Abstract
The effectiveness of mRNA vaccines largely depends on their lipid nanoparticle (LNP) component. Herein, we investigate the effectiveness of DLin-KC2-DMA (KC2) and SM-102-based LNPs for the intramuscular delivery of a plasmid encoding B.1.617.2 (Delta) spike fused with CD40 ligand. LNP encapsulation of this CD40L-adjuvanted DNA vaccine with either LNP formulation drastically enhanced antibody responses, enabling neutralization of heterologous Omicron variants. The DNA-LNP formulations provided excellent protection from homologous challenge, reducing viral replication, and preventing histopathological changes in the pulmonary tissues. Moreover, the DNA-LNP vaccines maintained a high level of protection against heterologous Omicron BA.5 challenge despite a reduced neutralizing response. In addition, we observed that DNA-LNP vaccination led to the pulmonary downregulation of interferon signaling, interleukin-12 signaling, and macrophage response pathways following SARS-CoV-2 challenge, shedding some light on the mechanisms underlying the prevention of pulmonary injury. These results highlight the potential combination of molecular adjuvants with LNP-based vaccine delivery to induce greater and broader immune responses capable of preventing inflammatory damage and protecting against emerging variants. These findings could be informative for the future design of both DNA and mRNA vaccines.
Keywords: SARS-CoV-2, lipid nanoparticle, DNA vaccine, nucleic acid, antibody, Omicron variants, CD40 ligand
Graphical abstract
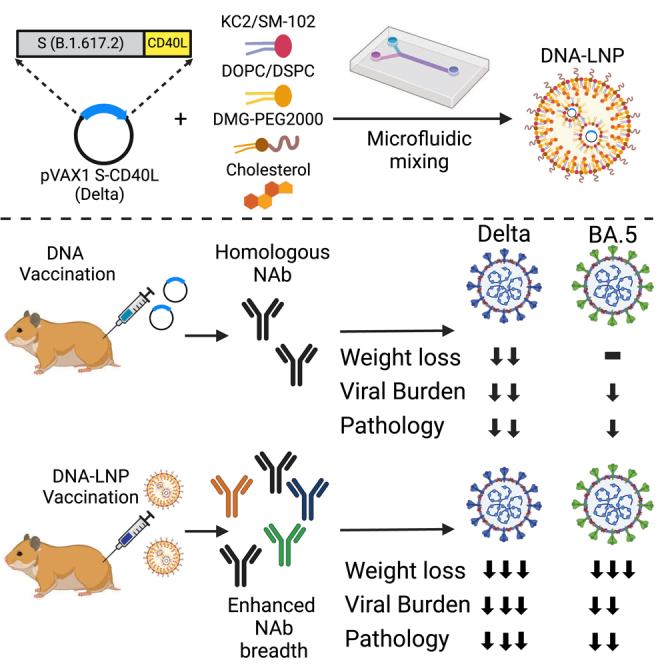
Tamming and colleagues demonstrate that the encapsulation of DNA vaccines within the same lipid nanoparticle formulations found in commercial mRNA vaccines greatly enhances immune responses generated against SARS-CoV-2. These enhanced responses enabled the neutralization of heterologous variants and significantly increased protection in Syrian hamster challenge models.
Introduction
Since the emergence of SARS-CoV-2 and the beginning of the COVID-19 pandemic, historic amounts of time, effort, and resources have been spent on vaccine research and development. Vaccines have proven to be highly effective at reducing adverse outcomes and hospitalizations associated with SARS-CoV-2 infection,1,2,3 and are estimated to have prevented millions of deaths throughout the pandemic.4 Spearheading global vaccination efforts have been several mRNA lipid nanoparticle (LNP) vaccines, which have shown tremendous efficacy.5,6,7,8 A key factor contributing to the successful implementation of these mRNA vaccines has been their LNP delivery system.9,10 LNPs contribute to vaccine efficacy by reducing degradation of the nucleic acid payload and improving its cellular uptake.11,12,13 Additionally, the ionizable lipid component of LNP vaccines has been shown to be immunogenic,14,15,16,17 which may contribute to improved immune responses. DNA vaccines are often presented as an alternative to mRNA vaccines, sharing many properties but with a lower cost of production and a decreased likelihood to induce undesirable ribosomal frameshifting during translation.18,19,20 However, DNA vaccine efficacy has historically been limited by poor cellular uptake, susceptibility to DNase degradation, and limited immunogenicity.21 In the wake of widespread LNP-mRNA vaccination during the COVID-19 pandemic, more research is essential to comprehend how effectively LNP delivery can be translated for use in DNA-based vaccination.
We previously described the immunogenicity and efficacy of pcDNA3.1 S.dTM.PP-CD40L, a plasmid DNA (pDNA)-vectored vaccine encoding a pre-fusion stabilized ancestral SARS-CoV-2 spike (S) ectodomain fused to the ectodomain of CD40 ligand (CD40L).22 Engagement with CD40L on the surface of activated T cells represents a key regulator of antigen-presenting cell activation.23 While the addition of CD40L to our pDNA-based vaccine improved humoral responses following intramuscular vaccination in Syrian hamsters and afforded significant protection from challenge with an ancestral SARS-CoV-2 isolate, the adjuvanted vaccine failed to completely prevent viral replication and lung pathology. We encapsulated an updated version of our CD40L-adjuvanted vaccine based on the B.1.617.2 Delta variant, pVAX1 S-CD40LB.1.617.2, in two different LNP formulations containing either DLin-KC2-DMA (KC2) or SM-102 ionizable lipids. We evaluated both the immunogenicity and efficacy of the LNP-encapsulated vaccines in comparison with unencapsulated pDNA in Syrian hamsters following B.1.617.2 and BA.5 SARS-CoV-2 challenge. Herein, we demonstrate that LNP encapsulation drastically improves the strength and breadth of humoral responses induced by DNA vaccination and provides superior protection upon homologous and heterologous challenge.
Results
LNP encapsulation enhances strength and breadth of DNA vaccine-induced humoral response
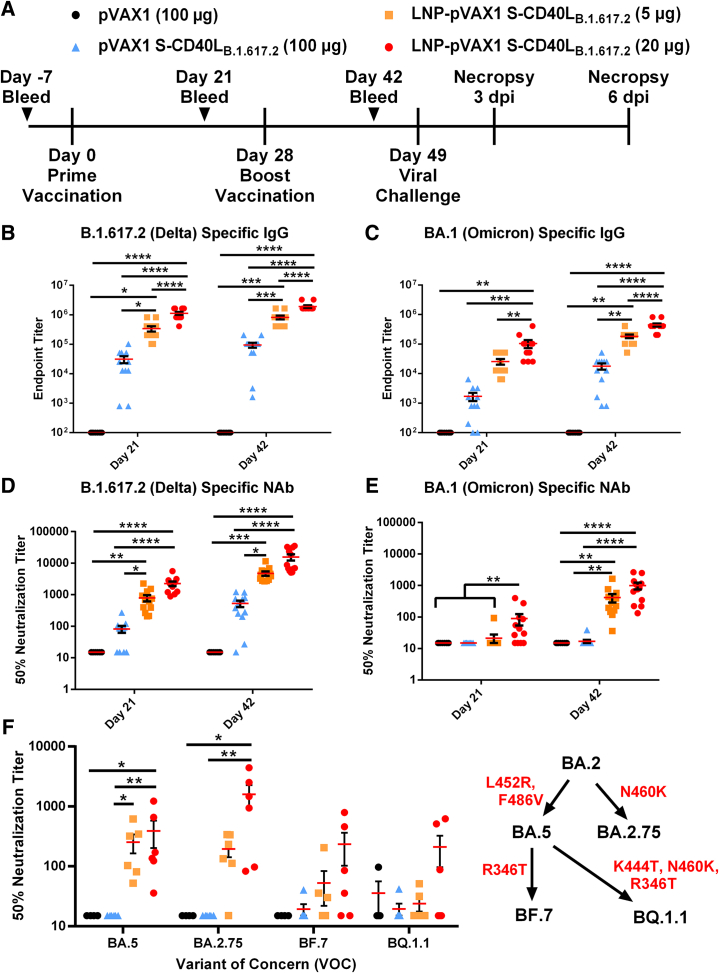
The B.1.617.2-based DNA construct (pVAX1 S-CD40LB.1.617.2) differs from our previous construct22 in that nine mutations found in the B.1.617.2 variant S protein were introduced into the vaccine antigen (Figure S1A) and the pVAX1 vector was used rather than pcDNA3.1. Due to the omission of non-essential elements, the pVAX1 vector is considerably smaller than pcDNA3.1, which should allow a greater number of plasmids to be administered in an equivalent dose improving antigen expression and vaccine efficiency.24 An in vitro CD40 signaling bioassay confirmed the fused CD40L domain remained functional (Figure S1B). To first assess DNA-LNP immunogenicity and efficacy, we immunized male Syrian hamsters intramuscularly at a 4-week interval with 5 or 20 μg of LNP-encapsulated pVAX1 S-CD40LB.1.617.2 (Figure 1A). Male hamsters were chosen as they experience more severe infection compared with females.25,26 Formulated with KC2 ionizable lipids, the DNA-LNPs had an average size distribution around 100 nm (Table S1). For comparison, hamsters were also immunized with a 100-μg dose of unencapsulated pVAX1 S-CD40LB.1.617.2, this dose of naked DNA having previously conferred significant yet incomplete protection.22 Both the 5- and 20-μg doses of KC2 LNP-encapsulated pVAX1 S-CD40LB.1.617.2 induced greater binding antibody responses than the significantly higher 100-μg dose of unencapsulated vaccine against homologous B.1.617.2 S (Figure 1B) and heterologous Omicron BA.1 S (Figure 1C). Furthermore, the magnitude of this increase appears to rely on the fusion of CD40L (Figure S1C). LNP encapsulation also substantially improved neutralizing antibody (NAb) responses. As shown in Figure 1D, unlike unencapsulated pVAX1 S-CD40LB.1.617.2, both 5- and 20-μg doses of the DNA-LNP induced significant NAb responses against B.1.617.2 pseudotyped-VSV after a single dose. Interestingly, despite having a strong binding Ab response, a single 5-μg dose of LNP-formulated pVAX1 S-CD40LB.1.617.2 failed to elicit a detectable neutralizing response against BA.1 pseudotyped-VSV (Figure 1E). However, following the second vaccination, both the 5- and 20-μg doses of LNP-formulated pVAX1 S-CD40LB.1.617.2 induced significant NAb responses against BA.1. In contrast, vaccination with naked pVAX1 S-CD40LB.1.617.2 failed to induce a detectable Omicron neutralizing response even after two vaccine doses. Notably, NAb responses following the boost vaccination were also measured against four other Omicron subvariants (Figure 1F). Both the 5-μg and 20-μg doses of the LNP-encapsulated vaccine induced a significant response against BA.5, while the higher dose also raised a significant response against BA.2.75. With respect to BF.7 and BQ.1, while not statistically significant relative to control groups, vaccination with 20-μg of the DNA-LNPs did raise some neutralizing activity against these newer variants.
Figure 1.
