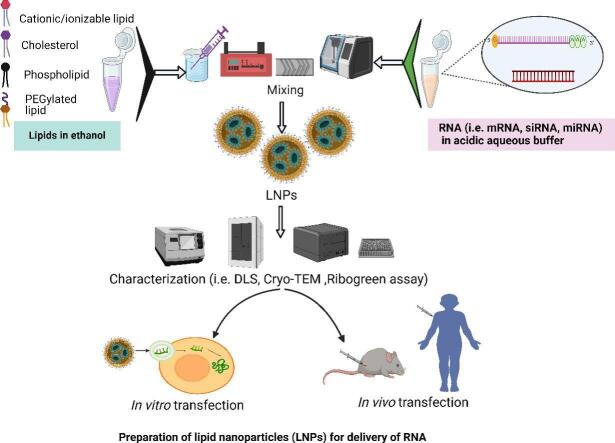
Abstract
Nucleic acid-based therapeutics are a common approach that is increasingly popular for a wide spectrum of diseases. Lipid nanoparticles (LNPs) are promising delivery carriers that provide RNA stability, with strong transfection efficiency, favorable and tailorable pharmacokinetics, limited toxicity, and established translatability. In this review article, we describe the lipid-based delivery systems, focusing on lipid nanoparticles, the need of their use, provide a comprehensive analysis of each component, and highlight the advantages and disadvantages of the existing manufacturing processes. We further summarize the ongoing and completed clinical trials utilizing LNPs, indicating important aspects/questions worth of investigation, and analyze the future perspectives of this significant and promising therapeutic approach.
Keywords: Lipid nanoparticles, mRNA, siRNA, Transfection, Microfluidics
Graphical abstract
Highlights
-
•
Groundbreaking success of using lipid nanoparticles (LNPs) in Covid vaccines opened a window for RNA therapeutics.
-
•
LNP’s components and physical properties have a role in nucleic acid stability, encapsulation, and transfection efficiency.
-
•
Different manufacturing set-ups for LNPs have their own merits, with studies focusing on their improvement.
-
•
Pre- clinical and clinical studies on LNPs pave the way for their applications in various diseases.
1. Introduction
Nucleic acids are a popular therapeutic approach against a plethora of diseases, including cancer and immune therapies, with increasing interest. As fundamental building blocks of life, nucleic acids present unique opportunities to target diseases at their fundamental origin, such as cancer. Nonetheless, nucleic acid-based therapeutics have presented some challenges associated with the successful delivery of these molecules. Nucleic acids are not stable in the human body if adequate protection is not provided, and they cannot penetrate cell membranes without additional help. Not surprisingly, nanotechnology emerged with solutions to these problems. In this review paper, we focus on one of the most prominent delivery carriers, lipid nanoparticles (LNPs), used in preclinical and clinical studies and translated to patient treatment for the delivery of RNAs. We attempt to summarize some of the most important perspectives of the LNP formulations, with potential advantages or drawbacks associated with these particles.
1.1. Historical background of RNA-based therapeutics
Significant advancements in gene sequencing technology have been made after the completion of the human genome project in 2003, which deepened our understanding of genetic causes for different diseases (Goodwin et al., 2016) and paved the way for the development of RNA-based therapeutics, which target disease-related genes that could not be targeted by conventional drugs, small molecules or protein-based drugs (Kaczmarek et al., 2017). RNA therapy includes the targeting of different cellular mechanisms of protein expression, including the use of mRNA and non-coding RNA, such as small interfering RNA (siRNA), micro-RNA (miRNA), antisense oligonucleotides (ASO), and RNA aptamers to facilitate the expression or silencing of desired genes via RNA interference (RNAi) (Kim, 2022). ASOs are single-stranded oligonucleotides, having 18–30 base-pairs complementary to a target RNA, to which they bind and impede translation through RNAse H-mediated hydrolysis of the mRNA strand or steric hindrance. ASO-based drugs, such as nusinersen, eteplirsen, and inotersen, have been approved for the treatment of spinal muscular atrophy, Duchenne muscular dystrophy, and familial amyloid polyneuropathy (Damase et al., 2021; Feng et al., 2021). Pegaptanib is a chemically modified RNA aptamer that can specifically recognize and bind the vascular endothelial growth factor (VEGF) to impede angiogenesis. It was approved for clinical use in age–related macular degeneration (AMD) (Zhou et al., 2012). siRNAs are double-stranded RNA molecules of 20–25 base pair length. These molecules present high specificity in targeting mRNA strands, inducing post-transcriptional gene silencing, mediated by the RNA-induced silencing complex (RISC) and the RNAi cell's mechanism (Laganà et al., 2015). Among different siRNA therapeutics, patisiran is the first siRNA-based drug against hereditary amyloidogenic transthyretin amyloidosis that received FDA approval, followed by givosiran, lumasiran and inclisiran (Hu et al., 2020; Sparmann and Vogel, 2023).
Even though miRNA-based therapeutics have not yet received FDA-approval, several products, such as cobomarsen and remlarsen, are in clinical trials. In general, miRNA-based therapeutics can be promising tools as miRNA mimics or inhibitors (Niazi, 2023). Finally, mRNA-based vaccines were recently used against COVID-19, utilizing mRNAs to induce the expression of the SARS-CoV-2 spike protein, which eventually is detected by the body's immune cells for protection against the disease (Curreri et al., 2023). The development of these mRNA-based vaccines also opened up a new avenue for using mRNA-based therapeutics for non-infectious diseases.
1.2. Challenges of RNA delivery
The incorporation of RNA-based treatments against genetically predisposed diseases, such as polyneuropathy of hereditary transthyretin-mediated amyloidosis (Patisiran), acute hepatic porphyria (Givosiran), primary hyperoxaluria type 1 (Lumasiran) and primary hyperlipidemia (Inclisiran), seems promising (Padda et al., 2024) and can lead to potential therapeutics. Nonetheless, the safe and effective delivery of RNA-based therapeutics is challenging. For example, the large molecular size (varying from a few to hundreds of kDa for ASOs, siRNAs, or mRNAs) and the negative charge of the nucleic acid constructs are significant impediments to internalizing these molecules across the negatively charged cell membranes. Furthermore, even though unmodified nucleic acids could be uptaken by endosomes, their release into the cytoplasm is not feasible, eventually leading to their degradation (Dowdy, 2017; Hossian et al., 2019; Labatut and Mattheolabakis, 2018; Lahooti et al., 2021; Poudel et al., 2021; Shrestha et al., 2023). This can be critical, as RNA-based molecules need to reach the cell's cytoplasm to exert their function. Another impediment associated with RNAs and their administration is that RNAs rapidly degrade by nucleases in body fluids (Poudel et al., 2021; Rhym and Anderson, 2022; Shrestha et al., 2023), while the exogenous RNA delivery should evade the innate immune system response mediated by toll-like receptors (TLR) and retinoic acid-inducible gene I (RIG-I), because they can be recognized by the pattern recognition receptors and trigger immune responses, reducing their translational capability (Chandela and Ueno, 2019; Rhym and Anderson, 2022; Zhu et al., 2022). In summary, considerations need to be taken for using RNA molecules as drugs regarding their bioavailability, elimination and degradation, biodistribution to target areas, renal clearance, uptake into the cellular component, and endosomal escape into the cytoplasm.
Thus, there has been significant effort in developing and characterizing appropriate delivery systems that can protect unmodified RNA molecules in the body for their subsequent delivery to the target area with minimal toxicity and immunogenicity (Niazi, 2023). Furthermore, the delivery systems should promote endosomal escape when taken up into cells via endocytosis (Hamilton et al., 2023; Yan et al., 2022). This poses a significant challenge, as the therapeutic cargo needs to escape this degradative pathway to remain effective. In this review article, we focus on lipid nanoparticles, presenting the different factors for their formulations and how they affect the final properties of the carriers.
2. Stating the expectations: Circulation and endosomal escape of the nucleic acids
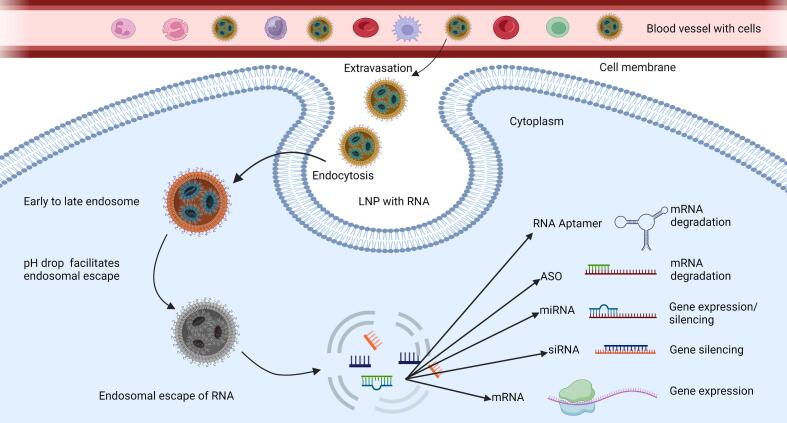
The physiological barriers to RNA delivery can be either extracellular or intracellular. Nucleic acid-loaded carriers must protect their load against degradation in plasma, while the carriers themselves should prevent detection and phagocytosis by the mononuclear phagocytic system (MPS) prior to reaching the targeted cells (Nitika Wei and Hui, 2022; Zelepukin et al., 2024). After being uptaken into cells via endocytosis (Fig. 1), the nanoparticles will experience an environment where the pH drops during endosomal maturation, which can reach values around pH 4–5. The nanoparticles will need to demonstrate specific properties in order to promote their escape from the endosomes. For example, the most commonly used nanoparticles for gene delivery rely on positively charged molecules, such as amines, where protonation is induced by these amine groups when pH reaches lower than their pKa. Even nanoparticles that do not rely on amines, such as poly-(lactide-co-glycolide) (PLGA) polymers, endosomal escape takes place during pH change due to a selective reversal of the particle's surface (Panyam et al., 2002). These properties cause the accumulation of protons and counterions, facilitating the movement of liquids from the cytoplasm to the endosomes. This osmotically-driven phenomenon causes swelling of the endosomes and destabilization of their membrane, compounded in some cases by the electrostatic interactions between the positively charged nanoparticles and the negatively charged membrane bilayer or fusion with the endosomal membrane. Eventually, this leads to the rupture of the endosomes, releasing the nanoparticles and their nucleic acid load to the cytoplasm. In a higher cytoplasmic pH, the binding interaction between the nanoparticles and the nucleic acids weakens, eventually promoting the release of the encapsulated molecules (Patel et al., 2021; Varkouhi et al., 2011; Zheng et al., 2023).
Fig. 1.
Cellular uptake and endosomal escape of nucleic acid carrying lipid nanoparticles into the cytosol to modulate gene expression. Image was created using BioRender.com
3. Delivery systems for RNA therapeutics
Different delivery vehicles based on viral and non-viral vectors have been explored over the decades to meet the aforementioned challenges for efficient RNA delivery. For example, viral vectors can naturally and efficiently deliver genetic materials into a host cell. Furthermore, they can present short-term and long-term transgene expression ability (Lundstrom, 2023). A wide variety of viral vectors have been employed in clinical applications, including adenovirus, adeno-associated virus, herpes simplex virus, retrovirus, and lentivirus for either in vivo or ex vivo gene therapies (Zhao et al., 2022), with retroviral and lentiviral vectors being exploited for delivering RNA-based payloads (Sung and Kim, 2019). However, potential inflammatory reactions, immunogenicity, and mutagenesis of host cells are significant drawbacks associated with viral vector-mediated delivery (Butt et al., 2022).
In contrast, non-viral vectors were developed to diminish immunogenic side effects observed by viral vectors potentially. Nonetheless, recognizing that viral and non-viral vectors have advantages and disadvantages, selecting a delivery system depends on the therapeutic target and the host's safety. Different types of non-viral nanocarriers for the delivery of RNA have been explored, including polymeric nanoparticles, lipid nanoparticles, inorganic nanoparticles, hybrid vector systems (i.e., nanoparticles with a combination of lipids, polymers and/or inorganic materials, such as polymer-lipid hybrid or inorganic-organic hybrid nanoparticles), and biomimetic nanoparticles (Yan et al., 2022). Here, among the different carriers currently researched for RNA delivery, we focus on lipid nanoparticles, a delivery system category that is associated with products that have translated to patient care and have attracted significant attention in recent years.
3.1. Lipid-based systems - Introduction
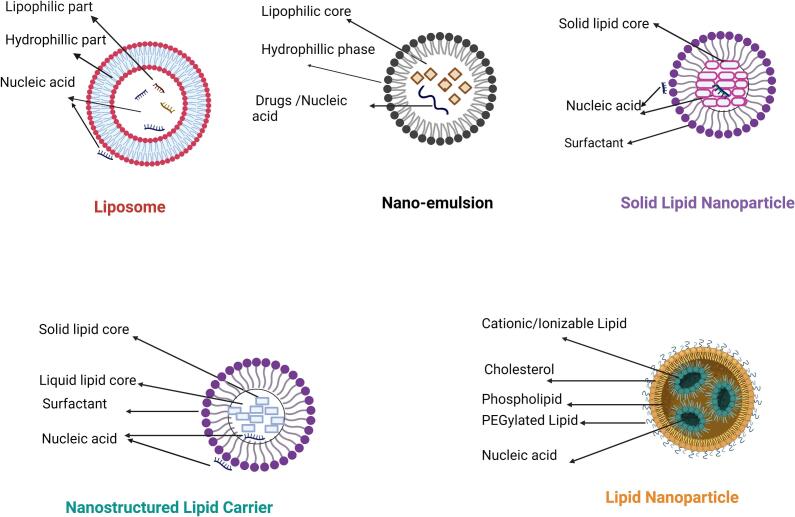
Initially, it is necessary to acknowledge that there are different types of lipid-based systems for nucleic acid delivery, and these carriers can be produced to nanosized dimensions. For example, lipid-based carriers include liposomes, lipid nanoparticles, solid lipid nanoparticles (including nanostructured lipid carriers), and nanoemulsions (Fig. 2), which have been valuable tools for RNA delivery due to properties common among lipid-based carriers. These properties include biocompatibility, versatility, ease of modification and scale up-production, strong internalization due to structural similarities of the cell membrane, and potentially diminished immune reactions compared to viral vectors (Ge et al., 2020; Xue et al., 2015). In fact, lipids used in one type of lipid-based nanocarrier could also be frequently used to formulate the other types. For example, DSPC, a commonly used lipid for liposomes is frequently used in formulating lipid nanoparticles as well. On the other hand, specific formulations, physical characteristics, and lipid differences among the different groups are the reasons for the different classifications. Before focusing on the lipid nanoparticles, we briefly describe representative lipid-based formulations.
Fig. 2.
Graphical illustration of lipid-based systems used for nucleic acid delivery. The image was created with BioRender.com
3.2. Liposomes
Liposomes are self-assembled, sphere-shaped vesicles comprised of lipid bilayers surrounding a single or multiple aqueous cores. Liposomes in the submicron range, i.e., in <200 nm diameter, such as small unilamellar vesicles and large unilamellar vesicles (Pattni et al., 2015), are commonly used in drug delivery applications. In fact, there are currently more than 15 approved liposomal-based drug formulations, though not for nucleic acids (Tsakiri et al., 2022). A “variant” of liposomes, called cationic liposomes, utilize positively charged lipids or molecules, which are positively charged molecules that the negatively nucleic acids can complex with through electrostatic interactions (Liu et al., 2020). In their study, Albakr et al. developed a liposomal delivery system for miR-1296 that sensitized MDA-MB-231 cells to cisplatin treatment (Albakr et al., 2021). Despite the promising attributes of the cationic liposomes for the delivery of nucleic acids, permanently positively charged cationic lipids have created undesired outcomes, such as toxicity, aggregation, stability, and rapid elimination from circulation, among others (Buck et al., 2019; Nsairat et al., 2023; Yi et al., 2000).
3.3. Solid Lipid Nanoparticles (SLNs)
SLNs are colloidal carriers comprised of physiological lipids, such as triglycerides, fatty acids, and waxes, that are dispersed in aqueous solution and stabilized by a surfactant, with particle size of 50 to 1000 nm. They may remain in a homogenous matrix structure, drug-enriched shell structure, or drug-enriched core structure. SLNs differ from liposomes, nano-emulsions, and polymeric nanoparticles in preparation method and use of organic solvents (Ganesan and Narayanasamy, 2017; Xu et al., 2022). DOTAP- and DODAP- based SLNs were explored for DNA and RNA delivery, and DOTAP SLNs presented improved long-term stability (Gomez-Aguado et al., 2020). As the electrostatic interaction of RNAs and lipids primarily takes place at the surface between the aqueous and the lipid phase of the SLNs, proper encapsulation of negatively charged hydrophilic RNAs within the lipophilic core matrix can be challenging and limits the uses of SLNs (Tsakiri et al., 2022).
3.4. Nanostructured Lipid Carriers (NLCs)
Nanostructured Lipid Carriers (NLCs) can be considered a variation or evolutionary product of SLNs. Inside NLC's core, a combination of solid and liquid lipids creates an unorganized environment, which permits better loading, less drug expulsion, and improved drug solubility compared to SLNs (Khan et al., 2023). Similarly, NLCs have also been explored for gene delivery. Representatively, Garbuzenko et al. demonstrated that a Luteinizing Hormone-Releasing Hormone decapeptide coated (LHRH)-NLC nanoparticles system that entrapped a pool of siRNAs against EGFR and paclitaxel enhanced the delivery and efficacy of the active compounds, siRNAs, and paclitaxel, against the cancer cells (Garbuzenko et al., 2019).
3.5. Lipid nanoemulsions
Nanoemulsions are dispersions of nanometer-sized droplets of one liquid, commonly an oil, within another immiscible liquid, i.e., water, stabilized by surfactants or emulsifying agents, such as lipids, also referred to as oil-in-water (o/w) emulsions (Souto et al., 2011). Commonly, these types of nanoemulsions have been explored for the administration of water-insoluble compounds, where the oil nanodroplets provide a hydrophobic environment for lipophilic drugs, while the medium remains aqueous (Hormann and Zimmer, 2016). Cationic nanoemulsions were developed to deliver nucleic acid products, using lipids that have also been utilized to develop cationic liposomes. For example, Yi et al. (Yi et al., 2000) reported on developing cationic lipid emulsions using soybean oil and DOTAP, as the cationic lipid, DOPE and PEG-PE, for complexation with a plasmid. The authors reported stability against DNAse I, while the system successfully transfected cells in vitro. In another example, Brito et al. (Brito et al., 2014) developed a nano emulsion-based carrier to deliver a self-amplifying mRNA vaccine using the cationic lipid DOTAP at Nitrogen to Phosphate (N/P) ratio of 7, and found that the vaccine-induced antibody and T cell response in vivo. As mentioned above, the oil phase of the nanoemulsions allows the incorporation of hydrophobic drugs. This property was utilized by Oh et al. (Oh et al., 2013) to incorporate into an iodinated poppy seed oil phase the hydrophobic anti-cancer compound paclitaxel. The objective was to entrap two active compounds, paclitaxel and siRNA, in the nanocarrier for synergistic dual treatment. Though in this case, the authors did not utilize cationic lipids, but a cationic polymer (i.e., polyethyleneimine) to complex with an siRNA against BCL-2, the nanoemulsion was formed with the two active compounds and was stabilized by PEGylated phospholipids and cholesterols. The authors reported a strong apoptotic activity for the combinatorial drug treatment, higher that the individual compounds (Oh et al., 2013). Even though nanoemulsions have advantages, such as the mixing of more than one drug in the same formulation or the use of natural oils that exhibit biocompatibility, there are concerns regarding their stability, particle size, and the use of permanently cationic molecules for entrapment of nucleic acids, that may cause similar side effects described above for cationic liposomes (Xue et al., 2015).
3.6. Lipid nanoparticles
The application of lipid nanoparticles (LNPs) for RNA delivery has been actively and extensively explored recently. LNPs have been instrumental in delivering groundbreaking therapeutics, such as the approval of Patisiran (siRNA-based therapeutic) and their use in the mRNA-based vaccinations during the COVID-19 pandemic (Suzuki and Ishihara, 2021). LNPs generally have a lower risk of immunogenicity compared to viral vectors, while they can carry a relatively larger payload of nucleic acids, and as the recent COVID-19 vaccinations indicated, their production can be scaled up. Not surprisingly, research and development on LNP-based therapeutics for the administration of nucleic acids have accelerated, and several clinical applications are being evaluated (Swetha et al., 2023). In fact, LNPs have been tested in clinical trials for different infectious diseases caused by the zika virus, the Chikungunya virus, and influenza (Han et al., 2023). The following sections also list clinical trials indicating which LNPs have been the cornerstone of this approach.
The versatility of LNPs, their ability to efficiently deliver various types of nucleic acids, and their biocompatibility make them a promising platform for advancing therapeutic interventions in genetic diseases, cancer, infectious diseases, and beyond (Jung et al., 2022; Pilkington et al., 2021). Finally, significant research is being conducted on developing ionizable lipids, which are major components for the LNP-based formulations and their potential. These molecules promote nucleic acid complexation and endosomal escape, and the synthesis of novel molecules with improved properties for transfection while minimizing toxicity compared to permanently charged cationic lipids has been a proliferative area of research (Swetha et al., 2023).
3.6.1. Fundamentals of LNPs
The therapeutic potential of LNPs relies on various key factors, including chemical composition, transfection capacity, physicochemical characteristics, biodegradability, and immunogenicity, which can be designed or tailored during the initial optimization of each formulation. Lipid components predominately have been the defining materials of LNPs. However, formulation and manufacturing processes can significantly impact the final products (Gyanani and Goswami, 2023).
Initially, LNP's characteristics result from the combination of the different lipid components, rather than being solely attributed to a single lipid, and a synergy of lipids is necessary.(Hald Albertsen et al., 2022). Lipids commonly used in nanoparticles are usually amphiphilic molecules that have both hydrophilic and hydrophobic parts, with the different types of lipids contributing to the formation of the core of a nanoparticle, and the development of structures, such as multilamellar vesicles or the development of a homogenous core-shell in the nanosize diameter to encapsulate nucleic acids (Viger-Gravel et al., 2018). However, it is important to note early on that LNP formulations used for the delivery of short nucleic acids, such as siRNAs or miRNAs, may not necessarily exhibit the same benefits, physical characteristics, or transfection capacity with larger molecular weight nucleic acids, such as mRNAs. Subsequently, structural differences, for example, mRNA is longer than siRNA, charge density, lipid used or stability of the nucleic acid products may necessitate optimization for each LNP formulation, as it is generally regarded that LNP formulations developed for short RNAs (i.e., siRNAs) would not be efficient for mRNAs or DNAs (Hald Albertsen et al., 2022; Kauffman et al., 2015a).
A prototypical LNP formulation comprises four main structural components: a cationic or ionizable lipid, cholesterol, a helper lipid, and a PEGylated lipid. The molar ratio of the lipids in the final lipid mixture can vary depending on the therapeutic targets (Cárdenas et al., 2023; Eygeris et al., 2022; Gyanani and Goswami, 2023; Hald Albertsen et al., 2022; Jung et al., 2022; Pilkington et al., 2021; Swetha et al., 2023; Zhang et al., 2021b).
The rationale for the different materials and their selection impacts stability, encapsulation and transfection capacity of the final products, among others. Briefly, positively charged ionizable or cationic lipids aid the entrapment of nucleic acids and help to destabilize the endosomal membrane to release nucleic acids into the cytoplasm. Ionizable lipids facilitate pH-dependent protonation, and they offer a neutral charge at physiological pH that can minimize potential cytotoxic effects associated with cationic lipids (Sun and Lu, 2023). Thus, ionizable lipids' pKa is preferred at values that allow the lipid to be predominantly neutral at physiological pH and protonated at endosomal pH for endosomal escape. On the other end of these lipids, the lipid tail impacts carrier stability and can affect the fusion of the lipid carrier with the endosomal membrane for nucleic acid release in the cytoplasm (Gyanani and Goswami, 2023). Similarly, cholesterol and a helper lipid build the lipid layer of the LNPs, and sustain the structural integrity of the carriers, while different helper lipids affect the transfection capacity of the LNPs. Finally, PEGylated lipids affect the size and circulation time of the nanoparticles in vivo (Zhang et al., 2024).
On the other hand, optimization of the molar ratio among the lipid components, the molar ratio between lipids and nucleic acids, and the mixing procedure are the remaining key determinants for encapsulation efficiency, particle size, stability and transfection efficiency of LNPs. Briefly, a sufficient amount of the cholesterol and the helper lipids is needed to produce the lipid envelop that will protect the nucleic acid content. Similarly, sufficient quantity of the ionizable lipid at the proper molar ratio to the nucleic acids is needed in order to encapsulate and sufficiently protect the nucleic acids. In the following sections, we delve into published research to present this complex interplay of materials, conditions, and formulation procedures for LNPs, providing the most commonly used molar ratios, lipids, and formulation methods, while providing examples of how these properties/materials affect the final products.
