Abstract
Background
Controlled trials have consistently demonstrated the efficacy of poly(ADP-ribose) polymerase inhibitors (PARPis) in patients with metastatic castration-resistant prostate cancer (mCRPC) and BRCA1 or BRCA2 alterations (BRCAalt). However, the reported efficacy of PARPi for alterations in other homologous recombination repair (HRR) genes is less consistent. We sought to evaluate the routine practice effectiveness of PARPi between and within these groups.
Design
Patient-level data from a deidentified nationwide (USA-based) cancer clinico-genomic database between January 2011 and September 2023 were extracted. Patients with mCRPC and comprehensive genomic profiling by liquid biopsy [circulating tumor DNA (ctDNA)] or tissue (tumor) biopsy and who received single-agent PARPi were included and grouped by BRCAalt, ATMalt, other HRR, or no HRR. We further subcategorized BRCAalt into homozygous loss (BRCAloss) and all other deleterious BRCAalt (otherBRCAalt).
Results
A total of 445 patients met inclusion criteria: 214 with tumor and 231 with ctDNA. BRCAalt had more favorable outcomes to PARPi compared with ATM, other HRR, and no HRR groups. Within the BRCAalt subgroup, compared with other BRCAalt, BRCAloss had a more favorable time to next treatment (median 9 versus 19.4 months, P = 0.005), time to treatment discontinuation (median 8 versus 14 months, P = 0.006), and routine practice overall survival (median 14.7 versus 19.4 months, P = 0.016). Tumor BRCAloss prevalence (3.1%) was similar to ctDNA prevalence in liquid biopsy specimens with high tumor fraction (>20%). BRCAloss was not detected in orthogonal germline testing.
Conclusions
PARPi routine practice effectiveness between groups mirrors prospective trials. Within the BRCAalt group, BRCAloss had the best outcomes. Unless the ctDNA tumor fraction is very high, somatic tissue testing (archival or metastatic) should be prioritized to identify patients who may benefit most from PARPi. When tissue testing is not clinically feasible, sufficient ctDNA tumor fraction levels for detection are enriched at clinical timepoints associated with tumor progression.
Key words: mCRPC, PARPi, BRCA, homozygous loss
Highlights
-
•
Homozygous loss of BRCA is present in the tumors of patients with mCRPC who derived the most durable benefit from PARPi.
-
•
Tumor tissue specimens should be prioritized to optimize the identification of those with BRCA homozygous loss.
-
•
Sufficient ctDNA tumor fraction for the detection of BRCA loss is enriched at timepoints associated with tumor progression.
Introduction
Metastatic castration-resistant prostate cancer (mCRPC) is a genetically heterogeneous disease and 20%-30% of men with mCRPC harbor somatic or germline loss-of-function mutations in genes involved in DNA damage repair.1,2 Deleterious mutations in homologous recombination repair (HRR) genes are often associated with distant metastases and worse overall survival (OS).3 HRR-deficient prostate cancers are dependent on alternative mechanisms to prevent deleterious accumulation of DNA damage. One such mechanism for maintaining genomic stability is mediated via poly(ADP-ribose) polymerases (PARPs), which are enzymes involved in the repair of single-strand DNA breaks. Inhibition of PARP in HRR-deficient cancer cells leads to the creation of DNA double-strand breaks and resultant synthetic lethality in prostate tumor cells with HRR deficiency.4 Randomized clinical trials have shown efficacy for the inhibition of PARP in men with HRR-deficient mCRPC.5,6 On the basis of these trials, rucaparib was approved by the United States Food and Drug Administration (FDA) as a single agent for men with mCRPC and BRCA1/2 mutations (BRCAalt) previously treated with next-generation androgen receptor directed therapy and prior taxane therapy.7 Olaparib was approved for men with mCRPC and any germline or somatic HRR mutations previously treated with next-generation androgen receptor directed therapy.8 More recently, talazoparib plus enzalutamide and niraparib or olaparib plus abiraterone have been approved by the FDA for patients with mCRPC with any HRR mutation (talazoparib) or BRCA1/2 mutations only (niraparib and olaparib) based on phase III data from TALAPRO-2, MAGNITUDE, and PROPEL, respectively.9, 10, 11
The varying results surrounding the importance of different HRR alterations in driving sensitivity to PARP inhibitor (PARPi) therapy12 leave open key questions surrounding the role of these treatments beyond the BRCAalt setting. This is reflected in the different criteria for olaparib and talazoparib use (any HRR mutation) compared with the criteria for rucaparib and niraparib (only BRCAalt). Although this may reflect differences in the drugs themselves, it also may be related to the study design in each of the pivotal trials.9,11,13 In addition, there is an emerging hypothesis that homozygous BRCA1 or BRCA2 loss may be associated with more favorable responses to PARPi compared with other types of BRCA alterations14,15 because deletions are irreversible16,17 which may result in prolonged patient benefit.18 Scar-based homologous recombination deficiency (HRD) biomarkers based on patterns of chromosomal instability, such as those approved in ovarian cancer, are hypothesized to have utility in identifying genomic subsets of prostate cancer most likely to respond to PARPi.4,19
The routine clinical use patterns of PARPi are not well described, nor is the routine practice effectiveness of PARPi across HRR subsets and BRCA alteration types. There is a need to more precisely define the molecular determinants that confer durable PARPi benefit to ensure appropriate patient selection for targeted PARPi therapy, as well as the mode of assessment: tissue biopsy or liquid biopsy. Certain types of alterations, such as copy number losses, are more challenging to reliably detect than mutations, not universally validated or reported by next-generation sequencing (NGS) assays,20 and when assessed with liquid biopsy, additionally require a higher level of circulating tumor DNA (ctDNA) tumor fraction to detect.21, 22, 23 Here we describe routine practice outcomes of patients with mCRPC undergoing PARPi therapy according to molecular subgroups stratified by specific HRR mutation status or a scar-based HRD signature (HRDsig). In addition, we sought to assess the association of specific BRCA alterations with PARPi effectiveness.
Methods
Study design and patient selection
The cohort consisted of patients with a confirmed diagnosis of mCRPC included in the USA-wide Flatiron Health–Foundation Medicine Inc. (FMI) deidentified clinico-genomic database between January 2011 and September 2023. All patients underwent genomic testing using Foundation Medicine comprehensive genomic profiling (CGP) assays (described later) with molecular data linked directly.
Deidentified clinical data originated from ∼280 United States cancer clinics (∼800 sites of care). Retrospective longitudinal clinical data were derived from electronic health records, comprising patient-level structured and unstructured data, curated via technology-enabled human abstraction of clinical notes and radiology/pathology reports, which were linked to genomic data derived from FMI testing by deidentified, deterministic matching.24,25
Clinical data included demographics, clinical and laboratory features, timing of treatment exposure, treatment progression, survival, and orthogonal unstructured somatic and germline molecular testing abstracted from PDF reports of non-Foundation Medicine testing.26 Lines of therapy in the database were oncology clinician-defined and rule-based. Patients with a birthyear of 1938 or earlier may have an adjusted birthyear in Flatiron datasets due to patient deidentification requirements. All data elements used in this study have undergone a rigorous quality assessment against published frameworks.27
Patients were included in this study if they received a first-line single-agent PARPi (olaparib, rucaparib, or niraparib) in the mCRPC setting and genomic profiling by tissue or liquid biopsy. Patients were grouped by genes with deleterious alterations detected: BRCA1/2 (BRCAalt), ATM, other HRR (ATR, ATRX, BAP1, BARD1, BRIP1, CHEK1, CHEK2, CDK12, FANCA, FANCL, MRE11, RAD51B, RAD51C, RAD51D, RAD54L, PALB2), or no HRR (if negative by tissue or liquid profiling). We further subclassified BRCAalt into BRCA homozygous loss (BRCAloss) and all other BRCA alterations (otherBRCA), including point mutations, small sequence deletions, and rearrangements. While homozygous loss is always biallelic, other BRCA alterations may be mono-allelic. Institutional Review Board approval of the study protocol was obtained before conducting the study and included a waiver of informed consent based on the observational, noninterventional nature of the study (WCG IRB, Protocol No. 20225562).
