Abstract
The crucial molecular factors that shape the interfaces of lipid-binding proteins with their target ligands and surfaces remain unknown due to the complex makeup of biological membranes. Cholesterol, the major modulator of bilayer structure in mammalian cell membranes, is recognized by various proteins, including the well-studied cholesterol-dependent cytolysins. Here, we use in vitro evolution to investigate the molecular adaptations that preserve the cholesterol specificity of perfringolysin O, the prototypical cholesterol-dependent cytolysin from Clostridium perfringens. We identify variants with altered membrane-binding interfaces whose cholesterol-specific activity exceeds that of the wild-type perfringolysin O. These novel variants represent alternative evolutionary outcomes and have mutations at conserved positions that can only accumulate when epistatic constraints are alleviated. Our results improve the current understanding of the biochemical malleability of the surface of a lipid-binding protein.
Keywords: directed evolution, ribosome display, lipid specificity, cholesterol specificity, pore forming toxins
Biological membranes serve as molecular hubs where coordinated and highly specific interactions between proteins and their cognate ligands control cellular responses to a changing microenvironment. The major modulator of the bilayer structure, fluidity, thickness, compressibility, and intrinsic membrane curvature of mammalian cell membranes is cholesterol (Chol) (1). Chol often coordinates functional events relevant to cell physiology by modulating the biophysical properties of the membrane (2). Heterogeneous Chol distribution allows some proteins to concentrate in Chol-rich membrane domains and thus controls the cellular response to environmental signals (3, 4). The rise of Chol and other sterols in the evolution of biological membranes has been proposed as a cellular strategy to restrict the diffusion of free oxygen after the appearance of atmospheric oxygen (5). Posterior probability analysis for a number of membrane proteins showed that the binding sites for lipid receptors (such as Chol) are subject to higher selective pressure than the rest of the membrane interface residues and tend to be conserved (6). Similarly, through the course of evolution, peripheral lipid-binding proteins have developed a range of motifs and domains that specifically recognize particular membrane lipids (7, 8, 9, 10). Among the Chol-binding peripheral proteins, members of the Chol-dependent cytolysins (CDCs) represent one of the best-described examples (11).
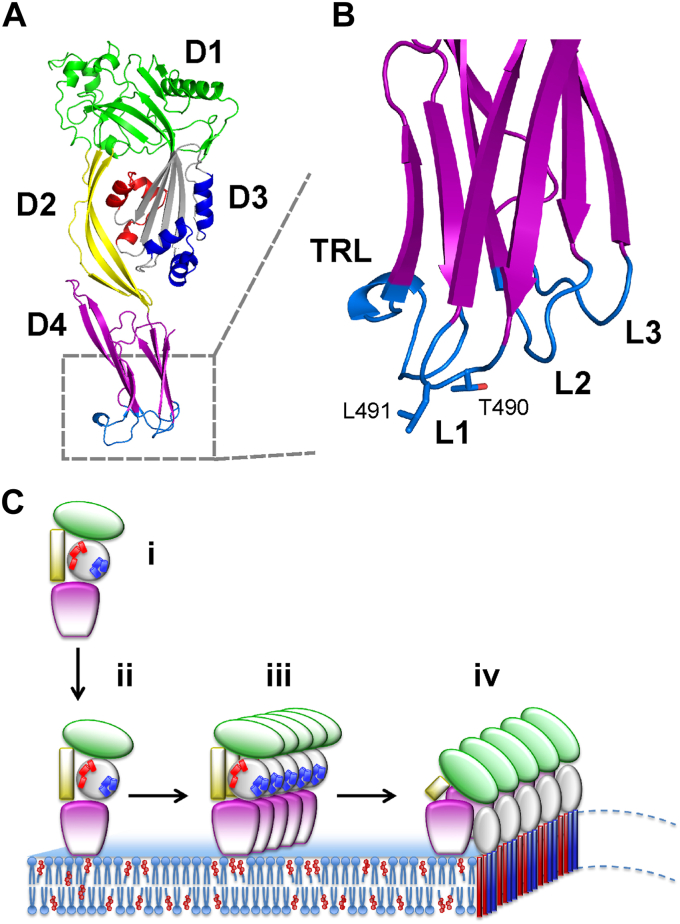
CDCs are potent toxins secreted predominantly by pathogenic Gram-positive bacteria (12). The archetypal CDC perfringolysin O (PFO) secreted by Clostridium perfringens, a bacterial pathogen that causes gastroenteritis associated with food poisoning in humans and animals, is the most extensively studied and the best described member of the family (13). CDC monomers have a four-domain architecture (domains D1-D4) (Fig. 1A) and exhibit 40 to 70% sequence identity (11, 14), with certain motifs in the membrane-binding domain (D4) being particularly conserved (Fig. 1B). These include amino acid residues in the tryptophan-rich loop (TRL) and loops L1 and L2, all located at the tip of the domain (12), while the remaining loop L3 on the far edge of the membrane-interacting D4 surface is the least conserved (11). The TRL is composed of the undecapeptide ECTGLAWEWWR and represents the characteristic signature motif for CDCs. D4 mediates the initial contacts of the monomers with Chol in a target membrane (15, 16) (Fig. 1B), and a threonine-leucine pair in loop L1 (T490-L491 in PFO, PDB ID: 1PFO), termed the cholesterol recognition motif (CRM), has been shown to be essential for Chol specificity (17). Binding of the monomers to the Chol-containing membrane enables oligomerization on the membrane and subsequent extensive structural changes between domains, especially in the distant D3, eventually leading to the membrane insertion of a large transmembrane β-barrel pore consisting of up to ∼30 protomers (11, 14). Even in the membrane-inserted pore, the contact of D4 with the membrane remains limited to interaction loops, whereas the rest of D4 projects from the membrane surface and retains the same conformation as found in solution prior to membrane binding (Fig. 1C) (15, 18).
Figure 1.
3D structure of PFO and mechanism of pore formation.A, a 3D model of the structure of PFO (PDB ID: 1PFO (29)). Different domains are labeled and colored. Two clusters of α-helices in D3 that participate in pore formation are labeled in red and blue. B, enlargement of the lower part of D4. Loops 1 to 3 (L1–L3) and the tryptophan-rich loop (TRL) comprising membrane-binding surface are labeled. The conserved residues Thr490-Leu491 implicated in cholesterol recognition (CRM) are shown in sticks. The view is rotated 90° clockwise in comparison to panel A. C, pore formation by PFO proceeds through several steps: binding of the monomer (i) to the lipid membrane (ii), oligomerization of the monomers in the plane of the membrane (iii) and insertion of the beta strands of D3 into the lipid membrane to form the final pore (iv). Labeling of different parts of a protein is as in (A). Images of the 3D models were created in Pymol (61).
CDCs provide an excellent model system for studying deeper evolutionary principles that shape the interacting surface of lipid-binding proteins. PFO pore formation can be influenced by the lipid composition of the membrane (19) and it has been shown that particular mutations in the D4 membrane-binding interface influence the activity and the Chol threshold needed for binding (20, 21). Unlike other CDCs domains, D4 is a discrete domain that is contiguous in its sequence. For this reason, it can be readily produced as an independent protein and, as a nontoxic product, has been used extensively as a research tool for the study of membrane Chol (22, 23, 24, 25, 26).
Here we address an important unresolved question about the evolution of lipid-binding proteins, in particular the molecular adaptations that enable the protein to recognize an important ligand Chol in the biochemically heterogeneous microenvironment of a lipid bilayer. To this end, we used a previously developed adaptation of the ribosome display technique to investigate the potential evolutionary pathways of the development of Chol specificity in PFO (27). By studying the distribution of variants’ activity across sequence space, we deciphered the particular evolutionary conserved positions, which need to be changed to allow the development of alternative evolutionary outcomes. We discovered that certain combinations of residues can support Chol specificity and that some of these variants possess activity that surpasses that of the wild-type (WT) protein. These variants represent alternative solutions for Chol-specific membrane binding that do not occur in nature. The selection of the variants with the Chol-specific activity is strongly influenced by epistasis, where mutations at certain positions affect the selective constraints on CRM. Our results, which reveal the surprising ability of PFO to accommodate mutations at strictly conserved positions and the major influence of epistasis on their occurrence, are of importance for the design of lipid-binding proteins.
