Summary
Background
Passive administration of SARS-CoV-2 neutralizing monoclonal antibodies (mAbs), such as CAS + IMD (Casirivimab + Imdevimab) antibody cocktail demonstrated beneficial effects on clinical outcomes in hospitalized patients with COVID-19 who were seronegative at baseline and outpatients. However, little is known about their impact on the host immunophenotypes.
Methods
We conducted an immunoprofiling study in 46 patients from a single site of a multi-site trial of CAS + IMD in hospitalized patients. We collected longitudinal samples during October 2020 ∼ April 2021, prior to the emergence of the Delta and Omicron variants and the use of COVID-19 vaccines. All collected samples were analyzed without exclusion and post-hoc statistical analysis was performed. We examined the dynamic interplay of CAS + IMD with host immunity applying dimensional reduction approach on plasma proteomics and high dimensional flow cytometry data.
Findings
Using an unbiased clustering method, we identified unique immunophenotypes associated with acute inflammation and disease resolution. Compared to placebo group, administration of CAS + IMD accelerated the transition from an acute inflammatory immunophenotype, to a less inflammatory or “resolving” immunophenotype, as characterized by reduced tissue injury, proinflammatory markers and restored lymphocyte/monocyte imbalance independent of baseline serostatus. Moreover, CAS + IMD did not impair the magnitude or the quality of host T cell immunity against SARS-CoV-2 spike protein.
Interpretation
Our results identified immunophenotypic changes indicative of a possible SARS-CoV-2 neutralizing antibodies-induced anti-inflammatory effect, without an evident impairment of cellular antiviral immunity, suggesting that further studies of Mabs effects on SAS-CoV-2 or other viral mediated inflammation are warranted.
Funding
Regeneron Pharmaceuticals Inc and federal funds from the Department of Health and Human Services; Administration for Strategic Preparedness and Response; Biomedical Advanced Research and Development Authority, under OT number: HHSO100201700020C.
Keywords: COVID-19, SARS-CoV-2 neutralizing antibodies, Host immunity, Longitudinal immunophenotyping, Plasma proteomics, High dimensional flow cytometry
Research in context.
Evidence before this study
Clinical trials have proved the safety and clinical benefits of neutralizing mAbs therapy for patients with COVID-19. Previous research has suggested that treatment with SARS-CoV-2 neutralizing mAbs may decrease the level of systemic inflammatory markers in certain patient groups. However, these earlier studies primarily focused on a limited number of markers, such as CRP and IL-6. The complex biology of COVID-19 makes it challenging to identify specific biomarkers that respond to therapeutics, including mAbs. Therefore, the precise effect of rapid virus clearance by mAbs on the host's immune and inflammatory responses is still not fully understood.
Added value of this study
We used global clustering method to deconvolute longitudinal blood proteomics and high dimensional flow cytometry data to find distinct immunophenotypes linked to the response to mAb treatment. Although limited to small sample size and potential biases due to patient heterogenicity, we found that administrating a mAb cocktail (CAS + IMD) to patients was associated with a more rapid shift from an acute inflammatory immune response to a calmer or “resolving” stage of immune response, with lower level of markers associated with tissue damage and inflammation, and restored the balance of lymphocytes and monocytes, regardless of the patient's baseline serostatus.
Implications of all the available evidence
Our study underscores the potential benefits of using SARS-CoV-2 neutralizing mAbs, some of which were previously unrecognized. These potential benefits, include the rapid resolution of inflammatory responses and reduced need for systemic corticosteroids. However, additional larger studies will need to be designed in order to validate any association between these findings with clinical outcomes. Despite this, our data suggest that mAbs could be a potential tool to treat inflammatory disease caused by SARS-CoV-2. Our study also shows that the use of an unbiased clustering approach with integrated immune profiling data can be a valuable approach to access the therapeutic effect of treatment intervention in diseases lacking pre-identified biomarkers.
Introduction
Monoclonal antibodies (mAbs) have emerged as a key treatment class for cancer and immune disorders.1 However, only a small number of mAbs are approved to treat or prevent infectious diseases. The COVID-19 pandemic has stimulated extensive efforts to develop neutralizing mAbs against SARS-CoV-2, providing an important component of strategies to combat COVID-19 as well as a boost to harness mAbs in the treatment of other infectious diseases. Clinical trials have demonstrated the beneficial effects of SARS-CoV-2 neutralizing mAbs cocktail therapy on mortality, clinical course, and viral load in hospitalized patients (seronegative at baseline) and outpatients with COVID-19, which led to emergency use authorization (EUA) from the U.S. Food and Drug Administration for several products, and subsequent clinical use in adults and children at risk for severe COVID-19.2, 3, 4
Although clinical trials have shown both safety and efficacy of neutralizing mAbs therapy for patients with COVID-19, questions remain regarding the impact of mAb on host immune and inflammatory responses. One of the questions is whether treatment with neutralizing mAbs during acute COVID-19 could impair host humoral and cellular adaptive immunity. Emerging data suggest that administration of neutralizing Abs prior to COVID-19 vaccination may not have drastic impact on the host mounting humoral antibody immune responses.5, 6, 7 One group did observe that patients receiving neutralizing antibodies for SARS-CoV-2 can alter epitopes of memory B cells induced by subsequent COVID-19 vaccination through antibody feedback mechanism.5 However, the impact of antiviral mAb therapies on host humoral immunity at cellular and molecular level in the context of active infection and prior vaccination remains largely unknown. Recent studies have shown that patients who received neutralizing Ab can still mount similar level of SARS-CoV-2-specific memory CD4/CD8 T cell responses in both vaccination and in acute infection setting in COVID-19 outpatients.8,9
Importantly, an unanswered question is whether rapid clearance of virus by mAbs would impact kinetics of host inflammatory responses, thus contributing to an improved clinical course. Studies have established that SARS-CoV-2 infection can mediate hyperinflammation responses in the host (increased proinflammatory cytokines or chemokines) associated with dysregulated adaptive and innate immunity through disruption of the myeloid and lymphoid lineage balance.10, 11, 12 These studies also highlighted that dysregulation in the myeloid and lymphoid compartments can be associated with severe disease. Excess circulating monocytes, particularly classical monocyte with immature phenotypes (HLA-DRlow) are well-documented in patients with COVID-19 and associated with severity and clinical outcome.10,13,14 Lymphopenia, characterized by drastically reduced number of lymphocytes (including T, B, and NK cells), is another prominent feature of COVID-19, with additional studies linking lymphopenia with disease severity, poor clinical outcomes and potential links to persistent symptoms.15, 16, 17
As part of this pandemic response effort, we developed a cocktail of two non-competing high-affinity neutralizing mAbs that bind to the receptor binding domain (RBD) of the spike protein, casirivimab (CAS, REGN10933, R10933) and imdevimab (IMD, REGN10987, R10987).18 It has demonstrated clinical benefits in reducing viral load and preventing hospitalization and death in SARS-CoV-2 infected individuals, as well as preventing COVID-19 when administrated as prophylaxis in individuals at high risk of COVID-19.2,19,20 CAS + IMD was previously authorized for treatment and post-exposure prophylaxis of COVID-19 in the United States (U.S.) and treatment and/or prevention of COVID-19 in other jurisdictions. Because more recent data revealed that CAS + IMD are unlikely to be active against SARS-CoV2 omicron-lineage variants, it was no longer authorized for use in any region of the U.S. as of January 24, 2022. We hypothesize that neutralizing Abs may impact host immunity (adaptive/innate) during active SARS-CoV-2 infection. To test this hypothesis, we recruited hospitalized patients with COVID-19 from a double-blind, placebo-controlled trial [NCT04426695] of CAS + IMD cocktail mAbs to perform immune profiling (plasma proteomics and peripheral blood flow cytometry analysis). Longitudinal blood samples were collected from a total 46 hospitalized patients at a single clinical site between October 2020 and April 2021 from one study site. Here, we present data using an unbiased global clustering approach to identify dynamic changes and/or unique immunophenotypes corresponding to the course of the disease and the impact of CAS + IMD treatment.
Methods
Ethics
Human samples used in this research were obtained from The Miriam Hospital, Providence, RI, USA, which is one of the study sites for a phase I/II/III, double-blind, placebo-controlled trial (NCT04426695).19 The trial was conducted in accordance with the principles of the Declaration of Helsinki International Council for Harmonisation Good Clinical Practice guidelines, and all applicable regulatory requirements. The Miriam Hospital review board and ethics committee oversaw the trial conduct documentation (IRB00004624). All the patients provided written informed consent before participating in the trial (Reference numbers: 20/EE/0101 and R10933-10987-COV-2066).2,19 Healthy donor samples were obtained from the New York Blood Center. Approval for donation and collection of blood from donors was attained by written consent. All donors were over 16 years of age.
