Abstract
To investigate the mechanism and functional significance of infection of CD8+ lymphocytes by human immunodeficiency virus type 1 (HIV-1) in vivo, we determined frequencies of infection, proviral conformation, and genetic relationships between HIV-1 variants infecting naive (CD45RA+) and memory (CD45RO+) peripheral blood CD4+ and CD8+ lymphocytes. Infection of CD3+ CD8+ CD45RA+ cells was detected in 9 of 16 study subjects at frequencies ranging from 30 to 1,400 proviral copies/106 cells, more frequently than CD3+ CD8+ lymphocytes expressing the RO isoform of CD45 (n = 2, 70 and 260 copies /106 cells). In agreement with previous studies, there was no evidence for a similar preferential infection of CD4+ naive lymphocytes. Proviral sequences in both CD4+ and CD8+ lymphocyte subsets were complete, as assessed by quantitation using primers from the long terminal repeat region spanning the tRNA primer binding site. In six of the seven study subjects investigated, variants infecting CD8+ lymphocytes were partially or completely genetically distinct in the V3 region from those recovered from CD4+ lymphocytes and showed a greater degree of compartmentalization than observed between naive and memory subsets of CD4+ lymphocytes. In two study subjects, arginine substitutions at position 306, associated with use of the chemokine coreceptor CXCR4, were preferentially found in CD4 lymphocytes. These population differences may have originated through different times of infection rather than necessarily indicating a difference in their biological properties. The preferential distribution of HIV-1 in naive CD8+ lymphocytes indeed suggests that infection occurred early in T-lymphocyte ontogeny, such as during maturation in the thymus. Destruction of cells destined to become CD8+ lymphocytes may be a major factor in the decline in CD8+ lymphocyte frequencies and function on disease progression and may contribute directly to the observed immunodeficiency in AIDS.
CD4+ lymphocytes are considered to be the principal target of human immunodeficiency virus type 1 (HIV-1), but more recently a number of studies have shown the in vivo infection of other lymphoid cell types including CD8+ T lymphocytes (12, 21, 31, 33, 51). CD8+ T cells are a major immunological defence against HIV-1 infection. In most individuals, HIV mediates a strong specific cytotoxic activity that eliminates productively infected cells. This response also blocks intracellular viral replication in CD4 cells by production and secretion of a number of soluble inhibitory factors such as macrophage inflammatory proteins 1α and 1β and RANTES (regulated upon activation, normal T-cell expressed and secreted), interleukin-16, and other, as yet unidentified factors (17, 32). During the course of infection, there is a gradual loss of CD4 cells; after an initial increase in the number of cytotoxic T cells, the CD8 cell number similarly falls during disease progression (29). The observed decline in both the number of CD8 cells and specific HIV cytotoxic activity coincides with an increase in viral load and may ultimately be a contributory factor to the eventual collapse of the immune system and the development of AIDS.
The mechanisms leading to CD8 T-cell dysfunction and depletion are still unclear. CD8 T-cell function may be the indirectly influenced by a defective HIV-1-specific CD4 T helper response that is necessary for the maturation and function of cytotoxic T cells. It has also been demonstrated after antigenic stimulation some CD8 T cells develop a state of unresponsiveness and eventual death is mediated via apoptosis (19, 30). However, it has been observed that productive HIV-1 infection of cytotoxic T lymphocytes in vitro and in vivo in animal models can occur (47). The peripheral interaction between CD4 and CD8 cells occurring in vivo as part of the immune response may allow direct transmission of infection to the CD8 lymphocytes. Alternatively, it is possible that damage to or deficiency of the thymus may also confer direct infection. HIV-1 may infect and destroy both intrathymic T progenitor cells (CD3− CD4+ CD8−), double-positive thymocytes (CD3+ CD4+ CD8+) (4, 49) and mature CD3hi CD8+ thymocytes (28). Recent studies alternatively suggest that infection of CD8 lymphocytes may occur by a conventional CD4-dependent mechanism, as CD4 expression is up-regulated following activation through the T-cell receptor complex (12, 49, 51).
Information on the mechanisms of CD8 lymphocyte infection can be obtained by analysis of relative frequencies of infection of CD8 lymphocytes expressing the CD45RA (naive) and CD45RO (memory/effector) isoforms, as has been previously described for CD4 lymphocyte infection (36). Abortive or noncytopathic infection of double-positive (CD4+ CD8+) thymocytes during maturation in the thymus would produce differentiated, naive CD8+ lymphocytes in the circulation that contained stably integrated HIV proviral sequences. However, if infection occurred during the phase of CD4 expression after antigenic stimulation of CD8 lymphocytes, then proviral sequences would be preferentially distributed in the memory/effector population, as the activation process would result in a change of phenotype from CD45RA+ to CD45RO+. In this study, we investigated the frequency of HIV infection in CD4 and CD8 naive and memory cell populations separated from peripheral blood mononuclear cells (PBMCs) of HIV-seropositive individuals by quantitative PCR for HIV-1 proviral sequences. We also investigated possible genetic differences between variants of HIV-1 isolated from each of the lymphocyte subsets that might relate to time of infection or determine differences in cellular tropism.
MATERIALS AND METHODS
Samples and clinical details of study patients.
Samples (20 to 30 ml) of whole blood anticoagulated with EDTA were collected from seropositive individuals attending the genitourinary medicine clinic or the infectious disease unit in Edinburgh. CD4 counts, viral load information, and risk groups from the patient group in whom distribution of HIV in naive and memory subsets of CD4 and CD8 lymphocytes were analyzed (Table 1). Plasma virus loads were determined by a commercially available PCR (Roche Monitor, Lewes, East Sussex, United Kingdom). Further samples were collected from five healthy, HIV-seronegative control for cell purity measurements.
TABLE 1.
Clinical profile and frequencies of infection of naive and memory subsets of CD4 and CD8 T lymphocytes.
| Study subject | Sexa | Risk groupb | Viral load (genome eq/ml) | CD4 count (μl) | Proviral copies/106 cells
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| CD4+ lymphocytes
|
CD8+ CD45RA+ (naive) cells
|
CD8+ CD45RO+ (memory) cells
|
||||||||
| CD45RA+ | CD45RO+ | Proviral load | CD4 contaminationc | Proviral load | CD4 contamination | |||||
| p1 | F | IDU | 14,000 | 10 | 98 | 834 | <10 | 0.9 | <10 | 11 |
| p2 | M | MH | 514,500 | 29 | 1,106 | 587 | 107 | 10 (9.4) | <10 | 8.0 |
| p3 | M | MH | 116,000 | 74 | 133 | 278 | 1,444 | 1.2 (0.1) | <10 | 3.8 |
| p4 | M | MH | 18,000 | 130 | 596 | 4,272 | <10 | 5.4 | <10 | 58 |
| p5 | M | MH | <400 | 232 | 686 | 457 | 422 | 6.2 (1.5) | 67 | 6.2 (17) |
| p6 | F | Het | 52,000 | 265 | 866 | 1,566 | <10 | 7.9 | <10 | 21 |
| p7 | M | IDU/Het | 48,000 | 267 | 95 | 520 | <10 | 0.9 | <10 | 7.1 |
| p8 | M | IDU | 460,000 | 280 | 4,307 | 2,137 | 22 | 39 (180) | 259 | 29 (21) |
| p9 | M | MH | >750,000 | 282 | 575 | 2,321 | 40 | 5.2 (13) | <10 | 32 |
| p10 | M | MH | 81,540 | 301 | 1,307 | 464 | 622 | 12 (1.9) | <10 | 6.3 |
| p11 | M | MH | 661,000 | 311 | 834 | 3,916 | <10 | 7.6 | 20 | 53 (270) |
| p12 | F | IDU | 64,680 | 329 | 55 | 115 | <10 | 0.5 | <10 | 1.6 |
| p13 | M | MH | 67,980 | 354 | 115 | 222 | 71 | 1.0 (1.5) | <10 | 3.0 |
| p14 | F | Het | <513 | 360 | 533 | 980 | 119 | 4.9 (4.1) | <10 | 13 |
| p15 | M | MH | 60,589 | 748 | 653 | Negative | 31 | 5.9 (19) | <10 | <0.1 |
| p16 | M | MH | 1,680 | 850 | 143 | 25 | <10 | 1.3 | <10 | 0.3 |
F, female; M, male.
Abbreviations: IDU, injecting drug user; MH, male homosexual; Het, infection acquired through heterosexual contact.
Estimated contribution of contaminating CD4 cells to proviral load (percent of total) detected in CD8 lymphocytes, using mean purity data from controls (Table 4).
Cell separations.
