Abstract
Pseudotyping retrovirus and lentivirus vectors with different viral fusion proteins is a useful strategy to alter the host range of the vectors. Although lentivirus vectors are efficiently pseudotyped by Env proteins from several different subtypes of murine leukemia virus (MuLV), the related protein from gibbon ape leukemia virus (GaLV) does not form functional pseudotypes. We have determined that this arises because of an inability of GaLV Env to be incorporated into lentivirus vector particles. By exploiting the homology between the GaLV and MuLV Env proteins, we have mapped the determinants of incompatibility in the GaLV Env. Three modifications that allowed GaLV Env to pseudotype human immunodeficiency virus type 1 particles were identified: removal of the R peptide (C-terminal half of the cytoplasmic domain), replacement of the whole cytoplasmic tail with the corresponding MuLV region, and mutation of two residues upstream of the R peptide cleavage site. In addition, we have previously proposed that removal of the R peptide from MuLV Env proteins enhances their fusogenicity by transmitting a conformational change to the ectodomain of the protein (Y. Zhao et al., J. Virol. 72:5392–5398, 1998). Our analysis of chimeric MuLV/GaLV Env proteins provides further evidence in support of this model and suggests that proper Env function involves both interactions within the cytoplasmic tail and more long-range interactions between the cytoplasmic tail, the membrane-spanning region, and the ectodomain of the protein.
Retrovirus vectors derived from murine leukemia virus (MuLV) are the most commonly used gene transfer vectors in current human gene therapy applications (reviewed in reference 5). Like those of their parental retroviruses, the host ranges of such vectors are influenced in large part by the properties of the fusion protein contained in the outer lipid envelope of the vector particle. An attractive property of these vectors is the relative ease with which different fusion proteins can be incorporated into particles in place of the native envelope (Env) protein, a process referred to as pseudotyping. This allows the vectors to transduce ranges of cells and tissues different from those that would be possible with just the native Env.
There are many examples of pseudotyping in the literature. Both MuLV and retrovirus vectors derived from it can be pseudotyped by the Env proteins from other type C mammalian retroviruses. These include proteins from the different subtypes of MuLV, such as amphotropic (7, 34, 46), polytropic (15, 29), and xenotropic (3, 46) subtypes and 10A1 (35), as well as the Env proteins from gibbon ape leukemia virus (GaLV) (36, 46, 63) and feline leukemia virus type B (59). In addition, Env proteins from more-distantly related retroviruses can also pseudotype MuLV particles, such as feline endogenous virus RD114 (46, 59), Jaagsiekte sheep retrovirus (48), human T-cell-lymphotropic virus type 1 (HTLV-1) (63), and simian immunodeficiency virus (21). Finally, MuLV-based vectors can also be pseudotyped by nonretrovirus fusion proteins, including the glycoproteins from vesicular stomatitis virus (VSV) (4, 11), rabies virus (38), lymphocytic choriomeningitis virus (LCMV) (32), and Ebola virus (64).
Despite the potentially wide host range conferred by the use of heterologous fusion proteins, retrovirus vectors still suffer from certain limitations. In particular, MuLV-based vectors are unable to transduce nondividing cells (37). In contrast, lentivirus vectors derived from human immunodeficiency virus type 1 (HIV-1) are able to transduce a variety of nondividing cells, including hematopoietic cells (39) and neurons (41, 42). HIV-1 and lentivirus vectors can also be pseudotyped by different viral fusion proteins, including the VSV protein G (VSV-G) (42), different MuLV subtypes (25, 30), HTLV-1 (25), and the rabies virus and Mokola virus G proteins (40).
Most current retrovirus and lentivirus vector protocols use fusion proteins with a broad host range, such as those from the humantropic MuLV subtypes (amphotropic, xenotropic, and polytropic subtypes and 10A1), the GaLV Env protein, and VSV-G. Although VSV-G has an extremely wide host range (4), its inherent cytotoxicity has made the establishment of stable producer cell lines difficult unless inducible systems are used (2, 65). In contrast, both the MuLV and GaLV Env proteins are able to form stable producer cell lines (7, 8, 29, 34, 36, 46), making them potentially more useful for the large-scale production of pseudotyped vectors.
Although the GaLV and amphotropic MuLV receptor proteins are widely expressed on human cells (reviewed in references 20 and 33), several side-by-side comparisons have demonstrated that GaLV Env is better at transducing certain human cell types than the amphotropic Env (6, 24, 57, 62). The GaLV and MuLVs are closely related type C mammalian retroviruses, and their entry pathways have common features (26). Their Env proteins contain two subunits, SU and TM, which are cleaved from a common precursor protein during transport to the cell surface. An additional feature of these Env proteins is that the C-terminal region of the cytoplasmic tail, the R peptide, is cleaved by the viral protease at, or shortly after, viral budding. R peptide cleavage is necessary to confer full activity to the Env protein (47, 50), although not all of the Env proteins in a virion are cleaved (13, 22). The processing of the cytoplasmic tail of Env is not unique to the mammalian type C retroviruses and has also been reported for more-distantly related retroviruses such as the Mason-Pfizer monkey virus (55) and equine infectious anemia virus (52).
Although the combined properties of the GaLV Env and HIV-1 cores would make lentivirus vectors pseudotyped with GaLV Env potentially useful for human gene therapy applications, our initial attempts to produce such vectors were unsuccessful. In agreement with a recent report (58), we have determined that the incompatibility between the GaLV Env and lentivirus vectors lies in the cytoplasmic tail of the Env protein. We here describe a detailed analysis of the mechanism of this incompatibility and describe three different strategies that allow functional pseudotypes to form. In addition, we discuss the implications that our findings have for Env protein function and Env-particle interactions in the retroviruses.
MATERIALS AND METHODS
Cell lines.
293T and HeLa cells were obtained from the American Type Culture Collection and were maintained in Dulbecco's modified Eagle's medium plus 10 mM glutamine (Norris Cancer Center cell culture core facility, University of Southern California) and 10% fetal bovine serum (Hyclone, Logan, Utah).
Production of retrovirus and lentivirus vectors.
Retrovirus vectors were produced by transient transfection of 293T cells, essentially as described previously (15, 56). Ten micrograms each of plasmids pCgp and pCnBg, together with 1 to 10 μg of an appropriate fusion protein expression plasmid, was cotransfected into 60 to 70% confluent 293T cells in 10-cm-diameter plates by calcium phosphate precipitation. Plasmid pCgp is a packaging construct expressing MuLV Gag-Pol (15), and pCnBg has a retrovirus vector genome carrying a nuclear β-galactosidase marker gene (14). Lentivirus vectors were generated in the same way using 10 μg each of HIV-1 packaging construct pCMVΔR8.2 (68) and a plasmid with the lentivirus vector genome, pHR′-CMVLacZ (42), together with 1 to 10 μg of a fusion protein expression plasmid. For experiments where both retrovirus and lentivirus vectors were produced from the same cell, we cotransfected 7 μg each of plasmids pCgp and pCMVΔR8.2, 5 μg each of pHanPuro and pHR′-CMVLacZ, and 7 μg of the expression plasmids for either amphotropic MuLV or GaLV Env proteins. pHanPuro has a retrovirus vector genome containing an internal simian virus 40 promoter driving expression of a puromycin resistance gene. For the control experiments where only pCgp or pCMVΔR8.2 was used, the total amount of DNA per transfection was normalized to 31 μg using plasmid pBluescript (Stratagene, La Jolla, Calif.).
Env and fusion protein expression vectors.