Samples and participants
Blood samples were collected from 46 patients hospitalized at The Miriam Hospital, Providence, RI, USA, who consented to participate in phase I/II/III, double-blind, placebo-controlled trial of CAS + IMD cocktail mAbs (part of the parent clinical trial NCT04426695, casirivimab and imdevimab, Regeneron Pharmaceuticals, Tarrytown, NY, USA) Key inclusion criteria include: 1) The participants were confirmed SARS-CoV-2 ≤ 72 h (by validated SARS-CoV-2 antigen, RT-PCR, or other molecular diagnostic assay, using an appropriate sample such as NP, nasal, oropharyngeal or saliva). 2) Has symptoms consistent with COVID-19 (e.g., Influenza-like illness with fever and muscle pain, or respiratory illness with cough and shortness of breath), as determined by investigator with onset ≤10 days from randomization. 3) Hospitalized for ≤72 h on low-flow or no supplemental oxygen. Key exclusion criteria: 1) Patients maintained O2 saturation >94% on room air. 2) In the opinion of the investigator, unlikely to survive for >48 h from screening. 3) Receiving extracorporeal membrane oxygenation (ECMO). 4) Has new-onset stroke or seizure disorder during hospitalization. 5) Initiated renal replacement therapy due to COVID-19. Additional detailed information with patient enrolment criteria can be found in parent study NCT04426695. Enrolled patients were randomized 1:1:1 to a single intravenous dose of 2.4 g CAS + IMD (1.2 g casirivimab and 1.2 g imdevimab), 8.0 g CAS + IMD (4.0 g casirivimab and 4.0 g imdevimab), or placebo.
Standard-of-care treatments for COVID-19 were permitted. Samples were collected from October 2020 to April 2021, before the widespread of Delta and Omicron and the use of COVID-19 vaccines. All participants provided written informed consent. Details of the trial design, and full inclusion and exclusion criteria can be found in earlier reports.2,19
SARS-CoV-2 viral load and serostatus determination
All patients were assessed prior to dosing for baseline viral load in nasopharyngeal swabs and anti-SARS-CoV-2 antibodies as described previously.2,19 Briefly, anti-spike (S1) immunoglobulin (Ig) A (EUROIMMUNE), anti-S1 IgG (EUROIMMUN), and anti-nucleocapsid IgG (Abbott) were accessed using the cut-offs for negative, positive, or borderline as defined per manufacturer's instructions for use. For patients with board line or missing data, they are categorized as “other” in this study.
PBMC isolation and handling
Whole blood was collected in Sodium Heparin tube (vendor) and stored at room temperature prior to processing for PBMC isolation. In brief, PBMC were isolated by density-gradient sedimentation using SepMate tube (STEM CELL Technology) following vendor's manual. Sterile technique was used to transfer the blood to sterile conical tubes. Whole blood was diluted 1:2 in room temperature RPMI (Corning, Manassas, VA, USA). Appropriate volume of room temperature Ficoll PAQUE plus (GE Healthcare Pharmacia) was added to the bottom of a SepMate tube. Diluted blood was gently overlayed on Ficoll layer and centrifuge for 10 min at 1200g at room temperature with the brake on. PBMC layer was collected and washed with RPMI. If red blood cells contamination was present, red blood cells were lysed using ACK lysis buffer (GIBCO, Grans Island, NY, USA). Isolated PBMC were then cryopreserved in Recovery Cell Culture Freezing Medium (Gibco), placed in Mr. Frosty freezing container (Thermo Scientific, USA) and frozen overnight at −80C, and then transferred to liquid nitrogen for further storage until used in the assays. All blood samples were handled in a BSL-2 laboratory with the use of appropriate personal protective equipment and safety precautions, in accordance with the blood processing protocol approved by the Regeneron Institutional Biosafety Committee.
Flow cytometry
Cells were added to a 96-well V-bottom plate (1 × 106 cells/well) and incubated in Human TruStain FcX and True-Stain Monocyte Blocker (Biolegend), followed by incubation at 4C for 30 min with staining antibodies. Flow cytometry antibodies used in the study are listed in Supplementary Table S2. Cells were then washed and fixed with BD Stabilizing Fixative and filtered through an AcroPrep Advance Filter Plate (PALL Corporation) prior to acquisition on a BD FACSymphony™ Cell Analyzers. Data were analysed by OMIQ (Dotmatics).
Plasma proteomics
The OLINK assay were performed using OLINK Explore 384 Inflammation panel (OLINK, Uppsala, Sweden). Plasma samples were inactivated with 1% TritonX-100 for 2 h at room temperature according to the virus inactivation protocol provide by OLINK. Plasma protein concentration was measured on the OLINK platform based on the so-called Proximity Extension Assay technology (PEA). The OLINK-generated data was preprocessed, and quality controlled using the platform-specific “OLINK NPX manager” software, which background corrects, log2 transforms and normalizes all samples to an arbitrary NPX scale. NPX (Normalized Protein eXpression) is Olink's arbitrary unit which is in Log2 scale. It is calculated from Ct values and data pre-processing (normalization) is performed to minimize both intra- and inter-assay variation. The NPX is a relative quantification unit where a difference of 1 NPX equates to a doubling of protein concentration. Proteomics data are provided in Supplementary Table S5.
Pathway enrichment analysis
Proteomic markers that are significantly enriched in Cluster 1 were subjected for pathway analysis using Metascape web-based portal which integrates over 40 independent knowledge bases, including the GO, KEGG, UniProt, and GWAS databases (https://metascape.org/).21 We used Metascape to identify and visualize the top statistically enriched terms of upregulated pathway in Cluster 1 (Supplementary Table S4).
Dimensional reduction analysis using integrated flow cytometry and proteomics data
Data collected from the same patient at one given timepoint using OLINK, flow cytometry platforms and clinical data were integrated as one data file, which were then uploaded to OMIQ cloud (Dotmatrics, www.omiq.ai, www.dotmatics.com) for dimensional reduction analysis. Clinical data include patient demographic information, antiviral treatment, days post-Ab infusion, viral load, disease duration, oxygen requirement, baseline serology, disease severity (baseline and highest), outcome. Integrated data were processed in Omiq platform, dimensional reduced data were visualized using Uniform Manifold Approximation and Projection (UMAP) algorithm (Neighbors = 60, Minimum Distance = 0.4, Components = 2, Euclidean metric, Epochs = 200, spectral embedding initialization, using total 482 features identified from proteomics and flow cytometry immune cell subsets/clusters). Phenograph algorithm was used to identify major clusters after dimensional reduction (K nearest neighbors = 20, Euclidean distance metric, using UMAP 1, 2 as features). Multivariant correlation analysis of flow cytometry and proteomics data was performed using JMP16 (SAS Institute Inc).
Activation induced cell marker (AIM) assay
For AIM assay, we adapt the protocol as previously described.22 Briefly, cryopreserved cells were thawed by diluting cells in 10 ml complete RPMI 1640 with 10% heat inactivated FBS (Gibco) in the presence of benzonase (0.5%). PBMC were cultured for 24 h in the presence of 1 ug/ml SARS-CoV-2 specific spike peptides in 96-well U bottomed plate at 1 × 106 cells per well. Three PepTivator SARS-CoV-2 pools (Prot_S, S1 and S+) were purchased from Miltenyi Biotech. The pools comprise 15 mer peptides overlapping by 11 amino acids and cover the entire sequences of the SARS-CoV-2 spike protein. Peptides were dissolved in DMSO and diluted in AIM culture medium for T cell stimulation. Negative (DMSO) and positive controls (anti-CD3/CD28 Ab stimulation, combined CD4 and CD8 specific peptides for CMV or EBV) were included for internal control (JPT, Berlin, Germany). Flow cytometry gating strategy is presented in Supplementary Fig. S2. Supernatants were harvested at 24 h post-stimulation for cytokine quantification by V-plex Human proinflammatory 10-plex kit following the manufacturer's instruction (Catalog No K15049D, Meso Scale Diagnostics).
Statistical analysis
Statistical analysis was performed using Prism software (Graphpad Version 9) or JMP (Version 16). There are no sample exclusion criteria used for this study. This was entirely a post-hoc analysis and no sample size/power calculations were conducted. Post-hoc power analysis does not provide information beyond the pathway analysis themselves. The pathway analysis is a hypothesis generating exercise and those result should be interpreted as it is. All analyses were performed as described and no specific adjustment for imbalance between the groups was performed. Nominal P values are shown in relevant interpretation are described. For multiple comparisons, FDR method of Benjamini–Hochberg was used for post-hoc test. Sample sizes (n) are indicated in corresponding figures or figure legends. Kruskal–Wallis test and followed by Dunn's test to access differences among groups. P value less than or equal to 0.05 were considered significant. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
Role of funders
This project has been funded in whole or in part by Regeneron Pharmaceuticals Inc and with federal funds from the Department of Health and Human Services; Administration for Strategic Preparedness and Response; Biomedical Advanced Research and Development Authority, under OT number: HHSO100201700020C. The findings and conclusions herein are those of the authors and do not necessarily represent the views of the Department of Health and Human Services or its components.
