Abstract
Background
Recent outbreaks of clade 2.3.4.4b highly pathogenic avian influenza (HPAI) H5N1 viruses in regions previously less affected since 2020 have raised global concerns. Implementing mass immunization or ring vaccination in poultry should be a countermeasure ready to contain disease outbreaks. This study focuses on developing a recombinant H5N2 vaccine based on virus-like particles (VLPs) against clade 2.3.4.4c, the predominant HPAI subclade in Taiwan since its emergence, leading to a large outbreak in 2015.
Methods
The study aimed to confirm the effectiveness of clade 2.3.4.4c H5N2 VLPs in protecting chickens and identify the best adjuvants for the VLP vaccine. We used Montanide 71VG-adjuvanted inactivated RG6 to establish the immunization protocol, followed by prime-boost H5N2-VLP immunizations. We compared adjuvants: 71VG, 71VG with VP3, and Alum with VP3. Serum samples were tested for antibodies against homologous vaccine antigens and cross-clade antigens by hemagglutination inhibition (HI) assays. Finally, we evaluated the protective efficacy by lethally challenging immunized chickens with H5 viruses from clade 1 or 2.3.4.4c.
Results
Poultry adjuvant 71VG significantly enhanced antibody responses in chickens with inactivated RG6 compared to unadjuvanted inactivated virus. While increasing antigen dosage enhanced 71VG adjuvanted RG6-induced antibody titers, the vaccine displayed minimal cross-reactivity against locally circulating HPAI H5N2. In contrast, H5N2-VLP containing the HA protein of clade 2.3.4.4c, adjuvanted with (FMDV) VP3 in 71VG, significantly promoted HI antibody responses. All H5N2-VLP immunized chickens survived lethal challenges with the local clade 2.3.4.4c H5 strain.
Conclusion
The study demonstrated the immunogenic potential of the VLP vaccine in chickens. Our findings offer insights for optimizing VLP vaccines, allowing the incorporation of the HA of currently circulating H5 viruses to effectively mitigate the impact of the rapidly evolving clade 2.3.4.4 H5 outbreaks.
Keywords: Highly pathogenic avian influenza (HAPI), H5N2 subtype, Chicken, Poultry vaccination, Virus-like particles
Introduction
Since the emergence of the highly pathogenic avian influenza (HPAI) A (H5) viruses, specifically the A/Goose/Gaundong/01/1996 (Gs/GD) lineage in 1996, they have undergone rapid evolution into ten phylogenetic clades (0 to 9) and multiple subclades [1]. The clade 2.3.4.4 H5 HPAI viruses have caused more than 541 million birds culled as of January 5, 2024, in response to the outbreak [2], [3], [4]. According to the World Health Organization (WHO), as of January 5, 2024, there have been 880 human infections and 460 deaths caused by H5N1 [5]. The onward and extensive transmission of HPAI H5Nx viruses among susceptible avian hosts poses a threat of novel HPAI virus emergence and raises concerns about human pandemic infection. In recent outbreaks, there were more than 3,663 H5 HPAI outbreaks across Europe, Asia, and Africa from 2020 to 2021, and nearly 5,212 H5 HPAI outbreaks from 2022 to 2024 occurred in unvaccinated countries in Europe, Asia, Africa, and North America [6].
Taiwan experienced its largest epidemic of H5 HPAI outbreak in poultry in history in 2015, impacting 944 poultry farms and resulting in the death or culling of more than 4.15 million geese [7], [8]. Given the rapid mutations and global distribution of clade 2.3.4.4 H5 viruses, there is an increasing need to implement mass immunization of poultry, including ring vaccination strategies, to effectively contain the virus's spread. However, significant concerns regarding the implementation of vaccines in poultry include virus escape due to incomplete sterilization of inactivated viral vaccines, viral survival from vaccine-induced herd immunity, and further evolution. Additionally, it is challenging to distinguish antibodies derived from vaccination or natural infection.
To address these concerns, we have developed a recombinant VLP platform that is devoid of the viral genome, convenient for updating the HA gene according to circulating virus strains, and amenable to the mass production of vaccines for a vast number of susceptible birds [9]. Using this flexible platform, conducted at the biosafety level one, we generated an H5N2-VLP vaccine containing the HA gene from the A/Goose/Taiwan/01004/2015 (clade 2.3.4.4c) strain [8]. We assessed its immunogenicity and cross-clade protection in chicken hosts. In addition, we compared the results with chickens immunized with inactivated H5N1 whole virus derived from IBCDC-RG6 (A/Anhui/1/2005), a candidate vaccine virus belonging to clade 2.3.4 [10]. The findings highlight the potential of VLPs as a vaccine platform for rapidly producing epidemic-strain matched vaccines against zoonotic influenza virus infections.
Materials & Methods
Cells and viruses
Epithelial Madin-Darby canine kidney (MDCK) cells (ATCC CCL-34) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10 % fetal bovine serum (FBS) and antibiotics. A monolayer of MDCK cells was infected with AIVs at a multiplicity of infection (MOI) of 0.001 in the presence of TPCK-trypsin (2 μg/mL). The culture supernatant containing the virus was harvested, aliquoted, and stored in DMEM with 10 % DMSO at −80 °C.
RG6 AIV (IBCDC-RG6) is a candidate vaccine virus for clade 2.3.4 H5N1 generated through the reassortment of the HA and NA genes from H5N1 (A/Anhui/1/2005) with PR8 virus using reverse genetics techniques [10]. The HPAI H5 viruses used in this study included clade 1 H5N1 (A/Vietnam/1203/2004), and local H5N2 viruses (A/Goose/Taiwan/GV20018//2016, A/Goose/Taiwan/GV20686/2017, A/Chicken/Taiwan/CV20623/2017, A/Chicken/Taiwan/CV20629/2017) which were isolated from surveillance study in Taiwan.
Phylogenetic analysis
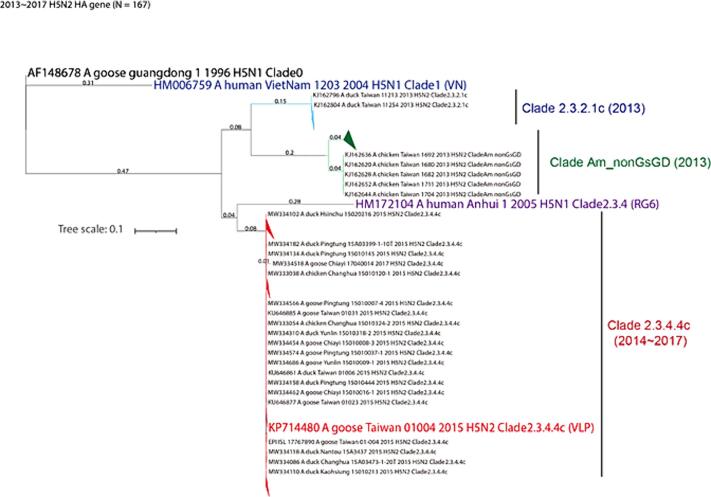
To address the phylogenetic relationship between the H5Nx viruses involved in this study and the H5N2 strains isolated in Taiwan during 2013–2017 (total of 164 strains), we retrieved the HA sequences from National Center for Biotechnology Information Influenza Virus Database (NCBI-IVD) [11] and Global Initiative on Sharing All Influenza Data (GISAID-Epiflu) [12]. Downloaded sequence information was processed by our previously developed software packages, FluConvert and IniFlu, for sequence alignment and clustering [13]. The phylogenetic tree of each of the HA sequences was analyzed using the Maximum Likelihood method based on the Tamura-Nei model, and evolutionary analyses were conducted using MEGA X [14]. The MEGA X program was automatically run by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances. Using the Maximum Composite Likelihood (MCL) approach, we selected the best tree with superior log-likelihood value, measured branch lengths within the number of substitutions per site, and plotted a phylogenetic tree based on these values.
