Abstract
Neuroimmune crosstalk participates in intestinal tissue homeostasis and host defense. However, the matrix of interactions between arrays of molecularly-defined neuron subsets and of immunocyte lineages remains unclear. We utilized a chemogenetic approach to activate eight distinct neuronal subsets, assessing effects by deep immunophenotyping, microbiome profiling and immunocyte transcriptomics in intestinal organs. Distinct immune perturbations followed neuronal activation: Nos1+ neurons regulated Th17-like cells and Chat+ neurons regulated neutrophils. Trpv1+ neurons elicited the broadest immunomodulation, inducing changes in innate lymphocytes, macrophages, and RORγ+ T regulatory (Treg) cells. Further neuroanatomical, genetic and pharmacological analysis showed that Trpv1+ neurons in dorsal root ganglia decreased Treg cell numbers via the neuropeptide CGRP. Given the role of these neurons in nociception, these data potentially link pain signaling with gut Treg function.
One Sentence Summary:
Chemogenetic screen for neuroimmune interactions identifies a role for Trpv1 neurons in controlling RORγ+ Treg cells via CGRP.
INTRODUCTION
The mammalian gastrointestinal (GI) tract is one of the body’s critical barrier sites, interfacing for information exchange between the host, nutrients and resident microbiota. A dynamic interplay between the immune system, the nervous system, and microbial ecosystem maintains normal gut physiology and tissue integrity (1-3). The immune and nervous systems both possess exquisitely specific sensing and effector capabilities, and there is great interest in how they can be combined to sense chemical, damaging or infectious threats at the GI frontier.
The GI tract is densely innervated by a complex network of sensory and autonomic neurons. Gut-innervating neurons include the enteric nervous system (ENS), a largely autonomous intrinsic system that coordinates motility and secretion. In addition, extrinsic sensory neurons from dorsal root ganglia (DRG) and nodose ganglia (NG), as well as autonomic neurons that reside in the brainstem and sympathetic ganglia, project to the gut, modulating the activity of the ENS (4-6). DRG and NG neurons function primarily as sensory neurons, possessing multiple receptors such as Piezo2, Transient receptor potential (TRP) channels, and G-protein-coupled receptors (GPCRs) to detect mechanical stretch, dietary and noxious stimuli. Gut-innervating neuronal populations are highly diverse, with many molecular categories identified by single-cell RNA-sequencing (scRNAseq) (7-9). The gut immune system is equally complex, with a representation of immunocytes from all lineages: cells expressing innate receptors that respond immediately to microbial patterns (macrophages (MF) or innate-like lymphocytes (ILCs)), adaptive T or B lymphocytes with tailored responses to antigens on micro-organisms and food, and cell populations whose final role is to dampen inflammation and promote peaceful coexistence with symbiotic microbiota while ensuring barrier integrity and tissue repair (T regulatory cells (Treg), dendritic cells (DCs)).
Several elegant studies in this field have targeted specific neuronal or immunologic subtypes and revealed specific neuro-immune interactions. For example, neurons expressing vasoactive intestinal peptide (VIP) have been shown to modulate the activity of ILC3s (10-12), and Neuromedin U (NMU) expressing enteric neurons amplify ILC2-driven type 2 immune responses (13-15). Sympathetic neurons mediate gut tissue protection by crosstalk with enteric macrophages (16, 17) and ILCs (18). However, beyond these one-to-one interactions, there is a need for a more systematic and integrated approach to identify specific neuro-immune interactions in the gut.
Chemogenetic approaches (19, 20) offer the potential for activation or inhibition of molecularly defined neurons and permit one to probe many such neuronal types in parallel. They rest on the targeted expression of DREADDs (Designer Receptor Exclusively Activated by Designer Drugs), which are engineered GPCRs that are inert except when activated by a synthetic ligand that has no counterpart or receptor in mammalian organisms. Neurons can thus be activated in a temporally controlled manner. If expressed as cell type-specific transgenes, or delivered by viral vectors with fine tropism, they also allow spatial and anatomical targeting. As examples, chemogenetics has allowed the identification of the neural basis of feeding and other behaviors (21-23).
Here, we leveraged an intersectional chemogenetic approach to determine how distinct neurons modulate the gut immune system. We delivered DREADDs to the peripheral nervous system using an adeno-associated virus (AAV) vector with neuron-specific tropism, whose expression was unleashed by Cre recombinase expression in a panel of eight Cre-transgenic mouse lines, chosen to target the major classes of neuronal types that innervate the gut. Following neuronal activation, we analyzed the effects on the gut immune system and microbiota by immunophenotyping, microbial profiling, and single-cell transcriptomics. Activation of distinct neuronal subsets led to discrete alterations in immunocytes. Among these changes, activation of Trpv1+ neurons had the broadest effects, in particular leading to a decrease in Treg cells in the cecum and colon. We identified the neuro-anatomical basis of this regulation and molecular pathways involved, introducing a new model for neuro-immune collaboration in non-self sensing at the organismal frontier.
RESULTS
Chemogenetic screen of neurons for modulatory capacities of the GI immune system and microbiome
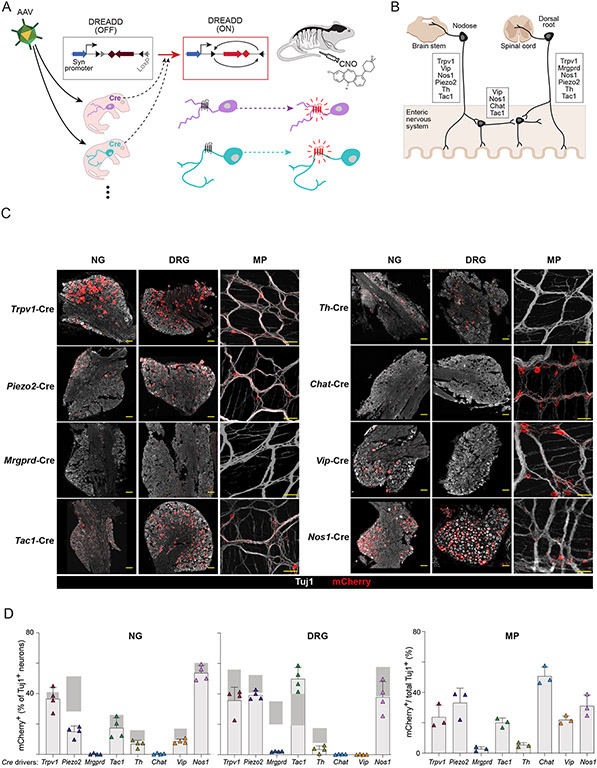
We set up a systematic screen to detect how activation of peripheral neuronal subtypes modulate the immune system and gut microbiome, based on targeted expression of DREADD molecules. Our overall strategy was to transduce via an adeno-associated virus (AAV) vector the DREADD-encoding sequence in an inactive configuration, which was unblocked by Cre recombinase expressed in specific neuronal subtypes, and finally activating these targeted neurons by injection of DREADD ligand (Fig. 1A). For expression, we used the AAV.PHP.S viral vector, which efficiently infects peripheral sensory and autonomic neurons in mice without boosting strong immune responses (24). It allows expression of a DREADD and a mCherry fluorescent protein under the human SYN1 promoter, but only after inversion by Cre recombinase. As a DREADD, we utilized hM3Dq, a muscarinic Gq protein-coupled receptor that had been engineered to respond to the synthetic ligand, clozapine N-oxide (CNO), leading to calcium influx and neuronal activation (25). CNO has a half-life of several hours in vivo, such that the neuronal activation was expected to last for several hours. High-titer AAV.PhP.S-hSyn-DIO-hM3Dq-mCherry stocks were injected into neonatal mice, an age which results in higher levels of neuronal labeling compared with adult injections, and avoids immunologic confounders from recent viral infection (26-28).
Fig 1. Screening neuronal subtypes for immunomodulatory capabilities using viral mediated DREADDs.
(A) Schematic of experimental procedures describing setup of DREADD-based chemogenetic screen for neuronal effects on the gut immune system.
(B) Diagram showing neuronal subtypes in different anatomical locations (dorsal root ganglia, nodose ganglia, enteric nervous system) that innervate the gut and markers expressed in each location.
(C) Representative images of mCherry (red) and Tuj1 (βIII-tubulin, gray) staining in nodose ganglia (NG) and dorsal root ganglia (DRG) and myenteric plexus (MP) of the ENS after AAV.PhP.S-hSyn-DIO-mCherry-DREADD labeling of eight Cre lines. Scale bars are 100μm.
(D) Quantification of mCherry expression patterns across the eight Cre lines in the DRG, NG, and MP. For DRG and NG, the proportion of mCherry+ cells out of total Tuj1+ cells were quantified (4-5 fields per mouse). For MP, the area labeled by mCherry out of total Tuj1 area was quantified (2 fields per mouse). Gray bars indicate the expected expression range for marker genes.
For neuronal targeting, we chose a panel of 8 transgenic mouse lines expressing Cre in major subsets of peripheral sensory and autonomic neurons that innervate the gut (Table S1). From published scRNAseq datasets (Fig. S1A) and well-defined classical markers of gut innervating neurons (29, 30), we chose the following driver genes: Chat, Nos1, Vip, Tac1, Trpv1, Mrgprd, Th, and Piezo2. These Cre lines (31-38) were chosen to label intrinsic neurons of the enteric nervous system (Chat-cre, Nos1-cre, Vip-cre, Tac1-cre), sensory neurons in the vagal jugular/nodose ganglia (NG, Trpv1-cre, Vip-cre, Nos1-cre, Piezo2-cre, Tac1-cre, Th-cre), and sensory neurons in the dorsal root ganglia (DRG, Trpv1-cre, Tac1-cre, Mrgprd-cre, Piezo2-cre, Th-cre, Nos1-cre) (Fig. 1B). The Th-cre and Chat-cre lines also target postganglionic sympathetic neurons and parasympathetic neurons, respectively. To confirm expression of the DREADD in neurons of the injected mice, we stained sections of the NG, DRG and the ENS myenteric plexus (MP) in the colon with antibodies against the pan-neuronal marker Beta-III Tubulin (Tuj1) and mCherry (Fig. 1C). The NG and DRG showed labeling of mCherry+ neuronal cell bodies, while the colon showed both ENS cell bodies and fibers that may represent either intrinsic or extrinsic innervation. Quantification of mCherry expression out of total Tuj1+ neurons in each Cre line showed expression in the DRG, NG or MP that fit expected patterns for the transgenic markers (Fig. 1D), with frequencies of labeling close to, but lower than those expected from known marker or reporter expression. The only exception was Mrgprd-creErT2 mice, which required tamoxifen injection for Cre induction and for which mCherry expression was distinctly present but less than expected, possibly because of low or mis-timed Cre expression. Overall, this intersectional chemogenetic approach eschewed non-neuronal DREADD expression and limited it to targeted expression within neurons, in a sizeable proportion but not necessarily in every neuron, which may be an advantage for the functional experiments.
DREADD-based activation of specific neurons elicits distinct changes in gut immunocyte pools
After confirming the specificity and efficiency of this AAV.PhP.ShM3Dq Cre-based activity, we investigated the changes within intestinal populations of immunocytes induced by chemogenetic activation of various neurons. Mice were infected neonatally with AAV.PhP.ShM3dq, and treated at adult age with CNO (hereafter abbreviated as “ADC” mice) every other day over a two week period. Lymphoid and myeloid cells from different intestinal locales (ileum, cecum, colon) were then analyzed by high-parameter flow cytometry (31 immunophenotypes altogether, staining panels and gating strategies in Fig. S2A). For robustness, each experiment included ADC mice and matched Cre-negative littermates also AAV-injected and CNO treated (hereafter CTRL).
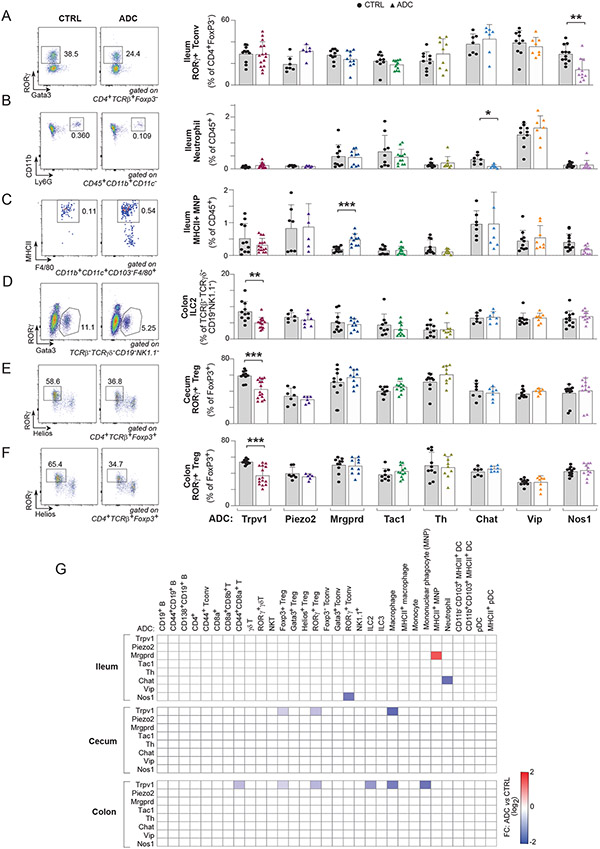
Overall, a number of neuro-immune relationships were detected by this screen, where the chemogenetic activation of a given neuronal subset led to distinct changes in immunophenotypes (illustrated in Figs 2A-F, tabulated in Fig. 2G). Some relationships were very exclusive. Nos1+ nitrergic neuron activation specifically downregulated RORγ+ CD4+ T conventional (Th17-like) cells in the ileum compared to CTRLs (Fig. 2A), whereas Chat+ cholinergic neuron activation downregulated ileal neutrophils (Fig. 2B). Mrgprd+ neuron activation led to increased ileal populations of MHCII+ mononuclear phagocytes (MNP) (Fig. 2C). These three effects were specific to the ileum, with no corresponding counterpart in the cecum or colon (Fig. S2B-D). By contrast, activation of Trpv1+ sensory neurons affected several cell populations: ILC2s in the colon (Fig. 2D), and RORγ+ Treg cells in both the colon and cecum (Fig. 2E, F), but also total colonic Tregs, colonic macrophages (significantly but more modestly) and the CD44+ fraction of colonic CD8+ T cells (Fig. 2G, Fig. S2E-G and Fig. S3). In contrast, some immunocyte populations were not affected by activation of any neuronal subtypes tested (B, γδT, and NK cells; Fig. 2G, Fig. S2H-J). Conversely, activation of some neurons (Piezo2, Th or VIP) seemed without influence on the homeostatic setting of gut immunocytes (Fig. 2G). Neuronal types that had effects on immunocyte populations were found among the various origins of intestinal innervation (NG, DRG or ENS – Fig. 1B). Thus, the screen uncovered a patchwork of discrete alterations induced by chemogenetic activation of distinct neuronal types, with Trpv1+ nociceptor neurons having the most widespread effects.
