SUMMARY
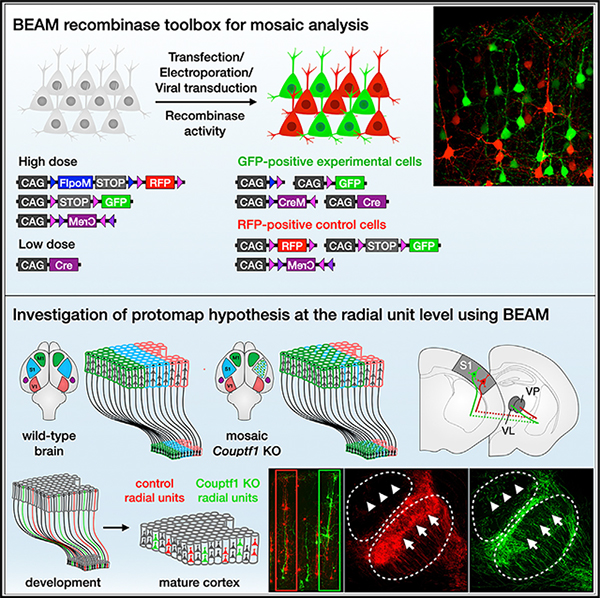
We describe a binary expression aleatory mosaic (BEAM) system, which relies on DNA delivery by transfection or viral transduction along with nested recombinase activity to generate two genetically distinct, non-overlapping populations of cells for comparative analysis. Control cells labeled with red fluorescent protein (RFP) can be directly compared with experimental cells manipulated by genetic gain or loss of function and labeled with GFP. Importantly, BEAM incorporates recombinase-dependent signal amplification and delayed reporter expression to enable sharper delineation of control and experimental cells and to improve reliability relative to existing methods. We applied BEAM to a variety of known phenotypes to illustrate its advantages for identifying temporally or spatially aberrant phenotypes, for revealing changes in cell proliferation or death, and for controlling for procedural variability. In addition, we used BEAM to test the cortical protomap hypothesis at the individual radial unit level, revealing that area identity is cell autonomously specified in adjacent radial units.
In brief
Greig et al. describe a dual-recombinase system for mosaic genetic analysis using DNA transfection or transduction termed BEAM. Nested recombinase activity generates two distinct populations of cells in a single experiment, labeled with either green or red fluorescent protein, enabling direct comparison of control and genetically manipulated cells.
Graphical Abstract
INTRODUCTION
Mosaic genetic analysis of individual mutant cells in an otherwise wild-type organism and local environment offers a powerful approach for investigating gene function with cellular resolution. It can circumvent embryonic lethality and avoid the confounding pleiotropic effects that often arise in the setting of global loss of gene function throughout an entire organism or even with conditional loss of gene function in a specific organ, tissue, or cell type.1–3 Through the combination of mosaic genetic analysis with distinct fluorescent labeling of mutant cells, direct identification of a broad range of abnormal phenotypes becomes possible. Phenotypic analysis can be facilitated further by labeling a second population of wild-type cells with a different fluorescent protein, enabling direct comparison of wild-type and mutant cells within the same tissue.
A number of genome-based strategies for mosaic analysis in mice have been developed, including mosaic analysis with double markers (MADM)4,5 and mosaic analysis with spatial and temporal regulation (MASTR).6 Although these methods have been used in recent years to make important discoveries across a broad range of scientific fields,7–11 they require time-consuming genetic crosses to enable experimentation and entail additional complexities and limitations. In contrast, genetic analysis by in vivo delivery of exogenous DNA enables versatile manipulation of gene expression with substantial potential to accelerate biological research in vertebrate model systems. However, the potential for considerable procedural variability inherent in the introduction of exogenous DNA can be a limiting drawback relative to genome-based methods. Procedural variability can cause substantial differences in the number, type, and spatial distribution of cells targeted, often increasing the difficulty of obtaining well-matched pairs of experimental and control specimens and, therefore, complicating data analysis and interpretation.
Importantly, these limitations can be circumvented by incorporating an internal control. Such an approach has been applied to study graded EPHA7/EPHRIN-A signaling in corticothalamic axon sorting within the ventrobasal thalamic nucleus.12 By electroporating a mix of a high concentration of red fluorescent protein (RFP) expression plasmid and low concentration of green fluorescent protein (GFP)/EphA7 bicistronic expression plasmid, the authors compared the spatial distribution of RFP-positive and RFP/GFP-double-positive axons within the same brain, eliminating the variability introduced by differences in the areal distribution of electroporated neurons across surgeries. A similar approach is to mix a high concentration of RFP expression plasmid and of a CRE-dependent GFP expression plasmid together with a low concentration of a Cre expression plasmid, resulting in a subset of cells expressing GFP.13 Both approaches result in control cells labeled with RFP and experimental cells double labeled with both RFP and GFP, but neither approach yields truly binary gene expression. Reporter constructs have been previously described that convert from RFP expression to GFP expression after CRE-mediated recombination.14,15 However, initial expression of RFP in all cells, and/or incomplete recombination, results in a large number of cells co-expressing both fluorescent proteins,16–19 thus producing potentially ambiguous results.
Here, we substantially extend and refine the range of tools available for mosaic genetic analysis in mice by developing a modular recombinase-based system for binary gene expression and mosaic analysis that can be delivered by transfection or transduction directly into wild-type or floxed mice, without the need for complex breeding schemes. The system relies on sparse recombinase activation, combined with recombinase-mediated signal amplification and delayed expression, to generate two genetically distinct fluorescently labeled populations of cells for comparative analysis. We have termed this method BEAM, for binary expression aleatory mosaic. Any gene of interest can be misexpressed or deleted in GFP-positive cells and compared with interspersed RFP-positive control cells. We applied BEAM to previously published loss- and gain-of-function phenotypes to illustrate its advantages for investigating cell autonomy, identifying temporally or spatially aberrant phenotypes, revealing changes in cell proliferation or death, and controlling for procedural variability. We also applied BEAM to investigate a long-standing hypothesis in the field of cortical development—that each neural progenitor and its progeny constitute an independent radial unit and that subsequent organization of the cerebral cortex into functional areas is determined in neural progenitors as a cell identity protomap that is then transferred to their progeny within a cortical column. These original experiments demonstrate that intermingled cortical columns can acquire divergent area identities through cell-autonomous transcriptional mechanisms.
RESULTS
Combinatorial recombinase activity can be used to generate genetically distinct populations of cells
Delivery of DNA by transfection, electroporation, and viral transduction is a stochastic process in which each cell can receive anywhere from 0 to more than 100 copies of the delivered DNA construct, depending on the plasmid concentration or viral titer used, with an approximately normal distribution of copy numbers across the entire population of cells.20 It is, therefore, not possible to use incompatible recombinase sites to generate fully distinct outcomes on a cell-by-cell basis,21 since each possible outcome of recombination is represented multiple times within most cells. However, the proportion of cells that are successfully transfected can be controlled by adjusting plasmid concentration or viral titer. When one plasmid or virus is introduced at a higher concentration and another at a lower concentration, most cells will receive both or only the one at higher concentration (while a much smaller number of cells will receive only the one at lower concentration). Here, we exploit this principle, in conjunction with the approximately all-or-none activity of recombinases, to generate binary gene expression outcomes in individual cells (Figure 1A).
Figure 1. A recombinase-based strategy for binary gene expression.
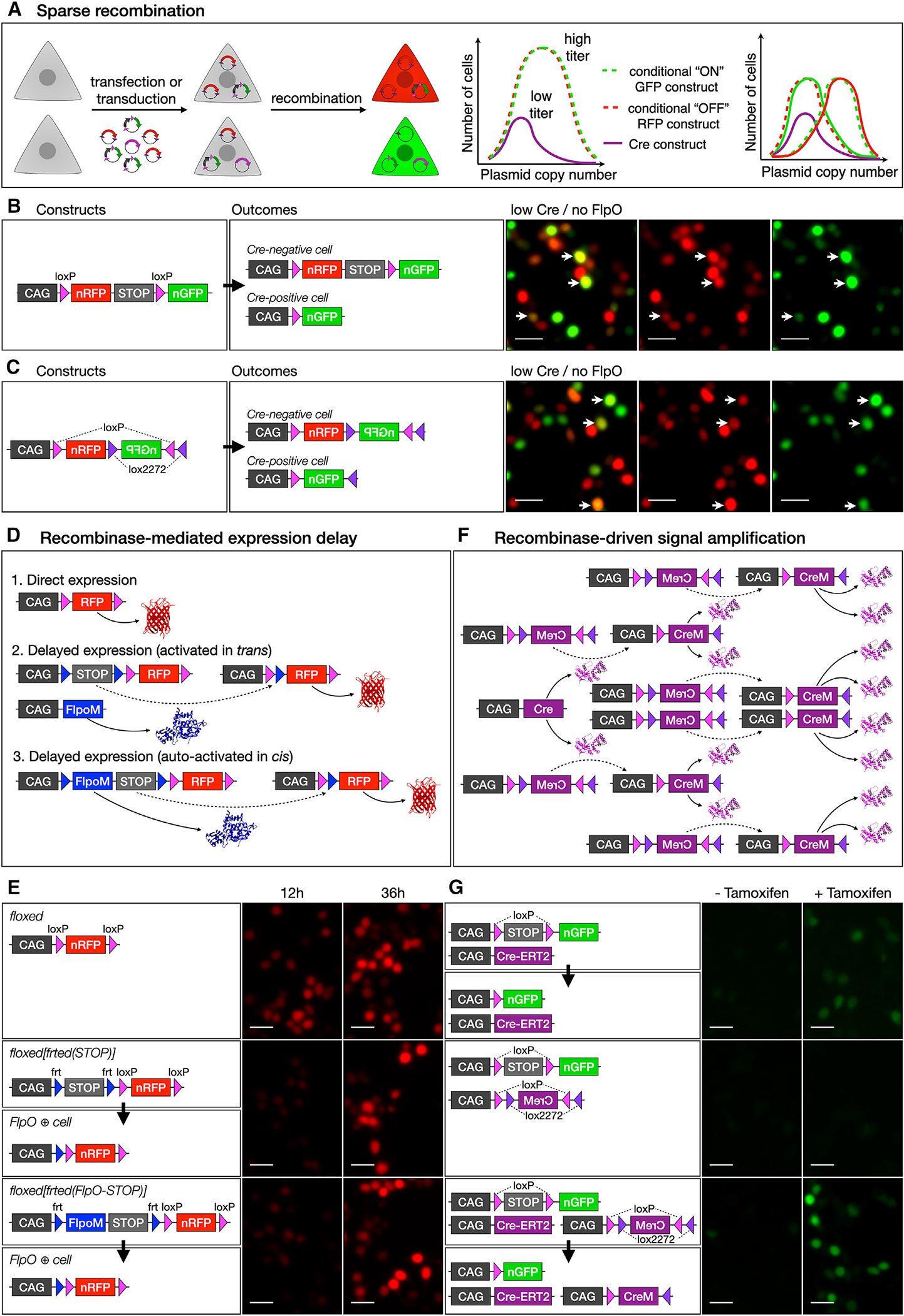
(A) Conceptual representation showing the frequency of plasmid copy number across a population of cells, which can be exploited to achieve binary gene expression by using sparse recombinase delivery in combination with conditional expression plasmids.
(B and C) Significant overlap of RFP and GFP (arrows, right) results with both a loxP-RFP-STOP-loxP-GFP construct (B) and an RFP-FLEX(GFP) construct (C).
(D and E) RFP can be directly expressed from a CAG promoter (D1) or be dependent on excision of an frt-flanked STOP cassette by FLPO, which can be expressed from a second construct, in trans (D2) or autoactivated in cis (D3). Constitutively active loxP-flanked RFP is expressed at 12 h post-transfection in 293T cells, with strong expression at 36 h (E, top). Expression can be delayed using an frt-flanked STOP cassette and FlpO expression in trans (E, middle) or in cis (E, bottom), resulting in lower expression levels at 12 h post-transfection but similar expression levels compared with direct expression at 36 h.
(F and G) CRE activity can be amplified in trans by transfecting a low-dose CAG-Cre that activates multiple copies of a FLEX-CreM construct. A low dose of tamoxifen was used to induce GFP expression in 293T cells transfected with a Cre-ERT2 expression construct and a loxP-STOP-loxP-GFP reporter, resulting in low levels of GFP (G, top). Transfection with a FLEX(CreM) construct did not result in significant autoactivation (G, middle). However, in the presence of CRE-ERT2 and tamoxifen, FLEX(CreM) substantially amplified recombination, increasing GFP levels (G, bottom). All scale bars, 10 μm. See also Figure S1.