Chicken immunization experiment
One-day-old White Leghorn chickens were purchased from the Animal Health Research Institute or JD-SPF Biotech, Taiwan, and were maintained in a specific-pathogen-free (SPF) facility at the Animal Research Center of National Taiwan University (NTU). At seven days of age, the chickens were vaccinated by subcutaneous injection with 150 μL of the vaccine. A booster dose of the same vaccine (300 μL) was administered either on day 7 (0/7 schedule) or day 14 (0/14 schedule) after the primary immunization. Serum samples were collected weekly for at least four weeks. The animal protocols (no. NTU105-EL-00161 and no. NTU106-EL-00135) were approved by the Institute Animal Care and Use Committee (IACUC) at NTU.
Inactivated RG6 (H5N1) vaccine
The culture supernatant from RG6 virus-infected MDCK cells was treated with 0.01 % formalin at 37 °C overnight, pelleted through ultracentrifugation at 29,000 rpm (103,800 × g) for 1.5 h at 4 °C and resuspended in PBS. The HA unit of the RG6 antigen was determined by hemagglutination of turkey red blood cells (RBCs) in an HA assay. HA contents were measured using single radial immunodiffusion (SRID) utilizing the WHO standard reagents or by densitometry of SDS-PAGE if the standard reagent was unavailable [15]. In this study, 16 HAU of RG6 contained 228 ng of HA protein.
H5N2-VLP vaccine
The H5N2-VLP was produced by a stable transfection clone of the 293F cell line as previously described with modifications [9]. Briefly, the human 293F cell line was stably transfected with a plasmid harboring an EF1a/eIF4g-pCI-TetR-P2A-BSR cassette and a tetracycline-inducible promoter (CMV/TO)-GFP reporter cassette flanked by two heterotypic FLP recombination target sites (FRT and F3) to trap a high-expression genome locus. Single-cell clones were isolated and characterized as clones with a single-copy reporter as founder cell lines. A founder cell line was selected for further transfection with FLPe flippase and the donor plasmid to swap GFP carrying the FRT/F3-flanked H5N2-VLPs gene cluster, comprising CMV/TO driven H5, N2, and M1-IRES-M2 genes linked in tandem. The HA, NA, and M genes were from clade 2.3.4.4c H5N2 influenza A virus (A/goose/Taiwan/01004/2015). Gene-swapped cells were enriched based on loss-of-GFP after doxycycline induction. Single-cell clones were isolated and characterized for their inducible expression of VLP genes and secretion of H5N2-VLP in the induced conditioned medium. VLP-producer cells were scaled up in suspension culture and induced by adding 1 µg/mL doxycycline in the medium. H5N2-VLP were harvested from the conditioned medium and purified by sucrose density gradient (30 % and then 40 %-60 %) ultracentrifugation. Purified H5N2-VLP (batch-1 and batch-2) were analyzed for their hemagglutination activity. The content of HA antigen in each batch of the H5N2-VLP was determined by ELISA using HA-specific monoclonal antibody F10, a gift from Dr. An-Suei Yang at Academia Sinica, Taiwan. Each dose of the H5N2-VLP vaccine was equivalent to 16 HAU (containing 30 ng of HA protein) or 64 HAU (containing 120 ng of HA protein) by HA assay [16], [17].
Vaccine adjuvant
The Montanide SeppicTM adjuvant 71VG (71VG) is a mineral oil-based adjuvant widely used in veterinary emulsion vaccines [18], [19]. A vaccine formulation was developed by combining 70 % of the adjuvant 71VG with 30 % of the vaccine antigen and emulsified by sonication before vaccination. VP3, a structural protein of the foot-and-mouth disease virus (FMDV), has been shown to exhibit agonistic properties toward TLR2/TLR1 and TLR2/TLR6 [20]. Our previous chicken vaccination observed that combining VP3 (utilizing a range of 25–75 μg) with Alum adjuvant (2 % Alhydrogel, Invivogen) resulted in the most significant immune-enhancing effects and optimal efficacy.
Study design of VLP immunizations
Aluminum salts are commonly used in vaccine formulations, including approved VLP vaccines [21]. Combining pattern recognition receptor (PRR) agonists with aluminum hydroxide has been shown to enhance the immune response in VLP vaccines [22], [23], [24]. Our previous research demonstrated that VLPs adjuvanted with Alum and combined with the FMDV VP3 structural protein significantly protected chickens from lethal challenges (unpublished results). In this study, our objective was not only to confirm the effectiveness of VLPs in protecting poultry but also to investigate how the use of appropriate adjuvants could reduce the required antigen dose without compromising the vaccine's protective effect. This approach has the potential to lower the manufacturing cost of recombinant VLP vaccines, making them more accessible to poultry farms.
The commercially available poultry adjuvant 71VG had never been tested for its immune-enhancing effects in VLP vaccines for chicken immunization. To establish the chicken immunization protocol, we first immunized chickens with 71VG-adjuvanted inactivated RG6 to demonstrate the antigen dosage-dependent antibody-enhancing effects of 71VG in chickens. Since the primary goal of our study was to investigate whether combining the appropriate amount of VP3 with the 71VG formulation could reduce the antigen dosage while achieving superior clinical benefits, we did not design the study to compare adjuvanted VLP vaccine with an unadjuvanted control. Instead, we focused on comparing the adjuvant groups 71VG, 71VG+VP3, and Alum + VP3 to evaluate their respective impacts on VLP vaccine-induced antibody responses.
Challenge experiment
The immunized chickens were infected with 10 LD50 of the HAPI Clade 1 A/Vietnam/1194/2004 H5N1 virus or infected with 10 LD50 of the HAPI local H5N2/GV20686/2017 H5N2 virus. Viral shedding in fecal samples was confirmed by measuring viral plaques in MDCK cells. Additionally, the number of days each animal survived was recorded for 9 days for further analysis. The experiments were conducted in a BSL3 laboratory at the Genomics Research Center at Academia Sinica (AS_IACUC Protocol no. 15–02-819).
Hemagglutination (HA) and hemagglutination inhibition (HI) assays
HA and HI assays used washed turkey RBCs. In the HA assay, 50 μl of a 2-fold serially diluted virus solution was combined with an equal volume of turkey RBCs (4 × 107 cells/mL) in a V-bottom 96-well microtiter plate. After 30 min incubation, the highest virus dilution that resulted in visible hemagglutination was recorded as 1 HAU. A standard 512 HAU of inactivated RG6 supernatant was used as a positive control, while turkey RBCs with PBS alone were the negative control.
