Abstract
Infections with high-risk human papillomaviruses (HPVs) are the major risk factor for the development of anogenital cancers. Viral E2 proteins are involved in viral DNA replication and regulation of transcription. Repression of the viral P97 promoter by E2 proteins has been implicated in the modulation of the immortalization capacity and DNA replication properties of high-risk HPVs. Analysis of the cis and trans requirements for repression of the HPV type 31 (HPV31) P97 promoter, however, revealed striking differences between the full-length E2 and the E8̂E2C fusion protein which were due to conserved residues W6 and K7 of the E8 domain. In contrast to E2, E8̂E2C completely inhibited the P97 promoter from a single promoter-distal E2 binding site. This novel long-distance repression activity of the E8 domain also enabled E8̂E2C to inhibit the HPV6a P2 promoter and minimal-promoter constructs containing E2 binding sites. Thus, E8̂E2C may represent the master repressor of viral gene expression during a high-risk HPV infection, and changes in the activity of E8̂E2C might contribute to the progression of high-risk HPV-induced lesions.
The replication cycle of human papillomaviruses (HPVs) can be divided into two stages. In the basal cell layer of the epidermis, which is composed of division-competent keratinocytes, HPVs establish a persistent, nonproductive infection in which viral DNA genomes are replicated as extrachromosomal elements at low levels and only early viral genes are transcribed. The productive viral replication cycle occurs upon differentiation of infected keratinocytes, which results in high-level replication of viral genomes, induction of late-gene transcription, and synthesis of infectious virions (22, 49).
Infections with a subset of HPV types dramatically increase the risk for the development of malignancies of the anogenital tract, and these types have been designated as high-risk HPVs (56, 57). Within this group, high-risk HPV type 16 (HPV16), HPV18, and HPV31 have been most intensively studied at the molecular level. Expression of the early E6 and E7 gene products of high-risk HPVs is sufficient to immortalize normal human keratinocytes (NHKs), the natural target cells for HPVs (34). This property is not shared by E6 and E7 genes from low-risk HPV6 and HPV11 and is therefore regarded as being relevant to the carcinogenic potential of high-risk HPVs (34). The exact molecular events that lead to malignant progression of high-risk HPV-induced lesions are still largely unknown.
Several lines of evidence suggest that transcriptional modulation of early viral gene expression is a central regulatory event. In the absence of viral gene products, HPV early-gene transcription is activated by a variety of host cell transcription factors, which interact with regulatory sequences located upstream of the major early promoter of high-risk HPV16, -18, and -31, designated P97 for HPV16 and -31 and P105 for HPV18 (22). The basal activity of HPV early promoters can be further modulated by viral E2 proteins, which are sequence-specific DNA binding proteins (22, 32). Target sequences for E2 are designated as E2 binding sites (E2BSs), four of which are located in highly conserved positions in the regulatory region of a large group of HPVs, including all high-risk types. Recognition of E2BSs is mediated by the C terminus of E2, which is also responsible for dimerization of E2 proteins (32). The N-terminal domain is required for activation of transcription and viral DNA replication (32). Interestingly, E2 can function as a transcriptional repressor of the high-risk HPV major early P97/P105 promoter (2, 42, 48, 54, 55). This appears to be mainly due to competition with cellular transcription factors. Binding of E2 to promoter-proximal E2BS4 interferes with the recognition of the neighboring TATA box by the TATA box binding protein (TBP), a subunit of the TFIID complex (14). In addition, E2 may affect the stability of the preassembled preinitiation complex after binding of TBP to DNA has occurred (21). Furthermore, binding of E2 to E2BS2 and -3 may contribute to promoter repression by competition with cellular transcription factors such as SP1, depending on the cell line and the HPV type being analyzed (8, 9, 12, 52).
Transcripts initiated at P97 are polycistronic, with the potential to encode oncoproteins E6 and E7 as well as replication proteins E1 and E2 (49), which implies that early-promoter repression by E2 may modulate viral DNA replication and immortalization of keratinocytes. In line with this, HPV31 genomes mutated in E2BS4 replicated to higher levels than wild-type genomes, which suggests that transcriptional repression by E2 is involved in regulating the extent of viral DNA replication (51). Furthermore, analysis of HPV16 genomes revealed that the ability of HPV16 to immortalize NHKs can be enhanced by mutations in an E2BS or the E2 gene (41). Further evidence that E2 repression modulates immortalization of cells by high-risk HPVs has come from cell lines containing integrated high-risk HPV DNA, in which overexpression of E2 resulted in decreased E6/E7 transcript levels (10, 15, 16, 25). It has also been noted that high-risk HPV DNA is often integrated into the host chromosomes in cervical carcinoma lesions in a way that disrupts the E2 gene, resulting in derepression of the HPV major early promoter (56, 57). Taken together, these findings gave rise to the hypothesis that early-promoter repression by E2 counteracts the development of cancer in vivo.
Aside from the full-length form of E2, two additional E2 proteins have been identified in bovine papillomavirus type 1 (BPV1)-infected cells, which have been named E2C and E8/E2 (7, 23, 27, 28). BPV1 E2C is an N-terminally truncated E2 protein which is generated from a promoter located within the E2 gene (28). E8/E2, a fusion protein in which parts of the E8 gene are linked to the C-terminal half of the E2 gene, is translated from an alternatively spliced viral transcript (7). Both proteins retain the DNA binding-dimerization domain of E2 and are therefore able to form homo- and heterodimers which specifically recognize E2BSs (1, 29, 33). Spliced viral transcripts comparable to the BPV1 E8/E2 mRNA that generate fusion proteins in which also the N-terminal domain of E2 is replaced with the small viral E8 gene have also been described for high-risk HPV16, -31, and -33 and low-risk HPV11 (13, 43, 46, 48). The respective proteins have been designated E8̂E2C, E2C, or sE2 depending on the virus type (13, 43, 46, 48). HPV31 genomes that were unable to express E8̂E2C replicated their DNA to much higher levels than wild-type HPV31 in short-term assays in undifferentiated keratinocytes (48). This indicated that E8̂E2C is a potent negative regulator of HPV DNA replication during the early phase of the viral life cycle. Overreplication of mutated HPV31 genomes may be primarily due to an increase in E2 activity since E8̂E2C (and other N-terminally truncated E2 proteins) have been demonstrated to inhibit E2 in DNA replication and transcription assays, which may be in part explained by competition at E2BSs (1, 4–7, 28–30, 33, 48). Therefore, the major role of HPV E8̂E2C is believed to be counteracting E2.
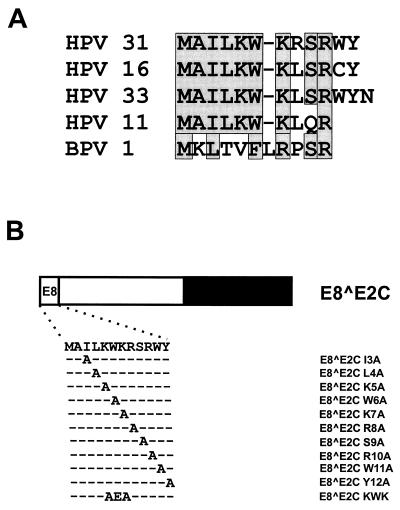
In addition to inhibiting E2, E8̂E2C may also control viral replication by modulating viral gene expression on its own, since it has been demonstrated that repression of the HPV major early promoter is not restricted to E2 but can also be achieved by E8̂E2C and E2 proteins lacking the activation domain (3, 6, 8, 12, 48, 54), which suggested that the hinge-DNA binding-dimerization domain is sufficient for promoter repression. However, we have noted that the E8 domain is conserved among HPVs (see Fig. 4a), making it likely that the E8 part of E8̂E2C is important for functions ascribed to the fusion protein.
FIG. 4.
Alanine-scanning mutagenesis of the conserved E8 domain. (A) Alignment of E8 domains from HPV11, -16, -31, and -33 and BPV1. Identical or similar residues are boxed. (B) Structure and sequences of mutated HPV31 E8̂E2C proteins. The different domains of the E8̂E2C protein are shown at the top. The amino acid sequence of E8 residues 1 to 12 is depicted below, and the mutated residues are indicated.
Our data indicate that the E8 domain is required for a novel transcriptional repression activity, which appears to be different from the mechanism of repression achieved by E2 or the E2 DNA binding-dimerization domain. In contrast to E2, E8̂E2C not only repressed the HPV31 P97 promoter from promoter-distal E2BSs but also inhibited the activity of the HPV6a P2 (or E7) promoter as well as that of synthetic transcription units containing E2BSs. Alanine-scanning mutagenesis of the E8 domain revealed that repression activity is largely dependent on amino acids W6 and K7, which are conserved among all HPV E8 genes described so far. The identification of the high-risk HPV31 E8̂E2C protein as a long-distance transcriptional repressor raises the intriguing possibility that E8̂E2C not only antagonizes E2's activity but serves as a master repressor for all viral promoters. E8̂E2C may therefore be involved in gene expression changes throughout the course of a high-risk HPV infection, and changes in its activity may contribute to the progression of high-risk HPV-induced lesions.