LNP encapsulation improves humoral response to DNA vaccination
(A) Male Syrian hamsters were randomly divided into four experimental groups (n = 8 for pVAX1, n = 12 for pVAX1 S-CD40LB.1.617.2) and immunized intramuscularly on days 0 and 28 with 100 μg of pVAX1, 100 μg of pVAX1 S-CD40LB.1.617.2, or 5 or 20 μg of LNP-encapsulated pVAX1 S-CD40LB.1.617.2. Animals were challenged intranasally with 1.67 × 105 TCID50 of a SARS-CoV-2 B.1.617.2 isolate on day 49 and euthanized 3 and 6 days post-infection (dpi). ELISA determination of total B.1.617.2 (B) and BA.1 (C) spike-specific IgG in the sera of immunized hamsters on days 21 and 42. The 50% neutralizing titer (NT50) of immunized hamster sera on days 21 and 42 was determined using (D) B.1.617.2 and (E) BA.1 pseudotyped-VSV. (F) The NT50 on day 42 was also determined using BA.5, BA.2.75, BF.7, and BQ.1.1 SARS-CoV-2 spike pseudotyped-VSV. Red indicates mutations acquired in the SARS-CoV-2 spike protein. Data shown are mean ± SEM; n = 8 for pVAX1, n = 12 for pVAX1 S-CD40LB.1.617.2 groups, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
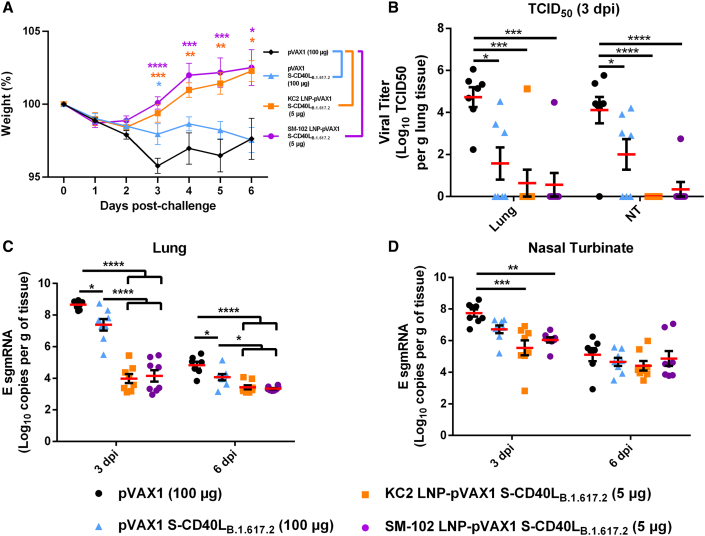
LNP encapsulation protects hamsters against homologous B.1.617.2 challenge
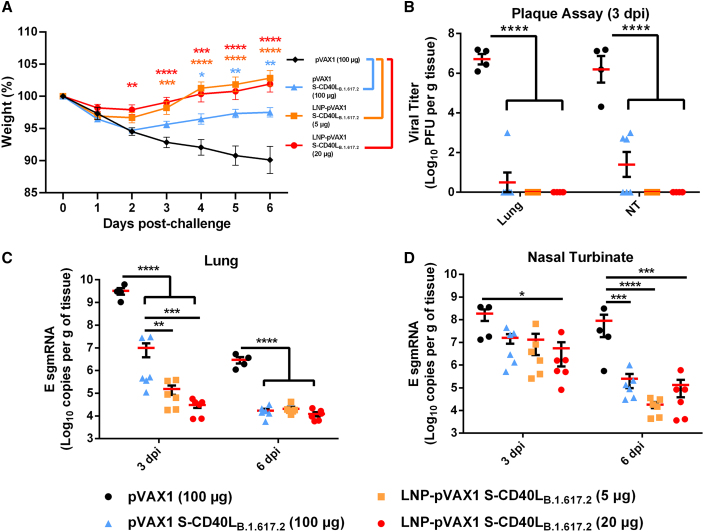
Three weeks after the second immunization, the Syrian hamsters were challenged intranasally with 1 × 105 plaque-forming units (PFU) of a SARS-CoV-2 B.1.617.2. Although all groups experienced slight body weight loss on the first day post-challenge, only pVAX1 control group continuously lost weight over the course of the infection, reaching a mean weight loss of 9.9% ± 4.2% on day 6 post-challenge (Figure 2A). In contrast, all three pVAX1 S-CD40LB.1.617.2 vaccinated groups began recovering from the initial weight loss. Importantly, however, hamsters vaccinated with 5 and 20 μg of the DNA-LNP consistently lost less body weight and recovered faster than those that received 100 μg of the naked DNA vaccine. These results reflect the viral burden in the lungs and nasal turbinates of these animals. As shown in Figure 2B, the vaccinated groups had significantly less viral burden in lungs and nasal turbinates, with no virus being detectable by plaque assay in the DNA-LNP groups. In addition to quantification of live virus, we used RT-PCR to quantify viral envelope (E) subgenomic mRNA (sgmRNA) expression in the same tissues, which revealed a similar observation. All vaccines significantly reduced E sgmRNA levels in the lung tissue relative to the pVAX1 vector control at both 3 and 6 days post-infection (dpi) (Figure 2C). Additionally, vaccination with the DNA-LNP also significantly reduced E sgmRNA expression in the lungs relative to unformulated pVAX1 S-CD40LB.1.617.2 3-dpi (Figure 2C). We also observed significant decreases in nasal turbinate E sgmRNA expression in vaccinated animals (Figure 2D). These data demonstrated that LNP-delivered DNA vaccines afford more effective protection than naked DNA administered at a much higher dose.
Figure 2.
LNP-formulated DNA vaccination greatly reduces Delta variant replication
(A) Syrian hamster body weight was measured for 6 days following intranasal challenge with 1.67 × 105 TCID50 of a SARS-CoV-2 B.1.617.2 isolate. (B) Plaque assays were used to determine the number of plaque-forming units (PFU) in the lung and nasal turbinate tissue collected 3 dpi. The number of SARS-CoV-2 E sgmRNA copies in lung (C) and nasal turbinate (D) tissues was determined via RT-qPCR 3 and 6 dpi and normalized per gram of tissue. Data shown are mean ± SEM; n = 4 for pVAX1, n = 6 for pVAX1 S-CD40LB.1.617.2 groups, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
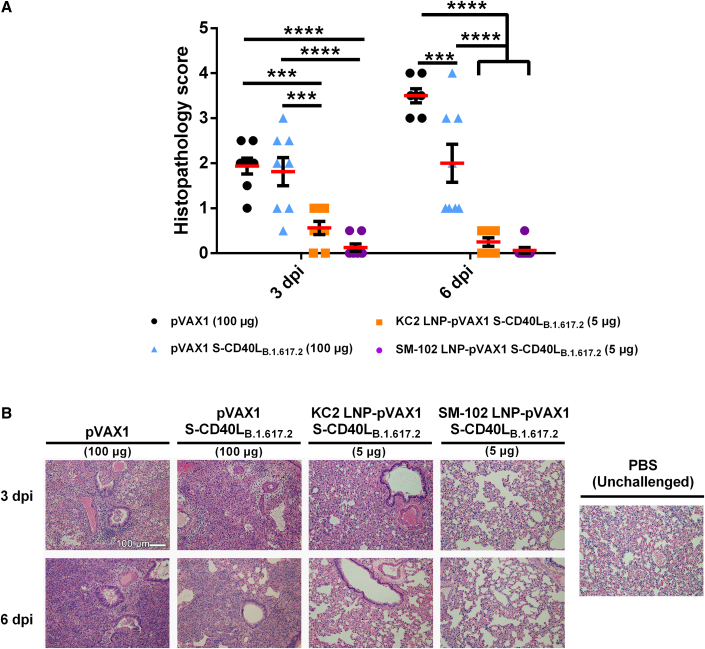
LNP encapsulation prevents lung pathology following homologous B.1.617.2 challenge
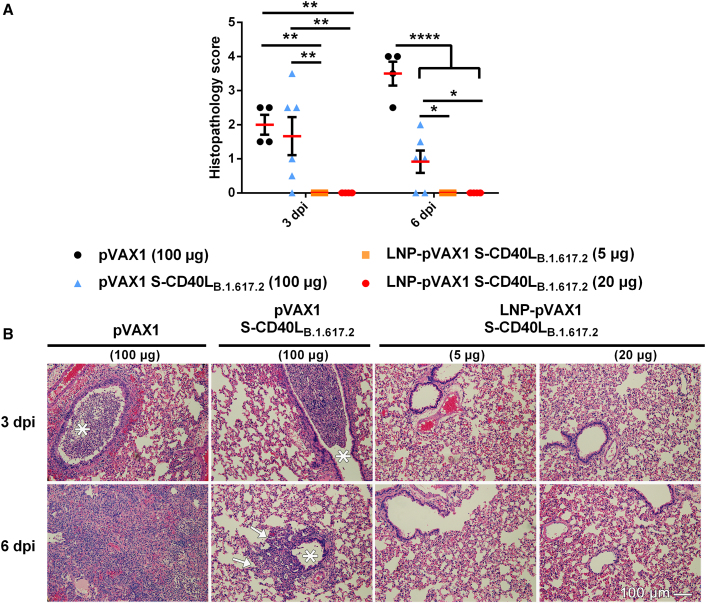
Next, lung samples collected from the challenged hamsters were analyzed for histopathological changes (Figure 3). At 3 dpi, hamsters immunized with the pVAX1 vector control or unencapsulated pVAX1 S-CD40LB.1.617.2 vaccine developed acute bronchitis, bronchiolitis, and interstitial pneumonia in their lungs. However, interstitial pneumonia in the area adjacent to the airways was generally milder in those immunized with unencapsulated pVAX1 S-CD40LB.1.617.2 than those immunized with the pVAX1 control vector. In contrast, hamsters immunized with either dose of LNP-formulated pVAX1 S-CD40LB.1.617.2 displayed no obvious airway or pulmonary inflammatory infiltrates or tissue damage. As anticipated, histopathological changes further progressed in the pVAX1 vector control group by 6 dpi, with the majority or entire lung tissues having become consolidated in most of the animals with very few aerated alveoli visible. At 6 dpi, the lungs of hamsters immunized with the LNP-formulated vaccines continued to display no overt histopathological changes while mild to moderate bronchopneumonia and interstitial pneumonia remained visible in some areas of lung tissues from hamsters vaccinated with unencapsulated pVAX1 S-CD40LB.1.617.2.
Figure 3.
LNP encapsulation prevents lung pathology following SARS-CoV-2 Delta variant challenge
Histopathological changes in the lungs of hamsters immunized with different vaccine formulations and euthanized at 3 and 6 days after being challenged intranasally with an isolate of SARS-CoV-2 B.1.617.2. (A) Summary of histopathological scores. Data shown are mean ± SEM; n = 4 for pVAX1, n = 6 for pVAX1 S-CD40LB.1.617.2 groups, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001. (B) Representative photomicrograph of H&E-stained lung tissue. ∗ Bronchial lumen. Peribronchiolar infiltration (arrows). Scale bar, 100 μm.