In the HI assays, chicken serum samples were pre-treated with a receptor-destroying enzyme (Sigma) overnight, using a volume ratio of 1:3, followed by heat inactivation at 56 °C for 30 min. Serially 2-fold diluted serum samples were incubated with 8 HAU of inactivated AIV at room temperature for 30 min., followed by another 30 min incubation with Turkey RBCs (50 μl). The highest serum dilution without hemagglutination determined the HI titer.
Statistical analysis
Survival curves after the lethal challenge of HPAI H5 viruses were generated using the Kaplan-Meier method, given that the data is not normally distributed. The Log-Rank test assessed significant differences between the sham-vaccinated and vaccinated groups. The plot and the statistical analysis were performed using GraphPad Prism version 7 software. The null hypothesis of identical survival outcomes was rejected when a p-value was less than 0.05.
Results
Phylogenetic analysis of the circulating H5 viruses for developing the VLP vaccine
The WHO reported significant diversity between the HA genes of clade 2.3.4 and clade 2.3.4.4 viruses in its February 2015 publication. Specifically, the antigenic differences were evident in the poor reactivity of antiserum raised against the reference antigen (A/Anhui/1/2015, clade 2.3.4) with the A(H5N2/N8) and A(H5N6) viruses of clade 2.3.4.4 [25]. The emergence of the HPAI H5N2 clade 2.3.4.4 in Taiwan occurred in late 2014, leading to a significant outbreak in 2015 [8]. To comprehend the phylogenetic relationship of H5N2 viruses in Taiwan between 2013 and 2017 for designing the VLP vaccine, we retrieve HA gene segments from 164 H5N2 for further analysis. Among the 26 strains collected before the 2015 large outbreak (2013–2014), 46 % (N=12) belonged to clade 2.3.2.1c, while 54 % (N=14) were classified as the American lineage non-A/goose/guangdong/1/1996/H5N1 Clade 0 (clade Am_nonGsGD). In contrast, of the total 138 strains collected from 2015 to 2017, only 4.3 % (N=6) were identified as clade Am_nonGsGD, while 95.7 % (N=132) belonged to clade 2.3.4.4c (Fig. 1 and Supplementary Fig. 1). Notably, the nucleotide sequences of the HA gene from H5N2 viruses (Goose/GV20018/2016 and Goose/GV20686/2017) obtained through environmental surveillance were characterized as clade 2.3.4.4c (data not shown). These analyses indicated that HAPI H5N2 clade 2.3.4.4c has become the predominant clade in Taiwan since its emergence in 2015 and persisted into 2017 [26]. Furthermore, these viruses were genetically away from two historical WHO-recommended human influenza H5N1 vaccine viruses (A/Vietnam/1203/2004, clade 1; and A/Anhui/1/2005 RG6, clade 2.3.4) (Fig. 1).
Fig. 1.
Phylogenetic analysis of HA proteins in H5N1 VN, RG6, and selected circulating H5N2 viruses (2013–2017) in Taiwan Maximum likelihood phylogenies of HA genes from H5Nx viruses in this study, including A/VietNam/1203/2004 H5N1 Human CDC RG (in blue), A/AnHui/1/2005 H5N1 Human IBCDC-RG6 (in purple), KP714479 A/goose/Taiwan/01004/2015 4 H5N2 goose VLP (in red) are depicted. This analysis encompasses 164 circulating H5N2 strains to illustrate the phylogenetic relationship of H5N2 viruses in Taiwan before and after the 2015 large outbreak. Bootstrap support and Bayesian posterior clade classifications are indicated in the phylogenetic tree. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Upon aligning the DNA sequences of the H5N2 VLP (H5 of the 2.3.4.4c H5N2 influenza A virus, A/goose/Taiwan/01004/2015) with the RG6 H5N1 strain, we identified 4 nucleotide variations in the signal peptide region, 89 in the HA1 region, and 35 in the HA2 region. These variations led to changes in amino acid residues: 2 in the signal peptide, 37 in the HA1 region, and 3 in the HA2 region, as detailed in the supplementary Table 1. As was shown in Fig. 3, serum samples from RG6 vaccine-immunized chickens exhibited limited cross-reactivity against HA antigens of local H5N2 viruses. Given the fundamental importance of antigenic matching for vaccine efficacy [27], we chose the HA gene of influenza A/goose/Taiwan/01004/2015 (H5N2) to design the H5N2-VLP vaccines (see Fig. 1).
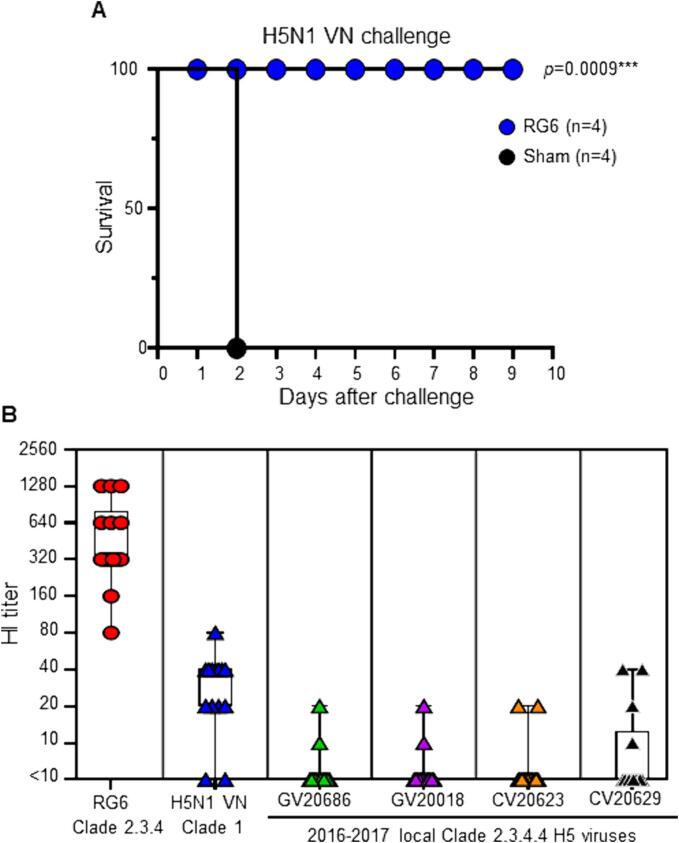
Fig. 3.
