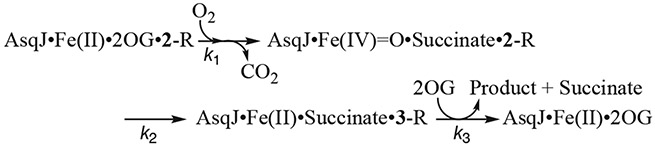Abstract
Iron and 2-oxoglutarate dependent (Fe/2OG) enzymes exhibit an exceedingly broad reaction repertoire. The most prevalent reactivity is hydroxylation, but many other reactivities have also been discovered in recent years, including halogenation, desaturation, epoxidation, endoperoxidation, epimerization, and cyclization. To fully explore the reaction mechanisms that support such a diverse reactivities in Fe/2OG enzyme, it is necessary to utilize a multi-faceted research methodology, consisting of molecular probe design and synthesis, in vitro enzyme assay development, enzyme kinetics, spectroscopy, protein crystallography, and theoretical calculations. By using such a multi-faceted research approach, we have explored reaction mechanisms of desaturation and epoxidation catalyzed by a bi-functional Fe/2OG enzyme, AsqJ. Herein, we describe the experimental protocols and computational workflows used in our studies.
1. Introduction
Non-heme mononuclear iron-containing (NHM-Fe) enzymes catalyze a broad set of chemical reactions, which in many cases constitute key chemical transformations to enable important biological functions, ranging from gene regulation to natural product biosynthesis (Abu-Omar, Loaiza, & Hontzeas, 2005; Buongiorno & Straganz, 2013; Costas, Mehn, Jensen, & Que, 2004; Gao, Naowarojna, Cheng, Liu, & Liu, 2018; Hausinger, 2015; Kovaleva & Lipscomb, 2008; Solomon et al., 2000; Song, Naowarojna, Cheng, Lopez, & Liu, 2019). As one of the biggest NHM-Fe subfamilies, iron and 2-oxoglutarate (Fe/2OG) dependent enzymes have been shown to catalyze chemically challenging reactions, including hydroxylation, halogenation, desaturation, epoxidation, endoperoxidation, epimerization, and cyclization (Bollinger et al., 2015; Gao et al., 2018; Guo, Chang, Li, & Davidson, 2021; Hausinger, 2015). This catalytic versatility has attracted intense research efforts in the past two decades to explore detailed reaction mechanisms of this group of NHM-Fe enzymes. These efforts have led to the discovery of a common high-valent iron reactive intermediate, the S = 2 oxyferryl (ferryl, oxoiron(IV), or Fe(IV)=O) intermediate, in Fe/2OG enzymes (Chang, Li, Lee, Cronican, & Guo, 2016; Krebs, Galonić Fujimori, Walsh, & Bollinger, 2007; Matthews, Krest, et al., 2009; Price, Barr, Tirupati, Bollinger, & Krebs, 2003), and yielded a large number of protein crystal structures, showing a highly conserved protein fold for these enzymes, the double stranded beta-helix (DSBH, also known as cupin) fold (Aik et al., 2015; Roach et al., 1995, 1997; Valegård et al., 1998; Zhang et al., 2000). These efforts have also revealed the reaction mechanism of Fe/2OG enzyme catalyzed hydroxylation (Kal & Que, 2017; Krebs et al., 2007; Solomon, DeWeese, & Babicz, 2021), which is by far the most prevalent reactivity demonstrated by these enzymes. In hydroxylation, the O2 activation is achieved by the oxidative decaboxylation of Fe(II) bound 2OG, which leads to the formation of the S = 2 oxyferryl intermediate together with succinate and CO2. Subsequently, the oxyferryl intermediate activates the designated C-H bond on the enzyme substrate via a hydrogen atom transfer (HAT) step, which leads to the generation of a carbon centered radical on the substrate and a hydroxyl-ferric (Fe(III)-OH) species. Finally, the hydroxyl (OH·) rebound enables C-O bond formation on the substrate radical to complete the hydroxylation reaction. But what is not fully understood is how this hydroxyl rebound paradigm can be applied to understand other reactivities demonstrated by Fe/2OG enzymes that do not rely on this rebound step.
To address this mechanistic challenge, a multi-faceted research strategy combining a wide range of research tools is needed. Spectroscopic methods, such as Mössbauer spectroscopy and electron paramagnetic resonance (EPR), are commonly used in the studies of Fe/2OG enzymes and other non-heme iron-containing enzymes (Cammack & Cooper, 1993; Krebs & Bollinger, 2009; Münck, Ksurerus, & Hendrich, 1993). These techniques are frequently combined with freeze quench techniques (Krebs & Bollinger, 2009) and presteady state kinetics (Bollinger & Krebs, 2006) to reveal the kinetics and the electronic structures of reactive intermediates. For example, the combined application of these techniques was crucial for the discovery of the first example of the S = 2 oxyferryl intermediate in taurine/2OG dioxygenase (Price, Barr, Hoffart, Krebs, & Bollinger, 2005; Price, Barr, Glass, Krebs, & Bollinger, 2003; Price, Barr, Tirupati, et al., 2003). In addition, substrate analog design and synthesis and enzyme assay development based on liquid chromatography coupled mass spectrometry (LC-MS) establish the reaction profile of a given Fe/2OG enzyme and provide evidence of reaction mechanism through the analysis of product distribution and isotope tracing experiments (some examples include (Cha et al., 2023; Chen et al., 2022; Dunham et al., 2018; Matthews, Neumann, et al., 2009)). X-ray protein crystallography provides crucial structural information of the overall protein, the iron center, and the substrate binding configuration, which can be used to correlate with spectroscopic and enzyme assay results to better derive reaction mechanisms (Aik et al., 2015; Bowman, Bridwell-Rabb, & Drennan, 2016). Theoretical calculations, such as density functional theory (DFT) calculations and full protein quantum mechanics/molecular mechanics (QM/MM) calculations have been commonly used in recent years to predict reaction coordinates and reaction energetics to differentiate different reaction pathways (Blomberg, Borowski, Himo, Liao, & Siegbahn, 2014; Proos Vedin & Lundberg, 2016; Quesne, Borowski, & de Visser, 2016; Ye & Neese, 2009; Zhang, Wu, Zhang, & Wang, 2023). Finally, molecular dynamics (MD) simulations (Karplus & McCammon, 2002) have shown great utilities in the studies of Fe/2OG enzymes (Li et al., 2020; Liao et al., 2018; Pang, Han, & Cui, 2013; Tang et al., 2021; Waheed et al., 2020), which reveal dynamic nature of protein-substrate interactions and complement to the protein crystallography analysis.
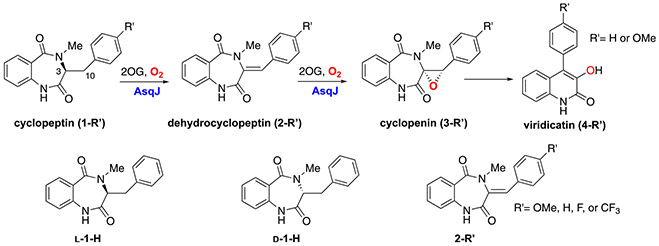
In recent years, we have applied the aforementioned research tools to study a bi-functional Fe/2OG enzyme, AsqJ (Chang et al., 2016; Li et al., 2020; Liao et al., 2018; Tang et al., 2021). AsqJ catalyzes a stepwise oxidation (desaturation and epoxidation) in the biosynthesis of a quinolone-type fungal alkaloid 4′-methoxy-viridicatin (Scheme 1) (Ishikawa et al., 2014). In this chapter, we document detailed experimental protocols and MD simulation methods used in our original publications (Chang et al., 2016; Li et al., 2020; Liao et al., 2018; Tang et al., 2021). Specifically, protocols for protein purification, in vitro enzymatic assays, kinetics, Mössbauer spectroscopy, and protein crystallography are described. Details of the procedures on performing MD simulations to predict protein dynamics and free energy calculations for structural transformations are also described. Finally, the overall reaction mechanisms of AsqJ-catalyzed desaturation and epoxidation derived from our studies are summarized.
Scheme 1.
Top: Overall reaction catalyzed by AsqJ; Bottom: Substrate analogs used in our original studies (Chang et al., 2016; Li et al., 2020; Liao et al., 2018).
2. Biochemical and spectroscopic characterization of AsqJ
2.1. Heterologous expression and purification of AsqJ
The DNA sequence that encodes AsqJ from Aspergillus nidulans was codon-optimized for over-expression in Escherichia coli, synthesized, and inserted into the expression vector pET-28a by GeneArt (Regensburg, Germany). This plasmid construct, which places the gene under the control of a T7 promoter, allows overproduction of the protein containing an N-terminal hexa-histidine (His6) tag. The protocol for protein over-expression used in one of our original studies (Chang et al., 2016) is listed below:
Inoculate and grow the pre-cultures of the E. coli BL21 (DE3) transformants at 37 °C in Luria-Bertani (LB) medium containing 20 mg/L kanamycin overnight.
The next morning, the pre-culture is used to inoculate 1 L (1 to 100 v/v ratio) of the same growth media in a shaker at 37 °C with constant agitation (225 r.p.m.) until the optical density at 600 nm (OD600) reaches 0.5–0.7.
Induce expression of protein with the addition of Isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM and keep cell growth at 27 °C and 225 r.p.m. for an additional 4 h.
Harvest the cells by centrifugation at 4 °C for 20 min at 6000 × g and subsequently store the cell pellets at −80 °C.
Thaw and resuspend the frozen cells in an ice-chilled buffer containing 50 mM Tris (hydroxymethyl) aminomethane (Tris) at pH 8.0, 5 mM imidazole, and 100 mM NaCl.
Disrupt the cells by sonication (3 × 60 s pulses) in the presence of the phenylmethylsulfonyl fluoride (PMSF) protease inhibitor, lysozyme, and benzonase nuclease. Centrifuge the resulting lysate for 20 min at 15,000 r.p.m. at 4 °C.
Transfer the supernatant to nickel-nitrilotriacetic acid (Ni-NTA) resin pre-equilibrated with 50 mM Tris buffer (pH 8.0) containing 5 mM imidazole and 100 mM NaCl.
Wash the protein-bound resin with 50 mM Tris buffer (pH 8.0) containing 10 mM imidazole and 100 mM NaCl. Perform this step using the solution buffer volume equivalent to 8 volumes of the resin used.
Elute the protein with 50 mM Tris buffer (pH 8.0) containing 250 mM imidazole and 100 mM NaCl.
Analyze the protein purity using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Elution fractions containing pure AsqJ protein were then pooled together and concentrated to a volume of approximately 20 mL by using centrifugal 10 kDa molecular weight cutoff (MWCO) filters.
