription factor (REST), and NRGN.[41] A significant increase was found in the expression of Aβ42, p-S396-tau, and p-T181-tau in exosomes derived from AD samples compared to healthy controls.[41] In contrast, the expression of REST and NRGN were significantly reduced in exosomes obtained from plasma samples of individuals with AD.[41] Thus, the ability to isolate exosomes and measure their protein and nucleic acids, such as miRNA content, may serve as a standard gold method to diagnose AD in the future.
Owing to the heterogeneity and low concentration of biomarkers in biofluidic samples, highly sensitive and reliable detection techniques that can overcome the limitations of traditional methods, such as the enzyme-linked immunosorbent assay (ELISA), are urgently needed. Therefore, designing innovative detection methods and diagnostic devices may ultimately enhance the field of AD biomarkers and diagnosis. The mounting attention to applying NPs to systems that can help in the diagnosis of a multifactorial disease like AD is due to their attractive chemical, physical, and optical properties. This interest in NPs has been sparked in the hope of designing detection strategies that are more adequate for high-throughput screening, low detection limits, and real-time or point-of-care analysis. The following discussion will address various biosensor technologies (e.g., electrochemical, optical, and colorimetric) that are engineered by using a variety of NPs and the impact these are having on the detection of AD biomarkers.
3. Nanoparticles in the Fabrication of Diagnostic Biosensors for Alzheimer’s Disease
3.1. Electrochemical Biosensor Designs for the Diagnosis of AD
NPs have been widely used in the fabrication of electrochemical biosensors because they are easily synthesized, their outer surface can be functionalized with several ligands, they possess tunable size, shape, and geometric parameters, and their high surface area to volume ratio allows for ideal detection. NPs, such as gold NPs (AuNPs), silver NPs (Ag-NPs), quantum dots (QDs), and magnetic NPs (MNPs), exhibit excellent conductivity and catalytic properties. The unique properties of these particles make them excellent candidates for the fabrication of electrochemical sensors and biosensors to enhance selectivity and limit of detection (LOD) of AD biomarkers for biosensing applications.
3.1.1. Gold Nanoparticles in Electrochemical Biosensor Fabrication
AuNPs have been widely used in the fabrication of electrochemical biosensors because they have excellent conductivity and catalytic properties. The unique properties of AuNPs make them excellent candidates for the fabrication of electrochemical sensors and biosensors to enhance the accuracy of detecting AD pathogenic proteins from biological samples. Several studies have utilized AuNPs to coat transducers due to their ability to conjugate the biomolecular recognition elements without affecting bioactivity and in a way that their binding sites are accessible for the target antigens, as well as their ability to enhance electron transference.
Diba et al. designed a quantitative electrochemical immunosensor based on a surface sandwich immunoassay approach. The biosensor was designed as follows: (I) AuNPs were deposited on a screen-printed carbon electrode (SPCE).[42] (II) The particles were conjugated with polyethylene glycol (PEG) and mercaptopropionic acid (MPA) to decrease non-specific protein adsorption to the outer surface of the particles and to functionalize the particles covalently with anti-Aβ (12F4).[42] After the Aβ42 antigen binds to anti-Aβ (12F4), another antibody, anti-Aβ (1E11), functionalized with alkaline phosphatase (ALP), binds to a second epitope of the antigen. The surface-bound ALP interacts with APP, which converts it to its dephosphorylated form, producing 4-aminophenol molecules. At −0.05 V versus Ag/AgCl, 4-aminophenol molecules are oxidized to 4-quinoneimine and generate an anodic peak response. The sensor was able to quantitatively measure the concentration of Aβ42, and there was a proportional relationship between the Aβ42 concentration and the anodic peak response while achieving a LOD of 100 fM.[42] The main challenge of sandwich assays is that the epitopes of the proteins have to be accessible to the antibodies; however, the agglomeration of protein, which is the main cause of AD, can lead to steric hindrance and epitope hiding.[43] As a result, the Aβ concentration in sandwich assays analysis may be underestimated and should be a source for further investigations. In this paper, AuNPs were used to covalently immobilize antibodies to the outer surface using MPA as a linker.
In another study, AuNPs were used in a label-free electrochemical immunosensor for Aβ42 detection.[44] The sensor was engineered by using a gold electrode covered by a self-assembled layer of MPA which was further decorated by another layer of AuNPs. Thiol groups were added to antibodies via a thiolation process to form covalent bonds with the outer surface of the AuNPs.[44] The authors stated that the designed sensor required only 10 minutes to detect Aβ42 with a concentration that lies in the range of 10–1000 pg, with a LOD and limit of quantitation (LOQ) of 5.2 pg mL−1 and 17.4 pg mL−1, respectively.[44]
In 2020, Iglesias-Mayor et al. utilized AuNPs due to their ability to immobilize antibodies and their unique electrocatalytic activity.[45] They designed a competitive electrochemical immunosensor platform consisting of an Au@Pt/Au core–shell system, which has the potential to detect and quantify conformationally altered p53 peptides as low as 66 nM.[45] Conformationally altered p53 was detected in a competitive manner between the Au@Pt/Au NPs/anti-p53 and magnetic beads (MBs) conjugated with p53 in a way that the absence of the p53 molecules led to the capture of Au@Pt/Au NPs/anti-p53 on the beads. This process enhanced the water oxidation reaction (WOR), which resulted in increasing current. The presence of the conformationally altered p53 leads to the attachment of Au@Pt/Au NPs/anti-p53 to the p53 molecules, blocking the WOR catalytic activity of Au@Pt/Au NPs, and decreasing the signal current (Figure 3A).[45] A spike and recovery experiment was performed to validate the ability and accuracy of the platform to detect conformationally altered p53 in real samples. Thus, plasma samples were collected from healthy individuals and spiked with 100, 500, and 1000 nM of conformationally altered p53 protein. The authors contend that their electrochemical immunosensor was not affected by the complexity of real samples and was able to accurately detect conformationally altered p53 with a recovery percentage of ≈90%.[45]
Figure 3.
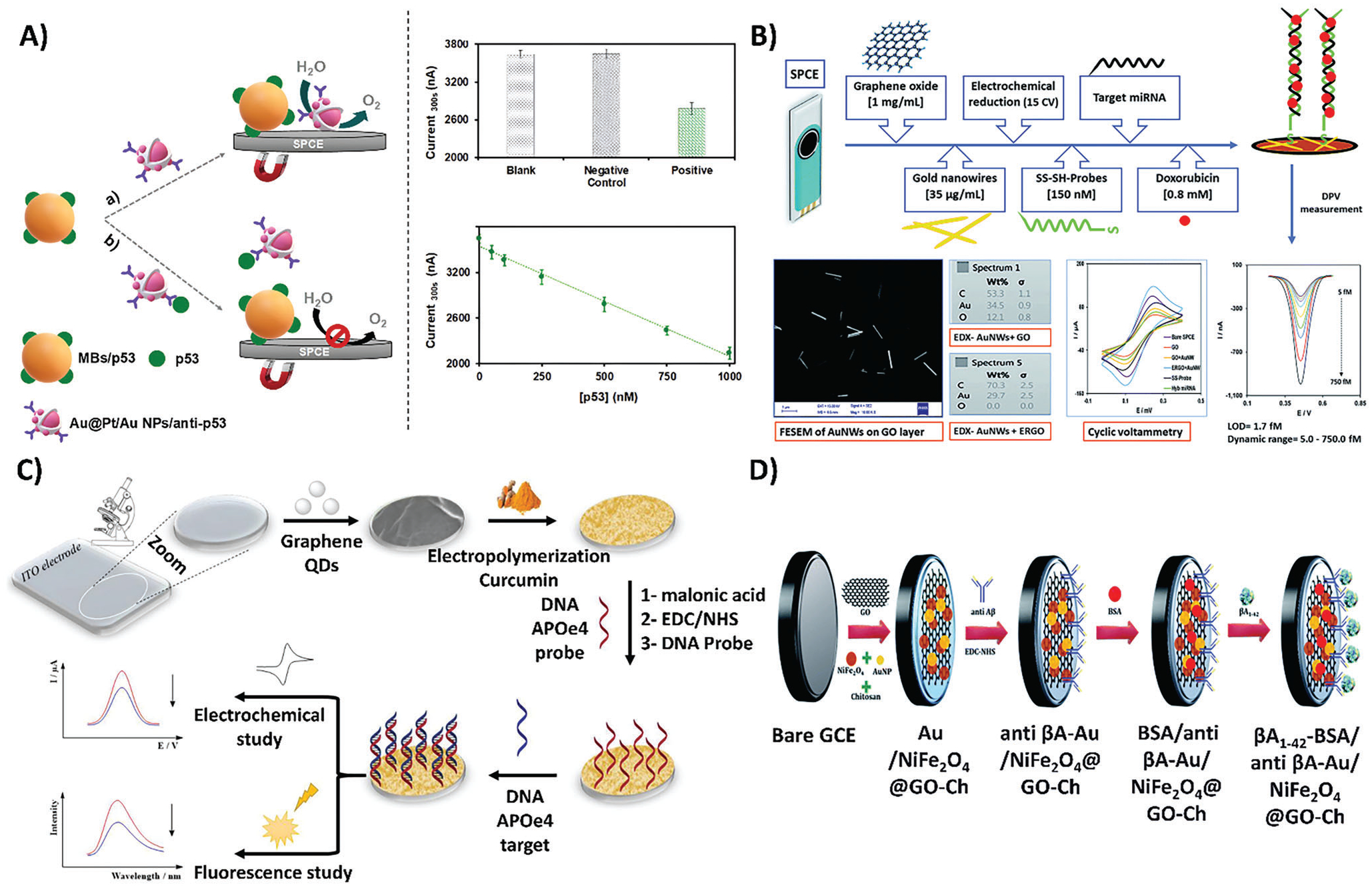
Schematic representations of various electrochemical biosensor designs, fabrication processes, and quantification techniques to detect AD biomarkers. A) Method of detection of an electrochemical biosensor for quantifying conformationally altered p53 through the use of a competitive immunoassay made of Au@Pt/Au NP tags. Reproduced with permission.[45] Copyright 2020, American Chemical Society. B) Outlined steps for the fabrication of an electrochemical biosensor composed of graphene oxide and gold nanowires, that quantifies miRNA-137. Reproduced with permission.[48] Copyright 2017, Royal Society of Chemistry. C) The steps utilized to design curcumin-graphene quantum dots (QDs) for a dual-mode sensing platform: fluorescence and electrochemical detection of apolipoprotein e4 (ApoE4). Reproduced with permission.[49] Copyright 2018, Elsevier. The labels on the original figure have been modified to improve readability. D) Fabrication of an electrochemical biosensor to capture and quantify Aβ. The biosensor was designed by modifying a glassy carbon electrode (GCE) with graphene oxide-chitosan nanocomposite functionalized with gold nanoparticle/nickel ferrite (AuNP/NiFe2O4) to immobilize anti- Aβ on the surface of the platform. Reproduced with permission.[50] Copyright 2020, Royal Society of Chemistry. The labels on the original figure have been modified to improve readability.
De Olivera et al., designed a disposable microfluidic platform integrated with an electrochemical immunosensor to detect and quantify a disintegrin and metalloprotease 10 (ADAM 10) biomarker in clinical samples, including plasma and CSF.[46] ADAM 10 is the α-secretase responsible for the cleavage of APP protein in the non-amyloidogenic pathway producing soluble amyloid precursor protein α2 (sAPPα 2). In fact, sAPPα plays a crucial role in neuroprotection and memory.[47] The platform was composed of eight working electrodes (WEs) coated with poly(diallyldimethylammonium chloride) (PDDA)/AuNPs to immobilize anti-ADAM10 antibodies on the surface of the electrodes.[46] ADAM 10 was first separated from the sample using MNPs conjugated with HRP and another ADAM10 antibody, then the HRP-MB-Ab2/ADAM10 was injected into the disposable microfluidic platform and captured by the antibodies immobilized on the surface of the electrodes resulting in a sandwich structure. The authors found that their designed platform was able to detect and quantify ADAM10 in blood samples collected from 10 healthy controls, 10 individuals diagnosed with MCI, and 25 individuals diagnosed with AD. Further, the results from the platform were in line with the results of ELISA, which indicates the good specificity and selectivity of the platform in detecting ADAM10 in human plasma samples. Also, the receiver operating characteristic (ROC) curve shows that the platform was able to detect ADAM10 from human plasma with 72% sensitivity, 100% specificity, and an area under the curve (AUC) of 0.888.[46]
Another study illustrated the detection and quantification of miRNA-137 with a LOD of 1.7 fM using an electrochemical biosensor composed of graphene oxide and gold nanowires.[48] The biosensor was designed as follows: (I) graphene oxide and gold nanowires were utilized to cover an SPCE to enhance the electron transference rate and conductivity; (II) A single-stranded DNA probe that binds explicitly to miRNA-137 was conjugated to Au nanowires; (III) Doxorubicin molecules were used due to their ability to bind, via the intercalation mechanism, to the double-stranded oligonucleotides formed after the target miRNA’s attachment to the single-stranded DNA probe (Figure 3B).[48] Results showed that the biosensor had the potential to definitively distinguish its target of interest, miRNA-137, from non-specific miRNAs even at high concentrations of non-specific oligos.[48] A summary of recent electrochemical biosensors for AD is detailed in Table 1.
Table 1.
Summary of electrochemical biosensors incorporating AuNPs in AD.