Vaccination resulted in reduced pulmonary expression of proinflammatory macromolecules
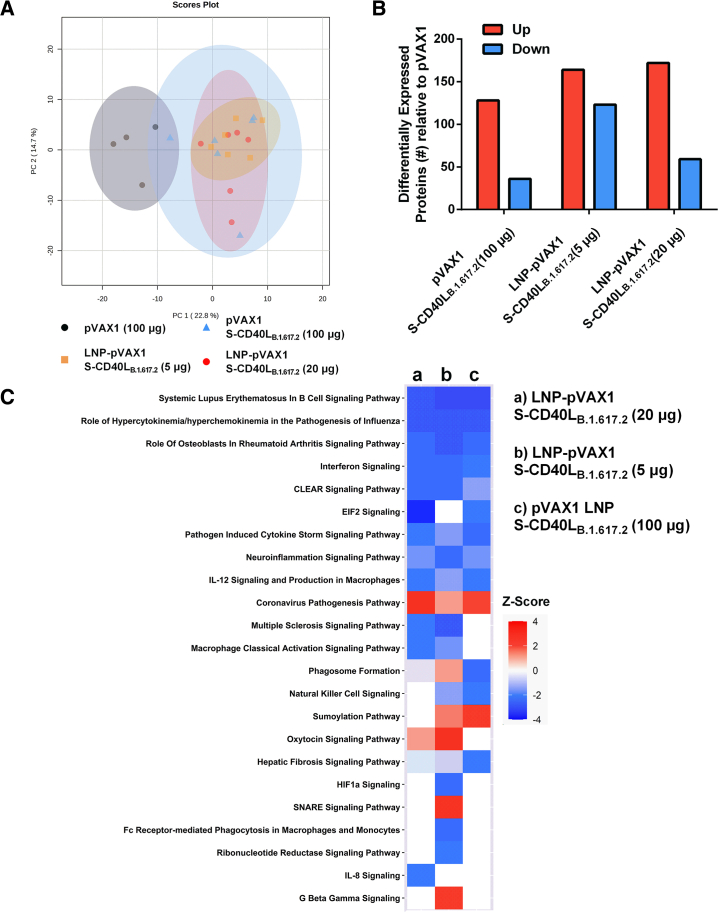
We next sought to characterize potential mechanisms underlying the prevention of lung pathology by conducting quantitative proteomic analysis of lung tissues 6 dpi, when the most severe histopathology was observed in control animals. In total, 3,651 protein groups were identified and quantified. Principal-component analysis (PCA) revealed a distinct separation between the protein expression profiles of the pVAX1 vector control-vaccinated animals and the three pVAX1 S-CD40LB.1.617.2-vaccinated groups (Figure 4A). The PCA showed no obvious overall proteome differences among the three vaccines, which was reflected by the absence of significantly differentially expressed protein (DEP) when comparing spike-vaccinated groups with one another (Table S3). In contrast, there were 164, 287, and 231 DEPs between the pVAX1 control group and the unencapsulated, 5-μg and 20-μg dose LNP pVAX1 S-CD40LB.1.617.2 groups, respectively (Figure 4B). There was considerable overlap in the three sets of DEPs (Figure S2). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of these DEPs revealed an enrichment of pathways related to immune responses and viral infection for all three comparisons (Figure S3; Table S4). The sets of DEPs from the LNP comparisons were additionally enriched with proteins related to other immune processes such as antigen processing, phagosomes, and cytosolic DNA sensing. Ingenuity pathway analysis (IPA) revealed downregulation of various immune-related response pathways including interferon signaling, EIF2 signaling interleukin (IL)-12 signaling, and macrophage responses in the pVAX1 S-CD40LB.1.617.2-vaccinated animal lungs (Figure 4C; Table S5). LNP-vaccinated lung tissues additionally had proteomic signatures linked to decreased macrophage activity, including the inhibition of classical activation signaling, Fc-receptor-mediated phagocytosis, and IL-8 signaling.
Figure 4.
Quantitative proteomic analysis of vaccinated hamster lung tissue post-challenge
(A) PCA score plot of the four experimental groups. The pVAX1 vector control group demonstrates a distinct pattern relative to pVAX1 S-CD40LB.1.617.2-vaccinated animals. (B) The number of differentially expressed proteins (DEPs) between the pVAX1 control and spike-vaccinated groups determined by pairwise comparison. Red indicates proteins with increased expression in spike-vaccinated groups, while blue shows proteins with decreased expression. (C) Heatmap of the predicted activation or inhibition of canonical signaling pathways based on DEPs in the three comparisons. Color refers to the Z score, where >2 indicates an activated pathway (red), while Z score < −2 means the pathway is inhibited (blue).
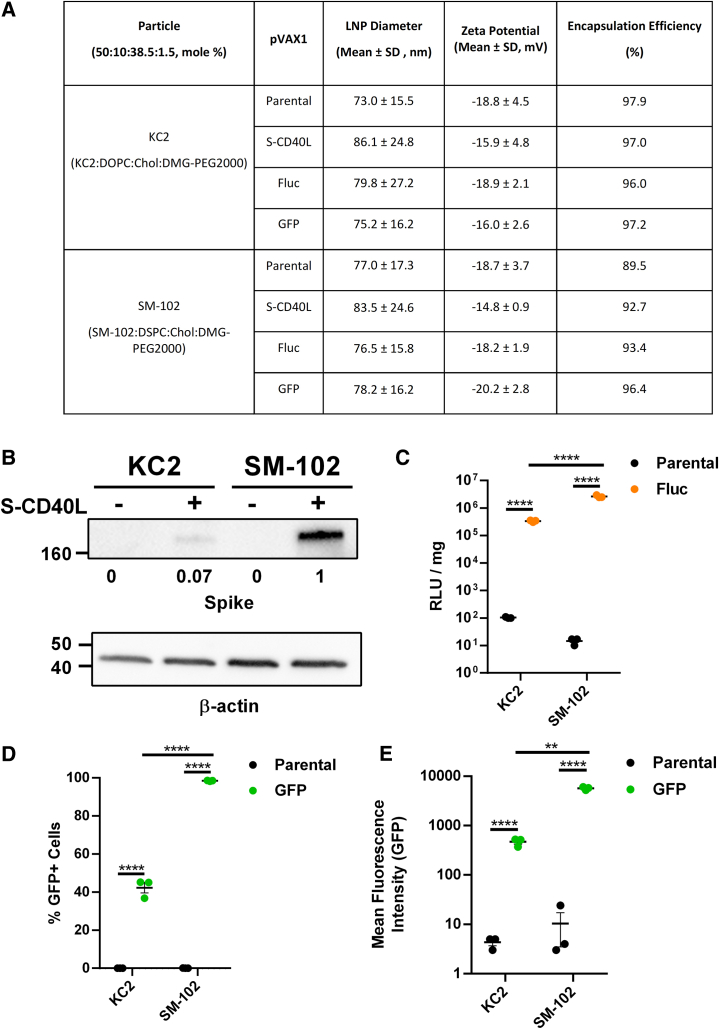
Characterization of DNA-LNP formulations in vitro
Having demonstrated the enhanced efficacy of DNA vaccination afforded by KC2-based LNPs, we next wanted to examine whether immunogenicity and protection would be altered when using LNPs formulated with SM-102, the ionizable lipid found in commercially approved Spikevax COVID-19 mRNA vaccines.7,27 To this end, the potency of these two DNA-LNP formulations were first evaluated and compared in vitro using HEK293T cells. Across tested plasmids, KC2 and SM-102 DNA-LNP formulations had similar size distributions and charge (Figure 5A). Relative to KC2, DNA-LNPs formulated using SM-102 induced approximately a 10-fold increase in expression of both S-CD40LB.1.617.2 (Figure 5B) and firefly luciferase (Fluc) (Figure 5C) 48 h after transfection, mirroring previous observations.28 Using flow cytometry to measure the proportion of green fluorescent protein-positive (GFP+) HEK293T cells, SM-102 formulated DNA-LNPs also showed a significantly greater transfection efficiency (Figure 5D). The intensity of GFP expression was also greater for SM-102 DNA-LNP-transfected cells (Figure 5E).
Figure 5.
In vitro characterization of DNA-LNPs
(A) Characterization of DNA-LNP formulations. Chol = cholesterol. HEK293T cells were transfected in triplicate with 500 ng of each formulation. (B) Western blotting was done to detect SARS-CoV-2 spike expression in HEK293T cells 48 h after transfection with pVAX1 S-CD40L or parental control. β-actin expression was used as a loading control and spike expression was normalized to SM-102 pVAX1 S-CD40L. (C) Relative light units (RLU) were measured 48 h after transfection with pVAX1 Fluc or parental control. RLU was standardized per mg of protein as quantified by Bicinchoninic Acid (BCA) assay. Quantification of the (D) percentage of GFP+ HEK293T cells and (E) mean fluorescence intensity (MFI) 48 h after transfection with pVAX1 GFP or parental control. Data shown are mean ± SEM; n = 3, ∗∗∗∗p < 0.0001.
LNP composition has minor effect on DNA vaccine immunogenicity
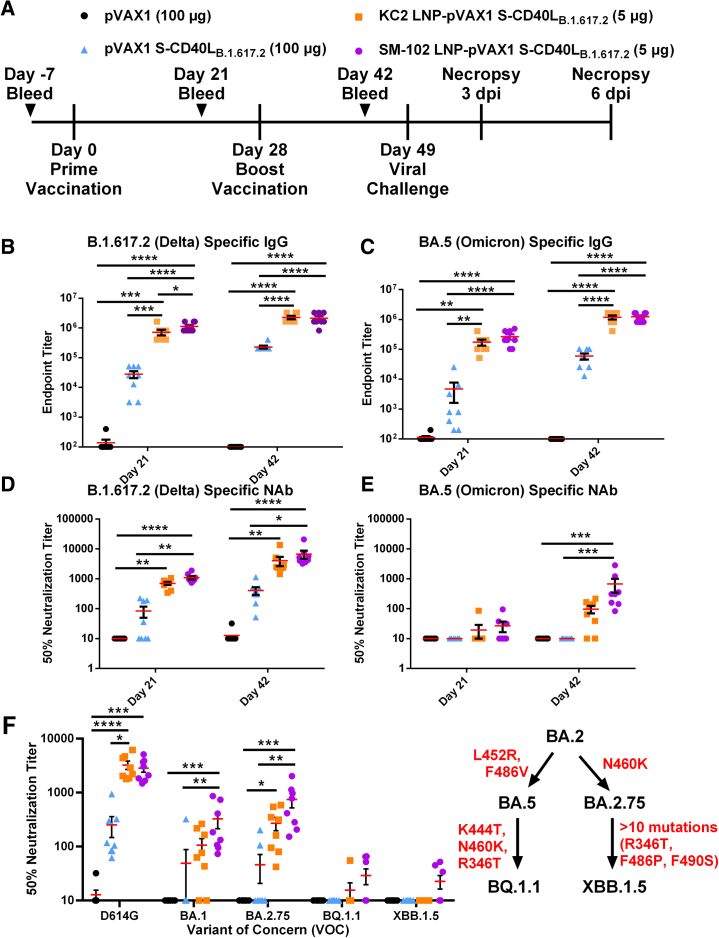
Syrian hamsters were vaccinated on days 0 and 28 with 5 μg of either KC2 or SM-102 LNP-encapsulated pVAX1 S-CD40LB.1.617.2 (Figure 6A). Control animals were again vaccinated with a higher dose of 100 μg of either unencapsulated pVAX1 S-CD40LB.1.617.2 or parental pVAX1. Nanoparticle tracking analysis (NTA) showed that both DNA-LNP formulations had similar mean size distributions and encapsulation efficiencies (Table S2). Compared with the naked DNA vaccine, both KC2 and SM-102 DNA-LNPs induced greater binding antibody responses against homologous B.1.617.2 (Figure 6B) and heterologous BA.5 (Figure 6C) S proteins. While the binding antibody responses induced by the two LNP formulations were quite similar, the SM-102 LNP formulation generated higher levels of B.1.617.2 S-specific immunoglobulin (Ig)G on day 21.In terms of neutralizing activity, both DNA-LNP vaccines induced a significant response against homologous B.1.617.2 (Figure 6D). Moreover, only the SM-102 DNA-LNPs induced a significant response relative to naked DNA (Figure 6D). Importantly, when assayed against heterologous BA.5, only the SM-102 formulation induced a significant NAb response despite the similar IgG levels (Figure 6E).