MATERIALS AND METHODS
Recombinant plasmids.
Luciferase reporter plasmids pGL31URR, 6aNCR-P1∗P2-luc, and p6×E2BS-luc have been described previously (39, 47). Mutations in the E2BSs of plasmid pGL31URR were introduced by overlap-extension PCR (20). The exact nucleotide changes in the mutated E2BSs have been described previously (51). PCR-generated fragments carrying mutations in E2BS2, -3, or -4 were digested with BstXI and HindIII and then used to replace the BstXI-HindIII fragment from pGL31URR, resulting in plasmids pGL31URR BS4MT, pGL31URR BS3,4MT, and pGL31URR BS2,3,4MT. Plasmids pGL31URR BS1,3,4MT and pGL31URR BS1,2,3,4MT were obtained by replacing the RsrII-SpeI fragment from pGL31URR BS3,4MT or pGL31URR BS2,3,4MT with the respective fragment from pHPV31-BS1 (51).
The introduction of mutations in the HPV6a E2BS of plasmid 6aNCR-P2luc has been described previously (39). Plasmid 6aNCR-P1∗P2-BS1mt-luc was constructed by ligating the SalI-DraI fragment from plasmid E2BS-1mt (39) and the DraI-KpnI fragment from 6aNCR-P1∗P2luc to SalI and KpnI-digested vector plasmid pALuc. Plasmid E2BS-2mt was used to clone plasmid 6aNCR-P1∗P2-BS2mt-luc as described for 6aNCR-P1∗P2-BS1mt. The SalI-BstXI fragment from 6aNCR-P1∗P2-BS1mt-luc was used to replace the corresponding fragment in 6aNCR-P1∗P2-BS2mt-luc, giving rise to plasmid 6aNCR-P1∗P2-BS1,2mt-luc. Plasmids 6aNCR-P1∗P2-BS3,4mt-luc and 6aNCR-P1∗P2-BS2,3,4mt-luc were constructed by replacing the MluI fragment from 6aNCR-P1∗P2-BS4mt-luc with the corresponding fragment from plasmid E2BS3/4mt or E2BS2/3/4mt (39), respectively. Plasmid 6aNCR-P1∗P2-BS1,2,3,4mt-luc was cloned by replacing the BstXI-HindIII fragment from 6aNCR-P1∗P2-BS2,3,4mt-luc with the corresponding fragment from 6aNCR-P1∗P2-BS1mt-luc. Plasmid pC18-SP1-luc (a kind gift of G. Steger, Institute of Virology, Cologne, Germany) consists of four synthetic E2 binding sites (5′-CTAGACCGAAAACGGTG-3′) and two synthetic SP1 binding sites (5′-GATCTAAACCCCGCCCAGCCG-3′) upstream of a minimal adenovirus major late promoter composed of the TATA box and the initiator element inserted into the luciferase reporter plasmid pALuc (G. Steger, unpublished data). Plasmid pC18-luc, which is comparable to plasmid pC18 (19), was constructed by removing the SP1 binding sites from pC18-SP1-luc by BamHI digestion. E2 and SP1 binding sites were deleted from plasmid pC18-luc by digesting with HindIII and BamHI, filling in the ends with Klenow polymerase, and religating the fragments. This plasmid, named pML44-luc, is comparable to pML-44 (19).
The eukaryotic expression vectors for HPV31 E2 (pSXE2) and HPV31 E8̂E2C (pSGE8̂E2C) are based on pSG5 (Stratagene) and have been described previously (47, 48). The plasmid originally called pSGE8̂E2C (48) was renamed pSGE8̂E2C-L and was then modified to facilitate the introduction of mutations. The coding region of E8̂E2C (HPV31 nucleotides [nt] 1259 to 1296 and 3295 to 3810) was amplified by PCR using plasmid pSGE8̂E2C-L as a template and an upstream primer with an EcoRI restriction site and an NcoI restriction site overlapping the ATG start codon (Table 1). The PCR-generated fragment was cloned into pSG5, giving rise to plasmid pSGE8̂E2C, which lacks the sequences upstream of the E8 start codon (HPV31 nt 1212 to 1258). Site-specific mutagenesis of pSGE8̂E2C was performed by PCR with the oligonucleotides shown in Table 1. Mutated fragments were used to replace the EcoRI fragment in pSGE8̂E2C, giving rise to plasmids pSGE8̂E2C-I3A, -L4A, -K5A, -W6A, -K7A, -R8A, -S9A, -R10A, -W11A, -Y12A, and -KWK and pSGE8̂E2C d3-12 (see Fig. 1 and 4B). All mutations were confirmed by DNA sequence analysis of the complete cloned fragment.
TABLE 1.
Oligonucleotide sequences used for the generation of wild-type and mutated E8̂E2C expression constructs
| Oligonucleotide | Sequencea | HPV31 nt |
|---|---|---|
| E8RI-WT | CCAGACGAATTCCATGGCAATAC | 1246–1268 |
| E8RI-I3A | CCAGACGAATTCCATGGCAGCACTGAAGTGG | 1246–1276 |
| E8RI-L4A | CCAGACGAATTCCATGGCAATAGCGAAGTGGAAAC | 1246–1280 |
| E8RI-K5A | CCAGACGAATTCCATGGCAATACTGCGTGGAAACGC | 1246–1282 |
| E8RI-W6A | CCAGACGAATTCCATGGCAATACTGAAGGCGAAACGC | 1246–1282 |
| E8RI-K7A | CCAGACGAATTCCATGGCCAATACTGAAGTGGGCACGCAGCAG | 1246–1287 |
| E8RI-R8A | CCAGACGAATTCCATGGCAATACTGAAGTGGAAAGCCAGCAGATG | 1246–1290 |
| E8RI-S9A | CCAGACGAATTCCATGGCAATACTGAAGTGGAAACGCGCCAGATGGTAC | 1246–1294 |
| E8RI-R10A | CCAGACGAATTCCATGGCAATACTGAAGTGGAAACGCAGCGCATGGTACAG | 1246–1296 |
| E8RI-W11A | CCAGACGAATTCCATGGCAATACTGAAGTGGAAACGCAGCAGAGCGTACAGC AGTG | 1246–1296̂ 3295–3299 |
| E8RI-Y12A | CCAGACGAATTCCATGGCAATACTGAAGTGGAAACGCAGCAGATGGGCCAGC AGTGAC | 1246–1296̂ 3295–3301 |
| E8RI-KWK | CCAGACGAATTCCATGGCAATACTGGCGGAGGCACGCAGCAG | 1246–1287 |
| E8RI-d3-12 | GGTTAAGAATTCCATGGCTAGTGACGAAATATCCTTTGCTGGG | 3296–3319 |
Sequences are shown in the 5′-3′ direction. Nucleotide changes resulting in amino acid exchanges are underlined. Sequences nonhomologous to HPV31 are shown in italics.
FIG. 1.
Schematic depiction of the HPV31 E2 proteins and the deletion mutant E8̂E2C d3-12. The E2 proteins consist of a variable N terminus and a common C-terminal hinge and DNA binding-dimerization domain. The deletion mutant E8̂E2C d3-12 retains only the hinge and dimerization-DNA-binding domain common to both E2 and E8̂E2C.
Generation and culture of human keratinocytes.
NHKs were isolated from human foreskin epithelium as described previously (44) and were maintained in keratinocyte growth medium (Clonetics). The RTS3b keratinocyte cell line was maintained in E medium without fibroblast feeder cells (38, 40).
Transient luciferase expression assay.
Approximately 105 RTS3b or NHK cells (passage 2 to 5) were seeded into 35-mm-diameter dishes. The next day, cells were cotransfected with 200 ng of luciferase reporters and 10 ng of pSG5 or the respective HPV31 expression vector DNA as indicated in the figure legends. Transfections were carried out with 5 μl of Lipofectamine (Life Technologies) in OptiMEM (Life Technologies) for RTS3b cell lines or in keratinocyte growth medium for NHKs in accordance with the manufacturer's recommendations. Luciferase assays were carried out 48 h after transfection. The cells were washed twice with cold phosphate-buffered saline (PBS) and then lysed by adding 150 μl of cold luciferase extraction buffer (0.1 M potassium phosphate [pH 7.8], 1% Triton X-100, 1 mM dithiothreitol [DTT]). Lysates were cleared by centrifugation (20,000 × g, 5 min, 4°C), and 20 to 80 μl of extract was subjected to luminometer analysis as described in the manufacturer's manual. Transient luciferase expression assays were repeated with different plasmid preparations at least four times to ensure reproducibility. NHKs from different donors were used to exclude donor-specific effects.