Comprehensive genomic profiling
Hybrid capture-based NGS assays were carried out on patient tumor or blood specimens in a Clinical Laboratory Improvement Amendments (CLIA)-certified, College of American Pathologists-accredited laboratory (FMI, Cambridge, MA). Foundationone and FoundationOne CDx assays report single-nucleotide variants, insertions/deletions, genomic rearrangements, copy number amplifications, and homozygous losses.28 FoundationOne Liquid CDx is an NGS panel assay interrogating 324 cancer-related genes, and reporting single-nucleotide variants, insertions/deletions, genomic rearrangements, copy number amplifications, and homozygous losses. Cell-free DNA was extracted from whole blood and CGP was carried out using hybridization-captured, adaptor ligation–based libraries.22
HRDsig is a machine learning algorithm developed to predict genomic scarring consistent with homologous repair deficiency.29 HRDsig utilizes a broad set of copy number features, including absolute modeled copy number, segment size, oscillation patterns, and breakpoints per chromosome arm with features examined genomewide and specifically within the telomeric and centromeric portions of chromosome arms.30 The broad set of copy number features was used as inputs into an extreme gradient boosting machine learning model. HRDsig is a continuous factor from 0 to 1 and a cut-off of 0.7 was prespecified for calling a sample HRDsig(+) based on 90% sensitivity to detect biallelic BRCA1/2 alterations in canonically BRCA-driven diseases (ovary, prostate, pancreas, and breast cancers).
Predominant genetic ancestry was assigned by training a random forest classifier to distinguish the five ancestral superpopulations of the 1000 Genomes Project,31 as previously described,32 and then determining the closest match for each specimen.
Time-to-event outcomes
Time to next treatment (TTNT) was calculated from the treatment start date to the start of the next treatment line (due to any cause) or death. Patients not yet reaching the next treatment line or death were censored at the date of the last clinical visit. Time to PARPi discontinuation (TTD) was calculated from the treatment start date to the cessation of PARPi use (due to any cause), or death. Patients still on PARPi were censored at the date of the last clinical visit or structured activity. OS was calculated from the start of PARPi to death from any cause, and patients with no record of mortality were right censored at the date of the last clinic visit. As patients cannot enter the database until a CGP report is delivered, OS risk intervals were left truncated to the date of the CGP report to account for immortal time. Truncation independence with censoring was evaluated with Kendall’s tau, with P < 0.05 considered acceptable. Flatiron Health database mortality information is a composite derived from three sources: documents within the electronic health records, the Social Security Death Index, and a commercial death dataset mining data from obituaries and funeral homes, with validations reported in comparison to the National Death Index.33
Prostate-specific antigen response
A line of PARPi therapy was eligible for prostate-specific antigen (PSA) response assessment if a PSA result was available within 60 days before PARPi initiation and a separate PSA result was available 1-180 days after. If multiple results were available, respective values most proximal to treatment initiation and 12 weeks of treatment were used. PSA response calculated as previously described34: (on treatment PSA – baseline PSA)/(baseline PSA + 0.01).
Statistical analysis
Differences in time-to-event outcomes were assessed using the log-rank test and Cox proportional hazard models. Chi-square tests and Wilcoxon rank-sum tests were used to assess differences between groups of categorical and continuous variables, respectively. Multiple comparison adjustments were not carried out; P values were reported to quantify the strength of association for biomarker and each outcome, not for null hypothesis significance testing, and interpretations adopted broadly considering the consistency of multiple outcome measures in concert (TTNT, TTD, PSA response, and OS), with no outcome measure standing on its own.
Missing values were handled by simple imputation with expected values determined using random forests with the R package ‘missForest.’ In subsequent analyses, imputed values were treated identically to measured values. R software (R Foundation, Vienna, Austria) was used for all statistical analyses.
Results
Characteristics of the analysis cohort
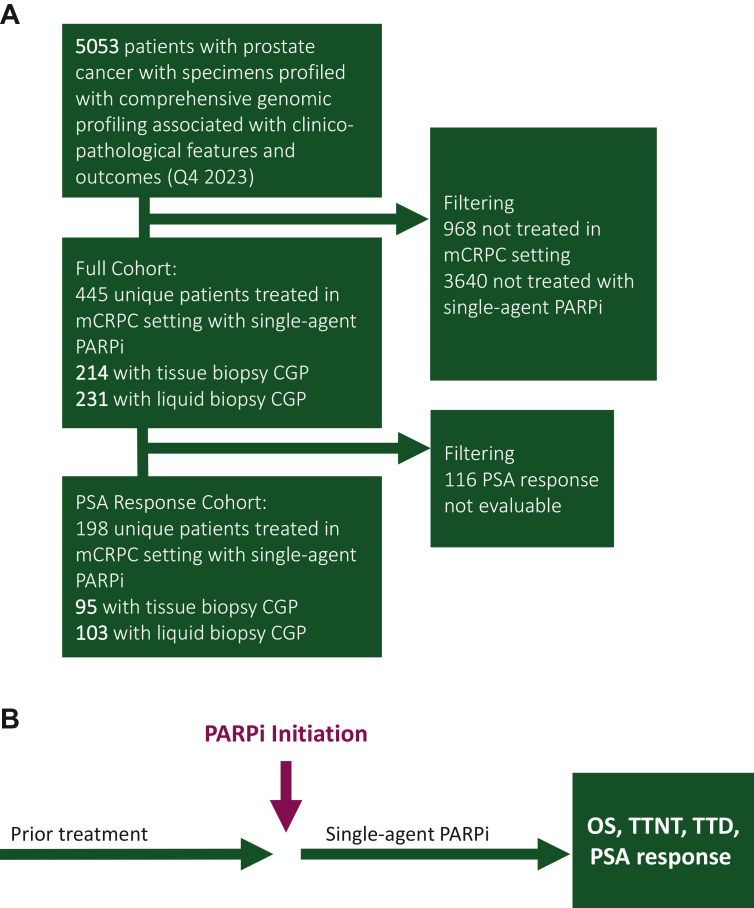
We included 4559 patients with mCRPC and CGP data, of whom 445 unique patients were treated with single-agent PARPi and met inclusion criteria (Figure 1). There were 214 patients who had tumor tissue CGP and 231 who had liquid biopsy CGP; 68 (15%) patients were predicted of majorly African ancestry, 337 (76%) of majorly European ancestry, and 40 (9%) other. Of these patients, 170 (38.2%) had BRCAalt, 110 (24.7%) had ATM mutations, 109 (24.5%) had other HRR mutations, and 56 (12.6%) had no known HRR alteration (Table 1). BRCAalt was more highly represented in the tissue CGP cohorts compared with liquid CGP (46.7% versus 30.3% respectively, P < 0.001). Timing of PARPi therapy was variable: it was administered as a first-line mCRPC treatment in 10.1% (45 patients), second-line in 28.3% (126 patients), third-line in 23.1% (103 patients), and fourth-line in 38.4% (171 patients). This was similar between men with tissue or liquid biopsy genomic profiling. The vast majority of patients (n = 390, 87.6%) had records of prior novel hormonal therapy, 225 (50.6%) had records of prior taxane therapy, and 29 (6.5%) had records of prior platinum therapy. Most patients were treated with olaparib (95.5%) and the remainder received rucaparib (4.0%) and niraparib (0.5%). Differences between molecular subgroups are listed in Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2024.103684.
Figure 1.
Cohort overview. (A) Cohort selection diagram and (B) overview diagram of outcome associations assessed from (A). CGP, comprehensive genomic profiling; mCRPC, metastatic castration-resistant prostate cancer; OS, overall survival; PARPi, poly(ADP-ribose) polymerase inhibitor; PSA, prostate-specific antigen (pre-PARPi PSA level); TTD, time to PARPi discontinuation; TTNT, time to next treatment.
Table 1.