Blood samples were diluted with an equal volume of phosphate-buffered saline and PBMCs were isolated by density centrifugation over a Ficoll-Hypaque gradient (Lymphoprep; Nycomed). T lymphocytes were purified from PBMCs using a negative selection pan-T isolation kit on an automated MACS cell sorter (Miltenyi Biotec). CD8+ and CD4+ lymphocytes were isolated from the purified T cells by positive selection using CD8- and CD4-conjugated MACS beads. Naive and memory subsets of CD4 and CD8 lymphocytes were separated from PBMCs by initial separation into CD4+ and CD8+ lymphocytes by negative selection (Miltenyi Biotec). Naive and memory subsets of isolated CD4 and CD8 lymphocytes by obtained by positive selection using CD45RA- and CD45RO-conjugated MACS beads.
Analysis of the purity of isolated subsets by flow cytometry.
For analyses in this report, the following combinations of labeled monoclonal antibodies were used (10 μl per 105 cells; 30 min at 4°C): (i) fluorescein isothiocyanate isomer 1 (FITC)-conjugated CD8, phycoerythrin (PE)-conjugated CD3, and phycoerythrin-Cy5 (Cy5)-conjugated CD4; (ii) FITC-CD45RA and PE-CD45RO; (iii) FITC-CD45RA, PE-CD45RO, and Cy5-CD4; (iv) FITC-CD45RO, PE-CD45RA, and Cy5-CD4; (v) FITC-CD45RA, PE-CD45RO, and Cy5-CD8; and (vi) FITC-CD45RO, PE-CD45RA, and Cy5-CD8 (DAKO, Glostrup, Denmark). Cells were washed twice in phosphate-buffered saline with 2% bovine serum albumin and fixed in 200 μl of 2% formalin. The purity of the isolated CD4+ CD45RA+, CD4+ CD45RO+, CD8+ CD45RA+, and CD8+ CD45RO+ cell populations was assessed on a Coulter Epics Elite after gating on lymphocytes based on their forward and side scatter characteristics. Each analysis was based on a minimum of 5,000 events.
Detection and quantitation of HIV sequences.
DNA was extracted from isolated cell subsets as previously described (44). HIV proviral sequences were quantified by limiting-dilution nested PCR (44, 54) using nested sets of highly conserved PCR primers from the long terminal repeat (LTR) region. Pan-LTR primers were 5′-GRAACCCACTGCTTAASSCTCAA-3′ (outer, sense) 5′-TGTTCGGGCGCCACTGCTAGAGA-3′ (outer, antisense), 5′-CTCAATAAAGCTTGCCTTGAG-3′ (inner, sense), and 5′-GAGGGATCTCTAGNYACCAGAGT-3′ (inner, antisense) (5′ base positions 506, 626, 524, and 578, respectively, in the HXB2 genome). Complete LTR (C-LTR) primers were 5′-ACTCTGGTRNCTAGAGATCCCTC-3′ (outer, sense), 5′-GGCGTACTCACCAGTCG CCG-3′ (outer, antisense), 5′-TCTCTAGCAGTGGCGCCCGAAC-3′ (inner, sense), and 5′-TCAGCAAGCCGAGTCCTG-3′ (inner, antisense) (5′ base positions 578, 735, 626, and 692, respectively, in the HXB2 genome). Both primary and secondary PCRs for pan-LTR and C-LTR primers were carried out using the following parameters: 94°C for 18 s, 55°C for 21 s, and 72°C for 1.5 min for 30 cycles, followed by a final extension step of 72°C for 6 min. PCR amplicons were run at 150 V for 30 min on 2% agarose gels containing 0.5 μg of ethidium bromide/ml and visualized under UV light.
Quantification was performed by limiting-dilution PCR as previously described (44). Nucleotide sequences from the V3 region (patient samples p2, p3, p4, p11, p13, p14, and p15) were amplified using previously described primers (24, 40, 43). To serve as negative controls, parallel separations, extractions, and amplifications were carried out with PBMCs isolated from buffy coats leukocytes derived from HIV-negative blood. HIV-1 DNA could be detected in CD4 T lymphocytes and CD8 T lymphocytes from HIV-seropositive individuals but not in any negative controls.
Cloning of the PCR products.
Amplified DNA was ligated into a plasmid vector prior to nucleotide sequencing using the pGEM-T vector system (Promega, Southampton, United Kingdom). The ligated product was transformed into competent cells (JM109; Promega) and plated on Luria-Bertani plates (200 μg of ampicillin/ml). The plasmid DNA was denatured by incubation with 1/10 volume of 2 M sodium hydroxide–2 mM EDTA at 37°C for 30 min. DNA was precipitated by addition of 1/10 volume of 3 M sodium acetate and 2 volumes of ethanol and incubation at −20°C overnight.
Nucleotide sequencing and analysis.
Dideoxynucleotide sequencing was carried out with a U.S. Biochemical Sequenase 2.0 kit (Amersham Life Science, Piscataway, N.J.) with [35S]dATP, and patient plasma samples were sequenced using a Thermosequenase radiolabeled terminator cycle sequencing kit according to the manufacturer's instructions.
Sequences were aligned and distances were estimated using the Simmonic 2000 sequence editor package. Phylogenetic analysis was carried out using the MEGA program (27). The nucleotide sequences from V1/V2 and V3 amplified from each of the study subjects were compared with each other and with a range of standard HIV-1 variants. Each set of sequences from the four study subjects was monophyletic in both genomic regions and distinct from those of the published sequences of subtype B: HIVSF2 (K02007), HIVRF (M17451), HIVOYI (M26727), HIVLAI (K02013), HIVJRFL (M74978), HIVYU2 (M93258), HIVCAM1 (D10112), HIVNY5CG (M38431), HIVHAN (U43131), HIVWMJ22 (M12507), and HIVSFAAA (M65024).
Nucleotide sequence accession numbers.
Nucleotide sequences obtained in this study have been submitted to GenBank and assigned accession numbers AF353734 through AF353941.
RESULTS
Detection of HIV proviral sequences in CD8 lymphocytes.
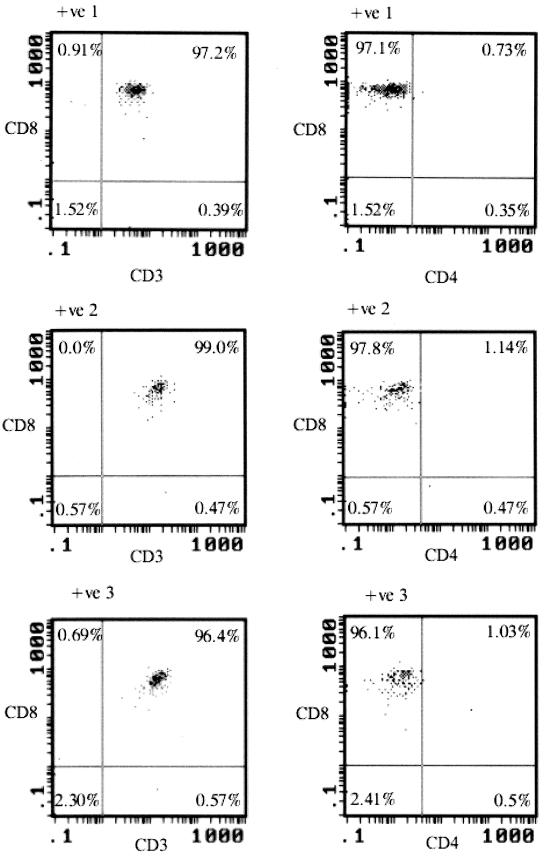
T lymphocytes were separated from PBMCs by negative selection using immobilized magnetic beads. Fluorescence-activated cell sorting analysis showed substantial purity of the separated T cells, with ≥94% of cells coexpressing CD3 and CD45 (mean, 97.4%) (Table 2). Monocyte contamination, detected by the expression of CD14 and CD45, was minimal (<3%; mean, 1.1%). Purified CD3+ lymphocytes were subsequently separated by positive selection for CD8 expression and, in a separate step carried on the remaining cells, for CD4. Analysis of the purity of selected CD8 lymphocytes revealed extremely low numbers of contaminating CD4 lymphocytes (Table 2; Fig. 1), in no case exceeding 0.5% of the total population. Knowledge of this frequency combined with quantitation of proviral load in purified CD8 and CD4 lymphocytes allowed calculation in worst-case situations (i.e., 0.5% CD4 contamination) of the contribution of CD4 lymphocytes to the proviral load detected in CD8 lymphocytes (see below).
TABLE 2.