Fig 2. Distinct gut immune changes after DREADD-mediated neuronal activation.
(A-F) Quantification of proportions of ileum RORγ+ Tconv (A), ileum neutrophil (B), ileum MHCII+ MNP (C), colon ILC2 (D), colon RORγ+ Treg (E), and cecum RORγ+ Treg (F) with representative flow cytometric plots after chemogenetic activation of different neuronal subsets in distinct ADC mice and controls (CTRL).
(G) Heatmap of average fold changes (relative to CTRL) for significantly changed immune cell populations in the ileum, cecum, and colon from each Cre line compared to control littermates after CNO treatment (P<0.05). White squares are where P>0.05.
Each symbol (A-F) represents an individual mouse; Error bars represent mean and standard deviation. *,P < 0.05; **, P < 0.01 ***; P < 0.001 (unpaired Student’s t test with Holm-Sidak correction for multiple comparison). Data are representative of ≥2 independent experiments. n = 7-12 mice/group.
DREADD-based neuronal activation induces modest changes in the gut microbiota
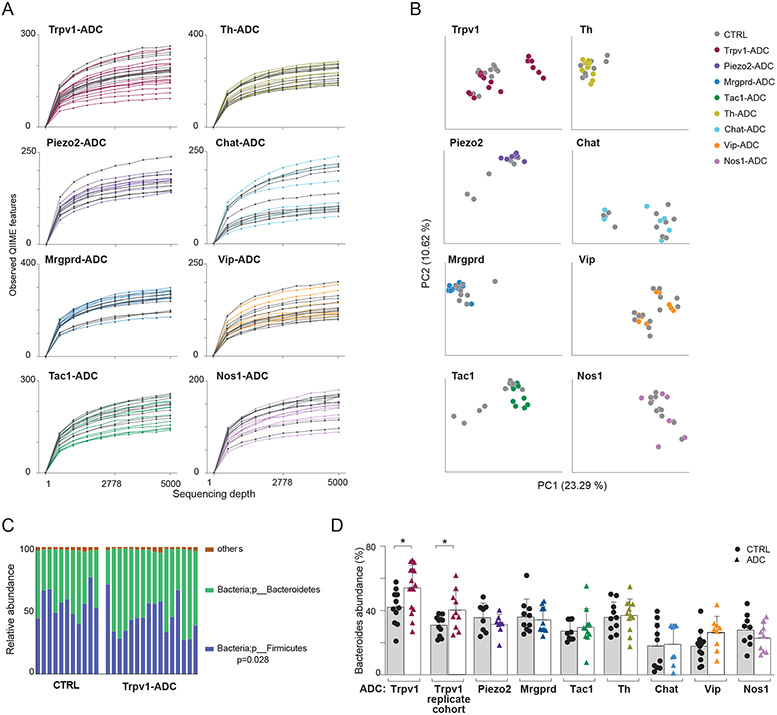
Many situations have been described in which alterations in gut microbes influence the peripheral nervous system (1, 39, 40), and a few cases where perturbations of gut-innervating neurons reflect on the intestinal microbiota (41, 42). We asked whether the specific neuronal activation we were eliciting in ADC mice provoked strong dysbiosis (which could be a confounder for the interpretation of the immunophenotypes observed), or rather more specific effects on a microbial family or genus. Bacterial populations were profiled in colonic content from ADC and CTRL littermates by 16S rDNA amplification and sequencing (testing littermate pairs to eschew cage-of-origin issues). There was no indication of strong dysbiosis in any of the mice, as α-diversity in ADC and CTRL littermates was generally comparable, except for a few of the Trpv1-ADC mice (Fig. 3A; these mice with lower diversity did not have particularly low colonic RORγ+ Tregs or other immunologic changes). Similarly, principal components analysis showed no marked deviation between ADC and CTRL littermates (both CNO-treated), except for some of the mice in the Trpv1-ADC and Tac1-ADC groups (Fig. 3B). Closer examination at the phylum level showed a modest but significant increase in the abundance of Bacteroidetes in Trpv1-ADC (with a corresponding decrease in Firmicutes (Fig. 3C), of interest because Zhang et al showed that disruption of intestinal Trpv1 innervation led to an increase in Bacteroidetes (42). This deviation in Bacteroidetes/Firmicute balance was confirmed in a replication cohort of Trpv1-ADC mice (Fig. 3D). On the other hand, analysis of finer taxonomic ranks failed to reveal significant changes in ADC mice beyond the level of experimental noise (estimated by iterative permutation) or that were reproducible in the replication cohort (the sole exception was a 25-fold increase in abundance of Clostridiaceae in Tac1-ADC mice; Table S3). Thus, activation of the wide range of intestinal neuronal types in our panel did not induce large reorganization of the gut microbiota, beyond modest shifts in phylum representation.
Fig 3. Microbiome changes upon specific neuronal activation.
(A) Alpha diversity rarefaction plots of microbial populations in different pairs of ADC mice and CTRL mice after CNO treatment.
(B) Principal coordinates analysis (PCoA) of unweighted UniFrac distance measurements in different ADC mice and CTRL mice after CNO treatment.
(C) Phylum-level analysis of the microbiome in the colon of Trpv1-ADC and CTRL mice.
(D) Quantification of proportions of Bacteroidetes (phylum-level) in the colon of different ADC mice and CTRL mice after CNO treatment.
Each symbol (C and E) represents an individual mouse; Error bars represent mean and standard deviation. *, P < 0.05 (unpaired Student’s t test with Holm-Sidak correction for multiple comparison). Data are representative of ≥2 independent experiments. n = 7-12 mice/group.
Trpv1+ neurons control the gut Treg niche
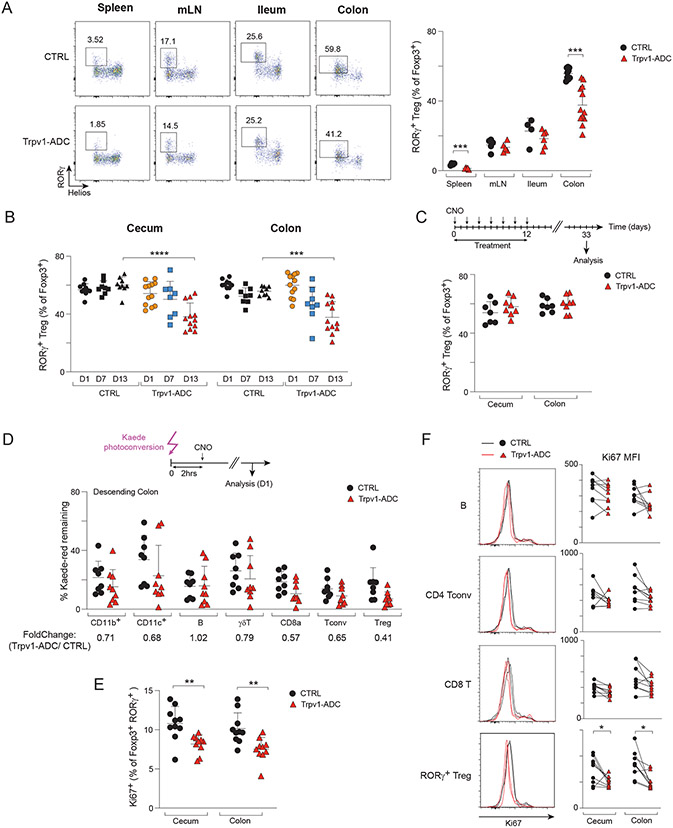
Among the different “hits” brought forth by our screen of immunologic changes following neuronal chemogenetic activation, we opted to focus deeper analyses on the reduction in RORγ+ Treg cells resulting from activation of Trpv1+ neurons, which mediate nociception of noxious heat and capsaicin (43) and mainly reside in the DRG and NG. This phenotype was robust and reproducible (Fig. 2), and RORγ+ Tregs are a particularly interesting population of immunoregulatory cells (44). Their frequencies are modulated by gut microbes, they help control tissue inflammation, oral tolerance, and maintain peaceful co-existence at the host/symbiont interface (44, 45). Most germane to the present study, we demonstrated previously that the control of RORγ+ Tregs by gut microbes is associated with perturbations of the ENS, proposing a triangular interaction between gut microbes, RORγ+ Tregs and the intestinal nervous system (46, 47), and a vagal brain-gut arc has also been proposed to maintain gut Tregs (48). RORγ+ Treg cells result, in a large part, from conversion of FoxP3-negative T conventional cells (Tconv) in the intestinal lymphoid tissues, but there is some debate whether this conversion takes place in the draining lymph nodes or in the lamina propria. Here, the effect of Trpv1+ neuron activation was visible in the lamina propria, but not in the mesenteric lymph nodes (mLN) (Fig. 4A), as was the reduction in total Treg cells, suggesting that the impact of neuronal activation was playing out in the intestinal wall itself, not in the draining lymph nodes. Interestingly, we also noted a significant reduction of RORγ+ Tregs in the spleen of Trpv1-ADC mice. Although far less abundant than in the colon, recent results support the notion that these small pools share many characteristics of RORγ+ Treg cells in the colon, and result from trafficking from the gut to the systemic lymphoid organs (49-51). Thus, the consequences of neuro-immune interactions in the gut seem to be reflected more systemically.
Fig 4. Trpv1+ neurons control the gut Treg cell niche.
(A) Representative flow cytometric plots and quantification of proportions of RORγ+ Tregs in the spleen, mLN, ileum and colon tissues from Trpv1-ADC and CTRL littermates.
(B) Quantification of proportions of RORγ+ Tregs in the cecum and colon of Trpv1-ADC and CTRL mice from indicated timepoints post-CNO treatment (day 1, day 7: CNO injected daily, day 13: CNO injected every other day).
(C) Quantification of proportions of RORγ+ Tregs in the cecum and colon of Trpv1-ADC and CTRL mice from indicated timepoint.
(D) Quantification of proportions of Kaede red+ immune cells in the descending colon from Trpv1-ADC-Kaede and littermate controls.
(E) Quantification of proportions of Ki67+ cells (of RORγ+ Tregs) in the cecum and colon from Trpv1-ADC and CTRL mice.
(F) Representative flow cytometric plots and quantification of geometric MFI (gMFI) of Ki67 (of B, CD4 Tconv, CD8 T, and RORγ+ Treg) in the cecum and colon from Trpv1-ADC and CTRL mice.
Each symbol (A-F) represents an individual mouse; Error bars represent mean and standard deviation. *; P < 0.05; **, P < 0.01 ***; P < 0.001; ****, P < 0.0001 (unpaired Student’s t test with Holm-Sidak correction for multiple comparison). Data are representative of ≥3 independent experiments. n = 5-12 mice/group.
A new class of unconventional antigen presenting cells, Thetis/Janus cells were identified recently and shown to control RORγ+ Treg frequencies (52, 53). In the mLNs of Trpv1-ADC mice, we did not detect significant changes in MHCII+IL7R-CXCR6− cells after activation (Fig. S4A), indicating that Thetis/Janus cells are not an intermediate for Trpv1-ADC effects. As a next step in elucidating the mechanism that connects Trpv1+ neuron activation and RORγ+ Tregs we determined the kinetics of the response: was the drop on RORγ+ Tregs a fast response, or did it represent a longer-lasting change in homeostatic setpoints? The frequencies of RORγ+ Tregs were not affected one day after CNO treatment, excluding a rapid response. Some Trpv1-ADC mice began showing a drop after one week, but the fully significant decre0ase in RORγ+ Tregs was not evident until two weeks of activation (Fig. 4B), suggesting a slow adaptation. On the other hand, the drop in RORγ+ Treg frequencies did not represent a stably altered setpoint: three weeks after cessation of treatment, the frequencies had reverted back to normal (Fig. 4C).