Results
Unbiased global clustering analysis of integrated proteomics and flow cytometry data identifies distinct immunophenotypes that correlate with the course of disease in hospitalized patients with COVID-19
To provide an unbiased characterization of the circulating immune cell compartment we used an integrated analysis of high-dimensional flow cytometry data from peripheral blood mononuclear cells (PBMC), extensive proteomics screening in plasma, and clinical information from patients included in the study (Fig. 1a). A total of 46 hospitalized patients with COVID-19 were recruited and sampled at baseline and at various time points post CAS + IMD treatment. Samples were collected between the period of October 2020 ∼ April 2021, prior to the emergence of the Delta and Omicron variants and the use of COVID-19 vaccines. Of those, 11 patients received 2.4 g of CAS + IMD, 18 patients received 8.0 g of CAS + IMD, and 17 patients received the placebo (Table 1). Baseline demographics and COVID-19 characterization were mostly similar among treatment groups and consistent with the larger clinical trial (NCT04426695).2,19 Placebo group has a higher percentage of seronegative patients at baseline than the 2.4 g group (9/16, 52.9% versus 3/11 27.3% respectively, P = 0.031). Viral load data were collected from these patients as part of the clinical study; however, the available viral load data are limited for statistical analysis (available viral load data are shown in Supplementary Fig. S1).
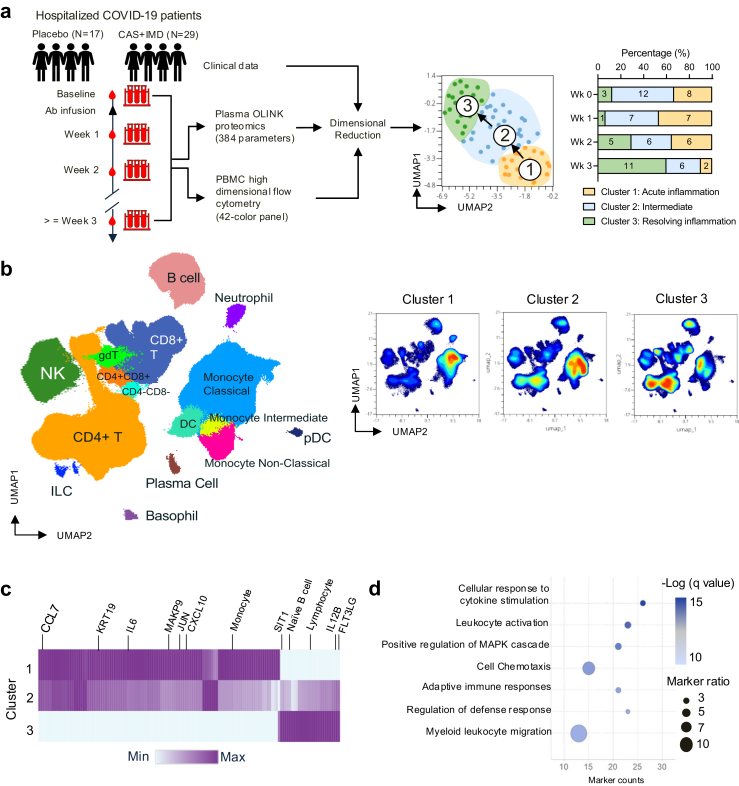
Fig. 1.
Identification of distinct immunophenotypes correlate with the course of disease using biomarker guided unbiased clustering analysis. (a) Workflow of longitudinal samples collection, analysis, and dimensional reduction. Percentage of Cluster 1, 2, 3 in longitudinal samples (binned in weeks). Numbers for individual samples from treatment group and immune clusters are labelled on the graph. (b) High dimensional flow cytometry analysis. UMAP plot of concatenated all longitudinal samples was overlaid with colour-coded and labelled major immune cell populations identified by FlowSOM (left); density UMAP plots showing concatenated samples from immunophenotype Cluster 1, 2 and 3. (c) Heatmap showing differentially expressing proteomic markers or enriched immune cell subsets (136 markers and immune cell populations) from each cluster. Selected top enriched markers are marked on the heatmap. Colour-coded based on min to max expression of each marker. (d) Quantitative result of top enriched pathway from plasma proteomics analysis. Colour gradient depicts the -Log (q value). The size of the dot corresponds to the significant representation of the indicated pathway as quantified by marker ratio. (Marker ratio = [the number of markers in the dataset/the number of expected markers from each pathway]∗100).
Table 1.
Characteristics of patients at baseline and indicated timepoints.
| Placebo |
CAS + IMD 2.4 g IV |
CAS + IMD 8.0 g IV |
P value Placebo vs 2.4 g | P value Placebo vs 8.0 g | P value 2.4 vs 8.0 g | |
|---|---|---|---|---|---|---|
| N = 17 (37%) | N = 11 (23.9%) | N = 18 (39.1%) | ||||
| Age | ||||||
| Mean (SD) | 61.5 (16.3) | 65.8 (17.8) | 64.4 (13.8) | 0.5058 | 0.6772 | 0.7659 |
| SEM | 3.9 | 5.4 | 3.3 | |||
| Age ≥ 50 | ||||||
| <50 | 3 (17.6%) | 2 (18.2%) | 4 (22.2%) | 0.685 | 0.9453 | 0.6379 |
| ≥50 | 14 (82.4%) | 9 (81.8%) | 14 (77.8%) | |||
| Sex | ||||||
| Female | 8 (47.1%) | 4 (36.4%) | 10 (55.6%) | 0.1443 | 0.7649 | 0.1058 |
| Male | 9 (52.9%) | 7 (63.6%) | 8 (44.4%) | |||
| Race | ||||||
| Black or African American | 2 (11.8%) | 2 (18.2%) | 0 | 0.9992 | >0.9999 | 0.9992 |
| Not reported | 0 | 0 | 1 (5.6%) | |||
| Unknown | 2 (11.8%) | 1 (9.1%) | 3 (16.7%) | |||
| White | 13 (76.5%) | 8 (72.7%) | 14 (77.8%) | |||
| BMI (kg/m2) | ||||||
| Mean (SD) | 32.1 (6.8) | 30.3 (7.4) | 31.8 (6.7) | 0.4958 | 0.8978 | 0.57 |
| SEM | 1.6 | 2.2 | 1.6 | |||
| BMI >30 (kg/m2) | ||||||
| ≤30 | 5 (29.4%) | 7 (63.6%) | 8 (44.4%) | 0.4093 | >0.9999 | 0.4093 |
| >30 | 12 (70.6%) | 4 (36.4%) | 9 (50%) | |||
| Missing data | 0 | 0 | 1 (5.6%) | |||
| Days of COVID-19 prior to baseline sampling | ||||||
| Mean (SD) | 5.6 (2.1) | 6 (2.6) | 5.8 (2.2) | 0.6461 | 0.7926 | 0.8163 |
| SEM | 0.5 | 0.8 | 0.5 | |||
| Baseline viral load (Log10 copies/mL) | ||||||
| Mean (SD) | 6.4 (2.4) | 7.4 (1.6) | 5.6 (2.8) | 0.3129 | 0.3551 | 0.0699 |
| SEM | 0.6 | 0.5 | 0.7 | |||
| Min:Max | 0 : 10 | 5 : 9.4 | 0 : 9.9 | |||
| Baseline serology | ||||||
| Negative | 9 (52.9%) | 3 (27.3%) | 8 (44.4%) | 0.3144 | 0.7838 | 0.4319 |
| Other (missing/board line data) | 1 (5.9%) | 3 (27.3%) | 2 (11.1%) | |||
| Positive | 7 (41.2%) | 5 (45.5%) | 8 (44.4%) | |||
| Neutralizing antibody titers for seropositive patients | ||||||
| N/A, Negative serology | 10 (58.8%) | 6 (54.5%) | 10 (55.6%) | 0.9121 | 0.9578 | 0.9541 |
| Negative | 2 (11.8%) | 0 | 1 (5.6%) | |||
| Positive | 5 (29.4%) | 4 (36.4%) | 6 (33.3%) | |||
| Unknown/Missing/Indeterminate | 0 | 1 (9.1%) | 1 (5.6%) | |||
| Oxygen cohort | ||||||
| Low flow oxygen | 13 (76.5%) | 8 (72.7%) | 10 (55.6%) | 0.534 | 0.9144 | 0.4736 |
| No supplemental oxygen | 4 (23.5%) | 3 (27.3%) | 8 (44.4%) | |||
| Comorbidities | ||||||
| Cardiovascular | ||||||
| Cardiovascular disease | 14 (82.4%) | 10 (90.9) | 11 (61.1%) | 0.7654 | 0.96 | 0.7285 |
| No history of disease | 3 (17.6%) | 1 (9.1%) | 7 (38.9%) | |||
| Autoimmune | ||||||
| Autoimmune disease | 2 (11.8%) | 1 (9.1%) | 1 (5.6%) | 0.6529 | 0.9391 | 0.6022 |
| No history of disease | 15 (88.2%) | 10 (90.9%) | 17 (94.4%) | |||
| Cancer | ||||||
| Cancer | 2 (11.8%) | 1 (9.1%) | 4 (22.2%) | 0.7207 | 0.9519 | 0.6772 |