The simplest version of a reporter construct for this purpose would comprise a CMV/β-actin hybrid promoter (CAG) and a loxP-flanked (floxed) RFP and transcriptional STOP cassette, followed by GFP.14,16 At baseline, the construct drives expression of RFP, but GFP is expressed only after Cre deletes RFP (Figure 1B). Alternatively, the same outcome can be achieved using a CAG-driven flip excision (FLEX) construct with inverted lox sites, in which the coding sequence for RFP is oriented in the sense direction and the coding sequence for GFP is oriented in the antisense direction.15,17 In this case, CRE permanently reverses the orientation of GFP and, in the process, excises the coding sequence for RFP (Figure 1C). By co-transfecting each of these constructs with a low dose of CAG-Cre, it is possible to generate cells that are primarily green or primarily red. However, substantial residual overlap in expression results from two main limitations: (1) these constructs drive expression of RFP until CRE mediates recombination and (2) low Cre expression results in incomplete recombination of the multiple copies of reporter plasmid in each cell.
To address the first limitation of these prior methods, we sought to delay initial expression of RFP. We designed a construct with a CAG promoter and an frt-flanked (frted) transcriptional STOP cassette followed by a floxed(RFP) (Figure 1D), resulting in minimal expression of RFP at baseline until it is activated by FLP-mediated excision of the frted(STOP) cassette. When cells are transfected with this construct together with a CAG-FlpO construct, RFP is expressed, but with a delay introduced by the requirement for FlpO transcription and translation followed by excision of the frted(STOP) cassette. We also generated a construct with an intron-modified FlpO (FlpoM) between the first frt site and the STOP cassette (Figure 1D). FlpoM contains a β-globin intron that interrupts its coding sequence and prevents FLPO production in bacteria, thereby avoiding recombination of frt sites in E. coli during plasmid production. Once introduced into eukaryotic cells, the plasmid drives transcription of FlpoM, the β-globin intron is spliced out, and FLPO protein is produced and excises its own coding sequence, along with the transcriptional STOP cassette from the plasmid, enabling expression of RFP. Therefore, the plasmid functions as a self-activating recombinase-operated delay switch.
In 293T cells transfected with FlpO-delayed expression constructs, RFP fluorescence is clearly reduced 12 h after transfection, compared to cells transfected with a direct expression construct, but reaches similar levels by 36 h after transfection (Figure 1E). When these plasmids are co-transfected with a CAG-Cre construct, there is direct competition between CRE-mediated excision of RFP and FLPO-mediated excision of the STOP cassette, leading to reduced RFP fluorescence in CRE-positive cells (Figure S1A). Therefore, this approach substantially mitigates one of the mechanisms that leads to overlap between GFP and RFP in a Cre-dependent binary expression system.
To address the second limitation of existing methods, incomplete recombination, we developed a strategy for recombinase-mediated signal amplification. We cloned an intron-modified Cre (CreM) into a CAG-driven FLEX cassette in the antisense orientation, thereby generating a CRE-activated Cre expression construct in which the enzymatic activity of CRE recursively activates expression of more Cre in a self-amplifying reaction (Figure 1F). We tested this approach by transfecting 293T cells with a Cre-ERT2 expression construct and a floxed(STOP)-GFP reporter and then inducing recombination with a low concentration of tamoxifen (Figure 1G). This results in recombination of only a subset of the available copies of floxed(STOP)-GFP in each cell and, therefore, low levels of GFP fluorescence. Co-transfecting FLEX(CreM) along with Cre-ERT2 results in higher levels of GFP fluorescence. Importantly, we do not observe leaky autoactivation of FLEX(CreM) in the absence of Cre-ERT2, indicating that it remains inert in mammalian cells in the absence of initiating CRE activity from a second source. Unlike Beatrix19, Cre amplification is decoupled from FLEX reporter activation, allowing for a floxed(STOP) reporter, which can be activated more efficiently (Figure S1B).
BEAM drives binary gene expression in vitro and in vivo
Combining both of these advances—FlpO-delayed RFP expression and amplification of CRE activity with FLEX(CreM)—dramatically reduced overlap between EGFP and RFP (Figures 2A and 2B). We used fluorescence-activated cell sorting (FACS) to quantify the extent to which each of these approaches can sharpen binary expression. By introducing FlpO-delayed RFP expression, we found a substantial reduction in the overlap of EGFP and RFP, from 26.7% to 23.1% (Figure 2C). By introducing Cre signal amplification, the overlap of EGFP and RFP was reduced to 13.7%. By combining FlpO-delayed RFP expression with Cre signal amplification, overlap between EGFP and RFP was reduced to only 8.3%. This combination of constructs forms the basis of our BEAM recombinase system and is used in subsequent experiments unless otherwise noted.
Figure 2. BEAM results in distinct expression of GFP or RFP, both in vitro by transfection of 293T cells and in vivo by electroporation of cortical progenitors.
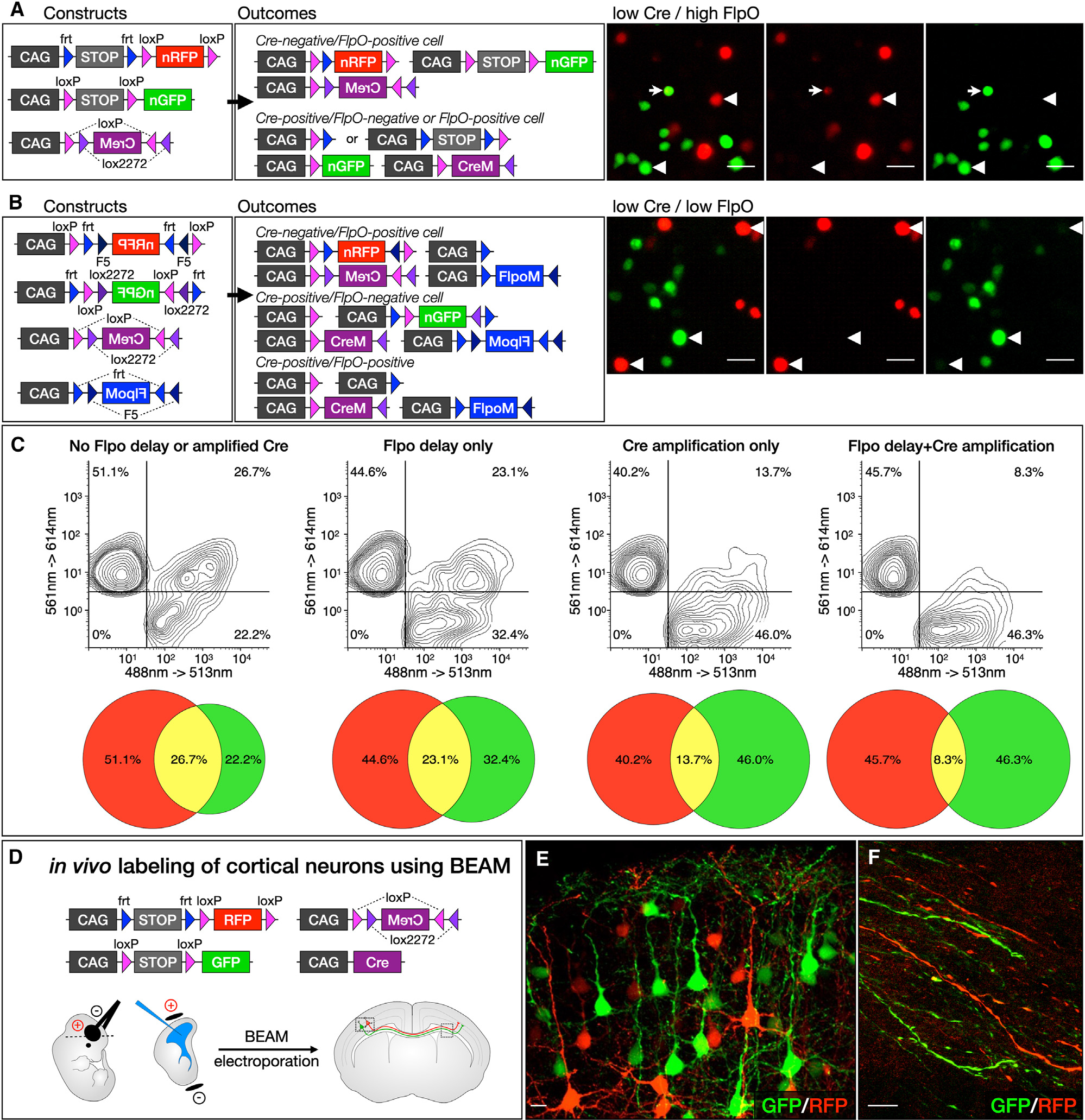
(A) Incorporating recombinase-mediated delayed expression of RFP and recombinase-driven signal amplification substantially reduces overlapping expression of GFP and RFP (arrowheads, right). Only rare CRE-positive cells maintain even low-level, residual RFP expression (arrows, right).
(B) FLEX cassettes produce recombination with minimal crossover between fluorophores (arrowheads, right).
(C) Combining delayed FlpO expression with Cre amplification in vivo dramatically reduces overlap between GFP- and RFP-expressing cells (right upper quadrant in plots).
(D–F) In vivo labeling of cortical neurons using BEAM at E14.5 (D). Electroporated neurons express either GFP or RFP, appear phenotypically normal (E), and extend axons across the corpus callosum (F). Scale bars, 10 μm. See also Figure S2.
We also generated equivalent reagents for mosaic genetic analysis using FLEX configurations, in which the coding sequence for GFP or RFP is initially oriented in the antisense direction and is flanked by inverted repeats of incompatible lox sites (FLEX) or frt sites (FREX). Therefore, CRE- or FLPO-mediated recombination will reverse the orientation of GFP or RFP, activating expression of the corresponding fluorescent protein (Figures S2A and S2B). Each cassette is additionally flanked by direct repeats of frt sites (frt-FLEX-frt) or loxP sites (loxP-FREX-loxP) so that, while one recombinase activates expression, the other recombinase excises the entire cassette, turning off expression. Sparse transfection with Cre and FlpO, along with the corresponding amplification constructs, results in cells that receive a single recombinase and activate expression of GFP or RFP and cells that receive both recombinases and express neither (Figure 2B). These FLEX constructs provide an alternative to transcriptional STOP cassettes, which allow low levels of readthrough, especially when placed in front of a strong promoter such as CAG. CRE-positive green cells have no history of any RFP expression, and FLPO-positive red cells have no history of any GFP expression. Because the two-step inversion and excision process is substantially less efficient than direct excision,22 little expression of GFP or RFP develops in double-positive cells. However, for the same reason, lower GFP and RFP expression levels are present in cells transfected with a single recombinase. In addition, fewer cells are labeled in total, because transfection with each recombinase is sparse, and double-positive cells remain unlabeled. Therefore, this approach is most useful for experiments in which even low levels of overlap would substantially complicate interpretation or in which sparse labeling is advantageous.
Finally, we compared the performance of BEAM with that of two previously published methods for binary gene expression, Double UP and Beatrix.18,19 We found that both methods result in nearly twice as many cells expressing both GFP and RFP compared to BEAM. Specifically, 12.5% of cells are both GFP and RFP positive with Double UP and 11.8% with Beatrix, compared to only 6.2% with BEAM (Figure S2C). We suspect that Double UP results in more overlap because it lacks Flpo-mediated delayed expression of RFP or Cre amplification of GFP expression and RFP inactivation. Although Beatrix utilizes Cre amplification, it does so within a FLEX context, which is known to be less efficient than direct excision (Figure S1B),22 and it also lacks Flpo-mediated delayed RFP expression.
To investigate whether BEAM can also be used successfully in vivo, we introduced the constructs into cerebral cortex progenitors in mice by in utero electroporation on embryonic day 14.5 (E14.5) and analyzed the resulting brains on post-natal day (P4) (Figure 2D). Similar to the results obtained in vitro, we observed only red cells in the absence of CAG-Cre and increasing numbers of green cells with escalating doses of CAG-Cre, enabling the large majority of cells to be labeled green (Figure S3). Importantly, green (CRE-positive) and red (CRE-negative) neurons both migrate into the cortex and reside intermingled with one another, with no indication that one population is otherwise phenotypically different from the other. High-magnification confocal imaging shows that both populations of neurons have normal pyramidal morphologies and long apical dendrites that arborize in layer I (Figure 2E). In addition, both red and green axons extend normally across the corpus callosum (Figure 2F).
