Abstract
Prymnesium parvum are harmful haptophyte algae that cause massive environmental fish-kills. Their polyketide polyether toxins, the prymnesins, are among the largest nonpolymeric compounds in nature and have biosynthetic origins that have remained enigmatic for more than 40 years. In this work we report the “PKZILLAs”, massive P. parvum polyketide synthase (PKS) genes, that have evaded previous detection. PKZILLA-1 and -2 encode giant protein products of 4.7 and 3.2 megadaltons that have 140 and 99 enzyme domains. Their predicted polyene product matches the proposed pre-prymnesin precursor of the 90-carbon-backbone A-type prymnesins. We further characterize the variant PKZILLA-B1, which is responsible for the shorter B-type analog prymnesin-B1 from P. parvum RCC3426 and thus establish a general model of haptophyte polyether biosynthetic logic. This work expands expectations of genetic and enzymatic size limits in biology.
One-Sentence Summary:
Fish-killing Prymnesium parvum algae use enzymes of unprecedented size to biosynthesize their large prymnesin polyketide toxins.
Large-scale fish deaths caused by harmful algal blooms are global health, environmental, and food security problems (1). Anthropogenic causes appear to be hastening the severity and frequency of toxic eukaryotic microalgal blooms in freshwater and marine ecosystems, including the massive fish kill along the Oder River in 2022 by the golden alga Prymnesium parvum (Haptophyta) that decimated half of the river’s fish population through Poland and Germany (2, 3). Its hemolytic poisons, including prymnesin-1 and prymnesin-B1, are regularly implicated in worldwide mass fish die offs (4, 5) (Fig. 1) and are notable for their giant, fused polycyclic polyether structures. Prymnesin-1 (6), at 90 carbon atoms in length, joins other massive microalgal polyketide biotoxins such as palytoxin (7) and maitotoxin (8) that are among the largest nonpolymeric carbon chain molecules in nature (9). Collectively, this group of microalgal polyketides pose serious human and environmental health risks as neurotoxins. Fundamental knowledge of microalgal polyketide biology is still poorly understood because no causal biosynthesis gene has yet been identified in a haptophyte, chlorophyte, or dinoflagellate.
Fig. 1. Prymnesin, its source PKZILLA polyketide synthases (PKSs), and other large proteins and PKS systems.

(A) Map of historical or ongoing major P. parvum or Prymnesium spp. fish-killing blooms: England (49, 50), Netherlands (51), Israel (6, 52, 53), Texas (5, 54, 55), Scandinavia (4), and Oder River (3, 37). (B) Molecular structure of prymnesin-1 (31). (C) Molecular structure of prymnesin-B1 (56). (D) Comparison of polypeptide and coding nucleotide sizes from representative PKSs or computationally summed PKS systems. Blue indicates PKZILLAs from P. parvum 12B1 (this work); [S] indicates computationally summed lengths for independent PKS proteins that participate in the same biosynthetic system; black dashed lines indicate divisions of PKS systems into independent proteins; red and * indicate the largest known protein (non-PKS) (18); gold indicates representative bacterial PKS systems, including the quinolidomicin (** indicates the previous largest known PKS system) (42) and erythromycin (57) PKSs; and green and *** indicate the previous largest genetically studied microalgal PKS (16). AA, amino acid.
The chemical structure of giant microalgal polyethers such as prymnesin-1 imply a biosynthetic assembly-line construction of two-carbon chain length iterations to a polyene intermediate that undergoes epoxidation followed by a nucleophilic reaction cascade to construct their distinctive trans-fused (“ladder-frame”) polyether frameworks (10, 11). However, the biosynthesis of these massive microalgal toxins has remained an enigma despite a wealth of intimate knowledge of polyketide biochemistry from decades of research in bacteria and fungi (12). Despite numerous transcriptomic studies that identified orphan biosynthetic gene candidates in multimodular type I polyketide synthases (PKSs) from toxic microalgae (13, 14), including numerous from diverse P. parvum strains (15), no PKS gene or fragment has yet been connected to a microalgal polyketide structure. The sheer size of microalgal polyether biotoxins presents experimental challenges. There is also a lack of methods to study their genetic origin. The model green alga Chlamydomonas reinhardtii, for instance, hosts a single large ~80–kilo–base pair (kbp) PKS, known as PKS1 (Fig. 1D). Genetic knockout experiments established that PKS1 participates in formation of the zygospore cell wall, but its polyketide product remains unknown (16). Furthermore, unlike bacteria and fungi that organize their PKS encoding genes into polycistrons and biosynthetic gene clusters, other eukaryotes typically use monocistronic mRNAs and infrequently functionally colocalize most genes, which thus greatly obfuscates gene discovery efforts (17).
Here, we report the application of a customized gene annotation strategy that enabled the discovery of two massive PKS genes, PKZILLA-1 and -2, from P. parvum strain 12B1 that we propose are responsible for the complete backbone assembly of its notorious ladder-frame polyether toxin, prymnesin-1. Not only are the two giant PKZILLA “gigasynthases” organized consistently with the long-anticipated polyene intermediate structure of a microalgal ladder-frame polyether, but PKZILLA-1 is larger than the exceptionally large human protein titin (18) (Fig. 1D).
Genomic and transcriptomic evidence for the PKZILLA gigasynthases
We selected the A-type prymnesin-1 (19) producing P. parvum strain 12B1 as a model system to resolve microalgal polyether biosynthesis, as its 116 Mbp genome and our recently published near-chromosome-level genomic assembly (20) makes strain 12B1 relatively tractable amongst microalgae and other toxic P. parvum strains. We first cataloged PKS genes potentially involved in prymnesin-1 biosynthesis within our automated gene annotation (20), identifying 44 PKS genes encoding relatively small proteins with 1–3 trans-acyltransferases (trans-AT) PKS modules. However, confirmatory tblastn queries using PKS domains unveiled three seemingly contiguous and strikingly large PKS “hotspot” loci that stood apart at 137, 93, and 74 kbp on pseudo-chromosomes 17, 7, and 10, respectively. These hotspots showed a high concentration of visible coding regions that were only partially captured by 25 fragmented PKS gene models, thus we hypothesized they represented massive and mis-annotated single-genes. Upon manual revision, we successfully constructed single-gene models from each hotspot, as described below, and dubbed the resulting genes PKZILLA-1, -2, and -3 (Fig. 1D, Fig. 2, table S1). At final count, we annotated 22 PKS genes distributed across 16 of 34 pseudo-chromosomes that dramatically ranged in size from 3 to 137 kbp (table S1).