3.6.2. Choice of cationic/ionizable lipids in LNPs
Choosing the appropriate cationic/ionizable lipid is crucial for LNP formulation. It is important to note that the cationic/ionizable lipids comprise the larger portion of the lipid combination for the LNP formulations. Cationic lipids comprise a positively charged amine group, a lipophilic tail, and a linker connecting the hydrophilic portion (amine group) to the hydrophobic tail. They can be classified as monovalent aliphatic lipids, multivalent aliphatic lipids, and cationic cholesterol derivatives (Lechanteur et al., 2018; Rietwyk and Peer, 2017). Uses of cationic lipids for gene delivery based on liposomal formulation gained prominence during 1980s, contributing to improved transfection efficiency, albeit with initial challenges mostly associated with the encapsulation of the payload (Sun and Lu, 2023). 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) and 1,2-di-O-octadecenyl-3-trimethylammonium propane (DOTMA) were among the earliest cationic lipids explored for gene delivery (Guéguen et al., 2024). Felgner et al. in 1987 prepared unilamellar liposomes with DOTMA that efficiently encapsulated and transfected plasmid DNA in vitro (Felgner et al., 1987). Cationic lipids were also effective in RNA delivery (Zhang et al., 2021b). Other commonly used cationic lipids include dioctadecylamidoglycylspermine (DOGS), 2,3-dioleyloxy-N-[2-(sperminecarboxamido)ethyl]-N,N-dimethyl-1-propanaminium (DOSPA), dimethyldioctadecylammonium bromide (DDAB), and 3β[N-(N′, N′-dimethylaminoethane)-carbamoyl] cholesterol (DC-Chol), among others (Balazs and Godbey, 2011; Lechanteur et al., 2018). Even though RNA entrapment depends on electrostatic interactions between positively charged cationic lipid and negatively charged RNA, permanently positive-charged lipids associate with cytotoxicity mostly due to the primary, secondary, tertiary, or quaternary ammonium headgroups (Rietwyk and Peer, 2017) and rapid clearance due to phagocytosis by the mononuclear phagocytosis system (Pilkington et al., 2021). DOTAP, a cationic lipid-based siRNA-LNP, was found to increase liver enzyme release, suggesting possible hepatoxicity and toll-like receptor-mediated inflammatory response (Kedmi et al., 2010; Landesman-Milo and Peer, 2014).
Such shortcomings necessitated the development of new lipids. To address toxicities with quaternary ammonium-based cationic lipids, such as the DOTAP molecule, headgroups of cationic lipids were modified using guanidinium, amidine, pyridinium, and imidazolium to delocalize the positive charge. Such lipids include diC14-amidine, AtuFECT01, Atu027, DODAG-9, DiLA2, C12ANHC18, and synthetic amphiphiles interdisciplinary (SAINT). Guanidinium functionalized lipids provide the advantages of their protonation capability in a wide range of pH and can be a good choice for RNA delivery (Zhang et al., 2021b).
A new group of lipids, namely ionizable lipids, were developed to improve RNA delivery further. The positive charge on cationic lipids remains relatively constant at various pH solutions, whereas an ionizable lipid poses no charge at neutral pH but becomes positively charged in relatively acidic conditions due to protonation (Jayaraman et al., 2012; Kowalski et al., 2019; Patel et al., 2021; Rietwyk and Peer, 2017; Whitehead et al., 2014). The ionizable lipids in the LNPs also assist in the interplay during cellular uptake and endosomal escape by interacting with the endosomal walls' negative charge, disrupting the endosomal membrane (Pilkington et al., 2021; Schlich et al., 2021; Sun and Lu, 2023).
The ionizable lipids have three structural segments: hydrophobic alkyl chains, one or more hydrophilic amines as the hydrophilic headgroup, and a linker (amide, ester, or ether) (Sun and Lu, 2023; Zou et al., 2022). The ability of the ionizable lipid to mediate efficient nucleic acid delivery is influenced by its capacity to facilitate cellular uptake and subsequent release of its load into the target cells, which is highly dependent on the pH-responsive characteristics conferred by the lipid's pKa value. It has been proposed that the optimal pKa value for an ionizable lipid would be at values between 6.2 and 6.5 (Jayaraman et al., 2012). In ionizable lipids, the ester linkage can be hydrolyzed under certain conditions, impacting the net charge and pKa of the respective lipid. The ionization state of the amino headgroup, which is often responsible for the pH-dependent charge of the lipid, may be modulated by the presence or absence of the ester group (Rietwyk and Peer, 2017). Sabnis et al. synthesized a set of ionizable lipids to prepare mRNA-LNPs. They concluded that alterations in the structure of the lipid tails, such as removing ester groups, can lead to a reduction in the pKa, while replacing the alcohol functionality in the headgroup with dimethylamine does not lead to a change in pKa. However, delivery efficiency was lost (Sabnis et al., 2018). In another research paper by Ni et al. (Ni et al., 2022), it was reported that specific modifications in the head group positively influenced mRNA transfection efficiency in Kupffer cells and spleen macrophages.
DODAP is regarded as the first ionizable lipid developed, which was used for nucleic acid delivery (Rietwyk and Peer, 2017). The presence of double bonds on the alkyl chains has been evaluated on the capacity of ionizable lipids for gene silencing (Heyes et al., 2005). Briefly, Heyes et al. synthesized a series of two-tailed dimethyl amino analogs of DODMA, which contains a single, double bond per alkyl chain, lipids with: a) no unsaturated bonds (DSDMA); b) two double bonds per chain (DLinDMA), and; c) three double bonds per chain (DLenDMA). The authors reported that ionizable lipids with two double bonds are more effective in gene silencing compared to those with three double bonds, followed by a single double bond. More importantly, ionizable lipids with saturated alkyl chains exhibited limited to negligible silencing ability. This was correlated to the capacity of the lipids to form a reversed hexagonal II phase, where the degree of unsaturation favors the shift from lamellar to reverse hexagonal II phase, facilitating fusogenicity (DLenDMA = DLinDMA >DODMA> DSDMA) (Heyes et al., 2005).
By introducing a branched lipid tail, Hajj et al. demonstrated a 10-fold increase in mRNA transfection in LNPs with branched chains compared to LNPs with unbranched ones (Hajj et al., 2019). In another study, 7-fold higher potency was found using a multiple-tailed ionizable lipid, C12–200, in LNP formulations for erythropoietin-mRNA delivery (Kauffman et al., 2015b). By independently varying the linker and headgroup of lipids based on DLinDMA, while keeping the other lipid components constant, Semple et al. prepared a series of lipid molecules and their respective formulations to deliver siRNA. The DLinDMA-based modified ionizable lipids were altered either in the linker, yielding the products DLinDAP, DLin-2-DMAP, DLin-C-DAP, Dlin-S-DMA, and DLin-K-DMA, or the headgroup, yielding the products DLin-K-MPZ, DLin-K-MA, DLin-K-TMA.Cl, DLin-K2-DMA, DLin-KC2-DMA, DLin-KC3-DMA and DLin-KC4-DMA. They observed that introducing ester, carbamate, or thioether linkages to the ionizable lipids led to a substantial reduction in the in vivo activity compared to LNPs containing DLinDMA. In contrast, LNPs with ketal ring linker, i.e., DLin-K-DMA, were approximately 2.5-fold more potent compared to the DLinDMA benchmark. Addition of a single additional methylene group into the headgroup of DLin-K-DMA, i.e., the DLin-KC2-DMA lipid, showed a significant increase in potency, whereas further extension with more methylene groups, i.e., the DLin-KC3-DMA and DLin-KC4-DMA lipids, resulted in a substantial decrease in activity, concluding that DLin-KC2-DMA was the best performing ionizable lipid (Semple et al., 2010). In a similar study, Lin et al. (Lin et al., 2013) depicted that the low potency of LNPs containing DLinDAP can be attributed to hydrolysis caused by endogenous lipases after cell internalization, while DLin-KC2-DMA, DLin-K-DMA, or DLin-DMA, which do not contain ester linkage, were less vulnerable to lipase digestion and facilitated better gene silencing when used in a GAPDH siRNA LNP formulation and RAW 264.7 cells. Furthermore, the researchers reported that DLin-KC2-DMA was more effective in gene silencing compared to the other lipids examined in the study.
DLin-MC3-DMA has been recognized as one of the most potent ionizable lipids used in LNPs. Jayaraman et al. did a comprehensive evaluation of 56 amino lipids for their LNP-mediated in vivo gene silencing activity targeting the Factor VII gene in mice, through alterations on their amine head group and the respective lipid's pKa values. The authors recognized DLin-MC3-DMA to demonstrate the most potent gene silencing with low effective doses in rodents and nonhuman primates (Jayaraman et al., 2012). DLin-MC3-DMA stands out as a key ionizable lipid that depicted robust hepatic gene silencing since its advent as part of the FDA-approved siRNA-LNP drug to treat hereditary amyloidogenic transthyretin amyloidosis (hATTR) (Adams et al., 2018; Akinc et al., 2019). In an interesting study by Nabhan et al. (Nabhan et al., 2016), the authors reported that the delivery of a frataxin (FXN) mRNA using LNPs formulated with DLin-MC3-DMA demonstrated efficient translation into mFXN protein in hepatocytes, maintaining increased protein expression after sever days following the intravenous administration of the formulation.
Although DLin-MC3-DMA has received extensive recognition for its transfection capacity, efforts are being made to improve this by developing derivatives of this molecule or new structures. For example, in one study, the authors attempted to improve the biodegradability of the DLin-MC3-DMA via modification on the aliphatic tails and by introducing ester linkages into them, which will result in the breakdown of the lipid tails to more hydrophilic components once internalized. One of the resulting products, L319, showed biodegradability and tolerability while maintaining in vivo potency on par with the DLin-MC3-DMA (Maier et al., 2013). Similarly, another molecule, 3-(dimethylamino)propyl(12Z,15Z)-3-[(9Z,12Z)-octadeca-9,12-dien-1-yl]henicosa-12,15-dienoate (DMAP-BLP) was used to develop LNPs with GRIN1 siRNA. The formulation efficiently silenced neuronal gene expression in vitro, while presenting selective reduction of synaptic NMDAR currents in the brains of mice following intracranial injection (Rungta et al., 2013). Finally, we need to point out two important lipids, ALC-0315, ((4-hydroxybutyl)azanediyl)bis(hexane-6,1-diyl)bis(2-hexyldecanoate), and SM-102, Heptadecan-9-yl 8-(2-hydroxyethyl)[6-oxo-6-(undecyloxy)hexyl]amino octanoate, which are the ionizable lipids used in the formulation of LNPs for mRNA delivery in the COVID-19 vaccines by Pfizer-BioNTech and Moderna, vaccines respectively (de Alwis et al., 2021; Sun and Lu, 2023).
3.6.3. Choice of cholesterol
Cholesterol is a naturally occurring component in cell membranes that contributes to membrane fluidity, stability, and permeability (Ercole et al., 2015; Raffy and Teissie, 1999; Zhang et al., 2019). Cholesterol affects the transition temperature and the phase behavior of lipid layers, which facilitates the release of mRNA from LNPs to the cytosol (Pilkington et al., 2021). As cholesterol helps to fill the spaces between phospholipids and increases the rigidity of the lipid membranes, it prevents leakage of the payload from LNPs, while it provides stability and promotes membrane fusion (Cheng and Lee, 2016; Kim et al., 2021; Kulkarni et al., 2019). The incorporation of cholesterol in lipid-based formulations, such as liposomes, reduces surface-bound proteins and opsonization. This effect improves circulation half-life, allowing the formulations to remain in circulation for extended periods and increasing their chances of reaching the target cells (Semple et al., 1996).
Cholesterol localization within the LNPs appears to depend on factors, such as the cholesterol's molecular ratio to other lipids or the type of ionizable lipid/other lipids. In an interesting study by Kulkarni et al. (Kulkarni et al., 2018), the authors described how the localization of cholesterol within the nanoparticles is affected by the solubility of the molecule in the ionizable lipid oil phase and the interaction with the helper lipid, in the case of the paper, DSPC. By extension, the molar ratio of the DSPC and cholesterol, in this case, needs to be considered or optimized for the proper formation of a complete external surface monolayer. Excess cholesterol can result in the formation of crystalline structures by the molecule and subsequent particle instability (Kulkarni et al., 2018), with similar observations presented by another group approximately at the same time (Yanez Arteta et al., 2018). Overall, the cholesterol content in LNP formulations has been predominately at ∼37–40 % of total molecular lipid content in multiple LNP formulations with different ionizable lipids (Ball et al., 2018a; Rungta et al., 2013; Yamamoto et al., 2015). Both Moderna and Pfizer-BioNTech used cholesterol in their LNP-based COVID-19 vaccine formulations (Batty et al., 2021).
Modifications in cholesterol molecules can affect their interactions with other components of the LNPs, potentially influencing the nanoparticles' ability to remain stable and/or target specific cells or tissues. Patel et al. (Patel et al., 2020) explored the role of different cholesterol analogues on LNPs and suggested that the hydroxyl group's polarity, sterol ring's flexibility to undergo conformational changes, and length of the alkyl tail in cholesterol are important factors for maintaining high transfection efficiency. In another research conducted by Paunovska et al. (Paunovska et al., 2018), the authors adopted six different cholesterol variants in LNP formulations and concluded that LNPs formulated with esterified cholesterol derivatives, such as cholesteryl oleate, demonstrated improved nucleic acid delivery compared to regular and oxidized cholesterols. Alternatively, oxidative modifications in the hydrocarbon tail of cholesterol were better tolerated than modifications in the B cholesterol ring during transfection in vivo, potentially leading to improved delivery (Paunovska et al., 2019). Eygeris et al. (Eygeris et al., 2020) studied the impact of different naturally occurring cholesterol analogues like β-sitosterol, fucosterol, campesterol, stigmasterol, and Vitamin D2 on mRNA-LNP delivery. They demonstrated that substituting cholesterol with phytosterols contributed to changes in morphology, crystallinity, lamellarity, lipid partitioning, thermal response, and gene transfection with different degrees of variations. LNPs formulated with β-sitosterol exhibited high lamellarity, fewer internal defects, and high mRNA transfection capacity in vitro compared to others. LNPs formulated with vitamin D2 were reported to have high fragility due to fluidity in the lipid membrane, which prevented effective crossing through the cell membrane.
3.6.4. Choice of phospholipid
Phospholipids are essential helper lipids in the formation and functionality of LNPs. Phospholipids are amphiphilic molecules that have hydrophilic and hydrophobic portions. The hydrophilic phosphate-linked head group can be modified with choline, ethanolamine, or serine to yield phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylserine (PS), respectively, among others (Nsairat et al., 2022). Their ability to spontaneously organize into lipid layers, contribute to endosomal escape, and provide membrane stability makes them key components in designing effective LNP-based drug or nucleic acid delivery systems (Eygeris et al., 2022; Granot and Peer, 2017; Zhang et al., 2021b). Commonly used helper lipids for nucleic acid delivery include DSPC, DPPC, HSPC, DOPC, POPC, SOPC, for phosphatidylcholines, and DOPE, POPE, SOPE, for phosphatidylethanolamines (Álvarez-Benedicto et al., 2022; Ball et al., 2018b; Jung et al., 2022; Semple et al., 2010; Zatsepin et al., 2016; Zuhorn et al., 2005).
Due to their more cylindrical molecular shapes, PCs have an inherent tendency to adopt a bilayer phase that is beneficial for the formation of stable lipid bilayers, while the cone shape associated with PE headgroups and tails promotes a cone shape and inverted hexagonal (HII) conformation (Li et al., 2015b). Adopting the inverted hexagonal (HII) phase by lipids in LNPs promotes endosomal membrane destabilization and fusion, facilitating the release of nucleic acids to the cytoplasm from endosomes. Using the ionizable lipid DLin-MC3-DMA, DSPC, cholesterol, and a PEG-lipid, Arteta et al. prepared mRNA-LNPs and evaluated their structure for different sizes. The authors identified the inverse hexagonal internal structure while mRNA was present and identified that DSPC is mainly on the surface of the mRNA-LNPs, with the particle size and surface composition affecting the protein production following transfection (Yanez Arteta et al., 2018). The geometrical structures that lipids assume in an aqueous environment reflects their amphiphilicity, the conformational positioning of the lipid tails, and their respective molar ratio in a formulation associated with their “packing parameter” (Barenholz and Thompson, 1999; Hsu et al., 2005). A helper lipid, such as DOPE and DSPE that present a packing parameter of >1, can contribute to the formation of the inverted hexagonal (HII) phase and the fusion with the endosomal membrane (Eygeris et al., 2022; Mukalel et al., 2019; Zhang et al., 2023b; Zhang et al., 2021b). However, in contrast to phosphatidylethanolamine (PE), which favors an inverted hexagonal (HII) phase, phosphatidylcholines (PC), which would present a packing parameter closer to 1, have been reported to favor lamellar structures (Barenholz and Thompson, 1999; Zhang et al., 2021b). Nonetheless, we need to point out that although the helper lipids, whether PE or PC, can contribute to the membrane fusion or the form of the lipid layer, their molar ratio in the formulation and the ionizable lipid used have a significant impact on the formation of inverted hexagonal (HII) phase in LNPs, as well as remind that even though DOPE has these benefiting properties, like fusogenicity and capacity to favor inverted hexagonal phase, DSPC has been favored in LNPs-siRNA systems (Heyes et al., 2005; Kulkarni et al., 2019). Nonetheless, increased fluidity of DOPE due to the unsaturated lipid tails might enhance interaction between LNPs and cellular membranes, facilitating internalization and intracellular release of the payload (Eygeris et al., 2022).
Li et al. (Li et al., 2015a) developed LNP nanoparticles using N1,N3,N5-tris(2-aminoethyl)benzene-1,3,5-tricarboxamide (TT) lipids. Their study evaluated how the helper lipid affected the transfection of the LNPs and compared DOPE, DSPC, and POPE. The researchers found superior potency of DOPE over DSPC and POPE for mRNA delivery. Similarly, Cheng Q et al. (Cheng et al., 2018) evaluated mRNA delivery in vivo using DOPE and DSPC in dendrimer-based LNP formulations. The authors concluded that DOPE was advantageous in delivering nucleic acids.
Interestingly, in an in vivo study comparing identical LNP formulations with either DOPE or DSPC as helper lipids, LNPs formulated with DOPE preferentially accumulated and exhibited better mRNA delivery to the liver compared to LNPs formulated with DSPC, which presented preferential accumulation to the spleen (Zhang et al., 2021a). In a study utilizing the ionizable lipid DLin-KC2-DMA, the authors evaluated how the encapsulation efficiency of a siRNA in LNPs is affected while altering the cationic lipid and PEG-c-DMA molar ratios, but having fixed ratios for either DSPC or DOPE. The siRNA encapsulation efficiency was greatly affected by the change of the DSPC to DOPE, whereas DOPE allowed a consistently high encapsulation efficiency with up to 70 % of ionizable lipid molar ratio, while the encapsulation efficiently greatly dropped under similar conditions while using DSPC (Leung et al., 2015). In their study, Kulkarni et al. (Kulkarni et al., 2017a) compared DSPC to unsaturated PCs (i.e. SOPC, DOPC) or DOPE in LNPs using the ionizable lipid Dlin-KC2-DMA. The authors concluded that pDNA-LNPs containing the unsaturated SOPC and DOPC demonstrated stronger transfection than LNPs containing DSPC, or DOPE, in HeLa cells in vitro, with DSPC mostly presenting the lowest transfection, while in vivo studies indicated DOPE to be the most potent for inducing transfection.
DSPC was used in the first FDA-approved RNAi, Patisiran (siRNA-LNP) formulations, followed by the COVID-19 vaccines mRNA-1273 (Moderna) and BNT162b2 (Pfizer-BioNTech) (Hajiaghapour Asr et al., 2023; Kulkarni et al., 2019; Zhang et al., 2020b). Hence, the choice of the helper lipids in the LNP formulation can significantly influence their physicochemical properties and interactions with biological components, affect nucleic acid encapsulation and transfection efficiency.
3.6.5. Choice of the PEGylated lipid
PEGylation involves the attachment of polyethylene glycol (PEG) chains to molecules, such as drugs, proteins, or nanoparticles to improve systemic circulation time and reduce immunogenicity. PEG-lipids create a ‘stealth’ effect by shielding the surface of nanoparticles to limit the adsorption of serum proteins onto the nanoparticle's surface and minimize their recognition by the immune system (Hossian et al., 2019; Labatut and Mattheolabakis, 2018; Mattheolabakis et al., 2014; Suk et al., 2016). The steric hindrance introduced by the brush-like PEG molecules from the surface of the nanoparticles can result in prolonged residence time in the circulation of the nanoparticles, and potential accumulation to specific tissues, such as tumors, due to the enhanced permeation and retention effect (EPR) (Lahooti et al., 2023). Similar to other lipids, PEG-lipids have a hydrophobic portion that is usually made of alkyl chains, which is integrated into the lipid layer, and a hydrophilic part that also contains the PEG, which extends from the surface of the LNPs. While PEGylation enhances circulation time and reduces immune recognition of nanoparticles, excessive PEGylation might hinder the efficient interaction of LNPs with target cells, leading to reduced therapeutic effects (Kumar et al., 2014), and fusion of the LNPs with the endosomal membrane due to steric hindrance, leading to therapeutic inefficacy (Aldosari et al., 2021; Kulkarni et al., 2019). More importantly, the PEG-lipids significantly impact LNP formulation and size (Kulkarni et al., 2019; Samaridou et al., 2020). In two studies by Belliveau et al. and Ryals et al., particle size decreases upon PEG lipid content increase in the formulations (Belliveau et al., 2012; Ryals et al., 2020).
Thus, choosing an appropriate PEG-lipid and PEG-lipid molar content for a formulation for proper PEG density is crucial for optimizing the performance of LNPs. In fact, the absence of PEG-lipids may result in larger or aggregated particles (Lokugamage et al., 2021), whereas as little as 0.5–2.5 mol% of a PEG-lipid may yield stable and homogeneous LNPs with around 80 nm (Kulkarni et al., 2019). Mui et al., (Mui et al., 2013) showed that, by using varying amounts and different types of PEG-lipids, 1.5 mol% of PEG-C16 and PEG-C18 is optimal for gene silencing by siRNA-LNPs, while above 3.5 mol% content for any of the PEG-lipids impaired gene silencing. In their study, Suzuki et al. (Suzuki et al., 2020) explored the accelerated blood clearance phenomenon associated with PEGylated lipids. They depicted that the choice of PEG-lipid and its shedding characteristics influence the immune response. LNPs with PEG-lipids that shed more rapidly are associated with reduced production of anti-PEG IgM antibodies compared to LNPs with PEG-lipids that shed more slowly. The results indicated that PEG-lipids with short acyl chain (DMG-PEG) used in the siFVII-DSG-LNP formulation showed greater gene silencing compared to the long acyl chain DSG-PEG (Suzuki et al., 2020). DMG-C-PEG2000 was used in onpattro siRNA-LNP (Wahane et al., 2020), while ALC-0159 was the PEGylated lipid component for Spikevax-and Comirnaty COVID-19 vaccines, respectively (Wang et al., 2023a).
3.6.6. Selection of appropriate molar ratio of lipid components
Apart from selecting the appropriate lipid combination, finding the proper mixing ratio, i.e., the molar ratio among the lipid components, is necessary. Current lipid-based RNA delivery systems, such as LNPs have drawn inspiration from conventional liposomal systems, notably the Doxil formulation, which was the first FDA-approved nanomedicine (Barenholz, 2012). Earlier liposomal formulations were primarily composed of higher cholesterol and phospholipid contents, such as HSPC: Cholesterol: PEG 2000-DSPE at 56:39:5 M ratio or without PEGylated lipid, with EPC: Cholesterol at 55:45 M ratio (Bulbake et al., 2017; Chang and Yeh, 2012). However, LNP formulations differ, as the ionizable/cationic lipid is the major component, along with the phospholipid, cholesterol, and the PEG-lipid. The portion of the ionizable and cationic lipids relative to the total lipids in the formulation is approximately 30–50 %, whereas cholesterol, phospholipid and PEGylated lipid stand at 20–50 %, 10–20 % and 0.5–5 %, respectively (Eygeris et al., 2022; Sun and Lu, 2023). Of note, the lipid mixture is usually prepared in ethanol. Although there is a difference in the selection of the individual lipid components, for example, choosing different ionizable and PEG lipid, there are commonalities in the lipid formulations of several mRNA-based COVID-19 vaccines (specifically BNT162b2 by BioNTech/Pfizer and mRNA-1273 by Moderna) and the first-ever approved siRNA-LNP therapeutic, Onpattro. Both mRNA-1273 vaccine and Onpattro used a molar ratio of 50:10:38.5:1.5 mol% for the lipid components (i.e., ionizable lipid: Cholesterol: helper lipid: PEG-lipid), while BNT162b2 used a 46.3:42.7:9.4:1.6 ratio (Kon et al., 2022). The lipid formulations of these therapeutics have similarities regarding the presence of tertiary amine groups in the ionizable lipids (Mendonça et al., 2023; Suzuki and Ishihara, 2021; Verbeke et al., 2021; Zhang et al., 2023a). Across several research papers, similar molar ratios of approximately 50:10:38.5:1.5 have been maintained for developing LNPs (Carrasco et al., 2021; Chander et al., 2023; Ge et al., 2020; Gyanani and Goswami, 2023; Jayaraman et al., 2012; Kauffman et al., 2015a; Whitehead et al., 2014). Nonetheless, the proper ratio might need to be evaluated ad-hoc in different settings, which necessitates optimization of these parameters across different mRNA and siRNA-LNP formulations for efficient nucleic acid delivery.