Dialyze the concentrated protein solution against 1 L of 50 mM Tris buffer (pH 8.0) containing 100 mM NaCl and 10 mM ethylenedia-minetetraacetic acid (EDTA) to remvoe imidazole and enzyme bound metals.
Dialyze the concentrated protein solution three times against 1 L of 50 mM Tris buffer (pH 8.0) containing 100 mM NaCl to remove EDTA introduced in the previous step.
Concentrate the dialyzed protein solution to 50–80 mg/mL by using centrifugal 10kDa MWCO filters.
Place the concentrated protein solution in a sealed flask on ice and connect the flask to a Schlenk line to remove O2 by exchanging the atmosphere in the flask to argon.
Transfer the degassed protein solution from the previous step into a glovebox maintained in N2 atmosphere, aliquot the solution to cryovials and freeze them using liquid nitrogen and store it at −80 °C.
2.2. In vitro assays of AsqJ reactions
To elucidate the reaction mechanism, liquid chromatography mass spectrometry (LC-MS) experiments were used to monitor the AsqJ catalyzed reactions. The experimental procedure for how to setup assays and LC-MS conditions used in our original studies (Chang et al., 2016; Liao et al., 2018) are described below.
All samples were prepared in a MBraun Unilab glove box filled with a nitrogen atmosphere. The high-performance liquid chromatography (HPLC) with detection by mass spectrometry (MS) was conducted on an Agilent Technologies (Santa Clara, CA) 1200 system coupled to an Agilent Technologies 6410 QQQ mass spectrometer.
2OG and ferrous ammonium sulfate solutions (with final concentrations of 100 and 50 mM respectively) were prepared in the box by dissolving them in buffer (100 mM Tris, pH 8.0). The substrate dissolved in DMSO was used directly. The protein samples were prepared by incubating anaerobic apo AsqJ (metal free) with ferrous ion, 2OG, and enzymes substrates with a ratio of 1:0.9:10:2. Here, the final concentration of enzyme is 100 μM. We chose this molar ratio to ensure saturation of the metal binding site of the enzyme with ferrous ion and the maximal formation of Asq•Fe(II)•2OĜsubstrate complex, which was based on the previous studies on the binding affinities of ferrous ion, 2OG, and substrate for an Fe/2OG enzyme, taurine:2OG dioxygenase (TauD).
Prepare an O2-saturated (~1.8 mM O2) buffer solution by bubbling O2 into a solution in a sealed 50 mL round bottom flask and keep the flask on ice.
For the chemical quench experiments, the anaerobic solution of AsqJ•FeII•2OG•substrate (e.g. 1-H, Scheme 1) was loaded onto a KinTek quench-flow instrument and rapidly mixed with oxygenated buffer at 5 °C. The reaction was halted by injecting the reaction mixture into an acetone solution in the volumetric ratio of 1:4 (reaction mixture:acetone) at different reaction times. The need for the chemical quench experiments is to provide a time-dependent product formation profile for the enzyme reactions. It is particularly useful for AsqJ catalysis because AsqJ has been proposed to carry out a sequential two-step reaction (desaturation followed by epoxidation). The time dependent product formation determined by chemical quench experiments provided strong experimental evidence to support this conclusion. Other enzyme reactions that do not require the chemical quench setup were prepared in a similar manner. However, after sample preparation, samples were removed from the glove box and exposed to air to initiate the reaction.
After a reaction mixture is prepared, spin down the solution by using centrifugation (14,000 g × 30 min) and transfer the supernatant to a vial.
Inject the supernatant into LC-MS. A C18 column (Agilent ZORBAX Extended 4.6 × 50 mm, 1.8 μm particle size). An isocratic system of 68% of solvent A (20 mM aqueous ammonium formate, pH 6.8) and 32% solvent B (acetonitrile) is used for this step. Detection is performed using electrospray ionization in positive mode (ESI+). Considering the polarity of the substrate and the products and to achieve separation with an effective running time for each sample, we chose to use a reversephase column, e.g., a C-18 column, and used acetonitrile/water as the solvent system. Additionally, we chose LC-MS (ESI+, positive mode of electrospray ionization) as our detection method because this method allows quantification/determination of products with good separation using a small amount of samples. It is worth noting that the substrate and products are more sensitive to detect by MS under the positive mode.
Analyze data using Agilent MassHunter software package.
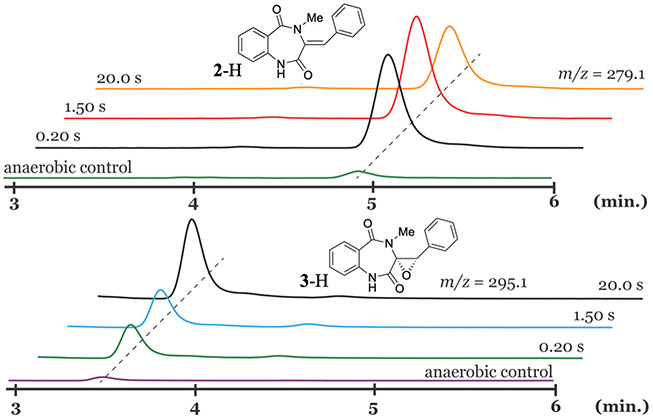
Fig. 1 shows typical LC-MS analysis of the AsqJ-catalyzed reaction by using 1-H as the substrate. The samples were prepared by reacting the anaerobic AsqJ-Fe(II)-2OG-1-H with O2 buffer at different time durations before quenched in acetone by using the chemical quench apparatus. The analysis revealed that the desaturated product, 2-H was initially formed at early reaction times, such as 0.2 and 1.5 s. At a longer time point, such as 20 s, 2-H started depleting while the epoxide product, 3-H, accumulated. This data set confirmed that AsqJ is a bi-functional enzyme, which reacts with 1-H to first produce 2-H and further converts 2-H to 3-H with additional 2OG and O2.
Fig. 1.
Chemical quench results of the AsqJ reaction with 1-H at different reaction times. The LC-MS analysis revealed the sequential formation of 2-H and 3-H. The figure is adapted with permission from Liao, H.-J., Li, J., Huang, J.-L., Davidson, M., Kurnikov, I., Lin, T.-S., … Chang, W. (2018). Insights into the desaturation of cyclopeptin and its C3 epimer catalyzed by a non-heme iron enzyme: Structural characterization and mechanism elucidation. Angewandte Chemie International Edition, 57(7), 1831–1835. https://doi.org/10.1002/anie.201710567 Copyright 2018 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
2.3. Stopped-flow absorption spectroscopy
To elucidate the reaction mechanisms of the AsqJ-catalyzed desaturation and epoxidation, transient kinetics experiments via stopped-flow absorption spectroscopy (SF-Abs) were used initially to reveal any optically detectable intermediates. The protocol for SF-Abs experiment used in our original studies (Chang et al., 2016; Li et al., 2020; Liao et al., 2018) is listed below:
All experiments were performed using an Applied Photophysics SX20 stopped-flow instrument (Applied Photophysics Inc., Charlotte, NC), operated within an MBraun Unilab glove box filled with a nitrogen atmosphere. The glovebox was maintained at an O2 level of <0.5 ppm throughout all SF-Abs experiments. An external water chiller was used to maintain the temperature of the reaction solution during kinetic analysis, while a photodiode array detector was used as the optical detection device.
Transfer solid powders of ferrous ammonium sulfate, 2OG, and substrate into the glovebox. Allow all materials to equilibrate with the glovebox atmosphere overnight.
Prepare an O2-saturated (~1.8 mM O2) buffer solution by bubbling O2 (g) into a solution of 50 mM Tris pH 8.0 containing 100 mM NaCl in a sealed 50 mL round bottom flask. Store the flask within an ice bath throughout the oxygenation step to maintain a temperature of ~4 °C. The buffer solution should be bubbled for at least 1 h to yield the final O2-saturated solution.
Prepare stock solutions of ferrous ion, 2OG, and substrate using anerobic 50 mM Tris pH 8.0 containing 100 mM NaCl. Then, prepare the AsqJ•Fe(II)•2OG•substrate quaternary complex solution wherein each component concentration is the following: 0.65 mM AsqJ apo enzyme, 0.58 mM Fe(II), 5.8 mM 2OG, and 1.16 mM substrate (e.g. 2-H or 2-OMe, Scheme 1). Here similar to that described in step 2 of Section 2.2, the chosen molar ratio of various components was to ensure the maximal formation of the AsqJ•Fe(II)•2OG•substrate complex.
Carefully load the enzyme quaternary complex solution, using a 1 mL syringe, into the SF-Abs instrument. Avoid introducing bubbles while loading the enzyme solution. The presence of bubbles can lead to deleterious baseline drifting in the optical traces which often produces misleading results.
Transfer the O2-saturated buffer into the glovebox and load ~1 mL of the buffer into the SF-Abs instrument. Again, avoid the introduction of bubbles.
Collect SF-Abs data by using the single mixing mode under various timescales to gain insight into the formation/decay kinetics of optical features possibly attributable to the enzyme quaternary complex, the oxyferryl species, or other intermediates. Absorbance scans from 270 to 720 nm were collected. The specific wavelengths monitored during our original studies were 310 nm, which reflects the optical absorption feature from the substrate 1-H or 1-OMe due to the presence of the aromatic moiety in the substrate structure, 330 nm, which is a part of absorption features belonging the oxyferryl intermediate (Matthews, Krest, et al., 2009; Price, Barr, Tirupati, et al., 2003; Srnec et al., 2016), and 470 nm, which reflects absorption feature of the AsqJ-Fe(II)-2OG complex metal-to-ligand charge transfer band (Chang et al., 2016).
Process the resulting data using any graphical interface software available which reads CSV files. The software packages used in our data processing were KaleidaGraph (Synergy Software, Reading, PA) and/or Spincount (Petasis & Hendrich, 2015) Quantitative kinetics simulations were accomplished with the software package KinTek Explorer (Johnson, Simpson, & Blom, 2009).
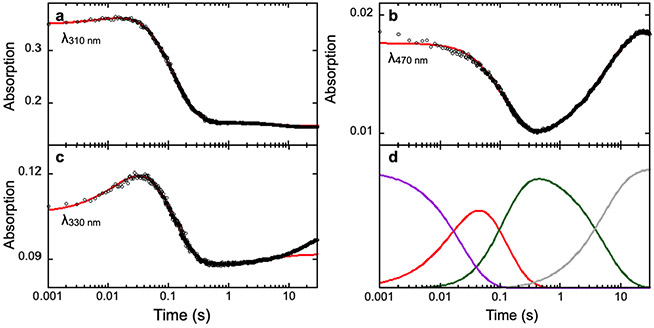
Fig. 2 shows typical SF-Abs data and analysis of the AsqJ-catalyzed epoxidation reaction by using 2-H as the substrate (Li et al., 2020) Three time dependent kinetic traces at different wavelengths are shown, which represent the kinetics of the consumption of the substrate, 2-H, at 310 nm, the kinetics of the formation and the decay of the oxyferryl intermediate at 330 nm, and the kinetics of the consumption and the reformation (due to depletion of O2 in the reaction solution) of the AsqJ•Fe(II)•2OG•2-H quaternary complex. The model used for the kinetics simulation is shown in Scheme 2 below:(Li et al., 2020).