Characteristics of patients included in the study, by method of genomic profiling
| Characteristics | LBx (n = 231) | TBx (n = 214) | Total (n = 445) | P value |
|---|---|---|---|---|
| Age, years | <0.001 | |||
| Median (Q1-Q3) | 75.0 (69.0-80.0) | 71.0 (65.0-77.0) | 73.0 (67.0-79.0) | |
| ECOG, n (%) | 0.14 | |||
| 1 | 73 (31.6) | 83 (38.8) | 156 (35.1) | |
| 2 | 44 (19.0) | 26 (12.1) | 70 (15.7) | |
| 3+ | 66 (28.6) | 66 (30.8) | 132 (29.7) | |
| Unknown | 48 (20.8) | 39 (18.2) | 87 (19.6) | |
| Treatment setting, n (%) | 0.51 | |||
| 1st line mCRPC | 21 (9.1) | 24 (11.2) | 45 (10.1) | |
| 2nd line mCRPC | 61 (26.4) | 65 (30.4) | 126 (28.3) | |
| 3rd line mCRPC | 59 (25.5) | 44 (20.6) | 103 (23.1) | |
| 4th+ line mCRPC | 90 (39.0) | 81 (37.9) | 171 (38.4) | |
| Biopsy site, n (%) | <0.001 | |||
| Blood | 231 (100.0) | 0 (0.0) | 231 (51.9) | |
| Metastatic | 0 (0.0) | 107 (50.0) | 107 (24.0) | |
| Prostate | 0 (0.0) | 107 (50.0) | 107 (24.0) | |
| HRR category, n (%) | <0.001 | |||
| No HRR alteration | 28 (12.1) | 28 (13.1) | 56 (12.6) | |
| ATMalt | 75 (32.5) | 35 (16.4) | 110 (24.7) | |
| BRCA1/2 alt | 70 (30.3) | 100 (46.7) | 170 (38.2) | |
| Other HRR alt | 58 (25.1) | 51 (23.8) | 109 (24.5) | |
| Pre-Tx PSA | 0.80 | |||
| Median (Q1-Q3) | 75.7 (17.9-293.5) | 79.3 (16.8-310.7) | 77.4 (17.1-307.2) | |
| N-Miss, n | 109 | 105 | 214 | |
| Pre-Tx albumin, n (%) | 0.90 | |||
| Below LLN | 30 (15.7) | 28 (16.2) | 58 (15.9) | |
| Normal | 161 (84.3) | 145 (83.8) | 306 (84.1) | |
| N-Miss, n | 40 | 41 | 81 | |
| Pre-Tx alkaline phosphatase, n (%) | 0.62 | |||
| Above ULN | 62 (32.5) | 57 (35.0) | 119 (33.6) | |
| Normal | 129 (67.5) | 106 (65.0) | 235 (66.4) | |
| N-Miss, n | 40 | 51 | 91 | |
| Pre-Tx hemoglobin, n (%) | 0.45 | |||
| Below LLN | 154 (81.5) | 147 (84.5) | 301 (82.9) | |
| Normal | 35 (18.5) | 27 (15.5) | 62 (17.1) | |
| N-Miss, n | 42 | 40 | 82 | |
| Practice type, n (%) | 0.66 | |||
| Academic | 47 (20.3) | 40 (18.7) | 87 (19.6) | |
| Community | 184 (79.7) | 174 (81.3) | 358 (80.4) | |
| Prior novel hormonal therapy, n (%) | 0.66 | |||
| No | 27 (11.7) | 28 (13.1) | 55 (12.4) | |
| Yes | 204 (88.3) | 186 (86.9) | 390 (87.6) | |
| Prior taxane, n (%) | 0.14 | |||
| No | 122 (52.8) | 98 (45.8) | 220 (49.4) | |
| Yes | 109 (47.2) | 116 (54.2) | 225 (50.6) | |
| Prior platinum, n (%) | 0.13 | |||
| No | 212 (91.8) | 204 (95.3) | 416 (93.5) | |
| Yes | 19 (8.2) | 10 (4.7) | 29 (6.5) | |
| Pre-Tx opioid use, n (%) | 0.76 | |||
| No evidence | 166 (71.9) | 151 (70.6) | 317 (71.2) | |
| Yes | 65 (28.1) | 63 (29.4) | 128 (28.8) | |
| Treatment received, n (%) | 0.32 | |||
| Niraparib | 2 (0.9) | 0 (0.0) | 2 (0.4) | |
| Olaparib | 221 (95.7) | 204 (95.3) | 425 (95.5) | |
| Rucaparib | 8 (3.5) | 10 (4.7) | 18 (4.0) | |
| PSA response, n (%) | 0.97 | |||
| Evaluable | 103 (44.6) | 95 (44.4) | 198 (44.5) | |
| Unevaluable | 128 (55.4) | 119 (55.6) | 247 (55.5) | |
| Genetic ancestry, n (%) | 0.21 | |||
| African | 42 (18.2) | 26 (12.1) | 68 (15.3) | |
| European | 169 (73.2) | 168 (78.5) | 337 (75.7) | |
| Other | 20 (8.7) | 20 (9.3) | 40 (9.0) |
ECOG, Eastern Cooperative Oncology Group; HRR, homologous recombination repair; LBx, liquid biopsy; LLN, lower limit of normal; mCRPC, metastatic castration-resistant prostate cancer; PSA, prostate-specific antigen; TBx, tissue biopsy; Tx, treatment; ULN, upper limit of normal.
Outcomes of PARPi stratified by molecular subgroup
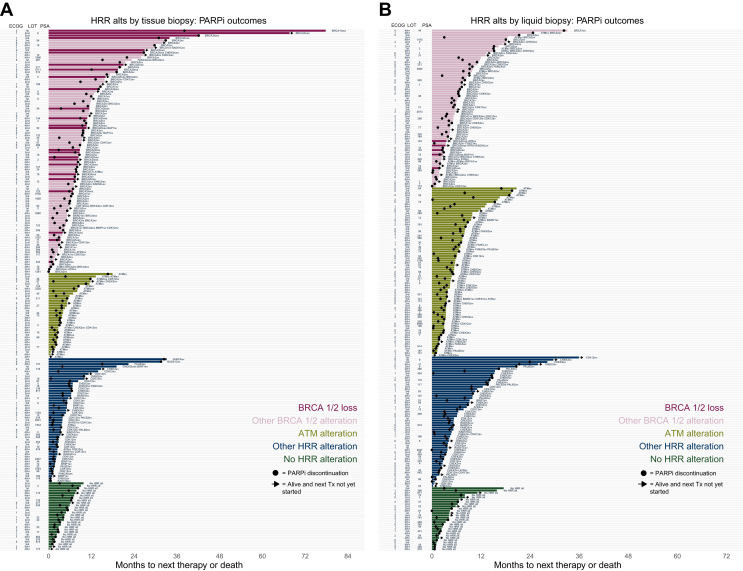
We next assessed TTNT, TTD, and OS stratified by molecular subgrouping. A granular analysis of gene alterations associated with outcomes of individual patients is shown with swimmer plots for TTNT for patients with tissue biopsy CGP (Figure 2A) and liquid biopsy CGP (Figure 2B). In the tissue cohort, several patients with the longest TTNT (>12 months) were patients with BRCA1 or BRCA2 homozygous loss, which were less commonly detected in the liquid cohort. Several patients with non-BRCA alterations (FANCA, RAD51C, PALB2, CHEK2, CDK12, and BRIP1) had >12 months’ TTNT. A small group of patients were administered PARPi without the detection of any HRR subgroup. No significant differences in PARPi outcomes were found between genetic ancestry groups.
Figure 2.
Outcomes on PARPi by biomarker group. Swimmer’s plots of TTNT per patient receiving single-agent PARPi and receiving genomic profiling via (A) tissue biopsy or (B) liquid biopsy. ECOG, Eastern Cooperative Oncology Group; HRR, homologous recombination repair; loss, homozygous loss; LOT, line of therapy in the mCRPC setting; mCRPC, metastatic castration-resistant prostate cancer; PARPi, poly(ADP-ribose) polymerase inhibitor; PSA, prostate-specific antigen (pre-PARPi PSA level); re, rearrangement; sv, short variant mutation; TTNT, time to next treatment; Tx, treatment.