In principle, this slowly unfolding drop in colon and cecum RORγ+ Tregs could be due to induced extravasation from the gut, cell death, and/or decreased proliferation from a reduced niche size. To assess the turnover of intestinal Tregs, we bred Trpv1-Cre mice with transgenic mice encoding the photoconvertible Kaede fluorescent protein (54). In Kaede mice, colon cells can be temporarily switched from green to red by colonoscopic illumination with violet light, and both their residence in the colon and emigration into systemic organs tracked over time (49, 50, 55). In Trpv1-ADC mice, CNO treatment slightly lowered the proportion of photoconverted cells remaining in the descending colon after 24 hours, a trend that affected essentially all cell-types, not specifically RORγ+ Tregs (Fig. 4D); CNO treatment did not alter photoconversion efficiency. This result indicated that Trpv1+ neuron activation slightly increased immunocyte turnover in the colon, but was unlikely to account for the specific effect on RORγ+ Tregs. Annexin-V staining showed that there was no increase in the proportion of dying cells as a result of neuronal activation (Fig. S4B). Finally, an effect on Treg proliferation was assessed by staining for the nuclear factor Ki67. Ki67 has a complex relationship with the cell cycle, being actively synthesized during S, G2 and M phases, then progressively degraded during G1 and G0, serving as a marker of the time since the last division (56). Here, Tregs in Trpv1-ADC mice showed slightly decreased proportion of Ki67hi RORγ+ Tregs as their CTRL littermates (Fig. 4E), and a shift was noted in the noncycling peak, which lost Ki67 intensity in RORγ+ Tregs but not in other lymphocytes (Fig. 4F). These results suggest that Trpv1+ nociceptor neuron activation decreases the RORγ+ Treg pool not by cell death but by curtailing the proportion of Tregs that are poised to enter the cell cycle.
Colonic Treg cells contribute to maintaining gut barrier integrity and to regulating immunopathology (44). Thus, we interrogated whether Treg downregulation induced by Trpv1+ neuron activation had functional consequences. To this end, at the end of the two-week activation period by CNO treatment, we fed mice Dextran Sodium Sulfate (DSS) to induce colitis. Trpv1-ADC mice displayed more severe weight loss than did their CTRL littermates, not during the initiation phase but during the recovery phase (Fig. S5A), with higher inflammation by histology (Fig. S5B). We also infected mice with Citrobacter rodentium, an attaching/effacing bacterial pathogen that induces colon inflammation and pathology in mice (57). Trpv1-ADC mice lost more body weight than the controls (Fig. S5C). In addition, even though their gut colonization with C. rodentium was comparable with that of controls (Fig. S5D), many of the Trpv1-ADC mice showed systemic dissemination of the bacterium to the spleen and liver (Fig. S5E). Thus, nociceptor neuron activation not only resulted in decreased levels of RORγ+ Tregs, but affected proper inflammatory responses in various models.
Transcriptome changes in immunocytes resulting from Trpv1+ neuron activation
Having observed the effects of sensory neuron activation on colonic Treg cells, and as a first step towards elucidating the functional connection, we explored more generally the changes in the intestinal ecosystem that might explain this observation. Overall, histological analysis revealed no striking change in the general architecture of the colon, with no inflammatory infiltrate or tissue destruction (Fig. S6A). The overall architecture of the MP did not show major distortion in Trpv1-ADC mice, as evidenced by Tuj1 staining (Fig. S6B). These results indicated that Trpv1+ neuron activation did not grossly affect the immunologic or neurologic architecture in the colon.
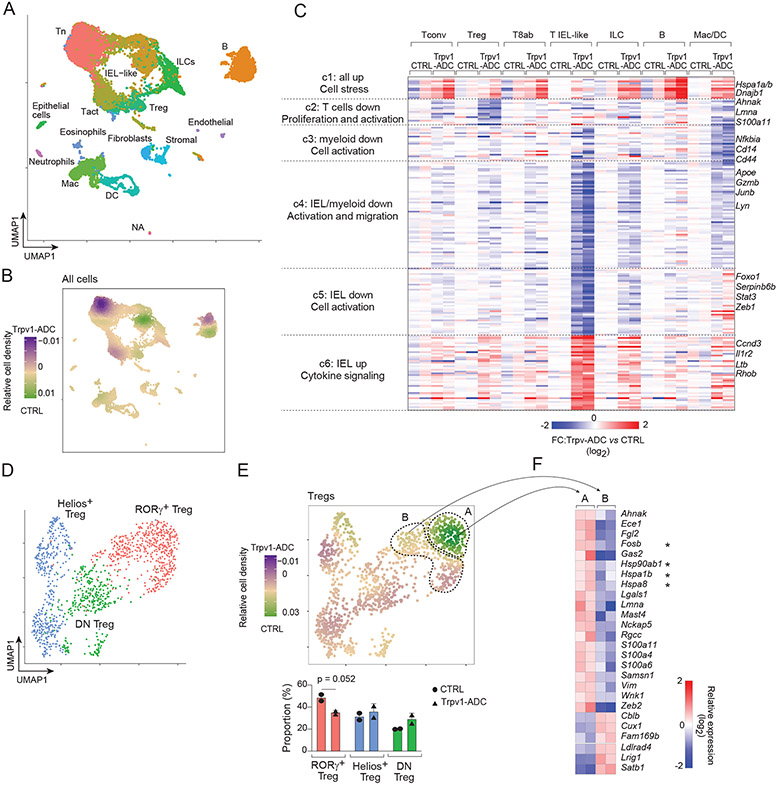
To investigate possible intermediate events between Trpv1+ neuron activation and changes in RORγ+ Tregs, we performed single-cell RNA sequencing (scRNAseq) on total CD45+ immunocytes from the cecum of Trpv1-ADC and CTRL mice (in biological duplicates). Dimensionality reduction and projection on a Uniform Manifold Approximation and Projection (UMAP) revealed all the common immunologic lineages expected in the cecum, including T and B lymphocytes, ILCs and myeloid cells (Fig. 5A), identifiable by the usual specific transcripts (Fig. S7A), with equivalent distributions in all four mice profiled (Fig. S7B). Comparing the relative density of cells from Trpv1-ADC and CTRL mice revealed shifts in some immunocyte populations, most markedly in lymphoid cells, less so in myeloid cells (Fig. 5B). To identify the immunocyte transcripts perturbed by Trpv1+ neuron activation, while avoiding those appearing differential merely because of altered cell-type representation, we performed Differential Gene Expression analysis separately on individual cell populations, tallying the resulting most affected 187 transcripts on the heatmap of Fig. 5C (Table S4; statistical significance of the clusters is shown on the volcano plots of Fig. S7C). These data illustrate a diversity of effects of Trpv1+ neuron activation on immunocytes, generally mild but broader than we initially realized from the immunophenotyping, many cutting across cell-types. This breadth was particularly true of gene cluster c1, induced in almost all cell-types, which was almost exclusively composed of heat-shock proteins and other stress response genes (Hspa1a/b, Hsp8, Hsp90, Dnajb1), suggesting that immunocyte stress resulted from Trpv1+ neuronal activation. To ask whether Hsp induction reflected a general cell stress, we analyzed the distribution of transcripts of a prototypical signature of cell damage (58). Only a minority of these transcripts was upregulated (Fig. S7D), indicating that the stress response to neuron activation had specificity, and was not merely uncontrolled cell damage, Two other clusters (c4, c5) were most prominently altered in intraepithelial lymphocyte (IEL)-like T cells, but also in other cell-types, and formed a network enriched (p=10−4 to 10−6) in cytokine receptors (Tnfrsf1, Il21r, Ifngr1), cell signaling (Lyn, Itk, Map3k1) and transcriptional regulators centered around Nfkb1, Stat3 and Stat4 (Fig.S7E), suggesting that neuronal activation might have tuned inter-cellular communication between immunocytes.
Fig 5. scRNAseq reveals cell stress and cell activation upon nociceptor neuron activation.
(A and B) Single-cell RNA sequencing analysis of cecum immunocytes from Trpv1-ADC and CTRL mice. UMAP plot (A). Differential density plot (B). Tn: naïve T cells; Tact: activated T cells.
(C) Heatmap of average expression fold changes (Trpv1-ADC vs. CTRL mice, log2) of differentially expressed genes (DEGs) extracted from 7 cell clusters as indicated. T8ab: CD8a+CD8b+ T, Mac/DC: Macrophages and dendritic cells.
(D and E) scRNAseq analysis of cecum Tregs from Trpv1-ADC and CTRL mice. UMAP plot (D). Proportions of different clusters and differential density plot (E).
(F) Heatmap of relative expression level (log2) of differential expressed genes (DEGs) extracted from the two RORγ+ Treg clusters as indicated in E from CTRL mice only.
Each symbol (E) represents an individual mouse; Error bars represent mean and standard deviation. n = 2 mice/group.
To analyze more closely the changes occurring in Treg cells of Trpv1-ADC mice, we recomputed a UMAP focused on Treg cells alone, which clearly identified RORγ+, Helios+ and DN Treg cells (Fig. 5D) substantiated by the expression of signature genes (59) (Fig. S7F). Many of the changes seen in Tregs in the wider analysis of Fig. 5C were observed in both RORγ+ and Helios Tregs (e.g. the induction of heat-shock proteins, or the downregulation of Lmna or S100a6 - Fig. S7G) suggesting that they may not themselves discriminate between Treg subsets. A comparative density plot revealed a focused reduction that affected only a segment of the RORγ+ Treg group (“A” cluster in Fig. 5E), but not the other (“B”). We asked what genes might distinguish, in CTRL mice devoid of induced neuronal activation, the “A” RORγ+ Tregs destined to be reduced electively by Trpv1+ neuron signaling, relative to their more resistant “B” counterparts (Fig. 5F). Interestingly, several of the differential transcripts revealed by this comparison within CTRL Treg subsets belonged to those further altered by Trpv1+ neuron activation (Hsp family, Table S5). This observation suggests that the programmatic changes in RORγ+ T reg cells that could be altered or induced by neuronal activation were already in play in SPF mice at steady state.
Trpv1+ spinal afferent neurons are responsible for altering the Treg niche
After characterizing the immunocyte changes induced by activation of Trpv1+ neurons, we next aimed to determine the neuronal mechanisms that led to Treg modulation. Trpv1 is expressed in two main anatomical locations, DRG neurons that centrally project to the spinal cord and NG neurons that project via the vagus nerve to the brainstem (8, 9) (Fig. S1B). Given that our chemogenetic strategy would label and activate all of these neurons, it was important to determine the relative contribution of the two Trpv1+ neuron locations in controlling gut Treg populations, adopting a strategy of AAV9 mediated delivery of Cre-dependent reporters into NG by intranodose injections or to the DRG via intrathecal injections (60, 61).
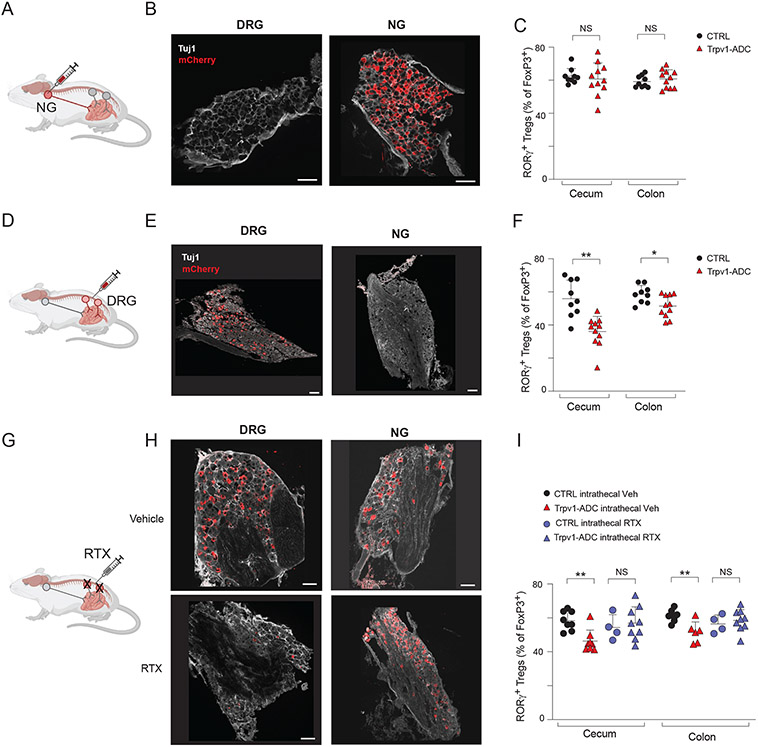
To target Trpv1+ neurons in the NG, we performed bilateral intranodose injections of AAV9hM3Dq virus in Trpv1-cre mice or control littermates (Fig. 6A). Intranodose injection yielded robust DREADD-mCherry expression in NG neurons, with no labeling of DRG neurons (Fig. 6B). In these mice, chemogenetic activation by CNO treatment did not elicit changes in colonic RORγ+ Tregs or any other immunocyte populations relative to CTRL littermates (Fig. 6C, S8A). We next performed intrathecal injections of AAV9hM3Dq virus into the lumbar spinal column of Trpv1-cre mice or control littermates (Fig. 6D). Intrathecal injection resulted in specific expression of DREADD-mCherry in DRG neurons but not vagal neurons (Fig. 6E). Chemogenetic activation of DRG Trpv1+ neurons by 2-weeks of CNO treatment resulted in a significant decrease in cecal and colonic RORγ+ Tregs and macrophages in intrathecally injected mice, suggesting that DRG Trpv1+ neurons modulated multiple immunocyte populations as we observed for whole body Trpv1+ neuron activation (Fig. 6F, S8B). Together, these data indicate that activation of sensory Trpv1+ neurons in the DRG, but not of vagal neurons, was sufficient to induce Treg population changes in the GI tract.
Fig 6. Trpv1+ DRG neurons, but not NG, regulate the gut Treg niche.
(A) Schematic of intranodose ganglionic injection into Trpv1-Cre and control mice for targeting of AAV9hM3Dq virus to nodose/jugular ganglia.
(B) Representative images of DRG and NG of mCherry (red) and Tuj1 (gray) after intranodose injection.
(C) Quantification of RORγ+ Treg cells in the cecum and colon from intranodose ganglionic injected Trpv1-Cre and control mice.