Fig. 2.
SF-Abs derived kinetic traces of the time-dependent absorption changes at various wavelengths in the AsqJ-catalyzed epoxidation by using 2-H as the substrate. Experimental data are shown in black dots, the kinetic simulations are shown in red curves. The resulting kinetics of individual species from the simulations are shown in panel d with the following color codes: purple, the AsqJ•Fe(II)•2OG•2-H quaternary complex; red, the oxyferryl intermediate; green, the AsqJ•Fe(II)•sucinnat•product complex; gray, the AsqJ•Fe(II)•2OG ternary complex. The figure is adapted with permission from Li, J., Liao, H.-J., Tang, Y., Huang, J.-L., Cha, L., Lin, T.-S., … Guo, Y. (2020). Epoxidation catalyzed by the nonheme iron(II)- and 2-oxoglutarate-dependent oxygenase, AsqJ: Mechanistic elucidation of oxygen atom transfer by a ferryl intermediate. Journal of the American Chemical Society, 142(13), 6268–6284. https://doi.org/10.1021/jacs.0c00484 Copyright 2020 American Chemical Society.
Scheme 2.
Three-step kinetic model used for analyzing AsqJ-catalyzed epoxidation.
2.4. Freeze-quench Mössbauer experiments
To further reveal the nature of the intermediates involved in AsqJ catalyzed reactions and validate the kinetic model derived from the SF-Abs studies, Mössbauer spectroscopy coupled with freeze-quench technique was used. The protocol for freeze-quench Mössbauer experiments used in our original studies (Chang et al., 2016; Li et al., 2020; Liao et al., 2018) is listed below:
Transfer pre-weighed amounts of 2OG and substrate into the glove box. Allow all materials to equilibrate with the box atmosphere overnight. Different from the previous section, a stock of 57Fe(II) solution is to be used to reconstitute the apo enzyme.
The following day, prepare an O2-saturated buffer solution in the same manner as described in the previous section.
Transfer a prepared anaerobic 57Fe(II) aliquot into the glove box. The 57Fe solution is prepared by dissolving 57Fe metal powders in 0.5 M sulfuric acid. The pH condition of this acidic 57Fe solution is adjusted back to the neutral condition by using an anaerobic buffer before using it to reconstitute apo AsqJ.
Prepare the anaerobic AsqJ•57Fe(II)•2OG•substrate complex with the following component concentrations: 1.44 mM enzyme, 1.3 mM 57Fe(II), 13 mM 2OG, and 2.6 mM substrate (e.g. 2-H or 2-OMe, Scheme 1) in 50 mM Tris buffer in pH 8.0 with 100 mM NaCl. Prior to addition to the enzyme, the 57Fe(II) should be diluted in 1 M Tris buffer in pH 8.0 to avoid crashing the enzyme out of solution due to the concentrated sulfuric acid.
Transfer a small volume (~300 μL) of the anaerobic AsqJ•57Fe(II)•2OG•substrate complex into a Mössbauer sample cup and freeze the sample in a pre-cooled metal block in the glovebox. This sample will serve as an anaerobic control.
Load the remaining volume of anaerobic AsqJ•57Fe(II)•2OG•substrate complex into an air-tight Hamilton syringe and seal the syringe.
Load the syringe with the anaerobic enzyme solution into a KinTek quench-flow instrument, which will be maintained at 4 °C using an external water chiller throughout the experiment.
Transfer the O2-saturated buffer into the KinTek quench flow instrument using another pre-chilled air-tight Hamilton syringe.
- Use the KinTek quench flow instrument to rapidly mix the O2-staurated buffer with the anaerobic enzyme solution in a 1:1 vol ratio. Set the instrument delay times accordingly to match the previously collected kinetic data by using SF-Abs.
- Delay time variability is produced using delay lines to increase/decrease the time the reaction solution incubates prior to freeze quench.
- Reaction quenching is accomplished by rapidly injecting the reaction solution, after a designated delay time, into a solution of liquid ethane maintained at 90 K, which is contained in a falcon tube. Such a freeze quench process will generate a frozen solution powder that is suspended in liquid ethane.
Use a vacuum system to carefully remove the liquid ethane from the container that holds the frozen solution sample powder. During liquid ethane removal, the sample powder was stored in a cold liquid ethane solution.
Pack the dried frozen sample powder carefully into a freeze quench-Mössbauer cup using a metal funnel and packing rod (both pre-cooled in liquid nitrogen) after the complete liquid ethane removal.
Record Mössbauer data on each sample using a Mössbauer spectrometer.
Perform Mössbauer spectral simulations using applicable graphical software, such as either WMOSS (See Co., Minneapolis, MN) or Spincount (Petasis & Hendrich, 2015).
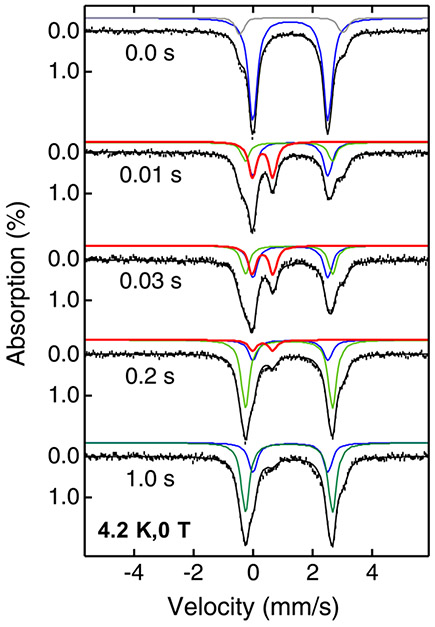
Fig. 3 shows the typical FQ-Mössbauer data and spectral simulations of the AsqJ-catalyzed epoxidation reaction by using 2-H as the substrate.(Chang et al., 2016) The spectral simulations reveal that the AsqJ•57Fe(II)•2OG•2-H quaternary complex exhibits a typical high-spin ferrous species with Mössbauer parameters of δ = 1.24 mm/s ∣ΔEQ∣ = 2.54 mm/s (the blue trace in Fig. 3). Upon reacting with O2, the spectral component representing the enzyme quaternary complex is quickly depleted. In the meantime, an oxyferryl intermediate with the Mössbauer parameters of δ = 0.31 mm/s ∣ΔEQ∣ = 0.68 mm/s is formed within 0.03 s and decayed after 0.2 s (the red trace in Fig. 3). This FQ-Mössbauer derived oxyferryl kinetics is fully consistent with that derived from SF-Abs analysis (see Fig. 2). The FQ-Mössbauer data also identify a AsqJ•57Fe(II)•2OG•product complex with the Mössbauer parameters of δ = 1.20 mm/s ∣ΔEQ∣ = 2.95 mm/s (the green trace in Fig. 3), which is accumulated to a maximal level at ~ 1s.
Fig. 3.
4.2 K zero-field Mossbauer spectra of the reaction of the AsqJ•57Fe(II)•2OG•2-H complex with O2 at different times. Black vertical bars mark experimental spectra, black solid lines represent overall spectral simulations, blue solid lines represent the spectral simulation of the AsqJ•57Fe(II)•2OG•2-H complex, red solid lines represent the spectral simulation of the oxyferryl intermediate, green solid lines represent the spectral simulations of the AsqJ•57Fe(II)•2OG•product complex. The figure is adapted with permission from Chang, W., Li, J., Lee, J. L., Cronican, A. A., & Guo, Y. (2016). Mechanistic investigation of a non-heme iron enzyme catalyzed epoxidation in (−)-4′-methoxycyclopenin biosynthesis. Journal of the American Chemical Society, 138(33), 10390–10393. https://doi.org/10.1021/jacs.6b05400, Copyright 2016 American Chemical Society.
3. X-ray protein crystallographic characterization of AsqJ
3.1. Protein crystallization
To elucidate the structural basis underlying the AsqJ-catalyzed desaturation and epoxidation, crystallographic analysis was employed to determine the binding modes of the substrates and the metal cofactor in the active site. The protocol for the expression and purification of AsqJ and the crystallization procedure has been previously described (Li et al., 2020; Liao et al., 2018; Liao, 2019) and is outlined below:
E. coli C43 (DE3) cells, harboring the expression plasmid, were cultured in LB medium supplemented with 30 mg/L kanamycin at 37 °C with shaking at 200 rpm.
Upon reaching an OD600 of 0.4–0.6, isopropyl β-d-1-thiogalacto-pyranoside (IPTG) was added to achieve a final concentration of 0.5 mM. Protein expression was then induced at 28 °C for 4 h. Bacteria were harvested by centrifugation and subsequently stored at −80 °C for future use.
For the purification of His6-tagged AsqJ samples that were suitable for crystallization, the bacteria were thawed and resuspended in Buffer A (100 mM Tris/HCl, pH 7.5; 500 mM NaCl; 20 mM Imidazole; 10% glycerol), supplemented with an EDTA-free protease inhibitor cocktail tablet (SIGMAFAST™). The cells were then disrupted by sonication. This buffer composition was chosen to maintain protein stability and integrity, essential for obtaining high-quality crystals, as opposed to the conditions used in Section 2 which were optimized for biochemical and spectroscopic analyses.
The crude cell lysate was clarified by centrifuging at 17,500 × g for an hour, repeated three times, at 4 °C. The supernatant was pooled and applied to a pre-equilibrated nickel-NTA resin column. The column was washed with eight column volumes of Buffer A, followed by an equal volume of Buffer A mixed with varying ratios of Buffer B (100 mM Tris/HCl, pH 7.5; 500 mM NaCl; 500 mM Imidazole; 10% glycerol), including 2%, 4%, and 6%.
His6-tagged AsqJ was subsequently eluted with Buffer A containing 50% Buffer B. The eluted fractions were pooled and dialyzed first against 1 L of buffer containing 50 mM Tris/HCl (pH 8.0), 100 mM NaCl, and 10 mM EDTA, and then twice against 1 L of EDTA-free buffer (50 mM Tris/HCl, pH 8.0; 100 mM NaCl). The resulting protein was concentrated and mixed with glycerol to achieve a final concentration of 50%, and then stored at −20 °C.
Before crystallization, the sample was further purified using a size-exclusion column (Hi-Load Superdex 200) with gel filtration buffer (20 mM Tris/HCl, pH 7.5; 100 mM sodium chloride; 5 mM β-mercaptoethanol). The purified protein was collected, concentrated to 1 mM for crystallization, and its concentration was determined by UV absorption at 280 nm using a calculated molar absorptivity of 28,082 M−1 cm−1, as obtained from the ProtParam web tool (https://web.expasy.org/protparam/).