Results
Single amino acid variants of PFO retain characteristic CDC identities
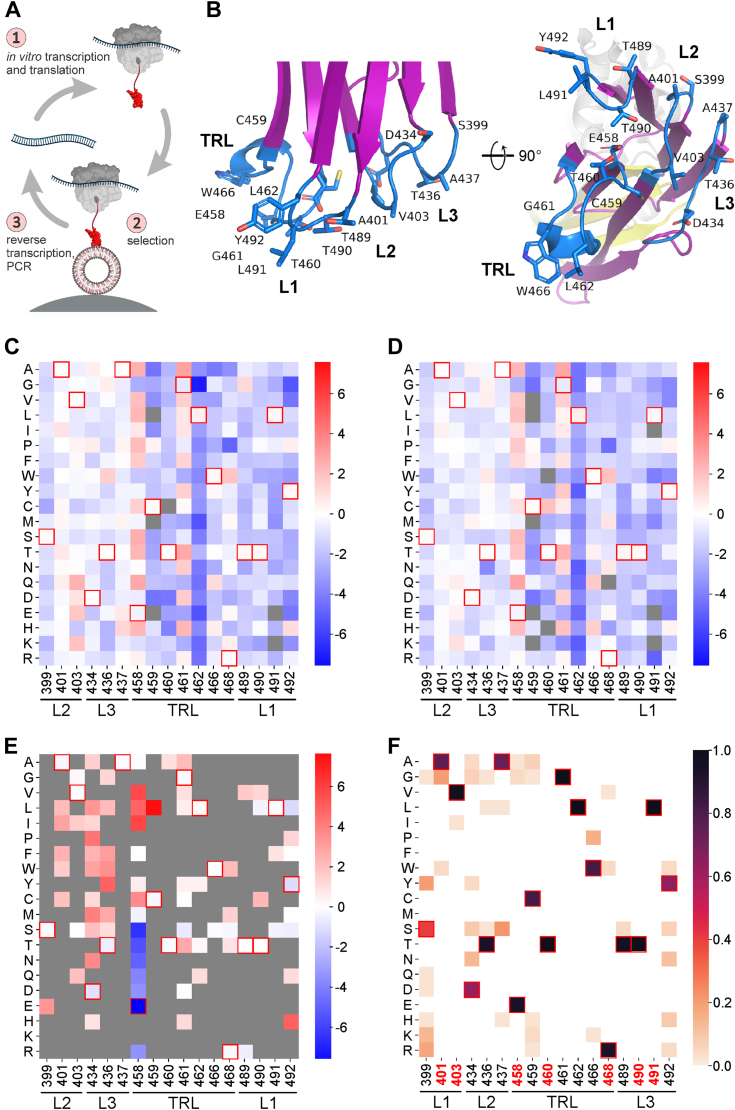
To gain insight into the potential biochemical variability of the Chol-specific interaction surface of the D4 domain of PFO, we used a ribosomal display approach to select variants based on their lipid-binding affinities (Fig. 2A) (27). We systematically mapped the sequence space underlying the membrane-binding interface using a site saturation library (SSL). The SSL contained single amino acid substitutions at nearly all positions surrounding the binding surface of D4 (Fig. 2B). Only the positions with glycine residues (G400 in L2 and G488 in L1) and those where the side chains face away from the plane of the membrane (Y402 in L2, K435, H438 in L3, A463, W464, E465, W467 in the TRL) were excluded from the analysis, as they are unlikely to contribute to initial membrane binding. A complete set of amino acid substitutions was allowed at 17 positions (Fig. 2B), resulting in a library that encodes 324 unique protein variants. To assess the contribution of individual amino acid substitutions to Chol-specific binding, we performed affinity selection using the SSL library against three groups of lipid vesicles: (i) vesicles without Chol consisting only of the phospholipid 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), (ii) POPC vesicles with 20 mol% Chol, which is below the Chol threshold required for the WT PFO binding to membranes (20), and (iii) POPC vesicles with 50 mol% Chol. After six rounds of selection, we subjected the input SSL and affinity-selected SSL libraries to deep sequencing. Analysis of the input library showed that complete sequence coverage was achieved, as all possible 324 protein variants were detected. The vast majority of sequences were found in nearly equal proportions, showing that the input library is unbiased, which is important when only positive selections are involved (28).
Figure 2.
Representation of amino acids in the membrane-binding region of PFO analyzed with the site saturation library.A, a ribosomal display was used to select and identify PFO variants against lipid vesicles with defined lipid composition. Figure adapted from (27) with permission. B, the structure of the tip of the D4 domain. Residues that were included in the analysis are highlighted. Labeling and coloring as in Figure 1B. C–E, enrichment of the SSL library at 17 randomized positions after selection against vesicles containing POPC (C), POPC vesicles containing 20 mol% Chol (D), and POPC vesicles containing 50 mol% Chol (E). The color scale represents log2 of amino acid residue frequencies normalized to the input SSL library. The gray color represents amino acid residues that were not detected after selection. The red boxes outline amino acid residues present in the WT PFO. F, relative abundance of amino acid residues at the selected positions of 46 members of the CDC family, highlighting their variability. Red numbers mark residues used in the combinatorial library (see below).
We calculated the frequency as the ratio of each variant with single substitution in the population of analyzed sequencing reads and the enrichment of affinity-selected variants as the ratio of the frequency of a single substitution in the selected population compared to the nonselected population. This enrichment value served as a quantifiable indicator of the impact of substitution on Chol binding. As expected, no clear enrichment was observed after selection against POPC vesicles (Fig. 2C) and POPC vesicles with 20 mol% Chol (Fig. 2D) and nearly all theoretically possible variants were detected in both libraries, specifically 318 after selection against POPC vesicles and 313 of 324 possible variants after selection against vesicles containing 20 mol% Chol. The occurrences of the top five variants selected against POPC vesicles ranged from 2.3% to 4.6%, comparable to 20 mol% Chol, where they ranged from 2.2% to 4.1%. When frequencies after selection were normalized to the frequencies in the library before selection, the enrichment of polar amino acids with longer side chains at positions 403, 458, and 461 when selecting against vesicles with 20% Chol or less was observed, which may indicate that such amino acids promote binding to Chol-free vesicles in a nonspecific manner.
Conversely, a clear pattern is observed in the enrichment of variants in the library selected against POPC vesicles with 50 mol% Chol. Out of 324 possible sequences, we detected only 92 indicating that nearly three quarters were eliminated during the affinity selection process (Fig. 2E) and the most enriched variant represented 45% of all analyzed sequencing reads. Overall, the most abundant variants possess amino acids found in members of the CDC family (Fig. 2F). Specifically, the 11 enriched residues located at 7 out of 17 positions analyzed correspond to those found in CDC family members. The greatest variability was observed at positions in the evolutionarily less conserved L3, while TRL and L1 remained relatively unchanged. Invariability and lack of tolerance to amino acid substitutions were particularly evident in T490 and L491 in the proposed CRM (17) and in V403 and T460, which is also consistent with their relative conservation in CDC representatives (Fig. 2F).
Interestingly, the most abundant sequences selected on POPC vesicles with 50 mol% Chol contained nonsynonymous substitutions at conserved position E458. Their total representation in the affinity-selected library was around 30%, which, given the low relative abundance in the input library (<0.5%), suggests 100-fold enrichment by the affinity selection. The most abundant sequences contain the hydrophobic residues valine, leucine, and isoleucine at position 458. In the WT protein, glutamic acid at position 458 forms a hydrogen bond with T460 that stabilizes the TRL loop (29). The preferential enrichment of residues with shorter side chains and the loss of hydrogen bonding potentially increases the flexibility of the TRL loop and thus may facilitate the first contact with the target membrane. We identified a similar trend of increasing surface hydrophobicity at position T436, where residues with aromatic side chains (phenylalanine, tyrosine, and tryptophan) dominate. The residue T436 is located in the loop L3, which is, during the initial contact with the target membrane, farther apart from the membrane surface compared to the L1 and the TRL (Fig. 2B). Replacement of the threonine side chain with bulkier aromatic side chains likely brings the L3 loop closer to the plane of the membrane, allowing for better binding. This is consistent with a previous report where the T436A substitution was shown to reduce the hemolytic activity of PFO by ∼20% (17). Similarly, we observed a pronounced enrichment of sequences with nonsynonymous substitutions at position D434 (Fig. 2E). The enrichment of proline, asparagine, serine, leucine, and aromatic residues indicates that the removal of the negative charge at this position contributes to better binding of D4. Indeed, the D434S substitution has been associated with a reduction in the membrane Chol concentration required for efficient PFO binding (20).
Concurrent D4 mutations reveal alternative evolutionary trajectories
To evaluate the broader adaptability of the D4 protein-membrane binding interface, we used a previously developed combinatorial library in which seven interfacial positions were randomized simultaneously (27). The residues at the selected positions, namely 401, 403 in L2, 458, 460 and 468 in TRL, and 490, 491 in L1, are almost completely conserved in CDCs (red residues in Fig. 2F) (27). They are all located within a narrow pocket formed by TRL, L1, and L2 loops with a solution-exposed side chains and are therefore likely to be involved in the initial contact with the target membrane.
The combinatorial library was selected against lipid vesicles composed of (i) POPC only, (ii) vesicles containing POPC and 50 mol% of Chol, and (iii) vesicles composed of POPC and 50 mol% of the Chol analog cholesteryl-acetate (Fig. 3A). The latter contains a modification of the hydroxyl group of Chol and does not promote binding of CDCs to lipid membranes (30, 31, 32). We subjected the libraries to deep sequencing before and after selection and analyzed the identity and abundance of each sequence in the population for each sample. The distribution of variants is heterogeneous and without biased enrichment when selected against POPC membranes (Fig. 3B). These variants, detected in samples selected against vesicles without Chol, probably represent background noise originating from D4 variants and/or mRNA molecules nonspecifically associated with lipid vesicles (33, 34, 35).
Figure 3.
Representation of amino acids in the membrane-binding region of D4 analyzed with the combinatorial library.A, occurrence of sequences in the combinatorial library and after selection against lipid vesicles with different lipid compositions as indicated. B–D, relative abundance of amino acid residues at randomized positions after selection against vesicles containing only POPC (B), POPC and 50 mol% Chol (C), and POPC and 50 mol% acetyl-Chol (D). The color scale represents log2 of amino acid residue frequencies normalized to the input combinatorial library. The amino acid residues in the boxes outlined in red represent WT PFO residues. E, sequence logos for all variants with alternative CRM. F, enrichment of nonsynonymous mutations observed at the indicated positions in the SSL and the combinatorial library. The sequence logos were generated with WebLogo 3 (62).