Figure 6.
DNA-LNPs formulated with SM-102 induce superior humoral responses
(A) Male Syrian hamsters were immunized intramuscularly on days 0 and 28 with 100 μg of pVAX1, 100 μg of pVAX1 S-CD40LB.1.617.2, or 5 μg of KC2 or SM-102 LNP-encapsulated pVAX1 S-CD40LB.1.617.2. Animals were challenged intranasally with a SARS-CoV-2 BA.5 isolate on day 49 and euthanized 3 and 6 days post-infection (dpi). ELISA determination of the total B.1.617.2 (B) and BA.5 (C) spike-specific IgG in the sera of immunized hamsters 21 and 42 days post-vaccination. The 50% neutralizing titer (NT50) of immunized hamster sera on days 21 and 42 was determined using (D) B.1.617.2 and (E) BA.5 pseudotyped-VSV. (F) The NT50 on day 42 was also determined using D614G, BA.1, BA.2.75, BQ.1.1, and XBB.1.5 SARS-CoV-2 spike pseudotyped-VSV. Red indicates mutations acquired in the SARS-CoV-2 spike protein. Data shown are mean ± SEM; n = 8, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
We next tested NAb responses at day 42 against a panel of select SARS-CoV-2 variants (Figure 6F). Both DNA-LNP formulations induced robust NAb responses against the ancestral D614G variant, comparable to their response against homologous B.1.617.2. However, only the SM-102 DNA-LNPs induced a significant response against both the Omicron BA.1 and BA.2.75 variants, while also inducing slightly higher, although not statistically significant, responses against BQ.1.1 and XBB.1.5.
LNP encapsulation enables DNA vaccine protection against heterologous Omicron challenge
Having characterized the humoral response induced by the two DNA-LNP formulations, we next determined the protection against heterologous Omicron challenge. Toward this, the vaccinated hamsters were challenged intranasally on day 49 with the SARS-CoV-2 BA.5 variant. pVAX1 control vector-vaccinated hamsters experienced mild weight loss, with a maximum mean weight reduction of 4.2% at 3 dpi (Figure 7A). Over the course of the infection, the naked DNA vaccine failed to prevent weight loss (Figure 7A). However, animals vaccinated with either the KC2 or SM-102 LNP-encapsulated vaccines began recovering weight at significant levels starting from 3 dpi. All three pVAX1 S-CD40LB.1.617.2 vaccines significantly reduced viral burden in both the lung and nasal turbinates 3 dpi (Figure 7B), although to a greater extent with the LNP-encapsulated vaccines. Notably, both DNA-LNP formulations significantly reduced E sgmRNA expression in the lungs relative to unencapsulated pVAX1 S-CD40LB.1.617.2 (Figure 7C). In the upper respiratory tract at 3 dpi, only the DNA-LNP vaccines significantly reduced E sgmRNA levels relative to the empty vector control (Figure 7D).
Figure 7.
DNA-LNP encapsulation improves protection against the BA.5 Omicron variant
Vaccinated Syrian hamsters were challenged intranasally with 1.67 × 105 median tissue culture infectious dose (TCID50) of a SARS-CoV-2 BA.5 isolate. (A) Syrian hamster body weight was measured for 6 days following challenge. (B) TCID50 was determined in the lung and nasal turbinate tissue collected 3 dpi. The number of SARS-CoV-2 E sgmRNA copies in lung (C) and nasal turbinate (D) tissues was determined via RT-qPCR 3 and 6 dpi and normalized per gram of tissue. Data shown are mean ± SEM; n = 8, ∗p < 0.05, ∗∗p 7 < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Last, we again conducted histopathological analysis of lung tissue sections collected 3 and 6 dpi (Figure 8). Consistent with the weight loss and viral burden, both DNA-LNP formulations reduced lung pathology relative to both the naked vaccine and control vector on both days 3 and 6 (Figure 8A). Notably, the lowest histopathology scores were observed following SM-102 vaccination, with there being no overt inflammatory responses or tissue damage observable even at 3 dpi (Figure 8B). Collectively, these data indicate that both DNA-LNP formulations afford effective protection against heterologous challenge, with SM-2 formulation performing slightly better than KC2, as demonstrated by level of neutralizing antibodies against mismatched Omicron strains and protection from BA.5 challenge.
Figure 8.
LNP encapsulation greatly reduces lung pathology following Omicron BA.5 challenge
Histopathological changes in the lungs of hamsters immunized with different vaccine formulations and euthanized at 3 and 6 days after being challenged intranasally with a SARS-CoV-2 BA.5 isolate. (A) Summary of histopathological scores. Data shown are mean ± SEM; n = 8, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001. (B) Representative photomicrograph of H&E-stained lung tissue. Scale bar, 100 μm.
Discussion
This current study centered on the LNP-mediated delivery of a DNA vaccine encoding SARS-CoV-2 Delta spike fused with the ectodomain of CD40L. Our approach differs from previous studies in several aspects. First, the Delta spike was chosen as the vaccine antigen. While the B.1617.2 variant was one of the most virulent,29,30 it lacks most of the spike mutations found within the Omicron lineage. As such, this vaccine would give us an opportunity to investigate the relationship between humoral responses and in vivo protection against significantly mismatched viral challenge. Second, we were able to directly compare the immunogenicity and efficacy of two distinct LNP formulations for DNA vaccine delivery, an approach that remains severely understudied relative to mRNA-LNP vaccination. Last, through the addition of CD40L, we were able to investigate the combination of a molecular adjuvant with LNP-mediated vaccine delivery, an avenue of exploration with boundless therapeutic potential for both DNA and mRNA vaccines.
The B.1.617.2 S protein does not contain many notable mutations such as K417N, N501Y, G446S, E484A, and G496S, which contribute to the Omicron lineage’s resistance to neutralizing antibodies.31,32,33,34 However, in our studies, encapsulation of the B.1.617.2 pDNA vaccine within KC2- or SM-102-based LNPs led to the induction of a significant NAb response against BA.1, BA.2, BA.2.75, and BA.5, albeit to a lower extent than against B.1.617.2. It is also of note that low but detectable NAb activities were observed against BF.7, BQ.1.1, and XBB.1.5, variants in which R346T, N640K, K444T, and F486P mutations are likely to further enhance neutralization resistance.35,36 These findings are largely in agreement with previous reports with respect to cross-neutralizing activities of human antisera.35,37
To compare the two different LNP formations, we chose to assess the level of protection afforded by our B.1.617.2-based vaccine against challenge with the Omicron BA.5 variant to which considerable but reduced NAb activities, relative to homologous B.1.617.2, were induced by both the KC-2 and SM-102 DNA-LNPs. Despite the reduced NAb response, immunization with either of the two DNA-LNP formulations led to excellent control of the heterologous BA.5 infection, comparable to that following homologous B.1.617.2 challenge, suggesting the reduced level of NAb induced by the DNA-LNP vaccines may have remained sufficient to neutralize viruses in vivo. Nonetheless, the future use of variants such as XBB.1.5 as a challenge strain, to which the B.1.617.2 vaccines failed to elicit a significant NAb response, could further delineate the functional roles of neutralizing vs. non-neutralizing antibodies, along with the importance of cell-mediated immune responses.
Immune escape by emerging variants has and will likely continue to be a recurring problem for SARS-CoV-2 vaccines.38,39 Research continues to develop methods of inducing broadly neutralizing and variant-proof immune responses.40 One factor affecting the breadth of humoral immunity following vaccination is germinal center (GC) formation and subsequent B cell somatic hypermutation.41,42,43 While previous studies have shown CD40-targeting vaccine adjuvants to increase T follicular helper cell counts and promotes GC formation,44,45 our unencapsulated CD40L-adjuvanted pDNA vaccine was unable to overcome the hurdle of neutralizing heterologous Omicron variants. It has been shown in recent studies that SARS-CoV-2 mRNA-LNP vaccines drive strong GC responses in lymph nodes following vaccination.16,46,47 The enhanced breadth of neutralization observed in our LNP-formulated pDNA vaccines could potentially be attributed to enhanced vaccine immunogenicity, subsequently driving superior GC formation and a broader humoral response.
Finally, our proteomic analysis of lung tissues collected 6 dpi demonstrated a general decrease in innate immune and infection-related response pathways in pVAX1 S-CD40LB.1.617.2-vaccinated groups relative to the pVAX1 control-vaccinated animals (Figure 5). Although the three vaccinated groups had similar proteomic profiles, DNA-LNP-vaccinated groups had additional DEP signatures linked to the inhibition of macrophage responses. Other transcriptomics and proteomics studies have correlated upregulated monocyte, macrophage, and neutrophil signatures with disease severity during SARS-CoV-2 infection.48,49,50,51,52 Stronger vaccine-induced adaptive immune responses should provide better protection during the early stages of viral infection, reducing the subsequent induction of innate immune responses. Clearly, effective vaccination can attenuate the upregulation of these innate immune and inflammatory response pathways following SARS-CoV-2 infection.52,53,54 Altogether, these findings support the histopathological absence of pulmonary inflammatory infiltrates observed in DNA-LNP-vaccinated lung tissues upon B.1.617.2 challenge.
Research demonstrating the potential therapeutic benefits of encapsulating pDNA vaccines within ionizable lipid nanoparticles is beginning to accumulate.13,55,56,57 During the preparation of this manuscript, Liao et al. reported results of a DNA-LNP vaccine encoding ancestral or BA.1 spike that mirror our own.58 Interestingly, they also found that DNA-LNPs provided superior antibody responses and protection than mRNA-LNPs. Their results appear to be at odds with our recently published study comparing mRNA- and DNA-LNPs, where we reported that mRNA-LNPs induce superior antigen expression and antibody responses.28 While this discrepancy remains to be understood, antigen selection, construct design, and nanoparticle fabrication could be among other unknown contributing factors. These questions, along with the rates of possible ribosomal skipping in DNA vaccines, await further investigation.