Because the purification of His6-tagged AsqJ using Ni-NTA resin may result in replacement of iron by Ni2+ (Bräuer, Beck, Hintermann, & Groll, 2016), FeCl2 (100 mM) was added to the purified AsqJ (0.57 mM) in gel filtration buffer (20 mM Tris/HCl, pH 7.5; 5 mM β-mercaptoethanol; 100 mM NaCl) to achieve a final concentrations of 0.85 mM for FeCl2 and 0.56 mM for AsqJ, respectively. After overnight incubation at 4 °C, the mixture was centrifuged at 21,500 × g for 30 min at 4 °C to remove any precipitated material, this step ensures that the AsqJ used for crystallization was predominantly in the iron-bound form.
Succinate (100 mM stock in water) and the primary substrate, 1-H (100 mM stock in DMSO) were added to the supernatant, adjusting the final concentrations of AsqJ, FeCl2, succinate, and 1-H to 0.5, 0.75, 1.0, and 1.0 mM, respectively. These molar ratios were to ensure the formation of the AsqJ•Fe•succinate•1-H complex. Similarly, for the 2-CF3-bound AsqJ complex, the final concentrations of AsqJ, FeCl2, succinate, and 2-CF3 were also adjusted to 0.5, 0.75, 1.0, and 1.0 mM, respectively.
The sitting-drop vapor diffusion method was used for crystallizing the AsqJ•Fe•succinate•1-H and AsqJ•Fe•succinate•2-CF3 complexes. Specifically, to crystallize the AsqJ•Fe•succinate•1-H complex, 0.1 μL of protein sample was mixed with an equal amount of reservoir solution (20% (w/v) PEG 1000, 100 mM imidazole/HCl, pH 8.0, 0.2 M calcium acetate) and 0.2 μL of ddH2O, then equilibrated against 25 μL of reservoir solution at 4 °C (Table 1, left column). For the crystallization of the AsqJ•Fe•succinate•2-CF3 complex, 0.1 μL of protein sample was mixed with an equal amount of reservoir solution (either 1.2 M potassium sodium tartrate tetrahydrate or 0.6 M potassium sodium tartrate tetrahydrate, both with 0.1 M Tris, pH 8.5), and then equilibrated against 25 μL of reservoir solution at 4 °C (Table 1, right column). Crystals of the AsqJ•Fe•succinate•1-H complex typically appeared in about 3 weeks, whereas those of the AsqJ•Fe•succinate•2-CF3 complex usually formed within about 6 weeks.
Crystals of AsqJ•Fe•2OG•1-H and AsqJ•Fe•2OG•D-1-H were obtained by soaking pre-grown crystals of AsqJ•Fe•succinate•1-H in substitute mother liquor containing 25% PEG 1000, 100 mM imidazole/HCl (pH 8.0), 10 mM 2OG, and 2mM of either L- or D-1-H, and the crystals were soaked for 24 h at 4 °C (Table 2).
To obtain crystals of AsqJ•Fe•2OG•2-H•O2, pre-grown crystals of AsqJ•Fe•succinate•1-H were soaked in a substitute mother liquor containing 25% PEG 1000, 100 mM imidazole/HCl (pH 8.0), 5 mM 2OG, and 1 mM of 2-H for 24 h at 4 °C (Table 3).
To acquire the crystal of AsqJ•Fe•2OG•3-H, the pre-grown crystals of AsqJ•Fe•succinate•1-H were soaked in a substitute mother liquor containing 25% PEG 1000, 100 mM imidazole/HCl (pH 8.0), 25 mM 2OG, and 20 mM 3-H for 24 h at 4 °C (Table 4).
The crystal structure representing a potential intermediate during the AsqJ catalyzed epoxidation reaction, the AsqJ•Fe•succinate•2-H (O-C10 bond formation) complex was obtained by soaking pre-grown crystals of AsqJ•Fe•succinate•1-H in a substitute mother liquor. This liquor contained 25% PEG 1000, 100 mM imidazole/HCl (pH 8.0), 5 mM 2OG, and varying concentrations of ascorbate (5, 0.1, 0.5, and 1 mM, respectively) for 24 h at 4°C (Table 5).
All AsqJ crystals mentioned above were harvested by first transferring them into paratone oil, then looping, and finally flash-freezing in liquid nitrogen for data collection.
Table 1.
Preparation of crystallization drops for AsqJ•Fe•succinate•1-H and AsqJ•Fe•2OG•2-CF3 complexes using the hanging drop method.
| AsqJ•Fe•succinate•1-H complex |
AsqJ•Fe•succinate•2-CF3 complex |
|
|---|---|---|
| AsqJ•Fe•succinate•1-H (protein sample, μL) | 0.1 | – |
| AsqJ•Fe•succinate•2-CF3 (protein sample, μL) | – | 0.1 |
| ddH2O (μL) | 0.2 | – |
| reservoir solution (20% (w/v) PEG 1000, 100 mM imidazole/HCl, pH 8.0, 0.2 M calcium acetate) (μL) | 0.1 | – |
| reservoir solution (either 1.2 M potassium sodium tartrate tetrahydrate or 0.6 M potassium sodium tartrate tetrahydrate, both with 0.1 M Tris, pH 8.5) (μL) | – | 0.1 |
| Total drop volume (μL) | 0.4 | 0.2 |
Table 2.
Preparation of AsqJ•Fe•2OG•1-H and AsqJ•Fe•2OG•D-1-H crystals by soaking pre-grown AsqJ•Fe•succinate•1-H crystals.
| AsqJ•Fe•2OG•1-H complex | AsqJ•Fe•2OG•D-1-H complex | |
|---|---|---|
| Pre-grown crystal | AsqJ•Fe•succinate•1-H | AsqJ•Fe•succinate•1-H |
| Soaking solution | 25% PEG 1000, 100 mM imidazole/HCl (pH 8.0), 10 mM 2OG, 2 mM 1-H | 25% PEG 1000, 100 mM imidazole/HCl (pH 8.0), 10 mM 2OG, 2 mM D-1-H |
Table 3.
Preparation of AsqJ•Fe•2OG•2-H•O2 crystals by soaking pre-grown AsqJ•Fe•succinate•1-H crystals.
| AsqJ•Fe•2OG•2-H•O2 complex | |
|---|---|
| Pre-grown crystal | AsqJ•Fe•succinate•1-H |
| Soaking solution | 25% PEG 1000, 100 mM imidazole/HCl (pH 8.0), 5 mM 2OG, 1 mM 2-H |
Table 4.
Preparation of AsqJ•Fe•2OG•3-H crystal by soaking pre-grown AsqJ•Fe•succinate•1-H crystal.
| AsqJ•Fe•2OG•3-H complex | |
|---|---|
| Pre-grown crystal | AsqJ•Fe•succinate•1-H |
| Soaking solution | 25% PEG 1000, 100 mM imidazole/HCl (pH 8.0), 25 mM 2OG, 20 mM 3-H |
Table 5.
Preparation of AsqJ crystals in different stages of the catalytic cycle by soaking pre-grown AsqJ•Fe•succinate•1-H crystals.
| Soaking solution | |
|---|---|
| AsqJ•Fe•succinate•2-H complex (O-C10 bond formation) | 25% PEG 1000, 100 mM imidazole/HCl (pH 8.0), 5 mM 2OG, 0.5 mM ascorbate |
3.2. Crystal structure determination
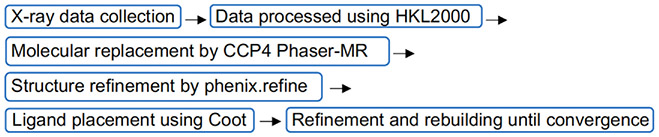
In this section, the general protocols and procedures for crystal structure determination used in our original studies (Li et al., 2020; Liao et al., 2018) are described below.
Collecting X-ray diffraction data sets of AsqJ•Fe•2OG•1-H and AsqJ•Fe•2OG•D-1-H crystals were at a synchrotron beamline. In our case, BL13C1 and BL15A1 at the National Synchrotron Radiation Research Center (NSRRC), Taiwan, were used.
Processing the raw diffraction data onsite using the HKL2000 program suite (Otwinowski & Minor, 1997).
Determining the structure of AsqJ•Fe•2OG•1-H by molecular replacement with CCP4 Phaser-MR (McCoy et al., 2007; Winn et al., 2011) using the AsqJ structure from the AsqJ•Ni•2OG•1-H complex (PDB ID: 5DAW, (Bräuer et al., 2016)) as the search model. This choice was made because the search model features the simultaneous presence of both metal cofactor and substrate within the active site, suggesting that the conformation of the catalytic site in the search model likely resembles that of the AsqJ•Fe•2OG•1-H complex to be determined and thereby suitable for conducting molecular replacement.
Performing initial structural refinement by using phenix.refine (Afonine et al., 2012). Upon convergence of refinement, the presence of iron, 2OG, and 1-H in the active site can be readily recognized in the Fo-Fc density map.
Manually docking the bound iron, 2OG, and 1-H into the initial unbiased Fo-Fc density maps using Coot (Emsley, Lohkamp, Scott, & Cowtan, 2010).
Conducting additional rounds of manual rebuilding by refining the positioning of ligands and amino acid residue side-chains based on features observed in subsequent Fo-Fc density maps.
The structure of AsqJ•Fe•2OG•D-1-H was determined following the same procedure. In both structures, continuous electron density can be observed between the mononuclear iron center and the side-chain Nε atoms of His134 and His211, a side-chain Oδ atom of Asp136, and two oxygen atoms from the α-ketoacid moiety of 2OG. A clear bulge in the iron’s electron density was interpreted as an iron-bound water molecule, completing the hexacoordinate iron center in an octahedral geometry.
Performing structural refinement by restraining the metal-ligand bond lengths and angles to averages observed in other non-heme iron-dependent dioxygenases.
The final atomic coordinates and structure factors were deposited in the Protein Data Bank under accession codes 5Y7R and 5Y7T for AsqJ•Fe•2OG•1-H and AsqJ•Fe•2OG•D-1-H, respectively (Fig. 4) (Liao et al., 2018).
The remaining AsqJ structures were determined by molecular replacement with CCP4 Phaser-MR, using the AsqJ structure from the AsqJ•Fe•2OG•1-H complex (PDB ID: 5Y7R, (Liao et al., 2018)) as the search model. These structures include the AsqJ•Fe•2OG•2-H•O2 complex (PDB ID: 6K0E, (Li et al., 2020)), the AsqJ•Fe•2OG•3-H complex (PDB ID: 6K0F, (Li et al., 2020)), the AsqJ•Fe•succinate• 2-CF3 complex (PDB ID: 6KD9 and 6K30, (Li et al., 2020)) and the AsqJ•Fe•succinate•2-H (O-C10 bond formation) complex (PDB ID: 6JZM, (Li et al., 2020)).