A population (n = 56) of patients received PARPi without a positive HRR alteration from Foundation Medicine (somatic) testing. Of these, half (n = 28) were assessed with tissue testing and half (n = 28) with liquid testing (Supplementary Figure S1A, available at https://doi.org/10.1016/j.esmoop.2024.103684). The group baseline PSA levels (median 155.5, interquartile range 22.2-242.2) were qualitatively higher than other groups, without other substantive differences in other baseline clinical features (Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2024.103684). As many as 37 patients in this group had orthogonal (germline or somatic) BRCA testing from laboratories other than Foundation Medicine. Of these, 12 had a BRCA mutation detected. The median TTNT of these three groups (BRCA mutation detected, no BRCA mutation detected, and no record of orthogonal testing) were 4.3, 4.4, and 4.4 months, respectively (log-rank P = 0.80; Supplementary Figure S1B, available at https://doi.org/10.1016/j.esmoop.2024.103684).
Using the no HRR group as a reference, we next carried out multivariable analyses for TTNT, TTD, and OS in the tissue and liquid CGP cohorts, adjusting for clinical factors that might confound time-to-event analyses [e.g. pre-tx PSA, Eastern Cooperative Oncology Group (ECOG) performance status]. In the tissue CGP cohort, BRCAalt was associated with more favorable TTNT compared with men with no HRR alterations [n = 100, hazard ratio (HR) 0.30, 95% confidence interval (CI) 0.18-0.51, P < 0.001]. ATMalt was not associated with more favorable TTNT (n = 35, HR 1.00, 95% CI 0.56-1.78, P = 0.999), consistent with prior reports,35 and similarly other HRR mutations (n = 51, HR 0.75, 95% CI 0.44-1.28, P = 0.296) were not associated with more favorable TTNT compared with the no HRR cohort (Supplementary Figure S2A, available at https://doi.org/10.1016/j.esmoop.2024.103684). OS was also more favorable in the BRCAalt group relative to no HRR (n = 100, HR 0.43, 95% CI 0.24-0.75, P = 0.003). ATM (n = 35, HR 0.73, 95% CI 0.38-1.41, P = 0.351) and other HRR (n = 51, HR 1.08, 95% CI 0.58-1.92, P = 0.854) cohorts were not statistically different than no HRR (Supplementary Figure S2C, available at https://doi.org/10.1016/j.esmoop.2024.103684).
In the liquid CGP cohort, adjusted TTNT was more favorable in the BRCAalt subgroup (n = 70, HR 0.46, 95% CI 0.26-0.82, P = 0.009) and similarly in the other HRR subgroup (n = 58, HR 0.51, 95% CI 0.29-0.88, P = 0.017) compared with no HRR. ATM was not statistically different than no HRR (Supplementary Figure S2D, available at https://doi.org/10.1016/j.esmoop.2024.103684). In addition, OS was similarly favorable in BRCAalt (n = 70, HR 0.50, 95% CI 0.25-1.01, P = 0.053) and other HRR subgroups (n = 58, HR 0.53, 95% CI 0.27-1.04, P = 0.064) compared with no HRR. No difference was observed in the ATM subgroup (n = 75, HR 0.95, 95% CI 0.51-1.78, P = 0.877; Supplementary Figure S2F, available at https://doi.org/10.1016/j.esmoop.2024.103684).
The multivariable model for TTD for both tissue and liquid cohorts was very similar to TTNT (Supplementary Figure S2C and D, available at https://doi.org/10.1016/j.esmoop.2024.103684). Unadjusted, univariable associations between HRR groups can be found in Supplementary Figure S3, available at https://doi.org/10.1016/j.esmoop.2024.103684.
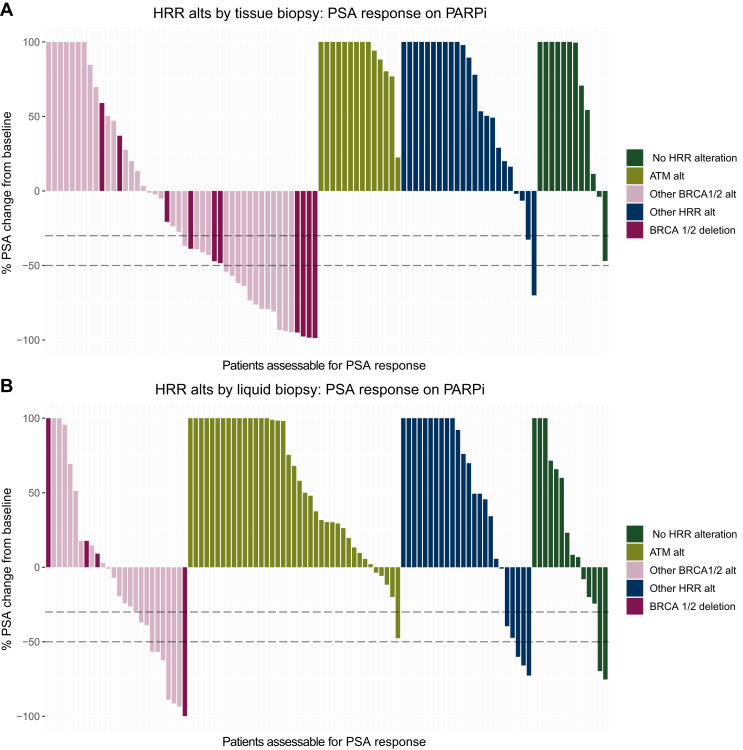
PSA response
Baseline and 12-week PSA responses were evaluable in 198 patients (n = 95 with tissue biopsy and n = 103 with liquid biopsy). In the tissue CGP cohort, 16 (34.7%) patients with BRCAalt had a >50% decline in PSA at 12 weeks after PARPi initiation. By contrast, 0 patients in the ATM alteration subgroup, 1 patient in the other HRR subgroup (4.2%), and 0 patients in the no HRR subgroup had a PSA decline of ≥50% (Figure 3A). Several of the exceptional responders had BRCAloss. In the liquid CGP cohort, 7 patients (26.9%) with BRCAalt had a 12-week 50% PSA response (Figure 3B). In this cohort, 0 patients with ATM alterations, 3 patients (12.5%) with other HRR alterations, and 2 patients (14.0%) with no HRR alterations had a 50% PSA response. Patient characteristics were broadly similar between those with an evaluable PSA response (n = 198) and those with an unevaluable for PSA response (n = 247; Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2024.103684).
Figure 3.
PSA response at week 12. The change in PSA from baseline to most proximal PSA assessment to 12-week timepoint on-therapy (see Methods) is shown for patients with evaluable baseline and on-therapy PSA with alterations identified via (A) tissue biopsy and (B) liquid biopsy. Dashed horizontal lines represent 30% and 50% PSA declines from the baseline. HRR, homologous recombination repair; PARPi, poly(ADP-ribose) polymerase inhibitor; PSA, prostate-specific antigen (pre-PARPi PSA level); TBx, tissue biopsy; LBx, liquid biopsy.