Purity of isolated pan-T and CD8+ lymphocytes
| Sample | T lymphocytes
|
CD8 lymphocytes
|
||
|---|---|---|---|---|
| % CD4+ CD45+ | % CD3+ CD45+ | % CD4+ CD8− | % CD3+ CD8+ | |
| HIV negative | ||||
| 1 | 2.6 | 98.1 | 0.2 | 99.0 |
| 2 | 0.5 | 96.4 | 0.2 | 98.2 |
| 3 | 0.1 | 99.8 | 0.3 | 96.0 |
| 4 | 0.1 | 96.7 | 0.4 | 97.3 |
| Mean | 0.6 | 98.2 | 0.3 | 97.3 |
| HIV positive | ||||
| 1 | 2.5 | 94.0 | 0.4 | 97.2 |
| 2 | 1.3 | 98.0 | 0.5 | 99.0 |
| 3 | 0.7 | 99.0 | 0.5 | 96.4 |
| Mean | 1.5 | 97.0 | 0.4 | 97.5 |
FIG. 1.
Analysis by three-color flow cytometry of CD8 lymphocytes purified by positive selection from PBMCs of three HIV-positive (+ve) individuals. Cells were labeled with monoclonal antibodies to CD3 (PE), CD8 (FITC), and CD4 (Cy5).
Having established the effectiveness of the cell separation method, we separated samples from seven HIV-infected individuals into CD4 and CD8 T lymphocytes and assayed them for HIV proviral DNA sequences using highly conserved primers from the LTR. These pan-LTR primers amplify DNA sequences immediately downstream from the tRNA primer binding site and are therefore potentially able to detect incomplete transcripts resulting from abortively infected cells (18, 52). Frequencies of infected cells ranged from 30 to 670 proviral copies/106 CD4 lymphocytes and from 8 to 500 /106 CD8 lymphocytes. The worst-case scenario, where CD4 lymphocytes comprise 0.5% of the CD8 population (see above), failed to account for the proviral sequences detected in CD8 lymphocytes in at least five of the seven samples (Table 3; Fig. 2A). Maximum contributions from contaminating CD4 lymphocytes ranged from <5% in these four samples and 27 and 33% in the remaining two. These latter samples contained low proviral loads in the CD8 lymphocyte subset (<10 copies/106 cells). Similar results were obtained on retesting proviral loads using a different set of primers (C-LTR) (Table 3). In this case, CD4 contamination accounted for ≤1% of proviral load detected in five samples and for 16 and 44% in the remaining two).
TABLE 3.
Frequencies of infection of CD4 and CD8 lymphocytes and contribution of contaminating CD4 lymphocytes to the proviral load detected in CD8 lymphocytes
| Study subject | CD4 (proviral copies/106 CD4 lymphocytes) | Proviral contaminationa
|
CD8 (proviral copies/106 CD8 lymphocytes) | % Mean (range) CD4 contributionb | ||
|---|---|---|---|---|---|---|
| Minimum | Mean | Maximum | ||||
| Pan-T LTR primers | ||||||
| p17 | 215 | 0.41 | 0.73 | 1.01 | 101 | 0.72 (0.40–1.00) |
| p18 | 309 | 0.59 | 1.05 | 1.45 | 23 | 4.55 (2.55–6.31) |
| p19 | 672 | 1.28 | 2.28 | 3.16 | 56 | 4.07 (2.28–5.64) |
| p20 | 579 | 1.10 | 1.96 | 2.72 | 10 | 19.6 (11.0–27.2) |
| p21 | 30 | 0.06 | 0.10 | 0.14 | 19 | 0.53 (0.30–0.74) |
| p22 | 563 | 1.07 | 1.91 | 2.65 | 8 | 23.8 (13.4–33.1) |
| p23 | 551 | 1.05 | 1.87 | 2.59 | 499 | 0.37 (0.21–0.52) |
| C-LTR primers | ||||||
| p17 | 216 | 0.41 | 0.73 | 1.01 | 505 | 0.14 (0.08–0.20) |
| p18 | 514 | 0.98 | 1.74 | 2.42 | 585 | 0.30 (0.17–0.41) |
| p19 | 52 | 0.10 | 0.18 | 0.24 | 56 | 0.31 (0.31–0.44) |
| p20 | 901 | 1.71 | 3.05 | 4.23 | 9.6 | 31.82 (17.8–44.1) |
| p21 | 63 | 0.12 | 0.21 | 0.30 | 29 | 0.74 (0.41–1.02) |
| p22 | 271 | 0.51 | 0.92 | 1.27 | 8 | 11.48 (6.4–15.92) |
Contribution of CD4 lymphocytes to proviral load detected in CD8 lymphocyte subset, corresponding to minimum (0.19%), mean (0.34%), and maximum (0.47%) contamination levels observed in control experiments (Table 2), expressed as copies per 106 cells.
Percentage of proviral load in CD8 lymphocyte subset attributable to CD4 contamination.
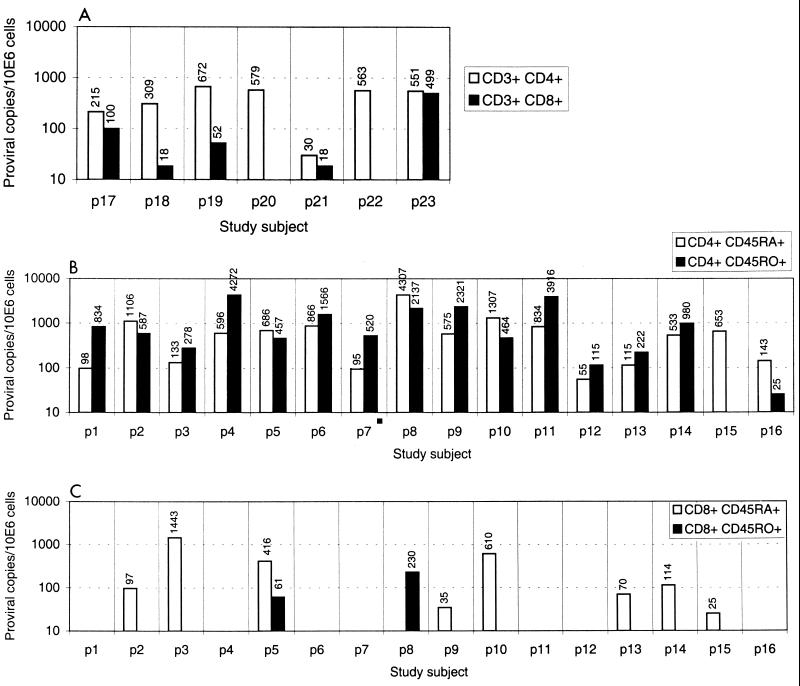
FIG. 2.
Comparison of net proviral load in CD4 and CD8 lymphocytes separated by positive selection (A), and in CD4 lymphocytes (B) and CD8 lymphocytes (C) separated by negative selection, followed by positive selection for CD45RA and CD45RO.
Detection of complete proviral transcripts.
To determine whether proviral sequences detected in CD4 and CD8 lymphocytes were incomplete or complete transcripts, DNA samples from each subset were quantified using a second set of primers spanning the region on either side of the primer binding site. Quantitation of the plasmid HXB2 demonstrated that pan-LTR and C-LTR primers had equal sensitivities (data not shown), and so the detection of greater proviral loads using the pan-LTR primers in CD4 or CD8 lymphocytes would indicate the presence of incomplete proviral transcripts. Proviral loads using the two sets of primers were comparable in each case, with no evidence for higher virus loads detected with the pan-LTR primers (median ratio of pan-LTR to C-LTR, 0.638; Z = 0.76 using Wilcoxon signed rank test; P = 0.445 [not significant]).
Distribution of HIV in naive and memory subsets of CD4 and CD8 lymphocytes.
To obtain phenotypically unchanged CD4+ and CD8+ T lymphocytes for separation of naive and memory subsets, it was necessary to use negative selection to isolate CD4 and CD8 lymphocytes prior to positive selection with CD45RA and CD45RO monoclonal antibodies. The purity of the isolated CD4+ CD45RA+, CD4+ CD45RO+, CD8+ CD45RA+, and CD8+ CD45RO+ cell populations from five HIV-1-negative individuals and one HIV-1-positive individual was determined (Table 4). CD4+ lymphocytes contained 0.9 to 1.3% contaminating CD8+ cells, similar to that found in the sample from the HIV-positive sample (0.7 to 1.2%). Contamination of the separated CD8+ lymphocytes by CD4+ cells was similarly low (0.8 to 1.7%). As described above for CD4 and CD8 lymphocytes separated by positive selection, the contribution of contaminating CD4+ lymphocytes could not account for the proviral load detected in the majority if CD8 lymphocytes samples (see below).
TABLE 4.