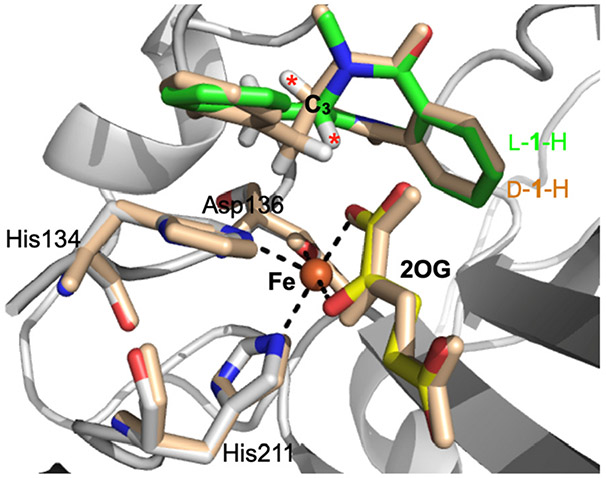
Fig. 4.
Crystal structure of AsqJ in the presence of L-1-H (multi-color) and D-1-H (wheat color) based on PDB IDs: 5Y7R and 5Y7T (Liao et al., 2018). The iron center, the iron bound 2OG and the protein residues are labeled accordingly. L-1-H is colored in green while D-1-H in wheat. The figure also highlights the C3 position and the opposite orientations of the C3 proton of L-1-H and D-1-H (labeled as red asterisks). This opposite orientations of the C3 protons provided a crucial structural information of the plausible desaturation reaction mechanism used by AsqJ (See Section 5 for further discussion).
The overall process and workflow of crystal structure determination is illustrated in Scheme 3 below and the structural overlay of the active centers from the crystal structures of the AsqJ•Fe•2OG•1-H and the AsqJ•Fe•2OG•D-1-H complexes are shown in Fig. 4.
Scheme 3.
The overall workflow of crystal structure determination as described in Section 3.2.
4. Molecular dynamics simulations
Molecular dynamics (MD) simulations can provide a detailed and quantitative description of molecular processes in biological systems and greatly augment information obtained with experimental techniques (Karplus & McCammon, 2002). MD gave an invaluable insight into catalytic mechanisms of numerous enzymes (Gao et al., 2006). MD was especially successful to discern the role of particular interactions, such as protein substrate interactions, that lower free energies of transition states and direct the reaction to a particular path.
Empirical analytical force fields allow to model molecular interactions involving biopolymers (e.g. AMBER; Case, Aktulga, Belfon, Ben-Shalom, et al., 2023) and CHARMM (Brooks et al., 2009) and general stable organic molecules (e.g. GAFF (Wang, Wolf, Caldwell, Kollman, & Case, 2004; Wang, Wang, Kollman, & Case, 2006) and CGenFF (Vanommeslaeghe et al., 2010)). To model chemical transformations, such as reaction coordinates calculations, a part of the simulated biocatalytic system is often described with a quantum chemical model (QM/MM method (Senn & Thiel, 2009)). However, QM/MM calculations can be computationally much more expensive than pure MD that employ empirical atom-atom interaction potentials, therefore, QM/MM generally has difficulties to adequately sample conformational space of the system studied. In our studies, we do not focus on reaction coordinate calculations, but rather focus on analyzing the protein-substrate interactions and dynamics of specific reaction intermediate states (Li et al., 2020; Liao et al., 2018). This information is important for understanding catalytic mechanism of the enzyme studied. Therefore, we applied pure MD simulations to our studies on AsqJ, keeping conformations of metal coordinating groups, not described by standard force fields, restrained to experimentally observed geometries. This method allowed us to run sufficiently long trajectories to adequately sample conformation space of the enzyme and compute relative free energies of reaction intermediates.
Several computationally efficient MD software packages have been developed that allow the routine modeling of proteins on a time scale of hundreds of nanoseconds and longer. Here we used freely accessible GROMACS package (Hess, Kutzner, van der Spoel, & Lindahl, 2008) to model AsqJ function. GROMACS is highly optimized for both CPU and GPU and implements modern biomolecular and general molecule force fields such as AMBER (Case, Aktulga, Belfon, Ben-Shalom, et al., 2023) and GAFF used in this work. GROMACS also provides facilities to perform free energy calculations for several types of alchemical transformations that we used to compute probabilities of distinct states of AsqJ/substrate complex and clarify the mechanism of AsqJ catalysis.
4.1. MD system setup
4.1.1. Initial structure preparation
A setup of a model for MD simulations of AsqJ with a substrate and a cosubstrate (2OG) bound involved additional steps compared to a setup of simulations of a protein without cofactors. The starting structure of the system was taken from AsqJ crystal structures, such as PDB database entry 6K0E, that contains Fe(II), 2OG and dehydrocyclopeptin (2-H, see Scheme 1 for structure). First, the protonation state of Histidine residues (δ-N of the imidazole ring) (HID residue), the ε-N of the imidazole ring (HIE), or at both nitrogen atoms (HIP) were determined using PROPKA website (Olsson, Søndergaard, Rostkowski, & Jensen, 2011). Next, hydrogen atoms were added to the protein and AMBER99SB-ILDN (Lindorff-Larsen et al., 2010) force field parameters were set to the protein residues with the GROMACS pdb2gmx utility. 2OG and substrate GAFF force field parameters were assigned using ACPYPE utility (Sousa da Silva & Vranken, 2012) that calls ANTECHAMBER program of AMBERTOOLS suite (Case, Aktulga, Belfon, Cerutti, et al., 2023). Geometry of the substrate molecule were optimized using DFT B3LYP functional and 6-311G basis set in the GAUSSIAN program (Frisch et al., 2016).
4.1.2. Iron center modeling
Since there are no published force field parameters to accurately describe the mononuclear iron center in Fe/2OG enzymes, we simulated non-heme iron centers in Fe/2OG enzymes by applying additional restraints that keep the geometry of the iron center close to that observed in the AsqJ X-ray structure. Here, we introduced specific restraints aimed at maintaining the correct bidentate configuration of 2OG and preventing the swing movement of imidazole ring of iron-bound histidine residues, which involves adjusting the distance restraints, angle restraints, and dihedral restraints within the active center. Fe2+ VdW parameters (ε = 0.0264 kcal/mol, σ = 2.4033 Å) were taken from the literature (Babu & Lim, 2006).
4.1.3. Solvated enzyme system preparation
After assigning force field parameters and initial geometry for the AsqJ complex with 2OG cofactor and the substrate, we solvated the system in the cubic box of TIP3P (Jorgensen, Chandrasekhar, Madura, Impey, & Klein, 1983; Neria, Fischer, & Karplus, 1996) water molecules using GROMACS editconf and solvate utilities. Sodium and chloride ions were added with GROMACS genion utility to ensure physiological ionic strength and overall neutrality of the simulated system.
4.2. Energy minimization and equilibration MD
Before starting MD simulations, the initial geometry of the solvated protein system was relaxed with Steepest Decent energy minimization method using GROMACS mdrun program. The minimization was stopped when the maximum atom energy gradient value fell below 1000 kJ/mol/nm. After energy minimization, the system attains a reasonably optimized initial structure, exhibiting improved geometry and solvent orientation.
The first MD equilibration step was performed in the NVT ensemble conditions. We gradually increase the temperature from 0 to 300 K using the Langevin thermostat with a timestep of 2 fs. Typically, 50–100 ps is sufficient for equilibration; however, for the Fe/2OG enzyme AsqJ system, we set it to 1 ns.
We then transitioned to an NPT ensemble for equilibration, employing the Langevin integrator with parameters 300 K, 20 ns relaxation time, 2 fs timestep, and Berendsen barostat (1 atm). In all preceding steps, positional restraints of 1000kJ/mol were maintained on all atoms of protein, the substrate and cofactor molecules, and the iron center, while allowing solvent molecules to move freely.
Next, staged relaxation followed, during which we gradually relaxed positional restraints: initially, controlling the entire protein 1000 kJ/mol/nm2, then limiting restraints only to Cα atoms, then further decreasing the restraint force constant to 400 kJ/mol, and finally to 200 kJ/mol. Each stage lasted for 20 ns, with a goal to achieve an enhanced stability of the simulated system. We monitored structural stability of the system by assessing the Root Mean Square Deviation (RMSD) of the protein atoms during the equilibration period.
4.3. MD production runs and analysis
4.3.1. Criteria during equilibration
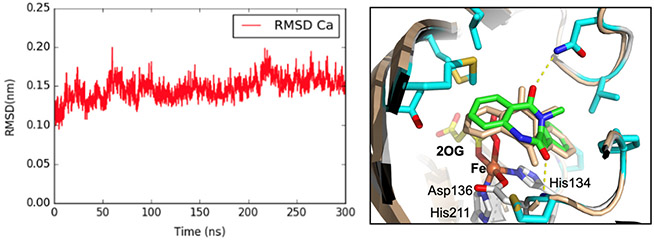
Our main NPT MD simulation, lasting 300 ns, proceeded with the release of positional restraints of all atoms, except for those on the Fe first coordination sphere. Before drawing any conclusions, it’s crucial to assess the quality of MD simulations. During these simulations, it's important to check the geometry stability of the iron, meanwhile the protein structure should maintain its integrity without significant unfolding or other abrupt alterations. GROMACS offers built-in functions tailored for processing trajectory files (.xtc). Initially, the trjconv function is used to address translational and rotational movements and resolve issues like proteins crossing periodic boundaries. Following this, the GROMACS rms function computes the root mean square deviation (RMSD) of the protein relative to the initial protein structure determined from X-ray crystallography (Fig. 5, left). Lastly, the GROMACS distance function is employed to determine the distance between atom pairs, such as between Fe and each atom in the first coordination shell. All these functions necessitate user interaction for making appropriate selections.
Fig. 5.
Left: RMSD Cα atoms of MD simulated AsqJ structure (relative to AsqJ crystal structure, PDB ID: 6K0E) during 300 ns production MD trajectories. Right: Structure overlay of MD simulated AsqJ structure in the presence of 2-H (wheat color) and the corresponding X-ray crystal structure (multi-color, PDB ID: 6K0E, (Li et al., 2020)). The figure highlights the overlay of the 2-H in the MD structure (wheat color) and in the crystal structure (green color).
4.3.2. Trajectory analysis
Finally, we can analyze the trajectory of the MD production. In this context, we focused on determining the distance between the Fe center and the C3 and C10 atoms of the substrate, 2-H. To calculate this distance, a separate index file (.ndx) is required to specify the pairs of atoms to be measured. Ultimately, the trajectory can be visualized in PyMOL (Schrödinger, 2020) as a dynamic movie.