Gel retardation analysis.
Approximately 5 × 105 NHKs were seeded into 60-mm-diameter dishes. The next day, cells were transfected with 2 μg of expression vector DNA in the presence of 15 μl of Lipofectamine (Life Technologies). Cells were harvested 48 h after transfection, and crude nuclear extracts were prepared as described previously (50). Briefly, cells were washed once with cold PBS, scraped in 1 ml of cold PBS into a microcentrifuge tube, and pelleted by centrifugation (20,000 × g, 30 s, 4°C). The cell pellet was incubated for 5 min on ice in 150 μl of lysis buffer (10 mM HEPES [pH 7.9], 300 mM saccharose, 50 mM NaCl, 0.25 mM EGTA, 0.5% [vol/vol] Igepal CA 630 [Sigma Aldrich], 1 mM EDTA, 1 mM DTT, 0.5 mM sodium orthovanadate, 50 mM NaF, protease inhibitor cocktail [Sigma Aldrich], and 10 μM N-acetyl-Leu-Leu-Nle-CHO [Calbiochem]). Nuclei were pelleted at 3,000 × g and 4°C for 5 min. The nuclear pellet was extracted on ice for 15 min with 30 μl of elution buffer (20% [vol/vol] glycerol, 10 mM HEPES [pH 7.9], 500 mM NaCl, 0.5 mM EDTA, 0.1 mM EGTA, 1 mM DTT, 50 mM NaF, 0.5 mM sodium orthovanadate, protease inhibitor cocktail, and 10 μM N-acetyl-Leu-Leu-Nle-CHO). The supernatant, representing the crude nuclear extract, was recovered by centrifugation (20,000 × g, 5 min, 4°C) in a microcentrifuge. Aliquots were snap frozen and stored at −80°C. Gel retardation analysis was carried out with 50,000 cpm of a 32P-end-labeled double-stranded oligonucleotide representing E2BS4 (HPV31 nt 45 to 70) or E2BS4 MT (51). Binding reactions were carried out for 10 min on ice in a final volume of 20 μl containing equal amounts of crude nuclear extract, the labeled oligonucleotide, and final concentrations of 10 mM HEPES (pH 7.9), 125 mM NaCl, 5 mM DTT, 10% glycerol, 50 μg of salmon sperm DNA/ml, and 75 μg of poly (dI-dC)/ml (Amersham Pharmacia). Complexes were separated in a native 7% polyacrylamide gel (55 parts acrylamide to 1 part bisacrylamide) containing 0.25× Tris-borate-EDTA. Gels were run at 200 V, dried, and autoradiographed with an intensifying screen or exposed to storage screens and than visualized with a Fuji BAS 1800 phosphorimager and AIDA software.
Western blot analysis.
A chicken polyclonal antiserum (82996) was generated against a peptide consisting of amino acids 58 to 75 of the HPV31 E8̂E2C protein and affinity purified (Research Genetics, Inc.). The antiserum specifically recognized bacterially expressed E8̂E2C proteins (data not shown). Transfected NHKs were lysed in sodium dodecyl sulfate-polyacrylamide sample buffer including protease inhibitors. The lysates were heated to 95°C for 5 min and then separated in a sodium dodecyl sulfate–15% polyacrylamide gel. Proteins were transferred to a nitrocellulose membrane (Protran; pore size, 0.2 μm; Schleicher & Schuell) in a buffer containing 10 mM 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS) (pH 10.3) and 10% methanol at 70 V for 1 h. The membrane was blocked with 5% nonfat dry milk in Tris-buffered saline–0.1% Tween 20 (MTBST) for 1 h. The membrane was incubated for 90 min at room temperature with primary antibody diluted 1:2,000 in MTBST. To detect bound antibody, donkey anti-chicken immunoglobulin Y antibody coupled to horseradish peroxidase (Jackson Immunochemicals) was added at a dilution of 1:2,500 in MTBST and the membrane was further incubated for 1 h. E8̂E2C proteins were detected using the chemiluminescence reagent ECL (Amersham Pharmacia).
RESULTS
Efficient repression of HPV31 P97 activity by E8̂E2C in NHKs requires only a single, promoter-distal E2BS.
Since the regulation of the HPV major early promoter is central to the extent of viral DNA replication and oncogene expression, we decided to investigate the cis and trans requirements for promoter repression by HPV31 E8̂E2C. We have previously reported that the HPV31 P97 promoter is weakly activated in the presence of small amounts of transfected HPV31 E2 expression vector, whereas large amounts result in a moderate repression of P97 by E2 in SCC13 keratinocytes. In contrast, cotransfection of an E8̂E2C expression vector repressed P97 activity at all concentrations of vector tested (48). To evaluate whether this difference could be ascribed to the different N-terminal domains of E2 and E8̂E2C, we constructed an E2 protein that retained only the linker-hinge domain and the DNA binding-dimerization domain (E8̂E2C d3-12), which is present in both E2 and E8̂E2C (Fig. 1). It has been reported that the requirements for HPV early-promoter repression by E2 proteins differ among established epithelial cell lines (8, 11, 39). We therefore decided to analyze the regulation of the HPV31 P97 promoter by E8̂E2C in NHKs, which represent the natural target cells for HPV and are a suitable tissue culture model for the complete HPV replication cycle (17, 18, 35).
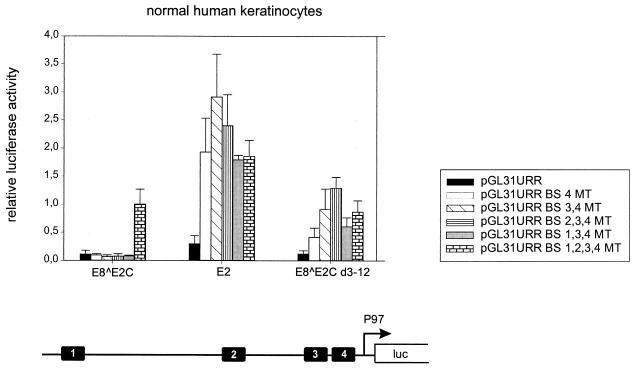
E2-mediated repression of early-promoter activity is mainly due to binding of E2 to promoter-proximal E2BS4 and is further enhanced by binding of E2 to E2BS3 and E2BS2 (8, 9, 12, 21, 39, 42, 52, 53). To delineate the contributions of individual E2BSs to E8̂E2C-mediated repression of P97 activity, we constructed a set of P97 reporter plasmids in which different combinations of mutated E2BSs were introduced. The respective mutations of individual HPV31 E2BSs have been previously shown to abolish E2 binding and also to influence the levels of transient replication of HPV31 genomes (51). Reporter plasmids (200 ng each) were transiently transfected into low-passage-number NHKs, isolated from two different donors, together with expression vectors for E2, E8̂E2C, or E8̂E2C d3-12. Cotransfection of 10 ng of the E2 expression vectors resulted in repression of the pGL31URR plasmid, but to slightly different extents (Fig. 2). Both E8̂E2C and E8̂E2C d3-12 inhibited P97 activity to 10% of the basal levels, whereas E2 repressed promoter activity to 25% (Fig. 2). Surprisingly, E8̂E2C repressed promoter activity not only from plasmid pGL31URR but also, and to the same extent (7 to 11%), from plasmids pGL31URR BS4MT, pGL31URR BS3,4MT, pGL31URR BS2,3,4MT, and pGL31URR BS1,3,4MT, which suggested that binding to neither promoter-proximal E2BS3 and -4 nor promoter-distal E2BS2 was required for efficient repression (Fig. 2). No repression was observed when all four E2BSs were inactivated by mutation (pGL31URR BS1,2,3,4MT), which demonstrated that sequence-specific recognition of either promoter-distal E2BS1 or -2 by E8̂E2C is necessary for complete inhibition of P97 activity (Fig. 2). In contrast to E8̂E2C, E2 no longer repressed transcription from pGL31URR BS4MT but instead weakly activated expression (1.9-fold) (Fig. 2). Additional mutation of E2BS1, -2, and -3 did not significantly change activation levels by E2, and therefore the weak activation does not appear to be binding site dependent (Fig. 2). The deletion mutant E8̂E2C d3-12 was able to inhibit P97 to the same extent as E8̂E2C only when E2BS4 was present, since mutation of E2BS4 (pGL31URR BS4MT) resulted in a 12 to 42% increase of the basal promoter activity (Fig. 2). Plasmid pGL31URR BS3,4MT could no longer be repressed by E8̂E2C d3-12, and the additional mutation of E2BS2 resulted even in a slight activation of promoter activity (130%). However, plasmid pGL31URR BS1,3,4MT was inhibited more strongly than pGL31URR BS2,3,4MT or pGL31URR BS1,2,3,4MT, which suggests that E2BS2, -3, and -4 contribute to repression by the hinge-DNA binding-dimerization domain (E8̂E2C d3-12), whereas binding to E2BS1 resulted in weak activation (Fig. 2).