| No history of disease | 15 (88.2%) | 10 (90.9%) | 14 (77.8%) | |||
| Diabetes | ||||||
| Diabetes | 5 (29.4%) | 4 (36.4%) | 1 (5.6%) | 0.7062 | 0.9492 | 0.6616 |
| No history of disease | 12 (70.6%) | 7 (63.6%) | 17 (94.4%) | |||
| Lung | ||||||
| Lung disease | 6 (35.3%) | 2 (18.2%) | 6 (33.3%) | 0.534 | 0.9144 | 0.4736 |
| No history of disease | 11 (64.7%) | 9 (81.8%) | 12 (66.7%) | |||
| Kidney | ||||||
| Kidney disease | 2 (11.8%) | 4 (36.4%) | 1 (5.6%) | 0.7476 | 0.9568 | 0.7083 |
| No history of disease | 15 (88.2%) | 7 (63.6%) | 17 (94.4%) | |||
| Concomitant medication | ||||||
| Systemic Corticosteroid | ||||||
| Dexamethasone | ||||||
| Pre-Treatment | 7 (41.2%) | 6 (54.5%) | 10 (55.6%) | 0.0936 | 0.7335 | 0.0629 |
| On Treatment | 8 (47.1%) | 1 (9.1%) | 5 (26.3%) | 0.0352 | 0.2132 | 0.2852 |
| Prednisone | ||||||
| Pre-Treatment | 0 | 1 (9.1%) | 0 | 0.1135 | >0.9999 | 0.1096 |
| On Treatment | 2 (11.8%) | 0 | 1 (5.6%) | 0.2324 | 0.4685 | 0.5661 |
| Methylprednisolone | ||||||
| Pre-Treatment | 0 | 0 | 0 | >0.9999 | >0.9999 | >0.9999 |
| On Treatment | 1 (5.9%) | 0 | 0 | 0.3099 | 0.2462 | >0.9999 |
| Systemic antiviral | ||||||
| Remdesivir | 15 (88.2%) | 9 (81.8%) | 14 (77.8%) | 0.7928 | 0.916 | 0.7143 |
| Systemic antibacterials | ||||||
| Systemic antibacterials | 7 (41.2%) | 6 (54.5%) | 8 (44.4%) | 0.0936 | 0.7335 | 0.0629 |
Longitudinal blood samples were collected from forty-six patients during the hospitalization and follow-up visits after discharge from twenty patients. Median blood sampling points for each patient is 2 (25% percentile = 1; 75% percentile = 3.25). Median hospitalization time is 7 days (25% percentile = 3.5, 75% percentile = 11). Median follow-up time is 28 days (25% percentile = 21.5, 75% percentile = 43). Most majority of the patients were recovered and discharged from the hospital with a total mortality of all causes 9.2% (7/46). We did not exclude any data collected from the available samples with sufficient material. We analysed 384 unique plasma proteins measured by proximity extension assay (PEA) using the OLINK platform (OLINK Explore 384 Inflammation panel). A 42-colour flow cytometry panel was developed and applied on frozen PBMC samples to identify and characterize the peripheral host immune cell populations. Using a manual gating strategy (Supplementary Fig. S2) and unsupervised clustering algorithm (Fig. 1b, Supplementary Fig. S3), we were able to identify major and rare immune subsets with unique phenotypes, including monocytes, T, B, NK, DC, ILC, γδT, plasma, and basophil subsets.
We then performed dimensional reduction of integrated flow cytometry (Fig. 1b), proteomics, and clinical data using paired (baseline and longitudinal) samples to characterize the global immunophenotype and interplay with clinical features (including patient demographic information, antiviral treatment, days post-Ab infusion, viral load, disease duration, oxygen requirement, and baseline serology) (Fig. 1a). Using this approach, we identified three major immunophenotypic clusters (Cluster 1, 2, 3) with differential expression levels of plasma protein marks, immune cell subsets and clinical features (Fig. 1a–c, d). A full list of markers that were significantly enriched in each cluster can be found in Supplementary Table S3 and a top enriched pathway analysis of upregulated plasma proteins in Cluster 1 is shown in Fig. 1d and Supplementary Table S4. Interestingly, we found that Cluster 1 and 2 are the dominant immunophenotypes correlating with the acute phase of the disease. Cluster 1 and 2 gradually decreased over time and transitioned to Cluster 3 (Fig. 1a, Supplementary Fig. S4a).
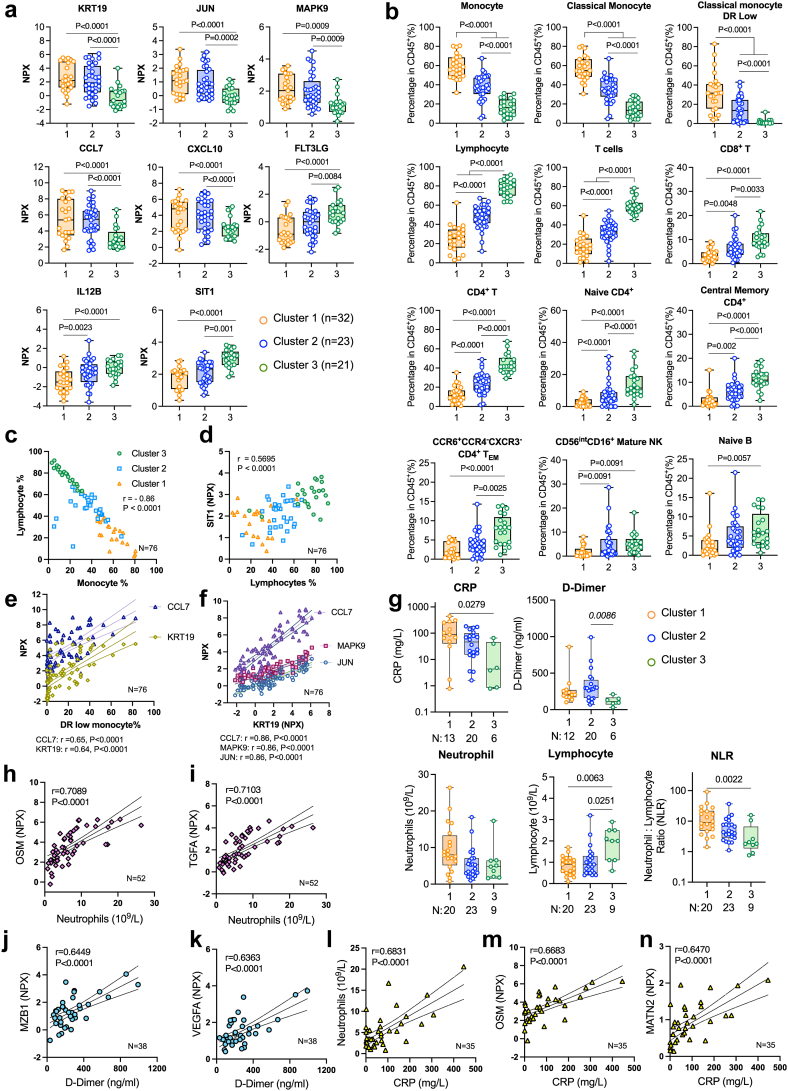
We found that samples in Cluster 1 or “Acute” immunophenotype highly express plasma markers associated with lung epithelial damaging (KRT19),23,24 cellular response to inflammation (JUN, MAPK9), leukocyte chemotaxis (CCL7, CXCL10), and show a high frequency of classical monocytes, monocytes with the immature phenotype (HLA-DRlow) and lower frequency of lymphocyte subsets, such as T, B, and NK cells (Figs. 1c and 2a, b). In contrast, samples in Cluster 3 expressed lower levels of proinflammatory markers and showed a reduction in the monocyte population, while significantly increased expression of plasma FLT3LG, IL12B and restored lymphocytes population including T, B, and NK cells, and a subset of CCR6+CCR4−CXCR3−CCR7-CD45RA−CD4+ effector memory T cells. Cluster 3 contains samples expressing a lowest level of inflammatory markers compared to Cluster 1 and 2, suggesting they are associated with the resolving phase of the inflammation; thus, we refer Cluster 3 as the “resolving” immunophenotype in this study. Notably, Cluster 2 shared features with both Cluster 1 and 3. Specifically, Cluster 2 had intermediate expression levels of markers that were enriched in either Cluster 1 or 3 (Figs. 1c and 2a, b). Moreover, there were no significant differences in the distribution of patients’ age, sex, baseline oxygen requirement, viral load, or serology across these three distinct clusters (Supplementary Fig. S4). Further, multivariant correlation analysis revealed a significant association between markers identified by proteomics and flow cytometry. Specifically, the frequency of total lymphocytes was reversely correlated with monocytes, and positively correlated with plasma SIT1 level (Fig. 2c and d), while HLA-DR low monocytes positively correlated with plasma level of KRT19 and CCL7 (Fig. 2e). Finally, top expressed proteomic inflammatory markers (CCL7, MAKP9, JUN) in “Acute” immunophenotype strongly correlated with lung injury marker KRT19 (Fig. 2f).