CD3, CD4, and CD8 expression on isolated CD4+ CD45RA+, CD4+ CD45RO+, CD8+ CD45RA+, and CD8+ CD45RO+ lymphocytesa
| Sample | CD3 expression (%)
|
CD4 expression (%)
|
CD8 expression (%)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD4+ RA+ | CD4+ RO+ | CD8+ RA+ | CD8+ RO+ | CD4+ RA+ | CD4+ RO+ | CD8+ RA+ | CD8+ RO+ | CD4+ RA+ | CD4+ RO+ | CD8+ RA+ | CD8+ RO+ | |
| HIV negative | ||||||||||||
| 1 | 79.1 | 98.7 | 98.7 | 68.5 | 73.4 | 93.9 | 2.5 | 2.4 | 2.3 | 1.9 | 93.1 | 60.9 |
| 2 | 97.2 | 93.8 | 99.2 | 97.6 | 95.6 | 90.9 | 0.4 | 1.3 | 0.6 | 0.3 | 98.4 | 97.5 |
| 3 | 98.9 | 99.9 | 98.4 | 91.8 | 96.1 | 97.1 | 0.9 | 2.6 | 1.6 | 1.3 | 92.6 | 79.2 |
| 4 | 99.4 | 99.2 | 88.4 | 96.8 | 95.2 | 96.5 | 0.1 | 0.3 | 0 | 0 | 84.2 | 91.0 |
| 5 | 99.2 | 97.6 | 88.8 | 50.2 | 98.1 | 95.3 | 0 | 0 | 1.7 | 0 | 80.9 | 45.6 |
| Mean | 97.6 | 97.6 | 94.7 | 81.0 | 91.7 | 86.5 | 7 | 1.6 | 1.3 | 0.9 | 83.3 | 70.1 |
| HIV positive | 94.8 | 97.8 | 94.5 | 92.8 | 95.1 | 94.7 | 0.8 | 1.3 | 1.2 | 0.7 | 89.8 | 78.8 |
RA+ and RO+, CD45RA+ and CD45RO+.
HIV sequences in DNA from each of the purified lymphocyte subsets of the 16 study subjects was quantified by limiting dilution (Table 1; Fig. 2B and C). HIV-1 proviral DNA was detected in the CD4 naive and CD4 memory subsets of all patients studied (between 55 and 4,307 provirus copies per 106 cells) except in the CD4 memory subset of p15 (Table 1). The frequency of infection and proportion of virus load contributed by naive and memory subsets of CD4 lymphocytes varied with absolute CD4 count. CD4 naive lymphocytes contributed the majority of infected cells in those with high CD4 counts (r = 0.703, P = 0.02), while memory cells were predominantly infected in individuals with low CD4 counts (r = −0.450, P = 0.08).
HIV DNA was also detected in nine samples of separated CD8+ CD45RA+ lymphocytes, and in three CD8+ CD45RO+ subsets from 10 of the 16 individuals. The contribution of contaminating CD4 lymphocytes to the measured virus loads was small or insignificant in 10 of these samples (Table 1), and these were considered to represent infection of CD8 lymphocytes. In eight individuals, HIV preferentially infected the naive subset of CD8 T lymphocytes, with proviral frequencies ranging from 25 to and 1,440 provirus copies per 106 cells). There was a tendency for higher frequencies of infected CD8+ CD45RA+ cells to be found in individuals with lower CD4 counts, although this difference did not reach statistical significance (P = 0.286). Frequencies of HIV-infected CD8 lymphocytes did not correlate with disease status, risk group, antiviral therapy, or total CD4 lymphocyte counts. There was no correlation between frequency of infection of CD8 cells with either CD4 subset.
Effect of antiviral therapy on CD4 and CD8 lymphocyte infection.
CD4 and CD8 naive and memory lymphocyte subsets were separated from pretreatment and sequential posttreatment samples collected from five individuals receiving combination antiviral treatment and then analyzed for proviral sequences (Table 5). Treated individuals showed rises in circulating CD4 counts and reductions in circulating viremia. Despite the effectiveness of the antiviral therapy, the distribution and frequency of infected cells in each of the subsets remained relatively stable, in all cases remaining within a ±1-log10 range of the levels detected in the initial samples. Infection of CD8 lymphocytes remained detectable in the three individuals whose first samples were positive, while the two individuals with negative CD8 lymphocytes in the pretreatment samples remained negative during treatment. There was evidence for a decline in the frequency of infected CD4+ CD45RA+ and CD4+ CD45RO+ lymphocytes over the course of treatment (r = −0.428; P = 0.098 and r = 0.545; P = 0.029 respectively). A similar decrease in the frequencies of infected CD8+ CD45RA+ cells was not observed, although fewer observations were made (r = 0.070; P = 0.870).
TABLE 5.
Infection of lymphocyte subsets of study subjects receiving combination antiviral therapy
| Patient | Therapya | Time (days) since start of treatment | CD4 count/μl | Circulating plasma virus load (genome eq of RNA/ml)b | Proviral copies/106 cells
|
|||
|---|---|---|---|---|---|---|---|---|
| CD4+
|
CD8+
|
|||||||
| CD45RA+ | CD45RO+ | CD45RA+ | CD45RO+ | |||||
| p1 | Pre | 0 | 10 | 14,000 | 98 | 834 | Neg | Neg |
| + | 27 | 150 | 1,700 | 34 | 239 | Neg | Neg | |
| p9 | Pre | 0 | 283 | 750,000 | 575 | 2,321 | 40 | Neg |
| + | 48 | 3,000 | 72 | 1,807 | 215 | Neg | ||
| p10 | Pre | 0 | 301 | 81,540 | ||||
| + | 49 | 7 | Neg | 1,307 | 464 | 622 | Neg | |
| + | 62 | 466 | Neg | 533 | 533 | 697 | Neg | |
| + | 100 | Neg | 878 | 364 | 1,049 | Neg | ||
| + | 115 | ± | 667 | 577 | 2,764 | Neg | ||
| + | 154 | 428 | ± | 1,498 | 1,390 | 489 | Neg | |
| Stop | 360 | 349 | 16,000 | 734 | 2,884 | 435 | Neg | |
| p12 | Pre | 0 | 329 | 64,680 | 55 | 115 | Neg | Neg |
| + | 14 | 477 | Neg | 15 | 188 | Neg | Neg | |
| + | 41 | 540 | Neg | 19 | 119 | Neg | Neg | |
| + | 100 | 442 | Neg | 9 | 48 | Neg | Neg | |
| + | 340 | 207 | Neg | 18 | 56 | Neg | Neg | |
| p14 | Pre | 0 | 360 | Neg | 533 | 980 | 119 | Neg |
| + | 27 | 418 | Neg | 158 | 1,307 | 203 | Neg | |
| + | 153 | 465 | Neg | 351 | 183 | 144 | Neg | |
Combination treatment over study period. Pre, sample collected pretreatment (tested in Table 1); +, sample affected while on treatment; stop, treatment stopped because of side effects.
Neg, <400 genome eq/ml; ±, equivocal result.
Sequence comparison of HIV infecting different lymphocyte subsets.
To investigate the possible genetic partitioning and differences in predicted phenotype of HIV infecting CD4 and CD8 lymphocytes, proviral sequences of the V3 hypervariable region of env were compared (Fig. 3 and 4). From seven study subjects, approximately 10 clones derived from amplified DNA extracted from the separated lymphocyte subsets were sequenced. The combined set of V3 sequences from each individual were monophyletic and distinct from those of previously published V3 region sequences (data not shown).