(D) Schematic of intrathecal injection into Trpv1-Cre and control mice for targeting of AAV9hM3Dq virus to DRG neurons.
(E) Representative images of DRG and NG of mCherry (red) and Tuj1 (gray) from intrathecally injected mice.
(F) Quantification of RORγ+ Tregs in the cecum and colon from intrathecally injected Trpv1-Cre and control mice.
(G) Schematic of resiniferatoxin (RTX) intrathecal injection for ablation of Trpv1+ DRG neurons.
(H) Representative images of DRG and NG of mCherry (red) and Tuj1 (gray) from Trpv1-ADC mice injected intrathecally with vehicle or RTX.
(I) Quantification of RORγ+ Tregs in the cecum and colon from Trpv1-ADC and CTRL mice with intrathecal injection of vehicle or RTX.
Each symbol (C, F, and I) represents an individual mouse; Error bars represent mean and standard deviation. *, P < 0.05; **, P < 0.01; NS, no significance (unpaired Student’s t test with Holm-Sidak correction for multiple comparison). Scale Bars: 100 μm in (B, E, and H). Data are representative of ≥2 independent experiments. n = 4-11 mice/group.
We next determined, in loss-of-function experiments, if DRG neurons were necessary for Trpv1+ neuronal regulation of Tregs. As above, we generated Trpv1-ADC mice, in which we performed targeted ablation of DRG Trpv1+ neurons by intrathecal injection of resiniferatoxin (RTX), a high affinity Trpv1 agonist that causes the loss of Trpv1+ neurons (62) (Fig. 6G). Mice treated with intrathecal injection of RTX had greatly diminished DREADD-mCherry expression in the DRG, while sparing those neurons in the NG (Fig. 6H). Following chemogenetic activation by CNO-treatment, Trpv1-ADC mice treated with RTX lost the downregulation of cecal and colonic RORγ+ Tregs observed in the Trpv1-ADC mice (Fig. 6I).
Taken together, we demonstrated that activation of Trpv1+ neurons in the DRG was necessary and sufficient to induce a decrease in RORγ+ Tregs. By contrast, activation of Trpv1+ vagal neurons did not affect this population. A previous study had reported that vagotomy and capsaicin injections into the vagus nerve altered colonic Treg populations and proposed a brain-liver-gut sensory-autonomic circuit (48). However, our data indicates that spinal DRG Trpv1+ neurons but not vagal neurons played a critical role in regulating RORγ+ Tregs in the context of repeated nociceptor activation.
Trpv1+ neurons regulate the gut Treg niche through a CGRP-Ramp1 axis
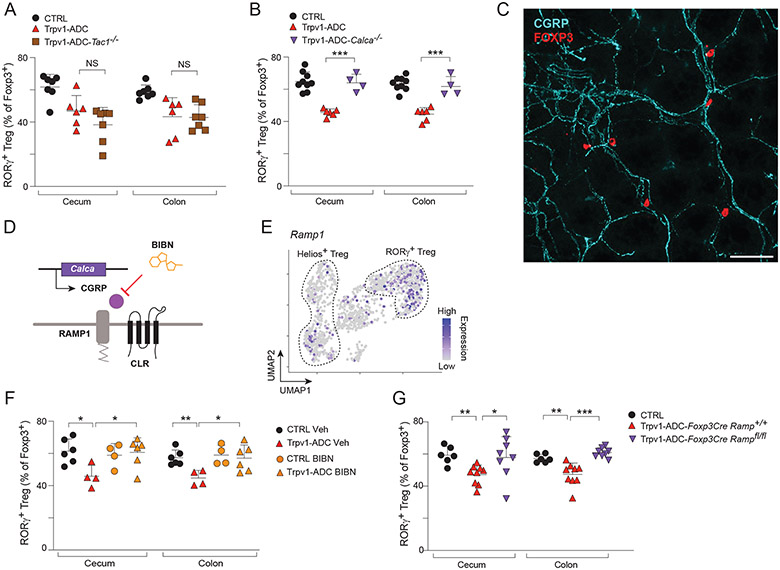
After determining the location of the Trpv1+ neurons responsible for decreasing RORγ+ Treg frequencies, we wanted to elucidate the mechanism through which these neurons signal to the Treg cells. In principle, this regulation could be via direct neuroimmune communication and/or indirectly via an intermediate cell-type. Trpv1+ neurons in the DRG are peptidergic (63), expressing neuropeptides including calcitonin-gene related peptide (CGRP) and Substance P (SP) (Fig. S9A - mousebrain.org). Given the role of these neuropeptides in regulating both vascular and immune responses in neurogenic inflammation (64), we hypothesized that Trpv1+ neurons might release neuropeptides upon activation that could signal to and impair RORγ+ Treg turnover. Substance P is encoded by Tac1, which can also be spliced to encode neurokinins A and B (65). We intercrossed Tac1−/− with Trpv1-cre mice to generate Tac1-deficient Trpv1-ADC mice. In these animals, the reduction in total RORγ+ Tregs in the colon following CNO treatment was similar to that of Trpv1-ADC mice (Fig. 7A), suggesting that SP and neurokinins were not required. Similarly, macrophage and ILC2 populations were still reduced in Trpv1-ADC mice (Fig. S9B), indicating that SP is not essential for effects of neuronal activation in this model. CGRP is expressed by two genes, Calca (encodes CGRPα) and Calcb (CGRPβ), whose products both bind to the same receptor complex and have similar functional activity (66). Calca is expressed at higher levels in DRG than enteric neurons, while Calcb is expressed in both DRG and enteric neurons (7, 66). Given our finding that DRG nociceptors regulated Tregs (Fig. 6), we next crossed Trpv1-Cre mice with Calca−/− mice to deplete CGRPα. Calca−/− Trpv1-ADC mice did not show the usual decrease in colonic and cecal RORγ+ Tregs compared with CTRL littermates treated with CNO (Fig. 7B), indicating an important role for CGRPα in nociceptor/Treg crosstalk. However, the macrophage compartment was still downregulated (Fig. S9C), denoting a specific involvement of CGRPα in neuronal regulation of Tregs.
Fig 7. Trpv1+ neurons regulate the gut Treg niche via CGRP.
(A) Quantification of RORγ+ Tregs in the cecum and colon from CTRL, Trpv1-ADC, and Trpv1-ADC-Tac1−/− mice.
(B) Quantification of RORγ+ Tregs in the cecum and colon from CTRL, Trpv1-ADC, and Trpv1-ADC-Calca−/− mice.
(C) Representative image of Foxp3+ Tregs (red) in proximity to CGRP fibers (cyan) from whole mount colon tissue.
(D) Schematic of CGRP signaling through its coreceptor complex formed by RAMP1 and CLR.
(E) UMAP of Ramp1 expression in Treg cell populations from CTRL mice.
(F) Quantification of RORγ+ Tregs in the cecum and colon from CTRL and Trpv1-ADC mice treated with vehicle or BIBN4096.
(G) Quantification of RORγ+ Tregs in the cecum and colon from CTRL, Trpv1-ADC-FoxP3CreRamp1+/+, and Trpv1-ADC-FoxP3CreRamp1fl/fl mice.
Each symbol (A-C, and E) represents an individual mouse; Error bars represent mean and standard deviation. *, P < 0.05; **, P < 0.01; ***; P < 0.001; NS, no significance (unpaired Student’s t test with Holm-Sidak correction for multiple comparison). Scale Bars: 100 μm in (D). Data are representative of ≥2 independent experiments. n = 4-10 mice/group.
CGRP has been reported as a mediator in neuro-immune interactions (67, 68) involving several immunocytes of the innate immune system like ILC2s (69-71), neutrophils (72, 73) or dendritic cells (74). These raised the possibility that the effect of Trpv1+ neuron activation was indirect, via one of these cells as intermediates (in line with observations in the injured muscle, where neuronal CGRP tunes IL-33 production by mesenchymal stromal cells, and hence the accumulation of muscle Tregs (75). Interestingly, immunofluorescence analysis on whole-mounts of normal colon tissue showed distinct proximity between FoxP3+ Treg cells and CGRP-positive fibers of sensory neurons in the lamina propria, consistent with the notion that these cells might directly interact (Fig. 7C). To further test this hypothesis, we asked whether the CGRP receptor on Treg cells was required for the Trpv1-ADC effect. CGRP acts on target cells by binding to a high-affinity receptor complex formed by CALCRL and modifying co-receptor RAMP1 (66) (Fig. 7D). In the scRNAseq data, Ramp1 was preferentially expressed in RORγ+ Tregs, relative to Helios+ Tregs, providing a possible explanation for a preferential effect of Trpv1-ADC on RORγ+ Tregs (Fig. 7E). To assess whether the RAMP1 receptor is indeed required for Trpv1+ neuronal signaling to Tregs, we treated mice with the RAMP1 antagonist BIBN4096 (76) or vehicle one hour before every CNO injection in Trpv1-ADC mice or CTRL littermates. This pre-treatment reverted the action of neuronal activation, showing that RAMP1 blockade rescued the decrease in RORγ+ Tregs resulting from Trpv1+ neuron activation (Fig. 7F). In addition, we generated Ramp1-deficient Tregs by crossing the Ramp1flox conditional allele with Foxp3-Cre mice (in addition to Trpv1-cre), to ablate Ramp1 in Treg cells (because of the two Cre transgenes in these mice, Ramp1 is also deleted in Trpv1+ neurons, which should be irrelevant). These Foxp3-creRamp1fl/fl Trpv1-ADC mice did not show the usual decrease in colonic RORγ+ Tregs following CNO treatment (Fig. 7G). Macrophage and ILC2 populations were still downregulated (Fig. S9D). These data together suggest that CGRPα released from Trpv1+ neurons upon neuronal activation bound to the RAMP1/CALCRL receptor complex on RORγ+ Tregs, affecting their homeostatic control. Thus, several independent clues demonstrated that CGRP was the mediator from activated neurons that exerted their negative control on RORγ+ Treg cells.
DISCUSSION
The nervous and immune systems jointly regulate tissue homeostasis and physiology at the main interface with the microbiota in the gut, each bringing into play wide constellations of cell-types of varied phenotypes and functions. Gut-extrinsic and intrinsic neurons influence the homeostasis of particular populations of immunocytes (47, 77-81), but we lacked an integrated understanding of how the panels of neurons and immunocytes interact. Here, we leveraged chemogenetic activation of distinct neuronal subsets to determine effects on immunocyte populations and the gut microbiota. The results highlight a number of individual neuroimmune connections. Trpv1+ neuron activation induced the most robust changes in immunocyte populations, in particular the reduction in Treg cells. We further investigated this interaction using neuroanatomical and molecular approaches, finding that spinal afferent Trpv1+ neurons mediated Treg changes via the neuropeptide CGRP. Given that these neurons mediate visceral pain, these data reveal a potential crosstalk between pain signaling and immune regulation in the gut.
Several recent studies have described the relationship between particular neuronal subsets and immunocytes in the gut (10-12, 14, 16-18, 71, 82-86). The present work extends by providing a more extensive and integrated view of neuroimmune interactions of relevance to the gut, by leveraging a panel of neuron subtype-specific DREADDs. We must acknowledge that there is a degree of artificiality in the approach, as it involves coordinated GPCR-induced activation, extending in a number of body locations. However, we chose our experimental design (repeated activation over a two-week period, instead of a single high-intensity burst) because it might reveal effects occurring over time, and mimic conditions encountered in diseases where peripheral neurons are chronically activated, such as in chronic visceral pain, autonomic imbalance, or GI dysmotility (87-89).
We uncovered several neuron-immunocyte interactions that pointed to a broader involvement of neuronal types than had been hitherto recognized, and will make for fascinating future explorations. Nitrergic (Nos1+) enteric neurons produce nitric oxide and mainly play an inhibitory role in peristalsis (90); their activation reduced the proportion of Th17-like RORγ+ Tconv cells in the ileum, possibly indicating a coordinated regulation of motor activity and Type-3/17 immune responses. Conversely, cholinergic (Chat+) excitatory motor neuron activation decreased neutrophils in the ileum. It remains to be determined if this finding relates to the cholinergic anti-inflammatory reflex, which was defined as acting through Chat+ vagal neurons that operate through macrophages and T cells in the spleen (91, 92). As these immunocytes are functionally related in host defense (e.g. Th17 and neutrophils), and these neuronal subsets coordinate motor function and peristalsis, one might speculate that motility and effector T cell responses might be coordinated. Mrgprd+ sensory neuron activation boosted the proportion of MHCIIhi MNPs in the ileum. Mrgprd+ neurons suppress mast cell activation through glutamate in the skin (93), but the role of Mrgprd+ neurons in the gut is less well understood.
Amidst this diversity, Trpv1+ nociceptors were clearly those whose activation elicited the broadest and most robust changes, not only altering the gut RORγ+ Treg niche, but also ILC2s, macrophages, and activated CD8+ T cells (Fig. 2G). Transcriptome profiling revealed changes in an even broader set of immunocytes resulting from Trpv1+ neuron activation. The most general effect was the increased expression of heat-shock and other stress response genes in most cell types, suggesting that signals from activated nociceptor neurons are interpreted as stress signals by immunocytes (Fig. 5C). Importantly, this induction did not include most of the transcripts of the generic van Oudenaarden signature of “cell suffering” (58), indicating some specificity to the stress signals involved, and arguing against experimental artefacts. In addition, the expression of this stress signature was more pronounced in normal mice at steady state in the targets of Trpv1 neurons, suggesting that this mode of regulation operates at baseline, independent of experimental manipulation.