Two doses of 71VG-adjuvanted RG6 confer protection against H5N1 VN clade 1 virus but exhibit limited cross-reactivity against clade 2.3.4.4 H5 viruses Chickens vaccinated with 2 doses of RG6 were challenged with a 10 × LD50 of H5N1 VN (Clade 1) after day 28 post-vaccination. The Kaplan-Meier method presents the survival curves. The significant difference in the survival outcome between sham-vaccinated (black circles) and RG6-vaccinated (blue circles) chickens was assessed by the Log-Rank test (p-value < 0.005). (B) The HI antibody titers against homologous RG6 antigen or against viral antigens from H5N1 VN Clade 1 and GV20686, GV20018, CV20623, and CV20629 (Clade 2.3.4.4c) were obtained for 12 antiserum samples from chickens vaccinated with RG6 (Clade 2.3.4) at day 28. Each symbol represents an individual animal. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Prime-boost immunization of 71VG-adjuvanted inactivated RG6 effectively induces dose-dependent antibody responses in chickens
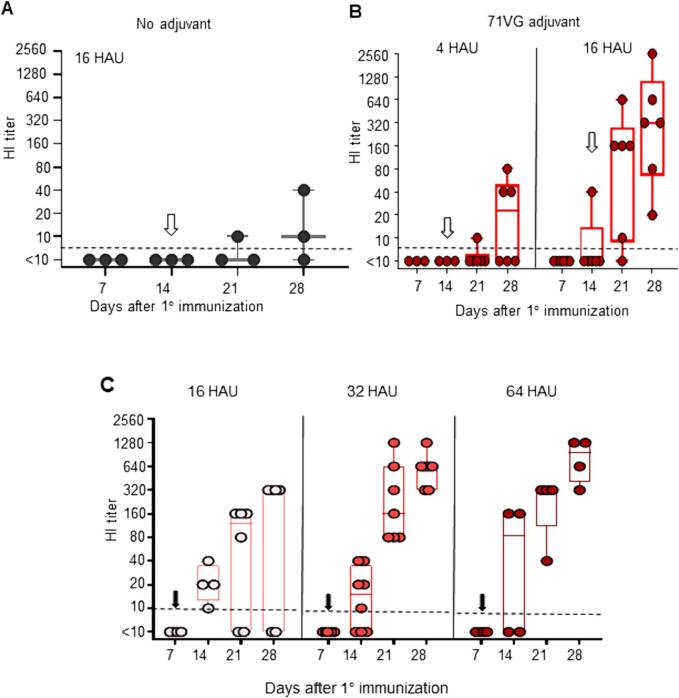
To establish a chicken immunization protocol using 71VG adjuvant, we first immunized chickens with inactivated RG6 by conducting standard prime-boost vaccination (0/14 days) compared to no adjuvant. Prime-boost immunization with two doses of adjuvant-free inactivated RG6 resulted in poor HI antibody responses (Fig. 2A). In contrast, 71VG-adjuvanted RG6 significantly increased HI antibody titers after booster immunization (Fig. 2B). The minimal effective dose of inactivated RG6 was 16 HAU, as vaccination with 4 HAU of 71VG-adjuvanted inactivated RG6 failed to induce a robust antibody response in chickens (Fig. 2B).
Fig. 2.
Prime-boost immunization of 71VG-adjuvanted inactivated RG6 effectively induces dose-dependent antibody responses in chickens Seven-day-old chickens were subcutaneously vaccinated with 16 HAU of inactivated RG6 without adjuvant (A) or adjuvanted with water-in-oil 71VG (B, C) in various amounts of HAU of RG6. After the primary vaccination, the same booster dose was administered on day 14 (indicated by an open arrowhead) (B) or on day 7 (C). Each circle represents an individual animal. HI titers were measured against RG6 antigen. An HI titer of ≥1:10 was defined as seroconversion (dashed line). The results show a pooled analysis of multiple studies employing the same immunization protocol.
Research has shown that the percentage of class-switched IgY-positive cells peaks on day 14 and declines on day 21 [28]. To align the second dose of inactivated RG6 with the time before the rise of immunoglobulin class switching, we changed the booster timing from day 14 to day 7. Early booster time did not enhance the seroconversion rate and the GMTs of HI antibodies in the 16 HAU group (Fig. 2C). In the 32 HAU and 64 HAU groups, seroconversion rates reached 100 % after two weeks of booster immunization. The GMTs on day 28 became 580 in the 32 HAU and 761 in the 64 HAU group (Fig. 2C). These results demonstrated that increasing the antigen dosage is required to achieve a high antibody titer in chicken vaccination when shortening the booster immunization interval.
Two doses of 71VG-adjuvanted RG6 confer protection against H5N1 VN clade 1 virus but exhibit limited cross-reactivity against 2016–2017 epidemic H5 strains
To assess the protective efficacy of inactivated RG6 vaccination in chickens, we conducted challenge experiments using lethal doses (10 × LD50) of H5N1 VN (clade 1). After the challenge infection with H5N1 VN, the age-matched naïve chickens succumbed to the infection, and all died by day 2. However, all of the RG6-vaccinated chickens survived the lethal challenge, demonstrating the potent protective efficacy of RG6 vaccination against clade 1 H5N1 VN (p = 0.0009, Fig. 3A).
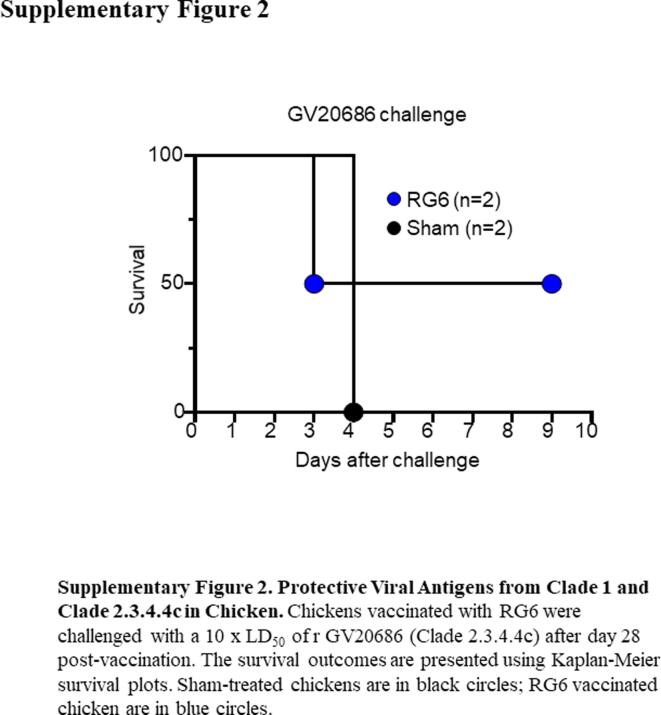
Next, we assessed the cross-reactivity of RG6-immunized chicken serum samples using HI assays against homologous (RG6 antigen) and heterologous antigens from clade 1 H5N1 VN or from local strains (GV20686, GV20018, CV20623, and CV20629), which were isolated after 2015 clade 2.3.4.4c H5 epidemics [8]. RG6-immunized antiserum samples exhibited reactivity against the homologous RG6 antigen as well as against the H5N1 VN antigen. However, they showed poor reactivity against GV20686, GV20018, CV20623, and CV20629 (Fig. 3B). These results indicated that while RG6 vaccination effectively induced antibody responses against the homologous antigen and clade 1 H5N1, their recognition of the clade 2.3.4.4 strains is limited. In parallel, we saved two RG6-vaccinated chickens for the GV20686 challenge. The unvaccinated chickens succumbed to the infection by day 4 post-challenge. Of two RG6 vaccinated chickens, one survived the infection, but the other succumbed to the challenge on day 3 (Supplementary Fig. 1). Although the sample size of the RG6 vaccinated group was small, these results were consistent with the findings on limited HI cross-reactivity against locally circulating H5 antigens observed (Fig. 3B), implicating new vaccines containing closely related epidemic H5 strains were needed.