FIG. 2.
Repression of HPV31 P97 activity by E8̂E2C in NHKs does not require promoter-proximal E2 binding sites. NHK were cotransfected with different HPV31 P97 luciferase reporter plasmids and eukaryotic expression vectors for E8̂E2C, E2 and E8̂E2C d3-12 or with the parental plasmid pSG5. The average relative luciferase activities were calculated with respect to the activity of each construct in the presence of the parental pSG5 expression vector, which was set to 1. Standard deviations are indicated by the vertical lines above the bars. The structure of the pGL31URR plasmid is shown below the graph. Conserved E2BS1 to -4 and the P97 RNA initiation site are indicated.
Taken together, these data suggested that repression of P97 by E2 and E8̂E2C proteins in NHKs is fundamentally different depending on the E2BS involved. In line with published data, we found that in the presence of all four E2BSs, all E2 proteins investigated were able to repress P97 activity. However, mutational analysis of individual E2BSs revealed that E2 repressed only in the presence of E2BS4, which is similar to E2-mediated repression of the HPV16 P97 and the HPV18 P105 promoters in C33A cells and of the HPV11 E6 promoter in in vitro transcription studies (8, 9, 21, 42, 54). The E8̂E2C d3-12 mutant protein inhibited promoter activity mainly through E2BS4, but E2BS2 and -3 seemed to contribute to repression, which confirms previously published data for HPV11 and -18 (12, 54). In striking contrast, E8̂E2C repressed P97 activity to almost identical levels as long as a single promoter-distal E2BS was present. These differences in repression activity between E8̂E2C and E8̂E2C d3-12 strongly suggest that the E8 domain is responsible for the novel repressor function of E8̂E2C.
The E8 domain is necessary for long-distance repression of the HPV6a P2 early promoter by E8̂E2C.
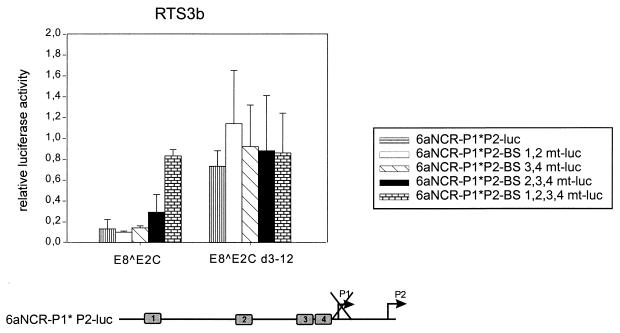
To address the issue of whether long-distance repression by E8̂E2C is restricted to the HPV31 P97 promoter, we analyzed the influence of E8̂E2C on the activity of the HPV6a P2 promoter (39, 45). The P2 (or E7) promoter is specific for low-risk HPV6 and -11, and transcription initiates at nt 270 within the HPV6a E6 gene (45). Based on the distance of E2BS1 to -4 from the P2 initiation site (200 to 660 nt), all E2BSs can be regarded as being in promoter-distal positions (Fig. 3). In contrast to the HPV6a P1 promoter, which is the equivalent of the P97/P105 promoters of high-risk HPVs, the P2 promoter is not repressed but instead is activated by full-length HPV6a E2 as well as by HPV31 E2 (reference 39 and data not shown). The reporter construct 6aNCR-P1∗P2-luc consists of the complete regulatory region of HPV6a and extends to nt 446 in the early region (Fig. 3). This plasmid contains the four conserved E2BSs (1 to 4), which are similar in sequence and location to their HPV31 counterparts. As previously described, luciferase mRNA is initiated only at the P2 promoter due to the mutational inactivation of the P1 TATA box (Fig. 3) (39). The HPV6a P2 reporter construct was transiently contransfected with empty expression vector pSG5, E8̂E2C, or E8̂E2C d3-12 into the RTS3b keratinocyte cell line, and luciferase activity was analyzed 48 h posttransfection (Fig. 3). E8̂E2C strongly inhibited promoter activity from 6aNCR-P1∗P2-luc, to 15% of the basal levels, which is similar to the repression levels obtained with the HPV31 P97 promoter. This inhibition was highly dependent on the presence of the E8 domain, since the deletion mutant E8̂E2C d3-12 repressed promoter activity to only 75%, suggesting that the long-distance inhibition activity of the E8 domain is not restricted to the HPV31 P97 promoter (Fig. 3). Since no changes in repression levels of the P97 promoter by E8̂E2C were observed as long as either E2BS1 or E2BS2 was present, it was possible that long-distance repression of HPV promoters is linked to the presence of E2BS1 or -2. We therefore analyzed the binding site requirements for P2 regulation by E8̂E2C with a set of reporter plasmids in which different combinations of E2BSs were mutated by site-directed mutagenesis. Reporter plasmids with mutations in E2BS1 and -2 (6aNCR-P1∗P2-BS1,2mt-luc) or E2BS3 and -4 (6aNCR-P1∗P2-E2BS3,4mt-luc) were inhibited to the same extent as the wild-type reporter plasmid by E8̂E2C, which suggested that long-distance repression is not specific for E2BS1 or -2 (Fig. 3). A slight loss of inhibition levels by E8̂E2C was seen with plasmid 6aNCR-P1∗P2-E2BS2,3,4mt-luc (Fig. 3). As with the HPV31 P97 promoter, repression of the P2 promoter is highly dependent on the presence of one intact E2BS, since plasmid 6aNCR-P1∗P2-BS1,2,3,4mt-luc could not be inhibited by E8̂E2C (Fig. 3). Taken together, the data obtained with the HPV31 P97 and HPV6a P2 reporter plasmids demonstrate that we have identified a novel repression activity of the E8̂E2C protein that is distinct from the repression mechanism of the HPV major early promoters by E2 or the DNA binding-dimerization domain of E2. The E8-specific inhibition works from promoter-distal E2BSs in the URR and requires amino acid residues 3 to 12 of the E8 domain.
FIG. 3.
The E8 domain is necessary for long-distance repression of the HPV6a P2 early promoter by E8̂E2C. RTS3b cells were cotransfected with expression vectors for E8̂E2C or E8̂E2C d3-12 and the 6aNCR-P1∗P2-luc luciferase reporter plasmids indicated on the right. The average relative luciferase activities were calculated with respect to the activity of each construct in the presence of the parental pSG5 expression vector, which was set to 1. Standard deviations are indicated by the vertical lines above the bars. The structure of the 6aNCR-P1∗P2-luc plasmid is shown below the graph. Conserved E2BS1 to -4 (gray boxes) and the initiation sites for the P1 and P2 promoters are indicated. No transcripts initiate at P1 because of a TATA box mutation.
Conserved tryptophan 6 and lysine 7 in the E8 domain are the major contributors to long-distance repression.