Why did Trpv1+ neuron activation cause the broadest immunocyte changes among the neurons subtypes we probed? Trpv1+ neurons, as a subset of nociceptors whose primary function is to detect and transmit noxious signals to the central nervous system, are sensitized by a variety of inflammatory mediators, and nociceptors can activate the immune system through neurogenic inflammation (64). Correspondingly, several reports have already described isolated interactions between immunocytes and Trpv1+ sensory neurons. In the skin, Trpv1+ neurons interact with IL23-producing DCs and are required for Imiquimod-induced dermatitis (94), and their optogenetic activation can trigger Th17 responses in the skin, setting the stage for defense against fungal and bacterial pathogens (95). Trpv1+ neurons have also been shown to have inhibitory influence over immunocytes. In bacterial skin and lung infections, Trpv1+ neurons suppress neutrophil recruitment and myeloid cell responses to Streptococcus pyogenes and Staphylococcus aureus (72, 73). Conversely, Th17 cells induced by skin commensals promote sensory neuron regeneration during wound healing through IL17 receptor expressed on these neurons (96), indicating that the interface is bidirectional. Overall, the dominance of Trpv1+ neurons in neuroimmune crosstalk makes sense, if one considers the nociceptive nervous system and immune systems as sensory organs that constantly monitor the environment for danger and mediate inflammation: tight communication between these sentinels would seem important.
Emerging evidence also connects Treg cells to pain modulation. CD4+ T cells can signal to nociceptors mediating an anti-nociceptive role. In inflammatory models of chronic somatic pain measured in the footpad (97-99), as well as in DSS-colitis-induced visceral pain in the gut (100), T cell depletion caused prolonged pain hypersensitivity. Treg specific depletion with anti-CD25 treatment (101) or in Foxp3-DTR mice (102) led to increased mechanical hypersensitivity and neuropathic pain in sciatic nerve injury. Thus, Tregs seem to play a protective role in pain, likely through release of anti-inflammatory cytokines such as IL10 and production of endogenous opioids including proenkephalin, which can act directly on nociceptors to block pain; inhibition of proinflammatory immune cells can also indirectly lead to decreased pain (101-104). In the present study, persistent Trpv1+ nociceptor activation induced a decrease of Tregs, which we speculate might represent an adaptation, the dampening of a negative feedback loop in the face of an unresolved noxious stimulus, leading to prolonged or enhanced chronic pain. Thus, nociceptors and Tregs may have bidirectional communication and imbalance of this communication may be important in the development and maintenance of chronic pain.
The biogeography and neural circuitry of neuroimmune interactions in the gut remains a poorly understood topic. Neural tracing studies have shown that vagal ganglia innervation decreases in the lower GI tract (29), whereas DRG spinal afferent neurons innervate the entire GI tract (4). Our data show that DRG but not vagal neurons regulate colonic and cecal Tregs. This signaling is likely through local secretion of CGRP, but we cannot rule out the possibility that Trpv1+ neurons also signal through a neural reflex arc through the CNS, although not through a previously proposed vagal route (48). DRG sensory neurons have central terminals and reflex circuits in the spinal cord, and more complex circuits in higher cortical areas. Recent work has mapped CNS regions linked to mouse models of gut inflammation including DSS induced colitis and food allergies (105-107). It would be interesting to determine if such brain regions are also connected with peripheral neuron subsets that actively regulate immunity in the gut.
Overall, our study used a DREADD-based in vivo neuronal activation system to discover a number of neuro-immune interactions that control and modify immunocyte populations in the gut, and demonstrate a role for Trpv1+ neurons in modulating gut Tregs via CGRP-Ramp1 signaling. These observations raise the intriguing possibility that the nervous and immune systems, with their molecularly different sensory modalities, cooperate towards a balanced response to potentially noxious challenges in the digestive tract.
Materials and Methods
Mice
We utilized aged matched 6- to 12-week-old littermate male and female mice for all experiments. C57BL/6J (B6), B6;129S6-Chattm2(cre)Lowl/J (Chat-Cre), B6.129-Trpv1tm1(cre)Bbm/J (Trpv1-Cre), B6.129-Nos1tm1(cre)Mgmj/J (Nos1Cre), Viptm1(cre)Zjh/J (Vip-Cre), B6.Cg-7630403G23RikTg(Thcre)1Tmd/J (Th-Cre), Mrgprd tm1.1(cre/ERT2)Wql/J (Mrgprd-CreER), B6; 129S-Tac1 tm1.1(cre)Hze/J (Tac1-Cre), B6(SJL)-Piezo2 tm1.1(cre)Apat/J (Piezo2-Cre), B6.129-Gt(ROSA)26Sortm1(CAG-CHRM4*,-mCitrine)Ute/J (R26-hM4Di/mCitrine), B6.129(Cg)-Foxp3tm4(YFP/icre)Ayr/J (FoxP3-CreYFP), and B6.Cg-Tac1tm1Bbm/J (Tac1−/−) mice were purchased and obtained from the Jackson Laboratory (Bar Harbor, Maine) and bred in the specific-pathogen free animal facility at Harvard Medical School (HMS). B6.129S6-Calcatm1Hku (Calca−/−) (108) mice were kindly provided by Vijay Kuchroo (Harvard Medical School). Ramp1tm1a(EUCOMM)Wtsi/H (Ramp1flox/flox) mice were purchased from the European Mouse Mutant Archive (EMMA). Kaede transgenic (Kaede) mice were originally obtained from O.Kanagawa (RIKEN, Wako, Japan). Trpv1-Cre, Chat-Cre, Nos1-Cre, Vip-Cre, Th-Cre, Mrgprd-CreER, Tac1-Cre, Piezo2-Cre mice were crossed with B6 mice to generate Trpv1-Cre+/−, Chat-Cre+/−, Nos1-Cre+/−, Vip-Cre+/−, Th-Cre+/−, Mrgprd-CreER+/−, Tac1-Cre+/−, Piezo2-Cre+/− and littermate control mice to use for subsequent AAV-based experiments outlined below. For analysis of immune migration, Trpv1-Cre+/− mice were crossed with Kaede mice to generate Trpv1-Cre+/−Kaede and Trpv1-Cre−/−Kaede mice. For assays related to the role of neuropeptides in immunity, Trpv1-Cre mice were crossed with Tac1−/− or Calca−/− to generate Trpv1-Cre+/− Tac1+/− or Trpv1-Cre+/− Calca+/− mice, and F1 heterozygotes further crossed to Tac1+/− or Calca+/− mice to generate Trpv1-Cre+/− Tac1−/− or Trpv1-Cre+/− Calca−/− mice and littermate controls, respectively. For Treg specific deletion of Ramp1, which is the co-receptor for CGRP, FoxP3-CreYFP mice were crossed with Ramp1flox/flox mice. FoxP3-CreYFP-Ramp1flox/flox mice were then crossed with Trpv1-Cre mice to generate Trpv1Cre+/−FoxP3-CreYFP-Ramp1flox/flox mice. Mice were bred and maintained in the animal facility at Harvard Medical School (HMS) under specific pathogen-free (SPF) conditions with food and water ad libitum and a 12 h dark/light cycle. All experiments with animals were approved by the Harvard Medical School Institutional Animal Use and Care Committee (protocols IS00001257 and IS00000054-6).
Systemic and targeted viral DREADD expression and chemogenetic activation
For systemic AAV-mediated delivery, we utilized the adeno-associated virus AAV.PhP.S-hSyn-DIO-hM3D(Gq)-mCherry (AAVhM3Dq) to mediate Cre-dependent expression of the designer receptor exclusively activated by designer drugs (DREADD) hM3Dq in neurons (24). Viruses were produced at the Boston Children’s Hospital (BCH) viral core facility. Neonatal pups (either Cre+ or Cre− littermates) at postnatal stage 1 (P1) were injected intraperitoneally with 10μL of AAVhM3Dq at a dose of 2x1011 vg per mouse.
For DRG-targeted AAV-mediated delivery, we utilized the AAV9-hSyn-DIO-hM3D(Gq)-mCherry (AAV9hM3Dq). Viruses were purchased from Addgene. 10 μL of AAV9hM3Dq virus was injected intrathecally on three consecutive days. Mice were briefly anesthetized with isoflurane and injected in the L5-L6 region. Mice were rested for 3-4 weeks before being used for experiments.
For vagal ganglia-targeted expression, the nodose/jugular ganglia were bilaterally injected as previously described (61). Briefly, adult mice were anesthetized and an incision was made along the ventral surface of the neck. The nodose/jugular ganglia were surgically exposed by blunt dissection and a micropipette containing the virus was inserted into the nodose ganglia. 150 nL of AAV9hM3Dq virus was injected using a Nanoinject II injector (Drummond). Mice were allowed to recover for 4 weeks before being used for experiments.
To stimulate neuronal activation in mice, 1mg/kg of clozapine-N-oxide (CNO, Tocris 4936) was injected intraperitoneally. Mice were administered CNO every other day for a 2-week period, or consecutively for 1 day or 7 days. Mice were sacrificed the day after the last CNO injection, unless otherwise specified in the Results section.
Trpv1+ DRG neuron ablation
For targeted ablation of Trpv1+ DRG neurons, 4-6 week old mice were intrathecally injected with Resiniferatoxin (RTX, Alamone labs R-400), (25 ng/mouse) or vehicle in 10ul of 0.25% DMSO/0.02% Tween-80/0.05% ascorbic acid/PBS in the L5-L6 region under isoflurane on two consecutive days as described (62, 109). Control mice were injected intrathecally with vehicle alone. Mice were allowed to rest for 3-4 weeks before being used for experiments. We confirmed the loss of Trpv1+ neurons in the DRG but not vagal ganglia by immunostaining (see section below) or reduced thermal responses to noxious heat during hot plate tests (55°C).
RAMP1 antagonist administration
For antagonist experiments, BIBN 4096 (Torcis, 4561) was injected intraperitoneally at the dose of 0.3mg/kg 1 hour before CNO injections.
DSS induced colitis and Citrobacter rodentium infection
For DSS-induced colitis in mice, 2.5% dextran sulfate sodium salt (DSS, Thermo Scientific) was dissolved in the drinking water and given to mice for 6 days, followed by normal drinking water for 4 days. Mice were monitored daily for morbidity (piloerection, lethargy), weight loss and rectal bleeding.
For Citrobacter rodentium infections in mice, C. rodentium strain DBS100 (ATCC, 51459) was used. Bacteria were grown overnight for 16-18 hours in Luria Bertani (LB) broth at 37°C at 250 rpm. The OD600 was determined to estimate bacterial density and serial plating was performed to quantify the infection dose by counting colony forming units (CFU). Mice were fasted overnight and then orally gavaged with 200μL of sterile PBS containing 2 x 109 CFU. Mice were monitored daily throughout the experiment. To detect C. rodentium colonization, fecal pellets, liver, or spleen were homogenized in 1 mL of PBS, serially diluted, and plated on MacConkey agar for counting. C. rodentium CFU were counted after overnight incubation at 37°C.
Photo-conversion procedures
Colon was photoconverted as previously described (55). Briefly, a custom-built fiberoptic endoscope (ZIBRA Corporation) was coupled to a handheld 405 nm blue purple laser (≤ 5mW) via an in-house custom-made connection device (fixed mounts from ThorLabs). Mice were anesthetized with ketamine:xylazine (10 mg/kg:2mg/kg i.p). After cleansing the colon of fecal pellets with PBS, the fiberoptic endoscope was inserted through the anus to a depth of 3 cm. The laser was switched on, exposing the inner colon to violet light (3.5 mm beam diameter). Subsequently, the endoscope was gently retracted, pausing at 2 mm increments for 30 second light pulses at each interval (for a total of up to 10 minutes).
Immunocyte isolation from tissues
Spleen:
Single-cell suspensions were prepared by mashing the splenic tissue through a 70μm cell strainer followed by washing in RPMI containing 5% fetal calf serum (FCS). Red blood cells in the spleen were lysed with ACK lysing buffer (Gibco, ref A10492-01).
Mesenteric lymph node (mLN)
mLNs were minced into small pieces and dissociated in collagenase solution (1 mg/mL collagenase VIII (Sigma), 0.1mg/mL DNase I (Sigma) and 2% FCS in RPMI) with constant shaking at 37°C for 30 minutes. Single-cell suspensions were filtered through a 40μm cell strainer and washed with RPMI containing 5% FCS.
Ileum, Cecum, Colon:
Intestines were cleaned (Peyer’s patches were removed in the case of the ileum), and treated with RPMI containing 1 mM DTT, 20 mM EDTA and 2% FCS at 37°C for 15 minutes to remove epithelial cells. Tissues were then minced and dissociated in collagenase solution (1.5 mg/mL collagenase II (Gibco), 0.5mg/mL dispase (Gibco) and 1% FCS in RPMI) with constant stirring at 37°C for 40 minutes. Single-cell suspensions were filtered through a 40μm cell strainer and washed with RPMI containing 5% FCS.
Flow cytometry
Single-cell suspensions from spleen and intestinal tissues were prepared as above.
The cells were stained with 2 constant panels of antibodies for consistency. The first panel (lymphoid panel) included surface markers for CD19, CD4, CD8a, CD8b, TCR-β, TCR-γδ, NK1.1, CD138, CD44, and intracellular markers for RORγ, FoxP3, Tbet, Gata3, Helios. The second panel (myeloid panel) included surface markers for CD45, CD19, CD11b, CD11c, Ly6c, Ly6g, PDCA-1, F4/80, CD103, and MHCII. The Thetis/Janus cell staining panel included surface markers for CD45, TCR-β, TCR-γδ, B220, CD11c, CD127, MHCII, CCR6, CXCR6, and intracellular markers for RORγ, FoxP3. The cells were stained with zombie live/dead dye (BioLegend) at 4°C for 20 minutes, followed by surface staining at 4°C for 30 minutes. For intracellular staining, cells were fixed in eBioscience Fix/Perm buffer at room temperature for 1 hour, followed by permeabilization in eBioscience permeabilization buffer at room temperature for 1 hour in the presence of antibodies. For Annexin-V staining, the cells were stained with antibody for Annexin-V in Annexin-V binding buffer at room temperature for 10 minutes after surface staining. Cells were acquired with a Symphony flow cytometer (BD Biosciences) and analysis was performed with FlowJo (Tree Star) software.