Fig. 2. Genomic, transcriptomic, and proteomic evidence for the PKZILLAs.

(A to C) Genomic PKS hotspot loci with gene models and PKS domain and module annotations for (A) PKZILLA-1 and (B) PKZILLA-2. (A, B) Red boxes denote chromosomal locations and relative sizes of the PKZILLA genes. The contiguous log-scale forward-stranded read coverage from the stranded rRNA-depletion RNA-seq (in gray) is shown across the PKZILLA gene models (in blue). Introns are highlighted with black arrows, and exons are numbered 1 to 17 for PKZILLA-1 and 1 to 12 for PKZILLA-2. See fig. S2 for an alternative view and figs. S3 and S4 for a detailed view of each intron. The numbered protein-coding exons are colored light blue, medium blue, or dark blue on the basis of whether supporting proteomic peptides from that exon were not detected, detected by protein-multimatch peptide matches alone, or detected by protein-unique plus exon-unique peptide matches, respectively (see section “Proteomic evidence for the PKZILLAs” in the text and fig. S10). Domain and module annotations (starting with the loading module (LM) and module 1 (M1) of PKZILLA-1 and ending with M56 of PKZILLA-2) are shown below the gene models, see key in (C). The bi- and trimodules are boxed in gray and categorized as S2/3M (saturating bi- and trimodule), PT2M (pass-through bimodule), and DH2M (dehydrating bimodule). See figs. S6 and S7 for non–length-normalized domains. KRn, N-terminal KR subdomain; KRc, C-terminal KR subdomain; KR*, catalytically novel or inactive KR.
Constructing the PKZILLA gene models from their candidate hotspots required several manual gene annotation interventions. Initial realignments of our Oxford Nanopore Technologies (ONT) long genomic DNA reads (20) to the PKZILLA hotspots revealed an assembly collapse of a tandem repetitive region within the coding N-terminus of PKZILLA-1 that was fixed by targeted reassembly. After that revision, we found no further PKZILLA assembly concerns (fig. S1). To test for and classify transcriptional activity at these hotspots, we next analyzed coverage from a P. parvum 12B1 oligo-dT mRNA enrichment/poly-A tail pulldown Illumina RNA sequencing (RNA-seq) dataset (21) to localize the putative PKZILLA mRNA 3’-ends. This dataset indicated one transcriptional termination site (TTS) per hotspot (fig. S2), however as established for poly-A pulldown RNA-Seq datasets, the coverage was negligible beyond ~10 kbp to the 5’-end and thus uninformative to 5’ transcriptional activity (fig. S2). To evaluate if the full PKZILLA hotspots were transcriptionally active and consistent with single genes, we applied mRNA length-unbiased Rrna-depletion Illumina RNA-seq by generating and sequencing two dUTP-stranded libraries from exponentially growing P. parvum cultures from the day and night phases. The low relative expression of the PKZILLAs combined with abundant rRNA reads from unoptimized rRNA depletion required four independent sequencing runs to accumulate sufficient coverage. Ultimately, we calculated that the PKZILLA transcript expression levels were uniformly low with transcripts per million (TPM) values of 1, 2, and 0.5 for PKZILLA-1, -2, and -3, respectively, in both day and night phases (table S1). These rRNA datasets further showed contiguous and sense-stranded transcriptional activity across the three PKZILLA gene models indicating one transcriptional start site (TSS) per PKZILLA hotspot (Fig. 2, fig. S2). Critically, the rRNA depletion data identified the presence and location of the 34 PKZILLA introns, all of which showed canonical eukaryotic GT-AG splice sites (table S2, fig. S3, S4, S5). The gene model-derived PKZILLA polypeptide sequences show near-contiguous sequence similarity to known PKS domains, with limited evidence for internal breaks (fig. S6, S7). Thus, we concluded that PKZILLA-1, -2, and -3 are single genes that each encode a single major transcriptional and translational product (Fig. 2). Remarkably, the calculated size of the PKZILLA-1 transcript at 136,071 nucleotides and the associated protein at 45,212 amino acids is about 25% larger than the mammalian muscle protein titin (18) (Fig. 1D, table S5).
Proteomic evidence for the PKZILLAs
To validate the PKZILLA proteins predicted by our gene models, we analyzed lyophilized P. parvum 12B1 biomass using an optimized bottom-up proteomics method. We identified and confidently validated 43 and 38 proteomic peptides from PKZILLA-1 and -2, respectively, yet none for PKZILLA-3 (fig. S8, table S6). Only 9 and 6 peptides from PKZILLA-1 and -2, respectively, were single-copy (single-match) within a single predicted PKZILLA polypeptide (protein-unique). Instead, most of the detected peptides were multimatch peptides present in multiple copies, either protein-unique to a given PKZILLA, or present in both PKZILLA-1 and -2 polypeptides (protein-multimatch) (fig. S9, S10). This high proportion of multimatch peptides highlights the internally repetitive nature of the “giga-modular” PKZILLAs, both within and across proteins. These peptides were only present in the PKZILLA gene models and were not found anywhere else in 6-frame translations of the 12B1 genome.
We next established which regions of the PKZILLA polypeptides were supported by proteomics. A complication is that most of the P. parvum proteomic data were multimatch peptides, which are rare in proteomic analyses of typical non-large, non-repetitive proteins. Since they cannot be unambiguously assigned to a single polypeptide region, multimatch peptides are often ignored in downstream analyses, in favor of simpler protein-unique single-match peptides (22). We judged that overlooking the multimatch peptides, although a simple solution, needlessly limited our analysis and discarded valuable data. We adapted to this challenge by sub-classifying each protein-unique yet multimatch peptide that also only arose from the translation product of a single exon as exon-unique (fig. S10), thus localizing proteomic support to the exon rather than the residue level. Of the 43 PKZILLA-1 proteomic peptides, 14 met both the protein-unique and exon-unique criteria (fig. S10), and thus established unambiguous proteomic support for translation of seven out of 17 PKZILLA-1 exons (41%), bounded upstream and downstream by exons 2 and 15, respectively (Fig. 2A). When considering the remaining 29 tryptic peptides despite their exon-multimatch or protein-multimatch ambiguity (fig. S10), we established increased proteomic support for 76% of the PKZILLA-1 exons (Fig. 2A). Applying the same criteria to PKZILLA-2, we measured proteomic support of 16 exon-unique peptides from three of the 12 exons (25%), bounded by exons 3 and 6, which increased to 75% of PKZILLA-2 exons after considering all PKZILLA matching peptides (Fig. 2B). Overall, these results confidently validate the translation of the PKZILLA-1/-2 transcripts into proteins and are consistent with a single translational product per gene.