Materials and methods
Cell lines and viruses
HEK293T, HEK293T-ACE2, and HEK-Blue CD40L cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Thermo Fisher) supplemented with 25 mM HEPES, 20 U/mL penicillin, 0.02 mg/mL streptomycin, and 10% heat-inactivated fetal bovine serum (FBS). HEK-Blue CD40L medium was additionally supplemented with 100 μg/mL of Normocin. Vero-TMPRSS2 cells were cultured in DMEM with L-glutamine supplemented with 1X non-essential amino acid, 1 mM sodium pyruvate, and 5% fetal bovine serum (FBS). SARS-CoV-2 viruses were obtained from BEI Resources. B.1.617.2 hCoV-19/USA/MD-HP05647/2021 (NR-55672) and BA.5 hCoV-19/South Africa/CERI-KRISP-K040013/2022 (NR-56798) were propagated and titered using Vero-TMPRSS2 cells and sequenced to confirm genetic fidelity. Passage four virus stocks were used in all subsequent experiments that required live virus.
Immunization
Animal experiments and procedures were approved by the National Research Council Canada (NRC) Human Health Therapeutics Animal Care Committee and performed by trained staff in accordance with regulations and guidelines set by the Canadian Council on Animal Care. All infectious work was carried out under ABSL-3 conditions at the NRC. Six- to 8-week-old male Syrian hamsters were purchased from Charles River Laboratories (Senneville, QC). Animals were randomly allocated into four experimental groups, being immunized with either 100 μg of pVAX1, 100 μg of pVAX1 S-CD40LB.1.617.2, and 5 or 20 μg of LNP-formulated pVAX1 S-CD40LB.1.617.2. Vaccines were suspended in 100 μL of PBS and administered intramuscularly in the hamster’s left tibialis anterior muscle with a needle syringe on day 0 and day 28. Serum was collected on days 21 and 42 post-vaccination. On day 49 post-vaccination the hamsters were intranasally challenged with 1.67 ×105 TCID50 of B.1.617.2 or BA.5. Challenge doses were found to induce significant illness in earlier dose-optimization experiments (data not shown). Animals were euthanized by CO2 either 3 or 6 days post-challenge, after which the nasal turbinate and lung tissues were collected for downstream experiments.
DNA vaccine design and synthesis
The DNA vaccine was designed and prepared as previously described with minor modifications.22 A DNA sequence encoding the SARS-CoV-2 B.1.617.2 spike ectodomain (T19R, G142D, E156G, Del 157/158, L452R, T478K, D614G, P681R, D950N) fused via “GSGG” glycine-serine linkers to a T4 fibritin foldon trimerization motif and the ectodomain of Mesocricetus auratus CD40L (GenBank: XM_005084522.4, residues 118–260) was commercially synthesized (Genscript). The spike sequence was mutated to contain a “GSAS” substitution at the furin cleavage site (residues 682–685) and pre-fusion stabilizing proline substitutions at residues 986 and 987.59 The vaccine was codon optimized for expression in Syrian hamsters and subcloned into the mammalian expression plasmid pVAX1 (Thermo Fisher) using KpnI and XhoI restriction enzymes. Bulk DNA vaccine preparations were prepared with plasmid gigaprep kits (Qiagen) and sequence validated with Sanger sequencing.
LNP generation
LNPs were synthesized as previously described within 3 days of vaccination and stored at 4°C.28,55 Briefly, pDNA-LNPs were prepared via the microfluidic mixing of an aqueous and organic phase. The aqueous phase was prepared by suspending pDNA in 25 mM acetate buffer (pH 4.0). The organic phase was prepared in ethanol and consisted of 2,2-dilinoleyl-4-dimethylaminoethyl-[1,3]-dioxolane (DLin-KC2-DMA, MedKoo Biosciences) or heptadecan-9-yl-8-((2-hydroxyethyl) (6-oxo-6-(undecyloxy)hexyl)amino)octanoate (SM-102, MedKoo Biosciences), 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC, Avanti Polar Lipids) or 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC, Avanti Polar Lipids), ovine cholesterol (Avanti Polar Lipids), and 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (DMG-PEG2000, Avanti Polar Lipids) at a ratio of 50:10:38.5:1.5 mol %, respectively. The two phases were mixed with a polymer amine (N = nitrogen) group to nucleic acid phosphate (P) group (N/P) ratio of 6:1 using either a NanoAssemblr BT instrument (Precision Nanosystems, Vancouver, BC) with a microfluidics cartridge containing a staggered herringbone mixing unit or a NanoAssemblr Ignite instrument with NxGen cartridges equipped with toroidal structures. LNPs were dialyzed against a 1,000-fold volume of phosphate-buffered saline (PBS) (pH 7.4) for 18 h at 4°C in a 10k MWCO cassette (Thermo Fisher), passed through a 0.22-μm filter and then concentrated using an Amicon Ultra 4 10 k MWCO centrifugal concentrator (Millipore Sigma). After concentration, the nanoparticle size was measured via nanoparticle tracking analysis (NTA) under static conditions (NanoSight, Malvern Panalytical, Westborough, MA, USA). Five separate tracking videos (1 min each) were taken consecutively and merged together to generate the NTA sizing data. Zeta potential was measured in 5 mM phosphate buffer (pH 7.4) using disposable capillary cells on a Zetasizer Ultra instrument (Malvern Panalytical, Westborough, MA, USA). pDNA-LNPs were stored at 4°C and administered within 48 h.
LNP encapsulation efficiency
Nucleic acid encapsulation efficiency and vaccine dose were determined as previously described.55 Briefly, LNPs were either untreated or disrupted with 1% Triton X-100 (Millipore Sigma) in a 96-well plate before the addition of SYBR Gold dye (Thermo Fisher). Fluorescence (Ex/Em: 495/537 nm) was measured in each sample using a Synergy MX plate reader (BioTek). Total pDNA concentration in each sample was determined by comparing the fluorescence relative to that of a standard curve of pDNA prepared in the same buffer. Untreated and disrupted samples were used to determine unencapsulated and total pDNA, respectively. The amount of encapsulated pDNA, calculated by subtracting the amount of unencapsulated pDNA from the total amount, was used for dose calculations. Last, the encapsulation efficiency was determined from the amount of encapsulated pDNA relative to the total amount (Tables S1 and S2).
In vitro transfection
In a 24-well plate, HEK293T cells were transfected in triplicate with 500 ng of DNA-LNPs, suspended in PBS (Thermo Fisher). The DNA-transfected cells were then incubated at 37°C, 5% CO2 for 48 h prior to quantification of S-CD40L, firefly luciferase (Fluc), or green fluorescent protein expression (GFP).
Western blotting
For determination of S-CD40L expression, transfected cells were washed with PBS and then lysed with radioimmunoprecipitation assay buffer (Thermo Fisher). Lysates were electrophoresed on a 4%–15% TGX stain-free SDS-PAGE gel (Bio-Rad) and then transferred to a polyvinylidene difluoride membrane. Membranes were blocked for 1 h at room temperature with tris-buffered saline (TBS) containing 0.5% Tween 20 (Sigma-Aldrich) (v/v) (TBS-T) and 5% (w/v) non-fat milk powder, then incubated overnight at 4°C in blocking buffer containing either polyclonal rabbit anti-SARS-CoV-2 spike antibody (1:3,000 dilution) (40591-T62, Sino Biological) or polyclonal rabbit anti-β-actin antibody (1:1,000 dilution) (#4967, Cell Signaling Technology). Membranes were then incubated for 1 h at room temperature with goat anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibody (1:75,000 dilution) (Thermo Fisher) in blocking buffer and developed using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher) and a ChemiDoc MP imaging system (Bio-Rad) (Figures S4 and S5).
Quantification of luciferase
Luciferase expression in cells transfected with pVAX1 Fluc DNA-LNPs was quantified as previously described.28 Briefly, media were aspirated and replaced with 200 μL of passive lysis buffer (Promega). Cells were then incubated for 30 min at room temperature on an orbital shaker at 50 rpm. Lysates were clarified via centrifugation at 15,000 × g for 5 min at room temperature and then added in triplicate, 100 μL per well, to a white Costar 96-well plate (Corning) followed by 100 μL of room temperature Bright-Glo Reagent (Promega). Luminescence was read within 5 min and expressed as relative light units/mg of protein (RLU/mg) after being normalized to total protein content as measured by a Bicinchoninic Acid (BCA) Kit (Millipore Sigma).
Flow cytometric analysis of GFP-positive cells
HEK293T cells transfected with pVAX1 GFP DNA-LNPs were analyzed by flow cytometry (BD FACSymphony A1). Briefly, cells were lifted with PBS supplemented with 0.5 mM EDTA (Thermo Fisher) for 10 min at 37°C. Data were acquired using the FACS DIVA software (version 9.0.2). GFP signal was measured off the 488-nm blue laser, using 505-nm long-pass and 530/30-nm band-pass filters. GFP signal was used to analyze the % GFP+ HEK293T cells and measure mean fluorescence intensity (MFI) (Figure S6).
CD40 ligand bioactivity assay
CD40L activity was assessed as previously described.22 Briefly, HEK293T cells were transfected in a 24-well plate with 1 μg of pVAX1 or pVAX1 S-CD40LB.1.617.2 using lipofectamine 3000 (Thermo Fisher). After a 24-h incubation at 37°C and 5% CO2, 100 μL of 0.45-μm filtered supernatant was added to a 96-well plate and mixed with 100 μL of HEK-Blue CD40L cells (InvivoGen) resuspended at 2.0 × 105 cells/mL. After 24 h, 20 μL of cell culture supernatant was mixed with 180 μL of QUANTI-Blue Reagent (InvivoGen) in a 96-well plate. The absorbance at 630 nm was measured after a 30-min incubation at 37°C using a Synergy 2 microplate reader (BioTek).
ELISA
Spike protein ectodomains were obtained from the National Research Council of Canada for the following strains: B.1.617.2 (PRO7604-10 [SmT1v3 (B.1.617.2)]), BA.1 (PRO7911-2 [SmT1v3-B.1.1.529]), and BA.5 (PRO8213 [SmT1v3 (BA.5)]), which were produced using previously described methods.60,61 ELISAs were conducted as described previously.22 Briefly, Nunc MaxiSorp flat-bottom 96-well plates (Thermo Fisher) were coated with 1 μg/mL of antigen diluted in PBS and incubated overnight at 4°C. Plates were washed with PBS containing 0.1% Tween 20 (PBS-T) before blocking with 3% (w/v) bovine serum albumin (IgG-free, protease-free) (Jackson Immuno Research) in PBS-T for 2 h at 37°C. Plates were washed again and 2-fold serial dilutions of hamster serum were prepared. After a 1-h incubation at 37°C the plates were washed again and Peroxidase AffiniPure Goat Anti-Syrian Hamster IgG (H + L) (Jackson Immuno Research) was added to each well at 1:4,000 and incubated at 37°C for 1 h. After a final wash, 100 μL of tetramethylbenzidine (TMB) substrate (Cell Signaling Technology) was added to each well. After a 2-min incubation at room temperature, 100 μL of 0.16-M sulfuric acid was added to terminate the reaction and absorbance was measured at 450 nm. Endpoint titers were expressed as the reciprocals of the final detectable dilution with an optical density above the cutoff value, which was defined as the average OD of the empty vector samples plus three standard deviations.