In contrast, the library selected against vesicles containing 50 mol% Chol shows a significant enrichment of certain sequences (Fig. 3C). The ten most abundant sequences in the population of variants selected against Chol-containing vesicles represent 68% of all identified sequences. Despite the large number of potential outcomes in the D4 interaction surface resulting from simultaneous substitutions allowed in the combinatorial library, the most abundant sequences after affinity selection against Chol-containing vesicles retain the WT residues A401, V403, T460, and R468, as well as the T490-L491 pair (Fig. 3C). On the other hand, and similar to the trend observed with the SSL, residue E458 emerges as an exception, where glutamic acid, chosen by natural selection, becomes replaced by hydrophobic residues, primarily valine, during in vitro evolution. These findings indicate that this residue, despite being nearly universally conserved, is tolerant to substitutions with hydrophobic amino acid residues (Fig. 3C). In contrast, after selection against control vesicles with cholesteryl-acetate, the output library shows no preference for a particular residue at any of the positions and is similar to the selection against vesicles without Chol (Fig. 3D).
Considering the T490-L491 pair as a CRM (17), and based on a previous result where we discovered a nonconventional CRM (27), we conducted an additional search within the population of variants identified after affinity selection against 50 mol% Chol-containing vesicles to identify rare variants lacking this motif. Intriguingly, we discovered a subset of variants that deviate markedly from D4, lacking the strictly conserved T490-L491 pair. These divergent variants contain the hydrophobic valine or alanine at position T490, whereas residues with aromatic side groups, namely tryptophan, proline, and phenylalanine occupy position L491. While V403 and T460 remain conserved in these variants, A401W is a common substitution. Notably, in all of these sequences, the position E458 is occupied by hydrophobic residues and not by the evolutionarily conserved glutamate (Fig. 3E).
Taken together, affinity selection using simplified model lipid membranes clearly shows the absence of distinct sequence enrichment after selection against Chol-free vesicles or control vesicles containing cholesteryl-acetate and the profound enrichment following selection against Chol-containing vesicles using the SSL and combinatorial libraries both indicate that Chol is the key evolutionary force that shaped the D4 interaction surface. Moreover, the strong enrichment of variants with nonsynonymous substitutions at E458 identified using both libraries and the identification of variants lacking the T490-L491 motif using the combinatorial library reveal extensive plasticity of the Chol-specific membrane-binding D4 interface and a wider sequence-space not explored by natural evolution. The low abundance of variants lacking the T490-L491 motif indicates that they are tolerated only in certain biochemical contexts, accompanied by other substitutions in the membrane-binding region. In addition, the significantly higher variability at all positions analyzed, especially A401 and V403, after affinity selection against Chol-containing vesicles using the combinatorial library compared to SSL (Fig. 3F) suggests that certain mutations become permissible when combined with mutations at other positions and that epistatic interactions between mutations thus have an important influence on the in vitro evolution of D4 variants. As these variants profoundly differ from those found in nature, they represent an alternative evolutionary history in the evolutionary trajectory of CDCs.
The activity of PFO variants correlates with their enrichment after affinity selection against Chol-containing vesicles
To examine whether the variants enriched in the affinity selection steps are indeed functional, we prepared a series of full-length PFO variants and tested their pore-forming activity using bovine erythrocytes. We named the PFO variants after the residues at the positions randomized in the combinatorial library, with those that differ from the WT underlined. Since the mutations are located at the tip of D4 and are far from other domains, we anticipated that the differences in activity likely reflect differences in membrane binding. However, changes in the D4 membrane-binding region might also be involved in other steps in the formation of a functional transmembrane pore. Positions V403 and T460 were identified as less tolerant to substitutions after affinity selection using both libraries. Therefore, we tested the hemolytic activity of full-length PFO with single substitutions at these two positions. In the context of full-length PFO, both positions show some tolerance to substitutions, with the exception of charged amino acids, which are strongly disfavored. For example, the activity of variants V403I, V403F, T460I, or T460L is almost identical to that of WT PFO, while substitutions to charged amino acids almost completely abolish hemolytic activity (Fig. 4, A and B).
Figure 4.
The hemolytic profile of PFO variants with mutations at invariable and variable positions. The observed hemolytic activities were normalized to the activity of the WT PFO. The hemolytic activity of the WT PFO (AVETRTL) is highlighted in red. A–D, relative hemolytic activity of variants with substitutions at positions 403 (A) and 460 (B), the most abundant sequences in the combinatorial library (C) and variants with alternative CRM (D). In (C) and (D), each variant is named after the amino acids it has at the positions studied by the combinatorial library. Underlined are the mutated positions. The plots show the mean values ± SD of three independent experiments. The c50 concentrations (the concentration of the protein that causes 50% of the maximal rate of the hemolysis) of the PFO variants were compared with the c50 of the WT PFO using a two-tailed unpaired t test (n.s. = p > 0.05; ∗p < 0.05, ∗∗p < 0.01; ∗∗∗p < 0.001). n.a., non-hemolytic variants.
The most frequent substitutions in both libraries selected against Chol-containing vesicles were A401 and E458, and to a lesser extent R468V (Fig. 3F). To compare the effects of isolated and concurrent mutations at these positions, we generated a series of single, double, and triple full-length PFO variants (Fig. 4C). A single mutation variant A401W (variant WVETRTL) had a two-fold higher hemolytic activity than the WT PFO (AVETRTL) (Fig. 4C). On the other hand, the activity of variants A401L (LVETRTL) and A401Y (YVETRTL) is somewhat lower, approximately five-fold and two-fold, respectively, consistent with less pronounced enrichment after affinity selection (Fig. 3E). Interestingly, the activity of the variant containing the most abundant substitution E458V (AVVTRTL) was found to be similar to the WT. However, the variant containing the combined substitutions A401W and E458V (WVVTRTL) had a nearly 40-fold improvement in activity compared to the WT PFO (Fig. 4C). The addition of the third substitution R468V (WVVTVTL) reduces this activity to approximately the level of the A401W single mutant. Similarly, the variant with the double mutation E458V and R468V (AVVTVTL) leads to about ten-fold lower hemolytic activity than the WT PFO. The comparison with the triple mutation variant A401W/T458V/R468V (WVVTVTL), whose activity is even slightly better than that of the WT PFO, shows that A401W compensates for the effects of the double mutation E458V and R468V. In conclusion, the hamolytic activity of the most enriched variants is comparable or even better than the activity of the WT PFO. Therefore, the amino acid substitutions at the positions defined as variable are indeed tolerant to substitutions.
Variants with alternative cholesterol recognition motifs retain functional properties of PFO
To validate the activity of PFO variants with alternative CRMs, we tested the hemolytic activity of the recombinantly produced identified full-length PFO variants (Fig. 4D). The results of the hemolytic assay confirm that mutations in the conserved pair T490-L491 can indeed be replaced. Interestingly, in almost all cases where the hemolytic activity was similar to the WT protein, changes in the T490-L491 pair were accompanied by mutations at other positions that have been identified as tolerant. For example, the variants WVATRIW and WVVTRAF with substitutions T490V/L491W and T490A/L491F in the CRM, respectively, exhibit even higher activity than WT PFO. The substitutions in the CRM are accompanied by A401W and a substitution of E458 to a small hydrophobic residue (alanine or valine) (Fig. 4D). The most active variants have a smaller hydrophobic residue at position 490 (isoleucine, valine, or alanine) and an aromatic residue at position 491 (tryptophan or phenylalanine). Mutation A401W appears to be beneficial but not essential for rescuing PFO activity, while E458V is present in all variants with the best performance, consistent with its relative abundance in the SSL and combinatorial libraries selected against Chol-containing vesicles. These results suggest that an epistatic effect exerted by substitutions at variable positions E458 and A401 shapes specific amino acid patterns that determine Chol specificity.
Among the variants with alternative Chol-binding motif, WVATRIW and WVVTRAF demonstrated higher hemolytic activity than the WT PFO (Fig. 4D). Therefore, we further focused on examining their Chol specificity. We detected a concentration-dependent binding of the variants and WT PFO only to Chol and not to POPC or cholesteryl-acetate (Fig. 5A). The Chol specificity of the two variants was also confirmed by the lipid overlay assay, in which the WT and both variants bound to spotted Chol but not to POPC or sphingomyelin (Fig. 5A inset) and in vesicles sedimentation assay (Fig. 5B). To investigate Chol-specific binding of the variants in the cellular context, we used mouse macrophage-derived RAW 264.7 cells treated or untreated with methyl-β-cyclodextrin (MβCD), which extracts Chol from cell membranes. Both WT EGFP-tagged D4 and the variants with an alternative CRM bound the RAW 264.7 cells in an MβCD concentration-dependent manner, confirming that binding is indeed Chol-dependent (Fig. 5C). Similarly, monitoring permeabilization of the plasma membrane of RAW 264.7 cells induced by the full-length PFO variants shows that the activity of the WVVTRAF is almost identical to that of the WT PFO (Fig. 5D). The WVATRIW variant achieves cell permeabilization at a slightly higher protein concentration. Pre-incubation of RAW 264.7 cells with MβCD resulted in a strong inhibition of the permeabilization potency of all PFO variants (Fig. 5D). Finally, we used cryo transmission electron microscopy to image the pores formed on the surface of the Chol-containing large unilamellar vesicles. We found that the pores of the WVATRIW and WVVTRAF variants are similar in shape and size of the inner diameter (26.8 ± 2.9 nm (n = 82) and 23.1 ± 1.6 nm (n = 114), respectively) to those of the WT PFO (21.0 ± 0.3 nm (n = 88)) (Fig. 5E). Thus, both variants retain functional properties of PFO and have full capacity to form pores similar in shape and size to the pores of the WT protein.
Figure 5.