| Electrochemical biosensor design | The role of AuNPs in the sensor | Targeted biomarker | Limit of detection | Sample type | Ref. |
|---|---|---|---|---|---|
| ITO is deposited by AuNPs in the presence or absence of PEG. | Enhance the electrical conductivity Enhance the sensor sensitivity to the biomarker | Aβ40 | 20.7 ng g−1 | Aβ samples extracted from rats’ brain | [59] |
| Immobilized oligonucleotides were used to capture ApoE4, and ferrocene functionalized with AuNPs modified with streptavidin to amplify the signal. | Enhance the electrochemical signal | ApoE4 | 0.1 pM | ApoE 4 DNA extracts from serum samples | [60] |
| A polycrystalline gold surface was deposited by a layer of microporous gold nanostructures. The gold nanostructures were conjugated with Aβ42 binding peptides. | Immobilize the Aβ42 binding peptide Improve electron transfer | Aβ42 | 0.2 pg mL−1 | Artificial CSF and spiked serum samples | [61] |
| Gold electrodes were modified with cellular prion protein (PrPC) that has the ability to specifically target AβO. | Improve electron transfer | AβO | 45 pM | Serum samples | [62] |
| Sandwich immunoassay was designed by conjugating SPCEs with AuNPs- polyamidoamine (PAMAM) dendrimer nanocomposite (3D-Au-PAMAM). Anti-tau antibodies were conjugated on the 3D-Au-PAMAM. After the protein was captured, the protein was sandwiched with a secondary Ab conjugated with horseradish peroxidase (HRP-DAb). | Immobilize tau antibody | tau | 1.7 pg mL−1 | Raw plasma Brain tissue extracts Postmortem diagnosis of AD | [63] |
3.1.2. Silver Nanoparticles in Electrochemical Biosensor Fabrication
Like AuNPs, AgNPs represent potent candidates to perform as electrochemical tags due to their ability to enhance the detection of electrochemical sensors due to their excellent electrochemical activity as well. An electrochemical biosensor was designed to detect Aβ oligomers (AβOs) based on the formation of AgNP aggregate tags.[51] Briefly, adamantine cellular prion protein (Ad-PrP 95−110) molecules were attached to AgNPs, and Ad-PrP 95−110/AgNPs were anchored to β-cyclodextrin (β-CD) molecules that were immobilized on the surface of an electrode through (host-guest) interactions. The electrochemical signal was produced, as follows: (I) in the absence of AβOs, Ad-PrP 95−110 interacted with AgNPs and triggered the formation of AgNPs aggregates, producing a high electrochemical signal. (II) In the presence of AβOs, the oligomers interfered in the Ad-PrP 95−110/AgNPs interaction and bound to Ad-PrP 95−110, which resulted in decreased formation of the AgNPs aggregates and led to a decrease in signal.[51] The authors contend that the sensor was able to detect AβOs as low as 8 pM. However, the sensor was not able to detect AβO in serum samples until the samples were diluted by more than 50-fold.[51] In a follow-up study, this electrochemical sensor was modified using cyclodextrins (CDs) to attach Ad-PrP 95−110/AgNPs to the surface of the electrode, and Ad-PrP 95−110/AgNPs complexed were immobilized directly to the electrochemical platform.[52] Using this technique, the LOD decreased to 6 pM instead of 8 pM using the previous sensor. Most importantly, this electrochemical sensor technique was able to detect AβOs in CSF and serum samples with recovery percentages in the range of 86% to 109%.[52] Another study used AgNPs in the fabrication of a sandwich-type electrochemical sensor to detect an AD biomarker, α−1 antitrypsin (AAT), to immobilize antibodies on the outer surface of the particles and enhance the electrochemical signal.[53] The sensor was designed as follows: (I) electrodes were covered by carbon nanotubes (CNTs); (II) alkaline phosphatase-labeled AAT antibodies were conjugated to AgNPs (ALP-AAT Ab-AgNPs).[53] In the presence of AAT, ALP molecules were anchored to the particles enzymatically using dephosphorylates 4-amino phenyl phosphate to 4-aminophenol leading to a high electrochemical signal. The biosensor was able to detect AAT as low as 0.01 pM, and it was successfully able to detect the biomarker in biological samples, such as serum.[53]
3.1.3. Quantum Dots in Electrochemical Biosensor Fabrication
QD-based electrochemical biosensors have been used to detect biomarkers via two approaches; first, by dissolving QDs as electrode modifier labels and releasing metal ions, and second, the direct detection using QDs as nanolabels.[54] To diagnose AD, QDs have primarily been utilized to modify electrodes. Medina-Sánchez et al. designed a QD-based electrochemical biosensor that was able to detect apolipoprotein e4 (ApoE4) in human plasma through the release of metal ions.[54] The suggested biosensor was made by cadmium-selenide/zinc-sulfide (CdSe/ZnS) QDs as labels and tosyl-activated MBs to modify a polydimethylsiloxane-polycarbonate microfluidic chip integrated into screen-printed electrodes. The detection procedure consisted of two parts; firstly, MBs were modified with antibodies and anchored to the electrode using a neodymium magnet. Next, ApoE4 was incorporated in different concentrations with the addition of biotinylated antibodies. The electrochemical measurement of Cd2+ reduction and re-oxidation was obtained by streptavidin-modified QDs. The linear range of ApoE4 was from 0.29 to 5.86 nM (10–200 ng mL−1) with a LOD of 0.37 Nm (12.5 ng mL−1).[54]
In another recent study, curcumin-graphene QDs were used as a dual electrochemical and fluorescence platform was used to develop indium tin oxide (ITO) electrodes capable of detecting ApoE4 in human blood plasma. This platform exhibits high performance, including repeatability, reproducibility, selectivity, and long-term storage stability.[49] The transparent electrode, ITO, was functionalized with graphene QDs, electro-polymerized with curcumin, then malonic acid and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide/N-hydroxysuccinimide (EDC/NHS) chemistry was used to immobilize an amino-substituted DNA probe. In the last step, ApoE4 was incorporated to test biosensor performance. The recorded quenching signals of curcumin were employed to quantify ApoE4 DNA, showing a linear decrease in the amperometric response with a LOD of 16.7 fM (Figure 3C).[49] Regarding Aβ detection, Bu et al. developed a quantitative cathodic photoelectrochemical aptasensor by means of black phosphorous QDs as a photoactive material, assisted by heme as an electron acceptor.[55] The transparent ITO electrode was coated by black phosphorous QDs, followed by the adsorption of positively charged poly-l-lysine (PLL) onto black phosphorous QDs through electronic interaction. The aptamer as the specific recognition element for Aβ was incorporated on the modified electrodes, yielding Aβ-heme complexes. Aβ was simultaneously captured by the aptamer on the electrode, resulting in an enhanced photocurrent response, which demonstrates a linear response range of Aβ from 1.0 fM to 100 nM with a LOD of 0.87 fM.[55]
3.1.4. Magnetic Nanoparticles in Electrochemical Biosensor Fabrication
MNPs have been used in the fabrication of several biosensors to identify various pathological biomarkers, such as p-tau and acetylcholinesterase (AChE). Da Silva et al. suggested a highly sensitive electrochemical biosensor to detect AChE.[56] The AChE was fixed on a poly (neutral red) (PNR) film, which was grown on Fe2O3 MNPs-modified GCE by potential cycling electropolymerization in ethaline deep eutectic solvent with an acid dopant.[56] The rate of PNR growth on MNPs has been determined in different acid dopant anions (NO3−, SO42−, Cl−, ClO4), and the PNREthaline-HNO3/Fe2O3 NP/GCE sensing platform was introduced as the best option for the AChE biosensor. The proposed advantage of the designed biosensor included good selectivity, reproducibility, stability, and high selectivity, with fast response and low LOD, 1.04 μM, which was successfully used for AChE detection in synthetic urine with good recoveries.[56]
Devi et al. engineered a label-free electrochemical immunosensor for the detection of Aβ using gold/nickel ferrite (NiFe2O4) NPs decorated with a graphene oxide-chitosan nanocomposite in combination with a GCE (Figure 3D). The incorporation of NiFe2O4 NPs on 2D graphene oxide nanosheets demonstrates an excellent platform for sensitive and selective sensing applications in the monitoring of AD. They employed differential pulse voltammetry to study the amperometric response of the developed immunosensor as a function of Aβ42 concentration. The results demonstrated a wide linear range from 1 pg mL−1 to 1 ng mL−1 with a LOD of 3.0 pg mL−1.[50]
Importantly, MNPs-based immunoassays could differentiate between dementia and prodromal stages of AD. In a recent study, the immunomagnetic reduction assay comprising MNPs coated with surfactants (e.g., dextran) was used to detect the blood-based biomarkers (Aβ and p-tau).[57] Three different kinds of reagents in phosphoryl buffer solutions (pH 7.2) were functionalized by immobilizing antibodies against Aβ and p-tau. The platform was used to examine the concentration of Aβ42, Aβ40, and tau in plasma samples collected from 3 different groups: (I) individuals with MCI due to AD, (II) individuals with AD dementia, (III) and healthy controls. The concentration of Aβ40 was found to decrease in the MCI group and AD dementia group compared to the healthy control group. However, the concentration of Aβ42 and tau was significantly higher in MCI compared to the healthy control group but lower than their concentration in the AD dementia group. According to the ROC results, the sensitivity and specificity of the platform in differentiating the concentration of the three biomarkers between the healthy control group from MCI and AD dementia groups were high. In contrast, the sensitivity and specificity were moderate when differentiating the MCI group from the AD dementia group. In fact, this platform was not only able to examine the concentration of the biomarkers in the clinical samples but also differentiate between the groups that participated in the study. This finding makes this platform promising for use as a diagnostic and staging tool.[57]
The screening of targeted AD biomarkers could be achieved in a short time using magnetic-based biosensors and bioassays. These approaches could be developed to be low-cost and user-friendly, with high sensitivity and specificity. However, there are still some challenges to MNP-based biosensing, including large-scale manufacture, multiplexing of biomarkers, standard detection limit, efficiency, and long-term stability, which must be considered for effective clinical translation.[58] Additionally, combining MNPs with other nanomaterials (e.g., AuNPs) in biosensors could enhance performance and dramatically increase functional features.
3.2. Non-Colorimetric Optical Biosensor Designs for the Diagnosis of AD
Optical biosensors are a unique class of biosensor modality that utilizes optical transduction, such as changes in fluorescence intensity or optical phenomenon, to distinguish concentrations of biomarkers in artificial or human samples. As such, optical biosensors are diverse and versatile in their clinical applications.[64] Unlike conventional diagnostic imaging techniques, optical biosensors can offer relevant diagnostic data on a patient’s condition in real-time and can be used for early-onset diagnosis of complex diseases.[64a] Therefore, by analyzing the optimization and characterization of nanomaterials in various optical biosensor fabrication techniques, the quantification and detection of target AD biomarkers can be maximized, which is the objective of the literature reviewed in this section.
3.2.1. Gold Nanoparticles in Non-Colorimetric Optical Biosensor Design
AuNPs are the most used NP in the development of nanomaterial-based optical biosensors. This is due to their intrinsic metallic properties, which enhance their optical behaviors. Specifically, AuNPs, being metallic by nature, exhibit optical properties that can be quantified through surface plasmon resonance (SPR).[65] SPR is a phenomenon that introduces an electromagnetic field to a nanomaterial, which causes the oscillation of the material’s metal-free surface electrons. This, in turn, results in either the absorption or scattering of light, creating resonance in the nanomaterial that is quantifiable by measuring the index of refraction of light as it interacts with the material. When these metallic nanomaterials are exposed to target biomarkers, their ability to absorb and refract light is altered, and through SPR, these changes can be quantified to detect the concentration of the introduced biomarker.[66]
In the case of AuNPs, there is an added advantage as each individual NP exhibits SPR, creating a localized SPR (LSPR) (see Figure 4A), which makes the detection even more specific.[65,67] In fact, LSPR is highly affected by the AuNP’s geometry and, as such, can be leveraged in biosensor design by optimizing the size and shape of the individual AuNPs. AuNPs can be tuned to be highly stable, biocompatible, and customizable for a specific biomarker [65] The LSPR phenomenon, alongside wide-range customizability, has fueled scientific and clinical interest in utilizing AuNPs to fabricate optical AD diagnostic techniques.
Figure 4.
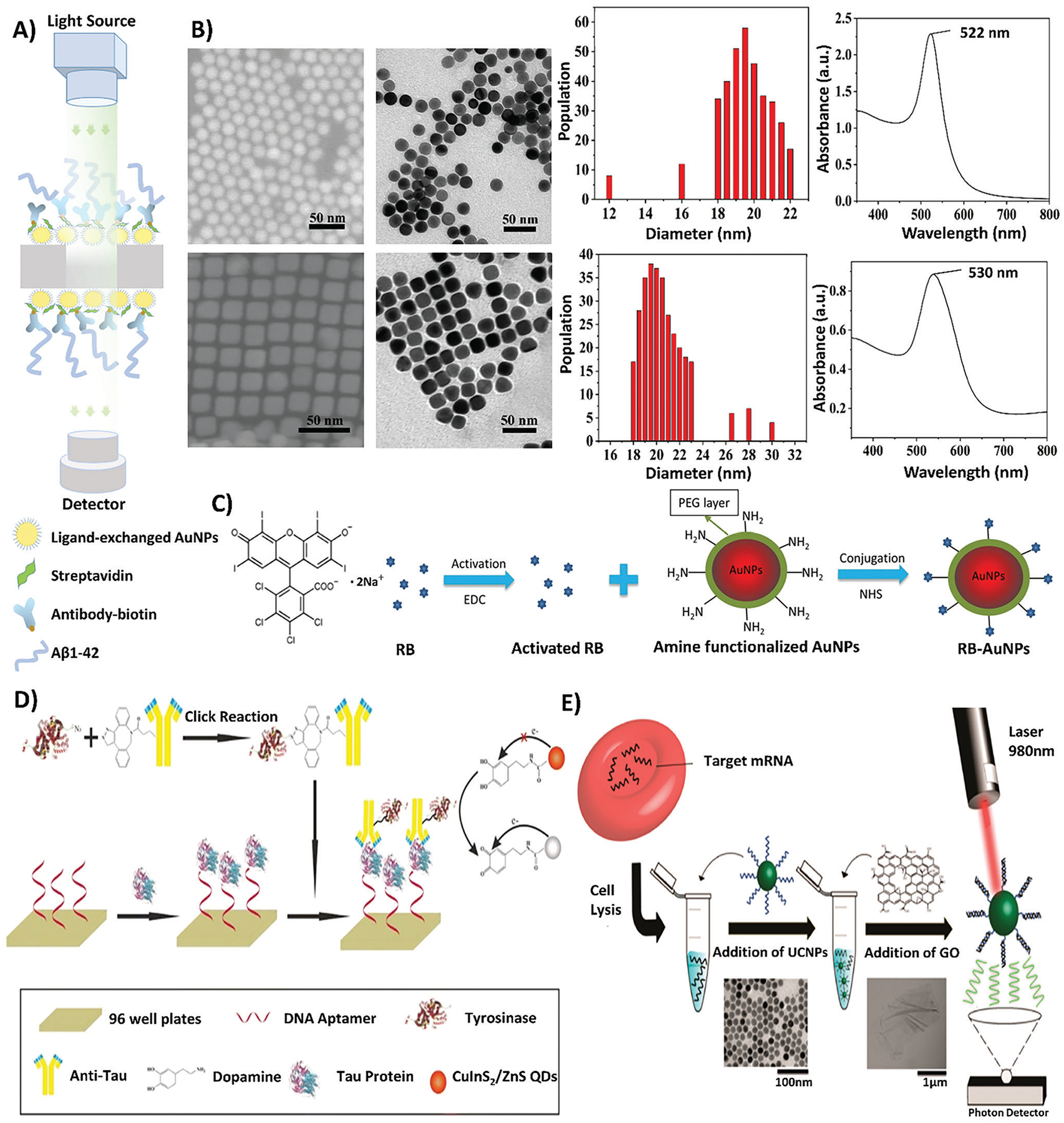
A schematic of various optical biosensing measurement and fabrication techniques. A) A schematic depicting the utilization of LSPR quantification techniques to measure the optical properties of AuNPs in the biosensing platform. Reproduced with permission.[65] Copyright 2020, Elsevier. B) Depicts the characterization of AuNSs and AuNCs in terms of shape and absorbance. Reproduced with permission.[69] Copyright 2019, American Chemical Society. C) A schematic showing the conjugation process of the F-SERS probe, where RB is activated using EDC and combined with PEGylated AuNPs and NHS for conjugation, forming RB-AuNP complexes. Reproduced with permission.[75a,b] Copyright 2019, Springer Nature. D) A schematic outlining the application of CuInS2/ZnS QDs within a sandwich immunoassay for the fabrication of an optical biosensor. The process of creating the sandwich immunoassay involves the immobilization of DNA aptamers to a 96-well plate substrate, followed by the introduction of tau protein, and ending in the binding of anti-tau antibody conjugated to tyrosinase. Through dopamine-functionalization induced by tyrosinase enzymatic activity, the quenching effects of CuInS2/ZnS QDs are induced. Reproduced with permission.[83] Copyright 2019, Springer. E) A depiction of UCNPs in the detection of AD miRNAs, in which target miRNAs are extracted from cells through cell lysis and combined with UCNPs. With the addition of graphene oxide and the utilization of a 980 nm laser, the excitability of UCNPs with graphene oxide can be detected by a photon detector. Reproduced with permission.[84] Copyright 2017, American Chemical Society. The labels on all original figures have been modified to improve readability.
One key benefit of leveraging LSPR is the ability to achieve low LODs simply by adjusting the geometric parameters and structure of the AuNPs (Figure 4A). This is demonstrated in a study by Ly et al. in which their AuNP biosensor is optimized for the detection of Aβ42, a key derivative of Aβ, one of the hallmark biomarkers of AD. Ly et al. fabricated AuNP films that coated polyethylene terephthalate substrates. By testing and optimizing the diameters of each AuNP, Ly et al. determined that a 9 nm NP diameter utilized in two film layers creates the ideal features for maximizing light absorption surrounding the AuNPs in the presence of Aβ42 (Figure 4A). As such, through the fine-tuning of AuNP size, it was possible to lower the LOD to 1 pg mL−1 in human CSF samples, making the overall biosensor more sensitive after optimization of AuNP size.[65]
Wang et al. further assessed the effects of the geometric optimization of AuNPs on the ability to detect and interact with Aβ40 during the fibrillation process, such as during the formation of beta-amyloid fibrils (AβFs) and aggregates, rather than just observing the concentration of Aβ40.[69] Specifically, Wang et al. looked to demonstrate the difference in functionality between gold nanospheres (AuNSs) and gold nanocubes (AuNCs), as shown in Figure 4B. To ensure that shape was the only factor change, the AuNSs, and AuNCs were both fabricated with the same traditional seed method and functionalized with cationic gemini surfactant (C12C6C12Br2).[69] A common cationic benzothiazole dye, thioflavin T (ThT), was utilized in a fluorescence assay to detect fluorescence intensity when the AuNPs were exposed and interacted with Aβ40 in the solution buffer.[67,69] In this assay, it was shown that despite similar synthesis techniques and the same value of 20 μM of Aβ40, AuNSs had higher ThT fluorescence intensity compared to AuNCs, which in turn signifies an increased induced interaction between AuNSs and Aβ40 during fibrillation. As such, Wang et al. demonstrated the effect of shape in AuNP interactions with Aβ40 fibrillation and concluded that AuNSs are a more promising avenue for biosensor fabrication.[69]
Another benefit in the optimization of AuNP geometric parameters includes the ability to detect multiple AD biomarkers simultaneously, by combining AuNPs of different shapes and sizes into one optical biosensing platform. Simultaneous detection of different AD biomarkers, or multiplex detection, is advantageous as it allows for more accurate diagnostic information to be gathered at once from a single sample rather than requiring multiple samples.[66a,68] Kim et al.’s study aimed to demonstrate the feasibility of designing a multiplex detection biosensing platform for multiple AD biomarkers. Here, three different AuNPs were optimized in terms of their shape and size to be specific to either Aβ40, Aβ, or tau protein in mimicked blood. The optimized shapes are nanospheres with a diameter of 50 nm for Aβ40, short nanorods for Aβ, and long nanorods for tau. These three distinct AuNPs were then combined to create a shape-code nanoplasmonic biosensor, which was able to detect Aβ40, Aβ, and tau simultaneously at extremely low LODs of 34.9, 26, and 23.6 fM, respectively.[68]
While AuNPs have many benefits in the detection of AD biomarkers and can be optimized for their specificity and sensitivity, AuNPs alone have limitations. Currently, they are only used for AD diagnosis through optical biosensing platforms in vitro. This is because AuNPs have the potential to interact in complex ways with biological processes and systems, and as such, their toxicity and safety for in vivo applications are not fully established. Further research to assess the appropriate administration routes, bioaccumulation rates, and adverse interactions would be needed for in vivo use of AuNPs.[70] At times, collecting human blood or CSF samples from a patient still poses challenges in terms of invasiveness, and as such, having a biosensing platform that can detect biomolecules in vivo can be more desirable as it can overcome these limitations.[69,71]
3.2.2. Dyes, Probes, and Agents for Non-Colorimetric Optical Biosensor Enhancement
While there is still limited research conducted on in vivo detection for AD diagnostics, approaches that utilize fluorescent labels or dyes can enhance the biocompatibility and detection ability of AuNPs in tissues and organs rather than through blood or CSF samples. Fluorescent dyes and labels emit light in a fluorescence spectrum that optimized AuNPs on their own cannot achieve, and this specialized fluorescence makes the signals from the dye-conjugated AuNPs detectible and distinguishable in tissues and organs. Distinguishing dye-conjugated AuNPs in tissue and organ samples can be done through various imaging modalities, including fluorescence microscopy, Fourier-transform infrared spectroscopy (FTIR), and ultraviolet-visible light (UV–vis) spectroscopy.[69,71,72] The applicability of dyes, labels, and agents is dependent on the target biomarker that the optical biosensing platform is being designed for. Fluorescent dyes can be derived from a wide range of natural and synthetic sources, including peptides and DNA or RNA strands, known as aptamers, which can be customized and designed for the detection of specific AD biomarkers of interest. As such, their high affinity and biocompatibility have been shown in ex vivo models, which aids in advancing the field of research toward a more in vivo approach.[71–73]
Jara-Guajardo et al. analyzed the feasibility of conjugating fluorescent peptide probes in combination with AuNPs ex vivo to detect AβFs in brain slices of transgenic mice with induced AD.[71] They began by synthesizing gold nanorods (AuNRs), which were then modified by PEG spacers (HS-PEG-OMe and HS-PEG-COOH) and functionalized with the D1 peptide to form AuNR-PEG-D1. The fluorescent peptide probe utilized in conjugation with this biosensing platform, CRANAD-2, was derived from curcumin and emitted fluorescence in the near-infrared (NIR) spectrum at 715 nm. This unique fluorescence band that CRANAD-2 exhibits, when co-incubated with the AuNR-PEG-D1 biosensor at a limit of 0.001 nM, was shown to be successful for ex vivo detection, especially in the AD brain tissue of mice.