Pseudovirus production and neutralization
The neutralizing activity of hamster sera was determined by using a VSV-based SARS-CoV-2 S pseudovirus reporter assay as described previously.22,62 Briefly, pseudotyped-VSV was generated by concurrently infecting HEK293T cells with G∗ΔG-VSV (Kerafast) and transfecting them with pLV vectors encoding SARS-CoV-2 S Δ19. Vectors encoding D614G, B.1.617.2, BA.1, and BA.5 spike were purchased commercially (Invivogen) while vectors encoding BA.2.75, BF.7, BQ.1.1, and XBB.1.5 were synthesized commercially (GenScript, Brockville, ON, Canada). Supernatant containing the pseudovirus was collected 48 and 72 h post-infection and passed through a 0.45-μm filter. In a 96-well plate, serum samples heat inactivated at 56°C for 30 min were serially diluted 3-fold and mixed with 1.3 × 104 TCID50 of pseudovirus. After a 1-h incubation at 37°C, 5% CO2, 2 × 104 HEK293T-ACE2 cells were added to each well. After a 24-h incubation at 37°C, 5% CO2, luminescence was measured using Bright-Glo luciferase reagent (Promega) and a Synergy 2 microplate reader (BioTek). The 50% neutralization titers (NT50) were measured as the reciprocal of the sample dilution at which a 50% reduction in relative light units (RLU) was observed relative to the average of the no-serum control wells.
Quantification of viral burden
Lung and nasal turbinate tissues were homogenized in PBS using a Precellys Evolution. Spin-clarified supernatants of the homogenates were then used for viral quantification. B.1.617.2 viral burden was determined by plaque assay as previously described.22 Briefly, a 1:10 serial dilution of clarified supernatant was prepared in infection media (DMEM supplemented with 1X non-essential amino acid, 20 U/mL penicillin, 0.02 mg/mL streptomycin, 1 mM sodium pyruvate, and 0.1% bovine serum albumin). Virus was adsorbed on Vero E6 cells at 37°C, 5% CO2 for 1 h before the inoculum was replaced with overlay media (1X infection media with 0.6% ultrapure, low-melting point agarose). Cells were incubated at 37°C, 5% CO2 for 72 h, then fixed with 10% formaldehyde and stained with crystal violet. Plaques were enumerated and PFU was determined per gram of tissue. BA.5 viral burden was determined by TCID50 assay on Vero-TMPRSS2 cells. Spin-clarified supernatants were serially diluted 1:10 in infection media (DMEM supplemented 1X non-essential amino acid, 1 mM sodium pyruvate, and 1% FBS). Dilutions were adsorbed on Vero-TMPRSS2 cells seeded in 96-well plates for 1 h at 37°C, 5% CO2. After adsorption, inoculum was removed and 100 mL of infection media was added to each well and incubated at 37°C, 5% CO2 for 5 days. Observed cytopathic effect indicates presence of infectious virus was recorded and TCID50/g of lung tissue was calculated using the Reed-Muench method.63
RNA extraction and quantitative reverse-transcription PCR (qRT-PCR)
Quantification of SARS-CoV-2 E subgenomic mRNA (sgmRNA) was completed as described previously.22 Briefly, lung and nasal turbinate tissues were placed into RNA shield buffer (Zymo Research) and incubated overnight at 4°C before freezing at −80°C. Viral RNA was extracted from the mechanically homogenized samples using a Quick-RNA Viral Kit (Zymo Research). Viral RNA expression was quantified using a one-step Fast Virus master mix (ThermoFisher) and E sgmRNA-specific primer/probe set.64 sgmRNA copy numbers were calculated by comparing sample Ct values to a standard curve of in vitro transcribed E sgmRNA prepared using a TranscriptAid T7 High Yield Transcription Kit (Thermo Fisher) and normalized by tissue weight. All RT-qPCR reactions were conducted in MicroAmp Fast Optical 96 wells with an Applied Biosystems 7500 Fast Real-time PCR instrument.
Histopathology
Histopathology analysis was conducted as described previously.22 Briefly, right lung lobes were fixed for 72 h in 10% neutral buffered formalin and processed by standard paraffin embedding methods.65 Four-micrometer-thick sections were stained with hematoxylin-eosin (H&E) and examined under microscopy. The severity and extent of pneumonia was scored blinded based on previously established criteria.66
Mass spectrometry sample preparation and LC-MS/MS acquisition
Lung tissue was mechanically homogenized in 4% SDS supplemented with Halt Protease Inhibitor Cocktail (Thermo Fisher) to inactivate SARS-CoV-2 virus and extract proteins. Extracted proteins were then purified using acetone precipitation, reduced using Pierce Premium-Grade TCEP HCl (Thermo Fisher), and alkylated using Iodoacetamide (Sigma-Aldrich). Proteins were digested with trypsin and labeled with isobaric tags (TMT 11plex, Thermo Fisher) using a dry TMT-based labeling approach as described previously.67 Briefly, samples were randomly assigned to tags that were pre-aliquoted dried in 12-tube strips. A pooled sample was included in each strip for normalization between strips. After quenching, the labeled peptides from each strip were combined and fractionated (Pierce High pH Reverse-Phase Peptide Fractionation Kit, Thermo Fisher) into eight fractions. Collected peptide samples were analyzed with an Orbitrap Fusion Lumos Tribrid Mass Spectrometer coupled to an Easy-nLC 1200 (Thermo Fisher). For each sample 2 μL (corresponding to approximately 500 ng of peptide) were analyzed by loading onto a NanoViper Acclaim pepmap 100-trap column (75-μm 20 mm with 3-μm beads) and desalting with 0.1% formic acid in water (solvent A) before separating on an Easyspray pepmap C18 reverse-phase analytical column (50-μm 150 mm with 2-μm beads). Chromatographic separation was achieved at a flow rate of 0.300 μL/min over 100 min in five linear steps as follows (solvent B was 0.1% formic acid in 80% acetonitrile): initial, 2% B; 80 min, 25% B; 90 min, 40% B; 95 min, 95% B; 100 min, 95% B. The eluting peptides were analyzed in data-dependent mode MSMS. An MS survey scan of 400–1600 m/z was performed in the Orbitrap at a resolution of 120,000, 50-ms maximum injection time and an AGC target of 4 × 105. The top speed mode was used to select ions for MS2 analysis with dynamic exclusion 20 s with a ±10 ppm window. During the MS2 analyses, precursors were isolated using a width of 0.7 m/z and fragmented by HCD at 42% collision energy, AGC target 1.25 × 105 followed by Orbitrap analysis at 50,000 resolutions.
Bioinformatics and statistical analysis pathway analysis
Raw MS data were subjected to protein identification and quantification using MaxQuant 1.6.3.3 against a Mesocricetus auratus (golden hamster) database that was downloaded from UniprotKB (https://www.uniprot.org/, downloaded 2021-08-31, 89281 entries). MaxQuant search was performed with reporter ion MS2 (TMT11) mode with the following parameters: fixed modification was set as carbamidomethyl cysteine, variable modifications set as oxidized methionine, protein N-term acetylation; enzyme was set as trypsin/P with maximum missing cleavage site of 2; minimal peptide length of 7; and an FDR (false discovery rate) of peptide-spectrum match (PSM) of 0.01. Isotopic impurities of each TMT channel were obtained according to the reagent lot and was used during database search to correct the purities of TMT channels for each set of TMT labeling data. MaxQuant output was then summarized and normalized using MSstatsTMT,68 using the open-source R software available on Bioconductor. Proteins in all groups were compared pairwise using a pairwise t test. Proteins with an absolute fold change >1.5 and an adjusted p value <0.05 were defined as differentially expressed proteins (DEPs). Principal-component analysis (PCA) was performed with Metaboanalyst (https://www.metaboanalyst.ca/) using K-Nearest Neighbors Algorithm to estimate missing values and all other parameters set to default. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed on the DAVID Bioinformatics database (https://david.ncifcrf.gov/) with all parameter settings at their default values. DEPs were imported into ingenuity pathway analysis (IPA) (QIAGEN) and used to predict the activation or inhibition of canonical pathways. An enrichment score of 1.3 was considered significant. The overall predicted activation state of the biological attributes was assigned a Z score (<0: inhibition, >0: activation) and those that gave a Z score >2 or < −2 were considered significant. The results of the KEGG and IPA analysis were plotted as heat maps using RStudio.
Quantification and statistical analysis
Statistical analyses were performed using GraphPad Prism 9. An unpaired two-tailed t test was used for comparisons of CD40L bioactivity. If possible, before pairwise comparison, normality of the data was assessed by a Shapiro-Wilk test (alpha-level = 0.05). Whenever data or their log transformations were deemed not of normal distribution, a non-parametric approach was adopted. A non-parametric Kruskal-Wallis H test with Dunn’s test for multiple comparisons was applied for pairwise (between-group) comparisons of spike-specific IgG endpoint titers, neutralizing antibody titers, and plaque assay viral burden. A one-way analysis of variance (ANOVA) with Bonferroni’s adjustment was applied for pairwise (between-group) comparisons of weight loss data by day, log subgenomic mRNA expression, RLU/mg, % GFP+ cells, GFP MFI, and lung histopathology. The significance level and n number for each test is indicated in the figure legends.
Data and code availability
All proteomics data along with the database search results were deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org) via the PRIDE partner repository with the identifier PXD: 049053. All additional data needed to evaluate the conclusions in the paper are present in the main text and supplementary materials or available from the corresponding author upon request.
Acknowledgments
We gratefully acknowledge the histology and staining services provided by the Louise Pelletier HCF at the University of Ottawa. We gratefully acknowledge the technical contribution of many members of the Mammalian Cell Expression Section of the NRC-HHT for spike antigens production. We also gratefully acknowledge Dr. Lu Huixin and Dr. Roger Tam for commenting on the manuscript and Greg Harris for preparing the photomicrographs. This work is supported by the Government of Canada (intramural funding from Health Canada) and in part by the Pandemic Response Challenge Program of the National Research Council Canada.
Author contributions
Conceptualization, L.T., A.T., A.M.H., D.S., J.C., L.W., S.S., M.R.-M., M.J.W.J., and X.L.; Methodology, L.T., A.T., C.L., E.E.F.F., M.C., A.S., M.S., X.Z.; Formal Analysis and Data Curation, L.T., E.E.F.F., M.C., A.S., X.Z., and J.B.; Investigation, L.T., D.D., A.T., C.L., G.F, J.W., M.C., S.R., E.L., W.Z., A.P., C.G., W.C., and M.S.; Writing – Original Draft, L.T., W.C., and X.L.; Writing - Review & Editing, L.T., A.T., C.L., G.F., S.R., W.Z., A.P., C.G., A.M.H., L.W., S.S., M.R.-M., M.J.W.J., J.B., and X.L.; Resources, A.T., C.L., C.G., M.C., and Y.D. Funding Acquisition, Project administration and Supervision, A.T., Y.D., D.S., J.C., L.W., S.S., M.R.-M., M.J.W.J., and X.L.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2024.101325.
Contributor Information
Anh Tran, Email: anh.tran@nrc-cnrc.gc.ca.
Michael J.W. Johnston, Email: michael.johnston@hc-sc.gc.ca.
Xuguang Li, Email: sean.li@hc-sc.gc.ca.
Supplemental information
Protein expression in Syrian hamster lung tissue samples was compared pairwise using a pairwise t test. Proteins with an absolute fold change >1.5 and an adjusted p value <0.05 were defined as differentially expressed proteins (DEPs).
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed on the DAVID Bioinformatics database with all parameter settings at their default values.