In this case, a representative structure of AsqJ, captured during the 300 ns MD trajectory (Fig. 5, right), was chosen based on its minimal deviation from the average RMSD. Analyzing various snapshots extracted from AsqJ’s MD trajectory, we noted the Fe coordination centers closely mirrored their positioning in the original crystal structure. Specifically, the distance from Fe to the substrate's C3 position stood at 4.6 Å, and the distance between Fe and the substrate's C10 atom was 4.2 Å. Although these measurements were slightly greater than those found in the crystal structure (Fe-C3 at 4.9 Å and Fe-C10 at 4.4 Å), they were sufficiently proximate to validate our model's accuracy.
4.4. Calculations of free energy of oxygen binding to Fe(II) center
MD simulation can be utilized to calculate free energies of different thermodynamic processes. Generally, two types of processes can be simulated. One is to calculate the free energy difference between two static states without the need to know the activation barrier that connects these two states. These calculations allow to estimate the relative equilibrium population of different states of the system. The other calculation type computes not only the free energy difference but also the activation barrier between the two states – thus allow evaluate the kinetic of transition between states. In this section, we describe MD simulations for the first type that use alchemical transformations and thermodynamic integration technique. We again use AsqJ as an example.
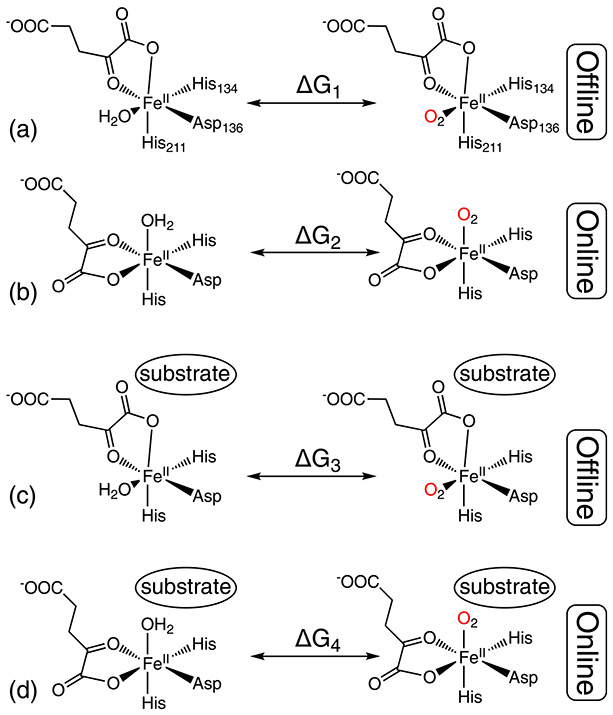
One key step during the AsqJ reaction is the O2 binding step at the iron center. Upon O2 binding, the iron bound water molecule will be replaced. However, it is well-known that 2OG can bind to the Fe(II) center of Fe/2OG enzymes in two orientations, the online and the offline orientation, which create two different O2 binding sites that are occupied by H2O molecule before the introduction of O2. For AsqJ, one of the binding sites (the 2OG offline orientation) points away from the substrate and the other site (the 2OG online orientation) locates right between the iron center and the substrate (Scheme 4). Interestingly, all the AsqJ crystal structures only capture the 2OG offline orientation. Thus, The MD simulation objective is to calculate the free energy difference between the H2O ligated state and the O2 ligated state for both the 2OG online and offline orientations to determine which orientation state would be thermodynamically favorable for O2 binding.
Scheme 4.
The four system configurations used for free energy calculations by alchemical transformations and thermodynamic integration via MD simulations. (A) and (C) represent the 2OG offline binding configuration in the absence (A) or the presence (C) of the substrate. (B) and (D) represent the 2OG online binding configuration in the absence (B) or the presence (D) of the substrate. For each system configuration free energy difference (ΔGi, i = 1–4) between the H2O bound state and the O2 bound state was calculated.
Though the real process of O2 addition may involve departure of H2O and approaching of O2, in-silica simulation enables us to construct a hypothetical reaction pathway by mutating the H2O to O2 in place. Thermodynamical analysis of such an alchemical process allow for calculation of the relative free energy differences between the two end states of the pathway (the H2O bound state and the O2 bound state), at different system configurations, in this case with online vs offline configuration and substrate bound and unbound states (a total of 4 system configurations, see Scheme 4). The mutation process is divided into small steps, during which the mutated molecule force field parameters are intermediate between those of H2O and O2 and the free energy of the total alchemical process is calculated by integrating, the free energy differences of all the steps. This is the essence of the thermodynamic integration (TI) technique.
The preparation of the system was similar to that of the previous sections. After the equilibrium step, a production MD step is executed for 10 steps of the gradual mutation (H2O to O2). Each step is indicated by a parameter λ. λ = 0 represents the initial state (the H2O bound state), and λ = 1 represents the end state (the O2 bound state). The transformation from H2O to O2 is achieved by mutating one H to O and the other H to a dummy atom. The mutation of an atom in MD simulation is through replacing the atom mass, non-bonded parameters, and bonded parameters. The bond length needs to change from O-H to O-O in the simulation, however, GROMACS LINCS algorithm treat the initial O-H bond as a constraint such that the bond length cannot be changed during the TI simulation. To avoid that issue, we performed the reverse process by transforming O2 to H2O. The free energy calculated for the reverse process is the negative of the original process.
One production MD would be produced for each lambda state with the corresponding lambda combinations. The parameters set the protocols as gradually changing the van-der-Waals interactions before the Coulomb interactions. This is necessary to ensure that the van-der-Waals interactions are already set to offset the Coulomb attractions between the opposite charges. Parameters for bonded interactions are changed gradually throughout all lambdas.
Finally, the free energy changes for an alchemical process were computed using the BAR (Bennett Acceptance Ratio) method (Bennett, 1976) as implemented in gmx bar utility of GROMACS. Simulations were performed for 4 system states (online or offline iron center configurations with bound or unbound DHCP substrate molecule) to investigate the effect of the substrate binding and Fe coordination geometry on the free energy Fe oxygen binding (bound water to oxygen substitution).
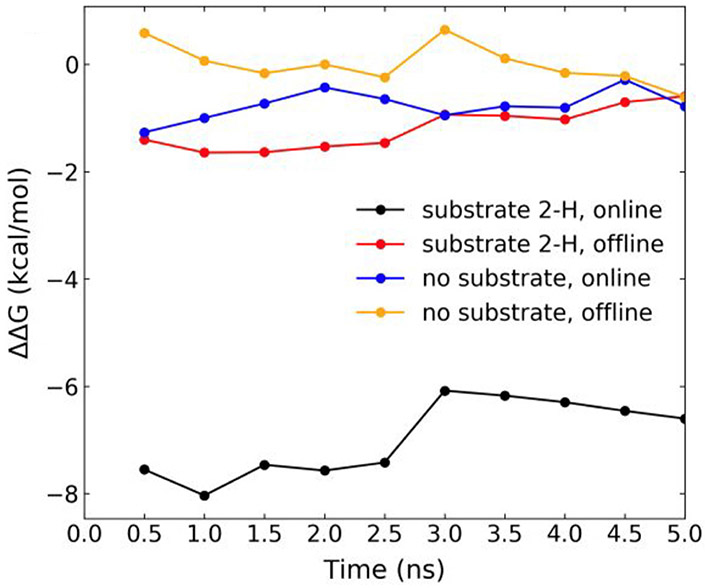
As can be seen from Fig. 6, the free energy of replacement of Fe-bound water to O2 is much more negative (by ~ 6 kcal/mol) in the online Fe center configuration with 2-H substrate bound than when the substrate is not bound or with the substrate bound but the system in the offline configuration. This result allows us to make a conclusion that reaction activating binding of O2 to iron occurs only when the substrate is present and Fe-2OG coordination configuration is in the online mode.
Fig. 6.
Free energy differences (ΔΔG) for H2O to O2 replacement at the Fe(II) center of AsqJ in two different 2OG binding configurations derived from MD simulations. Cumulative ΔΔG for four system configurations (see Scheme 3 for the illustration of the four system configurations) averaged over 0.5 ns MD simulation intervals by using the 2OG offline configuration without substrate as the reference state. Simulation for each lambda runs for 5 ns. The BAR method calculates free energy difference along the 5 ns trajectories. We plot the running average over 0.5 ns in the figure. For black trace, ΔΔG = ΔG4–ΔG1, for the red trace, ΔΔG = ΔG3–ΔG1, for the blue trace, ΔΔG = ΔG2–ΔG1, and for the orange trace, ΔΔG = ΔG1–ΔG1. See the definition of ΔGi (i = 1–4) in Scheme 4. The figure is adapted from Li, J., Liao, H.-J., Tang, Y., Huang, J.-L., Cha, L., Lin, T.-S., … Guo, Y. (2020). Epoxidation catalyzed by the nonheme iron(II)- and 2-oxo-glutarate-dependent oxygenase, AsqJ: Mechanistic elucidation of oxygen atom transfer by a ferryl intermediate. Journal of the American Chemical Society, 142(13), 6268–6284. https://doi.org/10.1021/jacs.0c00484 Copyright 2020 American Chemical Society.
5. Proposed reaction mechanism
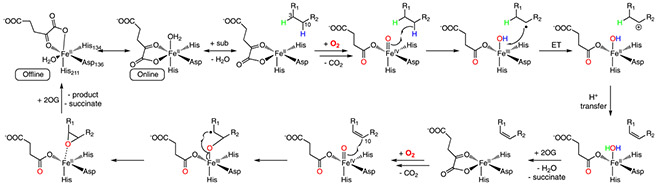
By using a multi-faceted approach including protein biochemistry, protein X-ray crystallography, substrate analog synthesis, pre-steady state kinetics, advanced spectroscopy, and molecular dynamics simulations, we have studied both desaturation and epoxidation reactions catalyzed by a bifunctional Fe/2OG enzymes, AsqJ. A proposed reaction mechanism based on all our studies is shown in Scheme 5 (Chang et al., 2016; Li et al., 2020; Liao et al., 2018; Tang et al., 2021).
Scheme 5.
Overall reaction mechanism of AsqJ-catalyzed desaturation and epoxidation derived based on our original studies (Chang et al., 2016; Li et al., 2020; Liao et al., 2018).