All fusion proteins were expressed from plasmids containing the human cytomegalovirus immediate-early promoter and the origin of replication from simian virus 40. The fusion proteins used were obtained from the amphotropic 4070A MuLV (14, 16), the polytropic mink cell focus-forming virus (15, 17), the xenotropic NZB MuLV (27), 10A1 MuLV (14, 49), the SEATO strain of GaLV (9), the Indiana strain of VSV (53), influenza A virus/fowl plague virus/Rostock/34 (61), and the Armstrong 53b strain of LCMV (10). Chimeric and truncated MuLV/GaLV Env proteins were generated by splice-overlap PCR (18), and the final constructs were fully sequenced.
Determination of vector titer.
Retrovirus and lentivirus vector titers were measured by transduction of 293T or HeLa cells. Serial dilutions of the vector supernatants were prepared, and 1 ml of each dilution was added to a well of a six-well plate seeded with 105 cells the previous day in the presence of 8 μg of Polybrene (Sigma, St. Louis, Mo.)/ml. For vectors carrying β-galactosidase markers, the titer was determined by X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining at 48 h posttransduction. Cells were fixed in 0.5% glutaraldehyde for 10 min, washed with phosphate-buffered saline for 10 min, and then incubated with staining solution (4 mM potassium ferricyanide, 4 mM potassium ferrocyanide, 2 mM MgCl2, 80 μg of X-Gal [Sigma]/ml) at 37°C overnight. Titer was determined by counting the blue colonies in each well under a light microscope and multiplying this number by the appropriate dilution factor. For experiments using vector pHanPuro, titer was determined as the number of puromycin-resistant colonies that grew out after 7 days of treatment with 2.5 μg of puromycin (Sigma)/ml. Titer was expressed as CFU per milliliter of viral supernatant.
Western analysis of retrovirus and lentivirus vector particles.
Vector particles were harvested from the supernatants of transiently transfected 293T cells and partially purified by centrifugation through 2 ml of 20% sucrose at 25,000 rpm at 4°C for 2 h using an SW41 rotor. The transmembrane (TM) subunit of the Env proteins was detected using rat monoclonal antibody 42/114 against AKR MuLV TM (45) at a 1:2,000 dilution, the capsid (CA) protein from the retrovirus vectors was detected using a goat anti-Rauscher MuLV p30 antiserum (Quality Biotech; lot 78S221) at a 1:5,000 dilution, and the lentivirus CA protein was detected using mouse anti-p24 monoclonal antibody 183-H12-5C (National Institutes of Health AIDS Research and Reference Reagent Program) at a 1:1,000 dilution. The secondary antibodies used were horseradish peroxidase (HRP)-conjugated rabbit anti-goat immunoglobulin G (IgG) (1:20,000), HRP-conjugated goat anti-rat IgG (1:10,000), and HRP-conjugated goat anti-mouse IgG (1:10,000) (Pierce, Rockford, Ill.). Specific proteins were visualized using the enhanced chemiluminescence detection system (Amersham International plc., Arlington Heights, Ill.).
To analyze the form of the Env proteins present in cell lysates, 293T cells were incubated in 200 μl of lysis buffer (20 mM Tris-HCl [pH 7.5], 1% Triton X-100, 0.05% sodium dodecyl sulfate [SDS], 5 mg of sodium deoxycholate/ml, 150 mM NaCl, 1 mM phenylethanolamine fluoride) for 10 min at 4°C, followed by centrifugation at 14,000 × g for 10 min to pellet nuclei. Fifteen microliters of the resulting supernatants was analyzed by SDS-polyacrylamide gel electrophoresis and Western blotting, as described above.
RESULTS
Ability of viral fusion proteins to pseudotype retrovirus or lentivirus vectors.
We examined the efficiencies with which a panel of different fusion proteins could pseudotype either retrovirus (MuLV) or lentivirus (HIV-1) vectors. We included the Env proteins from the different MuLV subtypes that have tropism for human cells (amphotropic, polytropic, and xenotropic subtypes and 10A1), as well as the related Env protein from GaLV. The heterologous viral fusion proteins that we used were a hemagglutinin (HA) protein from an avian pathogenic strain of influenza virus, VSV-G, and the glycoprotein (GP) from the Armstrong 53b strain of LCMV.
Retrovirus and lentivirus vectors were generated using a three-plasmid transient transfection system (56) in which the Gag-Pol and transfer vector components were kept constant but the fusion protein was varied accordingly. The vectors so generated were initially titered on human 293T cells to screen for the formation of functional pseudotypes (Table 1) and subsequently titered on a range of predictive cell lines to confirm the expected tropism of the vectors (data not shown). Although most of the vectors tested gave titers that were in the range of 104 to 106 CFU/ml, strikingly, the combination of the GaLV Env and lentivirus vector components did not result in any titer.
TABLE 1.
Titers of pseudotyped retrovirus and lentivirus vectorsa
| Vector type | Titer (CFU/ml) for:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Retrovirus Env proteinb:
|
Nonretrovirus proteinc:
|
|||||||
| Ampho | 10A1 | Poly | Xeno | GaLV | VSV-G | HA | LCMV-GP | |
| Retrovirus | 5 × 106 | 3 × 106 | 5 × 105 | 6 × 106 | 1 × 106 | 2 × 107 | 7 × 104 | 3 × 105 |
| Lentivirus | 4 × 106 | 4 × 106 | 4 × 104 | 4 × 105 | <50 | 3 × 106 | 8 × 104 | 1 × 104 |
Viral fusion proteins were coexpressed with retrovirus or lentivirus vector components and were titered on 293T cells. Titers are averages of at least three independent experiments.
MuLV Env proteins were derived from the amphotropic 4070A virus (ampho), 10A1, the polytropic mink cell focus-forming virus MCF247 (poly), and the xenotropic virus NZB (xeno); the GaLV Env protein was from the SEATO strain.
VSV-G is from the Indiana strain of VSV, HA is from the A/Rodstock/2/34 strain of avian pathogenic influenza virus, and LCMV-GP is from the Armstrong 53b strain.
GaLV Env does not pseudotype lentivirus vectors.
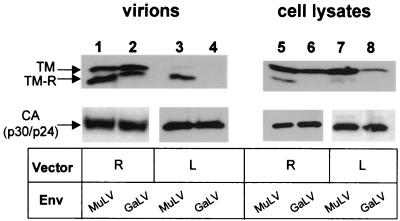
We wished to determine the basis for this lack of titer. As a first step, we examined the efficiency with which the GaLV and amphotropic MuLV Env proteins could be incorporated into retrovirus and lentivirus vector particles. Vector particles were harvested from the supernatants of transfected cells and analyzed by Western blotting, as were the producer cell lysates. For Env detection, we used an antibody raised against the MuLV TM protein that also cross-reacts with the corresponding GaLV protein.
Both the MuLV and GaLV TM proteins were readily detected in the pelleted retrovirus vector supernatants, indicating good Env incorporation (Fig. 1). We did, however, observe a difference in the ratios of the immature (TM) and processed (TM-R) forms of the two proteins, with the MuLV Env being more efficiently processed to the TM-R form. In contrast, examination of the lentivirus vector supernatants revealed that only the MuLV Env protein was present in these vector particles. The GaLV TM was not detected, even when the blot was overexposed (data not shown). Western analysis of cell lysates confirmed that the GaLV TM protein was expressed in cells transfected with the lentivirus components, although it was present at a lower level than that in cells expressing the retrovirus vectors (Fig. 1, compare lanes 6 and 8). From these results, it is therefore not apparent whether the lack of pseudotyping of lentivirus vectors by the GaLV Env arises because of a significant block to the incorporation of GaLV Env into HIV-1 particles or whether the effect is secondary to an inhibition of the steady-state levels of GaLV Env in cells producing lentivirus vectors.
FIG. 1.