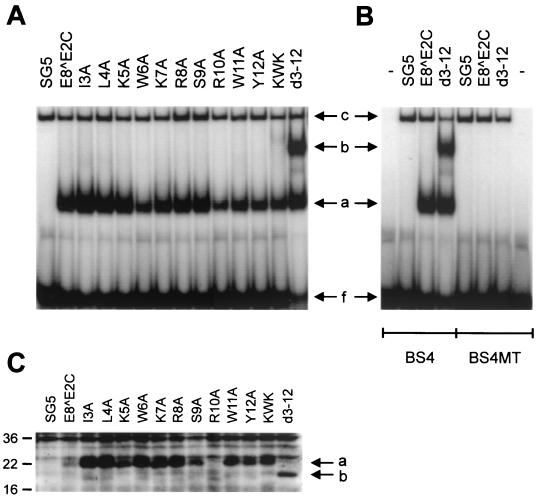
Sequence comparison revealed that the E8 gene is highly conserved among HPV11, -16, -31, and -33 (Fig. 4A), and the data presented so far indicated that E8 residues 3 to 12 are responsible for long-distance repression. We therefore investigated next what residue(s) of the HPV31 E8 domain was responsible for promoter repression. We performed an alanine-scanning mutagenesis of E8 residues 3 to 12 and also mutated a KWK motif (amino acids 5 to 7), which is part of a highly conserved stretch of charged amino acids in the central portion of the E8 domain (Fig. 4B). To control for the ability of the mutant proteins to specifically interact with DNA, NHKs were transfected with the expression vectors for E8̂E2C and mutant genes. Nuclear extracts were isolated 48 h posttransfection and were analyzed by gel retardation analysis with 32P-labeled oligonucleotides representing E2BS4 and E2BS4 MT (Fig. 5A and B). Extracts isolated from wild-type and mutant E8̂E2C-transfected cells revealed two complexes (complexes a and c) whose mobility differed from that of the unbound E2BS4 oligonucleotide (Fig. 5A, band f). In contrast to complex c, complex a was not present in pSG5-transfected cells and was abolished when E2BS4 MT was used, strongly indicating that complex a represents E8̂E2C proteins bound to DNA (Fig. 5A and B). Complex b appeared consistently only when extracts from E8̂E2C d3-12-transfected cells were used and displayed a DNA specificity identical to that of complex a (Fig. 5A and B). The appearance of two complexes with different mobilities may be due to either posttranslational modification or different protein conformations (Fig. 5A). A similar observation has been made for the full-length E2 protein of BPV1, which has been demonstrated to exist in different conformations when bound to DNA (46a). To control for expression levels of E8̂E2C proteins in transfected NHKs, immunoblot analyses were performed with cell extracts from transfected NHKs and a polyclonal antiserum directed against a peptide from E8̂E2C encompassing residues 58 to 75 (Fig. 5C). Unlike extracts from vector-transfected cells, in extracts from E8̂E2C- and mutant-transfected cells there was a specific band that corresponded to a protein with a molecular mass of approximately 22 kDa, which is similar to the calculated molecular mass (20.5 kDa) for E8̂E2C (Fig. 5C). Extracts from E8̂E2C d3-12-transfected cells displayed a band with a decreased molecular mass in line with the deletion of 10 amino acids from E8̂E2C (Fig. 5C). Mutant proteins, with the exception of R10A, were present at higher levels than the wild-type protein. In other experiments, the levels of the R10A mutant were similar to wild-type levels. We have consistently observed that the levels of mutant proteins E8̂E2C I3A, L4A, W6A, KWK, and d3-12 were increased compared to the wild-type E8̂E2C protein whereas the other mutants were present at levels similar to the wild type. These data suggested that the E8 domain not only is responsible for repression activity but also influences protein levels. Taken together, these data indicate that all mutant proteins are stably expressed and are able to specifically interact with E2BSs.
FIG. 5.
E8̂E2C mutant proteins are DNA binding competent and are stably expressed in transfected human keratinocytes. (A and B) Gel retardation analysis were performed with or without (−) nuclear extracts from NHK transfected with expression vectors as indicated above the lanes and a 32P-end-labeled double-stranded oligonucleotide representing HPV31 E2BS4 (A) or mutated E2BS4 (E2BS4 MT) (B). The mutagenesis changes the E2 recognition sequence from ACCGAAAACGGT to TTCGAAAACCCA (51). The position of the unbound oligonucleotide is indicated (f). Complexes with migration properties different from that of the oligonucleotide are labeled a to c. (C) Western blot analysis of extracts from transfected NHK. The positions of E8̂E2C proteins (a) and the E8̂E2C d3-12 (b) protein are indicated by arrows. Molecular masses are expressed in kilodaltons and are shown to the left.
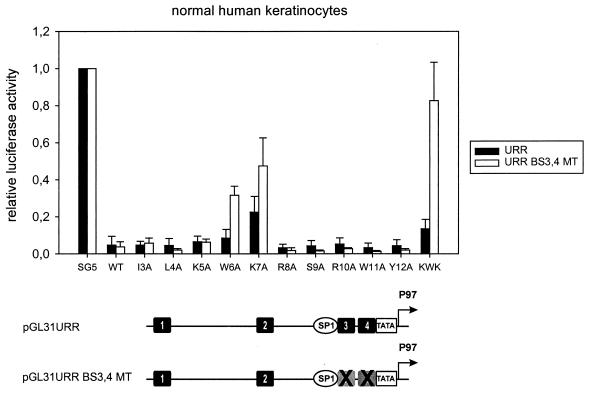
The ability of the E8̂E2C mutant proteins to repress transcription was analyzed with reporter plasmids pGL31URR and pGL31URR BS3,4MT in NHKs (Fig. 6). The activity from the wild-type reporter plasmid pGL31URR was repressed by all E8̂E2C proteins, providing further evidence that all mutant proteins were able to specifically interact with E2BSs in the nucleus (Fig. 6). E8̂E2C mutants K7A and KWK showed a slight decrease in repression activity and inhibited promoter activity to 23 and 14% of the basal activity, respectively. All other mutants behaved essentially as the wild-type E8̂E2C protein. When assayed with the pGL31URR BS3,4MT reporter plasmid, which only retains promoter-distal E2BS1 and -2, repression levels by the wild-type E8̂E2C protein and the single mutants I3A, L4A, K5A, R8A, S9A, R10A, W11A, and Y12A were not altered (Fig. 6). In contrast, repression by E8̂E2C mutants W6A and K7A decreased significantly, from 9 to 32% and from 23 to 48%, respectively (Fig. 6). An almost complete loss of promoter inhibition was observed with the E8̂E2C KWK mutant, which has mutations at positions 5, 6, and 7 (Fig. 6). In summary, these data provide evidence that the decreased long-distance repression activity of mutant proteins W6A, K7A, and KWK is not due to a loss of DNA-binding activity or decreased protein levels but strongly suggest that residues 6 and 7 are responsible for a novel, long-distance repression mechanism.
FIG. 6.
The conserved tryptophan 6 and lysine 7 residues of the E8 domain are required for long-distance repression. NHK were cotransfected with expression vectors for E8̂E2C or the respective E8̂E2C mutants and the pGL31URR (URR) or pGL31URR BS3,4MT (URR BS3,4 MT) luciferase reporter plasmids. The average relative luciferase activities were calculated with respect to the activity of each construct in the presence of the parental pSG5 expression vector, which was set to 1. Standard deviations are indicated by the vertical lines above the bars. The structure of the reporter plasmids is shown below the graph (see the legend to Fig. 2). The mutations of E2 BS3 and -4 are indicated by X's.
E8̂E2C acts as a repressor of synthetic minimal-promoter constructs containing E2BSs.
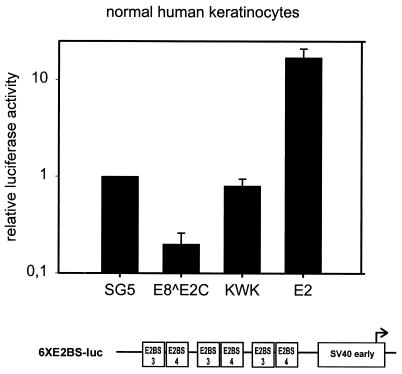
We next asked whether cis elements from the HPV URR aside from E2BSs were required for repression by E8̂E2C. To address this issue, we performed cotransfection experiments with a synthetic E2-responsive reporter plasmid which consists of the minimal simian virus 40 (SV40) early promoter and three copies of an oligonucleotide representing E2BS3 and -4 from HPV31 (47). E2 significantly enhanced luciferase expression from plasmid 6XE2BS-luc in NHKs (Fig. 7). In contrast, cotransfection of E8̂E2C repressed activity from the reporter plasmid to 20% of the basal level. No repression was observed when an expression vector for the E8̂E2C KWK mutant which is deficient for long-distance repression of the HPV31 P97 promoter, was cotransfected (Fig. 6). These data indicated that only E2BSs from the HPV31 URR are required for E8̂E2C-specific promoter repression. Furthermore, these data suggested that E2BS3 and -4 mediate repression by E2 and E8̂E2C KWK proteins (Fig. 2 and 6) only when located in close proximity to the P97 TATA box. This argues strongly that competition with cellular transcription factors interacting with E2BS3 and/or -4 does not account for promoter repression by E8̂E2C and strongly suggests that there exists a novel repression mechanism.
FIG. 7.
E8̂E2C specifically represses the SV40 early promoter. NHK were cotransfected with expression vectors for E2, E8̂E2C, or E8̂E2C KWK (KWK) and the 6×E2BS-luc luciferase reporter plasmid. The structure of the 6×E2BS-luc plasmid is shown below the graph. HPV31-specific E2BS3 and -4, the minimal SV40 early promoter (SV40 early), and the RNA initiation site (arrow) are indicated. The average relative luciferase activities were calculated with respect to the activity of each construct in the presence of the parental pSG5 expression vector, which was set to 1. Standard deviations are indicated by the vertical lines above the bars.