Certain properties, such as RNA encapsulation capacity, depend on the LNP preparation. For example, Ball et al. (Ball et al., 2018b) prepared five different formulations with varying all lipids' content, for the co-delivery of an siRNA and an mRNA. They found that a composition ionizable lipid: DSPC: DOPE: Cholesterol: C14-PEG at 38.8:3.6:10.9:44.5:2.25 mol%, which was in between the two tested extreme conditions of i) ionizable lipid: DSPC: DOPE: Cholesterol: C14-PEG at 35:0:16:46.5:2.5 mol% and ii) ionizable lipid: DSPC: DOPE: Cholesterol: C14-PEG 50:10:38.5:0:1.5 mol%, was the optimal for all tested applications, such as silencing, gene expression, size and entrapment. They observed that decreasing the ionizable lipid content, while increasing the cholesterol, helper lipid, and PEG-lipid contents promoted RNA encapsulation and gene delivery overall (Ball et al., 2018b). In another study, Sago et al. (Sago et al., 2018) compared LNPs and found the two best-performing LNPs having a molar ratio of 7C1:cholesterol:C14-PEG2000:18:1 Lyso PC at 50:23.5:6.5:20 and 7C1:cholesterol:C14-PEG2000:DOPE at 60:10:25:5, which is an indication that LNP formulations not only need to be optimized in terms of their lipids (here, helper lipid), but also for the respective molecular ratio/content. Similarly, Lam et al. (Lam et al., 2023) showed that altering the PEG-lipid content, from 1.6 % to 2.8 %, increased gene expression in vivo, while further PEG-lipid increase brought diminishing results. Roces et al. (Roces et al., 2020) successfully prepared LNPs using different cationic/ionizable lipids, namely DOTAP, DDAB and DLin-MC3-DMA at various molar ratios, to identify the optimal conditions for the preparation of the LNPs. In another study, Prakash et al. (Prakash et al., 2013) formulated LNPs at a molar ratio of 57.5:7.5:31.5:3.5 using DLin-KC2-DMA: DSPC: cholesterol: DMG-PEG to deliver single-stranded siRNA to mouse livers. All these studies are representative examples of how the optimization process for choosing lipids and their respective molar ratios is a necessary step during LNP formulations.
3.6.7. Selection of aqueous buffer/lipids for RNA and Nitrogen to Phosphate (N/P) ratio
The manufacturing process for LNPs involves using ethanol as a solvent for cationic/ionizable lipids, phospholipids, cholesterol, and PEG-lipids to form the lipid phase. This phase is subsequently mixed with a low-pH aqueous solution of the nucleic acids (Webb et al., 2022). As we focus on LNPs and ionizable lipids, which appear to be the most commonly studied, we will describe procedures associated with ionizable lipids. Selection of an appropriate buffer and pH is important for the protonation of an ionizable lipid. Across the literature, three buffers have been most commonly studied for the dissolution of nucleic acids. Briefly, representative examples include 1–100 mM citrate buffer at pH 3–6 (Alabi et al., 2013; Álvarez-Benedicto et al., 2022; Billingsley et al., 2020; Geall et al., 2012; Larson et al., 2022; Naderi Sohi et al., 2021; Patel et al., 2017; Prakash et al., 2013; Zhang et al., 2021a), 20–50 mM acetate buffer at pH 4–6 (Belliveau et al., 2012; Carrasco et al., 2021; Hassett et al., 2019; Jürgens et al., 2023) or 20 mM malic acid buffer at pH 3 (Tanaka et al., 2021) have been used for LNP preparation. Malic acid buffer at pH 3 has been proposed for encapsulating DNA molecules, while the citrate buffer at pH 4.5 is proposed for mRNA encapsulation, as a lower pH may lead to mRNA degradation (Bailey-Hytholt et al., 2021).
Optimizing the appropriate ratio between the ionizable lipid and RNA is crucial. The Nitrogen to Phosphate (N/P) ratio indicates the ratio between the positively charged lipids and the negatively charged nucleic acid molecules during LNP formation. Simply stated, it reflects the balance of positive and negative charges in the LNPs. Achieving an optimal N/P ratio is critical for ensuring effective complexation, stability, and delivery of nucleic acids. For example, Hassett et al. (Hassett et al., 2019) used for their formulation an N/P ratio of 5.67 to prepare mRNA-LNPs, while Jürgens et al. (Jürgens et al., 2023) used N/P ratio of 3 for siRNA and N/P ratio of 6 for mRNA to formulate the respective LNPs in their study. In an interesting study by Carrasco et al. (Carrasco et al., 2021), the authors prepared a series of LNPs with varying N/P ratios from 2 to 16, while fixing the molar lipid ratio at 50:10:38.5:1.5 (KC2: DSPC: Cholesterol: DMG-PEG). They found a decrease of mRNA encapsulation efficiency from 80 to 40 % while lowering the N/P ratio from 8 to 2. They also described that adjusting the N/P ratio affects the size of the LNPs and, by extension, the number of mRNA copies they can accommodate (Carrasco et al., 2021). In other study, Ball et al. (Ball et al., 2018b) used N/P ratio 8.4 for LNP-mediated siRNA and mRNA codelivery, while Roces et al. (Roces et al., 2020) in their study prepared mRNA -LNP with N/P ratio 8. Philipp et al. (Philipp et al., 2023) for mRNA-LNP preparation used N/P of 3, whereas 7.5 was optimal for Sanghani et al. (Sanghani et al., 2021) for siRNA-LNP. In a different study, Chen et al. (Chen et al., 2016a) concluded that increasing the N/P ratio from 2 to 12, a progressive improvement in potency occurred up to N/P of 6, beyond which there was little additional improvement. Finally, the N/P ratio of BNT162B2 (Pfizer) and mRNA-1273 (Moderna) vaccines has been estimated at 6, where for siRNA-LNP (Onpattro), the N/P has been reported at 3 (Schoenmaker et al., 2021b).
Alternatively, many researchers focus on RNA to ionizable lipid w/w ratio instead of N/P ratio. For single amine lipids, such as DOTAP, DLin-MC3-DMA or SM-102 and ALC-0315, the N/P ratio changes correspond to 1:1 proportional change to lipid:nucleic acid weight changes. Attention must only be paid for lipids with more than one amine, such as DOGS and DOSPA, or dendrimers, such as PAMAM. The lipid formulations of both Pfizer and Moderna mRNA COVID-19 vaccines used RNA to ionizable lipid w/w ratio of ∼0.05 (Verbeke et al., 2021). However, different studies show that the RNA-to-ionizable lipid w/w ratio varies from 0.04 to 0.2, considering individual lab optimization processes (Alabi et al., 2013; Belliveau et al., 2012; Kauffman et al., 2015a; Kumar et al., 2014; Whitehead et al., 2014). Hence, the choice of N/P ratio or RNA to ionizable lipid ratio (w/w) depends on various factors, including the specific properties of the lipids used or the structure of the RNA, and the desired characteristics of the resulting nanoparticles. Not surprisingly, and as can be seen from the examples above, researchers resort to optimizing each new formulation of LNPs and often experiment with different ratios to optimize the performance of the respective applications.
3.6.8. Manufacturing considerations of RNA delivery via lipid-based formulations
3.6.8.1. Thin film hydration
The thin film hydration method is one of the most commonly used approaches in the development of liposomes, and has been a methodology used for the development of cationic liposomes, while it has been less commonly utilized for the production of LNPs (Mattheolabakis et al., 2012; Wang et al., 2023b). The method relies on the development of a thin lipid film during evaporation of a volatile organic solvent, such as ethanol or chloroform, in which under hydration, the lipids detach from the solid phase and re-assemble into a liposomal structure, frequently multi-lamellar structures of microsized dimensions (Pattni et al., 2015). Subsequently, the particles are reduced in size via extrusion through membranes with specific pore sizes or sonication to produce unilameral vesicles in the nanometer dimensions (Vogelaar et al., 2023). Cationic lipids can substitute negatively charged lipids for the development of cationic liposomes. In traditional liposomal formulations, the drug encapsulation usually occurs during the hydration step, using a solution of the drug, which commonly results in lower encapsulation or post-loading or remote-loading based on pH gradient-dependent drug loading (Nambiar et al., 2024). In cationic liposomes, the cationic lipids are located on the lipid bilayer, including the surface, which allows the complexation of the cationic liposomes with nucleic acids to occur during hydration or after their formation. The drawback of this is that the nucleic acids may remain on the surface of the liposomes (Haghiralsadat et al., 2018; Luiz et al., 2022; Zhang et al., 2006), or the complexation between liposomes and nucleic acids can lead to aggregation and size increase at certain nucleic acid-to-lipid ratios (Pires et al., 1999), as well as present challenges in upscaling or reproducibility (McKenzie et al., 2023; Vogelaar et al., 2023).
3.6.8.2. Ethanol injection
The ethanol injection method has also been a commonly used technique for liposomal preparations. The original methodology has been adapted for the preparation of lipid nanoparticles. Focusing on ionizable lipids, the approach relies on the mixture under intensive stirring of an organic solvent that contains the lipids and a low pH buffer solution of the nucleic acids in excess. The miscible organic phase disperses in the aqueous phase rapidly due to intense stirring, causing the formation of lipid nanoparticles, where the ionized lipids complex with the nucleic acids and are entrapped inside the nanoparticles (Wagner et al., 2002).
For example, Khare et al. (Khare et al., 2021) prepared siRNA- LNP based on dropwise mixing of a lipid phase into a 10 mM citrate buffer pH 4. Though this methodology can directly lead to the formulation of nano-sized carriers and represents a simple and straightforward methodology, the reproducibility, scalability, and encapsulation efficiency are of concern.
3.6.8.3. T-Junction method
As an alternative approach to ethanol injection, the T-junction mixing method is a frequently used method to leverage controlled and more precise mixing of the alcoholic and buffer solutions with the help of pumps. With two inlet channels in a T-shaped mixer, each carrying a different liquid component, such as the ethanolic solution of the lipids and the low pH buffer solution with the nucleic acids, the chaotic and turbulent flow at the T-junction, where the two flows meet, facilitates rapid and thorough mixing of the two liquids and yielding lipid nanoparticles in the outlet (Jürgens et al., 2023; Li and Xu, 2023). Though this technique can achieve efficient encapsulation of nucleic acids in LNPs, frequently at or above 90 % encapsulation, larger particle size has also been reported compared to the methods described below. Furthermore, a higher flow rate is required to achieve proper mixing, which may not be convenient in a laboratory setup (Jürgens et al., 2023; Leung et al., 2014), affecting the size and polydispersity index of the produced formulations (Jürgens et al., 2023; Kulkarni et al., 2017b). Representatively, Crawford et al. (Crawford et al., 2011) prepared siRNA-LNP with an average particle size ranging from 63.3 nm to 120.1 nm, using T-junction mixing. Goswami et al. (Goswami et al., 2019) prepared LNP with the range of ∼140–155 nm and up to 88 % encapsulation efficiency for the delivery of self-amplifying mRNA in mannosylated LNPs. In a different study, Lazzaro et al. (Lazzaro et al., 2015) adopted T-junction mixing to prepare mRNA-LNPs, which were then delivered to CD8 T-cells to evaluate immune responses against the encoded antigen. Finally, Kumar et al. and Abrams et al. used T-junction mixing for preparing LNPs using the ionizable lipids DLin-MC3-DMA and CLinDMA, respectively (Abrams et al., 2010; Kumar et al., 2014).
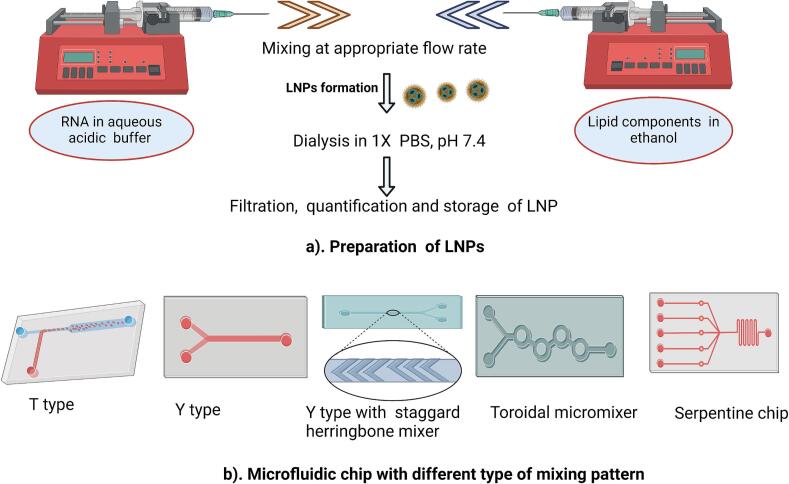
3.6.8.4. Microfluidic mixing
Microfluidic devices and technologies have gained attention for their role in producing LNPs. Microfluidic-based mixing provides precise control and improved mixing, resulting in smaller particle sizes with increased homogeneity in the particle sizes and consistent encapsulation efficiencies with reduced batch-to-batch variability. While initially developed for small-scale production of LNPs, microfluidic systems can be designed for scale-up, making them suitable for both research and potential industrial applications (Bezelya et al., 2023; Liu et al., 2019; Maeki et al., 2022; Thomas et al., 2018). The microfluidic architectures are designed in such a way that two solutions are mixed within the device under controlled conditions to generate LNPs. Similarly to the T-junction, one inlet of the microfluidic device is connected to a pumping system to deliver the lipid components dissolved in ethanol, while the other inlet is connected to the low pH buffer solution of the nucleic acids, most commonly RNA (Menon et al., 2022). During LNP preparation, the flow rate ratio (FRR) of mixing between the lipid and nucleic acid solutions is crucial for the final LNP characteristics and their particle size, with different studies utilizing FRR values that span between 1:1 and 5:1 (aqueous to lipid solution mix), but mostly commonly utilizing the ratio of 3:1 (Belliveau et al., 2012; Billingsley et al., 2020; Hassett et al., 2019; Jürgens et al., 2023; Roces et al., 2020; Walsh et al., 2014; Zhang et al., 2023a).
Types of microfluidic chips that can be used for mixing and subsequent nanoparticle formation include T type, Y type, serpentine, toroidal mixer, and staggered herringbone micromixer (SHM) (Belliveau et al., 2012; Chen et al., 2016b; Mendonça et al., 2023). In T- and Y-shaped microfluidic systems, the lipid and buffer solutions come into contact at a liquid-liquid interface within the microfluidic device, allowing controlled and gradual formation of LNPs at the liquid-liquid interface. Slow ethanol dilution is a characteristic feature of this method compared to other approaches, impacting the final size of the LNPs produced. Injection in both inlets is usually facilitated by pumps (Maeki et al., 2022; Mendonça et al., 2023). In contrast, the design of a serpentine microfluidic device is specifically tailored to enhance mixing. Chaotic convection is generated within the device, contributing to increased uniformity in the resulting nanoparticles (Niculescu et al., 2022). Alternatively, a sheath-flow-type microfluidic device design allows for a controlled and well-defined flow environment and strong ethanol-buffer mixing (Hood and DeVoe, 2015; Jahn et al., 2004), which may yield better particles than Y- or T-type devices.
On the other hand, the SHM microfluidic chip's unique pattern of V-shaped ridges promotes efficient mixing, enhances substance diffusion, and facilitates rapid and uniform reactions, making them well-suited for applications such as the production of LNPs (Hama et al., 2018). Li et al. (Li et al., 2017) compared two methods for forming transferrin-conjugated lipid nanoparticles (Tf-LNPs): a single-step microfluidic process and a conventional multi-step batch mixing method. Results suggested that microfluidic-formed LNPs were more effective than the multi-step one in delivering siRNA to tumor sites. Belliveau et al. (Belliveau et al., 2012) prepared siRNA-LNPs using SHM and observed that at higher flow rates, the PDI and size of siRNA-LNP decreased.
Several studies have portrayed the advantageous role of microfluidic mixing techniques (Fig. 3) for the development of LNPs, with several innovative designs of microfluidic systems being evaluated to improve the mixing and/or dilution of the lipid and nucleic acid solutions while achieving strong encapsulation efficiency and transfection (Billingsley et al., 2020; Chen et al., 2012; Chen et al., 2014; Fenton et al., 2017; Kauffman et al., 2015a; Leung et al., 2012; Philipp et al., 2023; Shepherd et al., 2021). For example, Chen at el. (Chen et al., 2012) prepared siRNA-LNPs with 90 % gene silencing in vivo using a microfluidic device created using PDMS and soft lithography, where the nucleic acid solution was initially mixed with the lipid solution and subsequently diluted with a buffer solution.
Fig. 3.
LNP formulation using microfluidic setups involves pump systems for regulating the flow of ethanolic and aqueous solutions for mixing in the microfluidic chip. (a) Representative formulation setup for LNPs; (b) Representative microfluidic chips used for mixing the two solutions and producing the LNPs. The image was created with BioRender.com
While the herringbone design and similar advanced mixing geometries in microfluidic systems are highly effective for controlled and efficient mixing, there are challenges when scaling up to commercial production with high-throughput requirements. One benefit of the microfluidic systems is their ability to be utilized in parallel, compared to previous methodologies, which can streamline their upscaling, although relatively slow flow rates are still necessitated in a reliable and reproducible manner per microfluidic chip. Significant efforts on microfluidic designs take place with the focus on improving these aspects. Among them, toroidal mixers have been demonstrated to be a promising technology to overcome these limitations. The circular flow pattern created within the tori helps to thoroughly blend and homogenize the components (Webb et al., 2020). Similarly, microfluidic platforms, i.e., systems designed to utilize the microfluid chips, are developed and are often equipped with computer-controlled pump systems that regulate the flow and mixture rate of solutions. Such examples include the NanoAssemblr benchtop systems by precision nanosystems, which allow precise adjustment of parameters, such as flow rates, concentrations, and reaction times (Prakash et al., 2022). Hence, such automated microfluidic systems are increasingly exploited for LNP preparations (Carrasco et al., 2021; Robinson et al., 2018; Sebastiani et al., 2021). Overall, using microfluidics in LNP preparations offered significant advantages in particle size control, homogeneity, and reproducibility for subsequent in vitro and in vivo evaluations.
In Table 1, we summarize the primary characteristics that are being evaluated during the preparation of the LNP nanoparticles and include representative examples for each category.
Table 1.
Preparation variables and materials for LNPs.
| Features of LNP preparation | Representative Examples |
|---|---|
| 1. Lipids | |
| Cationic/ionizable lipid | DOTAP, DODAB, DLin-MC3-DMA, C12–200, DLin-KC2, DMA, ALC-0315, SM-102, L319 |
| Cholesterol | Cholesterol, β-sitosterol |
| Helper lipids (Phospholipids) | DSPC, DOPE, DOPC, DSPE |
| PEGylated lipid | DMG-PEG, DSG-PEG, DMG-C-PEG2000, ALC-0159 |
| 2. Molar ratio of lipids | Typically, molar ratio of lipids (Cationic/ionizable lipid: Cholesterol: Phospholipid: PEG lipid) ranges from 30 to 50 %: 20–50 %:10–20 %: 0.5–5 % |
| 3. Nitrogen to phosphate (N/P) ratio | N/P ratio can vary from 2 to 16 |
| 4. Aqueous buffer | Sodium acetate, Sodium citrate and Malic acid buffer. pH 3–6. |
| 5. Mixing pattern | Hand mixing, T junction method, Microfluidic mixing. |
| 6. Flow rate | Total flow rate changes from 0.5 to 20 (ml/min) whereas flow rate ratio be 1:1 to 1:5 (lipid to aqueous). |
After the preparation of LNPs, qualitative and quantitative tests should be conducted to ensure that LNPs meet the desired encapsulation efficiency, stability, and integrity of RNA. In Table 2, representative characterization methods for LNPs are presented.
Table 2.
Methods for LNP characterization.
| Characterization criteria of LNPs | Implications | Assay methods |
|---|---|---|
| Particle size | To determine size of nanoparticles |
Dynamic light scattering (DLS) |
| Polydispersity index (PDI) | To assess homogeneity and size distribution | |
| Zeta potential | Gives idea about surface charge of nanoparticles | |
| Morphology & structure | To evaluate structural characteristics on the LNPs | Cryogenic transmission electron microscopy (Cryo-TEM), SAXS (small -angle x-ray scattering) |
| Encapsulation% | To quantify the payload inside LNPs. | Ribogreen/Picogreen assay |
| Integrity of the payload | To detect stability and integrity of entrapped nucleic acid, following encapsulation | Gel electrophoresis |
| Gene expression efficiency | Indicates effectiveness of LNP to deliver payload | In vitro/in vivo transfection |
3.6.9. Stability and storage of RNA-LNPs
As RNA molecules are prone to degradation, LNP preparation and long-term storage are crucial for successful internalization and functionality. Chemical modifications of siRNAs with phosphorothioate substitution, 2′ -O-methylation and fluorination (Choung et al., 2006), as modification of 5′ cap and 3′ poly-A tail in mRNA enhances their stability as well as their therapeutic efficiency (Kim et al., 2022; Shrestha et al., 2023). Encapsulation of such RNA payload inside LNPs ensures further protection from nuclease degradation. However, it does not guarantee long-term stability during storage and handling. Although there are commonalities in the LNP preparations among LNPs in Onpattro and the Moderna or Pfizer COVID-19 vaccines, there are differences in their shelf-life and storage conditions. Onpattro is stable for 3 years at 2–8 °C, whereas the Pfizer COVID-19 vaccine requires −80 to −60 °C to remain stable for few months (Schoenmaker et al., 2021b). In a study by Ball et al. (Ball et al., 2017), the authors prepared siRNA -LNPs using the lipidoid 306O13, and they studied the impact of pH (3, 7.4, 9) and temperature (−20, 2 and 25 °C) on LNPs. It was determined that when stored at the low temperature of 2 °C, LNPs remained stable for the longest period, at least 150 days, while the pH did not affect the formulation's stability. It has also been reported that preserving mRNA-LNP at −80 °C without a cryoprotectant decreases gene expression due to possible particle aggregation, while other external factors, such as vibrations and light, may also contribute to this outcome (Kamiya et al., 2022). Thus, to improve on LNP stability during storage without lyophilization, proper excipient selection, including the buffering agents, osmolytes and cryoprotectant, can minimize the degradation of the payload (Muralidhara et al., 2016; Schoenmaker et al., 2021b).
In contrast, lyophilization of suspensions has been explored to potentially prolong the stability of nanoparticles (Gatto and Najahi-Missaoui, 2023). Interestingly, Zhao et al. (Zhao et al., 2020) demonstrated that adding 20 % (w/v) sucrose or trehalose may stabilize nanoparticles' size and mRNA delivery efficiency in vitro, but the lyophilized nanoparticles did not exhibit efficiency in vivo. They concluded that adding 5 % (w/v) sucrose or trehalose may be optimal for long-term storage of mRNA in lipid-like nanoparticles in liquid nitrogen. In another study, Kim et al. (Kim et al., 2023) suggested that storing LNPs at −20 °C in phosphate-buffered saline (PBS) with 10 % sucrose effectively maintains LNPs stability and in vivo potency for one month. For reference, both Moderna and Pfizer COVID-19 vaccines included sucrose in their formulations (Schoenmaker et al., 2021b). Hence, a proper understanding of these factors helps establish optimal storage conditions for preserving the integrity of LNPs and their loads, ensuring that therapeutic efficacy is maintained throughout their shelf life.