H5N2-VLP adjuvanted with Alum plus FMDV VP3 or with 71VG is a potential chicken vaccination candidate
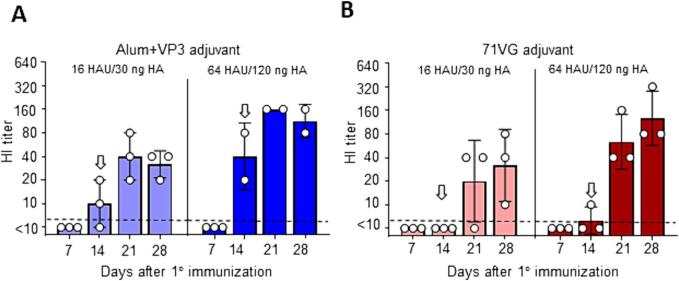
To explore a better vaccine to protect the avian hosts from novel clade 2.3.4.4c H5 viruses, we incorporated the HA of clade 2.3.4.4c H5N2 into the VLP (H5N2-VLP). FMDV VP3, known for its agonistic effect on chicken TLR2 and TLR6, can stimulate higher levels of IFN-γ and IL-18 to induce an enhanced cell-mediated immune response in chickens [20]. To assess the immunogenicity of H5N2-VLP in chickens, we conducted a standard prime-boost vaccination schedule (0/14 days) with the H5N2-VLP, using Alum + VP3 (25 μg) adjuvant and 71VG adjuvant.
In the Alum + VP3 adjuvant group, two doses of 16 HAU of H5N2-VLP effectively induced antibody responses and reached 100 % seroconversion by day 21. Increasing the antigen dosage to 64 HAU stimulated an early and complete seroconversion (100 %) by day 14. Two doses of the 64 HAU vaccines significantly enhanced the GMT to 113 compared to 31.7 with the 16 HAU vaccine on day 28 (Fig. 4A).
Fig. 4.
H5N2-VLP adjuvanted with Alum plus FMDV VP3 or 71VG is a potential candidate for chicken vaccination The immunogenicity of the clade 2.3.4.4c H5N2-VLP vaccine was assessed in chickens. The VLP vaccine was adjuvanted with either Alum and VP3 (25 μg) (A) or 71VG (B) at different doses (16 HAU or 64 HAU). After the primary vaccination, the same booster dose was administered on day 14 (indicated by an open arrow). The figure shows the HI antibody titers induced by each vaccine formulation with different adjuvants and antigen doses. Each circle represents an individual animal. A HI titer of ≥1:10 was defined as seroconversion (dash line).
In the 71VG adjuvant groups, two doses of 64 HAU H5N2-VLP stimulated a faster seroconversion and more prominent antibody responses than 16 HAU (Fig. 4B). Results from these experiments demonstrated the immunogenic potential of H5N2-VLP in chickens with the appropriate adjuvant and dosage.
Diverse impacts of adjuvant and booster strategies on H5N2-VLP vaccination in chickens
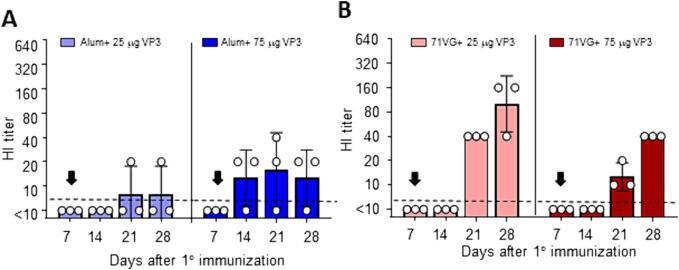
We next evaluated whether chickens gained benefit in generating a better antibody response from an early booster immunization with a reduced amount of the H5N2-VLP (16 HAU). This experiment included four different adjuvant conditions: Alum + 25 μg VP3, Alum + 75 μg VP3, 71VG+25 μg VP3, and 71VG+75 μg VP3. Chickens prime-boosted on day 7 with Alum/VP3-adjuvanted H5N2-VLP exhibited lower antibody titers and seroconversion rates, regardless of the VP3 protein amount (Fig. 5A). In contrast, early booster immunization with 71VG+25 μg VP3 adjuvanted 16 HAU of H5N2-VLP yielded significant benefits, displaying substantially higher HI antibody titers and seroconversion rates than the 71VG adjuvant plus 75 μg VP3 (Fig. 5B). Specifically, the GMTs of HI antibody titers were 40 on day 21 and 101 on day 28 in the 25 μg VP3 group, compared to the GMTs of 13 on day 21 and 40 on day 28 in the 75 μg VP3 group. These findings demonstrated that a prime-boost vaccination strategy with a 7-day interval, using 71VG+25 μg VP3 adjuvanted H5N2-VLP, effectively elicited an optimal antibody response in chickens.
Fig. 5.
Diverse impacts of adjuvant and booster strategies on H5N2-VLP vaccination in chickens Seven-day-old chickens were subcutaneously vaccinated with a minimal dose of the clade 2.3.4.4c H5N2 VLP vaccine, containing 30 ng of HA (16 HAU). The VLP vaccine was combined with either Alum (A) or 71VG (B) adjuvant, along with two different doses (25 μg or 75 μg) of VP3. The booster immunization was also administered on day 7 (indicated by a filled arrow) after the primary immunization instead of day 14. A HI titer of ≥1:10 was defined as seroconversion (dashed line).
H5N2-VLP vaccination induces cross-reactive antibody and protection against clade 2.3.4.4 H5 viruses
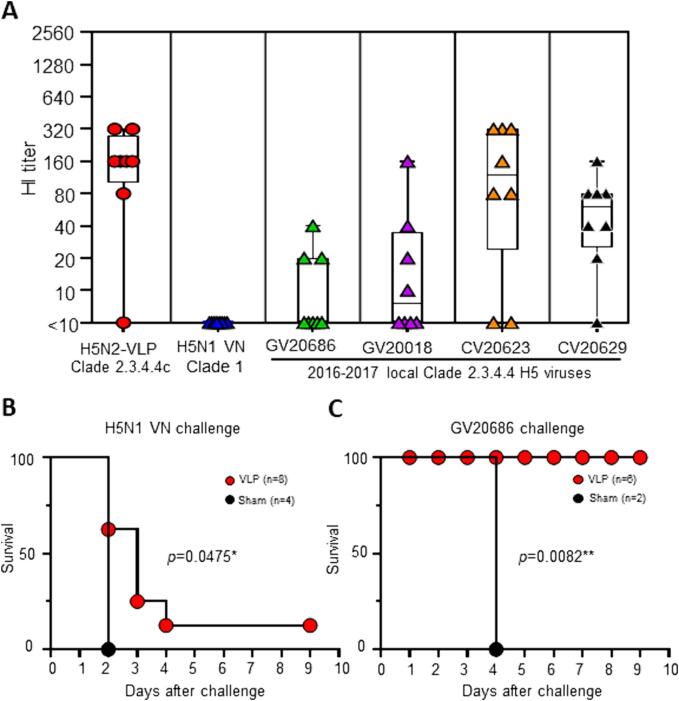
To investigate the potential cross-reactivity of antibodies induced by H5N2-VLP vaccination, we selected serum samples from chickens immunized with a 64 HAU of 71VG/VP3 adjuvanted H5N2-VLP and tested their HI antibody reactivity against antigens from clade 1 H5N1 VN and 2016–2017 local HPAI H5 viruses (clade 2.3.4.4c). H5N2-VLP vaccination effectively induced HI antibodies against the homologous H5N2-VLP antigen. However, none of the antibodies reacted to the H5N1 antigen from clade 1. They exhibited varied reactivity against the 2016–2017 H5 viral antigen. The GMTs of HI antibody titers against CV20623, CV20629, GV20018, and GV20686 were 73, 44, 13, and 9, respectively (Fig. 6A).