Annotation of PKZILLA domain & modular structures and their compatibility with prymnesin
With proteomics-validated PKZILLA gene models in hand, we next tested their possible role in prymnesin-1 biosynthesis by annotating their PKS domains and evaluating their modular-arrangement against the chemical structure of a retrobiosynthetically proposed pre-prymnesin biosynthetic precursor (PPBP; Fig. 3A). We identified 140 and 99 protein domains for PKZILLA-1 and -2, respectively (table S3, S4, S5), using InterProScan (23). We also cataloged 30 candidate domains of unknown function (cDUFs), however none of these cDUFs showed strong evidence of being unannotated enzyme domains (table S7, S8). The first two domains of PKZILLA-1, an Acyl-CoA synthetase/NRPS adenylation domain/Luciferase (ANL) superfamily ligase adjacent to an acyl-carrier-protein (ACP) domain (Fig. 2A), comprise an unconventional, yet precedented (24), loading module (LM) to initiate polyketide chain elongation. PKZILLA-2 lacked any recognizable N-terminal loading domains; however, it does possess a C-terminal thioesterase (TE) domain, consistent with polyketide chain termination. In the end, we organized the combined 239 domains into 56 trans-AT PKS modules, including module-34 (M34) split across the C-terminus of PKZILLA-1 (Fig. 2A) and the N-terminus of PKZILLA-2 (Fig. 2B, table S5, S9, S10).
Fig. 3. Alignment of PKZILLA PKS modules with the proposed prymnesin biosynthetic precursor.

(A to C) PKZILLA-1 snapshot assembly line reactions at (bi)modules M6, M21/M22, and M30/M31 that depict FLX-catalyzed α-hydroxylation, reducing nonelongation “pass-through”, and dual methylation-saturation reactions. Dotted lines indicate omitted contiguous PKZILLA protein sequence. (B) PKZILLA-2 snapshot assembly line reactions at modules M41, M50, and M56 that depict 2,4-dienoyl-ACP reduction, transamination, and decarboxylative desulfation reactions. (C) Proposed structures of the PKZILLA-compatible pre-prymnesin-1 biosynthetic precursor, its corresponding epoxy-pre-prymnesin-1 intermediate, and the prymnesin-1 aglycone (31). The unassigned starter R group in PPBP is presently unknown yet responsible for providing the terminal acetylene group. For simplicity, all oxidative modifications are depicted post assembly line. The snapshot reactions in panels (A) and (B) are correspondingly boxed in (C). See fig. S21 for further detail. Me, methyl.
Prymnesin-1 is devoid of standard polyketide termini and had an unknown direction of chain elongation. We resolved its biosynthetic directionality by first correlating the diagnostic polyol segment at C84–C76 with the five adjacent modules M3–M7 that contain a ketoreductase (KR) as the terminal reductive domain. Based on this observation, we could infer the directionality of biosynthesis, with the PKZILLA-1 LM initiating the polyketide biosynthetic pathway with a three-carbon carboxylate of yet unknown origin followed by chain extension with seven malonate molecules via M1–M7 with interceding KR reduction to produce a hexol intermediate (fig. S21). Although five of the “β”-hydroxyls originate from malonate extender units, the out-of-sequence “α”-hydroxy group at C77 is instead likely installed by the α-hydroxylase flavoprotein (FLX) domain (25, 26) contained in M6. PKZILLA-1 harbors two additional FLX-domain containing modules, M9 and M11, both of which align with the installation of the further α-hydroxylations at C71 and C67, respectively. The absence of domains corresponding to additional C–H oxygenations (C81 and C83) and halogenation (C85) is suggestive that these functionalities are installed by intermodular- or trans-acting enzymes during chain elongation or post polyketide assembly via oxidative enzymes (27).
The precyclized region spanning prymnesin-1’s polyether rings (C74–C20) coincides with a break from canonical trans-AT modular architecture with 11 non-elongating ketosynthase (KS0) containing modules interspersed amongst 16 canonical trans-AT PKS modules (Fig. 2, 3A, table S9, S10). Two of the KS0s are found as part of “dehydrating bimodules” (Fig. 2; M39/M40, M42/M43) first characterized in bacterial trans-AT pathways (28), wherein the first module (KS-KR(-ACPn)) catalyzes chain elongation and ketone reduction, and the second module, consisting of a (KS0-DH-ACP), performs the corresponding dehydration to yield an α,β-alkene thioester intermediate (fig. S21). Three other KS0s are integrated into simple “pass-through bimodules” (Fig. 2.; M21/M22, M24/M25, M36/M37), also found in bacterial trans-AT BGCs (29), whose minimal KS0-ACP architecture preserves the β-hydroxy group generated by the upstream module. The remaining KS0s reside in unprecedented “saturating” bi- and trimodules (Fig. 2; M13/M14, M18/M19, M27/M28, M30/M31, M33/M34, and M44/M45/M46). In these systems, the second and third modules contain the full complement of reductive domains which convert the β-hydroxy group to a saturated methylene at positions C64, C56, C44, C40, C36, and C22 (Fig. 3A, fig. S21). Notably, saturating bimodule M30/M31 also contains the only methyltransferase (MT) domain in the entire assembly line and is positioned to install prymnesin’s lone methyl group at C39 (Fig. 3, fig. S21). We further confirmed the module-to-precursor alignment throughout this C74–C20 region by applying trans-AT ketoreductase precedent (30) to bioinformatically predict the stereochemical outcome of reduction for each KR domain (table S12). These bioinformatic predictions matched with six of the seven previously assigned configurations from the most recent structure revision of prymnesin-1 (31), with the C32 hydroxyl as the exception (table S13, fig. S21). By extrapolating these predictions, we propose β-hydroxy stereochemical assignments for the yet unassigned C84–C76 region of prymnesin-1 (Fig. 3A, table S13).