To further identify cis elements required for E8̂E2C-specific promoter repression, we used reporter plasmid pC18-SP1-luc, which is composed of defined transcriptional elements such as four identical E2BSs, two SP1 binding sites, and the TATA box-initiator element from the adenovirus major late promoter (Fig. 8). To address the question of whether repression by E8̂E2C requires SP1 binding sites, which are present in all reporter plasmids used in this study, the SP1 sites were deleted from pC18-SP1-luc, giving rise to pC18-luc. The basal promoter activity of pC18-luc was reduced approximately 30-fold compared to that of pC18-SP1-luc, indicating that the SP1 sites contribute to promoter activity. However, the basal promoter activity was still 10-fold higher than background levels, making it possible to determine specific repression by E8̂E2C. Reporter plasmids (200 ng) were transiently transfected into NHK in the presence of 10 ng of pSG5 or expression vectors for E2, E8̂E2C, or E8̂E2C KWK. Analysis of luciferase expression revealed that pC18-SP1-luc and pC18-luc could be transactivated by E2 approximately 100-fold (Fig. 8). The basal activities from both pC18-SP1-luc and pC18-luc were inhibited by E8̂E2C to 10 and 30%, respectively (Fig. 8). In contrast, no inhibition of basal promoter activity by the E8̂E2C KWK mutant protein was observed, but instead a slight activation of plasmid pC18-luc was detected. As has been described for the natural HPV31 P97 and the HPV6a P2 promoters (Fig. 2 and 3), the effects of the different E2 proteins on reporter gene expression were highly dependent on the presence of E2BSs, since only minor effects were observed in cotransfection experiments with plasmid pML44-luc, which was derived from plasmid pC18-luc by deletion of the E2BS (Fig. 8). These data indicate that neither activation by E2 nor repression by E8̂E2C is highly dependent on SP1 binding sites. Taken together, our data suggest that repression activity by E8̂E2C is not restricted to the early HPV promoters tested. It also indicated that the only cis elements from the HPV regulatory region that are required for repression by E8̂E2C are E2BSs, and the most likely likely target for the repression activity seems to be the protein complexes formed on the TATA box-initiator region.
FIG. 8.
E8̂E2C represses a minimal promoter consisting of E2BS and the adenovirus major late TATA box-initiator elements. NHK were cotransfected with expression vectors for E2, E8̂E2C, or E8̂E2C KWK (KWK) and the pC18-SP1-luc, pC18-luc, or pML44-luc luciferase reporter plasmids, respectively. The average relative luciferase activities were calculated with respect to the activity of each construct in the presence of the parental pSG5 expression vector, which was set to 1. Standard deviations are indicated by the vertical lines above the bars. The structures of the luciferase reporter plasmids are shown below the graph. Transcriptional control elements representing E2BS, SP1 binding sites (SP1), the adenovirus major late promoter TATA box-initiator element (INR), and the RNA initiation site (arrow) are indicated.
The long-distance repression activity of E8̂E2C is not required for inhibition of E2-transactivated transcription.
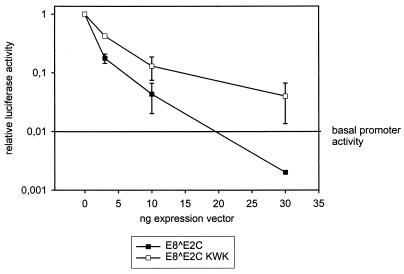
To investigate whether the E8 domain plays a role in the inhibition of E2-activated transcription by E8̂E2C, we cotransfected NHK with the E2-responsive reporter construct pC18-SP1-luc (Fig. 8), a fixed amount of the HPV31 E2 expression vector (10 ng), and increasing amounts of the expression vector for either E8̂E2C or the E8̂E2C KWK mutant, which is deficient in long-distance promoter repression (Fig. 6 to 8). Cotransfection of increasing amounts of the E8̂E2C expression vector inhibited E2-activated transcription in a concentration-dependent manner, as has been previously described for BPV1, HPV16, and HPV31 (3, 7, 48). At 30 ng of cotransfected E8̂E2C expression vector, luciferase expression levels dropped below the basal promoter activity level (Fig. 9), to levels similar to those obtained in the presence of the E8̂E2C expression vector only (Fig. 8). This indicated that E8̂E2C not only inhibits E2 transactivation but is able to repress basal promoter activity in the presence of E2. Cotransfection of increasing amounts of the E8̂E2C KWK mutant resulted also in a concentration-dependent decrease in E2-activated luciferase expression, which indicated that long-distance repression activity per se is not required for inhibition of E2 transactivation (Fig. 9). However, inhibition of E2-transactivated promoter activity by the E8̂E2C KWK mutant was slightly less efficient than that by the wild-type E8̂E2C protein, which suggests that the long-distance repression activity of E8̂E2C enhances inhibition of E2 transactivation (Fig. 9). In line with published reports, these data suggest that inhibition of E2's transactivation function by E8̂E2C could be due to formation of heterodimers and/or competition at E2BSs which requires the common C terminus of E2 (1, 4, 6, 7, 28, 33, 48).
FIG. 9.
Long-distance repression by E8̂E2C is not required for inhibition of E2-mediated transactivation of transcription. NHKs were cotransfected with 200 ng of the pC18-SP1-luc luciferase reporter plasmid and 10 ng of E2 expression vector (pSXE2) together with 0, 3, 10, or 30 ng of the E8̂E2C or E8̂E2C KWK expression vector. The total amount of expression vector was kept constant by adding the pSG5 plasmid. The average relative luciferase activity in the presence of 10 ng of pSXE2 was set to 1. The average basal promoter activity in the presence of the parental expression vector pSG5 was 0.01 and is indicated in the graph as a reference. Standard deviations are indicated by the vertical lines.
DISCUSSION
Repression of the major early promoter of high-risk HPVs by E2 proteins has been implicated in modulation of the immortalization capacity, viral DNA copy number, and extrachromosomal maintenance of these viruses (10, 15, 16, 25, 41, 51). In many carcinomas, high-risk HPV genomes are no longer extrachromosomally maintained but are present in the host genome, integrating in a way that disrupts the E2 gene (56, 57). This has led to the hypothesis that the loss of transcriptional regulation of the E6/E7 promoter by E2 proteins contributes to the development of cervical carcinomas in vivo.
The major finding of our work is that the E8 domain confers a novel transcriptional repression activity to the HPV31 E8̂E2C protein which enables the fusion protein to specifically repress promoters from promoter-distal E2BSs. However, this domain is not required for the ability of E8̂E2C to interfere with the transactivation by E2. In contrast to E2 or the truncated E8̂E2C d3-12 protein and the E8̂E2C KWK mutant protein, repression by E8̂E2C was not restricted to the HPV31 P97 promoter. E8̂E2C also inhibited basal promoter activity from the HPV6a P2 promoter and synthetic reporter constructs consisting of multimerized E2BSs and different minimal-promoter elements unrelated to HPV. Repression activity was mainly ascribed to residues W6 and K7 of the E8 domain. The respective mutant proteins (E8̂E2C W6A, K7A, KWK, and d3-12) were able to interfere with E2 transactivation and inhibited P97 activity in the presence of all four E2BSs, similar to the wild-type protein. These proteins, however, were greatly impaired in their ability to repress the P97 promoter and the HPV6a P2 promoter from distal E2BSs as well as several synthetic promoters. Therefore, we conclude that these mutations specifically interfere with the ability to repress transcription from a distance. Our data strongly suggest that sequence-specific recognition of at least one E2BS is necessary for repression by E8̂E2C. The inhibition of synthetic promoters by E8̂E2C suggests that repression does not require specific enhancer or promoter elements from the HPV regulatory region aside from E2BSs. Since a minimal promoter construct consisting of four E2BSs and the TATA box-initiator element from the adenovirus major late promoter was specifically repressed by E8̂E2C, an attractive model holds that the E8 domain interferes with the basal transcription machinery that assembles over the TATA box-initiator region, as has been described for other eukaryotic repressors (31).