Fig. 6.
H5N2-VLP vaccination induces cross-reactive antibody and protection against clade 2.3.4.4c H5 viruses HI titers against viral antigens from H5N1 VN clade 1 and GV20686, GV20018, CV20623, and CV20629 (Clade 2.3.4.4c) were measured for antiserum samples obtained from chickens vaccinated with clade 2.3.4.4c H5N2-VLP containing 120 ng of HA adjuvanted with 71VG+25 μg VP3 at day 28. Each symbol represents an individual animal. (B) Chickens were vaccinated with H5N2-VLP containing 30 ng HA combined with different adjuvant formulations. On day 28, they were pooled and challenged with 10 LD50 of H5N1 VN (Clade 1). Adjuvant groups: Alum + 25 μg VP3, n = 2; Alum + 75 μg VP3, n = 2; 71VG+25 μg VP3, n = 2; 71VG+75 μg VP3, n = 2. (C) Chickens were vaccinated with H5N2-VLP containing 120 ng HA and 71VG+25 μg VP3 (n = 6). On day 28, they were challenged with 10 LD50 of GV20686 HPAI H5N2 (Clade 2.3.4.4c). The Kaplan-Meier method presents the survival curves. The significant difference in the survival outcome between sham-vaccinated (black circles) and H5N2-VLP-vaccinated (red circles) chickens was assessed by the Log-Rank test (p-value < 0.005). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We subsequently assessed the protective efficacy of H5N2-VLP immunization in chickens by challenging the animals with a lethal dose of H5N1 VN or GV20686. In the H5N1 VN challenge group, all age-matched naïve chickens succumbed to the infection by day 2. Among the eight H5N2-VLP immunized chickens, only one animal survived the infection. Of the other seven animals, three chickens died by day 2, three died by day 3, and one died on day 4. The median survival time for the VLP-vaccinated group significantly extended to 3 days, as opposed to the 2 days observed in the unvaccinated group, with a statistically significant difference (p = 0.0475, Fig. 6B).
In contrast, in the GV20686 challenge group, the unvaccinated chickens succumbed to the infection by day 4 post-challenge. All of the H5N2-VLP immunized chickens survived the lethal challenge (p = 0.0082, Fig. 6C). The challenge experiments demonstrated the potent protective efficacy of antigenic matched H5N2-VLP vaccination against the clade 2.3.4.4c H5 virus.
Discussion
Unlike the earlier clade 2.3.4.4 H5 virus outbreaks that mainly occurred in Asia, the Middle East, and Africa between 2005 and 2019, the recent HPAI H5N1 outbreaks in Europe and North America since 2020 have raised global concerns. One primary concern is the desperate loss of over 200 million birds within a short period [3]. The other significant concern is that the health threat posed by clade 2.3.4.4 H5N1 viruses persists and has now impacted previously less affected regions. The successful use of H5/H7 bivalent inactivated vaccines in poultry to prevent HPAI infection in China in 2017 underscores the importance of reconsidering the application of poultry vaccines as a control measure in countries without mass vaccination programs [2], [3], [29].
In early 2015, a sizeable avian influenza H5 epidemic occurred in poultry farms in Taiwan, causing a severe loss of goose hosts [8]. The 2015 H5 of avian flu viruses belong to clade 2.3.4.4c and have persisted in Taiwan since its emergence [26]. The 2015 WHO report suggests the antigen diversity of 2.3.4.4 viruses compared to 2.3.4 viruses [25]. Although the inactive RG6 vaccine chicken showed 100 % protection against H5N1 VN Class 1 challenges, our study found that the inactivated RG6 of Clade 2.3.4 did not induce a serum-cross-reactivity against H5 viruses from the Taiwanese epidemic in 2016–2017, providing limited protection against lethal infections. In contrast, the H5N2-VLP vaccine containing HA of clade 2.3.4.4c exhibited reactivity in chicken serum samples against local H5 viral antigens. Moreover, chickens immunized with the H5N2-VLP demonstrated 100 % survival from a lethal challenge with the H5 virus strains that are currently spreading. These results suggest that clade 2.3.4.4c VLP-based vaccines offer adequate protection against locally circulating viruses in poultry.
The egg-based inactivated viral vaccines are the most commonly used type for AIV control because of low production cost. However, injecting the virus into the eggs, allowing replication, harvesting, inactivating, and purifying are time-consuming and often ineffective for HPAI viruses [30], [31]. Recombinant vaccines, such as H5N2-VLPs, though slightly more costly, can be developed and manufactured with a lower safety margin at the BSL-1 level and updated quickly to match antigens with pandemic/epidemic strains. The direct cost of manufacturing the H5N2-VLP used in this study was estimated to be 4.4 cents USD per dose, approaching the affordable price of commercial products. Our recent methodology optimization using the same HEK293F cell line can increase the VLP yield from the current 1.6 mg/L to 10–12 mg/L and will further reduce the cost of the VLP vaccine. On an industrial scale, using semi-continuous cultures of HEK293F cells in a stir-tank bioreactor can produce yields 2–4 times higher than those of a shake flask, which can economically make it for poultry vaccines.
The current study assessed the antibody responses of inactivated virus and VLP vaccines in chickens with different vaccine adjuvants, antigen doses/dosages, and booster timing. We found that an appropriate adjuvant combined with a minimally effective dosage of vaccine antigens was critical to significantly enhancing antibody responses in chickens. The H5N2-VLP effectively stimulated chicken antibody responses after prime-boost immunization with two doses of vaccine combined with Alum + VP3 adjuvant. Moreover, increasing the dosage of the 71VG-adjuvanted H5N2-VLP achieved a faster seroconversion and higher antibody GMTs than a lower dosage regimen, suggesting that the 71VG adjuvant might be more effective in boosting antibody production, particularly at higher vaccine dosages.
Alum facilitates strong antibody responses and promotes Th2-biased T-cell differentiation [32]. FMDV VP3 can enhance activation of the TLR2/TLR6 pathways and proinflammatory cytokine production [20]. The oligosaccharide oleate ester derived from mannitol and oleic acid esterification involves adjuvant activity in 71VG, possibly through the activation of TLR4 signaling [33]. As a vaccine emulsifier, the dispersion of oligosaccharide oleate ester forms a milky white suspension in water. It can absorb vaccine antigens to stimulate antigen presentation by dendritic cells or macrophages, activating antibody and cell-mediated immune responses in chickens [19], [34], [35].