Characterization of the cholesterol-specific functional and structural properties of the WVATRIW and WVVTRAF variants carrying alternative CRMs. The variants are named after the amino acids they contain at the positions studied by the combinatorial library. A, enzyme-linked immunosorbent assay. The wells of the microtiter plates were coated with POPC (black), Chol (red), or cholesteryl-acetate (blue). The mean ± SD of three independent experiments is shown. The inset shows the results of lipid overlay assay where binding of the whole-length proteins to POPC, sphingomyelin, and cholesterol spotted to nitrocellulose membrane was analyzed. B, sedimentation assay of whole-length proteins with multilamellar vesicles composed of POPC or POPC with 50 mol% Chol (Chol). The vesicle-free sample (-ves) was used as a control. C, binding of EGFP-tagged D4 to the plasma membrane of RAW 264.7 cells in the absence or presence of methyl-β-cyclodextrin (MβCD) at the indicated concentrations. The left panel for each protein shows flow cytometry data, and the graph on the right shows the green fluorescence of the cells (mean ± SD of three independent experiments). D, permeabilization of Raw 267.4 cells with whole-length WT PFO (red) and variants WVATRIW (blue) and WVVTRAF (green). Permeabilization is shown as the maximum rate of increase in fluorescence of the nuclear dye Sytox Green, which is not permeable to intact membranes. In each panel, responses in intact cells (the intense color) are compared to cells with MβCD-extracted Chol (pale color) (mean ± SD of four independent experiments). E, cryo transmission electron microscopy micrographs of whole-length WT and PFO variants bound to large unilamellar vesicles composed of POPC and 50 mol% Chol (top). Arrows indicate pores imaged as inserted rings. 2D class averages of top views of pores inserted in the vesicle lipid bilayer. Size bars (in nm) are depicted on micrographs and under the 2D class averages (right). P, pellet; RFU, relative fluorescence units; S, supernatant; T, total amount of protein used in the binding reaction.
Epistasis shapes the mutagenic pathways leading to alternative cholesterol-binding motifs
The results of our in vitro affinity selection indicate that epistasis plays an important role in shaping PFO specificity for membrane Chol (Figs. 3F and 4D). To examine epistatic constraints in the transition from WT PFO to two alternative variants, WVATRIW and WVVTRAF, we purified and analyzed the hemolytic activity of all mutation combinations at positions that differ in WT PFO and the two alternative variants (i.e., 401, 458, 490, and 491). The obtained c50 values, that is, the concentration of the variant that induces 50% of the maximum hemolysis rate, were normalized to the activity of the WT and used to construct the activity landscape in which the edges connect the intermediate variants separated by a single residue substitution (Fig. 6). The mutagenic pathway was defined as possible if the activity of the intermediate variants was comparable to or better than that of the WT PFO.
Figure 6.
A mutagenic pathway linking AVETRTL (WT) and variants WVATRIW and WVVTRAF.A, a mutagenic pathway for a WVVTRAF variant. The amino acid residues for the WT and the WVATRIW variant are indicated in the top right-hand corner. Residues that were mutated in the WT to reach the sequence of this variant are underlined. Each variant is presented as a node and the respective residues that differ in WT and a particular variant are indicated. Nodes are shaded according to their relative hemolytic activity. The possible mutagenic pathways are indicated by red lines. B, relative hemolytic activity of WVATRIW variants with additional mutations at conserved positions V403 and T460. C, a mutagenic pathway for a WVVTRAF variant. All details are as in (A).
When analyzing the mutagenic pathway from AVETRTL to WVATRIW, we found that the activity of the variants AVETRTW and AVETRIL, which describe stepwise substitutions in T490 or L491, is about 100 times lower. A similarly low activity is also characteristic of the AVETRIW variant (Fig. 6A), in which both amino acids have been replaced in the CRM. This suggests that a mutagenic pathway from AVETRTL to WVATRIW that begins with mutations in the CRM is not possible. On the other hand, the activity of the AVATRTL and WVETRTL variants that have substitutions at the tolerant positions is higher than that of WT PFO. For the continuation of the pathway and the final substitution in the T490 and L491 motif, a simultaneous change of amino acids at both positions tolerant to substitutions is a prerequisite, which then imparts the plasticity of the motif with a permissive T491I change. In addition, we confirmed that even in the amino acid context of the alternative CRM, substitution of V403 or T460 is not permissible, as the WAATRIW variant is about ten-fold less active than the WT, whereas no hemolytic activity is detected in the WVAARIW variant (Fig. 6B). Likewise, in the case of the mutagenic pathway from AVETRTL to WVVTRAF, changes at mutation-permissive positions (A401W and E458V) are required and consequently allow the development of the alternative CRM (Fig. 6C). In both cases, the most active variant was the one with substitutions at mutation-permissive positions 401 and 458.
The possible mutational pathway from the WT PFO motif to the alternative motif is therefore dictated by specific epistatic relationships, which manifest as a requirement for a specific order of amino acid substitutions. Our results clearly show that the plasticity of the Chol recognition motif is much greater than could be inferred by site-specific mutagenesis or by analysis of the natural variability of the CDCs orthologs. Amino acid interdependence in the D4 loops limits the greater diversification of the binding surface that could evolve during the process of natural selection, as it requires conditional prior changes at mutation-tolerant positions.
Discussion
The fitness landscape and general paradigm for the molecular adaptations of a protein that specifically recognizes a particular lipid in a biochemically diverse environment, such as cell membranes, is experimentally challenging to approach. This is partly due to the inherent challenges in the biochemical and structural interpretation of protein–lipid interactions and limitations of existing molecular simulation techniques. In addition, the membrane lipid can serve simultaneously as a solvent, receptor, and regulatory cofactor for the activity of a lipid-binding protein (36). Furthermore, once embedded in the membrane, the protein modulates its local lipid environment, resulting in an enrichment or depletion of specific lipid components (37). As a result, the specificity determinants of protein-membrane lipid interactions remain poorly defined.
In this study, we investigated the plasticity of a lipid-binding protein interface using in vitro evolution of PFO, the prototypical CDC. Through a comprehensive comparative analysis of amino acid variability in D4 loops, we examined the biochemical heterogeneity that can maintain the fundamental function of D4, namely the recognition of Chol-containing membranes. We show that in vitro evolution enables optimization beyond natural evolution by identifying PFO variants with an order of magnitude higher activity than the WT (Figs. 4 and 6). In addition to identifying mutation-tolerant and mutant-intolerant positions, we discovered variants with novel CRMs that represent alternative evolutionary outcomes.
Unlike protein–protein or protein–DNA interactions, in which a genetically encoded subset of residues from each interaction partner can co-evolve (38), the molecular adaptations required to maintain recognition of a molecule embedded in the lipid bilayer depend solely on the ability of the interacting protein to evolve. The initial interaction of CDCs with the lipid membrane is mediated by the four loops at the bottom of the molecule (Fig. 1B) (15, 17, 39). These loops also contain the CRM responsible for Chol recognition (17). Molecular dynamics simulations showed that the CRM and aromatic amino acids from the TRL interact directly and transiently with Chol in pneumolysin, a CDC of Streptococcus pneumoniae (40). However, the interaction of CDCs with Chol has not been fully elucidated, and the molecular details of the interaction of CDCs with Chol remain elusive (12, 41). Our results show that Chol specificity is maintained independent of profound changes in amino acid identities at the membrane-binding interface and CRM (Fig. 5). This suggests that the determinants of Chol specificity in CDCs are not exclusively defined by the identity of individual residues, that is, the CRM, in the binding surface.
The complexity of cholesterol–CDCs interactions can be further illustrated by the fact that some mutations in the least conserved L3 loop of PFO decrease the amount of Chol required for binding (20, 21, 23). Similarly, the binding of CDCs to membranes containing ergosterol, a structural analog of Chol, is significantly weaker than Chol (30, 41). Ergosterol differs from Chol mainly in the length of the hydrophobic tail, which may affect both the interaction interface with CDCs and the organization of the membrane bilayer, resulting in a different lipid environment and membrane properties despite the structural similarities between the two lipids.
The experimental design of our approach allowed us to study the protein–lipid interaction independently of the conformational changes that lead to the insertion of the protein into the membrane. This is due to the fact that the same element, namely the CDC undecapeptide, is involved in binding to the lipid membrane as well as in long-range signaling to the distal D3 domain, which forms a transmembrane pore following a conformational change (42, 43). A similar mechanism has been demonstrated for most other CDCs (44, 45, 46, 47, 48, 49, 50). Therefore, by employing the isolated D4 domain and selecting variants based on their affinity for Chol, we were able to uncouple Chol binding from the process of pore formation, which is important from several points of view. First, we were able to circumvent other epistatic interactions that occur at the level of full-length protein and are required for protein function (51). In full-length PFO, mutations in the Chol-binding site may result in proteins with altered activity, which may be due to changes in affinity for target membranes, but also due to changes in the interaction between monomers or protomers or to an effect on structural remodeling associated with pore formation. Each of these processes is likely to have an individual fitness landscape that can expand the sequence space of interest beyond the technical capabilities of current methods of directed evolution.
The loops in D4 also play an important role in oligomer formation by providing an interaction surface between the monomers in the oligomeric form (18). In particular, W433 in pneumolysin (W464 in PFO, which was not included in our screen) was shown to interact with T405 (T436 in PFO). According to the results with SSL, T436 can be replaced by several bulkier and hydrophobic amino acids (Fig. 2E), which indicates its important role in the initial interactions of D4 with the membrane and could only be revealed when working with isolated D4.
However, we cannot exclude the possibility that the effects of some important residues may be overlooked in this context and due to possible conformational differences in TRL between the protein in solution and when inserted in the membrane. A comparison of the crystal structures of PFO with other CDCs, namely pneumolysin (i.e., PDB ID: 5AOD (52)), shows that TRL adopts its extended form when bound to the membrane. As a result, residues 463 to 465 (PFO numbering) become placed in the membrane environment (18) but away from the CRM and therefore do not affect Chol recognition.