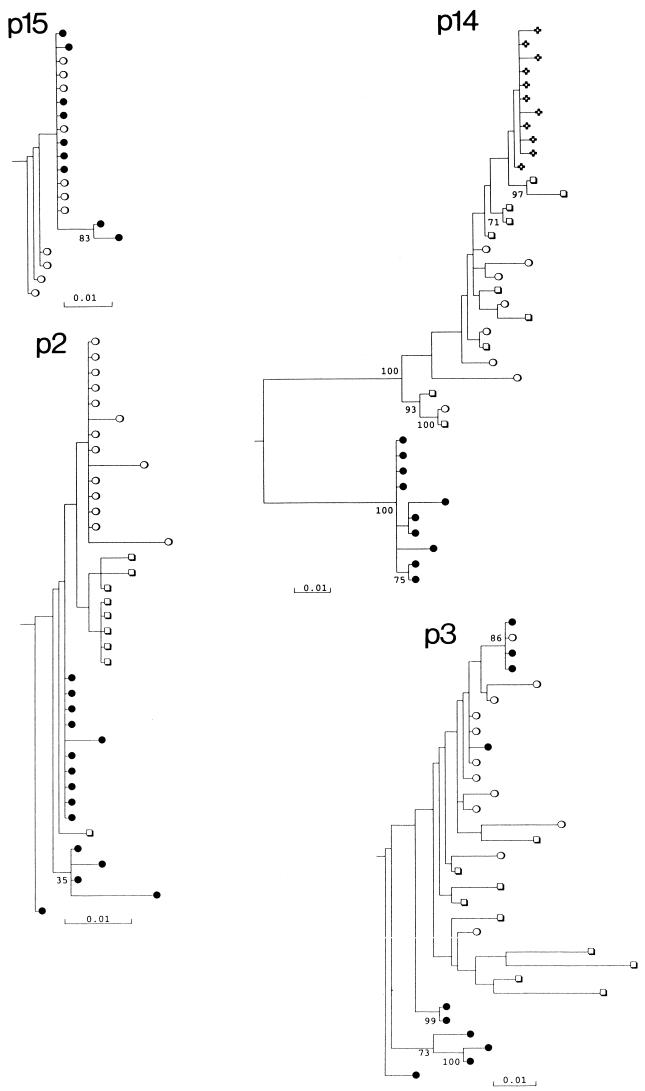
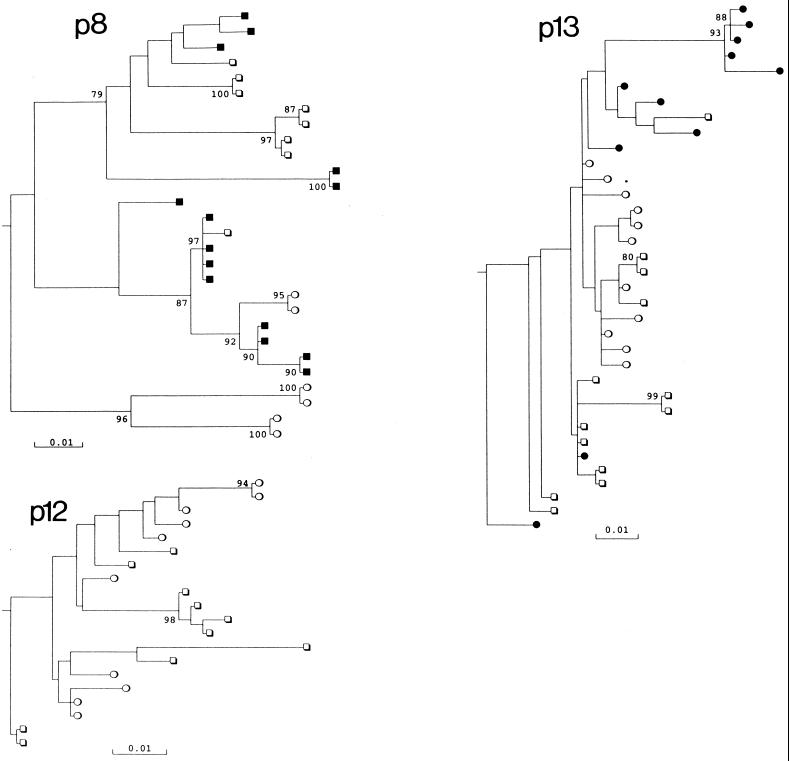
FIG. 3.
Phylogenetic analysis of V3 region sequences from different lymphocyte subsets of seven study subjects. Symbols: ●, CD8+ CD45RA+ (naive) cells; ○, CD4+ CD45RA+ cells; ■, CD8+ CD45RO+ cells; □, CD4+ CD45RO+ cells; ✜, cDNA sequences from plasma (p14). Trees were rooted using the HIV-1MN sequence; a scale bar indicated at the bottom. Clades with >70% bootstrap supported are indicated by numbers on branches.
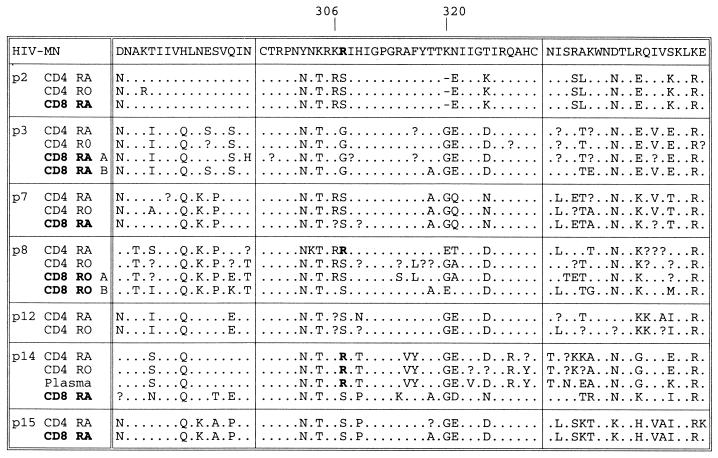
FIG. 4.
Consensus V3 and flanking region sequences of lymphocyte subsets from seven study subjects. Separate consensus sequences were calculated for the two phylogenetic groupings of CD8 sequences from p3 and p8. Amino acid residues are numbered according to HIV-1HXB2; positions 306 and 320 are indicated; basic residues are indicated in bold. Symbols: ., sequence identity with HIV-1MN; ?, variable sites without 75% majority consensus residue.
Sequence diversity within lymphocyte subsets and the degree of partitioning between them varied considerably between the study subjects. V3 sequences from the naive CD4 and CD8 subsets of p15 grouped together and showed mean diversities of 0.002 to 0.0064, similar to the mean pairwise distance between subsets (0.0071). In marked contrast, sequences from CD8 lymphocytes of p14 were distinct from all other cell types, forming a separate clade supported in 100% of bootstrap resamplings. In this individual, the mean pairwise distance between the CD8 and CD4 lymphocytes ranged from 0.113 (CD4+ CD45RA+) to 0.119 (CD4+ CD45RO+), much greater than the diversity within the CD8 population (0.0076). There was no correlation between sequence diversity in any of the subsets with disease progression, nor was there correlation with proviral load (data not shown).
Among the six study subjects from whom CD8 lymphocyte sequences were obtained, five showed either partial or complete separation from CD4 virus populations. In contrast, only one of six study subjects showed any evidence for separation of CD4 naive and memory subsets (p8) (Fig. 4). Most sequences contained neutral or acidic residues at positions 306 and 320. However, sequences of certain subsets of lymphocytes from p8 (CD4+ CD45RA+) and p14 (CD4+ lymphocytes and plasma) contained arginine residues at position 306 along with several other amino acid changes in V3 and flanking regions. These sequences formed phylogenetically distinct clades that grouped separately from variants found in other subsets.
DISCUSSION
The use of magnetic bead separation provides an effective method to isolate subsets of lymphocytes that can be assayed for HIV infection by conventional PCR. One of the problems with the technique when used to investigate infection of CD8 lymphocytes is the possibility that even low-level contamination by CD4 lymphocytes may produce false positive results. To verify that CD8 lymphocytes were infected, we combined purity measurements with calculations of the contribution to proviral load of contaminating CD4 lymphocytes. Using positive selection methods to isolate CD4 and CD8 lymphocytes, we demonstrated that CD4 contamination could not account for the proviral load detected in the separated CD8 lymphocytes from at least five of the seven samples tested (Table 3). A similar conclusion was drawn from comparison of frequencies of infected CD45RA+ and CD45RO+ CD8 lymphocytes with measurements of CD4 contamination (Table 1). The purity of T cells isolated by the pan-T negative selection method and in particular the lack of significant monocyte contamination (Table 2) provided evidence against a significant contribution to proviral load by other cell types in the PBMC population.
Further evidence was provided by lack of a correlation between frequencies of infected CD4 lymphocytes and detection of proviral sequences in either the CD3+, CD8+, CD8+ CD45RA+ or CD3+, CD8+, CD8+ CD45RO+ populations (Tables 1 and 3). For example, the sample with the highest frequency of infected CD4 lymphocytes (p4) contained undetectable frequencies of CD8 infection; similarly, the sample with the highest CD8 proviral load contained relative low frequencies of infection of both subsets of CD4 cells. Third, although there was considerable intersubject variability in the detection of CD8 lymphocyte infection, longitudinal sampling of the same individual over time revealed remarkable stability in proviral loads in each of the lymphocyte subsets. For example, pretreatment samples from p1 and p12 contained undetectable frequencies of infected CD8 lymphocytes, while samples subsequently remained negative. The remaining individuals with infected CD8 lymphocytes pretreatment remained PCR positive subsequently. The consistency of provirus detection in the lymphocyte subset argues against sporadic contamination by CD4 lymphocytes as a reason for our observations. Finally, CD4 contamination would not account for the frequent genetic differences observed in the V3 hypervariable regions between variants recovered from CD8 with those from the CD45RA+ and CD45RO+ subpopulations of CD4 lymphocytes (Fig. 3).