Nucleic acids, such as DNA and RNA, are also viable aptamers for conjugation with AuNPs and enhance the detection of AuNPs for clinical applications. For instance, Song et al. developed a DNA-encoded AuNP biosensing platform for the detection of various AD-associated exosome miRNA strands, with LODs ranging from 3.37–4.01 aM. A double-stranded DNA aptamer facilitated the formation of a novel programmable curved nanoarchitecture of the AuNPs, aiding in signal amplification and increased selectivity. This was shown in the ability of their optical biosensor to distinguish AD patient serum samples from patients with MCI and healthy controls. Through ROC analysis, the AUC values for the tested exosome miRNA showed a range of 0.936–0.957, with a sensitivity of 95.83%, making the detection of miRNA strands through nucleic acid aptamers conjugated to uniquely structured AuNPs highly effective in AD diagnostics.[74]
Alongside the use of fluorescence probes and aptamers, AuNP biosensing platforms can also be enhanced for clinical applications by leveraging the surface-enhanced Raman scattering (SERS) phenomenon.[75b] SERS is a phenomenon characterized by Raman scattering, which increases the overall effects of the intrinsic LSPR phenomenon of individual AuNPs, as well as other metallic nanomaterials. This can be tailored by introducing nanostructures as protruding additions to each AuNP and contributes to the excitability and Raman scattering of the AuNPs, which strengthen the existing magnetic field and LSPR effect. SERS nanostructures can be combined with common fluorescence probes to create fluorescent SERS (F-SERS) probes (see Figure 4C). F-SERS probes conjugated on AuNPs broaden the applicability of the AuNP-based biosensing platform by allowing for real-time data and image acquisition, as well as being able to use common dyes widely used in the market, including fluorescein isothiocyanate (FITC).[75a,b] F-SERS probes reduce the need to construct or fabricate specialized probes for detection by utilizing existing fluorescence probes and modalities that are more accessible.[75b]
In Xia et al.’s study, the application of F-SERS probes in an AuNP-based biosensor was shown to be both effective and superior to biosensing modalities containing only unconjugated AuNPs when detecting Aβ42 peptides. The F-SERS probe consisted of bifunctional AuNPs in conjugation with the widely available Rose Bengal (RB) dye (4,5,6,7-tetrachloro-2′,4′,5′,7′-tetraiodofluorescein), creating a multifunctional Raman and fluorescent biosensor, as shown in Figure 4C. The quality and detection ability of the F-SERS probe was assessed by comparing the Raman and fluorescence data of the RB-AuNP complexes with unconjugated AuNPs. In terms of Raman signaling, the RB-AuNPs showed peaks at 1490 and 1610 cm−1, while unconjugated AuNPs showed peaks at 200 to 400 cm−1, and free RB had little to no Raman signaling. Looking at fluorescent intensity, the RB-AuNP complexes showed the highest fluorescent intensity of the three conditions, demonstrating the advantages of an F-SERS probe AuNP biosensing platform. However, Xia et al. also noted the overall limitations of an F-SERS probe, as the SERS effect is most concentrated within 2 nm. As such, the detection of extremely large biomolecules, such as large proteins or peptides, is limited due to their size. In the case of the Aβ42 peptide, the LOD was 2 μM, which is less specific than other optical biosensing modalities. Therefore F-SERS probes have promising future applications in the detection of small biomolecules, such as small nucleotide miRNAs responsible for misregulated neuronal activity, rather than large proteins with hundreds of nucleotide strands and epitopes.[75b,76] Studies applying SERS in the detection of miRNAs for other diseases, hepatocellular carcinoma, further show the potential of translating similar SERS-tagging techniques for miRNAs in AD.[77]
Certain biomarkers have complex structures or are extremely stable, making the epitopes necessary for detection difficult to access, even with customized fluorescence or F-SERS probe conjugation on AuNPs.[78] This effect is amplified more in biological samples and tissues as there are additional proteins and molecules that may interact with the AD biomarker and prevent accessibility. For instance, in blood plasma, the AD biomarker tau protein has its essential epitopes hidden by hydrogen bonds in the blood plasma solution, as well as hydrophobic bonds between multiple tau proteins or other proteins in blood plasma. Due to these interactions, tau protein is difficult to detect as the epitopes are not easily accessible to the optical biosensor.[79]
The addition of a chaotropic agent alongside the use of the biosensor ensures that the epitopes used for detection are readily accessible. The chaotropic agent is responsible for weakening and inhibiting the interactions that can take place between the tau protein and its environment in a biological sample, thus making the optical biosensing platform more robust for clinical and medical applications.[78,79] Kim et al. tested and analyzed the effects and benefits of chaotropic agents, specifically guanidine hydrochloride (Gua-HCl), in combination with an AuNP optical biosensor for the detection of tau in human blood samples. The optical biosensing platform was created by fabricating AuNRs, conducting PEG treatment to remove non-specific attachment, and then conjugating the AuNRs to antibody immune complexes using functionalization with EDC/NHS. Once human blood samples were collected from patients with AD and healthy controls, 6 M Gua-HCl was used to weaken the interactions of the tau protein and expose its epitopes for detection. The detection of tau protein was analyzed through LSPR shifts. When Gua-HCl was used in combination with the optical biosensor, significantly higher LSPR shifts were detected compared to the use of the biosensor without a chaotropic agent. A significant enhancement and improvement in LOD were shown, reducing to 0.1 pM (or 100 fM) of tau protein, compared to 1 pM without the chaotropic agent. This work showed that utilizing a chaotropic agent alongside an optical biosensor enhanced the detection of AD biomarkers in human biological samples, creating a highly specific detection platform to distinguish AD human blood samples from healthy controls.[79]
While many advantages exist in the use of customized fluorescent probes, F-SERS probes, or chaotropic agents, these optical biosensing modalities are still limited in their specificity, especially as there is limited capability to distinguish the multiple stages of this complex disease process, such as the formation of senile plaques in the AD brain pathology.[80] Specifically, the ability to accurately distinguish the various stages and concentrations of Aβ in plaque formation, such as monomers (AβMs), oligomers (AβOs), and fibrils (AβFs), requires significant improvement.[81] This is because probes, being either peptide or aptamer-based, can interact with their environment in unexpected ways due to other elements and processes that take place in CSF, blood, and tissue. For instance, probes can potentially exhibit inhibitory effects or interference effects, as well as overlap with tissue autofluorescence.[67,81b,82] These potentially unwanted effects can, in turn, alter the optical fluorescence intensity signals quantified by introducing noise and reducing the strength of the signal measured, leading to lower signal-to-noise ratios and poor quantification ability.[82] As such, novel strategies to address these limitations are beneficial in designing AuNP biosensors that are more specific and accurate to various complex processes that are present in AD pathology.
3.2.3. Quantum Dots in Non-Colorimetric Optical Biosensor Design
A strategy that has gained traction in the literature of AuNP optical biosensing is the conjugation of AuNPs with QDs to address the limitations of fluorescent probes, F-SERS probes, and chaotropic agents.[89,95] QDs can either be incorporated with existing AuNP optical biosensors or can be designed as their own unique biosensing platform.[80,81,83,85] QDs are a feasible and viable option for optical biosensor design targeting Aβ peptides, as they can specifically aid in the detection of AβMs, AβOs, and AβFs.[80,85] QDs possess unique fluorescent and optical properties for the detection of biomarkers involved in complex processes, such as the formation of AβFs and plaques, deposition of these plaques onto neurons in the brain, and misregulation of gene expression in neurons, resulting in disease.[81,86] QDs can be composed of multiple materials, such as carbon or graphene, and can be functionalized and modified with DNA, antibodies, and enzymes.[80,81,83,85] As such, QDs can be designed to be highly specific to various stages of Aβ plaque formation characteristic of AD.
In various stages of plaque formation and Aβ aggregation, AβMs, AβOs, and AβFs all play a crucial role. As such, a study by Xia et al. focused on designing their QD AuNP biosensor to quantify plaque formation, specifically by detecting AβOs, the intermediate stage in the plaque formation process. They utilized the inner filter effect (IFE) of AuNPs on the fluorescence properties of cadmium telluride (CdTe) QDs, by quenching the fluorescence of CdTe QDs and, thus, reducing their fluorescent intensity. Upon specific binding and interactions of AβOs with the peptide PrP (95–110), interference occurs with the AuNPs IFE, allowing the AuNPs to aggregate around CdTe QDs and stop their IFE quenching effects. As such, in the presence of AβOs, the fluorescence of the CdTe QDs is restored and can be detected or quantified through optical analysis. For this AuNP and CdTe QDs optical fluorescence biosensing platform, the LOD is reported as 0.2 nM of AβOs in the solution buffer.[85]
Along with Aβ, QD-based fluorescent biosensors, and assays have been employed for the detection of other AD biomarkers, such as tau protein, without the use of chaotropic agents. In a study by Chen et al., dopamine-functionalized CuInS2/ZnS QDs were utilized for the sensitive detection of tau protein in human serum (see Figure 4D). The proposed redox-mediated fluorescence immunoassay was designed for detection caused by tyrosinase-induced interactions with QDs. The high luminescence CuInS2/ZnS core/shell QDs were modified with dopamine through amide conjugation and used to replace the conventional fluorophore modality. In the presence of functionalized QDs in the sandwich fluorescence immunoassay system, tyrosinase enzyme catalyzed the transformation of dopamine-to-dopamine quinone, serving as an effective electron acceptor and triggering fluorescence quenching. The engineered fluorescence immunoassay has a linear range for tau protein concentration and could detect tau protein at a concentration range of 10 pM to 200 nM with a LOD of 9.3 pM.[83]
Looking at the deposition of plaques in the AD brain, AChE is one of the key biomarkers in this process that can be utilized for the construction of QD-based optical biosensors. AchE’s key role in AD pathology includes facilitating the deposition of peptides, such as Aβ, into plaques that are insoluble in the AD brain, thus building and growing the size of these plaques and worsening AD pathology.[87] In Qian et al.’s study, a fluorometric assay was engineered to detect and monitor AchE with adequate sensitivity for human serum and seminal plasma samples by taking carbon QDs as the signal reporter. In the first step, copper (II) ions interacted and were bound with the carboxyl groups on the carbon QDs, resulting in quenching of their fluorescence. The presence of AchE then served to catalyze the hydrolysis of acetylthiocholine into thiocholine, inducing fluorescence recovery because of a stronger affinity between thiocholine and copper ions. Finally, in the presence of the inhibitor tacrine, AchE lost its catalytic ability for the hydrolysis of acetylthiocholine, and thus the fluorescence remains quenched. As a result, the activity of AchE with a concentration as low as 4.25 U L−1 and a broad linear scope ranging from 14.2 to 121.8 U L−1 was reported, thus demonstrating the ability to detect biomarkers of plaque deposition using QDs.[86a]
As mentioned, in AD pathology, miRNAs, which play a key role in gene expression and regulation in neurons, are misregulated or changed and result in inefficient neuronal activity and synapse function. miRNA misregulation is associated with the characteristic memory loss of AD.[88] As such, miRNAs are alsoa target of interest for optical biosensor applications. The combination of two miRNA biomarkers, namely miR-501-3p and miR-455-3p, can be employed to enhance AD diagnostic accuracy. To this end, Guo et al. designed a dual-signal DNA probe based on the fluorescent recovery of carbon QDs and the sulfo-cyanine5 dye to identify the associated miRNAs simultaneously. By taking advantage of duplex-specific nuclease (DSN) amplification of signals, the dynamic ranges were 0.01 to 4 pM for miR-501-3p and 0.01 to 5 pM for miR-455–3p in human serum, meeting the required sensitivities for clinical applications and relevance.[86b]
Overall, the benefits of QD on their own or in conjugation with AuNPs have been shown in their ability to replicate or enhance LOD for biomarkers in complex processes, such as AβO, tau protein (without the use of chaotropic agents), AChE, and miRNAs. However, the application and fabrication of QDs for biosensing are still relatively novel, and as such, challenges exist that need to be addressed to create scalable optical biosensors using QDs, such as shortening the time of fabrication, reducing chemical resources required, and simplifying the fabrication steps required.[66b] The optimization of QD parameters, geometry, and fabrication processes is a future avenue of research that can be investigated to enhance the applications of QDs in clinical settings.[66b,89]
3.2.4. Metal Oxide Nanoparticles in Non-Colorimetric Optical Biosensor Designs
While AuNPs still stand as the most common and widely researched modality in developing optical biosensors for AD early-onset diagnostics, recent research is beginning to emerge in which other metallic and non-metallic NPs show benefits in their applications for optical biosensing. A metal oxide platform used in developing optical biosensors includes ZnO NPs. ZnO is advantageous in optical biosensor design for its surface area and metal-enhanced fluorescence (MEF), allowing it to have lower LODs than other biosensing modalities.[90] ZnO has the added ability to be customizable, as it can be fabricated in a wider variety of shapes than previous biosensing fabrication techniques using AuNPs, depending on their application. These enhanced NP shapes and structures include nanowires, nanobelts, nanotubes, and nanoflowers.[90a]
Like AuNPs, ZnO can also be conjugated with fluorophores, labels, or dyes to increase fluorescent output in the presence of biomarkers for in vitro, ex vivo, and potential in vivo biosensing.[91] Lee et al., for example, conjugated nano-porous ZnO-NPs with a multivalent peptide probe, specifically, polyva-lent directed peptide polymer (PDPP). This nano-porous ZnO was chosen for its permeability as well as its high surface area, leading to enhanced target binding and detection, as seen by enhanced fluorescence. In mouse and human CSF samples, as well as mouse brain tissue, Lee et al. showed the ability to detect Aβ42 at lower LODs than current clinical standards. As such, this biosensing platform utilizing ZnO-NPs is very promising for clinical applications. In fact, the PDPP-conjugated nano-porous ZnO-NP biosensor was able to detect Aβ42 at 12 ag mL−1 in human CSF samples, whereas clinical practices have a typical LOD of 100 pg mL−1.[91] Thus, the ZnO-NPs biosensor created here was not only viable for AD biomarker detection but also enhanced the ability to detect biomarkers compared to current standard techniques.