Second, by focusing exclusively on the surface of D4 that is involved in membrane interactions, we were able to locate the permissive mutation A401W, which is at the beginning of the mutagenic pathways of the two PFO variants with altered Chol-binding motifs (Fig. 6). The permissive mutations allow mutations that confer new functions to occur. However, the permissive mutations themselves are not acquired by selection for the same function (51). Since the chance that the permissive mutation will be fixed by drift alone is quite low, the occurrence of specific permissive mutations is rare, which in turn regulates the relative abundance of PFO variants with multiple mutations in the final round of selection. The other permissive mutation, E458V, appears to have arisen through selection pressure, based on its prevalence in both SSL and combinatorial libraries. Natural membranes represent a unique lipid and protein environment that might contribute to evolutionarily conservation of the E458 among CDC members. In contrast, artificial Chol-containing model membranes, with their more hydrophobic surface due to absence of other membrane components, might enhance the selection of the hydrophobic residue such as valine at position E458 among variants affinity selected against Chol-containing lipid vesicles. Additionally, due to a simplified lipid composition and the absence of proteins in model membranes, the pH in the vesicle bilayer might not be the same as in the outer leaflet of the cell membrane. In the crystal structure of the PFO monomer (29), E458 is involved in a hydrogen bonding network with the side chains of the neighboring residues of the undecapeptide, which breaks off upon entry into the hydrophobic environment of the outer membrane leaflet and subsequent protonation of E458 (41). The protonated glutamate makes the binding surface of PFO less polar and thus increases affinity toward membranes. This is consistent with the observation that binding of PFO in lipid vesicles mimicking the outer membrane leaflet was highest at pH 5.5 to 6 (32, 53). Since binding of PFO decreases rapidly at pH above 7 and is almost absent at pH 8 (53), it is not surprising that the E458 variants dominate in the SSL library (Fig. 2E). Importantly, substitutions in evolutionarily conserved CRM residues within the membrane-binding interface clearly support the subsequent changes necessary for pore formation (Fig. 6). It is noteworthy that an evolutionary pathway has emerged under in vitro conditions that allow the binding surface of the PFO membrane to be altered without compromising its ability to form pores (Fig. 5), which is essential for its function.
In conclusion, the unique architecture makes CDCs an excellent model system for studying amino acid substitutions responsible for specific lipid–protein interactions, as initial lipid binding can be separated from the complex structural rearrangements that follow receptor recognition. Using isolated D4 and simplified membrane models provides a controlled environment to elucidate the effects of individual mutations and their interactions, offering insights into the mechanisms underlying mutational pathways. Our study revealed alternative CRM, which are not present among CDC family members. Thus, despite the numerous accessible alternative CRM, historically evolved variants remain highly conserved among present-day descendants. Systematic exploration of the epistasis on mutational pathways showed significant constraints on the mutational pathways D4 could follow through sequence space. Namely, the alternative CRM can be reached from WT only in the case of ordered amino acid substitutions. Additionally, the probabilities of amino acid transitions influenced by their codon requirements have likely limited evolutionarily exploration of the sequence space. Namely, the substitution A401W, which represents a prerequisite for the development of the alternative CRM from the WT, is evolutionarily less probable due to complex genetic code transitions from alanine (encoded by one of four codons GCT, GCC, GCA, GCG) to tryptophan (TGG). However, our approach also isolates the bacterial toxin from the plethora of other bacterial virulence factors. As a result, a PFO variant with increased activity may not confer a competitive advantage to the bacterium, if for example it leads to detrimental effects such as increased host damage or energy expenditure. Additionally, variants with increased activity may encounter reduced evolutionarily pressure for further adaptations. On the other hand, evolution may have progressed as it did due to stochastic events rather than deterministic processes (54). The specific evolutionary trajectory may not be the only possible outcome as alternative pathways might show different scenarios that were possible, but were not taken.
In vitro evolution also sheds light on the evolution of toxins and host–parasite interactions (55) as well as the evolution of potential antimicrobial targets based on different sterolomes (56). With advances in the analysis of lipid–protein interactions using native mass spectrometry (10), in vitro–directed evolution systems may become an invaluable aspect of studying the activity of membrane proteins in defined lipid environments (36).
Experimental procedures
Materials
The plasmid pRDV was provided by Dr Andreas Plückthun (57). The D4 gene libraries were obtained from Eurofins Genomics or GeneScript. All primers and synthetic gene fragments were purchased from Integrated DNA Technologies. Restriction enzymes were from New England Biolabs, while RNase inhibitor RiboLock, NTPs, T4 DNA ligase, T7 RNA polymerase, DNAse I, Ion Plus Fragment Library Kit, Ion PGMTM Hi-QTM OT2 kit, and Ion Sphere Quality Control kit were from Thermo Fisher Scientific. POPC, Chol, 1-oleoyl-2-(12-biotinyl (aminododecanoyl))-sn-glycero-3-phosphoethanolamine (Biotin-PE) were from Avanti Polar Lipids. Lissamine rhodamine B 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (rhodamine DHPE) was from Invitrogen. All other chemicals were from Sigma, unless stated otherwise.
Ribosome display and affinity selection
The SSL was prepared by combining the 17 synthetic DNA D4 libraries in equimolar ratio, each of which contained randomized codons (one per DNA molecule) at the following 17 positions (PDB ID: 1PFO): 399, 401, 403, 434, 436, 437, 458, 459, 460, 461, 462, 466, 468, 489, 490, 491, 492. The combinatorial library contained seven randomized codons per DNA molecule at positions 401, 403, 458, 460, 468, 490, 491 (27). Both libraries start with the codon for S386 (PDB ID: 1PFO) and contain randomized codons in the NNK scheme (N denotes any nucleotide and K represents G or T). Gene elements required for in vitro transcription and translation were added to D4 libraries by cloning them into the pRDV vector in a two-step PCR (27). In vitro transcription was performed for 3 h at 37 °C in T7 buffer (200 mM Hepes, 30 mM magnesium acetate, 2 mM spermidine, 40 mM DTT, pH 7.6) using 1 μg of template DNA, 1.75 mM of each nucleotide ATP, GTP, UTP, CTP, 10 mM spermidine, 1 U/μl RNase inhibitor RiboLock, and 16 U/μl T7 RNA polymerase. Next, the template DNA was cleaved with 0.08 U/μl DNase I, and the mRNA was purified by precipitation with 6 M LiCl and 3 M sodium acetate (58) and used immediately for in vitro translation in an S30 system from Escherichia coli strain MRE600 (ATCC 2941) purified as described previously (27). The in vitro translation reaction was performed in 50 mM Tris-acetate buffer (pH 7.4 at 4 °C) and contained 60% (v/v) S30 extract, 3 μg mRNA, 0.35 mM each amino acid, 3.6 μM anti-ssrA oligonucleotide (5′ TTAGCTGCTAAAGCGTAGTTTTCGTCGTTTGC 3′), 2 mM ATP, 0.5 mM GTP, 1 mM cAMP, 30 mM 3-phosphoglyceric acid, 0.5 mg/ml tRNA from E. coli MRE strain, 0.02 mg/ml folinic acid, magnesium acetate (from 11 mM to 14.4 mM depending on S30 batch), potassium glutamate (from 180 mM to 220 mM), and trehalose (3.5%–6% (w/v)). The translation was stopped after 9 min by cooling and five-fold dilution with ice-cold washing buffer (50 mM Tris-acetate, pH 7.5, 150 mM NaCl, 50 mM magnesium acetate, 0.1% bovine serum albumin (BSA), with added 2.5 mg/ml heparin). Next, the diluted reaction mixture was centrifuged at 13,000g for 5 min at 4 °C and then the supernatant was added to the immobilized target vesicles.
The target vesicles were immobilized on magnetic streptavidin beads (Dynabeads M-280, Thermo Fisher Scientific) as described previously (27). Briefly, the beads (20 μl/sample) were pre-washed with solution A (100 mM NaOH and 50 mM NaCl) and washing buffer (50 mM Tris-acetate, pH 7.5, 150 mM NaCl, 50 mM magnesium acetate, and 0.1% BSA). Then, 20 μl of vesicles composed of 1 mM appropriate lipids and 0.5 mol% Biotin-PE were added and gently shaken at 4 °C for 1 h. Bound vesicles were next treated with 0.5% BSA in washing buffer at 4 °C for 16 to 18 h. The vesicles were then washed with ice-cold washing buffer, and the translation mixture was added and incubated for 1 h at 4 °C with gentle shaking. The selection pressure was applied by successive washing with 100 μl of the ice-cold washing buffer as follows: 3 × 2 min for the first and second round, 2 × 2 min and 1 × 3 min for the third round, 3 × 3 min for the fourth, and finally 2 × 3 min, 1 × 4 min and 2 × 5 min for the fifth and sixth round. Next, the bound protein–mRNA–ribosome complexes were dissociated with the elution buffer (50 mM Tris-acetate, pH 7.5, 150 mM NaCl, 25 mM EDTA, and 50 μg/ml Saccharomyces cerevisiae RNA) and the eluted RNA was purified using High Pure RNA Isolation Kit (Roche Diagnostics). Immediately after isolation, the RNA was reversely transcribed by the High Capacity cDNA RT Kit (Life Technologies) and amplified using D4 library-specific primers. The amplified DNA was used to prepare the DNA template for the next round of selection or for next-generation sequencing (NGS) library preparation.
