Abstract
Objective
This study aims to investigate the association between antimicrobial resistance genes and virulence factors in ST11 and non-ST11 types of CR-KP in bloodstream infections in the intensive care unit, providing a theoretical basis for infection control and clinical diagnosis and treatment.
Methods
From January 2021 to June 2023, samples of Klebsiella pneumoniae from bloodstream infections were collected at our hospital, focusing on those resistant to carbapenems. The resistance genes, housekeeping genes, and virulence genes were identified through PCR and analyzed using the GrapeTree software to perform MLST-based minimum spanning tree typing.
Results
Among the 85 CR-KP cases, 61.18% were of the ST11 type, predominantly of the KL64 capsular type; non-ST11 types were mainly ST15, accounting for 25.88%, predominantly of the KL5 capsular type. The carriage rates of virulence genes such as rmpA2, entB, silS, kpn, iucA, peg-344, and terB were significantly higher in the ST11 group than in the non-ST11 group. The primary carbapenemase identified was class A enzyme blaKPC-2, with a higher carriage rate in the ST11 group. Drug susceptibility tests showed that the resistance rates for cefepime, ertapenem, nitrofurantoin, amikacin, and gentamicin were also higher in the ST11 group, consistent with the resistance genotype findings.
Conclusion
The study reveals that ST11 type CR-KP in intensive care unit bloodstream infections exhibits stronger resistance and higher virulence compared to non-ST11 types, posing significant challenges to clinical treatment. Thus, strict control over the use of carbapenem antibiotics is essential to prevent the spread of resistant plasmids.
Keywords: intensive care unit, bloodstream infections, ST11 CR-KP, Non-ST11 CR-KP, resistance genes, virulence genes
Introduction
Klebsiella pneumoniae (KP), a Gram-negative Enterobacteriaceae bacterium, colonizes the upper respiratory tract and intestines and is a significant pathogen in both hospital-acquired and community-acquired infections. It primarily affects individuals with compromised immune systems. According to Chen et al1 bloodstream infections caused by Klebsiella pneumoniae result in a threefold increase in the rate of septic shock compared to other infections, presenting significant challenges to clinical treatment.2 Carbapenems serve as the last line of defense against multidrug-resistant Gram-negative bacterial infections.3,4 However, the widespread transmission of carbapenem-resistant plasmids has led to an increasing detection rate of Carbapenem-resistant Klebsiella pneumoniae (CR-KP) in bloodstream infections.5,6 The high mortality rate associated with these infections poses a serious threat to public health safety.7,8 Qi et al9 conducted a screening of 95 CR-KP isolates from 5 provinces and 9 cities in China, collected from 13 hospitals, and identified the predominant clonal circulation in China as the ST11 type. The PFGE pattern similarity among ST11 CR-KP isolates exceeded 80%. Similarly, Liu et al10 observed that ST11 KPC-2-producing CR-KP was the most prevalent CR-KP in the Shanghai area. The majority of these isolates contained IncFII plasmids, contributing to the widespread dissemination of KPC-2 resistance plasmids. This suggests that current prevention and control strategies for CR-KP hospital infections need to be adjusted. Moreover, Elmanakhly et al11 discovered that Klebsiella pneumoniae not only developed resistance but also underwent adaptive changes in virulence genes as it acquired resistance, which differs from previous studies indicating that highly resistant Klebsiella pneumoniae were typically susceptible to commonly used antibiotics.12 Recently, Pu et al13 identified CR-KP strains of the ST11-KL64 type containing the iucABCD, iutA, and rmpA/rmpA2 genes, with their iron carrier production exceeding that of classical Klebsiella pneumoniae (cKp). An increasing number of studies are demonstrating the evolution of Klebsiella pneumoniae towards high virulence and resistance, posing greater challenges for clinical treatment.14–16 Infection by these highly virulent and resistant strains further complicates clinical management. Thus, to better serve clinical needs, our research group gathered carbapenem-resistant Klebsiella pneumoniae strains from bloodstream infections in the intensive care unit (ICU) of a hospital in Ningbo between January 2021 and June 2023, analyzing their resistance and virulence genes to provide a theoretical basis for the diagnosis and treatment of bloodstream infections caused by CR-KP.
Materials and Methods
Source of Bacterial Strains
We collected 85 strains of carbapenem-resistant Klebsiella pneumoniae from bloodstream infections in our hospital’s intensive care unit from January 2021 to June 2023. The selection method involved identifying the strains as Klebsiella pneumoniae using the EXS3600 microbial mass spectrometry analyzer. Subsequently, the collected strains were subjected to in vitro antimicrobial susceptibility testing using the VITEK-2 Compact automated microbiology system. The criteria for determining antimicrobial resistance were based on the Clinical and Laboratory Standards Institute (CLSI) M100-S33 standards, with a meropenem MIC value ≥4 mg/mL indicating resistance. Strains from the same patient were screened, and duplicates were excluded to identify the target experimental strains.
Instruments and Reagents
Microbial mass spectrometry analyzer EXS3600, purchased from Zhongyuan Huiji Company; VITEK-2 Compact automated microbiology system, acquired from bioMérieux, France; PCR amplification system, bought from Bio-Rad, USA; gel imaging system, also from Bio-Rad, USA; Gram-negative bacillus antimicrobial susceptibility test card N13, obtained from bioMérieux, France. Quality control strains Escherichia coli (ATCC25922) and Pseudomonas aeruginosa (ATCC27853) are preserved in our hospital’s biological sample strain repository.
Methods
Bacterial DNA Extraction
Retrieve the frozen strain from the −80°C freezer, inoculate it onto Columbia blood agar plates using a sterile inoculating loop, and incubate at 37°C in a CO2 incubator for 16 h. Subsequently, select a single colony and transfer it into a sterile EP tube containing 500μL of sterile distilled water. The samples were vortexed, then heated in a 100°C metal bath for 10 minutes. After centrifuging at high speed for 5 minutes, the supernatant was transferred to a new sterile Eppendorf tube and stored at −20°C for future use.