BEAM efficiently reports genomic recombination status
To determine how closely GFP and RFP expression tracks with the genomic recombination status of transfected cells in vivo, we electroporated the BEAM plasmids into the cerebral cortex of R26loxP(STOP)loxP-LacZ mice23 on E14.5. We collected the brains at P7, performed immunocytochemistry for β-GAL, and examined co-expression with GFP and RFP by confocal imaging (Figures 3A–3H). Highly effective BEAM operation would produce a few green cells that were β-GALnegative or red cells that were β-GAL positive. However, some β-GAL-positive cells that do not express GFP are expected, because the electroporated plasmids are epigenomic and are therefore gradually diluted in post-mitotic neurons after progenitors undergo multiple rounds of mitosis, while the recombined R26LacZ reporter allele undergoes genomic replication during each round of mitosis. Importantly, we found almost perfect co-localization between GFP and β-GAL immunolabeling (91.7% ± 5.5% of GFP-positive cells were also β-GAL positive), although some β-GAL-positive cells were GFP negative, as expected. Conversely, there was almost no overlap between RFP and β-GAL immunolabeling (4.0% ± 2.4% of RFP-positive cells were also β-GAL-positive). To determine whether BEAM could be delivered by viral transduction, we generated adeno-associated virus (AAV) constructs and packaged these into AAV 2–1 capsids for each BEAM plasmid. We injected BEAM AAVs in the cerebral cortex of R26loxP(STOP)loxP-LacZ mice at P1 and collected tissue for analysis at P14. We found that viral transduction is also compatible with mosaic analysis using BEAM, with effective segregation of GFP and RFP expression and high concordance with genomic recombination status (Figures S3J–S3N).
Figure 3. BEAM-driven expression of GFP and RFP accurately reflects genomic recombination status.
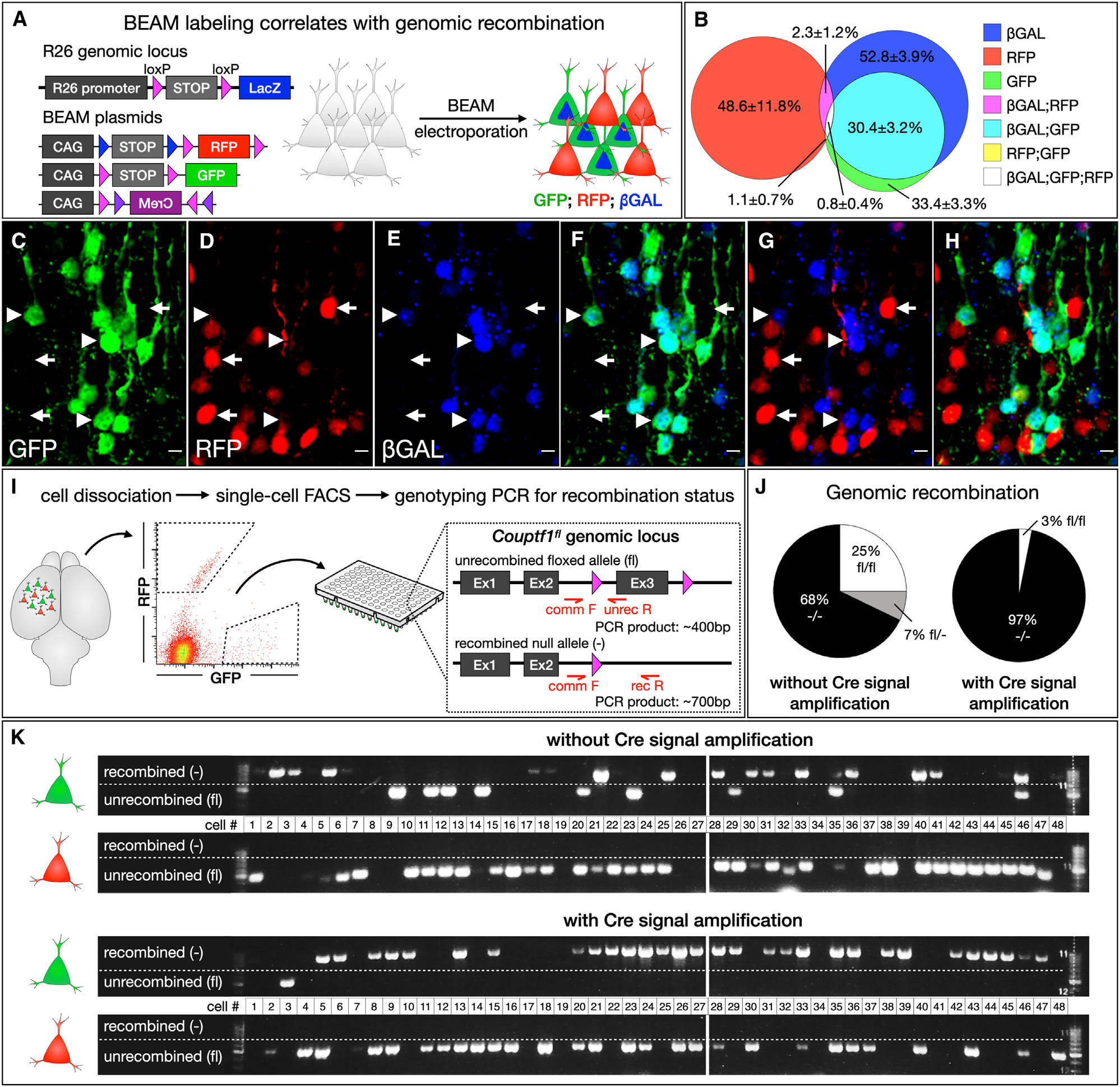
(A–H) BEAM electroporation into Rosa26loxP-STOP-loxP-LacZ mice (A). Most GFP-positive neurons are also β-GAL-positive (arrowheads in C–G), while most RFP-positive neurons lack β-GAL staining (arrows in C–G). Quantification (B, n = 3).
(I–K) BEAM electroporated neurons were dissociated, and single-cell sorting was performed into a 96-well plate (I). Single-cell genotyping PCR demonstrates that all RFP-positive neurons remain unrecombined. GFP-positive cells were recombined with considerably higher efficiency when FLEX(CreM) was used to amplify CRE activity (K). Quantification (J). Quantification shows mean ± SEM. Scale bars, 10 μm. See also Figure S3.
We also tested how closely excision of a floxed gene of interest correlates with fluorescent reporter expression by electroporating BEAM plasmids into the cerebral cortex of Couptf1fl/fl mice24,25 and analyzing the recombination status of GFP- and RFP-labeled cells (Figures 3I–3K). Electroporations were performed at E14.5, tissue was collected at P4 and dissociated, and individual neurons were then FACS purified into 96-well plates. Single-cell PCR was used to interrogate the recombination status of the Couptf1 locus in each isolated cell. Strikingly, we found that 96% of BEAM-electroporated green cells (31/32 cells) were fully recombined when recombinase amplification was used, while only 68% (19/28 cells) were fully recombined when the CAG-FLEX(CreM) plasmid was omitted, with 7/9 cells genotyping as unrecombined and 2/9 genotyping as having one unrecombined and one recombined copy of the allele. Importantly, all of the genotyped BEAM-electroporated red cells remained unrecombined (64/64 cells), indicating that there is no spontaneous CRE activity arising from the presence of CAG-FLEX-CreM in CRE-negative cells. Taken together, these data indicate that BEAM plasmids can be effectively used to investigate gene function by electroporation into genetically modified floxed mice.
Cell autonomy of gene function can be rigorously investigated using BEAM
As discussed above, dual-population mosaic analysis is a particularly powerful approach for investigating cell autonomy. When using BEAM, RFP-labeled (CRE-negative) cells provide an internal and spatially interspersed control for direct comparison to experimental GFP-labeled (CRE-positive) cells, eliminating pleiotropic effects present in global and even in tissue- or cell-type-specific mutants. As a proof-of-principle experiment, we reinvestigated the effect of Satb2 loss of function on the trajectory of callosal projection neuron (CPN) axons in Satb2fl/fl mice.26–28 In Satb2−/− mice, superficial-layer neuron axons fail to project toward the corpus callosum and project subcortically instead.29,30 In addition, these neurons abnormally activate expression of Ctip2, a transcriptional control over corticofugal projection neuron development that is specific to that population (though present later in interneurons at lower expression levels).29,31 Previous studies have proposed that these defects in axonal targeting are due to a cell-autonomous failure of callosal projection neuron subtype differentiation and excluded a number of alternative possibilities, including abnormalities in glial structures necessary for midline fusion.29
To further investigate the cell autonomy of this phenotype, BEAM plasmids were electroporated into the cerebral cortex of Satb2fl/fl mice at E14.5, and the resulting brains were examined at P7 (Figures 4A and S4A). We found that a number of GFP-labeled neurons undergo migrational arrest in the white matter (Figure S4K′), while their axons enter the internal capsule and fasciculate with other corticofugal axons (Figure S4L′), projecting into the thalamus and cerebral peduncle (Figures 4D′ and 4E′). Immediately adjacent RFP-positive control neurons migrate normally (Figure S4K) and do not project axons into the internal capsule (Figure S4L) or subcortical targets (Figures 4D and 4E). Electroporation of BEAM into wild-type mice resulted in GFP-positive neurons with unaltered phenotypes (Figures S4F–S4I). These results are similar to what has been observed in Fezf2 and Ctip2 overexpression experiments32,33 and reinforce that abnormal migration in Satb2−/− mice might result, at least in part, from aberrant and premature expression of Ctip2 and other molecular controls of subcerebral projection neuron differentiation, as previously proposed.29,32 Importantly, immunostaining for SATB2 and CTIP2 demonstrates that the majority of RFP-labeled neurons are SATB2 positive and CTIP2 negative (Figure 4G, quantification in 4I). Conversely, a large majority of GFP-labeled neurons lack SATB2 and have upregulated CTIP2 (Figures 4G and 4H, quantification in 4I). One surprising finding from these experiments is the large number of GFP-positive axons that project successfully across the corpus callosum in Satb2fl/fl brains (Figure S4M′, quantification in S4N). These data sharply contrast with the absolute failure of cortical neurons to project across the corpus callosum in Satb2−/− mice.29 Although recombination at the Satb2 locus is expected to fail in a small percentage of GFP-labeled neurons, these few escapees cannot account for the large number of GFP-labeled axons projecting across the corpus callosum. Therefore, it seems likely that axons of GFP-labeled Satb2−/− neurons, though impaired, are often able to fasciculate with the axons of unrecombined Satb2fl/fl neurons (both RFP positive and unelectroporated) to project successfully across the corpus callosum. These results further highlight the advantages of such mosaic genetic analysis via BEAM to elucidate cell-autonomous gene function.
Figure 4. BEAM enables investigation of cell-autonomous gene function by conditional deletion of floxed alleles and provides a platform for rapid in vivo screening of gene function by gain- and loss-of-function approaches.
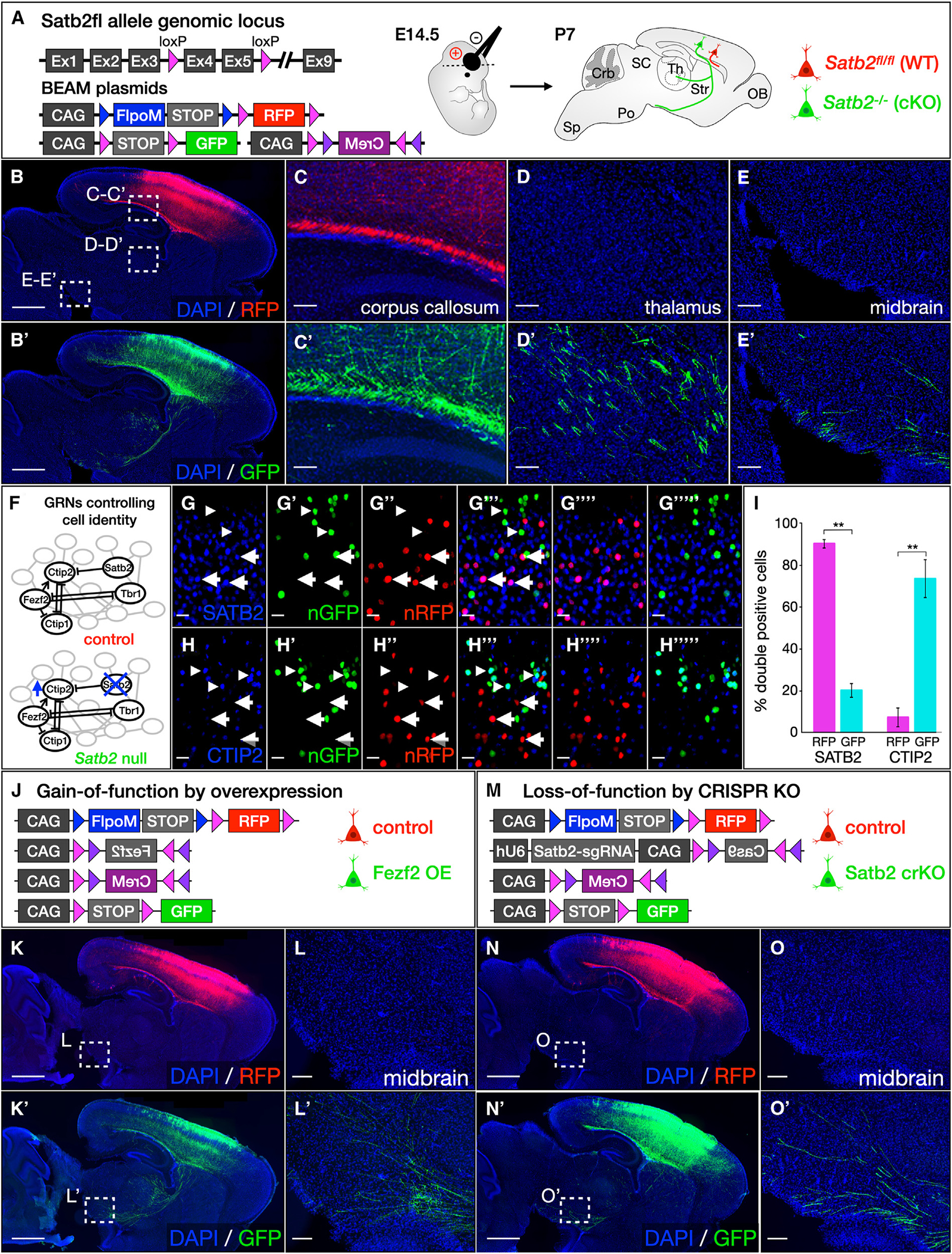
(A–E) Electroporation of BEAM into Satb2fl/fl mice demonstrates abnormal axon targeting by conditional-null neurons (A). Both RFP- and GFP-positive axons are present in the corpus callosum (C and C′), but only GFP-positive axons are redirected to the thalamus (D and D′) and midbrain (E and E′). These results were consistent across multiple replicates (n = 3 control, n = 3 Satb2fl/fl).