Library preparation and NGS
Libraries for NGS were prepared as previously described (27). Briefly, input D4 libraries and libraries after the sixth round of affinity selection amplified with D4-specific primers were purified and subjected to preparation of the NGS library using Ion Plus Fragment Library Kit. Adapter and barcode ligation, size selection, nick repair, five amplification cycles with 100 ng of the DNA template were performed according to the manufacturer’s instructions. Enrichment of NGS libraries (diluted to a 15 pM concentration and mixed in the equimolar ratio) was performed using the Ion OneTouch 2 (ThermoFisher Scientific) System and the Ion PGMTM Hi-QTM OT2 kit. The quality of the prepared NGS libraries was determined using the Ion Sphere Quality Control kit and the Qubit 3.0 fluorimeter (Thermo Fisher Scientific). The NGS libraries were sequenced with the Ion 530 Chip. Signal processing, base calling, and adapter sequence trimming were performed using Torrent Suite software (http://coolgenes.cahe.wsu.edu/ion-docs/Technical-Note---Analysis-Pipeline_6455567.html). The quality-filtered numbers of reads that were used in the analysis were between 92,773 and 958,851 for the input combinatorial library and 422,916 or 499,317 for the SSL, while the number of reads for the libraries after affinity selection were between 72,013 and 805,244.
Analysis of high-throughput sequencing data and variant identification
Quality-filtered sequencing reads were aligned to the reference (nucleotide sequence of the PFO D4) using LAST (59). The error rate and mutation frequency at each position in the nucleotide sequence were determined, and the alignments were further processed to account for any insertion or deletion artefacts that occurred during PCR amplification or sequencing. We analyzed only the sequencing reads spanning all randomized positions. Sequences were then translated in silico and amino acid frequency, calculated as a ratio of its occurrence in the population of detected NGS reads, was analyzed for each randomized position. The enrichment was calculated by normalizing the frequency (fa,p) of amino acid residue a at the position p to the frequency ( of amino acid residues in the input library.
Data processing, calculations, and visualization were performed using the Python packages NumPy, SciPy, Pandas, and Seaborn.
Preparation of multilamellar and unilamellar lipid vesicles
Multilamellar vesicles (MLVs) were prepared by dissolving lipids in chloroform to 10 mM solutions and mixing them in the desired molar ratio. The solvent was then evaporated, and the formed lipid film was hydrated with a suitable buffer depending on its further use. For vesicles used for affinity selection, the lipid film was hydrated in the washing buffer (50 mM Tris acetate, pH 7.5, 150 mM NaCl, 50 mM magnesium acetate, 0.1% (w/v) BSA), whereas for vesicles used for hemolysis inhibition assay or sedimentation assay, the lipid film was hydrated in 50 mM Tris-acetate, pH 7.5, and 150 mM NaCl. To prepare small unilamellar vesicles, MLVs were subjected to six freeze-thaw cycles and sonicated on ice for 25 min with 10-s pulses at 35% amplitude. The size distribution profile of the vesicles was measured using dynamic light scattering (Zetasizer Nano-ZS, Malvern Instruments).
Plasmid preparation and protein purification
The plasmid vector pHT-PFO wt (27) encoding the N-terminal 6× His-tag, the recognition site for the TEV protease, and the gene encoding PFO with a silent mutation leading to the formation of the BstBI restriction site at the beginning of the coding region for the D4 domain was used to prepare the constructs encoding PFO variants. The D4-coding region of the pHT-PFO wt was replaced with synthetic DNA fragments encoding D4 variants using the BstBI and MluI restriction sites. Similarly, the synthetic DNA fragments replaced the D4 WT coding sequence in the pHT-EGFP-D4 wt, which was prepared in the same expression system (pET8c) as pHT-PFO wt. In the pHT-EGFP-D4 wt, the EGFP coding sequence was inserted after the TEV-coding recognition site using XhoI and MluI restriction enzymes. AvrII restriction site created at 3′ end of the EGFP coding sequence (together with the flexible Gly-Ser linker) and MluI restriction site were used for insertion of the D4 gene or D4 gene variants. All resulting constructs were verified by Sanger sequencing.
Recombinant proteins were expressed in the E. coli BL21(DE3) pLysS cells cultivated at 37 °C in LB medium supplemented with 100 μg/ml ampicillin and 30 μg/ml chloramphenicol. The protein production was induced with 0.5 mM IPTG and the cells were harvested ∼20 h after induction at 20 °C. To purify the proteins, the cells were resuspended in the lysis buffer (50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole, pH 8.0) at a ratio of 10 ml/g wet bacterial mass, lysed by sonication and, after removal of the cell debris by centrifugation and supernatant filtration, subjected to nickel affinity chromatography (Ni-NTA Superflow, Qiagen) as described previously (27). Protein purity was confirmed by SDS-PAGE and Coomassie staining and proteins were dialyzed against Tris–HCl buffer (50 mM Tris–HCl, pH 7.4, and 150 mM NaCl) using Slide-a-Lyzer with MWCO 10 kDa (Thermo Fisher Scientific), flash frozen in liquid nitrogen, and stored at −20 °C until use.
Hemolytic activity of purified PFO variants
The hemolytic activity of PFO variants was measured using bovine erythrocytes as described previously (33). Briefly, the erythrocyte suspension was washed 3 to 5 times with the erythrocyte buffer (20 mM Tris–HCl, pH 7.4, and 140 mM NaCl) and diluted to the absorbance of ∼1 at 630 nm (A630). The prepared erythrocyte suspension was added to the equal volume of the two-fold serial dilutions of the proteins and A630 nm was measured at 25 °C immediately every 20 s for the next 20 min using the Synergy MX microplate reader (Biotek). The concentration of PFO variants (c50) that causes the half maximum hemolysis rate was determined from the data. The relative hemolytic activity of each variant was calculated as a reciprocal c50 value normalized to the reciprocal c50 value of the WT PFO.
Enzyme-linked immunosorbent assay assessment of protein variant binding to various lipids
Lipids (POPC, cholesteryl-acetate, and Chol) dissolved in chloroform at 2 mM were 100× diluted in ethanol, and 100 μl was used to coat the wells of the microtiter plate. After overnight solvent evaporation at 22 to 24 °C, 200 μl of 3% (w/v) BSA in Tris-buffered saline buffer (TBS; 10 mM Tris–HCl, pH 7.4, 150 mM NaCl) was used as a blocking reagent and incubated for 2 h at 22 to 24 °C. Then, the blocking reagent was removed and the wells were rinsed 3× with 200 μl TBS. Next, 100 μl of two-fold serial dilutions of proteins in a concentration range 0.2 to 200 nM prepared in TBS with 1% (w/v) BSA was added and incubated for 1 h at 22 to 24 °C. Unbound protein was removed by six washes with 200 μl of TBS, while bound proteins were detected with primary monoclonal mouse Penta-His antibodies (Qiagen) diluted at a ratio of 1:1000 and secondary anti-mouse antibodies conjugated with horseradish peroxidase diluted at a ratio of 1:5000. The detection of bound complexes was performed with 3,3′,5,5′-tetramethylbenzidine substrate (Thermo Fisher Scientific) and the intensity of the absorbance of the developed substrate color was measured at 490 nm with a reference of 630 nm with the Synergy MX microplate reader (Biotek).
Lipid overlay assay
One microliter of chloroform:methanol:water mixture (1:1:0.3, v:v:v) containing 1000 pmol of POPC, Chol, and sphingomyelin was spotted to the polyvinylidene difluoride membrane, treated with casein blocking buffer for 2 h, and incubated overnight with 10 μg/ml proteins. After removal of unbound proteins by washing in TBS containing 0.03% Tween-20, the bound proteins were detected by mouse anti-His IgG (Qiagen) and horseradish peroxidase–coupled rabbit anti-mouse antibodies. Detection was performed using chemiluminescence substrate (Millipore, Billerica) and captured with an iBright 1500 imaging system (Thermo Fisher Scientific).
Vesicles sedimentation assay
Proteins (2 μM) were incubated with a 2000× molar excess of MLVs prepared in 50 mM Tris-acetate and 150 mM NaCl, pH 7.4. Mixtures were incubated for 30 min at 22 to 24 °C and then centrifuged at 16,000g for 30 min at 4 °C. The supernatants were transferred to fresh microtubes, while unbound proteins were removed from the pellets by resuspension in 100 μl buffer solution and centrifugation under the same conditions. Afterward, unbound proteins in the supernatant and bound proteins in the pellet were subjected to SDS-PAGE analysis followed by protein detection using SimplyBlue SafeStain (Thermo Fisher Scientific).
Cryo transmission electron microscopy
Five micromolars of proteins were added to LUVs prepared from 2.5 mM POPC and 2.5 mM Chol. The reaction mixture was incubated at 22 to 24 °C for 20 min. Three microliters of the mixture was applied to glow-discharged Quantifoil 200 mesh R1.2/1.3 holey carbon grids (Quantifoil) and frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fischer Scientific). Micrographs were acquired on a Glacios transmission cryo transmission electron microscope (Thermo Fisher Scientific) operated at 200 kV and equipped with a Falcon III direct electron detector at 73,000× magnification (pixel size of 1.6 Å) with a total dose of 30 e−/Å2 and defocus of −3 or −4 μm. For each PFO variant, micrographs were CTF estimated and 114 to 384 top views of the PFO pores were manually picked for 2D classification in cryoSPARC (60). The inner pore diameter was determined by analyzing 82 pores of the variant WVATRIW, 144 of the WVVTRAF, and 88 of the WT PFO.
Cell lines and media
Mouse macrophage suspension cell line RAW 267.4 (ATCC, TIB-71) were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% heat inactivated fetal bovine serum (all from Sigma-Aldrich) and maintained at standard conditions of 37 °C, 5% CO2, and 90% humidity.