In this study, expression of the RA and RO isoforms of CD45 was used to select between two functionally different subsets of T lymphocytes (naive and memory /effector, respectively) in the peripheral circulation. The separation method had to be modified to avoid potential cellular activation and resulting change in CD45 expression that may occur during the two rounds of positive selection required by the original method (positive selection for CD8 or CD4, followed by bead removal and positive selection by CD45RA or CD45RO monoclonal antibodies). Evidence for the effectiveness of the second method is provided by the similarity in the combined frequencies of infected CD45RA+ and CD45RO+ CD8 and CD4 cells with those measured in CD8 and CD4 lymphocytes separated by positive selection in this (Fig. 2) and previous (31) studies. It is unlikely that CD8 lymphocytes were significantly contaminated with CD4 lymphocytes with Nef-induced down-regulation of CD4 expression. It has been long established that the proportion of provirus-positive PBMCs that are actively infected with HIV is extremely low. PBMCs expressing HIV mRNAs detectable by in situ hybridization were undetectable or present only at very low frequencies (16), frequencies considerably adrift from the numbers of PBMCs containing proviral DNA sequences (42, 44). More recently, quantitative PCR methods to detect multiply spliced mRNA transcripts (from which nef is translated) have been developed. They have confirmed that few, if any, cells in the peripheral circulation contain transcriptionally active HIV (2). In a variety of HIV-infected individuals, frequencies of multiply spliced mRNA ranged from 0 to 700 copies per 2 × 105 PBMCs. As a productively infected cell expresses at least 100 to 1,000 spliced transcripts, the expression detected in vivo can be accounted for by a few or even single virus-expressing cells in the sample. These frequencies are considerably adrift from the numbers of provirus-positive cells detected in the CD8 fraction purified by negative selection (Table 1).
Quantitation of proviral sequences in these two subsets indicated that the CD45RA+ (naive) subset was infected with HIV-1 in vivo, in contrast to the distribution of proviral sequences in the RO and RA subsets of CD4+ lymphocytes. Separate identification of HIV-1 infection in naive and memory/effector cells provides information on the time of infection of T cells relative to maturation and activation, which in turn provides indirect information on the likely mechanisms underlying infection of peripheral CD8 lymphocytes. While expression of CD45RA and CD45RO has been the most commonly used method for separating naive and memory/effector cells, expression of other markers in combination with CD45 has been used to more strictly define functionally distinct subsets. Markers of cellular activation such as CD38 expression and down-regulation of CD27 and CD28 have been used to identify effector from memory and naive CD8+ lymphocytes (8, 14, 15, 20, 35). Expression of CD62L in combination with CD45RA has been used to define purer populations of naive CD4 cells, as there is evidence that a proportion of memory CD4 cells of HIV-positive individuals express CD45RA (36, 39). Whether abnormal expression of CD45RA on the CD8 lymphocytes of HIV-positive lymphocytes also occurs has not been determined. Recently it has been reported that the isoform of CD45 expressed on memory cells may ultimately revert to RA, potentially producing a subset of CD45RA+ cells functionally distinct from the naive phenotype (3).
While it is possible that a proportion of the CD45RA+ CD4 and CD8 populations analyzed in our study may have originally been memory cells, the predominance of HIV-1 infection in the CD8 CD45RA+ subset argues empirically that the main target CD8 lymphocyte is the naive subpopulation. For example, it is unlikely that the infected CD8 cells detected are memory revertants, as it would be likely that a similar frequency of nonrevertant CD45RO+ lymphocytes would also be infected.
Infection of naive cells has been documented extensively in CD4 lymphocytes (36, 41, 45) and is confirmed in this study, although in contrast to our findings, frequencies of infected CD45RA+ have been reported to be lower than in the CD45RO+ effector/memory population. The latter cells are considered to be the main contributors to the pool of infected lymphocytes in vivo, where cellular activation on antigenic stimulation produces cells susceptible to infection and destruction by HIV-1 (9, 11, 48, 52, 53). Indeed, productive infection of naive CD4 lymphocytes in vitro by laboratory isolates of HIV-1 is demonstrably inefficient, most likely through lack of T-cell activation necessary for efficient reverse transcription and integration of the HIV genome after entry (9, 11, 39, 46, 48, 52, 53), and because they do not express the CCR5 chemokine coreceptor required for the entry of primary (non-syncytium-inducing) variants (5, 34). Hypotheses developed to account for the infection of naive CD4 lymphocytes may also be relevant for the infection of naive CD8 lymphocytes in vivo reported here.
There are several potential mechanisms for the infection of CD8 lymphocytes, although most would not predict the preferential distribution of infection in the naive subset. Peripheral CD8 lymphocytes express low levels of CD4 on mitogenic and antigenic stimulation in vitro, and it has been hypothesized that this allows productive infection of CD8 lymphocytes in vivo to occur (12, 25, 51). This hypothesis is supported by our recent finding that CD4 expression can be detected on the CD69+, activated subpopulation of CD8 lymphocytes in PBMCs of both HIV-negative and HIV-positive individuals (S. Imlach et al., unpublished data), but the requirement for cellular activation suggests that the CD45RO+ population would be the main reservoir of infected CD8 lymphocytes. The absence of any measurable excess of incomplete proviral transcripts in CD8 lymphocytes (Table 3) provides further evidence against active infection of this subset.
It is possible that genetic variants of HIV-1 evolve during persistent infection to infect cells by a non-CD4-dependent mechanism; high levels of virus replication and extensive depletion of the CD4 target population may contribute to this switch. While phenotypic characterization of HIV variants infecting CD4 and CD8 lymphocytes would clearly be of value, and the genetic differences observed in V3 provide evidence for a genetic difference between the CD4 and CD8 populations, CD4-independent infection would likely remain cell cycle-dependent, or perhaps become more so if based on expression of high levels of coreceptors, and again would preferentially target the memory/effector CD8 cell population.
HIV-1 infection of CD4+ CD8+ immature thymocytes destined to become CD8 lymphocytes during thymic maturation (26, 28) would provide both a plausible mechanistic explanation for their infection, and would also explain the presence of proviral sequences in the naive subsets of both CD4 and CD8 lymphocytes in peripheral blood. The thymus represents a major target for HIV-1 infection in vivo, and destruction of thymopoietic areas is observed on autopsy examination of AIDS cases. Destruction of CD8 precursor cells would also explain the eventual failure of CD8 homeostasis and decline in circulating numbers of first naive and then memory CD8 lymphocytes on disease progression (37, 38), as well as the recovery in numbers of naive CD8+ (and CD4+) lymphocytes generally observed after commencement of antiviral treatment (1, 6). Without biopsy material, it was not possible to directly observe the replication of HIV within thymocytes. Indeed, proviral sequences in naive cells in peripheral blood can be detected only in circumstances where thymocytes survive infection with HIV and mature into CD4 and CD8 lymphocytes. Survival of infected cells may occur because the infecting virus was defective or because the provirus integrated into a site in the host chromosome that prevented transcription. As documented for naive CD4 lymphocytes in the peripheral circulation, where frequencies of provirus-positive cells were approximately 100-fold greater than infectivity titers (36), it is likely that the vast majority of integrated proviral sequences detected in naive CD8 lymphocytes would also be defective. The presence of culturable virus from this subset therefore provides little information on the degree of CD8 destruction that occurs in the thymus.
Evidence for the quiescent nature of HIV-1 infection in CD8 naive lymphocytes, consistent with thymic infection, is provided by the observation of equivalence in proviral load in CD8 lymphocytes between the pan-LTR and C-LTR primers (Table 3), indicating that HIV proviruses were complete and potentially stably integrated into the cellular genome. Second, we observed a great stability in frequencies of infected CD8 population on antiretroviral therapy; combination treatment achieved a complete clearance of circulating viremia in the majority of study subjects (Table 5), while over the period of virus suppression ranging from 1 to 11 months, there was no consistent reduction in frequencies of infected CD8 lymphocytes. Over the same period, frequencies of infected naive and memory subsets of CD4 lymphocytes showed a modest decline, consistent with previous kinetic studies reporting half-lives of proviral sequences from 21 to 58 weeks (7, 22, 23, 50).
Sequences recovered from the CD8 lymphocytes were frequently distinct from those from CD4 lymphocytes, whereas the CD45RA+ and CD45RO+ subsets of the latter were generally undifferentiated from each other. Although there was great individual variation in the sequence relationships between different cell types, CD8 lymphocytes of p14 retained V3 sequences that lacked positively charged amino acid residues associated with a CXCR4-dependent phenotype (10, 13), when both subsets of CD4 lymphocytes and the plasma population contained variants with arginine residues at these sites and which were likely to have replaced the original population. This indicates a slower turnover of the CD8 population in this individual. Similar differences in rates of cell turnover may therefore underlie the genetic differences between CD4 and CD8 lymphocytes in other study subjects.