Fig. 2.
Characterization of three immunophenotypes. (a) Boxplots of differentially expressed plasma makers based on mean normalized protein expression unit (NPX) value across 3 clusters. False discovery rate (FDR)-adjusted P-values from Benjamini–Hochberg test are indicated. (b) Boxplots of selected immune cell subsets (% in CD45+ immune cells). (c, d) Multivariant Pearson correlation analysis showing selected pairs of immune cell subsets and plasma protein markers. Correlation coefficient (r) and P values are indicated on the figures. (n = 76) (c) Correlation of lymphocyte and monocyte frequency with colour-coded for samples from Cluster 1, 2, 3. (d) Correlation of lymphocyte frequency with plasma SIT1 level (NPX) colour-coded for samples from Cluster 1, 2, 3. (e) Correlation of monocyte frequency with plasma CCL7 and KRT19 level. (f) Correlation of plasma KRT19 with CCL7, MAPK9 and JUN. Adjusted FDR P-values are indicated. ∗∗∗∗P < 0.0001, ∗∗∗P < 0.001, ∗∗P < 0.01 and ∗P < 0.05. (g) Expression of clinical measurements (CRP, D-Dimer, neutrophil counts, lymphocyte counts and neutrophil to lymphocyte ratio) in immune clusters. (h-n) Multivariant Pearson correlation analysis showing selected pairs of clinical measurements with immune cell subsets or plasma protein markers. P values shown on the plots were determined by Knuskal–Wallies test and corrected for multiple comparisons by Dunn's test. Box and whisker plots, box extends from the 25th to 75th percentiles, whisker from min to max, show all points. Number of samples are indicated in each panel.
We obtained clinical measures (D-Dimer, C-reactive protein/CRP, blood counts) and analysed their level across clusters. Although we don't have matched clinical data for every timepoint that was used for longitudinal analysis, with the available dataset, we found that Cluster 1 has a higher CRP level than Cluster 3 and Cluster 2 has a higher D-Dimer level than Cluster 3. Both Cluster 1 and 2 have lower lymphocyte counts than Cluster 3. Cluster 1 also has a higher neutrophil-to-lymphocyte ratio (NLR) than Cluster 3 consistent with a more acute inflammatory immunophenotype (Fig. 2g). In addition, we performed multivariant correlation analysis and identified the top inflammatory markers associated with the clinical measures. We found that neutrophil counts are significantly associated with plasma transforming growth factor A (TGFA) and oncostatin M (OSM) levels (Fig. 2h and i). Top plasma proteomics markers associated with D-Dimer are Marginal Zone B And B1 Cell Specific Protein (MZB1) and Vascular endothelial growth factor A (VEGFA) (Fig. 2j and k). CRP is strongly associated with Neutrophil counts, plasma OSM, and an extracellular matrix protein matrilin-2 (MATN2) level (Fig. 2l–n). These data suggest that the immunophenotypes we identified are consistent with the established clinical measures for systemic inflammation and disease progression in COVID-19.
Treatment with CAS + IMD enhances the resolution of host inflammatory responses and restores lymphocyte/monocyte balance independently of the baseline serostatus
To examine the effect of monoclonal Abs on global host immune and inflammatory responses, we evaluated the kinetic distribution of these three immunophenotypic clusters in patients who received placebo versus CAS + IMD treatment. Importantly, upon CAS + IMD treatment there is a trend towards a faster switching to Cluster 3 as early as 1 ∼2-week post-treatment (Fig. 3a). At 2-wk post-treatment, % of patients in Cluster 3 was ∼2 fold higher in Ab-treated patients compared to the placebo group (Fig. 3a). At later time points (3 weeks or more), ∼60% of the subjects transitioned to Cluster 3 except for patients whose disease clinically progressed and eventually died. Those subjects remained in or relapsed back to Cluster 2 before death (Supplementary Fig. S5). For example, patient 40 received CAS + IMD on day 1 and showed a transient shifting immunophenotype to “resolving” at day 13 but by day 21 returned to the “acute” inflammatory phenotype before clinical progression (Supplementary Fig. S5).
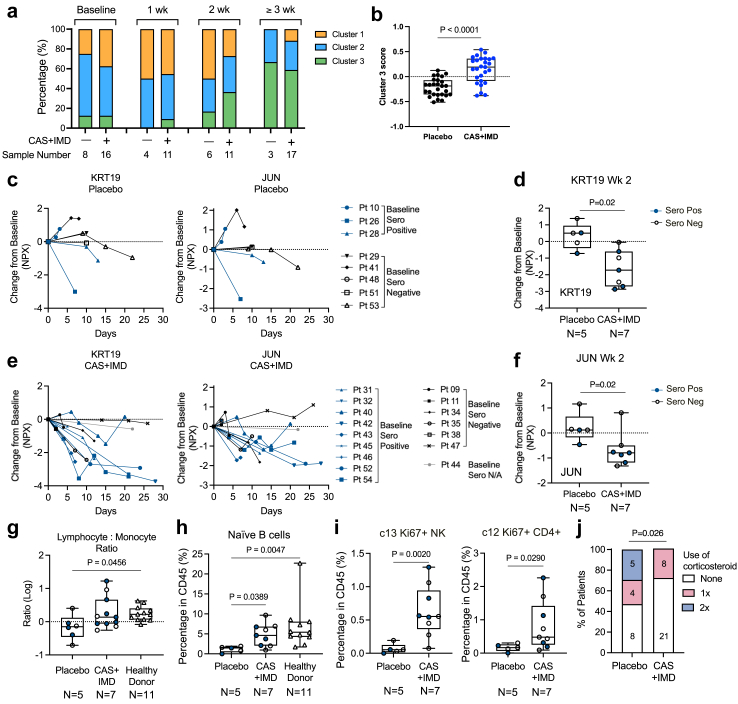
Fig. 3.
Patients received CAS + IMD rapidly reduced markers associated with tissue injury and inflammation and restore lymphocyte balance. (a) Kinetic distribution of immunophenotype clusters in patients receive placebo or CAS + IMD treatment. Percentage of each cluster in samples from indicated timepoint and treatment samples is shown. (b) z-score of Cluster 3 differentially expressed markers and immune cell subsets (n = 27 measurements, P value by non-parametric Mann Whitney t-test). Patient numbers used for analysis, Placebo n = 6, CAS + IMD n = 11. (c-f) Changes of plasma KRT19 and JUN level in paired longitudinal samples compared to baseline level. Blue symbols mark baseline seropositive patients. (c, e) Longitudinal changes with lines connecting paired samples from individual patients. (d, f) Boxplot showing changes from placebo or CAS + IMD treated patients at 2 weeks post-treatment. (d) Changes in KRT19 level. (f) Changes in JUN level (P value by unpaired t test). (g) Lymphocyte to monocyte ratio (as in Log scale) in patients received placebo or CAS + IMD at 2 weeks post-treatment (P value by One-way ANOVA, Dunn's multiple comparison test). (h) Percentage of naïve B cells in total CD45+ PBMC 1 week post placebo or CAS + IMD treatment. (P value by One-way ANOVA, Dunn's multiple comparison test). (I) Percentage of c13 (Ki67+ NK cells) and c12 (Ki67+CD4+ T cells) in PBMC 1 week post placebo or CAS + IMD treatment (P value by un-paired t test). (j) Percentage of patients underwent systemic corticosteroids during the trial in placebo or CAS + IMD group (P value by unpaired t test). Box and whisker plots, box extends from the 25th to 75th percentiles, whisker from min to max, show all points. Number of samples are indicated in each panel. Number of samples are indicated in the figure.