Incorporation of GaLV and MuLV Env proteins into retrovirus and lentivirus vectors. 293T cells were transiently transfected with either retrovirus (R) or lentivirus (L) vector components and the Env proteins indicated. Vector particles were partially purified from the supernatant by centrifugation through 20% sucrose, and both vector particles and cell lysates were subjected to Western analysis using antibodies raised against the MuLV TM protein that also recognize the GaLV TM. TM-R is the form of TM with the R peptide cleaved. The CA proteins from the vectors were used as loading controls and were detected using anti-p30 (MuLV) or anti-p24 (HIV-1) antibodies, respectively.
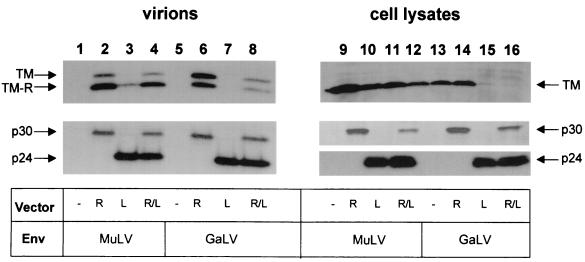
Coexpression of retrovirus and lentivirus vectors reveals that GaLV Env is available for incorporation into vector particles.
In order to distinguish between these two possibilities, we repeated our analyses using cells cotransfected with both retrovirus and lentivirus vector components. We used two different marker genes on the retrovirus and lentivirus transfer vectors (those for puromycin resistance and β-galactosidase, respectively), to enable us to distinguish between functional pseudotypes of the two vector types. Western analysis of vector supernatants and cell lysates confirmed that the presence of lentivirus vector components always inhibited the steady-state levels of GaLV Env in cell lysates, even when retrovirus vectors were also present (Fig. 2, lanes 15 and 16). However, as pelleted supernatants produced from cells transfected with both retrovirus and lentivirus vectors were found to contain the GaLV TM (lane 8), we conclude that there is still sufficient protein present to be incorporated into vector particles.
FIG. 2.
Coexpression of retrovirus and lentivirus vectors. 293T cells were transfected with the vector components and Env proteins shown, and both vector supernatants and cell lysates were subjected to Western analysis for the proteins indicated. R, retrovirus vectors alone; L, lentivirus vectors alone; R/L, retrovirus and lentivirus vectors cotransfected.
We next examined whether the GaLV TM present in Fig. 2, lane 8, was associated with retrovirus or lentivirus vectors. We performed titer assays on HeLa cells and measured the numbers of both puromycin-resistant and β-galactosidase-expressing colonies (Table 2). This revealed that the vector supernatant produced from the cotransfection of GaLV Env with both lentivirus and retrovirus vectors was not able to transfer β-galactosidase. In contrast, the same supernatant produced puromycin-resistant colonies at a titer similar to that obtained by the combination of the GaLV Env with retrovirus vectors alone. Taken together, these results suggest that the pelleted GaLV Env was exclusively associated with retrovirus vectors and that the inability of GaLV Env to pseudotype lentivirus vectors is not primarily due to a destabilization of the protein but arises because of some specific incompatibility between HIV-1 particles and GaLV Env.
TABLE 2.
Titers of coexpressed retrovirus and lentivirus vectorsa
| Env source | Packaging component | Vector titer (CFU/ml)b
|
||
|---|---|---|---|---|
| Retrovirus (HeLa) | Lentivirus (HeLa) | Lentivirus (293T) | ||
| MuLV | Retrovirus | (2.5 ± 0.7) × 104 | 0 | 0 |
| MuLV | Lentivirus | 0 | (5.0 ± 4.0) × 103 | (1.4 ± 0.9) × 106 |
| MuLV | Retrovirus/Lentivirus | (7.0 ± 1.3) × 103 | (1.5 ± 0.7) × 103 | (5.0 ± 0.2) × 106 |
| GaLV | Retrovirus | (9.0 ± 6.7) × 103 | 0 | 0 |
| GaLV | Lentivirus | 0 | 0 | (2.0 ± 0.7) × 101 |
| GaLV | Retrovirus/Lentivirus | (5.0 ± 1.7) × 103 | 0 | (2.0 ± 0.1) × 102 |
Amphotropic MuLV or GaLV Env proteins were coexpressed with retrovirus and/or lentivirus vector packaging (Gag-Pol) plasmids. In all cases, both, retrovirus and lentivirus transfer vector genomes were cotransfected.
Titers were measured on HeLa and 293T cells as either puromycin-resistant colonies (retrovirus-vectors) or β-galactosidase-expressing colonies (lentivirus-vectors) and are the averages of two independent experiments ± standard deviations.
We also performed β-galactosidase titer assays with the various vector supernatants on 293T cells (Table 2) but were unable to determine the corresponding retrovirus vector titers due to the difficulty in selecting for adherent puromycin-resistant 293T colonies. Surprisingly, we observed a reproducible 1-log-unit increase in the number of β-galactosidase-positive cells produced by the GaLV/retrovirus/lentivirus combination on 293T cells compared to the GaLV/lentivirus vectors alone, from an average of 20 to 200 CFU/ml. Control experiments ruled out the possibility that this increase was caused by encapsidation of the lentivirus vector genome by GaLV pseudotyped retrovirus vectors (data not shown). Although such an enhancement was not observed when the same supernatants were titered on HeLa cells, we cannot rule out the possibility that the presence of the retrovirus vectors in some way increases the ability of GaLV Env to pseudotype lentivirus vectors. At present, the mechanism of such an effect is not known.
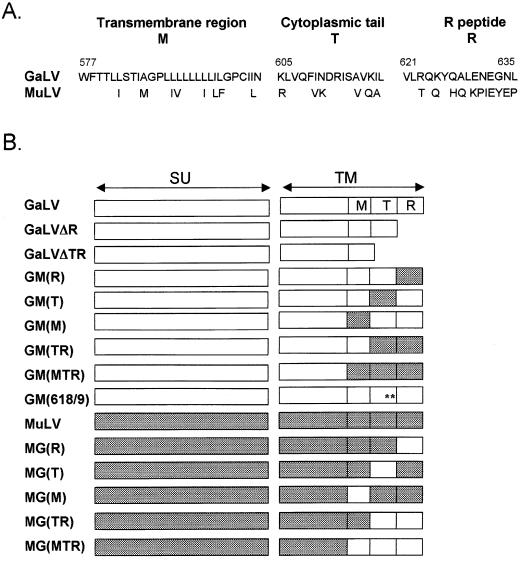
Construction of chimeric MuLV/GaLV Env proteins.
To further examine the basis for the incompatibility between the GaLV Env and lentivirus vectors, we took advantage of the homology between the GaLV and amphotropic MuLV Env proteins in order to construct chimeric proteins (Fig. 3). We concentrated in particular on the cytoplasmic domain, as previous reports of incompatibility between fusion proteins and retrovirus particles have implicated this region. For example, both the HIV-1 (63) and human foamy virus (40) Env proteins are unable to pseudotype MuLV cores, but modifying their cytoplasmic tails alleviates the problem (28, 31, 54). In addition, as we have previously shown that truncating the R peptide or the whole cytoplasmic tail from the ecotropic MuLV Env results in proteins that still retain some function (22), we also constructed truncated versions of the GaLV Env.
FIG. 3.
MuLV and GaLV Env proteins. (A) Comparison of the sequences of the transmembrane (M) and cytoplasmic tail regions (T and R) of the GaLV SEATO and amphotropic 4070A MuLV Env proteins. Numbering for GaLV Env is from the start of the mature protein after removal of the signal peptide (60). The C terminus of the cytoplasmic tail (R) is cleaved from Env during virion maturation, leaving 16 amino acids in the tail of the mature Env protein (T). (B) Schematic of the truncated and substituted GaLV (open boxes) and MuLV (shaded boxes) Env proteins used in this study. Construct GM(618/9) is the GaLV Env with the substitutions K618Q and I619A, ∗∗.