In summary, the evidence for infection of the CD45RA+ population of peripheral CD8 lymphocytes provides the basis for testing a number of competing theories for the mechanism CD8 lymphocyte infection in vivo. In the future, phenotypic characterization of variants infecting the two cell types should indicate whether CD4-independent entry of HIV-1 can occur. Further studies on the turnover of HIV-1-infected CD8 lymphocytes in vivo, such as the acquisition of antiviral resistance during treatment relapse, will provide information on the dynamics and consequences of CD8 infection and potentially on the mechanisms underlying the loss of CD8 lymphocytes on disease progression.
ACKNOWLEDGMENTS
We thank staff at the Genitourinary Medicine Clinic, Royal Infirmary of Edinburgh, and the Regional Infectious Diseases Unit, Western General Hospital, for collection of samples from the study subjects.
Grant support for this study was provided by the Medical Research Council (G9632414).
REFERENCES
- 1.Arno A, Ruiz L, Juan M, Zayat M K, Puig T, Balague M, Romeu J, Pujol R, OBrien W A, Clotet B. Impact on the immune system of undetectable plasma HIV-1 RNA for more than 2 years. AIDS. 1998;12:697–704. doi: 10.1097/00002030-199807000-00005. [DOI] [PubMed] [Google Scholar]
- 2.Bagnarelli P, Valenza A, Menzo S, Sampaolesi R, Varaldo P E, Butini L, Montroni M, Perno C F, Aquaro S, Mathez D, Leibowitch J, Balotta C, Clementi M. Dynamics and modulation of human immunodeficiency virus type 1 transcripts in vitro and in vivo. J Virol. 1996;70:7603–7613. doi: 10.1128/jvi.70.11.7603-7613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell E B, Sparshott S M, Bunce C. CD4+ T-cell memory, CD45R subsets and the persistence of antigen—a unifying concept. Immunol Today. 1998;19:60–64. doi: 10.1016/s0167-5699(97)01211-5. [DOI] [PubMed] [Google Scholar]
- 4.Blanpain C, Migeotte I, Lee B, Vakili J, Doranz B J, Govaerts C, Vassart G, Doms R W, Parmentier M. CCR5 binds multiple CC-chemokines: MCP-3 acts as a natural antagonist. Blood. 1999;94:1899–1905. [PubMed] [Google Scholar]
- 5.Bleul C C, Wu L J, Hoxie J A, Springer T A, Mackay C R. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohler T, Walcher J, Holzl-Wenig G, Geiss M, Buchholz B, Linde R, Debatin K M. Early effects of antiretroviral combination therapy on activation, apoptosis and regeneration of T cells in HIV-1-infected children and adolescents. AIDS. 1999;13:779–789. doi: 10.1097/00002030-199905070-00006. [DOI] [PubMed] [Google Scholar]
- 7.Bruisten S M, Reiss P, Loeliger A E, VanSwieten P, Schuurman R, Boucher C A B, Weverling G J, Huisman J G. Cellular proviral HIV type 1 DNA load persists after long-term RT-inhibitor therapy in HIV type 1 infected persons—short communication. AIDS Res Hum Retroviruses. 1998;14:1053–1058. doi: 10.1089/aid.1998.14.1053. [DOI] [PubMed] [Google Scholar]
- 8.Burgisser P, Hammann C, Kaufmann D, Battegay M, Rutschmann O T. Expression of CD28 and CD38 by CD8+ T lymphocytes in HIV-1 infection correlates with markers of disease severity and changes towards normalization under treatment. Clin Exp Immunol. 1999;115:458–463. doi: 10.1046/j.1365-2249.1999.00818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chun T W, Chadwick K, Margolick J, Siliciano R F. Differential susceptibility of naive and memory CD4+ T cells to the cytopathic effects of infection with human immunodeficiency virus type 1 strain LAI. J Virol. 1997;71:4436–4444. doi: 10.1128/jvi.71.6.4436-4444.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Jong J J, de Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol. 1992;66:6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farnet C M, Bushman F D. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 12.Flamand L, Crowley R W, Lusso P, Colombini-Hatch S, Margolis D M, Gallo R C. Activation of CD8+ T lymphocytes through the T cell receptor turns on CD4 gene expression: implications for HIV pathogenesis. Proc Natl Acad Sci USA. 1998;95:3111–3116. doi: 10.1073/pnas.95.6.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouchier R A M, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3138–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giorgi J V, Ho H N, Hirji K, Chou C C, Hultin L E, Orourke S, Park L, Margolick J B, Ferbas J, Phair J P, Saah A J, Armenian H, Farzadegan H, Graham N, Mcarthur J, Palenicek J, Munoz A, Hoover D, Galai N, Jacobson L P, Piantadosi S, Su S, Bauer K, Chmiel J S, Cohen B, Variakojis D, Wesch J, Wolinsky S M, Detels R, Visscher B R, Chen I, Dudley J, Fahey J L, Martinezmaza O, Nishanian P, Taylor J, Zack J, Rinaldo C R, Becker J T, Gupta P, Ho M, Kingsley L, Schrager L, Kaslow R A, Vanraden M J, Seminara D. CD8+ lymphocyte activation at human immunodeficiency virus type 1 seroconversion: development of HLA-DR+ CD38− CD8+ cells is associated with subsequent stable CD4+ cell levels. J Infect Dis. 1994;170:775–781. doi: 10.1093/infdis/170.4.775. [DOI] [PubMed] [Google Scholar]
- 15.Hamann D, Baars P, Rep M H G, Hooibrink B, Kerkhof-Garde S R, Klein M R, Vanlier R A W. Phenotypic and functional separation of memory and effector human CD8+ T cells. J Exp Med. 1997;186:1407–1418. doi: 10.1084/jem.186.9.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harper M E, Marselle L M, Gallo R C, Wong Staal F. Detection of lymphocytes expressing human T lymphotropic virus type III in lymph nodes and peripheral blood from infected individuals by in situ hybridisation. Proc Natl Acad Sci USA. 1986;83:772–776. doi: 10.1073/pnas.83.3.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haynes B F, Pantaleo G, Fauci A S. Toward an understanding of the correlates of protective immunity to HIV infection. Science. 1996;271:324–328. doi: 10.1126/science.271.5247.324. [DOI] [PubMed] [Google Scholar]
- 18.Helbert M R, Walter J, L'Age J, Beverley P C L. HIV infection of CD45RA+ and CD45RO+ CD4+ cells. Clin Exp Immunol. 1997;107:300–305. doi: 10.1111/j.1365-2249.1997.280-ce1170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herbein G, Mahlknecht U, Batliwalla F, Gregersen P, Pappas T, Butler J, O'Brien W A, Verdin E. Apoptosis of CD8+ T cells is mediated by macrophages through interaction of HIV gp120 with chemokine receptor CXCR4. Nature. 1998;395:189–194. doi: 10.1038/26026. [DOI] [PubMed] [Google Scholar]
- 20.Ho H N, Hultin L E, Mitsuyasu R T, Matud J L, Hausner M A, Bockstoce D, Chou C C, Orourke S, Taylor J M G, Giorgi J V. Circulating HIV-specific CD8+ cytotoxic T cells express CD38 and HLA-DR antigens. J Immunol. 1993;150:3070–3079. [PubMed] [Google Scholar]
- 21.Huang P L, Sun Y T, Chen H C, Kung H F, Leehuang S. Proteolytic fragments of anti-HIV and anti-tumor proteins MAP30 and GAP31 are biologically active. Biochem Biophys Res Commun. 1999;262:615–623. doi: 10.1006/bbrc.1999.1213. [DOI] [PubMed] [Google Scholar]
- 22.Ibanez A, Puig T, Elias J, Clotet B, Ruiz L, Martinez M A. Quantification of integrated and total HIV-1 DNA after long-term highly active antiretroviral therapy in HIV-1-infected patients. AIDS. 1999;13:1045–1049. doi: 10.1097/00002030-199906180-00007. [DOI] [PubMed] [Google Scholar]
- 23.Izopet J, Tamalet C, Pasquier C, Sandres K, Marchou B, Massip P, Puel J. Quantification of HIV-1 proviral DNA by a standardized colorimetric PCR-based assay. J Med Virol. 1998;54:54–59. doi: 10.1002/(sici)1096-9071(199801)54:1<54::aid-jmv8>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 24.Kaufmann G R, Duncombe C, Cunningham P, Beveridge A, Carr A, Sayer D, French M, Cooper D A. Treatment response and durability of a double protease inhibitor therapy with saquinavir and ritonavir in an observational cohort of HIV-1-infected individuals. AIDS. 1998;12:1625–1630. doi: 10.1097/00002030-199813000-00009. [DOI] [PubMed] [Google Scholar]