3.2.5. Non-Metallic Nanoparticles in Non-Colorimetric Optical Biosensor Designs
Non-metal-based NPs are also commonly used in literature and are diverse, promising avenues for optical biosensing to detect AD biomarkers. These include graphene oxide, Prussian blue NPs (PBNPs), carbon nanostructures, cyclic peptide NPs (c-PNPs), and SPR fibers.[92]
The benefits of using graphene oxide in optical biosensing include their higher characteristic affinity for target biomarkers, as well as their high solubility and large surface area, leading to more enhanced and efficient detection. For example, Vilela et al. utilized graphene oxide when developing an optical biosensor to detect AD-associated mRNA, BACE-1. This study created graphene oxide up-conversion NPs (UCNPs) (see Figure 4E). These graphene oxide UCNPs were shown to detect lower concentrations of biomarkers, as they required a minimum of 2 low-energy photons to become active for biomarker detection. In other words, the excitability of the graphene oxide UCNPs was exceptionally low, making them more efficient for biomarker detection. Graphene UCNPs in this biosensor were able to detect the short polyA sequences of BACE-1 mRNA at a LOD of 500 fM in blood plasma, with a range spanning the femto- and pico-scales, thus showing more specific detection compared to AuNPs.[84]
Furthermore, graphene oxide is advantageous for clinical use as it can be easily conjugated with other nanoparticle types, such as metallic and magnetic NPs. It can also be integrated with fluorescent dyes and probes and can leverage the SERS phenomenon. Yu et al., for instance, developed a SERS-based biosensing platform in which graphene oxide magnetic (Fe3O4@GOs) NPs were conjugated alongside tannin-capped AgNPs, achieving LODs of 1.62 fg mL−1 of Aβ1–42 and 5.74 fg mL−1 of tau protein, specifically P-tau-181. SERS spectra peaks were also observed at 1585 cm−1 and 1076 cm−1. In their study, not only were they able to determine the concentration of AD biomarkers in patient serum samples, but they also distinguished individuals with AD from individuals with non-AD dementia and healthy controls. By assessing a pool of 63 patient serum samples using an AUC-ROC analysis, tau protein was shown to have a higher AUC of 0.770 compared to Aβ, with an AUC of 0.383. As such, the detection of P-tau-181 protein was determined to be a more sensitive biomarker for AD, and can differentially diagnose dementia that originates from AD.[93]
Next, PBNPs are used in biosensing applications for their advantageous qualities, including biocompatibility and ease of preparation clinically. Chen et al. utilized PBNPs in their biosensing model for the detection of AβOs in human CSF samples. Using PBNPs, conjugated with carboxyl fluorescein (FAM) modified Aβ40O-targeting aptamer (FAM-AptAβ), showed increased stability in the overall biosensing complex, giving rise to increased fluorescence intensity and enhanced detection ability. In fact, the range of detection of this PBNP-based biosensor is on the nanoscale in human CSF, from 1 nM to 100 nM.[92c]
Looking at carbon structures, the main benefits of the detection of AD biomarkers include their versatility in structure, shape, and design. As carbon structures are fabricated in a layer-by-layer method, this allows for diversity in both shape and size. For instance, Lisi et al. created multi-walled carbon nanotubes (MWCNTs). This MWCNT biosensing assay for tau protein used enhancement of SPR to increase detection ability based on MWCNT conjugation with antibodies. The LODs were 7.8 nM in the solution buffer and 15.0 nM in artificial CSF when applied directly to the solution. In the context of a sandwich fluorescence immunoassay, the LOD for artificial CSF dropped to 2 nM, demonstrating the ability to change the limit based on different applications. However, limits exist in using MWCNTs for clinical applications, as it is still a novel technique in AD biomarker literature and requires more investigation to reach clinically relevant limits.[92b]
Not only can carbon be used to form an optical biosensing platform for AD, but peptides can also be leveraged. In fact, Sun et al. were able to utilize cyclic peptides in a negatively charged c-PNP model to detect AβF and AβO through negative charge interactions. The detection of Aβ was possible in human samples, such as human serum, at LODs of 15 μg mL−1 for AβF and 10 μg mL−1 for AβO. Human serum samples were collected from 24 subjects at various stages of the AD prognosis cycle (healthy, early-onset, late-stage). Fluorescence intensity imaging and analysis were used to distinguish between healthy, early-onset, and late-stage AD samples from subjects, where fluorescence intensity decreased 4-fold between healthy and early-onset AD samples, as well as decreasing 4-fold again between early-onset and late-stage AD samples. This illustrates that the c-PNP platform can distinguish between healthy individuals and different severities of AD in its clinical diagnosis.[92d]
Finally, SPR fibers, made from silica, can be used in biosensing platforms that utilize optic properties and may be promising avenues for point-of-care diagnosis of AD in clinical settings. SPR fibers effectively leverage SPR optical phenomena and do not require a label or dye. Nu et al. demonstrated the ability of an SPR fiber-based biosensor to detect tau protein in a population of 40 subjects, half of whom were clinically diagnosed with AD, and the other half were healthy controls. In the fabrication process, the multimode SPR fibers, composed of silica cores, were coated in Au film and mounted on polydimethylsiloxane (PDMS) ring-shaped flow cells. Human serum samples were collected from all subjects and the biosensor was used to detect both T-tau protein and p-tau protein. The LODs were 2.4 pg mL−1 and 1.6 pg mL−1 for T-tau protein and p-tau protein, respectively. Not only did this platform successfully differentiate between healthy controls and AD patients, but it was also able to show a 6-fold increase in T-tau protein and a 3-fold increase in p-tau protein amongst the AD samples relative to the healthy controls.[92a]
Through the studies shown and summarized in Table 2, both AuNP and other NP biosensing platforms have advantages when it comes to the detection of AD biomarkers for early diagnosis of AD. However, limitations in the use of non-colorimetric optical biosensors in clinical applications still arise, mainly in the measurement of fluorescent and optical signals. In non-colorimetric optical biosensing, the use of a microscope or measurement device is required to detect changes in biomarker concentration and biological processes, and as such, makes it challenging to integrate biosensing platforms in a quick, easy-to-interpret diagnostic tool. As such, considering other types of biosensing modalities can aid in addressing the limitations of optical biosensing, by reducing or eliminating the need for microscopes or measurement devices.[64a]
Table 2.
Optical biosensing platforms utilizing nanoparticles in their fabrication.
| Biosensor | AD Biomarker | Sample Used | Limit of Detection | Fabrication Method | Ref. |
|---|---|---|---|---|---|
| LSPR–based AuNPs | Aβ42 | Human CSF | 1 pg mL−1 | AuNP films coated on polyethylene terephthalate substrate | [65] |
| AuNSs and AuNCs | Aβ40 fibrils | Solution buffer | 20 μM | Traditional seed-method, functionalized with C12C6C12Br2, combined with ThT in solution buffer | [69] |
| Shape-code nanoplasmonic AuNPs | Aβ40, Aβ42, tau protein (multiplex detection) | Mimicked blood | Aβ40 at 34.9 fM Aβ42 at 26 fM tau protein at 23.6 fM | Spherical and rod-shaped (long and short) AuNPs are fabricated, functionalized with PEG, and immobilized on a glass slide | [68] |
| AuNRs conjugated with CRANAD-2 AuNPs conjugated with double-stranded DNA aptamers | AβFs Exosome miRNAs | Mouse brain tissue Human serum | 0.001 nM 3.37–4.01 aM | AuNRs modified with PEG spacers (HS-PEG-OMe and HS-PEG-COOH), functionalized with the D1 peptide AuNPs conjugated with double-stranded DNA aptamers, facilitating the formation of a programmable curved nanoarchitecture | [71] [74] |
| AuNPs conjugated with F-SERS probe | Aβ42 | Mouse brain tissue | 2 μM | Bifunctional AuNPs conjugated with Rose Bengal | [77] |
| AuNPs conjugated with Gua-HCl chaotropic agent | tau protein | Human blood | 0.1 pM (100fM) | PEG treatment, AuNRs to conjugated antibody immune complexes, functionalized with EDC/NHS. | [79] |
| AuNPs quenched by CdTe QDs | AβOs | Solution buffer | 0.2 nM | IFE utilized on CdTe QDs synthesized and combined with AuNPs | [85] |
| CuInS2/ZnS QDs | tau protein | Human serum | 9.3 pM | QDs functionalized with dopamine are structured in a redox-mediated fluorescence immunoassay | [83] |
| Carbon QDs | AChE | Human serum | 4.25 U L−1 | Copper (II) ions interact with carboxyl groups on carbon QDs, resulting in fluorescence quenching | [86a] |
| Carbon QDs | miR-501-3p and miR-455-3p | Human serum | 0.01 – 4 pM miR-501-3p, 0.01 – 5 pM miR-455-3p | Dual-signal DNA probe based on the fluorescent recovery of carbon QDs and sulfo-cyanine5 dye | [86b] |
| Graphene oxide UCNPs Graphene oxide magnetic NPs | BACE-1 Aβ1–42, P-tau-181 | Blood plasmaHuman Serum | 500 fM Aβ1–42 at 1.62 fg mL−1, P-tau-181 at 5.74 fg mL−1 | UCNPs functionalized with EDC/NHS and conjugated with target-specific oligomers Graphene oxide magnetic NPs (Fe3O4@GOs) conjugated with tannin-coated AgNPs in a SERS-based probe | [84] [93] |
| ZnO-NPs | Aβ42 | Human and mouse CSF, Mouse brain tissue | 12 ag mL−1 (Human CSF) | Nanoporous ZnO-NPs conjugated with PDPP | [91] |
| PBNPs | Aβ40O | Human CSF | 1 nM - 100 nM | PBNPs conjugated with FAM and modified with Aβ40O-targeting aptamer (FAM-AptAβ) | [92c] |
| MWCNT | tau protein | Solution buffer, artificial CSF | 7.8 nM in solution buffer, 15.0 nM in artificial CSF | Carbon structures synthesized and fabricated in layer-by-layer method, creating walls of CNTs | [92b] |
| Negatively charged c-PNP | AβF, AβO | Human serum | 15 μg mL−1 for AβF, 10 μg mL−1 for AβO | c-PNP synthesized using solid-phase peptide synthesis method | [92d] |
| SPR fiber-based | T-tau protein, p-tau protein | Human serum | 2.4 pg mL−1 for T-tau protein 1.6 pg mL−1 for p-tau protein | Multimode fiber coated in Au film and mounted on PDMS ring-shaped flow cell | [92a] |
3.3. Colorimetric Biosensor Designs for the Diagnosis of AD
Colorimetric biosensors detect biomarkers through physical and chemical interactions, but unlike optical biosensors, these interactions result in a visible color change or rapid fading of color.[111–113] Once the LOD is met, the color change or rapid fading occurs and is easily detectable with the naked eye, requiring no equipment.[94a,b,95] The reduced need for equipment makes colorimetric biosensors a promising avenue for AD diagnostics, as there is a corresponding reduced cost in their clinical applications compared to their electrochemical or optical counterparts.[94a,c] For instance, colorimetric biosensors in clinical practice can be manufactured as lateral flow or microfluidic devices, which are portable and can be easily integrated into current AD diagnostic practices.[94b,96] Their versatility and customizability to varying targets based on different color changes also make them candidates for designing multiplex detection or integrated biosensing devices.[68]
The color change characteristic of colorimetric biosensors is caused by a significant increase or shift in spectral wavelength peak upon exposure to the target biomarker. These increases or shifts are caused by significant chemical or physical changes, such as induced particle aggregation or enlargement for sensors utilizing NPs, altered chemical configuration, enzymatic activity, or induced interactions with molecules, ions, or proteins.[94b,97] In the initial development and testing stages, specialized quantification techniques and measuring tools detect the minimal amount of change, or threshold, needed to induce a color change, to further understand the biochemical mechanisms leading to these visual changes. For instance, if exposure to an AD biomarker causes NP enlargement, dynamic light scattering (DLS) can detect these subtle changes in particle size and size distribution, allowing for the LOD of the biomarker to be determined relative to particle size when testing novel colorimetric biosensors that have the potential for AD diagnostics.[98] Colorimetric biosensors are versatile as the color change is determined by the nanoscale features utilized as well as how these features are structured and functionalized.[68,94b,c,97]
3.3.1. Gold Nanoparticles in the Design of Colorimetric Biosensors
Just like optical biosensing, the use of AuNPs for biosensing applications is widespread in the field of colorimetric biosensor fabrication. In the context of colorimetric biosensing, the ability to incorporate AuNPs in typical detection assays, such as sandwich ELISA, is an essential component to consider when fabricating colorimetric biosensors.[94a,98] Furthermore, the use of AuNPs enhances the performance and clinical applicability of typical detection assays by tuning them to specialized target biomolecules for AD diagnostics, allowing for the robust use of colorimetric biosensors and bioassays in clinical practice.[98]
To demonstrate the versatility of AuNPs in typical assays, Hu et al. developed a colorimetric ELISA assay modified by conjugating antibodies to AuNPs. In the fabrication of the modified ELISA, both C and N-terminal antibodies were immobilized on AuNPs, the AuNPs were functionalized with bovine serum albumin (BSA), and interactions between the antibody-conjugated AuNPs and target Aβ42 induced color change. The cause of the color change was determined to be a combination of physical changes to the AuNPs, including enlargement where diameters increased from 15.4 nm to 32.6 nm, as well as enhanced aggregation upon exposure to Aβ42. In the solution buffer, this colorimetric modified ELISA had a LOD of 2.3 nM Aβ42, which induced a color change from red to blue. While this is not as specific as the standard ELISA that demonstrates a LOD in the picomolar range, the colorimetric biosensor could detect the presence of Aβ42 clearly and visually in a way that is quick and rapid, demonstrating the utility of the colorimetric biosensing modalities.[98,99]
When designing colorimetric biosensors for AD diagnostics, certain factors or components can be adjusted and optimized for a specific target biomarker. In AuNP colorimetric biosensors, like optical biosensor counterparts, the size, shape, and geometric parameters of the AuNPs can be optimized for certain biomarkers as these factors determine the unique wavelength scattering that the NP experiences. In the case of detecting Aβ, for example, different forms of Aβ exist depending on the stage of fibrillation they are in, where AβM is the smallest form of Aβ, followed by AβO, and AβF being the largest. AuNP size could be altered and adjusted to detect Aβ at multiple stages in fibrillation, where smaller AuNPs would facilitate AβM interaction and detection while large AuNPs would be more effective if AβF was the target biomarker. As such, the ability to alter size is beneficial to customize the colorimetric biosensor to the desired Aβ form, making it suitable for detecting and monitoring Aβ fibrillation.[100]
Alongside size, the shape of the AuNPs can be designed for target biomarkers and express unique colorimetric properties for detection. By combining multiple shapes on a single colorimetric biosensing platform, the ability to incorporate multiplex detection in biosensor design is possible. Multiplexed detection is advantageous in colorimetric biosensing as each AD biomarker will express and show a different color when in clinical use, making the detection of multiple biomarkers easily observable by simply determining which color is present. For instance, when fabricating their optical shape-code AuNP biosensor using AuNPs functionalized with PEG and immobilized on glass substrates, Kim et al. examined the option of incorporating colorimetric biosensing modalities by assessing the difference between the color spectrum of spherical particles and rod-shaped particles. It was determined that spherical AuNPs had wavelength properties in the green spectrum, with a spherical diameter of 50 nm having an olive-green hue, specifically. Rod-shaped AuNPs, on the other hand, possessed traits on the lower wavelengths of the color spectrum, depending on the length of the rod. As the length of the rod decreased, the wavelength increased. In other words, shorter rods appeared as orange, while longer rods were red.[68] After optimization of AuNPs size, shape, and colorimetric characteristics, the shape-code biosensor was utilized in the multiplex detection of Aβ40, Aβ42, and tau protein, at respective LODs of 34.9 fM, Aβ42 at 26 fM, and tau protein at 23.6 fM, through both the application of optical and colorimetric biosensing. As such, through their investigations on color spectrums of AuNPs optimized for size and shape, Kim et al. investigated and showed the simultaneous use of optical and colorimetric modalities for multiplex detection of AD biomarkers.[68]
In colorimetric biosensing platforms, there is still an emphasis on utilizing bioassays, and as such, investigating probes for conjugation with AuNPs is essential to enhancing detection ability. For example, probes such as aptamers, or short synthesized DNA strands, are exceptional in enhancing the performance of AuNPs as part of a colorimetric bioassay. This is because aptamers can be designed for high-level targeting of the specific AD biomarker of interest, which is difficult to achieve through optimizing AuNP geometric parameters alone, thus, enhancing the selectivity of the overall colorimetric biosensing assay. A colorimetric biosensor produced by fabricating AuNPs with specialized aptamers is referred to as a colorimetric aptasensor.[94a] Zhu et al. focused on demonstrating the benefits of utilizing aptamers in conjugation with AuNPs for colorimetric biosensing of AβO. Aptamers designed to be selective to AβO formed AβO-aptamer complexes, which alongside AuNPs in NaCl solution created a specialized AβO colorimetric aptasensor. The NaCl solution aided in stabilizing the AβO-aptamer complexes conjugated with AuNPs by salt-induced AuNP aggregation. The LOD of this colorimetric aptasensor was 0.56 nM in artificial CSF, resulting in a hypsochromic shift from a longer absorption wavelength to a shorter one, leading to a color change from deep red to blue or purple.[94a] Comparing the results of aptamer-conjugated AuNPs with unconjugated AuNPs, the LODs are lower and more specific for aptamer-conjugated AuNPs.[94a,98] As such, when considering the design of AuNP-based colorimetric biosensors for clinical applications and point-of-care use, the increased sensitivity of the colorimetric aptasensor with maintained functionality, is more beneficial in determining the presence of AD biomarkers in patient samples than the use of AuNPs alone.