Interestingly, when the 71VG adjuvant included FMDV VP3 (25 g), the VLP vaccine elicited higher seroconversion rates and antibody titers than when paired with Alum + VP3. Moreover, this regimen reduced the required HA amount in the VLP vaccine and shortened the booster timing. These findings underscore the VLP vaccination strategies for chickens against novel clade 2.3.4.4c H5 viruses. Due to limited space in the ABSL-3 facility, we could only conduct challenge studies with small sample sizes, which constrained our ability to include all possible adjuvant combinations in the challenge experiment. Despite this, we prioritized the most relevant groups based on our previous findings to demonstrate the effectiveness of VLP vaccines for combating rapidly evolving avian influenza viruses. Efficacy studies evaluating potential vaccine candidates identified in this article warrant further validation in larger sample sizes.
In the face of the ongoing threat posed by HPAI H5 viruses, which has become a global concern for poultry safety and human health, in addition to enhanced biosecurity measures, developing effective vaccines to combat HPAI outbreaks is a growing need. A recent study employed reverse genetic technology to produce recombinant PR8-based vaccines targeting H5Nx viruses (rgH5N1_2.3.4.4b, rgH5N8_2.3.4.4b, and rgH5N1_2.2.1.2) that have been co-circulating in Egypt since 2022 [36]. This investigation demonstrated that the generated candidate vaccines provided complete protection and significantly reduced viral shedding in experimental chickens against homologous circulating strains [37]. These findings highlight the importance of incorporating HA proteins that closely match the currently circulating H5 viruses for an effective AIV vaccine against the subclades of 2.3.4.4 H5 viruses.
Our study extensively investigated the factors crucial for enhancing antibody responses and protective efficacy elicited by VLP vaccines in chickens. This exploration encompassed adjuvant selection, antigen dosage, and booster timing. With advantages such as easy swapping of the HA gene to match circulating viral strains, incorporation of multiple HA genes from different viral strains, and avoidance of manufacturing the recombinant protein in a biosafety level-3 laboratory, the VLP platform serves as an alternative approach to generate poultry vaccines against the novel clade 2.3.4.4 highly pathogenic influenza viruses.
Conclusions
In conclusion, our findings emphasize that the VLP vaccine, when administered with the appropriate adjuvant and dosage, elicits robust antibody responses and protects immunized chicken from circulating clade 2.3.4.4c H5 viruses. The versatility demonstrated in our study underscores the potential of the VLP as a recombinant platform for AIV vaccine development, thereby offering a promising avenue to mitigate the impact of novel H5 virus outbreaks on poultry and human health.
Funding
This work was supported by the Sustainability Science Research Program at Academia Sinica (Grant number AS-SS-106- 04).
CRediT authorship contribution statement
Chia-Chi Ku: Writing – review & editing, Writing – original draft, Validation, Supervision, Investigation, Funding acquisition, Formal analysis, Conceptualization. Cheng-Yu Lin: Methodology, Investigation. Chin-Rur Yang: Methodology, Investigation, Formal analysis. Yu-Chih Yang: Investigation. Po-Ling Chen: Methodology, Investigation. Yi-Te Lin: Methodology, Investigation. Pei-Ru Wang: Investigation. Min-Shi Lee: Writing – review & editing, Validation, Conceptualization. Shu-Mei Liang: Methodology, Funding acquisition, Conceptualization. Pei-Wen Hsiao: Writing – review & editing, Writing – original draft, Methodology, Investigation, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the Sustainability Science Research Program at Academia Sinica (Grant number AS-SS-106-04) to support this research. We are grateful to Dr. Chia-Tsorng Chan at Genomic Research Center, Academia Sinica, for helping with challenge study; Ms. Yu-Ming Chen and Dr. Chwan-Chuen King at the Institute of Epidemiology and Preventive Medicine, College of Public Health, NTU for providing Taiwan local H5 viruses; Dr. Betty A. Wu-Hsieh for reading the manuscript. The authors thank veterinarians Hsin-Sun Huang and Siou-Hui Chen and the staff of Mr. Chuo-Ming Yu of the Animal Resource Center at NTU for excellent technique training, animal care, research protocols, and logistical support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jvacx.2024.100552.
Contributor Information
Chia-Chi Ku, Email: chiachiku@ntu.edu.tw.
Pei-Wen Hsiao, Email: pwhsiao@gate.sinica.edu.tw.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. 1.
Supplementary Fig. 2.
Data availability
No data was used for the research described in the article.
References
- 1.Lee D.H., Bertran K., Kwon J.H., Swayne D.E. Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx clade 2.3.4.4. J Vet Sci. 2017;18:269–280. doi: 10.4142/jvs.2017.18.S1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen J. Bird shots. Science. 2023;380:24–27. doi: 10.1126/science.adi1004. [DOI] [PubMed] [Google Scholar]
- 3.Shi J., Zeng X., Cui P., Yan C., Chen H. Alarming situation of emerging H5 and H7 avian influenza and effective control strategies. Emerg Microbes Infect. 2023;12:2155072. doi: 10.1080/22221751.2022.2155072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Organisation for Animal Health faO. https://wahiswoahorg/#/dashboards/qd-dashboard. Accessed on January 15, 2024.
- 5.World Organisation for Animal Health faO. https://cdnwhoint/media/docs/default-source/wpro---documents/emergency/surveillance/avian-influenza/ai_20240105pdf?sfvrsn=5f006f99_124. Accessed on January 5, 2024.
- 6.World Organisation for Animal Health faO. https://wahiswoahorg/#/event-management. Accessed on January 14, 2024.