Ability of MuLV/GaLV Env proteins to pseudotype retrovirus and lentivirus vectors.
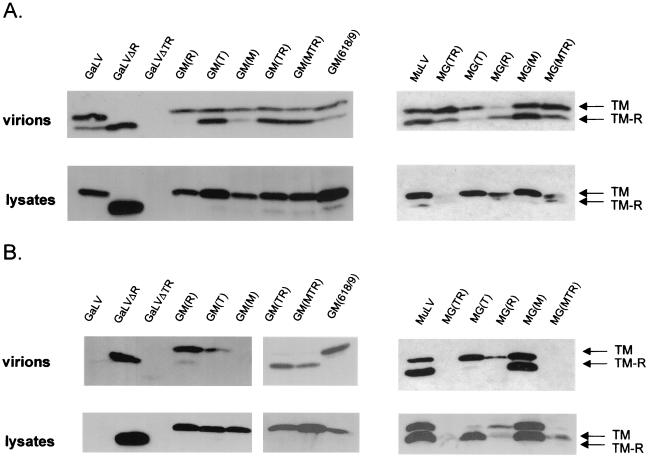
The truncated and chimeric Env proteins were assessed for their ability to pseudotype both retrovirus and lentivirus vectors. As before, vector particles were generated by transient transfection and both vector supernatants and cell lysates were analyzed by Western blotting. All of the Env proteins were found to be efficiently incorporated into the retrovirus vector particles, except for construct GaLVΔTR (Fig. 4A). Since this protein was only present at very low levels in cells transfected with either retrovirus or lentivirus vectors, it is likely that poor protein stability underlies its lack of incorporation.
FIG. 4.
Incorporation of chimeric and truncated Env proteins into vector particles. Retrovirus (A) or lentivirus (B) vectors were generated using the Env proteins indicated, and both partially purified vectors and cell lysates were subjected to Western analysis.
In contrast to the situation with the retrovirus vectors, the extent of incorporation of the various Env proteins into lentivirus vectors varied significantly (Fig. 4B). Moreover, the nature of the cytoplasmic tail sequences alone was sufficient to determine the incorporation pattern, as the reciprocal chimeras GM(TR) and MG(TR) displayed the phenotypes of the Env proteins from which the cytoplasmic tail was derived. The whole cytoplasmic tail was needed for this effect, as substitution of either the T or R regions alone did not fully alleviate the block to incorporation in the GaLV Env or confer total incompatibility to the MuLV Env. We noted that the presence of the GaLV R peptide in particular was associated with poor incorporation, as GM(R) was incorporated at higher levels than GM(T), while the converse was true for MG(R) and MG(T). Furthermore, the simple removal of the R peptide from GaLV Env allowed strong incorporation of GaLVΔR into lentivirus vectors. However, the incorporation-competent phenotype of mutant GM(618/9) argues against a model whereby the GaLV R peptide per se prevents incorporation and, instead, suggests that some feature of the complete cytoplasmic tail influences the association with HIV-1 particles.
We also examined the titers directed by the various Env proteins on 293T cells (Table 3). Interestingly, for the retrovirus vectors, the GM series of chimeras always gave titers that were lower than those obtained with either the MuLV or GaLV parental Env proteins. The effects on titer could not simply be attributed to reduced Env incorporation into vector particles. For example, construct GM(M) gave titers that were nearly 3 orders of magnitude lower than those for wild-type GaLV Env, despite reasonable levels of incorporation into retrovirus vectors. Instead, this suggests that even small changes in the GaLV Env protein can compromise its function.
TABLE 3.
Properties of Env proteins
| Env | Incorporationa
|
Titer (CFU/ml)b
|
||
|---|---|---|---|---|
| R | L | R | L | |
| GaLV | +++ | − | (1.4 ± 0.9) × 106 | <50 |
| GaLVΔR | +++ | +++ | (2.0 ± 1.0) × 106 | (1.7 ± 0.8) × 104 |
| GaLVΔTR | + | + | (1.4 ± 0.9) × 104 | (2.0 ± 0.8) × 102 |
| GM(R) | ++ | ++ | (5.8 ± 3.7) × 104 | <50 |
| GM(T) | +++ | + | (2.5 ± 0.7) × 105 | (2.9 ± 1.1) × 102 |
| GM(M) | ++ | − | (3.8 ± 1.6) × 103 | <50 |
| GM(TR) | +++ | +++ | (4.0 ± 1.0) × 105 | (1.2 ± 0.6) × 104 |
| GM(MTR) | +++ | +++ | (3.0 ± 1.0) × 105 | (4.8 ± 3.3) × 103 |
| GM(618/9) | +++ | +++ | (1.1 ± 0.8) × 105 | (6.2 ±1.8) × 103 |
| MuLV | +++ | +++ | (4.9 ± 1.4) × 106 | (3.5 ± 1.7) × 106 |
| MG(R) | ++ | + | (6.0 ± 4.6) × 106 | (5.0 ± 2.2) × 103 |
| MG(T) | ++ | ++ | (7.3 ± 2.1) × 105 | (3.8 ± 1.5) × 103 |
| MG(M) | +++ | +++ | (2.7 ± 1.2) × 106 | (1.3 ± 0.7) × 106 |
| MG(TR) | +++ | − | (4.7 ± 3.1) × 106 | <50 |
| MG(MTR) | +++ | − | (2.2 ± 1.4) × 106 | <50 |
GaLV/MuLV Env proteins were used to pseudotype either retrovirus (R) or lentivirus (L) vectors, and the relative efficiencies of incorporation of the Env proteins into the vector particles were assessed by Western blotting. +++, wild-type MuLV Env amount; ++, less than 50% of the wild-type level; +, trace amount detected; −, no Env.
Titers are averages ± standard deviations of at least three independent experiments.
Similarly, analysis of the titers directed by the incorporation-competent Env proteins on lentivirus vectors revealed that factors other than absolute incorporation levels affected titers. The four GaLV-based proteins that were well incorporated [GaLVΔR GM(TR), GM(MTR), and GM(618/9)] gave titers only in the range of 103 to 104 CFU/ml with lentivirus vectors, while retrovirus vectors pseudotyped with the same proteins gave titers in the range of 105 to 106 CFU/ml. This 2-order-of-magnitude difference in titers was not simply a property of the lentivirus vectors, as lentivirus vectors pseudotyped with the MuLV Env gave a titer of 3.5 × 106, which was comparable to those achieved with the retrovirus vectors. Instead, it appears that these GaLV Env derivatives have an additional block to full function that is only manifest when the proteins are present on lentivirus vectors.
R peptide cleavage patterns.
The above observations suggested that specific interactions between the vector particle and the pseudotyping Env protein could play a role in influencing the overall efficiency of transduction. A clear example of such Env-particle interactions is the fact that the viral protease is responsible for R peptide processing. Although complete R peptide removal is not necessary for Env function (67), the efficiency of processing has been shown to correlate with titer in at least one case (23). We were therefore interested to examine whether differences in the extent of R peptide processing could account for the differences in titer achieved with retrovirus and lentivirus vectors.
We have repeatedly observed that cleavage of the GaLV Env by MuLV cores is less efficient than processing of the MuLV Env (Fig. 1, 2, and 4). Typically two-thirds of the MuLV TM proteins were cleaved in pelleted vector particles, and the same degree of processing was observed for both the homologous MuLV particles and the heterologous HIV-1 particles. Although the GaLV Env was processed less efficiently, this was clearly not a problem for Env function as titers of 1.4 × 106 CFU/ml were achieved with retrovirus vectors. It is possible that this lower level of processing occurs naturally in GaLV virions and is sufficient to produce a fully functional Env. Indeed, the extreme fusogenicity and cytotoxicity of the R-less form of the GaLV Env (12) provide a rationale for such a situation.