Remarkably, at 2 weeks post-baseline collection, the expression of markers associated with Cluster 3 “resolving” immunophenotype was significantly higher in samples from patients that received CAS + IMD treatment versus placebo (Fig. 3b, data shown quantitative analysis using 27 markers enriched in Cluster 3, median z-score for the placebo group is −0.188, CAS + IMD is 0.192, P < 0.0001). Longitudinal analysis of the top inflammatory markers enriched in “acute” immunophenotype (Cluster 1) showed CAS + IMD treatment trends to have more reduction of inflammatory markers (KRT19, JUN) compared to the placebo group (Fig. 3c–e). To quantify the differences between treatment groups, we compared the changes from baseline at 2-wk post-treatment, at which timepoint we had the greatest number of paired patient samples (Placebo N = 5, CAS + IMD N = 7). We found that while KRT19 level continued to rise from baseline in 60% (3/5) of the patients from the placebo group, the majority (86%, 6/7) of the patients in the CAS + IMD mAb treated group showed reduction of KRT19 level and no further increase from baseline (Fig. 3d). Similarly, plasma level of JUN was decreased in 6 out of 7 patients receiving CAS + IMD mAb treatment, while only 1 out of 5 patients showed decrease in the placebo group (Fig. 3f). This reduction of plasma inflammatory markers (KRT19 and JUN) was observed irrespectively of serostatus, either seropositive or seronegative for SARS-Cov-2 virus, at baseline prior to mAb therapy (baseline serostatus is annotated in Fig. 3c–f).
The lymphocyte to monocyte ratio recovered to a level like healthy donors (collected pre-pandemic) at 2 weeks post-treatment (Fig. 3g) in patients that received CAS + IMD. However, patients in the placebo group still had significantly lower lymphocyte to monocyte ratio at the same time point (Fig. 3g). Interestingly, the peripheral naïve B cell subset was significantly higher in patients that received CAS + IMD and equivalent to that observed in healthy donors (Fig. 3h).
Using an unbiased clustering approach for high dimensional flow cytometry analysis, we identified minor populations of cell cycling lymphocytes which were significantly increased in the CAS + IMD treated subjects. Specifically, a proliferating NK cell subset (c13, CD3−CD19−CD14−CD56++CD16+CD38+CD57-Ki67+) and a CD4+ effector memory T cells subset (c12, CD3+CD56−CD19−CD14−CD45RA-CCR7−ICOS+PD1+CD28+CD27-Ki67+) were significantly higher at week one post CAS + IMD treatment (Fig. 3i) compared to the placebo group at the same time point. The global immunophenotyping and quantitative analysis indicated that CAS + IMD mAb treatment can rapidly resolve inflammatory responses, tissue damage and restore the lymphocyte/monocyte balance in this patient population regardless of the baseline seropositivity.
At baseline a similar percentage of subjects in each arm of the study had previously received systemic corticosteroids for COVID-19 prior to start Ab treatment (seven out of seventeen [41.1%] subjects in the placebo arm compared with seventeen out of twenty-nine subjects [58.6%] in the CAS + IMD group) (Supplementary Table S1). Notably, the overall use of systemic corticosteroids on study (as plotted in doses of corticosteroids per patient during study) was significantly lower in patients who received CAS + IMD (Fig. 3j).
CAS + IMD treatment does not impair or interfere with the development of host cellular immunity against SARS-CoV-2 in hospitalized patients with COVID-19
To understand the impact of CAS + IMD on host cellular immunity, we identified and quantified the SARS-CoV-2 specific T cell responses using the activation induced marker (AIM) assay (Supplementary Fig. S6), which has been previously used to identify virus-specific T cells including SARS-CoV-2-specific T cells.22 We found that all patients developed SARS-CoV-2 spike-specific CD4+ and CD8+ T cell responses regardless of the treatment assignment (Fig. 4). Circulating T cell responses against spike protein were broadly stable during the study and CAS + IMD treatment did not have a negative impact on the quantity of the T cell responses (Fig. 4a–d).
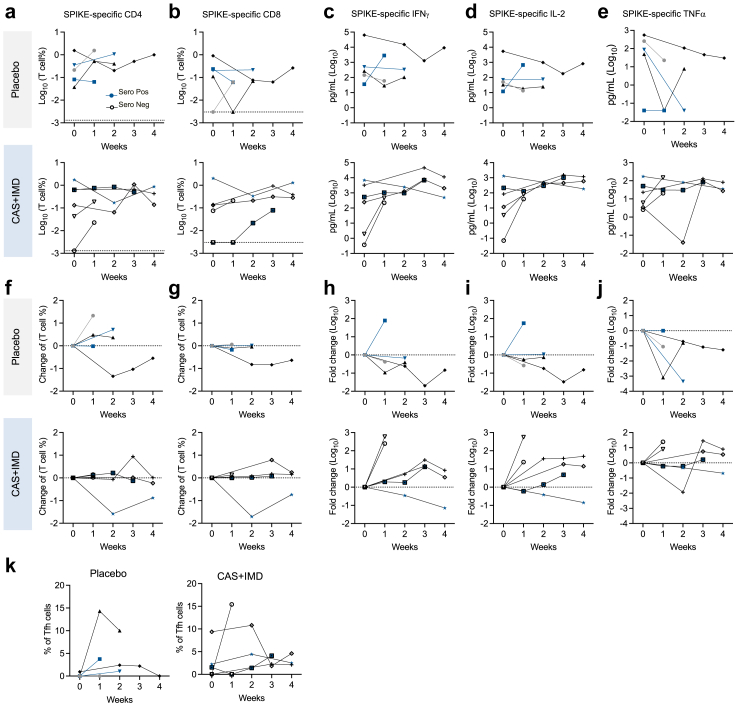
Fig. 4.
CAS + IMD treatment does not impair host T cell immunity against SARS-CoV-2. Activation induced cell marker (AIM) assay was performed in longitudinal PBMC samples to quantify viral-specific T cell responses. Cytokines were quantified by MSD assay in supernatant collected in AIM assay. (a-e). Longitudinal data showing magnitude of circulating SARS-CoV-2 spike-specific CD4+ and CD8+ T cells and cytokine (IFN-γ, IL-2, and TNF-α) production from individual with different serostatus and treatment. (f-j) Absolute changes from baseline. Longitudinal samples are binned by weeks. Serostatus is annotated under the graph. Lines connecting samples from the same individual. (K) Spike-specific TFH (T follicular helper) cell frequency in spike-specific CD4+ T cells.
To assess the functionality of the SARS-CoV-2 specific T cells, we collected the supernatant from the culture of the AIM assay for cytokine production analysis. Spike-specific Th1-skewed response, as quantified by IFN-γ, IL-2, and TNF-α production, was detected in most of the patients (Fig. 4e–j). We observed increases of IFN-γ, IL-2 and TNF-α production overtime compared to baseline (1/5 patients in placebo group; 5/6 in CAS + IMD group). Although at baseline seropositive patients showed higher T cell responses compared to seronegative patients (Supplementary Fig. S7), both seropositive (blue symbols, Fig. 4) and seronegative (black/grey symbols, Fig. 4) patients were able to develop detectable T cell responses over the course of the disease. In addition, both placebo and CAS + IMD treated patients were able to develop spike-specific T follicular helper T cells (Fig. 4k). Using available viral load data, we attempted to explore the effect of CAS + IMD on the dynamic relationship between the viral load and viral-specific T cell frequency in these patients. We found that CAS + IMD treatment does not seem to affect the dynamics and showed similar kinetics compared to the placebo group in relationship with viral load (Supplementary Fig. S8).
Discussion
Given the increased uses of mAb therapy for the treatment of infectious diseases and ongoing efforts to develop Ab therapies for omicron variants, understanding the dynamic impact of mAb therapy on host immune and inflammatory responses is of high interest and may deepen our knowledge of how mAb therapy works and subsequently influences its clinical applications. In acute COVID-19, morbidity, and mortality are associated with respiratory failure due to host hyperinflammatory responses, often independent of viral load.25 Although mAb therapies have been widely used in patients with COVID-19, there is a lack of understanding of how it may impact the resolution of host inflammatory responses, partly due to the high heterogenicity and lack of “universal” inflammatory marker(s). There is limited and inconsistent evidence suggesting that mAb therapies may have benefits in reducing systemic inflammation as measured by CRP and/or IL-6 without affecting clinical outcomes.26,27 Here we expanded clinical inflammatory markers to the integrated high-dimensional dataset and applied an unbiased global clustering approach to examine the dynamic impact of mAb therapy on host immune and inflammatory responses. The goal of our study is to identify unique immunophenotypes that may be associated with Ab therapy. Due to the limited sample size, the impact on clinical outcomes is beyond the scope of this study.
We performed immune profiling from the peripheral blood of forty-six hospitalized patients who had acute COVID-19 following infection with ancestral SARS-CoV-2 and who participated in a randomized, placebo-controlled clinical trial.3,19 We used high dimensional flow cytometry and plasma proteomics to perform deep immune profiling of immune subsets and inflammatory markers with temporal analysis of immune changes during infection. We combined these profiling data with clinical features to identify the immunophenotypes across samples and treatment arms. Using this approach, we made several key findings. First, we identified 3 major immunophenotypes (Cluster 1–3) associated with the course of the infection regardless of the treatment arms. Cluster 1, or “acute” phenotype, is enriched with samples expressing higher levels of plasma inflammatory markers. Pathway enrichment analysis revealed that many of these markers are involved in proinflammatory cytokine/chemokine and downstream signaling responses (e.g., MAPK/JUN), which are known to play a key role in COVID-19 immunopathology.28
Some of the top expressed markers have been previously identified and associated with tissue damage and disease severity e.g., KRT19 (keratin 19), which is an intracellular filament protein that is important for the structural integrity of lung epithelial cells.29 Notably, several studies showed that increase of KRT19 is associated with disease severity in patients with COVID-19.23,24 Other previously identified markers associated with acute and more severe disease, such as IL-6, CXCL10, CCL3, CCL7 (Supplementary Table S3), are also highly expressed in samples from Cluster 1.30,31 Interestingly, we observed a significant correlation between proinflammatory markers (CCL7, MAPK9 and JUN) and epithelial marker KRT19, suggesting a link between systemic inflammation and tissue damaging.