Serotyping and Virulence Gene Detection of Strains
PCR amplification was used to detect common capsular genes of Klebsiella pneumoniae, including KL1, KL2, KL5, KL20, KL54, KL57, and KL64; and 21 virulence genes such as rmpA, magA, aerobactin, entB, ybtS, kfu, ycfM, mrkD, fimH, kpn, allS, ureA, uge, wabG, wcaG, rmpA2, peg344, iroB, iucA, terB, silS. The primer sequences were designed and synthesized by Sangon Biotech, Shanghai. The PCR reaction system comprised 3μL of DNA template, 1μL each of the upstream and downstream primers, 10μL of Taq polymerase, and 25μL of double-distilled water (ddH2O). The reaction conditions included an initial denaturation at 95°C for 5 minutes, followed by denaturation at 95°C for 30 seconds, annealing at 55°C for 40 seconds, elongation at 72°C for 30 seconds; this cycle was repeated for a total of 35 cycles with a final elongation step at 72°C for an additional duration of 5 minutes.
Resistance Gene Detection
Using PCR amplification, common carbapenem resistance genes were detected, including Class A blaKPC enzymes, Class B metallo-beta-lactamases like blaNDM, and Class D blaOXA enzymes. The primer sequences were designed and synthesized by Sangon Biotech, Shanghai. The PCR Reaction System: 12.5μL of Tag enzyme, 1μL each of upstream and downstream primers, and 1μL of DNA template; deionized water is added to achieve a total volume of 25μL. Reaction conditions include an initial denaturation at 94°C for 5 minutes, followed by denaturation at 94°C for 45 seconds, annealing at 56°C for 45 seconds, and extension at 72°C for one minute. This cycle is repeated for a total of 30 cycles with a final extension step at 72°C lasting for ten minutes. All sequences of PCR primers can be found in the supplementary materials accompanying this article (Supplementary Table 1).
PCR Product Electrophoresis
Electrophoresis was conducted at 110 V for 20 minutes. Gel images were captured using a gel imaging system. The target bands were sent to Sangon Biotech, Shanghai, for sequencing verification.
MLST Homology Analysis
Using PCR amplification, seven housekeeping genes of Klebsiella pneumoniae (gapA, infB, mdh, pgi, phoE, tonB, ropB) were amplified for multi-locus sequence typing (MLST: http://pubmlst.org/). The PCR products were sent to Sangon Biotech, Shanghai, for sequencing and the sequences were submitted to the NCBI website in order to acquire alleles. Following the acquisition of the allele and its subsequent classification as ST type, the data were analyzed using the GrapeTree software (https://doi.org/10.1101/gr.232397.117) to perform MLST-based minimum spanning tree typing.
Statistical Analysis
Data were processed and analyzed using Excel and SPSS version 21.0 software. Count data were represented as number (n) or percentage (%), and quantitative data were analyzed using the chi-square (χ2) test. Where conditions for the χ2 test were not met, Fisher’s exact test was used. A P-value of less than 0.05 was considered statistically significant.
Results
Basic Clinical Data of the Strains
From January 2021 to June 2023, 85 strains of carbapenem-resistant Klebsiella pneumoniae (CR-KP) were collected from bloodstream infections in the intensive care unit of our hospital. Among the identified strains, the predominant capsule type of ST11 CR-KP was KL64 (27/52, 51.92%), followed by KL2 (6/52, 11.54%), KL20 (4/52, 7.69%) and KL54 (2/52, 3.85%), with 13 strains remaining unclassified; conversely, the primary capsule type of non-ST11 CR-KP was KL5 (19/33, 57.58%), succeeded by KL64 (5/33, 15.15%), KL20 (4/33, 12.12%) and KL1 (1/33, 3.03%) with an additional four strains unclassified. Notably, significant statistical differences were observed between the KL64 type of ST11 CR-KP and the KL5 type of non-ST11 CR-KP (P≤0.001). However, there was no statistical difference in the gender and age of the patients between the two groups (see Table 1).