Finally, the polyene segment (C19–C1) contains several distinguishing structural features, all of which align with the final ten modules of PKZILLA-2. The M49 dehydratase is positioned to catalyze a precedented (32) vinylogous dehydration to reconfigure the C19–C16 diene out of conjugation relative to the ACP-tethered thioester, and the unconventional configuration of six consecutively arranged pyridoxal 5’-phosphate (PLP) dependent aminotransferase (AMT) domains (33) in M50 are located at the precise location to incorporate the sole primary amine at C14 (Fig. 3C). Much like the initial modules in PKZILLA-1, the final modules in PKZILLA-2, M51–M55, possess standard trans-AT domain architecture and generate the C12–C7 triene and a transient C4–C3 alkene that must undergo further desaturation to give prymnesin’s observed alkyne. The final module, M56, is precedented by the terminal PKS module from curacin biosynthesis (34) wherein an unusual sulfotransferase (ST) domain sulfates the β-hydroxy, and TE-mediated offloading initiates simultaneous decarboxylation and sulfate elimination to give a terminal alkene (Fig. 3D). As prymnesin terminates in a vinyl chloride, an additional halogenase must act pre- or post-chain offloading to install the third and final chloride at C1 (35).
PKZILLA gene models establish broad prymnesin polyketide biosynthetic logic
We next predicted that A- and B-type-producing strains of P. parvum would differ at the PKZILLA-1 locus based on structural differences between their prymnesins (Fig. 1B,C) (36). Illumina gDNA reads from nine strains of diverse A- and B-type P. parvum strains (20) were aligned to the 12B1 genome and read coverage at the three PKZILLA loci were compared. Precise structural differences could not be determined from read mapping alone due to the abundance of multi-mapped reads across the highly repetitive PKZILLA loci (fig. S22). In general, regions of uniquely mapped reads in all three PKZILLA genes were conserved across A- and B-type strains, however, the four B-type strains showed reduced, uniquely mapped read coverage at modules 18 and 21 of PKZILLA-1 indicative of a deletion relative to the 12B1 sequence (fig. S22A). To better characterize this structural difference, we used ONT long-reads to create a de novo assembly of B-type strain RCC3426 and characterize its locus homologous to PKZILLA-1. Synteny analysis identified a single 15 kbp region in 12B1 PKZILLA-1 absent in RCC3426. The absent region was comprised of a small portion of M17, all M18–20, and over half M21 for PKZILLA-1 amongst other changes (Fig. 4A). Recently, a similar observation was reported in the B-type P. parvum ODER1 strain (37).
Fig. 4. Comparative genomics, biosynthesis, and chemistry of the A- and B-type prymnesins.
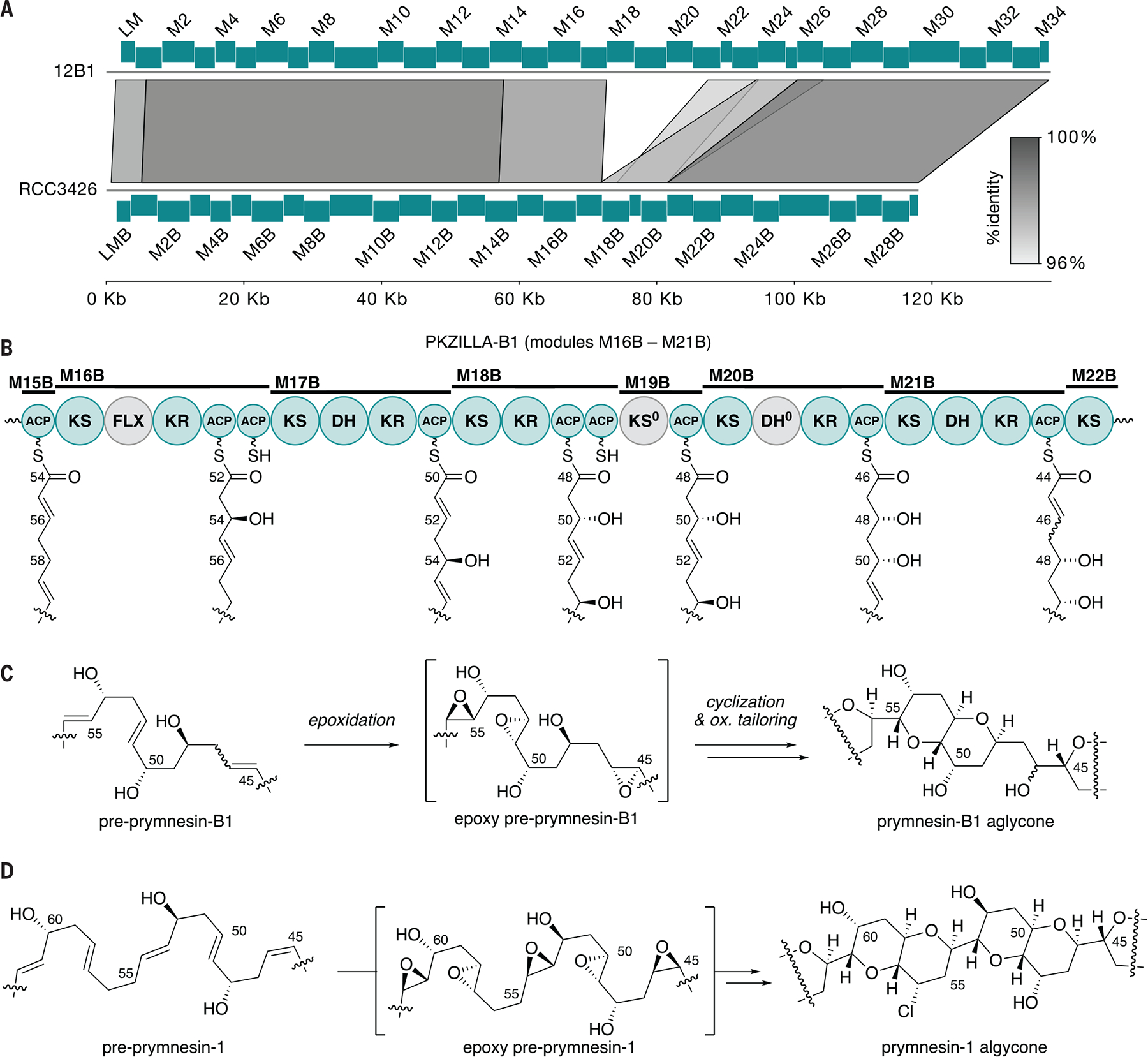
(A) Synteny plot of PKZILLA-1 from P. parvum 12B1 (top) and PKZILLA-B1 from P. parvum RCC3426 (bottom) showing a 15-kbp deletion and other modifications relative to the 12B1 sequence. (B) Domain organization of PKZILLA-B1 modules M16B to M21B and its assembly line biosynthesis of the differentiating C45–C55 fragment. Inactive domains are colored gray. (C) Partial structure and proposed conversion of pre–prymnesin-B1 via epoxy-pre–prymnesin-B1 to prymnesin-B1. Because the C46 stereocenter in prymnesin-B1 has not yet been established (56), the configuration of the C45-C46 alkene is not drawn. Full structures are shown in fig. S23. (D) Corresponding partial structures and biosynthesis of prymnesin-1 and intermediates highlighting the structure region that is distinct from the B1 series shown in (C). The omitted structure regions are identical between the A- and B-type prymnesins as supported by their respective PKZILLA gigasynthases. See Fig. 1 for full chemical structures.