Hyperinflammation can drive profound myeloid cells (such as neutrophils and monocytes) and lymphocytes alterations in severe COVID-19.13,14 Consistent with the current understanding, we also found that there is a significant reduction of lymphocytes subsets (T, B and NK cells) and excess monocytes, particularly immature monocytes, in Cluster 1. Multivariant correlation analysis identified CCL7 and KRT19 as the top plasma markers that are associated with immature monocytes subsets, further provide evidence supporting close relationship among systemic inflammation, myeloid dysregulation, and tissue injury.
In contrast to Cluster 1, we found Cluster 3 represents an immunophenotype mostly found in samples collected from patients later in the course of the infection, or “resolving” phase. Samples from Cluster 3 have lower levels of inflammatory plasma markers and monocyte subsets but higher lymphocytes, notably T, B and NK cell subsets, suggesting an immune cell balance restoration. Intriguingly, the top 2 elevated proteins in cluster 3 were FLT3LG and IL12B. FLT3LG is a key factor driving dendritic cell homeostasis, while IL12B is produced by dendritic cells driving cellular immunity. The increased expression of FLT3LG and IL12B in Cluster 3 suggested recovery from dendritic cells deficiency, which can persist over months post SARS-CoV-2 infection.32 Samples from Cluster 2 seem to have an “transitional” phenotype sharing many features with both Cluster 1 and 3.
A second key finding from our approach was the identification of the impact of mAb therapy on the host inflammatory responses during the infection. We found that patients receiving CAS + IMD mAb treatment showed an enhanced switch to “resolving” immunophenotype with significantly increased scores of the markers enriched in Cluster 3, and significantly attenuated plasma markers associated with tissue injury and inflammatory responses at 2 weeks treatment compared to placebo. In addition, mAb therapy accelerated the restoration of the lymphocyte/monocyte imbalance 2 weeks post-treatment. Interestingly, we observed a significant increase in circulating naïve B cell frequency in mAb treated patients, suggesting that rapid neutralization of virus with mAb likely relieves the pressure on host humoral immunity and prevents further loss of naïve B cells. It would be interesting to further examine if similar impact on host immunity can be mediated by other mAb therapies with different properties.
Further, we also observed an increase of proliferating NK and CD4+ T cells with mAb treatment, which may be linked to the enhanced dendritic cell homeostasis (FLT3LG, and IL12B as discussed earlier). The various immunophenotypes are comparable between mAb therapy and the placebo group at 3 weeks or later time point, which correlates with the resolution of the infection in most patients. Upon further examination of the data from the subject that failed to respond to mAb treatment, we found that there was a transient resolution of the inflammatory responses, consistent with the “resolving” immunophenotype from samples collected at day 13 post mAb treatment. However, the condition of the patient declined later and was re-presented with the “acute” inflammatory phenotype before death. Although this study was not designed or powered as a predictive biomarker study, our data suggest unbiased immunophenotyping, using integrated immune profiling together with clinical data may be a valuable strategy for diseases with highly diverse profiles. In addition, we explored if such rapid resolution of the host inflammation could lead to a sparing of systemic corticosteroid use, providing supportive evidence for the impact of mAb therapy on host immunity. Interestingly, in this study we observed a potential association between mAb therapy and reduced need for systemic corticosteroids.
We have also explored the effect of Mab on host inflammatory responses in patients with different baseline serostatus. Based on previous clinical studies (NCT04426695 and NCT04381936) efficacy of CAS + IMD is largely limited to seronegative patients. Interestingly, our immunoprofiling data suggest that the rapid reduction of host inflammatory responses mediated by CAS + IMD may not be limited to seronegative patients. CAS + IMD could help accelerate neutralizing viruses in the airway thus reducing the inflammation to some level. However, due to the small sample size, we cannot examine the direct impact of reduced inflammation on clinical outcomes in seropositive patients.
Although this study focused on understanding the impact of CAS + IMD therapy on host inflammatory responses with an unbiased clustering approach, we were also able to examine SARS-CoV-2 specific memory T cell responses in the same subject. From a limited number of samples with enough cells to perform T cell assay, we found that mAb treatment did not impair host mounting spike-specific T cell responses. This result is consistent with two previous studies in either vaccination or acute infection setting in COVID-19 outpatients.8,9 However, in comparison to previous studies, here we observed a trend of enhanced effector cytokine (IL-2, IFN-γ, and TNF-α) production in patients that received mAb therapy (1/5 patients in the placebo group; 5/6 in CAS + IMD group showed increased cytokines production compared to baseline). Several factors may have contributed to this difference among studies. First, the patient populations are different, our study was using samples from hospitalized patients versus outpatient or non-acute COVID-19 in previous studies. Second, the mAbs used for treatment were different across studies, which may contribute to unaware differences in antigen presentation upon viral neutralization through the immune complex. In addition, different viral clearance rates by different mAbs may also have contributed to host mounting cellular immunity. Third, the assays used to detect cytokine production were different between our study (supernatant MSD assay) versus previous studies (intracellular cytokine staining and flow cytometry analysis). We have also explored the kinetics and correlation between viral load and T-cell responses. Although the number of patients is low to perform statistical analysis, we found that our kinetic data agrees with earlier studies showing that T cell immunity typically peaks in line with effective viral clearance.33,34
The major limitation of our study is the small sample size which precludes analysis of the clinical significance (e.g., survival benefit) of the immunophenotype changes with respect to the progression towards severe COVID-19. The study was designed to test a hypothesis on whether neutralizing Abs can impact host immunity, which is challenging to perform in a large clinical trial due to the high cost, risk, and justification of large amount of resources. Although interesting associations were observed in our hypothesis-driven study, we need to be cautious of generalizing our conclusions, as a small sample size may produce false-positive results or overestimate the magnitude of the responses. A large confirmatory study is needed to solidify the findings. Further, due to the small sample size, the result can also be biased or affected by heterogeneous treatment with other antiviral (Remdesivir) and/or anti-inflammatory (corticosteroids) regimens. In addition, we were not able to differentiate the effect of low versus high doses. Largely due to the clinical efficacious dose being determined at 1.2 g dose, we do not think we would observe meaningful biological differences between the two doses. By combining samples from both dose groups, we have more statistical power. As the Mab doses were higher than the dose that was recommended for clinical use, it is unclear whether the effect on immunophenotypes would be same with a lower Mab dose. Further, due to the relatively short duration of this study, the effect of neutralizing Ab on long-term host immune and inflammatory response could not be assessed. It would be interesting to evaluate whether reduction of systemic immune and inflammatory responses could be associated with a lower risk of long COVID. In addition, our findings may not be applicable in patients with comorbidities that were not overly represented in the current study, for example, patients with immunodeficiency or immunocompromised conditions. Similarly, there may be different properties of mAbs that could impact the host immune responses differently which limits the generalization of our findings to other mAbs. Our study evaluated an Ab cocktail that has no longer sufficient antiviral activity against novel variants, thus our findings may not be generalizable to COVID-19 mediated by different SARS-CoV-2 variants. Lastly, there were challenges obtaining sufficient blood samples from each patient at planned sampling time points. The missing samples may cause additional biases. Mechanistic studies are not possible due to lack of patient sample availability. A larger validation study will be helpful to confirm and validate our results. Although it would be unlikely to validate these findings in a COVID-19 clinical setting, testing other passive immunotherapies with neutralizing Mabs in a different infectious disease setting would be helpful to extend and generalize these results.