All the available AsqJ crystal structures show an offline binding configuration for 2OG at the Fe(II) center with and without substrate (Scheme 5). Such a binding configuration may lead to an oxyferryl intermediate exhibiting an offline Fe(IV)=O structure, which would be far away from the substrate to enable facile substrate oxidation. However, our MD simulations together with free energy calculations by using thermodynamic integration suggest that in solution the O2 binding to the Fe(II) center is energetically favorable only when the 2OG binds to the iron center in an online configuration in the presence of enzyme substrate (Scheme 4 and Fig. 6). It is most likely because the presence of substrate generates a hydrophobic local environment to facilitate O2 binding while disfavor H2O coordination. Thus the oxyferryl intermediate formed based on the online binding configuration of 2OG will position the Fe(IV)=O moiety close to the substrate to enable facile reaction. We further hypothesize that all the AsqJ crystal structures only capture the offline 2OG binding mode, while in solution the two binding modes (offline vs. online) may interconvert in the absence of substrate (Scheme 5). Once the substrate presents close to the iron center, only the 2OG online binding mode leads to facile O2 addition and activation at the iron center.
For desaturation, the oxyferryl intermediate most likely attacks the C10-H bond of cyclopeptin (1-H) to initiate the reaction. This conclusion is based on the observation that AsqJ reacts with both L-cyclopeptin (1-H) and D-cyclopeptin (D-1-H) to produce the correct enzyme product, dehydroxycyclopeptin (2-H), with a similar efficiency determined by pre-steady state kinetic studies. Although both C3-H and C10-H of 1-H face the iron center and presumably available for C-H activation, only C10-H of D-1-H is available for C-H activation. Followed by C-H activation, a C10 radical species for both 1-H and D-1-H could form. Again, based on the fact that D-1-H can be metabolized by AsqJ to product the correct enzyme product, the C10 radical species may likely to convert to a cationic intermediate via a facile electron transfer to the Fe(III)-OH species. Finally, a deprotonation at C3 of 1-H or D-1-H completes the olefin group formation.
For epoxidation, an oxyferryl intermediate is observed to act as the key reactive intermediate. The initial C-O bond formation during the oxygen atom transfer step is likely to occur at C10 position of 2-H based on crystal structures, MD simulations, and enzyme kinetics when using different para-substituted substrate analogs. This initial C10-O bond formation is most likely followed by the generation of a Fe(III)-alkoxide species with a carbon radical located at the C3 position of the substrate. The subsequent C3-O bond formation completes the epoxide ring formation. The release of the epoxide product, cyclopenin (3-H), from the enzyme will further trigger a non-enzymatic rearrangement to finally generate viridicatin (4-H).
Acknowledgments
This work was supported by the National Institutes of Health (R01-GM125924 to M.G. K., W.-c. C., and Y. G.) and by the National Science and Technology Council-Taiwan (grant number: 111-2113-M-002-014-MY3 to N.-L. C.).
References
- Abu-Omar MM, Loaiza A, & Hontzeas N (2005). Reaction mechanisms of mononuclear non-heme iron oxygenases. Chemical Reviews, 105(6), 2227–2252. 10.1021/cr040653o. [DOI] [PubMed] [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, … Adams PD (2012). Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallographica. Section D, Biological Crystallography, 68(4), 352–367. 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aik WS, Chowdhury R, Clifton IJ, Hopkinson RJ, Leissing T, McDonough MA, … Walport LJ (2015). Introduction to structural studies on 2-oxoglutarate-dependent oxygenases and related enzymes. In Hausinger RP, & Schofield CJ (Eds.). 2-Oxoglutarate-dependent oxygenases (pp. 59–94). The Royal Society of Chemistry. 10.1039/9781782621959-00059. [DOI] [Google Scholar]
- Babu CS, & Lim C (2006). Empirical force fields for biologically active divalent metal cations in water. The Journal of Physical Chemistry. A, 110(2), 691–699. 10.1021/jp054177x. [DOI] [PubMed] [Google Scholar]
- Bennett CH (1976). Efficient estimation of free energy differences from Monte Carlo data. Journal of Computational Physics, 22(2), 245–268. 10.1016/0021-9991(76)90078-4. [DOI] [Google Scholar]
- Blomberg MRA, Borowski T, Himo F, Liao R-Z, & Siegbahn PEM (2014). Quantum chemical studies of mechanisms for metalloenzymes. Chemical Reviews, 114(7), 3601–3658. 10.1021/cr400388t. [DOI] [PubMed] [Google Scholar]
- Bollinger JM Jr., Chang W, Matthews ML, Martinie RJ, Boal AK, & Krebs C (2015). Mechanisms of 2-oxoglutarate-dependent oxygenases: The hydroxylation paradigm and beyond. In Hausinger R, & Schofield C (Eds.). 2-Oxoglutarate-dependent oxygenases (pp. 95–122). The Royal Society of Chemistry. 10.1039/9781782621959-00095. [DOI] [Google Scholar]
- Bollinger JM Jr., & Krebs C (2006). Stalking intermediates in oxygen activation by iron enzymes: Motivation and method. Journal of Inorganic Biochemistry, 100(4), 586–605. 10.1016/j.jinorgbio.2006.01.022. [DOI] [PubMed] [Google Scholar]
- Bowman SEJ, Bridwell-Rabb J, & Drennan CL (2016). Metalloprotein crystallography: More than a structure. Accounts of Chemical Research, 49(4), 695–702. 10.1021/acs.accounts.5b00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräuer A, Beck P, Hintermann L, & Groll M (2016). Structure of the dioxygenase AsqJ: Mechanistic insights into a one-pot multistep quinolone antibiotic biosynthesis. Angewandte Chemie International Edition, 55(1), 422–426. 10.1002/anie.201507835. [DOI] [PubMed] [Google Scholar]
- Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, … Im W, … Karplus M (2009). CHARMM: The biomolecular simulation program. Journal of Computational Chemistry, 30(10), 1545–1614. 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buongiorno D, & Straganz GD (2013). Structure and function of atypically coordinated enzymatic mononuclear non-heme-Fe(II) centers. Coordination Chemistry Reviews, 257(2), 541–563. 10.1016/j.ccr.2012.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammack R, & Cooper CE (1993). [12] Electron paramagnetic resonance spectroscopy of iron complexes and iron-containing proteins. Methods in Enzymology, 227, 353–384. 10.1016/0076-6879(93)27014-8. [DOI] [PubMed] [Google Scholar]
- Case DA, Aktulga HM, Belfon K, Cerutti DS, Cisneros GA, Cruzeiro VWD, … Merz KM (2023). AmberTools. Journal of Chemical Information and Modeling, 63(20), 6183–6191. 10.1021/acs.jcim.3c01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case DA, Aktulga HM, Belfon K, Ben-Shalom IY, Berryman JT, Brozell SR, … Kollman PA (2023). Amber 2023 (2023). San Francisco: University of California. [Google Scholar]
- Cha L, Paris JC, Zanella B, Spletzer M, Yao A, Guo Y, & Chang W (2023). Mechanistic studies of aziridine formation catalyzed by mononuclear non-heme iron enzymes. Journal of the American Chemical Society, 145(11), 6240–6246. 10.1021/jacs.2c12664. [DOI] [PubMed] [Google Scholar]
- Chang W, Li J, Lee JL, Cronican AA, & Guo Y (2016). Mechanistic investigation of a non-heme iron enzyme catalyzed epoxidation in (−)-4′-methoxycyclopenin biosynthesis. Journal of the American Chemical Society, 138(33), 10390–10393. 10.1021/jacs.6b05400. [DOI] [PubMed] [Google Scholar]
- Chen T-Y, Zheng Z, Zhang X, Chen J, Cha L, Tang Y, … Chang W (2022). Deciphering the reaction pathway of mononuclear iron enzyme-catalyzed N≡C triple bond formation in isocyanide lipopeptide and polyketide biosynthesis. ACS Catalysis, 12(4), 2270–2279. 10.1021/acscatal.1c04869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costas M, Mehn MP, Jensen MP, & Que L (2004). Dioxygen activation at mononuclear nonheme iron active sites: Enzymes, models, and intermediates. Chemical Reviews, 104(2), 939–986. 10.1021/cr020628n. [DOI] [PubMed] [Google Scholar]
- Dunham NP, Chang W, Mitchell AJ, Martinie RJ, Zhang B, Bergman JA, … Bollinger JM (2018). Two distinct mechanisms for C─C desaturation by iron(II)- and 2-(oxo)glutarate-dependent oxygenases: Importance of α-heteroatom assistance. Journal of the American Chemical Society, 140(23), 7116–7126. 10.1021/jacs.8b01933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, & Cowtan K (2010). Features and development of coot. Acta Crystallographica. Section D, Biological Crystallography, 66(4), 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, … Fox DJ (2016). Gaussian 16. Revision C.01. Gaussian, Inc. [Google Scholar]
- Gao J, Ma S, Major DT, Nam K, Pu J, & Truhlar DG (2006). Mechanisms and free energies of enzymatic reactions. Chemical Reviews, 106(8), 3188–3209. 10.1021/cr050293k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Naowarojna N, Cheng R, Liu X, & Liu P (2018). Recent examples of α-ketoglutarate-dependent mononuclear non-haem iron enzymes in natural product biosyntheses. Natural Product Reports, 35(8), 792–837. 10.1039/C7NP00067G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Chang W, Li J, & Davidson M (2021). Non-heme mono-iron enzymes: Co-substrate-dependent dioxygen activation. In Constable EC, Parkin G, & Que LBT-CCCIII (Eds.). Comprehensive coordination chemistry III (pp. 269–300). Elsevier. 10.1016/B978-0-12-409547-2.14820-6. [DOI] [Google Scholar]
- Hausinger RP (2015). Biochemical diversity of 2-oxoglutarate-dependent oxygenases. In Hausinger RP, & Schofield CJ (Eds.). 2-Oxoglutarate-dependent oxygenases (pp. 1–58). The Royal Society of Chemistry. 10.1039/9781782621959-00001. [DOI] [Google Scholar]
- Hess B, Kutzner C, van der Spoel D, & Lindahl E (2008). GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. Journal of Chemical Theory and Computation, 4(3), 435–447. 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Ishikawa N, Tanaka H, Koyama F, Noguchi H, Wang CCC, Hotta K, & Watanabe K (2014). Non-heme dioxygenase catalyzes atypical oxidations of 6,7-bicyclic systems to form the 6,6-quinolone core of viridicatin-type fungal alkaloids. Angewandte Chemie International Edition, 53(47), 12880–12884. 10.1002/anie.201407920. [DOI] [PubMed] [Google Scholar]
- Johnson KA, Simpson ZB, & Blom T (2009). Global kinetic explorer: A new computer program for dynamic simulation and fitting of kinetic data. Analytical Biochemistry, 387(1), 20–29. 10.1016/j.ab.2008.12.024. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, & Klein ML (1983). Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics, 79(2), 926–935. 10.1063/1.445869. [DOI] [Google Scholar]