Outcomes stratified by BRCA alteration type
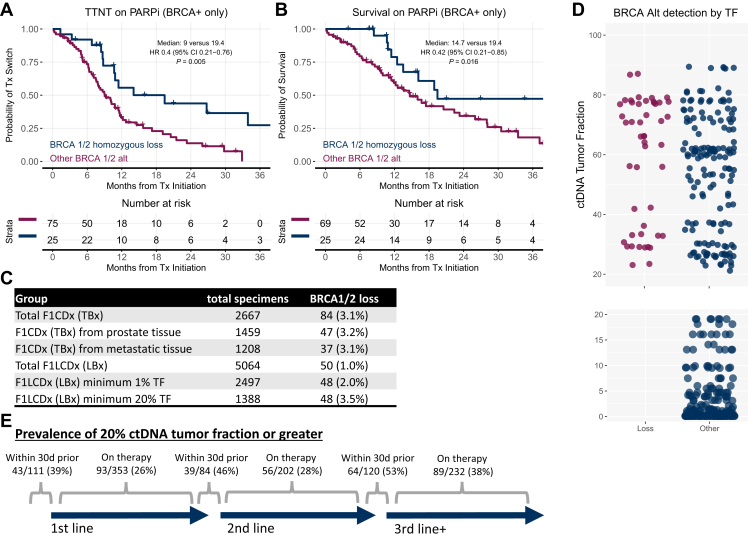
We next assessed if homozygous BRCAloss detected on tissue CGP is associated with more favorable outcomes to single-agent PARPi compared with other BRCAalt. For men with homozygous BRCAloss, there was more favorable TTNT (adjusted HR 0.4, 95% CI 0.21-0.76, P = 0.005) and OS (adjusted HR 0.42, 95% CI 0.21-0.85, P = 0.016) compared with other BRCAalt (Figure 4A and B). Time-to-treatment discontinuation associations were similar to TTNT (Supplementary Figure S4, available at https://doi.org/10.1016/j.esmoop.2024.103684), and multivariable assessments adjusting for standard baseline prognostic factors estimate a similar magnitude of effect as seen in univariable assessments (Supplementary Figure S5, available at https://doi.org/10.1016/j.esmoop.2024.103684), including when adjusting for co-occurring alterations in other commonly altered pathways in prostate cancer (Supplementary Figures S6 and S7, available at https://doi.org/10.1016/j.esmoop.2024.103684).
Figure 4.
Outcomes on PARPi by BRCA alteration type. Among the subset of patients with tissue biopsy and BRCA 1/2 alterations, the outcomes associated with BRCA 1/2 homozygous loss versus other alterations are shown for (A) TTNT and (B) OS. (C) Prevalence of BRCA 1/2 homozygous loss among FDA-approved assays included in the database. (D) Distribution of ctDNA tumor fraction associated with the detection of BRCA alterations in liquid biopsy. (E) Prevalence of ≥20% ctDNA tumor fraction by the timing of blood draw relative to the start of a line of therapy. CI, confidence interval; ctDNA, circulating tumor DNA; FDA, Food and Drug Administration; HR, hazard ratio; OS, overall survival; PARPi, poly(ADP-ribose) polymerase inhibitor; TF, ctDNA tumor fraction; TTNT, time to next treatment; Tx, treatment.
To further evaluate the hypothesis that BRCA homozygous loss is challenging to detect in liquid biopsy in the absence of high levels of ctDNA tumor fraction (TF), we explored the frequency of detection of BRCA homozygous loss in FDA-approved CGP assays for tissue and liquid biopsy, regardless of the treatment. We identified 2667 tissue specimens profiled with FoundationOne CDx. Among these, 3.1% have BRCA homozygous loss. Among 5064 liquid biopsy specimens profiled with FoundationOne Liquid CDx, 1.0% had homozygous BRCAloss. Considering only the specimens with a minimum of 1% TF and 20% TF, the prevalence is 2% and 3.5%, respectively (Figure 4C). A visualization of the TF associated with BRCA alteration types in liquid biopsy is also shown (Figure 4D). We further evaluated the prevalence of ≥20% ctDNA tumor fraction as determined by FoundationOne Liquid CDx, with respect to the timing of initiation of first, second, and third lines of therapy, and found the respective prevalences of 39%, 46%, and 53% when assessed within 30 days before when a new line of therapy began (Figure 4E). When ctDNA tumor fraction was assessed outside of these ranges, the respective prevalences were 26%, 28%, and 38%, respectively.
BRCAloss was highly associated with genomic scarring (HRDsig), consistent with it being biallelic loss (Supplementary Figure S8 and S9, available at https://doi.org/10.1016/j.esmoop.2024.103684). While the presence of HRDsig was significantly associated with more favorable outcomes in the tissue cohort (TTNT, TTD, and OS all P < 0.001) the associations with outcome within the BRCA altered, and separately, the non-BRCA altered groups, were less clear in this cohort.
As BRCA1 and BRCA2 knockout mice are embryonic lethal,36 it is hypothesized that BRCAloss in human cancers is a somatic-only event. We sought to evaluate if orthogonal germline testing would identify BRCAloss. Among the tissue cohort, 88 patients had germline BRCA testing results available (Supplementary Figure S10, available at https://doi.org/10.1016/j.esmoop.2024.103684). Of those with BRCA mutations or rearrangements in tumor tissue, 22 of 39 (56%) were positive for germline BRCA mutations. Notably, 0 of 9 (0%) patients with BRCAloss detected by Foundation Medicine tumor tissue testing had positive germline BRCA results.
Discussion
Multiple lines of level 1 evidence support PARPi therapy in the context of certain HRR alterations. However, questions have remained with regard to PARPi efficacy across the spectrum of alterations in HRR pathway genes and the use of tumor versus liquid CGP for the detection of alterations. In this routine practice cohort study, we show that men with mCRPC treated with single-agent PARPi were most likely to have favorable outcomes and durable benefits in the setting of homozygous BRCA1 or BRCA2 loss compared with those in other biomarker-defined subgroups (other BRCA alterations, ATM alterations, other HRR alterations, or no HRR alterations). These findings were robust across multiple endpoints—PSA response, TTNT, TTD, and OS—and are in line with previous reports: the greatest benefit was most consistently observed in patients whose tumors had evidence of deleterious BRCA1/2 alterations, and smaller cohorts reporting extreme benefit in the presence of homozygous BRCAloss.14,18 There is an existing body of biological evidence to suggest that reversion mutations are a common mechanism of resistance to PARPi37 and that complete loss of BRCA1 or BRCA2 confers an inability to evolve this resistance mechanism. We report and further validate this phenomenon in a larger cohort more reflective of routine clinical practice.
In contrast to prior evaluations of the effectiveness of PARPi in BRCA1- versus BRCA2-altered groups,38 we observed similar relative outcomes between these groups when additionally adjusting for routine clinical prognostic factors, alteration type (homozygous loss versus other), and TP53 status (Supplementary Figure S5, available at https://doi.org/10.1016/j.esmoop.2024.103684). This was also observed when further adjusting for additional commonly coaltered genes (Supplementary Figure S6, available at https://doi.org/10.1016/j.esmoop.2024.103684). A recent report suggested potential extreme benefit from the coalteration of SPOP and BRCA2 mutations,39 and while we did not observe this finding here (Supplementary Figure S6, available at https://doi.org/10.1016/j.esmoop.2024.103684), such inferences must be interpreted with caution given the small number with SPOP mutations in our cohort. Further exploratory assessments of non-HRR alterations (Supplementary Figure S7, available at https://doi.org/10.1016/j.esmoop.2024.103684) did not suggest an association with PARPi effectiveness for other non-HRR-altered genes or pathways, with the notable exception of TP53, which was consistently associated with less favorable outcomes. This was true both the overall cohort (Supplementary Figure S7, available at https://doi.org/10.1016/j.esmoop.2024.103684) and in just those with BRCA1 or BRCA2 alterations (Supplementary Figure S6, available at https://doi.org/10.1016/j.esmoop.2024.103684).
A surprising finding in our study was the non-insignificant (n = 56) population of patients who received PARPi without a positive Foundation Medicine test for HRR gene alteration. All BRCA results as available in the electronic health record are abstracted. As results are not always available in the electronic health record, it is possible that more patients than reflected received testing. A total of 37 patients had orthogonal testing results available: 12 had positive BRCA test results and 25 had negative BRCA results. The lack of TTNT differences observed between these groups (log-rank P = 0.80) is consistent with the possibility that these orthogonal test results might represent false-positive results.
While liquid biopsy CGP offers a less invasive alternative to tissue biopsy, liquid biopsy approaches may be more prone to false-negative and false-positive results. Clonal hematopoiesis (CH) is a well-described process that can lead to the accumulation of somatic mutations in blood cells, and these can confound the interpretation of ctDNA results. In a retrospective study of 69 patients with mCRPC, 7 had detectable CH mutations in HRR genes (5 ATM, 1 BRCA2, and 1 CHEK2).40 It is possible a subset of liquid CGP patients had CH interference which may also impact interpretation of PARPi outcomes.