(F–I) Transcription factor network in Satb2 wild-type and conditional-null neurons (F). Immunolabeling for SATB2 demonstrates that GFP-positive neurons do not express SATB2 (arrowheads, G–G‴), while RFP-positive neurons do (arrows). Conversely, immunolabeling for CTIP2 is absent from RFP-positive neurons (arrows, H–H%‴), but is present in GFP-positive cells (arrowheads). Quantification (I, p < 0.01, n = 3 control, n = 3 Satb2fl/fl).
(J–L) Overexpression of Fezf2 in callosal projection neurons using BEAM (J). CRE-positive GFP-labeled neurons ectopically express Fezf2 due to activation of the CAG-FLEX(Fezf2) construct, causing them to extend aberrant projections to the thalamus and brain stem (K′ and L′) instead of the contralateral hemisphere. Intermingled control RFP-labeled cells project exclusively across the corpus callosum (K and L). These results were consistent across multiple replicates (n =3 control, n = 3 Fezf2 overexpression).
(M–O) CRISPR-Cas9 ablation of Satb2 in callosal projection neurons using BEAM (M). Although all neurons express guide RNAs targeting the Satb2 gene, Cas9 expression from a CAG-FLEX(Cas9) construct is activated by CRE exclusively in GFP-labeled cells, introducing mutations that inactivate Satb2 function and redirecting their axons from the corpus callosum to the thalamus and brain stem (N′ and O′). In the absence of Cas9 expression, intermingled control RFP-labeled neurons are unaffected by the guide RNA and project normally across the corpus callosum (N and O). These results were consistent across multiple replicates (n = 3 control, n =3 Satb2 crKO). Quantification shows mean ± SEM. Scale bars, 10 μm in (G) and (H); 100 μm in (C)–(E), (L), and (O); and 1,000 μm in (B), (K), and (N). See also Figure S4.
Rapid screening of gene function using BEAM
Internally controlled experiments provide a particularly attractive platform for functional screening of genes predicted to regulate a biological process of interest. For such an approach to be optimally versatile, it cannot rely solely on the use of genetically modified conditional knockout mice. We therefore sought to determine whether BEAM would be compatible with existing gain- and loss-of-function strategies mediated by the delivery of exogenous DNA into wild-type mice by transfection or transduction.
The most straightforward and broadly used gain-of-function strategy is to misexpress a gene of interest in a population of cells from which it is normally absent, using a strong ubiquitous promoter. As proof of principle, we generated a CAG-driven FLEX construct for the transcription factor Fezf2, which is normally expressed during cortical development by early-born corticofugal projection neurons that extend axons to the thalamus and brain stem, but excluded from late-born callosal projection neurons that extend axons across the corpus callosum to the contralateral hemisphere.32,34,35 Misexpression of Fezf2 by late-born callosal projection neurons causes them to redirect their axons to the thalamus and brain stem.32 We electroporated the CAG-FLEX-Fezf2 construct, along with the previously described BEAM plasmids, at E14.5 (Figure 4J) and found that control RFP-labeled axons project exclusively across the corpus callosum (Figures 4K and 4L). However, many Fezf2-overexpressing GFP-labeled axons were misrouted to the thalamus and brain stem (Figures 4K′ and 4L′), providing striking confirmation of previously published results.
For proof-of-principle genetic loss-of-function experiments, we focused on CRISPR-Cas9-mediated gene editing. We generated a bicistronic construct with a U6 promoter driving expression of a guide RNA and a CAG promoter driving expression of Cas9 in a CRE-dependent manner using FLEX (Figure 4M). To enable direct comparison to the loss-of-function experiments with gene deletion using a floxed allele, we cloned guide RNAs targeting the coding sequence of Satb236 and electroporated these constructs into developing cortex of wild-type mice at E14.5. Successful CAS9-mediated gene deletion was apparent from the aberrant migration of Satb2 CRISPR KO (crKO) GFP-positive neurons (Figures S4O and S4P) and further confirmed by loss of SATB2 immunostaining (Figure S4Q). Consistent with previously reported results,36 CAS9-mediated ablation of Satb2 expression resulted in many GFP-labeled axons aberrantly projecting to the thalamus and brain stem (Figures 4N′ and 4O′), while RFP-labeled axons projected exclusively across the corpus callosum (Figure 4N). These findings again rigorously confirm the prior reports, with BEAM’s mosaic genetic analysis highlighting the cell autonomy of Satb2 function.
BEAM can identify abnormalities in the timing of developmental processes and the organization of cells into complex tissues
In addition to providing a rigorous experimental approach for investigating cell autonomy, dual-population mosaic analysis is particularly well suited to uncovering phenotypes that are subject to substantial intertrial variability, such as the timing of developmental events. To illustrate this application, we used BEAM to investigate regulation of neuronal migration by Satb2 in the developing cerebral cortex. Superficial-layer neuron migration is severely disrupted in Satb2−/− mice, and few neurons generated at later stages of cortical development are able to reach their appropriate laminar position.29,30 We electroporated BEAM plasmids carrying nuclear-targeted RFP and GFP into cortical progenitor cells at E14.5 and collected brains for analysis 3 days later, at E17.5 (Figure 5A). In wild-type embryos, both GFP- and RFP-labeled neurons migrated efficiently into the cortical plate (Figure 5B). In Satb2fl/fl embryos, control RFP-labeled neurons still reached the cortical plate normally, but Satb2-null GFP-labeled neurons became stalled in the progenitor and intermediate zones, with few reaching the cortical plate (Figure 5C). To investigate whether this represents a delay or a complete failure of migration, we repeated the experiments, but analyzed the brains at P7. Interestingly, although a subset of Satb2-null GFP-labeled neurons underwent migrational arrest in the white matter and never entered the cortical plate, the majority of them joined control RFP-labeled neurons in layers II/III of the cortex and, in fact, migrated past them to reside in aberrantly superficial positions (Figures S5A and S5B). These direct wild-type and mutant population comparisons of interspersed neurons enable identification of subtle dynamic alterations during development and maturation.
Figure 5. BEAM readily identifies abnormal timing of developmental processes and shifts in the spatial distribution of cells.
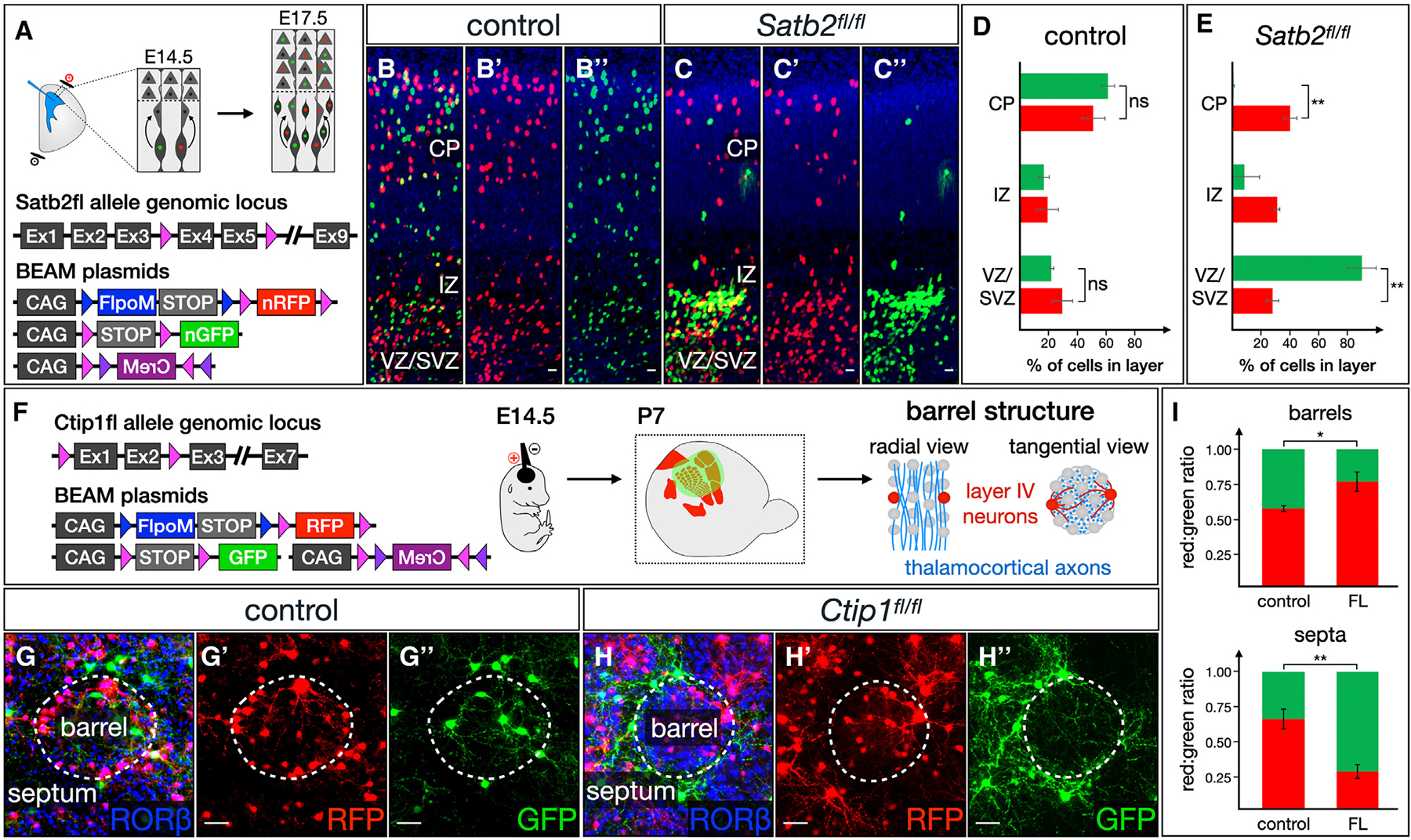
(A–E) BEAM reveals cell autonomous migrational delay of cortical neurons in the absence of Sabt2 function (A). While both RFP-positive and GFP-positive neurons migrate into the cortex at E17.5 following E14.5 electroporation in wild-type mice (B–B″), only RFP-positive control neurons migrate successfully into the cortex in Satb2fl/fl mice (C–C″). GFP-positive mutant neurons are stalled in the ventricular/subventricular zones (VZ/SVZ) and intermediate zone (IZ) in Satb2fl/fl mice (B″). (D and E) Quantification (p < 0.01, n = 3 control, n = 3 Satb2fl/fl).
(F–I) BEAM demonstrates abnormal spatial distribution of neurons lacking Ctip1 within the barrel field (F). Following electroporation at E14.5, both RFP- and GFP-labeled neurons integrate into barrels by P7 in wild-type mice, extending dendrites within the barrel to receive input from thalamocortical axons (G–G″). In Ctip1fl/fl mice, control RFP-labeled neurons adopt this normal configuration, while Ctip1-null GFP-positive neurons and their dendrites are excluded from barrels, instead taking up residence in the septa (H–H″). (I) Quantification (p < 0.05 for barrels, p < 0.01 for septa, n = 3 control, n =3 Ctip1fl/fl). Quantification shows mean ± SEM. Scale bars, 10 μm. See also Figure S5.