Table 1.
Basic Clinical Data of the Strains
| Categories | ST11 CR-KP | Non-ST11 CR-KP | χ2 value | P value |
|---|---|---|---|---|
| (n=52) | (n=33) | |||
| Gender | ||||
| Male | 40 | 24 | 0.191 | 0.662 |
| Female | 12 | 9 | 0.191 | 0.662 |
| Age | ||||
| <age 60 | 14 | 8 | 0.076 | 0.783 |
| ≥age 60 | 38 | 25 | 2.018 | 0.155 |
| Capsule type | ||||
| KL1 | 0 | 1 | 1.564 | 0.393 |
| KL2 | 6 | 0 | 4.097 | 0.077 |
| KL5 | 0 | 19 | 38.558 | <0.001 |
| KL20 | 4 | 4 | 0.465 | 0.705 |
| KL54 | 2 | 0 | 1.300 | 0.519 |
| KL57 | 0 | 0 | - | 1.000 |
| KL64 | 27 | 5 | 11.629 | 0.001 |
| Unknown capsule | 13 | 4 | 2.093 | 0.148 |
MLST Typing Results of Carbapenem-Resistant Klebsiella Pneumoniae
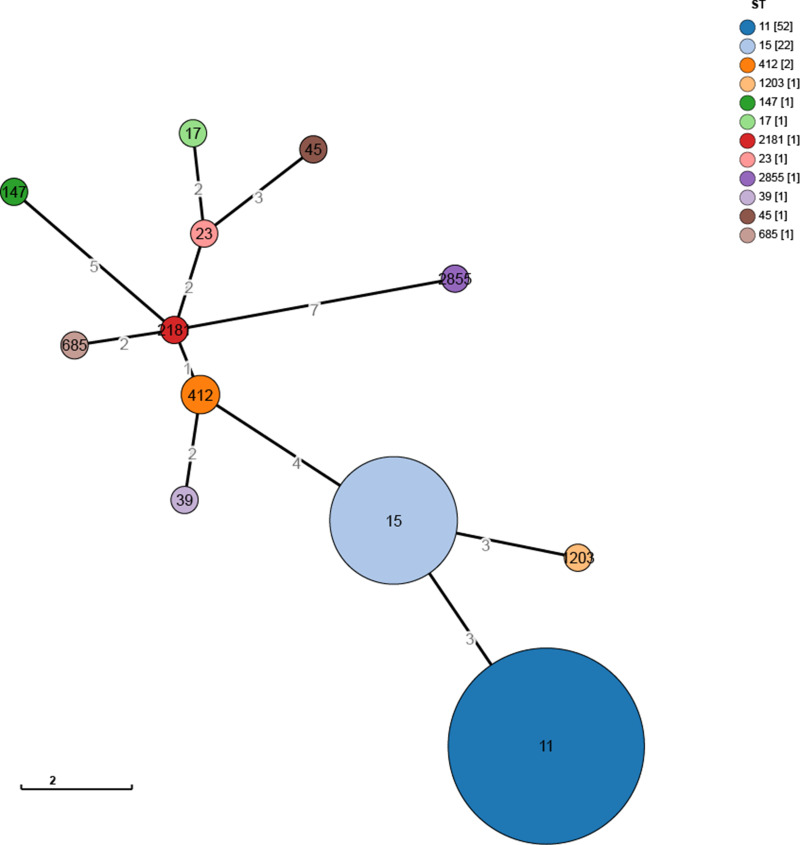
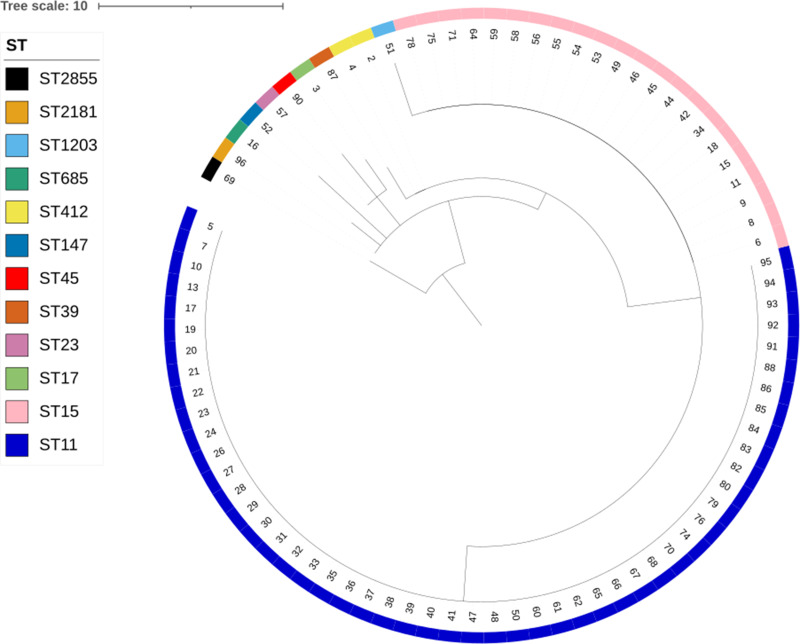
The results of the MLST-based minimum spanning tree showed that the predominant sequence type among CR-KP strains was ST11 (52 strains, accounting for 61.18%), followed by ST15 (22 strains, accounting for 25.88%), ST412 (2 strains, accounting for 2.35%), and one strain each of ST1203, ST147, ST17, ST2181, ST23, ST2855, ST39, ST45, and ST685, each accounting for 1.18% (see Figure 1). The hierarchical clustering tree from the MLST analysis indicated the following clusters: cluster1 (sample 63, ST2855, showing the greatest difference from other samples), cluster2 (samples 2, 13, 47, 52, 79, 85, each belonging to different ST types), cluster3 (samples 3, 1, 77), cluster4 (22 samples of ST type 15), and cluster5 (52 samples of ST type 11, see Figure 2).
Figure 1.
Minimum Spanning Tree of 85 CR-KP Strains. Constructed using GrapeTree software based on the MSTreeV2 algorithm.
Figure 2.
Hierarchical Clustering Tree of 85 CR-KP Strains. The tree file constructed with GrapeTree software was visualized using iTOL software (https://itol.embl.de/). Annotations on the outermost layer indicate the sequence type (ST) of each sample.
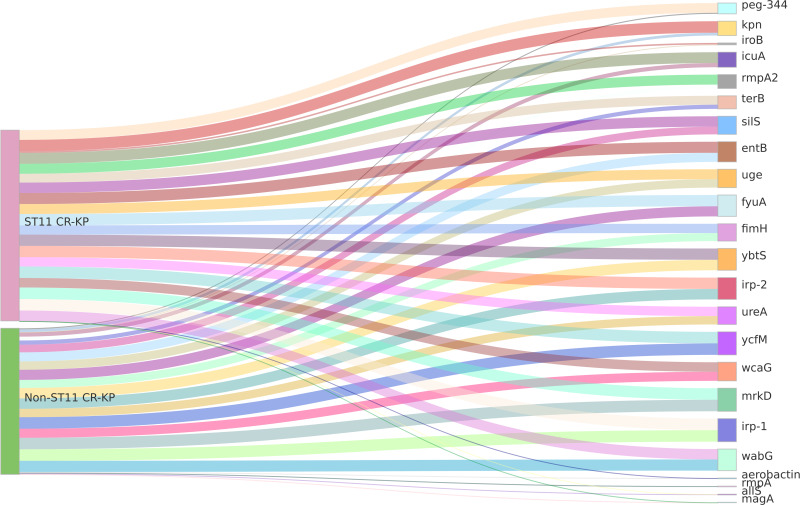
Detection Results of Virulence Genes in Carbapenem-Resistant Klebsiella Pneumoniae
Among the 23 virulence genes screened, the iron transporter-related gene irp-1 and the fimbrial attachment and adhesion-related genes ycfM and mrkD were identified in 100% of both ST11 type CR-KP and non-ST11 type CR-KP strains. Notably, all ST11 type CR-KP strains harbored the fimbrial attachment and adhesion-related gene kpn, with seven virulence genes (entB, ybtS, irp-2, fyuA, silS, wabG, iucA) exhibiting a carrying rate exceeding 90%. In contrast, the prevalence of virulence genes in non-ST11 type CR-KP was comparatively low; only three genes (ybtS, irp-2, wabG) demonstrated a carrying rate above 90%. And CR-KP strains of the ST11 type showed higher carriage rates of the following genes compared to non-ST11 types: the capsule synthesis-promoting gene rmpA2, iron-carrier related gene entB, silver ion resistance gene silS; fimbriae fixation and adhesion-related gene kpn, and pPLVPK-like virulence plasmid related genes iucA, peg-344, terB. The differences were statistically significant (P<0.05) (see Table 2). The visualization of these results can be seen in Figure 3.