Using the panel of chimeric GaLV/MuLV proteins, we were able to examine the factors that influenced the degree of R peptide processing by either retrovirus or lentivirus particles (Table 4). The viral protease recognition site includes at least seven residues spanning the actual cleavage site (residues P4 to P3′) (43), so that both the T and R regions of the tail could influence processing efficiency. However, for proteins present in retrovirus vectors, our analysis showed that only the nature of the T region influenced the extent of R peptide cleavage; any protein containing an MuLV T region was cleaved efficiently, while any protein containing a GaLV T region was not. This pattern held true whatever the absolute incorporation levels of the Env protein and is shown most clearly by the opposite patterns exhibited by MG(T) and MG(R) (Fig. 4A). However simply changing the GaLV residues in the T region that were part of the protease recognition site to the corresponding MuLV sequences, as occurs in construct GM(618/9), was not sufficient to produce the efficient MuLV pattern of cleavage, implying a contribution by more-upstream sequences in the GaLV T region. In contrast, for the lentivirus vectors, we observed that the presence of either the GaLV R or T regions reduced the levels of processing to the wild-type GaLV pattern and that both the T and R regions had to be replaced by the MuLV sequences before the efficient MuLV pattern could be observed.
TABLE 4.
R peptide cleavage patterns
| Env | R peptide cleavagea
|
|
|---|---|---|
| R | L | |
| GaLV | G | — |
| GaLVΔR | n.a. | n.a. |
| GaLVΔTR | n.a. | n.a. |
| GM(R) | G | G |
| GM(T) | M | G |
| GM(M) | G | — |
| GM(TR) | M | M |
| GM(MTR) | M | M |
| GM(618/9) | G | G |
| MuLV | M | M |
| MG(R) | M | G |
| MG(T) | G | G |
| MG(M) | M | M |
| MG(TR) | G | — |
| MG(MTR) | G | — |
Pattern of R peptide cleavage in retrovirus (R) or lentivirus (L) vectors is defined as GaLV-like (G) if less than 50% of the TM protein is truncated and MuLV-like (M) if greater than 50% is truncated. n.a., not applicable; —, insufficient Env incorporated.
In general, the same patterns of processing were observed for the chimeric Env proteins regardless of whether the proteins were incorporated into retrovirus or lentivirus vectors. However, a discrepancy between the two vector types was noted for constructs MG(R) and GM(T). Both of these chimeras displayed the MuLV pattern in retrovirus vectors but the GaLV pattern in lentivirus vectors. The feature uniquely shared by these two proteins is the combination of the MuLV T region and the GaLV R region. It is possible that the negative effect that the GaLV R peptide has on incorporation into lentivirus vectors precludes efficient R peptide removal, despite the presence of the MuLV T region. In contrast, as incorporation of Env proteins into retrovirus vectors is not affected by the presence of the GaLV R peptide, the extent of R peptide removal in these vectors was determined solely by the nature of the T region and was therefore MuLV-like. Finally, we note that, as construct GM(618/9) retained the GaLV pattern of R peptide cleavage on both vector types, we are able to separate the two properties conferred by the GaLV tail; while changing residues K618 and I619 to those of the MuLV sequence was sufficient to overcome the block to incorporation into lentivirus vectors, increasing the level of R peptide cleavage to the MuLV level required the substitution of the entire T region (retrovirus vectors) or both T and R regions combined (lentivirus vectors). Overall, this indicates that Env association with viral particles and the extent of R peptide cleavage, while related, are not absolutely correlated.
Factors affecting protein stability.
Retrovirus Env proteins are initially synthesized as a single polypeptide that is cleaved by a host cell protease to the SU and TM subunits during transport to the cell surface (44). Small amounts of TM in cell lysates, as well as poor ratios of TM to uncleaved Env, can result from reduced rates of processing and transport, as well as poor protein stability. Furthermore, the R-less form of TM is not always apparent on Western blots of cell lysates, reflecting the fact that this cleavage event may occur during or after the budding of the virion from the cell.
Analysis of the lysates of cells transfected with the various truncated and chimeric Env proteins identified certain proteins as having phenotypes that differed from those of either of the parental Env proteins. As previously mentioned, the GaLVΔTR TM signal was barely detectable in the presence of either vector type, suggesting an inherently low stability of this protein. In addition, the three MuLV/GaLV chimeras containing the GaLV R peptide [constructs MG(R), MG(TR), and MG(MTR)] gave relatively weak TM and TM-R signals in cell lysates and had relatively stronger bands of the uncleaved Env (data not shown), suggesting a problem in transport and/or processing. Despite this defect, all three constructs gave wild-type levels of incorporation and titer when expressed with retrovirus vectors (Table 2). Finally, the GaLV Env itself was notable in that it was the only Env protein that was significantly inhibited or destabilized by the coexpression in transfected cells of lentivirus vector components. Although the basis for this inhibition is currently unknown, the inhibition could be prevented in several different ways, including the separate replacement of the R, T, or M region and the substitution of residues K618 and I619. Taken together, these findings further suggest that overall interactions between the different regions of the GaLV Env protein contribute to its sensitivity to the presence of lentivirus vectors.
DISCUSSION
We have demonstrated that the GaLV Env protein is unable to pseudotype lentivirus vectors because certain features of its cytoplasmic tail are incompatible with incorporation into HIV-1 particles. We have identified three different strategies that enable such pseudotypes to form: (i) removal of the GaLV Env R peptide, (ii) replacement of the whole (R plus T regions) GaLV Env cytoplasmic tail with the corresponding MuLV sequences, but not either region alone, and (iii) the replacement of residues K618 and I619 by the corresponding MuLV residues. The resulting proteins, designated GaLVΔR, GM(TR), and GM(618/9), may have applications in human gene therapy, as such pseudotyped lentivirus vectors retain the GaLV Env host range (data not shown). In particular, GM(TR) and GM(618/9) may prove useful for the establishment of stable cell lines, as they are not toxic when expressed in cells (data not shown).
There are other examples in the literature of incompatibility between retrovirus particles and heterologous fusion proteins. For example, the Env proteins from HIV-1 (61), HIV-2 (19), Mason-Pfizer monkey virus, and simian retrovirus-1 (59) do not pseudotype MuLV. For the HIV-1 and HIV-2 Envs, the block resides in their long cytoplasmic tails and truncated versions of the proteins can be incorporated (19, 31, 54). Similarly, although the human foamy virus Env protein does not efficiently pseudotype MuLV, a chimera containing the cytoplasmic domain of MuLV Env shows improved incorporation (28). However, as the GaLV Env cytoplasmic tail is relatively short (29 amino acids) and has reasonable homology to the tail of the incorporation-compatible MuLV Env proteins, it was surprising to observe such incompatibility between the GaLV Env and lentivirus vectors.
Our initial attempts to understand why the GaLV Env was excluded from lentivirus vectors were hampered by the greatly reduced levels of GaLV TM that we observed in transfected 293T cells in the presence of lentivirus components. However, further experiments revealed that even the low level of protein that was present in these cells was sufficient to pseudotype coexpressed retrovirus vectors. This in turn suggested that the lack of GaLV Env in lentivirus vectors was not simply due to low steady-state levels of the protein in cells or at the cell surface but instead reflected a specific incompatibility between the Env and vector components. Although we do not yet understand why lentivirus vectors reduce the cellular levels of GaLV Env, we note that several different approaches can alleviate this effect. They include the individual replacement of either the M, T, or R region of the GaLV Env with MuLV sequences and just the substitution of residues K618 and I619. Finally, it is noteworthy that even good levels of expression of a protein in transfected cells are not sufficient to allow incorporation into lentivirus vectors, as can be seen most clearly for construct GM(M).