- 7.Huang P.Y., Lee C.D., Yip C.H., Cheung C.L., Yu G., Lam T.T., et al. Genetic characterization of highly pathogenic H5 influenza viruses from poultry in Taiwan, 2015. Infect Genet Evol. 2016;38:96–100. doi: 10.1016/j.meegid.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 8.Lee M.S., Chen L.H., Chen Y.P., Liu Y.P., Li W.C., Lin Y.L., et al. Highly pathogenic avian influenza viruses H5N2, H5N3, and H5N8 in Taiwan in 2015. Vet Microbiol. 2016;187:50–57. doi: 10.1016/j.vetmic.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Wu C.Y., Yeh Y.C., Yang Y.C., Chou C., Liu M.T., Wu H.S., et al. Mammalian expression of virus-like particles for advanced mimicry of authentic influenza virus. PLoS One. 2010;5:e9784. doi: 10.1371/journal.pone.0009784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong J., Matsuoka Y., Maines T.R., Swayne D.E., O'Neill E., Davis C.T., et al. Development of a new candidate H5N1 avian influenza virus for pre-pandemic vaccine production. Influenza Other Respir Viruses. 2009;3:287–295. doi: 10.1111/j.1750-2659.2009.00104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shu Y., McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017:22. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatcher E.L., Zhdanov S.A., Bao Y., Blinkova O., Nawrocki E.P., Ostapchuck Y., et al. Virus Variation Resource - improved response to emergent viral outbreaks. Nucleic Acids Res. 2017;45:D482–D490. doi: 10.1093/nar/gkw1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang C.R., King C.C., Liu L.D., Ku C.C. FluConvert and IniFlu: a suite of integrated software to identify novel signatures of emerging influenza viruses with increasing risk. BMC Bioinf. 2020;21:316. doi: 10.1186/s12859-020-03650-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen P.L., Tzeng T.T., Hu A.Y., Wang L.H., Lee M.S. Development and evaluation of vero cell-derived master donor viruses for influenza pandemic preparedness. Vaccines (Basel) 2020:8. doi: 10.3390/vaccines8040626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tung C.P., Chen I.C., Yu C.M., Peng H.P., Jian J.W., Ma S.H., et al. Discovering neutralizing antibodies targeting the stem epitope of H1N1 influenza hemagglutinin with synthetic phage-displayed antibody libraries. Sci Rep. 2015;5:15053. doi: 10.1038/srep15053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu C.Y., Kao S.E., Tseng Y.C., Lin Y.P., Hou J.T., Wu L.Y., et al. Pilot-scale production of inactivated monoglycosylated split H(1)N(1) influenza virus vaccine provides cross-strain protection against influenza viruses. Antiviral Res. 2023;216 doi: 10.1016/j.antiviral.2023.105640. [DOI] [PubMed] [Google Scholar]
- 18.Aucouturier J., Ascarateil S., Dupuis L. The use of oil adjuvants in therapeutic vaccines. Vaccine. 2006;24(Suppl 2) doi: 10.1016/j.vaccine.2005.01.116. pp. S2–44-5. [DOI] [PubMed] [Google Scholar]
- 19.Dupuis L., Ascarateil S., Aucouturier J., Ganne V. SEPPIC vaccine adjuvants for poultry. Ann N Y Acad Sci. 2006;1081:202–205. doi: 10.1196/annals.1373.024. [DOI] [PubMed] [Google Scholar]
- 20.Lin Y.T., Chen Y.P., Fang C.H., Huang P.Y., Liang S.M. Capsid proteins of foot-and-mouth disease virus interact with TLR2 and CD14 to induce cytokine production. Immunol Lett. 2020;223:10–16. doi: 10.1016/j.imlet.2020.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Jain N.K., Sahni N., Kumru O.S., Joshi S.B., Volkin D.B., Russell M.C. Formulation and stabilization of recombinant protein based virus-like particle vaccines. Adv Drug Deliv Rev. 2015;93:42–55. doi: 10.1016/j.addr.2014.10.023. [DOI] [PubMed] [Google Scholar]
- 22.Cimica V., Galarza J.M. Adjuvant formulations for virus-like particle (VLP) based vaccines. Clin Immunol. 2017;183:99–108. doi: 10.1016/j.clim.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martins K.A.O., Cooper C.L., Stronsky S.M., Norris S.L.W., Kwilas S.A., Steffens J.T., et al. Adjuvant-enhanced CD4 T Cell Responses are Critical to Durable Vaccine Immunity. EBioMedicine. 2016;3:67–78. doi: 10.1016/j.ebiom.2015.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nooraei S., Bahrulolum H., Hoseini Z.S., Katalani C., Hajizade A., Easton A.J., et al. Virus-like particles: preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers. J Nanobiotechnology. 2021;19:59. doi: 10.1186/s12951-021-00806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.(WAHIS) WOfAH. https://wwwwhoint/teams/global-influenza-programme/vaccines/who-recommendations. 2015.
- 26.Huang C.W., Chen L.H., Lee D.H., Liu Y.P., Li W.C., Lee M.S., et al. Evolutionary history of H5 highly pathogenic avian influenza viruses (clade 2.3.4.4c) circulating in Taiwan during 2015–2018. Infect Genet Evol. 2021;92 doi: 10.1016/j.meegid.2021.104885. [DOI] [PubMed] [Google Scholar]
- 27.Wong S.S., Webby R.J. Traditional and new influenza vaccines. Clin Microbiol Rev. 2013;26:476–492. doi: 10.1128/CMR.00097-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yasuda M., Kajiwara E., Ekino S., Taura Y., Hirota Y., Horiuchi H., et al. Immunobiology of chicken germinal center: I. Changes in surface Ig class expression in the chicken splenic germinal center after antigenic stimulation. Dev Comp Immunol. 2003;27:159–166. doi: 10.1016/s0145-305x(02)00066-6. [DOI] [PubMed] [Google Scholar]
- 29.Wille M., Barr I.G. Resurgence of avian influenza virus. Science. 2022;376:459–460. doi: 10.1126/science.abo1232. [DOI] [PubMed] [Google Scholar]
- 30.Rajao D.S., Perez D.R. Universal Vaccines and Vaccine Platforms to Protect against Influenza Viruses in Humans and Agriculture. Front Microbiol. 2018;9:123. doi: 10.3389/fmicb.2018.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swayne D.E., Spackman E., Pantin-Jackwood M. Success factors for avian influenza vaccine use in poultry and potential impact at the wild bird-agricultural interface. Ecohealth. 2014;11:94–108. doi: 10.1007/s10393-013-0861-3. [DOI] [PubMed] [Google Scholar]
- 32.McKee A.S., Marrack P. Old and new adjuvants. Curr Opin Immunol. 2017;47:44–51. doi: 10.1016/j.coi.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaeffler A., Gross P., Buettner R., Bollheimer C., Buechler C., Neumeier M., et al. Fatty acid-induced induction of Toll-like receptor-4/nuclear factor-kappaB pathway in adipocytes links nutritional signalling with innate immunity. Immunology. 2009;126:233–245. doi: 10.1111/j.1365-2567.2008.02892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jang S.I., Kim D.K., Lillehoj H.S., Lee S.H., Lee K.W., Bertrand F., et al. Evaluation of Montanide ISA 71 VG adjuvant during profilin vaccination against experimental coccidiosis. PLoS One. 2013;8:e59786. doi: 10.1371/journal.pone.0059786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Awate S., Babiuk L.A., Mutwiri G. Mechanisms of action of adjuvants. Front Immunol. 2013;4:114. doi: 10.3389/fimmu.2013.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mosaad Z., Elhusseiny M.H., Zanaty A., Fathy M.M., Hagag N.M., Mady W.H., et al. Emergence of Highly Pathogenic Avian Influenza A Virus (H5N1) of Clade 2.3.4.4b in Egypt, 2021–2022. Pathogens. 2023:12. doi: 10.3390/pathogens12010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahmoud S.H., Khalil A.A., Abo Shama N.M., El Sayed M.F., Soliman R.A., Hagag N.M., et al. Immunogenicity and Cross-Protective Efficacy Induced by an Inactivated Recombinant Avian Influenza A/H5N1 (Clade 2.3.4.4b) Vaccine against Co-Circulating Influenza A/H5Nx Viruses. Vaccines (Basel) 2023:11. doi: 10.3390/vaccines11091397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.