Table 2.
Carrying Status of Carbapenem-Resistant Klebsiella Pneumoniae Virulence Genes
| Virulence genes | ST11 CR-KP | Non-ST11 CR-KP | χ2 value |
P value |
||
|---|---|---|---|---|---|---|
| (n=52) | (n=33) | |||||
| Strains | Percentage (%) |
Strains | Percentage (%) |
|||
| (n) | (n) | |||||
| rmpA | 0 | 0.00 | 2 | 6.06 | 3.228 | 0.148 |
| magA | 0 | 0.00 | 1 | 3.03 | 1.595 | 0.388 |
| aerobactin | 0 | 0.00 | 2 | 6.06 | 3.228 | 0.148 |
| entB | 50 | 96.15 | 26 | 78.79 | 6.431 | 0.025 |
| ybtS | 51 | 98.08 | 30 | 90.91 | 2.313 | 0.294 |
| irp-1 | 52 | 100.00 | 33 | 100.00 | - | 1.000 |
| irp-2 | 51 | 98.08 | 30 | 90.91 | 2.313 | 0.294 |
| fyuA | 51 | 98.08 | 29 | 87.88 | 3.792 | 0.072 |
| silS | 47 | 90.38 | 22 | 66.67 | 7.432 | 0.006 |
| ycfM | 52 | 100.00 | 33 | 100.00 | - | 1.000 |
| mrkD | 52 | 100.00 | 33 | 100.00 | - | 1.000 |
| fimH | 42 | 80.77 | 23 | 69.70 | 1.376 | 0.241 |
| kpn | 52 | 100.00 | 8 | 24.24 | 54.600 | <0.001 |
| allS | 0 | 0.00 | 2 | 6.06 | 3.228 | 0.148 |
| ureA | 42 | 80.77 | 24 | 72.73 | 0.392 | 0.532 |
| uge | 45 | 86.54 | 24 | 72.73 | 1.798 | 0.180 |
| wabG | 47 | 90.38 | 32 | 96.97 | 1.334 | 0.398 |
| wcaG | 43 | 82.69 | 26 | 78.79 | 0.201 | 0.694 |
| rmpA2 | 45 | 86.54 | 11 | 33.33 | 25.425 | <0.001 |
| iroB | 7 | 13.46 | 1 | 3.03 | 2.577 | 0.143 |
| iucA | 49 | 94.23 | 12 | 36.36 | 33.363 | <0.001 |
| peg-344 | 44 | 84.62 | 3 | 9.09 | 46.573 | <0.001 |
| terB | 40 | 76.92 | 12 | 36.36 | 13.983 | <0.001 |
Figure 3.
Sankey Diagram of Virulence Genes in CR-KP. A Sankey diagram, also known as a Sankey energy flow chart, is used here. The left nodes represent ST11 and non-ST11 types of CR-KP, while the right nodes represent 23 common virulence genes. The diagram shows the flow from CR-KP types on the left to the virulence genes on the right, the width of the branches correlates with the number of virulence genes.
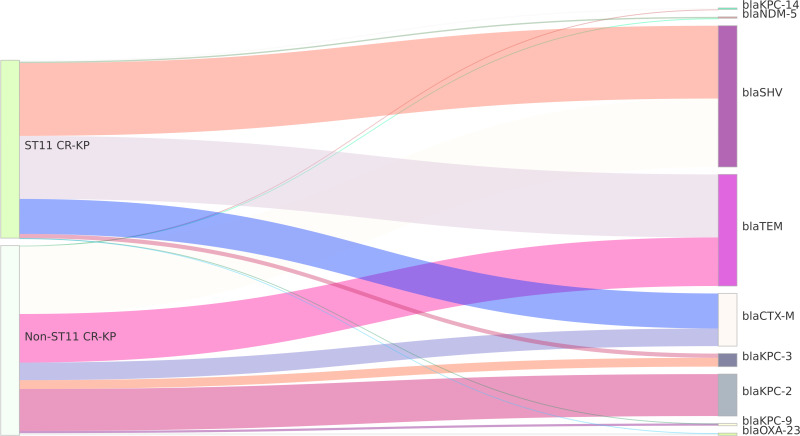
Detection Results of Resistance Genes in Carbapenem-Resistant Klebsiella Pneumoniae
The detection of carbapenemase resistance genes in CR-KP strains showed that the A-class enzyme blaKPC-2 subtype was predominantly carried, with a higher carriage rate in ST11 type CR-KP compared to non-ST11 type CR-KP, a statistically significant difference (P<0.05). Specifically, in the ST11 type CR-KP, one strain carrying the B-class metallo-beta-lactamase blaNDM-5 subtype was found, with no strains carrying blaVIM or blaIMP enzymes detected. In non-ST11 type CR-KP, one strain carrying the D-class enzyme blaOXA-23 subtype was identified. Four ST11 type and eight non-ST11 type CR-KP strains remained untyped. The detection of β-lactamase resistance genes revealed that the carriage rates of blaCTX-M and blaTEM were higher in ST11 type CR-KP compared to non-ST11 type CR-KP, with statistically significant differences (P<0.05) (see Table 3). The visualization of these results can be seen in Figure 4.
Table 3.
Carrying Status of Carbapenem-Resistant Klebsiella Pneumoniae Drug Resistance Genes
| Drug resistance genes |
ST11 CR-KP | Non -ST11 CR-KP | χ2 value |
P value |
||
|---|---|---|---|---|---|---|
| (n=52) | (n=33) | |||||
| Strains (n) |
Percentage (%) | Strains (n) |
Percentage (%) | |||
| Carbapenemases | ||||||
| blaKPC-2 | 43 | 82.69 | 19 | 57.58 | 6.453 | 0.011 |
| blaKPC-3 | 3 | 5.77 | 4 | 12.12 | 1.078 | 0.423 |
| blaKPC-9 | 0 | 0.00 | 1 | 3.03 | 1.595 | 0.388 |
| blaKPC-14 | 1 | 1.92 | 0 | 0.00 | 0.642 | 1.000 |
| blaVIM | 0 | 0.00 | 0 | 0.00 | - | - |
| blaIMP | 0 | 0.00 | 0 | 0.00 | - | - |
| blaOXA-23 | 0 | 0.00 | 1 | 3.03 | 1.595 | 0.388 |
| blaNDM-5 | 1 | 1.92 | 0 | 0.00 | 0.642 | 1.000 |
| β-lactamase | ||||||
| blaCTX-M | 25 | 48.08 | 8 | 24.24 | 4.829 | 0.028 |
| blaSHV | 52 | 100.00 | 31 | 93.94 | 3.228 | 0.148 |
| blaTEM | 45 | 86.54 | 22 | 66.67 | 4.776 | 0.029 |
Figure 4.