Liquid biopsy CGP assay sensitivity is dependent on the amount of ctDNA shed and measured by tumor fraction on liquid biopsy assays.21,22,41 Detection of homozygous loss is particularly challenging with the current generation of liquid biopsy technologies. In a post hoc analysis of the PROfound study, Chi et al. found that while the concordance of liquid CGP for the detection of nonsense, splice, and frameshift mutations on matched tissue CGP was high (each >85%), the concordance of homozygous loss was only 27%.42 Corroborating this, we observed a similar prevalence of BRCAloss detection between tissue biopsy (TBx) and liquid biopsy (LBx) when the ctDNA tumor fraction is very high (≥20%; Figure 4C). This is consistent with the differences observed between the cohort of outcomes associated with TBx and LBx detection of HRR alterations (Figure 2). At the face value, without consideration of confounders, in men who underwent liquid biopsy CGP, the BRCAalt group did not have more favorable outcomes compared with the other HRR alterations. Our interpretation is that, because BRCA homozygous losses are only detectable in liquid biopsy with a level of ctDNA tumor fraction that many patients in routine practice will not have, potential extreme responders are underrepresented in the liquid biopsy cohort presented here.
Our data support the use of tumor tissue for CGP for the detection of HRR alterations in mCRPC that can improve care. However, biopsy is not always feasible, and biopsies of bone metastases, in particular, have a high rate of failure of CGP.42,43 Previous studies suggest that HRR alterations are largely truncal, and would be present in an archival prostate tissue biopsy,44,45 suggesting that it would be valid to use archival specimens to guide decisions for PARPi use in metastatic settings. Corroborating this, we observed a similar prevalence of BRCAloss in specimens originating from the prostate and from other tissues (Figure 4C). In our cohorts of patients treated with PARPi, among the 214 with tissue biopsy, exactly half of the biopsies were obtained from the prostate and half from metastatic tissue (Table 1). We did not observe differences in outcomes associated with either group (Supplementary Figures S2 and S5-S7, available at https://doi.org/10.1016/j.esmoop.2024.103684). While prospective, longitudinal assessments of BRCAloss would be able to more definitively assess whether it can be acquired over the course of treatments, current data support a truncal nature and the appropriateness of using an archival prostate biopsy to assess BRCAloss status.
In cases where fresh biopsy is not feasible and archival tissue is not available, liquid biopsy is an important alternative. Certain liquid biopsy assays are validated to detect BRCAloss, but even when these tools are used, the lack of detection of BRCAloss should be interpreted as ‘indeterminate’ rather than ‘negative’ in the absence of very high ctDNA fraction, whereas properly validated tissue biopsy assays can definitively rule out the presence of BRCAloss. However, liquid biopsy assays have much higher sensitivity (requiring less ctDNA tumor fraction) to detect mutations and rearrangements. Detection of other types of BRCA alterations via liquid biopsy was associated with improved OS compared with control in a post hoc analysis of the PROfound study.46,47
Providers should be aware of the inherent limitations of liquid biopsy with regard to ctDNA tumor fraction and the detection of homozygous BRCAloss. Providers should also be aware that not all NGS tumor tissue assays have the ability to report copy number losses in BRCA1 and BRCA2.20 Lastly, it is also important to note that blood-based germline testing is also unlikely to detect homozygous BRCAloss; BRCA1 and BRCA2 knockout mice are embryonic lethal,36 suggesting a somatic-only origin. In this study, all patients (n = 9) with detected BRCAloss in tumor tissue who underwent orthogonal germline testing were negative for germline BRCA alterations (Supplementary Figure S10, available at https://doi.org/10.1016/j.esmoop.2024.103684).
Consistent with prior reports48 we observed that ctDNA tumor fraction was enriched in specimens obtained within 30 days before the initiation of a new line of therapy (Figure 4E), with prevalence rates of ≥20% tumor fraction present in 39%, 46%, and 53% of specimens, respectively, obtained within 30 days before first-, second-, and third-line treatment initiations in our cohort, consistent with the hypothesis that progressing tumors will shed more DNA into circulation compared with points in the patient journey where disease may be slower growing. This observation supports current treatment guideline recommendations to prioritize liquid biopsy CGP testing at times of progression.
Notably, there were no differences in clinical outcomes to PARPi for patients across genetic ancestry cohorts. Each registrational trial of PARPi enrolled <5% of patients of African descent. This routine care population with 15% of patients of African ancestry having similar treatment effects and outcomes to PARPi is reassuring for clinical practice, supporting the use of PARPi agents for patients of African descent.
The PARPi monotherapy has not shown a consistent clinical benefit for patients with ATM mutations,35 and our results are consistent with prior reports. Single-agent PARPi has shown some efficacy in a small subset of other HRR mutations. In the TRITON2 study, 6/11 patients with PALB2 alterations treated with rucaparib had a 50% PSA response.49 There were also occasional responses in patients with other HRR genes (RAD51B, FANCA, and BRIP1). Similarly, in the TOPARP-B trial, 4/7 patients with mCRPC and PALB2 mutations had either a radiographic or 50% PSA response to single-agent olaparib.50 A United States FDA pooled analysis of six trials of PARPi in mCRPC, which included studies of single-agent PARPi and dual PARPi plus androgen receptor pathway inhibitors, found that PARPi benefit was greatest for patients with BRCA, PALB2, and CDK12 mutations.51 In our study, the small fraction of men with non-BRCA HRR and durable TTNT and TTD had alterations in these genes as well. A total of nine patients in our cohort tested positive for HRDsig who were negative for BRCA alterations (Supplementary Figure S9, available at https://doi.org/10.1016/j.esmoop.2024.103684), and two of these had PALB2 mutations. While more favorable outcomes were not strongly enriched in this small cohort, larger cohorts may yield more insights.
Limitations
Observational and/or retrospective analyses are more prone to false discovery than prospective randomized trials, due to multiple hypothesis testing and potential imbalances between groups. We carried out rigorous adjustments of prognostic factors to reduce potential imbalances between groups that might confound time-to-event comparisons. However, these adjustments do not account for all potential imbalances, and there are some biases that are known that must be carefully considered to generalize results, such as BRCAloss being easier to detect with tissue biopsy. Results abstracted from unstructured data, such as orthogonal (non-Foundation Medicine) somatic and germline testing can be subject to degrees of human or machine reader error, especially around BRCAloss. We recognize that clinical annotation, bioinformatic pipelines, and reporting can vary between laboratories. Therefore the results from our study may not be generalizable to the biomarker performance of all NGS platforms.
Conclusions
In this analysis of the effectiveness of PARPi, patients with BRCA alterations identified on tissue biopsy CGP had more favorable outcomes across all endpoints relative to other patient subgroups in the setting of PARPi monotherapy. Patients with homozygous BRCAloss had significantly more favorable outcomes relative to all other BRCA alterations. While BRCAloss can be detected with validated liquid biopsy assays, many patients with mCRPC do not have high-enough ctDNA tumor fraction to rule out the presence of BRCAloss using liquid biopsy. However, when tumor tissue testing is not feasible, the greater signal in liquid biopsy is anticipated when tumors are progressing. No patients with BRCAloss detected in tumor tissue had germline BRCA alterations detected, consistent with the existing biological understanding of BRCAloss. Our results suggest that tumor tissue CGP should be prioritized when clinically feasible for the detection of alterations in HRR genes, especially for those associated with durable benefits from PARPi.
Acknowledgements
We thank the patients whose deidentified data made this research possible, the clinical and laboratory staff at Foundation Medicine, and the team at Flatiron Health. This work is additionally dedicated to precision medicine pioneer Bryce Olson, whose life was shortened by prostate cancer and is dearly missed.
Foundation Medicine, a wholly owned subsidiary of Roche, is a for-profit company and producer of FDA-regulated molecular diagnostics. Authors employed by Foundation Medicine were involved in the design and conduct of the study, analysis, interpretation of the data, preparation, review, and approval of the manuscript.