DEPs identified through pairwise t tests were imported into ingenuity pathway analysis (QIAGEN) and used to predict the activation or inhibition of canonical pathways.
References
- 1.Moghadas S.M., Vilches T.N., Zhang K., Wells C.R., Shoukat A., Singer B.H., Meyers L.A., Neuzil K.M., Langley J.M., Fitzpatrick M.C., Galvani A.P. The Impact of Vaccination on Coronavirus Disease 2019 (COVID-19) Outbreaks in the United States. Clin. Infect. Dis. 2021;73:2257–2264. doi: 10.1093/CID/CIAB079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feikin D.R., Higdon M.M., Abu-Raddad L.J., Andrews N., Araos R., Goldberg Y., Groome M.J., Huppert A., O’Brien K.L., Smith P.G., et al. Duration of effectiveness of vaccines against SARS-CoV-2 infection and COVID-19 disease: results of a systematic review and meta-regression. Lancet. 2022;399:924–944. doi: 10.1016/S0140-6736(22)00152-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bobrovitz N., Ware H., Ma X., Li Z., Hosseini R., Cao C., Selemon A., Whelan M., Premji Z., Issa H., et al. Protective effectiveness of previous SARS-CoV-2 infection and hybrid immunity against the omicron variant and severe disease: a systematic review and meta-regression. Lancet Infect. Dis. 2023;23:556–567. doi: 10.1016/S1473-3099(22)00801-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watson O.J., Barnsley G., Toor J., Hogan A.B., Winskill P., Ghani A.C. Global impact of the first year of COVID-19 vaccination: a mathematical modelling study. Lancet Infect. Dis. 2022;22:1293–1302. doi: 10.1016/S1473-3099(22)00320-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMOA2035389/SUPPL_FILE/NEJMOA2035389_DATA-SHARING.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMOA2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chalkias S., Harper C., Vrbicky K., Walsh S.R., Essink B., Brosz A., McGhee N., Tomassini J.E., Chen X., Chang Y., et al. A Bivalent Omicron-Containing Booster Vaccine against Covid-19. N. Engl. J. Med. 2022;387:1279–1291. doi: 10.1056/NEJMOA2208343/SUPPL_FILE/NEJMOA2208343_DATA-SHARING.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas S.J., Moreira E.D., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Polack F.P., Zerbini C., et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine through 6 Months. N. Engl. J. Med. 2021;385:1761–1773. doi: 10.1056/NEJMOA2110345/SUPPL_FILE/NEJMOA2110345_DATA-SHARING.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021;6:1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson B., Geetha K.M. Lipid nanoparticles in the development of mRNA vaccines for COVID-19. J. Drug Deliv. Sci. Technol. 2022;74:103553. doi: 10.1016/J.JDDST.2022.103553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blakney A.K., McKay P.F., Yus B.I., Aldon Y., Shattock R.J. Inside out: optimization of lipid nanoparticle formulations for exterior complexation and in vivo delivery of saRNA. Gene Ther. 2019;26:363–372. doi: 10.1038/s41434-019-0095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J., Eygeris Y., Gupta M., Sahay G. Self-assembled mRNA vaccines. Adv. Drug Deliv. Rev. 2021;170:83–112. doi: 10.1016/J.ADDR.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mucker E.M., Karmali P.P., Vega J., Kwilas S.A., Wu H., Joselyn M., Ballantyne J., Sampey D., Mukthavaram R., Sullivan E., et al. Lipid Nanoparticle Formulation Increases Efficiency of DNA-Vectored Vaccines/Immunoprophylaxis in Animals Including Transchromosomic Bovines. Sci. Rep. 2020;101:8764–8813. doi: 10.1038/s41598-020-65059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connors J., Joyner D., Mege N.J., Cusimano G.M., Bell M.R., Marcy J., Taramangalam B., Kim K.M., Lin P.J.C., Tam Y.K., et al. Lipid nanoparticles (LNP) induce activation and maturation of antigen presenting cells in young and aged individuals. Commun. Biol. 2023;61:1–13. doi: 10.1038/s42003-023-04555-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ndeupen S., Qin Z., Jacobsen S., Bouteau A., Estanbouli H., Igyártó B.Z. The mRNA-LNP platform’s lipid nanoparticle component used in preclinical vaccine studies is highly inflammatory. iScience. 2021;24:103479. doi: 10.1016/J.ISCI.2021.103479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alameh M.G., Tombácz I., Bettini E., Lederer K., Sittplangkoon C., Wilmore J.R., Gaudette B.T., Soliman O.Y., Pine M., Hicks P., et al. Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity. 2021;54:2877–2892.e7. doi: 10.1016/j.immuni.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee Y., Jeong M., Park J., Jung H., Lee H. Immunogenicity of lipid nanoparticles and its impact on the efficacy of mRNA vaccines and therapeutics. Exp. Mol. Med. 2023:2085–2096. doi: 10.1038/s12276-023-01086-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kafetzis K.N., Papalamprou N., McNulty E., Thong K.X., Sato Y., Mironov A., Purohit A., Welsby P.J., Harashima H., Yu-Wai-Man C., Tagalakis A.D. The Effect of Cryoprotectants and Storage Conditions on the Transfection Efficiency, Stability, and Safety of Lipid-Based Nanoparticles for mRNA and DNA Delivery. Adv. Healthc. Mater. 2023;12:2203022. doi: 10.1002/ADHM.202203022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu M.A. A Comparison of Plasmid DNA and mRNA as Vaccine Technologies. Vaccines. 2019;7:37. doi: 10.3390/VACCINES7020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mulroney T.E., Pöyry T., Puc Y.-, Carlos J., Rust M., Harvey R.F., Kalmar L., Horner E., Booth L., Ferreira A.P., et al. N1-methylpseudouridylation of mRNA causes +1 ribosomal frameshifting. Nature. 2023;2023:189–194. doi: 10.1038/s41586-023-06800-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melo A.R.da S., de Macêdo L.S., Invenção M.d.C.V., Invenção M.da C.V., da Gama M.A.T.M., Andrêssa I., Silva A.J.D., Batista M.V.d.A., Freitas A.C.d., Cristiane M.L., et al. Third-Generation Vaccines: Features of Nucleic Acid Vaccines and Strategies to Improve Their Efficiency. Genes. 2022;13:2287. doi: 10.3390/GENES13122287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamming L.A., Duque D., Tran A., Zhang W., Pfeifle A., Laryea E., Wu J., Raman S.N.T., Gravel C., Russell M.S., et al. DNA Based Vaccine Expressing SARS-CoV-2 Spike-CD40L Fusion Protein Confers Protection Against Challenge in a Syrian Hamster Model. Front. Immunol. 2021;12:785349. doi: 10.3389/FIMMU.2021.785349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elgueta R., Benson M.J., De Vries V.C., Wasiuk A., Guo Y., Noelle R.J. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol. Rev. 2009;229:152–172. doi: 10.1111/J.1600-065X.2009.00782.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harzandi N., Aghababa H., Khoramabadi N., Tabaraie T. Efficient Immunization of BALB/c Mice against Pathogenic Brucella melitensis and B. ovis: Comparing Cell-Mediated and Protective Immune Responses Elicited by pCDNA3.1 and pVAX1 DNA Vaccines Coding for Omp31 of Brucella melitensis. Iran. J. Biotechnol. 2021;19:40–47. doi: 10.30498/IJB.2021.2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan L., Zhu H., Zhou M., Ma J., Chen R., Chen Y., Chen L., Wu K., Cai M., Hong J., et al. Gender associates with both susceptibility to infection and pathogenesis of SARS-CoV-2 in Syrian hamster. Signal Transduct. Target. Ther. 2021;6:136–138. doi: 10.1038/s41392-021-00552-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castellan M., Zamperin G., Franzoni G., Foiani G., Zorzan M., Drzewnioková P., Mancin M., Brian I., Bortolami A., Pagliari M., et al. Host Response of Syrian Hamster to SARS-CoV-2 Infection including Differences with Humans and between Sexes. Viruses. 2023;15:428. doi: 10.3390/V15020428/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., Himansu S., Deterling J., Geilich B.M., Ketova T., et al. Optimization of Lipid Nanoparticles for Intramuscular Administration of mRNA Vaccines. Mol. Ther. Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W., Pfeifle A., Lansdell C., Frahm G., Cecillon J., Tamming L., Gravel C., Gao J., Thulasi Raman S.N., Wang L., et al. The Expression Kinetics and Immunogenicity of Lipid Nanoparticles Delivering Plasmid DNA and mRNA in Mice. Vaccines. 2023;11:1580. doi: 10.3390/VACCINES11101580/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saito A., Irie T., Suzuki R., Maemura T., Nasser H., Uriu K., Kosugi Y., Shirakawa K., Sadamasu K., Kimura I., et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature. 2022;602:300–306. doi: 10.1038/s41586-021-04266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 31.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., et al. COVID-19 Genomics UK COG-UK Consortium SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kudriavtsev A.V., Vakhrusheva A.V., Novoseletsky V.N., Bozdaganyan M.E., Shaitan K.V., Kirpichnikov M.P., Sokolova O.S. Immune Escape Associated with RBD Omicron Mutations and SARS-CoV-2 Evolution Dynamics. Viruses. 2022;14:1603. doi: 10.3390/V14081603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chakraborty C., Sharma A.R., Bhattacharya M., Lee S.S. A Detailed Overview of Immune Escape, Antibody Escape, Partial Vaccine Escape of SARS-CoV-2 and Their Emerging Variants With Escape Mutations. Front. Immunol. 2022;13:801522. doi: 10.3389/FIMMU.2022.801522/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang R., Ye Z.W., Ong C.P., Qin Z., Xie Y., Fan Y., Tang K., Poon V.K.M., Chan C.C.S., Yang X., et al. The spike receptor-binding motif G496S substitution determines the replication fitness of SARS-CoV-2 Omicron sublineage. Emerg. Microbes Infect. 2022;11:2093–2101. doi: 10.1080/22221751.2022.2111977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qu P., Evans J.P., Faraone J.N., Zheng Y.M., Carlin C., Anghelina M., Stevens P., Fernandez S., Jones D., Lozanski G., et al. Enhanced neutralization resistance of SARS-CoV-2 Omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2. Cell Host Microbe. 2023;31:9–17.e3. doi: 10.1016/j.chom.2022.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uraki R., Ito M., Furusawa Y., Yamayoshi S., Iwatsuki-Horimoto K., Adachi E., Saito M., Koga M., Tsutsumi T., Yamamoto S., et al. Humoral immune evasion of the omicron subvariants BQ.1.1 and XBB. Lancet Infect. Dis. 2023;23:30–32. doi: 10.1016/S1473-3099(22)00816-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qu P., Faraone J.N., Evans J.P., Zheng Y.M., Carlin C., Anghelina M., Stevens P., Fernandez S., Jones D., Panchal A.R., et al. Enhanced evasion of neutralizing antibody response by Omicron XBB.1.5, CH.1.1, and CA.3.1 variants. Cell Rep. 2023;42:112443. doi: 10.1016/J.CELREP.2023.112443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ao D., He X., Hong W., Wei X. The rapid rise of SARS-CoV-2 Omicron subvariants with immune evasion properties: XBB.1.5 and BQ.1.1 subvariants. MedComm. 