Most of the variation between the cytoplasmic tails of the GaLV and MuLV Env proteins is located in the R peptide. Although the good incorporation of the GaLVΔR protein into lentivirus vectors initially suggested that a steric block between the R peptide and HIV-1 virions caused their incompatibility, replacing the R peptide alone with the corresponding MuLV region was not sufficient to allow full incorporation. Furthermore, the ability of construct GM(618/9) to be incorporated into lentivirus vectors suggests that it was not the GaLV R peptide per se that was the problem but rather a more general structure in the whole GaLV Env tail. Overall, these findings suggest that interactions occur between the R and T regions in the cytoplasmic tails of these Env proteins that are important for the secondary structure of the whole of the tail and that the R peptide cleavage site itself may be a major determinant of this property.
One of the major differences that we noted between the GaLV and MuLV Env proteins was the different degrees of R peptide cleavage for the two proteins when present in retrovirus vectors. Since R peptide cleavage is performed by the viral protease, it presumably requires an intimate association between the Env protein and the viral core. We therefore asked whether R peptide cleavage rates would correlate well with efficiency of incorporation into lentivirus vectors. Interestingly, this was found not to be the case, and we were able to distinguish between the determinants that governed R peptide cleavage and those that controlled incorporation. The most extreme example is construct GM(618/9), where we observed good incorporation into lentivirus vectors without high levels of R peptide cleavage. Recently, an HIV-1 MA mutant that does not block MuLV Env incorporation into HIV-1 particles but that does prevent R peptide cleavage by the HIV-1 protease has been described (23). The phenotype of this mutant also suggests that incorporation into a lentivirus vector does not necessarily lead to a normal interaction with the HIV-1 core.
Our study has also produced evidence for specific Env-vector interactions that act to influence the overall rate of transduction. We observed a 2-order-of-magnitude difference in the titers directed by retrovirus and lentivirus vectors pseudotyped with the same incorporation-competent GaLV Env derivatives [constructs GM(TR), GM(MTR), GM(618/9), and GaLVΔR] that could not simply be attributed to differences in Env levels. One possible explanation is that the MuLV and HIV-1 virions are differentially sensitive to the entry pathway directed by the GaLV Env. A similar situation has been reported for HIV-1 virions produced in the presence or absence of the Nef protein, which showed a 10-fold difference in infectivity when pseudotyped with the amphotropic MuLV Env protein but not when the fusion protein used was VSV-G (1). However, if the GaLV entry pathway is indeed more productive for retrovirus vectors than lentivirus vectors, then it is an unexpected finding given that no such differences were seen for vectors pseudotyped with either the amphotropic or 10A1 Env proteins, which recognize similar or identical receptors. In addition, our studies with fusion proteins that direct entry pathways markedly different from those used by the mammalian type C retroviruses, including the pH-dependent proteins from VSV, influenza virus, and LCMV, did not provide evidence of any pathway-specific differences between retrovirus and lentivirus vectors.
An alternative explanation to account for these findings is that the MuLV or HIV-1 particles themselves in some way influence Env function and thereby affect the efficiency of transduction. Indeed, the process of R peptide cleavage is a clear precedent to indicate that retrovirus particles can influence Env protein function through an interaction with the cytoplasmic tail. Although our data show that the efficiency of R peptide processing was not a simple predictor of vector titer, it remains possible that an additional influence of viral particles on Env function exists that is so subtle that our current assays cannot detect it and that the Env-particle interactions that occur in the retroviruses are more complex than has previously been realized.
Our studies also have implications for understanding the mechanism of fusion enhancement of retrovirus Env proteins by R peptide cleavage. We (66, 67) and others (51) have previously reported that R peptide-truncated forms of MuLV Env can function in trans within an Env protein oligomer to stimulate Env fusogenicity. The data presented here further support this model, as even the low levels of R peptide truncation seen for the native GaLV Env protein were sufficient to give titers on retrovirus vectors as high as those obtained with the MuLV Env. Furthermore, even though replacing the tail of the MuLV Env with the corresponding GaLV domain in constructs MG(TR) and MG(MTR) reduced R peptide cleavage levels to the GaLV level, this did not reduce the titers obtained for pseudotyped retrovirus vectors. Overall, this indicates that even low levels of R peptide cleavage can confer full function to both the GaLV and MuLV Env proteins in the context of retrovirus vectors. Finally, we have previously proposed that R peptide cleavage enhances MuLV Env fusogenicity by transmitting a conformational change from the cytoplasmic tail of Env through to the ectodomain of the protein (67). Examination of the properties of the chimeric Env proteins lends further support to this hypothesis by suggesting the occurrence of long-range interactions between the different domains of the Env protein, including the cytoplasmic tail, the membrane-spanning region, and the ectodomain.
ACKNOWLEDGMENTS
We thank our colleagues in the Gene Therapy Laboratories for their support, in particular Maria Barcova, Celina Ngiam, and Kathleen Burke. We also thank French Anderson and Nori Kasahara (USC) for their thoughtful suggestions. The following reagent was obtained from the NIH AIDS Research and Reference Reagent Program: anti-p24 monoclonal antibody 183-H12-5C from Bruce Chesebro and Kathy Wehrly. We thank Maribeth Eiden (NIH) for providing the GaLV SEATO Env plasmid.
This work was supported by funding from Genetic Therapy, Inc./Novartis and Public Health Service grant CA-59318 from the National Cancer Institute.