Significantly, these changes in the B-type PKZILLA-1 gigasynthase (“PKZILLA-B1”) resulted in a smaller 29-modular synthase that corresponded to the 6-carbon shorter prymnesin-B1 molecule in which three elongating and two non-elongating modules were absent in relation to PKZILLA-1 (Fig. 4A). Domain predictions based upon the A-type PKZILLA model mapped seamlessly with the pre-prymnesin B1 polyene intermediate (Fig. 4B, fig. S23). Two key differences included the replacement of the M18/M19 saturating bimodule with a reducing M18B/M19B bimodule and the tetradomain reducing module M20B with a nonfunctional DH domain (table S15). These changes in module organization support the C50 and C48 hydroxyls in the pre-prymnesin-B1 intermediate (Fig. 4C) that, in addition to the loss of three extension modules, distinguish prymnesin-B1 from prymnesin-1 (Fig. 4D).
Although much of the prymnesin assembly line conforms to trans-AT PKS biochemistry, there are a few module-to-precursor alignments that may signal distinctive enzyme-catalyzed steps consistent across the A- and B-type PKZILLAs. In the case of M40 and its dienoyl intermediate, we propose that the adjacent M41 module elongates the growing polyketide chain to a 2,4-dienoyl-ACP intermediate before reduction (Fig. 3B, S21) by a phylogenetically distinct enoyl reductase “ER2” (table S10, fig. S15, S20) to generate a β,γ-alkene out of conjugation with the thioester carbonyl. This type of reduction has precedence in fatty acid biosynthesis on acyl-CoA intermediates (38). Similarly, module M48 is missing an explicit KS domain (Fig. 2, S7, table S10) and instead harbors the 186-residue sequence-unique cDUF9 in the analogous upstream position (table S8, S10) that may help recruit a restorative cis- or trans-acting KS enzyme (Fig. 3A). Finally, based on the currently revised structure of prymnesin-1 (31), our biosynthetic model requires a (Z)-ene at C45–C46 to accommodate the stereochemical outcome of the prymnesin aglycone, despite the M26-DH/KR pair predicting an (E)-ene product (table S12). Taken together, the sequence of the assembly line operations in the A- and B-type prymnesin systems strongly supports a causal role for PKZILLAs as the “gigasynthases” responsible for synthesis of the prymnesin backbone and suggests future focus areas to identify the remaining biosynthetic enzymes for the polyether cascade.
Discussion
The structural elucidation of brevetoxin B as the causative agent of the toxic red-tide dinoflagellate Karenia brevis over four decades ago (39), established microalgae as exquisite producers of polycyclic polyether toxins. Over 150 members with five- to nine-membered cyclic ethers, including maitotoxin as the largest with 32 fused rings, have since been discovered (40) and have helped shape the field of marine natural products due to their toxicity to both marine life and humans. One such marine polyether, halichondrin B, was even the inspiration of the synthetic derivative eribulin that has since been approved as an anticancer drug (41). The discovery and initial characterization of the prymnesin PKZILLA gigasynthases now sheds light on the long-standing question about how microalgae biosynthesize their giant polyketide polyether molecules.
The domain composition of the PKZILLA-1 and -2 gigasynthases are impeccably aligned with the cooperative assembly of the prymnesin carbon scaffold, and support decades-old hypotheses that diverse microalgal ladder-frame polyethers from haptophytes and dinoflagellates are constructed from all (E)- linear polyene intermediates (10, 11). Our discovery identified several unexpected features of the prymnesin assembly line. The sizes of the PKZILLA enzymes are stunning and surprisingly require just two modular trans-AT PKS proteins at 239 total domains for the construction of the 90-carbon long prymnesin molecule. In contrast, the longest known bacterial polyketide, quinolidomicin at 68 carbons (42), is assembled by 13 PKS gene products (Fig. 1D). The remarkable size of the PKZILLASs expands our imagination on the capabilities of enzymes in the construction of complex molecules. And finally, many non-elongating KS0s are featured in modules associated with the construction of the polycyclic interior of prymnesin, which may contribute to the timing and mechanism of polyether assembly.
No comparable PKS system has yet been identified from a toxic dinoflagellate (14); however, numerous studies have established that dinoflagellates encode large numbers of multimodular and single-domain type I PKSs (43), including a promising, yet reportedly incomplete 35-knt 7-module PKS transcript candidate from the ciguatoxin-producing Gambierdiscus polynesiensis (13). If dinoflagellates similarly encode giant PKSs reminiscent of the PKZILLAs, then common transcriptomic practices involving poly-A pulldown RNA sequencing may bias against giant transcripts and instead require length-unbiased rRNA-depletion RNA-seq alongside customized assembly and annotation as performed in this study. Notably, the PKZILLAs went unrecognized from recent P. parvum transcriptomic (15) and genomic (44) analyses, highlighting the challenges of assembling and annotating giant PKS genes with their highly repetitive sequences. Retrospective analysis of the recent transcriptomic study (15), with hindsight from knowledge of the full PKZILLA gene models, revealed that the 3’-ends of the PKZILLA-1 and -2 genes were indeed captured, covering modules M28–M34 (contig 7) and M54–56 (contig 6), respectively. These observations are similar to ours when viewed only by poly-A RNA-seq (fig. S2) and highlight the limitations of this common RNA-seq method when conducted on giant transcripts.
Dinoflagellate polyketides also share a distinctive biosynthetic feature involving the irregular incorporation of intact and C1-deleted acetate building blocks as illuminated by isotope labeling studies (45). The prymnesin biosynthetic model, on the other hand, supports the intact incorporation of 43 contiguous malonate units, which is standard in most bacterial systems. The α-hydroxylating FLX domains and “pass-through” modules found within the PKZILLA modules provide a tantalizing hypothesis for this yet to be described dinoflagellate PKS biochemistry: Assembly line oxidation to the α-ketone followed by transacylation by a KS0 may lead to excision of single carbon atoms by decarbonylation as precedented in the biosynthesis of marine polyketides enterocin (46) and barbamide (47).