Whole mount staining
Mice were euthanized and a small piece (3 mm x3 mm) of distal colon was collected for staining. The colons were opened and fixed in a Silgard dish with 4% PFA at 4°C for 2 hours. Then the tissues were washed with PBST (PBS with 0.5% Triton-X100) 6 times at room temperature for 20 minutes, followed by incubation with primary antibodies (Tuj1 and mCherry, or CGRP and FoxP3) in blocking buffer (20% DMSO, 5% donkey serum or goat serum in PBST) for 2-3 days and corresponding secondary antibodies in blocking buffer for 1-2 days. In subsets of experiments, tissue was dehydrated with serial (50%, 80%, 100%) methanol solution and cleared in BABB buffer (1 volume Benzyl Alcohol to 2 volume Benzyl Benzoate). The tissues were mounted on glass slides using vacuum grease for imaging. Three different fields of each tissue were imaged by Ti2 Spinning Disk microscope (Nikon) or a Stellaris 8 FALCON CFS system (Leica) and processed by ImageJ software. All images were maximum intensity projections of z-stacks. For quantification of the myenteric plexus in whole mount colon images, the percentage of mCherry+ area out of total Tuj1+ area was determined for each sample as an average of 2 fields per mouse and quantified by investigators blinded to genotypes.
Immunofluorescence and Microscopy
For immunostaining of extrinsic ganglia, tissues were placed in 4% PFA for 1-2 hours. Thoracic (T11-T13) DRG, lumbar DRG (L5-L6), and vagal ganglia were dissected, incubated overnight at 4°C in 30% sucrose, and embedded in OCT. Sections (14 μm) were cut, blocked with 10% NDS, 0.05% Tween-20 in PBS for 2 hours at room temperature, and stained with primary antibodies (goat anti-mCherry, rabbit anti-Tuj1) overnight at room temperature. Sections were washed in PBS, then stained with secondary antibodies (donkey anti-goat Alexa594, donkey anti-rabbit Alexa488) for 2 hours at room temperature. After washing with PBS, sections were mounted in VectaShield (Vector Labs). Sections were imaged on a Leica Stellaris 8 FALCON CFS confocal microscope at 20X magnification. Leica software (Leica Application Suite X) was used for image capture and post-processing. For quantification of extrinsic ganglia neurons, the percentage of mCherry+ neurons out of the total Tuj1+ neurons were determined for each sample as an average of 3-5 fields per mouse and quantified by investigators were blinded to genotypes.
Gut histopathology
Mice were euthanized and colon tissues were collected and fixed in 10% formalin for at least 24 hours prior to H&E (hematoxylin and eosin) staining. Whole cross-sections were scanned and imaged on a widefield microscope (Nikon) at either 10X or 20X magnitude. For H&E staining, images were analyzed using a scoring system as previously described (110). Briefly, cross-sections were randomly split into 8 sections and 4 random sections were scored qualitatively using a 0-5 point scale for immune infiltrate, goblet cell loss, crypt density, crypt hyperplasia, muscle thickening, submucosal infiltrate, ulceration, and abscess. Scores from each item were combined for a composite score for each mouse. Scores from each section were averaged for each mouse. Scoring was performed blinded.
Fecal DNA extraction, 16S rDNA sequencing, and data analysis
Feces from colons were collected and frozen at −80°C until use. Bacterial genomic DNA from fecal samples was extracted using phenol:chloroform:isoamyl alcohol and purified with the QIAquick PCR Purification kit (Qiagen). Purified DNA was quantified by Qubit dsDNA HS Assay (Thermo Fisher) and normalized to 6ng/μl for following amplification. Amplicons were quantified by Qubit dsDNA HS Assay and combined with equal mass to make a pooled library. The pooled library was purified and multiplexed sequenced (Illumina MiSeq, 251 nt x 2 pair-end reads with 12 nt index reads) through Harvard’s Biopolymers Facility. Raw sequencing data was processed with QIIME2. In brief, raw sequencing data was imported to QIIME2 and demultiplexed, then DADA2 was used for sequence quality control and feature table construction. The feature table was used for beta diversity analysis, taxonomic analysis, and differential abundance testing using QIIME2. Beta group significance was determined by permutational analysis of variance (PERMANOVA). Identification of taxa associated with different groups was determined using Analysis of Composition of Microbiomes (ANCOM).
Single-Cell RNA sequencing and data analysis
Sample preparation and sequencing:
Single-cell RNA sequencing (scRNAseq) experiments were performed and analyzed as previously described (111) Cecum cell suspensions were stained with surface markers for CD45, CD4, CD19, CD8a, CD11b, CD11c, TCR-γδ, NK1.1 and DAPI as a viability dye, along with hashtag antibodies (hashtag 1, CTRL#1; hashtag 3, CTRL#2; hashtag 7, Trpv1-ADC#1; hashtag 9, Trpv1-ADC#2). Cells were sorted as DAPI-CD45+. Additional sorting was performed for CD19−CD45+, NK1.1+CD45+, and TCR-γδ +CD45+ to enrich for these populations. All samples were pooled together, centrifuged, and resuspended in 0.04% BSA. Encapsulation was done on the 10X Chromium microfluidic instrument (10X Genomics). Libraries were prepared using Chromium Single cell 3’ reagents kit v2 according to manufacturer’s protocol. Hashtag oligonucleotide (HTO) libraries were prepared as described in (112). Libraries were sequenced together on the Illumina HiSeq X.
Data analysis:
scRNAseq data were processed using the standard CellRanger pipeline (10X Genomics). HTO counts were obtained using the CITE-seq-Count package (113). Data was analysed in R using the Seurat package (114). HTOs were assigned to cells using the HTODemux function, and doublets were eliminated from analysis. Cells with less than 700 UMIs or 500 genes and more than 2,500 UMIs, 10,000 genes and 5% of reads mapped to mitochondrial genes were also excluded from the analysis. Dimensionality reduction, visualization and clustering analysis were performed in Seurat using the NormalizeData, ScaleData, FindVariableGenes, RunPCA, FindNeighbours (dims=1:30), RunUMAP (dims=1:30) and FindClusters functions. Cluster identity was determined based on expression of key marker genes (Fig. S7A). The SubsetData function was used to gate individual clusters for further analysis.
Differentially expressed genes (DEGs) between Trpv1-ADC and CTRL mice were obtained using the FindMarkers function with the cutoff based on fold change and P value (logfc.threshold = 0.5 and adjusted P value < 0.05) on 7 populations including CD4+ Tconv cells (T4conv), CD4+ Treg cells (Treg), CD8a+CD8b+ T cells (T8ab), CD160+ IEL (IEL-like), B cells (B), ILCs (ILC), and selected myeloid cells (Macrophage and dendritic cells, MacDC), respectively. Then the DEGs from each population were collated non-redundantly. The average expression of these DEGs in each sample (2 pairs of Trpv1-ADC and CTRL mice) across each population were generated using AverageExpression function. Heatmaps of differentially expressed genes were generated using Morpheus (https://software.broadinstitute.org/morpheus).
Gene signatures:
The IEL gene signature was based on the expression of the following marker genes: Klra1, Klre1, Klra7, Itgae, Cd160, Klrk1, Fas1, Itgb7, Ccr9, Cd8a. The Helios Treg gene signature was on the expression of the following marker genes: Cd200r1, Cd83, Dgat2, Epas1, Fam46a, Gas2l3, Ikzf2, Il9r, Naip5, Nrp1, Ppp2r3a, Swap70, Foxp3. The Rorc Treg gene signature was on the expression of the following marker genes: Ccr1, Ccr2, Ccr5, Ccr9, Clic4, F2rl2, Gpr15, Havcr2, Itgb5, Marcks, Matn2, Nr1d1, Prg4, Rorc.
Data analysis and statistics
Data are represented as mean and standard deviation where n represents the number of mice, unless indicated otherwise. Statistical tests used were unpaired Student’s t test with Holm-Sidak correction for multiple comparison, or Two-way repeated measures ANOVA as indicated in the figure legends. Differences were considered significant if p < 0.05. Statistics were performed in GraphPad Prism.
Supplementary Material
Table S1. List of 8 Cre transgenic lines used in this study.
Table S3. Analysis of bacterial abundance at the family level by 16S rDNA sequencing of colonic feces from ADC and CTRL mice for each Cre line.
Table S2. Immunophenotyping results of ileum, cecum, and colon from ADC and CTRL mice for each Cre line.
Table S4. The fold change (log2) of 187 Differentially expressed genes from analysis of 7 individual cell populations in Trpv1-ADC vs. CTRL mice.
Table S5. Relative expression of genes differentially expressed in subsets A and B of RORγ+ Treg cells, in control mice (corresponds to Fig. 5F).
Fig. S1. Subtypes of neurons labeled by the eight Cre lines utilized for the screen.
(A) Heatmap showing expression pattern of genetic markers chosen in this study that correspond with Cre lines to mark neuronal subsets and their patterns in published scRNAseq datasets from 3 studies: gut-innervating DRG neurons (8) (Top portion of heatmap), nodose/jugular vagal ganglia (9) (middle portion of heat-map), and myenteric plexus enteric neurons in the small intestine (7) (Bottom portion of heatmap). Row names were demarcated by original RNAseq databases.
Fig. S2. Immunophenotyping results upon specific neuronal activation.
(A) Representative flow cytometric plots of gating strategies for lymphoid and myeloid staining panels.
(B-J) Quantification of proportions of indicated cell populations from Figure.2G, showing cecum RORγ+ Tconv (B), colon neutrophil (C), colon MHCII+ MNP (D), colon FoxP3+ Treg (E), colon macrophage (F), colon activated CD8 T cell (G), cecum B cell (H), colon RORγ+ γδT cell (I), and colon NK cell (J).
Each symbol (B-J) represents an individual mouse; Error bars represent mean and standard deviation. *, P < 0.05; **, P < 0.01; ****, P < 0.0001 (unpaired Student’s t test with Holm-Sidak correction for multiple comparison). Data are representative of ≥2 independent experiments. n = 7-12 mice/group.
Fig. S3. Immunophenotyping results upon Trpv1+ neuron activation.
(A-C) Quantification of proportions of indicated cell populations (of CD45+) in the ileum (A), cecum (B), and colon (C) from Trpv1-ADC and CTRL mice.
Each symbol (A-C) represents an individual mouse; Error bars represent mean and standard deviation. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (unpaired Student’s t test with Holm-Sidak correction for multiple comparison). Data are representative of ≥3 independent experiments. n = 9-12 mice/group.
Fig. S4. Activation of Trpv1+ neurons does not affect Thetis/Janus cells.
(A) Representative flow cytometric plots of gating strategy for Thetis/Janus cells (Top) and quantification of proportions of MHCII+IL7R-CXCR6− Thetis/Janus cells (of CD45+) in the mLN from Trpv1-ADC and CTRL mice.
(B) Quantification of proportions of Annexin-V+ cells (of RORγ+ Tregs) in the cecum and colon from Trpv1-ADC and CTRL mice. Each symbol (A and B) represents an individual mouse; Error bars represent mean and standard deviation. (unpaired Student’s t test with Holm-Sidak correction for multiple comparison). Data are representative of ≥2 independent experiments. n = 5-6 mice/group.
Fig. S5. Trpv1+ neuron activation alters susceptibility to colitis and C. rodentium infection.
(A-B) Schematic diagram of DSS treatment and body weight loss (A), representative images of H&E staining and histology scores (B) of Trpv1-ADC and CTRL mice after DSS treatment as in A.
(C-E) Schematic diagram of C. rodentium infection and body weight loss (C), bacterial load in the feces (D), spleen and liver (E) of Trpv1-ADC and CTRL mice after C. rodentium infection as in C.
Each symbol (A-E) represents an individual mouse; Error bars represent mean and standard deviation. *, P < 0.05; **, P < 0.01 (Two-way repeated measures ANOVA) Scale Bars: 100 μm in (B). Data are representative of ≥2 independent experiments. n = 5-6 mice/group.
Fig. S6. No global histological changes or neuron structure difference upon Trpv1+ neuron activation.
(A) Representative images of H&E staining from colons of Trpv1-ADC and CTRL mice.
(B) Representative images of whole-mount staining of Tuj1 of colons from AAVhM3Dq-infected Trpv1-Cre mice treated with DMSO or CNO.
Scale Bars: 50 μm in (A), 100 μm in (B).
Fig. S7. Global transcriptional changes in immune populations upon Trpv1+ neuron activation.
(A-G) scRNAseq analysis of cecal immunocytes from Trpv1-ADC and CTRL mice. Feature plots of select markers (A). UMAP plots of cecal immunocytes from each mouse (B). Volcano plots of different cell populations as in Fig. 5C, red or blue, genes from c2-c6 clusters as in Fig. 5C (C). Volcano plots of all cells, blue, gene signature of cell damage (58) (D). String cluster plot of genes from c4 and c5 clusters of Fig. 5C (E). UMAP plots of cecal Tregs from Trpv1-ADC and CTRL mice with RORγ+ Treg and Helios+ Treg signature (F). Heatmap of average expression fold changes (Trpv1-ADC vs. CTRL mice, log2) of DEGs extracted from RORγ+ Treg and Helios+ Treg clusters (G). n = 2 mice/group.