Previous analyses provided evidence that repression by E2 and N-terminally truncated E2 proteins (comparable to HPV31 E8̂E2C d3-12) of the P97 promoter (and its equivalents from other HPV types) is mainly due to binding site competition with cellular transcription factors at specific E2BSs. In line with these data, we found that E2BS4 is required for P97 repression by HPV31 E2 and is involved in repression by E8̂E2C d3-12 in NHK. E2BS4 has an important role in regulation of the HPV16 P97 and HPV18 P105 promoters as previously determined in transfection experiments with the cervical carcinoma cell line C33A, and of the HPV11 E6 promoter, as evidenced by in vitro transcription analyses (8, 9, 21, 42, 54). The close proximity of E2BS4 to the early-promoter TATA box may account for its unique role in repression, and there is evidence that binding of E2 interferes with recognition of the TATA box by TBP, leading to promoter repression (14). A recent study suggested that binding of HPV11 E2 and HPV11 E1-E2 fusion proteins to E2BS4 additionally affects the stability of the assembled preinitiation complex, which contributes to repression when measured by in vitro transcription assays (21). Transient-transfection analyses of HPV11 E6 promoter repression by HPV11 E2 and HPV11 E2C (comparable to HPV31 E8̂E2C d3-12, since the construct used lacks the HPV11 E8 sequence) in C33A cells suggested that in addition to E2BS4, E2BS2 and -3 contribute to repression (12). A similar conclusion was drawn from studies of the HPV18 P105 promoter in HeLa and HaCat cell lines (8). The underlying mechanism seems to be binding site competition with cellular transcription factors binding to the GT-1 motif and the SP1 binding site. In our hands, repression by HPV31 E2 is completely dependent on the presence of E2BS4 in NHKs. It is still possible that E2 and E8̂E2C d3-12 compete with transcription factors at E2BS2 and -3, but this may contribute to repression only when the E2 transactivation domain is absent. Some evidence that E2 may functionally replace certain transcription factors has come from studies of HPV11 and HPV18 (8, 9, 12). However, our data strongly indicate that binding site competition does not account for repression of the HPV6a P2 promoter or 6×E2BS-luc or pC18 plasmid, since E8̂E2C d3-12 and E8̂E2C KWK, which are both DNA binding competent and are expressed at high levels (Fig. 5), were unable to inhibit these promoters, in contrast to the wild-type P97 promoter. Therefore, our data suggest that E8-specific long-distance repression is mediated by a different mechanism, which may involve specific binding to cellular proteins as has been described for several transcriptional repressors (26, 31).
Our data imply that the repression of basal promoter activity by E8̂E2C is not restricted to the P97 promoter and that it also occurs in the presence of E2. It is therefore very likely that other viral early promoters, and possibly the HPV31 major late promoter P742, are negatively regulated by E8̂E2C during a viral infection cycle (24, 36, 37). In this case, E8̂E2C may represent the master repressor of HPV transcription and may thus be involved in the establishment of a latent or persistent infection and/or in the differentiation-dependent early-to-late switch during a productive infection. It will also be of great interest to determine whether changes in the levels or activity of high-risk HPV E8̂E2C proteins contribute to the development of malignant lesions.
ACKNOWLEDGMENTS
We thank G. Steger and L. A. Laimins for providing reagents.
This work was funded by a grant from the Deutsche Forschungsgemeinschaft to F.S. (DFG Stu 218/2-1).
REFERENCES
- 1.Barsoum J, Prakash S S, Han P, Androphy E J. Mechanism of action of the papillomavirus E2 repressor: repression in the absence of DNA binding. J Virol. 1992;66:3941–3945. doi: 10.1128/jvi.66.6.3941-3945.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernard B A, Bailly C, Lenoir M-C, Darmon M, Thierry F, Yaniv M. The human papillomavirus type 18 (HPV18) E2 gene product is a repressor of the HPV18 regulatory region in human keratinocytes. J Virol. 1989;63:4317–4324. doi: 10.1128/jvi.63.10.4317-4324.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouvard V, Storey A, Pim D, Banks L. Characterization of the human papillomavirus E2 protein: evidence of trans-activation and trans-repression in cervical keratinocytes. EMBO J. 1994;13:5451–5459. doi: 10.1002/j.1460-2075.1994.tb06880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiang C-M, Broker T R, Chow L T. An E1M̂E2C fusion protein encoded by human papillomavirus type 11 is a sequence-specific transcription repressor. J Virol. 1991;65:3317–3329. doi: 10.1128/jvi.65.6.3317-3329.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiang C-M, Dong G, Broker T R, Chow L T. Control of human papillomavirus type 11 origin of replication by the E2 family of transcription regulatory proteins. J Virol. 1992;66:5224–5231. doi: 10.1128/jvi.66.9.5224-5231.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chin M T, Hirochika R, Hirochika H, Broker T R, Chow L T. Regulation of human papillomavirus type 11 enhancer and E6 promoter by activating and repressing proteins from the E2 open reading frame: functional and biochemical studies. J Virol. 1988;62:2994–3002. doi: 10.1128/jvi.62.8.2994-3002.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choe J, Vaillancourt P, Stenlund A, Botchan M. Bovine papillomavirus type 1 encodes two forms of a transcriptional repressor: structural and functional analysis of new viral cDNAs. J Virol. 1989;63:1743–1755. doi: 10.1128/jvi.63.4.1743-1755.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demeret C, Desaintes C, Yaniv M, Thierry F. Different mechanisms contribute to the E2-mediated transcriptional repression of human papillomavirus type 18 viral oncogenes. J Virol. 1997;71:9343–9349. doi: 10.1128/jvi.71.12.9343-9349.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demeret C, Yaniv M, Thierry F. The E2 transcriptional repressor can compensate for SP1 activation of the human papillomavirus type 18 early promoter. J Virol. 1994;68:7075–7082. doi: 10.1128/jvi.68.11.7075-7082.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desaintes C, Demeret C, Goyat S, Yaniv M, Thierry F. Expression of the papillomavirus E2 protein in HeLa cells leads to apoptosis. EMBO J. 1997;16:504–514. doi: 10.1093/emboj/16.3.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dollard S C, Broker T R, Chow L T. Regulation of the human papillomavirus type 11 E6 promoter by viral and host transcription factors in primary human keratinocytes. J Virol. 1993;67:1721–1726. doi: 10.1128/jvi.67.3.1721-1726.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong G, Broker T R, Chow L T. Human papillomavirus type 11 E2 proteins repress the homologous E6 promoter by interfering with the binding of host transcription factors to adjacent elements. J Virol. 1994;68:1115–1127. doi: 10.1128/jvi.68.2.1115-1127.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doorbar J, Parton A, Hartley K, Banks L, Crook T, Stanley M, Crawford L. Detection of novel splicing patterns in a HPV16-containing keratinocyte cell line. Virology. 1990;178:254–262. doi: 10.1016/0042-6822(90)90401-c. [DOI] [PubMed] [Google Scholar]
- 14.Dostatni N, Lambert P F, Sousa R, Ham J, Howley P M, Yaniv M. The functional BPV-1 E2 trans-activating protein can act as a repressor by preventing formation of the initiation complex. Genes Dev. 1991;5:1657–1671. doi: 10.1101/gad.5.9.1657. [DOI] [PubMed] [Google Scholar]
- 15.Dowhanick J J, McBride A A, Howley P M. Suppression of cellular proliferation by the papillomavirus E2 protein. J Virol. 1995;69:7791–7799. doi: 10.1128/jvi.69.12.7791-7799.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francis D A, Schmid S I, Howley P M. Repression of the integrated papillomavirus E6/E7 promoter is required for growth suppression of cervical cancer cells. J Virol. 2000;74:2679–2686. doi: 10.1128/jvi.74.6.2679-2686.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frattini M G, Lim H B, Doorbar J, Laimins L A. Induction of human papillomavirus type 18 late gene expression and genomic amplification in organotypic cultures from transfected DNA templates. J Virol. 1997;71:7068–7072. doi: 10.1128/jvi.71.9.7068-7072.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frattini M G, Lim H B, Laimins L A. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc Natl Acad Sci USA. 1996;93:3062–3067. doi: 10.1073/pnas.93.7.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ham J, Dostatni N, Arnos F, Yaniv M. Several different upstream promoter elements can potentiate transactivation by the BPV-1 E2 protein. EMBO J. 1991;10:2931–2940. doi: 10.1002/j.1460-2075.1991.tb07843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higuchi R, Krummel B, Saiki R K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou S Y, Wu S-Y, Zhou T, Thomas M C, Chiang C-M. Alleviation of human papillomavirus E2-mediated transcriptional repression via formation of a TATA binding protein (or TFIID)-TFIIB-RNA polymerase II-TFIIF preinitiation complex. Mol Cell Biol. 2000;20:113–125. doi: 10.1128/mcb.20.1.113-125.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howley P M. Papillomavirinae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2045–2076. [Google Scholar]