In addition to the modification of the geometric parameters of AuNPs, the medium in which AuNPs are suspended can also be fine-tuned to induce further color changes.[94a] Specifically, metals or salts in solution can increase or enhance the aggregation kinetics of AuNPs, making them more sensitive to the presence of target AD biomarkers and leading to more rapid and enhanced color change. In terms of experimental prototyping and bioassay construction, adjusting the surrounding medium is beneficial as it reduces the steps required in the fabrication process.[94a,c] Zhu et al., after assessing aptamer-conjugation, demonstrated the effects of fine-tuning AuNP performance and induced color change using salt ions (Na+) in artificial CSF. To optimize the conditions of the artificial CSF medium, various concentrations, and pH levels were tested, and the ideal conditions were determined to be 50 mM of Na+, resulting in a 5.0 pH solution. At these optimized Na+ concentration and pH levels, the LOD for detecting AβO was 0.56 nM and a purple hypsochromic shift was observed, as previously mentioned.[94a]
A current limitation in AuNP-based colorimetric biosensors is the reliance on existing conventional bioassays, such as double-antibody sandwich immunoassays or ELISAs, which require multiple fabrication steps and complex conjugation of probes, antibodies, and dyes.[96d] To make colorimetric biosensors suitable for scalable point-of-care device manufacturing and clinical use, the number of fabrication steps needs to be reduced and simplified to make the manufacturing of colorimetric biosensors streamlined and efficient. A novel strategy to reduce or eliminate the need for existing bioassay structures is by combining colorimetric principles with the optical application of the SERS effect using Raman dyes, creating a dual-modal biosensor that does not require the use of a bioassay. Zhang et al. aimed to introduce the SERS effect, while also leveraging the benefits of a colorimetric aptasensor model, for the multiplex detection of Aβ42 oligomers (Aβ42O) and tau protein. Initially, poly A aptamers were conjugated onto the AuNPs to induce stabilization and selectivity between AuNPs, either for Aβ42O or tau protein. Upon poly A aptasensor conjugation, increased aggregation and AuNP enlargement from 53 nm to 69 nm were observed, which led to color changes from red to blue. Then, to show the advantages of SERS in the colorimetric aptasensor, the Raman dyes 4-acetamidothiophenol (AATP) and 5,5′-dithio-bis (2-nitrobenzoic acid) (DTNB) were added for enhanced specificity of Aβ42O and tau protein, respectively. This demonstrated that SERS-based colorimetric aptasensors are highly accurate in reaching clinically relevant LODs of 3.7 × 10−2 nM for Aβ42O and 4.2 × 10−4 pM for tau protein. To investigate the ability of the colorimetric aptasensor to simultaneously detect Aβ42O and tau protein amongst other proteins in artificial CSF, Zhang et al. measured and quantified the Raman spectral peaks. In artificial CSF containing multiple proteins and peptides, specific Raman peaks were still detected at 1073 cm−1 for Aβ42O and 1327 cm−1 tau protein, as shown in Figure 5A. As such, the SERS-based colorimetric aptasensor demonstrated not only the ability to detect Aβ42O and tau protein at clinically relevant LODs but also the ability to distinguish Aβ42O and tau protein from other proteins in artificial CSF, thus making a colorimetric biosensor suitable for clinical use. By utilizing Raman dyes, the number of fabrication steps for the colorimetric aptasensor was reduced (Figure 5A), as there was a reduced need to conjugate additional antibodies and fluorescent dyes like in a typical bioassay, making the fabrication of SERS-based biosensors more scalable. However, new challenges arise in the development stages of SERS-based colorimetric aptasensors as the specialized Raman dyes require additional measurement and quantification steps to determine and analyze the Raman spectral peaks, for the calibration of the aptasensor before scaling and implementation. As a result, by incorporating SERS and Raman dyes, the development stages are prolonged before the SERS-based colorimetric aptasensor is ready to be scaled for manufacturing and clinical applications.[101] Evidently, when designing point-of-care AD diagnostic devices using colorimetric biosensor techniques, there is a trade-off between increasing manufacturing scalability for long-term benefits and clinical use, at the expense of prolonging the necessary development and initial testing stages of the colorimetric biosensor to ensure accuracy and functionality. Therefore, the ability to design a device that can rapidly detect AD biomarkers clinically, while also being easy to develop and manufacture, is important to consider for future research that is conducted on colorimetric biosensing for AD diagnostics.
Figure 5.
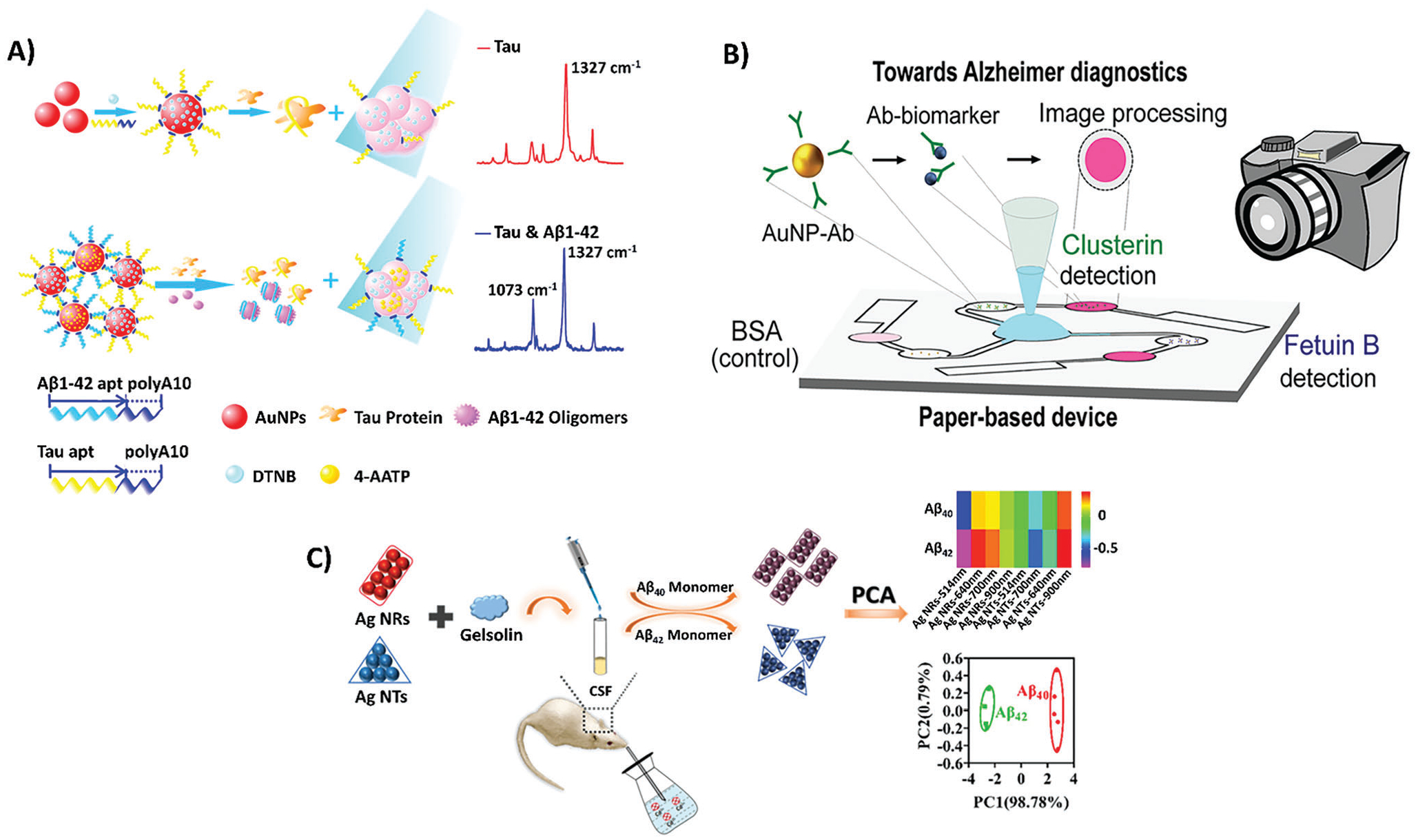
A schematic of various colorimetric biosensing measurement and fabrication techniques. A) A schematic showing the application of the F-SERS platform in colorimetric biosensing, where Aβ42O and tau protein biomarker-specific aptamers and respective 4-AATP and DTNB Raman dyes are conjugated on AuNPs, resulting in unique Raman peaks of 1073 cm−1 in the presence of Aβ42O and 1327 cm−1 in the presence of tau protein. Reproduced with permission.[101] Copyright 2019, American Chemical Society. The labels on the original figure have been modified to improve readability. B) Image depicting the design of colorimetric multi-channel μPAD lateral flow assay where AuNPs conjugated with biomarker specific antibodies are passed through a paper-based device containing individual microchannels for clusterin and fetuin B, which induce color change from white to pink in simple and easy to use design for clinical use. Reproduced with permission.[96a] Copyright 2019, American Chemical Society. C) A schematic showing the conjugation of AgNTs and AgNRs to gelsolin, followed by in vivo administration of AgNPs in mice induced with AD from exposure to Cd+, in which sampling of CSF for UV–vis spectroscopy absorbance depicts subsequent color changes if Aβ40 and Aβ42 are present. Reproduced with permission.[104] Copyright 2020, American Chemical Society. The labels on the original figure have been modified to improve readability.
Additionally, it is essential that clinical diagnostic biosensors designed for AD diagnostics are viable in detecting extremely low concentrations of target biomarkers. As such, another method to further reduce the LOD of colorimetric biosensors to make them clinically relevant for AD diagnostics and device manufacturing is promising in furthering biosensor platform technologies. For example, rather than utilizing AuNPs on their own, AuNPs can be immobilized on treated surfaces and combined with other NP platforms, such as treated nanobody (Nb) structures as well as titanium dioxide NPs (TiO2 NPs). Ren et al. proposed an Nbs colorimetric biosensor, composed of 3-aminopropyltrimethoxysilane (APTMS) and glutaraldehyde (GA), for the detection of genetic AD risk factor ApoE, which results in amyloid plaque formations. The biosensor was fabricated by taking APTMS-GA complexes and modifying them to produce Nbs. The APTMS-GA Nbs foundation was then assembled and formulated through a layer-by-layer method, and conjugated further with AuNP-coated TiO2 NPs, to create a colorimetric biosensor. After exposure to ApoE, the initial red of the structural APTMS-GA complex faded to a clear or transparent color. Through this novel strategy utilizing APTMS-GA Nbs, a LOD of 0.42 pg mL−1 in human serum was achieved, showing the enhancement of LOD from a nanomolar range to a reduced picomolar range and making it more effective for early-onset AD diagnosis applications.[102]
In the investigation of the manufacturing and clinical applications for colorimetric biosensors, the ability to create a portable point-of-care device for AD is beneficial for non-invasive yet accurate early diagnosis of AD.[96a] These portable point-of-care colorimetric devices are one way of addressing some of the challenges in biosensor design for clinical applications by incorporating easy-to-use device design that can distinguish different biomarkers through color differentiation. While these point-of-care devices can utilize multiple modalities for detection, AuNP-based colorimetric biosensors are the most advantageous as they provide an easy-to-read output of a color change, which is interpretable by the naked eye.[68,85,96a] One example is microfluidic paper-based analytical devices (μPADs), which are microfluidic devices utilizing colorimetric biosensor detection. Leveraging microfluidics, μPADs can also be designed for multiplex detection, as there are multiple channels for biomarker detection.[94b,96a] Brazaca et al., for instance, developed a μPAD specifically for multiplex detection of AD biomarkers in human blood, such as fetuin B and clusterin, at LODs of 0.24 and 0.12 nmol L−1, respectively. In their multi-channel μPAD design, each channel was specific to its own AD biomarker. Once a sample of blood was placed on the absorption pad, the sample was able to disperse through multiple channels in the device, one for each biomarker being detected, plus a control. In each channel, a lateral flow assay was performed, where conjugation occurs between Ab-AuNPs and the biomarkers, and these complexes underwent color change if the biomarker was present in the human blood sample. The detection and color change were then displayed on the test zone if the biomarker was present, while flow moved on to an absorption pad at the end of the lateral flow assay for final absorption and completion of the lateral flow assay. This makes it simple and quick to read the results through observation of whether the color change occurred in the test zone or not. μPAD and portable lateral flow assays are This is promising for point-of-care AD diagnostics, as they demonstrate the feasibility of developing a point-of-care device that is viable in a clinical setting (Figure 5B).[96a] AuNPs are versatile and can be effectively incorporated in colorimetric biosensor design, whether that be in existing bioassay detection models or novel SERS-based techniques.[94a,98] As well, simply by optimizing AuNP geometry or medium of suspension and assessing color spectrums of different shapes, sizes, and geometric parameters, AuNPs have the potential to combine both optical and colorimetric biosensor modalities in one platform for AD detection.[68] Evidently, through the work done on developing portable point-of-care diagnostic devices for AD diagnosis, AuNPs have contributed greatly and are typically used as the basis for colorimetric detection.[96a] However, key limitations exist with AuNPs in colorimetric biosensing, especially regarding their longevity and stability. For a color change to be induced, interactions between AuNPs and target AD biomarkers must elicit a physical or chemical change, most commonly by inducing aggregation or enlargement of AuNPs. These changes take the stable AuNPs and make them unstable, which prevents the color change from lasting longer than a few hours. Over a relatively short period of time, the overall color can fade, making it difficult to quantify the effectiveness of the AuNPs in their detection ability. While probes can aid in temporary stability, they do not address the longevity of the AuNP color shifts. Similarly, in the development and initial testing stages of colorimetric biosensor design, AuNPs require additional and sometimes extensive data analysis and spectroscopy techniques for assessing their functionality before they can be scaled for manufacturing and clinical use. As such, the short timeframe of the induced color change and limitations pose challenges in the development stages of AuNP-based colorimetric biosensors, as the quantification and data analysis would need to be done in the short time frame for the most accurate representation of AD detection data, despite typical data analysis techniques for AuNPs being extensive and lengthy.[101,103]
3.3.2. Silver Nanoparticles in the Design of Colorimetric Biosensors
To address the limitations of AuNPs in colorimetric biosensing, the short lifetime of AuNP color change, and prolonged development processes, recent studies propose novel NP strategies for AD biomarker detection without using AuNPs. These studies instead investigate other metallic NPs, such as AgNPs.[102,104,105]
Of the novel strategies presented, AgNPs show the most promise in detecting AD biomarkers, with high specificity and selectivity in colorimetric biosensing.[105b] AgNPs have a wider range of possible color shifts, becoming more effective and diverse based on distance. More specifically, the colorimetric properties and absorption shifts of AgNPs are enhanced and diversified the closer they are to their target biomarker.[104,105b] As well, AgNPs have superior longevity, based on higher extinction coefficients compared to AuNPs, which allows color changes to last longer during the development stages of biosensor design.[105b] As such, while AgNPs are a more novel approach when compared to AuNPs, AgNPs show promise in outperforming AuNPs in colorimetric biosensing platforms and models.[104,105b] For instance, Hu et al. demonstrated the applicability of Aβ(40/42) antibody-conjugated AgNPs (Ab-AgNPs) in colorimetric biosensing, which were designed to detect Aβ(40/42) in human blood samples. Initially, the Ab-AgNPs possessed absorption wavelengths of 393 nm, a distinctive yellow color. In the presence of Cu+, the Ab-AgNPs experienced enhanced aggregation after binding to the ion, which made colorimetric detection possible. In fact, there was a significant color change from yellow to red, or a jump from 393 nm to 520 nm, once Aβ(40/42) was added. The LOD observed for Aβ(40/42) was 86 pM, where significant color change occurred, making this AgNP-based colorimetric biosensor viable for clinical application. AgNP-based colorimetric biosensors are here proven to be specific and selective for the detection of Aβ(40/42), revealing the viability of using AgNPs for future biosensor development.[105b]
AgNPs show huge benefits and applicability in colorimetric biosensing, as well as viability in the use of clinical biosensors using in vitro human samples.[104,105b] Recent studies also propose the potential use of AgNP colorimetric biosensors for in vivo clinical applications. Specifically, Liu et al. propose the use of AgNP colorimetric biosensors in vivo by investigating and demonstrating the effectiveness in detecting Aβ40 and Aβ42 in CSF and in vivo monitoring of mouse brain AD models. AgNPs can be modified and functionalized with proteins found in CSF and brain tissue, such as the secretory protein gelsolin, making monitoring possible (see Figure 5C). After functionalization with gelsolin, AgNP shapes were optimized, creating nanotriangles (AgNTs) and nanorods (AgNRs). Upon exposure to Aβ40 and Aβ42, both the AgNTs and AgNRs aggregated in the CSF samples and brain tissue. As in vivo applications of AgNPs are relatively novel and most AgNP-based colorimetric biosensors are still in the design and developmental stages of biosensor fabrication, additional signal analysis, mainly UV–vis spectroscopy, was utilized to quantify the changes in absorbance wavelength spectrum. Specifically, UV–vis aimed to measure peaks or spikes in wavelength absorbance that occurred once the AD biomarker was presented, and an observable color change or color fade was seen with the naked eye. This color change or color fade was a result of the subsequent light that was not absorbed. For the AgNTs, rather than a color change, there was a visible fading of color, corresponding to a change in color intensity from dark blue to a light blue or grey tone. While there was no direct color change, the change in color intensity was observable, and a new absorbance peak also occurred at 900 nm for the wavelength spectrum of the AgNTs. Meanwhile, AgNRs experienced a new absorbance peak at 700 nm when AD biomarkers were introduced. As 700 nm, corresponding to red, was absorbed, a subsequent direct color change from red to blue or purple was seen. During initial in vivo testing of the colorimetric biosensor, exposure to Cd+, known to alter brain activity and lead to neurodegeneration, was able to induce AD in an in vivo mouse model. The resulting LODs in CSF were 20 nM for Aβ40 and Aβ42, and colorimetric in vivo monitoring of Aβ40 and Aβ42 in the hippocampus, prefrontal cortex and striatum was able to distinguish early-onset AD induced by Cd+ compared to controls that did not experience induced early-onset AD. As such, this investigation demonstrated not only the benefits of using AgNPs in colorimetric biosensing but also the potential of using AgNPs in in vivo applications, which aids in real-time monitoring and tracking of AD biomarkers in early-onset AD (Figure 5C).[104]
Nonetheless, AgNPs, like AuNPs, are still limited by their LODs. While AgNPs have lower LODs than AuNPs, it is still difficult to achieve LODs that are reliable and accurate for repeatable clinical applications. This is because previous studies have shown the LODs for metallic NPs only reach the nanomolar range, or – high picomolar range and none of the studies previously mentioned were able to achieve a LOD below 10 pg mL−1 or in the femtomolar range unless a combination of optical and colorimetric biosensing was used.[68] In human samples, having a colorimetric biosensor with reduced LOD is important for more accurate and repeatable color changes, which would function even when the biomarker is presented at lower concentrations in human blood, serum, or tissue. Being able to induce color change at lower picomolar or femtomolar levels can ensure early-onset detection of biomarkers, thus informing clinical diagnostic and monitoring of patient health earlier on.[102,105a]
Colorimetric biosensors are diverse and variable when creating biosensing platforms to detect AD biomarkers, as shown in the studies outlined in Table 3. The versatility in design and customization, as well as the easy-to-read color change indicative of biomarker presence, gives colorimetric biosensors an advantage over other biomarker modalities. While AuNPs are a great foundation for colorimetric biosensing in AD, novel strategies to address their limitations are emerging and worth investigating, such as the use of APTMS-GA Nbs and AgNPs.