- 25.Kitchen S G, Korin Y D, Roth M D, Landay A, Zack J A. Costimulation of naive CD8+ lymphocytes induces CD4 expression and allows human immunodeficiency virus type 1 infection. J Virol. 1998;72:9054–9060. doi: 10.1128/jvi.72.11.9054-9060.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitchen S G, Uittenbogaart C H, Zack J A. Mechanism of human immunodeficiency virus type 1 localization in CD4-negative thymocytes: differentiation from a CD4-positive precursor allows productive infection. J Virol. 1997;71:5713–5722. doi: 10.1128/jvi.71.8.5713-5722.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar S, Tamura K, Nei M. MEGA: Molecular Evolutionary Genetics Analysis, version 1.0. University Park, Pa: The Pennsylvania State University; 1993. [Google Scholar]
- 28.Lee S, Goldstein H, Baseler M, Adelsberger J, Golding H. Human immunodeficiency virus type 1 infection of mature CD3hi CD8+ thymocytes. J Virol. 1997;71:6671–6676. doi: 10.1128/jvi.71.9.6671-6676.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy J A. Pathogenesis of human immunodeficiency virus infection. Microbiol Rev. 1993;57:183–289. doi: 10.1128/mr.57.1.183-289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis D E, Tang D S N, Aduoppong A, Schober W, Rodgers J R. Anergy and apoptosis in CD8+ T cells from HIV-infected persons. J Immunol. 1994;153:412–420. [PubMed] [Google Scholar]
- 31.Livingstone W J, Moore M, Innes D, Bell J E, Simmonds P, Whitelaw J, Wyld R, Robertson J R, Brettle R P. Frequent infection of peripheral blood CD8-positive T-lymphocytes with HIV-1. Lancet. 1996;348:649–654. doi: 10.1016/s0140-6736(96)02091-0. [DOI] [PubMed] [Google Scholar]
- 32.McMichael A J, O'Callaghan C A. A new look at T cells. J Exp Med. 1998;187:1367–1371. doi: 10.1084/jem.187.9.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mercure L, Phaneuf D, Wainberg M A. Detection of unintegrated human immunodeficiency virus type-1 DNA in persistently infected CD8+ cells. J Gen Virol. 1993;74:2077–2083. doi: 10.1099/0022-1317-74-10-2077. [DOI] [PubMed] [Google Scholar]
- 34.Mo H M, Monard S, Pollack H, Ip J, Rochford G, Wu L J, Hoxie J, Borkowsky W, Ho D D, Moore J P. Expression patterns of the HIV type 1 coreceptors CCR5 and CXCR4 on CD4+ T cells and monocytes from cord and adult blood. AIDS Res Hum Retroviruses. 1998;14:607–617. doi: 10.1089/aid.1998.14.607. [DOI] [PubMed] [Google Scholar]
- 35.Ogg G S, Jin X, Bonhoeffer S, Moss P, Nowak M A, Monard S, Segal J P, Cao Y, Rowland-Jones S L, Hurley A, Markowitz M, Ho D D, McMichael A J, Nixon D F. Decay kinetics of human immunodeficiency virus-specific effector cytotoxic T lymphocytes after combination antiretroviral therapy. J Virol. 1999;73:797–800. doi: 10.1128/jvi.73.1.797-800.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ostrowski M A, Chun T W, Justement S J, Motola I, Spinelli M A, Adelsberger J, Ehler L A, Mizell S B, Hallahan C W, Fauci A S. Both memory and CD45RA+ CD62L+ naive CD4+ T cells are infected in human immunodeficiency virus type 1-infected individuals. J Virol. 1999;73:6430–6435. doi: 10.1128/jvi.73.8.6430-6435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roederer M. T cell dynamics of immunodeficiency. Nat Med. 1996;1:621–622. doi: 10.1038/nm0795-621. [DOI] [PubMed] [Google Scholar]
- 38.Roederer M, Dubs J G, Anderson M T, Raju P A, Herzenberg L A. CD8 naive T cell counts decrease progressively in HIV-infected adults. J Clin Investig. 1995;95:2061–2066. doi: 10.1172/JCI117892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roederer M, Raju P A, Mitra D K, Herzenberg L A. HIV does not replicate in naive CD4 T cells stimulated with CD3/CD28. J Clin Investig. 1997;99:1555–1564. doi: 10.1172/JCI119318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanchez X, Cousins-Hodges B, Aguilar T, Gosselink P, Lu Z J, Navarro J. Activation of HIV-1 coreceptor (CXCR4) mediates myelosuppression. J Biol Chem. 1997;272:27529–27531. doi: 10.1074/jbc.272.44.27529. [DOI] [PubMed] [Google Scholar]
- 41.Schnittman S M, Lane H C, Greenhouse J, Justement J S, Baseler M, Fauci A S. Preferential infection of CD4+ memory T cells by human immunodeficiency virus type 1: evidence for a role in the selective T-cell functional defects observed in infected individuals. Proc Natl Acad Sci USA. 1990;87:6058–6062. doi: 10.1073/pnas.87.16.6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schnittman S M, Psallidopoulos M C, Lane H C, Thompson L, Baseler M, Massari F, Fox C H, Salzman N P, Fauci A S. The reservoir for HIV-1 in human peripheral blood is a T cell that maintains expression of CD4. Science. 1989;245:305–308. doi: 10.1126/science.2665081. [DOI] [PubMed] [Google Scholar]
- 43.Simmonds P, Balfe P, Ludlam C A, Bishop J O, Leigh Brown A J. Analysis of sequence diversity in hypervariable regions of the external glycoprotein of human immunodeficiency virus type 1. J Virol. 1990;64:5840–5850. doi: 10.1128/jvi.64.12.5840-5850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simmonds P, Balfe P, Peutherer J F, Ludlam C A, Bishop J O, Leigh Brown A J. Human immunodeficiency virus-infected individuals contain provirus in small numbers of peripheral mononuclear cells and at low copy numbers. J Virol. 1990;64:864–872. doi: 10.1128/jvi.64.2.864-872.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sleasman J W, Aleixo L F, Morton A, SkodaSmith S, Goodenow M M. CD4+ memory T cells are the predominant population of HIV-1-infected lymphocytes in neonates and children. AIDS. 1996;10:1477–1484. doi: 10.1097/00002030-199611000-00004. [DOI] [PubMed] [Google Scholar]
- 46.Spina C A, Prince H E, Richman D D. Preferential replication of HIV-1 in the CD45RO memory cell subset of primary CD4 lymphocytes in vitro. J Clin Investig. 1997;99:1774–1785. doi: 10.1172/JCI119342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stanley S K, McCune J M, Kaneshima H, Justement J S, Sullivan M, Boone E, Baseler M, Adelsberger J, Bonyhadi M, Orenstein J, Fox C H, Fauci A S. Human immunodeficiency virus infection of the human thymus and disruption of the thymic microenvironment in the SCID-hu mouse. J Exp Med. 1993;178:1151–1163. doi: 10.1084/jem.178.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stevenson M, Stanwick T L, Dempsey M P, Lamonica C A. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 1990;9:1551–1560. doi: 10.1002/j.1460-2075.1990.tb08274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Su L, Kaneshima I, Bonyhadi M, Salimi S, Kraft D, Rabin L, McCue J M. HIV-1-induced thymocyte depletion is associated with indirect cytopathicity and infection of progenitor cells in vivo. Immunity. 1995;2:25–36. doi: 10.1016/1074-7613(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 50.vanHarmelen J, Wood R, Lambrick M, Rybicki E P, Williamson A L, Williamson C. An association between HIV-1 subtypes and mode of transmission in Cape Town, South Africa. AIDS. 1997;11:81–87. doi: 10.1097/00002030-199701000-00012. [DOI] [PubMed] [Google Scholar]
- 51.Yang L P, Riley J L, Carroll R G, June C H, Hoxie J, Patterson B K, Ohshima Y, Hodes R J, Delespesse G. Productive infection of neonatal CD8+ T lymphocytes by HIV-1. J Exp Med. 1998;187:1139–1144. doi: 10.1084/jem.187.7.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 53.Zack J A, Haislip A M, Krogstad P, Chen I S Y. Incompletely reverse transcribed human immunodeficiency virus type 1 genomes in quiescent cells can function as intermediates in the retroviral life cycle. J Virol. 1992;66:1717–1725. doi: 10.1128/jvi.66.3.1717-1725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang L Q, Simmonds P, Ludlam C A, Leigh Brown A J. Detection, quantification and sequencing of HIV-1 from the plasma of seropositive individuals and from factor VIII concentrates. AIDS. 1991;5:675–681. doi: 10.1097/00002030-199106000-00006. [DOI] [PubMed] [Google Scholar]