In conclusion, our study demonstrates that use of an unbiased clustering approach with integrated immune profiling data can be a valuable approach to access therapeutic effect of treatment intervention in disease lacking pre-identified biomarkers. The overall approach may be generalized to broader applications for translational immunology purposes. Applying such an approach, we found that SARS-CoV2 neutralizing mAb therapy was associated with a faster transition from an acute inflammation immunophenotype to an inflammation resolving phase with reduced tissue injury, proinflammatory markers and restored lymphocyte/monocyte imbalance independent of baseline serostatus. We identified effects that might be beneficial in the resolution of COVID-19-associated inflammatory processes. The increased rate of recovery from inflammation of Mab recipients may provide a mechanistic framework to understand the benefit of a more rapid antibody response (e.g., in seronegative individuals) and/or antibody-based therapies in patients in whom they are not expected to work.35 Our findings may provide a rationale for the use of specific (e.g., Mab-based), rather than non-specific (e.g., corticosteroids, IL-6 inhibitors) immune modulators for COVID-19. Although our result can't be generalized to other virus variants and Mab treatment modalities, our findings evaluating the effect of Ab treatment on host immunity should be interesting for general audiences in the field. Moreover, CAS + IMD did not impair the magnitude or the quality of host T cell immunity against SARS-CoV-2 spike protein. Our results indicate that administration of SARS-CoV-2 neutralizing antibodies may provide previously unknown benefit by rapidly resolving inflammatory responses without impairing cellular immunity.
Contributors
All authors read and approved the final version of the manuscript. B.W., M.F.W, E.C. S.C.H. and E.M (Eleftherios Mylonakis) have accessed and verified the underlying data.
Conceptualization: B.W., E.M., D.S. Conceived, designed, and analysed experiments: B.W., E.O., J.G., E.M., G.D.K., and D.S.
Designed clinical protocols: B.W. D.S., G.D.K., S.K., F.S., E.M.
Processing samples, data and performed experiments: B.W., E.O., P.P., Q.W., C.L., J.G., F.S., M.K, E.K.M, L.F., B.M.
Biostatistical analysis: M.F.W. E.C., S.C.H., B.J.M.
Project administration: A.T.H., J.D.H.
Writing: B.W, D.S. and E.M. wrote the manuscript with input from G.D.K. C.D.P and M.A.S.
Data sharing statement
All data are available in the main text or the supplementary materials.
Declaration of interests
B.W., J.G., P.P, Q.W., C.L., M.F.W., E.C., S.C.H., A.T.H., S.K., G.D.K., B.J.M., C.D.P., J.D.H., M.A.S., D.S. are employees of Regeneron Pharmaceuticals, Inc. E.M.O is former employee of Regeneron. E.M. (Eleftherios Mylonakis) received fundings from Regeneron Pharmaceuticals, SciClone Pharmaceuticals, Pfizer, Chemic Labs/KODA Therapeutics, Cidara, and Leidos Biomedical Research Inc./NCI, NIH/NIAID, NIH/NIGMS, and BARDA.
Acknowledgements
We thank all study participants who devoted their time to our research. Ben Daniel for flow cytometry panel consultation. We would also like to thank all study staff and investigators of COV-2071 and COV-2066 for their contributions.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105334.
Appendix A. Supplementary data
References
- 1.Pantaleo G., Correia B., Fenwick C., Joo V.S., Perez L. Antibodies to combat viral infections: development strategies and progress. Nat Rev Drug Discov. 2022;21:676–696. doi: 10.1038/s41573-022-00495-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Group R.C. Casirivimab and imdevimab in patients admitted to hospital with COVID-19 (RECOVERY) a randomised, controlled, open-label, platform trial. Lancet. 2022 doi: 10.1016/s0140-6736(22)00163-5. published online Feb 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinreich D.M., Sivapalasingam S., Norton T., et al. REGEN-COV antibody combination and outcomes in outpatients with covid-19. N Engl J Med. 2021 doi: 10.1056/nejmoa2108163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li G., Hilgenfeld R., Whitley R., Clercq E.D. Therapeutic strategies for COVID-19: progress and lessons learned. Nat Rev Drug Discov. 2023;22:449–475. doi: 10.1038/s41573-023-00672-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schaefer-Babajew D., Wang Z., Muecksch F., et al. Antibody feedback regulates immune memory after SARS-CoV-2 mRNA vaccination. Nature. 2022:1–3. doi: 10.1038/s41586-022-05609-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benschop R.J., Tuttle J.L., Zhang L., et al. The anti-SARS-CoV-2 monoclonal antibody, bamlanivimab, minimally impacts the endogenous immune response to COVID-19 vaccination. Sci Transl Med. 2022;14 doi: 10.1126/scitranslmed.abn3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L., Poorbaugh J., Dougan M., et al. Endogenous antibody responses to SARS-CoV-2 in patients with mild or moderate COVID-19 who received bamlanivimab alone or bamlanivimab and etesevimab together. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.790469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rotundo S., Vecchio E., Abatino A., et al. Spike-specific T-cell responses in patients with COVID-19 successfully treated with neutralizing monoclonal antibodies against SARS-CoV-2. Int J Infect Dis. 2022;124:55–64. doi: 10.1016/j.ijid.2022.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramirez S.I., Grifoni A., Weiskopf D., et al. Bamlanivimab therapy for acute COVID-19 does not blunt SARS-CoV-2-specific memory T cell responses. JCI Insight. 2022;7 doi: 10.1172/jci.insight.163471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silvin A., Chapuis N., Dunsmore G., et al. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020;182:1–45. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vabret N., Britton G.J., Gruber C., et al. Immunology of COVID-19: current state of the Science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lucas C., Klein J., Sundaram M.E., et al. Delayed production of neutralizing antibodies correlates with fatal COVID-19. Nat Med. 2021:1–9. doi: 10.1038/s41591-021-01355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulte-Schrepping J., Reusch N., Paclik D., et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1–64. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilk A.J., Rustagi A., Zhao N.Q., et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020;20:533. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan L., Wang Q., Zhang D., et al. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct Target Ther. 2020;5:33. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen G., Wu D., Guo W., et al. Clinical and immunologic features in severe and moderate Coronavirus Disease 2019. J Clin Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varghese J., Sandmann S., Ochs K., et al. Persistent symptoms and lab abnormalities in patients who recovered from COVID-19. Sci Rep. 2021;11 doi: 10.1038/s41598-021-91270-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen J., Baum A., Pascal K.E., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Somersan-Karakaya S., Mylonakis E., Menon V.P., et al. Casirivimab and imdevimab for the treatment of hospitalized patients with COVID-19. J Infect Dis. 2022 doi: 10.1093/infdis/jiac320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hooper A.T., Somersan-Karakaya S., McCarthy S.E., et al. Casirivimab and imdevimab treatment reduces viral load and improves clinical outcomes in seropositive hospitalized COVID-19 patients with nonneutralizing or borderline neutralizing antibodies. mBio. 2022;13 doi: 10.1128/mbio.01699-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y., Zhou B., Pache L., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grifoni A., Weiskopf D., Ramirez S.I., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filbin M.R., Mehta A., Schneider A.M., et al. Longitudinal proteomic analysis of plasma from patients with severe COVID-19 reveal patient survival-associated signatures, tissue-specific cell death, and cell-cell interactions. Cell Rep Med. 2021 doi: 10.1016/j.xcrm.2021.100287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gisby J., Clarke C.L., Medjeral-Thomas N., et al. Longitudinal proteomic profiling of dialysis patients with COVID-19 reveals markers of severity and predictors of death. Elife. 2021;10 doi: 10.7554/eLife.64827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chew K.W., Moser C., Daar E.S., et al. Antiviral and clinical activity of bamlanivimab in a randomized trial of non-hospitalized adults with COVID-19. Nat Commun. 2022;13:4931. doi: 10.1038/s41467-022-32551-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen T.O., Grandits G.A., Jain M.K., et al. Effect of neutralizing monoclonal antibody treatment on early trajectories of virologic and immunologic biomarkers in patients hospitalized with COVID-19. J Infect Dis. 2023;229:671–679. doi: 10.1093/infdis/jiad446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrison A.G., Lin T., Wang P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 2020;41:1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saha S.K., Choi H.Y., Kim B.W., et al. KRT19 directly interacts with β-catenin/RAC1 complex to regulate NUMB-dependent NOTCH signaling pathway and breast cancer properties. Oncogene. 2017;36:332–349. doi: 10.1038/onc.2016.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valle D.M.D., Kim-Schulze S., Huang H.-H., et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. The Cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pérez-Gómez A., Vitallé J., Gasca-Capote C., et al. Dendritic cell deficiencies persist seven months after SARS-CoV-2 infection. Cell Mol Immunol. 2021;18:2128–2139. doi: 10.1038/s41423-021-00728-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moss P. The T cell immune response against SARS-CoV-2. Nat Immunol. 2022;23:186–193. doi: 10.1038/s41590-021-01122-w. [DOI] [PubMed] [Google Scholar]
- 34.Notarbartolo S., Ranzani V., Bandera A., et al. Integrated longitudinal immunophenotypic, transcriptional, and repertoire analyses delineate immune responses in patients with COVID-19. Sci Immunol. 2021;6 doi: 10.1126/sciimmunol.abg5021. [DOI] [PubMed] [Google Scholar]
- 35.Misset B., Piagnerelli M., Hoste E., et al. Convalescent plasma for covid-19–induced ARDS in mechanically ventilated patients. N Engl J Med. 2023;389:1590–1600. doi: 10.1056/NEJMoa2209502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.