Sankey Diagram of Resistance Genes in CR-KP. A Sankey diagram, also known as a Sankey energy flow chart, is used here. The left nodes represent non-ST11 and ST11 types of CR-KP, while the right nodes represent 11 detected resistance genes. The diagram illustrates the flow from CR-KP types on the left to the resistance genes on the right, the width of the branches correlates with the number of resistance genes.
Antimicrobial Susceptibility of Carbapenem-Resistant Klebsiella Pneumoniae
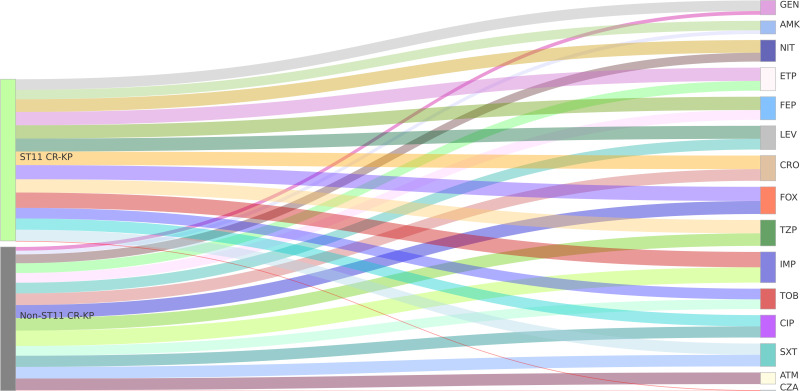
The results of the antibiotic susceptibility testing indicated that CR-KP, carbapenem-resistant Klebsiella pneumoniae, exhibited significant resistance to a range of clinical antibiotics, with the exception of ceftazidime/avibactam. Antibiotics such as ceftriaxone, cefepime, and imipenem classified respectively as cephalosporins and carbapenems exhibit a resistance rate of up to 100%. The resistance rates of ST11 type CR-KP were higher than those of non-ST11 type CR-KP. Specifically, the carbapenem ertapenem, the nitrofuran antibiotic nitrofurantoin, the aminoglycoside amikacin, and the aminoglycoside gentamicin showed higher resistance rates in ST11 type CR-KP compared to non-ST11 type, with statistically significant differences (see Table 4). The visualization of these results can be seen in Figure 5. Overall, the resistance situation is quite severe.
Table 4.
Antimicrobial Susceptibility of Carbapenem-Resistant Klebsiella Pneumoniae
| Antimicrobial Agent |
MIC Breakpoints (μg/mL) |
ST11 CR-KP | Non-ST11 CR-KP | χ2 value | P value | ||||
|---|---|---|---|---|---|---|---|---|---|
| (n=52) | (n=33) | ||||||||
| S | I | R | Resistant strains (n) |
Percentage (%) | Resistant strains (n) |
Percentage (%) | |||
| ATM | ≤4 | 8 | ≥16 | 43 | 82.69 | 25 | 75.76 | 0.607 | 0.436 |
| FOX | ≤8 | 16 | ≥32 | 48 | 92.31 | 28 | 84.85 | 1.187 | 0.300 |
| CRO | ≤1 | 2 | ≥4 | 52 | 100.00 | 33 | 100.00 | - | 1.000 |
| FEP | ≤2 | - | ≥16 | 52 | 100.00 | 33 | 100.00 | - | 1.000 |
| CIP | ≤0.25 | 0.5 | ≥1 | 38 | 73.08 | 24 | 72.73 | 0.001 | 0.972 |
| LEV | ≤0.5 | 1 | ≥2 | 44 | 84.62 | 23 | 69.70 | 2.692 | 0.101 |
| IMP | ≤1 | 2 | ≥4 | 52 | 100.00 | 33 | 100.00 | - | 1.000 |
| ETP | ≤0.5 | 1 | ≥2 | 45 | 86.54 | 21 | 63.64 | 6.101 | 0.014 |
| TZP | ≤8/4 | - | ≥32/4 | 45 | 86.54 | 27 | 81.82 | 0.347 | 0.556 |
| SXT | ≤2/38 | - | ≥4/76 | 38 | 73.08 | 25 | 75.76 | 0.076 | 0.783 |
| NIT | ≤32 | 64 | ≥128 | 43 | 82.69 | 19 | 57.58 | 6.453 | 0.011 |
| AMK | ≤4 | 8 | ≥16 | 32 | 61.54 | 8 | 24.24 | 11.272 | 0.001 |
| GEN | ≤2 | 4 | ≥8 | 36 | 69.23 | 8 | 24.24 | 16.364 | 0.001 |
| TOB | ≤2 | 4 | ≥8 | 36 | 69.23 | 21 | 63.64 | 0.286 | 0.593 |
| CZA | ≤8/4 | - | ≥16/4 | 0 | 0.00 | 1 | 3.03 | - | - |
Abbreviations: ATM, aztreonam; FOX, cefoxitin; CRO, ceftriaxone; FEP, cefepime; CIP, ciprofloxacin; LEV, levofloxacin; IMP, imipenem; ETP, ertapenem; TZP, piperacillin/tazobactam; SXT, trimethoprim/sulfamethoxazole; NIT, nitrofurantoin; AMK, amikacin; GEN, gentamicin; TOB, tobramycin; CZA, ceftazidime-avibactam.
Figure 5.