We next applied BEAM to investigate the extension of axons across the midline and into the contralateral hemisphere by callosal projection neurons, which has been reported to be delayed in the absence of Couptf1 function.37 BEAM plasmids were electroporated at E14.5, and brains were collected for analysis at P0, when the axons of late-born superficial-layer callosal projection neurons are still en route to the contralateral hemisphere. GFP-labeled axons behaved identically to control RFP-labeled axons in wild-type brains, crossing the corpus callosum and invading the contralateral hemisphere (Figures S5E–S5G). In Couptf1fl/fl brains, however, GFP-labeled mutant axons reached the corpus callosum, but were absent from the contralateral hemisphere (Figures S5H–S5J). The presence of an internal control in the same brains rules out the possibility of even subtly mismatched developmental stages across surgeries.
Another broad category of phenotypes that can be advantageously investigated using BEAM are those relating to the spatial distribution of cells. As proof of principle, we used BEAM to examine the organization of layer IV neurons in the somatosensory cortex of Ctip1fl/fl mice.38 In the vibrissal barrel field, thalamocortical input from each whisker condenses into distinct clusters, each relaying information from a single whisker to a cytoarchitecturally distinct cluster of layer IV neurons known as a barrel (Figure 5F). Ctip1 is necessary for proper differentiation of layer IV neurons, and their ability to organize into barrels is severely impaired in its absence.39 Electroporation of BEAM into wild-type cortex at E14.5 results in both RFP- and GFP-labeled neurons adopting an unbiased distribution intermingled across the barrel cortex, with higher density of neurons in the barrel walls and most dendrites extending toward the center of each barrel (Figure 5G). Electroporation of BEAM into Ctip1fl/fl brains leads to a similar overall distribution of control RFP-labeled cells. Strikingly, however, Ctip1 conditional-null GFP-labeled cells and their dendrites position themselves preferentially between barrels, segregating themselves in the septa (Figure 5H). The mosaic results highlight the dynamic nature of cellular migration and multicellular assembly into structurally complex tissues whose function depends on their precise histological organization.
BEAM controls for procedural variability enabling efficient investigation of cell proliferation and survival
Experiments to investigate cellular proliferation and survival can be difficult to interpret due to significant variability in the number of cells labeled across trials. To investigate whether BEAM can provide more robust measures of these phenotypes by enabling normalization across trials, we performed experiments involving manipulation of cortical progenitor proliferation. Previous studies have shown that the Wnt signaling pathway regulates the cell cycle in cortical progenitors, so that overexpression of β-catenin stimulates proliferation of cortical progenitors,40 while a dominant-negative mutant version of Tcf4 (Tcf4-DN) inhibits proliferation.41 We generated CAG-driven FLEX constructs for the expression of β-catenin-2A-H2B-EGFP or Tcf4-DN-2A-H2B-EGFP (Figure 6A). Using BEAM, we found a 25% ± 9% increase in the ratio of GFP:RFP-labeled cells in β-catenin electroporations relative to control experiments (Figures 6C and 6D, quantification in 6B), indicating increased proliferation or survival of cells overexpressing β-catenin. Conversely, the ratio of GFP:RFP-labeled cells in Tcf4-DN electroporations was decreased by 62% ± 15% relative to control experiments (Figures 6D and 6E, quantification in 6B), indicating decreased proliferation or survival of cells overexpressing Tcf4-DN. Importantly, without normalization, the trends toward more cells with overexpression of β-catenin and fewer cells with overexpression of Tcf4-DN were still present, but the differences across conditions were no longer statistically significant. Normalization to an internal control eliminates an important source of experimental variability, substantially increasing statistical power and enabling smaller changes to be detected without a large sample size.
Figure 6. BEAM enables unequivocal investigation of changes in mitotic activity and cell survival.
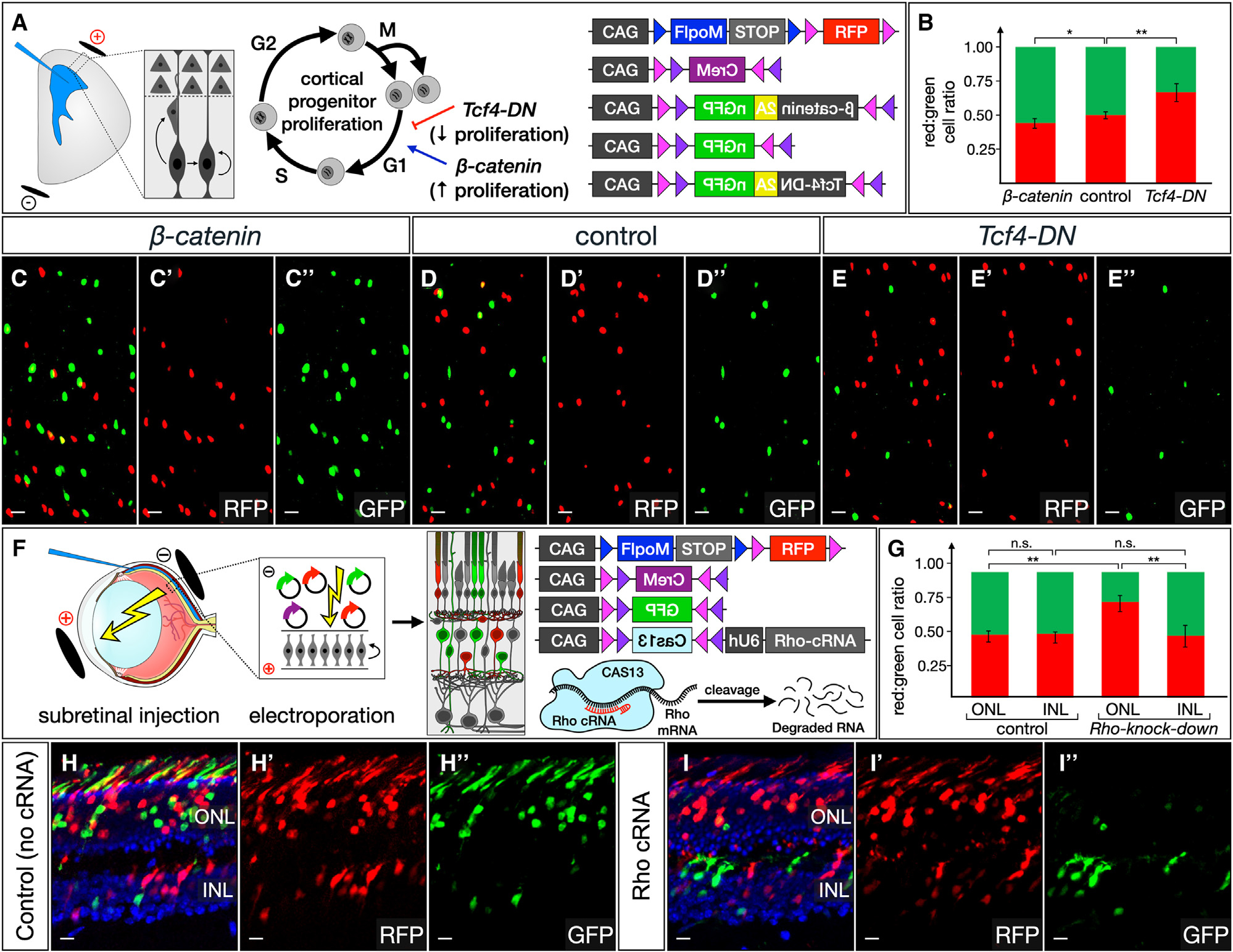
(A–E) Manipulation of cortical progenitor cell-cycle dynamics was carried out using BEAM and overexpression of either β-catenin or Tcf-DN (A). The relative ratios of control RFP- and genetically manipulated GFP-labeled cells after electroporation at E12.5 and analysis at E17.5 were quantified (B, p < 0.05 for β-catenin, p < 0.01 for Tcf4-DN, n = 3 control, n =3 β-catenin, n =3 TCF4-DN). Compared with control experiments (D–D″), β-catenin overexpression increases the relative number of GFP-labeled cells (C–C″), while a dominant-negative Tcf4 mutant decreases the relative number of GFP-labeled cells (E–E″).
(F–I) BEAM reveals selective cell death due to photoreceptor degeneration (F). Approximately equal numbers of RFP- and GFP-positive cells are present in control retinas in both the INL and the ONL. CAS13-mediated knockdown of Rho does not affect survival of cells in the INL; however, there is a striking decrease in GFP-positive cells in the ONL. Note also the complete absence of GFP-positive outer segments in Rho knockdown (I″), while there is robust GFP labeling of outer segments in control experiments (H″). Quantification (G, p < 0.01, n = 3 control, n =3 b-Rho knockdown). Quantification shows mean ± SEM. Scale bars, 10 μm. See also Figure S6.
Procedural variability in the number of cells labeled across experiments similarly complicates efforts to investigate cell survival. Here, we demonstrate that CAS13-mediated knockdown can be used to recapitulate a photoreceptor degeneration phenotype in the retina (Figure 6F). We focused on retinitis pigmentosa, the most common Mendelian degenerative retinopathy, which is caused by mutations in one or more genes important for photoreceptor development and function leading to progressive loss of rods and decreased vision. Rhodopsin-null mice were one of the earliest published mouse models of retinitis pigmentosa.42 In Rho−/− mice, rods fail to elaborate outer segments during development, and most photoreceptors are lost by 3 months of age. We first tested whether Cas13,43 together with an array of three guide RNAs, could induce efficient knockdown of Rho. In contrast to the extensive overlap of RHO immunostaining with the outer segments of electroporated photoreceptors in control experiments (Figures S6A and S6B), we found a complete lack of overlap when guide RNAs targeting Rho were used, indicating efficient knockdown (Figures S6C and S6D). In BEAM experiments, approximately equal ratios of RFP- and GFP-positive cells were present in the inner nuclear layer (INL), where electroporated bipolar cells, Müller glia, and amacrine cells reside, and in the outer nuclear layer (ONL), where photoreceptors reside (R:G ratio 1 in the INL and 0.94 in the ONL, no statistically significant difference; Figure 6H, quantification in 6G). Although in Rho-knockdown experiments the ratio of RFP- to GFP-positive cells remained approximately equal in the INL (R:G ratio 1.09, no statistically significant difference compared to control INL; Figure 6I, quantification in 6G), the ratio was strikingly changed in favor of RFP-positive cells in the ONL (R:G ratio 3.37, p < 0.01; Figure 6I, quantification in 6G), indicating a relative depletion of GFP-positive photoreceptors. Importantly, the few remaining GFP-positive cells in the ONL generally exhibit low levels of fluorescence, and no GFP-positive outer segments can be discerned in Rho-knockdown retinas. These results are consistent with the death of photoreceptors due to loss of Rho expression and, together with our experiments manipulating cell-cycle dynamics in cortical progenitors, illustrate the power of mosaic analysis using BEAM to investigate cell proliferation and survival.
Evaluating the protomap hypothesis of cortical area development at the level of individual radial units using BEAM
The protomap hypothesis of cortical development proposes that area identity is specified in cortical progenitors and that information is transferred to post-mitotic neurons in each radial unit.44,45 Manipulating morphogen activity or transcription factor function is sufficient to change the relative size and position of cortical areas.25,46,47 For instance, loss of Couptf1 function throughout the cerebral cortex results in a dramatic expansion of motor areas and a reciprocal reduction in the size of sensory areas (Figure 7A). However, it remains unknown whether area identity is independently programmed within each radial unit or is interdependent across adjacent radial units. To provide insights into this long-standing debate in the field of cortical development, we took advantage of the sophisticated investigation of cell autonomy made possible by BEAM by examining the arealization of adjacent control and Couptf1-null cortical columns.
Figure 7. Investigating the protomap hypothesis at the level of individual radial units reveals that area identity is independently specified in each progenitor cell and its progeny.