4. Clinical trials on RNA-LNP drug candidates
The number of clinical trials using LNP formulations is rapidly increasing, with applications based on administering RNAs against a plethora of diseases, including the respiratory syncytial virus, SARS-CoV-2, influenza virus, Zika virus, among others, and life-threatening genetic disorders, such as cancer. Pioneering the LNP-encapsulated formulation of siRNA against Transthyretin-mediated amyloidosis, Alnylam Pharmaceutics successfully developed a first-of-its-kind Onpattro that marked the advent of RNA delivery through LNPs (Adams et al., 2018). The LNP composition comprised of (6Z,9Z,28Z,31Z)-hepatatriaconta-6,9,28,31-tetraen-19-yl-4(dimethylaminio)-butanoate (DLin-MC3-DMA) lipid, DSPC, cholesterol and PEG-DMG (Hald Albertsen et al., 2022). This drug first entered its clinical trial phase in 2010 and received FDA approval in 2020. This marked the advent of the new era of nucleic acid delivery through LNPs. The significance of LNP formulations for nucleic acid was further accentuated during the COVID-19 pandemic when the search for appropriate drug delivery carriers for nucleic acid therapeutics increased. Another breakthrough for LNP-encapsulated mRNA vaccines was during the global pandemic of SARS-CoV2, when Moderna and BioNTech/Pfizer resorted to lipid nanoparticles as carrier to successfully deliver mRNA against the COVID-19 virus (Polack et al., 2020a; Sahly et al., 2021).
In August 2021, the FDA approved the groundbreaking BNT162b2 COVID-19 vaccine by BioNTech/Pfizer known now as Comirnaty (Lamb, 2021). The LNP formulation was comprised of a proprietary ionizable cationic lipid ((4-hydroxybutyl)azanediyl)bis(hexane-6,1-diyl)bis(2-hexyl decanoate), DSPC, cholesterol and PEG-lipid (Shi et al., 2022). In January 2022, Moderna's Spikevax also received the FDA's Emergency Use Authorization (EUA) within one year during the COVID-19 pandemic. Spikevax consists of mRNA-1273 encapsulated with LNP formulated with SM-102 (ionizable lipid), proprietary helper lipids, cholesterol, DSPC, and PEG2000-DMG (Shi et al., 2022).
Similarly, LNP-encapsulated mRNA-1345 which encodes for prefusion F glycoprotein was administered prophylactically against respiratory syncytial virus infection (Wilson et al., 2023). Currently, it is undergoing a Phase 2/3 randomized study in adults ≥60 years of age (NCT05127434), to assess its safety and tolerability. Additionally, a Phase 3 clinical trial for LNP-mRNA-1345 co-administered with a quadrivalent influenza vaccine (Afluria® Quadrivalent; NCT05330975) in adults above 50 years old is also underway. The mRNA-1010 LNP vaccine against seasonal influenza A (H1N1/H3N2) and influenza B (Yamagata- and Victoria lineages) was evaluated in clinical trials (NCT04956575) and demonstrated no vaccine-related serious adverse effects or deaths while eliciting a potent immune response (Ananworanich et al., 2024; Lee et al., 2023a). mRNA-1647 LNP vaccine against cytomegalovirus (CMV), known as CMVictory, is undergoing Phase 3 study (NCT05085366) in women of childbearing age. In a previous clinical study (Phase 2), the vaccine demonstrated safety, tolerability, and immunogenic responses in both CMV -seropositive and -seronegative populations (Panther et al., 2023). Similarly, some other ongoing clinical studies on mRNA-LNP drug candidates for other infectious diseases are mRNA-1893 (Phase 2, NCT04917861) against Zika virus, mRNA-1215 (Phase 1, NCT05398796) against Nipah virus, H1ssF-3928 (Phase 1, NCT05755620) against influenza, mRNA-1189 (Phase 1, NCT05164094) against Epstein Virus, among others.
Additionally, LNP-encapsulated mRNAs are under clinical trials against various cancer types. For instance, mRNA-2752 encodes OX40L T-cell co-stimulator, IL-23, and IL36γ proinflammatory cytokines and is undergoing Phase I clinical study against relapsed or refractory solid tumor (NCT03739931) (Deng et al., 2022; Manish et al., 2021). Similarly, mRNA-4157 (V940) in combination with Pembrolizumab (NCT03897881) is currently in phase 2 clinical study for treatment against melanoma (Weber et al., 2024). Some other examples of mRNA-LNP drug candidates in clinical studies for cancer treatments are MEDI1191 (Phase 1, NCT03946800), SAR441000 (Phase 1, NCT03871348), mRNA-2416 (Phase 1/2, NCT03323398), mRNA-5671 (Phase 1, NCT03948763), NCI-4650 (Phase 1/2, NCT03480152), among others. Similarly, mRNA-LNP vaccines targeting genetic disorders also include the mRNA-3704 (NCT03810690) and mRNA-3705 (NCT05295433) against methylmalonic acidemia, mRNA-3927 (NCT05130437) against propionic acidemia, MRT5201 (NCT03767270) and ARCT-810 (NCT04442347) against ornothine transcarbylase deficiency, and MRT5005 (NCT03375047) against cystic fibrosis, among others. The LNP-encapsulated mRNA-0184 is undergoing phase 1 clinical trial (NCT05659264) as a novel investigational mRNA drug candidate to treat chronic heart failure.
In different approaches, Intellia Therapeutics has developed the very first CRISPR/CAS9 single guide (sg)RNA encapsulated LNP, NTLA-2001 (NCT04601051) against transthyretin (hATTR)-amyloidosis (Gillmore et al., 2021; Lee et al., 2023b). Similarly, for drug candidate development, Verve Therapeutics' VERVE-102 encapsulated CRISPR-Cas9 guide RNA targeting PCSK-9 gene in LNPs for evaluation in a Phase 1 trial (NCT06164730). siRNA-LNPs are also gaining momentum in the treatment of various cancer types. Briefly, NBF-006 (NCT03819387) are LNPs with encapsulated siRNAs against glutathione S-transferase Pi (GSTP) and evaluated in its Phase 1 clinical study against colorectal cancer (Xie and Wang, 2022).
In Table 3, we present past and on-going clinical trials of LNPs as a carrier system for mRNA, siRNA, and sgRNA against various diseases. It is evident that LNPs are a sought-after lipid-based, non-viral delivery carrier system for effective RNA delivery that has found extensive applicability.
Table 3.
Summary of ongoing or completed clinical trials utilizing LNPs. Formulation information is provided anywhere possible. Additional sources used in the construction of this table: https://clinicaltrials.gov; https://classic.clinicaltrials.gov; https://www.cdek.liu.edu. N/A: Formulation information not found, or any information identified were not peer-reviewed and not included.
| Nuclei Acid Candidate | Clinical Trial study ID (NCT) | Phase | Formulation/Description | Lipid Composition | Disease/Conditions | Route of Administration | Ref. | Sponsor | Status |
|---|---|---|---|---|---|---|---|---|---|
| mRNA-LNP | |||||||||
| Cancer | |||||||||
| mRNA-2752/ Durvalumab | NCT03739931 | Phase 1 (2018–2026) | Novel mRNA-based therapeutic agent encoding OX40L T cell co-stimulator, IL-23 and IL-36γ pro-inflammatory cytokines |
DLin-MC3-DMA, DSPC, Cholesterol, DSPE-PEG2000 | Relapsed/refractory solid tumor malignancies or lymphoma | Intratumoral | (Deng et al., 2022; Manish et al., 2021) | ModernaTX, Inc. | Active, Not Recruiting |
| MEDI1191/Durvalumab | NCT03946800 | Phase 1 (2019–2023) | LNP formulation therapy of IL-12 mRNA to induce a potent TH1-mediated anti-tumor response | N/A | Solid Tumors | Intratumoral/IV | (Hamid et al., 2021) | MediImmune LLC | Completed |
| mRNA-4157/Pembrolizumab |
NCT03313778 NCT03897881 |
Phase 1 (2017–2025) Phase 2 (2019–2029) |
LNP-encapsulated mRNA-4157 that encodes 34 different patient-specific neoantigens in combination with Pembrolizumab |
N/A | Solid tumors including melanoma, bladder carcinoma, HPV-neg HNSCC, NSCLC, SCLC, MSI-high, TMB-high cancers | IM/IV | (Burris et al., 2019; Julie et al., 2020; Weber et al., 2024) | ModernaTX, Inc. | Recruiting |
| SAR441000 | NCT03871348 | Phase 1 (2019–2024) | A mixture of four mRNAs encoding IL-12sc, IFN-α-2b, GM-CSF and IL-15sushi as monotherapy and in combination with cemiplimab | N/A | Advanced solid tumors | Intratumoral/IV | (Oliver et al., 2020) | Sanofi | Active, Not recruiting |
| mRNA-2416 | NCT03323398 | Phase1/2 (2017-2021) | LNP-encapsulated mRNA encoding human OX40L alone or in combination with Duvulamab | N/A | Relapsed/Refractory Solid Tumor Malignancies or Lymphoma and Ovarian Cancer | Intratumoral | (Jimeno et al., 2020) | ModernaTX, Inc. | Terminated |
| mRNA-5671/ V941 and Pembrolizumab | NCT03948763 | Phase 1 (2019–2022) | LNP-encapsulated mRNA-5671 that encodes for most common KRAS substitutions (G12d, G12V, G13D, G12C) either alone or in combination with Pembrolizumab | N/A | Neoplasms | IM/IV | (Barbier et al., 2022) | Merck Sharp & Dohme LLC | Completed |
| NCI-4650 | NCT03480152 | Phase 1/2 (2018–2019) | A mRNA based personalized cancer vaccine that targets up to 15 tumor-associated antigens | N/A | Melanoma, Colon Cancer, Gastrointestinal Cancer, Genitourinary cancer, hepatocellular cancer | IM | (Cafri et al., 2020) | National Cancer Institute | Terminated |
| Cardiovascular disease | |||||||||
| mRNA-0184 | NCT05659264 | Phase 1 (2022–2025) | LNP-encapsulated mRNA-0184 that encodes for relaxin hormone | N/A | Chronic Heart Failure | IV | (Soroudi et al., 2024) | ModernaTX, Inc. | Recruiting |
| Viral diseases/Influenza | |||||||||
| mRNA-1345/ Afluria®Quadrivalent/mRNA-1273.214 | NCT05330975 | Phase 3 (2022–2024) | mRNA-1345 co-administered with seasonal influenza vaccine (Afluria) to evaluate the impact of co-administration on immune respose of RSV-A and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) | N/A | Respiratory Syncytial Virus in adults over 50 and SARS-COV2 | IM | (Li et al., 2023; Wilson et al., 2023) | ModernaTX, Inc. | Active, not recruiting |
| mRNA-1345 | NCT05127434 | Phase 2/3 2021–2025 |
LNPs with encapsulated mRNA encoding for a stabilized prefusion F glycoprotein | N/A | Respiratory Syncytial Virus (RSV) | IM | Moderna TX, Inc | Active, not recruiting | |
| mRNA-1273 mRNA-1010 mRNA-1345 mRNA −1647 FLUAD® |
NCT05397223 | Phase 1 (2022–2026) | LNPs encapsulating mRNA-1273, mRNA-1010, mRNA-1345, and mRNA-1647 to observe systemic reactogenicity, adverse effects and adverse reactions | N/A | Respiratory Syncytial Virus in healthy adults, SARS-CoV-2, Cytomegalovirus | IM | (Lee et al., 2023a) | ModernaTX, Inc. | Active, not recruiting |
| mRNA-1273 mRNA-1273.351 |
NCT04405076 | Phase 2 (2020−2021) | LNP-encapsulated mRNA-1273 vaccine encoding a pre-fusion stabilized form of the SARS-CoV-2 spike protein (S—2P). | N/A | SARS-COV-2 Booster dose |
IM | (Choi et al., 2021) | ModernaTX, Inc. | Completed |
| Spikevax mRNA-1273 |
NCT04283461 NCT04470427 |
Phase 1 (2020−2022) Phase 3 (2020–2022) |
LNP-mRNA-1273 is a lipid-encapsulated mRNA-based vaccine that encodes for a full-length, prefusion stabilized spike (S) protein of SARS-CoV-2 | SM-102, and 3 commercially available lipids, cholesterol, DSPC, and PEG2000 DMG. | SARS-CoV-2 | IM | (Baden et al., 2021) | National Institute of Allergy and Infectious Diseases (NIAID) | Completed |
| mRNA-1283 | NCT05137236 | Phase 2 (2021−2023) | Next generation SARS-CoV2 vaccine composed of LNP encapsulated mRNA-1283 | N/A | SARS-CoV-2 | IM | (Yassini et al., 2023) | ModernaTX, Inc. | Completed |
| Comirnaty (BNT162b2) | NCT04368728 | Phase 1/2/3 (2020−2023) | LNP-encapsulated nucleoside-modified mRNA that encodes membrane-anchored, full-length SARS-CoV-19 S-protein | ALC-0315, ALC-0519, DSPC Cholesterol | SARS-CoV-2 | IM | (Polack et al., 2020b) (Lamb, 2021) | BioNTech SE | Completed |
| CV-NCOV | NCT04449276 | Phase 1 (2020–2021) | LNP-formulated SARS-CoV-2 vaccine containing mRNA encoding perfusion conformation-stabilized full-length SARS-CoV-2 spike protein | DSPC, cationic lipid, PEG-lipid conjugate, cholesterol | SARS-CoV-2 | IM | (Kremsner et al., 2021) | CureVac | Completed |
| ARCoV | NCT04847102 | Phase 3 (2021–2023) | mRNA-LNP that encodes the receptor binding domain of the S-protein of SARS-CoV2 | Ionizable lipid (Lipid 9001), (1,2-DSPC, cholesterol and PEG-lipid | SARS-CoV-2 | IM | (Zhang et al., 2020a) (Chen et al., 2022) | Walvax Biotechnology Co., Ltd. | Unknown Status |
| mRNA-1010 | NCT04956575 | Phase 1/2 (2021−2022) | LNP encapsulating quadrivalent seasonal influenza vaccine encoding membrane-bound HA surface glycoproteins of four influenza strains (A/H1N1, A/H3N2, B/Victoria, and B/Yamagata) recommended by the WHO for cell- or recombinant vaccines | N/A | Seasonal influenza A: H1N1 and H3N2 Influenza B: strains including Victoria-lineage and Yamagata lineage |
IM | (Lee et al., 2023a) | ModernaTX, Inc. | Completed |
| mRNA −1647 mRNA-1443 |
NCT03382405 |
Phase 1 (2017–2020) |
LNP encapsulated mRNA-1647 comprises six mRNAs encoding for CMV antigens given along with LNP-encapsulated mRNA-1443 encoding pp65 from T cells of the CMV antibody |
N/A | Cytomegalovirus (CMV) | IM | (Jung et al., 2022) | ModernaTX, Inc. | Completed |
| mRNA-1647 | NCT04232280 | Phase 2 (2020–2023) | LNP encapsulating mRNA-1647 cytomegalovirus vaccine in CMV-seronegative and CMV-seropositive healthy adults | N/A | IM | (Panther et al.) | Completed | ||
| NCT05085366 | Phase 3 (2021–2026) | Active, not recruiting | |||||||
| mRNA-1653 | NCT04144348 | Phase 1 (2019–2022) | LNP encapsulated bivalent vaccine comprised of nucleoside-modified mRNA encoding full-length membrane-bound fusion proteins of hMPV and PIV3 | N/A | Human Metapneumovirus and Human parainfluenza virus | IM | (August et al., 2022; Nelson et al., 2020; Schnyder Ghamloush et al., 2024) | ModernaTX, Inc. | Completed |
| mRNA-1440 (VAL-506440) | NCT03076385 | Phase 1 (2015–2018) | LNP-formulated modified mRNA-based vaccine encoding hemagglutinin (HA)proteins of H10N8 or H7N9 influenza strain | N/A | Influenza A (H10N8 and H7N9) | IM | (Bahl et al., 2017; Feldman et al., 2019) | ModernaTX, Inc. | Completed |
| mRNA-1851 (VAL-339851) | NCT03345043 | Phase 1 (2016–2018) | LNP encapsulated mRNA-1851 that encodes for the HA protein of H7N9 | N/A | Influenza A virus H7N9 subtype | IM | (Shi et al., 2022) | ModernaTX, Inc. | Completed |
| CV7202 | NCT03713086 | Phase 1 (2018–2021) | RABV-G mRNA using initial formulation CV7201 antigen with cationic protein protamine encapsulated in LNP | Ionizable amino lipid, DSPC, PEG-2000-DMG, Cholesterol | Rabies virus glycoprotein (RABV-G) | IM | (Aldrich et al., 2021; Shi et al., 2022) | CureVac | Completed |
| mRNA-1325 | NCT03014089 | Phase 1 (2016–2019) | LNP-encapsulated modified mRNA vaccine encoding premembrane and envelope E structural proteins (prME) from a Micronesia 2007 Zika virus isolate | Proprietary ionizable lipids, DSPC, cholesterol, and PEG-lipid | Zika Virus | IM | (Richner et al., 2017) (Bollman et al., 2023) | ModernaTX, Inc. | Completed |
| mRNA-1944 | NCT03829384 | Phase 1 (2019–2021) | LNP-encapsulated mRNA-1944 that encodes for the heavy and light chains of CHKV-24 antibody | Proprietary high purity PEG-2000 stearate monoester, IAL (proprietary ionizable amino lipid), cholesterol, DSPC | Chikungunya virus | IV | (August et al., 2021) | ModernaTX, Inc. | Completed |
| mRNA-1893 | NCT04917861 | Phase 2 (2021–2024) | LNP-encapsulated mRNA-1893 vaccine encoding the envelope E structural proteins (prME) from the RIO-U1 Zika virus isolate | Proprietary ionizable lipid, DSPC, cholesterol, and PEG lipid | Zika virus | IV | (Bollman et al., 2023; Essink et al., 2023; Li et al., 2024) | ModernaTX, Inc. | Active, not recruiting |
| mRNA-1215 | NCT05398796 | Phase 1 (2022–2024) | LNP encapsulated mRNA-1215 that encodes for Nipah perfusion F protein and G protein | N/A | Nipah Virus (NiV) Infection | IM | (Rodrigue et al., 2024; Wang et al., 2023c) | ModernaTX, Inc. | Active, not recruiting |
| H1ssF-3928 |
NCT03814720 NCT05755620 |
Phase 1 (2019–2021) Phase 1 (2023–2025) |
VRC H1ssF 3928 mRNA-LNP vaccine encoding influenza H1 hemagglutinin stem | N/A | Influenza | IM | (Andrews et al., 2023; Widge et al., 2023) | National Institute of Allergy and Infectious Diseases (NIAID) | Completed Recruiting |
| DCVC H1 HA | NCT05945485 | Phase 1 (2023–2024) | mRNA-LNP vaccine encoding full length H1 HA of influenza A/California/07/2009 (H1N1) | N/A | Influenza A/H1N1 | IM | National Institute of Allergy and Infectious Diseases (NIAID) | Recruiting | |
| AVX502 | NCT00440362 | Phase 1/2 (2007) | Alphavirus Replicon Vaccine Expressing Influenza HA protein | N/A | Influenza | IM/SC | AlphaVax, Inc. | Completed | |
| mRNA-1189 | NCT05164094 | Phase 1 (2021–2025) | LNP-encapsulated mRNAs that encode Epstein-Barr Virus (EBV) envelope glycoproteins gp42, gp220, gH and gL | N/A | Epstein-Barr Virus (EBV) | IM | (Zhong et al., 2022) | ModernaTX, Inc | Active, Not Recruiting |
| Genetic disorders | |||||||||
| mRNA-3704 | NCT03810690 | Phase 1/2 (2019–2020) | LNP-encapsulated mRNA encoding human methylmalonyl-CoA mutase (hMUT) | N/A | Methylmalonic acidemia | IV | (Hou et al., 2021) | ModernaTX, Inc. | Withdrawn |
| mRNA-3705 | NCT05295433 | Phase 1/2 (2022–2034) | LNP-encapsulated mRNA encoding hMUT | SM-86, DSPC, cholesterol, and OL-56 [polyethylene glycol-lipid conjugate] | Methylmalonic Acidemia | IV | (Baek et al., 2024; Suzuki et al., 2023) | ModernaTX, Inc | Recruiting |
| mRNA-3927 |
NCT04159103 NCT05130437 |
Phase1/2 (2021−2031) | LNP-encapsulated dual mRNA therapy that encodes for (propionic-CoA mutase) PCC-A and PCC-B subunit proteins restoring PCC enzyme in liver | SM-86, DSPC, cholesterol, and OL-56 [polyethylene glycol-lipid conjugate] | Propionic Acidemia | IV | (Attarwala et al., 2023; Baek et al., 2024) | ModernaTX, Inc. | Recruiting |
| MRT5201 | NCT03767270 | Phase 1/2 (2019–2022) | LNP-encapsulated codon-optimized human OTC mRNA | N/A | Ornithine Transcarbamylase Deficiency | IV | Translate Bio, Inc. | Withdrawn | |
| MRT5005 | NCT03375047 | Phase 1/2 (2018–2021) | Aerosolized LNP-encapsulated a codon-optimized CFTR mRNA | N/A | Cystic Fibrosis | Nebulization | (Barbier et al., 2018; Hou et al., 2021; Rowe et al., 2023) | Translate Bio, Inc. | Unknown Status |
| ARCT-810 | NCT04442347 | Phase 1 (2020–2023) | Human ornithine transcarbamylase (hOTC) mRNA-LNP | N/A | Ornothine Transcarbamylase Deficiency | IV | (Yamazaki et al., 2023) | Arcturus Therapeutics, Inc. | Active, not recruiting |
| NCT05526066 | Phase 2 (2022–2024) | Recruiting | |||||||
| siRNA | |||||||||
| ALN-TTR01 | NCT01148953 | Phase 1 (2010−2012) | First gen formulation of LNPs to deliver siRNAs | DLin-MC3- DMA, DSPC, PEG2000-C-DMG, Cholesterol | Transthyretin (TTR) Mediated amyloidosis (ATTR) | IV | (Coelho et al., 2013; Schoenmaker et al., 2021a; Zatsepin et al., 2016) | Alnylam Pharmaceuticals | Completed |
| ALN-TTR02 | NCT01559077 | Phase 1 (2012) | Second gen formulation of LNPs to deliver siRNAs | DLin-MC3- DMA, DSPC, PEG2000-C-DMG, Cholesterol | Transthyretin (TTR) Mediated amyloidosis (ATTR) | IV | (Adams et al., 2018; Coelho et al., 2020; Coelho et al., 2013; Schmidt et al., 2022; Suhr et al., 2015; Zatsepin et al., 2016) | Alnylam Pharmaceuticals | Completed |
| NCT01617967 | Phase 2 (2012–2014) | Completed | |||||||
| NCT01961921 | Phase 2 (2013–2016) | Completed | |||||||
| NCT01960348 | Phase 3 (2013–2017) | Completed | |||||||
| NCT03862807 | Phase 3 (2019–2020) | Completed | |||||||
| NCT02510261 | Phase 3 (2015–2022) | Completed | |||||||
| DCR-MYC | NCT02110563 | Phase 1 (2014–2016) | DCR-MYC is a LNP-formulated Dicer substrate siRNA (DsiRNA) that silences MYC mRNA | N/A | Solid tumors, multiple myeloma, non-Hodgkins lymphoma | IV | (Chipumuro et al., 2016; Tolcher et al., 2015) | Dicerna Pharmaceuticals | Terminated |
| NCT02314052 | Phase 1/2 (2015–2016) | Hepatocellular carcinoma | Terminated | ||||||
| ND-L02-s0201 | NCT01858935 | Phase 1 (2013–2014) | LNP-encapsulated anti- hsp47 siRNA formulation designed to reversibly inhibit the expression of HSP47 | Six key lipid components including cationic, helper and targeting lipids (names of lipids not specified) | Safety, Tolerability and Pharmacokinetics in Healthy Normal Subjects | IV | (Liu et al., 2021) | Bristol-Myers Squibb | Completed |
| NCT02227459 | Phase 1/2 (2014–2016) | Moderate to Extensive Hepatic Fibrosis | Bristol-Myers Squibb | Completed | |||||
| NCT03538301 | Phase 2 (2018–2022) | Idiopathic Pulmonary Fibrosis | Nitto Denko Corporation | Completed | |||||
| TKM-080301 | NCT01437007 | Phase 1 (2011−2012) | LNP containing siRNA against PLK1 gene products | Four lipid components | Hepatocellular carcinoma | Intra-arterial /IV | (Ramanathan et al., 2013) | National Cancer Institute (NCI) | Completed |
| NCT01262235 | Phase 1/2 (2010–2015) | Adrenocortical carcinoma | (Demeure et al., 2016) | Arbutus Biopharma Corporation | Completed | ||||
| NCT02191878 | Phase 1/2 (2014–2016) | Hepatocellular carcinoma | (El Dika et al., 2019) | Arbutus Biopharma Corporation | Completed | ||||
| NBF-006 | NCT03819387 | Phase 1 (2019–2024) | LNP encapsulating siRNA targeting glutathione-S-transferase Pi | N/A | NSCLC, Pancreatic, Colorectal cancer | IV | (Hattab et al., 2021) | Nitto BioPharma., | Completed |
| BMS-986263 | NCT03142165 | Phase 1 (2017) | BMS-986263, a retinoid-conjugated lipid nanoparticle delivering small interfering RNA designed to target heat shock protein (HSP)-47 mRNA, for the treatment of advanced fibrosis. | N/A | Hepatic Cirrhosis -Hepatic Impairment | IV | (Qosa et al., 2023a; Qosa et al., 2023b) | Bristol-Myers Squibb | Completed |
| NCT03420768 | Phase 2 (2018–2019) | Completed | |||||||
| NCT04225936 | Phase 1 (2020–2021) | Completed | |||||||
| NCT04267393 | Phase 2 (2021–2024) | Terminated | |||||||
| CRISPR/Cas9 (sgRNA) | |||||||||
| NTLA-2001 |
NCT04601051 NCT06128629 |
Phase 1 (2020–2026) Phase 3 (2023–2028) |
LNP-encapsulated single guide RNA (sgRNA) targeting human TTR and a human-codon optimized mRNA sequence of S. pyogens Cas9 protein | N/A | Transthyretin (ATTR) amyloidosis with cardiomyopathy | IV | (Gillmore et al., 2021) | Intellia Therapeutics | Active, not recruiting Recruiting |
| VERVE-101 | NCT05398029 | Phase 1 (2022–2024) | LNPs for a CRISPR-based gene editing that inactivates the PCSK9 gene | N/A | Heterozygous Familial Hypercholesteremia and Cardiovascular Disease | IV | (Lee et al., 2023b) | Verve Therapeutics | Recruiting |
| VERVE-102 | NCT06164730 | Phase 1 (2024–2026) | LNP-encapsulated CRISPR-based treatment using a guide RNA targeting PCSK-9 gene | N/A | Heterozygous Familial Hypercholesteremia or Premature Coronary Artery Disease | IV | Verve Therapeutics | Not yet recruiting | |
5. Future perspectives
LNPs have emerged as promising tools for the delivery of RNA products and have attracted significant attention in recent years in preclinical and clinical studies. Not surprisingly, the intensive research taking place focuses not only on the applications that the LNPs permit, i.e., the use of siRNAs or mRNAs vs. different disease types, but also on the LNPs themselves. For example, research on ionizable lipids produced the most popular lipid, DLin-MC3-DMA. Further research will provide additional insights on improving transfection efficiency across different types of cells, including types of nucleic acids, i.e., plasmid vs RNA. For example, as mentioned above, modification in the head group influenced transfection efficiency in Kupffer cells and spleen macrophages, and demonstrated how the helper lipid could influence the fate of the LNPs (Ni et al., 2022; Zhang et al., 2021a). Thus, the type of cell targeting, tissue accumulation or immune responses can potentially be affected or regulated by the ionizable lipid and the LNP composition, which indicates further research could potentially provide important information. Another consideration is the safety of LNPs. Though efforts in the evaluation of the biocompatibility and biodegradability of the ionizable lipids and the other components take place, work remains to ensure that there are minimal or non-existent side effects, especially if taken into consideration that these particles are intended for the delivery of nucleic acids, the building blocks of life. Nonetheless, there should be an underlying understanding that the nucleic acid delivered with LNPs are prominently mRNAs or siRNAs, which do not associate with genomic changes. Finally, it should be noted that long-term stability of LNPs and their ability to freeze-dry consists of another research area that should or could be addressed in the years to come. The COVID-19 vaccines and the concerns about their proper shipping and storage presented how real-life applications of these systems have room for improvement, though the technology is still feasible. Finally, LNPs, by definition, describe a formulation based on lipids. Nonetheless, there have been important drug delivery approaches, some with established FDA-approved applications, such as protein-based carriers (i.e., albumin), that may also be able to minimize any potential side effects. Although mRNAs are unsuitable to be delivered without any carrier protection, ideally, nucleic acids with limited requirements for protection or carrier for transfection should be the ultimate goal.