Though PKZILLA-1 and -2 are responsible for the construction of the majority of the prymnesin molecule, additional enzymes (acyltransferases, desaturases, hydroxylases, chlorinases, epoxidases, glycosyltransferases) are needed to complete the full biosynthetic pathway and install prymnesin’s remaining functional groups and sugar moieties. In contrast to the small alkaloid domoic acid with its clustered causal genes (48), the distribution of the PKZILLA-1 and -2 genes across separate pseudo-chromosomes indicates prymnesin biosynthesis is not encoded within a single biosynthetic gene cluster, and its tailoring enzymes may also not be clustered. The discovery of the PKZILLAs and their role in prymnesin biosynthesis lays the foundation for the development and implementation of alternative linked ‘omics approaches to fully uncover the complete suite of prymnesin biosynthetic enzymes. Moreover, PKZILLAs offer the opportunity to dissect the enzymology of ladder-frame polyether biosynthesis and will serve as a model to capture and dissect giant genes, transcripts, and proteins in specialized metabolism.
Supplementary Material
Acknowledgments:
The authors acknowledge helpful discussions and feedback from members of the Moore laboratory, and from Prof. April Lukowski (UCSD). This publication includes data generated at the UC San Diego IGM Genomics Center utilizing an Illumina NovaSeq 6000 that was purchased with funding from a National Institutes of Health SIG grant (#S10 OD026929). Large language models (LLMs), including code generation by GitHub Copilot (Microsoft, Redmond, WA, USA) within Visual Studio Code (Microsoft, Redmond, WA, USA), and code generation and limited text rephrasing by the Claude 3 Opus application programming interface (API) (Anthropic PBC, San Francisco, CA, USA), were used in this work. The authors reviewed all LLM generated text and rejected any inaccuracies.
Funding:
National Institutes of Health grant F32-ES032276 (TRF)
National Institutes of Health grant F32-GM145146 (VVS)
National Institutes of Health grant R01-GM085770 (BSM)
National Science Foundation grant DEB-1831493 (JHW)
Footnotes
Competing interests: Authors declare that they have no competing interests.
Data and materials availability:
The P. parvum 12B1 v1.1 genome assembly, PKZILLA gene models, and updated gene annotation have been deposited to the National Center for Biotechnology Information (NCBI) Whole Genome Shotgun (WGS) database (accession number JBGBPQ010000000). Raw rRNA depletion RNA-seq have been deposited to the NCBI Sequence Read Archive (SRA) archive (BioProject PRJNA936443). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium through the PRIDE (58) partner repository under the dataset identifier PXD044632 (59). Other extended datasets and analysis code (scripts) are available on Zenodo and/or Github and are both cited in-line throughout the manuscript and listed in table S16.
References and Notes
- 1.Hallegraeff GM, Anderson DM, Davidson K, Gianella F, Hansen P, Fish-Killing Marine Algal Blooms: Causative Organisms, Ichthyotoxic Mechanisms, Impacts and Mitigation. (UNESCO, Paris, France, 2023; 10.25607/OBP-1964)IOC Manuals and Guides. [DOI] [Google Scholar]
- 2.Griffith AW, Gobler CJ, Harmful algal blooms: A climate change co-stressor in marine and freshwater ecosystems. Harmful Algae 91, 101590 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Sobieraj J, Metelski D, Insights into Toxic Prymnesium parvum Blooms as a Cause of the Ecological Disaster on the Odra River. Toxins 15, 403 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karlson B, Andersen P, Arneborg L, Cembella A, Eikrem W, John U, West JJ, Klemm K, Kobos J, Lehtinen S, Lundholm N, Mazur-Marzec H, Naustvoll L, Poelman M, Provoost P, De Rijcke M, Suikkanen S, Harmful algal blooms and their effects in coastal seas of Northern Europe. Harmful Algae 102, 101989 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Patiño R, Christensen VG, Graham JL, Rogosch JS, Rosen BH, Toxic Algae in Inland Waters of the Conterminous United States—A Review and Synthesis. Water 15, 2808 (2023). [Google Scholar]
- 6.Igarashi T, Satake M, Yasumoto T, Structures and Partial Stereochemical Assignments for Prymnesin-1 and Prymnesin-2: Potent Hemolytic and Ichthyotoxic Glycosides Isolated from the Red Tide Alga Prymnesium parvum. J. Am. Chem. Soc 121, 8499–8511 (1999). [Google Scholar]
- 7.Moore RE, Bartolini G, Structure of palytoxin. J. Am. Chem. Soc 103, 2491–2494 (1981). [Google Scholar]
- 8.Murata M, Naoki H, Iwashita T, Matsunaga S, Sasaki M, Yokoyama A, Yasumoto T, Structure of maitotoxin. J. Am. Chem. Soc 115, 2060–2062 (1993). [Google Scholar]
- 9.Nicolaou KC, Frederick MO, Aversa RJ, The Continuing Saga of the Marine Polyether Biotoxins. Angew. Chem. Int. Ed 47, 7182–7225 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakanishi K, The chemistry of brevetoxins: A review. Toxicon 23, 473–479 (1985). [DOI] [PubMed] [Google Scholar]
- 11.Vilotijevic I, Jamison TF, Epoxide-Opening Cascades Promoted by Water. Science 317, 1189–1192 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nivina A, Yuet KP, Hsu J, Khosla C, Evolution and Diversity of Assembly-Line Polyketide Synthases. Chem. Rev 119, 12524–12547 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dolah FMV, Morey JS, Milne S, Ung A, Anderson PE, Chinain M, Transcriptomic analysis of polyketide synthases in a highly ciguatoxic dinoflagellate, Gambierdiscus polynesiensis and low toxicity Gambierdiscus pacificus, from French Polynesia. PLOS ONE 15, e0231400 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verma A, Barua A, Ruvindy R, Savela H, Ajani PA, Murray SA, The Genetic Basis of Toxin Biosynthesis in Dinoflagellates. Microorganisms 7, 222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anestis K, Kohli GS, Wohlrab S, Varga E, Larsen TO, Hansen PJ, John U, Polyketide synthase genes and molecular trade-offs in the ichthyotoxic species Prymnesium parvum. Sci. Total Environ 795, 148878 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Heimerl N, Hommel E, Westermann M, Meichsner D, Lohr M, Hertweck C, Grossman AR, Mittag M, Sasso S, A giant type I polyketide synthase participates in zygospore maturation in Chlamydomonas reinhardtii. Plant J. 95, 268–281 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Medema MH, de Rond T, Moore BS, Mining genomes to illuminate the specialized chemistry of life. Nat. Rev. Genet 22, 553–571 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bang M-L, Centner T, Fornoff F, Geach AJ, Gotthardt M, McNabb M, Witt CC, Labeit D, Gregorio CC, Granzier H, Labeit S, The Complete Gene Sequence of Titin, Expression of an Unusual ≈700-kDa Titin Isoform, and Its Interaction With Obscurin Identify a Novel Z-Line to I-Band Linking System. Circ. Res 89, 1065–1072 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Binzer SB, Svenssen DK, Daugbjerg N, Alves-de-Souza C, Pinto E, Hansen PJ, Larsen TO, Varga E, A-, B- and C-type prymnesins are clade specific compounds and chemotaxonomic markers in Prymnesium parvum. Harmful Algae 81, 10–17 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Wisecaver JH, Auber RP, Pendleton AL, Watervoort NF, Fallon TR, Riedling OL, Manning SR, Moore BS, Driscoll WW, Extreme genome diversity and cryptic speciation in a harmful algal-bloom-forming eukaryote. Curr. Biol 33, 2246–2259.e8 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.National Center for Biotechnology Information (NCBI), Sequence Read Archive (SRA), Prymnesium parvum strain 12B Illumina reads − SRA PRJNA201451, SRR1685644. https://www.ncbi.nlm.nih.gov/sra/?term=SRR1685644.