Fig. S8. Immunophenotyping results from intranodose or intrathecally injected Trpv1-ADC and CTRL mice.
(A-B) Quantification of proportions of indicated cell populations (of CD45+) in the colon from intranodose injected Trpv1-Cre and CTRL mice (A), or intrathecally injected Trpv1-Cre and CTRL mice (B).
Each symbol (A, B) represents an individual mouse; Error bars represent mean and standard deviation. *, P < 0.05 (unpaired Student’s t test with Holm-Sidak correction for multiple comparison). Data are representative of ≥2 independent experiments. n = 9-11 mice/group.
Fig. S9. Substance P does not regulate macrophage or ILC2 populations downstream of Trpv1+ neuronal activation.
(A) Expression of CGRP and substance P genes in Trpv1+ DRG neurons (7) Graphic was made from mousebrain.org.
(B) Quantification of proportions of ILC2s and macrophages in the cecum and colon from CTRL, Trpv1-ADC, and Trpv1-ADC-Tac1−/− mice.
(C) Quantification of proportions of macrophages in the cecum and colon from CTRL, Trpv1-ADC, and Trpv1-ADC-Calca−/− mice.
(D) Quantification of proportions of ILC2s and macrophages in the cecum and colon from CTRL, Trpv1-ADC-FoxP3CreRamp1+/+, and Trpv1-ADC-FoxP3CreRamp1fl/fl mice.
Each symbol (B-D) represents an individual mouse; Error bars represent mean and standard deviation. *, P < 0.05; **, P < 0.01; ****, P < 0.0001; NS, no significance (unpaired Student’s t test with Holm-Sidak correction for multiple comparison). Data are representative of ≥2 independent experiments. n = 4-9 mice/group.
Acknowledgements:
We thank Drs. Dan Littman, Viviana Gradinaru, Meng Wu, Meena Rao, and members of Chiu and Mathis/Benoist lab for insightful discussion and advice, L. Yang and D. Mallah for help with computational analyses, K. Hattori and S. Choi for help with mice, Dr H. Basu for help with image analysis, Dr P. Montero-Llopis and MicRoN facility for microscopy, the Boston Children's Hospital Viral Core for virus stocks, and Ian Magill for single-cell sequencing.
Funding:
This work was supported by National Institutes of Health (NIH) grants AI125603 and AI150686 and the JPB Foundation to CB&DM; RO1DK127257, Kenneth Rainin Foundation, and the Food Allergy Science Initiative to IMC. KAM was supported by NIH fellowship F32DK137456, SG-P by a fellowship from European Molecular Biology Organisation (ALTF 547-2019).
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: Data newly reported in this paper are available at the NCBI GEO and SRA repositories under accession number GSE240982.
References and Notes
- 1.Jacobson A, Yang D, Vella M, Chiu IM, The intestinal neuro-immune axis: crosstalk between neurons, immune cells, and microbes. Mucosal. Immunol 14, 555–565 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang H, Foong JPP, Harris NL, Bornstein JC, Enteric neuroimmune interactions coordinate intestinal responses in health and disease. Mucosal. Immunol 15, 27–39 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cryan JF, Dinan TG, Gut microbiota: Microbiota and neuroimmune signalling-Metchnikoff to microglia. Nat Rev Gastroenterol. Hepatol 12, 494–496 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Blackshaw LA, Brookes SJ, Grundy D, Schemann M, Sensory transmission in the gastrointestinal tract. Neurogastroenterol. Motil 19, 1–19 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Costa M, Brookes SJ, Hennig GW, Anatomy and physiology of the enteric nervous system. Gut. 47 Suppl 4, iv15–iv19 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCorry LK, Physiology of the autonomic nervous system. Am. J Pharm. Educ 71, 78 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeisel A et al. , Molecular architecture of the mouse nervous system. Cell. 174, 999–1014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hockley JRF et al. , Single-cell RNAseq reveals seven classes of colonic sensory neuron. Gut. 68, 633–644 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kupari J et al. , An Atlas of Vagal Sensory Neurons and Their Molecular Specialization. Cell Rep. 27, 2508–2523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Talbot J et al. , Feeding-dependent VIP neuron-ILC3 circuit regulates the intestinal barrier. Nature. 579, 575–580 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seillet C et al. , The neuropeptide VIP confers anticipatory mucosal immunity by regulating ILC3 activity. Nat Immunol. 21, 168–177 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Pascal M et al. , The neuropeptide VIP potentiates intestinal innate type 2 and type 3 immunity in response to feeding. Mucosal. Immunol 15, 629–641 (2022). [DOI] [PubMed] [Google Scholar]
- 13.Wallrapp A et al. , The neuropeptide NMU amplifies ILC2-driven allergic lung inflammation. Nature. 549, 351–356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cardoso V et al. , Neuronal regulation of type 2 innate lymphoid cells via neuromedin U. Nature. 549, 277–281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klose CSN et al. , The neuropeptide neuromedin U stimulates innate lymphoid cells and type 2 inflammation. Nature. 549, 282–286 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabanyi I et al. , Neuro-immune interactions drive tissue programming in intestinal macrophages. Cell. 164, 378–391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matheis F et al. , Adrenergic Signaling in Muscularis Macrophages Limits Infection-Induced Neuronal Loss. Cell. 180, 64–78 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moriyama S et al. , β2-adrenergic receptor-mediated negative regulation of group 2 innate lymphoid cell responses. Science. 359, 1056–1061 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Urban DJ, Roth BL, DREADDs (designer receptors exclusively activated by designer drugs): chemogenetic tools with therapeutic utility. Annu Rev Pharmacol. Toxicol 55, 399–417 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Roth BL, DREADDs for Neuroscientists. Neuron. 89, 683–694 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krashes MJ et al. , Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J Clin Invest. 121, 1424–1428 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atasoy D, Betley JN, Su HH, Sternson SM, Deconstruction of a neural circuit for hunger. Nature. 488, 172–177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoo ES et al. , Galphαi/o-coupled Htr2c in the paraventricular nucleus of the hypothalamus antagonizes the anorectic effect of serotonin agents. Cell Rep. 37, 109997 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan KY et al. , Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat Neurosci. 20, 1172–1179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armbruster BN et al. , Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci U S A. 104, 5163–5168 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogura T et al. , Utility of intraperitoneal administration as a route of AAV serotype 5 vector-mediated neonatal gene transfer. J Gene Med. 8, 990–997 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Foust KD et al. , Neonatal intraperitoneal or intravenous injections of recombinant adeno-associated virus type 8 transduce dorsal root ganglia and lower motor neurons. Hum. Gene Ther 19, 61–70 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Machida A et al. , Intraperitoneal administration of AAV9-shRNA inhibits target gene expression in the dorsal root ganglia of neonatal mice. Mol. Pain 9, 36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips RJ, Powley TL, Innervation of the gastrointestinal tract: patterns of aging. Auton. Neurosci 136, 1–19 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uesaka T, Young HM, Pachnis V, Enomoto H, Development of the intrinsic and extrinsic innervation of the gut. Dev. Biol 417, 158–167 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Woo SH et al. , Piezo2 is required for Merkel-cell mechanotransduction. Nature. 509, 622–626 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris JA et al. , Anatomical characterization of Cre driver mice for neural circuit mapping and manipulation. Front Neural Circuits. 8, 76 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olson W et al. , Sparse genetic tracing reveals regionally specific functional organization of mammalian nociceptors. eLife. 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Savitt JM et al. , Bcl-x is required for proper development of the mouse substantia nigra. J Neurosci. 25, 6721–6728 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cavanaugh DJ et al. , Trpv1 reporter mice reveal highly restricted brain distribution and functional expression in arteriolar smooth muscle cells. J Neurosci. 31, 5067–5077 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossi J et al. , Melanocortin-4 receptors expressed by cholinergic neurons regulate energy balance and glucose homeostasis. Cell Metab. 13, 195–204 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leshan RL et al. , Leptin action through hypothalamic nitric oxide synthase-1-expressing neurons controls energy balance. Nat Med. 18, 820–823 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taniguchi H et al. , A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 71, 995–1013 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gershon MD, Margolis KG, The gut, its microbiome, and the brain: connections and communications. J. Clin. Invest 131, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cryan JF et al. , The Microbiota-Gut-Brain Axis. Physiol Rev. 99, 1877–2013 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Yang D et al. , Nociceptor neurons direct goblet cells via a CGRP-RAMP1 axis to drive mucus production and gut barrier protection. Cell. 185, 4190–4205 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang W et al. , Gut-innervating nociceptors regulate the intestinal microbiota to promote tissue protection. Cell. 185, 4170–4189 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caterina MJ et al. , The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 389, 816–824 (1997). [DOI] [PubMed] [Google Scholar]
- 44.Ramanan D et al. , Regulatory T cells in the face of the intestinal microbiota. Nat Rev Immunol. In press, (2023). [DOI] [PubMed] [Google Scholar]
- 45.Traxinger BR, Richert-Spuhler LE, Lund JM, Mucosal tissue regulatory T cells are integral in balancing immunity and tolerance at portals of antigen entry. Mucosal. Immunol 15, 398–407 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yissachar N et al. , An intestinal organ culture system uncovers a role for the nervous system in microbe-immune crosstalk. Cell. 168, 1135–1148 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan Y et al. , Interleukin-6 produced by enteric neurons regulates the number and phenotype of microbe-responsive regulatory T cells in the gut. Immunity. 54, 499–513 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teratani T et al. , The liver-brain-gut neural arc maintains the Treg cell niche in the gut. Nature. 585, 591–596 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Galván-Peña S et al. , A dynamic atlas of immunocyte migration from the gut. bioRxiv. 10.1101/2022.11.16.516757 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanna BS et al. , The gut microbiota promotes distal tissue regeneration via RORγ+ regulatory T cell emissaries. Immunity. 56, 829–846 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chowdhary K et al. , An interwoven network of transcription factors, with divergent influences from FoxP3, underlies Treg diversity. bioRxiv. DOI 10.1101/2023.05.18.541358 (2023). [DOI] [Google Scholar]
- 52.Kedmi R et al. , A RORγ+ cell instructs gut microbiota-specific Treg cell differentiation. Nature. 610, 737–743 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Akagbosu B et al. , Novel antigen presenting cell imparts Treg-dependent tolerance to gut microbiota. Nature. 610, 752–760 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tomura M et al. , Monitoring cellular movement in vivo with photoconvertible fluorescence protein "Kaede" transgenic mice. Proc Natl Acad Sci U S A. 105, 10871–10876 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morton AM et al. , Endoscopic photoconversion reveals unexpectedly broad leukocyte trafficking to and from the gut. Proc Natl Acad Sci U S A. 111, 6696–6701 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller I et al. , Ki67 is a Graded Rather than a Binary Marker of Proliferation versus Quiescence. Cell Rep. 24, 1105–1112 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crepin VF, Collins JW, Habibzay M, Frankel G, Citrobacter rodentium mouse model of bacterial infection. Nat Protoc. 11, 1851–1876 (2016). [DOI] [PubMed] [Google Scholar]
- 58.van den Brink SC et al. , Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods 14, 935–936 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Ramanan D et al. , Homeostatic, repertoire and transcriptional relationships between colon T regulatory cell subsets. bioRxiv. DOI 10.1101/2023.05.17.541199 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Skorput AGJ et al. , Targeting the somatosensory system with AAV9 and AAV2retro viral vectors. PLoS One. 17, e0264938 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang RB et al. , Vagal Sensory Neuron Subtypes that Differentially Control Breathing. Cell. 161, 622–633 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mishra SK, Hoon MA, Ablation of TrpV1 neurons reveals their selective role in thermal pain sensation. Mol. Cell Neurosci 43, 157–163 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Price TJ, Flores CM, Critical evaluation of the colocalization between calcitonin gene-related peptide, substance P, transient receptor potential vanilloid subfamily type 1 immunoreactivities, and isolectin B4 binding in primary afferent neurons of the rat and mouse. J Pain. 8, 263–272 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chiu IM, Von Hehn CA, Woolf CJ, Neurogenic inflammation and the peripheral nervous system in host defense and immunopathology. Nat Neurosci. 15, 1063–1067 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cao YQ et al. , Primary afferent tachykinins are required to experience moderate to intense pain. Nature. 392, 390–394 (1998). [DOI] [PubMed] [Google Scholar]