- Kal S, & Que L (2017). Dioxygen activation by nonheme iron enzymes with the 2-his-1-carboxylate facial triad that generate high-valent oxoiron oxidants. Journal of Biological Inorganic Chemistry, 22(2–3), 339–365. 10.1007/s00775-016-1431-2. [DOI] [PubMed] [Google Scholar]
- Karplus M, & McCammon JA (2002). Molecular dynamics simulations of biomolecules. Nature Structural Biology, 9(9), 646–652. 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- Kovaleva EG, & Lipscomb JD (2008). Versatility of biological non-heme Fe(II) centers in oxygen activation reactions. Nature Chemical Biology, 4(3), 186–193. 10.1038/nchembio.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs C, & Bollinger JM (2009). Freeze-quench 57Fe-Mössbauer spectroscopy: Trapping reactive intermediates. Photosynthesis Research, 102(2), 295–304. 10.1007/s11120-009-9406-6. [DOI] [PubMed] [Google Scholar]
- Krebs C, Galonić Fujimori D, Walsh CT, & Bollinger JM (2007). Non-heme Fe (IV)–oxo intermediates. Accounts of Chemical Research, 40(7), 484–492. 10.1021/ar700066p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liao H-J, Tang Y, Huang J-L, Cha L, Lin T-S, … Guo Y (2020). Epoxidation catalyzed by the nonheme iron(II)- and 2-oxoglutarate-dependent oxygenase, AsqJ: Mechanistic elucidation of oxygen atom transfer by a ferryl intermediate. Journal of the American Chemical Society, 142(13), 6268–6284. 10.1021/jacs.0c00484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H-J, Li J, Huang J-L, Davidson M, Kurnikov I, Lin T-S, … Chang W (2018). Insights into the desaturation of cyclopeptin and its C3 epimer catalyzed by a non-heme iron enzyme: Structural characterization and mechanism elucidation. Angewandte Chemie International Edition, 57(7), 1831–1835. 10.1002/anie.201710567. [DOI] [PubMed] [Google Scholar]
- Liao H-J (2019). Structural insight into the mechanism of AsqJ-catalyzed consecutive oxidation reactions. National Taiwan University. http://tdr.lib.ntu.edu.tw/jspui/handle/123456789/73532. [Google Scholar]
- Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, & Shaw DE (2010). Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins: Structure, Function, and Bioinformatics, 78(8), 1950–1958. 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews ML, Krest CM, Barr EW, Vaillancourt FH, Walsh CT, Green MT, … Bollinger JM (2009). Substrate-triggered formation and remarkable stability of the C─H bond-cleaving chloroferryl intermediate in the aliphatic halogenase, SyrB2. Biochemistry, 48(20), 4331–4343. 10.1021/bi900109z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews ML, Neumann CS, Miles L. a, Grove TL, Booker SJ, Krebs C, … Bollinger JM (2009). Substrate positioning controls the partition between halogenation and hydroxylation in the aliphatic halogenase, SyrB2. Proceedings of the National Academy of Sciences, 106(42), 17723–17728. 10.1073/pnas.0909649106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, & Read RJ (2007). Phaser crystallographic software. Journal of Applied Crystallography, 40(4), 658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münck E, Ksurerus K, & Hendrich MP (1993). [17] Combining Mössbauer spectroscopy with integer spin electron paramagnetic resonance. Methods in Enzymology, 227, 463–479. 10.1016/0076-6879(93)27019-D. [DOI] [PubMed] [Google Scholar]
- Neria E, Fischer S, & Karplus M (1996). Simulation of activation free energies in molecular systems. The Journal of Chemical Physics, 105(5), 1902–1921. 10.1063/1.472061. [DOI] [Google Scholar]
- Olsson MHM, Søndergaard CR, Rostkowski M, & Jensen JH (2011). PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. Journal of Chemical Theory and Computation, 7(2), 525–537. 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, & Minor W (1997). [20] Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology, 276, 307–326. 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pang X, Han K, & Cui Q (2013). A simple but effective modeling strategy for structural properties of non-heme Fe(II) sites in proteins: Test of force field models and application to proteins in the AlkB family. Journal of Computational Chemistry, 34(19), 1620–1635. 10.1002/jcc.23305. [DOI] [PubMed] [Google Scholar]
- Petasis DT, & Hendrich MP (2015). Quantitative interpretation of multifrequency multimode EPR spectra of metal containing proteins, enzymes, and biomimetic complexes. Methods in Enzymology, 563, 171–208. 10.1016/bs.mie.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JC, Barr EW, Glass TE, Krebs C, & Bollinger JMJ (2003). Evidence for hydrogen abstraction from C1 of taurine by the high-spin Fe(IV) intermediate detected during oxygen activation by taurine:α-ketoglutarate dioxygenase (TauD). Journal of the American Chemical Society, 125, 13008–13009. [DOI] [PubMed] [Google Scholar]
- Price JC, Barr EW, Hoffart LM, Krebs C, & Bollinger JM (2005). Kinetic dissection of the catalytic mechanism of taurine:α-ketoglutarate dioxygenase (TauD) from Escherichia coli. Biochemistry, 44(22), 8138–8147. 10.1021/bi050227c. [DOI] [PubMed] [Google Scholar]
- Price JC, Barr EW, Tirupati B, Bollinger JM, & Krebs C (2003). The first direct characterization of a high-valent iron intermediate in the reaction of an α-ketoglutarate-dependent dioxygenase: A high-spin Fe(IV) complex in taurine/α-ketoglutarate dioxygenase (TauD) from Escherichia coli. Biochemistry, 42(24), 7497–7508. 10.1021/bi030011f. [DOI] [PubMed] [Google Scholar]
- Proos Vedin N, & Lundberg M (2016). Protein effects in non-heme iron enzyme catalysis: Insights from multiscale models. JBIC Journal of Biological Inorganic Chemistry, 21(5–6), 645–657. 10.1007/s00775-016-1374-7. [DOI] [PubMed] [Google Scholar]
- Quesne MG, Borowski T, & de Visser SP (2016). Quantum mechanics/molecular mechanics modeling of enzymatic processes: Caveats and breakthroughs. Chemistry – A European Journal, 22(8), 2562–2581. 10.1002/chem.201503802. [DOI] [PubMed] [Google Scholar]
- Roach PL, Clifton IJ, Fülöp V, Harlos K, Barton GJ, Hajdu J, … Baldwin JE (1995). Crystal structure of isopenicillin N synthase is the first from a new structural family of enzymes. Nature, 375(6533), 700–704. 10.1038/375700a0. [DOI] [PubMed] [Google Scholar]
- Roach PL, Clifton IJ, Hensgens CM, Shibata N, Schofield CJ, Hajdu J, & Baldwin JE (1997). Structure of isopenicillin N synthase complexed with substrate and the mechanism of penicillin formation. Nature, 387(6635), 827–830. 10.1038/42990. [DOI] [PubMed] [Google Scholar]
- Schrödinger L (2020). The {PyMOL} molecular graphics system, Version~2.0. http://www.pymol.org/pymol. [Google Scholar]
- Senn HM, & Thiel W (2009). QM/MM methods for biomolecular systems. Angewandte Chemie International Edition, 48(7), 1198–1229. 10.1002/anie.200802019. [DOI] [PubMed] [Google Scholar]
- Solomon EI, Brunold TC, Davis MI, Kemsley JN, Lee S-K, Lehnert N, … Zhou J (2000). Geometric and electronic structure/function correlations in non-heme iron enzymes. Chemical Reviews, 100(1), 235–350. 10.1021/cr9900275. [DOI] [PubMed] [Google Scholar]
- Solomon EI, DeWeese DE, & Babicz JT (2021). Mechanisms of O2 activation by mononuclear non-heme iron enzymes. Biochemistry, 60(46), 3497–3506. 10.1021/acs.biochem.1c00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Naowarojna N, Cheng R, Lopez J, & Liu P (2019). Non-heme iron enzyme-catalyzed complex transformations. Advances in Protein Chemistry and Structural Biology, 1–61. 10.1016/bs.apcsb.2019.06.002. [DOI] [PubMed] [Google Scholar]
- Sousa da Silva AW, & Vranken WF (2012). ACPYPE - AnteChamber PYthon Parser interfacE. BMC Research Notes, 5(1), 367. 10.1186/1756-0500-5-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srnec M, Wong SD, Matthews ML, Krebs C, Bollinger JM, & Solomon EI (2016). Electronic structure of the ferryl intermediate in the α-ketoglutarate dependent non-heme iron halogenase SyrB2: Contributions to H atom abstraction reactivity. Journal of the American Chemical Society, 138(15), 5110–5122. 10.1021/jacs.6b01151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Tang Y, Kurnikov IV, Liao H-J, Chan N-L, Kurnikova MG, … Chang W (2021). Harnessing the substrate promiscuity of dioxygenase AsqJ and developing efficient chemoenzymatic synthesis for quinolones. ACS Catalysis, 11(12), 7186–7192. 10.1021/acscatal.1c01150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valegård K, Terwisscha van Scheltinga AC, Lloyd MD, Hara T, Ramaswamy S, Perrakis A, … Andersson I (1998). Structure of a cephalosporin synthase. Nature, 394, 805–809. [DOI] [PubMed] [Google Scholar]
- Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, … Mackerell AD (2010). CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. Journal of Computational Chemistry, 31(4), 671–690. 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waheed SO, Ramanan R, Chaturvedi SS, Lehnert N, Schofield CJ, Christov CZ, & Karabencheva-Christova TG (2020). Role of structural dynamics in selectivity and mechanism of non-heme Fe(II) and 2-oxoglutarate-dependent oxygenases involved in DNA repair. ACS Central Science, 6(5), 795–814. 10.1021/acscentsci.0c00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang W, Kollman PA, & Case DA (2006). Automatic atom type and bond type perception in molecular mechanical calculations. Journal of Molecular Graphics and Modelling, 25(2), 247–260. 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Wang J, Wolf RM, Caldwell JW, Kollman PA, & Case DA (2004). Development and testing of a general amber force field. Journal of Computational Chemistry, 25(9), 1157–1174. 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, … Wilson KS (2011). Overview of the CCP 4 suite and current developments. Acta Crystallographica. Section D, Biological Crystallography, 67(4), 235–242. 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S, & Neese F (2009). Quantum chemical studies of C─H activation reactions by high-valent nonheme iron centers. Current Opinion in Chemical Biology, 13(1), 89–98. 10.1016/j.cbpa.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Ren J, Stammers DK, Baldwin JE, Harlos K, & Schofield CJ (2000). Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase. Nature Structural Biology, 7(2), 127–133. 10.1038/72398. [DOI] [PubMed] [Google Scholar]
- Zhang J, Wu P, Zhang X, & Wang B (2023). Coordination dynamics of iron is a key player in the catalysis of non-heme enzymes. Chembiochem: A European Journal of Chemical Biology, 24(14), 10.1002/cbic.202300119. [DOI] [PubMed] [Google Scholar]