- 22.Dou Y, Liu Y, Yi X, Olsen LK, Zhu H, Gao Q, Zhou H, Zhang B, SEPepQuant enhances the detection of possible isoform regulations in shotgun proteomics. Nat. Commun 14, 5809 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong S-Y, Lopez R, Hunter S, InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hemmerling F, Lebe KE, Wunderlich J, Hahn F, An Unusual Fatty Acyl:Adenylate Ligase (FAAL)–Acyl Carrier Protein (ACP) Didomain in Ambruticin Biosynthesis. ChemBioChem 19, 1006–1011 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Hemmerling F, Meoded RA, Fraley AE, Minas HA, Dieterich CL, Rust M, Ueoka R, Jensen K, Helfrich EJN, Bergande C, Biedermann M, Magnus N, Piechulla B, Piel J, Modular Halogenation, α-Hydroxylation, and Acylation by a Remarkably Versatile Polyketide Synthase. Angew. Chem. Int. Ed 61, e202116614 (2022). [DOI] [PubMed] [Google Scholar]
- 26.Winter AJ, Khanizeman RN, Barker-Mountford AMC, Devine AJ, Wang L, Song Z, Davies JA, Race PR, Williams C, Simpson TJ, Willis CL, Crump MP, Structure and Function of the α-Hydroxylation Bimodule of the Mupirocin Polyketide Synthase. Angew. Chem. Int. Ed 62, e202312514 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helfrich EJN, Piel J, Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep 33, 231–316 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Wagner DT, Zeng J, Bailey CB, Gay DC, Yuan F, Manion HR, Keatinge-Clay AT, Structural and Functional Trends in Dehydrating Bimodules from trans-Acyltransferase Polyketide Synthases. Structure 25, 1045–1055.e2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masschelein J, Sydor PK, Hobson C, Howe R, Jones C, Roberts DM, Ling Yap Z, Parkhill J, Mahenthiralingam E, Challis GL, A dual transacylation mechanism for polyketide synthase chain release in enacyloxin antibiotic biosynthesis. Nat. Chem 11, 906–912 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helfrich EJN, Ueoka R, Dolev A, Rust M, Meoded RA, Bhushan A, Califano G, Costa R, Gugger M, Steinbeck C, Moreno P, Piel J, Automated structure prediction of trans-acyltransferase polyketide synthase products. Nat. Chem. Biol 15, 813–821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasaki M, Takeda N, Fuwa H, Watanabe R, Satake M, Oshima Y, Synthesis of the JK/LM-ring model of prymnesins, potent hemolytic and ichthyotoxic polycyclic ethers isolated from the red tide alga Prymnesium parvum: confirmation of the relative configuration of the K/L-ring juncture. Tetrahedron Lett. 47, 5687–5691 (2006). [Google Scholar]
- 32.Taft F, Brünjes M, Knobloch T, Floss HG, Kirschning A, Timing of the Δ10,12-Δ11,13 Double Bond Migration During Ansamitocin Biosynthesis in Actinosynnema pretiosum. J. Am. Chem. Soc 131, 3812–3813 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Aron ZD, Dorrestein PC, Blackhall JR, Kelleher NL, Walsh CT, Characterization of a New Tailoring Domain in Polyketide Biogenesis: The Amine Transferase Domain of MycA in the Mycosubtilin Gene Cluster. J. Am. Chem. Soc 127, 14986–14987 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Gu L, Wang B, Kulkarni A, Gehret JJ, Lloyd KR, Gerwick L, Gerwick WH, Wipf P, Håkansson K, Smith JL, Sherman DH, Polyketide Decarboxylative Chain Termination Preceded by O-Sulfonation in Curacin A Biosynthesis. J. Am. Chem. Soc 131, 16033–16035 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang Y, Kim A, Olive C, Lewis JC, Selective C−H Halogenation of Alkenes and Alkynes Using Flavin-Dependent Halogenases. Angew. Chem. Int. Ed 63, e202317860 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rasmussen SA, Andersen AJC, Andersen NG, Nielsen KF, Hansen PJ, Larsen TO, Chemical diversity, origin, and analysis of phycotoxins. J. Nat. Prod 79, 662–673 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Kuhl H, Strassert JFH, Čertnerova D, Varga E, Kreuz E, Lamatsch DK, Wuertz S, Köhler J, Monaghan MT, Stöck M, The haplotype-resolved Prymnesium parvum (type B) microalga genome reveals the genetic basis of its fish-killing toxins. bioRxiv [Preprint] (2024). 10.1101/2024.03.27.587007. [DOI] [PubMed] [Google Scholar]
- 38.Hua T, Wu D, Ding W, Wang J, Shaw N, Liu Z-J, Studies of Human 2,4-Dienoyl CoA Reductase Shed New Light on Peroxisomal β-Oxidation of Unsaturated Fatty Acids. J. Biol. Chem 287, 28956–28965 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin Yong-Yeng, Risk Martin, Ray Sammy M., Donna Van Engen Jon Clardy, Golik Jerzy, James John C., Nakanishi Koji, Isolation and structure of brevetoxin B from the “red tide” dinoflagellate Ptychodiscus brevis (Gymnodinium breve). J. Am. Chem. Soc 103, 6773–6775 (1981). [Google Scholar]