Funding
This work was supported by the National Institutes of Health (NIH) through the National Institute of General Medical Science Grants P20 GM103424-21 and R&D, Research Competitiveness Subprogram (RCS) of the Louisiana Board of Regents through the Board of Regents Support Fund (Grant Number LEQSF(2021-24)-RD-A-23).
CRediT authorship contribution statement
Md. Anamul Haque: Writing – review & editing, Writing – original draft, Visualization, Conceptualization. Archana Shrestha: Writing – review & editing, Writing – original draft, Conceptualization. Constantinos M. Mikelis: Writing – review & editing, Writing – original draft, Conceptualization. George Mattheolabakis: Writing – review & editing, Writing – original draft, Visualization, Conceptualization.
Declaration of competing interest
The authors declare no competing interest.
Data availability
No data was used for the research described in the article.
References
- Abrams M.T., Koser M.L., Seitzer J., Williams S.C., DiPietro M.A., Wang W., Shaw A.W., Mao X., Jadhav V., Davide J.P., Burke P.A., Sachs A.B., Stirdivant S.M., Sepp-Lorenzino L. Evaluation of efficacy, biodistribution, and inflammation for a potent siRNA nanoparticle: effect of dexamethasone co-treatment. Mol. Ther. 2010;18:171–180. doi: 10.1038/mt.2009.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams D., Gonzalez-Duarte A., O’Riordan W.D., Yang C.C., Ueda M., Kristen A.V., Tournev I., Schmidt H.H., Coelho T., Berk J.L., Lin K.P., Vita G., Attarian S., Planté-Bordeneuve V., Mezei M.M., Campistol J.M., Buades J., Brannagan T.H., 3rd, Kim B.J., Oh J., Parman Y., Sekijima Y., Hawkins P.N., Solomon S.D., Polydefkis M., Dyck P.J., Gandhi P.J., Goyal S., Chen J., Strahs A.L., Nochur S.V., Sweetser M.T., Garg P.P., Vaishnaw A.K., Gollob J.A., Suhr O.B. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 2018;379:11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- Akinc A., Maier M.A., Manoharan M., Fitzgerald K., Jayaraman M., Barros S., Ansell S., Du X., Hope M.J., Madden T.D., Mui B.L., Semple S.C., Tam Y.K., Ciufolini M., Witzigmann D., Kulkarni J.A., van der Meel R., Cullis P.R. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019;14:1084–1087. doi: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- Alabi C.A., Love K.T., Sahay G., Yin H., Luly K.M., Langer R., Anderson D.G. Multiparametric approach for the evaluation of lipid nanoparticles for siRNA delivery. Proc. Natl. Acad. Sci. USA. 2013;110:12881–12886. doi: 10.1073/pnas.1306529110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albakr L., Alqahtani F.Y., Aleanizy F.S., Alomrani A., Badran M., Alhindas H., Al-Mohanna F. Improved delivery of miR-1296 loaded cationic nanoliposomes for effective suppression of triple negative breast cancer. Saudi Pharmaceut. J. 2021;29:446–455. doi: 10.1016/j.jsps.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldosari B.N., Alfagih I.M., Almurshedi A.S. Lipid Nanoparticles as delivery Systems for RNA-Based Vaccines. Pharmaceutics. 2021;13 doi: 10.3390/pharmaceutics13020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldrich C., Leroux-Roels I., Huang K.B., Bica M.A., Loeliger E., Schoenborn-Kellenberger O., Walz L., Leroux-Roels G., von Sonnenburg F., Oostvogels L. Proof-of-concept of a low-dose unmodified mRNA-based rabies vaccine formulated with lipid nanoparticles in human volunteers: a phase 1 trial. Vaccine. 2021;39:1310–1318. doi: 10.1016/j.vaccine.2020.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Benedicto E., Farbiak L., Márquez Ramírez M., Wang X., Johnson L.T., Mian O., Guerrero E.D., Siegwart D.J. Optimization of phospholipid chemistry for improved lipid nanoparticle (LNP) delivery of messenger RNA (mRNA) Biomater. Sci. 2022;10:549–559. doi: 10.1039/d1bm01454d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananworanich J., Lee I.T., Ensz D., Carmona L., Schaefers K., Avanesov A., Stadlbauer D., Choi A., Pucci A., McGrath S., Kuo H.H., Henry C., Chen R., Huang W., Nachbagauer R., Paris R. Healthy Adults: Final Results from a Phase 1/2 Randomized Trial. J Infect Dis; 2024. Safety and immunogenicity of mRNA-1010, an investigational seasonal influenza vaccine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S.F., Cominsky L.Y., Shimberg G.D., Gillespie R.A., Gorman J., Raab J.E., Brand J., Creanga A., Gajjala S.R., Narpala S., Cheung C.S.F., Harris D.R., Zhou T., Gordon I., Holman L., Mendoza F., Houser K.V., Chen G.L., Mascola J.R., Graham B.S., Kwong P.D., Widge A., Dropulic L.K., Ledgerwood J.E., Kanekiyo M., McDermott A.B. An influenza H1 hemagglutinin stem-only immunogen elicits a broadly cross-reactive B cell response in humans. Sci. Transl. Med. 2023;15:eade4976. doi: 10.1126/scitranslmed.ade4976. [DOI] [PubMed] [Google Scholar]
- Attarwala H., Lumley M., Liang M., Ivaturi V., Senn J. Translational Pharmacokinetic/Pharmacodynamic Model for mRNA-3927, an Investigational Therapeutic for the Treatment of Propionic Acidemia. Nucleic Acid Ther. 2023;33:141–147. doi: 10.1089/nat.2022.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- August A., Attarwala H.Z., Himansu S., Kalidindi S., Lu S., Pajon R., Han S., Lecerf J.-M., Tomassini J.E., Hard M., Ptaszek L.M., Crowe J.E., Zaks T. A phase 1 trial of lipid-encapsulated mRNA encoding a monoclonal antibody with neutralizing activity against Chikungunya virus. Nat. Med. 2021;27:2224–2233. doi: 10.1038/s41591-021-01573-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- August A., Shaw C.A., Lee H., Knightly C., Kalidindia S., Chu L., Essink B.J., Seger W., Zaks T., Smolenov I., Panther L. Safety and immunogenicity of an mRNA-based human metapneumovirus and parainfluenza virus Type 3 combined vaccine in healthy adults. Open Forum Infect. Dis. 2022;9:ofac206. doi: 10.1093/ofid/ofac206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., McGettigan J., Khetan S., Segall N., Solis J., Brosz A., Fierro C., Schwartz H., Neuzil K., Corey L., Gilbert P., Janes H., Follmann D., Marovich M., Mascola J., Polakowski L., Ledgerwood J., Graham B.S., Bennett H., Pajon R., Knightly C., Leav B., Deng W., Zhou H., Han S., Ivarsson M., Miller J., Zaks T., Group, C.S Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek R., Coughlan K., Jiang L., Liang M., Ci L., Singh H., Zhang H., Kaushal N., Rajlic I.L., Van L., Dimen R., Cavedon A., Yin L., Rice L., Frassetto A., Guey L., Finn P., Martini P.G.V. Characterizing the mechanism of action for mRNA therapeutics for the treatment of propionic acidemia, methylmalonic acidemia, and phenylketonuria. Nat. Commun. 2024;15:3804. doi: 10.1038/s41467-024-47460-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl K., Senn J.J., Yuzhakov O., Bulychev A., Brito L.A., Hassett K.J., Laska M.E., Smith M., Almarsson Ö., Thompson J., Ribeiro A., Watson M., Zaks T., Ciaramella G. Preclinical and Clinical Demonstration of Immunogenicity by mRNA Vaccines against H10N8 and H7N9 Influenza Viruses. Mol. Ther. 2017;25:1316–1327. doi: 10.1016/j.ymthe.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Hytholt C.M., Ghosh P., Dugas J., Zarraga I.E., Bandekar A. Formulating and characterizing lipid nanoparticles for gene delivery using a microfluidic mixing platform. J. Vis. Exp. 2021;(168) doi: 10.3791/62226. [DOI] [PubMed] [Google Scholar]
- Balazs D.A., Godbey W. Liposomes for use in gene delivery. J. Drug Deliv. 2011;2011 doi: 10.1155/2011/326497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball R.L., Bajaj P., Whitehead K.A. Achieving long-term stability of lipid nanoparticles: examining the effect of pH, temperature, and lyophilization. Int. J. Nanomedicine. 2017;12:305–315. doi: 10.2147/IJN.S123062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball R.L., Bajaj P., Whitehead K.A. Oral delivery of siRNA lipid nanoparticles: Fate in the GI tract. Sci. Rep. 2018;8:2178. doi: 10.1038/s41598-018-20632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball R.L., Hajj K.A., Vizelman J., Bajaj P., Whitehead K.A. Lipid Nanoparticle Formulations for Enhanced Co-delivery of siRNA and mRNA. Nano Lett. 2018;18:3814–3822. doi: 10.1021/acs.nanolett.8b01101. [DOI] [PubMed] [Google Scholar]
- Barbier A., Rosa F., Karve S., Smith L., Askew K., Kaza N., Shei R.-J., Stanford D., Heartlein M., Rowe S. 2018. In vitro and in vivo evaluation of an MRNA therapeutic for the treatment of patients with cystic fibrosis. [Google Scholar]
- Barbier A.J., Jiang A.Y., Zhang P., Wooster R., Anderson D.G. The clinical progress of mRNA vaccines and immunotherapies. Nat. Biotechnol. 2022;40:840–854. doi: 10.1038/s41587-022-01294-2. [DOI] [PubMed] [Google Scholar]
- Barenholz Y. Doxil®--the first FDA-approved nano-drug: lessons learned. J. Control. Release. 2012;160:117–134. doi: 10.1016/j.jconrel.2012.03.020. [DOI] [PubMed] [Google Scholar]
- Barenholz Y., Thompson T.E. Sphingomyelin: biophysical aspects. Chem. Phys. Lipids. 1999;102:29–34. doi: 10.1016/s0009-3084(99)00072-9. [DOI] [PubMed] [Google Scholar]
- Batty C.J., Heise M.T., Bachelder E.M., Ainslie K.M. Vaccine formulations in clinical development for the prevention of severe acute respiratory syndrome coronavirus 2 infection. Adv. Drug Deliv. Rev. 2021;169:168–189. doi: 10.1016/j.addr.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belliveau N.M., Huft J., Lin P.J., Chen S., Leung A.K., Leaver T.J., Wild A.W., Lee J.B., Taylor R.J., Tam Y.K., Hansen C.L., Cullis P.R. Microfluidic Synthesis of Highly Potent Limit-size Lipid Nanoparticles for in Vivo delivery of siRNA. Mol. Thera. Nucleic Acids. 2012;1 doi: 10.1038/mtna.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezelya A., Küçüktürkmen B., Bozkır A. Microfluidic Devices for Precision Nanoparticle Production. Micro. 2023;3:822–866. [Google Scholar]
- Billingsley M.M., Singh N., Ravikumar P., Zhang R., June C.H., Mitchell M.J. Ionizable lipid nanoparticle-mediated mrna delivery for human CAR T cell engineering. Nano Lett. 2020;20:1578–1589. doi: 10.1021/acs.nanolett.9b04246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollman B., Nunna N., Bahl K., Hsiao C.J., Bennett H., Butler S., Foreman B., Burgomaster K.E., Aleshnick M., Kong W.-P., Fisher B.E., Ruckwardt T.J., Morabito K.M., Graham B.S., Dowd K.A., Pierson T.C., Carfi A. An optimized messenger RNA vaccine candidate protects non-human primates from Zika virus infection. npj Vaccines. 2023;8:58. doi: 10.1038/s41541-023-00656-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito L.A., Chan M., Shaw C.A., Hekele A., Carsillo T., Schaefer M., Archer J., Seubert A., Otten G.R., Beard C.W., Dey A.K., Lilja A., Valiante N.M., Mason P.W., Mandl C.W., Barnett S.W., Dormitzer P.R., Ulmer J.B., Singh M., O'Hagan D.T., Geall A.J. A cationic nanoemulsion for the delivery of next-generation RNA vaccines. Mol. Ther. 2014;22:2118–2129. doi: 10.1038/mt.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck J., Grossen P., Cullis P.R., Huwyler J., Witzigmann D. Lipid-based DNA Therapeutics: Hallmarks of Non-Viral Gene delivery. ACS Nano. 2019;13:3754–3782. doi: 10.1021/acsnano.8b07858. [DOI] [PubMed] [Google Scholar]
- Bulbake U., Doppalapudi S., Kommineni N., Khan W. Liposomal formulations in clinical use: an updated review. Pharmaceutics. 2017;9 doi: 10.3390/pharmaceutics9020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burris H.A., Patel M.R., Cho D.C., Clarke J.M., Gutierrez M., Zaks T.Z., Frederick J., Hopson K., Mody K., Binanti-Berube A., Robert-Tissot C., Goldstein B., Breton B., Sun J., Zhong S., Pruitt S.K., Keating K., Meehan R.S., Gainor J.F. A phase I multicenter study to assess the safety, tolerability, and immunogenicity of mRNA-4157 alone in patients with resected solid tumors and in combination with pembrolizumab in patients with unresectable solid tumors. J. Clin. Oncol. 2019;37:2523. [Google Scholar]
- Butt M.H., Zaman M., Ahmad A., Khan R., Mallhi T.H., Hasan M.M., Khan Y.H., Hafeez S., Massoud E.E.S., Rahman M.H., Cavalu S. Appraisal for the potential of viral and nonviral vectors in gene therapy: a review. Genes (Basel) 2022;13 doi: 10.3390/genes13081370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafri G., Gartner J.J., Zaks T., Hopson K., Levin N., Paria B.C., Parkhurst M.R., Yossef R., Lowery F.J., Jafferji M.S., Prickett T.D., Goff S.L., McGowan C.T., Seitter S., Shindorf M.L., Parikh A., Chatani P.D., Robbins P.F., Rosenberg S.A. mRNA vaccine-induced neoantigen-specific T cell immunity in patients with gastrointestinal cancer. J. Clin. Invest. 2020;130:5976–5988. doi: 10.1172/JCI134915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas M., Campbell R.A., Yanez Arteta M., Lawrence M.J., Sebastiani F. Review of structural design guiding the development of lipid nanoparticles for nucleic acid delivery. Curr. Opin. Colloid Interface Sci. 2023;66 [Google Scholar]
- Carrasco M.J., Alishetty S., Alameh M.-G., Said H., Wright L., Paige M., Soliman O., Weissman D., Cleveland T.E., Grishaev A., Buschmann M.D. Ionization and structural properties of mRNA lipid nanoparticles influence expression in intramuscular and intravascular administration. Commun. Biol. 2021;4:956. doi: 10.1038/s42003-021-02441-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandela A., Ueno Y. Systemic delivery of Small Interfering RNA Therapeutics: Obstacles and advances. Rev. Agricult. Sci. 2019;7:10–28. [Google Scholar]
- Chander N., Basha G., Yan Cheng M.H., Witzigmann D., Cullis P.R. Lipid nanoparticle mRNA systems containing high levels of sphingomyelin engender higher protein expression in hepatic and extra-hepatic tissues. Mol. Ther. Meth. Clin. Dev. 2023;30:235–245. doi: 10.1016/j.omtm.2023.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.I., Yeh M.K. Clinical development of liposome-based drugs: formulation, characterization, and therapeutic efficacy. Int. J. Nanomedicine. 2012;7:49–60. doi: 10.2147/IJN.S26766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Love K.T., Chen Y., Eltoukhy A.A., Kastrup C., Sahay G., Jeon A., Dong Y., Whitehead K.A., Anderson D.G. Rapid discovery of potent siRNA-containing lipid nanoparticles enabled by controlled microfluidic formulation. J. Am. Chem. Soc. 2012;134:6948–6951. doi: 10.1021/ja301621z. [DOI] [PubMed] [Google Scholar]
- Chen S., Tam Y.Y., Lin P.J., Leung A.K., Tam Y.K., Cullis P.R. Development of lipid nanoparticle formulations of siRNA for hepatocyte gene silencing following subcutaneous administration. J. Contro. Release. 2014;196:106–112. doi: 10.1016/j.jconrel.2014.09.025. [DOI] [PubMed] [Google Scholar]
- Chen S., Tam Y.Y.C., Lin P.J.C., Sung M.M.H., Tam Y.K., Cullis P.R. Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA. J. Contro. Release. 2016;235:236–244. doi: 10.1016/j.jconrel.2016.05.059. [DOI] [PubMed] [Google Scholar]
- Chen Z.Z., Gao Z.M., Zeng D.P., Liu B., Luan Y., Qin K.R. A Y-shaped microfluidic device to study the combined effect of wall shear stress and ATP signals on intracellular calcium dynamics in vascular endothelial cells. Micromachines. 2016;7 doi: 10.3390/mi7110213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.L., Li X.F., Dai X.H., Li N., Cheng M.L., Huang Z., Shen J., Ge Y.H., Shen Z.W., Deng Y.Q., Yang S.Y., Zhao H., Zhang N.N., Zhang Y.F., Wei L., Wu K.Q., Zhu M.F., Peng C.G., Jiang Q., Cao S.C., Li Y.H., Zhao D.H., Wu X.H., Ni L., Shen H.H., Dong C., Ying B., Sheng G.P., Qin C.F., Gao H.N., Li L.J. Safety and immunogenicity of the SARS-CoV-2 ARCoV mRNA vaccine in Chinese adults: a randomised, double-blind, placebo-controlled, phase 1 trial. The Lancet. Microbe. 2022;3:e193–e202. doi: 10.1016/S2666-5247(21)00280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Lee R.J. The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv. Drug Deliv. Rev. 2016;99:129–137. doi: 10.1016/j.addr.2016.01.022. [DOI] [PubMed] [Google Scholar]
- Cheng Q., Wei T., Jia Y., Farbiak L., Zhou K., Zhang S., Wei Y., Zhu H., Siegwart D.J. Dendrimer-based lipid nanoparticles deliver therapeutic FAH mRNA to normalize liver function and extend survival in a mouse model of hepatorenal tyrosinemia Type I. Adv. Materials (Deerfield Beach, Fla.) 2018;30 doi: 10.1002/adma.201805308. [DOI] [PubMed] [Google Scholar]
- Chipumuro E., Siddiquee Z., Ganesh S., Shui S., Shah A., Kim B., Chen D., Pandya P., Storr R., Wang W., Dudek H., Lai C., Abrams M., Brown B. Abstract 2925: Anti-tumor activity of a MYC-targeting dicer substrate siRNA in combination with BRD4/CDK7 inhibitors. Cancer Res. 2016;76:2925. [Google Scholar]
- Choi A., Koch M., Wu K., Chu L., Ma L., Hill A., Nunna N., Huang W., Oestreicher J., Colpitts T., Bennett H., Legault H., Paila Y., Nestorova B., Ding B., Montefiori D., Pajon R., Miller J.M., Leav B., Carfi A., McPhee R., Edwards D.K. Safety and immunogenicity of SARS-CoV-2 variant mRNA vaccine boosters in healthy adults: an interim analysis. Nat. Med. 2021;27:2025–2031. doi: 10.1038/s41591-021-01527-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choung S., Kim Y.J., Kim S., Park H.O., Choi Y.C. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem. Biophys. Res. Commun. 2006;342:919–927. doi: 10.1016/j.bbrc.2006.02.049. [DOI] [PubMed] [Google Scholar]
- Coelho T., Adams D., Silva A., Lozeron P., Hawkins P.N., Mant T., Perez J., Chiesa J., Warrington S., Tranter E., Munisamy M., Falzone R., Harrop J., Cehelsky J., Bettencourt B.R., Geissler M., Butler J.S., Sehgal A., Meyers R.E., Chen Q., Borland T., Hutabarat R.M., Clausen V.A., Alvarez R., Fitzgerald K., Gamba-Vitalo C., Nochur S.V., Vaishnaw A.K., Sah D.W., Gollob J.A., Suhr O.B. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 2013;369:819–829. doi: 10.1056/NEJMoa1208760. [DOI] [PubMed] [Google Scholar]
- Coelho T., Adams D., Conceição I., Waddington-Cruz M., Schmidt H.H., Buades J., Campistol J., Berk J.L., Polydefkis M., Wang J.J., Chen J., Sweetser M.T., Gollob J., Suhr O.B. A phase II, open-label, extension study of long-term patisiran treatment in patients with hereditary transthyretin-mediated (hATTR) amyloidosis. Orphanet J. Rare Dis. 2020;15:179. doi: 10.1186/s13023-020-01399-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford R., Dogdas B., Keough E., Haas R.M., Wepukhulu W., Krotzer S., Burke P.A., Sepp-Lorenzino L., Bagchi A., Howell B.J. Analysis of lipid nanoparticles by Cryo-EM for characterizing siRNA delivery vehicles. Int. J. Pharm. 2011;403:237–244. doi: 10.1016/j.ijpharm.2010.10.025. [DOI] [PubMed] [Google Scholar]
- Curreri A., Sankholkar D., Mitragotri S., Zhao Z. RNA therapeutics in the clinic. Bioeng. Transl. Med. 2023;8 doi: 10.1002/btm2.10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damase T.R., Sukhovershin R., Boada C., Taraballi F., Pettigrew R.I., Cooke J.P. The Limitless Future of RNA Therapeutics. Front. Bioeng. Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.628137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alwis R., Gan E.S., Chen S., Leong Y.S., Tan H.C., Zhang S.L., Yau C., Low J.G.H., Kalimuddin S., Matsuda D., Allen E.C., Hartman P., Park K.J., Alayyoubi M., Bhaskaran H., Dukanovic A., Bao Y., Clemente B., Vega J., Roberts S., Gonzalez J.A., Sablad M., Yelin R., Taylor W., Tachikawa K., Parker S., Karmali P., Davis J., Sullivan B.M., Sullivan S.M., Hughes S.G., Chivukula P., Ooi E.E. A single dose of self-transcribing and replicating RNA-based SARS-CoV-2 vaccine produces protective adaptive immunity in mice. Mol. Ther. 2021;29:1970–1983. doi: 10.1016/j.ymthe.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeure M.J., Armaghany T., Ejadi S., Ramanathan R.K., Elfiky A., Strosberg J.R., Smith D.C., Whitsett T., Liang W.S., Sekar S., Carpten J.D., Fredlund P., Niforos D., Dye A., Gahir S., Semple S.C., Kowalski M.M. A phase I/II study of TKM-080301, a PLK1-targeted RNAi in patients with adrenocortical cancer (ACC) J. Clin. Oncol. 2016;34:2547. [Google Scholar]
- Deng Z., Yang H., Tian Y., Liu Z., Sun F., Yang P. An OX40L mRNA vaccine inhibits the growth of hepatocellular carcinoma. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.975408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdy S.F. Overcoming cellular barriers for RNA therapeutics. Nat. Biotechnol. 2017;35:222–229. doi: 10.1038/nbt.3802. [DOI] [PubMed] [Google Scholar]
- El Dika I., Lim H.Y., Yong W.P., Lin C.C., Yoon J.H., Modiano M., Freilich B., Choi H.J., Chao T.Y., Kelley R.K., Brown J., Knox J., Ryoo B.Y., Yau T., Abou-Alfa G.K. An Open-Label, Multicenter, phase I, Dose Escalation Study with phase II expansion Cohort to Determine the Safety, Pharmacokinetics, and preliminary Antitumor activity of Intravenous TKM-080301 in Subjects with Advanced Hepatocellular Carcinoma. Oncologist. 2019;24:747–e218. doi: 10.1634/theoncologist.2018-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercole F., Whittaker M.R., Quinn J.F., Davis T.P. Cholesterol Modified Self-Assemblies and their Application to Nanomedicine. Biomacromolecules. 2015;16:1886–1914. doi: 10.1021/acs.biomac.5b00550. [DOI] [PubMed] [Google Scholar]