Disclosure
RPG, RWM, OG, HT, JSR, JCFQ, and GL are employees of Foundation Medicine, a wholly owned subsidiary of Roche, and have equity interests in Roche. HHC reports research funds to the institution from Clovis Oncology, Color Genomics, Janssen, Medivation, Promontory Pharmaceutics, and Sanofi; served as a consultant for AstraZeneca (2021); and royalties from UpToDate. AJZ reports research funding from Pfizer, Astellas, X4 Pharma, Infinity, Merck, ABX, Curium, Clarity; consulting fees from Pfizer, Astellas, Incyte, Bayer, Exelixis, Dendreon, Astra Zeneca; and speaker’s fees/travel support from Pfizer, Janssen, HIKMA, Amedco, CancerNet, McKesson Specialty Health. QQ reports consulting for Exelixis and has received an honorarium from MJH Life Sciences. TZ reports research funds to the institution from CPRIT, Merck, Janssen, AstraZeneca, Pfizer, Astellas, Eli Lilly, Tempus, ALX Oncology, Janux Therapeutics, and Exelixis; and consulting/advisory relationships with Merck, Exelixis, Sanofi-Aventis, Janssen, AstraZeneca, Pfizer, Amgen, BMS, Seagen, Eisai, Aveo, Eli Lilly, Bayer, Gilead, Novartis, EMD Serono, Aravive, MJH Associates, Vaniam, Aptitude Health, and PeerView. MC reports research funds to the institution from Bristol Meyers Squibb. NA has received honorarium before May 2021 and during his lifetime for consulting to Astellas, AstraZeneca, Aveo, Bayer, Bristol Myers Squibb, Calithera, Clovis, Eisai, Eli Lilly, EMD Serono, Exelixis, Foundation Medicine, Genentech, Gilead, Janssen, Merck, MEI Pharma, Nektar, Novartis, Pfizer, Pharmacyclics, and Seattle Genetics; and has received research funding during his lifetime (to his institution) from Arvinas, Astellas, AstraZeneca, Bavarian Nordic, Bayer, Bristol Meyers Squibb, Calithera, Celldex, Clovis, CRISPR Therapeutics, Eisai, Eli Lilly, EMD Serono, Exelixis, Genentech, Gilead, Glaxo Smith Kline, Immunomedics, Janssen, Lava, Medivation, Merck, Nektar, Neoleukin, New Link Genetics, Novartis, Oric, Pfizer, Prometheus, Rexahn, Roche, Sanofi, Seattle Genetics, Takeda, and Tracon Pharmaceuticals. ZRR reports institutional/research funding from AstraZeneca; and performs personal advisory roles in Janssen and AstraZeneca. JM has served as an advisor for AstraZeneca, Amunix/Sanofi, Daiichi-Sankyo, Janssen, MSD, Pfizer, and Roche; is member of the Scientific Board for Nuage Therapeutics; and also the principal investigator of grants funded by AstraZeneca, Amgen, and Pfizer to VHIO (institution), but none of these are related to this work. TMM serves on the advisory boards of Foundation Medicine and Tempus. All other authors have declared no conflicts of interest.
Funding
This work was supported by Foundation Medicine, a wholly owned subsidiary of Roche (no grant number).
Data sharing
The data that support the findings of this study were originated by Flatiron Health, Inc. and Foundation Medicine, Inc. Requests for data sharing by license or by permission for the specific purpose of replicating results in this manuscript can be submitted to PublicationsDataaccess@flatiron.com and cgdb-fmi@flatiron.com.
Supplementary data
References
- 1.Pritchard C.C., Mateo J., Walsh M.F., et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375(5):443–453. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson D., Van Allen E.M., Wu Y.-M., et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castro E., Goh C., Olmos D., et al. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J Clin Oncol. 2013;31(14):1748–1757. doi: 10.1200/JCO.2012.43.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.von Werdt A., Brandt L., Schärer O.D., Rubin M.A. PARP inhibition in prostate cancer with homologous recombination repair alterations. JCO Precis Oncol. 2021;5:1639–1649. doi: 10.1200/PO.21.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Bono J., Mateo J., Fizazi K., et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med. 2020;382(22):2091–2102. doi: 10.1056/NEJMoa1911440. [DOI] [PubMed] [Google Scholar]
- 6.Abida W., Patnaik A., Campbell D., et al. Rucaparib in men with metastatic castration-resistant prostate cancer harboring a BRCA1 or BRCA2 gene alteration. J Clin Oncol. 2020;38(32):3763–3772. doi: 10.1200/JCO.20.01035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.US Food & Drug Administration FDA Grants Accelerated Approval to Rucaparib for BRCA-Mutated Metastatic Castration-Resistant Prostate Cancer. https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-accelerated-approval-rucaparib-brca-mutated-metastatic-castration-resistant-prostate Available at:
- 8.US Food & Drug Administration FDA Approves Olaparib for HRR Gene-Mutated Metastatic Castration-Resistant Prostate Cancer. https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-olaparib-hrr-gene-mutated-metastatic-castration-resistant-prostate-cancer Available at:
- 9.Agarwal N., Azad A.A., Carles J., et al. Talazoparib plus enzalutamide in men with first-line metastatic castration-resistant prostate cancer (TALAPRO-2): a randomised, placebo-controlled, phase 3 trial. Lancet. 2023;402(10398):291–303. doi: 10.1016/S0140-6736(23)01055-3. [DOI] [PubMed] [Google Scholar]
- 10.Chi K.N., Rathkopf D., Smith M.R., et al. Niraparib and abiraterone acetate for metastatic castration-resistant prostate cancer. J Clin Oncol. 2023;41(18):3339–3351. doi: 10.1200/JCO.22.01649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saad F., Clarke N.W., Oya M., et al. Olaparib plus abiraterone versus placebo plus abiraterone in metastatic castration-resistant prostate cancer (PROpel): final prespecified overall survival results of a randomised, double-blind, phase 3 trial. Lancet Oncol. 2023;24:1094–1108. doi: 10.1016/S1470-2045(23)00382-0. [DOI] [PubMed] [Google Scholar]
- 12.Akbıyık I., Ürün Y. Determining magnitude of benefit from poly(ADP-ribose) polymerase inhibitors in prostate cancer. Future Oncol. 2023;19(39):2585–2591. doi: 10.2217/fon-2023-0550. [DOI] [PubMed] [Google Scholar]
- 13.Murai J., Huang S.N., Das B.B., et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 2012;72(21):5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carreira S., Porta N., Arce-Gallego S., et al. Biomarkers associating with PARP inhibitor benefit in prostate cancer in the TOPARP-B trial. Cancer Discov. 2021;11(11):2812–2827. doi: 10.1158/2159-8290.CD-21-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.VanderWeele D.J., Paner G.P., Fleming G.F., Szmulewitz R.Z. Sustained complete response to cytotoxic therapy and the PARP inhibitor veliparib in metastatic castration-resistant prostate cancer – a case report. Front Oncol. 2015;5:169. doi: 10.3389/fonc.2015.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodall J., Mateo J., Yuan W., et al. Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov. 2017;7(9):1006–1017. doi: 10.1158/2159-8290.CD-17-0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards S.L., Brough R., Lord C.J., et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451(7182):1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 18.Mateo J., de Bono J.S., Fizazi K., et al. Olaparib for the treatment of patients with metastatic castration-resistant prostate cancer and alterations in BRCA1 and/or BRCA2 in the PROfound trial. J Clin Oncol. 2023;42(5):571–583. doi: 10.1200/JCO.23.00339. [DOI] [PubMed] [Google Scholar]