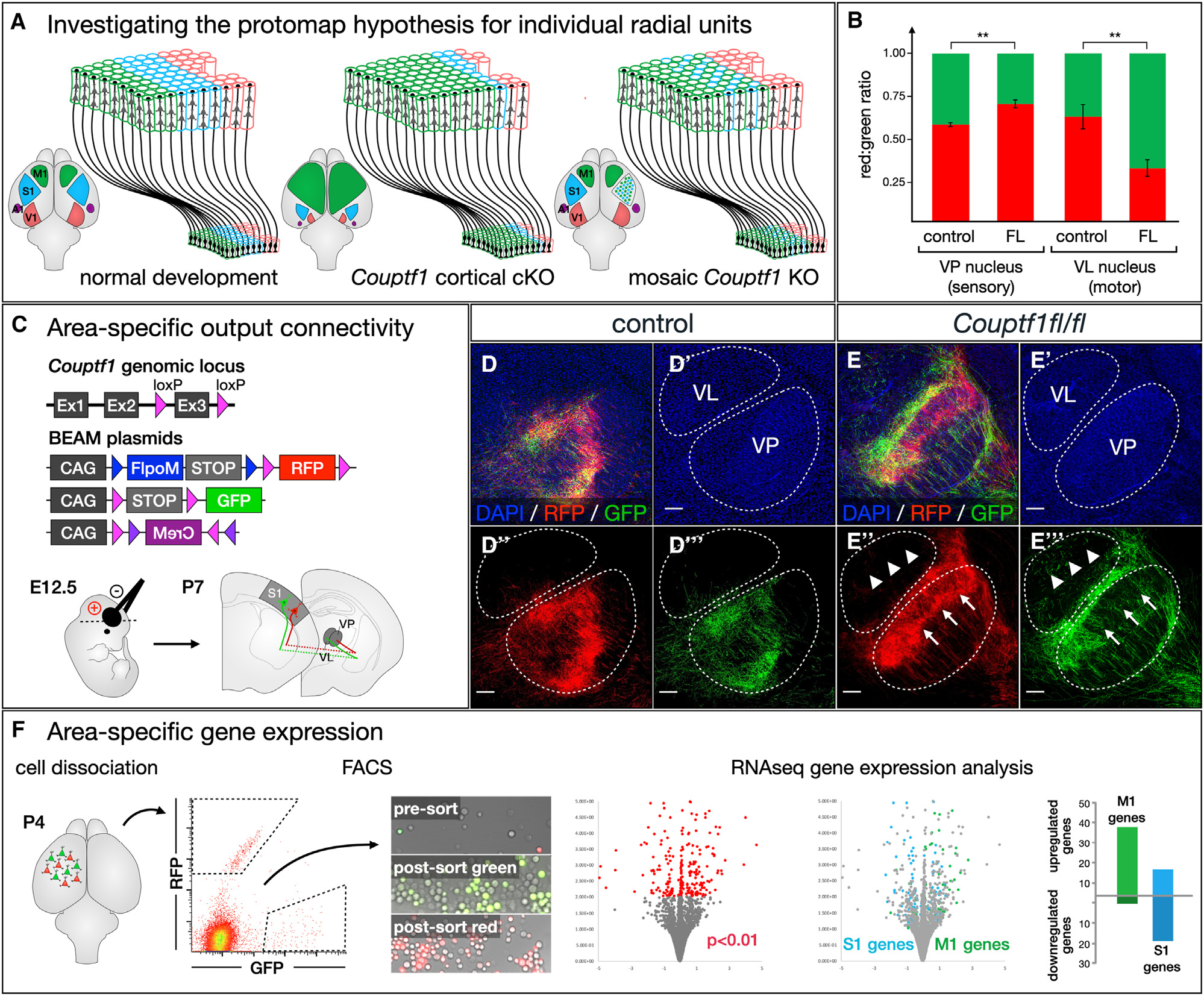
(A) The protomap hypothesis proposes that individual cortical progenitors acquire specific area identities and transfer this information to post-mitotic neurons generated by each radial unit. Cortex-wide loss of transcription factors that specify area identity leads to relative changes in the size and position of cortical areas. It is not well understood whether area specification occurs independently within each radial unit or whether there are mechanisms driving interdependent area specification of adjacent radial units.
(B–E) Each cortical area establishes axonal projections to specific thalamic nuclei, and this area-specific connectivity can be experimentally interrogated by electroporation of corticothalamic neurons at E12.5 (C). In control brains, both RFP- and GFP-labeled axons originating from somatosensory radial units project primarily to the ventral posterior (VP) sensory nucleus (D” and D‴). In Couptf1fl/fl brains, RFP-labeled axons from unrecombined radial units similarly arborize within the VP (E″, arrows), but GFP-labeled axons originating from intermingled Couptf1-null radial units avoid arborizing within the VP (E‴, arrows), instead entering and arborizing within the ventral lateral (VL) motor nucleus (E‴, arrowheads). Quantification (B, p < 0.01, n = 3 control, n = 3 Couptf1fl/fl).
(F) Couptf1fl/fl brains were electroporated at E12.5, then cortical tissue was collected from the somatosensory cortex at P4 and dissociated. Control RFP-labeled cells and conditional-null GFP-labeled cells were purified by FACS, and gene expression was analyzed by RNA-seq. Among differentially expressed genes, motor-specific genes were upregulated, while sensory-specific genes were downregulated. Quantification shows mean ± SEM. Scale bars, 100 μm. See also Figure S7 and Data S1.
We first investigated output connectivity. Each cortical area establishes projections to specific thalamic nuclei, with somatosensory cortex projecting mainly to the ventral posterior nucleus (VP) and motor cortex projecting primarily to the ventral anterior and ventral lateral nuclei (VA and VL). We electroporated BEAM into somatosensory cortex of wild type and Couptf1fl/fl mice at E12.5, when corticothalamic projection neurons are being generated (Figure 7C). In control electroporations, both RFP- and GFP-labeled axons projected to the VP and arborized with a similar distribution (Figure 7D, quantification in 7B). Electroporation into Couptf1fl/fl brains also resulted in RFP-labeled axons that projected and arborized primarily within the VP, while GFP-labeled axons arising from intermingled Couptf1 conditional-null radial units largely avoided arborizing within the VP and shifted toward arborizing in the VL instead (Figure 7E, quantification in 7B).
Another defining feature of neurons in different cortical areas is their unique gene expression pattern. In parallel experiments, we microdissected BEAM-electroporated Couptf1fl/fl somatosensory cortex at P3, dissociated the tissue, and FACS purified RFP- and GFP-labeled cells arising from intermingled radial units in the same brain. We then performed RNA-sequencing (RNA-seq) gene expression analysis and identified differentially expressed transcripts. By using previously published datasets of motor- and somatosensory-specific genes (Figure S7A),39 we showed that 74 of 201 differentially expressed genes in these BEAM Couptf1 loss-of-function experiments are area specific, with overall upregulation of motor-specific genes and downregulation of sensory-specific genes (Figure 7F and Data S1). Gene ontology indicated that genes differentially expressed between Couptf1 wild-type and conditional-null neurons regulate developmental processes critical for controlling axon targeting and circuit assembly (Figure S7B). Taken together, these data demonstrate that area-specific output connectivity and gene expression are independently determined in neighboring radial units.
These results highlight that BEAM not only flexibly enables new investigations with added discovery power, rigor, ease, and comparative insight, but also enables elucidation of prior work and observations at more detailed levels of mechanism and cell autonomy. Even subtle effects from molecular and cellular manipulations are placed in direct contrast by comparison with fully intermingled cells in exactly the same area of any tissue or organ system under investigation.
DISCUSSION
BEAM comprises a flexible and powerful approach for molecular functional analyses that relies on combinatorial recombinase activation to generate two genetically distinct, fully interspersed, fluorescently labeled populations of cells for comparative analysis in the same spatial domain of the same tissue and animal. For many experimental paradigms, BEAM offers critical advantages over genome-based tools (Table S1). In addition, compared with existing plasmid-based tools, BEAM enables sharper and earlier delineation of control and experimental cells, it is validated for a broad range of experimental manipulations (including floxed alleles, overexpression, Cas9-mediated knockout, and Cas13-mediated knockdown), and it has been shown to be compatible with multiple DNA-delivery strategies (chemical and physical transfection as well as viral transduction).
BEAM enables improved reproducibility of genetic manipulation experiments
Reproducibility and replicability are critical for rigorous interpretation of experimental data and have been an area of significant interest and concern for the scientific community for many years. More recently, the US National Institutes of Health (NIH) have developed action plans to address these concerns. As one notable example, studies of spinal cord injury have suffered from a lack of replicability, a problem brought into sharp focus by the NINDS-sponsored FORE-SCI project.48 One likely explanation for failures to replicate across studies, as well as the resultant, often erroneous, conclusions, is the variability across trials in the severity of experimentally induced injuries and in the level of genetic or viral manipulation. The ability to analyze experimental and control cells in the same animal enables control for variable severity of injury, degree of genetic or viral manipulation, timing, and other experimental variables. Although we focus here on developmental phenotypes, we demonstrate that application of BEAM for internally controlled experiments substantially reduces procedural variability and significantly enhances the ability to detect or rule out genuine effects. Therefore, application of BEAM would enable more rigorous evaluation of therapeutic benefits in spinal cord injury regeneration studies and increase reproducibility more broadly across a variety of fields.
Applications of BEAM beyond analysis of gene function
Here, we focus on characterizing the BEAM system for investigations of gene function. However, introducing different effector molecules into two distinct cell populations creates opportunities for many other experimental paradigms. Application of channel rhodopsins (ChRs)49,50 or designer receptors exclusively activated by designer drugs (DREADDs)51,52 can allow independent stimulation or silencing of specific proportions of neurons in sequence or simultaneously. Anterograde or retrograde synaptic tracing using tetanus toxin C-terminal fragment (TTC)53,54 or wheat germ agglutinin (WGA)55,56 tagged with distinct epitopes or fluorescent proteins would enable comparative connectomics. In several regenerative paradigms, it might be desirable to instruct production of multiple distinct cell types from a single population of endogenous progenitors, and BEAM could facilitate programming subsets of progenitors toward divergent cell-type-specification pathways. Finally, although this current iteration of the BEAM system is not well suited to clonal analysis, this application could be enabled by combining BEAM with transposon-based genomic integration and activation, for instance, using a leak-proof integration-coupled On (LiOn) system.57
Limitations of the study
Although mosaic analysis eliminates multiple sources of experimental variability, it is important to remain mindful of biases inherent in the generation of genetically distinct populations of cells using the BEAM recombinase system. Whether any individual cell becomes a control cell or an experimental cell depends on the presence or absence of Cre, which is a stochastic event on a cell-by-cell level. However, on a population level, cells that receive a higher copy number of plasmids or viral genomes are also more likely to receive a copy of Cre, which is introduced at a lower dose. The average number of plasmids or viral genomes present in experimental CRE-positive cells is therefore higher than the average number present in control CRE-negative cells (Figure 1A). Moreover, which cells take up a higher copy number of plasmids or viral genomes is unlikely to be entirely random. For example, some cortical progenitor subtypes might be electroporated more efficiently, based on proximity to the lateral ventricle, cell morphology, or expression of specific cell-membrane proteins. Viral transduction efficiency varies widely across different neuron and glial cell types, as determined by the tropism of particular viral serotypes. Cells carrying higher copy numbers of plasmid or viral genomes might respond to the additional foreign DNA with stronger activation of immune cell signaling pathways. In addition, high levels and prolonged Cre expression have in some instances been reported to cause toxicity.58 With these considerations in mind, we recommend that, before transitioning to internally controlled experiments, investigators first establish baseline controls to exclude any significant differences between CRE-negative and CRE-positive cells in the absence of any other manipulations. We do so in all of our discovery experiments and have not found any differences between CRE-negative and CRE-positive cells with respect to any of the phenotypes analyzed in this paper.
In summary, we have developed and validated a highly flexible system for mosaic genetic analysis in mice that will be useful for studying a broad range of biological questions in diverse fields of study. The BEAM system is ideal for investigation of cell autonomy, as demonstrated here by our analysis of multiple cellular phenotypes controlled by transcription factors during cortical development. The sophistication and precision of mosaic genetic analysis enabled by BEAM allowed us to directly test the long-standing cortical protomap hypothesis, revealing that radial unit identity is specified cell autonomously at the level of individual radial units. In addition, BEAM should bring exceptional clarity to subtle biological questions that are difficult to address definitively with existing methods due to experimental variability. This work lays the foundation for a new generation of progressively more sophisticated recombinase-based tools for mosaic genetic analysis in mice that will significantly increase capabilities for discovery and experimental manipulation across a wide range of experimental systems and fields of study.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jeffrey Macklis (jeffreymacklis@fas.harvard.edu).
Materials availability
Plasmids generated in this study will be deposited to Addgene.
Data and code availability
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
| Mouse anti-SATB2 | Abcam | RRID:AB_882455 |
| Chicken anti-B-GAL | Abcam | RRID:AB_10899635 |
| Rabbit anti-GFP | Molecular Probes | RRID:AB_221569 |
| Rat anti-CTIP2 | Abcam | RRID:AB_2064130 |
| Rabbit anti-RORB | H. Stunnenberg | RRID:AB_2566817 |
|
Critical commercial assays | ||
| SMART-Seq v4 Ultra Low Input RNA kit | Takara | 634888 |
| RNeasy kit | Qiagen | 74104 |
|
Deposited data | ||
| Couptf1 BEAM RNAseq was deposited in GEO data repository | This paper | Accession # GSE271632 |
|
Experimental models: cell lines | ||
| HEK-293T | ATCC | CRL-3216 |
|
Experimental models: organisms/strains | ||
| Satb2fl/fl | Savarese et al.26 | RRID:MGI_6363495 |
| Couptf1fl/fl | Armentano et al.24,25 | RRID:MGI_5523158 |
| Ctip1 fl/fl | Sankaran et al.38 | RRID:MGI_4358088 |
| Rosa26-loxP-STOP-loxP-LacZ | JAX | RRID:MGI_2164639 |
|
Oligonucleotides | ||
| Satb2 sgRNA: GTCTCGCACAAAGATCTCTTTGG |
Shinmyo et al.36 | N/A |
| Rho Cas13 guide 1: GGTCTCCGTCTTGGAAGCGGTGGCAGAGGC |
This paper | N/A |
| Rho Cas13 guide 2: GTCGTCATCTCCCAGTGGATTCTTGCCGCA |
This paper | N/A |
| Rho Cas13 guide 3: GCACAGCGTGGTGAGCATACAGTTCCGGAA |
This paper | N/A |
|
Recombinant DNA | ||
| pBeatrix | Addgene | Plasmid #167528 |
| pDouble-Up | Addgene | Plasmid #125134 |
| BEAM plasmids will be deposited with Addgene | This paper | Plasmid # pending |
|
Software and algorithms | ||
| ImageJ | Schneider et al.59 | N/A |
| Tuxedo tools | Trapnell et al.60 | N/A |
| PANTHER overrepresentation test | Mi et al.61 | N/A |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
All mouse studies were approved by the Harvard University IACUC, and were performed in accordance with institutional and federal guidelines. The date of vaginal plug detection was designated E0.5, and the day of birth as P0. The influence and association with sex was not analyzed because these studies are focused on method development rather than biological discovery. Moreover, the genetic pathways studied relate to basic mechanisms of brain development that are unlikely to exhibit sexual dimorphism as they regulate specification of cell identities and circuit formation that are equivalent in males and females.