- Essink B., Chu L., Seger W., Barranco E., Le Cam N., Bennett H., Faughnan V., Pajon R., Paila Y.D., Bollman B., Wang S., Dooley J., Kalidindi S., Leav B. The safety and immunogenicity of two Zika virus mRNA vaccine candidates in healthy flavivirus baseline seropositive and seronegative adults: the results of two randomised, placebo-controlled, dose-ranging, phase 1 clinical trials. Lancet Infect. Dis. 2023;23:621–633. doi: 10.1016/S1473-3099(22)00764-2. [DOI] [PubMed] [Google Scholar]
- Eygeris Y., Patel S., Jozic A., Sahay G. Deconvoluting Lipid Nanoparticle Structure for Messenger RNA delivery. Nano Lett. 2020;20:4543–4549. doi: 10.1021/acs.nanolett.0c01386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eygeris Y., Gupta M., Kim J., Sahay G. Chemistry of Lipid Nanoparticles for RNA delivery. Acc. Chem. Res. 2022;55:2–12. doi: 10.1021/acs.accounts.1c00544. [DOI] [PubMed] [Google Scholar]
- Feldman R.A., Fuhr R., Smolenov I., Ribeiro A., Panther L., Watson M., Senn J.J., Smith M., Almarsson Ӧ., Pujar H.S., Laska M.E., Thompson J., Zaks T., Ciaramella G. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine. 2019;37:3326–3334. doi: 10.1016/j.vaccine.2019.04.074. [DOI] [PubMed] [Google Scholar]
- Felgner P.L., Gadek T.R., Holm M., Roman R., Chan H.W., Wenz M., Northrop J.P., Ringold G.M., Danielsen M. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. USA. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng R., Patil S., Zhao X., Miao Z., Qian A. RNA Therapeutics - Research and Clinical Advancements. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.710738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton O.S., Kauffman K.J., Kaczmarek J.C., McClellan R.L., Jhunjhunwala S., Tibbitt M.W., Zeng M.D., Appel E.A., Dorkin J.R., Mir F.F., Yang J.H., Oberli M.A., Heartlein M.W., DeRosa F., Langer R., Anderson D.G. Synthesis and biological evaluation of ionizable lipid materials for the in vivo delivery of messenger RNA to B lymphocytes. Adv. Materials (Deerfield Beach, Fla.) 2017;29 doi: 10.1002/adma.201606944. [DOI] [PubMed] [Google Scholar]
- Ganesan P., Narayanasamy D. Lipid nanoparticles: different preparation techniques, characterization, hurdles, and strategies for the production of solid lipid nanoparticles and nanostructured lipid carriers for oral drug delivery. Sustain. Chem. Pharm. 2017;6:37–56. [Google Scholar]
- Garbuzenko O.B., Kuzmov A., Taratula O., Pine S.R., Minko T. Strategy to enhance lung cancer treatment by five essential elements: inhalation delivery, nanotechnology, tumor-receptor targeting, chemo- and gene therapy. Theranostics. 2019;9:8362–8376. doi: 10.7150/thno.39816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto M.S., Najahi-Missaoui W. Lyophilization of nanoparticles, does it really work? Overview of the current status and challenges. Int. J. Mol. Sci. 2023;24 doi: 10.3390/ijms241814041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Chen L., Zhao B., Yuan W. Rationale and application of PEGylated lipid-based system for advanced target delivery of siRNA. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.598175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geall A.J., Verma A., Otten G.R., Shaw C.A., Hekele A., Banerjee K., Cu Y., Beard C.W., Brito L.A., Krucker T., O'Hagan D.T., Singh M., Mason P.W., Valiante N.M., Dormitzer P.R., Barnett S.W., Rappuoli R., Ulmer J.B., Mandl C.W. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. USA. 2012;109:14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillmore J.D., Gane E., Taubel J., Kao J., Fontana M., Maitland M.L., Seitzer J., O’Connell D., Walsh K.R., Wood K., Phillips J., Xu Y., Amaral A., Boyd A.P., Cehelsky J.E., McKee M.D., Schiermeier A., Harari O., Murphy A., Kyratsous C.A., Zambrowicz B., Soltys R., Gutstein D.E., Leonard J., Sepp-Lorenzino L., Lebwohl D. CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N. Engl. J. Med. 2021;385:493–502. doi: 10.1056/NEJMoa2107454. [DOI] [PubMed] [Google Scholar]
- Gomez-Aguado I., Rodriguez-Castejon J., Vicente-Pascual M., Rodriguez-Gascon A., Pozo-Rodriguez A.D., Solinis Aspiazu M.A. Nucleic acid delivery by solid lipid nanoparticles containing switchable lipids: plasmid DNA vs. messenger RNA. Molecules. 2020;25 doi: 10.3390/molecules25245995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S., McPherson J.D., McCombie W.R. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami R., Chatzikleanthous D., Lou G., Giusti F., Bonci A., Taccone M., Brazzoli M., Gallorini S., Ferlenghi I., Berti F., O’Hagan D.T., Pergola C., Baudner B.C., Adamo R. Mannosylation of LNP results in improved potency for self-amplifying RNA (SAM) vaccines. ACS Infect. Dis. 2019;5:1546–1558. doi: 10.1021/acsinfecdis.9b00084. [DOI] [PubMed] [Google Scholar]
- Granot Y., Peer D. Delivering the right message: challenges and opportunities in lipid nanoparticles-mediated modified mRNA therapeutics-an innate immune system standpoint. Semin. Immunol. 2017;34:68–77. doi: 10.1016/j.smim.2017.08.015. [DOI] [PubMed] [Google Scholar]
- Guéguen C., Ben Chimol T., Briand M., Renaud K., Seiler M., Ziesel M., Erbacher P., Hellal M. Evaluating how cationic lipid affects mRNA-LNP physical properties and biodistribution. Eur. J. Pharm. Biopharm. 2024;195:114077. doi: 10.1016/j.ejpb.2023.08.002. ISSN 0939-6411. [DOI] [PubMed] [Google Scholar]
- Gyanani V., Goswami R. Key Design Features Of Lipid Nanoparticles And Electrostatic Charge-Based Lipid Nanoparticle Targeting. Pharmaceutics. 2023;15 doi: 10.3390/pharmaceutics15041184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghiralsadat F., Amoabediny G., Naderinezhad S., Forouzanfar T., Helder M.N., Zandieh-Doulabi B. Preparation of PEGylated cationic nanoliposome-siRNA complexes for cancer therapy. Artif Cells Nanomed. Biotechnol. 2018;46:684–692. doi: 10.1080/21691401.2018.1434533. [DOI] [PubMed] [Google Scholar]
- Hajiaghapour Asr M., Dayani F., Saedi Segherloo F., Kamedi A., Neill A.O., MacLoughlin R., Doroudian M. Lipid nanoparticles as promising carriers for mRNA vaccines for viral lung infections. Pharmaceutics. 2023;15 doi: 10.3390/pharmaceutics15041127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajj K.A., Ball R.L., Deluty S.B., Singh S.R., Strelkova D., Knapp C.M., Whitehead K.A. Branched-tail lipid nanoparticles potently deliver mRNA in vivo due to enhanced ionization at endosomal pH. Small (Weinheim an der Bergstrasse, Germany) 2019;15 doi: 10.1002/smll.201805097. [DOI] [PubMed] [Google Scholar]
- Hald Albertsen C., Kulkarni J.A., Witzigmann D., Lind M., Petersson K., Simonsen J.B. The role of lipid components in lipid nanoparticles for vaccines and gene therapy. Adv. Drug Deliv. Rev. 2022;188 doi: 10.1016/j.addr.2022.114416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama B., Mahajan G., Fodor P.S., Kaufman M., Kothapalli C.R. Evolution of mixing in a microfluidic reverse-staggered herringbone micromixer. Microfluid. Nanofluid. 2018;22:54. [Google Scholar]
- Hamid O., Hellman M., Carneiro B., Marron T., Subbiah V., Mehmi I., Eyles J., Dubois V., Ridgway B., Hamid O., Gasco Hernandez A. 19O preliminary safety, antitumor activity and pharmacodynamics results of HIT-IT MEDI1191 (mRNA IL-12) in patients with advanced solid tumours and superficial lesions. Ann. Oncol. 2021;32:S9. [Google Scholar]
- Hamilton A.G., Swingle K.L., Mitchell M.J. Biotechnology: Overcoming biological barriers to nucleic acid delivery using lipid nanoparticles. PLoS Biol. 2023;21 doi: 10.1371/journal.pbio.3002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Lim J., Wang C.J., Han J.H., Shin H.E., Kim S.N., Jeong D., Lee S.H., Chun B.H., Park C.G., Park W. Lipid nanoparticle-based mRNA delivery systems for cancer immunotherapy. Nano Convergence. 2023;10:36. doi: 10.1186/s40580-023-00385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., Himansu S., Deterling J., Geilich B.M., Ketova T., Mihai C., Lynn A., McFadyen I., Moore M.J., Senn J.J., Stanton M.G., Almarsson Ö., Ciaramella G., Brito L.A. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Thera. Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattab D., Gazzali A.M., Bakhtiar A. Clinical advances of siRNA-based nanotherapeutics for cancer treatment. Pharmaceutics. 2021;13 doi: 10.3390/pharmaceutics13071009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyes J., Palmer L., Bremner K., MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Control. Release. 2005;107:276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Hood R.R., DeVoe D.L. High-throughput continuous flow production of nanoscale liposomes by microfluidic vertical flow focusing. Small (Weinheim an der Bergstrasse, Germany) 2015;11:5790–5799. doi: 10.1002/smll.201501345. [DOI] [PubMed] [Google Scholar]
- Hormann K., Zimmer A. Drug delivery and drug targeting with parenteral lipid nanoemulsions - a review. J. Control. Release. 2016;223:85–98. doi: 10.1016/j.jconrel.2015.12.016. [DOI] [PubMed] [Google Scholar]
- Hossian A., Mackenzie G.G., Mattheolabakis G. miRNAs in gastrointestinal diseases: can we effectively deliver RNA-based therapeutics orally? Nanomedicine (London) 2019;14:2873–2889. doi: 10.2217/nnm-2019-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021;6:1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu W.L., Chen H.L., Liou W., Lin H.K., Liu W.L. Mesomorphic complexes of DNA with the mixtures of a cationic surfactant and a neutral lipid. Langmuir. 2005;21:9426–9431. doi: 10.1021/la051863e. [DOI] [PubMed] [Google Scholar]
- Hu B., Zhong L., Weng Y., Peng L., Huang Y., Zhao Y., Liang X.-J. Therapeutic siRNA: state of the art. Signal Transduct. Target. Ther. 2020;5:101. doi: 10.1038/s41392-020-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn A., Vreeland W.N., Gaitan M., Locascio L.E. Controlled vesicle self-assembly in microfluidic channels with hydrodynamic focusing. J. Am. Chem. Soc. 2004;126:2674–2675. doi: 10.1021/ja0318030. [DOI] [PubMed] [Google Scholar]
- Jayaraman M., Ansell S.M., Mui B.L., Tam Y.K., Chen J., Du X., Butler D., Eltepu L., Matsuda S., Narayanannair J.K., Rajeev K.G., Hafez I.M., Akinc A., Maier M.A., Tracy M.A., Cullis P.R., Madden T.D., Manoharan M., Hope M.J. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int. Ed. Eng. 2012;51:8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno A., Gupta S., Sullivan R., Do K.T., Akerley W.L., Wang D., Teoh D., Schalper K., Zacharek S.J., Sun J., Laino A.S., Frederick J., Zhou H., Randolph W., Pascarella S., Johansen L., Cohen P.S., Meehan R.S., Bauer T.M. Abstract CT032: a phase 1/2, open-label, multicenter, dose escalation and efficacy study of mRNA-2416, a lipid nanoparticle encapsulated mRNA encoding human OX40L, for intratumoral injection alone or in combination with durvalumab for patients with advanced malignancies. Cancer Res. 2020;80 CT032-CT032. [Google Scholar]
- Julie B., Howard B., Jeffrey C., Manish P., Daniel C., Martin G., Ricklie J., Aaron S., Pamela C., Joshua F., Celine R.-T., Honghong Z., Kinjal M., Karen K., Robert M., Justin G. 798 Safety, tolerability, and immunogenicity of mRNA-4157 in combination with pembrolizumab in subjects with unresectable solid tumors (KEYNOTE-603): an update. J. Immunother. Cancer. 2020;8:A477. [Google Scholar]
- Jung H.N., Lee S.Y., Lee S., Youn H., Im H.J. Lipid nanoparticles for delivery of RNA therapeutics: current status and the role of in vivo imaging. Theranostics. 2022;12:7509–7531. doi: 10.7150/thno.77259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens D.C., Deßloch L., Porras-Gonzalez D., Winkeljann J., Zielinski S., Munschauer M., Hörner A.L., Burgstaller G., Winkeljann B., Merkel O.M. Lab-scale siRNA and mRNA LNP manufacturing by various microfluidic mixing techniques – an evaluation of particle properties and efficiency. OpenNano. 2023;12 [Google Scholar]
- Kaczmarek J.C., Kowalski P.S., Anderson D.G. Advances in the delivery of RNA therapeutics: from concept to clinical reality. Genome Med. 2017;9:60. doi: 10.1186/s13073-017-0450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya M., Matsumoto M., Yamashita K., Izumi T., Kawaguchi M., Mizukami S., Tsurumaru M., Mukai H., Kawakami S. Stability study of mRNA-lipid nanoparticles exposed to various conditions based on the evaluation between physicochemical properties and their relation with protein expression ability. Pharmaceutics. 2022;14 doi: 10.3390/pharmaceutics14112357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman K.J., Dorkin J.R., Yang J.H., Heartlein M.W., DeRosa F., Mir F.F., Fenton O.S., Anderson D.G. Optimization of lipid nanoparticle formulations for mRNA delivery in vivo with fractional factorial and definitive screening designs. Nano Lett. 2015;15:7300–7306. doi: 10.1021/acs.nanolett.5b02497. [DOI] [PubMed] [Google Scholar]
- Kauffman K.J., Dorkin J.R., Yang J.H., Heartlein M.W., DeRosa F., Mir F.F., Fenton O.S., Anderson D.G. Optimization of Lipid Nanoparticle Formulations for mRNA delivery in vivo with fractional factorial and definitive screening designs. Nano Lett. 2015;15:7300–7306. doi: 10.1021/acs.nanolett.5b02497. [DOI] [PubMed] [Google Scholar]
- Kedmi R., Ben-Arie N., Peer D. The systemic toxicity of positively charged lipid nanoparticles and the role of Toll-like receptor 4 in immune activation. Biomaterials. 2010;31:6867–6875. doi: 10.1016/j.biomaterials.2010.05.027. [DOI] [PubMed] [Google Scholar]
- Khan S., Sharma A., Jain V. An overview of nanostructured lipid carriers and its application in drug delivery through different routes. Adv Pharm Bull. 2023;13:446–460. doi: 10.34172/apb.2023.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare P., Dave K.M., Kamte Y.S., Manoharan M.A., O’Donnell L.A., Manickam D.S. Development of lipidoid nanoparticles for siRNA delivery to neural cells. AAPS J. 2021;24:8. doi: 10.1208/s12248-021-00653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.K. RNA therapy: rich history, various applications and unlimited future prospects. Exp. Mol. Med. 2022;54:455–465. doi: 10.1038/s12276-022-00757-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Eygeris Y., Gupta M., Sahay G. Self-assembled mRNA vaccines. Adv. Drug Deliv. Rev. 2021;170:83–112. doi: 10.1016/j.addr.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.C., Sekhon S.S., Shin W.R., Ahn G., Cho B.K., Ahn J.Y., Kim Y.H. Modifications of mRNA vaccine structural elements for improving mRNA stability and translation efficiency. Mol. Cell. Toxicol. 2022;18:1–8. doi: 10.1007/s13273-021-00171-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B., Hosn R.R., Remba T., Yun D., Li N., Abraham W., Melo M.B., Cortes M., Li B., Zhang Y., Dong Y., Irvine D.J. Optimization of storage conditions for lipid nanoparticle-formulated self-replicating RNA vaccines. J. Control. Release. 2023;353:241–253. doi: 10.1016/j.jconrel.2022.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon E., Elia U., Peer D. Principles for designing an optimal mRNA lipid nanoparticle vaccine. Curr. Opin. Biotechnol. 2022;73:329–336. doi: 10.1016/j.copbio.2021.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski P.S., Rudra A., Miao L., Anderson D.G. Delivering the Messenger: advances in Technologies for Therapeutic mRNA delivery. Mol. Ther. 2019;27:710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremsner P.G., Mann P., Kroidl A., Leroux-Roels I., Schindler C., Gabor J.J., Schunk M., Leroux-Roels G., Bosch J.J., Fendel R., Kreidenweiss A., Velavan T.P., Fotin-Mleczek M., Mueller S.O., Quintini G., Schönborn-Kellenberger O., Vahrenhorst D., Verstraeten T., Alves de Mesquita M., Walz L., Wolz O.O., Oostvogels L. Safety and immunogenicity of an mRNA-lipid nanoparticle vaccine candidate against SARS-CoV-2: A phase 1 randomized clinical trial. Wien. Klin. Wochenschr. 2021;133:931–941. doi: 10.1007/s00508-021-01922-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni J.A., Myhre J.L., Chen S., Tam Y.Y.C., Danescu A., Richman J.M., Cullis P.R. Design of lipid nanoparticles for in vitro and in vivo delivery of plasmid DNA. Nanomedicine. 2017;13:1377–1387. doi: 10.1016/j.nano.2016.12.014. [DOI] [PubMed] [Google Scholar]
- Kulkarni J.A., Tam Y.Y.C., Chen S., Tam Y.K., Zaifman J., Cullis P.R., Biswas S. Rapid synthesis of lipid nanoparticles containing hydrophobic inorganic nanoparticles. Nanoscale. 2017;9:13600–13609. doi: 10.1039/c7nr03272b. [DOI] [PubMed] [Google Scholar]
- Kulkarni J.A., Darjuan M.M., Mercer J.E., Chen S., van der Meel R., Thewalt J.L., Tam Y.Y.C., Cullis P.R. On the formation and morphology of lipid nanoparticles containing ionizable cationic lipids and siRNA. ACS Nano. 2018;12:4787–4795. doi: 10.1021/acsnano.8b01516. [DOI] [PubMed] [Google Scholar]
- Kulkarni J.A., Witzigmann D., Leung J., Tam Y.Y.C., Cullis P.R. On the role of helper lipids in lipid nanoparticle formulations of siRNA. Nanoscale. 2019;11:21733–21739. doi: 10.1039/c9nr09347h. [DOI] [PubMed] [Google Scholar]
- Kumar V., Qin J., Jiang Y., Duncan R., Brigham B., Fishman S., Nair J.K., Akinc A., Barros S.A., Kasperkovitz P.V. Shielding of lipid nanoparticles for siRNA delivery: impact on physicochemical properties, cytokine induction, and efficacy. molecular therapy. Nucleic acids. 2014;3 doi: 10.1038/mtna.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labatut A.E., Mattheolabakis G. Non-viral based miR delivery and recent developments. Eur. J. Pharm. Biopharm. 2018;128:82–90. doi: 10.1016/j.ejpb.2018.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganà A., Veneziano D., Russo F., Pulvirenti A., Giugno R., Croce C.M., Ferro A. Computational design of artificial RNA molecules for gene regulation. Meth. Mol. Biol. (Clifton, N.J.) 2015;1269:393–412. doi: 10.1007/978-1-4939-2291-8_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahooti B., Poudel S., Mikelis C.M., Mattheolabakis G. MiRNAs as anti-angiogenic adjuvant therapy in cancer: synopsis and potential. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.705634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahooti B., Akwii R.G., Zahra F.T., Sajib M.S., Lamprou M., Alobaida A., Lionakis M.S., Mattheolabakis G., Mikelis C.M. Targeting endothelial permeability in the EPR effect. J. Control. Release. 2023;361:212–235. doi: 10.1016/j.jconrel.2023.07.039. [DOI] [PubMed] [Google Scholar]
- Lam K., Schreiner P., Leung A., Stainton P., Reid S., Yaworski E., Lutwyche P., Heyes J. Optimizing lipid nanoparticles for delivery in primates. Adv. Materials (Deerfield Beach, Fla.) 2023;35:e2211420. doi: 10.1002/adma.202211420. [DOI] [PubMed] [Google Scholar]
- Lamb Y.N. BNT162b2 mRNA COVID-19 Vaccine: first Approval. Drugs. 2021;81:495–501. doi: 10.1007/s40265-021-01480-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landesman-Milo D., Peer D. Toxicity profiling of several common RNAi-based nanomedicines: a comparative study. Drug Deliv. Transl. Res. 2014;4:96–103. doi: 10.1007/s13346-013-0158-7. [DOI] [PubMed] [Google Scholar]
- Larson N.R., Hu G., Wei Y., Tuesesca A., Forrest M.L., Middaugh C.R. pH-Dependent phase behavior and stability of cationic lipid-mRNA nanoparticles. J. Pharm. Sci. 2022;111(3):690–698. doi: 10.1016/j.xphs.2021.11.004. [DOI] [PubMed] [Google Scholar]
- Lazzaro S., Giovani C., Mangiavacchi S., Magini D., Maione D., Baudner B., Geall A.J., De Gregorio E., D'Oro U., Buonsanti C. CD8 T-cell priming upon mRNA vaccination is restricted to bone-marrow-derived antigen-presenting cells and may involve antigen transfer from myocytes. Immunology. 2015;146:312–326. doi: 10.1111/imm.12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechanteur A., Sanna V., Duchemin A., Evrard B., Mottet D., Piel G. Cationic liposomes carrying siRNA: impact of lipid composition on physicochemical properties, cytotoxicity and endosomal escape. Nanomaterials (Basel) 2018:8. doi: 10.3390/nano8050270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I.T., Nachbagauer R., Ensz D., Schwartz H., Carmona L., Schaefers K., Avanesov A., Stadlbauer D., Henry C., Chen R., Huang W., Schrempp D.R., Ananworanich J., Paris R. Safety and immunogenicity of a phase 1/2 randomized clinical trial of a quadrivalent, mRNA-based seasonal influenza vaccine (mRNA-1010) in healthy adults: interim analysis. Nat. Commun. 2023;14:3631. doi: 10.1038/s41467-023-39376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.G., Mazzola A.M., Braun M.C., Platt C., Vafai S.B., Kathiresan S., Rohde E., Bellinger A.M., Khera A.V. Efficacy and safety of an investigational single-course CRISPR Base-editing therapy targeting PCSK9 in nonhuman primate and mouse models. Circulation. 2023;147:242–253. doi: 10.1161/CIRCULATIONAHA.122.062132. [DOI] [PubMed] [Google Scholar]
- Leung A.K., Hafez I.M., Baoukina S., Belliveau N.M., Zhigaltsev I.V., Afshinmanesh E., Tieleman D.P., Hansen C.L., Hope M.J., Cullis P.R. Lipid nanoparticles containing siRNA synthesized by microfluidic mixing exhibit an electron-dense nanostructured core. J. Phys. Chem. C. Nanomater. Interfaces. 2012;116:18440–18450. doi: 10.1021/jp303267y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A.K., Tam Y.Y., Cullis P.R. Lipid nanoparticles for short interfering RNA delivery. Adv. Genet. 2014;88:71–110. doi: 10.1016/B978-0-12-800148-6.00004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A.K., Tam Y.Y., Chen S., Hafez I.M., Cullis P.R. Microfluidic mixing: a general method for encapsulating macromolecules in lipid nanoparticle systems. J. Phys. Chem. B. 2015;119:8698–8706. doi: 10.1021/acs.jpcb.5b02891. [DOI] [PubMed] [Google Scholar]