Flow cytometry measurement
RAW 264.7 cells (106 cells/ml) were incubated for 1 h in DMEM or in DMEM containing 1 mM or 2.5 mM MβCD. After incubation, cells (5 × 106) were washed with DMEM, resuspended in 200 μl PBS, and purified proteins, namely 0.5 μM EGFP-D4 AVETRTL (the WT), 2 μM EGFP-D4 WVATRIW, and 2 μM EGFP-D4 WVVTRAF were added. After 15 min incubation at 22 to 24 °C, protein binding was monitored on a Cytec Aurora flow cytometer (Cytek Biosciences) using a 488 nm laser and forward and side light scattering (FS/SS). The amount of fluorescently labeled protein on the cells was monitored on the FL1 (B1-A) channel, which detects green fluorescence. The results were analyzed using the Flowjo software version 10.9.0 (BD Biosciences).
Cell membrane permeabilization assay
RAW267.4 cells (106/ml) were washed twice in serum-free DMEM and incubated for 150 min with 1 mM mβCD prepared in DMEM or DMEM only. After incubation, cells were washed twice with DMEM and 25,000 cells were transferred to each well of the 384-microtiter plate containing 5 μM Sytox Green and serially twofold-diluted PFO variants at concentrations ranging from 6.25 to 100 nM. RAW267.4 cells incubated only with DMEM instead of 1 mM mβCD served as controls. Fluorescence of DNA-bound Sytox Green was excited at 504 nm and emission was measured at 523 nm every minute for 30 min using FLUOstar Galaxy (BMG Lab Technologies), immediately after the proteins were added to the cells. The permeabilization rate for each protein concentration was determined from the fluorescence kinetics curve as the maximum increase in fluorescence per minute. The value was determined by curve fitting using Origin 8.1 software (OriginLab).
Data availability
All data are contained within the manuscript and can be shared upon request to corresponding authors.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors thank Magdalena Kulma for inspiring discussions.
Author contributions
A. S., N. K., N. O., S. A., A. K., M. K., A. B. Z., S. C., R. J. C. G., M. P., and G. A. formal analysis; A. S., N. K., N. O., S. A., A. K., M. K., A. B. Z., and S. C. investigation; A. S., N. K., N. O., S. A., A. K., M. K., A. B. Z., and S. C. methodology; A. S., A. C., and G. A. visualization; A. S., N. K., A. C., and G. A. writing–original draft; A. S., N. K., N. O., S. A., A. K., M. K., A. B. Z., S. C., R. J. C. G., M. P., A. C., and G. A. writing–review and editing; M. P., A. C., and G. A. funding acquisition; G. A. conceptualization; G. A. project administration; G. A. supervision.
Funding and additional information
The work was supported by the programme grant of the Slovenian Research Agency (Molecular Interactions P1-0391). Part of this work was performed at the National Institute of Chemistry Cryo-EM Facility, supported by Slovenian Research Agency Infrastructure Programme IO-0003. A. C. acknowledges funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No. 896849.
Reviewed by members of the JBC Editorial Board. Edited by Karen Fleming
Contributor Information
Ana Crnković, Email: ana.crnkovic@ki.si.
Gregor Anderluh, Email: gregor.anderluh@ki.si.
References
- 1.Redondo-Morata L., Losada-Pérez P., Giannotti M.I. In: Current Topics in Membranes. Levitan I., Trache A., editors. Academic Press; San Diego, CA: 2020. Chapter one - lipid bilayers: phase behavior and nanomechanics; pp. 1–55. [DOI] [PubMed] [Google Scholar]
- 2.Zhang T., Yuan D., Xie J., Lei Y., Li J., Fang G., et al. Evolution of the cholesterol biosynthesis pathway in animals. Mol. Biol. Evol. 2019;36:2548–2556. doi: 10.1093/molbev/msz167. [DOI] [PubMed] [Google Scholar]
- 3.Corradi V., Sejdiu B.I., Mesa-Galloso H., Abdizadeh H., Noskov S.Y., Marrink S.J., et al. Emerging diversity in lipid-protein interactions. Chem. Rev. 2019;119:5775–5848. doi: 10.1021/acs.chemrev.8b00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 5.Galea A.M., Brown A.J. Special relationship between sterols and oxygen: were sterols an adaptation to aerobic life? Free Radic. Biol. Med. 2009;47:880–889. doi: 10.1016/j.freeradbiomed.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 6.Adamian L., Naveed H., Liang J. Lipid-binding surfaces of membrane proteins: evidence from evolutionary and structural analysis. Biochim. Biophys. Acta. 2011;1808:1092–1102. doi: 10.1016/j.bbamem.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderluh G., Lakey J.H. Disparate proteins use similar architectures to damage membranes. Trends Biochem. Sci. 2008;33:482–490. doi: 10.1016/j.tibs.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Fantini J., Epand R.M., Barrantes F.J. In: Direct Mechanisms in Cholesterol Modulation of Protein Function. Rosenhouse-Dantsker A., Bukiya A.N., editors. Springer International Publishing; Cham: 2019. Cholesterol-recognition motifs in membrane proteins; pp. 3–25. [Google Scholar]
- 9.Lee A.G. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 10.Bolla J.R., Agasid M.T., Mehmood S., Robinson C.V. Membrane protein-lipid interactions probed using mass spectrometry. Annu. Rev. Biochem. 2019;88:85–111. doi: 10.1146/annurev-biochem-013118-111508. [DOI] [PubMed] [Google Scholar]
- 11.Tweten R.K., Hotze E.M., Wade K.R. The unique molecular choreography of giant pore formation by the cholesterol-dependent cytolysins of gram-positive bacteria. Annu. Rev. Microbiol. 2015;69:323–340. doi: 10.1146/annurev-micro-091014-104233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnstone B.A., Joseph R., Christie M.P., Morton C.J., McGuiness C., Walsh J.C., et al. Cholesterol-dependent cytolysins: the outstanding questions. IUBMB Life. 2022;74:1169–1179. doi: 10.1002/iub.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verherstraeten S., Goossens E., Valgaeren B., Pardon B., Timbermont L., Haesebrouck F., et al. Perfringolysin O: the underrated Clostridium perfringens toxin? Toxins (Basel) 2015;7:1702–1721. doi: 10.3390/toxins7051702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert R. Structural features of cholesterol dependent cytolysins and comparison to other MACPF-domain containing proteins. Subcell. Biochem. 2014;80:47–62. doi: 10.1007/978-94-017-8881-6_4. [DOI] [PubMed] [Google Scholar]
- 15.Ramachandran R., Heuck A.P., Tweten R.K., Johnson A.E. Structural insights into the membrane-anchoring mechanism of a cholesterol-dependent cytolysin. Nat. Struct. Biol. 2002;9:823–827. doi: 10.1038/nsb855. [DOI] [PubMed] [Google Scholar]
- 16.Shimada Y., Maruya M., Iwashita S., Ohno-Iwashita Y. The C-terminal domain of perfringolysin O is an essential cholesterol-binding unit targeting to cholesterol-rich microdomains. Eur. J. Biochem. 2002;269:6195–6203. doi: 10.1046/j.1432-1033.2002.03338.x. [DOI] [PubMed] [Google Scholar]
- 17.Farrand A.J., LaChapelle S., Hotze E.M., Johnson A.E., Tweten R.K. Only two amino acids are essential for cytolytic toxin recognition of cholesterol at the membrane surface. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4341–4346. doi: 10.1073/pnas.0911581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Pee K., Neuhaus A., D'Imprima E., Mills D.J., Kuhlbrandt W., Yildiz O. CryoEM structures of membrane pore and prepore complex reveal cytolytic mechanism of Pneumolysin. Elife. 2017;6 doi: 10.7554/eLife.23644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Q., London E. Altering hydrophobic sequence lengths shows that hydrophobic mismatch controls affinity for ordered lipid domains (rafts) in the multitransmembrane strand protein perfringolysin O. J. Biol. Chem. 2013;288:1340–1352. doi: 10.1074/jbc.M112.415596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson B.B., Moe P.C., Wang D., Rossi K., Trigatti B.L., Heuck A.P. Modifications in perfringolysin O domain 4 alter the cholesterol concentration threshold required for binding. Biochemistry. 2012;51:3373–3382. doi: 10.1021/bi3003132. [DOI] [PubMed] [Google Scholar]
- 21.Johnson B.B., Brena M., Anguita J., Heuck A.P. Mechanistic insights into the cholesterol-dependent binding of perfringolysin O-based probes and cell membranes. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-14002-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohno-Iwashita Y., Shimada Y., Waheed A.A., Hayashi M., Inomata M., Nakamura M., et al. Perfringolysin O, a cholesterol-binding cytolysin, as a probe for lipid rafts. Anaerobe. 2004;10:125–134. doi: 10.1016/j.anaerobe.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Liu S.L., Sheng R., Jung J.H., Wang L., Stec E., O'Connor M.J., et al. Orthogonal lipid sensors identify transbilayer asymmetry of plasma membrane cholesterol. Nat. Chem. Biol. 2017;13:268–274. doi: 10.1038/nchembio.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lasič E., Lisjak M., Horvat A., Božič M., Šakanović A., Anderluh G., et al. Astrocyte specific remodeling of plasmalemmal cholesterol composition by ketamine indicates a new mechanism of antidepressant action. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-47459-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rituper B., Guček A., Lisjak M., Gorska U., Šakanović A., Bobnar S.T., et al. Vesicle cholesterol controls exocytotic fusion pore. Cell Calcium. 2022;101 doi: 10.1016/j.ceca.2021.102503. [DOI] [PubMed] [Google Scholar]