REFERENCES
- 1.Aiken C. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J Virol. 1997;71:5871–5877. doi: 10.1128/jvi.71.8.5871-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arai T, Matsumoto K, Saitoh K, Ui M, Ito T, Murakami M, Kanegae Y, Saito I, Cosset F L, Takeuchi Y, Iba H. A new system for stringent, high-titer vesicular stomatitis virus G protein-pseudotyped retrovirus vector induction by introduction of Cre recombinase into stable prepackaging cell lines. J Virol. 1998;72:1115–1121. doi: 10.1128/jvi.72.2.1115-1121.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battini J L, Danos O, Heard J M. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J Virol. 1995;69:713–719. doi: 10.1128/jvi.69.2.713-719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burns J C, Friedmann T, Driever W, Burrascano M, Yee J K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannon P M, Anderson W F. Retroviral vectors for human gene therapy. In: Templeton N S, Lasic D D, editors. Gene therapy: therapeutic mechanisms and strategies. New York, N.Y: Marcel Dekker, Inc; 2000. pp. 1–16. [Google Scholar]
- 6.Chuah M K L, Brems H, Vanslembrouck V, Collen D, Vandendriessche T. Bone marrow stromal cells as targets for gene therapy of hemophilia A. Hum Gene Ther. 1998;9:353–365. doi: 10.1089/hum.1998.9.3-353. [DOI] [PubMed] [Google Scholar]
- 7.Cone R D, Mulligan R. High-efficiency gene transfer into mammalian cells: generation of helper-free recombinant retrovirus with broad mammalian host range. Proc Natl Acad Sci USA. 1984;81:6349–6353. doi: 10.1073/pnas.81.20.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danos O, Mulligan R C. Safe and efficient generation of recombinant retroviruses with amphotropic and ecotropic host ranges. Proc Natl Acad Sci USA. 1988;85:6460–6464. doi: 10.1073/pnas.85.17.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Paoli A, Johnsen D O, Noll W W. Granulocytic leukemia in whitehanded gibbons. J Am Vet Med Assoc. 1973;163:624–628. [PubMed] [Google Scholar]
- 10.Dutko F J, Oldstone M B A. Genomic and biological variation among commonly used lymphocytic choriomeningitis virus strains. J Gen Virol. 1983;64:1689–1698. doi: 10.1099/0022-1317-64-8-1689. [DOI] [PubMed] [Google Scholar]
- 11.Emi N, Friedmann T, Yee J K. Pseudotype formation of murine leukemia virus with the G protein of vesicular stomatitis virus. J Virol. 1991;65:1202–1207. doi: 10.1128/jvi.65.3.1202-1207.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fielding A K, Chapel-Fernandez S, Chadwick M P, Bullough F J, Cosset F L, Russell S J. A hyperfusogenic gibbon ape leukemia envelope glycoprotein: targeting of a cytotoxic gene by ligand display. Hum Gene Ther. 2000;11:817–826. doi: 10.1089/10430340050015437. [DOI] [PubMed] [Google Scholar]
- 13.Gray K D, Roth M J. Mutational analysis of the envelpe gene of Moloney murine leukemia virus. J Virol. 1993;67:3489–3496. doi: 10.1128/jvi.67.6.3489-3496.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han J Y, Cannon P M, Lai K M, Zhao Y, Eiden M V, Anderson W F. Identification of envelope protein residues required for the expanded host range of 10A1 murine leukemia virus. J Virol. 1997;71:8103–8108. doi: 10.1128/jvi.71.11.8103-8108.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han J Y, Zhao Y, Anderson W F, Cannon P M. Role of variable regions A and B in the receptor binding domain of amphotropic murine leukemia virus envelope protein. J Virol. 1998;72:9101–9108. doi: 10.1128/jvi.72.11.9101-9108.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartley J W, Rowe W P. Naturally occurring murine leukemia viruses in wild mice: characterization of a new “amphotropic” class. J Virol. 1976;19:19–25. doi: 10.1128/jvi.19.1.19-25.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartley J W, Wolford N K, Old L J, Rowe W P. A new class of murine leukemia virus associated with development of spontaneous lymphomas. Proc Natl Acad Sci USA. 1977;74:789–792. doi: 10.1073/pnas.74.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 19.Höhne M, Thaler S, Dudda J C, Groner B, Schnierle B S. Truncation of the human immunodeficiency virus-type-2 envelope glycoprotein allows efficient pseudotyping of murine leukemia virus retroviral vector particles. Virology. 1999;261:70–78. doi: 10.1006/viro.1999.9847. [DOI] [PubMed] [Google Scholar]
- 20.Hunter E. Viral entry and receptors. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 71–119. [PubMed] [Google Scholar]
- 21.Indracollo S, Minuzzo S, Feroli F, Mammano F, Calderazzo F, Chieco-Bianchi L, Amadori A. Pseudotyping of Moloney leukemia virus-based retroviral vectors with simian immunodeficiency virus leads to targeted infection of human CD4+ lymphoid cells. Gene Ther. 1998;5:209–217. doi: 10.1038/sj.gt.3300603. [DOI] [PubMed] [Google Scholar]
- 22.Januszeski M M, Cannon P M, Chen D, Rozenberg Y, Anderson W F. Functional analysis of the cytoplasmic tail of Moloney murine leukemia virus envelope protein. J Virol. 1997;71:3613–3619. doi: 10.1128/jvi.71.5.3613-3619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiernan R E, Freed E O. Cleavage of the murine leukemia virus transmembrane Env protein by human immunodeficiency virus type 1 protease: transdominant inhibition by matrix mutations. J Virol. 1998;72:9621–9627. doi: 10.1128/jvi.72.12.9621-9627.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam J S, Reeves M E, Cowherd R, Rosenberg S A, Hwu P. Improved gene transfer into human lymphocytes using retroviruses with the gibbon ape leukemia virus envelope. Hum Gene Ther. 1996;7:1415–1422. doi: 10.1089/hum.1996.7.12-1415. [DOI] [PubMed] [Google Scholar]
- 25.Landau N R, Page K A, Littman D R. Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range. J Virol. 1991;65:162–169. doi: 10.1128/jvi.65.1.162-169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lavillette D, Ruggieri A, Russell S J, Cosset F L. Activation of a cell entry pathway common to type C mammalian retroviruses by soluble envelope fragments. J Virol. 2000;74:295–304. doi: 10.1128/jvi.74.1.295-304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy J A, Pincus T. Demonstration of biological activity of a murine leukemia virus of New Zealand Black mice. Science. 1970;170:326–327. doi: 10.1126/science.170.3955.326. [DOI] [PubMed] [Google Scholar]
- 28.Lindemann D, Bock M, Schweizer M, Rethwilm A. Efficient pseudotyping of murine leukemia virus particles with chimeric human foamy virus envelope proteins. J Virol. 1997;71:4815–4820. doi: 10.1128/jvi.71.6.4815-4820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loiler S A, Difronzo N L, Holland C A. Gene transfer to human cells using retrovirus vectors produced by a new polytropic packaging cell line. J Virol. 1997;71:4825–4828. doi: 10.1128/jvi.71.6.4825-4828.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lusso P, Di Marzo Veronese F, Ensoli B, Franchini G, Jemma C, DeRocco S E, Kalyanaraman V S, Gallo R C. Expanded HIV-1 cellular tropism by phenotypic mixing with murine endogenous retroviruses. Science. 1990;247:848–852. doi: 10.1126/science.2305256. [DOI] [PubMed] [Google Scholar]
- 31.Mammano F, Salvatori F, Indracollo S, De Rossi A, Chieco-Bianchi L, Göttlinger H G. Truncation of the human immunodeficiency virus type 1 envelope glycoprotein allows efficient pseudotyping of Moloney murine leukemia virus particles and gene transfer into CD4+ cells. J Virol. 1997;71:3341–3345. doi: 10.1128/jvi.71.4.3341-3345.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miletic H, Bruns M, Tsiakas K, Vogt B, Rezai R, Baum C, Kuhlke K, Cosset F L, Ostertag W, Lother H, von Laer D. Retroviral vectors pseudotyped with lymphocytic choriomeningitis virus. J Virol. 1999;73:6114–6116. doi: 10.1128/jvi.73.7.6114-6116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller A D. Cell-surface receptors for retroviruses and implications for gene transfer. Proc Natl Acad Sci USA. 1996;93:11407–11413. doi: 10.1073/pnas.93.21.11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller A D, Buttimore C. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol Cell Biol. 1986;8:2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller A D, Chen F. Retrovirus packaging cells based on 10A1 murine leukemia virus for production of vectors that use multiple receptors for cell entry. J Virol. 1996;70:5564–5571. doi: 10.1128/jvi.70.8.5564-5571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller A D, Garcia J V, von Shur N, Lynch C M, Wilson C, Eiden M V. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller D G, Adam M A, Miller A D. Gene transfer by retrovirus vectors occurs in cells that are actively replicating at the time of infection. Mol Cell Biol. 1990;10:4239–4242. doi: 10.1128/mcb.10.8.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitrophanous K A, Yoon S, Rohll J B, Patil D, Wilkes F J, Kim V N, Kingsman S M, Kingsman A J, Mazarakis N D. Stable gene transfer to the nervous system using a non-primate lentiviral vector. Gene Ther. 1999;6:1808–1818. doi: 10.1038/sj.gt.3301023. [DOI] [PubMed] [Google Scholar]