Table 3.
Colorimetric biosensing platforms utilizing nanoparticles in their fabrication.
| Biosensor | AD Biomarker | Sample Used | Limit of Detection | Fabrication Method | Color Shifts/Properties | Ref |
|---|---|---|---|---|---|---|
| AuNP-based sandwich ELISA immunoassay | Aβ42 | Solution buffer | 2.3 nM | Conjugation with C-/N-terminus of Aβ42 specific antibodies, functionalization with BSA | Red-to-blue | [98] |
| Shape-code nanoplasmonic | Aβ40, Aβ42, tau protein (multiplex detection) | Mimicked blood | Aβ40 at 34.9 fM, Aβ42 at 26 fM, and tau protein at 23.6 fM | Spherical and rod-shaped (long and short) AuNPs are fabricated, functionalized with PEG, and immobilized on a glass slide | Green for spheres, orange for short rods, red for long rods | [68] |
| Colorimetric aptasensor | AβO | Artificial CSF | 0.56 nM | AuNPs conjugated with AβO specific DNA aptamer and suspended in Na+ medium | Deep red to Blue/purple (hypsochromic) | [94a] |
| SERS-based colorimetric aptasensor | Aβ42O, tau protein (multiplex) | Solution buffer, artificial CSF | 3.7 × 10−2 nM for Aβ42O, 4.2 × 10−4 pM for tau protein | AuNPs conjugated with specific DNA aptamer, then modified and conjugated with Raman dyes specific to target biomarkers | Red-to-blue | [101] |
| APTMS-GA Nb complexes | ApoE | Human serum | 0.42 pg mL−1 | APTMS-GA complexes fabricated as Nb, nanostructures are assembled and formulated in a layer-by-layer method with immobilized TiO2 NPs coated in AuNPs on nanostructure | Red to clear (fade in color) | [97] |
| Multi-channel μPAD lateral flow assay | Clusterin, fetuin B | Human blood | 0.12 nmol L−1 for clusterin, 0.24 nmol L−1 for fetuin B | μPAD designed with multi-channel, each containing AuNPs conjugated to different antibodies specific to a certain biomarker | White to pink | [96a] |
| AgNP-based immunoassay | Aβ(40/42) | Human serum | 86 pM | Conjugation with C-terminus Aβ(40/42) specific antibodies | Yellow to red | [105b] |
| AgNPs aggregation-based colorimetric sensor array | Aβ40, Aβ42 (multiplex detection) | Mouse brain tissue (in vivo) and CSF | 20 nM | AgNPs in triangle and rod shapes fabricated and modified with gelsolin | Color fade for triangles, red to blue/purple for rods | [104] |
3.4. Nanoparticles as Contrast Agent Carriers for AD Diagnostic Applications
Due to its noninvasive nature and high spatial resolution, MRI is one of the most common techniques used to diagnose AD. There are two types of contrast agents (CAs) typically used for this modality. First, positive CAs that enhance the MR signal intensity, such as gadolinium-based compounds. Second, negative Cas, such as MNPs, decrease the MR signal intensity of the targeted regions, making the targeted regions appear darker in the image.[106] In the advanced stages of AD, the accumulation of iron ions in the plaque tissue is relatively high; therefore, the Aβ plaques can be recognized directly through MRI without any CAs.[107] However, only plaques with diameters above 50 μm using very long acquisition times (≈7 h) and very high magnetic fields (>7T) can be detected this way. In addition, the iron accumulation in Aβ plaques is not homogenous in all brain regions.[108] Moreover, due to the ionic Ga3+ compound, the toxic effect of gadolinium-based CAs is significant.[107] As an alternative to these methods, we turn to the second type of CA mentioned, negative CAs. The capability of using MRI for early detection of AD could be enhanced by employing NPs, in particular MNPs, as negative CAs. Functionalized MNPs have great potential for use as CAs in MRI, given their ability to cross the BBB, enhancement in selectivity and target-specific binding to disease hallmark, non-toxicity, and proper clearing from the body.[109] The nanocrystal structure of MNPs with thousands of paramagnetic centers leads to higher sensitivity, which provides imaging at lower concentrations than Gd3+ chelates, creating a greater relaxation enhancement per entity.[107] Conjugate material can also be added to the surface of CA in order to mediate specific binding to Aβ plaques and enhance plaque detection. Zeng et al. developed mixed ferrite MNPs (MnZnFe2O4), functionalized with Pittsburgh compound B to bind specifically to Aβ plaques.[110] The engineered 100 nm MNPs demonstrate excellent negative CA and demonstrate success in vitro binding to Aβ plaques without toxicity.[110] In another in vivo study, curcumin-functionalized superparamagnetic iron oxide nanoparticles (SPIONs) coated with PEG-polylactic acid were used to target Aβ plaques for visualization with MRI. Curcumin improves the targeting efficiency of MNPs, and coating agents enhance the bypass of BBB, as well as MNPs circulation half-life.[111] The coating agent used on MNPs plays a significant role in CA and enhances the MRI contrast. As another instance, core–shell MNPs made using SPIONs as the magnetic core, coated with three different inorganic or organic modified silica-based sol-gels, were investigated in Zebrafish (Danio rerio) animal model. The effect of core–shell MNPs was quantified in a set of MRIs of agar phantoms obtained at a 7 T magnetic field and with an imaging gradient field of 1.6 T m−1. As a result, the efficiency of negative CAs for MRI was strongly dependent on the SPIONs coating.[112] For MRI, negative CA efficiency is quantified using the ratio of transverse (r2) and longitudinal (r1) relaxivities, r2/r1, with efficient negative CAs having high ratios. In this study, the three types of SPIONs produced r2/r1 values of 6767, 863, and 554, significantly outperforming the commercial CA with an r2/r1 value of 82.7 under the same conditions.
MNPs can also be functionalized by different antibodies for AD detection. For instance, Fernandez and co-workers designed a novel functionalized MNP-conjugated anti-ferritin antibody, which after intravenous injection in vivo, could recognize and bind specifically to the ferritin protein accumulated in areas with high deposition of Aβ plaques.[109c] In another in vivo study, a new CA based on a nanoconjugate composed of MNPs bound to an anti-cholesterol antibody was used to detect abnormal cholesterol deposits in senile plaques. After intravenous administration, the senile plaques and vascular Aβ deposits were successfully detected in vivo by MRI.[113] In favor of improving lesion detectability in specific pathologies and early diagnosis, molecular imaging of Aβ could employ MNPs. Vectorized CA binds to the specific Aβ, which significantly increases sensitivity and specificity. In an interesting in vivo study, the vectorized ultra-small iron oxide NPs functionalized with different peptides were utilized to target Aβ plaques.[114] The authors demonstrate an affinity of the engineered MNPs for Aβ plaques, which enhanced the labeling potential of the CA. Moreover, the introduced non-toxic CAs can be cleared from the body, and the elimination half-life is ≈3 h. They can cross BBB without any facilitating strategy.[114] To track the senile plaques, Zhou et al. engineered SPIONs as a molecular imaging nanoprobe, functionalized with a small hydrophobic lipophilic fluorescent probe (1,1-dicyano-2-[6-(dimethylamino) naphthalene-2-yl] propene carboxyl derivative) to mark the senile plaques and identify the plaque regions.[109a] The in vitro experiment shows that the proposed MNPs can cross BBB and demonstrate a great candidate for MRI CA, which enables senile plaque tracking.[109a]
Due to their unique optical features, QDs have recently been used as promising fluorescence and MRI CAs for AD detection. Quan et al. developed a novel QD-based CA that provides strong red-emitting fluorescence, multivalence binding, decreased background signal, and nonspecific binding to detect Aβ in artificial CSF, where the signal was four times higher than conventional ThT derivative dyes.[115] The engineered nanoprobes contain a red-emitting fluorescent QD core coated with a PEG shell having benzotriazole targeting molecules on the surface. These demonstrated highly efficient results for AD diagnosis, particularly in samples with a very low concentration of analytes, which are not detectable using the conventional ThT-based fluorescence.[115] Yousaf et al. developed graphene QDs functionalized with bovine-serum-albumin-capped fluorine as a multimodal fluorescence CA and MRI-based probe, which detects AβMs/AβOs through in vivo labeled Aβ in the brains of mouse models for AD and enable real-time monitoring of the dynamics of AβMs into amyloid fibrillation.[116] The suggested QD-based CA demonstrates higher sensitivity and affinity to bind with the AβMs and crosses the BBB, leading to the detection of Aβ plaques in vivo by MRI imaging with higher contrast, compared with conventional dye ThT.[116]
Despite these recent advances, it is essential to develop novel engineered MNPs and QDs as CAs that can efficiently detect other hallmarks associated with AD in the early stages of the disease. Moreover, despite the high accuracy and sensitivity, the disadvantages of MRI compared with bioassays and biosensors are the interpretation, complexity, time-consuming, and high-cost instruments.
4. Conclusion
Currently, the ability to diagnose AD mainly depends on the presentation and documentation of patient symptoms, such as mental decline and cognitive impairment. Targeting AD at early stages before irreversible brain damage has occurred could significantly help physicians to slow the progression of the disease and mitigate the symptoms. To this end, there have been tremendous efforts and remarkable advances in early AD detection through its associated biomarkers found in blood or saliva. While many of these novel approaches have demonstrated good sensitivity and specificity, including the recent work by Gonzalez-Ortiz et al. to establish a new blood-based tau biomarker,[28] establishing a reliable and cost-effective biosensing system with good precision and accuracy has remained a challenge. Among AD-specific biomarkers, Aβ and tau have been widely utilized as promising biomarkers in different electrochemical, non-colorimetric optical, as well as colorimetric biosensing applications. Recently, other biomarker candidates, such as AD-associated miRNAs and exosomes have also shown great potential in highly sensitive detection of AD.
Although the exercise of these biomarkers can greatly enhance the specificity of AD diagnostic sensors, the sensitivity and LOD of such devices are not satisfactory in many cases. The use of NPs as nano-biomaterials in different optical and electrochemical platforms has proven to be an effective strategy for increasing the sensitivity and precision of AD diagnosis. This is mainly due to the increased surface area provided by the NPs for bioreceptor immobilization, thereby augmenting the achieved signal in the immunoassays. AuNPs are undoubtedly the most frequently used NPs in such systems, owing to their easy synthesis, tuneability, biofunctionalization, good stability, and biocompatibility, as well as excellent conductivity. Alternatively, AgNPs, have also been used in electrochemical and optical detection techniques. Considering their wider range of possible color shifts as well as higher extinction coefficients, AgNPs could outperform AuNPs in many colorimetric-based AD biosensors. MNPs and other NPs, such as Fe2O3, ferrite composites, and ZnO, have also shown great stability, biocompatibility, and strong adsorption, making them a good substrate for AD detection. Nonetheless, the long-term stability and multiplexing of NPs are still problematic for the scale-up of these biosensor platforms. In general, the LODs of metallic NPs are limited to nanomolar or high picomolar ranges. Thus, the utilization of QDs and non-metallic NPs such as graphene oxide UCNPs, carbon-based NPs, cyclic peptides, APTMS-GA, and Prussian blue has attracted more attention in recent studies. QDs made of different materials such as carbon, CuInS2/ZnS, CdSe/ZnS, and curcumin-graphene possess unique optical properties and can be functionalized with various biomolecules such as DNA, antibodies, and enzymes. The use of QDs in AD-related assays could significantly enhance the LOD and help distinguish the state of the disease due to their high specificity. QDs have also been employed for electrode modification in electrochemical sensing. However, the complexity of the fabrication of these materials and the lack of information on their biofunctionality hinder their application for mass-production. Carbon-based NPs, such as graphene oxide, fullerene, and MWCNTs, have also shown high performance in optical biosensing due to their large surface area and high solubility, low excitability, and diversity in both shape and size.
Typically, electrochemical biosensing of AD provides excellent sensitivity and substantially lower LODs. Further, electrochemical biosensors are usually much faster than optical strategies, especially in label-free electrochemical immunosensors eliminating the need for the addition of multiple antibodies and proteins required in sandwich-based assays. The sophistication in fabrication and instrumentation of these techniques, however, sometimes limits their applicability in AD point-of-care diagnostics. Optical sensors for the detection of AD biomarkers, on the other hand, are often more straightforward to produce but lack sufficient precision and clinically relevant LODs. Implementation of SPR, fiber-based SPR, LSPR, SERS, and F-SERS probes could substantially improve the sensitivity and LOD in optical biosensing; however, they require further instrumentation and well-trained technicians. The run-time of fluorescence-based immunoassays is also usually longer than electrochemical sensors, making them not suitable for point-of-care diagnostic applications. Colorimetric detection of AD biomarkers can easily be conducted using the naked eye and usually does not require further equipment, which is very promising for point-of-care diagnosis of AD. Despite ongoing challenges with low LOD in typical colorimetric devices and ongoing difficulties with multiplex detection of different biomarkers in AD, it is evident that this platform presents the most clinically user-friendly option for rapid diagnostic purposes. Indeed, the utilization of μPADs in the colorimetric detection of AD appears to be one of the best strategies to address the simplicity and sensitivity issues.[96a] Continued optimization with combinations of different optical tests, various biomarkers, and NPs made of different materials as well as different sizes and shapes have shown high capacity in boosting the sensitivity, multiplexity, and specificity in the diagnosis of different states of AD disease,[68,85] which promise to pave the way for colorimetric platforms capable of detection at clinically relevant concentrations.
To this end, the functionalization of NPs as nano-biomaterials for the immobilization of bio-entities such as antibodies and aptamers is an important factor determining the sensitivity and robustness of biosensors. In future work, it is suggested that novel covalent immobilization techniques capable of preserving the preferred orientation and compactness of antibodies can be leveraged to prevent steric hindrance and contribute to higher sensitivity, reproducibility, and shelf life of the biosensors.[117] Further, another important parameter in the sensitivity of AD biosensors is blocking the non-specific attachment of biomolecules to the surface. Due to the presence of many proteins, cells, and antibodies in complex biofluids such as blood as well as the high surface-to-volume ratio of NPs, non-specific adsorption of biomolecules to NPs and sensing platforms is very probable. This could decrease electrode sensitivity and hinder the attachment of AD biomarkers to the bioreceptors in electrochemical sensors. Similarly, in optical approaches, non-specific adsorption results in higher background noise and consequently diminished sensitivity. Recently, innovative strategies such as the creation of hierarchical topography on micro- and nanoparticles, as well as rendering the particles omniphobic through lubricant-infused surface technology and other polymer grafting methods, have been shown to inhibit non-specific adsorption, increase the specificity, and lower LODs in blood samples.[118] Together, these techniques could drastically improve the performance of AD biosensors and revolutionize AD diagnosis.