- Li H., Xu D. An overview of fluids mixing in T-shaped mixers. Theor. Appl. Mech. Lett. 2023;13 [Google Scholar]
- Li B., Luo X., Deng B., Wang J., McComb D.W., Shi Y., Gaensler K.M., Tan X., Dunn A.L., Kerlin B.A., Dong Y. An orthogonal array optimization of lipid-like nanoparticles for mRNA delivery in vivo. Nano Lett. 2015;15:8099–8107. doi: 10.1021/acs.nanolett.5b03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang X., Zhang T., Wang C., Huang Z., Luo X., Deng Y. A review on phospholipids and their main applications in drug delivery systems. Asian J. Pharma. Sci. 2015;10:81–98. [Google Scholar]
- Li Y., Lee R.J., Huang X., Li Y., Lv B., Wang T., Qi Y., Hao F., Lu J., Meng Q., Teng L., Zhou Y., Xie J., Teng L. Single-step microfluidic synthesis of transferrin-conjugated lipid nanoparticles for siRNA delivery. Nanomedicine. 2017;13:371–381. doi: 10.1016/j.nano.2016.09.014. [DOI] [PubMed] [Google Scholar]
- Li D.F., Liu Q.S., Yang M.F., Xu H.M., Zhu M.Z., Zhang Y., Xu J., Tian C.M., Yao J., Wang L.S., Liang Y.J. Nanomaterials for mRNA-based therapeutics: challenges and opportunities. Bioeng. Transl. Med. 2023;8 doi: 10.1002/btm2.10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Qi J., Wang J., Hu W., Zhou W., Wang Y., Li T. Nanoparticle technology for mRNA: delivery strategy, clinical application and developmental landscape. Theranostics. 2024;14:738–760. doi: 10.7150/thno.84291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P.J., Tam Y.Y., Hafez I., Sandhu A., Chen S., Ciufolini M.A., Nabi I.R., Cullis P.R. Influence of cationic lipid composition on uptake and intracellular processing of lipid nanoparticle formulations of siRNA. Nanomedicine. 2013;9:233–246. doi: 10.1016/j.nano.2012.05.019. [DOI] [PubMed] [Google Scholar]
- Liu C., Feng Q., Sun J. Lipid nanovesicles by microfluidics: manipulation, synthesis, and drug delivery. Adv. Materials (Deerfield Beach, Fla.) 2019;31:e1804788. doi: 10.1002/adma.201804788. [DOI] [PubMed] [Google Scholar]
- Liu C., Zhang L., Zhu W., Guo R., Sun H., Chen X., Deng N. Barriers and strategies of cationic liposomes for cancer gene therapy. Mol. Ther. Meth. Clin Dev. 2020;18:751–764. doi: 10.1016/j.omtm.2020.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu J., Quimbo A., Xia F., Yao J., Clamme J.P., Zabludoff S., Zhang J., Ying W. Anti-HSP47 siRNA lipid nanoparticle ND-L02-s0201 reverses interstitial pulmonary fibrosis in preclinical rat models. ERJ Open Res. 2021;7 doi: 10.1183/23120541.00733-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage M.P., Vanover D., Beyersdorf J., Hatit M.Z.C., Rotolo L., Echeverri E.S., Peck H.E., Ni H., Yoon J.K., Kim Y., Santangelo P.J., Dahlman J.E. Optimization of lipid nanoparticles for the delivery of nebulized therapeutic mRNA to the lungs. Nat. Biomed. Eng. 2021;5:1059–1068. doi: 10.1038/s41551-021-00786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luiz M.T., Dutra J.A.P., Tofani L.B., de Araujo J.T.C., Di Filippo L.D., Marchetti J.M., Chorilli M. Targeted liposomes: a nonviral gene delivery system for cancer therapy. Pharmaceutics. 2022;14 doi: 10.3390/pharmaceutics14040821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundstrom K. Viral vectors in gene therapy: where Do we stand in 2023? Viruses. 2023;15 doi: 10.3390/v15030698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeki M., Uno S., Niwa A., Okada Y., Tokeshi M. Microfluidic technologies and devices for lipid nanoparticle-based RNA delivery. J. Contro. Release. 2022;344:80–96. doi: 10.1016/j.jconrel.2022.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier M.A., Jayaraman M., Matsuda S., Liu J., Barros S., Querbes W., Tam Y.K., Ansell S.M., Kumar V., Qin J., Zhang X., Wang Q., Panesar S., Hutabarat R., Carioto M., Hettinger J., Kandasamy P., Butler D., Rajeev K.G., Pang B., Charisse K., Fitzgerald K., Mui B.L., Du X., Cullis P., Madden T.D., Hope M.J., Manoharan M., Akinc A. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol. Ther. 2013;21:1570–1578. doi: 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manish P., Antonio J., Ding W., Salomon S., Todd B., Randy S., Ravit G., Shivaani K., Patrick R., Ruth P., Patricia L., Shilpa G., Sima Z., Andressa L., Oleg M., Josh F., Sheryl C., Stephanie P., William R., Praveen A., Lisa J., Khanh D., Robert M., Ryan S. 539 Phase 1 study of mRNA-2752, a lipid nanoparticle encapsulating mRNAs encoding human OX40L/IL-23/IL-36γ, for intratumoral (ITu) injection +/- durvalumab in advanced solid tumors and lymphoma. J. Immunother. Cancer. 2021;9:A569. [Google Scholar]
- Mattheolabakis G., Nie T., Constantinides P.P., Rigas B. Sterically stabilized liposomes incorporating the novel anticancer agent phospho-ibuprofen (MDC-917): preparation, characterization, and in vitro/in vivo evaluation. Pharm. Res. 2012;29:1435–1443. doi: 10.1007/s11095-011-0619-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattheolabakis G., Wong C.C., Sun Y., Amella C.A., Richards R., Constantinides P.P., Rigas B. Pegylation improves the pharmacokinetics and bioavailability of small-molecule drugs hydrolyzable by esterases: a study of phospho-Ibuprofen. J. Pharmacol. Exp. Ther. 2014;351:61–66. doi: 10.1124/jpet.114.217208. [DOI] [PubMed] [Google Scholar]
- McKenzie R.E., Minnell J.J., Ganley M., Painter G.F., Draper S.L. mRNA Synthesis and encapsulation in ionizable lipid nanoparticles. Curr. Protoc. 2023;3 doi: 10.1002/cpz1.898. [DOI] [PubMed] [Google Scholar]
- Mendonça M.C.P., Kont A., Kowalski P.S., O’Driscoll C.M. Design of lipid-based nanoparticles for delivery of therapeutic nucleic acids. Drug Discov. Today. 2023;28 doi: 10.1016/j.drudis.2023.103505. [DOI] [PubMed] [Google Scholar]
- Menon I., Zaroudi M., Zhang Y., Aisenbrey E., Hui L. Fabrication of active targeting lipid nanoparticles: challenges and perspectives. Mater. Today Adv. 2022;16 [Google Scholar]
- Mui B.L., Tam Y.K., Jayaraman M., Ansell S.M., Du X., Tam Y.Y., Lin P.J., Chen S., Narayanannair J.K., Rajeev K.G., Manoharan M., Akinc A., Maier M.A., Cullis P., Madden T.D., Hope M.J. Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Molecular therapy. Nucleic acids. 2013;2 doi: 10.1038/mtna.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukalel A.J., Riley R.S., Zhang R., Mitchell M.J. Nanoparticles for nucleic acid delivery: applications in cancer immunotherapy. Cancer Lett. 2019;458:102–112. doi: 10.1016/j.canlet.2019.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidhara B.K., Baid R., Bishop S.M., Huang M., Wang W., Nema S. Critical considerations for developing nucleic acid macromolecule based drug products. Drug Discov. Today. 2016;21:430–444. doi: 10.1016/j.drudis.2015.11.012. [DOI] [PubMed] [Google Scholar]
- Nabhan J.F., Wood K.M., Rao V.P., Morin J., Bhamidipaty S., LaBranche T.P., Gooch R.L., Bozal F., Bulawa C.E., Guild B.C. Intrathecal delivery of frataxin mRNA encapsulated in lipid nanoparticles to dorsal root ganglia as a potential therapeutic for Friedreich’s ataxia. Sci. Rep. 2016;6:20019. doi: 10.1038/srep20019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naderi Sohi A., Kiani J., Arefian E., Khosrojerdi A., Fekrirad Z., Ghaemi S., Zim M.K., Jalili A., Bostanshirin N., Soleimani M. Development of an mRNA-LNP vaccine against SARS-CoV-2: evaluation of immune response in mouse and rhesus macaque. Vaccines. 2021;9 doi: 10.3390/vaccines9091007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambiar N.R., Gaur S., Ramachandran G., Pandey R.S., Nath L.R., Dutta T., Sudheesh M.S. Remote loading in liposome: a review of current strategies and recent developments. J. Liposome Res. 2024:1–13. doi: 10.1080/08982104.2024.2315449. [DOI] [PubMed] [Google Scholar]
- Nelson J., Sorensen E.W., Mintri S., Rabideau A.E., Zheng W., Besin G., Khatwani N., Su S.V., Miracco E.J., Issa W.J., Hoge S., Stanton M.G., Joyal J.L. Impact of mRNA chemistry and manufacturing process on innate immune activation. Sci. Adv. 2020;6:eaaz6893. doi: 10.1126/sciadv.aaz6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni H., Hatit M.Z.C., Zhao K., Loughrey D., Lokugamage M.P., Peck H.E., Cid A.D., Muralidharan A., Kim Y., Santangelo P.J., Dahlman J.E. Piperazine-derived lipid nanoparticles deliver mRNA to immune cells in vivo. Nat. Commun. 2022;13:4766. doi: 10.1038/s41467-022-32281-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niazi S.K. RNA Therapeutics: a healthcare paradigm shift. Biomedicines. 2023;11 doi: 10.3390/biomedicines11051275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculescu A.G., Mihaiescu D.E., Grumezescu A.M. A review of microfluidic experimental designs for nanoparticle synthesis. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms23158293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitika Wei J., Hui A.M. The delivery of mRNA vaccines for therapeutics. Life (Basel, Switzerland) 2022;12 doi: 10.3390/life12081254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nsairat H., Khater D., Sayed U., Odeh F., Al Bawab A., Alshaer W. Liposomes: structure, composition, types, and clinical applications. Heliyon. 2022;8 doi: 10.1016/j.heliyon.2022.e09394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nsairat H., Alshaer W., Odeh F., Esawi E., Khater D., Bawab A.A., El-Tanani M., Awidi A., Mubarak M.S. Recent advances in using liposomes for delivery of nucleic acid-based therapeutics. OpenNano. 2023;11 [Google Scholar]
- Oh M.H., Kim J.S., Lee J.Y., Park T.G., Nam Y.S. Radio-opaque theranostic nanoemulsions with synergistic anti-cancer activity of paclitaxel and Bcl-2 siRNA. RSC Adv. 2013;3:14642–14651. [Google Scholar]
- Oliver B., Jochen U., Jean-Francois B., Christophe M., Ugur S., Evelyna D., Marie-Laure O., Rahul M., Esteban-Rodrigo I., Nicolas A., Carmen L. 391 A first-in-human study of intratumoral SAR441000, an mRNA mixture encoding IL-12sc, interferon alpha2b, GM-CSF and IL-15sushi as monotherapy and in combination with cemiplimab in advanced solid tumors. J. Immunother. Cancer. 2020;8:A237. [Google Scholar]
- Padda I.S., Mahtani A.U., Patel P., Parmar M. Mayur Parmar declares no relevant financial relationships with ineligible companies. 2024. Small Interfering RNA (siRNA) Therapy, StatPearls, Treasure Island (FL) ineligible companies. Disclosure: Arun Mahtani declares no relevant financial relationships with ineligible companies. Disclosure: Preeti Patel declares no relevant financial relationships with ineligible companies. Disclosure. [Google Scholar]
- Panther L., Basnet S., Fierro C., Brune D., Leggett R., Peterson J., Pickrell P., Lin J., Wu K., Lee H., Hasselbeck R., Natenshon A., Miller J. 2892. Safety and immunogenicity of mRNA-1647, an mRNA-based cytomegalovirus vaccine in healthy adults: results of a phase 2, randomized, observer-blind, placebo-controlled, dose-finding trial. Open Forum Infect. Dis. 2023;10(Suppl. 2) doi: 10.1093/ofid/ofad500.2475. ofad500.2475. eCollection 2023 Dec. [DOI] [Google Scholar]
- Panyam J., Zhou W.Z., Prabha S., Sahoo S.K., Labhasetwar V. Rapid endo-lysosomal escape of poly(DL-lactide-co-glycolide) nanoparticles: implications for drug and gene delivery. FASEB J. 2002;16:1217–1226. doi: 10.1096/fj.02-0088com. [DOI] [PubMed] [Google Scholar]
- Patel S., Ashwanikumar N., Robinson E., DuRoss A., Sun C., Murphy-Benenato K.E., Mihai C., Almarsson Ö., Sahay G. Boosting intracellular delivery of lipid nanoparticle-encapsulated mRNA. Nano Lett. 2017;17:5711–5718. doi: 10.1021/acs.nanolett.7b02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S., Ashwanikumar N., Robinson E., Xia Y., Mihai C., Griffith J.P., 3rd, Hou S., Esposito A.A., Ketova T., Welsher K., Joyal J.L., Almarsson Ö., Sahay G. Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nat. Commun. 2020;11:983. doi: 10.1038/s41467-020-14527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel P., Ibrahim N.M., Cheng K. The importance of apparent pKa in the development of nanoparticles encapsulating siRNA and mRNA. Trends Pharmacol. Sci. 2021;42:448–460. doi: 10.1016/j.tips.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattni B.S., Chupin V.V., Torchilin V.P. New developments in liposomal drug delivery. Chem. Rev. 2015;115:10938–10966. doi: 10.1021/acs.chemrev.5b00046. [DOI] [PubMed] [Google Scholar]
- Paunovska K., Gil C.J., Lokugamage M.P., Sago C.D., Sato M., Lando G.N., Gamboa Castro M., Bryksin A.V., Dahlman J.E. Analyzing 2000 in vivo drug delivery data points reveals cholesterol structure impacts nanoparticle delivery. ACS Nano. 2018;12:8341–8349. doi: 10.1021/acsnano.8b03640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paunovska K., Da Silva Sanchez A.J., Sago C.D., Gan Z., Lokugamage M.P., Islam F.Z., Kalathoor S., Krupczak B.R., Dahlman J.E. Nanoparticles containing oxidized cholesterol deliver mRNA to the liver microenvironment at clinically relevant doses. Adv. Materials (Deerfield Beach, Fla.) 2019;31:e1807748. doi: 10.1002/adma.201807748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipp J., Dabkowska A., Reiser A., Frank K., Krzysztoń R., Brummer C., Nickel B., Blanchet C.E., Sudarsan A., Ibrahim M., Johansson S., Skantze P., Skantze U., Östman S., Johansson M., Henderson N., Elvevold K., Smedsrød B., Schwierz N., Lindfors L., Rädler J.O. pH-dependent structural transitions in cationic ionizable lipid mesophases are critical for lipid nanoparticle function. Proc. Natl. Acad. Sci. 2023;120 doi: 10.1073/pnas.2310491120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilkington E.H., Suys E.J.A., Trevaskis N.L., Wheatley A.K., Zukancic D., Algarni A., Al-Wassiti H., Davis T.P., Pouton C.W., Kent S.J., Truong N.P. From influenza to COVID-19: Lipid nanoparticle mRNA vaccines at the frontiers of infectious diseases. Acta Biomater. 2021;131:16–40. doi: 10.1016/j.actbio.2021.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires P., Simoes S., Nir S., Gaspar R., Duzgunes N., Pedroso de Lima M.C. Interaction of cationic liposomes and their DNA complexes with monocytic leukemia cells. Biochim. Biophys. Acta. 1999;1418:71–84. doi: 10.1016/s0005-2736(99)00023-1. [DOI] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Marc G.P., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Hammitt L.L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D.B., Mather S., Dormitzer P.R., Şahin U., Jansen K.U., Gruber W.C. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Jr., Hammitt L.L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D.B., Mather S., Dormitzer P.R., Şahin U., Jansen K.U., Gruber W.C. Safety and Efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poudel S., Napit P.R., Briski K.P., Mattheolabakis G. Oral delivery of nucleic acids with passive and active targeting to the intestinal tissue using polymer-based nanocarriers. Pharmaceutics. 2021;13 doi: 10.3390/pharmaceutics13071075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash T.P., Lima W.F., Murray H.M., Elbashir S., Cantley W., Foster D., Jayaraman M., Chappell A.E., Manoharan M., Swayze E.E., Crooke S.T. Lipid nanoparticles improve activity of single-stranded siRNA and gapmer antisense oligonucleotides in animals. ACS Chem. Biol. 2013;8:1402–1406. doi: 10.1021/cb4001316. [DOI] [PubMed] [Google Scholar]
- Prakash G., Shokr A., Willemen N., Bashir S.M., Shin S.R., Hassan S. Microfluidic fabrication of lipid nanoparticles for the delivery of nucleic acids. Adv. Drug Deliv. Rev. 2022;184 doi: 10.1016/j.addr.2022.114197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qosa H., de Oliveira C., Cizza G., Lawitz E.J., Colletti N., Wetherington J., Charles E.D., Tirucherai G.S. Pharmacokinetics, safety, and tolerability of BMS-986263, a lipid nanoparticle containing HSP47 siRNA, in participants with hepatic impairment. Clin. Transl. Sci. 2023;16:1791–1802. doi: 10.1111/cts.13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qosa H., de Oliveira C.H.M.C., Cizza G., Lawitz E.J., Colletti N., Wetherington J., Charles E.D., Tirucherai G.S. Pharmacokinetics, safety, and tolerability of BMS-986263, a lipid nanoparticle containing HSP47 siRNA, in participants with hepatic impairment. Clin. Transl. Sci. 2023;16:1791–1802. doi: 10.1111/cts.13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffy S., Teissie J. Control of lipid membrane stability by cholesterol content. Biophys. J. 1999;76:2072–2080. doi: 10.1016/S0006-3495(99)77363-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan R.K., Hamburg S.I., Borad M.J., Seetharam M., Kundranda M.N., Lee P., Fredlund P., Gilbert M., Mast C., Semple S.C., Judge A.D., Crowell B., Vocila L., MacLachlan I., Northfelt D.W. Abstract LB-289: a phase I dose escalation study of TKM-080301, a RNAi therapeutic directed against PLK1, in patients with advanced solid tumors. Cancer Res. 2013;73 LB-289-LB-289. [Google Scholar]
- Rhym L.H., Anderson D.G. Nanoscale delivery platforms for RNA therapeutics: challenges and the current state of the art. Med. 2022;3:167–187. doi: 10.1016/j.medj.2022.02.001. [DOI] [PubMed] [Google Scholar]
- Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C., Ciaramella G., Diamond M.S. Modified mRNA vaccines protect against Zika Virus infection. Cell. 2017;168:1114–1125.e1110. doi: 10.1016/j.cell.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietwyk S., Peer D. Next-generation lipids in RNA interference therapeutics. ACS Nano. 2017;11:7572–7586. doi: 10.1021/acsnano.7b04734. [DOI] [PubMed] [Google Scholar]
- Robinson E., MacDonald K.D., Slaughter K., McKinney M., Patel S., Sun C., Sahay G. Lipid nanoparticle-delivered chemically modified mRNA restores chloride secretion in cystic fibrosis. Mol. Ther. 2018;26:2034–2046. doi: 10.1016/j.ymthe.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roces C.B., Lou G., Jain N., Abraham S., Thomas A., Halbert G.W., Perrie Y. Manufacturing considerations for the development of lipid nanoparticles using microfluidics. Pharmaceutics. 2020;12 doi: 10.3390/pharmaceutics12111095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue V., Gravagna K., Yao J., Nafade V., Basta N.E. Current progress towards prevention of Nipah and Hendra disease in humans: a scoping review of vaccine and monoclonal antibody candidates being evaluated in clinical trials. Trop. Med. Int. Health. 2024;29:354–364. doi: 10.1111/tmi.13979. [DOI] [PubMed] [Google Scholar]
- Rowe S.M., Zuckerman J.B., Dorgan D., Lascano J., McCoy K., Jain M., Schechter M.S., Lommatzsch S., Indihar V., Lechtzin N., McBennett K., Callison C., Brown C., Liou T.G., MacDonald K.D., Nasr S.Z., Bodie S., Vaughn M., Meltzer E.B., Barbier A.J. Inhaled mRNA therapy for treatment of cystic fibrosis: Interim results of a randomized, double-blind, placebo-controlled phase 1/2 clinical study. J. Cyst. Fibros. 2023;22:656–664. doi: 10.1016/j.jcf.2023.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rungta R.L., Choi H.B., Lin P.J., Ko R.W., Ashby D., Nair J., Manoharan M., Cullis P.R., Macvicar B.A. Lipid nanoparticle delivery of siRNA to silence neuronal gene expression in the brain. Mol. Ther. Nucleic Acids. 2013;2 doi: 10.1038/mtna.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals R.C., Patel S., Acosta C., McKinney M., Pennesi M.E., Sahay G. The effects of PEGylation on LNP based mRNA delivery to the eye. PLoS One. 2020;15 doi: 10.1371/journal.pone.0241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabnis S., Kumarasinghe E.S., Salerno T., Mihai C., Ketova T., Senn J.J., Lynn A., Bulychev A., McFadyen I., Chan J., Almarsson Ö., Stanton M.G., Benenato K.E. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther. 2018;26:1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sago C.D., Lokugamage M.P., Paunovska K., Vanover D.A., Monaco C.M., Shah N.N., Gamboa Castro M., Anderson S.E., Rudoltz T.G., Lando G.N., Munnilal Tiwari P., Kirschman J.L., Willett N., Jang Y.C., Santangelo P.J., Bryksin A.V., Dahlman J.E. High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing. Proc. Natl. Acad. Sci. USA. 2018;115:E9944–e9952. doi: 10.1073/pnas.1811276115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahly H.M.E., Baden L.R., Essink B., Doblecki-Lewis S., Martin J.M., Anderson E.J., Campbell T.B., Clark J., Jackson L.A., Fichtenbaum C.J., Zervos M., Rankin B., Eder F., Feldman G., Kennelly C., Han-Conrad L., Levin M., Neuzil K.M., Corey L., Gilbert P., Janes H., Follmann D., Marovich M., Polakowski L., Mascola J.R., Ledgerwood J.E., Graham B.S., August A., Clouting H., Deng W., Han S., Leav B., Manzo D., Pajon R., Schödel F., Tomassini J.E., Zhou H., Miller J. Efficacy of the mRNA-1273 SARS-CoV-2 vaccine at completion of blinded phase. N. Engl. J. Med. 2021;385:1774–1785. doi: 10.1056/NEJMoa2113017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaridou E., Heyes J., Lutwyche P. Lipid nanoparticles for nucleic acid delivery: current perspectives. Adv. Drug Deliv. Rev. 2020;154-155:37–63. doi: 10.1016/j.addr.2020.06.002. [DOI] [PubMed] [Google Scholar]