METHOD DETAILS
Construct assembly
All constructs for constitutive mammalian expression were derived by subcloning into the XhoI and StuI sites of CBIG (gift of C. Lois), which contains an abelson murine leukemia virus (A-MuLV) Psi packaging element, a CMV/beta-actin promoter, a woodchuck hepatitis virus post-transcriptional regulatory element (WPRE), and an A-MuLV long terminal repeat (LTR). Various configurations of loxP, lox2272, frt and F5 sites were designed in silico, produced by GeneArt using gene synthesis, and subcloned into the CBIG backbone. The coding sequences for EGFP or tdTomato were then subcloned, as appropriate. The coding sequence for CreM was obtained from Addgene (Kaczmarczyk and Green JE, 2001; Addgene plasmid #8395). FlpOM was generated by introducing the human b-globin intron from CreM into a codon optimized Flp (Raymond and Soriano, 2007; Addgene plasmid # 13792), interrupting the coding sequence between amino acids 159 and 160. Cas9, the U6 promoter and sgRNA sequences were subcloned from the pX330 plasmid (Addgene #42230)62 and the Satb2-specific guides have been previously described36. Cas13, the U6 promoter and guide arrays were subcloned from pXR plasmids (Addgene plasmids # 109049 and 10905443). All plasmids generated for this study will be shared on Addgene.
Mice
All mouse studies were approved by the Harvard University IACUC, and were performed in accordance with institutional and federal guidelines. The date of vaginal plug detection was designated E0.5, and the day of birth as P0. Satb2fl/fl mice were generated by Grosschedl and colleagues26,27, and were generously provided by Susan McConnell. Couptf1fl/fl mice were generated and generously shared by Studer and colleagues25,63. Ctip1fl/fl mice were generated by Tucker and colleagues38 and generously shared by Orkin and colleagues. Rosa26loxP-STOP-loxP-LacZ mice (stock number 003474) were purchased from Jackson Laboratories.
Immunocytochemistry
Mice were transcardially perfused with 4% paraformaldehyde, and brains were dissected and post-fixed at 4°C overnight in 4% paraformaldehyde. Tissue was sectioned at 50um on a vibrating microtome (Leica). Non-specific binding was blocked by incubating tissue and antibodies in 8% goat serum/0.3% bovine serum albumin in phosphate-buffered saline.
Imaging
For epifluorescence microscopy, tissue sections were imaged using an Eclipse 90i microscope (Nikon Instruments) with a mounted CCD camera (ANDOR Technology). For confocal microscopy, cells were imaged on an inverted LSM 880 microscope (Zeiss) at the Harvard Center for Biological Imaging. Quantification of fluorescence intensity was carried out using ImageJ59.
Cell culture and transfection
Human embryonic kidney (HEK) 293T cells were obtained from the ATCC and grown in DMEM (Gibco) containing 10% fetal bovine serum (Seradigm), 5000U/ml penicillin, and 5g/ml streptomycin (Invitrogen). Transfection was performed using Lipofectamine 2000 (Invitrogen) following the protocol provided by the manufacturer.
In vivo electroporation
Surgeries were performed as previously described32. Briefly, plasmids were microinjected into the lateral ventricle of developing embryos under ultrasound guidance, and electroporated into cortical progenitors using a CUY21EDIT electroporator (Nepa Gene, Japan) set to deliver five 30V pulses of 50ms, separated by 950ms intervals. Retina electroporations were also carried out as previously described 64. Briefly, plasmids were microinjected subretinally under direct visualization using a FemtoJet 4i microinjector, and electroporated into retinal progenitors using an NEPA21 electroporator (Nepa Gene, Japan) set to deliver five 50ms pulses of 80V each at 950ms intervals. For BEAM electroporation experiments all plasmids were used at 1ug/ul each, except for CAG-Cre, which was used at 75–125ng/ul (the exact concentration necessary for approximately equal numbers of GFP and RFP labeled cells varies slightly across DNA preps).
AAV labeling
All virus work was approved by the Harvard Committee on Microbiological Safety, and conducted according to institutional guidelines. We obtained a pAAV-CAG-EGFP construct from the MGH Virus Core, which contains the following elements flanked by AAV2 ITRs: a CMV/beta-actin promoter, the coding sequence for EGFP, the woodchuck hepatitis virus post-transcriptional regulatory element (WPRE), a bovine GH pA signal, and an SV40 pA signal. Mosaic expression pAAV constructs were generated by replacing the EGFP coding sequence in pAAV-CAG-EGFP with the various BEAM modules described above. Constructs were packaged and serotyped with the AAV2/1 capsid protein by the MGH Virus Core. Neurons were labeled by pressure injection of virus under ultrasound guidance at P1, and brains were collected for analysis at P14. All AAV plasmids generated for this study will be shared on Addgene.
Fluorescence activated cell sorting
Flow cytometry analysis and FACS purification were performed on a MoFlo Astrios cell sorter (Beckman Coulter) at the Bauer Flow Cytometry Core Facility. Cells were initially gated based on forward and side scatter. Fluorescence gates were set relative to negative controls. Plots in Figure 2C were downsampled to display 1000 fluorescently-labeled cells in each condition, in order to enable direct comparison across samples.
RNA sequencing
Parietal (somatosensory) cortex was dissected from BEAM-electroporated Couptf1fl/fl brains, acutely dissociated as previously described65, and FACS purified on a MoFlo Astrios cell sorter at the Harvard Bauer flow cytometry core. Three replicates consisting of 10,000 RFP- or GFP-labeled cells each were obtained from independently electroporated brains. RNA was isolated using RNeasy kit (Qiagen), and libraries were prepared using the SMART-Seq v4 Ultra Low Input RNA kit (TaKaRa). Sequencing was performed on an Illumina NextSeq at the Harvard Bauer core facility. Data were analyzed using Tuxedo tools60. Gene ontology category enrichment analysis was performed using the PANTHER overrepresentation test61.
QUANTIFICATION AND STATISTICAL ANALYSIS
Littermate pairs of experimental and control mice were collected at the age indicated in the figure and processed as for immunocytochemistry. Anatomically matched sections from each mouse were selected, and single confocal slices were imaged. For quantification of cells positive for specific markers, each marker was counted within a box of predefined size spanning the radial thickness of cortex, with the same size of box applied to control and experimental images. For quantification of axonal projections, fluorescence intensity was quantified using ImageJ with the same size of box applied to control and experimental images. Graphs show the mean value with error bars denoting the standard error of the mean. Statistical analyses were conducted using unpaired two-tailed t tests in Microsoft Excel, with a significance threshold of p < 0.05.
Supplementary Material
Highlights.
BEAM analysis reliably, sharply, and rapidly delineates control and experimental cells
BEAM controls for cell autonomy, proliferation/survival, and procedural variability
BEAM is validated with floxed alleles, overexpression, Cas9 knockout, and Cas13 knockdown
BEAM testing of the protomap hypothesis reveals area specification in individual radial units
ACKNOWLEDGMENTS
We thank S. Dymecki, S. Ross, R. Grosschedl, S. McConnell, and M. Studer for generous sharing of mice and reagents; P. Davis, E. Gillis-Buck, M. Wettstein, and B. Wall for technical assistance; J. Flanagan, S. Dymecki, L. Goodrich, and H. Padmanabhan for scientific discussions; and members of the Macklis lab for helpful suggestions. This work was supported by National Institutes of Health grant R21 NS104733 to J.D.M., with additional infrastructure support from NIH DP1 NS106665, NS075672, NS045523, NS104055, and NS049553; the Max and Anne Wien Professor of Life Sciences fund; and the Emily and Robert Pearlstein Fund for Nervous System Repair. L.C.G. was partially supported by the Harvard Medical Scientist Training Program, NIH individual predoctoral National Research Service Award NS080343, and the DEARS Foundation (to J.D.M.). M.B.W. was partially supported by NIH individual predoctoral National Research Service Award NS064730 and the DEARS Foundation (to J.D.M.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2024.114650.