- 26.Gay A., Rye D., Radhakrishnan A. Switch-like responses of two cholesterol sensors do not require protein oligomerization in membranes. Biophys. J. 2015;108:1459–1469. doi: 10.1016/j.bpj.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Šakanović A., Kranjc N., Omersa N., Podobnik M., Anderluh G. More than one way to bind to cholesterol: atypical variants of membrane-binding domain of perfringolysin O selected by ribosome display. RSC Adv. 2020;10:38678–38682. doi: 10.1039/d0ra06976k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang M.S., Brunner S.F., Huguenin-Dezot N., Liang A.D., Schmied W.H., Rogerson D.T., et al. Biosynthesis and genetic encoding of phosphothreonine through parallel selection and deep sequencing. Nat. Methods. 2017;14:729–736. doi: 10.1038/nmeth.4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossjohn J., Feil S.C., McKinstry W.J., Tweten R.K., Parker M.W. Structure of a cholesterol-binding, thiol-activated cytolysin and a model of its membrane form. Cell. 1997;89:685–692. doi: 10.1016/s0092-8674(00)80251-2. [DOI] [PubMed] [Google Scholar]
- 30.Bavdek A., Gekara N.O., Priselac D., Gutierrez Aguirre I., Darji A., Chakraborty T., et al. Sterol and pH interdependence in the binding, oligomerization, and pore formation of Listeriolysin O. Biochemistry. 2007;46:4425–4437. doi: 10.1021/bi602497g. [DOI] [PubMed] [Google Scholar]
- 31.Kozorog M., Sani M.A., Lenarčič Živkovič M., Ilc G., Hodnik V., Separovic F., et al. 19F NMR studies provide insights into lipid membrane interactions of listeriolysin O, a pore forming toxin from Listeria monocytogenes. Sci. Rep. 2018;8:6894. doi: 10.1038/s41598-018-24692-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson L.D., Johnson A.E., London E. How interaction of perfringolysin O with membranes is controlled by sterol structure, lipid structure, and physiological low pH - insights into the origin of perfringolysin O-lipid raft interaction. J. Biol. Chem. 2008;283:4632–4642. doi: 10.1074/jbc.M709483200. [DOI] [PubMed] [Google Scholar]
- 33.Whitehead T.A., Chevalier A., Song Y., Dreyfus C., Fleishman S.J., De Mattos C., et al. Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat. Biotechnol. 2012;30:543–548. doi: 10.1038/nbt.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu N.C., Grande G., Turner H.L., Ward A.B., Xie J., Lerner R.A., et al. In vitro evolution of an influenza broadly neutralizing antibody is modulated by hemagglutinin receptor specificity. Nat. Commun. 2017;8 doi: 10.1038/ncomms15371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishikawa T., Sunami T., Matsuura T., Ichihashi N., Yomo T. Construction of a gene screening system using giant unilamellar liposomes and a fluorescence-activated cell sorter. Anal. Chem. 2012;84:5017–5024. doi: 10.1021/ac300678w. [DOI] [PubMed] [Google Scholar]
- 36.Levental I., Lyman E. Regulation of membrane protein structure and function by their lipid nano-environment. Nat. Rev. Mol. Cell Biol. 2023;24:107–122. doi: 10.1038/s41580-022-00524-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corradi V., Mendez-Villuendas E., Ingolfsson H.I., Gu R.X., Siuda I., Melo M.N., et al. Lipid-protein interactions are unique fingerprints for membrane proteins. ACS Cent. Sci. 2018;4:709–717. doi: 10.1021/acscentsci.8b00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szurmant H., Weigt M. Inter-residue, inter-protein and inter-family coevolution: bridging the scales. Curr. Opin. Struct. Biol. 2018;50:26–32. doi: 10.1016/j.sbi.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soltani C.E., Hotze E.M., Johnson A.E., Tweten R.K. Structural elements of the cholesterol-dependent cytolysins that are responsible for their cholesterol-sensitive membrane interactions. Proc. Natl. Acad. Sci. U. S. A. 2007;104:20226–20231. doi: 10.1073/pnas.0708104105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vogele M., Bhaskara R.M., Mulvihill E., van Pee K., Yildiz O., Kuhlbrandt W., et al. Membrane perforation by the pore-forming toxin pneumolysin. Proc. Natl. Acad. Sci. U. S. A. 2019;116:13352–13357. doi: 10.1073/pnas.1904304116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Savinov S.N., Heuck A.P. Interaction of cholesterol with perfringolysin O: what have we learned from functional analysis? Toxins (Basel) 2017;9:381. doi: 10.3390/toxins9120381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dowd K.J., Farrand A.J., Tweten R.K. The cholesterol-dependent cytolysin signature motif: a critical element in the allosteric pathway that couples membrane binding to pore assembly. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iwamoto M., Ohno-Iwashita Y., Ando S. Role of the essential thiol group in the thiol-activated cytolysin from Clostridium perfringens. Eur. J. Biochem. 1987;167:425–430. doi: 10.1111/j.1432-1033.1987.tb13355.x. [DOI] [PubMed] [Google Scholar]
- 44.Polekhina G., Giddings K.S., Tweten R.K., Parker M.W. Insights into the action of the superfamily of cholesterol-dependent cytolysins from studies of intermedilysin. Proc. Natl. Acad. Sci. U. S. A. 2005;102:600–605. doi: 10.1073/pnas.0403229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saunders F.K., Mitchell T.J., Walker J.A., Andrew P.W., Boulnois G.J. Pneumolysin, the thiol-activated toxin of Streptococcus pneumoniae, does not require a thiol group for in vitro activity. Infect. Immun. 1989;57:2547–2552. doi: 10.1128/iai.57.8.2547-2552.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pinkney M., Beachey E., Kehoe M. The thiol-activated toxin streptolysin O does not require a thiol group for cytolytic activity. Infect. Immun. 1989;57:2553–2558. doi: 10.1128/iai.57.8.2553-2558.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michel E., Reich K.A., Favier R., Berche P., Cossart P. Attenuated mutants of the intracellular bacterium Listeria monocytogenes obtained by single amino acid substitutions in listeriolysin O. Mol. Microbiol. 1990;4:2167–2178. doi: 10.1111/j.1365-2958.1990.tb00578.x. [DOI] [PubMed] [Google Scholar]
- 48.Sekino-Suzuki N., Nakamura M., Mitsui K.i., Ohno-Iwashita Y. Contribution of individual tryptophan residues to the structure and activity of θ-toxin (perfringolysin O), a cholesterol-binding cytolysin. Eur. J. Biochem. 1996;241:941–947. doi: 10.1111/j.1432-1033.1996.00941.x. [DOI] [PubMed] [Google Scholar]
- 49.Korchev Y.E., Bashford C.L., Pederzolli C., Pasternak C.A., Morgan P.J., Andrew P.W., et al. A conserved tryptophan in pneumolysin is a determinant of the characteristics of channels formed by pneumolysin in cells and planar lipid bilayers. Biochem. J. 1998;329:571–577. doi: 10.1042/bj3290571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Billington S.J., Songer J.G., Jost B.H. The variant undecapeptide sequence of the Arcanobacterium pyogenes haemolysin, pyolysin, is required for full cytolytic activity. Microbiology (Reading) 2002;148:3947–3954. doi: 10.1099/00221287-148-12-3947. [DOI] [PubMed] [Google Scholar]
- 51.Starr T.N., Thornton J.W. Epistasis in protein evolution. Protein Sci. 2016;25:1204–1218. doi: 10.1002/pro.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Pee K., Mulvihill E., Muller D.J., Yildiz O. Unraveling the pore-forming steps of pneumolysin from Streptococcus pneumoniae. Nano Lett. 2016;16:7915–7924. doi: 10.1021/acs.nanolett.6b04219. [DOI] [PubMed] [Google Scholar]
- 53.McGuinness C., Walsh J.C., Bayly-Jones C., Dunstone M.A., Christie M.P., Morton C.J., et al. Single-molecule analysis of the entire perfringolysin O pore formation pathway. Elife. 2022;11 doi: 10.7554/eLife.74901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Starr T.N., Picton L.K., Thornton J.W. Alternative evolutionary histories in the sequence space of an ancient protein. Nature. 2017;549:409–413. doi: 10.1038/nature23902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bucciarelli G.M., Alsalek F., Kats L.B., Green D.B., Shaffer H.B. Toxic relationships and arms-race coevolution revisited. Annu. Rev. Anim. Biosci. 2022;10:63–80. doi: 10.1146/annurev-animal-013120-024716. [DOI] [PubMed] [Google Scholar]
- 56.Haubrich B.A. Microbial sterolomics as a chemical biology tool. Molecules. 2018;23:2768. doi: 10.3390/molecules23112768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Binz H.K., Amstutz P., Kohl A., Stumpp M.T., Briand C., Forrer P., et al. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat. Biotechnol. 2004;22:575–582. doi: 10.1038/nbt962. [DOI] [PubMed] [Google Scholar]
- 58.Zahnd C., Amstutz P., Pluckthun A. Ribosome display: selecting and evolving proteins in vitro that specifically bind to a target. Nat. Methods. 2007;4:269–279. doi: 10.1038/nmeth1003. [DOI] [PubMed] [Google Scholar]
- 59.Kielbasa S.M., Wan R., Sato K., Horton P., Frith M.C. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011;21:487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 61.Schrodinger L.L.C. 2015. The PyMOL Molecular Graphics System, Version 1.8. [Google Scholar]
- 62.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the manuscript and can be shared upon request to corresponding authors.