- 39.Miyoshi H, Smith K A, Mosier D E, Verma I M, Torbett B E. Transduction of human CD34+ cells that mediate long-term engraftment of NOD/SCID mice by HIV vectors. Science. 1999;283:682–686. doi: 10.1126/science.283.5402.682. [DOI] [PubMed] [Google Scholar]
- 40.Mochizuki H, Schwartz J P, Tanaka K, Brady R O, Reiser J. High-titer human immunodeficiency virus type 1-based vector systems for gene delivery into nondividing cells. J Virol. 1998;72:8873–8883. doi: 10.1128/jvi.72.11.8873-8883.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naldini L, Blömer U, Gage F H, Trono D, Verma I M. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci USA. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 43.Pettit S C, Simsic J, Loeb D D, Everitt L, Hutchison III C A, Swanstrom R. Analysis of retroviral protease cleavage sites reveals two types of cleavage sites and the structural requirements of the P1 amino acid. J Biol Chem. 1991;266:14539–14547. [PubMed] [Google Scholar]
- 44.Pinter A, Honnen W J. Topography of murine leukemia virus envelope proteins: characterization of transmembrane components. J Virol. 1983;46:1056–1060. doi: 10.1128/jvi.46.3.1056-1060.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinter A, Honnen W J, Tung J S, O'Donnell P V, Hammerling U. Structural domains of endogenous murine leukemia virus gp70s containing specific antigenic determinants defined by monoclonal antibodies. Virology. 1982;116:499–516. doi: 10.1016/0042-6822(82)90143-x. [DOI] [PubMed] [Google Scholar]
- 46.Porter C D, Collins M K, Tailor C S, Parkar M H, Cosset F L, Weiss R A, Takeuchi Y. Comparison of efficiency of infection of human gene therapy target cells via four different retroviral receptors. Hum Gene Ther. 1996;7:913–919. doi: 10.1089/hum.1996.7.8-913. [DOI] [PubMed] [Google Scholar]
- 47.Ragheb J A, Anderson W F. pH-independent Moloney murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM domain in viral entry. J Virol. 1994;68:3220–3231. doi: 10.1128/jvi.68.5.3220-3231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rai S K, DeMartini J C, Miller A D. Retrovirus vectors bearing Jaagsiekte sheep retrovirus Env transduce human cells by using a new receptor localized to chromosome 3p21.3. J Virol. 2000;74:4698–4704. doi: 10.1128/jvi.74.10.4698-4704.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rasheed S, Pal B K, Gardner M B. Characterization of a highly oncogenic murine leukemia virus from wild mice. Intl J Cancer. 1982;29:345–350. doi: 10.1002/ijc.2910290319. [DOI] [PubMed] [Google Scholar]
- 50.Rein A, Mirro J, Haynes J G, Ernst S M, Nagashima K. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J Virol. 1994;68:1773–1781. doi: 10.1128/jvi.68.3.1773-1781.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rein A, Yang C, Haynes J A, Mirro J, Compans R. Evidence for cooperation between murine leukemia virus Env molecules in mixed oligomers. J Virol. 1998;72:3432–3435. doi: 10.1128/jvi.72.4.3432-3435.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rice N R, Henderson L E, Soweder R C, Copeland T D, Oroszlan S, Edwards J F. Synthesis and processing of the transmembrane envelope protein of equine infectious anemia virus. J Virol. 1990;64:3770–3778. doi: 10.1128/jvi.64.8.3770-3778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rose J K, Shafferman A. Conditional expression of the vesicular stomatitis virus glycoprotein gene in Escherichia coli. Proc Natl Acad Sci USA. 1981;78:6670–6674. doi: 10.1073/pnas.78.11.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schnierle B S, Stitz J, Bosch V, Nocken F, Merget-Millitzer H, Engelstädter M, Kurth R, Groner B, Cichutek K. Pseudotyping of murine leukemia virus with the envelope glycoproteins of HIV generates a retroviral vector with specificity of infection for CD4-expressing cells. Proc Natl Acad Sci USA. 1997;94:8640–8645. doi: 10.1073/pnas.94.16.8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sommerfelt M A, Petteway S R, Jr, Dreyer G B, Hunter E. Effect of retroviral proteinase inhibitors on Mason-Pfizer monkey virus maturation and transmembrane glycoprotein cleavage. J Virol. 1992;66:4220–4227. doi: 10.1128/jvi.66.7.4220-4227.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soneoka Y, Cannon P M, Ramsdale E E, Griffiths J C, Romano G, Kingsman S M, Kingsman A J. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song J J, Kim J H, Lee H, Kim E, Kim J, Park Y S, Ahn J, Yoo N C, Roh J K, Kim B S. Enhancement of gene transfer efficiency into human cancer cells by modification of retroviral vectors and addition of chemicals. Oncol Rep. 2000;7:119–124. doi: 10.3892/or.7.1.119. [DOI] [PubMed] [Google Scholar]
- 58.Stitz J, Buchholz C J, Engelstadter M, Uckert W, Bloemer U, Schmitt I, Cichutek K. Lentiviral vectors pseudotyped with envelope glycoproteins derived from gibbon ape leukemia virus and murine leukemia virus 10A1. Virology. 2000;273:16–20. doi: 10.1006/viro.2000.0394. [DOI] [PubMed] [Google Scholar]
- 59.Takeuchi Y, Simpson G, Vile R G, Weiss R A, Collins M K L. Retroviral pseudotypes produced by rescue of Moloney murine leukemia virus vector by C-type, but not D-type, retroviruses. Virology. 1992;186:792–794. doi: 10.1016/0042-6822(92)90049-u. [DOI] [PubMed] [Google Scholar]
- 60.Ting Y T, Wilson C A, Farrell K B, Chaudry G J, Eiden M V. Simian sarcoma-associated virus fails to infect Chinese hamster cells despite the presence of functional gibbon ape leukemia virus receptors. J Virol. 1998;72:9453–9458. doi: 10.1128/jvi.72.12.9453-9458.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vey M, Orlich M, Alder S, Klenk H D, Rott R, Garten W. Hemagglutinin activation of pathogenic avian influenza viruses of serotype H7 requires the protease recognition motif R-X-K/R-R. Virology. 1992;188:408–413. doi: 10.1016/0042-6822(92)90775-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.von Kalle C, Kiem H P, Goehle S, Darovsky B, Heimfeld S, Dorok-Storb B, Storb R, Shuening F G. Increased gene transfer into human hematopoietic progenitor cells by extended in vitro exposure to a pseudotyped retroviral vector. Blood. 1994;84:2890–2897. [PubMed] [Google Scholar]
- 63.Wilson C M, Reitz S, Okayma H, Eiden M V. Formation of infectious hybrid virions with gibbon ape leukemia virus and human T-cell leukemia virus retroviral envelope glycoproteins and the Gag and Pol proteins of Moloney murine leukemia virus. J Virol. 1989;63:2374–2378. doi: 10.1128/jvi.63.5.2374-2378.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wool-Lewis R J, Bates P. Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. J Virol. 1998;72:3155–3160. doi: 10.1128/jvi.72.4.3155-3160.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang Y, Vanin E F, Whitt M A, Fornerod M, Zwart R, Schneiderman R D, Grosveld G, Nienhuis A W. Inducible, high-level production of infectious murine leukemia retroviral vector particles pseudotyped with vesicular stomatitis virus G envelope protein. Hum Gene Ther. 1995;6:1203–1213. doi: 10.1089/hum.1995.6.9-1203. [DOI] [PubMed] [Google Scholar]
- 66.Zhao Y, Lee S, Anderson W F. Functional interactions between monomers of the retroviral envelope protein complex. J Virol. 1997;71:6967–6972. doi: 10.1128/jvi.71.9.6967-6972.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao Y, Zhu L, Benedict C A, Chen D, Anderson W F, Cannon P M. Functional domains in the retroviral transmembrane protein. J Virol. 1998;72:5392–5398. doi: 10.1128/jvi.72.7.5392-5398.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zufferey R, Nagy D, Mandel R J, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]