- Sanghani A., Kafetzis K.N., Sato Y., Elboraie S., Fajardo-Sanchez J., Harashima H., Tagalakis A.D., Yu-Wai-Man C. Novel PEGylated lipid nanoparticles have a high encapsulation efficiency and effectively deliver MRTF-B siRNA in conjunctival FIBROBLASTS. Pharmaceutics. 2021;13 doi: 10.3390/pharmaceutics13030382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlich M., Palomba R., Costabile G., Mizrahy S., Pannuzzo M., Peer D., Decuzzi P. Cytosolic delivery of nucleic acids: the case of ionizable lipid nanoparticles. Bioeng. Transl. Med. 2021;6 doi: 10.1002/btm2.10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H.H., Wixner J., Planté-Bordeneuve V., Muñoz-Beamud F., Lladó L., Gillmore J.D., Mazzeo A., Li X., Arum S., Jay P.Y., Adams D. Patisiran treatment in patients with hereditary transthyretin-mediated amyloidosis with polyneuropathy after liver transplantation. Am. J. Transplant. Off. J. Am. Soc. Transplant. Am. Soc. Transplant Surg. 2022;22:1646–1657. doi: 10.1111/ajt.17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnyder Ghamloush S., Essink B., Hu B., Kalidindi S., Morsy L., Egwuenu-Dumbuya C., Kapoor A., Girard B., Dhar R., Lackey R., Snape M.D., Shaw C.A. Safety and immunogenicity of an mRNA-based hMPV/PIV3 combination vaccine in seropositive children. Pediatrics. 2024;153 doi: 10.1542/peds.2023-064748. [DOI] [PubMed] [Google Scholar]
- Schoenmaker L., Witzigmann D., Kulkarni J.A., Verbeke R., Kersten G., Jiskoot W., Crommelin D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021;601 doi: 10.1016/j.ijpharm.2021.120586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenmaker L., Witzigmann D., Kulkarni J.A., Verbeke R., Kersten G., Jiskoot W., Crommelin D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021;601 doi: 10.1016/j.ijpharm.2021.120586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani F., Yanez Arteta M., Lerche M., Porcar L., Lang C., Bragg R.A., Elmore C.S., Krishnamurthy V.R., Russell R.A., Darwish T., Pichler H., Waldie S., Moulin M., Haertlein M., Forsyth V.T., Lindfors L., Cárdenas M. Apolipoprotein E binding drives structural and compositional rearrangement of mRNA-containing lipid nanoparticles. ACS Nano. 2021;15:6709–6722. doi: 10.1021/acsnano.0c10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple S.C., Chonn A., Cullis P.R. Influence of cholesterol on the association of plasma proteins with liposomes. Biochemistry. 1996;35:2521–2525. doi: 10.1021/bi950414i. [DOI] [PubMed] [Google Scholar]
- Semple S.C., Akinc A., Chen J., Sandhu A.P., Mui B.L., Cho C.K., Sah D.W., Stebbing D., Crosley E.J., Yaworski E., Hafez I.M., Dorkin J.R., Qin J., Lam K., Rajeev K.G., Wong K.F., Jeffs L.B., Nechev L., Eisenhardt M.L., Jayaraman M., Kazem M., Maier M.A., Srinivasulu M., Weinstein M.J., Chen Q., Alvarez R., Barros S.A., De S., Klimuk S.K., Borland T., Kosovrasti V., Cantley W.L., Tam Y.K., Manoharan M., Ciufolini M.A., Tracy M.A., de Fougerolles A., MacLachlan I., Cullis P.R., Madden T.D., Hope M.J. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- Shepherd S.J., Warzecha C.C., Yadavali S., El-Mayta R., Alameh M.G., Wang L., Weissman D., Wilson J.M., Issadore D., Mitchell M.J. Scalable mRNA and siRNA Lipid nanoparticle production using a parallelized microfluidic device. Nano Lett. 2021;21:5671–5680. doi: 10.1021/acs.nanolett.1c01353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Huang M.W., Lu Z.D., Du X.J., Shen S., Xu C.F., Wang J. Delivery of mRNA for regulating functions of immune cells. J. Control. Release. 2022;345:494–511. doi: 10.1016/j.jconrel.2022.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha A., Haque M.A., Mattheolabakis G. Pulmonary delivery for miRs: present and Future potential. Processes. 2023;11:1788. [Google Scholar]
- Soroudi S., Jaafari M.R., Arabi L. Lipid nanoparticle (LNP) mediated mRNA delivery in cardiovascular diseases: advances in genome editing and CAR T cell therapy. J. Control. Release. 2024;372:113–140. doi: 10.1016/j.jconrel.2024.06.023. [DOI] [PubMed] [Google Scholar]
- Souto E.B., Nayak A.P., Murthy R.S. Lipid nanoemulsions for anti-cancer drug therapy. Pharmazie. 2011;66:473–478. [PubMed] [Google Scholar]
- Sparmann A., Vogel J. RNA-based medicine: from molecular mechanisms to therapy. EMBO J. 2023;42 doi: 10.15252/embj.2023114760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhr O.B., Coelho T., Buades J., Pouget J., Conceicao I., Berk J., Schmidt H., Waddington-Cruz M., Campistol J.M., Bettencourt B.R., Vaishnaw A., Gollob J., Adams D. Efficacy and safety of patisiran for familial amyloidotic polyneuropathy: a phase II multi-dose study. Orphanet J. Rare Dis. 2015;10:109. doi: 10.1186/s13023-015-0326-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suk J.S., Xu Q., Kim N., Hanes J., Ensign L.M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 2016;99:28–51. doi: 10.1016/j.addr.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Lu Z.R. Structure and function of cationic and ionizable lipids for nucleic acid delivery. Pharm. Res. 2023;40:27–46. doi: 10.1007/s11095-022-03460-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung Y.K., Kim S.W. Recent advances in the development of gene delivery systems. Biomate. Res. 2019;23:8. doi: 10.1186/s40824-019-0156-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Ishihara H. Difference in the lipid nanoparticle technology employed in three approved siRNA (Patisiran) and mRNA (COVID-19 vaccine) drugs. Drug Metab. Pharmacokinet. 2021;41 doi: 10.1016/j.dmpk.2021.100424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Suzuki Y., Hihara T., Kubara K., Kondo K., Hyodo K., Yamazaki K., Ishida T., Ishihara H. PEG shedding-rate-dependent blood clearance of PEGylated lipid nanoparticles in mice: Faster PEG shedding attenuates anti-PEG IgM production. Int. J. Pharm. 2020;588 doi: 10.1016/j.ijpharm.2020.119792. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Katsurada Y., Hyodo K. Differences and similarities of the intravenously administered lipid nanoparticles in three clinical trials: potential linkage between lipid nanoparticles and extracellular vesicles. Mol. Pharm. 2023;20:4883–4892. doi: 10.1021/acs.molpharmaceut.3c00547. [DOI] [PubMed] [Google Scholar]
- Swetha K., Kotla N.G., Tunki L., Jayaraj A., Bhargava S.K., Hu H., Bonam S.R., Kurapati R. Recent Advances in the Lipid Nanoparticle-Mediated Delivery of mRNA Vaccines. Vaccines (Basel) 2023:11. doi: 10.3390/vaccines11030658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Miyama R., Sakurai Y., Tamagawa S., Nakai Y., Tange K., Yoshioka H., Akita H. Improvement of mRNA delivery Efficiency to a T Cell Line by Modulating PEG-Lipid Content and Phospholipid Components of Lipid Nanoparticles. Pharmaceutics. 2021;13 doi: 10.3390/pharmaceutics13122097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A., De Souza R.A.G., Ouellet E., Tharmarajah G., Reichert D., Ordobadi M., Ip S., Ramsay E.C. Microfluidic production and application of lipid nanoparticles for nucleic acid transfection. Meth. Mol. Biol. (Clifton, N.J.) 1792. 2018:193–203. doi: 10.1007/978-1-4939-7865-6_14. [DOI] [PubMed] [Google Scholar]
- Tolcher A.W., Papadopoulos K.P., Patnaik A., Rasco D.W., Martinez D., Wood D.L., Fielman B., Sharma M., Janisch L.A., Brown B.D., Bhargava P., Ratain M.J. Safety and activity of DCR-MYC, a first-in-class Dicer-substrate small interfering RNA (DsiRNA) targeting MYC, in a phase I study in patients with advanced solid tumors. J. Clin. Oncol. 2015;33:11006. [Google Scholar]
- Tsakiri M., Zivko C., Demetzos C., Mahairaki V. Lipid-based nanoparticles and RNA as innovative neuro-therapeutics. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.900610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkouhi A.K., Scholte M., Storm G., Haisma H.J. Endosomal escape pathways for delivery of biologicals. J. Control. Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Verbeke R., Lentacker I., De Smedt S.C., Dewitte H. The dawn of mRNA vaccines: the COVID-19 case. J. Control. Release. 2021;333:511–520. doi: 10.1016/j.jconrel.2021.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viger-Gravel J., Schantz A., Pinon A.C., Rossini A.J., Schantz S., Emsley L. Structure of Lipid Nanoparticles Containing siRNA or mRNA by Dynamic Nuclear Polarization-Enhanced NMR Spectroscopy. J. Phys. Chem. B. 2018;122:2073–2081. doi: 10.1021/acs.jpcb.7b10795. [DOI] [PubMed] [Google Scholar]
- Vogelaar A., Marcotte S., Cheng J., Oluoch B., Zaro J. Use of Microfluidics to Prepare Lipid-based Nanocarriers. Pharmaceutics. 2023;15 doi: 10.3390/pharmaceutics15041053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A., Vorauer-Uhl K., Kreismayr G., Katinger H. The crossflow injection technique: an improvement of the ethanol injection method. J. Liposome Res. 2002;12:259–270. doi: 10.1081/lpr-120014761. [DOI] [PubMed] [Google Scholar]
- Wahane A., Waghmode A., Kapphahn A., Dhuri K., Gupta A., Bahal R. Role of lipid-based and polymer-based non-viral vectors in nucleic acid delivery for next-generation gene therapy. Molecules (Basel, Switzerland) 2020;25 doi: 10.3390/molecules25122866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C., Ou K., Belliveau N.M., Leaver T.J., Wild A.W., Huft J., Lin P.J., Chen S., Leung A.K., Lee J.B., Hansen C.L., Taylor R.J., Ramsay E.C., Cullis P.R. Microfluidic-based manufacture of siRNA-lipid nanoparticles for therapeutic applications. Meth. Mol. Biol. (Clifton, N.J.) 2014;1141:109–120. doi: 10.1007/978-1-4939-0363-4_6. [DOI] [PubMed] [Google Scholar]
- Wang H., Wang Y., Yuan C., Xu X., Zhou W., Huang Y., Lu H., Zheng Y., Luo G., Shang J., Sui M. Polyethylene glycol (PEG)-associated immune responses triggered by clinically relevant lipid nanoparticles in rats. npj Vaccines. 2023;8:169. doi: 10.1038/s41541-023-00766-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Liu S., Sun Y., Yu X., Lee S.M., Cheng Q., Wei T., Gong J., Robinson J., Zhang D., Lian X., Basak P., Siegwart D.J. Preparation of selective organ-targeting (SORT) lipid nanoparticles (LNPs) using multiple technical methods for tissue-specific mRNA delivery. Nat. Protoc. 2023;18:265–291. doi: 10.1038/s41596-022-00755-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.S., Kumari M., Chen G.H., Hong M.H., Yuan J.P., Tsai J.L., Wu H.C. mRNA-based vaccines and therapeutics: an in-depth survey of current and upcoming clinical applications. J. Biomed. Sci. 2023;30:84. doi: 10.1186/s12929-023-00977-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb C., Forbes N., Roces C.B., Anderluzzi G., Lou G., Abraham S., Ingalls L., Marshall K., Leaver T.J., Watts J.A., Aylott J.W., Perrie Y. Using microfluidics for scalable manufacturing of nanomedicines from bench to GMP: a case study using protein-loaded liposomes. Int. J. Pharm. 2020;582 doi: 10.1016/j.ijpharm.2020.119266. [DOI] [PubMed] [Google Scholar]
- Webb C., Ip S., Bathula N.V., Popova P., Soriano S.K.V., Ly H.H., Eryilmaz B., Nguyen Huu V.A., Broadhead R., Rabel M., Villamagna I., Abraham S., Raeesi V., Thomas A., Clarke S., Ramsay E.C., Perrie Y., Blakney A.K. Current Status and Future Perspectives on MRNA Drug Manufacturing. Mol. Pharm. 2022;19:1047–1058. doi: 10.1021/acs.molpharmaceut.2c00010. [DOI] [PubMed] [Google Scholar]
- Weber J.S., Carlino M.S., Khattak A., Meniawy T., Ansstas G., Taylor M.H., Kim K.B., McKean M., Long G.V., Sullivan R.J., Faries M., Tran T.T., Cowey C.L., Pecora A., Shaheen M., Segar J., Medina T., Atkinson V., Gibney G.T., Luke J.J., Thomas S., Buchbinder E.I., Healy J.A., Huang M., Morrissey M., Feldman I., Sehgal V., Robert-Tissot C., Hou P., Zhu L., Brown M., Aanur P., Meehan R.S., Zaks T. Individualised neoantigen therapy mRNA-4157 (V940) plus pembrolizumab versus pembrolizumab monotherapy in resected melanoma (KEYNOTE-942): a randomised, phase 2b study. Lancet. 2024;403:632–644. doi: 10.1016/S0140-6736(23)02268-7. [DOI] [PubMed] [Google Scholar]
- Whitehead K.A., Dorkin J.R., Vegas A.J., Chang P.H., Veiseh O., Matthews J., Fenton O.S., Zhang Y., Olejnik K.T., Yesilyurt V., Chen D., Barros S., Klebanov B., Novobrantseva T., Langer R., Anderson D.G. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat. Commun. 2014;5:4277. doi: 10.1038/ncomms5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widge A.T., Hofstetter A.R., Houser K.V., Awan S.F., Chen G.L., Burgos Florez M.C., Berkowitz N.M., Mendoza F., Hendel C.S., Holman L.A., Gordon I.J., Apte P., Liang C.J., Gaudinski M.R., Coates E.E., Strom L., Wycuff D., Vazquez S., Stein J.A., Gall J.G., Adams W.C., Carlton K., Gillespie R.A., Creanga A., Crank M.C., Andrews S.F., Castro M., Serebryannyy L.A., Narpala S.R., Hatcher C., Lin B.C., O'Connell S., Freyn A.W., Rosado V.C., Nachbagauer R., Palese P., Kanekiyo M., McDermott A.B., Koup R.A., Dropulic L.K., Graham B.S., Mascola J.R., Ledgerwood J.E., team, V.R.C.s An influenza hemagglutinin stem nanoparticle vaccine induces cross-group 1 neutralizing antibodies in healthy adults. Sci. Transl. Med. 2023;15 doi: 10.1126/scitranslmed.ade4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson E., Goswami J., Baqui A.H., Doreski P.A., Perez-Marc G., Zaman K., Monroy J., Duncan C.J.A., Ujiie M., Ramet M., Perez-Breva L., Falsey A.R., Walsh E.E., Dhar R., Wilson L., Du J., Ghaswalla P., Kapoor A., Lan L., Mehta S., Mithani R., Panozzo C.A., Simorellis A.K., Kuter B.J., Schodel F., Huang W., Reuter C., Slobod K., Stoszek S.K., Shaw C.A., Miller J.M., Das R., Chen G.L., Conquer R.S.V.S.G. Efficacy and safety of an mRNA-based RSV PreF vaccine in older adults. N. Engl. J. Med. 2023;389:2233–2244. doi: 10.1056/NEJMoa2307079. [DOI] [PubMed] [Google Scholar]
- Xie J., Wang S. Small interfering RNA in colorectal cancer liver metastasis therapy. Technol. Cancer Res. Treat. 2022;21 doi: 10.1177/15330338221103318. 15330338221103318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Wang X., Liu Y., Yang G., Falconer R.J., Zhao C.-X. Lipid nanoparticles for drug delivery. Adv. NanoBiomed Res. 2022;2:2100109. [Google Scholar]
- Xue H.Y., Guo P., Wen W.C., Wong H.L. Lipid-based nanocarriers for RNA delivery. Curr. Pharm. Des. 2015;21:3140–3147. doi: 10.2174/1381612821666150531164540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Lin P.J., Beraldi E., Zhang F., Kawai Y., Leong J., Katsumi H., Fazli L., Fraser R., Cullis P.R., Gleave M. siRNA Lipid nanoparticle potently silences clusterin and delays progression when combined with androgen receptor cotargeting in enzalutamide-resistant prostate cancer. Clin. Cancer Res. 2015;21:4845–4855. doi: 10.1158/1078-0432.CCR-15-0866. [DOI] [PubMed] [Google Scholar]
- Yamazaki K., Kubara K., Ishii S., Kondo K., Suzuki Y., Miyazaki T., Mitsuhashi K., Ito M., Tsukahara K. Lipid nanoparticle-targeted mRNA formulation as a treatment for ornithine-transcarbamylase deficiency model mice. Mol. Thera. Nucleic Acids. 2023;33:210–226. doi: 10.1016/j.omtn.2023.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Liu X.Y., Lu A., Wang X.Y., Jiang L.X., Wang J.C. Non-viral vectors for RNA delivery. J. Control. Release. 2022;342:241–279. doi: 10.1016/j.jconrel.2022.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez Arteta M., Kjellman T., Bartesaghi S., Wallin S., Wu X., Kvist A.J., Dabkowska A., Szekely N., Radulescu A., Bergenholtz J., Lindfors L. Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. Proc. Natl. Acad. Sci. USA. 2018;115:E3351–E3360. doi: 10.1073/pnas.1720542115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassini P., Hutchens M., Paila Y.D., Schoch L., Aunins A., Siangphoe U., Paris R. Interim analysis of a phase 1 randomized clinical trial on the safety and immunogenicity of the mRNA-1283 SARS-CoV-2 vaccine in adults. Hum. Vaccin. Immunother. 2023;19:2190690. doi: 10.1080/21645515.2023.2190690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi S.W., Yune T.Y., Kim T.W., Chung H., Choi Y.W., Kwon I.C., Lee E.B., Jeong S.Y. A cationic lipid emulsion/DNA complex as a physically stable and serum-resistant gene delivery system. Pharm. Res. 2000;17:314–320. doi: 10.1023/a:1007553106681. [DOI] [PubMed] [Google Scholar]
- Zatsepin T.S., Kotelevtsev Y.V., Koteliansky V. Lipid nanoparticles for targeted siRNA delivery - going from bench to bedside. Int. J. Nanomedicine. 2016;11:3077–3086. doi: 10.2147/IJN.S106625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelepukin I.V., Shevchenko K.G., Deyev S.M. Rediscovery of mononuclear phagocyte system blockade for nanoparticle drug delivery. Nat. Commun. 2024;15:4366. doi: 10.1038/s41467-024-48838-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.S., Liu F., Conwell C.C., Tan Y., Huang L. Mechanistic studies of sequential injection of cationic liposome and plasmid DNA. Mol. Ther. 2006;13:429–437. doi: 10.1016/j.ymthe.2005.08.021. [DOI] [PubMed] [Google Scholar]
- Zhang J., Li Q., Wu Y., Wang D., Xu L., Zhang Y., Wang S., Wang T., Liu F., Zaky M.Y., Hou S., Liu S., Zou K., Lei H., Zou L., Zhang Y., Liu H. Cholesterol content in cell membrane maintains surface levels of ErbB2 and confers a therapeutic vulnerability in ErbB2-positive breast cancer. Cell Commun. Signal. 2019;17:15. doi: 10.1186/s12964-019-0328-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N.N., Li X.F., Deng Y.Q., Zhao H., Huang Y.J., Yang G., Huang W.J., Gao P., Zhou C., Zhang R.R., Guo Y., Sun S.H., Fan H., Zu S.L., Chen Q., He Q., Cao T.S., Huang X.Y., Qiu H.Y., Nie J.H., Jiang Y., Yan H.Y., Ye Q., Zhong X., Xue X.L., Zha Z.Y., Zhou D., Yang X., Wang Y.C., Ying B., Qin C.F. A thermostable mRNA vaccine against COVID-19. Cell. 2020;182:1271–1283.e1216. doi: 10.1016/j.cell.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Goel V., Robbie G.J. Pharmacokinetics of Patisiran, the first Approved RNA Interference Therapy in patients with Hereditary Transthyretin-Mediated Amyloidosis. J. Clin. Pharmacol. 2020;60:573–585. doi: 10.1002/jcph.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., El-Mayta R., Murdoch T.J., Warzecha C.C., Billingsley M.M., Shepherd S.J., Gong N., Wang L., Wilson J.M., Lee D., Mitchell M.J. Helper lipid structure influences protein adsorption and delivery of lipid nanoparticles to spleen and liver. Biomater. Sci. 2021;9:1449–1463. doi: 10.1039/d0bm01609h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Sun C., Wang C., Jankovic K.E., Dong Y. Lipids and Lipid Derivatives for RNA delivery. Chem. Rev. 2021;121:12181–12277. doi: 10.1021/acs.chemrev.1c00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., More K.R., Ojha A., Jackson C.B., Quinlan B.D., Li H., He W., Farzan M., Pardi N., Choe H. Effect of mRNA-LNP components of two globally-marketed COVID-19 vaccines on efficacy and stability. npj Vaccines. 2023;8:156. doi: 10.1038/s41541-023-00751-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Hai L., Gao Y., Yu G., Sun Y. Lipid nanomaterials-based RNA therapy and cancer treatment. Acta Pharm. Sin. B. 2023;13:903–915. doi: 10.1016/j.apsb.2022.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Yin H., Li Y., Yang H., Ge K., Zhang J., Yuan Q., Dai X., Naeem A., Weng Y., Huang Y., Liang X.J. Optimized lipid nanoparticles (LNPs) for organ-selective nucleic acids delivery in vivo. iScience. 2024;27 doi: 10.1016/j.isci.2024.109804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Hou X., Yan J., Du S., Xue Y., Li W., Xiang G., Dong Y. Long-term storage of lipid-like nanoparticles for mRNA delivery. Bioactive Materials. 2020;5:358–363. doi: 10.1016/j.bioactmat.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Anselmo A.C., Mitragotri S. Viral vector-based gene therapies in the clinic. Bioeng. Transl. Med. 2022;7 doi: 10.1002/btm2.10258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Bandara S.R., Tan Z., Leal C. Lipid nanoparticle topology regulates endosomal escape and delivery of RNA to the cytoplasm. Proc. Natl. Acad. Sci. USA. 2023;120 doi: 10.1073/pnas.2301067120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L., Krummenacher C., Zhang W., Hong J., Feng Q., Chen Y., Zhao Q., Zeng M.S., Zeng Y.X., Xu M., Zhang X. Urgency and necessity of Epstein-Barr virus prophylactic vaccines. NPJ Vaccines. 2022;7:159. doi: 10.1038/s41541-022-00587-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Bobbin M.L., Burnett J.C., Rossi J.J. Current progress of RNA aptamer-based therapeutics. Front. Genet. 2012;3:234. doi: 10.3389/fgene.2012.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Zhu L., Wang X., Jin H. RNA-based therapeutics: an overview and prospectus. Cell Death Dis. 2022;13:644. doi: 10.1038/s41419-022-05075-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y., Zhou Q., Zhao Y., Zhi D., Chen H., Wang R., Ju B., Zhang S. Structure-activity relationships of pH-responsive and ionizable lipids for gene delivery. Int. J. Pharm. 2022;617 doi: 10.1016/j.ijpharm.2022.121596. [DOI] [PubMed] [Google Scholar]
- Zuhorn I.S., Bakowsky U., Polushkin E., Visser W.H., Stuart M.C.A., Engberts J.B.F.N., Hoekstra D. Nonbilayer phase of lipoplex–membrane mixture determines endosomal escape of genetic cargo and transfection efficiency. Mol. Ther. 2005;11:801–810. doi: 10.1016/j.ymthe.2004.12.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.