- 23.Hubbert N L, Schiller J T, Lowy D R, Androphy E J. Bovine papilloma virus-transformed cells contain multiple E2 proteins. Proc Natl Acad Sci USA. 1988;85:5864–5868. doi: 10.1073/pnas.85.16.5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hummel M, Hudson J B, Laimins L A. Differentiation-induced and constitutive transcription of human papillomavirus type 31b in cell lines containing viral episomes. J Virol. 1992;66:6070–6080. doi: 10.1128/jvi.66.10.6070-6080.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang E-S, Riese II D J, Settleman J, Nilson L A, Honig J, Flynn S, DiMaio D. Inhibition of cervical carcinoma cell line proliferation by the introduction of a bovine papillomavirus regulatory gene. J Virol. 1993;67:3720–3729. doi: 10.1128/jvi.67.7.3720-3729.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knoepfler P S, Eisenman R N. Sin meets NuRD and other tails of repression. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- 27.Lambert P F, Hubbert N L, Howley P M, Schiller J T. Genetic assignment of multiple E2 gene products in bovine papillomavirus-transformed cells. J Virol. 1989;63:3151–3154. doi: 10.1128/jvi.63.7.3151-3154.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambert P F, Spalholz B A, Howley P M. A transcriptional repressor encoded by BPV-1 shares a common carboxy-terminal domain with the E2 transactivator. Cell. 1987;50:69–78. doi: 10.1016/0092-8674(87)90663-5. [DOI] [PubMed] [Google Scholar]
- 29.Lim D A, Gossen M, Lehman C W, Botchan M R. Competition for DNA binding sites between the short and long forms of E2 dimers underlies repression in bovine papillomavirus type 1 DNA replication control. J Virol. 1998;72:1931–1940. doi: 10.1128/jvi.72.3.1931-1940.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J S, Kuo S R, Broker T R, Chow L T. The functions of human papillomavirus type 11 E1, E2, and E2C proteins in cell-free DNA replication. J Biol Chem. 1995;270:27283–27291. doi: 10.1074/jbc.270.45.27283. [DOI] [PubMed] [Google Scholar]
- 31.Maldonado E, Hampsey M, Reinberg D. Repression: targeting the heart of the matter. Cell. 1999;99:455–458. doi: 10.1016/s0092-8674(00)81533-0. [DOI] [PubMed] [Google Scholar]
- 32.McBride A A, Romanczuk H, Howley P M. The papillomavirus E2 regulatory proteins. J Biol Chem. 1991;266:18411–18414. [PubMed] [Google Scholar]
- 33.McBride A A, Byrne J C, Howley P M. E2 polypeptides encoded by bovine papillomavirus type 1 form dimers through the common carboxyl-terminal domain: transactivation is mediated by the conserved amino-terminal domain. Proc Natl Acad Sci USA. 1989;86:510–514. doi: 10.1073/pnas.86.2.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDougall J K. Immortalization and transformation of human cells by human papillomavirus. Curr Top Microbiol Immunol. 1994;186:101–119. doi: 10.1007/978-3-642-78487-3_6. [DOI] [PubMed] [Google Scholar]
- 35.Meyers C, Mayer T J, Ozbun M A. Synthesis of infectious human papillomavirus type 18 in differentiating epithelium transfected with viral DNA. J Virol. 1997;71:7381–7386. doi: 10.1128/jvi.71.10.7381-7386.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozbun M A, Meyers C. Temporal usage of multiple promoters during the life cycle of human papillomavirus type 31b. J Virol. 1998;72:2715–2722. doi: 10.1128/jvi.72.4.2715-2722.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ozbun M A, Meyers C. Two novel promoters in the upstream regulatory region of human papillomavirus type 31b are negatively regulated by epithelial differentiation. J Virol. 1999;73:3505–3510. doi: 10.1128/jvi.73.4.3505-3510.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purdie K J, Sexton C J, Proby C M, Glover M T, Williams A T, Stables J N, Leigh I M. Malignant transformation of cutaneous lesions in renal allograft patients: a role for human papillomavirus. Cancer Res. 1993;53:5328–5333. [PubMed] [Google Scholar]
- 39.Rapp B, Pawellek A, Kraetzer F, Schaefer M, May C, Purdie K, Grassmann K, Iftner T. Cell-type-specific separate regulation of the E6 and E7 promoters of human papillomavirus type 6a by the viral transcription factor E2. J Virol. 1997;71:6956–6966. doi: 10.1128/jvi.71.9.6956-6966.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rheinwald J G, Beckett M A. Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultures from human squamous cell carcinomas. Cancer Res. 1981;41:1657–1663. [PubMed] [Google Scholar]
- 41.Romanczuk H, Howley P M. Disruption of either the E1 or the E2 regulatory gene of human papillomavirus type 16 increases viral immortalization capacity. Proc Natl Acad Sci USA. 1992;89:3159–3163. doi: 10.1073/pnas.89.7.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romanczuk H, Thierry F, Howley P M. Mutational analysis of cis elements involved in E2 modulation of human papillomavirus type 16 P97 and type 18 P105 promoters. J Virol. 1990;64:2849–2859. doi: 10.1128/jvi.64.6.2849-2859.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rotenberg M O, Chow L T, Broker T R. Characterization of rare human papillomavirus type 11 mRNAs coding for regulatory and structural proteins, using the polymerase chain reaction. Virology. 1989;172:489–497. doi: 10.1016/0042-6822(89)90191-8. [DOI] [PubMed] [Google Scholar]
- 44.Ruesch M N, Stubenrauch F, Laimins L A. Activation of papillomavirus late gene transcription and genome amplification upon differentiation in semisolid medium is coincident with expression of involucrin and transglutaminase but not keratin-10. J Virol. 1998;72:5016–5024. doi: 10.1128/jvi.72.6.5016-5024.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smotkin D, Prokoph H, Wettstein F O. Oncogenic and nononcogenic human genital papillomaviruses generate the E7 mRNA by different mechanisms. J Virol. 1989;63:1441–1447. doi: 10.1128/jvi.63.3.1441-1447.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Snijders P J F, van den Brule A J C, Schrijnemakers H F J, Raaphorst P M C, Meijer C J L M, Walboomers J M M. Human papillomavirus type 33 in a tonsillar carcinoma generates its putative E7 mRNA via two E6∗ transcript species which are terminated at different early region poly(A) sites. J Virol. 1992;66:3172–3178. doi: 10.1128/jvi.66.5.3172-3178.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46a.Steger G, Ham J, Lefebvre O, Yaniv M. The bovine papillomavirus 1 E2 protein contains two activation domains: one that interacts with TBP and another that functions after TBP binding. EMBO J. 1995;14:329–340. doi: 10.1002/j.1460-2075.1995.tb07007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stubenrauch F, Colbert A M E, Laimins L A. Transactivation by the E2 protein of oncogenic human papillomavirus type 31 is not essential for early and late viral functions. J Virol. 1998;72:8115–8123. doi: 10.1128/jvi.72.10.8115-8123.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stubenrauch F, Hummel M, Iftner T, Laimins L A. The E8̂E2C protein, a negative regulator of viral transcription and replication, is required for extrachromosomal maintenance of human papillomavirus type 31 in keratinocytes. J Virol. 2000;74:1178–1186. doi: 10.1128/jvi.74.3.1178-1186.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stubenrauch F, Laimins L A. Human papillomavirus life cycle: active and latent phases. Semin Cancer Biol. 1999;9:379–386. doi: 10.1006/scbi.1999.0141. [DOI] [PubMed] [Google Scholar]
- 50.Stubenrauch F, Leigh I M, Pfister H. E2 represses the late gene promoter of human papillomavirus type 8 at high concentrations by interfering with cellular factors. J Virol. 1996;70:119–126. doi: 10.1128/jvi.70.1.119-126.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stubenrauch F, Lim H B, Laimins L A. Differential requirements for conserved E2 binding sites in the life cycle of oncogenic human papillomavirus type 31. J Virol. 1998;72:1071–1077. doi: 10.1128/jvi.72.2.1071-1077.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan S H, Gloss B, Bernard H-U. During negative regulation of the human papillomavirus-16 E6 promoter, the viral E2 protein can displace Sp1 from a proximal promoter element. Nucleic Acids Res. 1992;20:251–256. doi: 10.1093/nar/20.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan S-H, Leong L E-C, Walker P A, Bernard H-U. The human papillomavirus type 16 E2 transcription factor binds with low cooperativity to two flanking sites and represses the E6 promoter through displacement of Sp1 and TFIID. J Virol. 1994;68:6411–6420. doi: 10.1128/jvi.68.10.6411-6420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thierry F, Howley P M. Functional analysis of E2-mediated repression of the HPV18 P105 promoter. New Biol. 1991;3:90–100. [PubMed] [Google Scholar]
- 55.Thierry F, Yaniv M. The BPV1–E2 trans-acting protein can be either an activator or a repressor of the HPV18 regulatory region. EMBO J. 1987;6:3391–3397. doi: 10.1002/j.1460-2075.1987.tb02662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.zur Hausen H. Papillomavirus infections—a major cause of human cancers. Biochim Biophys Acta. 1996;1288:F55–F78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]
- 57.zur Hausen H. Molecular pathogenesis of cancer of the cervix and its causation by specific human papillomavirus types. In: zur Hausen H, editor. Human pathogenic papillomaviruses. Berlin, Germany: Springer-Verlag KG; 1994. pp. 131–156. [DOI] [PubMed] [Google Scholar]