Sankey Diagram of Antimicrobial Susceptibility of CR-KP. A Sankey diagram, also known as a Sankey energy flow chart, is used here. The left nodes represent ST11 and non-ST11 types of CR-KP, while the right nodes represent 15 detected antibiotics. The diagram illustrates the flow from CR-KP types on the left to the antibiotics on the right, the width of the branches correlates with the number of resistant strains.
Discussion
In recent years, the excessive utilization of carbapenem antibiotics has led to an escalating emergence of CR-KP resistant plasmids, exemplified by the identification of blaNDM-117 in South Africa in 2012 and blaNDM-518 in Japan in 2014. The prevalence shift from blaOXA-2319 to the current global clonal dissemination of blaKPC-2 highlights a progressive rise in CR-KP resistance.20–22 The Chinese Bacterial Resistance Monitoring Network (www.chinets.com) has found that the resistance rate of Klebsiella pneumoniae to imipenem has risen from 3% in 2005 to 24.8% in 2023, especially the detection rate of Carbapenem-resistant Klebsiella pneumoniae (CR-KP) in bloodstream infections has risen from 7.0% in 2014 to 19.6% in 2019.6 Notably, research by Chen et al23 found that the ST11 type CR-KP, in its evolution to the KL64 type, tends to carry more virulence genes, indicating a severe situation in the control of highly virulent, carbapenem-resistant Klebsiella pneumoniae in China. Additionally, the in-hospital mortality rate and 14-day mortality rate for bloodstream infections caused by Klebsiella pneumoniae are as high as 58.3% and 41.7%,24 respectively. Furthermore, 31.4% of patients with Klebsiella pneumoniae bacteremia are infected with hypervirulent Klebsiella pneumoniae (hvKP).25 Therefore, studying the resistance and virulence genes of Klebsiella pneumoniae in intensive care unit bloodstream infections is urgent.
Our research group collected 85 strains of CR-KP from intensive care unit bloodstream infections, predominantly of the ST11 type, with the ST11 type CR-KP primarily exhibiting the KL64 capsular genotype. This distribution across China is consistent with findings reported by Wang et al26 and further studies by Qi et al9 have shown that ST11 type CR-KP possesses greater resistance and faster transmission capabilities. The non-ST11 type CR-KP primarily includes the ST15 type with the KL5 capsular gene, which differs from the findings of Feng et al, who reported that the ST15 type CR-KP predominantly had the KL112 capsular genotype at 59.2%,27 suggesting a relation to the capsular gene polymorphism in Klebsiella pneumoniae.
In terms of virulence gene detection, the ST11 type CR-KP, compared to non-ST11 types, showed higher carriage rates of the capsular synthesis-promoting gene rmpA2, iron-carrier related gene entB, silver ion resistance gene silS, fimbriae fixation and adhesion-related gene kpn, and pPLVPK-like virulence plasmid related genes iucA, peg-344, and terB, with significant differences (P<0.05). Research confirms that the rmpA2 gene, with over 95% specificity, regulates the synthesis of capsular polysaccharides and serves as a marker for screening hypervirulent Klebsiella pneumoniae.28 The entB gene enhances KP’s adaptability to environments by mediating trivalent iron uptake,29 while the silS gene, part of heavy metal resistance, boosts CR-KP’s environmental resilience.30 The kpn gene promotes biofilm formation and host tissue adhesion, decreasing the host’s clearance rate.31 Yang et al32 found that the presence of the rmpA2, iroB, iucA, peg344, and terB virulence genes on a pLVPK virulence plasmid is associated with more severe infections, particularly noting the peg344 gene’s role in encoding an inner membrane transporter, a hallmark of hvKP.33 This study revealed that the prevalence of the peg344 gene in ST11 type CR-KP reached 84.62%, significantly surpassing that observed in non-ST11 type CR-KP (P < 0.01). This finding indicates a heightened pathogenicity associated with ST11 type CR-KP.
In resistance gene detection, bloodstream infection CR-KP primarily carried the A-type enzyme blaKPC-2 subtype, with a higher carriage rate in ST11 type CR-KP, consistent with Du et al’s findings.34 A blaNDM-5 subtype was also found in non-ST11 type CR-KP, naturally resistant to novel beta-lactam drugs like ceftazidime/avibactam, indicating the need to monitor the spread of the blaNDM-5 resistance plasmid.35 The extended-spectrum β-lactamases blaCTX-M and blaTEM were more prevalent in ST11 type CR-KP, aligning with drug susceptibility tests indicating higher resistance rates to cefepime. Other results showed higher resistance rates in ST11 type CR-KP to ertapenem, nitrofurantoin, amikacin, and gentamicin compared to non-ST11 types, complicating clinical first-line antibiotic choices and underscoring the need for rigorous monitoring and control of hospital-acquired ST11 type CR-KP infections.13
Overall, this study isolated 85 strains of CR-KP from ICU bloodstream infections, predominantly of the ST11 type, which accounted for 61.18% of the cases, indicating that ST11 is the dominant clonal group in our hospital. The study found that all 85 CR-KP strains exhibited high levels of resistance. Specifically, the ST11 type CR-KP group showed higher resistance rates to cephalosporin antibiotics such as cefepime, carbapenem antibiotics such as ertapenem, nitrofuran antibiotics such as nitrofurantoin, and aminoglycoside antibiotics such as amikacin and gentamicin compared to the non-ST11 type CR-KP group, with statistically significant differences. Additionally, the ST11 type had higher carriage rates of the capsule synthesis-promoting gene rmpA2, iron-carrier related gene entB, silver ion resistance gene silS, fimbriae fixation and adhesion related gene kpn, and pPLVPK-like virulence plasmid related genes iucA, peg-344, and terB, exhibiting more potent virulence traits. Both groups carried the blaKPC-2 carbapenem resistance gene, but the ST11 type had higher carriage rates of the extended-spectrum β-lactamase genes blaCTX-M and blaTEM. These results show that the ST11 type CR-KP displays stronger resistance, higher virulence characteristics, and greater survivability compared to the non-ST11 type CR-KP, posing a severe threat to hospital infection control and clinical treatment. This necessitates stricter antimicrobial stewardship to prevent the spread of resistant plasmids.