Acknowledgements
S.A.A. and L.N. contributed equally to this work. K.D. was funded by the National Institute of General Medical Sciences of the National Institutes of Health under award number 1 R16GM149496-01, the North Carolina A&T State University KL2 Scholar Award from the National Center for Advancing Translational Sciences, National Institutes of Health, Grant KL2TR002490, and start-up funds from the Joint School of Nanoscience and Nanoengineering, North Carolina A&T State University. This research was undertaken, in part, thanks for Canada Research Chairs Program to T.F.D. T.F.D was funded by Natural Sciences and Engineering Research Council (NSERC) of Canada Discovery Grant and Ontario Early Researcher Award. The content was solely the responsibility of the authors and did not necessarily represent the official views of the NIH.
Biographies

Saqer Al Abdullah is currently a Ph.D. student in Nanoengineering, at the Joint School of Nanoscience and Nanoengineering at North Carolina A&T State University. Previously, Saqer graduated with a B.Sc. in Pharmacy in 2016 from Jordan University of Science and Technology (JUST), Jordan, and an M.Sc. in Drug Design and Discovery in 2019 from the University of Salford, Manchester, UK. His research focuses on designing diagnostic tools and drug delivery systems for multifactorial diseases, such as Alzheimer’s and cancer.

Lubna Najm is an Accelerated Master of Biomedical Engineering (MASc) student at McMaster University. She graduated from the inaugural cohort of the Health, Engineering Science, and Entrepreneurship (HESE) program at McMaster University. Currently, she is leading a research team to develop optimized contact and non-contact bioprinting techniques used in the fabrication of optical biosensors for early-onset diagnostics.

Liane Ladouceur is a Clinical Engineer at Toronto General Hospital, part of the University Health Network. Liane was part of the Didar Lab at McMaster University for 4 years, having completed her MASc in Biomedical Engineering before moving into a Research Associate role for the lab. Her research ranged from fabrication and testing of antimicrobial surfaces to development of novel biosensors. Liane received a B.Sc. from Carleton University in Neuroscience and a BEng from McMaster University in Electrical and Biomedical Engineering. She is passionate about innovation and advancement in the healthcare field.

Farbod Ebrahimi is currently pursuing a Ph.D. degree in Nanoengineering at the Joint School of Nanoscience and Nanoengineering at North Carolina A&T State University. He graduated with an M.Sc. from the University of Duisburg-Essen Nanoengineering. Currently, his research focus includes designing and characterizing functional nanoparticles and modifying existing nanomaterials for diagnostics and delivery regarding multifactorial diseases.

Amid Shakeri received his B.S. degree (2010) and M.Sc. degree (2013) in materials science and engineering from the University of Tehran. He gained his Ph.D. degree in 2020 from McMaster University, Department of Mechanical Engineering. During his Ph.D., he mainly worked on the Development of biofunctional interfaces for different applications such as self-cleaning surfaces, lab-on-a-chip systems, as well as biosensors and diagnostics. Currently, he is a postdoctoral fellow at the University of Toronto. His interests include microfluidics and lab-on-a-chip technologies for biomedical applications and tissue engineering as well as fabrication of smart bio-interfaces for biosensors and point-of-care diagnostic

Nadine Al-Jabouri is a fourth-year B.H.Sc. student in the Biomedical Discovery and Commercialization program at McMaster University. She is interested in exploring how human health intersects with technology and nanobiotechnology. She is currently involved in microfluidics research, working on developing biosensors for clinical diagnostics.

Tohid F. Didar is an Associate Professor in the Department of Mechanical Engineering, School of Biomedical Engineering, and a member of the Institute for Infectious Disease Research (IIDR) at McMaster University. He is also Canada Research Chair (CRC) in Nano-Biomaterials. Before joining McMaster in 2016, Dr. Didar was a postdoctoral fellow at the Wyss Institute for Biologically Inspired Engineering at Harvard University. Dr. Didar’s lab is focused on developing smart nano-biomaterials for applications in diagnostics, therapeutics, and food safety.

Kristen Dellinger is an Assistant Professor in the Department of Nanoengineering, Joint School of Nanoscience and Nanoengineering at North Carolina A&T State University. Before joining A&T in 2019, Dr. Dellinger was a research associate at the University of North Carolina at Greensboro and obtained her Ph.D. in biomedical engineering at McGill University. Dr. Dellinger’s lab is focused on developing functional nanomaterials and devices for molecular bio-sensing with applications in multifactorial diseases and environmental testing.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
The ORCID identification number(s) for the author(s) of this article can be found under https://doi.org/10.1002/adfm.202302673
Contributor Information
Saqer Al Abdullah, Department of Nanoengineering, Joint School of Nanoscience and Nanoengineering, North Carolina A&T State University, 2907 East Gate City Boulevard, Greensboro, NC 27401, USA.
Lubna Najm, School of Biomedical Engineering, McMaster University, 1280 Main Street West, Hamilton, ON L8S 4L8, Canada.
Liane Ladouceur, Department of Mechanical Engineering, McMaster University, 1280 Main Street West, Hamilton, ON L8S 4L7, Canada.
Farbod Ebrahimi, Department of Nanoengineering, Joint School of Nanoscience and Nanoengineering, North Carolina A&T State University, 2907 East Gate City Boulevard, Greensboro, NC 27401, USA.
Amid Shakeri, Department of Mechanical Engineering, McMaster University, 1280 Main Street West, Hamilton, ON L8S 4L7, Canada.
Nadine Al-Jabouri, Department of Biochemistry and Biomedical Sciences, McMaster University, 1280 Main Street West, Hamilton, ON L8S 4K1, Canada.
Tohid F. Didar, School of Biomedical Engineering, McMaster University, 1280 Main Street West, Hamilton, ON L8S 4L8, Canada Department of Mechanical Engineering, McMaster University, 1280 Main Street West, Hamilton, ON L8S 4L7, Canada; Institute for Infectious Disease Research (IIDR), 1280 Main St W, McMaster University, Hamilton, ON L8S 4L8, Canada.
Kristen Dellinger, Department of Nanoengineering, Joint School of Nanoscience and Nanoengineering, North Carolina A&T State University, 2907 East Gate City Boulevard, Greensboro, NC 27401, USA.
References
- [1].Dementia, https://www.who.int/news-room/fact-sheets/detail/dementia, accessed: Febreuary, 2022.
- [2].Kepp KP, Chem. Rev 2012, 112, 5193. [DOI] [PubMed] [Google Scholar]
- [3].Livingston G, Sommerlad A, Orgeta V, Costafreda SG, Huntley J, Ames D, Ballard C, Banerjee S, Burns A, Cohen-Mansfield J, Cooper C, Fox N, Gitlin LN, Howard R, Kales HC, Larson EB, Ritchie K, Rockwood K, Sampson EL, Samus Q, Schneider LS, Selbæk G, Teri L, Mukadam N, Lancet 2017, 390, 2673. [DOI] [PubMed] [Google Scholar]
- [4].Weller J, Budson A, F1000Res 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mantzavinos V, Alexiou A, Curr. Alzheimer Res 2017, 14, 1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Snyder HM, Carrillo MC, Grodstein F, Henriksen K, Jeromin A, Lovestone S, Mielke MM, O’Bryant S, Sarasa M, Sjøgren M, Soares H, Teeling J, Trushina E, Ward M, West T, Bain LJ, Shineman DW, Weiner M, Fillit HM, Alzheimers Dement. 2014, 10, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].a) Naseri NN, Wang H, Guo J, Sharma M, Luo W, Neurosci. Lett 2019, 705, 183; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hajipour MJ, Santoso MR, Rezaee F, Aghaverdi H, Mahmoudi M, Perry G, Trends Biotechnol. 2017, 35, 937; [DOI] [PubMed] [Google Scholar]; c) Munshe F, Khan MA, Nanosci. Nanotechnol. - Asia 2022, 12, 10. [Google Scholar]
- [8].Lloret A, Esteve D, Lloret M-A, Cervera-Ferri A, Lopez B, Nepomuceno M, Monllor P, Int. J. Mol. Sci 2019, 20, 5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Laske C, Sohrabi HR, Frost SM, López-de-Ipiña K, Garrard P, Buscema M, Dauwels J, Soekadar SR, Mueller S, Linnemann C, Bridenbaugh SA, Kanagasingam Y, Martins RN, O’Bryant SE, Alzheimers Dement. 2015, 11, 561. [DOI] [PubMed] [Google Scholar]
- [10].Delenclos M, Jones DR, McLean PJ, Uitti RJ, Parkinsonism Relat. Disord 2016, 22, S106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Murphy MP, LeVine H 3rd, J Alzheimers Dis 2010, 19, 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Karran E, Mercken M, Strooper BD, Nat. Rev. Drug Discovery 2011, 10, 698. [DOI] [PubMed] [Google Scholar]
- [13].Selkoe DJ, Hardy J, EMBO Mol. Med 2016, 8, 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cheignon C, Tomas M, Bonnefont-Rousselot D, Faller P, Hureau C, Collin F, Redox Biol. 2018, 14, 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Motter R, Vigo-Pelfrey C, Kholodenko D, Barbour R, Johnson-Wood K, Galasko D, Chang L, Miller B, Clark C, Green R, Olson D, Southwick P, Wolfert R, Munroe B, Lieberburg I, Seubert P, Schenk D, Ann. Neurol 1995, 38, 643. [DOI] [PubMed] [Google Scholar]
- [16].a) Maia LF, Kaeser SA, Reichwald J, Hruscha M, Martus P, Staufenbiel M, Jucker M, Sci. Transl. Med 2013, 5, 194re2; [DOI] [PubMed] [Google Scholar]; b) Sunderland T, Linker G, Mirza N, Putnam KT, Friedman DL, Kimmel LH, Bergeson J, Manetti GJ, Zimmermann M, Tang B, Bartko JJ, Cohen RM, JAMA, J. Am. Med. Assoc 2003, 289, 2094. [DOI] [PubMed] [Google Scholar]
- [17].Funke SA, Birkmann E, Willbold D, Curr. Alzheimer Res 2009, 6, 285. [DOI] [PubMed] [Google Scholar]
- [18].Peña-Bautista C, Vigor C, Galano J-M, Oger C, Durand T, Ferrer I, Cuevas A, López-Cuevas R, Baquero M, López-Nogueroles M, Vento M, Hervás D, García-Blanco A, Cháfer-Pericás C, Free Radic Biol Med 2018, 124, 388. [DOI] [PubMed] [Google Scholar]
- [19].Janelidze S, Stomrud E, Palmqvist S, Zetterberg H, van Westen D, Jeromin A, Song L, Hanlon D, Tan Hehir CA, Baker D, Blennow K, Hansson O, Sci. Rep 2016, 6, 26801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hansson O, Zetterberg H, Vanmechelen E, Vanderstichele H, Andreasson U, Londos E, Wallin A, Minthon L, Blennow K, Neurobiol Aging 2010, 31, 357. [DOI] [PubMed] [Google Scholar]
- [21].García-Ayllón M-S, Lopez-Font I, Boix CP, Fortea J, Sánchez-Valle R, Lleó A, Molinuevo J-L, Zetterberg H, Blennow K, Sáez-Valero J, Sci. Rep 2017, 7, 2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wu G, Sankaranarayanan S, Wong J, Tugusheva K, Michener MS, Shi X, Cook JJ, Simon AJ, Savage MJ, J. Neurosci. Res 2012, 90, 2247. [DOI] [PubMed] [Google Scholar]
- [23].Naseri NN, Wang H, Guo J, Sharma M, Luo W, Neurosci. Lett 2019, 705, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bibow S, Ozenne V, Biernat J, Blackledge M, Mandelkow E, Zweckstetter M, J. Am. Chem. Soc 2011, 133, 15842. [DOI] [PubMed] [Google Scholar]
- [25].Mattsson N, Zetterberg H, Janelidze S, Insel PS, Andreasson U, Stomrud E, Palmqvist S, Baker D, Hehir CAT, Jeromin A, Neurology 2016, 87, 1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Suárez-Calvet M, Karikari TK, Ashton NJ, Lantero Rodríguez J, Milà-Alomà M, Gispert JD, Salvadó G, Minguillon C, Fauria K, Shekari M, Grau-Rivera O, Arenaza-Urquijo EM, Sala-Vila A, Sánchez-Benavides G, González-de-Echávarri JM, Kollmorgen G, Stoops E, Vanmechelen E, Zetterberg H, Blennow K, Molinuevo JL, A. S. for the, EMBO Mol. Med 2020, 12, 12921. [Google Scholar]
- [27].Dugger BN, Whiteside CM, Maarouf CL, Walker DG, Beach TG, Sue LI, Garcia A, Dunckley T, Meechoovet B, Reiman EM, Roher AE, J Alzheimers Dis 2016, 51, 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gonzalez-Ortiz F, Turton M, Kac PR, Smirnov D, Premi E, Ghidoni R, Benussi L, Cantoni V, Saraceno C, Rivolta J, Ashton NJ, Borroni B, Galasko D, Harrison P, Zetterberg H, Blennow K, Karikari TK, Brain 2022, 146, 1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lanni C, Uberti D, Racchi M, Govoni S, Memo M, J Alzheimers Dis 2007, 12, 93. [DOI] [PubMed] [Google Scholar]
- [30].Khalil M, Teunissen CE, Otto M, Piehl F, Sormani MP, Gattringer T, Barro C, Kappos L, Comabella M, Fazekas F, Petzold A, Blennow K, Zetterberg H, Kuhle J, Nat. Rev. Neurol 2018, 14, 577. [DOI] [PubMed] [Google Scholar]
- [31].Gaetani L, Blennow K, Calabresi P, Di Filippo M, Parnetti L, Zetterberg H, J. Neurol. Neurosurg. Psychiatry 2019, 90, 870. [DOI] [PubMed] [Google Scholar]
- [32].Mattsson N, Cullen NC, Andreasson U, Zetterberg H, Blennow K, JAMA Neurol 2019, 76, 791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jin M, Cao L, Dai Y.-p., Front Aging Neurosci 2019, 11, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nagaraj S, Zoltowska KM, Laskowska-Kaszub K, Wojda U, Ageing Res. Rev 2019, 49, 125. [DOI] [PubMed] [Google Scholar]
- [35].a) Jia L, Zhu M, Yang J, Pang Y, Wang Q, Li Y, Li T, Li F, Wang Q, Li Y, Wei Y, BMC Med. 2021, 19, 264; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Denk J, Oberhauser F, Kornhuber J, Wiltfang J, Fassbender K, Schroeter ML, Volk AE, Diehl-Schmid J, Prudlo J, Danek A, Landwehrmeyer B, Lauer M, Otto M, Jahn H, for the FTLDc study group, PLoS One 2018, 13, 0197329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tan L, Yu J-T, Liu Q-Y, Tan M-S, Zhang W, Hu N, Wang Y-L, Sun L, Jiang T, Tan L, J Neurol Sci 2014, 336, 52. [DOI] [PubMed] [Google Scholar]