REFERENCES
- 1.Adams D, Baldock R, Bhattacharya S, Copp AJ, Dickinson M, Greene NDE, Henkelman M, Justice M, Mohun T, Murray SA, et al. (2013). Bloomsbury report on mouse embryo phenotyping: recommendations from the impc workshop on embryonic lethal screening. Dis. Model. Mech. 6, 571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han KA, Woo D, Kim S, Choii G, Jeon S, Won SY, Kim HM, Heo WD, Um JW, and Ko J (2016). Neurotrophin-3 regulates synapse development by modulating trkc-ptps synaptic adhesion and intracellular signaling pathways. J. Neurosci. 36, 4816–4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Villalba A, Amberg N, and Hippenmeyer S (2023). Interplay of cell-autonomous gene function and tissue-wide mechanisms regulating radial glial progenitor lineage progression. Neocortical Neurogenesis in Development and Evolution, 169–191. 10.1002/9781119860914. [DOI] [Google Scholar]
- 4.Zong H, Espinosa JS, Su HH, Muzumdar MD, and Luo L (2005). Mosaic analysis with double markers in mice. Cell 121, 479–492. [DOI] [PubMed] [Google Scholar]
- 5.Contreras X, Amberg N, Davaatseren A, Hansen AH, Sonntag J, Andersen L, Bernthaler T, Streicher C, Heger A, Johnson RL, et al. (2021). A genome-wide library of madm mice for single-cell genetic mosaic analysis. Cell Rep. 35, 109274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lao Z, Raju GP, Bai CB, and Joyner AL (2012). Mastr: a technique for mosaic mutant analysis with spatial and temporal control of recombination using conditional floxed alleles in mice. Cell Rep. 2, 386–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu H-T, Han Z, Gao P, He S, Li Z, Shi W, Kodish O, Shao W, Brown KN, Huang K, and Shi SH (2014). Distinct lineage-dependent structural and functional organization of the hippocampus. Cell 157, 1552–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao P, Postiglione MP, Krieger TG, Hernandez L, Wang C, Han Z, Streicher C, Papusheva E, Insolera R, Chugh K, et al. (2014). Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell 159, 775–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu C, Sage JC, Miller MR, Verhaak RGW, Hippenmeyer S, Vogel H, Foreman O, Bronson RT, Nishiyama A, Luo L, and Zong H (2011). Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell 146, 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hippenmeyer S, Youn YH, Moon HM, Miyamichi K, Zong H, Wynshaw-Boris A, and Luo L (2010). Genetic mosaic dissection of lis1 and ndel1 in neuronal migration. Neuron 68, 695–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Espinosa JS, Wheeler DG, Tsien RW, and Luo L (2009). Uncoupling dendrite growth and patterning: single-cell knockout analysis of nmda receptor 2b. Neuron 62, 205–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torii M, Rakic P, and Levitt P (2013). Role of epha/ephrin-a signaling in the development of topographic maps in mouse corticothalamic projections. J. Comp. Neurol. 521, 626–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galazo MJ, Emsley JG, and Macklis JD (2016). Corticothalamic projection neuron development beyond subtype specification: Fog2 and intersectional controls regulate intraclass neuronal diversity. Neuron 91, 90–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang YS, and Hughes TE (2001). Cre stoplight: a red/green fluorescent reporter of cre recombinase expression in living cells. Biotechniques 31, 1036–1041. [DOI] [PubMed] [Google Scholar]
- 15.Schnütgen F, Doerflinger N, Calléja C, Wendling O, Chambon P, and Ghyselinck NB (2003). A directional strategy for monitoring cre-mediated recombination at the cellular level in the mouse. Nat. Biotechnol. 21, 562–565. [DOI] [PubMed] [Google Scholar]
- 16.Muzumdar MD, Tasic B, Miyamichi K, Li L, and Luo L (2007). A global double-fluorescent cre reporter mouse. genesis 45, 593–605. [DOI] [PubMed] [Google Scholar]
- 17.Franco SJ, Gil-Sanz C, Martinez-Garay I, Espinosa A, Harkins-Perry SR, Ramos C, and Müller U (2012). Fate-restricted neural progenitors in the mammalian cerebral cortex. Science 337, 746–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor RJ, Carrington J, Gerlach LR, Taylor KL, Richters KE, and Dent EW (2020). Double up: A dual color, internally controlled platform for in utero knockdown or overexpression. Front. Mol. Neurosci. 13, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trovato F, Parra R, Pracucci E, Landi S, Cozzolino O, Nardi G, Cruciani F, Pillai V, Mosti L, Cwetsch AW, et al. (2020). Modelling genetic mosaicism of neurodevelopmental disorders in vivo by a cre-amplifying fluorescent reporter. Nat. Commun. 11, 6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen RN, van der Aa MAEM, Macaraeg N, Lee AP, and Szoka FC Jr. (2009). Quantification of plasmid dna copies in the nucleus after lipoplex and polyplex transfection. J. Control. Release 135, 166–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, and Lichtman JW (2007). Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62. [DOI] [PubMed] [Google Scholar]
- 22.Sohal VS, Zhang F, Yizhar O, and Deisseroth K (2009). Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature 459, 698–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soriano P (1999). Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 24.Armentano M, Filosa A, Andolfi G, and Studer M (2006). Coup-tfi is required for the formation of commissural projections in the forebrain by regulating axonal growth. Development 133, 4151–4162. [DOI] [PubMed] [Google Scholar]
- 25.Armentano M, Chou S-J, Tomassy GS, Leingärtner A, O’Leary DD, and Studer M (2007). Coup-tfi regulates the balance of cortical patterning between frontal/motor and sensory areas. Nat. Neurosci. 10, 1277–1286. [DOI] [PubMed] [Google Scholar]
- 26.Savarese F, Dávila A, Nechanitzky R, De La Rosa-Velazquez I, Pereira CF, Engelke R, Takahashi K, Jenuwein T, Kohwi-Shigematsu T, Fisher AG, and Grosschedl R (2009). Satb1 and satb2 regulate embryonic stem cell differentiation and nanog expression. Genes Dev. 23, 2625–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srinivasan K, Leone DP, Bateson RK, Dobreva G, Kohwi Y, Kohwi-Shigematsu T, Grosschedl R, and McConnell SK (2012). A network of genetic repression and derepression specifies projection fates in the developing neocortex. Proc. Natl. Acad. Sci. USA 109, 19071–19078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leone DP, Heavner WE, Ferenczi EA, Dobreva G, Huguenard JR, Grosschedl R, and McConnell SK (2015). Satb2 regulates the differentiation of both callosal and subcerebral projection neurons in the developing cerebral cortex. Cereb. Cortex 25, 3406–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Fariñas I, Grosschedl R, and McConnell SK (2008). Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron 57, 364–377. [DOI] [PubMed] [Google Scholar]
- 30.Britanova O, de Juan Romero C, Cheung A, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, Mitkovski M, Agoston D, et al. (2008). Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron 57, 378–392. [DOI] [PubMed] [Google Scholar]
- 31.Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, and Macklis JD (2005). Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron 45, 207–221. [DOI] [PubMed] [Google Scholar]
- 32.Molyneaux BJ, Arlotta P, Hirata T, Hibi M, and Macklis JD (2005). Fezl is required for the birth and specification of corticospinal motor neurons. Neuron 47, 817–831. [DOI] [PubMed] [Google Scholar]
- 33.Chen B, Wang SS, Hattox AM, Rayburn H, Nelson SB, and McConnell SK (2008). The fezf2–ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc. Natl. Acad. Sci. USA 105, 11382–11387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen B, Schaevitz LR, and McConnell SK (2005). Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc. Natl. Acad. Sci. USA 102, 17184–17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J-G, Rašin M-R, Kwan KY, and Šestan N (2005). Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc. Natl. Acad. Sci. USA 102, 17792–17797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shinmyo Y, Tanaka S, Tsunoda S, Hosomichi K, Tajima A, and Kawasaki H (2016). Crispr/cas9-mediated gene knockout in the mouse brain using in utero electroporation. Sci. Rep. 6, 20611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alfano C, Viola L, Heng JI-T, Pirozzi M, Clarkson M, Flore G, De Maio A, Schedl A, Guillemot F, and Studer M (2011). Coup-tfi promotes radial migration and proper morphology of callosal projection neurons by repressing rnd2 expression. Development 138, 4685–4697. [DOI] [PubMed] [Google Scholar]
- 38.Sankaran VG, Xu J, Ragoczy T, Ippolito GC, Walkley CR, Maika SD, Fujiwara Y, Ito M, Groudine M, Bender MA, et al. (2009). Developmental and species-divergent globin switching are driven by bcl11a. Nature 460, 1093–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greig LC, Woodworth MB, Greppi C, and Macklis JD (2016). Ctip1 controls acquisition of sensory area identity and establishment of sensory input fields in the developing neocortex. Neuron 90, 261–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chenn A, and Walsh CA (2003). Increased neuronal production, enlarged forebrains and cytoarchitectural distortions in β-catenin overexpressing transgenic mice. Cereb. Cortex 13, 599–606. [DOI] [PubMed] [Google Scholar]
- 41.Woodhead GJ, Mutch CA, Olson EC, and Chenn A (2006). Cell-autonomous β-catenin signaling regulates cortical precursor proliferation. J. Neurosci. 26, 12620–12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Humphries MM, Rancourt D, Farrar GJ, Kenna P, Hazel M, Bush RA, Sieving PA, Sheils DM, McNally N, Creighton P, et al. (1997). Retinopathy induced in mice by targeted disruption of the rhodopsin gene. Nat. Genet. 15, 216–219. [DOI] [PubMed] [Google Scholar]
- 43.Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, and Hsu PD (2018). Transcriptome engineering with rna-targeting type vi-d crispr effectors. Cell 173, 665–676.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rakic P (1988). Specification of cerebral cortical areas. Science 241, 170–176. [DOI] [PubMed] [Google Scholar]
- 45.Rakic P, Ayoub AE, Breunig JJ, and Dominguez MH (2009). Decision by division: making cortical maps. Trends Neurosci. 32, 291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fukuchi-Shimogori T, and Grove EA (2001). Neocortex patterning by the secreted signaling molecule fgf8. Science 294, 1071–1074. [DOI] [PubMed] [Google Scholar]
- 47.O’Leary DD, Chou S-J, and Sahara S (2007). Area patterning of the mammalian cortex. Neuron 56, 252–269. [DOI] [PubMed] [Google Scholar]
- 48.Steward O, Popovich PG, Dietrich WD, and Kleitman N (2012). Replication and reproducibility in spinal cord injury research. Exp. Neurol. 233, 597–605. [DOI] [PubMed] [Google Scholar]
- 49.Berndt A, Lee SY, Ramakrishnan C, and Deisseroth K (2014). Structure-guided transformation of channelrhodopsin into a light-activated chloride channel. Science 344, 420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deisseroth K (2015). Optogenetics: 10 years of microbial opsins in neuroscience. Nat. Neurosci. 18, 1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Conklin BR, Hsiao EC, Claeysen S, Dumuis A, Srinivasan S, Forsayeth JR, Guettier J-M, Chang WC, Pei Y, McCarthy KD, et al. (2008). Engineering gpcr signaling pathways with rassls. Nat. Methods 5, 673–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Armbruster BN, Li X, Pausch MH, Herlitze S, and Roth BL (2007). Evolving the lock to fit the key to create a family of g protein-coupled receptors potently activated by an inert ligand. Proc. Natl. Acad. Sci. USA 104, 5163–5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sakurai T, Nagata R, Yamanaka A, Kawamura H, Tsujino N, Muraki Y, Kageyama H, Kunita S, Takahashi S, Goto K, et al. (2005). Input of orexin/hypocretin neurons revealed by a genetically encoded tracer in mice. Neuron 46, 297–308. [DOI] [PubMed] [Google Scholar]
- 54.Maskos U, Kissa K, St. Cloment, C., and Brûlet, P. (2002). Retrograde trans-synaptic transfer of green fluorescent protein allows the genetic mapping of neuronal circuits in transgenic mice. Proc. Natl. Acad. Sci. USA 99, 10120–10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshihara Y, Mizuno T, Nakahira M, Kawasaki M, Watanabe Y, Kagamiyama H, Jishage K, Ueda O, Suzuki H, Tabuchi K, et al. (1999). A genetic approach to visualization of multisynaptic neural pathways using plant lectin transgene. Neuron 22, 33–41. [DOI] [PubMed] [Google Scholar]
- 56.Tsai NY, Wang F, Toma K, Yin C, Takatoh J, Pai EL, Wu K, Matcham AC, Yin L, Dang EJ, et al. (2022). Trans-seq maps a selective mammalian retinotectal synapse instructed by nephronectin. Nat. Neurosci. 25, 659–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumamoto T, Maurinot F, Barry-Martinet R, Vaslin C, Vandormael-Pournin S, Le M, Lerat M, Niculescu D, Cohen-Tannoudji M, Rebsam A, et al. (2020). Direct readout of neural stem cell transgenesis with an integration-coupled gene expression switch. Neuron 107, 617–630.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Loonstra A, Vooijs M, Beverloo HB, Allak BA, van Drunen E, Kanaar R, Berns A, and Jonkers J (2001). Growth inhibition and dna damage induced by cre recombinase in mammalian cells. Proc. Natl. Acad. Sci. USA 98, 9209–9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schneider CA, Rasband WS, and Eliceiri KW (2012). Nih image to imagej: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, and Pachter L (2012). Differential gene and transcript expression analysis of rna-seq experiments with tophat and cufflinks. Nat. Protoc. 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mi H, Muruganujan A, Casagrande JT, and Thomas PD (2013). Large-scale gene function analysis with the panther classification system. Nat. Protoc. 8, 1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, and Zhang F (2013). Multiplex genome engineering using crispr/cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tomassy GS, De Leonibus E, Jabaudon D, Lodato S, Alfano C, Mele A, Macklis JD, and Studer M (2010). Area-specific temporal control of corticospinal motor neuron differentiation by coup-tfi. Proc. Natl. Acad. Sci. USA 107, 3576–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsuda T, and Cepko CL (2007). Controlled expression of transgenes introduced by in vivo electroporation. Proc. Natl. Acad. Sci. USA 104, 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Catapano LA, Arnold MW, Perez FA, and Macklis JD (2001). Specific neurotrophic factors support the survival of cortical projection neurons at distinct stages of development. J. Neurosci. 21, 8863–8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
| Mouse anti-SATB2 | Abcam | RRID:AB_882455 |
| Chicken anti-B-GAL | Abcam | RRID:AB_10899635 |
| Rabbit anti-GFP | Molecular Probes | RRID:AB_221569 |
| Rat anti-CTIP2 | Abcam | RRID:AB_2064130 |
| Rabbit anti-RORB | H. Stunnenberg | RRID:AB_2566817 |
|
Critical commercial assays | ||
| SMART-Seq v4 Ultra Low Input RNA kit | Takara | 634888 |
| RNeasy kit | Qiagen | 74104 |
|
Deposited data | ||
| Couptf1 BEAM RNAseq was deposited in GEO data repository | This paper | Accession # GSE271632 |
|
Experimental models: cell lines | ||
| HEK-293T | ATCC | CRL-3216 |
|
Experimental models: organisms/strains | ||
| Satb2fl/fl | Savarese et al.26 | RRID:MGI_6363495 |
| Couptf1fl/fl | Armentano et al.24,25 | RRID:MGI_5523158 |
| Ctip1 fl/fl | Sankaran et al.38 | RRID:MGI_4358088 |
| Rosa26-loxP-STOP-loxP-LacZ | JAX | RRID:MGI_2164639 |
|
Oligonucleotides | ||
| Satb2 sgRNA: GTCTCGCACAAAGATCTCTTTGG |
Shinmyo et al.36 | N/A |
| Rho Cas13 guide 1: GGTCTCCGTCTTGGAAGCGGTGGCAGAGGC |
This paper | N/A |
| Rho Cas13 guide 2: GTCGTCATCTCCCAGTGGATTCTTGCCGCA |
This paper | N/A |
| Rho Cas13 guide 3: GCACAGCGTGGTGAGCATACAGTTCCGGAA |
This paper | N/A |
|
Recombinant DNA | ||
| pBeatrix | Addgene | Plasmid #167528 |
| pDouble-Up | Addgene | Plasmid #125134 |
| BEAM plasmids will be deposited with Addgene | This paper | Plasmid # pending |
|
Software and algorithms | ||
| ImageJ | Schneider et al.59 | N/A |
| Tuxedo tools | Trapnell et al.60 | N/A |
| PANTHER overrepresentation test | Mi et al.61 | N/A |