Conclusion
In summary, compared to non-ST11 CR-KP, this study indicated that ST11 CR-KP was more common and that the KL64 type of ST11 CR-KP was more prevalent in bloodstream infections in the intensive care unit (ICU). Additionally, drug susceptibility testing revealed greater resistance rates and ST11 CR-KP carried more virulence and resistance genes than non-ST11 CR-KP. For these reasons, the use of antibiotics for bloodstream infections caused by ST11 CR-KP in the ICU should be done with caution. The hospital must strictly control the dosage and frequency of controlled antibiotics (Imipenem and Meropenem) usage. Dynamic management of antibiotic prescriptions is required. For example, if the patient has no obvious indicators of infection, the antibiotic prescription cannot be approved. If the attending physician insists on prescribing it, he/she must contact the relevant department (the infection control department) to file a statement explaining the reasons for the use, the course of treatment, and the criteria for discontinuation. The relevant department strictly monitors such use, and conducts a quarterly statistical analysis of which doctors have prescribed antibiotics without clear indications of infection. Regular in-house inquiries are conducted for doctors who frequently prescribe antibiotics without clear indications of infection, and severe cases may result in a one-week suspension of the doctor’s antibiotic prescription rights. Finally to enhance the understanding of the molecular epidemiological characteristics and resistance mechanisms associated with CR-KP bloodstream infections in the Ningbo area, we will continue to collect bacterial strain samples from local hospitals for a multi-center study in this region.
Acknowledgments
We thank all the staff of the Clinical Laboratory of Ningbo Medical Centre Lihuili Hospital for their significant contribution in this study and the Home for Researchers editorial team (www.home-for-researchers.com) for their assistance with language editing services.
Funding Statement
This work was supported by the research grants from Zhejiang Medical and Health Science and Technology Plan (NO.2023KY1042) and Ningbo Municipal Natural Science Fund (No. 2022J255).
Data Sharing Statement
The nucleotide sequences of blaKPC-2, blaKPC-3, blaKPC-9, blaKPC-14, blaOXA-23 and blaNDM-5 were deposited in the GenBank database under accession number PQ268540-PQ268601, PQ268533-PQ268539, PQ268530, PQ268531, PQ268532 and CP146099 respectively. Due to data sensitivity, the datasets generated in this study can be released after the article is published and indexed.
Ethical Approval
The authors are accountable for all aspects of the work and ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The present study received approval from the Ethics Committee of Ningbo Medical Centre Lihuili Hospital (No. KY2022SL390-01), and informed consent was obtained from the study participants prior to its implementation. The investigation was conducted in accordance with the guidelines of the Declaration of Helsinki.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Chen J, Li J, Huang F, et al. Clinical characteristics, risk factors and outcomes of Klebsiella pneumoniae pneumonia developing secondary Klebsiella pneumoniae bloodstream infection. BMC Pulmonary Medicine Mar. 2023;23(1):102. doi: 10.1186/s12890-023-02394-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imai K, Ishibashi N, Kodana M, et al. Clinical characteristics in blood stream infections caused by Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae: a comparative study. BMC Infect Dis. 2019;19(1):946. doi: 10.1186/s12879-019-4498-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong T, Fenn SJ, Hardie KR. JMM Profile: carbapenems: a broad-spectrum antibiotic. J Med Microbiol. 2021;70(12). doi: 10.1099/jmm.0.001462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu J, Qing Y, Dong N, et al. Effectiveness of a double-carbapenem combinations against carbapenem-resistant Gram-negative bacteria. Saudi Pharmaceutical j. 2022;30(6):849–855. doi: 10.1016/j.jsps.2022.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang J, Ye L, Guo L, et al. A nosocomial outbreak of KPC-2-producing Klebsiella pneumoniae in a Chinese hospital: dissemination of ST11 and emergence of ST37, ST392 and ST395. Clin Microbiol Infect. 2013;19(11):E509–15. doi: 10.1111/1469-0691.12275 [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Ji J, Ying C, et al. Blood bacterial resistant investigation collaborative system (BRICS) report: a national surveillance in China from 2014 to 2019. Antimicrob Resist Infect Control. 2022;11(1):17. doi: 10.1186/s13756-022-01055-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soares de Moraes L, Gomes Magalhaes GL, Material Soncini JG, Pelisson M, Eches Perugini MR, Vespero EC. High mortality from carbapenem-resistant Klebsiella pneumoniae bloodstream infection. Microbial Pathogenesis Jun. 2022;167:105519. doi: 10.1016/j.micpath.2022.105519 [DOI] [PubMed] [Google Scholar]
- 8.Ernst CM, Braxton JR, Rodriguez-Osorio CA, et al. Adaptive evolution of virulence and persistence in carbapenem-resistant Klebsiella pneumoniae. Nature Med. 2020;26(5):705–711. doi: 10.1038/s41591-020-0825-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrol Chemotherap. 2011;66(2):307–312. doi: 10.1093/jac/dkq431 [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Yu J, Chen F, et al. Emergence and establishment of KPC-2-producing ST11 Klebsiella pneumoniae in a general hospital in Shanghai, China. European j Clin Microbiol Infect Dise. 2018;37(2):293–299. doi: 10.1007/s10096-017-3131-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elmanakhly AR, Bendary MM, Safwat NA, et al. Carbapenem-Resistant Klebsiella pneumoniae: diversity, Virulence, and Antimicrobial Resistance. Infect Drug Resist. 2022;15:6177–6187. doi: 10.2147/idr.s387742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kocsis B. Hypervirulent Klebsiella pneumoniae: an update on epidemiology, detection and antibiotic resistance. Acta microbiologica et immunologica Hungarica. 2023;70(4):278–287. doi: 10.1556/030.2023.02186 [DOI] [PubMed] [Google Scholar]