- 66.Russell FA et al. , Calcitonin gene-related peptide: physiology and pathophysiology. Physiol Rev. 94, 1099–1142 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Assas BM, Pennock JI, Miyan JA, Calcitonin gene-related peptide is a key neurotransmitter in the neuro-immune axis. Front Neurosci. 8, 23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baral P, Udit S, Chiu IM, Pain and immunity: implications for host defence. Nat Rev Immunol. 19, 433–447 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nagashima H et al. , Neuropeptide CGRP limits group 2 innate lymphoid cell responses and constrains type 2 inflammation. Immunity. 51, 682–695 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wallrapp A et al. , Calcitonin gene-related peptide negatively regulates alarmin-driven type 2 innate lymphoid cell responses. Immunity. 51, 709–723 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu H et al. , Transcriptional atlas of intestinal immune cells reveals that neuropeptide α-CGRP modulates group 2 innate lymphoid cell responses. Immunity. 51, 696–708 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pinho-Ribeiro FA et al. , Blocking neuronal nignaling to immune cells treats streptococcal invasive infection. Cell. 173, 1083–1097 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baral P et al. , Nociceptor sensory neurons suppress neutrophil and gammadelta T cell responses in bacterial lung infections and lethal pneumonia. Nat Med. 24, 417–426 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hanc P et al. , Multimodal control of dendritic cell functions by nociceptors. Science. 379, eabm5658 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang K et al. , Neuronal, stromal, and T-regulatory cell crosstalk in murine skeletal muscle. Proc. Natl. Acad. Sci. U. S. A 117, 5402–5408 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miller PS et al. , Non-peptidic antagonists of the CGRP receptor, BIBN4096BS and MK-0974, interact with the calcitonin receptor-like receptor via methionine-42 and RAMP1 via tryptophan-74. Biochem. Biophys. Res. Commun 391, 437–442 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Veiga-Fernandes H, Mucida D, Neuro-immune interactions at barrier surfaces. Cell. 165, 801–811 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Klose CS, Artis D, Neuronal regulation of innate lymphoid cells. Curr. Opin. Immunol 56, 94–99 (2019). [DOI] [PubMed] [Google Scholar]
- 79.Muller PA, Matheis F, Mucida D, Gut macrophages: key players in intestinal immunity and tissue physiology. Curr. Opin. Immunol 62, 54–61 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vanuytsel T, Bercik P, Boeckxstaens G, Understanding neuroimmune interactions in disorders of gut-brain interaction: from functional to immune-mediated disorders. Gut. 72, 787–798 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Klein Wolterink RGJ, Wu GS, Chiu IM, Veiga-Fernandes H, Neuroimmune Interactions in Peripheral Organs. Annu. Rev. Neurosci 45, 339–360 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Matteoli G et al. , A distinct vagal anti-inflammatory pathway modulates intestinal muscularis resident macrophages independent of the spleen. Gut. 63, 938–948 (2014). [DOI] [PubMed] [Google Scholar]
- 83.Muller PA et al. , Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell. 158, 300–313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dalli J, Colas RA, Arnardottir H, Serhan CN, Vagal Regulation of Group 3 Innate Lymphoid Cells and the Immunoresolvent PCTR1 Controls Infection Resolution. Immunity. 46, 92–105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu HB et al. , Vasoactive intestinal peptide promotes host defense against enteric pathogens by modulating the recruitment of group 3 innate lymphoid cells. Proc Natl Acad Sci U S A. 118, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Viola MF et al. , Dedicated macrophages organize and maintain the enteric nervous system. Nature. 618, 818–826 (2023). [DOI] [PubMed] [Google Scholar]
- 87.Kuner R, Kuner T, Cellular Circuits in the Brain and Their Modulation in Acute and Chronic Pain. Physiol Rev. 101, 213–258 (2021). [DOI] [PubMed] [Google Scholar]
- 88.Kay MW, Jain V, Panjrath G, Mendelowitz D, Targeting Parasympathetic Activity to Improve Autonomic Tone and Clinical Outcomes. Physiology. (Bethesda. ). 37, 39–45 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Singh R, Zogg H, Ghoshal UC, Ro S, Current Treatment Options and Therapeutic Insights for Gastrointestinal Dysmotility and Functional Gastrointestinal Disorders. Front Pharmacol. 13, 808195 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bódi N, Szalai Z, Bagyanszki M, Nitrergic Enteric Neurons in Health and Disease-Focus on Animal Models. Int. J Mol. Sci 20, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tracey KJ, The inflammatory reflex. Nature. 420, 853–859 (2002). [DOI] [PubMed] [Google Scholar]
- 92.Kelly MJ, Breathnach C, Tracey KJ, Donnelly SC, Manipulation of the inflammatory reflex as a therapeutic strategy. Cell Rep Med. 3, 100696 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang S et al. , Nonpeptidergic neurons suppress mast cells via glutamate to maintain skin homeostasis. Cell. 184, 2151–2166 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Riol-Blanco L et al. , Nociceptive sensory neurons drive interleukin-23-mediated psoriasiform skin inflammation. Nature. 510, 157–161 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cohen JA et al. , Cutaneous TRPV1+ neurons trigger protective innate type 17 anticipatory immunity. Cell. 178, 919–932 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Enamorado M et al. , Immunity to the microbiota promotes sensory neuron regeneration. Cell. 186, 607–620 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Krukowski K et al. , CD8+ T cells and endogenous IL-10 are required for resolution of chemotherapy-induced neuropathic pain. J Neurosci. 36, 11074–11083 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Laumet G et al. , Cisplatin educates CD8+ T cells to prevent and resolve chemotherapy-induced peripheral neuropathy in mice. Pain. 160, 1459–1468 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Basso L et al. , Endogenous analgesia mediated by CD4+ T lymphocytes is dependent on enkephalins in mice. J Neuroinflammation. 13, 132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Boué J et al. , Endogenous regulation of visceral pain via production of opioids by colitogenic CD4(+) T cells in mice. Gastroenterology. 146, 166–175 (2014). [DOI] [PubMed] [Google Scholar]
- 101.Austin PJ, Kim CF, Perera CJ, Moalem-Taylor G, Regulatory T cells attenuate neuropathic pain following peripheral nerve injury and experimental autoimmune neuritis. Pain. 153, 1916–1931 (2012). [DOI] [PubMed] [Google Scholar]
- 102.Lees JG, Duffy SS, Perera CJ, Moalem-Taylor G, Depletion of Foxp3+ regulatory T cells increases severity of mechanical allodynia and significantly alters systemic cytokine levels following peripheral nerve injury. Cytokine. 71, 207–214 (2015). [DOI] [PubMed] [Google Scholar]
- 103.Liu XJ et al. , Nociceptive neurons regulate innate and adaptive immunity and neuropathic pain through MyD88 adapter. Cell Res. 24, 1374–1377 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Duffy SS et al. , Regulatory T cells and their derived cytokine, Interleukin-35, reduce pain in experimental autoimmune encephalomyelitis. J Neurosci. 39, 2326–2346 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Koren T et al. , Insular cortex neurons encode and retrieve specific immune responses. Cell. 184, 5902–5915 (2021). [DOI] [PubMed] [Google Scholar]
- 106.Florsheim EB et al. , Immune sensing of food allergens promotes avoidance behaviour. Nature. (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Plum T et al. , Mast cells link immune sensing to antigen-avoidance behaviour. Nature. (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Oh-hashi Y et al. , Elevated sympathetic nervous activity in mice deficient in alphaCGRP. Circ. Res 89, 983–990 (2001). [DOI] [PubMed] [Google Scholar]
- 109.Guptill V et al. , Disruption of the transient receptor potential vanilloid 1 can affect survival, bacterial clearance, and cytokine gene expression during murine sepsis. Anesthesiology. 114, 1190–1199 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Asseman C et al. , An essential role for interleukin 10 in the function of regulatory T cells that inhibit intestinal inflammation. J Exp Med. 190, 995–1004 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kiner E et al. , Gut CD4+ T cell phenotypes are a continuum molded by microbes, not by TH archetypes. Nat. Immunol (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stoeckius M et al. , Cell hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 19, 224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stoeckius M et al. , Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 14, 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Butler A et al. , Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of 8 Cre transgenic lines used in this study.
Table S3. Analysis of bacterial abundance at the family level by 16S rDNA sequencing of colonic feces from ADC and CTRL mice for each Cre line.
Table S2. Immunophenotyping results of ileum, cecum, and colon from ADC and CTRL mice for each Cre line.
Table S4. The fold change (log2) of 187 Differentially expressed genes from analysis of 7 individual cell populations in Trpv1-ADC vs. CTRL mice.
Table S5. Relative expression of genes differentially expressed in subsets A and B of RORγ+ Treg cells, in control mice (corresponds to Fig. 5F).
Fig. S1. Subtypes of neurons labeled by the eight Cre lines utilized for the screen.
(A) Heatmap showing expression pattern of genetic markers chosen in this study that correspond with Cre lines to mark neuronal subsets and their patterns in published scRNAseq datasets from 3 studies: gut-innervating DRG neurons (8) (Top portion of heatmap), nodose/jugular vagal ganglia (9) (middle portion of heat-map), and myenteric plexus enteric neurons in the small intestine (7) (Bottom portion of heatmap). Row names were demarcated by original RNAseq databases.
Fig. S2. Immunophenotyping results upon specific neuronal activation.
(A) Representative flow cytometric plots of gating strategies for lymphoid and myeloid staining panels.
(B-J) Quantification of proportions of indicated cell populations from Figure.2G, showing cecum RORγ+ Tconv (B), colon neutrophil (C), colon MHCII+ MNP (D), colon FoxP3+ Treg (E), colon macrophage (F), colon activated CD8 T cell (G), cecum B cell (H), colon RORγ+ γδT cell (I), and colon NK cell (J).
Each symbol (B-J) represents an individual mouse; Error bars represent mean and standard deviation. *, P < 0.05; **, P < 0.01; ****, P < 0.0001 (unpaired Student’s t test with Holm-Sidak correction for multiple comparison). Data are representative of ≥2 independent experiments. n = 7-12 mice/group.
Fig. S3. Immunophenotyping results upon Trpv1+ neuron activation.
(A-C) Quantification of proportions of indicated cell populations (of CD45+) in the ileum (A), cecum (B), and colon (C) from Trpv1-ADC and CTRL mice.
Each symbol (A-C) represents an individual mouse; Error bars represent mean and standard deviation. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (unpaired Student’s t test with Holm-Sidak correction for multiple comparison). Data are representative of ≥3 independent experiments. n = 9-12 mice/group.
Fig. S4. Activation of Trpv1+ neurons does not affect Thetis/Janus cells.
(A) Representative flow cytometric plots of gating strategy for Thetis/Janus cells (Top) and quantification of proportions of MHCII+IL7R-CXCR6− Thetis/Janus cells (of CD45+) in the mLN from Trpv1-ADC and CTRL mice.
(B) Quantification of proportions of Annexin-V+ cells (of RORγ+ Tregs) in the cecum and colon from Trpv1-ADC and CTRL mice. Each symbol (A and B) represents an individual mouse; Error bars represent mean and standard deviation. (unpaired Student’s t test with Holm-Sidak correction for multiple comparison). Data are representative of ≥2 independent experiments. n = 5-6 mice/group.
Fig. S5. Trpv1+ neuron activation alters susceptibility to colitis and C. rodentium infection.
(A-B) Schematic diagram of DSS treatment and body weight loss (A), representative images of H&E staining and histology scores (B) of Trpv1-ADC and CTRL mice after DSS treatment as in A.
(C-E) Schematic diagram of C. rodentium infection and body weight loss (C), bacterial load in the feces (D), spleen and liver (E) of Trpv1-ADC and CTRL mice after C. rodentium infection as in C.
Each symbol (A-E) represents an individual mouse; Error bars represent mean and standard deviation. *, P < 0.05; **, P < 0.01 (Two-way repeated measures ANOVA) Scale Bars: 100 μm in (B). Data are representative of ≥2 independent experiments. n = 5-6 mice/group.
Fig. S6. No global histological changes or neuron structure difference upon Trpv1+ neuron activation.
(A) Representative images of H&E staining from colons of Trpv1-ADC and CTRL mice.
(B) Representative images of whole-mount staining of Tuj1 of colons from AAVhM3Dq-infected Trpv1-Cre mice treated with DMSO or CNO.
Scale Bars: 50 μm in (A), 100 μm in (B).
Fig. S7. Global transcriptional changes in immune populations upon Trpv1+ neuron activation.
(A-G) scRNAseq analysis of cecal immunocytes from Trpv1-ADC and CTRL mice. Feature plots of select markers (A). UMAP plots of cecal immunocytes from each mouse (B). Volcano plots of different cell populations as in Fig. 5C, red or blue, genes from c2-c6 clusters as in Fig. 5C (C). Volcano plots of all cells, blue, gene signature of cell damage (58) (D). String cluster plot of genes from c4 and c5 clusters of Fig. 5C (E). UMAP plots of cecal Tregs from Trpv1-ADC and CTRL mice with RORγ+ Treg and Helios+ Treg signature (F). Heatmap of average expression fold changes (Trpv1-ADC vs. CTRL mice, log2) of DEGs extracted from RORγ+ Treg and Helios+ Treg clusters (G). n = 2 mice/group.
Fig. S8. Immunophenotyping results from intranodose or intrathecally injected Trpv1-ADC and CTRL mice.
(A-B) Quantification of proportions of indicated cell populations (of CD45+) in the colon from intranodose injected Trpv1-Cre and CTRL mice (A), or intrathecally injected Trpv1-Cre and CTRL mice (B).
Each symbol (A, B) represents an individual mouse; Error bars represent mean and standard deviation. *, P < 0.05 (unpaired Student’s t test with Holm-Sidak correction for multiple comparison). Data are representative of ≥2 independent experiments. n = 9-11 mice/group.
Fig. S9. Substance P does not regulate macrophage or ILC2 populations downstream of Trpv1+ neuronal activation.
(A) Expression of CGRP and substance P genes in Trpv1+ DRG neurons (7) Graphic was made from mousebrain.org.
(B) Quantification of proportions of ILC2s and macrophages in the cecum and colon from CTRL, Trpv1-ADC, and Trpv1-ADC-Tac1−/− mice.
(C) Quantification of proportions of macrophages in the cecum and colon from CTRL, Trpv1-ADC, and Trpv1-ADC-Calca−/− mice.
(D) Quantification of proportions of ILC2s and macrophages in the cecum and colon from CTRL, Trpv1-ADC-FoxP3CreRamp1+/+, and Trpv1-ADC-FoxP3CreRamp1fl/fl mice.
Each symbol (B-D) represents an individual mouse; Error bars represent mean and standard deviation. *, P < 0.05; **, P < 0.01; ****, P < 0.0001; NS, no significance (unpaired Student’s t test with Holm-Sidak correction for multiple comparison). Data are representative of ≥2 independent experiments. n = 4-9 mice/group.