- 40.Jiang Z-P, Sun S-H, Yu Y, Mándi A, Luo J-Y, Yang M-H, Kurtán T, Chen W-H, Shen L, Wu J, Discovery of benthol A and its challenging stereochemical assignment: opening up a new window for skeletal diversity of super-carbon-chain compounds. Chem. Sci 12, 10197–10206 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cortes J, Schöffski P, Littlefield BA, Multiple modes of action of eribulin mesylate: Emerging data and clinical implications. Cancer Treat. Rev 70, 190–198 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Hashimoto T, Hashimoto J, Kozone I, Amagai K, Kawahara T, Takahashi S, Ikeda H, Shin-ya K, Biosynthesis of Quinolidomicin, the Largest Known Macrolide of Terrestrial Origin: Identification and Heterologous Expression of a Biosynthetic Gene Cluster over 200 kb. Org. Lett 20, 7996–7999 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Dolah FMV, Kohli GS, Morey JS, Murray SA, Both modular and single-domain Type I polyketide synthases are expressed in the brevetoxin-producing dinoflagellate, Karenia brevis (Dinophyceae). J. Phycol 53, 1325–1339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jian J, Wu Z, Silva-Núñez A, Li X, Zheng X, Luo B, Liu Y, Fang X, Workman CT, Larsen TO, Hansen PJ, Sonnenschein EC, Long-read genome sequencing provides novel insights into the harmful algal bloom species Prymnesium parvum. Sci. Total Environ, 168042 (2023). [DOI] [PubMed] [Google Scholar]
- 45.Van Wagoner RM, Satake M, Wright JLC, Polyketide biosynthesis in dinoflagellates: what makes it different? Nat. Prod. Rep, 37 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Teufel R, Miyanaga A, Michaudel Q, Stull F, Louie G, Noel JP, Baran PS, Palfey B, Moore BS, Flavin-mediated dual oxidation controls an enzymatic Favorskii-type rearrangement. Nature 503, 552–556 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo S, Sang Y, Zheng C, Xue X-S, Tang Z, Liu W, Enzymatic α-Ketothioester Decarbonylation Occurs in the Assembly Line of Barbamide for Skeleton Editing. J. Am. Chem. Soc 145, 5017–5028 (2023). [DOI] [PubMed] [Google Scholar]
- 48.Brunson JK, McKinnie SMK, Chekan JR, McCrow JP, Miles ZD, Bertrand EM, Bielinski VA, Luhavaya H, Oborník M, Smith GJ, Hutchins DA, Allen AE, Moore BS, Biosynthesis of the neurotoxin domoic acid in a bloom-forming diatom. Science 361, 1356–1358 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holdway PA, Watson RA, Moss B, Aspects of the ecology of Prymnesium parvum (Haptophyta) and water chemistry in the Norfolk Broads, England. Freshw. Biol 8, 295–311 (1978). [Google Scholar]
- 50.Hems ES, “Synthesis of carbohydrate based tools to explore the biosynthesis of and develop detection methods for prymnesin toxins,” thesis, University of East Anglia, Norwich: (2017). [Google Scholar]
- 51.Liebert F, Deerns WM, Nederland Nationaal Archief - Inventaris van het archief van de Hoofinspecteur der Visserijen, (1907) 1910–1922 (1924) - Item 195: Verslag van een onderzoek naar de oorzaken van de vissterfte in de polder Workumer-Nieuwland nabij Workum, door N.W. Deerns van het Rijksinstituut voor Hydrografisch Visserijonderzoek, houdende een beschrijving van visdodende flagellaten (planktonorganismen). 1920 (1920). https://www.nationaalarchief.nl/onderzoeken/archief/2.11.12.
- 52.Reich K, Aschner M, Mass development and control of the phytoflagellate Prymnesium parvum in fish ponds in Palestine. Palest. J. Bot 4 (1947). [Google Scholar]
- 53.Shilo M, Rosenberger RF, STUDIES ON THE TOXIC PRINCIPLES FORMED BY THE CHRYSOMONAD PRYMNESIUM PARVUM CARTER. Ann. N. Y. Acad. Sci 90, 866–876 (1960). [Google Scholar]
- 54.James TL, De La Cruz A, PRYMNESIUM-PARVUM CARTER (CHRYSOPHYCEAE) AS A SUSPECT OF MASS MORTALITIES OF FISH AND SHELLFISH COMMUNITIES IN WESTERN TEXAS. Tex. J. Sci 41, 429–430 (1989). [Google Scholar]
- 55.Roelke DL, Grover JP, Brooks BW, Glass J, Buzan D, Southard GM, Fries L, Gable GM, Schwierzke-Wade L, Byrd M, Nelson J, A decade of fish-killing Prymnesium parvum blooms in Texas: roles of inflow and salinity. J. Plankton Res 33, 243–253 (2011). [Google Scholar]
- 56.Rasmussen SA, Meier S, Andersen NG, Blossom HE, Duus JØ, Nielsen KF, Hansen PJ, Larsen TO, Chemodiversity of Ladder-Frame Prymnesin Polyethers in Prymnesium parvum. J. Nat. Prod 79, 2250–2256 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Donadio S, Staver M, McAlpine J, Swanson S, Katz L, Modular organization of genes required for complex polyketide biosynthesis. Science 252, 675–679 (1991). [DOI] [PubMed] [Google Scholar]
- 58.Perez-Riverol Y, Bai J, Bandla C, García-Seisdedos D, Hewapathirana S, Kamatchinathan S, Kundu DJ, Prakash A, Frericks-Zipper A, Eisenacher M, Walzer M, Wang S, Brazma A, Vizcaíno JA, The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fallon T, Label-free bottom-up proteomics of Prymnesium pavum 12B1, PRIDE (2024); 10.6019/PXD044632. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The P. parvum 12B1 v1.1 genome assembly, PKZILLA gene models, and updated gene annotation have been deposited to the National Center for Biotechnology Information (NCBI) Whole Genome Shotgun (WGS) database (accession number JBGBPQ010000000). Raw rRNA depletion RNA-seq have been deposited to the NCBI Sequence Read Archive (SRA) archive (BioProject PRJNA936443). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium through the PRIDE (58) partner repository under the dataset identifier PXD044632 (59). Other extended datasets and analysis code (scripts) are available on Zenodo and/or Github and are both cited in-line throughout the manuscript and listed in table S16.