- 13.Pu D, Zhao J, Lu B, et al. Within-host resistance evolution of a fatal ST11 hypervirulent carbapenem-resistant Klebsiella pneumoniae. Interl j Antimicrol Agen. 2023;61(4):106747. doi: 10.1016/j.ijantimicag.2023.106747 [DOI] [PubMed] [Google Scholar]
- 14.Chen J, Zhang H, Liao X. Hypervirulent Klebsiella pneumoniae. Infect Drug Resist. 2023;16:5243–5249. doi: 10.2147/idr.S418523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choby JE, Howard-Anderson J, Weiss DS. Hypervirulent Klebsiella pneumoniae - clinical and molecular perspectives. J Intern Med. 2020;287(3):283–300. doi: 10.1111/joim.13007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendes G, Santos ML, Ramalho JF, Duarte A, Caneiras C. Virulence factors in carbapenem-resistant hypervirulent Klebsiella pneumoniae. Front Microbiol. 2023;14:1325077. doi: 10.3389/fmicb.2023.1325077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowman W, Sriruttan C, Nana T, et al. NDM-1 has arrived: first report of a carbapenem resistance mechanism in South Africa. S Afr Med J. 2011;101(12):873–875. [PubMed] [Google Scholar]
- 18.Nakano R, Nakano A, Hikosaka K, et al. First report of metallo-β-lactamase NDM-5-producing Escherichia coli in Japan. Antimicrob Agen Chemother. 2014;58(12):7611–7612. doi: 10.1128/aac.04265-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans BA, Amyes SG. OXA β-lactamases. Clinical microbiology reviews. Apr. 2014;27(2):241–263. doi: 10.1128/cmr.00117-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Labarca J, Poirel L, Ozdamar M, Turkoglü S, Hakko E, Nordmann P. KPC-producing Klebsiella pneumoniae, finally targeting Turkey. New Microbes New Infect. 2014;2(2):50–51. doi: 10.1002/nmi2.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wendt C, Schütt S, Dalpke AH, et al. First outbreak of Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae in Germany. European j Clinic Microbiol Infectious Dise. 2010;29(5):563–570. doi: 10.1007/s10096-010-0896-0 [DOI] [PubMed] [Google Scholar]
- 22.Bedenić B, Mazzariol A, Plečko V, et al. First report of KPC-producing Klebsiella pneumoniae in Croatia. J Chemother. 2012;24(4):237–239. doi: 10.1179/1973947812y.0000000017 [DOI] [PubMed] [Google Scholar]
- 23.Chen T, Wang Y, Zhou Y, et al. Recombination Drives Evolution of Carbapenem-Resistant Klebsiella pneumoniae Sequence Type 11 KL47 to KL64 in China. Microbiology Spectrum. 2023:e0110722. doi: 10.1128/spectrum.01107-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neuner EA, Yeh JY, Hall GS, et al. Treatment and outcomes in carbapenem-resistant Klebsiella pneumoniae bloodstream infections. Diagnostic microbiology and infectious disease. Apr. 2011;69(4):357–362. doi: 10.1016/j.diagmicrobio.2010.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu YM, Li BB, Zhang YY, et al. Clinical and molecular characteristics of emerging hypervirulent Klebsiella pneumoniae bloodstream infections in mainland China. Antimicrob Agen Chemother. 2014;58(9):5379–5385. doi: 10.1128/aac.02523-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Feng Y, Zong Z. The Origins of ST11 KL64 Klebsiella pneumoniae: a Genome-Based Study. Microbiology Spectrum. 2023;e0416522. doi: 10.1128/spectrum.04165-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng L, Zhang M, Fan Z. Population genomic analysis of clinical ST15 Klebsiella pneumoniae strains in China. Front Microbiol. 2023;14:1272173. doi: 10.3389/fmicb.2023.1272173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altayb HN, Elbadawi HS, Baothman O, et al. Genomic Analysis of Multidrug-Resistant Hypervirulent (Hypermucoviscous) Klebsiella pneumoniae Strain Lacking the Hypermucoviscous Regulators (rmpA/rmpA2). Antibiotics. 2022;11(5). doi: 10.3390/antibiotics11050596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han R, Niu M, Liu S, Mao J, Yu Y, Du Y. The effect of siderophore virulence genes entB and ybtS on the virulence of Carbapenem-resistant Klebsiella pneumoniae. Microb Pathogenesis. 2022;171:105746. doi: 10.1016/j.micpath.2022.105746 [DOI] [PubMed] [Google Scholar]
- 30.Woolley CA, Sutton JM, Wand ME. Mutations in SilS and CusS/OmpC represent different routes to achieve high level silver ion tolerance in Klebsiella pneumoniae. BMC microbiology. Apr. 2022;22(1):113. doi: 10.1186/s12866-022-02532-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo MY, Liu Y, Fei B, et al. Research progress on virulence factors of hypervirulent Klebsiella pneumoniae]. Zhonghua yu fang yi xue za zhi. 2021;55(11):1357–1363. doi: 10.3760/cma.j.cn112150-20210730-00732 [DOI] [PubMed] [Google Scholar]
- 32.Yang X, Xie M, Xu Q, et al. Transmission of pLVPK-like virulence plasmid in Klebsiella pneumoniae mediated by an Incl1 conjugative helper plasmid. iScience. 2022;25(6):104428. doi: 10.1016/j.isci.2022.104428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bulger J, MacDonald U, Olson R, Beanan J, Russo TA. Metabolite Transporter PEG344 Is Required for Full Virulence of Hypervirulent Klebsiella pneumoniae Strain hvKP1 after Pulmonary but Not Subcutaneous Challenge. Infect Immun. 2017;85(10). doi: 10.1128/iai.00093-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du FL, Huang QS, Wei DD, et al. Prevalence of Carbapenem-Resistant Klebsiella pneumoniae Co-Harboring blaKPC-Carrying Plasmid and pLVPK-Like Virulence Plasmid in Bloodstream Infections. Front Cell Infect Microbiol. 2020;10:556654. doi: 10.3389/fcimb.2020.556654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mezcord V, Traglia GM, Pasteran F, et al. Characterization of cefiderocol-resistant spontaneous mutant variants of Klebsiella pneumoniae producing NDM-5 with a single mutation in cirA. Int j Antimicrobial Agen. 1(2024):107131. doi: 10.1016/j.ijantimicag.2024.107131 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The nucleotide sequences of blaKPC-2, blaKPC-3, blaKPC-9, blaKPC-14, blaOXA-23 and blaNDM-5 were deposited in the GenBank database under accession number PQ268540-PQ268601, PQ268533-PQ268539, PQ268530, PQ268531, PQ268532 and CP146099 respectively. Due to data sensitivity, the datasets generated in this study can be released after the article is published and indexed.