- [37].Siedlecki-Wullich D, Català-Solsona J, Fábregas C, Hernández I, Clarimon J, Lleó A, Boada M, Saura CA, Rodríguez-Álvarez J, Miñano-Molina AJ, Alzheimer’s Res. Ther 2019, 11, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Swarbrick S, Wragg N, Ghosh S, Stolzing A, Mol. Neurobiol 2019, 56, 6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Saeedi S, Israel S, Nagy C, Turecki G, Transl Psychiatry 2019, 9, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Dong Z, Gu H, Guo Q, Liang S, Xue J, Yao F, Liu X, Li F, Liu H, Sun L, Zhao K, Mol. Neurobiol 2021, 58, 3084. [DOI] [PubMed] [Google Scholar]
- [41].Winston CN, Goetzl EJ, Akers JC, Carter BS, Rockenstein EM, Galasko D, Masliah E, Rissman RA, Alzheimers Dement (Amst) 2016, 3, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Diba FS, Kim S, Lee HJ, Catal. Today 2017, 295, 41. [Google Scholar]
- [43].Janssen L, Sobott F, De Deyn PP, Van Dam D, MethodsX 2015, 2, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Carneiro P, Loureiro J, Delerue-Matos C, Morais S, do Carmo Pereira M, Sens. Actuators, B 2017, 239, 157. [Google Scholar]
- [45].Iglesias-Mayor A, Amor-Gutierrez O, Novelli A, Fernandez-Sanchez MT, Costa-Garcia A, de la Escosura-Muniz A, Anal. Chem 2020, 92, 7209. [DOI] [PubMed] [Google Scholar]
- [46].De Oliveira TR, Erbereli CR, Manzine PR, Magalhães TNC, Balthazar MLF, Cominetti MR, Faria RC, ACS Sens. 2020, 5, 1010. [DOI] [PubMed] [Google Scholar]
- [47].Peron R, Vatanabe IP, Manzine PR, Camins A, Cominetti MR, Pharmaceuticals 2018, 11, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Azimzadeh M, Nasirizadeh N, Rahaie M, Naderi-Manesh H, RSC Adv 2017, 7, 55709. [Google Scholar]
- [49].Mars A, Hamami M, Bechnak L, Patra D, Raouafi N, Anal. Chim. Acta 2018, 1036, 141. [DOI] [PubMed] [Google Scholar]
- [50].Devi R, Gogoi S, Dutta HS, Bordoloi M, Sanghi SK, Khan R, Nanoscale Adv 2020, 2, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Xia N, Wang X, Zhou B, Wu Y, Mao W, Liu L, ACS Appl. Mater. Interfaces 2016, 8, 19303. [DOI] [PubMed] [Google Scholar]
- [52].Xing Y, Feng XZ, Zhang L, Hou J, Han GC, Chen Z, Int J Nanomedicine 2017, 12, 3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zhu G, Lee HJ, Biosens. Bioelectron 2017, 89, 959. [DOI] [PubMed] [Google Scholar]
- [54].Medina-Sánchez M, Miserere S, Morales-Narváez E, Merkoçi A, Biosens. Bioelectron 2014, 54, 279. [DOI] [PubMed] [Google Scholar]
- [55].Bu Y, Zhang M, Fu J, Yang X, Liu S, Anal. Chim. Acta 2022, 1189, 339200. [DOI] [PubMed] [Google Scholar]
- [56].da Silva W, Brett CM, J. Electroanal. Chem 2020, 872, 114050. [Google Scholar]
- [57].Chiu M-J, Chen T-F, Hu C-J, Yan S-H, Sun Y, Liu B-H, Chang Y-T, Yang C-C, Yang S-Y, Nanomed.: Nanotechnol. Biol. Med 2020, 28, 102182. [DOI] [PubMed] [Google Scholar]
- [58].a) Tripathy A, Nine MJ, Silva FS, Adv. Colloid Interface Sci 2021, 290, 102380; [DOI] [PubMed] [Google Scholar]; b) Khan AU, Chen L, Ge G, Inorg. Chem. Commun 2021, 134, 108995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].El-Said WA, Abd El-Hameed K, Abo El-Maali N, Sayyed HG, Electroanalysis 2017, 29, 748. [Google Scholar]
- [60].Lu H, Wu L, Wang J, Wang Z, Yi X, Wang J, Wang N, Mikrochim Acta 2018, 185, 549. [DOI] [PubMed] [Google Scholar]
- [61].Negahdary M, Heli H, Mikrochim Acta 2019, 186, 766. [DOI] [PubMed] [Google Scholar]
- [62].Xia N, Wang X, Yu J, Wu Y, Cheng S, Xing Y, Liu L, Sens. Actuators, B 2017, 239, 834. [Google Scholar]
- [63].Razzino CA, Serafín V, Gamella M, Pedrero M, Montero-Calle A, Barderas R, Calero M, Lobo AO, Yáñez-Sedeño P, Campuzano S, Pingarrón JM, Biosens. Bioelectron 2020, 163, 112238. [DOI] [PubMed] [Google Scholar]
- [64].a) Damborský P, Švitel J, Katrlík J, Essays Biochem 2016, 60, 91; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yousefi H, Ali MM, Su H-M, Filipe CDM, Didar TF, ACS Nano 2018, 12, 3287. [DOI] [PubMed] [Google Scholar]
- [65].Ly TN, Park S, J Ind Eng Chem 2020, 91, 182. [Google Scholar]
- [66].a) Song S, Lee JU, Jeon MJ, Kim S, Sim SJ, Biosens. Bioelectron 2022, 199, 113864; [DOI] [PubMed] [Google Scholar]; b) Kim D, Yoo S, Chemosensors 2021, 9, 318. [Google Scholar]
- [67].Elbassal EA, Morris C, Kent TW, Lantz R, Ojha B, Wojcikiewicz EP, Du D, J. Phys. Chem. C 2017, 121, 20007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kim H, Lee JU, Song S, Kim S, Sim SJ, Biosens. Bioelectron 2018, 101, 96. [DOI] [PubMed] [Google Scholar]
- [69].Wang W, Han Y, Fan Y, Wang Y, Langmuir 2019, 35, 2334. [DOI] [PubMed] [Google Scholar]
- [70].Jia Y-P, Ma B-Y, Wei X-W, Qian Z-Y, Chin. Chem. Lett 2017, 28, 691. [Google Scholar]
- [71].Jara-Guajardo P, Cabrera P, Celis F, Soler M, Berlanga I, Parra-Muñoz N, Acosta G, Albericio F, Guzman F, Campos M, Alvarez A, Morales-Zavala F, Kogan MJ, Nanomaterials 2020, 10, 690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Zhao J, Chang W, Liu L, Xing X, Zhang C, Meng H, Gopinath SCB, Lakshmipriya T, Chen Y, Liu Y, J. Immunol. Methods 2021, 489, 112942. [DOI] [PubMed] [Google Scholar]
- [73].Yousefi H, Su H-M, Ali M, Filipe CDM, Didar TF, Adv. Mater. Interfaces 2018, 5, 1800659. [Google Scholar]
- [74].Song S, Lee JU, Jeon MJ, Kim S, Lee C-N, Sim SJ, Biosens. Bioelectron 2023, 230, 115269. [DOI] [PubMed] [Google Scholar]
- [75].a) Yang SJ, Lee JU, Jeon MJ, Sim SJ, Anal. Chim. Acta 2022, 1195, 339445; [DOI] [PubMed] [Google Scholar]; b) Xia Y, Padmanabhan P, Sarangapani S, Gulyás B, Vadakke Matham M, Sci. Rep 2019, 9, 8497; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Suga K, Lai Y-C, Faried M, Umakoshi H, Int J Anal Chem 2018, 2018, 2571808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Klyucherev TO, Olszewski P, Shalimova AA, Chubarev VN, Tarasov VV, Attwood MM, Syvänen S, Schiöth HB, Transl Neurodegener 2022, 11, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Wu J, Zhou X, Li P, Lin X, Wang J, Hu Z, Zhang P, Chen D, Cai H, Niessner R, Haisch C, Sun P, Zheng Y, Jiang Z, Zhou H, Anal. Chem 2021, 93, 8799. [DOI] [PubMed] [Google Scholar]
- [78].Salvi G, De Los Rios P, Vendruscolo M, Proteins 2005, 61, 492. [DOI] [PubMed] [Google Scholar]
- [79].Kim H, Lee JU, Kim S, Song S, Sim SJ, ACS Sens. 2019, 4, 595. [DOI] [PubMed] [Google Scholar]
- [80].Luo T, Bu L, Peng S, Zhang Y, Zhou Z, Li G, Huang J, Talanta 2019, 205, 120117. [DOI] [PubMed] [Google Scholar]
- [81].a) Pi J, Long Y, Huang N, Cheng Y, Zheng H, Talanta 2016, 146, 10; [DOI] [PubMed] [Google Scholar]; b) Huang H, Li P, Zhang M, Yu Y, Huang Y, Gu H, Wang C, Yang Y, Nanoscale 2017, 9, 5044. [DOI] [PubMed] [Google Scholar]
- [82].Quan L, Wu J, Lane LA, Wang J, Lu Q, Gu Z, Wang Y, Bioconjug Chem 2016, 27, 809. [DOI] [PubMed] [Google Scholar]
- [83].Chen L, Lin J, Yi J, Weng Q, Zhou Y, Han Z, Li C, Chen J, Zhang Q, Anal. Bioanal. Chem 2019, 411, 5277. [DOI] [PubMed] [Google Scholar]
- [84].Vilela P, El-Sagheer A, Millar TM, Brown T, Muskens OL, Kanaras AG, ACS Sens. 2017, 2, 52. [DOI] [PubMed] [Google Scholar]
- [85].Xia N, Zhou B, Huang N, Jiang M, Zhang J, Liu L, Biosens. Bioelectron 2016, 85, 625. [DOI] [PubMed] [Google Scholar]
- [86].a) Qian Z, Chai L, Tang C, Huang Y, Chen J, Feng H, Sens. Actuators, B 2016, 222, 879; [Google Scholar]; b) Guo Y, Liu D, Yang Q, Shi W, Yang Z, Chen J, Xiang J, Anal. Lett 2022, 55, 1453. [Google Scholar]
- [87].Rees TM, Brimijoin S, Drugs Today (Barc) 2003, 39, 75. [DOI] [PubMed] [Google Scholar]
- [88].Silvestro S, Bramanti P, Mazzon E, Int. J. Mol. Sci 2019, 20, 3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Chen W, Lv G, Hu W, Li D, Chen S, Dai Z, Nanotechnol. Rev 2018, 7, 157. [Google Scholar]
- [90].a) Akhtar N, Metkar SK, Girigoswami A, Girigoswami K, Mater. Sci. Eng., C 2017, 78, 960; [DOI] [PubMed] [Google Scholar]; b) Lee S-C, Park H-H, Kim S-H, Koh S-H, Han S-H, Yoon M-Y, Anal. Chem 2019, 91, 5573; [DOI] [PubMed] [Google Scholar]; c) Adesoye S, Dellinger K, Sens Biosensing Res 2022, 37, 100499; [Google Scholar]; d) Adesoye S, Al Abdullah S, Nowlin K, Dellinger K, Nanomaterials 2022, 12, 3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Materón EM, Miyazaki CM, Carr O, Joshi N, Picciani PHS, Dalmaschio CJ, Davis F, Shimizu FM, Appl. Surf. Sci. Adv 2021, 6, 100163. [Google Scholar]
- [92].a) Vu Nu TT, Tran NHT, Nam E, Nguyen TT, Yoon WJ, Cho S, Kim J, Chang K-A, Ju H, RSC Adv. 2018, 8, 7855; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lisi S, Scarano S, Fedeli S, Pascale E, Cicchi S, Ravelet C, Peyrin E, Minunni M, Biosens. Bioelectron 2017, 93, 289; [DOI] [PubMed] [Google Scholar]; c) Chen W, Gao G, Jin Y, Deng C, Talanta 2020, 216, 120930; [DOI] [PubMed] [Google Scholar]; d) Sun L, Liu D, Fu D, Yue T, Scharre D, Zhang L, Chem. Eng. J 2021, 405, 126733. [Google Scholar]
- [93].Yu D, Yin Q, Wang J, Yang J, Chen Z, Gao Z, Huang Q, Li S, Int J Nanomedicine 2021, 16, 1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].a) Zhu X, Zhang N, Zhang Y, Liu B, Chang Z, Zhou Y, Hao Y, Ye B, Xu M, Anal. Methods 2018, 10, 641; [Google Scholar]; b) Pashchenko O, Shelby T, Banerjee T, Santra S, ACS Infect. Dis 2018, 4, 1162; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Khan ZA, Park S, Talanta 2022, 237, 122946. [DOI] [PubMed] [Google Scholar]
- [95].Yousefi H, Su H-M, Imani SM, Alkhaldi K, Filipe CDM, Didar TF, ACS Sens. 2019, 4, 808. [DOI] [PubMed] [Google Scholar]
- [96].a) Brazaca LC, Moreto JR, Martín A, Tehrani F, Wang J, Zucolotto V, ACS Nano 2019, 13, 13325; [DOI] [PubMed] [Google Scholar]; b) Didar TF, Tabrizian M, Lab Chip 2012, 12, 4363; [DOI] [PubMed] [Google Scholar]; c) Shakeri A, Imani SM, Chen E, Yousefi H, Shabbir R, Didar TF, Lab Chip 2019, 19, 3104; [DOI] [PubMed] [Google Scholar]; d) Imani SM, Badv M, Shakeri A, Yousefi H, Yip D, Fine C, Didar TF, Lab Chip 2019, 19, 3228. [DOI] [PubMed] [Google Scholar]
- [97].Carneiro P, Morais S, Pereira MC, Nanomaterials 2019, 9, 1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Hu T, Lu S, Chen C, Sun J, Yang X, Sens. Actuators, B 2017, 243, 792. [Google Scholar]
- [99].Xia W, Yang T, Shankar G, Smith IM, Shen Y, Walsh DM, Selkoe DJ, Arch. Neurol 2009, 66, 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].a) Zhao J, Xu N, Yang X, Ling G, Zhang P, Colloid Interface Sci. Commun 2022, 46, 100579; [Google Scholar]; b) Kim Y, Park J-H, Lee H, Nam J-M, Sci. Rep 2016, 6, 19548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Zhang X, Liu S, Song X, Wang H, Wang J, Wang Y, Huang J, Yu J, ACS Sens. 2019, 4, 2140. [DOI] [PubMed] [Google Scholar]
- [102].Ren X, Yan J, Wu D, Wei Q, Wan Y, ACS Sens. 2017, 2, 1267. [DOI] [PubMed] [Google Scholar]
- [103].Guo L, Xu Y, Ferhan AR, Chen G, Kim D-H, J. Am. Chem. Soc 2013, 135, 12338. [DOI] [PubMed] [Google Scholar]
- [104].Liu C, You X, Lu D, Shi G, Deng J, Zhou T, ACS Appl. Bio Mater 2020, 3, 7965. [DOI] [PubMed] [Google Scholar]
- [105].a) Lyu Z, Ding S, Zhang N, Zhou Y, Cheng N, Wang M, Xu M, Feng Z, Niu X, Cheng Y, Zhang C, Du D, Lin Y, Research 2020, 2020, 4724505; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hu T, Chen C, Huang G, Yang X, Sens. Actuators, B 2016, 234, 63. [Google Scholar]
- [106].Amiri H, Saeidi K, Borhani P, Manafirad A, Ghavami M, Zerbi V, ACS Chem. Neurosci 2013, 4, 1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Salerno M, S Domingo Porqueras D, Coord. Chem. Rev 2016, 327–328, 27. [Google Scholar]
- [108].Wadghiri YZ, Li J, Wang J, Hoang DM, Sun Y, Xu H, Tsui W, Li Y, Boutajangout A, Wang A, PLoS One 2013, 8, 57097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].a) Zhou J, Fa H, Yin W, Zhang J, Hou C, Huo D, Zhang D, Zhang H, Mater Sci Eng C Mater Biol Appl 2014, 37, 348; [DOI] [PubMed] [Google Scholar]; b) Sillerud LO, Solberg NO, Chamberlain R, Orlando RA, Heidrich JE, Brown DC, Brady CI, Vander Jagt TA, Garwood M, Vander Jagt DL, J Alzheimers Dis 2013, 34, 349; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Fernández T, Martínez-Serrano A, Cussó L, Desco M, Ramos-Gómez M, ACS Chem. Neurosci 2018, 9, 912. [DOI] [PubMed] [Google Scholar]
- [110].Zeng J, Wu J, Li M, Wang P, Arch Med Res 2018, 49, 282. [DOI] [PubMed] [Google Scholar]
- [111].Cheng KK, Chan PS, Fan S, Kwan SM, Yeung KL, Wang Y-XJ, Chow AHL, Wu EX, Baum L, Biomaterials 2015, 44, 155. [DOI] [PubMed] [Google Scholar]
- [112].Carvalho A, Domingues I, Gonçalves MC, Mater. Chem. Phys 2015, 168, 42. [Google Scholar]
- [113].Fernández-Cabada T, Ramos-Gómez M, Nanoscale Res. Lett 2019, 14, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Ansciaux E, Burtea C, Laurent S, Crombez D, Nonclercq D, Vander Elst L, Muller RN, Contrast Media Mol. Imaging 2015, 10, 211. [DOI] [PubMed] [Google Scholar]
- [115].Quan L, Wu J, Lane LA, Wang J, Lu Q, Gu Z, Wang Y, Bioconjugate Chem. 2016, 27, 809. [DOI] [PubMed] [Google Scholar]
- [116].Yousaf M, Ahmad M, Bhatti IA, Nasir A, Hasan M, Jian X, Kalantar-Zadeh K, Mahmood N, ACS Sens. 2019, 4, 200. [DOI] [PubMed] [Google Scholar]
- [117].a) Shakeri A, Khan S, Didar TF, Lab Chip 2021, 21, 3053; [DOI] [PubMed] [Google Scholar]; b) Shakeri A, Jarad NA, Khan S, Didar TF, Anal. Chim. Acta 2021, 1209, 339283; [DOI] [PubMed] [Google Scholar]; c) Okyem S, Awotunde O, Ogunlusi T, Riley MB, Driskell JD, Bioconjugate Chem. 2021, 32, 1753; [DOI] [PubMed] [Google Scholar]; d) Gao S, Guisán JM, Rocha-Martin J, Anal. Chim. Acta 2022, 1189, 338907. [DOI] [PubMed] [Google Scholar]
- [118].a) Shakeri A, Yousefi H, Jarad NA, Kullab S, Al-Mfarej D, Rottman M, Didar TF, Sci. Rep 2022, 12, 14486; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Shakeri A, Jarad NA, Terryberry J, Khan S, Leung A, Chen S, Didar TF, Small 2020, 16, 2003844; [DOI] [PubMed] [Google Scholar]; c) Ladouceur L, Shakeri A, Khan S, Rincon AR, Kasapgil E, Weitz JI, Soleymani L, Didar TF, ACS Appl. Mater. Interfaces 2022, 14, 3864; [DOI] [PubMed] [Google Scholar]; d) Khan S, Jarad NA, Ladouceur L, Rachwalski K, Bot V, Shakeri A, Maclachlan R, Sakib S, Weitz JI, Brown ED, Small 2022, 18, 2108112; [DOI] [PubMed] [Google Scholar]; e) Imani SM, Maclachlan R, Chan Y, Shakeri A, Soleymani L, Didar TF, Small 2020, 16, 2004886. [DOI] [PubMed] [Google Scholar]