- 19.Sztupinszki Z., Diossy M., Krzystanek M., et al. Detection of molecular signatures of homologous recombination deficiency in prostate cancer with or without BRCA1/2 mutations. Clin Cancer Res. 2020;26(11):2673–2680. doi: 10.1158/1078-0432.CCR-19-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vandekerkhove G., Giri V.N., Halabi S., et al. Toward informed selection and interpretation of clinical genomic tests in prostate cancer. JCO Precis Oncol. 2024;8 doi: 10.1200/PO.23.00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herberts C., Wyatt A.W. Technical and biological constraints on ctDNA-based genotyping. Trends Cancer. 2021;7(11):995–1009. doi: 10.1016/j.trecan.2021.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Woodhouse R., Li M., Hughes J., et al. Clinical and analytical validation of foundation one liquid CDx, a novel 324-gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS One. 2020;15(9) doi: 10.1371/journal.pone.0237802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Husain H., Pavlick D.C., Fendler B.J., et al. Tumor fraction correlates with detection of actionable variants across > 23,000 circulating tumor DNA samples. JCO Precis Oncol. 2022;6 doi: 10.1200/PO.22.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singal G., Miller P.G., Agarwala V., et al. Association of patient characteristics and tumor genomics with clinical outcomes among patients with non-small cell lung cancer using a clinicogenomic database. JAMA. 2019;321(14):1391–1399. doi: 10.1001/jama.2019.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liede A., Hernandez R.K., Roth M., Calkins G., Nicacio L., Larrabee K. Validation of international classification of diseases coding for bone metastases in electronic health records using technology-enabled abstraction. Clin Epidemiol. 2015;7:441. doi: 10.2147/CLEP.S92209. 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abernethy A.P., Gippetti J., Parulkar R., Revol C. Use of electronic health record data for quality reporting. J Oncol Pract. 2017;13(8):530–534. doi: 10.1200/JOP.2017.024224. [DOI] [PubMed] [Google Scholar]
- 27.Castellanos E.H., Wittmershaus B.K., Chandwani S. Raising the bar for real-world data in oncology: approaches to quality across multiple dimensions. JCO Clin Cancer Inform. 2024;8 doi: 10.1200/CCI.23.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milbury C.A., Creeden J., Yip W.-K., et al. Clinical and analytical validation of FoundationOne®CDx, a comprehensive genomic profiling assay for solid tumors. PLoS One. 2022;17(3) doi: 10.1371/journal.pone.0264138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore J.A., Chen K.-T., Madison R., et al. Pan-cancer analysis of copy-number features identifies recurrent signatures and a homologous recombination deficiency biomarker to predict poly (ADP-Ribose) polymerase inhibitor response. JCO Precis Oncol. 2023;7 doi: 10.1200/PO.23.00093. [DOI] [PubMed] [Google Scholar]
- 30.Macintyre G., Goranova T.E., De Silva D., et al. Copy number signatures and mutational processes in ovarian carcinoma. Nat Genet. 2018;50(9):1262–1270. doi: 10.1038/s41588-018-0179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Auton A., Abecasis G.R., Altshuler D.M., et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mata D.A., Rotenstein L.S., Ramos M.A., Jena A.B. Disparities according to genetic ancestry in the use of precision oncology assays. N Engl J Med. 2023;388(3):281–283. doi: 10.1056/NEJMc2213457. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Q., Gossai A., Monroe S., Nussbaum N.C., Parrinello C.M. Validation analysis of a composite real-world mortality endpoint for patients with cancer in the United States. Health Serv Res. 2021;56(6):1281–1287. doi: 10.1111/1475-6773.13669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graf R.P., Fisher V., Weberpals J., et al. Comparative effectiveness of immune checkpoint inhibitors vs chemotherapy by tumor mutational burden in metastatic castration-resistant prostate cancer. JAMA Netw Open. 2022;5(3):e225394. doi: 10.1001/jamanetworkopen.2022.5394. e225394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marshall C.H., Sokolova A.O., McNatty A.L., et al. Differential response to olaparib treatment among men with metastatic castration-resistant prostate cancer harboring BRCA1 or BRCA2 versus ATM mutations. Eur Urol. 2019;76(4):452–458. doi: 10.1016/j.eururo.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hakem R., Pompa JLD La, Mak T.W. Developmental studies of Brca1 and Brca2 knock-out mice. J Mammary Gland Biol Neoplasia. 1998;3(4):431–445. doi: 10.1023/a:1018792200700. [DOI] [PubMed] [Google Scholar]
- 37.Loehr A., Hussain A., Patnaik A., et al. Emergence of BRCA reversion mutations in patients with metastatic castration-resistant prostate cancer after treatment with rucaparib. Eur Urol. 2023;83(3):200–209. doi: 10.1016/j.eururo.2022.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taza F., Holler A.E., Fu W., et al. Differential activity of PARP inhibitors in BRCA1-versus BRCA2-altered metastatic castration-resistant prostate cancer. JCO Precis Oncol. 2021;5:1200–1220. doi: 10.1200/PO.21.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orme J.J., Taza F., De Sarkar N., et al. Co-occurring BRCA2/SPOP mutations predict exceptional poly (ADP-ribose) polymerase inhibitor sensitivity in metastatic castration-resistant prostate cancer. Eur Urol Oncol. 2024;7(4):877–887. doi: 10.1016/j.euo.2023.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jensen K., Konnick E.Q., Schweizer M.T. Association of clonal hematopoiesis in DNA repair genes with prostate cancer plasma cell-free DNA testing interference. JAMA Oncol. 2021;7(1):107–110. doi: 10.1001/jamaoncol.2020.5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reichert Z.R., Morgan T.M., Li G., et al. Prognostic value of plasma circulating tumor DNA fraction across four common cancer types: a real-world outcomes study. Ann Oncol. 2023;34(1):111–120. doi: 10.1016/j.annonc.2022.09.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chi K.N., Barnicle A., Sibilla C., et al. Detection of BRCA1, BRCA2, and ATM alterations in matched tumor tissue and circulating tumor DNA in patients with prostate cancer screened in PROfound. Clin Cancer Res. 2023;29(1):81–91. doi: 10.1158/1078-0432.CCR-22-0931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abida W., Armenia J., Gopalan A., et al. Prospective genomic profiling of prostate cancer across disease states reveals germline and somatic alterations that may affect clinical decision making. JCO Precis Oncol. 2017;1:1–16. doi: 10.1200/PO.17.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zurita A.J., Graf R.P., Villacampa G., et al. Genomic biomarkers and genome-wide loss-of-heterozygosity scores in metastatic prostate cancer following progression on androgen-targeting therapies. JCO Precis Oncol. 2022;6 doi: 10.1200/PO.22.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schweizer M.T., Sivakumar S., Tukachinsky H., et al. Concordance of DNA repair gene mutations in paired primary prostate cancer samples and metastatic tissue or cell-free DNA. JAMA Oncol. 2021;7(9):1378–1382. doi: 10.1001/jamaoncol.2021.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tukachinsky H., Madison R.W., Chung J.H., et al. Genomic analysis of circulating tumor DNA in 3,334 patients with advanced prostate cancer identifies targetable BRCA alterations and AR resistance mechanisms. Clin Cancer Res. 2021;27(11):3094–3105. doi: 10.1158/1078-0432.CCR-20-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsubara N., de Bono J., Olmos D., et al. Olaparib efficacy in patients with metastatic castration-resistant prostate cancer and BRCA1, BRCA2, or ATM alterations identified by testing circulating tumor DNA. Clin Cancer Res. 2023;29(1):92–99. doi: 10.1158/1078-0432.CCR-21-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Antonarakis E.S., Tierno M., Fisher V., et al. Clinical and pathological features associated with circulating tumor DNA content in real-world patients with metastatic prostate cancer. Prostate. 2022;82(7):867–875. doi: 10.1002/pros.24331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abida W., Campbell D., Patnaik A., et al. Rucaparib for the treatment of metastatic castration-resistant prostate cancer associated with a DNA damage repair gene alteration: final results from the phase 2 TRITON2 study. Eur Urol. 2023;84(3):321–330. doi: 10.1016/j.eururo.2023.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mateo J., Porta N., Bianchini D., et al. Olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair gene aberrations (TOPARP-B): a multicentre, open-label, randomised, phase 2 trial. Lancet Oncol. 2020;21(1):162–174. doi: 10.1016/S1470-2045(19)30684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fallah J., Xu J., Weinstock C., et al. Efficacy of poly(ADP-ribose) polymerase inhibitors by individual genes in homologous recombination repair gene-mutated metastatic castration resistant prostate cancer: a US Food and Drug Administration pooled analysis. J Clin Oncol. 2024;42(14):1687–1698. doi: 10.1200/JCO.23.02105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.