2023;4:e239. doi: 10.1002/MCO2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willett B.J., Grove J., MacLean O.A., Wilkie C., De Lorenzo G., Furnon W., Cantoni D., Scott S., Logan N., Ashraf S., et al. SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat. Microbiol. 2022;7:1161–1179. doi: 10.1038/s41564-022-01143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou J., Liu Z., Zhang G., Xu W., Xing L., Lu L., Wang Q., Jiang S. Development of variant-proof severe acute respiratory syndrome coronavirus 2, pan-sarbecovirus, and pan-β-coronavirus vaccines. J. Med. Virol. 2023;95:e28172. doi: 10.1002/JMV.28172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim W., Zhou J.Q., Horvath S.C., Schmitz A.J., Sturtz A.J., Lei T., Liu Z., Kalaidina E., Thapa M., Alsoussi W.B., et al. Germinal centre-driven maturation of B cell response to mRNA vaccination. Nature. 2022;604:141–145. doi: 10.1038/s41586-022-04527-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inoue T., Shinnakasu R., Kawai C., Yamamoto H., Sakakibara S., Ono C., Itoh Y., Terooatea T., Yamashita K., Okamoto T., et al. Antibody feedback contributes to facilitating the development of Omicron-reactive memory B cells in SARS-CoV-2 mRNA vaccinees. J. Exp. Med. 2023;220:e20221786. doi: 10.1084/JEM.20221786/213745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lenart K., Hellgren F., Ols S., Yan X., Cagigi A., Cerveira R.A., Winge I., Hanczak J., Mueller S.O., Jasny E., et al. A third dose of the unmodified COVID-19 mRNA vaccine CVnCoV enhances quality and quantity of immune responses. Mol. Ther. Methods Clin. Dev. 2022;27:309–323. doi: 10.1016/j.omtm.2022.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwa S., Lai L., Gangadhara S., Siddiqui M., Pillai V.B., Labranche C., Yu T., Moss B., Montefiori D.C., Robinson H.L., et al. CD40L-Adjuvanted DNA/Modified Vaccinia Virus Ankara Simian Immunodeficiency Virus SIV239 Vaccine Enhances SIV-Specific Humoral and Cellular Immunity and Improves Protection against a Heterologous SIVE660 Mucosal Challenge. J. Virol. 2014;88:9579–9589. doi: 10.1128/JVI.00975-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hashem A.M., Gravel C., Chen Z., Yi Y., Tocchi M., Jaentschke B., Fan X., Li C., Rosu-Myles M., Pereboev A., et al. CD40 Ligand Preferentially Modulates Immune Response and Enhances Protection against Influenza Virus. J. Immunol. 2014;193:722–734. doi: 10.4049/JIMMUNOL.1300093. [DOI] [PubMed] [Google Scholar]
- 46.Lau J.J., Cheng S.M.S., Leung K., Lee C.K., Hachim A., Tsang L.C.H., Yam K.W.H., Chaothai S., Kwan K.K.H., Chai Z.Y.H., et al. Real-world COVID-19 vaccine effectiveness against the Omicron BA.2 variant in a SARS-CoV-2 infection-naive population. Nat. Med. 2023;29:348–357. doi: 10.1038/s41591-023-02219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turner J.S., O’Halloran J.A., Kalaidina E., Kim W., Schmitz A.J., Zhou J.Q., Lei T., Thapa M., Chen R.E., Case J.B., et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature. 2021;596:109–113. doi: 10.1038/s41586-021-03738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aid M., Busman-Sahay K., Vidal S.J., Maliga Z., Bondoc S., Starke C., Terry M., Jacobson C.A., Wrijil L., Ducat S., et al. Vascular Disease and Thrombosis in SARS-CoV-2-Infected Rhesus Macaques. Cell. 2020;183:1354–1366.e13. doi: 10.1016/j.cell.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wendisch D., Dietrich O., Mari T., von Stillfried S., Ibarra I.L., Mittermaier M., Mache C., Chua R.L., Knoll R., Timm S., et al. SARS-CoV-2 infection triggers profibrotic macrophage responses and lung fibrosis. Cell. 2021;184:6243–6261.e27. doi: 10.1016/J.CELL.2021.11.033/ATTACHMENT/F61754D9-B3D7-4A0E-9D20-2067CC9C519F/MMC6.XLSX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nouailles G., Wyler E., Pennitz P., Postmus D., Vladimirova D., Kazmierski J., Pott F., Dietert K., Muelleder M., Farztdinov V., et al. Temporal omics analysis in Syrian hamsters unravel cellular effector responses to moderate COVID-19. Nat. Commun. 2021;12:4869. doi: 10.1038/S41467-021-25030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suresh V., Mohanty V., Avula K., Ghosh A., Singh B., Reddy R.K., Parida D., Suryawanshi A.R., Raghav S.K., Chattopadhyay S., et al. Quantitative proteomics of hamster lung tissues infected with SARS-CoV-2 reveal host factors having implication in the disease pathogenesis and severity. FASEB J. 2021;35:e21713. doi: 10.1096/FJ.202100431R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aid M., Vidal S.J., Piedra-Mora C., Ducat S., Chan C.N., Bondoc S., Colarusso A., Starke C.E., Nekorchuk M., Busman-Sahay K., et al. Ad26.COV2.S prevents upregulation of SARS-CoV-2 induced pathways of inflammation and thrombosis in hamsters and rhesus macaques. PLoS Pathog. 2022;18:e1009990. doi: 10.1371/JOURNAL.PPAT.1009990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waickman A.T., Victor K., Newell K., Li T., Friberg H., Foulds K.E., Roederer M., Bolton D.L., Currier J.R., Seder R. mRNA-1273 vaccination protects against SARS-CoV-2–elicited lung inflammation in nonhuman primates. JCI Insight. 2022;7:e160039. doi: 10.1172/JCI.INSIGHT.160039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meyer M., Wang Y., Edwards D., Smith G.R., Rubenstein A.B., Ramanathan P., Mire C.E., Pietzsch C., Chen X., Ge Y., et al. Attenuated activation of pulmonary immune cells in mRNA-1273–vaccinated hamsters after SARS-CoV-2 infection. J. Clin. Invest. 2021;131:e148036. doi: 10.1172/JCI148036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pfeifle A., Thulasi Raman S.N., Lansdell C., Zhang W., Tamming L., Cecillon J., Laryea E., Patel D., Wu J., Gravel C., et al. DNA lipid nanoparticle vaccine targeting outer surface protein C affords protection against homologous Borrelia burgdorferi needle challenge in mice. Front. Immunol. 2023;14:1020134. doi: 10.3389/FIMMU.2023.1020134/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Algarni A., Pilkington E.H., Suys E.J.A., Al-Wassiti H., Pouton C.W., Truong N.P. In vivo delivery of plasmid DNA by lipid nanoparticles: the influence of ionizable cationic lipids on organ-selective gene expression. Biomater. Sci. 2022;10:2940–2952. doi: 10.1039/D2BM00168C. [DOI] [PubMed] [Google Scholar]
- 57.Guimaraes L.C., Carvalho P.A., Júnior S., Aluotto S.R., Ferreira H.A.S., Braga A.C.S., Oliveira de, Camilo L., Figueiredo M.M., Shepherd S., et al. Nanoparticle-based DNA vaccine protects against SARS-CoV-2 variants in female preclinical models. Nat. Commun. 2024;15:1–19. doi: 10.1038/s41467-024-44830-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao H.-C., Shen K.-Y., Huang M.S., Yang C.H., Chiu F.F., Chiang C.Y., Chai K.M., Huang W.C., Ho H.M., Chen Y.H., et al. Lipid nanoparticle-encapsulated DNA vaccine robustly induce superior immune responses to the mRNA vaccine in Syrian hamsters. Mol. Ther. Methods Clin. Dev. 2024;32:101169. doi: 10.1016/J.OMTM.2023.101169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/SCIENCE.ABB2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stuible M., Gervais C., Lord-Dufour S., Perret S., L'Abbé D., Abbé D., Schrag J., St-Laurent G., Durocher Y. Rapid, high-yield production of full-length SARS-CoV-2 spike ectodomain by transient gene expression in CHO cells. J. Biotechnol. 2021;326:21–27. doi: 10.1016/J.JBIOTEC.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stark F.C., Akache B., Deschatelets L., Tran A., Stuible M., Durocher Y., McCluskie M.J., Agbayani G., Dudani R., Harrison B.A., et al. Intranasal immunization with a proteosome-adjuvanted SARS-CoV-2 spike protein-based vaccine is immunogenic and efficacious in mice and hamsters. Sci. Rep. 2022;12:9772–9811. doi: 10.1038/s41598-022-13819-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M., et al. Quantification of SARS-CoV-2 neutralizing antibody by a pseudotyped virus-based assay. Nat. Protoc. 2020;15:3699–3715. doi: 10.1038/s41596-020-0394-5. [DOI] [PubMed] [Google Scholar]
- 63.Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938;27:493–497. doi: 10.1093/OXFORDJOURNALS.AJE.A118408/2/27-3-493.PDF.GIF. [DOI] [Google Scholar]
- 64.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;2020:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 65.Harris G., Holbein B.E., Zhou H., Xu H.H., Chen W. Potential mechanisms of mucin-enhanced acinetobacter baumannii virulence in the mouse model of intraperitoneal infection. Infect. Immun. 2019;87 doi: 10.1128/IAI.00591-19/SUPPL_FILE/IAI.00591-19-S0001.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lien C.E., Lin Y.J., Chen C., Lian W.C., Kuo T.Y., Campbell J.D., Traquina P., Lin M.Y., Liu L.T.C., Chuang Y.S., et al. CpG-adjuvanted stable prefusion SARS-CoV-2 spike protein protected hamsters from SARS-CoV-2 challenge. Sci. Rep. 2021;11:8761–8767. doi: 10.1038/s41598-021-88283-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Creskey M., Li L., Ning Z., Fekete E.E.F., Mayne J., Walker K., Ampaw A., Ben R., Zhang X., Figeys D. An economic and robust TMT labeling approach for high throughput proteomic and metaproteomic analysis. Proteomics. 2023;23:2200116. doi: 10.1002/PMIC.202200116. [DOI] [PubMed] [Google Scholar]
- 68.Huang T., Choi M., Tzouros M., Golling S., Pandya N.J., Banfai B., Dunkley T., Vitek O. MSstatsTMT: Statistical Detection of Differentially Abundant Proteins in Experiments with Isobaric Labeling and Multiple Mixtures. Mol. Cell. Proteomics. 2020;19:1706–1723. doi: 10.1074/mcp.RA120.002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein expression in Syrian hamster lung tissue samples was compared pairwise using a pairwise t test. Proteins with an absolute fold change >1.5 and an adjusted p value <0.05 were defined as differentially expressed proteins (DEPs).
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed on the DAVID Bioinformatics database with all parameter settings at their default values.
DEPs identified through pairwise t tests were imported into ingenuity pathway analysis (QIAGEN) and used to predict the activation or inhibition of canonical pathways.
Data Availability Statement
All proteomics data along with the database search results were deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org) via the PRIDE partner repository with the identifier PXD: 049053. All additional data needed to evaluate the conclusions in the paper are present in the main text and supplementary materials or available from the corresponding author upon request.